m6A Regulator Information
General Information of the m6A Regulator (ID: REG00007)
| Regulator Name | Methyltransferase-like 3 (METTL3) | ||||
|---|---|---|---|---|---|
| Synonyms |
N6-adenosine-methyltransferase catalytic subunit; hMETTL3; N6-adenosine-methyltransferase 70 kDa subunit; MT-A70; MTA70
Click to Show/Hide
|
||||
| Gene Name | METTL3 | ||||
| Sequence |
MSDTWSSIQAHKKQLDSLRERLQRRRKQDSGHLDLRNPEAALSPTFRSDSPVPTAPTSGG
PKPSTASAVPELATDPELEKKLLHHLSDLALTLPTDAVSICLAISTPDAPATQDGVESLL QKFAAQELIEVKRGLLQDDAHPTLVTYADHSKLSAMMGAVAEKKGPGEVAGTVTGQKRRA EQDSTTVAAFASSLVSGLNSSASEPAKEPAKKSRKHAASDVDLEIESLLNQQSTKEQQSK KVSQEILELLNTTTAKEQSIVEKFRSRGRAQVQEFCDYGTKEECMKASDADRPCRKLHFR RIINKHTDESLGDCSFLNTCFHMDTCKYVHYEIDACMDSEAPGSKDHTPSQELALTQSVG GDSSADRLFPPQWICCDIRYLDVSILGKFAVVMADPPWDIHMELPYGTLTDDEMRRLNIP VLQDDGFLFLWVTGRAMELGRECLNLWGYERVDEIIWVKTNQLQRIIRTGRTGHWLNHGK EHCLVGVKGNPQGFNQGLDCDVIVAEVRSTSHKPDEIYGMIERLSPGTRKIELFGRPHNV QPNWITLGNQLDGIHLLDPDVVARFKQRYPDGIISKPKNL Click to Show/Hide
|
||||
| Family | MT-A70-like family | ||||
| Function |
The METTL3-METTL14 heterodimer forms a N6-methyltransferase complex that methylates adenosine residues at the N(6) position of some RNAs and regulates various processes such as the circadian clock, differentiation of embryonic and hematopoietic stem cells, cortical neurogenesis, response to DNA damage, differentiation of T-cells and primary miRNA processing. In the heterodimer formed with METTL14, METTL3 constitutes the catalytic core. N6-methyladenosine (m6A), which takes place at the 5'-[AG]GAC-3' consensus sites of some mRNAs, plays a role in mRNA stability, processing, translation efficiency and editing. In embryonic stem cells (ESCs), m6A methylation of mRNAs encoding key naive pluripotency-promoting transcripts results in transcript destabilization, promoting differentiation of ESCs. Involved in the response to DNA damage: in response to ultraviolet irradiation, METTL3 rapidly catalyzes the formation of m6A on poly(A) transcripts at DNA damage sites, leading to the recruitment of POLK to DNA damage sites. M6A is also required for T-cell homeostasis and differentiation: m6A methylation of transcripts of SOCS family members (SOCS1, SOCS3 and CISH) in naive T-cells promotes mRNA destabilization and degradation, promoting T-cell differentiation. Inhibits the type I interferon response by mediating m6A methylation of IFNB. Mediates m6A methylation of Xist RNA, thereby participating in random X inactivation: m6A methylation of Xist leads to target YTHDC1 reader on Xist and promote transcription repression activity of Xist. M6A also regulates cortical neurogenesis. METTL3 mediates methylation of pri-miRNAs, marking them for recognition and processing by DGCR8. Acts as a positive regulator of mRNA translation independently of the methyltransferase activity. Its overexpression in a number of cancer cells suggests that it may participate in cancer cell proliferation by promoting mRNA translation.
Click to Show/Hide
|
||||
| Gene ID | 56339 | ||||
| Uniprot ID | |||||
| Regulator Type | WRITER ERASER READER | ||||
| Mechanism Diagram | Click to View the Original Diagram | ||||
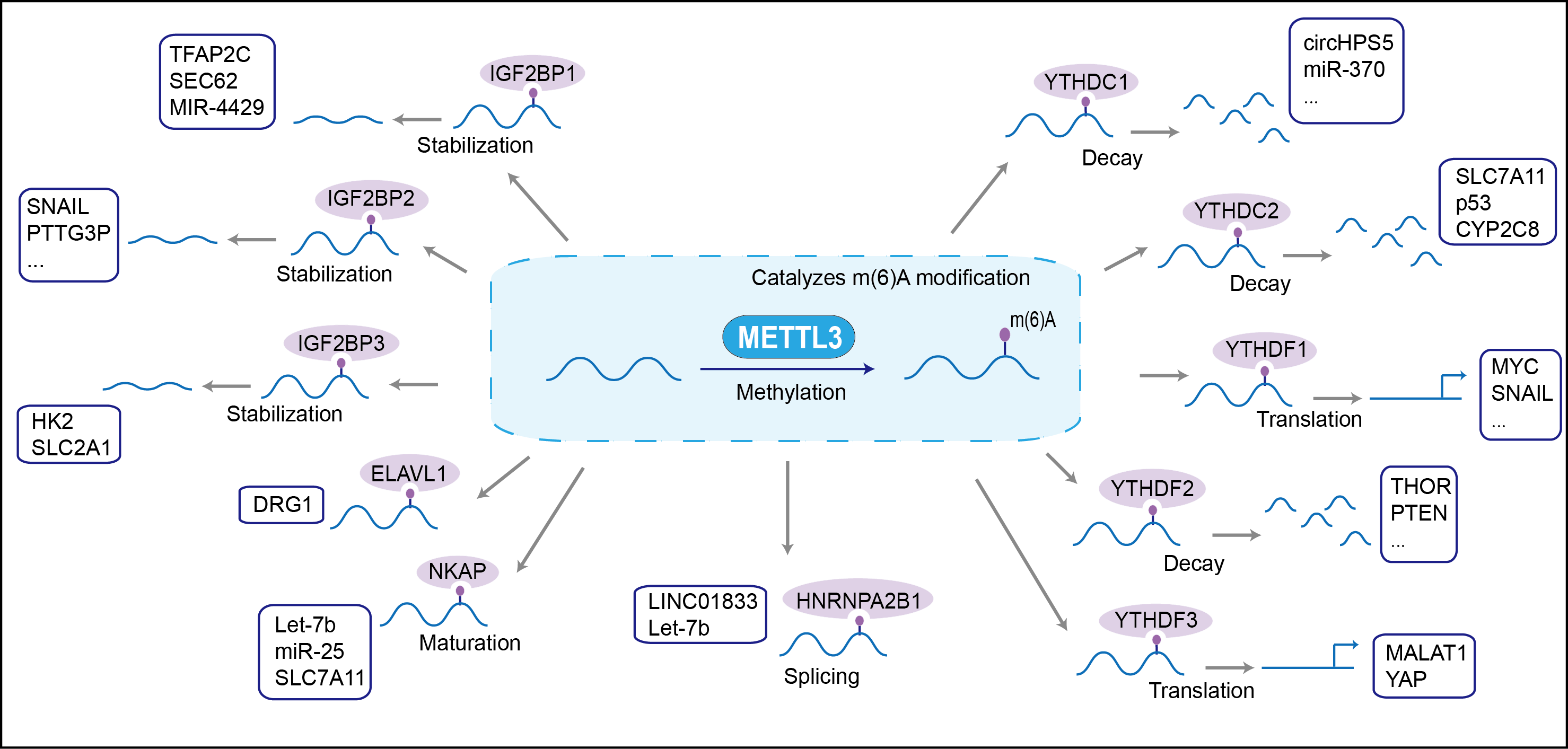
|
|||||
| Target Genes | Click to View Potential Target Genes of This Regulator | ||||
Full List of Target Gene(s) of This m6A Regulator and Corresponding Disease/Drug Response(s)
METTL3 can regulate the m6A methylation of following target genes, and result in corresponding disease/drug response(s). You can browse corresponding disease or drug response(s) resulted from the regulation of certain target gene.
Browse Target Gene related Disease
Browse Target Gene related Drug
72 kDa type IV collagenase (MMP2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
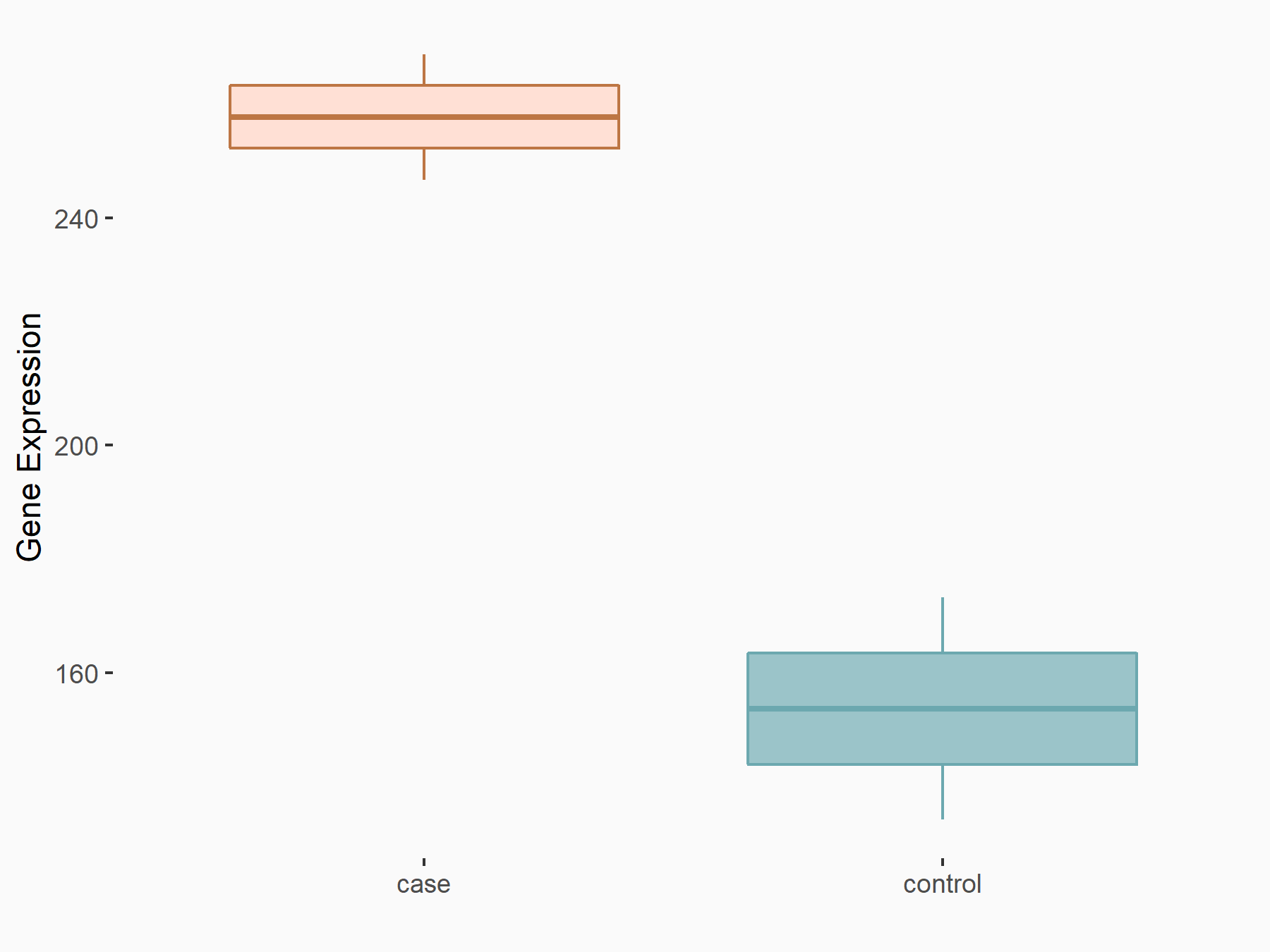  |
logFC: 7.46E-01 p-value: 1.39E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.08E+00 | GSE60213 |
Melanoma [ICD-11: 2C30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Melanoma [ICD-11: 2C30] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell invasion/migration | |||
In-vitro Model |
451Lu | Cutaneous melanoma | Homo sapiens | CVCL_6357 |
| A-375 | Amelanotic melanoma | Homo sapiens | CVCL_0132 | |
| A375-MA2 | Amelanotic melanoma | Homo sapiens | CVCL_X495 | |
| MeWo | Cutaneous melanoma | Homo sapiens | CVCL_0445 | |
| SK-MEL-2 | Melanoma | Homo sapiens | CVCL_0069 | |
| WM164 | Cutaneous melanoma | Homo sapiens | CVCL_7928 | |
| WM3211 | Acral lentiginous melanoma | Homo sapiens | CVCL_6797 | |
| WM3918 | Melanoma | Homo sapiens | CVCL_C279 | |
| WM793 | Melanoma | Homo sapiens | CVCL_8787 | |
| Response Summary | METTL3 is upregulated in human melanoma and plays a role in invasion/migration through MMP2. METTL3 overexpression promotes accumulation of 72 kDa type IV collagenase (MMP2) and N-cadherin in melanoma cells. | |||
Retinopathy [ICD-11: 9B71]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [264] | |||
| Responsed Disease | Retinopathy [ICD-11: 9B71] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | Mouse pups, along with their nursing mothers, were exposed to 75 ± 2% O2 in an incubator between postnatal day (P) 7 and P12 and were then returned to room air. | |||
Abnormal spindle-like microcephaly-associated protein (ASPM)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Mouse testis | Mus musculus |
|
Treatment: Mettl3 knockout mouse testis
Control: Mouse testis
|
GSE99771 | |
| Regulation |
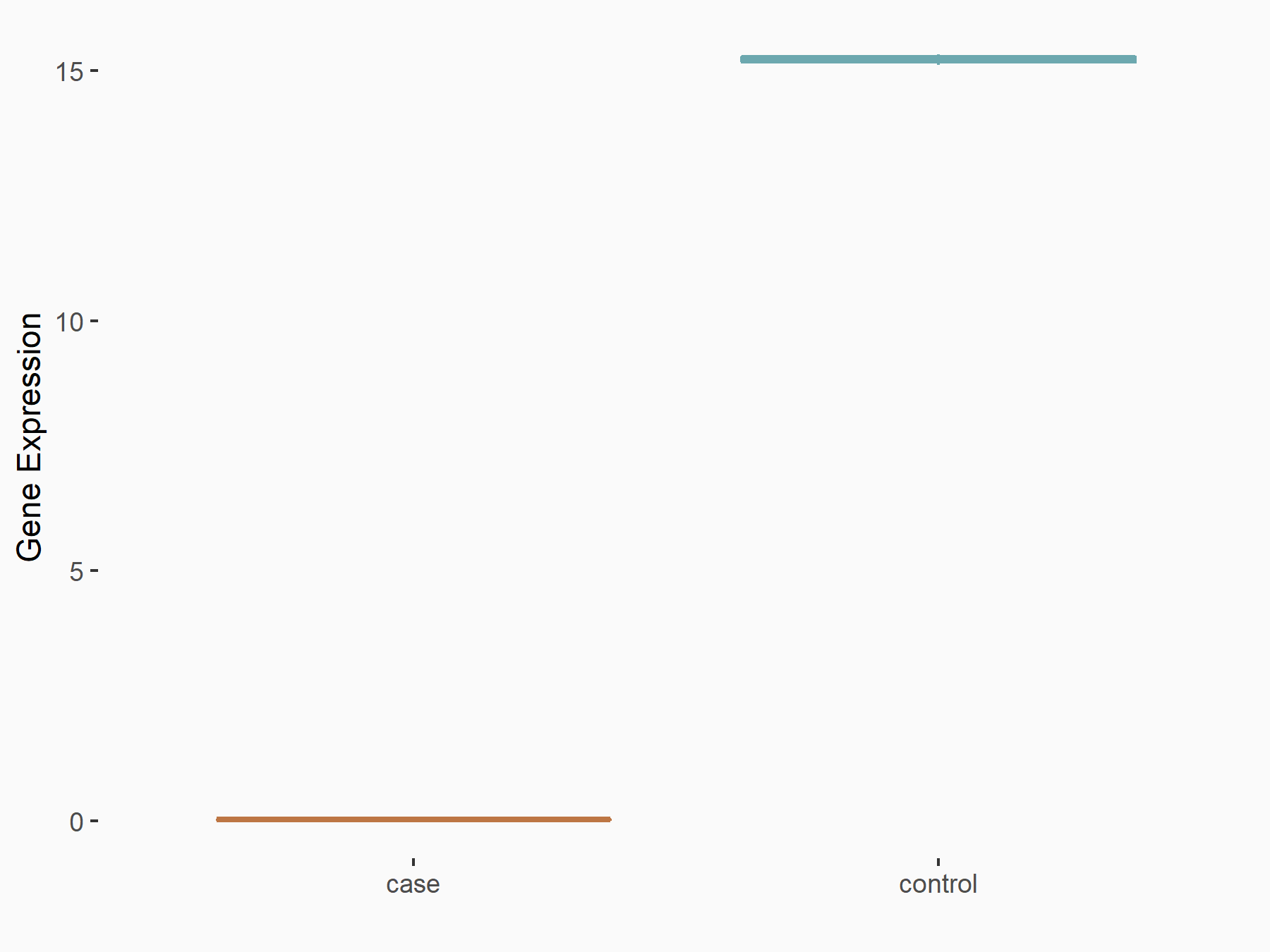 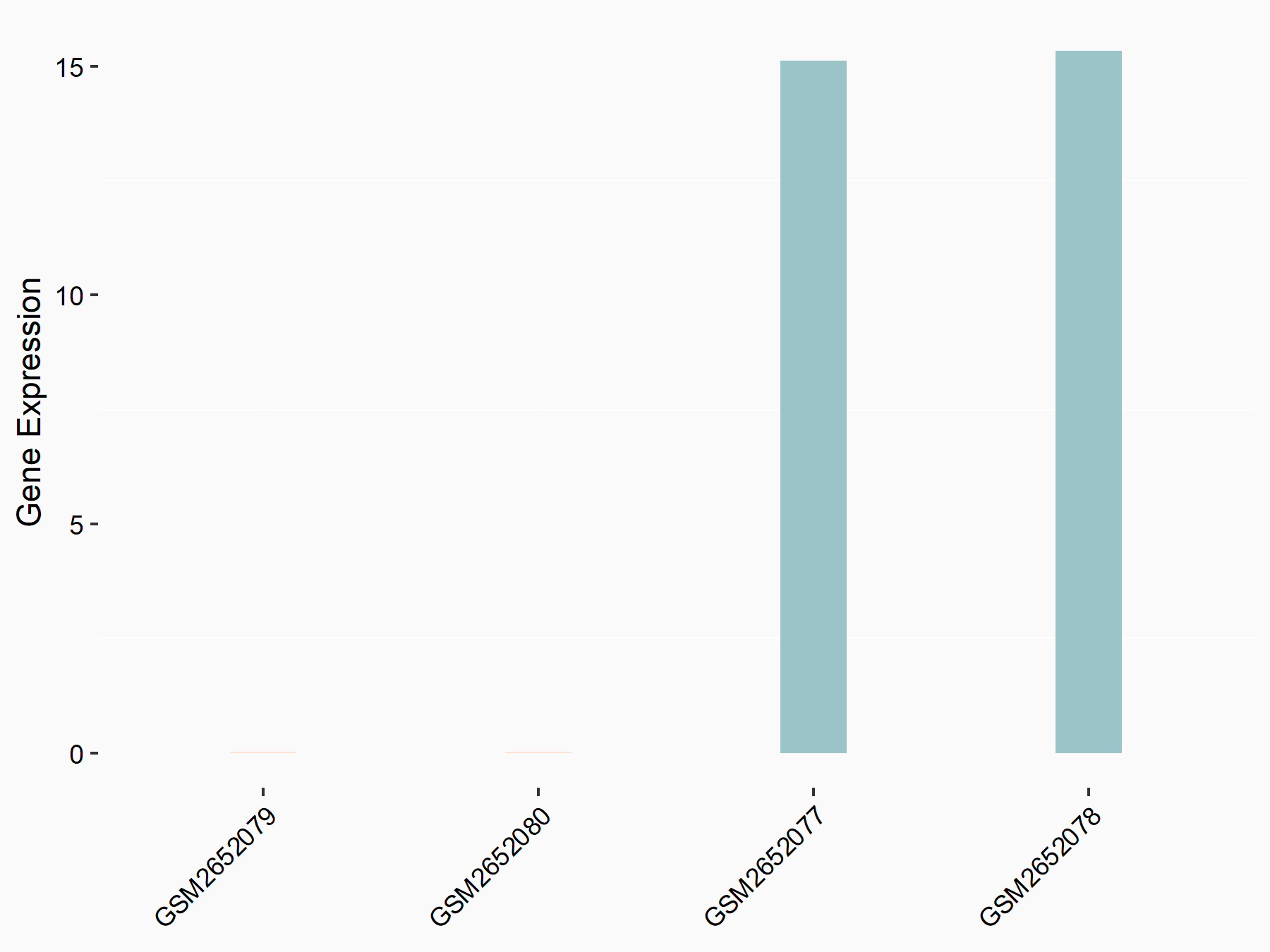 |
logFC: -3.99E+00 p-value: 5.74E-06 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.27E+00 | GSE60213 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [2] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cells growth | |||
| Cell metastasis | ||||
In-vitro Model |
SNU-449 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0454 |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hepg3b (Hepg3b were purchased from the American Type Culture Collection (ATCC, USA)) | ||||
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Response Summary | The N6-methyladenosine (m6A) modification of ASPM mRNA mediated by METTL3 promoted its expression in liver hepatocellular carcinoma. | |||
Adenomatous polyposis coli protein (APC)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
 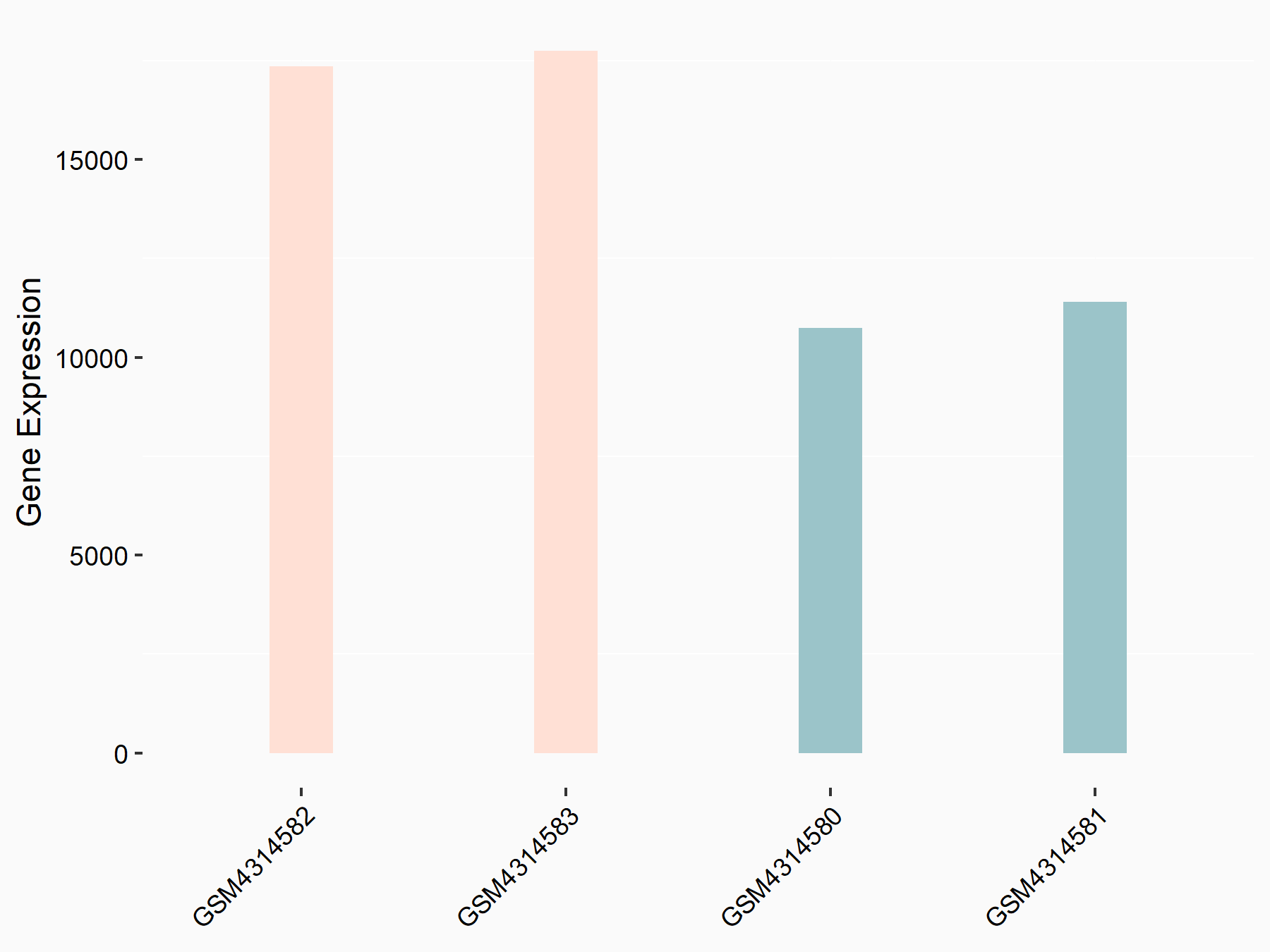 |
logFC: 6.65E-01 p-value: 8.92E-49 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.14E+00 | GSE60213 |
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [3] | |||
| Responsed Disease | Esophageal cancer [ICD-11: 2B70] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell cycle | hsa04110 | |||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glycolysis | |||
In-vitro Model |
TE-10 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1760 |
| TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 | |
| KYSE-70 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1356 | |
| KYSE-450 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1353 | |
| KYSE-410 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1352 | |
| KYSE-30 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1351 | |
| KYSE-180 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1349 | |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| KYSE-140 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1347 | |
| HET-1A | Normal | Homo sapiens | CVCL_3702 | |
| In-vivo Model | For the subcutaneous implantation model, 1 × 106 cells were injected subcutaneously into the flank regions of female BALB/c nude mice (4-5 weeks). | |||
| Response Summary | m6A-RNA immunoprecipitation sequencing revealed that METTL3 upregulates the m6A modification of Adenomatous polyposis coli protein (APC), which recruits YTHDF for APC mRNA degradation. Our findings reveal a mechanism by which the Wnt/Bete-catenin pathway is upregulated in ESCC via METTL3/YTHDF-coupled epitranscriptomal downregulation of APC. | |||
AF4/FMR2 family member 4 (AFF4)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 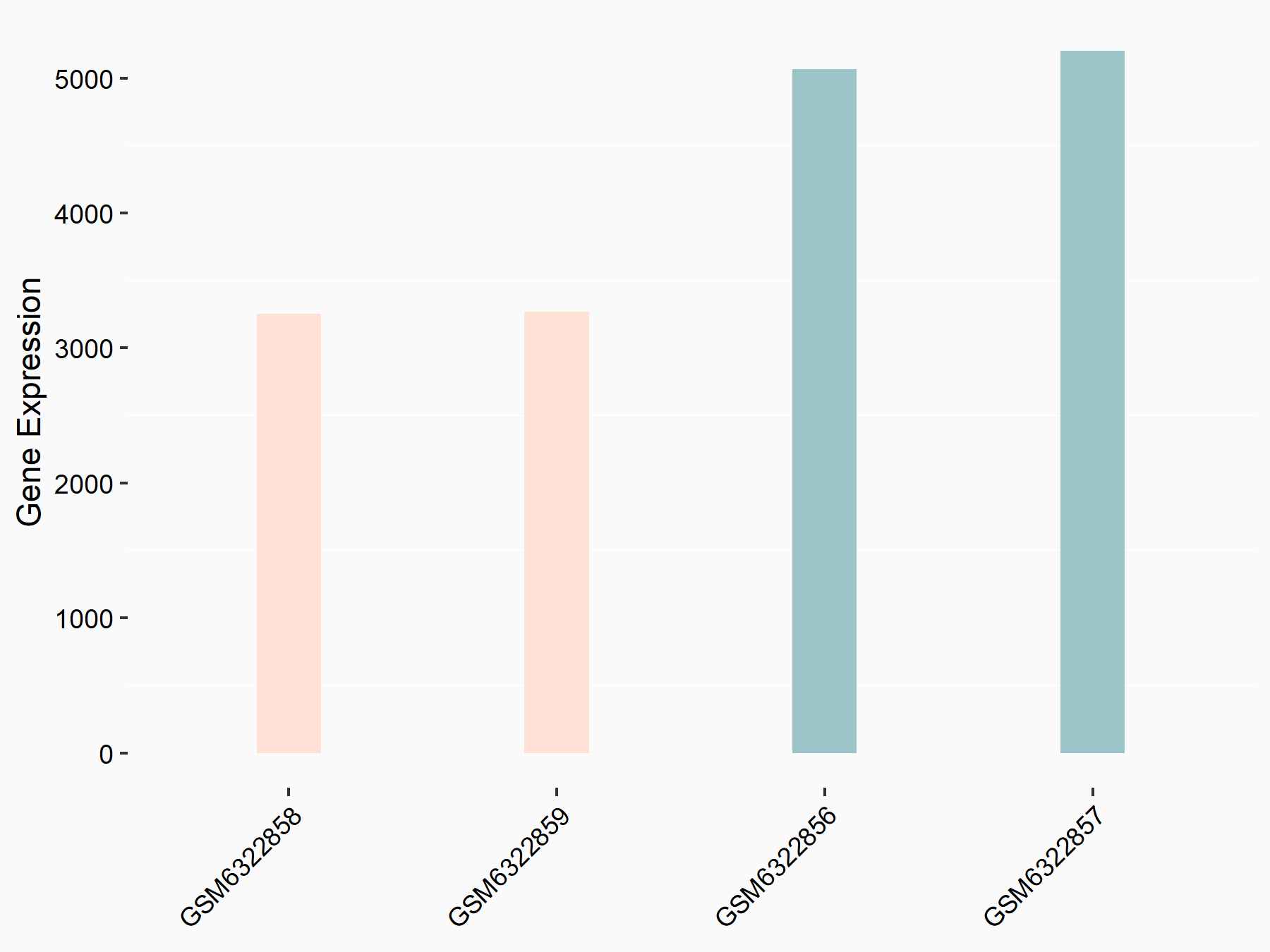 |
logFC: -6.53E-01 p-value: 1.79E-17 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.64E+00 | GSE60213 |
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Glucose metabolism | |||
| Response Summary | AF4/FMR2 family member 4 (AFF4), two key regulators of NF-Kappa-B pathway (IKBKB and RELA) and MYC were further identified as direct targets of METTL3-mediated m6A modification.overexpression of METTL3 significantly promoted Bladder cancer cell growth and invasion. | |||
Angiopoietin-1 receptor (TEK)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
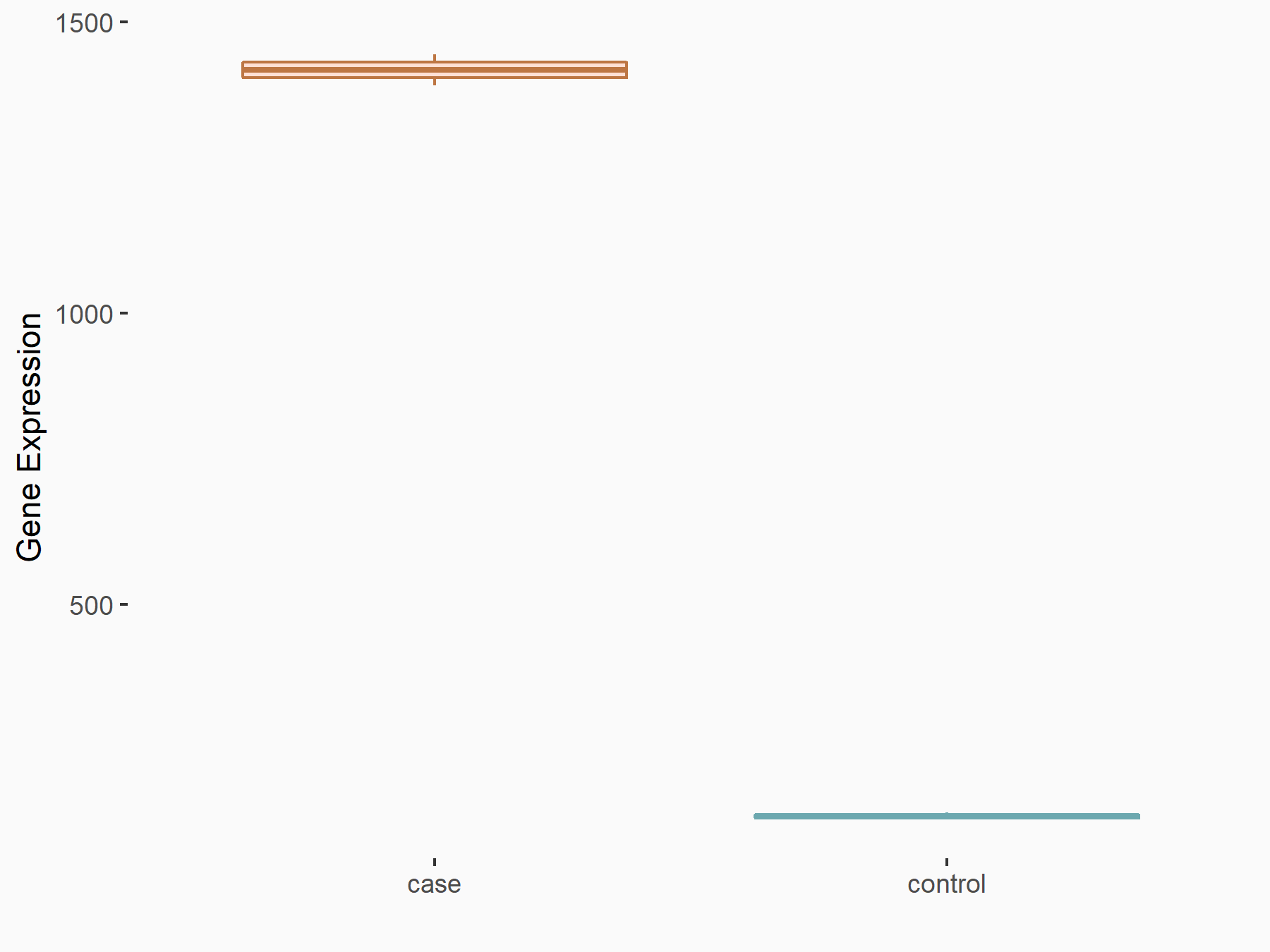 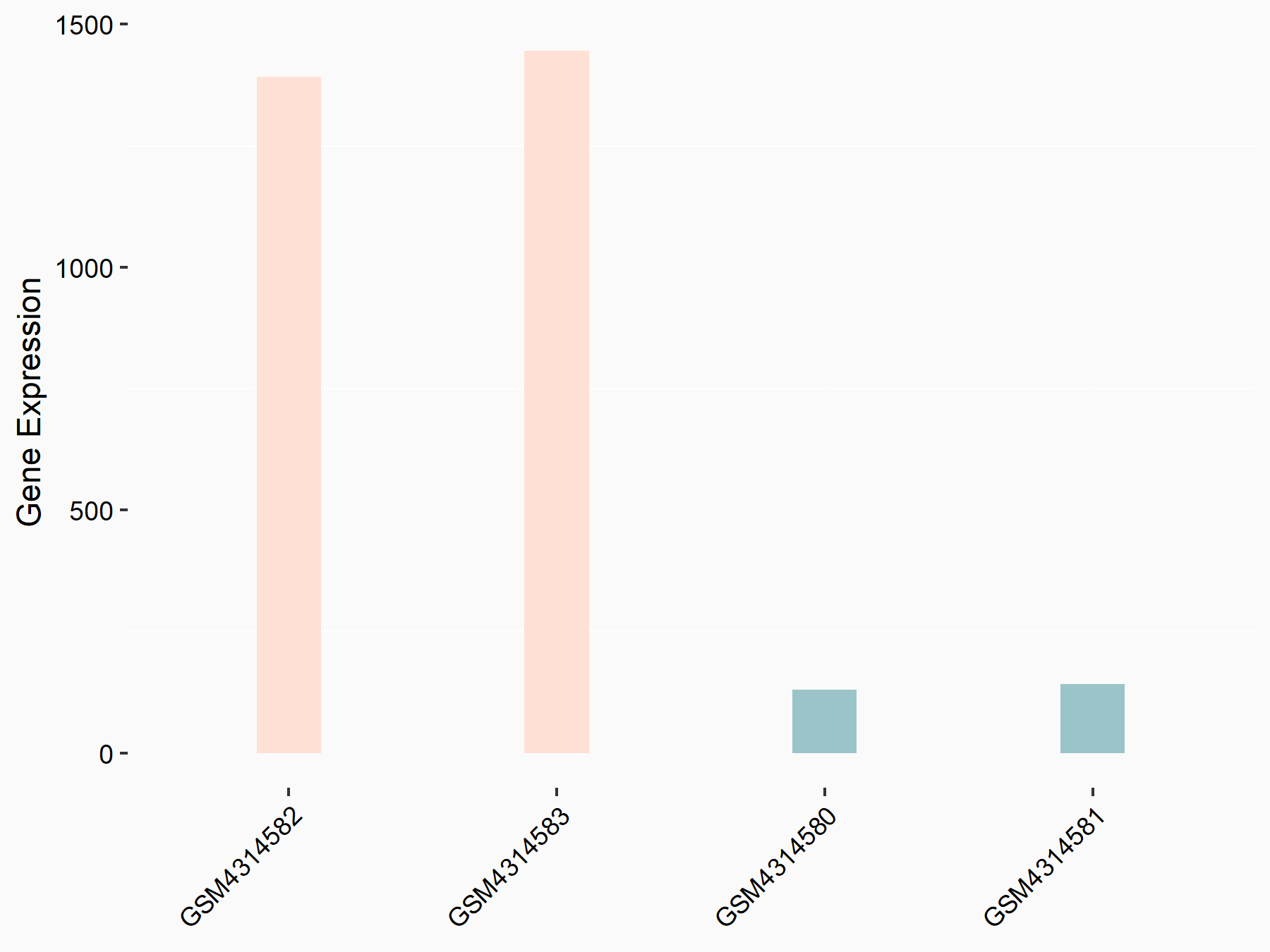 |
logFC: 3.39E+00 p-value: 6.45E-138 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 3.75E+00 | GSE60213 |
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cellular proliferation and survival | |||
In-vitro Model |
UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| In-vivo Model | For induction of BCa, 6-8-week-old mice were treated with drinking water containing 500 ug/ml BBN for 16 weeks and then given normal water for another 10 weeks. Tamoxifen was intraperitonelly injected to the mice with 0.08 mg/g of body weight each day for 3 days in order to inductively knock out the target gene. | |||
| Response Summary | Deletion of Mettl3 leads to the suppression of Angiopoietin-1 receptor (TEK) and VEGF-A,ablation of Mettl3 in bladder urothelial attenuates the oncogenesis and tumor angiogenesis of bladder cancer. | |||
Retinopathy [ICD-11: 9B71]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [264] | |||
| Responsed Disease | Retinopathy [ICD-11: 9B71] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | Mouse pups, along with their nursing mothers, were exposed to 75 ± 2% O2 in an incubator between postnatal day (P) 7 and P12 and were then returned to room air. | |||
Apoptosis regulator BAX (BAX)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Embryonic stem cells | Mus musculus |
|
Treatment: METTL3 knockout mESCs
Control: Wild type mESCs
|
GSE156481 | |
| Regulation |
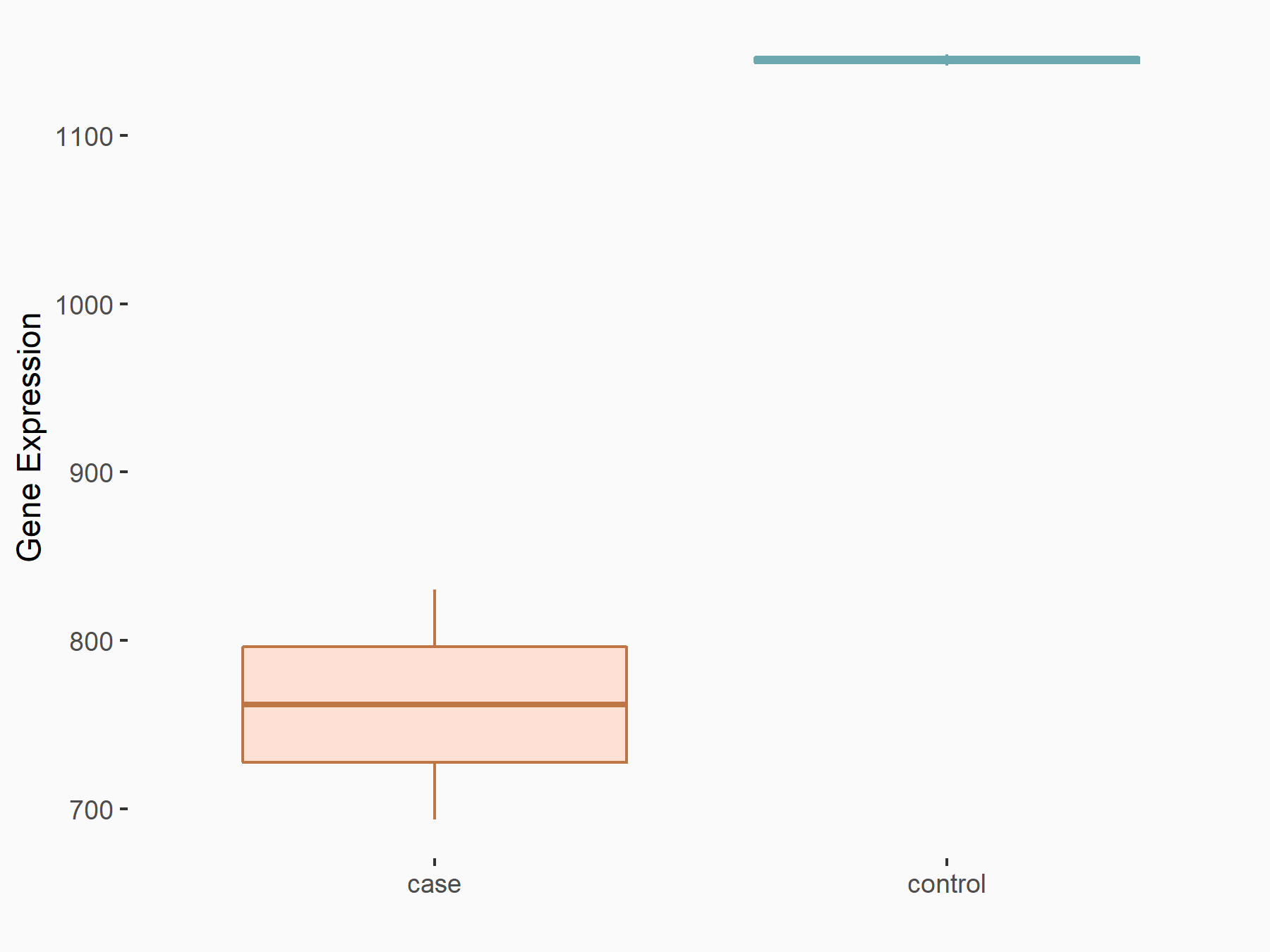 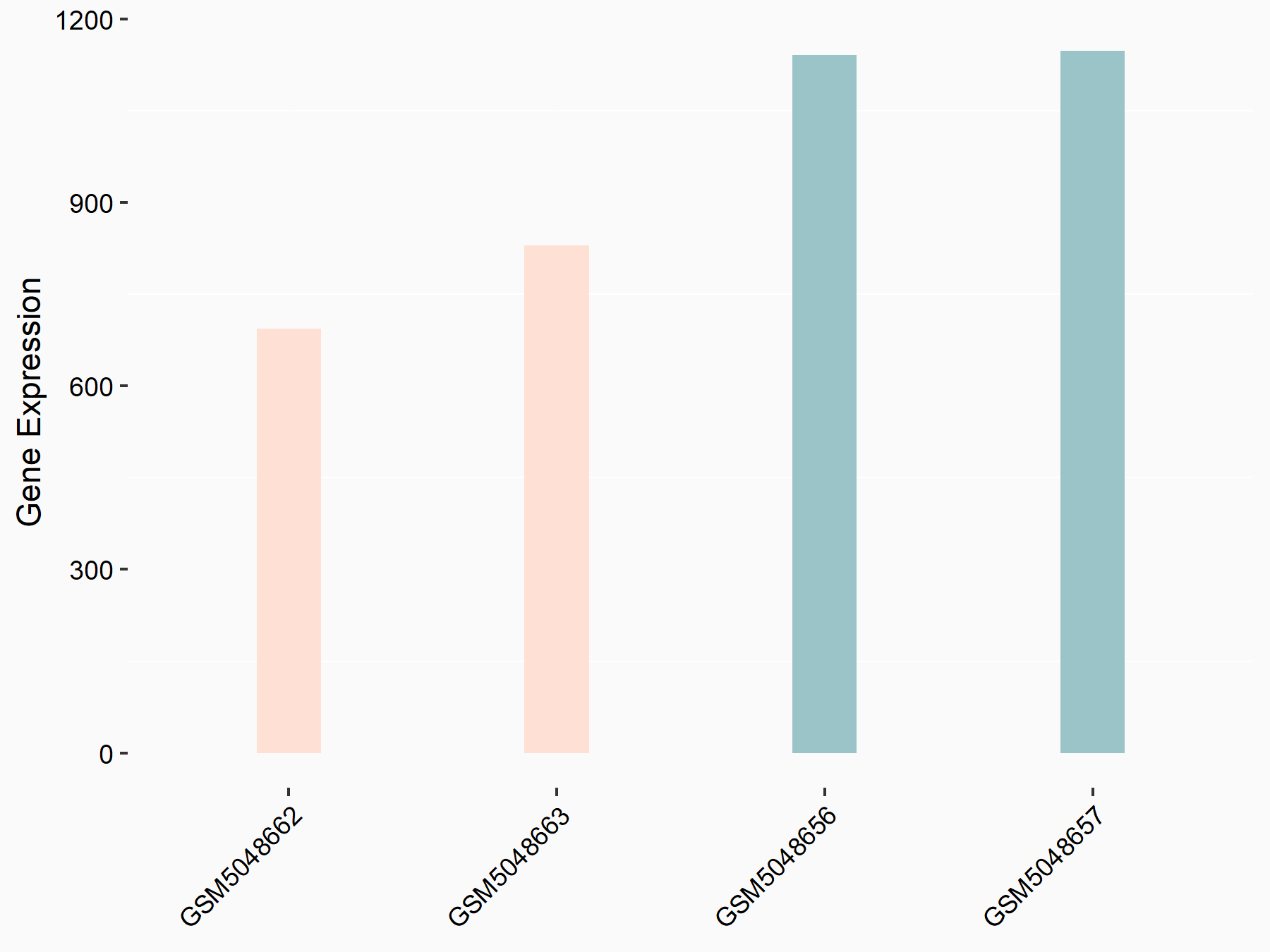 |
logFC: -5.89E-01 p-value: 1.41E-07 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.02E+01 | GSE60213 |
Enterovirus [ICD-11: 1A2Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Enterovirus [ICD-11: 1A2Y] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| Cell autophagy | ||||
In-vitro Model |
Schwann cells (A type of glial cell that surrounds neurons) | |||
| Response Summary | Knocking down METTL3 prevented Enterovirus 71-induced cell death and suppressed Enterovirus 71-induced expression of Apoptosis regulator BAX (BAX) while rescuing Bcl-2 expression after Enterovirus 71 infection. Knocking down METTL3 inhibited Enterovirus 71-induced expression of Atg5, Atg7 and LC3 II. Knocking down METTL3 inhibited Enterovirus 71-induced apoptosis and autophagy. | |||
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [8] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| Response Summary | Down-regulation of METTL3 inhibits the proliferation and mobility of human gastric cancer cells and leads to inactivation of the AKT signaling pathway, suggesting that METTL3 is a potential target for the treatment of human gastric cancer. METTL3 knockdown decreased Bcl2 and increased Apoptosis regulator BAX (BAX) and active Caspase-3 in gastric cancer cells, which suggested the apoptotic pathway was activated. METTL3 led to inactivation of the AKT signaling pathway in human gastric cancer cells, including decreased phosphorylation levels of AKT and expression of down-stream effectors p70S6K and Cyclin D1. | |||
Apoptosis regulator Bcl-2 (BCL2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Th1 cell line | Mus musculus |
|
Treatment: METTL3 knockout splenic Th1 cells
Control: Wild type splenic Th1 cells
|
GSE129648 | |
| Regulation |
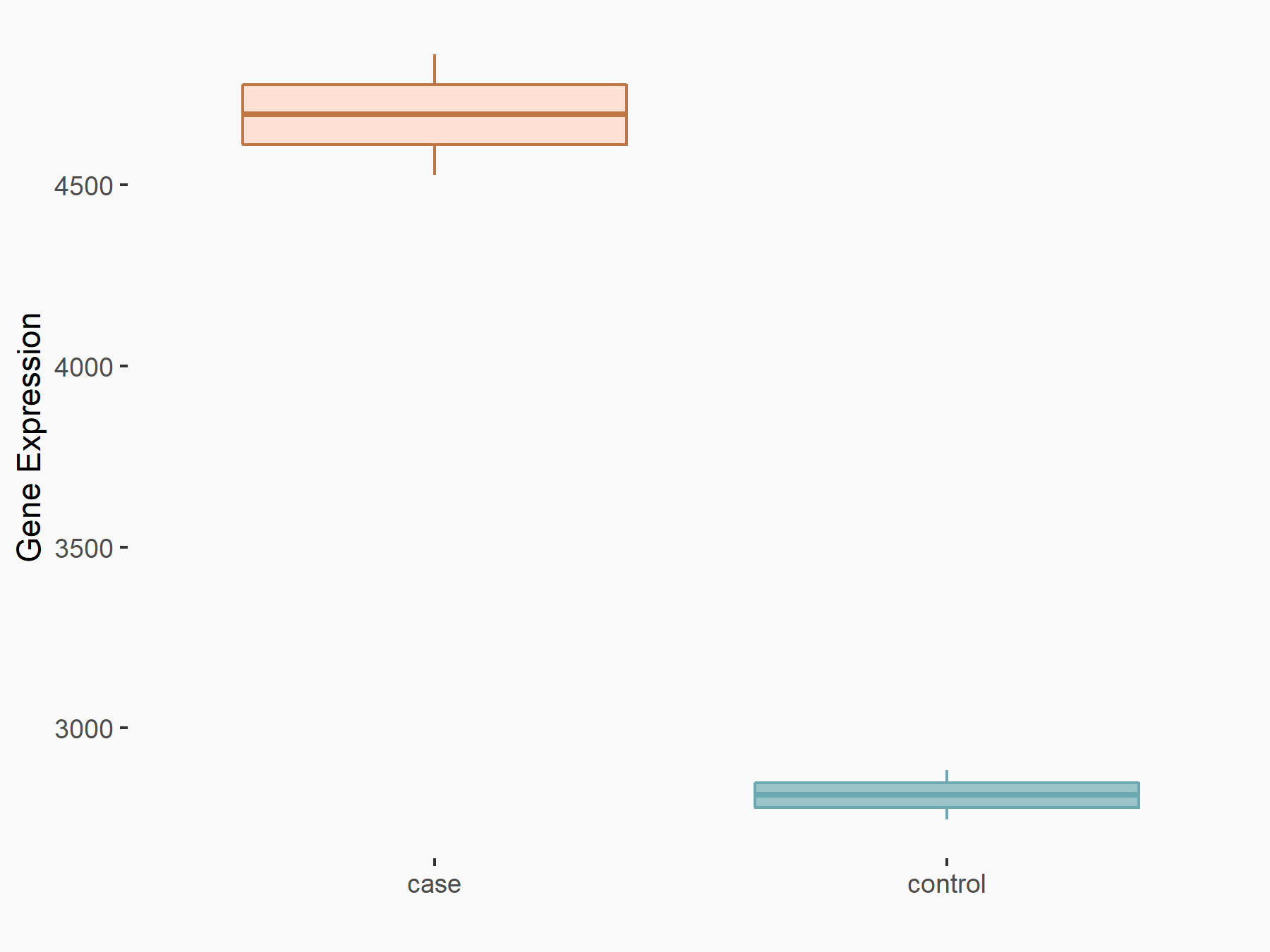 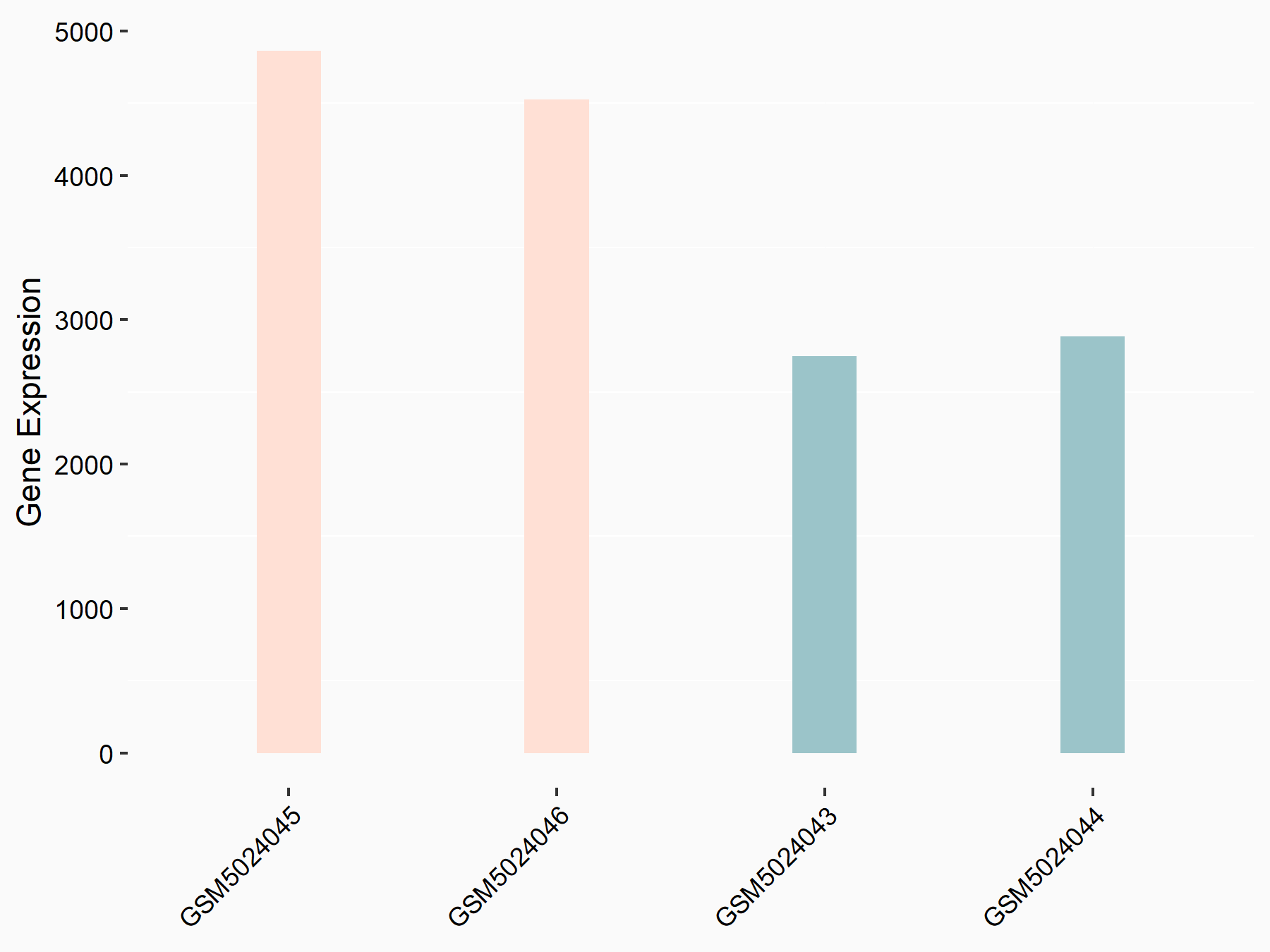 |
logFC: 7.38E-01 p-value: 3.55E-06 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.70E+00 | GSE60213 |
Enterovirus [ICD-11: 1A2Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Enterovirus [ICD-11: 1A2Y] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| Cell autophagy | ||||
In-vitro Model |
Schwann cells (A type of glial cell that surrounds neurons) | |||
| Response Summary | Knocking down METTL3 prevented Enterovirus 71-induced cell death and suppressed Enterovirus 71-induced expression of BAX while rescuing Apoptosis regulator Bcl-2 (BCL2) expression after Enterovirus 71 infection. Knocking down METTL3 inhibited Enterovirus 71-induced expression of Atg5, Atg7 and LC3 II. Knocking down METTL3 inhibited Enterovirus 71-induced apoptosis and autophagy. | |||
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [9] | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell differentiation and apoptosis | |||
In-vitro Model |
HSPC (Human hematopoietic stem cell) | |||
| In-vivo Model | 500,000 selected cells were injected via tail vein or retro-orbital route into female NSG (6-8 week old) recipient mice that had been sublethally irradiated with 475 cGy one day before transplantation. | |||
| Response Summary | METTL3 depletion in human myeloid leukemia cell lines induces cell differentiation and apoptosis and delays leukemia progression in recipient mice in vivo. Single-nucleotide-resolution mapping of m6A coupled with ribosome profiling reveals that m6A promotes the translation of c-MYC, Apoptosis regulator Bcl-2 (BCL2) and PTEN mRNAs in the human acute myeloid leukemia MOLM-13 cell line. Moreover, loss of METTL3 leads to increased levels of phosphorylated AKT. | |||
B-cell lymphomas [ICD-11: 2A86]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [10] | |||
| Responsed Disease | B-cell lymphomas [ICD-11: 2A86] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
In-vitro Model |
ATDC-5 | Mouse teratocarcinoma | Mus musculus | CVCL_3894 |
| In-vivo Model | For MIA + SAH control, S-adenosylhomocysteine (SAH), Mettl3 inhibitor (10 mg/kg) (MCE, NJ, USA) was injected intraperitoneally before MIA injection and maintained twice a week until mice were sacrificed. | |||
| Response Summary | Mettl3 inhibitor, S-adenosylhomocysteine promoted the apoptosis and autophagy of chondrocytes with inflammation in vitro and aggravated the degeneration of chondrocytes and subchondral bone in monosodium iodoacetate (MIA) induced temporomandibular joint osteoarthritis mice in vivo. Bcl2 protein interacted with Beclin1 protein in chondrocytes induced by TNF-alpha stimulation. Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through m6A/Ythdf1/Apoptosis regulator Bcl-2 (BCL2) signal axis which provides promising therapeutic strategy for temporomandibular joint osteoarthritis. | |||
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [8] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| Response Summary | Down-regulation of METTL3 inhibits the proliferation and mobility of human gastric cancer cells and leads to inactivation of the AKT signaling pathway, suggesting that METTL3 is a potential target for the treatment of human gastric cancer. METTL3 knockdown decreased Apoptosis regulator Bcl-2 (BCL2) and increased Bax and active Caspase-3 in gastric cancer cells, which suggested the apoptotic pathway was activated. METTL3 led to inactivation of the AKT signaling pathway in human gastric cancer cells, including decreased phosphorylation levels of AKT and expression of down-stream effectors p70S6K and Cyclin D1. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [11] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | The mice were housed with filtered air, 12 h light/dark cycle, constant temperature (25℃), relative humidity (50±5%) and free access to food and water. In order to establish a human NSCLC xenograft model, 5×106 H1299, sh-METTL3-H1299 and METTL3 stably overexpressed H1299 cells (2×106 per mouse) were subcutaneously injected into mice. Tumor growth was observed daily. Tumor volume was calculated as follows: 0.5× (length × width2). At 24 days post-inoculation, the maximum diameter exhibited by a single subcutaneous tumor was 15 mm and mice were anesthetized by intraperitoneal administration of sodium pentobarbital (50 mg/kg), then sacrificed by cervical dislocation. | |||
| Response Summary | METTL3 regulated cellular growth, survival and migration in non-small cell lung cancer. METTL3 promoted non-small cell lung cancer progression by modulating the level of Apoptosis regulator Bcl-2 (BCL2). | |||
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [12] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
MCF-10A | Normal | Homo sapiens | CVCL_0598 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| In-vivo Model | Mice were maintained at 22 ± 2 ℃ with a humidity of 35 ± 5% under a 12 h light and 12 h dark cycle, with free access to water and food. For the HFD experiment, female control (Ftoflox/flox) and adipose-selective fto knockout (Fabp4-Cre Ftoflox/flox, fto-AKO) mice were fed with high-fat diet (60% fat in calories; Research Diets, D12492) for the desired periods of time, and food intake and body weight were measured every week after weaning (at 3 weeks of age). | |||
| Response Summary | Apoptosis regulator Bcl-2 (BCL2) acted as the target of METTL3, thereby regulating the proliferation and apoptosis of breast cancer. | |||
Dentofacial anomalies [ICD-11: DA0E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [10] | |||
| Responsed Disease | Temporomandibular joint disorders [ICD-11: DA0E.8] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
In-vitro Model |
ATDC-5 | Mouse teratocarcinoma | Mus musculus | CVCL_3894 |
| In-vivo Model | For MIA + SAH control, S-adenosylhomocysteine (SAH), Mettl3 inhibitor (10 mg/kg) (MCE, NJ, USA) was injected intraperitoneally before MIA injection and maintained twice a week until mice were sacrificed. | |||
| Response Summary | Mettl3 inhibitor, S-adenosylhomocysteine promoted the apoptosis and autophagy of chondrocytes with inflammation in vitro and aggravated the degeneration of chondrocytes and subchondral bone in monosodium iodoacetate (MIA) induced temporomandibular joint osteoarthritis mice in vivo. Bcl2 protein interacted with Beclin1 protein in chondrocytes induced by TNF-alpha stimulation. Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through m6A/Ythdf1/Apoptosis regulator Bcl-2 (BCL2) signal axis which provides promising therapeutic strategy for temporomandibular joint osteoarthritis. | |||
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [267] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Responsed Drug | Celastrol | Preclinical | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| In-vivo Model | AsPC-1 cells suspended in 100 μl of PBS (2 × 106 cells/100 μl) were injected subcutaneously into the lateral flank of the mice, which were randomly divided into solvent group (n = 6), 1.0 mg/kg of celastrol group (n = 6) and 3.0 mg/kg of celastrol group (n = 6). The administration of celastrol was performed by intraperitoneal injection into tumor-bearing mice every 2 day after 10 day inoculation. The tumor sizes were monitored with calipers every 5 days, and the tumor volume was calculated with the formula: Volume (mm3) = 1/2 × length × width2. | |||
Injuries of spine or trunk [ICD-11: ND51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [268] | |||
| Responsed Disease | Spinal cord injury [ICD-11: ND51.2] | |||
| Responsed Drug | STM2457 | Investigative | ||
| Target Regulation | Down regulation | |||
In-vitro Model |
PC12 | Rat adrenal gland pheochromocytoma | Rattus norvegicus | CVCL_0481 |
| In-vivo Model | Rats were anesthetized with intraperitoneal injection of 1% sodium pentobarbital (20 mg/kg). For constructing a rat spinal cord hemisection model, the spinal colon was marked on the T9 spinous process and the skin was incised until exposing the T9-10 spinous process. The spinal cord was completely exposed by biting the vertebral plate with biting forceps. The spinal cord was cut on one side with ophthalmic scissors centered on the central canal of the spinal cord. | |||
Aspartate--tRNA ligase, cytoplasmic (DARS)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3 knockout liver
Control: Wild type liver cells
|
GSE198513 | |
| Regulation |
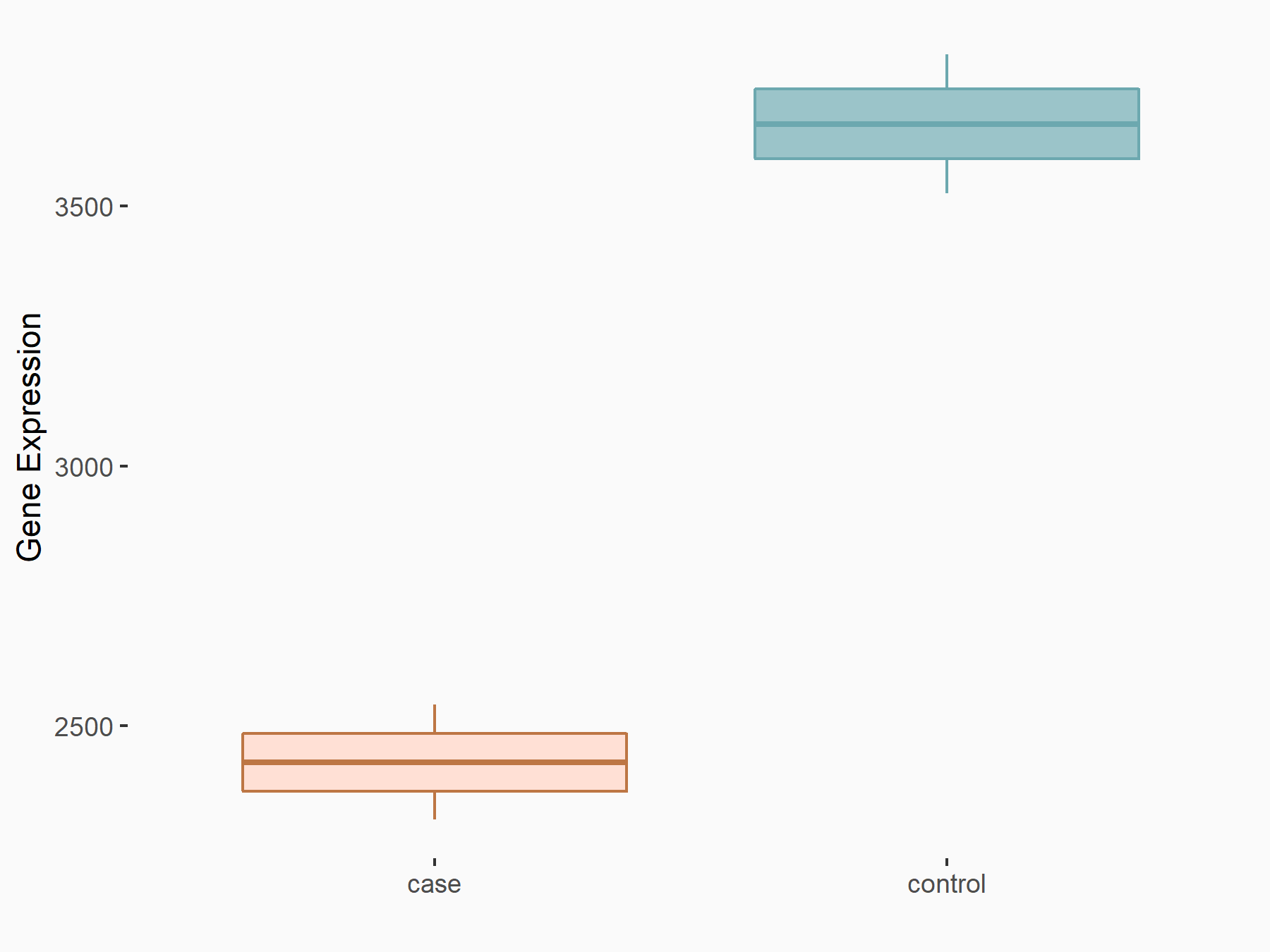 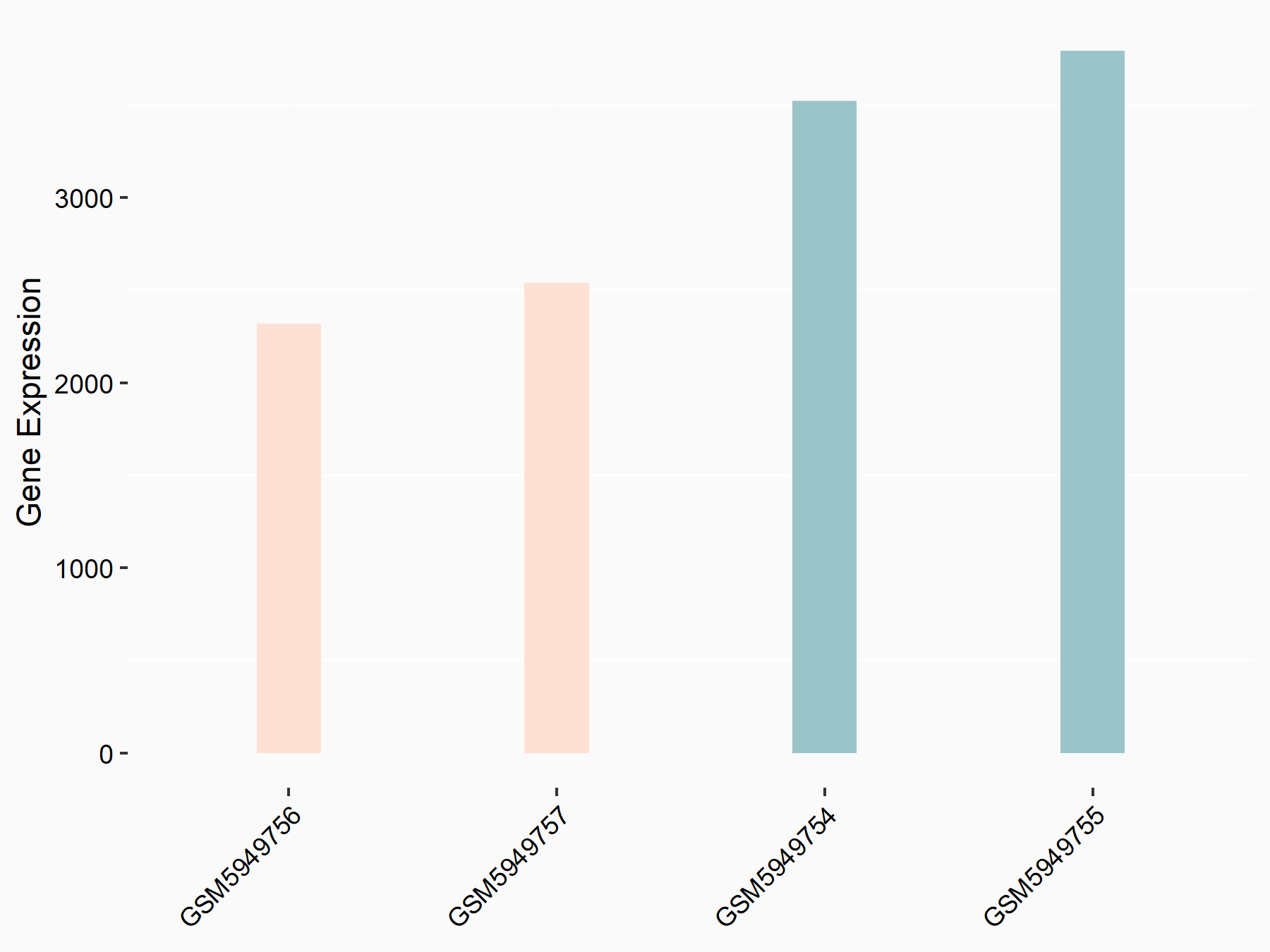 |
logFC: -5.90E-01 p-value: 7.45E-13 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.22E+00 | GSE60213 |
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [13] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell autophagy | |||
In-vitro Model |
SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| End1/E6E7 | Normal | Homo sapiens | CVCL_3684 | |
| DoTc2 4510 | Cervical carcinoma | Homo sapiens | CVCL_1181 | |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| Response Summary | DARS-AS1 was validated to facilitate DARS translation via recruiting METTL3 and METTL14, which bound with DARS mRNA Aspartate--tRNA ligase, cytoplasmic (DARS) mRNA 5' untranslated region (5'UTR) and promoting its translation. The present study demonstrated that the 'HIF1-Alpha/DARS-AS1/DARS/ATG5/ATG3' pathway regulated the hypoxia-induced cytoprotective autophagy of cervical cancer(CC) and is a promising target of therapeutic strategies for patients afflicted with CC. | |||
ATP-binding cassette sub-family C member 9 (ABCC9)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
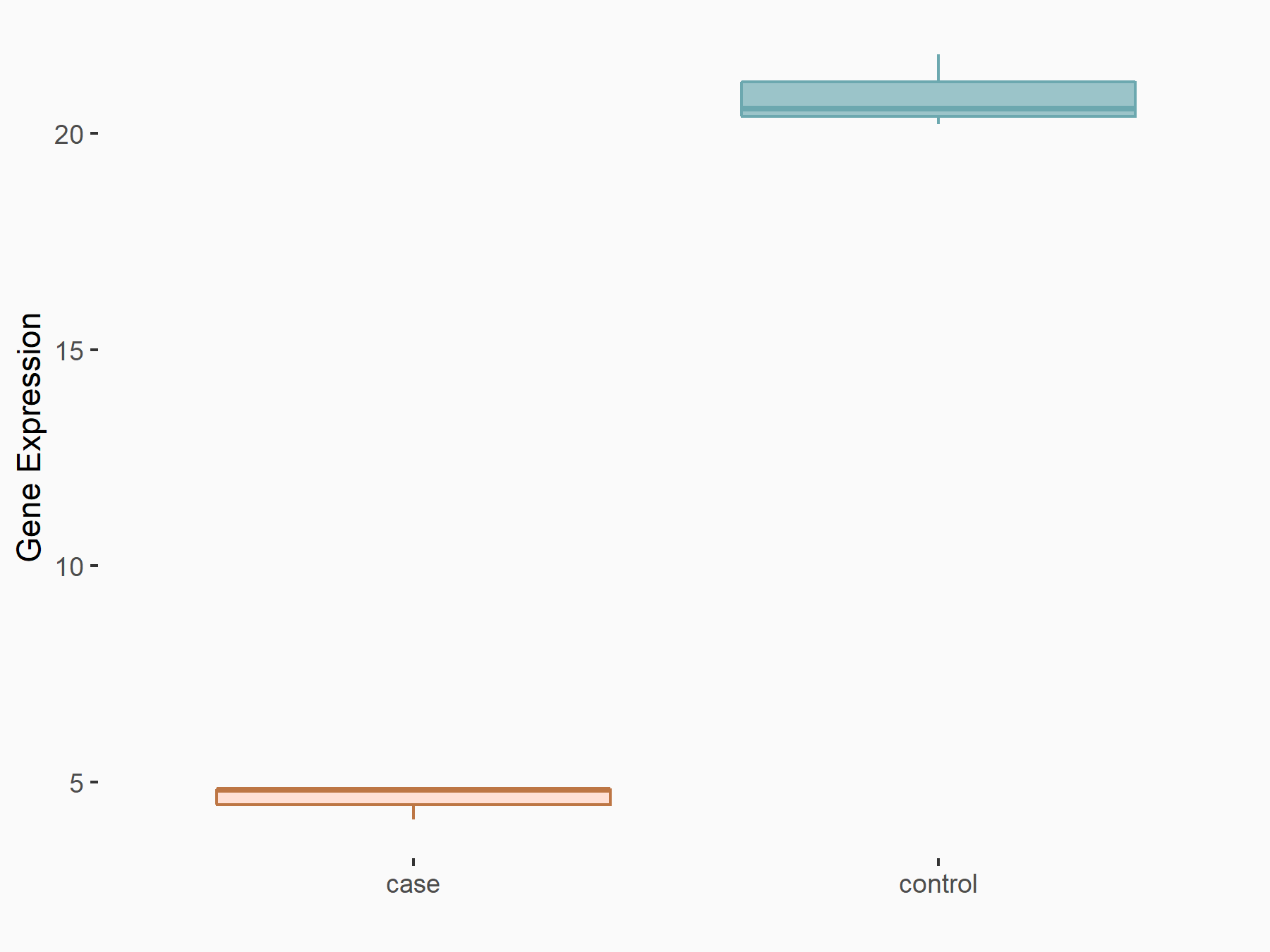  |
logFC: -1.97E+00 p-value: 5.10E-08 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.69E+00 | GSE60213 |
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [14] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | ABC transporters | hsa02010 | ||
| Wnt signaling pathway | hsa04310 | |||
| Ubiquitin mediated proteolysis | hsa04120 | |||
| Cell Process | Ubiquitination degradation | |||
In-vitro Model |
CNE-1 | Normal | Homo sapiens | CVCL_6888 |
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| In-vivo Model | A total of 2 × 106 cells was mixed with 0.2 ml PBS (pH 7.4) and 30% (v/v) Matrigel matrix (BD Biosciences). | |||
| Response Summary | TRIM11 regulates nasopharyngeal carcinoma drug resistance by positively modulating the Daple/beta-catenin/ATP-binding cassette sub-family C member 9 (ABCC9) signaling pathway. TRIM11 enhanced the multidrug resistance in NPC by inhibiting apoptosis in vitro and promoting cisplatin (DDP) resistance in vivo. METTL3-mediated m6A modification caused the upregulation of TRIM11 via IGF2BP2 in NPC drug-resistant cells. | |||
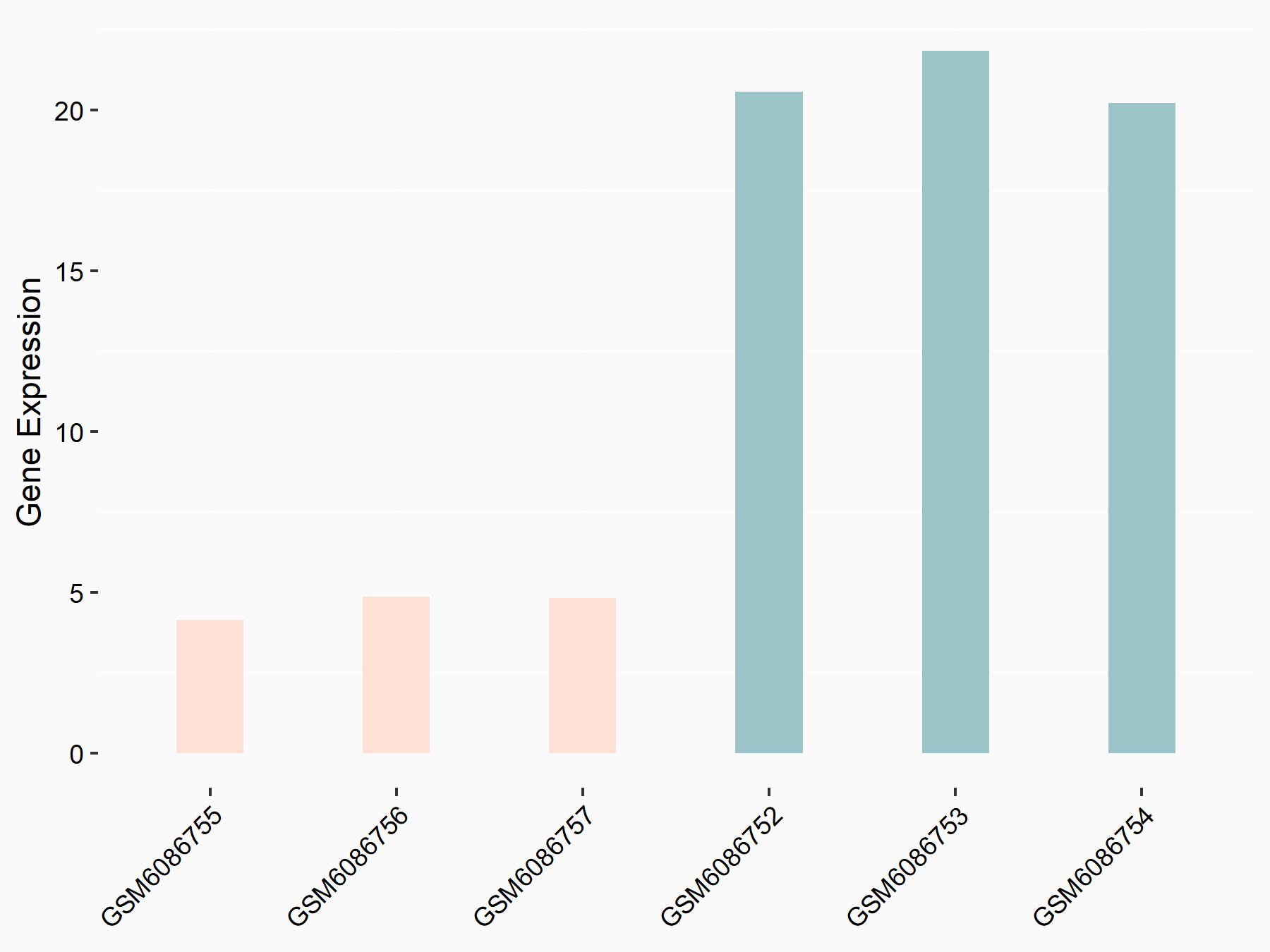
ATP-binding cassette sub-family D member 1 (ABCD1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shMETTL3 MOLM13 cells
Control: MOLM13 cells
|
GSE98623 | |
| Regulation |
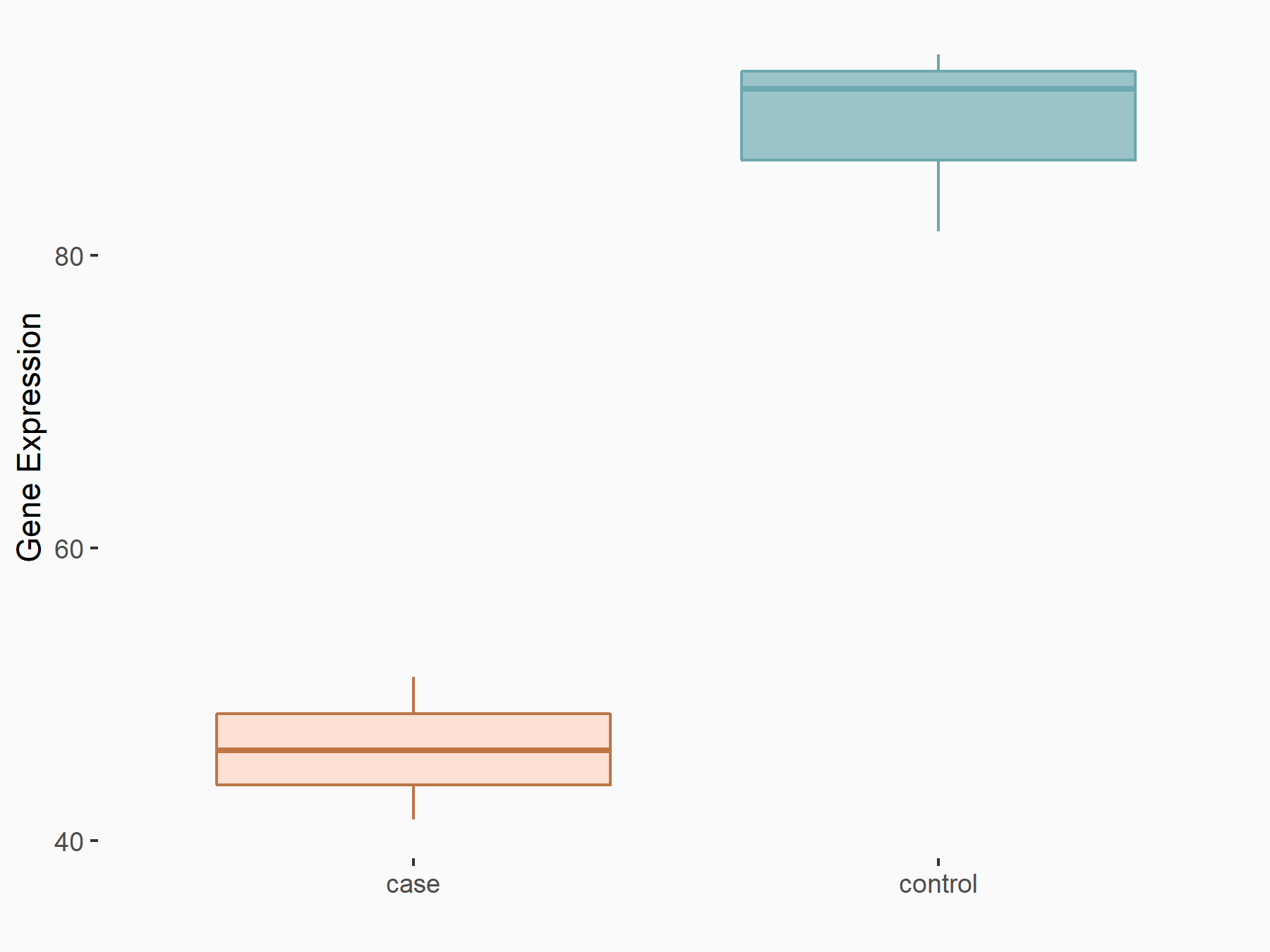 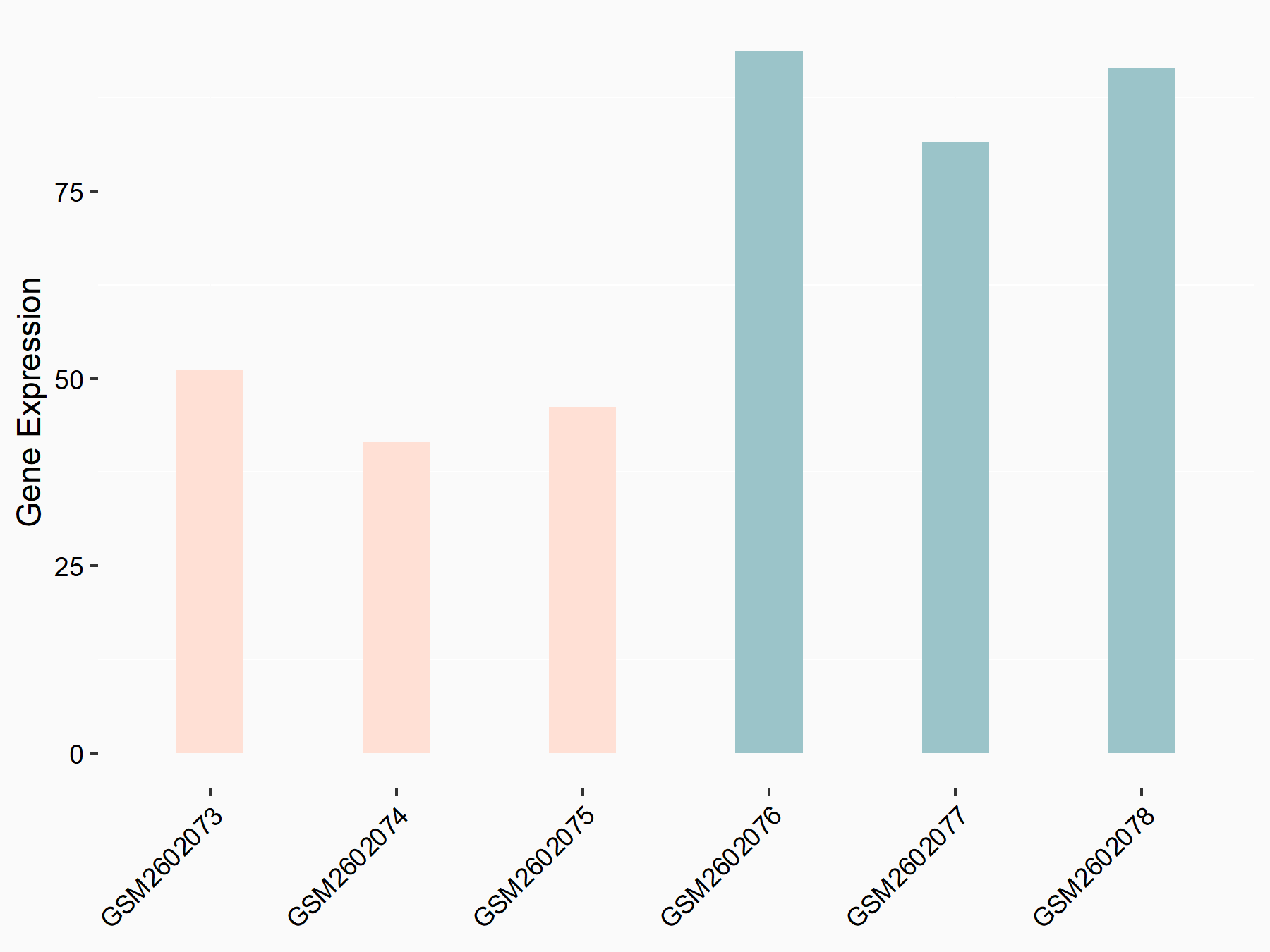 |
logFC: -9.42E-01 p-value: 1.69E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 8.82E+00 | GSE60213 |
Renal cell carcinoma [ICD-11: 2C90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [15] | |||
| Responsed Disease | Renal cell carcinoma of kidney [ICD-11: 2C90.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | ABC transporters | hsa02010 | ||
| Cell Process | Cell migration and spheroid formation | |||
In-vitro Model |
786-O | Renal cell carcinoma | Homo sapiens | CVCL_1051 |
| A-498 | Renal cell carcinoma | Homo sapiens | CVCL_1056 | |
| In-vivo Model | A498 cells (1 × 106 cells) were resuspended in 100 uL of PBS and subcutaneously injected into the axillary fossa of nude mice (BALB/c-nude, 4 weeks old). | |||
| Response Summary | Knockdown of METTL3 in clear cell renal cell carcinoma cell line impaired both cell migration capacity and tumor spheroid formation in soft fibrin gel, a mechanical method for selecting stem-cell-like tumorigenic cells. METTL3 knockdown cells and functional studies confirmed that translation of ATP-binding cassette sub-family D member 1 (ABCD1). | |||
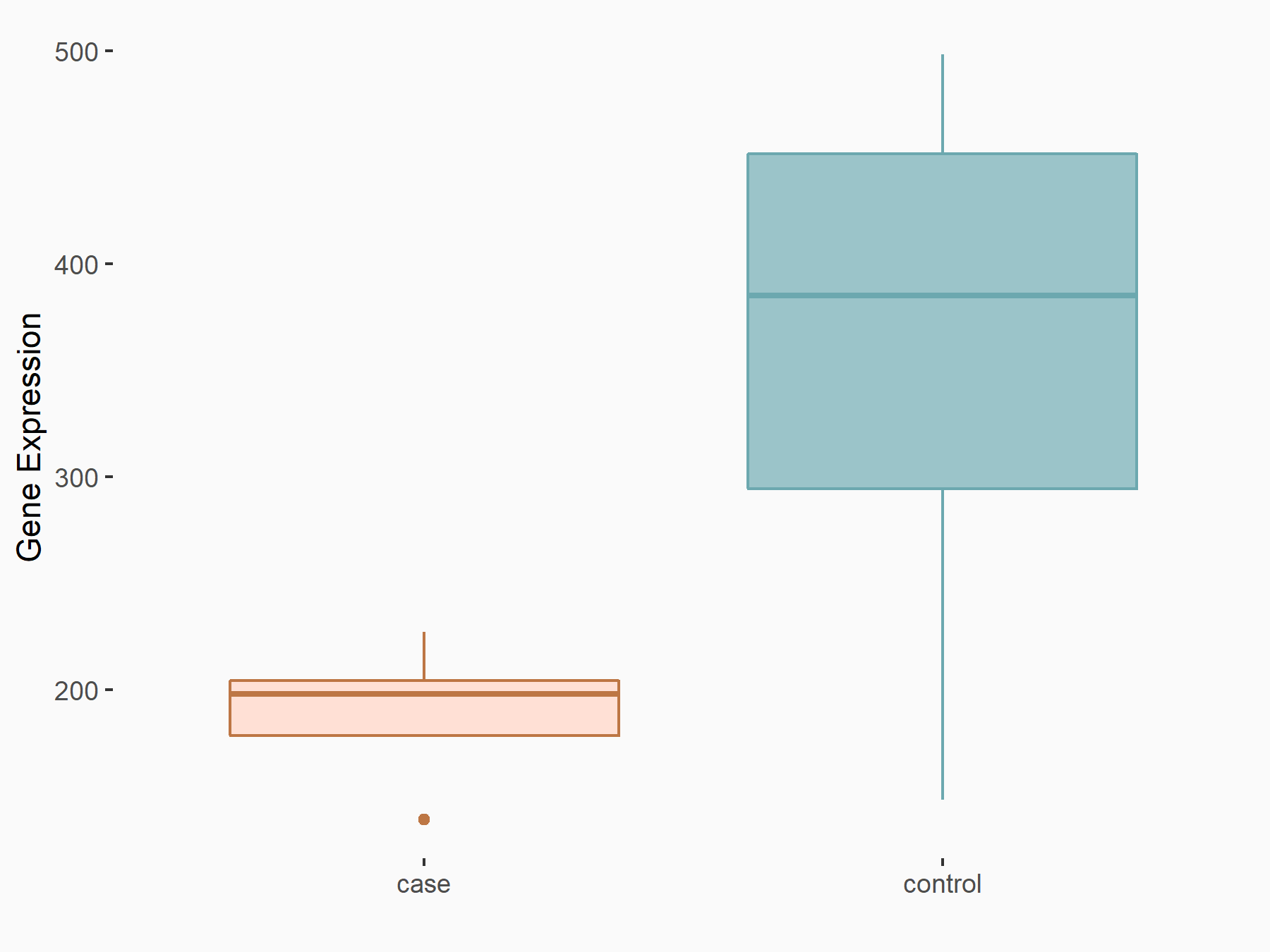
ATPase family AAA domain-containing protein 2 (ATAD2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | DKO-1 cell line | Homo sapiens |
|
Treatment: METTL3 knockdown DKO-1 cell
Control: DKO-1 cell
|
GSE182382 | |
| Regulation |
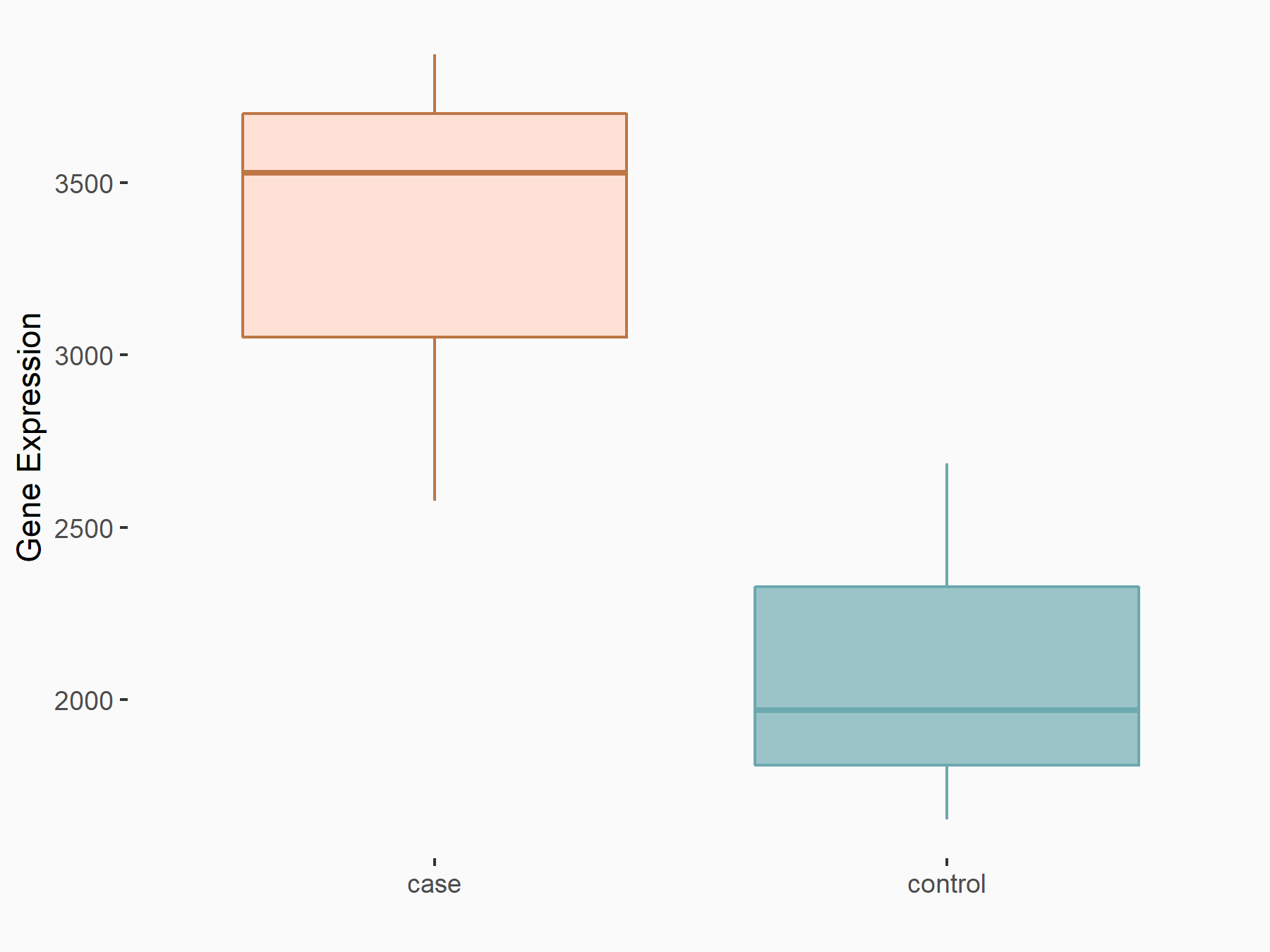 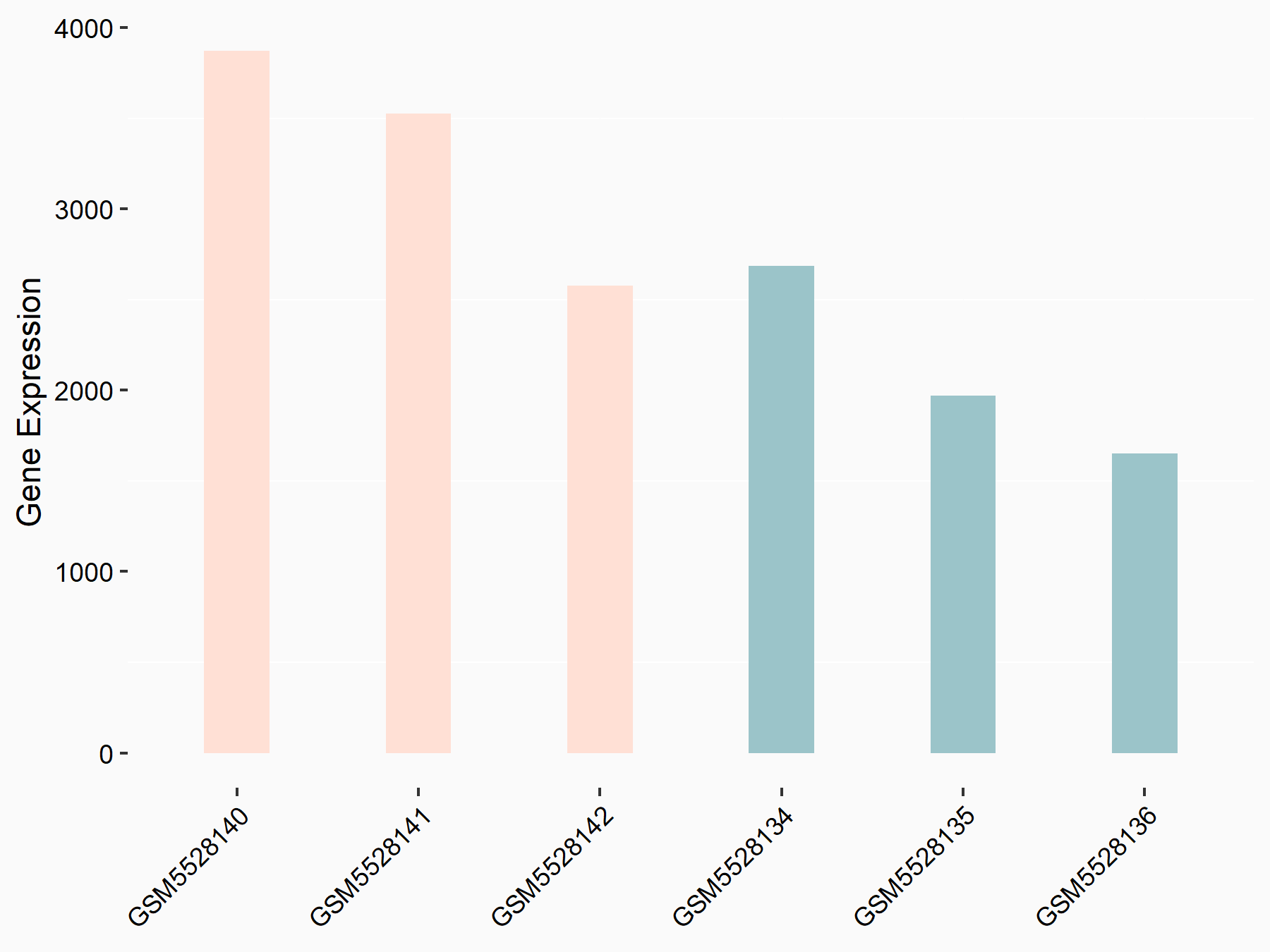 |
logFC: 6.62E-01 p-value: 6.71E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.75E+00 | GSE60213 |
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [16] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation and invasion | |||
| Cell apoptosis | ||||
In-vitro Model |
HOS | Osteosarcoma | Homo sapiens | CVCL_0312 |
| MG-63 | Osteosarcoma | Homo sapiens | CVCL_0426 | |
| SaOS-2 | Osteosarcoma | Homo sapiens | CVCL_0548 | |
| U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 | |
| Response Summary | METTL3 functions as an oncogene in the growth and invasion of osteosarcoma by regulating ATPase family AAA domain-containing protein 2 (ATAD2), suggesting a potential therapeutic target for osteosarcoma treatment. | |||
Autophagy protein 5 (ATG5)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | CT26 cell line | Mus musculus |
|
Treatment: METTL3 knockout CT26 cells
Control: CT26 cells
|
GSE142589 | |
| Regulation |
 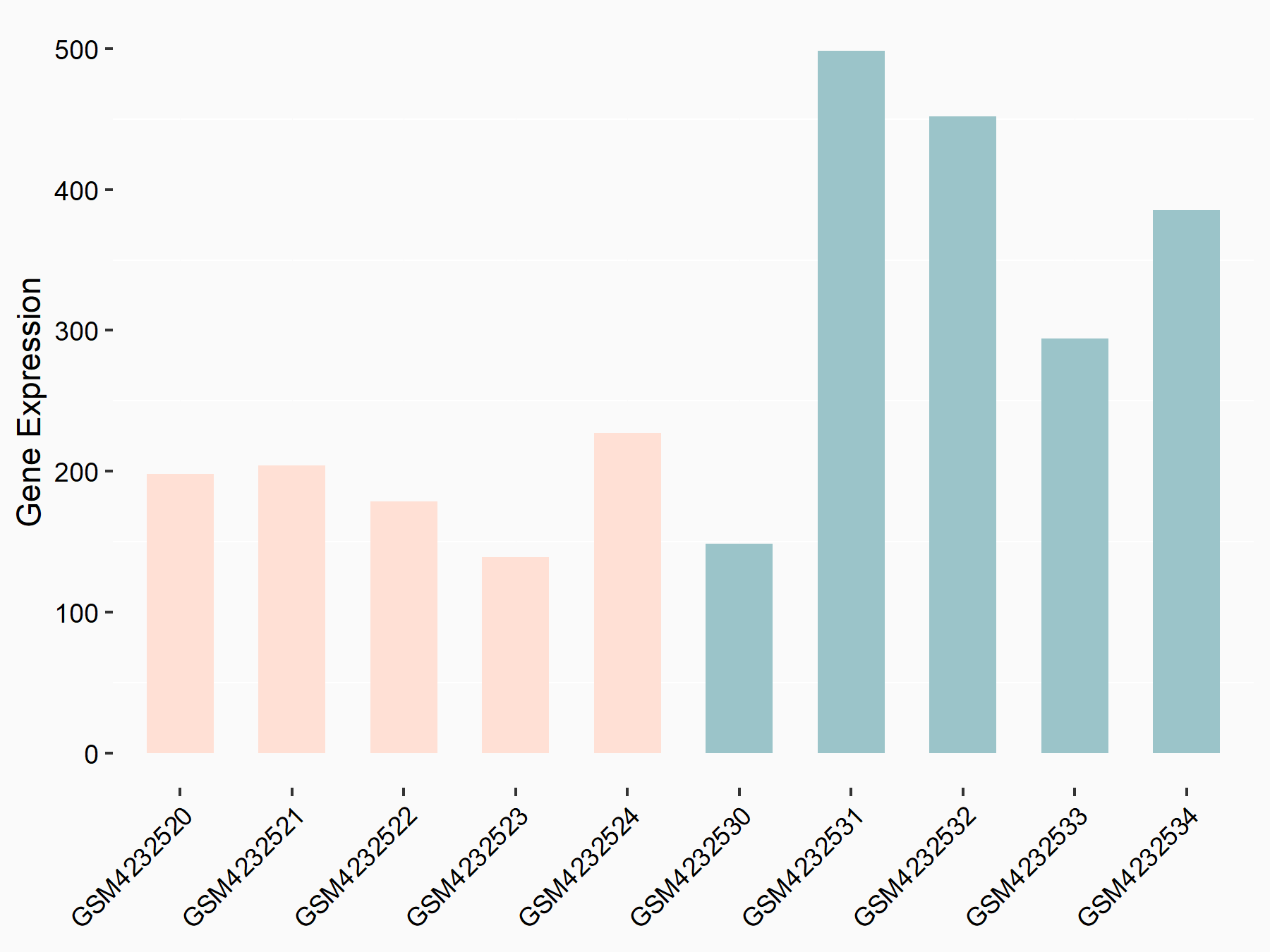 |
logFC: -9.09E-01 p-value: 4.91E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 8.30E+00 | GSE60213 |
Enterovirus [ICD-11: 1A2Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Enterovirus [ICD-11: 1A2Y] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| Cell autophagy | ||||
In-vitro Model |
Schwann cells (A type of glial cell that surrounds neurons) | |||
| Response Summary | Knocking down METTL3 prevented Enterovirus 71-induced cell death and suppressed Enterovirus 71-induced expression of Bax while rescuing Bcl-2 expression after Enterovirus 71 infection. Knocking down METTL3 inhibited Enterovirus 71-induced expression of Autophagy protein 5 (ATG5), Atg7 and LC3 II. Knocking down METTL3 inhibited Enterovirus 71-induced apoptosis and autophagy. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [17] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, Autophagy protein 5 (ATG5), ATG12, and ATG16L1. | |||
Lung cancer [ICD-11: 2C25]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Chloroquine | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
In-vitro Model |
Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as Autophagy protein 5 (ATG5), ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Gefitinib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
In-vitro Model |
Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as Autophagy protein 5 (ATG5), ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Experiment 3 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Beta-Elemen | Phase 3 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
In-vitro Model |
Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as Autophagy protein 5 (ATG5), ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
Testicular cancer [ICD-11: 2C80]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [19] | |||
| Responsed Disease | Testicular cancer [ICD-11: 2C80] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular Processes | |||
| Cellular Transport | ||||
| Cellular catabolism | ||||
| Cell autophagy | ||||
In-vitro Model |
Tcam-2/DDP (Cisplatin-resistant TCam-2 cell line) | |||
| TCam-2 | Testicular seminoma | Homo sapiens | CVCL_T012 | |
| Response Summary | m6A methyltransferase METTL3 regulates autophagy and sensitivity to cisplatin by targeting Autophagy protein 5 (ATG5) in seminoma. The use of autophagy inhibitors 3-MA could reverse the protective effect of METTL3 on TCam-2 cells. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [19] | |||
| Responsed Disease | Testicular cancer [ICD-11: 2C80] | |||
| Responsed Drug | 3-Methyladenine | Investigative | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular Processes | |||
| Cellular Transport | ||||
| Cellular catabolism | ||||
| Cell autophagy | ||||
In-vitro Model |
Tcam-2/DDP (Cisplatin-resistant TCam-2 cell line) | |||
| TCam-2 | Testicular seminoma | Homo sapiens | CVCL_T012 | |
| Response Summary | m6A methyltransferase METTL3 regulates autophagy and sensitivity to cisplatin by targeting Autophagy protein 5 (ATG5) in seminoma. The use of autophagy inhibitors 3-MA could reverse the protective effect of METTL3 on TCam-2 cells. | |||
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [270] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Responsed Drug | Spautin 1 | Preclinical | ||
| Target Regulation | Up regulation | |||
| In-vivo Model | For tumor growth assay, 2 × 106 OS cells in 100 μL medium were injected into the nude mice subcutaneously. For tumor metastasis assay, we injected medium containing 2 × 106 cells through the caudal vein. An IVIS200 imaging system (Caliper Life Science, USA) was used to image and assess the OS metastasis. For pharmacological inhibition of USP13, mice bearing xenografts were treated with Spautin-1 (40 mg/kg/day i.p.) or vehicle for 2 weeks. | |||
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [271] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
Autophagy-related protein 16-1 (ATG16L1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | CT26 cell line | Mus musculus |
|
Treatment: METTL3 knockout CT26 cells
Control: CT26 cells
|
GSE142589 | |
| Regulation |
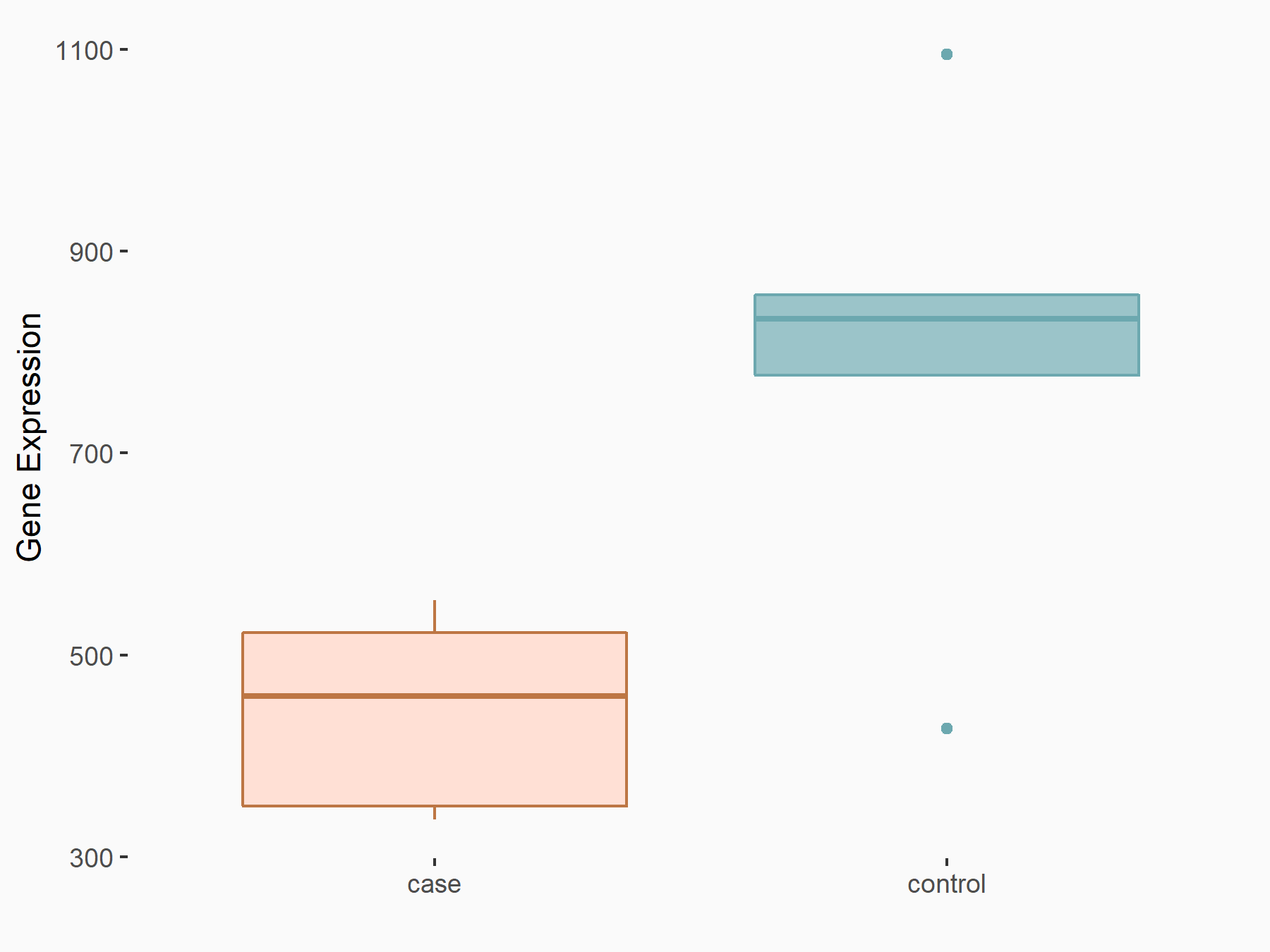 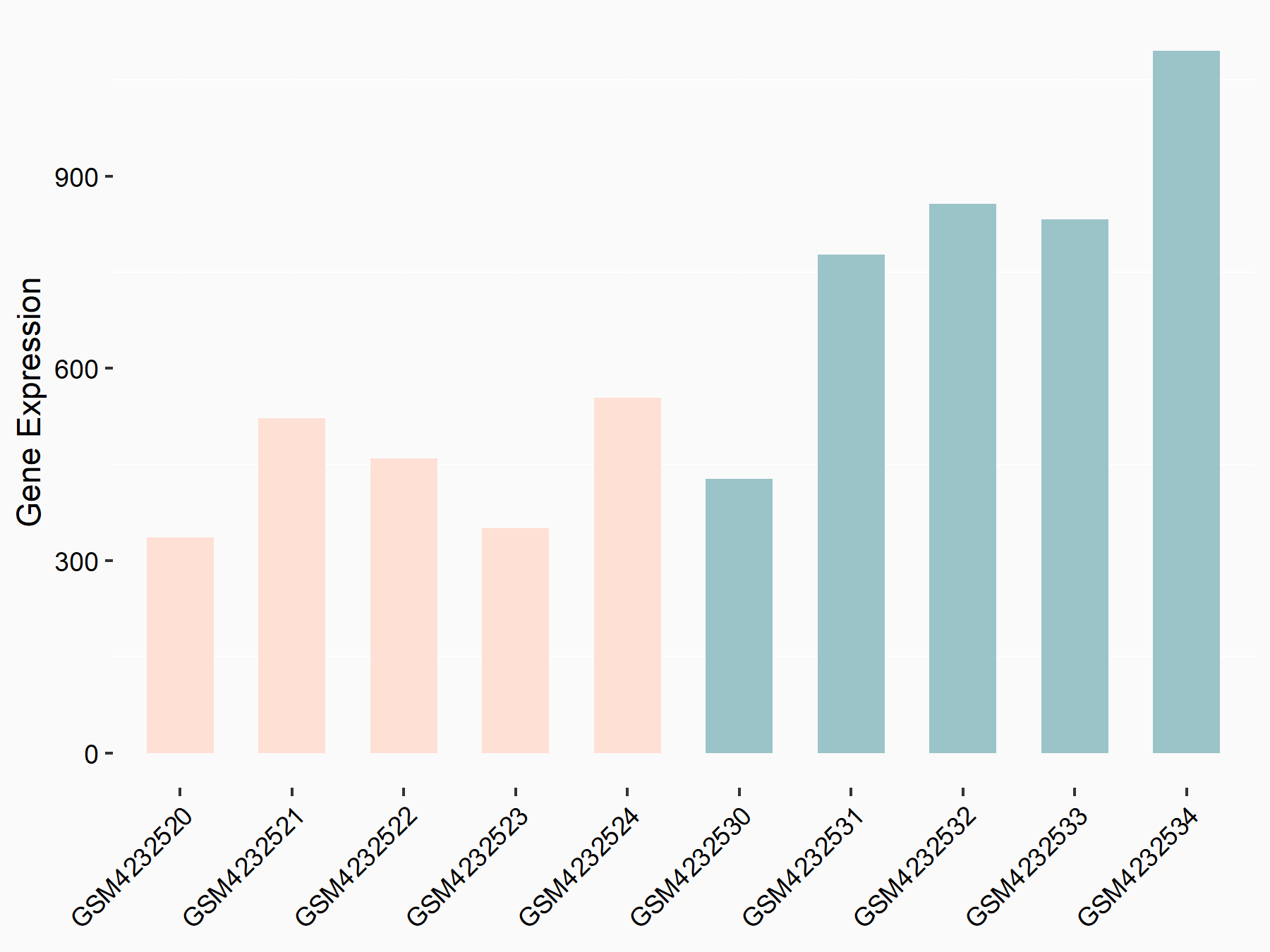 |
logFC: -8.44E-01 p-value: 3.07E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.03E+00 | GSE60213 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [17] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, ATG7, ATG12, and Autophagy-related protein 16-1 (ATG16L1). | |||
Basic leucine zipper transcriptional factor ATF-like 2 (BATF2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
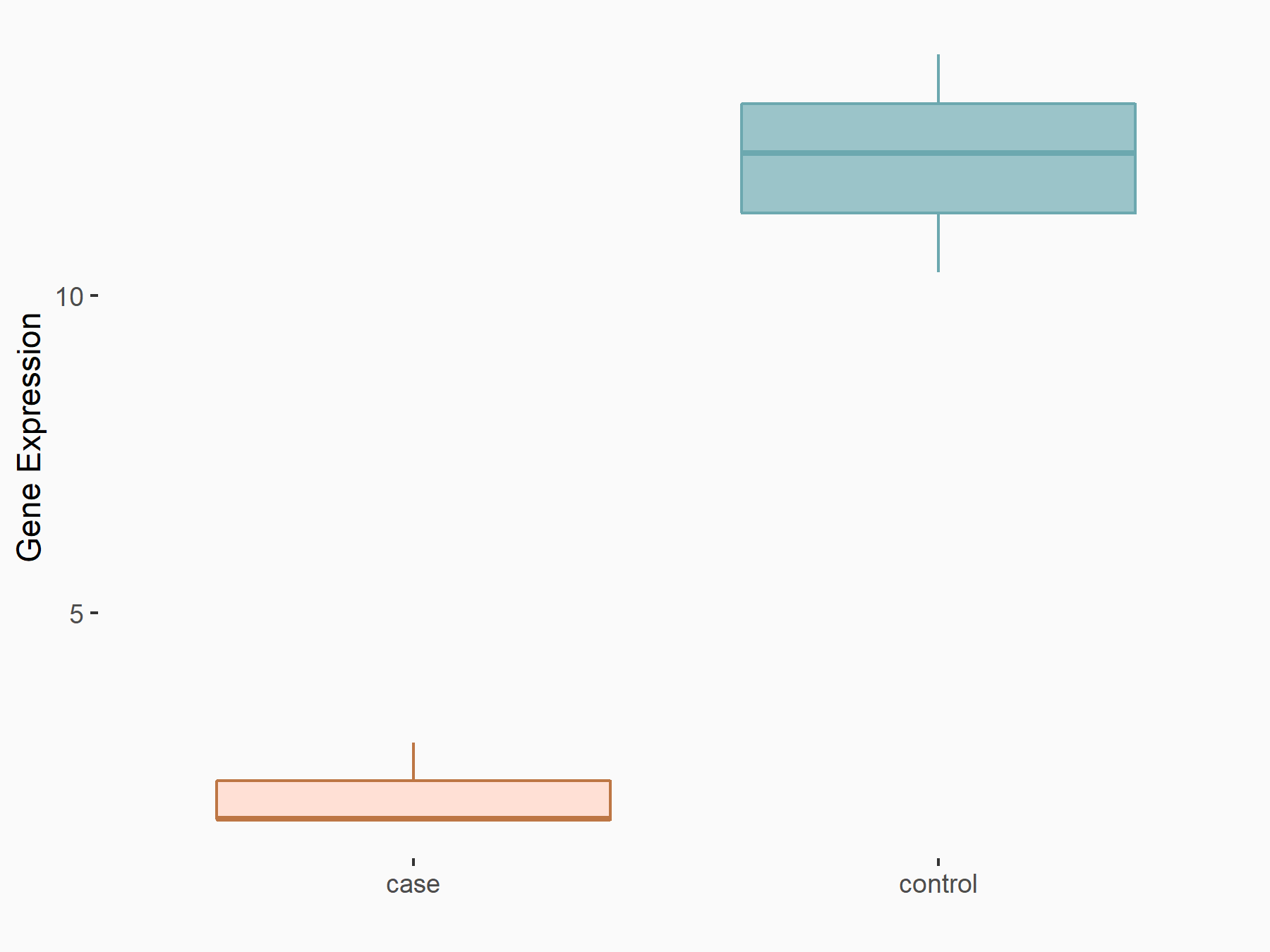 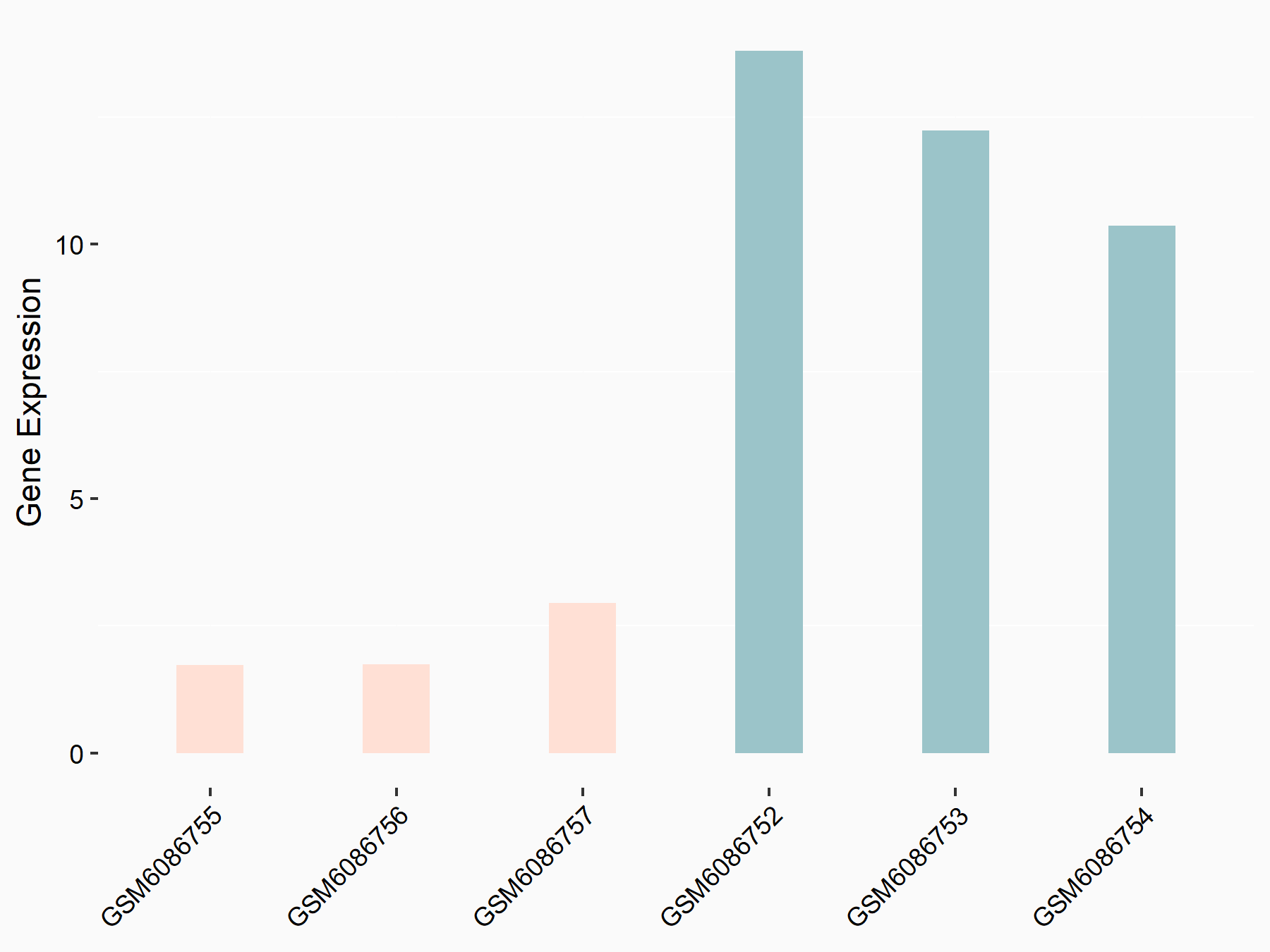 |
logFC: -2.08E+00 p-value: 6.32E-06 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.91E+00 | GSE60213 |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [20] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
In-vitro Model |
SNU-216 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_3946 |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| In-vivo Model | A total of 5 × 106 stably transfected HGC-27 cells were subcutaneously injected into the right axillary fossa of nude mice. Tumor volume was measured every 3 days and calculated with the following formula: V = (L × W2)/2 cm2 (V, tumor volume; L, length; W, width). The mice were sacrificed at 3-4 weeks after injection, and the tumors were weighed. For the lung metastasis model, 5 × 106 stably transfected HGC-27 cells were injected into the tail veins of nude mice. Forty-five days later, the mice were sacrificed, and the lungs were dissected to examine the histopathological metastatic loci. The peritoneal dissemination ability of GC cells was evaluated via intraperitoneal injection. A total of 5 × 106 stably transfected HGC-27 cells in 500 uL of PBS were injected into the peritoneal cavity of BALB/c nude mice. Mice were carefully monitored until they were killed at 4 weeks, at which point peritoneal metastases were examined and recorded. | |||
| Response Summary | N6-methyladenosine (m6A) modification of Basic leucine zipper transcriptional factor ATF-like 2 (BATF2) mRNA by METTL3 repressed its expression in gastric cancer. | |||
Beta-catenin-interacting protein 1 (CTNNBIP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
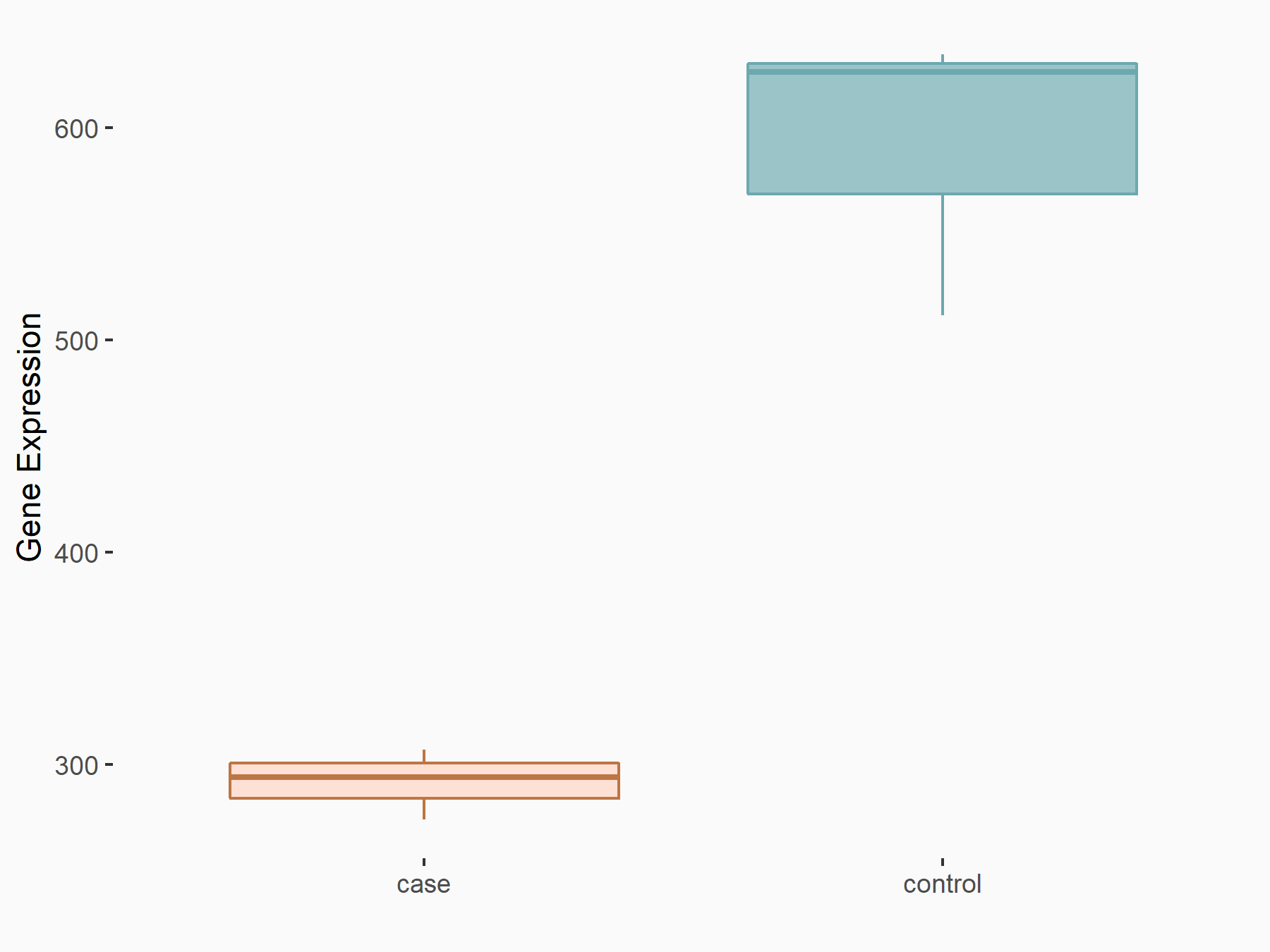 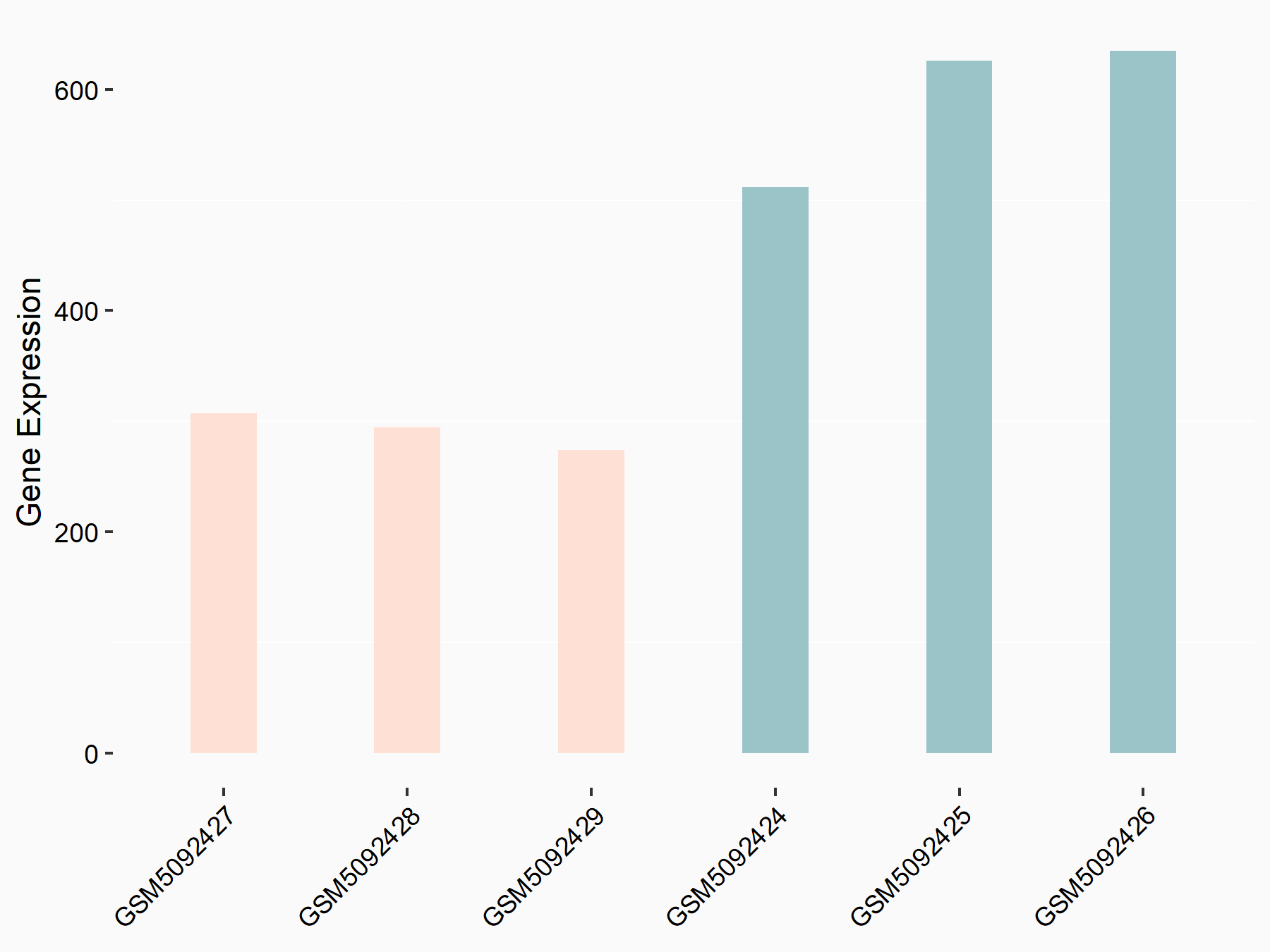 |
logFC: -1.02E+00 p-value: 3.20E-17 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.17E+00 | GSE60213 |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [21] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
T98G | Glioblastoma | Homo sapiens | CVCL_0556 |
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| LN-18 | Glioblastoma | Homo sapiens | CVCL_0392 | |
| HEB (human normal glial cell line HEB were obtained from Tongpai (Shanghai) biotechnology co., LTD (Shanghai, China)) | ||||
| A-172 | Glioblastoma | Homo sapiens | CVCL_0131 | |
| Response Summary | METTL3-mediated m6A modification upregulated circDLC1 expression, and circDLC1 promoted Beta-catenin-interacting protein 1 (CTNNBIP1) transcription by sponging miR-671-5p, thus repressing the malignant proliferation of glioma. | |||
Broad substrate specificity ATP-binding cassette transporter ABCG2 (BCRP/ABCG2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
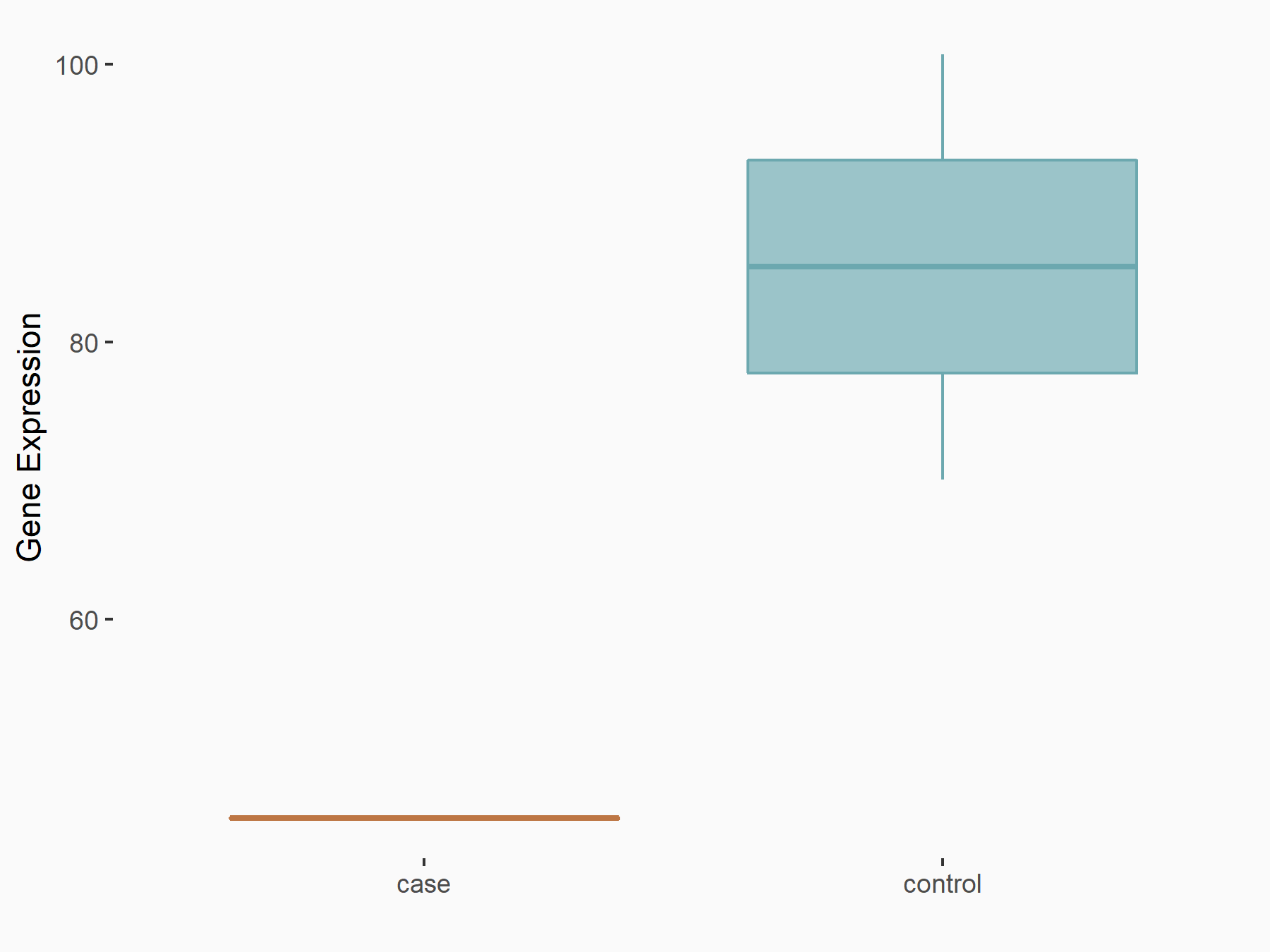 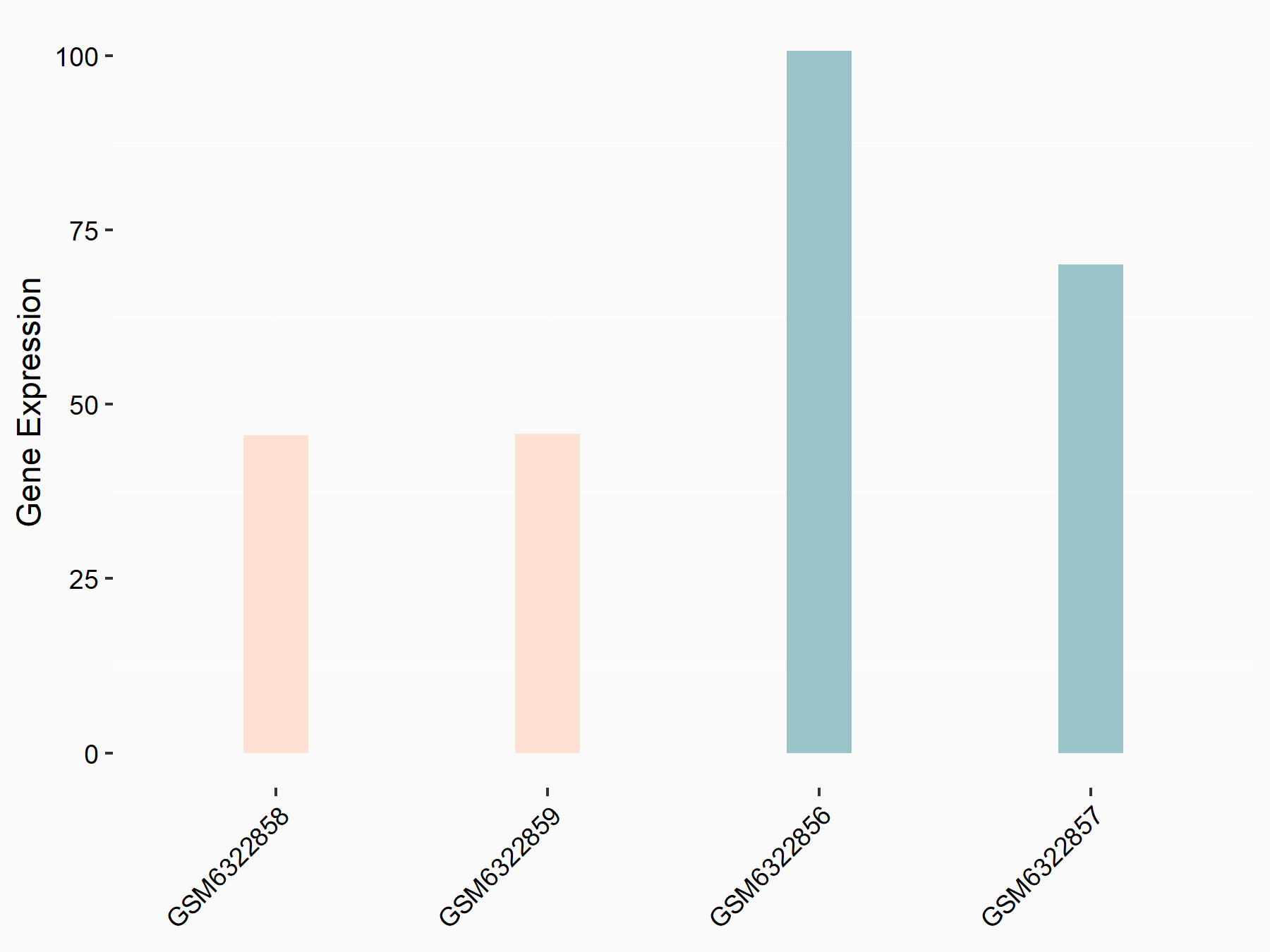 |
logFC: -9.03E-01 p-value: 2.29E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 4.88E+00 | GSE60213 |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [22] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Doxil | Approved | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell growth and death | |||
| Cell apoptosis | ||||
In-vitro Model |
ADR-resistant MCF-7 (MCF-7/ADR) cells (Human breast cancer doxorubicin-resistant cell line) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| In-vivo Model | Cell suspensions (2 × 106 cells/mL) made with MCF-7/ADR cells stably expressing METTL3 and/or miR-221-3p inhibitor were subcutaneously implanted into each mouse. One week later, xenografted mice were injected with 0.1 mL ADR (25 mg/kg, intraperitoneal injection) twice a week. | |||
| Response Summary | METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. METTL3 knockdown was shown to reduce the expression of miR-221-3p by reducing pri-miR-221-3p m6A mRNA methylation, reducing the expression of MDR1 and Broad substrate specificity ATP-binding cassette transporter ABCG2 (BCRP/ABCG2), and inducing apoptosis. Identified the METTL3/miR-221-3p/HIPK2/Che-1 axis as a novel signaling event that will be responsible for resistance of BC cells to ADR. | |||
Cadherin-1 (CDH1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
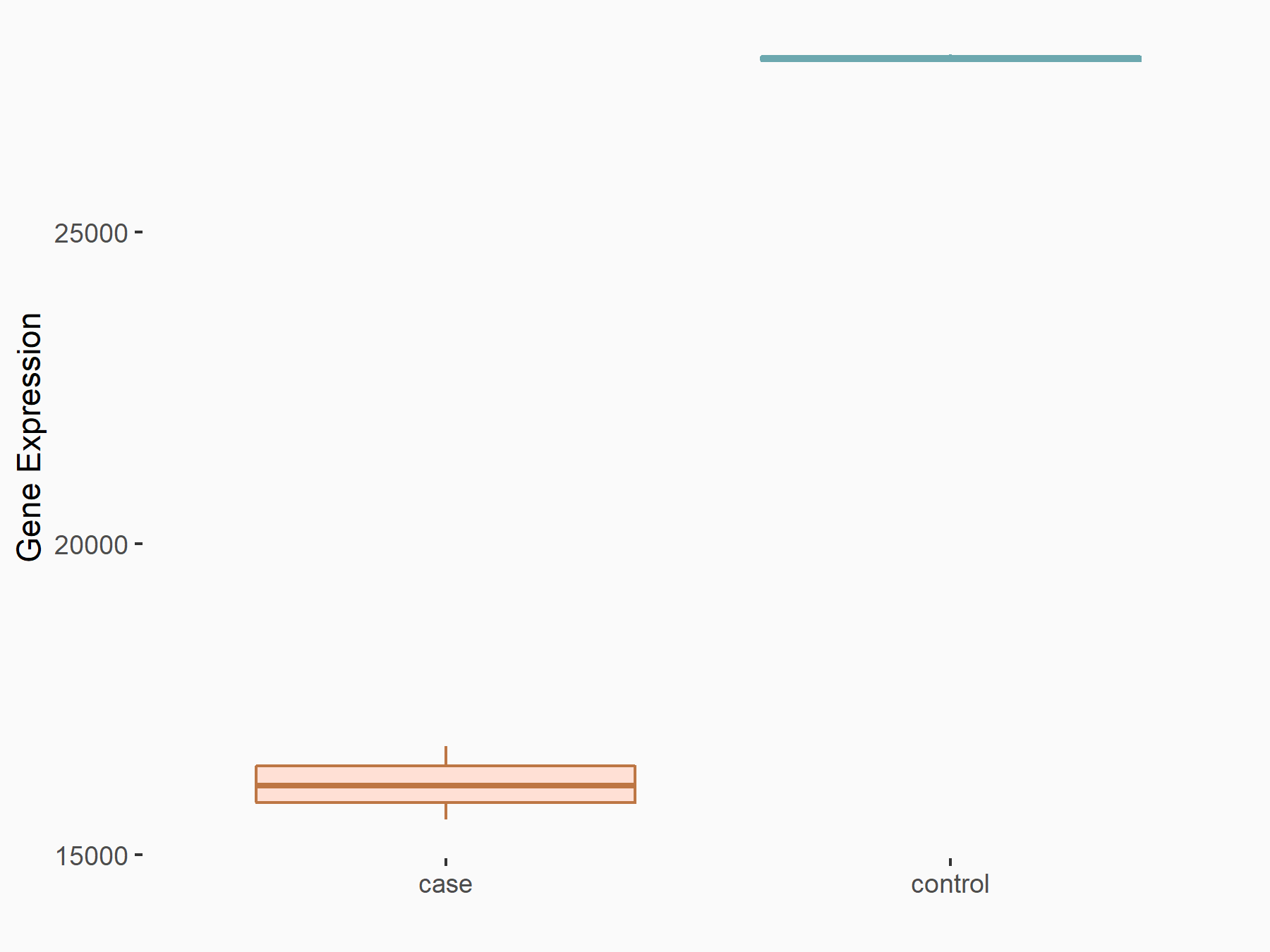 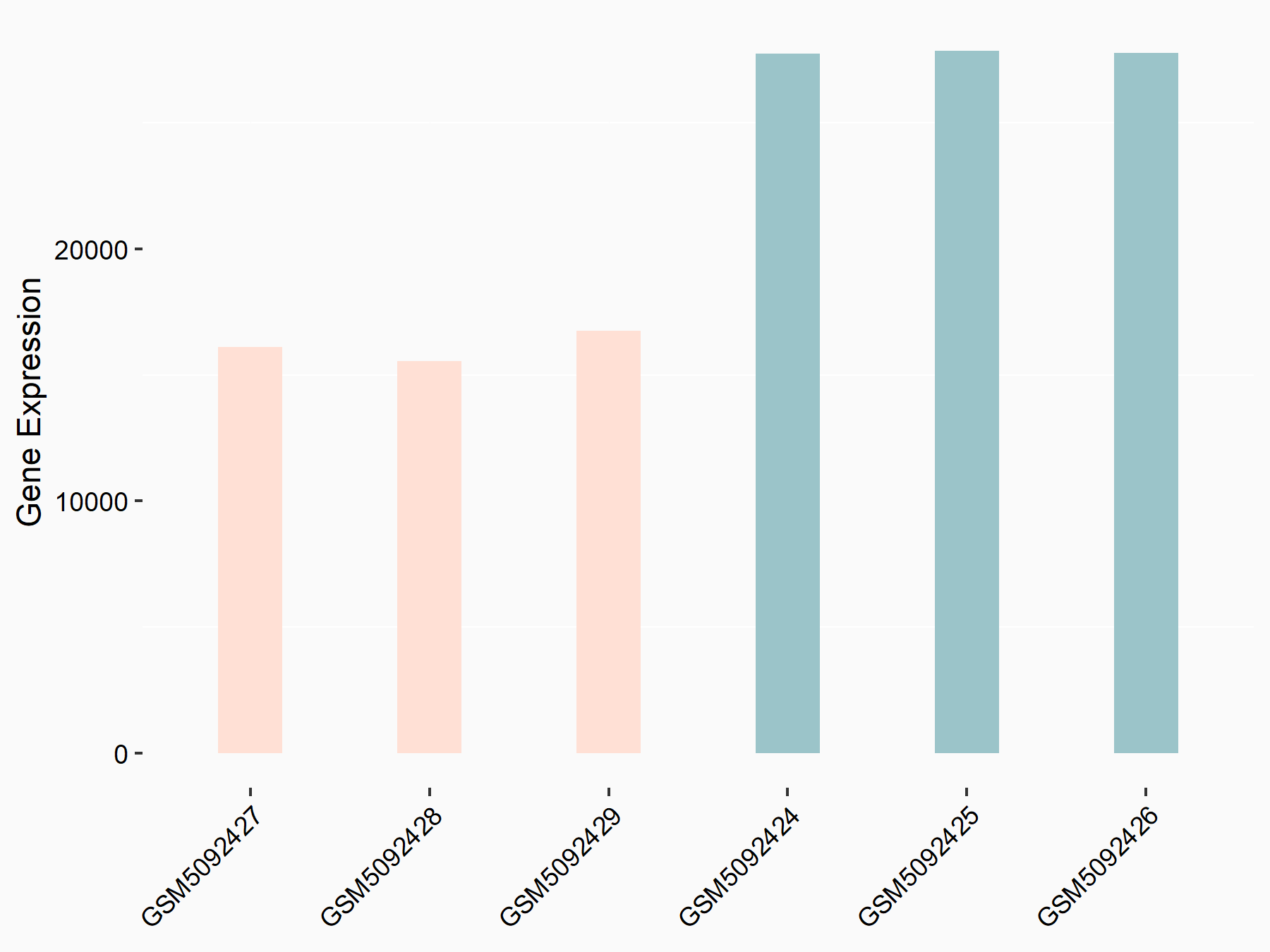 |
logFC: -7.84E-01 p-value: 1.59E-117 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 4.00E+00 | GSE60213 |
Idiopathic interstitial pneumonitis [ICD-11: CB03]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [23] | |||
| Responsed Disease | Pulmonary Fibrosis [ICD-11: CB03.4] | |||
| Pathway Response | Adherens junction | hsa04520 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Response Summary | PM2.5 exposure increased the levels of METTL3-mediated m6A modification of Cadherin-1 (CDH1) mRNA. PM2.5 exposure triggered EMT progression to promote the pulmonary fibrosis via miR-494-3p/YTHDF2 recognized and METTL3 mediated m6A modification. | |||
Cadherin-2 (CDH2/N-cadherin)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
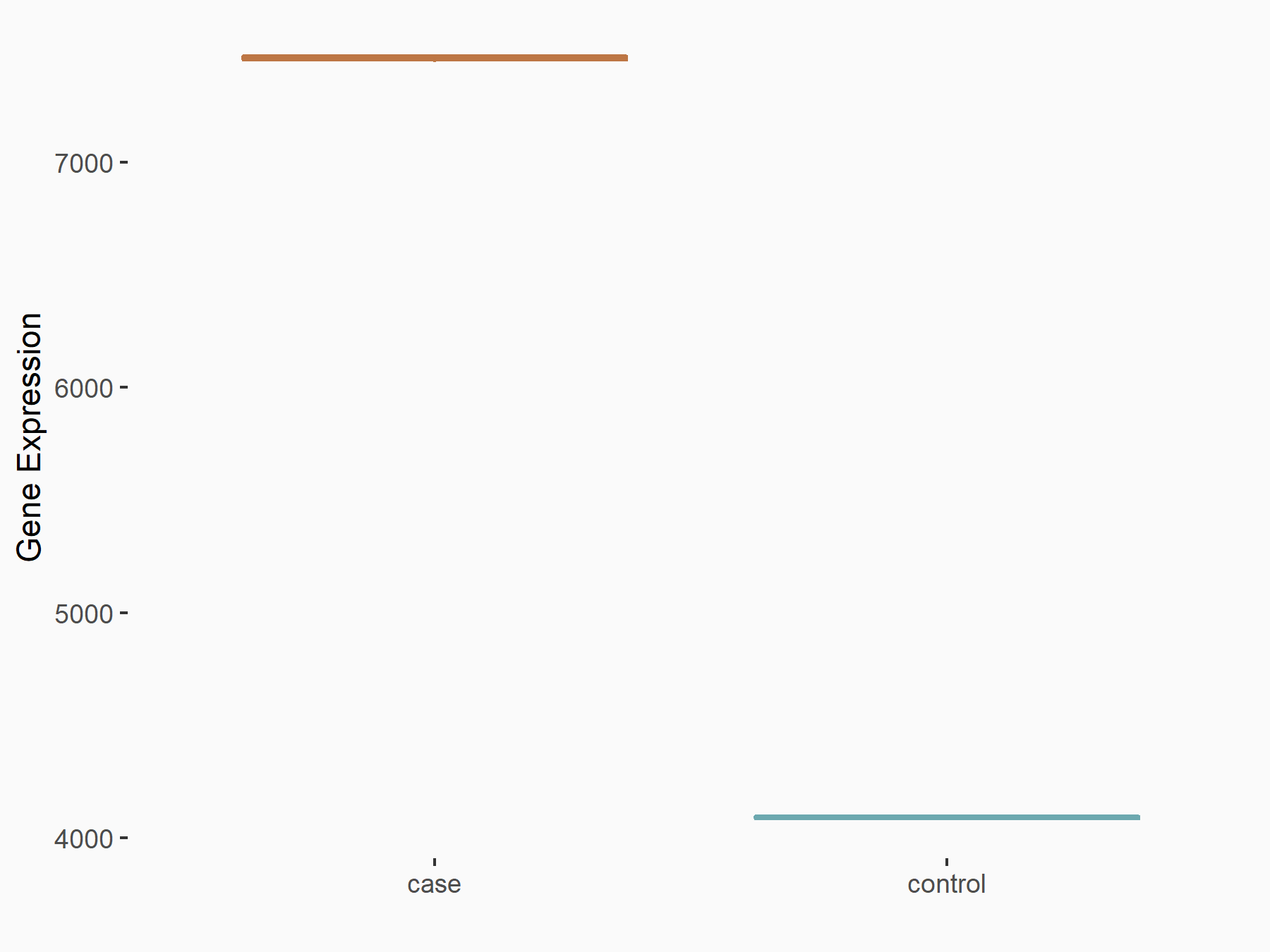 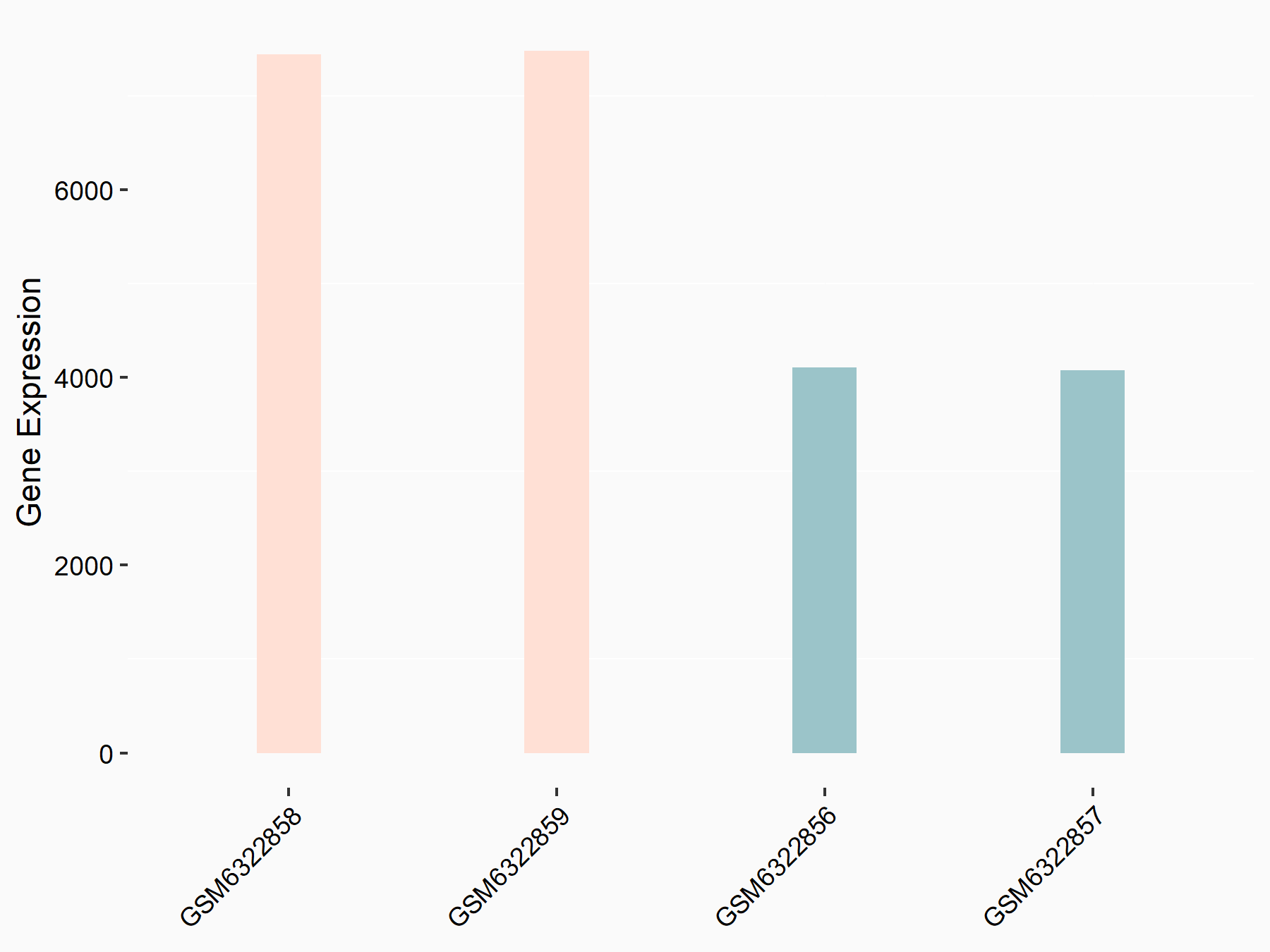 |
logFC: 8.67E-01 p-value: 3.90E-33 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 4.52E+00 | GSE60213 |
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [24] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
Neural progenitor cells (NPCs) (The progenitor cells of the CNS) | |||
| NP69 (A human immortalized nasopharyngeal epithelial) | ||||
| HNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_FA07 | |
| HNE-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_0308 | |
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| CNE-1 | Normal | Homo sapiens | CVCL_6888 | |
| In-vivo Model | 1 × 105 HNE2 cells (with or without METTL3 knockdown) were labeled with luciferase gene and injected into the tail vein of the nude mice. | |||
| Response Summary | METTL3 activated the luciferase activity of TOPflash (a reporter for beta-catenin/TCF signaling), and downregulation of METTL3 inhibited the expression of beta-catenin/TCF target genes vimentin and Cadherin-2 (CDH2/N-cadherin), which are two regulators of epithelial-mesenchymal transition. METTL3 silencing decreased the m6A methylation and total mRNA levels of Tankyrase, a negative regulator of axin. METTL3 is a therapeutic target for NPC. | |||
Melanoma [ICD-11: 2C30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Melanoma [ICD-11: 2C30] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell invasion/migration | |||
In-vitro Model |
451Lu | Cutaneous melanoma | Homo sapiens | CVCL_6357 |
| A-375 | Amelanotic melanoma | Homo sapiens | CVCL_0132 | |
| A375-MA2 | Amelanotic melanoma | Homo sapiens | CVCL_X495 | |
| MeWo | Cutaneous melanoma | Homo sapiens | CVCL_0445 | |
| SK-MEL-2 | Melanoma | Homo sapiens | CVCL_0069 | |
| WM164 | Cutaneous melanoma | Homo sapiens | CVCL_7928 | |
| WM3211 | Acral lentiginous melanoma | Homo sapiens | CVCL_6797 | |
| WM3918 | Melanoma | Homo sapiens | CVCL_C279 | |
| WM793 | Melanoma | Homo sapiens | CVCL_8787 | |
| Response Summary | METTL3 is upregulated in human melanoma and plays a role in invasion/migration through MMP2. METTL3 overexpression promotes accumulation of MMP2 and Cadherin-2 (CDH2/N-cadherin) in melanoma cells. | |||
Catenin beta-1 (CTNNB1/Beta-catenin)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Embryonic stem cells | Mus musculus |
|
Treatment: METTL3 knockout mESCs
Control: Wild type mESCs
|
GSE156481 | |
| Regulation |
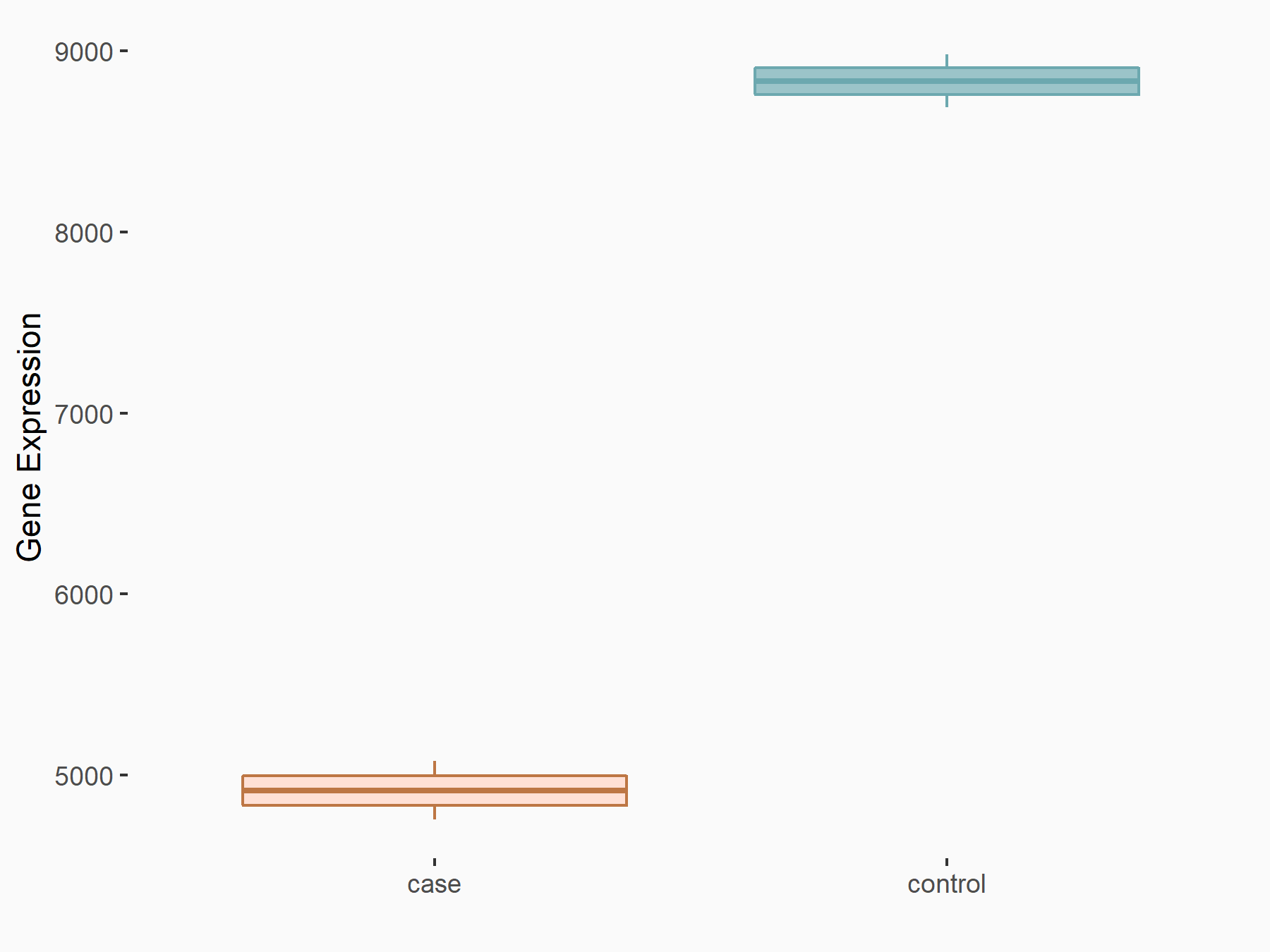 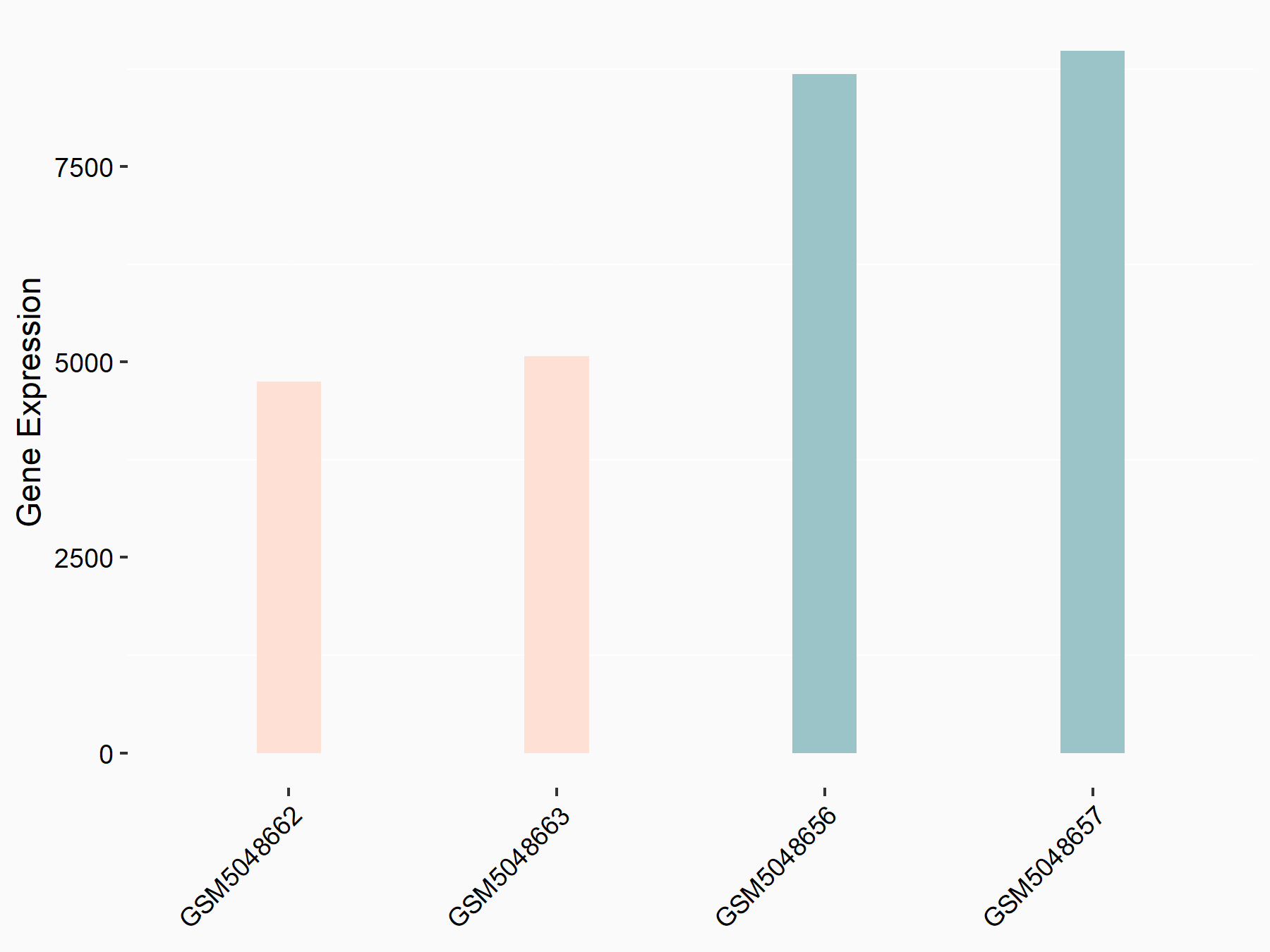 |
logFC: -8.46E-01 p-value: 2.97E-40 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.37E+00 | GSE60213 |
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [24] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
Neural progenitor cells (NPCs) (The progenitor cells of the CNS) | |||
| NP69 (A human immortalized nasopharyngeal epithelial) | ||||
| HNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_FA07 | |
| HNE-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_0308 | |
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| CNE-1 | Normal | Homo sapiens | CVCL_6888 | |
| In-vivo Model | 1 × 105 HNE2 cells (with or without METTL3 knockdown) were labeled with luciferase gene and injected into the tail vein of the nude mice. | |||
| Response Summary | METTL3 activated the luciferase activity of TOPflash (a reporter for beta-catenin/TCF signaling), and downregulation of METTL3 inhibited the expression of Catenin beta-1 (CTNNB1/Beta-catenin)/TCF target genes vimentin and N-cadherin, which are two regulators of epithelial-mesenchymal transition. METTL3 silencing decreased the m6A methylation and total mRNA levels of Tankyrase, a negative regulator of axin. METTL3 is a therapeutic target for NPC. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [25] | |||
| Responsed Disease | Hepatoblastoma [ICD-11: 2C12.01] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| QSG-7701 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6944 | |
| In-vivo Model | 5 × 106 cells were subcutaneously injected into the left or right flank of each mouse. | |||
| Response Summary | METTL3 is significantly up-regulated in Hepatoblastoma(HB) and promotes HB development.m6A mRNA methylation contributes significantly to regulate the Wnt/beta-catenin pathway. Reduced m6A methylation can lead to a decrease in expression and stability of the Catenin beta-1 (CTNNB1/Beta-catenin). | |||
Spina bifida [ICD-11: LA02]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [26] | |||
| Responsed Disease | Neural tube defect [ICD-11: LA02.Z] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
HT22 | Normal | Mus musculus | CVCL_0321 |
| In-vivo Model | The mice were maintained on a 12-h light/dark cycle (lights on from 8:00 a.m. to 8:00 p.m.). On day 7.5 of pregnancy (E7.5), ethionine (Sigma-Aldrich, USA) was intraperitoneally injected only once at a dose of 500 mg/kg to establish the NTDs embryo model. And SAM (MedChemExpress, USA) was intraperitoneally injected only once at a dose of 30 mg/kg. The same dose was intraperitoneally injected to the pregnant mice for control group. | |||
| Response Summary | SAM not only played a compensatory role, but also led to m6A modification changes in neural tube development and regulation. Ethionine affected m6A modification by reducing SAM metabolism. METTL3 is enriched in HT-22 cells, and METTL3 knockdown reduces cell proliferation and increases apoptosis through suppressing Wnt/Catenin beta-1 (CTNNB1/Beta-catenin) signaling pathway. Overexpression of ALKBH5 can only inhibit cell proliferation, but cannot promote cell apoptosis. | |||
Chronic kidney disease [ICD-11: GB61]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [276] | |||
| Responsed Disease | Chronic kidney disease [ICD-11: GB61] | |||
In-vitro Model |
HK-2 [Human kidney] | Normal | Homo sapiens | CVCL_0302 |
| In-vivo Model | Mice were injected with empty vector (pcDNA3) or Flag-tagged METTL3 expression vector (pFlag-METTL3) at 2 days after UUO or 4 days after UIRI, respectively. The expression of transgene was validated by Western blotting or immunostaining for Flag-tagged METTL3 fusion protein. | |||
Cellular tumor antigen p53 (TP53/p53)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
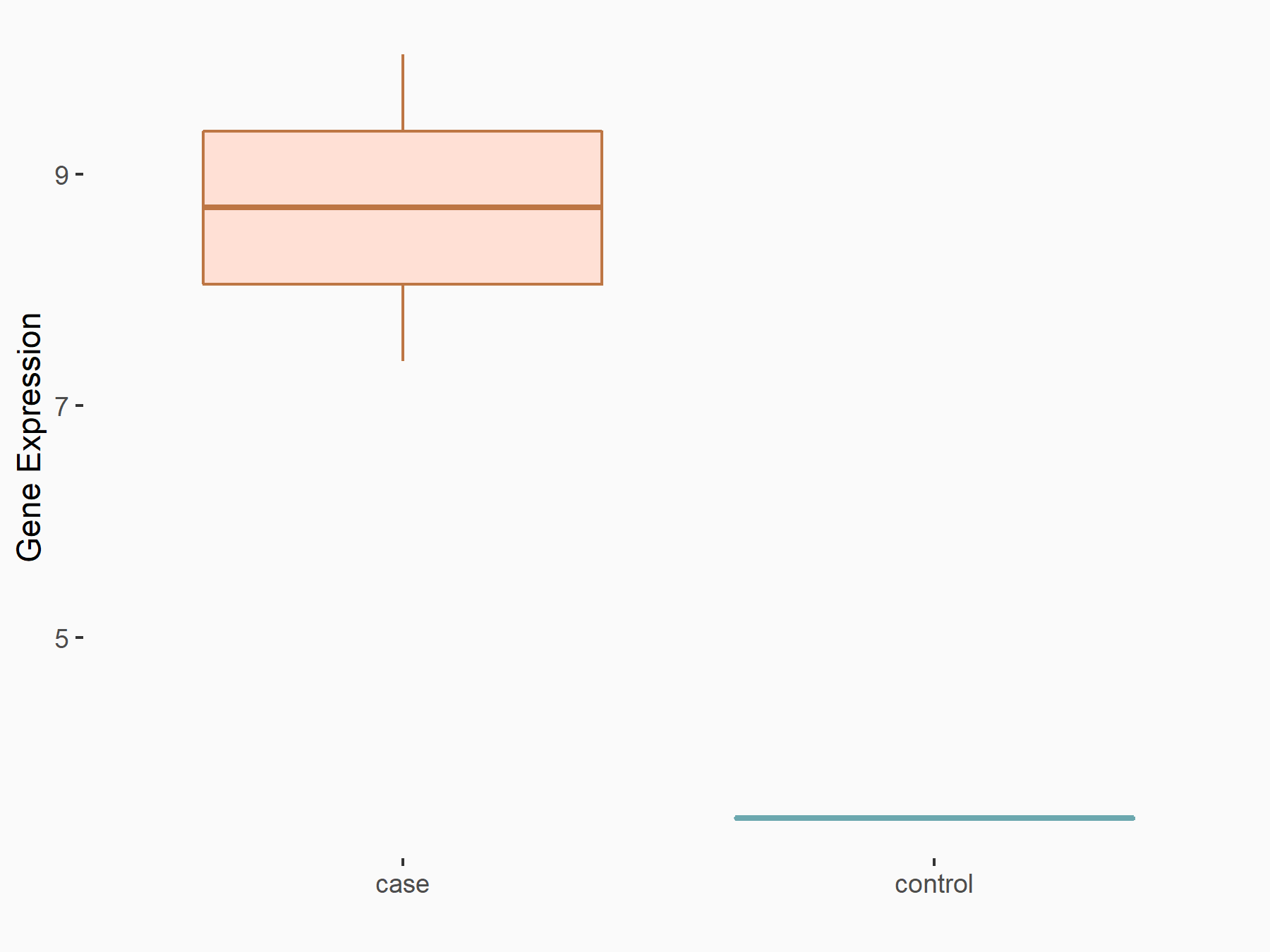 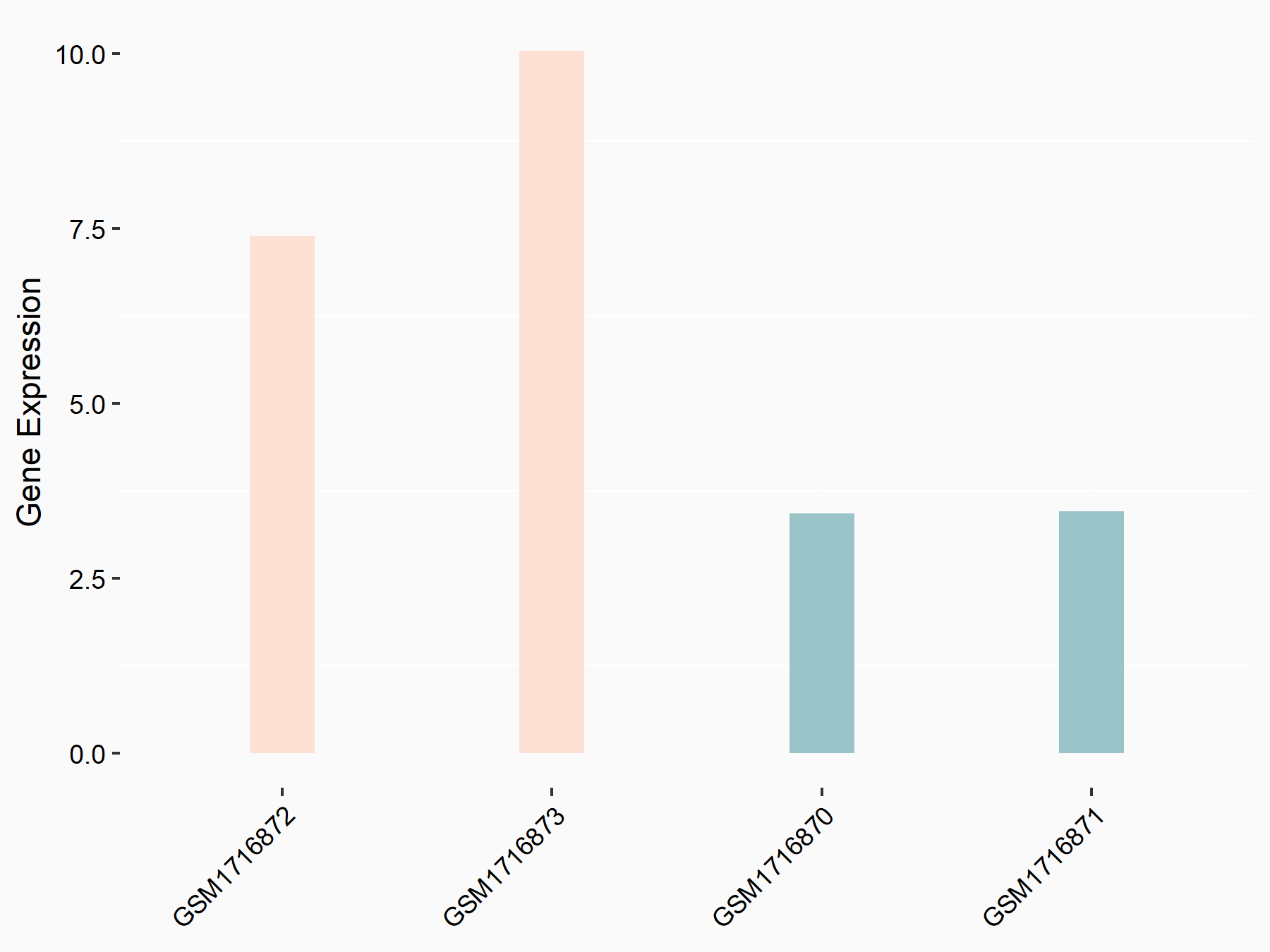 |
logFC: 1.12E+00 p-value: 1.48E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.05E+00 | GSE60213 |
Solid tumour/cancer [ICD-11: 2A00-2F9Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [27] | |||
| Responsed Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Responsed Drug | Arsenite | Phase 2 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
In-vitro Model |
HaCaT | Normal | Homo sapiens | CVCL_0038 |
| Response Summary | METTL3 significantly decreased m6A level, restoring Cellular tumor antigen p53 (TP53/p53) activation and inhibiting cellular transformation phenotypes in the arsenite-transformed cells. m6A downregulated the expression of the positive p53 regulator, PRDM2, through the YTHDF2-promoted decay of PRDM2 mRNAs. m6A upregulated the expression of the negative p53 regulator, YY1 and MDM2 through YTHDF1-stimulated translation of YY1 and MDM2 mRNA. This study further sheds light on the mechanisms of arsenic carcinogenesis via RNA epigenetics. | |||
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [28] | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Cell cycle | hsa04110 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell apoptosis | |||
| Cells in G3/M phase decreased | ||||
In-vitro Model |
THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| MOLT-4 | Adult T acute lymphoblastic leukemia | Homo sapiens | CVCL_0013 | |
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 | |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| CCRF-CEM C7 | T acute lymphoblastic leukemia | Homo sapiens | CVCL_6825 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| Response Summary | METTL3 and METTL14 play an oncogenic role in acute myeloid leukemia(AML) by targeting mdm2/Cellular tumor antigen p53 (TP53/p53) signal pathway. The knockdown of METTL3 and METTL14 in K562 cell line leads to several changes in the expression of p53 signal pathway, including the upregulation of p53, cyclin dependent kinase inhibitor 1A (CDKN1A/p21), and downregulation of mdm2. | |||
Colon cancer [ICD-11: 2B90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [29] | |||
| Responsed Disease | Colon cancer [ICD-11: 2B90] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Cell Process | Protein signaling | |||
In-vitro Model |
SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 |
| WiDr | Colon adenocarcinoma | Homo sapiens | CVCL_2760 | |
| Response Summary | The produced p53 R273H mutant protein resulted in acquired multidrug resistance in colon cancer cells. Either silencing METTL3 expression by using small interfering RNA (siRNA) or inhibiting RNA methylation with neplanocin A suppressed m6A formation in Cellular tumor antigen p53 (TP53/p53) pre-mRNA, and substantially increased the level of phosphorylated p53 protein (Ser15) and its function in cells heterozygously carrying the R273H mutation, thereby re-sensitizing these cells to anticancer drugs. | |||
Liver cancer [ICD-11: 2C12]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [30] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Apatinib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
QGY-7701 | Human papillomavirus-related endocervical adenocarcinoma | Homo sapiens | CVCL_6859 |
| HHL-5 | Normal | Homo sapiens | CVCL_S956 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | Nude mice (4-6 week-old) were administered sterile water and feed in a specific pathogen-free barrier. Using a 1-mL syringe, 1 × 107 HEPG2 cells were subcutaneously inoculated into the right axilla of nude mice to build the HCC xenograft model. When the tumor volume reached 50 mm3, the nude mice were randomly divided into 1 control (n = 4) and 3 treatment groups (n = 4 each). RG7112, apatinib, and RG7112 + apatinib were administered to the treatment groups and an equal volume of dimethyl sulfoxide to the control group by daily gavage for 14 d. The tumor length (L) and width (W) were measured on alternate days using vernier calipers. The following formula was used to calculate the tumor volume: volume (mm3) = 0.5 × L × W × W. At the end of the experiment, the nude mice were killed by CO2 overdose anesthesia. The tumors were dissected and weighed using a precision balance, and the tumor tissue was stored in liquid nitrogen for further analysis. | |||
| Response Summary | Cellular tumor antigen p53 (TP53/p53) n6-methyladenosine (m6A) played a decisive role in regulating Hepatocellular carcinoma(HCC) sensitivity to chemotherapy via the p53 activator RG7112 and the vascular endothelial growth factor receptor inhibitor apatinib. p53 mRNA m6A modification blockage induced by S-adenosyl homocysteine or siRNA-mediated METTL3 inhibition enhanced HCC sensitivity to chemotherapy. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [30] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | RG7112 | Phase 1 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
QGY-7701 | Human papillomavirus-related endocervical adenocarcinoma | Homo sapiens | CVCL_6859 |
| HHL-5 | Normal | Homo sapiens | CVCL_S956 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | Nude mice (4-6 week-old) were administered sterile water and feed in a specific pathogen-free barrier. Using a 1-mL syringe, 1 × 107 HEPG2 cells were subcutaneously inoculated into the right axilla of nude mice to build the HCC xenograft model. When the tumor volume reached 50 mm3, the nude mice were randomly divided into 1 control (n = 4) and 3 treatment groups (n = 4 each). RG7112, apatinib, and RG7112 + apatinib were administered to the treatment groups and an equal volume of dimethyl sulfoxide to the control group by daily gavage for 14 d. The tumor length (L) and width (W) were measured on alternate days using vernier calipers. The following formula was used to calculate the tumor volume: volume (mm3) = 0.5 × L × W × W. At the end of the experiment, the nude mice were killed by CO2 overdose anesthesia. The tumors were dissected and weighed using a precision balance, and the tumor tissue was stored in liquid nitrogen for further analysis. | |||
| Response Summary | Cellular tumor antigen p53 (TP53/p53) n6-methyladenosine (m6A) played a decisive role in regulating Hepatocellular carcinoma(HCC) sensitivity to chemotherapy via the p53 activator RG7112 and the vascular endothelial growth factor receptor inhibitor apatinib. p53 mRNA m6A modification blockage induced by S-adenosyl homocysteine or siRNA-mediated METTL3 inhibition enhanced HCC sensitivity to chemotherapy. | |||
Renal cell carcinoma [ICD-11: 2C90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [277] | |||
| Responsed Disease | Renal cell carcinoma [ICD-11: 2C90] | |||
| Responsed Drug | Erianin | Investigative | ||
| Target Regulation | Down regulation | |||
Acute ischemic stroke [ICD-11: 8B11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [278] | |||
| Responsed Disease | Acute ischemic stroke [ICD-11: 8B11] | |||
| Responsed Drug | Remimazolam | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 |
Senescent cell [ICD-11: MG2A]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [279] | |||
| Responsed Disease | Senescent cell [ICD-11: MG2A] | |||
In-vitro Model |
WI-38 | Normal | Homo sapiens | CVCL_0579 |
| In-vivo Model | 1 μg of labeled RNA was incubated with 500 μg of cell lysates from WI-38 cells at the indicated PDL. | |||
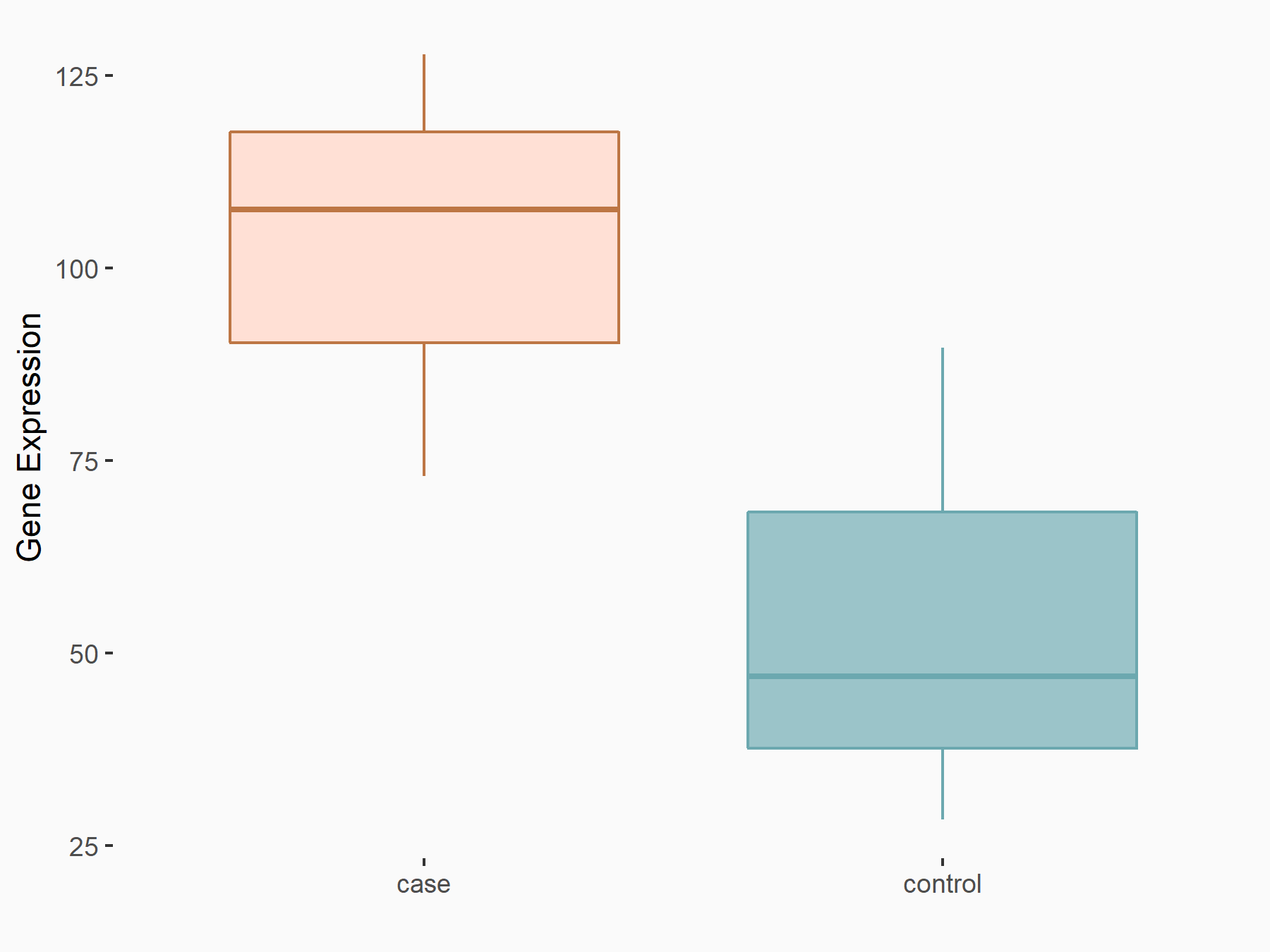
Collagen alpha-1 (III) chain (COL3A1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
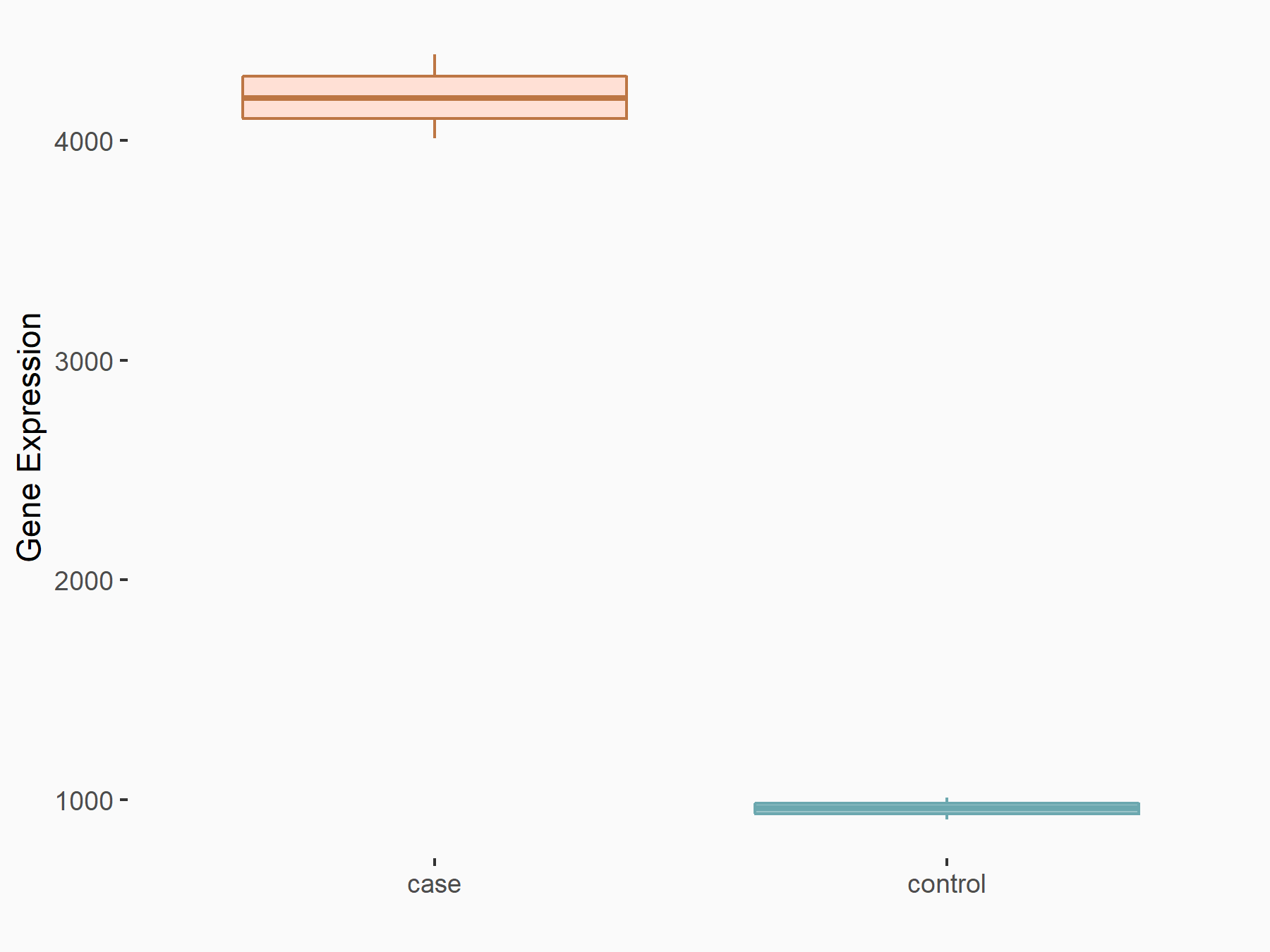 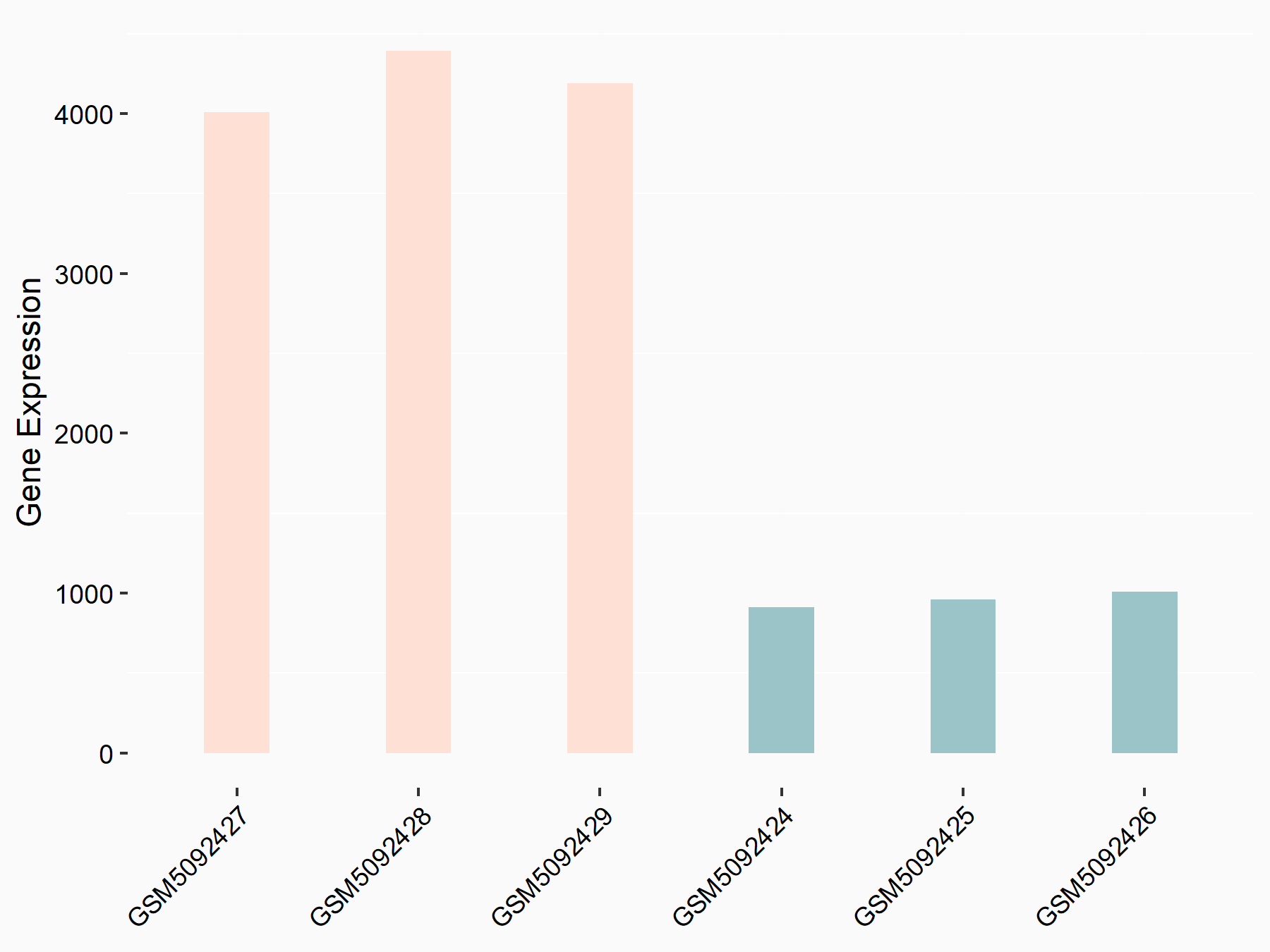 |
logFC: 2.13E+00 p-value: 6.01E-268 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 3.39E+00 | GSE60213 |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [31] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| Response Summary | METTL3 could down-regulate the expression of Collagen alpha-1 (III) chain (COL3A1) by increasing its m6A methylation, ultimately inhibiting the metastasis of TNBC cells. | |||
Collagenase 3 (MMP13)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | DKO-1 cell line | Homo sapiens |
|
Treatment: METTL3 knockdown DKO-1 cell
Control: DKO-1 cell
|
GSE182382 | |
| Regulation |
 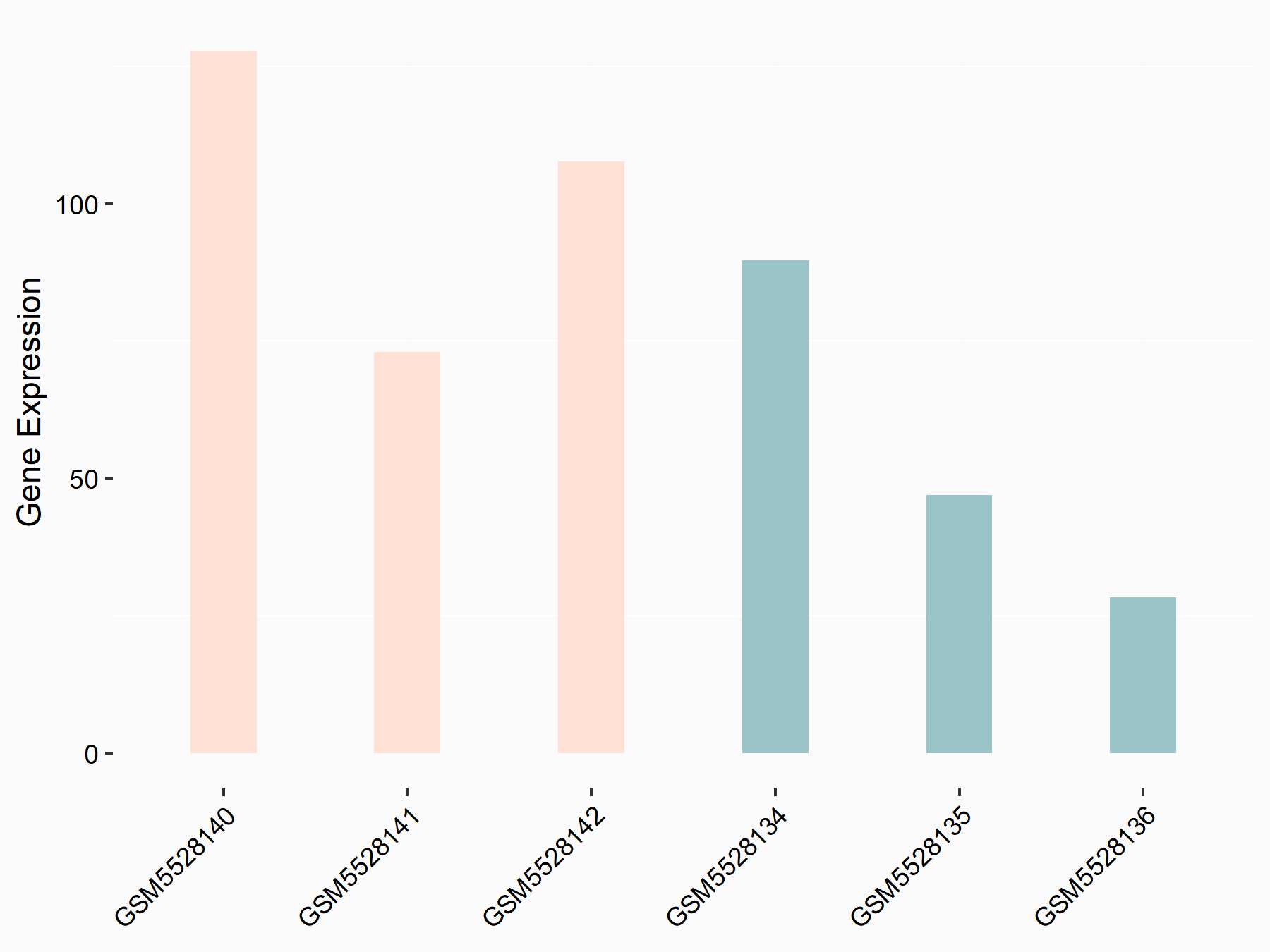 |
logFC: 9.33E-01 p-value: 3.02E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 4.73E+00 | GSE60213 |
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [32] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Inflammatory response and apoptosis | |||
In-vitro Model |
ATDC-5 | Mouse teratocarcinoma | Mus musculus | CVCL_3894 |
| In-vivo Model | The right knee joint of each OA mouse was injected with 1U of type VII collagenase over two consecutive days to obtain experimental OA joint, and the control mice received the equal volume of physiological saline. | |||
| Response Summary | METTL3 has a functional role in mediates osteoarthritis progression by regulating NF-Kappa-B signaling and ECM synthesis in chondrocytes that shed insight on developing preventive and curative strategies for OA by focusing on METTL3 and mRNA methylation. Silencing of METTL3 promotes degradation of extracellular matrix (ECM) by reducing the expression of Collagenase 3 (MMP13) and Coll X, elevating the expression of Aggrecan and Coll II. | |||
CUB domain-containing protein 1 (CDCP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
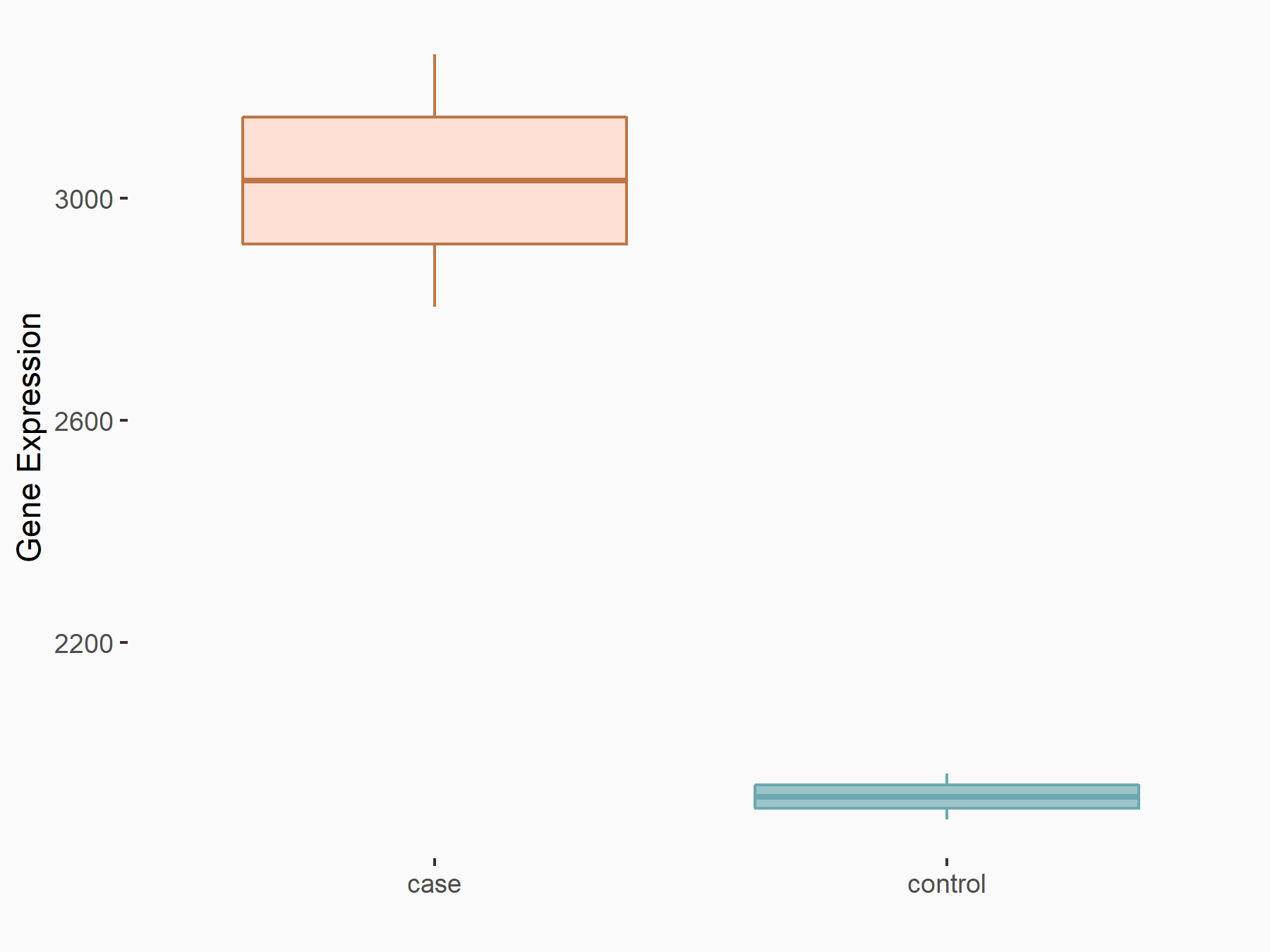 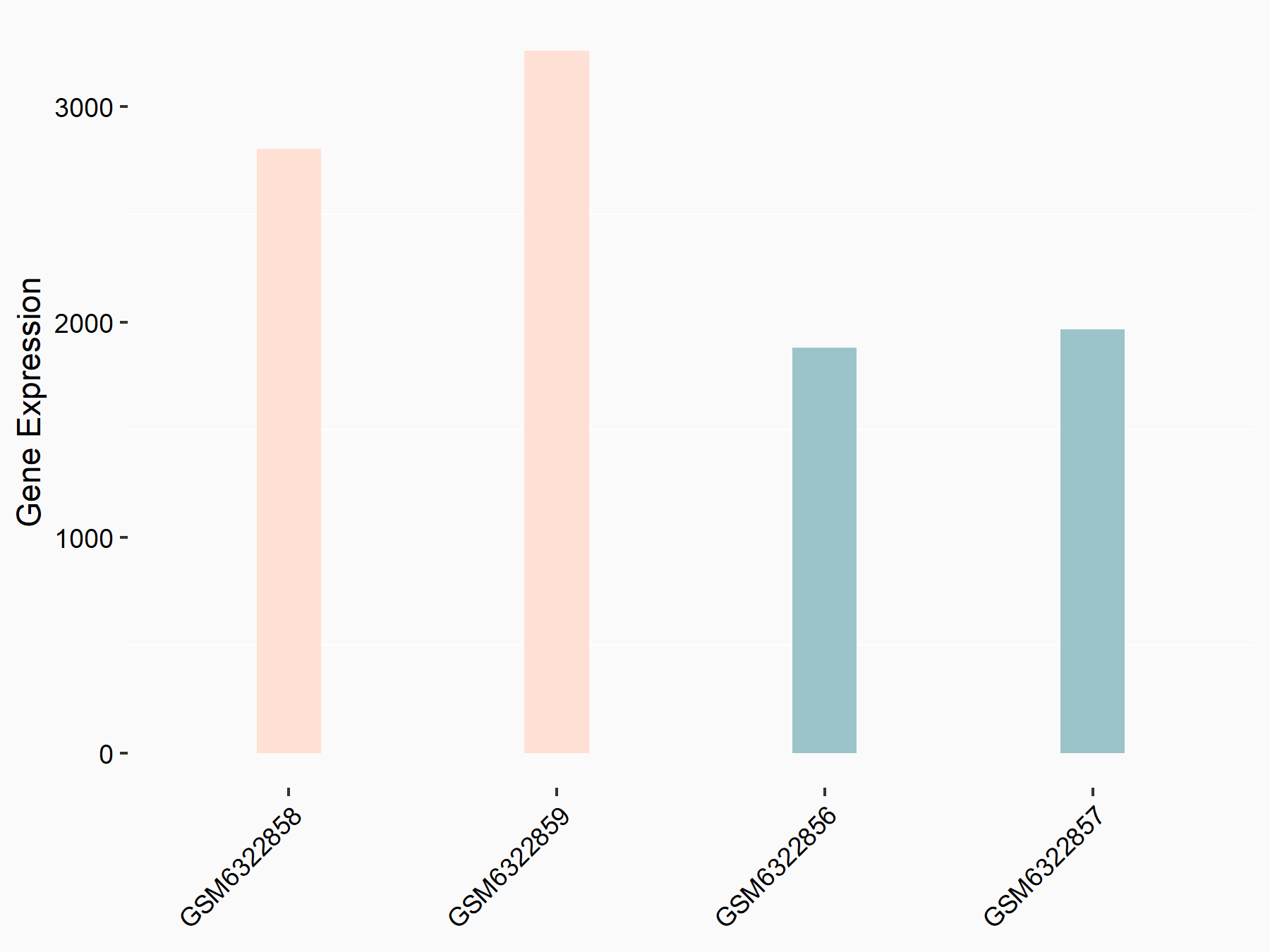 |
logFC: 6.56E-01 p-value: 5.99E-11 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.49E+00 | GSE60213 |
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [34] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| NSTC2 (Nickel-induced transformation of human cells) | ||||
| MC-SV-HUC T-2 | Ureteral tumor cell | Homo sapiens | CVCL_6418 | |
| 16HBE14o- | Normal | Homo sapiens | CVCL_0112 | |
| In-vivo Model | To test for malignant transformation, 1×107 cells were inoculated subcutaneously in the dorsal thoracic midline of ten NOD/SCID mice (Weitong Lihua Experimental Animal Technology Co. Ltd). Tumor formation and growth were assessed every 3 days. | |||
| Response Summary | m6A methyltransferase METTL3 and demethylases ALKBH5 mediate the m6A modification in 3'-UTR of CDCP1 mRNA. METTL3 and CUB domain-containing protein 1 (CDCP1) are upregulated in the bladder cancer patient samples and the expression of METTL3 and CDCP1 is correlated with the progression status of the bladder cancers. | |||
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [281] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
In-vitro Model |
HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
Cyclin-dependent kinase inhibitor 1 (CDKN1A)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
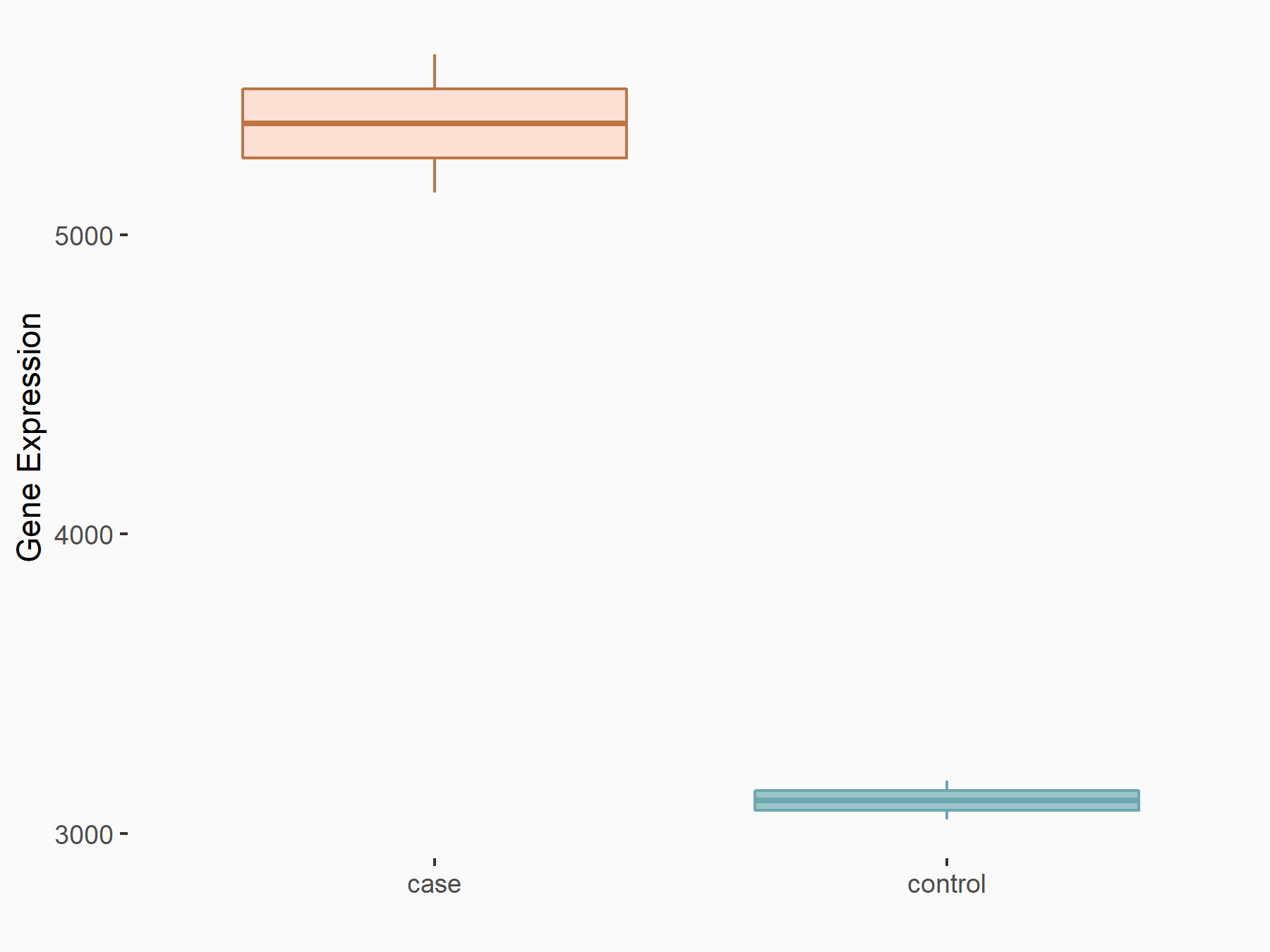 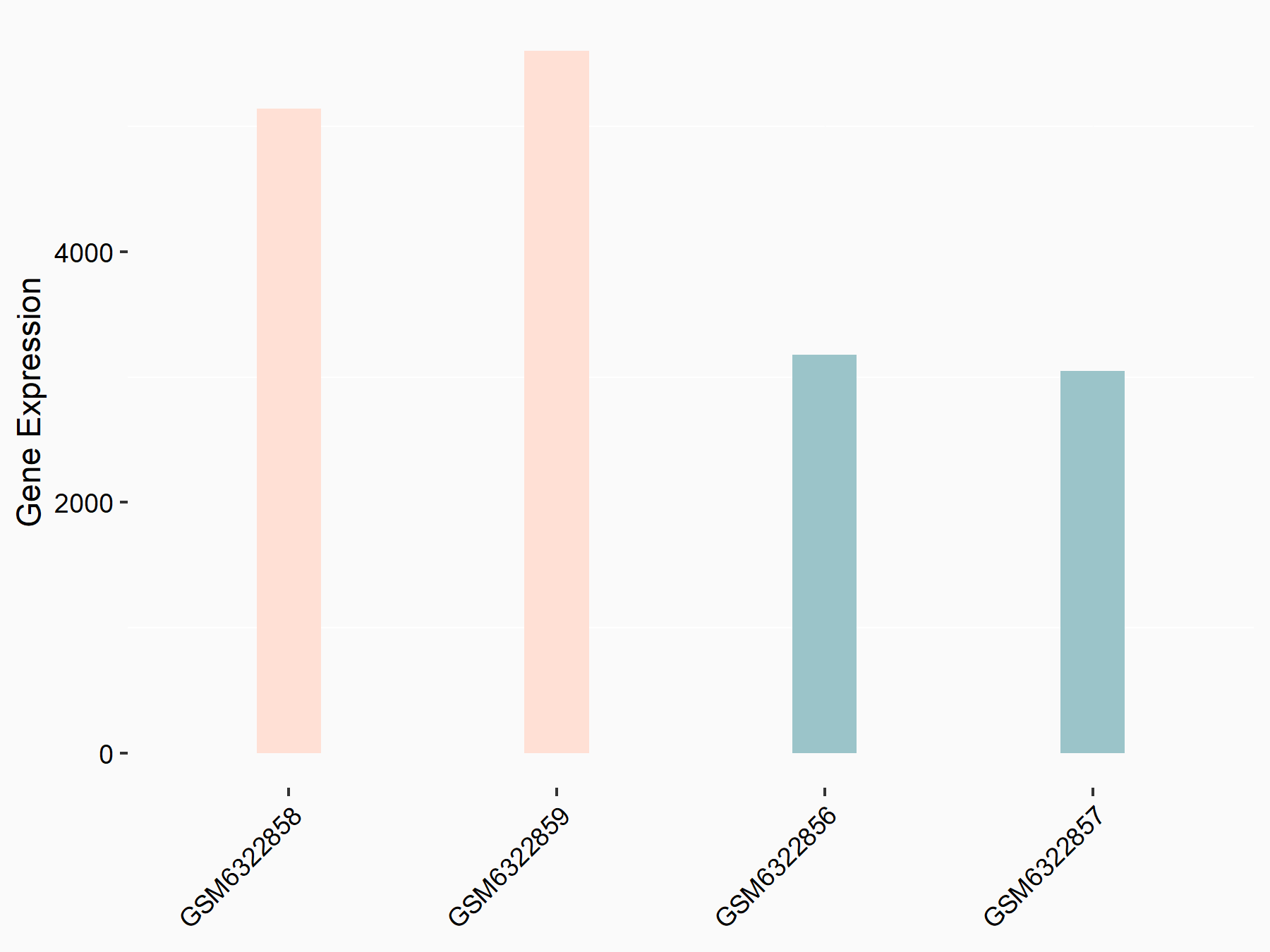 |
logFC: 7.88E-01 p-value: 3.07E-21 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 9.39E+00 | GSE60213 |
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [28] | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Cell apoptosis | |||
| Cells in G4/M phase decreased | ||||
In-vitro Model |
THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| MOLT-4 | Adult T acute lymphoblastic leukemia | Homo sapiens | CVCL_0013 | |
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 | |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| CCRF-CEM C7 | T acute lymphoblastic leukemia | Homo sapiens | CVCL_6825 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| Response Summary | METTL3 and METTL14 play an oncogenic role in acute myeloid leukemia(AML) by targeting mdm2/p53 signal pathway. The knockdown of METTL3 and METTL14 in K562 cell line leads to several changes in the expression of p53 signal pathway, including the upregulation of p53, cyclin dependent kinase inhibitor 1A (CDKN1A/Cyclin-dependent kinase inhibitor 1 (CDKN1A)), and downregulation of mdm2. | |||
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [36] | |||
| Responsed Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.1] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Arrest cell cycle at G2/M phase | |||
In-vitro Model |
TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| In-vivo Model | Totally 5 × 106 ESCC cells after treatment were administered to randomized animals by the subcutaneous route (right flank) in 200 uL DMEM. Tumor measurement was performed every other day. At study end (at least 3 weeks), the animals underwent euthanasia, and the tumors were extracted for histology. | |||
| Response Summary | METTL3 modulates the cell cycle of Esophageal squamous cell carcinoma(ESCC) cells through a Cyclin-dependent kinase inhibitor 1 (CDKN1A)-dependent pattern. METTL3-guided m6A modification contributes to the progression of ESCC via the p21-axis. | |||
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [37] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| SUM1315MO2 | Invasive breast carcinoma of no special type | Homo sapiens | CVCL_5589 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HBL-100 | Normal | Homo sapiens | CVCL_4362 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| In-vivo Model | About 1 × 107 stable METTL3 overexpression and negative control SUM-1315 cells were injected subcutaneously into the axilla of the female BALB/C nude mice (4-6 weeks old, 18-20 g, 10 mice/group). One week after injection, the two groups, METTL3 OE and NC, were then randomly allocated into the control group and experimental group (5 mice/group), which were treated with PBS or metformin (250 mg/kg/dose, respectively). PBS or metformin was administered every 2 days via intraperitoneal injection. | |||
| Response Summary | METTL3 is able to promote breast cancer cell proliferation by regulating the Cyclin-dependent kinase inhibitor 1 (CDKN1A) expression by an m6A-dependent manner. Metformin can take p21 as the main target to inhibit such effect. | |||
Ageing-related disease [ICD-11: 9B10-9B60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [283] | |||
| Responsed Disease | Ageing-related disease [ICD-11: 9B10-9B60] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HCT 116 TP53(-/-) | Colon carcinoma | Homo sapiens | CVCL_HD97 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
Cytochrome P450 1B1 (CYP1B1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
 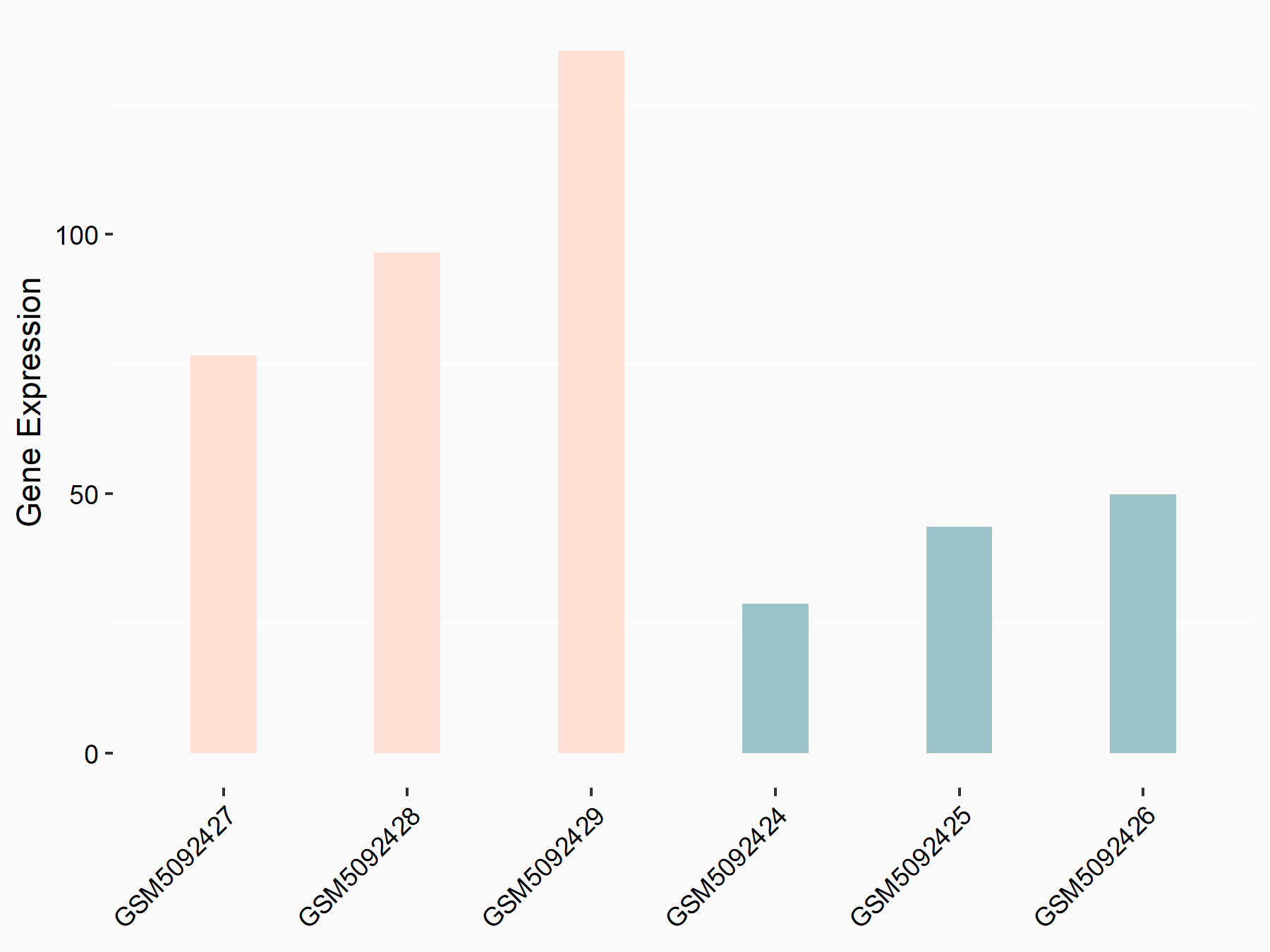 |
logFC: 1.33E+00 p-value: 1.93E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.72E+00 | GSE60213 |
Urinary/pelvic organs injury [ICD-11: NB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [38] | |||
| Responsed Disease | Injury of kidney [ICD-11: NB92.0] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell apoptosis | ||||
In-vitro Model |
NRK-52E | Normal | Rattus norvegicus | CVCL_0468 |
| In-vivo Model | Rats were anesthetized and incised through the midline of the abdomen, and the left renal vertebral arch and arteries were blocked for 45 min, thereby resulting in left kidney ischemia. At the same time, the right kidney was removed, further aggravating the degree of left kidney injury. | |||
| Response Summary | METTL3 contributes to renal ischemia-reperfusion injury by regulating Foxd1 methylation. When METTL3 was inhibited, m6A levels were accordingly decreased and cell apoptosis was suppressed in the H/R in vitro model. Based on MeRIP sequencing, transcription factor activating enhancer binding protein 2-alpha (tfap2a), Cytochrome P450 1B1 (CYP1B1), and forkhead box D1 (foxd1) were significantly differentially expressed, as was m6A, which is involved in the negative regulation of cell proliferation and kidney development. | |||
Death-associated protein kinase 2 (DAPK2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
 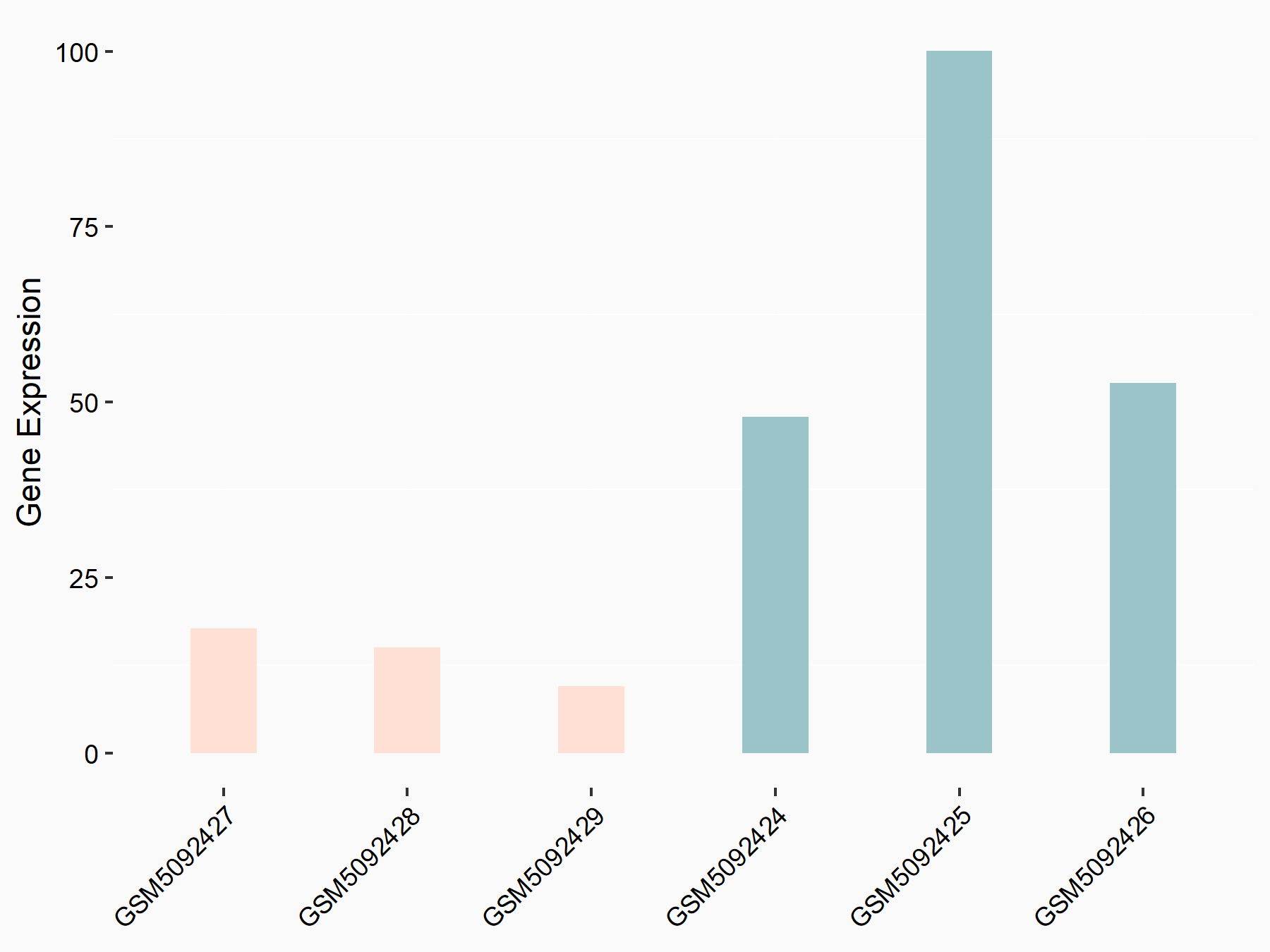 |
logFC: -2.25E+00 p-value: 1.26E-07 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.83E+00 | GSE60213 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [39] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | The nude mice were maintained under pathogen-free conditions and kept under timed lighting conditions mandated by the committee with food and water provided ad libitum. For xenograft experiments, nude mice were injected subcutaneously with 5 × 106 cells resuspended in 0.1 mL PBS. When a tumor was palpable, it was measured every 3 days. | |||
| Response Summary | Cigarette smoking induced aberrant N6-methyladenosine modification of Death-associated protein kinase 2 (DAPK2), which resulted in decreased DAPK2 mRNA stability and expression of its mRNA and protein. This modification was mediated by the m6A "writer" METTL3 and the m6A "reader" YTHDF2. BAY 11-7085, a NF-Kappa-B signaling selective inhibitor, was shown to efficiently suppressed downregulation of DAPK2-induced oncogenic phenotypes of NSCLC cells. | |||
Developmentally-regulated GTP-binding protein 1 (DRG1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Pancreatic islets | Mus musculus |
|
Treatment: Mettl3 knockout mice
Control: Mettl3 flox/flox mice
|
GSE155612 | |
| Regulation |
 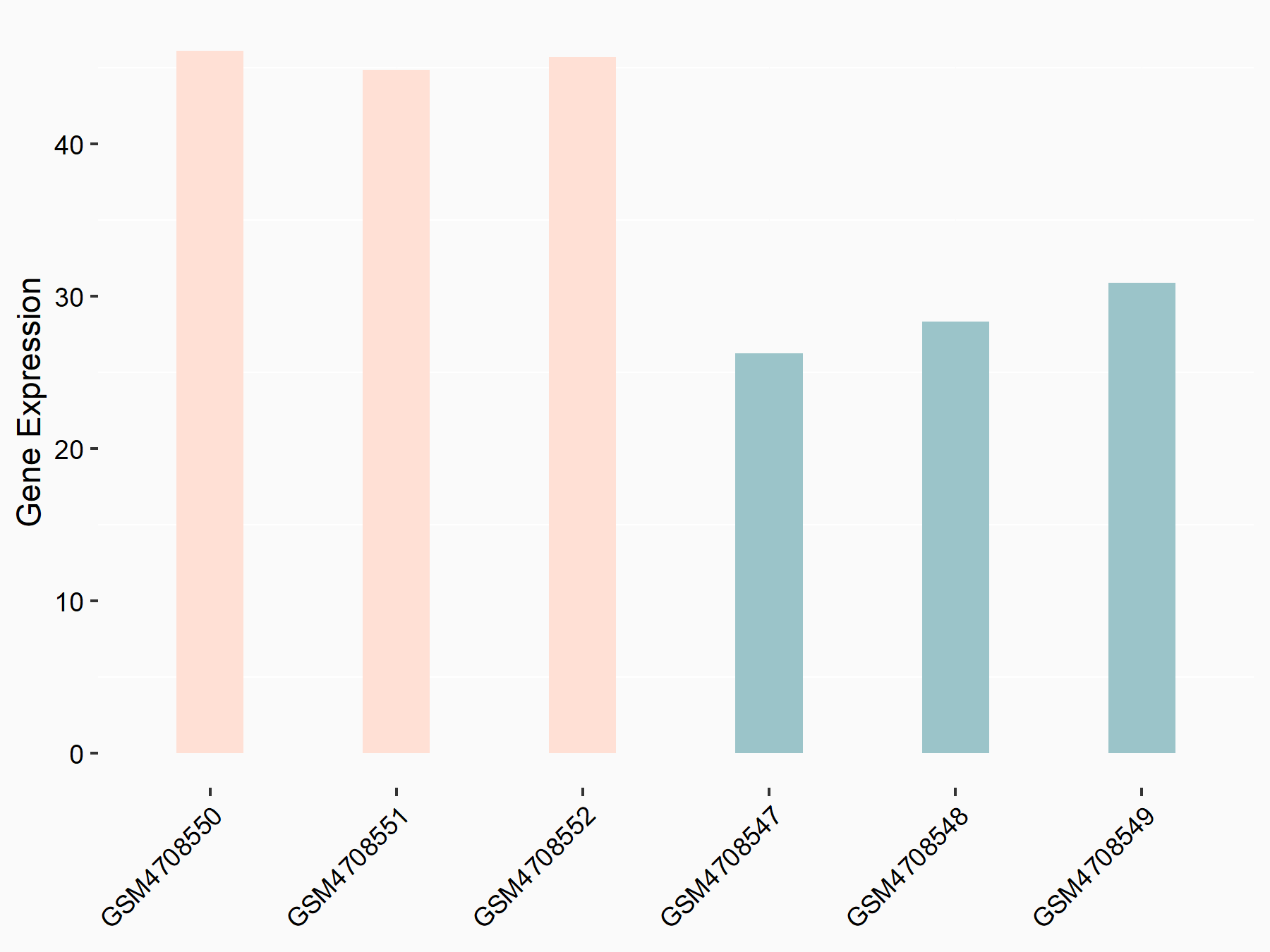 |
logFC: 6.62E-01 p-value: 3.32E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.01E+01 | GSE60213 |
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [40] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell cycle | |||
| Cell apoptosis | ||||
In-vitro Model |
143B | Osteosarcoma | Homo sapiens | CVCL_2270 |
| hFOB 1.19 | Normal | Homo sapiens | CVCL_3708 | |
| MG-63 | Osteosarcoma | Homo sapiens | CVCL_0426 | |
| SaOS-2 | Osteosarcoma | Homo sapiens | CVCL_0548 | |
| U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 | |
| Response Summary | ELAVL1 knockdown impaired the stability of DRG1 mRNA, thereby reducing both the mRNA and protein levels of Developmentally-regulated GTP-binding protein 1 (DRG1). In all, DRG1 exerted tumorigenic effects in osteosarcoma, and the up-regulation of DRG1 in OS was induced by METTL3 and ELAVL1 in an m6A-dependent manner. | |||
DNA polymerase kappa (Pol Kappa/POLK)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
 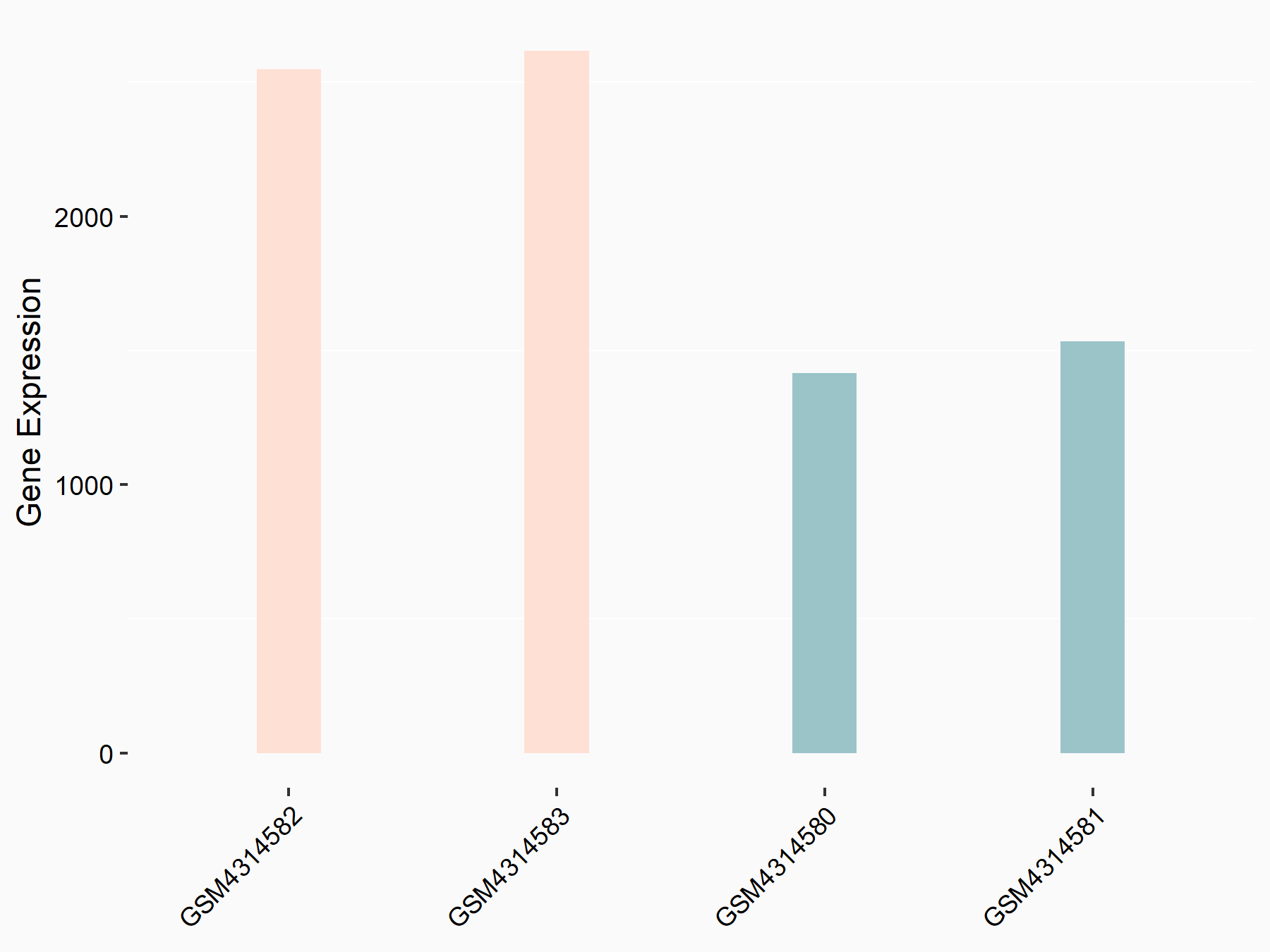 |
logFC: 8.09E-01 p-value: 1.19E-26 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.90E+00 | GSE60213 |
Exposure to radiation [ICD-11: PH73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [41] | |||
| Responsed Disease | Exposure to radiation [ICD-11: PH73] | |||
| Target Regulation | Up regulation | |||
| Cell Process | DNA repair | |||
| Nucleotide excision repair (hsa03420) | ||||
In-vitro Model |
A-375 | Amelanotic melanoma | Homo sapiens | CVCL_0132 |
| U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| MEF (Mouse embryonic fibroblasts) | ||||
| U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 | |
| Response Summary | Methylation at the 6 position of adenosine (m6A) in RNA is rapidly (within 2 min) and transiently induced at DNA damage sites in response to ultraviolet irradiation. This modification occurs on numerous poly(A)+ transcripts and is regulated by the methyltransferase METTL3 and the demethylase FTO. DNA DNA polymerase kappa (Pol Kappa/POLK), which has been implicated in both nucleotide excision repair and trans-lesion synthesis, required the catalytic activity of METTL3 for immediate localization to ultraviolet-induced DNA damage sites. | |||
DNA replication licensing factor MCM6 (MCM6)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3-deficient liver
Control: Wild type liver cells
|
GSE197800 | |
| Regulation |
 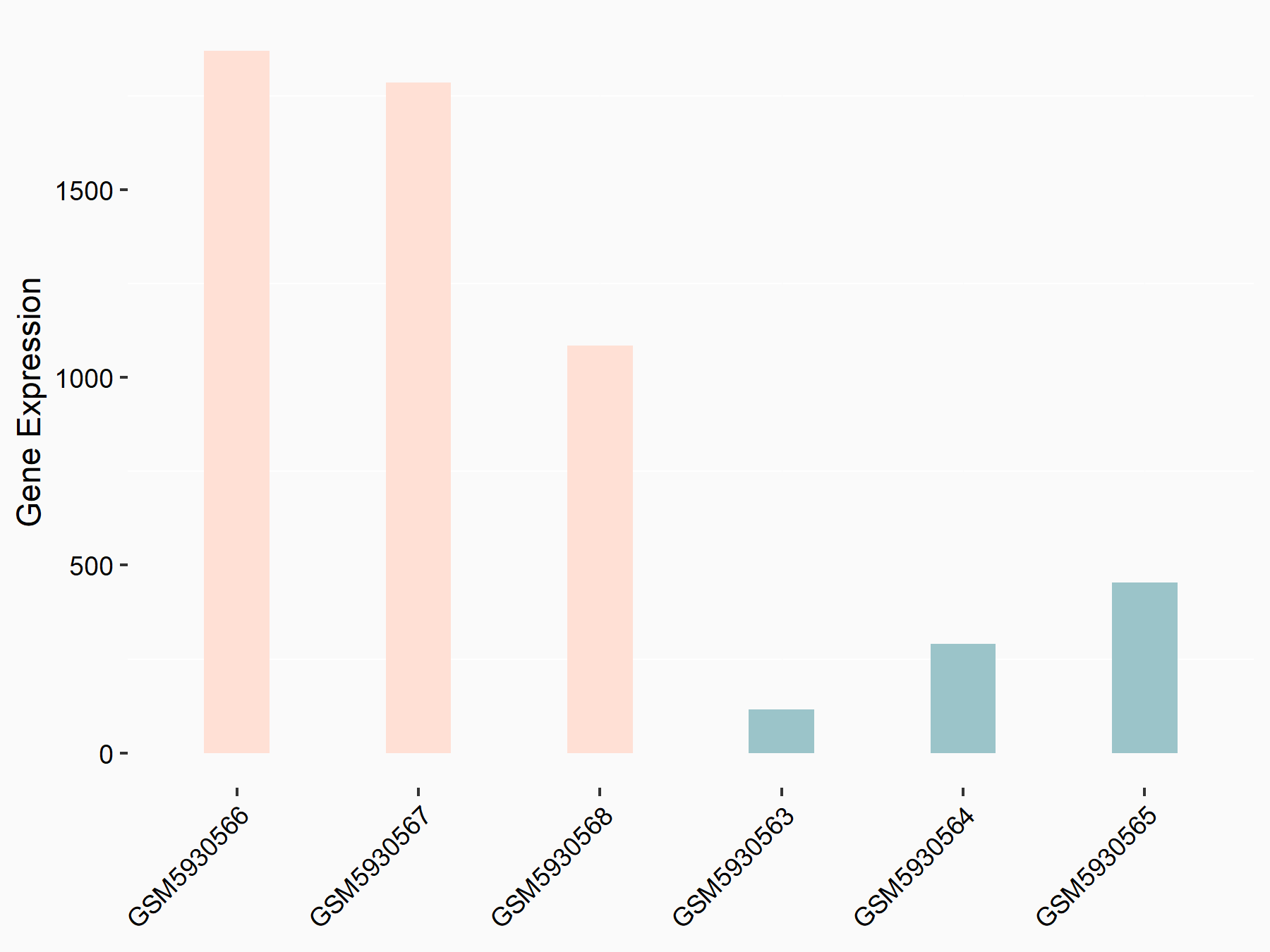 |
logFC: 2.47E+00 p-value: 3.65E-08 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.92E+00 | GSE60213 |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [42] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
GES-1 | Normal | Homo sapiens | CVCL_EQ22 |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MKN74 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_2791 | |
| pGCC (Primary GC cells) | ||||
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| In-vivo Model | A total of 2 × 106 GC cells were injected into the flank of nude mice in a 1:1 suspension of BD Matrigel (BD Biosciences) in phosphate-buffered saline (PBS) solution. | |||
| Response Summary | In gastric cancer, several component molecules (e.g., MCM5, DNA replication licensing factor MCM6 (MCM6), etc.) of MYC target genes were mediated by METTL3 via altered m6A modification. | |||
E3 ubiquitin-protein ligase TRIM7 (TRIM7)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
 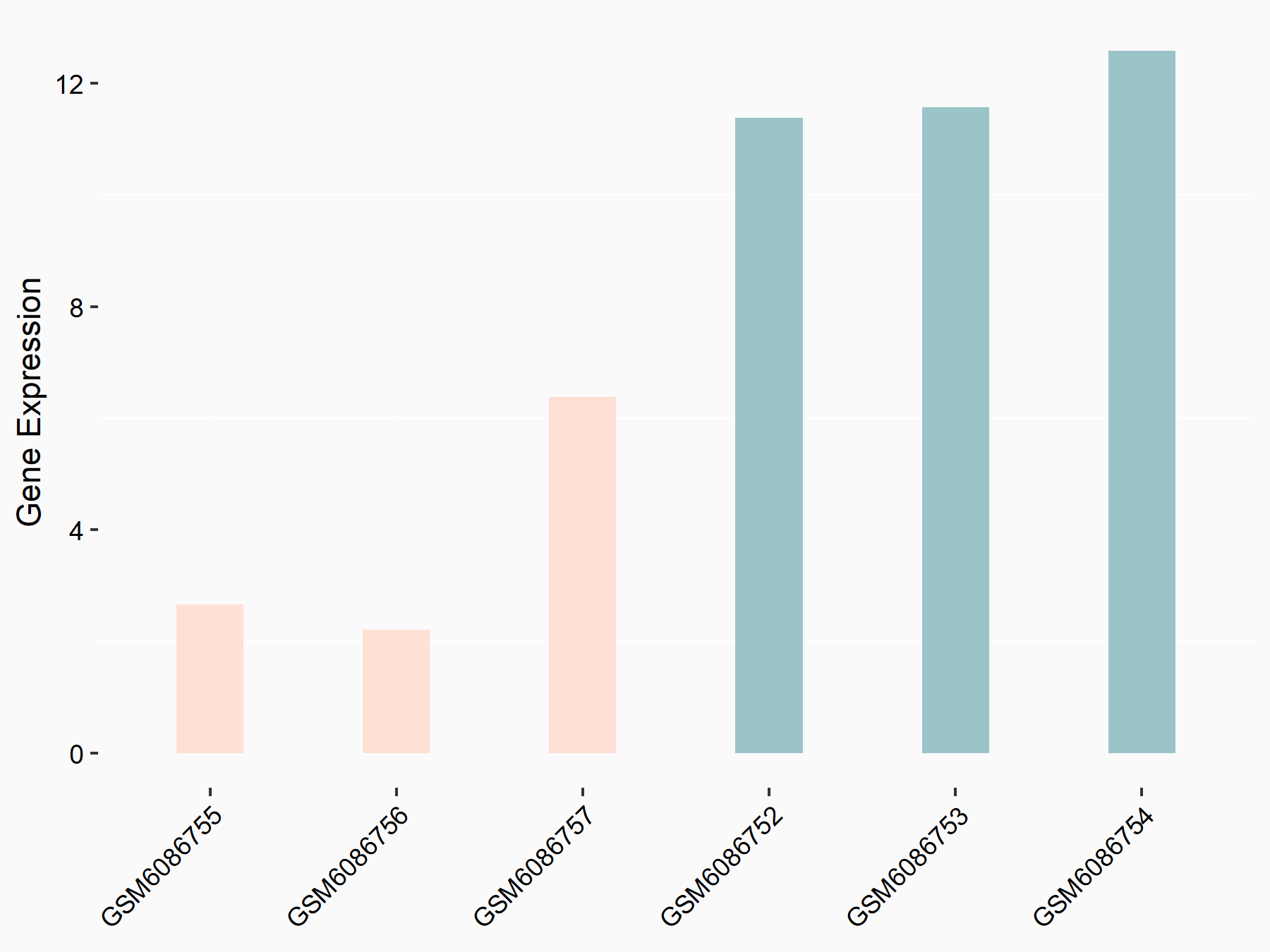 |
logFC: -1.54E+00 p-value: 1.28E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.18E+00 | GSE60213 |
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [43] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Ubiquitin mediated proteolysis | hsa04120 | ||
| Cell Process | Proteasome pathway degradation | |||
In-vitro Model |
U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 |
| SaOS-2 | Osteosarcoma | Homo sapiens | CVCL_0548 | |
| MG-63 | Osteosarcoma | Homo sapiens | CVCL_0426 | |
| HOS | Osteosarcoma | Homo sapiens | CVCL_0312 | |
| hFOB 1.19 | Normal | Homo sapiens | CVCL_3708 | |
| In-vivo Model | MG63 cells transduced with lentivirus expressing shTRIM7 or shNC, and SAOS2 cells transduced with lentivirus expressing TRIM7, BRMS1, TRIM7 plus BRMS1 or control vector, were injected via the tail vein into the nude mice (1 × 106 cells/mouse) (n = 11 per group). | |||
| Response Summary | E3 ubiquitin-protein ligase TRIM7 (TRIM7) mRNA stability was regulated by the METTL3/14-YTHDF2-mRNA in a decay-dependent manner. TRIM7 plays a key role in regulating metastasis and chemoresistance in osteosarcoma through ubiquitination of BRMS1. | |||
eIF4E-binding protein 1 (4EBP1/EIF4EBP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shMETTL3 MOLM13 cells
Control: MOLM13 cells
|
GSE98623 | |
| Regulation |
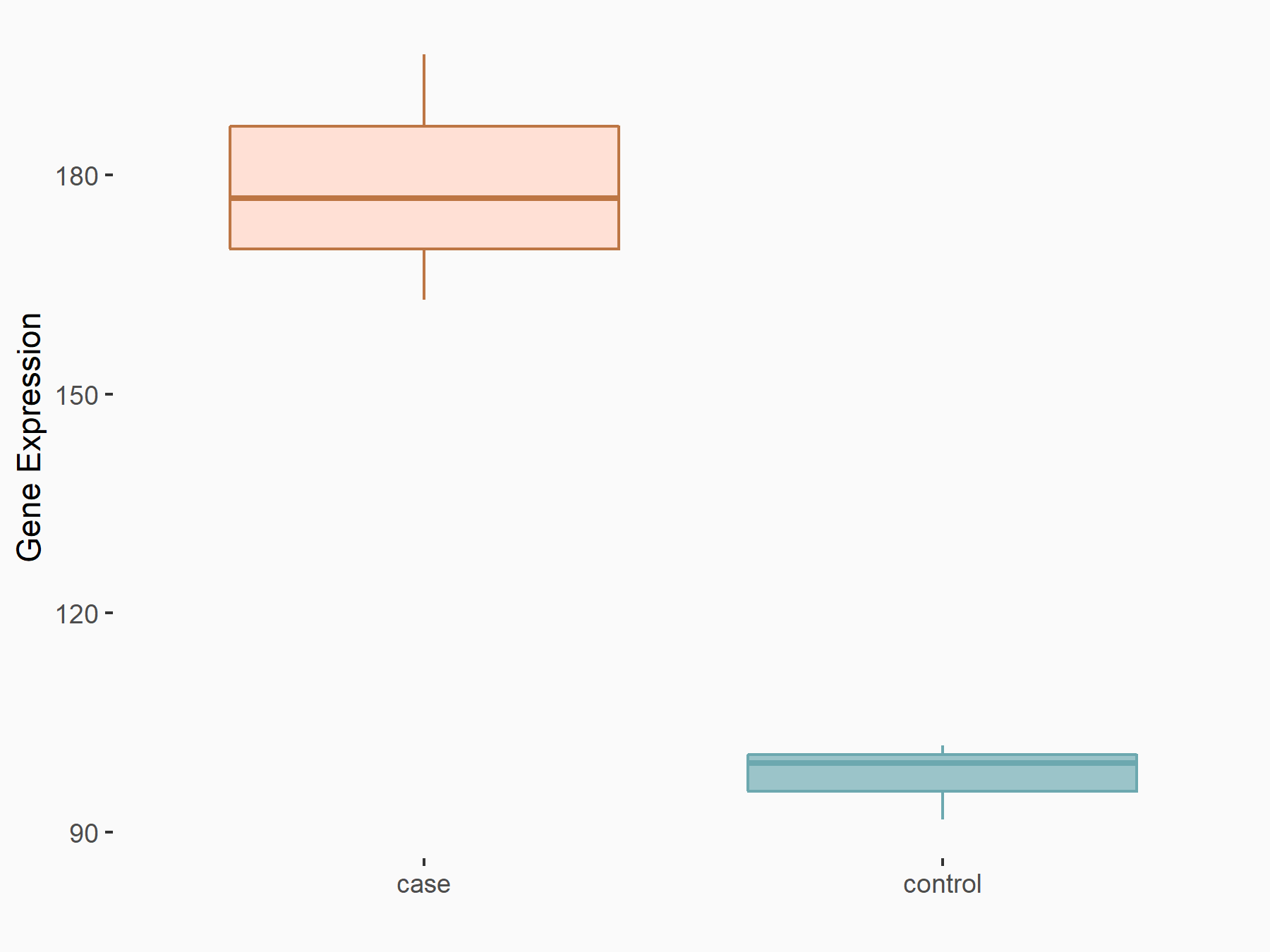 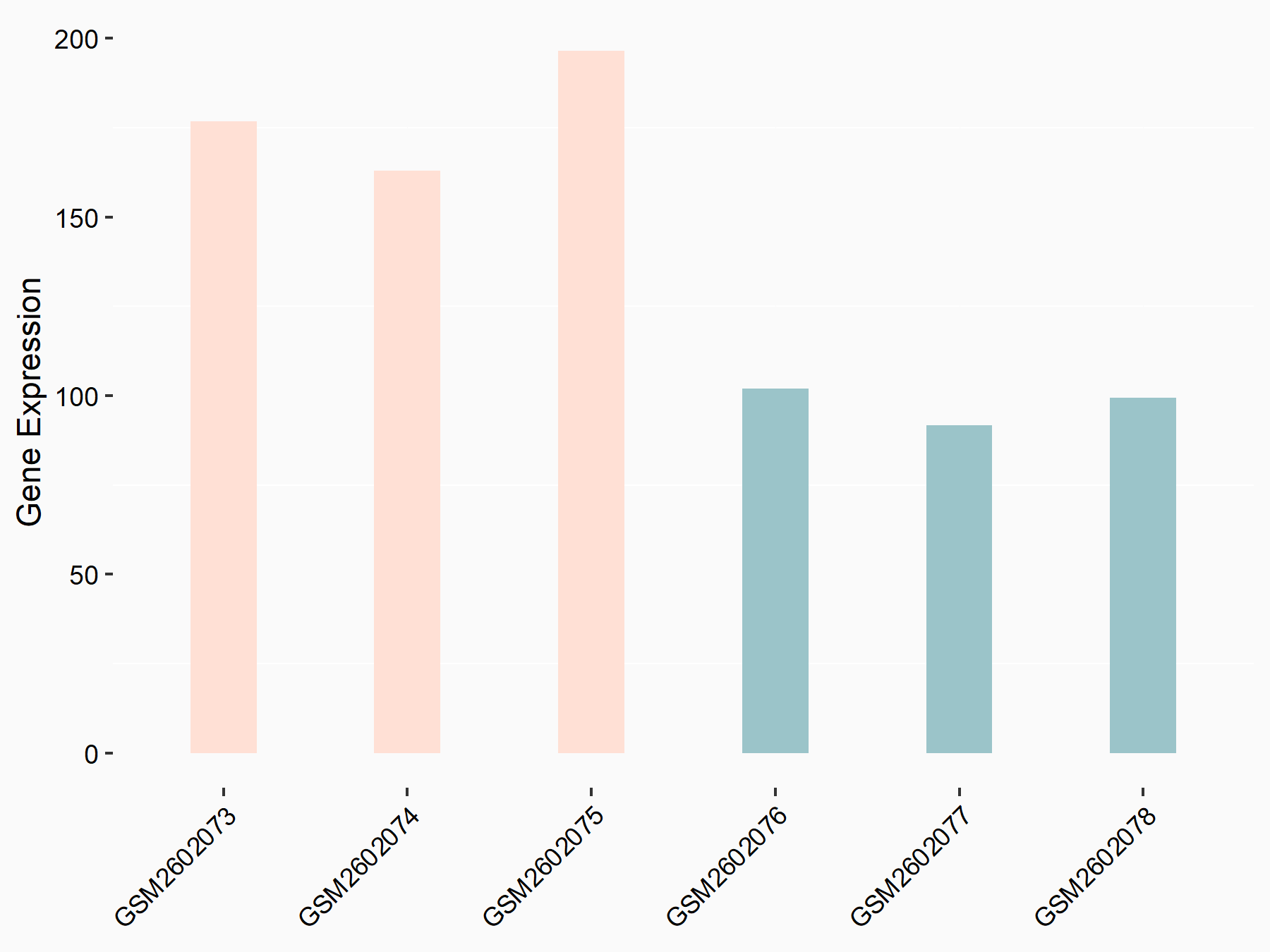 |
logFC: 8.71E-01 p-value: 3.32E-08 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 9.71E+00 | GSE60213 |
Retina cancer [ICD-11: 2D02]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [44] | |||
| Responsed Disease | Retinoblastoma [ICD-11: 2D02.2] | |||
| Responsed Drug | Rapamycin | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
In-vitro Model |
WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through PI3K/AKT/mTOR pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-P70S6K/eIF4E-binding protein 1 (4EBP1/EIF4EBP1) pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
Endoplasmic reticulum chaperone BiP (Grp78)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
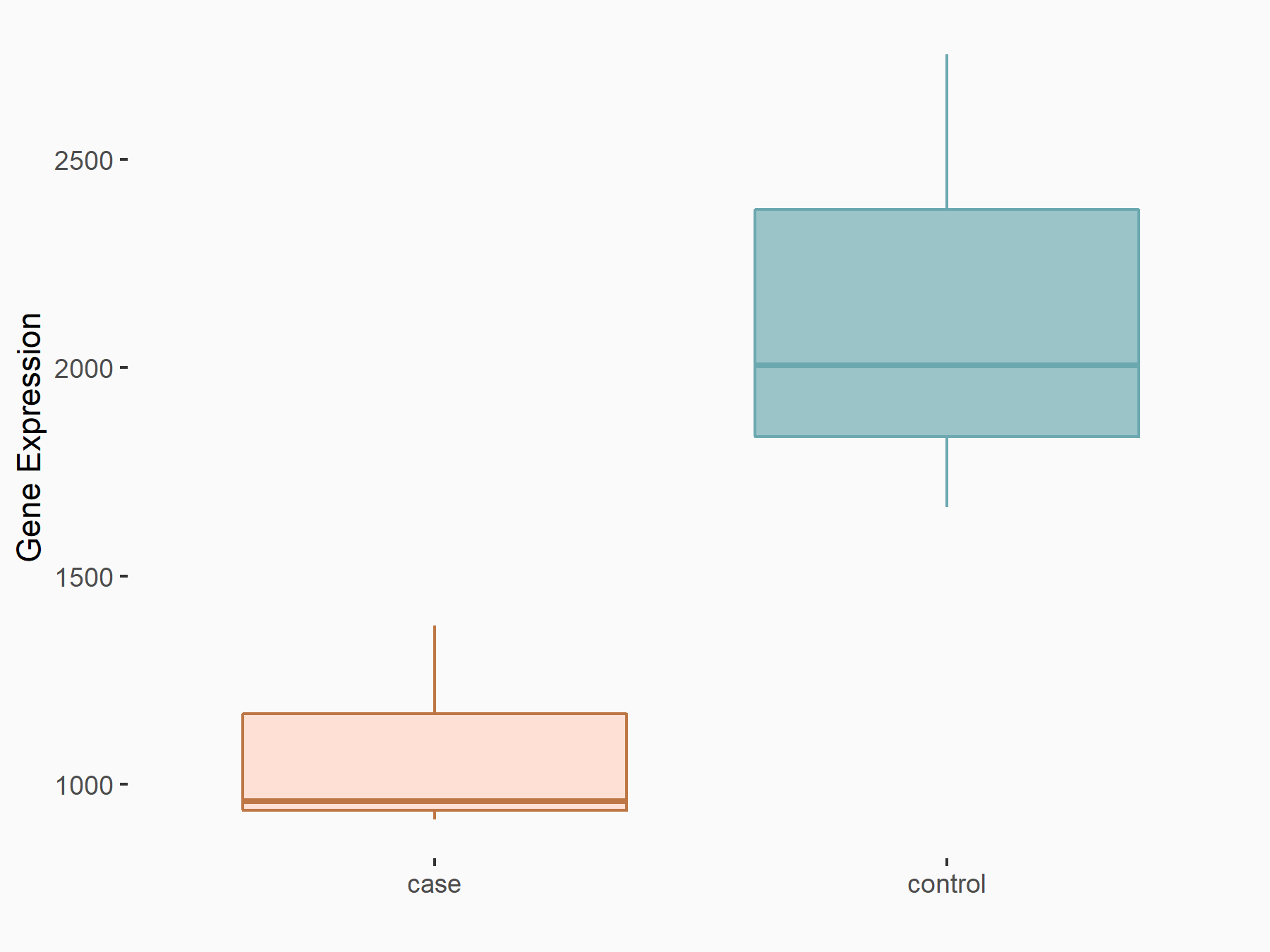 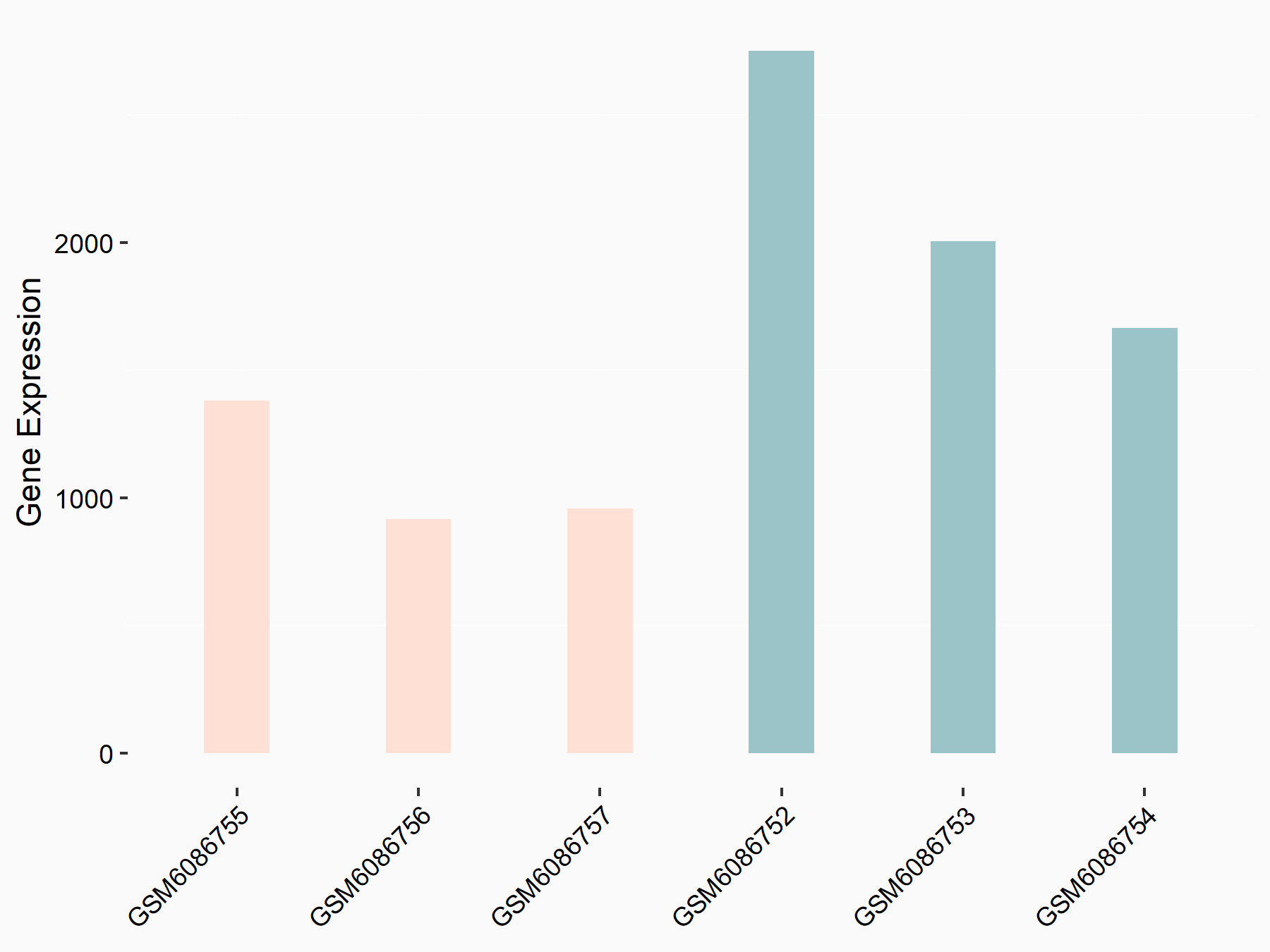 |
logFC: -9.73E-01 p-value: 3.50E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.24E+00 | GSE60213 |
Diseases of the musculoskeletal system [ICD-11: FC0Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [45] | |||
| Responsed Disease | Diseases of the musculoskeletal system [ICD-11: FC0Z] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
MC3T3-E1 | Normal | Mus musculus | CVCL_0409 |
| Response Summary | METTL3 knockdown enhanced Endoplasmic reticulum chaperone BiP (Grp78) expression through YTHDF2-mediated RNA degradation, which elicited ER stress, thereby promoting osteoblast apoptosis and inhibiting cell proliferation and differentiation under LPS-induced inflammatory condition. | |||
Ephrin type-A receptor 2 (EphA2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
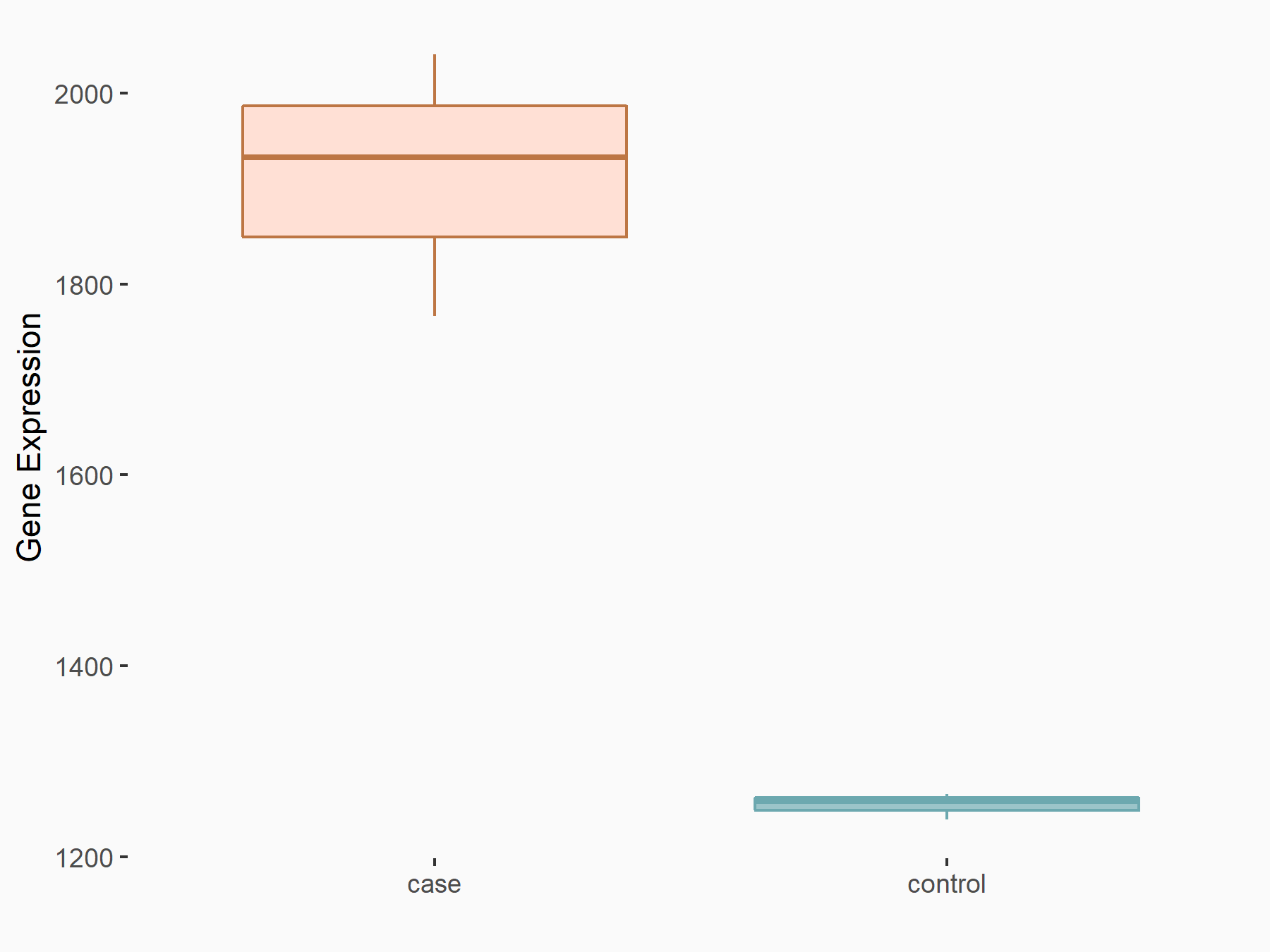 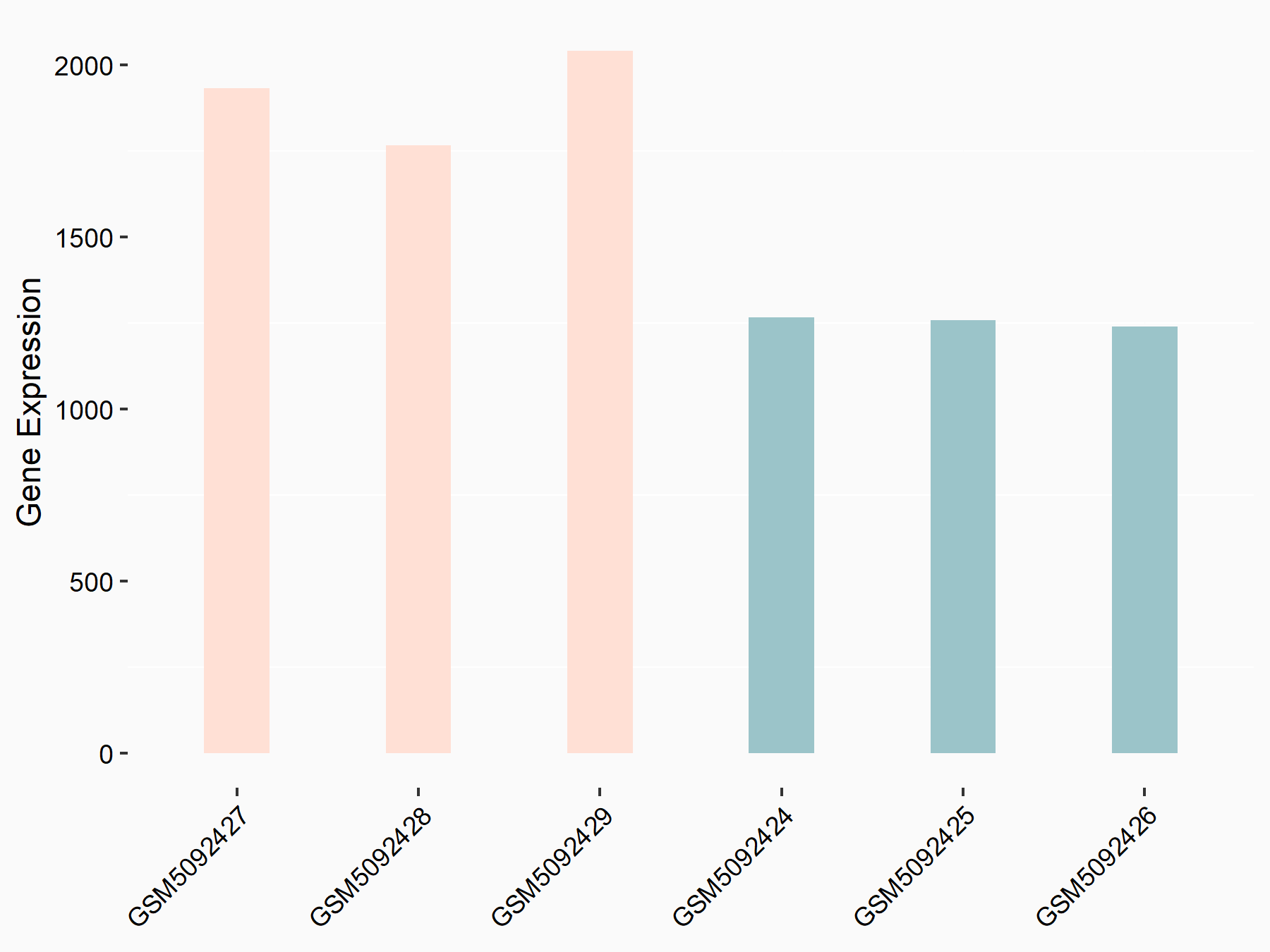 |
logFC: 6.09E-01 p-value: 1.09E-18 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.66E+00 | GSE60213 |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [46] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
In-vitro Model |
SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| In-vivo Model | A total of 8 × 106 wild-type (WT) or METTL3-knockdown cells were injected into the dorsal flanks of 6-week-old nude mice. Seven mice were randomly selected to calculate the volume according to the following formula: V = (width2 × length)/2. Mice were euthanized three weeks after injection and tumors removed, weighed, fixed, and embedded for immunohistochemical analysis. | |||
| Response Summary | Ephrin type-A receptor 2 (EphA2) and VEGFA targeted by METTL3 via different IGF2BP-dependent mechanisms were found to promote vasculogenic mimicry (VM) formation via PI3K/AKT/mTOR and ERK1/2 signaling in CRC. | |||
Far upstream element-binding protein 1 (FUBP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | DKO-1 cell line | Homo sapiens |
|
Treatment: METTL3 knockdown DKO-1 cell
Control: DKO-1 cell
|
GSE182382 | |
| Regulation |
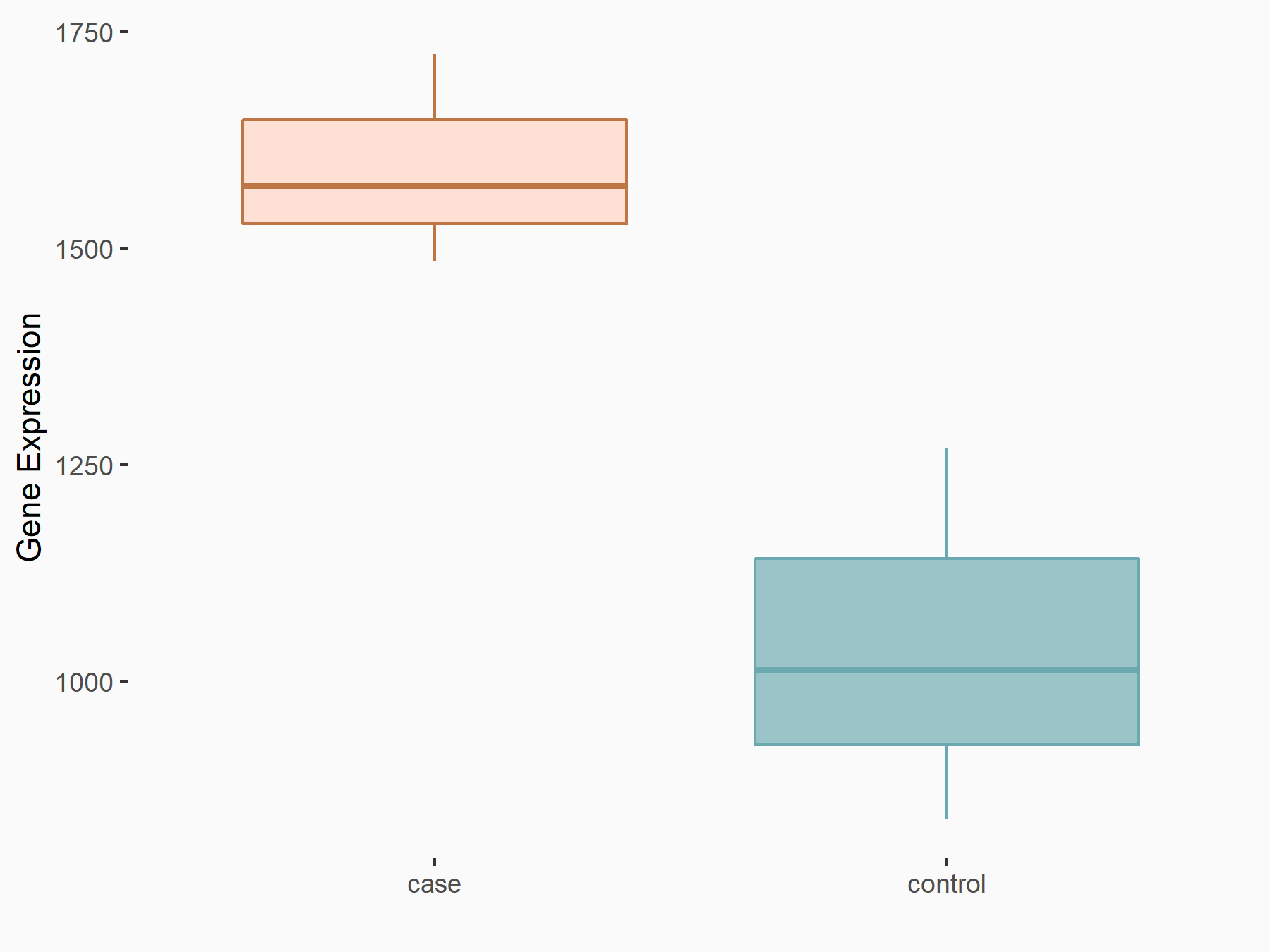 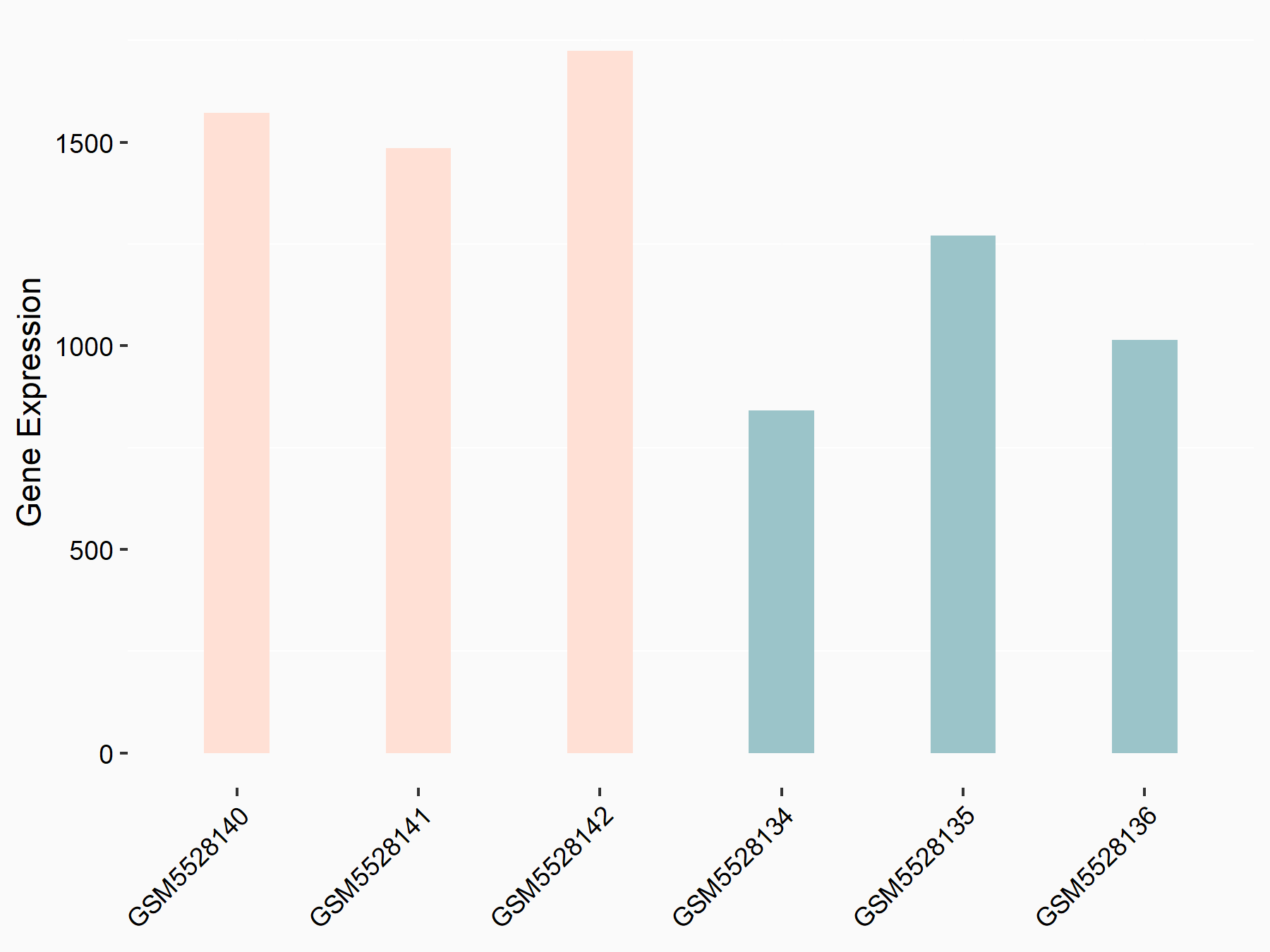 |
logFC: 6.14E-01 p-value: 4.72E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.31E+00 | GSE60213 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [47] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0608 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HOP-62 | Lung adenocarcinoma | Homo sapiens | CVCL_1285 | |
| In-vivo Model | For the in vivo tumorigenicity assay, female BALB/c nude mice (ages 4-5 weeks) were randomly divided into two groups (n = 6/group). Calu1 cells (4 × 106) that had been stably transfected with sh-LCAT3 or scramble were implanted subcutaneously into the nude mice. Tumor growth was measured after one week, and tumor volumes were calculated with the following formula: Volume (cm3) = (length × width2)/2. After four weeks, the mice were euthanized, and the tumors were collected and weighed. For the in vivo tumor invasion assay, 1.2 × 106 scramble or shLCAT3 cells were injected intravenously into the tail vein of nude mice (n = 6/group). 1.5 mg luciferin (Gold Biotech, St Louis, MO, USA) was administered once a week for 4 weeks, to monitor metastases using an IVIS@ Lumina II system (Caliper Life Sciences, Hopkinton, MA, USA). | |||
| Response Summary | LCAT3 upregulation is attributable to N6-methyladenosine (m6A) modification mediated by methyltransferase like 3 (METTL3), leading to LCAT3 stabilization. LCAT3 as a novel oncogenic lncRNA in the lung, and validated the LCAT3-Far upstream element-binding protein 1 (FUBP1)-MYC axis as a potential therapeutic target for lung adenocarcinomas. | |||
Fatty acid synthase (FASN)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LNCaP cell line | Homo sapiens |
|
Treatment: shMETTL3 LNCaP cells
Control: shControl LNCaP cells
|
GSE147884 | |
| Regulation |
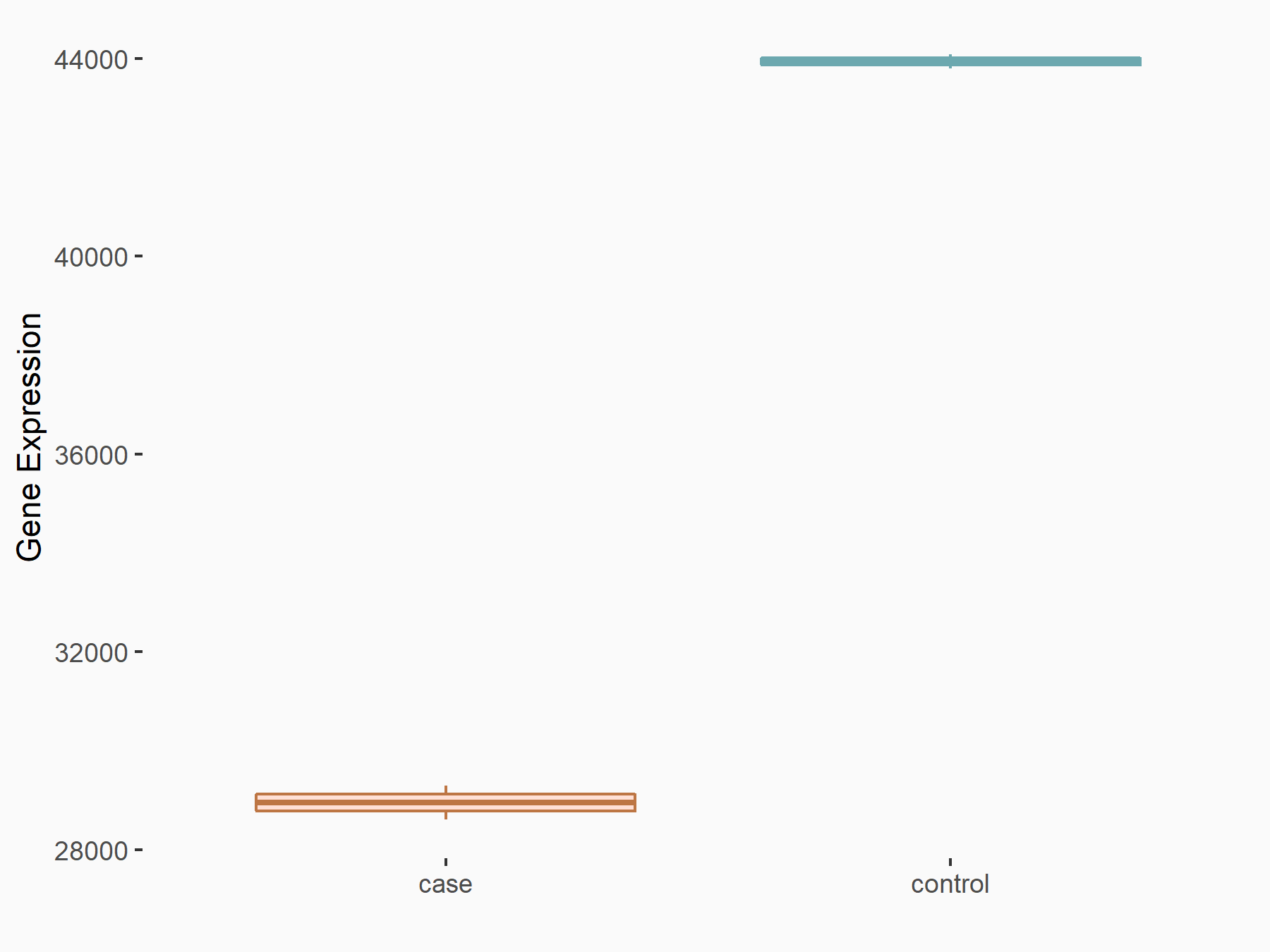 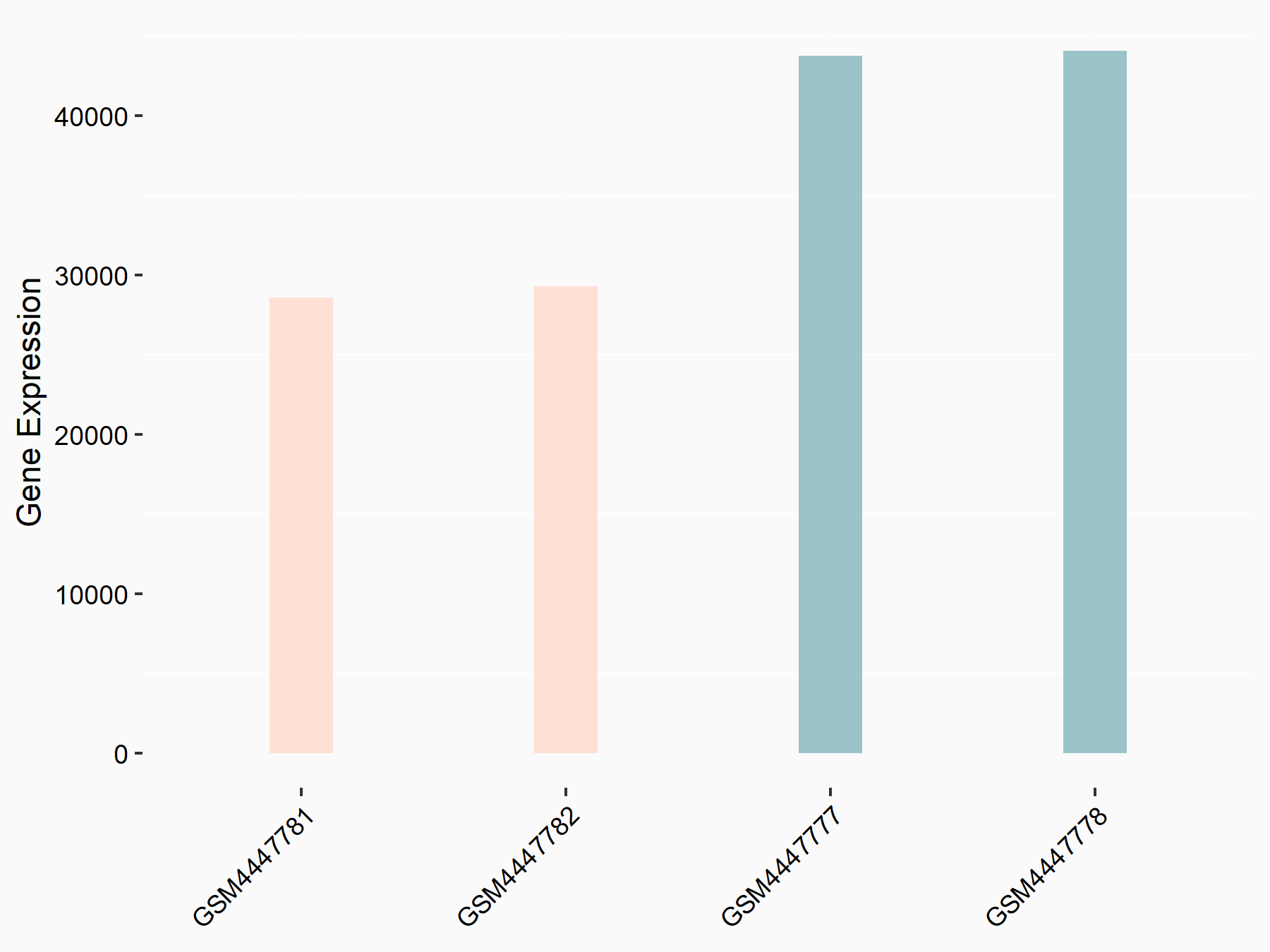 |
logFC: -6.03E-01 p-value: 2.01E-249 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.26E+00 | GSE60213 |
Type 2 diabetes mellitus [ICD-11: 5A11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [48] | |||
| Responsed Disease | Type 2 diabetes mellitus [ICD-11: 5A11] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Insulin resistance | hsa04931 | ||
| Cell Process | Lipid metabolism | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | Hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were generated by crossing mice with TBG-Cre Tg mice. METTL3 flox (METTL3 fl/fl) and hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were used for experiments. | |||
| Response Summary | Type 2 diabetes (T2D) is characterized by lack of insulin, insulin resistance and high blood sugar. METTL3 silence decreased the m6A methylated and total mRNA level of Fatty acid synthase (FASN), subsequently inhibited fatty acid metabolism. The expression of Acc1, Acly, Dgat2, Ehhadh, Fasn, Foxo, Pgc1a and Sirt1, which are critical to the regulation of fatty acid synthesis and oxidation were dramatically decreased in livers of hepatocyte-specific METTL3 knockout mice. | |||
Forkhead box protein D1 (FOXD1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
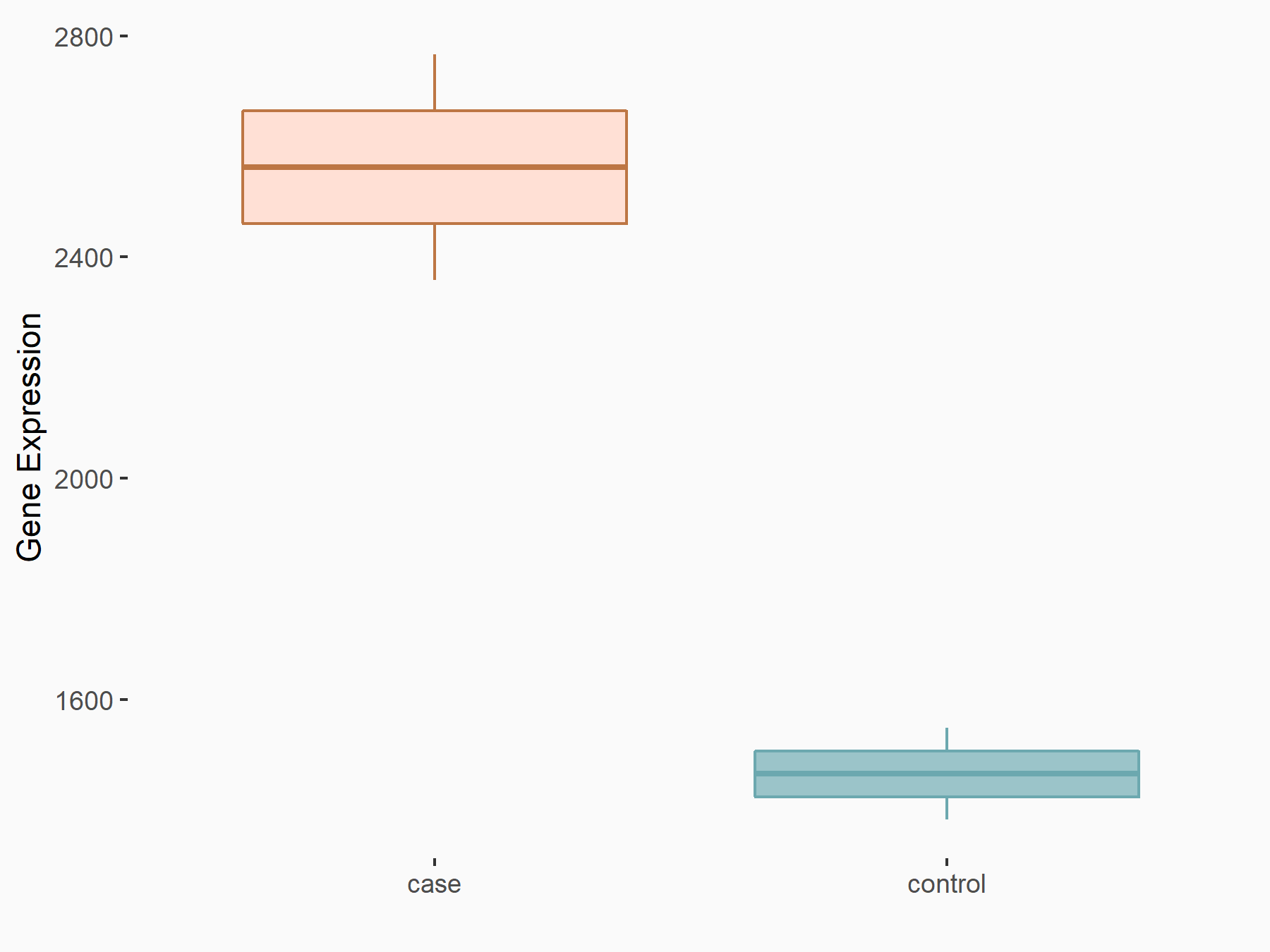 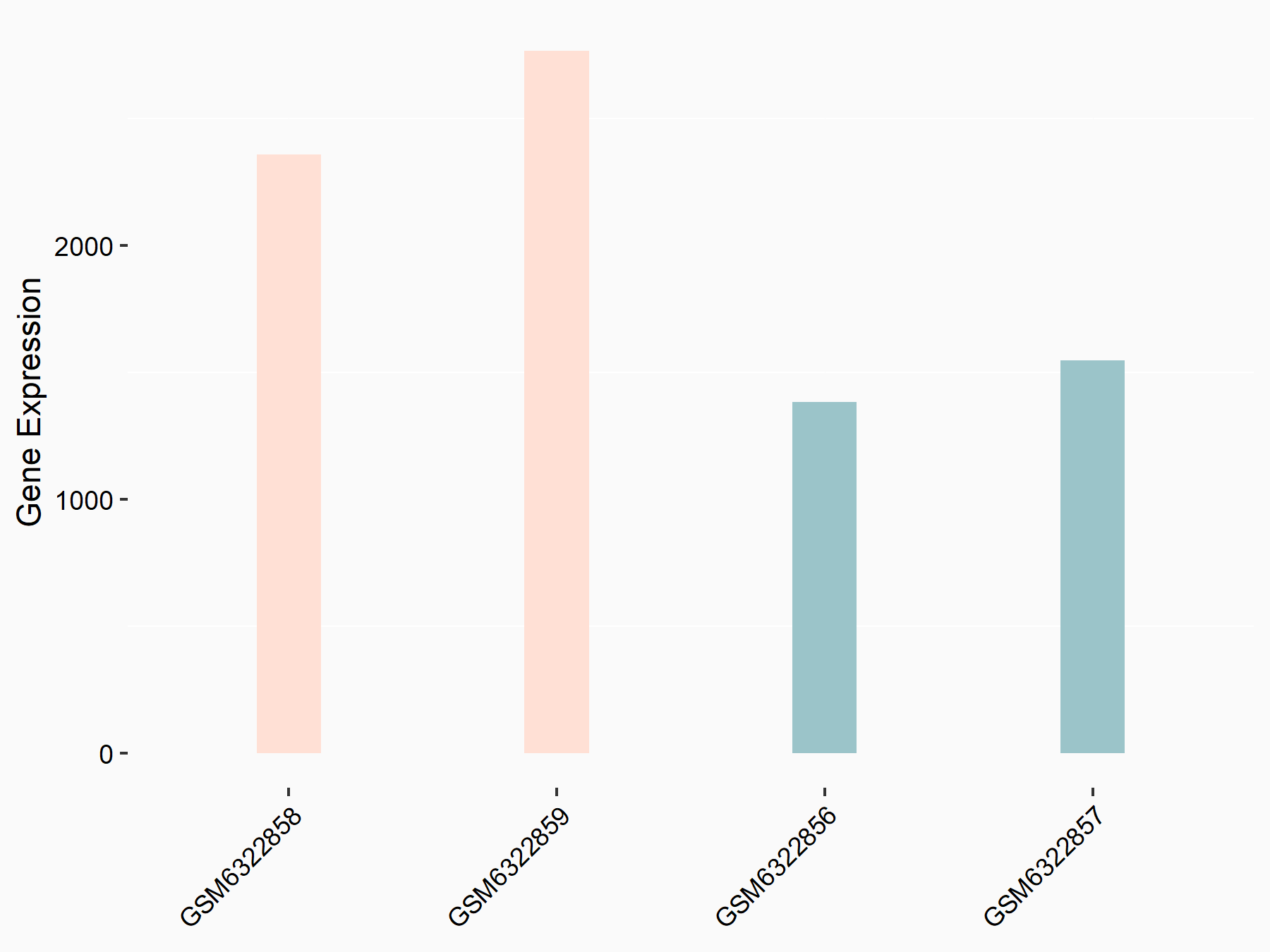 |
logFC: 8.07E-01 p-value: 2.88E-13 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.40E+00 | GSE60213 |
Urinary/pelvic organs injury [ICD-11: NB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [38] | |||
| Responsed Disease | Injury of kidney [ICD-11: NB92.0] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell proliferation | |||
| Cell apoptosis | ||||
In-vitro Model |
NRK-52E | Normal | Rattus norvegicus | CVCL_0468 |
| In-vivo Model | Rats were anesthetized and incised through the midline of the abdomen, and the left renal vertebral arch and arteries were blocked for 45 min, thereby resulting in left kidney ischemia. At the same time, the right kidney was removed, further aggravating the degree of left kidney injury. | |||
| Response Summary | METTL3 contributes to renal ischemia-reperfusion injury by regulating Forkhead box protein D1 (FOXD1) methylation. When METTL3 was inhibited, m6A levels were accordingly decreased and cell apoptosis was suppressed in the H/R in vitro model. Based on MeRIP sequencing, transcription factor activating enhancer binding protein 2-alpha (tfap2a), cytochrome P-450 1B1 (cyp1b1), and forkhead box D1 (foxd1) were significantly differentially expressed, as was m6A, which is involved in the negative regulation of cell proliferation and kidney development. | |||
G1/S-specific cyclin-E1 (CCNE1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
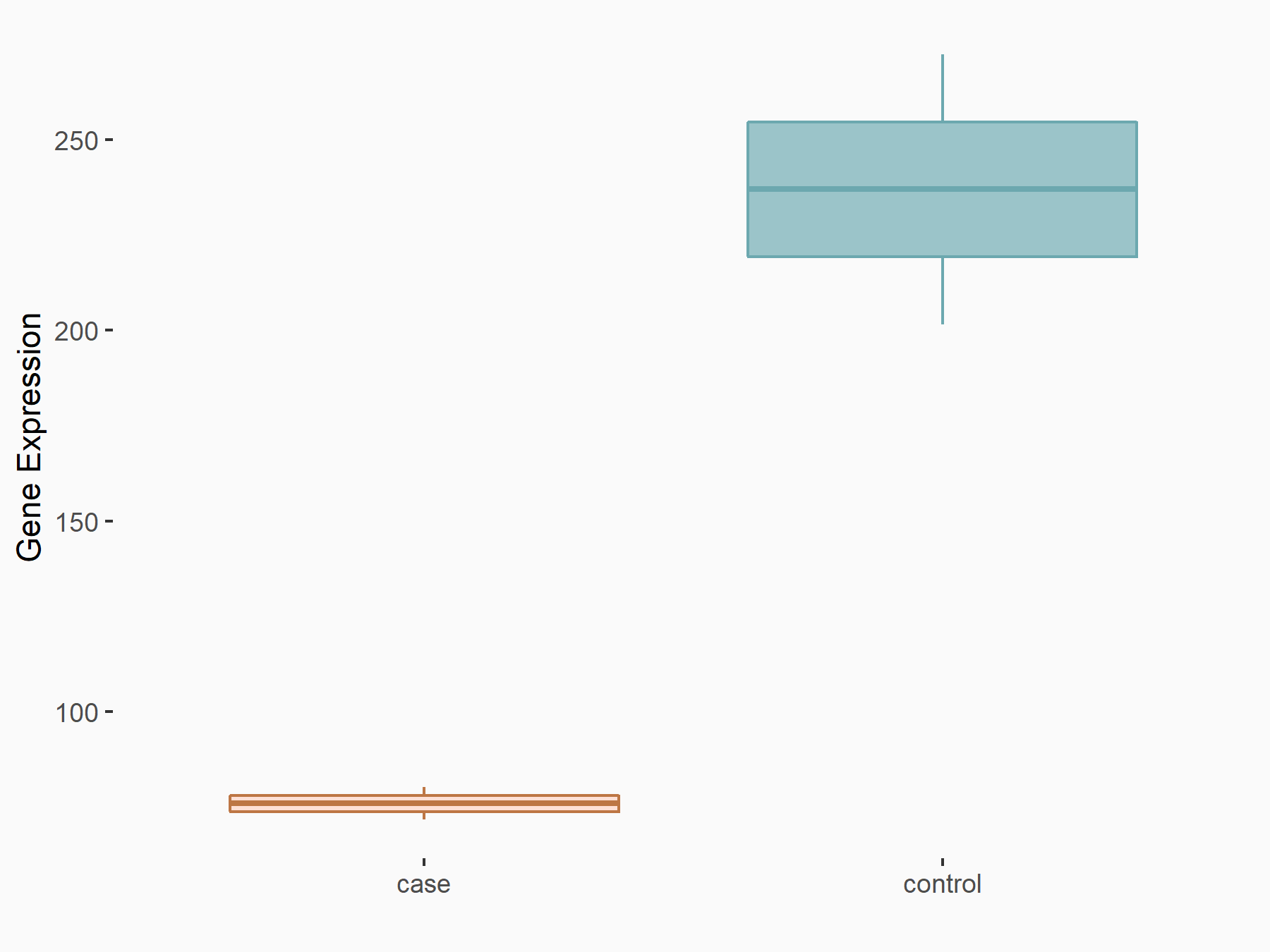 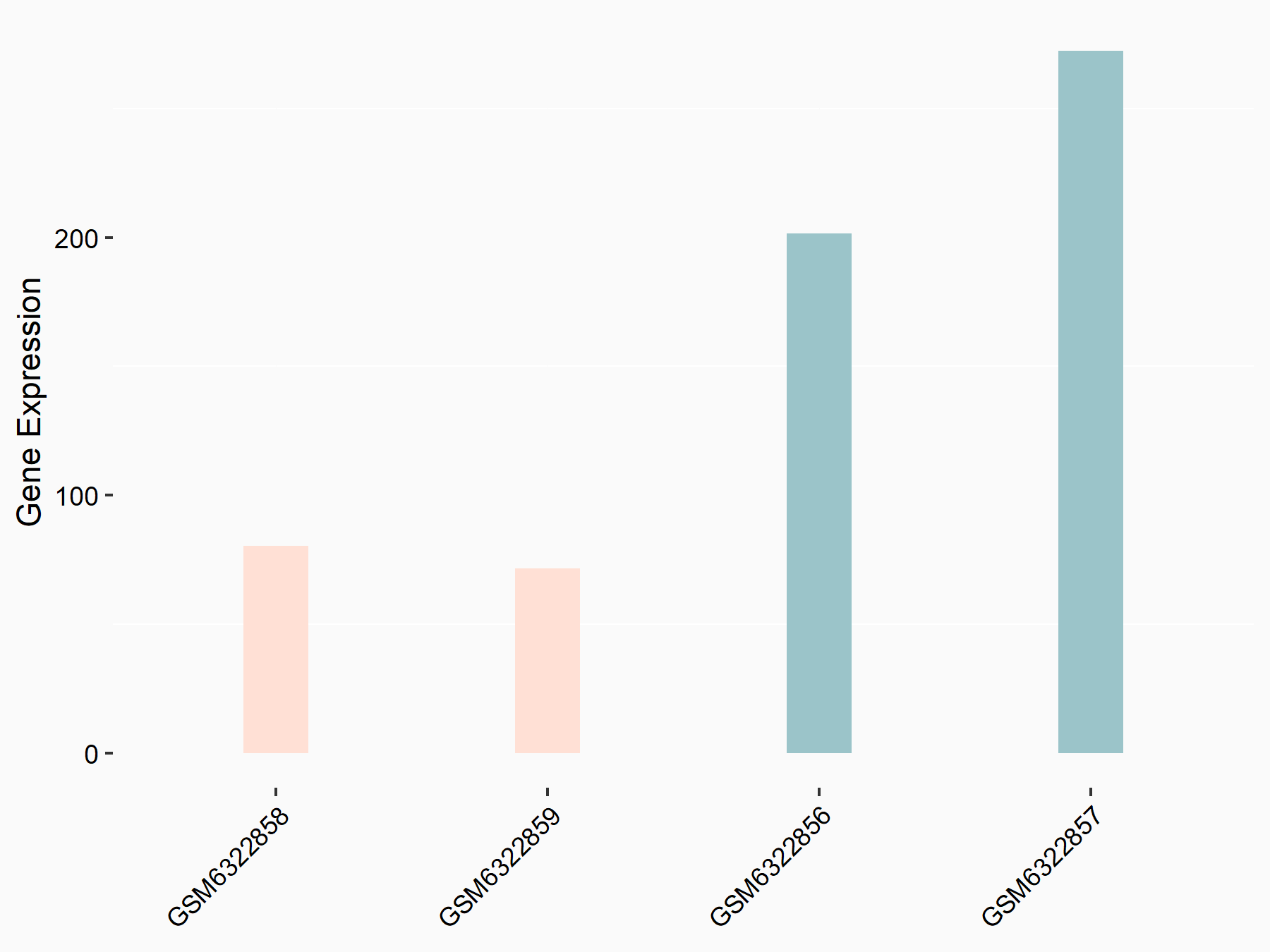 |
logFC: -1.64E+00 p-value: 3.22E-09 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.94E+00 | GSE60213 |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [49] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Cell proliferation | |||
| Arrest cell cycle at G1 phase | ||||
In-vitro Model |
HT29 | Colon cancer | Mus musculus | CVCL_A8EZ |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| In-vivo Model | LoVo cells (5 × 106 cells/ 200 uL PBS) stably transfected with METTL3 knockdown lentiviral vector or control vector were respectively injected subcutaneously into the left flank of each mouse. | |||
| Response Summary | METTL3 promotes colorectal cancer proliferation by stabilizing G1/S-specific cyclin-E1 (CCNE1) mRNA in an m6A-dependent manner, representing a promising therapeutic strategy for the treatment of CRC. | |||
H/ACA ribonucleoprotein complex subunit DKC1 (DKC1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Pancreatic islets | Mus musculus |
|
Treatment: Mettl3 knockout mice
Control: Mettl3 flox/flox mice
|
GSE155612 | |
| Regulation |
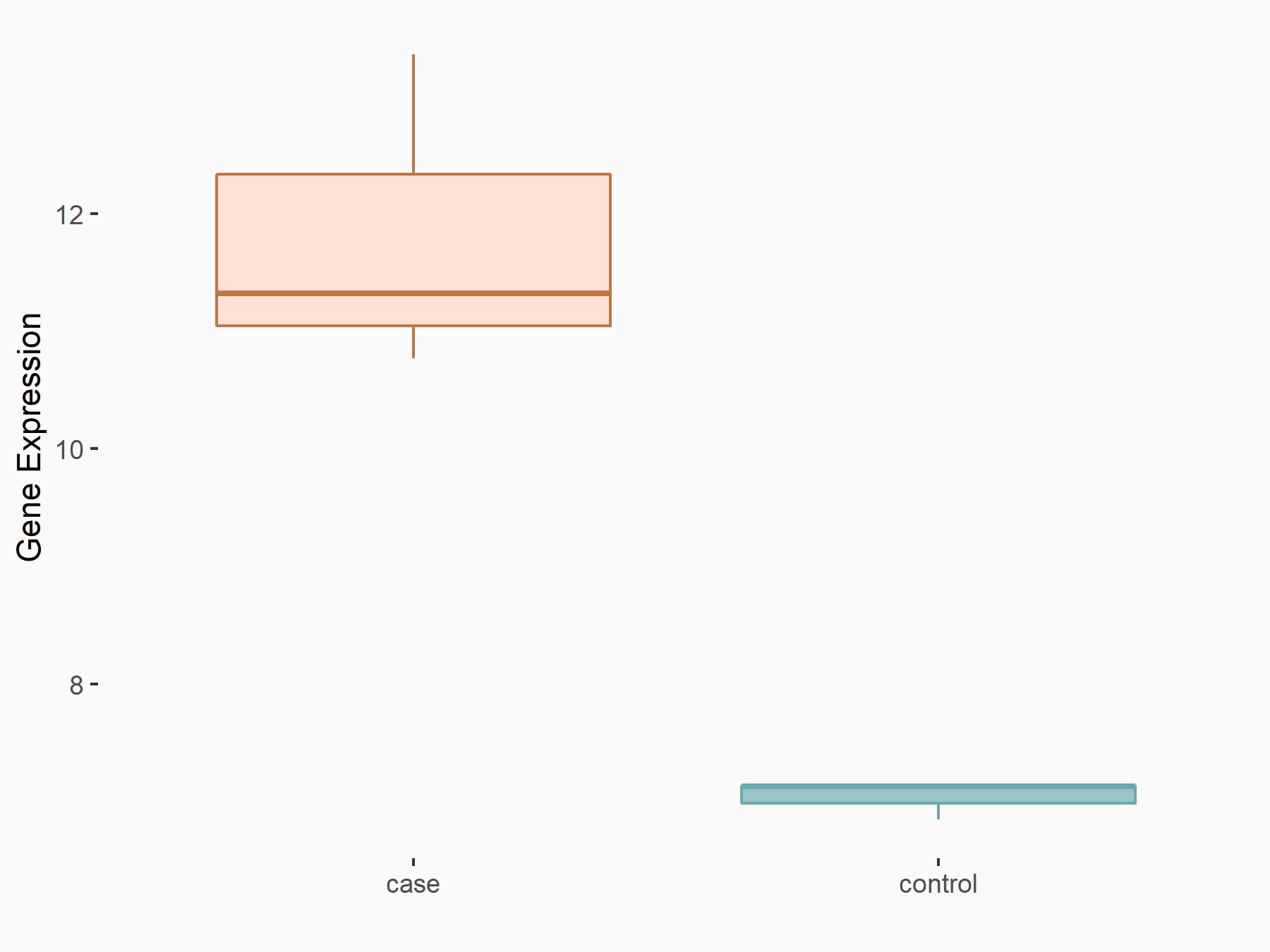 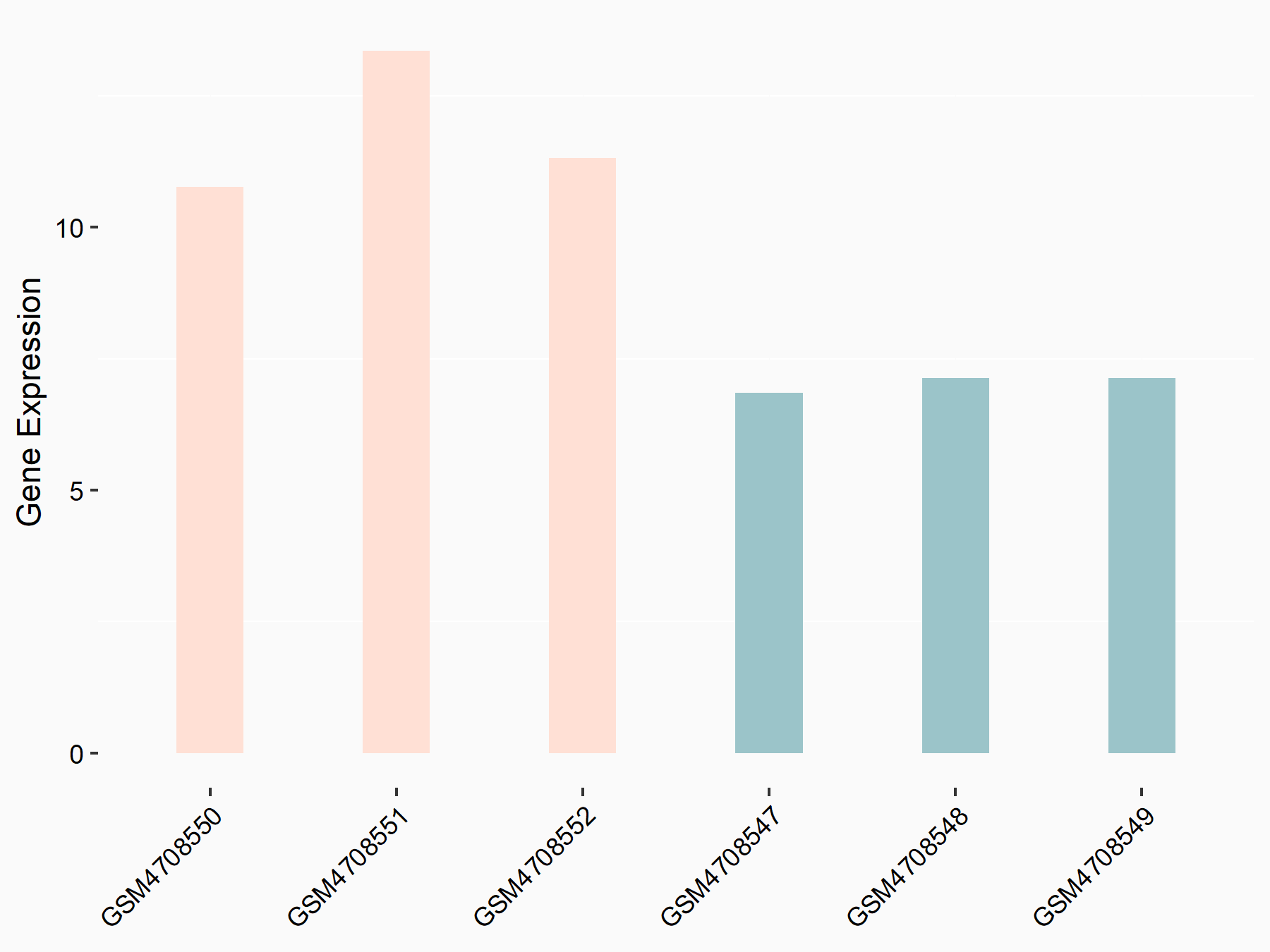 |
logFC: 6.68E-01 p-value: 1.04E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.61E+00 | GSE60213 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [50] | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 |
| In-vivo Model | Athymic BALB/c mice were injected with LCSC cells at the armpit area subcutaneously. The mice were then sacrificed and the tumors recovered. | |||
| Response Summary | CircMEG3 inhibits the expression of m6A methyltransferase METTL3 dependent on HULC. Moreover, CircMEG3 inhibits the expression of H/ACA ribonucleoprotein complex subunit DKC1 (DKC1), a component of telomere synthetase H/ACA ribonucleoprotein (RNP; catalyst RNA pseudouracil modification) through METTL3 dependent on HULC. These observations provide important basic information for finding effective liver cancer therapeutic targets. | |||
Heat shock protein HSP 90-alpha (HSP90/HSP90AA1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HUVEC cell line | Homo sapiens |
|
Treatment: shMETTL3 HUVEC cells
Control: shScramble HUVEC cells
|
GSE157544 | |
| Regulation |
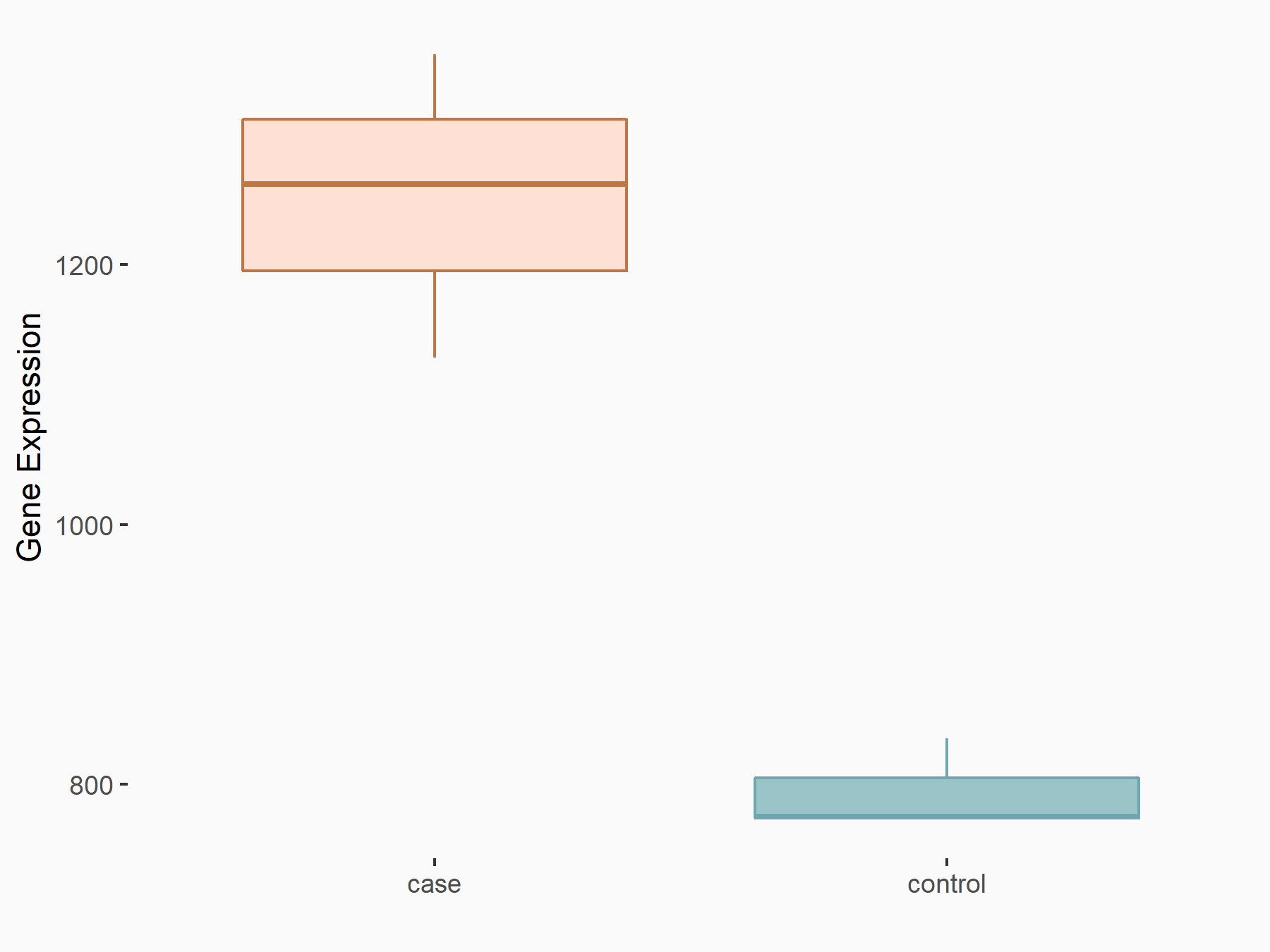 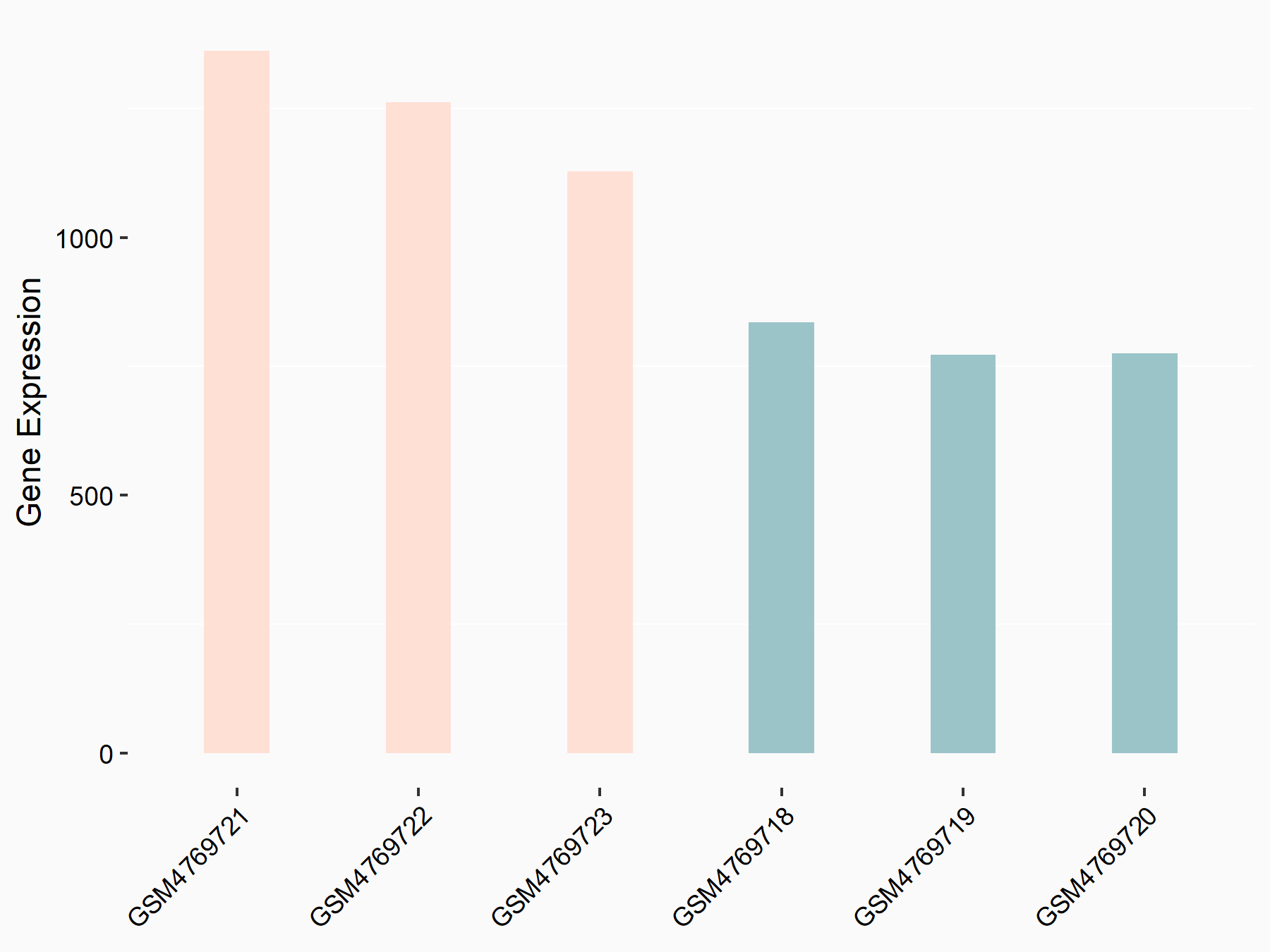 |
logFC: 6.51E-01 p-value: 1.12E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.19E+00 | GSE60213 |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [51] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell migration and proliferation | |||
In-vitro Model |
U251 (Fibroblasts or fibroblast like cells) | |||
| Response Summary | m6A regulated cell proliferation by influencing apoptosis of U251 cells through regulating Heat shock protein HSP 90-alpha (HSP90/HSP90AA1) expression. m6A level was decreased in glioma tissue, which was caused by decreased METTL3 and increased FTO levels. | |||
Hepatocyte nuclear factor 1-alpha (HNF1A/TCF1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
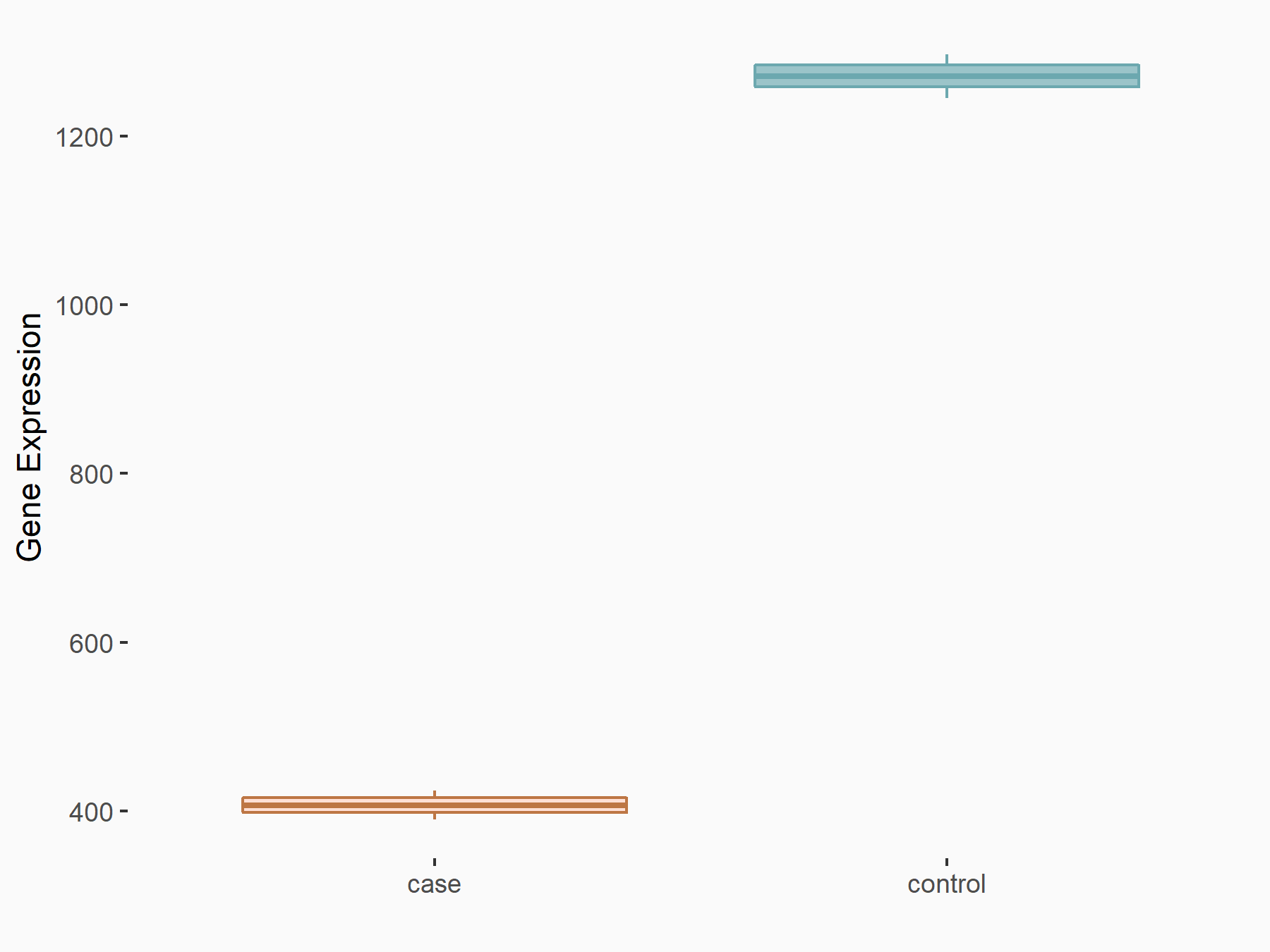 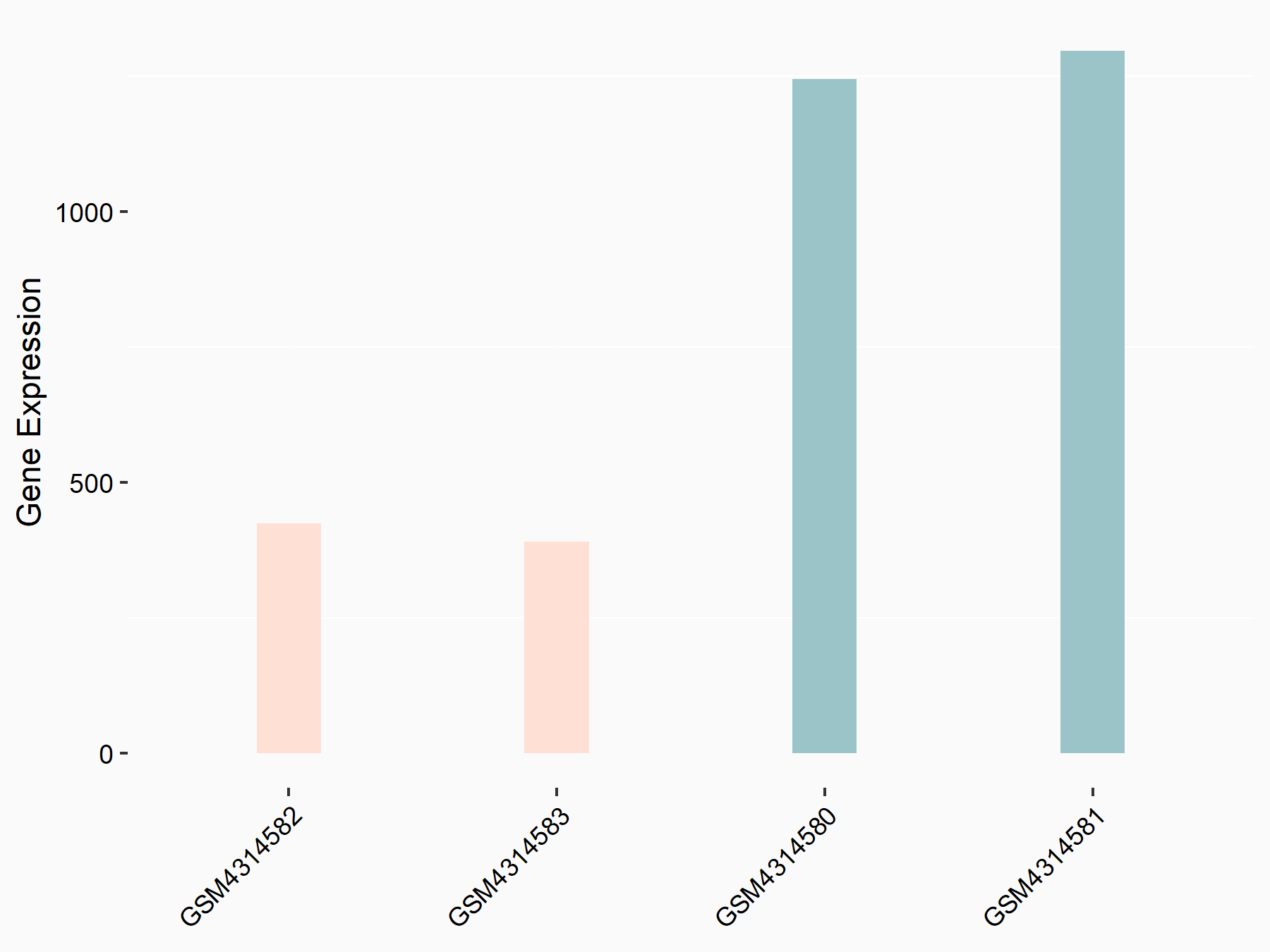 |
logFC: -1.64E+00 p-value: 4.47E-50 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.55E+00 | GSE60213 |
Thyroid Cancer [ICD-11: 2D10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [52] | |||
| Responsed Disease | Thyroid Cancer [ICD-11: 2D10] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Cell migratory | |||
In-vitro Model |
B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 |
| Nthy-ori 3-1 | Normal | Homo sapiens | CVCL_2659 | |
| TPC-1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_6298 | |
| Response Summary | Silence of METTL3 inhibited migratory ability and Wnt activity in TPC-1 cells. METTL3 positively regulated the enrichment abundance of Hepatocyte nuclear factor 1-alpha (HNF1A/TCF1) in anti-IGF2BP2. TCF1 was responsible for METTL3-regulated thyroid carcinoma progression via the m6A methylation. | |||
Hepatoma-derived growth factor (HDGF)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 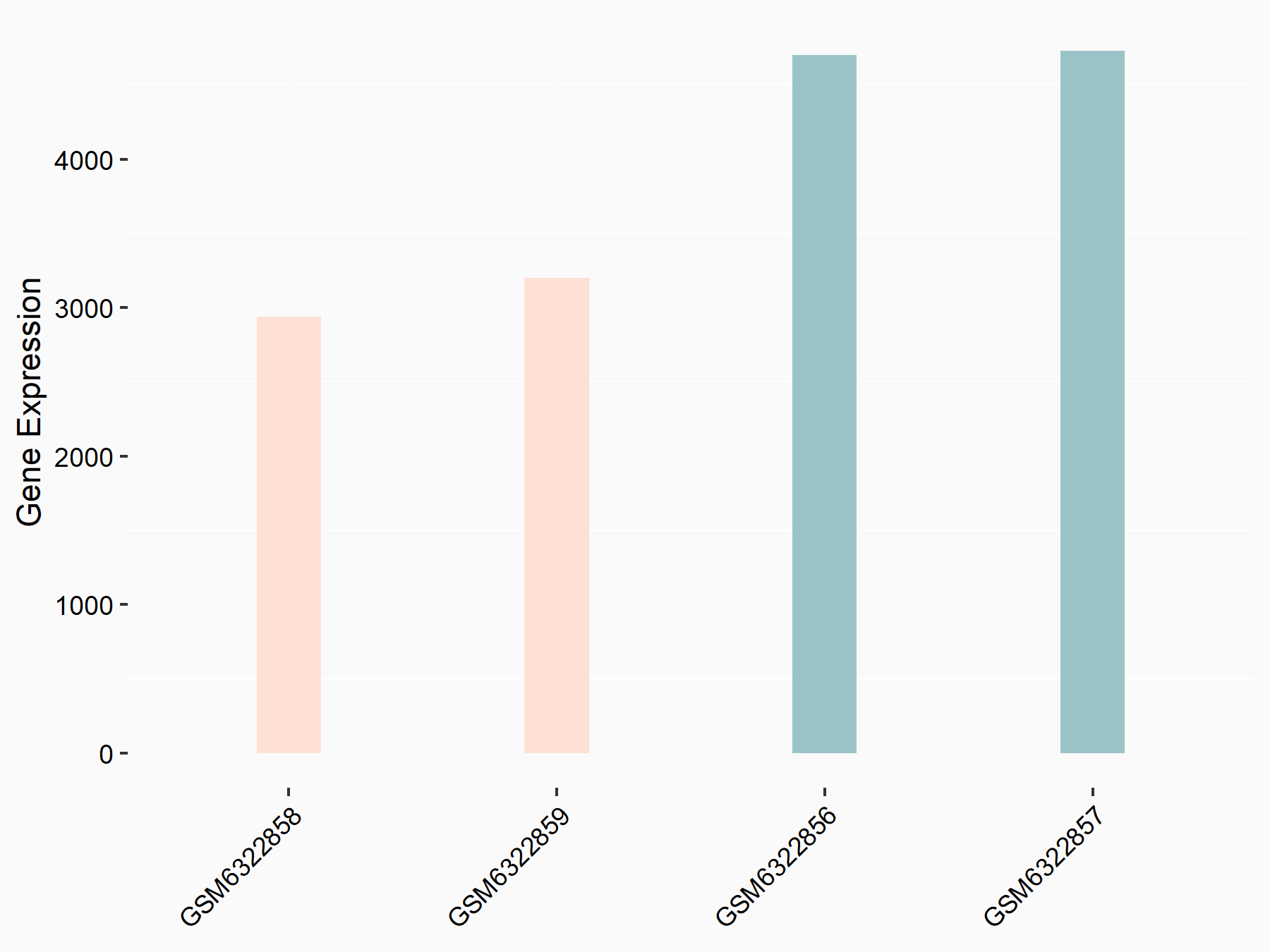 |
logFC: -6.20E-01 p-value: 6.85E-14 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.03E+00 | GSE60213 |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [53] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glycolysis | |||
In-vitro Model |
HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 |
| NCI-N87 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_1603 | |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| In-vivo Model | Mice 8 weeks after splenic portal vein injection of BGC823 cells with METTL3 overexpression or vector-transfected cells. | |||
| Response Summary | Elevated METTL3 expression promotes tumour angiogenesis and glycolysis in Gastric cancer. P300-mediated H3K27 acetylation activation in the promoter of METTL3 induced METTL3 transcription, which stimulated m6A modification of Hepatoma-derived growth factor (HDGF) mRNA, and the m6A reader IGF2BP3 then directly recognised and bound to the m6A site on HDGF mRNA and enhanced HDGF mRNA stability. | |||
Hexokinase-2 (HK2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 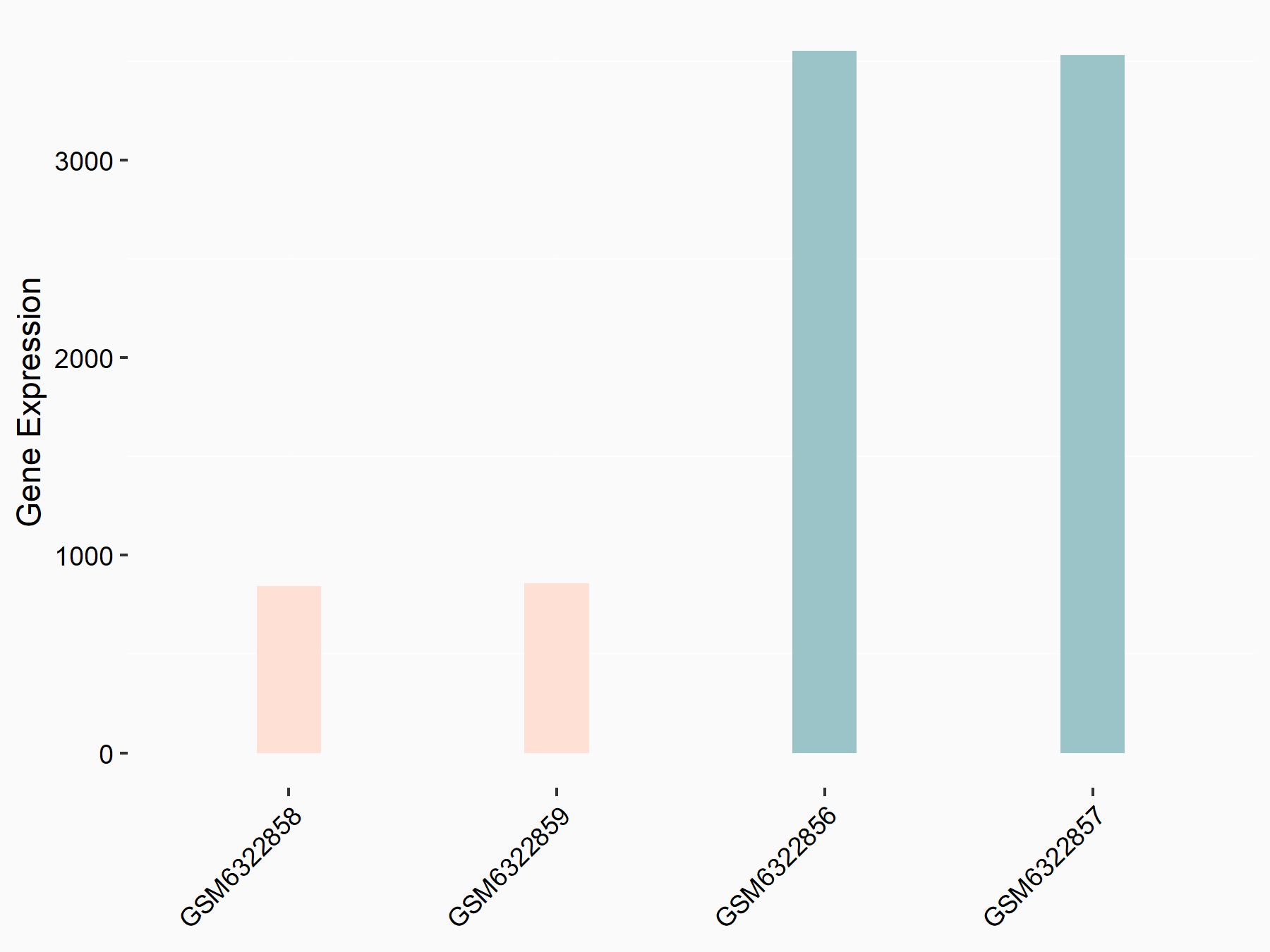 |
logFC: -2.06E+00 p-value: 3.79E-111 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.17E+00 | GSE60213 |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [54] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glucose metabolism | |||
| Response Summary | METTL3 stabilizes Hexokinase-2 (HK2) and SLC2A1 (GLUT1) expression in colorectal cancer through an m6A-IGF2BP2/3- dependent mechanism. | |||
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [55] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010), Central carbon metabolism in cancer | ||
| Cell Process | Glycolysis | |||
In-vitro Model |
SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| HT-3 | Cervical carcinoma | Homo sapiens | CVCL_1293 | |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| In-vivo Model | Five-week-old male nude BALB/C mice were applied for this animal studies and fed with certified standard diet and tap water ad libitum in a light/dark cycle of 12 h on/12 h off.The assay was performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Stable transfection of METTL3 knockdown (sh-METTL3) or negative control (sh-blank) in SiHa cells (5 × 106 cells per 0.1 mL) were injected into the flank of mice. | |||
| Response Summary | METTL3 enhanced the Hexokinase-2 (HK2) stability through YTHDF1-mediated m6A modification, thereby promoting the Warburg effect of CC, which promotes a novel insight for the CC treatment. | |||
High mobility group protein HMGI-C (HMGA2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
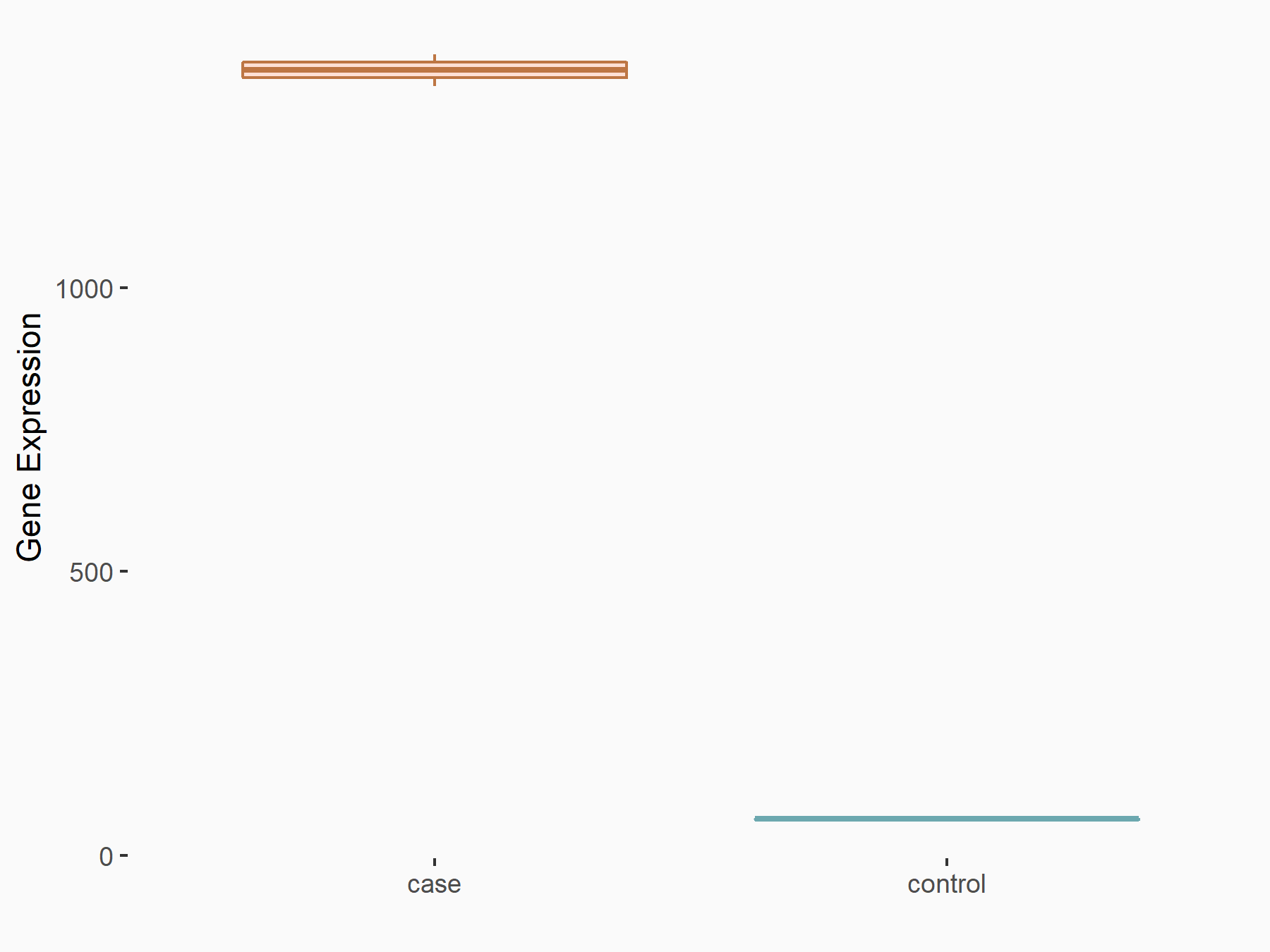 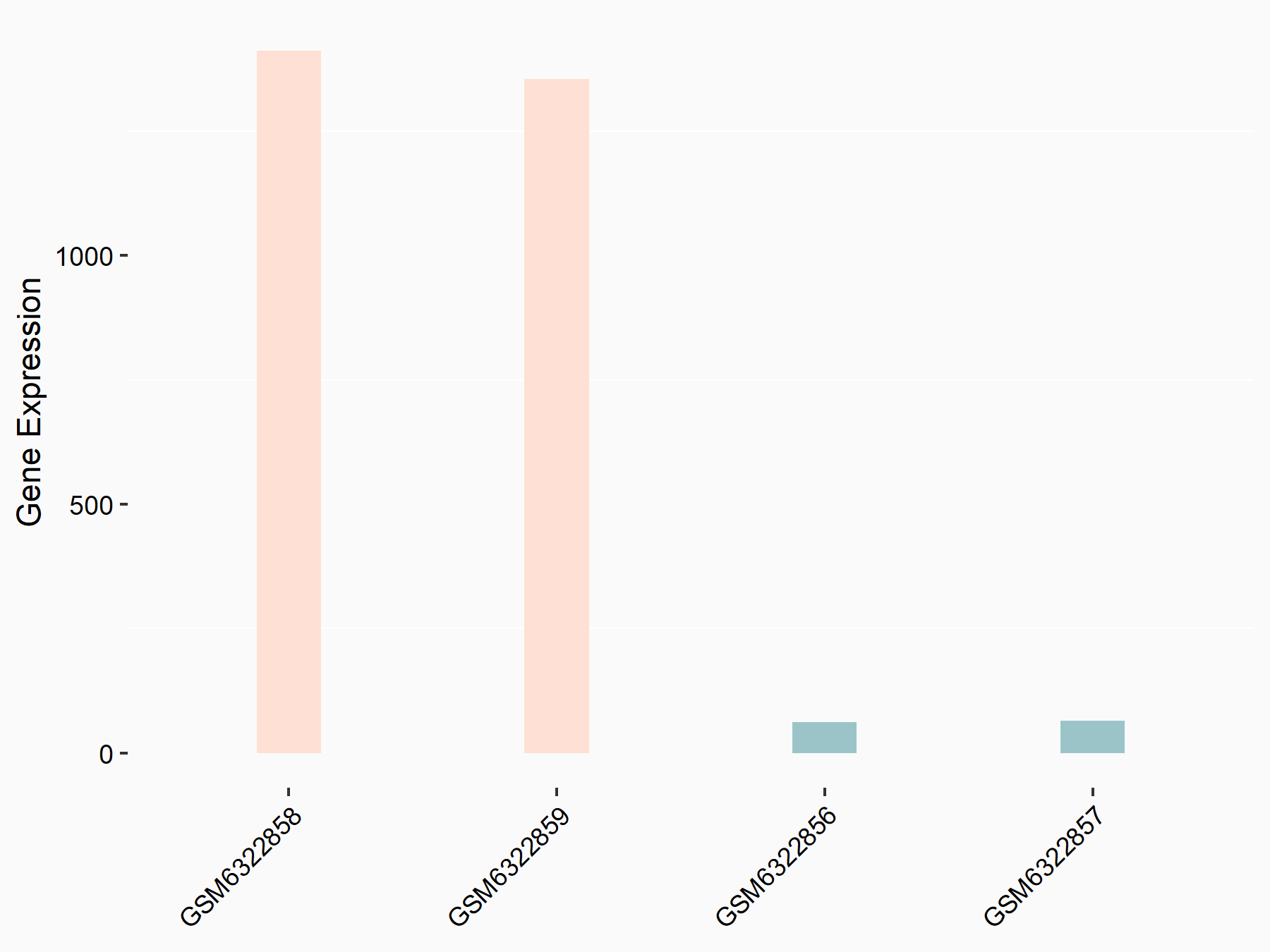 |
logFC: 4.44E+00 p-value: 5.85E-138 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.68E+00 | GSE60213 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [56] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Transcriptional misregulation in cancer | hsa05202 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell autophagy | ||||
In-vitro Model |
Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| In-vivo Model | To create the xenograft neoplasm system, 40 male BALB/c nude mice aged 5 weeks were randomly separated into sh-NC, sh-circHPS5, sh-circHPS5+CTRL, and sh-circHPS5+SAH groups (n = 5 for each group). HCC cells were subcutaneously injected into the axilla of the nude mice. | |||
| Response Summary | In hepatocellular carcinoma, METTL3 could direct the formation of circHPS5, and specific m6A controlled the accumulation of circHPS5. YTHDC1 facilitated the cytoplasmic output of circHPS5 under m6A modification. CircHPS5 can act as a miR-370 sponge to regulate the expression of High mobility group protein HMGI-C (HMGA2) and further accelerate hepatocellular carcinoma cell tumorigenesis. | |||
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [57] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| In-vivo Model | Eighteen BALB/C female nude mice aged 4-5 weeks and weighing 15-18 g were randomly assigned into three groups of six mice. The MCF-7 cell lines stably transfected with sh-NC + oe-NC, sh-METTL3 + oe-NC and sh-METTL3 + oe-HMGA2 were selected for subcutaneous establishment of the BC cell line MCF-7 as xenografts in the nude mice. For this purpose, MCF-7 cell lines in the logarithmic growth stage were prepared into a suspension with a concentration of about 1 × 107 cells/ml. The prepared cell suspension was injected into the left armpit of the mice, and the subsequent tumor growth was recorded. | |||
| Response Summary | Silencing METTL3 down-regulate MALAT1 and High mobility group protein HMGI-C (HMGA2) by sponging miR-26b, and finally inhibit EMT, migration and invasion in BC, providing a theoretical basis for clinical treatment of BC. | |||
Macular disorders [ICD-11: 9B75]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [296] | |||
| Responsed Disease | Macular disorders [ICD-11: 9B75.04] | |||
| Target Regulation | Up regulation | |||
Histone-lysine N-methyltransferase EZH2 (EZH2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
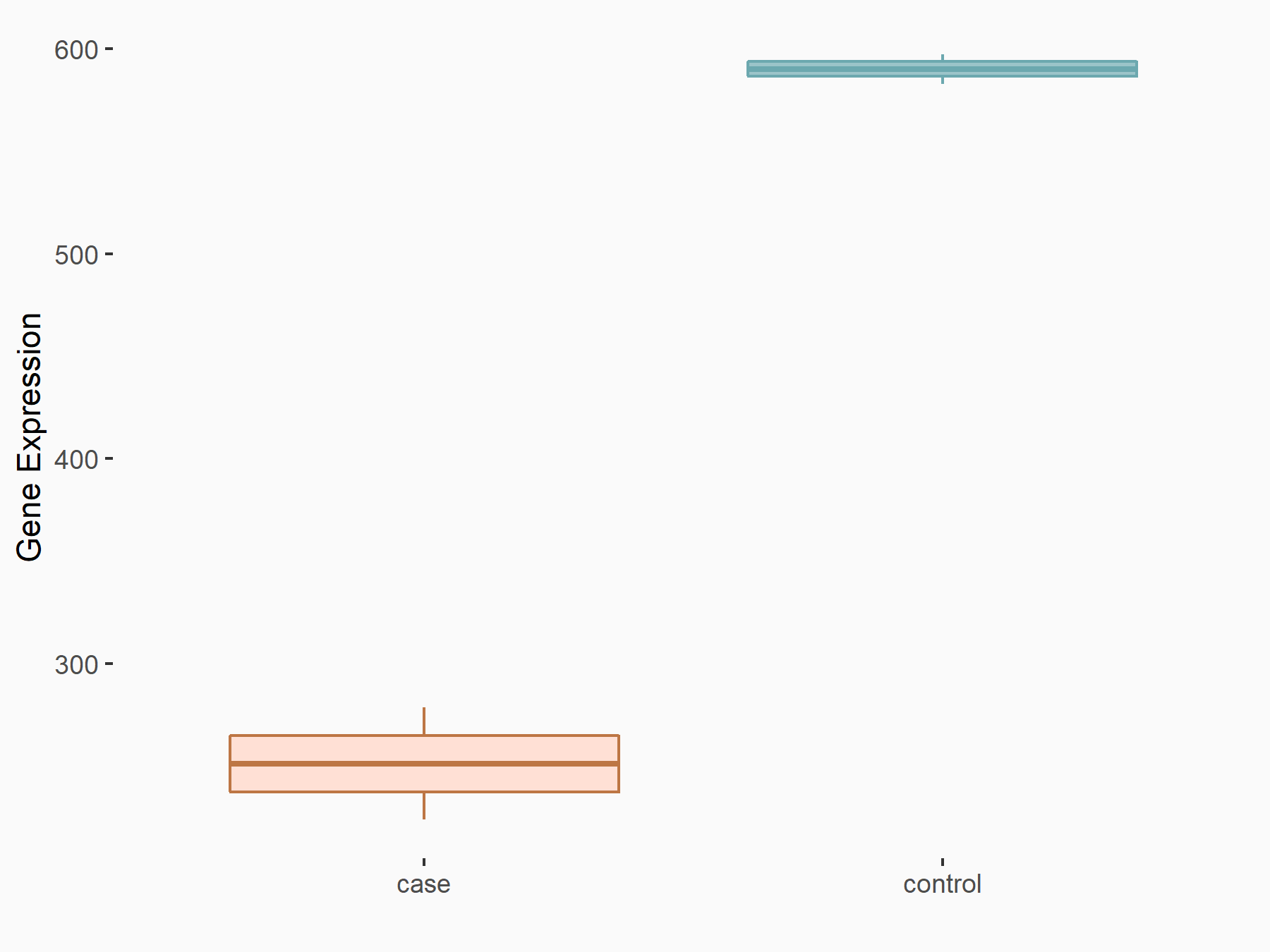 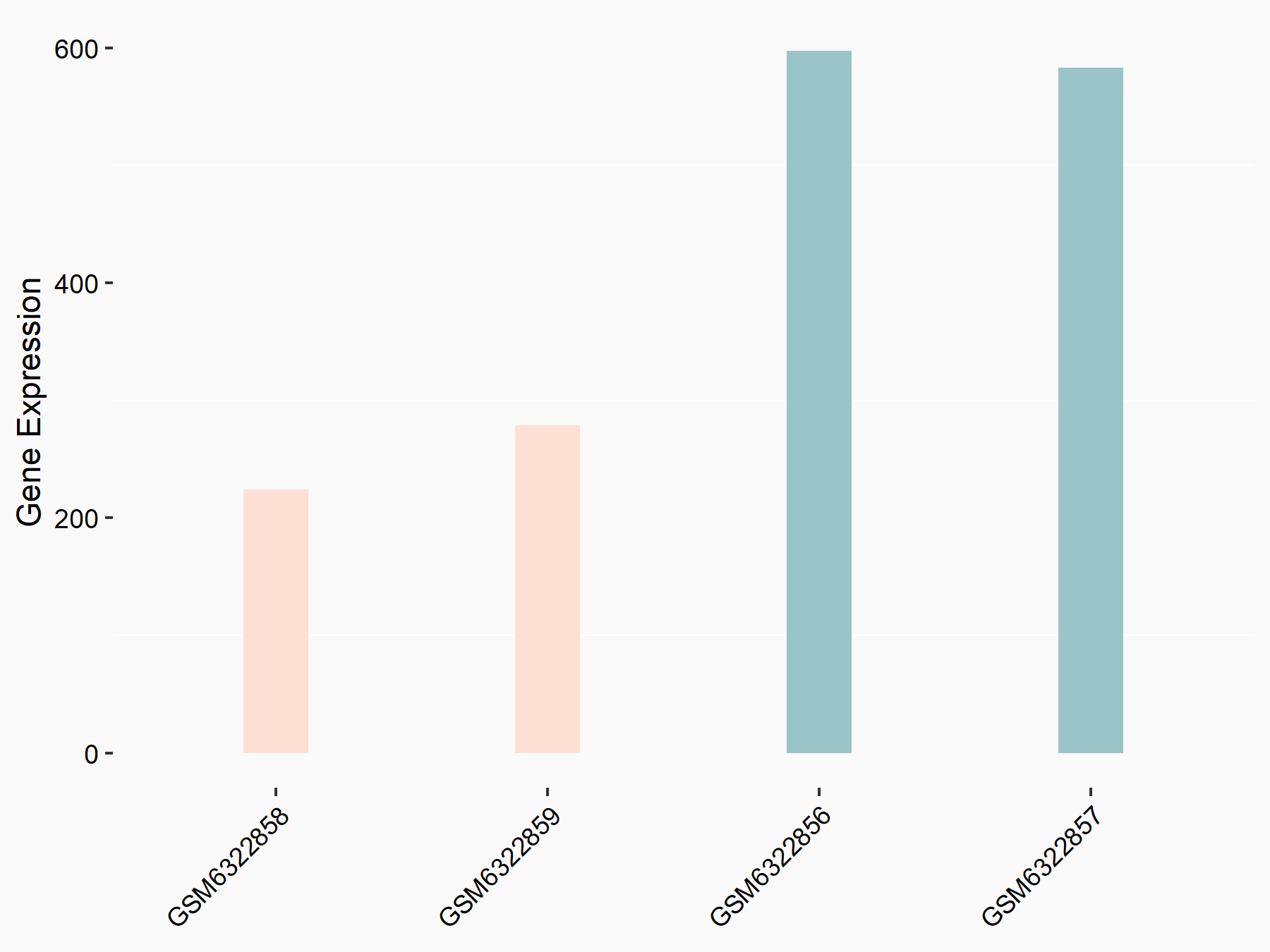 |
logFC: -1.23E+00 p-value: 1.10E-12 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.01E+00 | GSE60213 |
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [58] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell viability | |||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| C666-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_7949 | |
| SUNE1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6946 | |
| Response Summary | METTL3 was highly expressed in nasopharyngeal carcinoma tissues, which inhibited Histone-lysine N-methyltransferase EZH2 (EZH2) expression by mediating m6A modification of EZH2 mRNA. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [59] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Response Summary | Simvastatin induces METTL3 down-regulation in lung cancer tissues, which further influences EMT via m6A modification on Histone-lysine N-methyltransferase EZH2 (EZH2) mRNA and thus inhibits the malignant progression of lung cancer. | |||
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [60] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Adherens junction | hsa04520 | ||
In-vitro Model |
MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| Response Summary | METTL3 is upregulated in Breast Cancer. It could regulate the protein level of Histone-lysine N-methyltransferase EZH2 (EZH2) through m6A modification to promote EMT and metastasis in BCa cells, thereafter aggravating the progression of BCa. | |||
Congenital pneumonia [ICD-11: KB24]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [61] | |||
| Responsed Disease | Congenital pneumonia [ICD-11: KB24] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
| Cell Process | Inflammation | |||
In-vitro Model |
HPBM (Human Peripheral Blood Monocytes) | |||
| Response Summary | METTL3 promotes inflammation and cell apoptosis in a pediatric pneumonia model by regulating Histone-lysine N-methyltransferase EZH2 (EZH2). | |||
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [298] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Responsed Drug | Temozolomide | Approved | ||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | mRNA decay | |||
In-vitro Model |
U251 (Fibroblasts or fibroblast like cells) | |||
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| In-vivo Model | Used to inject 10 uL of the U87 MG-TMZ_R-luc cell suspension in the striatum at a depth of 3 mm from the dural surface. One week after the injection of the tumor cells, 40 mg/kg/day of TMZ in saline was administered for over 2 weeks by intraperitoneal injection. | |||
| Response Summary | Uncover the fundamental mechanisms underlying the interplay of m6 A RNA modification and histone modification in Temozolomide resistance and emphasize the therapeutic potential of targeting the SOX4/EZH2/METTL3 axis in the treatment of TMZ-resistant glioblastoma. | |||
Homeobox protein Nkx-3.1 (NKX3-1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
  |
logFC: 1.12E+00 p-value: 7.44E-16 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.11E+00 | GSE60213 |
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [62] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Oxidative phosphorylation | hsa00190 | ||
In-vitro Model |
VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| In-vivo Model | Approximately 2 × 106 PCa cells (PC-3 shNC, shYTHDF2, shMETTL3 cell lines) per mouse suspended in 100 uL PBS were injected in the flank of male BALB/c nude mice (4 weeks old). During the 40-day observation, the tumor size (V = (width2×length ×0.52)) was measured with vernier caliper. Approximately 1.5 × 106 PCa cells suspended in 100 uL of PBS (PC-3 shNC, shYTHDF2, and shMETTL3 cell lines) per mouse were injected into the tail vein of male BALB/c nude mice (4 weeks old). The IVIS Spectrum animal imaging system (PerkinElmer) was used to evaluate the tumor growth (40 days) and whole metastasis conditions (4 weeks and 6 weeks) with 100 uL XenoLight D-luciferin Potassium Salt (15 mg/ml, Perkin Elmer) per mouse. Mice were anesthetized and then sacrificed for tumors and metastases which were sent for further organ-localized imaging as above, IHC staining and hematoxylin-eosin (H&E) staining. | |||
| Response Summary | Knock-down of YTHDF2 or METTL3 significantly induced the expression of LHPP and Homeobox protein Nkx-3.1 (NKX3-1) at both mRNA and protein level with inhibited phosphorylated AKT. YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent way to regulate AKT phosphorylation-induced tumor progression in prostate cancer. | |||
Homeodomain-interacting protein kinase 2 (HIPK2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 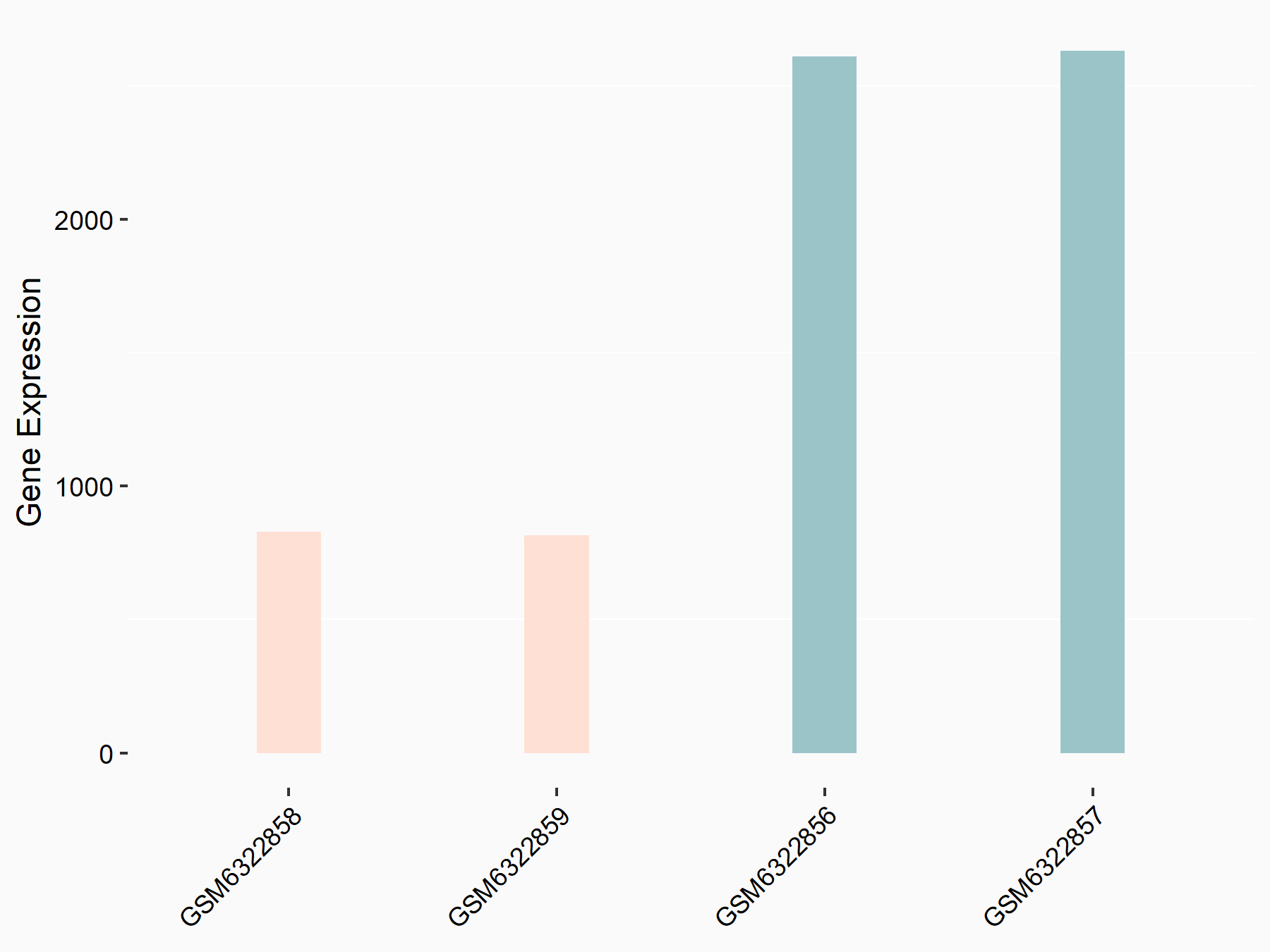 |
logFC: -1.67E+00 p-value: 2.62E-66 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.33E+00 | GSE60213 |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [22] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Doxil | Approved | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell growth and death | |||
| Cell apoptosis | ||||
In-vitro Model |
ADR-resistant MCF-7 (MCF-7/ADR) cells (Human breast cancer doxorubicin-resistant cell line) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| In-vivo Model | Cell suspensions (2 × 106 cells/mL) made with MCF-7/ADR cells stably expressing METTL3 and/or miR-221-3p inhibitor were subcutaneously implanted into each mouse. One week later, xenografted mice were injected with 0.1 mL ADR (25 mg/kg, intraperitoneal injection) twice a week. | |||
| Response Summary | METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. METTL3 knockdown was shown to reduce the expression of miR-221-3p by reducing pri-miR-221-3p m6A mRNA methylation, reducing the expression of MDR1 and BCRP, and inducing apoptosis. Identified the METTL3/miR-221-3p/Homeodomain-interacting protein kinase 2 (HIPK2)/Che-1 axis as a novel signaling event that will be responsible for resistance of BC cells to ADR. | |||
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [299] | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Responsed Drug | Verbascoside | Investigative | ||
In-vitro Model |
SCC-9 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1685 |
| UM1 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_VH00 | |
Insulin-like growth factor I (IGF1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | DKO-1 cell line | Homo sapiens |
|
Treatment: METTL3 knockdown DKO-1 cell
Control: DKO-1 cell
|
GSE182382 | |
| Regulation |
  |
logFC: -2.21E+00 p-value: 2.40E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.27E+00 | GSE60213 |
Ageing-related disease [ICD-11: 9B10-9B60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [63] | |||
| Responsed Disease | Ageing-related disease [ICD-11: 9B10-9B60] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | DNA repair and mitochondrial stress | |||
In-vitro Model |
Mouse fibroblasts (Major cellular components of loose connective tissue) | |||
| Response Summary | The long-lived endocrine mutants - Snell dwarf, growth hormone receptor deletion and pregnancy-associated plasma protein-A knockout - all show increases in the N6-adenosine-methyltransferases (METTL3/14) that catalyze 6-methylation of adenosine (m6A) in the 5' UTR region of select mRNAs. In addition, these mice have elevated levels of YTHDF1, which recognizes m6A and promotes translation by a cap-independent mechanism. Augmented translation by cap-independent pathways facilitated by m6A modifications contribute to the stress resistance and increased healthy longevity of mice with diminished GH and Insulin-like growth factor I (IGF1) signals. | |||
Integrin subunit beta 3 (ITGB3)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 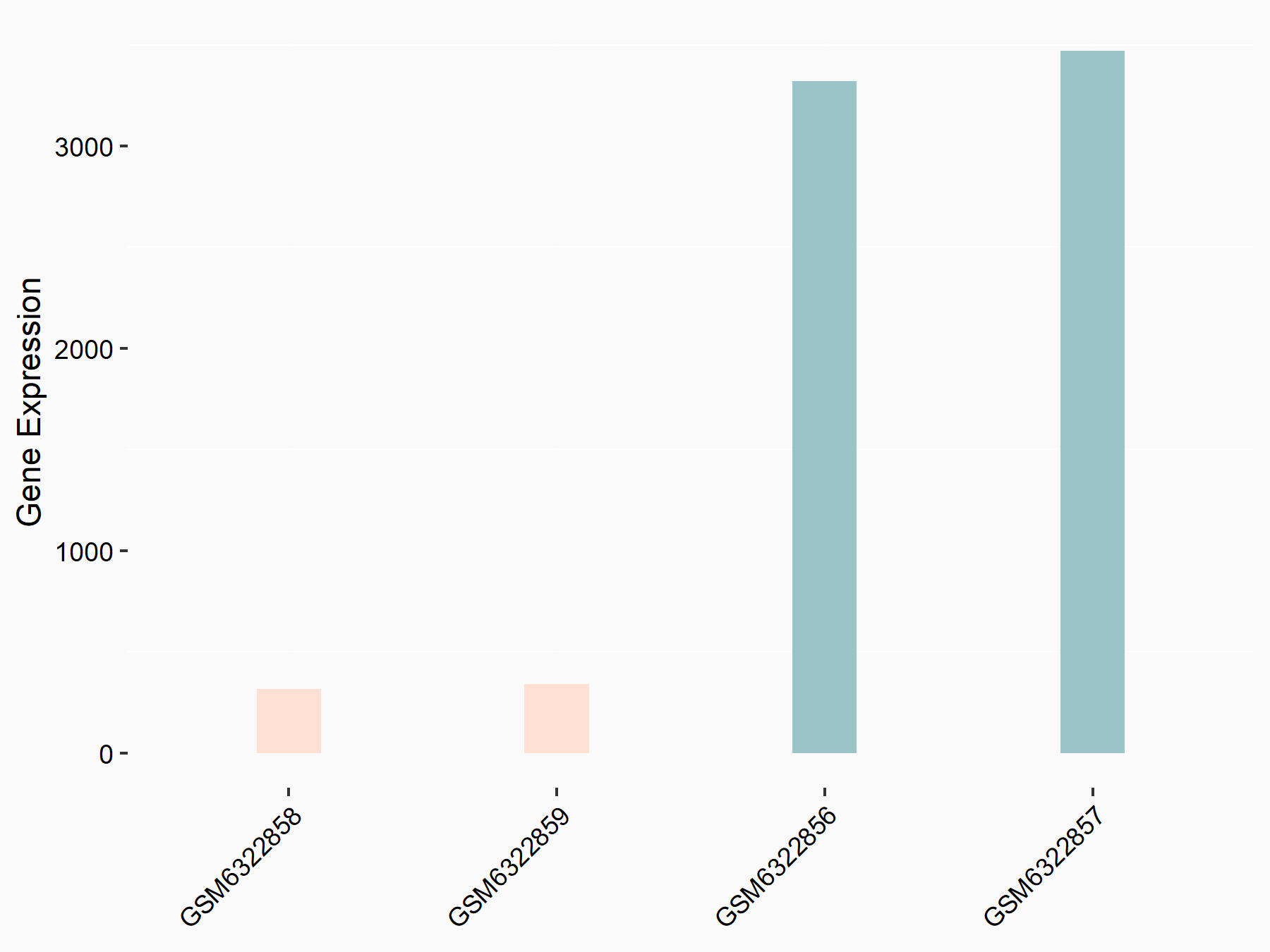 |
logFC: -3.38E+00 p-value: 2.43E-215 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.57E+00 | GSE60213 |
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [64] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell proliferation and metastasis | |||
In-vitro Model |
5-8F | Nasopharyngeal carcinoma | Homo sapiens | CVCL_C528 |
| 6-10B | Nasopharyngeal carcinoma | Homo sapiens | CVCL_C529 | |
| C666-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_7949 | |
| CNE-1 | Normal | Homo sapiens | CVCL_6888 | |
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| HK1 | Nasopharyngeal carcinoma | Acipenser baerii | CVCL_YE27 | |
| HNE-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_0308 | |
| HONE-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_8706 | |
| N2Tert (The human immortalized nasopharyngeal epithelial cell lines) | ||||
| NP69 (A human immortalized nasopharyngeal epithelial) | ||||
| S18 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_B0U9 | |
| S26 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_B0UB | |
| SUNE1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6946 | |
| In-vivo Model | For the tumor growth model, 1 × 106 HNE1-Scrambled or HNE1-shFAM2225A 2# cells were injected into the axilla of the mice, and the tumor size was measured every 3 days. | |||
| Response Summary | FAM225A functioned as a competing endogenous RNA (ceRNA) for sponging miR-590-3p and miR-1275, leading to the upregulation of their target Integrin subunit beta 3 (ITGB3), and the activation of FAK/PI3K/Akt signaling to promote Nasopharyngeal carcinoma cell proliferation and invasion. FAM225A showed lower RNA stability after silencing of METTL3. | |||
Interstitial collagenase (MMP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
 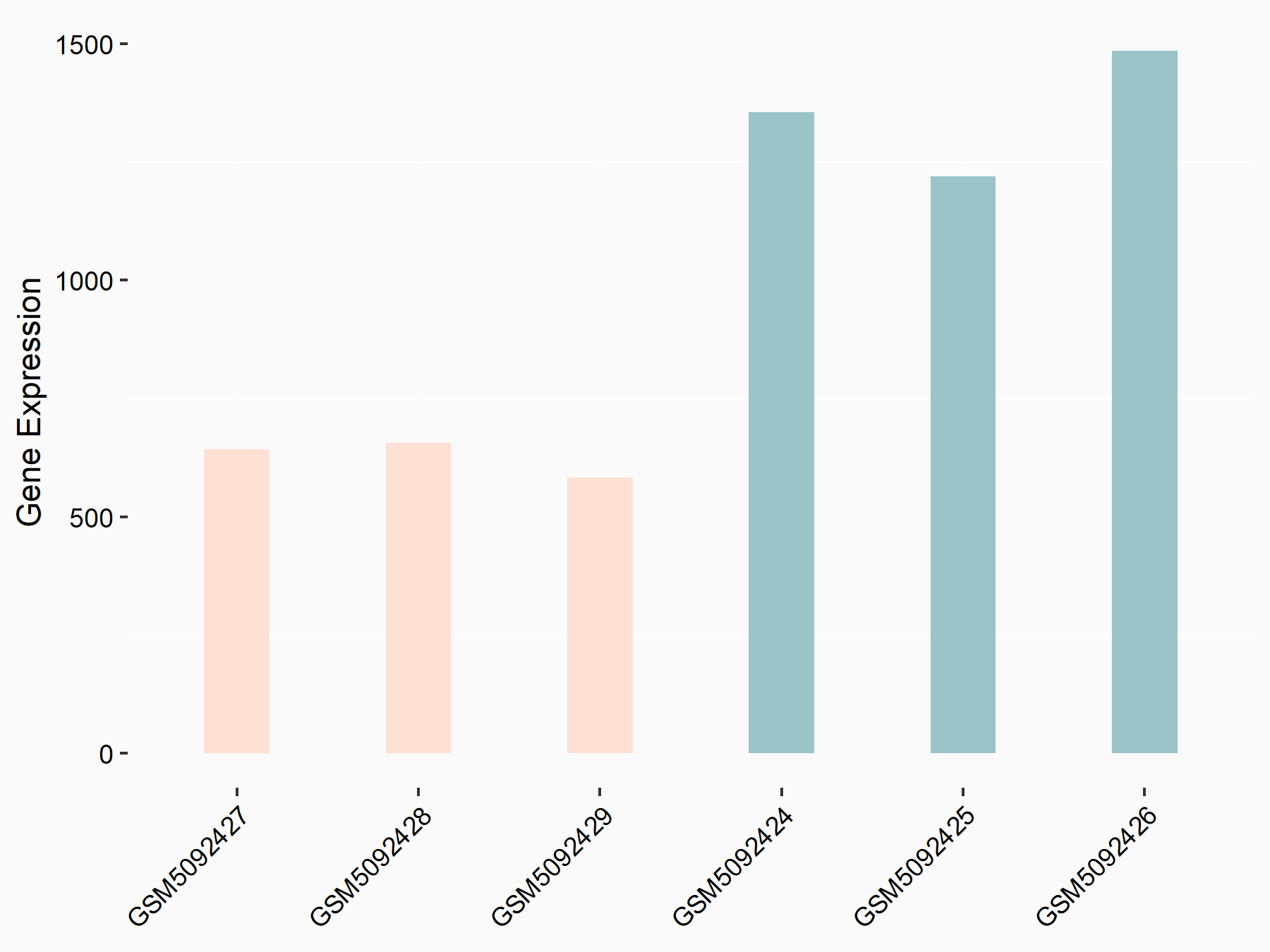 |
logFC: -1.11E+00 p-value: 1.30E-35 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 8.91E+00 | GSE60213 |
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [33] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Inflammatory response | |||
In-vitro Model |
SW1353 | Primary central chondrosarcoma | Homo sapiens | CVCL_0543 |
| Response Summary | METTL3 is involved in OA probably by regulating the inflammatory response. METTL3 overexpression affects extracellular matrix degradation in OA by adjusting the balance between TIMPs and Interstitial collagenase (MMP1). | |||
Kelch-like ECH-associated protein 1 (KEAP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
 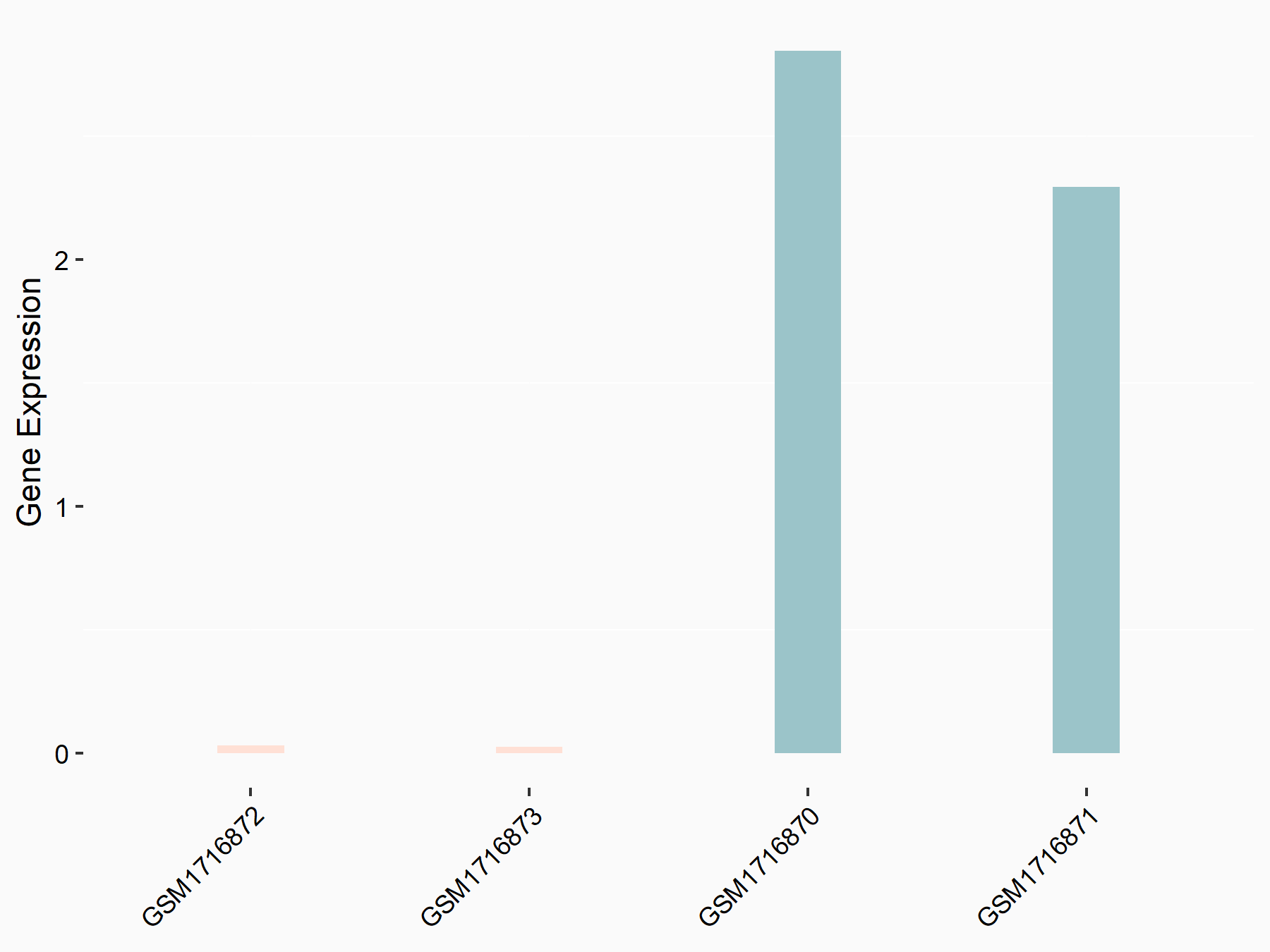 |
logFC: -1.79E+00 p-value: 1.23E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.17E+00 | GSE60213 |
Diseases of the urinary system [ICD-11: GC2Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [65] | |||
| Responsed Disease | Diseases of the urinary system [ICD-11: GC2Z] | |||
| Responsed Drug | Colistin | Approved | ||
| Target Regulation | Down regulation | |||
| Cell Process | Oxidative stress | |||
| Cell apoptosis | ||||
| In-vivo Model | The 60 female Kunming mice were divided into two groups (n = 30): control group (injection of physiological saline through the caudal vein) and colistin group (injection of 15 mg/kg colistin, twice a day, with an eight-hour interval). | |||
| Response Summary | m6A methylation was involved in oxidative stress-mediated apoptosis in the mechanism of colistin nephrotoxicity. METTL3-mediated m6A methylation modification is involved in colistin-induced nephrotoxicity through apoptosis mediated by Kelch-like ECH-associated protein 1 (KEAP1)/Nrf2 signaling pathway. | |||
Leukocyte surface antigen CD47 (CD47)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
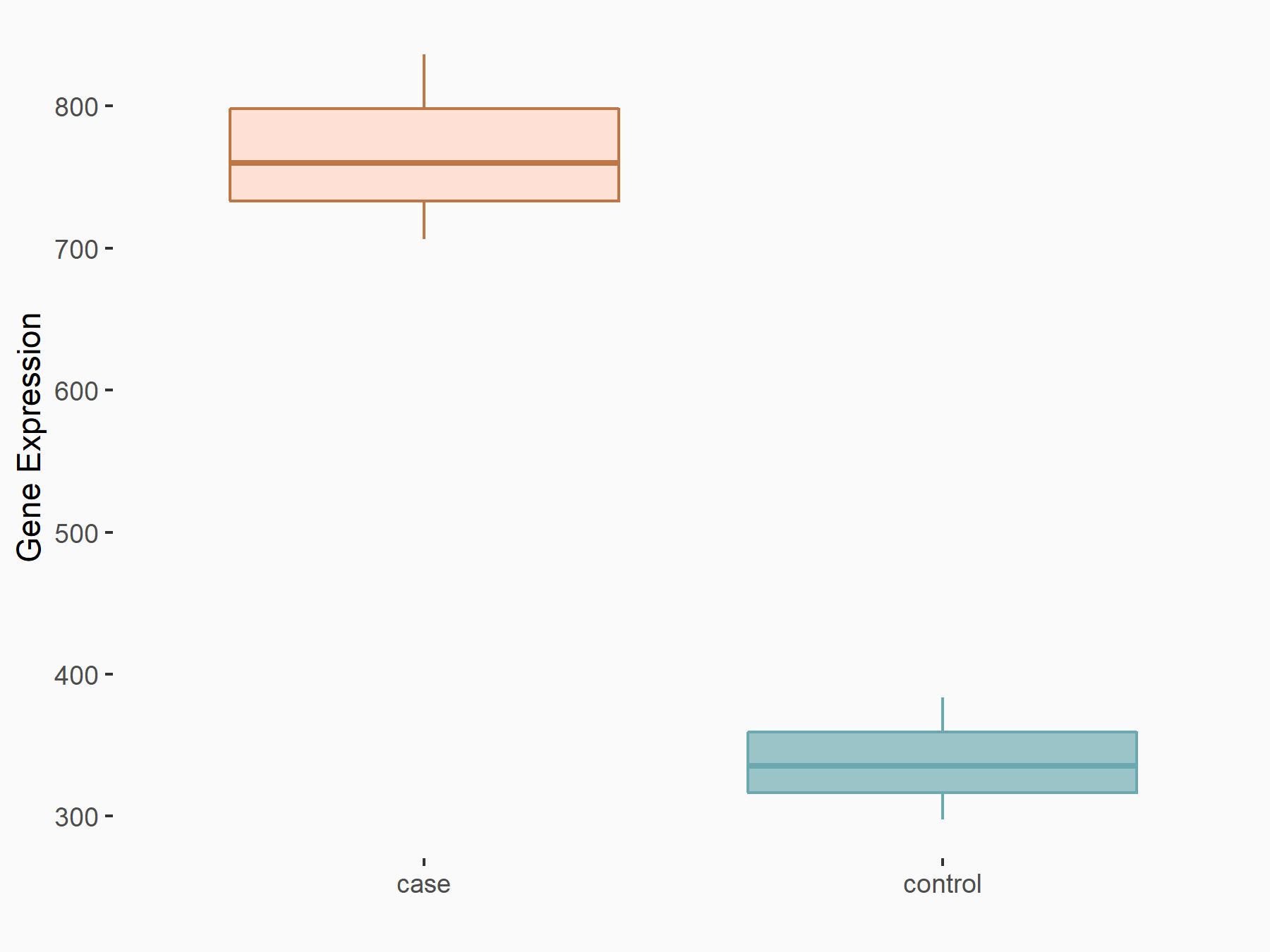 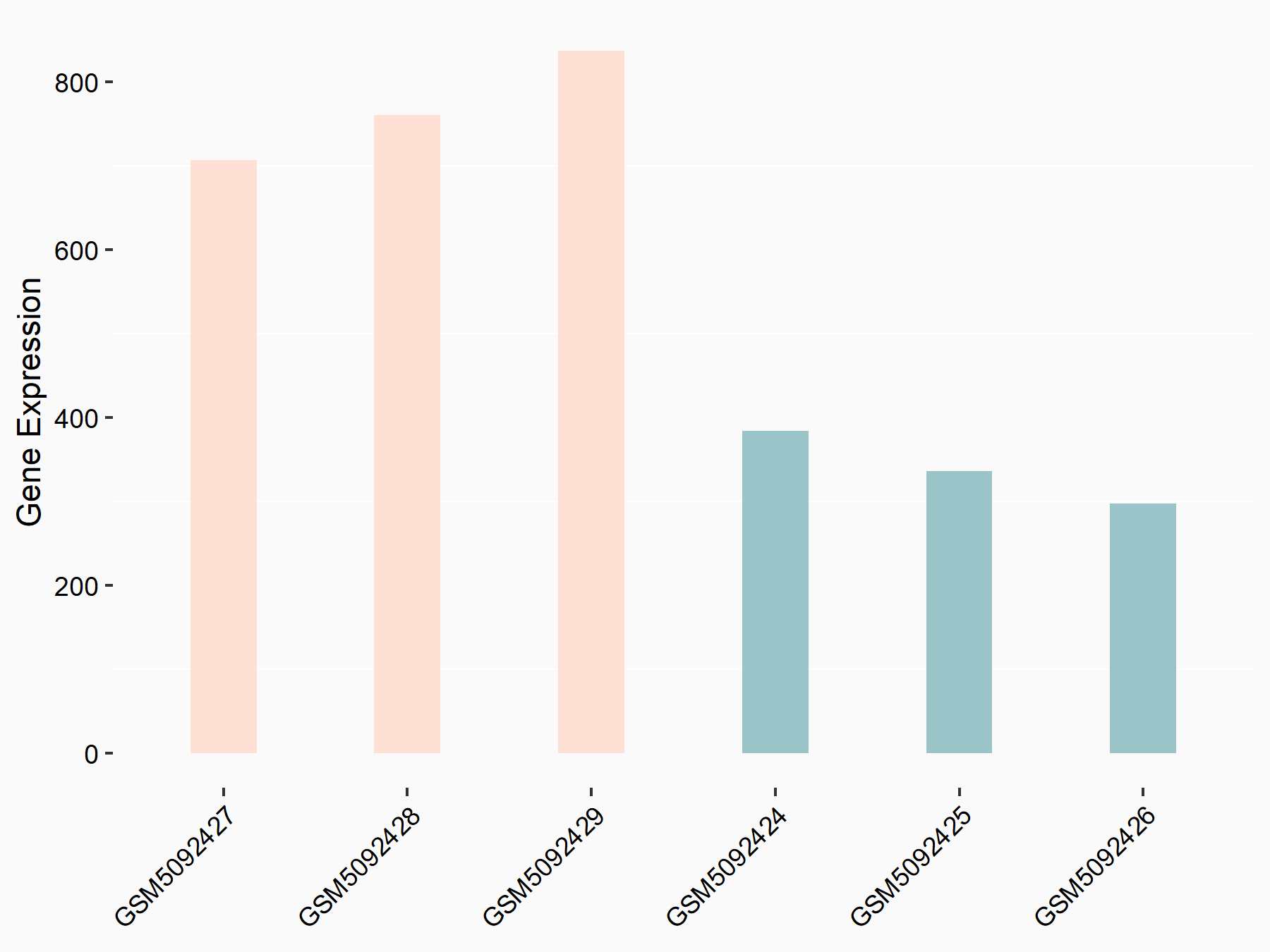 |
logFC: 1.18E+00 p-value: 9.07E-25 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 4.98E+00 | GSE60213 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [66] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| Response Summary | METTL3/IGF2BP1/Leukocyte surface antigen CD47 (CD47) mediated EMT transition contributes to the incomplete ablation induced metastasis in HCC cells. | |||
MAP kinase signal-integrating kinase 2 (MNK2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
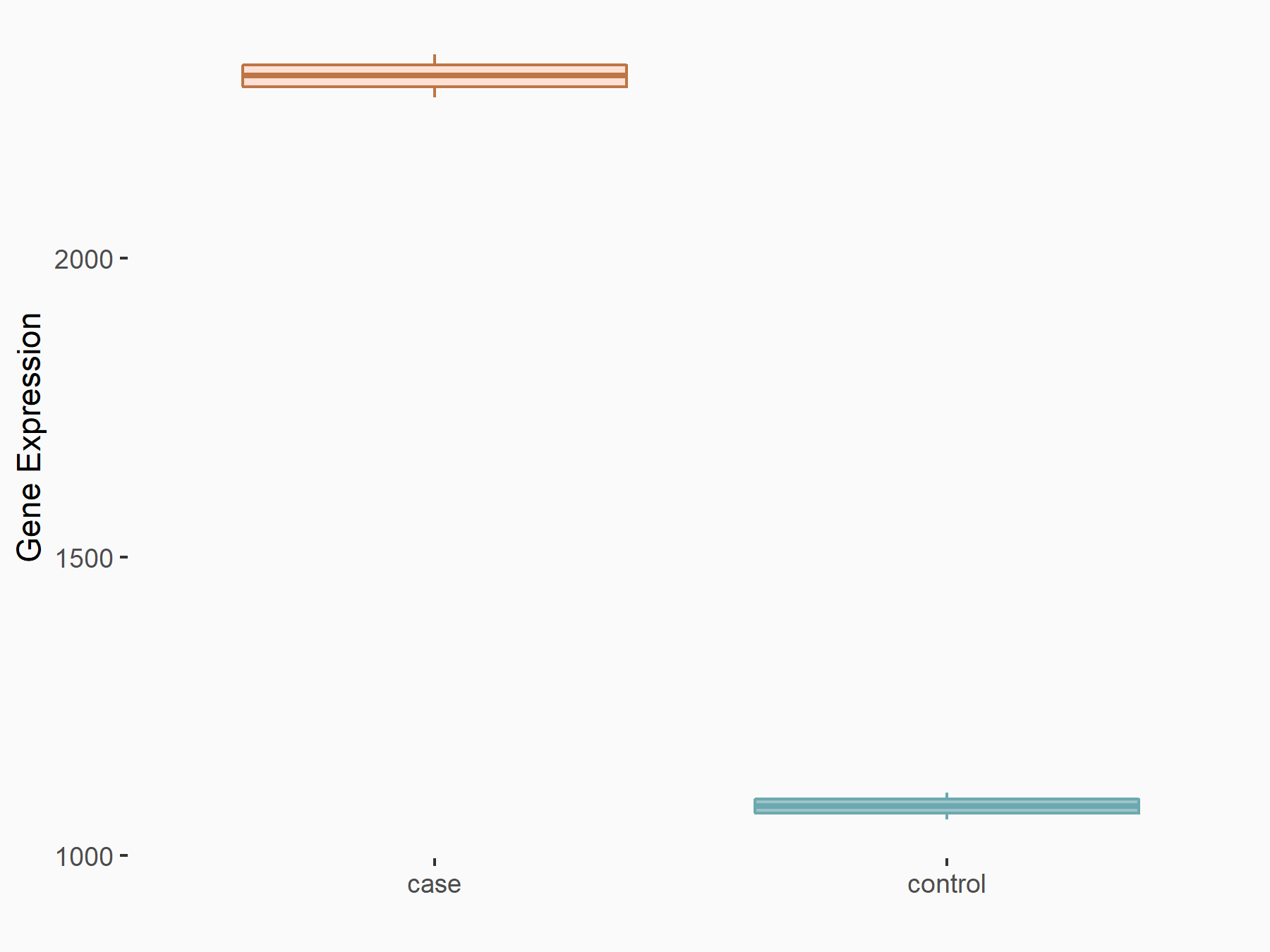 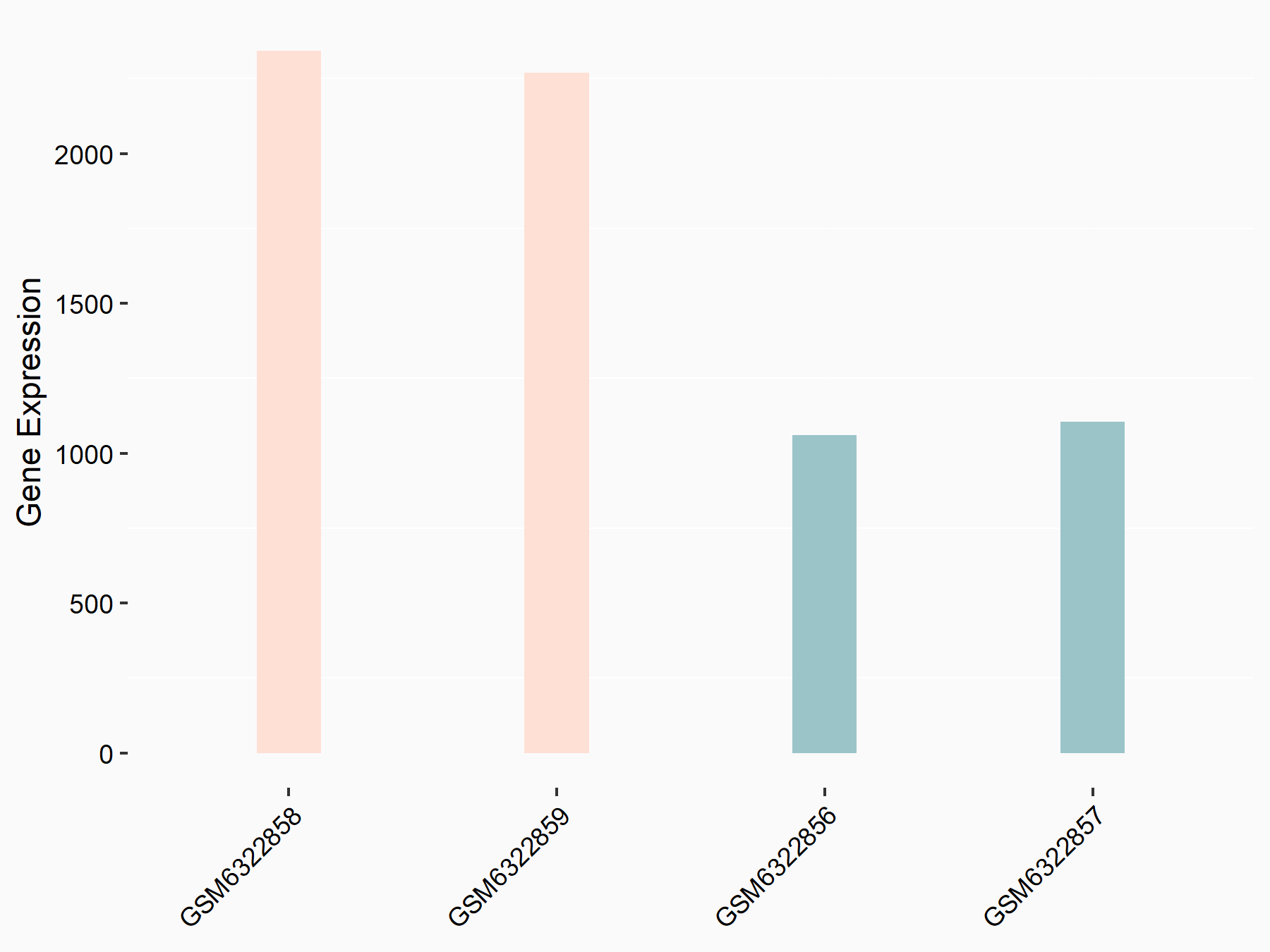 |
logFC: 1.09E+00 p-value: 4.06E-29 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.25E+00 | GSE60213 |
Muscular dystrophies [ICD-11: 8C70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [67] | |||
| Responsed Disease | Muscular dystrophies [ICD-11: 8C70] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| C2C12 | Normal | Mus musculus | CVCL_0188 | |
| In-vivo Model | For mouse muscle injury and regeneration experiment, tibialis anterior (TA) muscles of 6-week-old male mice were injected with 25 uL of 10 uM cardiotoxin (CTX, Merck Millipore, 217503), 0.9% normal saline (Saline) were used as control. The regenerated muscles were collected at day 1, 3, 5, and 10 post-injection. TA muscles were isolated for Hematoxylin and eosin staining or frozen in liquid nitrogen for RNA and protein extraction. | |||
| Response Summary | m6A writers METTL3/METTL14 and the m6A reader YTHDF1 orchestrate MAP kinase signal-integrating kinase 2 (MNK2) expression posttranscriptionally and thus control ERK signaling, which is required for the maintenance of muscle myogenesis and contributes to regeneration. | |||
Metalloproteinase inhibitor 1 (TIMP-1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 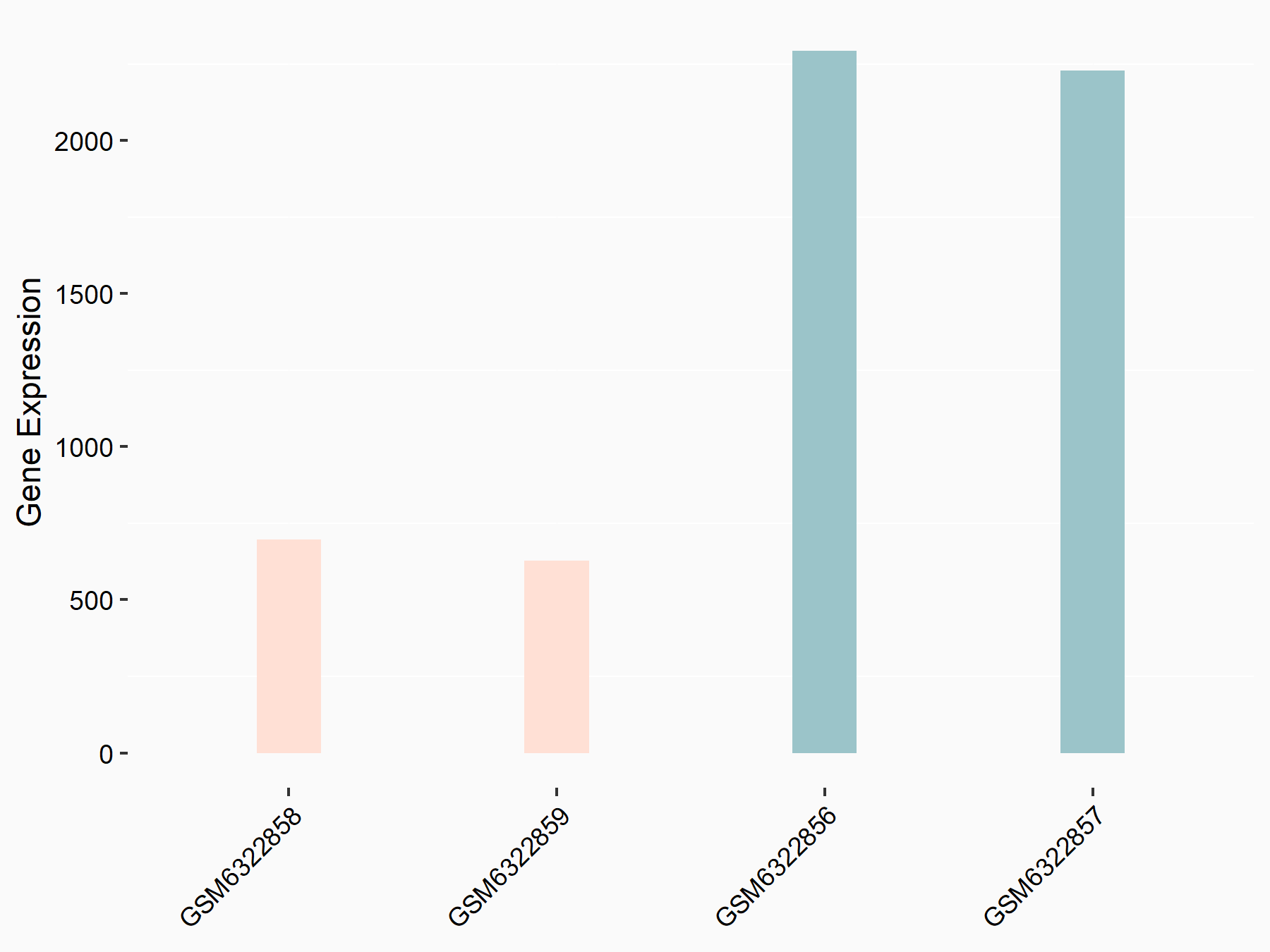 |
logFC: -1.77E+00 p-value: 2.62E-61 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.26E+00 | GSE60213 |
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [33] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Inflammatory response | |||
In-vitro Model |
SW1353 | Primary central chondrosarcoma | Homo sapiens | CVCL_0543 |
| Response Summary | METTL3 is involved in OA probably by regulating the inflammatory response. METTL3 overexpression affects extracellular matrix degradation in OA by adjusting the balance between Metalloproteinase inhibitor 1 (TIMP-1) and MMPs. | |||
Vascular disorders of the liver [ICD-11: DB98]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [307] | |||
| Responsed Disease | Vascular disorders of the liver [ICD-11: DB98.8] | |||
Metalloproteinase inhibitor 2 (TIMP2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 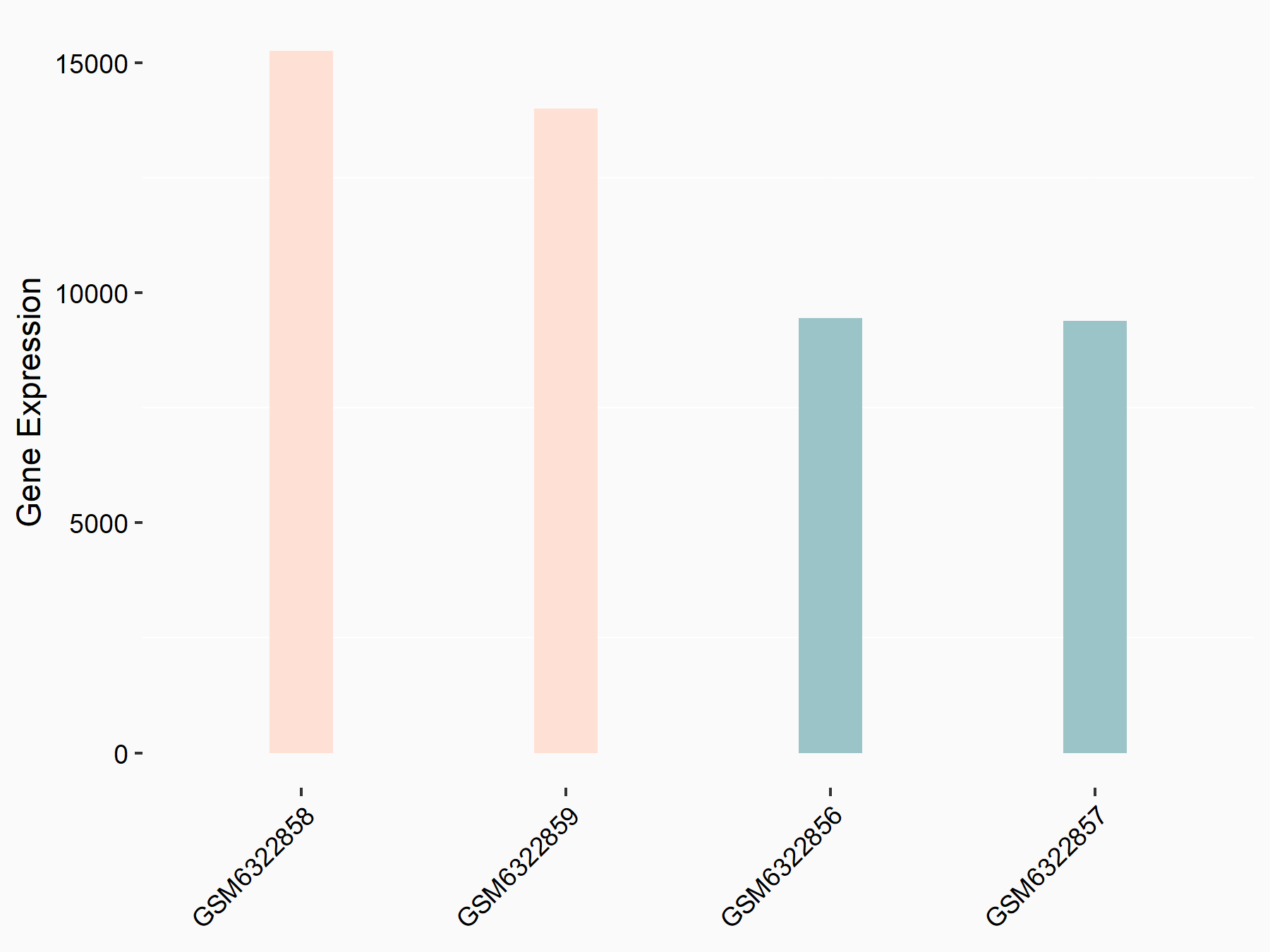 |
logFC: 6.36E-01 p-value: 3.68E-18 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.22E+00 | GSE60213 |
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [33] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Inflammatory response | |||
In-vitro Model |
SW1353 | Primary central chondrosarcoma | Homo sapiens | CVCL_0543 |
| Response Summary | METTL3 is involved in OA probably by regulating the inflammatory response. METTL3 overexpression affects extracellular matrix degradation in OA by adjusting the balance between Metalloproteinase inhibitor 2 (TIMP2) and MMPs. | |||
Chronic kidney disease [ICD-11: GB61]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [68] | |||
| Responsed Disease | Chronic kidney disease [ICD-11: GB61.Z] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Notch signaling pathway | hsa04330 | ||
In-vitro Model |
MPC-5 | Normal | Mus musculus | CVCL_AS87 |
| In-vivo Model | After 7 days of acclimatization, STZ, dissolved in 0.1 M citrate buffer, was intraperitoneally administered daily to mice at a dose of 50 mg/kg after 12 h of food deprivation each day for 5 consecutive days. The type 1 mice diabetic mice were randomly separated into four groups (n = 5-8): AAV9-scramble-control group; AAV9-scramble-STZ; AAV9-shRNA-control; and AAV9-shRNA-STZ group (Hanbio Biotechnology, China). The 50 UL titer of 1 × 1012 virus was injected into the renal pelvis using an insulin needle and maintained there for 2 to 3 s. The type 2 diabetic mice were randomly separated into four groups (n = 5-8): AAV9-scramble-db/m group; AAV9-scramble-db/db group; AAV9-shRNA-db/m group; and AAV9-shRNA-db/db group (Hanbio Biotechnology, China). The 100 UL titer of 1 × 1012 virus was injected into the tail vein using an insulin needle and maintained there for 2 to 3 s. Blood glucose was monitored weekly in mice. At the end of 12 weeks, the 24-h urine samples were collected from the mice kept in metabolic cages. | |||
| Response Summary | METTL3 modulated Notch signaling via the m6A modification of Metalloproteinase inhibitor 2 (TIMP2) in IGF2BP2-dependent manner and exerted pro-inflammatory and pro-apoptotic effects. This study suggested that METTL3-mediated m6A modification is an important mechanism of podocyte injury in DN. | |||
Methylated-DNA--protein-cysteine methyltransferase (MGMT)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
  |
logFC: -1.79E+00 p-value: 2.16E-20 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 5.15E+00 | GSE60213 |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [69] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Responsed Drug | Temozolomide | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | DNA repair | |||
In-vitro Model |
U251 (Fibroblasts or fibroblast like cells) | |||
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| In-vivo Model | Subcutaneously injected shMETTL3 or shNC-expressing U87-MG-TMZ cells into BALB/c NOD mice. After confirmation of GBM implantation, mice were treated with TMZ (66 mg/kg/d, 5 d per week, for 3 cycles). | |||
| Response Summary | Two critical DNA repair genes (Methylated-DNA--protein-cysteine methyltransferase (MGMT) and APNG) were m6A-modified by METTL3, whereas inhibited by METTL3 silencing or DAA-mediated total methylation inhibition, which is crucial for METTL3-improved temozolomide resistance in glioblastoma cells. | |||
Glioblastoma multiforme [ICD-11: XH0MB1]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [287] | |||
| Responsed Disease | Glioblastoma multiforme [ICD-11: XH0MB1] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
GSC11 | Glioblastoma | Homo sapiens | CVCL_DR55 |
| GSC23 | Glioblastoma | Homo sapiens | CVCL_DR59 | |
| In-vivo Model | To establish the xenograft model of GSCs in mice, GSCs or METTL3 knocking-down GSCs (v/v = 1:1) were subcutaneously inoculated into the right posterior limb of BALB/c nude mice (5 × 106 cells/mice, 4-week-old, female, body weight of 20 g). Tumor volume was measured with calipers every 3 days. The tumor size was calculated using the formula: (a2 × b)/2 (a: width in mm, b: length in mm). After about 27 days, all mice were sacrificed under general anesthesia and the xenografts were harvested for further pathological study. | |||
Methylcytosine dioxygenase TET1 (TET1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 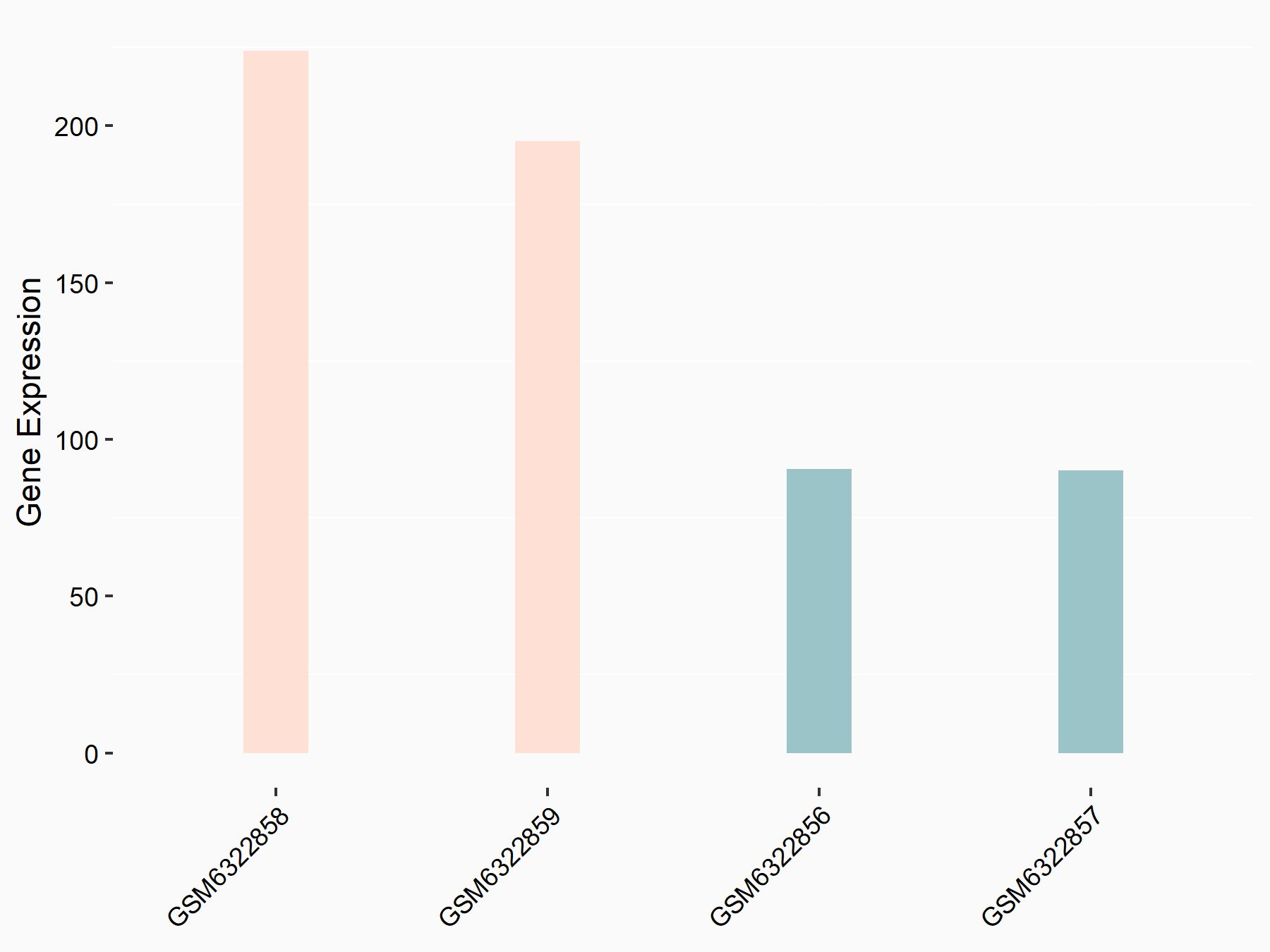 |
logFC: 1.21E+00 p-value: 3.76E-06 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.94E+00 | GSE60213 |
Chronic pain [ICD-11: MG30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [70] | |||
| Responsed Disease | Chronic pain [ICD-11: MG30] | |||
| Target Regulation | Down regulation | |||
| Response Summary | Downregulated spinal cord METTL3 coordinating with YTHDF2 contributes to the modulation of inflammatory pain through stabilizing upregulation of Methylcytosine dioxygenase TET1 (TET1) in spinal neurons. | |||
Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 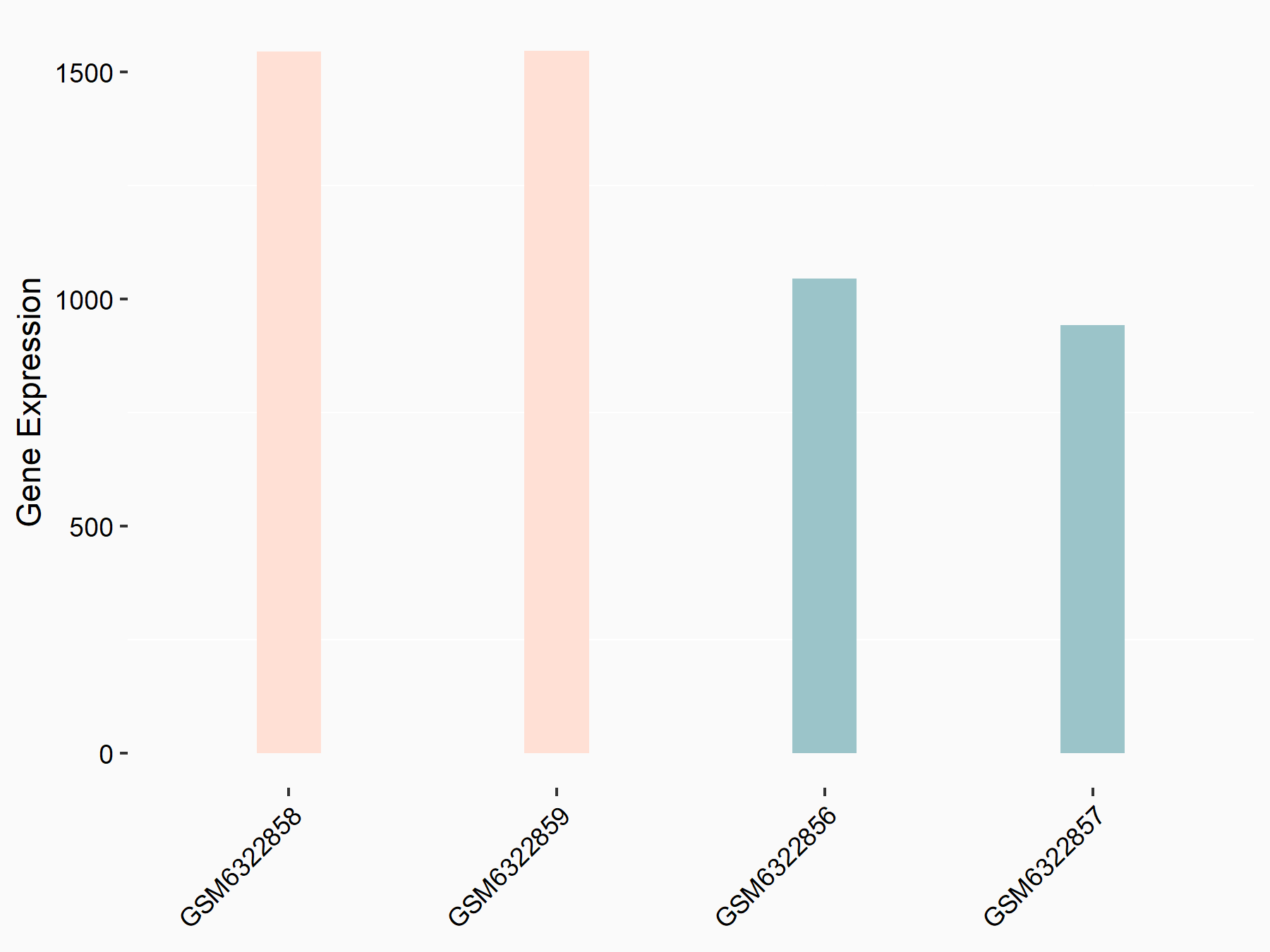 |
logFC: 6.38E-01 p-value: 4.93E-09 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.34E+00 | GSE60213 |
Enterovirus [ICD-11: 1A2Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Enterovirus [ICD-11: 1A2Y] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| Cell autophagy | ||||
In-vitro Model |
Schwann cells (A type of glial cell that surrounds neurons) | |||
| Response Summary | Knocking down METTL3 prevented Enterovirus 71-induced cell death and suppressed Enterovirus 71-induced expression of Bax while rescuing Bcl-2 expression after Enterovirus 71 infection. Knocking down METTL3 inhibited Enterovirus 71-induced expression of Atg5, Atg7 and Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II). Knocking down METTL3 inhibited Enterovirus 71-induced apoptosis and autophagy. | |||
Lung cancer [ICD-11: 2C25]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Chloroquine | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
In-vitro Model |
Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II). beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Gefitinib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
In-vitro Model |
Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II). beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Experiment 3 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Beta-Elemen | Phase 3 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
In-vitro Model |
Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II). beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
Mitogen-activated protein kinase 14 (p38/MAPK14)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
 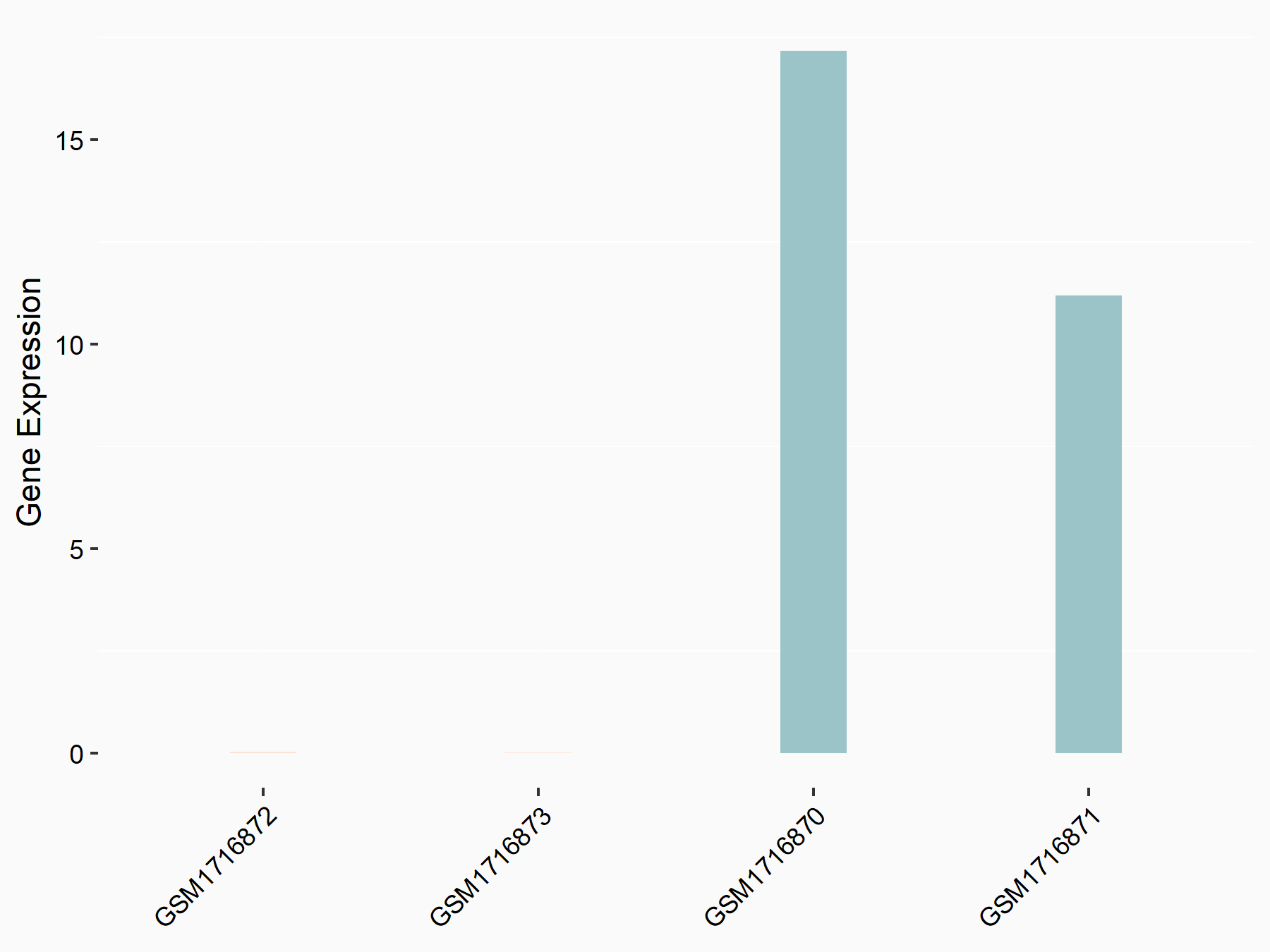 |
logFC: -3.87E+00 p-value: 1.84E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.30E+00 | GSE60213 |
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [71] | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SCC-25 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1682 |
| SCC-15 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1681 | |
| In-vivo Model | Male BALB/c-nu/nu mice, 9-10 weeks of age, were acclimatized for a week prior to the experiments. Nude mice (6 each group) were subcutaneously inoculated with 4 × 106 SCC-25 or SCC-15 cells through an injection into the center of the back, which consistently caused tumor formation within 1 week of inoculation. To monitor the initial tumor appearance, animals were observed every day. After tumor appeared, measurements were made every week with a caliper. After 28 days, mice were sacrificed and tumors were dissected out to be weighed. | |||
| Response Summary | High METTL3 expression was positively correlated with more severe clinical features of OSCC tumors. Furthermore, METTL3-KD and cycloleucine, a methylation inhibitor, decreased m6A levels and down-regulated Mitogen-activated protein kinase 14 (p38/MAPK14) expression in OSCC cells. | |||
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [72] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| KM12 | Colon carcinoma | Homo sapiens | CVCL_1331 | |
| Response Summary | METTL3 played a tumor-suppressive role in Colorectal cancer cell proliferation, migration and invasion through Mitogen-activated protein kinase 14 (p38/MAPK14)/ERK pathways, which indicated that METTL3 was a novel marker for CRC carcinogenesis, progression and survival. | |||
Mutated in multiple advanced cancers 1 (PTEN)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 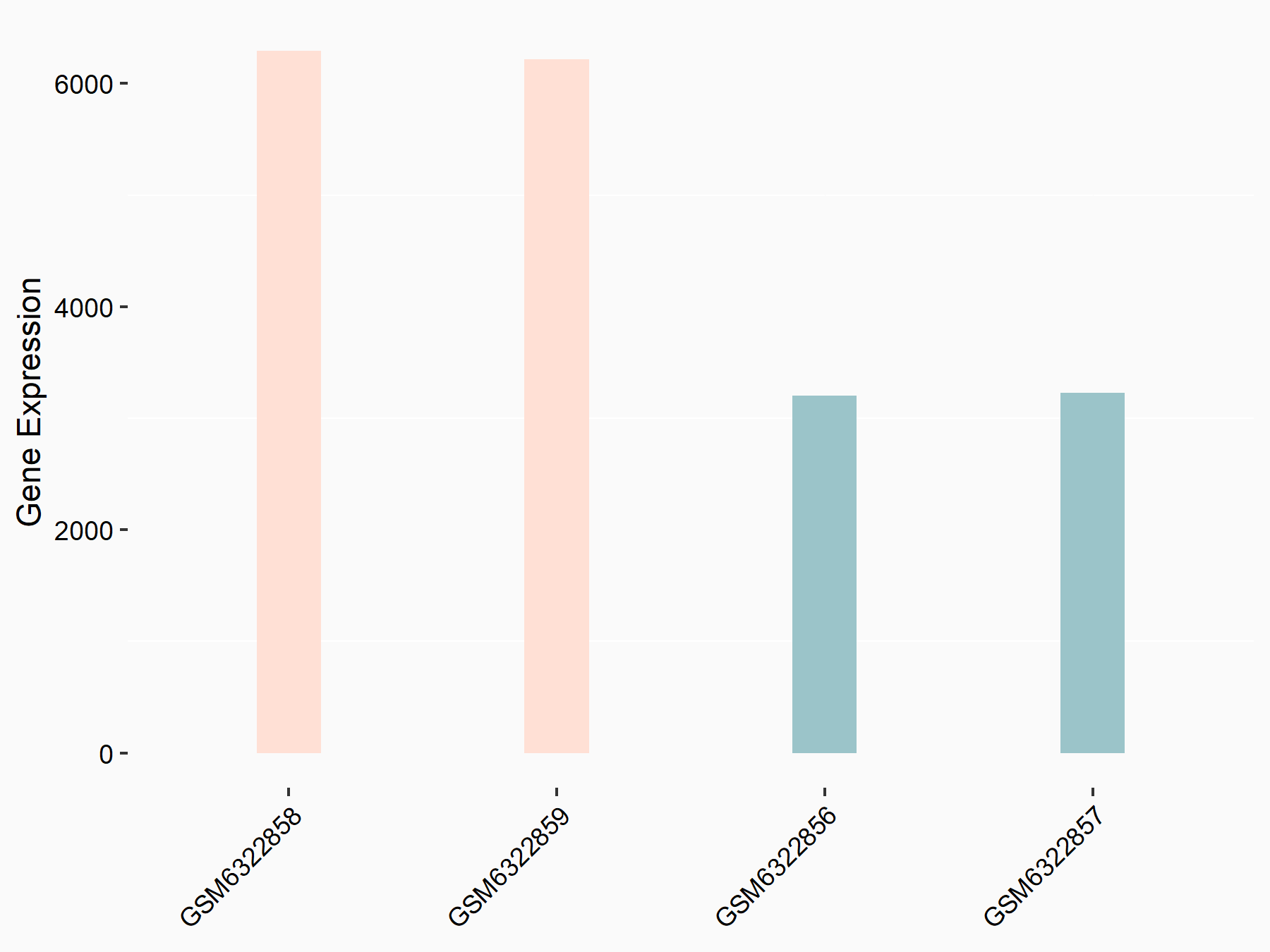 |
logFC: 9.60E-01 p-value: 1.46E-37 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.25E+00 | GSE60213 |
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [9] | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell differentiation and apoptosis | |||
| Apoptosis (hsa04210) | ||||
In-vitro Model |
HSPC (Human hematopoietic stem cell) | |||
| In-vivo Model | 500,000 selected cells were injected via tail vein or retro-orbital route into female NSG (6-8 week old) recipient mice that had been sublethally irradiated with 475 cGy one day before transplantation. | |||
| Response Summary | METTL3 depletion in human myeloid leukemia cell lines induces cell differentiation and apoptosis and delays leukemia progression in recipient mice in vivo. Single-nucleotide-resolution mapping of m6A coupled with ribosome profiling reveals that m6A promotes the translation of c-MYC, BCL2 and Mutated in multiple advanced cancers 1 (PTEN) mRNAs in the human acute myeloid leukemia MOLM-13 cell line. Moreover, loss of METTL3 leads to increased levels of phosphorylated AKT. | |||
Malignant haematopoietic neoplasm [ICD-11: 2B33]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [73] | |||
| Responsed Disease | Chronic myeloid leukaemia [ICD-11: 2B33.2] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | RNA stability | |||
| Cell autophagy | ||||
In-vitro Model |
K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 |
| KCL-22 | Chronic myelogenous leukemia | Homo sapiens | CVCL_2091 | |
| In-vivo Model | In the control mice or ADR mice group, the parental or chemo-resistant K562 cells were infected with LV-shCtrl. In the ADR + shLINC00470 group, the chemo-resistant K562 cells were infected with LV- shLINC00470. These cells were injected, respectively, into these 5-week-old mice subcutaneously. | |||
| Response Summary | The molecular mechanism underlying the effect of LINC00470 on chronic myelocytic leukaemia by reducing the Mutated in multiple advanced cancers 1 (PTEN) stability via RNA methyltransferase METTL3, thus leading to the inhibition of cell autophagy while promoting chemoresistance in CML. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [74] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | Four-week-old BALB/c nude mice were randomly divided into three groups: (1) vector group, (2) vector + Bete-elemene group, and (3) Bete-elemene + METTL3 group. Nude mice were raised in an SPF level animal house and were free to eat and drink. Mice in the vector group were subcutaneously injected with lung cancer cells transfected with empty vector and did not receive Bete-elemene administration, and this group was implemented as the negative control. Following establishing orthotopic xenografts by using A549 or H1299 cells transfected with empty vector, mice in the vector + Bete-elemene group underwent intraperitoneal injection with Bete-elemene once a day. For the subcutaneous transplanted model, A549 or H1299 cells transfected with METTL3-overexpressing vector were inoculated into mice from the Bete-elemene + METTL3 group. Then, mice were intraperitoneally administrated with Bete-elemene once a day. Three weeks later, all the animals were euthanized with CO2. Xenografts were removed and weighted after mice were euthanatized. | |||
| Response Summary | Bete-elemene exerted the restrictive impacts on the cell growth of lung cancer in vivo and in vitro through targeting METTL3. Bete-elemene contributed to the augmented PTEN expression via suppressing its m6A modification. | |||
Pulmonary hypertension [ICD-11: BB01]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [75] | |||
| Responsed Disease | Pulmonary hypertension due to lung disease or hypoxia [ICD-11: BB01.2] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
PASMC cell line (Pulmonary artery smooth muscle cell) | |||
| In-vivo Model | 10 rats were divided into control and HPH group. In detail, 5 rats of the hypoxia groups were exposed to hypoxia (10%O2) chamber (AiPu XBS-02B, China) for 4 weeks. In addition, 5 rats of control group were kept under normoxic conditions (21% O2) for 4 weeks. Rats were housed in standard polypropylene cages under controlled photocycle (12 h light/12 h dark) under 22-25 ℃ temperature. | |||
| Response Summary | METTL3/YTHDF2/Mutated in multiple advanced cancers 1 (PTEN) axis exerts a significant role in hypoxia induced PASMCs proliferation, providing a novel therapeutic target for hypoxic pulmonary hypertension. | |||
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [241] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Down regulation | |||
Myc proto-oncogene protein (MYC)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LNCaP cell line | Homo sapiens |
|
Treatment: shMETTL3 LNCaP cells
Control: shControl LNCaP cells
|
GSE147884 | |
| Regulation |
 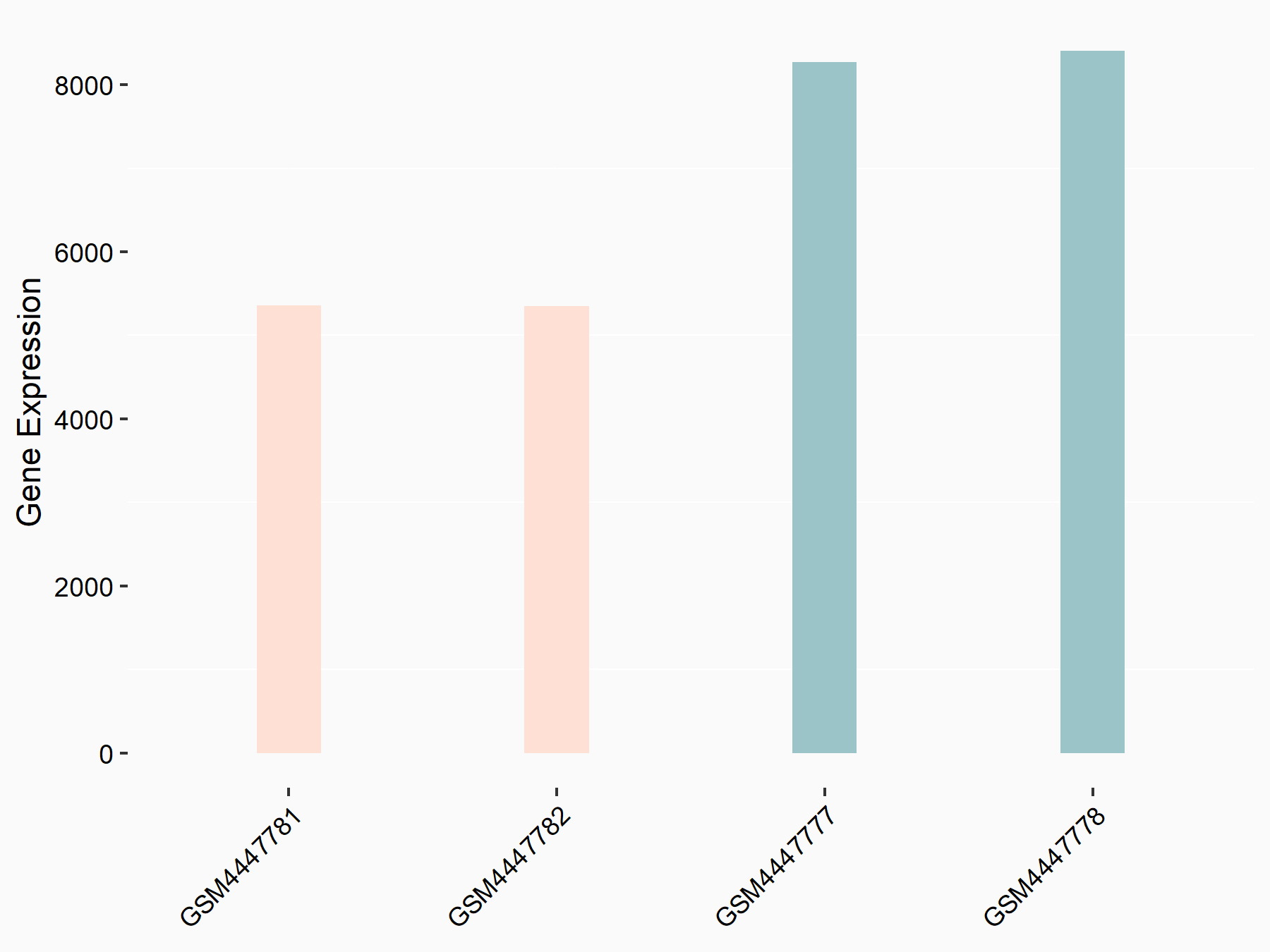 |
logFC: -6.39E-01 p-value: 4.30E-103 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.10E+00 | GSE60213 |
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [9] | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell differentiation and apoptosis | |||
| Apoptosis (hsa04210) | ||||
In-vitro Model |
HSPC (Human hematopoietic stem cell) | |||
| In-vivo Model | 500,000 selected cells were injected via tail vein or retro-orbital route into female NSG (6-8 week old) recipient mice that had been sublethally irradiated with 475 cGy one day before transplantation. | |||
| Response Summary | METTL3 depletion in human myeloid leukemia cell lines induces cell differentiation and apoptosis and delays leukemia progression in recipient mice in vivo. Single-nucleotide-resolution mapping of m6A coupled with ribosome profiling reveals that m6A promotes the translation of Myc proto-oncogene protein (MYC), BCL2 and PTEN mRNAs in the human acute myeloid leukemia MOLM-13 cell line. Moreover, loss of METTL3 leads to increased levels of phosphorylated AKT. | |||
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [77] | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 |
| NHOK (Normal oral keratinocytes) | ||||
| SCC-15 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1681 | |
| SCC-25 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1682 | |
| TSCCa | Endocervical adenocarcinoma | Homo sapiens | CVCL_VL15 | |
| In-vivo Model | The stable transfection of SCC25 cells (1 × 107 cells in 0.1 mL) with lenti-sh-METTL3 or blank vectors was injected subcutaneously into BALB/c nude mice. | |||
| Response Summary | In oral squamous cell carcinoma, YTH N6-methyladenosine RNA binding protein 1 (YTH domain family, member 1 [YTHDF1]) mediated the m6A-increased stability of Myc proto-oncogene protein (MYC) mRNA catalyzed by METTL3. | |||
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [42] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
GES-1 | Normal | Homo sapiens | CVCL_EQ22 |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MKN74 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_2791 | |
| pGCC (Primary GC cells) | ||||
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| In-vivo Model | A total of 2 × 106 GC cells were injected into the flank of nude mice in a 1:1 suspension of BD Matrigel (BD Biosciences) in phosphate-buffered saline (PBS) solution. | |||
| Response Summary | In gastric cancer, several component molecules (e.g., MCM5, MCM6, etc.) of Myc proto-oncogene protein (MYC) target genes were mediated by METTL3 via altered m6A modification. | |||
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [79] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | METTL3 stable knockdown or overexpression HCT116 cells were collected and resuspended at a density of 5 × 106 or 3 × 106 cells per 150 uL PBS. | |||
| Response Summary | METTL3 exerted its function through enhancing Myc proto-oncogene protein (MYC) expression, at least partially in an m6A-IGF2BP1-dependent manner. Knockdown of METTL3 suppressed colorectal cancer cell proliferation in vitro and in vivo. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [47] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0608 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HOP-62 | Lung adenocarcinoma | Homo sapiens | CVCL_1285 | |
| In-vivo Model | For the in vivo tumorigenicity assay, female BALB/c nude mice (ages 4-5 weeks) were randomly divided into two groups (n = 6/group). Calu1 cells (4 × 106) that had been stably transfected with sh-LCAT3 or scramble were implanted subcutaneously into the nude mice. Tumor growth was measured after one week, and tumor volumes were calculated with the following formula: Volume (cm3) = (length × width2)/2. After four weeks, the mice were euthanized, and the tumors were collected and weighed. For the in vivo tumor invasion assay, 1.2 × 106 scramble or shLCAT3 cells were injected intravenously into the tail vein of nude mice (n = 6/group). 1.5 mg luciferin (Gold Biotech, St Louis, MO, USA) was administered once a week for 4 weeks, to monitor metastases using an IVIS@ Lumina II system (Caliper Life Sciences, Hopkinton, MA, USA). | |||
| Response Summary | LCAT3 upregulation is attributable to N6-methyladenosine (m6A) modification mediated by methyltransferase like 3 (METTL3), leading to LCAT3 stabilization. LCAT3 as a novel oncogenic lncRNA in the lung, and validated the LCAT3-FUBP1-Myc proto-oncogene protein (MYC) axis as a potential therapeutic target for lung adenocarcinomas. | |||
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [81] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
LNCaP C4-2 | Prostate carcinoma | Homo sapiens | CVCL_4782 |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| Response Summary | METTL3 enhanced Myc proto-oncogene protein (MYC) expression by increasing m6A levels of MYC mRNA transcript, leading to oncogenic functions in prostate cancer. | |||
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Glucose metabolism | |||
| Response Summary | AF4/FMR2 family member 4 (AFF4), two key regulators of NF-Kappa-B pathway (IKBKB and RELA) and Myc proto-oncogene protein (MYC) were further identified as direct targets of METTL3-mediated m6A modification.overexpression of METTL3 significantly promoted Bladder cancer cell growth and invasion. | |||
Polycystic kidney disease [ICD-11: GB81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [82] | |||
| Responsed Disease | Polycystic kidney disease [ICD-11: GB81] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
mIMCD-3 | Normal | Mus musculus | CVCL_0429 |
| In-vivo Model | The clone, with one wild-type Mettl3 allele and one L1L2_Bact_P cassette inserted allele, was injected into C57BL/6 blastocysts. Mettl3-targeted mouse line was established from a germline-transmitting chimera. The chimeric mouse was crossed to C57BL/6 Flp mice to excise the neomycin resistance system. | |||
| Response Summary | Mettl3 activates the cyst-promoting c-Myc and cAMP pathways through enhanced Myc proto-oncogene protein (MYC) and Avpr2 mRNA m6A modification and translation. Thus, Mettl3 promotes Autosomal dominant polycystic kidney disease and links methionine utilization to epitranscriptomic activation of proliferation and cyst growth. | |||
Diffuse large B-cell lymphomas [ICD-11: 2A81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [311] | |||
| Responsed Disease | Diffuse large B-cell lymphomas [ICD-11: 2A81] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SU-DHL-4 | Diffuse large B-cell lymphoma | Homo sapiens | CVCL_0539 |
| DB | Diffuse large B-cell lymphoma germinal center B-cell type | Homo sapiens | CVCL_1168 | |
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [312] | |||
| Responsed Disease | Esophageal Squamous Cell Carcinoma [ICD-11: 2B70.1] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Eca-109 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_6898 |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| In-vivo Model | BALB/c-nu mice (5-6 weeks) were purchased from Sun Yat-sen University (SYSU) Animal Center. The mice were housed in a pathogen-free environment at the SYSU Animal Center. To establish a human ESCC xenograft model, 20 BALB/c-nu mice were randomly assigned to four groups, with each group consisting of five mice. Two groups of mice received a subcutaneous injection of either 2.0 × 106 Eca109 cells or 2.0 × 106 sh-METTL3-Eca109 cells. Tumor development was monitored daily. After one week, each mice was subjected to intratumoral injection of 10 μl PBS or Fn (1.0 × 107 CFU) once every two days, three times a week. The mice were sacrifices at the end of two weeks after the injection, and subsequently, the tumor, liver, and lungs were surgically removed. The following formula was used to determine tumor volume: 0.5× (length × width2). | |||
Myeloid differentiation primary response protein MyD88 (MYD88)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shMETTL3 MOLM13 cells
Control: MOLM13 cells
|
GSE98623 | |
| Regulation |
 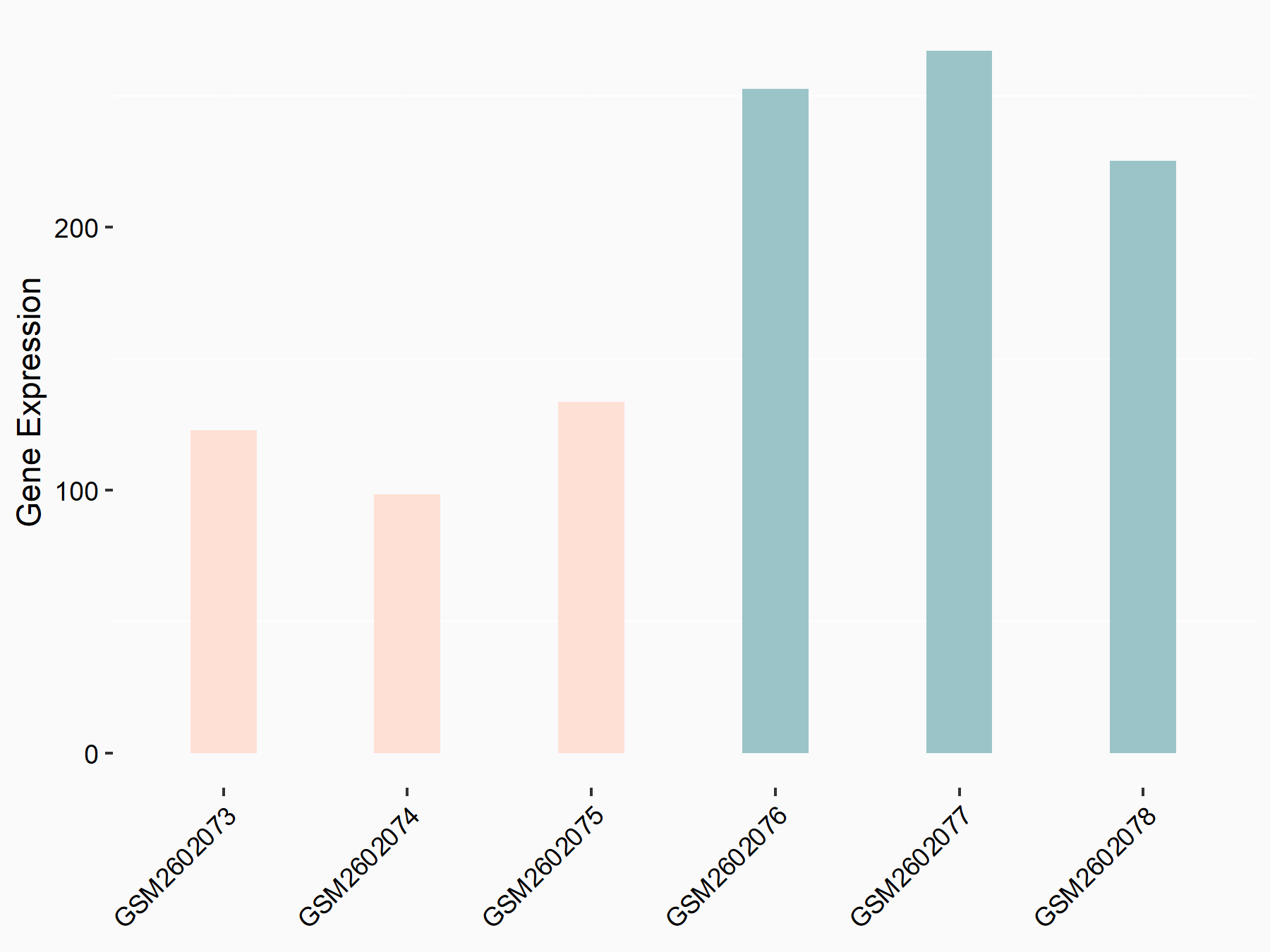 |
logFC: -1.07E+00 p-value: 1.83E-13 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.16E+00 | GSE60213 |
Pulpitis [ICD-11: DA09]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [83] | |||
| Responsed Disease | Pulpitis [ICD-11: DA09] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | Alternative splicing | |||
| Response Summary | Knocked down METTL3 and demonstrated that METTL3 depletion decreased the expression of inflammatory cytokines and the phosphorylation of IKK-alpha/beta, p65 and IKappa-B-alpha in the NF-Kappa-B signalling pathway as well as p38, ERK and JNK in the MAPK signalling pathway in LPS-induced HDPCs. METTL3 inhibits the LPS-induced inflammatory response of HDPCs by regulating alternative splicing of Myeloid differentiation primary response protein MyD88 (MYD88). | |||
Skeletal anomaly [ICD-11: LD24]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [84] | |||
| Responsed Disease | Skeletal anomaly [ICD-11: LD24] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Central carbon metabolism in cancer | hsa05230 | ||
| Cell Process | Glucose metabolism | |||
In-vitro Model |
Mesenchymal stem cell line (NP tissues were used to isolate NP cells) | |||
| Response Summary | METTL3 positively regulates expression of Myeloid differentiation primary response protein MyD88 (MYD88), a critical upstream regulator of NF-Kappa-B signaling, by facilitating m6A methylation modification to MYD88-RNA, subsequently inducing the activation of NF-Kappa-B which is widely regarded as a repressor of osteogenesis and therefore suppressing osteogenic progression. The METTL3-mediated m6A methylation is found to be dynamically reversed by the demethylase ALKBH5. | |||
NACHT, LRR and PYD domains-containing protein 3 (NLRP3)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 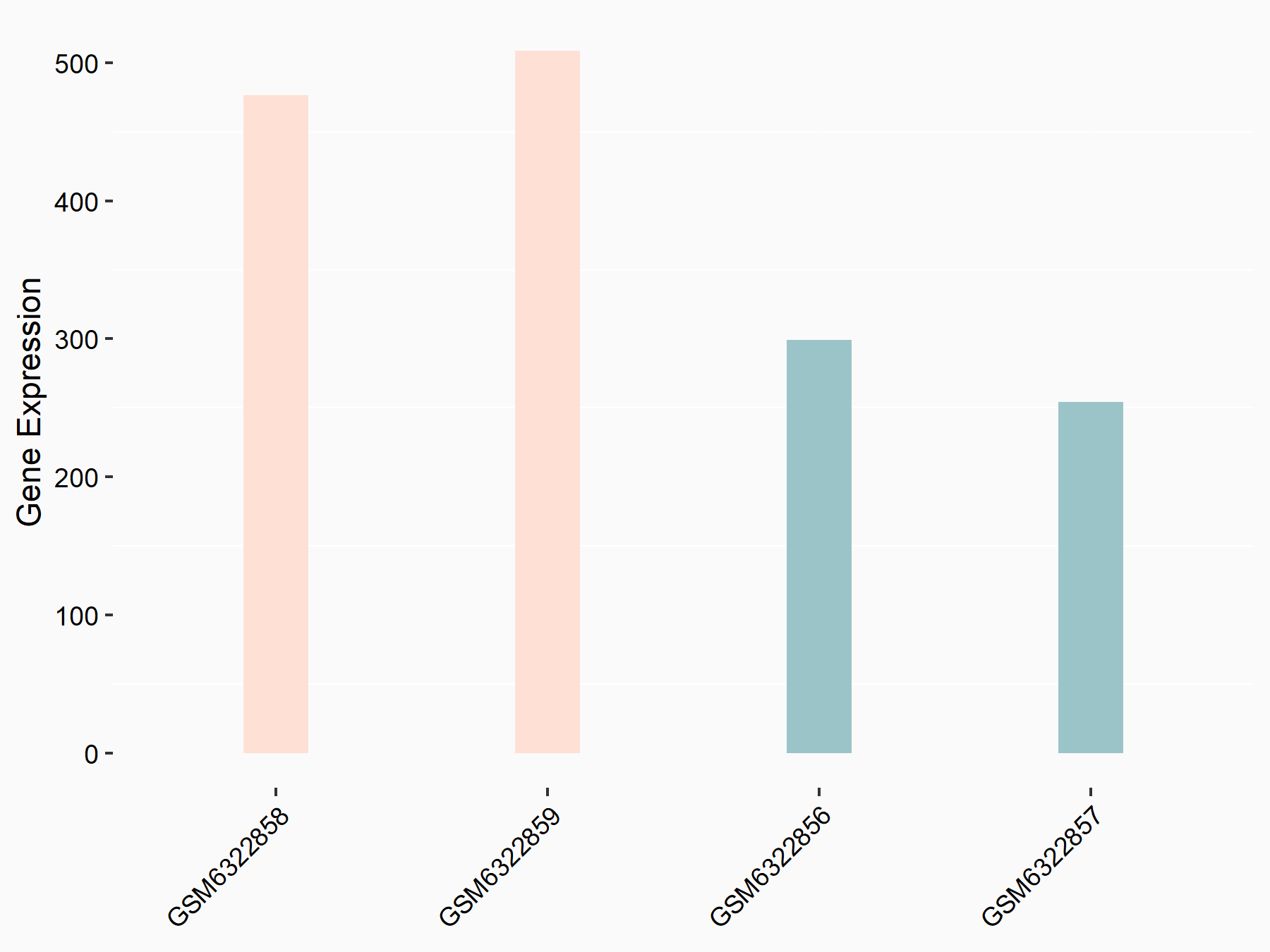 |
logFC: 8.32E-01 p-value: 1.76E-06 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.81E+00 | GSE60213 |
Atherosclerosis [ICD-11: BD40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [85] | |||
| Responsed Disease | Atherosclerosis [ICD-11: BD40.Z] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
MAEC | Normal | Mus musculus | CVCL_U411 |
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
| Response Summary | In the in vivo atherosclerosis model,partial ligation of the carotid artery led to plaque formation and up-regulation of METTL3 and NLRP1, with down-regulation of KLF4; knockdown of METTL3 via repetitive shRNA administration prevented the atherogenic process, NACHT, LRR and PYD domains-containing protein 3 (NLRP3) up-regulation, and KLF4 down-regulation. Collectively, it has demonstrated that METTL3 serves a central role in the atherogenesis induced by oscillatory stress and disturbed blood flow. | |||
Chronic kidney disease [ICD-11: GB61]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [86] | |||
| Responsed Disease | Chronic kidney disease [ICD-11: GB61.Z] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
In-vitro Model |
MPC-5 | Normal | Mus musculus | CVCL_AS87 |
| Response Summary | TFA could ameliorate HG-induced pyroptosis and injury in podocytes by targeting METTL3-dependent m6A modification via the regulation of NACHT, LRR and PYD domains-containing protein 3 (NLRP3)-inflammasome activation and PTEN/PI3K/Akt signaling. This study provides a better understanding of how TFA can protect podocytes in diabetic kidney disease (DKD). | |||
Lung cancer [ICD-11: 2C25]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [316] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Responsed Drug | Gefitinib | Approved | ||
In-vitro Model |
PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| PC-9/GR2 | Lung adenocarcinoma | Homo sapiens | CVCL_DI29 | |
|
HCC827/GR
|
N.A. | Homo sapiens | CVCL_E7R9 | |
| In-vivo Model | To establish xenograft models, five-week-old male mice were orthotopically injected with the same number of the PC9/GR cells. Tumours had developed after 4 days, at which time the xenografted mice were randomly divided into the following two experimental groups (each group with 6 mice): (1) the control group and (2) the gefitinib group. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [316] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Up regulation | |||
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [317] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
Neuroblast differentiation-associated protein AHNAK (AHNAK)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shMETTL3 MOLM13 cells
Control: MOLM13 cells
|
GSE98623 | |
| Regulation |
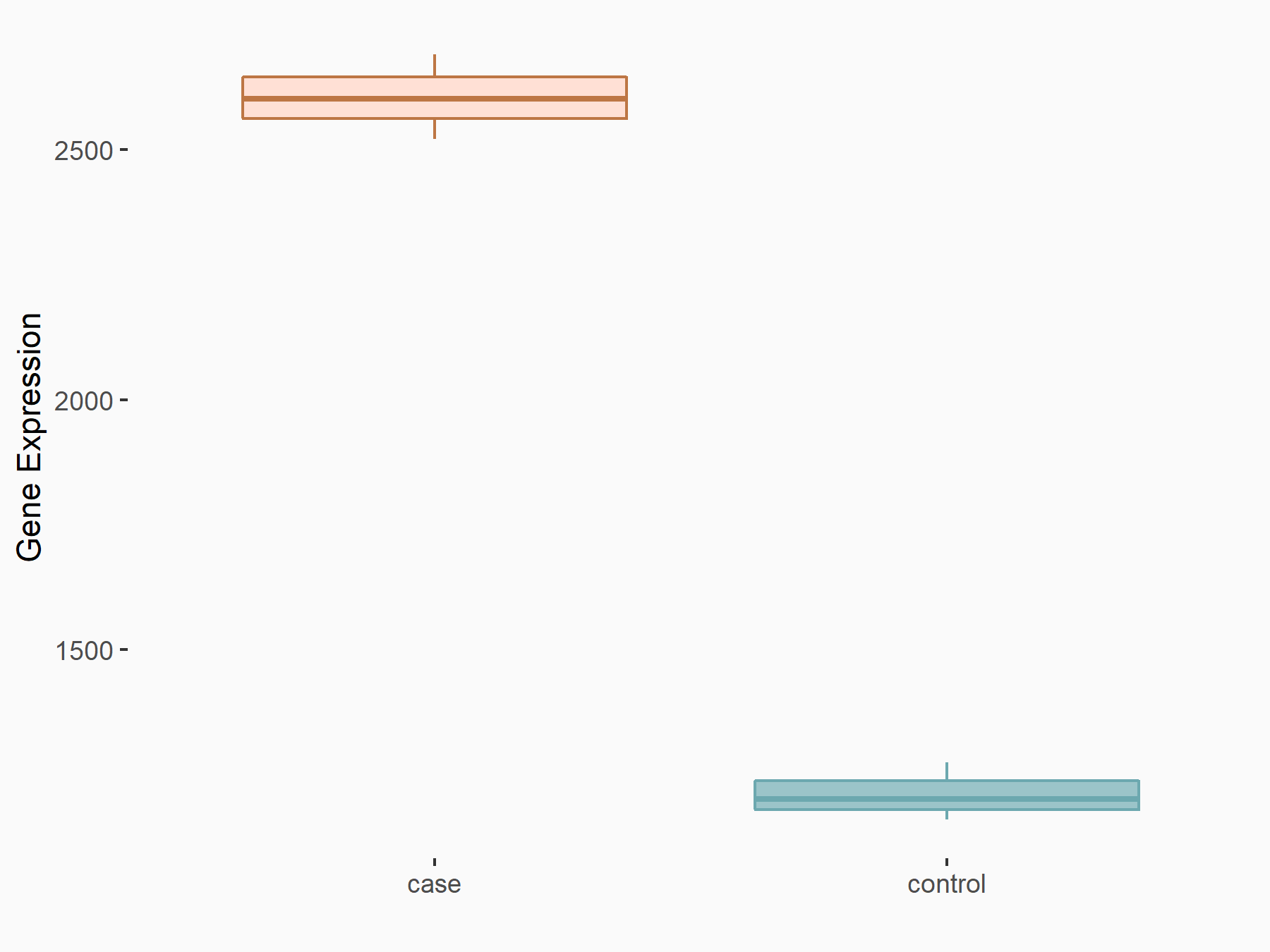 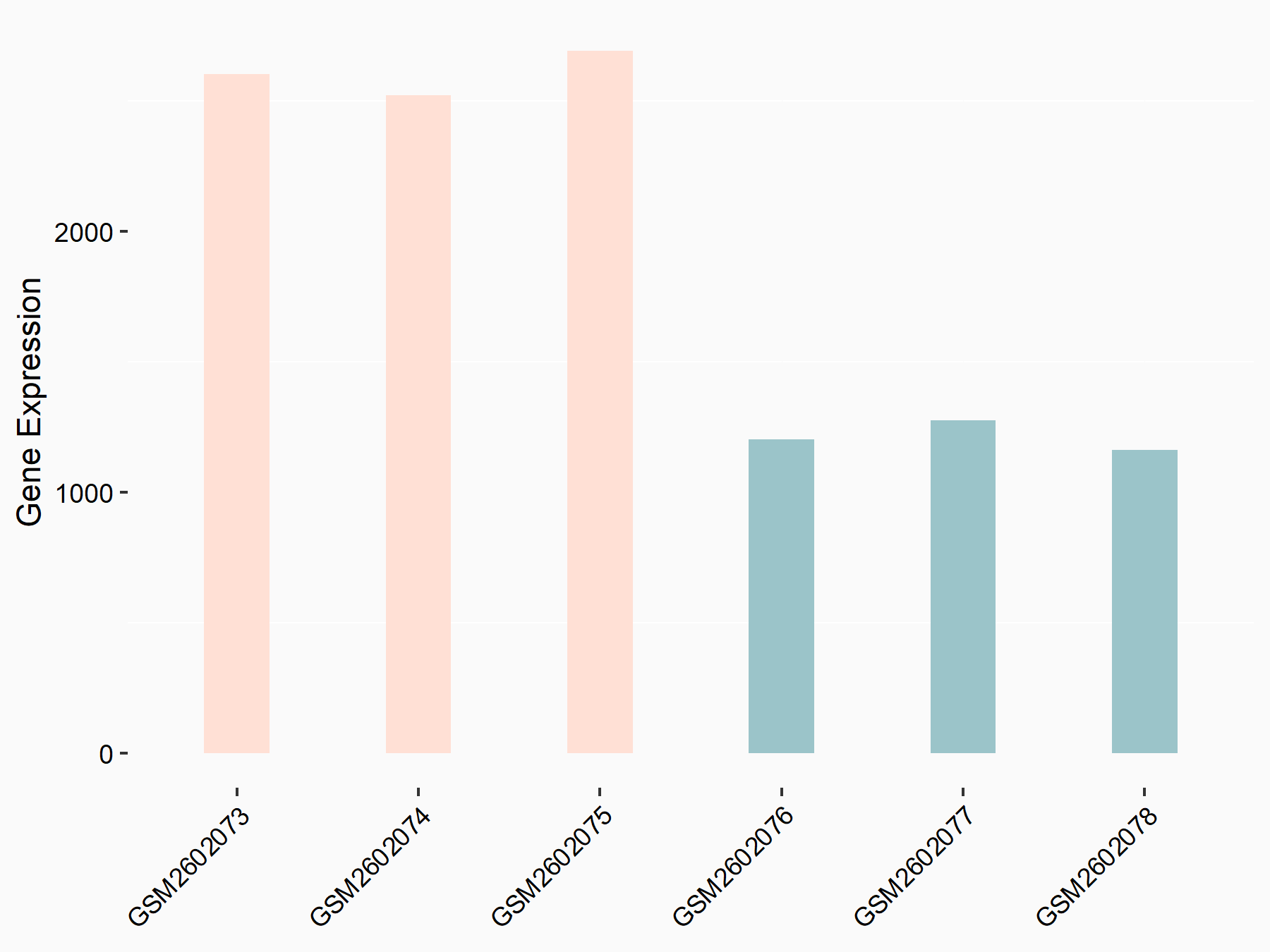 |
logFC: 1.10E+00 p-value: 5.50E-75 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.20E+00 | GSE60213 |
Corneal injury [ICD-11: NA06]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [87] | |||
| Responsed Disease | Corneal injury [ICD-11: NA06.4] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
In-vitro Model |
CGC (Conjunctival goblet cells) | |||
| In-vivo Model | Mettl3fl/wt mice were generated as previously described. Mettl3fl/wt mice were crossed with K14CreER mice to obtain K14creER/Mettl3fl/fl (cKO) mice. Mettl3 cKO and control mice were injected with tamoxifen and then were subjected to corneal alkali burn treatment. The right eye was the experimental eye, and the left eye was the control eye. The mice were sacrificed at 24 hours, 7 days, 14 days, 35 days, and 56 days after injury. Six mice were taken from each period. Both eyes were removed, frozen in OCT (n = 4), fixed in 4% paraformaldehyde, and embedded in conventional paraffin (n = 2). | |||
| Response Summary | METTL3 knockout in the limbal stem cells promotes the in vivo cell proliferation and migration, leading to the fast repair of corneal injury. In addition, m6A modification profiling identified stem cell regulatory factors Neuroblast differentiation-associated protein AHNAK (AHNAK) and DDIT4 as m6A targets. | |||
Neurocalcin-delta (NCALD)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LNCaP cell line | Homo sapiens |
|
Treatment: shMETTL3 LNCaP cells
Control: shControl LNCaP cells
|
GSE147884 | |
| Regulation |
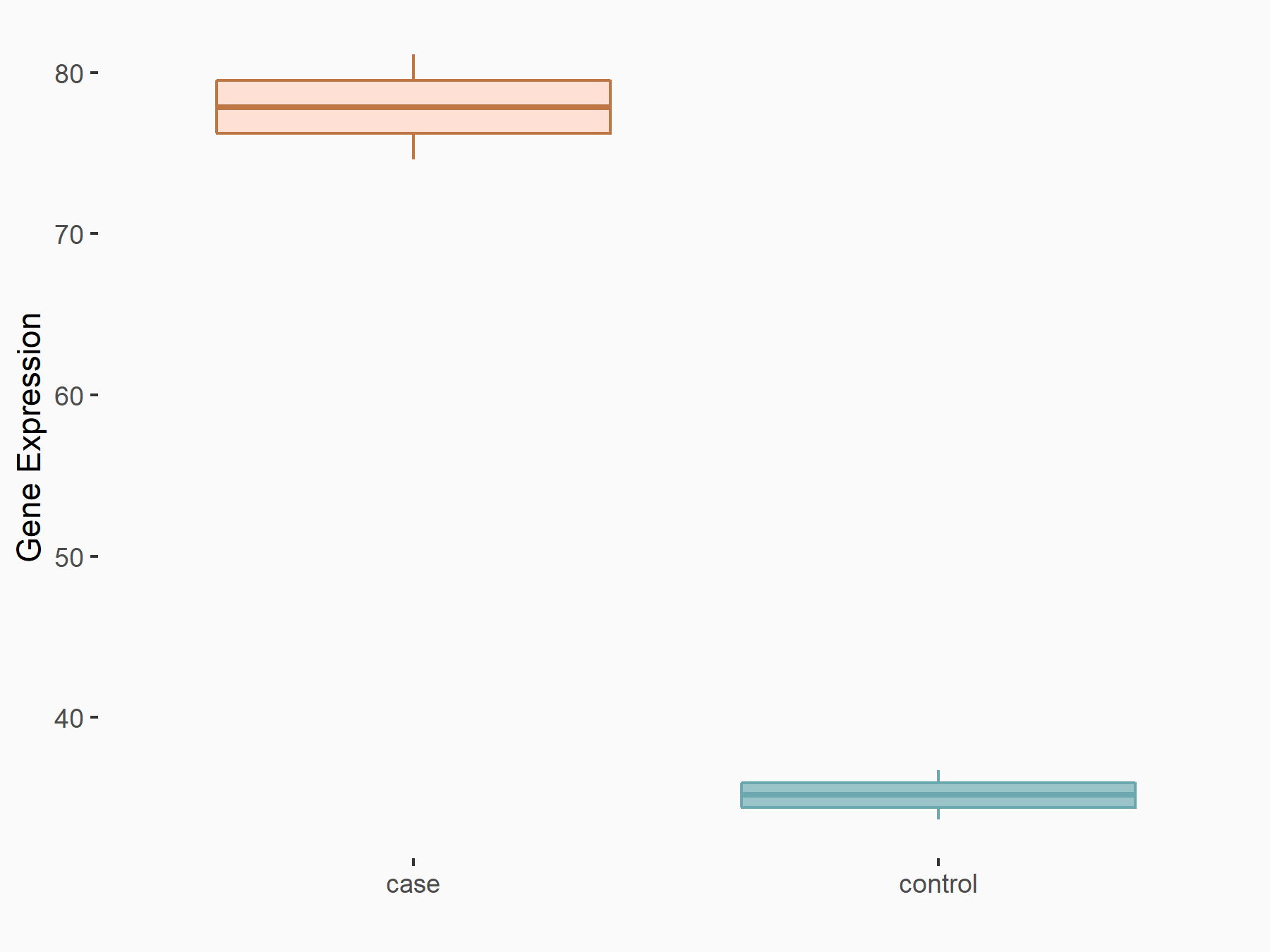 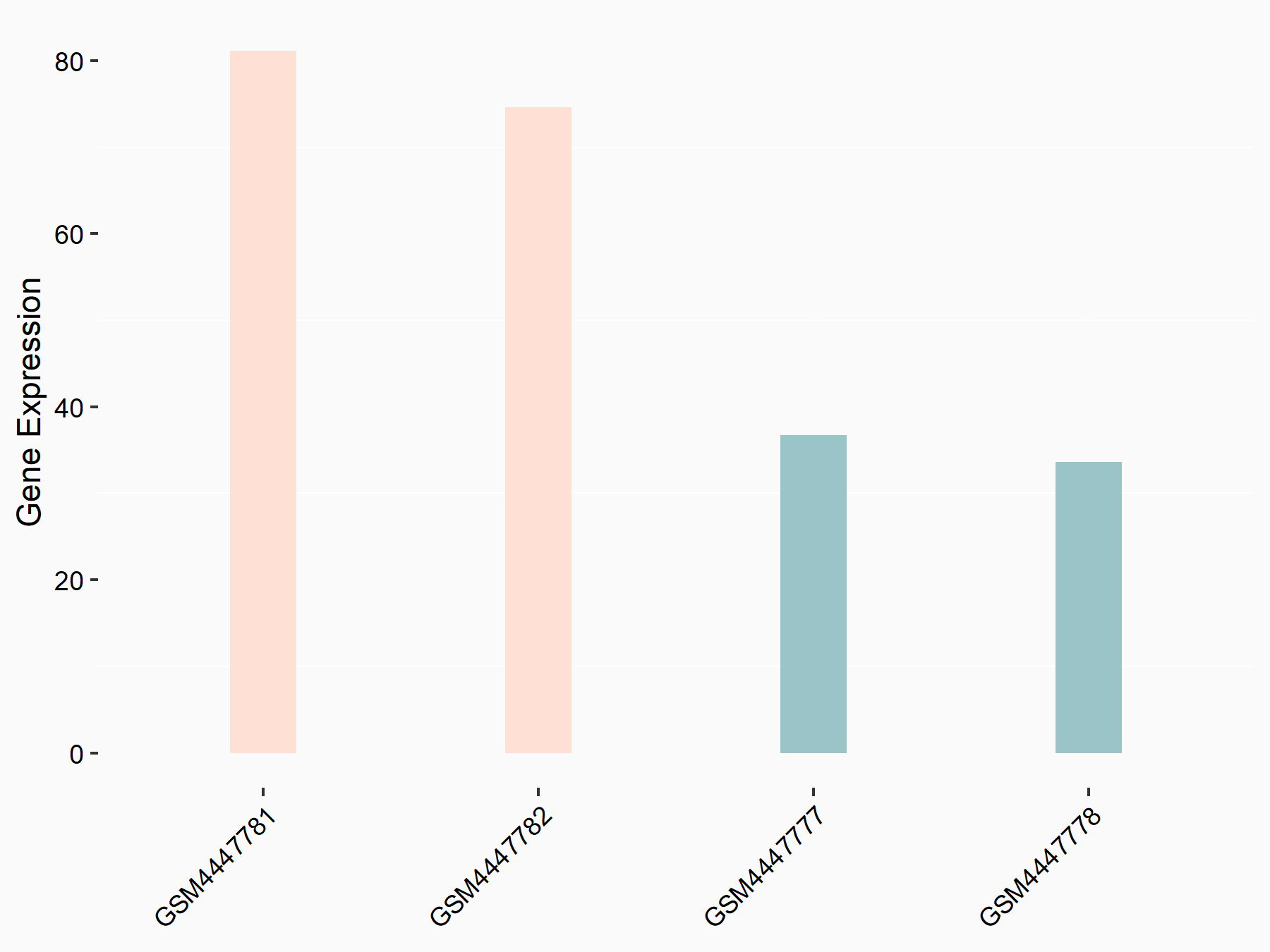 |
logFC: 1.15E+00 p-value: 3.16E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 8.03E+00 | GSE60213 |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [88] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Fluorouracil | Approved | ||
| Target Regulation | Down regulation | |||
In-vitro Model |
HT29 | Colon cancer | Mus musculus | CVCL_A8EZ |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| In-vivo Model | A tumor-bearing model was established by subcutaneously injecting 100 ul HT29 cells (5×106) followed by an intravenous injection of CAFs-derived exosomes (50 ug/mouse every three days) into the tail vein of the mice. An intraperitoneal injection of 5-FU (50 mg/kg, every week) was administered on day 12. | |||
| Response Summary | METTL3 dependent m6A methylation was upregulated in CRC to promote the processing of miR 181d 5p by DGCR8. This led to increased miR 181d 5p expression, which inhibited the 5 FU sensitivity of CRC cells by targeting Neurocalcin-delta (NCALD). | |||
Nuclear receptor-interacting protein 1 (NRIP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | DKO-1 cell line | Homo sapiens |
|
Treatment: METTL3 knockdown DKO-1 cell
Control: DKO-1 cell
|
GSE182382 | |
| Regulation |
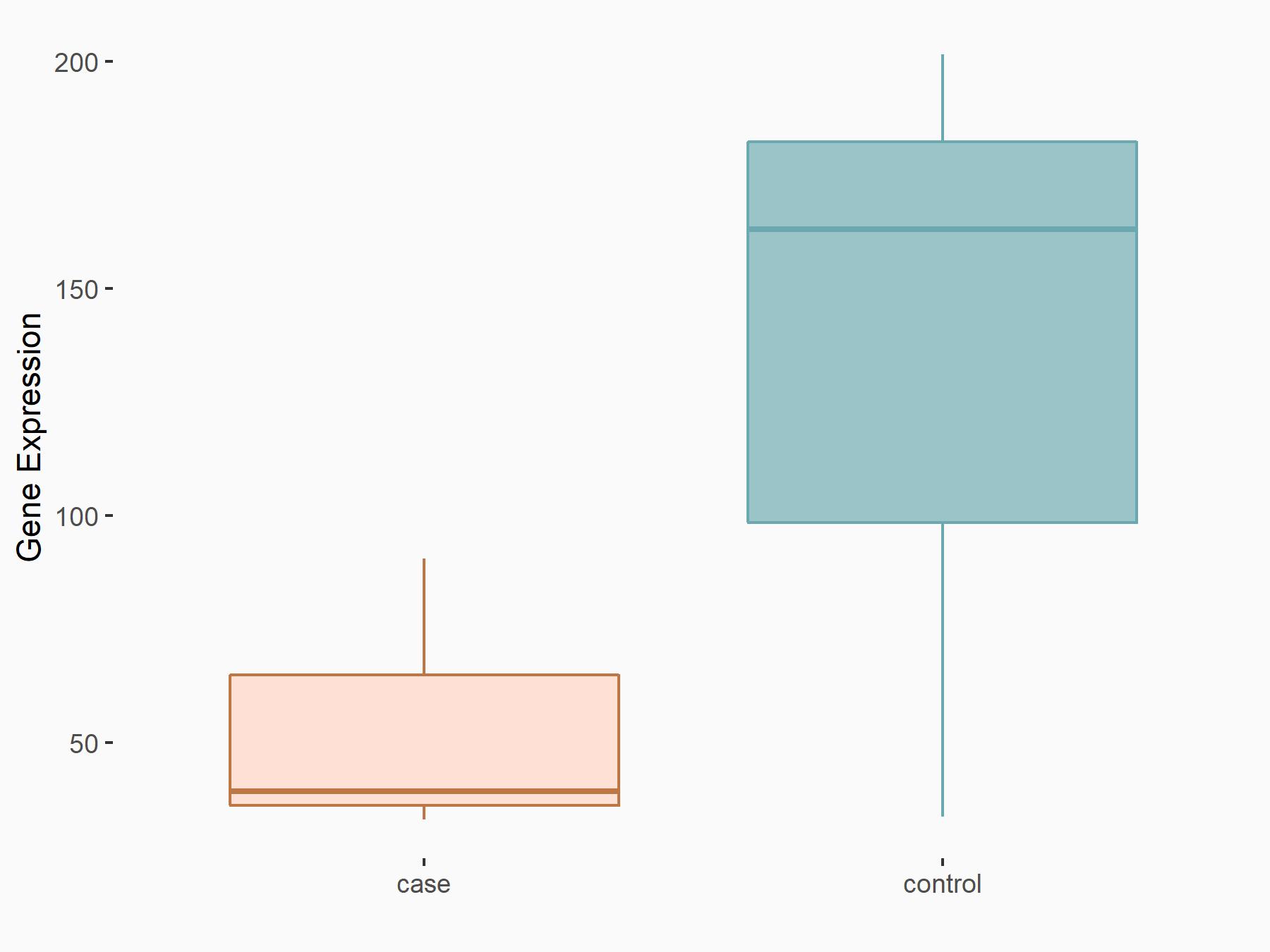 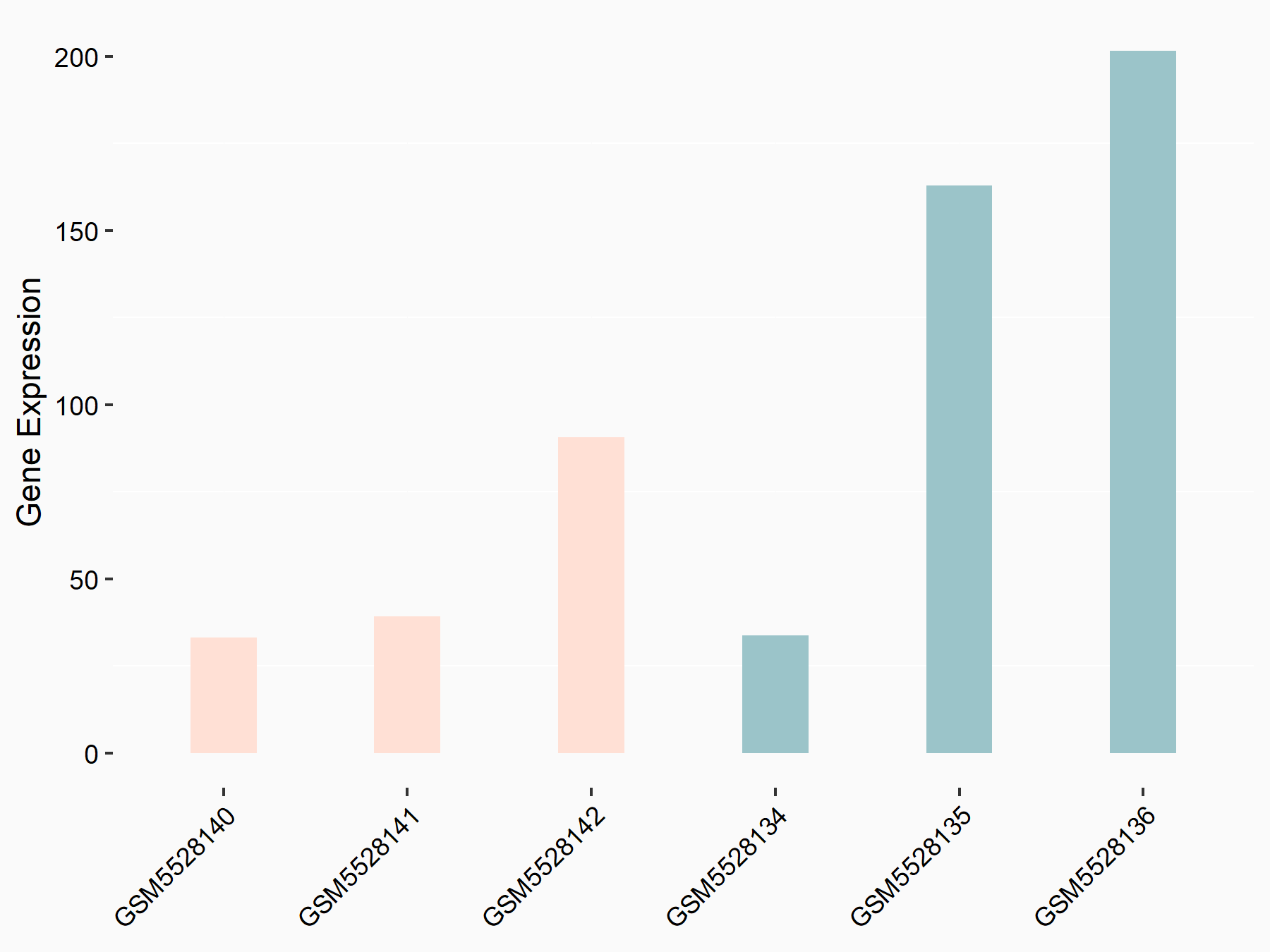 |
logFC: -1.29E+00 p-value: 1.52E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.30E+00 | GSE60213 |
Down syndrome [ICD-11: LD40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [89] | |||
| Responsed Disease | Complete trisomy 21 [ICD-11: LD40.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HT22 | Normal | Mus musculus | CVCL_0321 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| Response Summary | The NRIP1 mRNA increased in fetal brain tissues of DS, whereas the m6A modification of the NRIP1 mRNA significantly decreased. METTL3 knockdown reduced the m6A modification of NRIP1 mRNA and increased its expression, and an increase in NRIP1 m6A modification and a decrease in its expression were observed in METTL3-overexpressed cells. | |||
Parathyroid hormone 1 receptor (PTH1R)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
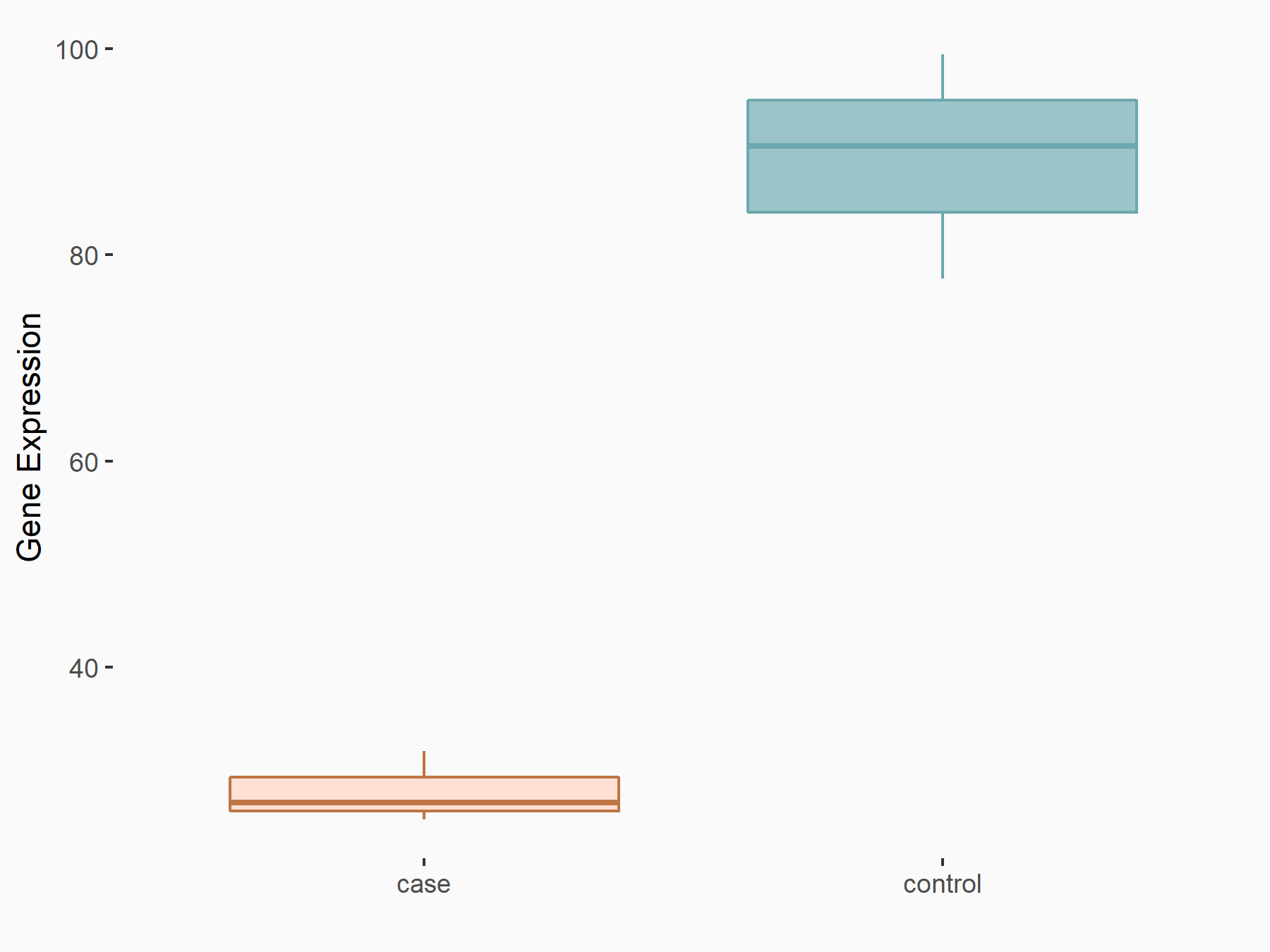 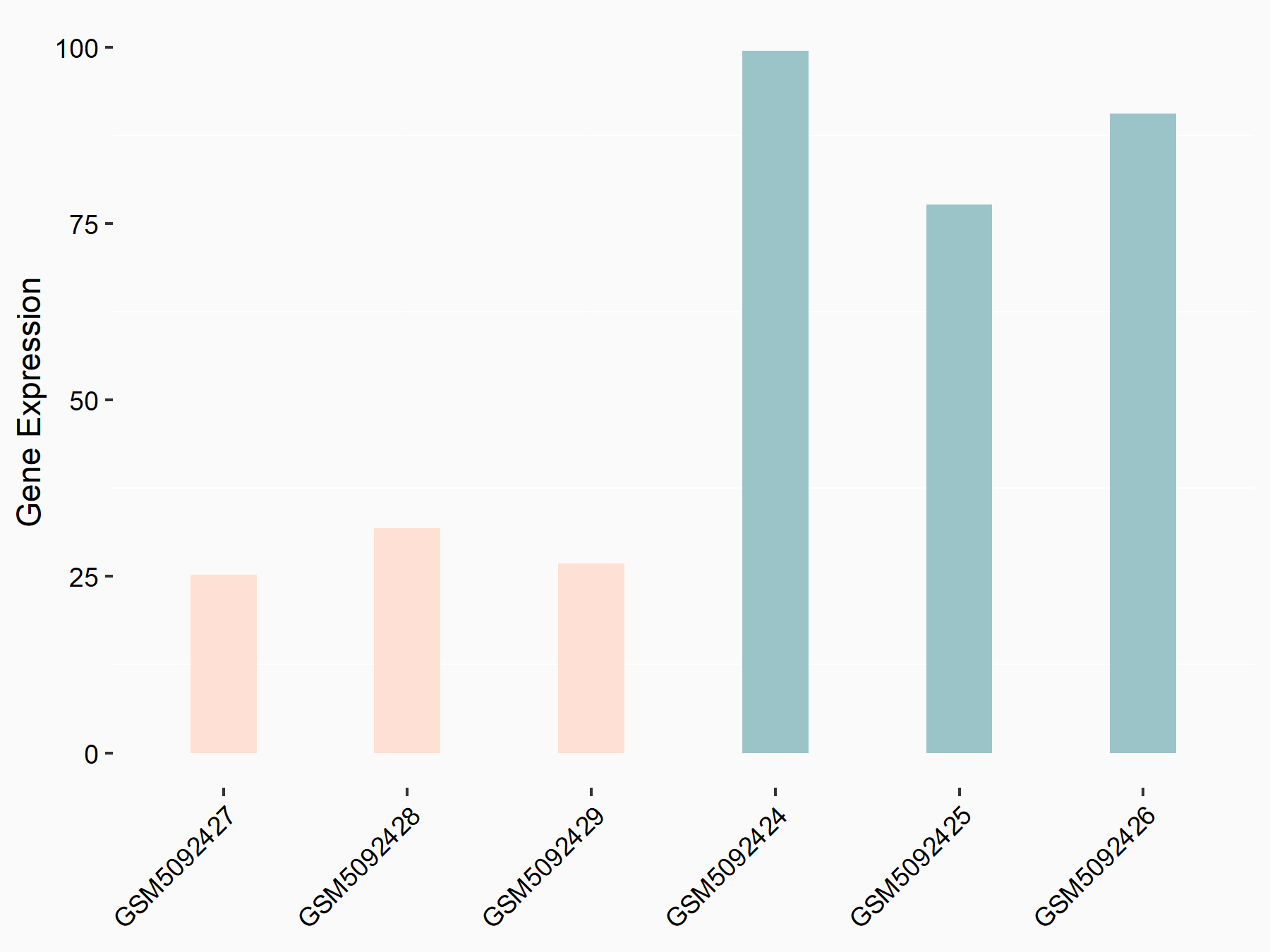 |
logFC: -1.67E+00 p-value: 4.51E-08 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.67E+00 | GSE60213 |
Low bone mass disorder [ICD-11: FB83]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [90] | |||
| Responsed Disease | Osteoporosis [ICD-11: FB83.1] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Parathyroid hormone synthesis, secretion and action | hsa04928 | ||
| Cell Process | Adipogenesis | |||
| In-vivo Model | Mettl3fl/+ mice with C57BL6/J background were generated using CRISPR-Cas9 systems. | |||
| Response Summary | Knockout of Mettl3 reduces the translation efficiency of MSCs lineage allocator Parathyroid hormone 1 receptor (PTH1R), and disrupts the PTH-induced osteogenic and adipogenic responses in vivo. | |||
Paternally-expressed gene 3 protein (PEG3)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LNCaP cell line | Homo sapiens |
|
Treatment: shMETTL3 LNCaP cells
Control: shControl LNCaP cells
|
GSE147884 | |
| Regulation |
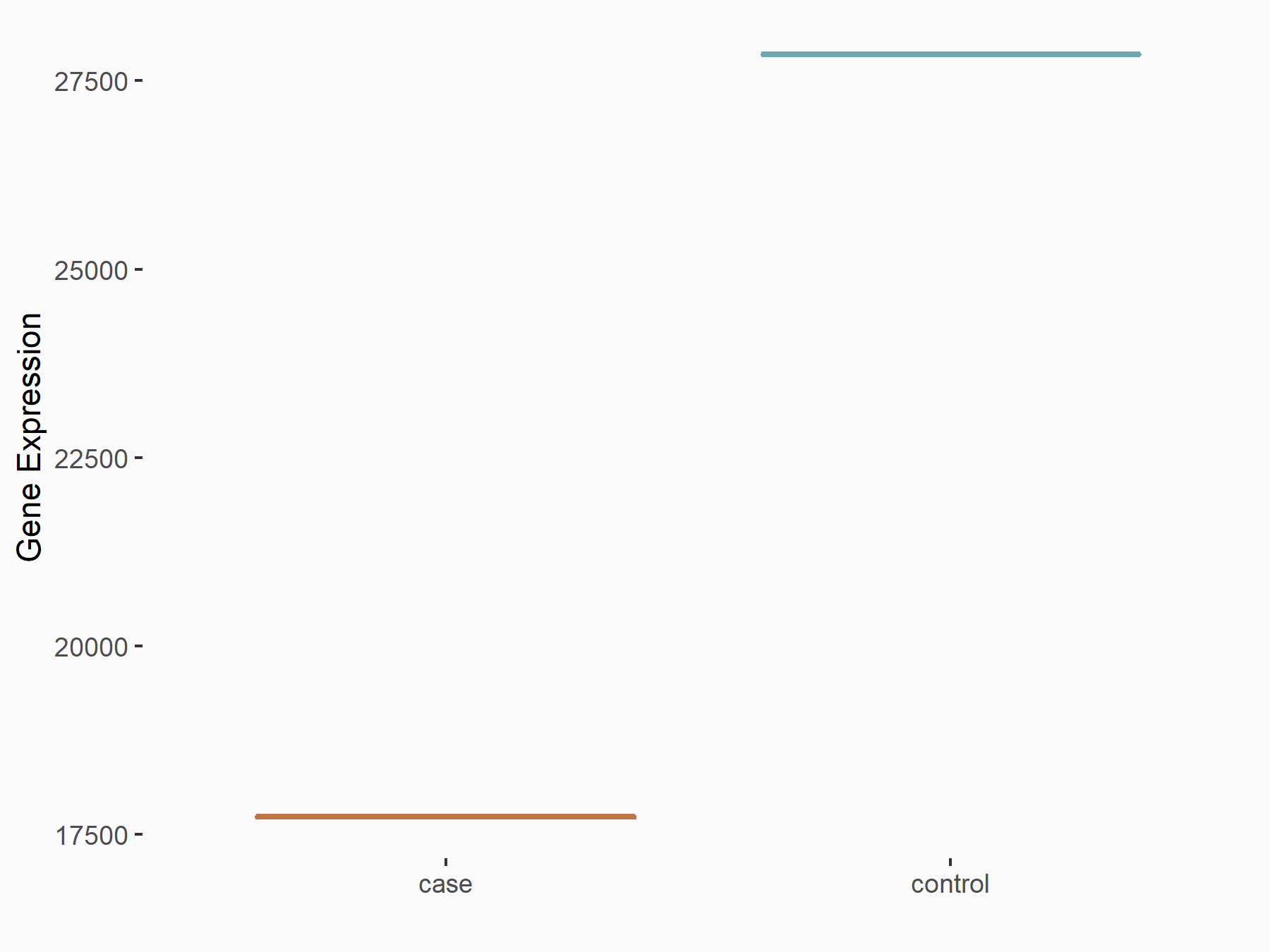 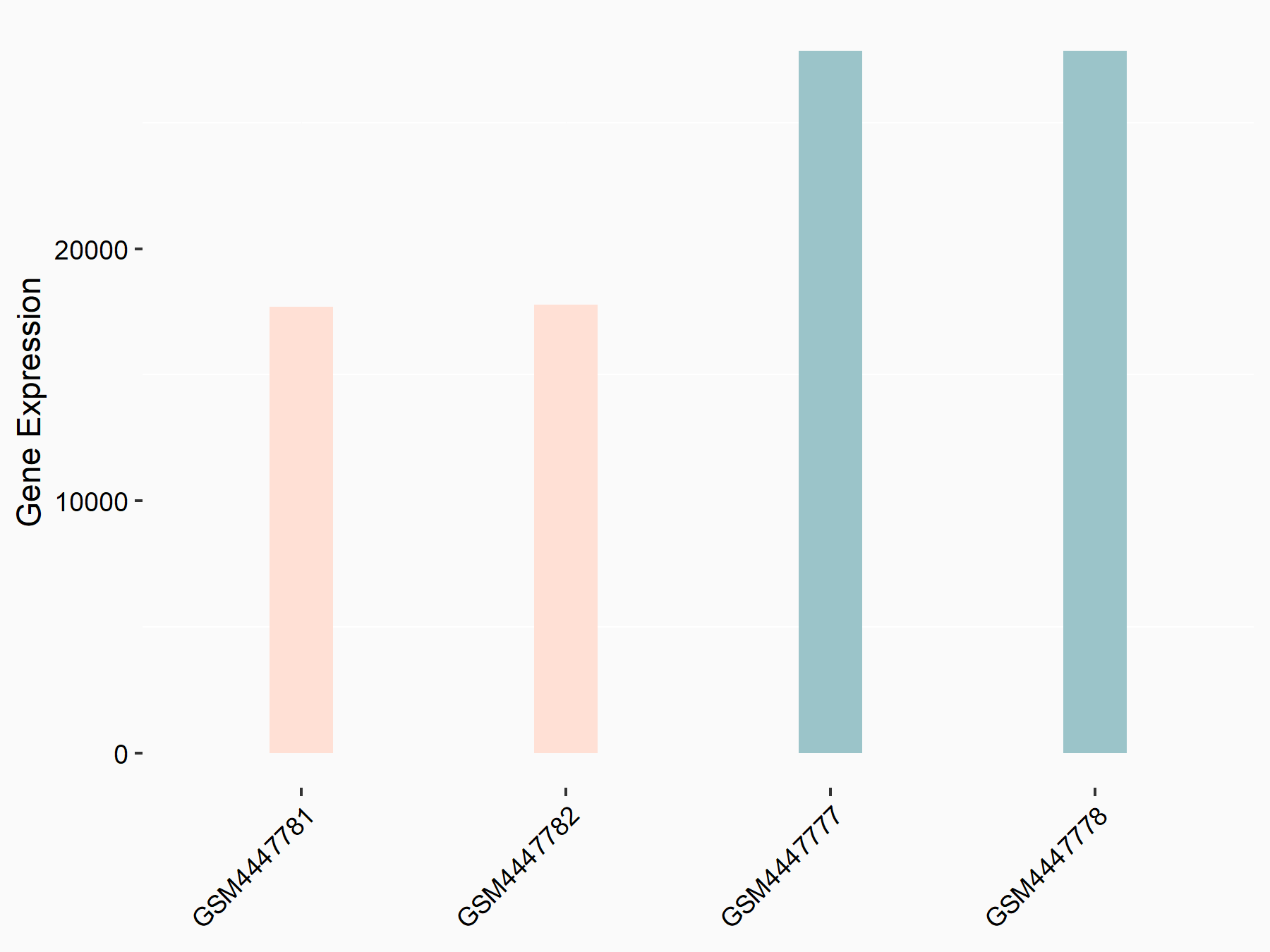 |
logFC: -6.51E-01 p-value: 1.16E-264 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 5.95E+00 | GSE60213 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [91] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | The 40 nude mice of each large group were classified into 8 small groups (n = 5) and subcutaneously injected with transfected cell suspension in the back (5 × 106 cells/mL/mouse). The length and width of tumors were recorded every 4 days, and the tumor volume = (length × width2)/2. The tumor growth curve was thereby graphed. On the 28th day of injection, mice were euthanized by neck dislocation, and the xenografts were harvested, photographed, and weighed. | |||
| Response Summary | METTL3 regulates the m6A modification to promote the maturation of miR-1246, which targets Paternally-expressed gene 3 protein (PEG3) to participate in occurrence and development of non-small cell lung cancer. | |||
Peroxisomal bifunctional enzyme (EHHADH)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
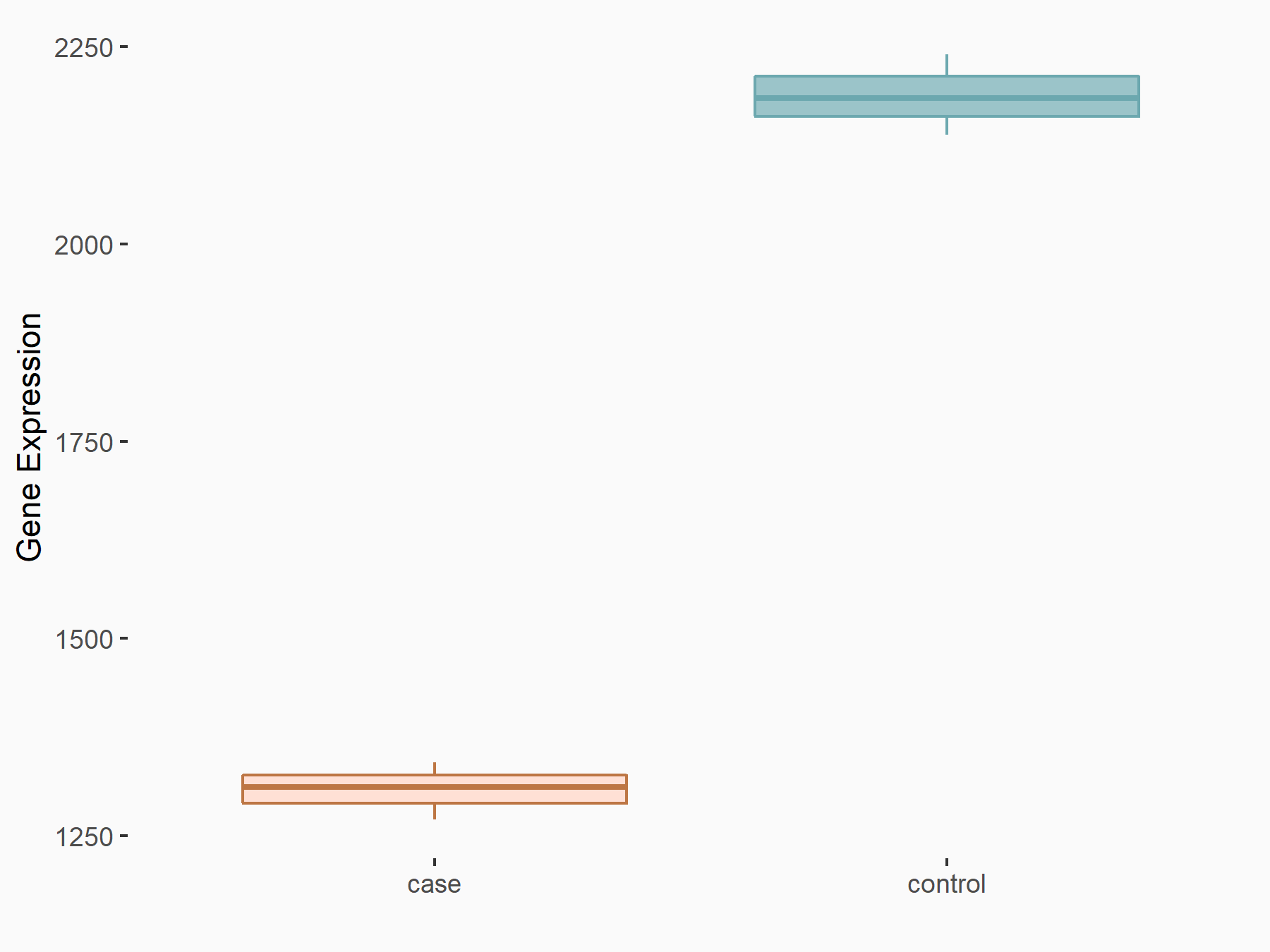 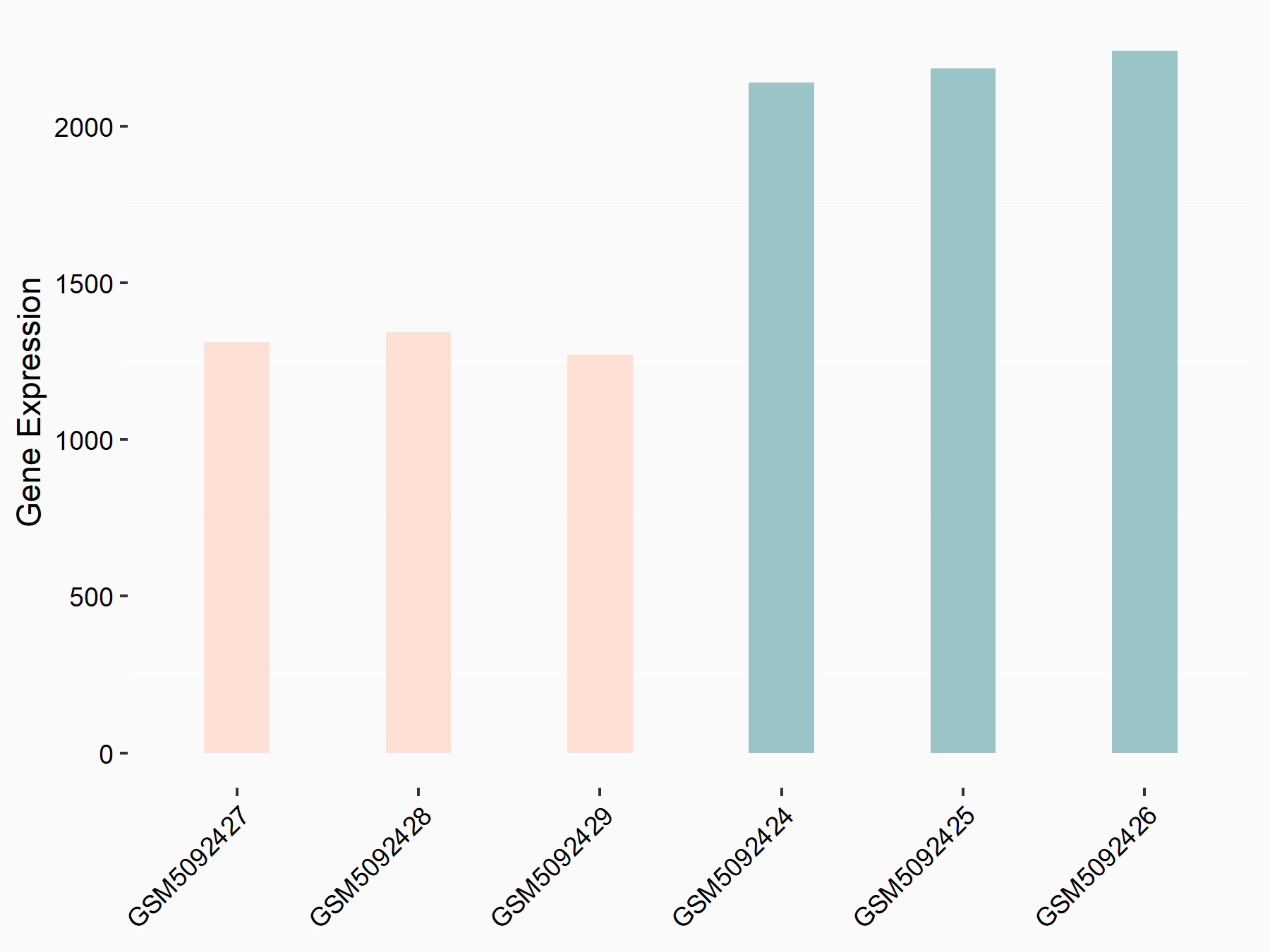 |
logFC: -7.43E-01 p-value: 3.92E-35 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.66E+00 | GSE60213 |
Type 2 diabetes mellitus [ICD-11: 5A11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [48] | |||
| Responsed Disease | Type 2 diabetes mellitus [ICD-11: 5A11] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Insulin resistance | hsa04931 | ||
| Cell Process | Lipid metabolism | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | Hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were generated by crossing mice with TBG-Cre Tg mice. METTL3 flox (METTL3 fl/fl) and hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were used for experiments. | |||
| Response Summary | Type 2 diabetes (T2D) is characterized by lack of insulin, insulin resistance and high blood sugar. METTL3 silence decreased the m6A methylated and total mRNA level of Fatty acid synthase (Fasn), subsequently inhibited fatty acid metabolism. The expression of Acc1, Acly, Dgat2, Peroxisomal bifunctional enzyme (EHHADH), Fasn, Foxo, Pgc1a and Sirt1, which are critical to the regulation of fatty acid synthesis and oxidation were dramatically decreased in livers of hepatocyte-specific METTL3 knockout mice. | |||
Peroxisome proliferator-activated receptor alpha (PPARalpha/PPARA)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
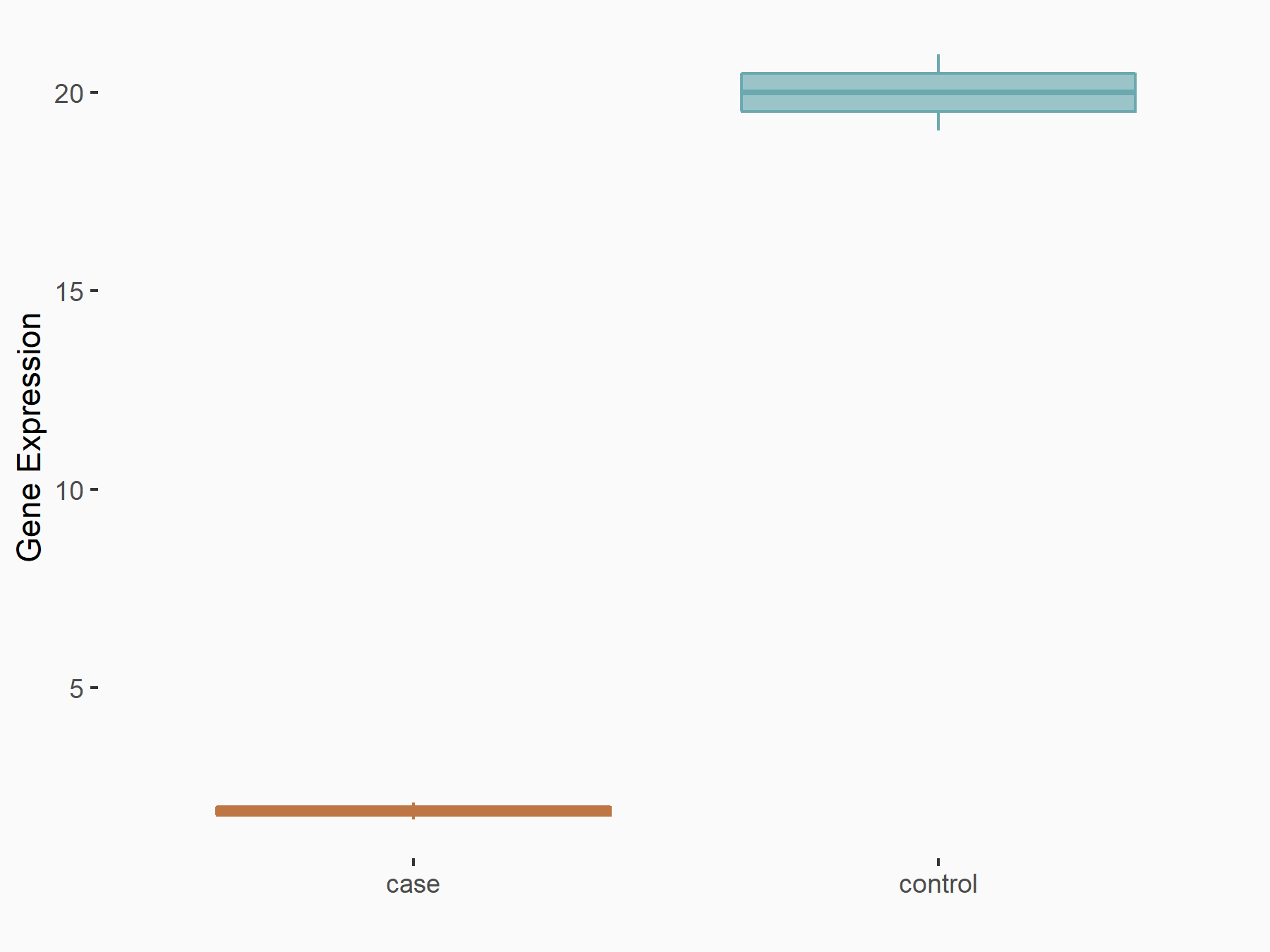 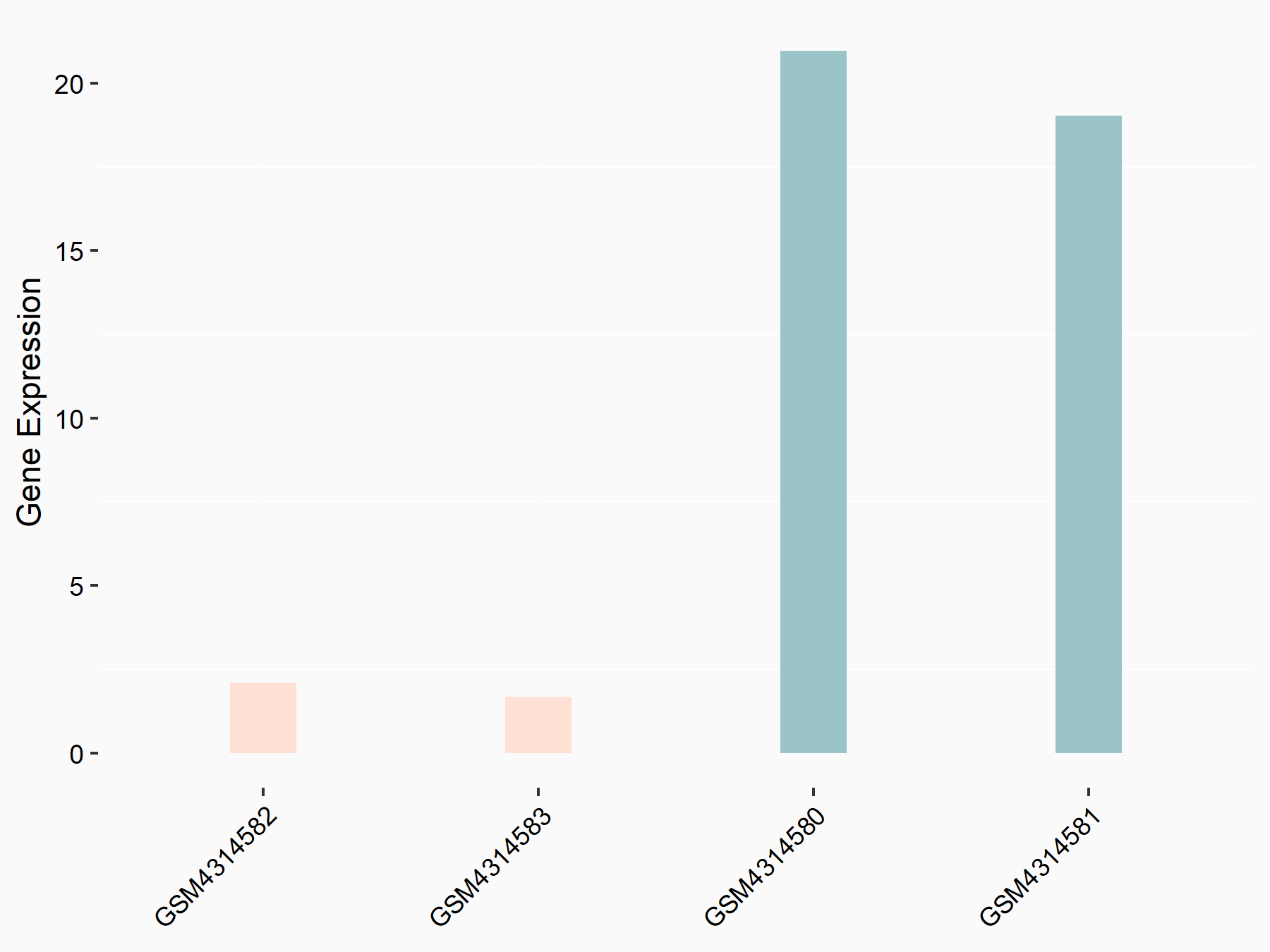 |
logFC: -3.42E+00 p-value: 1.41E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 8.50E+00 | GSE60213 |
Metabolic disorders [ICD-11: 5D2Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [92] | |||
| Responsed Disease | Metabolic disorders [ICD-11: 5D2Z] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PPAR signaling pathway | hsa03320 | ||
| Adipocytokine signaling pathway | hsa04920 | |||
| Cell Process | Llipid metabolism | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| Hepa 1-6 | Hepatocellular carcinoma of the mouse | Mus musculus | CVCL_0327 | |
| In-vivo Model | Liver-specific Bmal1f/f-AlbCre-knockout mice were purchased from Jackson Laboratory. C57BI/6J or Bmal1f/f-AlbCre-knockout male mice were maintained under a 12 hr light/12 hr dark (LD) cycle (ZT0 = 6 AM) and fed ad libitum with normal rodent chow (2018 Global 18% Protein diet, Envigo) and water. At 10-14 weeks of age, 10 male mice per group were sacrificed via CO2 asphyxiation at Zeitgeber Time (ZT) 0,2,6,10,12,14,18,22. In order to induce high levels of ROS in the liver, WT male mice were fasted 12 h and followed by intraperitoneal injection with 300 mg/kg APAP dissolved in PBS and re-fed. | |||
| Response Summary | PPaRalpha to mediate its mRNA stability to regulate lipid metabolism. Hepatic deletion of Bmal1 increases m6A mRNA methylation, particularly of Peroxisome proliferator-activated receptor alpha (PPARalpha/PPARA). Inhibition of m6A methylation via knockdown of m6A methyltransferase METTL3 decreases PPaR-Alpha m6A abundance and increases PPaRalpha mRNA lifetime and expression, reducing lipid accumulation in cells in vitro. YTHDF2 binds to PPaRalpha to mediate its mRNA stability to regulate lipid metabolism. Transcriptional regulation of circadian rhythms is essential for lipid metabolic homeostasis, disruptions of which can lead to metabolic diseases. | |||
Pescadillo homolog (PES1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
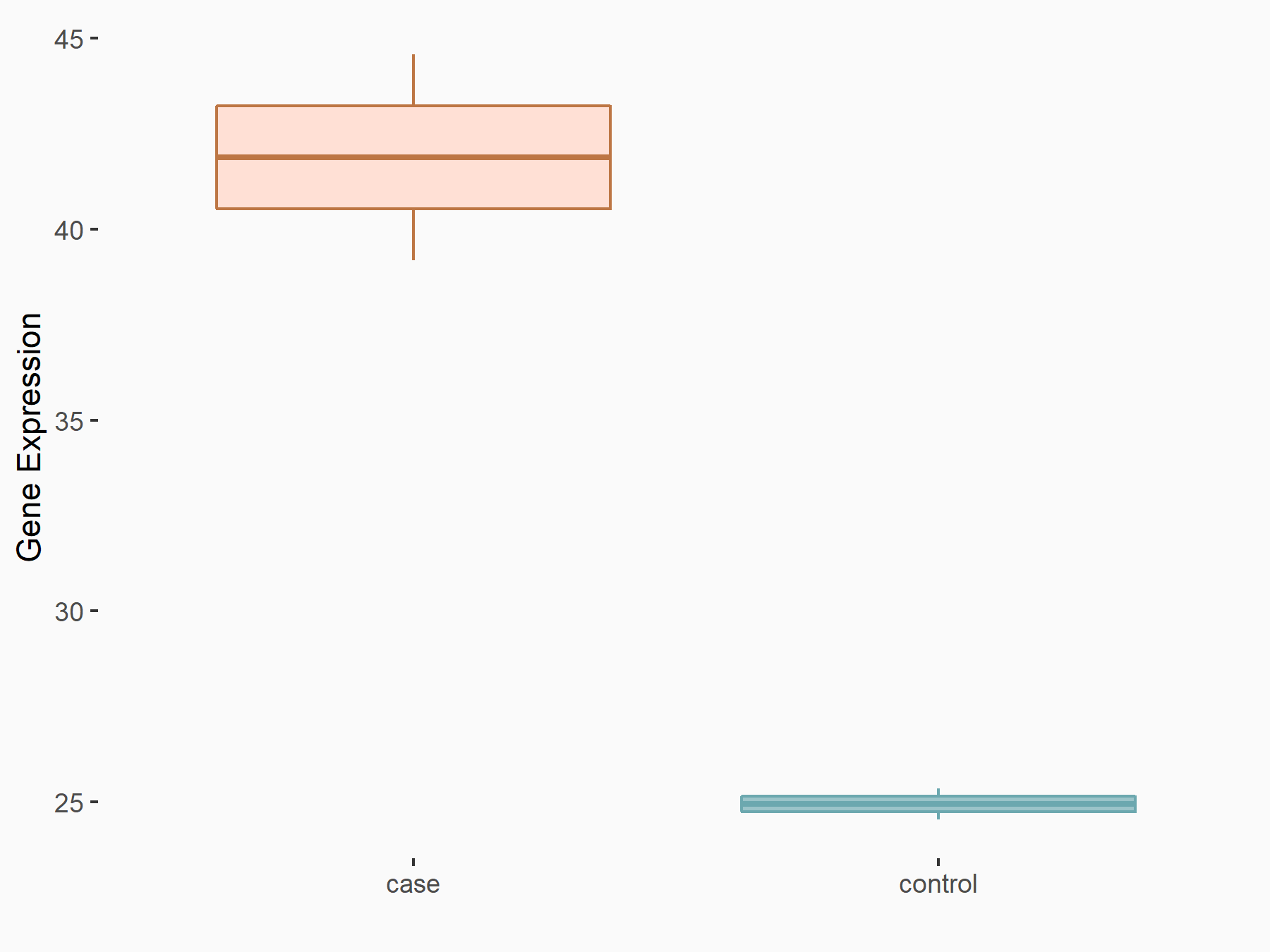 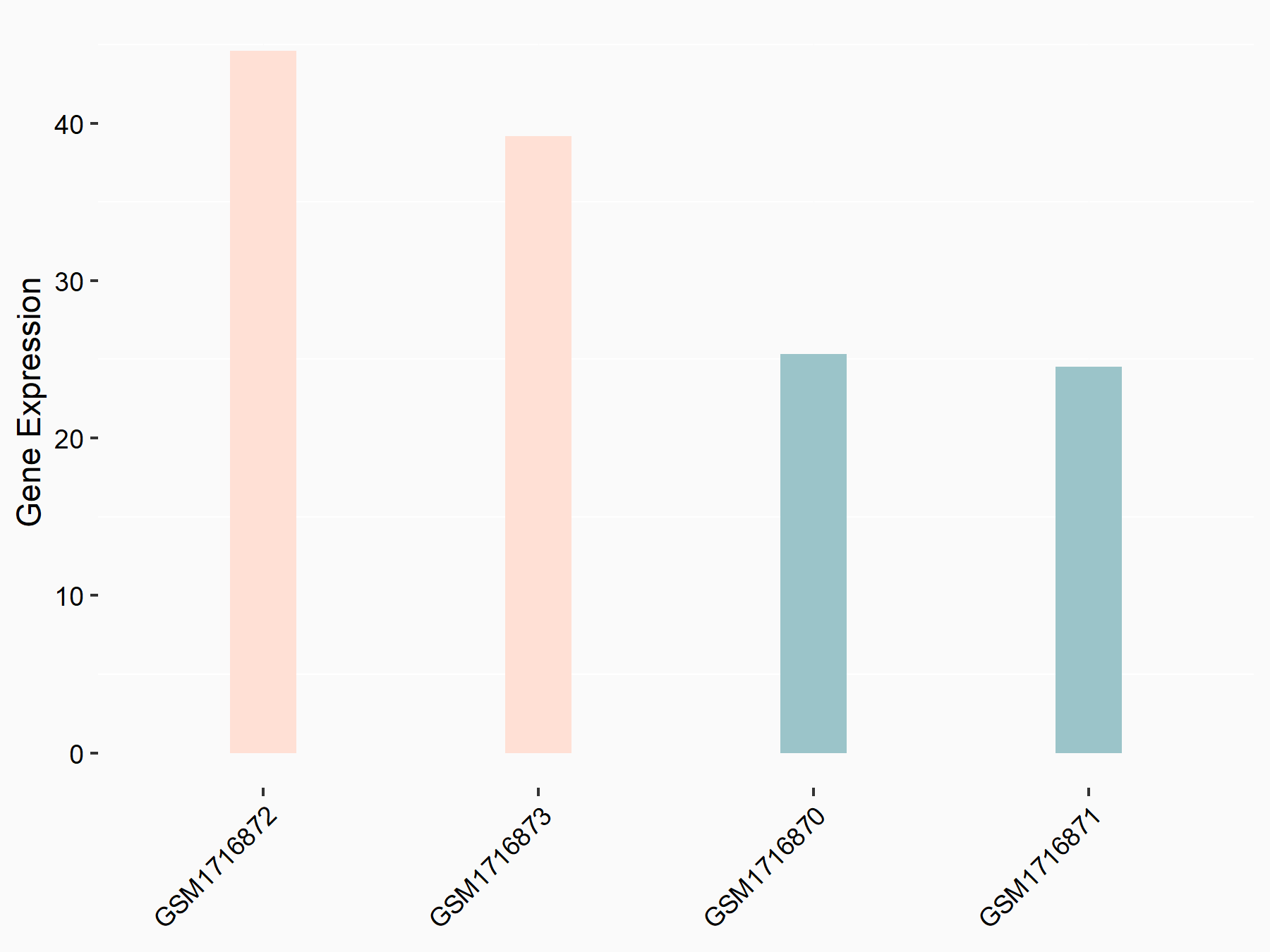 |
logFC: 7.23E-01 p-value: 7.36E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.38E+00 | GSE60213 |
Malignant haematopoietic neoplasm [ICD-11: 2B33]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [93] | |||
| Responsed Disease | Chronic myeloid leukaemia [ICD-11: 2B33.2] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Decrease of S phase | |||
In-vitro Model |
U-937 | Adult acute monocytic leukemia | Homo sapiens | CVCL_0007 |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| LAMA-84 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0388 | |
| KCL-22 | Chronic myelogenous leukemia | Homo sapiens | CVCL_2091 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 | |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| Response Summary | m6A methyltransferase complex METTL3/METTL14 is upregulated in CML patients and that is required for proliferation of primary CML cells and CML cell lines sensitive and resistant to the TKI imatinib. METTL3 directly regulates the level of Pescadillo homolog (PES1) protein identified as an oncogene in several tumors. These results point to METTL3 as a novel relevant oncogene in CML and as a promising therapeutic target for TKI resistant CML. | |||
PHLPP-like (PHLPP2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
|
Treatment: METTL3 knockdown MDA-MB-231 cells
Control: MDA-MB-231 cells
|
GSE70061 | |
| Regulation |
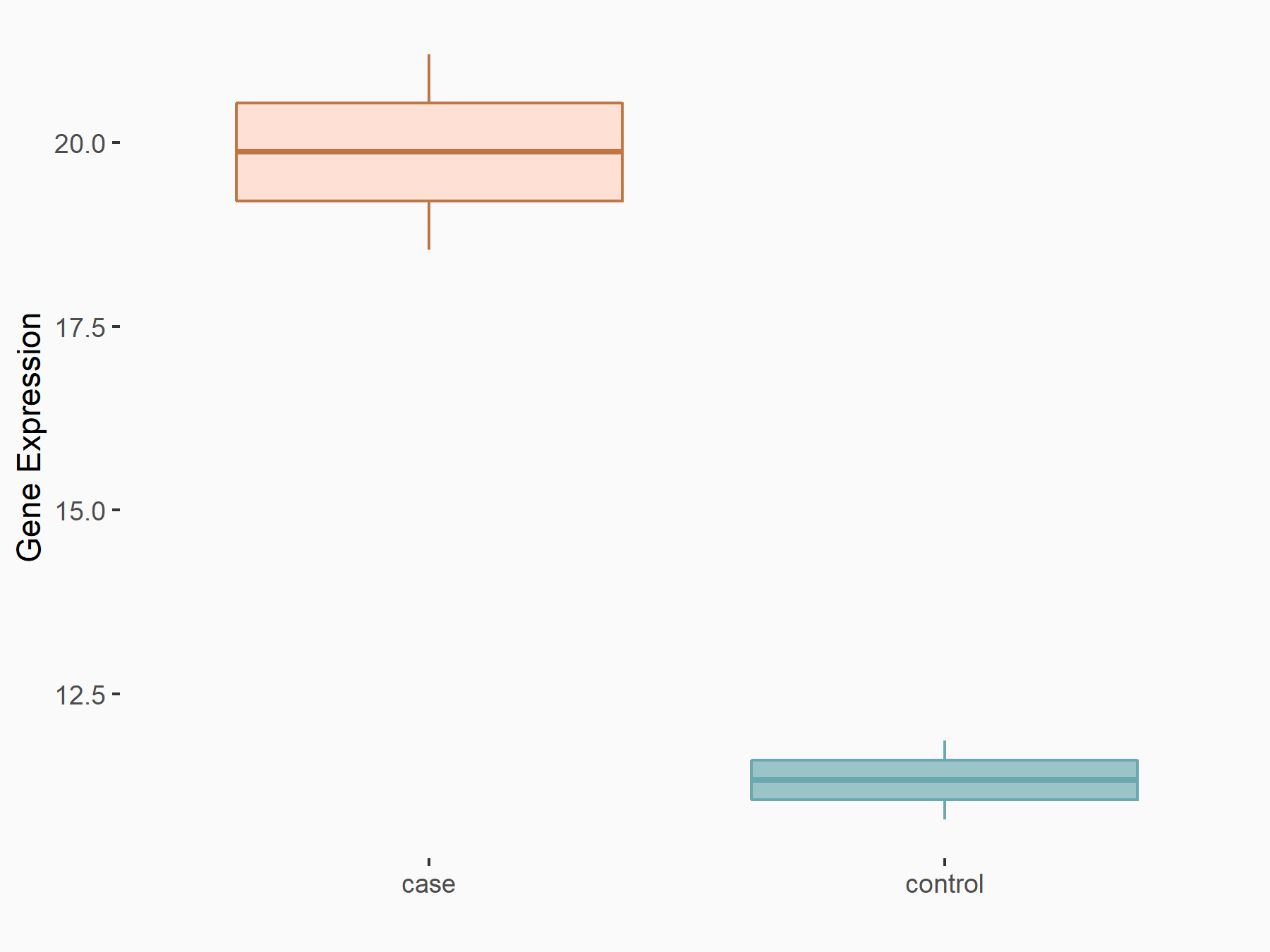 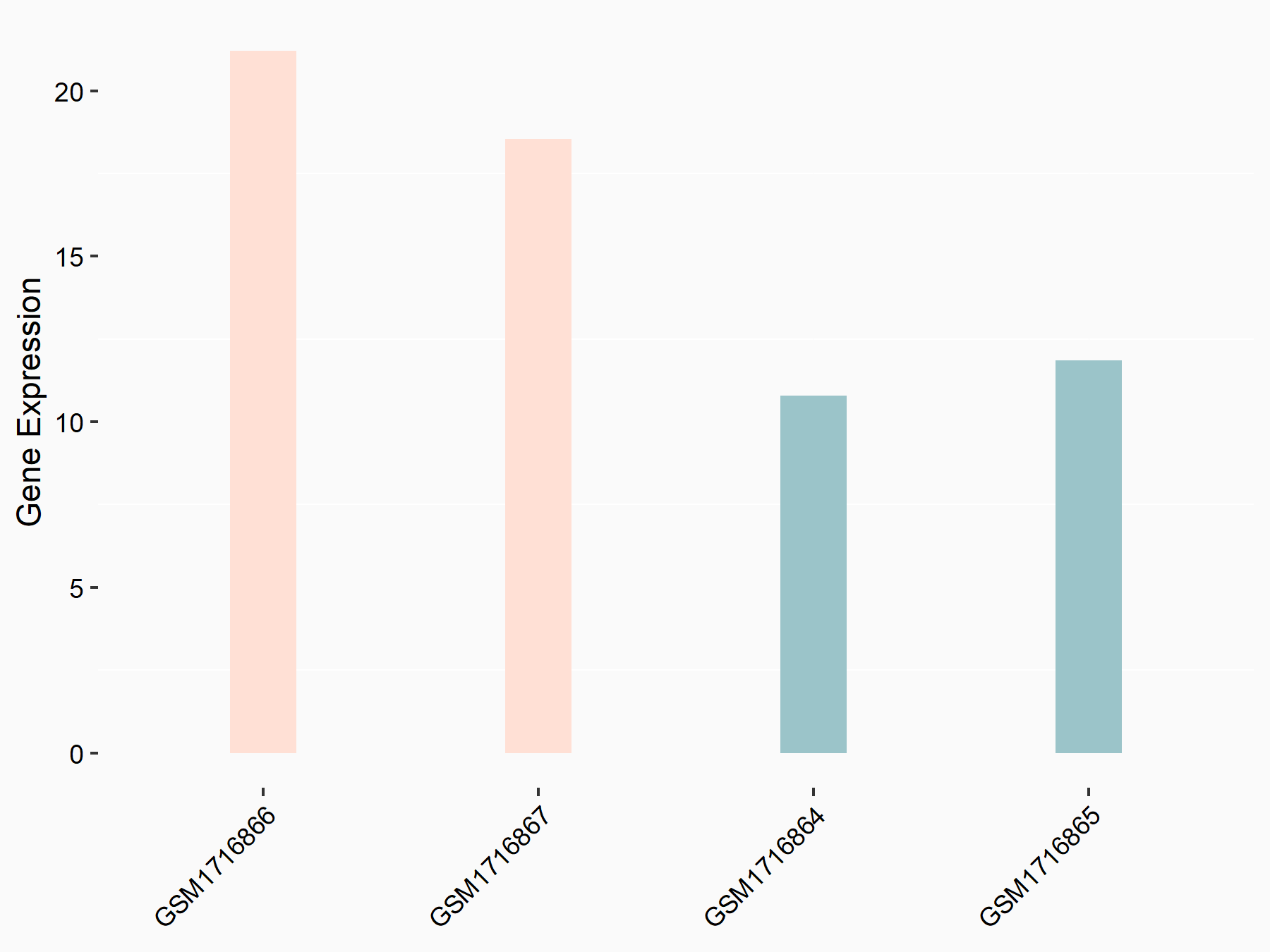 |
logFC: 7.59E-01 p-value: 9.18E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.40E+00 | GSE60213 |
Endometrial cancer [ICD-11: 2C76]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [94] | |||
| Responsed Disease | Endometrial cancer [ICD-11: 2C76] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell proliferation and tumorigenicity | |||
In-vitro Model |
HEC-1-A | Endometrial adenocarcinoma | Homo sapiens | CVCL_0293 |
| RL95-2 | Endometrial adenosquamous carcinoma | Homo sapiens | CVCL_0505 | |
| T HESCs | Normal | Homo sapiens | CVCL_C464 | |
| In-vivo Model | 4×106 HEC-1-A endometrial cancer cells (shCtrl, shMETTL3, wild-type, METTL14+/-, or METTL14+/- rescued with wild-type or mutant METTL14) were injected intraperitoneally into 5 week old female athymic nude mice (Foxn1nu, Harlan; n=10 per group). | |||
| Response Summary | About 70% of endometrial tumours exhibit reductions in m6A methylation that are probably due to either this METTL14 mutation or reduced expression of METTL3. Reductions in m6A methylation lead to decreased expression of the negative AKT regulator PHLPP-like (PHLPP2) and increased expression of the positive AKT regulator mTORC2. these results reveal reduced m6A mRNA methylation as an oncogenic mechanism in endometrial cancer and identify m6A methylation as a regulator of AKT signalling. | |||
Phosphatidylinositol 3-kinase regulatory subunit beta (PI3K-p85/PIK3R2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
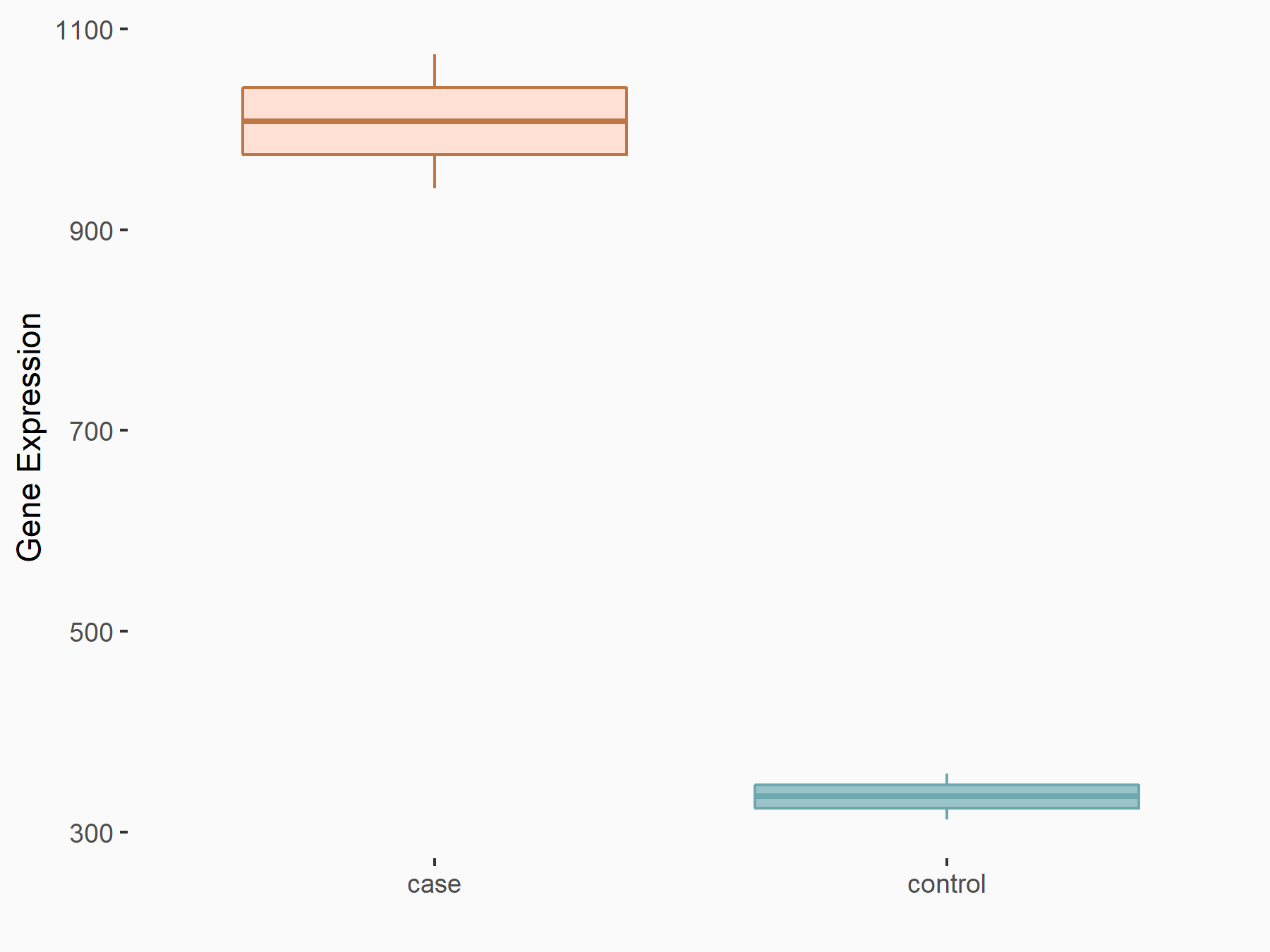 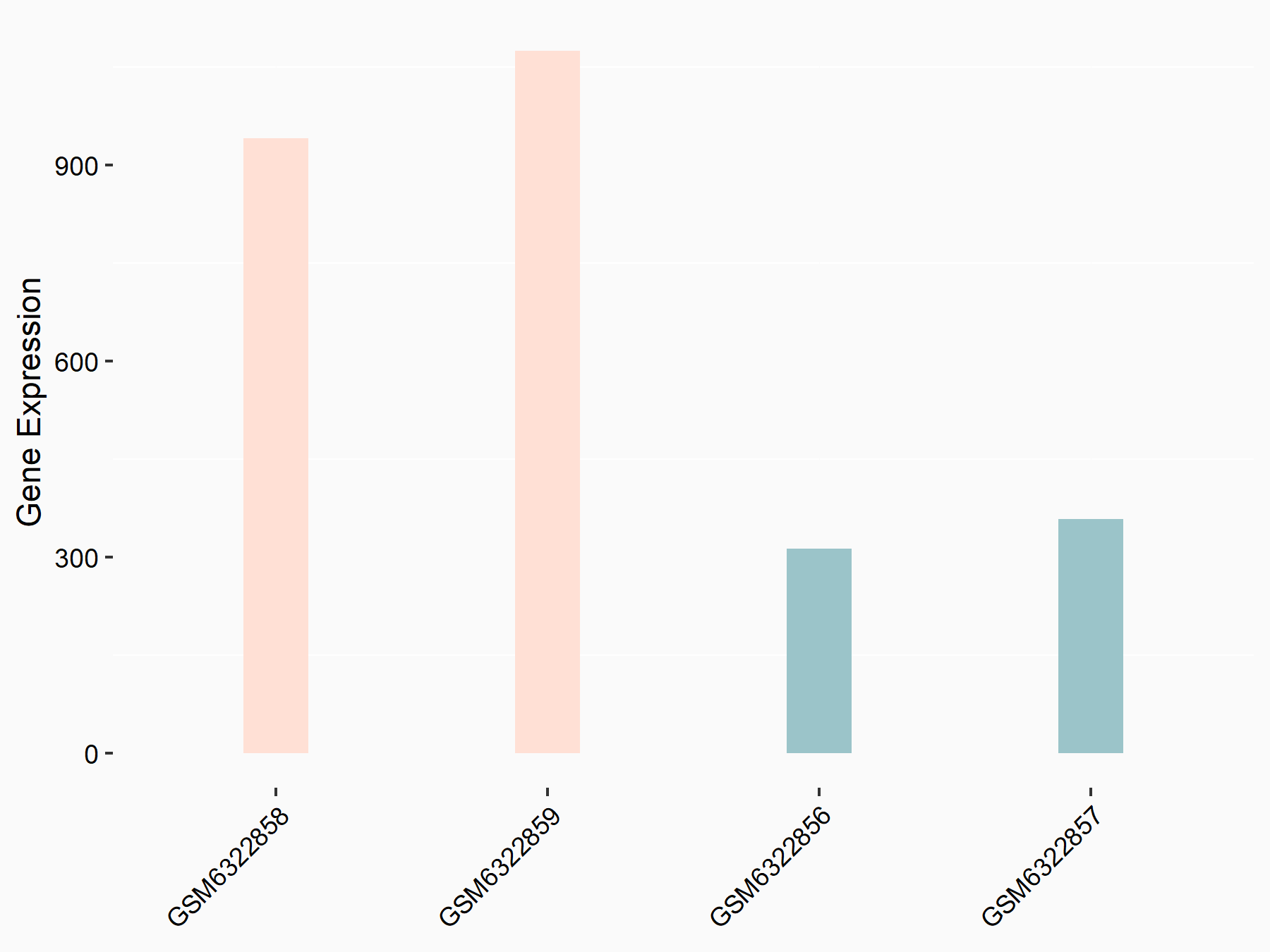 |
logFC: 1.59E+00 p-value: 3.69E-27 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.42E+00 | GSE60213 |
Retina cancer [ICD-11: 2D02]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [44] | |||
| Responsed Disease | Retinoblastoma [ICD-11: 2D02.2] | |||
| Responsed Drug | Rapamycin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
In-vitro Model |
WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through Phosphatidylinositol 3-kinase regulatory subunit beta (PI3K-p85/PIK3R2)/AKT/mTOR pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-P70S6K/4EBP1 pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
Phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
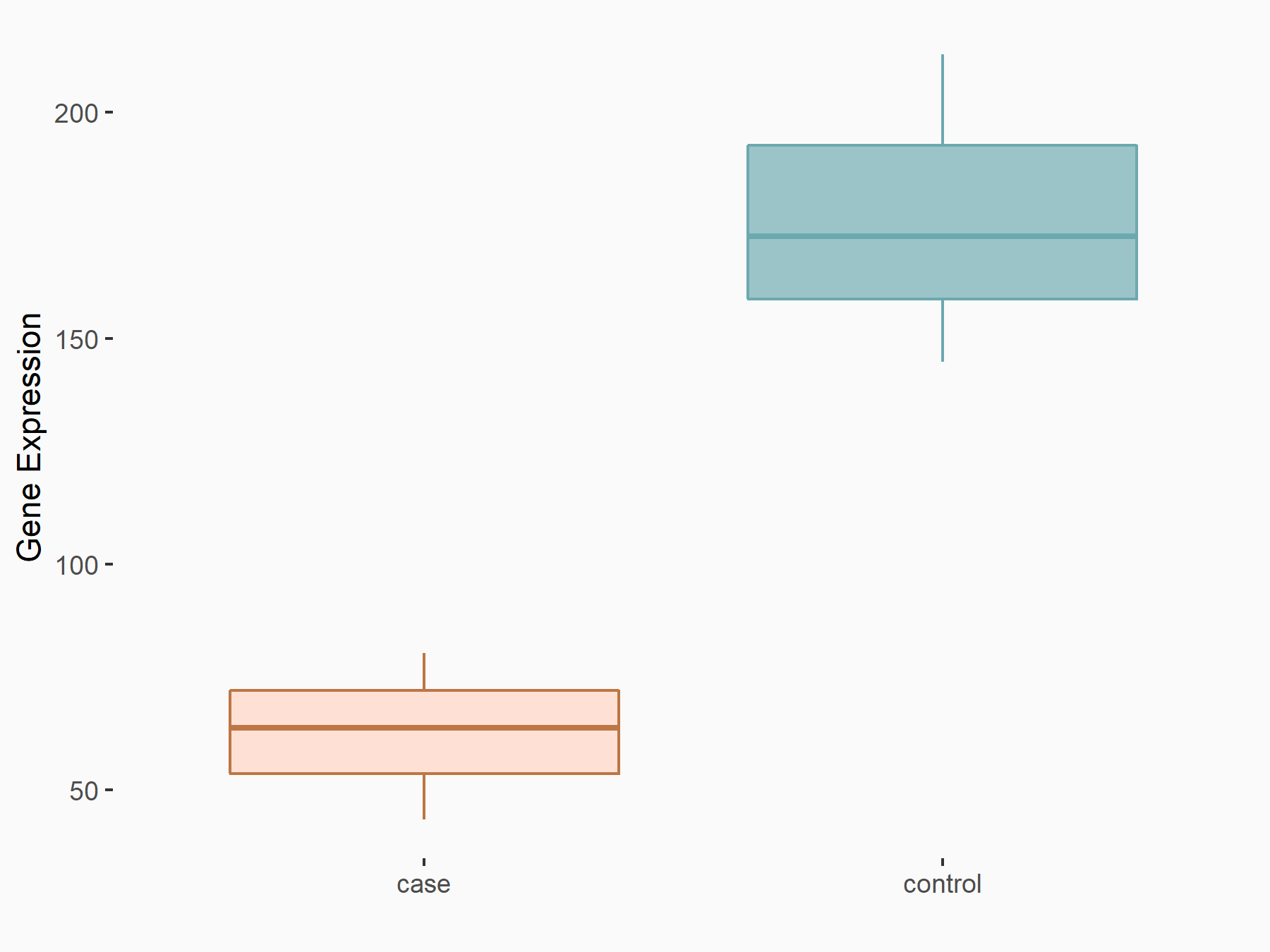 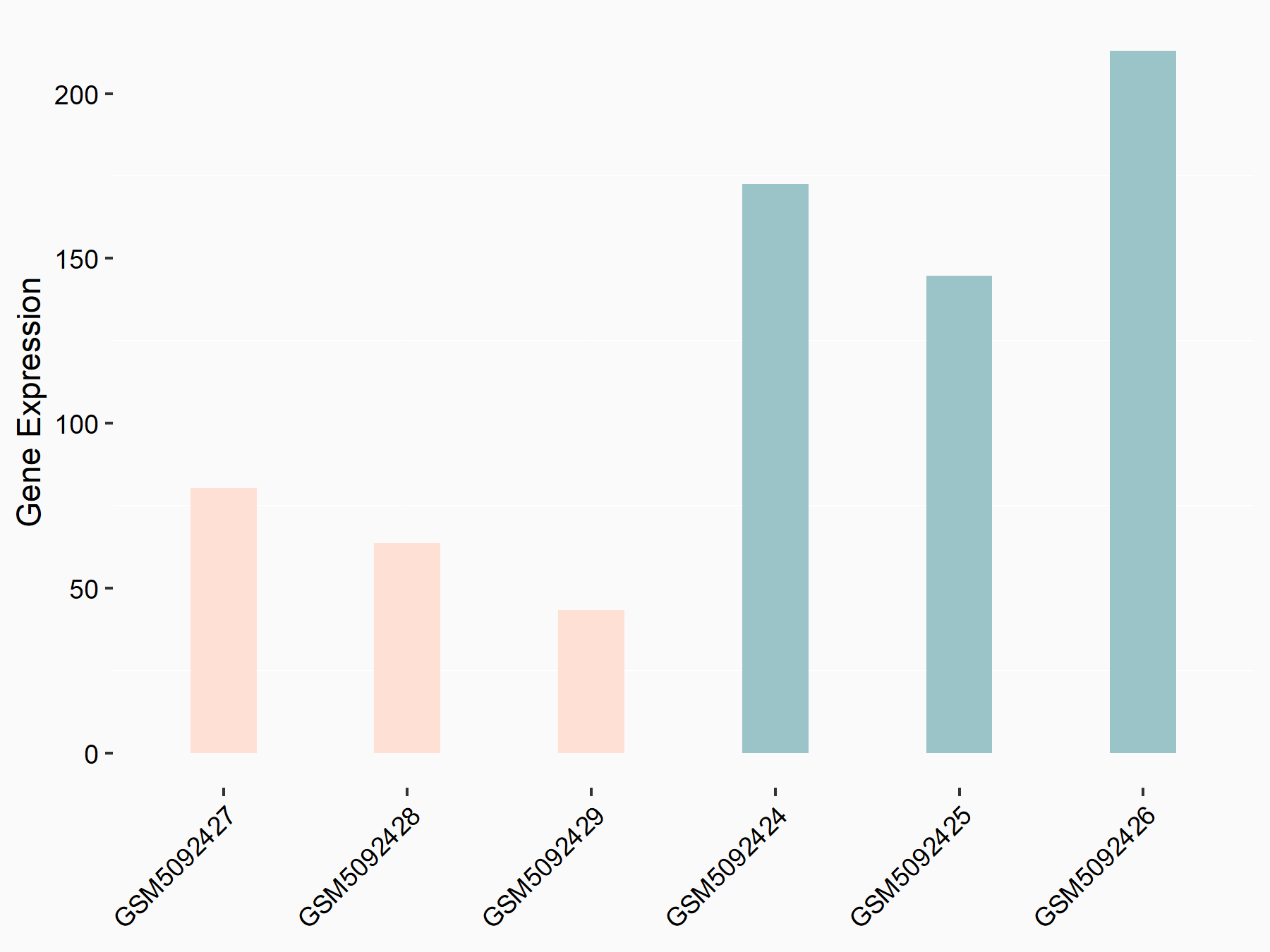 |
logFC: -1.51E+00 p-value: 1.12E-09 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 8.41E+00 | GSE60213 |
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [62] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Oxidative phosphorylation | hsa00190 | ||
In-vitro Model |
VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| In-vivo Model | Approximately 2 × 106 PCa cells (PC-3 shNC, shYTHDF2, shMETTL3 cell lines) per mouse suspended in 100 uL PBS were injected in the flank of male BALB/c nude mice (4 weeks old). During the 40-day observation, the tumor size (V = (width2×length ×0.52)) was measured with vernier caliper. Approximately 1.5 × 106 PCa cells suspended in 100 uL of PBS (PC-3 shNC, shYTHDF2, and shMETTL3 cell lines) per mouse were injected into the tail vein of male BALB/c nude mice (4 weeks old). The IVIS Spectrum animal imaging system (PerkinElmer) was used to evaluate the tumor growth (40 days) and whole metastasis conditions (4 weeks and 6 weeks) with 100 uL XenoLight D-luciferin Potassium Salt (15 mg/ml, Perkin Elmer) per mouse. Mice were anesthetized and then sacrificed for tumors and metastases which were sent for further organ-localized imaging as above, IHC staining and hematoxylin-eosin (H&E) staining. | |||
| Response Summary | Knock-down of YTHDF2 or METTL3 significantly induced the expression of Phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP) and NKX3-1 at both mRNA and protein level with inhibited phosphorylated AKT. YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent way to regulate AKT phosphorylation-induced tumor progression in prostate cancer. | |||
Pigment epithelium-derived factor (PEDF)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
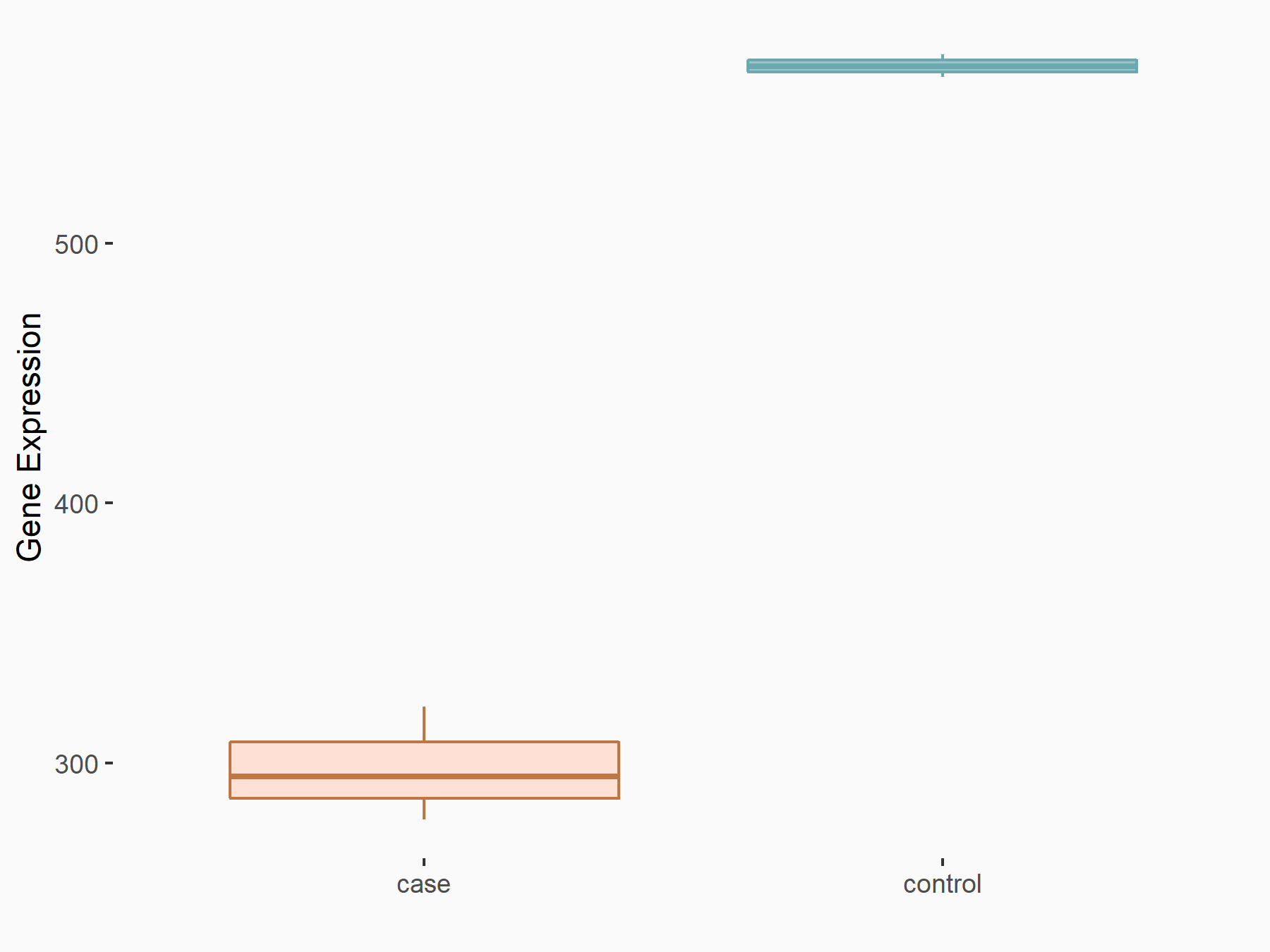 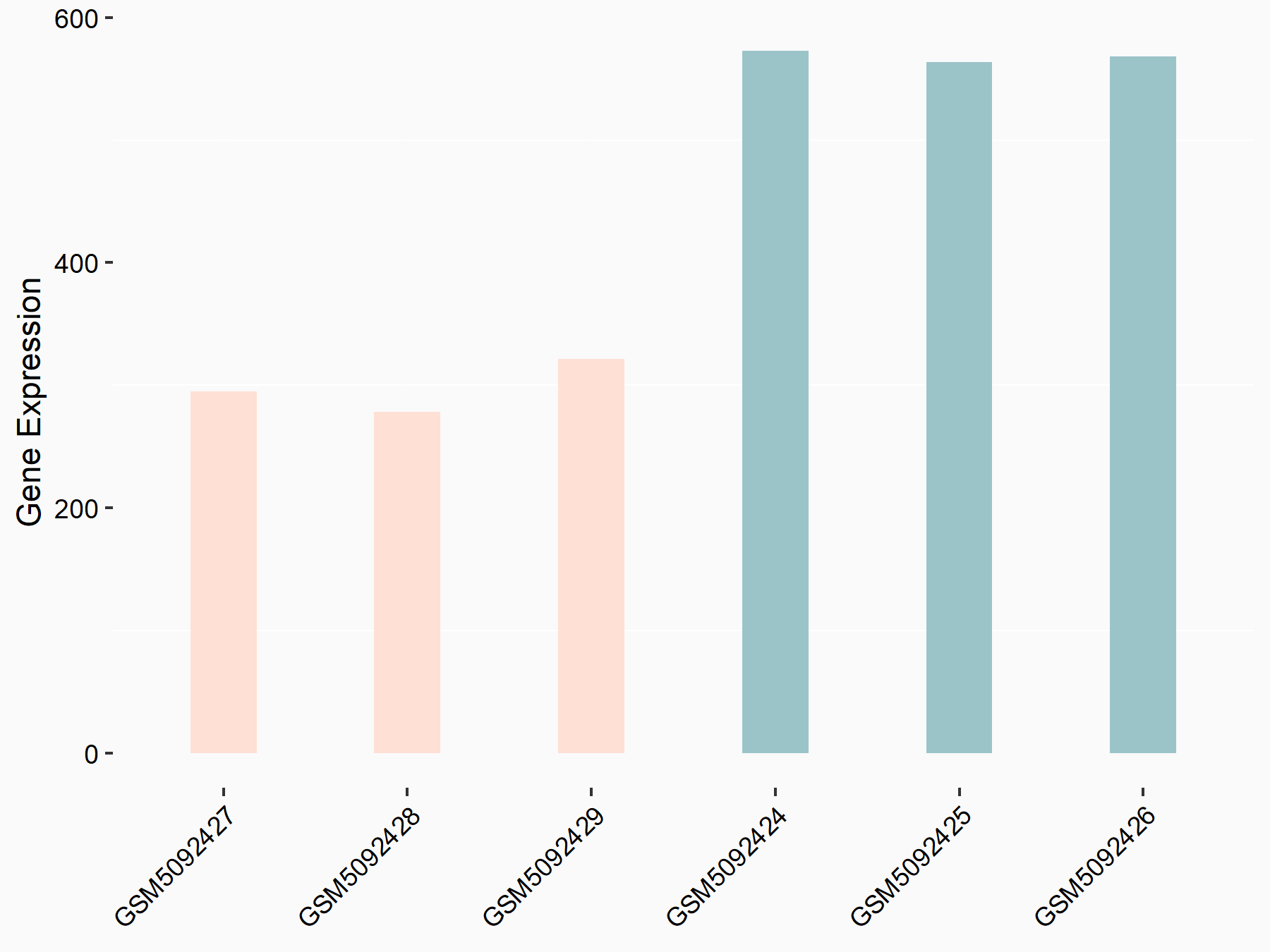 |
logFC: -9.30E-01 p-value: 1.13E-16 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 8.75E+00 | GSE60213 |
Diffuse large B-cell lymphomas [ICD-11: 2A81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [95] | |||
| Responsed Disease | Diffuse large B-cell lymphomas [ICD-11: 2A81] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
U-2932 | Diffuse large B-cell lymphoma | Homo sapiens | CVCL_1896 |
| SU-DHL-4 | Diffuse large B-cell lymphoma | Homo sapiens | CVCL_0539 | |
| OCI-Ly10 | Diffuse large B-cell lymphoma | Homo sapiens | CVCL_8795 | |
| HBL-1 [Human diffuse large B-cell lymphoma] | Diffuse large B-cell lymphoma | Homo sapiens | CVCL_4213 | |
| GM12878 | Human B lymphocytes | Homo sapiens | CVCL_7526 | |
| Farage | Diffuse large B-cell lymphoma | Homo sapiens | CVCL_3302 | |
| Response Summary | Overexpressed Pigment epithelium-derived factor (PEDF) abrogates the inhibition of cell proliferation in DLBCL cells that is caused by METTL3 silence. | |||
Platelet-derived growth factor receptor alpha (PDGFRA)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
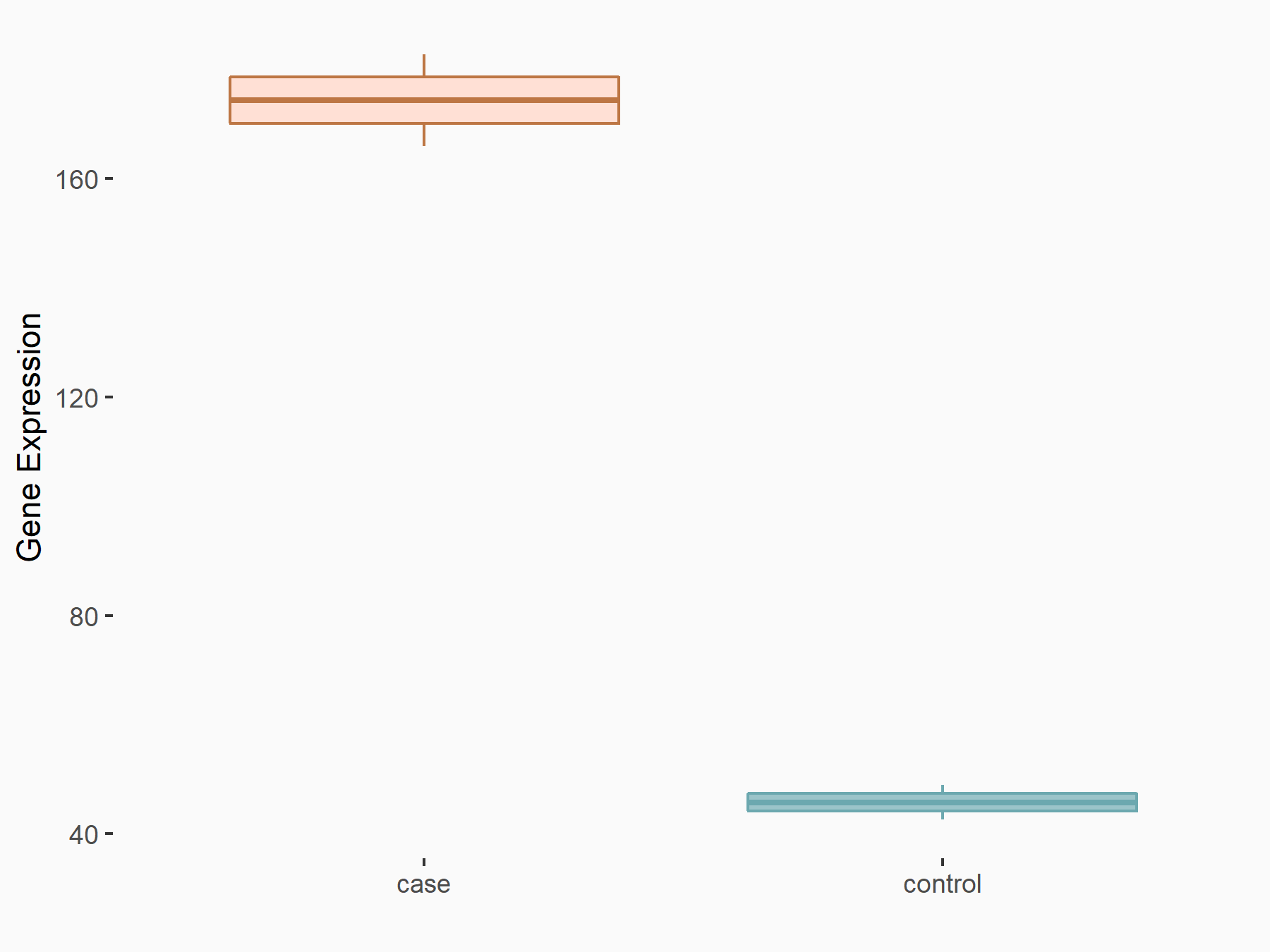 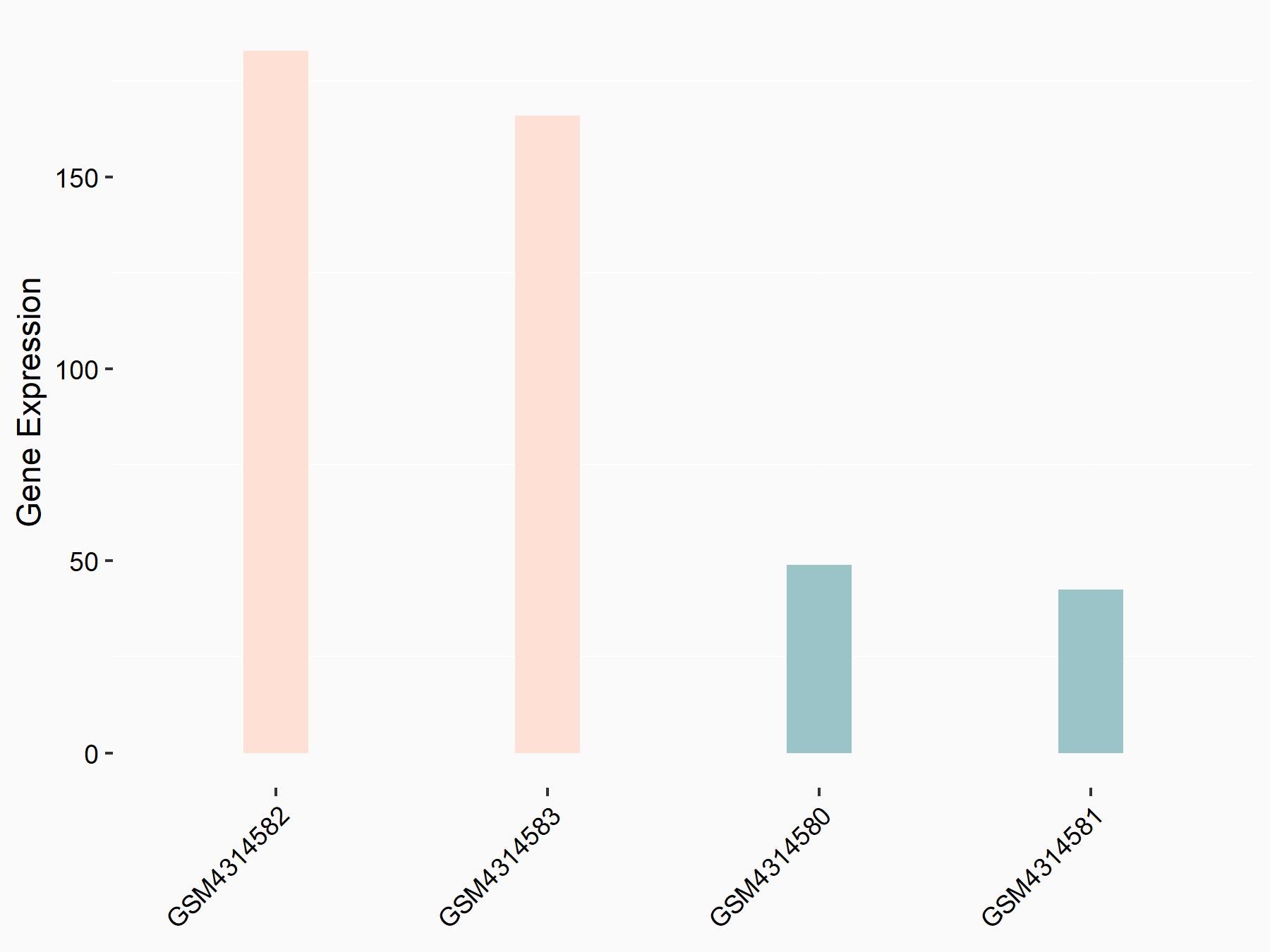 |
logFC: 1.93E+00 p-value: 8.84E-11 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 4.83E+00 | GSE60213 |
Diseases of arteries or arterioles [ICD-11: BD5Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [96] | |||
| Responsed Disease | Diseases of arteries or arterioles [ICD-11: BD5Y] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
ACBRI-183 (Human retinal pericytes (ACBRI-183) was obtained from Cell Systems Corp. (CSC, USA)) | |||
| In-vivo Model | Mettl3 floxed mice were purchased from GemPharmatech Co. Ltd (Nanjing, China). Pdgfr-Beta-Cre mice were purchased from Beijing Biocytogen Co. Ltd (Beijing, China) generated on C57BL/6J background. Mettl3 flox/flox mice were crossed with Pdgfr-Beta-Cre mice to generate pericyte-specific Mettl3 knockout mice. All mice were bred under the specific-pathogen free condition with free access to diet and water or their nursing mothers with alternating 12/12 light-dark cycle (lights on at 08:00 and off at 20:00). | |||
| Response Summary | Specific depletion of METTL3 in pericytes suppressed diabetes-induced pericyte dysfunction and Microvascular complication in vivo. METTL3 overexpression impaired pericyte function by repressing PKC-Eta, FAT4, and Platelet-derived growth factor receptor alpha (PDGFRA) expression, which was mediated by YTHDF2-dependent mRNA decay. | |||
Poly [ADP-ribose] polymerase tankyrase-1 (Tankyrase)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Pancreatic islets | Mus musculus |
|
Treatment: Mettl3 knockout mice
Control: Mettl3 flox/flox mice
|
GSE155612 | |
| Regulation |
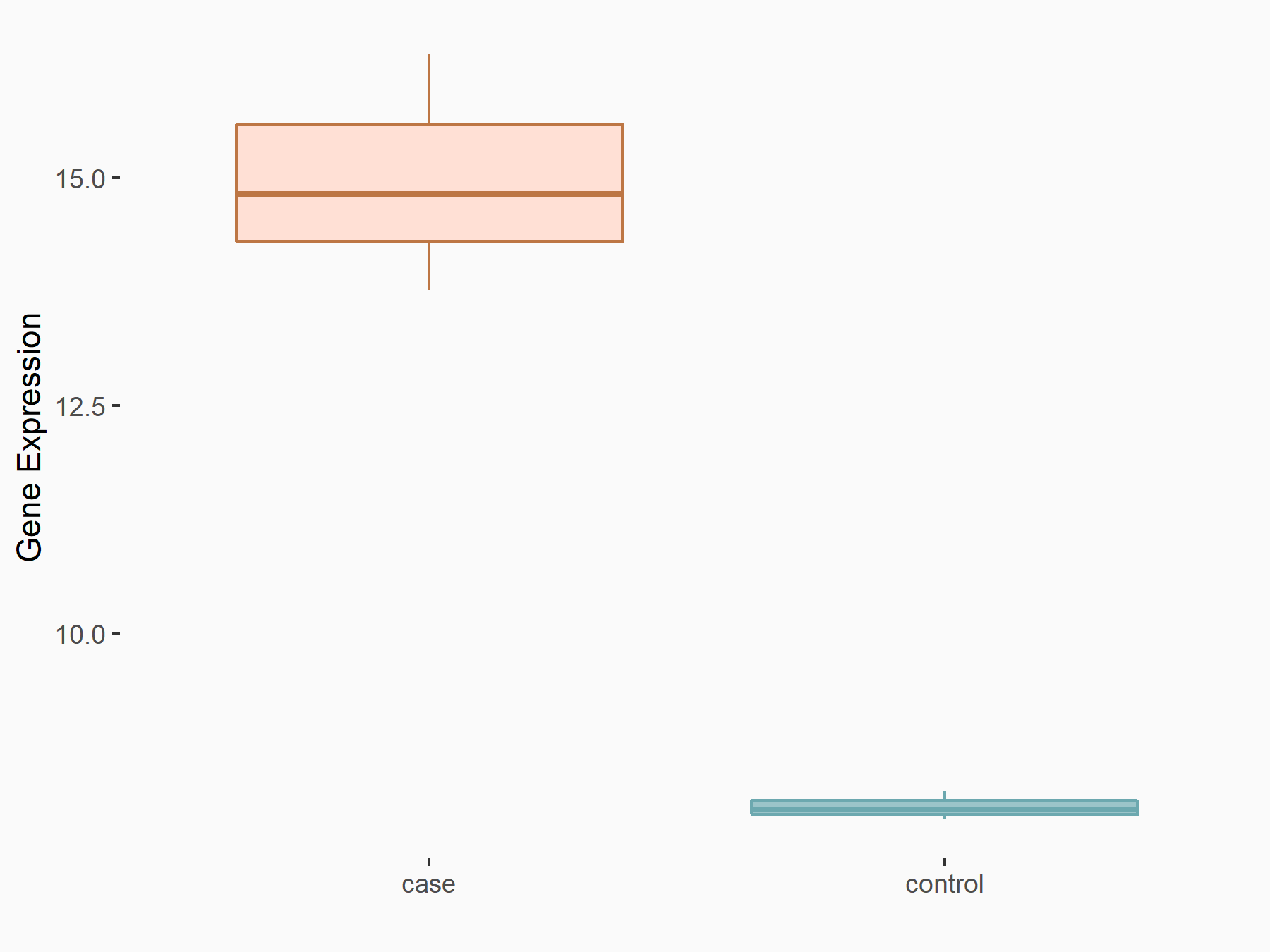 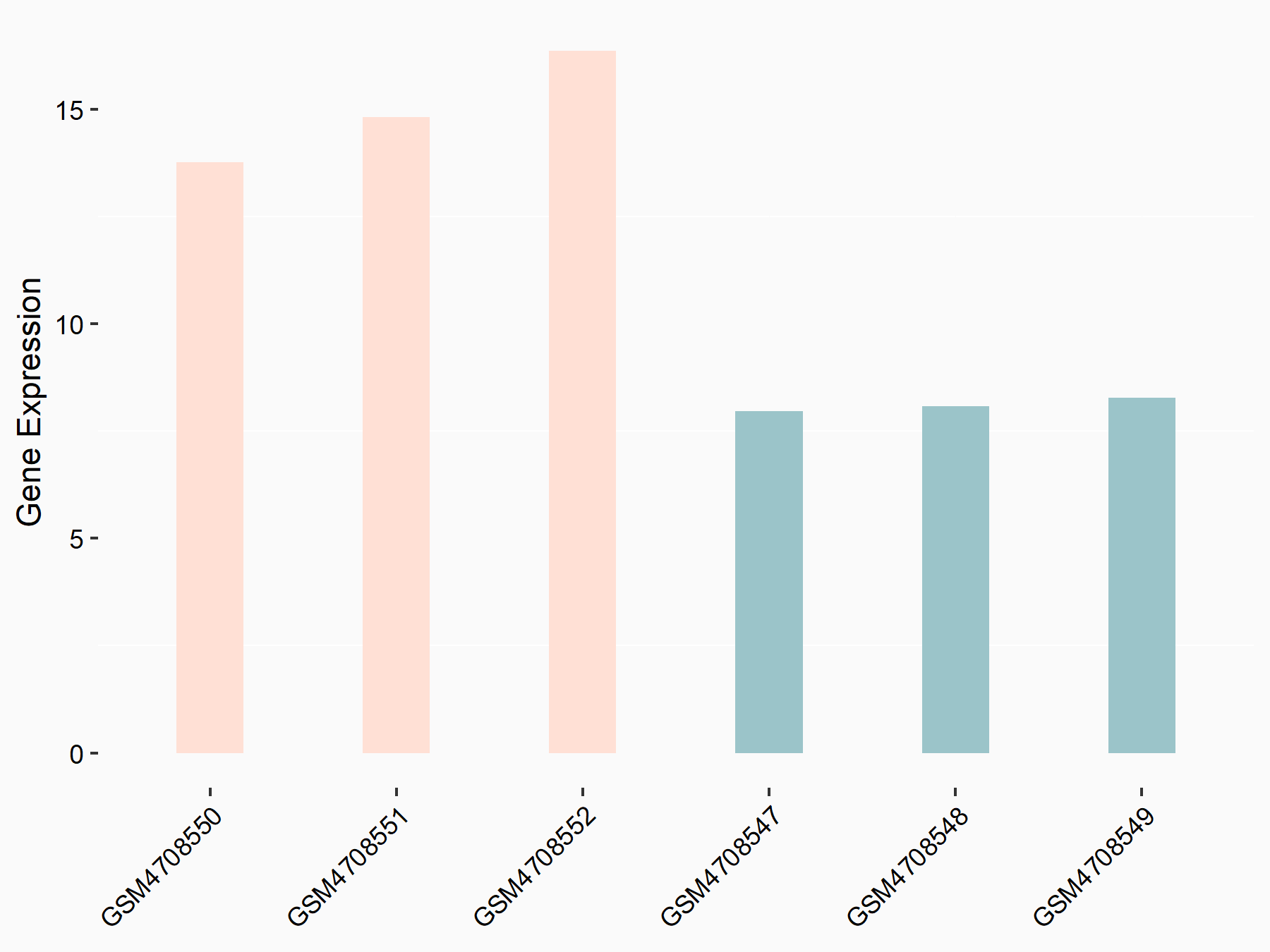 |
logFC: 8.10E-01 p-value: 1.68E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.44E+00 | GSE60213 |
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [24] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
Neural progenitor cells (NPCs) (The progenitor cells of the CNS) | |||
| NP69 (A human immortalized nasopharyngeal epithelial) | ||||
| HNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_FA07 | |
| HNE-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_0308 | |
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| CNE-1 | Normal | Homo sapiens | CVCL_6888 | |
| In-vivo Model | 1 × 105 HNE2 cells (with or without METTL3 knockdown) were labeled with luciferase gene and injected into the tail vein of the nude mice. | |||
| Response Summary | METTL3 activated the luciferase activity of TOPflash (a reporter for beta-catenin/TCF signaling), and downregulation of METTL3 inhibited the expression of beta-catenin/TCF target genes vimentin and N-cadherin, which are two regulators of epithelial-mesenchymal transition. METTL3 silencing decreased the m6A methylation and total mRNA levels of Poly [ADP-ribose] polymerase tankyrase-1 (Tankyrase), a negative regulator of axin. METTL3 is a therapeutic target for NPC. | |||
Potassium voltage-gated channel subfamily H member 6 (KCNH6)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Embryonic stem cells | Mus musculus |
|
Treatment: METTL3 knockout mESCs
Control: Wild type mESCs
|
GSE156481 | |
| Regulation |
 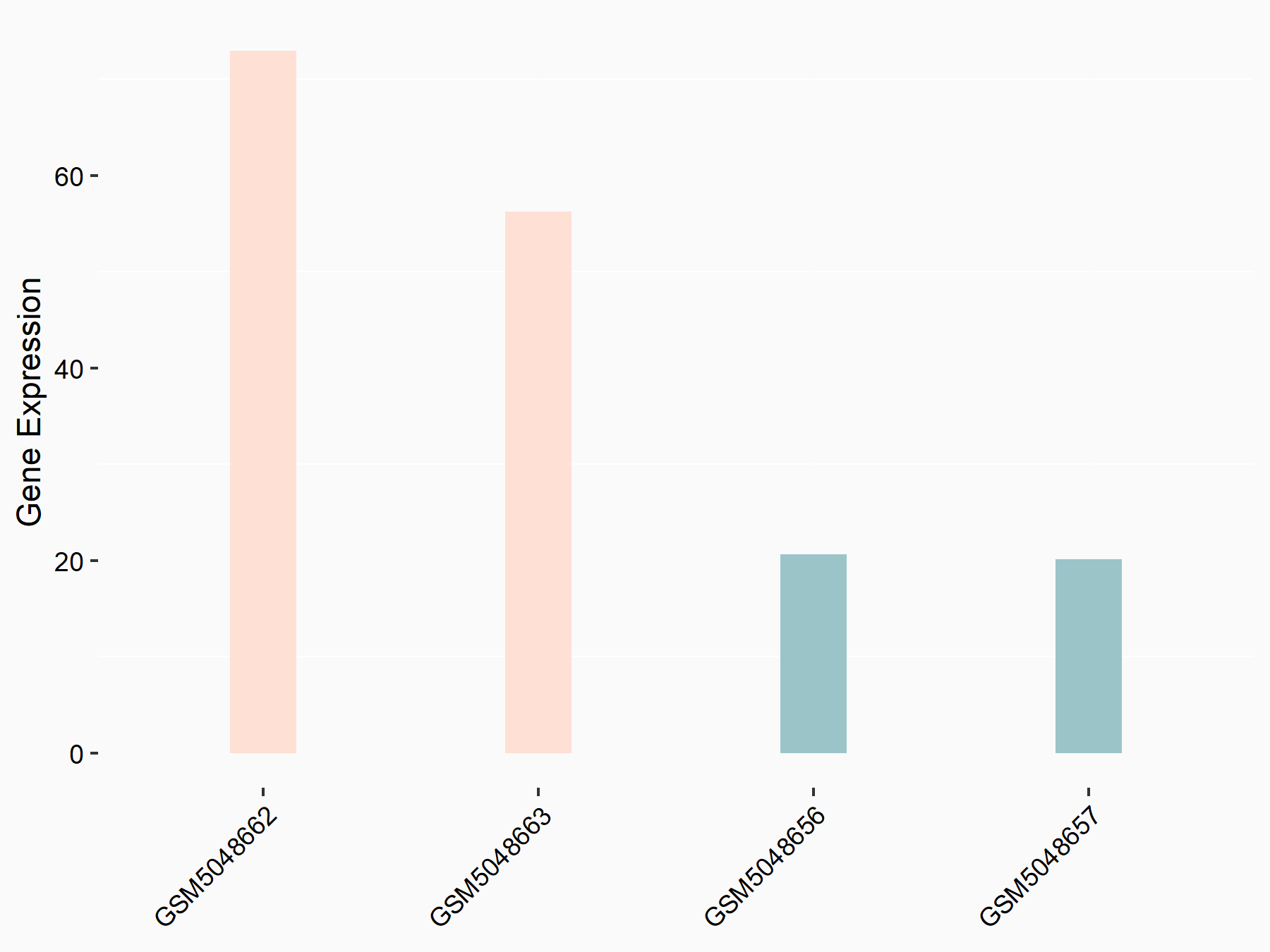 |
logFC: 1.67E+00 p-value: 2.01E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 4.49E+00 | GSE60213 |
Idiopathic interstitial pneumonitis [ICD-11: CB03]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [97] | |||
| Responsed Disease | Idiopathic pulmonary fibrosis [ICD-11: CB03.4] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
WI-38 | Normal | Homo sapiens | CVCL_0579 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | Animals were bred and housed in the pathogen-free facility of the Laboratory Animal Center of Shanghai General Hospital (Shanghai, China). All lungs were collected 4 weeks after BLM treatment for histology and further study. Lung microsections (5 uM) were applied to Masson's trichrome and Sirius red staining to visualize fibrotic lesions. | |||
| Response Summary | Lowering m6A levels through silencing METTL3 suppresses the FMT process in vitro and in vivo. m6A modification regulates EMT by modulating the translation of Potassium voltage-gated channel subfamily H member 6 (KCNH6) mRNA in a YTHDF1-dependent manner. Manipulation of m6A modification through targeting METTL3 becomes a promising strategy for the treatment of idiopathic pulmonary fibrosis. | |||
Pro-epidermal growth factor (EGF)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
 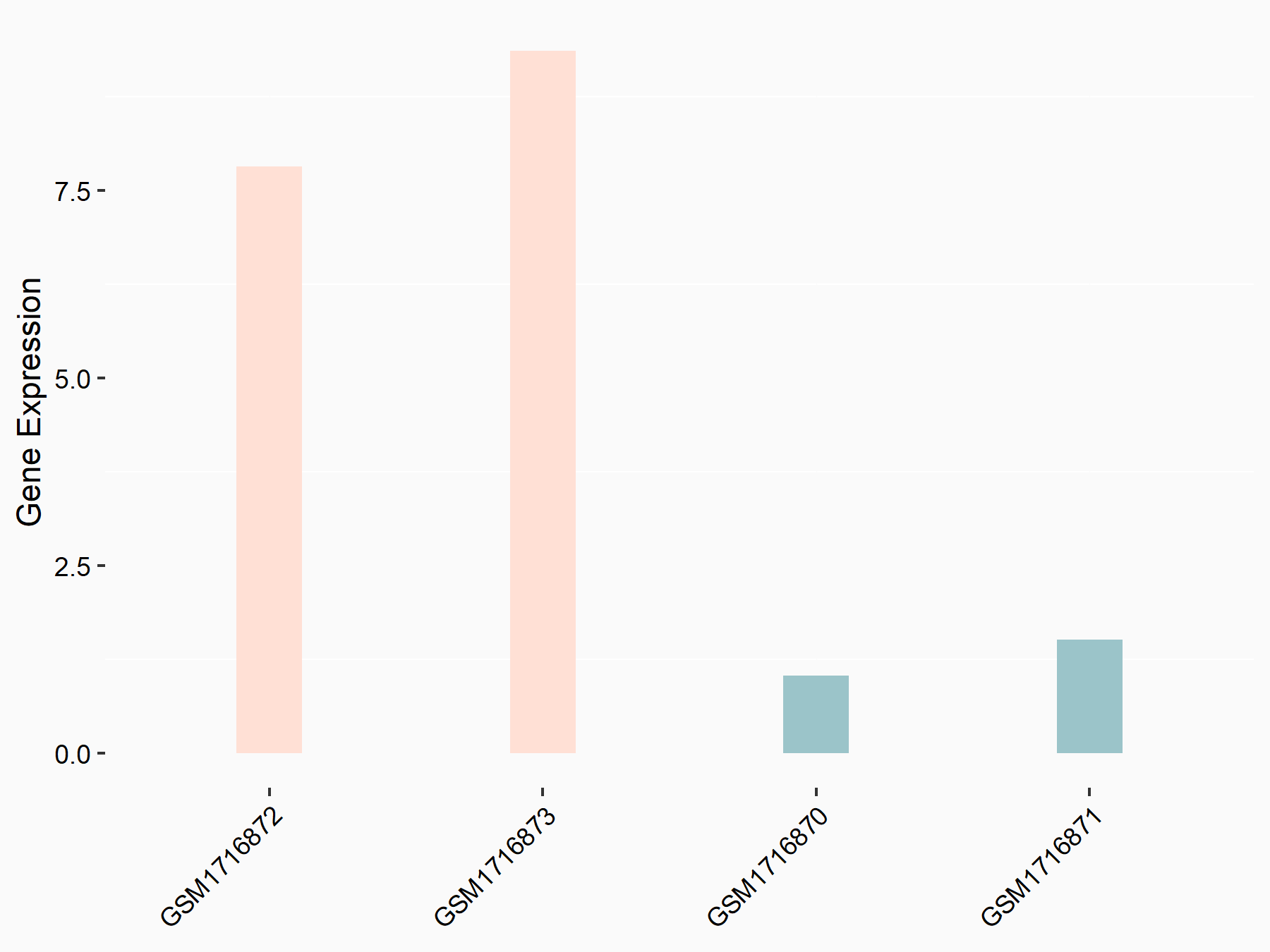 |
logFC: 2.08E+00 p-value: 3.08E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.98E+00 | GSE60213 |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [98] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Doxil | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Homologous recombination | hsa03440 | ||
| Cell Process | Homologous recombination repair | |||
In-vitro Model |
MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | Cells were trypsinized and resuspended in DMEM at a consistence of 1 × 107 cells/ml. A total of 1 × 106 cells were injected into flank of mice. 27 days after injection, tumors were removed for paraffin-embedded sections. | |||
| Response Summary | Knockdown of METTL3 sensitized these breast cancer cells to Adriamycin (ADR; also named as doxorubicin) treatment and increased accumulation of DNA damage. Mechanically, we demonstrated that inhibition of METTL3 impaired HR efficiency and increased ADR-induced DNA damage by regulating m6A modification of Pro-epidermal growth factor (EGF)/RAD51 axis. METTL3 promoted EGF expression through m6A modification, which further upregulated RAD51 expression, resulting in enhanced HR activity. | |||
Programmed cell death 1 ligand 1 (CD274/PD-L1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 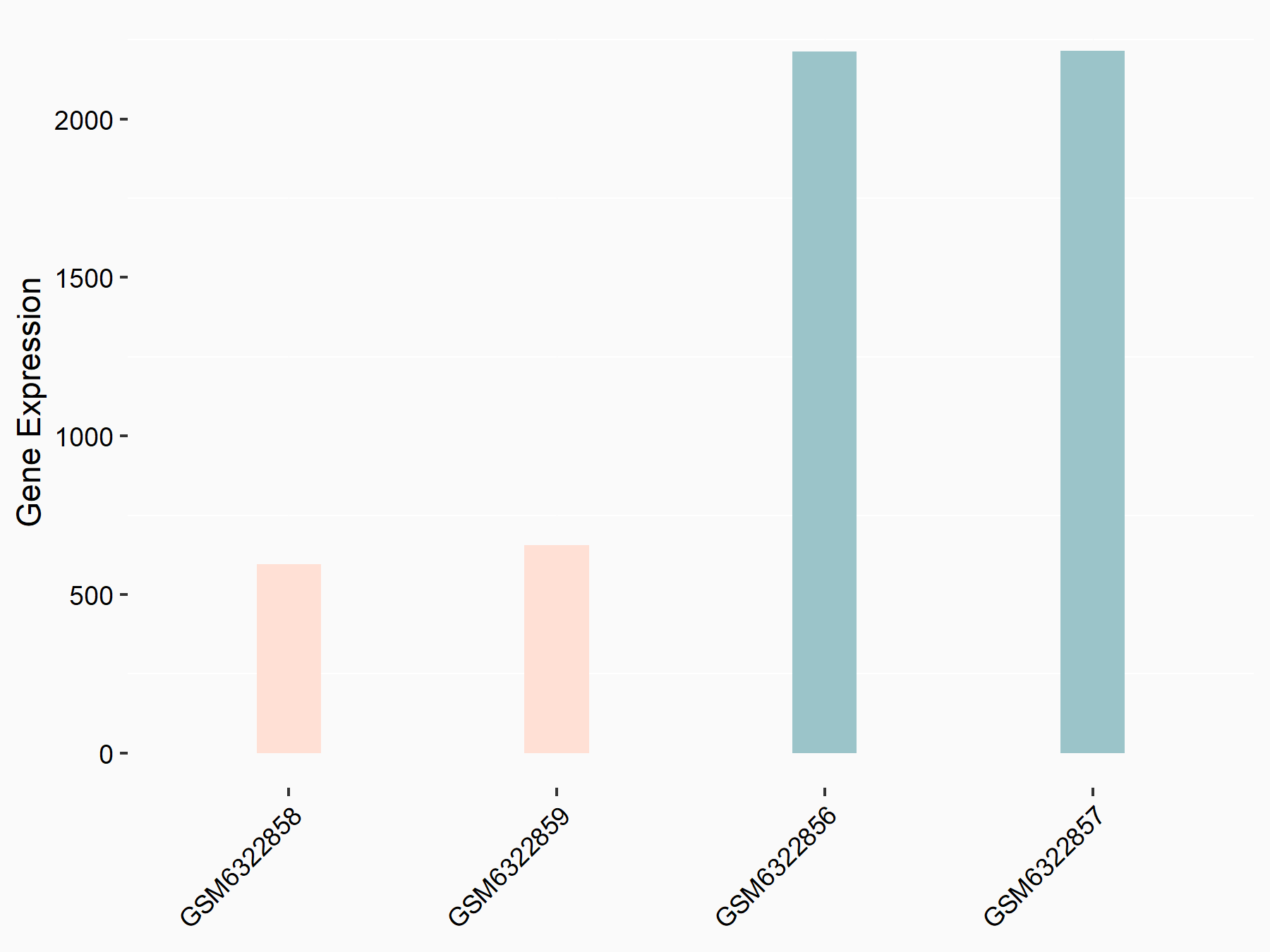 |
logFC: -1.82E+00 p-value: 4.68E-65 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.97E+00 | GSE60213 |
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [99] | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
In-vitro Model |
SCC-9 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1685 |
| SCC-4 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1684 | |
| SCC-25 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1682 | |
| CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 | |
| In-vivo Model | Six-week-old nude mice were randomly divided into two groups (three mice per group) and cultured with continuous access to sterile food and water in pathogen-free sterile conditions. To establish the OSCC xenograft model, we subcutaneously injected 5 × 106 SCC-9 cells stably transfected with METTL3 shRNA or sh-NC vectors into nude mice. | |||
| Response Summary | METTL3 intensified the metastasis and proliferation of OSCC by modulating the m6A amounts of PRMT5 and Programmed cell death 1 ligand 1 (CD274/PD-L1). | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [80] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | |||
| Response Summary | This study revealed that m6A methylation is closely related to the poor prognosis of non-small cell lung cancer patients via interference with the TIME, which suggests that m6A plays a role in optimizing individualized immunotherapy management and improving prognosis. The expression levels of METTL3, FTO and YTHDF1 in non-small cell lung cancer were changed. Patients in Cluster 1 had lower immunoscores, higher Programmed cell death 1 ligand 1 (CD274/PD-L1) expression, and shorter overall survival compared to patients in Cluster 2. The hallmarks of the Myelocytomatosis viral oncogene (MYC) targets, E2 transcription Factor (E2F) targets were significantly enriched. | |||
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [100] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
| Cell Process | Tumor immune escape | |||
In-vitro Model |
SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HCC38 | Breast ductal carcinoma | Homo sapiens | CVCL_1267 | |
| 4T1 | Normal | Mus musculus | CVCL_0125 | |
| In-vivo Model | For subcutaneous xenograft experiments in B-NDG mice, approximately 1 × 106 MDA-MB-231 and there was subcutaneous injection of the cells that resuspended in 100 uL PBS into the left flank of the mice and were divided into 11 groups randomly (each containing 5 mice). After the treatment Atezolizumab (Selleck, Shanghai, China) or corresponding iso control antibody (Selleck, Shanghai, China) was injected intratumorally on day 3, 6, 9, 12, 15 post-MDA-MB-231 inoculations, and 5 × 106 cytokine-induced killer (CIK) cells were injected in the tail vein on day 7, 14, 21. | |||
| Response Summary | Programmed cell death 1 ligand 1 (CD274/PD-L1) was a downstream target of METTL3-mediated m6A modification in breast cancer cells. METTL3-mediated PD-L1 mRNA activation was m6A-IGF2BP3-dependent. PD-L1 expression was also positively correlated with METTL3 and IGF2BP3 expression in breast cancer tissues. | |||
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [101] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
In-vitro Model |
UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| J82 | Bladder carcinoma | Homo sapiens | CVCL_0359 | |
| 5637 | Bladder carcinoma | Homo sapiens | CVCL_0126 | |
| In-vivo Model | Male C57BL/6J mice (6 weeks old) were given drinking water containing 0.05% (w/v) BBN (TCI, catalog no. B0938) for 20 weeks. After the BBN administration, mice were given normal drinking water and injected with 10% DMSO (as a control) or 20 mg/kg SP600125 (Selleck, catalog no. 129-56-6) i.p. every 3 days. After seven injections, mice were euthanized for tissue retrieval. For the bladder cancer cell-derived xenograft mouse model, male C57BL/6J mice (6 weeks old) were injected subcutaneously with 1 × 106 MB49 cells. One week after bladder cancer cell injection, 10% DMSO or 20 mg/kg SP600125 were injected i.p. every 3 days. | |||
| Response Summary | METTL3 was essential for bladder cancer cells to resist the cytotoxicity of CD8+ T cells by regulating Programmed cell death 1 ligand 1 (CD274/PD-L1) expression. Additionally, JNK signaling contributed to tumor immune escape in a METTL3-dependent manner both in vitro and in vivo. | |||
Inflammatory response [ICD-11: MG46]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [322] | |||
| Responsed Disease | Inflammatory response [ICD-11: MG46] | |||
Protein arginine N-methyltransferase 5 (PRMT5)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Pancreatic islets | Mus musculus |
|
Treatment: Mettl3 knockout mice
Control: Mettl3 flox/flox mice
|
GSE155612 | |
| Regulation |
 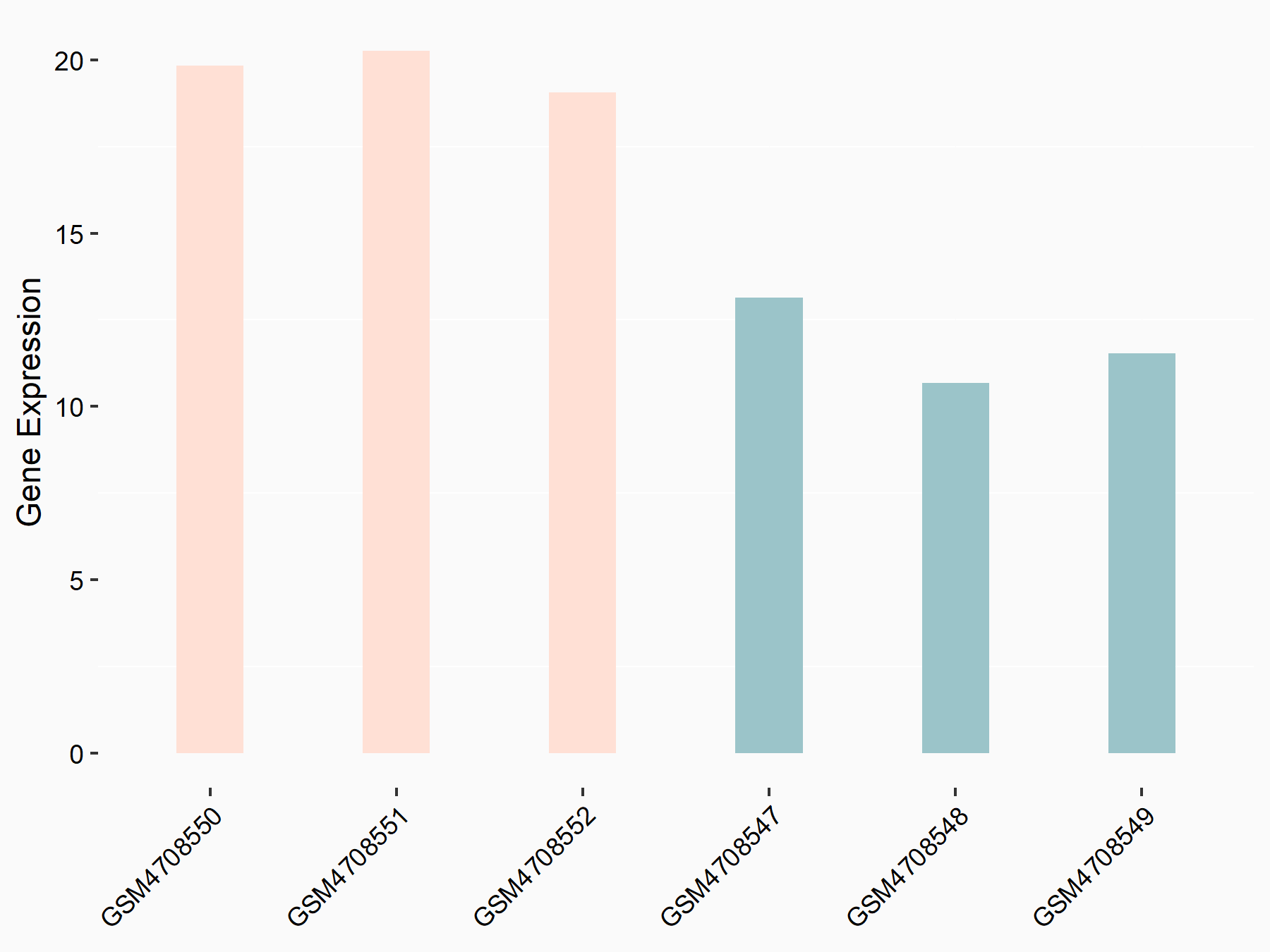 |
logFC: 7.02E-01 p-value: 6.96E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 5.94E+00 | GSE60213 |
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [99] | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
In-vitro Model |
SCC-9 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1685 |
| SCC-4 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1684 | |
| SCC-25 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1682 | |
| CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 | |
| In-vivo Model | Six-week-old nude mice were randomly divided into two groups (three mice per group) and cultured with continuous access to sterile food and water in pathogen-free sterile conditions. To establish the OSCC xenograft model, we subcutaneously injected 5 × 106 SCC-9 cells stably transfected with METTL3 shRNA or sh-NC vectors into nude mice. | |||
| Response Summary | METTL3 intensified the metastasis and proliferation of OSCC by modulating the m6A amounts of Protein arginine N-methyltransferase 5 (PRMT5) and PD-L1. | |||
Protein SET (SET)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
  |
logFC: 5.30E+00 p-value: 1.65E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.53E+00 | GSE60213 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [102] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | TNF signaling pathway | hsa04668 | ||
| Cell Process | Cell migration | |||
| Cell invasion | ||||
| Epithelial-mesenchymal transition | ||||
In-vitro Model |
NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| Response Summary | MiR-1915-3p expression was regulated by METTL3/YTHDF2 m6A axis through transcription factor KLF4. miR-1915-3p function as a tumor suppressor by targeting Protein SET (SET) and has an anti-metastatic therapeutic potential for lung cancer treatment. | |||
Proto-oncogene c-Rel (c-Rel)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Pancreatic islets | Mus musculus |
|
Treatment: Mettl3 knockout mice
Control: Mettl3 flox/flox mice
|
GSE155612 | |
| Regulation |
 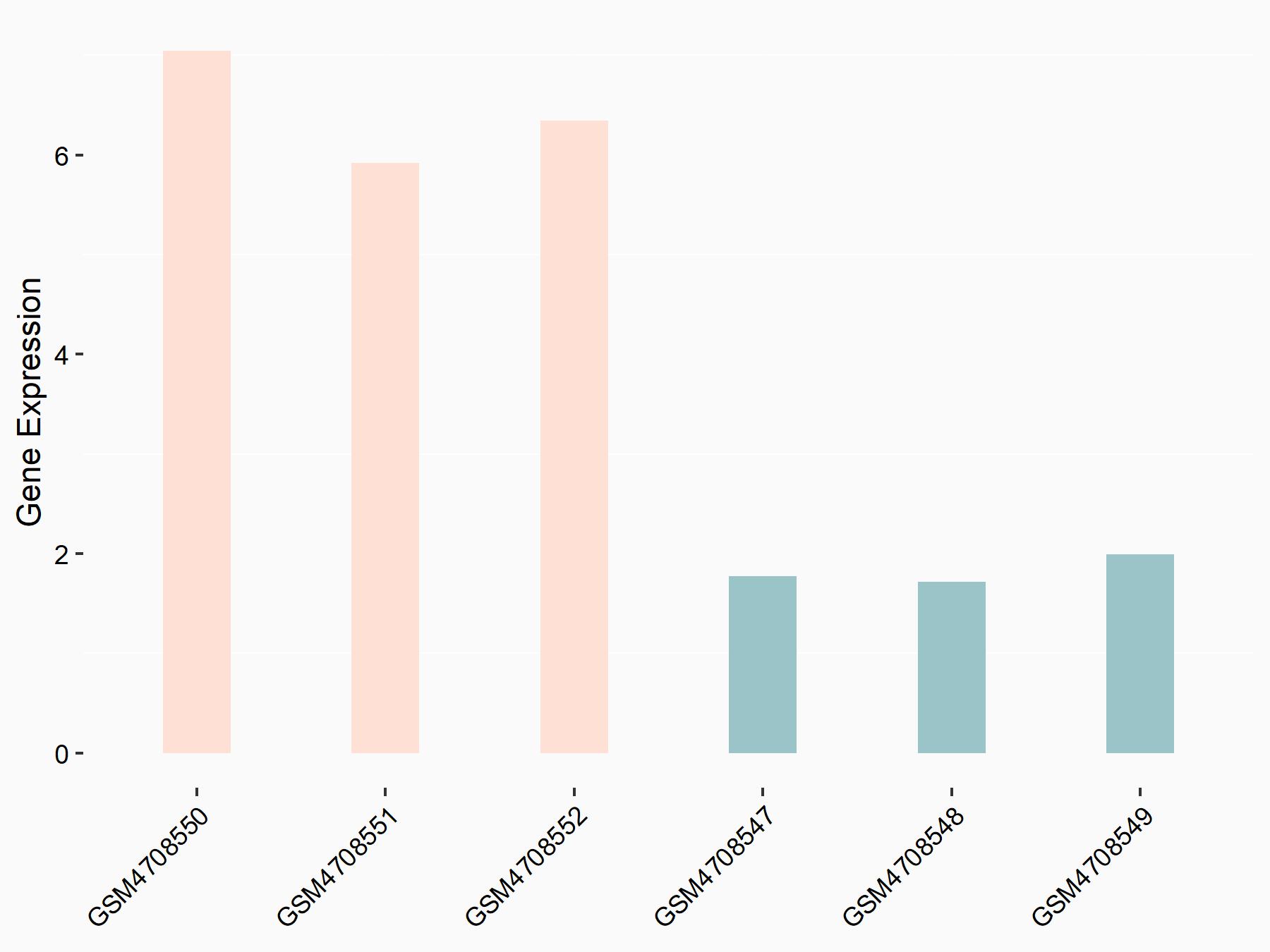 |
logFC: 1.39E+00 p-value: 2.50E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.02E+00 | GSE60213 |
Thyroid Cancer [ICD-11: 2D10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [103] | |||
| Responsed Disease | Papillary thyroid cancer [ICD-11: 2D10.1] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | NF-kappa B signaling pathway | hsa04064 | ||
In-vitro Model |
TPC-1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_6298 |
| Nthy-ori 3-1 | Normal | Homo sapiens | CVCL_2659 | |
| KTC-1 | Thyroid carcinoma | Homo sapiens | CVCL_6300 | |
| B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 | |
| In-vivo Model | For xenograft models, 5 × 106 BCPAP or KTC-1 cells from each group were injected subcutaneously into the flanks of female BALB/c nude mice (4-6 weeks old, Shanghai SLAC Laboratory Animal, China, n = 5 per group) in a volume of 150 uL PBS. Tumor growth was measured with a digital caliper every 4 days and calculated using the following formula: (length × width2)/2. To study the effect of IL-8 on tumor growth in vivo, scramble or shMETTL3 BCPAP cells were implanted hypodermically into BALB/c nude mice (2 × 106 cells in 150 uL PBS, n = 10 per group). When palpable tumors formed on day 14, mice were treated with DMSO or the IL-8 inhibitor SB225002 (10 mg/kg) by intraperitoneal injection 3 times per week for 3 weeks. Six weeks post-injection, the mice were sacrificed, and the tumors were collected to analyze the frequency of TANs by flow cytometry. For the lung metastasis model, BCPAP and KTC-1 cells (2 × 106 cells in 100 uL PBS) with the corresponding vectors were injected into the tail veins of BALB/c nude mice. Eight weeks after injection, the mice were euthanized, and metastatic lung nodules were analyzed (n = 5 for each group). | |||
| Response Summary | METTL3 played a pivotal tumor-suppressor role in papillary thyroid cancer carcinogenesis through Proto-oncogene c-Rel (c-Rel) and RelA inactivation of the nuclear factor Kappa-B (NF-Kappa-B) pathway by cooperating with YTHDF2 and altered TAN infiltration to regulate tumor growth. | |||
Pyruvate dehydrogenase kinase isoform 4 (PDK4)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
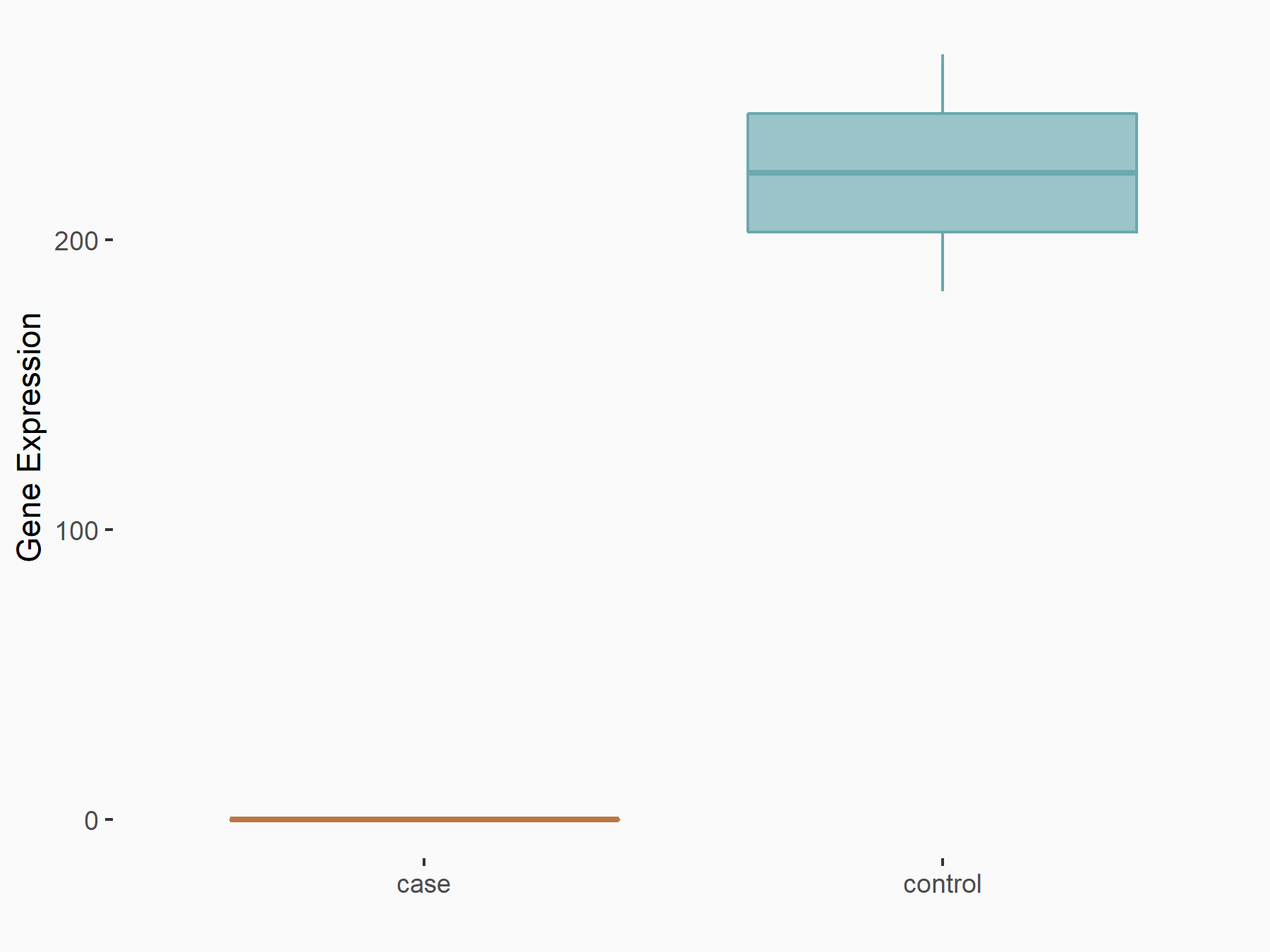 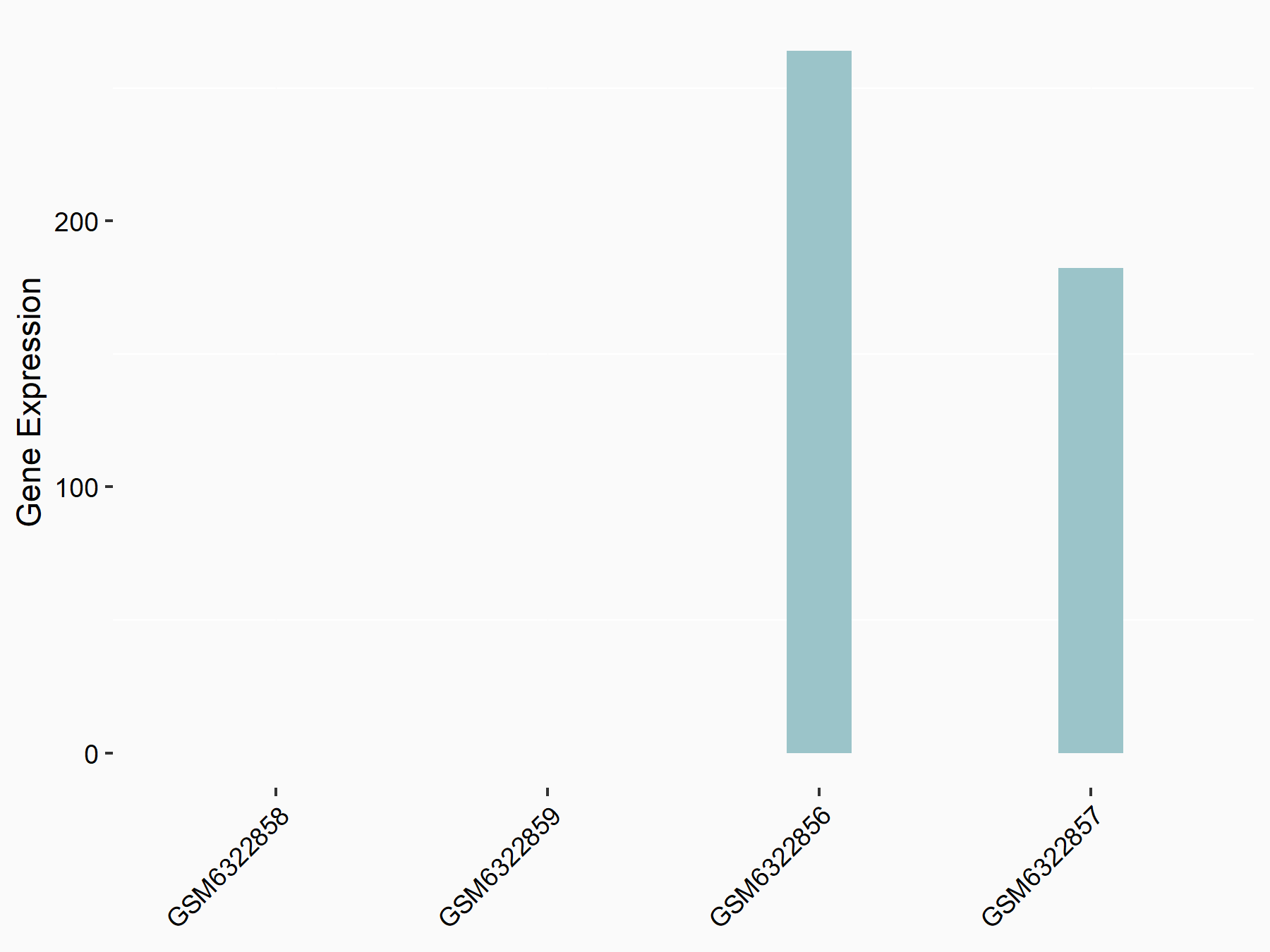 |
logFC: -1.03E+01 p-value: 3.59E-12 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 5.02E+00 | GSE60213 |
Solid tumour/cancer [ICD-11: 2A00-2F9Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [104] | |||
| Responsed Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycerolipid metabolism | hsa00561 | ||
| Cell Process | Glycolysis | |||
In-vitro Model |
SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| In-vivo Model | As to the subcutaneous transplanted model, WT Vec, Mettl3 Mut/-+vec, WTPDK4, Mettl3 Mut/-+vecPDK4 cells (2 × 10 6 per mouse, n = 10 for each group) were diluted in 200 uL normal medium + 200 uL Matrigel (BD Biosciences) and subcutaneously injected into immunodeficient mice to investigate tumor growth. | |||
| Response Summary | m6A regulates glycolysis of cancer cells through Pyruvate dehydrogenase kinase isoform 4 (PDK4). Knockdown of Mettl3 significantly attenuated m6A antibody enriched PDK4 mRNA in Huh7 cells. It reveals that m6A regulates glycolysis of cancer cells through PDK4. | |||
Ribosomal protein S6 kinase beta-1 (RPS6KB1/p70S6K)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | CT26 cell line | Mus musculus |
|
Treatment: METTL3 knockout CT26 cells
Control: CT26 cells
|
GSE142589 | |
| Regulation |
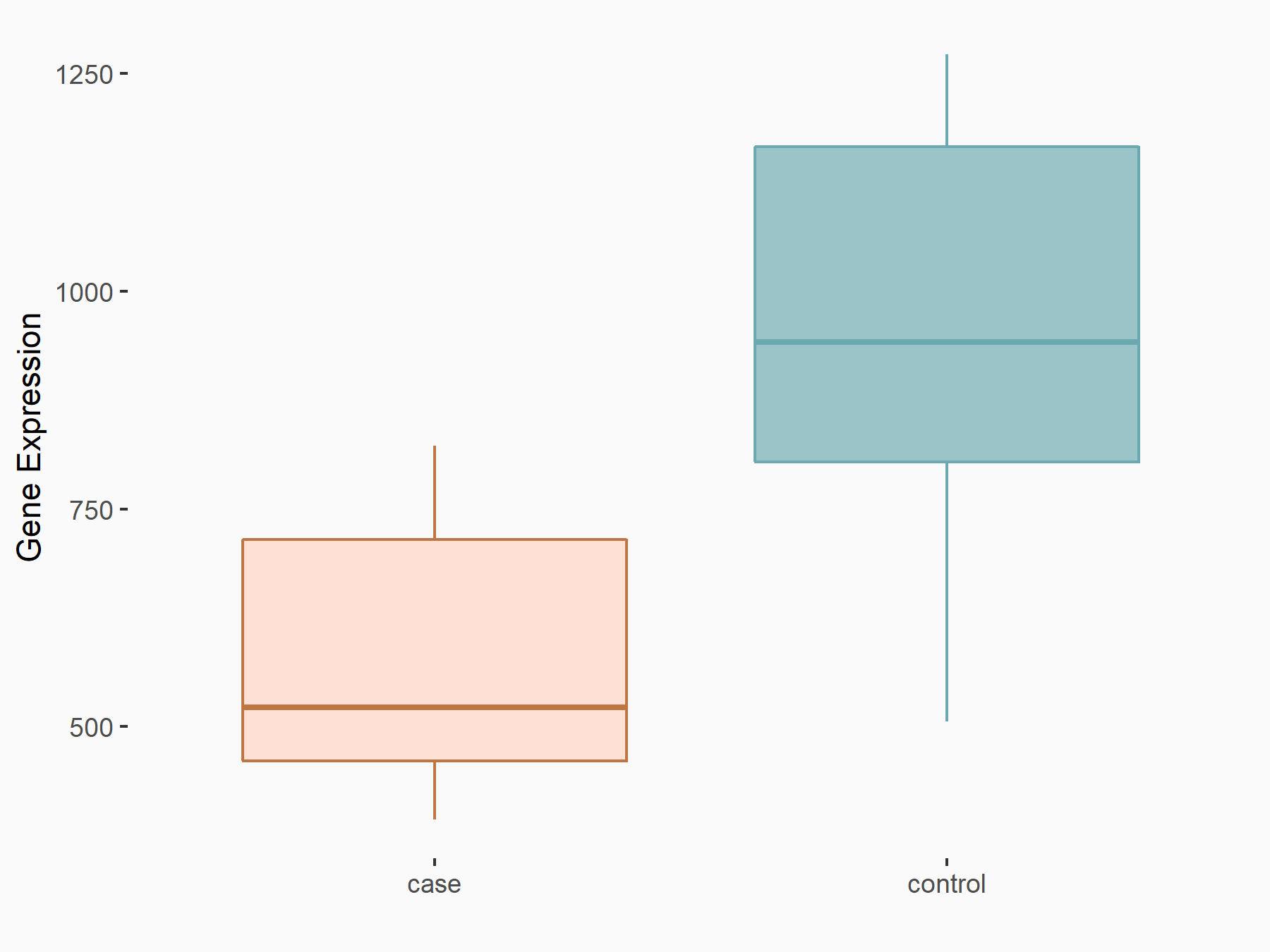 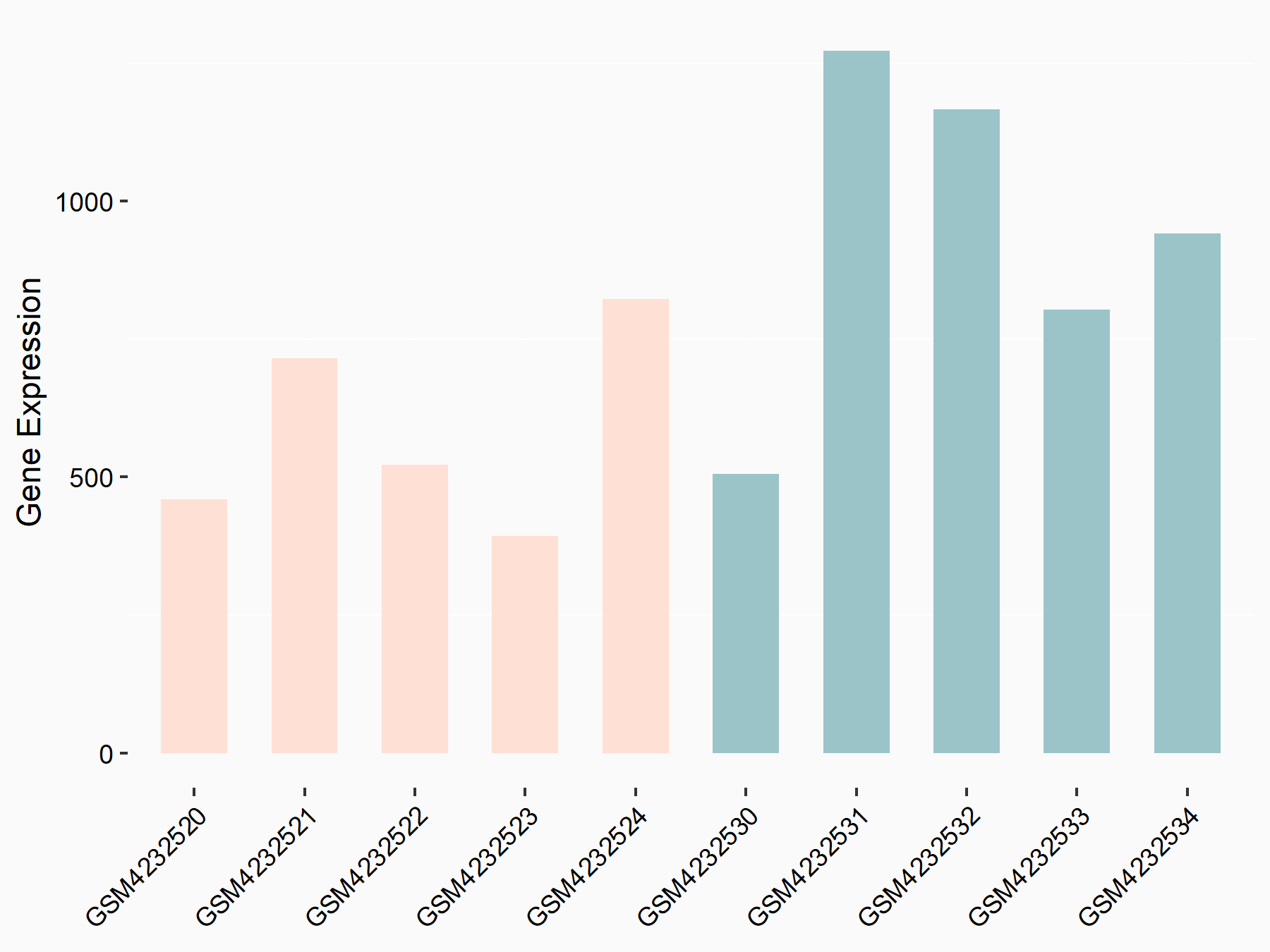 |
logFC: -6.86E-01 p-value: 2.67E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.70E+00 | GSE60213 |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [8] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| Response Summary | Down-regulation of METTL3 inhibits the proliferation and mobility of human gastric cancer cells and leads to inactivation of the AKT signaling pathway, suggesting that METTL3 is a potential target for the treatment of human gastric cancer. METTL3 knockdown decreased Bcl2 and increased Bax and active Caspase-3 in gastric cancer cells, which suggested the apoptotic pathway was activated. METTL3 led to inactivation of the AKT signaling pathway in human gastric cancer cells, including decreased phosphorylation levels of AKT and expression of down-stream effectors Ribosomal protein S6 kinase beta-1 (RPS6KB1/p70S6K) and Cyclin D1. | |||
Retina cancer [ICD-11: 2D02]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [44] | |||
| Responsed Disease | Retinoblastoma [ICD-11: 2D02.2] | |||
| Responsed Drug | Rapamycin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
In-vitro Model |
WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through PI3K/AKT/mTOR pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-Ribosomal protein S6 kinase beta-1 (RPS6KB1/p70S6K)/4EBP1 pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
RIG-I-like receptor 1 (RIG-I)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
  |
logFC: -1.14E+00 p-value: 3.48E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.75E+00 | GSE60213 |
Acute viral hepatitis [ICD-11: 1E50]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [105] | |||
| Responsed Disease | Acute hepatitis B [ICD-11: 1E50.1] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | RIG-I-like receptor signaling pathway | hsa04622 | ||
In-vitro Model |
Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Response Summary | METTL3 and METTL14 leads to an increase in viral RNA recognition by RIG-I-like receptor 1 (RIG-I), thereby stimulating type I interferon production. The obvious advantage is that m6A deficiency in HBV and HCV induces a higher IFN synthesis and in turn enhance adaptive immunity. | |||
COVID-19 [ICD-11: RA01]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [106] | |||
| Responsed Disease | COVID-19 [ICD-11: RA01] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | RIG-I-like receptor signaling pathway | hsa04622 | ||
| Cell Process | Immune responses | |||
In-vitro Model |
Calu-3 | Lung adenocarcinoma | Homo sapiens | CVCL_0609 |
| Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 | |
| HEK293-FT | Normal | Homo sapiens | CVCL_6911 | |
| Response Summary | In SARS-CoV-2 infection, depletion of the host cell m6A methyltransferase METTL3 decreases m6A levels in SARS-CoV-2 and host genes, and m6A reduction in viral RNA increases RIG-I-like receptor 1 (RIG-I) binding and subsequently enhances the downstream innate immune signaling pathway and inflammatory gene expression. | |||
Sequestosome-1 (SQSTM1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HUVEC cell line | Homo sapiens |
|
Treatment: shMETTL3 HUVEC cells
Control: shScramble HUVEC cells
|
GSE157544 | |
| Regulation |
 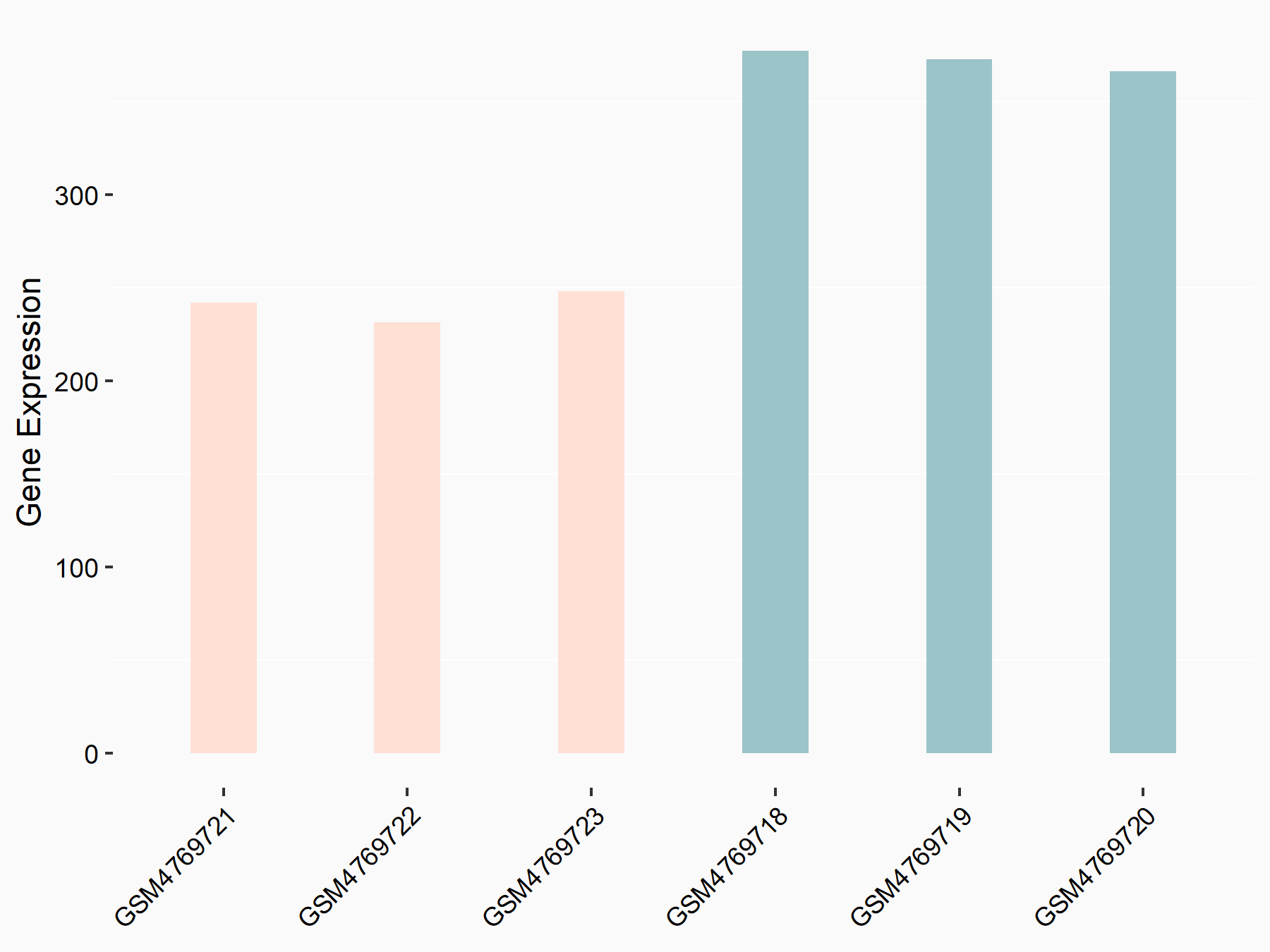 |
logFC: -6.28E-01 p-value: 1.94E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.03E+00 | GSE60213 |
Lung cancer [ICD-11: 2C25]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Chloroquine | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
In-vitro Model |
Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and Sequestosome-1 (SQSTM1). beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Gefitinib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
In-vitro Model |
Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and Sequestosome-1 (SQSTM1). beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Experiment 3 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Beta-Elemen | Phase 3 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
In-vitro Model |
Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and Sequestosome-1 (SQSTM1). beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
Breast cancer [ICD-11: 2C60]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [331] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Paclitaxel | Approved | ||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | The experimental mice were housed in an SPF-grade animal laboratory at a temperature of approximately 25 °C, a humidity range of 50%, and an average daylight duration of 12 h. BALB/C female mice at 5-6 weeks of age were taken and prepared for tumor-bearing. After the mice were sacrificed, their tissues were collected, weighed and immediately snap-frozen in liquid nitrogen. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [331] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | STM2457 | Investigative | ||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | The experimental mice were housed in an SPF-grade animal laboratory at a temperature of approximately 25 °C, a humidity range of 50%, and an average daylight duration of 12 h. BALB/C female mice at 5-6 weeks of age were taken and prepared for tumor-bearing. After the mice were sacrificed, their tissues were collected, weighed and immediately snap-frozen in liquid nitrogen. | |||
Serine/threonine-protein kinase 4 (STK4)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
 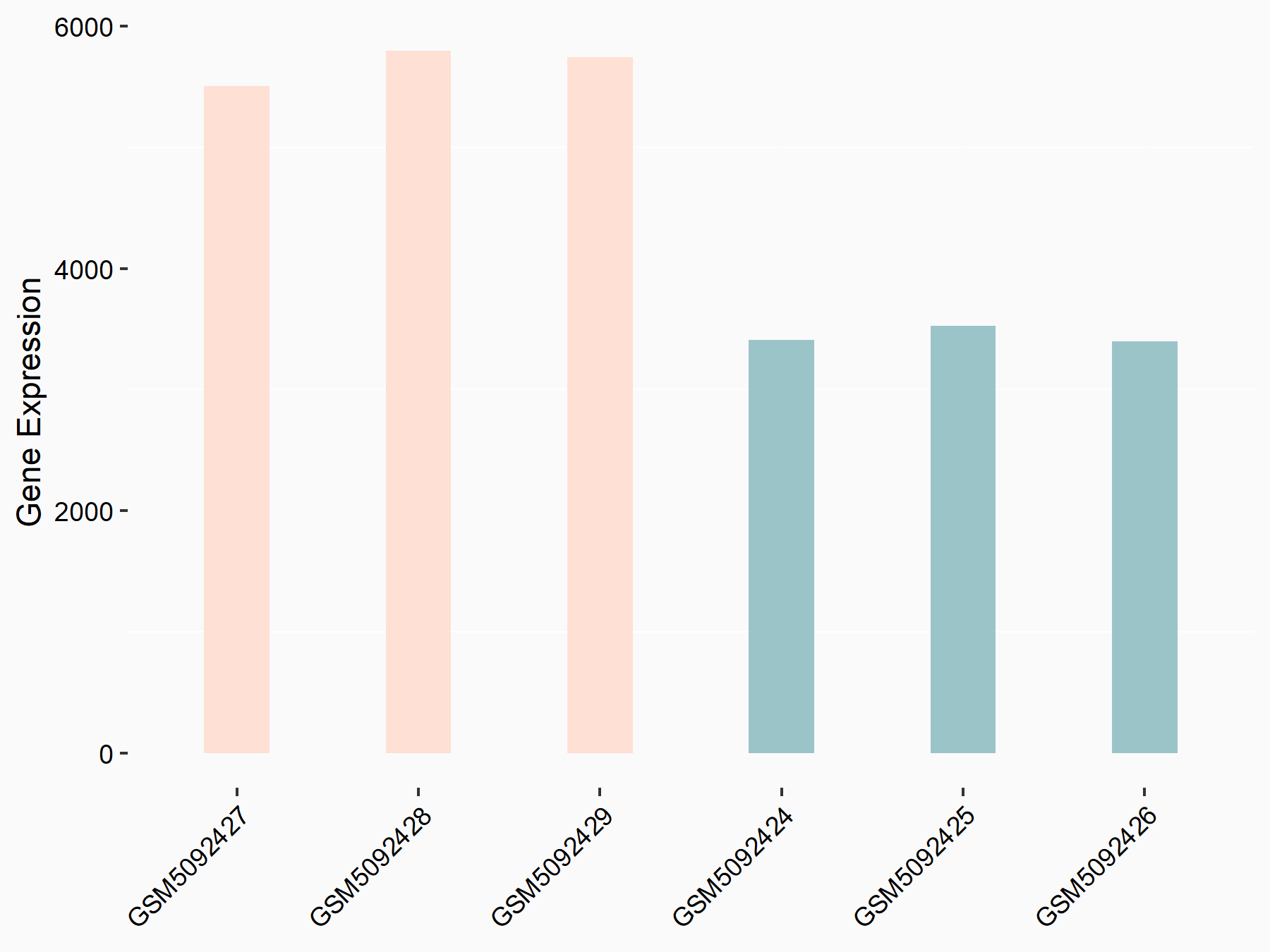 |
logFC: 7.22E-01 p-value: 1.68E-60 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.45E+00 | GSE60213 |
Thyroid Cancer [ICD-11: 2D10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [107] | |||
| Responsed Disease | Thyroid Cancer [ICD-11: 2D10] | |||
| Target Regulation | Down regulation | |||
| Response Summary | Silencing METTL3 suppresses miR-222-3p expression and thus stimulates Serine/threonine-protein kinase 4 (STK4) expression, thereby repressing the malignancy and metastasis of Thyroid Carcinoma. | |||
Serine/threonine-protein kinase mTOR (MTOR)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3 knockout liver
Control: Wild type liver cells
|
GSE198512 | |
| Regulation |
 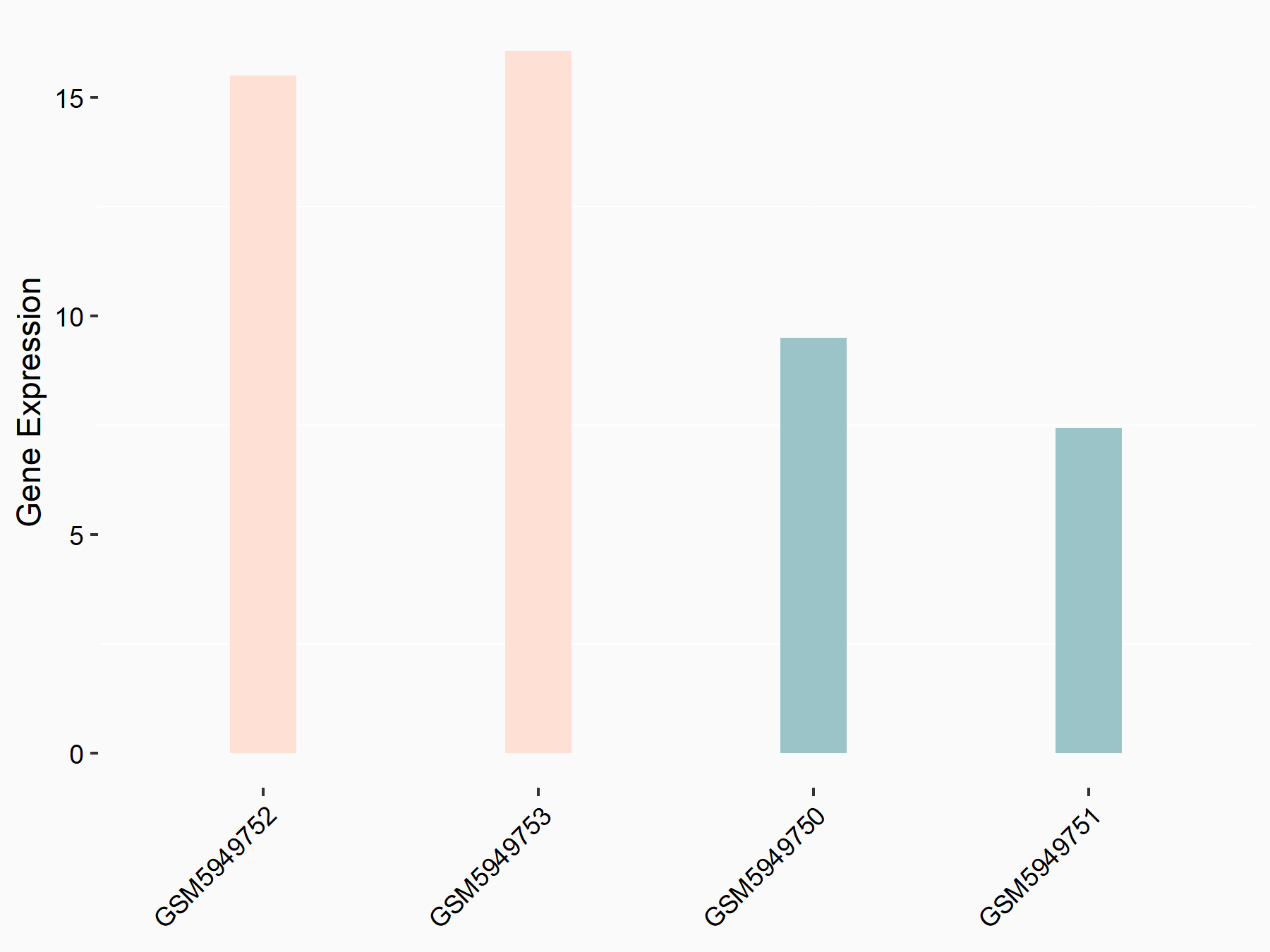 |
logFC: 8.34E-01 p-value: 1.58E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.47E+00 | GSE60213 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [108] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| In-vivo Model | 5 × 106 A549 cells overexpressing METTL3 (Lv-METTL3) or control (Lv-Ctrl) were suspended in 100 uL phosphate-buffered saline (PBS), and were subcutaneously injected into mouse lower right flank. Drug treatment started in the Lv-METTL3 group when the tumour volume reached around 100 mm3. Mice were randomly divided into three groups to receive vehicle, GSK2536771 (30 mg/kg) or rapamycin (1 mg/kg). Drugs were administrated daily through intraperitoneal injection for 18 days. Treatment conditions were chosen as previously reported. | |||
| Response Summary | METTL3-mediated m6 A methylation promotes lung cancer progression via activating PI3K/AKT/Serine/threonine-protein kinase mTOR (MTOR) pathway. | |||
Renal cell carcinoma [ICD-11: 2C90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [109] | |||
| Responsed Disease | Renal cell carcinoma [ICD-11: 2C90] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Epithelial-to-mesenchymal transition | |||
| Arrest cell cycle at G0/G1 phase | ||||
In-vitro Model |
ACHN | Papillary renal cell carcinoma | Homo sapiens | CVCL_1067 |
| Caki-1 | Clear cell renal cell carcinoma | Homo sapiens | CVCL_0234 | |
| Caki-2 | Papillary renal cell carcinoma | Homo sapiens | CVCL_0235 | |
| HK2 | Normal | Acipenser baerii | CVCL_YE28 | |
| In-vivo Model | Cells (5×106 cells in 200 uL) were suspended with 100 uL PBS and 100 uL Matrigel Matrix, and injected subcutaneously into the left armpit of each mouse. | |||
| Response Summary | Knockdown of METTL3 could obviously promote cell proliferation, migration and invasion function, and induce G0/G1 arrest,METTL3 acts as a novel marker for tumorigenesis, development and survival of RCC. Knockdown of METTL3 promoted changes in pI3K/AKT/Serine/threonine-protein kinase mTOR (MTOR) markers' expression with a gain in p-PI3k, p-AKT, p-mTOR and p-p70, and a loss of p-4EBP1. | |||
Retina cancer [ICD-11: 2D02]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [44] | |||
| Responsed Disease | Retinoblastoma [ICD-11: 2D02.2] | |||
| Responsed Drug | Rapamycin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
In-vitro Model |
WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through PI3K/AKT/Serine/threonine-protein kinase mTOR (MTOR) pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-P70S6K/4EBP1 pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [110] | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| HIF-1 signaling pathway | hsa04066 | |||
| In-vivo Model | The 8-10 weeks old mice were fed either a high fat diet or HF-CDAA , ad lib for 6-12 weeks. Chow diet was used as control for HFD.The mouse liver was perfused with PBS through portal vein, and liver tissue was cut into small pieces by a scissor. The single cell was made using syringe plunger to mull the tissue, and passed through a 40 uM cell strainer. | |||
| Response Summary | The contribution of METTL3-mediated m6A modification of Ddit4 mRNA to macrophage metabolic reprogramming in non-alcoholic fatty liver disease and obesity. In METTL3-deficient macrophages, there is a significant downregulation of Serine/threonine-protein kinase mTOR (MTOR) and nuclear factor Kappa-B (NF-Kappa-B) pathway activity in response to cellular stress and cytokine stimulation, which can be restored by knockdown of DDIT4. | |||
Signal transducer and activator of transcription 2 (STAT2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
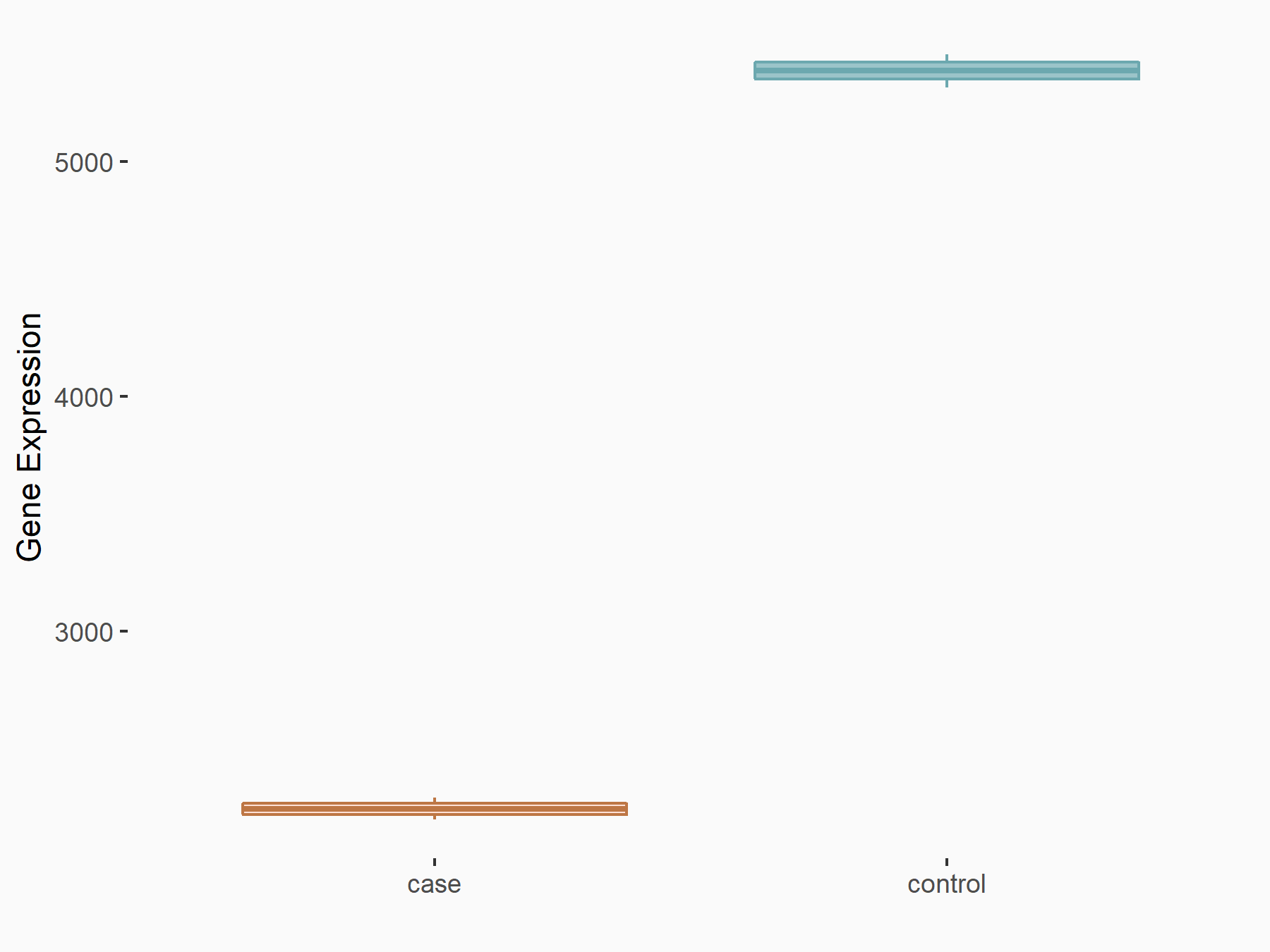 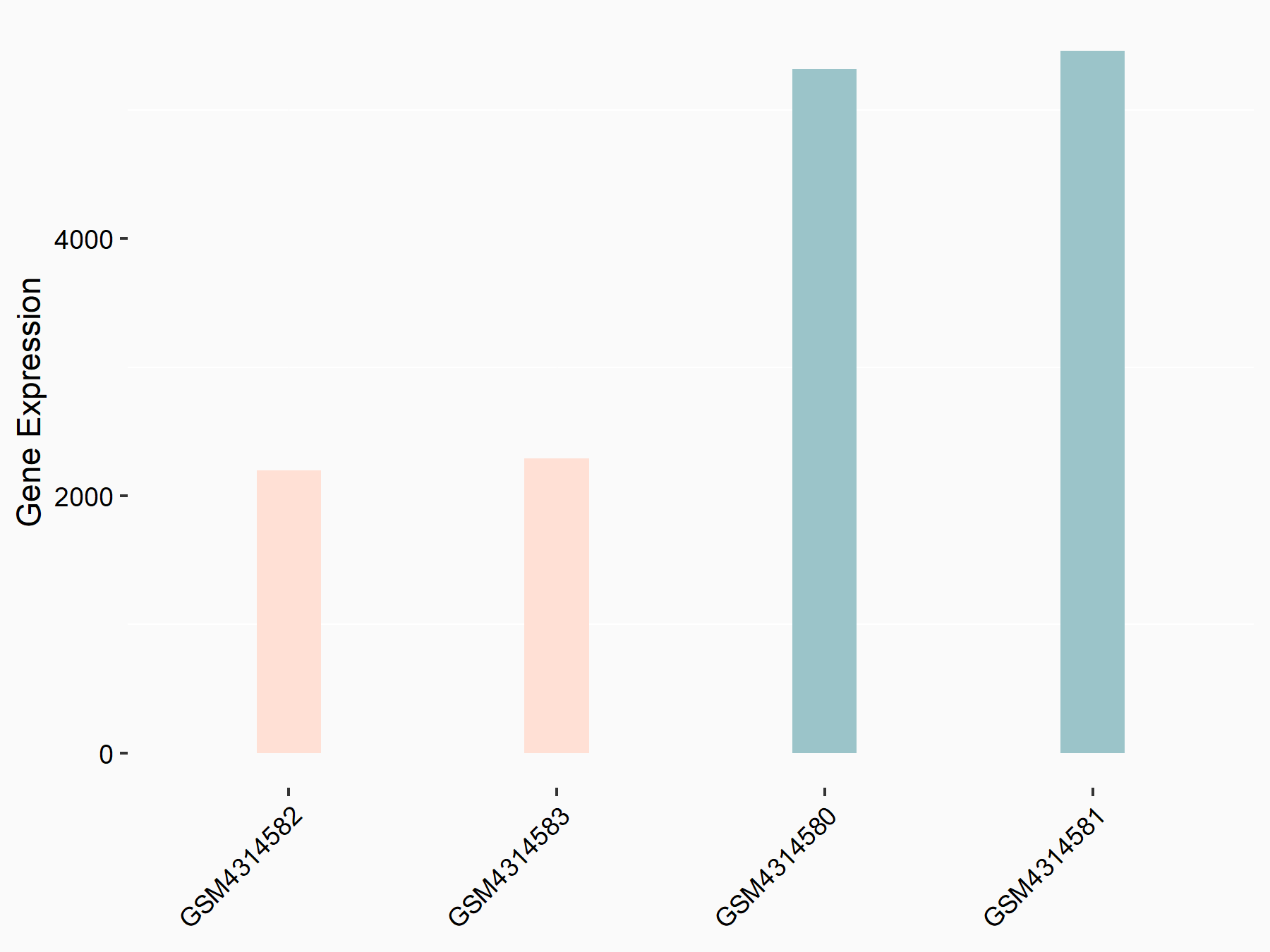 |
logFC: -1.27E+00 p-value: 1.09E-100 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.27E+00 | GSE60213 |
Congenital pneumonia [ICD-11: KB24]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [111] | |||
| Responsed Disease | Congenital pneumonia [ICD-11: KB24] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
WI-38 | Normal | Homo sapiens | CVCL_0579 |
| In-vivo Model | All mice were housed under a 12 h light/dark cycle with constant temperature about 25 ℃ and relative humidity approximating 55 %. The mice had free access to food and water for 10 days prior to the experiment. Forty mice were randomly selected and divided into four groups of 10 mice each. After 10 days, mice received an intraperitoneal injection of 22 mg / mL sodium pentobarbital (diluted in saline) followed by 167 uM LPS (60 uL) Saline solution was instilled into the oral cavity through the posterior pharyngeal wall. Pinch the nares quickly and hold for 30 s, model is successful when all fluid is absorbed into the nasal cavity, and slight tracheal rales appear. Lentiviral vectors containing pcDNA-SNHG4 (150 uM) or pcDNA-3.1 were intratracheally injected into mice. Twenty-one days after establishing the model, mice were intraperitoneally injected with 3% sodium pentobarbital and euthanized by overdose anesthesia at a dose of 90 mL/Kg, and organs and tissues were removed for follow-up studies. | |||
| Response Summary | SNHG4 was downregulated in the neonatal pneumonia patient serum and its overexpression could inhibit LPS induced inflammatory injury in human lung fibroblasts and mouse lung tissue. The molecular mechanism underlying this protective effect was achieved by suppression of METTL3-mediated m6A modification levels of YTHDF1-dependent Signal transducer and activator of transcription 2 (STAT2) mRNA. | |||
Stearoyl-CoA desaturase (SCD)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
|
Treatment: METTL3 knockdown MDA-MB-231 cells
Control: MDA-MB-231 cells
|
GSE70061 | |
| Regulation |
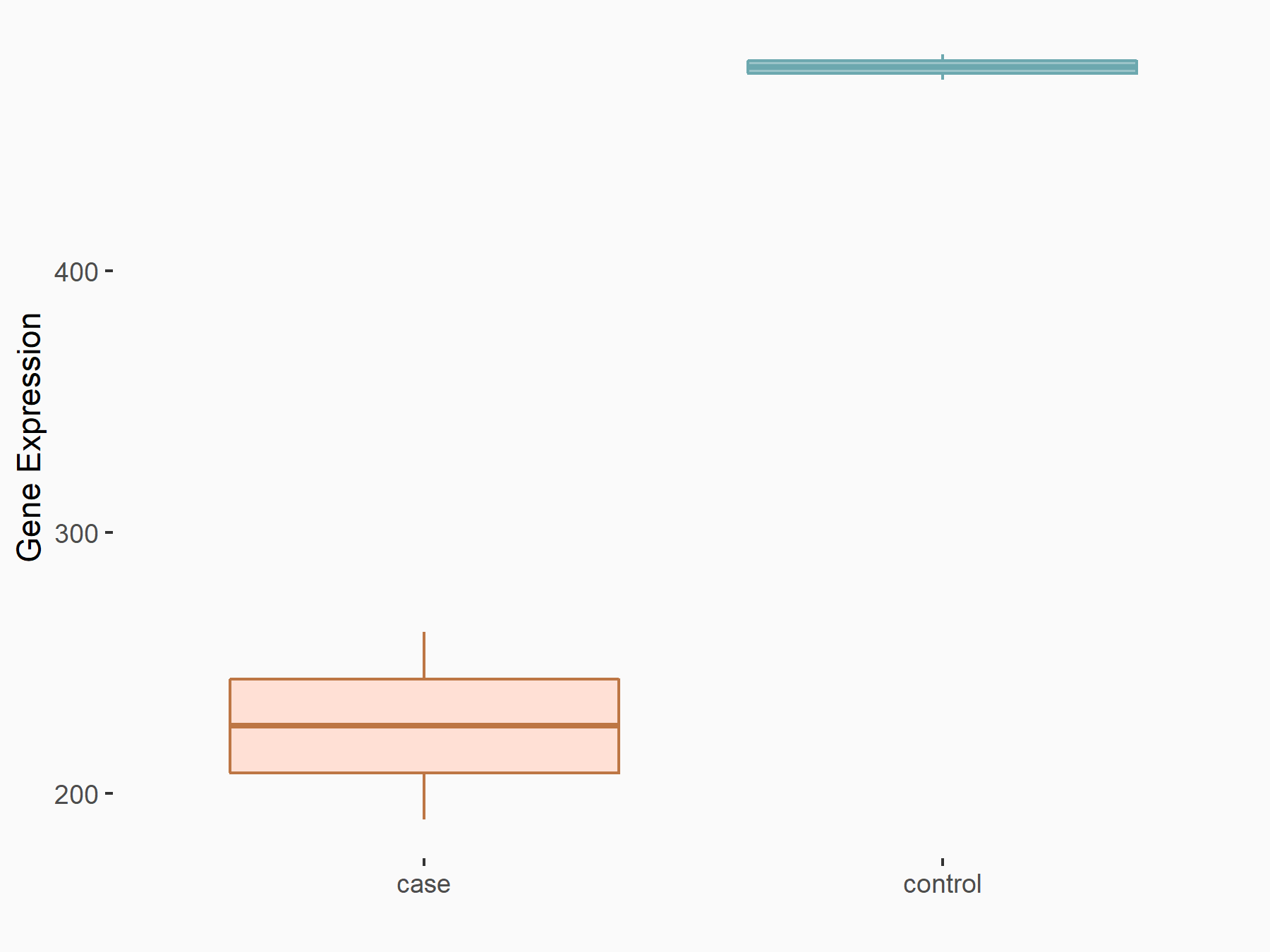 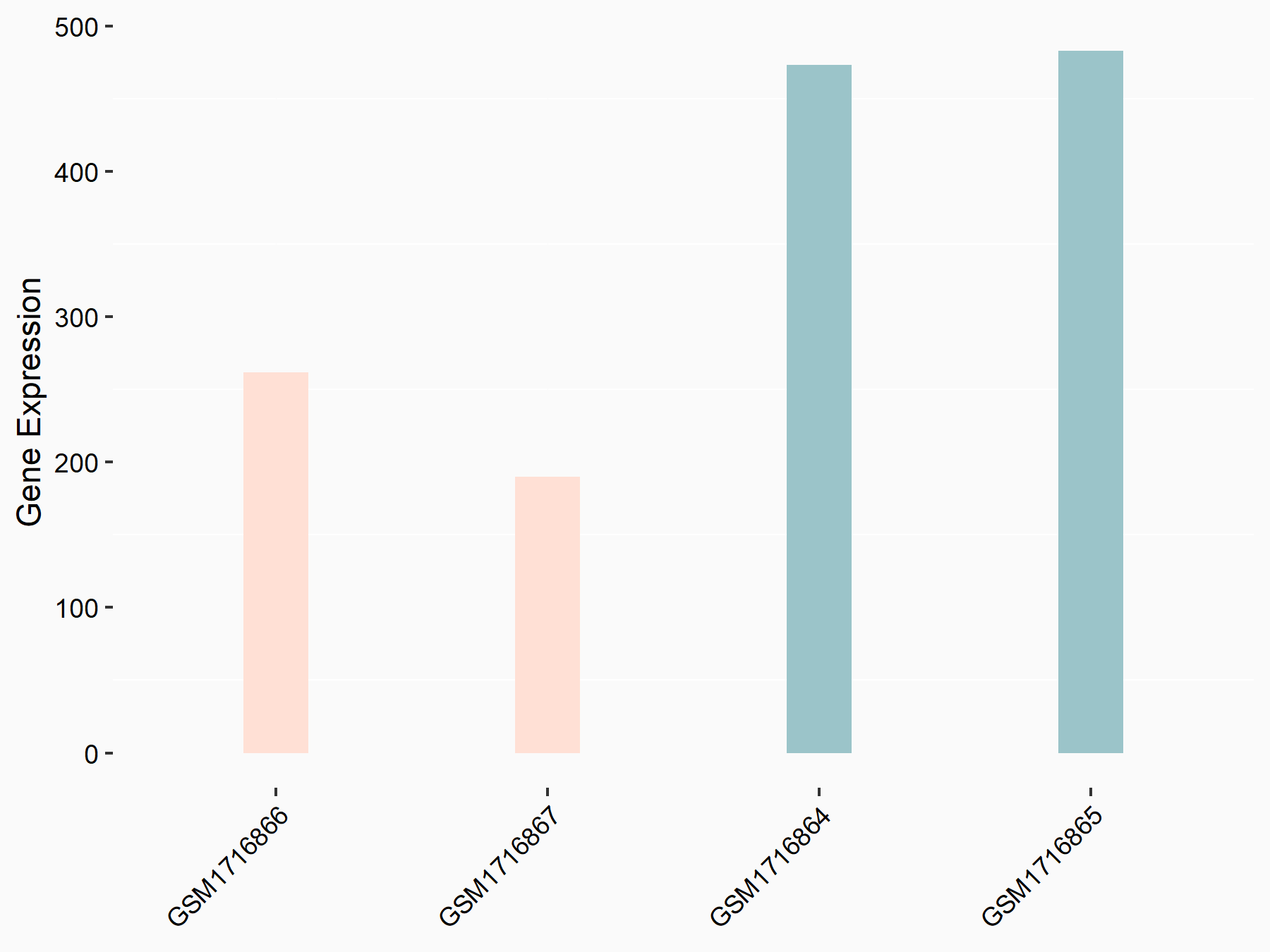 |
logFC: -1.10E+00 p-value: 2.11E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.17E+00 | GSE60213 |
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [112] | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycerolipid metabolism | hsa00561 | ||
| Cell Process | Lipid metabolism | |||
In-vitro Model |
LM3 | Malignant neoplasms | Mus musculus | CVCL_D269 |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| In-vivo Model | Mice with a Tmem30a deletion specifically in pancreatic beta cells were generated as previously described. Mice developed with NAFLD were named for Tmem30a-associated NAFLD (TAN) mice. The littermate mice with genotypes of Tmem30aloxP/loxP were used as controls. | |||
| Response Summary | Targeting METTL3/14 in vitro increases protein level of ACLY and Stearoyl-CoA desaturase (SCD) as well as triglyceride and cholesterol production and accumulation of lipid droplets. These findings demonstrate a new NAFLD mouse model that provides a study platform for DM2-related NAFLD and reveals a unique epitranscriptional regulating mechanism for lipid metabolism via m6A-modified protein expression of ACLY and SCD1. | |||
TNF receptor-associated factor 5 (TRAF5)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 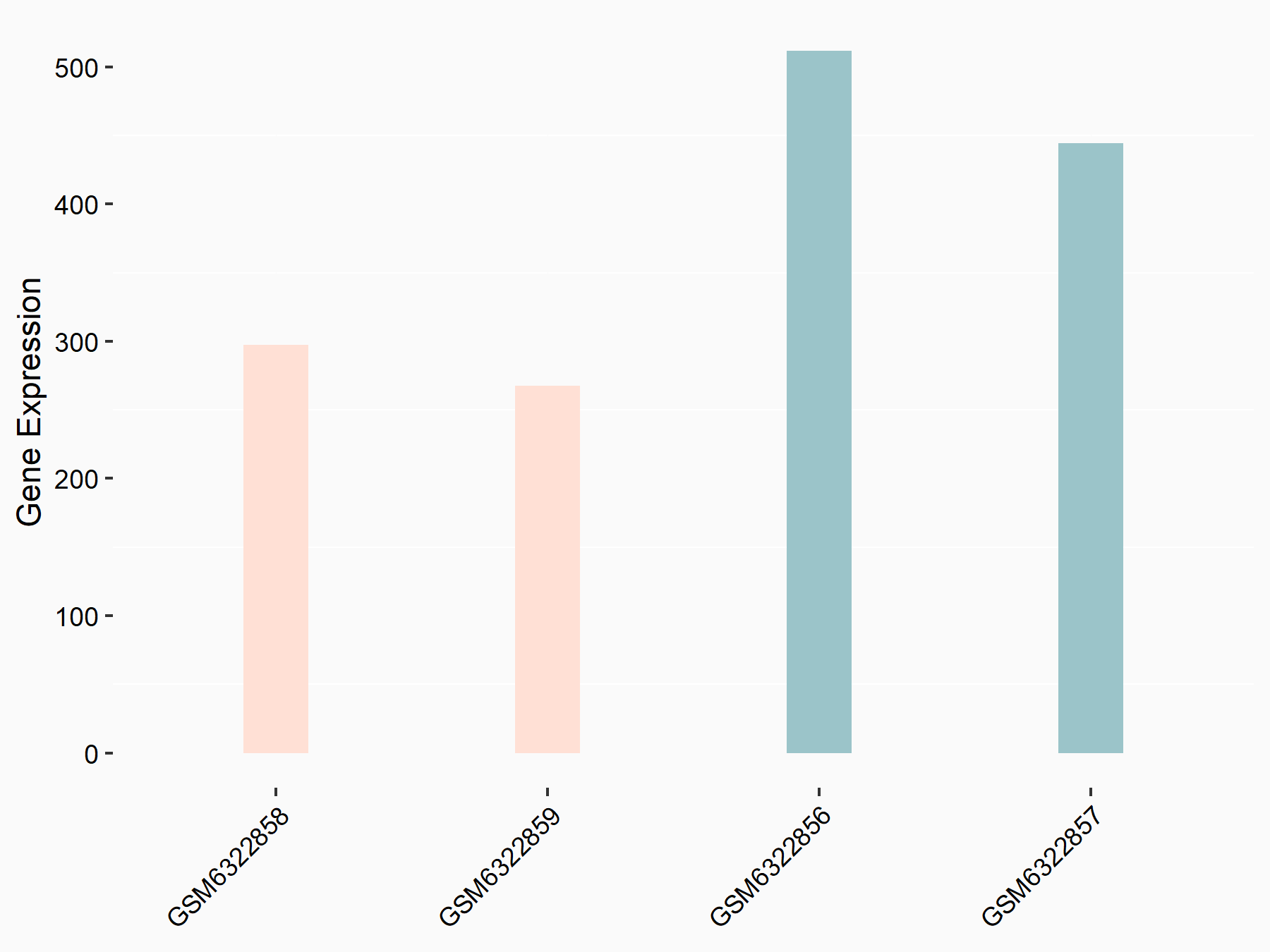 |
logFC: -7.59E-01 p-value: 1.50E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.24E+00 | GSE60213 |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [113] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Oxaliplatin | Approved | ||
| Target Regulation | Down regulation | |||
In-vitro Model |
LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| In-vivo Model | HCT-116 cells (3 × 105 cells in 200 uL of saline) were subcutaneously injected into the nude mice to establish xenograft tumors. After 10 days, 10 mg/kg OX or saline was intraperitoneally injected (n = 5 for each group). Si-METTL3 or si-TRAF5 (10 nmol/20 g body weight) was injected twice intratumorally before the start of OX treatment. The mice were examined every 2 days and sacrificed 4 weeks after the OX treatment. | |||
| Response Summary | 2-polarized tumor-associated macrophages enabled the oxaliplatin resistance via the elevation of METTL3-mediated m6A modification in Colorectal Cancer cells. Furthermore, they found that TNF receptor-associated factor 5 (TRAF5) contributes to the METTL3-triggered OX resistance in CRC cells. | |||
TNF receptor-associated factor 6 (TRAF6)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
 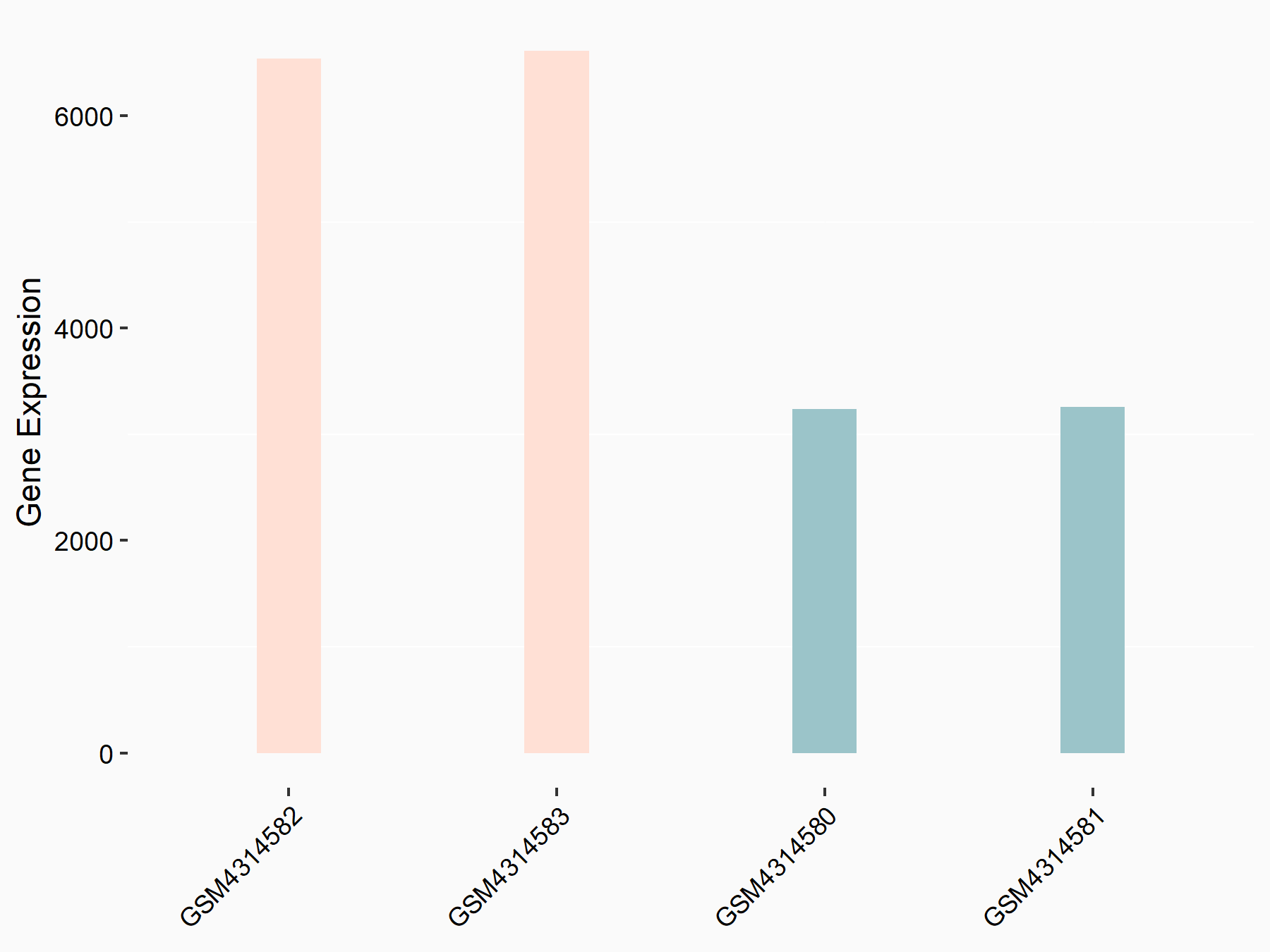 |
logFC: 1.02E+00 p-value: 2.31E-81 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.04E+00 | GSE60213 |
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [114] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 |
| SaOS-2 | Osteosarcoma | Homo sapiens | CVCL_0548 | |
| NHOst (Normal human osteoblast cells) | ||||
| MG-63 | Osteosarcoma | Homo sapiens | CVCL_0426 | |
| HOS | Osteosarcoma | Homo sapiens | CVCL_0312 | |
| In-vivo Model | For mouse xenograft tumor model, 1×106 Saos2 or HOS cells were mixed in 200 ul PBS and injected subcutaneously into 6- to 8-week-old C57BL/6 mice.umor size of mice was measured by vernier calipers every week after injection. The mice were euthanized after 28 days by intraperitoneal injection of 1% pentobarbital sodium (120 mg/kg), and the tumor tissues were removed from the mice. | |||
| Response Summary | METTL3 is highly expressed in OS and enhances TNF receptor-associated factor 6 (TRAF6) expression through m6A modification, thereby promoting the metastases of OS cells. | |||
Acute myocardial infarction [ICD-11: BA41]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [347] | |||
| Responsed Disease | Acute myocardial infarction [ICD-11: BA41] | |||
| Target Regulation | Up regulation | |||
Transcription factor AP-2 gamma (TFAP2C)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- mESCs
Control: Wild type ESCs
|
GSE147849 | |
| Regulation |
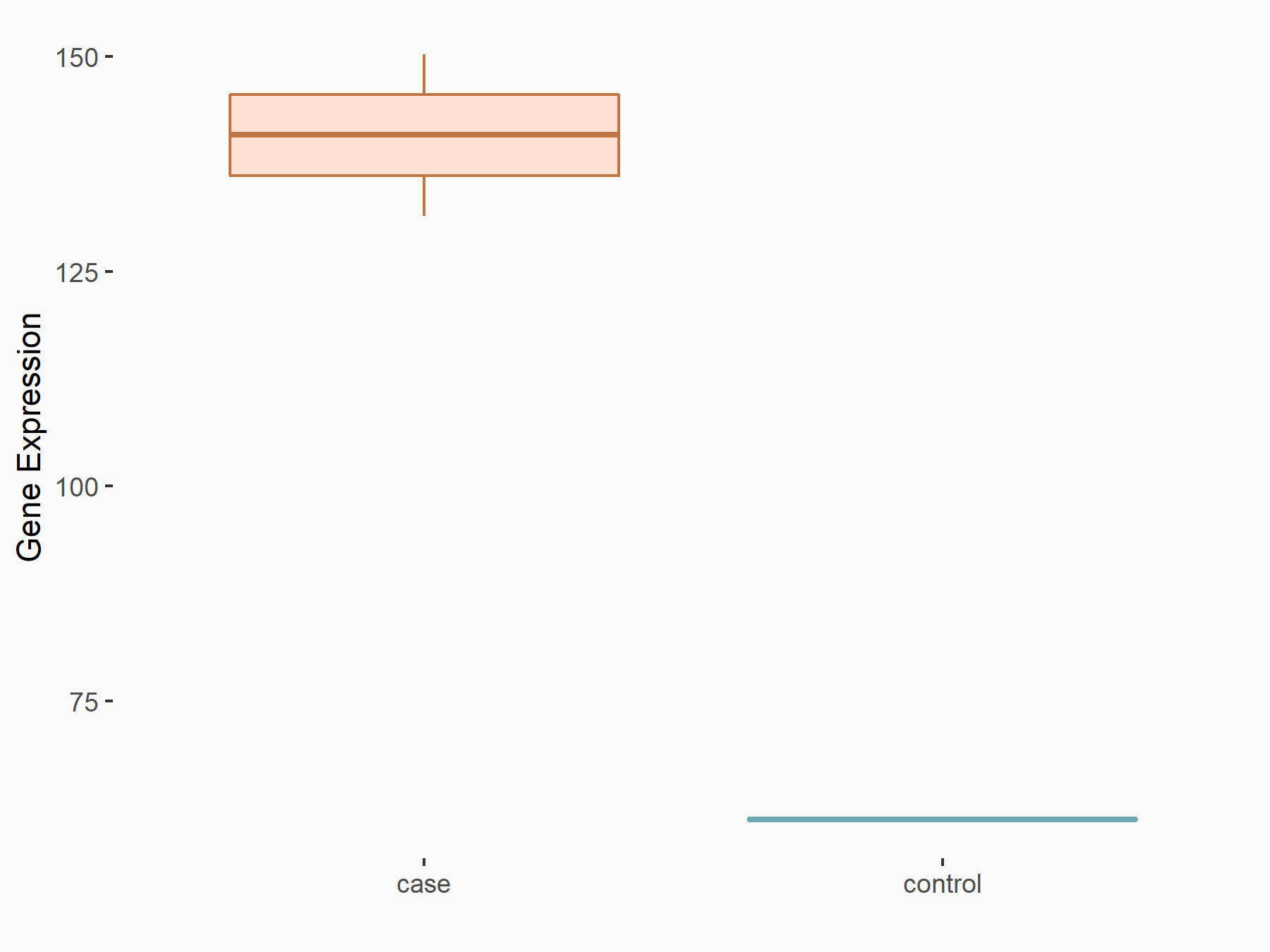 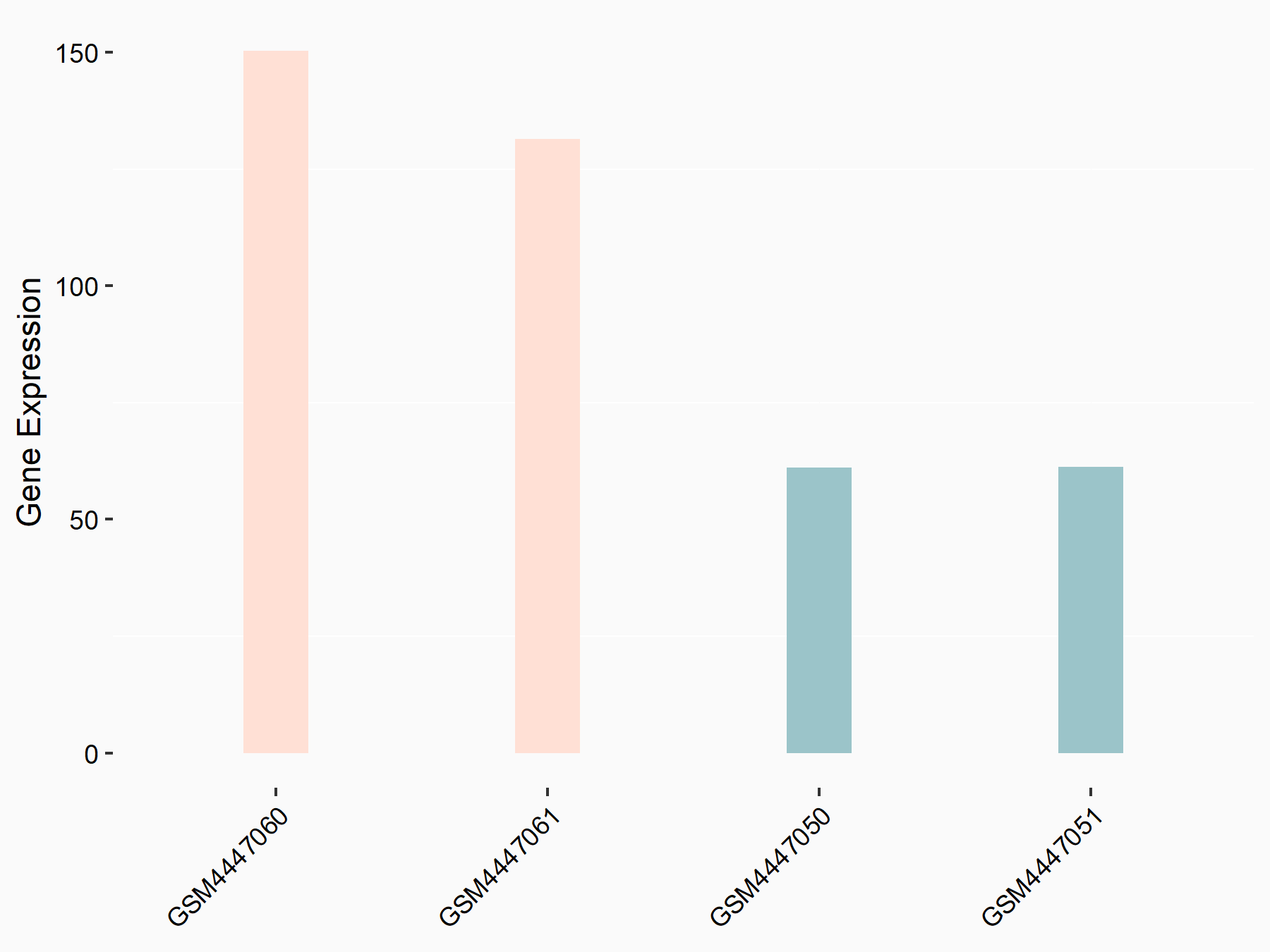 |
logFC: 1.20E+00 p-value: 4.47E-08 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.28E+00 | GSE60213 |
Testicular cancer [ICD-11: 2C80]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [115] | |||
| Responsed Disease | Testicular cancer [ICD-11: 2C80] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | DNA repair | |||
In-vitro Model |
TCam-2 | Testicular seminoma | Homo sapiens | CVCL_T012 |
| In-vivo Model | Male mice were subcutaneously injected with tumour cells near the limbs to establish xenografts (1 × 106/mouse, 0.2 mL for each injection site; METTL3-overexpressing TCam-2/CDDP cells were inoculated once at the initial time and IGF2BP1-inhibited TCam-2/CDDP cells were inoculated every 3 days). | |||
| Response Summary | METTL3 potentiates resistance to cisplatin through m6A modification of Transcription factor AP-2 gamma (TFAP2C) in seminoma. Enhanced stability of TFAP2C mRNA promoted seminoma cell survival under cisplatin treatment burden probably through up-regulation of DNA repair-related genes. IGF2BP1 binds to TFAP2C and enhances TFAP2C mRNA stability. | |||
Transcription factor E2F1 (E2F1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
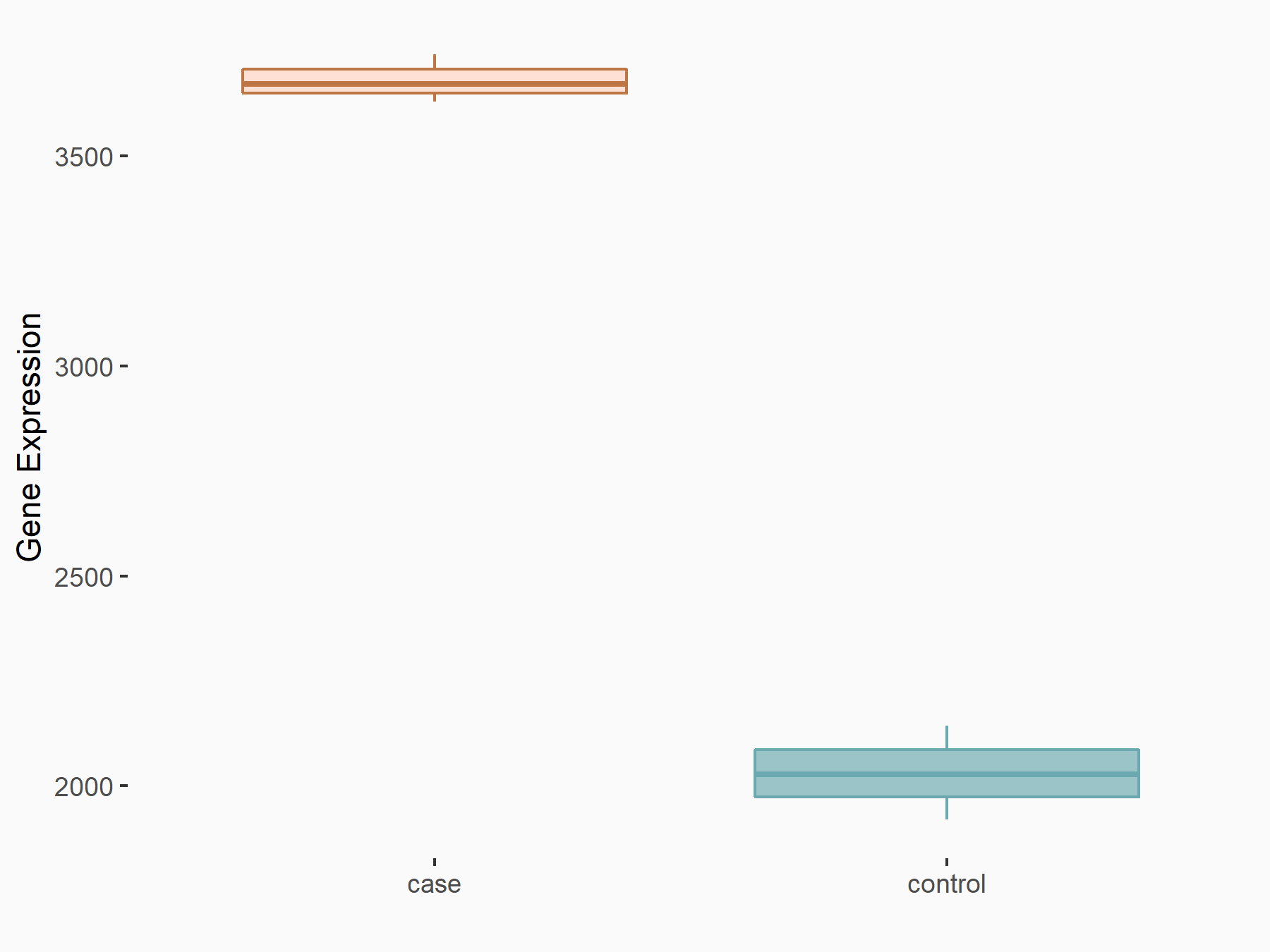 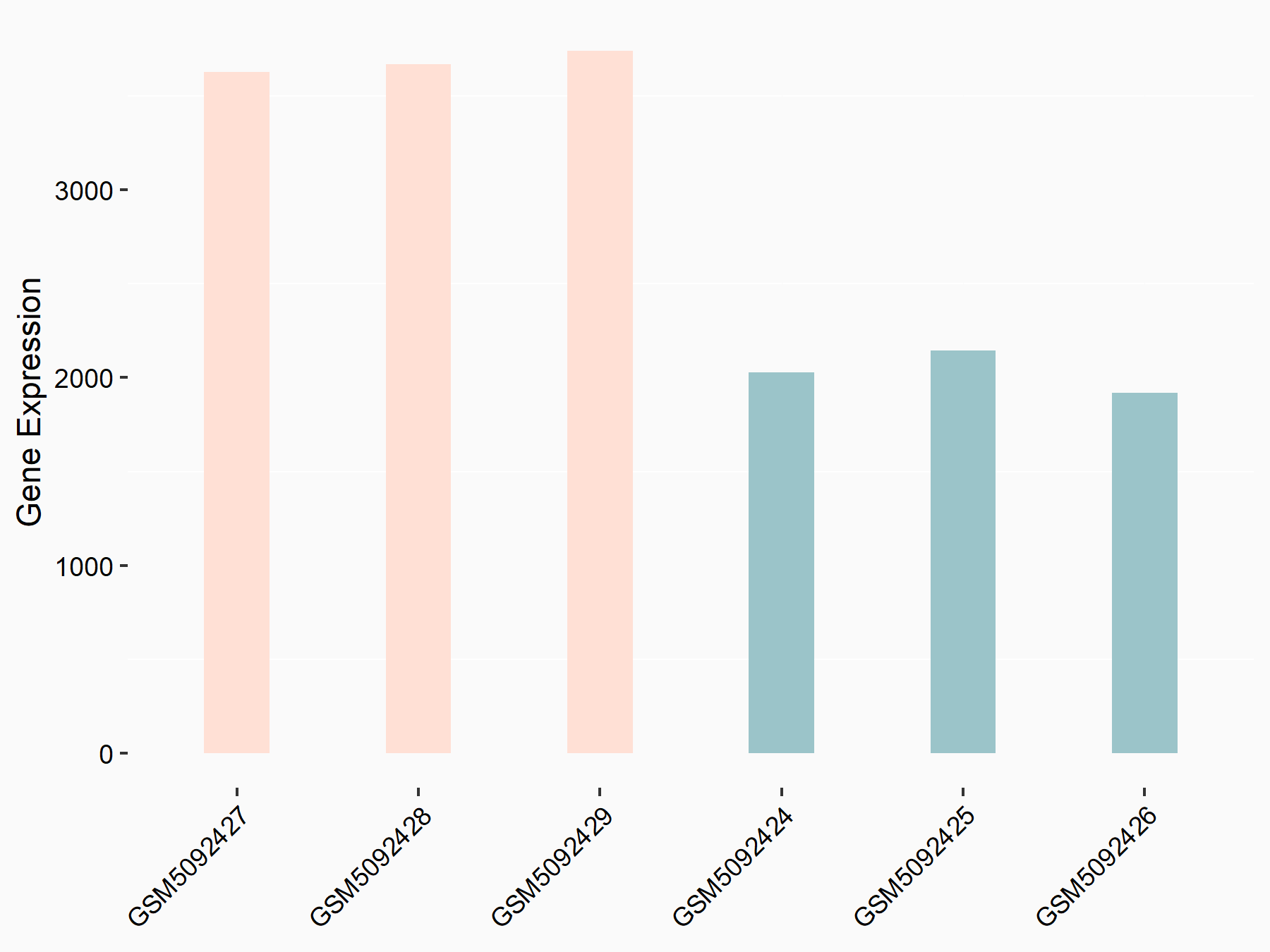 |
logFC: 8.59E-01 p-value: 2.73E-57 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.01E+00 | GSE60213 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [80] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | |||
| Response Summary | This study revealed that m6A methylation is closely related to the poor prognosis of non-small cell lung cancer patients via interference with the TIME, which suggests that m6A plays a role in optimizing individualized immunotherapy management and improving prognosis. The expression levels of METTL3, FTO and YTHDF1 in non-small cell lung cancer were changed. Patients in Cluster 1 had lower immunoscores, higher programmed death-ligand 1 (PD-L1) expression, and shorter overall survival compared to patients in Cluster 2. The hallmarks of the Myelocytomatosis viral oncogene (MYC) targets, Transcription factor E2F1 (E2F1) targets were significantly enriched. | |||
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [116] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Doxil | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
MCF7-DoxR (Adriamycin-resistant cell line MCF7-DoxR) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| In-vivo Model | Once the tumor volume increased to about 1 cm3, six groups of MCF7 bearing mice (n = 10 in each group) were injected with PBS (0.1 ml, caudal vein) and adriamycin (0.1 ml, 10 mg/kg), respectively. When the tumor reached 1.5 cm in any direction (defined as event-free survival analysis), 10 mice in each group were selected to measure the tumor size and weight on the 12th day after adriamycin injection. | |||
| Response Summary | METTL3 can regulate the expression of MALAT1 through m6A, mediate the Transcription factor E2F1 (E2F1)/AGR2 axis, and promote the adriamycin resistance of breast cancer. | |||
Transcription factor EB (TFEB)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LNCaP cell line | Homo sapiens |
|
Treatment: shMETTL3 LNCaP cells
Control: shControl LNCaP cells
|
GSE147884 | |
| Regulation |
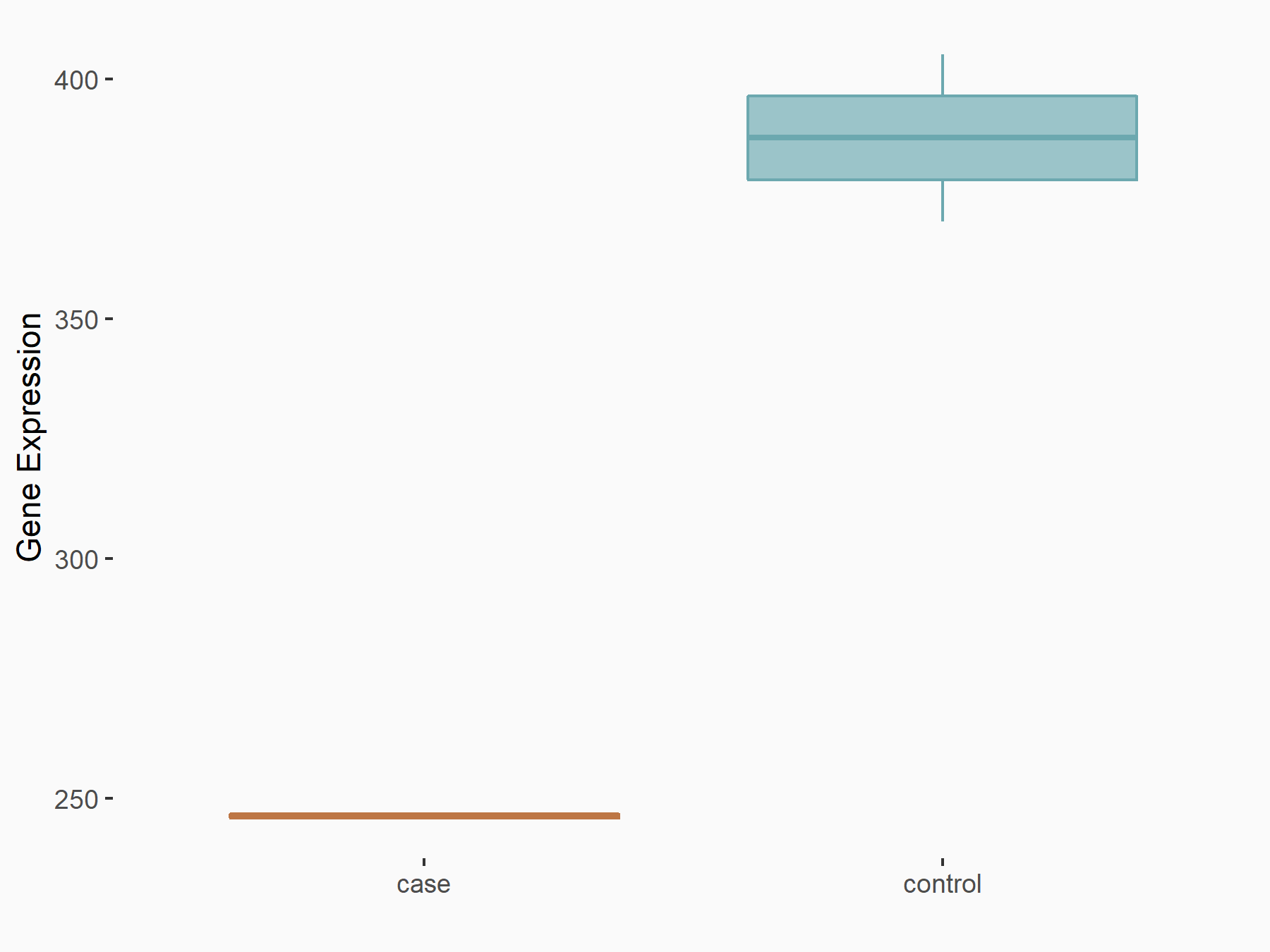 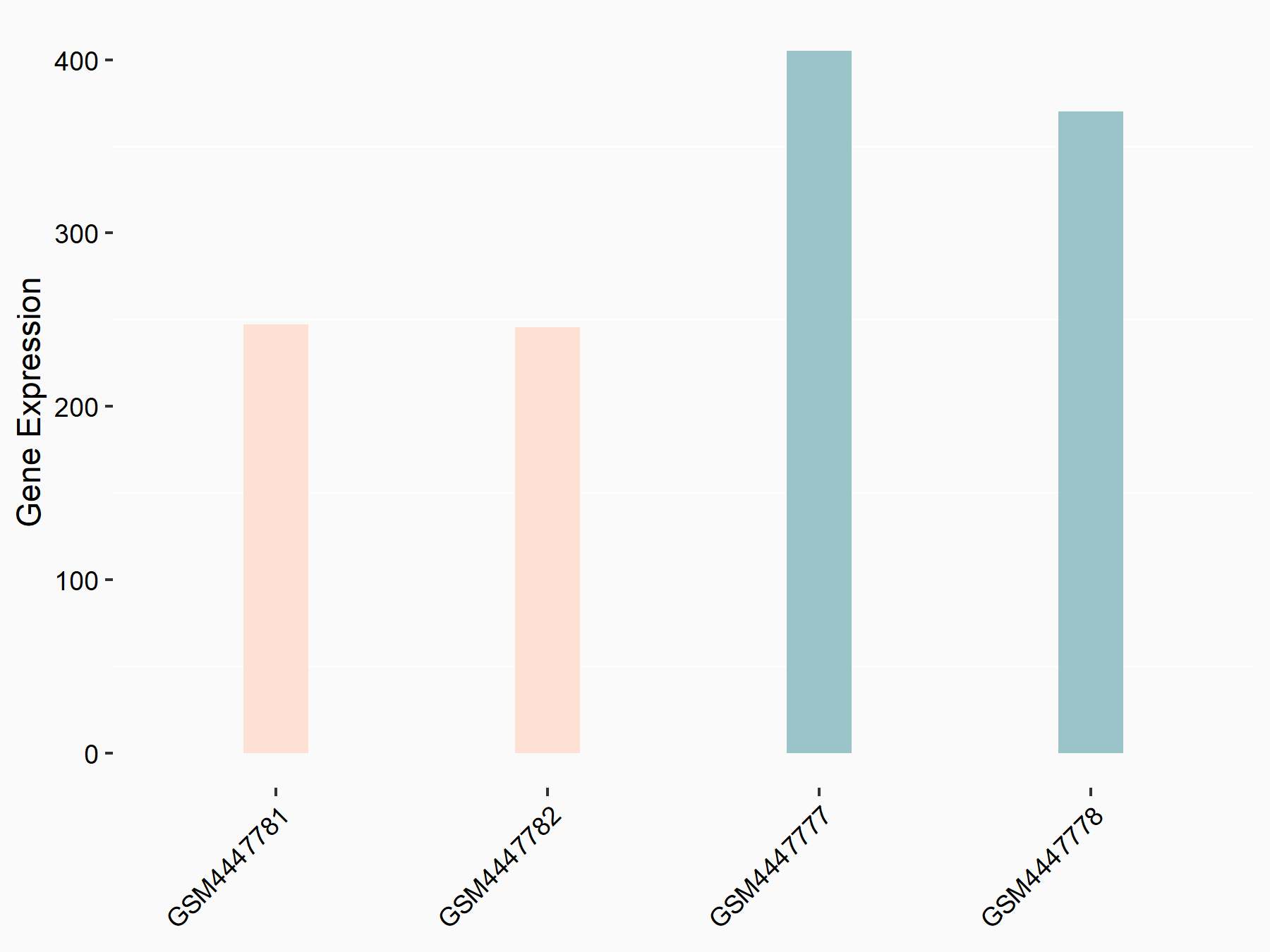 |
logFC: -6.54E-01 p-value: 8.08E-07 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 8.22E+00 | GSE60213 |
Ischemic heart disease [ICD-11: BA40-BA6Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [117] | |||
| Responsed Disease | Ischemic heart disease [ICD-11: BA40-BA6Z] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Apoptosis | hsa04210) | ||
| Cell Process | Cell proliferation | |||
In-vitro Model |
H9c2(2-1) | Normal | Rattus norvegicus | CVCL_0286 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | To cause I/R injury, mice were subjected to 30 min of LAD ischemia followed by 60 min of reperfusion. | |||
| Response Summary | METTL3 methylates Transcription factor EB (TFEB), a master regulator of lysosomal biogenesis and autophagy genes, at two m6A residues in the 3'-UTR, which promotes the association of the RNA-binding protein HNRNPD with TFEB pre-mRNA and subsequently decreases the expression levels of TFEB. METTL3-ALKBH5 and autophagy, providing insight into the functional importance of the reversible mRNA m6A methylation and its modulators in ischemic heart disease. | |||
Transcription factor ISGF-3 components p91/p84 (Stat1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Raw 264.7 cell line | Mus musculus |
|
Treatment: METTL3 knockout Raw 264.7 cells
Control: Wild type Raw 264.7 cells
|
GSE162248 | |
| Regulation |
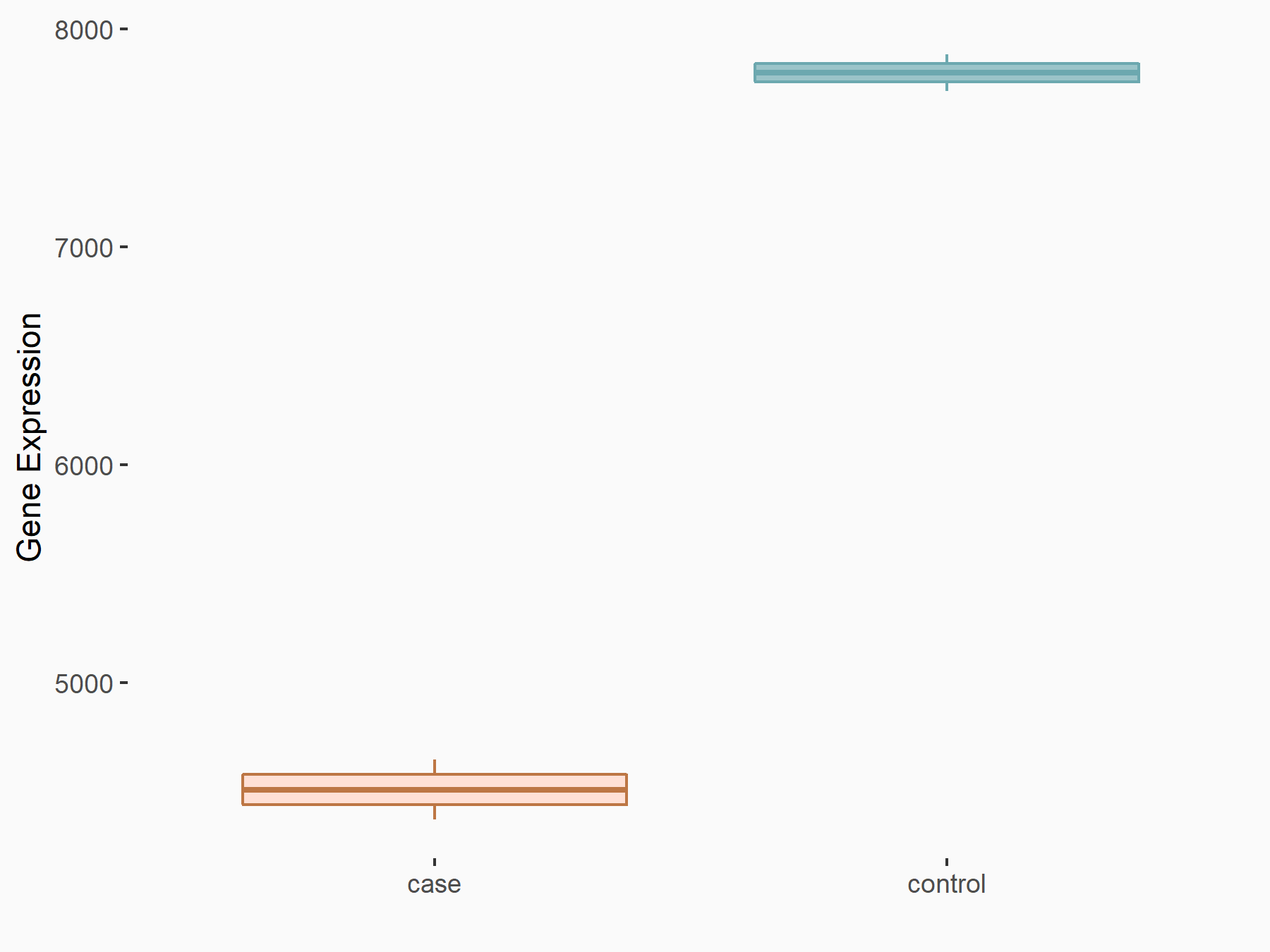 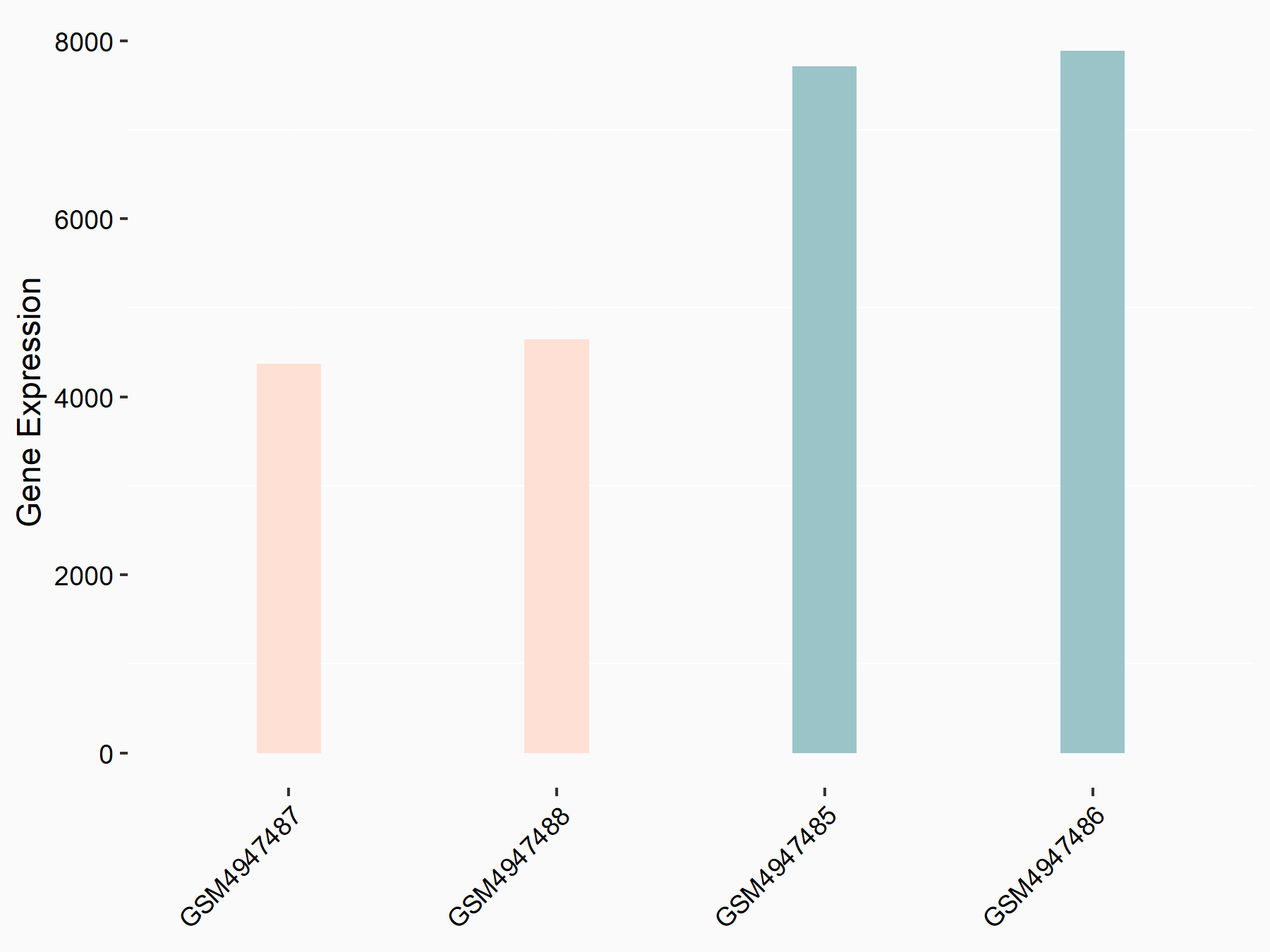 |
logFC: -7.90E-01 p-value: 1.53E-25 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.34E+00 | GSE60213 |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [118] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
| Cell Process | Immunity | |||
In-vitro Model |
CT26 | Mouse colon adenocarcinoma | Mus musculus | CVCL_7254 |
| B16-GM-CSF (B16-GM-CSF cell line was a kind gift from Drs. Glenn Dranoff and Michael Dougan (Dana-Farber/Harvard Cancer Center)) | ||||
| B16-F10 | Mouse melanoma | Mus musculus | CVCL_0159 | |
| In-vivo Model | 2 × 106 CT26 cells with knockout of Mettl3, Mettl14, Mettl3/Stat1, Mettl3/Irf1, Mettl14/Stat1, or Mettl14/Irf1 and control were suspended in 200 uL of PBS/Matrigel (Corning) (1:1) and then subcutaneously inoculated into flank of each mouse. | |||
| Response Summary | In colorectal cancer, Mettl3- or Mettl14-deficient tumors increased cytotoxic tumor-infiltrating CD8+ T cells and elevated secretion of IFN-gamma, Cxcl9, and Cxcl10 in tumor microenvironment in vivo. Mechanistically, Mettl3 or Mettl14 loss promoted IFN-gamma-Stat1-Irf1 signaling through stabilizing the Transcription factor ISGF-3 components p91/p84 (Stat1) and Irf1 mRNA via Ythdf2. | |||
Psoriasis [ICD-11: EA90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [348] | |||
| Responsed Disease | Psoriasis [ICD-11: EA90] | |||
| Target Regulation | Up regulation | |||
Transcription factor JunB (JUNB)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
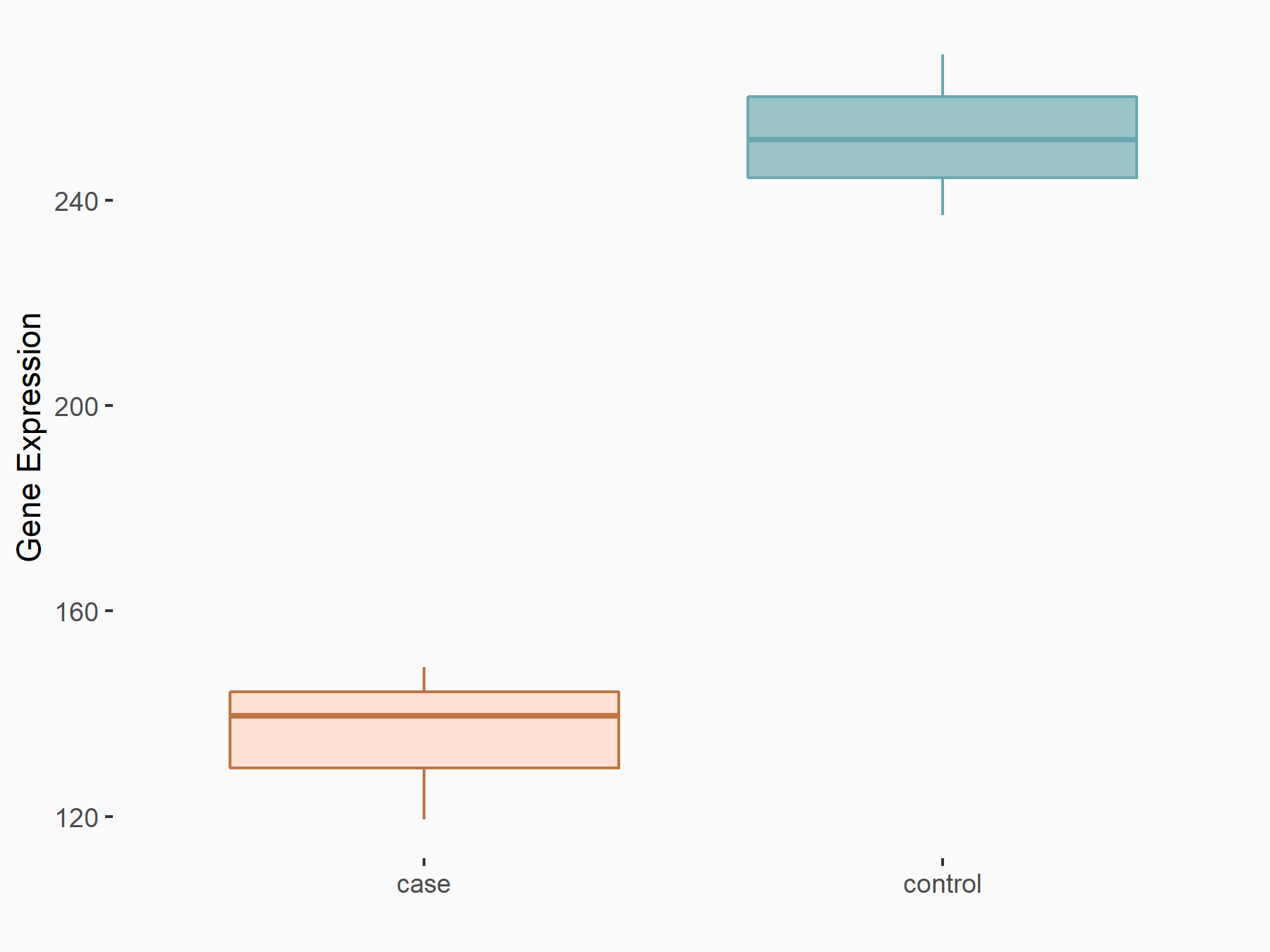 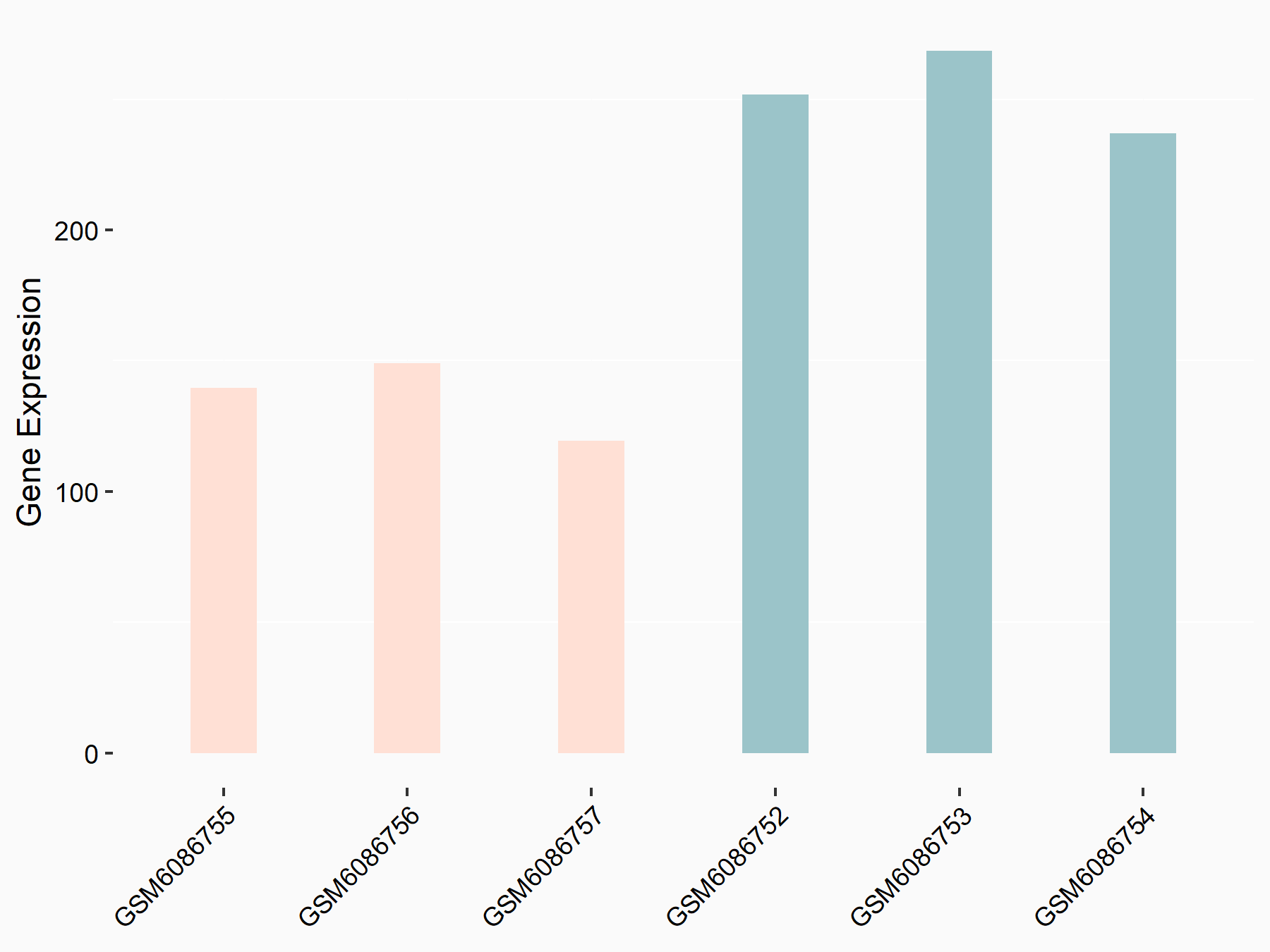 |
logFC: -8.92E-01 p-value: 4.99E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.95E+00 | GSE60213 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [119] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Cytokine-cytokine receptor interaction | hsa04060 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| LC-2/ad | Lung adenocarcinoma | Homo sapiens | CVCL_1373 | |
| Response Summary | m6A methyltransferase METTL3 is indispensable for TGF-beta-induced EMT of lung cancer cells through the regulation of Transcription factor JunB (JUNB). | |||
Transcription factor SOX-2 (SOX2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
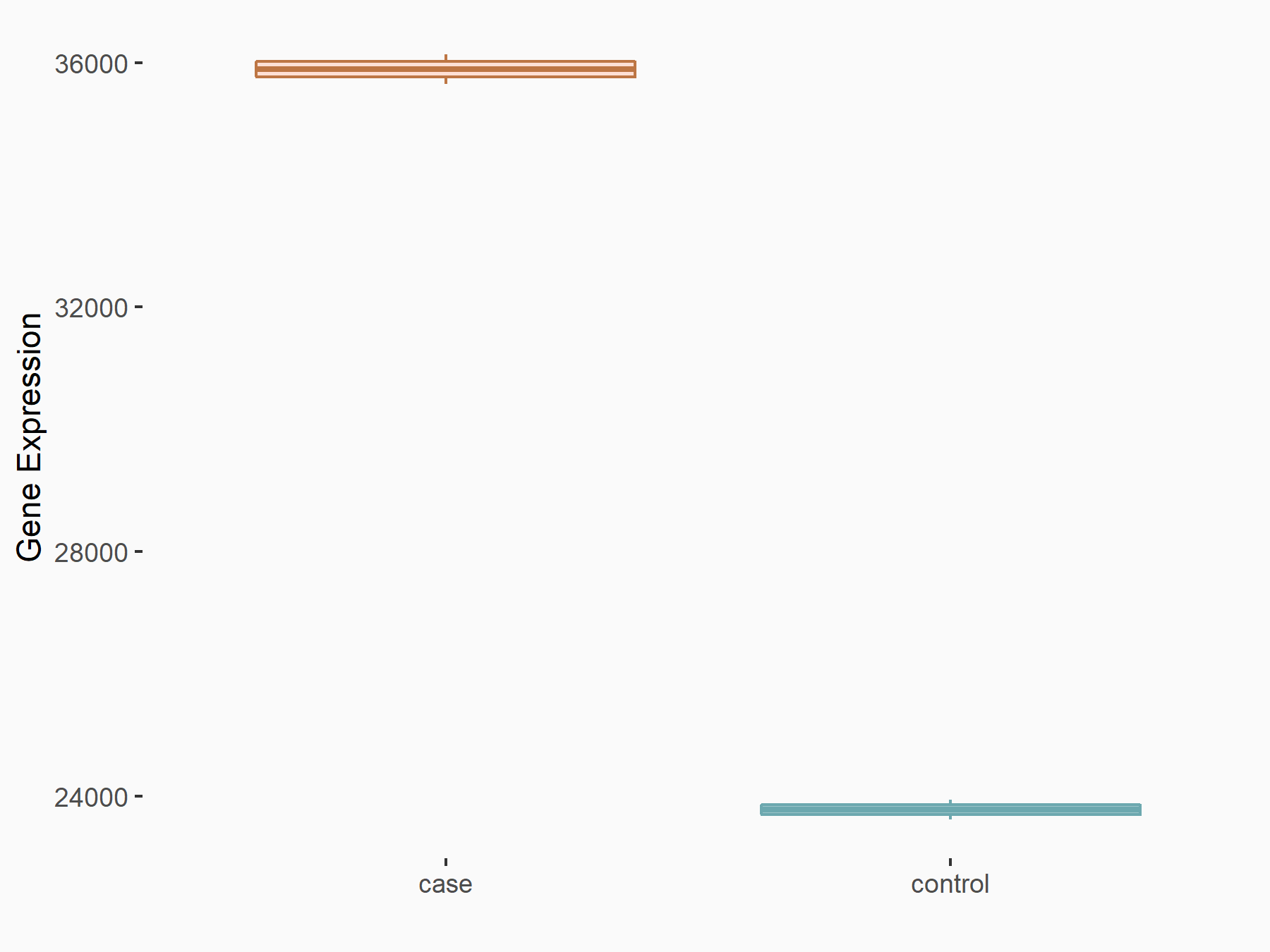 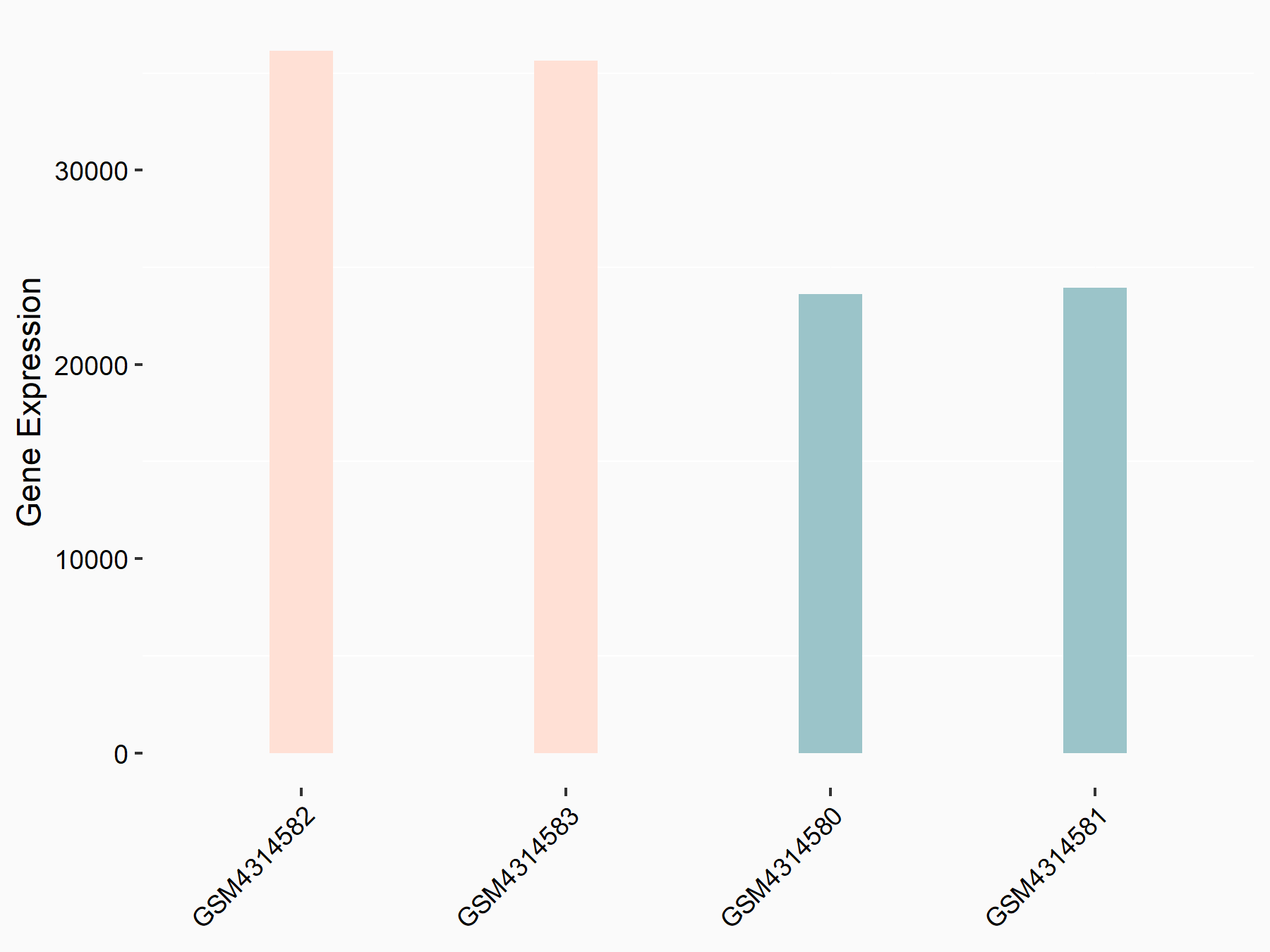 |
logFC: 5.94E-01 p-value: 9.57E-57 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.08E+00 | GSE60213 |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [120] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Signaling pathways regulating pluripotency of stem cells | hsa04550 | ||
| Cell Process | DNA repair | |||
| Nucleotide excision repair (hsa03420) | ||||
In-vitro Model |
Mouse immortalized astrocytes (A type of glial cell) | |||
| Response Summary | GBM tumors have elevated levels of METTL3 transcripts and silencing METTL3 in U87/TIC inhibited tumor growth in an intracranial orthotopic mouse model with prolonged mice survival. The exogenous overexpression of 3'UTR-less Transcription factor SOX-2 (SOX2) significantly alleviated the inhibition of neurosphere formation observed in METTL3 silenced GSCs. | |||
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [121] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Signaling pathways regulating pluripotency of stem cells | hsa04550 | ||
| Cell Process | Cell self-renewal | |||
| Stem cell frequency | ||||
| Cell migration | ||||
In-vitro Model |
CCD-112CoN | Normal | Homo sapiens | CVCL_6382 |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HCT 15 | Colon adenocarcinoma | Homo sapiens | CVCL_0292 | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| LS174T | Colon adenocarcinoma | Homo sapiens | CVCL_1384 | |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| Response Summary | METTL3, acting as an oncogene, maintained Transcription factor SOX-2 (SOX2) expression through an m6A-IGF2BP2-dependent mechanism in CRC cells, and indicated a potential biomarker panel for prognostic prediction in Colorectal carcinoma. | |||
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [122] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| Response Summary | Knockdown of METTL3 downregulated protein levels of Transcription factor SOX-2 (SOX2), CD133 and CD44 in MCF-7 cells. METTL3 is upregulated in breast cancer, and it promotes the stemness and malignant progression of BCa through mediating m6A modification on SOX2 mRNA. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [349] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
Cognitive disorders [ICD-11: 6E0Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [350] | |||
| Responsed Disease | Cognitive disorders [ICD-11: 6E0Z] | |||
| Responsed Drug | LPS | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
Transcriptional coactivator YAP1 (YAP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LNCaP cell line | Homo sapiens |
|
Treatment: shMETTL3 LNCaP cells
Control: shControl LNCaP cells
|
GSE147884 | |
| Regulation |
 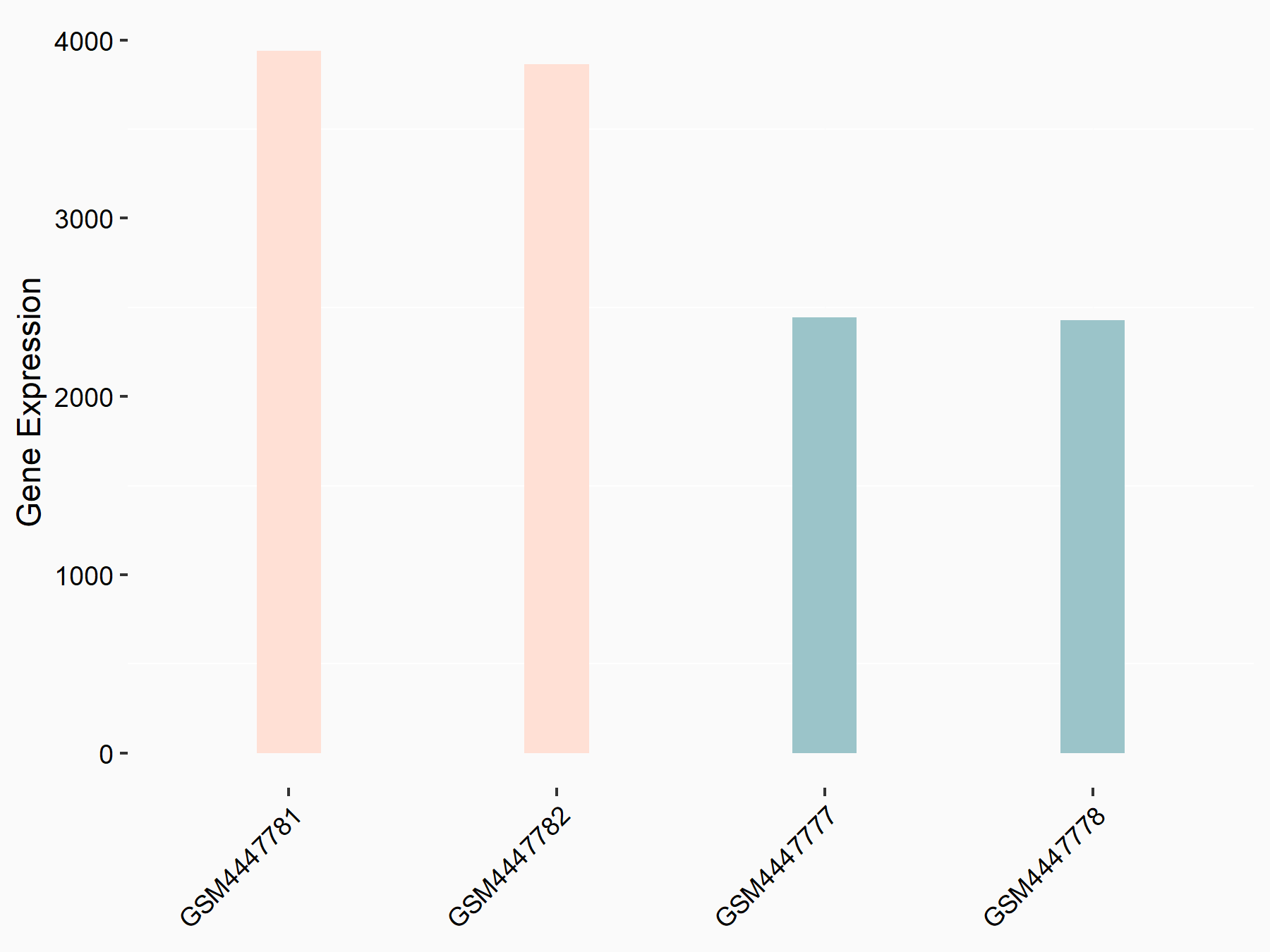 |
logFC: 6.80E-01 p-value: 6.99E-58 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.22E+00 | GSE60213 |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [123] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell metastasis | ||||
In-vitro Model |
MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| Response Summary | The expression of m6A and METTL3 was upregulated in human gastric cancer tissues and gastric cancer cell lines. m6A methyltransferase METTL3 promoted the proliferation and migration of gastric cancer cells through the m6A modification of Transcriptional coactivator YAP1 (YAP1). | |||
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [124] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Cell invasion | |||
| Cell migration | ||||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| In-vivo Model | BALB/c nude mice (4 weeks old) were acquired from Vital River Laboratory (Beijing, China). HCT116 cells with stable circ1662 expression (2 × 106 in 100 L of PBS) were injected via the tail vein. After 45 days, the mice were sacrificed. The lung metastatic carcinoma specimens were processed into paraffin-embedded sections for subsequent H&E staining and IHC. | |||
| Response Summary | METTL3-induced circ1662 promoted colorectal cancer cell invasion and migration by accelerating Transcriptional coactivator YAP1 (YAP1) nuclear transport. Circ1662 enhanced CRC invasion and migration depending on YAP1 and SMAD3. This result implies that circ1662 is a new prognostic and therapeutic marker for CRC metastasis. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [125] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Cell migration and invasion | |||
In-vitro Model |
Homo sapiens (SK-HEP-1-Luc (luciferase labeled) cells were obtained from OBIO (Shanghai, China).) | |||
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | 1 × 107 SK-HEP-1-Luc-shControl or SK-HEP-1-Luc-shMETTL3 stable cells were suspended in 300 uL of PBS and injected orthotopically into the left liver lobe of nude mice. | |||
| Response Summary | m6A methylation plays a key role in VM formation in HCC. METTL3 and Transcriptional coactivator YAP1 (YAP1) could be potential therapeutic targets via impairing VM formation in anti-metastatic strategies. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [126] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Metabolic | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-6 | Lung adenocarcinoma | Homo sapiens | CVCL_0236 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | Mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then administrated with PBS or DDP (3 mg/kg per day) for three times a week, and tumor volume were measured every second day. | |||
| Response Summary | METTL3, YTHDF3, YTHDF1, and eIF3b directly promoted YAP translation through an interaction with the translation initiation machinery. METTL3 knockdown inhibits tumor growth and enhances sensitivity to DDP in vivo.m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-Transcriptional coactivator YAP1 (YAP1) axis to induce Non-small cell lung cancer drug resistance and metastasis. | |||
Triple-negative breast cancer [ICD-11: 2C6Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [351] | |||
| Responsed Disease | Triple-negative breast cancer [ICD-11: 2C6Z] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 |
|
WI-38 VA13 subline 2RA
|
N.A. | Homo sapiens | CVCL_2759 | |
| CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Injuries of spine or trunk [ICD-11: ND51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [352] | |||
| Responsed Disease | Spinal cord injury [ICD-11: ND51.2] | |||
| Target Regulation | Up regulation | |||
Translocation protein SEC62 (SEC62)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | CT26 cell line | Mus musculus |
|
Treatment: METTL3 knockout CT26 cells
Control: CT26 cells
|
GSE142589 | |
| Regulation |
 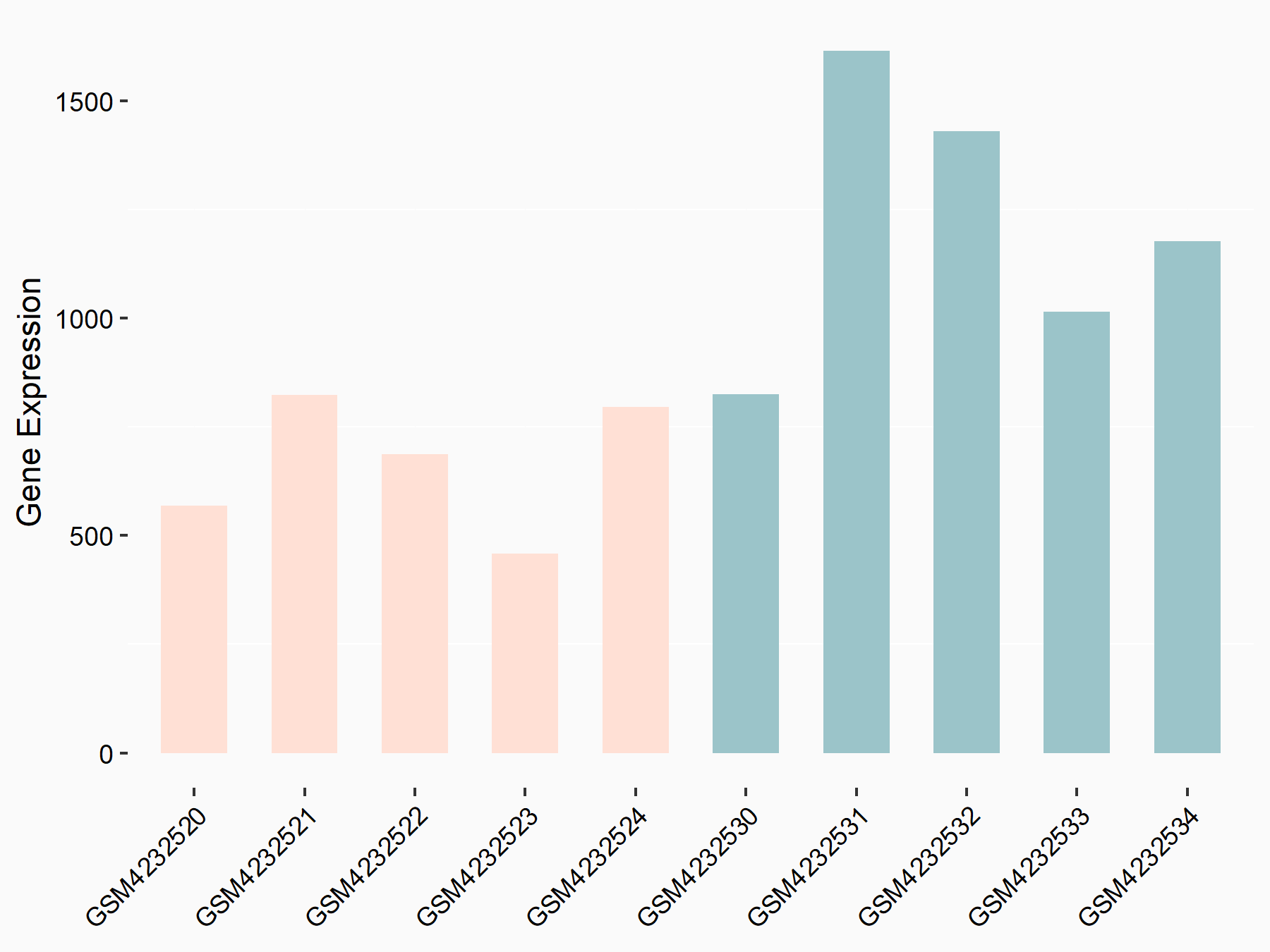 |
logFC: -8.61E-01 p-value: 1.21E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.99E+00 | GSE60213 |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [127] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Protein processing in endoplasmic reticulum | hsa04141 | ||
| Cell Process | RNA stability | |||
| Cell apoptosis | ||||
In-vitro Model |
GES-1 | Normal | Homo sapiens | CVCL_EQ22 |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| Response Summary | MiR-4429 prevented gastric cancer progression through targeting METTL3 to inhibit m6A-caused stabilization of Translocation protein SEC62 (SEC62), indicating miR-4429 as a promising target for treatment improvement for Gastric cancer. METTL3 interacted with SEC62 to induce the m6A on SEC62 mRNA, therefore facilitated the stabilizing effect of IGF2BP1 on SEC62 mRNA. | |||
Colorectal cancer [ICD-11: 2B91]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [128] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Fluorouracil | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Protein degradation | |||
In-vitro Model |
DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | DLD-1 cells were subcutaneously implanted into 4-6 weeks old female nude mice. When tumors reached a size of about 50 mm3, the nude mice were randomly divided into 6 groups. | |||
| Response Summary | Translocation protein SEC62 (SEC62) upregulated by the METTL3-mediated m6A modification promotes the stemness and chemoresistance of colorectal cancer by binding to beta-catenin and enhancing Wnt signalling. Depletion of Sec62 sensitized the CRC cells to 5-Fu or oxaliplatin treatment. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [128] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Oxaliplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Protein degradation | |||
In-vitro Model |
DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | DLD-1 cells were subcutaneously implanted into 4-6 weeks old female nude mice. When tumors reached a size of about 50 mm3, the nude mice were randomly divided into 6 groups. | |||
| Response Summary | Translocation protein SEC62 (SEC62) upregulated by the METTL3-mediated m6A modification promotes the stemness and chemoresistance of colorectal cancer by binding to beta-catenin and enhancing Wnt signalling. Depletion of Sec62 sensitized the CRC cells to 5-Fu or oxaliplatin treatment. | |||
Tumor necrosis factor (TNF/TNF-alpha)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Raw 264.7 cell line | Mus musculus |
|
Treatment: METTL3 knockout Raw 264.7 cells
Control: Wild type Raw 264.7 cells
|
GSE162248 | |
| Regulation |
 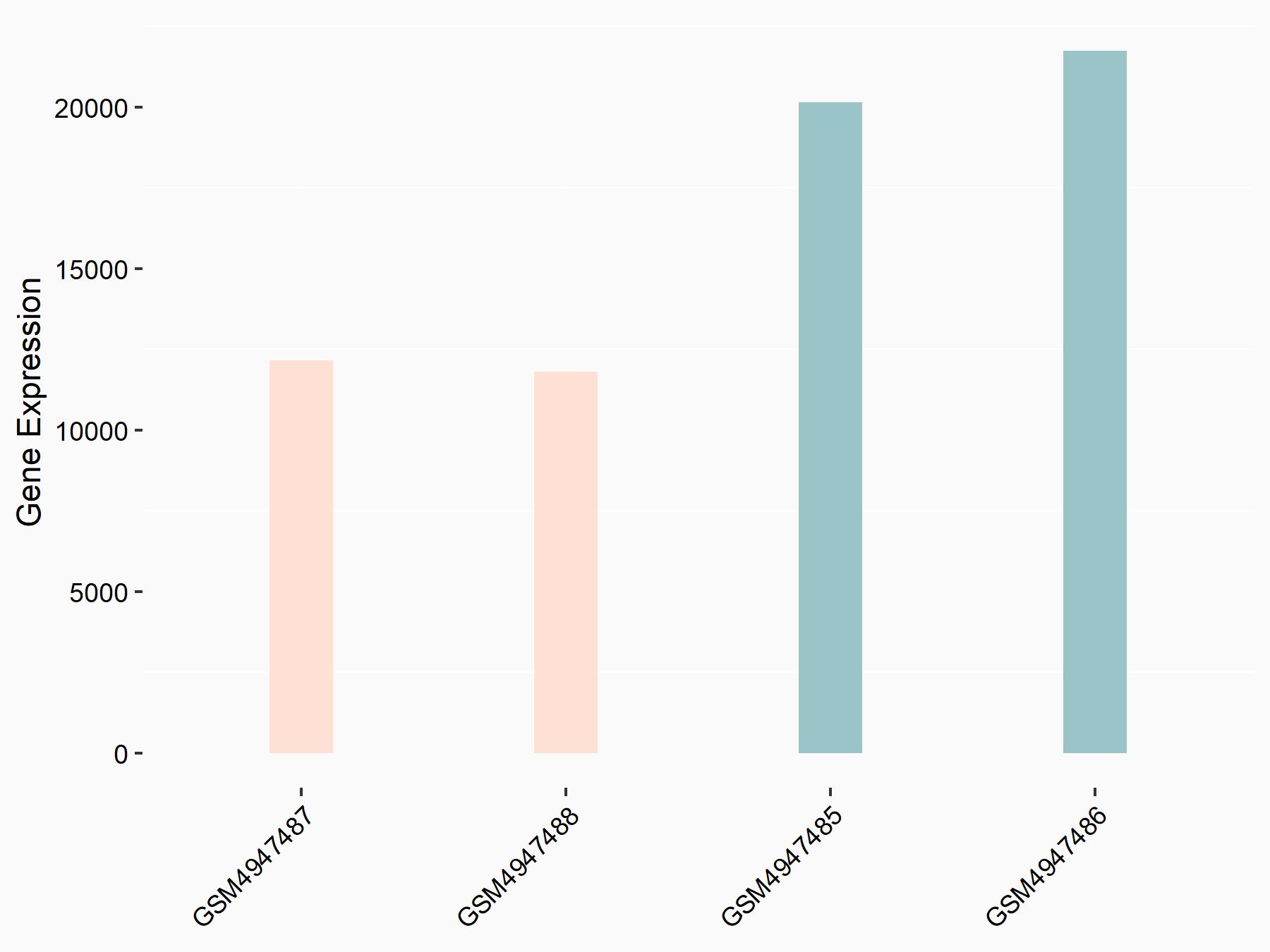 |
logFC: -8.06E-01 p-value: 1.88E-29 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 4.17E+00 | GSE60213 |
B-cell lymphomas [ICD-11: 2A86]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [10] | |||
| Responsed Disease | B-cell lymphomas [ICD-11: 2A86] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
In-vitro Model |
ATDC-5 | Mouse teratocarcinoma | Mus musculus | CVCL_3894 |
| In-vivo Model | For MIA + SAH control, S-adenosylhomocysteine (SAH), Mettl3 inhibitor (10 mg/kg) (MCE, NJ, USA) was injected intraperitoneally before MIA injection and maintained twice a week until mice were sacrificed. | |||
| Response Summary | Mettl3 inhibitor, S-adenosylhomocysteine promoted the apoptosis and autophagy of chondrocytes with inflammation in vitro and aggravated the degeneration of chondrocytes and subchondral bone in monosodium iodoacetate (MIA) induced temporomandibular joint osteoarthritis mice in vivo. Bcl2 protein interacted with Beclin1 protein in chondrocytes induced by Tumor necrosis factor (TNF/TNF-alpha) stimulation. Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through m6A/Ythdf1/Bcl2 signal axis which provides promising therapeutic strategy for temporomandibular joint osteoarthritis. | |||
Dentofacial anomalies [ICD-11: DA0E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [10] | |||
| Responsed Disease | Temporomandibular joint disorders [ICD-11: DA0E.8] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
In-vitro Model |
ATDC-5 | Mouse teratocarcinoma | Mus musculus | CVCL_3894 |
| In-vivo Model | For MIA + SAH control, S-adenosylhomocysteine (SAH), Mettl3 inhibitor (10 mg/kg) (MCE, NJ, USA) was injected intraperitoneally before MIA injection and maintained twice a week until mice were sacrificed. | |||
| Response Summary | Mettl3 inhibitor, S-adenosylhomocysteine promoted the apoptosis and autophagy of chondrocytes with inflammation in vitro and aggravated the degeneration of chondrocytes and subchondral bone in monosodium iodoacetate (MIA) induced temporomandibular joint osteoarthritis mice in vivo. Bcl2 protein interacted with Beclin1 protein in chondrocytes induced by Tumor necrosis factor (TNF/TNF-alpha) stimulation. Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through m6A/Ythdf1/Bcl2 signal axis which provides promising therapeutic strategy for temporomandibular joint osteoarthritis. | |||
Tumor protein 63 (TP63)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
 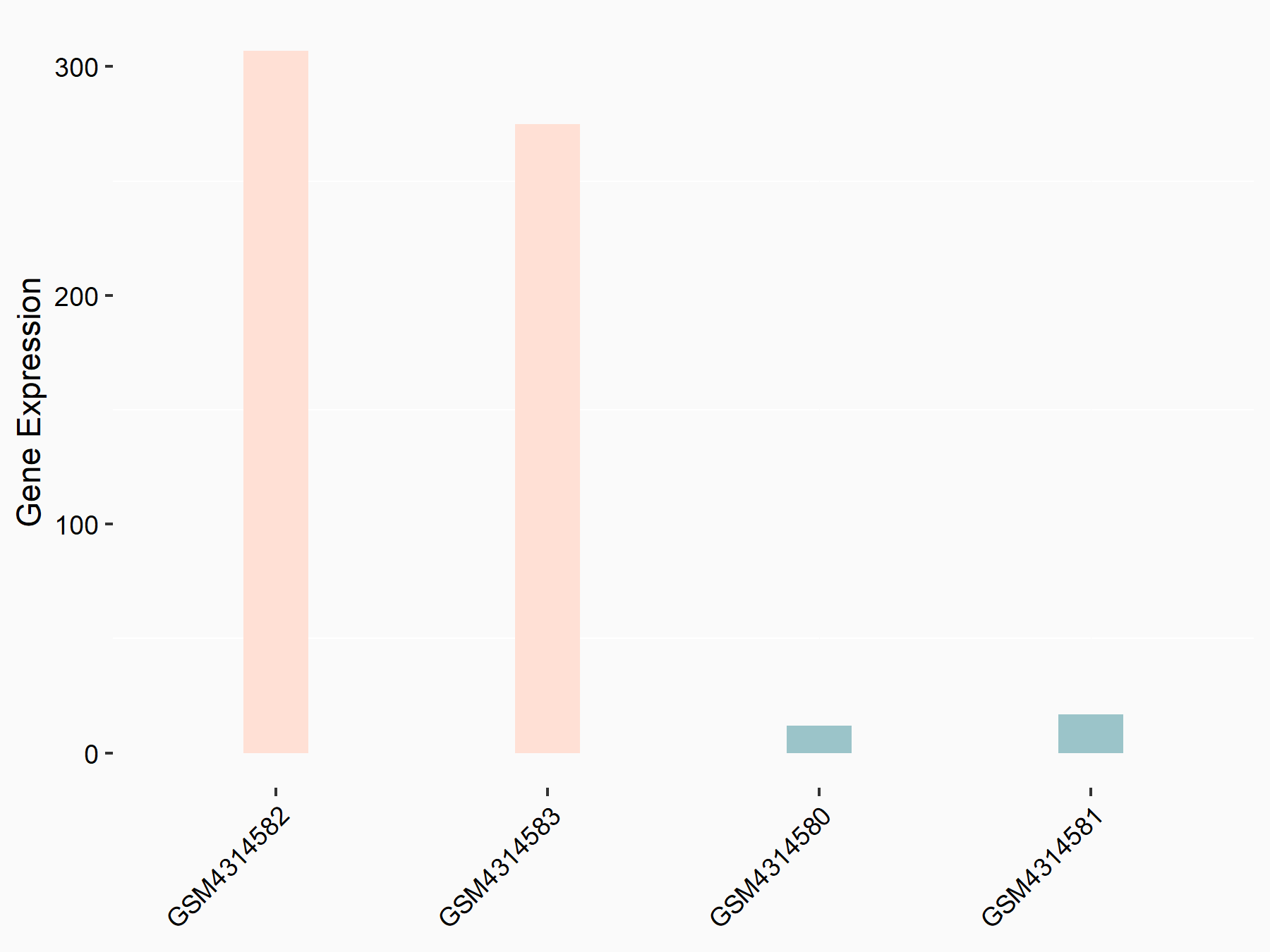 |
logFC: 4.35E+00 p-value: 1.15E-34 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.67E+00 | GSE60213 |
Cutaneous squamous cell carcinoma [ICD-11: 2C31]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [129] | |||
| Responsed Disease | Cutaneous squamous cell carcinoma [ICD-11: 2C31.Z] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Signaling pathways regulating pluripotency of stem cells | hsa04550 | ||
| Cell Process | Cell proliferation | |||
In-vitro Model |
A-431 | Skin squamous cell carcinoma | Homo sapiens | CVCL_0037 |
| HSC-1 | Skin squamous cell carcinoma | Homo sapiens | CVCL_2807 | |
| In-vivo Model | 2 × 106 cells were suspended in 150 uL DMEM. The A431 cells (scrambled group and shRNA1 group) were injected subcutaneously into the flanks of nude mice. | |||
| Response Summary | METTL3 knock down and methylation inhibitor cycloleucine could decrease the m6A levels and the expression of DeltaNp63 in Cutaneous squamous cell carcinoma. | |||
Tyrosine-protein kinase JAK2 (JAK2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Raw 264.7 cell line | Mus musculus |
|
Treatment: METTL3 knockout Raw 264.7 cells
Control: Wild type Raw 264.7 cells
|
GSE162248 | |
| Regulation |
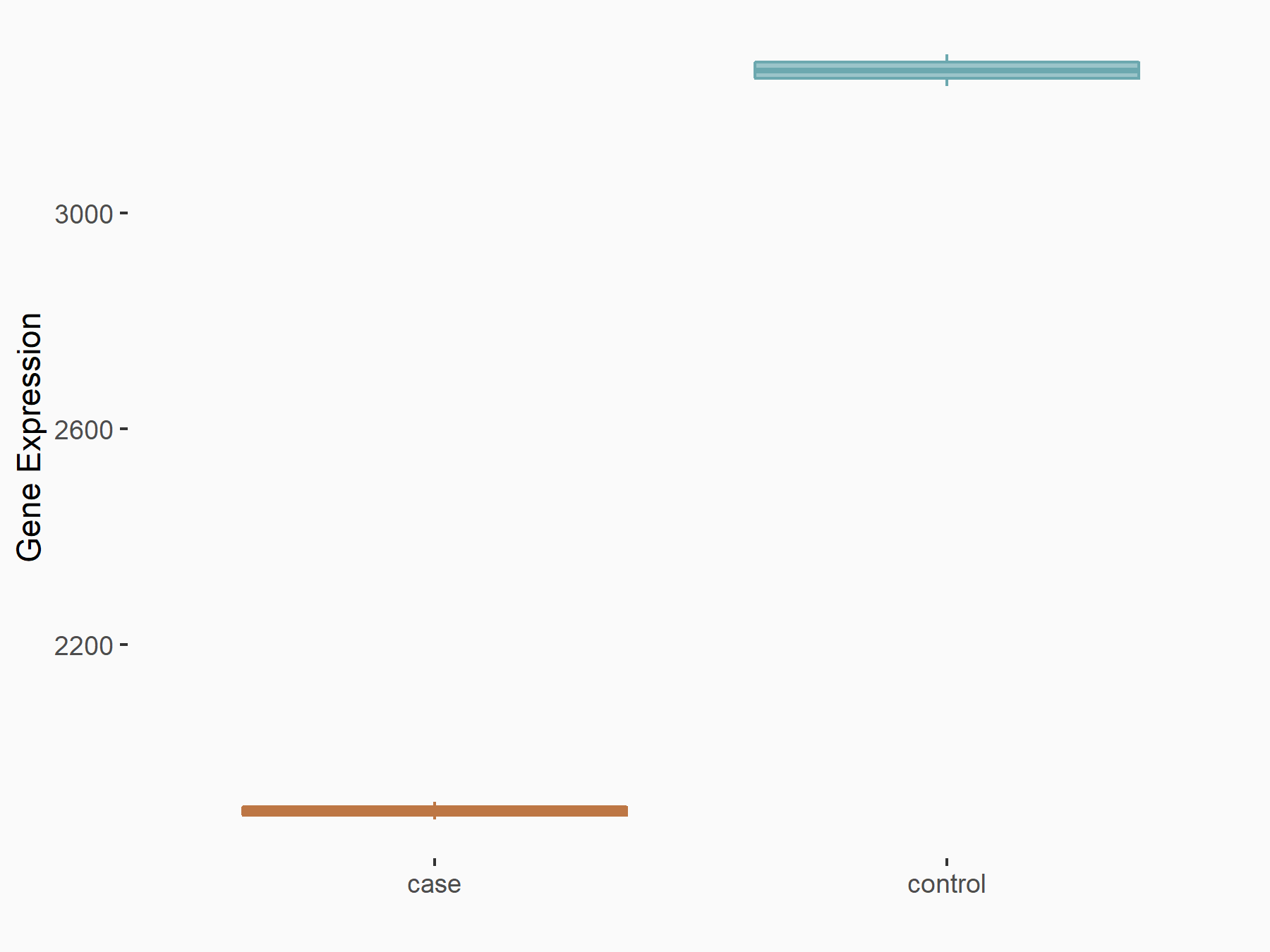 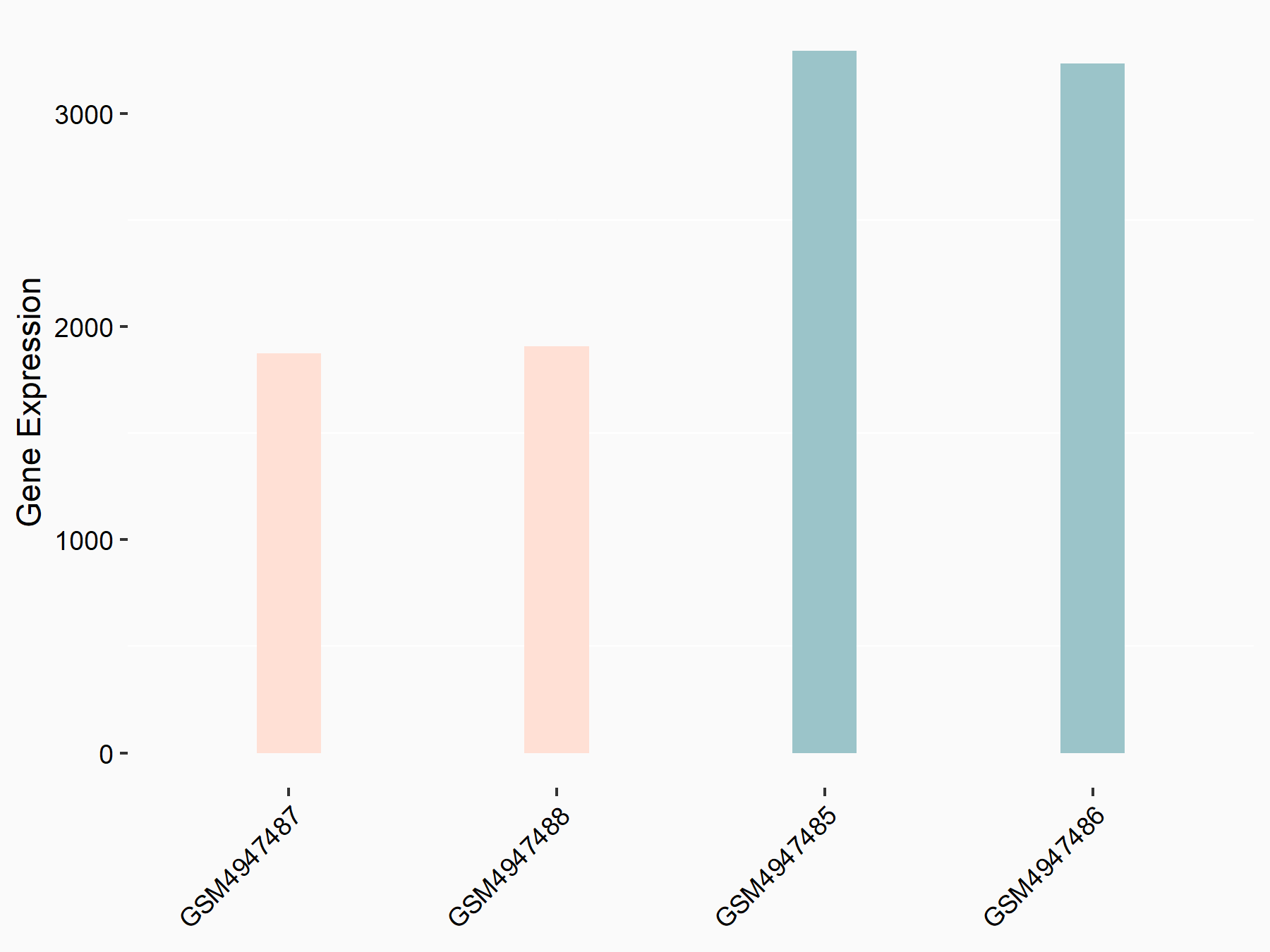 |
logFC: -7.87E-01 p-value: 4.26E-20 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.70E+00 | GSE60213 |
Atherosclerosis [ICD-11: BD40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [130] | |||
| Responsed Disease | Atherosclerosis [ICD-11: BD40.Z] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
| Cell Process | Cell proliferation and migration | |||
In-vitro Model |
HUVEC-C | Normal | Homo sapiens | CVCL_2959 |
| In-vivo Model | The adeno-associated viruses (AAV) that could silence METTL3 (sh-METTL3) and the negative control adeno-associated viruses (sh-NC) were obtained from WZ Biosciences Inc. (Jinan, China). APOE-/- mice were randomly divided into AS + sh-NC and AS + sh-METTL3 groups. Each group contains five mice. Mice were fed with the standard diet for 1 week to acclimatize. After 1 week of acclimation, mice were challenged with a high-fat and high-cholesterol feed H10540 (Beijing HFK BIOSCIENCE Co., Ltd., Beijing, China). The formula of the H10540 feed was shown in Supplementary File S1. After 8 weeks of HFD feeding, sh-NC or sh-METTL3 adeno-associated virus serotype 9 (AAV9, 1012 viral genome copies per mouse) were respectively delivered into mice in AS + sh-NC or AS + sh-METTL3 group through tail vein injection. At 14 weeks after HDF feeding, mice fasted overnight. | |||
| Response Summary | METTL3 knockdown prevented Atherosclerosis progression by inhibiting Tyrosine-protein kinase JAK2 (JAK2)/STAT3 pathway via IGF2BP1. | |||
Ubiquitin-like modifier-activating enzyme ATG7 (ATG7)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
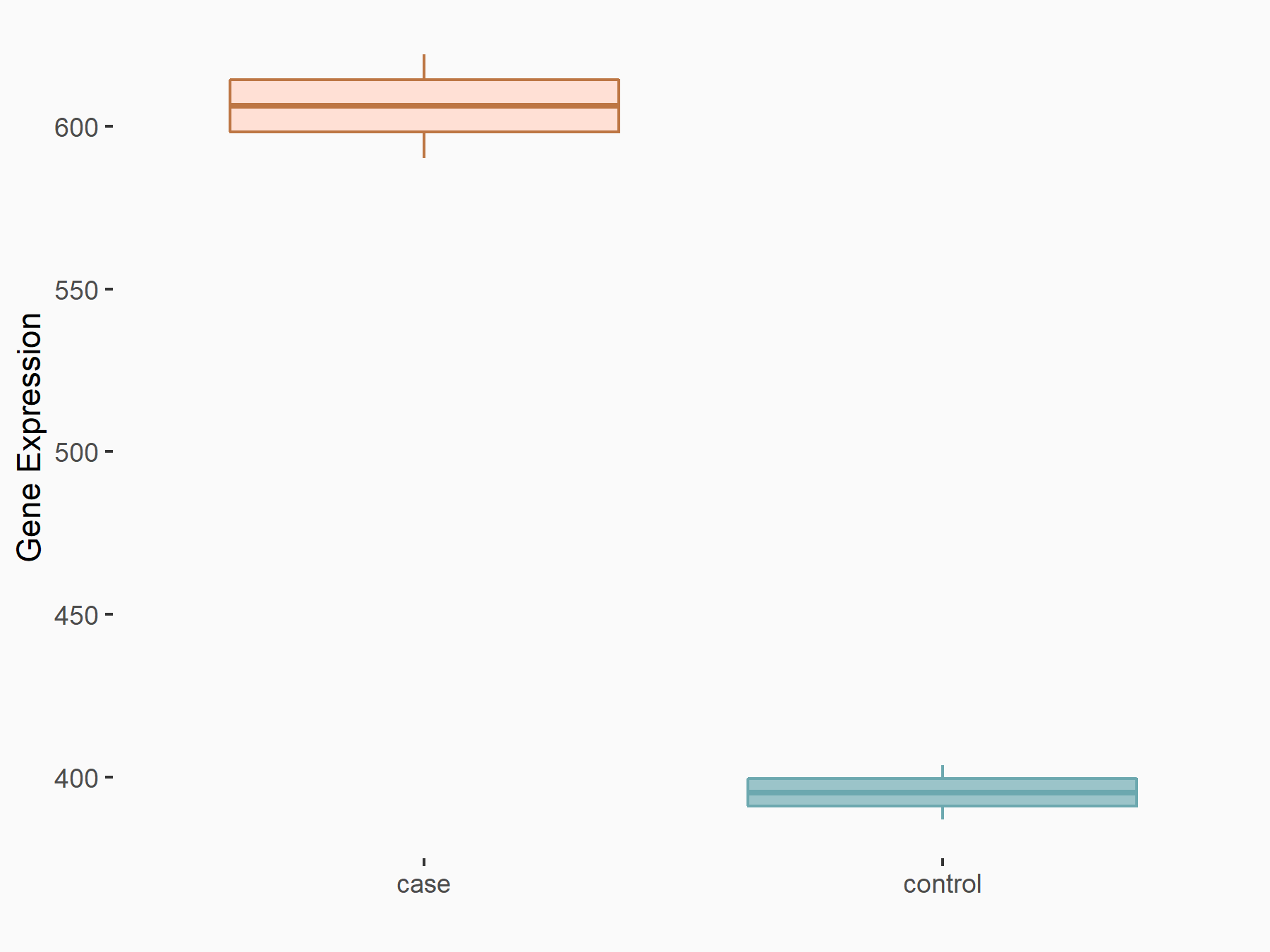 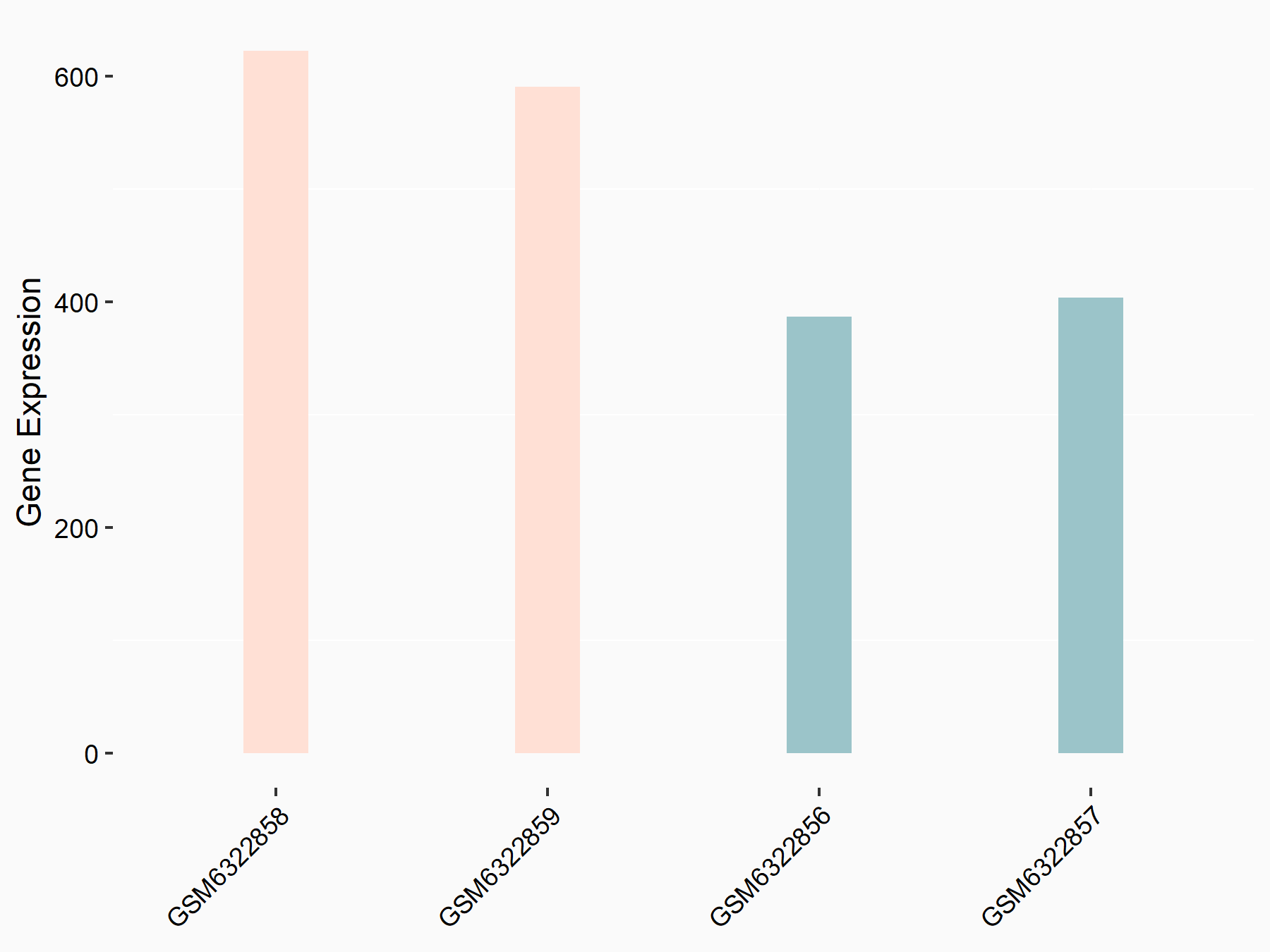 |
logFC: 6.17E-01 p-value: 3.67E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 4.77E+00 | GSE60213 |
Enterovirus [ICD-11: 1A2Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Enterovirus [ICD-11: 1A2Y] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| Cell autophagy | ||||
In-vitro Model |
Schwann cells (A type of glial cell that surrounds neurons) | |||
| Response Summary | Knocking down METTL3 prevented Enterovirus 71-induced cell death and suppressed Enterovirus 71-induced expression of Bax while rescuing Bcl-2 expression after Enterovirus 71 infection. Knocking down METTL3 inhibited Enterovirus 71-induced expression of Atg5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7) and LC3 II. Knocking down METTL3 inhibited Enterovirus 71-induced apoptosis and autophagy. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [17] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), ATG12, and ATG16L1. | |||
Lung cancer [ICD-11: 2C25]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Chloroquine | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
In-vitro Model |
Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Gefitinib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
In-vitro Model |
Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Experiment 3 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Beta-Elemen | Phase 3 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
In-vitro Model |
Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [131] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular senescence | |||
| Cell autophagy | ||||
In-vitro Model |
C-28/I2 | Normal | Homo sapiens | CVCL_0187 |
| FLS (Rat fibroblast synovial cell line) | ||||
| In-vivo Model | Mice were anaesthetized with isoflurane supplied in a mouse anaesthesia apparatus, followed with joint surgery on the right joint by sectioning the medial meniscotibial ligament. | |||
| Response Summary | In osteoarthritis METTL3-mediated m6A modification decreased the expression of autophagy-related 7, an E-1 enzyme crucial for the formation of autophagosomes, by attenuating its RNA stability. Silencing METTL3 enhanced autophagic flux and inhibited Ubiquitin-like modifier-activating enzyme ATG7 (ATG7) expression in OA-FLS. | |||
Ubiquitin-like protein ATG12 (ATG12)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Mouse testis | Mus musculus |
|
Treatment: Mettl3 knockout mouse testis
Control: Mouse testis
|
GSE99771 | |
| Regulation |
  |
logFC: -2.94E+00 p-value: 6.14E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.85E+00 | GSE60213 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [17] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, ATG7, Ubiquitin-like protein ATG12 (ATG12), and ATG16L1. | |||
Zinc finger and BTB domain-containing protein 4 (ZBTB4)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 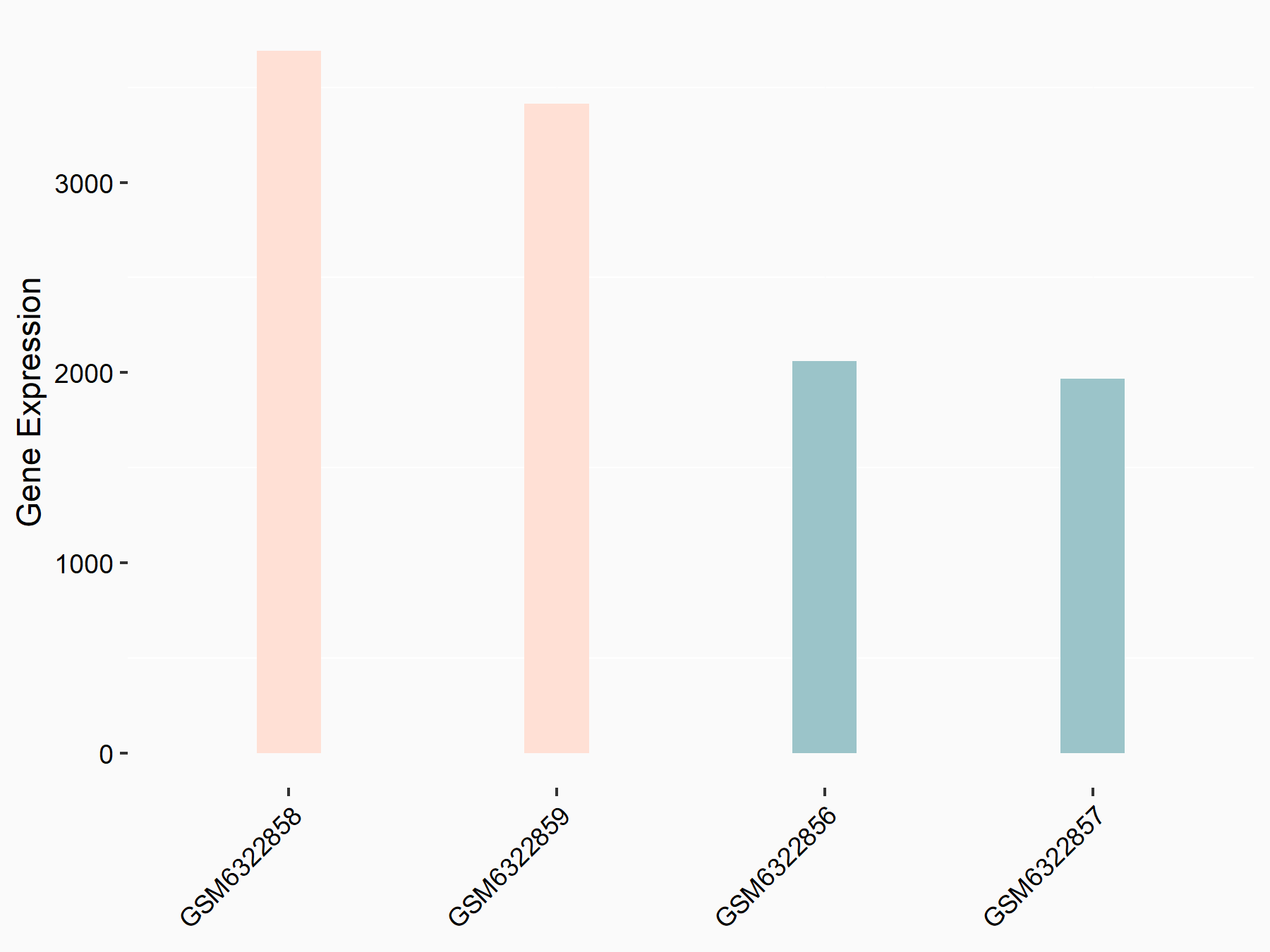 |
logFC: 8.19E-01 p-value: 3.59E-20 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.32E+00 | GSE60213 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [132] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
HBE (Human bronchial epithelial cell line) | |||
| In-vivo Model | Male BALB/c mice were grouped into control, CS, CS+AAV-ShMETTL3, and CS+AAV-NC shRNA groups, n = 6 animals per group. Mice in the AAV-ShMETTL3 and AAV-NC shRNA (GeneChem, China) groups were dosed by intranasal instillation after CS exposure for 4 weeks. | |||
| Response Summary | METTL3-mediated m6A modification of Zinc finger and BTB domain-containing protein 4 (ZBTB4) via EZH2 is involved in the CS-induced EMT and in lung cancer. | |||
Zinc finger MYM-type protein 1 (ZMYM1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3 knockout liver
Control: Wild type liver cells
|
GSE198513 | |
| Regulation |
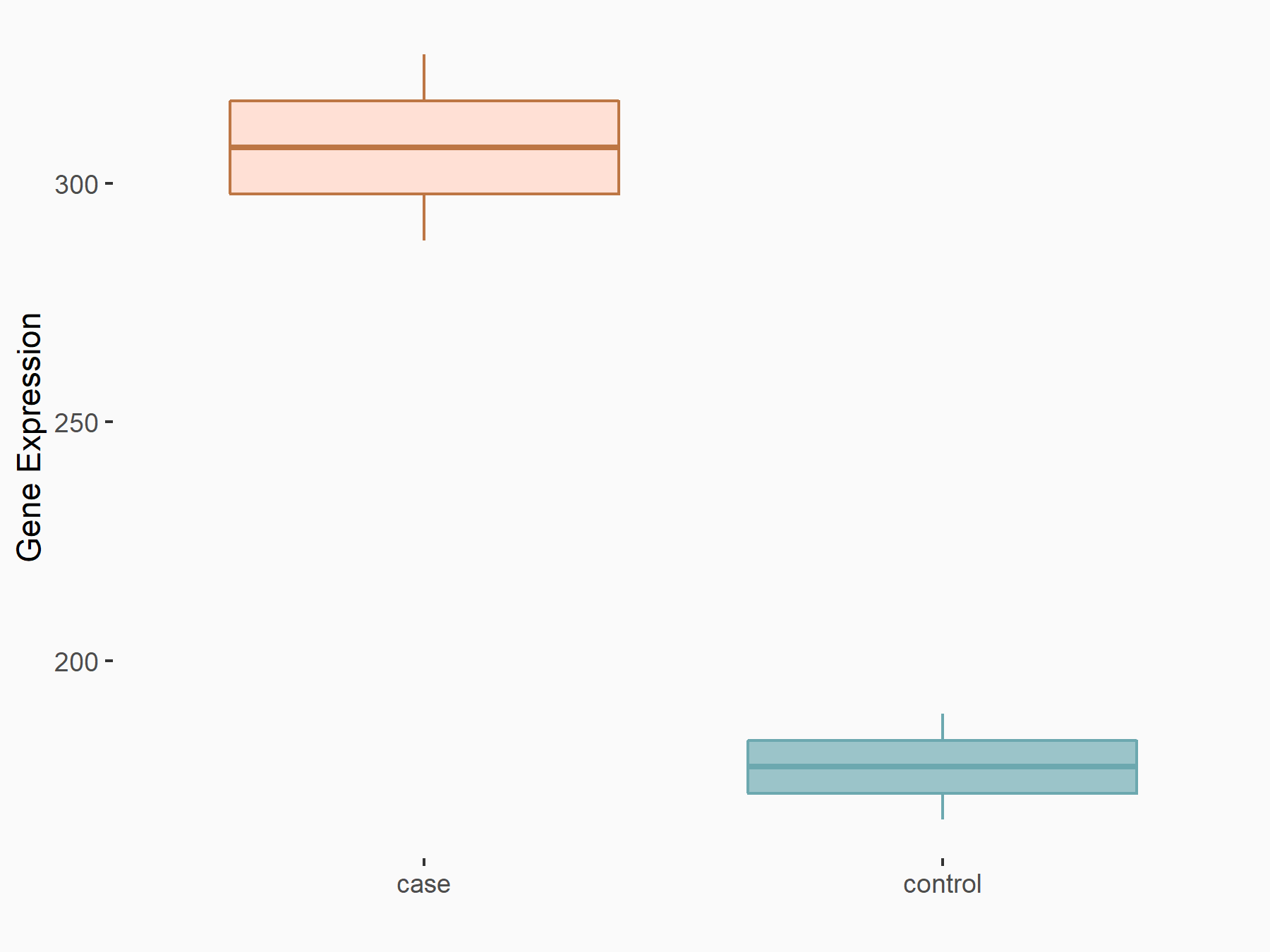 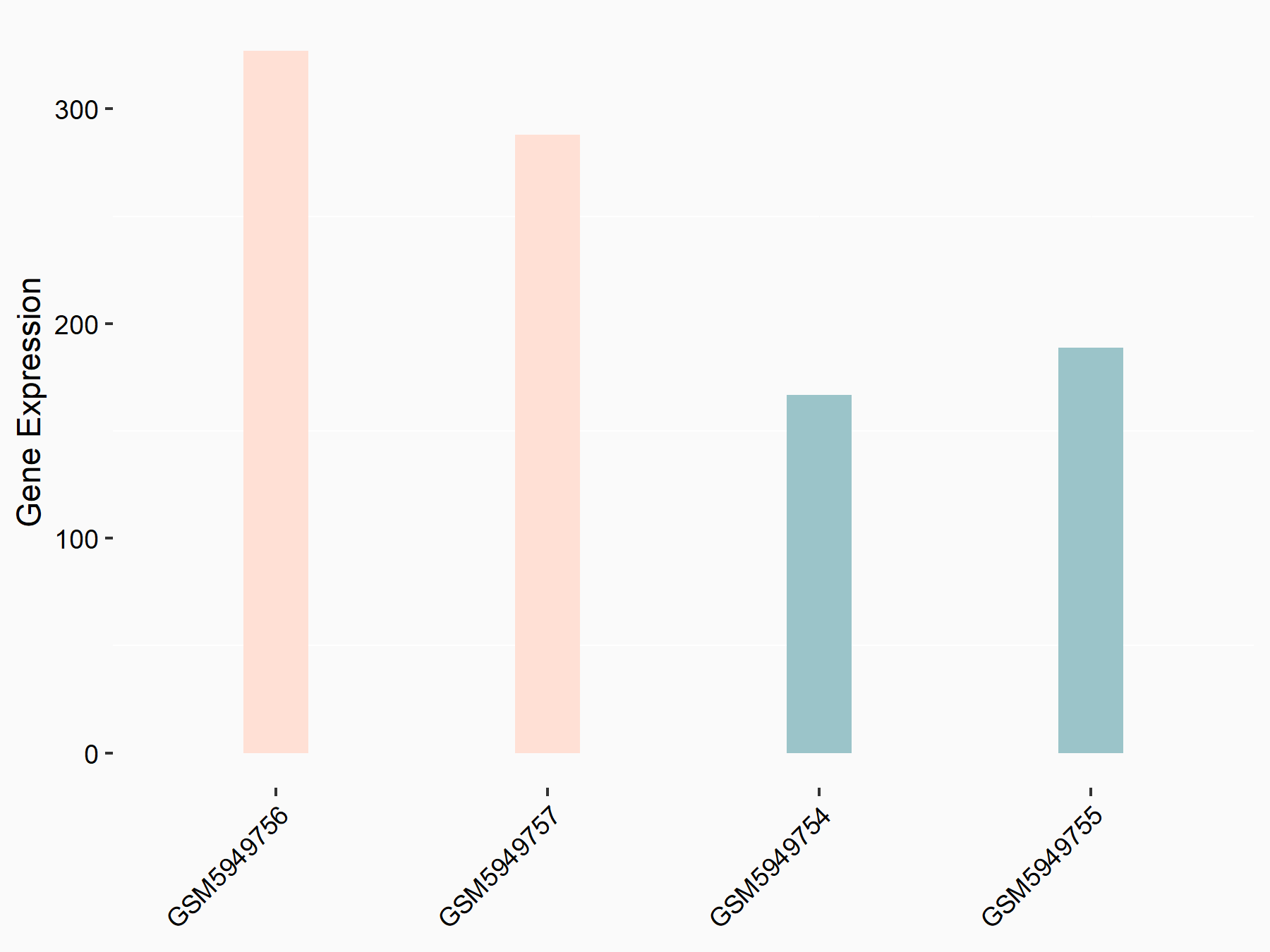 |
logFC: 7.91E-01 p-value: 6.45E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.20E+00 | GSE60213 |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [133] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Cytokine-cytokine receptor interaction | hsa04060 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| MKN28 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_1416 | |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| In-vivo Model | The luciferase signal intensity from days 7 to 42 is on equivalent scales in the models. Bioluminescent flux (photons/s/cm2/steradian) was determined for the lung metastases. | |||
| Response Summary | The m6A modification of Zinc finger MYM-type protein 1 (ZMYM1) mRNA by METTL3 enhanced its stability relying on the "reader" protein HuR (also known as ELAVL1) dependent pathway.The study uncover METTL3/ZMYM1/E-cadherin signaling as a potential therapeutic target in anti-metastatic strategy against Gastric cancer. | |||
Zinc finger protein 750 (ZNF750)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3-deficient liver
Control: Wild type liver cells
|
GSE197800 | |
| Regulation |
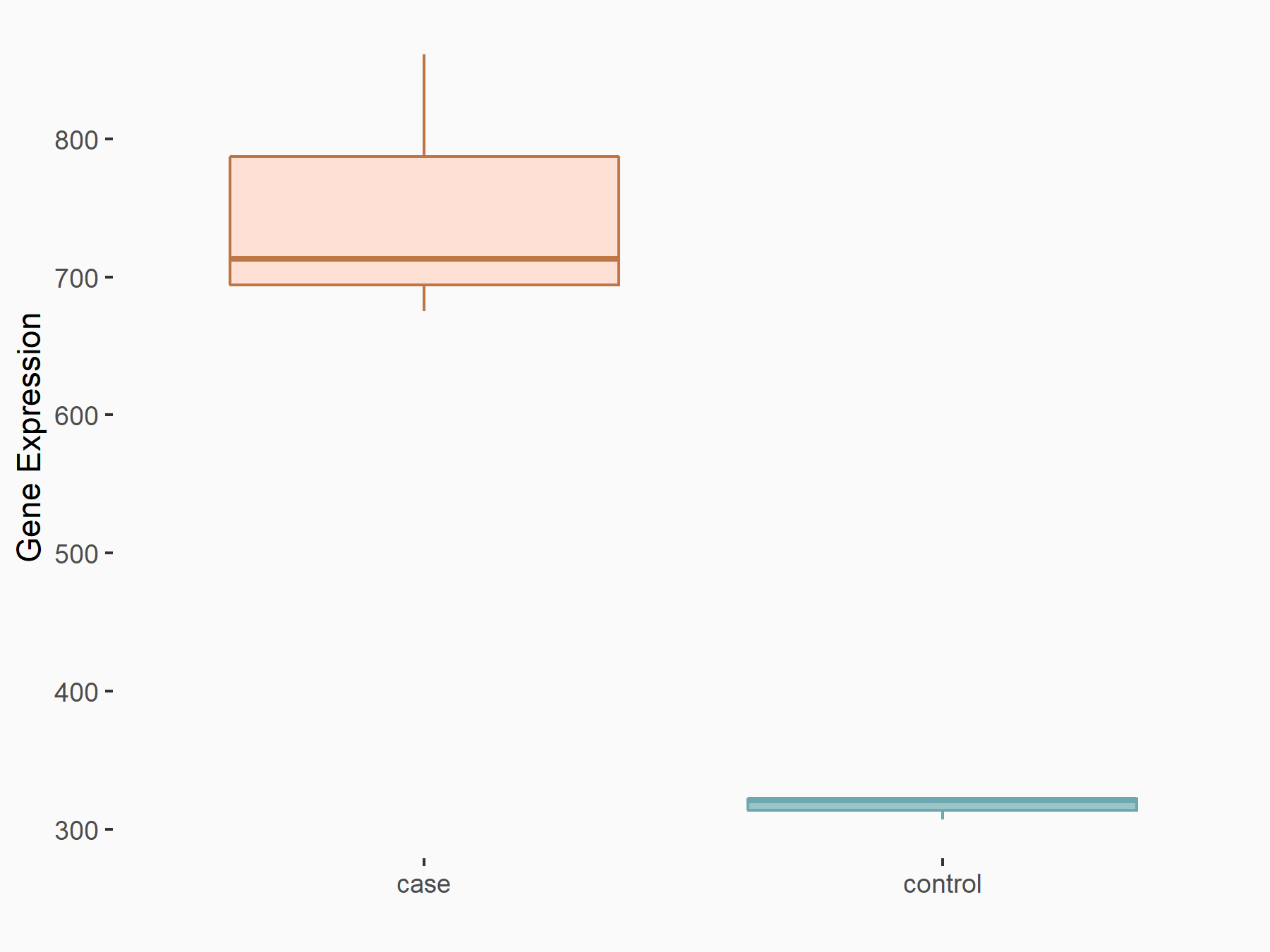 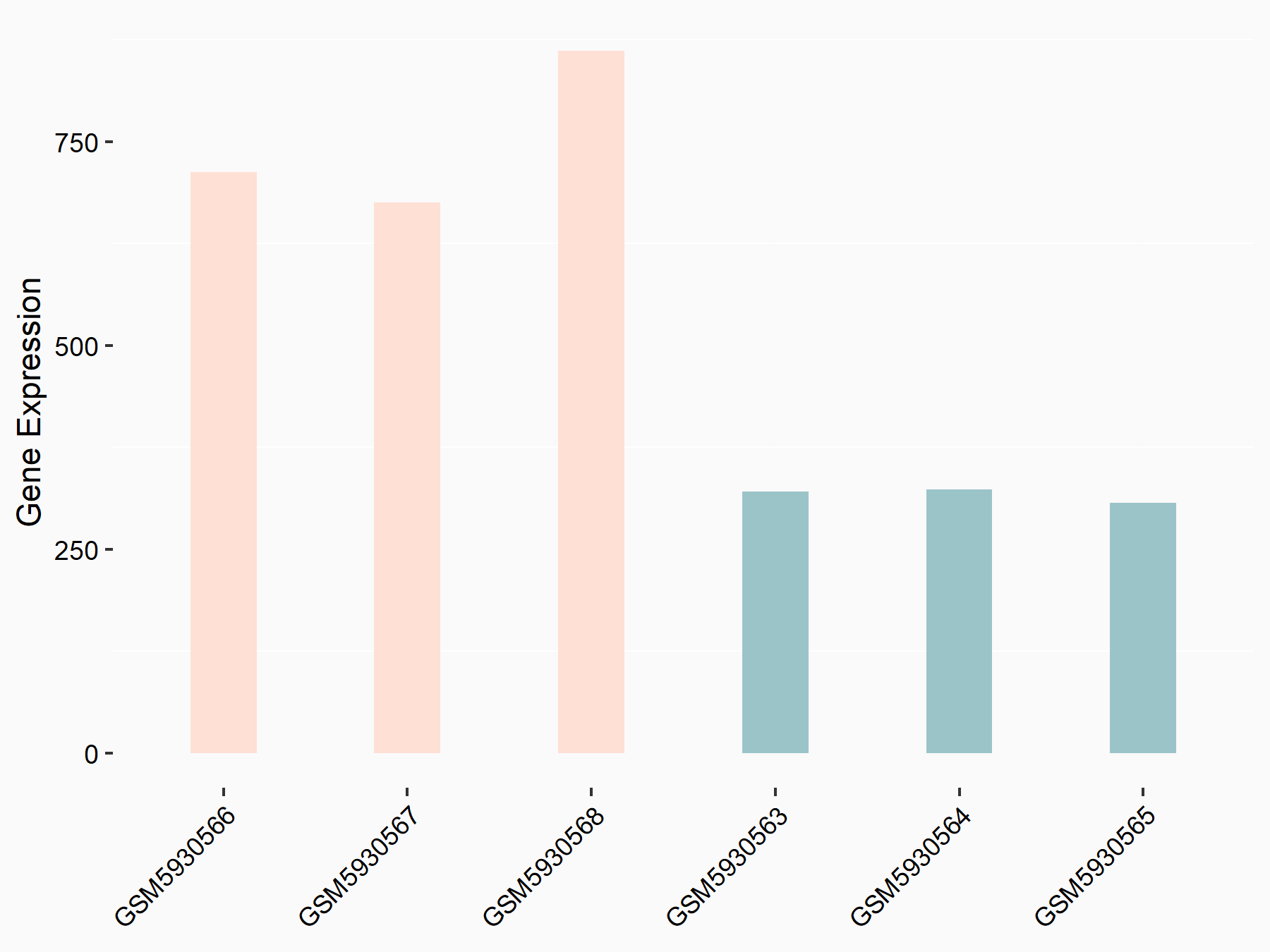 |
logFC: 1.24E+00 p-value: 8.99E-10 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 5.92E+00 | GSE60213 |
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [134] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
5-8F | Nasopharyngeal carcinoma | Homo sapiens | CVCL_C528 |
| 6-10B | Nasopharyngeal carcinoma | Homo sapiens | CVCL_C529 | |
| C666-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_7949 | |
| HNE-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_0308 | |
| HONE-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_8706 | |
| NP69 (A human immortalized nasopharyngeal epithelial) | ||||
| S18 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_B0U9 | |
| S26 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_B0UB | |
| SUNE1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6946 | |
| In-vivo Model | BALB/c-nu mice (4-6 weeks old, female) were purchased from Charles River Laboratories (Beijing, China), and SUNE1-vector-luciferase or SUNE1-ZNF750-luciferase cells (1 × 106) were subcutaneously injected into the dorsal or ventral flank. | |||
| Response Summary | m6A modifications maintained the low expression level of ZNF750 in NPC. Knocking down METTL3 stimulated endogenous Zinc finger protein 750 (ZNF750) expression, and vice versa. | |||
Zinc finger protein GLI1 (GLI1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HUVEC cell line | Homo sapiens |
|
Treatment: shMETTL3 HUVEC cells
Control: shScramble HUVEC cells
|
GSE157544 | |
| Regulation |
 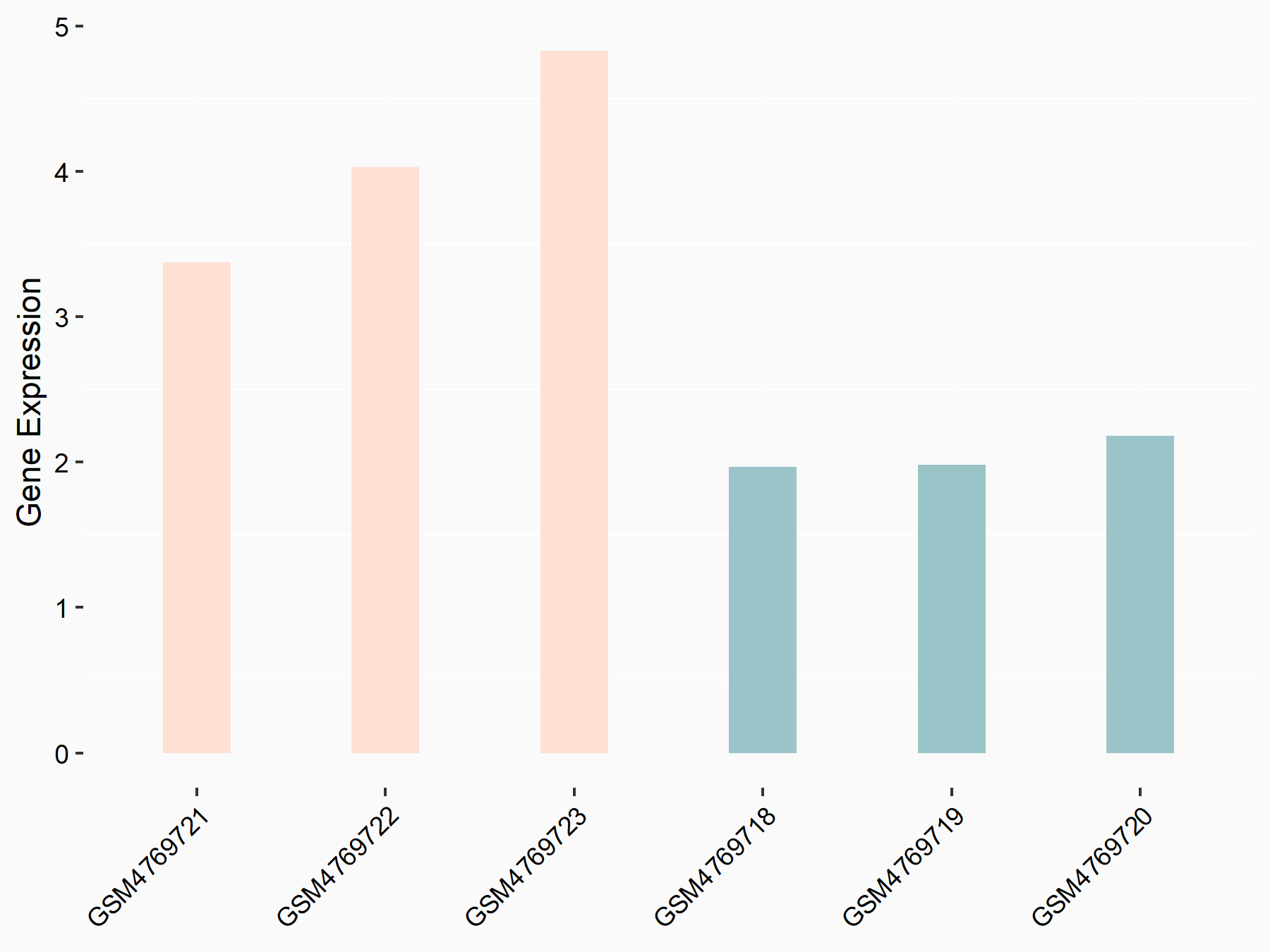 |
logFC: 7.30E-01 p-value: 2.91E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.23E+00 | GSE60213 |
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [135] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Hedgehog signaling pathway | hsa04340 | ||
| Cell Process | Cell proliferation | |||
| Cell survival | ||||
| Cell colony formation | ||||
| Cell invasion | ||||
In-vitro Model |
LNCaP C4-2 | Prostate carcinoma | Homo sapiens | CVCL_4782 |
| LNCaP C4-2B | Prostate carcinoma | Homo sapiens | CVCL_4784 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| In-vivo Model | Equal number of PC-3 cells was injected subcutaneously into right flank. | |||
| Response Summary | METTL3 silence decreased the m6A modification and expression of Zinc finger protein GLI1 (GLI1), an important component of hedgehog pathway, which led to cell apoptosis.the m6A methyltransferase METTL3 promotes the growth and motility of prostate cancer cells by regulating hedgehog pathway. | |||
FOXD2 adjacent opposite strand RNA 1 (FOXD2-AS1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
 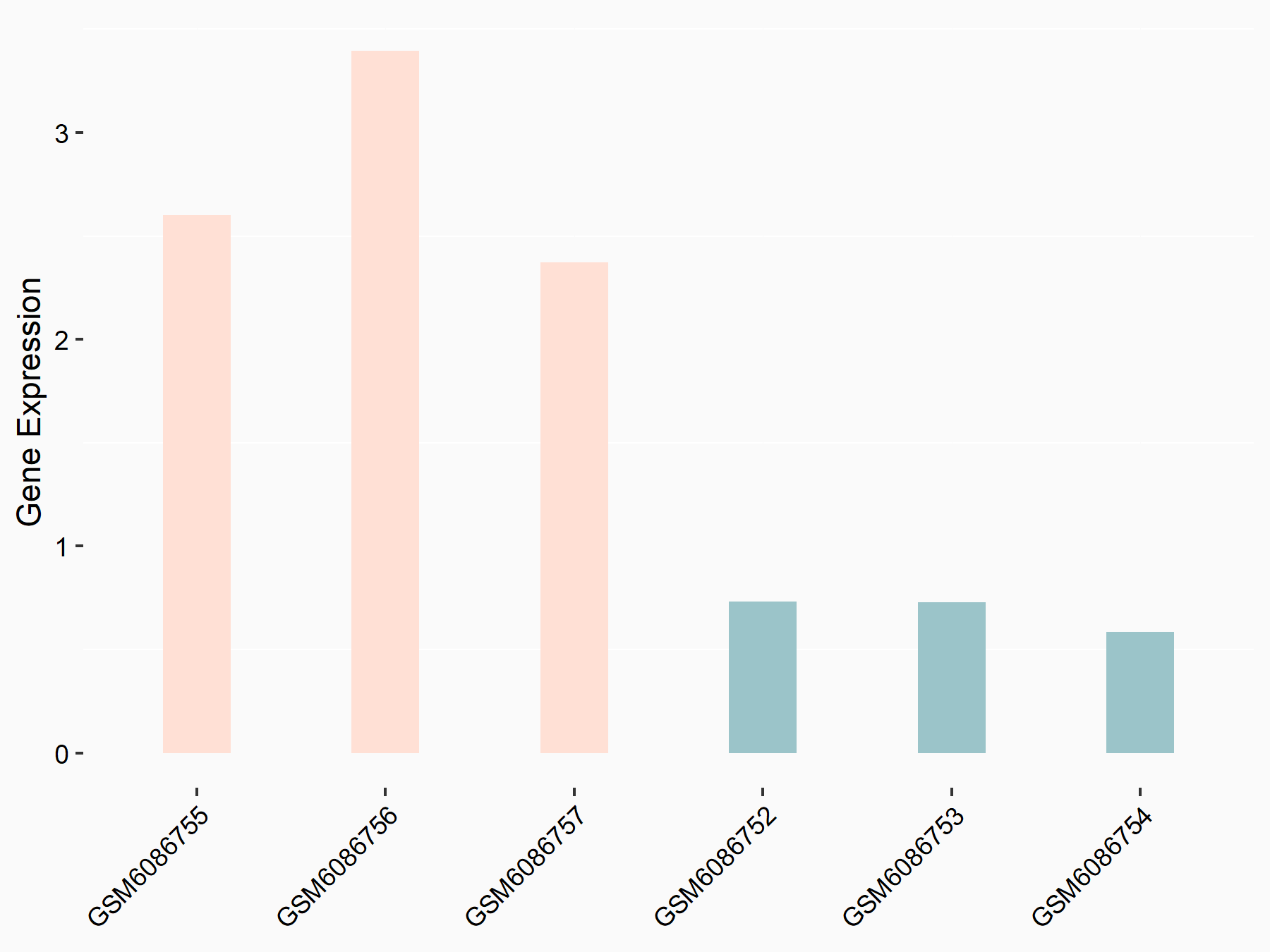 |
logFC: 1.16E+00 p-value: 1.63E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 5.78E+00 | GSE60213 |
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [136] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell migration and proliferation | |||
In-vitro Model |
C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| HaCaT | Normal | Homo sapiens | CVCL_0038 | |
| HT-3 | Cervical carcinoma | Homo sapiens | CVCL_1293 | |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| In-vivo Model | A total 5 × 106 stably transfected SiHa cells were subcutaneously injected into the flank of nude mice. | |||
| Response Summary | METTL3/FOXD2 adjacent opposite strand RNA 1 (FOXD2-AS1) accelerates cervical cancer progression via a m6A-dependent modality, which serves as a potential therapeutic target for cervical cancer. | |||
KRT7-AS
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HUVEC cell line | Homo sapiens |
|
Treatment: shMETTL3 HUVEC cells
Control: shScramble HUVEC cells
|
GSE157544 | |
| Regulation |
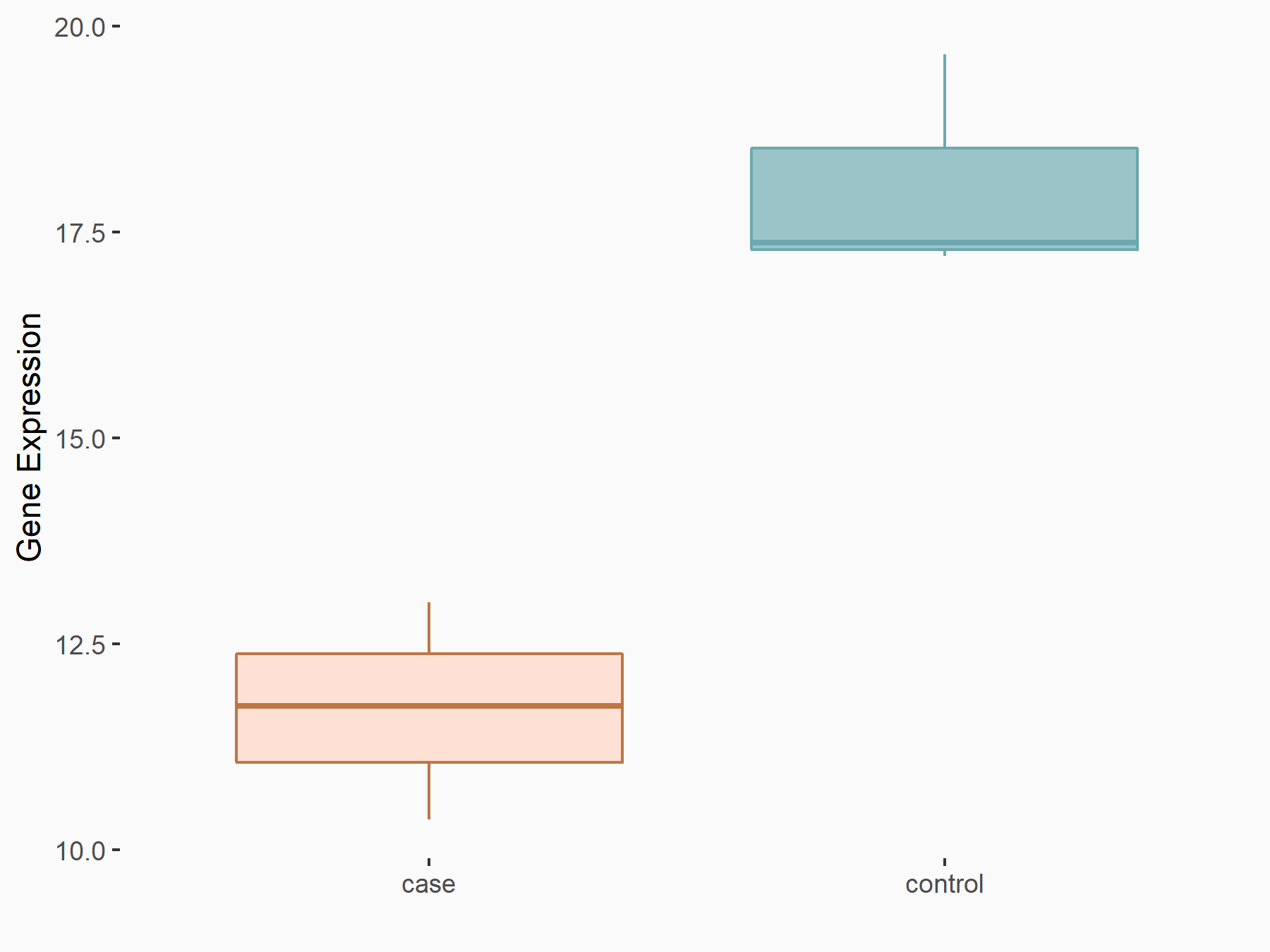 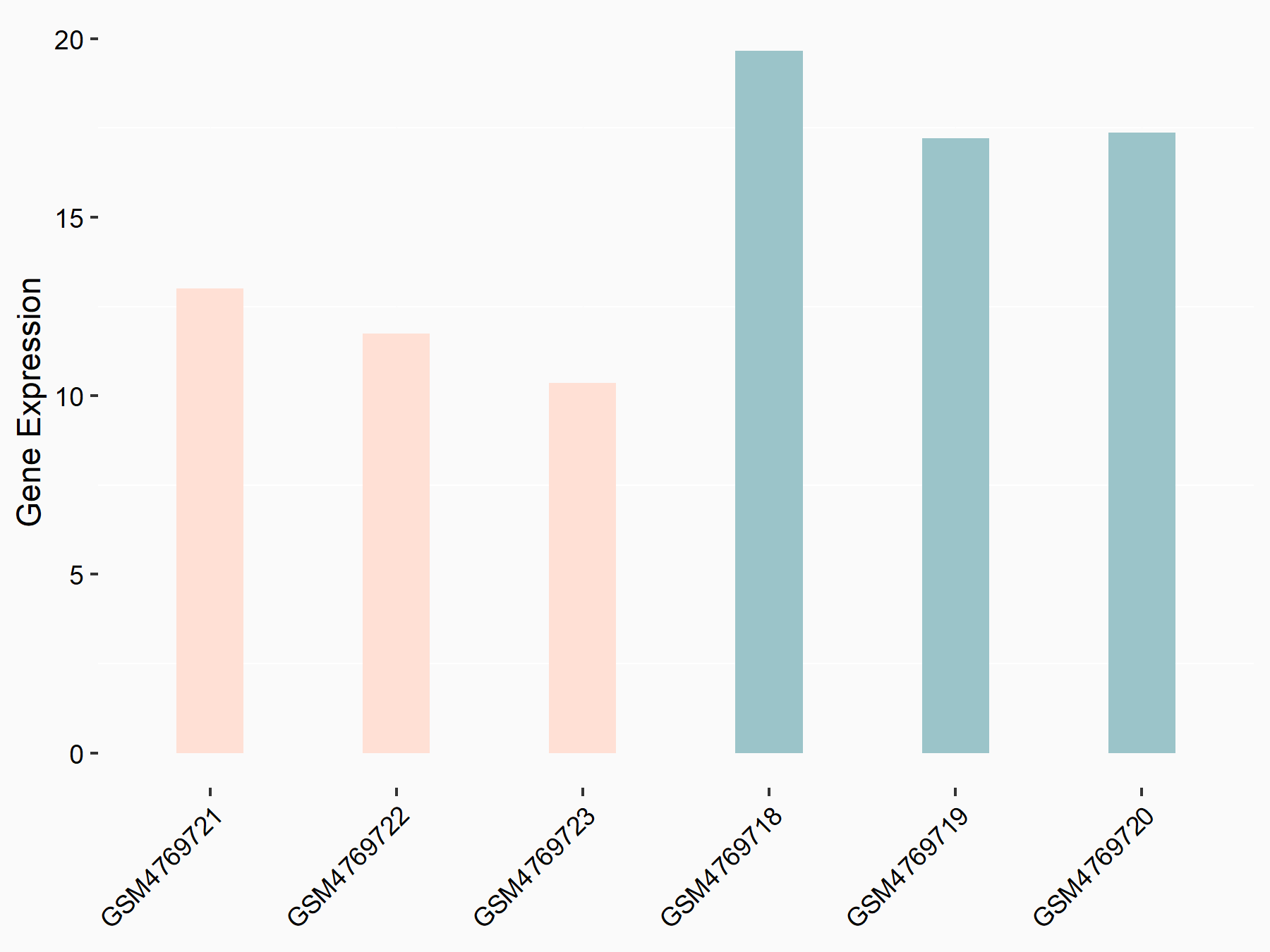 |
logFC: -5.89E-01 p-value: 3.54E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 8.95E+00 | GSE60213 |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [137] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Lung Metastasis | |||
In-vitro Model |
MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| In-vivo Model | First, subcutaneous transplanted model was used to evaluate the growth of BT-549LMF3 and BT-549 cells. Cells (5 × 106 per mouse, n = 5 for each group) were diluted in 200 ul PBS + 200 ul Matrigel (BD Biosciences) and subcutaneously injected into immunodeficient female mice. Second, subcutaneous transplanted model was used to evaluate the metastasis potential of BT-549LMF3 and BT-549 cells. Cells (5 × 106 per mouse, n = 5 for each group) were diluted in 200 ul PBS + 200 ul Matrigel (BD Biosciences) and subcutaneously injected into immunodeficient female mice. Third, the in vivo lung metastasis model was established by injecting with BT-549, BT-549LMF3, FTO stable BT-549LMF3, sh-METTL3 BT-549LMF3, and sh-KRT7 BT-549LMF3 stable cells (1 × 106 per mouse, n = 5 for each group). | |||
| Response Summary | Specifically, increased METTL3 methylated KRT7-AS at A877 to increase the stability of a KRT7-AS/KRT7 mRNA duplex via IGF2BP1/HuR complexes. m6A promotes breast cancer lung metastasis by increasing the stability of a KRT7-AS/KRT7 mRNA duplex and translation of KRT7. | |||
LBX2 antisense RNA 1 (LBX2-AS1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
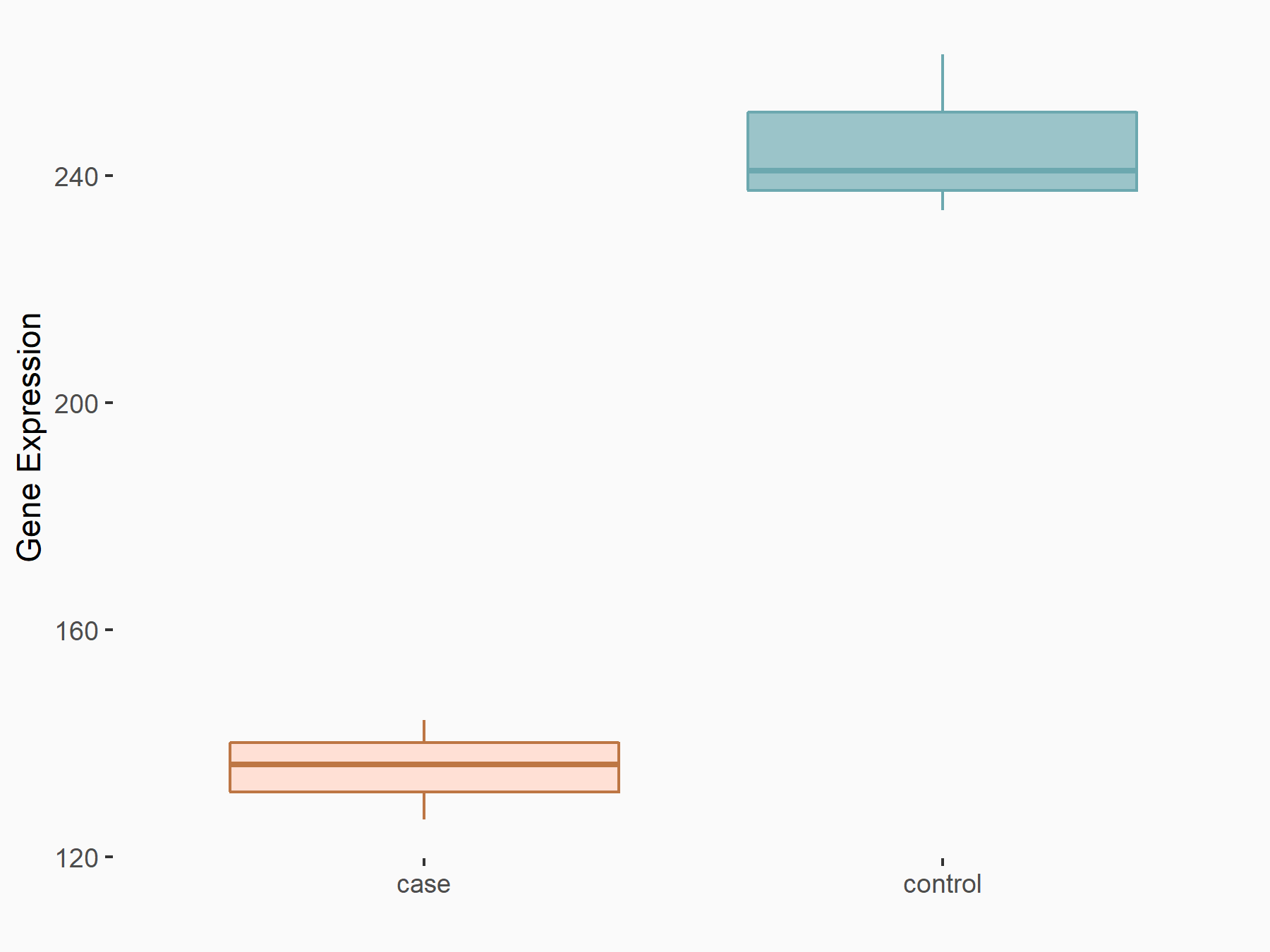 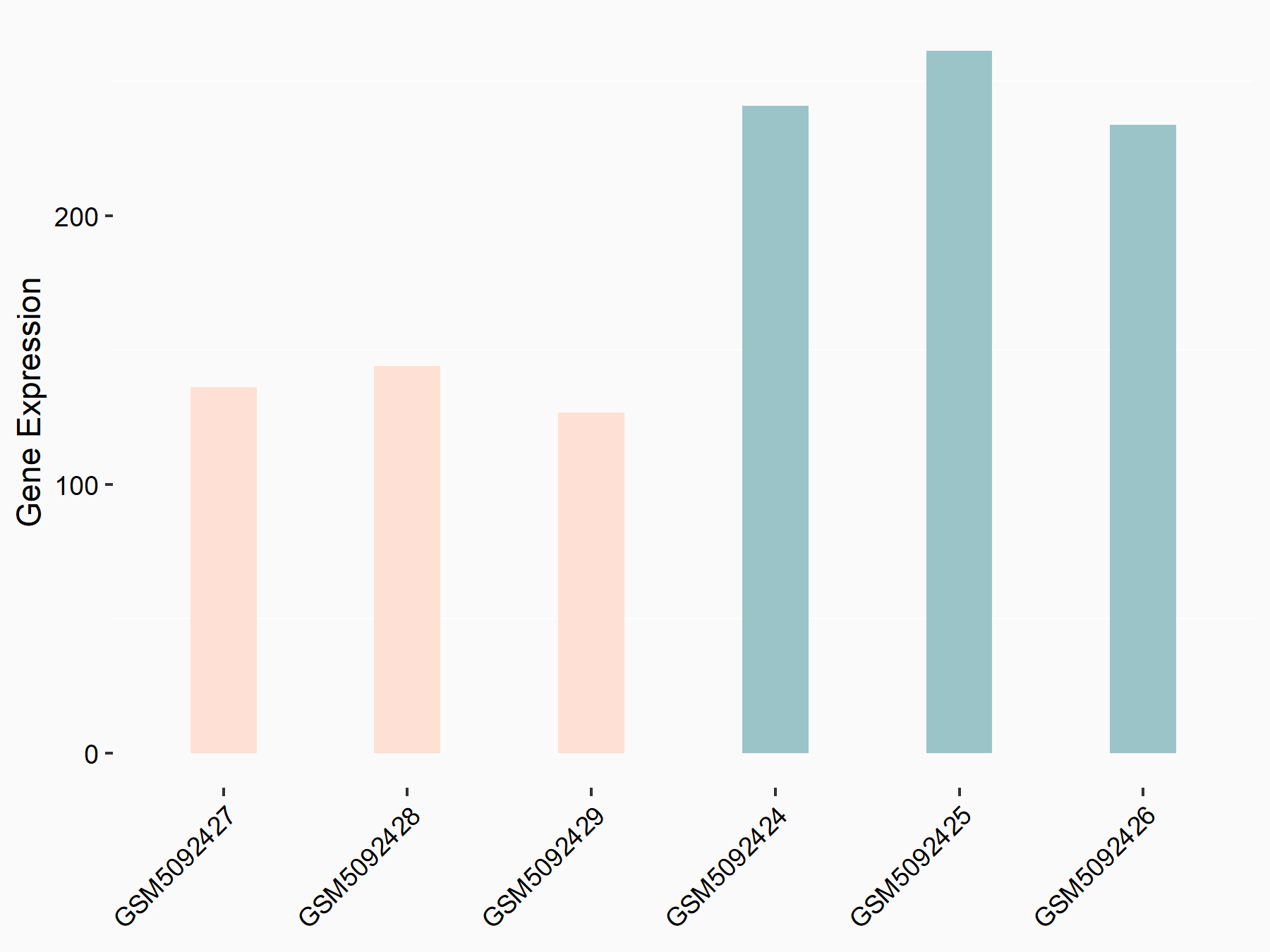 |
logFC: -8.54E-01 p-value: 2.13E-07 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.38E+00 | GSE60213 |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [138] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Fluorouracil | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | The increased LBX2 antisense RNA 1 (LBX2-AS1) in CRC was mediated by METTL3-dependent m6A methylation. LBX2-AS1 serves as a therapeutic target and predictor of 5-FU benefit in colorectal cancer patients. | |||
LncRNA activating regulator of DKK1 (LNCAROD)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
 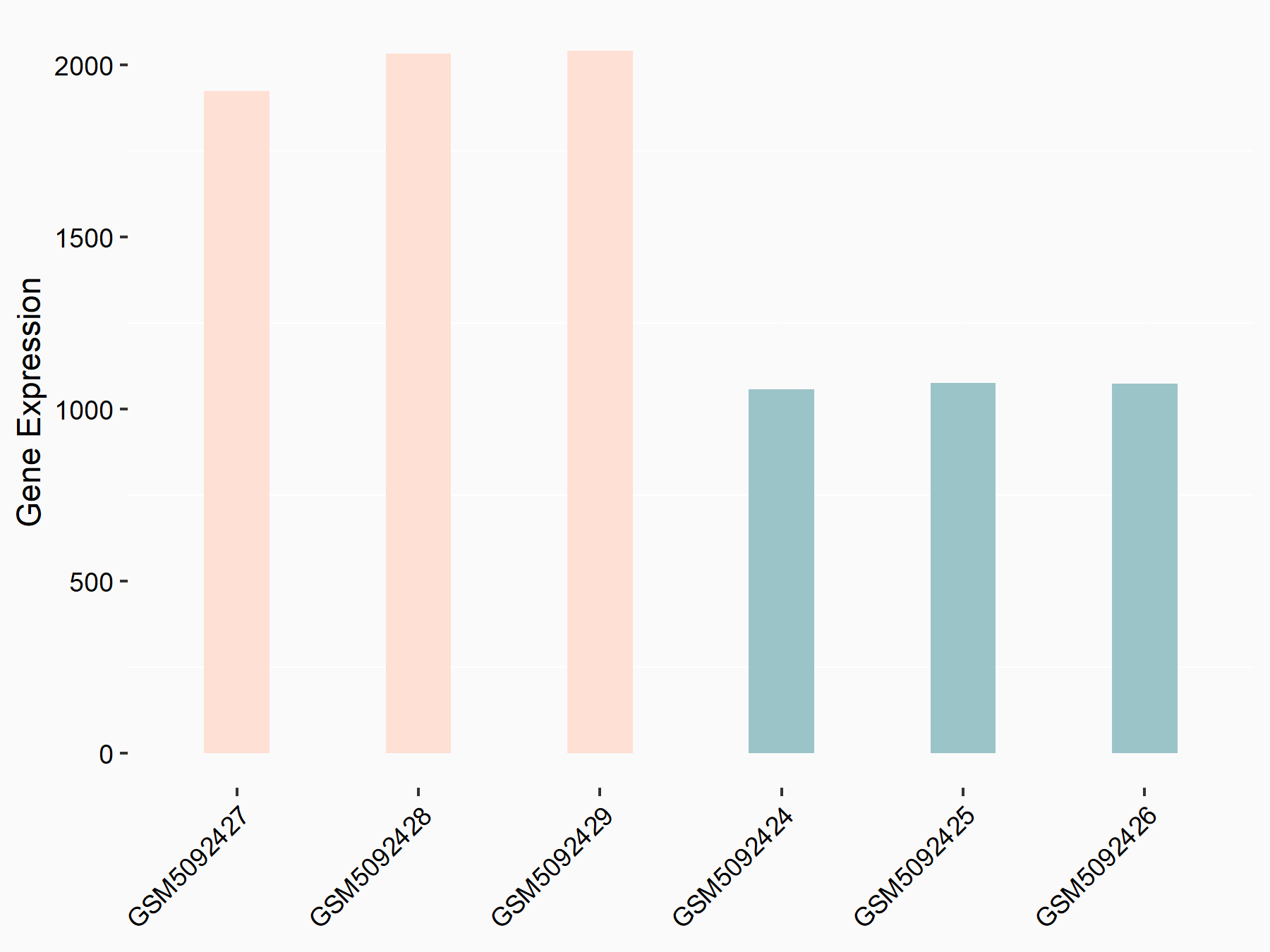 |
logFC: 9.04E-01 p-value: 2.47E-45 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 5.11E+00 | GSE60213 |
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [139] | |||
| Responsed Disease | Head and neck squamous carcinoma [ICD-11: 2B6E] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Proteasome | hsa03050 | ||
| Cell Process | Proteasomal degradation | |||
In-vitro Model |
C666-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_7949 |
| CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 | |
| FaDu | Hypopharyngeal squamous cell carcinoma | Homo sapiens | CVCL_1218 | |
| HK1 | Nasopharyngeal carcinoma | Acipenser baerii | CVCL_YE27 | |
| NP69 (A human immortalized nasopharyngeal epithelial) | ||||
| Tca8113 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6851 | |
| Response Summary | The N6-methyladenosine (m6A) modification mediated by METTL3 and METTL14 enhanced the stability of LncRNA activating regulator of DKK1 (LNCAROD) in head and neck squamous cell carcinoma cells. LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma. | |||
Long intergenic non-protein coding RNA 1833 (LINC01833)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
 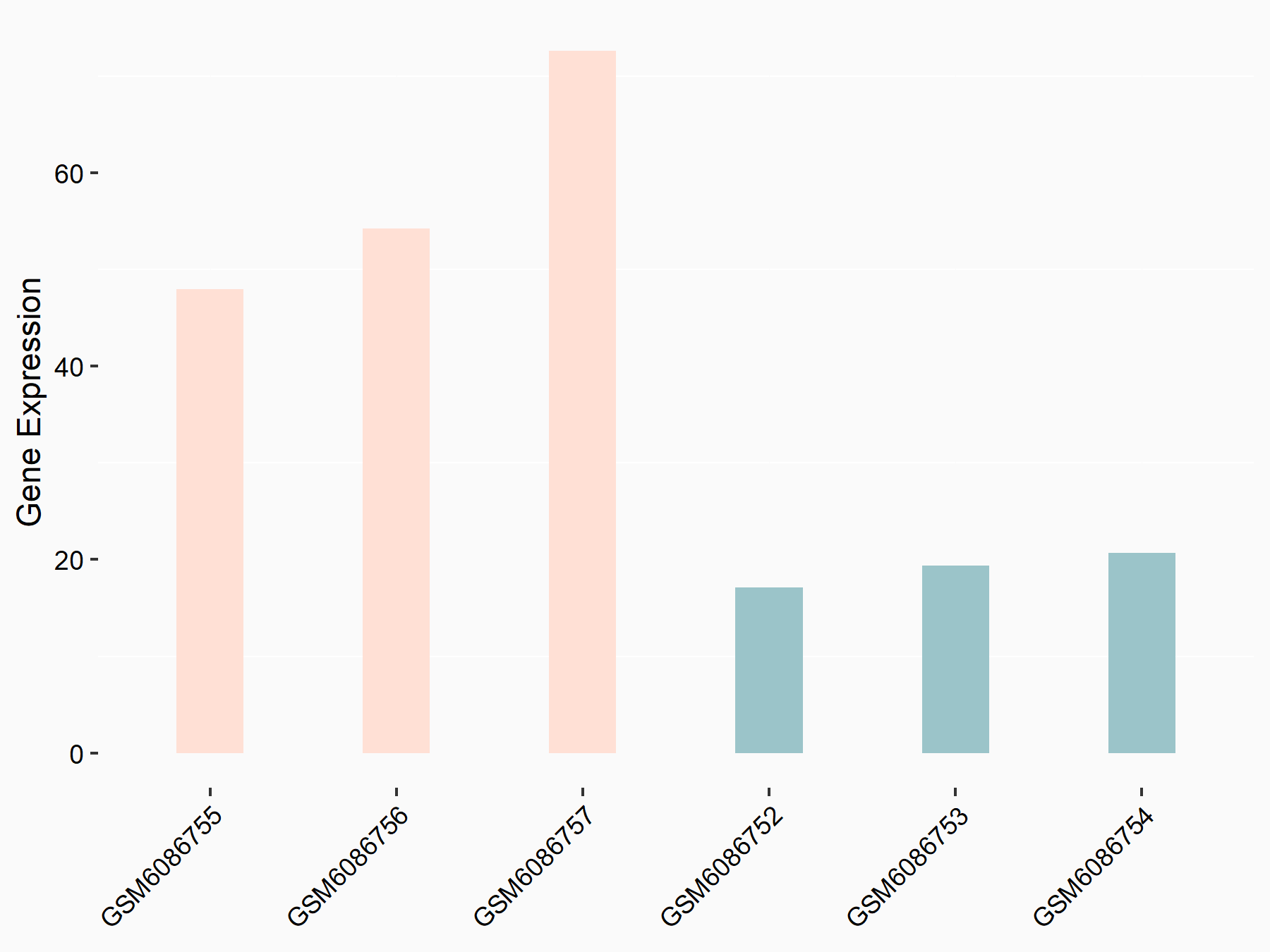 |
logFC: 1.55E+00 p-value: 2.63E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.09E+00 | GSE60213 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [140] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | For the tumorigenicity studies, 3 × 106 HCC827 cells stably expressing sh-LINC01833 or sh-NC were injected subcutaneously into the ventral side of male BALB/c nude mice in the according groups with five mice in each group. Five mice were sampled each time. Tumor size and weight were examined at 28 days after injection. | |||
| Response Summary | m6A transferase METTL3-induced Long intergenic non-protein coding RNA 1833 (LINC01833) m6A methylation promotes NSCLC progression through modulating HNRNPA2B1 expression. | |||
Long intergenic non-protein coding RNA 958 (LINC00958)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HUVEC cell line | Homo sapiens |
|
Treatment: shMETTL3 HUVEC cells
Control: shScramble HUVEC cells
|
GSE157544 | |
| Regulation |
  |
logFC: -6.47E-01 p-value: 2.79E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 5.60E+00 | GSE60213 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [141] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| Cell Process | RNA stability | |||
| Lipogenesis | ||||
In-vitro Model |
FOCUS | Adult hepatocellular carcinoma | Homo sapiens | CVCL_7955 |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| HEp-2 | Endocervical adenocarcinoma | Homo sapiens | CVCL_1906 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| QSG-7701 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6944 | |
| Response Summary | Long intergenic non-protein coding RNA 958 (LINC00958) sponged miR-3619-5p to upregulate hepatoma-derived growth factor (HDGF) expression, thereby facilitating Hepatocellular carcinoma lipogenesis and progression. METTL3-mediated N6-methyladenosine modification led to LINC00958 upregulation through stabilizing its RNA transcript. | |||
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [142] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| Cell Process | RNA transcript stability | |||
In-vitro Model |
BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| In-vivo Model | MCF-7 cells transfected with sh-LINC00958 or empty vector were resuspended at 2 × 107 cells/mL. | |||
| Response Summary | m6A methyltransferase-like 3 (METTL3) gave rise to the upregulation of Long intergenic non-protein coding RNA 958 (LINC00958) by promoting its RNA transcript stability in breast cancer. | |||
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | DKO-1 cell line | Homo sapiens |
|
Treatment: METTL3 knockdown DKO-1 cell
Control: DKO-1 cell
|
GSE182382 | |
| Regulation |
 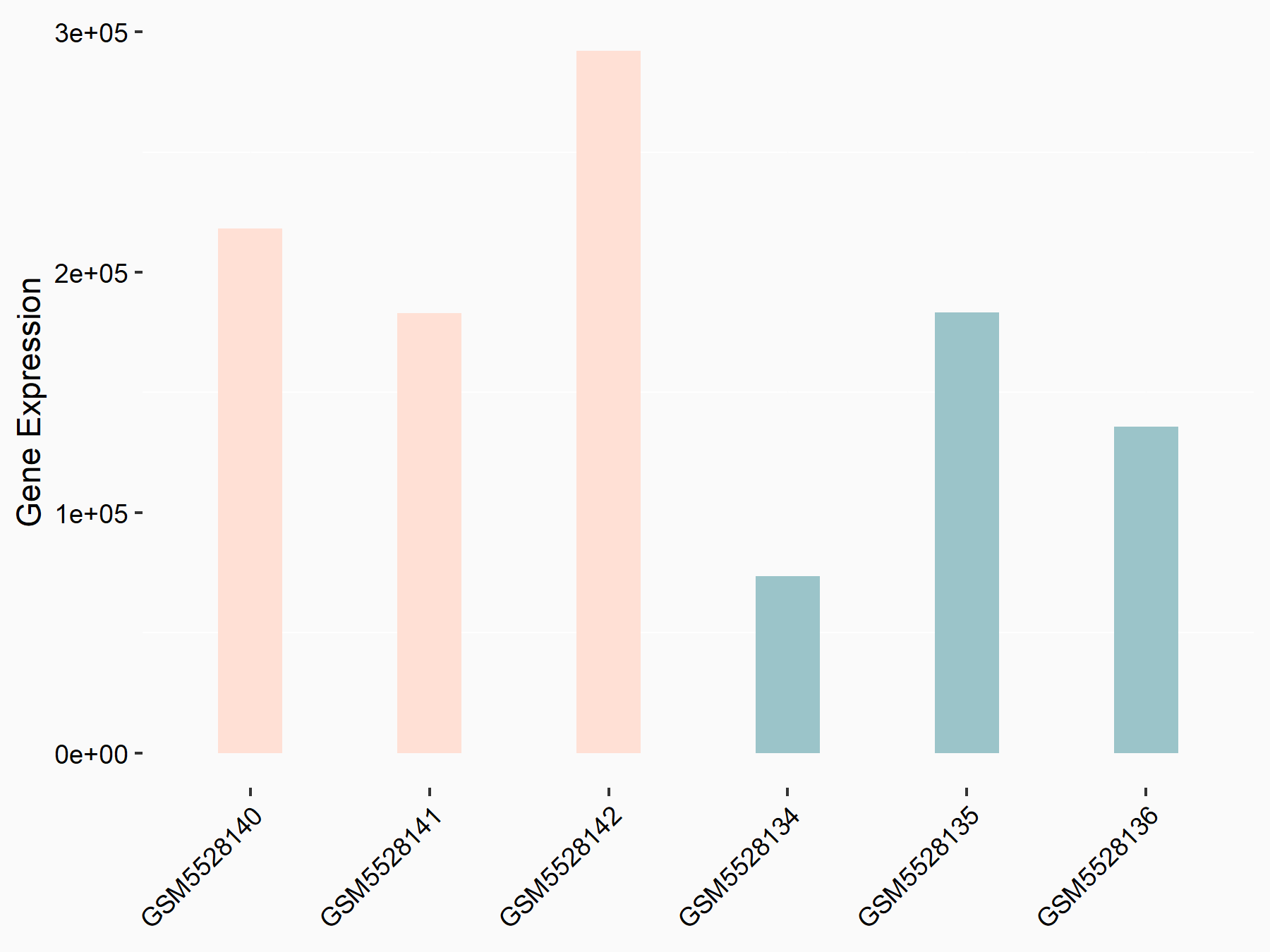 |
logFC: 8.20E-01 p-value: 5.93E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.28E+00 | GSE60213 |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [143] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | TNF signaling pathway | hsa04668 | ||
| Cell Process | Cell proliferation and metastasis | |||
In-vitro Model |
H4 | Astrocytoma | Homo sapiens | CVCL_1239 |
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| U87 (A primary glioblastoma cell line) | ||||
| In-vivo Model | U87 cells (5 × 105) transfected with an empty vector, METTL3 shRNA, or METTL3 overexpression vector were inoculated into the right frontal node of nude mice. | |||
| Response Summary | METTL3 promoted the malignant progression of IDH-wildtype gliomas and revealed important insight into the upstream regulatory mechanism of Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) and NF-Kappa-B with a primary focus on m6A modification. | |||
Lung cancer [ICD-11: 2C25]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [126] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Metabolic | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-6 | Lung adenocarcinoma | Homo sapiens | CVCL_0236 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | Mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then administrated with PBS or DDP (3 mg/kg per day) for three times a week, and tumor volume were measured every second day. | |||
| Response Summary | METTL3, YTHDF3, YTHDF1, and eIF3b directly promoted YAP translation through an interaction with the translation initiation machinery. METTL3 knockdown inhibits tumor growth and enhances sensitivity to DDP in vivo.m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)-miR-1914-3p-YAP axis to induce Non-small cell lung cancer drug resistance and metastasis. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [370] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
Thymoma [ICD-11: 2C27]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [144] | |||
| Responsed Disease | Thymic epithelial tumors [ICD-11: 2C27.Y] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Cellular senescence | hsa04218 | ||
| Cell Process | Cell viability and proliferation | |||
In-vitro Model |
T1889 | Thymic undifferentiated carcinoma | Homo sapiens | CVCL_D024 |
| Response Summary | This study highlighted METTL3 as a tumor promoter in Thymic tumors and c-MYC as a promising target to be exploited for the treatment of TET. High expression of c-MYC protein is enabled by lncRNA Metastasis associated lung adenocarcinoma transcript 1 (MALAT1), which is methylated and delocalized by METTL3. Silencing of METTL3 combined with cisplatin or c-MYC inhibitor induces cell death in TET cells. Blocking of c-MYC by using JQ1 inhibitor cooperates with METTL3 depletion in the inhibition of proliferation and induction of cell death. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [144] | |||
| Responsed Disease | Thymic epithelial tumors [ICD-11: 2C27.Y] | |||
| Responsed Drug | JQ-1 | Phase 1 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Cellular senescence | hsa04218 | ||
| Cell Process | Cell viability and proliferation | |||
In-vitro Model |
T1889 | Thymic undifferentiated carcinoma | Homo sapiens | CVCL_D024 |
| Response Summary | This study highlighted METTL3 as a tumor promoter in Thymic tumors and c-MYC as a promising target to be exploited for the treatment of TET. High expression of c-MYC protein is enabled by lncRNA Metastasis associated lung adenocarcinoma transcript 1 (MALAT1), which is methylated and delocalized by METTL3. Silencing of METTL3 combined with cisplatin or c-MYC inhibitor induces cell death in TET cells. Blocking of c-MYC by using JQ1 inhibitor cooperates with METTL3 depletion in the inhibition of proliferation and induction of cell death. | |||
Breast cancer [ICD-11: 2C60]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [116] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Doxil | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
MCF7-DoxR (Adriamycin-resistant cell line MCF7-DoxR) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| In-vivo Model | Once the tumor volume increased to about 1 cm3, six groups of MCF7 bearing mice (n = 10 in each group) were injected with PBS (0.1 ml, caudal vein) and adriamycin (0.1 ml, 10 mg/kg), respectively. When the tumor reached 1.5 cm in any direction (defined as event-free survival analysis), 10 mice in each group were selected to measure the tumor size and weight on the 12th day after adriamycin injection. | |||
| Response Summary | METTL3 can regulate the expression of Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) through m6A, mediate the E2F1/AGR2 axis, and promote the adriamycin resistance of breast cancer. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [57] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| Response Summary | Silencing METTL3 down-regulate Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) and HMGA2 by sponging miR-26b, and finally inhibit EMT, migration and invasion in breast cancer, providing a theoretical basis for clinical treatment of breast cancer. | |||
Chronic kidney disease [ICD-11: GB61]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [145] | |||
| Responsed Disease | Chronic kidney disease [ICD-11: GB61] | |||
| Responsed Drug | Artenimol | Approved | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
HK-2 [Human kidney] | Normal | Homo sapiens | CVCL_0302 |
| HK2 | Normal | Acipenser baerii | CVCL_YE28 | |
| In-vivo Model | For the unilateral ureteral obstruction (UUO) model, male C57BL/6J mice at 8 weeks of age (20-22 g body weight) were first anaesthetized with pentobarbital sodium (50 mg/kg) via intraperitoneal injection. Then, the left ureter was ligated using 3-0 silk and a left lateral incision. | |||
| Response Summary | Renal fibrosis is a key factor in chronic kidney disease (CKD). Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)/miR-145/FAK pathway was involved in the effect of dihydroartemisinin (DHA) on TGF-beta1-induced renal fibrosis in vitro and in vivo. | |||
Kidney failure [ICD-11: GB6Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [146] | |||
| Responsed Disease | Kidney failure [ICD-11: GB6Z] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
NIT-1 | Insulin tumor | Mus musculus | CVCL_3561 |
| Response Summary | m6A modification is co-regulated by METTL3 and FTO in cadmium-treated cells. Metastasis associated lung adenocarcinoma transcript 1 (MALAT1), LncRNA-PVT1 and m6A modification could be key nodes for cadmium-induced oxidative damage, and highlight their importance as promising preventive and therapeutic targets in cadmium toxicity. | |||
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [368] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
hFOB 1.19 | Normal | Homo sapiens | CVCL_3708 |
| MG-63 | Osteosarcoma | Homo sapiens | CVCL_0426 | |
| U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 | |
Ewing's sarcoma [ICD-11: 2B52]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [369] | |||
| Responsed Disease | Ewing's sarcoma [ICD-11: 2B52] | |||
| Target Regulation | Up regulation | |||
Prostate cancer associated transcript 6 (PCAT6)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
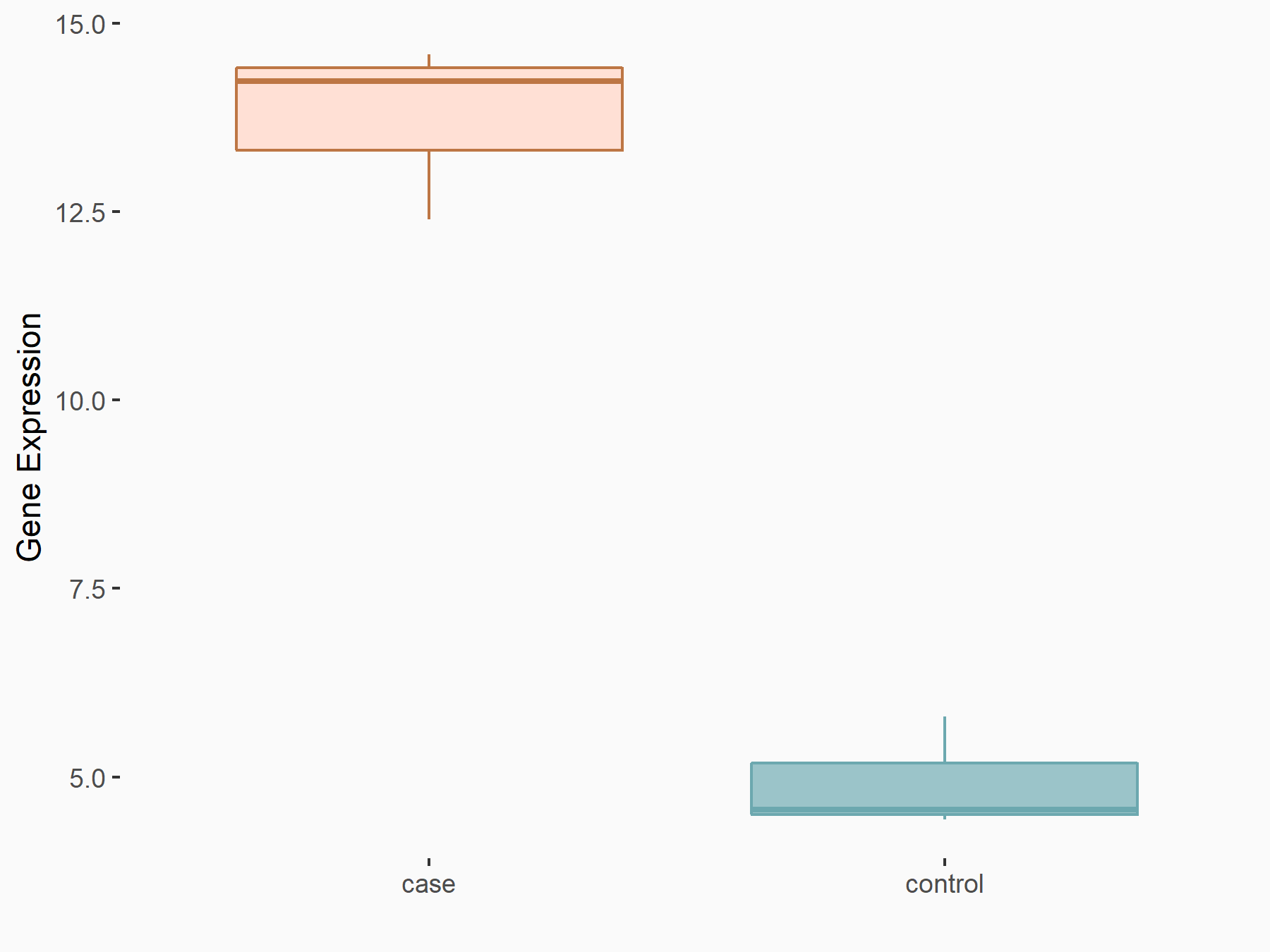 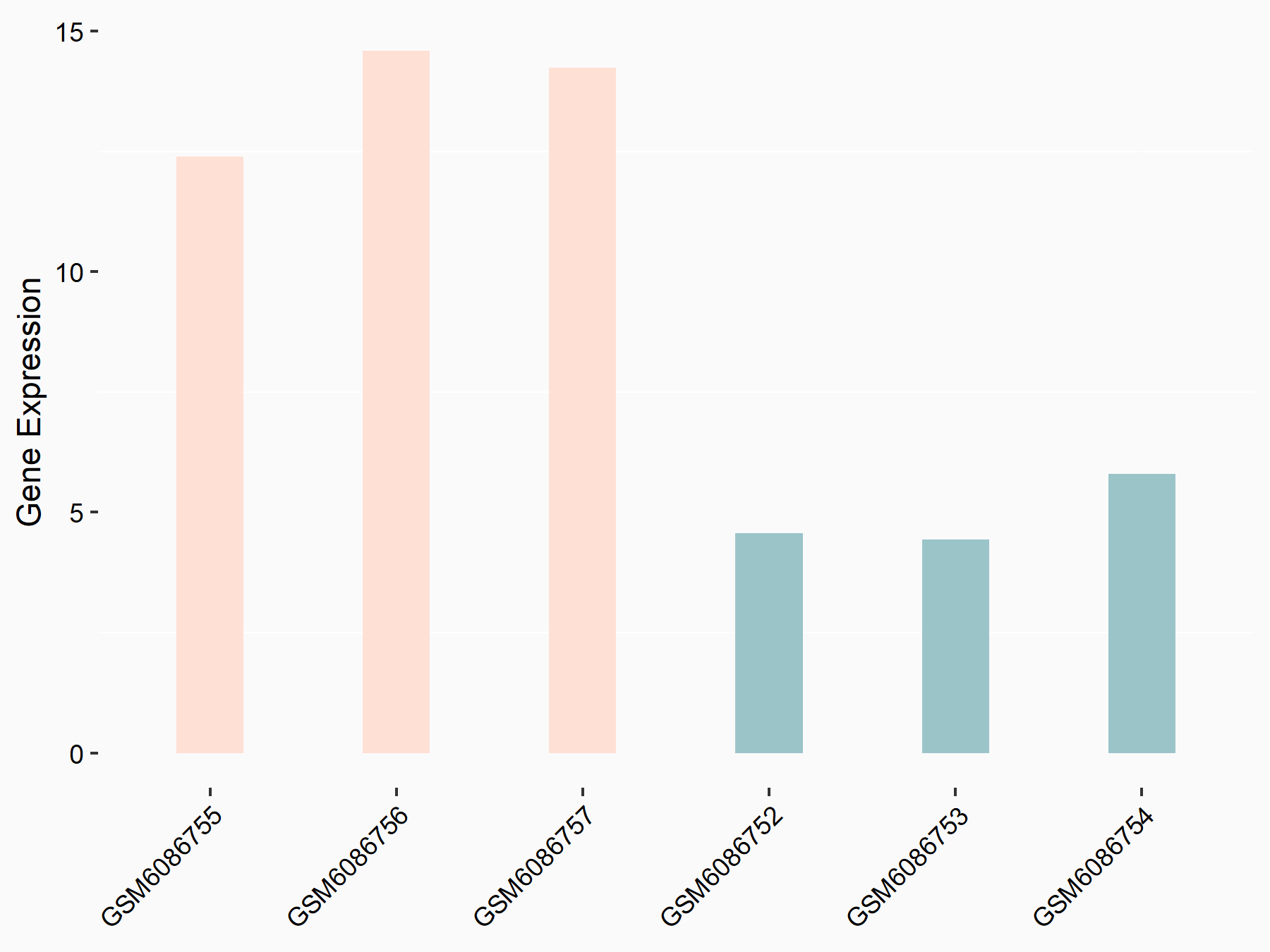 |
logFC: 1.32E+00 p-value: 7.57E-06 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.42E+00 | GSE60213 |
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [147] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
| Cell Process | RNA stability | |||
In-vitro Model |
PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 |
| LNCaP C4-2B | Prostate carcinoma | Homo sapiens | CVCL_4784 | |
| In-vivo Model | At 1 week post-injection with PC-3 cells, mice were randomly assigned to three groups (n = 8 per group): the ASO-NC group (injection with ASO negative control targeting unknown sequence, 5 nmol in 100 uL PBS for each mouse), the ASO-L group (injection with low-dose ASO targeting PCAT6, 5 nmol in 100 uL PBS for each mouse), and the ASO-H group (injection with high-dose ASO targeting PCAT6, 10 nmol in 100 uL PBS for each mouse). | |||
| Response Summary | METTL3-mediated m6A modification contributed to Prostate cancer associated transcript 6 (PCAT6) upregulation in an IGF2BP2-dependent manner. Furthermore, PCAT6 upregulated IGF1R expression by enhancing IGF1R mRNA stability through the PCAT6/IGF2BP2/IGF1R RNA-protein three-dimensional complex. The m6 A-induced PCAT6/IGF2BP2/IGF1R axis promotes PCa bone metastasis and tumor growth, suggesting that PCAT6 serves as a promising prognostic marker and therapeutic target against bone-metastatic PCa. | |||
Small nucleolar RNA host gene 1 (SNHG1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
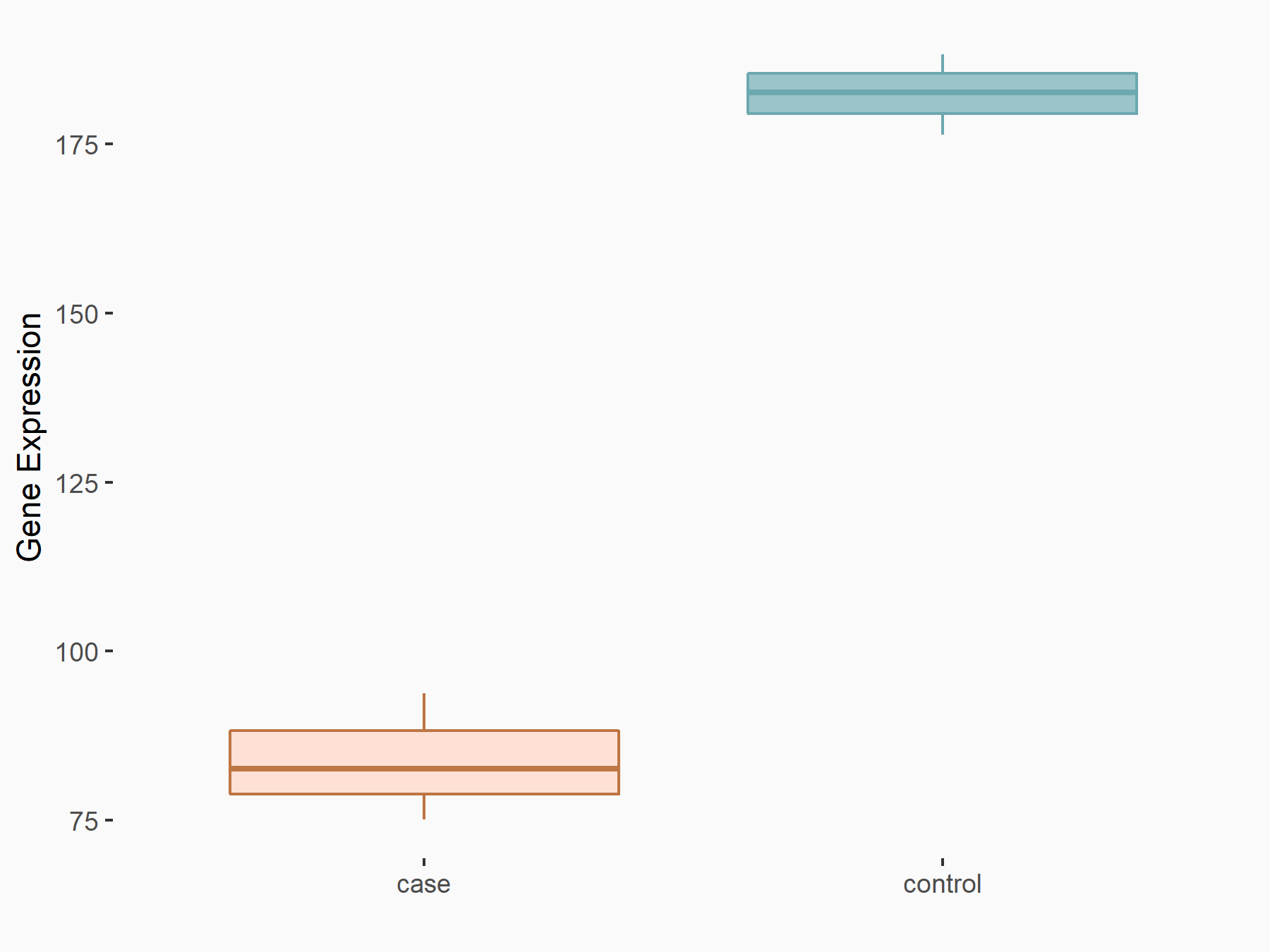 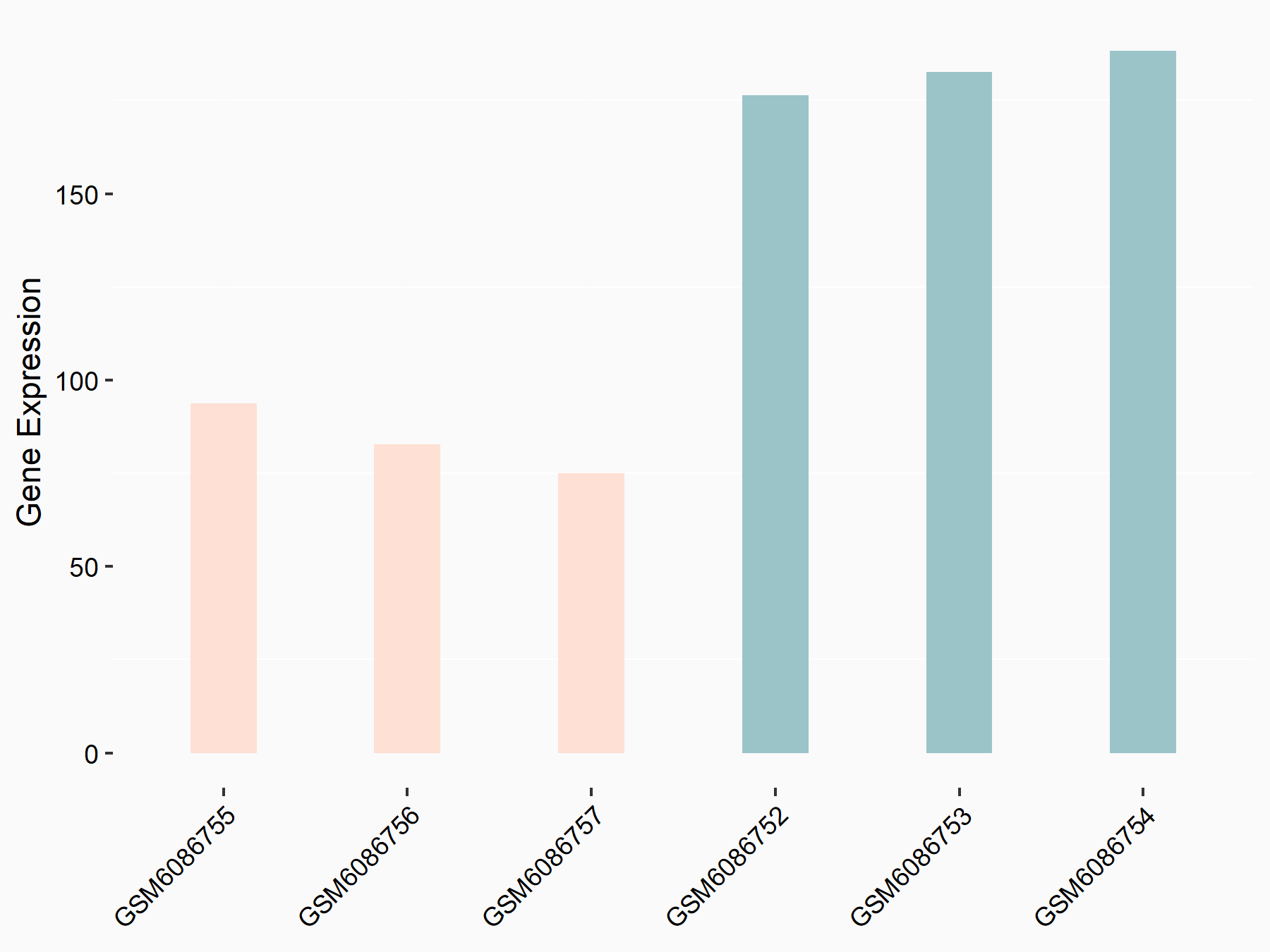 |
logFC: -1.12E+00 p-value: 7.04E-06 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.90E+00 | GSE60213 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [148] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| Response Summary | METTL3/Small nucleolar RNA host gene 1 (SNHG1)/miR-140-3p/UBE2C axis plays a crucial role in cancer progression and the immune response in non-small cell lung cancer. | |||
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [374] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
NCM460 | Normal | Homo sapiens | CVCL_0460 |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
THAP7 antisense RNA 1 (THAP7-AS1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | DKO-1 cell line | Homo sapiens |
|
Treatment: METTL3 knockdown DKO-1 cell
Control: DKO-1 cell
|
GSE182382 | |
| Regulation |
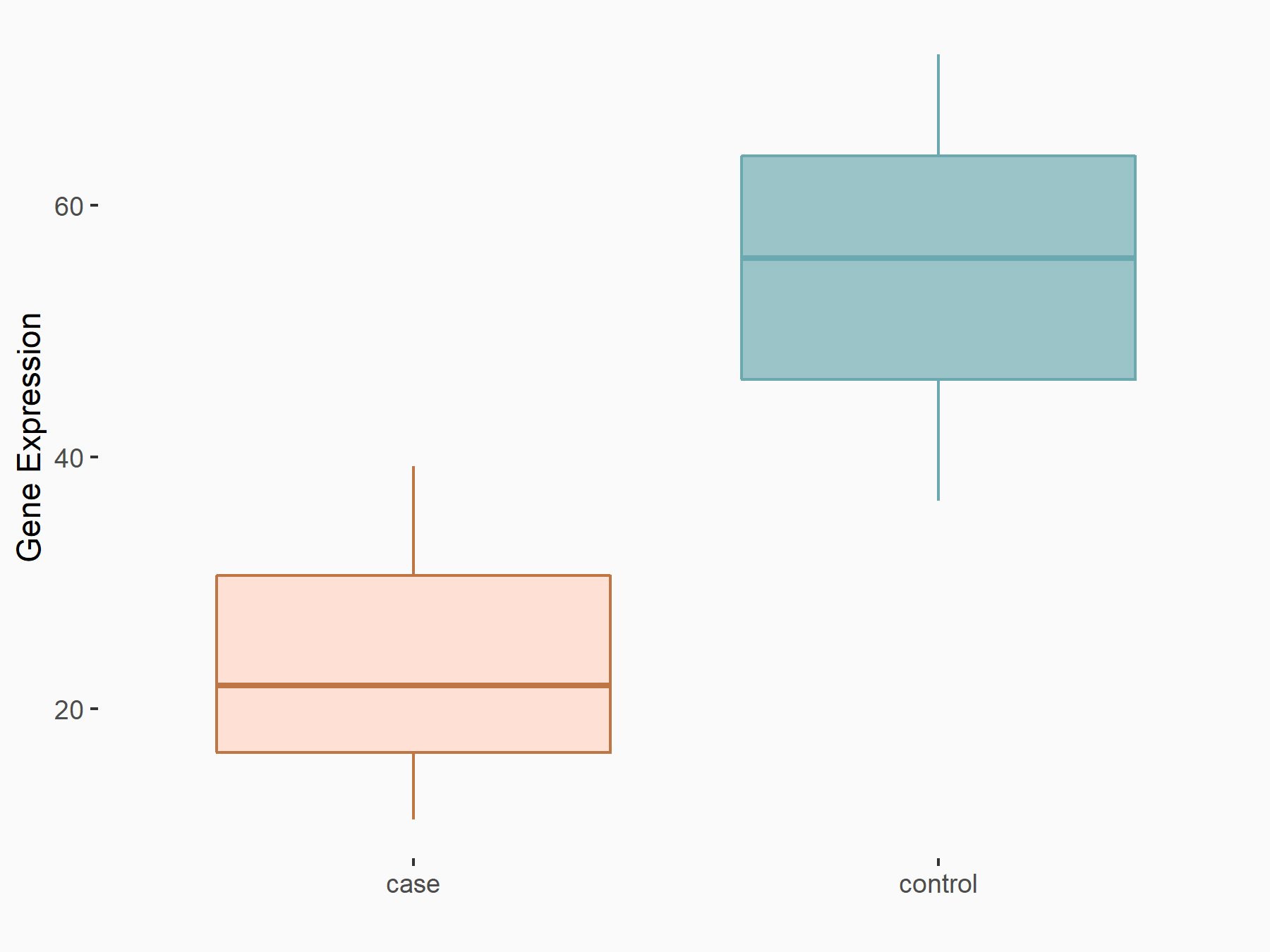 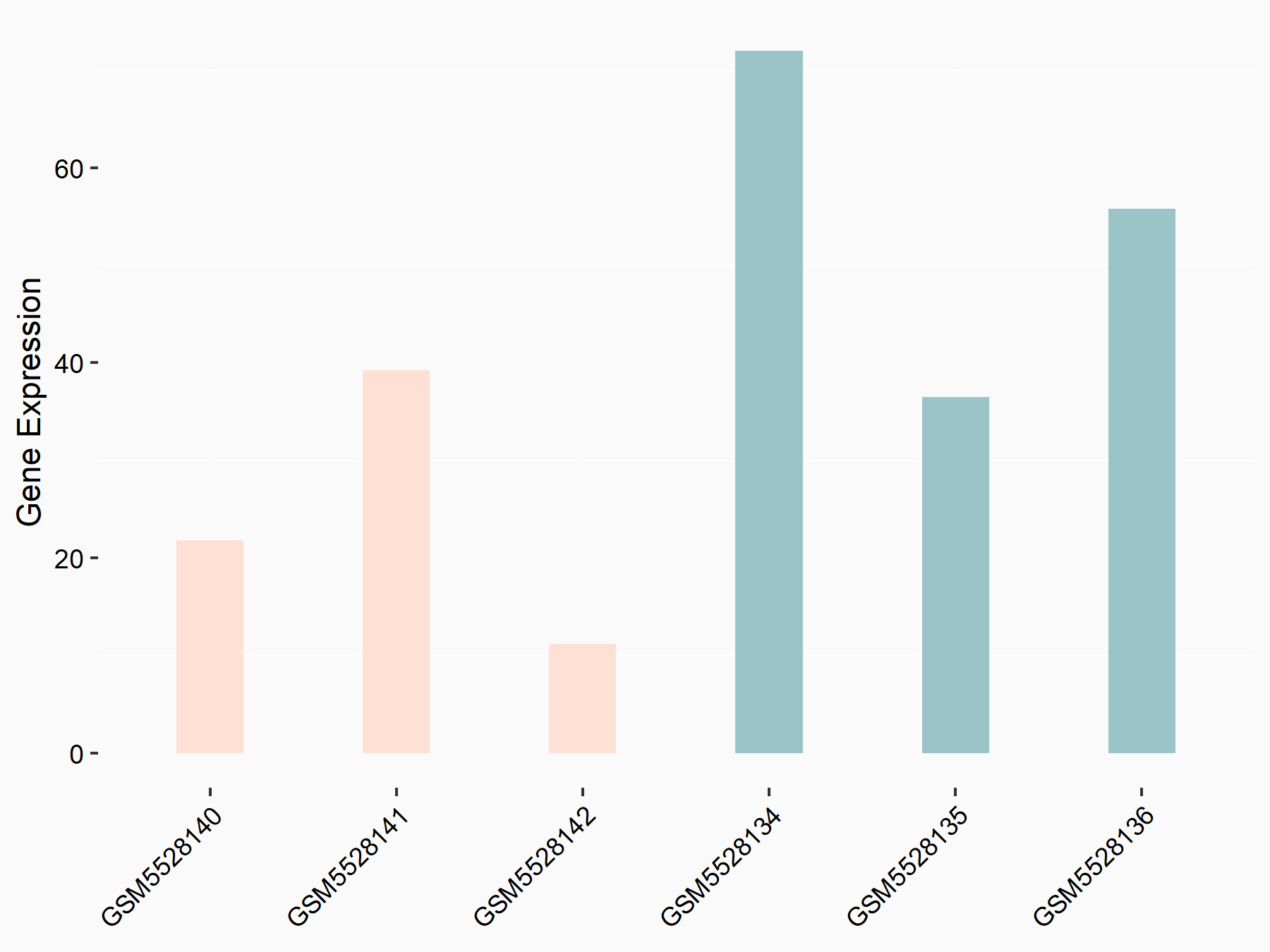 |
logFC: -1.23E+00 p-value: 1.77E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 5.66E+00 | GSE60213 |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [149] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell growth | |||
| Cell invasion | ||||
| Cell metastasis | ||||
| Response Summary | LV-sh-THAP7 antisense RNA 1 (THAP7-AS1) treatment could suppress gastric cancer growth. THAP7-AS1, transcriptionally activated by SP1 and then modified by METTL3-mediated m6A, exerts oncogenic functions, by promoting interaction between NLS and importin alpha-1 and then improving the CUL4B protein entry into the nucleus to repress the transcription of miR-22-3p and miR-320a. | |||
A disintegrin and metalloproteinase with thrombospondin motifs 9 (ADAMTS9)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HUVEC cell line | Homo sapiens |
|
Treatment: shMETTL3 HUVEC cells
Control: shScramble HUVEC cells
|
GSE157544 | |
| Regulation |
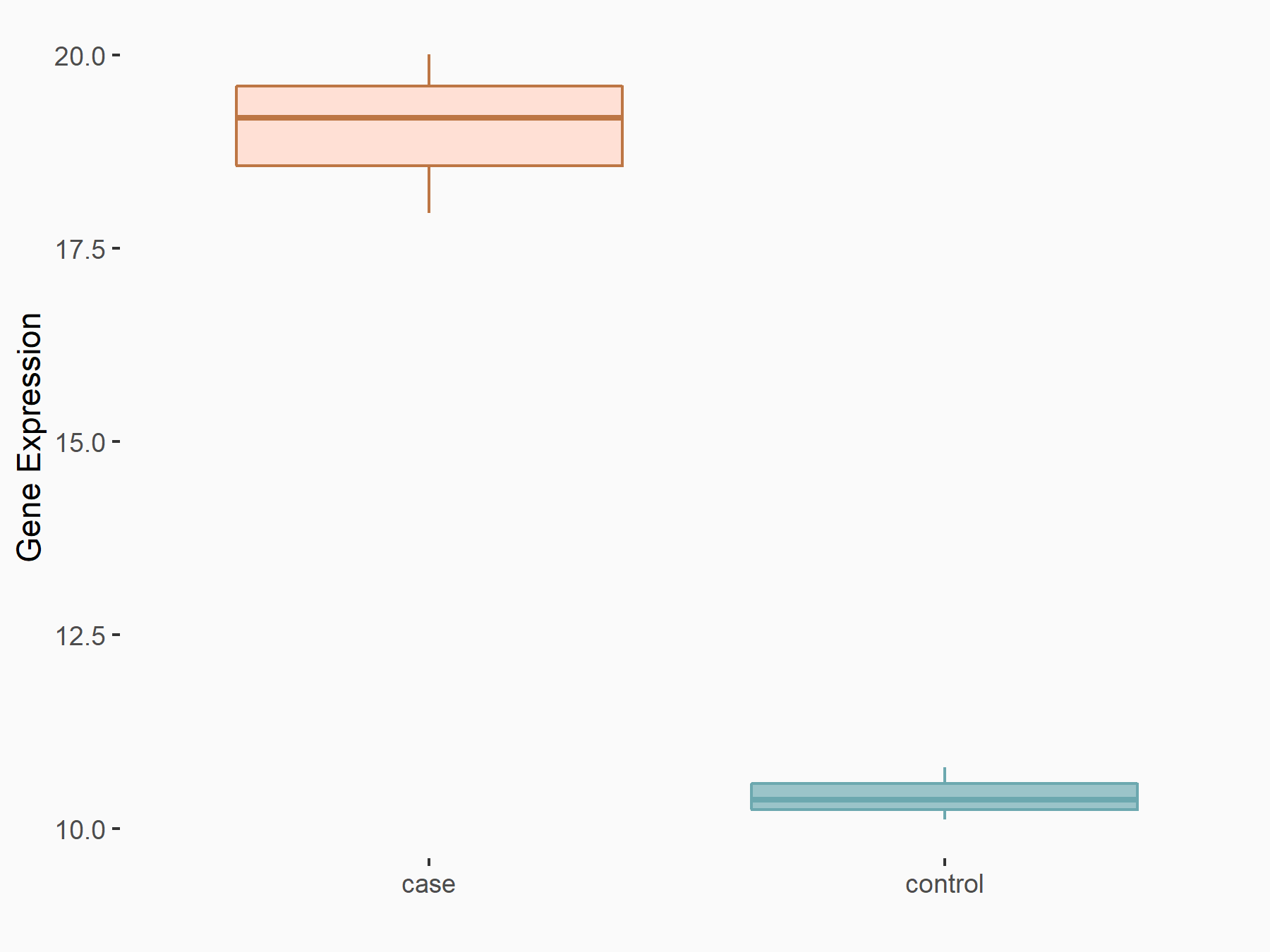 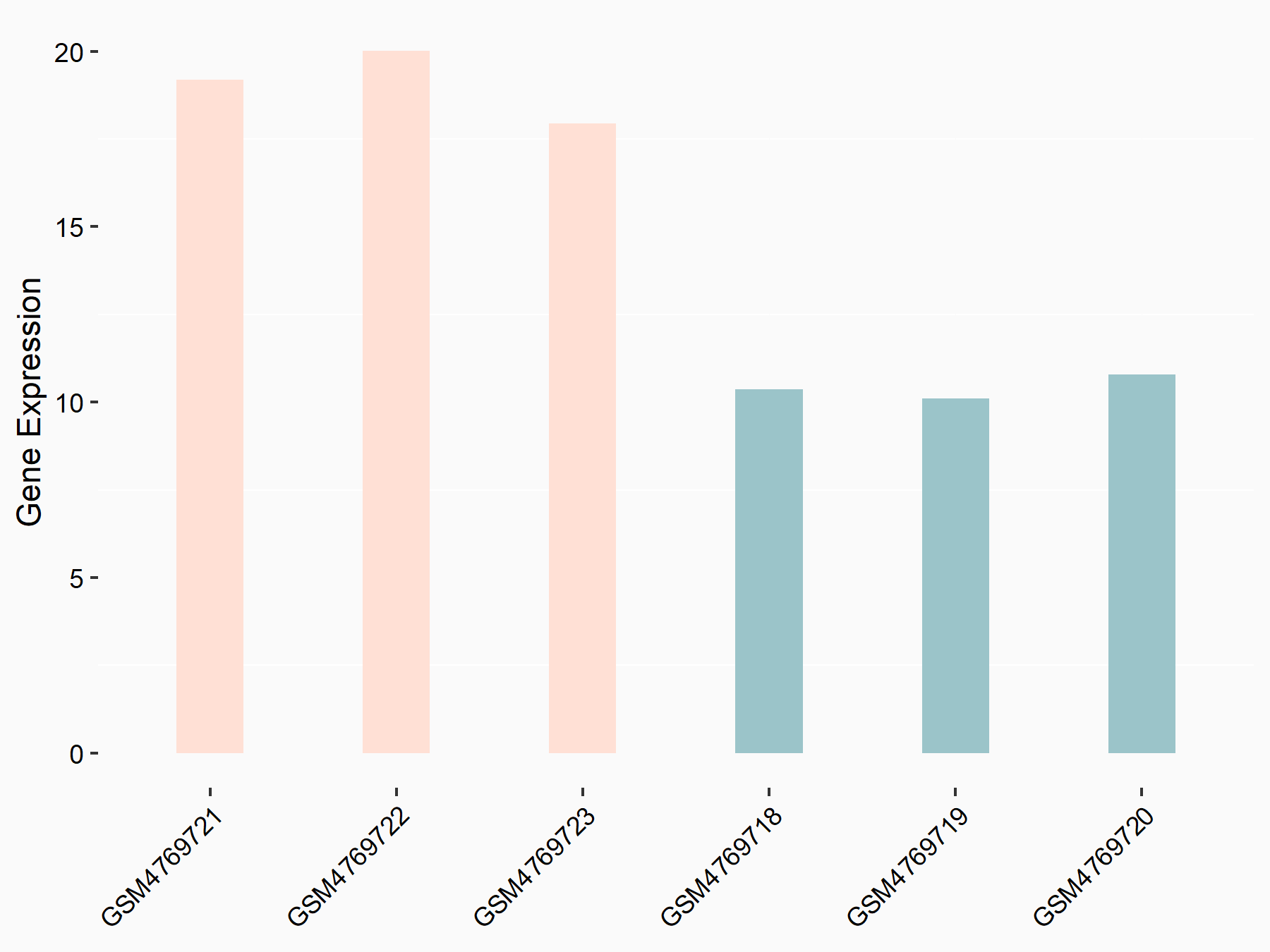 |
logFC: 8.11E-01 p-value: 4.51E-05 |
| More Results | Click to View More RNA-seq Results | |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [150] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | For the formation of xenograft tumors, 5 × 106 AGS cells mixed in Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) were subcutaneously injected into BALB/c nude mice (5-week-old male). | |||
| Response Summary | METTL3 facilitates GC progression through the A disintegrin and metalloproteinase with thrombospondin motifs 9 (ADAMTS9)-mediated PI3K/AKT pathway. | |||
Acetyl-CoA carboxylase 1 (ACC1/ACACA)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
|
Treatment: METTL3 knockdown MDA-MB-231 cells
Control: MDA-MB-231 cells
|
GSE70061 | |
| Regulation |
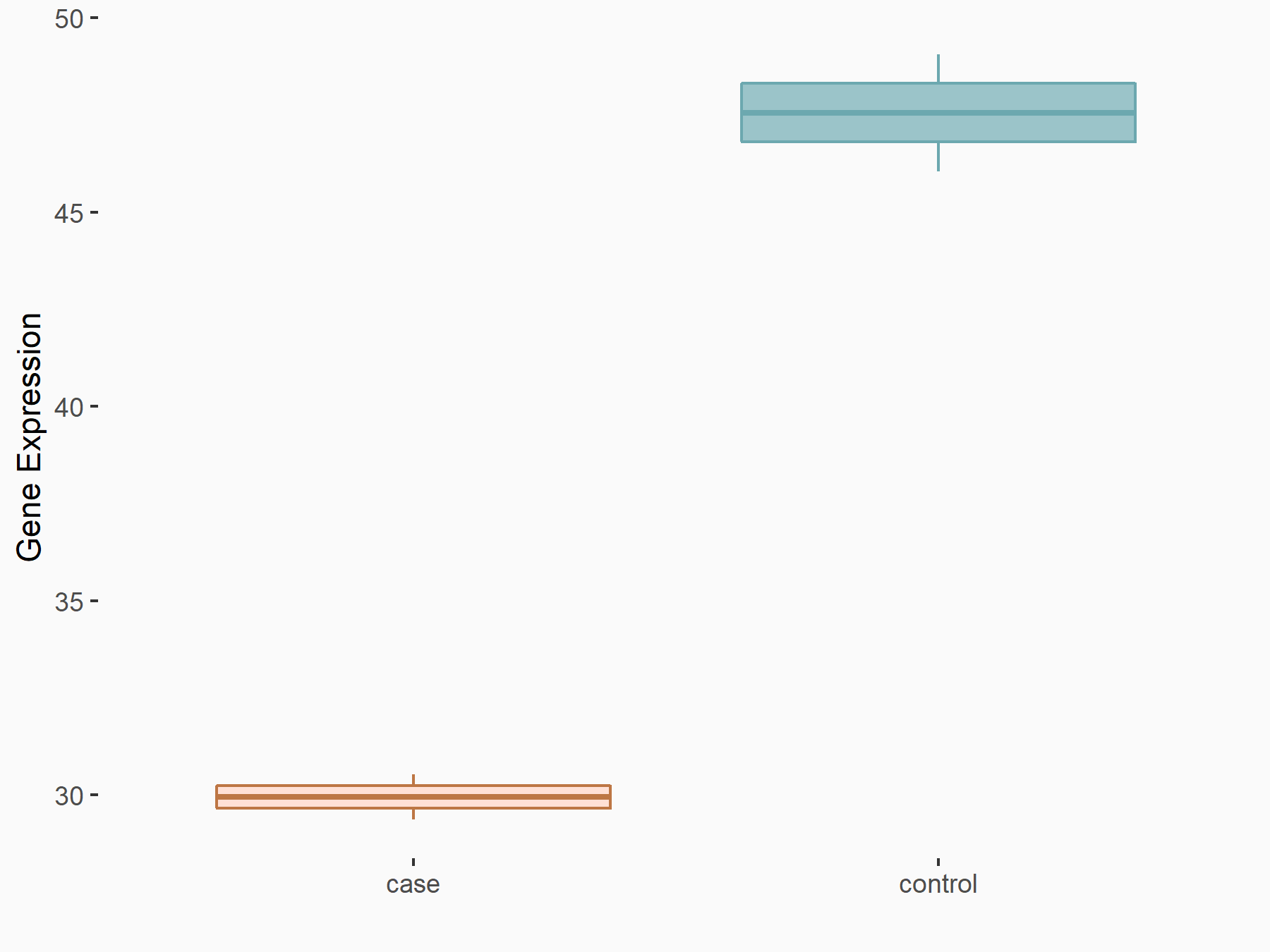 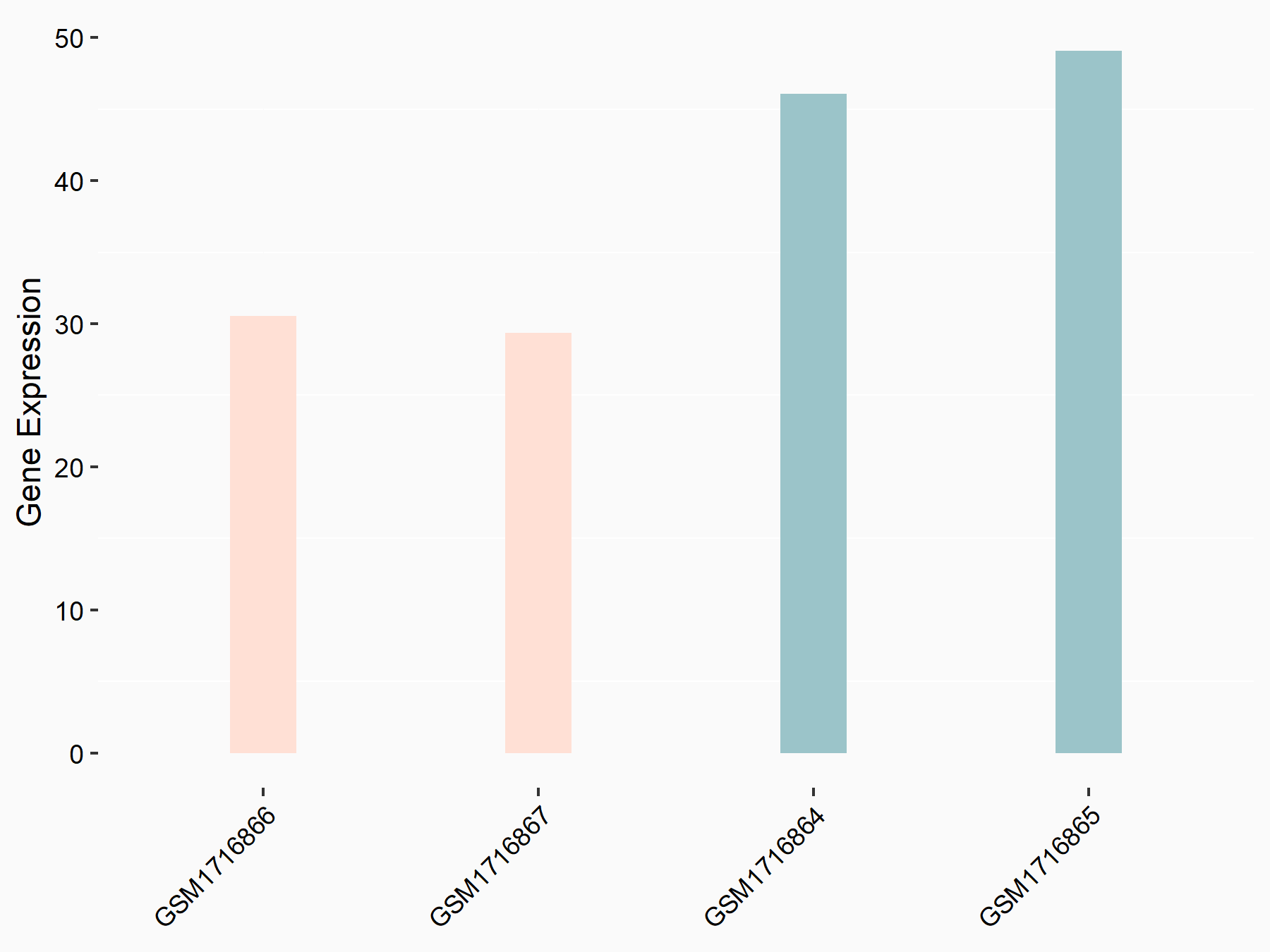 |
logFC: -6.50E-01 p-value: 2.31E-03 |
| More Results | Click to View More RNA-seq Results | |
Type 2 diabetes mellitus [ICD-11: 5A11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [48] | |||
| Responsed Disease | Type 2 diabetes mellitus [ICD-11: 5A11] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Insulin resistance | hsa04931 | ||
| Cell Process | Lipid metabolism | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | Hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were generated by crossing mice with TBG-Cre Tg mice. METTL3 flox (METTL3 fl/fl) and hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were used for experiments. | |||
| Response Summary | Type 2 diabetes (T2D) is characterized by lack of insulin, insulin resistance and high blood sugar. METTL3 silence decreased the m6A methylated and total mRNA level of Fatty acid synthase (Fasn), subsequently inhibited fatty acid metabolism. The expression of Acetyl-CoA carboxylase 1 (ACC1/ACACA), Acly, Dgat2, Ehhadh, Fasn, Foxo, Pgc1a and Sirt1, which are critical to the regulation of fatty acid synthesis and oxidation were dramatically decreased in livers of hepatocyte-specific METTL3 knockout mice. | |||
Actin, aortic smooth muscle (ACTA2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
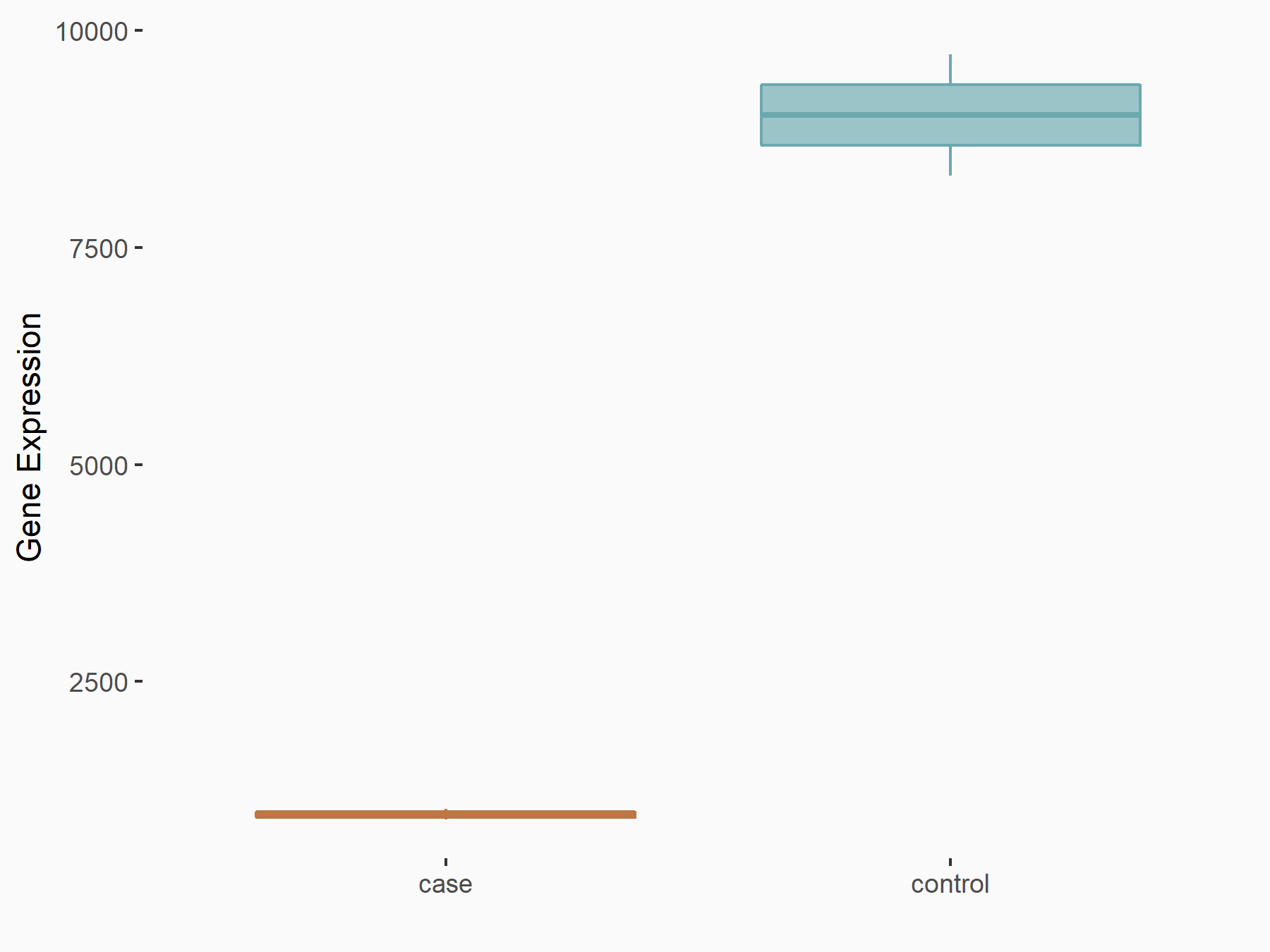 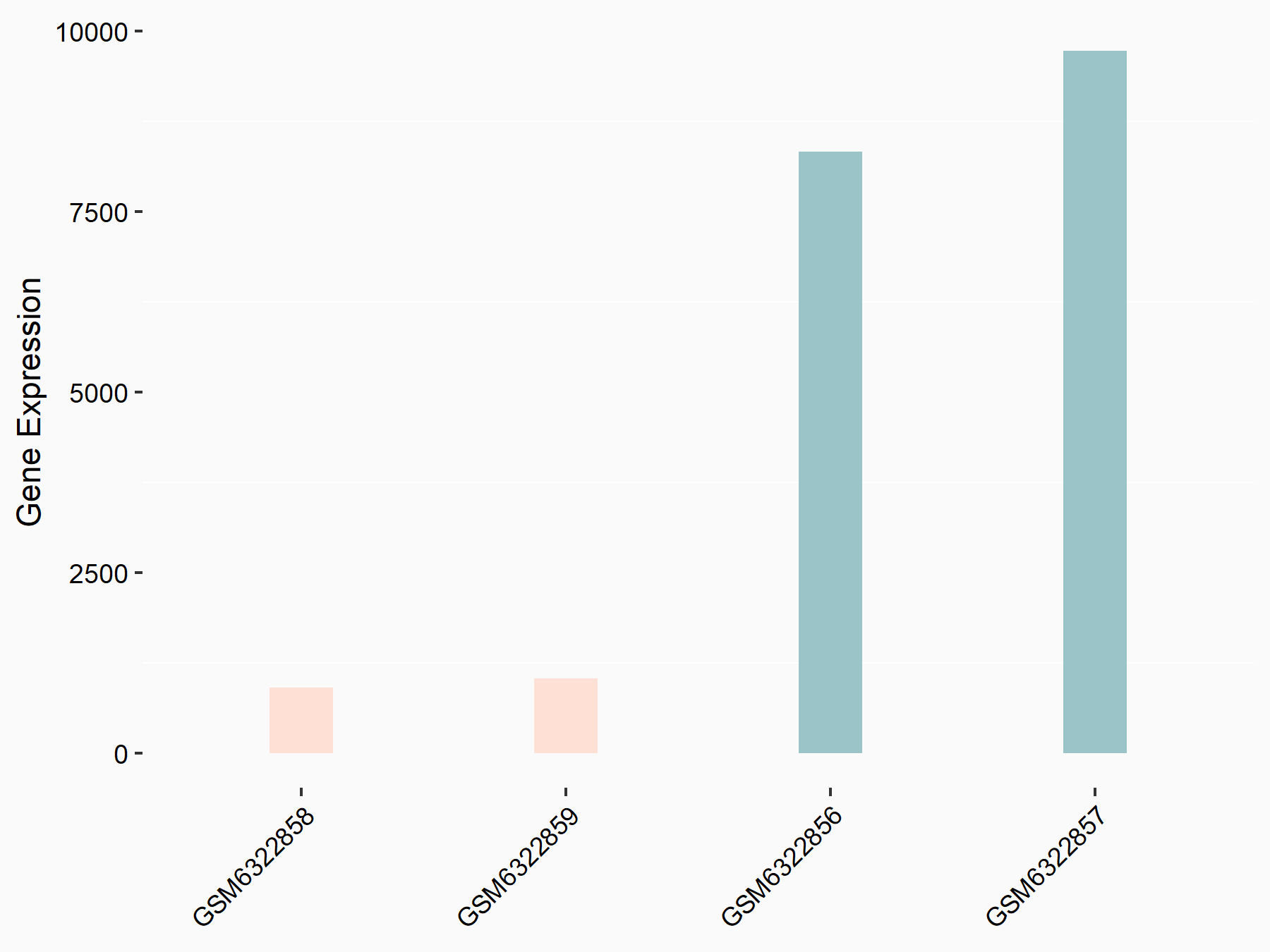 |
logFC: -3.22E+00 p-value: 6.14E-220 |
| More Results | Click to View More RNA-seq Results | |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [151] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
In-vitro Model |
BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| Response Summary | METTL3 knockdown decreased Actin, aortic smooth muscle (ACTA2) muscle actin. Taken together, our finding revealed that m6A methylation writer METTL3 serve as an oncogene in tumorigenesis of Gastric cancer. | |||
Adenylate kinase 4, mitochondrial (AK4)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
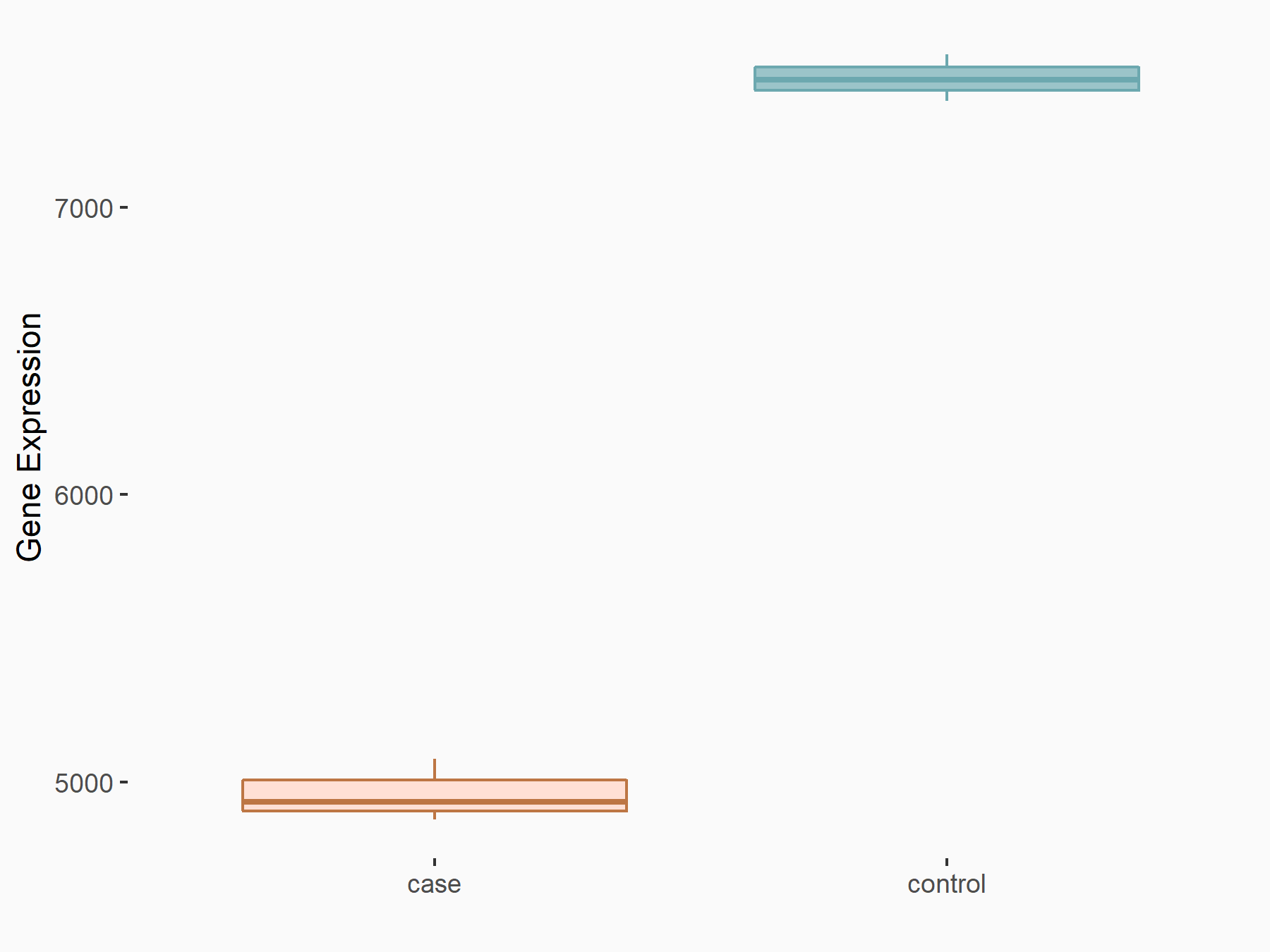 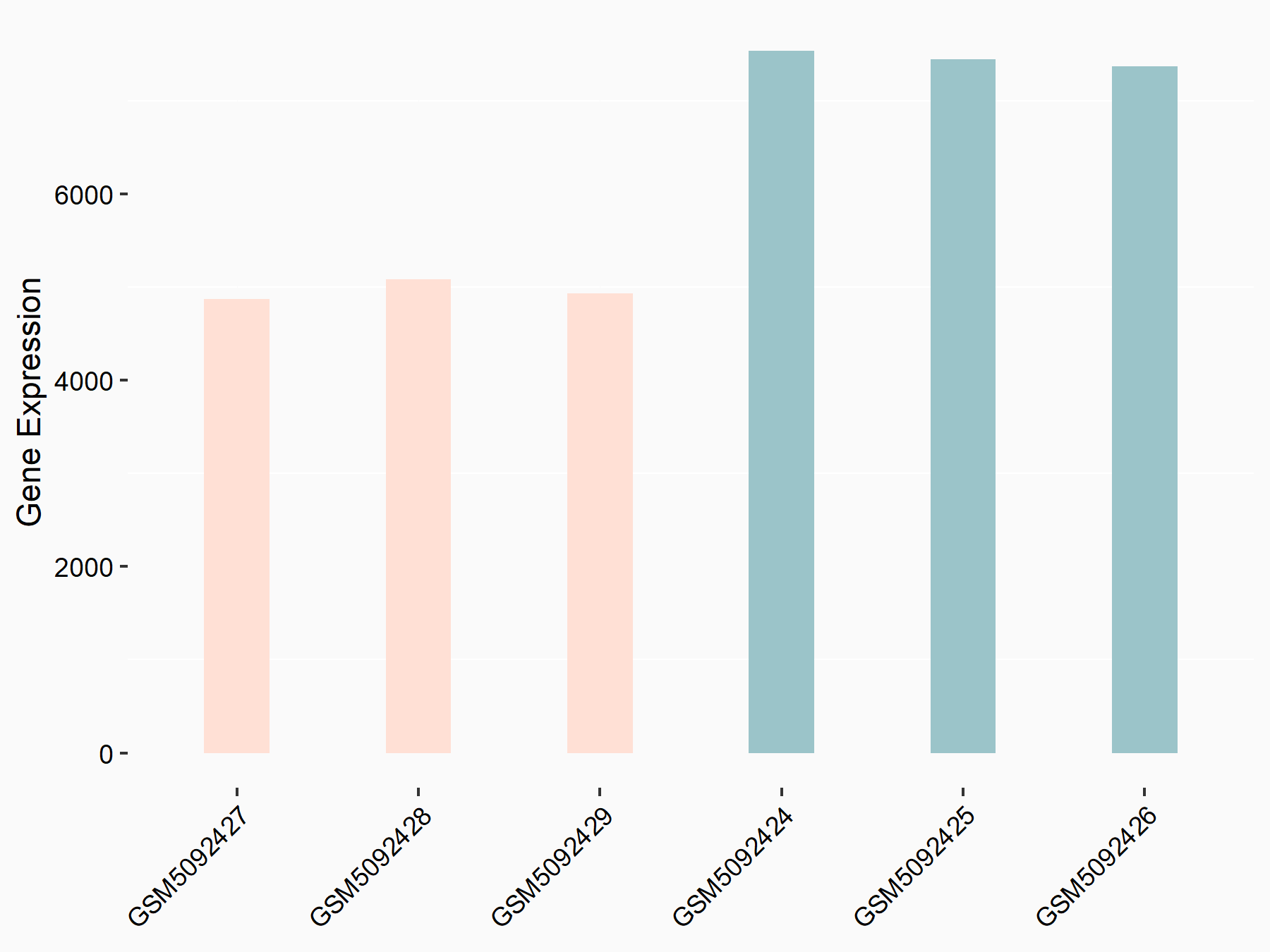 |
logFC: -5.87E-01 p-value: 2.40E-51 |
| More Results | Click to View More RNA-seq Results | |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [152] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Tamoxifen | Approved | ||
| Target Regulation | Up regulation | |||
| Cell Process | Mitochondrial apoptosis | |||
In-vitro Model |
MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| MCF7-TamR | Invasive breast carcinoma | Homo sapiens | CVCL_EG55 | |
| Response Summary | Adenylate kinase 4 modulates the resistance of breast cancer cells to tamoxifen through an m6A-based epitranscriptomic mechanism. Genetic depletion of METTL3 in TamR MCF-7 cells led to a diminished Adenylate kinase 4, mitochondrial (AK4) protein level and attenuated resistance to tamoxifen. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [265] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
In-vitro Model |
L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| BEL-7404 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6568 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
Alpha-enolase (ENO1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
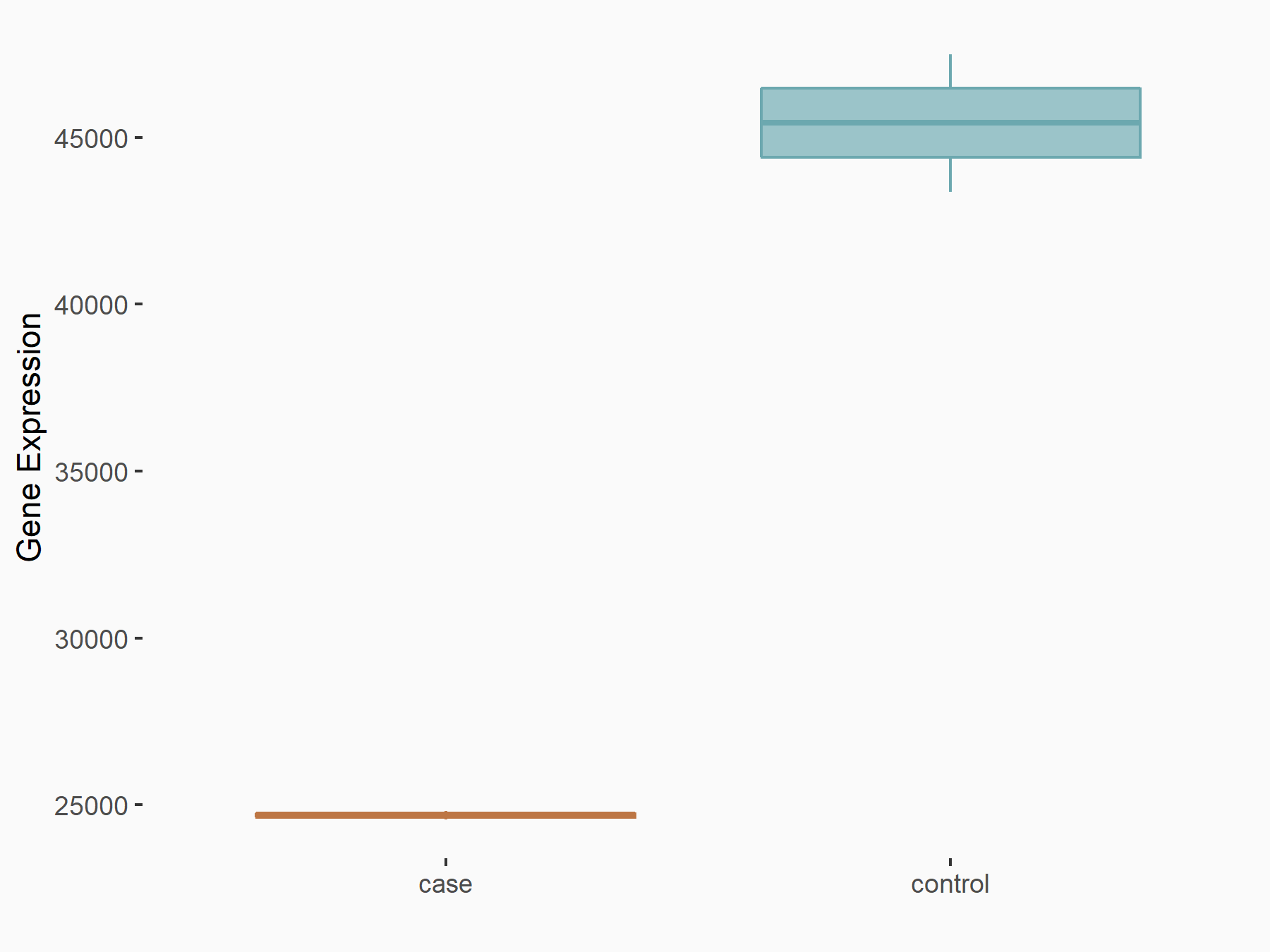 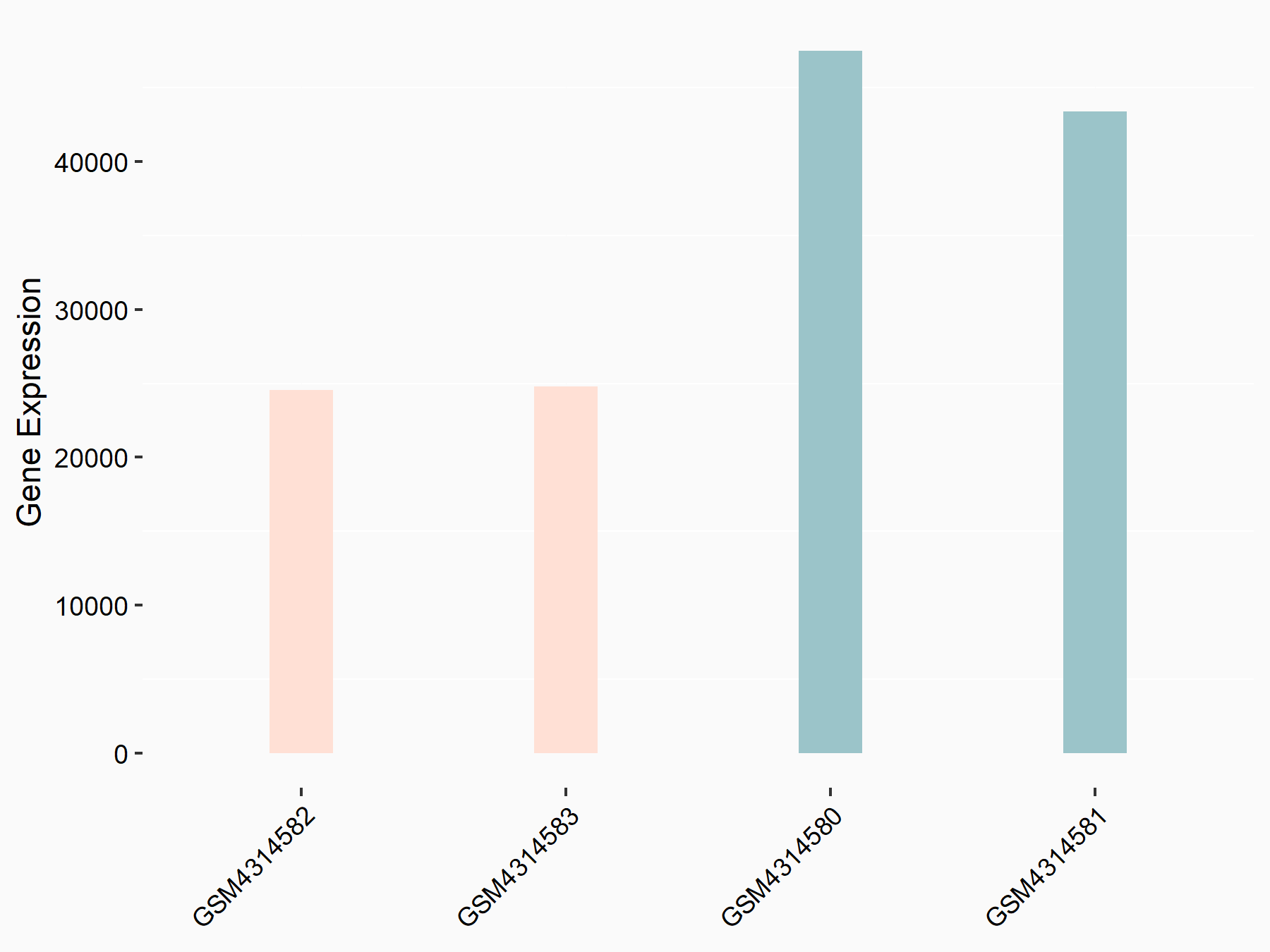 |
logFC: -8.80E-01 p-value: 3.03E-81 |
| More Results | Click to View More RNA-seq Results | |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [153] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Carbon metabolism | hsa01200 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glycolysis | |||
In-vitro Model |
PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| NCI-H441 | Lung papillary adenocarcinoma | Homo sapiens | CVCL_1561 | |
| NCI-H292 | Lung mucoepidermoid carcinoma | Homo sapiens | CVCL_0455 | |
| NCI-H2030 | Lung adenocarcinoma | Homo sapiens | CVCL_1517 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| 3LL | Malignant tumors of the mouse pulmonary system | Mus musculus | CVCL_5653 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| Calu-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0608 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| A-427 | Lung adenocarcinoma | Homo sapiens | CVCL_1055 | |
| 16HBE14o- | Normal | Homo sapiens | CVCL_0112 | |
| In-vivo Model | KP and Mettl3-/- mice were bred to generate KPM-/- mice. Afterwards, the KP and KPM-/- mice were intranasally infected under anesthesia with adeno-associated virus type 5 (AAV5) expressing Cre to initiate lung tumorigenesis along with ALKBH5-expressing AAV5 or Empty AAV5 to generate KPE, KPA, KPEM-/- and KPAM-/- spontaneous LUAD mouse models. For generation of LLC-based intra-pulmonary tumor mouse models, 1 × 107 LLC cells were injected into C57BL/6 mice via the tail vein.For cell-derived xenograft (CDX) mouse models, 1.0 × 107 H1299 or 1.5 × 107 H1975 cells were subcutaneously injected into 4-6-week-old athymic nude mice. The tumors were monitored at indicated time points and isolated for further analysis after sacrifice. | |||
| Response Summary | Alpha-enolase (ENO1) positively correlated with METTL3 and global m6A levels, and negatively correlated with ALKBH5 in human Lung adenocarcinoma(LUAD). In addition, m6A-dependent elevation of ENO1 was associated with LUAD progression. | |||
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [266] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 |
| 5637 | Bladder carcinoma | Homo sapiens | CVCL_0126 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| RT-4 | Bladder carcinoma | Homo sapiens | CVCL_0036 | |
| J82 | Bladder carcinoma | Homo sapiens | CVCL_0359 | |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | For the subcutaneous implantation model, 5 × 106 cells (n = 6 per group) were subcutaneously injected into the flank regions of 4-5 weeks female BALB/c nude mice. Tumor volume was calculated as 0.5 × W2 × L (where W and L represent a tumor's width and length, respectively). After xenografts were generated, DMSOand SB431542 (10 mg/kg, #301836-41-9, SANTA CRUZ BIOTECHNOLOGY), or ENOblock (#1177827-73-4, 8 mg/kg, MCE) were administered daily. All animal experiments were approved by The Affiliated Hospital of Qingdao University Committee on Animal Care. | |||
AMPK subunit alpha-1 (AMPK/PRKAA1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
 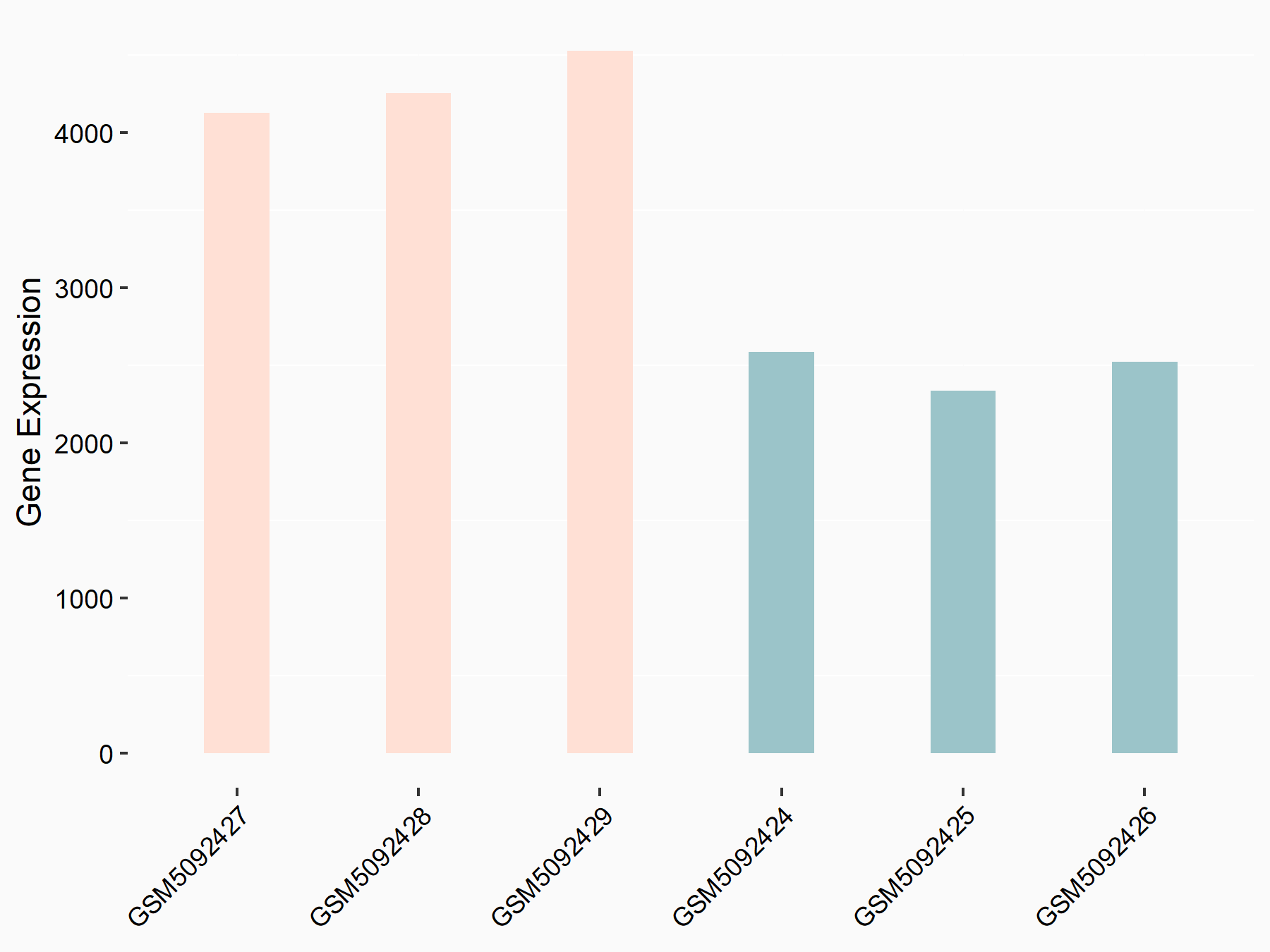 |
logFC: 7.94E-01 p-value: 1.12E-47 |
| More Results | Click to View More RNA-seq Results | |
Atherosclerosis [ICD-11: BD40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [154] | |||
| Responsed Disease | Atherosclerosis [ICD-11: BD40.Z] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | AMPK signaling pathway | hsa04152 | ||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
hTERT-RPE1 | Normal | Homo sapiens | CVCL_4388 |
| iHAEC | Normal | Homo sapiens | CVCL_C0EQ | |
| Response Summary | m6A driven machinery in virus-induced vascular endothelium damage and highlight the significance of vitamin D3 in the intervention of HCMV-induced atherosclerosis. METTL3 methylates mitochondrial calcium uniporter (MCU), the main contributor to HCMV-induced apoptosis of vascular endothelial cells, at three m6A residues in the 3'-UTR. Vitamin D3 downregulated the METTL3 by inhibiting the activation of AMPK subunit alpha-1 (AMPK/PRKAA1), thereby inhibiting the m6A modification of MCU and cell apoptosis. | |||
Anterior gradient protein 2 homolog (AGR2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
 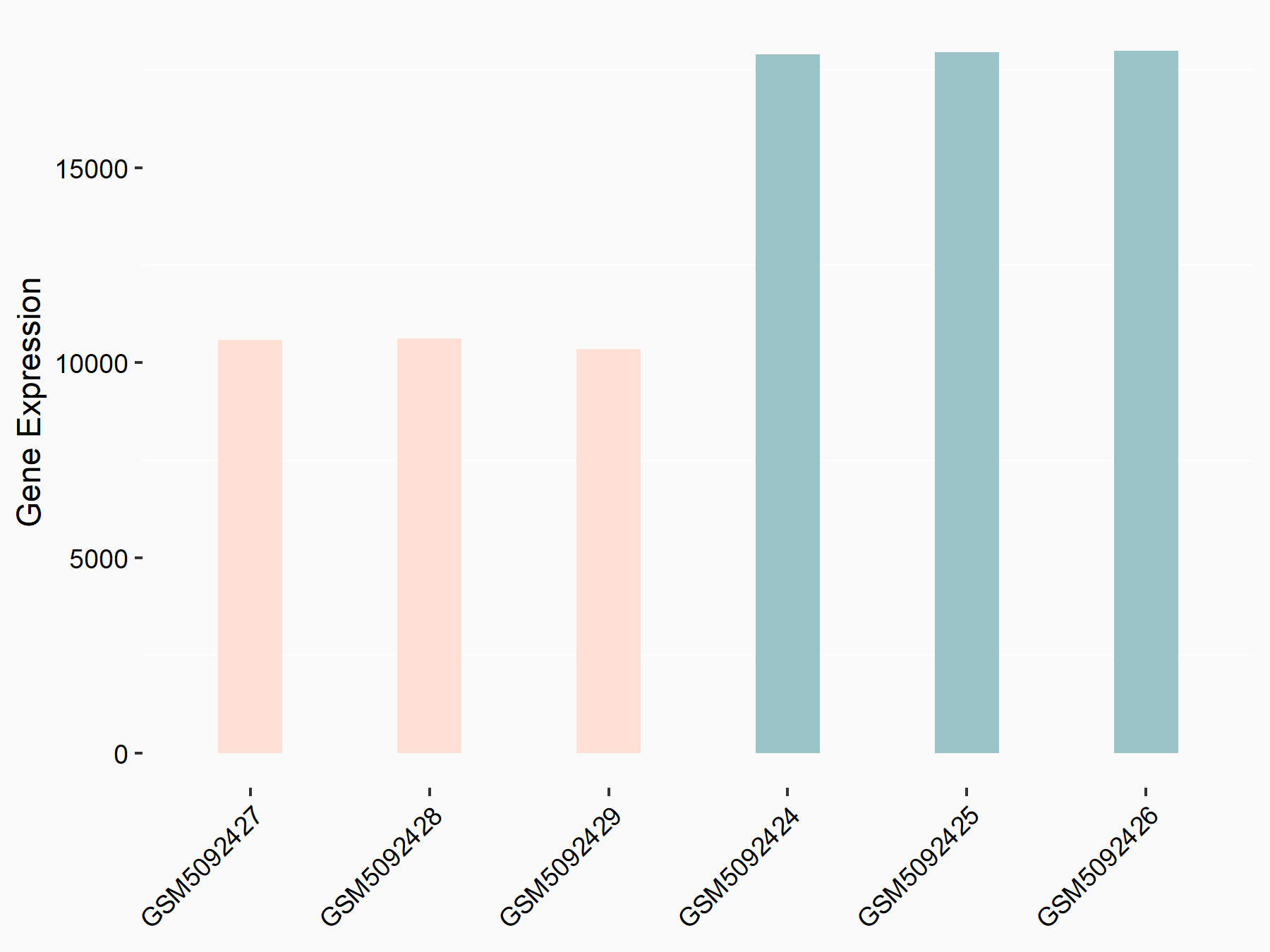 |
logFC: -7.71E-01 p-value: 1.08E-130 |
| More Results | Click to View More RNA-seq Results | |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [116] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Doxil | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
MCF7-DoxR (Adriamycin-resistant cell line MCF7-DoxR) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| In-vivo Model | Once the tumor volume increased to about 1 cm3, six groups of MCF7 bearing mice (n = 10 in each group) were injected with PBS (0.1 ml, caudal vein) and adriamycin (0.1 ml, 10 mg/kg), respectively. When the tumor reached 1.5 cm in any direction (defined as event-free survival analysis), 10 mice in each group were selected to measure the tumor size and weight on the 12th day after adriamycin injection. | |||
| Response Summary | METTL3 can regulate the expression of MALAT1 through m6A, mediate the E2F1/Anterior gradient protein 2 homolog (AGR2) axis, and promote the adriamycin resistance of breast cancer. | |||
ATP-citrate synthase (ACLY)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3 knockout liver
Control: Wild type liver cells
|
GSE198513 | |
| Regulation |
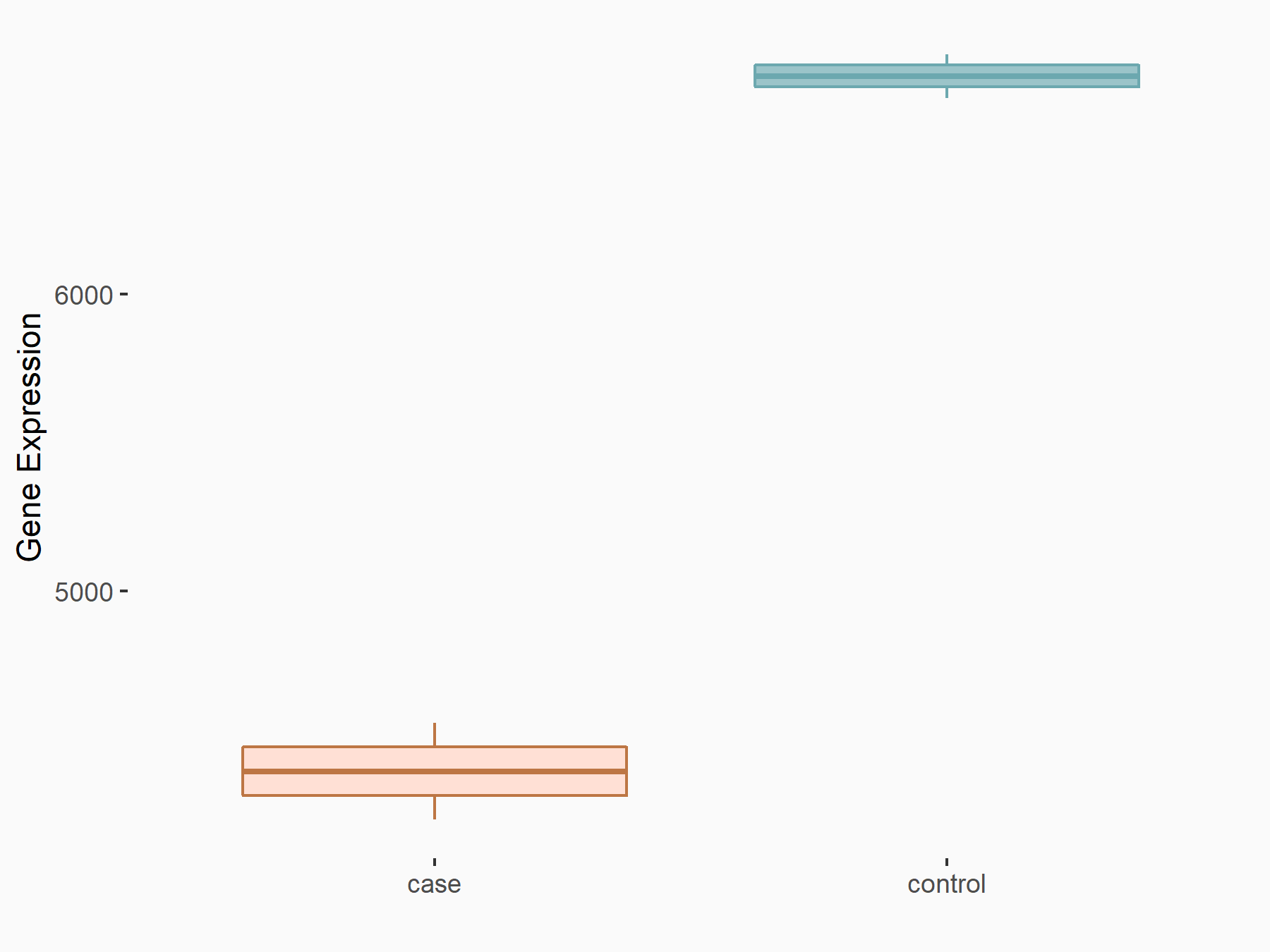 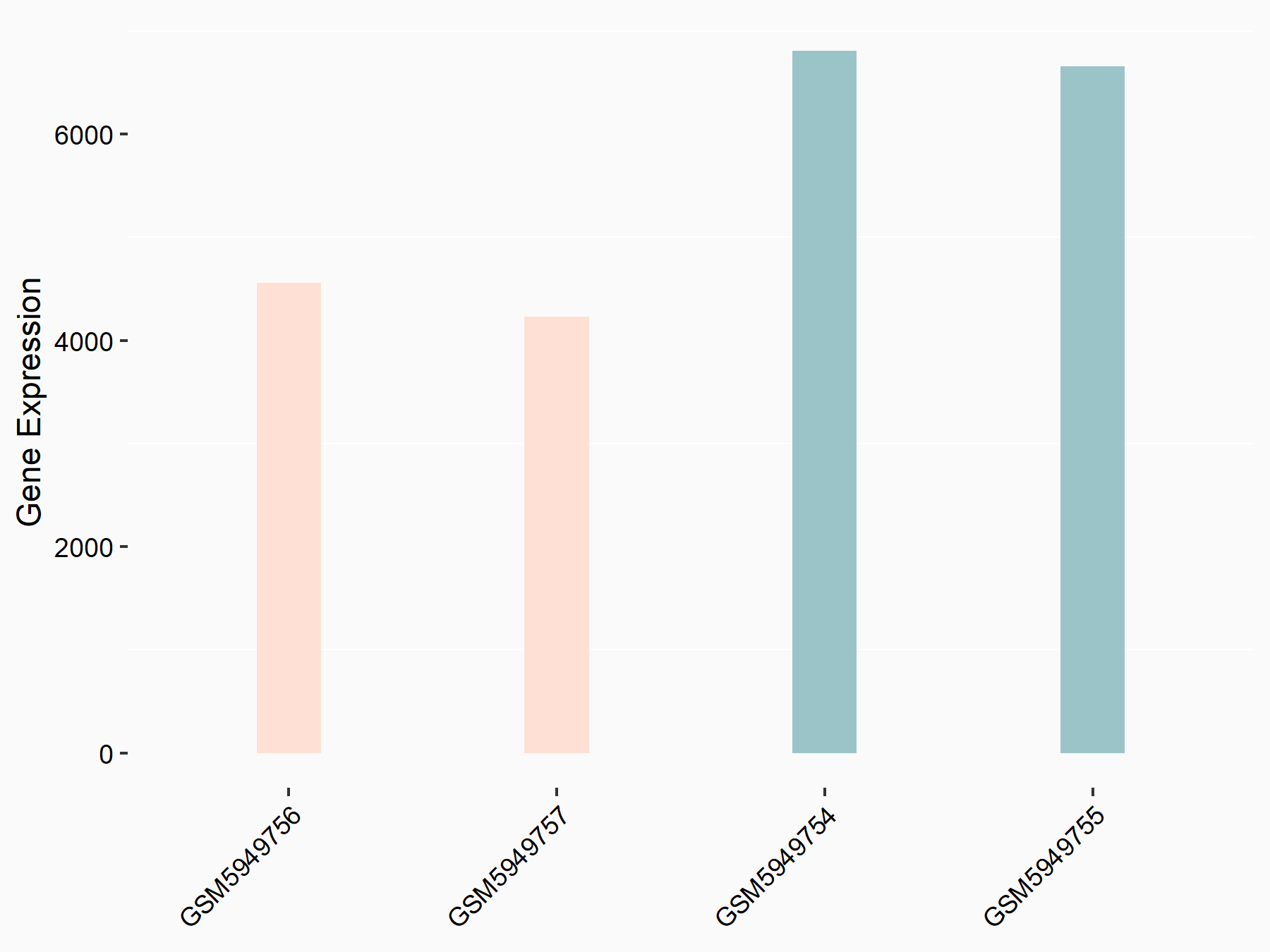 |
logFC: -6.16E-01 p-value: 1.78E-18 |
| More Results | Click to View More RNA-seq Results | |
Type 2 diabetes mellitus [ICD-11: 5A11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [48] | |||
| Responsed Disease | Type 2 diabetes mellitus [ICD-11: 5A11] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Insulin resistance | hsa04931 | ||
| Cell Process | Lipid metabolism | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | Hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were generated by crossing mice with TBG-Cre Tg mice. METTL3 flox (METTL3 fl/fl) and hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were used for experiments. | |||
| Response Summary | Type 2 diabetes (T2D) is characterized by lack of insulin, insulin resistance and high blood sugar. METTL3 silence decreased the m6A methylated and total mRNA level of Fatty acid synthase (Fasn), subsequently inhibited fatty acid metabolism. The expression of Acc1, ATP-citrate synthase (ACLY), Dgat2, Ehhadh, Fasn, Foxo, Pgc1a and Sirt1, which are critical to the regulation of fatty acid synthesis and oxidation were dramatically decreased in livers of hepatocyte-specific METTL3 knockout mice. | |||
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [112] | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycerolipid metabolism | hsa00561 | ||
| Cell Process | Lipid metabolism | |||
In-vitro Model |
LM3 | Malignant neoplasms | Mus musculus | CVCL_D269 |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| In-vivo Model | Mice with a Tmem30a deletion specifically in pancreatic beta cells were generated as previously described. Mice developed with NAFLD were named for Tmem30a-associated NAFLD (TAN) mice. The littermate mice with genotypes of Tmem30aloxP/loxP were used as controls. | |||
| Response Summary | Targeting METTL3/14 in vitro increases protein level of ATP-citrate synthase (ACLY) and SCD1 as well as triglyceride and cholesterol production and accumulation of lipid droplets. These findings demonstrate a new NAFLD mouse model that provides a study platform for DM2-related NAFLD and reveals a unique epitranscriptional regulating mechanism for lipid metabolism via m6A-modified protein expression of ACLY and SCD1. | |||
ATP-dependent translocase ABCB1 (ABCB1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
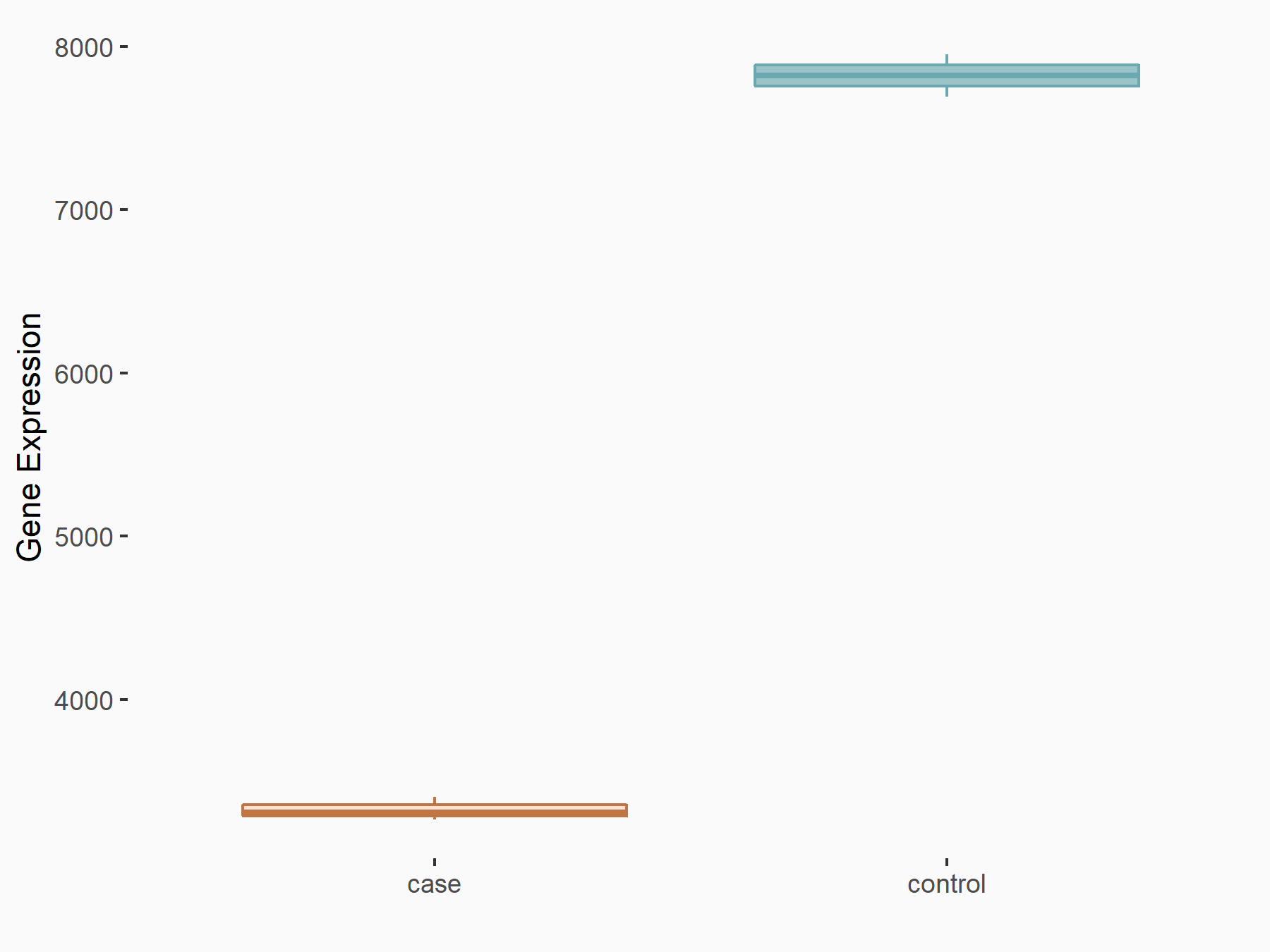 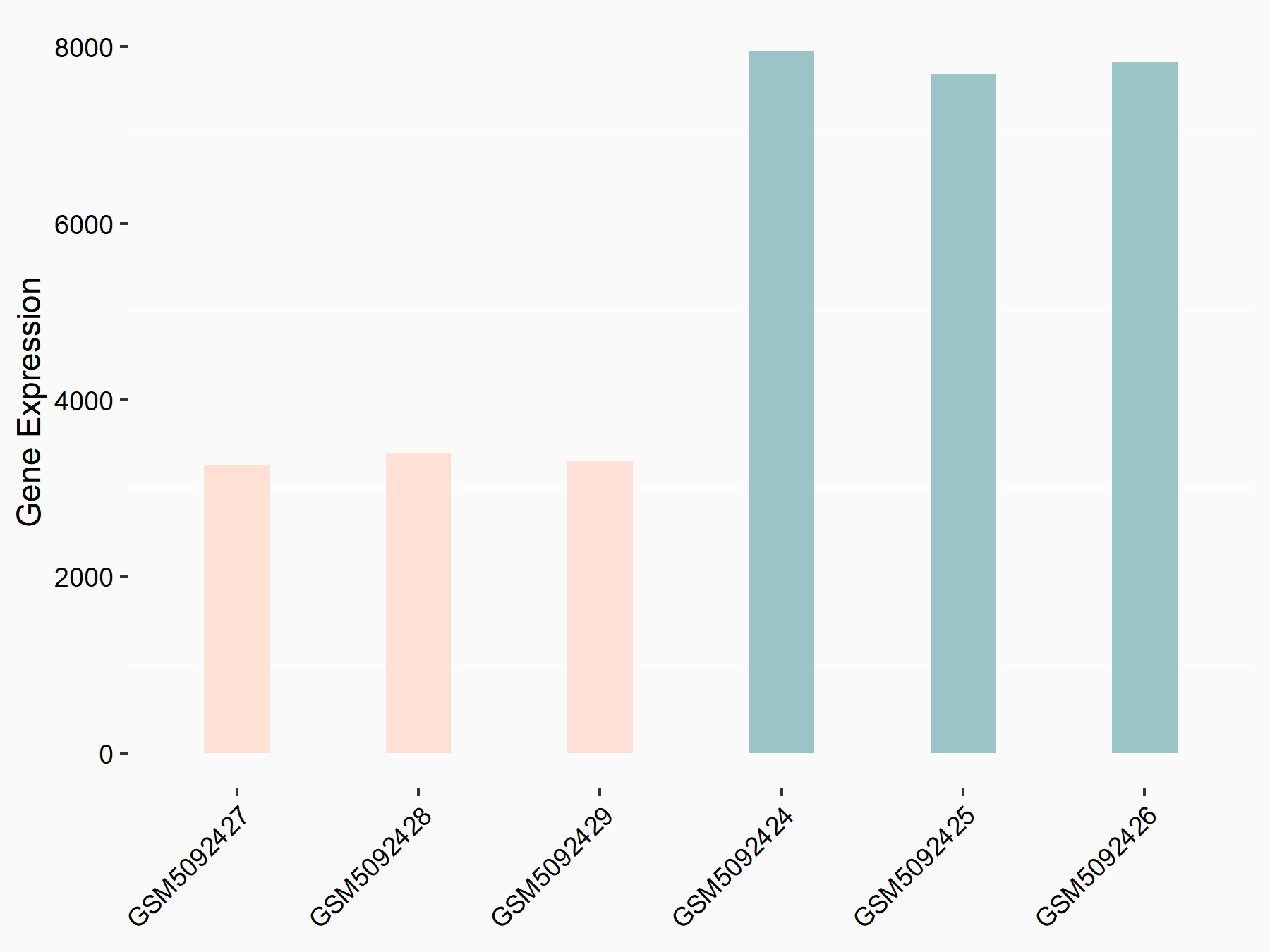 |
logFC: -1.24E+00 p-value: 2.14E-202 |
| More Results | Click to View More RNA-seq Results | |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [22] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Doxil | Approved | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell growth and death | |||
| Cell apoptosis | ||||
In-vitro Model |
ADR-resistant MCF-7 (MCF-7/ADR) cells (Human breast cancer doxorubicin-resistant cell line) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| In-vivo Model | Cell suspensions (2 × 106 cells/mL) made with MCF-7/ADR cells stably expressing METTL3 and/or miR-221-3p inhibitor were subcutaneously implanted into each mouse. One week later, xenografted mice were injected with 0.1 mL ADR (25 mg/kg, intraperitoneal injection) twice a week. | |||
| Response Summary | METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. METTL3 knockdown was shown to reduce the expression of miR-221-3p by reducing pri-miR-221-3p m6A mRNA methylation, reducing the expression of ATP-dependent translocase ABCB1 (ABCB1) and BCRP, and inducing apoptosis. Identified the METTL3/miR-221-3p/HIPK2/Che-1 axis as a novel signaling event that will be responsible for resistance of BC cells to ADR. | |||
Beclin-1 associated RUN domain containing protein (RUBCN/Rubicon)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
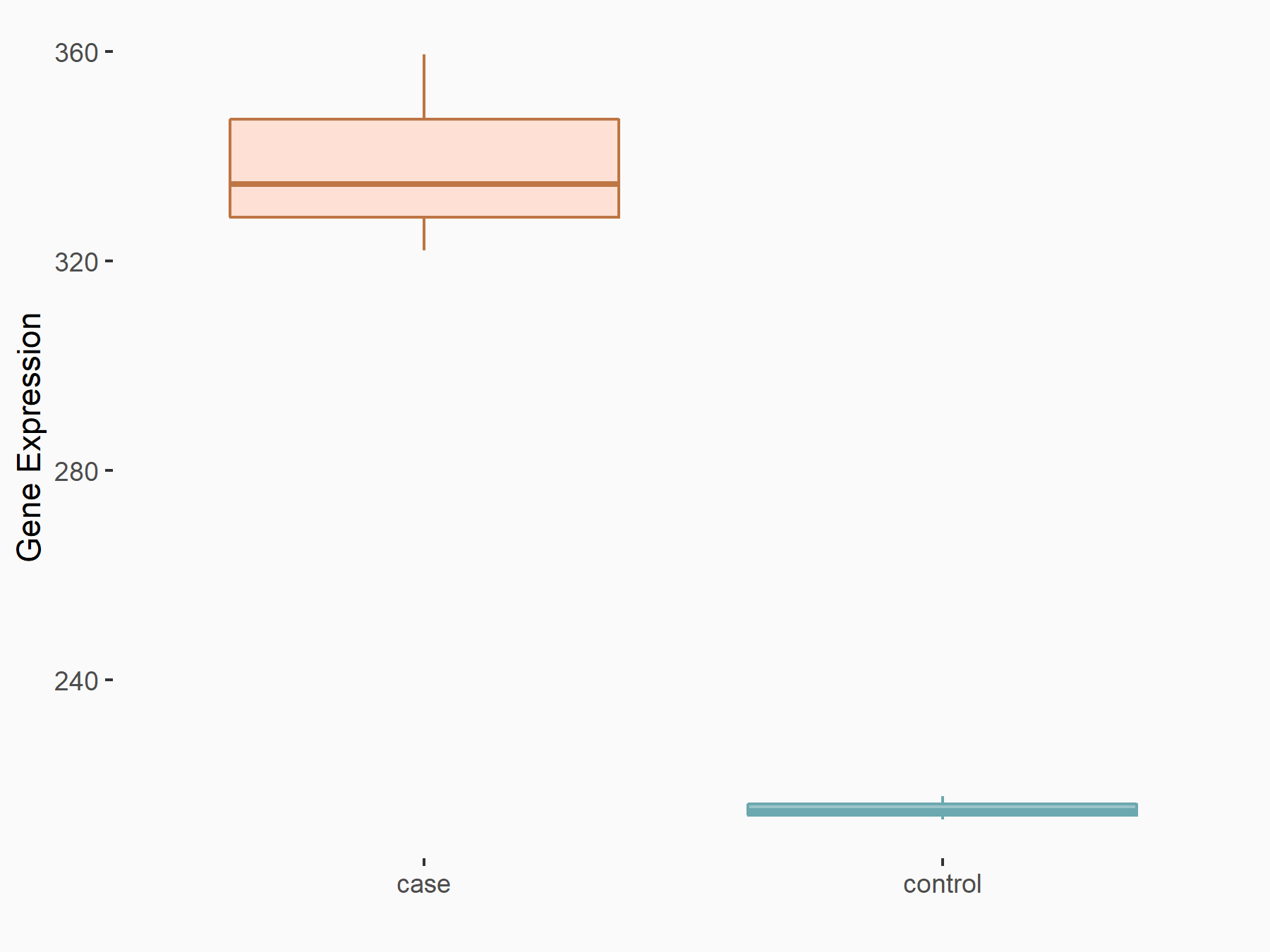 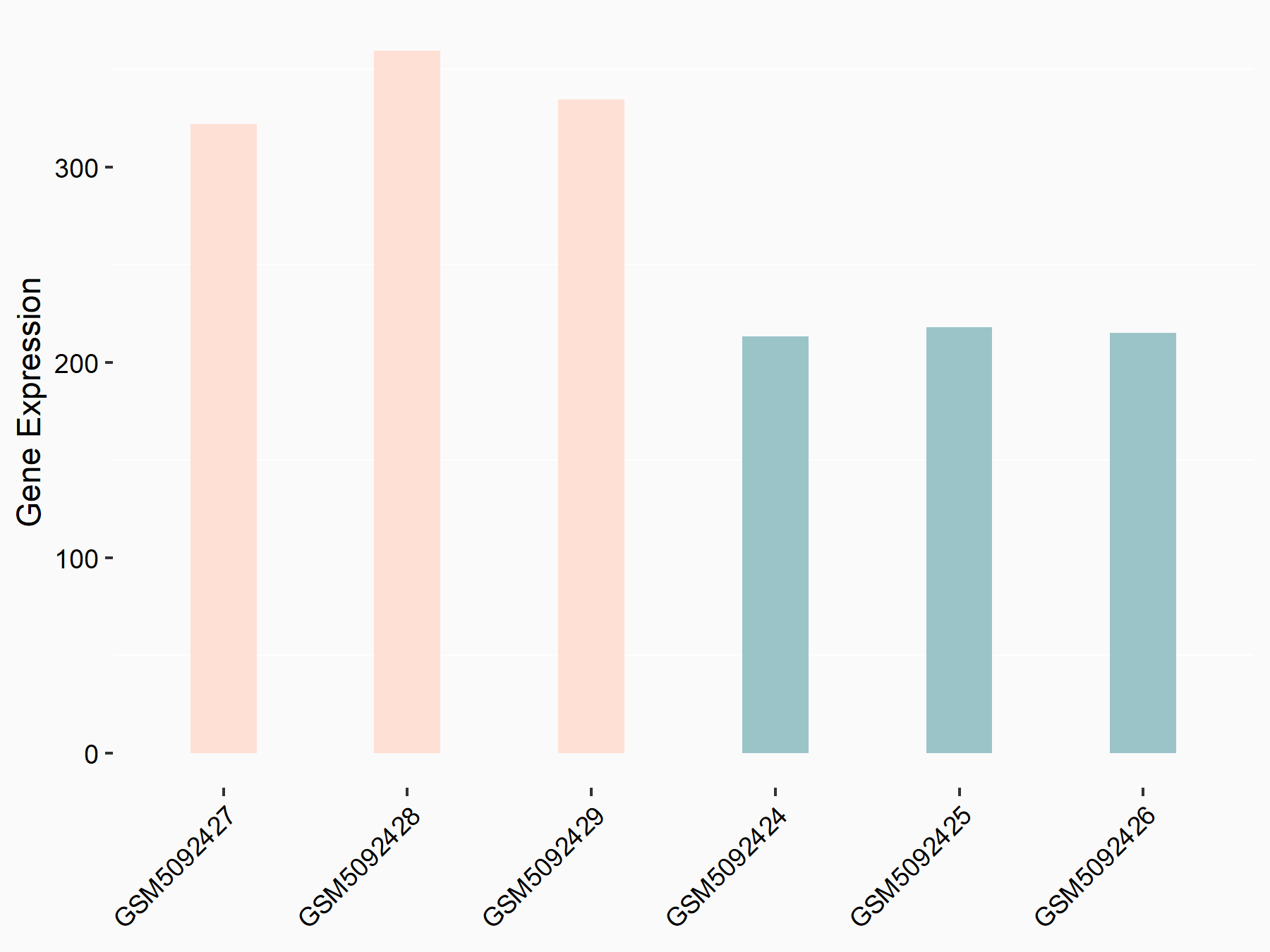 |
logFC: 6.54E-01 p-value: 1.80E-06 |
| More Results | Click to View More RNA-seq Results | |
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [155] | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell autophagy | |||
In-vitro Model |
AML12 | Normal | Mus musculus | CVCL_0140 |
| Hepa 1-6 | Hepatocellular carcinoma of the mouse | Mus musculus | CVCL_0327 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | All mice were housed in specific pathogen-free conditions in the animal facility with constant temperature and humidity under a 12-h light/12-h dark cycle, with free access to water and food. After a week of adaptation, they were then divided into two groups randomly and fed with a HFD (60 kcal% fat, D12492, Research Diets) or standard normal chow diet (CD) for 16 weeks, respectively. | |||
| Response Summary | The upregulation of METTL3 and YTHDF1 induced by lipotoxicity contributes to the elevated expression level of Beclin-1 associated RUN domain containing protein (RUBCN/Rubicon) in an m6A-dependent manner, which inhibits the fusion of autophagosomes and lysosomes and further suppresses the clearance of LDs via lysosomes in nonalcoholic fatty liver disease. | |||
Brain and muscle ARNT-like 1 (Bmal1/ARNTL)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
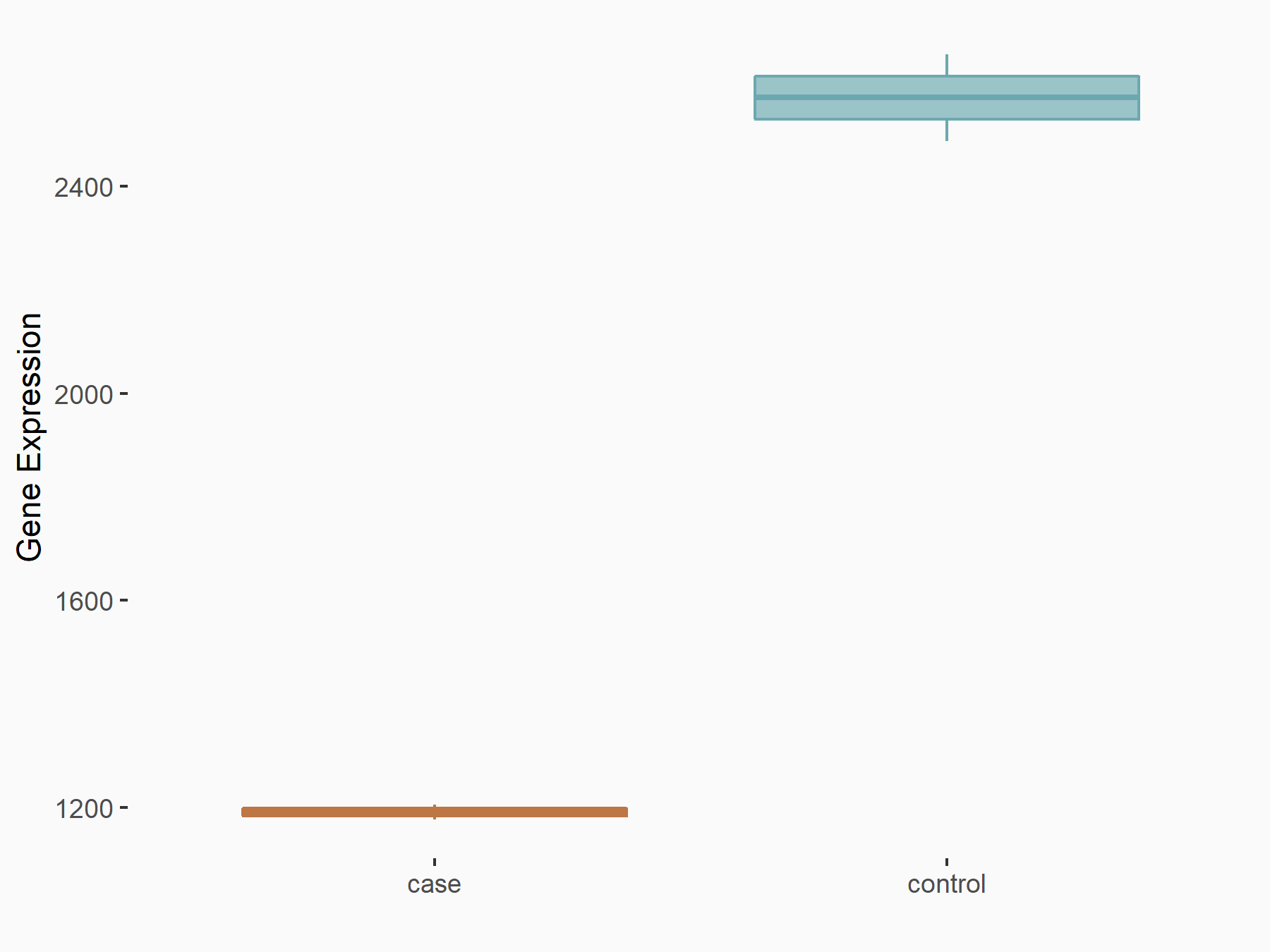 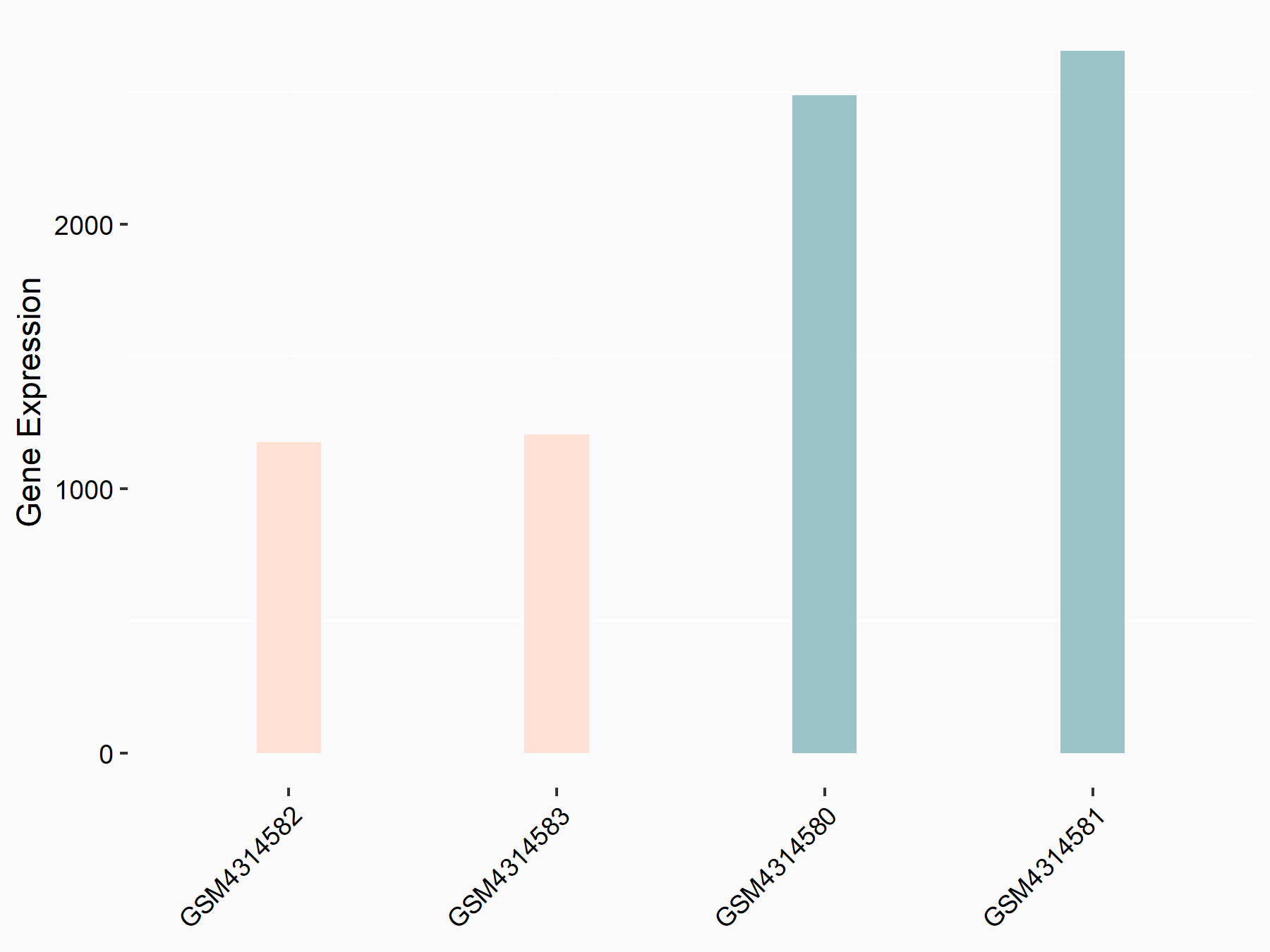 |
logFC: -1.11E+00 p-value: 1.53E-46 |
| More Results | Click to View More RNA-seq Results | |
Metabolic disorders [ICD-11: 5D2Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [92] | |||
| Responsed Disease | Metabolic disorders [ICD-11: 5D2Z] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Adipocytokine signaling pathway | hsa04920 | ||
| Cell Process | Llipid metabolism | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| Hepa 1-6 | Hepatocellular carcinoma of the mouse | Mus musculus | CVCL_0327 | |
| In-vivo Model | Liver-specific Bmal1f/f-AlbCre-knockout mice were purchased from Jackson Laboratory. C57BI/6J or Bmal1f/f-AlbCre-knockout male mice were maintained under a 12 hr light/12 hr dark (LD) cycle (ZT0 = 6 AM) and fed ad libitum with normal rodent chow (2018 Global 18% Protein diet, Envigo) and water. At 10-14 weeks of age, 10 male mice per group were sacrificed via CO2 asphyxiation at Zeitgeber Time (ZT) 0,2,6,10,12,14,18,22. In order to induce high levels of ROS in the liver, WT male mice were fasted 12 h and followed by intraperitoneal injection with 300 mg/kg APAP dissolved in PBS and re-fed. | |||
| Response Summary | PPaRalpha to mediate its mRNA stability to regulate lipid metabolism. Hepatic deletion of Brain and muscle ARNT-like 1 (Bmal1/ARNTL) increases m6A mRNA methylation, particularly of PPaRalpha. Inhibition of m6A methylation via knockdown of m6A methyltransferase METTL3 decreases PPaR-Alpha m6A abundance and increases PPaRalpha mRNA lifetime and expression, reducing lipid accumulation in cells in vitro. YTHDF2 binds to PPaRalpha to mediate its mRNA stability to regulate lipid metabolism. Transcriptional regulation of circadian rhythms is essential for lipid metabolic homeostasis, disruptions of which can lead to metabolic diseases. | |||
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [156] | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | M3LKO (Mettl3fl/fl; Alb-Cre) mice were generated by crossing Mettl3fl/fl mice (provided by Dr. Jacob Hanna) with albumin-Cre mice (Jackson Laboratories, Bar Harbor, ME), and genotypes were confirmed by tail-DNA PCR using primers as previously described. | |||
| Response Summary | Liver-specific Mettl3 knockout mice exhibited global decrease in m6A on polyadenylated RNAs and pathologic features associated with nonalcoholic fatty liver disease. Studies in the M3LKO model indicated that METTL3 exhibits pleotropic function to maintain liver homeostasis by deregulating m6A profile and expression of the liver transcriptome. A significant decrease in total Brain and muscle ARNT-like 1 (Bmal1/ARNTL) and Clock mRNAs but an increase in their nuclear levels were observed in M3LKO livers, suggesting impaired nuclear export. | |||
C-X-C motif chemokine 10 (Cxcl10)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | CT26 cell line | Mus musculus |
|
Treatment: METTL3 knockout CT26 cells
Control: CT26 cells
|
GSE142589 | |
| Regulation |
 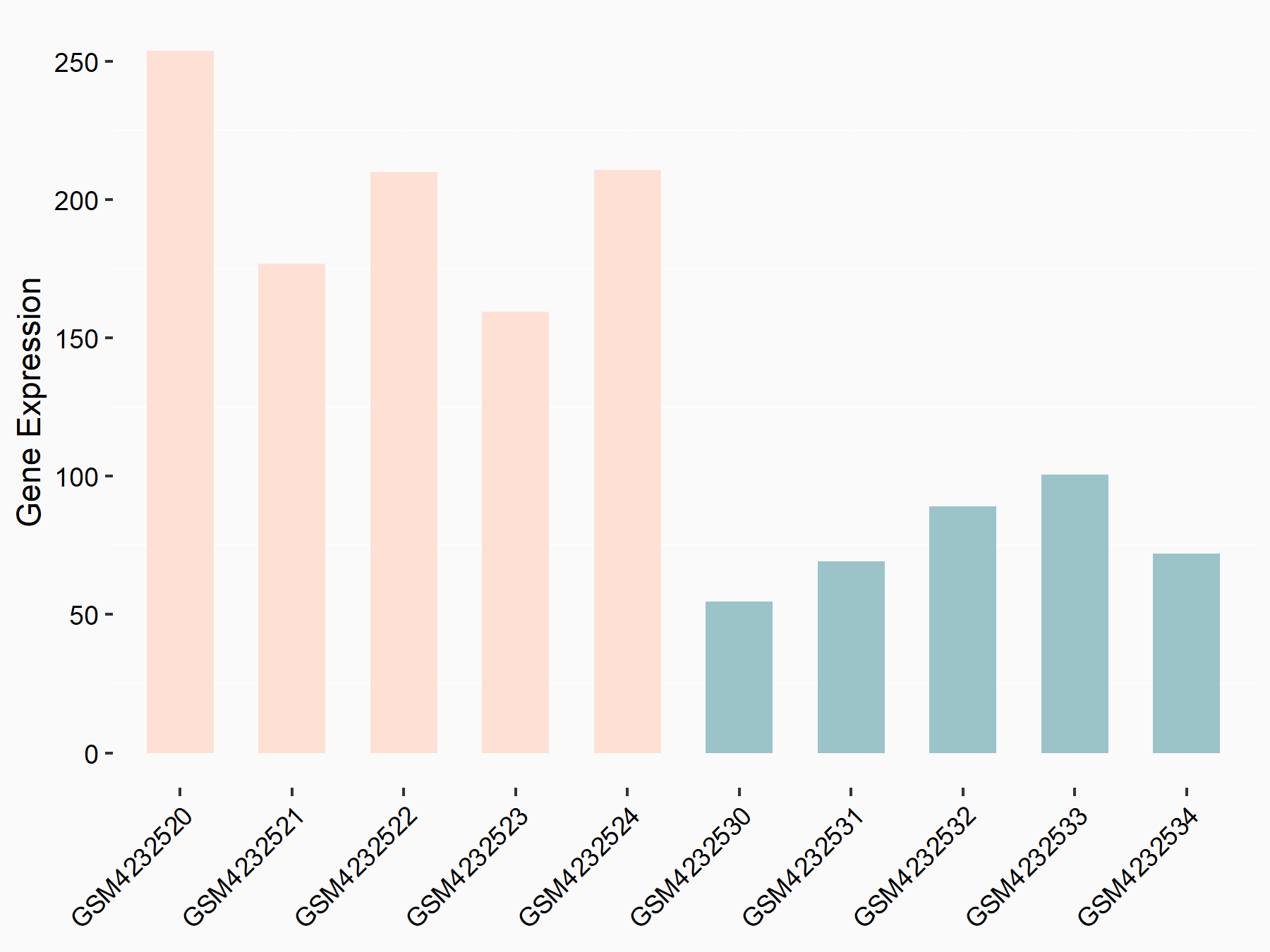 |
logFC: 1.38E+00 p-value: 1.00E-07 |
| More Results | Click to View More RNA-seq Results | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [118] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
| Cell Process | Immunity | |||
In-vitro Model |
CT26 | Mouse colon adenocarcinoma | Mus musculus | CVCL_7254 |
| B16-GM-CSF (B16-GM-CSF cell line was a kind gift from Drs. Glenn Dranoff and Michael Dougan (Dana-Farber/Harvard Cancer Center)) | ||||
| B16-F10 | Mouse melanoma | Mus musculus | CVCL_0159 | |
| In-vivo Model | 2 × 106 CT26 cells with knockout of Mettl3, Mettl14, Mettl3/Stat1, Mettl3/Irf1, Mettl14/Stat1, or Mettl14/Irf1 and control were suspended in 200 uL of PBS/Matrigel (Corning) (1:1) and then subcutaneously inoculated into flank of each mouse. | |||
| Response Summary | In colorectal cancer, Mettl3- or Mettl14-deficient tumors increased cytotoxic tumor-infiltrating CD8+ T cells and elevated secretion of IFN-gamma, Cxcl9, and C-X-C motif chemokine 10 (Cxcl10) in tumor microenvironment in vivo. Mechanistically, Mettl3 or Mettl14 loss promoted IFN-gamma-Stat1-Irf1 signaling through stabilizing the Stat1 and Irf1 mRNA via Ythdf2. | |||
C-X-C motif chemokine 9 (Cxcl9)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
 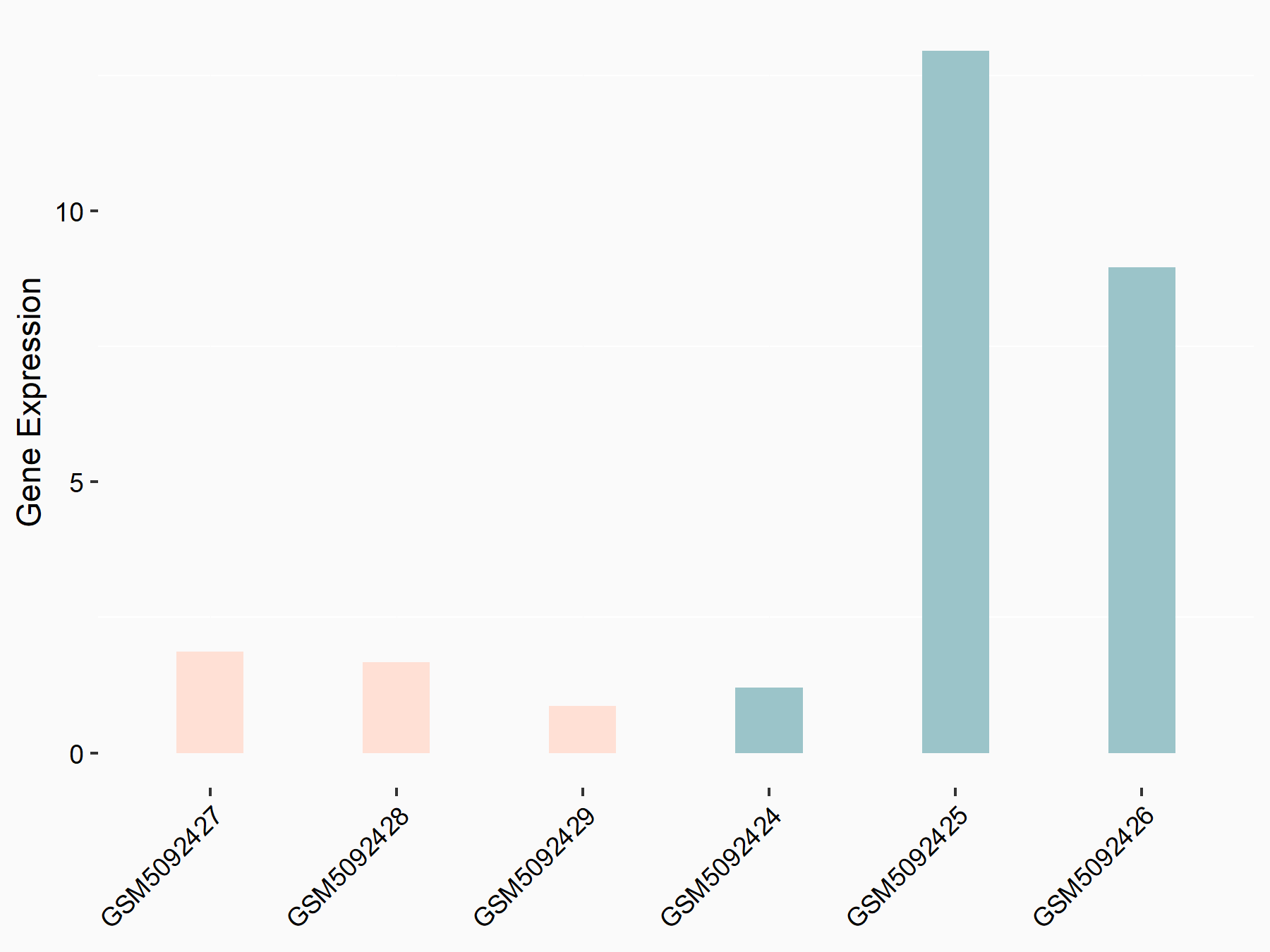 |
logFC: -2.40E+00 p-value: 4.57E-02 |
| More Results | Click to View More RNA-seq Results | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [118] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
| Cell Process | Immunity | |||
In-vitro Model |
CT26 | Mouse colon adenocarcinoma | Mus musculus | CVCL_7254 |
| B16-GM-CSF (B16-GM-CSF cell line was a kind gift from Drs. Glenn Dranoff and Michael Dougan (Dana-Farber/Harvard Cancer Center)) | ||||
| B16-F10 | Mouse melanoma | Mus musculus | CVCL_0159 | |
| In-vivo Model | 2 × 106 CT26 cells with knockout of Mettl3, Mettl14, Mettl3/Stat1, Mettl3/Irf1, Mettl14/Stat1, or Mettl14/Irf1 and control were suspended in 200 uL of PBS/Matrigel (Corning) (1:1) and then subcutaneously inoculated into flank of each mouse. | |||
| Response Summary | In colorectal cancer, Mettl3- or Mettl14-deficient tumors increased cytotoxic tumor-infiltrating CD8+ T cells and elevated secretion of IFN-gamma, C-X-C motif chemokine 9 (Cxcl9), and Cxcl10 in tumor microenvironment in vivo. Mechanistically, Mettl3 or Mettl14 loss promoted IFN-gamma-Stat1-Irf1 signaling through stabilizing the Stat1 and Irf1 mRNA via Ythdf2. | |||
Caspase-3 (CASP3)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
 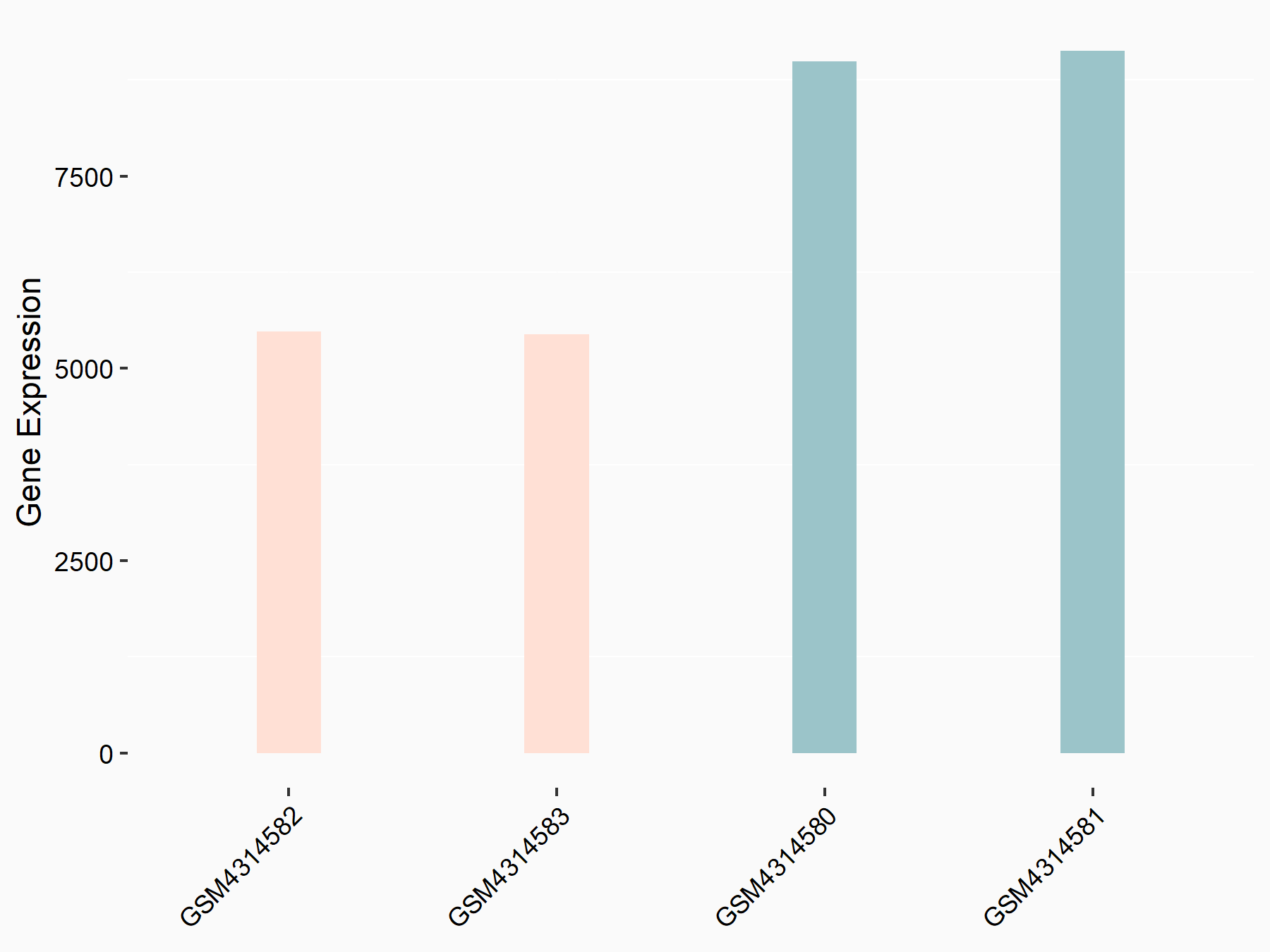 |
logFC: -7.30E-01 p-value: 2.00E-53 |
| More Results | Click to View More RNA-seq Results | |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [8] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| Response Summary | Down-regulation of METTL3 inhibits the proliferation and mobility of human gastric cancer cells and leads to inactivation of the AKT signaling pathway, suggesting that METTL3 is a potential target for the treatment of human gastric cancer. METTL3 knockdown decreased Bcl2 and increased Bax and active Caspase-3 (CASP3) in gastric cancer cells, which suggested the apoptotic pathway was activated. METTL3 led to inactivation of the AKT signaling pathway in human gastric cancer cells, including decreased phosphorylation levels of AKT and expression of down-stream effectors p70S6K and Cyclin D1. | |||
CD44 antigen (CD44)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 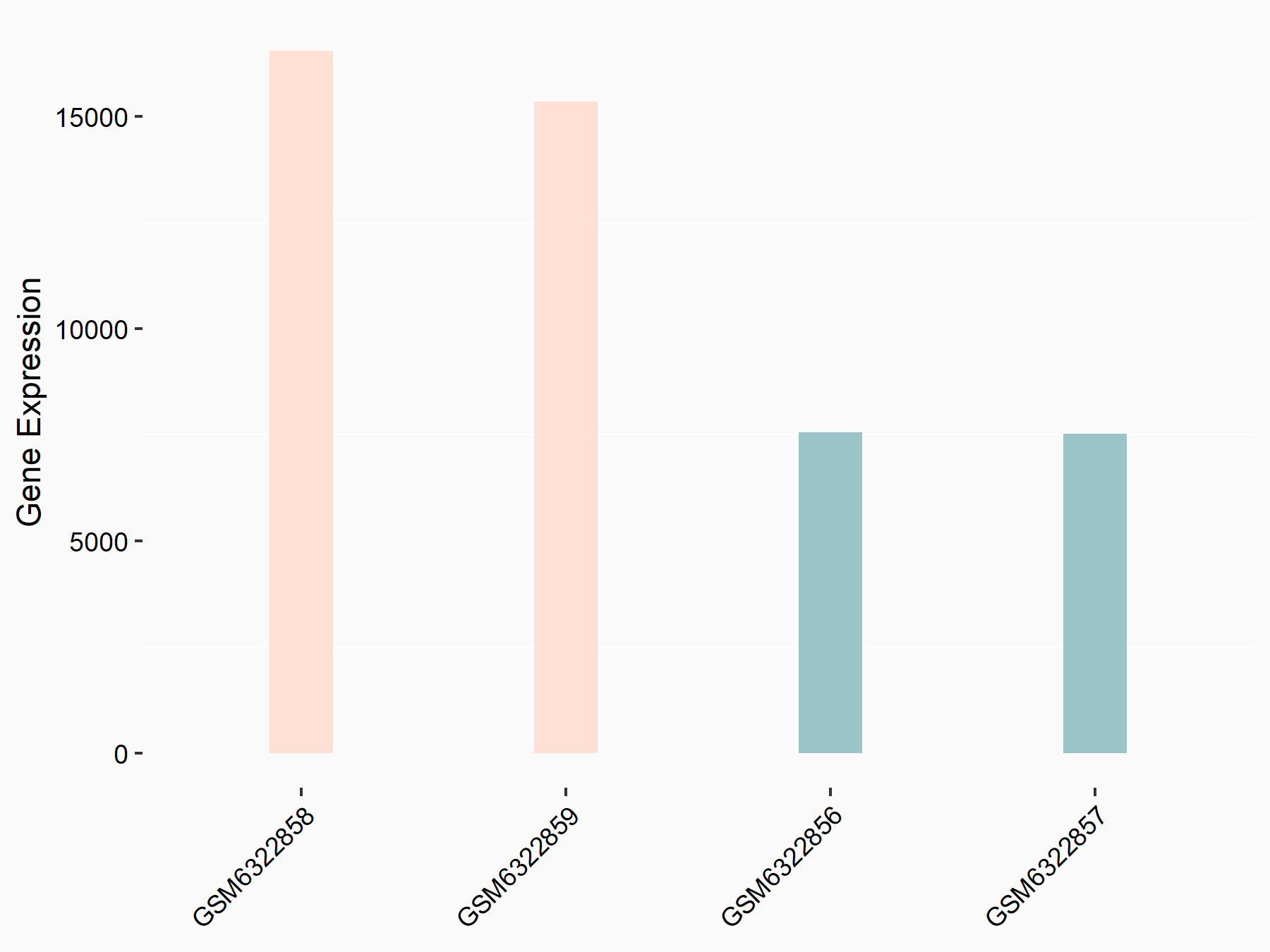 |
logFC: 1.08E+00 p-value: 6.88E-51 |
| More Results | Click to View More RNA-seq Results | |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [122] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| Response Summary | Knockdown of METTL3 downregulated protein levels of SOX2, CD133 and CD44 antigen (CD44) in MCF-7 cells. METTL3 is upregulated in breast cancer, and it promotes the stemness and malignant progression of BCa through mediating m6A modification on SOX2 mRNA. | |||
Circadian locomoter output cycles protein kaput (CLOCK)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
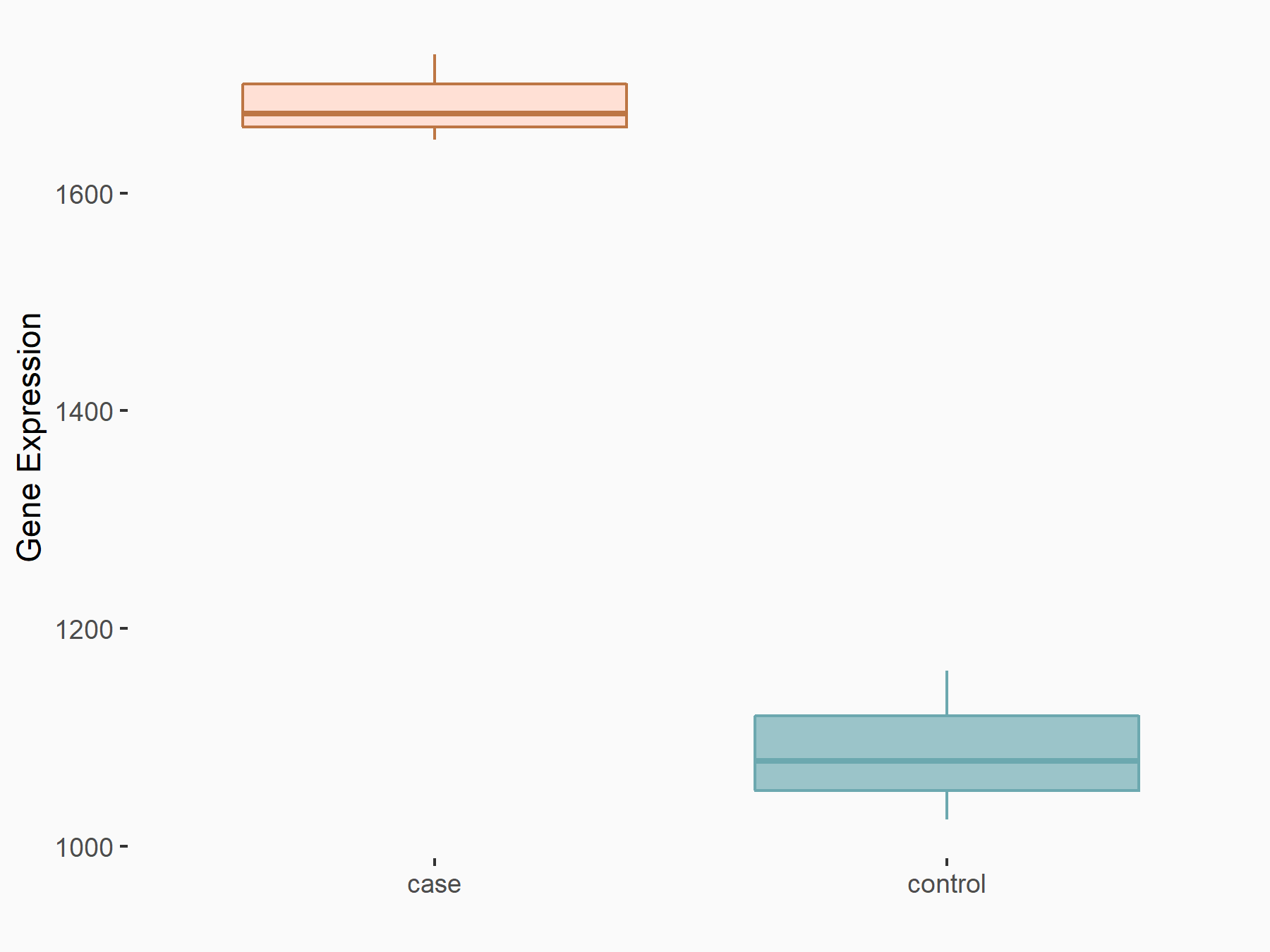 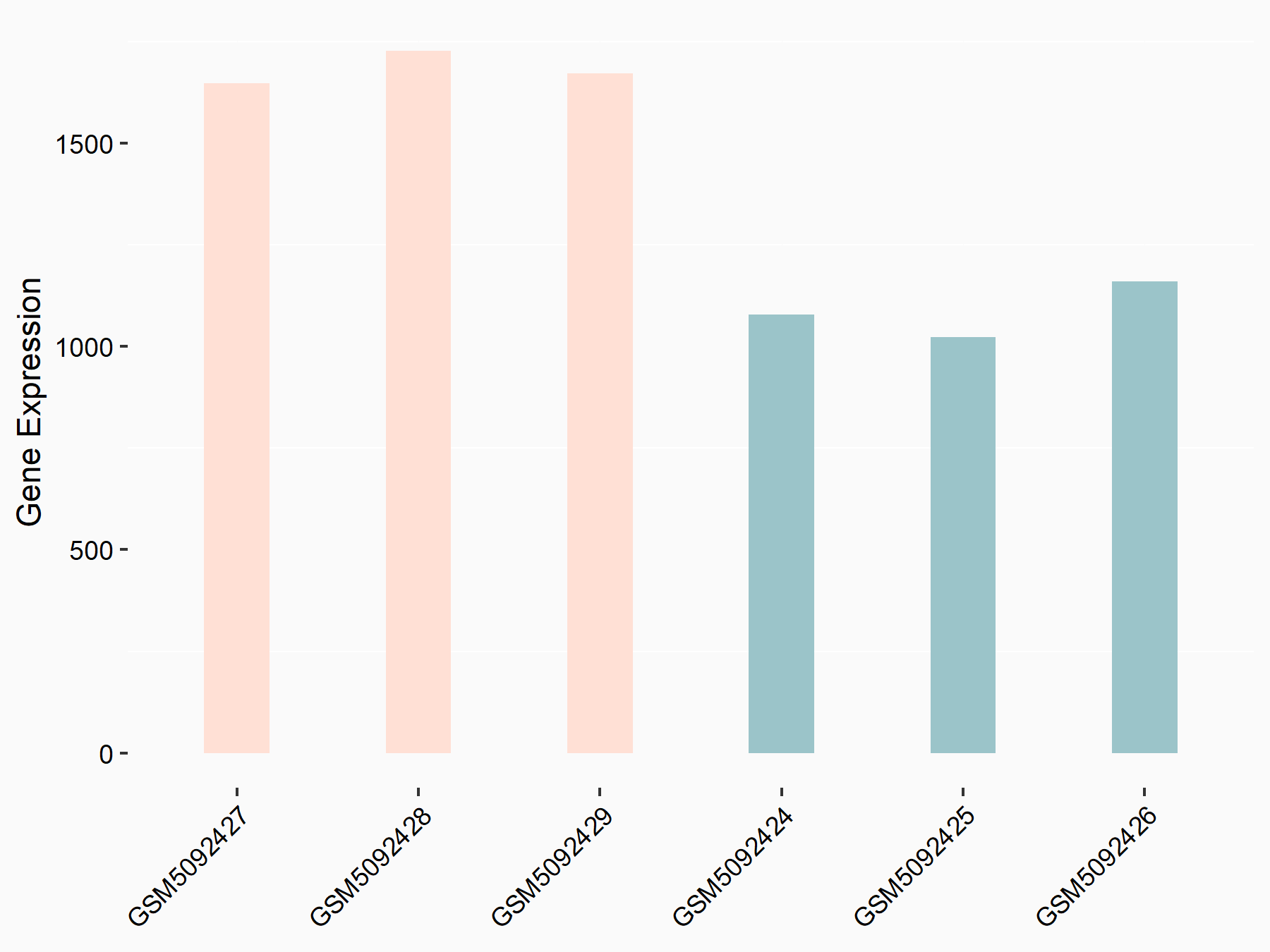 |
logFC: 6.28E-01 p-value: 1.95E-19 |
| More Results | Click to View More RNA-seq Results | |
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [156] | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | M3LKO (Mettl3fl/fl; Alb-Cre) mice were generated by crossing Mettl3fl/fl mice (provided by Dr. Jacob Hanna) with albumin-Cre mice (Jackson Laboratories, Bar Harbor, ME), and genotypes were confirmed by tail-DNA PCR using primers as previously described. | |||
| Response Summary | Liver-specific Mettl3 knockout mice exhibited global decrease in m6A on polyadenylated RNAs and pathologic features associated with nonalcoholic fatty liver disease. Studies in the M3LKO model indicated that METTL3 exhibits pleotropic function to maintain liver homeostasis by deregulating m6A profile and expression of the liver transcriptome. A significant decrease in total Bmal1 and Circadian locomoter output cycles protein kaput (CLOCK) mRNAs but an increase in their nuclear levels were observed in M3LKO livers, suggesting impaired nuclear export. | |||
Cyclin-dependent kinase 1 (CDK1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3-deficient liver
Control: Wild type liver cells
|
GSE197800 | |
| Regulation |
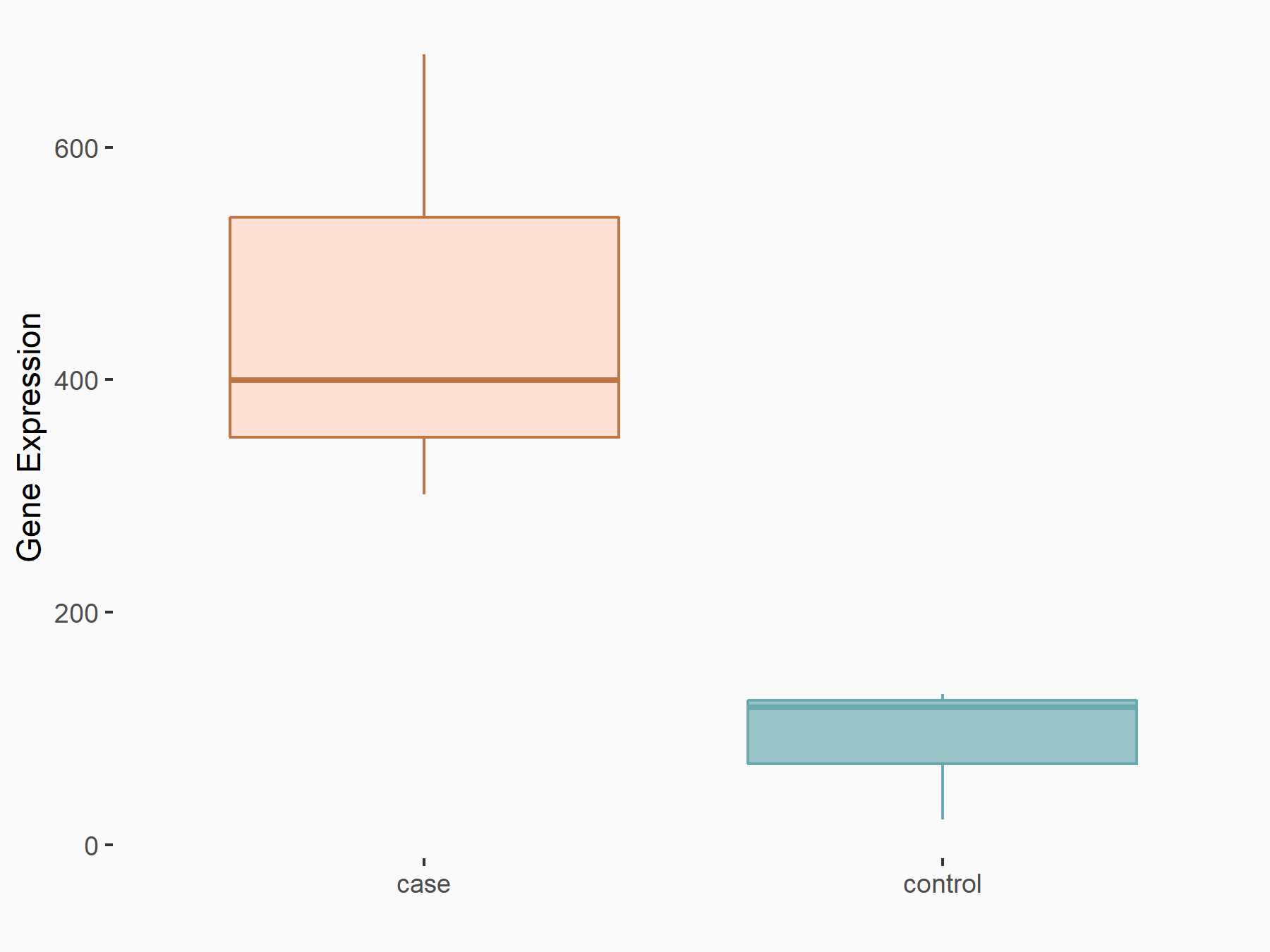 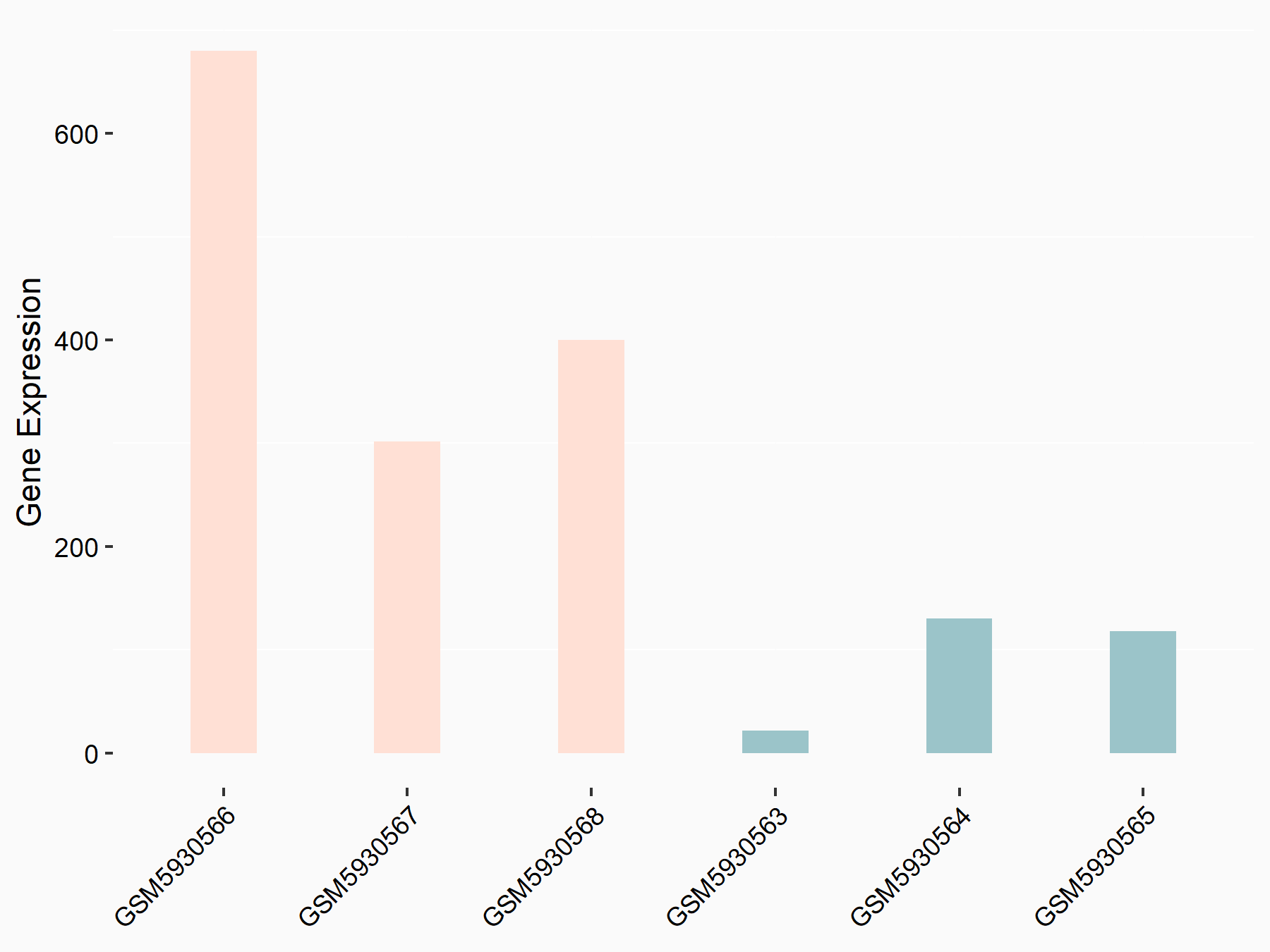 |
logFC: 2.36E+00 p-value: 3.11E-05 |
| More Results | Click to View More RNA-seq Results | |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [157] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Cell cycle | |||
In-vitro Model |
BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 |
| HCC1806 | Breast squamous cell carcinoma | Homo sapiens | CVCL_1258 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 | |
| In-vivo Model | Twelve female BALB/c nude mice (aged 4 weeks, 18-22g) were randomly divided into 2 groups. Stable circMETTL3-expression SUM1315 cells or control cells (1×106 cells in 0.1 mL PBS) was subcutaneously injected into mammary fat pads of the mice and the growth of tumors was followed up every week. Tumor volume was measured every week using a caliper, calculated as (length × width2)/2. After 4 weeks, mice were sacrificed and checked for final tumor weight. | |||
| Response Summary | CircMETTL3 promotes breast cancer progression through circMETTL3/miR-31-5p/Cyclin-dependent kinase 1 (CDK1) axis. | |||
Cystine/glutamate transporter (SLC7A11)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shMETTL3 MOLM13 cells
Control: MOLM13 cells
|
GSE98623 | |
| Regulation |
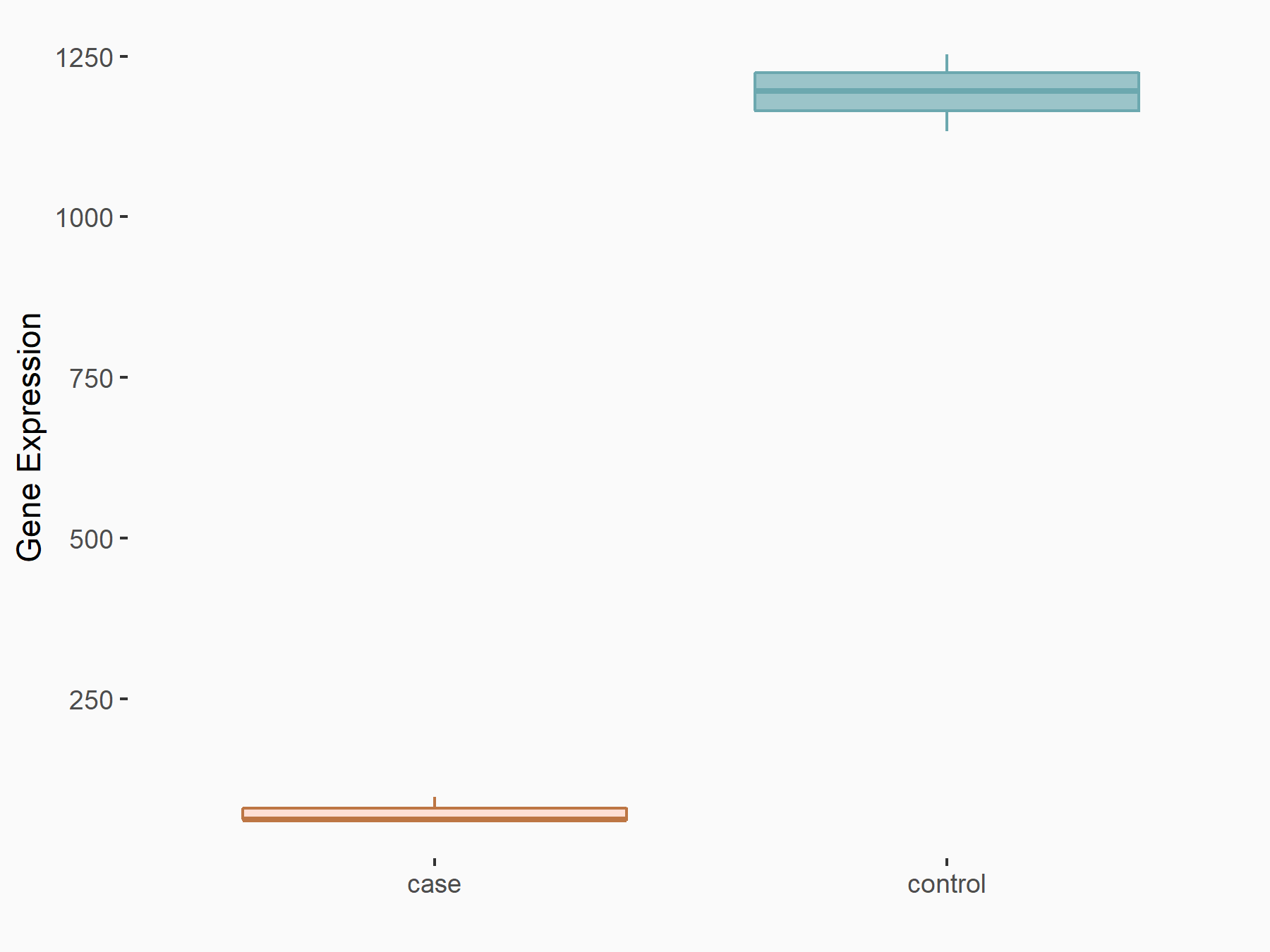 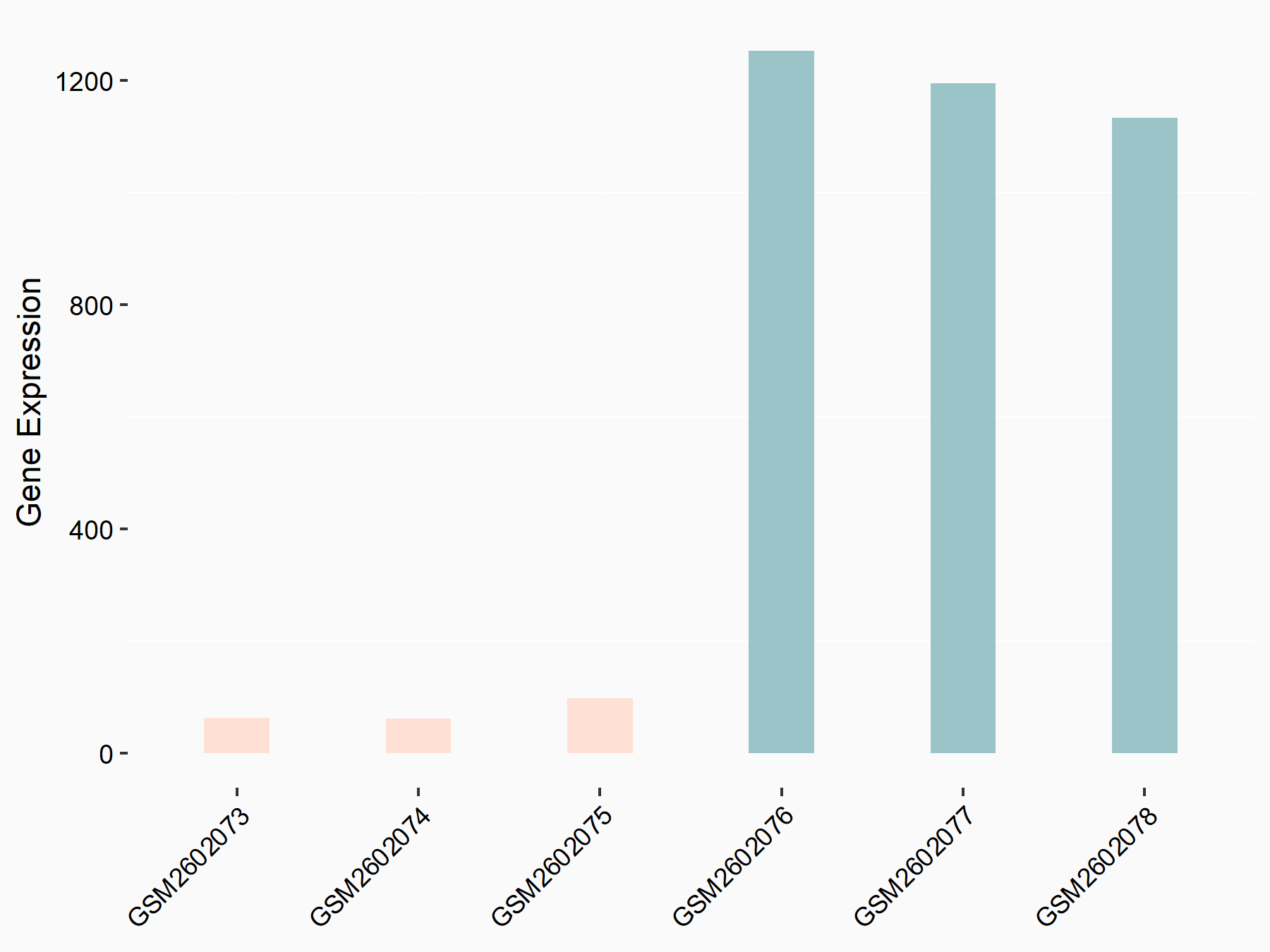 |
logFC: -4.02E+00 p-value: 2.26E-108 |
| More Results | Click to View More RNA-seq Results | |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [158] | |||
| Responsed Disease | Hepatoblastoma [ICD-11: 2C12.01] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Ferroptosis | hsa04216 | ||
| Cell Process | Ferroptosis | |||
In-vitro Model |
HuH-6 | Hepatoblastoma | Homo sapiens | CVCL_4381 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| Response Summary | METTL3-mediated Cystine/glutamate transporter (SLC7A11) m6A modification enhances hepatoblastoma ferroptosis resistance. The METTL3/IGF2BP1/m6A modification promotes SLC7A11 mRNA stability and upregulates its expression by inhibiting the deadenylation process. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [159] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Ferroptosis | hsa04216 | ||
| Cell Process | Ferroptosis | |||
In-vitro Model |
SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| NCI-H322 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1556 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | For the subcutaneous xenograft model, PC9 cells stably transfected with METTL3 knockdown (shMETTL3) or negative control (shNC) shRNA (5 × 106 cells per mouse, n = 6) were suspended in 200 uL PBS with 50% Matrigel matrix (Corning, USA, 354234) and then injected into one side of the axilla of nude mice. | |||
| Response Summary | METTL3-mediated m-6A modification could stabilize Cystine/glutamate transporter (SLC7A11) mRNA and promote its translation, thus promoting LUAD cell proliferation and inhibiting cell ferroptosis, a novel form of programmed cell death. | |||
Injury of heart [ICD-11: NB31]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [284] | |||
| Responsed Disease | Myocardial injury [ICD-11: NB31.Z] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
H9c2(2-1) | Normal | Rattus norvegicus | CVCL_0286 |
Diacylglycerol O-acyltransferase 2 (DGAT2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shMETTL3 MOLM13 cells
Control: MOLM13 cells
|
GSE98623 | |
| Regulation |
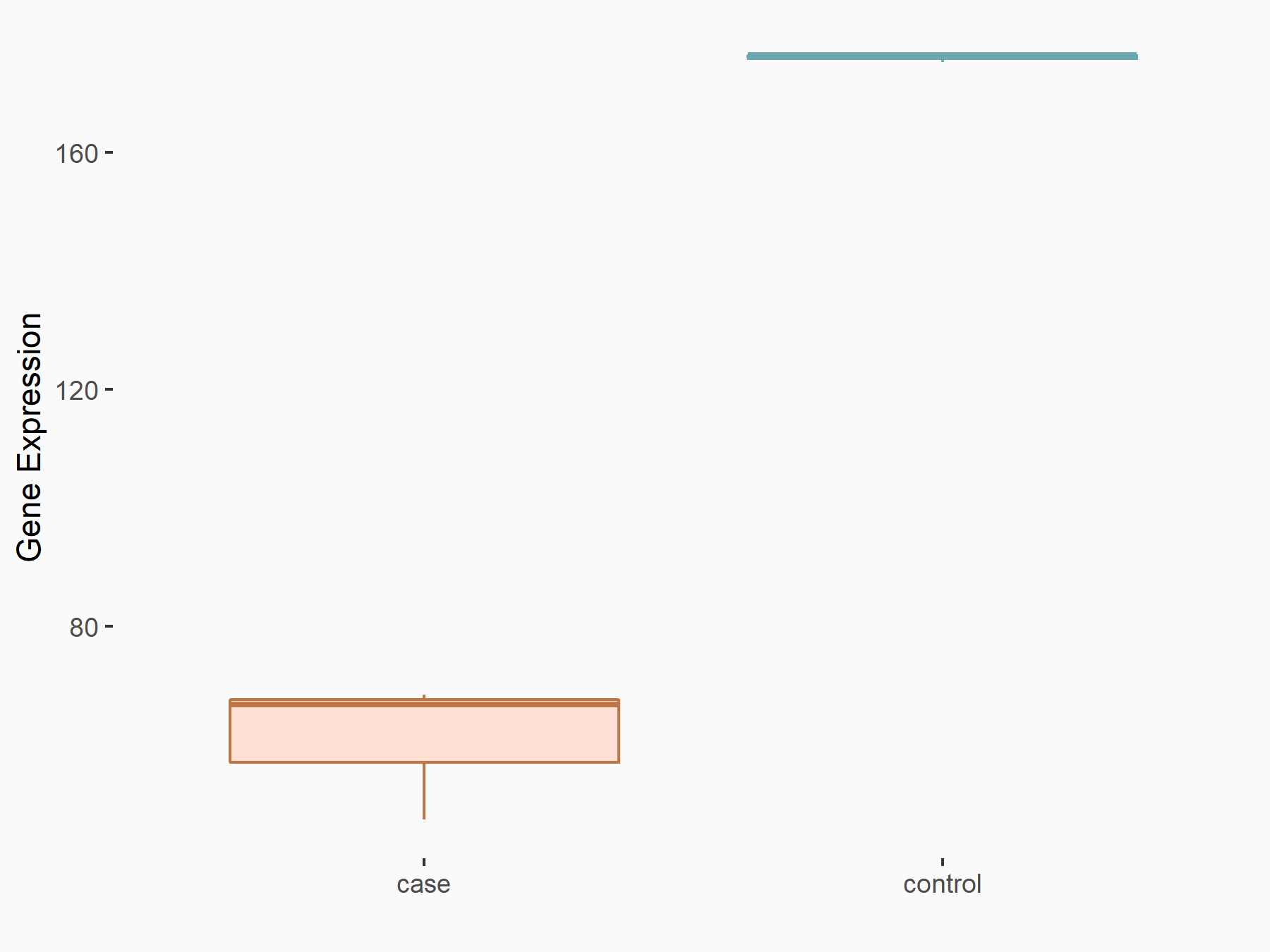 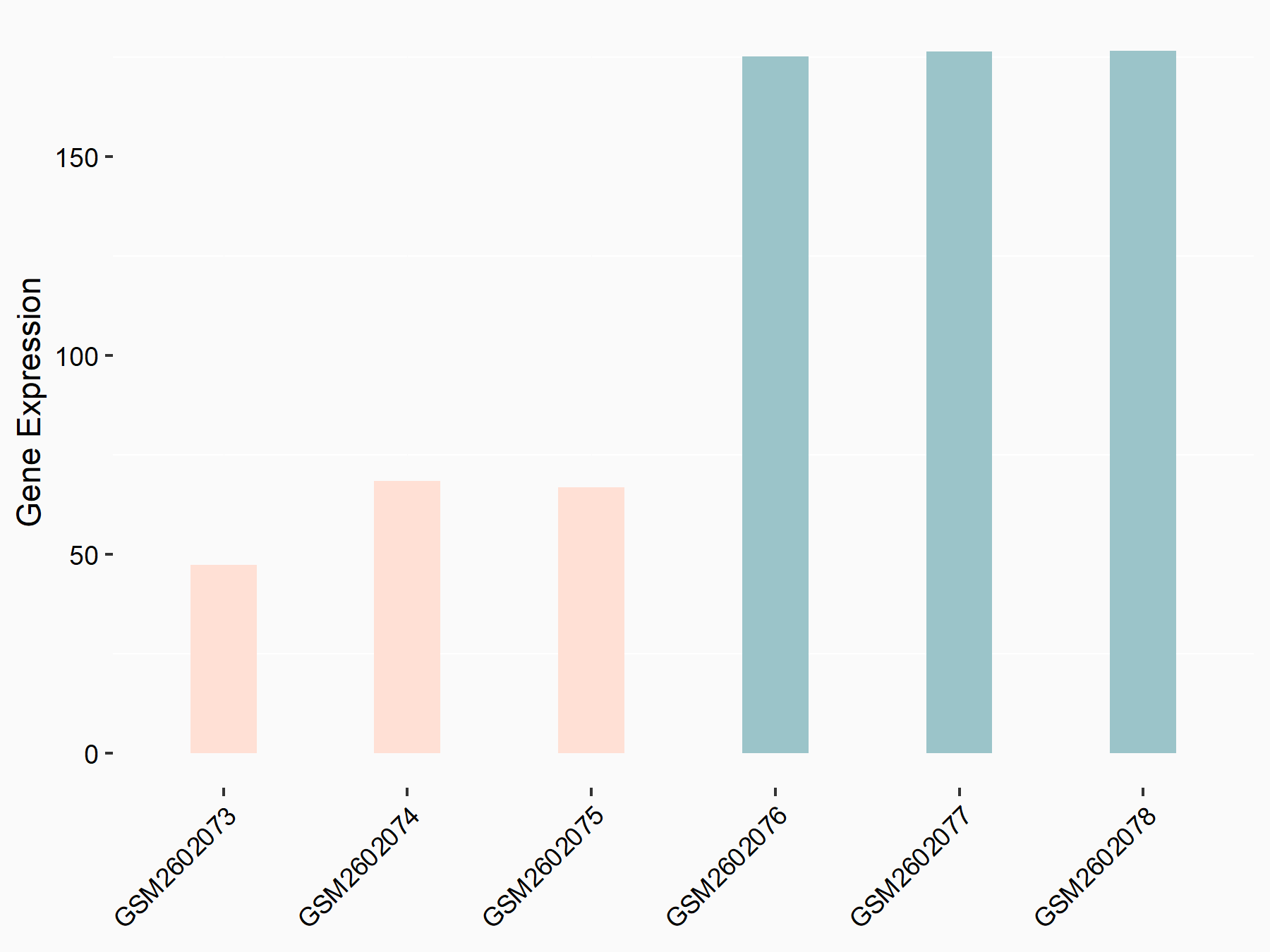 |
logFC: -1.53E+00 p-value: 7.36E-18 |
| More Results | Click to View More RNA-seq Results | |
Type 2 diabetes mellitus [ICD-11: 5A11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [48] | |||
| Responsed Disease | Type 2 diabetes mellitus [ICD-11: 5A11] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Insulin resistance | hsa04931 | ||
| Cell Process | Lipid metabolism | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | Hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were generated by crossing mice with TBG-Cre Tg mice. METTL3 flox (METTL3 fl/fl) and hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were used for experiments. | |||
| Response Summary | Type 2 diabetes (T2D) is characterized by lack of insulin, insulin resistance and high blood sugar. METTL3 silence decreased the m6A methylated and total mRNA level of Fatty acid synthase (Fasn), subsequently inhibited fatty acid metabolism. The expression of Acc1, Acly, Diacylglycerol O-acyltransferase 2 (DGAT2), Ehhadh, Fasn, Foxo, Pgc1a and Sirt1, which are critical to the regulation of fatty acid synthesis and oxidation were dramatically decreased in livers of hepatocyte-specific METTL3 knockout mice. | |||
DNA damage-inducible transcript 4 protein (DDIT4)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 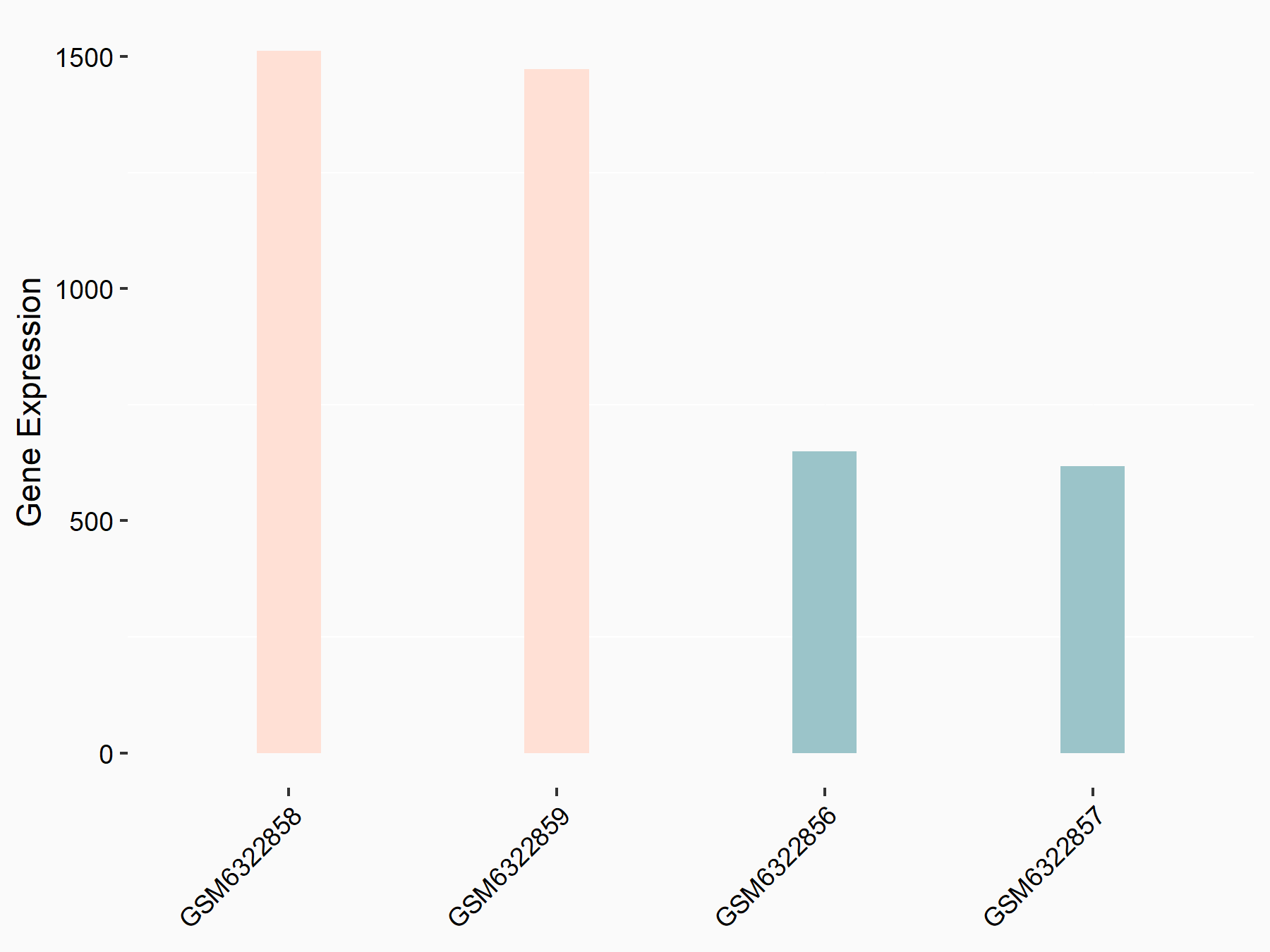 |
logFC: 1.24E+00 p-value: 1.11E-27 |
| More Results | Click to View More RNA-seq Results | |
Obesity [ICD-11: 5B81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [110] | |||
| Responsed Disease | Obesity [ICD-11: 5B81] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| HIF-1 signaling pathway | hsa04066 | |||
| In-vivo Model | The 8-10 weeks old mice were fed either a high fat diet or HF-CDAA , ad lib for 6-12 weeks. Chow diet was used as control for HFD.The mouse liver was perfused with PBS through portal vein, and liver tissue was cut into small pieces by a scissor. The single cell was made using syringe plunger to mull the tissue, and passed through a 40 uM cell strainer. | |||
| Response Summary | The contribution of METTL3-mediated m6A modification of DNA damage-inducible transcript 4 protein (DDIT4) mRNA to macrophage metabolic reprogramming in non-alcoholic fatty liver disease and obesity. In METTL3-deficient macrophages, there is a significant downregulation of mammalian target of rapamycin (mTOR) and nuclear factor Kappa-B (NF-Kappa-B) pathway activity in response to cellular stress and cytokine stimulation, which can be restored by knockdown of DDIT4. | |||
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [110] | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| HIF-1 signaling pathway | hsa04066 | |||
| In-vivo Model | The 8-10 weeks old mice were fed either a high fat diet or HF-CDAA , ad lib for 6-12 weeks. Chow diet was used as control for HFD.The mouse liver was perfused with PBS through portal vein, and liver tissue was cut into small pieces by a scissor. The single cell was made using syringe plunger to mull the tissue, and passed through a 40 uM cell strainer. | |||
| Response Summary | The contribution of METTL3-mediated m6A modification of DNA damage-inducible transcript 4 protein (DDIT4) mRNA to macrophage metabolic reprogramming in non-alcoholic fatty liver disease and obesity. In METTL3-deficient macrophages, there is a significant downregulation of mammalian target of rapamycin (mTOR) and nuclear factor Kappa-B (NF-Kappa-B) pathway activity in response to cellular stress and cytokine stimulation, which can be restored by knockdown of DDIT4. | |||
Corneal injury [ICD-11: NA06]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [87] | |||
| Responsed Disease | Corneal injury [ICD-11: NA06.4] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
In-vitro Model |
CGC (Conjunctival goblet cells) | |||
| In-vivo Model | Mettl3fl/wt mice were generated as previously described. Mettl3fl/wt mice were crossed with K14CreER mice to obtain K14creER/Mettl3fl/fl (cKO) mice. Mettl3 cKO and control mice were injected with tamoxifen and then were subjected to corneal alkali burn treatment. The right eye was the experimental eye, and the left eye was the control eye. The mice were sacrificed at 24 hours, 7 days, 14 days, 35 days, and 56 days after injury. Six mice were taken from each period. Both eyes were removed, frozen in OCT (n = 4), fixed in 4% paraformaldehyde, and embedded in conventional paraffin (n = 2). | |||
| Response Summary | METTL3 knockout in the limbal stem cells promotes the in vivo cell proliferation and migration, leading to the fast repair of corneal injury. In addition, m6A modification profiling identified stem cell regulatory factors AHNAK and DNA damage-inducible transcript 4 protein (DDIT4) as m6A targets. | |||
DNA repair protein RAD51 homolog 1 (RAD51)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
 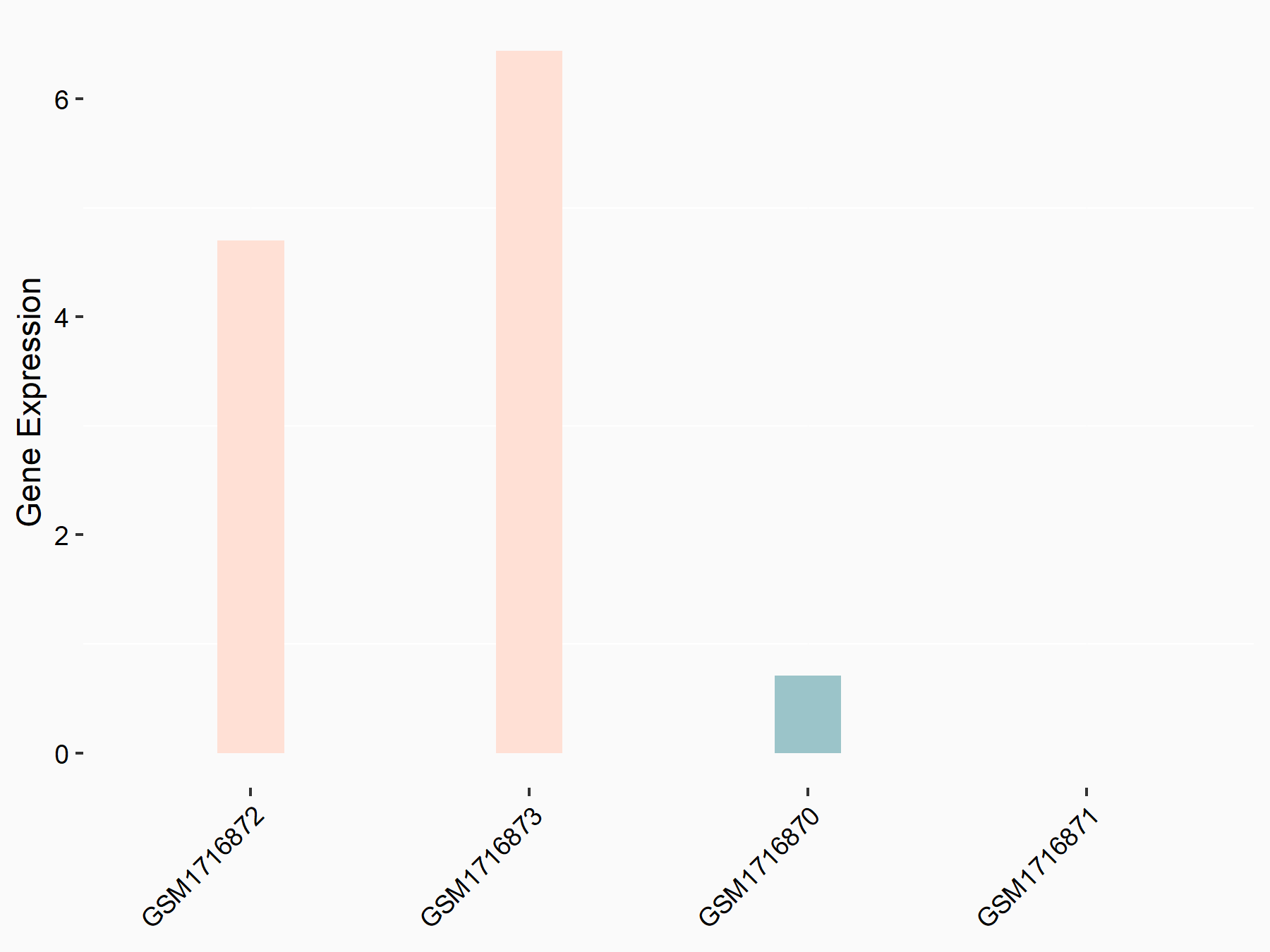 |
logFC: 2.32E+00 p-value: 1.62E-02 |
| More Results | Click to View More RNA-seq Results | |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [98] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Doxil | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Homologous recombination | hsa03440 | ||
| Cell Process | Homologous recombination repair | |||
In-vitro Model |
MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | Cells were trypsinized and resuspended in DMEM at a consistence of 1 × 107 cells/ml. A total of 1 × 106 cells were injected into flank of mice. 27 days after injection, tumors were removed for paraffin-embedded sections. | |||
| Response Summary | Knockdown of METTL3 sensitized these breast cancer cells to Adriamycin (ADR; also named as doxorubicin) treatment and increased accumulation of DNA damage. Mechanically, we demonstrated that inhibition of METTL3 impaired HR efficiency and increased ADR-induced DNA damage by regulating m6A modification of EGF/RAD51 axis. METTL3 promoted EGF expression through m6A modification, which further upregulated DNA repair protein RAD51 homolog 1 (RAD51) expression, resulting in enhanced HR activity. | |||
Genotoxicity [ICD-11: XE2PA]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [286] | |||
| Responsed Disease | Genotoxicity [ICD-11: XE2PA] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
GC-2spd(ts)
|
N.A. | Mus musculus | CVCL_6633 |
| In-vivo Model | Corn oil (control group), 1 mg/kg/day DEHP (low-dose: D1 group), 250 mg/kg/day DEHP (middle-dose: D250 group), and 500 mg/kg/day DEHP (high-dose: D500 group). | |||
DNA replication licensing factor MCM5 (MCM5)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
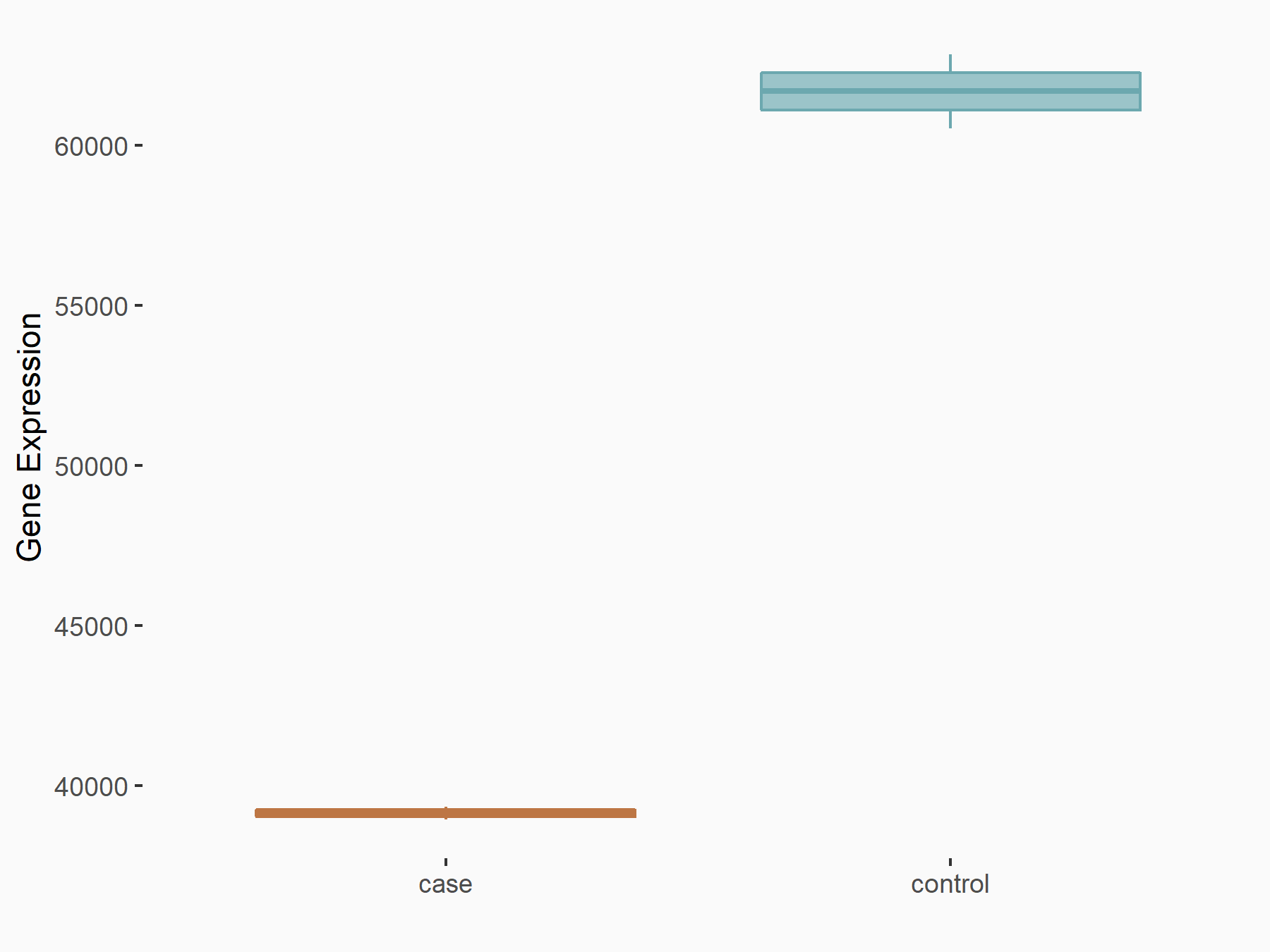 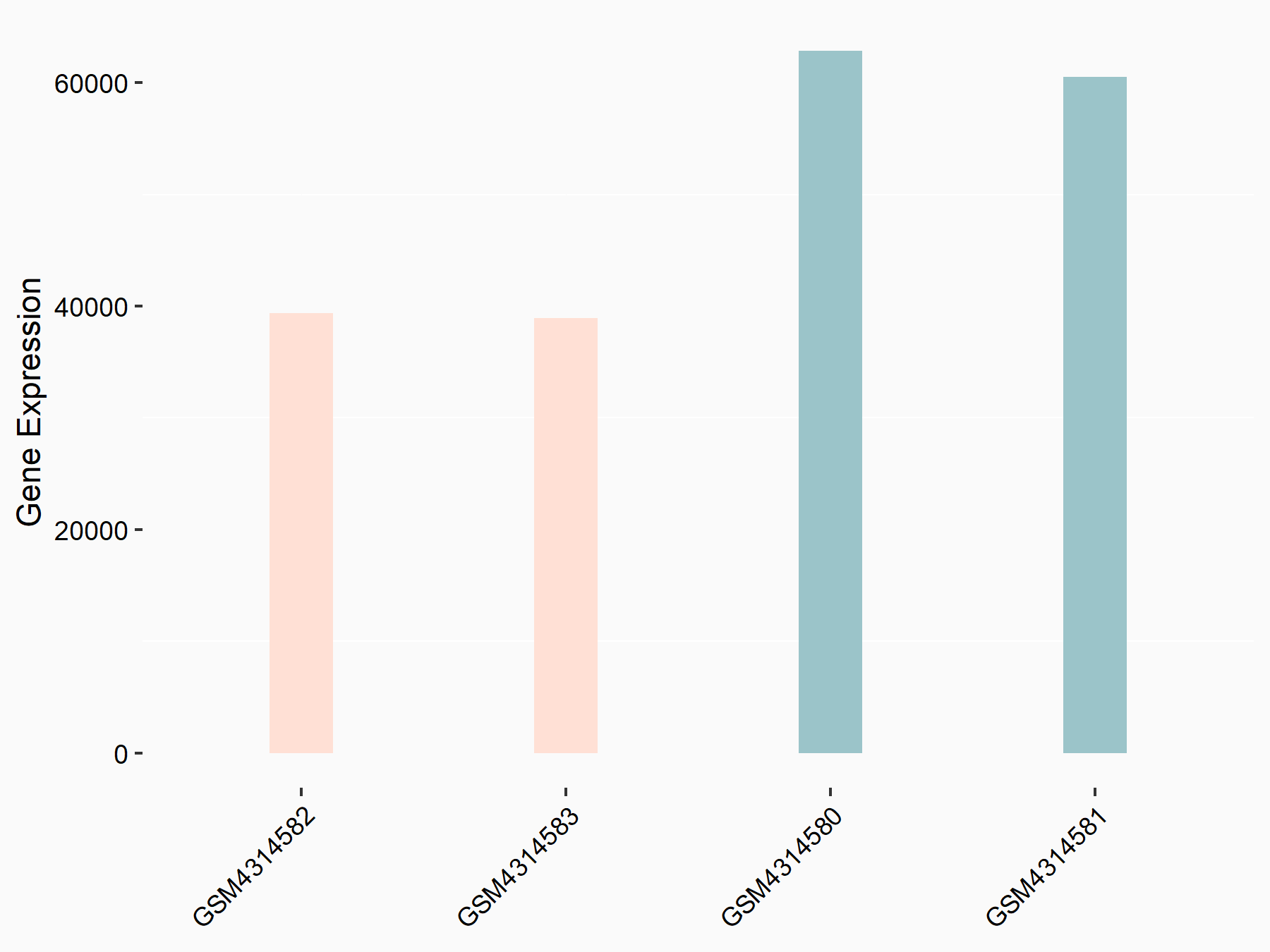 |
logFC: -6.56E-01 p-value: 6.81E-68 |
| More Results | Click to View More RNA-seq Results | |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [42] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
GES-1 | Normal | Homo sapiens | CVCL_EQ22 |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MKN74 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_2791 | |
| pGCC (Primary GC cells) | ||||
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| In-vivo Model | A total of 2 × 106 GC cells were injected into the flank of nude mice in a 1:1 suspension of BD Matrigel (BD Biosciences) in phosphate-buffered saline (PBS) solution. | |||
| Response Summary | In gastric cancer, several component molecules (e.g., DNA replication licensing factor MCM5 (MCM5), MCM6, etc.) of MYC target genes were mediated by METTL3 via altered m6A modification. | |||
DNA-3-methyladenine glycosylase (ANPG/MPG)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
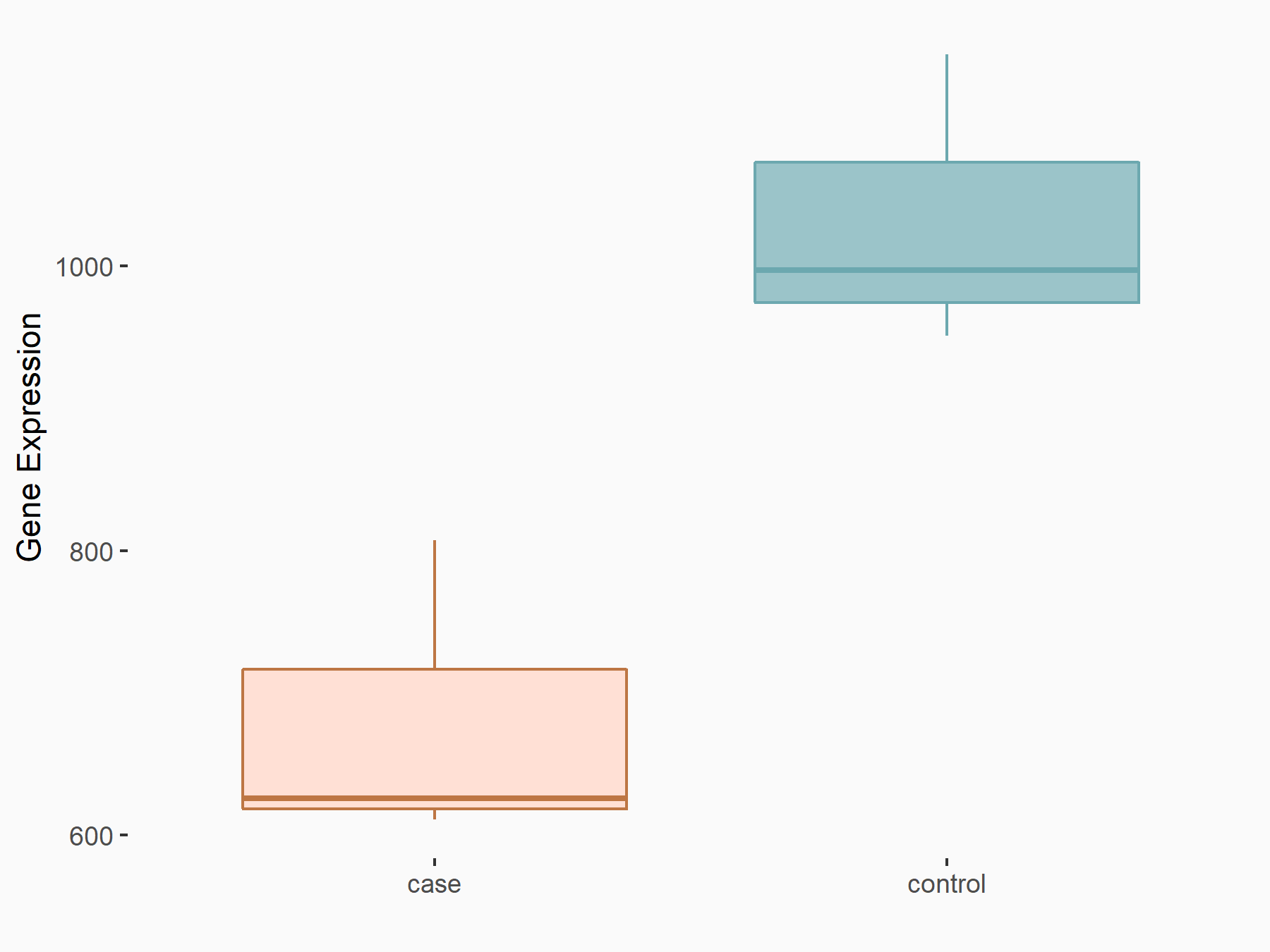 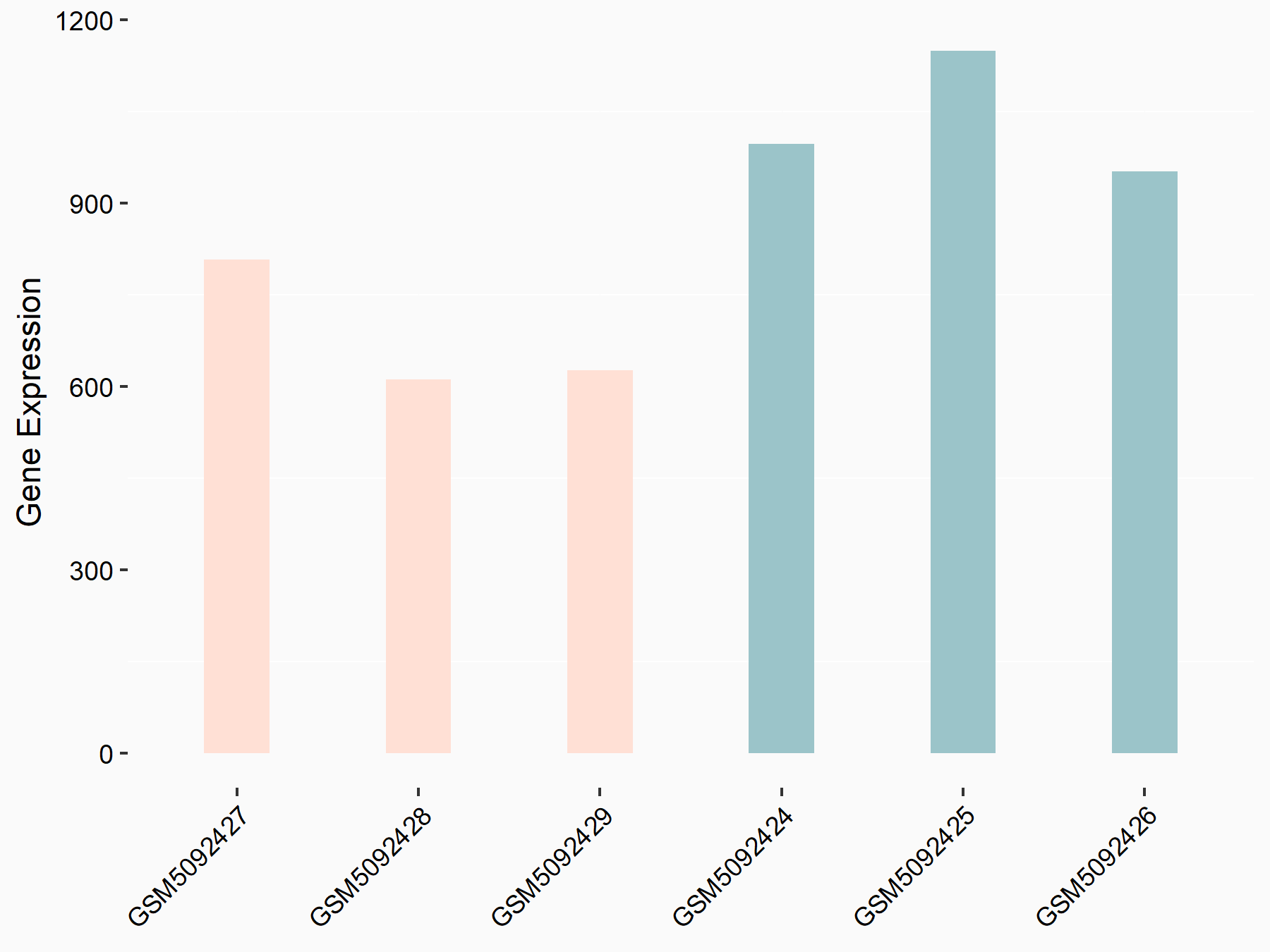 |
logFC: -6.00E-01 p-value: 3.04E-08 |
| More Results | Click to View More RNA-seq Results | |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [69] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Responsed Drug | Temozolomide | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | DNA repair | |||
In-vitro Model |
U251 (Fibroblasts or fibroblast like cells) | |||
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| In-vivo Model | Subcutaneously injected shMETTL3 or shNC-expressing U87-MG-TMZ cells into BALB/c NOD mice. After confirmation of GBM implantation, mice were treated with TMZ (66 mg/kg/d, 5 d per week, for 3 cycles). | |||
| Response Summary | Two critical DNA repair genes (MGMT and DNA-3-methyladenine glycosylase (ANPG/MPG)) were m6A-modified by METTL3, whereas inhibited by METTL3 silencing or DAA-mediated total methylation inhibition, which is crucial for METTL3-improved temozolomide resistance in glioblastoma cells. | |||
Glioblastoma multiforme [ICD-11: XH0MB1]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [287] | |||
| Responsed Disease | Glioblastoma multiforme [ICD-11: XH0MB1] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
GSC11 | Glioblastoma | Homo sapiens | CVCL_DR55 |
| GSC23 | Glioblastoma | Homo sapiens | CVCL_DR59 | |
| In-vivo Model | To establish the xenograft model of GSCs in mice, GSCs or METTL3 knocking-down GSCs (v/v = 1:1) were subcutaneously inoculated into the right posterior limb of BALB/c nude mice (5 × 106 cells/mice, 4-week-old, female, body weight of 20 g). Tumor volume was measured with calipers every 3 days. The tumor size was calculated using the formula: (a2 × b)/2 (a: width in mm, b: length in mm). After about 27 days, all mice were sacrificed under general anesthesia and the xenografts were harvested for further pathological study. | |||
Dual specificity protein phosphatase 2 (DUSP2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
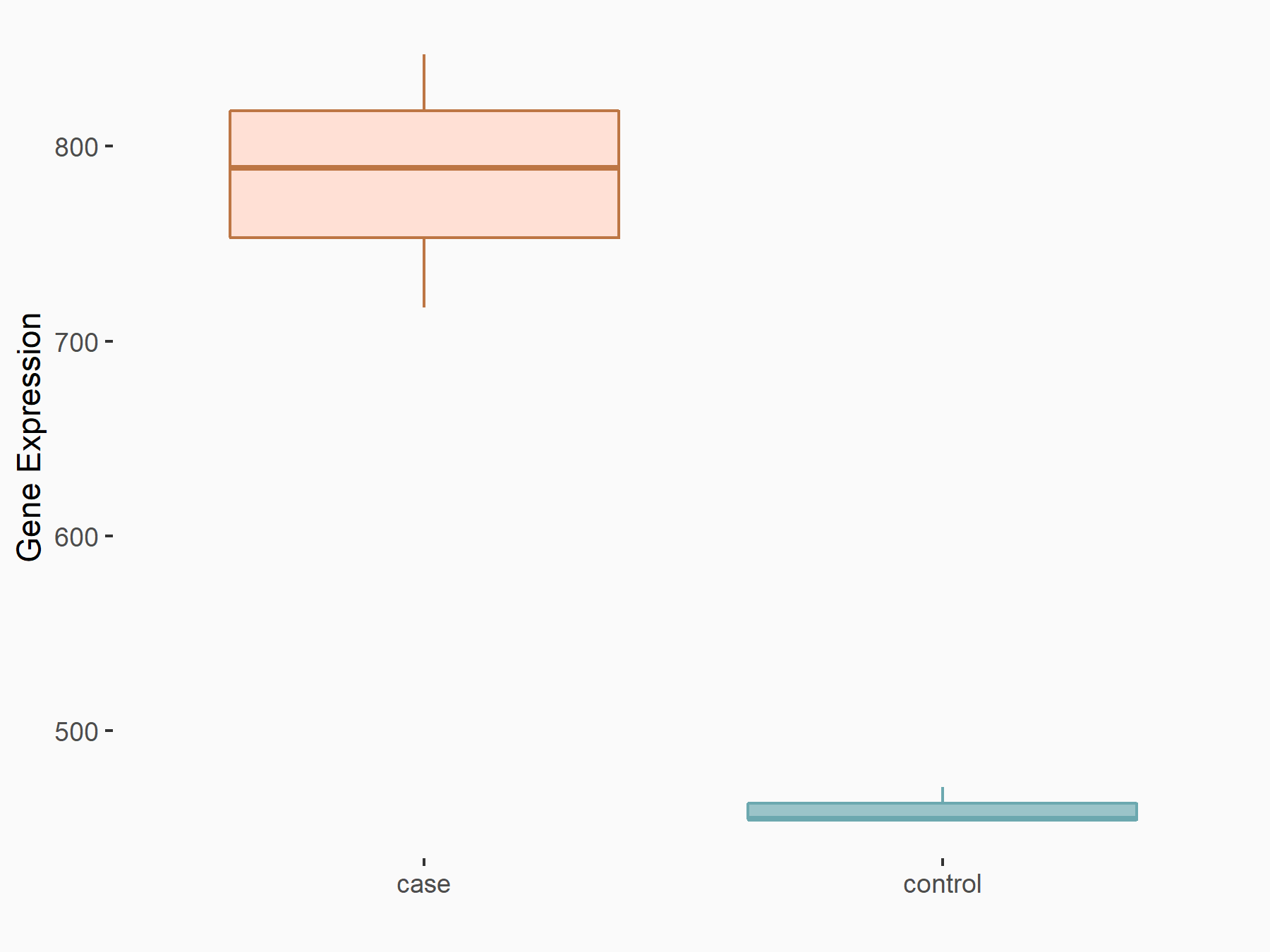 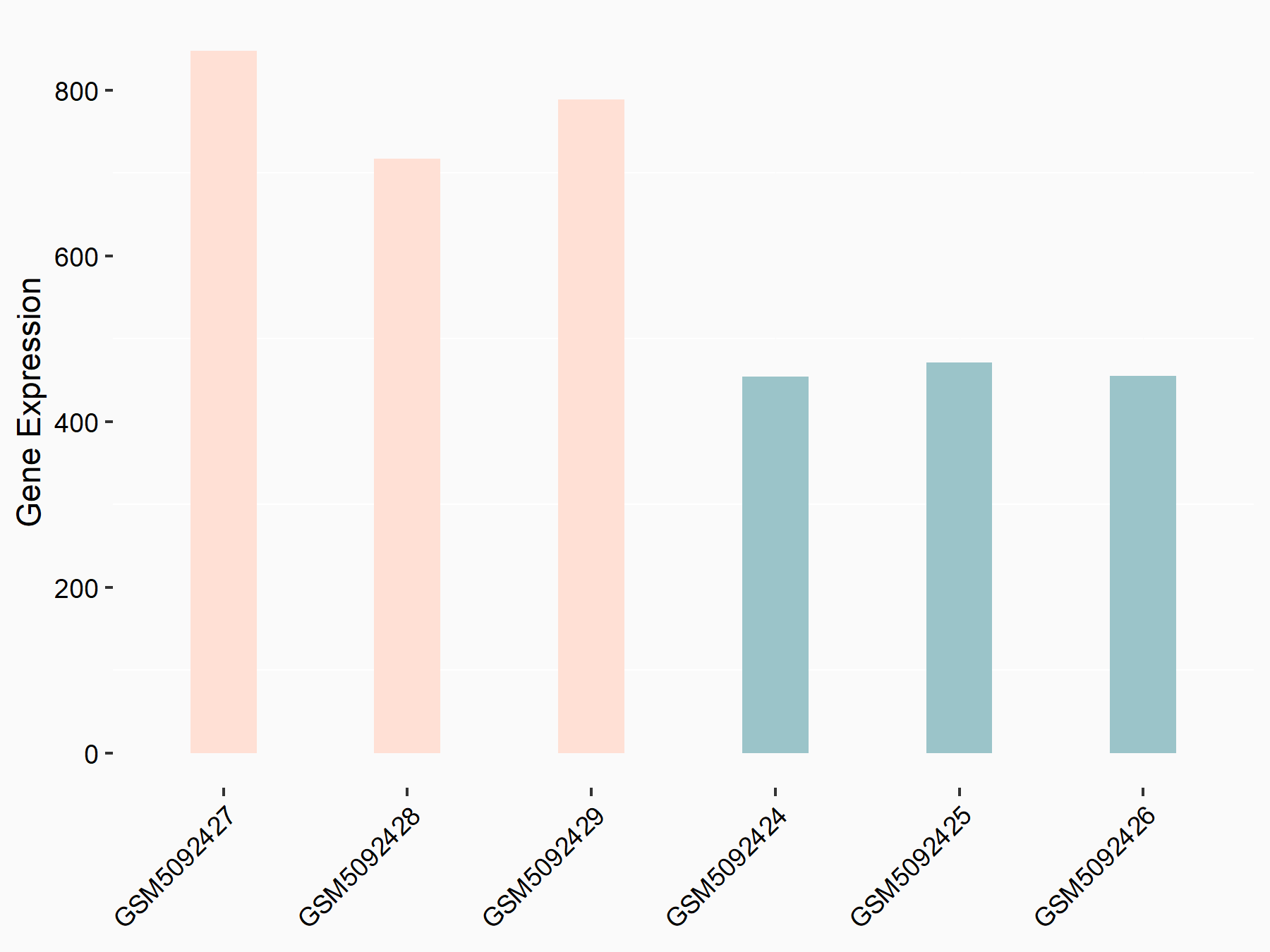 |
logFC: 7.70E-01 p-value: 1.05E-14 |
| More Results | Click to View More RNA-seq Results | |
Emphysema [ICD-11: CA21]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [160] | |||
| Responsed Disease | Emphysema [ICD-11: CA21] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Response Summary | METTL3-mediated formation of EV miR-93, facilitated by m6A, is implicated in the aberrant cross-talk of epithelium-macrophages, indicating that this process is involved in the smoking-related emphysema. EV miR-93 was used as a novel risk biomarker for CS-induced emphysema. MiR-93 activated the JNK pathway by targeting Dual specificity protein phosphatase 2 (DUSP2), which elevated the levels of matrix metalloproteinase 9 (MMP9) and matrix metalloproteinase 12 (MMP12) and induced elastin degradation, leading to emphysema. | |||
Dual specificity protein phosphatase 5 (DUSP5)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LNCaP cell line | Homo sapiens |
|
Treatment: shMETTL3 LNCaP cells
Control: shControl LNCaP cells
|
GSE147884 | |
| Regulation |
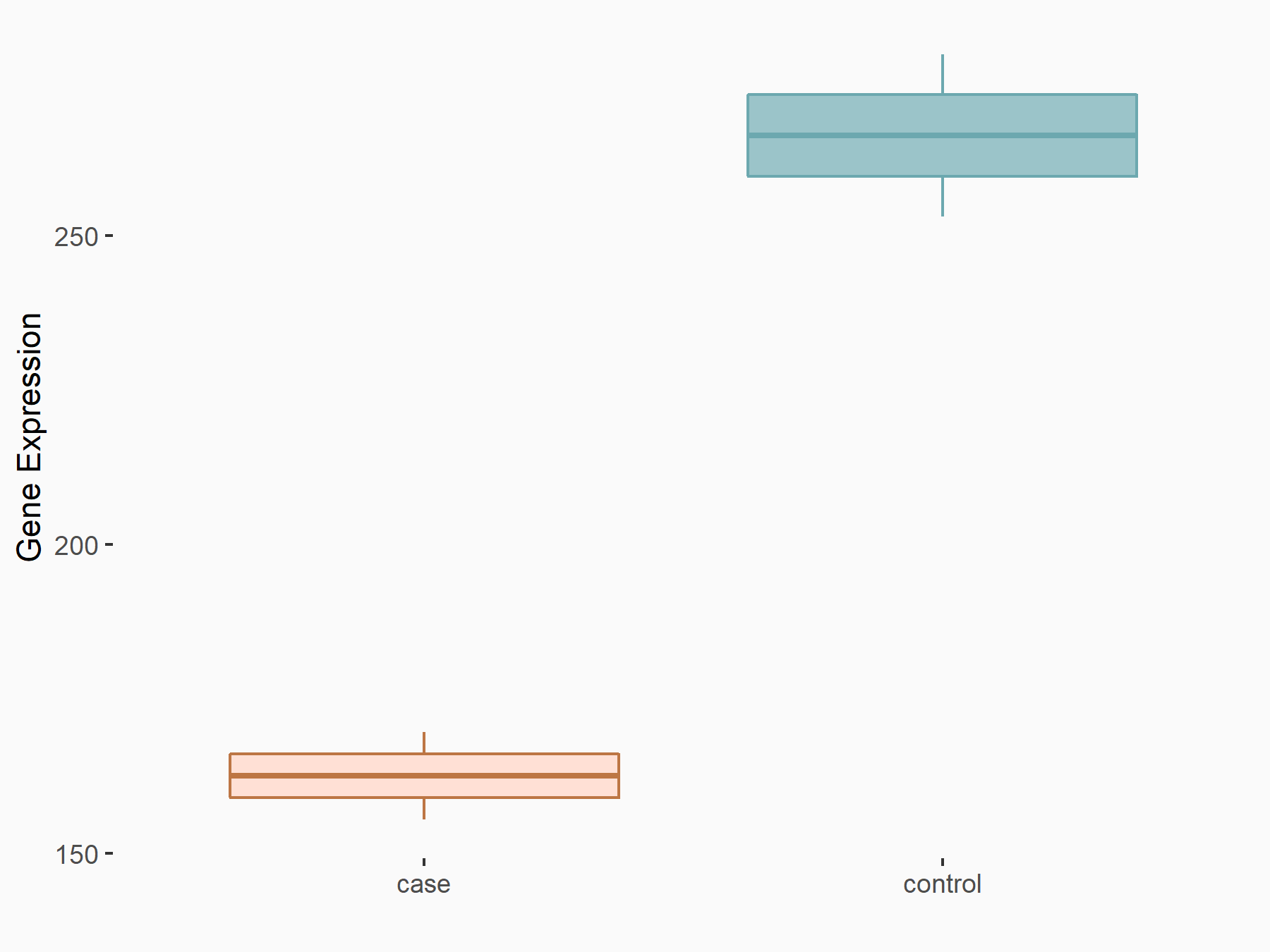 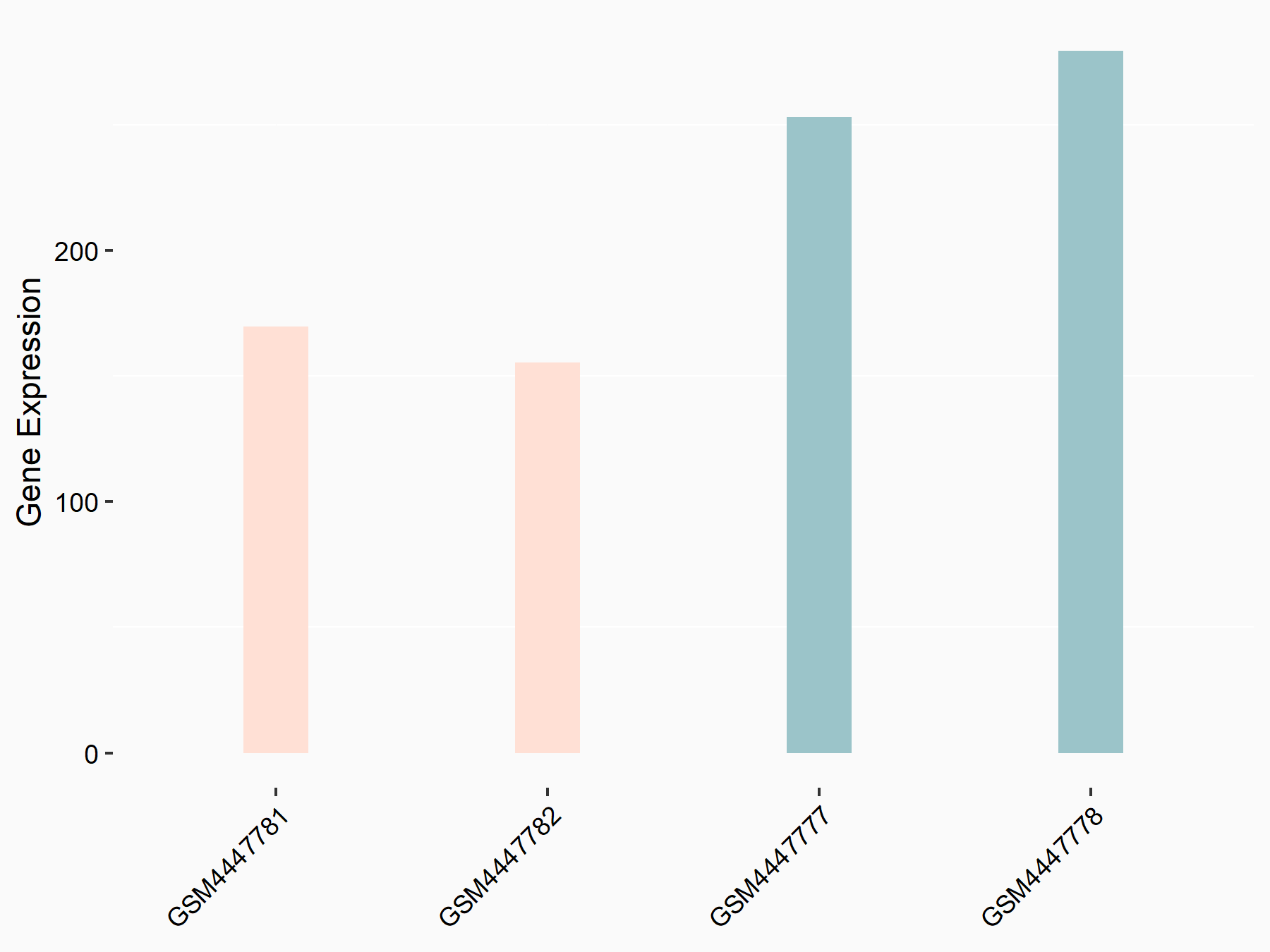 |
logFC: -7.11E-01 p-value: 1.15E-05 |
| More Results | Click to View More RNA-seq Results | |
Gallbladder cancer [ICD-11: 2C13]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [161] | |||
| Responsed Disease | Gallbladder cancer [ICD-11: 2C13] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell proliferation | |||
| Cell invasion | ||||
| Cell migration | ||||
In-vitro Model |
NOZ | Gallbladder carcinoma | Homo sapiens | CVCL_3079 |
| GBC-SD | Human gallbladder | Homo sapiens | CVCL_6903 | |
| Response Summary | METTL3-mediated m6A-modification profile in gallbladder-cancer cells and identified Dual specificity protein phosphatase 5 (DUSP5) as the downstream gene of METTL3. METTL3 promoted the degradation of DUSP5 mRNA in a YTHDF2-dependent manner. | |||
E3 ubiquitin-protein ligase Mdm2 (Mdm2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 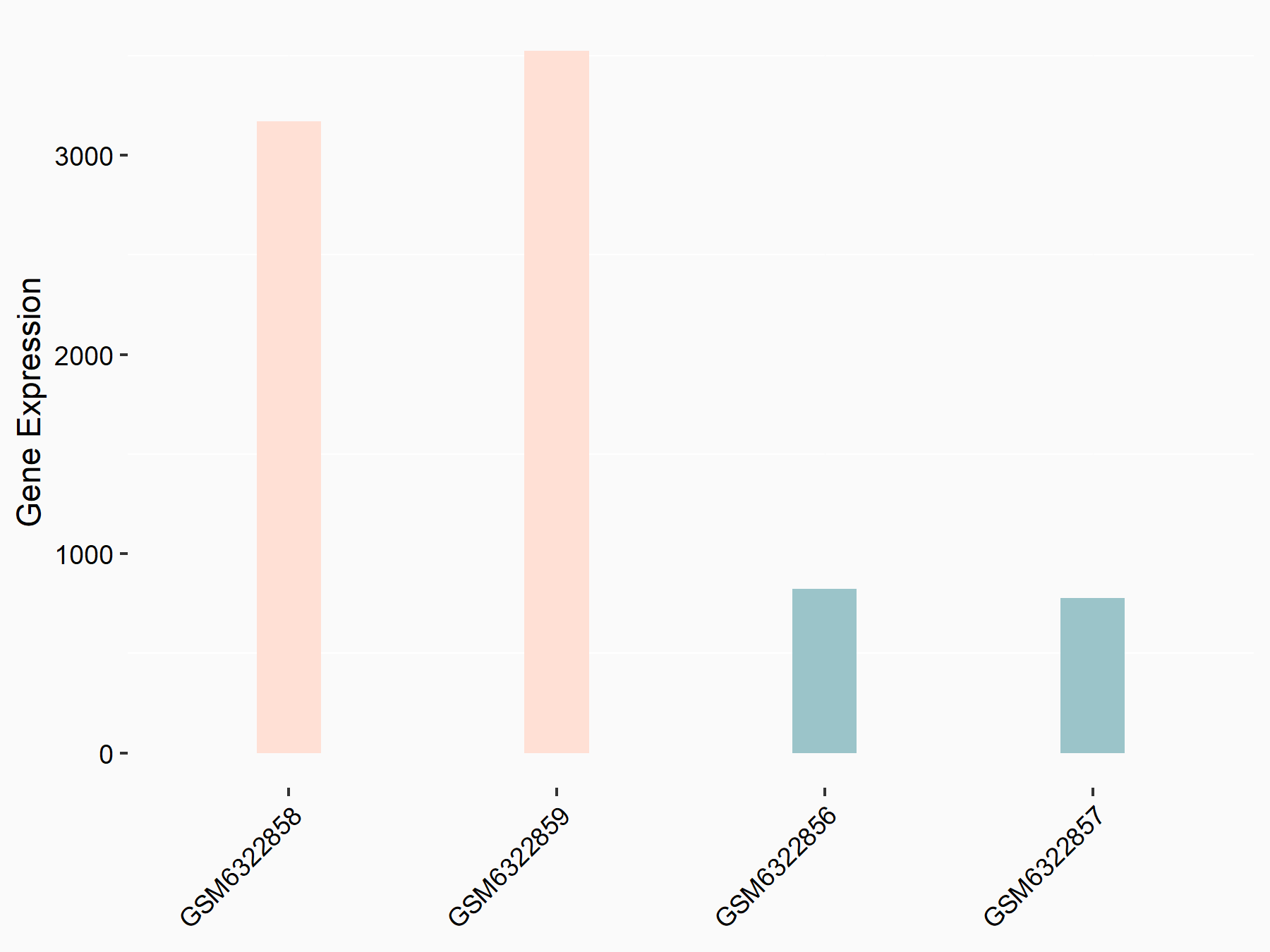 |
logFC: 2.07E+00 p-value: 7.73E-92 |
| More Results | Click to View More RNA-seq Results | |
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [28] | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Cell apoptosis | |||
| Cells in G2/M phase decreased | ||||
In-vitro Model |
THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| MOLT-4 | Adult T acute lymphoblastic leukemia | Homo sapiens | CVCL_0013 | |
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 | |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| CCRF-CEM C7 | T acute lymphoblastic leukemia | Homo sapiens | CVCL_6825 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| Response Summary | METTL3 and METTL14 play an oncogenic role in acute myeloid leukemia(AML) by targeting E3 ubiquitin-protein ligase Mdm2 (Mdm2)/p53 signal pathway. The knockdown of METTL3 and METTL14 in K562 cell line leads to several changes in the expression of p53 signal pathway, including the upregulation of p53, cyclin dependent kinase inhibitor 1A (CDKN1A/p21), and downregulation of mdm2. | |||
Acute kidney failure [ICD-11: GB60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [289] | |||
| Responsed Disease | Acute kidney failure [ICD-11: GB60] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HK-2 [Human kidney] | Normal | Homo sapiens | CVCL_0302 |
E3 ubiquitin-protein ligase SMURF1 (SMURF1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
 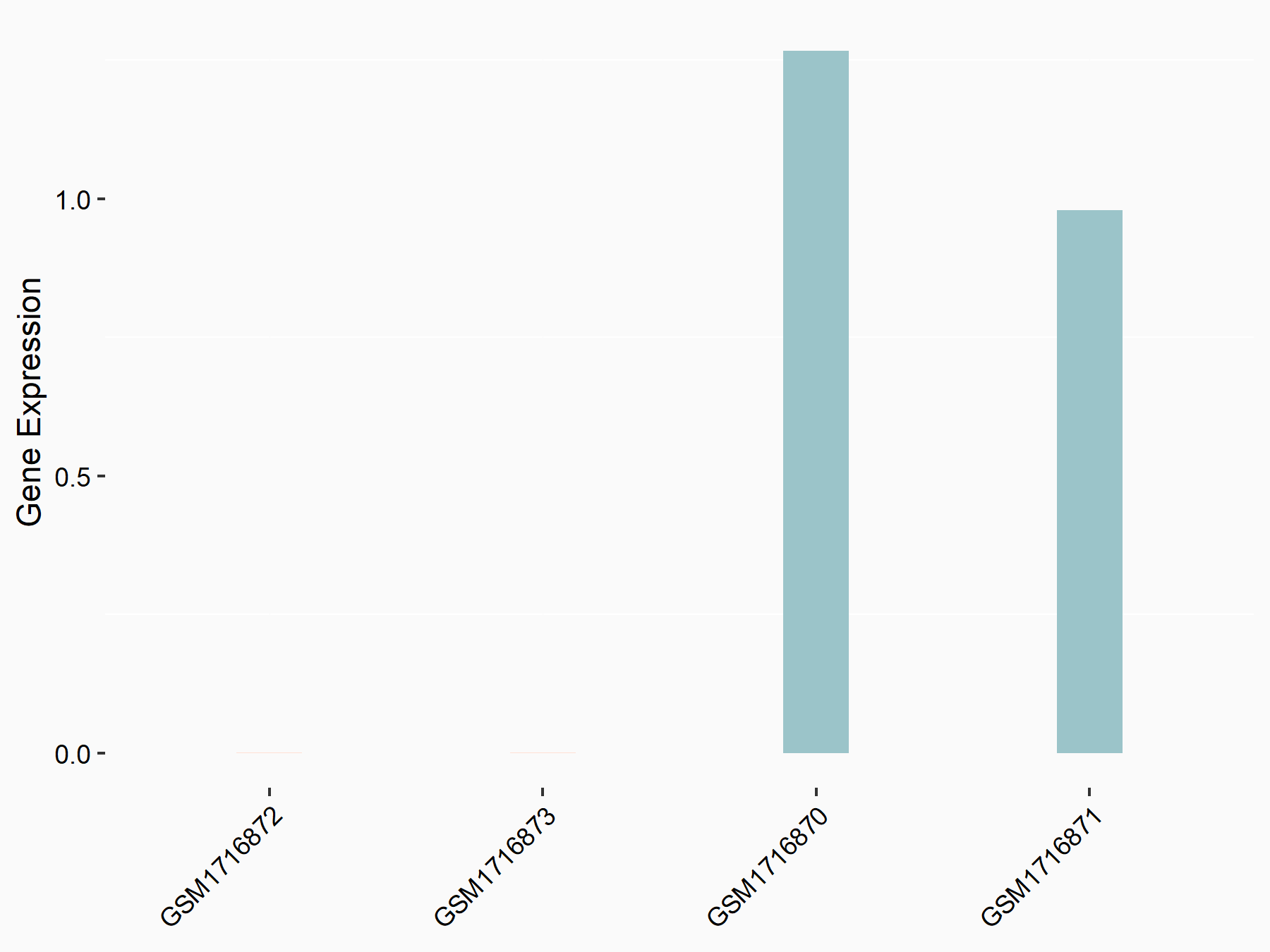 |
logFC: -1.08E+00 p-value: 3.04E-03 |
| More Results | Click to View More RNA-seq Results | |
Inflammatory response [ICD-11: MG46]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [162] | |||
| Responsed Disease | Inflammatory response [ICD-11: MG46] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
MC3T3-E1 | Normal | Mus musculus | CVCL_0409 |
| Response Summary | METTL3 knockdown inhibits osteoblast differentiation and Smad-dependent signaling by stabilizing Smad7 and E3 ubiquitin-protein ligase SMURF1 (SMURF1) mRNA transcripts via YTHDF2 involvement and activates the inflammatory response by regulating MAPK signaling in LPS-induced inflammation. | |||
ELAV-like protein 1 (HuR/ELAVL1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Embryonic stem cells | Mus musculus |
|
Treatment: METTL3 knockout ESCs
Control: Wild type ESCs
|
GSE146466 | |
| Regulation |
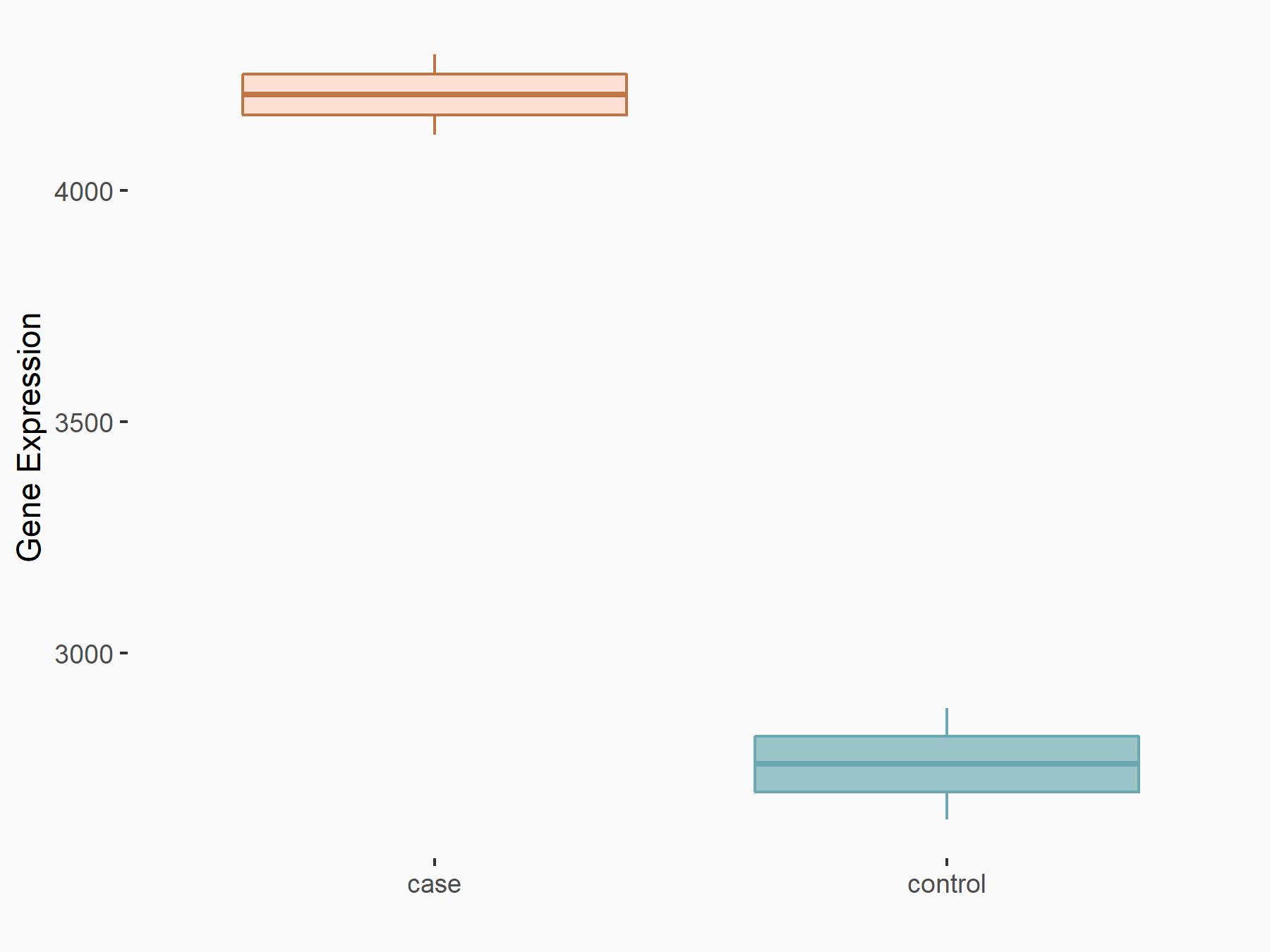 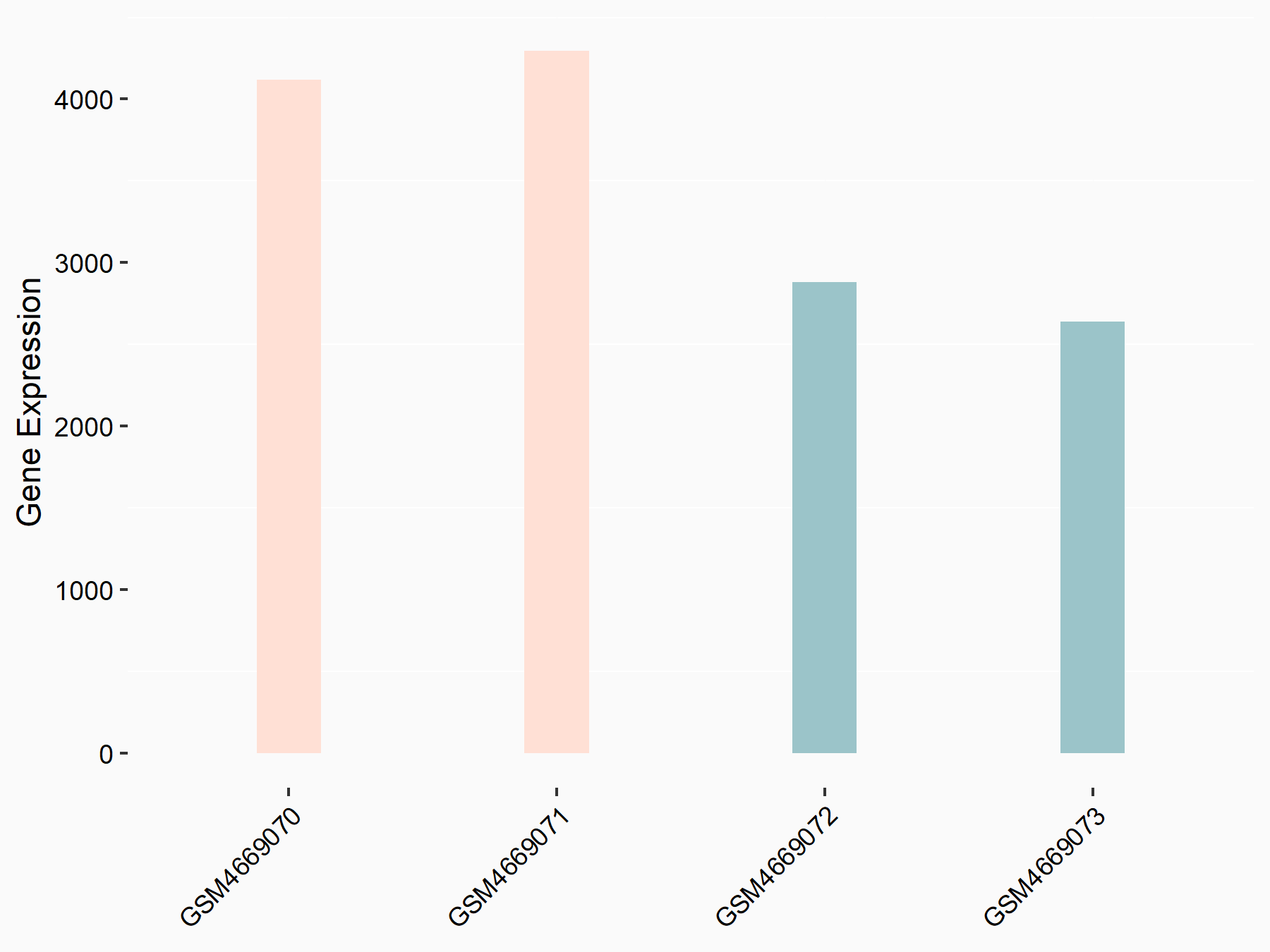 |
logFC: 6.09E-01 p-value: 1.50E-08 |
| More Results | Click to View More RNA-seq Results | |
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [163] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| In-vivo Model | A total of 1 × 106 PC3 cells or DU145 cells suspended in a mixture of 100 uL PBS and Matrigel were subcutaneously injected into BALB/c nude mice. Tumor weight were measured 2 months after the engraftment. To evaluate the role of METTL3 in tumor metastasis, PC3 cells with or without knockdown of METTL3 were injected into SCID mice through the tail vein (1 × 106 cells per mouse). After eight weeks, mice were sacrificed and their lung tissues were collected for subsequent analyses. | |||
| Response Summary | m6A modification levels were markedly upregulated in human PCa tissues due to increased expression of METTL3. METTL3 mediates m6A modification of USP4 mRNA at A2696, and m6A reader protein YTHDF2 binds to and induces degradation of USP4 mRNA by recruiting RNA-binding protein HNRNPD to the mRNA. Decrease of USP4 fails to remove the ubiquitin group from ELAVL1 protein, resulting in a reduction of ELAVL1 protein. Lastly, downregulation of ELAV-like protein 1 (HuR/ELAVL1) in turn increases ARHGDIA expression, promoting migration and invasion of PCa cells. | |||
Ephrin type-B receptor 2 (ERK/EPHB2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
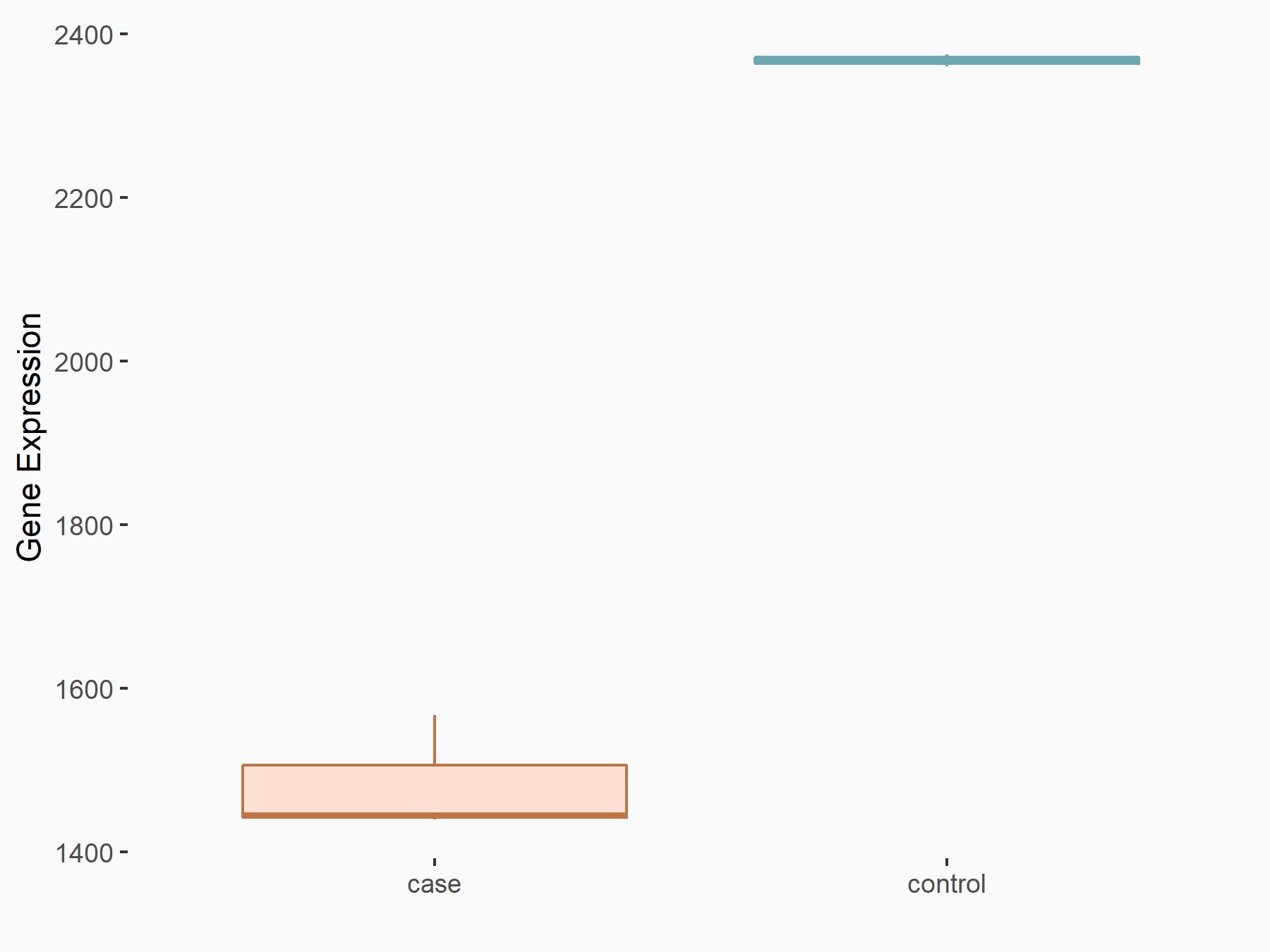 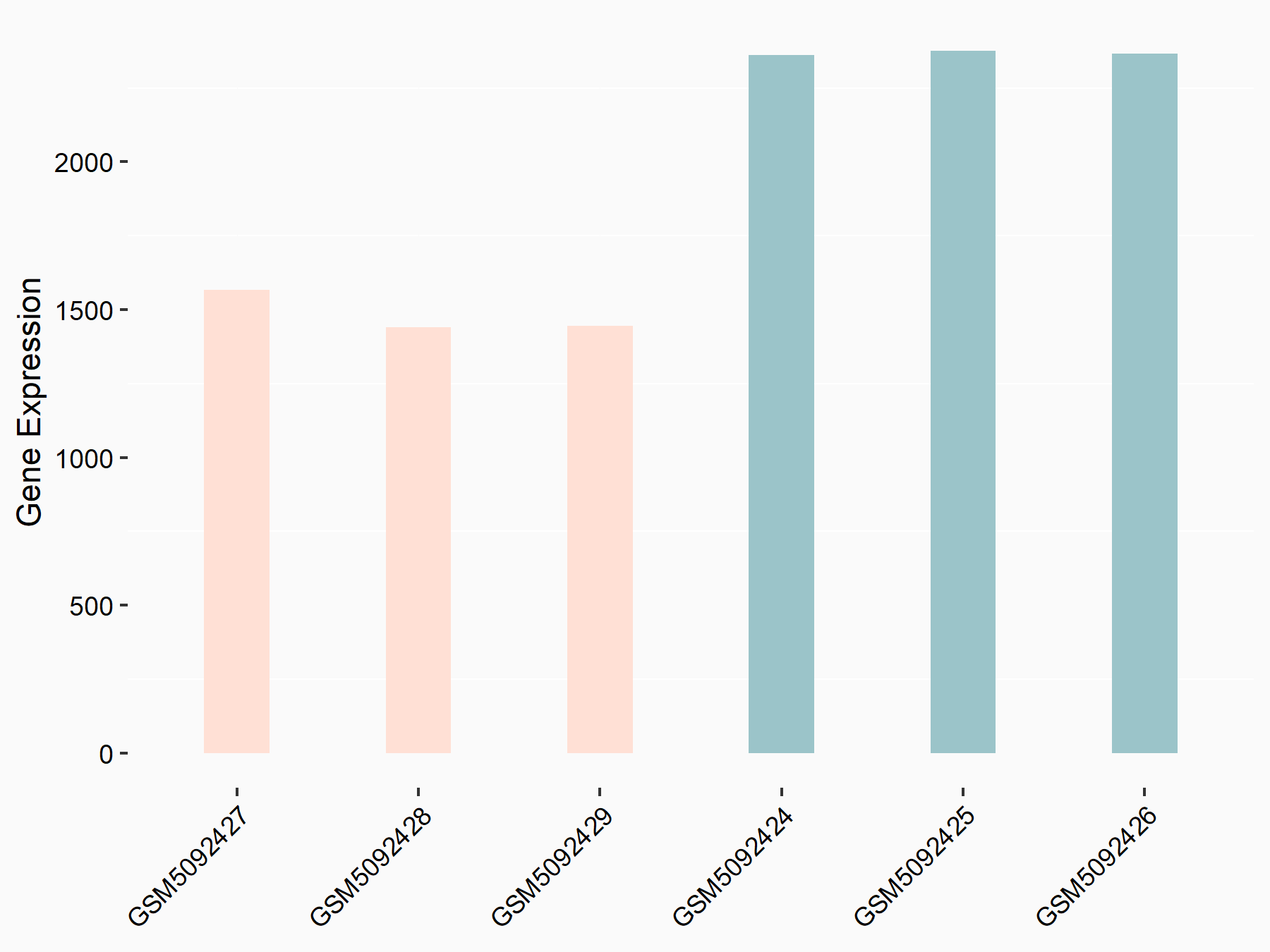 |
logFC: -6.74E-01 p-value: 5.11E-30 |
| More Results | Click to View More RNA-seq Results | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [72] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| KM12 | Colon carcinoma | Homo sapiens | CVCL_1331 | |
| Response Summary | METTL3 played a tumor-suppressive role in Colorectal cancer cell proliferation, migration and invasion through p38/Ephrin type-B receptor 2 (ERK/EPHB2) pathways, which indicated that METTL3 was a novel marker for CRC carcinogenesis, progression and survival. | |||
Muscular dystrophies [ICD-11: 8C70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [67] | |||
| Responsed Disease | Muscular dystrophies [ICD-11: 8C70] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| C2C12 | Normal | Mus musculus | CVCL_0188 | |
| In-vivo Model | For mouse muscle injury and regeneration experiment, tibialis anterior (TA) muscles of 6-week-old male mice were injected with 25 uL of 10 uM cardiotoxin (CTX, Merck Millipore, 217503), 0.9% normal saline (Saline) were used as control. The regenerated muscles were collected at day 1, 3, 5, and 10 post-injection. TA muscles were isolated for Hematoxylin and eosin staining or frozen in liquid nitrogen for RNA and protein extraction. | |||
| Response Summary | m6A writers METTL3/METTL14 and the m6A reader YTHDF1 orchestrate MNK2 expression posttranscriptionally and thus control Ephrin type-B receptor 2 (ERK/EPHB2) signaling, which is required for the maintenance of muscle myogenesis and contributes to regeneration. | |||
Epidermal growth factor receptor (EGFR)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
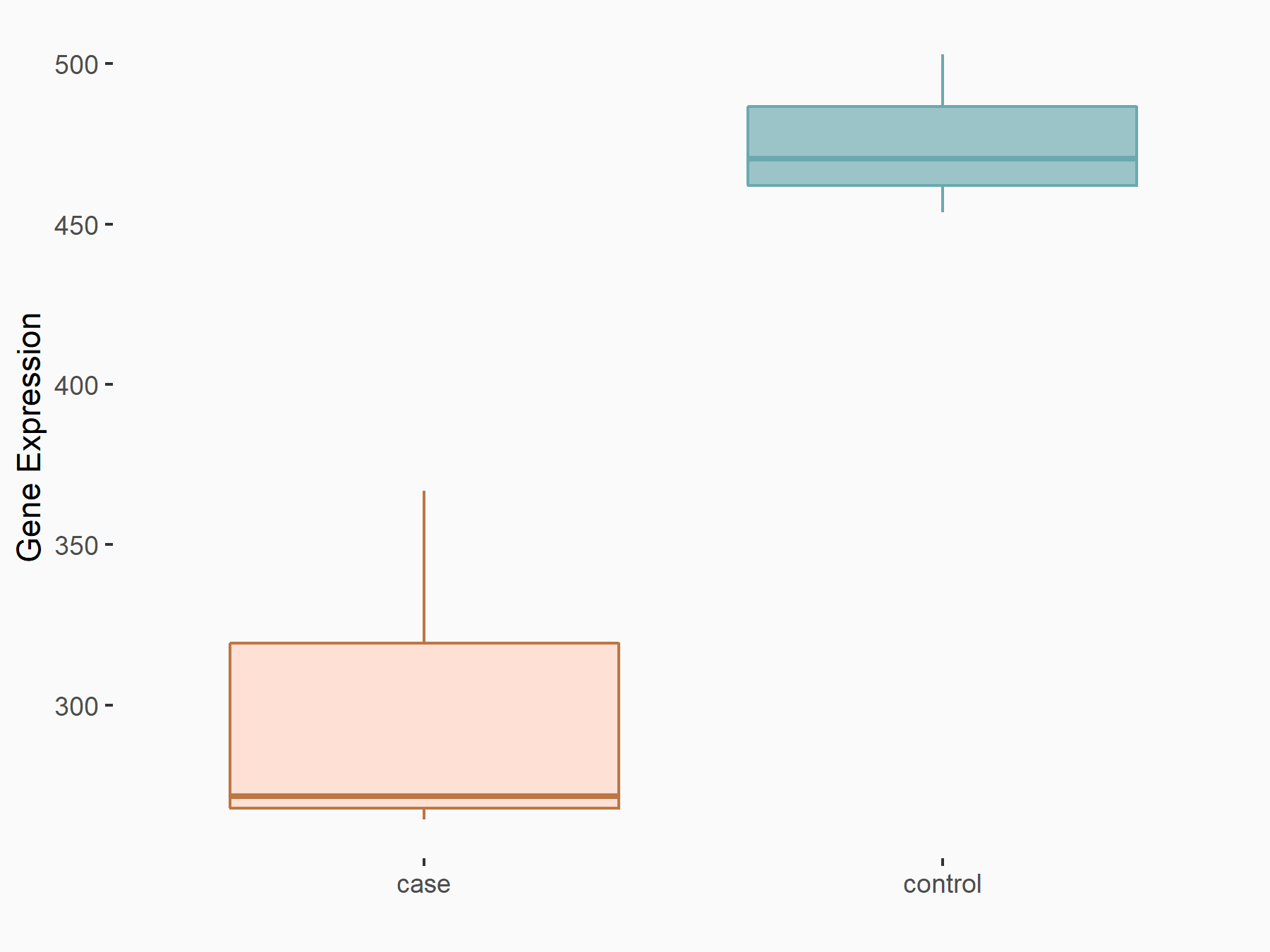 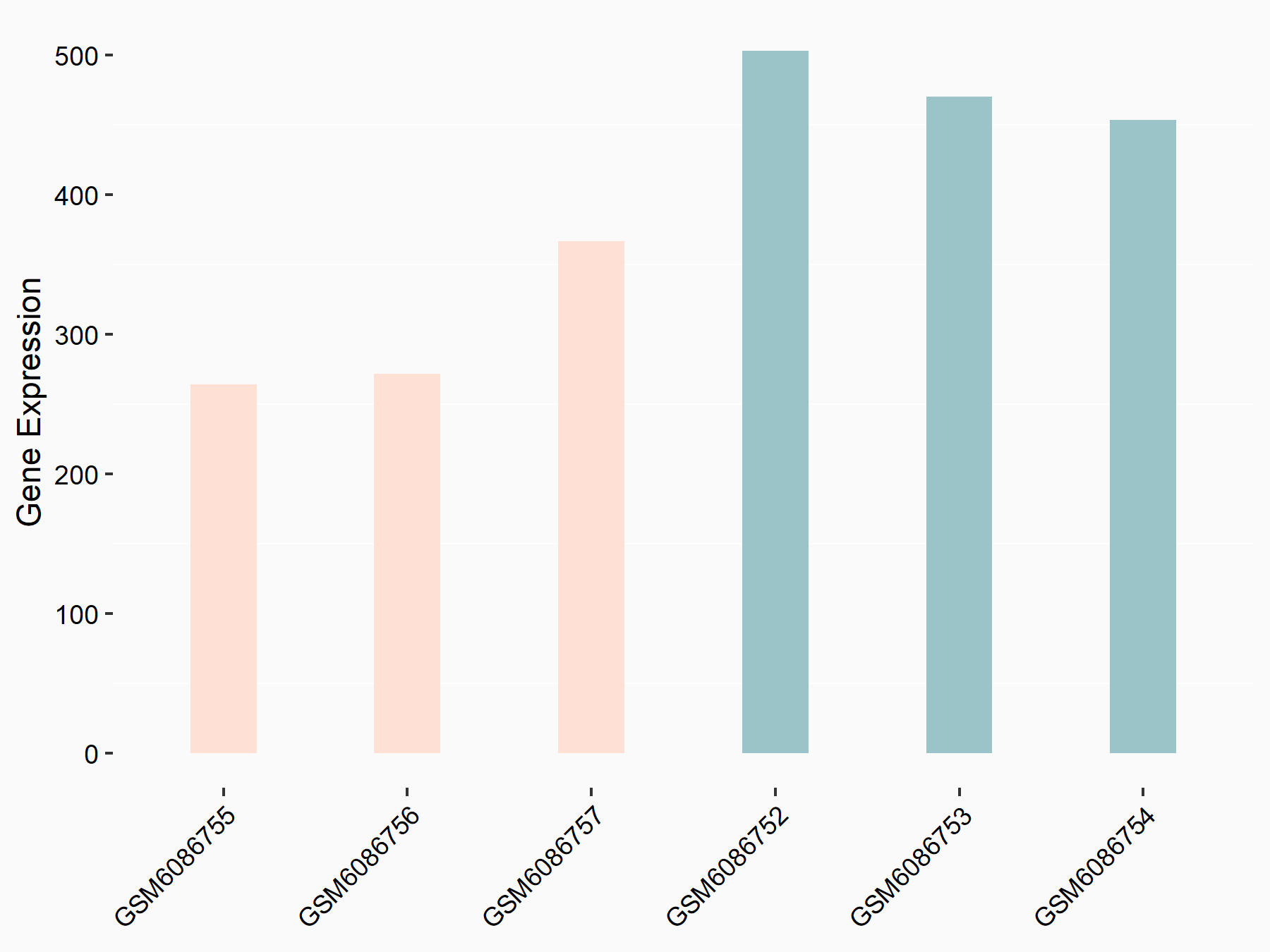 |
logFC: -6.74E-01 p-value: 1.53E-03 |
| More Results | Click to View More RNA-seq Results | |
Melanoma [ICD-11: 2C30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [164] | |||
| Responsed Disease | Melanoma [ICD-11: 2C30] | |||
| Responsed Drug | PLX4032 | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | EGFR tyrosine kinase inhibitor resistance | hsa01521 | ||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
A375-R | Amelanotic melanoma | Homo sapiens | CVCL_6234 |
| Response Summary | METTL3 increased the m6A modification of Epidermal growth factor receptor (EGFR) mRNA in A375R cells, which promoted its translation efficiency. Inhibiting METTL3 function to restore PLX4032 sensitivity in patients with melanoma. | |||
Liver cancer [ICD-11: 2C12]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [290] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Lenvatinib | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 |
| In-vivo Model | Tumor cells derived xenograft (MHCC97H cells) from subcutaneous mouse model were cut into 1 mm3 pieces and implanted into the left lobe of the livers of six-week-old male NCG mice. Once the tumors were established and reached approximately 100 mm3, mice were randomly assigned to daily treatment with vehicle, lenvatinib (4 mg/kg, oral gavage), STM2457 (50 mg/kg, intraperitoneal injection) or a drug combination in which each compound was administered at the same dose and duration as the single agent. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [290] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | STM2457 | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [291] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
F-box/WD repeat-containing protein 7 (FBXW7)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
|
Treatment: METTL3 knockdown MDA-MB-231 cells
Control: MDA-MB-231 cells
|
GSE70061 | |
| Regulation |
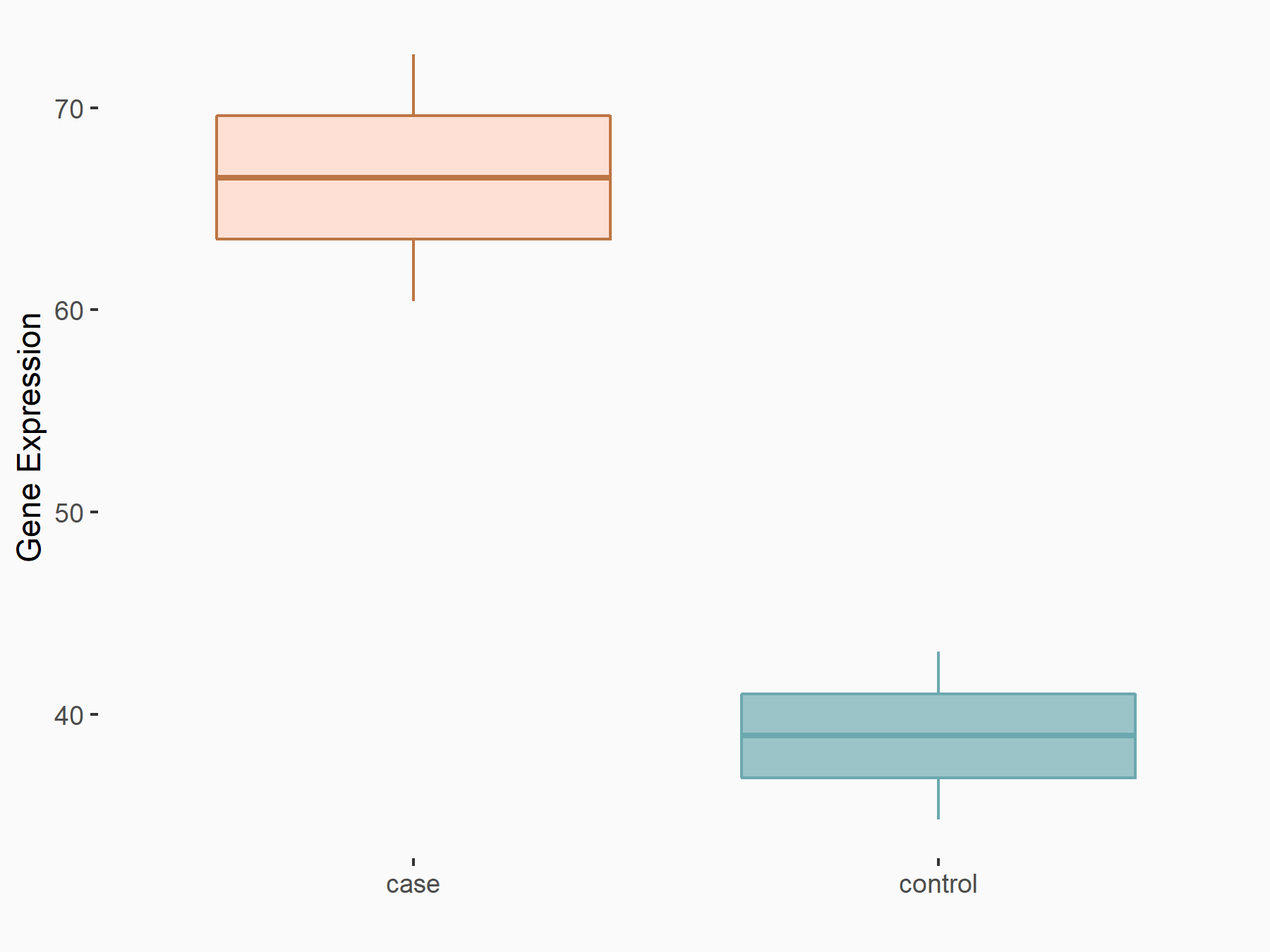 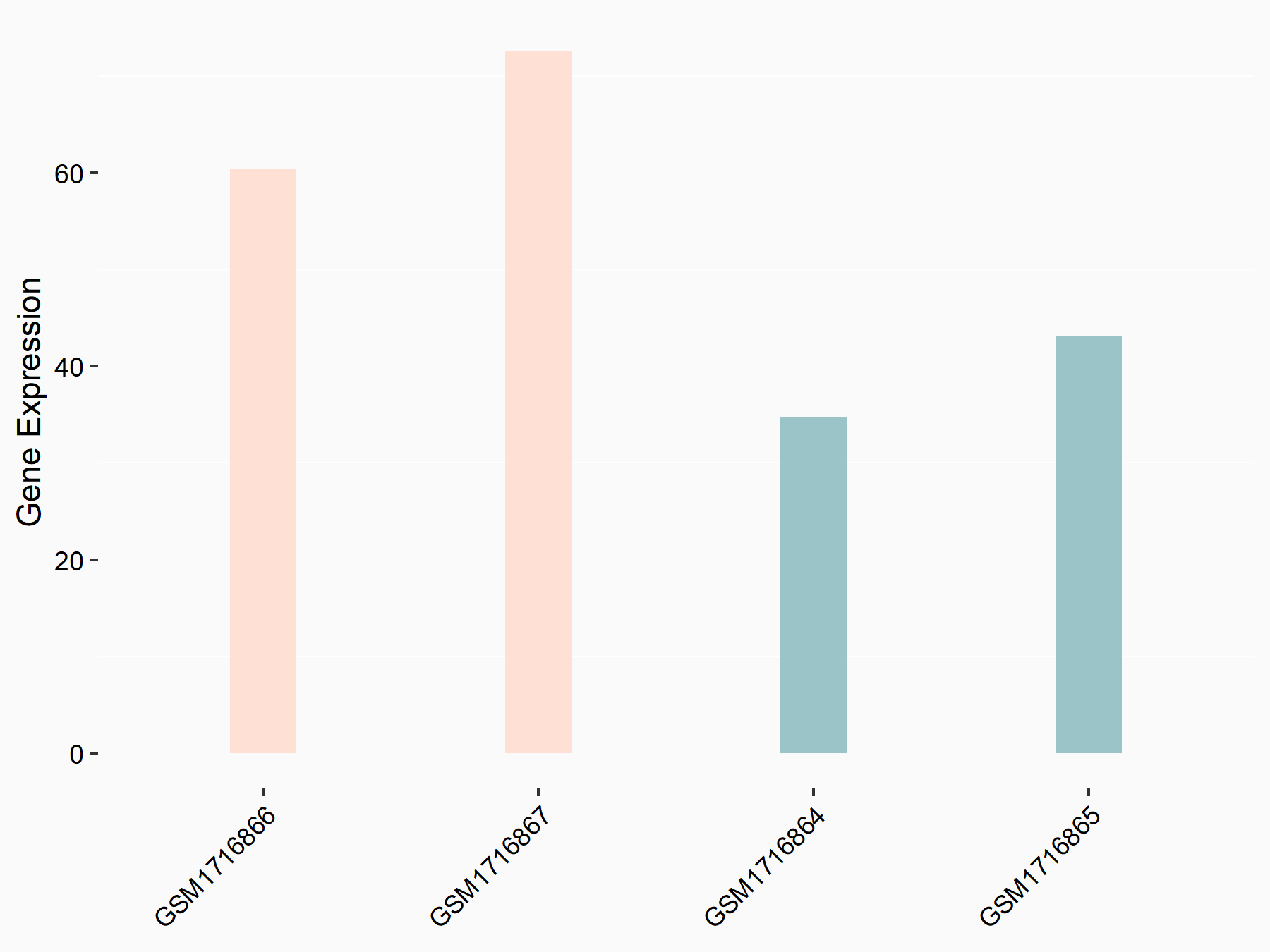 |
logFC: 7.60E-01 p-value: 3.50E-02 |
| More Results | Click to View More RNA-seq Results | |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [165] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulation | Down regulation | |||
| Cell Process | RNA stability | |||
| Cell apoptosis | ||||
In-vitro Model |
HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | The mice were randomly divided into groups before subcutaneous injection with cells (107) suspended in 200 uL of phosphate-buffered saline. Tumors were measured on day 7 and then once every 7 days until 28 days after injection. | |||
| Response Summary | METTL3 positively regulates F-box/WD repeat-containing protein 7 (FBXW7) expression and confirm the tumor-suppressive role of m6A-modified FBXW7, thus providing insight into its epigenetic regulatory mechanisms in lung adenocarcinoma initiation and development. | |||
Forkhead box protein C2 (FOXC2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Mesenchymal stem cell line | Mus musculus |
|
Treatment: Mettl3 knockout mesenchymal stem cells
Control: Wild type mesenchymal stem cells
|
GSE114933 | |
| Regulation |
 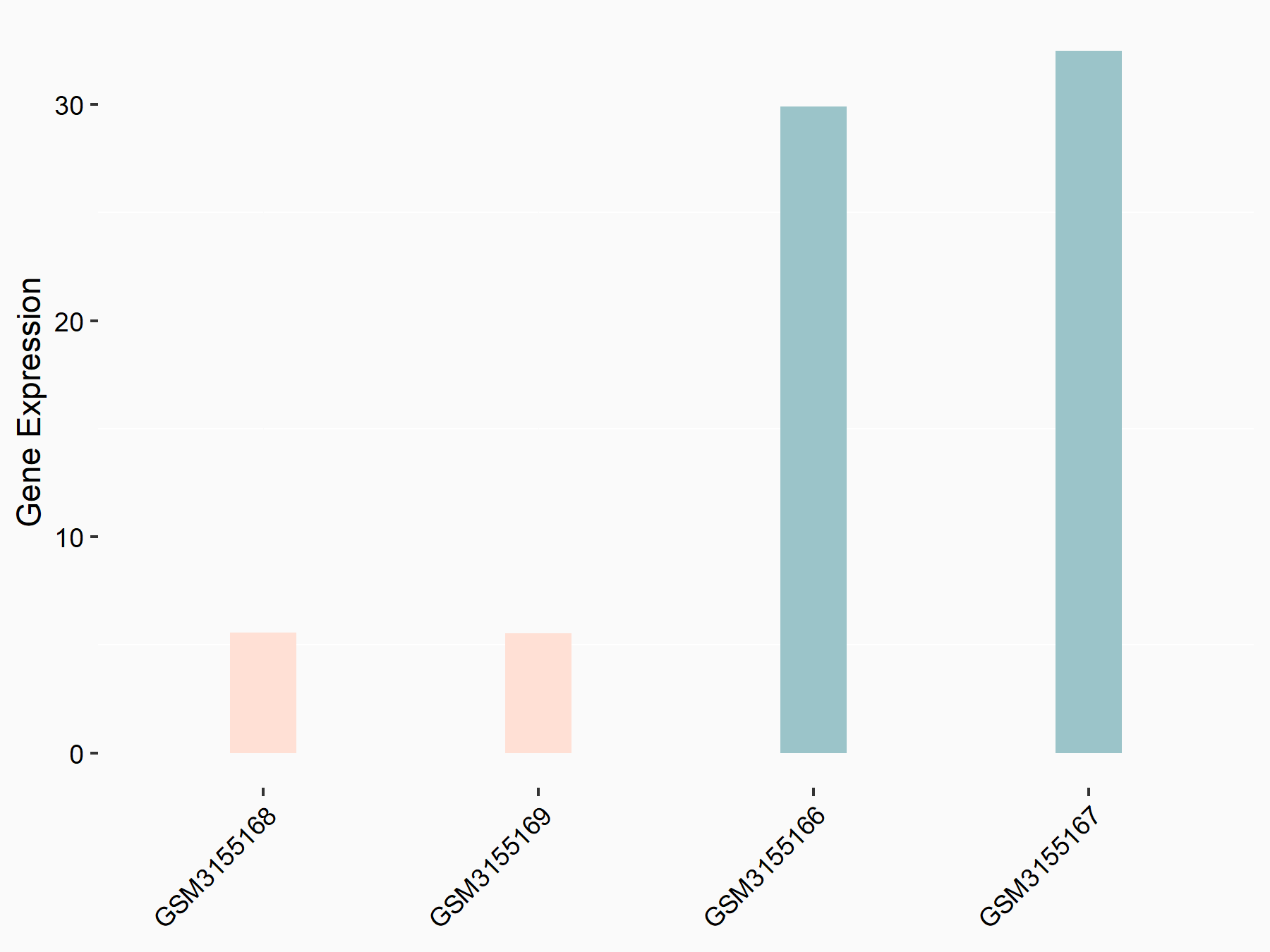 |
logFC: -2.30E+00 p-value: 6.85E-05 |
| More Results | Click to View More RNA-seq Results | |
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [166] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
hFOB 1.19 | Normal | Homo sapiens | CVCL_3708 |
| MG-63 | Osteosarcoma | Homo sapiens | CVCL_0426 | |
| U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 | |
| Response Summary | METTL3-mediated circNRIP1 exhibits oncogenic roles in osteosarcoma by regulating Forkhead box protein C2 (FOXC2) via sponging miR-199a, which provides new ideas for the treatment of osteosarcoma. | |||
Forkhead box protein O1 (FOXO1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
 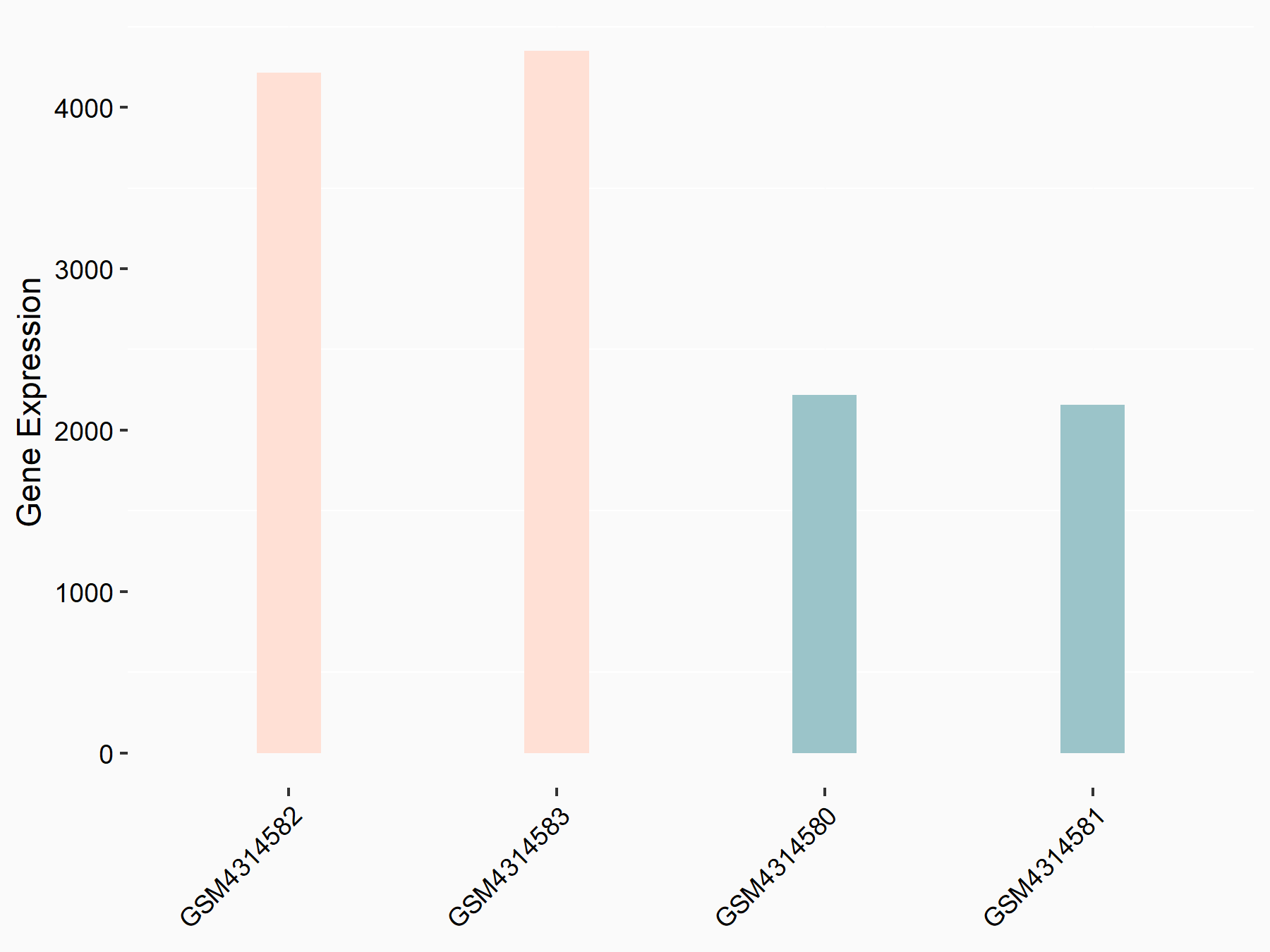 |
logFC: 9.70E-01 p-value: 7.07E-55 |
| More Results | Click to View More RNA-seq Results | |
Type 2 diabetes mellitus [ICD-11: 5A11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [48] | |||
| Responsed Disease | Type 2 diabetes mellitus [ICD-11: 5A11] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Insulin resistance | hsa04931 | ||
| Cell Process | Lipid metabolism | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | Hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were generated by crossing mice with TBG-Cre Tg mice. METTL3 flox (METTL3 fl/fl) and hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were used for experiments. | |||
| Response Summary | Type 2 diabetes (T2D) is characterized by lack of insulin, insulin resistance and high blood sugar. METTL3 silence decreased the m6A methylated and total mRNA level of Fatty acid synthase (Fasn), subsequently inhibited fatty acid metabolism. The expression of Acc1, Acly, Dgat2, Ehhadh, Fasn, Forkhead box protein O1 (FOXO1), Pgc1a and Sirt1, which are critical to the regulation of fatty acid synthesis and oxidation were dramatically decreased in livers of hepatocyte-specific METTL3 knockout mice. | |||
Female infertility [ICD-11: GA31]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [293] | |||
| Responsed Disease | Female infertility [ICD-11: GA31] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
T HESCs | Normal | Homo sapiens | CVCL_C464 |
| HTR-8/SVneo | Normal | Homo sapiens | CVCL_7162 | |
| HTR-8 | Normal | Homo sapiens | CVCL_D728 | |
| In-vivo Model | Five female mice were subjected to a normal pregnancy assay. Uterine tissues of 14 female donor mice were cut up and injected into the abdominal cavity of 28 female recipient mice. After 21 d, 10 male C57 mice were mated with the recipient mice, and the next day, when vaginal plugs were observed, was regarded as day 1. Eight recipient mice were euthanized by cervical dislocation after deep pentobarbital anesthesia on day 8, and the number of blastocysts in the uterus was counted. At the night of day 3, 10 μL of STM2457, a METTL3 inhibitor (Sellcek, S9870, 10 μM) and 10 μL dimethyl sulfoxide (DMSO) were individually injected into the uterine horns of 20 recipient mice. The mice were euthanized on day 8, and the number of blastocysts in the uterus was counted. The uterine tissues were collected and fixed in 4% (w/v) paraformaldehyde for histological and IHC analyses. | |||
Forkhead box protein O3 (FOXO3)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
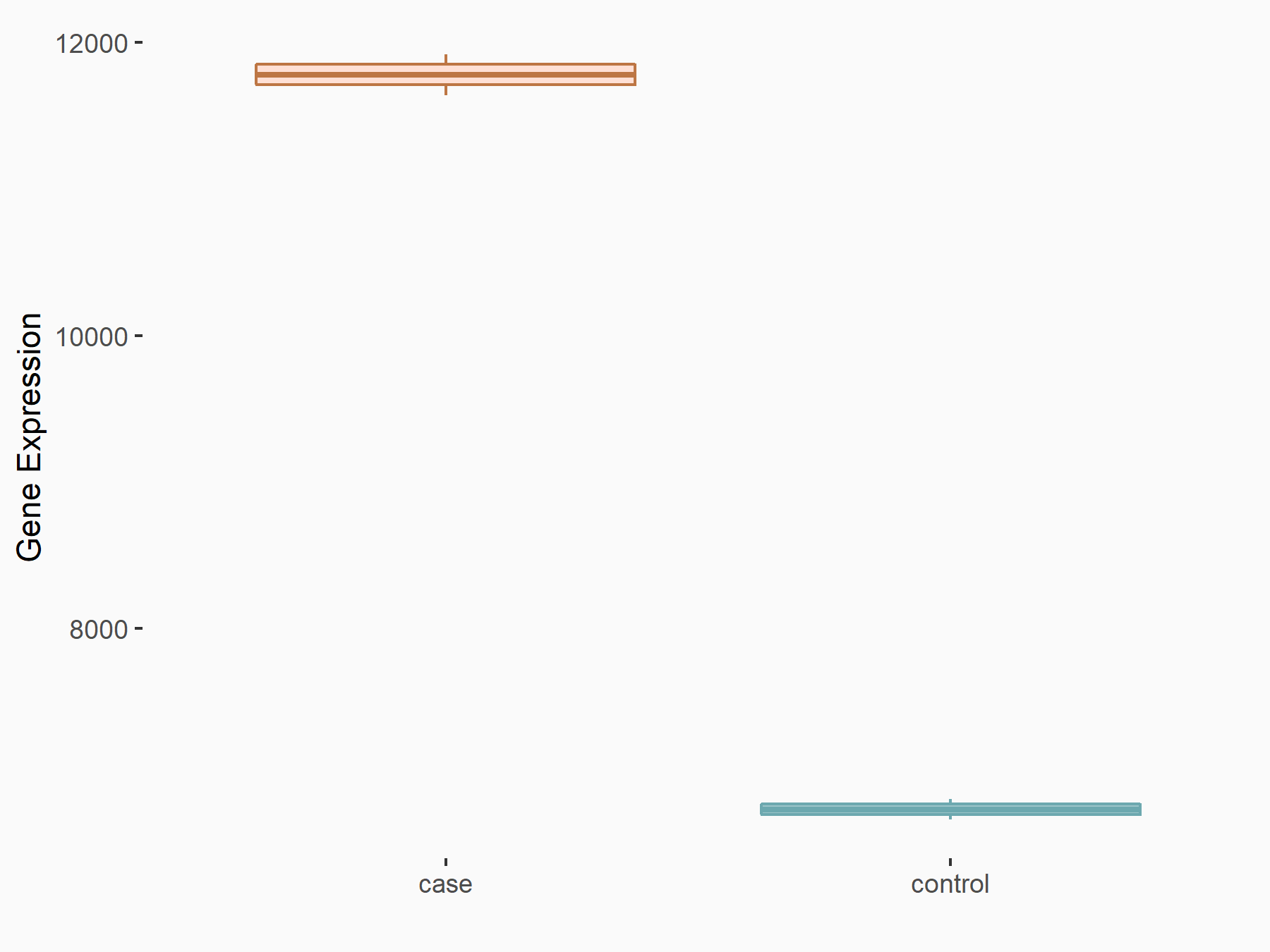 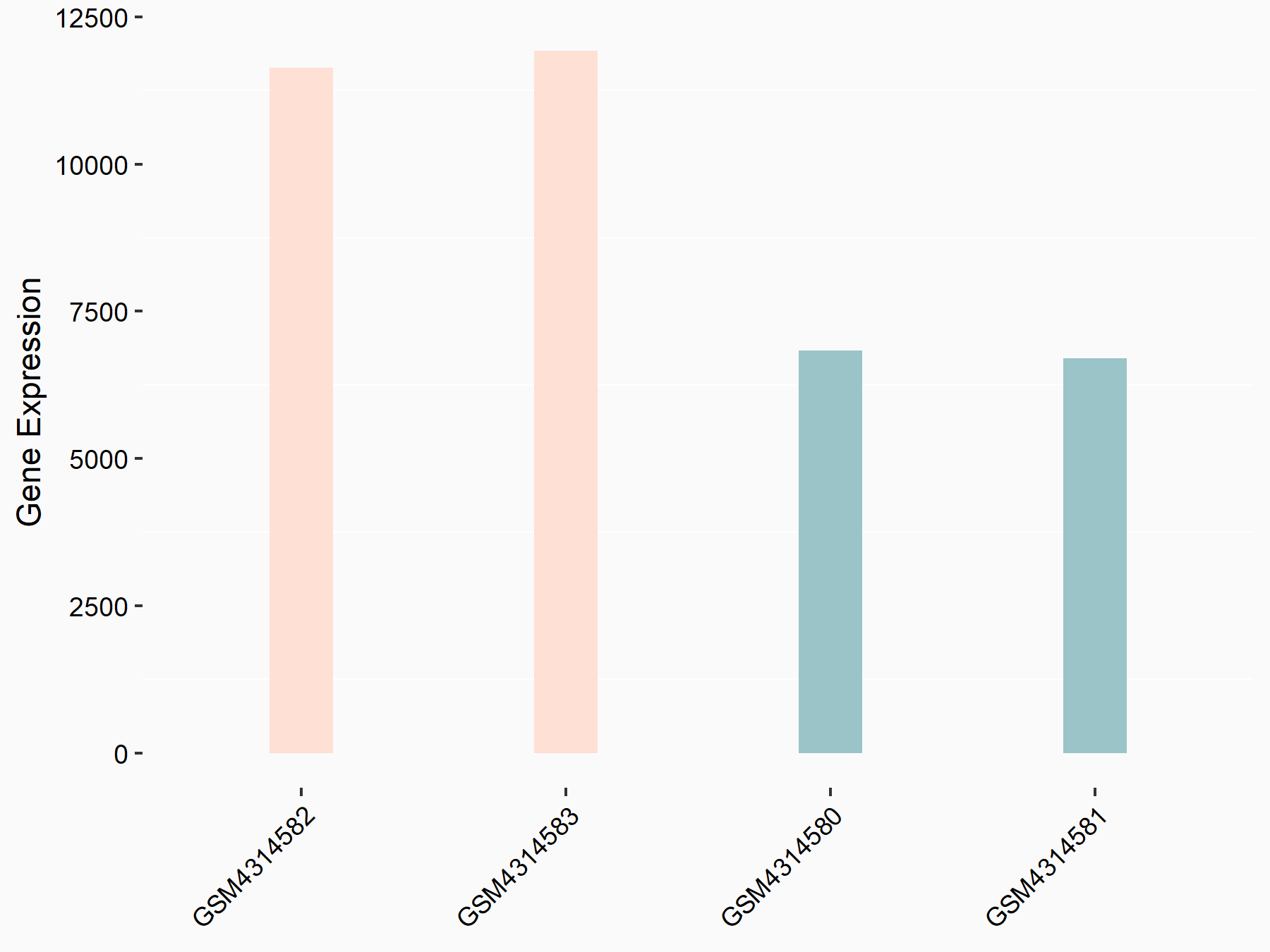 |
logFC: 8.00E-01 p-value: 1.28E-68 |
| More Results | Click to View More RNA-seq Results | |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [167] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Cell Process | Cell Transport | |||
| Cell catabolism | ||||
| Cell autophagy | ||||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| Hepa 1-6 | Hepatocellular carcinoma of the mouse | Mus musculus | CVCL_0327 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
| WRL 68 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0581 | |
| In-vivo Model | For the drug-resistant subcutaneous tumor models, drug administration was adopted when the tumors reached about 50 mm3 in size, at which point mice were randomized for treatment with DMSO(intraperitoneally) or sorafenib (50 mg/kg/every 2 days, intraperitoneally). For the patient-derived tumor xenograft model, drug administration began 4 weeks after tumors reached about 100 mm3 in size with sorafenib (50 mg/kg/every 3 days, intraperitoneally) or siCtrl/siMETTL3 intratumor injection. | |||
| Response Summary | METTL3 and Forkhead box protein O3 (FOXO3) levels are tightly correlated in hepatocellular carcinoma patients. In mouse xenograft models, METTL3 depletion significantly enhances sorafenib resistance of HCC by abolishing the identified METTL3-mediated FOXO3 mRNA stabilization, and overexpression of FOXO3 restores m6 A-dependent sorafenib sensitivity. | |||
G1/S-specific cyclin-D1 (CCND1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LNCaP cell line | Homo sapiens |
|
Treatment: shMETTL3 LNCaP cells
Control: shControl LNCaP cells
|
GSE147884 | |
| Regulation |
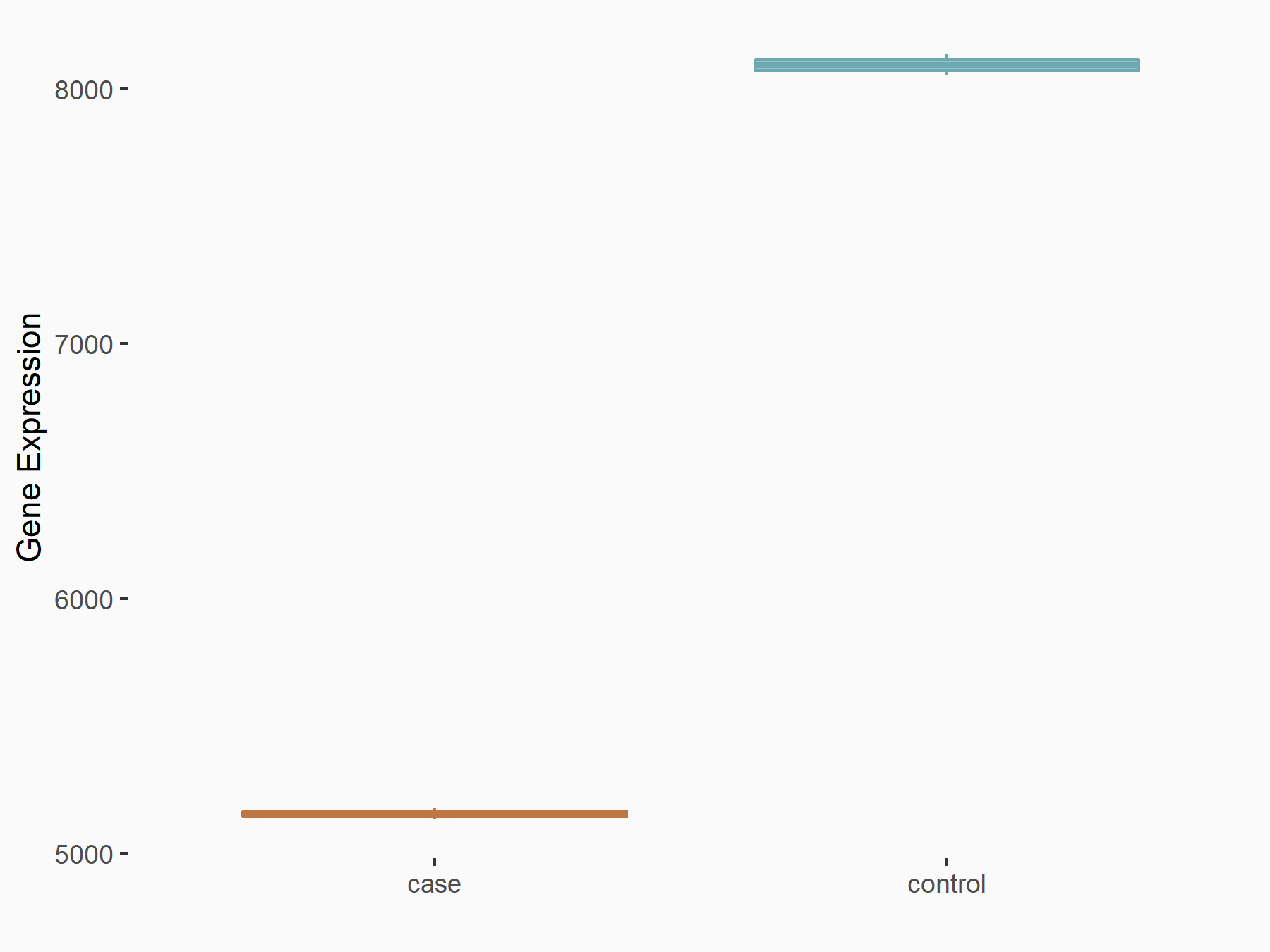 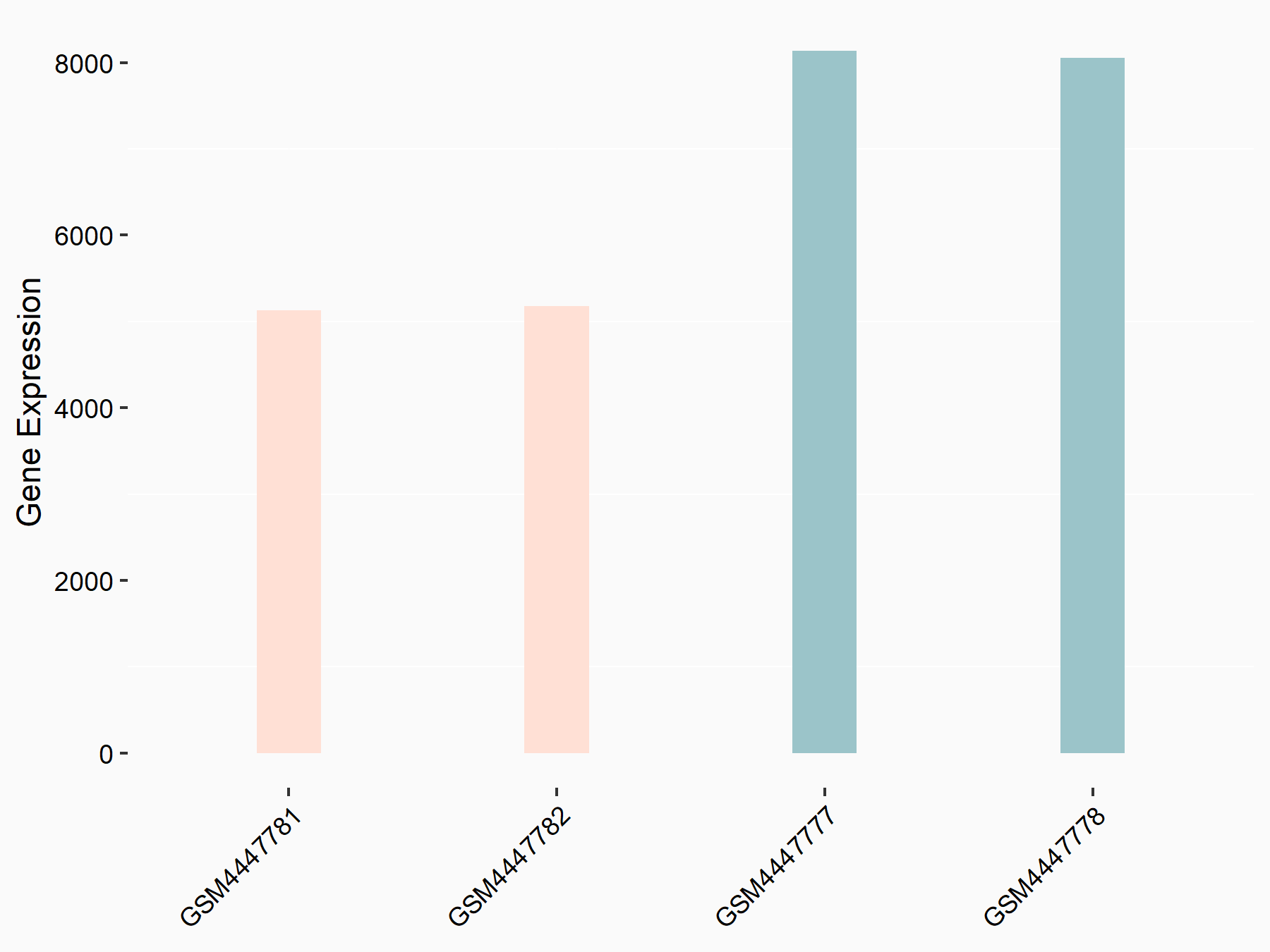 |
logFC: -6.51E-01 p-value: 1.46E-104 |
| More Results | Click to View More RNA-seq Results | |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [8] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| Response Summary | Down-regulation of METTL3 inhibits the proliferation and mobility of human gastric cancer cells and leads to inactivation of the AKT signaling pathway, suggesting that METTL3 is a potential target for the treatment of human gastric cancer. METTL3 knockdown decreased Bcl2 and increased Bax and active Caspase-3 in gastric cancer cells, which suggested the apoptotic pathway was activated. METTL3 led to inactivation of the AKT signaling pathway in human gastric cancer cells, including decreased phosphorylation levels of AKT and expression of down-stream effectors p70S6K and G1/S-specific cyclin-D1 (CCND1). | |||
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [168] | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell cycle | |||
| Cell apoptosis | ||||
In-vitro Model |
OVCAR-3 | Ovarian serous adenocarcinoma | Homo sapiens | CVCL_0465 |
| SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 | |
| Response Summary | METTL3 knockdown downregulated the phosphorylation levels of AKT and the expression of the downstream effector G1/S-specific cyclin-D1 (CCND1) in ovarian cancer. | |||
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [169] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| In-vivo Model | Mice were divided into two groups (n = 4/group) randomly. 3×106 cells suspended in 200 uL PBS were administered via subcutaneous injection over the right flank region of nude mice. After the development of palpable tumors (average volume, 50 mm3), intratumoral injection of synthetic miR-193b, or negative control complexed with siPORT Amine transfection reagent (Ambion, USA) was given 6 times at a 4-day interval. | |||
| Response Summary | METTL3 modulates miR-193b mature process in an m6A-dependent manner. Reintroduction of miR-193b profoundly inhibits tumorigenesis of cervical cancer cells both in vivo and in vitro through G1/S-specific cyclin-D1 (CCND1) targeting. | |||
Obesity [ICD-11: 5B81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [170] | |||
| Responsed Disease | Obesity [ICD-11: 5B81] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Mitotic clonal | |||
| Prolonged G1/S transition | ||||
In-vitro Model |
3T3-L1 | Normal | Mus musculus | CVCL_0123 |
| Response Summary | Obesity is becoming a global problem. ZFP217 knockdown-induced adipogenesis inhibition was caused by G1/S-specific cyclin-D1 (CCND1), which was mediated by METTL3 and YTHDF2 in an m6A-dependent manner. | |||
Glucose transporter type 1 (GLUT1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
  |
logFC: 8.54E-01 p-value: 1.08E-85 |
| More Results | Click to View More RNA-seq Results | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [54] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glucose metabolism | |||
| Response Summary | METTL3 stabilizes HK2 and Glucose transporter type 1 (SLC2A1) (GLUT1) expression in colorectal cancer through an m6A-IGF2BP2/3- dependent mechanism. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [294] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
Heat shock 70 kDa protein 1A (HSPA1A)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
 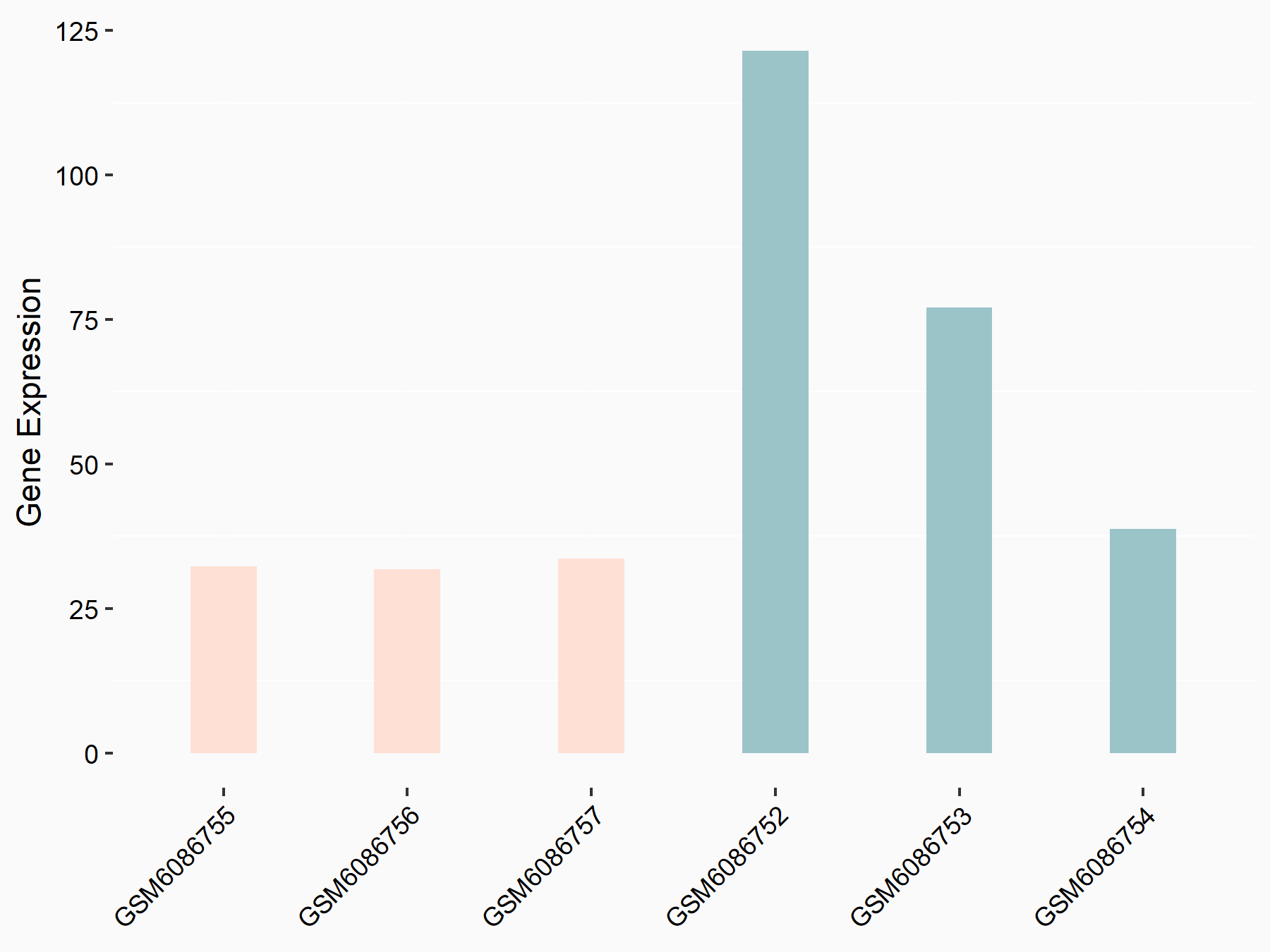 |
logFC: -1.11E+00 p-value: 1.96E-02 |
| More Results | Click to View More RNA-seq Results | |
Pre-eclampsia [ICD-11: JA24]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [172] | |||
| Responsed Disease | Pre-eclampsia [ICD-11: JA24] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| mTOR signaling pathway | hsa04150 | |||
| Response Summary | Findings show that METTL3 and METTL14 were up-regulated in preeclampsia(PE). Heat shock 70 kDa protein 1A (HSPA1A) is involved in the pathophysiology of PE as its mRNA and protein expression is regulated by m6A modification. | |||
Hepatocyte growth factor receptor (c-Met/MET)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 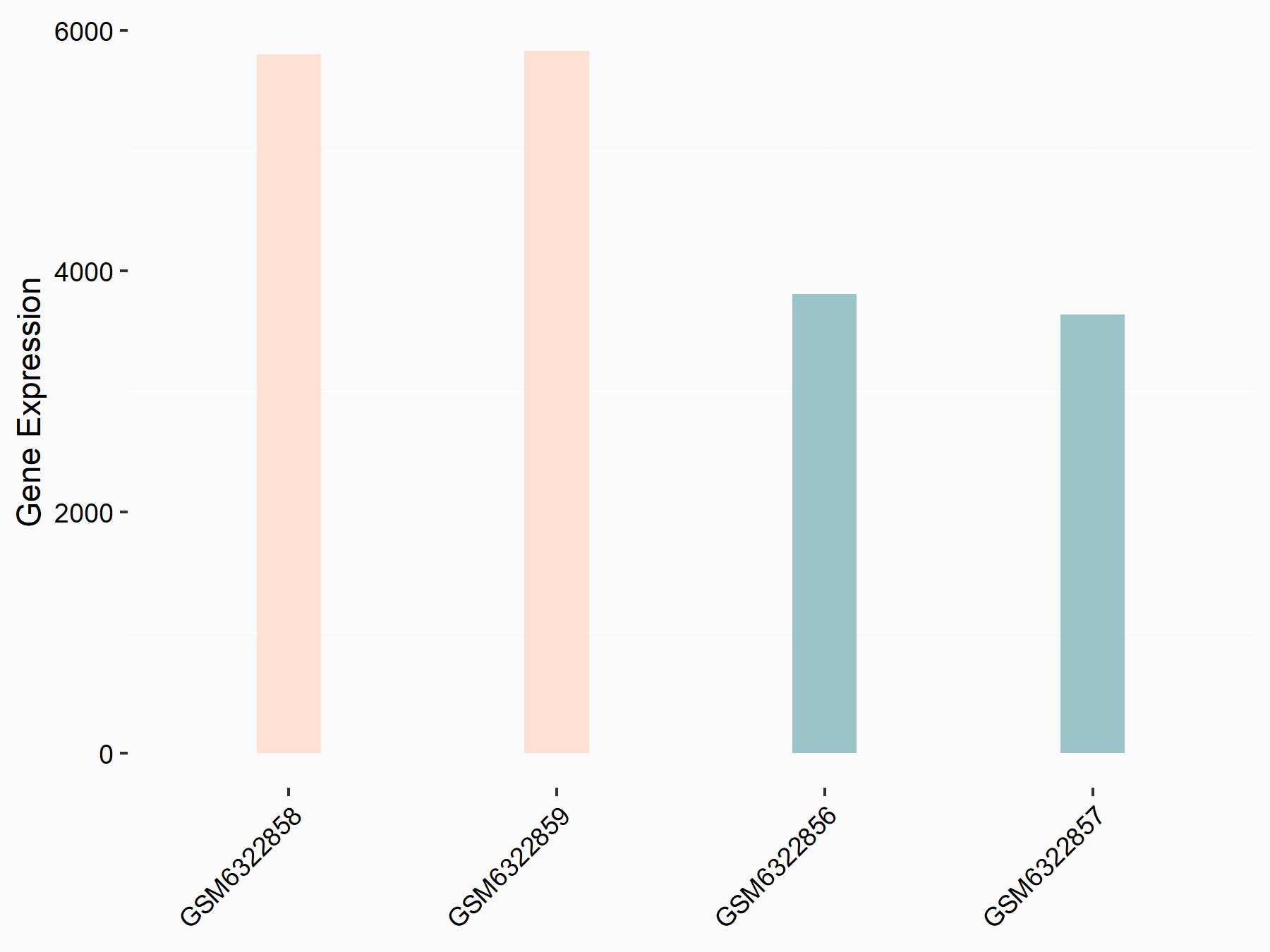 |
logFC: 6.42E-01 p-value: 3.45E-17 |
| More Results | Click to View More RNA-seq Results | |
Lung cancer [ICD-11: 2C25]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [173] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Crizotinib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | EGFR tyrosine kinase inhibitor resistance | hsa01521 | ||
In-vitro Model |
HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 |
| NCI-H661 | Lung large cell carcinoma | Homo sapiens | CVCL_1577 | |
| NCI-H596 | Lung adenosquamous carcinoma | Homo sapiens | CVCL_1571 | |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| NCI-H358 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1559 | |
| NCI-H292 | Lung mucoepidermoid carcinoma | Homo sapiens | CVCL_0455 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1395 | Lung adenocarcinoma | Homo sapiens | CVCL_1467 | |
| EBC-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_2891 | |
| Calu-3 | Lung adenocarcinoma | Homo sapiens | CVCL_0609 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | HCC827 (3×106) cells suspended in 100 uL of PBS were injected into the left inguen of female Balb/c nude mice (body weight 18-20 g; age 6 weeks; Beijing Huafukang Bioscience Co., Inc.). When the tumor volumes reached 50-100 mm3 on the 10th posttransplantation day, the mice were randomized into four groups (10 mice per group) and were intragastrically administered vehicle (normal saline), crizotinib (25 mg/kg body weight), chidamide (5 mg/kg), or the combination of the two drugs daily for 21 days. The tumor volumes and body weights of the mice were measured every 3 days. | |||
| Response Summary | Chidamide could decrease Hepatocyte growth factor receptor (c-Met/MET) expression by inhibiting mRNA N6-methyladenosine (m6A) modification through the downregulation of METTL3 and WTAP expression, subsequently increasing the crizotinib sensitivity of NSCLC cells in a c-MET-/HGF-dependent manner. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [174] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Responsed Drug | Gefitinib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
In-vitro Model |
PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| NCI-H3255 | Lung adenocarcinoma | Homo sapiens | CVCL_6831 | |
| Response Summary | METTL3 combines with Hepatocyte growth factor receptor (c-Met/MET) and causes the PI3K/AKT signalling pathway to be manipulated, which affects the sensitivity of lung cancer cells to gefitinib. METTL3 knockdown promotes apoptosis and inhibits proliferation of lung cancer cells. | |||
Melanoma of uvea [ICD-11: 2D0Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [175] | |||
| Responsed Disease | Melanoma of uvea [ICD-11: 2D0Y] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Arrest cell cycle at G1 phase | ||||
In-vitro Model |
M17 (Neuroblastoma cells) | |||
| M21 | Melanoma | Homo sapiens | CVCL_D031 | |
| M23 | Cutaneous melanoma | Homo sapiens | CVCL_RT32 | |
| SP-6.5 | Uveal melanoma | Homo sapiens | CVCL_7997 | |
| Response Summary | METTL3-mediated m6A RNA methylation modulates uveal melanoma cell proliferation, migration, and invasion by targeting Hepatocyte growth factor receptor (c-Met/MET). Cycloleucine (Cyc) was used to block m6 A methylation in UM cells. | |||
High mobility group protein HMG-I/HMG-Y (HMGA1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
 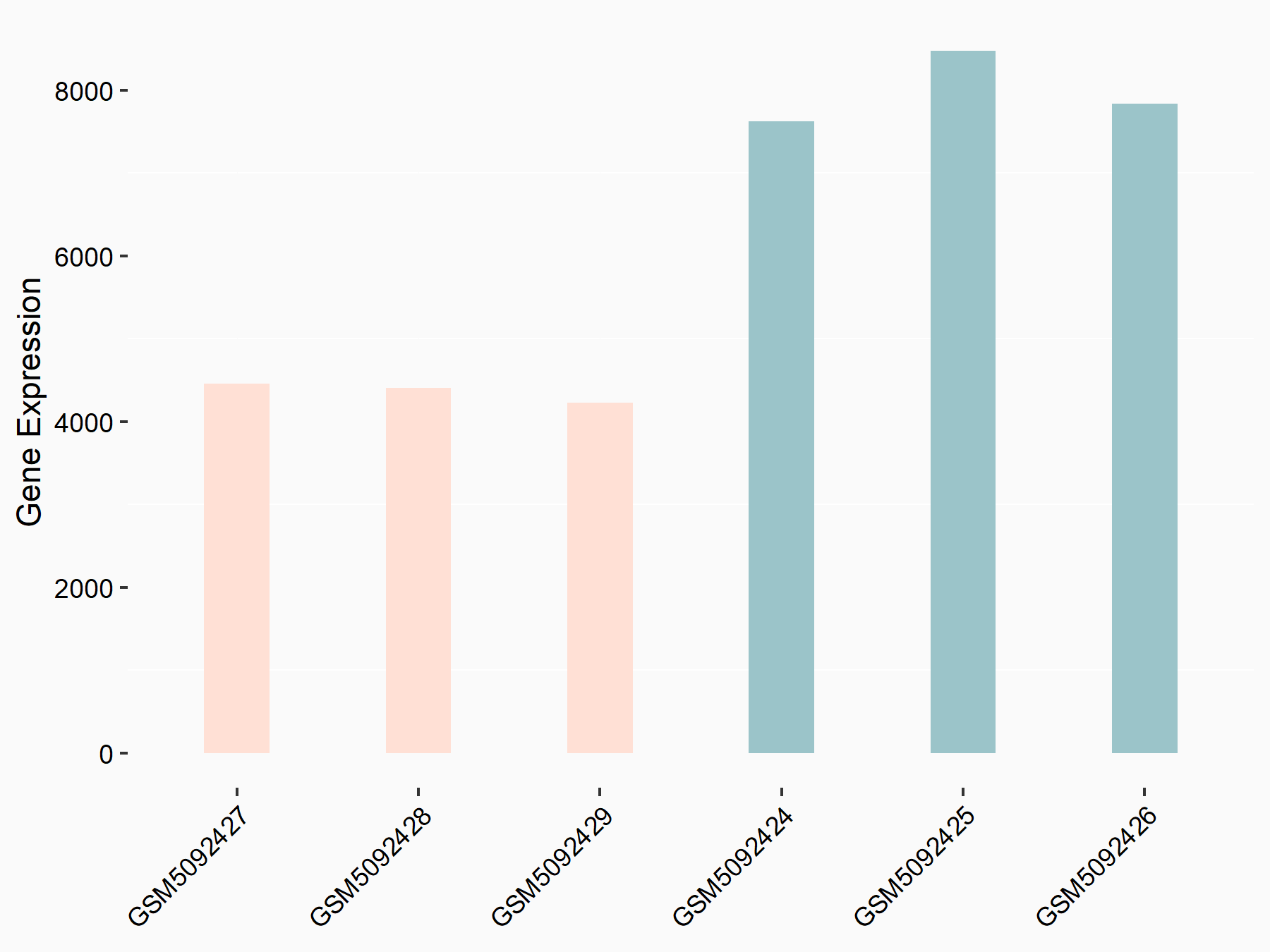 |
logFC: -8.70E-01 p-value: 2.47E-77 |
| More Results | Click to View More RNA-seq Results | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [176] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| Cell Process | mRNA stability | |||
| Epithelial-mesenchymal transition | ||||
In-vitro Model |
DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| FHC | Normal | Homo sapiens | CVCL_3688 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | Groups of HCT116-Luc-shCtrl, HCT116-Luc-shLINC00460, and HCT116-Luc-shLINC00460 + HMGA1 cells (5 × 106) were injected subcutaneously into the flanks of mice correspondingly. | |||
| Response Summary | LINC00460 is a novel oncogene of colorectal cancer through interacting with IGF2BP2 and DHX9 and bind to the m6A modified High mobility group protein HMG-I/HMG-Y (HMGA1) mRNA to enhance the HMGA1 mRNA stability. The N6-methyladenosine (m6A) modification of HMGA1 mRNA by METTL3 enhanced HMGA1 expression in CRC. | |||
Histone deacetylase 5 (HDAC5)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
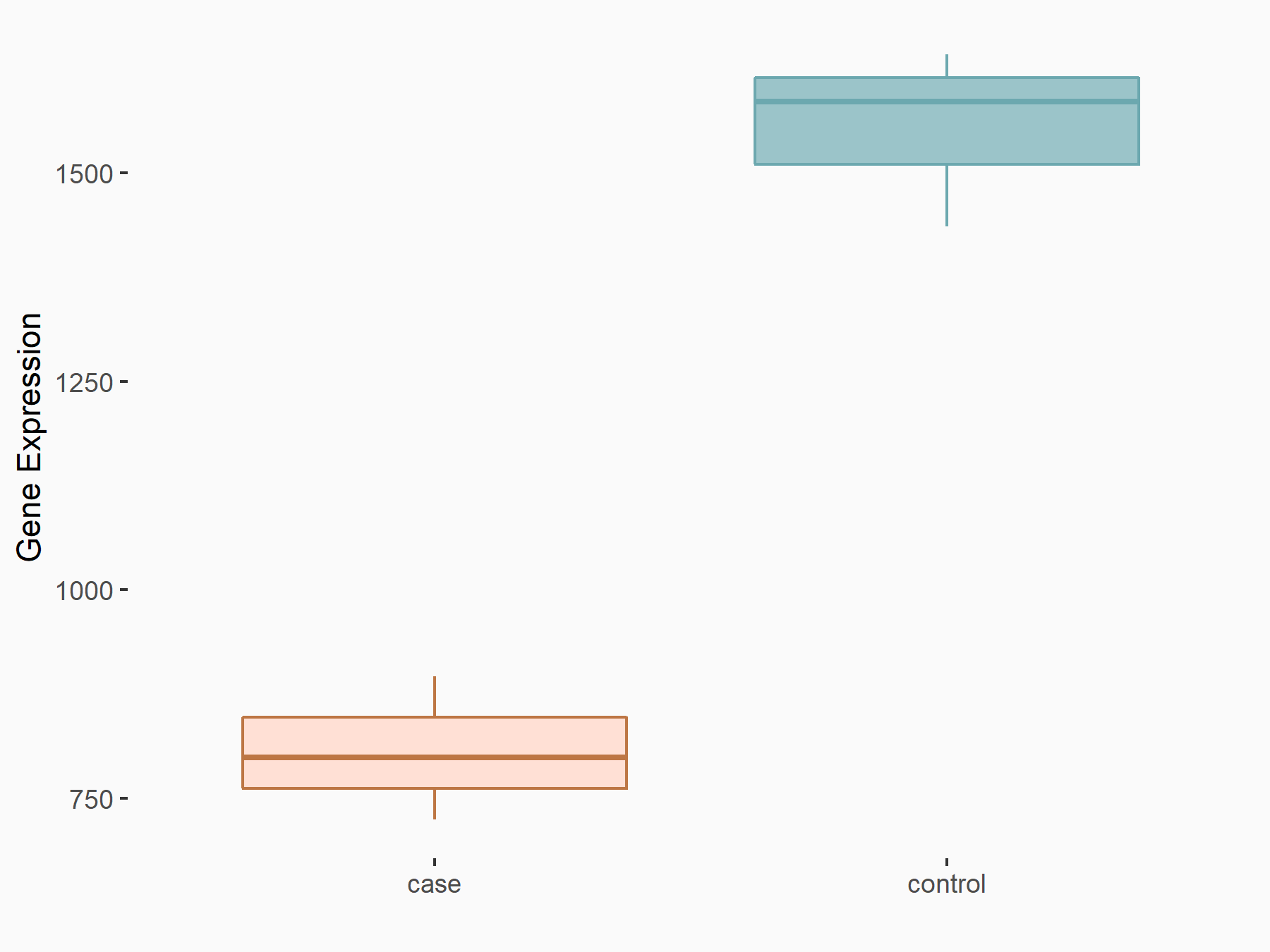 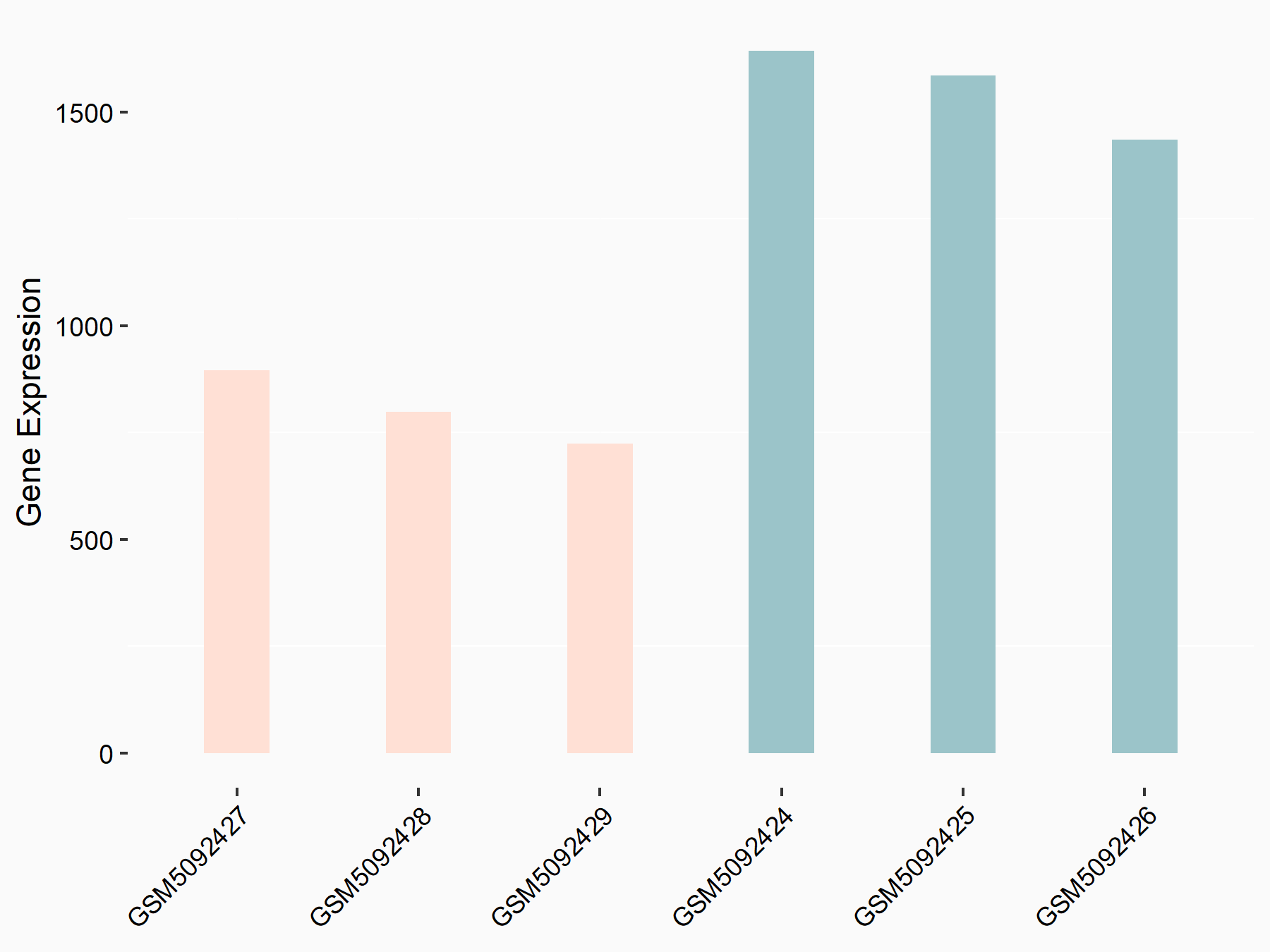 |
logFC: -9.48E-01 p-value: 2.87E-28 |
| More Results | Click to View More RNA-seq Results | |
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [177] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 |
| SaOS-2 | Osteosarcoma | Homo sapiens | CVCL_0548 | |
| HOS | Osteosarcoma | Homo sapiens | CVCL_0312 | |
| hFOB 1.19 | Normal | Homo sapiens | CVCL_3708 | |
| In-vivo Model | U2OS cells with stable METTL3 expression were screened using puromycin for subsequent experiments. U2OS cells (1 × 106) were injected subcutaneously to the right side of each mouse (the mice were numbered according to their weight, and the experimenters randomly divided the nude mice into 12 groups by the random number method, with 12 mice in each group). The status of mice was detected every 2 days. Tumor growth and volume were measured once a week.Tumor volume was measured: Volume = Width2 × Length/2. On the 35th day after the operation, nude mice were euthanized by intraperitoneal injection of ≥100 mg/kg pentobarbital sodium. Tumor resection was performed and tumor weight was recorded. Weight loss >10% of the body weight or the maximum diameter of the tumor >1.5 cm was the humane end point. | |||
| Response Summary | Higher METTL3 expression indicated poorer prognosis. METTL3 upregulated Histone deacetylase 5 (HDAC5) expression in osteosarcoma cells by increasing the m6A level. | |||
Histone-lysine N-methyltransferase SETD7 (SETD7)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
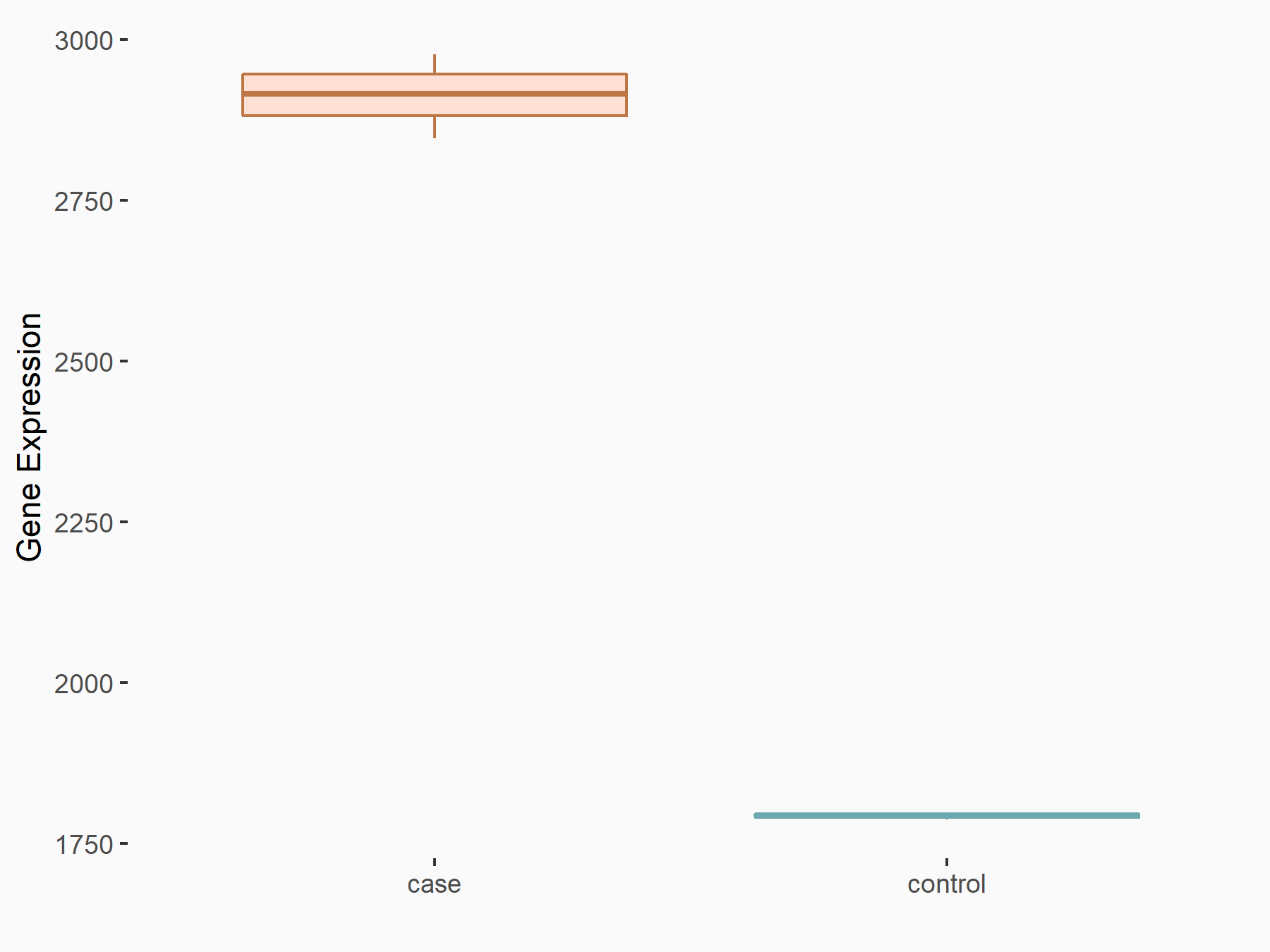 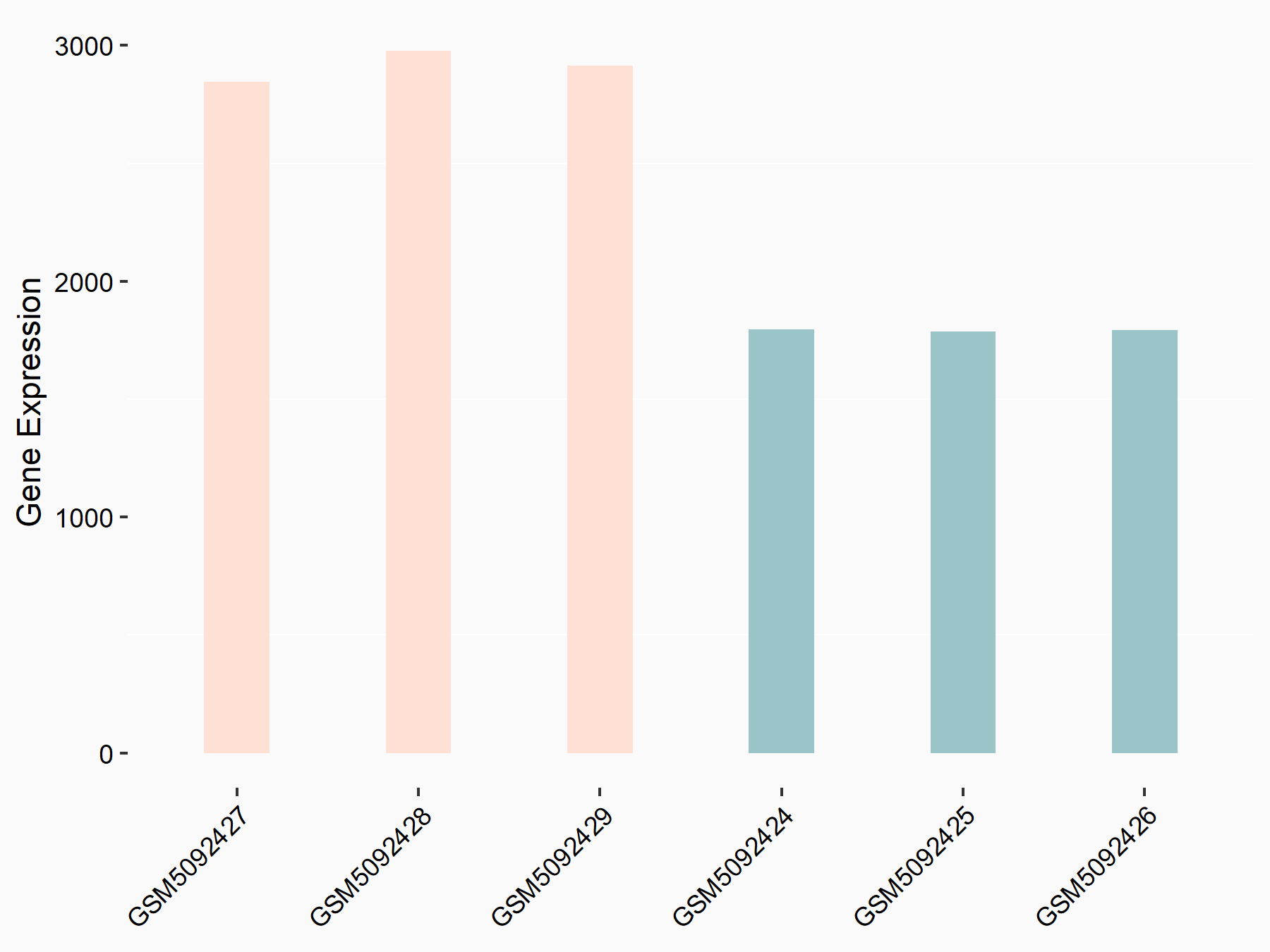 |
logFC: 7.00E-01 p-value: 6.39E-40 |
| More Results | Click to View More RNA-seq Results | |
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [178] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cancer proliferation | |||
| Cancer metastasis | ||||
In-vitro Model |
SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| In-vivo Model | For the subcutaneous implantation model, UM-UC-3 cells (2 × 106 cells per mouse) stably METTL3 knocked down (shMETTL3-1, shMETTL3-2) were injected into the flanks of mice. | |||
| Response Summary | METTL3/YTHDF2/Histone-lysine N-methyltransferase SETD7 (SETD7)/KLF4 m6 A axis provide the insight into the underlying mechanism of carcinogenesis and highlight potential therapeutic targets for bladder cancer. | |||
Homeobox protein NANOG (NANOG)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Soma | Mus musculus |
|
Treatment: Mettl3-/- soma
Control: Wild type soma
|
GSE171199 | |
| Regulation |
  |
logFC: 2.74E+00 p-value: 4.57E-02 |
| More Results | Click to View More RNA-seq Results | |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [179] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell migration | |||
| Cell invasion | ||||
| Epithelial-mesenchymal transition initiation | ||||
| Response Summary | In breast cancer, ZNF217 could upregulate Homeobox protein NANOG (NANOG) by reducing N6-methyladenosine levels via methyltransferase-like 13 (METTL3). | |||
Hypoxia-inducible factor 1-alpha (HIF-1-Alpha/HIF1A)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 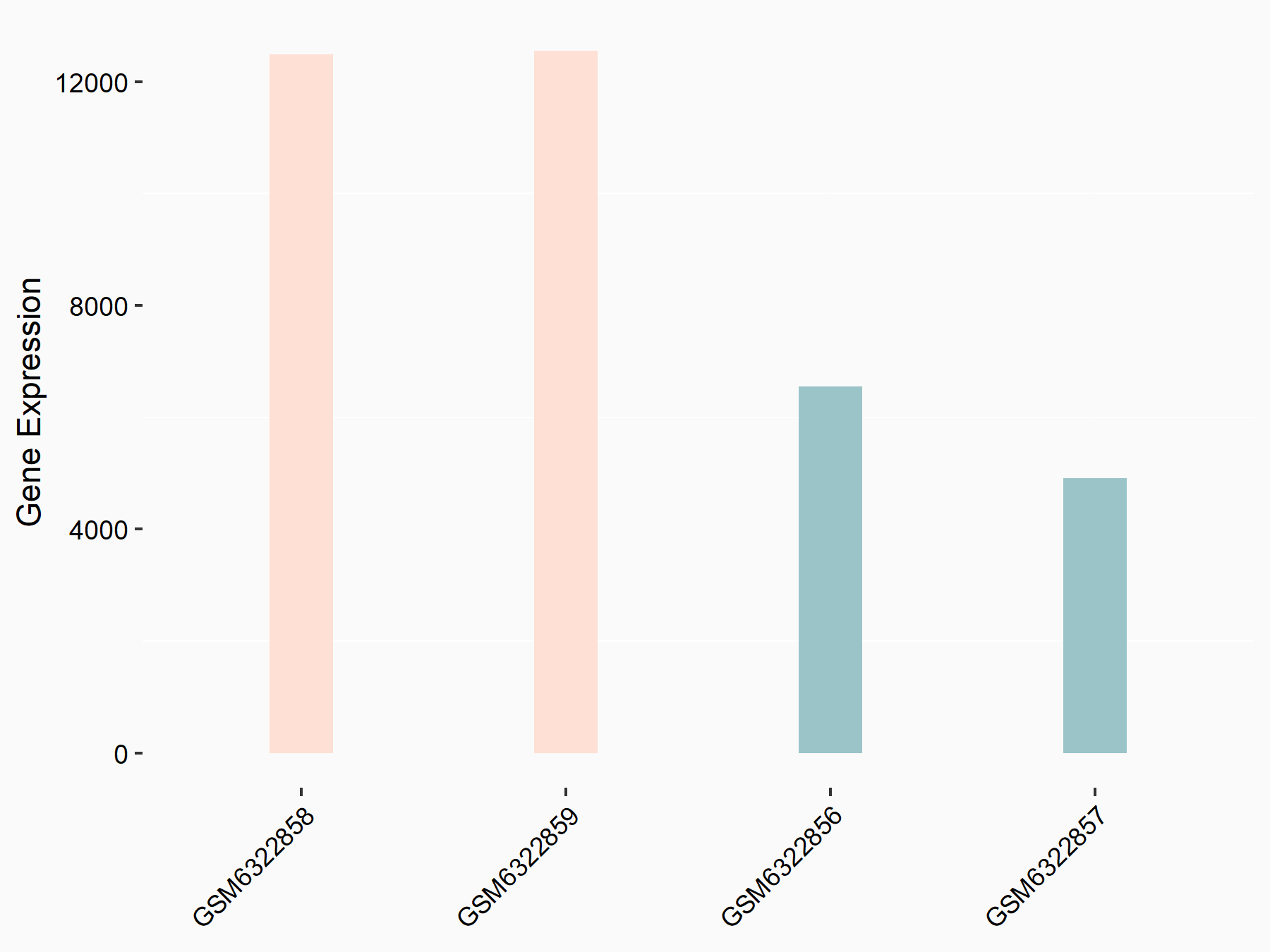 |
logFC: 1.13E+00 p-value: 4.17E-08 |
| More Results | Click to View More RNA-seq Results | |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [180] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | HIF-1 signaling pathway | hsa04066 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| Cell Process | Glycolysis | |||
| Glutaminolysis | ||||
In-vitro Model |
BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| MHCC97 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4971 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| Response Summary | HBXIP drives metabolic reprogramming in hepatocellular carcinoma cells via METTL3-mediated m6A modification of Hypoxia-inducible factor 1-alpha (HIF-1-Alpha/HIF1A). Highly expressed HBXIP and METTL3 are associated with dismal oncologic outcomes of patients with hepatocellular carcinoma. The positive relation between HBXIP and METTL3 can activate reprogramming of HCC cell metabolism by inducing m6A modification of HIF-1-alpha, which is unraveled to enhance aggressive biological behaviors of HCC cells. | |||
Injury of other or unspecified intrathoracic organs [ICD-11: NB32]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [300] | |||
| Responsed Disease | Injury of other or unspecified intrathoracic organs [ICD-11: NB32.3] | |||
| Target Regulation | Up regulation | |||
Inhibitor of nuclear factor kappa-B kinase subunit beta (IKBKB)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
  |
logFC: 3.01E+00 p-value: 5.15E-06 |
| More Results | Click to View More RNA-seq Results | |
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Glucose metabolism | |||
| Response Summary | AF4/FMR2 family member 4 (AFF4), two key regulators of NF-Kappa-B pathway (Inhibitor of nuclear factor kappa-B kinase subunit beta (IKBKB) and RELA) and MYC were further identified as direct targets of METTL3-mediated m6A modification.overexpression of METTL3 significantly promoted Bladder cancer cell growth and invasion. | |||
Insulin-like growth factor 1 receptor (IGF1R)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 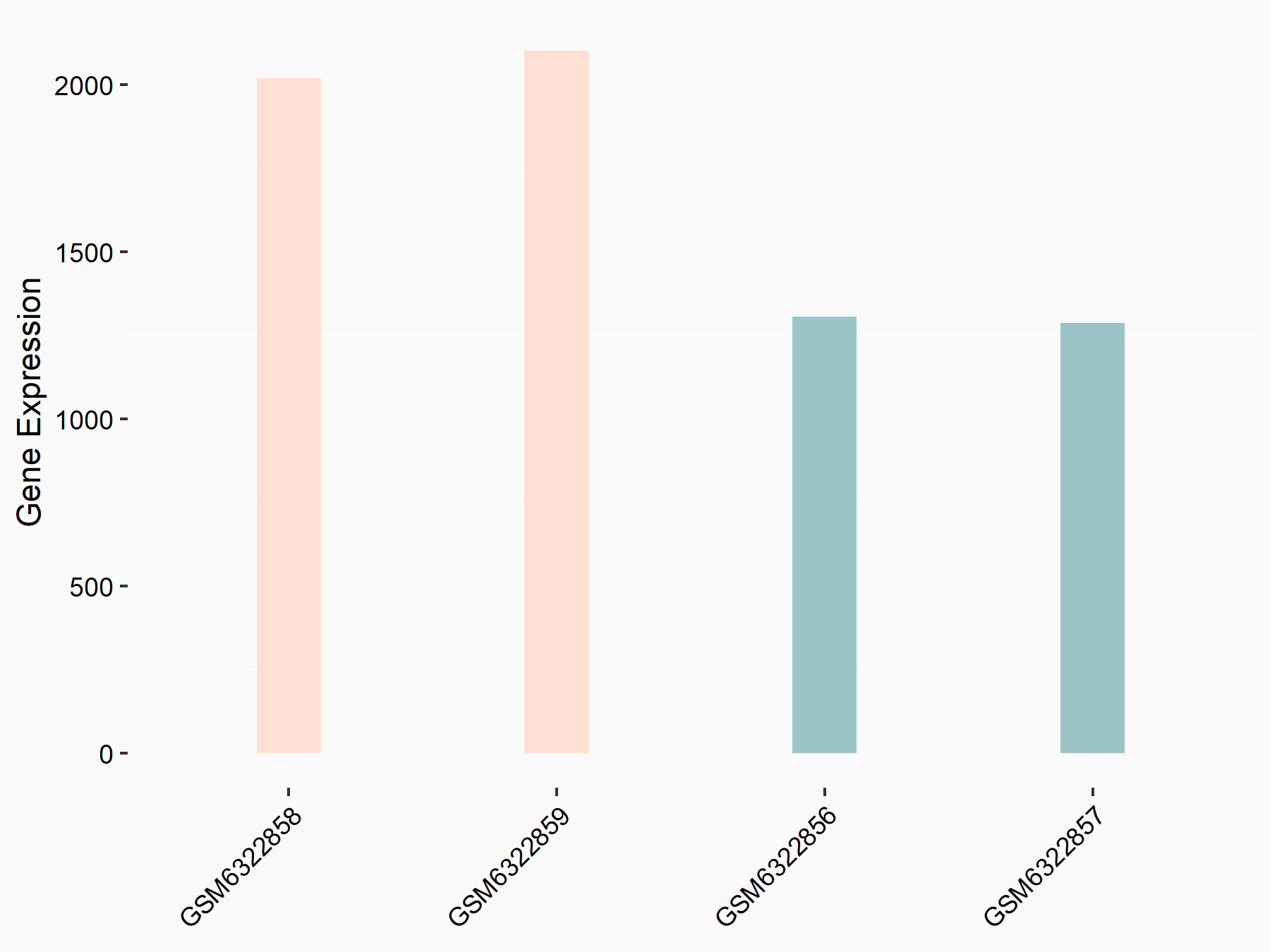 |
logFC: 6.69E-01 p-value: 4.12E-12 |
| More Results | Click to View More RNA-seq Results | |
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [147] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
| Cell Process | RNA stability | |||
In-vitro Model |
PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 |
| LNCaP C4-2B | Prostate carcinoma | Homo sapiens | CVCL_4784 | |
| In-vivo Model | At 1 week post-injection with PC-3 cells, mice were randomly assigned to three groups (n = 8 per group): the ASO-NC group (injection with ASO negative control targeting unknown sequence, 5 nmol in 100 uL PBS for each mouse), the ASO-L group (injection with low-dose ASO targeting PCAT6, 5 nmol in 100 uL PBS for each mouse), and the ASO-H group (injection with high-dose ASO targeting PCAT6, 10 nmol in 100 uL PBS for each mouse). | |||
| Response Summary | METTL3-mediated m6A modification contributed to PCAT6 upregulation in an IGF2BP2-dependent manner. Furthermore, PCAT6 upregulated Insulin-like growth factor 1 receptor (IGF1R) expression by enhancing IGF1R mRNA stability through the PCAT6/IGF2BP2/IGF1R RNA-protein three-dimensional complex. The m6 A-induced PCAT6/IGF2BP2/IGF1R axis promotes PCa bone metastasis and tumor growth, suggesting that PCAT6 serves as a promising prognostic marker and therapeutic target against bone-metastatic PCa. | |||
Integrin alpha-6 (ITGA6)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shMETTL3 MOLM13 cells
Control: MOLM13 cells
|
GSE98623 | |
| Regulation |
 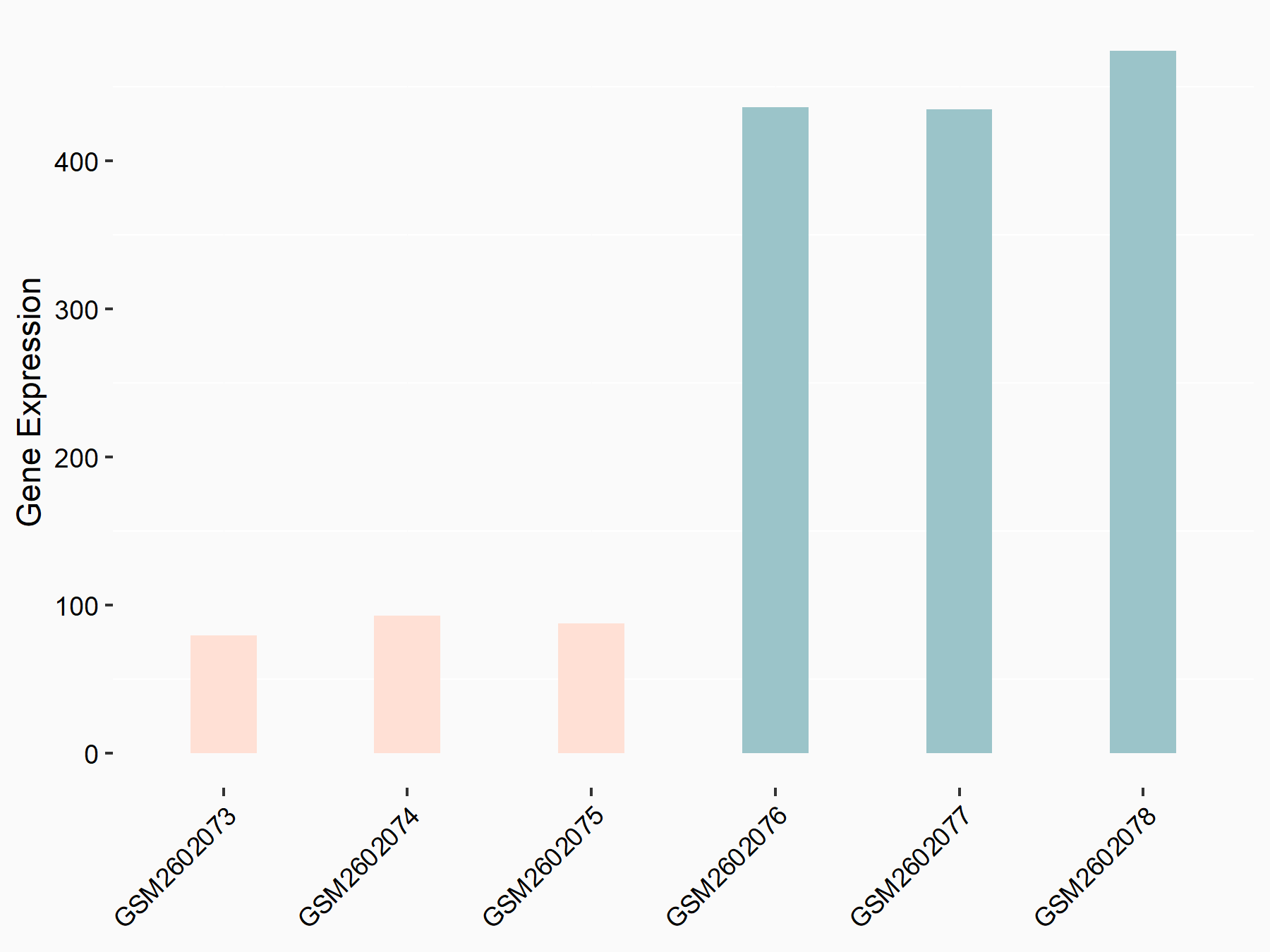 |
logFC: -2.38E+00 p-value: 1.75E-72 |
| More Results | Click to View More RNA-seq Results | |
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [181] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Cell adhesion molecules | hsa04514 | ||
| Cell Process | Cell adhesion | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
5637 | Bladder carcinoma | Homo sapiens | CVCL_0126 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| J82 | Bladder carcinoma | Homo sapiens | CVCL_0359 | |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| In-vivo Model | For the subcutaneous implantation model, 1 × 107 cells were subcutaneously implanted into 5-week-old BALB/cJNju-Foxn1nu/Nju nude mice. | |||
| Response Summary | m6A writer METTL3 and eraser ALKBH5 altered cell adhesion by regulating Integrin alpha-6 (ITGA6) expression in bladder cancer cells. m6A is highly enriched within the ITGA6 transcripts, and increased m6A methylations of the ITGA6 mRNA 3'UTR promotes the translation of ITGA6 mRNA via binding of the m6A readers YTHDF1 and YTHDF3. Inhibition of ITGA6 results in decreased growth and progression of bladder cancer cells in vitro and in vivo. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [301] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | Or tumor growth assessment, 104 Huh7/CSC infected with different lentiviral vectors were mixed with Matrigel at a 1:1 ratio, and the mixture was injected subcutaneously into nude mice (n = 6). For mouse survival assessment, a mixture of 105 infected Huh7/CSC cells with Matrigel was administered to the liver of nude mice. | |||
Integrin beta-1 (ITGB1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 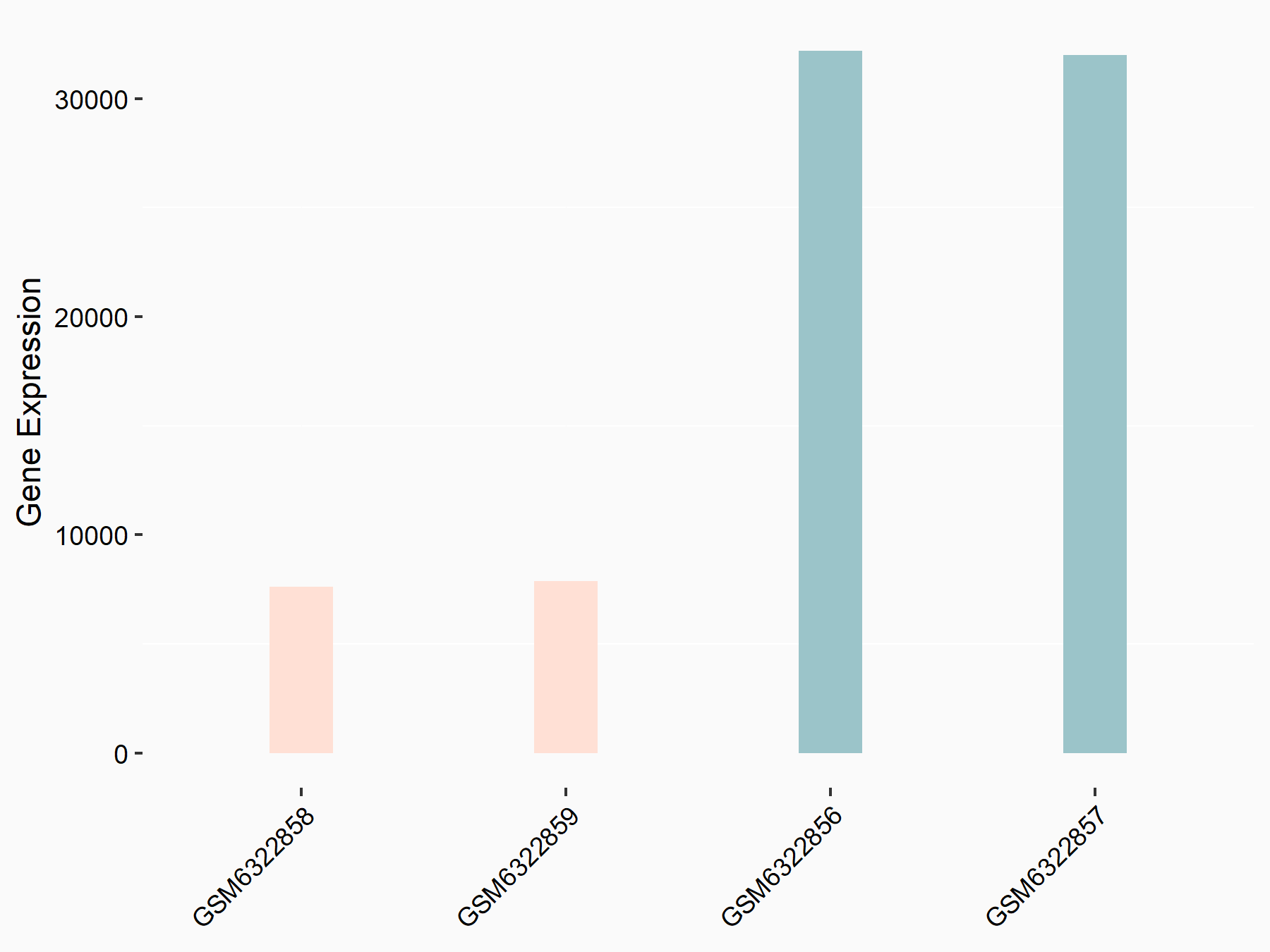 |
logFC: -2.05E+00 p-value: 1.83E-215 |
| More Results | Click to View More RNA-seq Results | |
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [182] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Cell adhesion molecules | hsa04514 | ||
| Cell Process | Cell adhesion | |||
In-vitro Model |
LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| In-vivo Model | All the mice were randomly divided into five groups of four mice each, including PC3-shNC, PC3-shMETTL3-1/2, LNCaP-vector and LNCaP-METTL3 group. | |||
| Response Summary | METTL3 regulates the expression of Integrin beta-1 (ITGB1) through m6A-HuR-dependent mechanism, which affects the binding of ITGB1 to Collagen I and tumor cell motility, so as to promote the bone metastasis of prostate cancer. | |||
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [303] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
In-vitro Model |
HPDE | Normal | Homo sapiens | CVCL_4376 |
| Capan-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0237 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| HPC-Y5 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_6918 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| In-vivo Model | Total of 27 BALB/c nude mice were chosen and divided into three groups (n = 6): Control group (injected with PANC-1 cells), si-NC group, and si-NNT-AS1 group. For the si-NC and si-NNT-AS1 groups, PANC-1 cells transfected with si-NC and si-NNT-AS1, respectively, were injected into mice. The back of each mouse was injected with above cell suspension (2 × 104 cells in 0.2 mL). The average volume of the tumor was measured three times in each 7 days. The mice were euthanized on the 28th day, and tumor tissues were collected for related analysis. Immunohistochemistry (IHC) and terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) staining were performed. The remaining nude mice were divided into three groups (n = 3): Control, si-NC, and si-NNT-AS1 groups. Cell suspension containing 2 × 104 cells was then injected into the back of each mouse. After 7 days, mice were administered a 50-μL suspension containing 5 × 106 CD8 T cells via peritumoral injection every 3 days for a total of five injections. After the last injection, mice were euthanized on the 28th day, and sera were collected. All animal studies were approved by the Ethics Committee of Xiangya Hospital, Central South University. | |||
Intercellular adhesion molecule 1 (ICAM1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
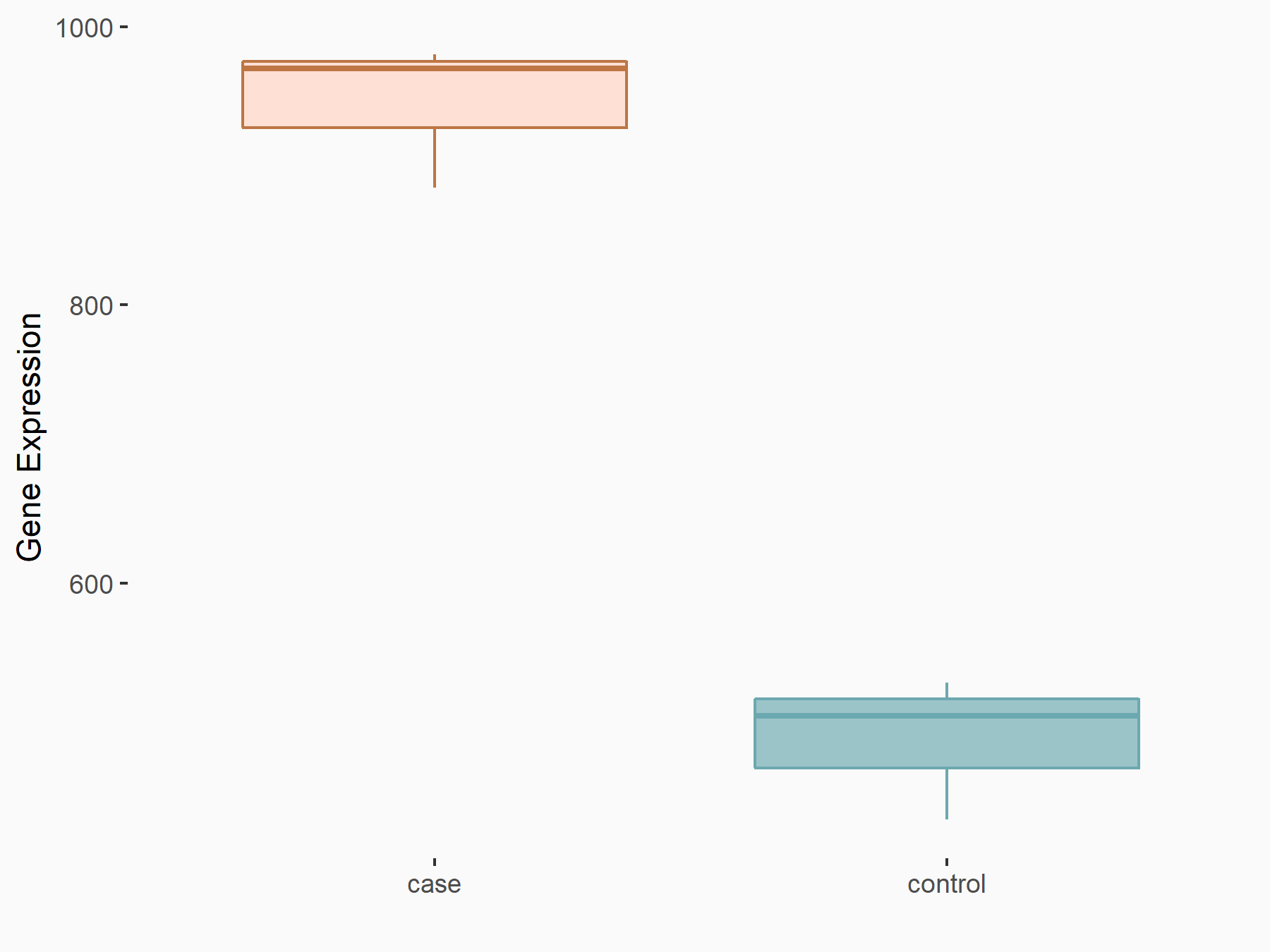 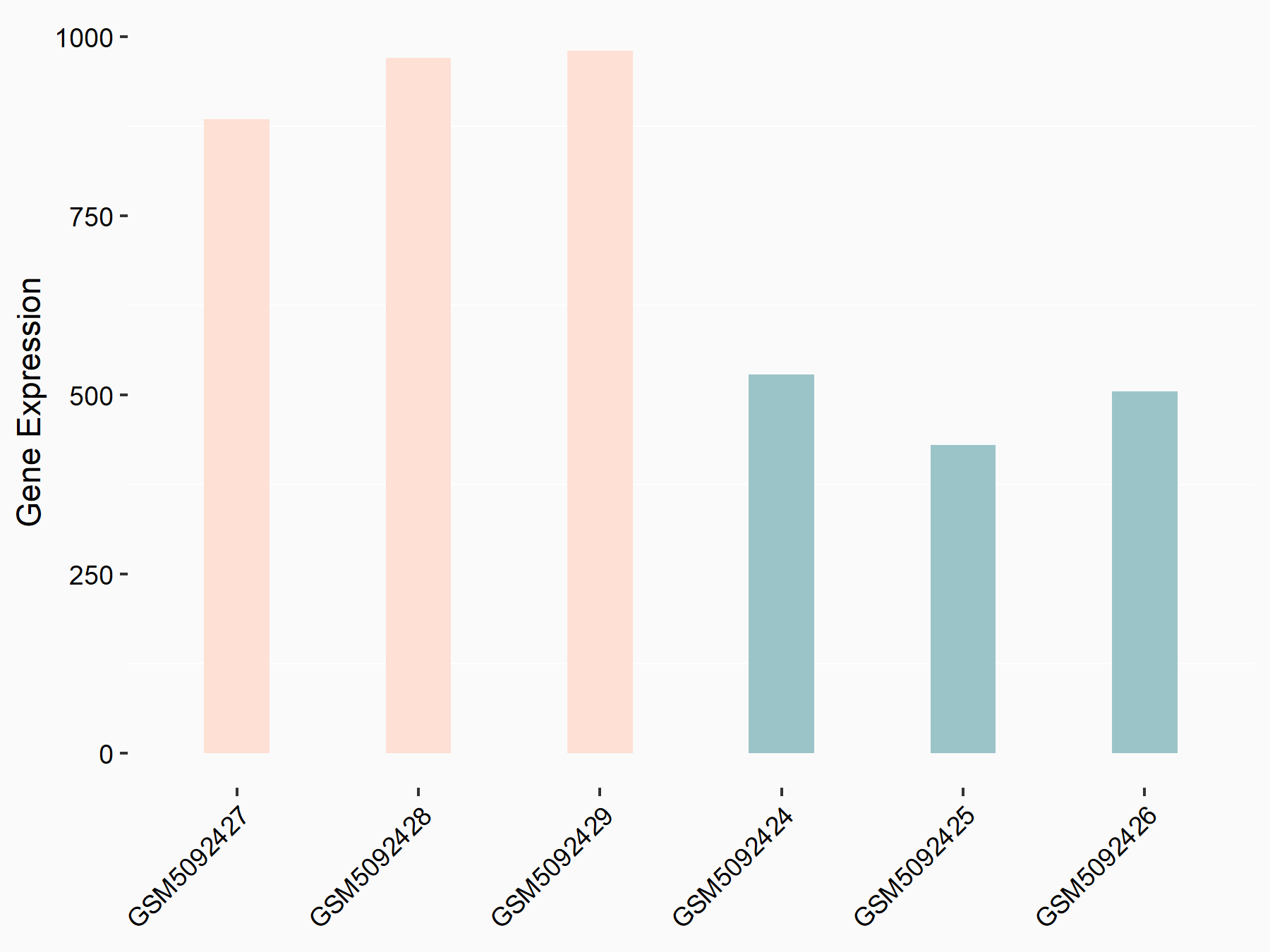 |
logFC: 9.55E-01 p-value: 2.64E-22 |
| More Results | Click to View More RNA-seq Results | |
Cataract [ICD-11: 9B10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [183] | |||
| Responsed Disease | Diabetic cataract [ICD-11: 9B10.21] | |||
| Target Regulation | Up regulation | |||
| Cell Process | RNA stability | |||
| Cell apoptosis | ||||
In-vitro Model |
B-3 | Normal | Homo sapiens | CVCL_6367 |
| Response Summary | METTL3 modulates the proliferation and apoptosis of lens epithelial cells in diabetic cataract. METTL3 targets the 3' UTR of Intercellular adhesion molecule 1 (ICAM1) to stabilize mRNA stability. | |||
Interferon gamma (IFN-gamma)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Th1 cell line | Mus musculus |
|
Treatment: METTL3 knockout splenic Th1 cells
Control: Wild type splenic Th1 cells
|
GSE129648 | |
| Regulation |
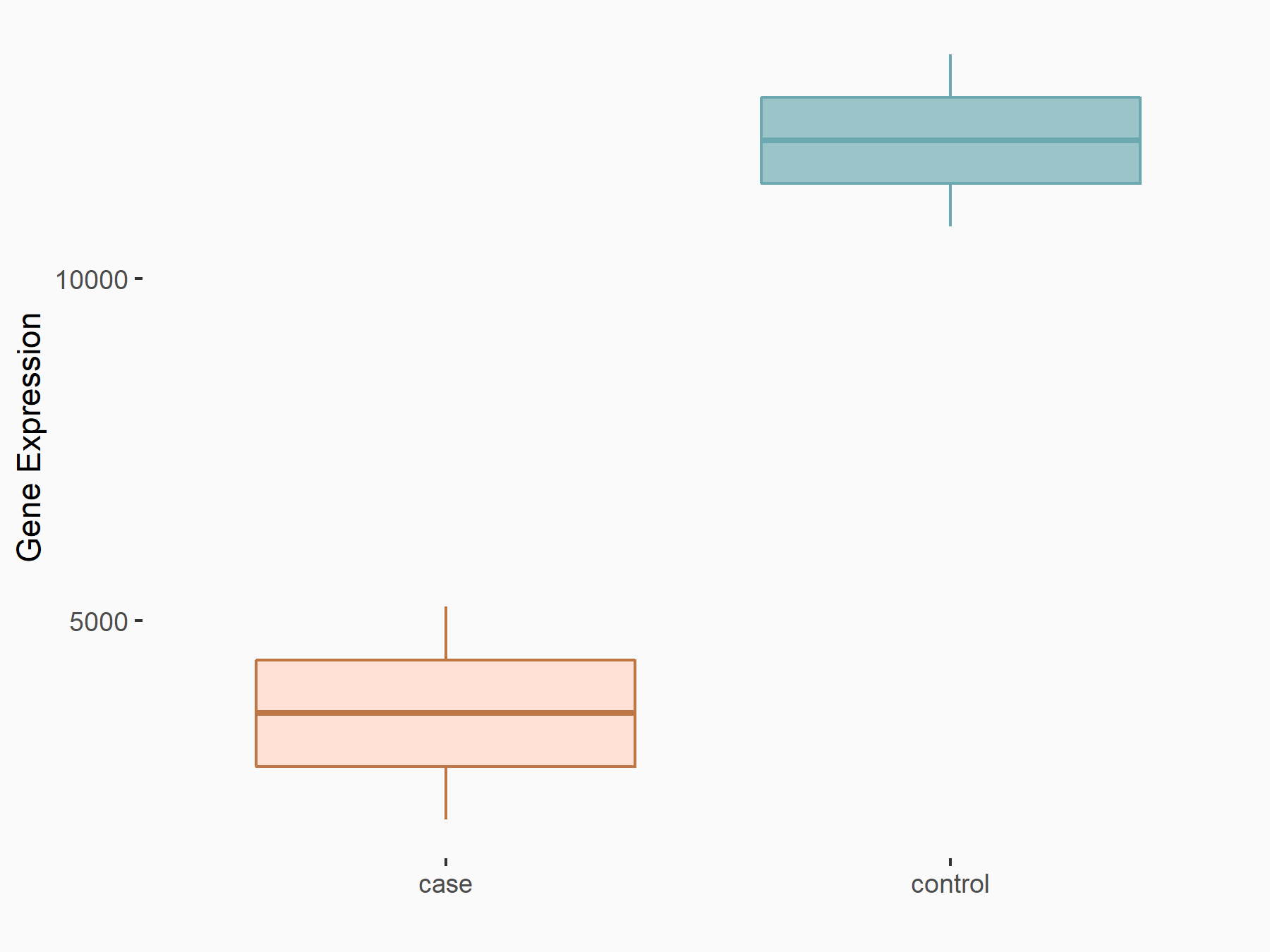 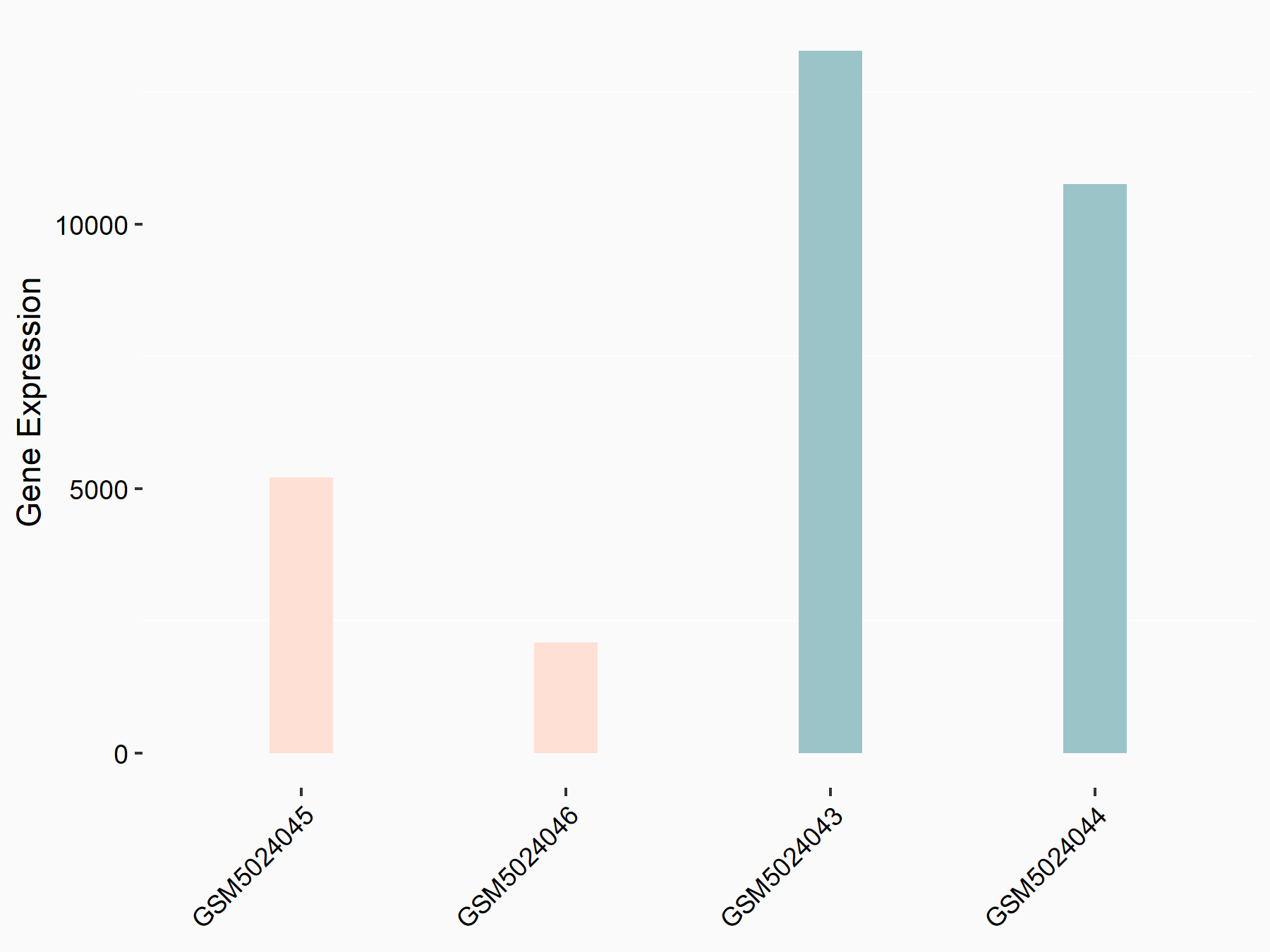 |
logFC: -1.72E+00 p-value: 6.66E-03 |
| More Results | Click to View More RNA-seq Results | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [118] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
| Cell Process | Immunity | |||
In-vitro Model |
CT26 | Mouse colon adenocarcinoma | Mus musculus | CVCL_7254 |
| B16-GM-CSF (B16-GM-CSF cell line was a kind gift from Drs. Glenn Dranoff and Michael Dougan (Dana-Farber/Harvard Cancer Center)) | ||||
| B16-F10 | Mouse melanoma | Mus musculus | CVCL_0159 | |
| In-vivo Model | 2 × 106 CT26 cells with knockout of Mettl3, Mettl14, Mettl3/Stat1, Mettl3/Irf1, Mettl14/Stat1, or Mettl14/Irf1 and control were suspended in 200 uL of PBS/Matrigel (Corning) (1:1) and then subcutaneously inoculated into flank of each mouse. | |||
| Response Summary | In colorectal cancer, Mettl3- or Mettl14-deficient tumors increased cytotoxic tumor-infiltrating CD8+ T cells and elevated secretion of Interferon gamma (IFN-gamma), Cxcl9, and Cxcl10 in tumor microenvironment in vivo. Mechanistically, Mettl3 or Mettl14 loss promoted IFN-gamma-Stat1-Irf1 signaling through stabilizing the Stat1 and Irf1 mRNA via Ythdf2. | |||
Interferon regulatory factor 1 (Irf1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
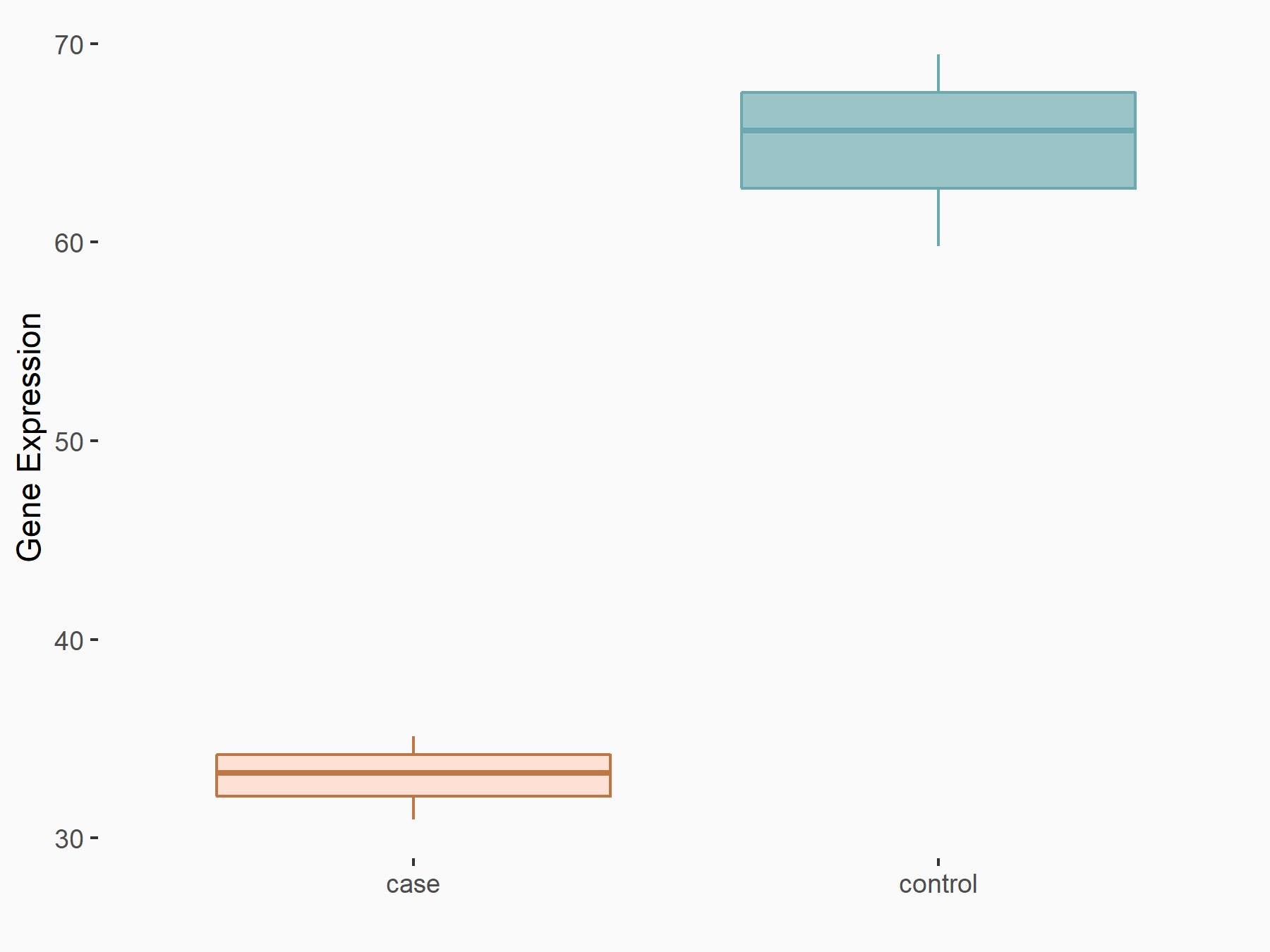 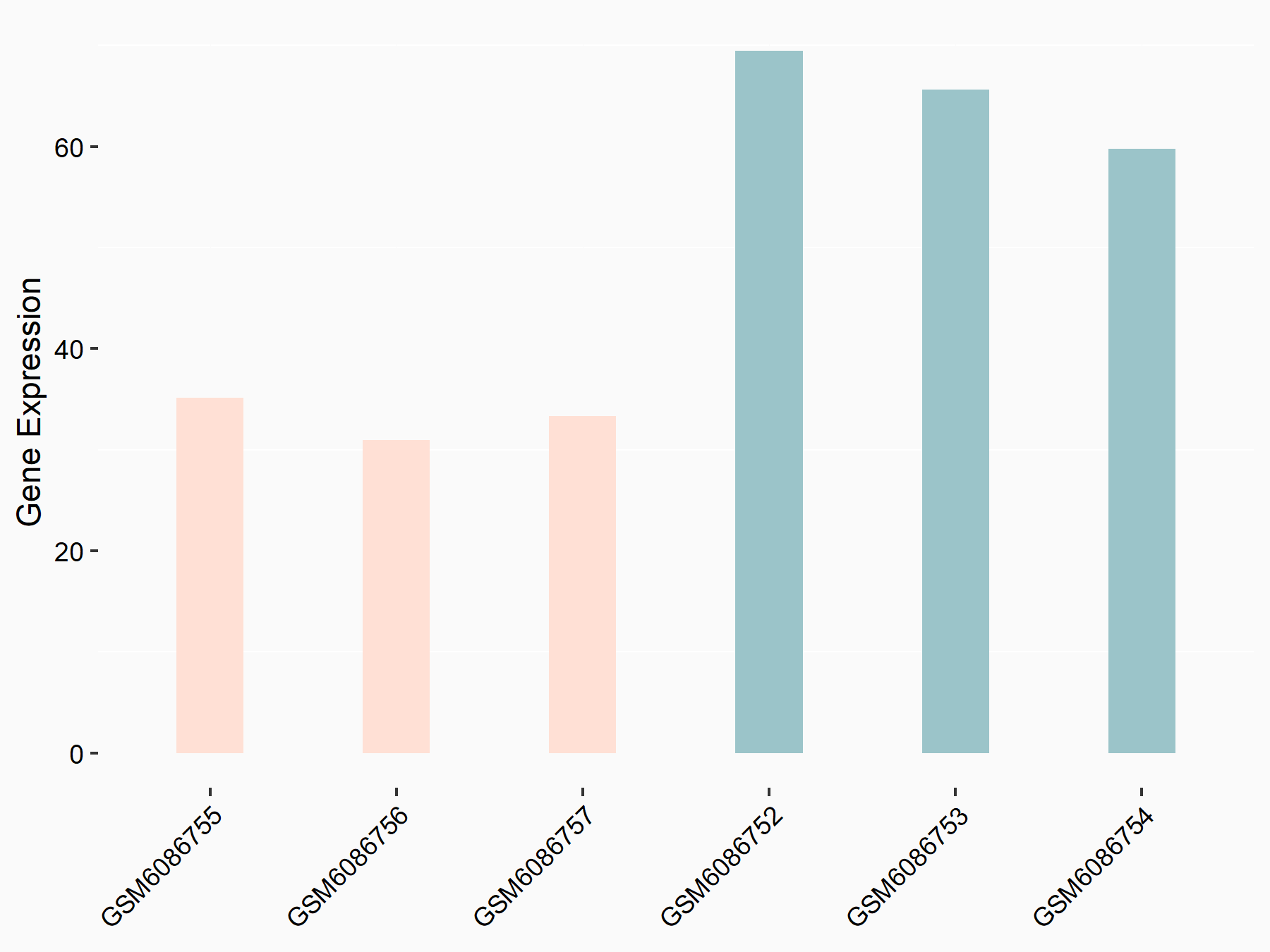 |
logFC: -9.51E-01 p-value: 1.06E-05 |
| More Results | Click to View More RNA-seq Results | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [118] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
| Cell Process | Immunity | |||
In-vitro Model |
CT26 | Mouse colon adenocarcinoma | Mus musculus | CVCL_7254 |
| B16-GM-CSF (B16-GM-CSF cell line was a kind gift from Drs. Glenn Dranoff and Michael Dougan (Dana-Farber/Harvard Cancer Center)) | ||||
| B16-F10 | Mouse melanoma | Mus musculus | CVCL_0159 | |
| In-vivo Model | 2 × 106 CT26 cells with knockout of Mettl3, Mettl14, Mettl3/Stat1, Mettl3/Irf1, Mettl14/Stat1, or Mettl14/Irf1 and control were suspended in 200 uL of PBS/Matrigel (Corning) (1:1) and then subcutaneously inoculated into flank of each mouse. | |||
| Response Summary | In colorectal cancer, Mettl3- or Mettl14-deficient tumors increased cytotoxic tumor-infiltrating CD8+ T cells and elevated secretion of IFN-gamma, Cxcl9, and Cxcl10 in tumor microenvironment in vivo. Mechanistically, Mettl3 or Mettl14 loss promoted IFN-gamma-Stat1-Irf1 signaling through stabilizing the Stat1 and Interferon regulatory factor 1 (Irf1) mRNA via Ythdf2. | |||
Interferon-induced 54 kDa protein (IFIT2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LNCaP cell line | Homo sapiens |
|
Treatment: shMETTL3 LNCaP cells
Control: shControl LNCaP cells
|
GSE147884 | |
| Regulation |
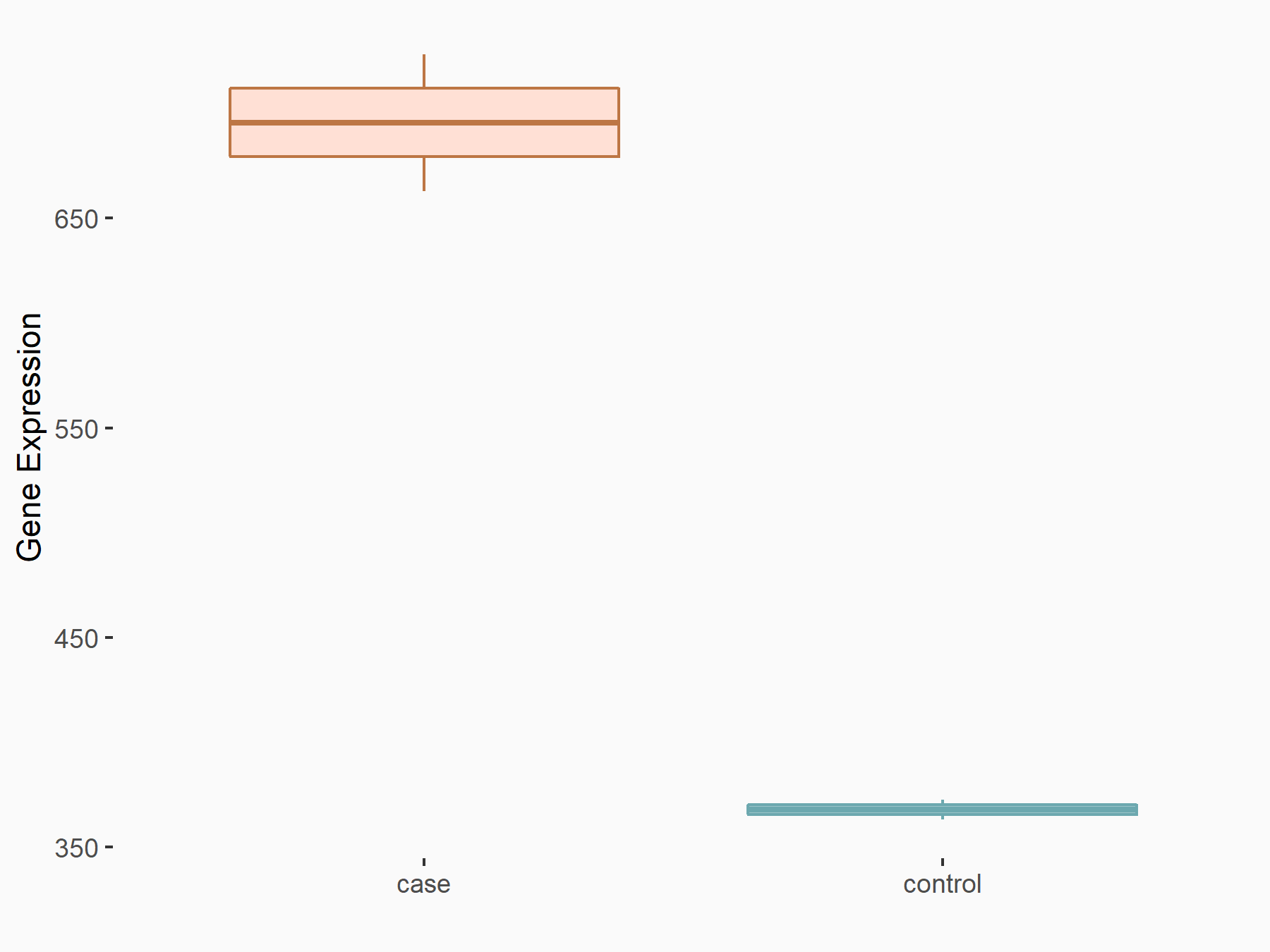 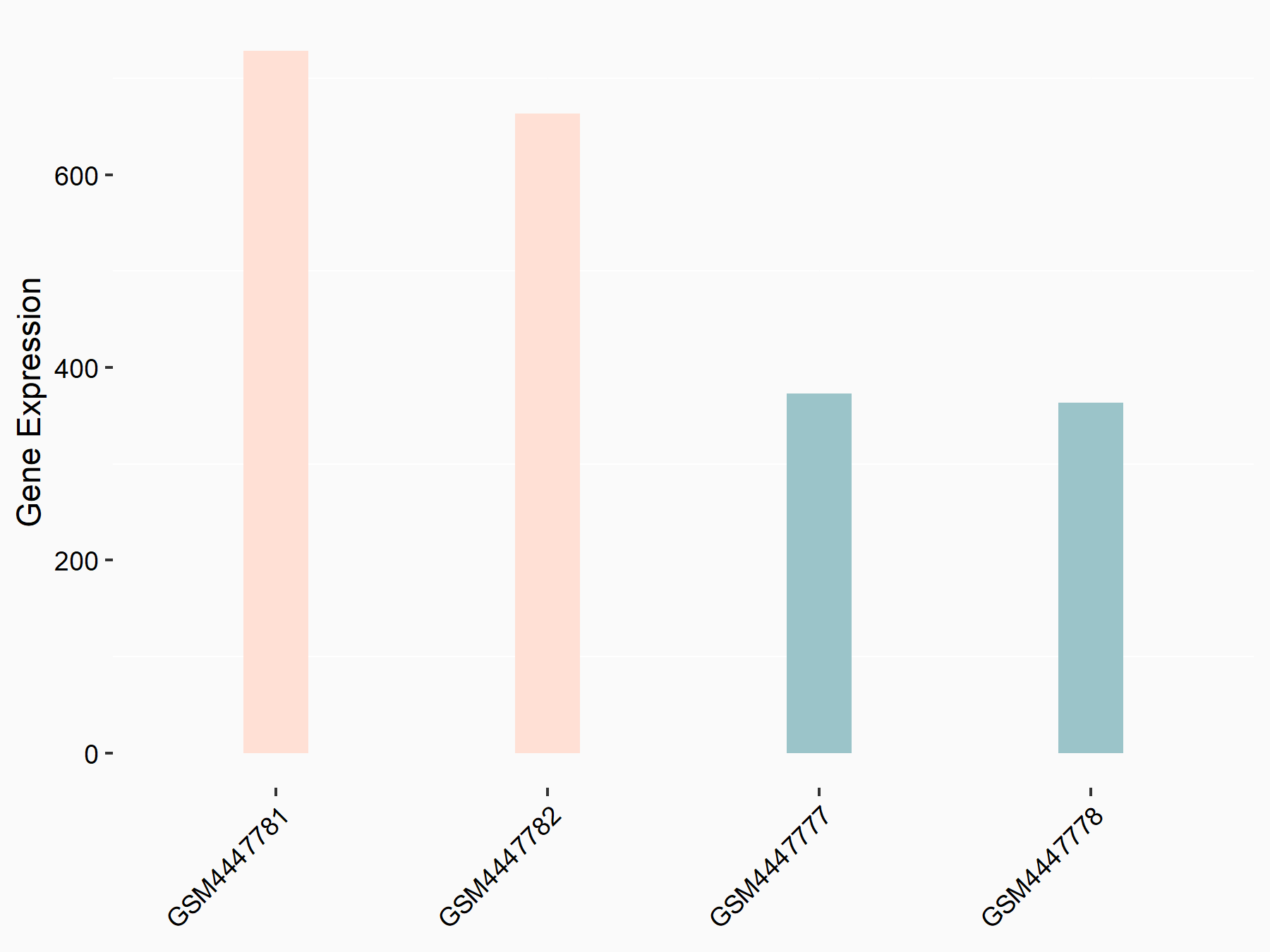 |
logFC: 9.21E-01 p-value: 1.94E-18 |
| More Results | Click to View More RNA-seq Results | |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [184] | |||
| Responsed Disease | Intrahepatic cholangiocarcinoma [ICD-11: 2C12.10] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HuCC-T1 | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_0324 |
| HCCC-9810 | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_6908 | |
| In-vivo Model | 4-week-old female BALB/c nude mice were used for HuCC-T1 tumor xenograft models and 4-week-old female B-NDG mice (Biocytogen, Beijing, China) were used for HCCC-9810 tumor xenograft models. 1 × 107 HuCC-T1 or HCCC-9810 cells were resuspended in 100 ul PBS with Matrigel (1:1), and injected into the right flank of mice (n = 6/group). | |||
| Response Summary | METTL3 was upregulated and predicted poor prognosis of patients with intrahepatic cholangiocarcinoma(ICC). H3K4me3 activation-driven METTL3 transcription promotes ICC progression by YTHDF2-mediated Interferon-induced 54 kDa protein (IFIT2) mRNA degradation. | |||
Interferon-inducible protein 4 (ADAR1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | E14.5 LSK cell line | Mus musculus |
|
Treatment: METTL3 knockout E14.5 LSK cells
Control: Wild type E14.5 LSK cells
|
GSE148882 | |
| Regulation |
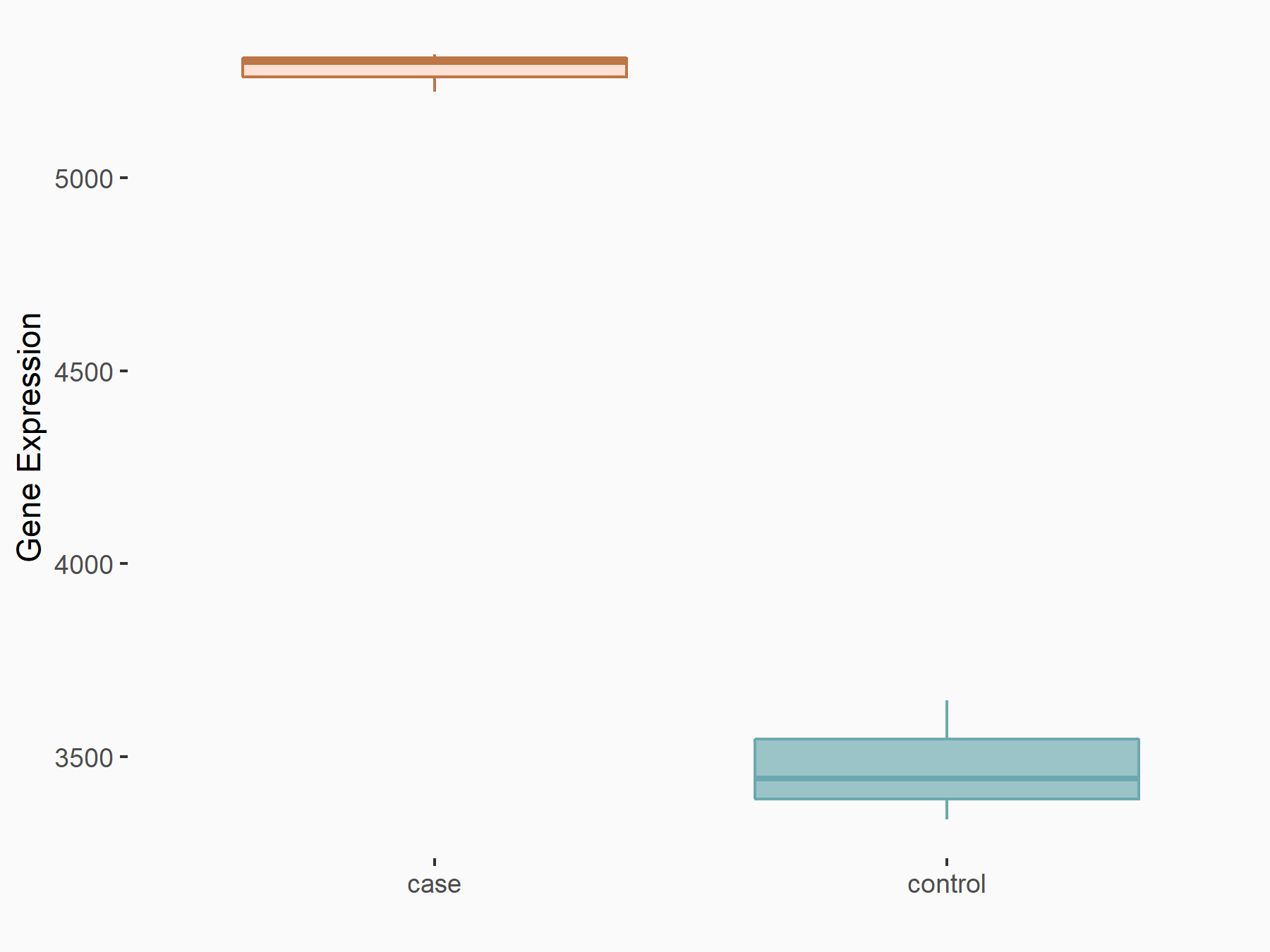 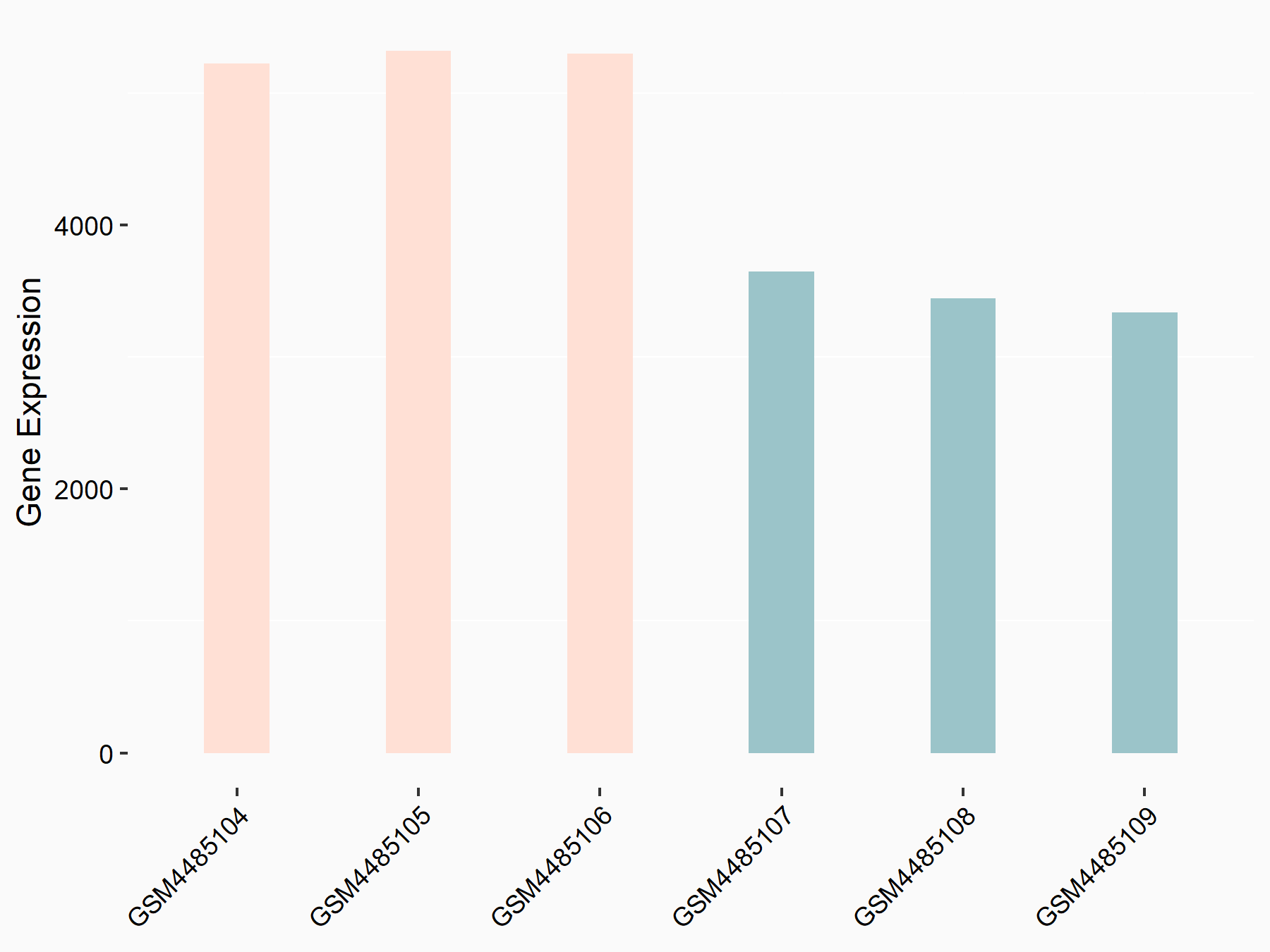 |
logFC: 6.03E-01 p-value: 6.54E-18 |
| More Results | Click to View More RNA-seq Results | |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [185] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| RNA degradation | hsa03018 | |||
| Cell Process | RNA stability | |||
In-vitro Model |
U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 |
| U-118MG | Astrocytoma | Homo sapiens | CVCL_0633 | |
| T98G | Glioblastoma | Homo sapiens | CVCL_0556 | |
| A-172 | Glioblastoma | Homo sapiens | CVCL_0131 | |
| In-vivo Model | 2 × 106 U87MG cells already expressing shscr or shADAR1 were subcutaneously injected in the flank of 6-week-old nude mice (nu/nu, Charles River, Wilmington, MA, USA). | |||
| Response Summary | METTL3, upregulated in glioblastoma, methylates Interferon-inducible protein 4 (ADAR1) mRNA and increases its protein level leading to a pro-tumorigenic mechanism connecting METTL3, YTHDF1, and ADAR1. | |||
Interleukin enhancer-binding factor 3 (ILF3)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
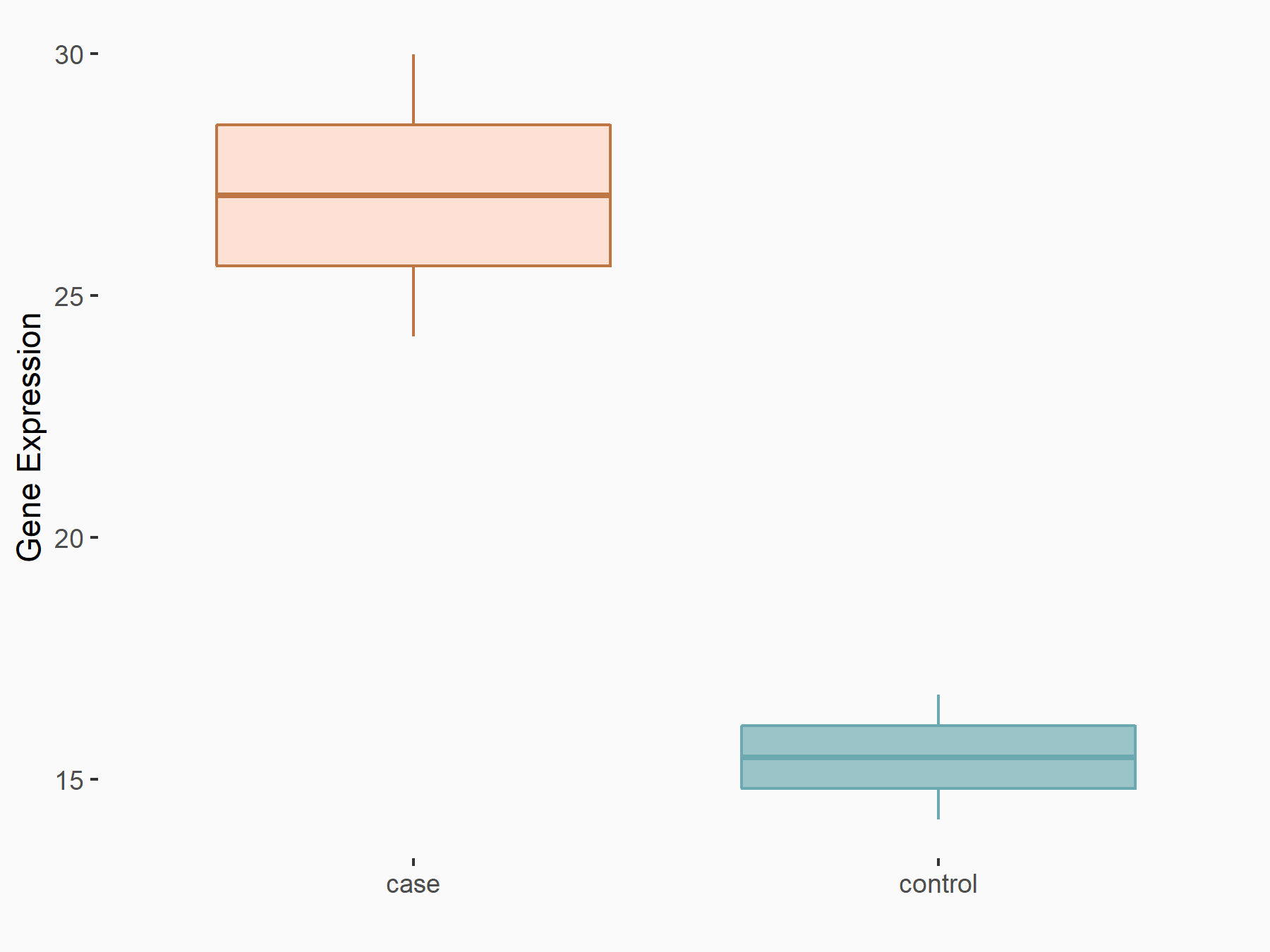 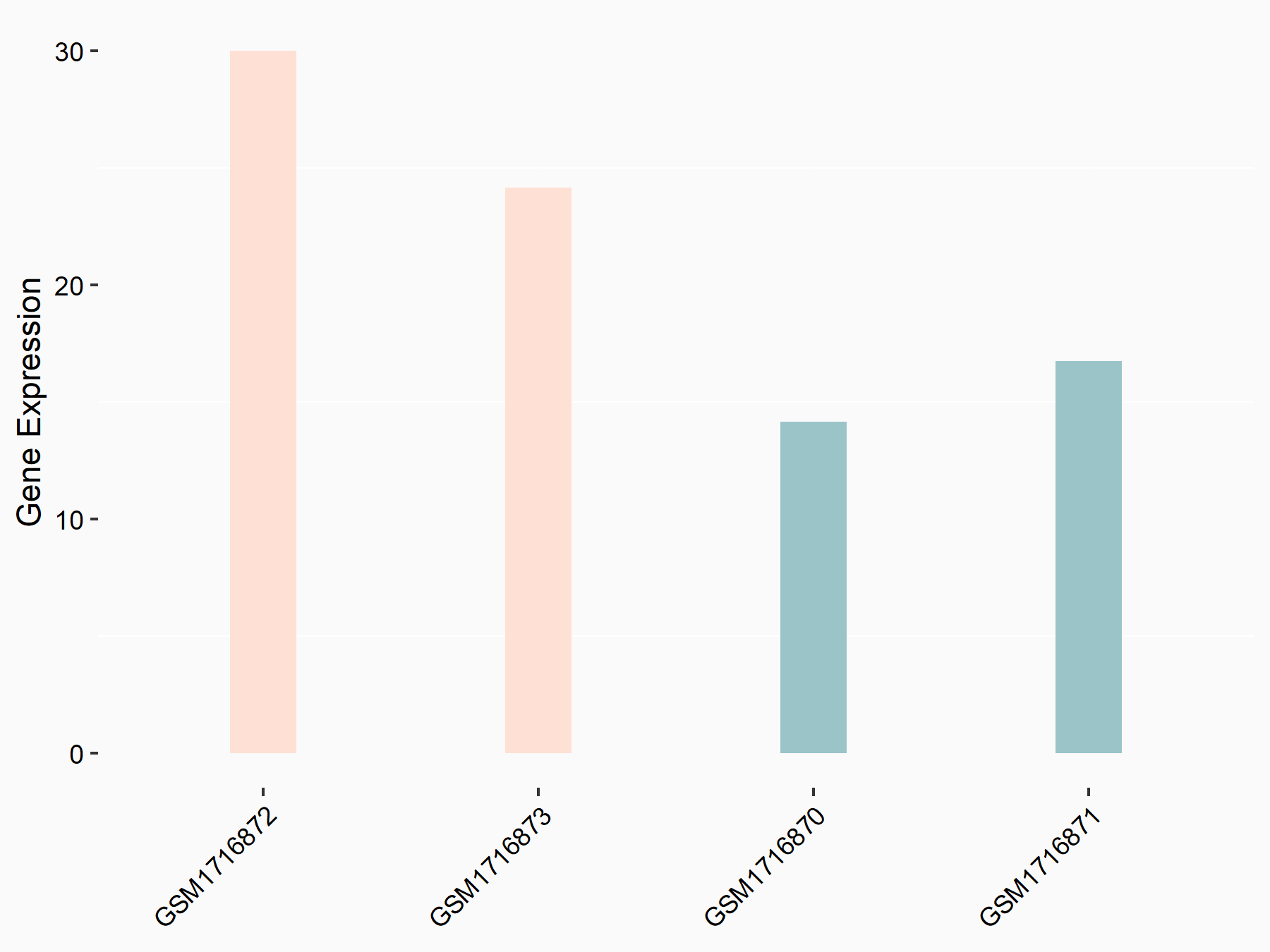 |
logFC: 7.68E-01 p-value: 3.12E-02 |
| More Results | Click to View More RNA-seq Results | |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [186] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | Approximately 5 × 106 control and ILF3-AS1 silencing Huh7 cells were subcutaneously implanted into the right flank of nude mice.Xenograft size was measured every 7 days and calculated using the equation V(mm3)=(length×width2)/2. 35 days later, the mice were sacrificed, and the tumor tissues were isolated and weighed. | |||
| Response Summary | ILF3-AS1 expression was significantly elevated in HCC tissues,mechanistically, ILF3-AS1 associated with Interleukin enhancer-binding factor 3 (ILF3) mRNA and inhibited its degradation. ILF3-AS1 increased ILF3 m6A level via recruiting N6-methyladenosine (m6A) RNA methyltransferase METTL3. Moreover, IFL3-AS1 enhanced the interaction between ILF3 mRNA and m6A reader IGF2BP1. | |||
Interleukin-6 (IL-6)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
 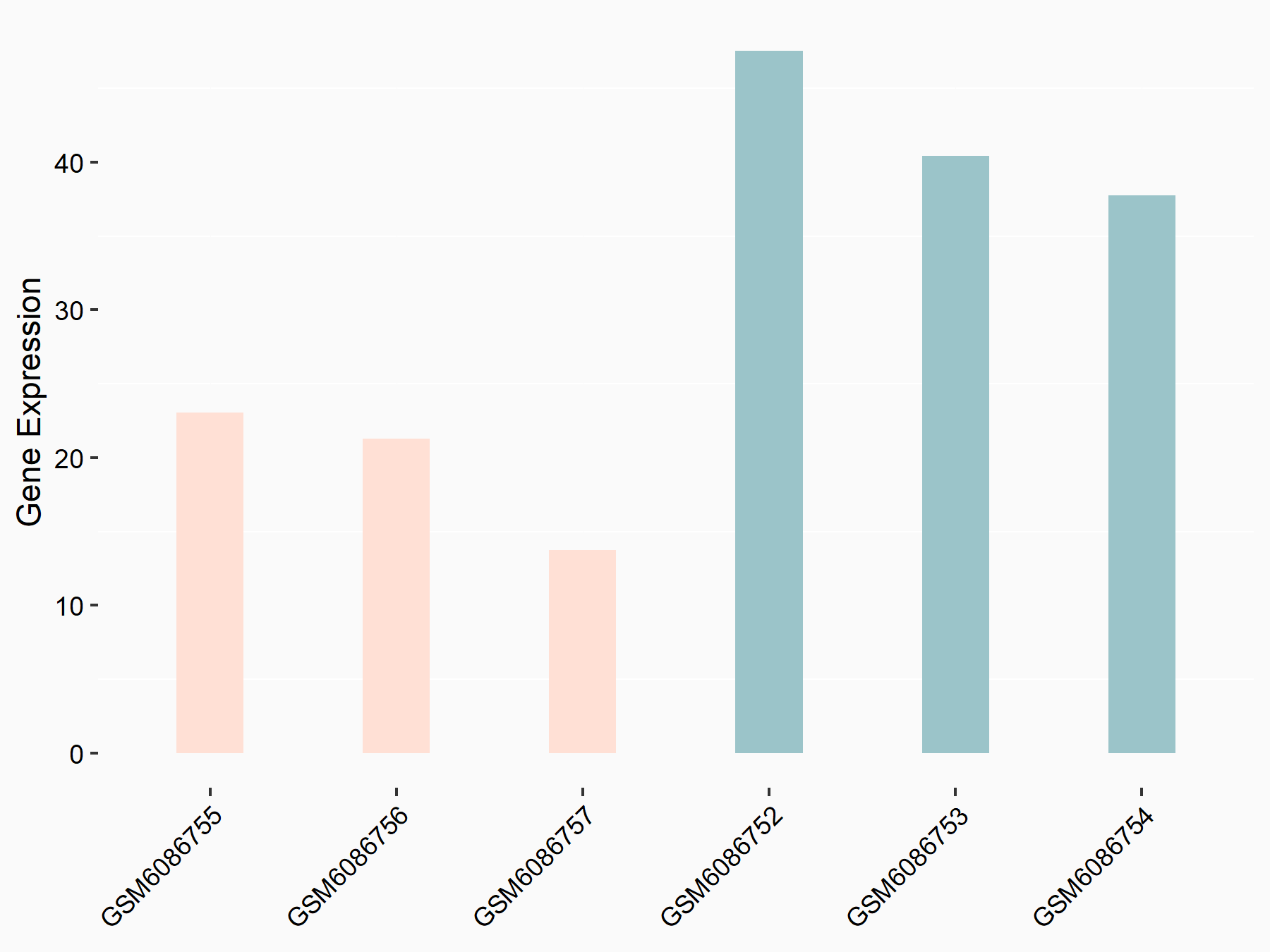 |
logFC: -1.10E+00 p-value: 7.68E-04 |
| More Results | Click to View More RNA-seq Results | |
Rheumatoid arthritis [ICD-11: FA20]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [187] | |||
| Responsed Disease | Rheumatoid arthritis [ICD-11: FA20] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Inflammatory response | |||
In-vitro Model |
FLS (Rat fibroblast synovial cell line) | |||
| In-vivo Model | To establish the adjuvant-induced arthritis (AIA) model, the rats were given complete Freund's adjuvant (CFA; Chondrex, Inc.) on the left paw of 0.1 ml per 100 g of body weight. Additionally, the rats were injected with normal saline to create the negative control (NC) group. | |||
| Response Summary | METTL3 knockdown suppressed Interleukin-6 (IL-6), matrix metalloproteinase (MMP)-3, and MMP-9 levels in human RA-FLSs and rat AIA-FLSs. | |||
Intersectin-2 (ITSN2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Pancreatic islets | Mus musculus |
|
Treatment: Mettl3 knockout mice
Control: Mettl3 flox/flox mice
|
GSE155612 | |
| Regulation |
 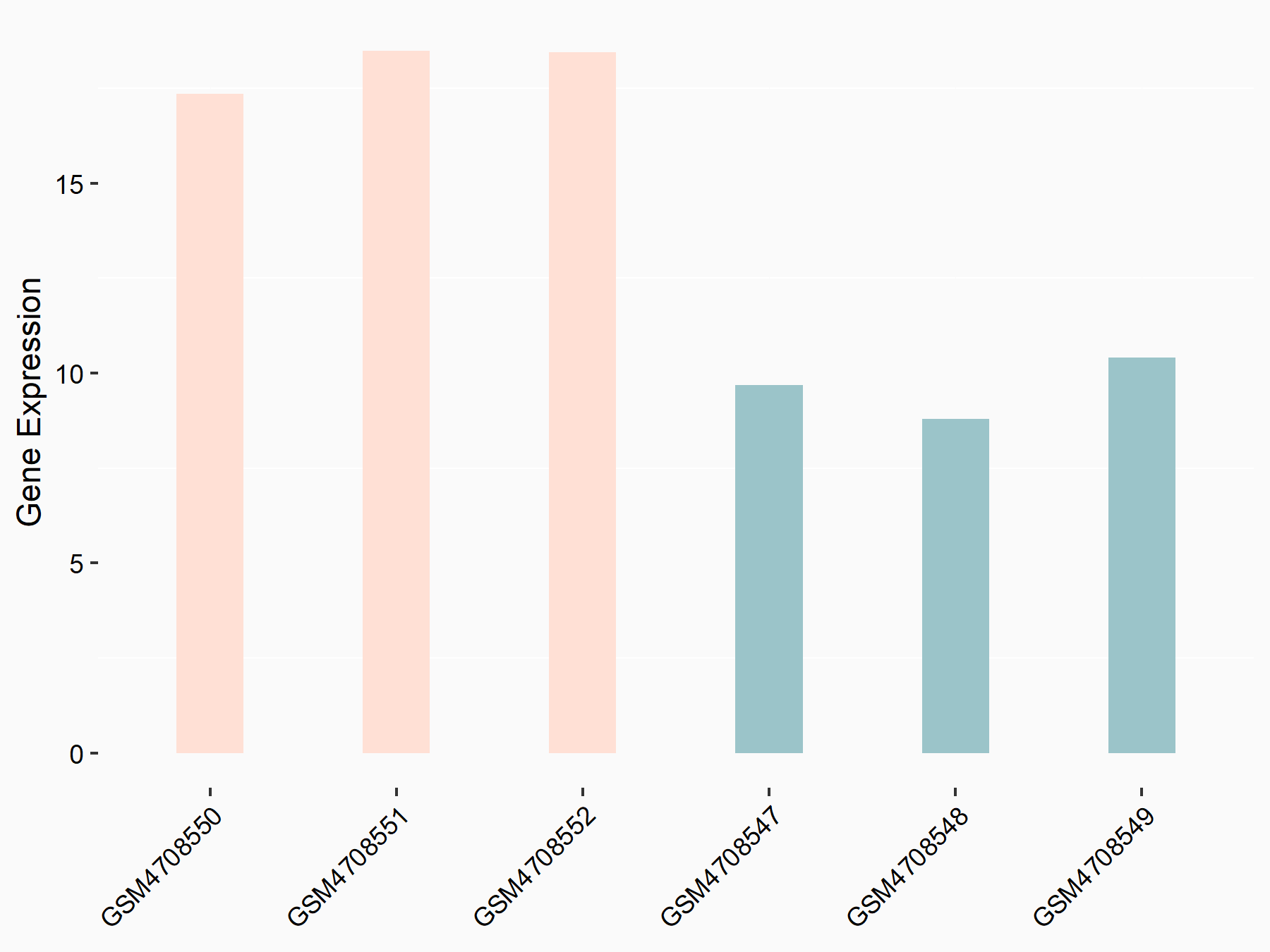 |
logFC: 8.48E-01 p-value: 1.42E-04 |
| More Results | Click to View More RNA-seq Results | |
Ovarian dysfunction [ICD-11: 5A80]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [188] | |||
| Responsed Disease | Ovarian dysfunction [ICD-11: 5A80] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | All mice described above were maintained on the C57BL/6 J background. Mice lacking Mettl3 in oocytes (referred to as Mettl3Gdf9 cKO) were generated by crossing Mettl3flox/flox mice with Gdf9-Cre mice. The Mettl3flox/flox female mice were used as the control group (referred to as WT). For the fertility test, six pairs of 6 weeks Mettl3flox/flox and Mettl3Gdf9 cKO female mice were randomly selected and continually mated to Mettl3flox/flox male mice which have been confirmed fertility for 5 months. The number of pups and litter size from each female was recorded. | |||
| Response Summary | METTL3 targets Intersectin-2 (ITSN2) for m6A modification and then enhances its stability to influence the oocytes meiosis. mRNA m6A modification in follicle development and coordination of RNA stabilization during oocyte growth. | |||
Female infertility [ICD-11: GA31]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [188] | |||
| Responsed Disease | Female infertility [ICD-11: GA31] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | All mice described above were maintained on the C57BL/6 J background. Mice lacking Mettl3 in oocytes (referred to as Mettl3Gdf9 cKO) were generated by crossing Mettl3flox/flox mice with Gdf9-Cre mice. The Mettl3flox/flox female mice were used as the control group (referred to as WT). For the fertility test, six pairs of 6 weeks Mettl3flox/flox and Mettl3Gdf9 cKO female mice were randomly selected and continually mated to Mettl3flox/flox male mice which have been confirmed fertility for 5 months. The number of pups and litter size from each female was recorded. | |||
| Response Summary | METTL3 targets Intersectin-2 (ITSN2) for m6A modification and then enhances its stability to influence the oocytes meiosis. mRNA m6A modification in follicle development and coordination of RNA stabilization during oocyte growth. | |||
Kinesin-like protein KIF3C (KIF3C)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 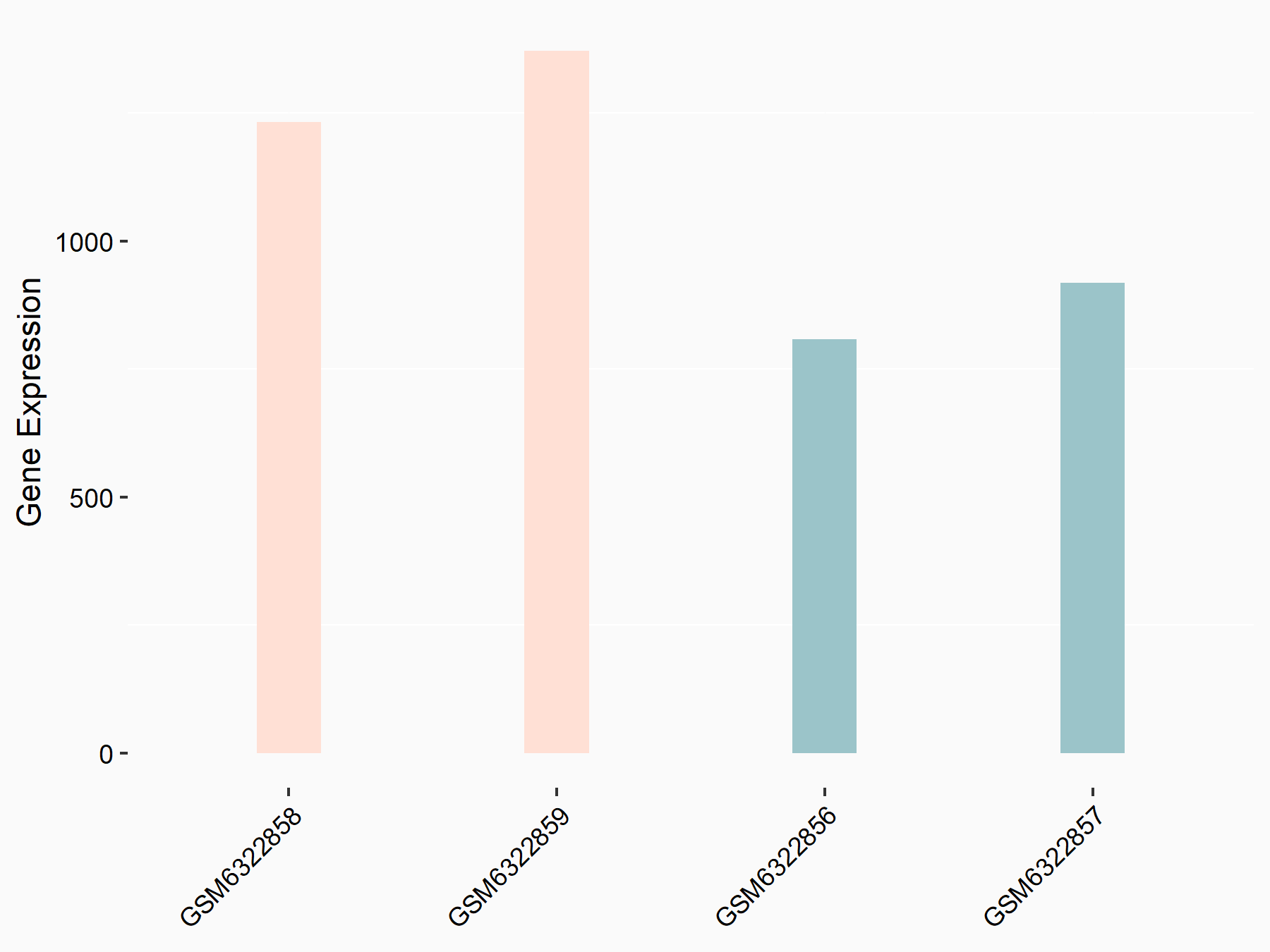 |
logFC: 5.93E-01 p-value: 1.16E-06 |
| More Results | Click to View More RNA-seq Results | |
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [189] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell migration | |||
| Cell invasion | ||||
In-vitro Model |
VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| LNCaP C4-2B | Prostate carcinoma | Homo sapiens | CVCL_4784 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| In-vivo Model | For the proliferation assays, C4-2B cells of KIF3C knockdown, negative control (1×106/200uL) were subcutaneously injected into BALB/c nude mice. The tumors were dissected and weighed (4-6 weeks old, male). | |||
| Response Summary | METTL3 induced m6A modification on Kinesin-like protein KIF3C (KIF3C), promoting the stabilization of KIF3C-mRNA by IGF2BP1. KIF3C was overexpressed in prostate cancer, promoting its growth migration and invasion was induced by miR-320d/METTL3 in an m6A dependent process. | |||
Krueppel-like factor 2 (KLF2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 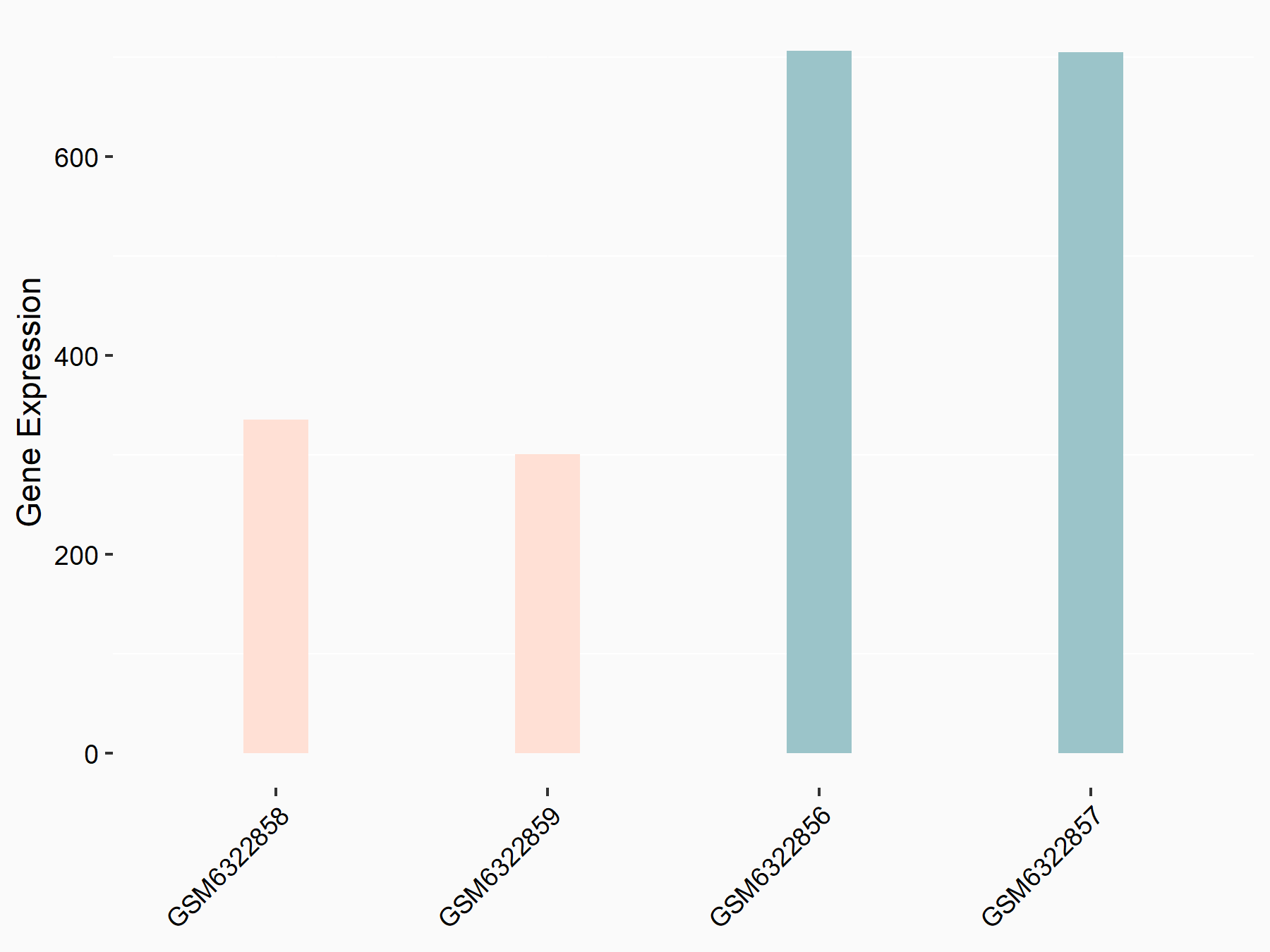 |
logFC: -1.15E+00 p-value: 3.23E-14 |
| More Results | Click to View More RNA-seq Results | |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [190] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Response Summary | METTL3 promotes translation of SPHK2 mRNA via an m6A-YTHDF1-dependent manner. Functionally, SPHK2 facilitates GC cell proliferation, migration and invasion by inhibiting Krueppel-like factor 2 (KLF2) expression. | |||
Krueppel-like factor 4 (KLF4)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HUVEC cell line | Homo sapiens |
|
Treatment: shMETTL3 HUVEC cells
Control: shScramble HUVEC cells
|
GSE157544 | |
| Regulation |
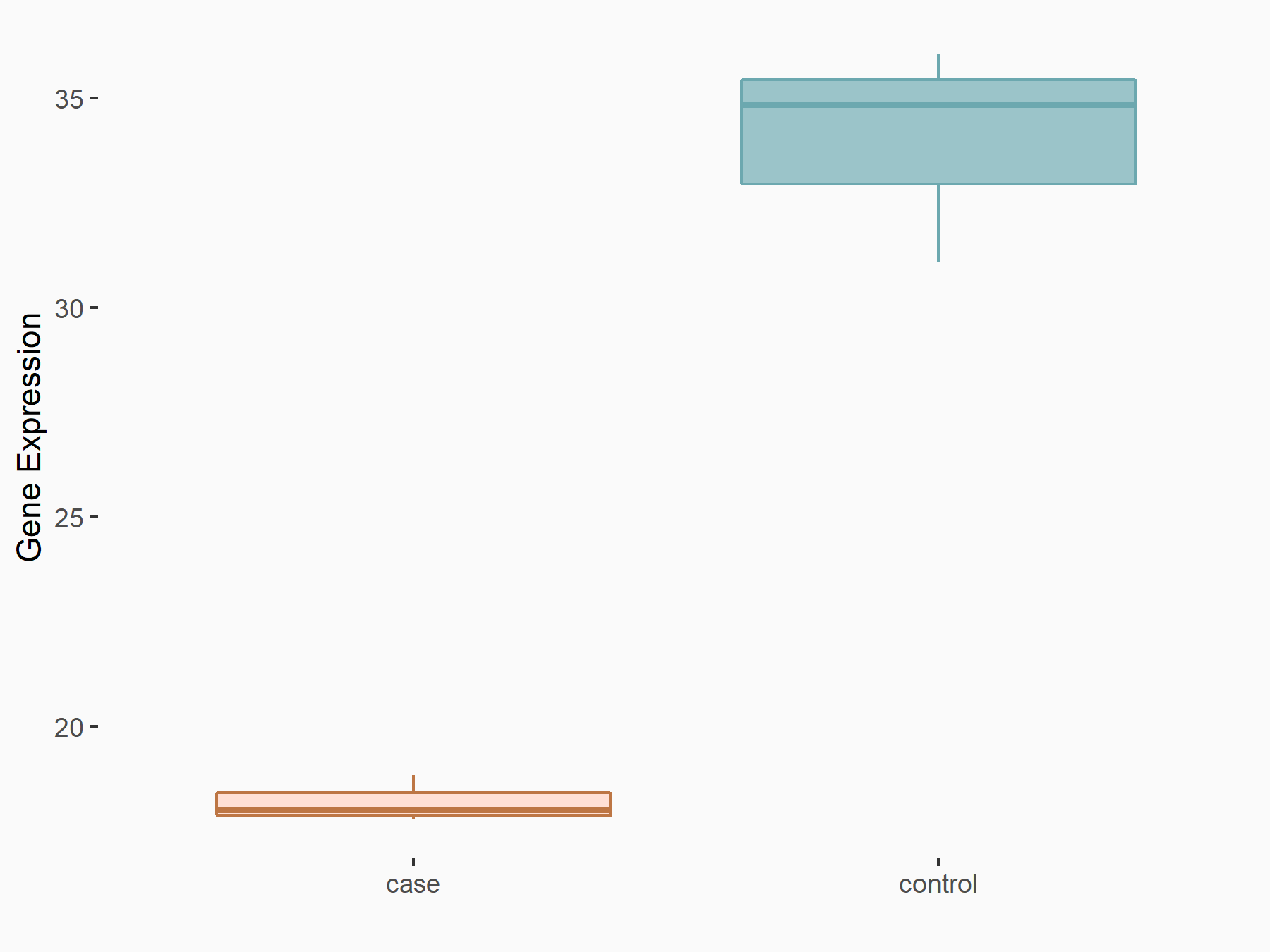 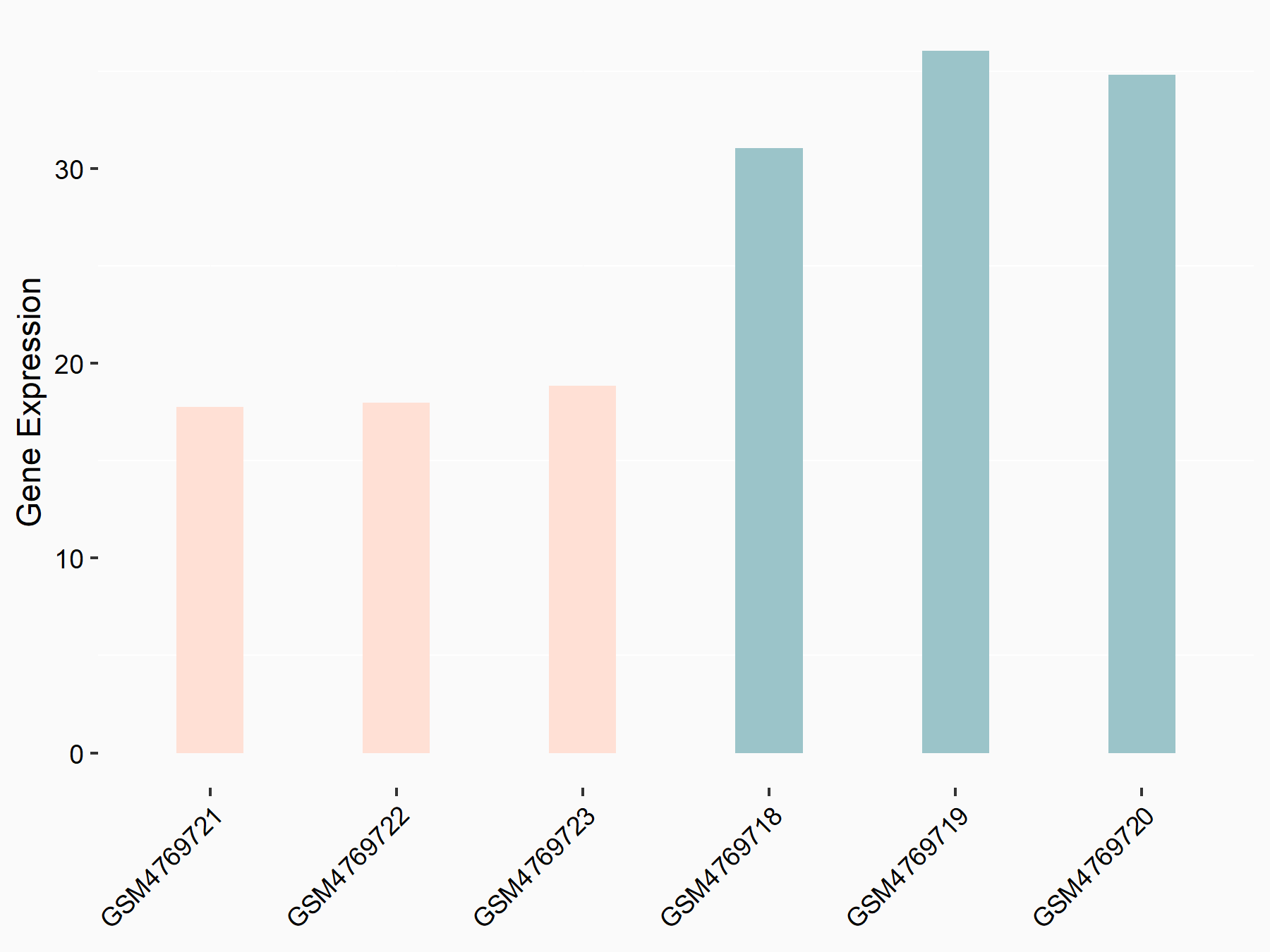 |
logFC: -8.63E-01 p-value: 1.22E-04 |
| More Results | Click to View More RNA-seq Results | |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [102] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | TNF signaling pathway | hsa04668 | ||
| Cell Process | Cell migration | |||
| Cell invasion | ||||
| Epithelial-mesenchymal transition | ||||
In-vitro Model |
NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| Response Summary | MiR-1915-3p expression was regulated by METTL3/YTHDF2 m6A axis through transcription factor Krueppel-like factor 4 (KLF4). miR-1915-3p function as a tumor suppressor by targeting SET and has an anti-metastatic therapeutic potential for lung cancer treatment. | |||
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [178] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cancer proliferation | |||
| Cancer metastasis | ||||
In-vitro Model |
SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| In-vivo Model | For the subcutaneous implantation model, UM-UC-3 cells (2 × 106 cells per mouse) stably METTL3 knocked down (shMETTL3-1, shMETTL3-2) were injected into the flanks of mice. | |||
| Response Summary | METTL3/YTHDF2/SETD7/Krueppel-like factor 4 (KLF4) m6 A axis provide the insight into the underlying mechanism of carcinogenesis and highlight potential therapeutic targets for bladder cancer. | |||
Atherosclerosis [ICD-11: BD40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [85] | |||
| Responsed Disease | Atherosclerosis [ICD-11: BD40.Z] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
MAEC | Normal | Mus musculus | CVCL_U411 |
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
| Response Summary | In the in vivo atherosclerosis model,partial ligation of the carotid artery led to plaque formation and up-regulation of METTL3 and NLRP1, with down-regulation of KLF4; knockdown of METTL3 via repetitive shRNA administration prevented the atherogenic process, NLRP1 up-regulation, and Krueppel-like factor 4 (KLF4) down-regulation. Collectively, it has demonstrated that METTL3 serves a central role in the atherogenesis induced by oscillatory stress and disturbed blood flow. | |||
Lymphoid enhancer-binding factor 1 (LEF1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
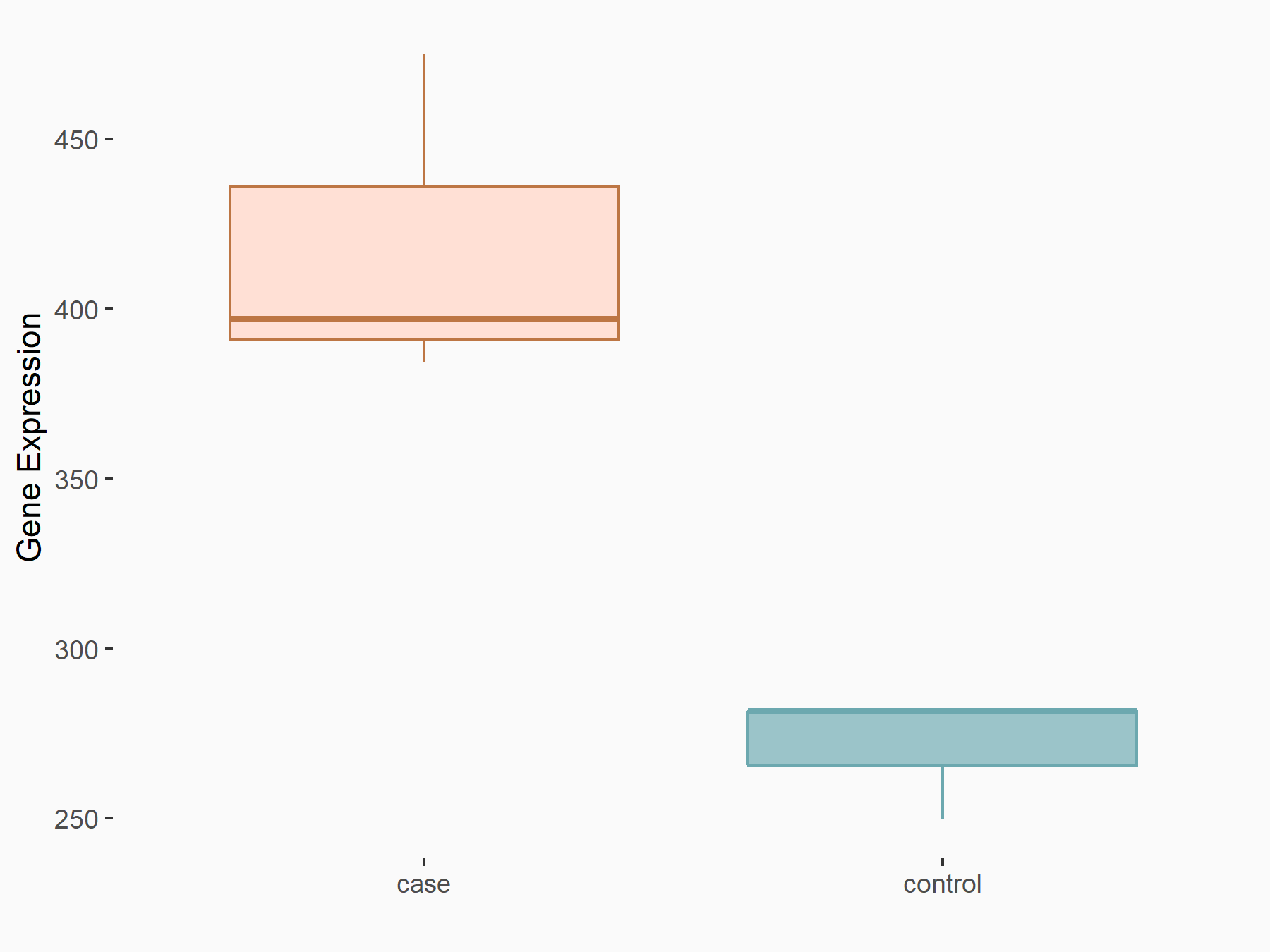 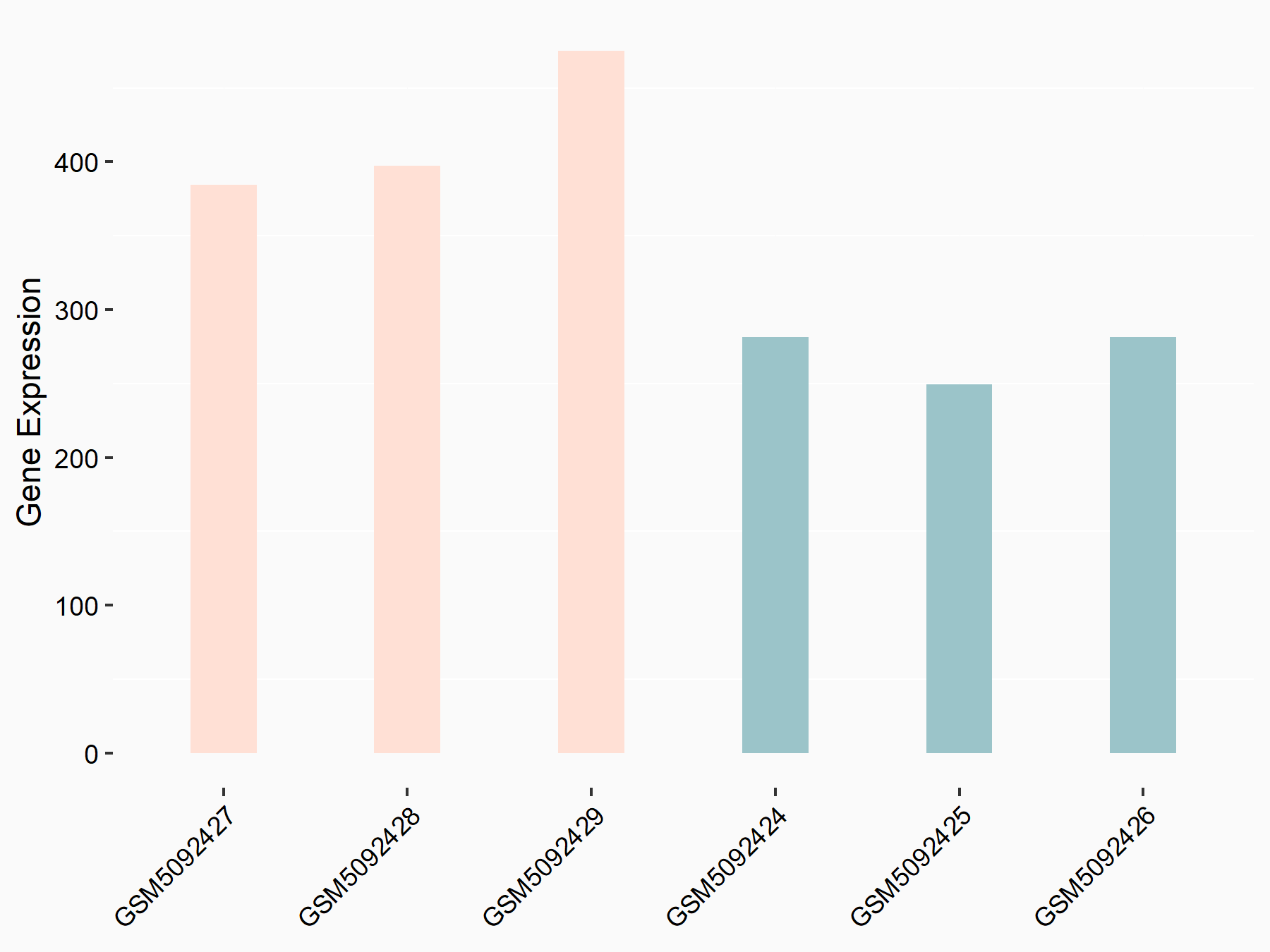 |
logFC: 6.28E-01 p-value: 2.52E-06 |
| More Results | Click to View More RNA-seq Results | |
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [191] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
HOS | Osteosarcoma | Homo sapiens | CVCL_0312 |
| MG-63 | Osteosarcoma | Homo sapiens | CVCL_0426 | |
| NHOst (Normal human osteoblast cells) | ||||
| SaOS-2 | Osteosarcoma | Homo sapiens | CVCL_0548 | |
| U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 | |
| In-vivo Model | The Metastatic Bone Tumor Model was established via injecting HOS cells into the right tibia of each animal. | |||
| Response Summary | METTL3 silence decreased the m6A methylation and total mRNA level of Lymphoid enhancer-binding factor 1 (LEF1),the m6A methyltransferase METTL3 promotes osteosarcoma cell progression by regulating the m6A level of LEF1 and activating Wnt/beta-catenin signaling pathway. | |||
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [192] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Cell activity and migratory | |||
In-vitro Model |
PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 |
| Response Summary | METTL3 influences the activity of the Wnt pathway through m6A methylation on Lymphoid enhancer-binding factor 1 (LEF1) mRNA, thereafter, promoting the progression of prostate cancer. | |||
M-phase inducer phosphatase 2 (CDC25B)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
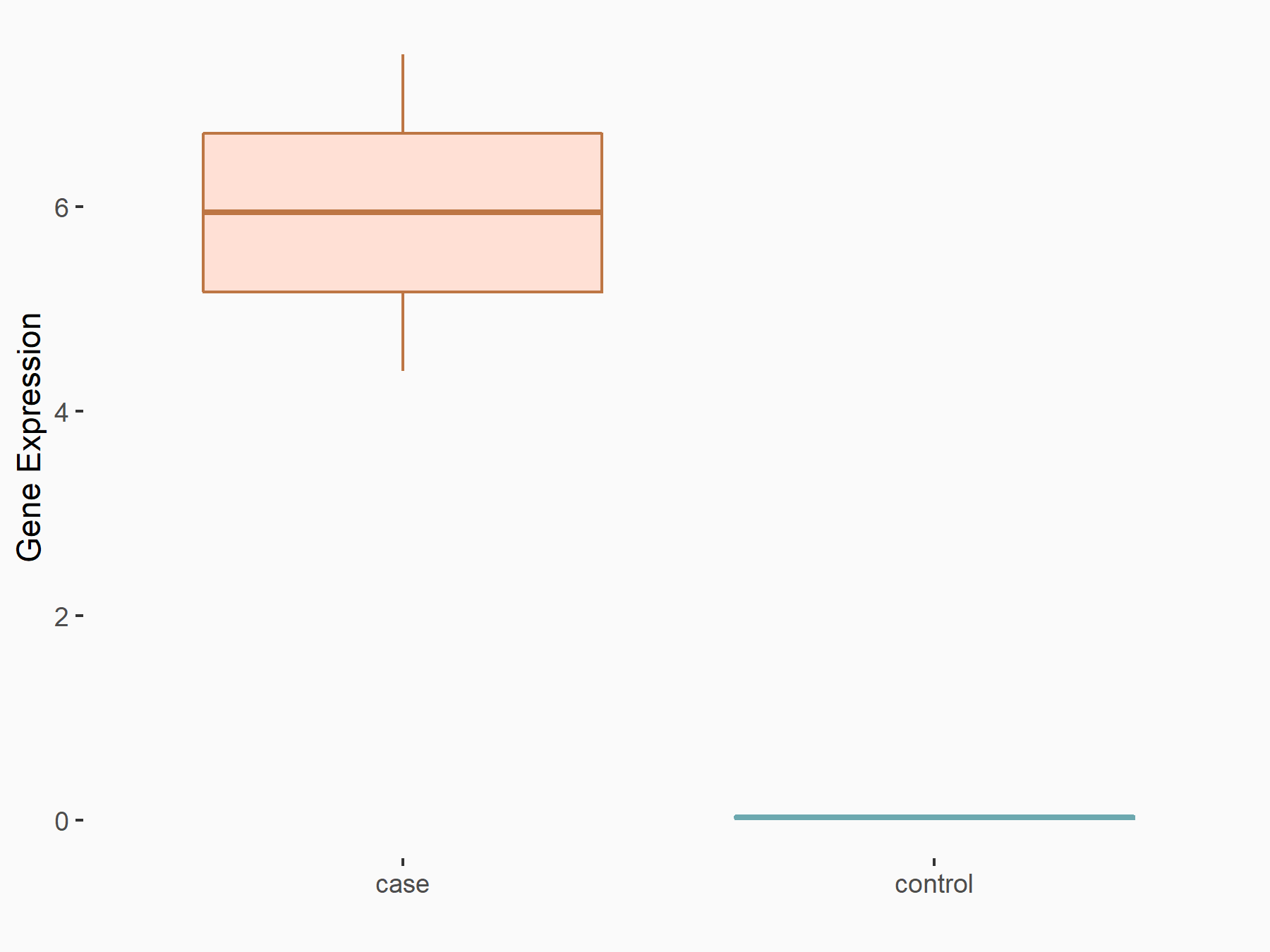  |
logFC: 2.72E+00 p-value: 5.89E-03 |
| More Results | Click to View More RNA-seq Results | |
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [193] | |||
| Responsed Disease | Head and neck squamous carcinoma [ICD-11: 2B6E] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
Tu 686 | Laryngeal squamous cell carcinoma | Homo sapiens | CVCL_4916 |
| Tu 212 | Head and neck squamous cell carcinoma | Homo sapiens | CVCL_4915 | |
| SAS | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1675 | |
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
| HEp-2 | Endocervical adenocarcinoma | Homo sapiens | CVCL_1906 | |
| FaDu | Hypopharyngeal squamous cell carcinoma | Homo sapiens | CVCL_1218 | |
| In-vivo Model | Tumour xenograft models were established in nude mice bearing: SAS cells cells stably transfected with METTL3-shRNA and the corresponding control vector. The different HNSCC cells (5 × 106) were subcutaneously injected into the right axilla of nude mice (n = 6 per group). | |||
| Response Summary | METTL3 enhanced the m6A modification of M-phase inducer phosphatase 2 (CDC25B) mRNA, which maintained its stability and upregulated its expression, thereby activating G2/M phase of cell cycle and leading to HNSCC malignant progression. | |||
Macrophage metalloelastase (MMP12)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Raw 264.7 cell line | Mus musculus |
|
Treatment: METTL3 knockout Raw 264.7 cells
Control: Wild type Raw 264.7 cells
|
GSE162248 | |
| Regulation |
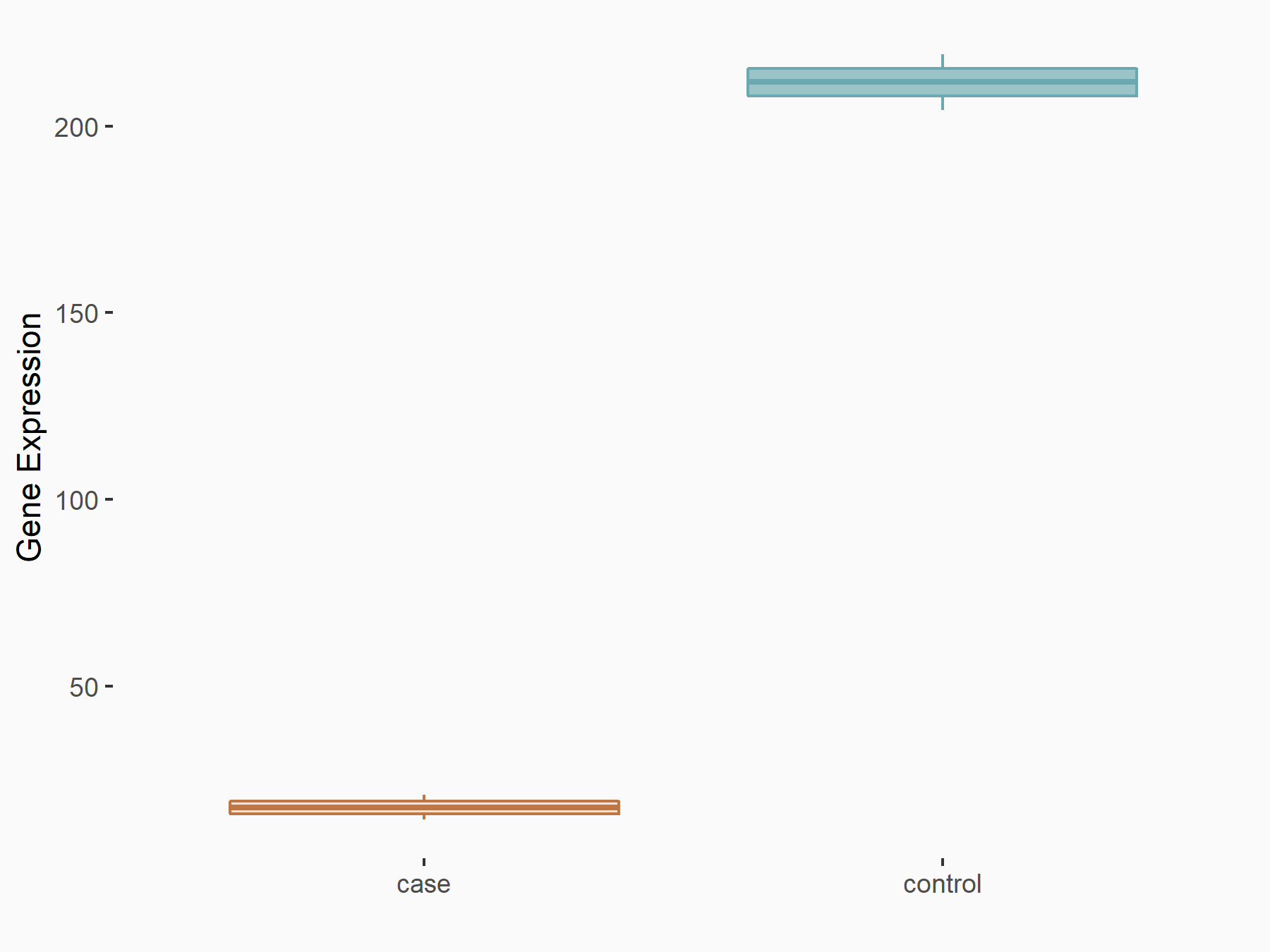 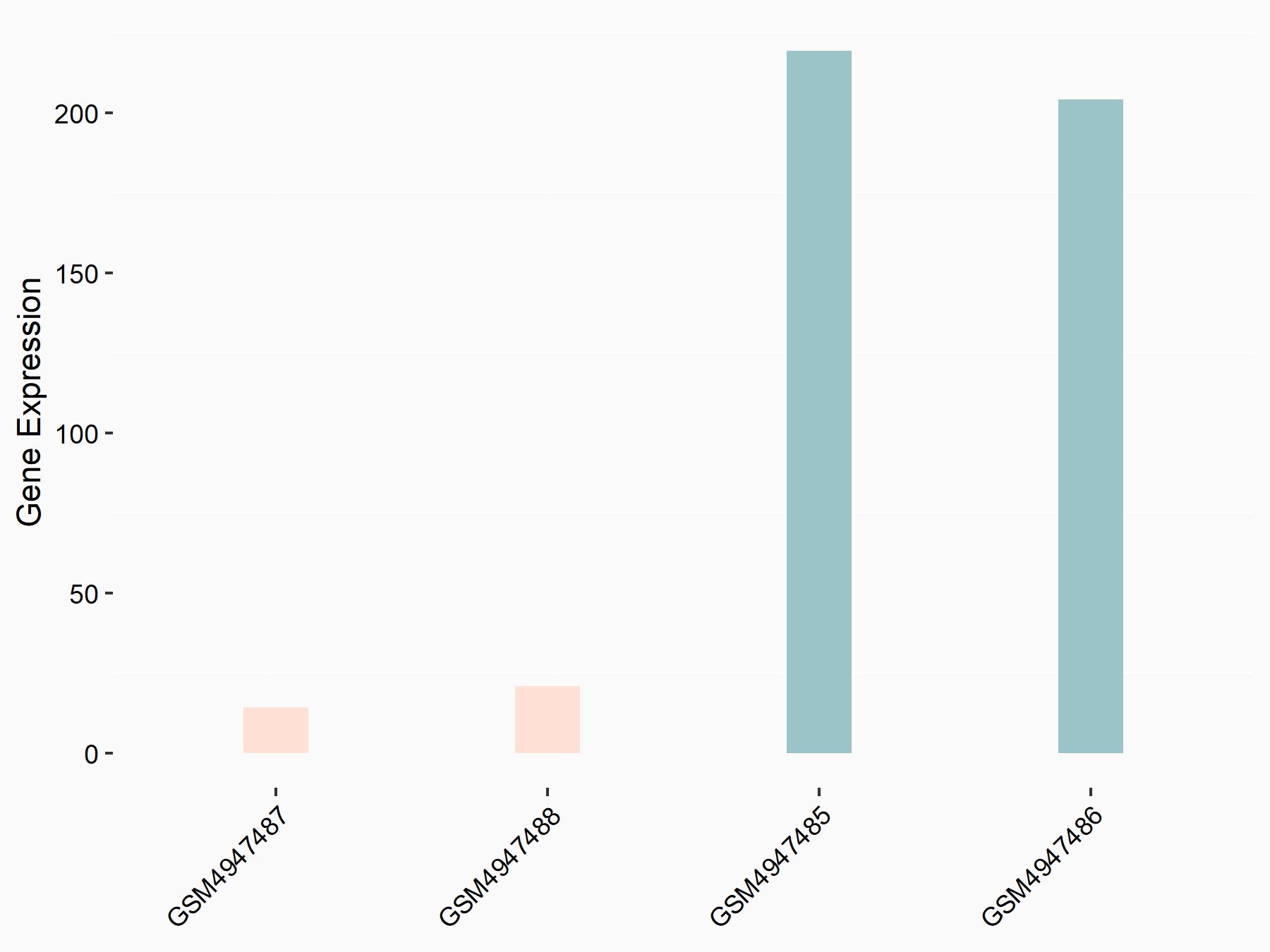 |
logFC: -3.62E+00 p-value: 1.51E-23 |
| More Results | Click to View More RNA-seq Results | |
Emphysema [ICD-11: CA21]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [160] | |||
| Responsed Disease | Emphysema [ICD-11: CA21] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Response Summary | METTL3-mediated formation of EV miR-93, facilitated by m6A, is implicated in the aberrant cross-talk of epithelium-macrophages, indicating that this process is involved in the smoking-related emphysema. EV miR-93 was used as a novel risk biomarker for CS-induced emphysema. MiR-93 activated the JNK pathway by targeting dual-specificity phosphatase 2 (DUSP2), which elevated the levels of matrix metalloproteinase 9 (MMP9) and Macrophage metalloelastase (MMP12) and induced elastin degradation, leading to emphysema. | |||
Matrix metalloproteinase-9 (MMP9)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 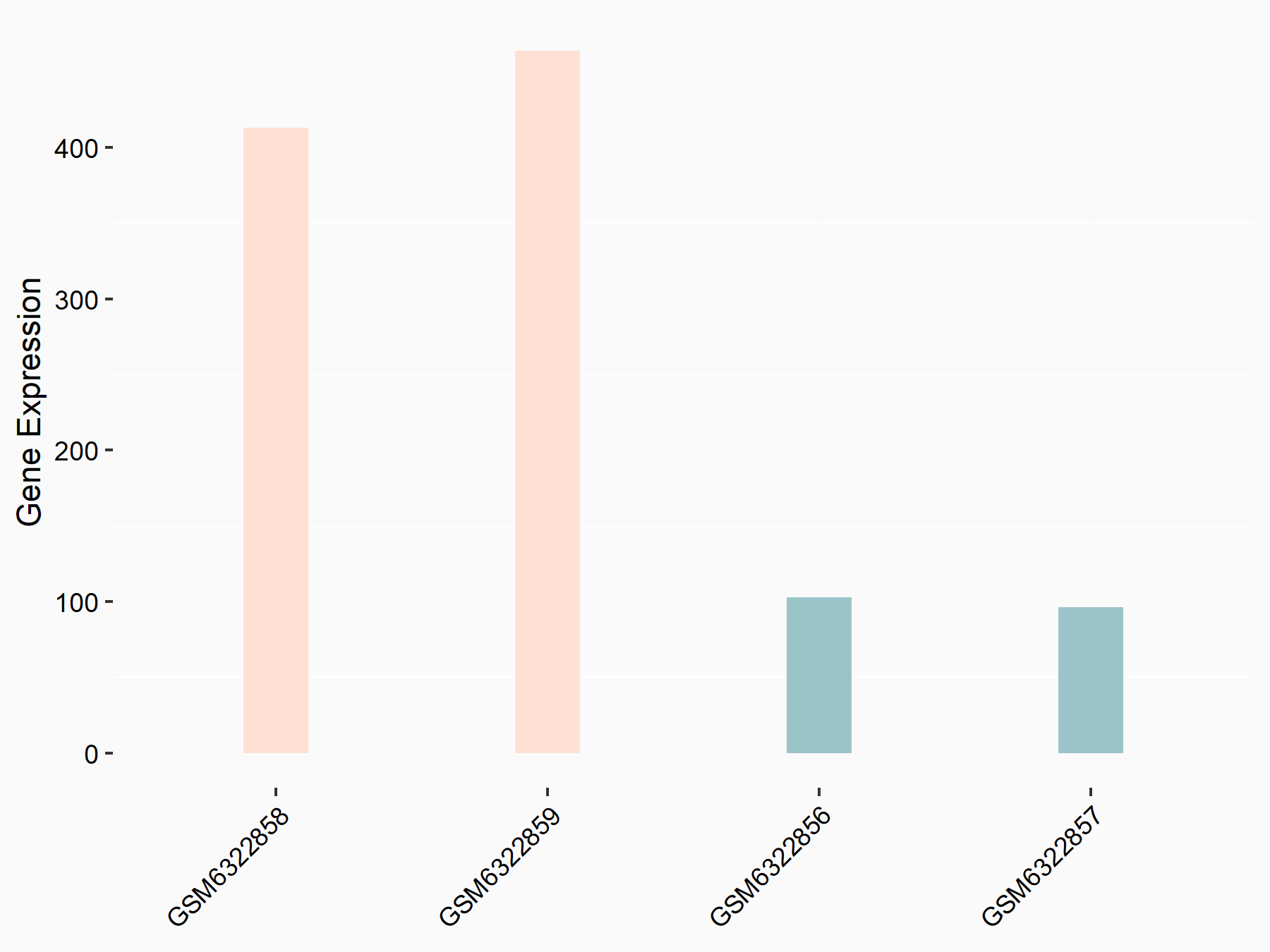 |
logFC: 2.14E+00 p-value: 4.12E-24 |
| More Results | Click to View More RNA-seq Results | |
Emphysema [ICD-11: CA21]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [160] | |||
| Responsed Disease | Emphysema [ICD-11: CA21] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Response Summary | METTL3-mediated formation of EV miR-93, facilitated by m6A, is implicated in the aberrant cross-talk of epithelium-macrophages, indicating that this process is involved in the smoking-related emphysema. EV miR-93 was used as a novel risk biomarker for CS-induced emphysema. MiR-93 activated the JNK pathway by targeting dual-specificity phosphatase 2 (DUSP2), which elevated the levels of Matrix metalloproteinase-9 (MMP9) and matrix metalloproteinase 12 (MMP12) and induced elastin degradation, leading to emphysema. | |||
Rheumatoid arthritis [ICD-11: FA20]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [187] | |||
| Responsed Disease | Rheumatoid arthritis [ICD-11: FA20] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Inflammatory response | |||
In-vitro Model |
FLS (Rat fibroblast synovial cell line) | |||
| In-vivo Model | To establish the adjuvant-induced arthritis (AIA) model, the rats were given complete Freund's adjuvant (CFA; Chondrex, Inc.) on the left paw of 0.1 ml per 100 g of body weight. Additionally, the rats were injected with normal saline to create the negative control (NC) group. | |||
| Response Summary | METTL3 knockdown suppressed interleukin (IL)-6, matrix metalloproteinase (MMP)-3, and Matrix metalloproteinase-9 (MMP9) levels in human RA-FLSs and rat AIA-FLSs. | |||
Meltrin-beta (ADAM19)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HUVEC cell line | Homo sapiens |
|
Treatment: shMETTL3 HUVEC cells
Control: shScramble HUVEC cells
|
GSE157544 | |
| Regulation |
 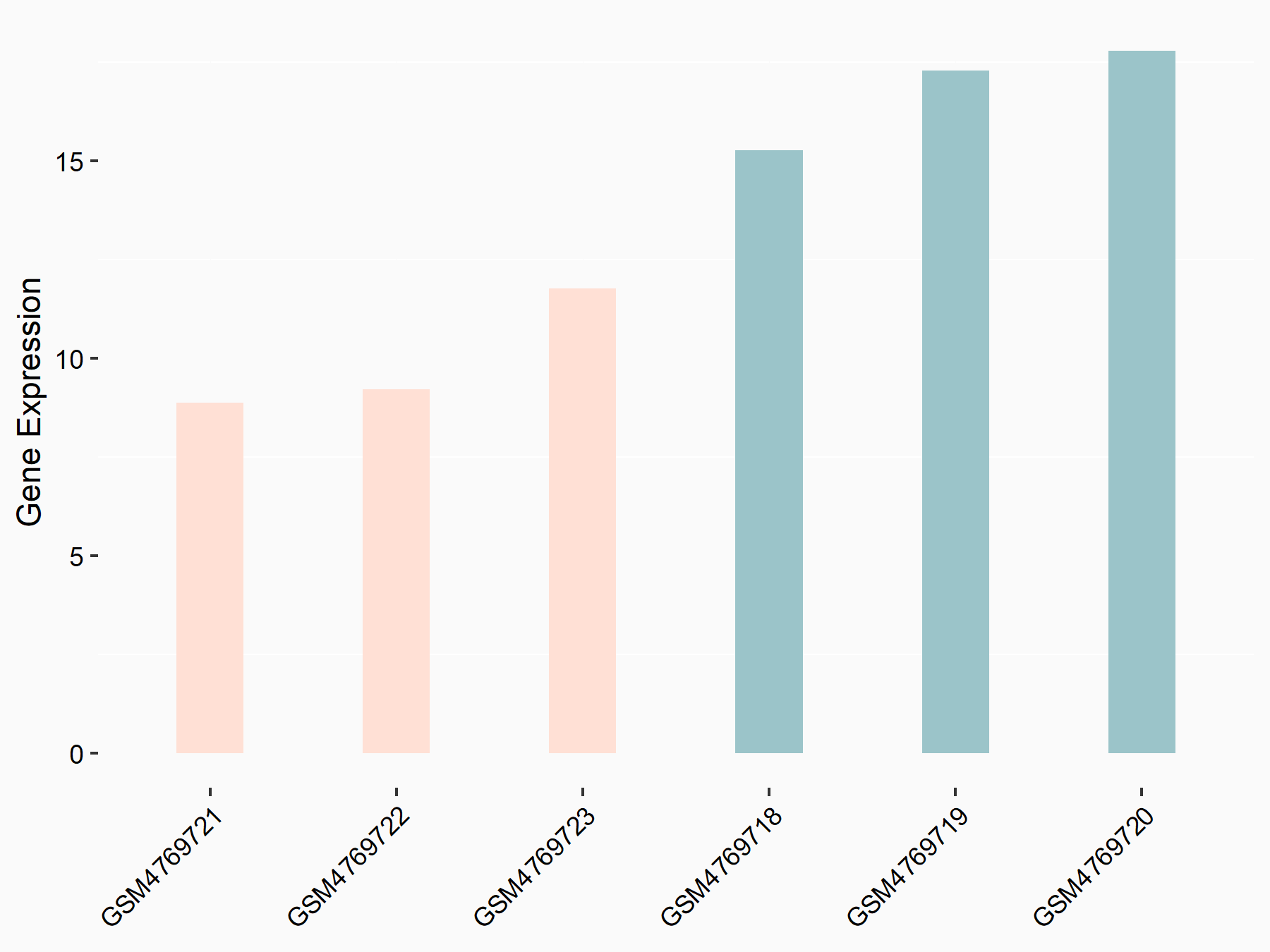 |
logFC: -7.06E-01 p-value: 4.35E-03 |
| More Results | Click to View More RNA-seq Results | |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [194] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Responsed Drug | Ethyl ester form of meclofenamic acid | Approved | ||
| Target Regulation | Down regulation | |||
| Cell Process | Cells growth | |||
| Cells self-renewal | ||||
| Tumorigenesis | ||||
| MicroRNAs in cancer (hsa05206) | ||||
In-vitro Model |
GSC | Glioma | Epinephelus akaara | CVCL_M752 |
| In-vivo Model | 2 × 105 dissociated cells in 2 uL PBS were injected into the following site (anteroposterior [AP] +0.6 mm, mediolateral [ML] +1.6 mm, and dorsoventricular [DV] 2.6 mm) with a rate of 1 uL/min. | |||
| Response Summary | Knockdown of METTL3 or METTL14 induced changes in mRNA m6A enrichment and altered mRNA expression of genes (e.g., Meltrin-beta (ADAM19)) with critical biological functions in GSCs. Treatment with MA2, a chemical inhibitor of FTO, dramatically suppressed GSC-induced tumorigenesis and prolonged lifespan in GSC-grafted animals. | |||
Metalloreductase STEAP2 (STEAP2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
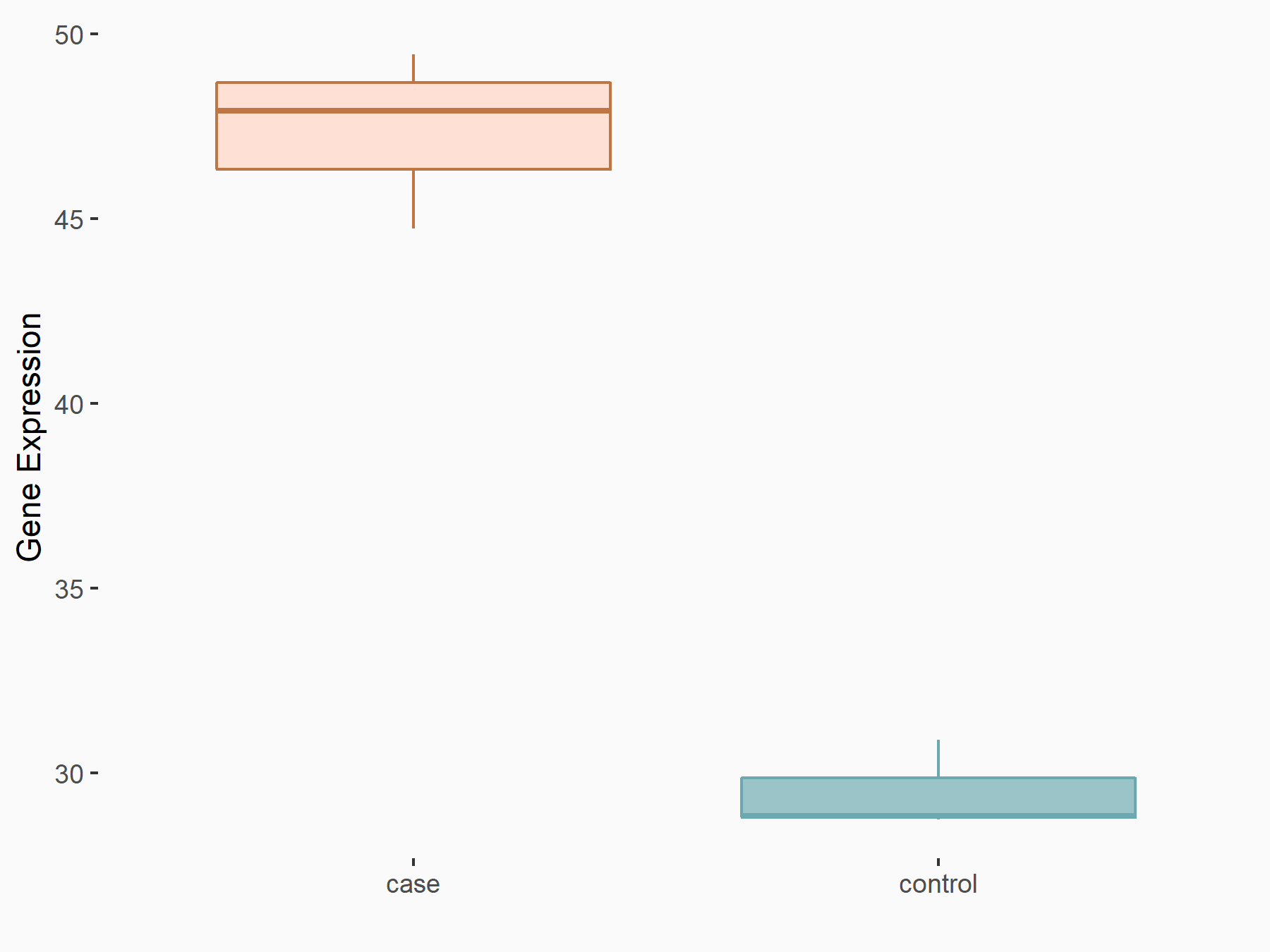 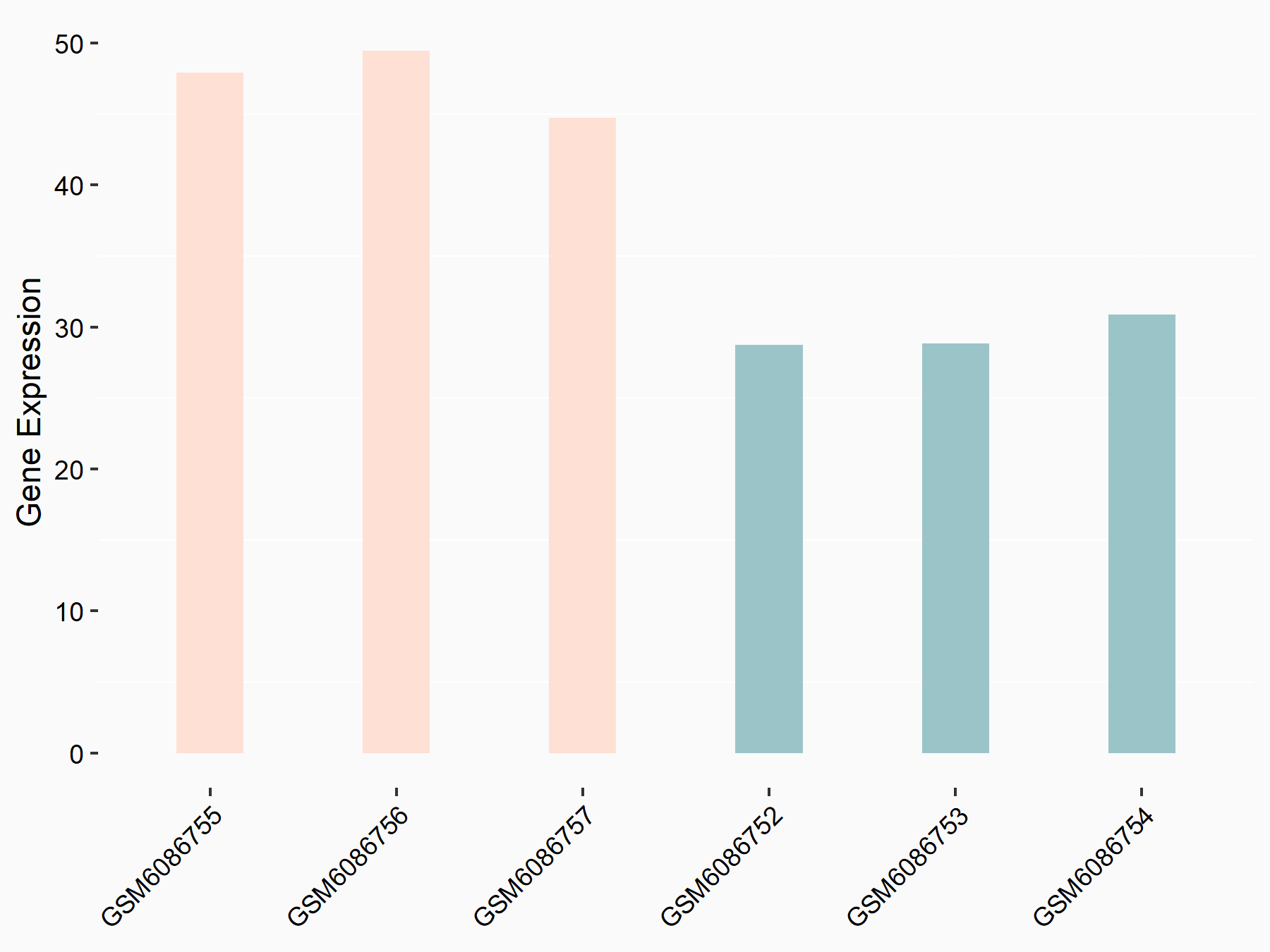 |
logFC: 6.65E-01 p-value: 3.16E-05 |
| More Results | Click to View More RNA-seq Results | |
Thyroid Cancer [ICD-11: 2D10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [195] | |||
| Responsed Disease | Papillary thyroid cancer [ICD-11: 2D10.1] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Hedgehog signaling pathway | hsa04340 | ||
| Cell Process | Epithelial-to-mesenchymal transition | |||
| Cell proliferation | ||||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
TPC-1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_6298 |
| KTC-1 | Thyroid carcinoma | Homo sapiens | CVCL_6300 | |
| B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 | |
| In-vivo Model | BCPAP cells (5×106) were introduced into the mice by means of subcutaneous injection through the flank area. STEAP2-saRNA or NC-saRNA (n = 6 for each group) was given by intratumoral multipoint injection at an interval of 3 days (5 injections in total) using an in vivo transfection reagent (Entranster -in vivo, Engreen, China) as per the vendor-provided protocol. Tumor volume (V) was monitored and calculated as follows: V = (L×W2)/2. For the in vivo tumor metastasis assay, BCPAP cells (5×106 cells) were administrated into mice through the tail vein. STEAP2-saRNA or NC-saRNA (n = 6 for each group) was given via tail vein injection at an interval of 3 days (8 injections in total). | |||
| Response Summary | Metalloreductase STEAP2 (STEAP2) overexpression inhibited papillary thyroid cancer cell proliferation, migration, and invasion in vitro and inhibited lung metastasis and tumorigenicity in vivo. METTL3 stabilized STEAP2 mRNA and regulated STEAP2 expression positively in an m6A-dependent manner. | |||
Microprocessor complex subunit DGCR8 (DGCR8)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3 knockout liver
Control: Wild type liver cells
|
GSE198513 | |
| Regulation |
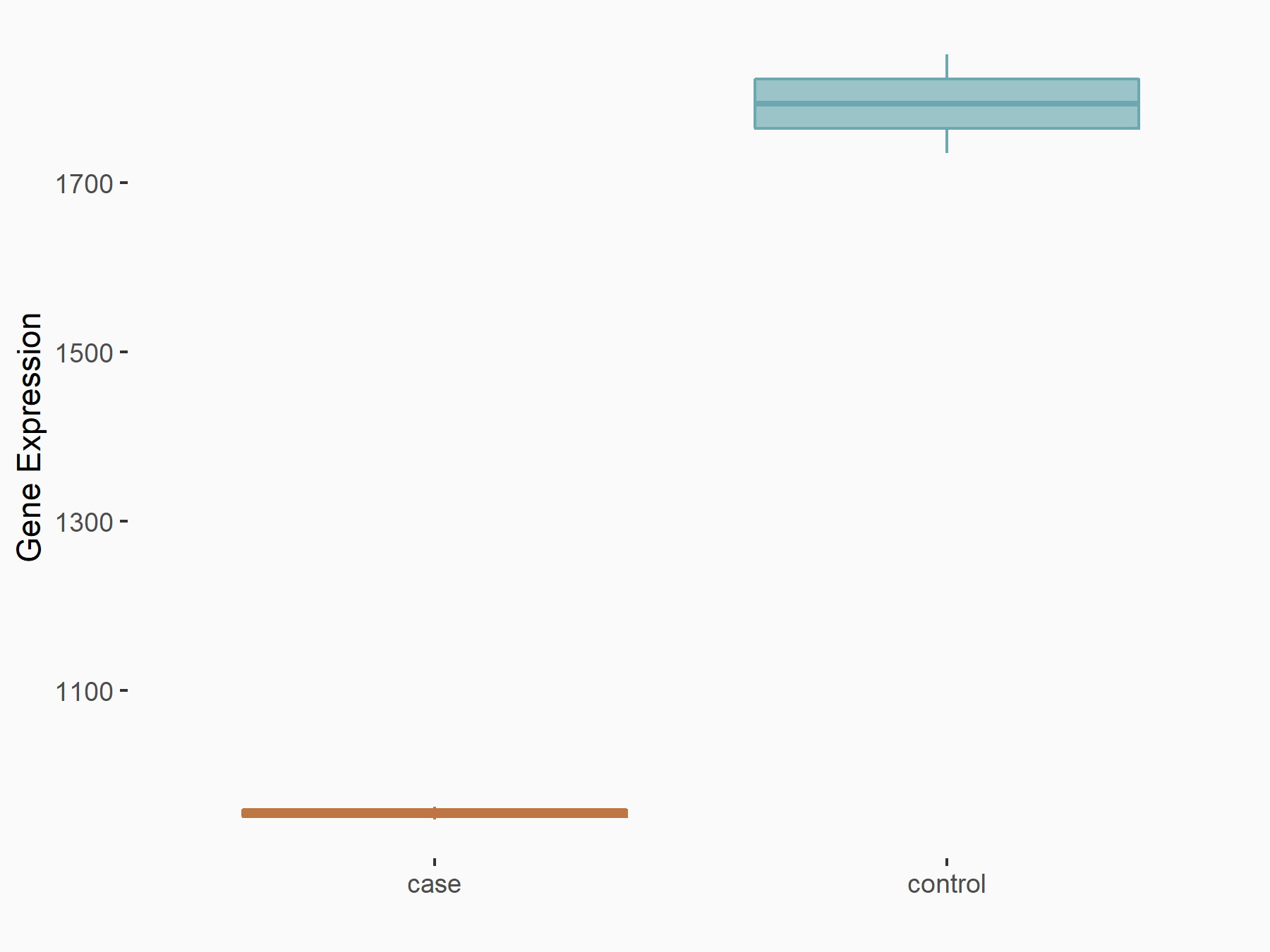 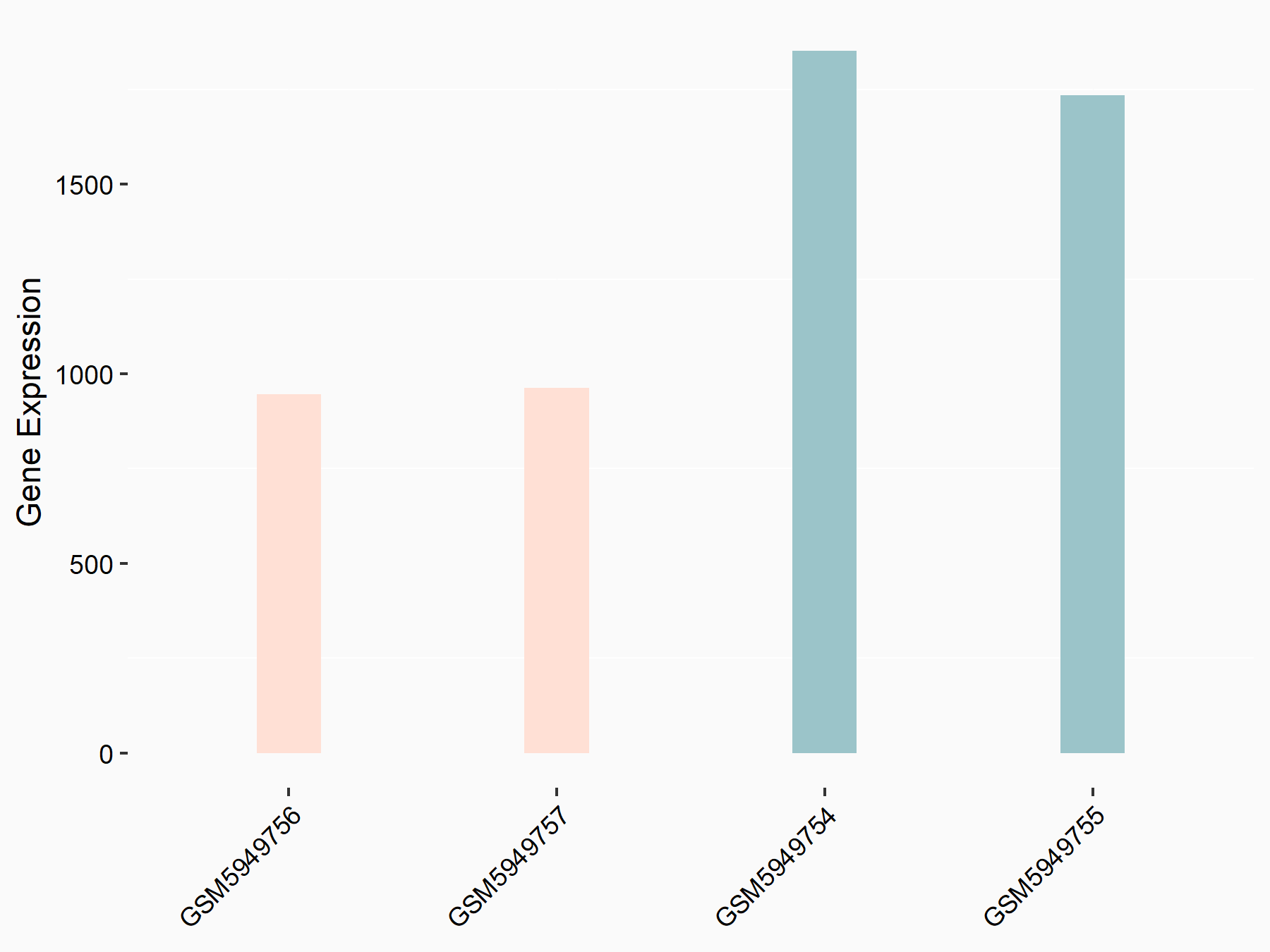 |
logFC: -9.09E-01 p-value: 2.99E-21 |
| More Results | Click to View More RNA-seq Results | |
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [196] | |||
| Responsed Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.1] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Epithelial-mesenchymal transition | ||||
In-vitro Model |
TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 |
| KYSE-30 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1351 | |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| HET-1A | Normal | Homo sapiens | CVCL_3702 | |
| CVCL_E307 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_E307 | |
| Eca-109 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_6898 | |
| In-vivo Model | Luciferase-labeled KYSE150 cells (5 × 106) were inoculated into the footpads of BALB/c nude mice (4-5 weeks old, 18-20 g) to establish the popliteal lymphatic metastasis model. | |||
| Response Summary | METTL3 could interact with Microprocessor complex subunit DGCR8 (DGCR8) protein and positively modulate pri-miR-320b maturation process in an N6-methyladenosine (m6A)-dependent manner. Therefore, our findings uncover a VEGF-C-independent mechanism of exosomal and intracellular miR-320b-mediated LN metastasis and identify miR-320b as a novel predictive marker and therapeutic target for LN metastasis in ESCC. | |||
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [197] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
WPMY-1 | Normal | Homo sapiens | CVCL_3814 |
| VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| Response Summary | METTL3 promoted cell proliferation, migration, invasion and tumorigenesis in PCa. METTL3 upregulating the level of m6A, and interacted with Microprocessor complex subunit DGCR8 (DGCR8) to recognize the m6A modification of pre-miR-182 to regulate its splicing and maturation and promote the high expression of miRNA. | |||
Chondropathies [ICD-11: FB82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [198] | |||
| Responsed Disease | Chondropathies [ICD-11: FB82] | |||
| Target Regulation | Up regulation | |||
| Cell Process | RNA mature | |||
In-vitro Model |
Cartilage cells (From the cartilage tissue samples from patients) | |||
| Response Summary | Interleukin 1-beta (IL-1-beta) is an important inducer of cartilage degeneration that can induce an inflammatory cascade reaction in chondrocytes and inhibit the normal biological function of cells. METTL3 could regulate miR-126-5p maturation, we first confirmed that METTL3 can bind the key protein underlying pri-miRNA processing, Microprocessor complex subunit DGCR8 (DGCR8). Additionally, when METTL3 expression was inhibited, the miR-126-5p maturation process was blocked. | |||
Mothers against decapentaplegic homolog 3 (SMAD3)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
  |
logFC: 1.25E+00 p-value: 1.46E-105 |
| More Results | Click to View More RNA-seq Results | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [124] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Cell invasion | |||
| Cell migration | ||||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| In-vivo Model | BALB/c nude mice (4 weeks old) were acquired from Vital River Laboratory (Beijing, China). HCT116 cells with stable circ1662 expression (2 × 106 in 100 L of PBS) were injected via the tail vein. After 45 days, the mice were sacrificed. The lung metastatic carcinoma specimens were processed into paraffin-embedded sections for subsequent H&E staining and IHC. | |||
| Response Summary | METTL3-induced circ1662 promoted colorectal cancer cell invasion and migration by accelerating YAP1 nuclear transport. Circ1662 enhanced CRC invasion and migration depending on YAP1 and Mothers against decapentaplegic homolog 3 (SMAD3). This result implies that circ1662 is a new prognostic and therapeutic marker for CRC metastasis. | |||
Liver cancer [ICD-11: 2C12]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [309] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Lenvatinib | Approved | ||
In-vitro Model |
Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| In-vivo Model | When the tumors in the CDX and PDX mice grew to about 80-150 mm3, forty tumor-bearing mice were randomly divided into four groups (ten mice in each group) according to the size of the transplanted tumor and the principle of consistency between groups. The four groups were: as follows: solvent control group, Len (3 mg kg-1) administration group, Res (0.375 mg kg-1) administration group, and Len (3 mg kg-1) plus Res (0.375 mg kg-1) administration group. The administration mode of all groups was intragastric and the volume was 0.2 ml/20 g. Mouse body weight and xenograft tumor volume were measured every other day. Tumor volume was measured weekly and calculated as V = (length × width2)/2. Three weeks after the onset of treatment, the tumors were excised and weighed. After the experiment, the heart, liver, spleen, lung and kidney tissues of the mice were completely removed and weighed. Then the organ index was calculated as organ index = weight of each organ/body weight × 100 %. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [309] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Reserpine | Approved | ||
In-vitro Model |
Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| In-vivo Model | When the tumors in the CDX and PDX mice grew to about 80-150 mm3, forty tumor-bearing mice were randomly divided into four groups (ten mice in each group) according to the size of the transplanted tumor and the principle of consistency between groups. The four groups were: as follows: solvent control group, Len (3 mg kg-1) administration group, Res (0.375 mg kg-1) administration group, and Len (3 mg kg-1) plus Res (0.375 mg kg-1) administration group. The administration mode of all groups was intragastric and the volume was 0.2 ml/20 g. Mouse body weight and xenograft tumor volume were measured every other day. Tumor volume was measured weekly and calculated as V = (length × width2)/2. Three weeks after the onset of treatment, the tumors were excised and weighed. After the experiment, the heart, liver, spleen, lung and kidney tissues of the mice were completely removed and weighed. Then the organ index was calculated as organ index = weight of each organ/body weight × 100 %. | |||
Mothers against decapentaplegic homolog 7 (SMAD7)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
 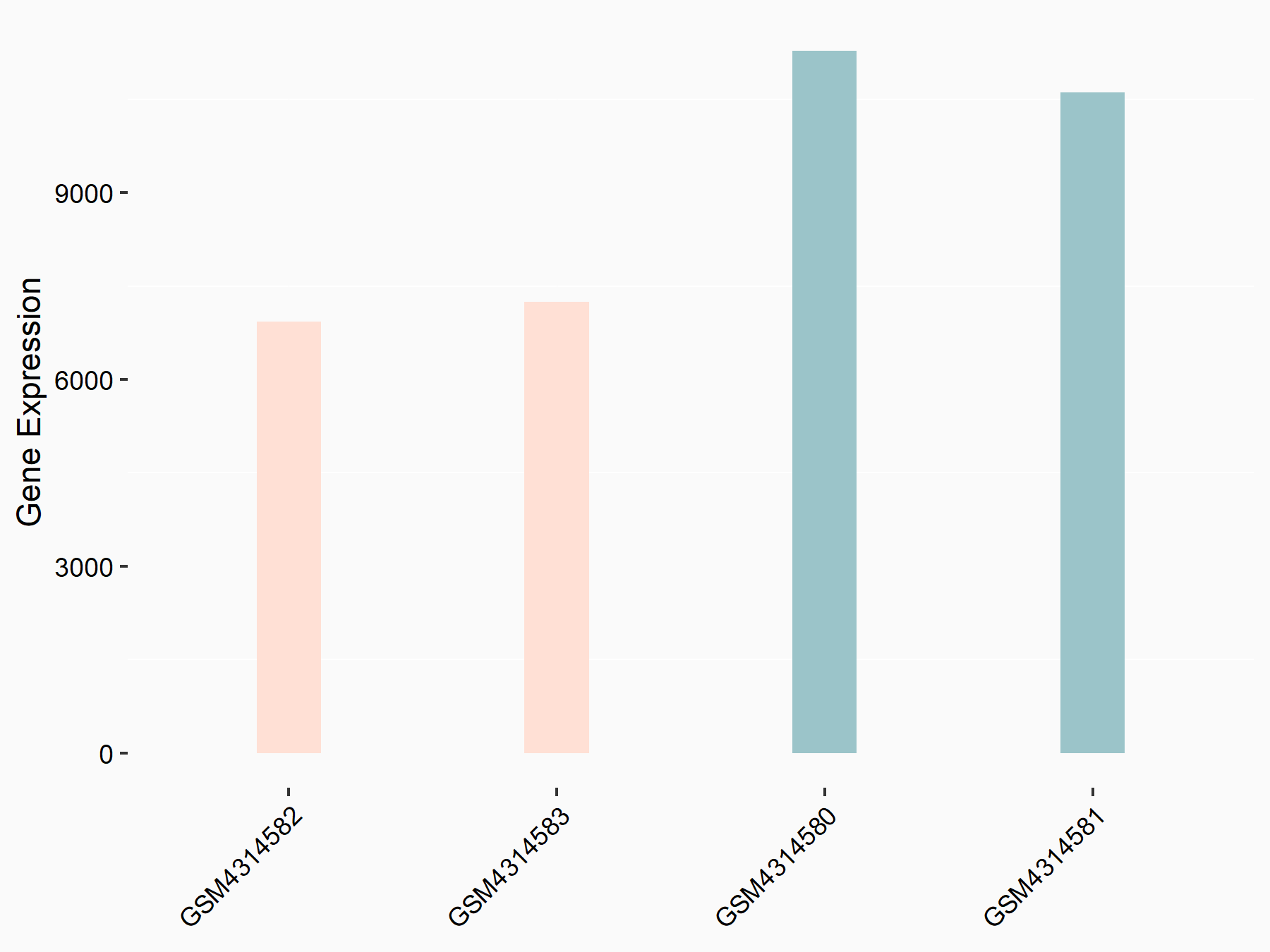 |
logFC: -6.27E-01 p-value: 8.80E-36 |
| More Results | Click to View More RNA-seq Results | |
Inflammatory response [ICD-11: MG46]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [162] | |||
| Responsed Disease | Inflammatory response [ICD-11: MG46] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
MC3T3-E1 | Normal | Mus musculus | CVCL_0409 |
| Response Summary | METTL3 knockdown inhibits osteoblast differentiation and Smad-dependent signaling by stabilizing Mothers against decapentaplegic homolog 7 (SMAD7) and Smurf1 mRNA transcripts via YTHDF2 involvement and activates the inflammatory response by regulating MAPK signaling in LPS-induced inflammation. | |||
NACHT/LRR/PYD domains-containing protein 1 (NLRP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HUVEC cell line | Homo sapiens |
|
Treatment: shMETTL3 HUVEC cells
Control: shScramble HUVEC cells
|
GSE157544 | |
| Regulation |
 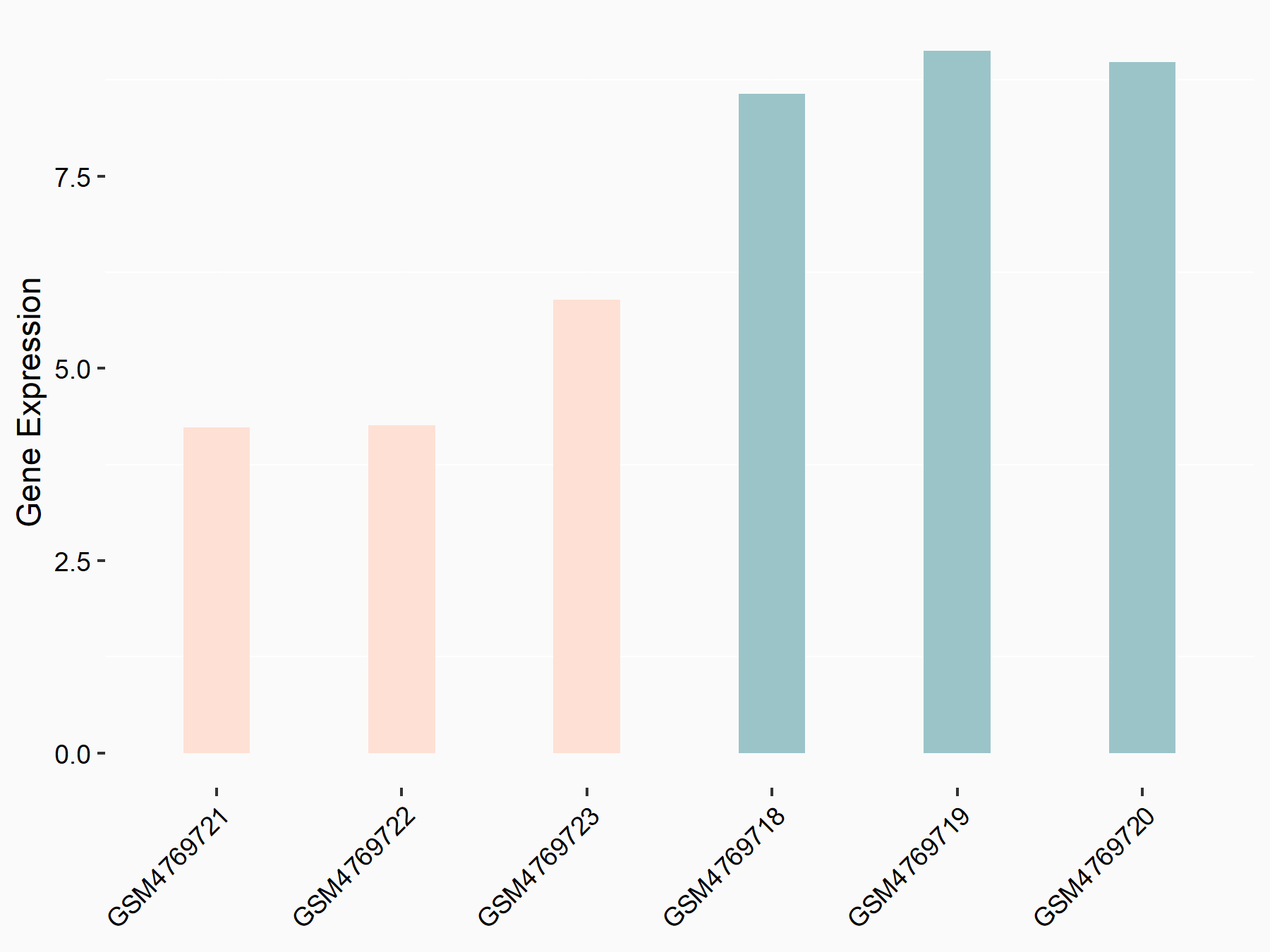 |
logFC: -7.83E-01 p-value: 2.96E-03 |
| More Results | Click to View More RNA-seq Results | |
Atherosclerosis [ICD-11: BD40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [85] | |||
| Responsed Disease | Atherosclerosis [ICD-11: BD40.Z] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
MAEC | Normal | Mus musculus | CVCL_U411 |
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
| Response Summary | In the in vivo atherosclerosis model,partial ligation of the carotid artery led to plaque formation and up-regulation of METTL3 and NACHT/LRR/PYD domains-containing protein 1 (NLRP1), with down-regulation of KLF4; knockdown of METTL3 via repetitive shRNA administration prevented the atherogenic process, NLRP3 up-regulation, and KLF4 down-regulation. Collectively, it has demonstrated that METTL3 serves a central role in the atherogenesis induced by oscillatory stress and disturbed blood flow. | |||
NAD-dependent protein deacetylase sirtuin-1 (SIRT1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Embryonic stem cells | Mus musculus |
|
Treatment: METTL3 knockout mESCs
Control: Wild type mESCs
|
GSE156481 | |
| Regulation |
 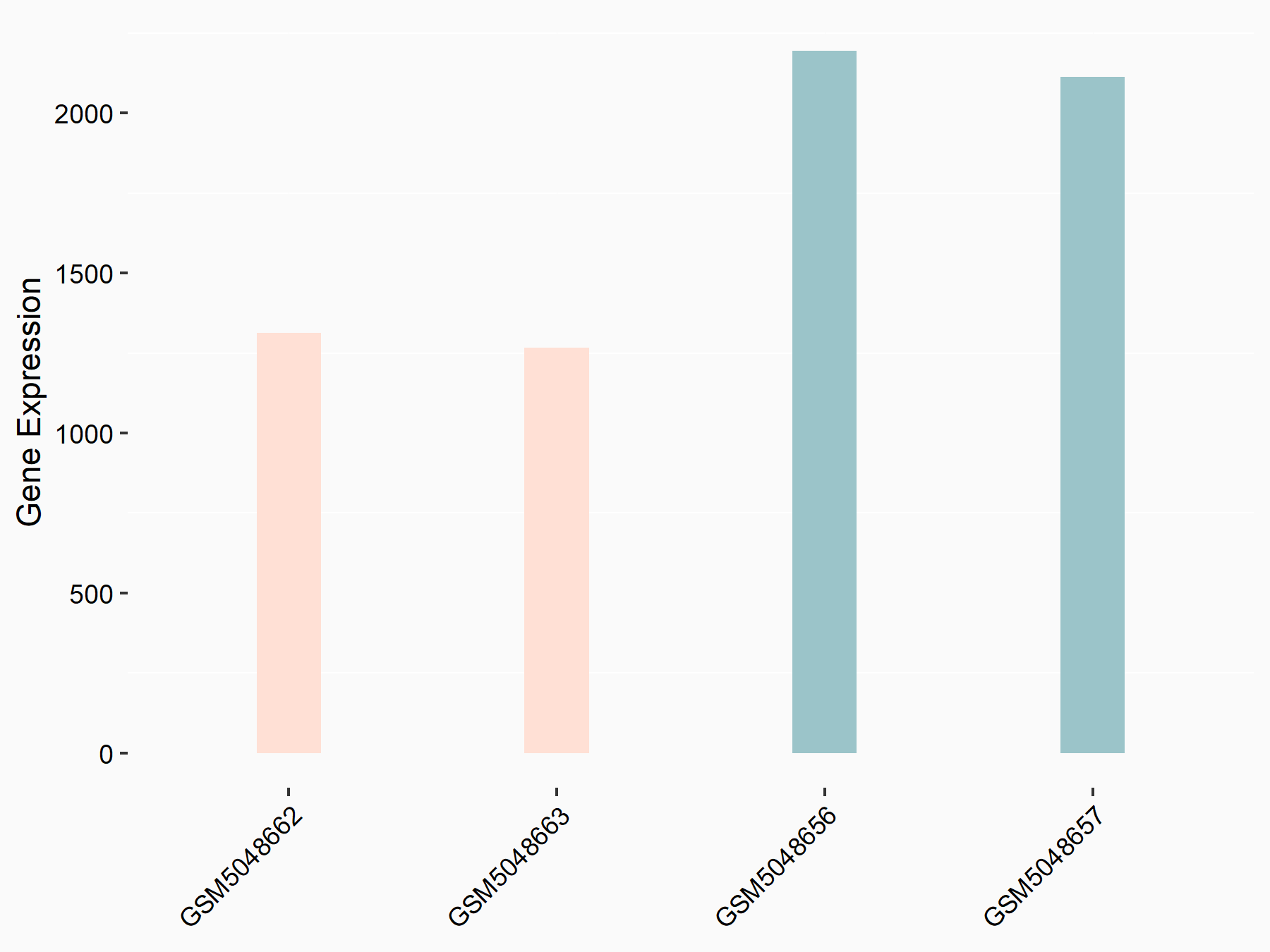 |
logFC: -7.40E-01 p-value: 1.00E-18 |
| More Results | Click to View More RNA-seq Results | |
Type 2 diabetes mellitus [ICD-11: 5A11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [48] | |||
| Responsed Disease | Type 2 diabetes mellitus [ICD-11: 5A11] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Insulin resistance | hsa04931 | ||
| Cell Process | Lipid metabolism | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | Hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were generated by crossing mice with TBG-Cre Tg mice. METTL3 flox (METTL3 fl/fl) and hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were used for experiments. | |||
| Response Summary | Type 2 diabetes (T2D) is characterized by lack of insulin, insulin resistance and high blood sugar. METTL3 silence decreased the m6A methylated and total mRNA level of Fatty acid synthase (Fasn), subsequently inhibited fatty acid metabolism. The expression of Acc1, Acly, Dgat2, Ehhadh, Fasn, Foxo, Pgc1a and NAD-dependent protein deacetylase sirtuin-1 (SIRT1), which are critical to the regulation of fatty acid synthesis and oxidation were dramatically decreased in livers of hepatocyte-specific METTL3 knockout mice. | |||
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [199] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Chondrocytes (Chondrocytes were isolated from human cartilage and cultured) | |||
| Response Summary | m6A-mediated LINC00680 regulates the proliferation and ECM degradation of chondrocytes through LINC00680/m6A/NAD-dependent protein deacetylase sirtuin-1 (SIRT1) mRNA axis. METTL3-mediated LINC00680 accelerates osteoarthritis(OA) progression, which provides novel understanding of the role of m6A and lncRNA in OA. | |||
Cataract [ICD-11: 9B10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [318] | |||
| Responsed Disease | Diabetic cataract [ICD-11: 9B10.21] | |||
In-vitro Model |
B-3 | Normal | Homo sapiens | CVCL_6367 |
| In-vivo Model | Male C57BL/6 mice (22-25 g; Animal Center of the Second Affiliated Hospital of Harbin Medical University) were housed under a 12-h light/dark cycle at a temperature of 22-24 °C, humidity between 50 ± 10%, and with ad libitum access to food and water. The mice were acclimatized for at least 1 week before the experiments. Type 1 DM was induced by intraperitoneally injecting streptozotocin (STZ) at a dose of 55 mg/kg for 5 days consecutively. Mice that received STZ injections were considered diabetic when their blood glucose levels consistently surpassed 16.7 mmol/L (300 mg/dL) for two days consecutively. Diabetic mice were used 4 months post-last STZ injection, with blood glucose levels above 16.7 mmol/L. Any mice that did not exhibit DM development were eliminated from the experiment. The control group of mice (CON) was administered a comparable dosage of citric acid buffer .Four weeks after successful STZ induction, AAV2sh-METTL3 (2 μL; 1 × 1013 vg/mL; WZ Biosciences Inc., China) was injected into the vitreous humor of the right eyes of these mice using a 30-gauge needle and a microinjector (Hamilto Sigma-Aldrich, St.Louis, MO, USA), forming the 'STZ + AAV2sh-METTL3' group. Mice injected with AAV2sh-NC were designated as the 'STZ + AAV2sh-NC' group. At the same time, the left eye was administered 2μL saline and was designated as the 'STZ' group. The eyes of control mice were administered the same saline dosage and were designated as the 'Control-sham' group. Mice were euthanized at 4 months, and their eyes were removed. The Second Affiliated Hospital Ethics Committee of Harbin Medical University authorized the animal experiments (YJSDW2023-088). | |||
Presbycusis [ICD-11: AB54]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [319] | |||
| Responsed Disease | Presbycusis [ICD-11: AB54] | |||
| Target Regulation | Up regulation | |||
Endometriosis [ICD-11: GA10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [320] | |||
| Responsed Disease | Endometriosis [ICD-11: GA10] | |||
| Target Regulation | Down regulation | |||
| In-vivo Model | For in vivo studies, the shMETTL3 sequence was subcloned into adeno-associated virus (AAV) construct 9 (AAV9). Recombinant AAV9 was manufactured by GenePharma (Shanghai, China). The ectopic endometrium was obtained from a 28-year-old woman who had undergone an operation for ovarian endometriosis. The tissue sample was washed twice with PBS and cut into three 3- to 5-mm pieces, and suspended by PBS.he animals were administered intraperitoneal injections of 30 mg/kg 17-oestradiol every three days after the endometrial injections. Xenografts were generated under bilateral axillae after 14 days with tumour volumes of approximately 10 mm3. | |||
Neurogenic locus notch homolog protein 1 (NOTCH1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
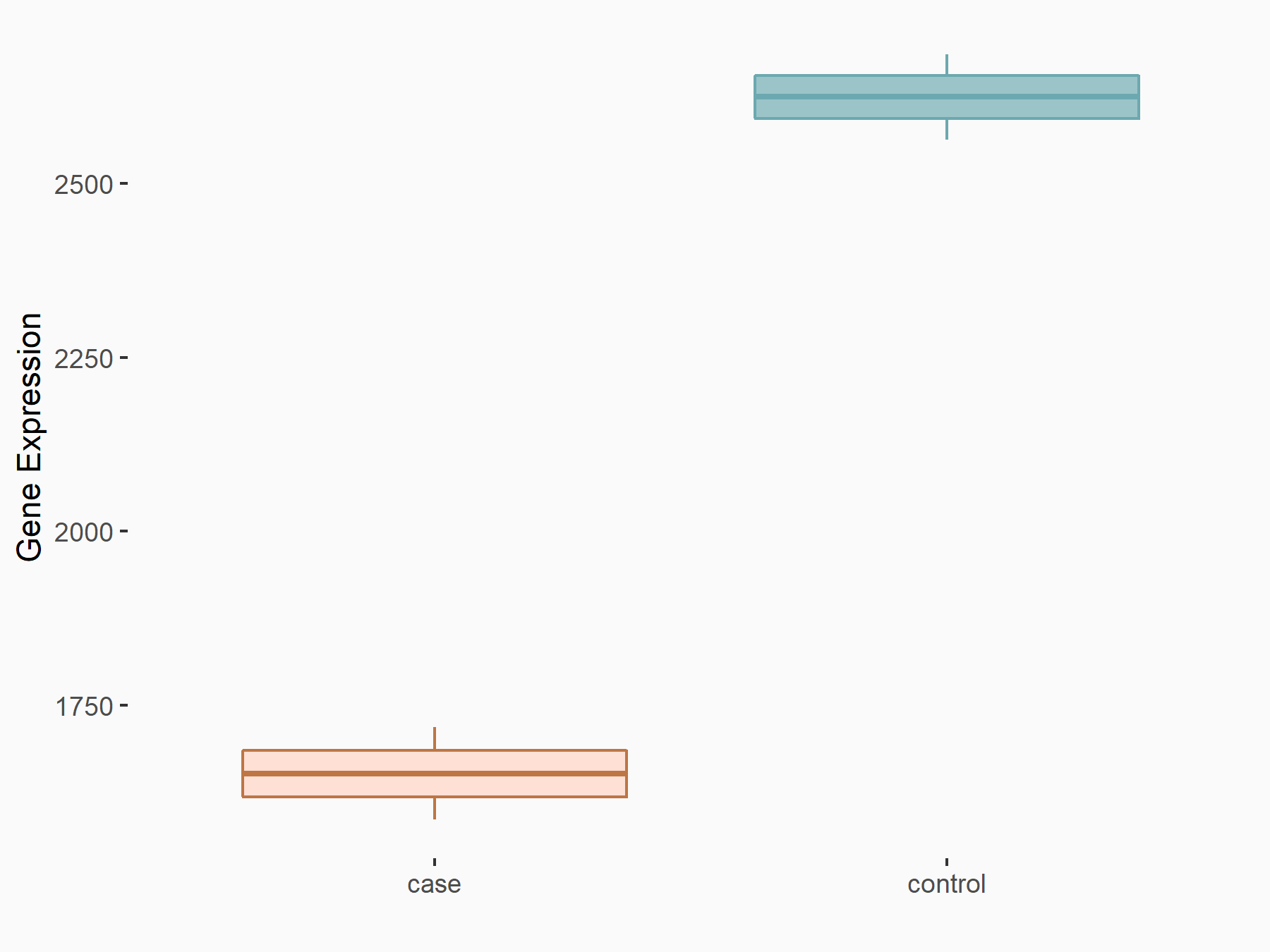 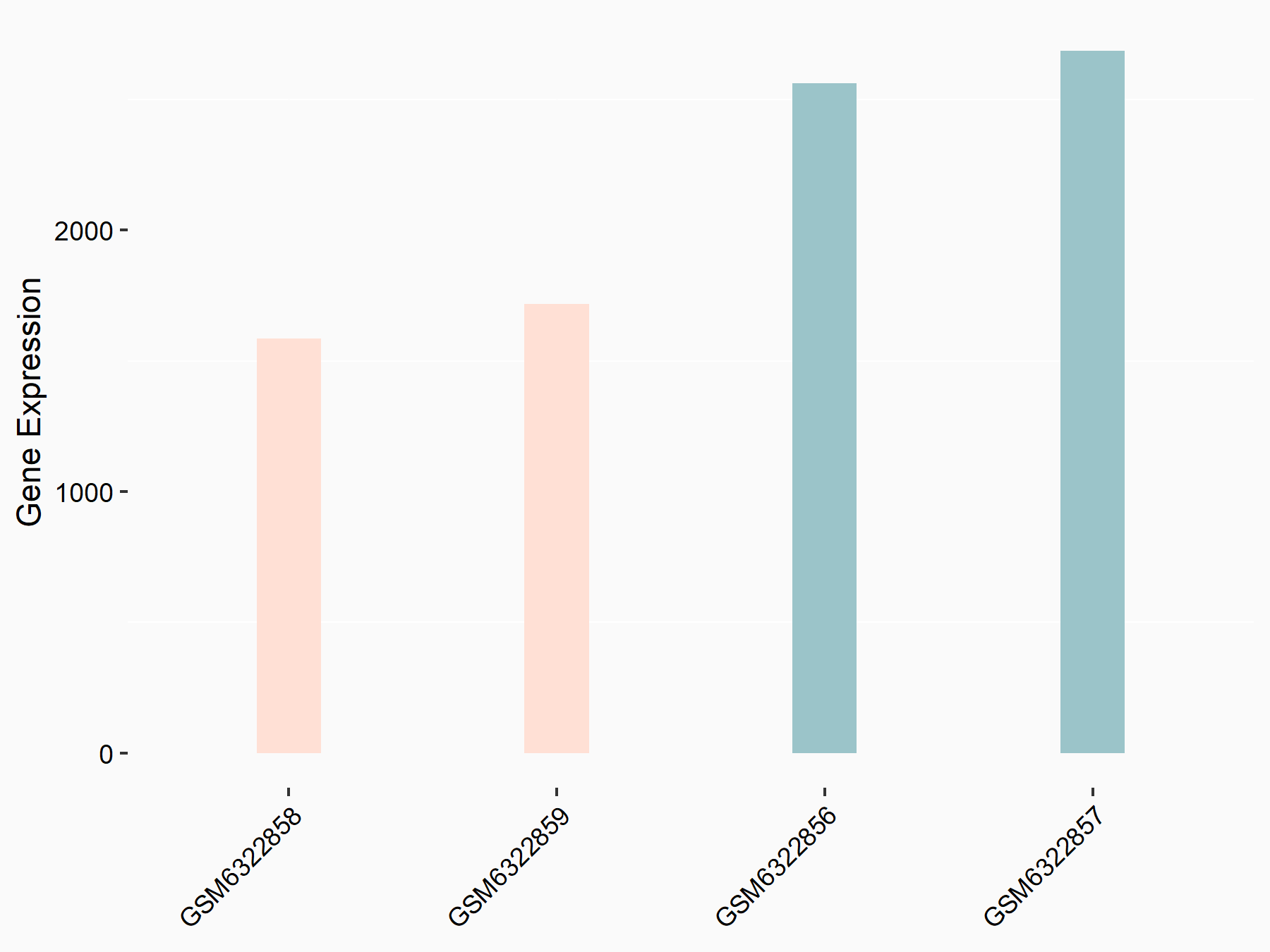 |
logFC: -6.68E-01 p-value: 1.32E-12 |
| More Results | Click to View More RNA-seq Results | |
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [200] | |||
| Responsed Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.1] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Notch signaling pathway | hsa04330 | ||
| Cell Process | Cell migration | |||
| Cell invasion | ||||
In-vitro Model |
TE-9 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1767 |
| KYSE-30 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1351 | |
| In-vivo Model | For induction of ESCC, 4-week-old mice were treated with drinking water containing 50 ug/mL 4NQO (Sigma-Aldrich, USA) for 16 weeks and then given normal drinking water for another 4-5 weeks. Cre was activated by the intraperitoneal injection of tamoxifen (Sigma-Aldrich, USA) at a dose of 9 mg per 40 g body weight every other day for a total of three injections. For tumor measurement, mice were sacrificed, and the esophagus was dissected immediately. The surface areas of tumors were measured as described previously. | |||
| Response Summary | METTL3-catalyzed m6A modification promotes Neurogenic locus notch homolog protein 1 (NOTCH1) expression and the activation of the Notch signaling pathway. Forced activation of Notch signaling pathway successfully rescues the growth, migration, and invasion capacities of METTL3-depleted ESCC cells. | |||
Aortic aneurysm or dissection [ICD-11: BD50]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [321] | |||
| Responsed Disease | Thoracic aortic dissection [ICD-11: BD50.3] | |||
| Target Regulation | Down regulation | |||
Neurogenic locus notch homolog protein 2 (NOTCH2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
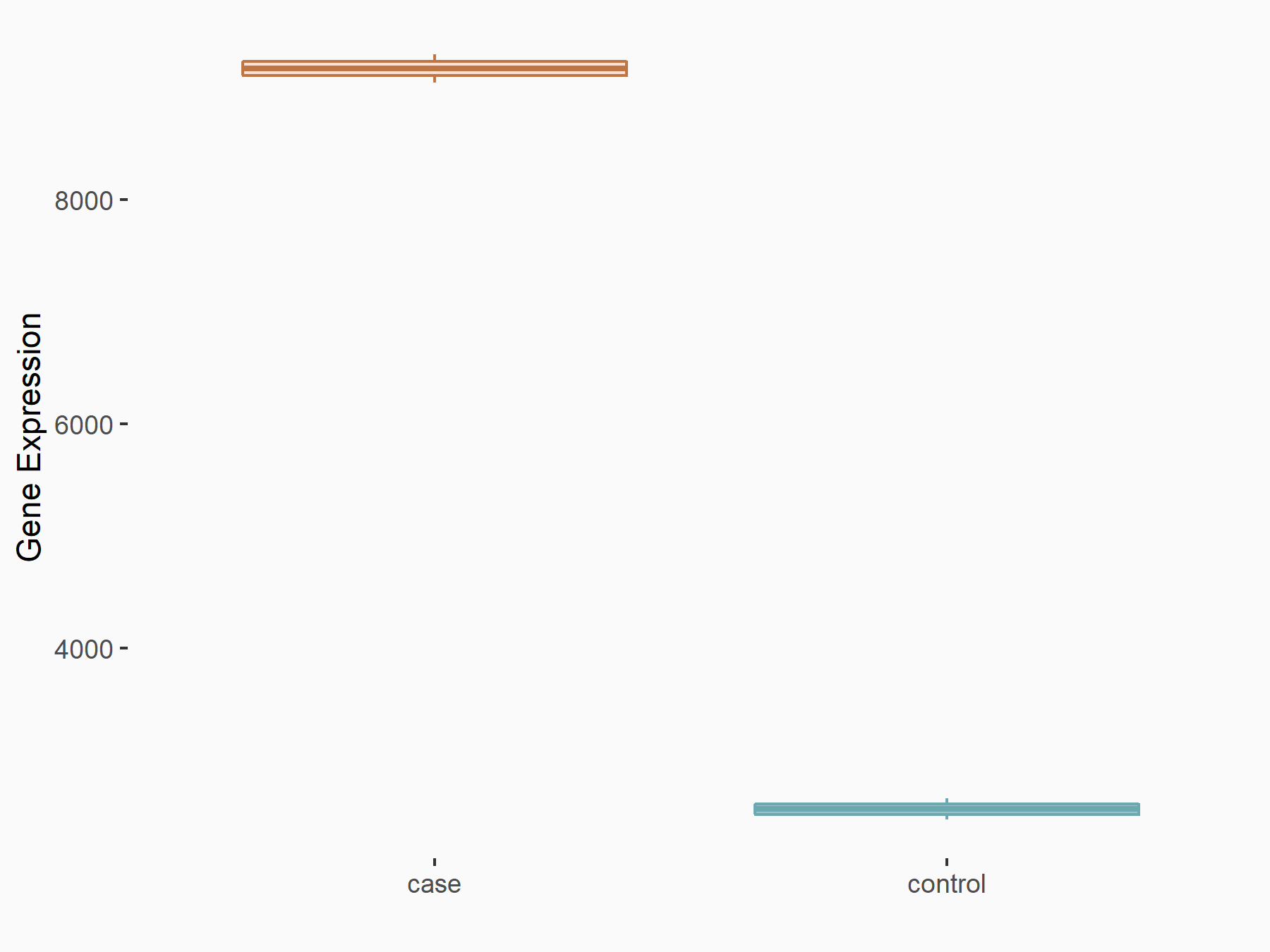 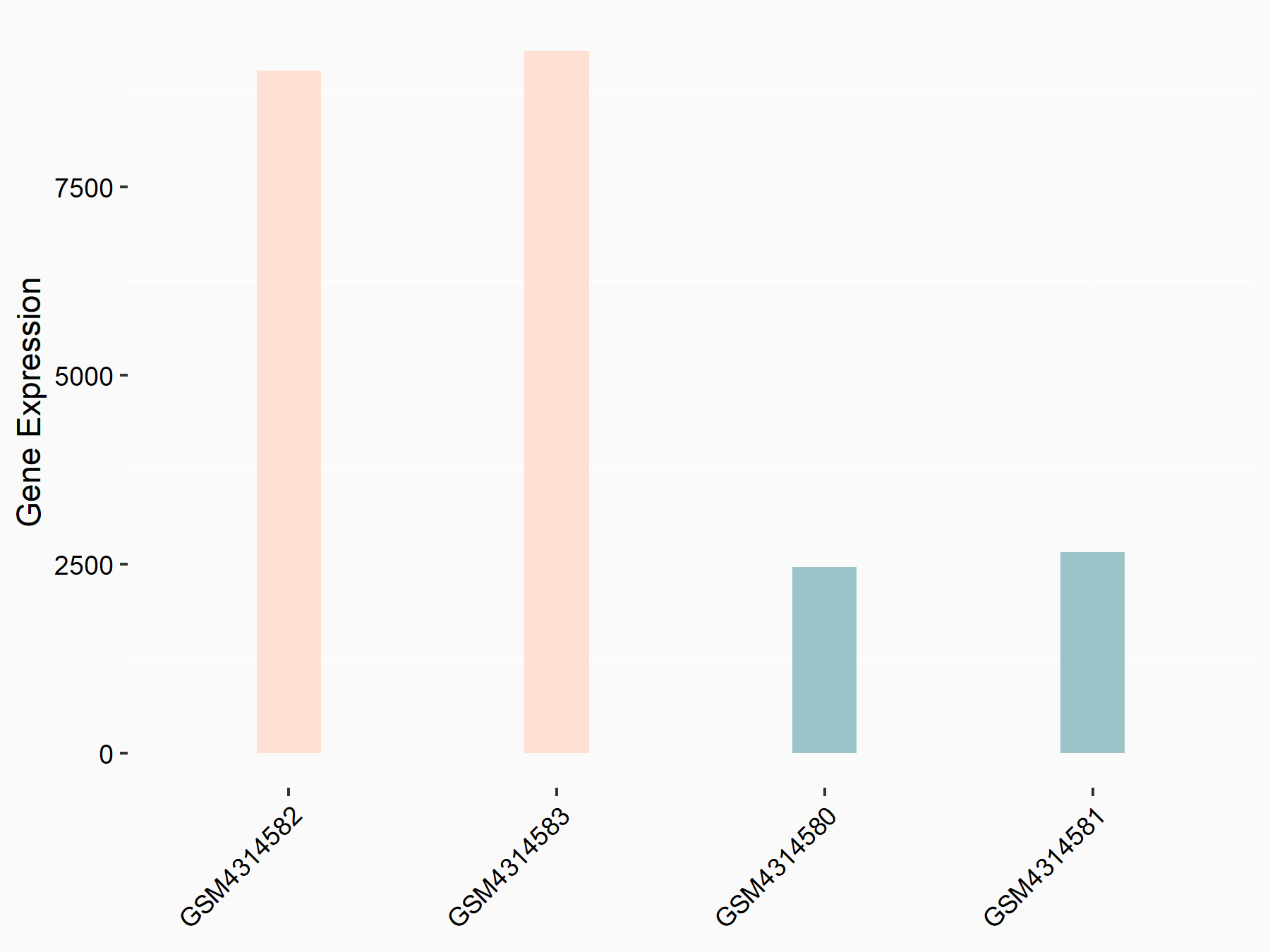 |
logFC: 1.84E+00 p-value: 2.61E-238 |
| More Results | Click to View More RNA-seq Results | |
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [201] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Responsed Drug | Melittin | Investigative | ||
| Target Regulation | Down regulation | |||
| Cell Process | miRNA maturation | |||
| Cell apoptosis | ||||
In-vitro Model |
T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| EJ (Human bladder cancer cells) | ||||
| BIU-87 | Human bladder cancer cells | Homo sapiens | CVCL_6881 | |
| In-vivo Model | For melittin treatment study, 4-week-old female BALB/c nude mice were subcutaneously injected with 1 × 107 T24 or BIU87 cells. | |||
| Response Summary | METTL3 acts as a fate determinant that controls the sensitivity of bladder cancer cells to melittin treatment. Moreover, METTL3/miR-146a-5p/NUMB/Neurogenic locus notch homolog protein 2 (NOTCH2) axis plays an oncogenic role in bladder cancer pathogenesis and could be a potential therapeutic target for recurrent bladder cancer treatment. | |||
Nuclear factor erythroid 2-related factor 2 (NFE2L2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
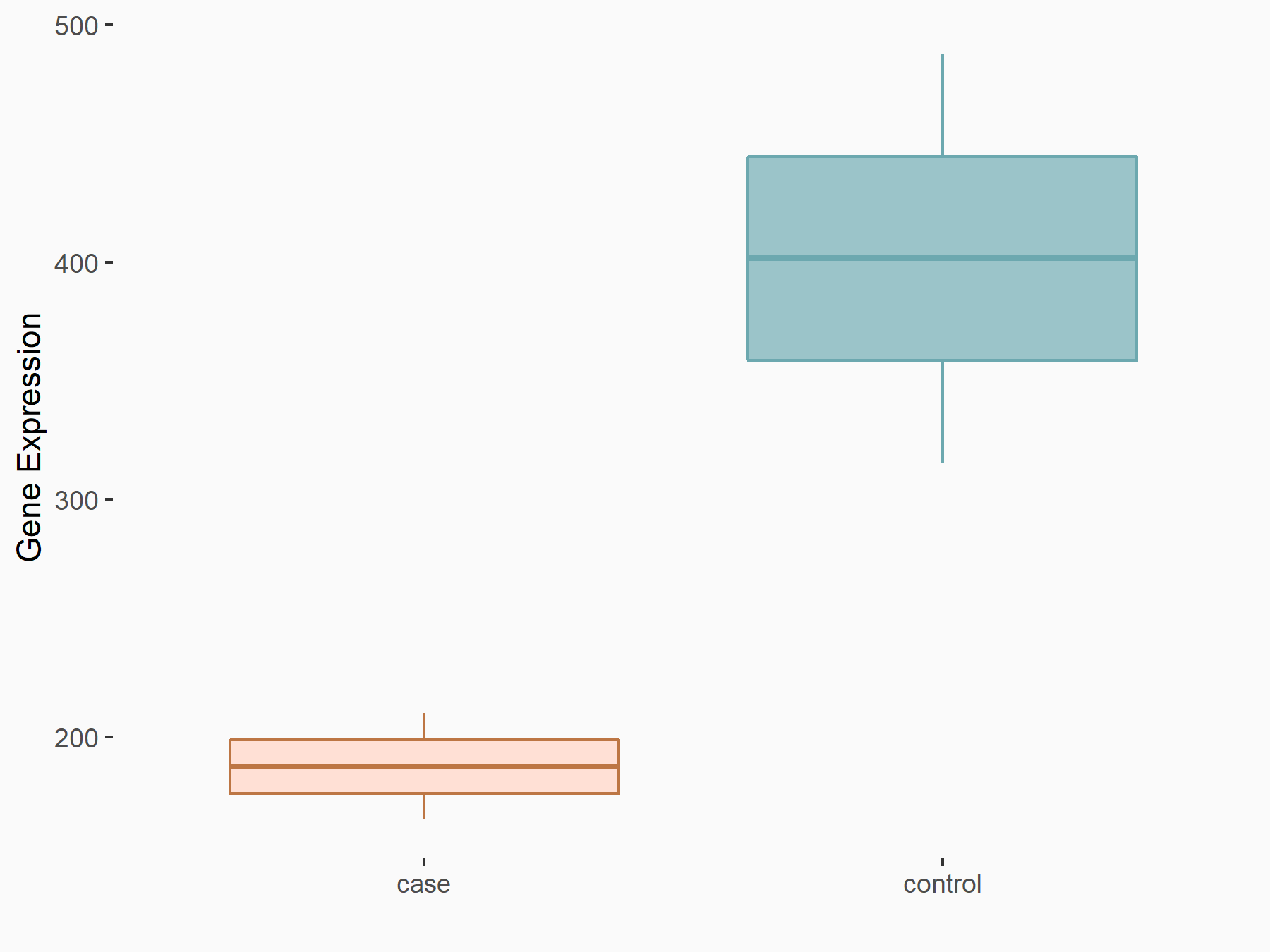 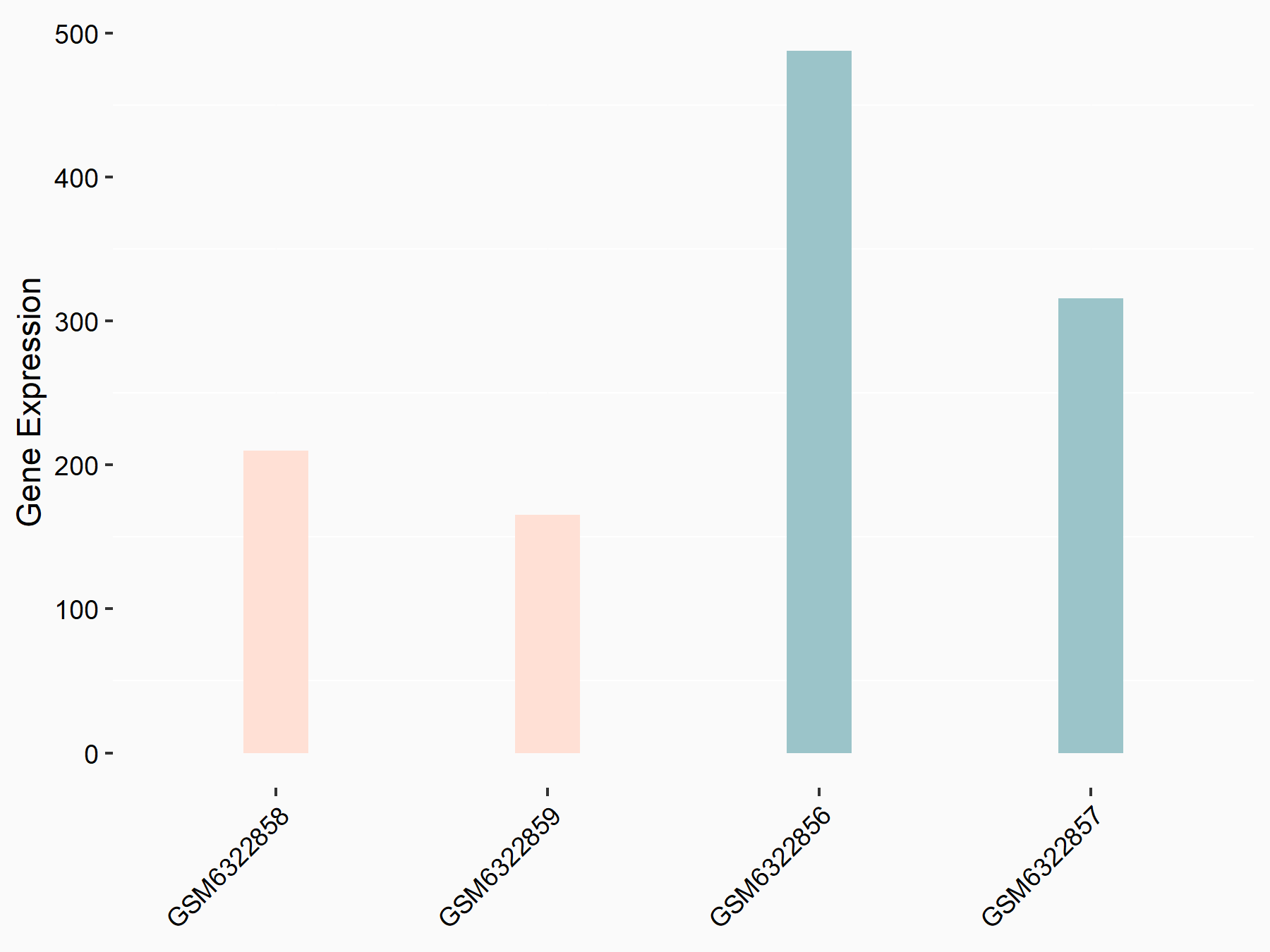 |
logFC: -1.10E+00 p-value: 4.72E-06 |
| More Results | Click to View More RNA-seq Results | |
Diseases of the urinary system [ICD-11: GC2Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [65] | |||
| Responsed Disease | Diseases of the urinary system [ICD-11: GC2Z] | |||
| Responsed Drug | Colistin | Approved | ||
| Target Regulation | Up regulation | |||
| Cell Process | Oxidative stress | |||
| Cell apoptosis | ||||
| In-vivo Model | The 60 female Kunming mice were divided into two groups (n = 30): control group (injection of physiological saline through the caudal vein) and colistin group (injection of 15 mg/kg colistin, twice a day, with an eight-hour interval). | |||
| Response Summary | m6A methylation was involved in oxidative stress-mediated apoptosis in the mechanism of colistin nephrotoxicity. METTL3-mediated m6A methylation modification is involved in colistin-induced nephrotoxicity through apoptosis mediated by Keap1/Nuclear factor erythroid 2-related factor 2 (NFE2L2) signaling pathway. | |||
Nuclear factor NF-kappa-B p105 subunit (NF-Kappa-B/NFKB1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
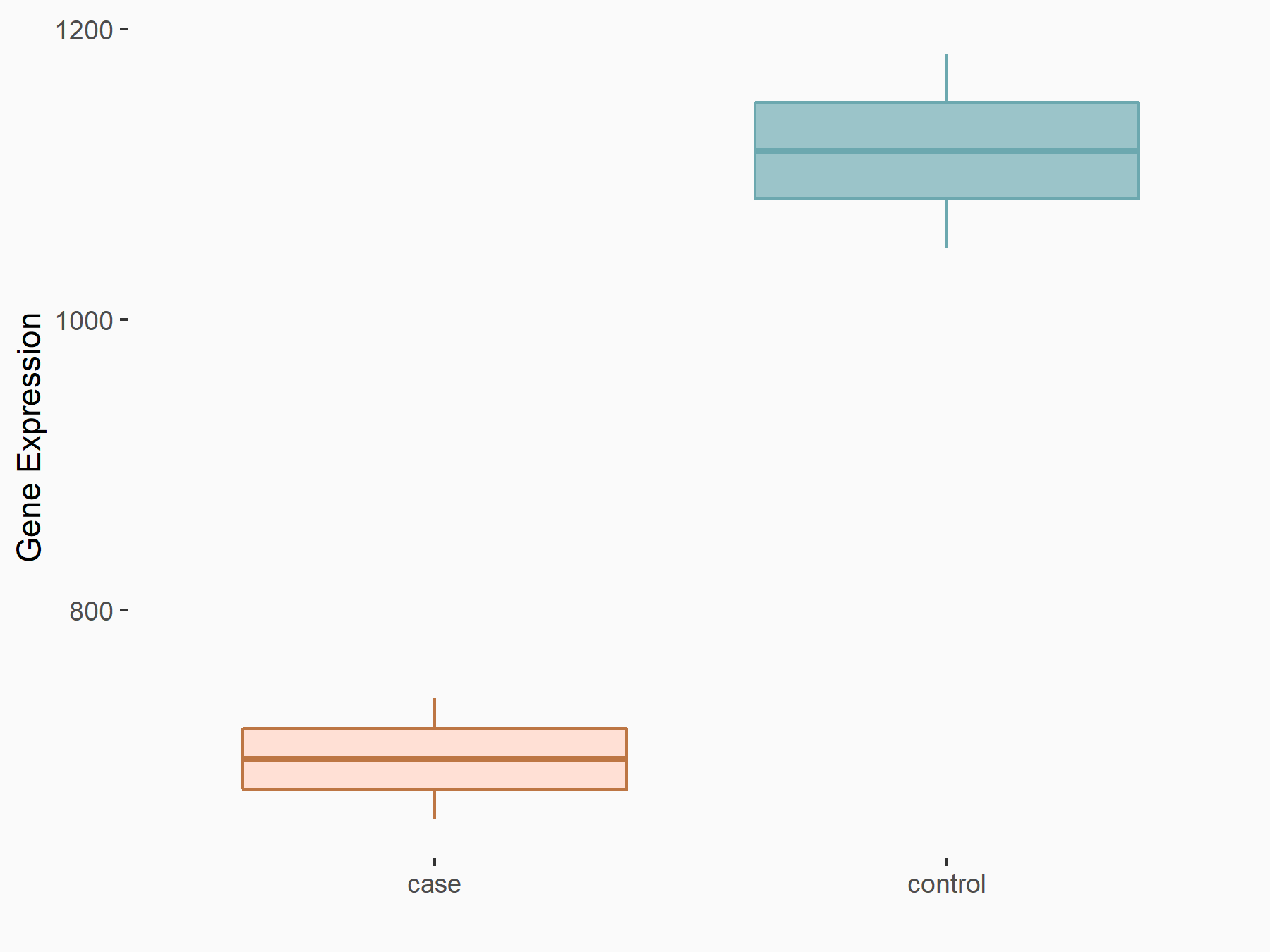 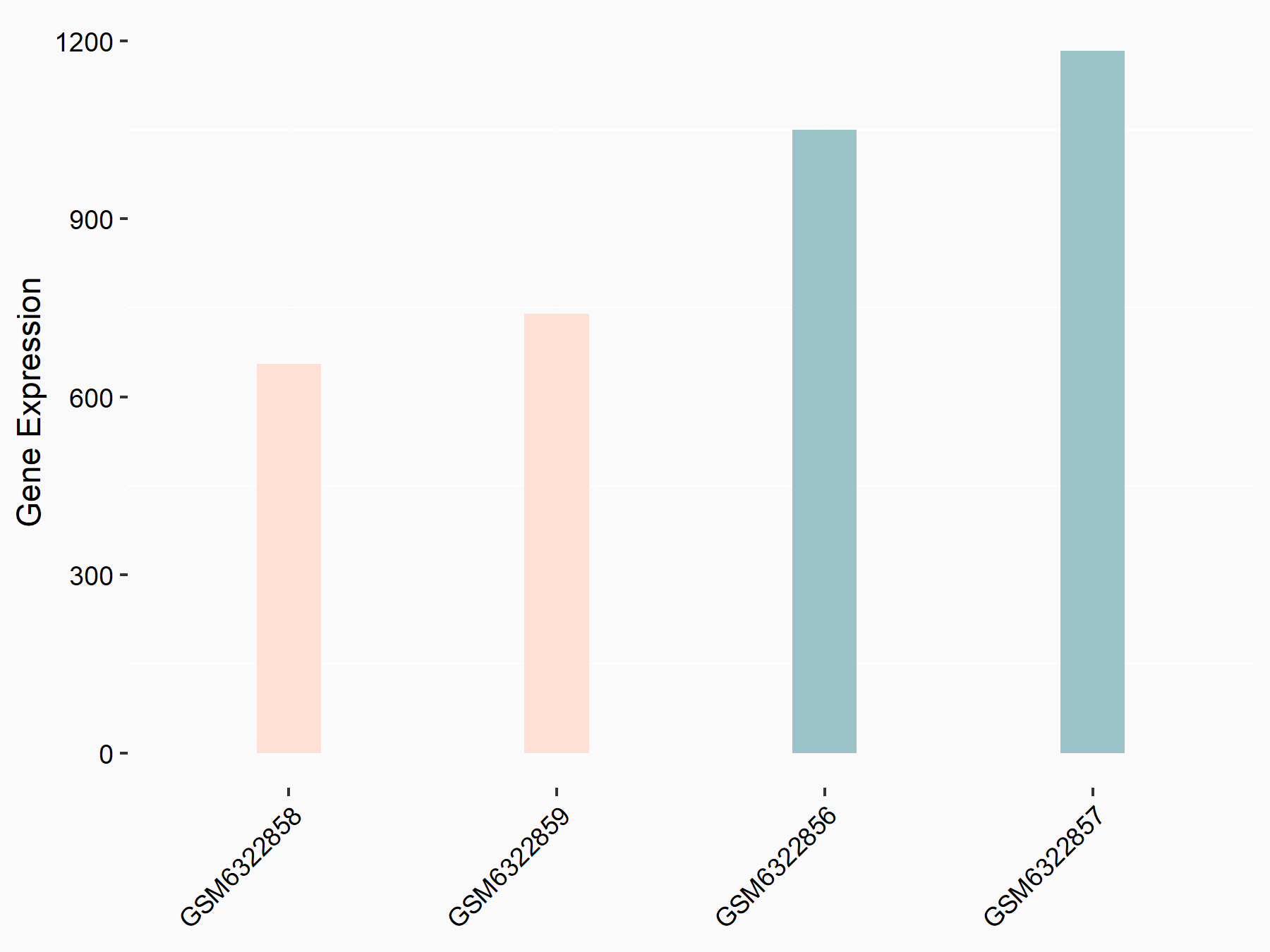 |
logFC: -6.78E-01 p-value: 1.44E-07 |
| More Results | Click to View More RNA-seq Results | |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [202] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | NF-kappa B signaling pathway | hsa04064 | ||
In-vitro Model |
U87 (A primary glioblastoma cell line) | |||
| N33 (The GBM patient-derived cell line) | ||||
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| H4 | Astrocytoma | Homo sapiens | CVCL_1239 | |
| In-vivo Model | Five-week-old female BALB/c nude mice (Charles Rivers, Beijing, China) were selected for the experiments. U87 cells (5 × 105) transfected with an empty vector, YTHDF2 overexpression, or METTL3 overexpression vectors were suspended in PBS and injected into the right frontal node of nude mice. The inoculation position was 2 mm lateral and 2 mm posterior to the anterior fontanel. Tumor size was estimated from luciferase volume measurements and MRI. The mice were sacrificed when they exhibited disturbed activity or convulsion. The brain was then harvested and embedded in paraffin. | |||
| Response Summary | YTHDF2 accelerated UBXN1 mRNA degradation via METTL3-mediated m6A, which, in turn, promoted Nuclear factor NF-kappa-B p105 subunit (NF-Kappa-B/NFKB1) activation. YTHDF2 promotes the malignant progression of gliomas and revealed important insight into the upstream regulatory mechanism of NF-Kappa-B activation via UBXN1 with a primary focus on m6A modification. | |||
Obesity [ICD-11: 5B81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [110] | |||
| Responsed Disease | Obesity [ICD-11: 5B81] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| HIF-1 signaling pathway | hsa04066 | |||
| In-vivo Model | The 8-10 weeks old mice were fed either a high fat diet or HF-CDAA , ad lib for 6-12 weeks. Chow diet was used as control for HFD.The mouse liver was perfused with PBS through portal vein, and liver tissue was cut into small pieces by a scissor. The single cell was made using syringe plunger to mull the tissue, and passed through a 40 uM cell strainer. | |||
| Response Summary | The contribution of METTL3-mediated m6A modif ication of Ddit4 mRNA to macrophage metabolic reprogramming in non-alcoholic fatty liver disease and obesity. In METTL3-deficient macrophages, there is a significant downregulation of mammalian target of rapamycin (mTOR) and Nuclear factor NF-kappa-B p105 subunit (NF-Kappa-B/NFKB1) pathway activity in response to cellular stress and cytokine stimulation, which can be restored by knockdown of DDIT4. | |||
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [203] | |||
| Responsed Disease | Nonalcoholic steatohepatitis [ICD-11: DB92.1] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
KCs (Mouse Kupffer cells (BeNa Culture Collection, Beijing, China; BNCC340733)) | |||
| In-vivo Model | At 8 weeks of age, METTL14 cKO and WT mice were challenged with LPS (Sigma-Aldrich, St. Louis, MO; L2880, single intraperitoneal injection at 5 mg/kg, n = 3) or CCl4 (10%, Macklin, Shanghai, China; C805332, intraperitoneal injection at 5 mL/kg diluted with corn oil, twice per week for 4 weeks, n = 3). The corresponding control groups were treated with single intraperitoneal injection of saline (n = 3) or intraperitoneal injection of corn oil twice per week for 4 weeks (n = 3), respectively. Two hours after LPS injection and 4 weeks after CCl4 treatment, METTL14 cKO and WT mice were etherized and the primary KCs were isolated from liver according to a previously published method. | |||
| Response Summary | Nuclear factor NF-kappa-B p105 subunit (NF-Kappa-B/NFKB1) acts as transcription factor to transactivate METTL3/METTL14 genes upon LPS challenge, leading to global RNA m6A hypermethylation. m6A modification in TGF-beta1 upregulation, which helps to shed light on the molecular mechanism of nonalcoholic steatohepatitis(NASH) progression. | |||
Nucleobindin-1 (NUCB1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | DKO-1 cell line | Homo sapiens |
|
Treatment: METTL3 knockdown DKO-1 cell
Control: DKO-1 cell
|
GSE182382 | |
| Regulation |
 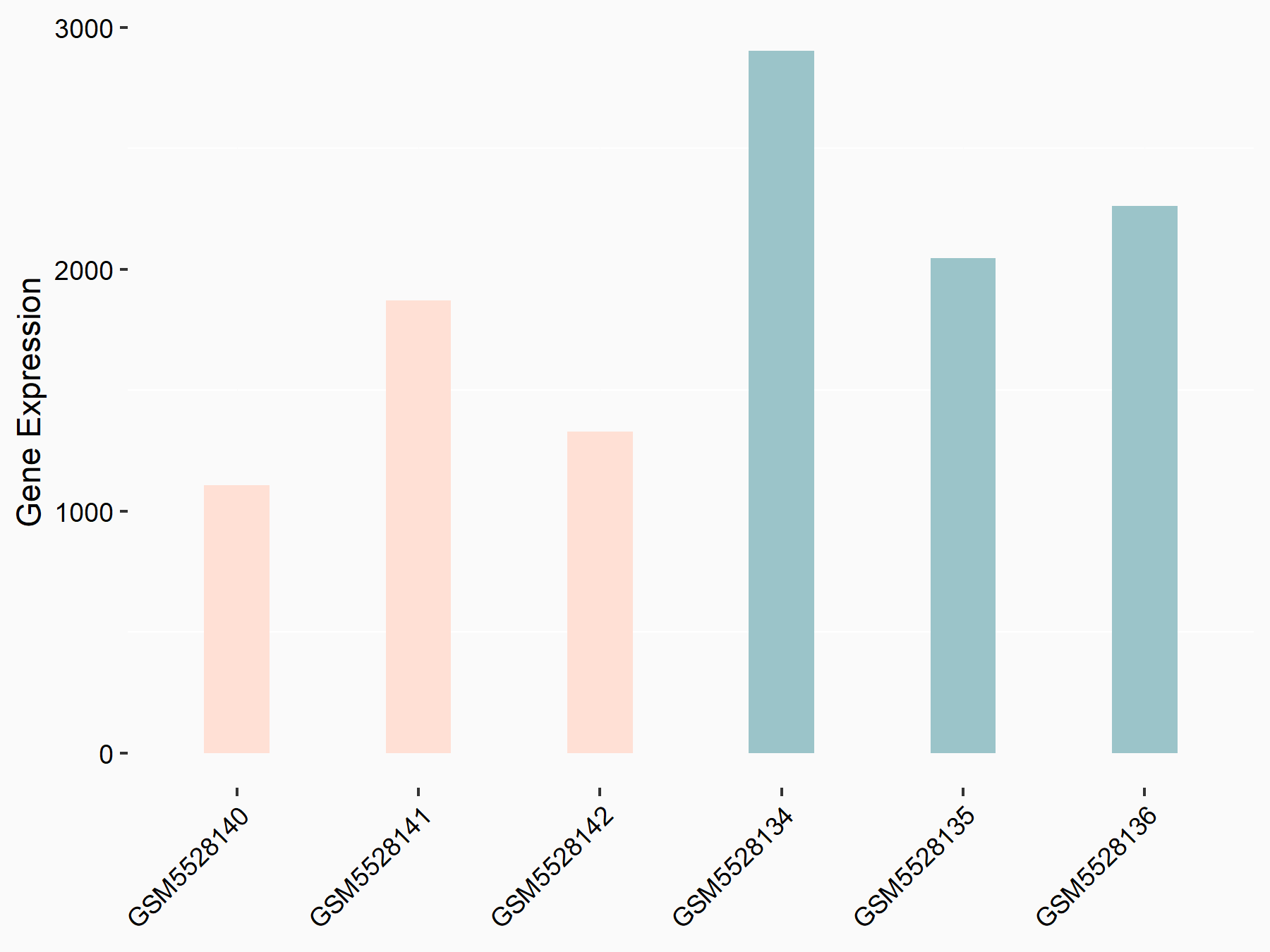 |
logFC: -7.45E-01 p-value: 2.41E-03 |
| More Results | Click to View More RNA-seq Results | |
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [204] | |||
| Responsed Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Responsed Drug | Gemcitabine | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell proliferation | |||
| Cell autophagy | ||||
In-vitro Model |
SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| In-vivo Model | 5 × 106 SW1990 cells expressing NUCB1 (oeNUCB1) or control vector (oeNC) were injected subcutaneously. | |||
| Response Summary | METTL3-mediated m6A modification on Nucleobindin-1 (NUCB1) 5'UTR via the reader YTHDF2 as a mechanism for NUCB1 downregulation in PDAC. This study revealed crucial functions of NUCB1 in suppressing proliferation and enhancing the effects of gemcitabine in pancreatic cancer cells. | |||
Nucleolar protein 3 (ARC)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Embryonic stem cells | Mus musculus |
|
Treatment: METTL3 knockout ESCs
Control: Wild type ESCs
|
GSE146466 | |
| Regulation |
 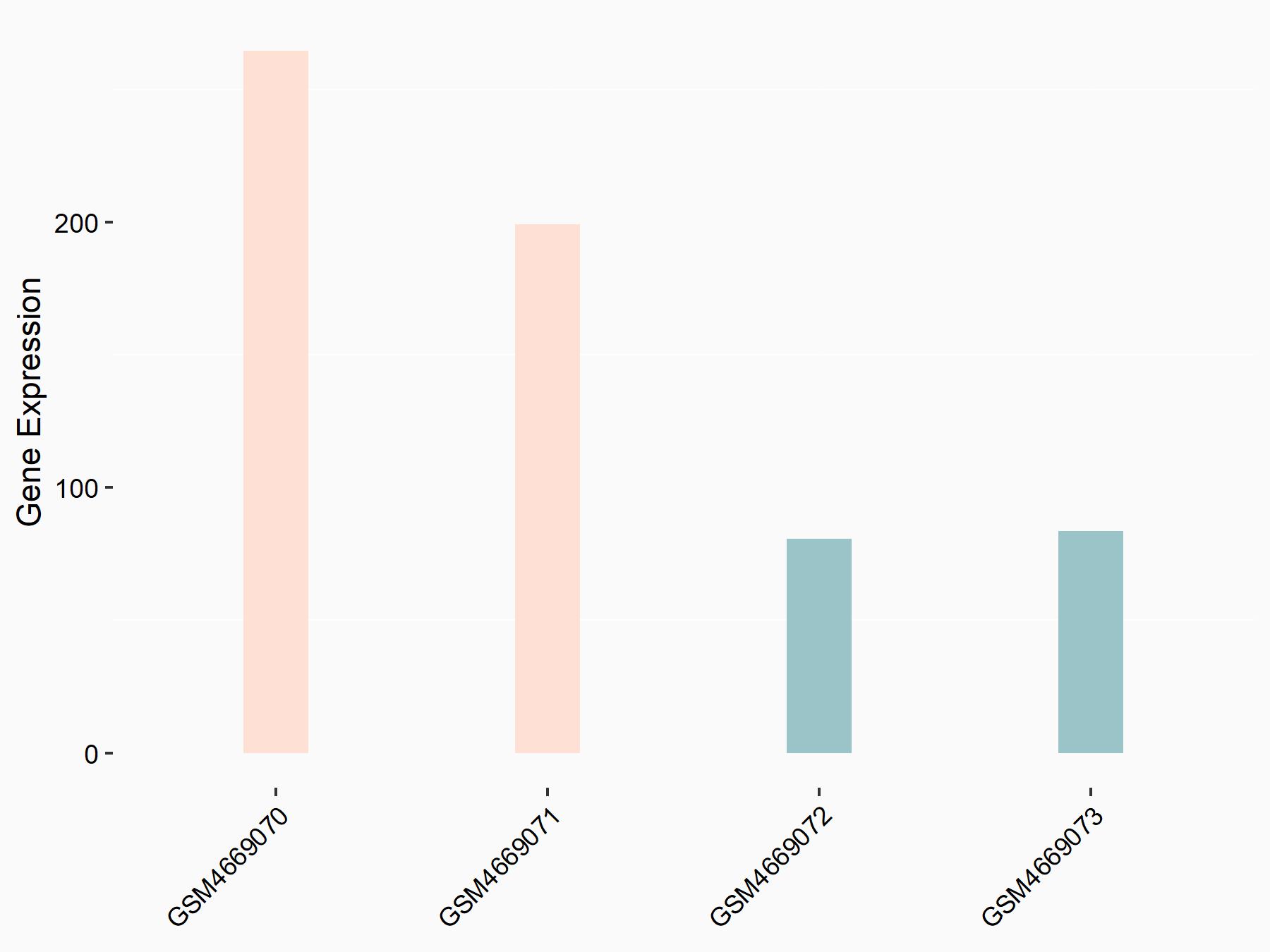 |
logFC: 1.50E+00 p-value: 1.12E-06 |
| More Results | Click to View More RNA-seq Results | |
Alzheimer disease [ICD-11: 8A20]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [205] | |||
| Responsed Disease | Alzheimer disease [ICD-11: 8A20] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
| Response Summary | METTL3 rescues the A-Bete-induced reduction of Nucleolar protein 3 (ARC) expression via YTHDF1-Dependent m6A modification, which suggests an important mechanism of epigenetic alteration in AD. | |||
Oxidative stress-induced growth inhibitor 1 (OSGIN1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3 knockout liver
Control: Wild type liver cells
|
GSE198513 | |
| Regulation |
 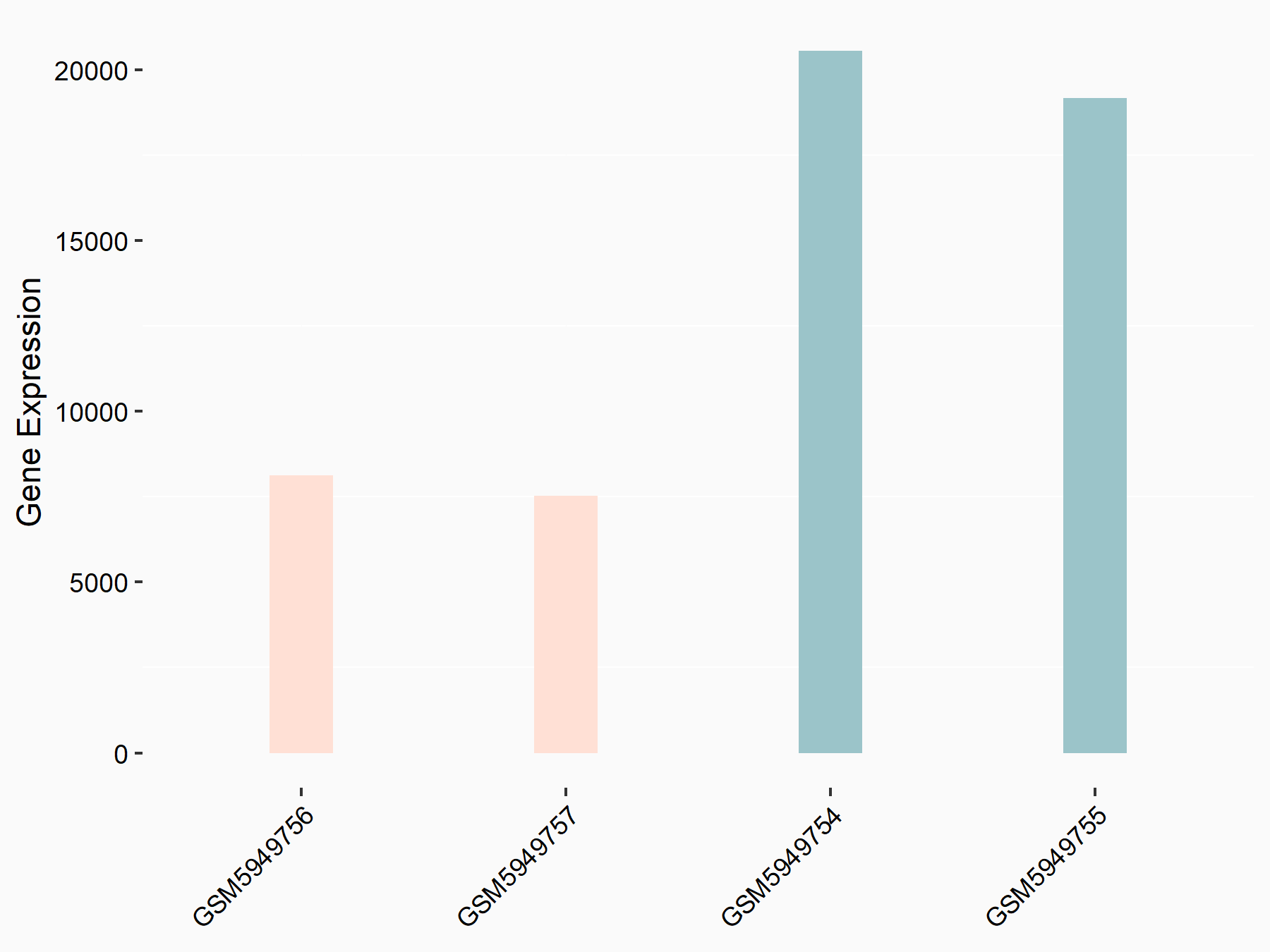 |
logFC: -1.34E+00 p-value: 4.00E-87 |
| More Results | Click to View More RNA-seq Results | |
Trachea cancer [ICD-11: 2C24]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [206] | |||
| Responsed Disease | Trachea cancer [ICD-11: 2C24] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell injury | |||
| Cell apoptosis | ||||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| HBE (Human bronchial epithelial cell line) | ||||
| Response Summary | METTL3 regulates PM 2.5-induced cell injury by targeting Oxidative stress-induced growth inhibitor 1 (OSGIN1) in human airway epithelial cells. | |||
PI3-kinase subunit alpha (PI3k/PIK3CA)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HULEC-5a cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HULEC-5a cells
Control: HULEC-5a cells
|
GSE200649 | |
| Regulation |
 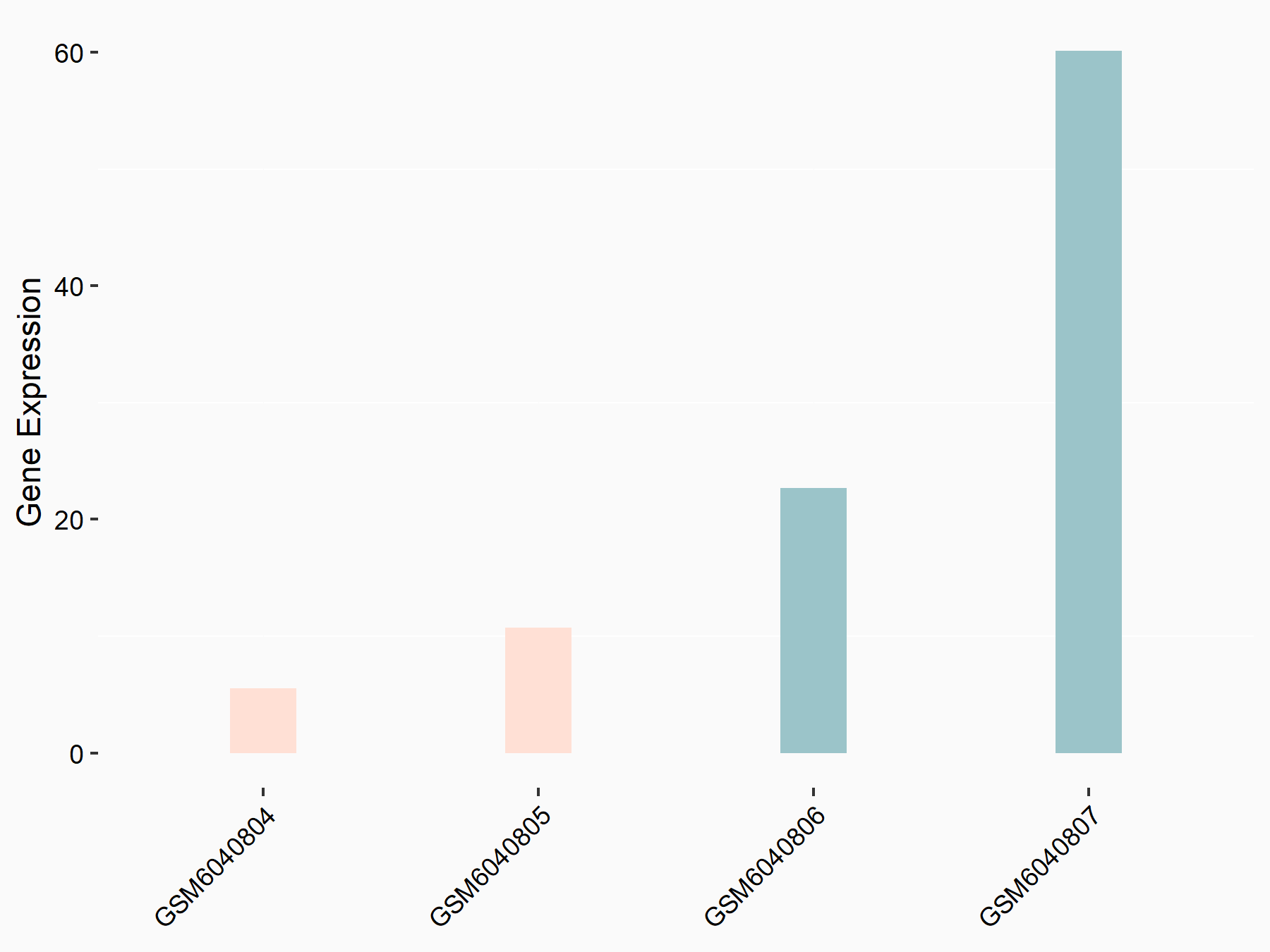 |
logFC: -2.36E+00 p-value: 7.04E-03 |
| More Results | Click to View More RNA-seq Results | |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [207] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell proliferation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| Response Summary | MiR-600 inhibited lung cancer via down-regulating METTL3 expression, and knockdown of METTL3 was used as a novel strategy for lung cancer therapy. The PI3-kinase subunit alpha (PI3k/PIK3CA)/Akt pathway is implicated in cell growth and survival and we also observed that knockdown of METTL3 changed the expression and phosphorylation of proteins of PI3K signaling pathway members. | |||
Renal cell carcinoma [ICD-11: 2C90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [109] | |||
| Responsed Disease | Renal cell carcinoma [ICD-11: 2C90] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Epithelial-to-mesenchymal transition | |||
| Arrest cell cycle at G0/G1 phase | ||||
In-vitro Model |
ACHN | Papillary renal cell carcinoma | Homo sapiens | CVCL_1067 |
| Caki-1 | Clear cell renal cell carcinoma | Homo sapiens | CVCL_0234 | |
| Caki-2 | Papillary renal cell carcinoma | Homo sapiens | CVCL_0235 | |
| HK2 | Normal | Acipenser baerii | CVCL_YE28 | |
| In-vivo Model | Cells (5×106 cells in 200 uL) were suspended with 100 uL PBS and 100 uL Matrigel Matrix, and injected subcutaneously into the left armpit of each mouse. | |||
| Response Summary | Knockdown of METTL3 could obviously promote cell proliferation, migration and invasion function, and induce G0/G1 arrest,METTL3 acts as a novel marker for tumorigenesis, development and survival of RCC. Knockdown of METTL3 promoted changes in PI3-kinase subunit alpha (PI3k/PIK3CA)/AKT/mTOR markers' expression with a gain in p-PI3k, p-AKT, p-mTOR and p-p70, and a loss of p-4EBP1. | |||
Poly [ADP-ribose] polymerase 1 (PARP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shMETTL3 MOLM13 cells
Control: MOLM13 cells
|
GSE98623 | |
| Regulation |
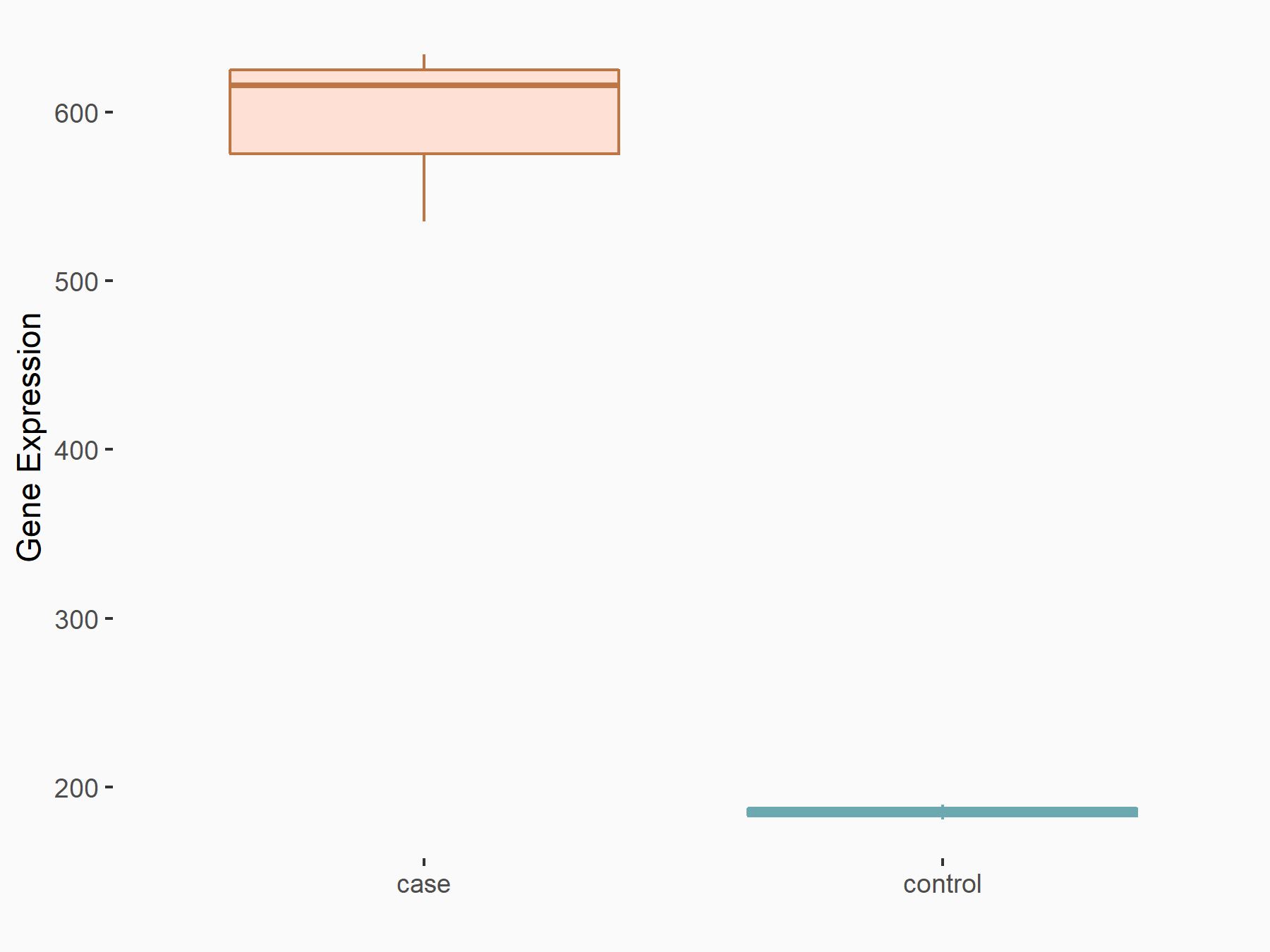 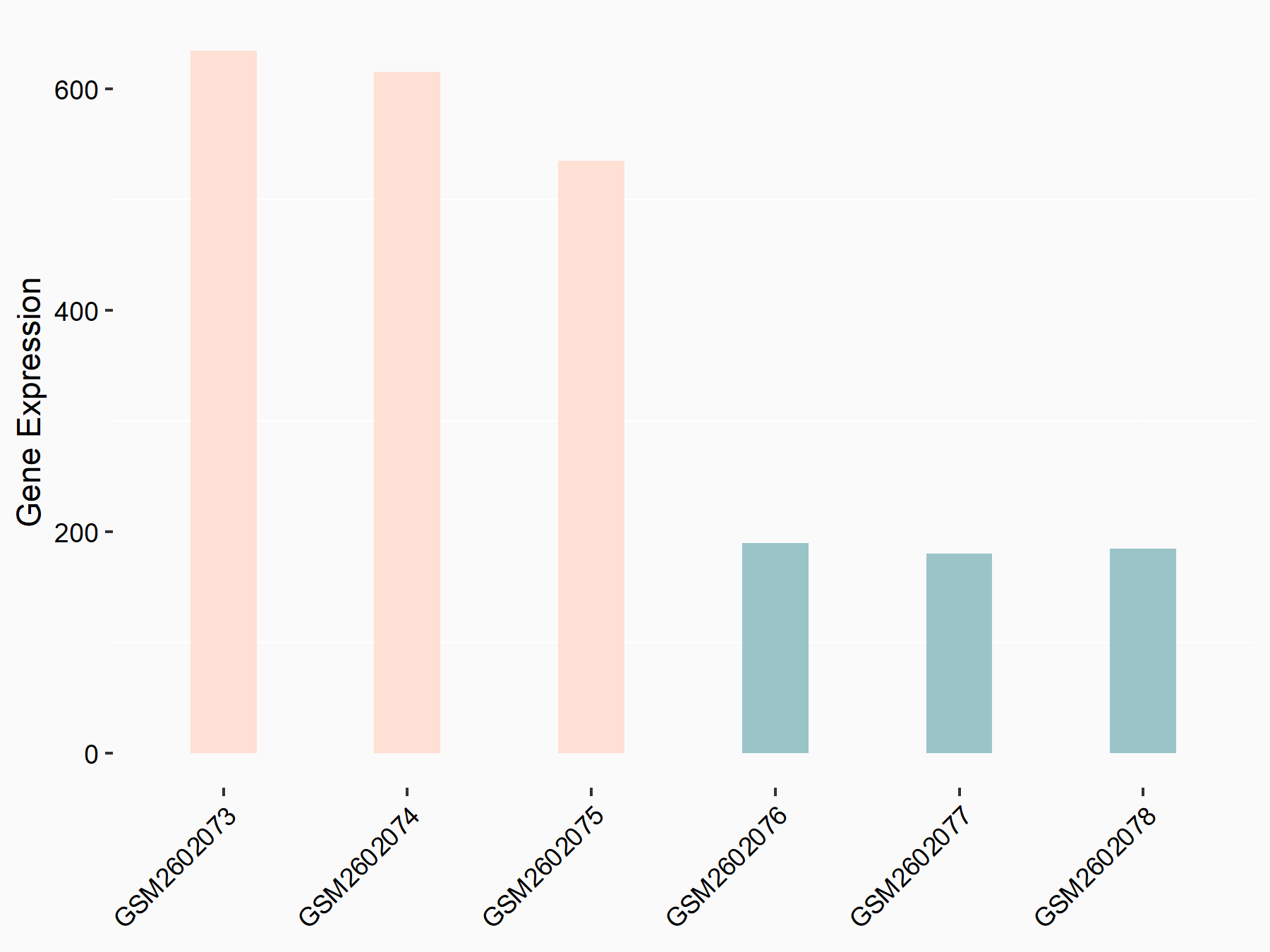 |
logFC: 1.69E+00 p-value: 1.05E-54 |
| More Results | Click to View More RNA-seq Results | |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [208] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Responsed Drug | Oxaliplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | |||
| Cell Process | RNA stability | |||
| Excision repair | ||||
In-vitro Model |
SNU-719 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_5086 |
| MKN74 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_2791 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | 100,000 pLKO and PARP1-sh1 (PT1 and PT2) cells were mixed with matrix gel and inoculate into BALB/C nude mice, respectively. After 25 days, 6 organoid transplanted tumor mice were treated with oxaliplatin (Sellekchem, s1224) twice a week for 4 weeks at a dose of 5 mg/kg. | |||
| Response Summary | m6A methyltransferase METTL3 facilitates oxaliplatin resistance in CD133+ gastric cancer stem cells by Promoting Poly [ADP-ribose] polymerase 1 (PARP1) mRNA stability which increases base excision repair pathway activity. METTTL3 enhances the stability of PARP1 by recruiting YTHDF1 to target the 3'-untranslated Region (3'-UTR) of PARP1 mRNA. | |||
Polycomb complex protein BMI-1 (BMI1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Pancreatic islets | Mus musculus |
|
Treatment: Mettl3 knockout mice
Control: Mettl3 flox/flox mice
|
GSE155612 | |
| Regulation |
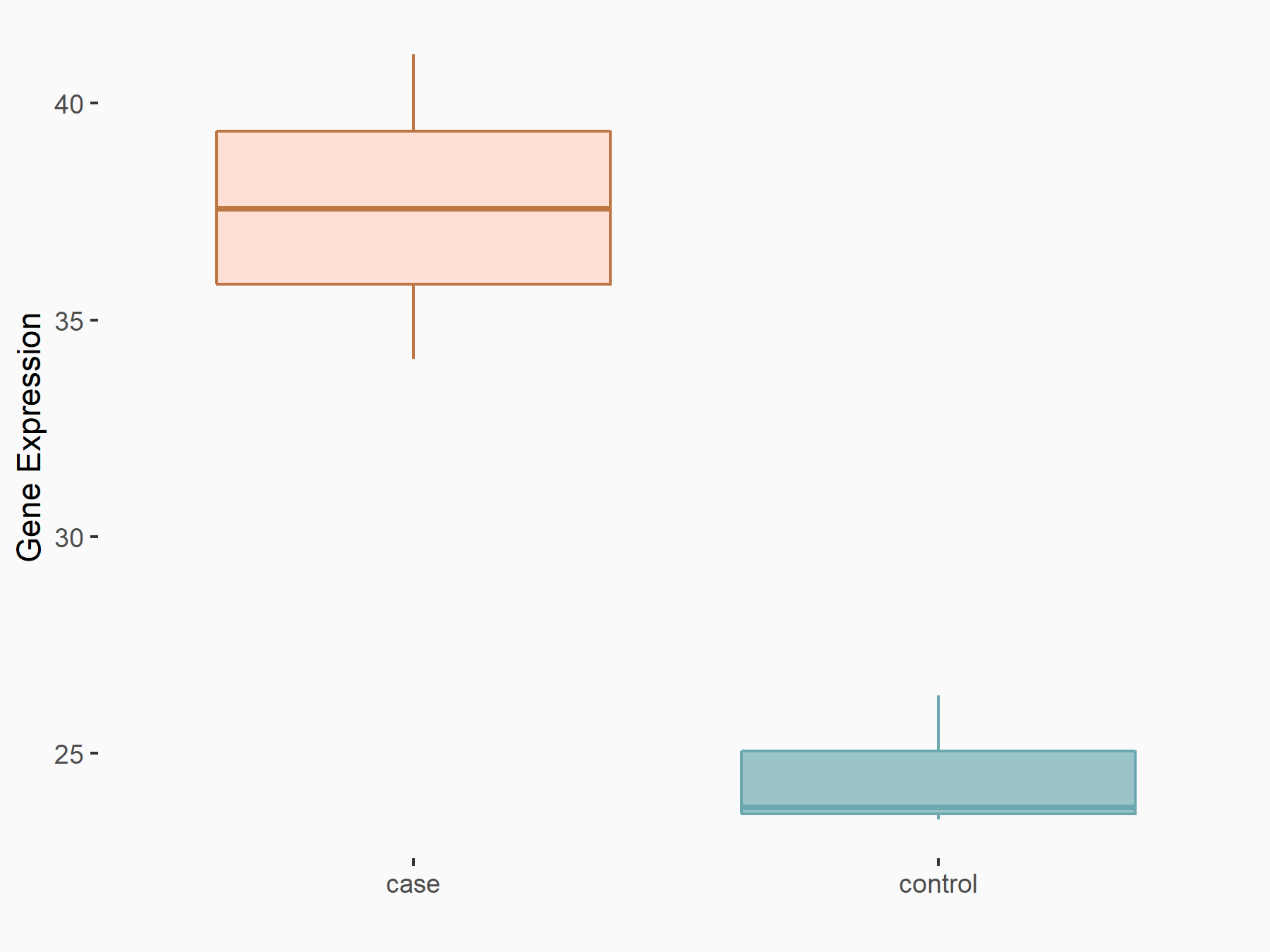 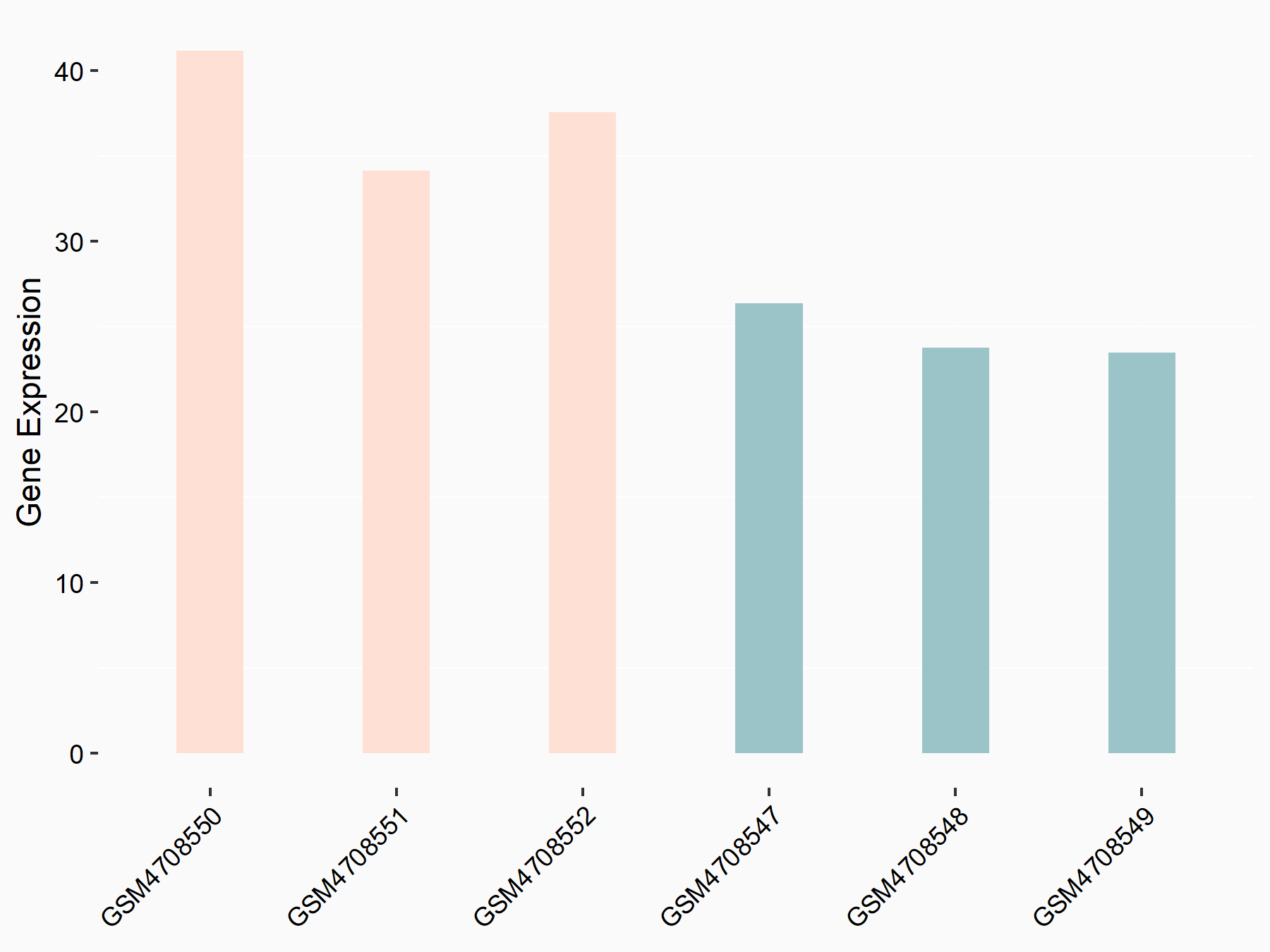 |
logFC: 5.95E-01 p-value: 1.91E-03 |
| More Results | Click to View More RNA-seq Results | |
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [209] | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
UM1 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_VH00 |
| SCC-9 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1685 | |
| SCC-25 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1682 | |
| SCC-15 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1681 | |
| HSC-3 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1288 | |
| HOK | Normal | Hexagrammos otakii | CVCL_YE19 | |
| In-vivo Model | To construct the subcutaneous tumorigenesis model, the cells were suspended in 100 uL of PBS and Matrigel matrix (BD Biosciences, USA) (1:1) and injected into the right flanks of 6-week-old female BALB/c nude mice.To construct the lymph node metastasis model, we injected 1 × 105/50 uL stably infected SCC9 cells into the left hind footpads of BALB/c mice. | |||
| Response Summary | METTL3 promotes Polycomb complex protein BMI-1 (BMI1) translation in OSCC under the cooperation with m6A reader IGF2BP1. And the study revealed that METTL3 promotes OSCC proliferation and metastasis through BMI1 m6A methylation. | |||
PPAR-gamma coactivator 1-alpha (PGC-1a/PPARGC1A)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shMETTL3 MOLM13 cells
Control: MOLM13 cells
|
GSE98623 | |
| Regulation |
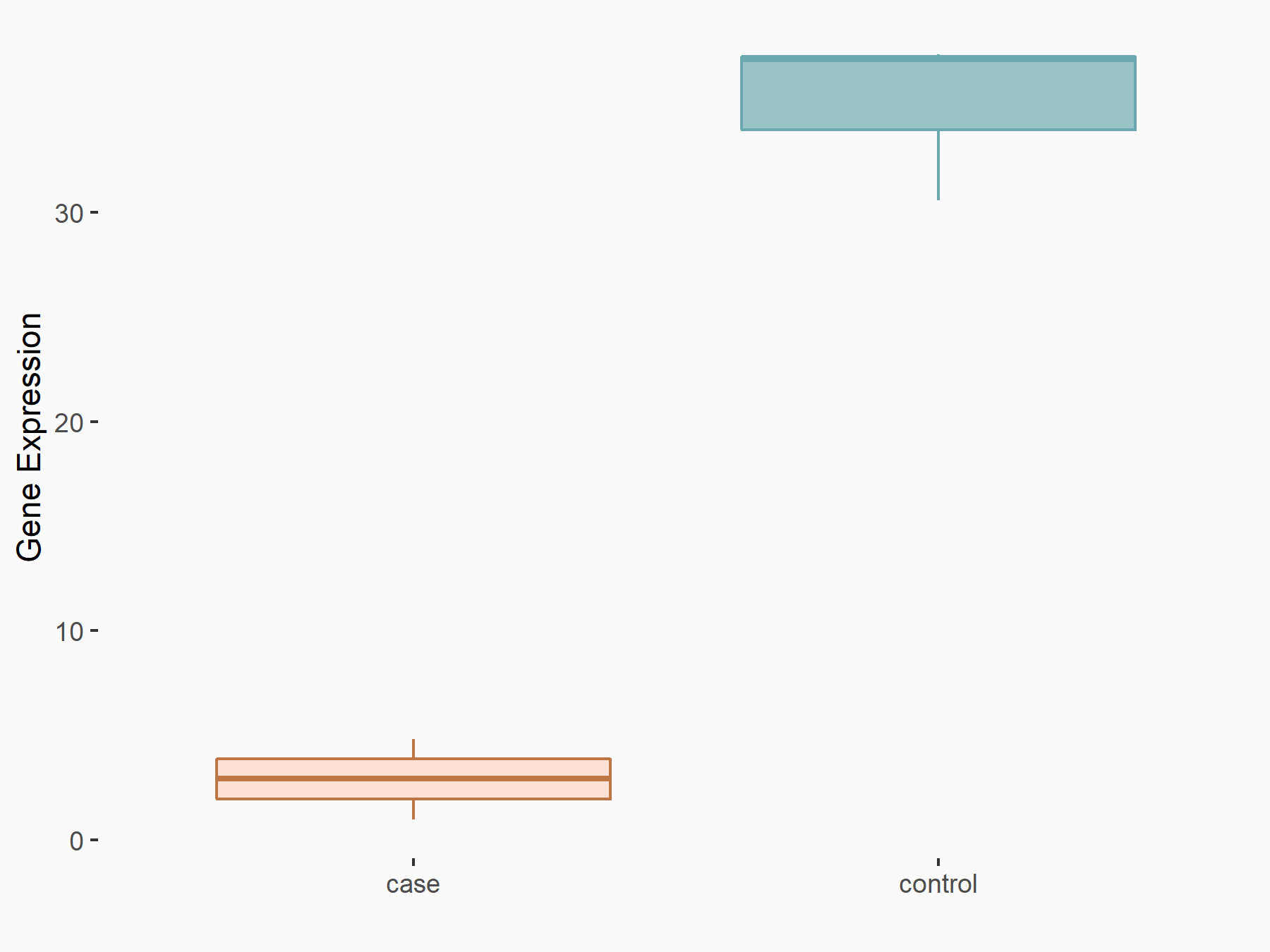 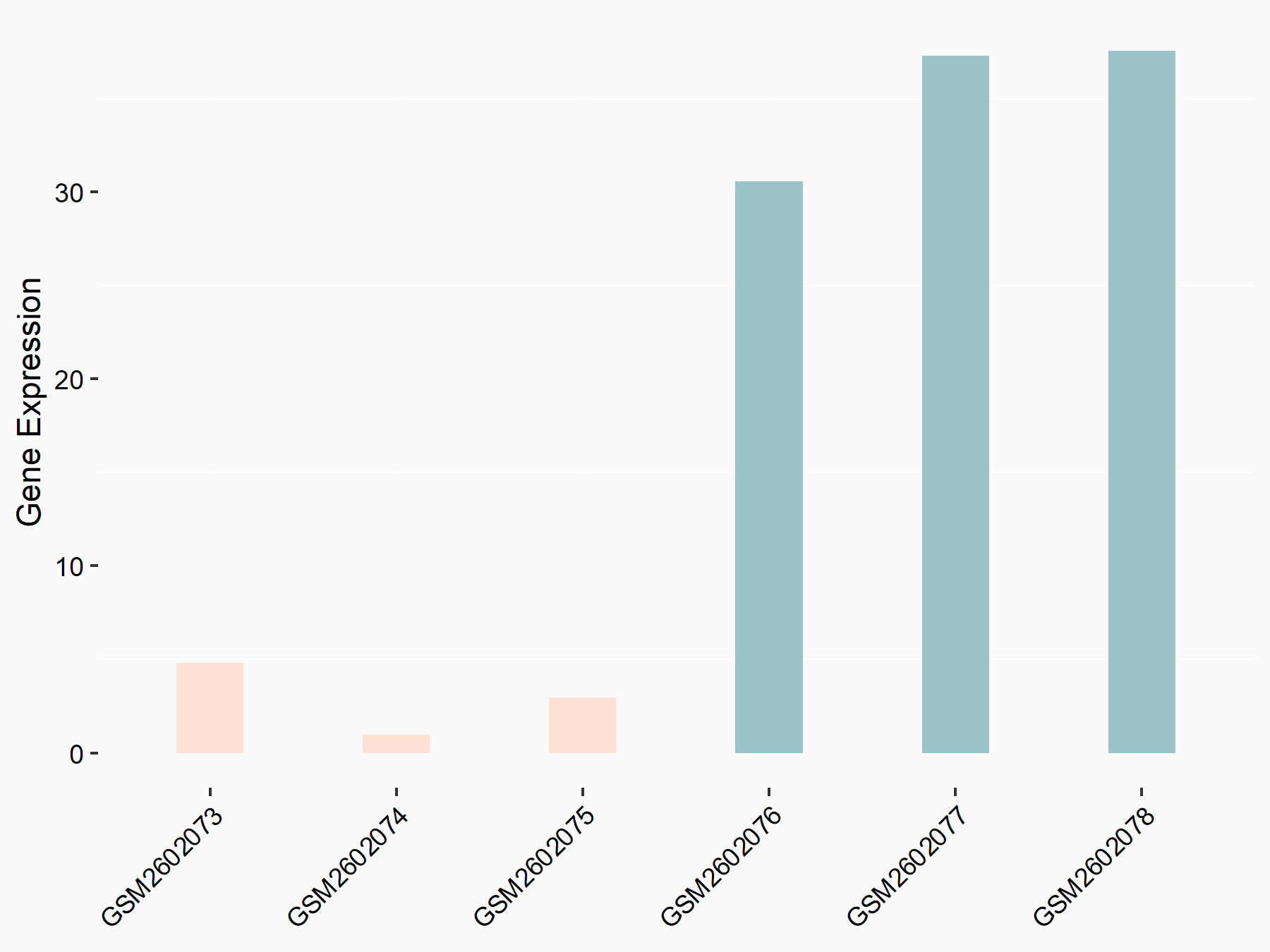 |
logFC: -3.59E+00 p-value: 8.55E-10 |
| More Results | Click to View More RNA-seq Results | |
Type 2 diabetes mellitus [ICD-11: 5A11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [48] | |||
| Responsed Disease | Type 2 diabetes mellitus [ICD-11: 5A11] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Insulin resistance | hsa04931 | ||
| Cell Process | Lipid metabolism | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | Hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were generated by crossing mice with TBG-Cre Tg mice. METTL3 flox (METTL3 fl/fl) and hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were used for experiments. | |||
| Response Summary | Type 2 diabetes (T2D) is characterized by lack of insulin, insulin resistance and high blood sugar. METTL3 silence decreased the m6A methylated and total mRNA level of Fatty acid synthase (Fasn), subsequently inhibited fatty acid metabolism. The expression of Acc1, Acly, Dgat2, Ehhadh, Fasn, Foxo, PPAR-gamma coactivator 1-alpha (PGC-1a/PPARGC1A) and Sirt1, which are critical to the regulation of fatty acid synthesis and oxidation were dramatically decreased in livers of hepatocyte-specific METTL3 knockout mice. | |||
PR domain zinc finger protein 2 (PRDM2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Pancreatic islets | Mus musculus |
|
Treatment: Mettl3 knockout mice
Control: Mettl3 flox/flox mice
|
GSE155612 | |
| Regulation |
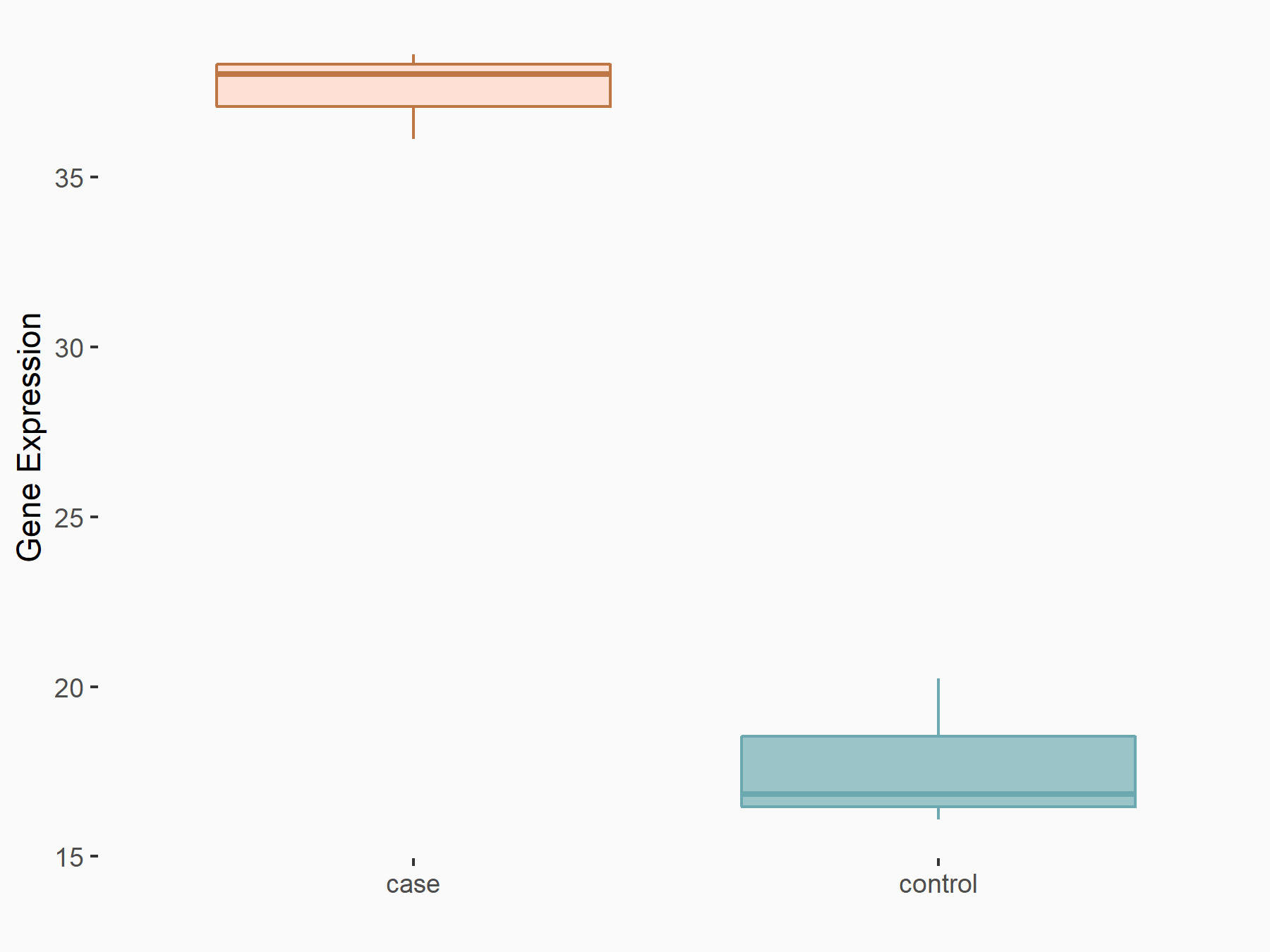 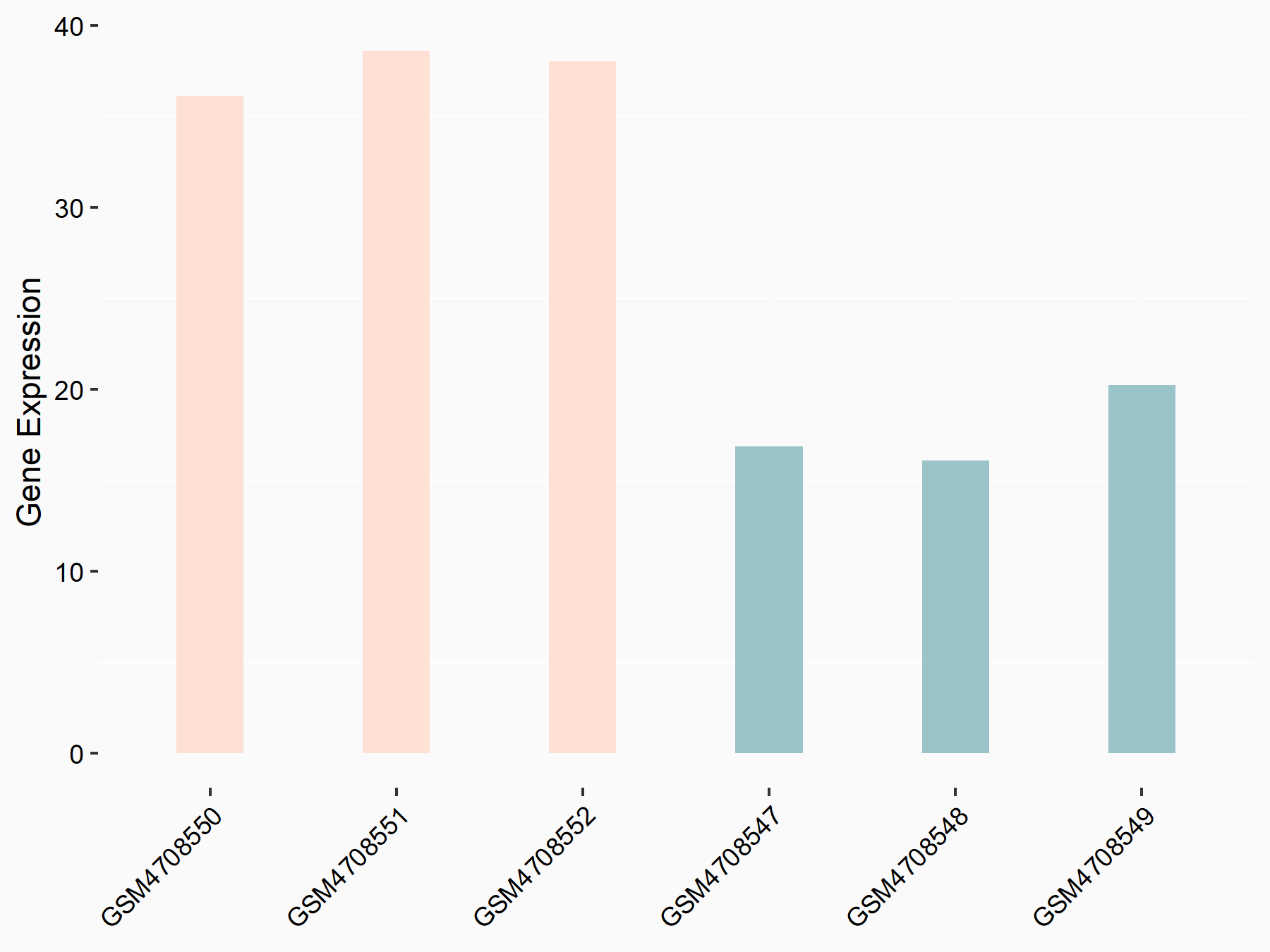 |
logFC: 1.05E+00 p-value: 2.68E-04 |
| More Results | Click to View More RNA-seq Results | |
Solid tumour/cancer [ICD-11: 2A00-2F9Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [27] | |||
| Responsed Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Responsed Drug | Arsenite | Phase 2 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
In-vitro Model |
HaCaT | Normal | Homo sapiens | CVCL_0038 |
| Response Summary | METTL3 significantly decreased m6A level, restoring p53 activation and inhibiting cellular transformation phenotypes in the arsenite-transformed cells. m6A downregulated the expression of the positive p53 regulator, PR domain zinc finger protein 2 (PRDM2), through the YTHDF2-promoted decay of PRDM2 mRNAs. m6A upregulated the expression of the negative p53 regulator, YY1 and MDM2 through YTHDF1-stimulated translation of YY1 and MDM2 mRNA. This study further sheds light on the mechanisms of arsenic carcinogenesis via RNA epigenetics. | |||
Prominin-1 (CD133)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
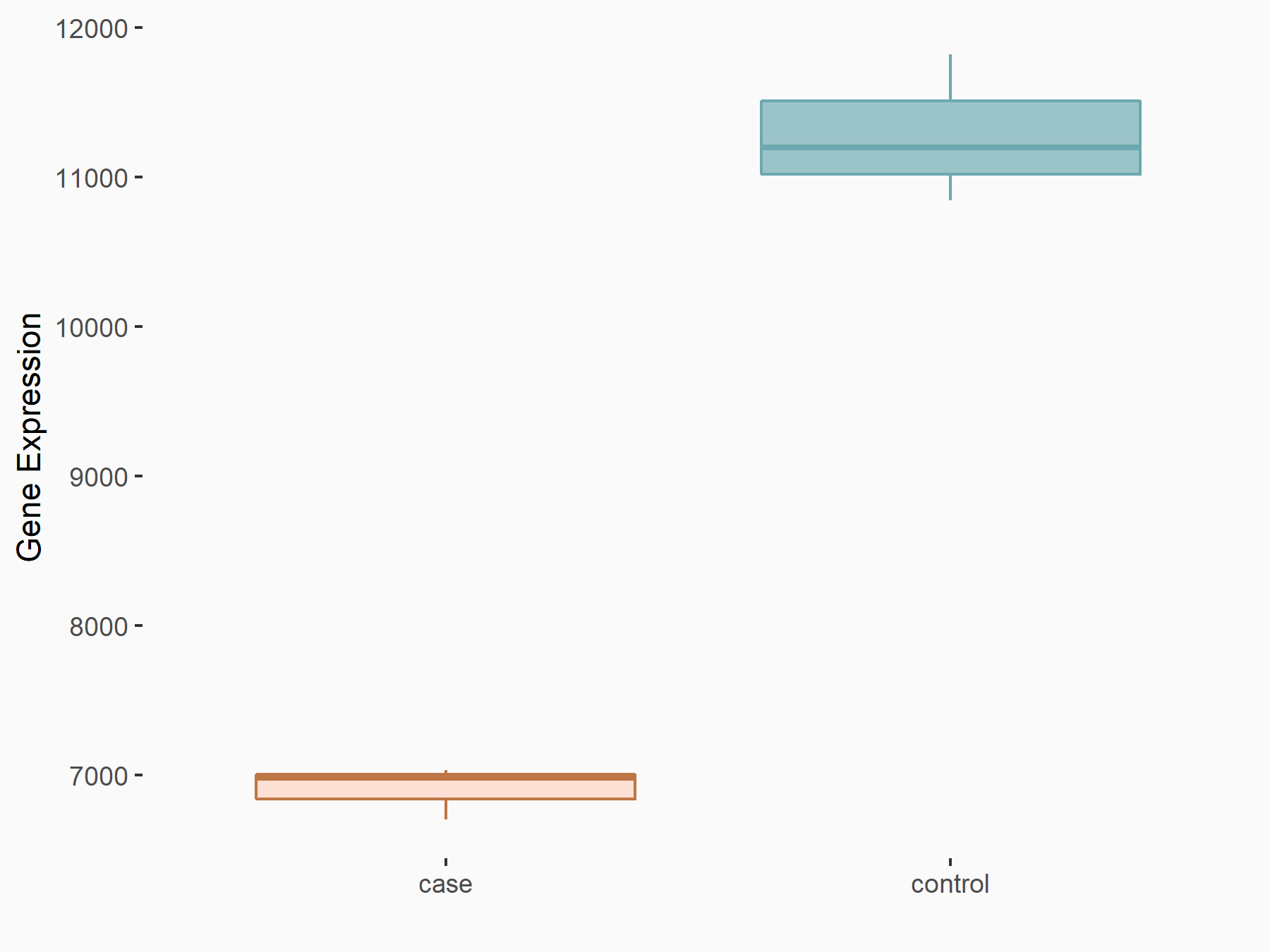 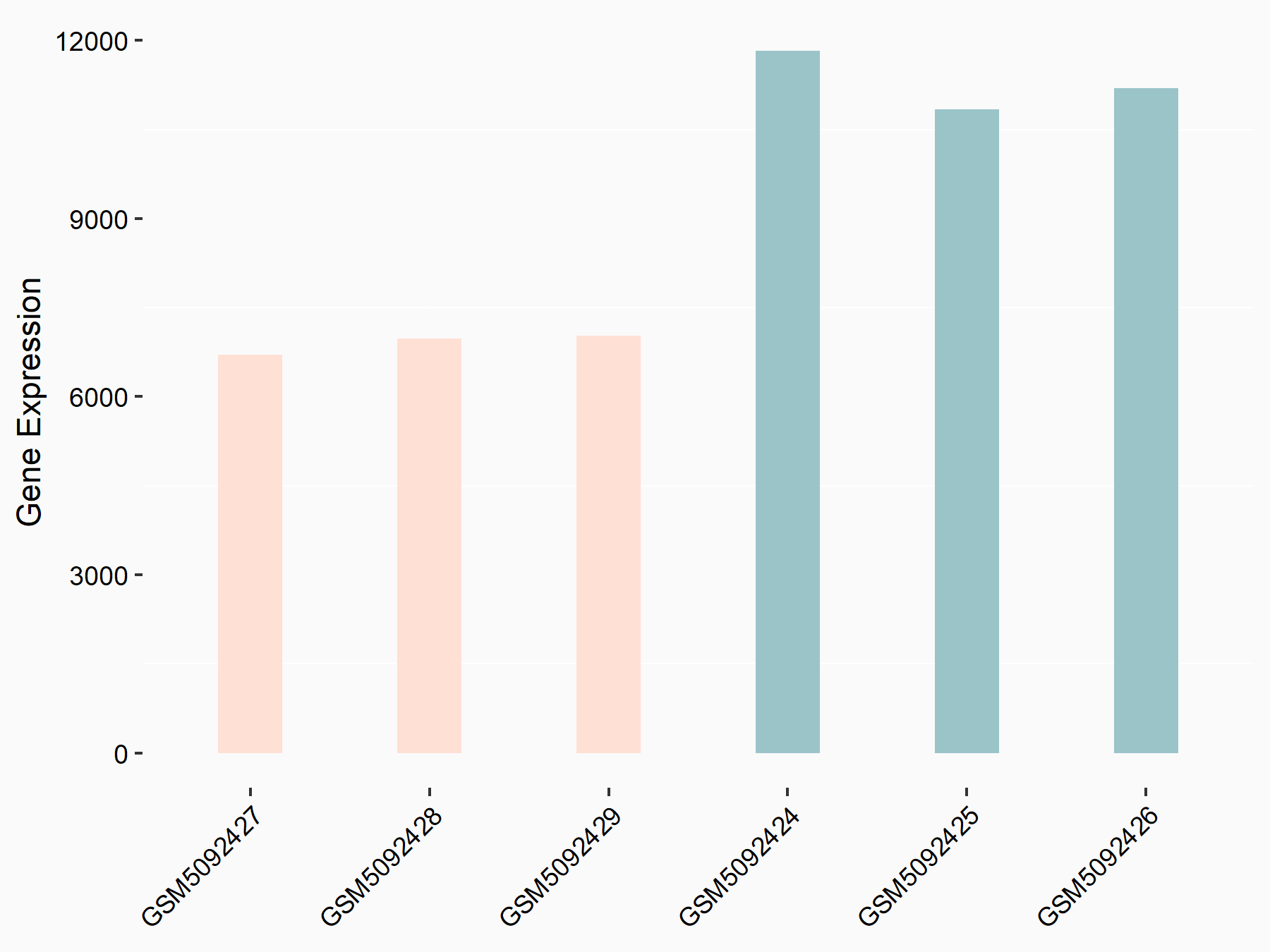 |
logFC: -7.09E-01 p-value: 5.71E-66 |
| More Results | Click to View More RNA-seq Results | |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [122] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| Response Summary | Knockdown of METTL3 downregulated protein levels of SOX2, Prominin-1 (CD133) and CD44 in MCF-7 cells. METTL3 is upregulated in breast cancer, and it promotes the stemness and malignant progression of BCa through mediating m6A modification on SOX2 mRNA. | |||
Prostaglandin G/H synthase 2 (Coll X/PTGS2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shMETTL3 MOLM13 cells
Control: MOLM13 cells
|
GSE98623 | |
| Regulation |
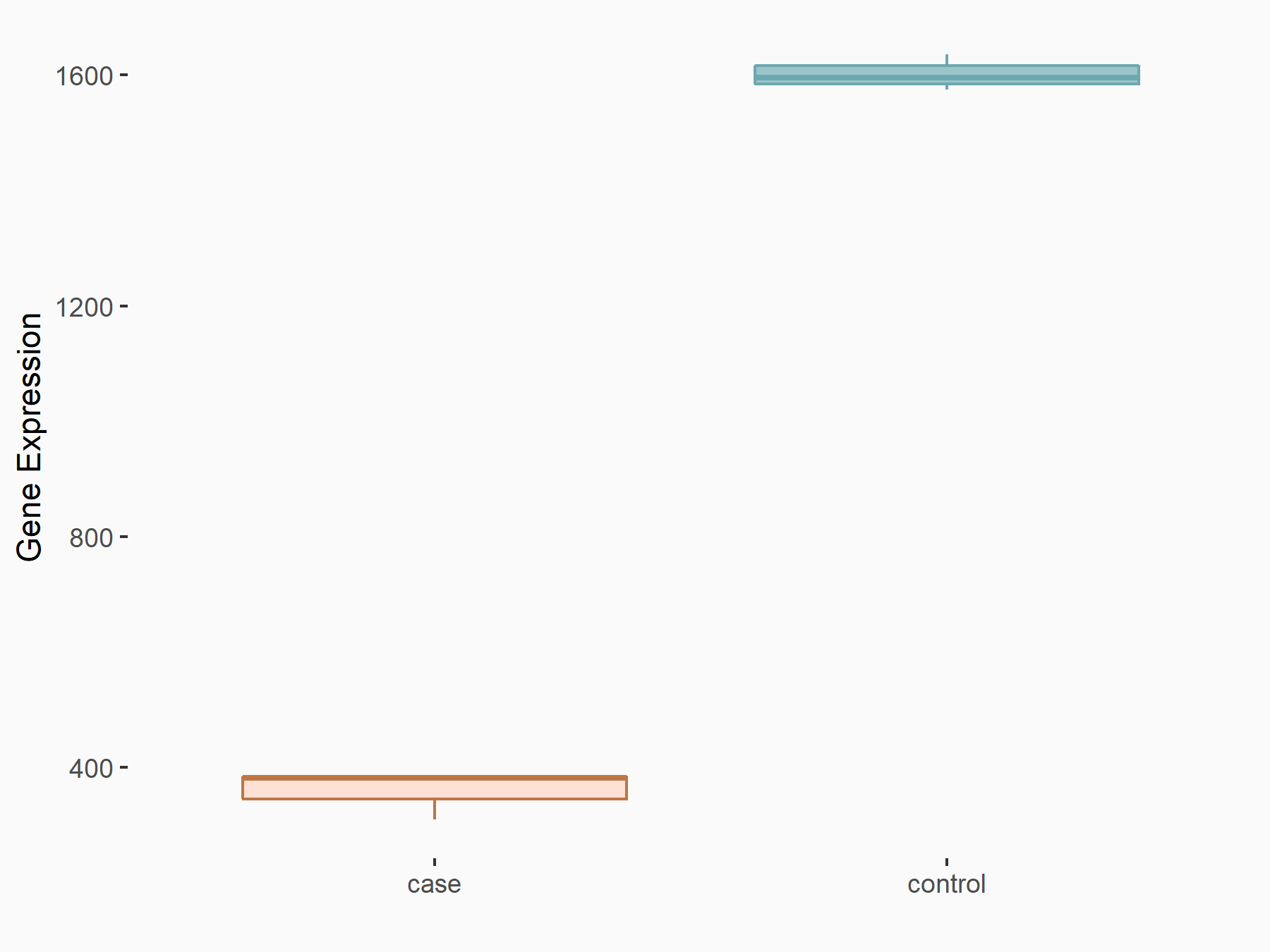 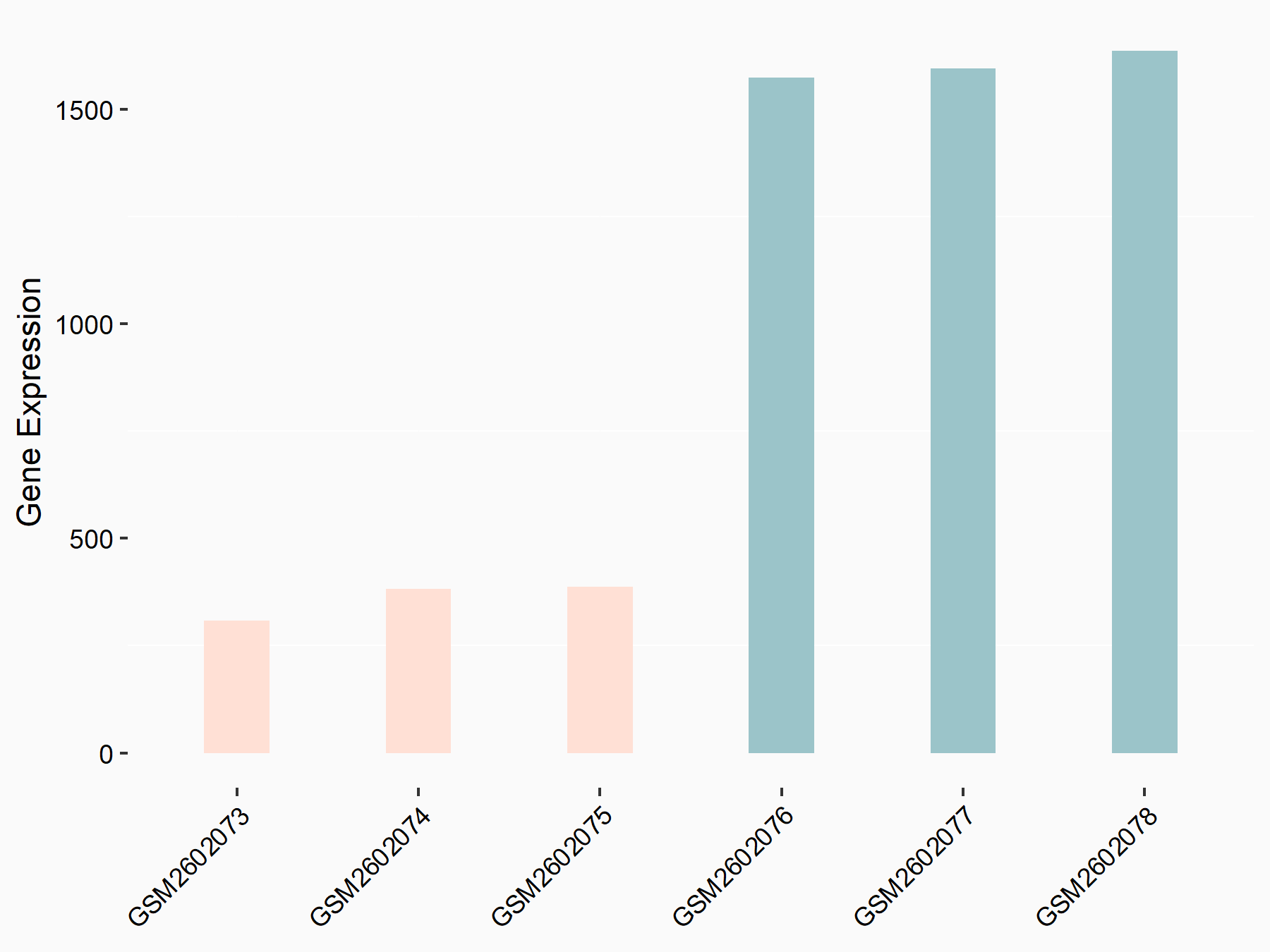 |
logFC: -2.16E+00 p-value: 5.15E-154 |
| More Results | Click to View More RNA-seq Results | |
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [32] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Inflammatory response and apoptosis | |||
In-vitro Model |
ATDC-5 | Mouse teratocarcinoma | Mus musculus | CVCL_3894 |
| In-vivo Model | The right knee joint of each OA mouse was injected with 1U of type VII collagenase over two consecutive days to obtain experimental OA joint, and the control mice received the equal volume of physiological saline. | |||
| Response Summary | METTL3 has a functional role in mediates osteoarthritis progression by regulating NF-Kappa-B signaling and ECM synthesis in chondrocytes that shed insight on developing preventive and curative strategies for OA by focusing on METTL3 and mRNA methylation. Silencing of METTL3 promotes degradation of extracellular matrix (ECM) by reducing the expression of MMP-13 and Prostaglandin G/H synthase 2 (Coll X/PTGS2), elevating the expression of Aggrecan and Coll II. | |||
Protein AATF (AATF/CHE1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3 knockout liver
Control: Wild type liver cells
|
GSE198513 | |
| Regulation |
 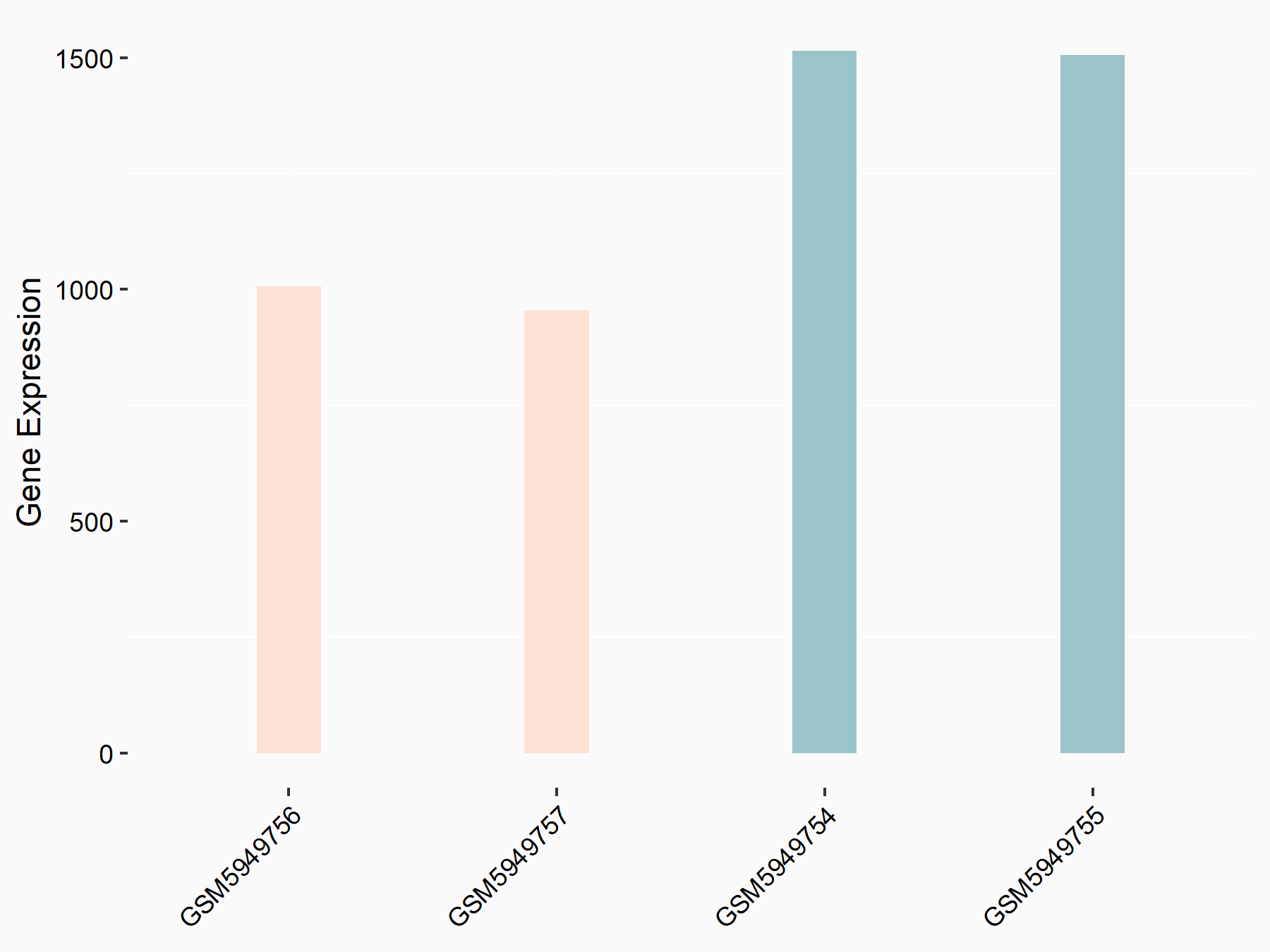 |
logFC: -6.23E-01 p-value: 2.02E-10 |
| More Results | Click to View More RNA-seq Results | |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [22] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Doxil | Approved | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell growth and death | |||
| Cell apoptosis | ||||
In-vitro Model |
ADR-resistant MCF-7 (MCF-7/ADR) cells (Human breast cancer doxorubicin-resistant cell line) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| In-vivo Model | Cell suspensions (2 × 106 cells/mL) made with MCF-7/ADR cells stably expressing METTL3 and/or miR-221-3p inhibitor were subcutaneously implanted into each mouse. One week later, xenografted mice were injected with 0.1 mL ADR (25 mg/kg, intraperitoneal injection) twice a week. | |||
| Response Summary | METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. METTL3 knockdown was shown to reduce the expression of miR-221-3p by reducing pri-miR-221-3p m6A mRNA methylation, reducing the expression of MDR1 and BCRP, and inducing apoptosis. Identified the METTL3/miR-221-3p/HIPK2/Protein AATF (AATF/CHE1) axis as a novel signaling event that will be responsible for resistance of BC cells to ADR. | |||
Protein kinase C eta type (PKC-eta)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
 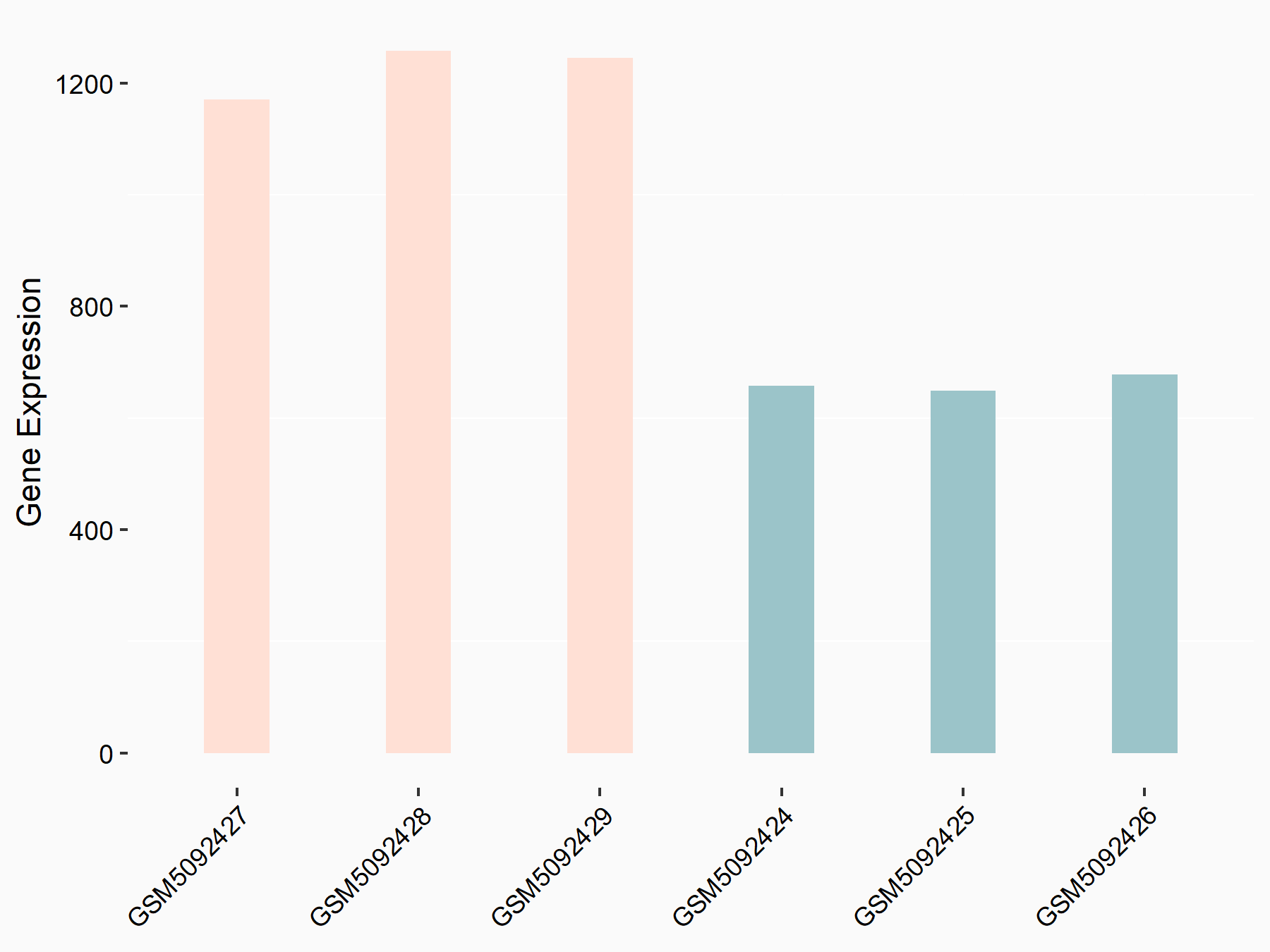 |
logFC: 8.88E-01 p-value: 2.37E-29 |
| More Results | Click to View More RNA-seq Results | |
Diseases of arteries or arterioles [ICD-11: BD5Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [96] | |||
| Responsed Disease | Diseases of arteries or arterioles [ICD-11: BD5Y] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
ACBRI-183 (Human retinal pericytes (ACBRI-183) was obtained from Cell Systems Corp. (CSC, USA)) | |||
| In-vivo Model | Mettl3 floxed mice were purchased from GemPharmatech Co. Ltd (Nanjing, China). Pdgfr-Beta-Cre mice were purchased from Beijing Biocytogen Co. Ltd (Beijing, China) generated on C57BL/6J background. Mettl3 flox/flox mice were crossed with Pdgfr-Beta-Cre mice to generate pericyte-specific Mettl3 knockout mice. All mice were bred under the specific-pathogen free condition with free access to diet and water or their nursing mothers with alternating 12/12 light-dark cycle (lights on at 08:00 and off at 20:00). | |||
| Response Summary | Specific depletion of METTL3 in pericytes suppressed diabetes-induced pericyte dysfunction and Microvascular complication in vivo. METTL3 overexpression impaired pericyte function by repressing Protein kinase C eta type (PKC-eta), FAT4, and PDGFRA expression, which was mediated by YTHDF2-dependent mRNA decay. | |||
Protein numb homolog (NUMB)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
|
Treatment: METTL3 knockdown MDA-MB-231 cells
Control: MDA-MB-231 cells
|
GSE70061 | |
| Regulation |
  |
logFC: 9.75E-01 p-value: 1.24E-03 |
| More Results | Click to View More RNA-seq Results | |
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [201] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Responsed Drug | Melittin | Investigative | ||
| Target Regulation | Down regulation | |||
| Cell Process | miRNA maturation | |||
| Cell apoptosis | ||||
In-vitro Model |
T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| EJ (Human bladder cancer cells) | ||||
| BIU-87 | Human bladder cancer cells | Homo sapiens | CVCL_6881 | |
| In-vivo Model | For melittin treatment study, 4-week-old female BALB/c nude mice were subcutaneously injected with 1 × 107 T24 or BIU87 cells. | |||
| Response Summary | METTL3 acts as a fate determinant that controls the sensitivity of bladder cancer cells to melittin treatment. Moreover, METTL3/miR-146a-5p/Protein numb homolog (NUMB)/NOTCH2 axis plays an oncogenic role in bladder cancer pathogenesis and could be a potential therapeutic target for recurrent bladder cancer treatment. | |||
Protein yippee-like 5 (YPEL5)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
 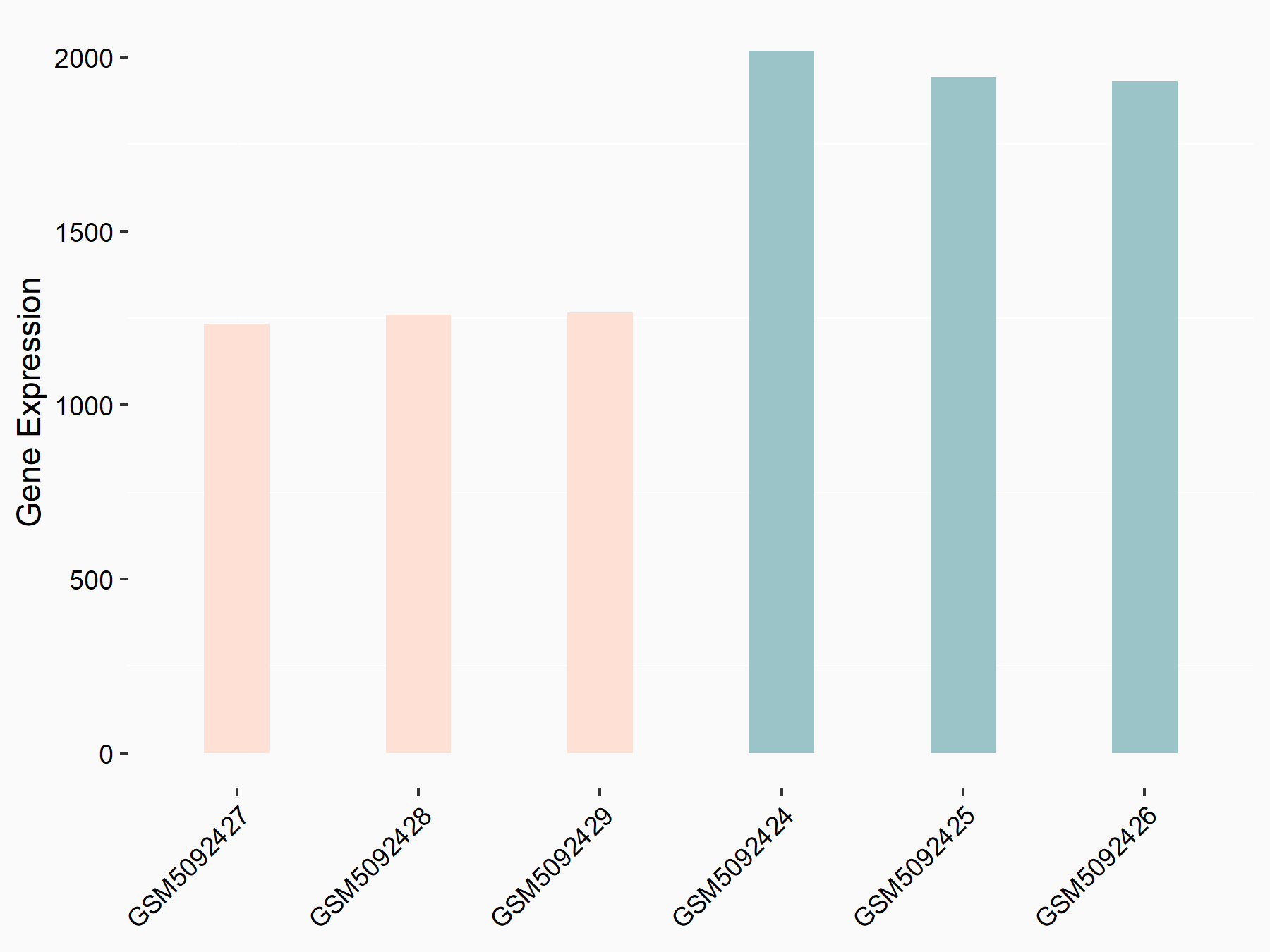 |
logFC: -6.48E-01 p-value: 2.75E-26 |
| More Results | Click to View More RNA-seq Results | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [210] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| In-vivo Model | For the xenograft model, METTL3 stable overexpressed SW620 cells (1 × 107) or control cells were subcutaneously injected into the right axilla of the female anesthetized BALB/C nude mice (4-6 weeks old, 18-20 g, four mice per group), respectively. The body weight and tumor volumes (length × width2 × 0.5) were measured twice a week. After 21 days, all mice were sacrificed and tumors were surgically removed for hematoxylin-eosin (H&E) staining.For the metastasis model, MTTL3 stable overexpressed SW620 cells (1 × 106) or control cells were injected into the exposed spleen of the anesthetized BALB/C nude mice, respectively. After 21 days, liver metastases were carefully detected using a fluorescent stereoscope and embedded for H&E staining. | |||
| Response Summary | METTL3-catalyzed m6A modification in CRC tumorigenesis, wherein it facilitates CRC tumor growth and metastasis through suppressing Protein yippee-like 5 (YPEL5) expression in an m6A-YTHDF2-dependent manner. | |||
Protocadherin Fat 4 (FAT4)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HUVEC cell line | Homo sapiens |
|
Treatment: shMETTL3 HUVEC cells
Control: shScramble HUVEC cells
|
GSE157544 | |
| Regulation |
 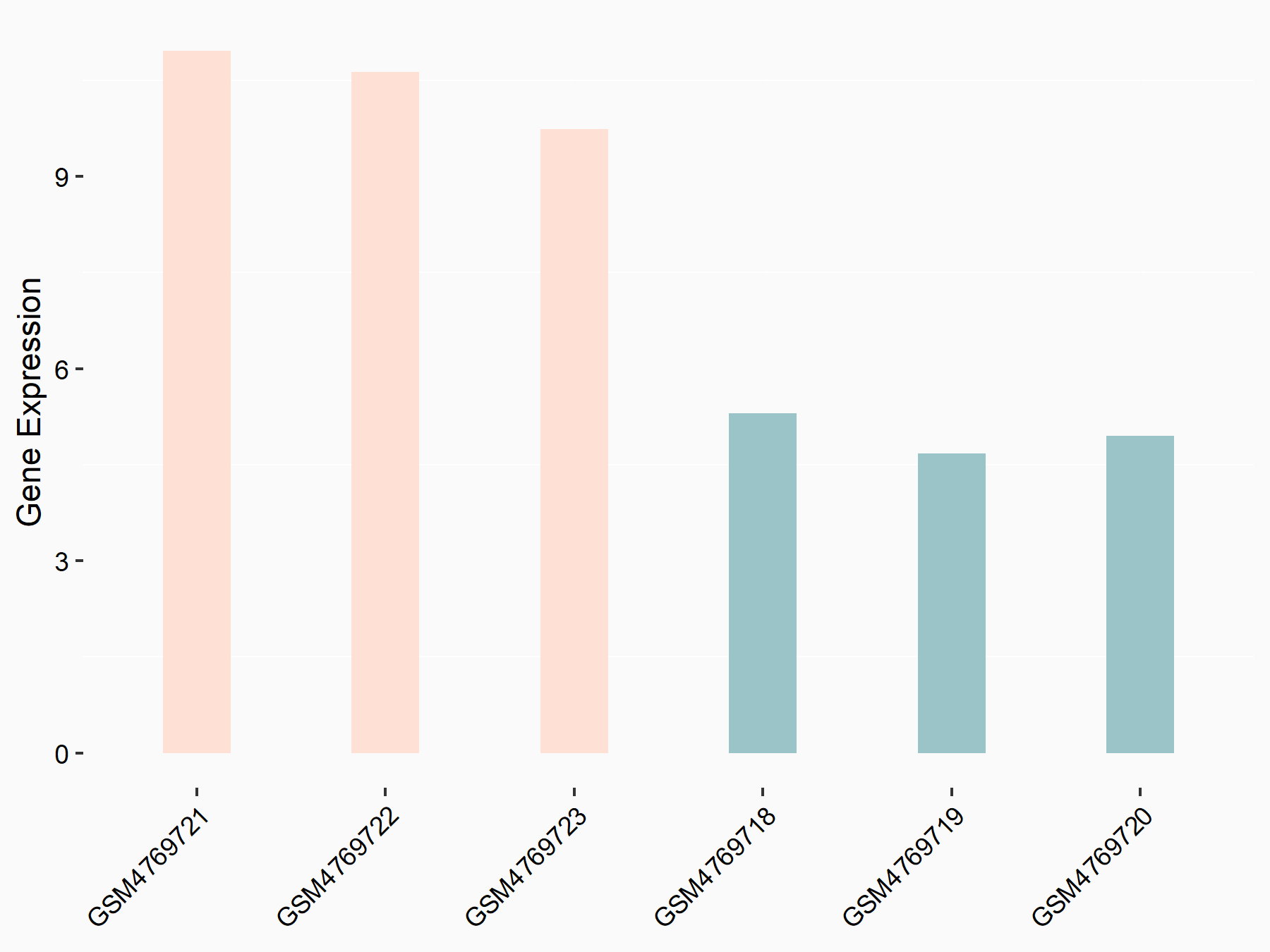 |
logFC: 9.37E-01 p-value: 7.13E-05 |
| More Results | Click to View More RNA-seq Results | |
Diseases of arteries or arterioles [ICD-11: BD5Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [96] | |||
| Responsed Disease | Diseases of arteries or arterioles [ICD-11: BD5Y] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
ACBRI-183 (Human retinal pericytes (ACBRI-183) was obtained from Cell Systems Corp. (CSC, USA)) | |||
| In-vivo Model | Mettl3 floxed mice were purchased from GemPharmatech Co. Ltd (Nanjing, China). Pdgfr-Beta-Cre mice were purchased from Beijing Biocytogen Co. Ltd (Beijing, China) generated on C57BL/6J background. Mettl3 flox/flox mice were crossed with Pdgfr-Beta-Cre mice to generate pericyte-specific Mettl3 knockout mice. All mice were bred under the specific-pathogen free condition with free access to diet and water or their nursing mothers with alternating 12/12 light-dark cycle (lights on at 08:00 and off at 20:00). | |||
| Response Summary | Specific depletion of METTL3 in pericytes suppressed diabetes-induced pericyte dysfunction and Microvascular complication in vivo. METTL3 overexpression impaired pericyte function by repressing PKC-Eta, Protocadherin Fat 4 (FAT4), and PDGFRA expression, which was mediated by YTHDF2-dependent mRNA decay. | |||
RAC-alpha serine/threonine-protein kinase (AKT1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3 knockout liver
Control: Wild type liver cells
|
GSE198512 | |
| Regulation |
 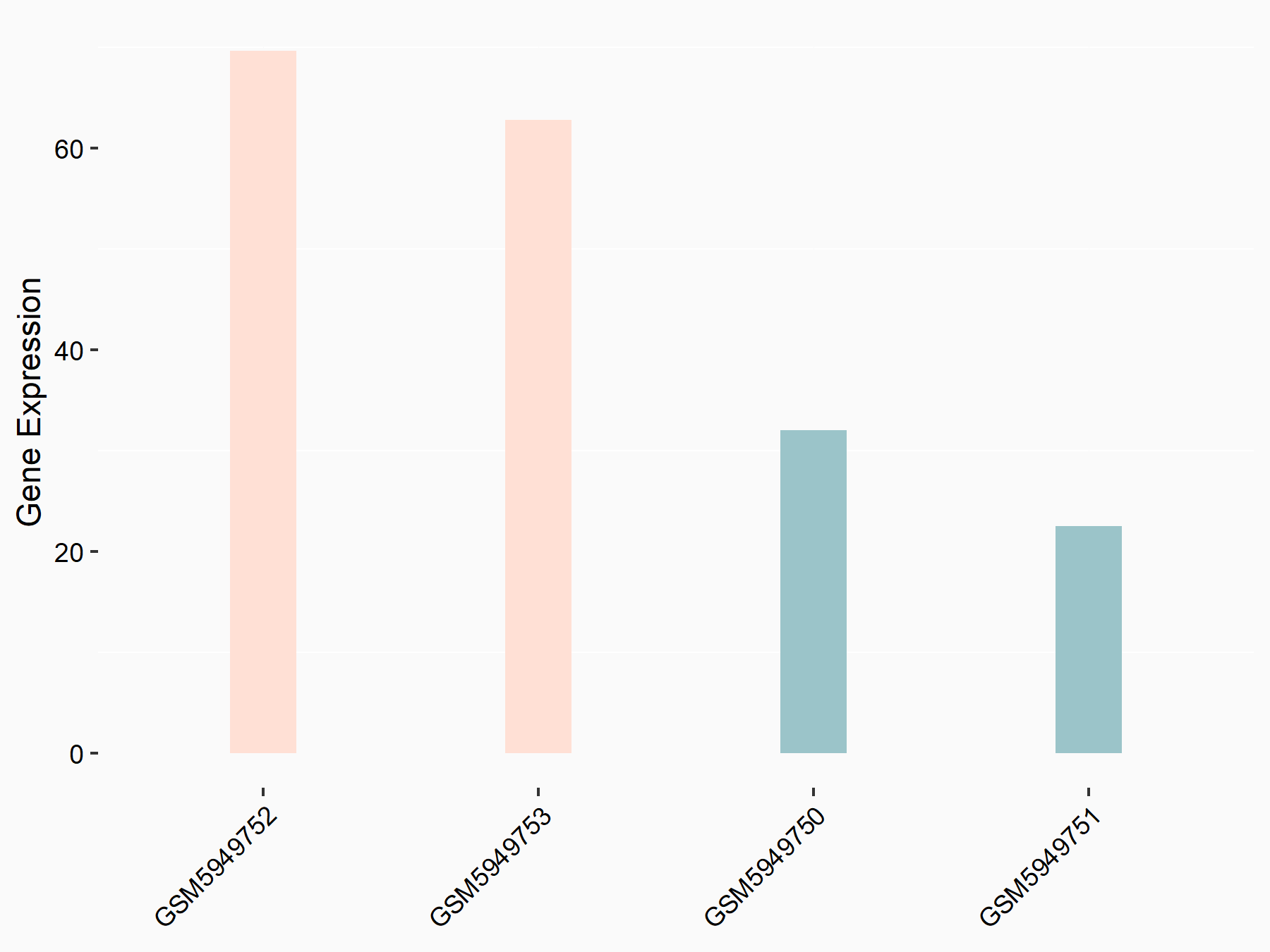 |
logFC: 1.27E+00 p-value: 1.77E-02 |
| More Results | Click to View More RNA-seq Results | |
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [9] | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell differentiation and apoptosis | |||
In-vitro Model |
HSPC (Human hematopoietic stem cell) | |||
| In-vivo Model | 500,000 selected cells were injected via tail vein or retro-orbital route into female NSG (6-8 week old) recipient mice that had been sublethally irradiated with 475 cGy one day before transplantation. | |||
| Response Summary | METTL3 depletion in human myeloid leukemia cell lines induces cell differentiation and apoptosis and delays leukemia progression in recipient mice in vivo. Single-nucleotide-resolution mapping of m6A coupled with ribosome profiling reveals that m6A promotes the translation of c-MYC, BCL2 and PTEN mRNAs in the human acute myeloid leukemia MOLM-13 cell line. Moreover, loss of METTL3 leads to increased levels of phosphorylated RAC-alpha serine/threonine-protein kinase (AKT1). | |||
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [212] | |||
| Responsed Disease | Esophageal cancer [ICD-11: 2B70] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Wnt signaling pathway | hsa04310 | |||
| mTOR signaling pathway | hsa04150 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell proliferation and invasion | |||
| Cell apoptosis | ||||
In-vitro Model |
Eca-109 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_6898 |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| Normal esophageal epithelial cell line (HEEC) (Isolated from the human esophagus) | ||||
| Response Summary | METTL3 plays a carcinogenic role in human EC progression partially through RAC-alpha serine/threonine-protein kinase (AKT1) signaling pathways, suggesting that METTL3 serves as a potential therapeutic target for esophageal cancer therapy. A double-effect inhibitor (BEZ235) inhibited AKT and mTOR phosphorylation and hindered the effect of METTL3 overexpression on the proliferation and migration of Eca-109 and KY-SE150 cells. | |||
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [8] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| Response Summary | Down-regulation of METTL3 inhibits the proliferation and mobility of human gastric cancer cells and leads to inactivation of the AKT signaling pathway, suggesting that METTL3 is a potential target for the treatment of human gastric cancer. METTL3 knockdown decreased Bcl2 and increased Bax and active Caspase-3 in gastric cancer cells, which suggested the apoptotic pathway was activated. METTL3 led to inactivation of the AKT signaling pathway in human gastric cancer cells, including decreased phosphorylation levels of RAC-alpha serine/threonine-protein kinase (AKT1) and expression of down-stream effectors p70S6K and Cyclin D1. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [213] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
| Response Summary | METTL3 contributes to the progression and chemoresistance of NSCLC by promoting RAC-alpha serine/threonine-protein kinase (AKT1) protein expression through regulating AKT1 mRNA m6A levels, and provides an efficient therapeutic intervention target for overcoming chemoresistance in NSCLC. | |||
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [168] | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell cycle | |||
| Cell apoptosis | ||||
In-vitro Model |
OVCAR-3 | Ovarian serous adenocarcinoma | Homo sapiens | CVCL_0465 |
| SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 | |
| Response Summary | METTL3 knockdown downregulated the phosphorylation levels of RAC-alpha serine/threonine-protein kinase (AKT1) and the expression of the downstream effector Cyclin D1 in ovarian cancer. | |||
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [62] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Oxidative phosphorylation | hsa00190 | ||
In-vitro Model |
VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| In-vivo Model | Approximately 2 × 106 PCa cells (PC-3 shNC, shYTHDF2, shMETTL3 cell lines) per mouse suspended in 100 uL PBS were injected in the flank of male BALB/c nude mice (4 weeks old). During the 40-day observation, the tumor size (V = (width2×length ×0.52)) was measured with vernier caliper. Approximately 1.5 × 106 PCa cells suspended in 100 uL of PBS (PC-3 shNC, shYTHDF2, and shMETTL3 cell lines) per mouse were injected into the tail vein of male BALB/c nude mice (4 weeks old). The IVIS Spectrum animal imaging system (PerkinElmer) was used to evaluate the tumor growth (40 days) and whole metastasis conditions (4 weeks and 6 weeks) with 100 uL XenoLight D-luciferin Potassium Salt (15 mg/ml, Perkin Elmer) per mouse. Mice were anesthetized and then sacrificed for tumors and metastases which were sent for further organ-localized imaging as above, IHC staining and hematoxylin-eosin (H&E) staining. | |||
| Response Summary | Knock-down of YTHDF2 or METTL3 significantly induced the expression of LHPP and NKX3-1 at both mRNA and protein level with inhibited phosphorylated RAC-alpha serine/threonine-protein kinase (AKT1). YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent way to regulate AKT phosphorylation-induced tumor progression in prostate cancer. | |||
Renal cell carcinoma [ICD-11: 2C90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [109] | |||
| Responsed Disease | Renal cell carcinoma [ICD-11: 2C90] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Epithelial-to-mesenchymal transition | |||
| Arrest cell cycle at G0/G1 phase | ||||
In-vitro Model |
ACHN | Papillary renal cell carcinoma | Homo sapiens | CVCL_1067 |
| Caki-1 | Clear cell renal cell carcinoma | Homo sapiens | CVCL_0234 | |
| Caki-2 | Papillary renal cell carcinoma | Homo sapiens | CVCL_0235 | |
| HK2 | Normal | Acipenser baerii | CVCL_YE28 | |
| In-vivo Model | Cells (5×106 cells in 200 uL) were suspended with 100 uL PBS and 100 uL Matrigel Matrix, and injected subcutaneously into the left armpit of each mouse. | |||
| Response Summary | Knockdown of METTL3 could obviously promote cell proliferation, migration and invasion function, and induce G0/G1 arrest,METTL3 acts as a novel marker for tumorigenesis, development and survival of RCC. Knockdown of METTL3 promoted changes in pI3K/RAC-alpha serine/threonine-protein kinase (AKT1)/mTOR markers' expression with a gain in p-PI3k, p-AKT, p-mTOR and p-p70, and a loss of p-4EBP1. | |||
Retina cancer [ICD-11: 2D02]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [44] | |||
| Responsed Disease | Retinoblastoma [ICD-11: 2D02.2] | |||
| Responsed Drug | Rapamycin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
In-vitro Model |
WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through PI3K/RAC-alpha serine/threonine-protein kinase (AKT1)/mTOR pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-P70S6K/4EBP1 pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
Ragulator complex protein LAMTOR5 (LAMTOR5/HBXIP)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Th1 cell line | Mus musculus |
|
Treatment: METTL3 knockout splenic Th1 cells
Control: Wild type splenic Th1 cells
|
GSE129648 | |
| Regulation |
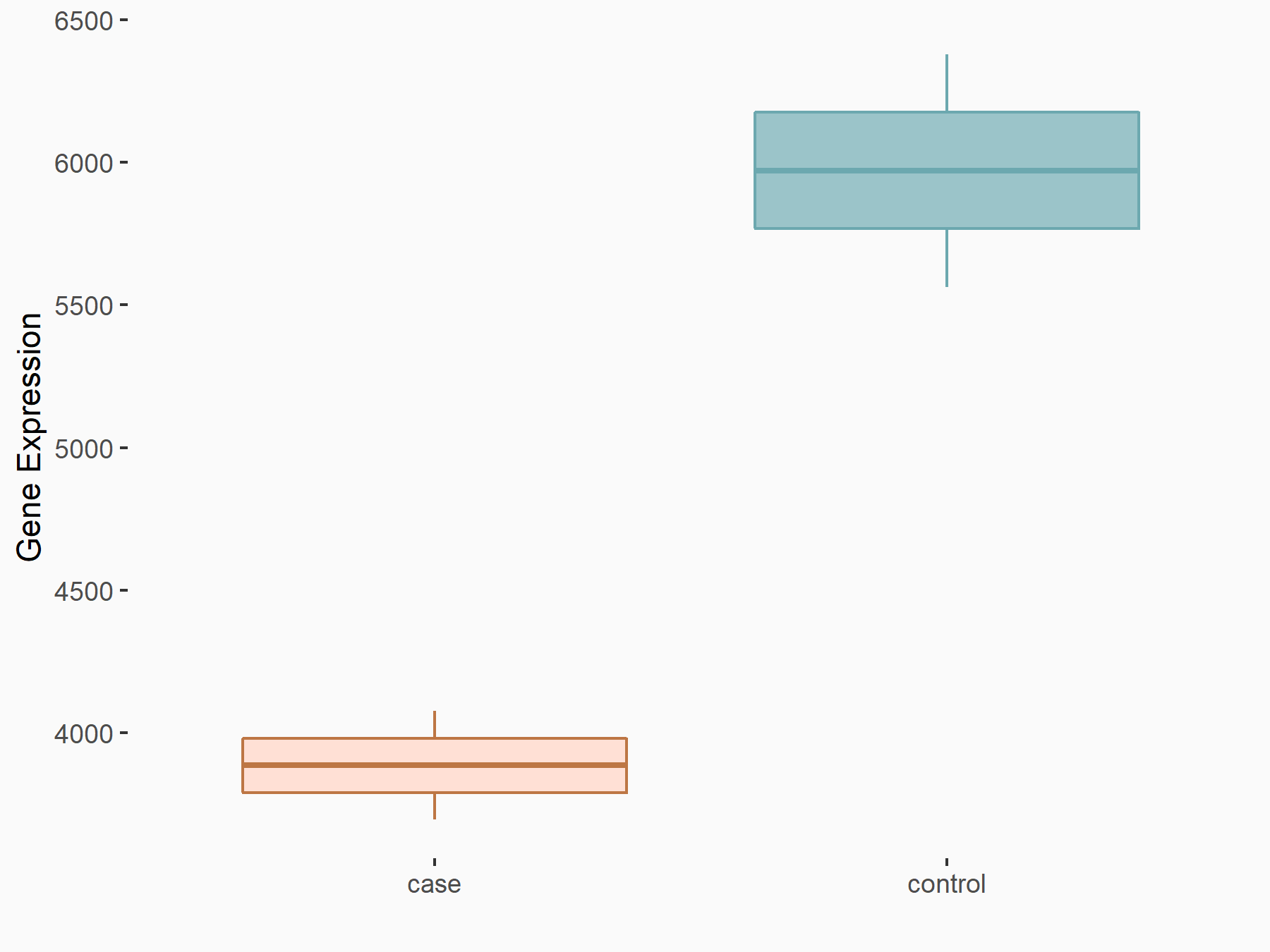 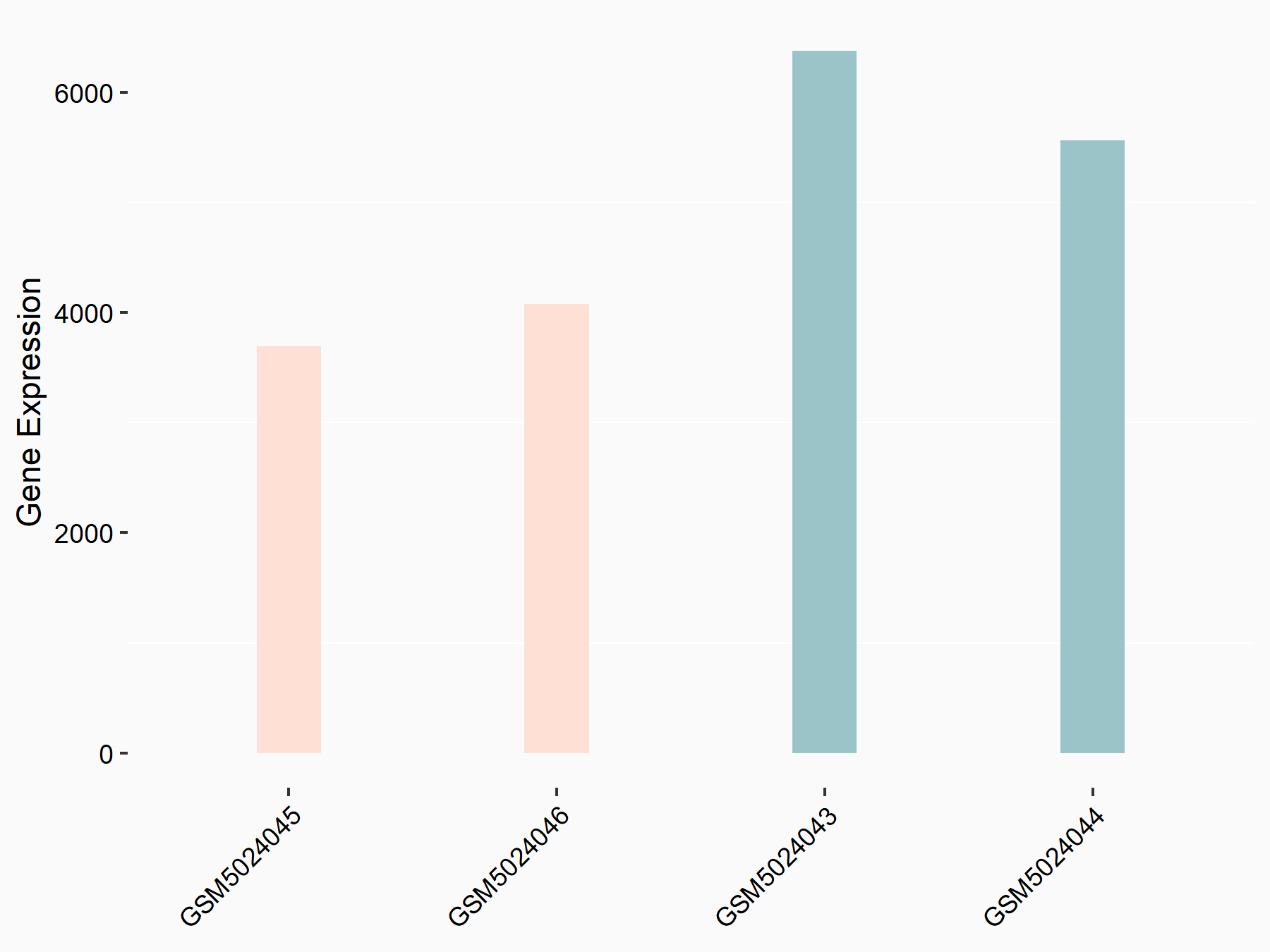 |
logFC: -6.20E-01 p-value: 1.93E-04 |
| More Results | Click to View More RNA-seq Results | |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [214] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell differentiation and apoptosis | |||
| Glutamine metabolism | ||||
| Apoptosis (hsa04210) | ||||
| Response Summary | Ragulator complex protein LAMTOR5 (LAMTOR5/HBXIP) up-regulates METTL3 by suppressing let-7g, in which METTL3 increased HBXIP expression forming a positive feedback loop of HBXIP/let-7g/METTL3/HBXIP, leading to accelerated cell proliferation in breast cancer. | |||
Rho GTPase activating protein 5 (ARHGAP5)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Pancreatic islets | Mus musculus |
|
Treatment: Mettl3 knockout mice
Control: Mettl3 flox/flox mice
|
GSE155612 | |
| Regulation |
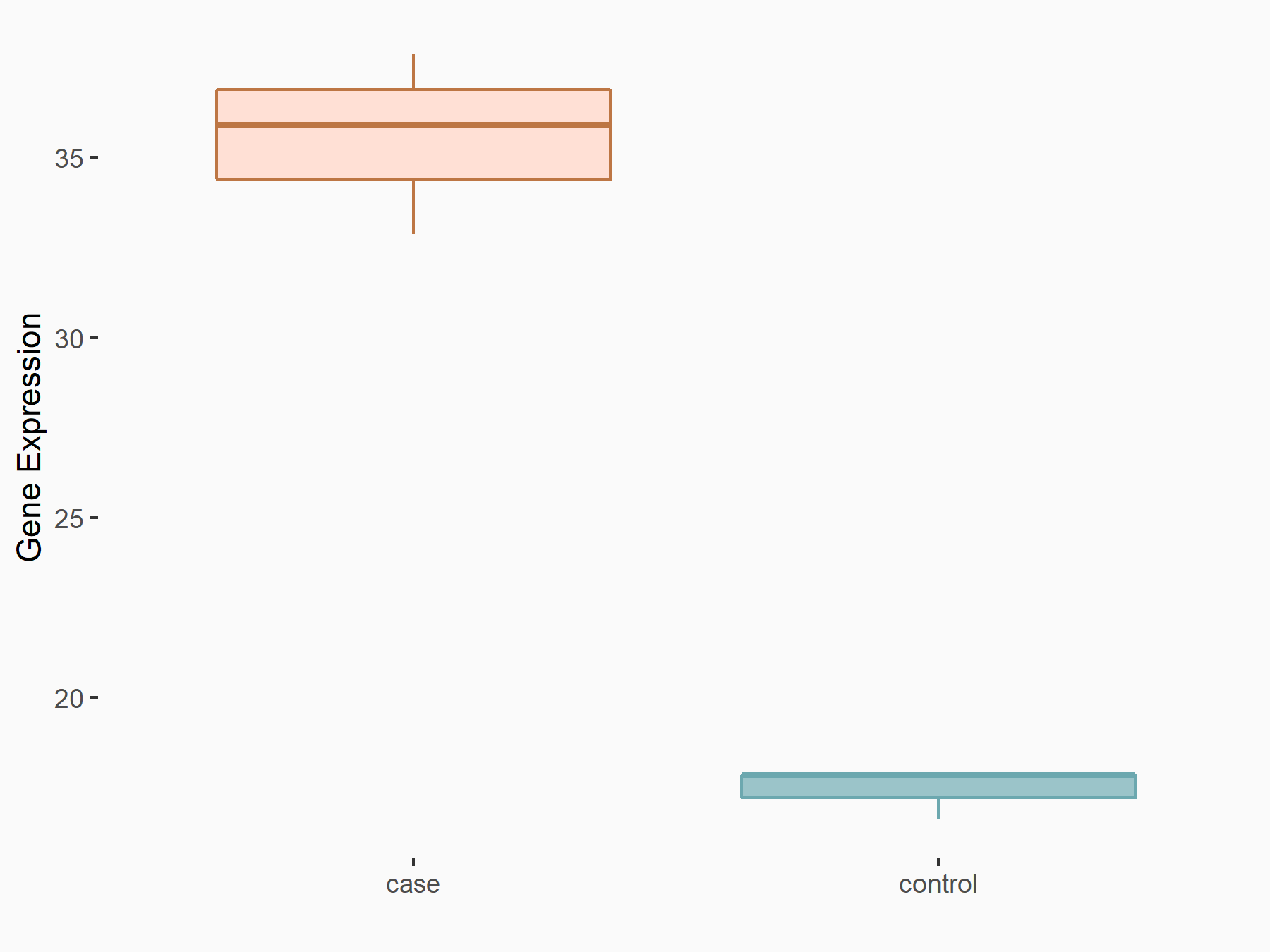 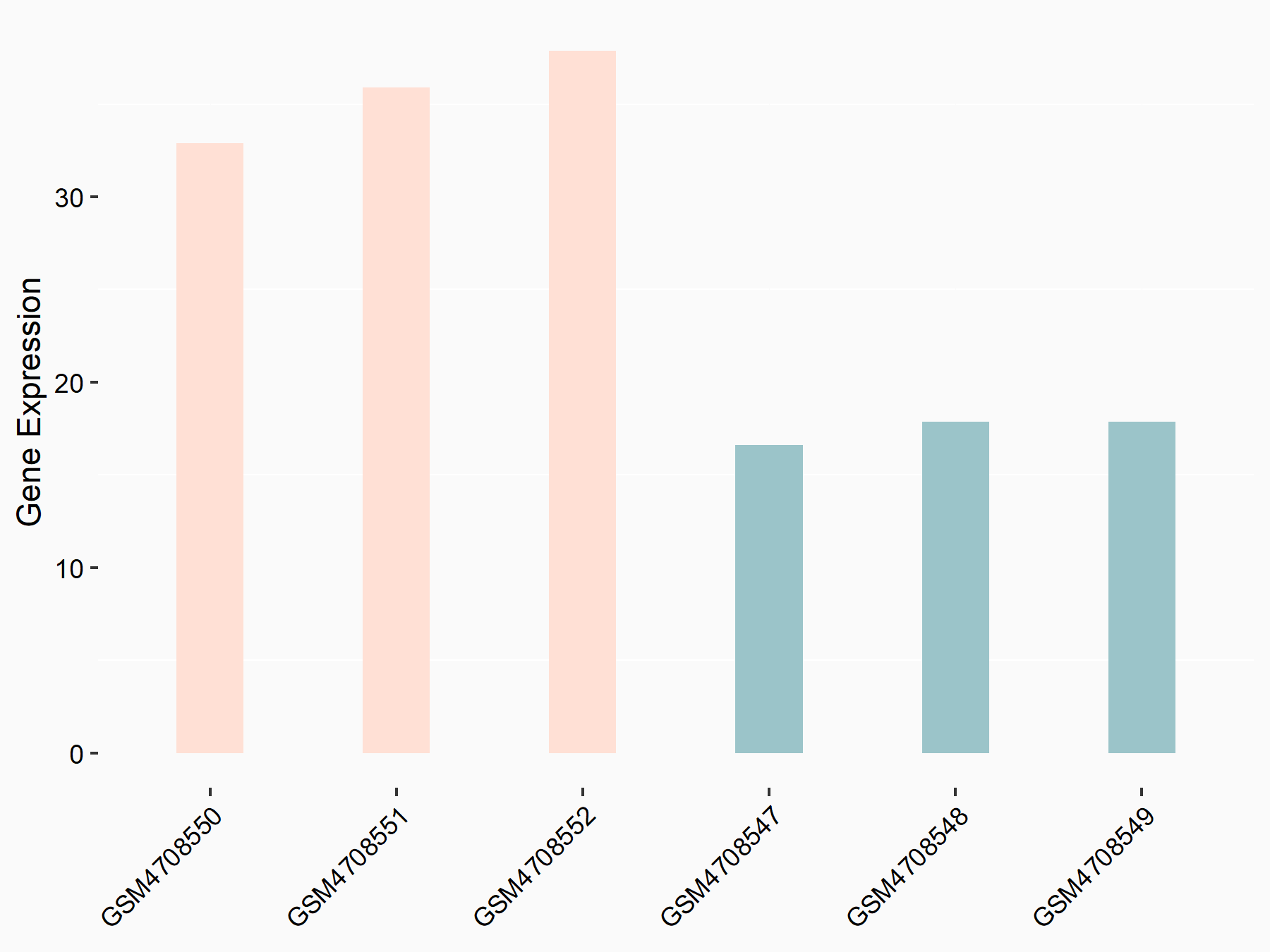 |
logFC: 9.85E-01 p-value: 6.23E-05 |
| More Results | Click to View More RNA-seq Results | |
Gastric cancer [ICD-11: 2B72]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [215] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular Processes | |||
| Transport and catabolism | ||||
In-vitro Model |
BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| Response Summary | ARHGAP5-AS1 also stabilized ARHGAP5 mRNA in the cytoplasm by recruiting METTL3 to stimulate m6A modification of Rho GTPase activating protein 5 (ARHGAP5) mRNA. As a result, ARHGAP5 was upregulated to promote chemoresistance and its upregulation was also associated with poor prognosis in gastric cancer. downregulation of ARHGAP5-AS1 in resistant cells evidently reversed the resistance to chemotherapeutic drugs including cisplatin (DDP), ADM, and 5-FU. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [215] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Responsed Drug | Fluorouracil | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular processes | |||
| Cellular transport | ||||
| Cellular catabolism | ||||
In-vitro Model |
BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| Response Summary | ARHGAP5-AS1 also stabilized ARHGAP5 mRNA in the cytoplasm by recruiting METTL3 to stimulate m6A modification of Rho GTPase activating protein 5 (ARHGAP5) mRNA. As a result, ARHGAP5 was upregulated to promote chemoresistance and its upregulation was also associated with poor prognosis in gastric cancer. downregulation of ARHGAP5-AS1 in resistant cells evidently reversed the resistance to chemotherapeutic drugs including cisplatin (DDP), ADM, and 5-FU. | |||
| Experiment 3 Reporting the m6A-centered Disease Response of This Target Gene | [215] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Responsed Drug | Adriamycin | Phase 3 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular processes | |||
| Cellular transport | ||||
| Cellular catabolism | ||||
In-vitro Model |
BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| Response Summary | ARHGAP5-AS1 also stabilized ARHGAP5 mRNA in the cytoplasm by recruiting METTL3 to stimulate m6A modification of Rho GTPase activating protein 5 (ARHGAP5) mRNA. As a result, ARHGAP5 was upregulated to promote chemoresistance and its upregulation was also associated with poor prognosis in gastric cancer. downregulation of ARHGAP5-AS1 in resistant cells evidently reversed the resistance to chemotherapeutic drugs including cisplatin (DDP), ADM, and 5-FU. | |||
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [328] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| In-vivo Model | Roughly 1 × 107 of MCF-7 or MDA-MB-231 cells stably transfected with ADAR1 sgRNA or Ctrl sgRNA (as described above) were injected subcutaneously into the thighs of the female athymic nude mice (5 weeks old, 17-18 g, three mice per group). Tumor growth was measured by measuring the width (W) and length (L) with calipers, and the volume (V) of the tumor was figured using the criterion V = (W2 × L)/2. At 42 days after injection, the mice were euthanized and tumors were removed and weighed. The tumor samples were further analyzed with IHC and Western blot assay. The animal studies were performed according to the institutional ethics guidelines for animal experiments and approved by the Chongqing Medical University Animal Care and Use Committee. | |||
Serine-protein kinase ATM (ATM)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
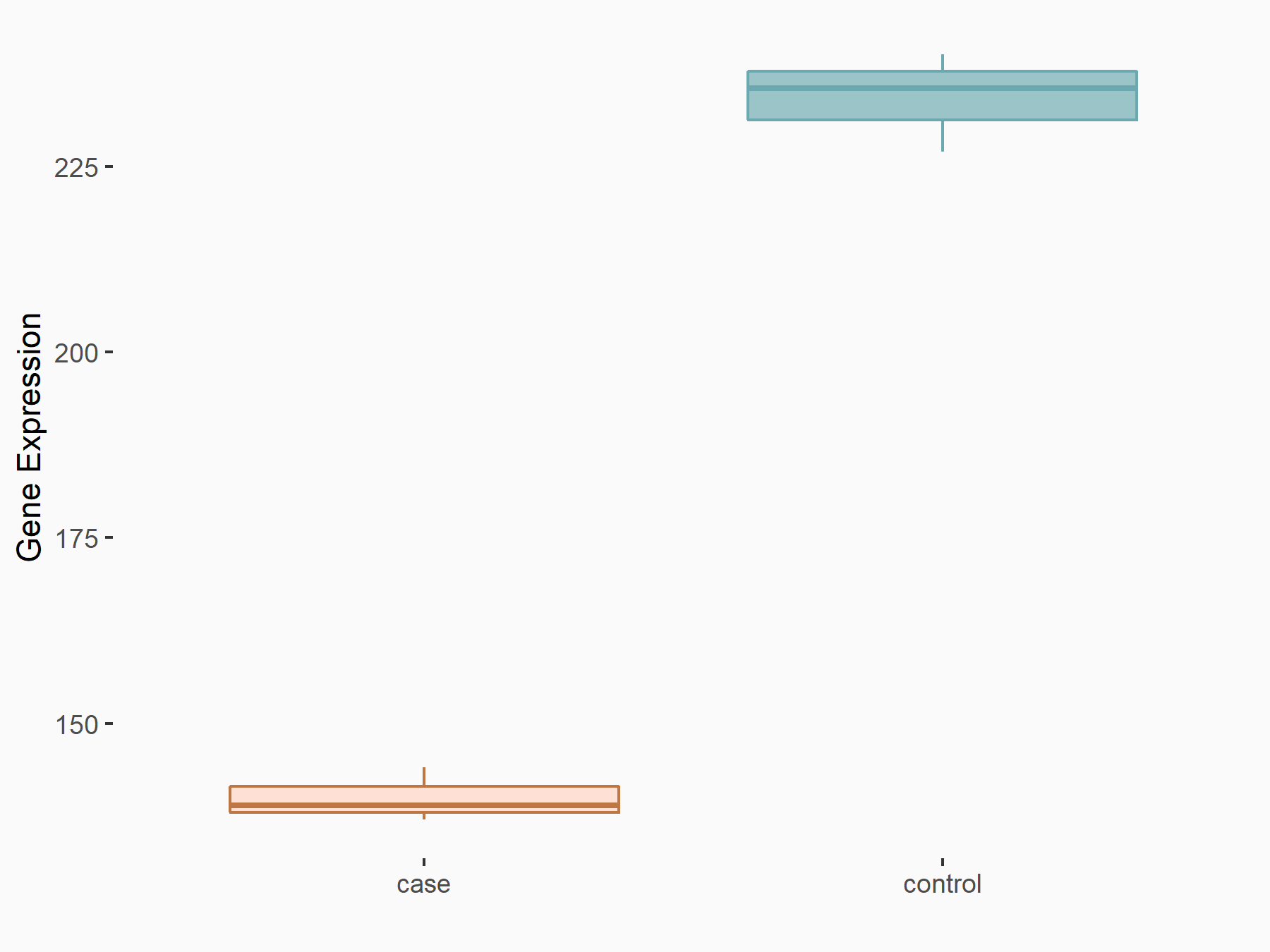 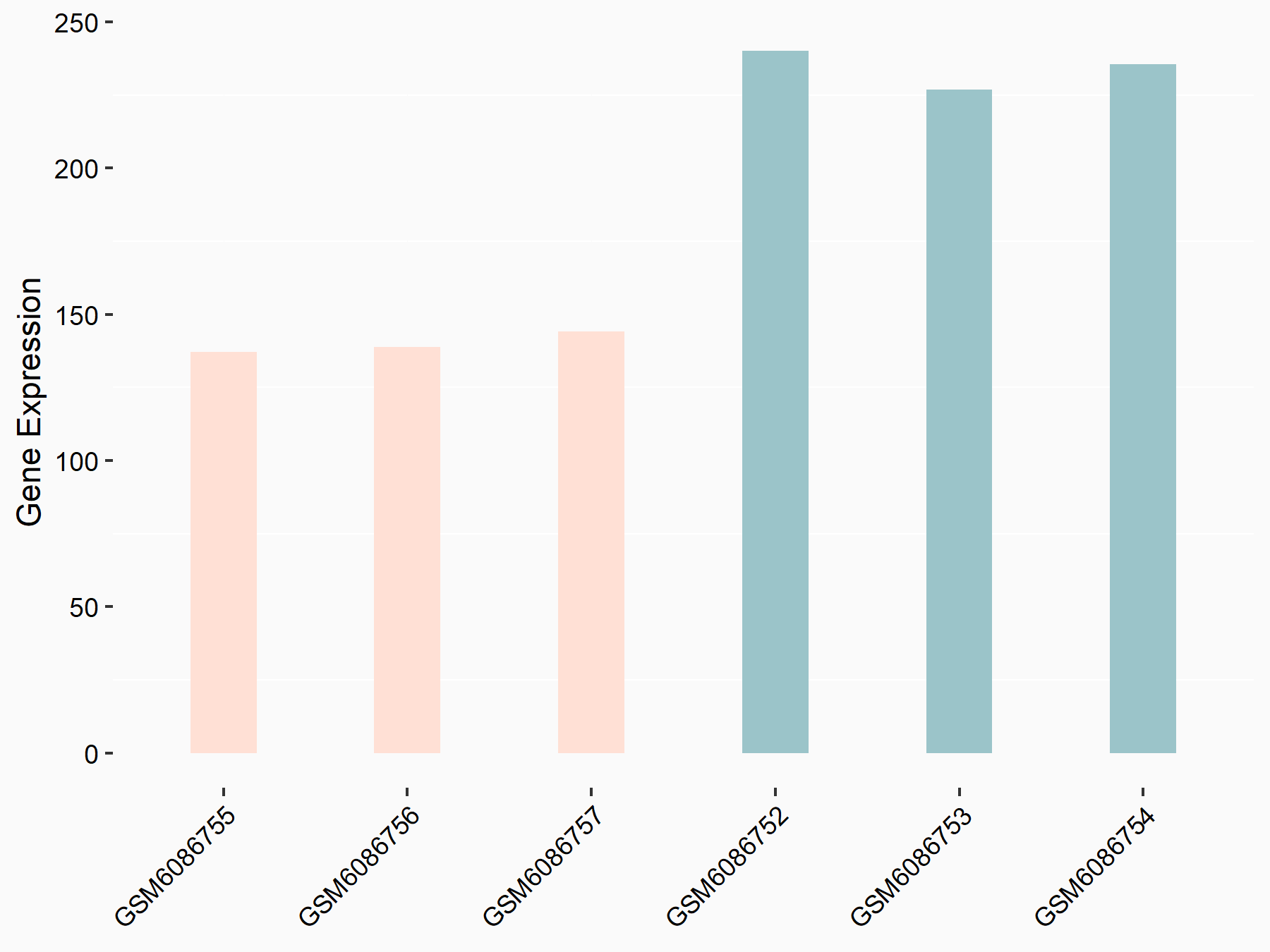 |
logFC: -7.38E-01 p-value: 7.12E-06 |
| More Results | Click to View More RNA-seq Results | |
Solid tumour/cancer [ICD-11: 2A00-2F9Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [216] | |||
| Responsed Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Homologous recombination | hsa03440 | ||
| p53 signaling pathway | hsa04115 | |||
In-vitro Model |
BJAB | Burkitt lymphoma | Homo sapiens | CVCL_5711 |
| Raji | EBV-related Burkitt lymphoma | Homo sapiens | CVCL_0511 | |
| Response Summary | m6A modification regulates ATM mRNA metabolism and ATM downstream signaling, which illustrates the importance of m6A modification-related molecules for being used as therapeutic targets in DNA damage-related diseases. METTL3 disrupts the ATM stability via m6A modification, thereby affecting the DNA-damage response. | |||
Serine/arginine-rich splicing factor 11 (SRSF11)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3 knockout liver
Control: Wild type liver cells
|
GSE198512 | |
| Regulation |
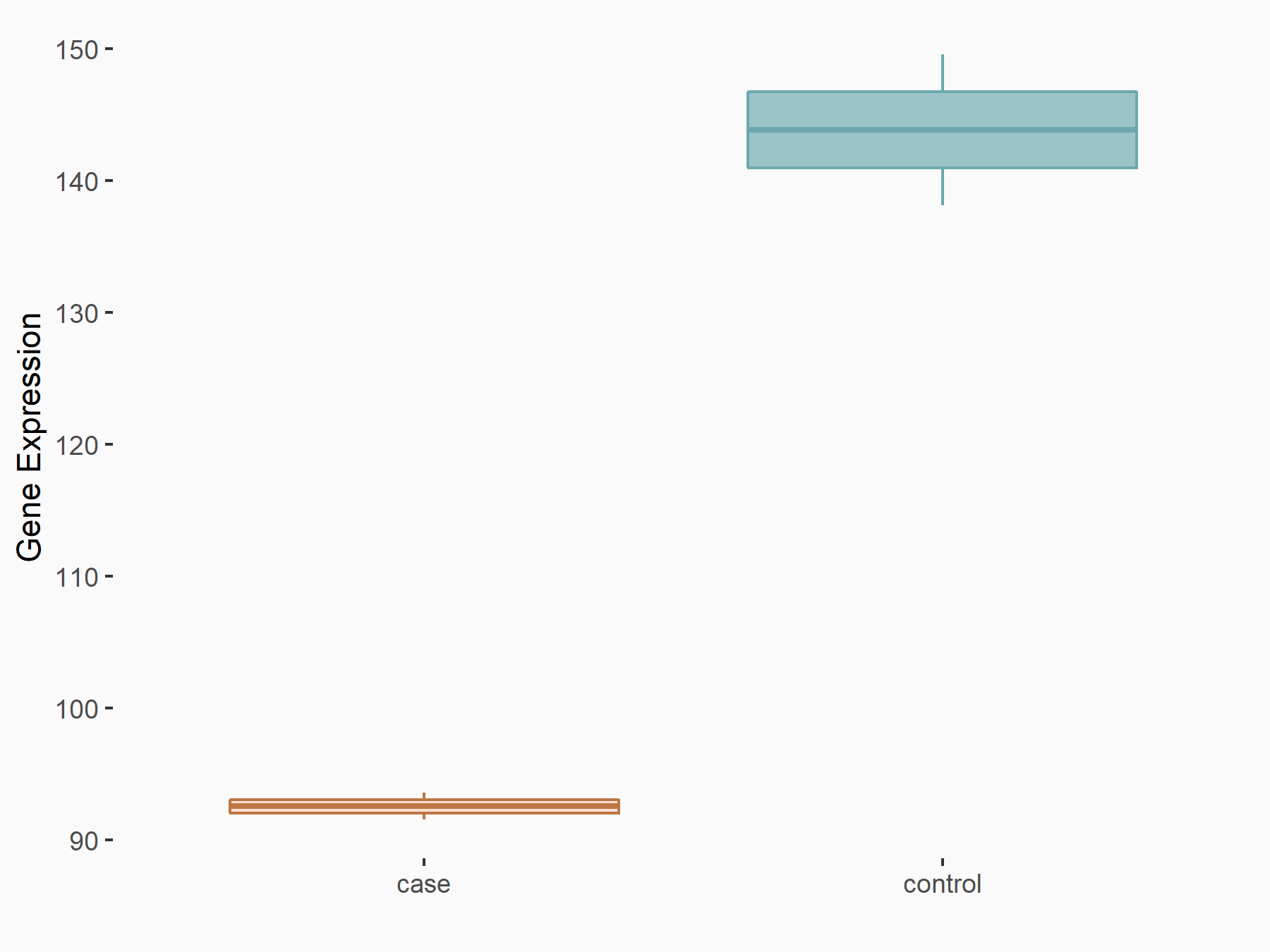 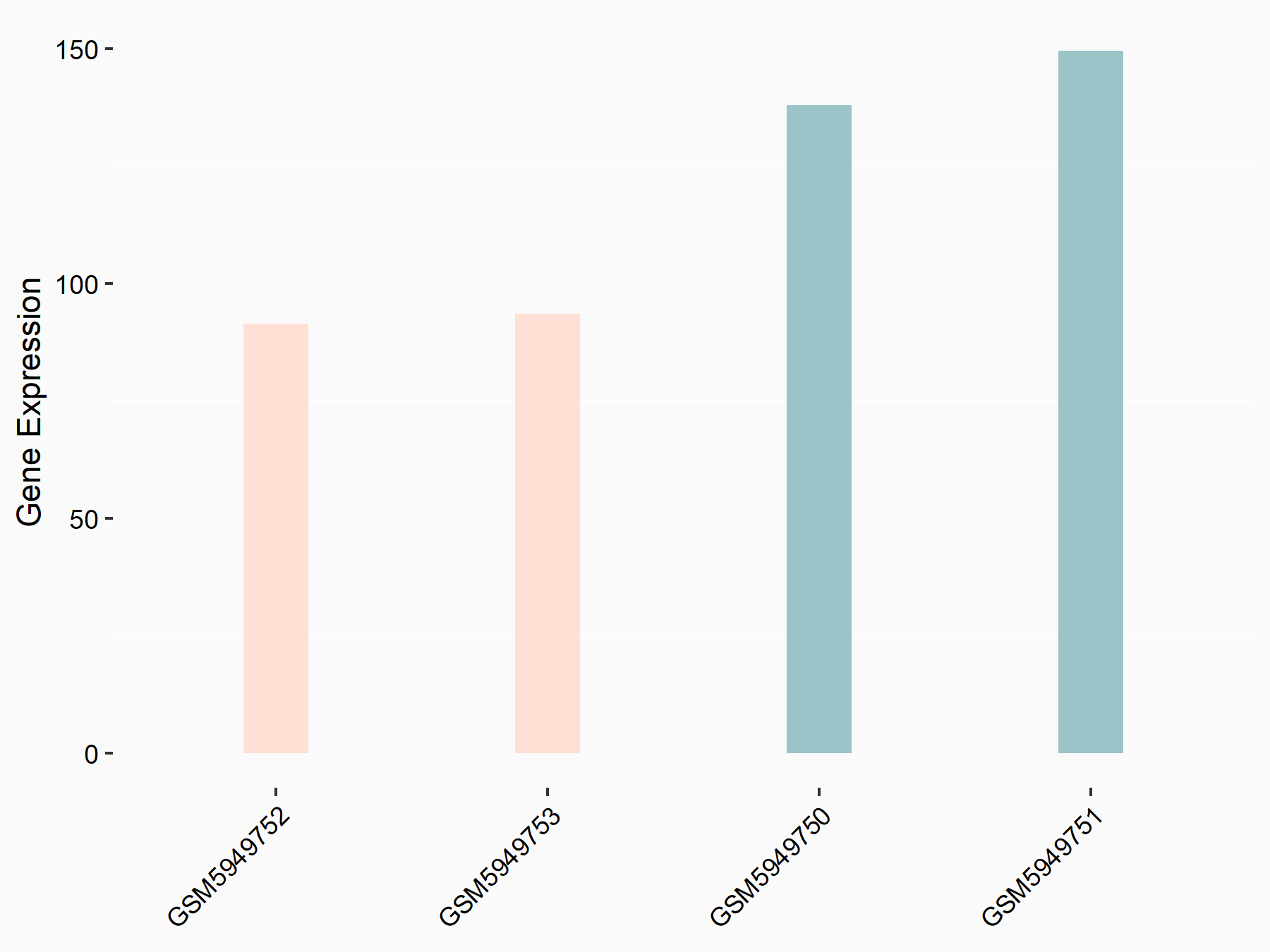 |
logFC: -6.30E-01 p-value: 2.77E-03 |
| More Results | Click to View More RNA-seq Results | |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [217] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | mRNA decay | |||
In-vitro Model |
U251 (Fibroblasts or fibroblast like cells) | |||
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| In-vivo Model | For subcutaneous tumor model, each mouse was injected subcutaneously in the right flank with 2 × 106 U87MG cells (METTL3-KD or control) in 100 uL PBS. | |||
| Response Summary | Silencing METTL3 or overexpressing dominant-negative mutant METTL3 suppressed the growth and self-renewal of Glioblastoma cells. Integrated transcriptome and MeRIP-seq analyses revealed that downregulating the expression of METTL3 decreased m6A modification levels of Serine/arginine-rich splicing factor 11 (SRSF11), which led to YTHDC1-dependent NMD of SRSF transcripts and decreased SRSF protein expression. | |||
Serine/arginine-rich splicing factor 3 (SRSF3)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LNCaP cell line | Homo sapiens |
|
Treatment: shMETTL3 LNCaP cells
Control: shControl LNCaP cells
|
GSE147884 | |
| Regulation |
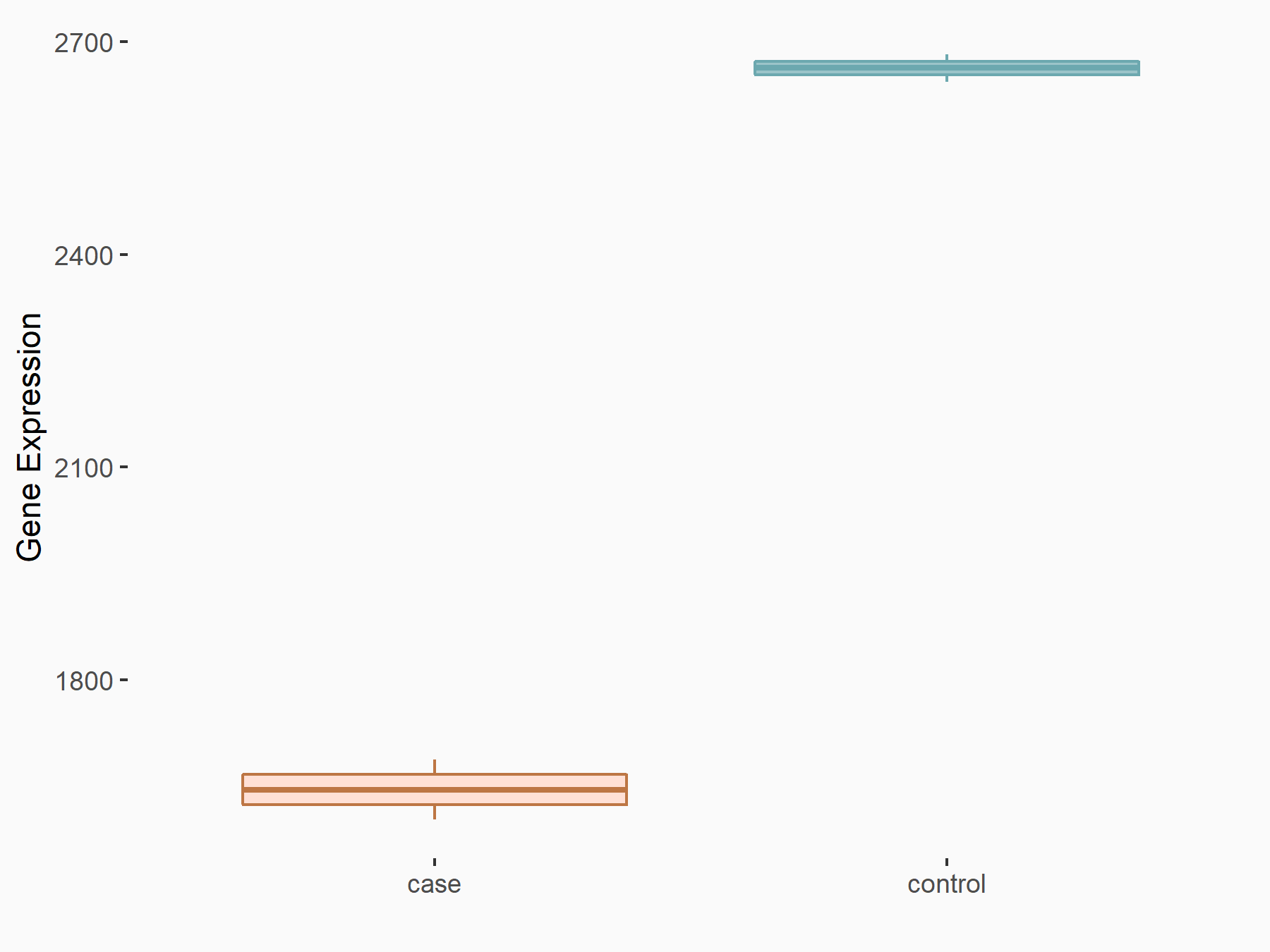 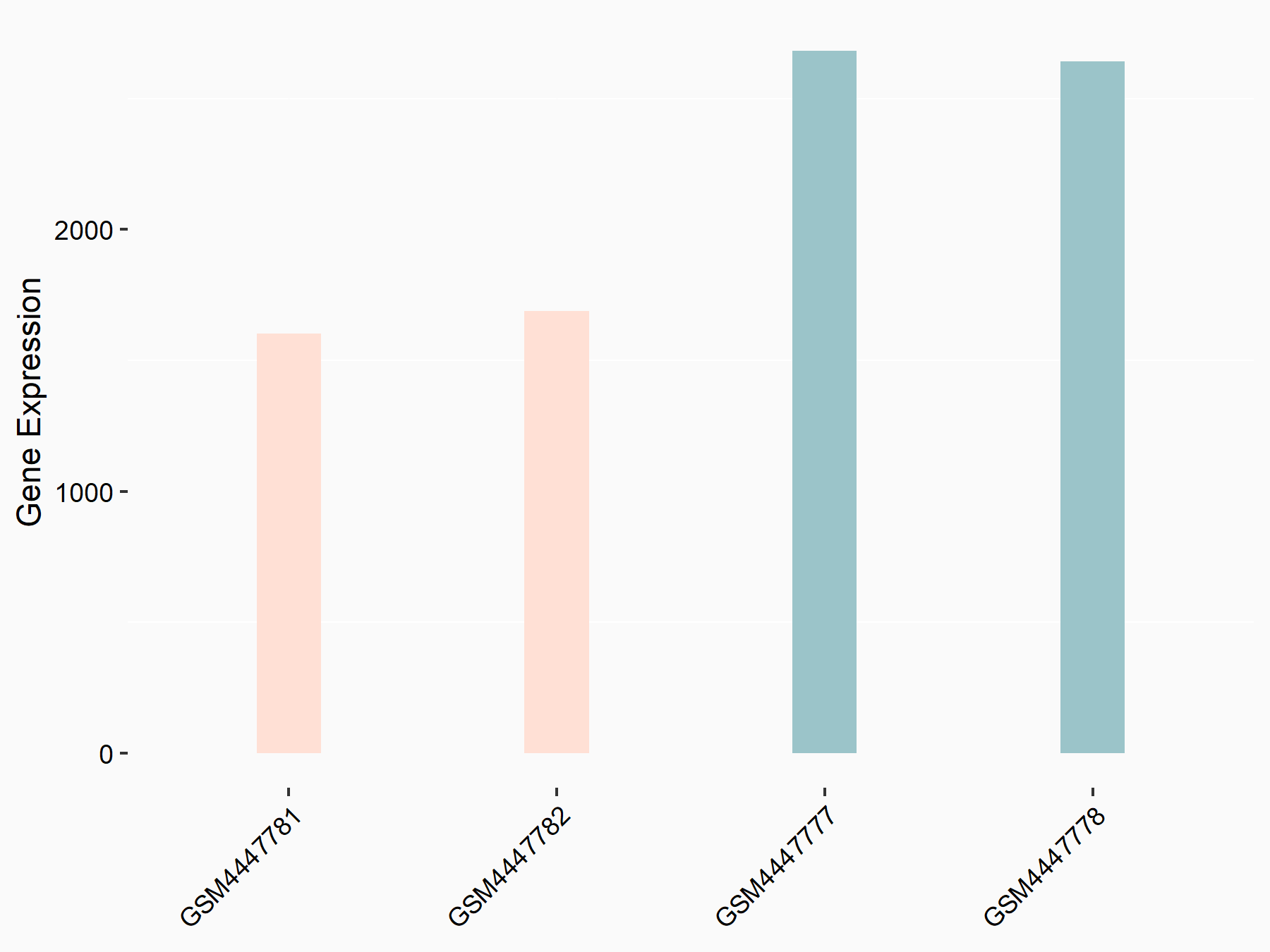 |
logFC: -6.96E-01 p-value: 4.00E-41 |
| More Results | Click to View More RNA-seq Results | |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [217] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | mRNA decay | |||
In-vitro Model |
U251 (Fibroblasts or fibroblast like cells) | |||
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| In-vivo Model | For subcutaneous tumor model, each mouse was injected subcutaneously in the right flank with 2 × 106 U87MG cells (METTL3-KD or control) in 100 uL PBS. | |||
| Response Summary | Silencing METTL3 or overexpressing dominant-negative mutant METTL3 suppressed the growth and self-renewal of Glioblastoma cells. Integrated transcriptome and MeRIP-seq analyses revealed that downregulating the expression of METTL3 decreased m6A modification levels of Serine/arginine-rich splicing factor 3 (SRSF3), which led to YTHDC1-dependent NMD of SRSF transcripts and decreased SRSF protein expression. | |||
Serine/arginine-rich splicing factor 6 (SRSF6)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
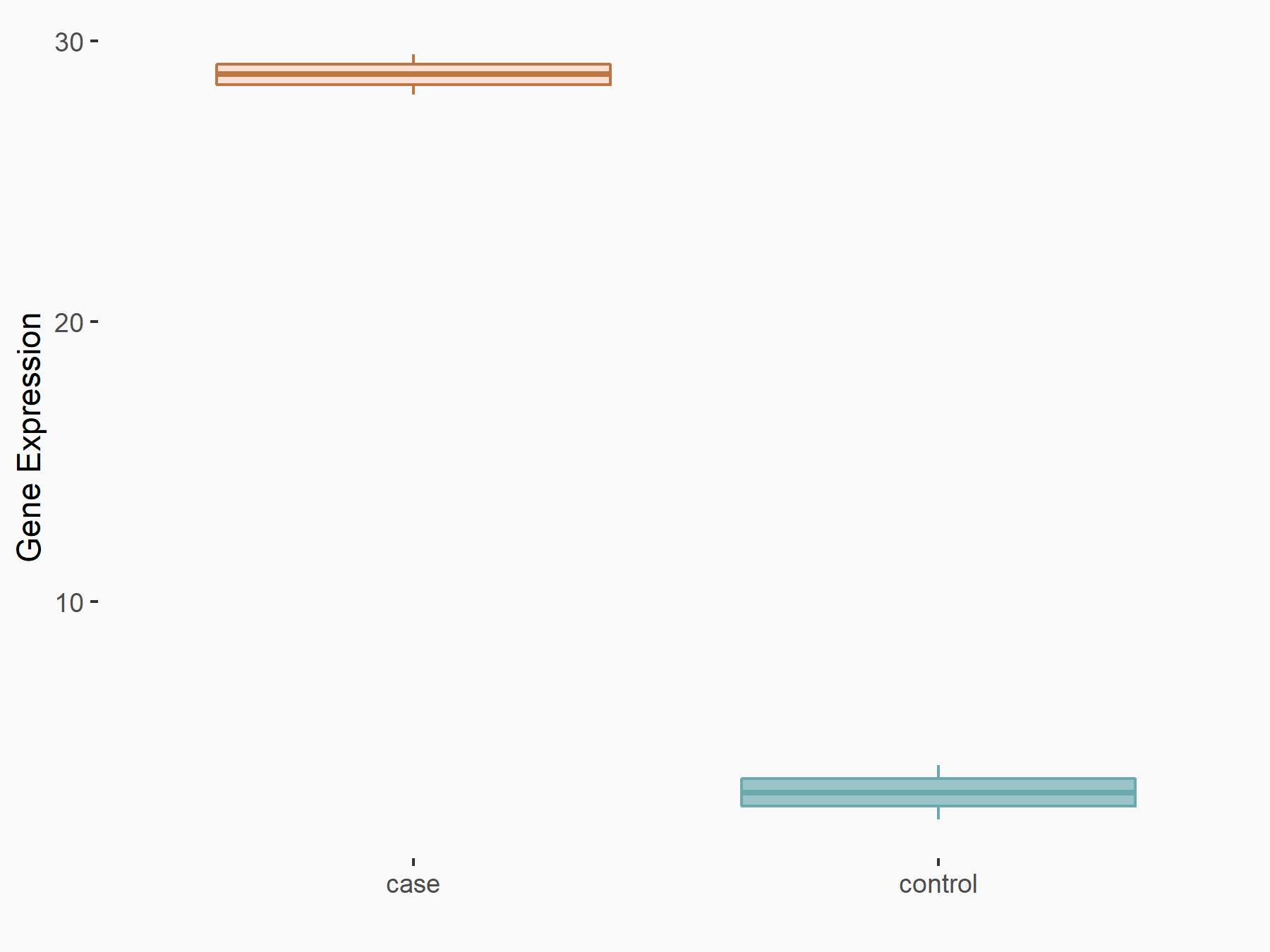 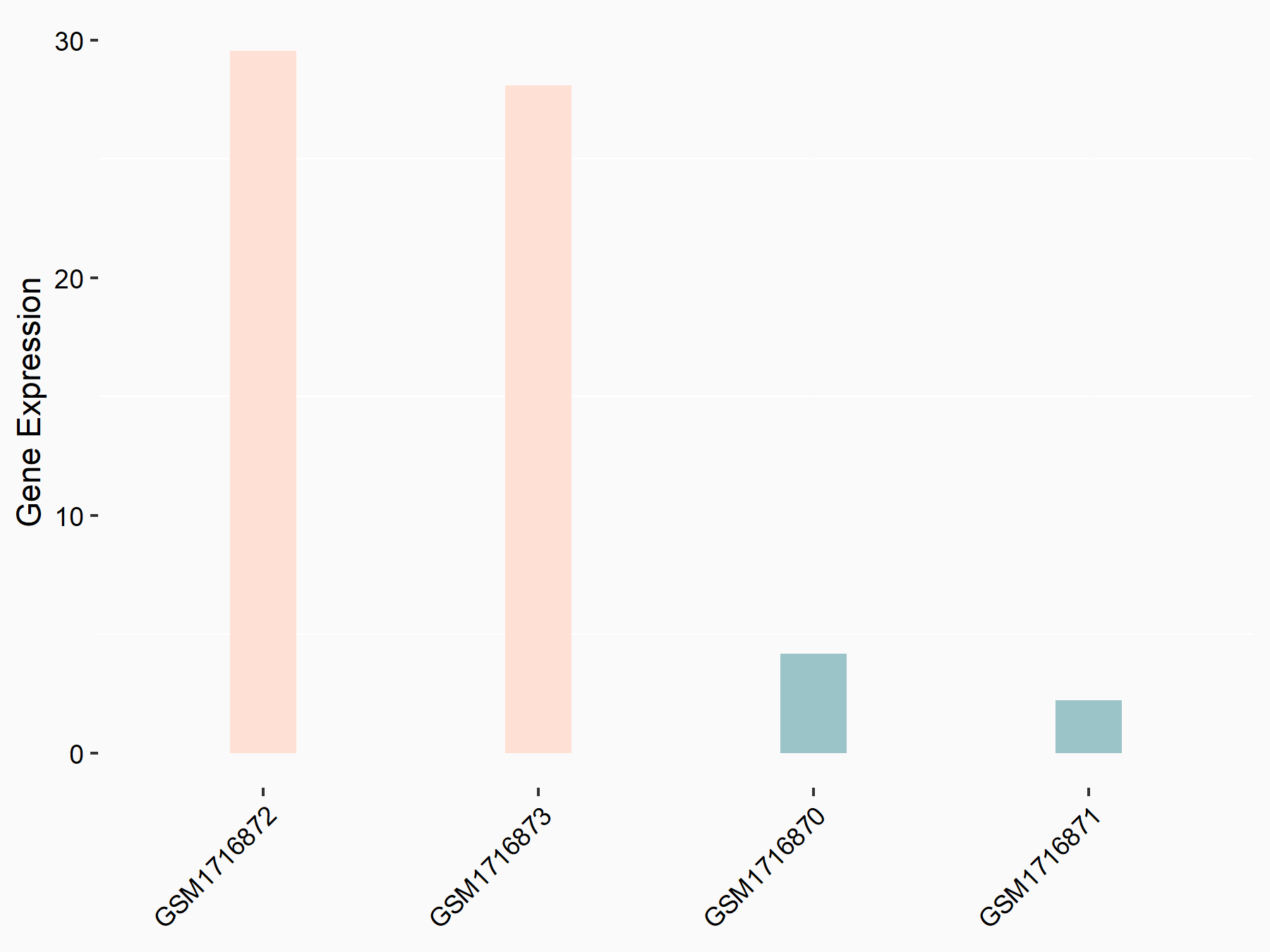 |
logFC: 2.87E+00 p-value: 5.80E-03 |
| More Results | Click to View More RNA-seq Results | |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [217] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | mRNA decay | |||
In-vitro Model |
U251 (Fibroblasts or fibroblast like cells) | |||
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| In-vivo Model | For subcutaneous tumor model, each mouse was injected subcutaneously in the right flank with 2 × 106 U87MG cells (METTL3-KD or control) in 100 uL PBS. | |||
| Response Summary | Silencing METTL3 or overexpressing dominant-negative mutant METTL3 suppressed the growth and self-renewal of Glioblastoma cells. Integrated transcriptome and MeRIP-seq analyses revealed that downregulating the expression of METTL3 decreased m6A modification levels of Serine/arginine-rich splicing factor 6 (SRSF6), which led to YTHDC1-dependent NMD of SRSF transcripts and decreased SRSF protein expression. | |||
Serine/threonine-protein kinase PLK1 (PLK1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
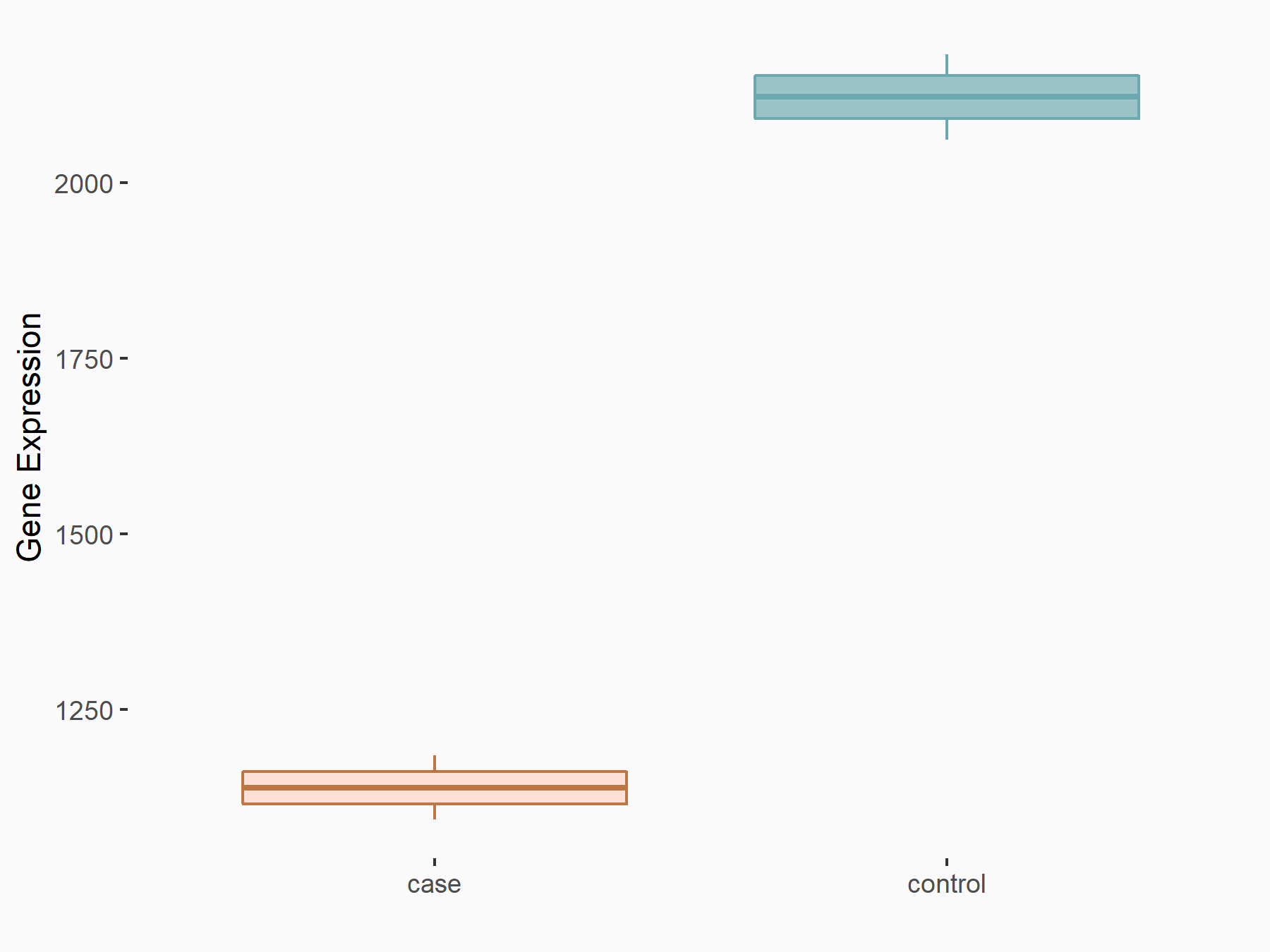 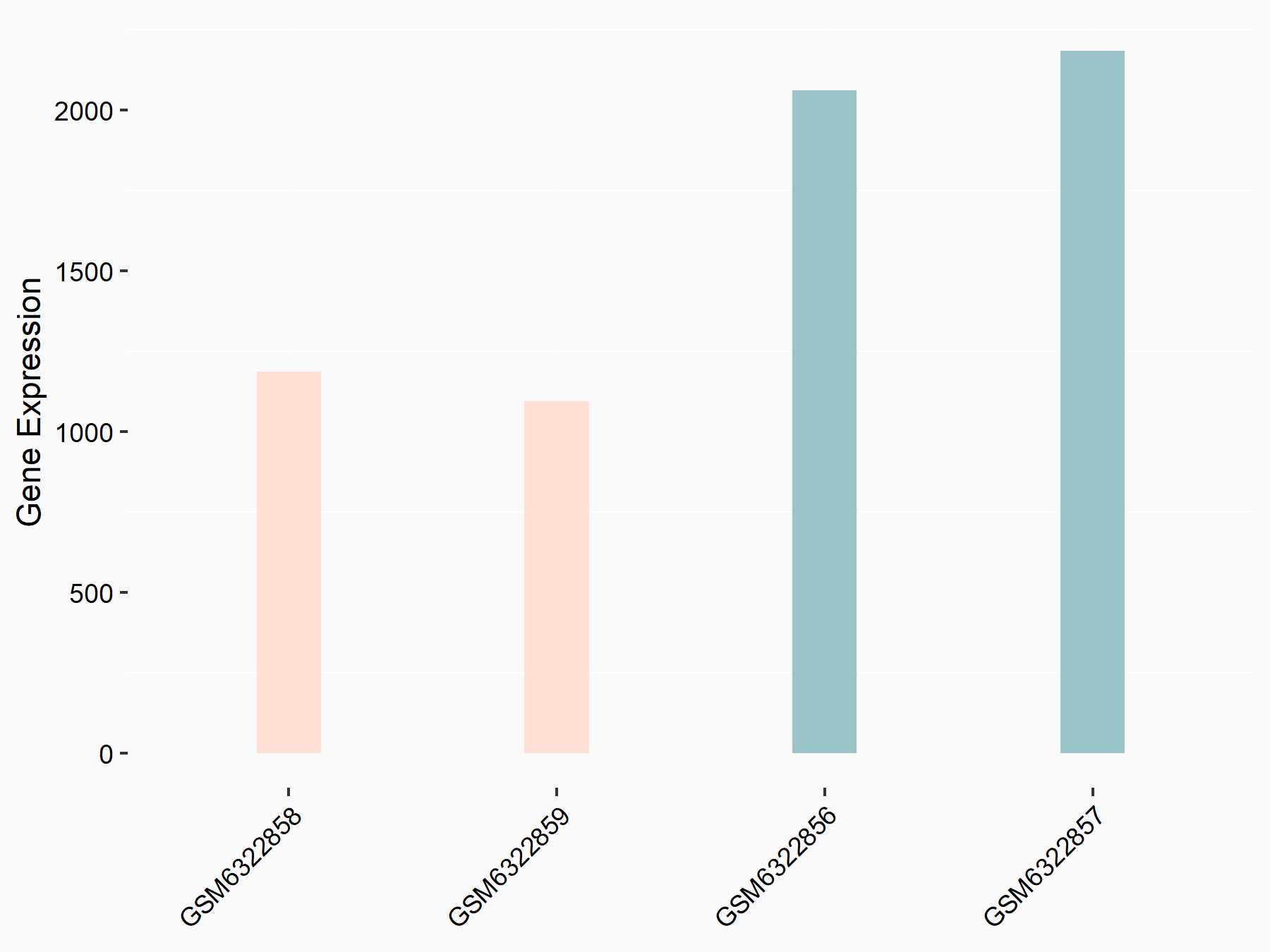 |
logFC: -8.98E-01 p-value: 1.15E-18 |
| More Results | Click to View More RNA-seq Results | |
Pulpitis [ICD-11: DA09]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [218] | |||
| Responsed Disease | Pulpitis [ICD-11: DA09] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell senescence | |||
In-vitro Model |
DPSC | Normal | Homo sapiens | CVCL_AV90 |
| Response Summary | Impaired METTL3 expression in dental pulp stem cells led to increasing cell senescence and apoptosis by interfering with the mitotic cell cycle in a m6A-dependent manner. The protein interaction network of differentially expressed genes identified Serine/threonine-protein kinase PLK1 (PLK1), a critical cycle modulator, as the target of METTL3-mediated m6A methylation in DPSCs. | |||
Signal transducer and activator of transcription 3 (STAT3)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Embryonic stem cells | Mus musculus |
|
Treatment: METTL3 knockout mESCs
Control: Wild type mESCs
|
GSE156481 | |
| Regulation |
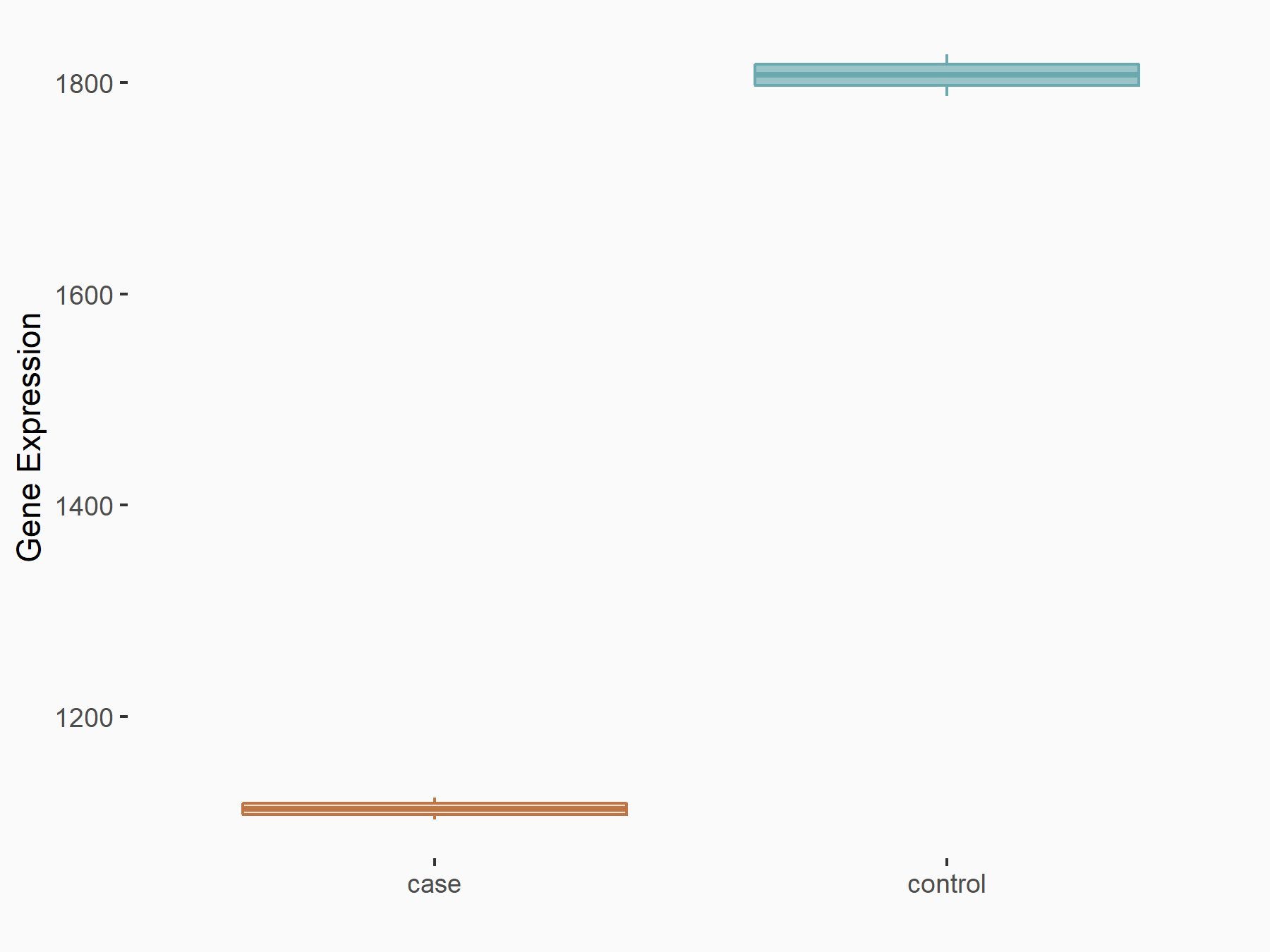 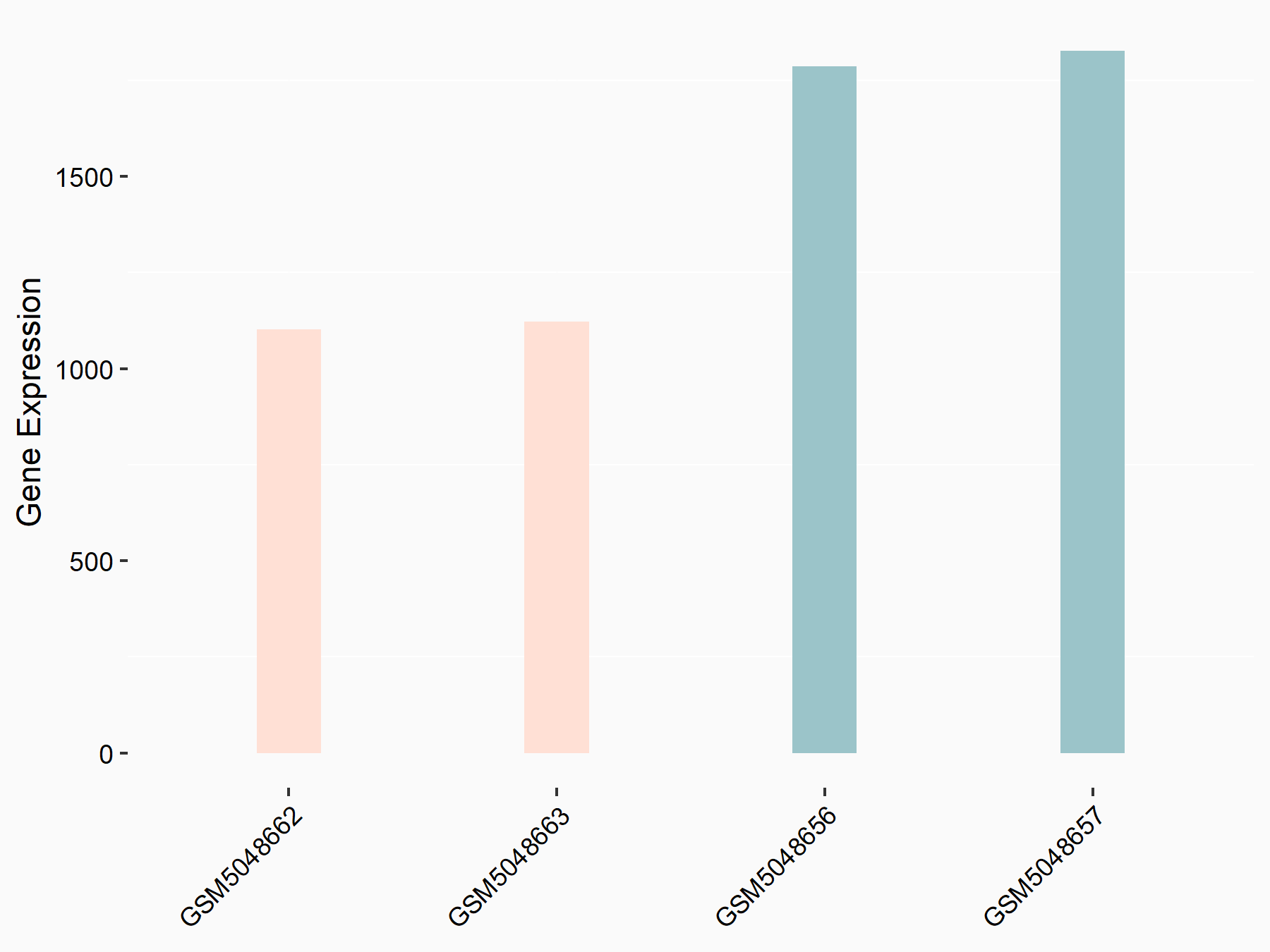 |
logFC: -6.99E-01 p-value: 1.19E-15 |
| More Results | Click to View More RNA-seq Results | |
Atherosclerosis [ICD-11: BD40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [130] | |||
| Responsed Disease | Atherosclerosis [ICD-11: BD40.Z] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
| Cell Process | Cell proliferation and migration | |||
In-vitro Model |
HUVEC-C | Normal | Homo sapiens | CVCL_2959 |
| In-vivo Model | The adeno-associated viruses (AAV) that could silence METTL3 (sh-METTL3) and the negative control adeno-associated viruses (sh-NC) were obtained from WZ Biosciences Inc. (Jinan, China). APOE-/- mice were randomly divided into AS + sh-NC and AS + sh-METTL3 groups. Each group contains five mice. Mice were fed with the standard diet for 1 week to acclimatize. After 1 week of acclimation, mice were challenged with a high-fat and high-cholesterol feed H10540 (Beijing HFK BIOSCIENCE Co., Ltd., Beijing, China). The formula of the H10540 feed was shown in Supplementary File S1. After 8 weeks of HFD feeding, sh-NC or sh-METTL3 adeno-associated virus serotype 9 (AAV9, 1012 viral genome copies per mouse) were respectively delivered into mice in AS + sh-NC or AS + sh-METTL3 group through tail vein injection. At 14 weeks after HDF feeding, mice fasted overnight. | |||
| Response Summary | METTL3 knockdown prevented Atherosclerosis progression by inhibiting JAK2/Signal transducer and activator of transcription 3 (STAT3) pathway via IGF2BP1. | |||
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [334] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
In-vitro Model |
GES-1 | Normal | Homo sapiens | CVCL_EQ22 |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| In-vivo Model | Male nude mice (age: 4 weeks) were obtained from Charles River (Hangzhou, Zhejiang, China). For tumorigenesis analysis, AGS cells (1 × 106) with stable knockdown of AGAP2-AS1 or scramble, were injected into mice. Next, we detected and measured the tumor volume each week. The weight of the tumor in each nude mouse was also measured at 4 weeks after injection. Immunohistochemistry (IHC) was used to detect Ki67- and caspase-3- positive cells in the tumor. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [335] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | The mice were housed according to the guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The corresponding cells (5 × 106) with luciferase signal were prepared and resuspended in 150 μl PBS/Matrigel (BD Biosciences, 1:1 mixture). The mice were randomly divided into 3 groups (n = 5 per group). The cells were injected into the mice's hepatic lobe. After 2 weeks, the mice were anesthetized with isoflurane and intraperitoneally injected with 150 mg/kg D-Luciferin once a week to evaluate tumor metastasis. | |||
Spectrin beta, non-erythrocytic 2 (SPTBN2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
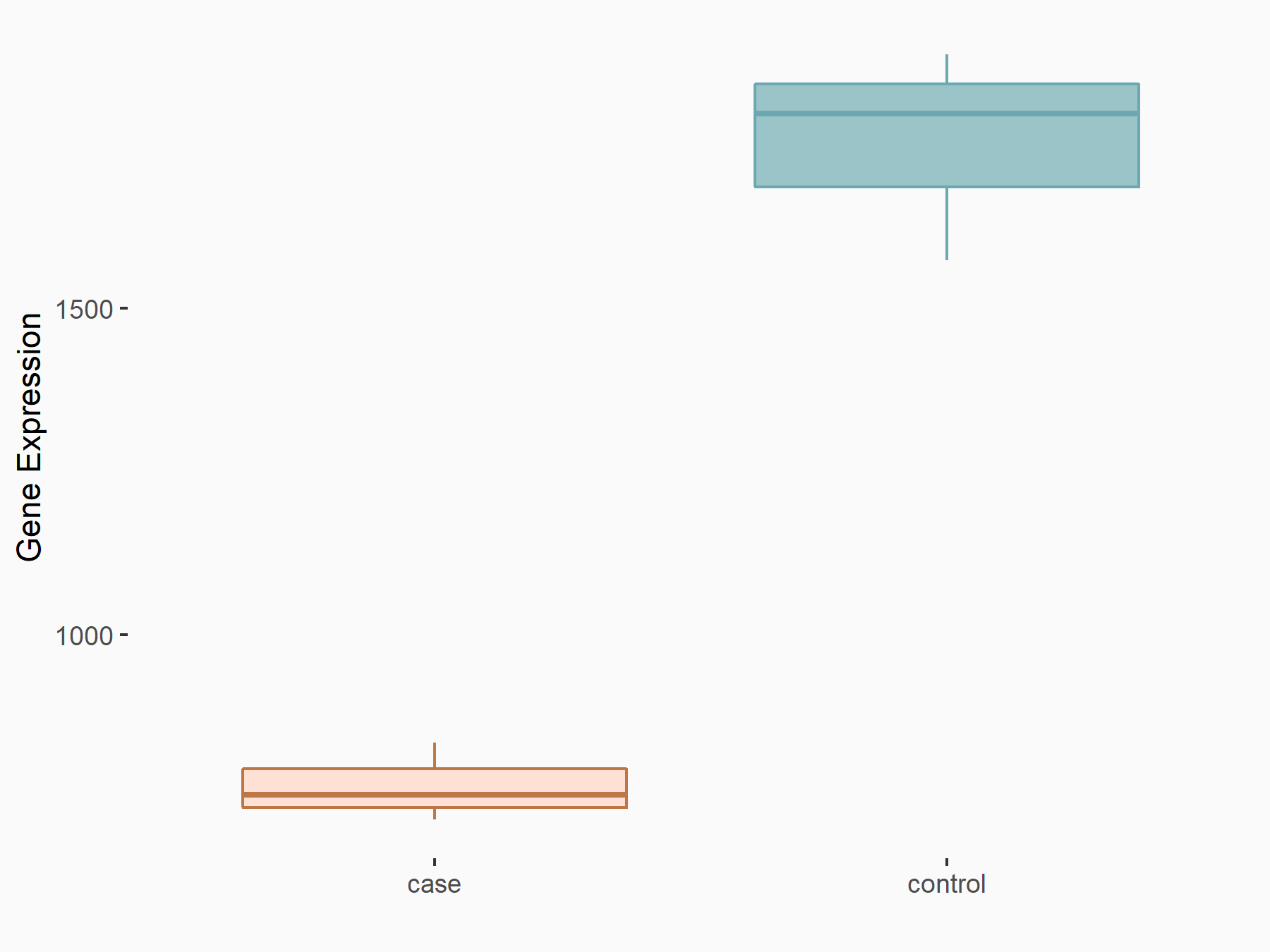 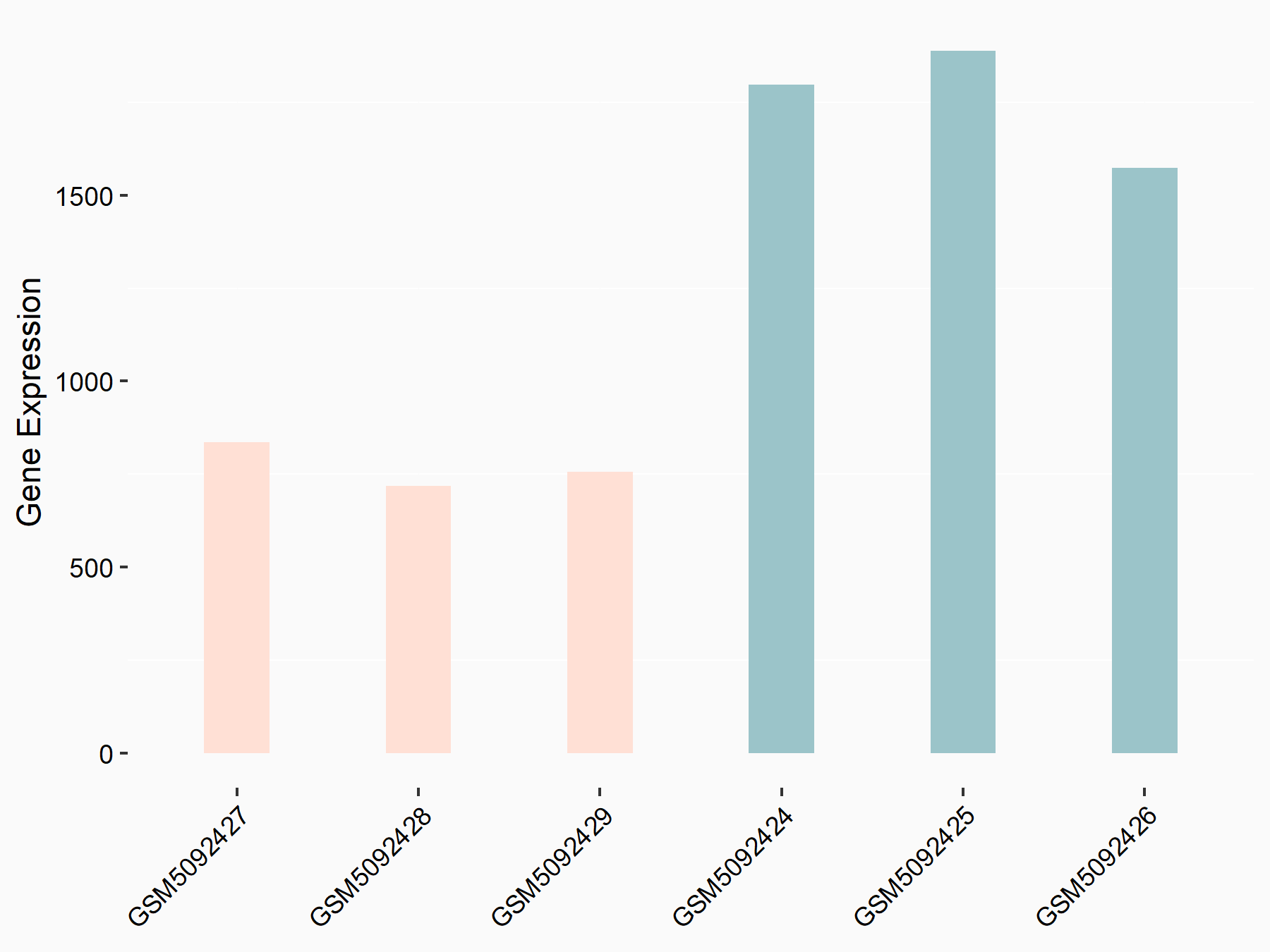 |
logFC: -1.19E+00 p-value: 2.92E-45 |
| More Results | Click to View More RNA-seq Results | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [219] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
In-vitro Model |
Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| In-vivo Model | The growth of CC cells in vivo was examined by subcutaneous injection of LoVo cells or Caco-2 cells into NSG mice, while the metastatic ability of cells in vivo by intracardiac injection into mice. | |||
| Response Summary | Overexpression of LINC01605, regulated by SMYD2-EP300-mediated H3K27ac and H3K4me3 modifications, bound to METTL3 protein to promote m6A modification of Spectrin beta, non-erythrocytic 2 (SPTBN2) mRNA, leading to the development of colorectal cancer. | |||
Sprouty-related, EVH1 domain-containing protein 2 (SPRED2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Embryonic stem cells | Mus musculus |
|
Treatment: METTL3 knockout mESCs
Control: Wild type mESCs
|
GSE156481 | |
| Regulation |
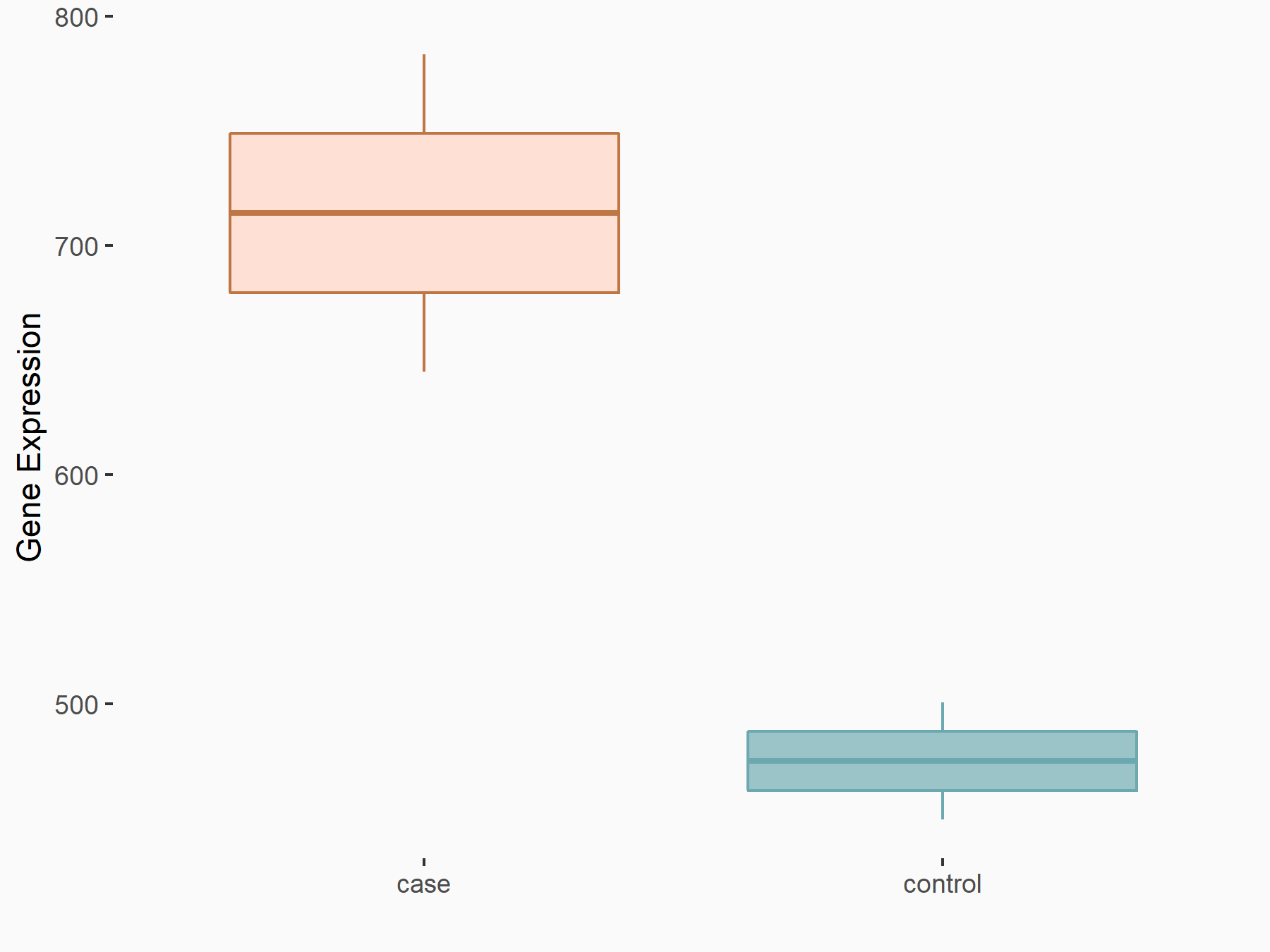 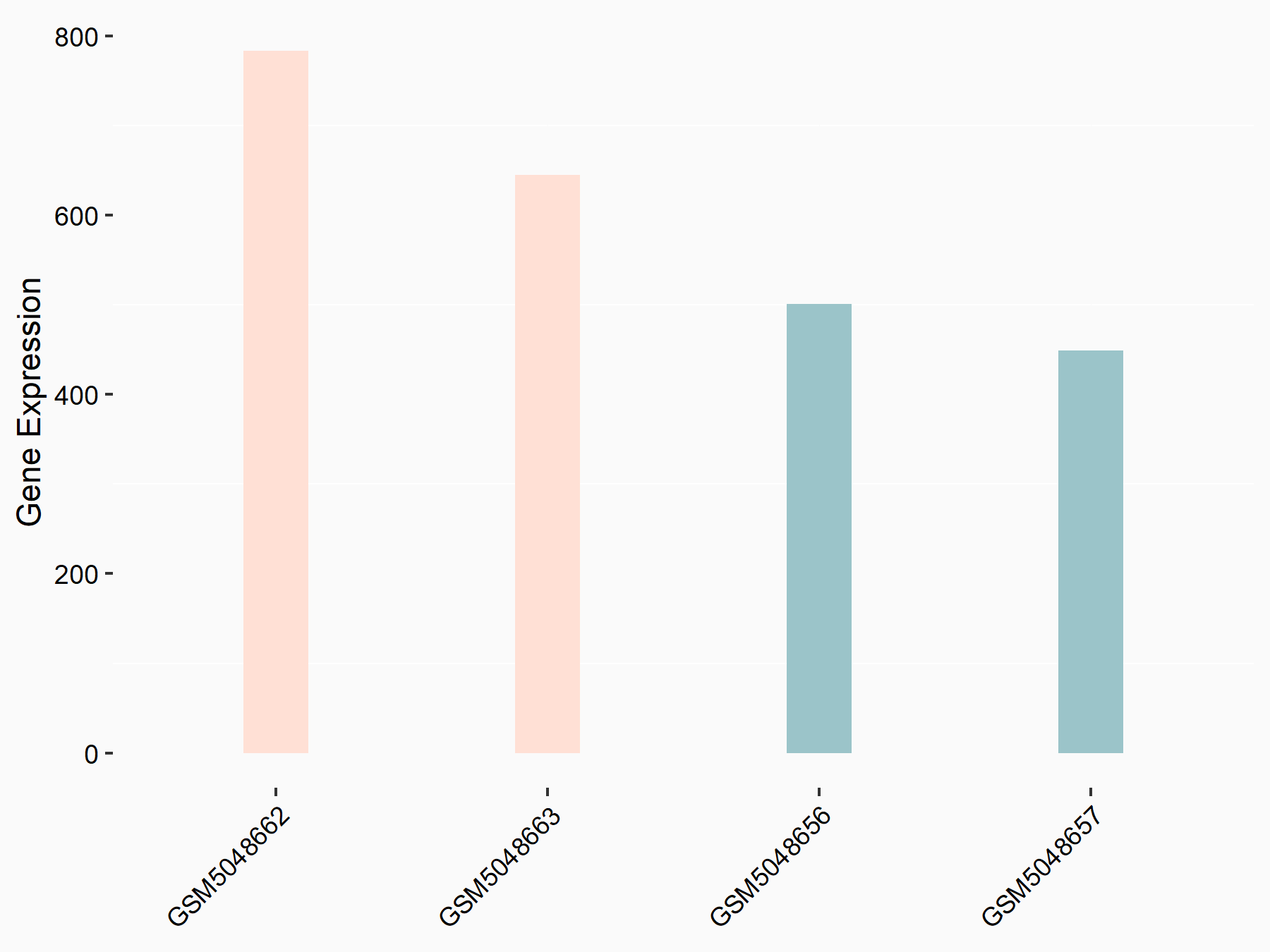 |
logFC: 5.88E-01 p-value: 1.67E-05 |
| More Results | Click to View More RNA-seq Results | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [220] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | Cell migration and invasion | |||
In-vitro Model |
Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| In-vivo Model | The spleen in the upper left lateral abdomen of the anesthetized mice were exposed, 106 cells suspended in 20 uL phosphate-buffered saline (PBS) were injected into the distal tip of the spleen. After injection, replacing the spleen, and closing the incision. | |||
| Response Summary | METTL3/miR-1246/Sprouty-related, EVH1 domain-containing protein 2 (SPRED2) axis plays an important role in tumor metastasis and provides a new m6A modification pattern in Colorectal cancer development. | |||
Stromelysin-1 (MMP-3)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HUVEC cell line | Homo sapiens |
|
Treatment: shMETTL3 HUVEC cells
Control: shScramble HUVEC cells
|
GSE157544 | |
| Regulation |
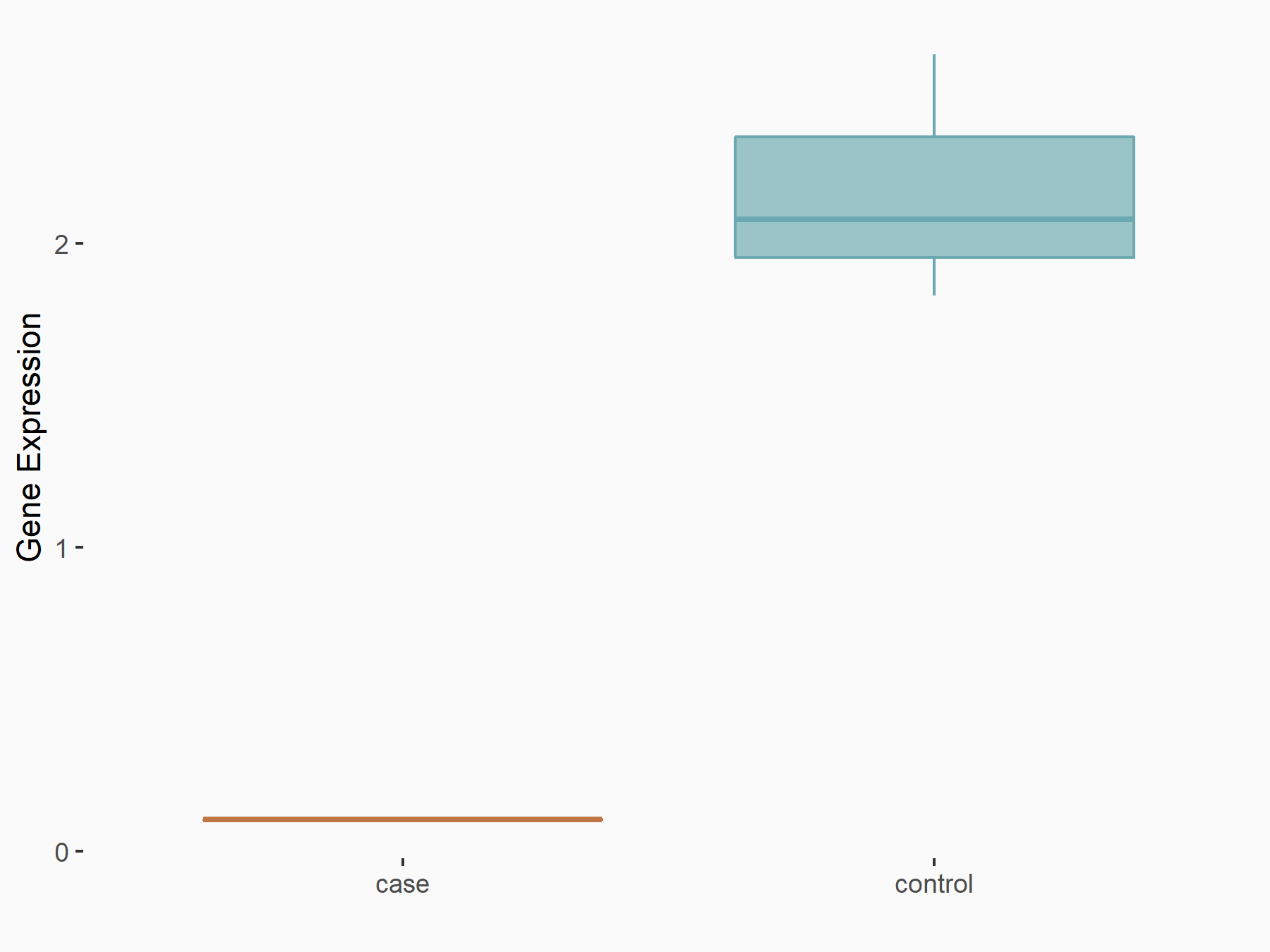 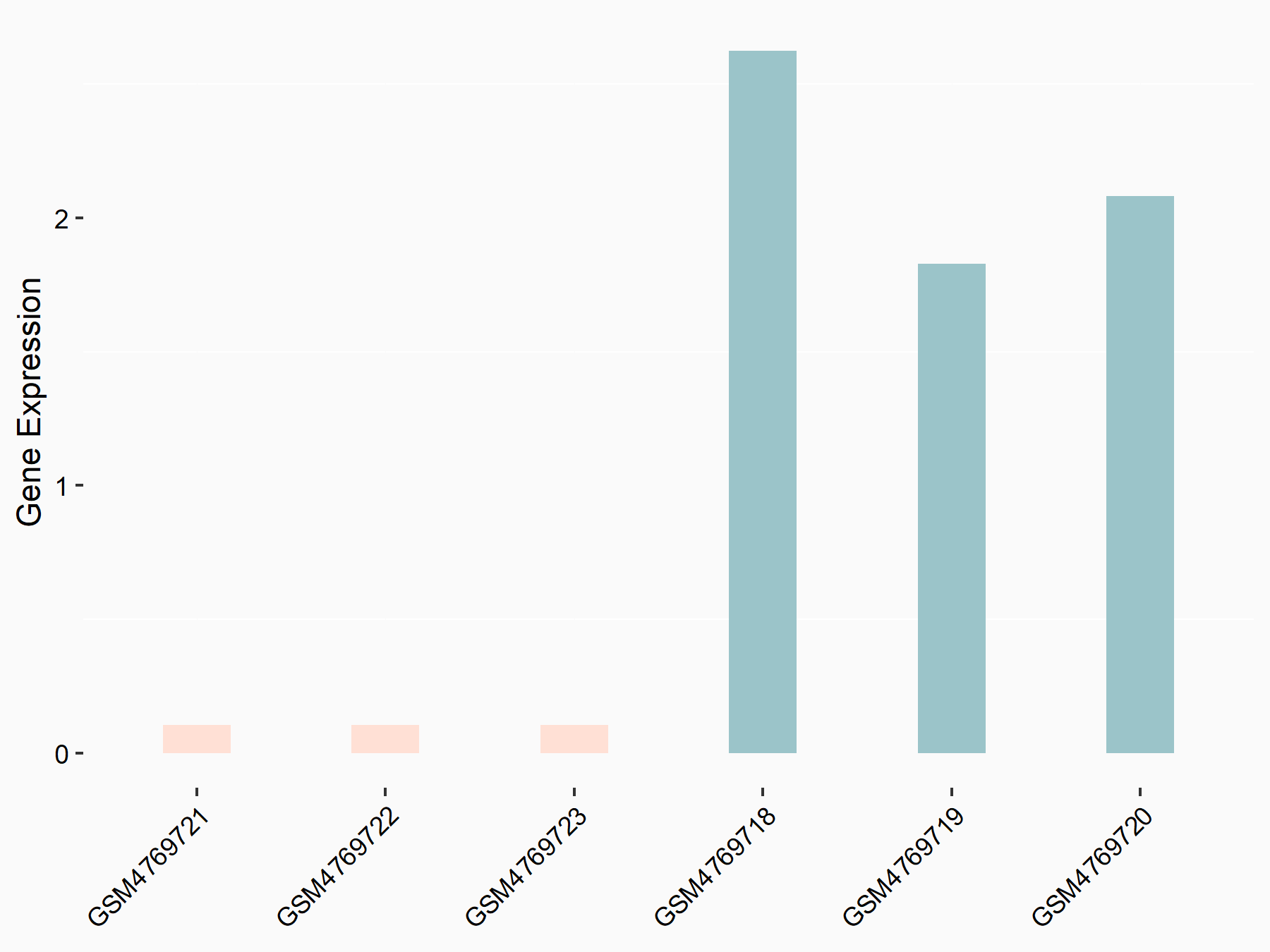 |
logFC: -1.52E+00 p-value: 7.27E-05 |
| More Results | Click to View More RNA-seq Results | |
Rheumatoid arthritis [ICD-11: FA20]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [187] | |||
| Responsed Disease | Rheumatoid arthritis [ICD-11: FA20] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Inflammatory response | |||
In-vitro Model |
FLS (Rat fibroblast synovial cell line) | |||
| In-vivo Model | To establish the adjuvant-induced arthritis (AIA) model, the rats were given complete Freund's adjuvant (CFA; Chondrex, Inc.) on the left paw of 0.1 ml per 100 g of body weight. Additionally, the rats were injected with normal saline to create the negative control (NC) group. | |||
| Response Summary | METTL3 knockdown suppressed interleukin (IL)-6, matrix metalloproteinase Stromelysin-1 (MMP-3), and MMP-9 levels in human RA-FLSs and rat AIA-FLSs. | |||
Suppressor of cytokine signaling 2 (SOCS2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
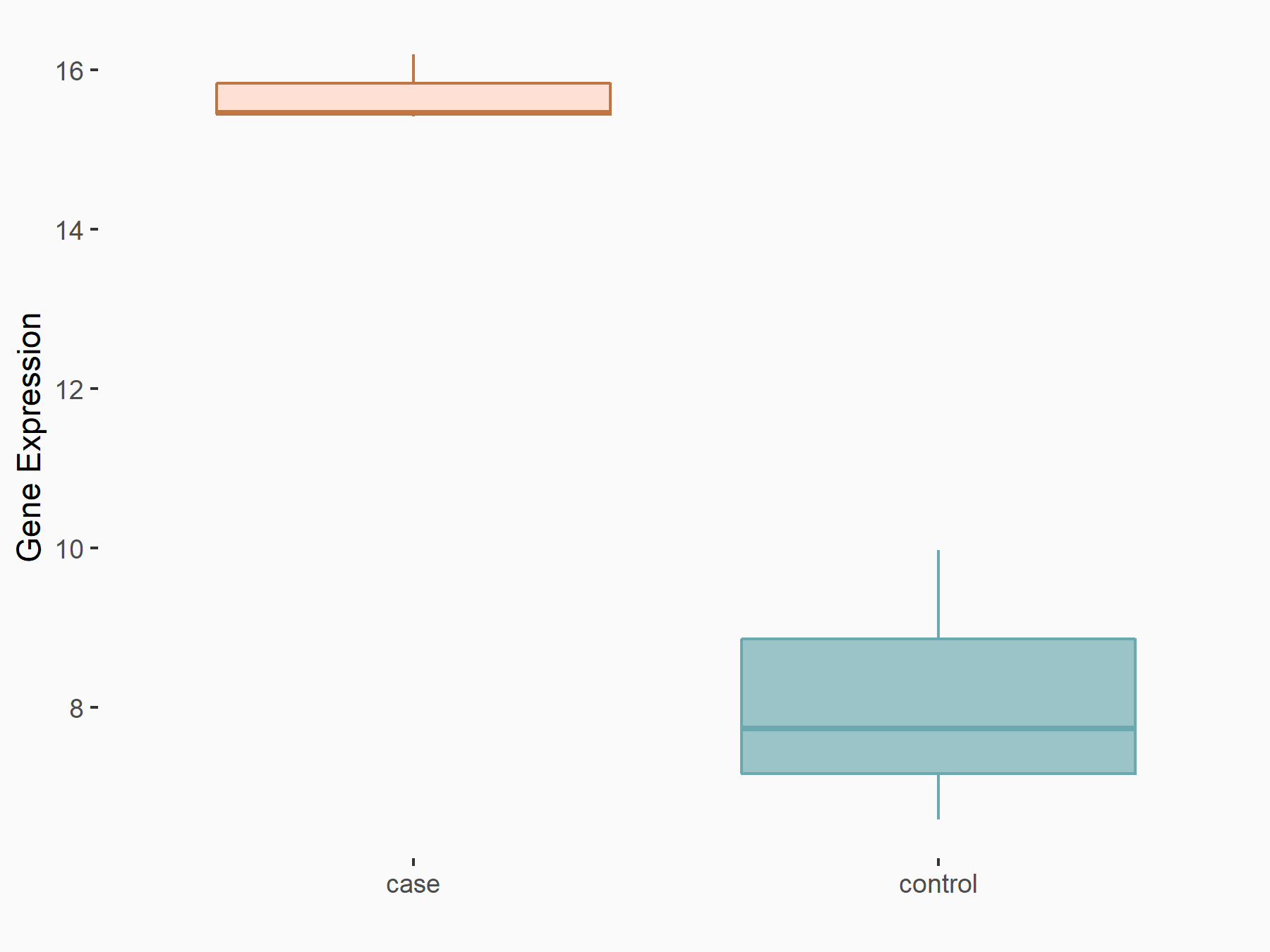 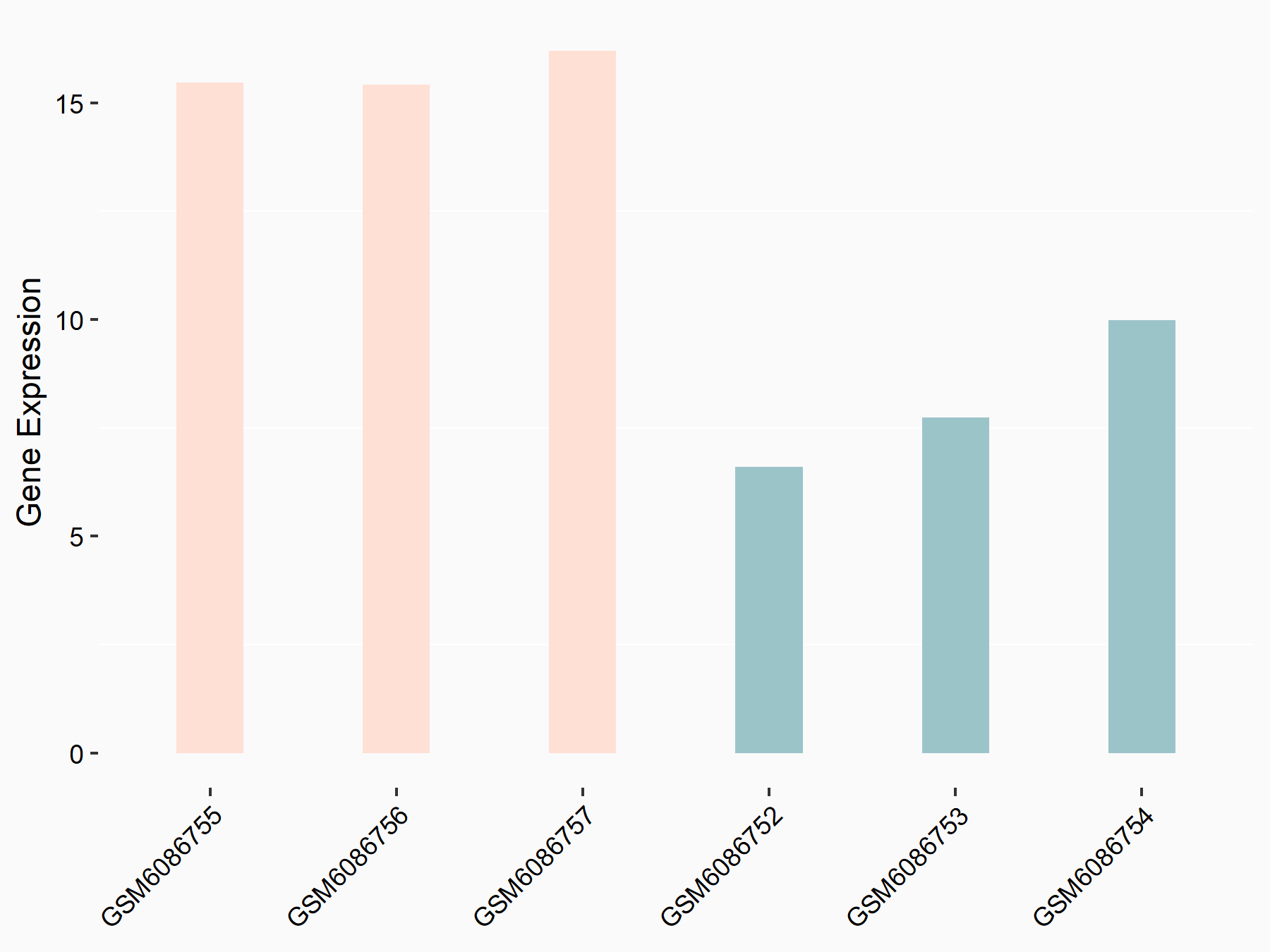 |
logFC: 8.91E-01 p-value: 2.94E-04 |
| More Results | Click to View More RNA-seq Results | |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [221] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Transcriptional misregulation in cancer | hsa05202 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 |
| LN-18 | Glioblastoma | Homo sapiens | CVCL_0392 | |
| HEB (human normal glial cell line HEB were obtained from Tongpai (Shanghai) biotechnology co., LTD (Shanghai, China)) | ||||
| In-vivo Model | Mice were subcutaneously injected with 2.5 × 106 U251 cells stably infected with OE-NC, OE-JMJD1C, OE-NC and sh-NC, OE-JMJD1C and sh-NC or OE-JMJD1C and sh-SOCS2 (n = 10 in each group) in 0.1 ml PBS. | |||
| Response Summary | MiR-302a was identified to target METTL3, which could inhibit Suppressor of cytokine signaling 2 (SOCS2) expression via m6A modification in glioma. | |||
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [222] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| Response Summary | METTL3-KO in gastric cancer cells resulted in the suppression of cell proliferation by inducing Suppressor of cytokine signaling 2 (SOCS2), suggesting a potential role of elevated METTL3 expression in gastric cancer progression. | |||
Colon cancer [ICD-11: 2B90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [223] | |||
| Responsed Disease | Colon cancer [ICD-11: 2B90] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell proliferation | |||
In-vitro Model |
SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 |
| Response Summary | An increased level of METTL3 may maintain the tumorigenicity of colon cancer cells by suppressing Suppressor of cytokine signaling 2 (SOCS2). | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [224] | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cells proliferation | |||
| Cells migration | ||||
| Cells invasion | ||||
| RNA degradation (hsa03018) | ||||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| In-vivo Model | For the subcutaneous implantation model, 2 × 106 METTL3 stable knockdown Huh-7 cells or METTL3 overexpression MHCC97L cells were injected subcutaneously into BABL/cAnN-nude mice. For orthotopic implantation, wild-type and METTL3 knockout Huh-7 cells were luciferase labelled, and 2 × 106 cells were then injected orthotopically into the left liver lobe of nude mice. | |||
| Response Summary | METTL3 is frequently up-regulated in human HCC and contributes to HCC progression. METTL3 represses Suppressor of cytokine signaling 2 (SOCS2) expression in HCC through an m6A-YTHDF2-dependent mechanism. | |||
Thyrotoxicosis [ICD-11: 5A02]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [225] | |||
| Responsed Disease | Graves disease [ICD-11: 5A02.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
PBMCs (Human peripheral blood mononuclear cells (PBMCs) are isolated from peripheral blood and identified as any blood cell with a round nucleus) | |||
| Response Summary | METTL3 knock-down experiment revealed that expressions of SOCS family members SOCS1, Suppressor of cytokine signaling 2 (SOCS2), SOCS4, SOCS5, and SOCS6 were increased after METTL3 knock-down. It indicated that METTL3 is involved in the development of Graves' disease (GD) by inducing mRNA m6A methylation modification of SOCS family members. | |||
Thioredoxin (TXN/SASP)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
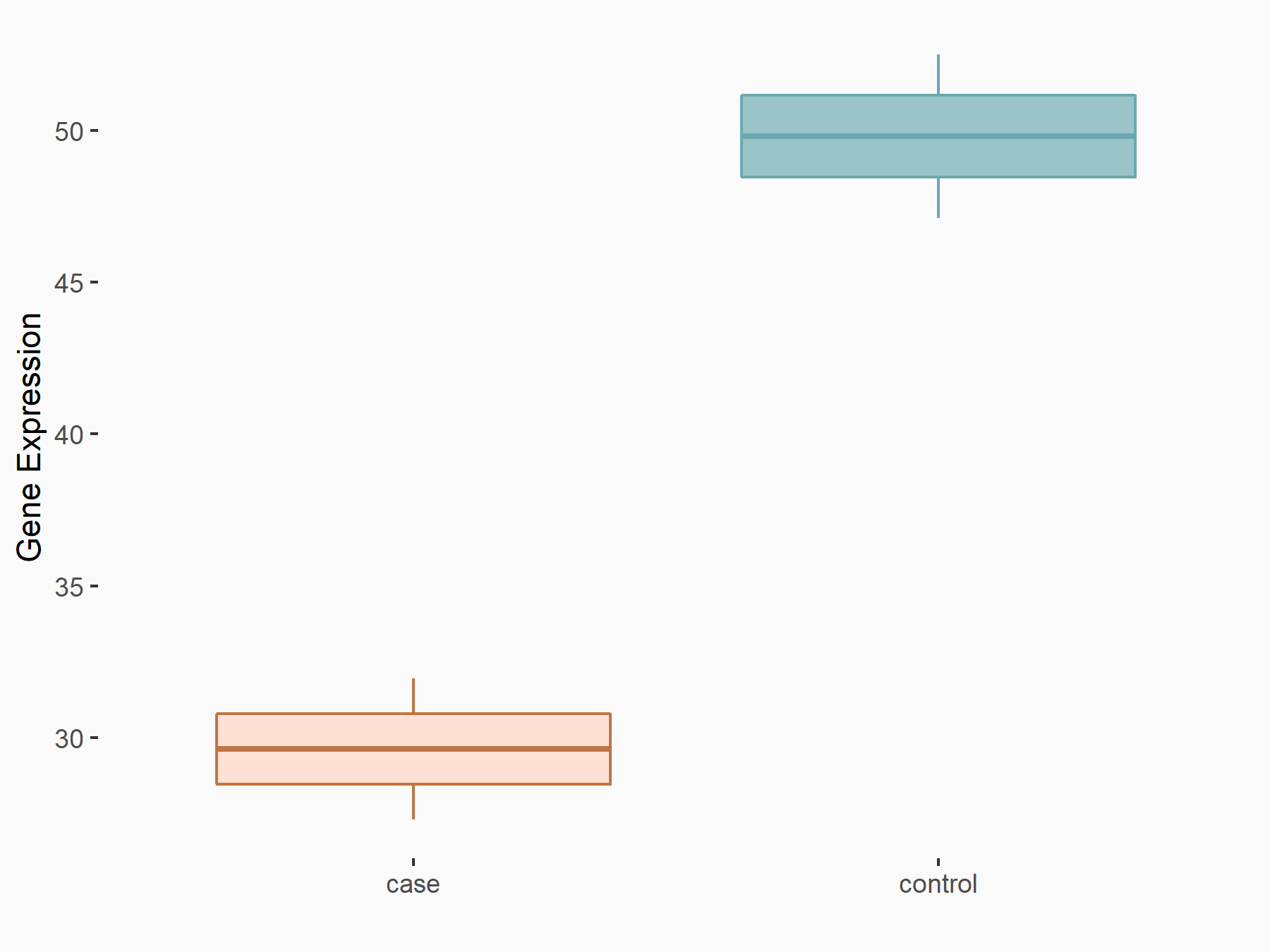 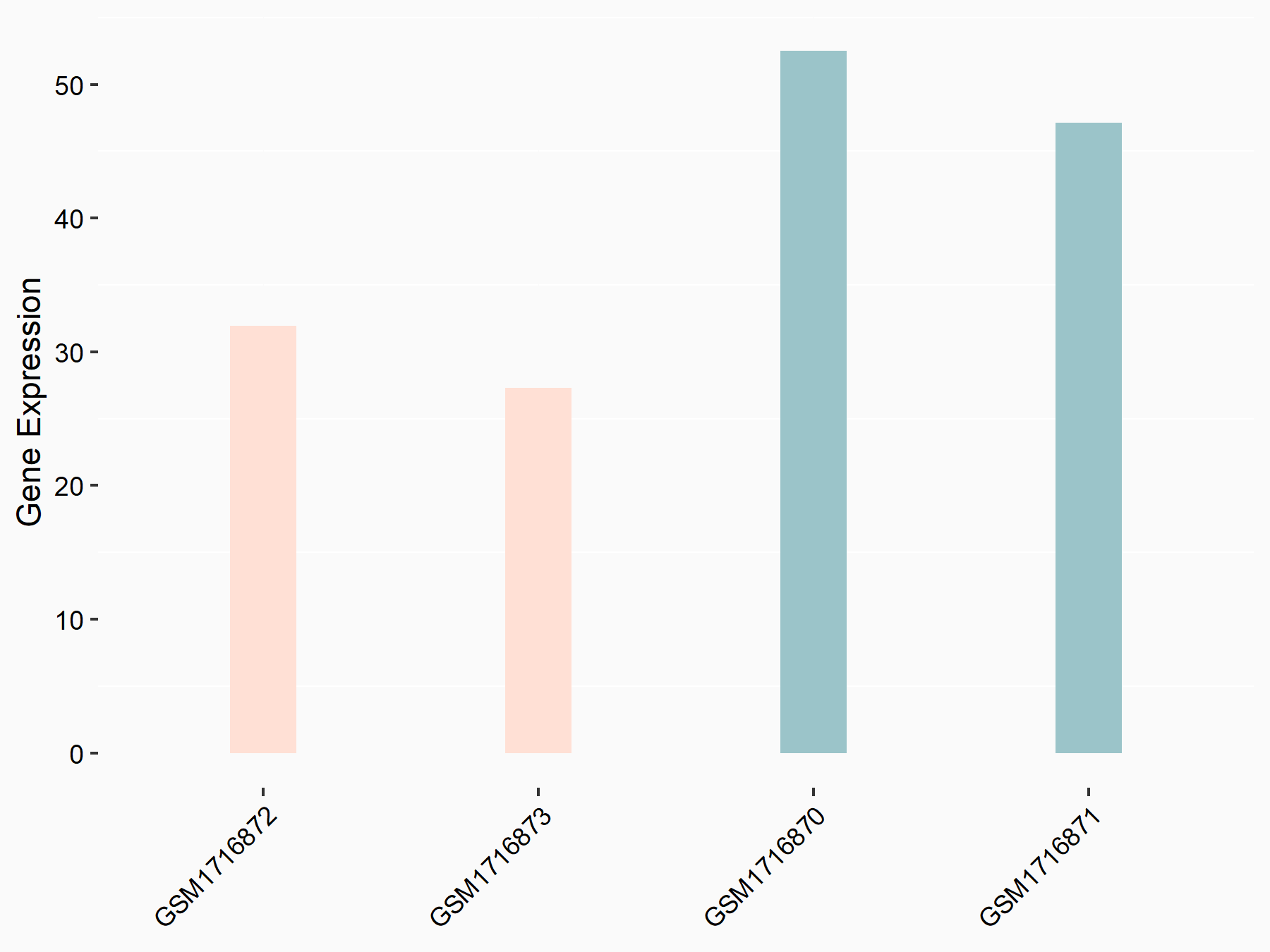 |
logFC: -7.32E-01 p-value: 1.61E-02 |
| More Results | Click to View More RNA-seq Results | |
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [131] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular senescence | |||
| Cell autophagy | ||||
In-vitro Model |
C-28/I2 | Normal | Homo sapiens | CVCL_0187 |
| FLS (Rat fibroblast synovial cell line) | ||||
| In-vivo Model | Mice were anaesthetized with isoflurane supplied in a mouse anaesthesia apparatus, followed with joint surgery on the right joint by sectioning the medial meniscotibial ligament. | |||
| Response Summary | In osteoarthritis METTL3-mediated m6A modification decreased the expression of Thioredoxin (TXN/SASP), an E-1 enzyme crucial for the formation of autophagosomes, by attenuating its RNA stability. Silencing METTL3 enhanced autophagic flux and inhibited SASP expression in OA-FLS. | |||
Thioredoxin domain-containing protein 5 (TXNDC5)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
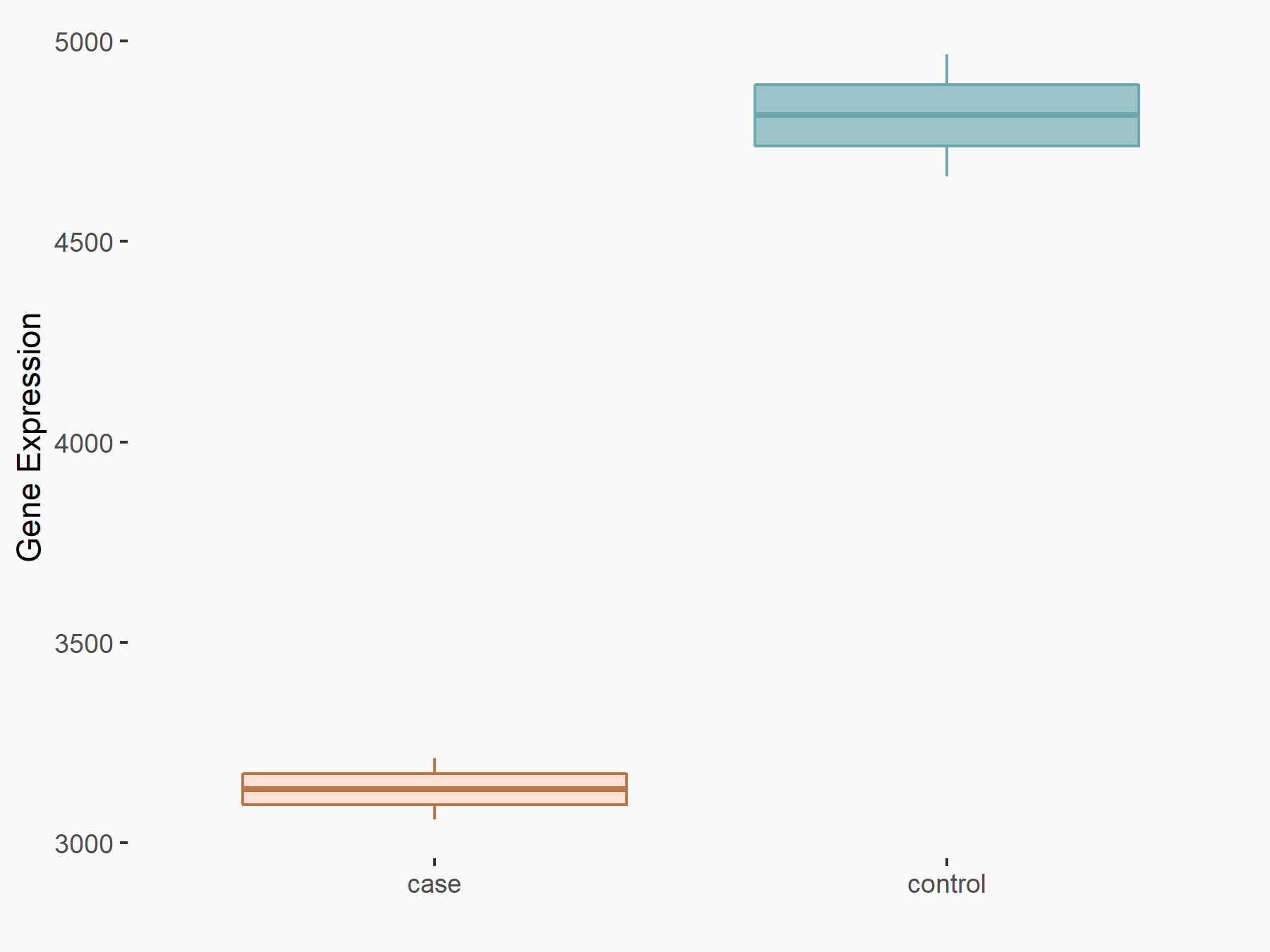 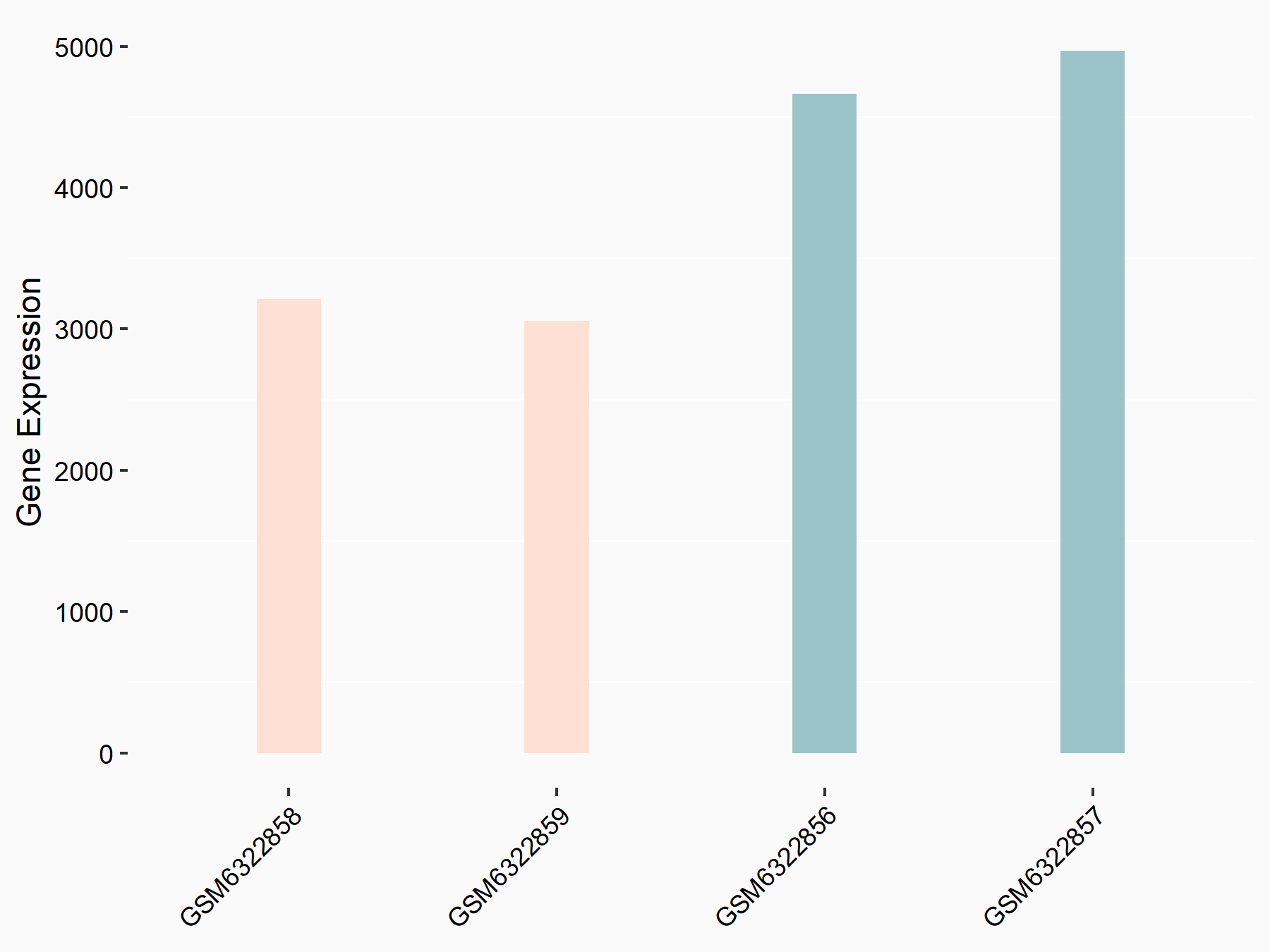 |
logFC: -6.19E-01 p-value: 4.11E-14 |
| More Results | Click to View More RNA-seq Results | |
Melanoma [ICD-11: 2C30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [226] | |||
| Responsed Disease | Melanoma [ICD-11: 2C30] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| cell migration | ||||
| cell invasion | ||||
In-vitro Model |
SK-MEL-28 | Cutaneous melanoma | Homo sapiens | CVCL_0526 |
| MV3 | Amelanotic melanoma | Homo sapiens | CVCL_W280 | |
| HMY-1 | Melanoma | Homo sapiens | CVCL_2950 | |
| HEMa (The normal human epidermal melanocyte (HEMa) was extracted from fresh foreskin tissue donated after circumcision in adults and cultured in melanocyte medium with 5% fetal bovine serum (Sciencell, USA)) | ||||
| A-875 | Melanoma | Homo sapiens | CVCL_4733 | |
| A375-MA2 | Amelanotic melanoma | Homo sapiens | CVCL_X495 | |
| A2058 | Amelanotic melanoma | Homo sapiens | CVCL_1059 | |
| In-vivo Model | Sixteen BALB/c nude mice (male, 6-week-old) were raised under pathogen-free conditions and randomly divided into two groups. A total of 2 × 106 A375 NTC or sh-METTL3#2 cells were subcutaneously inoculated into the right hind flank. Body weight and tumor size were measured every other day. The tumors were harvested at the end of the observation period, and tumor weight and gross images were recorded (11 days after inoculation). The tumors were embedded in RNA later and 10% formalin for further detection. | |||
| Response Summary | METTL3 regulates certain m6A-methylated transcripts, Thioredoxin domain-containing protein 5 (TXNDC5), targeting the m6A dependent-METTL3 signaling pathway serves as a promising therapeutic strategy for management of patients of acral melanomas. | |||
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [346] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
Transcription factor AP-2-alpha (TFAP2A)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 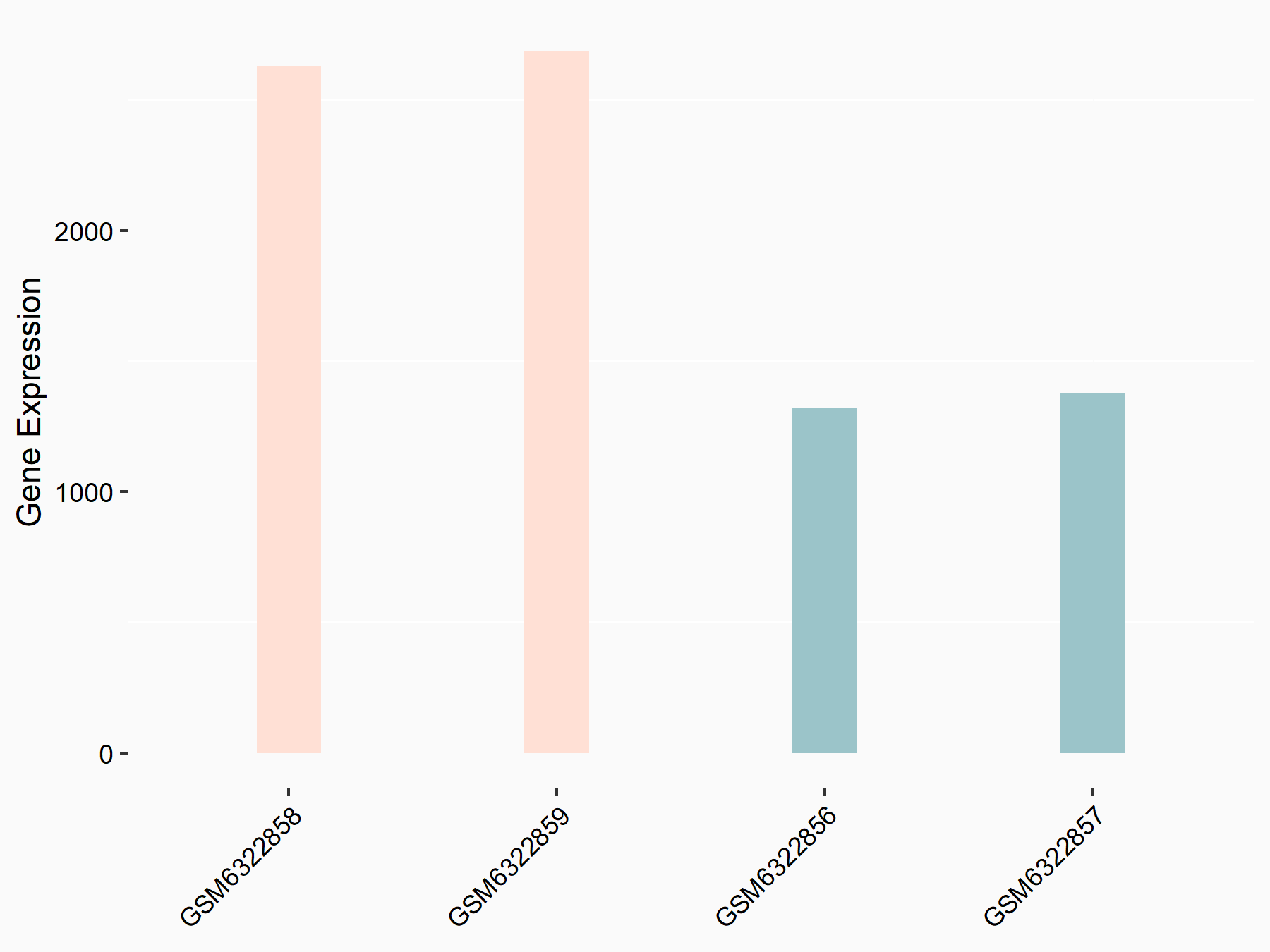 |
logFC: 9.80E-01 p-value: 2.54E-26 |
| More Results | Click to View More RNA-seq Results | |
Urinary/pelvic organs injury [ICD-11: NB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [38] | |||
| Responsed Disease | Injury of kidney [ICD-11: NB92.0] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell apoptosis | ||||
In-vitro Model |
NRK-52E | Normal | Rattus norvegicus | CVCL_0468 |
| In-vivo Model | Rats were anesthetized and incised through the midline of the abdomen, and the left renal vertebral arch and arteries were blocked for 45 min, thereby resulting in left kidney ischemia. At the same time, the right kidney was removed, further aggravating the degree of left kidney injury. | |||
| Response Summary | METTL3 contributes to renal ischemia-reperfusion injury by regulating Foxd1 methylation. When METTL3 was inhibited, m6A levels were accordingly decreased and cell apoptosis was suppressed in the H/R in vitro model. Based on MeRIP sequencing, Transcription factor AP-2-alpha (TFAP2A), cytochrome P-450 1B1 (cyp1b1), and forkhead box D1 (foxd1) were significantly differentially expressed, as was m6A, which is involved in the negative regulation of cell proliferation and kidney development. | |||
Transcription factor p65 (RELA)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
 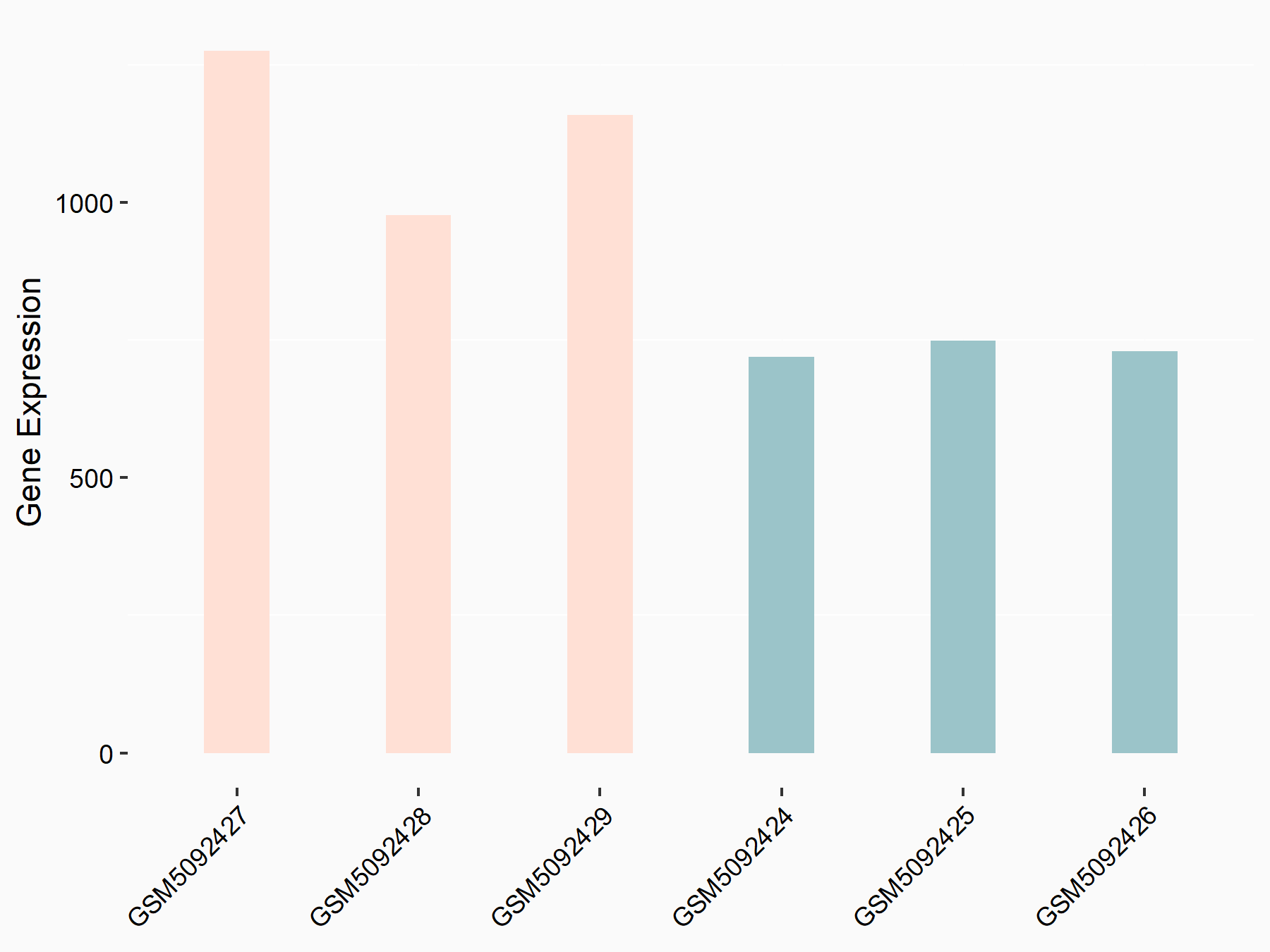 |
logFC: 6.33E-01 p-value: 3.93E-11 |
| More Results | Click to View More RNA-seq Results | |
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Glucose metabolism | |||
| Response Summary | AF4/FMR2 family member 4 (AFF4), two key regulators of NF-Kappa-B pathway (IKBKB and Transcription factor p65 (RELA)) and MYC were further identified as direct targets of METTL3-mediated m6A modification.overexpression of METTL3 significantly promoted Bladder cancer cell growth and invasion. | |||
Thyroid Cancer [ICD-11: 2D10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [103] | |||
| Responsed Disease | Papillary thyroid cancer [ICD-11: 2D10.1] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | NF-kappa B signaling pathway | hsa04064 | ||
In-vitro Model |
TPC-1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_6298 |
| Nthy-ori 3-1 | Normal | Homo sapiens | CVCL_2659 | |
| KTC-1 | Thyroid carcinoma | Homo sapiens | CVCL_6300 | |
| B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 | |
| In-vivo Model | For xenograft models, 5 × 106 BCPAP or KTC-1 cells from each group were injected subcutaneously into the flanks of female BALB/c nude mice (4-6 weeks old, Shanghai SLAC Laboratory Animal, China, n = 5 per group) in a volume of 150 uL PBS. Tumor growth was measured with a digital caliper every 4 days and calculated using the following formula: (length × width2)/2. To study the effect of IL-8 on tumor growth in vivo, scramble or shMETTL3 BCPAP cells were implanted hypodermically into BALB/c nude mice (2 × 106 cells in 150 uL PBS, n = 10 per group). When palpable tumors formed on day 14, mice were treated with DMSO or the IL-8 inhibitor SB225002 (10 mg/kg) by intraperitoneal injection 3 times per week for 3 weeks. Six weeks post-injection, the mice were sacrificed, and the tumors were collected to analyze the frequency of TANs by flow cytometry. For the lung metastasis model, BCPAP and KTC-1 cells (2 × 106 cells in 100 uL PBS) with the corresponding vectors were injected into the tail veins of BALB/c nude mice. Eight weeks after injection, the mice were euthanized, and metastatic lung nodules were analyzed (n = 5 for each group). | |||
| Response Summary | METTL3 played a pivotal tumor-suppressor role in papillary thyroid cancer carcinogenesis through c-Rel and Transcription factor p65 (RELA) inactivation of the nuclear factor Kappa-B (NF-Kappa-B) pathway by cooperating with YTHDF2 and altered TAN infiltration to regulate tumor growth. | |||
Transcription factor Sp1 (SP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Mouse testis | Mus musculus |
|
Treatment: Mettl3 knockout mouse testis
Control: Mouse testis
|
GSE99771 | |
| Regulation |
  |
logFC: -5.88E+00 p-value: 2.22E-06 |
| More Results | Click to View More RNA-seq Results | |
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [227] | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Cell Process | Cell cycle | |||
In-vitro Model |
EoL-1 | Chronic eosinophilic leukemia | Homo sapiens | CVCL_0258 |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 | |
| Jurkat | T acute lymphoblastic leukemia | Homo sapiens | CVCL_0065 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| KG-1 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0374 | |
| Loucy | Adult T acute lymphoblastic leukemia | Homo sapiens | CVCL_1380 | |
| MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| RN2c (Acute myeloid leukemia cell line) | ||||
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| In-vivo Model | Performed an ex vivo genome wide CRISPR dropout screen (Screen 1) using Cas9-expressing mouse primary leukaemia cells driven by an MLL-AF9 fusion gene and a FLT3 internal tandem duplication. | |||
| Response Summary | Genes regulated by METTL3 in this way are necessary for acute myeloid leukaemia. Two genes expressing the transcription factors Transcription factor Sp1 (SP1) and SP2, which have promoters occupied by METTL3. | |||
Transcription factor Sp2 (SP2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Pancreatic islets | Mus musculus |
|
Treatment: Mettl3 knockout mice
Control: Mettl3 flox/flox mice
|
GSE155612 | |
| Regulation |
 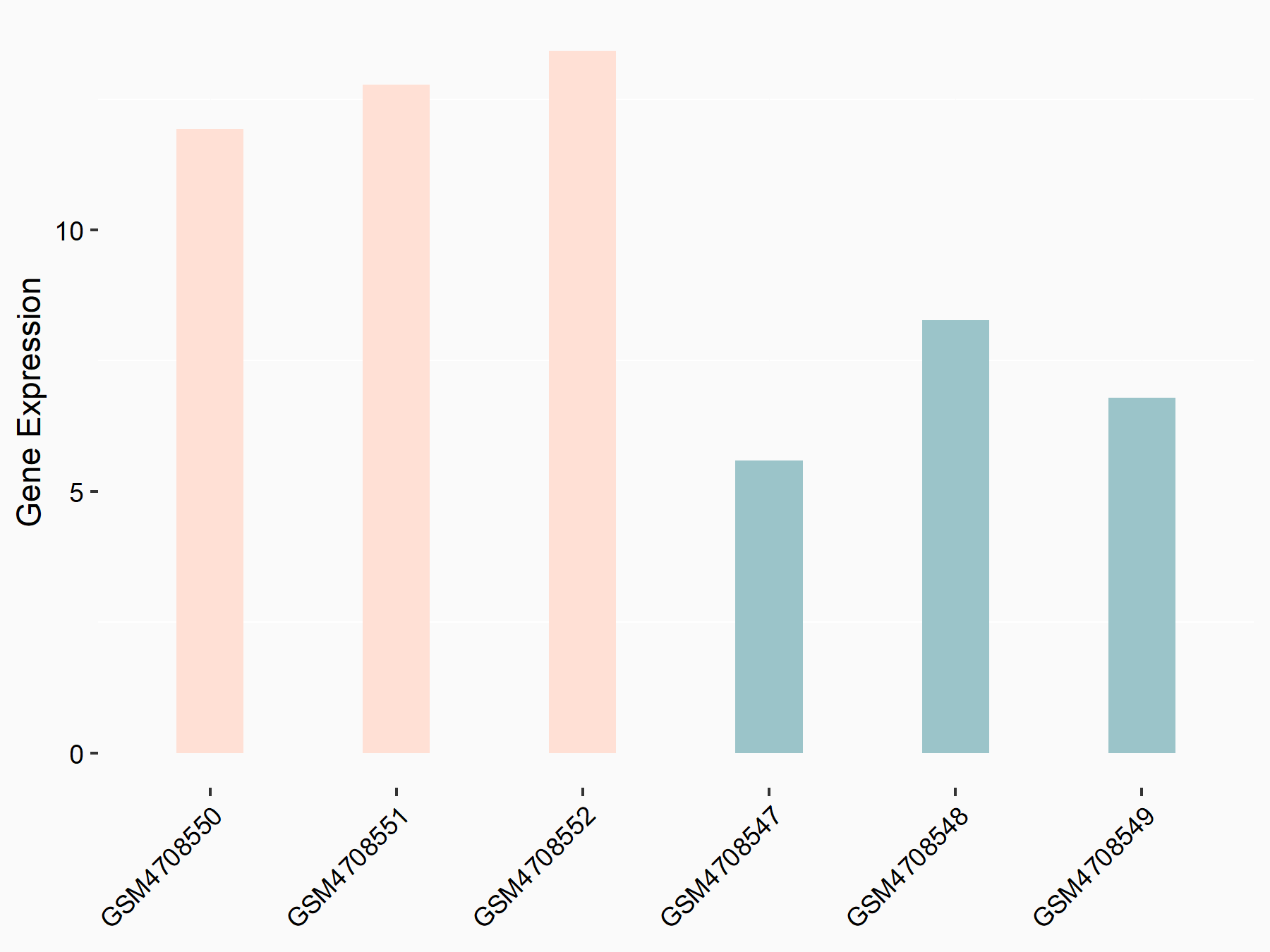 |
logFC: 8.11E-01 p-value: 3.84E-03 |
| More Results | Click to View More RNA-seq Results | |
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [227] | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Cell Process | Cell cycle | |||
In-vitro Model |
EoL-1 | Chronic eosinophilic leukemia | Homo sapiens | CVCL_0258 |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 | |
| Jurkat | T acute lymphoblastic leukemia | Homo sapiens | CVCL_0065 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| KG-1 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0374 | |
| Loucy | Adult T acute lymphoblastic leukemia | Homo sapiens | CVCL_1380 | |
| MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| RN2c (Acute myeloid leukemia cell line) | ||||
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| In-vivo Model | Performed an ex vivo genome wide CRISPR dropout screen (Screen 1) using Cas9-expressing mouse primary leukaemia cells driven by an MLL-AF9 fusion gene and a FLT3 internal tandem duplication. | |||
| Response Summary | Genes regulated by METTL3 in this way are necessary for acute myeloid leukaemia. Two genes expressing the transcription factors SP1 and Transcription factor Sp2 (SP2), which have promoters occupied by METTL3. | |||
Transcriptional repressor protein YY1 (YY1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Pancreatic islets | Mus musculus |
|
Treatment: Mettl3 knockout mice
Control: Mettl3 flox/flox mice
|
GSE155612 | |
| Regulation |
 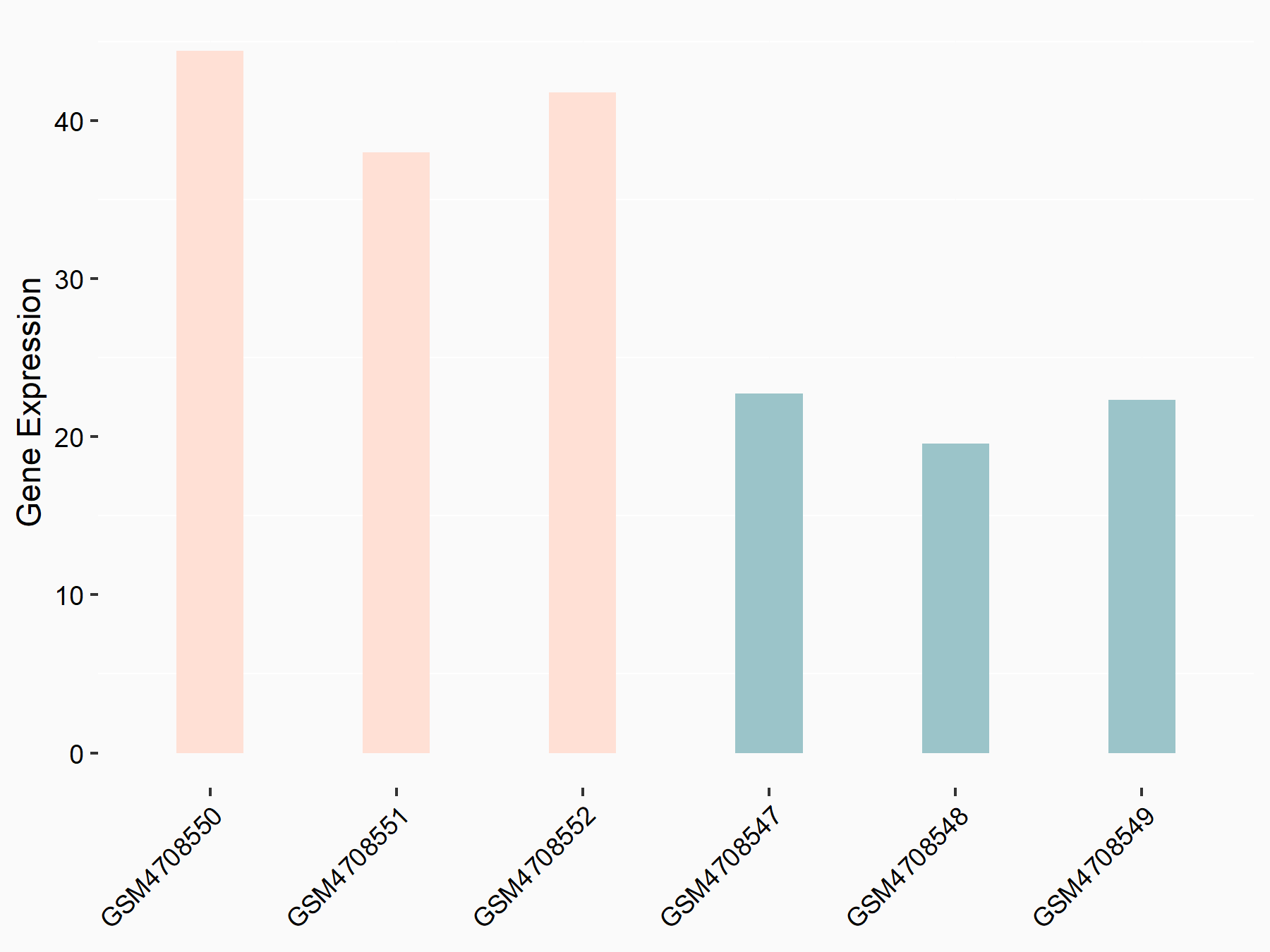 |
logFC: 9.11E-01 p-value: 3.29E-04 |
| More Results | Click to View More RNA-seq Results | |
Multiple myeloma [ICD-11: 2A83]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [228] | |||
| Responsed Disease | Multiple myeloma [ICD-11: 2A83.1] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
U266 (Human multiple myeloma cells) | |||
| RPMI-8226 | Plasma cell myeloma | Homo sapiens | CVCL_0014 | |
| NCI-H929 | Plasma cell myeloma | Homo sapiens | CVCL_1600 | |
| MM1.S | Plasma cell myeloma | Homo sapiens | CVCL_8792 | |
| In-vivo Model | BALB/C nude mice (5 weeks old, weighing 18-22 g) were fed in specific pathogen-free facilities and subcutaneously inoculated with U266 cells (1 × 106). The mice were randomly divided into 3 groups with 6 mice per group, when the tumor was measurable. Then, miR-27a-3p mimic or sh-METTL3 was injected intratumorally at an interval of 4 days a total of 4 times. Tumor volume was measured using a digital caliper every week and calculated using the formula V = 1/2 (width2 × length). | |||
| Response Summary | METTL3 affected the growth, apoptosis, and stemness of MM cells through accelerating the stability of Transcriptional repressor protein YY1 (YY1) mRNA and the maturation of primary-miR-27a-3p in vitro and in vivo. | |||
Tyrosine-protein kinase receptor UFO (AXL)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
 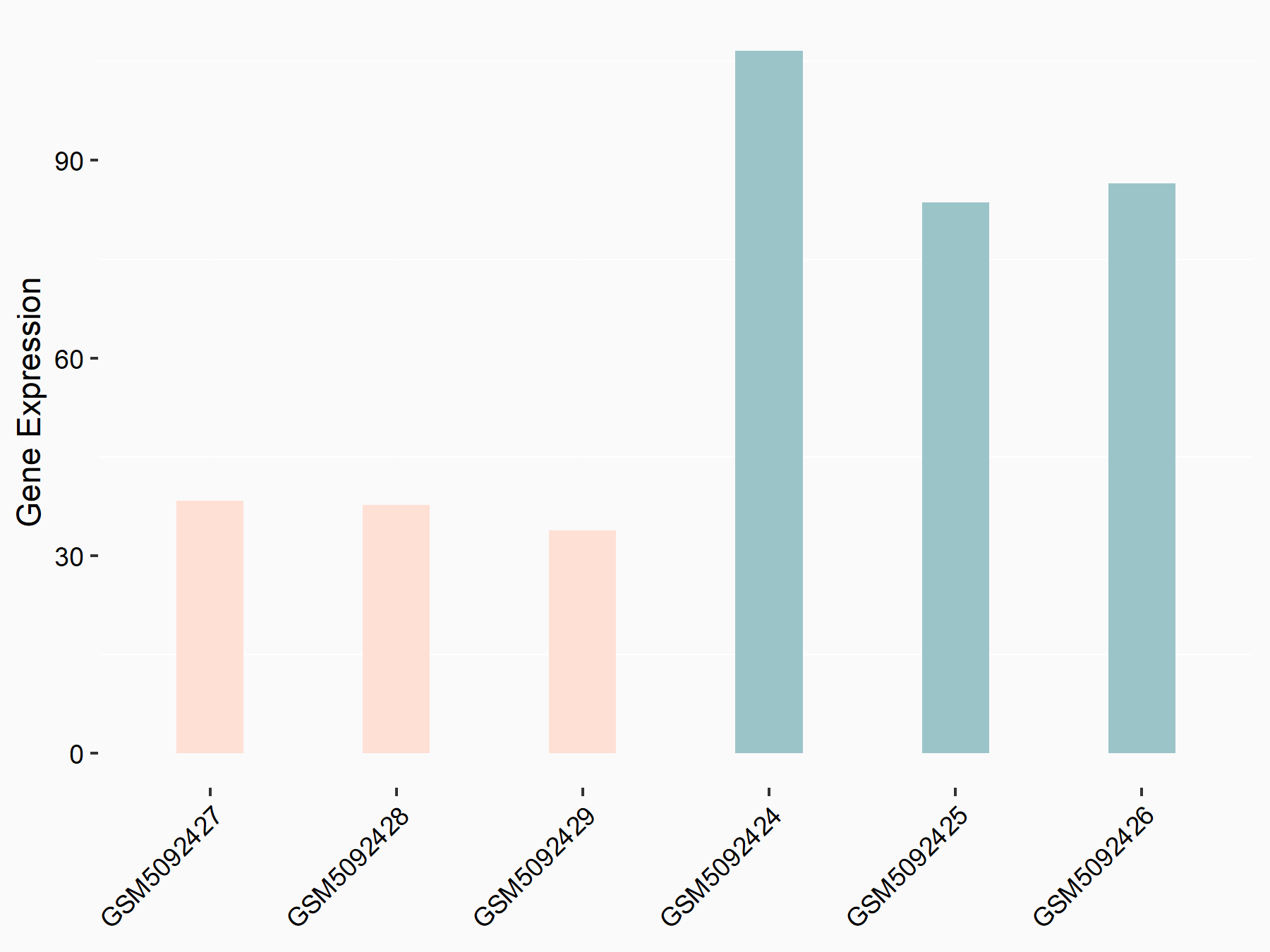 |
logFC: -1.33E+00 p-value: 3.11E-06 |
| More Results | Click to View More RNA-seq Results | |
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [229] | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 |
| COV504 | Ovarian carcinoma | Homo sapiens | CVCL_2424 | |
| ES2 | Ewing sarcoma | Homo sapiens | CVCL_AX39 | |
| HO-8910 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6868 | |
| OVCAR-3 | Ovarian serous adenocarcinoma | Homo sapiens | CVCL_0465 | |
| SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 | |
| In-vivo Model | 2 × 106 tumor cells (OVCAR3-METTL3 and OVCAR3-Ctrl) or 1 × 106 tumor cells (SKOV3-shMETTL3-1, SKOV3-shMETTL3-2 and SKOV3-shNC) were suspended in 200 uL of RPMI 1640 complete culture medium with 25% Matrigel (BD Biosciences) and inoculated subcutaneously into the right flank of the nude mice. | |||
| Response Summary | METTL3 promoted epithelial-mesenchymal transition (EMT) by upregulating the receptor tyrosine kinase Tyrosine-protein kinase receptor UFO (AXL) and that METTL3 serves as a novel prognostic and/or therapeutic target of interest in ovarian cancer. | |||
Ubiquitin carboxyl-terminal hydrolase 4 (USP4)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Mouse testis | Mus musculus |
|
Treatment: Mettl3 knockout mouse testis
Control: Mouse testis
|
GSE99771 | |
| Regulation |
  |
logFC: -6.58E+00 p-value: 2.47E-06 |
| More Results | Click to View More RNA-seq Results | |
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [163] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| In-vivo Model | A total of 1 × 106 PC3 cells or DU145 cells suspended in a mixture of 100 uL PBS and Matrigel were subcutaneously injected into BALB/c nude mice. Tumor weight were measured 2 months after the engraftment. To evaluate the role of METTL3 in tumor metastasis, PC3 cells with or without knockdown of METTL3 were injected into SCID mice through the tail vein (1 × 106 cells per mouse). After eight weeks, mice were sacrificed and their lung tissues were collected for subsequent analyses. | |||
| Response Summary | m6A modification levels were markedly upregulated in human PCa tissues due to increased expression of METTL3. METTL3 mediates m6A modification of Ubiquitin carboxyl-terminal hydrolase 4 (USP4) mRNA at A2696, and m6A reader protein YTHDF2 binds to and induces degradation of USP4 mRNA by recruiting RNA-binding protein HNRNPD to the mRNA. Decrease of USP4 fails to remove the ubiquitin group from ELAVL1 protein, resulting in a reduction of ELAVL1 protein. Lastly, downregulation of ELAVL1 in turn increases ARHGDIA expression, promoting migration and invasion of PCa cells. | |||
Multiple myeloma [ICD-11: 2A83]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [356] | |||
| Responsed Disease | Multiple myeloma [ICD-11: 2A83.1] | |||
| Responsed Drug | Metformin | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
NCI-H929 | Plasma cell myeloma | Homo sapiens | CVCL_1600 |
| MM1.S | Plasma cell myeloma | Homo sapiens | CVCL_8792 | |
| In-vivo Model | 5-week-old male athymic BALB/C nude mice were obtained from Experimental animal center of Fujian Medical University, and subsequently randomly divided into four groups (5 mice per group), including control, metformin, pcDNA-METTL3 and pcDNA-METTL3 + metformin. For the control or metformin group, to induce tumors, 5 × 105 H929 cells were suspended in 0.2 ml phosphate buffered saline and inoculated subcutaneously into the peritoneum of mice. Once the subcutaneous nodules grown to a rice grain size (required approximately a week), the subcutaneous xenograft model of MM in nude mice was successfully constructed. One week later, PBS or Metformin was diluted in drinking water (2 mg/ml) and administered orally throughout the indicated time periods. For the pcDNA-METTL3 group and pcDNA-METTL3 + metformin group, firstly, we transfected pcDNA-METTL3 into H929 cells to overexpress METTL3, and then 5 × 105 H929 cells were suspended in 0.2 ml phosphate-buffered saline and inoculated subcutaneously into the peritoneum of mice. One week later, PBS or Metformin was diluted in drinking water (2 mg/ml) and administered orally throughout the indicated time periods. All nude mice were closely monitored for tumor growth, skin condition, and behavior daily and any tumor ulceration or irritation was noted. Tumor length and width were calculated with vernier calipers every 7 days. After 35 days, the mice were humanely sacrificed, and the subcutaneous tumors were excised and removed. | |||
Ubiquitin carboxyl-terminal hydrolase 7 (USP7)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3 knockout liver
Control: Wild type liver cells
|
GSE198513 | |
| Regulation |
 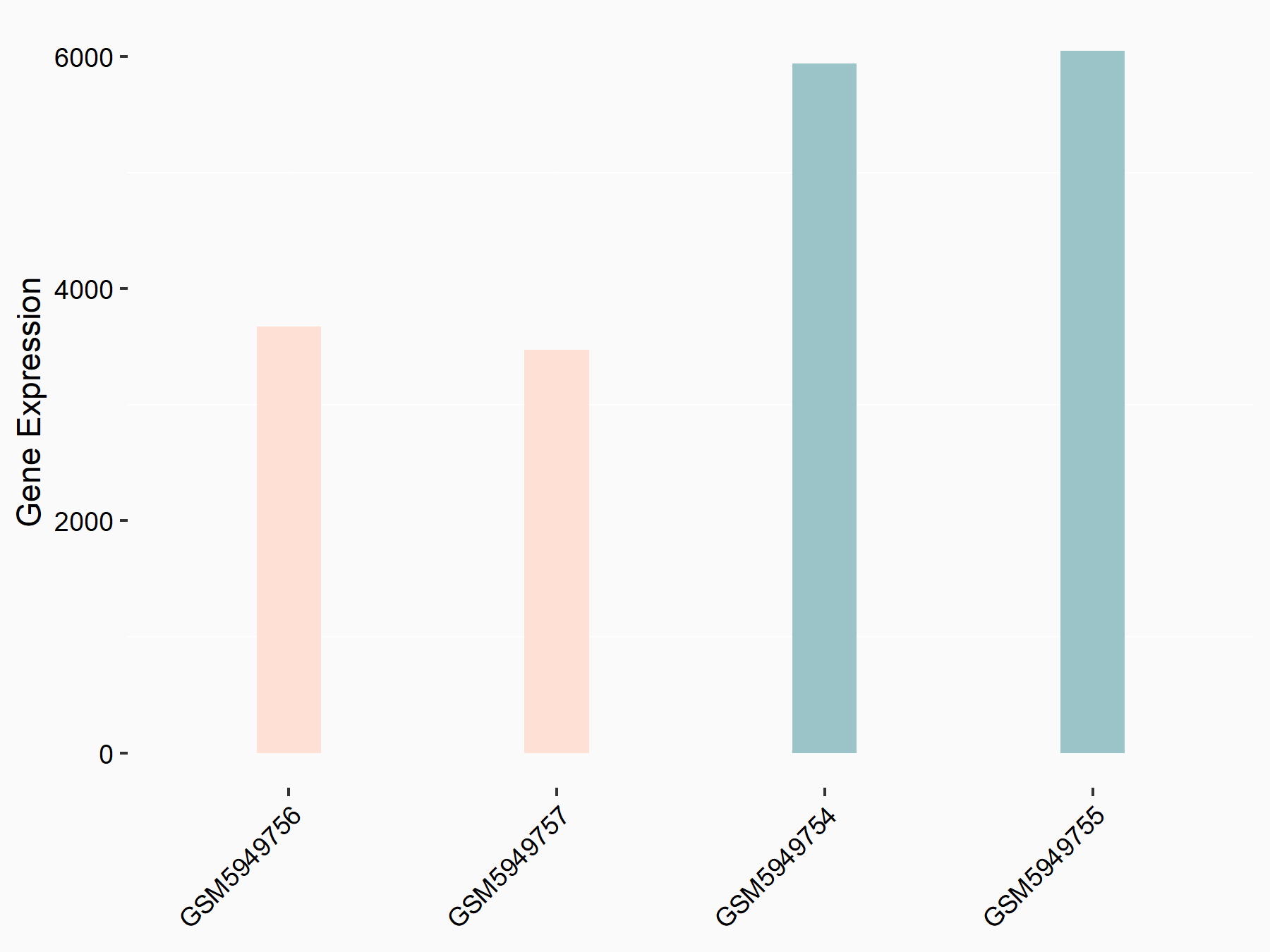 |
logFC: -7.46E-01 p-value: 2.02E-26 |
| More Results | Click to View More RNA-seq Results | |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [230] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell invasion | |||
| Cell migration | ||||
| Cell proliferation | ||||
In-vitro Model |
MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| In-vivo Model | Male nu/nu mice between 4 and 6 weeks of age received subcutaneous injections of equivalent Hep3B cells expressing either LV-shMETTL3 or LV-USP7 within 30 min of harvesting on the right and left flanks. The tumor was weighed after approximately 4 weeks, and the volume was measured every 5 days. | |||
| Response Summary | METTL3 regulates the expression of Ubiquitin carboxyl-terminal hydrolase 7 (USP7) through m6A methylation and facilitate the invasion, migration and proliferation of HCC cells. Besides, the elevated METTL3 expression was related to worse overall survival. | |||
Ubiquitin-like-conjugating enzyme ATG3 (ATG3)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3-deficient liver
Control: Wild type liver cells
|
GSE197800 | |
| Regulation |
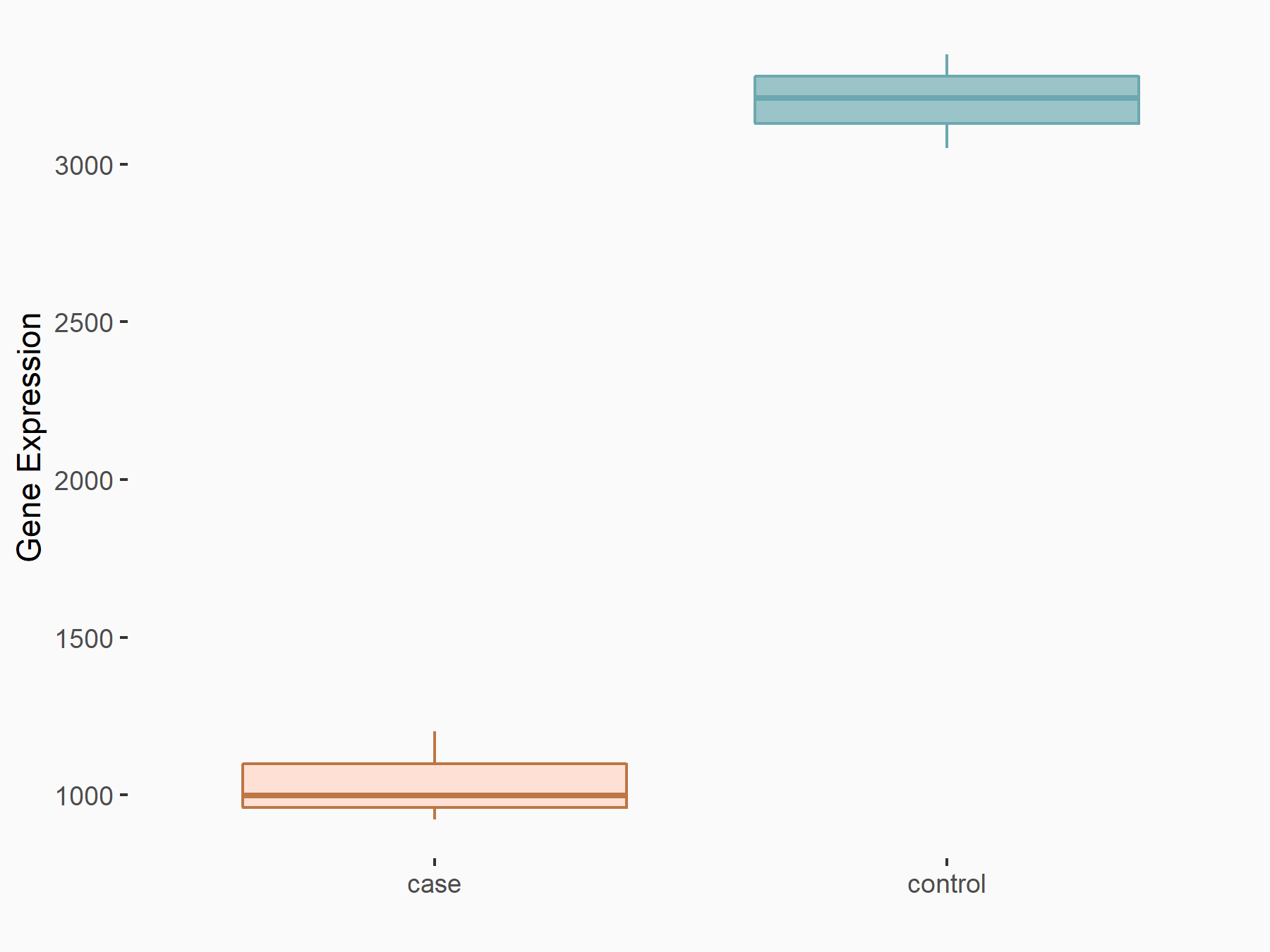 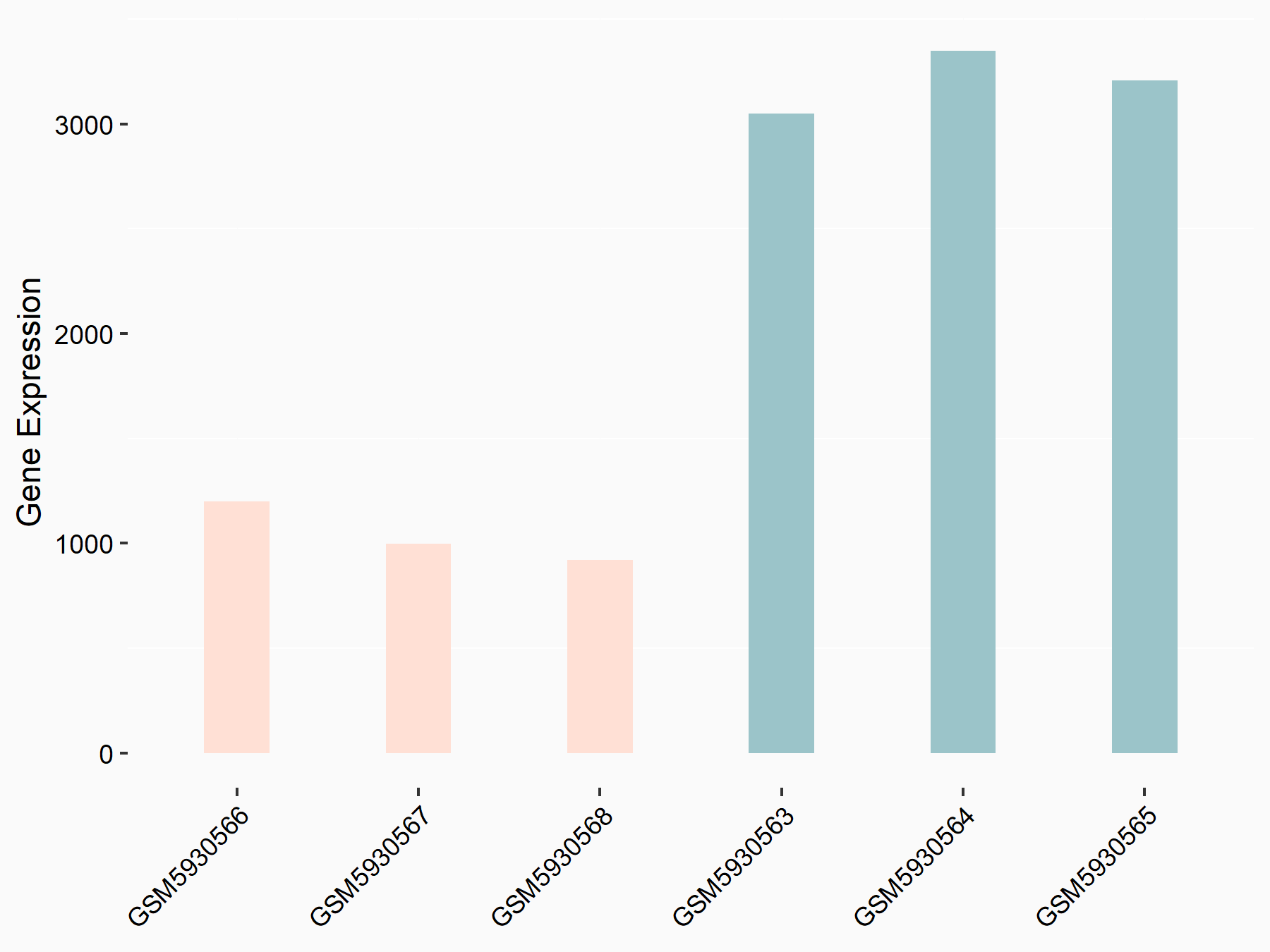 |
logFC: -1.62E+00 p-value: 2.55E-18 |
| More Results | Click to View More RNA-seq Results | |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [17] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including Ubiquitin-like-conjugating enzyme ATG3 (ATG3), ATG5, ATG7, ATG12, and ATG16L1. | |||
UBX domain-containing protein 1 (UBXN1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Embryonic stem cells | Mus musculus |
|
Treatment: METTL3 knockout mESCs
Control: Wild type mESCs
|
GSE156481 | |
| Regulation |
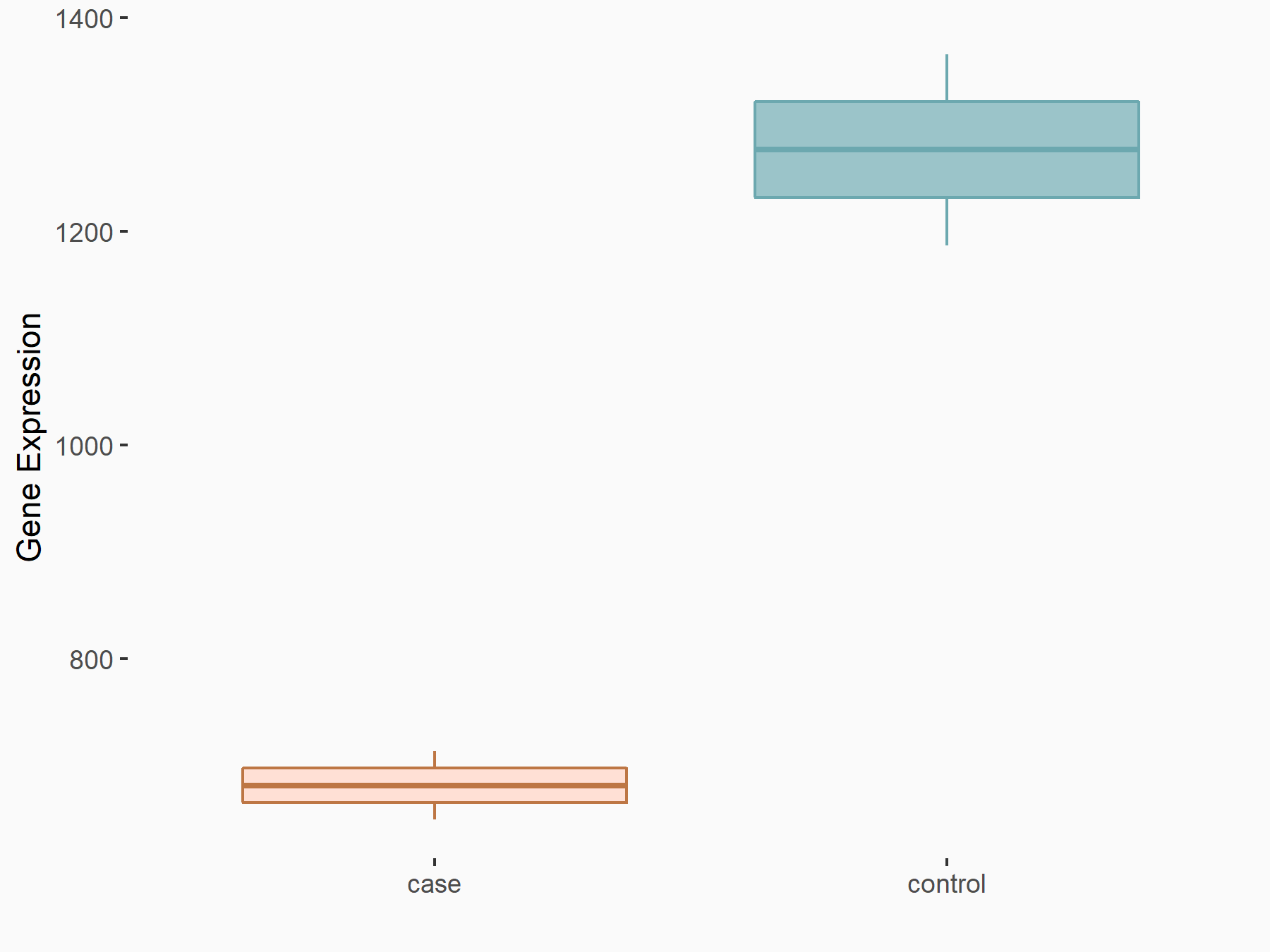 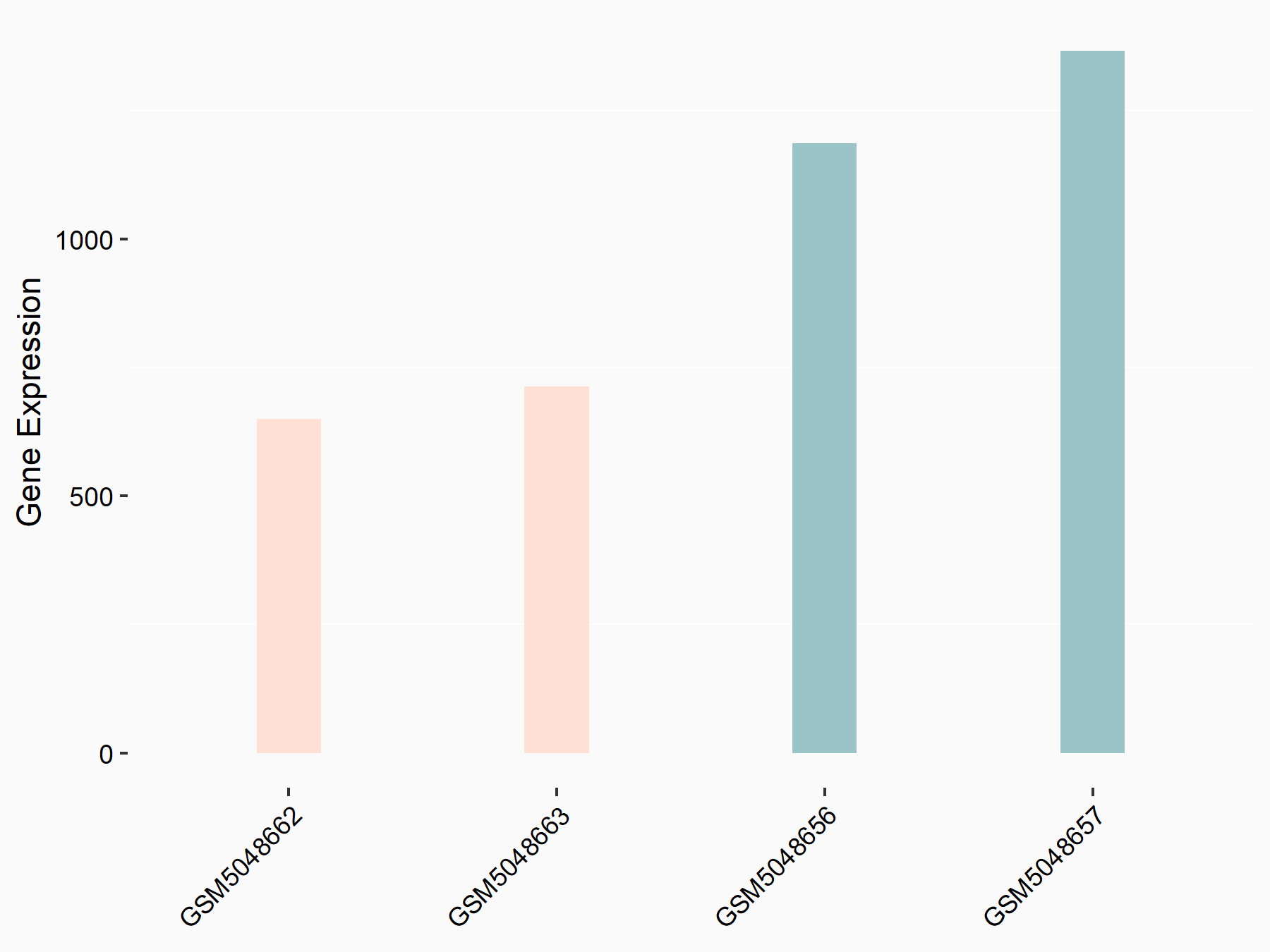 |
logFC: -9.05E-01 p-value: 4.00E-16 |
| More Results | Click to View More RNA-seq Results | |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [202] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | NF-kappa B signaling pathway | hsa04064 | ||
In-vitro Model |
U87 (A primary glioblastoma cell line) | |||
| N33 (The GBM patient-derived cell line) | ||||
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| H4 | Astrocytoma | Homo sapiens | CVCL_1239 | |
| In-vivo Model | Five-week-old female BALB/c nude mice (Charles Rivers, Beijing, China) were selected for the experiments. U87 cells (5 × 105) transfected with an empty vector, YTHDF2 overexpression, or METTL3 overexpression vectors were suspended in PBS and injected into the right frontal node of nude mice. The inoculation position was 2 mm lateral and 2 mm posterior to the anterior fontanel. Tumor size was estimated from luciferase volume measurements and MRI. The mice were sacrificed when they exhibited disturbed activity or convulsion. The brain was then harvested and embedded in paraffin. | |||
| Response Summary | YTHDF2 accelerated UBX domain-containing protein 1 (UBXN1) mRNA degradation via METTL3-mediated m6A, which, in turn, promoted NF-Kappa-B activation. YTHDF2 promotes the malignant progression of gliomas and revealed important insight into the upstream regulatory mechanism of NF-Kappa-B activation via UBXN1 with a primary focus on m6A modification. | |||
Uridine-cytidine kinase 2 (UCK2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
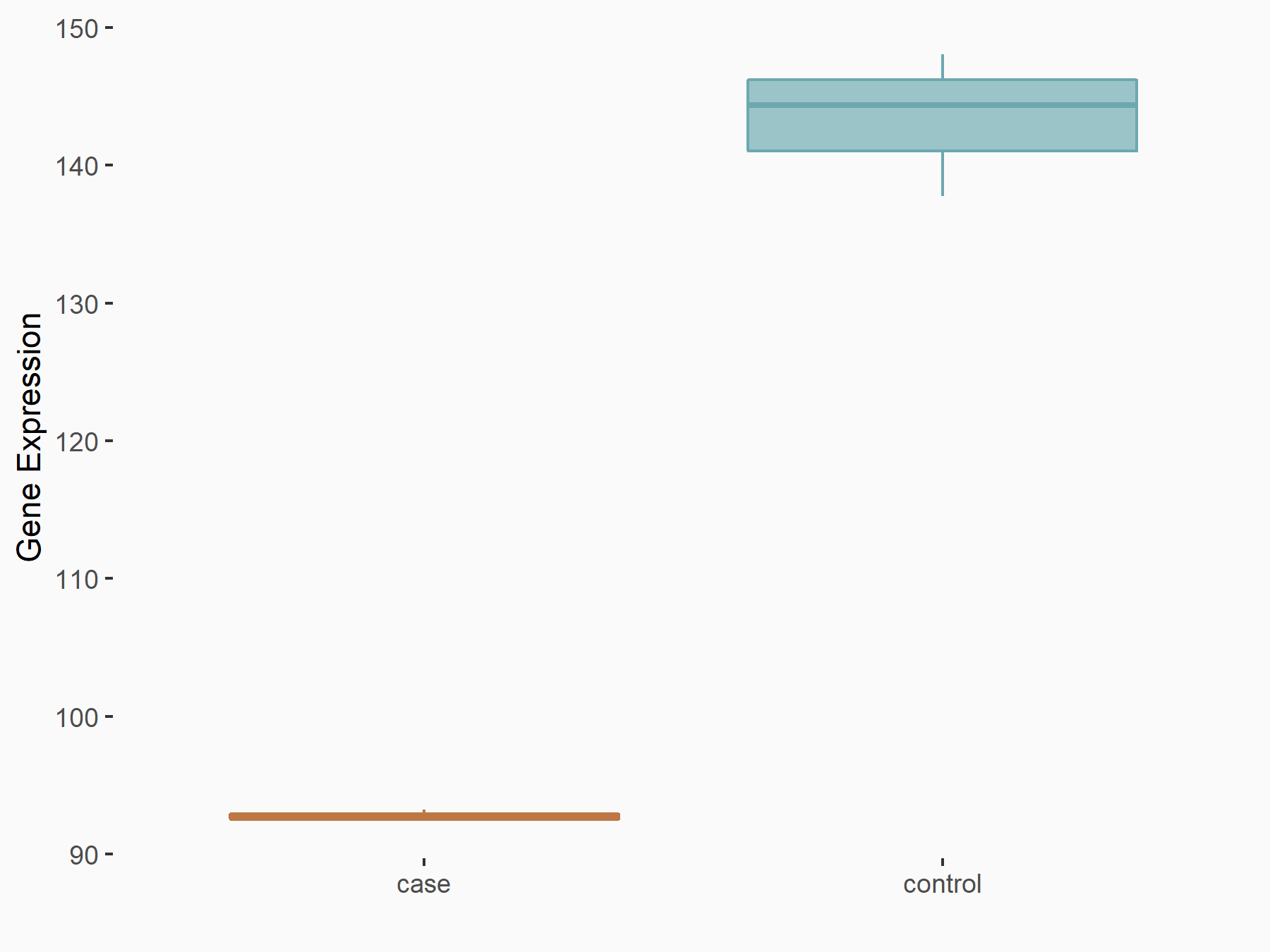 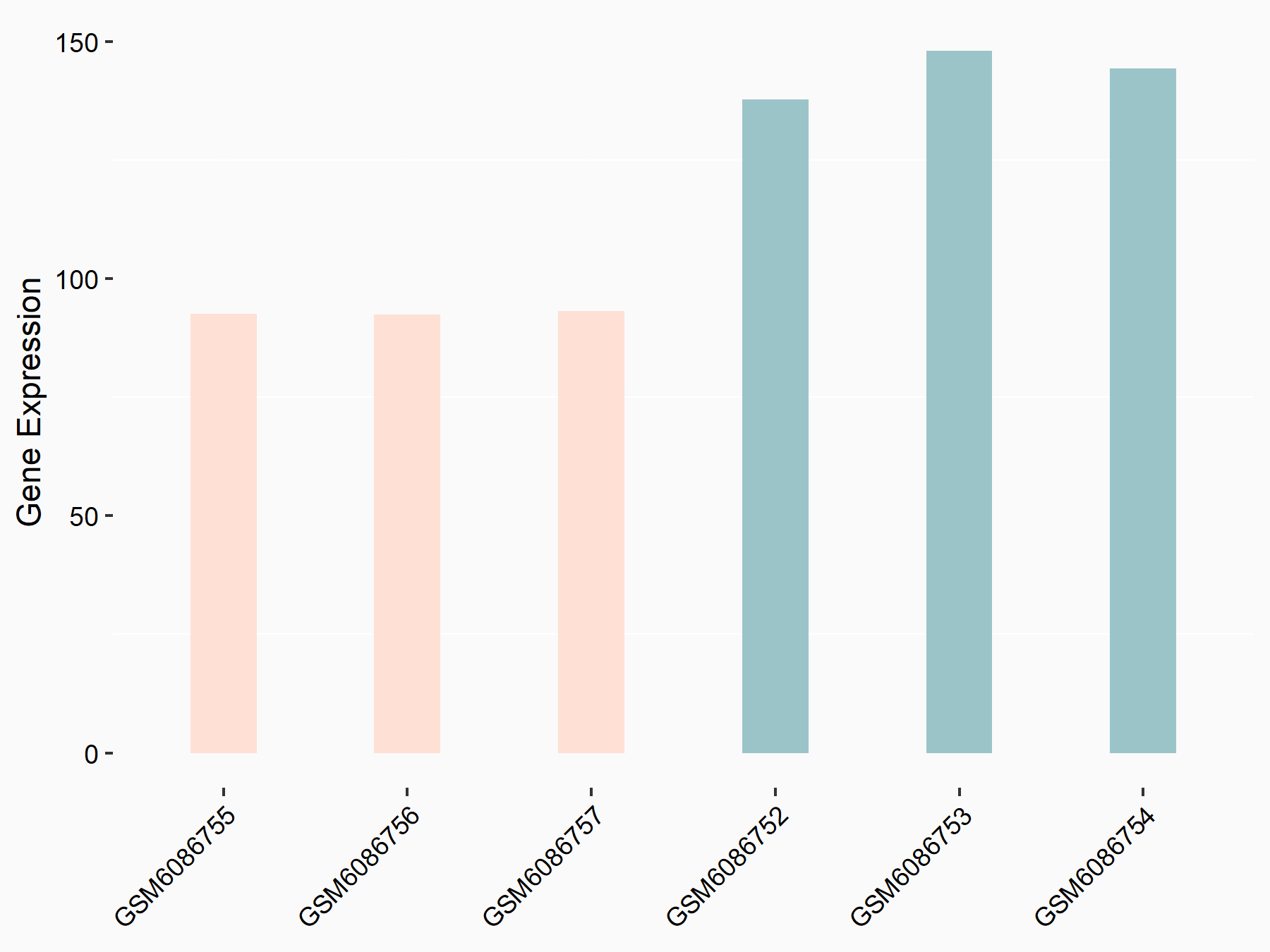 |
logFC: -6.22E-01 p-value: 2.07E-05 |
| More Results | Click to View More RNA-seq Results | |
Melanoma [ICD-11: 2C30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [232] | |||
| Responsed Disease | Melanoma [ICD-11: 2C30] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
In-vitro Model |
Hs 294T | Melanoma | Homo sapiens | CVCL_0331 |
| Response Summary | m6A-METTL3 axis induced abnormal Uridine-cytidine kinase 2 (UCK2) expression plays a role in melanoma metastasis by enhancing the Wnt/Bete-catenin pathway, which provided new clues for melanoma metastasis. It also provides a potential target for the prevention and treatment of melanoma. | |||
Urokinase-type plasminogen activator (PLAU)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
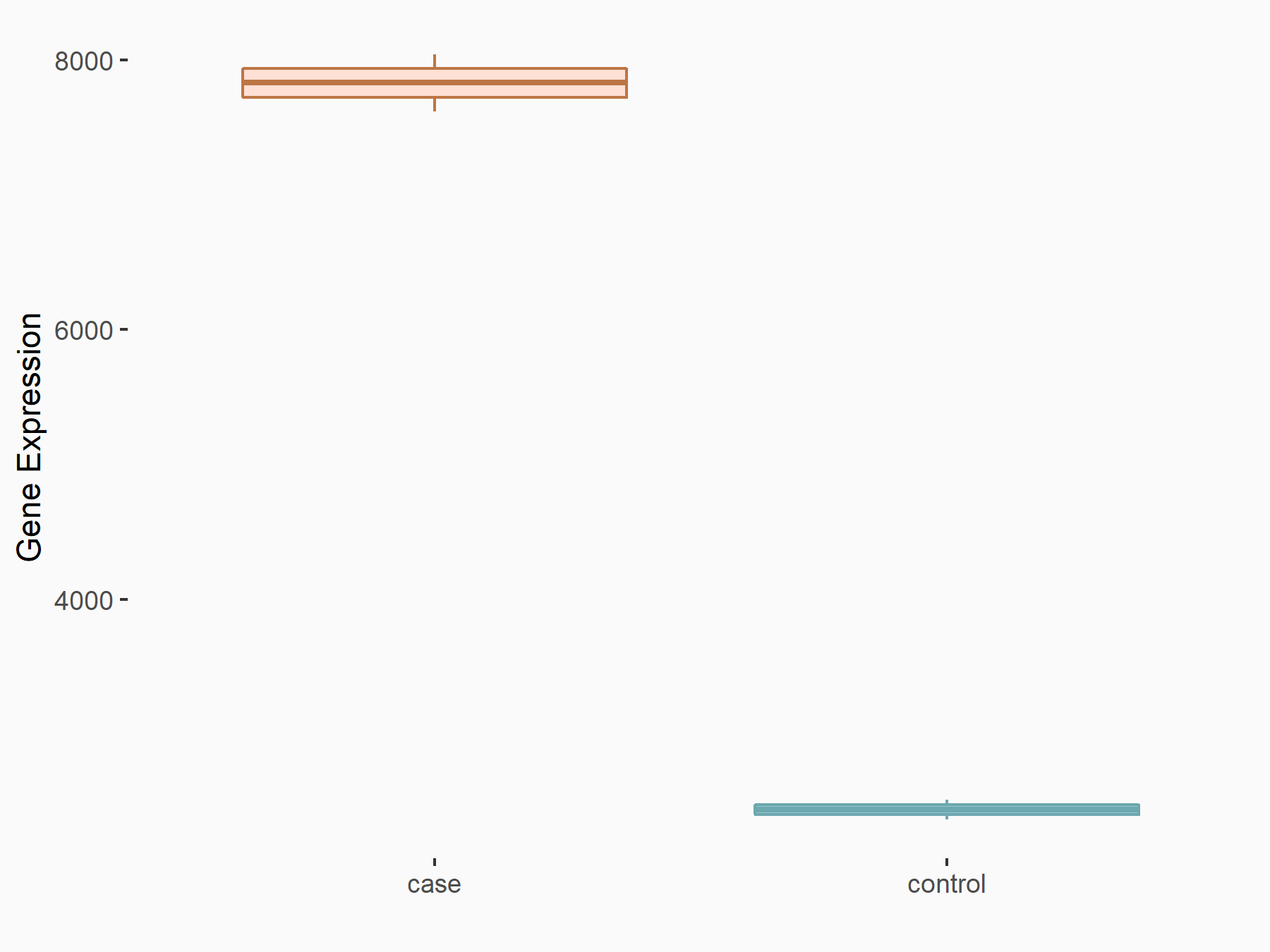 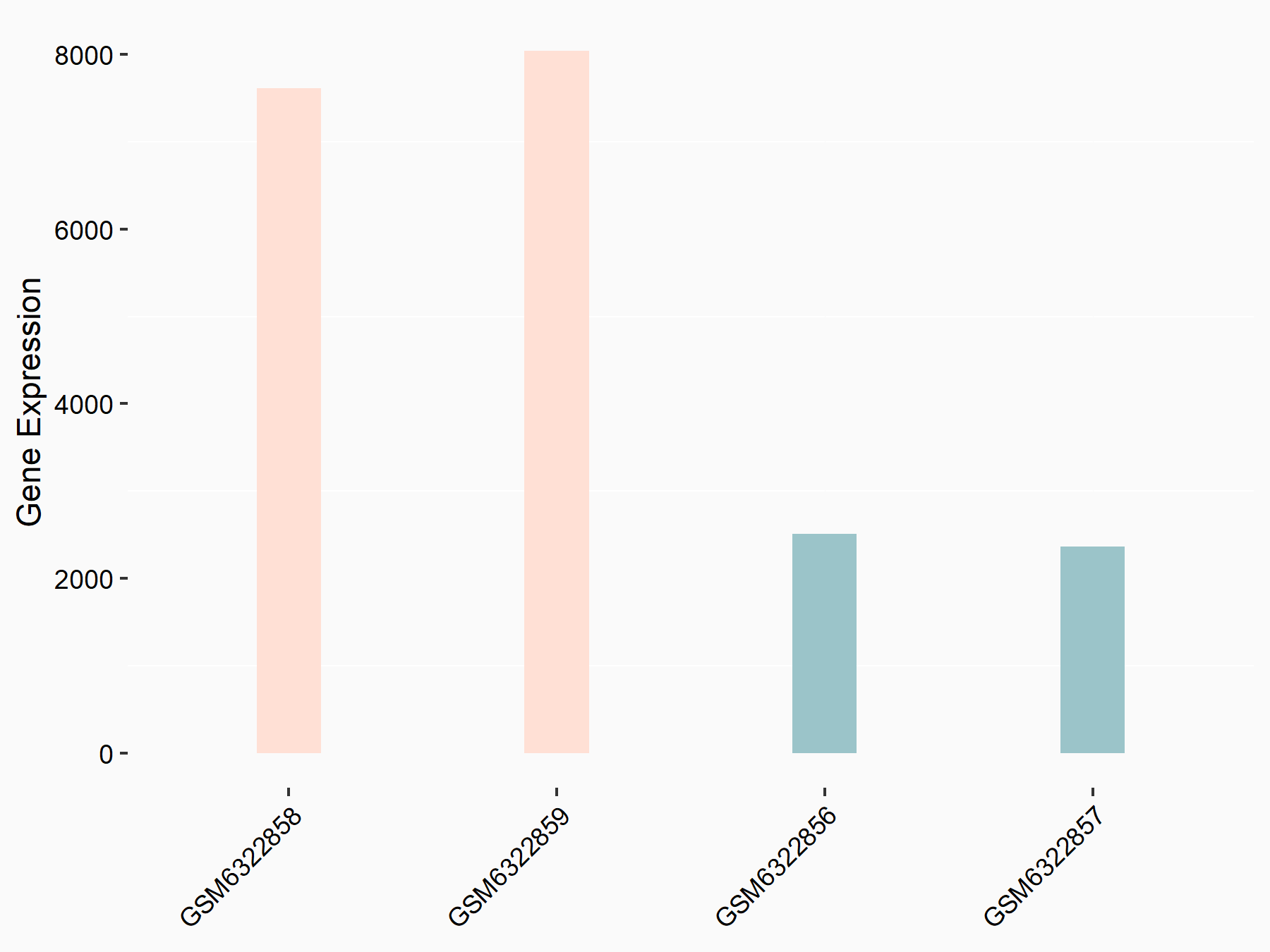 |
logFC: 1.68E+00 p-value: 4.01E-99 |
| More Results | Click to View More RNA-seq Results | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [233] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
In-vitro Model |
LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | 1 × 106 cells in 100 uL PBS (shMETTL3-1 or shNC) were respectively injected into each mouse through the tail vein. Pulmonary metastases were monitored after fourteen days using the imaging system (IVIS) Spectrum (PerkinElmer, USA). | |||
| Response Summary | METTL3 upregulated Urokinase-type plasminogen activator (PLAU) mRNA in an m6A-dependent manner, and then participated in MAPK/ERK pathway to promote angiogenesis and metastasis in CRC. | |||
Vascular endothelial growth factor A (VEGFA)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shMETTL3 MOLM13 cells
Control: MOLM13 cells
|
GSE98623 | |
| Regulation |
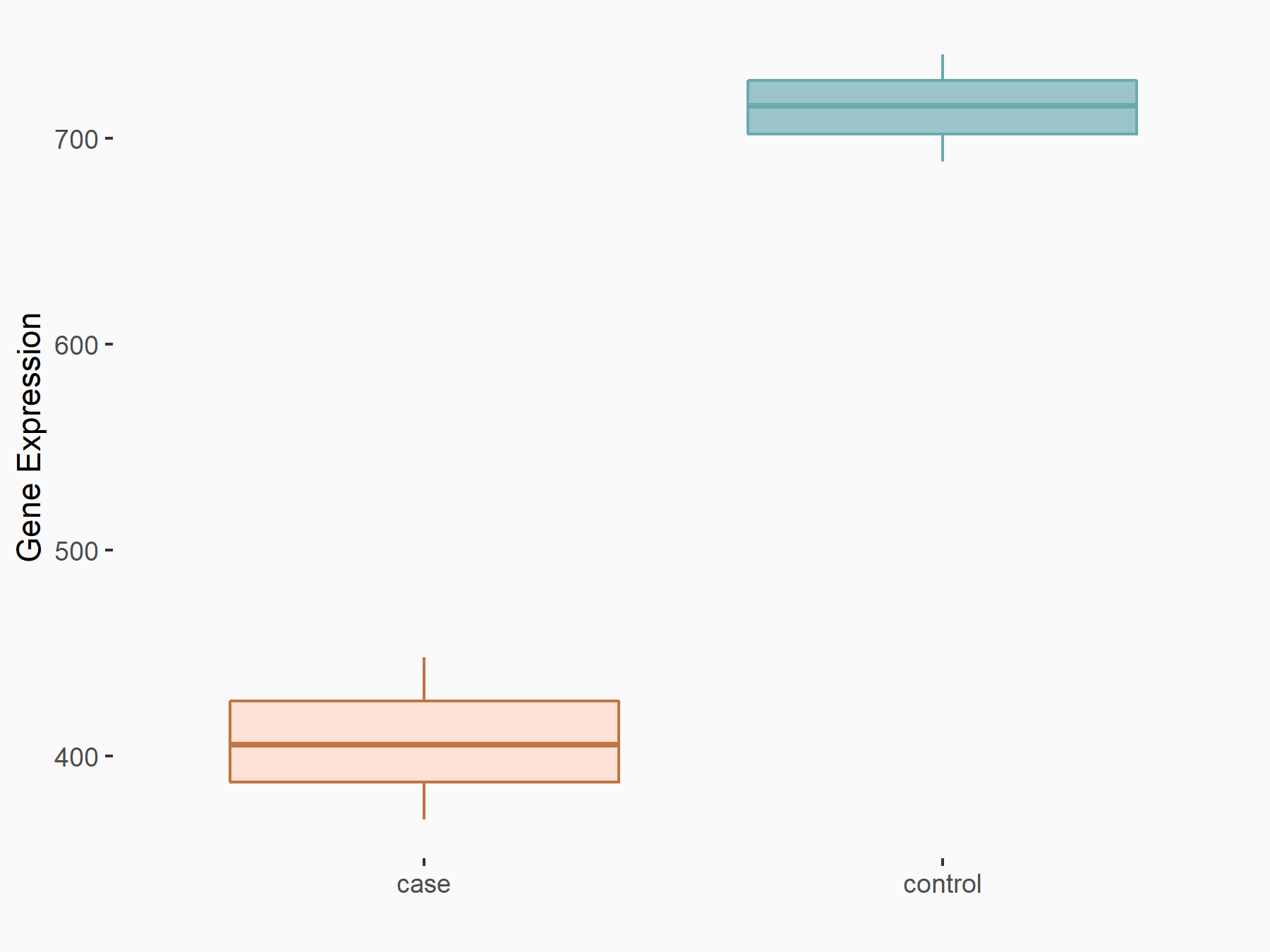 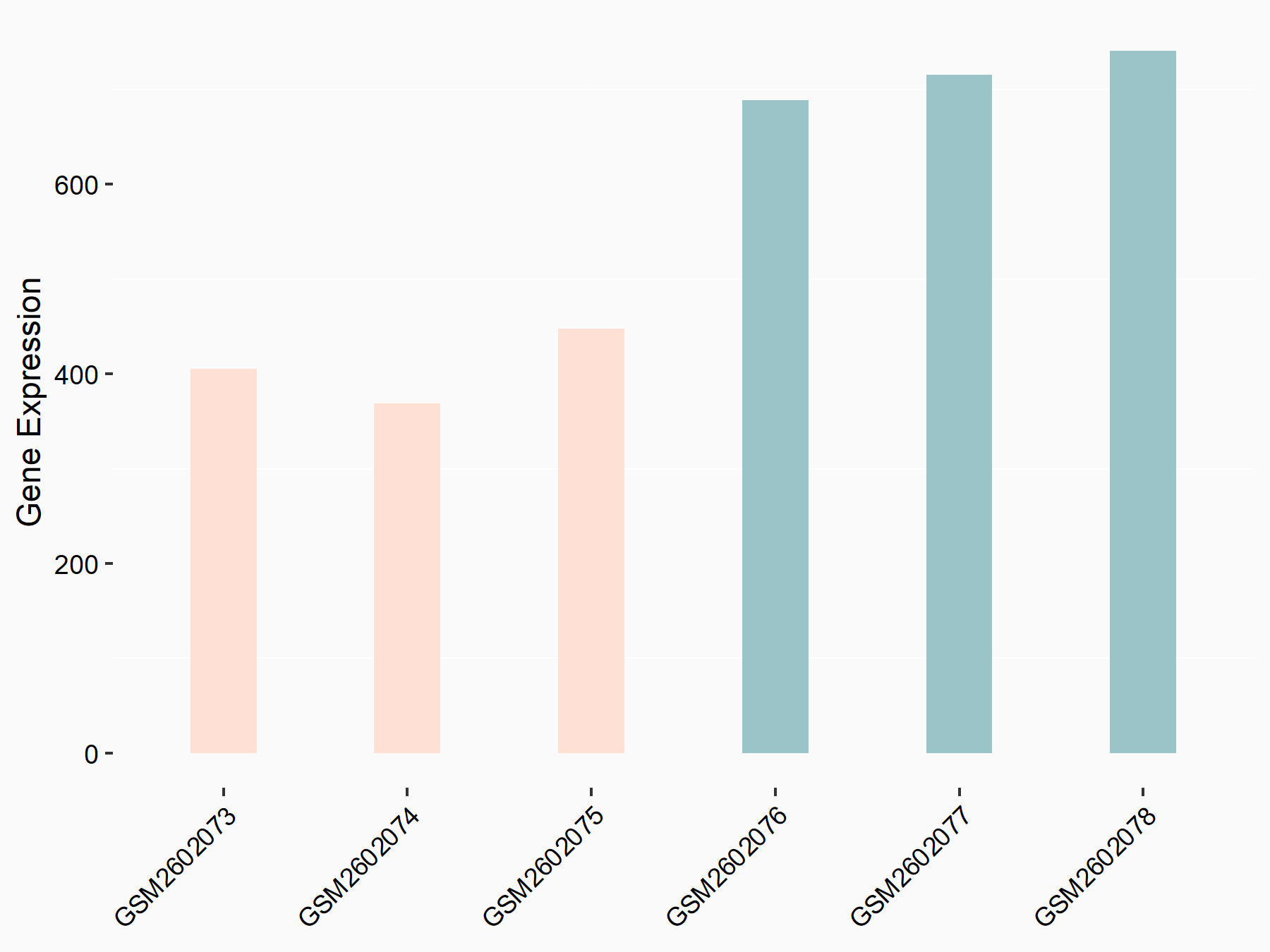 |
logFC: -8.11E-01 p-value: 1.38E-19 |
| More Results | Click to View More RNA-seq Results | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [46] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
In-vitro Model |
SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| In-vivo Model | A total of 8 × 106 wild-type (WT) or METTL3-knockdown cells were injected into the dorsal flanks of 6-week-old nude mice. Seven mice were randomly selected to calculate the volume according to the following formula: V = (width2 × length)/2. Mice were euthanized three weeks after injection and tumors removed, weighed, fixed, and embedded for immunohistochemical analysis. | |||
| Response Summary | EphA2 and Vascular endothelial growth factor A (VEGFA) targeted by METTL3 via different IGF2BP-dependent mechanisms were found to promote vasculogenic mimicry (VM) formation via PI3K/AKT/mTOR and ERK1/2 signaling in CRC. | |||
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cellular proliferation and survival | |||
In-vitro Model |
UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| In-vivo Model | For induction of BCa, 6-8-week-old mice were treated with drinking water containing 500 ug/ml BBN for 16 weeks and then given normal water for another 10 weeks. Tamoxifen was intraperitonelly injected to the mice with 0.08 mg/g of body weight each day for 3 days in order to inductively knock out the target gene. | |||
| Response Summary | Deletion of Mettl3 leads to the suppression of TEK and Vascular endothelial growth factor A (VEGFA),ablation of Mettl3 in bladder urothelial attenuates the oncogenesis and tumor angiogenesis of bladder cancer. | |||
Diseases of the musculoskeletal system [ICD-11: FC0Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [234] | |||
| Responsed Disease | Diseases of the musculoskeletal system [ICD-11: FC0Z] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Alternative splicing | |||
In-vitro Model |
BMSCs (BMSCs were obtained from the femurs and tibias of 2-3-week-old Sprague-Dawley male rats (Animal Center of Sun Yat-sen University)) | |||
| Response Summary | Mettl3 knockdown not only reduced the expression of Vascular endothelial growth factor A (VEGFA) but also decreased the level of its splice variants, vegfa-164 and vegfa-188, in Mettl3-deficient BMSCs. These findings contribute to novel progress in understanding the role of epitranscriptomic regulation in the osteogenic differentiation of BMSCs. | |||
Herpes infection [ICD-11: 1F00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [357] | |||
| Responsed Disease | Herpes simplex keratitis [ICD-11: 1F00.10] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HUVEC-CS
|
N.A. | Homo sapiens | CVCL_0F27 |
|
3T3-Swiss albino
|
N.A. | Mus musculus | CVCL_0120 | |
Vimentin (vimentin)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
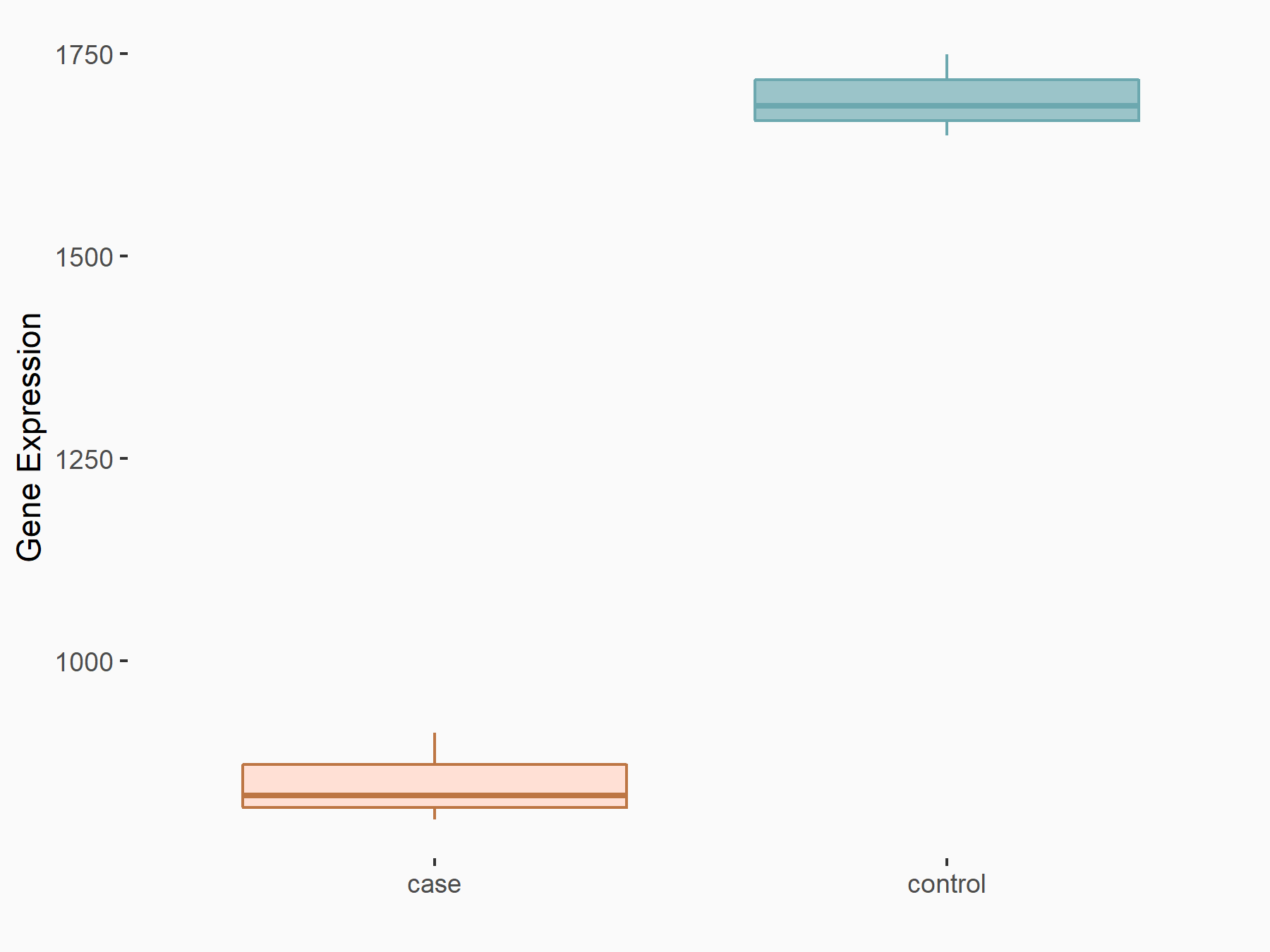 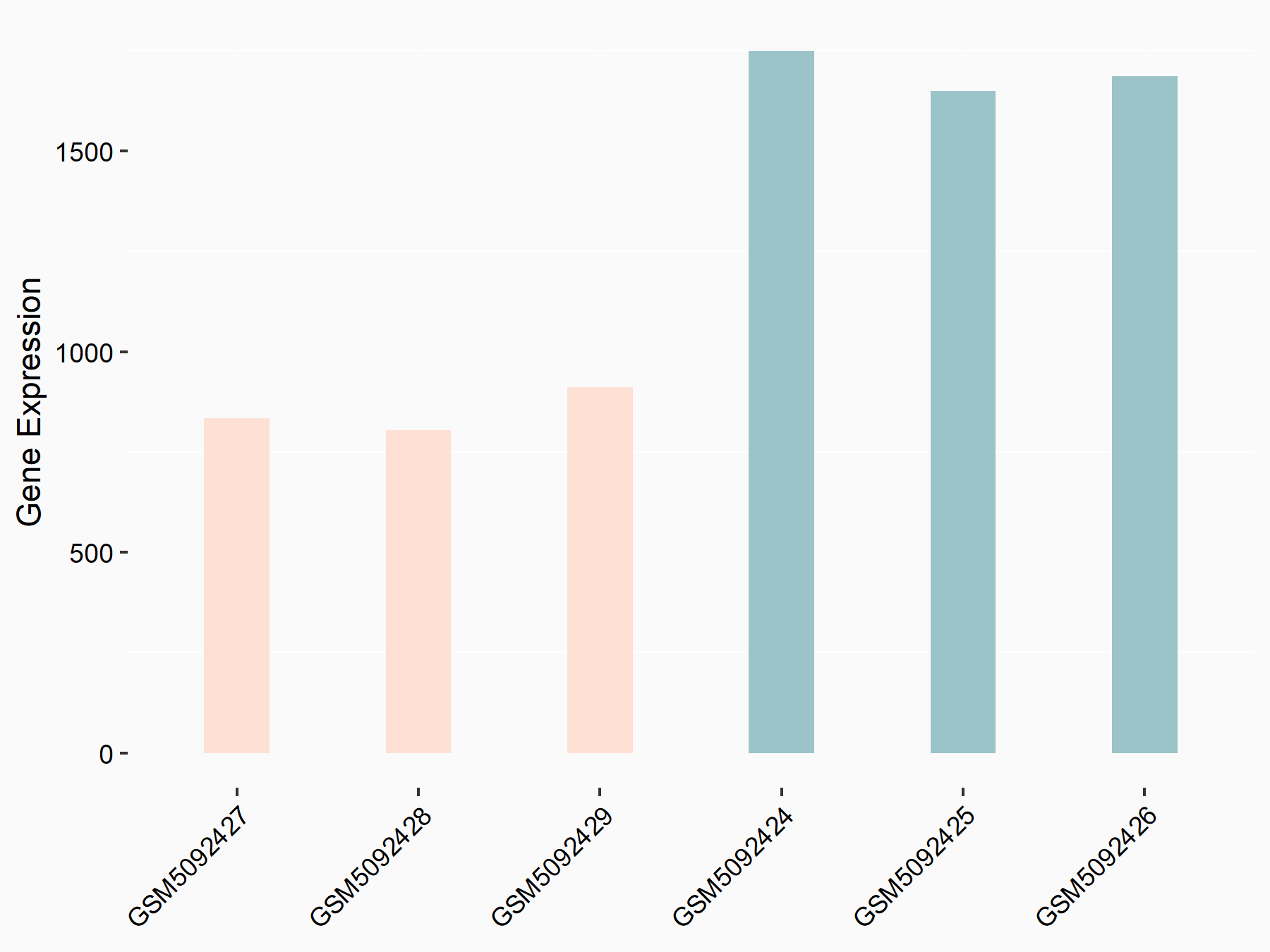 |
logFC: -9.97E-01 p-value: 8.40E-43 |
| More Results | Click to View More RNA-seq Results | |
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [24] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
Neural progenitor cells (NPCs) (The progenitor cells of the CNS) | |||
| NP69 (A human immortalized nasopharyngeal epithelial) | ||||
| HNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_FA07 | |
| HNE-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_0308 | |
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| CNE-1 | Normal | Homo sapiens | CVCL_6888 | |
| In-vivo Model | 1 × 105 HNE2 cells (with or without METTL3 knockdown) were labeled with luciferase gene and injected into the tail vein of the nude mice. | |||
| Response Summary | METTL3 activated the luciferase activity of TOPflash (a reporter for beta-catenin/TCF signaling), and downregulation of METTL3 inhibited the expression of beta-catenin/TCF target genes Vimentin (vimentin) and N-cadherin, which are two regulators of epithelial-mesenchymal transition. METTL3 silencing decreased the m6A methylation and total mRNA levels of Tankyrase, a negative regulator of axin. METTL3 is a therapeutic target for NPC. | |||
Zinc finger protein SNAI1 (SNAI1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
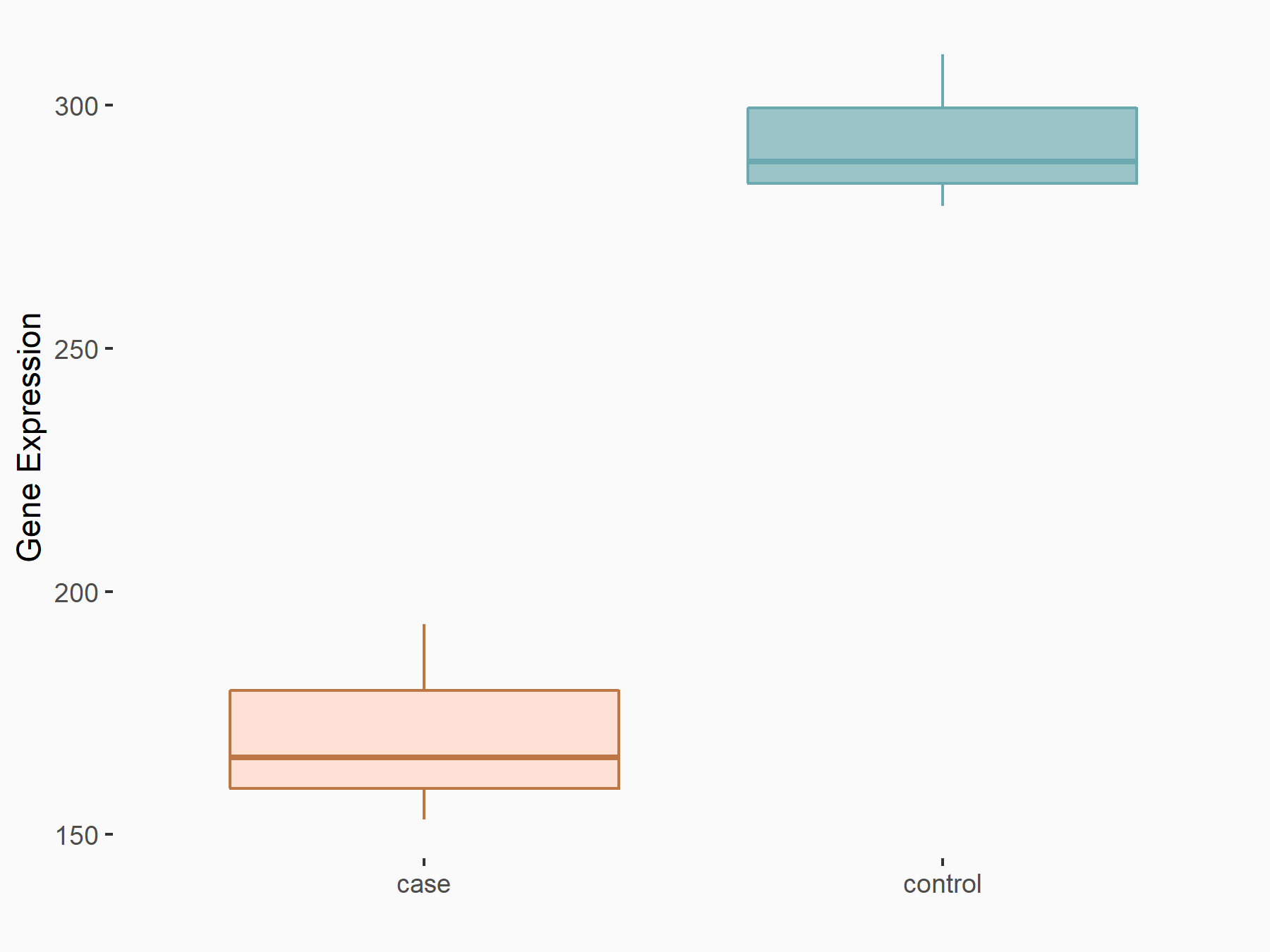 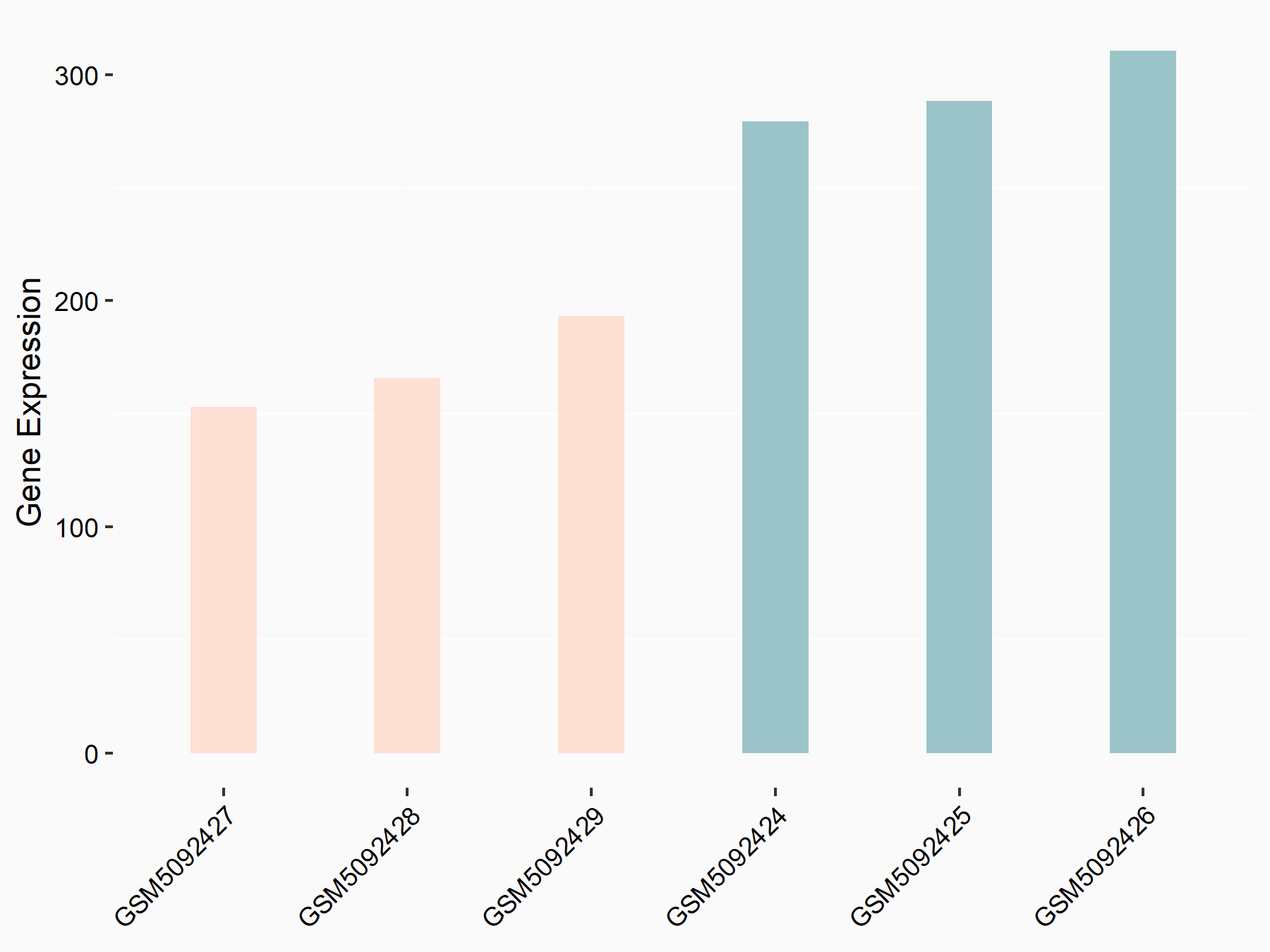 |
logFC: -7.78E-01 p-value: 5.01E-07 |
| More Results | Click to View More RNA-seq Results | |
Solid tumour/cancer [ICD-11: 2A00-2F9Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [235] | |||
| Responsed Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Adherens junction | hsa04520 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| Response Summary | The expression of TGFbeta1 was up-regulated, while self-stimulated expression of TGFbeta1 was suppressed in METTL3Mut/- cells. m6A performed multi-functional roles in TGFbeta1 expression and EMT modulation, suggesting the critical roles of m6A in cancer progression regulation. Zinc finger protein SNAI1 (SNAI1), which was down-regulated in Mettl3Mut/- cells, was a key factor responding to TGF-Beta-1-induced EMT. | |||
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [236] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
SUNE1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6946 |
| Response Summary | Overexpression of Zinc finger protein SNAI1 (SNAI1) partially reversed the regulatory effects of METTL3 on EMT-related gene expressions and metastatic abilities in nasopharyngeal carcinoma. Knockdown of METTL3 decreased the enrichment abundance of Snail in anti-IGF2BP2. | |||
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [237] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Adherens junction | hsa04520 | ||
| Cell Process | Cell proliferation | |||
| Cell invasion | ||||
In-vitro Model |
SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| HIEC-6 | Normal | Homo sapiens | CVCL_6C21 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| Response Summary | METTL3 acts as a critical m6A methyltransferase capable of facilitating colorectal cancer(CRC) progression, and revealed a novel mechanism by which METTL3 promotes CRC cell proliferation and invasion via stabilizing Zinc finger protein SNAI1 (SNAI1) mRNA in an m6A-dependent manner. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [238] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Epithelial-mesenchymal transition | |||
| cell migration | ||||
| invasion | ||||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | All animal experiments complied with Zhongshan School of Medicine Policy on Care and Use of Laboratory Animals. For subcutaneous transplanted model, sh-control and sh-METTL3 Huh7 cells (5 × 106 per mouse, n = 5 for each group) were diluted in 200 uL of PBS + 200 uL Matrigel (BD Biosciences) and subcutaneously injected into immunodeficient female mice to investigate tumor growth. | |||
| Response Summary | The upregulation of METTL3 and YTHDF1 act as adverse prognosis factors for overall survival (OS) rate of liver cancer patients. m6A-sequencing and functional studies confirm that Zinc finger protein SNAI1 (SNAI1), a key transcription factor of EMT, is involved in m6A-regulated EMT. | |||
Family with sequence similarity 225 member A (FAM225A)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
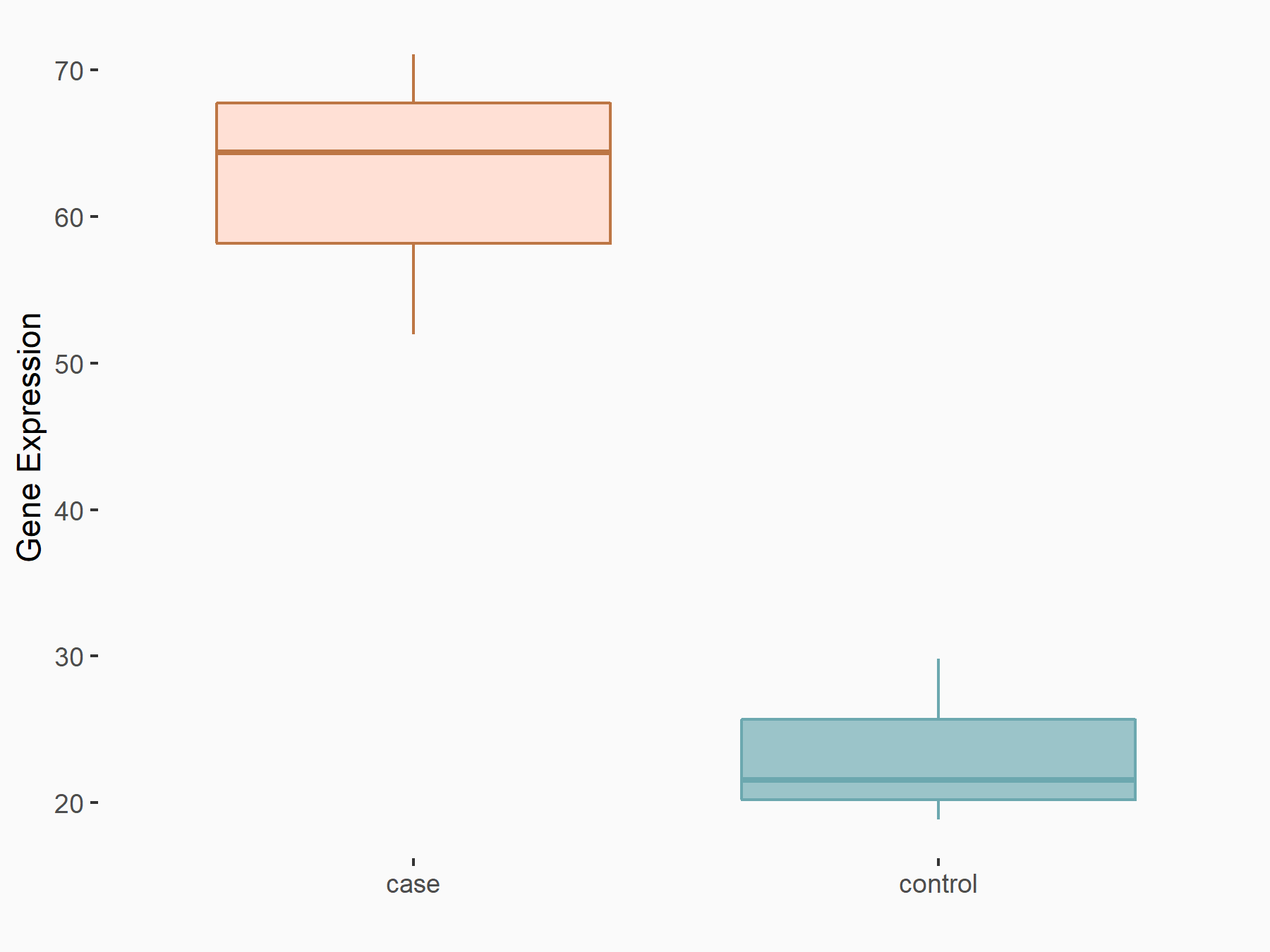 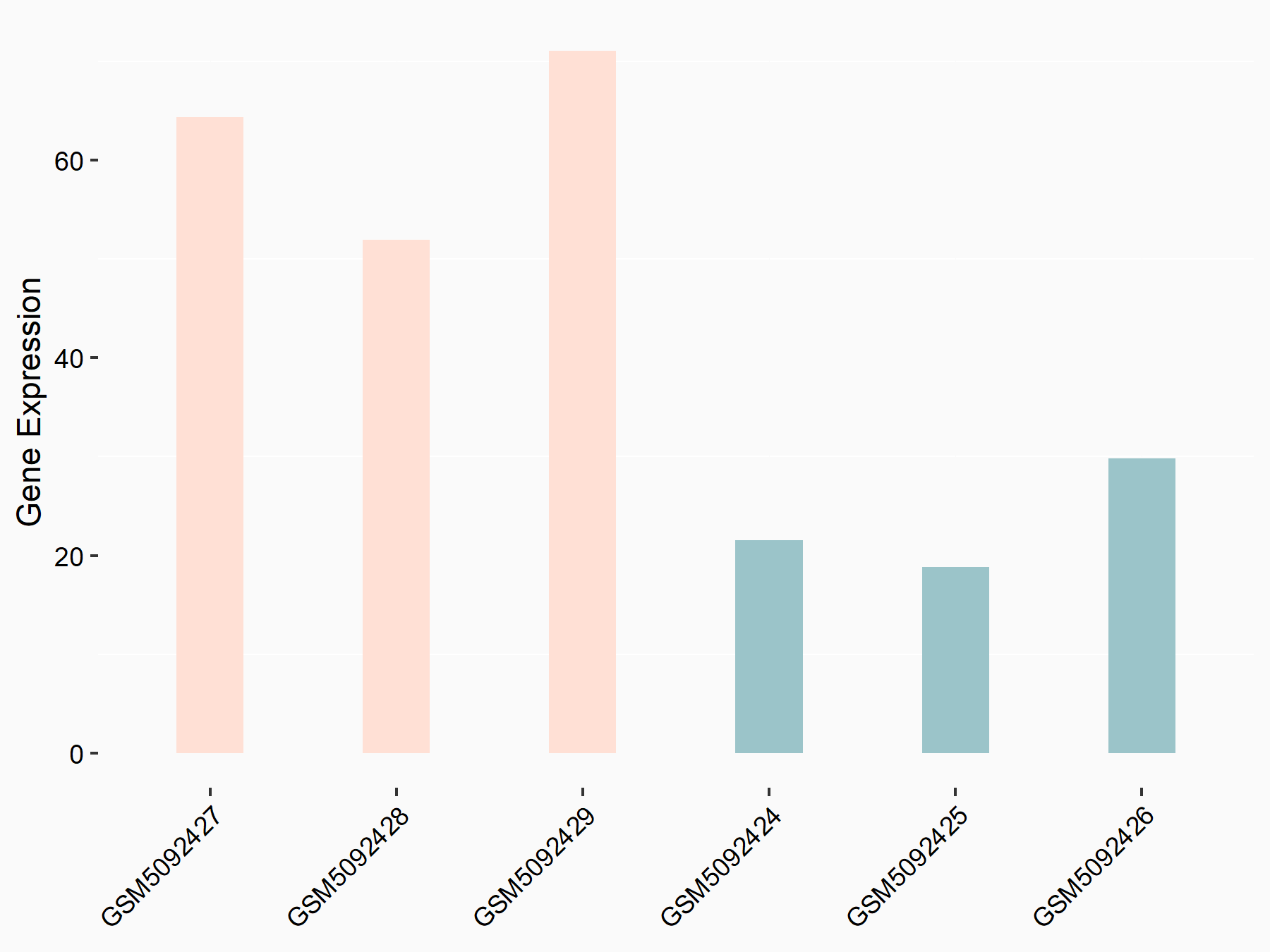 |
logFC: 1.41E+00 p-value: 1.18E-04 |
| More Results | Click to View More RNA-seq Results | |
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [64] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell proliferation and metastasis | |||
In-vitro Model |
5-8F | Nasopharyngeal carcinoma | Homo sapiens | CVCL_C528 |
| 6-10B | Nasopharyngeal carcinoma | Homo sapiens | CVCL_C529 | |
| C666-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_7949 | |
| CNE-1 | Normal | Homo sapiens | CVCL_6888 | |
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| HK1 | Nasopharyngeal carcinoma | Acipenser baerii | CVCL_YE27 | |
| HNE-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_0308 | |
| HONE-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_8706 | |
| N2Tert (The human immortalized nasopharyngeal epithelial cell lines) | ||||
| NP69 (A human immortalized nasopharyngeal epithelial) | ||||
| S18 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_B0U9 | |
| S26 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_B0UB | |
| SUNE1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6946 | |
| In-vivo Model | For the tumor growth model, 1 × 106 HNE1-Scrambled or HNE1-shFAM2225A 2# cells were injected into the axilla of the mice, and the tumor size was measured every 3 days. | |||
| Response Summary | Family with sequence similarity 225 member A (FAM225A) functioned as a competing endogenous RNA (ceRNA) for sponging miR-590-3p and miR-1275, leading to the upregulation of their target integrin beta-3 (ITGB3), and the activation of FAK/PI3K/Akt signaling to promote Nasopharyngeal carcinoma cell proliferation and invasion. FAM225A showed lower RNA stability after silencing of METTL3. | |||
H19 imprinted maternally expressed transcript (H19)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | DKO-1 cell line | Homo sapiens |
|
Treatment: METTL3 knockdown DKO-1 cell
Control: DKO-1 cell
|
GSE182382 | |
| Regulation |
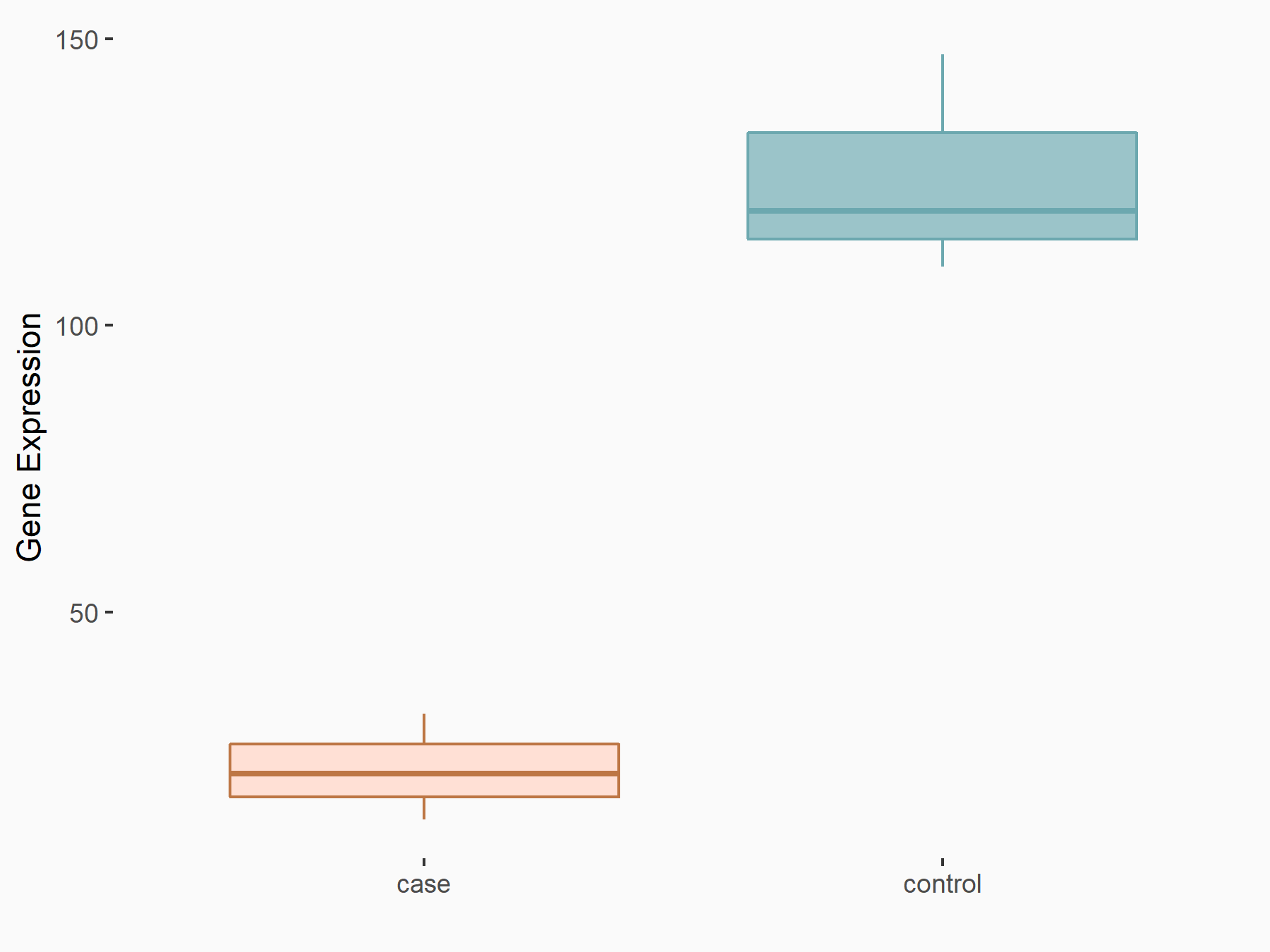 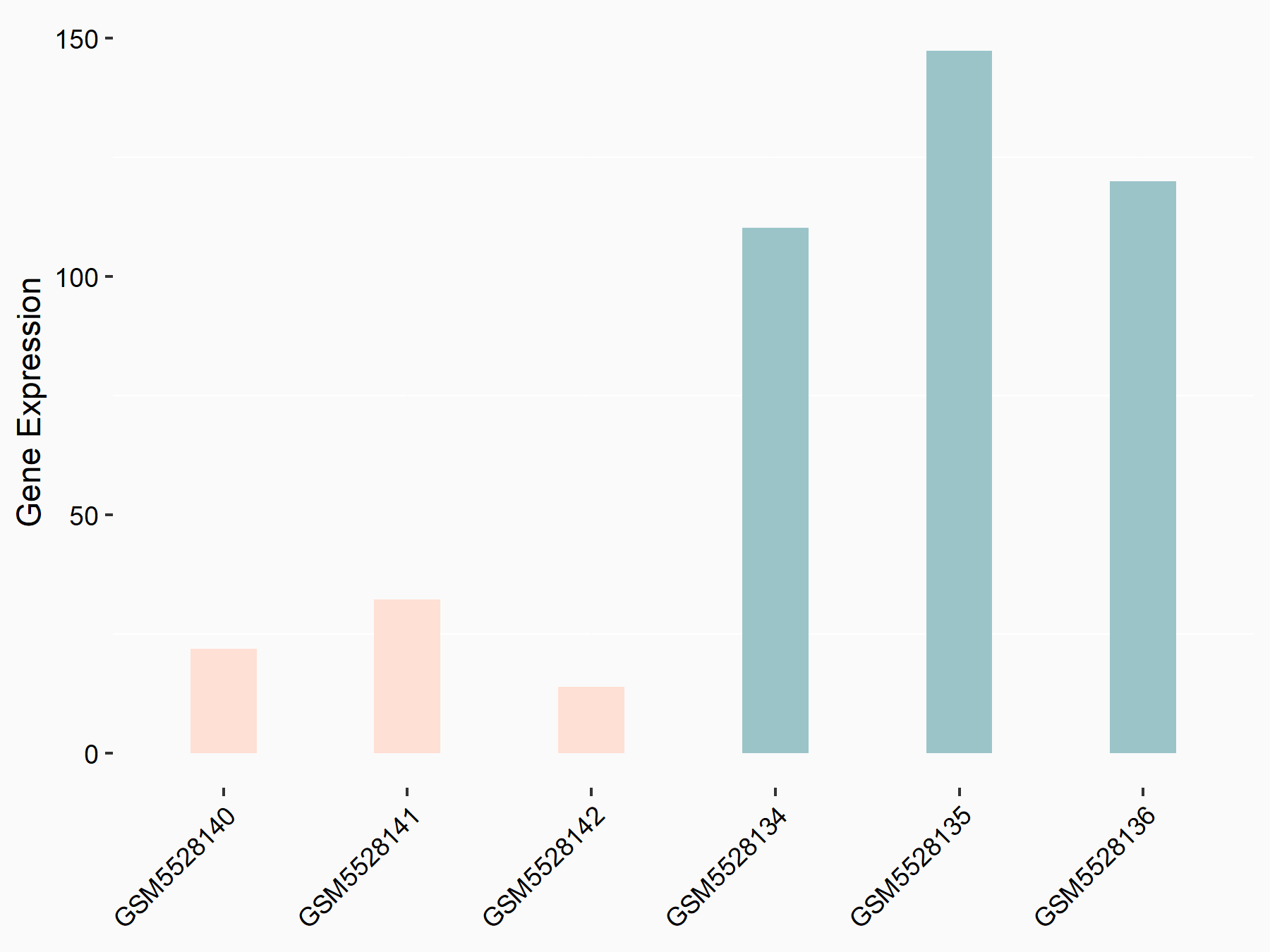 |
logFC: -2.53E+00 p-value: 9.95E-11 |
| More Results | Click to View More RNA-seq Results | |
Abnormalities of breathing [ICD-11: MD11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [240] | |||
| Responsed Disease | Abnormalities of breathing [ICD-11: MD11] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell viability | |||
| Cell apoptosis | ||||
In-vitro Model |
H9c2(2-1) | Normal | Rattus norvegicus | CVCL_0286 |
| In-vivo Model | In vivo myocardial I/R injury was performed by 60 min of ligation of LAD followed by 5 h of reperfusion. | |||
| Response Summary | Either knockdown of METTL3 or METTL14 notably reversed the hypoxic preconditioning-induced enhancement of cell viability, anti-apoptosis ability, and H19 imprinted maternally expressed transcript (H19) expression. | |||
Long intergenic non-protein coding RNA 470 (LINC00470)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
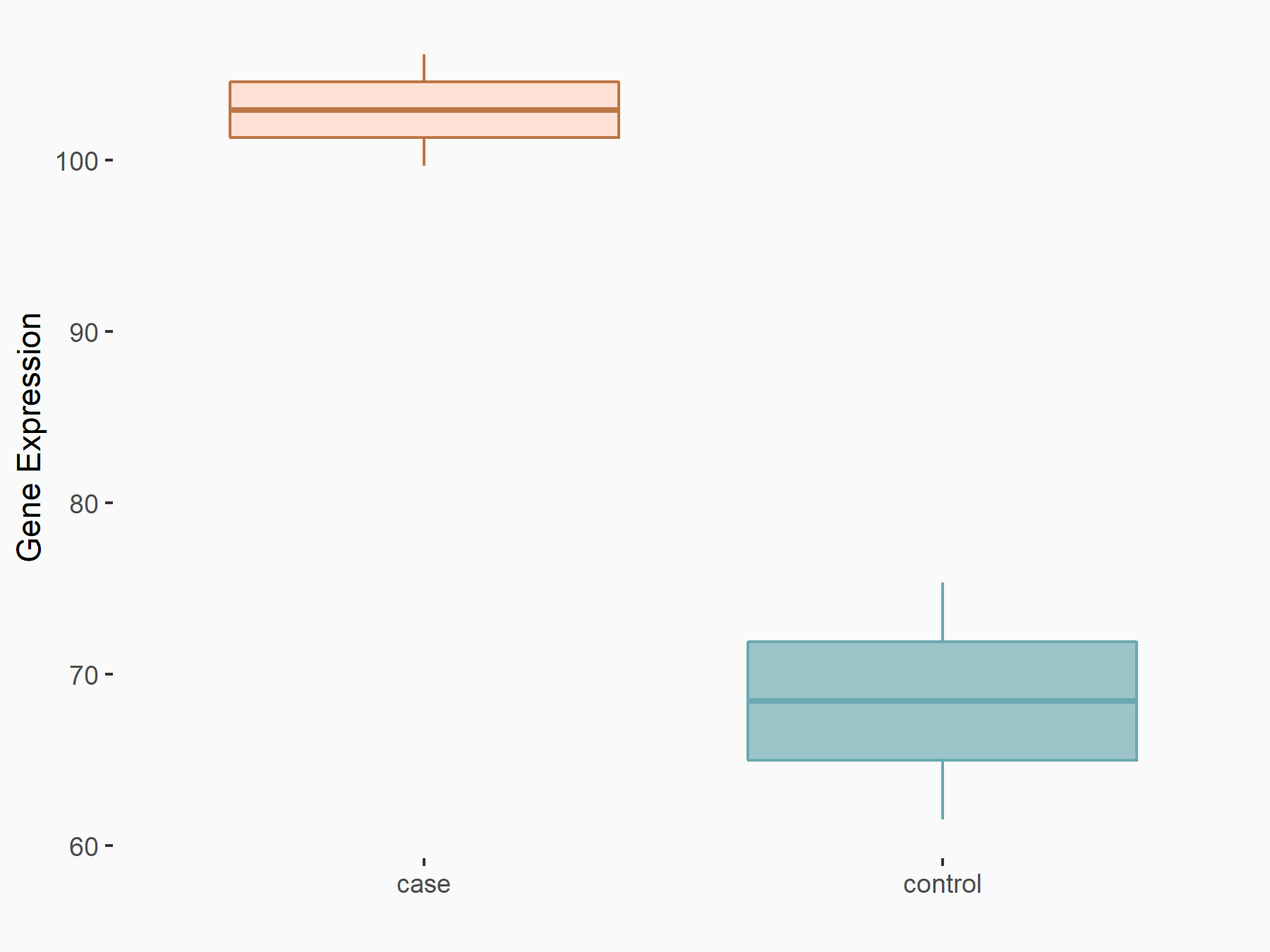 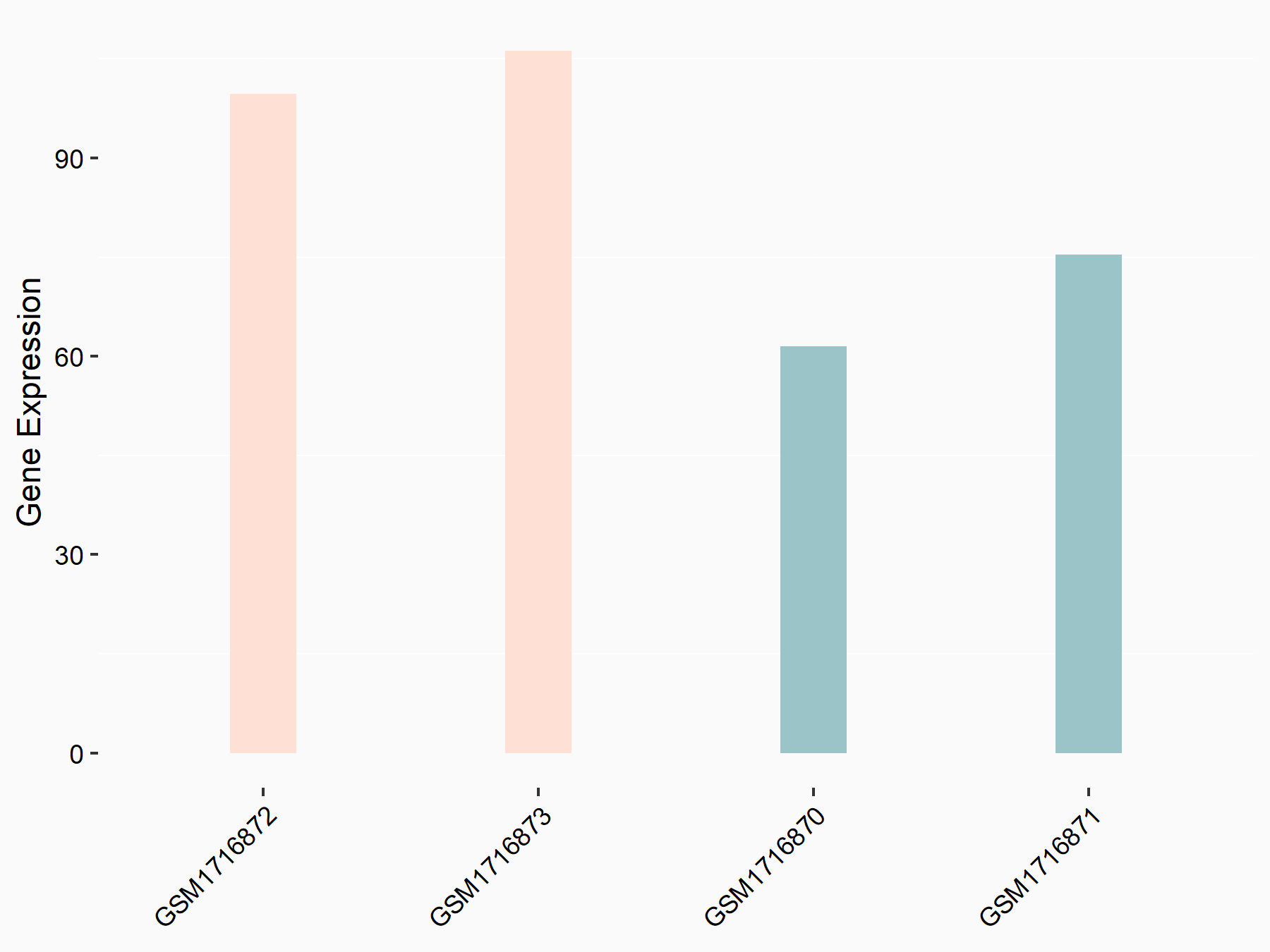 |
logFC: 5.89E-01 p-value: 3.48E-02 |
| More Results | Click to View More RNA-seq Results | |
Malignant haematopoietic neoplasm [ICD-11: 2B33]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [73] | |||
| Responsed Disease | Chronic myeloid leukaemia [ICD-11: 2B33.2] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | RNA stability | |||
| Cell autophagy | ||||
In-vitro Model |
K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 |
| KCL-22 | Chronic myelogenous leukemia | Homo sapiens | CVCL_2091 | |
| In-vivo Model | In the control mice or ADR mice group, the parental or chemo-resistant K562 cells were infected with LV-shCtrl. In the ADR + shLINC00470 group, the chemo-resistant K562 cells were infected with LV- shLINC00470. These cells were injected, respectively, into these 5-week-old mice subcutaneously. | |||
| Response Summary | The molecular mechanism underlying the effect of Long intergenic non-protein coding RNA 470 (LINC00470) on chronic myelocytic leukaemia by reducing the PTEN stability via RNA methyltransferase METTL3, thus leading to the inhibition of cell autophagy while promoting chemoresistance in CML. | |||
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [241] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Gastric cancer | hsa05226 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| Response Summary | Long intergenic non-protein coding RNA 470 (LINC00470)-METTL3-mediated PTEN mRNA degradation relied on the m6A reader protein YTHDF2-dependent pathway.LINC00470 served as a therapeutic target for Gastric cancer patients. | |||
Nuclear paraspeckle assembly transcript 1 (NEAT1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 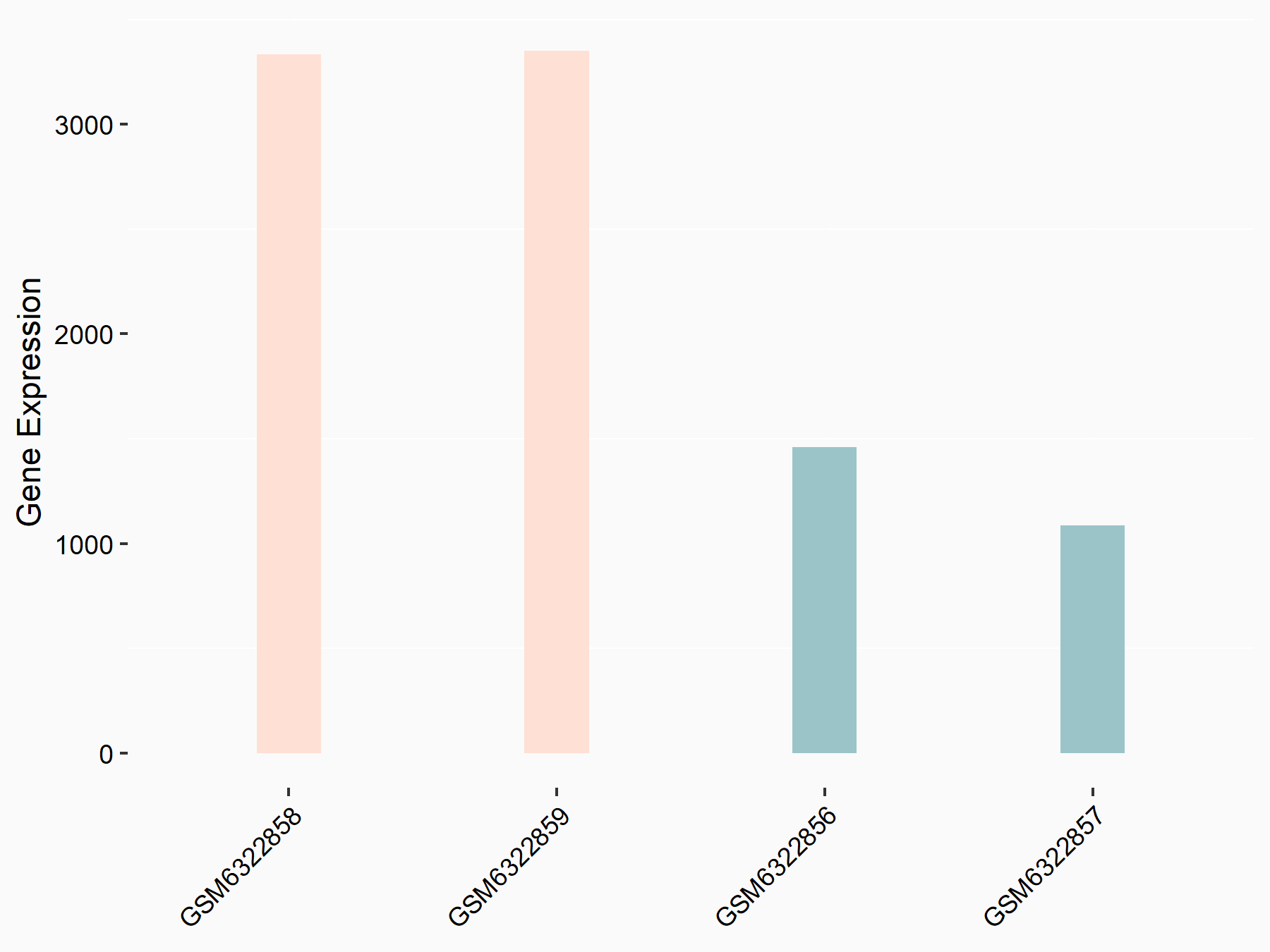 |
logFC: 1.39E+00 p-value: 3.32E-29 |
| More Results | Click to View More RNA-seq Results | |
Malignant haematopoietic neoplasm [ICD-11: 2B33]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [242] | |||
| Responsed Disease | Chronic myeloid leukaemia [ICD-11: 2B33.2] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell viability and apoptosis | |||
In-vitro Model |
BV-173 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0181 |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| KCL-22 | Chronic myelogenous leukemia | Homo sapiens | CVCL_2091 | |
| MEG-01 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0425 | |
| Response Summary | METTL3-mediated m6A modification induced the aberrant expression of Nuclear paraspeckle assembly transcript 1 (NEAT1) in chronic myelocytic leukemia. Overexpression of NEAT1 inhibited cell viability and promoted the apoptosis of chronic myelocytic leukemia cells. miR-766-5p was upregulated in CML PBMCs and abrogated the effects of NEAT1 on cell viability and apoptosis of the chronic myelocytic leukemia cells. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [371] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| In-vivo Model | BALB/c male nude mice aged 6 weeks were randomly divided into two groups (n = 5 / group). A549 cells with transfection of sh-NC or sh-NEAT1 were injected into mice. After 28 days, the mice were euthanized and the tumor tissues were collected and measured. | |||
microRNA 186 (MIR186)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
 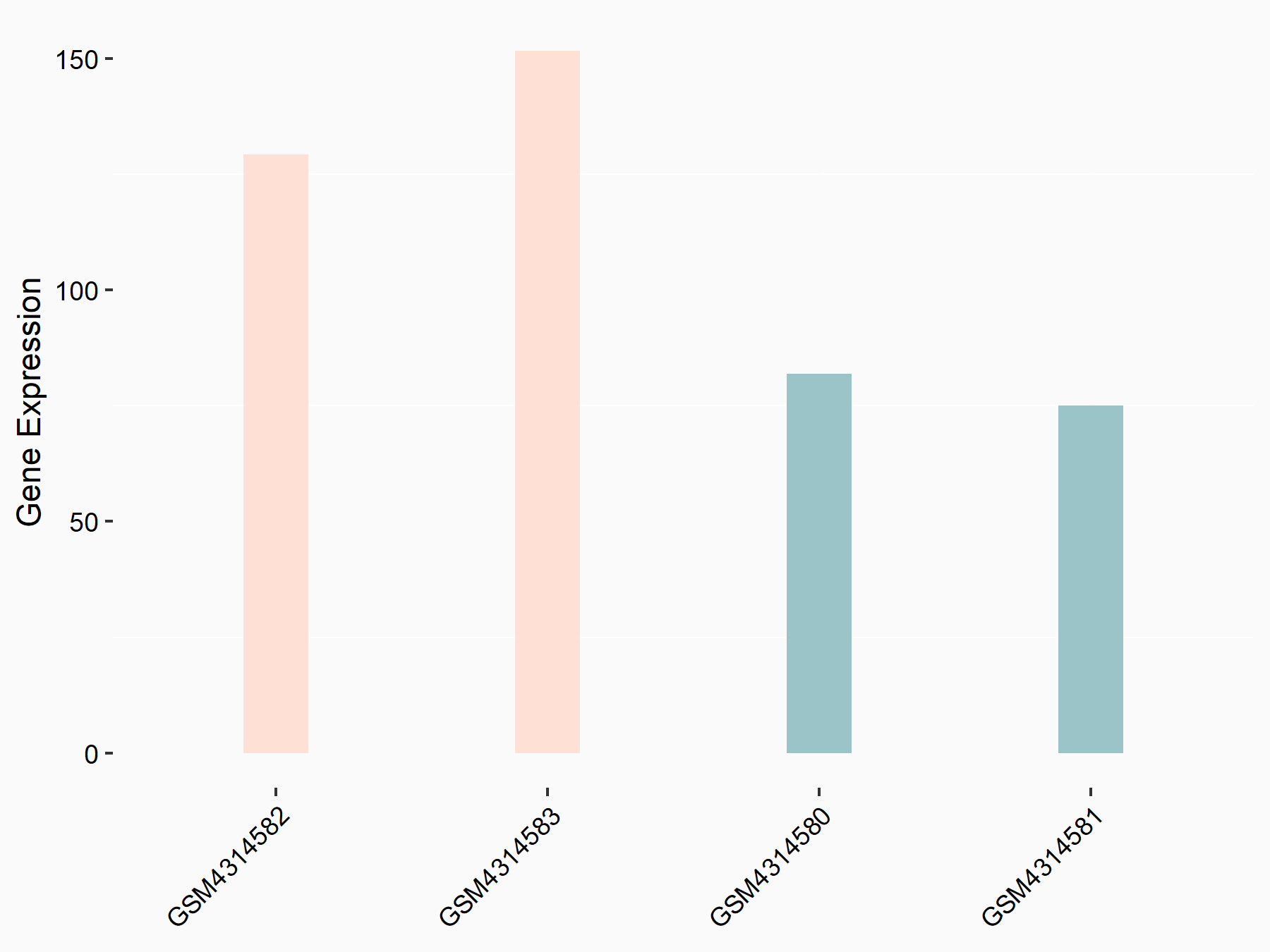 |
logFC: 8.42E-01 p-value: 2.96E-03 |
| More Results | Click to View More RNA-seq Results | |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [243] | |||
| Responsed Disease | Hepatoblastoma [ICD-11: 2C12.01] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Cell aggressive | |||
In-vitro Model |
HCCLM9 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_A5CU |
| HEK293 | Normal | Homo sapiens | CVCL_0045 | |
| Hepa 1-6 | Hepatocellular carcinoma of the mouse | Mus musculus | CVCL_0327 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| HuH-6 | Hepatoblastoma | Homo sapiens | CVCL_4381 | |
| In-vivo Model | Cells transfected with miR-186 overexpression lentivirus (Lenti-miR-186), miR-186 inhibitor (Lenti-anti-miR-186), Lenti-miR-186 & METTL3 overexpression plasmid (Lenti-METTL3), Lenti-anti-miR-186 & METTL3 shRNA (sh-METTL3) or empty lentivirus control (Lenti-NC) were subcutaneously injected into the lower flank of nude mice. | |||
| Response Summary | microRNA 186 (MIR186)/METTL3 axis contributed to the progression of Hepatoblastoma via the Wnt/beta-catenin signalling pathway. | |||
microRNA 221 (MIR221)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | DKO-1 cell line | Homo sapiens |
|
Treatment: METTL3 knockdown DKO-1 cell
Control: DKO-1 cell
|
GSE182382 | |
| Regulation |
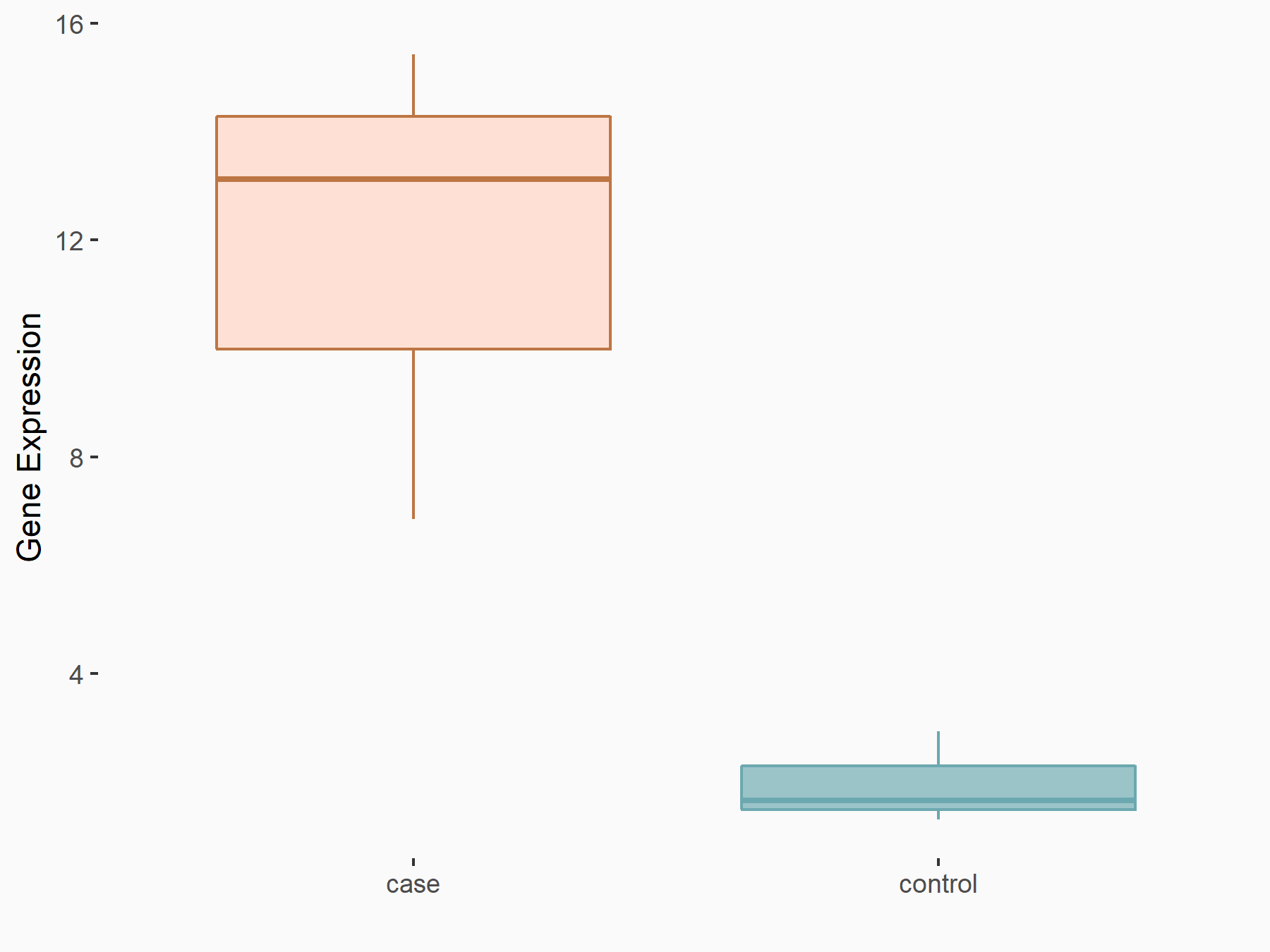 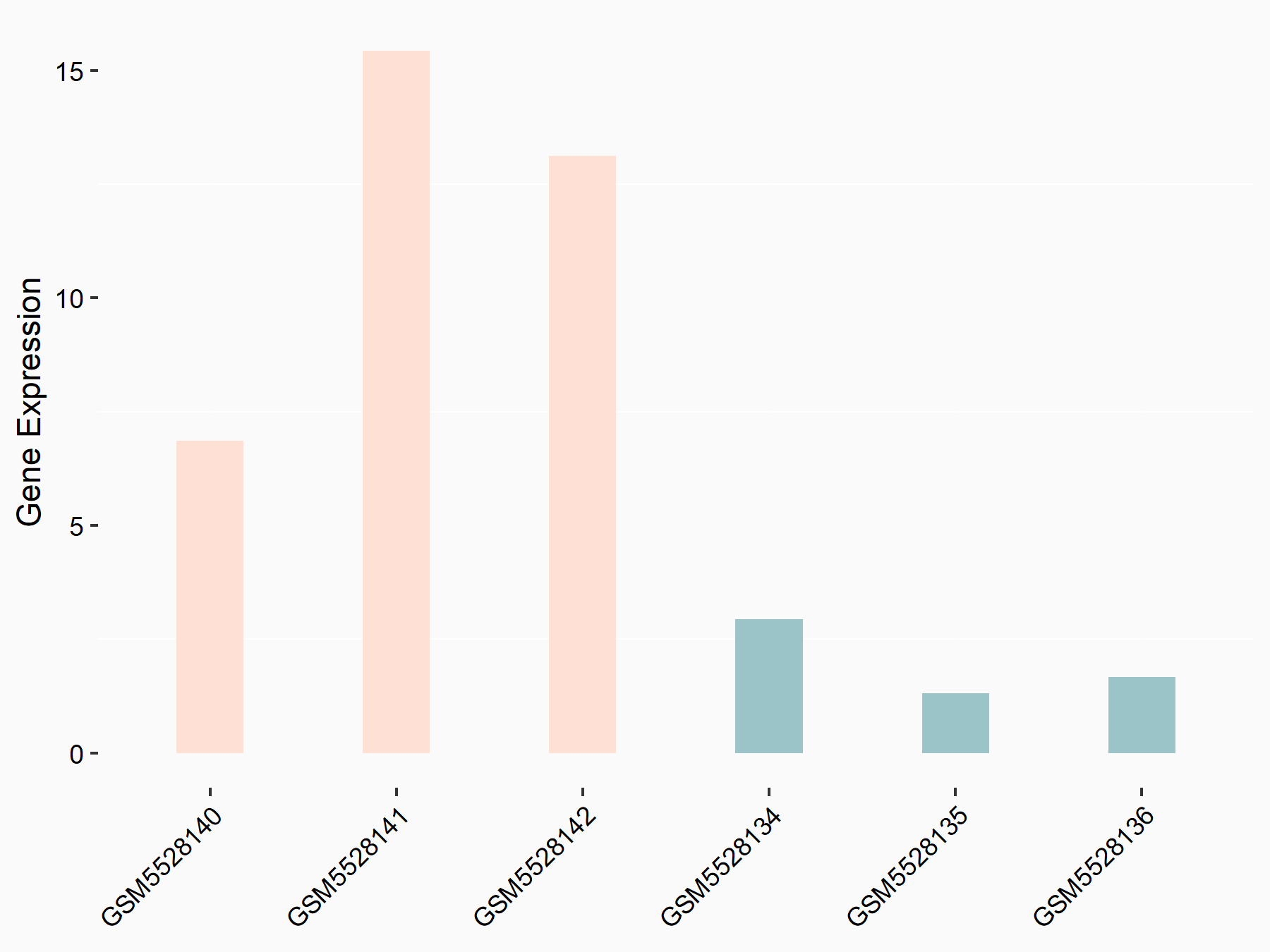 |
logFC: 2.60E+00 p-value: 1.63E-02 |
| More Results | Click to View More RNA-seq Results | |
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [244] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
In-vitro Model |
EJ (Human bladder cancer cells) | |||
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| In-vivo Model | About 1× 107 cells were injected subcutaneously into the axilla of the female athymic BALB/C nude mice (4-6 weeks old, 18-22 g, five mice per group). | |||
| Response Summary | METTL3 has an oncogenic role in bladder cancer through interacting with the microprocessor protein DGCR8 and positively modulating the microRNA 221 (MIR221) process in an m6A-dependent manner. | |||
Cardiomegaly [ICD-11: BC45]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [381] | |||
| Responsed Disease | Cardiomegaly [ICD-11: BC45] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
In-vitro Model |
NRCMs (Primary neonatal rat cardiomyocytes (NRCMs)NRCMs were prepared from the hearts of 2- to 3-day-old SD rats according to the following protocol) | |||
| In-vivo Model | The pump was prefilled with Ang-II or saline and then incubated in sterile saline at 37 ℃ for 48 h. After the mice were anesthetized with 3.0% isoflurane mixed with oxygen, an incision was made on the back skin of the mice, and the pump was implanted into the subcutaneous area, followed by suturing the incision. After the operation, the mice were given buprenorphine (0.1 mg/kg) to reach analgesia. Finally, the regaining consciousness mice were returned to cages and fed until the end of the experiment. | |||
| Response Summary | METTL3 positively modulates the pri-miR-221/222 maturation process in an m6A-dependent manner and subsequently activates Wnt/Beta-catenin signaling by inhibiting DKK2, thus promoting Ang-II-induced cardiac hypertrophy. | |||
microRNA 335 (MIR335)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Mouse testis | Mus musculus |
|
Treatment: Mettl3 knockout mouse testis
Control: Mouse testis
|
GSE99771 | |
| Regulation |
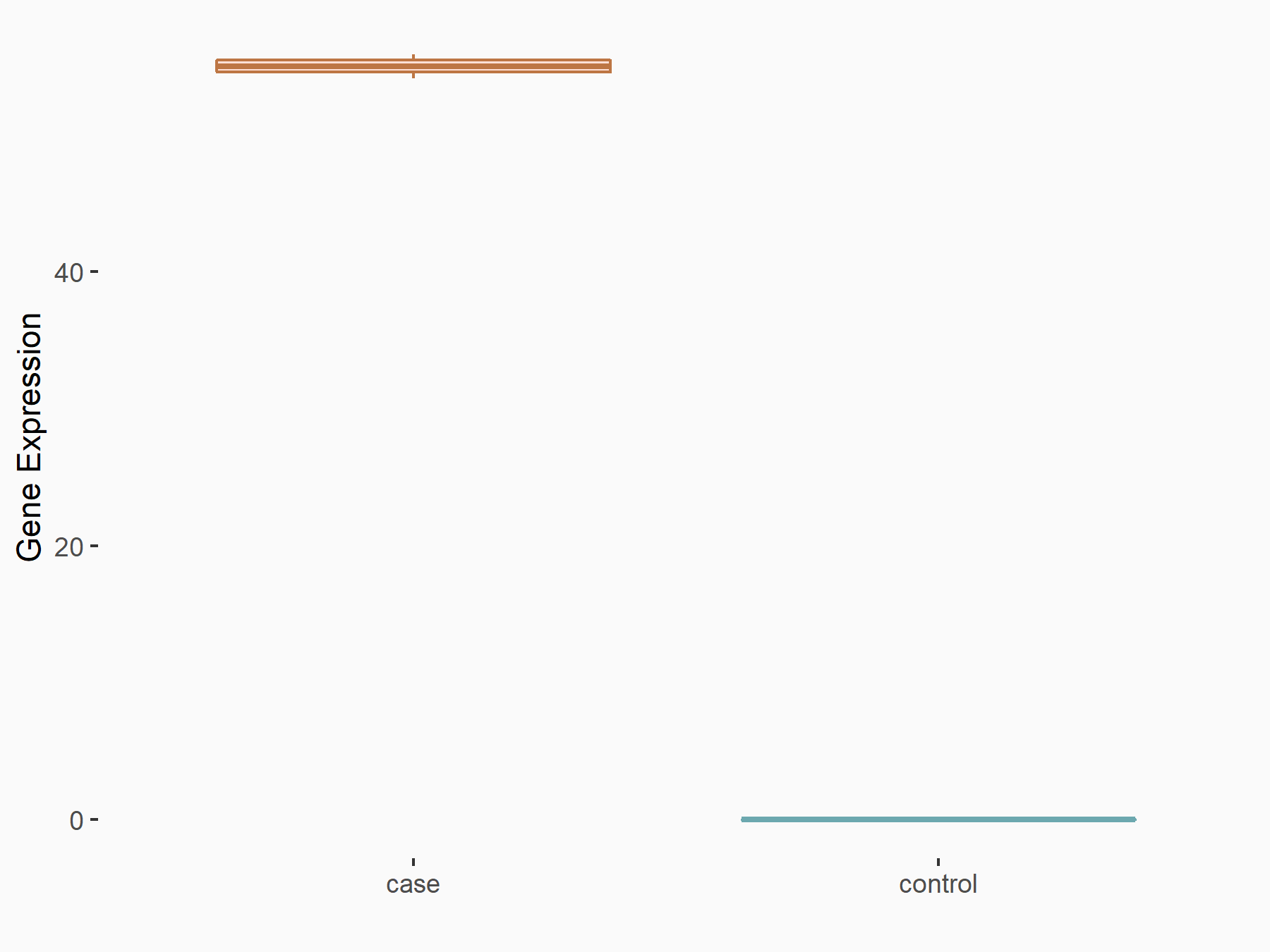 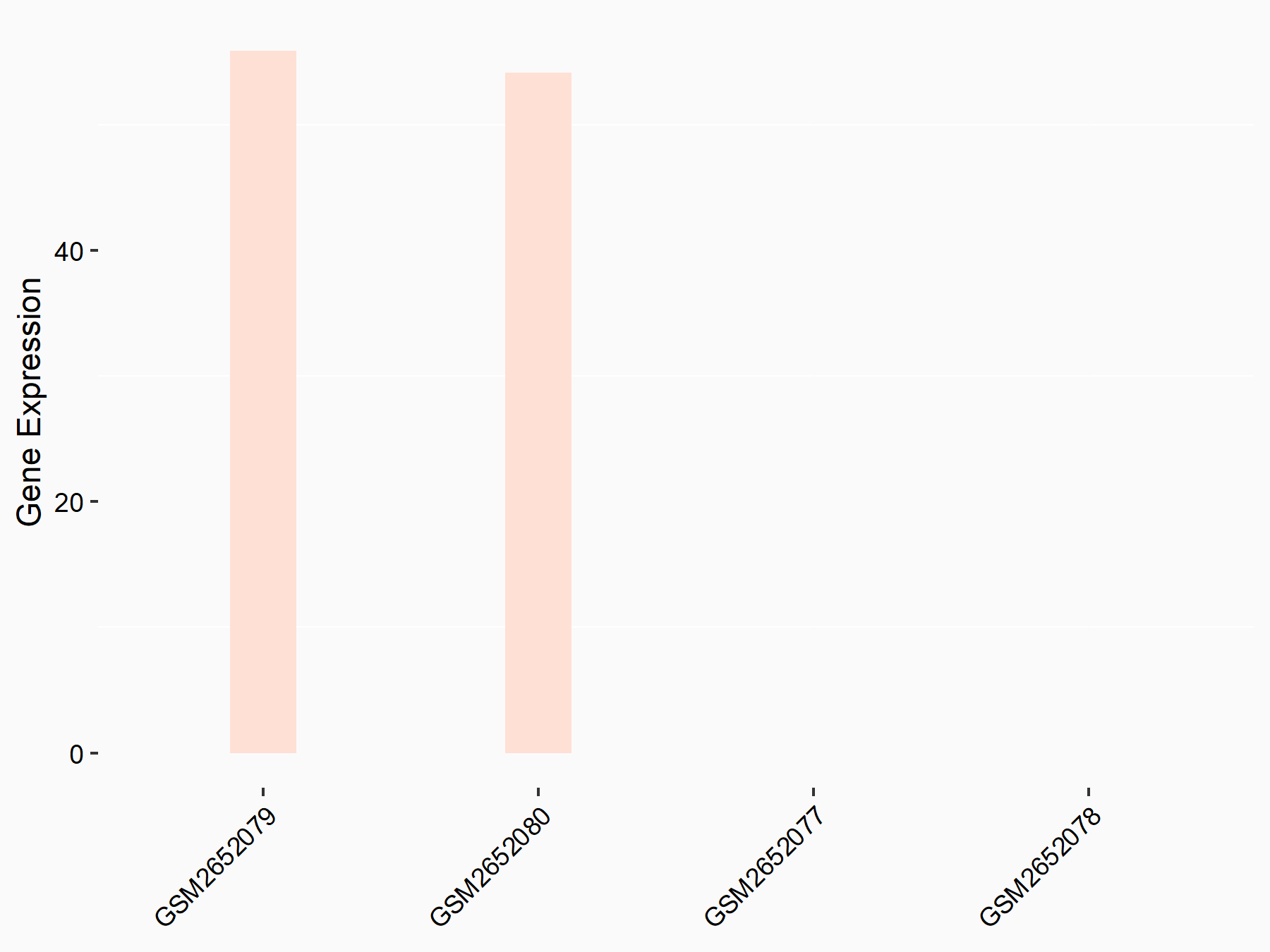 |
logFC: 5.81E+00 p-value: 4.09E-06 |
| More Results | Click to View More RNA-seq Results | |
Acute ischemic stroke [ICD-11: 8B11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [245] | |||
| Responsed Disease | Acute ischemic stroke [ICD-11: 8B11] | |||
| Target Regulation | Up regulation | |||
| Cell Process | RNA maturation | |||
| Cell apoptosis | ||||
In-vitro Model |
PC-12 | Lung papillary adenocarcinoma | Homo sapiens | CVCL_S979 |
| In-vivo Model | After a median incision on the neck, the left common carotid artery (CCA), internal carotid artery (ICA), and external carotid artery (ECA) were isolated. The left CCA and the ECA were ligated. | |||
| Response Summary | METTL3-mediated m6A methylation increases the maturation of microRNA 335 (MIR335), which promotes SG formation and reduces the apoptosis level of injury neurons and cells, and provides a potential therapeutic strategy for acute ischemic stroke. | |||
microRNA 370 (MIR370)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Embryonic stem cells | Mus musculus |
|
Treatment: METTL3 knockout mESCs
Control: Wild type mESCs
|
GSE156481 | |
| Regulation |
 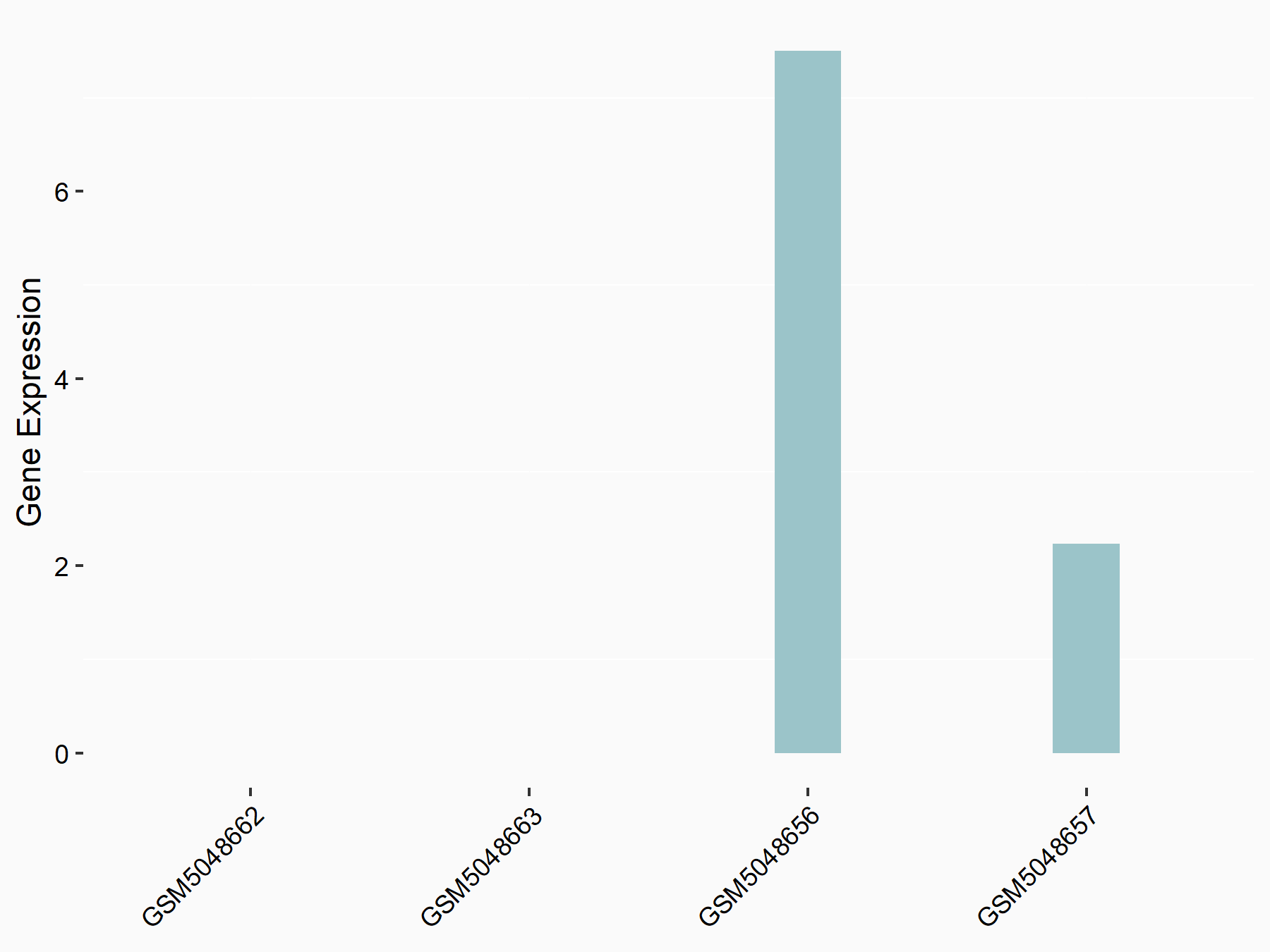 |
logFC: -4.78E+00 p-value: 3.06E-02 |
| More Results | Click to View More RNA-seq Results | |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [56] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Pathway Response | Transcriptional misregulation in cancer | hsa05202 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell autophagy | ||||
In-vitro Model |
Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| In-vivo Model | To create the xenograft neoplasm system, 40 male BALB/c nude mice aged 5 weeks were randomly separated into sh-NC, sh-circHPS5, sh-circHPS5+CTRL, and sh-circHPS5+SAH groups (n = 5 for each group). HCC cells were subcutaneously injected into the axilla of the nude mice. | |||
| Response Summary | In hepatocellular carcinoma, METTL3 could direct the formation of circHPS5, and specific m6A controlled the accumulation of circHPS5. YTHDC1 facilitated the cytoplasmic output of circHPS5 under m6A modification. CircHPS5 can act as a microRNA 370 (MIR370) sponge to regulate the expression of HMGA2 and further accelerate hepatocellular carcinoma cell tumorigenesis. | |||
miR-182
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
 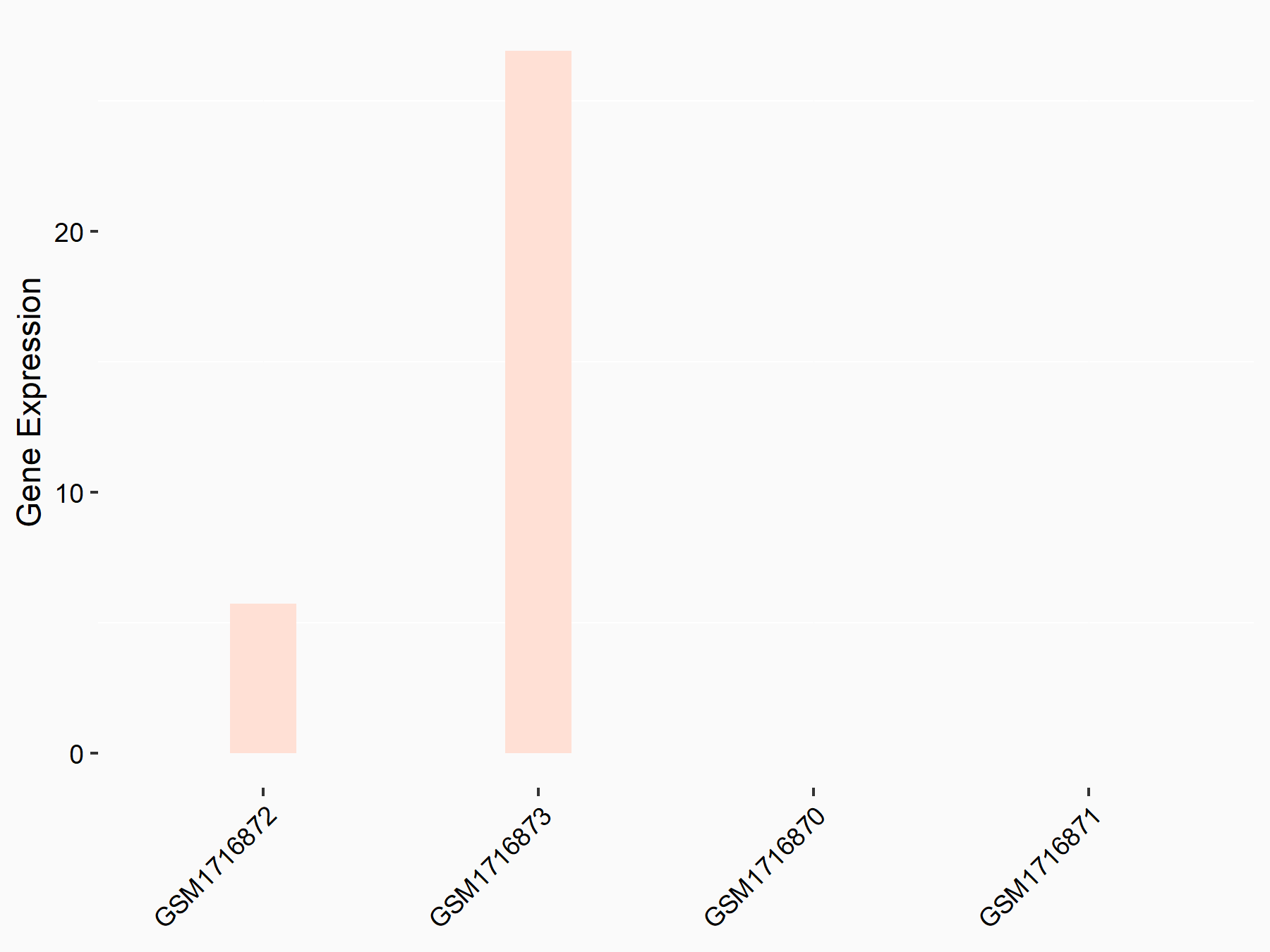 |
logFC: 3.78E+00 p-value: 3.86E-02 |
| More Results | Click to View More RNA-seq Results | |
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [197] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
WPMY-1 | Normal | Homo sapiens | CVCL_3814 |
| VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| Response Summary | METTL3 promoted cell proliferation, migration, invasion and tumorigenesis in PCa. METTL3 upregulating the level of m6A, and interacted with DGCR8 to recognize the m6A modification of pre-miR-182 to regulate its splicing and maturation and promote the high expression of miRNA. | |||
miR-503
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
|
Treatment: METTL3 knockdown MDA-MB-231 cells
Control: MDA-MB-231 cells
|
GSE70061 | |
| Regulation |
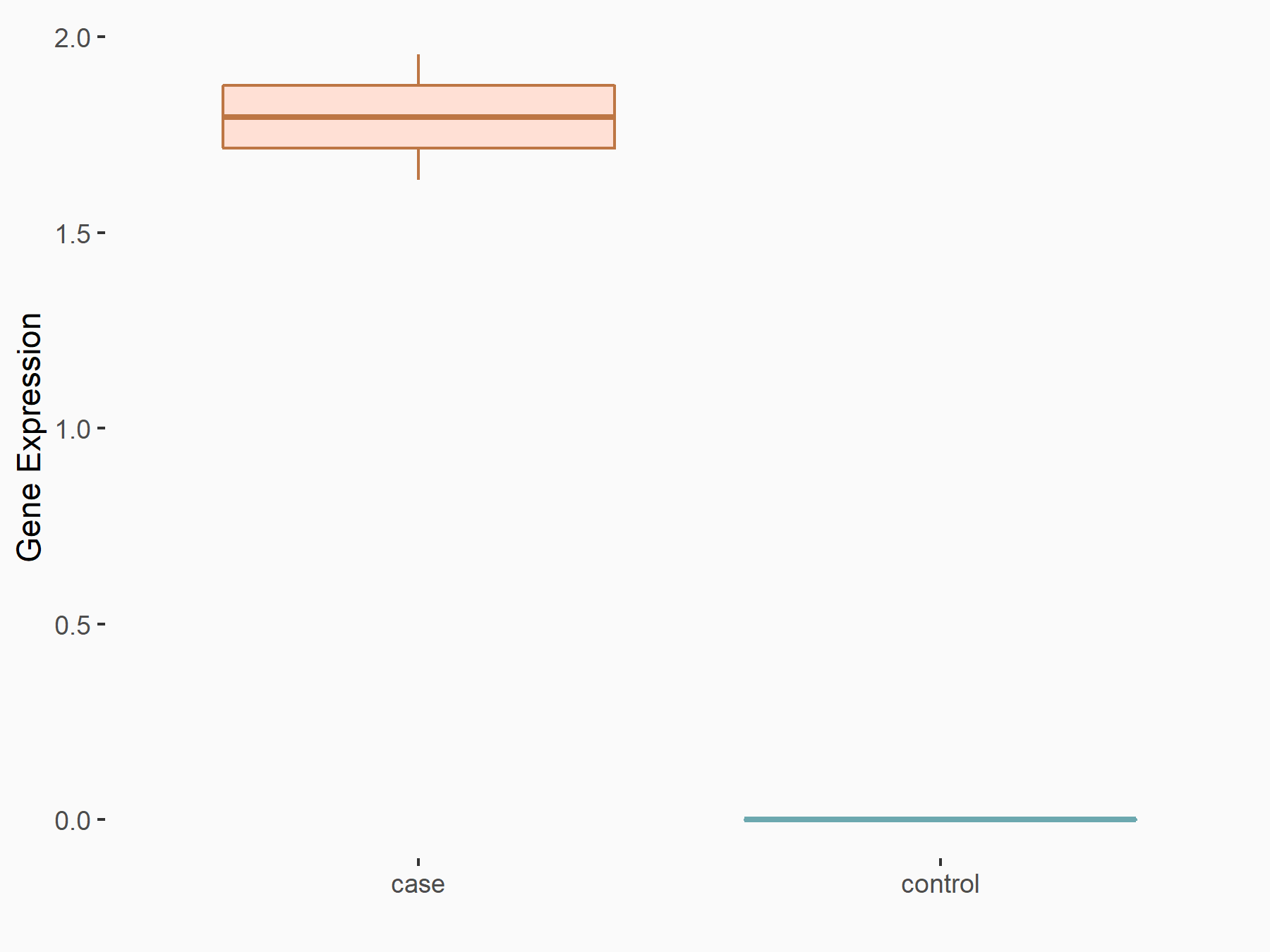 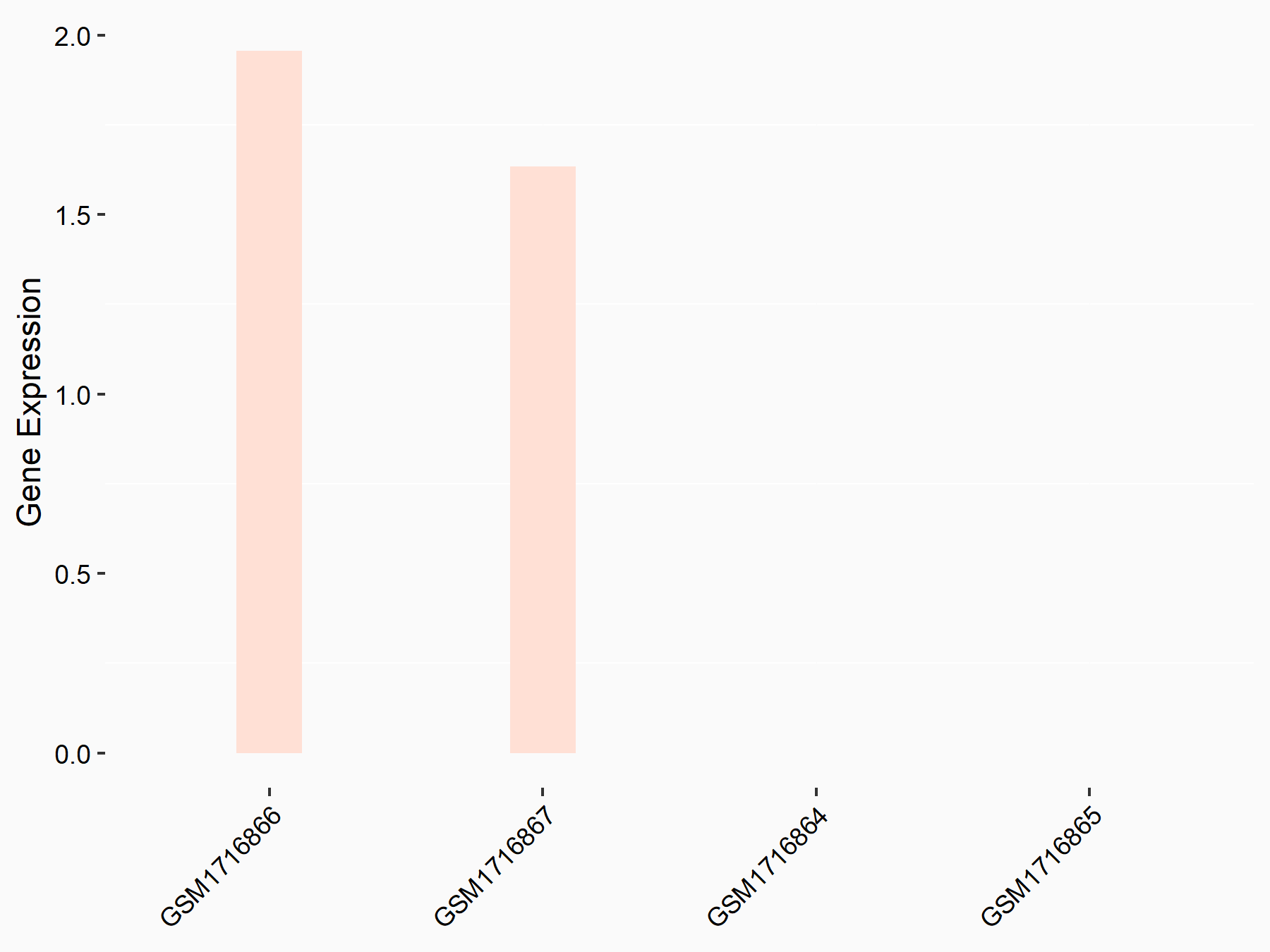 |
logFC: 1.48E+00 p-value: 8.87E-04 |
| More Results | Click to View More RNA-seq Results | |
Injury of heart [ICD-11: NB31]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [246] | |||
| Responsed Disease | Myocardial injury [ICD-11: NB31.Z] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Mitochondrial metabolic dysfunction | |||
| In-vivo Model | To generate an AMI mouse model, mice were anesthetised by intraperitoneal injection of sterile pentobarbital sodium at 50 mg/kg body weight. | |||
| Response Summary | Hypoxia induced rapid H3K4 methylation of the promoter of the methyltransferase-like 3 gene (METTL3) and resulted in its overexpression. METTL3 overexpression evokes N6-methyladenosine (m6A)-dependent miR-503 biogenesis in endothelial cells. In summary, this study highlights a novel endogenous mechanism wherein EVs aggravate myocardial injury during the onset of AMI via endothelial cell-secreted miR-503 shuttling. | |||
hsa-miR-26b
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Mouse testis | Mus musculus |
|
Treatment: Mettl3 knockout mouse testis
Control: Mouse testis
|
GSE99771 | |
| Regulation |
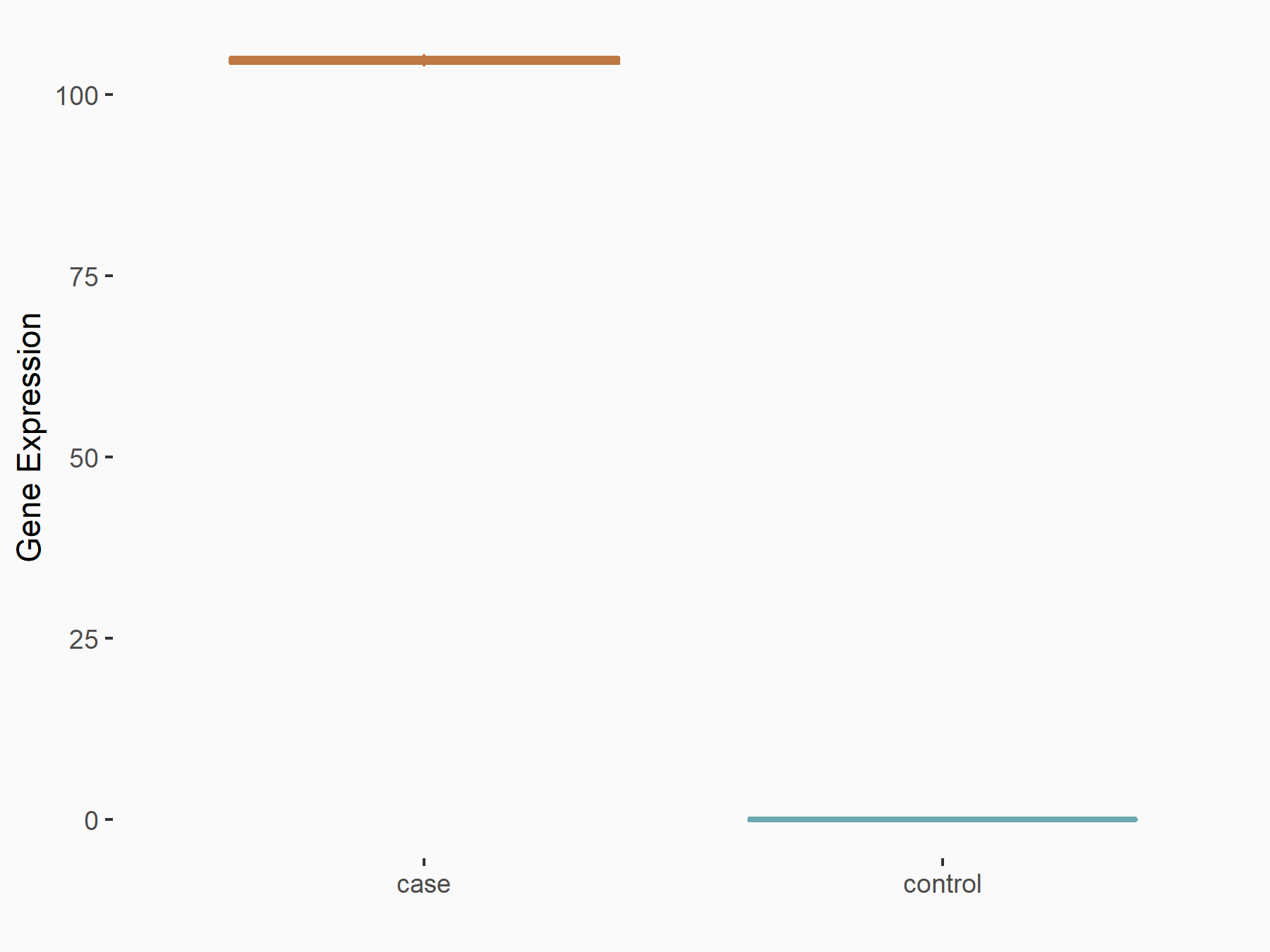 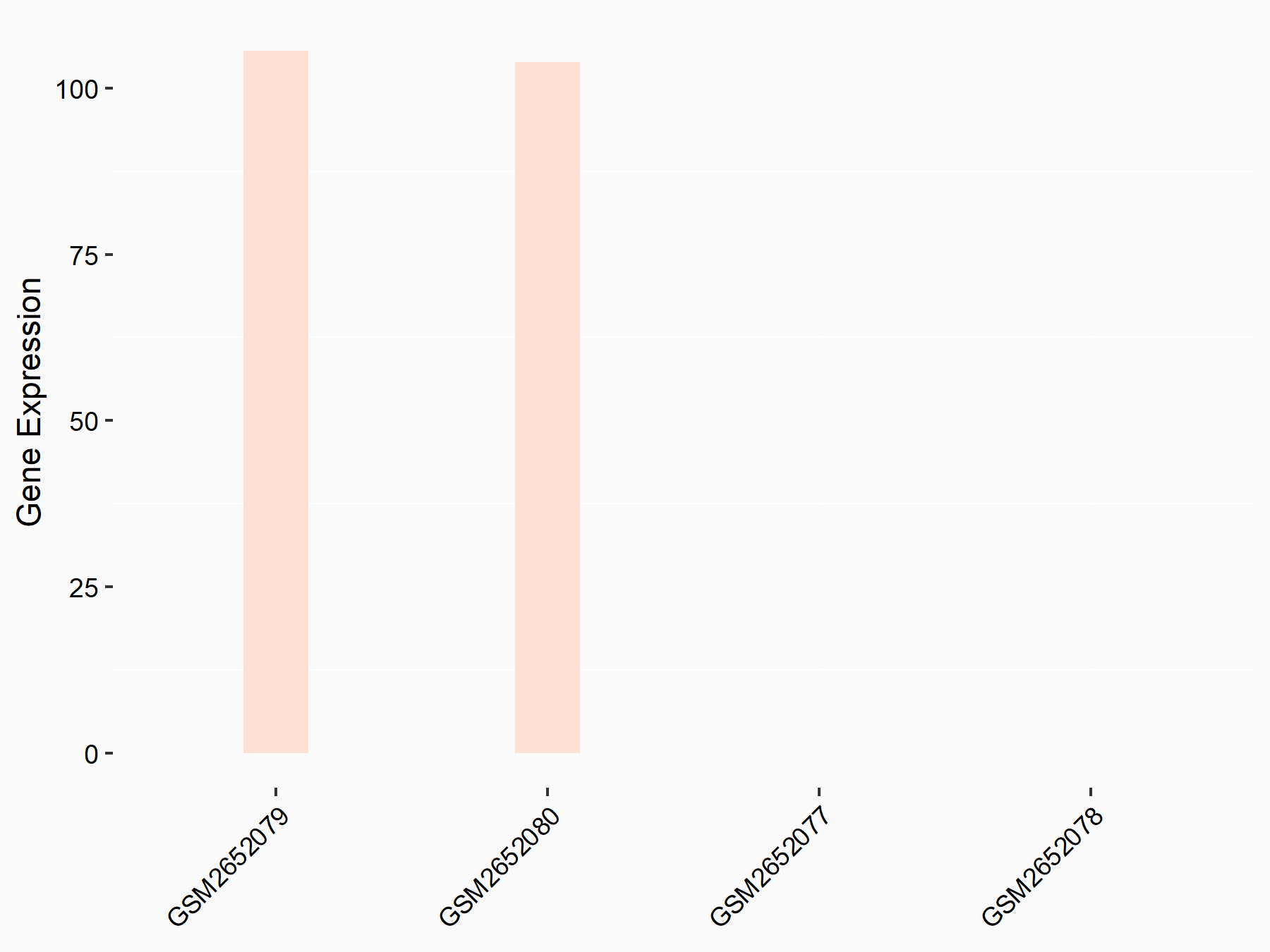 |
logFC: 6.72E+00 p-value: 1.75E-06 |
| More Results | Click to View More RNA-seq Results | |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [57] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| In-vivo Model | Eighteen BALB/C female nude mice aged 4-5 weeks and weighing 15-18 g were randomly assigned into three groups of six mice. The MCF-7 cell lines stably transfected with sh-NC + oe-NC, sh-METTL3 + oe-NC and sh-METTL3 + oe-HMGA2 were selected for subcutaneous establishment of the BC cell line MCF-7 as xenografts in the nude mice. For this purpose, MCF-7 cell lines in the logarithmic growth stage were prepared into a suspension with a concentration of about 1 × 107 cells/ml. The prepared cell suspension was injected into the left armpit of the mice, and the subsequent tumor growth was recorded. | |||
| Response Summary | Silencing METTL3 down-regulate MALAT1 and HMGA2 by sponging hsa-miR-26b, and finally inhibit EMT, migration and invasion in BC, providing a theoretical basis for clinical treatment of BC. | |||
Cyclin-dependent kinase inhibitor 2A (CDKN2A)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.42E+00 | GSE60213 |
Mature T-cell lymphoma [ICD-11: 2A90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [247] | |||
| Responsed Disease | Mature T-cell lymphoma [ICD-11: 2A90] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Response Summary | The decline in METTL3 levels was responsible for CTCL cell proliferation and migration,Cyclin-dependent kinase inhibitor 2A (CDKN2A) was a key regulator during this process in vitro and in vivo, and insufficient methylation modification blocked the interaction between CDKN2A and m6A reader IGF2BP2, resulting in mRNA degradation. | |||
Cytochrome P450 2C8 (CYP2C8)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 4.67E+00 | GSE60213 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [248] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Drug metabolism - cytochrome P450 | hsa00982 | ||
| Cell Process | Drug-metabolizing | |||
In-vitro Model |
HepaRG | Hepatitis C infection | Homo sapiens | CVCL_9720 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Response Summary | In the Hepatocellular carcinoma cells YTHDC2 promotes CYP2C8 mRNA degradation via recognizing the m6A in CYP2C8 mRNA, which is installed by METTL3/14 and removed by FTO. | |||
Differentiation antagonizing non-protein coding RNA (DANCR)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 9.97E+00 | GSE60213 |
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [249] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 |
| SJSA-1 | Osteosarcoma | Homo sapiens | CVCL_1697 | |
| SaOS-2 | Osteosarcoma | Homo sapiens | CVCL_0548 | |
| MG-63 | Osteosarcoma | Homo sapiens | CVCL_0426 | |
| HOS | Osteosarcoma | Homo sapiens | CVCL_0312 | |
| hFOB 1.19 | Normal | Homo sapiens | CVCL_3708 | |
| Response Summary | METTL3 contributes to OS progression by increasing Putative uncharacterized protein DANCR (DANCR) mRNA stability via m6A modification, meaning that METTL3 is a promising therapeutic target for OS treatment. | |||
Keratin, type II cytoskeletal 7 (KRT7)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.21E+00 | GSE60213 |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [137] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Lung Metastasis | |||
In-vitro Model |
MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| In-vivo Model | First, subcutaneous transplanted model was used to evaluate the growth of BT-549LMF3 and BT-549 cells. Cells (5 × 106 per mouse, n = 5 for each group) were diluted in 200 ul PBS + 200 ul Matrigel (BD Biosciences) and subcutaneously injected into immunodeficient female mice. Second, subcutaneous transplanted model was used to evaluate the metastasis potential of BT-549LMF3 and BT-549 cells. Cells (5 × 106 per mouse, n = 5 for each group) were diluted in 200 ul PBS + 200 ul Matrigel (BD Biosciences) and subcutaneously injected into immunodeficient female mice. Third, the in vivo lung metastasis model was established by injecting with BT-549, BT-549LMF3, FTO stable BT-549LMF3, sh-METTL3 BT-549LMF3, and sh-KRT7 BT-549LMF3 stable cells (1 × 106 per mouse, n = 5 for each group). | |||
| Response Summary | Specifically, increased METTL3 methylated KRT7-AS at A877 to increase the stability of a KRT7-AS/Keratin, type II cytoskeletal 7 (KRT7) mRNA duplex via IGF2BP1/HuR complexes. m6A promotes breast cancer lung metastasis by increasing the stability of a KRT7-AS/KRT7 mRNA duplex and translation of KRT7. | |||
L-glutamine amidohydrolase (GLS2)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 5.89E+00 | GSE60213 |
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [250] | |||
| Responsed Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.1] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell migration | |||
| Cell invasion | ||||
In-vitro Model |
HEEC cell line (Normal esophageal epithelial cell line) | |||
| TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 | |
| TE-13 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_4463 | |
| Eca-109 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_6898 | |
| EC/CUHK1 | Esophageal carcinoma | Homo sapiens | CVCL_RY08 | |
| Response Summary | L-glutamine amidohydrolase (GLS2) as a downstream target of METTL3. These findings uncover METTL3/GLS2 signaling as a potential therapeutic target in antimetastatic strategies against esophageal Squamous Cell Carcinoma(ESCC). | |||
Monocyte chemotactic and activating factor (CCL2)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 4.91E+00 | GSE60213 |
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [251] | |||
| Responsed Disease | Nonalcoholic steatohepatitis [ICD-11: DB92.1] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| In-vivo Model | Mettl3flox/flox and Mettl3-HKO mice were fasted overnight and then injected intraperitoneally with 20 uM BODIPY FL C16 in 200 ul saline for 20 min. | |||
| Response Summary | Mechanistically, METTL3 directly binds to the promoters of the Cd36 and Monocyte chemotactic and activating factor (CCL2) genes and recruits HDAC1/2 to induce deacetylation of H3K9 and H3K27 in their promoters, thus suppressing Cd36 and Ccl2 transcription. METTL3 negatively regulates hepatic Cd36 and Ccl2 gene transcription via a histone modification pathway for protection against Nonalcoholic steatohepatitis(NASH) progression. | |||
Multidrug resistance-associated protein 1 (MRP1/ABCC1)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.47E+00 | GSE60213 |
Gastrointestinal stromal tumor [ICD-11: 2B5B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [252] | |||
| Responsed Disease | Gastrointestinal stromal tumour [ICD-11: 2B5B] | |||
| Responsed Drug | Imatinib | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| In-vivo Model | For tumor growth assay, 4 × 106 logarithmically growing GIST cells were transfected with T1S-vector, T1S-METTL3, 882S-vector, or 882S-METTL3 constructs, and subcutaneously injected in 100 ul of PBS into the flank of female nude mice(4-week-old). Mice were then randomly divided into 8 groups (n = 5 in each group): (1) injected with T1S-vector-harboring cells, and treated with imatinib (600 mg/l in drinking water); (2) injected with T1S-vector-harboring cells, and treated with imatinib (600 mg/l in drinking water) and MRP1 inhibitor (100 mg/l in drinking water); (3) injected with T1S-METTL3-harboring cells, and treated with imatinib (600 mg/l in drinking water); (4) injected with T1S-METTL3-harboring cells, and treated with imatinib (600 mg/l in drinking water) and MRP1 inhibitor (100 mg/l in drinking water); (5) injected with 882S-vector-harboring cells and treated with imatinib (600 mg/l in drinking water); (6) injected with 882S-vector-harboring cells, and treated with imatinib (600 mg/l in drinking water) and MRP1 inhibitor (100 mg/l in drinking water); (7) injected with 882S-METTL3-harboring cells, and treated with imatinib (600 mg/l in drinking water); and (8) injected with 882S-METTL3-harboring cells, and treated with imatinib (600 mg/l in drinking water) and MRP1 inhibitor (100 mg/l in drinking water). | |||
| Response Summary | METTL3, and the YTHDF1/eEF-1 complex mediate the translation of Multidrug resistance-associated protein 1 (MRP1/ABCC1) mRNA in an m6A-dependent manner to regulate the intracellular concentration of imatinib and drug resistance of gastrointestinal stromal tumor (GIST). | |||
Protein kinase C epsilon (PRKCE)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 3.25E+00 | GSE60213 |
Ischemic heart disease [ICD-11: BA40-BA6Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [253] | |||
| Responsed Disease | Ischemic heart disease [ICD-11: BA40-BA6Z] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Pyroptosis | |||
In-vitro Model |
H9c2(2-1) | Normal | Rattus norvegicus | CVCL_0286 |
| In-vivo Model | The thoracic cavity of rats was exposed, and the left anterior descending coronary artery was ligated with a 6-0 silk thread. Successfully surgical MI could be observed, with myocardium color fading and pulse weakening. After 30 min of ischemia, the blood flow was restored by releasing the slipknot, and then 120-min perfusion was performed. Afterward, the thoracic cavity of rats was sutured. The rats were assigned into 4 groups, with 12 rats in each group. Lentivirus packaged short hairpin (sh)-negative control (NC) and sh-METTL3 (GenePharma, Shanghai, China) were injected into the rats via tail vein 24 h before operation. The titer of lentivirus was 1 × 109 TU/mL, and the injection rate was 0.2 ul/min for 10 min. Blood samples were collected 24 h after reperfusion. | |||
| Response Summary | METTL3 promoted DGCR8 binding to pri-miR-143-3p through m6A modification, thus enhancing miR-143-3p expression to inhibit Protein kinase C epsilon (PRKCE) transcription and further aggravating cardiomyocyte pyroptosis and MI/R injury. | |||
Sphingosine kinase 2 (SPHK2)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.82E+00 | GSE60213 |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [190] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Response Summary | METTL3 promotes translation of Sphingosine kinase 2 (SPHK2) mRNA via an m6A-YTHDF1-dependent manner. Functionally, SPHK2 facilitates GC cell proliferation, migration and invasion by inhibiting KLF2 expression. | |||
Suppressor of cytokine signaling 4 (SOCS4)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.78E+00 | GSE60213 |
Thyrotoxicosis [ICD-11: 5A02]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [225] | |||
| Responsed Disease | Graves disease [ICD-11: 5A02.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
PBMCs (Human peripheral blood mononuclear cells (PBMCs) are isolated from peripheral blood and identified as any blood cell with a round nucleus) | |||
| Response Summary | METTL3 knock-down experiment revealed that expressions of SOCS family members SOCS1, SOCS2, Suppressor of cytokine signaling 4 (SOCS4), SOCS5, and SOCS6 were increased after METTL3 knock-down. It indicated that METTL3 is involved in the development of Graves' disease (GD) by inducing mRNA m6A methylation modification of SOCS family members. | |||
Suppressor of cytokine signaling 6 (SOCS6)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.24E+00 | GSE60213 |
Thyrotoxicosis [ICD-11: 5A02]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [225] | |||
| Responsed Disease | Graves disease [ICD-11: 5A02.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
PBMCs (Human peripheral blood mononuclear cells (PBMCs) are isolated from peripheral blood and identified as any blood cell with a round nucleus) | |||
| Response Summary | METTL3 knock-down experiment revealed that expressions of SOCS family members SOCS1, SOCS2, SOCS4, SOCS5, and Suppressor of cytokine signaling 6 (SOCS6) were increased after METTL3 knock-down. It indicated that METTL3 is involved in the development of Graves' disease (GD) by inducing mRNA m6A methylation modification of SOCS family members. | |||
Transforming growth factor beta-1 proprotein (TGFB1)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.56E+00 | GSE60213 |
Solid tumour/cancer [ICD-11: 2A00-2F9Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [235] | |||
| Responsed Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Adherens junction | hsa04520 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| Response Summary | The expression of Transforming growth factor beta-1 proprotein (TGFB1) was up-regulated, while self-stimulated expression of TGFbeta1 was suppressed in METTL3Mut/- cells. m6A performed multi-functional roles in TGFbeta1 expression and EMT modulation, suggesting the critical roles of m6A in cancer progression regulation. Snail, which was down-regulated in Mettl3Mut/- cells, was a key factor responding to TGF-Beta-1-induced EMT. | |||
Vasopressin V2 receptor (Avpr2/V2R)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 4.52E+00 | GSE60213 |
Polycystic kidney disease [ICD-11: GB81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [82] | |||
| Responsed Disease | Polycystic kidney disease [ICD-11: GB81] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
mIMCD-3 | Normal | Mus musculus | CVCL_0429 |
| In-vivo Model | The clone, with one wild-type Mettl3 allele and one L1L2_Bact_P cassette inserted allele, was injected into C57BL/6 blastocysts. Mettl3-targeted mouse line was established from a germline-transmitting chimera. The chimeric mouse was crossed to C57BL/6 Flp mice to excise the neomycin resistance system. | |||
| Response Summary | Mettl3 activates the cyst-promoting c-Myc and cAMP pathways through enhanced c-Myc and Vasopressin V2 receptor (Avpr2/V2R) mRNA m6A modification and translation. Thus, Mettl3 promotes Autosomal dominant polycystic kidney disease and links methionine utilization to epitranscriptomic activation of proliferation and cyst growth. | |||
LINC00035 (ABHD11-AS1)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 5.94E+00 | GSE60213 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [254] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Central carbon metabolism in cancer | hsa05230 | ||
In-vitro Model |
NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | H1299 cells were transfected with sh-ABHD11-AS1 and harvested from six-well plates, then resuspended at a density 1 × 107 cells/ml. Mice were subsequently injected with 100 uL suspension at the right flank. After injection, tumor weight and length were examined every 3 days. | |||
| Response Summary | LINC00035 (ABHD11-AS1) was upregulated in NSCLC tissue specimens and cells and the ectopic overexpression was closely correlated with unfavorable prognosis of NSCLC patients.METTL3 installed the m6 A modification and enhanced ABHD11-AS1 transcript stability to increase its expression. | |||
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [364] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
In-vitro Model |
HCoEpiC (Healthy colon epithelial HCoEpiC cells) | |||
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| HT-29 | Colon adenocarcinoma | Homo sapiens | CVCL_0320 | |
| In-vivo Model | Harvested cells were resuspended in PBS and each side of mouse was injected about 1 × 106 cells. Tumor volume was estimated every four days and calculated as 0.5 × length × width2. | |||
Acute ischemic stroke [ICD-11: 8B11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [365] | |||
| Responsed Disease | Acute ischemic stroke [ICD-11: 8B11] | |||
| Target Regulation | Up regulation | |||
LOC10013.776 (AGAP2-AS1)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.67E+00 | GSE60213 |
Psoriasis [ICD-11: EA90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [255] | |||
| Responsed Disease | Psoriasis [ICD-11: EA90] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| PI3K-Akt signaling pathway | hsa04151), | |||
| Cell Process | Cell proliferation | |||
In-vitro Model |
HaCaT | Normal | Homo sapiens | CVCL_0038 |
| NHEK (Normal human epithelial keratinocytes) | ||||
| Response Summary | METTL3 resulted in the upregulation of AGAP2-AS1 in psoriasis. LOC10013.776 (AGAP2-AS1) is upregulated in the skin tissue of psoriasis patients and m6A methylation was involved in its upregulation. | |||
Long intergenic non-protein coding RNA 1273 (LINC01273)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.26E+00 | GSE60213 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [256] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Down regulation | |||
In-vitro Model |
SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Response Summary | Long intergenic non-protein coding RNA 1273 (LINC01273) was modified with m6A, METTL3 increased LINC01273 m6A modification, followed by LINC01273 decay in the presence of YTHDF2, a m6A 'reader'. And LINC01273 plays a key role in sorafenib resistant HCC cells. | |||
Long intergenic non-protein coding RNA 460 (LINC00460)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.69E+00 | GSE60213 |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [176] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| Cell Process | mRNA stability | |||
| Epithelial-mesenchymal transition | ||||
In-vitro Model |
DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| FHC | Normal | Homo sapiens | CVCL_3688 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | Groups of HCT116-Luc-shCtrl, HCT116-Luc-shLINC00460, and HCT116-Luc-shLINC00460 + HMGA1 cells (5 × 106) were injected subcutaneously into the flanks of mice correspondingly. | |||
| Response Summary | Long intergenic non-protein coding RNA 460 (LINC00460) is a novel oncogene of colorectal cancer through interacting with IGF2BP2 and DHX9 and bind to the m6A modified HMGA1 mRNA to enhance the HMGA1 mRNA stability. The N6-methyladenosine (m6A) modification of HMGA1 mRNA by METTL3 enhanced HMGA1 expression in CRC. | |||
Maternally expressed 3 (MEG3)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.64E+00 | GSE60213 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [257] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| THLE-3 | Normal | Homo sapiens | CVCL_3804 | |
| Response Summary | Maternally expressed 3 (MEG3) regulates the expression of BTG2 through miR-544b, thus affecting the malignant behavior of hepatocellular carcinoma. METTL3 regulates the m6A modification of MEG3 and its expression. | |||
NIFK antisense RNA 1 (NIFK-AS1)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.43E+00 | GSE60213 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [258] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| THLE-3 | Normal | Homo sapiens | CVCL_3804 | |
| In-vivo Model | For the PDX model, fresh patient HCC tissues were cut into fragments with a volume of 3 × 3 mm3 and then implanted subcutaneously into the flanks of nude mice. The mice were given sorafenib (30 mg/kg) or vehicle orally twice a week for 24 days. This procedure was approved by the Ethics Committee of Jinling Hospital. | |||
| Response Summary | Identified the lncRNA NIFK antisense RNA 1 (NIFK-AS1) as being highly expressed in hepatocellular carcinoma tissues and cells, promotes disease progression and sorafenib resistance, and showed this up-regulation resulted from METTL3-dependent m6A methylation. | |||
Pvt1 oncogene (PVT1)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.16E+01 | GSE60213 |
Kidney failure [ICD-11: GB6Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [146] | |||
| Responsed Disease | Kidney failure [ICD-11: GB6Z] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
NIT-1 | Insulin tumor | Mus musculus | CVCL_3561 |
| Response Summary | m6A modification is co-regulated by METTL3 and FTO in cadmium-treated cells. LncRNA-MALAT1, Pvt1 oncogene (PVT1) and m6A modification could be key nodes for cadmium-induced oxidative damage, and highlight their importance as promising preventive and therapeutic targets in cadmium toxicity. | |||
Small nucleolar RNA host gene 3 (SNHG3)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.17E+00 | GSE60213 |
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [259] | |||
| Responsed Disease | Esophageal cancer [ICD-11: 2B70] | |||
| Responsed Drug | Pt | Investigative | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cellular Processes | |||
| Cell growth and death | ||||
| Cell apoptosis | ||||
In-vitro Model |
Eca-9706 (Esophageal carcinoma cell line) | |||
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| In-vivo Model | Used 1 × 106 SNHG3 knocked down KY-SE150 cells and NC lentivirus to inject into the right flank of mice to generate xenografts. | |||
| Response Summary | Platinum can increase the overall m6A level of esophageal cancer. Small nucleolar RNA host gene 3 (SNHG3)/miR-186-5p, induced by platinum, was involved in regulating m6A level by targeting METTL3. miR-186-5p binds to the 3'UTR of METTL3 to inhibit its expression. Our manuscript has provided clues that regulating m6A level was a novel way to enhance the platinum efficacy. | |||
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [375] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
Melanoma [ICD-11: 2C30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [376] | |||
| Responsed Disease | Melanoma [ICD-11: 2C30] | |||
In-vitro Model |
A-375 | Amelanotic melanoma | Homo sapiens | CVCL_0132 |
| SK-MEL-28 | Cutaneous melanoma | Homo sapiens | CVCL_0526 | |
| In-vivo Model | A375 cells (1 × 107) transfected with sh-METTL3 or shNC lentiviruses were subcutaneously injected into the right flank of the mice. After 33 days, the mice were sacrificed and the xenografted tumors were collected for immunohistochemistry analysis. Animal experiments were approved by the Ethics Committee of Shanghai East Hospital (K-KYSB-2020-0). | |||
SVIL antisense RNA 1 (SVIL-AS1)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 9.54E+00 | GSE60213 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [260] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | A549 cells (1 × 106 cells) were intraperitoneally injected into the abdomen of nude mice. Tumor length and width were measured and recorded every 4 days after inoculation. Tumor volume was calculated as 1/2 × length × width2. The tumor weight was weighed and recorded 20 days after inoculation. | |||
| Response Summary | The reduction in cell proliferation induced by SVIL antisense RNA 1 (SVIL-AS1) overexpression could be rescued by E2F1 overexpression or METTL3 knockdown. In conclusion, METTL3-induced SVIL-AS1 in LUAD, which connects m6A and lncRNA in lung cancer carcinogenesis. | |||
X inactive specific transcript (XIST)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.31E+00 | GSE60213 |
Ossification of spinal ligaments [ICD-11: FA83]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [261] | |||
| Responsed Disease | Ossification of spinal ligaments [ICD-11: FA83] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Ubiquitin mediated proteolysis | hsa04120 | ||
| Cell Process | Ubiquitination degradation | |||
In-vitro Model |
Human primary ligament fibroblasts (Ligaments were dissected from a non-ossified site) | |||
| Response Summary | METTL3 regulates ossification of the posterior longitudinal ligament via the lncRNA X inactive specific transcript (XIST)/miR-302a-3p/USP8 axis. | |||
ZNFX1 antisense RNA 1 (ZFAS1)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 3.61E+00 | GSE60213 |
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [262] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell autophagy | ||||
In-vitro Model |
SUNE1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6946 |
| S26 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_B0UB | |
| S18 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_B0U9 | |
| NP69 (A human immortalized nasopharyngeal epithelial) | ||||
| N2Tert (The human immortalized nasopharyngeal epithelial cell lines) | ||||
| HONE-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_8706 | |
| HNE-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_0308 | |
| NPC/HK1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_7084 | |
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| CNE-1 | Normal | Homo sapiens | CVCL_6888 | |
| C666-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_7949 | |
| 6-10B | Nasopharyngeal carcinoma | Homo sapiens | CVCL_C529 | |
| In-vivo Model | Female BALB/c nude mice (ages 4-5 weeks, 18-20 g) were purchased from the Charles River Laboratories. For the tumor growth model, 1 × 106 HONE-1 sh-NC or sh-ZFAS1 cells were injected into the axilla of the mice, and the tumor size was measured every 3 days. On day 30, the mice were killed, and the tumors were dissected and weighed. | |||
| Response Summary | Knockout of METTL3 can reduce the total expression of ZNFX1 antisense RNA 1 (ZFAS1). ZFAS1 is used as an oncogenic lncRNA, which can promote NPCcell proliferation, migration and tumor growth. | |||
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [263] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| In-vivo Model | Nude mice were subjected to a subcutaneous injection of 5× 106 control and ZFAS1 silencing CaSki cells suspended in 0.2 mL DMEM medium. | |||
| Response Summary | ZNFX1 antisense RNA 1 (ZFAS1) sequestered miR-647, and this RNA-RNA interaction is regulated by METLL3-mediated m6A modification in cervical cancer. | |||
Apoptotic chromatin condensation inducer in the nucleus (ACIN1)
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [269] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| RNA degradation | hsa03018 | |||
| Cell Process | RNA stability | |||
In-vitro Model |
SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| End1/E6E7 | Normal | Homo sapiens | CVCL_3684 | |
| In-vivo Model | 2 × 106 stably transfected HeLa cells were subcutaneously inoculated into the left flank of mice. | |||
| Response Summary | METTL3 interacts with IGF2BP3 to promote the mRNA stability of Apoptotic chromatin condensation inducer in the nucleus (ACIN1), the overexpression of which induces the aggressiveness of CC cells. | |||
Axin-1 (AXIN1)
Disorders of refraction [ICD-11: 9D00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [272] | |||
| Responsed Disease | Disorders of refraction [ICD-11: 9D00.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
RF/6A
|
N.A. | Macaca mulatta | CVCL_4552 |
Bax inhibitor 1 (TMBIM6)
Pre-eclampsia [ICD-11: JA24]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [273] | |||
| Responsed Disease | Pre-eclampsia [ICD-11: JA24] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HTR-8/SVneo | Normal | Homo sapiens | CVCL_7162 |
BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 (BNIP3)
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [271] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
Bone morphogenetic protein 2 (BMP2)
Ossification of spinal ligaments [ICD-11: FA83]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [274] | |||
| Responsed Disease | Ossification of spinal ligaments [ICD-11: FA83] | |||
| Target Regulation | Up regulation | |||
Low bone mass disorder [ICD-11: FB83]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [275] | |||
| Responsed Disease | Osteoporosis [ICD-11: FB83.1] | |||
Class E basic helix-loop-helix protein 41 (BHLHE41)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [280] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Cytokine-cytokine receptor interaction | hsa04060 | ||
| Chemokine signaling pathway | hsa04062 | |||
| Cell Process | Cell immunity | |||
| Cell growth | ||||
| Cell migration | ||||
In-vitro Model |
CT26 | Mouse colon adenocarcinoma | Mus musculus | CVCL_7254 |
| MC-38 | Mouse colon adenocarcinoma | Mus musculus | CVCL_B288 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | METTL3 knockout or control CT26 and MC38 cells were injected subcutaneously into the dorsal flank of each 4- to 6-week-old male immunocompetent BALB/c and C57BL/6 mice, respectively. Anti-Gr-1 (BE0075; Bio-X-Cell, Lebanon, NH) or immunoglobulin (Ig)G isotype control (BE0090, Bio-XCell) was given every other day via intraperitoneal injection (150 ug/mouse). SB-265610 (Tocris, Bristol, UK) or phosphate-buffered saline was administrated through intraperitoneal injections at a dosage of 2 mg/kg per day. Tumor sizes were measured every other day. To establish an orthotopic mouse model of CRC, 5- to 6-week-old male C57BL/6 mice were treated with 1.7% dextran sodium sulfate in drinking water for 5 days, and then allowed to recover for 3 days. After 24 hours of fasting, METTL3 knockout or control MC38 cells suspended in 50 ul of 1 mg/mL Matrigel-phosphate-buffered saline (Corning, Corning, NY) were instilled into the colon lumen of anesthetized mice, coated sparingly with Vaseline. | |||
| Response Summary | METTL3 promoted Class E basic helix-loop-helix protein 41 (BHLHE41) expression in m6A-dependent manner, which subsequently induced CXCL1 transcription to enhance MDSC migration in vitro. METTL3 as a potential therapeutic target for CRC immunotherapy whose inhibition reverses immune suppression through m6A-BHLHE41-CXCL1 axis. | |||
Cytokine-responsive protein CR6 (GADD45-Gamma)
Endocrine glands benign cancer [ICD-11: 2F37]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [285] | |||
| Responsed Disease | Endocrine glands benign cancer [ICD-11: 2F37] | |||
| Target Regulation | Up regulation | |||
| Response Summary | METTL3 promotes tumor growth and hormone secretion by increasing expression of GNAS and Cytokine-responsive protein CR6 (GADD45-Gamma) in a m6A-dependent manner. Thus, METTL3 and the methylated RNAs constitute suitable targets for clinical treatment of Pituitary growth hormone-secreting-pituitary adenomas(GH-PAs). | |||
DNA-binding protein inhibitor ID-2 (ID2)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [288] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
In-vitro Model |
PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 |
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| HPDE6c7 | Normal | Homo sapiens | CVCL_0P38 | |
| Panc02 | Mouse pancreatic ductal adenocarcinoma | Mus musculus | CVCL_D627 | |
| In-vivo Model | For the subcutaneous tumor model, cells from the treatment group and the control group (1 × 107) were subcutaneously injected into the right groin of BALB/c nude mice (4-6 weeks old, 18-20 g, 4 mice each). Both the treatment group and control group had 5 BALB/c nude mice each. After 21 days, all mice were sacrificed and tumor volume and weight were measured. For the pancreatic cancer in situ tumor model, mice were anesthetized with Avertin at a dose of 0.2 ml/10 g body weight, and 5 × 105 panc2 treatment group cells and control group cells were injected into the pancreas of mice respectively, and the abdomen was closed for disinfection. Both the treatment group and control group had 5 C57 mice each. After 21 days, the mice were sacrificed, the in-situ tumor was removed, and the volume and weight were measured for subsequent experiments. | |||
E3 ubiquitin-protein ligase TRIM11 (TRIM11)
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [14] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | ABC transporters | hsa02010 | ||
| Wnt signaling pathway | hsa04310 | |||
| Ubiquitin mediated proteolysis | hsa04120 | |||
| Cell Process | Ubiquitination degradation | |||
In-vitro Model |
CNE-1 | Normal | Homo sapiens | CVCL_6888 |
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| In-vivo Model | A total of 2 × 106 cells was mixed with 0.2 ml PBS (pH 7.4) and 30% (v/v) Matrigel matrix (BD Biosciences). | |||
| Response Summary | TRIM11 regulates nasopharyngeal carcinoma drug resistance by positively modulating the Daple/beta-catenin/E3 ubiquitin-protein ligase TRIM11 (TRIM11) signaling pathway. TRIM11 enhanced the multidrug resistance in NPC by inhibiting apoptosis in vitro and promoting cisplatin (DDP) resistance in vivo. METTL3-mediated m6A modification caused the upregulation of TRIM11 via IGF2BP2 in NPC drug-resistant cells. | |||
Eukaryotic translation initiation factor 2 subunit 1 (EIF2S1/eIF2-Alpha)
Pulmonary hypertension [ICD-11: BB01]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [292] | |||
| Responsed Disease | Pulmonary arterial hypertension [ICD-11: BB01.0] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | Adult male Sprague-Dawley (150-200 g in body weight) rats were randomly divided into control or MCT/vehicle rats. Experimental rats were administered an intraperitoneal injection of monocrotaline (60 mg/kg, Sigma, 315-22-0), and their littermates were injected with saline. For GSK2606414 treatment, rats that underwent monocrotaline treatment were treated either with vehicle or GSK2606414 (10 mg/kg, Sigma, 1,337,531-89-1) by intraperitoneal injection per day. GSK2606414 was dissolved in a mixture of dimethyl sulfoxide (DMSO): polyethylene glycol (PEG) 400: distilled water (1: 4: 5). After 4 weeks, RV systolic blood pressure (RVSP) was measured with pressure transducers under anesthesia. The RV hypertrophy was analyzed as a ratio of RV to left ventricular and septal weight. Left lung tissues were fixed in 4% paraformaldehyde solution for the following histology staining; right lung tissues and pulmonary arteries were excised and immediately frozen in liquid nitrogen for other experiments. | |||
GTPase KRas (KRAS)
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [295] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| In-vivo Model | Subcutaneous xenograft model in nude mice: Each group of HeLa cells was prepared as a cell suspension at a concentration of 2 × 107/mL. | |||
Guanine nucleotide-binding protein G (GNAS)
Endocrine glands benign cancer [ICD-11: 2F37]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [285] | |||
| Responsed Disease | Endocrine glands benign cancer [ICD-11: 2F37] | |||
| Target Regulation | Up regulation | |||
| Response Summary | METTL3 promotes tumor growth and hormone secretion by increasing expression of Guanine nucleotide-binding protein G (GNAS) and GADD45-Gamma in a m6A-dependent manner. Thus, METTL3 and the methylated RNAs constitute suitable targets for clinical treatment of Pituitary growth hormone-secreting-pituitary adenomas(GH-PAs). | |||
Histone H2AX (H2AX)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [297] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| In-vivo Model | A549 cells (3 × 105 cells in 200 μl PBS) were subcutaneously injected into the nude mice to establish tumors. When the tumors had grown to almost 100 mm3, a 5-GyE dosage of carbon-ion irradiation was applied and the mice were examined every 3 days, then sacrificed 30 days after the injection. | |||
Interleukin-1 beta (IL1B)
Abortion [ICD-11: JA00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [304] | |||
| Responsed Disease | Abortion [ICD-11: JA00.09] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Swan 71
|
N.A. | Homo sapiens | CVCL_D855 |
| HTR-8/SVneo | Normal | Homo sapiens | CVCL_7162 | |
| In-vivo Model | Pregnant C57/BL6 were randomly assigned into four groups (each n = 10): (1) corn oil group, (2) BaP group, (3) BaP +AS-NC group, (4) BaP +AS-hz06 group. | |||
Lactate dehydrogenase A (LDHA)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [305] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | HIF-1 signaling pathway | hsa04066 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glycolysis | |||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | For subcutaneous transplanted model, sh-control and sh-METTL3 HCT-116/5-FU cells (5 × 106 per mouse) were diluted in 100ul PBS + 100 ul Matrigel (BD Biosciences, San Jose, CA, USA) and injected subcutaneously in the rear flank fat pad of the nude mice. | |||
| Response Summary | METTL3 enhances the expression of Lactate dehydrogenase A (LDHA), which catalyzes the conversion of pyruvate to lactate, to trigger glycolysis and 5-FU resistance. METTL3/LDHA axis-induced glucose metabolism is a potential therapy target to overcome 5-FU resistance in CRC cells. | |||
Male infertility [ICD-11: GB04]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [306] | |||
| Responsed Disease | Male infertility [ICD-11: GB04] | |||
| Responsed Drug | MICROCYSTIN-LR | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
TM4
|
N.A. | Mus musculus | CVCL_4327 |
Mammalian target of rapamycin complex 2 (mTORC2)
Endometrial cancer [ICD-11: 2C76]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [94] | |||
| Responsed Disease | Endometrial cancer [ICD-11: 2C76] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell proliferation and tumorigenicity | |||
In-vitro Model |
HEC-1-A | Endometrial adenocarcinoma | Homo sapiens | CVCL_0293 |
| RL95-2 | Endometrial adenosquamous carcinoma | Homo sapiens | CVCL_0505 | |
| T HESCs | Normal | Homo sapiens | CVCL_C464 | |
| In-vivo Model | 4×106 HEC-1-A endometrial cancer cells (shCtrl, shMETTL3, wild-type, METTL14+/-, or METTL14+/- rescued with wild-type or mutant METTL14) were injected intraperitoneally into 5 week old female athymic nude mice (Foxn1nu, Harlan; n=10 per group). | |||
| Response Summary | About 70% of endometrial tumours exhibit reductions in m6A methylation that are probably due to either this METTL14 mutation or reduced expression of METTL3. Reductions in m6A methylation lead to decreased expression of the negative AKT regulator PHLPP2 and increased expression of the positive AKT regulator Mammalian target of rapamycin complex 2 (mTORC2). these results reveal reduced m6A mRNA methylation as an oncogenic mechanism in endometrial cancer and identify m6A methylation as a regulator of AKT signalling. | |||
Mitogen-activated protein kinase 1 (MAPK/ERK2/MAPK1)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [308] | |||
| Responsed Disease | Brain cancer [ICD-11: 2A00] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 |
| U-251MG | Astrocytoma | Homo sapiens | CVCL_0021 | |
Platelet glycoprotein 4 (CD36)
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [251] | |||
| Responsed Disease | Nonalcoholic steatohepatitis [ICD-11: DB92.1] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| In-vivo Model | Mettl3flox/flox and Mettl3-HKO mice were fasted overnight and then injected intraperitoneally with 20 uM BODIPY FL C16 in 200 ul saline for 20 min. | |||
| Response Summary | Mechanistically, METTL3 directly binds to the promoters of the Platelet glycoprotein 4 (CD36) genes and recruits HDAC1/2 to induce deacetylation of H3K9 and H3K27 in their promoters, thus suppressing Cd36 and Ccl2 transcription. METTL3 negatively regulates hepatic Cd36 and Ccl2 gene transcription via a histone modification pathway for protection against Nonalcoholic steatohepatitis(NASH) progression. | |||
Protein crumbs homolog 3 (CRB3)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [323] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
FHC | Normal | Homo sapiens | CVCL_3688 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| Response Summary | m6A and METTL3 levels were substantially elevated in colorectal carcinoma(CRC) tissues, METTL3 knockdown substantially reduced the m6A level of Protein crumbs homolog 3 (CRB3), and inhibited the degradation of CRB3 mRNA to increase CRB3 expression. METTL3 regulated the progression of CRC by regulating the m6A-CRB3-Hippo pathway. | |||
Protein FAM83D (FAM83D)
Triple-negative breast cancer [ICD-11: 2C6Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [324] | |||
| Responsed Disease | Triple-negative breast cancer [ICD-11: 2C6Z] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
In-vitro Model |
MCF-10A | Normal | Homo sapiens | CVCL_0598 |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Protein patched homolog 1 (PTCH1)
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [325] | |||
| Responsed Disease | Esophageal cancer [ICD-11: 2B70] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
TE1
|
N.A. | Mus musculus | CVCL_C6K3 |
| KYSE-30 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1351 | |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| KYSE-410 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1352 | |
| CVCL_E307 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_E307 | |
| HET-1A | Normal | Homo sapiens | CVCL_3702 | |
| In-vivo Model | Nude mice were assigned into four groups (control, sh-NC + LV-NC, sh-METTL3 + LV-NC, and sh-METTL3 + LV-PTCH1) by the random number table method with six mice per group. After nude mice were anesthetized by intraperitoneal injection of sodium pentobarbital (60 mg/kg), the right dorsal skin was routinely disinfected with iodophor. The control mice were injected with untreated KYSE-30 cell suspensions (1 × 106 cells), and the remaining three groups were injected with equal amounts of transfected KYSE-30 cell suspensions (1 × 106 cells). After injection, the syringe was slowly withdrawn and the injection site was gently pressed with an alcohol cotton ball for 10 s to prevent cell suspension from flowing out. Ten days later, tumor volumes were measured every 3 days. | |||
Putative pituitary tumor-transforming gene 3 protein (PTTG3P)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [326] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Cell proliferation and suppression of apoptosis | |||
In-vitro Model |
FHC | Normal | Homo sapiens | CVCL_3688 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | Indicated cells (1 × 107) were subcutaneously injected into 4-week-old male nude mice. Tumor volume was measured every 5 days. | |||
| Response Summary | In colorectal cancer, n6-methyladenosine (m6A) subunit METTL3 increased PTTG3P expression by influencing its stability, while insulin-like growth factor 2 mRNA binding protein 2 (IGF2BP2) could identify Putative pituitary tumor-transforming gene 3 protein (PTTG3P) m6A methylation status and bind to it. | |||
RAD51-associated protein 1 (RAD51AP1)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [327] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Fluorouracil | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 |
| Response Summary | METTL3 augmented 5 FU induced DNA damage and overcame 5 FU resistance in HCT 8R cells, which could be mimicked by inhibition of RAD51-associated protein 1 (RAD51AP1). The present study revealed that the METTL3/RAD51AP1 axis plays an important role in the acquisition of 5 FU resistance in CRC. | |||
Runt-related transcription factor 2 (Runx2)
Maxillofacial bone defect is a critical obstacle for maxillofacial tumors and periodontal diseases. [ICD-11: NA02]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [329] | |||
| Responsed Disease | Maxillofacial bone defect is a critical obstacle for maxillofacial tumors and periodontal diseases. [ICD-11: NA02] | |||
| Target Regulation | Up regulation | |||
Secreted frizzled-related protein 2 (SFRP2)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [330] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Target Regulation | Down regulation | |||
Serine/threonine-protein kinase PINK1, mitochondrial (PINK1)
Chronic kidney disease [ICD-11: GB61]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [332] | |||
| Responsed Disease | Diabetic nephropathy [ICD-11: GB61.Z] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HK-2 [Human kidney] | Normal | Homo sapiens | CVCL_0302 |
| In-vivo Model | Five week old male C57bl/6 mice (20 ± 2 g) were divided into the DKD model group and the control group. The fasting blood glucose of all the mice was measured before the experiment, and was required to be less than 7 mmol/L. The mice in the DKD model group were fed with a high-fat and high-sugar diet for 8 weeks. Then, 1 day after restoring the high-fat and high-sugar diet, the mice in the DKD model group were fasted for 16 h and injected with STZ at 50 mg/kg per day for 5 days. The fasting blood glucose was measured 2 weeks after the last injection. If the fasting blood glucose was greater than 16 mmol/L, the medium-term diabetes model had been established successfully. Then, the rat urine was collected and the ratio of urinary albumin to urinary muscle intoxication (AIb/Cr) was measured. The AIb/Cr was greater than 30 mg/g, indicating a diagnosis of DKD. The mice in the control group received standard chow and were injected with the same amount of normal saline. Subsequently, all DKD mice were randomly divided into the sh-NC group and the sh-METTL3 group. After establishment of the DKD model, the lentiviruses carrying sh-NC or sh-METTL3 (MOI = 50) were injected into the caudal vein at a dose of 1 μg/g according to the weight of mice, once a week for 8 weeks. | |||
Serum response factor (SRF)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [333] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Responsed Drug | Tislelizumab | Approved | ||
In-vitro Model |
U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 |
Staphylococcal nuclease domain-containing protein 1 (SND1)
Nasal-type natural killer/T-cell lymphoma [ICD-11: XH3400]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [336] | |||
| Responsed Disease | Nasal-type natural killer/T-cell lymphoma [ICD-11: XH3400] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
NK-92 | Natural killer cell lymphoblastic leukemia/lymphoma | Homo sapiens | CVCL_2142 |
| YTS | Lymphoblastic leukemia/lymphoma | Homo sapiens | CVCL_D324 | |
| SNT-8 | Nasal type extranodal NK/T-cell lymphoma | Homo sapiens | CVCL_A677 | |
| SNK-6 | Nasal type extranodal NK/T-cell lymphoma | Homo sapiens | CVCL_A673 | |
| In-vivo Model | A total of 32 nude mice were randomly divided into four groups with eight mice in each group. Nude mice were subcutaneously injected with 2 × 106 cells (suspended in 100 μl of PBS) transfected with NC shRNA or SND1 shRNA into the back flank of mice. In DDP administration group, DDP (3 mg/kg per week for 2 weeks) was intraperitoneally injected into mice every 3 days. The control group received 200 μl of 0.1% DMSO. The tumor volume was measured every week for a total of 4 weeks. The mice were sacrificed after 4 weeks, then the tumors were harvested for weight measurement and other analysis. The tumor volume (mm3) was estimated with the formula (0.5 × length × width2). | |||
Sterol regulatory element-binding protein 1 (SREBF1)
Acute kidney failure [ICD-11: GB60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [338] | |||
| Responsed Disease | Acute kidney failure [ICD-11: GB60] | |||
In-vitro Model |
HK-2 [Human kidney] | Normal | Homo sapiens | CVCL_0302 |
Suppressor of cytokine signaling 1 (SOCS1)
Thyrotoxicosis [ICD-11: 5A02]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [225] | |||
| Responsed Disease | Graves disease [ICD-11: 5A02.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
PBMCs (Human peripheral blood mononuclear cells (PBMCs) are isolated from peripheral blood and identified as any blood cell with a round nucleus) | |||
| Response Summary | METTL3 knock-down experiment revealed that expressions of SOCS family members Suppressor of cytokine signaling 1 (SOCS1), SOCS2, SOCS4, SOCS5, and SOCS6 were increased after METTL3 knock-down. It indicated that METTL3 is involved in the development of Graves' disease (GD) by inducing mRNA m6A methylation modification of SOCS family members. | |||
Pain disorders [ICD-11: 8E43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [340] | |||
| Responsed Disease | Neuropathic pain [ICD-11: 8E43.0] | |||
Suppressor of cytokine signaling 3 (SOCS3)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [341] | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Responsed Drug | Colivelin | Investigative | ||
Diabetic [ICD-11: 5A14]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [342] | |||
| Responsed Disease | Diabetic [ICD-11: 5A14] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HUVEC-C | Normal | Homo sapiens | CVCL_2959 |
Paraneoplastic or autoimmune disorders of the nervous system [ICD-11: 8E4A]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [343] | |||
| Responsed Disease | Paraneoplastic or autoimmune disorders of the nervous system [ICD-11: 8E4A.1] | |||
| Target Regulation | Down regulation | |||
Chronic obstructive pulmonary disease [ICD-11: CA22]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [344] | |||
| Responsed Disease | Chronic obstructive pulmonary disease [ICD-11: CA22] | |||
| Responsed Drug | Cycloleucine | Investigative | ||
| Target Regulation | Down regulation | |||
Human skin lesions [ICD-11: ME60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [345] | |||
| Responsed Disease | Human skin lesions [ICD-11: ME60] | |||
| Responsed Drug | Arsenite | Phase 2 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
In-vitro Model |
HaCaT | Normal | Homo sapiens | CVCL_0038 |
Suppressor of cytokine signaling 5 (SOCS5)
Thyrotoxicosis [ICD-11: 5A02]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [225] | |||
| Responsed Disease | Graves disease [ICD-11: 5A02.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
PBMCs (Human peripheral blood mononuclear cells (PBMCs) are isolated from peripheral blood and identified as any blood cell with a round nucleus) | |||
| Response Summary | METTL3 knock-down experiment revealed that expressions of SOCS family members SOCS1, SOCS2, SOCS4, Suppressor of cytokine signaling 5 (SOCS5), and SOCS6 were increased after METTL3 knock-down. It indicated that METTL3 is involved in the development of Graves' disease (GD) by inducing mRNA m6A methylation modification of SOCS family members. | |||
Transferrin receptor protein 1 (TFRC)
Intracerebral hemorrhage [ICD-11: 8B00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [354] | |||
| Responsed Disease | Intracerebral hemorrhage [ICD-11: 8B00] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
PC12 | Rat adrenal gland pheochromocytoma | Rattus norvegicus | CVCL_0481 |
| In-vivo Model | For ICH model establishment, the mice in the ICH group were intraperitoneally injected with 2% sodium pentobarbital (0.2 mL/100 g body weight). Subsequently, the mice were routinely sterilized, skin prepared and fixed on a brain stereotaxic apparatus. Then 0.2 units of collagenase 4 were injected into the left caudate putamen (0.8 mm posterior to the bregma, 3.5 mm lateral to the left, and 5.5 mm deep). The sham-operated control group was injected with an equal volume of normal saline. For METTL3 knockdown, the mice were intravenously injected with sh-METTL3 or sh-NC (1×109 plaque forming unit). | |||
Tumor necrosis factor receptor superfamily member 6 (FAS)
Preeclampsia [ICD-11: JA23]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [355] | |||
| Responsed Disease | Preeclampsia [ICD-11: JA23] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HTR-8/SVneo | Normal | Homo sapiens | CVCL_7162 |
Wnt inhibitory factor 1 (WIF1)
Endometriosis [ICD-11: GA10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [358] | |||
| Responsed Disease | Endometriosis [ICD-11: GA10] | |||
| Target Regulation | Up regulation | |||
DBH antisense RNA 1 (Lnc_DBH-AS1)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [359] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Responsed Drug | Gemcitabine | Approved | ||
In-vitro Model |
MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 |
| HPDE | Normal | Homo sapiens | CVCL_4376 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| Canpan-2 (Pancreatic cancer cell line) | ||||
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| Response Summary | DBH antisense RNA 1 (Lnc_DBH-AS1) expression in pancreatic cancer(PC) was found to be linked to the METTL3-dependent m6A methylation of the lncRNA. MechanisticallyDBH-AS1 was able to increase PC cell sensitivity to gemcitabine by sequestering miR-3163 and thus upregulating USP44 in these tumor cells. | |||
Growth arrest specific 5 (GAS5)
Vascular disorders of the liver [ICD-11: DB98]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [360] | |||
| Responsed Disease | Vascular disorders of the liver [ICD-11: DB98.8] | |||
| Target Regulation | Down regulation | |||
| In-vivo Model | The in vivo assay was approved by the Animal Care and Use Committee of Anhui Medical University. And all experimental procedures and animal care were in accordance with the institutional ethics guidelines for animal experiments. The C57BL/6 mice (male, 8-10 weeks old) were housed (six per cage) in a specific and opportunistic pathogen-free facility and maintained on a 12-hlight-dark cycle with casually access to food and water. Detailed descriptions of the cell culture, cardiac fibrosis model, and treatment with lentiviral, were given in the Supplementary methods online. | |||
HOXD antisense growth-associated long non-coding RNA (HAGLR)
Pre-eclampsia [ICD-11: JA24]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [361] | |||
| Responsed Disease | Pre-eclampsia [ICD-11: JA24] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HTR-8/SVneo | Normal | Homo sapiens | CVCL_7162 |
KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1)
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [362] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Doxil | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
MCF-10A | Normal | Homo sapiens | CVCL_0598 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
LIFR antisense RNA 1 (LIFR-AS1)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [363] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell proliferation and metastasis | |||
| Response Summary | METTL3 induced m6A hyper-methylation on the 3' UTR of LIFR antisense RNA 1 (LIFR-AS1) to enhance its mRNA stability and LIFR-AS1 could directly interact with miR-150-5p, thereby indirectly up-regulating VEGFA expressions within cells. A noval m6A-LIFR-AS1 axis promotes pancreatic cancer progression at least in part via regulation of the miR-150-5p/VEGFA axis, indicating that this regulatory axis can be a viable clinical target for the treatment of pancreatic cancer. | |||
Long intergenic non-protein coding RNA 2598 (LINC02598/RP11)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [366] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Ubiquitin mediated proteolysis | hsa04120 | ||
| Cell Process | Ubiquitination degradation | |||
In-vitro Model |
HCT 15 | Colon adenocarcinoma | Homo sapiens | CVCL_0292 |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| Response Summary | Overexpression of METTL3 upregulates Long intergenic non-protein coding RNA 2598 (LINC02598/RP11) expression in colorectal cancer cells. Overexpression of ALKBH5 downregulates RP11 expression. | |||
Long intergenic non-protein coding RNA 680 (LINC00680)
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [199] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Chondrocytes (Chondrocytes were isolated from human cartilage and cultured) | |||
| Response Summary | m6A-mediated Long intergenic non-protein coding RNA 680 (LINC00680) regulates the proliferation and ECM degradation of chondrocytes through LINC00680/m6A/SIRT1 mRNA axis. METTL3-mediated LINC00680 accelerates osteoarthritis(OA) progression, which provides novel understanding of the role of m6A and lncRNA in OA. | |||
Small nucleolar RNA host gene (SNHG7)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [372] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
GES-1 | Normal | Homo sapiens | CVCL_EQ22 |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [373] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Central carbon metabolism in cancer | hsa05230 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glycolysis | |||
In-vitro Model |
VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| Response Summary | METTL3-stabilized lncRNA Small nucleolar RNA host gene (SNHG7) accelerates glycolysis in PCa via SRSF1/c-Myc axis and inspires the understanding of m6A roles in lncRNA metabolism and tumor progression. | |||
microRNA 1246 (MIR1246)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [220] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | Cell migration and invasion | |||
In-vitro Model |
Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| In-vivo Model | The spleen in the upper left lateral abdomen of the anesthetized mice were exposed, 106 cells suspended in 20 uL phosphate-buffered saline (PBS) were injected into the distal tip of the spleen. After injection, replacing the spleen, and closing the incision. | |||
| Response Summary | METTL3/microRNA 1246 (MIR1246)/SPRED2 axis plays an important role in tumor metastasis and provides a new m6A modification pattern in Colorectal cancer development. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [91] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | The 40 nude mice of each large group were classified into 8 small groups (n = 5) and subcutaneously injected with transfected cell suspension in the back (5 × 106 cells/mL/mouse). The length and width of tumors were recorded every 4 days, and the tumor volume = (length × width2)/2. The tumor growth curve was thereby graphed. On the 28th day of injection, mice were euthanized by neck dislocation, and the xenografts were harvested, photographed, and weighed. | |||
| Response Summary | METTL3 regulates the m6A modification to promote the maturation of microRNA 1246 (MIR1246), which targets PEG3 to participate in occurrence and development of non-small cell lung cancer. | |||
microRNA 126 (MIR126)
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [377] | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Mono-Mac-6 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1426 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | Cells were collected from the CD45.2 Ythdf2fl/flMx1-Cre mice, enriched for lin-population, and transduced with retroviral MA9 alone or retroviral MA9 + MSCV-pre-miR-126/MSCV-pre-miR-Control by spinoculation as described before. Clonal cells from CFA assays were washed and suspended together with healthy mouse BM cells (0.5 million CFA cells + 1 million healthy BM per mouse) with ice-cold PBS, and subsequently injected via the tail vein into lethally (900 rads) irradiated 8 to 10-week-old B6.SJL (CD45.1) recipient mice. Ten days after the injection, knockout was induced by i.p. delivering of poly I:C (428750 R&D Systems) at 300 μg each time per mice every other day for five times. Mice were observed regularly after cell injection and samples were collected at the end point. | |||
Endometriosis [ICD-11: GA10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [378] | |||
| Responsed Disease | Endometriosis [ICD-11: GA10] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HESC (Human endometrial stromal cells) | |||
| Response Summary | The METTL3/m6A/miR126 pathway, whose inhibition contributes to endometriosis development by enhancing cellular migration and invasion. | |||
microRNA 1305 (MIR1305)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [379] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Reverse splicing and biogenesis | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | Mice were randomly divided into three groups, and subcutaneously injected with control, circ-ARL3-silenced and circ-ARL3&miR-1305-silenced HepG2.2.15 cells. | |||
| Response Summary | Circ-ARL3 is a critical regulator in HBV-related HCC, targeting the axis of circ-ARL3/microRNA 1305 (MIR1305) can be a promising treatment for HBV+ HCC patients. HBx protein upregulated N6 -methyladenosine (m6A) methyltransferases METTL3 expression, increasing the m6A modification of circ-ARL3; then, m6A reader YTHDC1 bound to m6 A-modified of circ-ARL3 and favored its reverse splicing and biogenesis. Furthermore, circ-ARL3 was able to sponge miR-1305, antagonizing the inhibitory effects of miR-1305 on a cohort of target oncogenes, thereby promoting HBV+ HCC progression. | |||
microRNA 150 (MIR150)
Pain disorders [ICD-11: 8E43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [380] | |||
| Responsed Disease | Neuropathic Pain [ICD-11: 8E43.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
RN-sc (The rat neuron cell line RN-sc was purchased from ScienCell) | |||
| In-vivo Model | Based on our study design, the enrolled animals were blindingly grouped and received the following treatments: (1)Sham-NC vectors, (2)Sham-sh-METTL3, (3)Sham-sh-METTL3 + miR-150, (4)Sham-sh-METTL3 + Lv-YTHDF2, (5)Sham-sh-METTL3 + sh-BDNF, (6)Sham-anti-miR150, (7)Sham-anti-miR150+sh-BDNF, (8)SNI-Lv-METTL3, (9)SNI-Lv-METTL3 + anti-miR-150, (10)SNI-Lv-METTL3 + sh-YTHDF2, (11)SNI-Lv-METTL3 + Lv-BDNF, (12)SNI-miR150, (13)SNI-miR150+Lv-BDNF. The pain behaviors were detected in the respective time points and the expressions of METTL3 and miR-150 were determined at day 14 after the rats were sacrificed. | |||
| Response Summary | Enhanced METTL3 promoted the m6A methylation in total RNAs and inhibited neuropathic pain (NP) progression. Mechanistically, METTL3 accelerated microRNA 150 (MIR150) maturation via mediating m6A methylation of primiR-150 at locus 498, cooperating with the "m6A reader" YTHDF2. Therefore, the METTL3/miR-150/BDNF pathway is a promising therapeutic target for NP patients. | |||
microRNA 222 (MIR222)
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [244] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
In-vitro Model |
EJ (Human bladder cancer cells) | |||
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| In-vivo Model | About 1× 107 cells were injected subcutaneously into the axilla of the female athymic BALB/C nude mice (4-6 weeks old, 18-22 g, five mice per group). | |||
| Response Summary | METTL3 has an oncogenic role in bladder cancer through interacting with the microprocessor protein DGCR8 and positively modulating the microRNA 222 (MIR222) process in an m6A-dependent manner. | |||
Cardiomegaly [ICD-11: BC45]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [381] | |||
| Responsed Disease | Cardiomegaly [ICD-11: BC45] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
In-vitro Model |
NRCMs (Primary neonatal rat cardiomyocytes (NRCMs)NRCMs were prepared from the hearts of 2- to 3-day-old SD rats according to the following protocol) | |||
| In-vivo Model | The pump was prefilled with Ang-II or saline and then incubated in sterile saline at 37 ℃ for 48 h. After the mice were anesthetized with 3.0% isoflurane mixed with oxygen, an incision was made on the back skin of the mice, and the pump was implanted into the subcutaneous area, followed by suturing the incision. After the operation, the mice were given buprenorphine (0.1 mg/kg) to reach analgesia. Finally, the regaining consciousness mice were returned to cages and fed until the end of the experiment. | |||
| Response Summary | METTL3 positively modulates the pri-miR221/pri-miR-222 maturation process in an m6A-dependent manner and subsequently activates Wnt/Beta-catenin signaling by inhibiting DKK2, thus promoting Ang-II-induced cardiac hypertrophy. | |||
microRNA 25 (MIR25)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [382] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell proliferation | |||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| HPDE6c7 | Normal | Homo sapiens | CVCL_0P38 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| In-vivo Model | Mice (five in each group) were injected subcutaneously with 0.1 ml of cell suspension containing 2 × 106 cells in the back flank. | |||
| Response Summary | Cigarette smoke-induced microRNA 25 (MIR25) excessive maturation via m6A modification promotes the development and progression of pancreatic cancer. This modification is catalyzed by overexpressed methyltransferase-like 3 (METTL3) due to hypomethylation of the METTL3 promoter also caused by CSC. | |||
microRNA 675 (MIR675)
Gastrointestinal stromal tumor [ICD-11: 2B5B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [383] | |||
| Responsed Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B] | |||
In-vitro Model |
GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| In-vivo Model | The GIST-T1 cells were injected into the back flanks of the C57BL/6 female mice (Age six-week-old) at the concentration of 1 × 106 cells diluting in 200 μl PBS buffer solution. The GIST-T1 cells were allowed to grow in mice for a total of 25 days, the mice were sacrificed at day 25 and the tumors were obtained and weighed to evaluate the tumorigenesis abilities of the GIST-T1 cells in vivo. | |||
microRNA 93 (MIR93)
Emphysema [ICD-11: CA21]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [160] | |||
| Responsed Disease | Emphysema [ICD-11: CA21] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Response Summary | METTL3-mediated formation of EV microRNA 93 (MIR93), facilitated by m6A, is implicated in the aberrant cross-talk of epithelium-macrophages, indicating that this process is involved in the smoking-related emphysema. EV miR-93 was used as a novel risk biomarker for CS-induced emphysema. MiR-93 activated the JNK pathway by targeting dual-specificity phosphatase 2 (DUSP2), which elevated the levels of matrix metalloproteinase 9 (MMP9) and matrix metalloproteinase 12 (MMP12) and induced elastin degradation, leading to emphysema. | |||
microRNA let-7b (MIRLET7B)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [384] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Responsed Drug | Osimertinib | Approved | ||
| Pathway Response | Notch signaling pathway | hsa04330 | ||
In-vitro Model |
NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| H1975OR (Osimertinib resistant H1975 cells) | ||||
| HCC827OR (Osimertinib resistant HCC827 cells) | ||||
| Response Summary | The participation of Metformin decreased the bindings of DNMT3a/b to the METTL3 promoter with the help of the readers of NKAP and HNRNPA2B1.the mediation of m6A formation on pri-Let-7b processing increased the mature microRNA let-7b (MIRLET7B), whose key role is to suppress the Notch signaling and to re-captivate the Osimertinib treatment.The findings open up future drug development, targeting this pathway for lung cancer patients. | |||
microRNA let-7g (MIRLET7G)
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [214] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell differentiation and apoptosis | |||
| Glutamine metabolism | ||||
| Apoptosis (hsa04210) | ||||
| Response Summary | HBXIP up-regulates METTL3 by suppressing microRNA let-7g (MIRLET7G), in which METTL3 increased HBXIP expression forming a positive feedback loop of HBXIP/let-7g/METTL3/HBXIP, leading to accelerated cell proliferation in breast cancer. | |||
hsa-miR-126-5p
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [385] | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Response Summary | METTL3 promoted the maturation of hsa-miR-126-5p via the m6A modification of pri-miR-126-5p. Finally, in vitro and in vivo experiments substantiated that silencing of METTL3 impeded the progression and tumorigenesis of ovarian cancer by impairing the miR-126-5p-targeted inhibition of PTEN and thus blocking the PI3K/Akt/mTOR pathway. | |||
Chondropathies [ICD-11: FB82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [198] | |||
| Responsed Disease | Chondropathies [ICD-11: FB82] | |||
| Target Regulation | Up regulation | |||
| Cell Process | RNA mature | |||
In-vitro Model |
Cartilage cells (From the cartilage tissue samples from patients) | |||
| Response Summary | Interleukin 1-beta (IL-1-beta) is an important inducer of cartilage degeneration that can induce an inflammatory cascade reaction in chondrocytes and inhibit the normal biological function of cells. METTL3 could regulate hsa-miR-126-5p maturation, we first confirmed that METTL3 can bind the key protein underlying pri-miRNA processing, DGCR8. Additionally, when METTL3 expression was inhibited, the miR-126-5p maturation process was blocked. | |||
hsa-miR-140-3p
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [148] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| Response Summary | METTL3/SNHG1/hsa-miR-140-3p/UBE2C axis plays a crucial role in cancer progression and the immune response in non-small cell lung cancer. | |||
hsa-miR-143-3p
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [386] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA splicing | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| In-vivo Model | For subcutaneous transplanted model, control and miR-143-3p stable A549 cells (5 × 106 per mouse, n = 5 for each group) were diluted in 200 uL PBS + 200 uL Matrigel (BD Biosciences) and subcutaneously injected into immunodeficient mice to investigate tumor growth. | |||
| Response Summary | m6A methyltransferase Mettl3 can increase the splicing of precursor hsa-miR-143-3p to facilitate its biogenesis. The miR-143-3p/VASH1 axis in BM of lung cancers and suggests their critical roles in lung cancer pathogenesis. | |||
Ischemic heart disease [ICD-11: BA40-BA6Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [253] | |||
| Responsed Disease | Ischemic heart disease [ICD-11: BA40-BA6Z] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Pyroptosis | |||
In-vitro Model |
H9c2(2-1) | Normal | Rattus norvegicus | CVCL_0286 |
| In-vivo Model | The thoracic cavity of rats was exposed, and the left anterior descending coronary artery was ligated with a 6-0 silk thread. Successfully surgical MI could be observed, with myocardium color fading and pulse weakening. After 30 min of ischemia, the blood flow was restored by releasing the slipknot, and then 120-min perfusion was performed. Afterward, the thoracic cavity of rats was sutured. The rats were assigned into 4 groups, with 12 rats in each group. Lentivirus packaged short hairpin (sh)-negative control (NC) and sh-METTL3 (GenePharma, Shanghai, China) were injected into the rats via tail vein 24 h before operation. The titer of lentivirus was 1 × 109 TU/mL, and the injection rate was 0.2 ul/min for 10 min. Blood samples were collected 24 h after reperfusion. | |||
| Response Summary | METTL3 promoted DGCR8 binding to pri-miR-143-3p through m6A modification, thus enhancing hsa-miR-143-3p expression to inhibit PRKCE transcription and further aggravating cardiomyocyte pyroptosis and MI/R injury. | |||
Asthma [ICD-11: CA23]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [387] | |||
| Responsed Disease | Asthma [ICD-11: CA23] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
16HBE14o- | Normal | Homo sapiens | CVCL_0112 |
hsa-miR-146a-5p
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [201] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Responsed Drug | Melittin | Investigative | ||
| Target Regulation | Up regulation | |||
| Cell Process | miRNA maturation | |||
| Cell apoptosis | ||||
In-vitro Model |
T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| EJ (Human bladder cancer cells) | ||||
| BIU-87 | Human bladder cancer cells | Homo sapiens | CVCL_6881 | |
| In-vivo Model | For melittin treatment study, 4-week-old female BALB/c nude mice were subcutaneously injected with 1 × 107 T24 or BIU87 cells. | |||
| Response Summary | METTL3 acts as a fate determinant that controls the sensitivity of bladder cancer cells to melittin treatment. Moreover, METTL3/hsa-miR-146a-5p/NUMB/NOTCH2 axis plays an oncogenic role in bladder cancer pathogenesis and could be a potential therapeutic target for recurrent bladder cancer treatment. | |||
hsa-miR-181d-5p
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [88] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Fluorouracil | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
HT29 | Colon cancer | Mus musculus | CVCL_A8EZ |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| In-vivo Model | A tumor-bearing model was established by subcutaneously injecting 100 ul HT29 cells (5×106) followed by an intravenous injection of CAFs-derived exosomes (50 ug/mouse every three days) into the tail vein of the mice. An intraperitoneal injection of 5-FU (50 mg/kg, every week) was administered on day 12. | |||
| Response Summary | METTL3 dependent m6A methylation was upregulated in CRC to promote the processing of miR 181d 5p by DGCR8. This led to increased hsa-miR-181d-5p expression, which inhibited the 5 FU sensitivity of CRC cells by targeting NCALD. | |||
hsa-miR-186-5p
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [259] | |||
| Responsed Disease | Esophageal cancer [ICD-11: 2B70] | |||
| Responsed Drug | Pt | Investigative | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cellular Processes | |||
| Cell growth and death | ||||
| Cell apoptosis | ||||
In-vitro Model |
Eca-9706 (Esophageal carcinoma cell line) | |||
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| In-vivo Model | Used 1 × 106 SNHG3 knocked down KY-SE150 cells and NC lentivirus to inject into the right flank of mice to generate xenografts. | |||
| Response Summary | Platinum can increase the overall m6A level of esophageal cancer. SNHG3/hsa-miR-186-5p, induced by platinum, was involved in regulating m6A level by targeting METTL3. miR-186-5p binds to the 3'UTR of METTL3 to inhibit its expression. Our manuscript has provided clues that regulating m6A level was a novel way to enhance the platinum efficacy. | |||
hsa-miR-1914-3p
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [126] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Metabolic | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-6 | Lung adenocarcinoma | Homo sapiens | CVCL_0236 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | Mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then administrated with PBS or DDP (3 mg/kg per day) for three times a week, and tumor volume were measured every second day. | |||
| Response Summary | METTL3, YTHDF3, YTHDF1, and eIF3b directly promoted YAP translation through an interaction with the translation initiation machinery. METTL3 knockdown inhibits tumor growth and enhances sensitivity to DDP in vivo.m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-hsa-miR-1914-3p-YAP axis to induce Non-small cell lung cancer drug resistance and metastasis. | |||
hsa-miR-1915-3p
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [102] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | TNF signaling pathway | hsa04668 | ||
| Cell Process | Cell migration | |||
| Cell invasion | ||||
| Epithelial-mesenchymal transition | ||||
In-vitro Model |
NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| Response Summary | hsa-miR-1915-3p expression was regulated by METTL3/YTHDF2 m6A axis through transcription factor KLF4. miR-1915-3p function as a tumor suppressor by targeting SET and has an anti-metastatic therapeutic potential for lung cancer treatment. | |||
hsa-miR-193b
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [169] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| In-vivo Model | Mice were divided into two groups (n = 4/group) randomly. 3×106 cells suspended in 200 uL PBS were administered via subcutaneous injection over the right flank region of nude mice. After the development of palpable tumors (average volume, 50 mm3), intratumoral injection of synthetic miR-193b, or negative control complexed with siPORT Amine transfection reagent (Ambion, USA) was given 6 times at a 4-day interval. | |||
| Response Summary | METTL3 modulates miR-193b mature process in an m6A-dependent manner. Reintroduction of hsa-miR-193b profoundly inhibits tumorigenesis of cervical cancer cells both in vivo and in vitro through CCND1 targeting. | |||
hsa-miR-199a-5p
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [166] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
hFOB 1.19 | Normal | Homo sapiens | CVCL_3708 |
| MG-63 | Osteosarcoma | Homo sapiens | CVCL_0426 | |
| U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 | |
| Response Summary | METTL3-mediated circNRIP1 exhibits oncogenic roles in osteosarcoma by regulating FOXC2 via sponging hsa-miR-199a-5p, which provides new ideas for the treatment of osteosarcoma. | |||
hsa-miR-21-5p
Kidney disorders [ICD-11: GB90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [388] | |||
| Responsed Disease | Kidney disorders [ICD-11: GB90] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | NF-kappa B signaling pathway | hsa04064 | ||
In-vitro Model |
HK2 | Normal | Acipenser baerii | CVCL_YE28 |
| In-vivo Model | Each mouse was anaesthetized with inhaled isoflurane, and the left proximal ureter was exposed. Then, the ureter was ligated with 6-0 silk thread and severed. In the sham operation group, the left ureters of mice were exposed, but not ligated or severed. The 3rd, 7th and 14th days after surgery were the time points for killing. At each time point, a total of 10 mice in the UUO group were executed, and a total of 10 mice in the sham group were also executed to serve as controls. | |||
| Response Summary | METTL3-m6A-miR-21-5p-SPRY1/ERK/NF-kB axis in obstructive renal fibrosis and provides a deeper understanding of renal fibrosis. | |||
hsa-miR-221-3p
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [22] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Doxil | Approved | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell growth and death | |||
| Cell apoptosis | ||||
In-vitro Model |
ADR-resistant MCF-7 (MCF-7/ADR) cells (Human breast cancer doxorubicin-resistant cell line) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| In-vivo Model | Cell suspensions (2 × 106 cells/mL) made with MCF-7/ADR cells stably expressing METTL3 and/or miR-221-3p inhibitor were subcutaneously implanted into each mouse. One week later, xenografted mice were injected with 0.1 mL ADR (25 mg/kg, intraperitoneal injection) twice a week. | |||
| Response Summary | METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating hsa-miR-221-3p maturation in a m6A-dependent manner. METTL3 knockdown was shown to reduce the expression of miR-221-3p by reducing pri-miR-221-3p m6A mRNA methylation, reducing the expression of MDR1 and BCRP, and inducing apoptosis. Identified the METTL3/miR-221-3p/HIPK2/Che-1 axis as a novel signaling event that will be responsible for resistance of BC cells to ADR. | |||
hsa-miR-222-3p
Thyroid Cancer [ICD-11: 2D10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [107] | |||
| Responsed Disease | Thyroid Cancer [ICD-11: 2D10] | |||
| Target Regulation | Up regulation | |||
| Response Summary | Silencing METTL3 suppresses hsa-miR-222-3p expression and thus stimulates STK4 expression, thereby repressing the malignancy and metastasis of Thyroid Carcinoma. | |||
hsa-miR-25-3p
Diabetes [ICD-11: 5A10-5A14]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [389] | |||
| Responsed Disease | Diabetes [ICD-11: 5A10-5A14] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell viability | |||
| Cell apoptosis | ||||
In-vitro Model |
ARPE-19 | Normal | Homo sapiens | CVCL_0145 |
| Response Summary | METTL3 involves in the pathogenesis of diabetic retinopathy (DR). Both METTL3 mRNA and hsa-miR-25-3p were low-expressed in the peripheral venous blood samples of diabetes mellitus (DM) patients compared to normal volunteers, and high-glucose inhibited METTL3 and miR-25-3p expressions in RPE cells. Overexpression of METTL3 attenuates high-glucose induced RPE cell pyroptosis by regulating miR-25-3p/PTEN/Akt signaling cascade through DGCR8. | |||
Retinopathy [ICD-11: 9B71]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [389] | |||
| Responsed Disease | Diabetic retinopathy [ICD-11: 9B71.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell viability | |||
| Cell apoptosis | ||||
In-vitro Model |
ARPE-19 | Normal | Homo sapiens | CVCL_0145 |
| Response Summary | METTL3 involves in the pathogenesis of diabetic retinopathy (DR). Both METTL3 mRNA and hsa-miR-25-3p were low-expressed in the peripheral venous blood samples of diabetes mellitus (DM) patients compared to normal volunteers, and high-glucose inhibited METTL3 and miR-25-3p expressions in RPE cells. Overexpression of METTL3 attenuates high-glucose induced RPE cell pyroptosis by regulating miR-25-3p/PTEN/Akt signaling cascade through DGCR8. | |||
hsa-miR-29a-3p
Acute respiratory distress syndrome [ICD-11: CB00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [390] | |||
| Responsed Disease | Acute respiratory distress syndrome [ICD-11: CB00] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| In-vivo Model | All the mice were randomly divided into three groups: the normal saline group (NS), the lipopolysaccharide + negative control group (LPS + NC), and the LPS + miR-29a-3p agomir group (LPS + Agomir). | |||
| Response Summary | The knockout of methyltransferase 3 (N6-adenosine-methyltransferase complex catalytic subunit) removed the m6A modification of hsa-miR-29a-3p and reduced miR-29a-3p expression. These findings suggest that miR-29a-3p is a potential target that can be manipulated for ALI/ARDS. | |||
hsa-miR-31-5p
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [157] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Cell cycle | |||
In-vitro Model |
BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 |
| HCC1806 | Breast squamous cell carcinoma | Homo sapiens | CVCL_1258 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 | |
| In-vivo Model | Twelve female BALB/c nude mice (aged 4 weeks, 18-22g) were randomly divided into 2 groups. Stable circMETTL3-expression SUM1315 cells or control cells (1×106 cells in 0.1 mL PBS) was subcutaneously injected into mammary fat pads of the mice and the growth of tumors was followed up every week. Tumor volume was measured every week using a caliper, calculated as (length × width2)/2. After 4 weeks, mice were sacrificed and checked for final tumor weight. | |||
| Response Summary | CircMETTL3 promotes breast cancer progression through circMETTL3/hsa-miR-31-5p/CDK1 axis. | |||
hsa-miR-320b
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [196] | |||
| Responsed Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.1] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Epithelial-mesenchymal transition | ||||
In-vitro Model |
TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 |
| KYSE-30 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1351 | |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| HET-1A | Normal | Homo sapiens | CVCL_3702 | |
| CVCL_E307 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_E307 | |
| Eca-109 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_6898 | |
| In-vivo Model | Luciferase-labeled KYSE150 cells (5 × 106) were inoculated into the footpads of BALB/c nude mice (4-5 weeks old, 18-20 g) to establish the popliteal lymphatic metastasis model. | |||
| Response Summary | METTL3 could interact with DGCR8 protein and positively modulate pri-miR-320b maturation process in an N6-methyladenosine (m6A)-dependent manner. Therefore, our findings uncover a VEGF-C-independent mechanism of exosomal and intracellular hsa-miR-320b-mediated LN metastasis and identify miR-320b as a novel predictive marker and therapeutic target for LN metastasis in ESCC. | |||
hsa-miR-34a
Aortic aneurysm or dissection [ICD-11: BD50]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [391] | |||
| Responsed Disease | Abdominal aortic aneurysm [ICD-11: BD50.4] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
VSMC (Human aortic vascular smooth muscle cells) | |||
| In-vivo Model | Male C57BL/6J mice were anesthetized with an intraperitoneal injection of pentobarbital (40 mg/kg). The abdominal aorta between the renal arteries and the bifurcation of the iliac arteries was disassociated from the surrounding structures. Video microscopy was used to assay the diameter of the aorta in triplicate. After the measurements were taken, a small piece of gauze dipped in 0.5 mol/L CaCl2 was spread perivascularly onto the aortic passage for 15 min. Control mice received substitute treatment with NaCl (0.9%)-soaked gauze for 15 min. Then, the aorta was rinsed with 0.9% sterile saline, and the incision was sutured. After 3 or 6 weeks, all the animals were sacrificed, and the aortas were harvested for further analysis. | |||
| Response Summary | METTL3/m6A-mediated hsa-miR-34a maturation in AAA formation and provide a novel therapeutic target and diagnostic biomarker for AAA treatment. | |||
hsa-miR-375-3p
Atherosclerosis [ICD-11: BD40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [392] | |||
| Responsed Disease | Atherosclerosis [ICD-11: BD40.Z] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Focal adhesion | hsa04510 | ||
In-vitro Model |
MOVAS (Mouse aortic vascular smooth muscle cell lines) | |||
| In-vivo Model | The normal group consisted of C57BL/6J mice on normal diet and the HFD group consisted of ApoE C57BL/6J mice on HFD (containing 10% lard oil, 4% milk powder, 2% cholesterol, and 0.5% sodium cholate). Four weeks after HFD feeding, the mice were injected with 200 ul lentivirus containing 1 × 10-/--/-8 TU (lentivirus carrying sh-METTL3 or sh-NC; designed and constructed by GENCHEM (Shanghai, China)) via tail vein. Six weeks after transfection, all mice were euthanized by a tail vein injection of 200 mg/kg pentobarbital sodium. | |||
| Response Summary | Silencing METTL3 inhibited m6A level and decreased the binding of DGCR8 to pri-miR-375 and further limited hsa-miR-375-3p expression. METTL3-mediated m6A modification promoted VSMC phenotype transformation and made Atherosclerosis (AS) plaques more vulnerable via the miR-375-3p/PDK1 axis. | |||
hsa-miR-380-3p
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [393] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
HPDE | Normal | Homo sapiens | CVCL_4376 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| Capan-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0026 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| In-vivo Model | The PC cell line PANC1 was subcutaneously injected into the dorsal flank of the mice at the concentration of 1 × 106 cells per mouse. | |||
| Response Summary | hsa-miR-380-3p was enriched with m6A modifications, and elimination of m6A modifications by deleting METTL3 and METTL14 synergistically suppressed miR-380-3p expressions in pancreatic cancer cells. | |||
hsa-miR-422a
Unspecific body region injury [ICD-11: ND56]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [394] | |||
| Responsed Disease | Unspecific body region injury [ICD-11: ND56] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Neuronal cell apoptosis | |||
In-vitro Model |
SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
| Response Summary | Oxygen glucose deprivation/re-oxygenation (OGD/R) induces neuronal injury via mechanisms that are believed to mimic the pathways associated with brain ischemia. METTL3 shRNA reversed OGD/R-induced Lnc-D63785 m6A methylation to decrease hsa-miR-422a accumulation. | |||
hsa-miR-582-3p
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [395] | |||
| Responsed Disease | Intrahepatic cholangiocarcinoma [ICD-11: 2C12.10] | |||
In-vitro Model |
TFK-1 | Cholangiocarcinoma | Homo sapiens | CVCL_2214 |
| RBE | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_4896 | |
| HuH-28 | Cholangiocarcinoma | Homo sapiens | CVCL_2955 | |
| HuCC-T1 | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_0324 | |
| HIBEpic (Human intrahepatic bile duct epithelial cells) | ||||
| CC-LP-1 | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_0205 | |
| In-vivo Model | To detect the effect of NKILA on CAA growth, 5 × 106 control and NKILA-depleted HuCCT1 cells (n = 4 per group) were subcutaneously injected into male BALB/c nude mice (4-6 weeks old). | |||
| Response Summary | NKILA physically interacted with and suppressed hsa-miR-582-3p, which was regulated by METTL3-mediated N6 -methyladenosine (m6A) modification. YAP1 was a target of NKILA via miR-582-3p and NKILA functioned partially via YAP1 in CCA. | |||
hsa-miR-589-5p
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [396] | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Adherens junction | hsa04520 | ||
| Cell Process | Cell migration | |||
| Cell invasion | ||||
In-vitro Model |
THLE-2 | Normal | Homo sapiens | CVCL_3803 |
| SNU-423 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0366 | |
| SNU-387 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0250 | |
| SNU-182 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0090 | |
| SK-HEP-1 | Liver and intrahepatic bile duct epithelial neoplasm | Homo sapiens | CVCL_0525 | |
| PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | The mice were fed in an SPF environment (cycle of 12-h light and 12-h dark) with a free diet. All the mice were adaptively fed for 5 days before experiments and randomly divided into the following four groups: the siNC+MC group (n = 10), the METTL3 siRNA1+MC group (n = 10), siNC+M group (n = 10) and the METTL3 siRNA1+M group (n = 10). Then, the right forelimb of mice in the siNC+MC group was subcutaneously injected with 100 uL PBS containing 1 × 106 SK-Hep1 cells co-transfected with siNC and MC; the right forelimb of mice in the METTL3 siRNA1+MC group was subcutaneously injected with 100 uL PBS containing 1 × 106 SK-Hep1 cells co-transfected with METTL3 siRNA1 and MC; the right forelimb of mice in the siNC+M group was subcutaneously injected with 100 uL PBS containing 1 × 106 SK-Hep1 cells co-transfected with siNC and M; the right forelimb of mice in the METTL3 siRNA1+M group was subcutaneously injected with 100 uL PBS containing 1 × 106 SK-Hep1 cells co-transfected with METTL3 siRNA1 and M. | |||
| Response Summary | METTL3 up-regulated the expression of hsa-miR-589-5p and promoted the maturation of miR-589-5p. Overexpressed miR-589-5p and METTL3 promoted the viability, migration and invasion of liver cancer cells. | |||
hsa-miR-671-5p
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [21] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
T98G | Glioblastoma | Homo sapiens | CVCL_0556 |
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| LN-18 | Glioblastoma | Homo sapiens | CVCL_0392 | |
| HEB (human normal glial cell line HEB were obtained from Tongpai (Shanghai) biotechnology co., LTD (Shanghai, China)) | ||||
| A-172 | Glioblastoma | Homo sapiens | CVCL_0131 | |
| Response Summary | METTL3-mediated m6A modification upregulated circDLC1 expression, and circDLC1 promoted CTNNBIP1 transcription by sponging hsa-miR-671-5p, thus repressing the malignant proliferation of glioma. | |||
hsa-miR-766-5p
Malignant haematopoietic neoplasm [ICD-11: 2B33]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [242] | |||
| Responsed Disease | Chronic myeloid leukaemia [ICD-11: 2B33.2] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell viability and apoptosis | |||
In-vitro Model |
BV-173 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0181 |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| KCL-22 | Chronic myelogenous leukemia | Homo sapiens | CVCL_2091 | |
| MEG-01 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0425 | |
| Response Summary | METTL3-mediated m6A modification induced the aberrant expression of NEAT1 in chronic myelocytic leukemia. Overexpression of NEAT1 inhibited cell viability and promoted the apoptosis of chronic myelocytic leukemia cells. hsa-miR-766-5p was upregulated in CML PBMCs and abrogated the effects of NEAT1 on cell viability and apoptosis of the chronic myelocytic leukemia cells. | |||
hsa-miR-873-5p
Diseases of the urinary system [ICD-11: GC2Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [397] | |||
| Responsed Disease | Diseases of the urinary system [ICD-11: GC2Z] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Oxidative stress and apoptosis | |||
In-vitro Model |
mRTEC (Mouse renal tubular epithelial cells) | |||
| Response Summary | METTL3 interacts with the microprocessor protein DGCR8 and positively modulates hsa-miR-873-5p mature process in an m6A-dependent manner.METTL3/m6A in colistin-induced nephrotoxicity and provide a new insight on m6A modification in drug induced toxicity. | |||
hsa-miR-99a-5p
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [398] | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HOK | Normal | Hexagrammos otakii | CVCL_YE19 |
| CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 | |
| SCC-9 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1685 | |
| SCC-15 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1681 | |
| UM2 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_VH01 | |
hsa_circ_0000677
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [399] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
NCM460 | Normal | Homo sapiens | CVCL_0460 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HCT 15 | Colon adenocarcinoma | Homo sapiens | CVCL_0292 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 | |
| In-vivo Model | For the tumor xenograft, mice were randomly divided into three groups with four mice for each group. Then, 1 × 106 cells after indicated treatment were harvested and resuspended in 50 ul of PBS. Then the cells were subcutaneously injected into the right front flank of each mouse. | |||
| Response Summary | hsa_circ_0000677 and its downstream target ABCC1 were upregulated in CRC cells, induced by the METTL3-mediated m6 A modification of circ_0000677 and SUMO1-mediated SUMOylation of METTL3. This work provided a new strategy for the therapeutic treatment of CRC. | |||
mmu-miR-365-3p
Chronic pain [ICD-11: MG30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [400] | |||
| Responsed Disease | Chronic pain [ICD-11: MG30] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MicroRNAs in cancer | hsa05206 | ||
| Cell Process | RNA stability | |||
| Response Summary | METTL3 positively modulated the mmu-miR-365-3p processing in a microprocessor protein DiGeorge critical region 8-dependent manner.METTL3-mediated m6A modification in nociceptive sensitization and provide a novel perspective on m6A modification in the development of Inflammatory pain. | |||
mmu-miR-7212-5p
Unspecific body region injury [ICD-11: ND56]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [401] | |||
| Responsed Disease | Unspecific body region injury [ICD-11: ND56] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell differentiation | |||
In-vitro Model |
MC3T3-E1 | Normal | Mus musculus | CVCL_0409 |
| In-vivo Model | A longitudinal incision was made on the skin and the muscles were separated to expose the femur. A transverse osteotomy was performed in the mid-diaphysis of the femur, and the bones were stabilized by inserting a 23-gauge intramedullary needle. Equal amounts (100 uL) of phosphate-buffered saline (PBS), plasmid METTL3 and agomiR-7212-5p (10 mg/kg body weight) were locally injected into the femoral fracture site. | |||
| Response Summary | Down-regulation of METTL3 promotes osteogenic processes both in vitro and in vivo, and this effect is recapitulated by the suppression of mmu-miR-7212-5p maturation. miR-7212-5p inhibits osteoblast differentiation in MC3T3-E1 cells by targeting FGFR3. | |||
hsa_circ_0004771 (circ_NRIP1)
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [166] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
hFOB 1.19 | Normal | Homo sapiens | CVCL_3708 |
| MG-63 | Osteosarcoma | Homo sapiens | CVCL_0426 | |
| U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 | |
| Response Summary | METTL3-mediated hsa_circ_0004771 (circNRIP1) exhibits oncogenic roles in osteosarcoma by regulating FOXC2 via sponging miR-199a, which provides new ideas for the treatment of osteosarcoma. | |||
hsa_circ_0008399 (Circ_RBM3)
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [402] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Protein export | hsa03060 | ||
| Cell Process | Eukaryotic translation | |||
| Cell apoptosis | ||||
In-vitro Model |
5637 | Bladder carcinoma | Homo sapiens | CVCL_0126 |
| RT-4 | Bladder carcinoma | Homo sapiens | CVCL_0036 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| In-vivo Model | Chose 4-week-old female BALB/c nude mice for tumor xenograft experiments, which randomly were divided into four groups (n = 5 per group). Bladder cancer cells (3 × 106) were subcutaneously injected into the right axilla of the nude mice. | |||
| Response Summary | Circ0008399 bound WTAP to promote formation of the WTAP/METTL3/METTL14 m6A methyltransferase complex, reduce cisplatin sensitivity in bladder cancer, implicating the potential therapeutic value of targeting this axis. | |||
hsa_circ_0008542 (Circ_PHF12)
Diseases of the musculoskeletal system [ICD-11: FC0Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [403] | |||
| Responsed Disease | Diseases of the musculoskeletal system [ICD-11: FC0Z] | |||
| Target Regulation | Up regulation | |||
| Cell Process | RNA stability | |||
In-vitro Model |
RAW 264.7 | Mouse leukemia | Mus musculus | CVCL_0493 |
| MC3T3-E1 | Normal | Mus musculus | CVCL_0409 | |
| In-vivo Model | Circ_0008542, where rats were injected with exosomes containing circ_0008542 into the tail vein for 8 weeks. | |||
| Response Summary | METTL3 acts on the m6A functional site of 1956 bp in hsa_circ_0008542. RNA demethylase ALKBH5 inhibits the binding of circ_0008542 with miRNA-185-5p to correct the bone resorption process. | |||
hsa_circ_0021427 (circ_HPS5)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [56] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Transcriptional misregulation in cancer | hsa05202 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell autophagy | ||||
In-vitro Model |
Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| In-vivo Model | To create the xenograft neoplasm system, 40 male BALB/c nude mice aged 5 weeks were randomly separated into sh-NC, sh-circHPS5, sh-circHPS5+CTRL, and sh-circHPS5+SAH groups (n = 5 for each group). HCC cells were subcutaneously injected into the axilla of the nude mice. | |||
| Response Summary | In hepatocellular carcinoma, METTL3 could direct the formation of hsa_circ_0021427 (circHPS5), and specific m6A controlled the accumulation of circHPS5. YTHDC1 facilitated the cytoplasmic output of circHPS5 under m6A modification. CircHPS5 can act as a miR-370 sponge to regulate the expression of HMGA2 and further accelerate hepatocellular carcinoma cell tumorigenesis. | |||
hsa_circ_0029589 (Circ_CHFR)
Acute coronary syndrome [ICD-11: BA4Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [404] | |||
| Responsed Disease | Acute coronary syndrome [ICD-11: BA4Z] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Macrophage pyroptosis | |||
In-vitro Model |
CD14 + cells (CD14+ cells from human peripheral blood) | |||
| Response Summary | The relative RNA expression level of hsa_circ_0029589 in macrophages was decreased, whereas the N6-methyladenosine (m6A) level of hsa_circ_0029589 and the expression of m6A methyltransferase METTL3 were validated to be significantly elevated in macrophages in patients with acute coronary syndrome. | |||
hsa_circ_0058493 (Circ_RHBDD1)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [405] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Hepatocellular carcinoma | hsa05225 | ||
| Cell Process | Cell growth and metastasis | |||
In-vitro Model |
BEL-7404 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6568 |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| SK-HEP-1 | Liver and intrahepatic bile duct epithelial neoplasm | Homo sapiens | CVCL_0525 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| In-vivo Model | Groups of HCT116-Luc-shCtrl, HCT116-Luc-shLINC00460, and HCT116-Luc-shLINC00460 + HMGA1 cells (5 × 106) were injected subcutaneously into the flanks of mice correspondingly. | |||
| Response Summary | In hepatocellular carcinoma, hsa_circ_0058493 contained m6A methylation sites and that METTL3 mediated the degree of methylation modification of hsa_circ_0058493. YTHDC1 could bind to hsa_circ_0058493 and promote its intracellular localization from the nucleus to the cytoplasm. | |||
hsa_circ_0081609 (circ_CUX1)
Hypopharyngeal cancer [ICD-11: 2B6D]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [406] | |||
| Responsed Disease | Hypopharyngeal squamous cell carcinoma [ICD-11: 2B6D.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
CLA-27 cell line (The head and neck tumor cell lines) | |||
| HOK | Normal | Hexagrammos otakii | CVCL_YE19 | |
| SAS | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1675 | |
| SCC-15 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1681 | |
| SCC-4 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1684 | |
| SCC-9 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1685 | |
| Tca8113 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6851 | |
| Response Summary | METTL3-mediated m6A modification plays a key role in stabilizing the expression of hsa_circ_0081609 (circCUX1), thereby inhibiting the expression of caspase 1 and conferring the radiotherapy resistance of hypopharyngeal squamous cell carcinoma. | |||
hsa_circ_0092493 (circ_ARL3)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [379] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Reverse splicing and biogenesis | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | Mice were randomly divided into three groups, and subcutaneously injected with control, circ-ARL3-silenced and circ-ARL3&miR-1305-silenced HepG2.2.15 cells. | |||
| Response Summary | hsa_circ_0092493 (circ_ARL3) is a critical regulator in HBV-related HCC, targeting the axis of circ-ARL3/miR-1305 can be a promising treatment for HBV+ HCC patients. HBx protein upregulated N6 -methyladenosine (m6A) methyltransferases METTL3 expression, increasing the m6A modification of circ-ARL3; then, m6A reader YTHDC1 bound to m6 A-modified of circ-ARL3 and favored its reverse splicing and biogenesis. Furthermore, circ-ARL3 was able to sponge miR-1305, antagonizing the inhibitory effects of miR-1305 on a cohort of target oncogenes, thereby promoting HBV+ HCC progression. | |||
1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-2 (PLCG2)
Nasal-type natural killer/T-cell lymphoma [ICD-11: XH3400]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [407] | |||
| Responsed Disease | Nasal-type natural killer/T-cell lymphoma [ICD-11: XH3400] | |||
| Target Regulation | Up regulation | |||
2,4-dienoyl-CoA reductase [ (DECR1)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [408] | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Down regulation | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| SNU-449 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0454 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
|
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 | |
| In-vivo Model | For subcutaneous HCC mice xenograft model, the cell suspension was injected into the right flank of the mice (n = 5 per group). | |||
AC026356.1
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [409] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
THLE-2 | Normal | Homo sapiens | CVCL_3803 |
| SNU-398 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0077 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| In-vivo Model | Indicated HCC cells were intrasplenically injected into 5-week-old male BALB/C athymic nude mice (SpePharm Biotechnology, Beijing, China). After being fed in specific pathogen free condition for 35 days, the mice were euthanized and the livers were resected and subjected to HE staining. | |||
Acyl-CoA synthetase short-chain family member 3, mitochondrial (ACSS3)
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [410] | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
Alpha-actinin-4 (ACTN4)
SARS-CoV-2 [ICD-11: XN109]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [411] | |||
| Responsed Disease | SARS-CoV-2 [ICD-11: XN109] | |||
In-vitro Model |
A549-ACE2 | Lung adenocarcinoma | Homo sapiens | CVCL_C0Q5 |
| A549-ACE2 | Lung adenocarcinoma | Homo sapiens | CVCL_C0Q5 | |
|
Vero C1008
|
N.A. | Chlorocebus sabaeus | CVCL_0574 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Alpha-aminoadipic semialdehyde dehydrogenase (ALDH7A1)
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [412] | |||
| Responsed Disease | Head and neck squamous carcinoma [ICD-11: 2B6E] | |||
In-vitro Model |
SCC-9 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1685 |
| In-vivo Model | Bmi1CreER and RosatdTomato mice were obtained from The Jackson Laboratory, while Mettl3flox/flox and Mettl3KI/KI mice were provided from the laboratory of Prof. Quan Yuan at the West China Hospital of Stomatology. As previously described, we treated mice with 100 μg/mL 4-nitroquinoline 1-oxide (4NQO) (Sigma, N8141) to induce HNSCC formation. In brief, 6-week-old mice were fed with 4NQO for 16 weeks and then normal water for another 10 weeks. To lineage tracing and conditional knockout and knockin mouse model, tamoxifen (1 mg/mouse/day for 3 days; Sigma, T5648-1G) was injected intraperitoneally after 4NQO treatment. Then, we collected tongue samples, made frozen sections, and observed sections under a fluorescence microscope to calculate the positive percentage of BMI1 cells.+. | |||
Amphiregulin (AREG)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [413] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 |
| PancTu-II | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_4013 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| Human Pancreatic Nestin-Expressing cells (Human Pancreatic Nestin-Expressing cells) | ||||
Angiopoietin-related protein 3 (ANGPTL3)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [414] | |||
| Responsed Disease | Stomach adenocarcinoma [ICD-11: 2B72.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| In-vivo Model | For in vivo tumor formation, AGS cells (5 × 107) stably infected with sh-METTL3 lentivirus or sh-NC control cells were injected subcutaneously into nude mice (n = 3). The volume and size of the tumors were observed and recorded every three days from the third day after inoculation, calculated as 0.5×(length×width2). | |||
Anillin (ANLN)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [415] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
In-vitro Model |
Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 | |
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
| RAW 264.7 | Mouse leukemia | Mus musculus | CVCL_0493 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| In-vivo Model | Using a 100 μl Hamilton Microliter syringe, all the groups of 1 × 107 luciferase-labeled Huh-7 cells in 50 μl 1 x phosphate-buffered saline (PBS) was injected intracardially in the left ventricle of nu/nu, female 4-6-week-old nude mice. | |||
Annexin A2 (ANXA2)
Acute ischemic stroke [ICD-11: 8B11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [416] | |||
| Responsed Disease | Acute ischemic stroke [ICD-11: 8B11] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
Apolipoprotein C-III (APOC3)
COVID-19 [ICD-11: RA01]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [417] | |||
| Responsed Disease | COVID-19 [ICD-11: RA01] | |||
Aryl hydrocarbon receptor (AHR)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [333] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Responsed Drug | Tislelizumab | Approved | ||
In-vitro Model |
U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 |
AT-rich interactive domain-containing protein 1B (ARID1B)
Disorders of refraction [ICD-11: 9D00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [272] | |||
| Responsed Disease | Disorders of refraction [ICD-11: 9D00.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
RF/6A
|
N.A. | Macaca mulatta | CVCL_4552 |
ATP-dependent zinc metalloprotease YME1L1 (YME1L1)
Acute kidney failure [ICD-11: GB60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [338] | |||
| Responsed Disease | Acute kidney failure [ICD-11: GB60] | |||
In-vitro Model |
HK-2 [Human kidney] | Normal | Homo sapiens | CVCL_0302 |
Autophagy related 8 (ATG8)
Chronic respiratory disease originating in the perinatal period [ICD-11: KB29]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [418] | |||
| Responsed Disease | Chronic respiratory disease originating in the perinatal period [ICD-11: KB29.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | Neonatal C57BL/6J, B6.129S4-Ccr2tm1Ifc/J (METTL3-/-), and the respective Wild-Type (WT) control mice were purchased from the Experimental Animal Center of Nanjing Medical University. Mice were divided into three groups: 1) C57BL/6J mice were exposed to 21% oxygen (normoxia, NRMX) for 14 days; 2) C57BL/6J, METTL3-/- and WT control mice were exposed to 80% O2 oxygen (hyperoxia, HYRX) for 28 days; 3) Mice were injected with 15 μg of 3-MA 30 min before hyperoxia exposure. The procedures were performed in accordance with ARRIVE guidelines. | |||
B-cell lymphoma 6 protein (BCL6)
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [419] | |||
| Responsed Disease | Esophageal Squamous Cell Carcinoma [ICD-11: 2B70.1] | |||
| Cell Process | Transcription | |||
| Cell proliferation | ||||
| Cell migration | ||||
| Cell invasion | ||||
Baculoviral IAP repeat-containing protein 5 (BIRC5)
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [420] | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 |
| A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 | |
| In-vivo Model | Four weeks old female BALB/c nude mice were purchased from SJA Laboratory Animal Co, Ltd (Changsha, China), and acclimated in a specific pathogen free environment for 1 week. SKOV3/DDP cells treated with shBIRC5, or shBIRC5 combined with pcDNA-IGF2BP1 plasmid (IGF2BP1) or corresponding negative control were suspended in 100 μL McCoy's 5A medium and implanted subcutaneously into the left frank of mice in 1 × 107 cells/mice. After 7 days incubation, three groups of mice received 10 mg/kg cisplatin respectively, sacrificing and tumor collecting after 30 days. | |||
BDNF/NT-3 growth factors receptor (NTRK2)
Single episode depressive disorder, severe, without psychotic symptoms [ICD-11: 6A70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [421] | |||
| Responsed Disease | Major depressive disorder [ICD-11: 6A70.3] | |||
In-vitro Model |
SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
| In-vivo Model | Rats were housed at 23°C and 55% humidity and were given ad libitum food and water. During acclimatization (1 week), rats were placed randomly (3/cage); however, after initial behavioral testing, they were grouped according to their behavioral phenotypes. All experiments were performed under a light cycle (8:00 AM and 10:00 AM). The protocol to induce LH behavior was approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham. The animal study also adhered to the international guidelines for the use and care of laboratory animals. | |||
BRAF-activated non-protein coding RNA (BANCR)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [422] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
Calcium-binding and coiled-coil domain-containing protein 1 (CALCOCO1)
Breast cancer [ICD-11: 2C60]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [331] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Paclitaxel | Approved | ||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | The experimental mice were housed in an SPF-grade animal laboratory at a temperature of approximately 25 °C, a humidity range of 50%, and an average daylight duration of 12 h. BALB/C female mice at 5-6 weeks of age were taken and prepared for tumor-bearing. After the mice were sacrificed, their tissues were collected, weighed and immediately snap-frozen in liquid nitrogen. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [331] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | STM2457 | Investigative | ||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | The experimental mice were housed in an SPF-grade animal laboratory at a temperature of approximately 25 °C, a humidity range of 50%, and an average daylight duration of 12 h. BALB/C female mice at 5-6 weeks of age were taken and prepared for tumor-bearing. After the mice were sacrificed, their tissues were collected, weighed and immediately snap-frozen in liquid nitrogen. | |||
Caspase-1 (CASP1)
Inflammatory response [ICD-11: MG46]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [322] | |||
| Responsed Disease | Inflammatory response [ICD-11: MG46] | |||
Cathepsin K (CTSK)
Certain specified inflammatory arthropathies [ICD-11: FA27]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [423] | |||
| Responsed Disease | Certain specified inflammatory arthropathies [ICD-11: FA27.0] | |||
| In-vivo Model | Two-to-three-week-old male C57BL/6 mice were randomly divided into five groups based on their body weight: control (deionised water), solvent control (0.5 % CMC Na), T-2 (200 ng/g T-2 toxin), treatment (administered a specially formulated methionine diet one month after exposure), and prevention groups (fed a specially formulated methionine diet). Each group consisted of six mice that received treatment for 8 weeks. The T-2 toxin gavage solution was prepared using 0.5 % CMC Na solution. The standard diet contained 0.57 % methionine, whereas the specially formulated diet contained 1.2 % methionine. | |||
CCN family member 2 (CTGF)
Chronic kidney disease [ICD-11: GB61]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [424] | |||
| Responsed Disease | Chronic kidney disease [ICD-11: GB61] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SV40 MES 13
|
N.A. | Mus musculus | CVCL_5368 |
| NRK-52E | Normal | Rattus norvegicus | CVCL_0468 | |
Other disorders of bladder [ICD-11: GC01]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [425] | |||
| Responsed Disease | Other disorders of bladder [ICD-11: GC01.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 |
| In-vivo Model | A 26 G pediatric venous indwelling catheter (0.6 mm outer diameter) was used for urethral catheterization. To create intravesical obstruction, a 6-0 nonabsorbable polypropylene suture was gently ligatured. Then, after the suture knot was made, the catheter was carefully removed. Rats in the sham-operated group underwent all of the same procedures, except for the urethral ligation step. At the end of the experiments, the rats were killed humanely. | |||
CD40 ligand (CD40L)
Immune-related diseases [ICD-11: 4B4Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [426] | |||
| Responsed Disease | Immune-related diseases [ICD-11: 4B4Z] | |||
| Target Regulation | Down regulation | |||
CD70 antigen (CD70)
Thyroid Cancer [ICD-11: 2D10]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [427] | |||
| Responsed Disease | Papillary thyroid cancer [ICD-11: 2D10.1] | |||
| Responsed Drug | JNJ-74494550 | Phase 2 | ||
| Target Regulation | Down regulation | |||
In-vitro Model |
Nthy-ori 3-1 | Normal | Homo sapiens | CVCL_2659 |
| B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 | |
| K1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_2537 | |
| C-643 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_5969 | |
| BHT-101 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_1085 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| In-vivo Model | NCG mice (female, 4-5 weeks old, purchased from Jiangsu GemPharmatech) were acclimated for 1 week and were selected randomly to subcutaneously (s.c.) injected with 100 μl suspensions of 1 × 106 thyroid cancer cell lines. All mice were housed in animal facility under specific pathogen-free conditions. After 7 day, 5 × 106 huPBMCs were intravenously (i.v.) inoculated into mice. Mice were treated intravenously with anti-PD-1 mAb (nivolumab, 200 μg per injection), Combination (nivolumab + anti-CD70 mAb cusatuzumab, 200 μg per injection each), or PBS once a week. The treatment with M2-TAMs EVs or PBS continued in mice at 2ug EVs per injection at the tumor site twice a week [24]. The body weight of mice and the length (mm) and width (mm) of tumors were monitored every 3 or 4 days. Tumor volume (mm3) was calculated as Length × Width2 × 1/2. Tumor growth analyses were limited to 3 to 4 weeks because following this period mice started to show signs of xenograft-versus-host disease (xGVHD). Therefore, after 14 days or 21 days of PBMCs implantation, the mice were euthanized. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [427] | |||
| Responsed Disease | Papillary thyroid cancer [ICD-11: 2D10.1] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
Nthy-ori 3-1 | Normal | Homo sapiens | CVCL_2659 |
| B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 | |
| K1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_2537 | |
| C-643 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_5969 | |
| BHT-101 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_1085 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| In-vivo Model | NCG mice (female, 4-5 weeks old, purchased from Jiangsu GemPharmatech) were acclimated for 1 week and were selected randomly to subcutaneously (s.c.) injected with 100 μl suspensions of 1 × 106 thyroid cancer cell lines. All mice were housed in animal facility under specific pathogen-free conditions. After 7 day, 5 × 106 huPBMCs were intravenously (i.v.) inoculated into mice. Mice were treated intravenously with anti-PD-1 mAb (nivolumab, 200 μg per injection), Combination (nivolumab + anti-CD70 mAb cusatuzumab, 200 μg per injection each), or PBS once a week. The treatment with M2-TAMs EVs or PBS continued in mice at 2ug EVs per injection at the tumor site twice a week [24]. The body weight of mice and the length (mm) and width (mm) of tumors were monitored every 3 or 4 days. Tumor volume (mm3) was calculated as Length × Width2 × 1/2. Tumor growth analyses were limited to 3 to 4 weeks because following this period mice started to show signs of xenograft-versus-host disease (xGVHD). Therefore, after 14 days or 21 days of PBMCs implantation, the mice were euthanized. | |||
Cell division cycle-associated protein 7 (CDCA7)
Colon cancer [ICD-11: 2B90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [428] | |||
| Responsed Disease | Colon adenocarcinoma [ICD-11: 2B90.Y] | |||
In-vitro Model |
FHC | Normal | Homo sapiens | CVCL_3688 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| In-vivo Model | Under a specific pathogen-free environment with a 12 h light/dark cycle, BALB/C male nude mice (Slaike Jingda Laboratory, Hunan, China) were randomly divided into two groups. Meanwhile, all mice had free access to fresh water and feed. For the COAD model, 5×106 DLD1 cells sh-NC or sh-CDCA7 were suspended in 100 μL PBS and subcutaneously injected into mice(n=5 for each group). During this period, the volume of tumors was continuously examined every week to generate the tumor growth curve. After injection for five weeks, all mice were euthanized, and tumors were subjected to image, weight, and western blot analysis. Meanwhile, Immunohistochemical (IHC) staining was used for the detection of CDCA7-positive expression in dissected tumor tissues. | |||
Centromere protein F (CENPF)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [429] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
| In-vivo Model | Stably transfected GC cells (1 × 106) with sh-CENPF or CENPF overexpression were mixed in 0.1 mL PBS, and then were injected subcutaneously into the groin of 5-week-old BALB/C nude mice (n = 5 each group). After four weeks, the nude mice were sacrificed, and tumor tissues were dissected. Finally, the weight and volume of the tumors were measured. Tumor volume = length × width2 × 1/2.A total of 1 × 106 luciferase labeled GC cells were injected into the spleen of 5-week-old BALB/C nude mice. After 4-5 weeks, bioluminescence signals of liver metastases were detected via an IVIS imaging system (PerkinElmer, Norwalk, Connecticut, USA). Liver tissues were removed for hematoxylin-eosin (HE) (Beyotime, Shanghai, China) stained sections to evaluate metastatic liver lesions. | |||
Centrosomal protein of 170 kDa (CEP170)
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [430] | |||
| Responsed Disease | Esophageal cancer [ICD-11: 2B70] | |||
In-vitro Model |
KYSE-450 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1353 |
chromosome 2 open reading frame 92 (C2orf92)
Thyroid Cancer [ICD-11: 2D10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [431] | |||
| Responsed Disease | Papillary thyroid cancer [ICD-11: 2D10.1] | |||
| Target Regulation | Up regulation | |||
Circ_1662
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [124] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Cell invasion | |||
| Cell migration | ||||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| In-vivo Model | BALB/c nude mice (4 weeks old) were acquired from Vital River Laboratory (Beijing, China). HCT116 cells with stable circ1662 expression (2 × 106 in 100 L of PBS) were injected via the tail vein. After 45 days, the mice were sacrificed. The lung metastatic carcinoma specimens were processed into paraffin-embedded sections for subsequent H&E staining and IHC. | |||
| Response Summary | METTL3-induced Circ_1662 promoted colorectal cancer cell invasion and migration by accelerating YAP1 nuclear transport. Circ1662 enhanced CRC invasion and migration depending on YAP1 and SMAD3. This result implies that circ1662 is a new prognostic and therapeutic marker for CRC metastasis. | |||
Circ_ABCC4
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [432] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
Circ_ASK1
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [433] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Responsed Drug | Gefitinib | Approved | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 |
| SK-LU-1 | Lung adenocarcinoma | Homo sapiens | CVCL_0629 | |
| NCI-H1993 | Lung adenocarcinoma | Homo sapiens | CVCL_1512 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| 16HBE14o- | Normal | Homo sapiens | CVCL_0112 | |
| In-vivo Model | Established a xenograft model in BALB/c nude mice by inoculating HCC827-GR cells transfected with the constructs for circASK1 silencing, ASK1-272a.a overexpression and ASK1-272a.a overexpression/circASK1 knockdown. | |||
| Response Summary | Increased YTHDF2-mediated endoribonucleolytic cleavage of m6A-modified Circ_ASK1 accounts for its downregulation in gefitinib-resistant cells. Either METTL3 silencing or YTHDF2 silencing suppressed the decay of circASK1 in HCC827-GR cells. This study provides a novel therapeutic target to overcome gefitinib resistance in LUAD patients. | |||
Circ_ASXL1
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [434] | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 |
| OVCAR-3 | Ovarian serous adenocarcinoma | Homo sapiens | CVCL_0465 | |
| ES-2 | Ovarian clear cell adenocarcinoma | Homo sapiens | CVCL_3509 | |
| HEY | Ovarian serous adenocarcinoma | Homo sapiens | CVCL_0297 | |
| A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 | |
| COV362 | Ovarian serous adenocarcinoma | Homo sapiens | CVCL_2420 | |
| In-vivo Model | SKOV3 cells were treated with sh-NC or sh-circASXL1. 100 μL of PBS containing 1 × 106 cells were implanted into mice. Tumor volume was measured every 5 days. After 30 days, mice were euthanized. The tumor was excised and weighed. | |||
Circ_DLC1
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [21] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
T98G | Glioblastoma | Homo sapiens | CVCL_0556 |
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| LN-18 | Glioblastoma | Homo sapiens | CVCL_0392 | |
| HEB (human normal glial cell line HEB were obtained from Tongpai (Shanghai) biotechnology co., LTD (Shanghai, China)) | ||||
| A-172 | Glioblastoma | Homo sapiens | CVCL_0131 | |
| Response Summary | METTL3-mediated m6A modification upregulated Circ_DLC1 expression, and circDLC1 promoted CTNNBIP1 transcription by sponging miR-671-5p, thus repressing the malignant proliferation of glioma. | |||
Circ_EPHB4
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [435] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulation | Up regulation | |||
Circ_IGF2BP3
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [436] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
| Cell Process | Immunity | |||
In-vitro Model |
SW900 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1731 |
| SK-MES-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0630 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1703 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1490 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| 16HBE14o- | Normal | Homo sapiens | CVCL_0112 | |
| In-vivo Model | A total of 106 LLC cells transfected with the following constructs were injected subcutaneously into C57BL/6 mice. | |||
| Response Summary | In non-small-cell lung carcinoma, METTL3 mediates the N6-methyladenosine (m6A) modification of Circ_IGF2BP3 and promotes its circularization in a manner dependent on the m6A reader protein YTHDC1. | |||
Circ_MIRLET7BHG
BackgroundAllergic rhinitis [ICD-11: CA08]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [437] | |||
| Responsed Disease | BackgroundAllergic rhinitis [ICD-11: CA08.0] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | Briefly, the mice were intraperitoneally injected with 25 μg OVA mixed with 1 mg aluminum hydroxide gel was intraperitoneally injected into mice once a week for three weeks. Subsequently, nostril challenge with 500 μg OVA was performed once a day for one week. The control mice were exposed to PBS (vehicle). LV-sh-NC or LV-sh-circMIRLET7BHG (1 × 107 TU/mL, GenePharma) were injected into mice via the tail vein two days before the nostril challenge. | |||
Circ_MYO1C
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [438] | |||
| Responsed Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 |
| Capan-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0026 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| In-vivo Model | Male BALB/c nude mice (5-6 weeks) were obtained from Slac Laboratory Animal Center (Shanghai, China) and maintained under pathogen-free conditions. PANC-1 cells (2 × 106 cells suspended in 100 μl PBS) transfected with circMYO1C knockdown (sh-circMYO1C) or controls (sh-NC) were subcutaneously injected into the flank of nude mice. One week later, the tumor size was measured every three days. | |||
Circ_PRKAR1B
Crohn disease [ICD-11: DD70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [439] | |||
| Responsed Disease | Crohn disease [ICD-11: DD70.3] | |||
| In-vivo Model | Twelve-week-old wild-type (WT) and IL-10-KO mice were allocated randomly to four groups (n = 5 per group): (1) the WT group; (2) the IL-10 KO + si-control group (administrated saline as a placebo via enema once); (3) the IL-10 KO + si-circPRKAR1B group (administrated 200 μL of 1e11 vg adeno-associated virus (AAV) vector carrying si-circPRKAR1B via enema once); and (4) the IL-10 KO + si-METTL3 group (administrated 200 μL of 1e11 vg AAV vector carrying si-METTL3 via enema once). | |||
Circ_RNF13
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [440] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| C-4-I | Cervical squamous cell carcinoma | Homo sapiens | CVCL_2253 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| In-vivo Model | Stably transfected cell lines were created by silencing circRNF13 in CC SiHa cells. Once xenografts were established, the tumors reached an approximate volume of 200 mm3. A single dose of 15 Gy irradiation was administered to female BALB/c nude mice (4-5 weeks old) in the murine model. The tumor volume was measured and recorded using vernier calipers every five days after irradiation. After 30 days, the mice were euthanized under anesthesia, and tumor tissue was collected for further investigations. | |||
Circ_SETD2
Pre-eclampsia [ICD-11: JA24]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [441] | |||
| Responsed Disease | Pre-eclampsia [ICD-11: JA24] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | Pregnant rats were randomly assigned to 5 groups: normal pregnancy (normal) group, PE group, circSETD2 overexpression (PE + oe-circSETD2) group, METTL3 overexpression (PE + oe-METTL3) group, overexpression control (PE + oe-NC) group, joint treatment (PE + oe-METTL3 + si-circSETD2) group, and joint treatment control (PE + oe-METTL3 + si-NC) group, with 6 rats in each group. After continuous injection of L-NAME for 4 days, the rats in the overexpression and overexpression control groups were further injected with 30 μl lentivirus vector oe-circSETD2, oe-METTL3 or oe-NC solution (GenePharma, Shanghai, China) through the tail vein; after continuous injection of L-NAME for 4 days, the rats in the PE + oe-METTL3 + si-circSETD2 group were further injected with 30 μl lentiviral vector oe-circSETD2 and si-circSETD2 (GenePharma) at the virus titer of 5 × 107 PFU/mL. The rats were euthanized on the 16th day of pregnancy. Then, the chorionic trophoblast tissues were isolated from the placenta of pregnant rats and divided into 2 parts, with 1 part for extraction and detection of tissue RNA, and the other part fixed in the buffer containing 10% formalin, embedded in paraffin, and made into 5 μm sections. | |||
Circ_UHRF2
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [442] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| In-vivo Model | To create a xenograft model, 1 × 107 SW620 cells stably transfected shMETTL3 and shcircUHRF2 were subcutaneously injected into the nude mice. Tumor sizes were calculated by measuring the length and width (V = length × width2/2). Mice were euthanized four weeks later, and tumors were weighed. For in vivo liver metastasis assay, 1 × 106 SW620 cells stably transfected shMETTL3 and shcircUHRF2 were injected into the distal tip of the spleen of mice according to previous studies. | |||
Claspin (CLSPN)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [267] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Responsed Drug | Celastrol | Preclinical | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| In-vivo Model | AsPC-1 cells suspended in 100 μl of PBS (2 × 106 cells/100 μl) were injected subcutaneously into the lateral flank of the mice, which were randomly divided into solvent group (n = 6), 1.0 mg/kg of celastrol group (n = 6) and 3.0 mg/kg of celastrol group (n = 6). The administration of celastrol was performed by intraperitoneal injection into tumor-bearing mice every 2 day after 10 day inoculation. The tumor sizes were monitored with calipers every 5 days, and the tumor volume was calculated with the formula: Volume (mm3) = 1/2 × length × width2. | |||
Cohesin subunit SA-3 (STAG3)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [443] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| In-vivo Model | Oe-STAG3 and sh-METTL3 were transfected into HCT116 cells. Trypsin digestion and cell counting were performed on each set of cells after they had reached around 80% confluence. Nude mice were subcutaneously injected with 200 μL cells (2 × 106) and randomly assigned to 5 groups (n = 6 in each group): control group (mice were injected with cells without transfection plasmid), oe-NC group (mice were injected with cells transfected with oe-NC), oe-STAG3 group (mice were injected with cells transfected with oe-STAG3), oe-STAG3 + sh-NC group (mice were injected with cells transfected with oe-STAG3 and sh-NC) and oe-STAG3 + sh-METTL3 group. | |||
Collagen alpha-1 (COL1A1)
Keloid [ICD-11: EE60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [444] | |||
| Responsed Disease | Keloid [ICD-11: EE60] | |||
| Target Regulation | Up regulation | |||
| Cell Process | mRNA stability | |||
In-vitro Model |
CHSE-EC
|
N.A. | Oncorhynchus tshawytscha | CVCL_DG46 |
| In-vivo Model | Six- to eight-week-old C57BL/6 mice were purchased from Cyagen Biosciences, Inc. (Cyagen, Suzhou, China). Alkbh3-/- C57BL/6 mice were generated via the CRISPR/Cas9 system. Single-guide RNAs (sgRNAs) were designed to target exons 3 to 5 of Alkbh3 and were coinjected with Cas9 into the zygotes. The pups obtained were genotyped by PCR. After genotyping, the F0 mice were subjected to serial mating to generate homozygous mutant offspring. | |||
Collagen alpha-2(I) chain (COL1A2)
Vascular disorders of the liver [ICD-11: DB98]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [307] | |||
| Responsed Disease | Vascular disorders of the liver [ICD-11: DB98.8] | |||
Complement C1q subcomponent subunit A (C1QA)
Diffuse large B-cell lymphomas [ICD-11: 2A81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [445] | |||
| Responsed Disease | Diffuse large B-cell lymphomas [ICD-11: 2A81] | |||
| Responsed Drug | Rituximab | Approved | ||
| Target Regulation | Down regulation | |||
| In-vivo Model | Approximately 2 × 106 Farage/R or Farage/S cells stably transfected with shNC, shC1qA or shMETTL3 were subcutaneously injected into the left flank of each mouse. When the tumor volume reached ~100 mm3, each mouse received an intraperitoneal injection of Rituximab (20 mg/kg) every 4 days for a total of 5 injections. The diameter of each tumor was examined every 4 days using a caliper, and tumor volume was calculated as follows: (length × width2)/2. At 28 days after xenograft, the mice were sacrificed and the tumors were weighed and collected. | |||
CX3C chemokine receptor 1 (CX3CR1)
Inflammatory response [ICD-11: MG46]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [322] | |||
| Responsed Disease | Inflammatory response [ICD-11: MG46] | |||
Cyclic AMP-responsive element-binding protein 1 (CREB1)
Single episode depressive disorder, severe, without psychotic symptoms [ICD-11: 6A70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [421] | |||
| Responsed Disease | Major depressive disorder [ICD-11: 6A70.3] | |||
In-vitro Model |
SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
| In-vivo Model | Rats were housed at 23°C and 55% humidity and were given ad libitum food and water. During acclimatization (1 week), rats were placed randomly (3/cage); however, after initial behavioral testing, they were grouped according to their behavioral phenotypes. All experiments were performed under a light cycle (8:00 AM and 10:00 AM). The protocol to induce LH behavior was approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham. The animal study also adhered to the international guidelines for the use and care of laboratory animals. | |||
Cyclin-dependent kinase 2-associated protein 2 (CDK2AP2)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [446] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
Cysteine methyltransferase DNMT3A (DNMT3A)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [447] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [291] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
Alzheimer disease [ICD-11: 8A20]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [448] | |||
| Responsed Disease | Alzheimer disease [ICD-11: 8A20] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
Cysteine protease ATG4A (ATG4A)
Intervertebral disc degeneration [ICD-11: FA80]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [449] | |||
| Responsed Disease | Intervertebral disc degeneration [ICD-11: FA80] | |||
| Target Regulation | Down regulation | |||
Cytochrome c oxidase subunit 4 isoform 1, mitochondrial (COX4I1)
Asthma [ICD-11: CA23]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [450] | |||
| Responsed Disease | Asthma [ICD-11: CA23] | |||
| Target Regulation | Down regulation | |||
Cytochrome P450 2B6 (CYP2B6)
Insulin resistance [ICD-11: 5A44]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [451] | |||
| Responsed Disease | Insulin resistance [ICD-11: 5A44] | |||
| Responsed Drug | STM2457 | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | C57/BL6 male mice (eight per group) were randomly divided into four groups: basal CD group, HFD + 0.9% NaCl injection group, HFD + DMSO injection group, and HFD + STM2457 (Catalog No. 2499663-01-1, Selleck Chemicals, Houston, Texas) injection group. When the mean body weight of mice in HFD group was statistically different from that of mice in the control group [(mean body weight in HFD:mean body weight in CD)/mean body weight in CD > 30%], the treatment was started once a day for one or two weeks. STM2457 was administered at a dose of 50 mg/kg and weighed prior to each injection. DMSO or saline was also administered at an equal volume. Body weight was recorded every day. Injection of STM2457 was conducted daily and totally for one or two weeks. | |||
D-3-phosphoglycerate dehydrogenase (PHGDH)
Liver cancer [ICD-11: 2C12]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [452] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Lenvatinib | Approved | ||
In-vitro Model |
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| HepG3 | Hepatoblastoma | Homo sapiens | CVCL_Y479 | |
| In-vivo Model | For the subcutaneous injection model, 3 × 106 MHCC97L cells were washed with PBS thrice and resuspended in 100 μL 1:1 Matrixgel and DMEM-HG. HDTVi was performed as previously described.44 Plasmids were extracted using an EndoFree Plasmid extraction kit (QIAGEN). Plasmid sequence was confirmed using Sanger sequencing. A 2 mL mixture of 20 μg sgTp53 plasmids, 10 μg Myc overexpression plasmids, and 2 μg SB13 plasmids was dissolved in saline of 10% mice body weight and was tail-vein injected into C57BL/6 mice weighing at least 25 g. Plasmids were co-injected in the lateral tail vein to induce spontaneous HCC formation. Three weeks after HDTVi, mice were randomly subjected to vehicle or STM2457 treatment. STM2457 (DC chemicals) was dissolved in 20% (w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) and injected intraperitoneally. Sorafenib powder (Selleckchem) was dissolved in distilled water and administrated via oral gavage. A 5 days on/3 days off treatment regimen was applied. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [452] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
In-vitro Model |
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | For the subcutaneous injection model, 3 × 106 MHCC97L cells were washed with PBS thrice and resuspended in 100 μL 1:1 Matrixgel and DMEM-HG. HDTVi was performed as previously described.44 Plasmids were extracted using an EndoFree Plasmid extraction kit (QIAGEN). Plasmid sequence was confirmed using Sanger sequencing. A 2 mL mixture of 20 μg sgTp53 plasmids, 10 μg Myc overexpression plasmids, and 2 μg SB13 plasmids was dissolved in saline of 10% mice body weight and was tail-vein injected into C57BL/6 mice weighing at least 25 g. Plasmids were co-injected in the lateral tail vein to induce spontaneous HCC formation. Three weeks after HDTVi, mice were randomly subjected to vehicle or STM2457 treatment. STM2457 (DC chemicals) was dissolved in 20% (w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) and injected intraperitoneally. Sorafenib powder (Selleckchem) was dissolved in distilled water and administrated via oral gavage. A 5 days on/2 days off treatment regimen was applied. | |||
| Experiment 3 Reporting the m6A-centered Disease Response of This Target Gene | [452] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | STM2457 | Investigative | ||
In-vitro Model |
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | For the subcutaneous injection model, 3 × 106 MHCC97L cells were washed with PBS thrice and resuspended in 100 μL 1:1 Matrixgel and DMEM-HG. HDTVi was performed as previously described.44 Plasmids were extracted using an EndoFree Plasmid extraction kit (QIAGEN). Plasmid sequence was confirmed using Sanger sequencing. A 2 mL mixture of 20 μg sgTp53 plasmids, 10 μg Myc overexpression plasmids, and 2 μg SB13 plasmids was dissolved in saline of 10% mice body weight and was tail-vein injected into C57BL/6 mice weighing at least 25 g. Plasmids were co-injected in the lateral tail vein to induce spontaneous HCC formation. Three weeks after HDTVi, mice were randomly subjected to vehicle or STM2457 treatment. STM2457 (DC chemicals) was dissolved in 20% (w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) and injected intraperitoneally. Sorafenib powder (Selleckchem) was dissolved in distilled water and administrated via oral gavage. A 5 days on/4 days off treatment regimen was applied. | |||
D26496
Injury of nerves at hip or thigh level [ICD-11: NC74]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [453] | |||
| Responsed Disease | Injury of nerves at hip or thigh level [ICD-11: NC74.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
Schwann cells (A type of glial cell that surrounds neurons) | |||
| In-vivo Model | Rats were randomly divided into 4 groups: sham group (6 rats), SNI model group (24 rats), SNI + vector group (6 rats), and SNI + pcDNA-D26496 group (6 rats). D26496 overexpressed Adeno associated virus (AAVs, virus titer: 1.88E + 13) and vector (OBiO Technology Corp., Shanghai, China) were injected into the epineurium of sciatic nerve of rats (5 μL for each) in the SNI + vector group and SNI + pcDNA-D26496 group, respectively. | |||
Discoidin domain-containing receptor 2 (DDR2)
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [454] | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 |
| In-vivo Model | BALB nude mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. SKOV3 cells were transfected with sh-METTL3, sh-DDR2, DDR2 over-expression plasmid (oe-DDR2) or negative control. About one week later, the above SKOV3 cells were digested into single cells and subcutaneously injected to nude mice in different groups. After a duration of 4 weeks, the mice were euthanized by intraperitoneal injection of pentobarbital at a dosage 120 mg/kg. Finally, the subcutaneous tumors were removed to measure the size and volume (calculated according to volume = L (longest diameter) x W2 (shorter diameter)/2). | |||
DNA (cytosine-5)-methyltransferase 1 (DNMT1)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [455] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| In-vivo Model | To establishment the xenograftmodels, twenty mice were randomly divided into four groups, each of which contained five nude mice. In the sh-NC group, 4 × 106 A549 cells infected with LV-sh-NC were diluted in 100 μl of PBS and injected subcutaneously into nude mice. In the sh-DNMT1 group, 4 × 106 A549 cells infected with LV-sh-DNMT1 were diluted in 100 μl of PBS and injected subcutaneously into nude mice. In the sh-DNMT1 + sh-FOXO3a group, 4 × 106 A549 cells infected with LV-sh-DNMT1 and LV-sh-NC were diluted in 100 μl of PBS and injected subcutaneously into nude mice. | |||
DNA-binding protein SATB2 (SATB2)
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [419] | |||
| Responsed Disease | Esophageal Squamous Cell Carcinoma [ICD-11: 2B70.1] | |||
| Cell Process | Transcription | |||
| Cell proliferation | ||||
| Cell migration | ||||
| Cell invasion | ||||
Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2 (RPN2)
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [456] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
In-vitro Model |
5637 | Bladder carcinoma | Homo sapiens | CVCL_0126 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
Dynamin-1-like protein (DRP1)
Acute myocardial infarction [ICD-11: BA41]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [457] | |||
| Responsed Disease | Acute myocardial infarction [ICD-11: BA41] | |||
| Target Regulation | Up regulation | |||
E3 ubiquitin-protein ligase CHIP (STUB1)
Alzheimer disease [ICD-11: 8A20]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [458] | |||
| Responsed Disease | Alzheimer disease [ICD-11: 8A20] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
| HT22 | Normal | Mus musculus | CVCL_0321 | |
eIF5-mimic protein 1 (BZW2)
Multiple myeloma [ICD-11: 2A83]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [459] | |||
| Responsed Disease | Multiple myeloma [ICD-11: 2A83.1] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
NCI-H929 | Plasma cell myeloma | Homo sapiens | CVCL_1600 |
| MM1.S | Plasma cell myeloma | Homo sapiens | CVCL_8792 | |
| U266B1 | Plasma cell myeloma | Homo sapiens | CVCL_0566 | |
| INA-6 | Plasma cell myeloma | Homo sapiens | CVCL_5209 | |
| OPM-2 | Plasma cell myeloma | Homo sapiens | CVCL_1625 | |
| In-vivo Model | MM1S cells infected with sh-METTL3/sh-NC (selected by 2 μg/mL puromycin) were subcutaneously injected into the left abdominal flanks of NOD-SCID mice (SPF Biotechnology Co., Ltd., Beijing, China) (n = 6/group). Tumor volume was measured every 7 days using the calipers. After 28 days, the mice were sacrificed, and tumor tissues were collected, weighed, and photographed. One part of the tissue was used to detect the expression of METTL3 and BZW2 using qRT-PCR and WB analysis, and the other part was prepared into paraffin sections and then performing Ki-67 immumohistochemical (IHC) staining according to the SP kit (Solarbio, Beijing, China) instructions. | |||
Ena/VASP-like protein (EVL)
Macroscopic changes of size of the kidney [ICD-11: MF54]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [460] | |||
| Responsed Disease | Macroscopic changes of size of the kidney [ICD-11: MF54.0] | |||
| Responsed Drug | Isoforsythiaside | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
HEK293 | Normal | Homo sapiens | CVCL_0045 |
| HK-2 [Human kidney] | Normal | Homo sapiens | CVCL_0302 | |
| In-vivo Model | For the unilateral ureteral obstruction (UUO)-induced ON model, mice were subjected to permanent ureteral ligation of the left kidney ureter under aseptic and anaesthetic conditions, whereas sham-operated mice merely lacked ligation under equivalent conditions. The mice were sacrificed under anaesthesia at 3, 7 and 14 days postoperatively. For the ischemia/reperfusion (I/R)-induced renal fibrosis model, mice were recovered with blood supply after 42 min of bilateral renal artery clamping under both aseptic and anaesthetic conditions. These mice were sacrificed under anaesthesia at 7 and 14 days postoperatively. For drug treatment, isoforsythiaside was injected intraperitoneally at concentrations of 10, 25 and 50 mg/kg per day when the model was established. Similarly, the sham-operated group received an equivalent volume of saline intraperitoneally as a control. The harvested kidneys have been primarily used for morphological and histopathological observations and molecular biology studies. | |||
EN_42575
Change in bowel habit [ICD-11: ME05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [461] | |||
| Responsed Disease | Change in bowel habit [ICD-11: ME05.1] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
IPEC-J2
|
N.A. | Sus scrofa | CVCL_2246 |
F-box only protein 43 (FBXO43)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [462] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
Ferroptosis suppressor protein 1 (AIFM2)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [463] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Responsed Drug | iFSP1 | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
U-251MG | Astrocytoma | Homo sapiens | CVCL_0021 |
| In-vivo Model | Briefly, the mice were anesthetized and placed in a stereotactic frame. A burr hole was drilled into the skull (1.0 mm posterior and 3.0 mm lateral to the bregma). A 50 μL microinjector was used to inject 10 μL of 1 × 106 U251 cells suspended in the serum-free DMEM. The injection was performed slowly within 10 min and stayed for 10 min. After that, mice in cages and a cat were placed in the same room for fear stress induction . | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [464] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
FGD5 antisense RNA 1 (FGD5-AS1)
Endometrial cancer [ICD-11: 2C76]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [465] | |||
| Responsed Disease | Endometrial cancer [ICD-11: 2C76] | |||
| Responsed Drug | Paclitaxel | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
Ishikawa | Endometrial adenocarcinoma | Homo sapiens | CVCL_2529 |
| HHUA | Endometrial adenocarcinoma | Homo sapiens | CVCL_3866 | |
Fibronectin (FN1)
Keloid [ICD-11: EE60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [444] | |||
| Responsed Disease | Keloid [ICD-11: EE60] | |||
| Target Regulation | Up regulation | |||
| Cell Process | mRNA stability | |||
In-vitro Model |
CHSE-EC
|
N.A. | Oncorhynchus tshawytscha | CVCL_DG46 |
| In-vivo Model | Six- to eight-week-old C57BL/6 mice were purchased from Cyagen Biosciences, Inc. (Cyagen, Suzhou, China). Alkbh3-/- C57BL/6 mice were generated via the CRISPR/Cas9 system. Single-guide RNAs (sgRNAs) were designed to target exons 3 to 5 of Alkbh3 and were coinjected with Cas9 into the zygotes. The pups obtained were genotyped by PCR. After genotyping, the F0 mice were subjected to serial mating to generate homozygous mutant offspring. | |||
Forkhead box protein P3 (FOXP3)
Lupus erythematosus [ICD-11: 4A40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [466] | |||
| Responsed Disease | Systemic lupus erythematosus [ICD-11: 4A40.0] | |||
| Target Regulation | Up regulation | |||
Fos-related antigen 2 (FOSL2)
Ovarian dysfunction [ICD-11: 5A80]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [467] | |||
| Responsed Disease | Polycystic ovary syndrome [ICD-11: 5A80.1] | |||
| Responsed Drug | Short-chain fatty acid-butyric acid | Phase 4 | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
KGN | Ovarian granulosa cell tumor | Homo sapiens | CVCL_0375 |
| In-vivo Model | Specific-pathogen-free (SPF) grade C57/B6 mice (6-8 weeks old) were purchased from Weitong Lihua Co. (Beijing, China). The mice were housed at constant temperature and humidity, and 12 h light/dark cycle a day and provided with adequate food and water. | |||
Glucocorticoid receptor (NR3C1)
Single episode depressive disorder, severe, without psychotic symptoms [ICD-11: 6A70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [421] | |||
| Responsed Disease | Major depressive disorder [ICD-11: 6A70.3] | |||
In-vitro Model |
SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
| In-vivo Model | Rats were housed at 23°C and 55% humidity and were given ad libitum food and water. During acclimatization (1 week), rats were placed randomly (3/cage); however, after initial behavioral testing, they were grouped according to their behavioral phenotypes. All experiments were performed under a light cycle (8:00 AM and 10:00 AM). The protocol to induce LH behavior was approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham. The animal study also adhered to the international guidelines for the use and care of laboratory animals. | |||
Glucose-6-phosphate exchanger SLC37A2 (SLC37A2)
Inflammatory bowel disease [ICD-11: DD7Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [468] | |||
| Responsed Disease | Inflammatory bowel disease [ICD-11: DD7Z] | |||
In-vitro Model |
RAW 264.7 | Mouse leukemia | Mus musculus | CVCL_0493 |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| In-vivo Model | Mice were randomly allocated into three groups (n = 5/group): the negative control (NEG) group, the DSS-induced colitis (DSS) group, and the hucMSC-Ex-treated colitis (hucMSC-Ex) group. Mice in the NEG group were fed autoclaved water during the experiment, while mice in the DSS and hucMSC-Ex groups were fed autoclaved water containing 3% DSS (MP Biomedicals, USA). A total of 1 mg of hucMSC-Ex was administered by caudal vein to mice in the hucMSC-Ex group on the 3rd, 6th, and 9th day of modeling, while mice in other groups were injected with PBS. All mice were sacrificed when severe hematochezia and weight loss were observed in the DSS group. The disease progression of mice in the different groups was evaluated by weight change, DAI score, colorectal length, and weight ratio. After the mice were sacrificed, the general view of the spleen and colorectum of the mice was observed. The colorectal mucosa and spleen tissues of the mice in each group were separated, and RNA and protein of the tissues were extracted for subsequent experiments. | |||
glucosylceramidase beta 1 like, pseudogene (GBA1LP)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [469] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | In vivo tumor growth assay, 1 × 107 Hep3B-GBAP1 or MHCC97H-shGBAP1 subclones and the corresponding control cells were transplanted into the body of 6-week-old BALB/c nude mice (Slac Laboratory Animal Center, Shanghai, China) via subcutaneous injection. Tumor size was measured every 3 days. Twenty-one days later, tumors were removed from mice in different groups. Tumor weight was calculated after dissection. Permission of conducting animal study was obtained from the Research Ethics Committee of Xi'an Jiaotong University. In vivo lung metastasis model, 1 × 106 cells were intravenously injected into the lateral tail vein of nude mice (n = 5 mice/group). | |||
Glutaredoxin-1 (GLRX)
Parkinson disease [ICD-11: 8A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [470] | |||
| Responsed Disease | Parkinson disease [ICD-11: 8A00] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
BV-2 | Normal | Mus musculus | CVCL_0182 |
Glycogen phosphorylase, brain form (PYGB)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [471] | |||
| Responsed Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
In-vitro Model |
BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| HPDE | Normal | Homo sapiens | CVCL_4376 | |
| In-vivo Model | Six BALB/c nude mice (male, 4 weeks) were randomly divided into 2 groups and inoculated with BXPC-3 cells (1 × 106) transfected with shPYGB or shNC by subcutaneous injection. Tumor volume was recorded every 7 days and mice were sacrificed after 6 weeks. To evaluate metastasis in vivo, transfected cells (5 × 106) were injected into nude mice through the tail vein. | |||
Guanine nucleotide-binding protein G (GNAO1)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [472] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Responsed Drug | Temozolomide | Approved | ||
Heme oxygenase 1 (HMOX1)
Acute ischemic stroke [ICD-11: 8B11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [473] | |||
| Responsed Disease | Acute ischemic stroke [ICD-11: 8B11] | |||
| Target Regulation | Up regulation | |||
Diseases of the female genital system [ICD-11: GA6Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [474] | |||
| Responsed Disease | Diseases of the female genital system [ICD-11: GA6Z] | |||
| Target Regulation | Down regulation | |||
| In-vivo Model | Daily vaginal smears were collected between 4 and 6 PM, and rats showing four consecutive 4-day estrus cycles were chosen for constructing the endometrial injury model. Anesthesia was induced by injecting 3% mebumalnatrium (dose: 1.3 mL/kg) into the lumbar region, followed by a low midline abdominal incision for uterus exposure. | |||
Histone acetyltransferase KAT2A (KAT2A)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [475] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Target Regulation | Down regulation | |||
Histone deacetylase 9 (HDAC9)
Acute laryngitis or tracheitis [ICD-11: CA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [476] | |||
| Responsed Disease | Acute laryngitis or tracheitis [ICD-11: CA05] | |||
In-vitro Model |
TPC-1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_6298 |
| B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 | |
Histone-lysine N-methyltransferase ASH1L (ASH1L)
Uveitis [ICD-11: 9A96]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [477] | |||
| Responsed Disease | Uveitis [ICD-11: 9A96] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
TH1
|
N.A. | Homo sapiens | CVCL_5J51 |
Histone-lysine N-methyltransferase NSD2 (NSD2)
Chronic kidney disease [ICD-11: GB61]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [478] | |||
| Responsed Disease | Diabetic nephropathy [ICD-11: GB61.Z] | |||
| Target Regulation | Up regulation | |||
Histone-lysine N-methyltransferase SETD2 (SETD2)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [479] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
|
HBE1
|
N.A. | Homo sapiens | CVCL_0287 | |
Histone-lysine N-methyltransferase SETMAR (SETMAR)
Thyroid Cancer [ICD-11: 2D10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [480] | |||
| Responsed Disease | Thyroid Cancer [ICD-11: 2D10] | |||
| Target Regulation | Up regulation | |||
Histone-lysine N-methyltransferase SUV39H2 (SUV39H2)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [481] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | The nude mice were inoculated in the inguinal region subcutaneously with 1 × 106 stably transfected GC cells which were suspended in 100 μL PBS. After one week, preestablished tumor xenografts were treated with DMSO or OTS186935 (50 mg/kg, 2 × /wk × 3). | |||
Homeobox protein DLX-3 (DLX3)
Structural developmental anomalies of teeth and periodontal tissues [ICD-11: LA30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [482] | |||
| Responsed Disease | Structural developmental anomalies of teeth and periodontal tissues [ICD-11: LA30.0] | |||
Homeobox protein Hox-C10 (HOXC10)
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [483] | |||
| Responsed Disease | Head and neck squamous carcinoma [ICD-11: 2B6E] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
In-vitro Model |
Tu 686 | Laryngeal squamous cell carcinoma | Homo sapiens | CVCL_4916 |
| FaDu | Hypopharyngeal squamous cell carcinoma | Homo sapiens | CVCL_1218 | |
|
HuLa-PC
|
N.A. | Homo sapiens | CVCL_UC74 | |
| In-vivo Model | Animals had free access to standard rodent chow and water. For the subcutaneous xenograft tumor model, transfected cells (5×106) were injected on the sides of the flanks of mice (n=5 per group). Subsequently, 30 days later, the mice were euthanized by cervical dislocation following anesthesia induced by intraperitoneal injection of 0.3% sodium pentobarbital (30 mg/kg). Euthanasia was confirmed by verifying respiratory and cardiac arrest, along with pupil dilation, for a minimum of 10 min. Tumor size and tumor weight were measured (max tumor diameter, 9.6 mm; max area, 82.56 mm2; max volume, 355.01 mm3). For the pulmonary metastasis model, transfected cells (2×106) were injected into the mouse tail veins (n=5 per group). Subsequently, 60 days later, the mice were sacrificed, and lungs were obtained. Finally, lung and tumor tissues were available for H&E staining at room temperature for 5 min or immunohistochemistry staining. | |||
Homeobox protein Hox-C6 (HOXC6)
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [484] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
In-vitro Model |
VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| HNC PC3 | Retromolar trigone squamous cell carcinoma | Homo sapiens | CVCL_C8XA | |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
HOXA transcript antisense RNA, myeloid-specific 1 (HOTAIRM1)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [485] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 |
| In-vivo Model | BALB/c nude mice (4 weeks old, male) were purchased from Vital River Laboratory Animals and maintained under specific pathogen-free conditions. For in vivo tumor xenograft model establishment, 10 mice were randomly chosen and divided into two groups: the sh-NC and sh-HOTAIRM1 groups. Next, U87 cells (approximately 1 × 106/200 μl PBS) transfected with shRNAs (sh-HOTAIRM1 or sh-NC) were subcutaneously inoculated into the right frontal node of nude mice. Tumor growth was monitored once weekly. Mice were killed by cervical dislocation until the fifth week (35 days), and tumors were resected and measured for weight and volume. | |||
hsa-miR-148a-3p
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [486] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
RWPE-1 | Normal | Homo sapiens | CVCL_3791 |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 | |
| LAPC-4 | Prostate carcinoma | Homo sapiens | CVCL_4744 | |
| In-vivo Model | Sixteen SPF female BALB/c nude mice (6 weeks old, weighing 20-22 g) (SLAC Laboratory Animal, Shanghai, China) were randomly arranged into the sh-NC group and the sh-METTL3 group, with 8 mice per group. After transfected with sh-NC and sh-METTL3, respectively, cells were rinsed with PBS to remove excess medium, detached with 0.25% trypsin, and detachment was terminated with complete medium, followed by centrifugation to collect cell precipitates. Appropriate amounts of physiological saline were added and gently pipetted into single-cell suspensions for cell counting. A total of 3 × 106 cells were resuspended in 50 μL normal saline, mixed with 50 μL 25% Matrigel (1:1), and inoculated subcutaneously into the right inguinal of nude mice. Tumor growth curves were plotted with 7, 14, 21, and 28 days as time points, and the short diameter (a) and long diameter (b) of the tumor were determined using a vernier caliper to estimate the tumor volume V = (a2b)/2. Tumor mass was weighed on a scale. Following 4 weeks, nude mice were euthanized by an intraperitoneal injection of 150 mg/kg pentobarbital (P3761, Sigma). The skin near the tumor was cut short using surgical scissors, and the tumor was separated using ball pointed pliers. The tumors were partially embedded with paraffin for histological examination, and the remaining part was kept at -80°C. | |||
hsa-miR-181a-5p
Ageing-related disease [ICD-11: 9B10-9B60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [487] | |||
| Responsed Disease | Ageing-related disease [ICD-11: 9B10-9B60] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HK-2 [Human kidney] | Normal | Homo sapiens | CVCL_0302 |
| In-vivo Model | The mice were randomly separated into four groups (n = 3-5): young mice group (YNG); naturally aging mice group (OLD); PBS-treated mice group (Con); D-gal-treated aging mice group (D-gal). YNG group was the directly purchased mice (8-week-old) actually. OLD group: mice bought continued to be raised in a specific pathogen-free facility (SPF) individually for 24 months. Mice in Con group were injected subcutaneously with phosphate buffered saline (PBS) solution formulated from powder, with the same volume as the D-gal group. Mice from D-gal group were made by injecting (D-gal, 500 mg/kg body weight) daily for 8 weeks, subcutaneously. All animals were kept under a standard environmental condition (25 ± 2 °C; 12-h light-dark cycle) and allowed free access to water and regular sterile chow diets. All animal studies were designed in accordance with ARRIVE guidelines and experiment protocols followed NIH guidelines. | |||
hsa-miR-1908-5p
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [488] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
NP69SV40T
|
N.A. | Homo sapiens | CVCL_F755 |
| C666-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_7949 | |
| In-vivo Model | Male BALB/c-nude mice (5-week-old) were used in this study. First, the Antigo miR-1908-5p (Antigo-miR) for knocking down miR-1908-5p as well as its negative control (Antigo-NC) were acquired from GenePharma (China). Antigo-miR and Antigo-NC were each introduced into C666-1 cells which were then delivered through the tail of every mouse (n = 3 per group). Thereafter, all mice were raised well, and the long diameter and short diameter of tumors were measured every week. The tumor volume was calculated as tumor volume (mm3) = (short diameter)2 × long diameter/2. After 5 weeks, the mice were euthanized, and their tumors were weighed. All animal protocols were followed in accordance with the Institutional Animal Care and Use Committee of our hospital. | |||
hsa-miR-196b-5p
Structural developmental anomalies of teeth and periodontal tissues [ICD-11: LA30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [489] | |||
| Responsed Disease | Structural developmental anomalies of teeth and periodontal tissues [ICD-11: LA30.5] | |||
| Target Regulation | Down regulation | |||
| In-vivo Model | A total of 2.0 × 106 SCAPs were mixed with 20 mg HA/tricalcium phosphate (Engineering Research Center for Biomaterials, Sichuan University, Chengdu, China) at 37°C for 2 h, and then, the mixture was subcutaneously transplanted into the back of nude mice (10-week-old female, nu/nu). | |||
hsa-mir-19a
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [490] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Responsed Drug | Everolimus | Approved | ||
| Target Regulation | Up regulation | |||
hsa-mir-19b-1
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [490] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Responsed Drug | Everolimus | Approved | ||
| Target Regulation | Up regulation | |||
hsa-miR-27a-3p
Infantile hemangioma [ICD-11: 2E81]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [491] | |||
| Responsed Disease | Infantile hemangioma [ICD-11: 2E81] | |||
| Responsed Drug | Propranolol | Approved | ||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [491] | |||
| Responsed Disease | Infantile hemangioma [ICD-11: 2E81] | |||
| Responsed Drug | Oxymatrine | Investigative | ||
hsa_circ_0000677 (Circ_ABCC1)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [219] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
hsa_circ_0007905 (Circ_STX6)
Cataract [ICD-11: 9B10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [492] | |||
| Responsed Disease | Age-related cataracts [ICD-11: 9B10.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
B-3 | Normal | Homo sapiens | CVCL_6367 |
hsa_circ_0072380 (Circ_ZNF131)
Gestational diabetes mellitus [ICD-11: JA63]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [493] | |||
| Responsed Disease | Gestational diabetes mellitus [ICD-11: JA63] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
JEG-3 | Gestational choriocarcinoma | Homo sapiens | CVCL_0363 |
hsa_circ_0124554
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [494] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| In-vivo Model | HCT116 cells stably expressing sh-NC or sh-circ_0124554 were diluted to 2 × 106 cells in 0.2 mL phosphate buffer solution and injected into the mice, followed by 6 Gy irradiation once per day for 5 days. Ten days later, the tumor volume was calculated every 5 days for 5 cycles. Mice were sacrificed after 35 days, and the tumors were dissected for further analysis. | |||
hsa_circ_0136959
Thyroid Cancer [ICD-11: 2D10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [495] | |||
| Responsed Disease | Papillary thyroid cancer [ICD-11: 2D10.1] | |||
Hypoxia up-regulated protein 1 (HYOU1)
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [496] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Doxil | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
Insulin-like Growth Factor Binding Protein 7 Overlapping Transcript
Oosteoarthritis [ICD-11: FA0Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [497] | |||
| Responsed Disease | Oosteoarthritis [ICD-11: FA0Z] | |||
| In-vivo Model | 8-week-old male C57BL/6 mice were randomly divided into four groups: Ctrl+AAV-NC group (n = 6), Ctrl+AAV-OT group (n = 6), MIA+AAV-NC group (n = 8), and MIA+ AAV-OT group (n = 8). For the induction of OA, mice were given an intra-articular injection of MIA (Sigma-Aldrich) in the knee. Control mice were given normal saline in the same volume. | |||
Insulin-like growth factor-binding protein 4 (IGFBP4)
Endometrial cancer [ICD-11: 2C76]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [498] | |||
| Responsed Disease | Endometrial cancer [ICD-11: 2C76] | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| NF-kappa B signaling pathway | hsa04064 | |||
In-vitro Model |
HEC-1-A | Endometrial adenocarcinoma | Homo sapiens | CVCL_0293 |
| In-vivo Model | Ten specific pathogen-free (SPF) BALB/c nude mice (4-6 weeks old, 16 ± 2 g) were purchased from Shanghai Experimental Animal Center, Chinese Academy of Sciences. All experimental animals were kept in a SPF-level aseptic layer at 22-26 °C and 55 ± 5% humidity. Nude mice were randomly divided into sh-NC and sh-METTL3 groups, and these mice were anesthetized by intraperitoneal injection of pentobarbital sodium (60 mg/kg). After anesthesia, the right back skin of the nude mice was disinfected with conventional iodine. The mice in the sh-NC group were injected with 0.2 mL HEC-1-A cell suspension (1 × 106), and those in the sh-METTL3 group were injected with an equal volume of HEC-1-A cells that were stably transfected with sh-METTL3. The injector was slowly withdrawn, and the injection site was pressed for about 10 s to prevent an outflow of cell suspension. Three weeks later, the nude mice were euthanized by cervical dislocation, and subcutaneous tumors were separated and weighted. Tumor volume was measured as follows: tumor volume = 1/2 × length × width2. | |||
Inter-alpha-trypsin inhibitor heavy chain H1 (ITIH1)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [499] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | LY2157299 | Investigative | ||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| In-vivo Model | HCC cells stably transfected with a firefly luciferase expression vector (1 × 106 cells 30 μL) were injected into the left liver lobe of 5-week-old male BALB/c nude mice. Luciferase bioluminescence in each mouse was measured at week 6 after injection. Mice in all the groups were sacrificed 8 weeks after injection. ii) For the lung metastasis model, we slowly injected HCC cells (1 × 106cells 100 μL) into 5-week-old male BALB/c nude mice via the tail vein. Six weeks after injection, luciferase bioluminescence was measured, and the mice were then sacrificed. iii) As described in the previous study, 6-8-week-old male wild-type C57BL/6J mice were used for hydrodynamic tail vein injection (HTVi). The pT3-EF1alpha-myr-AKT-HA (human), pT3-EF1alpha-N90-beta-catenin (human), and pT3-EF1alpha-ITIH1 (human) plasmids were constructed by using the pT3-EF1alpha vector. The pooled pT3 plasmids and the Sleeping Beauty transposon plasmid (pCMV-SB) were mixed at a ratio of 25:1 and 20 μg of each pT3 plasmid was used for each mouse. | |||
Interferon regulatory factor 4 (IRF4)
Lupus erythematosus [ICD-11: 4A40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [500] | |||
| Responsed Disease | Systemic lupus erythematosus [ICD-11: 4A40.0] | |||
Interleukin-12 subunit beta (IL12B)
Inflammatory response [ICD-11: MG46]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [501] | |||
| Responsed Disease | Inflammatory response [ICD-11: MG46] | |||
| Cell Process | RNA decay | |||
Interleukin-13 (IL13)
Allergy [ICD-11: 4A8Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [502] | |||
| Responsed Disease | Allergy [ICD-11: 4A8Z] | |||
Asthma [ICD-11: CA23]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [502] | |||
| Responsed Disease | Asthma [ICD-11: CA23] | |||
Interleukin-17A (IL17A)
Psoriasis [ICD-11: EA90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [503] | |||
| Responsed Disease | Psoriasis [ICD-11: EA90] | |||
| Target Regulation | Up regulation | |||
Intraflagellar transport protein 80 homolog (IFT80)
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [504] | |||
| Responsed Disease | Head and neck squamous carcinoma [ICD-11: 2B6E] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HN4 | Clear cell renal cell carcinoma | Homo sapiens | CVCL_IS30 |
| HN-6 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_8129 | |
| WSU-HN30 | Pharyngeal squamous cell carcinoma | Homo sapiens | CVCL_5525 | |
| SCC-4 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1684 | |
| SCC-9 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1685 | |
| SCC-25 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1682 | |
| CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 | |
| In-vivo Model | To determine whether lnc-H2AFV-1 overexpression could enhance tumorigenicity in vivo, 1 × 106 HN6 cells stably transduced with LV-lnc-H2AFV-1 or LV-NC were subcutaneously injected into the right and left flanks of six mice. | |||
Kelch-like protein 5 (KLHL5)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [505] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
In-vitro Model |
GES-1 | Normal | Homo sapiens | CVCL_EQ22 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| In-vivo Model | Twenty nude mice were used to establish the transfer model. KLHL5 or normal SGC-7901 cells were knocked out using luciferase labeling (Genechem, China) and injected through the tail vein to construct tumor models. After 3 weeks, the tumor growth in mice was observed by small animal imaging technology. After the mice were euthanized, lung tissue was taken to observe tumor metastasis. In addition, eight nude mice were subcutaneously implanted with high/low KLHL5-expressing mass tissues from human subjects to evaluate the effect of KLHL5 on xenograft tumor proliferation. | |||
Krueppel-like factor 6 (KLF6)
Injury of heart [ICD-11: NB31]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [506] | |||
| Responsed Disease | Myocardial injury [ICD-11: NB31.Z] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HL-1 | Normal | Mus musculus | CVCL_0303 |
Krueppel-like factor 9 (KLF9)
Obesity [ICD-11: 5B81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [507] | |||
| Responsed Disease | Obesity [ICD-11: 5B81] | |||
| Responsed Drug | Methyl piperidine-3-carboxylate | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
3T3-L1 | Normal | Mus musculus | CVCL_0123 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
|
AAV-293
|
N.A. | Homo sapiens | CVCL_6871 | |
| In-vivo Model | For compound treatment, mice were intraperitoneally injected with methyl piperidine-3-carboxylate (MP3C) (Shanghai Aladdin Biochemical Technology) at 2.5 mg/kg every 3 days for 2 weeks. | |||
LCAT3
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [47] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0608 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HOP-62 | Lung adenocarcinoma | Homo sapiens | CVCL_1285 | |
| In-vivo Model | For the in vivo tumorigenicity assay, female BALB/c nude mice (ages 4-5 weeks) were randomly divided into two groups (n = 6/group). Calu1 cells (4 × 106) that had been stably transfected with sh-LCAT3 or scramble were implanted subcutaneously into the nude mice. Tumor growth was measured after one week, and tumor volumes were calculated with the following formula: Volume (cm3) = (length × width2)/2. After four weeks, the mice were euthanized, and the tumors were collected and weighed. For the in vivo tumor invasion assay, 1.2 × 106 scramble or shLCAT3 cells were injected intravenously into the tail vein of nude mice (n = 6/group). 1.5 mg luciferin (Gold Biotech, St Louis, MO, USA) was administered once a week for 4 weeks, to monitor metastases using an IVIS@ Lumina II system (Caliper Life Sciences, Hopkinton, MA, USA). | |||
| Response Summary | LCAT3 upregulation is attributable to N6-methyladenosine (m6A) modification mediated by methyltransferase like 3 (METTL3), leading to LCAT3 stabilization. LCAT3 as a novel oncogenic lncRNA in the lung, and validated the LCAT3-FUBP1-MYC axis as a potential therapeutic target for lung adenocarcinomas. | |||
Lithostathine-1-alpha (REG1A)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [508] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
NCM460 | Normal | Homo sapiens | CVCL_0460 |
| FHC | Normal | Homo sapiens | CVCL_3688 | |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| In-vivo Model | For in vivo lung metastases model, a total of 2 × 106 CRC cells with REG1alpha-overexpression or REG1alpha-knockdown were injected into nude mice via lateral tail vein. Six weeks after injection, the mice were sacrificed and the lung tissues were collected. | |||
Lnc668
Silicosis [ICD-11: CA60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [509] | |||
| Responsed Disease | Pulmonary fibrosis [ICD-11: CA60] | |||
| Target Regulation | Up regulation | |||
Lnc_D63785
Unspecific body region injury [ICD-11: ND56]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [394] | |||
| Responsed Disease | Unspecific body region injury [ICD-11: ND56] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Neuronal cell apoptosis | |||
In-vitro Model |
SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
| Response Summary | Oxygen glucose deprivation/re-oxygenation (OGD/R) induces neuronal injury via mechanisms that are believed to mimic the pathways associated with brain ischemia. METTL3 shRNA reversed OGD/R-induced Lnc_D63785 m6A methylation to decrease miR-422a accumulation. | |||
Long intergenic non-protein coding RNA 1638 (LINC01638)
Periodontitis [ICD-11: DA0C]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [510] | |||
| Responsed Disease | Periodontitis [ICD-11: DA0C] | |||
| Target Regulation | Up regulation | |||
long intergenic non-protein coding RNA 294 (LINC00294)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [294] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
Long intergenic non-protein coding RNA 426 (LINC00426)
Cervical cancer [ICD-11: 2C77]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [511] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Responsed Drug | Bleomycin | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| In-vivo Model | HeLa cells were stably transfected with LINC00426 overexpression lentivirus. Subsequently, for subcutaneously implanted tumor model, mice were randomly divided into two groups, and cells (1 × 107/100 μl) mixed with the same volume of matrix gel were injected subcutaneously into the right abdomen of mice. One month later the mice were executed and the tumors were stripped and weighed, measured for volume, and used for further analysis. For tumor metastasis assay, mice were randomly grouped, and 5 × 105/100 μl HeLa cells transfected with NC and LINC00426 overexpression lentivirus were intravenously injected into the tail vein of BALB/c nude mice which were sacrificed after 1 month. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [511] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| In-vivo Model | HeLa cells were stably transfected with LINC00426 overexpression lentivirus. Subsequently, for subcutaneously implanted tumor model, mice were randomly divided into two groups, and cells (1 × 107/100 μl) mixed with the same volume of matrix gel were injected subcutaneously into the right abdomen of mice. One month later the mice were executed and the tumors were stripped and weighed, measured for volume, and used for further analysis. For tumor metastasis assay, mice were randomly grouped, and 5 × 105/100 μl HeLa cells transfected with NC and LINC00426 overexpression lentivirus were intravenously injected into the tail vein of BALB/c nude mice which were sacrificed after 1 month. | |||
| Experiment 3 Reporting the m6A-centered Disease Response of This Target Gene | [511] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Responsed Drug | Imatinib | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| In-vivo Model | HeLa cells were stably transfected with LINC00426 overexpression lentivirus. Subsequently, for subcutaneously implanted tumor model, mice were randomly divided into two groups, and cells (1 × 107/100 μl) mixed with the same volume of matrix gel were injected subcutaneously into the right abdomen of mice. One month later the mice were executed and the tumors were stripped and weighed, measured for volume, and used for further analysis. For tumor metastasis assay, mice were randomly grouped, and 5 × 105/100 μl HeLa cells transfected with NC and LINC00426 overexpression lentivirus were intravenously injected into the tail vein of BALB/c nude mice which were sacrificed after 1 month. | |||
Long intergenic non-protein coding RNA 662 (LINC00662)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [512] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| COLO 357 | Pancreatic adenosquamous carcinoma | Homo sapiens | CVCL_0221 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| Human Pancreatic Nestin-Expressing cells (Human Pancreatic Nestin-Expressing cells) | ||||
| In-vivo Model | For the tumor growth study, nude mice were subcutaneously inoculated into the flanks with PC cells (3 × 106) suspended in 0.1 mL PBS. Tumor volumes were calculated as 1/2 (length × width2) weekly. After 4 weeks, the xenograft tumors were harvested for analysis. To examine the role of the FAK inhibitor in tumor growth, nude mice were inoculated subcutaneously into the flanks with 3 × 106 stable cells suspended in 0.1 mL PBS. After 1 week, pre-established tumor xenografts were treated with Y15 (0.5 mg/kg, 2 ×/week × 3) (MCE, New Jersey, China). After another 3 weeks, the xenograft tumors were harvested for analysis. In the metastasis model, PC cells (2 × 106) in 0.1 mL PBS were injected into nude mice through their tail veins. Six weeks later, the lungs were removed for the evaluation of lung metastasis using hematoxylin and eosin (H&E) staining. | |||
Triple-negative breast cancer [ICD-11: 2C6Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [513] | |||
| Responsed Disease | Triple-negative breast cancer [ICD-11: 2C6Z] | |||
| Responsed Drug | Docetaxel | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
Long-chain-fatty-acid--CoA ligase 4 (ACSL4)
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [514] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
| Target Regulation | Up regulation | |||
Injury of blood vessels of thorax [ICD-11: NB30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [515] | |||
| Responsed Disease | Sepsis-associated lung injury [ICD-11: NB30.4] | |||
| Responsed Drug | STM2457 | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
MLE-12
|
N.A. | Mus musculus | CVCL_3751 |
| In-vivo Model | To investigate the effect of lactate on sepsis-induced lung injury, sodium oxalate (0.75 g/kg body weight, Selleck, S6871) [18] or 2-DG (0.5 g/kg body weight, Selleck, S4701) [19] was injected intraperitoneally 3 h before CLP or sham operation to inhibit lactate production. To suppress METTL3 levels, mice were injected with STM2457 (30 mg/kg body weight, MedChemExpress, HY-134836) 12 h before CLP or sham surgery, with a supplemental injection of STM2457 before surgery. | |||
Lysine-specific demethylase 6B (KDM6B)
Inflammatory response [ICD-11: MG46]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [516] | |||
| Responsed Disease | Inflammatory response [ICD-11: MG46] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
m7GpppN-mRNA hydrolase (DCP2)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [517] | |||
| Responsed Disease | Small cell lung cancer [ICD-11: 2C25.1] | |||
| Responsed Drug | STM2457 | Investigative | ||
| Target Regulation | Down regulation | |||
In-vitro Model |
H-69
|
N.A. | Homo sapiens | CVCL_8121 |
| H69AR | Lung small cell carcinoma | Homo sapiens | CVCL_3513 | |
| H446 | Lung small cell carcinoma | Homo sapiens | CVCL_1562 | |
|
HBE1
|
N.A. | Homo sapiens | CVCL_0287 | |
| In-vivo Model | SCLC cells were collected, and cell suspensions were prepared at a concentration of 1 × 106 cells per 100 μL PBS and injected subcutaneously into nude mice. When the tumour volume reached 60 mm3, mice were randomly divided into control and treatment groups with five mice in each group. The mice in the treatment group were treated regularly, and chemotherapy drugs (CDDP 3 mg/kg and VP16 2 mg/kg) were administered via intraperitoneal injection. | |||
MAP kinase-activated protein kinase 2 (MAPKAPK2)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [291] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
Methyltransferase-like protein 1 (METTL1)
Cutaneous squamous cell carcinoma [ICD-11: 2C31]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [518] | |||
| Responsed Disease | Neck squamous cell carcinoma [ICD-11: 2C31.Z] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
In-vitro Model |
SCC-4 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1684 |
miR-17-92
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [490] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Responsed Drug | Everolimus | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | miRNA maturation | |||
In-vitro Model |
MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | For subcutaneous xenograft models, 0.1 mL of cell suspension containing 106 cells were injected subcutaneously into the right flank of mice (n = 6 for each group). | |||
| Response Summary | In gastric cancer, m6A facilitated processing of pri-miR-17-92 into the miR-17-92 cluster through an m6A/DGCR8-dependent mechanism. METTL3-high tumors showed preferred sensitivity to an mTOR inhibitor, everolimus. | |||
Mu-type opioid receptor (OPRM1)
Pain disorders [ICD-11: 8E43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [520] | |||
| Responsed Disease | Neuropathic pain [ICD-11: 8E43.0] | |||
| In-vivo Model | Male Sprague-Dawley rats (8-10 weeks old) weighing 200-250 g were purchased from Guangdong Medical Experimental Animal Cente [License No. SCXX (Guangdong) 2018-0002]. Animals were hosted in a centralized location in the Experimental Animal Center of the Third People's Hospital of Shenzhen [License No. SYXK (Guangdong) 2017-0120], with a standardized circadian cycle of 12 h for light and darkness at 20-25 °C and 55-60% humidity. Food pellets and water were provided ad libitum in specific-pathogen-free cages. All rats entered the experiment after 7 days of quarantine in the animal room without any abnormalities (normal activity and weight, normal gait, etc.). Thirty-four rats were randomly divided into 4 groups: 1 Sham operation group (Sham group, n = 7): only the right sciatic nerve trunk was exposed without ligation; 2 Chronic constriction injury (CCI) model group (NPP group, n = 7): the right sciatic nerve was exposed and ligated; 3 Intrathecal injection of AAV-METTL3 shRNA + CCI model group( M3 + NPP group, n = 10): AAV-METTL3 shRNA was intrathecally injected, and the right sciatic nerve was ligated after 19 days; 4 Intrathecal injection of negative control virus + CCI model group (Scr + NPP group, n = 10): AAV-scrambled shRNA was intrathecally injected, and the right sciatic nerve was ligated 19 days later. This experiment has been approved by the Animal Ethics Committee of the Third People's Hospital of Shenzhen (IACUC: 2019019), and strictly follows the 3 R principle of reduction, refinement and replacement in animal experiments. | |||
Myosin-3 (MYH3)
Cardiomyopathy [ICD-11: BC43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [521] | |||
| Responsed Disease | Cardiomyopathy [ICD-11: BC43] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
H9c2(2-1) | Normal | Rattus norvegicus | CVCL_0286 |
| In-vivo Model | Eight-week-old male SD rats were purchased from SPF (Beijing) biotechnology co., LTD (Animal certificate number: 1103212101024214414). After the animals were kept for 7 days for environmental adaptation, the production of animal models was started. The rats were divided into 4 groups, control group, sepsis group, siMETTL3 group and sepsis+siMETTL3 group respectively. siMETTL3 was given at a dose of 5 nmol/ time/animal by tail vein injection. Doses were given every 3 days for a total of 18 days (i.e., 6 doses). The sepsis model was established by intraperitoneal injection of LPS (5 mg/kg) 24 h after the end of administration, the survival rate of LPS-treated mice was 75 % [[47], [48], [49]]. The control group was intraperitoneally injected with the same volume of normal saline. Echocardiography was performed 6 h after LPS injection. The animals were anesthetized by isoflavane induction and cardiac functions were evaluated by transthoracic echocardiography with an ultrasound machine (VisualSonics USA vevo 2100). The serum was stored in the refrigerator at 4 °C for 1 h and centrifuged for 3000r for 15 min. The packaged erum was stored at -80 °C for subsequent detection. The apical tissue was cut into 1 mm × 1 mm tissue blocks and placed in the electron microscope liquid at 4 °C for electron microscope detection. | |||
N-myc proto-oncogene protein (MYCN)
Developmental anomalies [ICD-11: LD9Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [522] | |||
| Responsed Disease | Developmental anomalies [ICD-11: LD9Z] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
E14 | Lung carcinoma | Homo sapiens | CVCL_B4P5 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
N-terminal Xaa-Pro-Lys N-methyltransferase 1 (NTMT1)
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [523] | |||
| Responsed Disease | Head and neck squamous carcinoma [ICD-11: 2B6E] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 |
| FaDu | Hypopharyngeal squamous cell carcinoma | Homo sapiens | CVCL_1218 | |
NEDD4 family-interacting protein 1 (NDFIP1)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [524] | |||
| Responsed Disease | Intrahepatic cholangiocarcinoma [ICD-11: 2C12.10] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HIBEpic (Human intrahepatic bile duct epithelial cells) | |||
| HuCC-T1 | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_0324 | |
| RBE | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_4896 | |
| HCCC-9810 | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_6908 | |
| In-vivo Model | Mice were housed in a specific pathogen-free animal facility with a 12-h light-dark cycle. Lentivirus-infected HUCCT1 or HCCC-9810 cells (4 × 106) were suspended in 0.1 mL of phosphate-buffered saline and were subcutaneously injected into the right flank of nude mice. Tumor size was monitored every 4 days for a total of 28 days, and tumor volume was calculated as length × width2/2. After 4 weeks, the mice were euthanized by intraperitoneal injection of 200 mg/kg sodium pentobarbital. Then, tumor tissues were excised and weighed for further analysis. | |||
Neuronal acetylcholine receptor subunit alpha-4 (CHRNA4)
Cognitive disorders [ICD-11: 6E0Z]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [525] | |||
| Responsed Disease | Cognitive disorders [ICD-11: 6E0Z] | |||
| Responsed Drug | Bisphenol A | Investigative | ||
| Target Regulation | Up regulation | |||
| In-vivo Model | Rats were tested in the Morris Water Maze (MWM) after PND 60. The experimental device was a circular tank with a diameter of 120 cm and depth of 40 cm, containing water held at a constant 23 ± 1 °C. Rats were allowed to swim to a settled platform (a round, clear acrylic panel) for no more than 90 s with its top surface submerged 1.5 cm below the water level. Caramel was dissolved in water to ensuring that rats cannot directly observe the platform position. Rats were released from different positions in the water maze, and each rat performed four trials daily for five days. The escape latency was automatically recorded. On the sixth day, the rats were given a 90-s retention trial in which the platform was removed, and the mean speed, relative time in zone 2 and relative distance in zone 2 were recorded. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [525] | |||
| Responsed Disease | Cognitive disorders [ICD-11: 6E0Z] | |||
| Responsed Drug | STM2457 | Investigative | ||
| Target Regulation | Up regulation | |||
| In-vivo Model | Rats were tested in the Morris Water Maze (MWM) after PND 60. The experimental device was a circular tank with a diameter of 120 cm and depth of 40 cm, containing water held at a constant 23 ± 1 °C. Rats were allowed to swim to a settled platform (a round, clear acrylic panel) for no more than 90 s with its top surface submerged 1.5 cm below the water level. Caramel was dissolved in water to ensuring that rats cannot directly observe the platform position. Rats were released from different positions in the water maze, and each rat performed four trials daily for five days. The escape latency was automatically recorded. On the sixth day, the rats were given a 90-s retention trial in which the platform was removed, and the mean speed, relative time in zone 2 and relative distance in zone 2 were recorded. | |||
Neurosecretory protein VGF [Cleaved into: Neuroendocrine regulatory peptide-1 (VGF)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [479] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| In-vivo Model | For the in vivo tumor formation assay, transfected A549 cells (1 × 107) were injected subcutaneously into the right thigh of female BALBc nude mice. One month after transplantation, the mice were sacrificed and the tissue samples were measured, weighed, and immunohistochemically analyzed. The tumor volume was calculated by the formula 1/2 × length × width2. Female BALB/c nude mice were purchased from Beijing SPF (Beijing, China) and were grouped randomly (5 or 6 mice per group). | |||
Neurotrophic factor BDNF precursor form (BDNF)
Endometrial cancer [ICD-11: 2C76]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [526] | |||
| Responsed Disease | Endometrial cancer [ICD-11: 2C76] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
KLE | Endometrial adenocarcinoma | Homo sapiens | CVCL_1329 |
| AN3-CA | Endometrial adenocarcinoma | Homo sapiens | CVCL_0028 | |
| In-vivo Model | The mice were randomly divided into four groups (n=5 per group), labeled as OE-vector (caudal vein injection of 2×106 AN3CA cells), OE-SLERT (caudal vein injection of 2×106 SLERT-overexpressing AN3CA cells), OE-SLERT+ASO-BDNF (caudal vein injection of 2×106 SLERT-overexpressing AN3CA cells transfected with ASO-BDNF), and OE-SLERT+k252a (caudal vein injection of 2×106 SLERT-overexpressing AN3CA cells and intraperitoneal injection of 25μg/kg k252a). After 4 weeks, the lung tissues were dissected and weighted, followed by H&E staining. | |||
Single episode depressive disorder, severe, without psychotic symptoms [ICD-11: 6A70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [421] | |||
| Responsed Disease | Major depressive disorder [ICD-11: 6A70.3] | |||
In-vitro Model |
SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
| In-vivo Model | Rats were housed at 23°C and 55% humidity and were given ad libitum food and water. During acclimatization (1 week), rats were placed randomly (3/cage); however, after initial behavioral testing, they were grouped according to their behavioral phenotypes. All experiments were performed under a light cycle (8:00 AM and 10:00 AM). The protocol to induce LH behavior was approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham. The animal study also adhered to the international guidelines for the use and care of laboratory animals. | |||
NPC1-like intracellular cholesterol transporter 1 (NPC1L1)
Atherosclerosis [ICD-11: BD40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [527] | |||
| Responsed Disease | Atherosclerosis [ICD-11: BD40.Z] | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
In-vitro Model |
HUVEC-CS
|
N.A. | Homo sapiens | CVCL_0F27 |
| In-vivo Model | Eight-week-old male ApoE-/- and C57BL/6J mice were purchased from GemPharmatech Co., Ltd. All mice were housed in a 12-h light-dark environment with free access to food and water. ApoE-/- mice were fed a high-fat diet (21% crude fat, 0.15% cholesterol, 19.5% casein) for 8 weeks to establish an animal model of AS. C57BL/6J mice in control group were kept on a normal diet. Next, the modeled animals were randomly divided into 3 groups (n = 10): (1) mice in the AS group were fed with HFD for 8 weeks; (2) mice in the AS + sh-NC group were given Ad-sh-NC (2 × 109 pfu/mouse) injection every 14 days through the tail vein for 8 weeks, during which HFD was used to feed mice; (3) mice in the AS + sh-NPC1L1 group were given Ad-sh-NPC1L1 (2 × 109 pfu/mouse) injection every 14 days through the tail vein for 8 weeks, during which HFD was used to feed mice. | |||
Nuclear factor of activated T-cells 5 (NFAT5)
Liver cancer [ICD-11: 2C12]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [528] | |||
| Responsed Disease | Intrahepatic cholangiocarcinoma [ICD-11: 2C12.10] | |||
| Responsed Drug | Gemcitabine | Approved | ||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [528] | |||
| Responsed Disease | Intrahepatic cholangiocarcinoma [ICD-11: 2C12.10] | |||
| Responsed Drug | STM2457 | Investigative | ||
Nuclear pore complex protein Nup214 (NUP214)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [529] | |||
| Responsed Disease | Brain cancer [ICD-11: 2A00] | |||
| Target Regulation | Up regulation | |||
Nuclear receptor subfamily 4 group A member 2 (NR4A2)
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [530] | |||
| Responsed Disease | Esophageal Squamous Cell Carcinoma [ICD-11: 2B70.1] | |||
| Responsed Drug | Celecoxib | Approved | ||
In-vitro Model |
KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 |
| Eca-109 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_6898 | |
| KYSE-450 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1353 | |
| KYSE-30 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1351 | |
| HET-1A | Normal | Homo sapiens | CVCL_3702 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
NUTM2B antisense RNA 1 (NUTM2B-AS1)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [531] | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Target Regulation | Up regulation | |||
OIP5 antisense RNA 1 (OIP5-AS1)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [532] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
In-vitro Model |
MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | Female BALB/c nude mice aged 4 weeks old were adopted. In the subcutaneous xenograft tumor, GC cells (2 × 106/200 μl) were subcutaneously injected into the mice flank. | |||
Optineurin (OPTN)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [533] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulation | Down regulation | |||
Oxidized low-density lipoprotein receptor 1 (OLR1)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [534] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Responsed Drug | Temozolomide | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
U-343MG | Glioblastoma | Homo sapiens | CVCL_S471 |
| U-251MG | Astrocytoma | Homo sapiens | CVCL_0021 | |
p-P70
Renal cell carcinoma [ICD-11: 2C90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [109] | |||
| Responsed Disease | Renal cell carcinoma [ICD-11: 2C90] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Epithelial-to-mesenchymal transition | |||
| Arrest cell cycle at G0/G1 phase | ||||
In-vitro Model |
ACHN | Papillary renal cell carcinoma | Homo sapiens | CVCL_1067 |
| Caki-1 | Clear cell renal cell carcinoma | Homo sapiens | CVCL_0234 | |
| Caki-2 | Papillary renal cell carcinoma | Homo sapiens | CVCL_0235 | |
| HK2 | Normal | Acipenser baerii | CVCL_YE28 | |
| In-vivo Model | Cells (5×106 cells in 200 uL) were suspended with 100 uL PBS and 100 uL Matrigel Matrix, and injected subcutaneously into the left armpit of each mouse. | |||
| Response Summary | Knockdown of METTL3 could obviously promote cell proliferation, migration and invasion function, and induce G0/G1 arrest,METTL3 acts as a novel marker for tumorigenesis, development and survival of RCC. Knockdown of METTL3 promoted changes in pI3K/AKT/mTOR markers' expression with a gain in p-PI3k, p-AKT, p-mTOR and p-P70, and a loss of p-4EBP1. | |||
Paired box protein Pax-8 (PAX8)
Thyroid Cancer [ICD-11: 2D10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [535] | |||
| Responsed Disease | Thyroid Cancer [ICD-11: 2D10] | |||
| Responsed Drug | Sodium iodide I 131 | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 |
| KTC-1 | Thyroid carcinoma | Homo sapiens | CVCL_6300 | |
| TPC-1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_6298 | |
| K1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_2537 | |
| CAL-62 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_1112 | |
| C-643 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_5969 | |
| Nthy-ori 3-1 | Normal | Homo sapiens | CVCL_2659 | |
| In-vivo Model | A concentration of 1 × 107 CAL62 cells stably transfected with OE METTL3 or OE METTL3/shPAX8-transfected CAL-62 cells were injected subcutaneously into the dorsal flanks of the mice (five mice per group) to establish a xenograft model. | |||
Pappalysin-1 (PAPPA)
Endometrial cancer [ICD-11: 2C76]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [498] | |||
| Responsed Disease | Endometrial cancer [ICD-11: 2C76] | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| NF-kappa B signaling pathway | hsa04064 | |||
In-vitro Model |
HEC-1-A | Endometrial adenocarcinoma | Homo sapiens | CVCL_0293 |
| In-vivo Model | Ten specific pathogen-free (SPF) BALB/c nude mice (4-6 weeks old, 16 ± 2 g) were purchased from Shanghai Experimental Animal Center, Chinese Academy of Sciences. All experimental animals were kept in a SPF-level aseptic layer at 22-26 °C and 55 ± 5% humidity. Nude mice were randomly divided into sh-NC and sh-METTL3 groups, and these mice were anesthetized by intraperitoneal injection of pentobarbital sodium (60 mg/kg). After anesthesia, the right back skin of the nude mice was disinfected with conventional iodine. The mice in the sh-NC group were injected with 0.2 mL HEC-1-A cell suspension (1 × 106), and those in the sh-METTL3 group were injected with an equal volume of HEC-1-A cells that were stably transfected with sh-METTL3. The injector was slowly withdrawn, and the injection site was pressed for about 10 s to prevent an outflow of cell suspension. Three weeks later, the nude mice were euthanized by cervical dislocation, and subcutaneous tumors were separated and weighted. Tumor volume was measured as follows: tumor volume = 1/2 × length × width2. | |||
Phosphoenolpyruvate carboxykinase [GTP], mitochondrial (PCK2)
Hepatic inflammation [ICD-11: DB97]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [536] | |||
| Responsed Disease | Hepatic inflammation [ICD-11: DB97] | |||
| Target Regulation | Up regulation | |||
Phosphoglycerate kinase 1 (PGK1)
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [484] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
In-vitro Model |
VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| HNC PC3 | Retromolar trigone squamous cell carcinoma | Homo sapiens | CVCL_C8XA | |
|
RWPE-2
|
N.A. | Homo sapiens | CVCL_3792 | |
Phospholipid hydroperoxide glutathione peroxidase GPX4 (GPX4)
Ovarian dysfunction [ICD-11: 5A80]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [537] | |||
| Responsed Disease | Polycystic ovary syndrome [ICD-11: 5A80.1] | |||
| In-vivo Model | PCOS mice model was induced by the continuous subcutaneous injection of dehydroepiandrosterone (DHEA, GAOYUANbio, China) with a concentration of 60 mg/kg every day for 3 weeks. Meanwhile, mice in control group were subcutaneously injected with an equal volume of sesame oil. After three weeks, PCOS mice were divided into sh-NC and sh-METTL3 groups at random (n = 10). The ovary of mice in sh-NC group or sh-METTL3 group were subcutaneously injected with sh-NC or sh-METTL3 lentivirus at the concentration of 5 times; 108 PFU/mL synthesized by Shanghai GenePharma, Ltd. The estrus cycle, ovarian weight and follicular development were recorded 14 days following lentivirus infection. The ovary samples were collected for analysis. | |||
Phosphoserine aminotransferase (PSAT1)
Liver cancer [ICD-11: 2C12]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [452] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Lenvatinib | Approved | ||
In-vitro Model |
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | For the subcutaneous injection model, 3 × 106 MHCC97L cells were washed with PBS thrice and resuspended in 100 μL 1:1 Matrixgel and DMEM-HG. HDTVi was performed as previously described.44 Plasmids were extracted using an EndoFree Plasmid extraction kit (QIAGEN). Plasmid sequence was confirmed using Sanger sequencing. A 2 mL mixture of 20 μg sgTp53 plasmids, 10 μg Myc overexpression plasmids, and 2 μg SB13 plasmids was dissolved in saline of 10% mice body weight and was tail-vein injected into C57BL/6 mice weighing at least 25 g. Plasmids were co-injected in the lateral tail vein to induce spontaneous HCC formation. Three weeks after HDTVi, mice were randomly subjected to vehicle or STM2457 treatment. STM2457 (DC chemicals) was dissolved in 20% (w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) and injected intraperitoneally. Sorafenib powder (Selleckchem) was dissolved in distilled water and administrated via oral gavage. A 5 days on/6 days off treatment regimen was applied. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [452] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
In-vitro Model |
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | For the subcutaneous injection model, 3 × 106 MHCC97L cells were washed with PBS thrice and resuspended in 100 μL 1:1 Matrixgel and DMEM-HG. HDTVi was performed as previously described.44 Plasmids were extracted using an EndoFree Plasmid extraction kit (QIAGEN). Plasmid sequence was confirmed using Sanger sequencing. A 2 mL mixture of 20 μg sgTp53 plasmids, 10 μg Myc overexpression plasmids, and 2 μg SB13 plasmids was dissolved in saline of 10% mice body weight and was tail-vein injected into C57BL/6 mice weighing at least 25 g. Plasmids were co-injected in the lateral tail vein to induce spontaneous HCC formation. Three weeks after HDTVi, mice were randomly subjected to vehicle or STM2457 treatment. STM2457 (DC chemicals) was dissolved in 20% (w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) and injected intraperitoneally. Sorafenib powder (Selleckchem) was dissolved in distilled water and administrated via oral gavage. A 5 days on/5 days off treatment regimen was applied. | |||
| Experiment 3 Reporting the m6A-centered Disease Response of This Target Gene | [452] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | STM2457 | Investigative | ||
In-vitro Model |
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | For the subcutaneous injection model, 3 × 106 MHCC97L cells were washed with PBS thrice and resuspended in 100 μL 1:1 Matrixgel and DMEM-HG. HDTVi was performed as previously described.44 Plasmids were extracted using an EndoFree Plasmid extraction kit (QIAGEN). Plasmid sequence was confirmed using Sanger sequencing. A 2 mL mixture of 20 μg sgTp53 plasmids, 10 μg Myc overexpression plasmids, and 2 μg SB13 plasmids was dissolved in saline of 10% mice body weight and was tail-vein injected into C57BL/6 mice weighing at least 25 g. Plasmids were co-injected in the lateral tail vein to induce spontaneous HCC formation. Three weeks after HDTVi, mice were randomly subjected to vehicle or STM2457 treatment. STM2457 (DC chemicals) was dissolved in 20% (w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) and injected intraperitoneally. Sorafenib powder (Selleckchem) was dissolved in distilled water and administrated via oral gavage. A 5 days on/7 days off treatment regimen was applied. | |||
Phosphoserine phosphatase (PSPH)
Liver cancer [ICD-11: 2C12]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [452] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Lenvatinib | Approved | ||
In-vitro Model |
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | For the subcutaneous injection model, 3 × 106 MHCC97L cells were washed with PBS thrice and resuspended in 100 μL 1:1 Matrixgel and DMEM-HG. HDTVi was performed as previously described.44 Plasmids were extracted using an EndoFree Plasmid extraction kit (QIAGEN). Plasmid sequence was confirmed using Sanger sequencing. A 2 mL mixture of 20 μg sgTp53 plasmids, 10 μg Myc overexpression plasmids, and 2 μg SB13 plasmids was dissolved in saline of 10% mice body weight and was tail-vein injected into C57BL/6 mice weighing at least 25 g. Plasmids were co-injected in the lateral tail vein to induce spontaneous HCC formation. Three weeks after HDTVi, mice were randomly subjected to vehicle or STM2457 treatment. STM2457 (DC chemicals) was dissolved in 20% (w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) and injected intraperitoneally. Sorafenib powder (Selleckchem) was dissolved in distilled water and administrated via oral gavage. A 5 days on/9 days off treatment regimen was applied. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [452] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
In-vitro Model |
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | For the subcutaneous injection model, 3 × 106 MHCC97L cells were washed with PBS thrice and resuspended in 100 μL 1:1 Matrixgel and DMEM-HG. HDTVi was performed as previously described.44 Plasmids were extracted using an EndoFree Plasmid extraction kit (QIAGEN). Plasmid sequence was confirmed using Sanger sequencing. A 2 mL mixture of 20 μg sgTp53 plasmids, 10 μg Myc overexpression plasmids, and 2 μg SB13 plasmids was dissolved in saline of 10% mice body weight and was tail-vein injected into C57BL/6 mice weighing at least 25 g. Plasmids were co-injected in the lateral tail vein to induce spontaneous HCC formation. Three weeks after HDTVi, mice were randomly subjected to vehicle or STM2457 treatment. STM2457 (DC chemicals) was dissolved in 20% (w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) and injected intraperitoneally. Sorafenib powder (Selleckchem) was dissolved in distilled water and administrated via oral gavage. A 5 days on/8 days off treatment regimen was applied. | |||
| Experiment 3 Reporting the m6A-centered Disease Response of This Target Gene | [452] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | STM2457 | Investigative | ||
In-vitro Model |
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | For the subcutaneous injection model, 3 × 106 MHCC97L cells were washed with PBS thrice and resuspended in 100 μL 1:1 Matrixgel and DMEM-HG. HDTVi was performed as previously described.44 Plasmids were extracted using an EndoFree Plasmid extraction kit (QIAGEN). Plasmid sequence was confirmed using Sanger sequencing. A 2 mL mixture of 20 μg sgTp53 plasmids, 10 μg Myc overexpression plasmids, and 2 μg SB13 plasmids was dissolved in saline of 10% mice body weight and was tail-vein injected into C57BL/6 mice weighing at least 25 g. Plasmids were co-injected in the lateral tail vein to induce spontaneous HCC formation. Three weeks after HDTVi, mice were randomly subjected to vehicle or STM2457 treatment. STM2457 (DC chemicals) was dissolved in 20% (w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) and injected intraperitoneally. Sorafenib powder (Selleckchem) was dissolved in distilled water and administrated via oral gavage. A 5 days on/9 days off treatment regimen was applied. | |||
Pirin (PIR)
Melanoma of uvea [ICD-11: 2D0Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [538] | |||
| Responsed Disease | Melanoma of uvea [ICD-11: 2D0Y] | |||
| Target Regulation | Up regulation | |||
Pituitary homeobox 3 (PTX3)
Asthma [ICD-11: CA23]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [539] | |||
| Responsed Disease | Allergic asthma [ICD-11: CA23.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| In-vivo Model | Briefly, the Mettl3 targeting vector was designed to flank exon 2-3 with LoxP sites and electroporated into C57BL/6 embryonic stem cells (ES). Positive ES clones were confirmed by PCR and sequencing, and injected into C57BL/6 blastocysts to generate chimeric offspring. Chimeric mice were mated with C57BL/6 mice to obtain Mettl3flox/flox mice. These Mettl3flox/flox mice were crossed with Lyz2-Cre mice (The Jackson Laboratory) to generate Mettl3fl/fl (WT) and Mettl3fl/flLyz2Cre/+ (KO) mice. | |||
PncRNA-D
Liposarcoma [ICD-11: 2B59]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [540] | |||
| Responsed Disease | Liposarcoma [ICD-11: 2B59] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Cell cycle | |||
In-vitro Model |
HAP1 | Chronic myelogenous leukemia | Homo sapiens | CVCL_Y019 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Response Summary | Knockdown of METTL3 prolonged the half-life of PncRNA-D, and among the known m6A recognition proteins, YTHDC1 was responsible for binding m6A of pncRNA-D Knockdown of METTL3 or YTHDC1 also enhanced the interaction of pncRNA-D with TLS, and results from RNA pulldown assays implicated YTHDC1 in the inhibitory effect on the TLS-pncRNA-D interaction. | |||
Polyamine-transporting ATPase 13A3 (ATP13A3)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [541] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HCoEpiC (Healthy colon epithelial HCoEpiC cells) | |||
| In-vivo Model | The nude mice were randomly assigned to two groups consisting of six mice each. We injected transformed cells (P0 and P40) into the flank of each mouse in 0.1 mL of sterile PBS to form xenograft tumors. The tumor volume was measured every 2 or 3 days (volume = length × width2 × 1/2). The tumors were resected, imaged, and weighed after the mice were sacrificed. One piece of each tumor tissue was fixed in 4% (v/v) paraformaldehyde for hematoxylin and eosin (H&E) and immunohistochemistry (IHC) staining, and the remaining tissue was stored at -80 °C. | |||
Polyunsaturated fatty acid lipoxygenase ALOX12 (ALOX12)
Renal cell carcinoma [ICD-11: 2C90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [277] | |||
| Responsed Disease | Renal cell carcinoma [ICD-11: 2C90] | |||
| Responsed Drug | Erianin | Investigative | ||
| Target Regulation | Down regulation | |||
pre-miR-665
Structural developmental anomalies of teeth and periodontal tissues [ICD-11: LA30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [482] | |||
| Responsed Disease | Structural developmental anomalies of teeth and periodontal tissues [ICD-11: LA30.0] | |||
pri-miR-143
Acute myocardial infarction [ICD-11: BA41]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [542] | |||
| Responsed Disease | Acute myocardial infarction [ICD-11: BA41] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
neonatal ventricular myocytes (Mouse hearts were enzymatically digested to acquire the primary neonatal ventricular myocytes) | |||
| AC16 [Human hybrid cardiomyocyte] | Normal | Homo sapiens | CVCL_4U18 | |
| Response Summary | METTL3 deficiency contributes to heart regeneration after MI via METTL3-pri-miR-143-(miR-143)-Yap/Ctnnd1 axis. | |||
pri-miR-196b
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [543] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 | |
pri-miR-27
Multiple myeloma [ICD-11: 2A83]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [228] | |||
| Responsed Disease | Multiple myeloma [ICD-11: 2A83.1] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
U266 (Human multiple myeloma cells) | |||
| RPMI-8226 | Plasma cell myeloma | Homo sapiens | CVCL_0014 | |
| NCI-H929 | Plasma cell myeloma | Homo sapiens | CVCL_1600 | |
| MM1.S | Plasma cell myeloma | Homo sapiens | CVCL_8792 | |
| In-vivo Model | BALB/C nude mice (5 weeks old, weighing 18-22 g) were fed in specific pathogen-free facilities and subcutaneously inoculated with U266 cells (1 × 106). The mice were randomly divided into 3 groups with 6 mice per group, when the tumor was measurable. Then, miR-27a-3p mimic or sh-METTL3 was injected intratumorally at an interval of 4 days a total of 4 times. Tumor volume was measured using a digital caliper every week and calculated using the formula V = 1/2 (width2 × length). | |||
| Response Summary | METTL3 affected the growth, apoptosis, and stemness of MM cells through accelerating the stability of YY1 mRNA and the maturation of pri-miR-27 in vitro and in vivo. | |||
pri-miR-935
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [544] | |||
| Responsed Disease | Intrahepatic cholangiocarcinoma [ICD-11: 2C12.10] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
JEG-3 | Gestational choriocarcinoma | Homo sapiens | CVCL_0363 |
| JAR | Gestational choriocarcinoma | Homo sapiens | CVCL_0360 | |
| HTR-8/SVneo | Normal | Homo sapiens | CVCL_7162 | |
|
HUVEC-CS
|
N.A. | Homo sapiens | CVCL_0F27 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | The animals were first injected with JAR cells for 7 days, and the mice in AgomiR-935 group, AgomiR-NC group, AntagomiR-935 group, and AntagomiR-NC group were injected via tail vein with miR-935 agomiR (2 nmoL, Ribobio), miR-935 antagomiR (5 nmoL, Ribobio), or same dosage of negative controls (Ribobio) every 3 days for 2 weeks. | |||
Procathepsin L (CTSL)
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [545] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| HEK293-FT | Normal | Homo sapiens | CVCL_6911 | |
| In-vivo Model | Female BALB/c nude mice (4-5 weeks old) were purchased from the Center of Experimental Animals of Guangdong. To establish a tail vein metastasis model, 2 × 106 SiHa cells in 200 μl PBS were injected into the tail vein of each mouse (n = 6 for both METTL3-overexpressing and empty vector groups). The mice were killed at approximately 8 weeks, and lung tissues were isolated and embedded in paraffin. Hematoxylin and eosin staining was then used to determine the number of lung metastasis nodules. To establish the popliteal lymph node metastasis model, 1 × 106 SiHa cells in 50 μl PBS were injected subcutaneously into the footpad of each mouse (n = 6 for both groups). Cells from the experimental and control groups were inoculated under the right and left footpads of each mouse, respectively. After 8 weeks, the popliteal lymph nodes were excised. | |||
Prostaglandin E2 receptor EP4 subtype (PTGER2)
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [546] | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Responsed Drug | Carboplatin | Approved | ||
| Target Regulation | Up regulation | |||
Protein E6 (E6)
Malignant neoplasms of tonsil [ICD-11: 2B69]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [547] | |||
| Responsed Disease | Malignant neoplasms of tonsil [ICD-11: 2B69] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
WSU-HN26 | Squamous cell carcinoma of the oral cavity | Homo sapiens | CVCL_5523 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| C33A2 (The C33A2 cell line originates from C33A and has the subgenomic HPV16 plasmid pBELsLuc stably integrated into the genome) | ||||
| Response Summary | Overexpression of the ALKBH5 promoted production of intron retention on the human papillomavirus type 16 (HPV16) E6 mRNAs thereby promoting Protein E6 (E6) mRNA production.METLL3 induced production of intron-containing HPV16 E1 mRNAs over spliced E2 mRNAs and altered HPV16 L1 mRNA splicing in a manner opposite to ALKBH5. Overexpression of YTHDC1, enhanced retention of the E6-encoding intron and promoted E6 mRNA production. HPV16 mRNAs are m6A-methylated in tonsillar cancer cells. | |||
Protein mono-ADP-ribosyltransferase PARP10 (PARP10)
Cardiomegaly [ICD-11: BC45]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [548] | |||
| Responsed Disease | Cardiomegaly [ICD-11: BC45] | |||
| Target Regulation | Down regulation | |||
Protein NLRC5
Endometrial cancer [ICD-11: 2C76]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [549] | |||
| Responsed Disease | Endometrial cancer [ICD-11: 2C76] | |||
| Target Regulation | Up regulation | |||
Protein SON
Inflammatory response [ICD-11: MG46]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [550] | |||
| Responsed Disease | Inflammatory response [ICD-11: MG46] | |||
| Target Regulation | Up regulation | |||
Protein-tyrosine kinase 2-beta (PYK2)
Inflammatory response [ICD-11: MG46]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [551] | |||
| Responsed Disease | Inflammatory response [ICD-11: MG46] | |||
In-vitro Model |
RAW 264.7 | Mouse leukemia | Mus musculus | CVCL_0493 |
Pseudorabies Virus (PRV)
Rabies [ICD-11: 1C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [552] | |||
| Responsed Disease | Rabies [ICD-11: 1C82] | |||
| Responsed Drug | 3-deazidenosine | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
PK-15
|
N.A. | Sus scrofa | CVCL_2160 |
Putative microRNA 17 host gene protein (MIR17HG)
Endometriosis [ICD-11: GA10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [553] | |||
| Responsed Disease | Endometriosis [ICD-11: GA10] | |||
| Target Regulation | Up regulation | |||
Ras-related C3 botulinum toxin substrate 3 (RAC3)
Cancer-associated fibroblasts [ICD-11: XH45F3]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [554] | |||
| Responsed Disease | Cancer-associated fibroblasts [ICD-11: XH45F3] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| NF-kappa B signaling pathway | hsa04064 | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H661 | Lung large cell carcinoma | Homo sapiens | CVCL_1577 | |
| In-vivo Model | Nude mice were assigned randomly into two groups. For the first group, 2.5×106 A549 cells were injected subcutaneously into the right flank of mice, and 2.5×106 A549 cells mixed with 5×106 CAFs were injected into the left flank. For the second group, 2.5×106 A549-METTL3-KD cells were injected into the right flank, and A549-METTL3-KD cells mixed with 5×106 CAFs were injected into the left flank. Tumor volumes were measured every week. | |||
Ras-related protein R-Ras (RRAS)
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [555] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| J82 | Bladder carcinoma | Homo sapiens | CVCL_0359 | |
| TCCSUP | Bladder carcinoma | Homo sapiens | CVCL_1738 | |
| 5637 | Bladder carcinoma | Homo sapiens | CVCL_0126 | |
| In-vivo Model | A total of 16 male, 4- to 6-week-old BALB/c nude mice weighing 18-26 g, were purchased from the Institute of Model Animals, Nanjing University and raised in a specific pathogen-free (SPF) environment for the xenograft model. The mice were raised in animal individually ventilated cage (IVC cages) with a room temperature of 24°C and relative humidity of 70% and the air exchange rate was 15 times/h. The mice could drink filtered tap water and commercial feed ad libitum under a strict 12-h light/dark cycle. The animal laboratory was cleaned twice one day and sterilized with ultraviolet light for 1 h each week. Each mouse was injected with 100 μl PBS containing 1×106 T24 cell lines expressing either LV-NC or LV-shMETTL3 subcutaneously under the right armpit. Tumor volumes were recorded on day 7 after the injection and then measured each week. At day 28, the mice were sacrificed by cervical dislocation following the inhalation of 3% isoflurane anesthesia and the death of the mice was confirmed by respiratory arrest. Tumor weights were also compared. The data from each group of mice are expressed as the mean ± standard deviation (SD). | |||
Receptor-interacting serine/threonine-protein kinase 3 (RIP3)
Aortic aneurysm or dissection [ICD-11: BD50]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [556] | |||
| Responsed Disease | Abdominal aortic aneurysm [ICD-11: BD50.4] | |||
| Target Regulation | Up regulation | |||
Receptor-interacting serine/threonine-protein kinase 4 (RIPK4)
Malignant mixed epithelial mesenchymal tumour [ICD-11: 2B5D]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [557] | |||
| Responsed Disease | Malignant mixed epithelial mesenchymal tumour of ovary [ICD-11: 2B5D.0] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | NF-kappa B signaling pathway | hsa04064 | ||
In-vitro Model |
SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 |
| A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 | |
| In-vivo Model | Animals were purchased from Changzhou Cavens Laboratory Animal Company (Changzhou, China) and maintained under specific pathogen-free (SPF) conditions. All animal experiments were performed in accordance with the guidelines of the Laboratory Animal Health Committee of Jiangsu University. Approximately 1 × 107 cells were injected subcutaneously into the dorsal surface of female BALB/c nude mice (n = 5 for each group, 4 weeks old, 15 ± 2 g). When the xenograft volume reached approximately 60 mm3 (after 1 week), DDP was intraperitoneally injected three times per week for 3 weeks. On Day 28, the mice were sacrificed and the tumors were frozen at -80 °C for follow-up experiments. | |||
Retinaldehyde dehydrogenase 3 (ALDH1A3)
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [412] | |||
| Responsed Disease | Head and neck squamous carcinoma [ICD-11: 2B6E] | |||
In-vitro Model |
SCC-9 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1685 |
| In-vivo Model | Bmi1CreER and RosatdTomato mice were obtained from The Jackson Laboratory, while Mettl3flox/flox and Mettl3KI/KI mice were provided from the laboratory of Prof. Quan Yuan at the West China Hospital of Stomatology. As previously described, we treated mice with 100 μg/mL 4-nitroquinoline 1-oxide (4NQO) (Sigma, N8141) to induce HNSCC formation. In brief, 6-week-old mice were fed with 4NQO for 16 weeks and then normal water for another 10 weeks. To lineage tracing and conditional knockout and knockin mouse model, tamoxifen (1 mg/mouse/day for 3 days; Sigma, T5648-1G) was injected intraperitoneally after 4NQO treatment. Then, we collected tongue samples, made frozen sections, and observed sections under a fluorescence microscope to calculate the positive percentage of BMI1 cells.+. | |||
Retinoic Acid Receptor Alpha-Retinoic Acid Receptor Alpha (PML-RARalpha)
Malignant haematopoietic neoplasm [ICD-11: 2B33]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [558] | |||
| Responsed Disease | Blood malignancies [ICD-11: 2B33.Y] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell differentiation | |||
Ribose-5-phosphate isomerase (RPIA)
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [419] | |||
| Responsed Disease | Esophageal Squamous Cell Carcinoma [ICD-11: 2B70.1] | |||
| Cell Process | Transcription | |||
| Cell proliferation | ||||
| Cell migration | ||||
| Cell invasion | ||||
Ribosome-binding protein 1 (RRBP1)
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [559] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
In-vitro Model |
HNC PC3 | Retromolar trigone squamous cell carcinoma | Homo sapiens | CVCL_C8XA |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 | |
| In-vivo Model | Approximately 1 × 106 PCa cells (PC3-shNC, PC3-sh812 cell lines) or 2 × 106 DU145 cells (DU145-shNC, DU145-sh812, DU145-OENC, DU145-OEMETTL3-WT, DU145-OEMETTL3-Mut cell lines) per injection were suspended in a mixture of 100 μL PBS and Matrigel (1:1), and then injected subcutaneously into male BALB/c nude mice aged around 6 ~ 7 weeks old. Tumor volume was monitored at regular intervals and the volume was calculated as (length × width2)/2. Finally, the mice were sacrificed and the tumors were dissected and weighed. | |||
RNA component of 7SK nuclear ribonucleoprotein (RN7SK)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [561] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H23 | Lung adenocarcinoma | Homo sapiens | CVCL_1547 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| NCI-H2009 | Lung adenocarcinoma | Homo sapiens | CVCL_1514 | |
| MRC-5 | Normal | Homo sapiens | CVCL_0440 | |
RNA-binding protein 25 (RBM25)
Multiple myeloma [ICD-11: 2A83]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [356] | |||
| Responsed Disease | Multiple myeloma [ICD-11: 2A83.1] | |||
| Responsed Drug | Metformin | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
NCI-H929 | Plasma cell myeloma | Homo sapiens | CVCL_1600 |
| MM1.S | Plasma cell myeloma | Homo sapiens | CVCL_8792 | |
| In-vivo Model | 5-week-old male athymic BALB/C nude mice were obtained from Experimental animal center of Fujian Medical University, and subsequently randomly divided into four groups (5 mice per group), including control, metformin, pcDNA-METTL3 and pcDNA-METTL3 + metformin. For the control or metformin group, to induce tumors, 5 × 105 H929 cells were suspended in 0.2 ml phosphate buffered saline and inoculated subcutaneously into the peritoneum of mice. Once the subcutaneous nodules grown to a rice grain size (required approximately a week), the subcutaneous xenograft model of MM in nude mice was successfully constructed. One week later, PBS or Metformin was diluted in drinking water (2 mg/ml) and administered orally throughout the indicated time periods. For the pcDNA-METTL3 group and pcDNA-METTL3 + metformin group, firstly, we transfected pcDNA-METTL3 into H929 cells to overexpress METTL3, and then 5 × 105 H929 cells were suspended in 0.2 ml phosphate-buffered saline and inoculated subcutaneously into the peritoneum of mice. One week later, PBS or Metformin was diluted in drinking water (2 mg/ml) and administered orally throughout the indicated time periods. All nude mice were closely monitored for tumor growth, skin condition, and behavior daily and any tumor ulceration or irritation was noted. Tumor length and width were calculated with vernier calipers every 7 days. After 35 days, the mice were humanely sacrificed, and the subcutaneous tumors were excised and removed. | |||
RNF32 divergent transcript (RNF32-DT)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [562] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| In-vivo Model | Before subcutaneous injection, A549 cells were seeded into T-75 flask (Corning, USA) and transfected with vectors (control vector, LINC01006 overexpression and METTL3 overexpression, 10 μg respectively) or siRNA pools (LINC01006 and METTL3, 100 nM respectively) by Lipofectamine 2000 (Invitrogen, USA) based on the provided protocol when cell confluence reached 80%. After 48 h, counted 1x107 transfected A549 cells and suspended in 100 ul PBS. The suspended cells were injected into the same side armpit of mice. | |||
Sal-like protein 4 (SALL4)
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [563] | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
In-vitro Model |
SCC-9 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1685 |
| SCC-15 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1681 | |
| CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 | |
Semaphorin-3F (SEMA3F)
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [564] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
In-vitro Model |
RWPE-1 | Normal | Homo sapiens | CVCL_3791 |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| LNCaP C4-2B | Prostate carcinoma | Homo sapiens | CVCL_4784 | |
| VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | All the animal studies and protocols followed the institutional guidelines of the First Affiliated Hospital, School of Medicine, Zhejiang University. | |||
Serine/threonine-protein kinase B-raf (BRAF)
Atherosclerosis [ICD-11: BD40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [565] | |||
| Responsed Disease | Atherosclerosis [ICD-11: BD40.Z] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
RAW 264.7 | Mouse leukemia | Mus musculus | CVCL_0493 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Serine/threonine-protein kinase LATS1 (LATS1)
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [566] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | All animal experiments were approved by the Institutional Animal Care and Use Committee of Southern Medical University. Breast cancer cells (5 × 106) were implanted into the subcutaneous axilla of the forelimb of 3-4-week-old BALB/c nude mice. Seven days after transplantation, the diameter of the tumors was measured, and the tumors were removed after three weeks. | |||
Serine/threonine-protein kinase Nek2 (NEK2)
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [567] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
In-vitro Model |
HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
Serine/threonine-protein kinase Nek6 (NEK6)
Traumatic brain injury induced by controlled cortical impact injury [ICD-11: NA07]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [568] | |||
| Responsed Disease | Traumatic brain injury induced by controlled cortical impact injury [ICD-11: NA07] | |||
| Responsed Drug | Rapamycin | Approved | ||
In-vitro Model |
SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
Serine/threonine-protein kinase Nek7 (NEK7)
Intracerebral hemorrhage [ICD-11: 8B00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [569] | |||
| Responsed Disease | Intracerebral hemorrhage [ICD-11: 8B00] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HT22 | Normal | Mus musculus | CVCL_0321 |
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [570] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
| In-vivo Model | Twenty male healthy C57BL/6J mice (10 weeks) were purchased from the National Rodent Laboratory Animal Resources Center (Shanghai, China). All mice were raised and treated in accordance with the procedures set by the Animal Resource Center of our School of Medicine. We referred to the experiment of Xu and Xu. 32 Fifteen mice were randomly selected for medial meniscus (DMM) resection to establish mice models of OA knee joint. Sham operation was performed on mice in sham group. Two weeks after DMM operation, 10 mice were randomly selected and divided into 2 groups. Mice in both groups were injected weekly with 5 μg sh-METTL3 or sh-NC (GenePharma). Six weeks after surgery, knee joint specimens were collected for analysis. | |||
SET-binding protein (SETBP1)
Myelodysplastic neoplasms [ICD-11: 2A4Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [571] | |||
| Responsed Disease | Myelodysplastic neoplasms [ICD-11: 2A4Z] | |||
| In-vivo Model | In the first in vivo experiment focused on investigating the role of METTL14 in MDS cell proliferation in vivo, the MDS-L-luc cells were transduced with the Dox-inducible shMETTL14 (shMETTL14_Tet-on). After treatment with 2 μg/mL puromycin for a duration of 4 days, a total of 2.5 × 106 selected cells were injected via the tail vein into irradiated female NCG-M mice aged 8-10 weeks (GemPharmatech, China) to establish cell line-derived xenograft (CDX) models. On day 14 post transplantation, 20 mice were randomly assigned to two groups and treated with either Dox or vehicle. A total of 2 mg Dox was dissolved in water and administered by gastric lavage once a day. Five mice from each group underwent in vivo chemiluminescence imaging on day 14, 21, and 28 after receiving intraperitoneal injection of luciferin (Promega, USA). Their overall survival (OS) was also observed and documented. The remaining five mice from each group were utilized to assess the proportions of human CD45+cells in bone marrow and peripheral blood on day 28 through flow cytometry analysis. | |||
SH3 domain-binding protein 5 (SH3BP5)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [572] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
NCI-H522 | Lung adenocarcinoma | Homo sapiens | CVCL_1567 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| WI-38 | Normal | Homo sapiens | CVCL_0579 | |
| IMR-90 | Normal | Homo sapiens | CVCL_0347 | |
Signal transducer and activator of transcription 5B (STAT5B)
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [573] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
In-vitro Model |
MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| HEK293 | Normal | Homo sapiens | CVCL_0045 | |
| In-vivo Model | The mice were randomly allocated to four groups, with the specific number of mice detailed in each experiment. Subsequently, cells were injected into the abdominal mammary glands of the mice (2 × 106 cells in 100 μL) and allowed to grow until tumor formation occurred within 16 days. The tumor volume at the time of sacrifice was calculated using the formula, Volume = 0.5 × [(largest diameter) × (smallest diameter)^2]. The study protocol was approved by the Animal Experiments Committee of Chosun University (CIACUC 2020-S0022). | |||
SLC7A11 antisense RNA 1 (SLC7A11-AS1)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [574] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
SREBF2 antisense RNA 1 (SREBF2-AS1)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [575] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| SNU-398 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0077 | |
| THLE-2 | Normal | Homo sapiens | CVCL_3803 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
Sterile alpha motif domain-containing protein 9-like (SAMD9L)
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [419] | |||
| Responsed Disease | Esophageal Squamous Cell Carcinoma [ICD-11: 2B70.1] | |||
| Cell Process | Transcription | |||
| Cell proliferation | ||||
| Cell migration | ||||
| Cell invasion | ||||
Tafazzin (TAFAZZIN)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [291] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
Taurine up-regulated 1 protein (TUG1)
Diabetic [ICD-11: 5A14]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [576] | |||
| Responsed Disease | Diabetic [ICD-11: 5A14] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
GC-1 spg | Normal | Mus musculus | CVCL_8872 |
| In-vivo Model | STZ-induced diabetic mice were further divided into three groups: STZ, STZ+pcDNA3.1, and STZ+METTL3. Animals in the STZ+METTL3 group were intravenously injected with pAd-METTL3 vectors (109 plaque forming unit per mouse), while those in the STZ+pcDNA3.1 group were injected with pAd-NC vectors (109 plaque forming unit per mouse). The STZ group was administered with phosphate-buffered saline following the same procedure. After another 4 weeks, all mice were euthanized. Their serum and testicular tissues were collected for further analysis. | |||
Tenascin (TNC)
Acute myocardial infarction [ICD-11: BA41]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [577] | |||
| Responsed Disease | Acute myocardial infarction [ICD-11: BA41] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HL-1 | Normal | Mus musculus | CVCL_0303 |
| AC16 [Human hybrid cardiomyocyte] | Normal | Homo sapiens | CVCL_4U18 | |
| In-vivo Model | METTL3 and TNC overexpression model was achieved by in situ injection of overexpression AAV9 into the heart of C57/BL mice, and the injected virus titers were 1.8 × 1012 PFU/mL. The stable overexpression of METTL3 and overexpression of TNC could be obtained after 3 weeks. The overexpression AAV9 was purchased from Hanheng Biotechnology Co., Ltd. (Shanghai, China). The vector was HBAAV2/9-CMV-m-METTL3-3xflag-Null and HBAAV2/9-CMV-m-TNC-3xflag-Null. | |||
Thyroid hormone receptor-associated protein 3 (THRAP3)
Multiple myeloma [ICD-11: 2A83]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [356] | |||
| Responsed Disease | Multiple myeloma [ICD-11: 2A83.1] | |||
| Responsed Drug | Metformin | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
NCI-H929 | Plasma cell myeloma | Homo sapiens | CVCL_1600 |
| MM1.S | Plasma cell myeloma | Homo sapiens | CVCL_8792 | |
| In-vivo Model | 5-week-old male athymic BALB/C nude mice were obtained from Experimental animal center of Fujian Medical University, and subsequently randomly divided into four groups (5 mice per group), including control, metformin, pcDNA-METTL3 and pcDNA-METTL3 + metformin. For the control or metformin group, to induce tumors, 5 × 105 H929 cells were suspended in 0.2 ml phosphate buffered saline and inoculated subcutaneously into the peritoneum of mice. Once the subcutaneous nodules grown to a rice grain size (required approximately a week), the subcutaneous xenograft model of MM in nude mice was successfully constructed. One week later, PBS or Metformin was diluted in drinking water (2 mg/ml) and administered orally throughout the indicated time periods. For the pcDNA-METTL3 group and pcDNA-METTL3 + metformin group, firstly, we transfected pcDNA-METTL3 into H929 cells to overexpress METTL3, and then 5 × 105 H929 cells were suspended in 0.2 ml phosphate-buffered saline and inoculated subcutaneously into the peritoneum of mice. One week later, PBS or Metformin was diluted in drinking water (2 mg/ml) and administered orally throughout the indicated time periods. All nude mice were closely monitored for tumor growth, skin condition, and behavior daily and any tumor ulceration or irritation was noted. Tumor length and width were calculated with vernier calipers every 7 days. After 35 days, the mice were humanely sacrificed, and the subcutaneous tumors were excised and removed. | |||
Toll-like receptor 2 (TLR2)
Intervertebral disc degeneration [ICD-11: FA80]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [578] | |||
| Responsed Disease | Intervertebral disc degeneration [ICD-11: FA80] | |||
| In-vivo Model | Rats were anesthetized with 40 mg/kg sodium pentobarbital. A 21G needle was used to puncture discs 7-8 (Co7-8) from the back through the skin of the tail. The length of the needle was predetermined to ensure a puncture depth of approximately 5 mm. After penetrating the annulus, the needle was rotated 360° and held for 30 seconds to injure the annulus. The sham group underwent the same procedure but without puncture injury to the caudal disc. | |||
Transcription factor GATA-6 (GATA6)
Injury of other or unspecified intrathoracic organs [ICD-11: NB32]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [579] | |||
| Responsed Disease | Injury of other or unspecified intrathoracic organs [ICD-11: NB32.3] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | To perform in vivo plasmid transfection, the researchers mixed the pcDNA3.1-EGFP-METTL3 plasmid with in vivo jetPEI (Polyplus-transfection; Illkirch) per the manufacturer's guidelines. They mixed 40 μg of plasmid nucleic acid in 5% glucose (25 μl) with 6.4 μl of jetPEI solution in 5% glucose (25 μl) to obtain a final nitrogen and phosphorus ratio of 8. The mixture was incubated at room temperature for 30 minutes to form stable complexes before tracheal intubation of mice. | |||
Transcription factor SOX-6 (SOX6)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [580] | |||
| Responsed Disease | Intrahepatic cholangiocarcinoma [ICD-11: 2C12.10] | |||
| Target Regulation | Up regulation | |||
Transcription factor SOX-9 (SOX9)
Rheumatoid arthritis [ICD-11: FA20]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [581] | |||
| Responsed Disease | Rheumatoid arthritis [ICD-11: FA20] | |||
Transcriptional enhancer factor TEF-1 (TEAD1)
Liver disease [ICD-11: DB9Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [582] | |||
| Responsed Disease | Liver disease [ICD-11: DB9Z] | |||
| Target Regulation | Up regulation | |||
Transient receptor potential cation channel subfamily M member 7 (TRPM7)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [583] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 |
| SK-MES-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0630 | |
| Calu-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0608 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| In-vivo Model | The BALB/c nude mice (male, 4-6 weeks) were provided via Vital River Laboratory Animal Technology (Beijing, China). For xenograft tumor growth assay, 15 mice were randomly classified into 3 groups (n = 5/group), including control, sh-NC or sh-DGUOK-AS1 group. In sh-NC or sh-DGUOK-AS1 group, mice were subcutaneously injected with SK-MES-1 cells (3 × 106) stably transfected with sh-NC or sh-DGUOK-AS1; Moreover, mice were subcutaneously injected with non-transfected SK-MES-1 cells (3 × 106) as control group. The tumors were monitored every 7 days, and size was calculated by (length × width2)/2. Mice were euthanized after 4 weeks, and the tumors were collected and weighed, followed via collection for further analyses. Ki67 and CD31 levels were measured through immunohistochemistry staining using Ki67 (ab15580, 1:2000 dilution, Abcam) or CD31 (ab18298, 1:3000 dilution, Abcam) according to the protocols. For pulmonary metastatic assay, 3 × 106 SK-MES-1 cells stably transfected with sh-NC or sh-DGUOK-AS1 or non-transfected SK-MES-1 cells were intravenously injected into mice. Mice were sacrificed after 8 weeks, and lung specimens were collected, followed by hematoxylin-eosin (HE) staining assay for metastatic lesions in lung tissues. All experiments were approved via the Animal Ethics Committee of The Second Hospital of Shandong University. | |||
Transmembrane protein 127 (TMEM127)
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [584] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Rapamycin | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | Approximately 1 × 106 viable MDA-MB-231 breast cancer cells were resuspended in 1:1 ratio in 50 μl medium and 50 μl matrigel (Corning, 354234) and injected orthotopically into the fourth mammary fat pad of each mouse. After injection, tumor size was measured twice a week using an electronic caliper. Tumor volumes were calculated with the formula: volume = (L × W2)/2, where L is the tumor length and W is the tumor width measured in millimeters. | |||
Tribbles homolog 3 (TRIB3)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [585] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
Tumor-associated calcium signal transducer 2 (TROP2)
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [586] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Up regulation | |||
Ubiquitin carboxyl-terminal hydrolase 13 (USP13)
Gastrointestinal stromal tumor [ICD-11: 2B5B]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [587] | |||
| Responsed Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B] | |||
| Responsed Drug | 3-Methyladenine | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 | |
| In-vivo Model | 6 × 106 logarithmically growing GIST cells resuspended in 100 μL PBS were injected subcutaneously into the flank of 6-week-old female nude mice for tumor growth assays. The following treatments were initiated when the tumor volume reached approximately 300 mm3. (1) IM-sensitive GIST cell lines and IM-resistant GIST cell lines were used to establish a subcutaneous xenograft model of GIST (n = 6 mice/group). When the tumors grew to the required size, mice were treated with IM via drinking water; (2) GIST cells were pretreated with USP13 lentivirus or USP13 control lentivirus. In addition, IM-resistant GIST cells were pretreated with USP13 inhibitors or control. These lentivirus-transfected cells were implanted into subcutaneous mice tumors to construct the USP13 positive, USP13-negative, and control groups (n = 6 mice/group). When the transplanted tumor grew to the required volume, mice in each group were treated with IM via drinking water. (3) IM-resistant GIST cells-xenografted mice and control mice were treated with USP13 inhibitors or with USP13 inhibitors and autophagy inhibitors in combination therapy to assess reversal of IM resistance (n = 6 mice/group). Mice were treated with IM (45 mg/kg/day) and with or without 3-MA (15 mg/kg/day) via drinking water when the tumors grew to the required size. The USP13 inhibitor Spautin-1 was used at 20 mg/kg/day when required. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [587] | |||
| Responsed Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B] | |||
| Responsed Drug | Spautin 1 | Preclinical | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 | |
| In-vivo Model | 6 × 106 logarithmically growing GIST cells resuspended in 100 μL PBS were injected subcutaneously into the flank of 6-week-old female nude mice for tumor growth assays. The following treatments were initiated when the tumor volume reached approximately 300 mm3. (1) IM-sensitive GIST cell lines and IM-resistant GIST cell lines were used to establish a subcutaneous xenograft model of GIST (n = 6 mice/group). When the tumors grew to the required size, mice were treated with IM via drinking water; (2) GIST cells were pretreated with USP13 lentivirus or USP13 control lentivirus. In addition, IM-resistant GIST cells were pretreated with USP13 inhibitors or control. These lentivirus-transfected cells were implanted into subcutaneous mice tumors to construct the USP13 positive, USP13-negative, and control groups (n = 6 mice/group). When the transplanted tumor grew to the required volume, mice in each group were treated with IM via drinking water. (3) IM-resistant GIST cells-xenografted mice and control mice were treated with USP13 inhibitors or with USP13 inhibitors and autophagy inhibitors in combination therapy to assess reversal of IM resistance (n = 6 mice/group). Mice were treated with IM (45 mg/kg/day) and with or without 3-MA (15 mg/kg/day) via drinking water when the tumors grew to the required size. The USP13 inhibitor Spautin-1 was used at 20 mg/kg/day when required. | |||
Wnt family member 7B (WNT7B)
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [419] | |||
| Responsed Disease | Esophageal Squamous Cell Carcinoma [ICD-11: 2B70.1] | |||
| Cell Process | Transcription | |||
| Cell proliferation | ||||
| Cell migration | ||||
| Cell invasion | ||||
X-ray repair cross-complementing protein 5 (XRCC5)
Genotoxicity [ICD-11: XE2PA]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [286] | |||
| Responsed Disease | Genotoxicity [ICD-11: XE2PA] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
GC-2spd(ts)
|
N.A. | Mus musculus | CVCL_6633 |
| In-vivo Model | Corn oil (control group), 1 mg/kg/day DEHP (low-dose: D1 group), 250 mg/kg/day DEHP (middle-dose: D250 group), and 500 mg/kg/day DEHP (high-dose: D500 group). | |||
Zinc finger and BTB domain-containing protein 7C (ZBTB7C)
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [588] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Responsed Drug | STM2457 | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
MNNG/HOS Cl #5 | Osteosarcoma | Homo sapiens | CVCL_0439 |
| MG-63 | Osteosarcoma | Homo sapiens | CVCL_0426 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| hFOB 1.19 | Normal | Homo sapiens | CVCL_3708 | |
| In-vivo Model | (1) ZBTB7C functional assay in vivo, 2 × 106 MNNG/HOS cells were stably transfected with lentivirus (vector control and si-ZBTB7C, n = 5), and then subcutaneously incubated in BALB/c nude mice; (2) STM2457 intervention. MNNG/HOS cells were implanted subcutaneously first. When tumors reached approximately 80-100 mm3, PBS or STM2457 (30 mg/kg (STM-30), 60 mg/kg (STM-60), n = 4) were administered every 2 days; (3) ZBTB7C effects of STM2457 assay. Mice were assigned to 4 groups (n = 5 per group). Two groups had MNNG/HOS cells stably transfected with lentiviral vectors (Ctrl) and 2 with lentivirus-mediated ZBTB7C overexpression (ZBTB7C-OE), and then mice were treated with control vehicle or STM2457 (60 mg/kg), respectively. The experiments went on for 22 days, and the xenografted tumors were measured, weighted, and characterized by H&E and IHC staining. | |||
Zinc transporter ZIP9 (SLC39A9)
Periodontitis [ICD-11: DA0C]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [589] | |||
| Responsed Disease | Periodontitis [ICD-11: DA0C] | |||
| Target Regulation | Down regulation | |||
Unspecific Target Gene
Viral infections [ICD-11: 1D9Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [591] | |||
| Responsed Disease | Viral infections [ICD-11: 1D9Y] | |||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| Response Summary | m6A is the most abundant internal modification described in eukaryotic mRNA and several viral RNA including human respiratory syncytial virus (HRSV) infection. METTL3/METTL14 m6A writer complex plays a negative role in HRSV infections protein synthesis and viral titers, while m6A erasers FTO and ALKBH5 had the opposite effect. | |||
Herpes infection [ICD-11: 1F00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [592] | |||
| Responsed Disease | Herpes infection [ICD-11: 1F00] | |||
In-vitro Model |
HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| RD | Embryonal rhabdomyosarcoma | Homo sapiens | CVCL_1649 | |
| Response Summary | METTL3 knockdown suppressed the HSV-1 intermediate early and early genes (ICP0, ICP8 and UL23) and late genes (VP16, UL44, UL49 and ICP47). The components of m6A modification machinery, particularly m6A initiator METTL3 and reader YTHDF3, would be potential important targets for combating herpes virus type 1 (HSV-1) infections. | |||
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [593] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Cell Process | RNA editing | |||
In-vitro Model |
GSC | Glioma | Epinephelus akaara | CVCL_M752 |
| MGG8 (The primary tumor) | ||||
| Response Summary | METTL3 plays a vital role in many steps of RNA processing and orchestrates successful execution of oncogenic pathways in Glioma. | |||
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [595] | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
In-vitro Model |
EoL-1 | Chronic eosinophilic leukemia | Homo sapiens | CVCL_0258 |
| HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 | |
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| In-vivo Model | 6- to 10-week-old female Rosa26Cas9/+ mice were treated daily for two weeks with either vehicle or 50 mg kg-1 STM2457 (STORM). Four weeks after treatment, bone marrow cells from these mice were freshly dissected (as mentioned above) and blocked with anti-mouse CD16/32 (BD Pharmigen, cat. no. 553142) and 10% mouse serum (Sigma). | |||
| Response Summary | Inhibition of METTL3 by STM2457 targets key stem cell populations of acute myeloid leukaemia and reverses the AML phenotype, preventing or slowing the development of AML in re-transplantation experiments. | |||
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [596] | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
In-vitro Model |
HOK | Normal | Hexagrammos otakii | CVCL_YE19 |
| HN-6 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_8129 | |
| HN-5 | Squamous cell carcinoma of the oral cavity | Homo sapiens | CVCL_8128 | |
| HN-15 | Squamous cell carcinoma of the oral cavity | Homo sapiens | CVCL_W297 | |
| Response Summary | METTL3 and METTL14 are overexpressed in OSCC tissues and in the HN6 OSCC cell line that promotes cell proliferation. Overexpressed METTL3 or METTL14 is found to be an independent prognostic factor for short overall survival in patients with OSCC. | |||
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [597] | |||
| Responsed Disease | Esophageal Squamous Cell Carcinoma [ICD-11: 2B70.1] | |||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| KYSE-30 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1351 | |
| KYSE-510 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1354 | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [598] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
In-vitro Model |
SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| Response Summary | The results revealed a negative functional regulation of the LINC01559/miR-106b-5p/PTEN axis in CRC progression and explored a new mechanism of METTL3-mediated m6A modification on LINC01559. | |||
Pancreatic cancer [ICD-11: 2C10]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [599] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Responsed Drug | Gemcitabine | Approved | ||
| Pathway Response | Adipocytokine signaling pathway | hsa04920 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| HDE-CT cell line (A normal human pancreatic cell line) | ||||
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| Response Summary | Lasso regression identified a six-m6A-regulator-signature prognostic model (KIAA1429, HNRNPC, METTL3, YTHDF1, IGF2BP2, and IGF2BP3). Gene set enrichment analysis revealed m6A regulators (KIAA1429, HNRNPC, and IGF2BP2) were related to multiple biological behaviors in pancreatic cancer, including adipocytokine signaling, the well vs. poorly differentiated tumor pathway, tumor metastasis pathway, epithelial mesenchymal transition pathway, gemcitabine resistance pathway, and stemness pathway. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [600] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Cell Process | Mitogen-activated protein kinase cascades | |||
| Ubiquitin-dependent process | ||||
| RNA splicing | ||||
In-vitro Model |
MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 |
| Response Summary | METTL3 was associated with mitogen-activated protein kinase cascades, ubiquitin-dependent process and RNA splicing and regulation of cellular process, suggesting functional roles and targets of METTL3. METTL3 is a potent target for enhancing therapeutic efficacy in patients with pancreatic cancer. | |||
Liver cancer [ICD-11: 2C12]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [602] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Lenvatinib | Approved | ||
In-vitro Model |
Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | To increase the accuracy of the experimental results in vivo, we subcutaneously transplanted tumor tissues from HCC patients into BALB/c nude mice and then treated the tumor-bearing mice with lenvatinib to establish a Lenvatinib-resistant PDX mode. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [603] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Response Summary | METTL3 and YTHDF1 expression were found to be correlated with an increased risk and were included in an m6A-related gene signature for predicting prognosis of hepatocellular carcinoma. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [291] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Cell Process | RNA stability | |||
| RNA degradation (hsa03018) | ||||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| Response Summary | MIR33A can attenuate NSCLC cells proliferation via targeting to the 3'UTR of METTL3 mRNA. | |||
Melanoma [ICD-11: 2C30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [606] | |||
| Responsed Disease | Melanoma [ICD-11: 2C30] | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Cell cycle | |||
| Cell apoptosis | ||||
| Cell colony formation | ||||
| Cell migration | ||||
| Cell invasion | ||||
| Response Summary | Hsa-miR-302a-3p targets METTL3 and plays tumour suppressive roles in the proliferation, apoptosis, invasion, and migration of melanoma cells. | |||
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [607] | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
CRL-11731D cell line (Human ovarian cancer cell) | |||
| TOV-112D | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_3612 | |
| Response Summary | METTL3 can regulate m6A methylation independently of METTL14 and WTAP in endometrioid epithelial ovarian cancer. | |||
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [608] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
In-vitro Model |
HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| Response Summary | CD33+ MDSC expansion is linked to high levels of METTL3 and that METTL3 and CD33+ MDSCs are independent prognostic factors in CC. | |||
Renal cell carcinoma [ICD-11: 2C90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [609] | |||
| Responsed Disease | Renal cell carcinoma of kidney [ICD-11: 2C90.0] | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Cell Process | Adipogenesis | |||
| Response Summary | It is plausible that VHL-HIF-METTL3/14 pathways are involved in the m6A regulation in clear cell renal cell carcinoma cells, and PI3K-mTOR as well as p53 signaling pathways are possible downstream targets of m6A in ccRCC. | |||
Hematological disorders [ICD-11: 3C0Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [611] | |||
| Responsed Disease | Hematological disorders [ICD-11: 3C0Z] | |||
| Response Summary | This reviewed summarize and discuss recent findings regarding the biological functions and underlying mechanisms of m6A modification(i.e., the METTL3/METTL14/WTAP complex and other cofactor proteins) and the associated machinery in normal hematopoiesis and the initiation, progression, and drug response of acute myeloid leukemia (AML), a major subtype of leukemia usually associated with unfavorable prognosis. | |||
Obesity [ICD-11: 5B81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [612] | |||
| Responsed Disease | Obesity [ICD-11: 5B81] | |||
| Cell Process | Adipogenesis | |||
In-vitro Model |
3T3F442A | Normal | Mus musculus | CVCL_0122 |
| 3T3-L1 | Normal | Mus musculus | CVCL_0123 | |
| COS (From the African green monkey cell line (CV-1).) | ||||
| MEF (Mouse embryonic fibroblasts) | ||||
| In-vivo Model | Mice were anesthetized after 24 h of fasting, and 5 U of human insulin (Humalin R; Eli Lilly) was injected into the inferior vena cava. After 5 min, the liver and hind limb muscles were dissected and immediately frozen in liquid nitrogen. | |||
| Response Summary | WTAP, coupled with METTL3 and METTL14, is increased and distributed in nucleus by the induction of adipogenesis dependently on RNA in vitro Knockdown of each of these three proteins leads to cell cycle arrest and impaired adipogenesis associated with suppression of cyclin A2 upregulation during MCE, whose knockdown also impairs adipogenesis. | |||
Alzheimer disease [ICD-11: 8A20]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [613] | |||
| Responsed Disease | Alzheimer disease [ICD-11: 8A20] | |||
| Cell Process | Synaptic or neuron development and growth | |||
| Response Summary | The alterations of m6A RNA methylation in alzheimer's disease and in C57BL/6 mice were investigated using high-throughput sequencing. The expression of the m6A methyltransferase METTL3 was elevated and that of the m6A demethylase FTO was decreased in AD mice. | |||
Acute ischemic stroke [ICD-11: 8B11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [616] | |||
| Responsed Disease | Acute ischemic stroke [ICD-11: 8B11] | |||
| In-vivo Model | 36 male C57BL/6J mice (20-22 g, 2-month-old) were purchased from SPF (Beijing) biotechnology co., Ltd (Beijing, China) and maintained in the specific pathogen-free (SPF) animal laboratory with a 12/12 h light/dark cycle with free access to food and water. The mice were randomly assigned into six groups (n = 6 per group): (1) sham-operated group (Sham), (2) MCAO 6 h, (3) MCAO 12 h, (4) MCAO 1 d, (5) MCAO 3 d, and (6) MCAO 7 d. | |||
Cerebrovascular diseases [ICD-11: 8B22]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [617] | |||
| Responsed Disease | Arteriovenous malformation of cerebral vessels [ICD-11: 8B22.40] | |||
| Pathway Response | Notch signaling pathway | hsa04330 | ||
In-vitro Model |
HUVEC-C | Normal | Homo sapiens | CVCL_2959 |
| Response Summary | The expression level of METTL3 was reduced in the larger pathological tissues of cerebral arteriovenous malformation (AVM). Moreover, knockdown of METTL3 significantly affected angiogenesis of the human endothelial cells. | |||
Muscular dystrophies [ICD-11: 8C70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [618] | |||
| Responsed Disease | Muscular dystrophies [ICD-11: 8C70] | |||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| C2C12 | Normal | Mus musculus | CVCL_0188 | |
| In-vivo Model | For mouse muscle injury and regeneration experiment, tibialis anterior (TA) muscles of six-week-old male mice were injected with 25 uL of 10 uM cardiotoxin (CTX, Merck millipore, 217 503), then the C2C12 cells, which stably overexpressed METTL3 or GFP control, were injected to the injury site of mice on the following day. The regenerated muscles were collected at day 7 post-injection, frozen in liquid nitrogen for RNA and protein extraction. | |||
| Response Summary | Muscle specific miRNAs are essential for skeletal muscle differentiation. METTL3 regulated the expressions of muscle specific miRNAs by multiple mechanisms. | |||
Cortical development [ICD-11: 8E4Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [619] | |||
| Responsed Disease | Cortical development [ICD-11: 8E4Y] | |||
| In-vivo Model | Homologous recombination was used, cas9 mRNA, gRNA, and donor vector were microinjected into the fertilized eggs of C57BL/6J mice to obtain F0 generation mice. | |||
| Response Summary | Knockout of Mettl3 leads to a more severe disruption of translational regulation of mRNAs than deletion of Fto and results in altered translation of crucial genes in cortical radial glial cells and intermediate progenitors. Uncover a profound role of Mettl3 in regulating translation of major mRNAs that control proper cortical development. | |||
Disorders of the retina [ICD-11: 9B70-9C0Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [620] | |||
| Responsed Disease | Disorders of the retina [ICD-11: 9B70-9C0Z] | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
ARPE-19 | Normal | Homo sapiens | CVCL_0145 |
| In-vivo Model | The vitreous cavity of the right eye of each rat was injected either with ARPE-19 cells (1 × 105 per uL in PBS [pH 7.4]) overexpressing METTL3 or control vector cells or with an equal volume of PBS (pH 7.4). | |||
| Response Summary | METTL3 is involved in the PVR process, and METTL3 overexpression inhibits the EMT of ARPE-19 cells in vitro and suppresses the PVR process in vivo. | |||
Retinopathy [ICD-11: 9B71]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [621] | |||
| Responsed Disease | Retinopathy [ICD-11: 9B71] | |||
| Cell Process | Cell viability | |||
| Cell proliferation | ||||
| Cell migration | ||||
In-vitro Model |
HUVEC-C | Normal | Homo sapiens | CVCL_2959 |
| In-vivo Model | Mettl3flox/flox mice were crossed with the transgenic Cdh5-CreERT2 mice to generate the Mettl3-ecKO mice. Cdh5-Cre Mettl3flox/flox mice received an intragastric injection of 50 ul tamoxifen (1 mg/mL) at P1-P3 and P5 for Cre activation and Mettl3 knockout. After Mettl3 knockout, the mouse pups and their nursing mothers were exposed to 75% oxygen (hyperoxia) from P7 to P12 in an incubator chamber. Then, the pups were returned to normal oxygen conditions (normoxia). | |||
| Response Summary | METTL3 knockout in vivo decreased avascular area and pathological neovascular tufts in an oxygen-induced retinopathy model and inhibited alkali burn-induced corneal neovascularization. | |||
Presbycusis [ICD-11: AB54]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [622] | |||
| Responsed Disease | Presbycusis [ICD-11: AB54] | |||
Cardiomegaly [ICD-11: BC45]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [623] | |||
| Responsed Disease | Cardiomegaly [ICD-11: BC45] | |||
| Responsed Drug | A939572 | Investigative | ||
| In-vivo Model | Slack Laboratory Animal Co., Ltd. (Shanghai, China) supplied the six-week-old C57BL/6 male mice. Animals were reared at constant temperature (21 ± 1 °C) and maintained in a room with water and food during a 12 h dark/light cycle. After acclimatizing for a week, the animals were randomly assigned to two groups: model (TAC) and control (sham). In the TACgroup, the aortic arch of the mice was constricted using a 27 G needle and 6.0 sutures. The Sham group was operated in the same way as the TAC group without ligation of the aorta. All mice were housed for four weeks for further research. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [624] | |||
| Responsed Disease | Cardiomegaly [ICD-11: BC45] | |||
| Cell Process | Heart failure | |||
In-vitro Model |
Neonatal rat ventricular cardiomyocytes (Primary myocyte cells) | |||
| In-vivo Model | Mettl3 fl/fl mice were crossed with mice expressing cre recombinase under the control of the cardiac-specific Myh7 promoter (Beta-myosin heavy chain [Beta-MHC]) to obtain heart-restricted deletion of Mettl3 (METTL3-cKO; cKO). | |||
| Response Summary | Inhibition of METTL3 completely abrogated the ability of cardiomyocytes to undergo hypertrophy when stimulated to grow, whereas increased expression of the m6A RNA methylase METTL3 was sufficient to promote cardiomyocyte hypertrophy both in vitro and in vivo. | |||
Asthma [ICD-11: CA23]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [625] | |||
| Responsed Disease | Allergic asthma [ICD-11: CA23.0] | |||
| In-vivo Model | Six-week-old male BABL/c mice were purchased from Hunan SJA Laboratory Animal Co., Ltd. The mice were initially divided into four groups, each containing 6 mice: Control group, Control+S. boulardii group, Model group, and Model+S. boulardii group. With reference to the previous OVA method [27,28], the animal model of asthma was established. Mice in Control+S. boulardii and Model+S. boulardii groups were administered 300 μL/day of 1 × 107 CFU/d S. boulardiithrough oral gavage, starting from 6 days prior to the first OVA sensitization and continuing until day 21 [12,29]. The mice in Control and Control+S. boulardiigroups were injected with an equivalent volume of PBS. After the final OVA injection, the mice were challenged with 10 μL of 5 % OVA via intranasal inhalation for one week (day 15 to day 21), with 10 μL introduced into each nostril. The mice in Control and Control+S. boulardii groups were injected with an equivalent volume of PBS. Mice were sacrificed with pentobarbital sodium (150 mg/kg, intraperitoneal injection). On day 22, bronchoalveolar lavage fluid (BALF), intestinal tissue, and lung tissue were collected from all mice. | |||
Liver disease [ICD-11: DB9Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [626] | |||
| Responsed Disease | Liver disease [ICD-11: DB9Z] | |||
| Pathway Response | PPAR signaling pathway | hsa03320 | ||
| Cell Process | Fatty degeneration | |||
| In-vivo Model | A total of 24 male mice were randomly allocated to LFD (low-fat diet), LFDR (low-fat diet + resveratrol), HFD (high-fat diet), and HFDR (high-fat diet + resveratrol) groups for 12 weeks (n = 6/group). | |||
| Response Summary | The beneficial effect of resveratrol on lipid metabolism disorder under HFD is due to a decrease of m6A RNA methylation and an increase of PPARalpha mRNA, providing mechanistic insights into the function of resveratrol in alleviating the disturbance of lipid metabolism in mice. The resveratrol in HFD increased the transcript levels of methyltransferase like 3 (METTL3), alkB homolog 5 (ALKBH5), fat mass and obesity associated protein (FTO), and YTH domain family 2 (YTHDF2), whereas it decreased the level of YTH domain family 3 (YTHDF3) and m6A abundance in mice liver. | |||
Rheumatoid arthritis [ICD-11: FA20]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [627] | |||
| Responsed Disease | Rheumatoid arthritis [ICD-11: FA20] | |||
| Responsed Drug | STM2457 | Investigative | ||
| In-vivo Model | DBA/1 mice were injected intradermally with 100 μg of bovine type II collagen (CII) emulsified in 50 μL of complete Freund's adjuvant at a 1:1 ratio (vol/vol) as primary immunization on day 0. On day 21, the mice were boosted with CII emulsified at a 1:1 ratio in incomplete Freund's adjuvant. The weight, arthritis score, and thickness of the paws were observed and recorded once every 3 days. Starting on day 21, the CIA mice were administered an equal volume of methotrexate (MTX, 1 mg/kg) and METTL3 inhibitor STM2457 (50 mg/kg), which were intraperitoneally injected with twice a week. All mice were sacrificed and specimens were harvested on day 63. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [628] | |||
| Responsed Disease | Rheumatoid arthritis [ICD-11: FA20] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | TNF signaling pathway | hsa04668 | ||
| Cell Process | Inflammation in macrophages | |||
In-vitro Model |
THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| Response Summary | LPS could enhance the expression and biological activity of METTL3 in macrophages, while overexpression of METTL3 significantly attenuated the inflammatory response induced by LPS in macrophages.This study firstly demonstrates the critical role of METTL3 in RA, which provides novel insights into recognizing the pathogenesis of Rheumatoid Arthritis(RA) and a promising biomarker for RA. | |||
Low bone mass disorder [ICD-11: FB83]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [629] | |||
| Responsed Disease | Osteoporosis [ICD-11: FB83.1] | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Bone formation | |||
| In-vivo Model | OP rats were fixed prone, the skin was prepared and the skull was disinfected, resulting in a full-thickness defect of 8mm. BCP incubated with OP-BMSCs was implanted in the skull defect area. After 8 weeks, skull specimens were taken after the rats were euthanized. | |||
| Response Summary | Overexpression of Mettl3 could partially rescue the decreased bone formation ability of OP-BMSCs by the canonical Wnt signalling pathway. Therefore, Mettl3 can be a key targeted gene for bone generation and therapy of bone defects in OP patients. | |||
Male infertility [ICD-11: GB04]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [630] | |||
| Responsed Disease | Male infertility [ICD-11: GB04] | |||
| Responsed Drug | Ethyl ester form of meclofenamic acid | Approved | ||
| Cell Process | Cell cycle | |||
| Cell proliferation | ||||
In-vitro Model |
GC-1 spg | Normal | Mus musculus | CVCL_8872 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | Mouse GC-1 spg cells were treated with the ester form of meclofenamic acid (MA2) to inhibit the demethylase activity of FTO. | |||
| Response Summary | METTL3, METTL14, ALKBH5 and YTHDC2 are involved in the regulation of spermatogenesis and oogenesis. MA2 affected CDKs expression through the m6A-dependent mRNA degradation pathway, and thus repressed spermatogonial proliferation. Additionally, mutation of the predicted m6A sites in the Cdk2-3'UTR could mitigated the degradation of CDK2 mRNA after MA2 treatment. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [631] | |||
| Responsed Disease | Male infertility [ICD-11: GB04] | |||
| Cell Process | SSC proliferation/differentiation | |||
In-vitro Model |
Spermatogenic cells (Prepared from undifferentiated mouse spermatogonial cells) | |||
| In-vivo Model | All mice described above were maintained on the C57BL/6J (B6) background. Mettl3- and Mettl14-floxed mice (Mettl3flox/flox and Mettl14flox/flox) were then bred with germ cell-specific expressed Cre mice including Vasa-Cre mouse line (Jackson Laboratory, Bar Harbor, Maine, USA) and Stra8-GFPCre mouse line for excising the loxP-flanked exon 4 and exon 2 to generate germ cell-specific Mettl3 and Mettl14KO mice, respectively. Germ cell-specific Mettl3 and Mettl14 double KO mice were obtained by crossing Mettl3flox/floxMettl14flox/flox with Mettl3flox/+Mettl14flox/+ or Mettl3flox/+Mettl14flox/flox carried germ cell-specific expressed Cre mice. | |||
| Response Summary | Combined deletion of Mettl3 and Mettl14 in advanced germ cells with Stra8-GFPCre disrupts spermiogenesis, whereas mice with single deletion of either Mettl3 or Mettl14 in advanced germ cells show normal spermatogenesis. | |||
Structural developmental anomalies of large intestine [ICD-11: LB16]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [632] | |||
| Responsed Disease | Structural developmental anomalies of large intestine [ICD-11: LB16.1] | |||
CNS anomalies syndrome [ICD-11: LD20]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [633] | |||
| Responsed Disease | Pontocerebellar hypoplasia [ICD-11: LD20.01] | |||
| Cell Process | Extended RNA half-lives and aberrant splicing | |||
| Cell apoptosis | ||||
In-vitro Model |
CGC (Conjunctival goblet cells) | |||
| Neural stem cell (NSC) lines (A group of ectodermal progenitor cells) | ||||
| In-vivo Model | All pups were genotyped and two Mettl3flox/+ founder mice were obtained. | |||
| Response Summary | METTL3 depletion-induced loss of m6A modification causes extended RNA half-lives and aberrant splicing events, consequently leading to dysregulation of transcriptome-wide gene expression and premature CGC death.Mettl3 in mouse nervous system causes severe developmental defects in the brain. | |||
Senescent cell [ICD-11: MG2A]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [634] | |||
| Responsed Disease | Senescent cell [ICD-11: MG2A] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | nonhomologous end joining | NHEJ) pathwa | ||
Inflammatory response [ICD-11: MG46]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [516] | |||
| Responsed Disease | Inflammatory response [ICD-11: MG46] | |||
In-vitro Model |
THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Injury of liver [ICD-11: NB91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [635] | |||
| Responsed Disease | Liver injury [ICD-11: NB91.1] | |||
| Responsed Drug | BHBA | Investigative | ||
Exposure to radiation [ICD-11: PH73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [636] | |||
| Responsed Disease | Exposure to radiation [ICD-11: PH73] | |||
In-vitro Model |
MRC-5 | Normal | Homo sapiens | CVCL_0440 |
| Response Summary | m6A RNA methylation has been defined and appears to affect DNA repair by modulation of translation. | |||
COVID-19 [ICD-11: RA01]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [637] | |||
| Responsed Disease | COVID-19 [ICD-11: RA01] | |||
In-vitro Model |
Vero E6 (African green monkey kidney cell Vero E6 cells (ATCC CLR-1586)) | |||
| MRC-5 | Normal | Homo sapiens | CVCL_0440 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| Response Summary | Replication of SARS-CoV-2, the agent responsible for the COVID-19 pandemic, and a seasonal human Bete-coronavirus HCoV-OC43, can be suppressed by depletion of METTL3 or cytoplasmic m6A reader proteins YTHDF1 and YTHDF3 and by a highly specific small molecule METTL3 inhibitor. | |||
ATP-binding cassette sub-family C member 9 (ABCC9)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
  |
logFC: -1.97E+00 p-value: 5.10E-08 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.69E+00 | GSE60213 |
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [14] | |||
| Responsed Disease | Nasopharyngeal carcinoma | ICD-11: 2B6B | ||
| Target Regulation | Down regulation | |||
| Pathway Response | ABC transporters | hsa02010 | ||
| Wnt signaling pathway | hsa04310 | |||
| Ubiquitin mediated proteolysis | hsa04120 | |||
| Cell Process | Ubiquitination degradation | |||
| In-vitro Model | CNE-1 | Normal | Homo sapiens | CVCL_6888 |
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| In-vivo Model | A total of 2 × 106 cells was mixed with 0.2 ml PBS (pH 7.4) and 30% (v/v) Matrigel matrix (BD Biosciences). | |||
| Response Summary | TRIM11 regulates nasopharyngeal carcinoma drug resistance by positively modulating the Daple/beta-catenin/ATP-binding cassette sub-family C member 9 (ABCC9) signaling pathway. TRIM11 enhanced the multidrug resistance in NPC by inhibiting apoptosis in vitro and promoting cisplatin (DDP) resistance in vivo. METTL3-mediated m6A modification caused the upregulation of TRIM11 via IGF2BP2 in NPC drug-resistant cells. | |||
Autophagy protein 5 (ATG5)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | CT26 cell line | Mus musculus |
|
Treatment: METTL3 knockout CT26 cells
Control: CT26 cells
|
GSE142589 | |
| Regulation |
 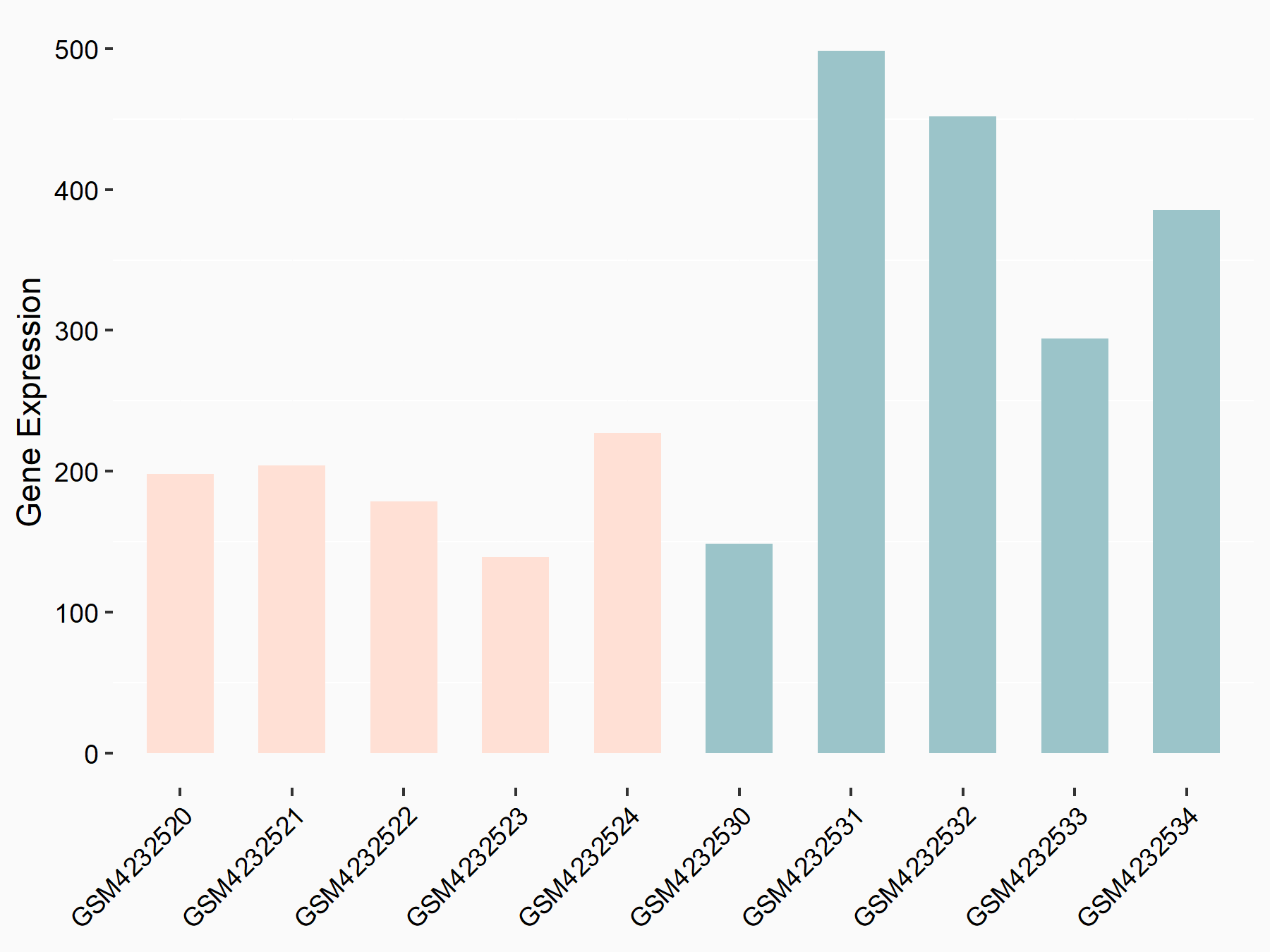 |
logFC: -9.09E-01 p-value: 4.91E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 8.30E+00 | GSE60213 |
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [17] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, Autophagy protein 5 (ATG5), ATG12, and ATG16L1. | |||
Chloroquine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as Autophagy protein 5 (ATG5), ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
Gefitinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as Autophagy protein 5 (ATG5), ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
Beta-Elemen
[Phase 3]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as Autophagy protein 5 (ATG5), ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [19] | |||
| Responsed Disease | Testicular cancer | ICD-11: 2C80 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular Processes | |||
| Cellular Transport | ||||
| Cellular catabolism | ||||
| Cell autophagy | ||||
| In-vitro Model | Tcam-2/DDP (Cisplatin-resistant TCam-2 cell line) | |||
| TCam-2 | Testicular seminoma | Homo sapiens | CVCL_T012 | |
| Response Summary | m6A methyltransferase METTL3 regulates autophagy and sensitivity to cisplatin by targeting Autophagy protein 5 (ATG5) in seminoma. The use of autophagy inhibitors 3-MA could reverse the protective effect of METTL3 on TCam-2 cells. | |||
3-Methyladenine
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [19] | |||
| Responsed Disease | Testicular cancer | ICD-11: 2C80 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular Processes | |||
| Cellular Transport | ||||
| Cellular catabolism | ||||
| Cell autophagy | ||||
| In-vitro Model | Tcam-2/DDP (Cisplatin-resistant TCam-2 cell line) | |||
| TCam-2 | Testicular seminoma | Homo sapiens | CVCL_T012 | |
| Response Summary | m6A methyltransferase METTL3 regulates autophagy and sensitivity to cisplatin by targeting Autophagy protein 5 (ATG5) in seminoma. The use of autophagy inhibitors 3-MA could reverse the protective effect of METTL3 on TCam-2 cells. | |||
Spautin 1
[Preclinical]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [270] | |||
| Responsed Disease | Osteosarcoma | ICD-11: 2B51 | ||
| Target Regulation | Up regulation | |||
| In-vivo Model | For tumor growth assay, 2 × 106 OS cells in 100 μL medium were injected into the nude mice subcutaneously. For tumor metastasis assay, we injected medium containing 2 × 106 cells through the caudal vein. An IVIS200 imaging system (Caliper Life Science, USA) was used to image and assess the OS metastasis. For pharmacological inhibition of USP13, mice bearing xenografts were treated with Spautin-1 (40 mg/kg/day i.p.) or vehicle for 2 weeks. | |||
Autophagy-related protein 16-1 (ATG16L1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | CT26 cell line | Mus musculus |
|
Treatment: METTL3 knockout CT26 cells
Control: CT26 cells
|
GSE142589 | |
| Regulation |
 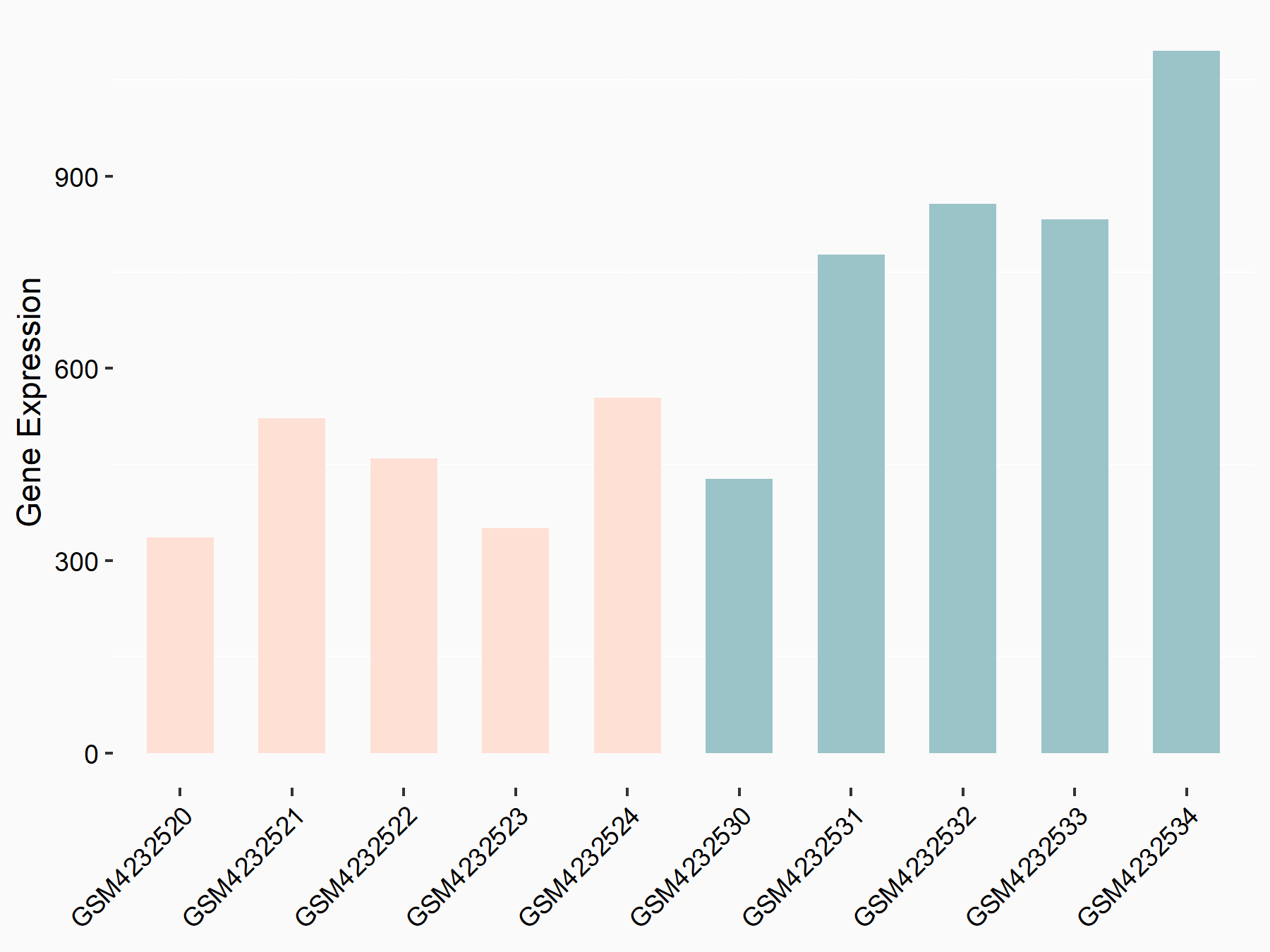 |
logFC: -8.44E-01 p-value: 3.07E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.03E+00 | GSE60213 |
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [17] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, ATG7, ATG12, and Autophagy-related protein 16-1 (ATG16L1). | |||
Broad substrate specificity ATP-binding cassette transporter ABCG2 (BCRP/ABCG2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
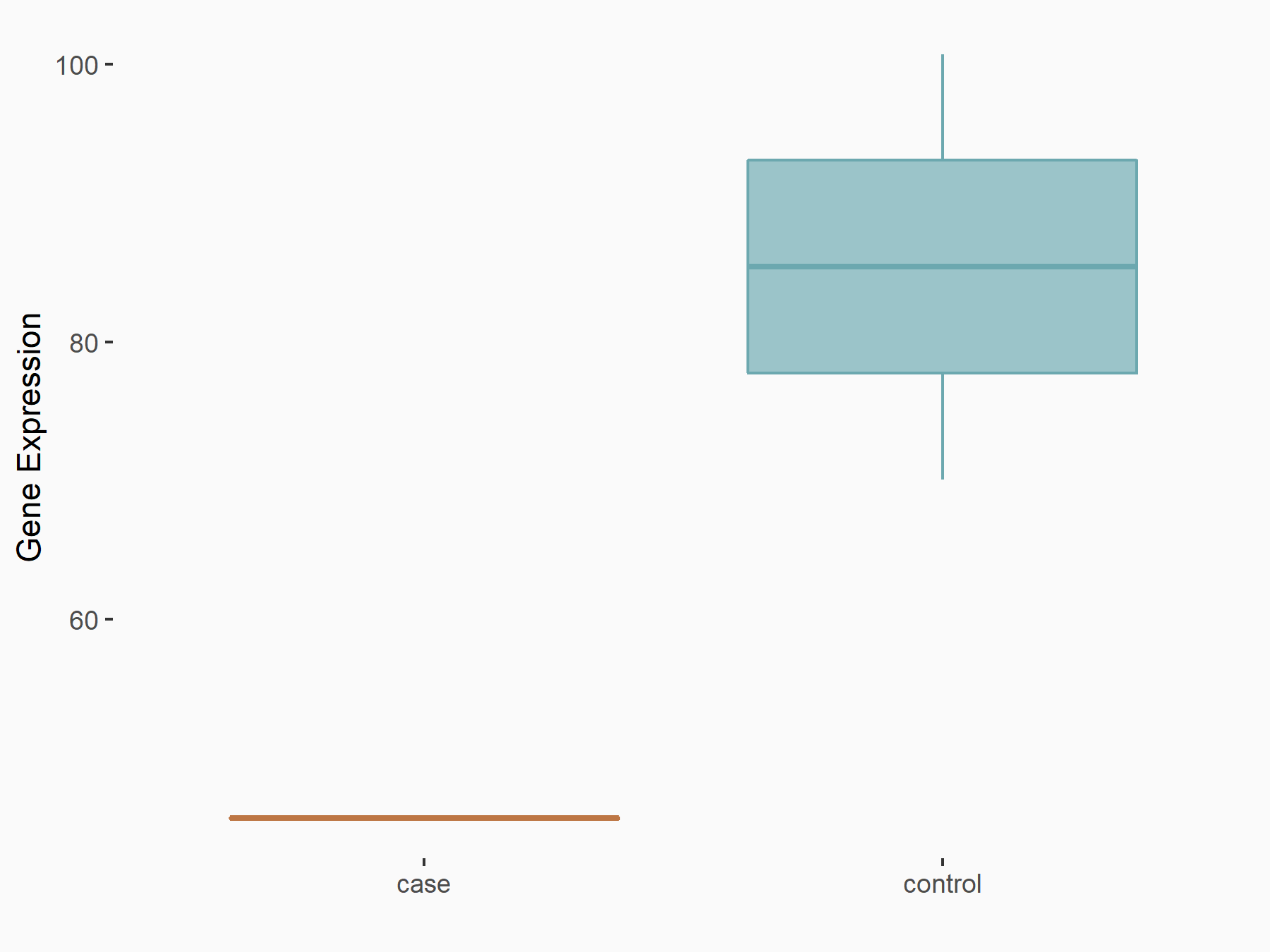 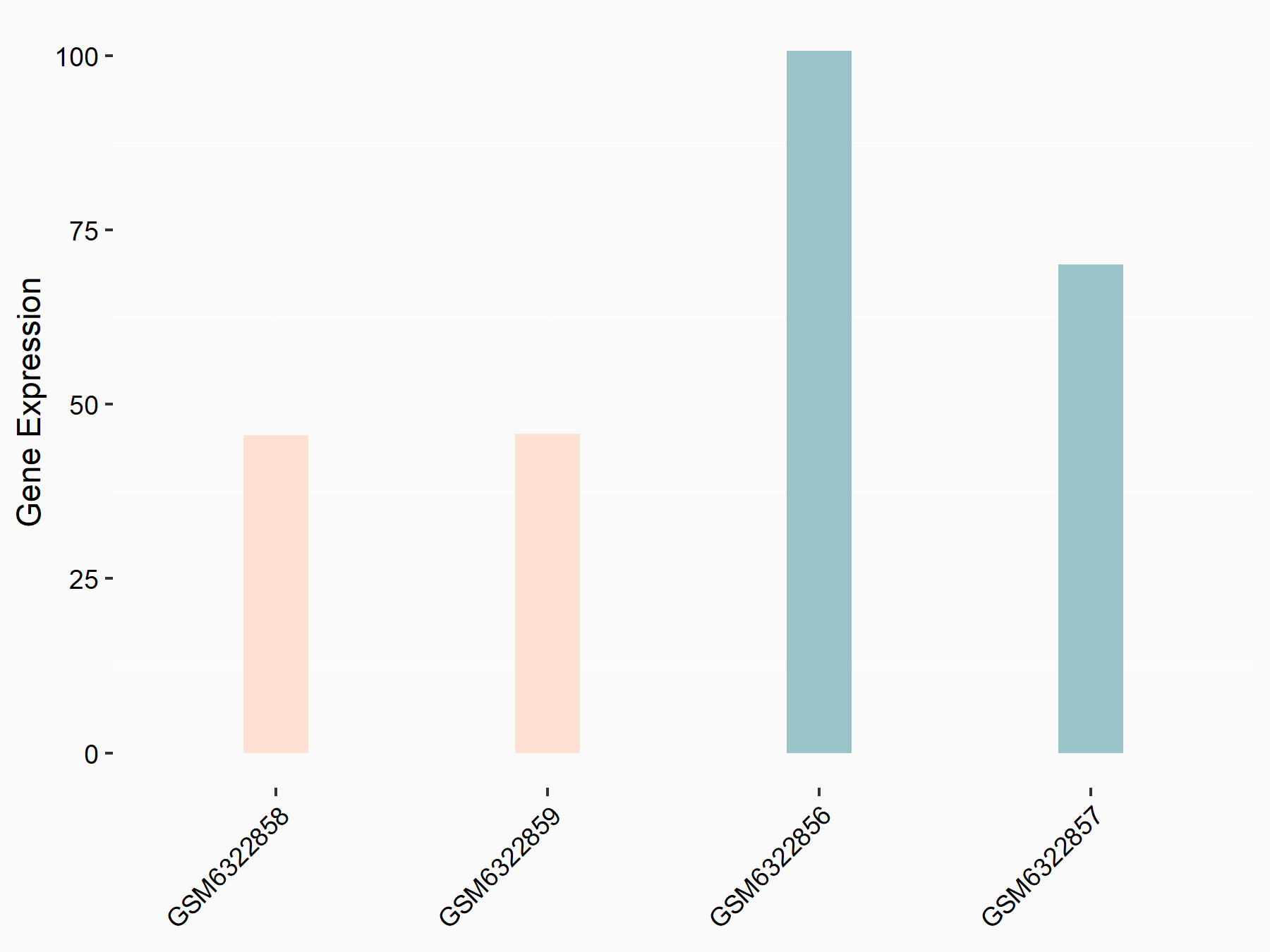 |
logFC: -9.03E-01 p-value: 2.29E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 4.88E+00 | GSE60213 |
Doxil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [22] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell growth and death | |||
| Cell apoptosis | ||||
| In-vitro Model | ADR-resistant MCF-7 (MCF-7/ADR) cells (Human breast cancer doxorubicin-resistant cell line) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| In-vivo Model | Cell suspensions (2 × 106 cells/mL) made with MCF-7/ADR cells stably expressing METTL3 and/or miR-221-3p inhibitor were subcutaneously implanted into each mouse. One week later, xenografted mice were injected with 0.1 mL ADR (25 mg/kg, intraperitoneal injection) twice a week. | |||
| Response Summary | METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. METTL3 knockdown was shown to reduce the expression of miR-221-3p by reducing pri-miR-221-3p m6A mRNA methylation, reducing the expression of MDR1 and Broad substrate specificity ATP-binding cassette transporter ABCG2 (BCRP/ABCG2), and inducing apoptosis. Identified the METTL3/miR-221-3p/HIPK2/Che-1 axis as a novel signaling event that will be responsible for resistance of BC cells to ADR. | |||
Cellular tumor antigen p53 (TP53/p53)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
 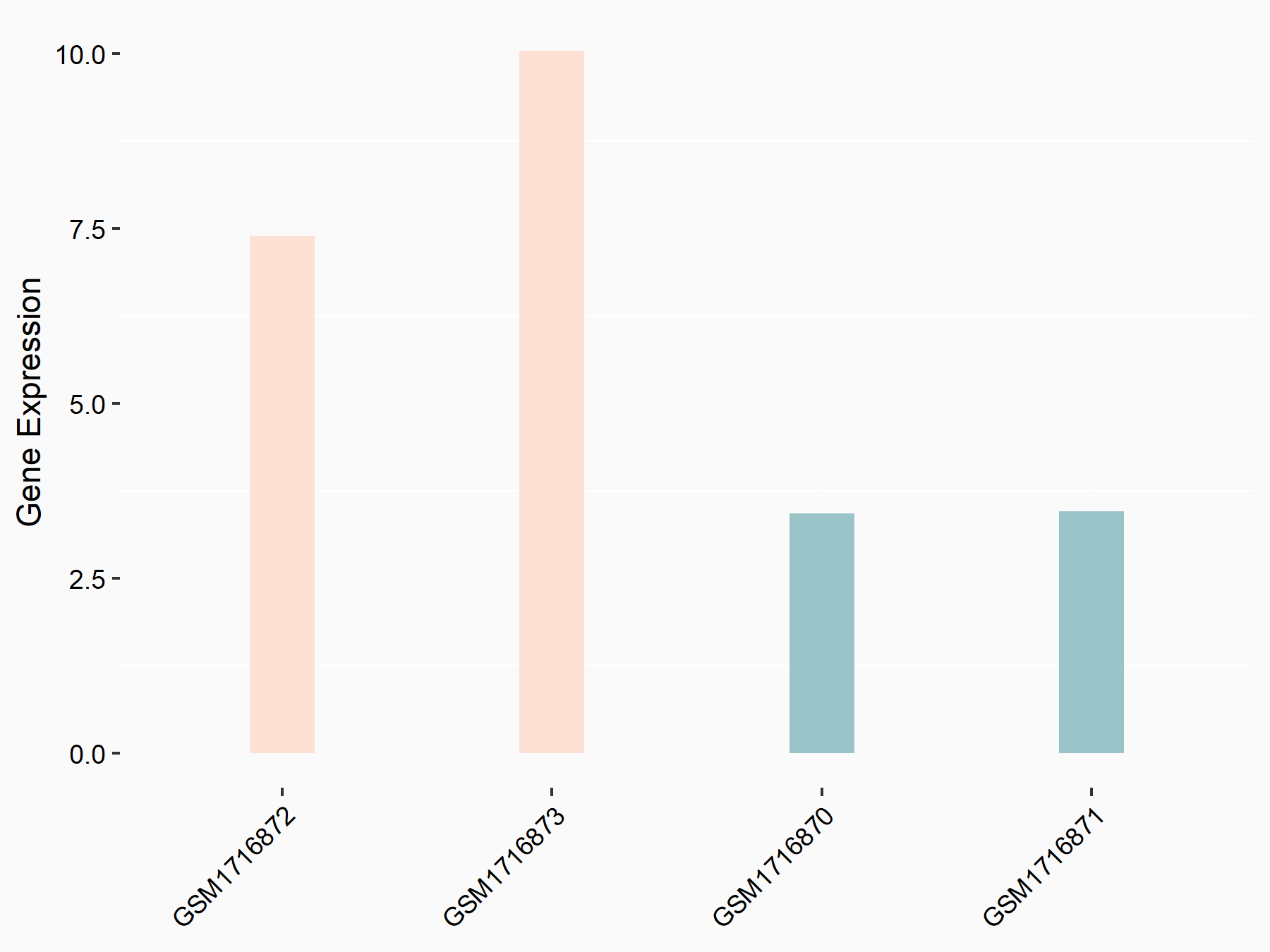 |
logFC: 1.12E+00 p-value: 1.48E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.05E+00 | GSE60213 |
Arsenite
[Phase 2]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [27] | |||
| Responsed Disease | Solid tumour/cancer | ICD-11: 2A00-2F9Z | ||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
| Response Summary | METTL3 significantly decreased m6A level, restoring Cellular tumor antigen p53 (TP53/p53) activation and inhibiting cellular transformation phenotypes in the arsenite-transformed cells. m6A downregulated the expression of the positive p53 regulator, PRDM2, through the YTHDF2-promoted decay of PRDM2 mRNAs. m6A upregulated the expression of the negative p53 regulator, YY1 and MDM2 through YTHDF1-stimulated translation of YY1 and MDM2 mRNA. This study further sheds light on the mechanisms of arsenic carcinogenesis via RNA epigenetics. | |||
Apatinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [30] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | QGY-7701 | Human papillomavirus-related endocervical adenocarcinoma | Homo sapiens | CVCL_6859 |
| HHL-5 | Normal | Homo sapiens | CVCL_S956 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | Nude mice (4-6 week-old) were administered sterile water and feed in a specific pathogen-free barrier. Using a 1-mL syringe, 1 × 107 HEPG2 cells were subcutaneously inoculated into the right axilla of nude mice to build the HCC xenograft model. When the tumor volume reached 50 mm3, the nude mice were randomly divided into 1 control (n = 4) and 3 treatment groups (n = 4 each). RG7112, apatinib, and RG7112 + apatinib were administered to the treatment groups and an equal volume of dimethyl sulfoxide to the control group by daily gavage for 14 d. The tumor length (L) and width (W) were measured on alternate days using vernier calipers. The following formula was used to calculate the tumor volume: volume (mm3) = 0.5 × L × W × W. At the end of the experiment, the nude mice were killed by CO2 overdose anesthesia. The tumors were dissected and weighed using a precision balance, and the tumor tissue was stored in liquid nitrogen for further analysis. | |||
| Response Summary | Cellular tumor antigen p53 (TP53/p53) n6-methyladenosine (m6A) played a decisive role in regulating Hepatocellular carcinoma(HCC) sensitivity to chemotherapy via the p53 activator RG7112 and the vascular endothelial growth factor receptor inhibitor apatinib. p53 mRNA m6A modification blockage induced by S-adenosyl homocysteine or siRNA-mediated METTL3 inhibition enhanced HCC sensitivity to chemotherapy. | |||
RG7112
[ Phase 1]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [30] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | QGY-7701 | Human papillomavirus-related endocervical adenocarcinoma | Homo sapiens | CVCL_6859 |
| HHL-5 | Normal | Homo sapiens | CVCL_S956 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | Nude mice (4-6 week-old) were administered sterile water and feed in a specific pathogen-free barrier. Using a 1-mL syringe, 1 × 107 HEPG2 cells were subcutaneously inoculated into the right axilla of nude mice to build the HCC xenograft model. When the tumor volume reached 50 mm3, the nude mice were randomly divided into 1 control (n = 4) and 3 treatment groups (n = 4 each). RG7112, apatinib, and RG7112 + apatinib were administered to the treatment groups and an equal volume of dimethyl sulfoxide to the control group by daily gavage for 14 d. The tumor length (L) and width (W) were measured on alternate days using vernier calipers. The following formula was used to calculate the tumor volume: volume (mm3) = 0.5 × L × W × W. At the end of the experiment, the nude mice were killed by CO2 overdose anesthesia. The tumors were dissected and weighed using a precision balance, and the tumor tissue was stored in liquid nitrogen for further analysis. | |||
| Response Summary | Cellular tumor antigen p53 (TP53/p53) n6-methyladenosine (m6A) played a decisive role in regulating Hepatocellular carcinoma(HCC) sensitivity to chemotherapy via the p53 activator RG7112 and the vascular endothelial growth factor receptor inhibitor apatinib. p53 mRNA m6A modification blockage induced by S-adenosyl homocysteine or siRNA-mediated METTL3 inhibition enhanced HCC sensitivity to chemotherapy. | |||
Erianin
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [277] | |||
| Responsed Disease | Renal cell carcinoma | ICD-11: 2C90 | ||
| Target Regulation | Down regulation | |||
Remimazolam
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [278] | |||
| Responsed Disease | Acute ischemic stroke | ICD-11: 8B11 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 |
eIF4E-binding protein 1 (4EBP1/EIF4EBP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shMETTL3 MOLM13 cells
Control: MOLM13 cells
|
GSE98623 | |
| Regulation |
 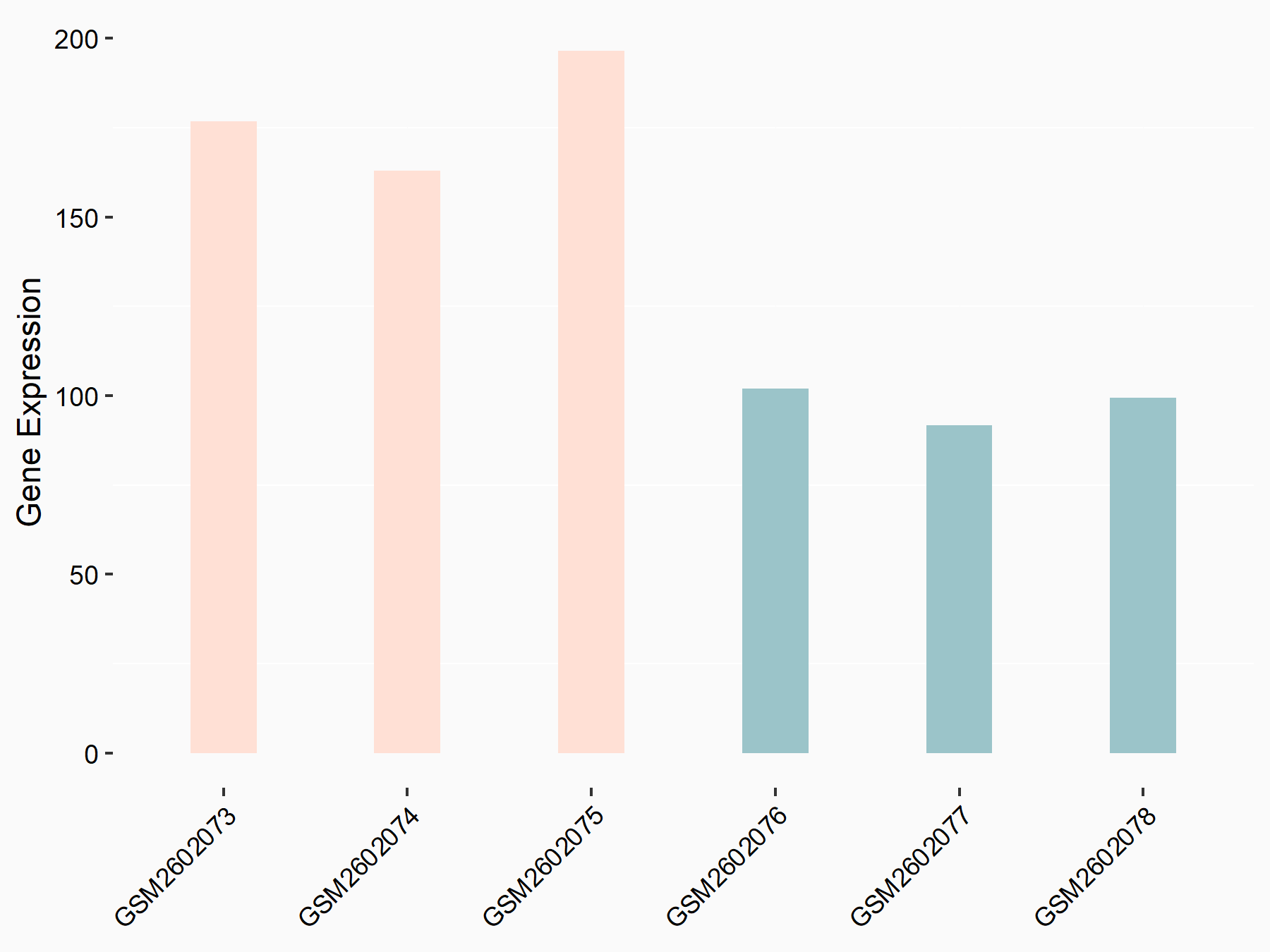 |
logFC: 8.71E-01 p-value: 3.32E-08 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 9.71E+00 | GSE60213 |
Rapamycin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [44] | |||
| Responsed Disease | Retinoblastoma | ICD-11: 2D02.2 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| In-vitro Model | WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through PI3K/AKT/mTOR pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-P70S6K/eIF4E-binding protein 1 (4EBP1/EIF4EBP1) pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
Homeodomain-interacting protein kinase 2 (HIPK2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
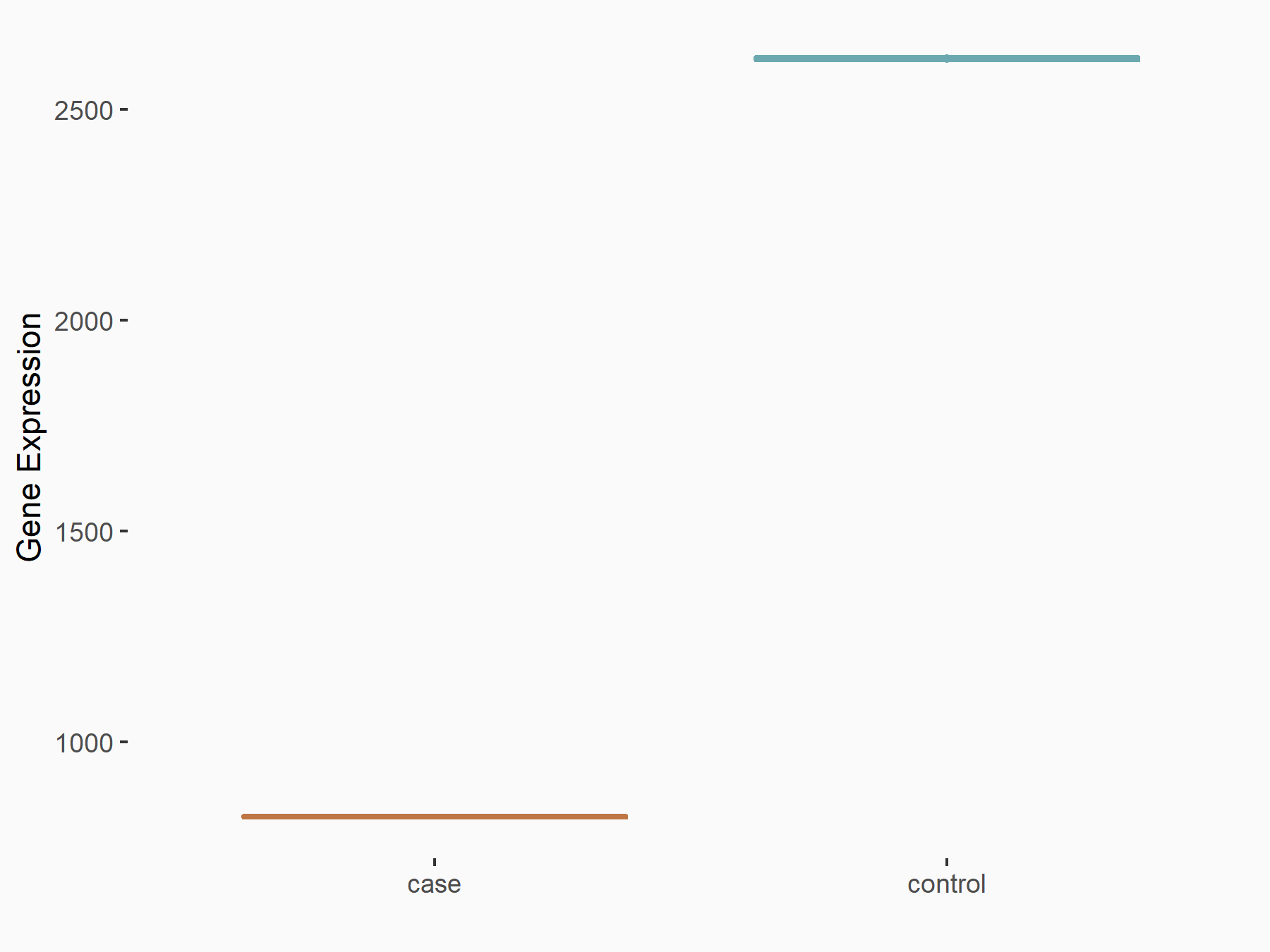 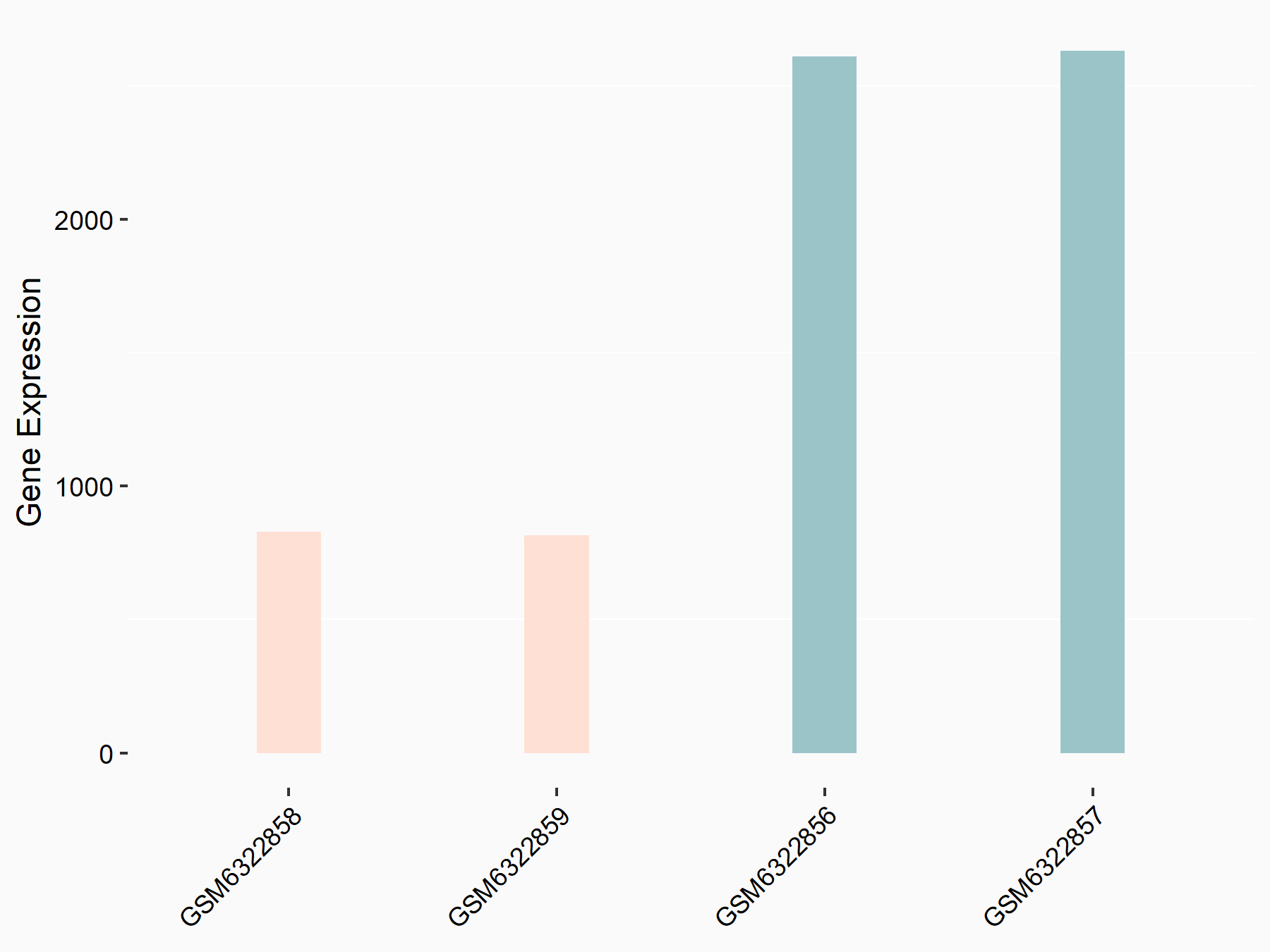 |
logFC: -1.67E+00 p-value: 2.62E-66 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.33E+00 | GSE60213 |
Doxil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [22] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell growth and death | |||
| Cell apoptosis | ||||
| In-vitro Model | ADR-resistant MCF-7 (MCF-7/ADR) cells (Human breast cancer doxorubicin-resistant cell line) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| In-vivo Model | Cell suspensions (2 × 106 cells/mL) made with MCF-7/ADR cells stably expressing METTL3 and/or miR-221-3p inhibitor were subcutaneously implanted into each mouse. One week later, xenografted mice were injected with 0.1 mL ADR (25 mg/kg, intraperitoneal injection) twice a week. | |||
| Response Summary | METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. METTL3 knockdown was shown to reduce the expression of miR-221-3p by reducing pri-miR-221-3p m6A mRNA methylation, reducing the expression of MDR1 and BCRP, and inducing apoptosis. Identified the METTL3/miR-221-3p/Homeodomain-interacting protein kinase 2 (HIPK2)/Che-1 axis as a novel signaling event that will be responsible for resistance of BC cells to ADR. | |||
Verbascoside
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [299] | |||
| Responsed Disease | Oral squamous cell carcinoma | ICD-11: 2B6E.0 | ||
| In-vitro Model | SCC-9 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1685 |
| UM1 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_VH00 | |
Kelch-like ECH-associated protein 1 (KEAP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
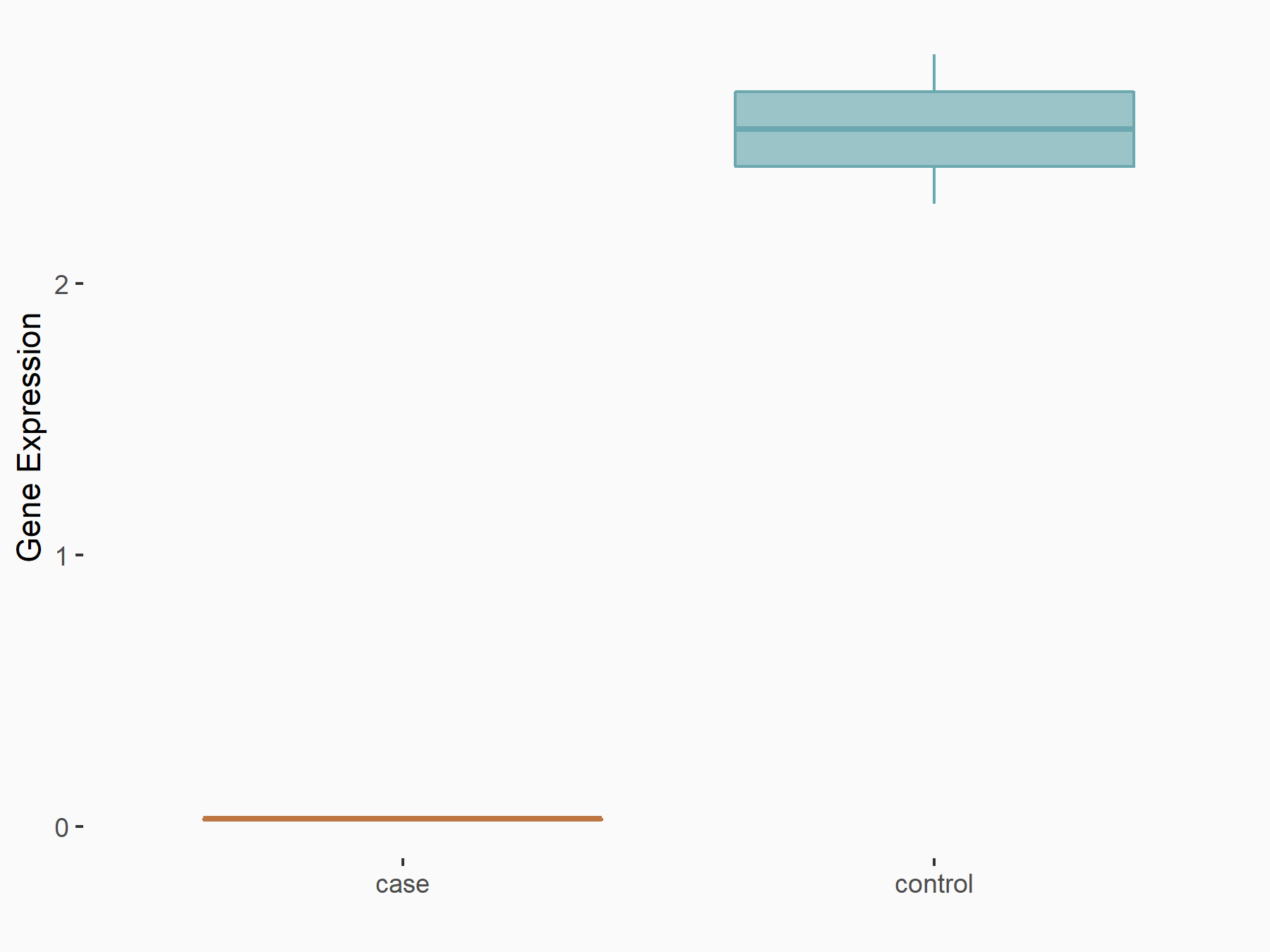 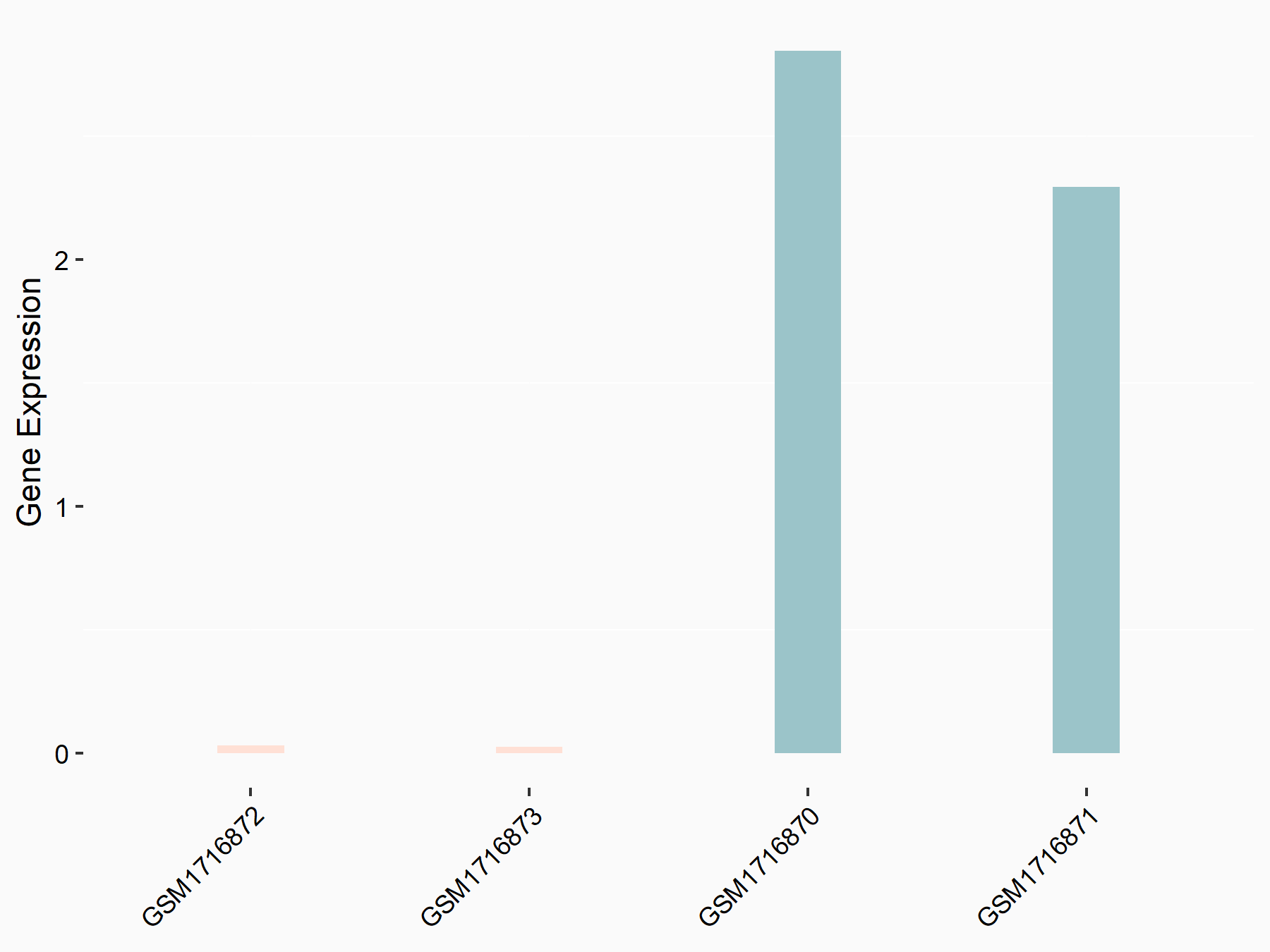 |
logFC: -1.79E+00 p-value: 1.23E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.17E+00 | GSE60213 |
Colistin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [65] | |||
| Responsed Disease | Diseases of the urinary system | ICD-11: GC2Z | ||
| Target Regulation | Down regulation | |||
| Cell Process | Oxidative stress | |||
| Cell apoptosis | ||||
| In-vivo Model | The 60 female Kunming mice were divided into two groups (n = 30): control group (injection of physiological saline through the caudal vein) and colistin group (injection of 15 mg/kg colistin, twice a day, with an eight-hour interval). | |||
| Response Summary | m6A methylation was involved in oxidative stress-mediated apoptosis in the mechanism of colistin nephrotoxicity. METTL3-mediated m6A methylation modification is involved in colistin-induced nephrotoxicity through apoptosis mediated by Kelch-like ECH-associated protein 1 (KEAP1)/Nrf2 signaling pathway. | |||
Methylated-DNA--protein-cysteine methyltransferase (MGMT)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
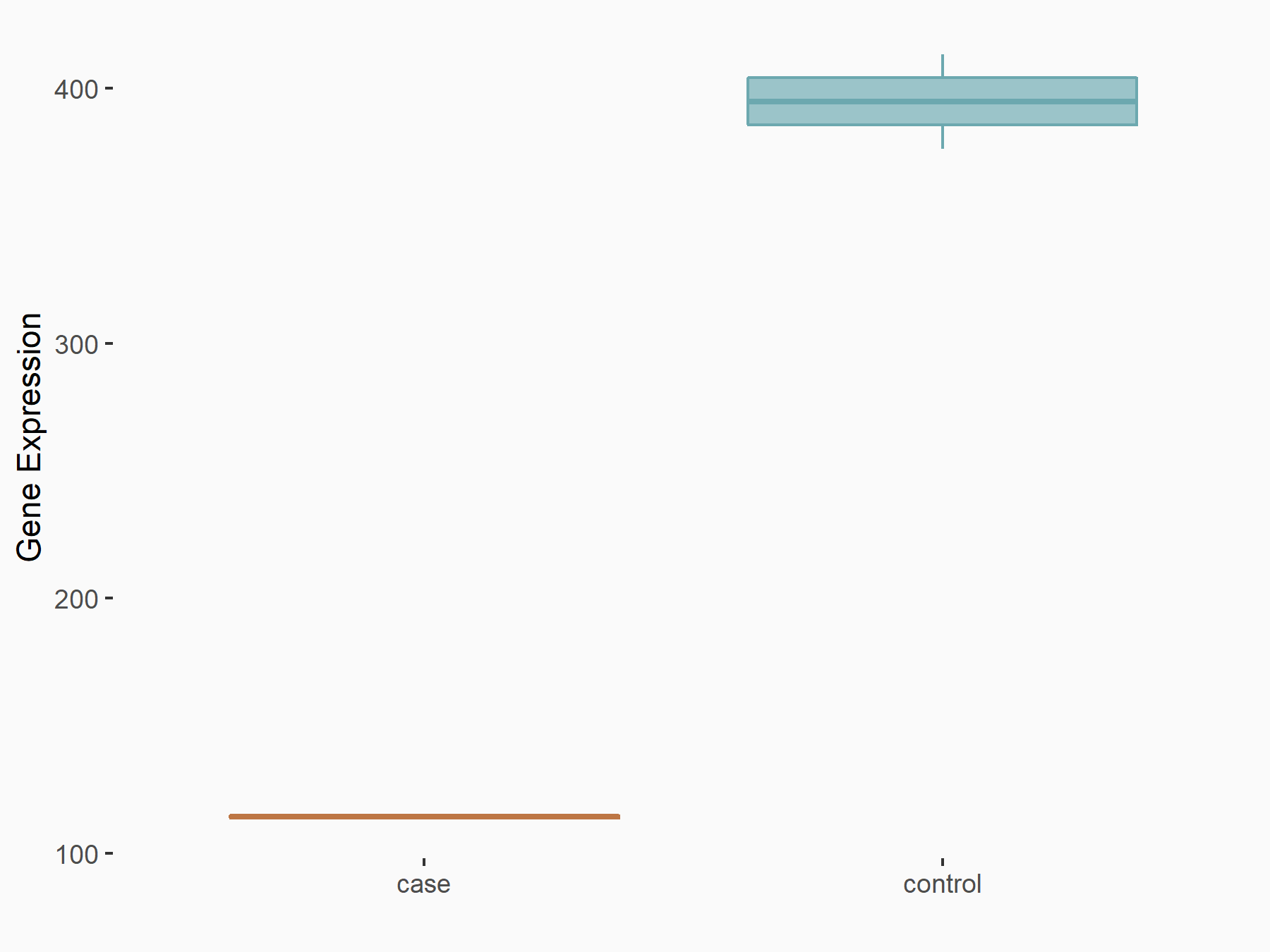  |
logFC: -1.79E+00 p-value: 2.16E-20 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 5.15E+00 | GSE60213 |
Temozolomide
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [69] | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | DNA repair | |||
| In-vitro Model | U251 (Fibroblasts or fibroblast like cells) | |||
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| In-vivo Model | Subcutaneously injected shMETTL3 or shNC-expressing U87-MG-TMZ cells into BALB/c NOD mice. After confirmation of GBM implantation, mice were treated with TMZ (66 mg/kg/d, 5 d per week, for 3 cycles). | |||
| Response Summary | Two critical DNA repair genes (Methylated-DNA--protein-cysteine methyltransferase (MGMT) and APNG) were m6A-modified by METTL3, whereas inhibited by METTL3 silencing or DAA-mediated total methylation inhibition, which is crucial for METTL3-improved temozolomide resistance in glioblastoma cells. | |||
Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
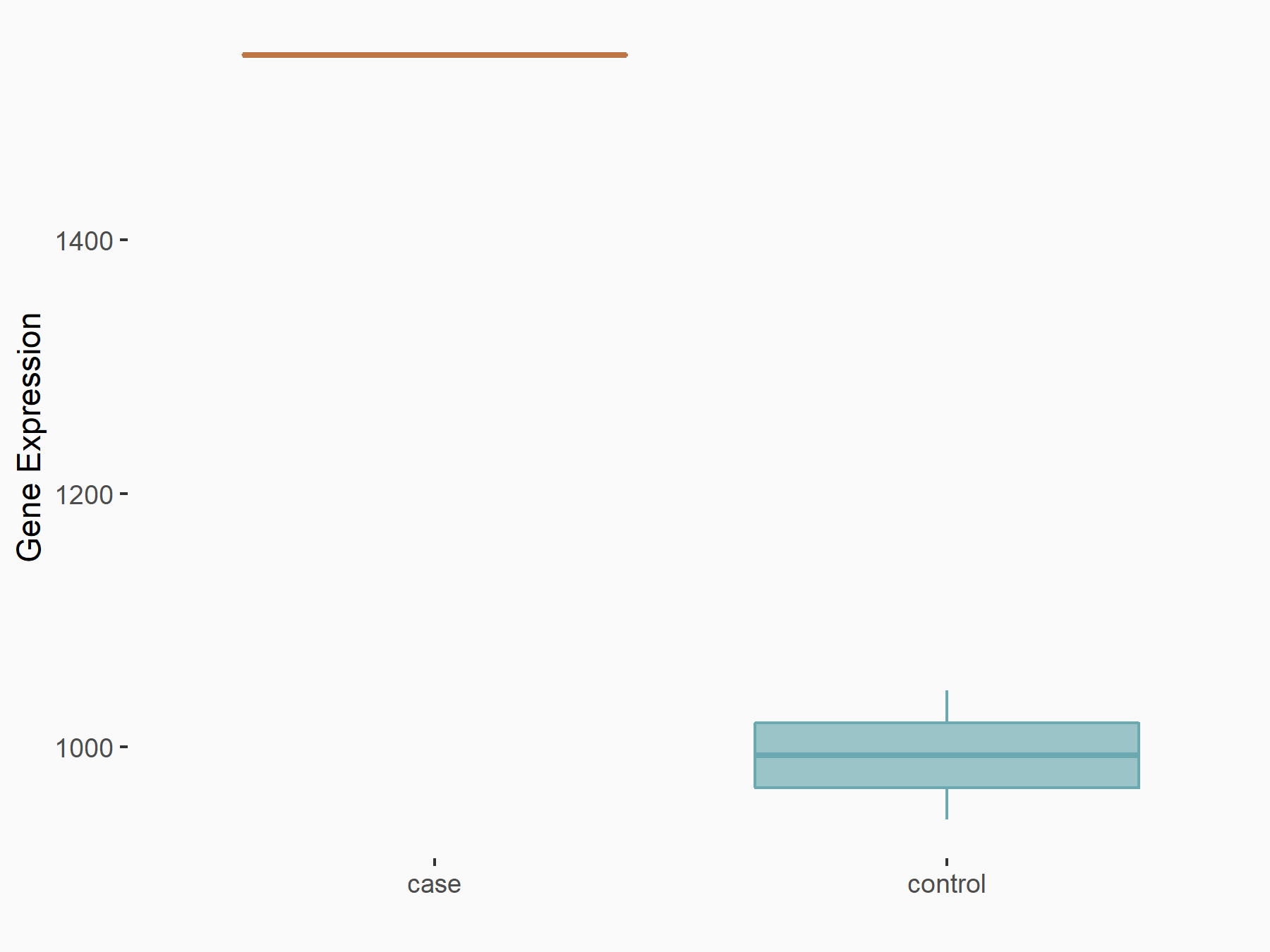  |
logFC: 6.38E-01 p-value: 4.93E-09 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.34E+00 | GSE60213 |
Chloroquine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II). beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
Gefitinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II). beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
Beta-Elemen
[Phase 3]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II). beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
Neurocalcin-delta (NCALD)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LNCaP cell line | Homo sapiens |
|
Treatment: shMETTL3 LNCaP cells
Control: shControl LNCaP cells
|
GSE147884 | |
| Regulation |
 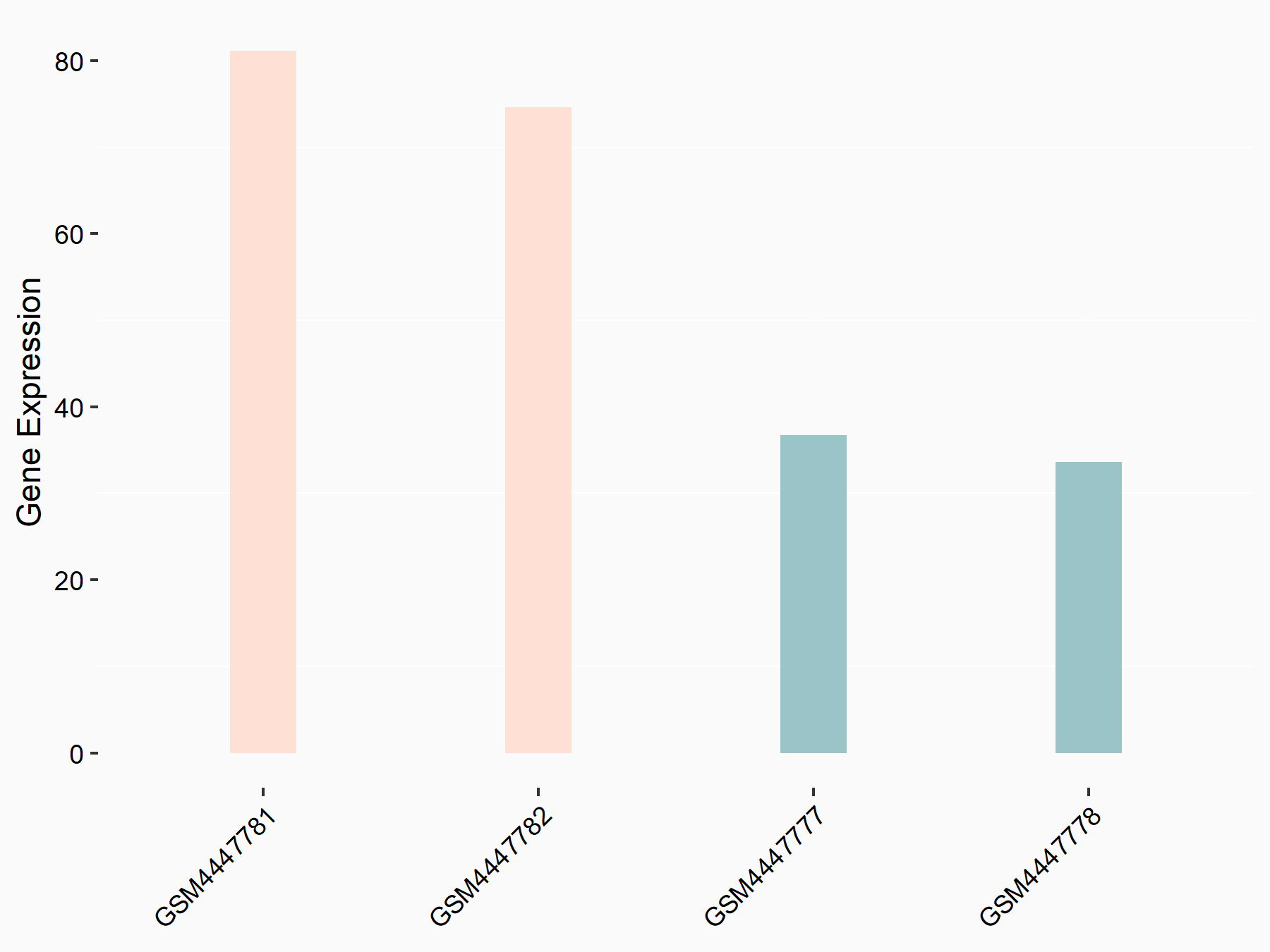 |
logFC: 1.15E+00 p-value: 3.16E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 8.03E+00 | GSE60213 |
Fluorouracil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [88] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | HT29 | Colon cancer | Mus musculus | CVCL_A8EZ |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| In-vivo Model | A tumor-bearing model was established by subcutaneously injecting 100 ul HT29 cells (5×106) followed by an intravenous injection of CAFs-derived exosomes (50 ug/mouse every three days) into the tail vein of the mice. An intraperitoneal injection of 5-FU (50 mg/kg, every week) was administered on day 12. | |||
| Response Summary | METTL3 dependent m6A methylation was upregulated in CRC to promote the processing of miR 181d 5p by DGCR8. This led to increased miR 181d 5p expression, which inhibited the 5 FU sensitivity of CRC cells by targeting Neurocalcin-delta (NCALD). | |||
Phosphatidylinositol 3-kinase regulatory subunit beta (PI3K-p85/PIK3R2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 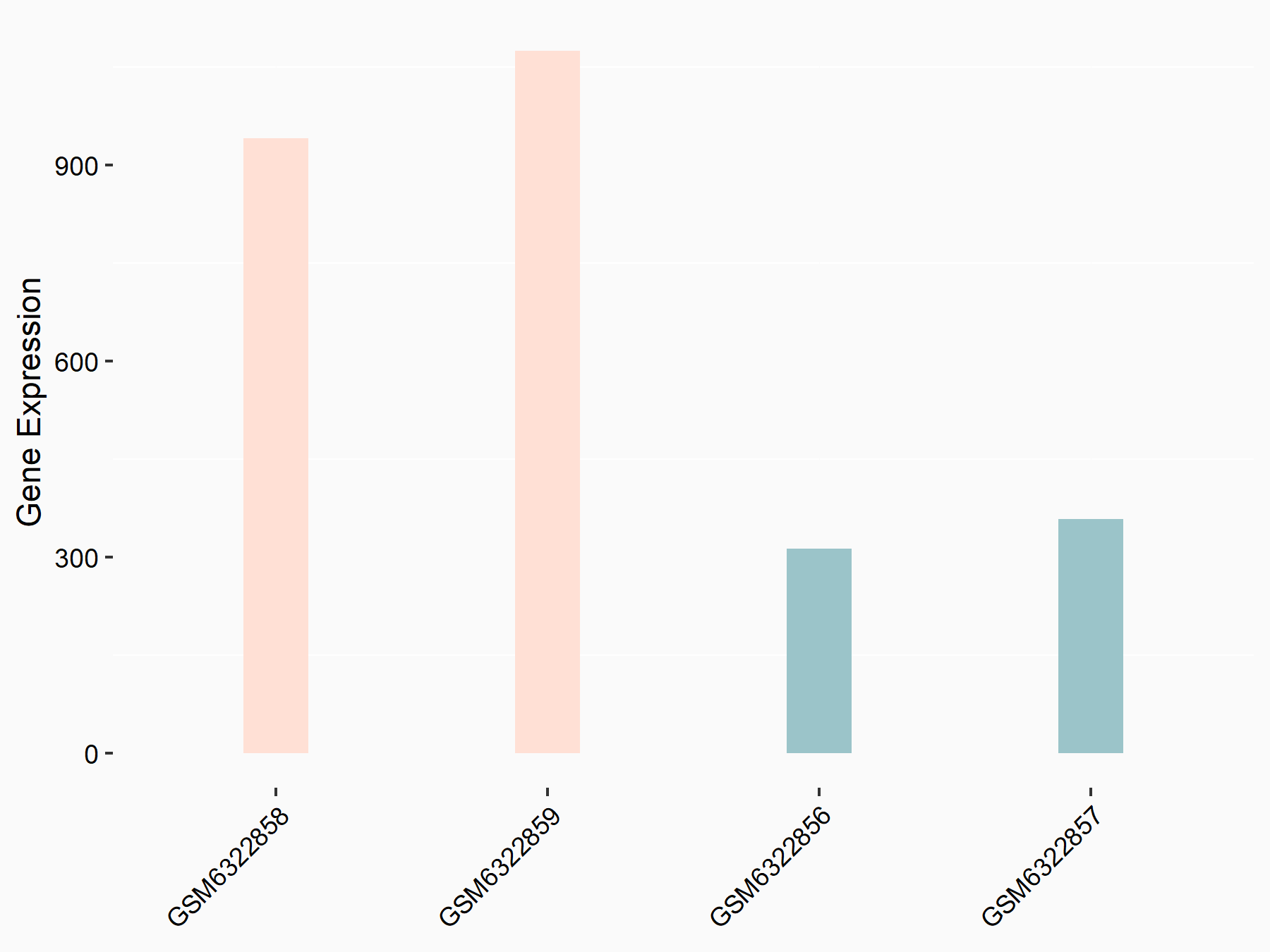 |
logFC: 1.59E+00 p-value: 3.69E-27 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.42E+00 | GSE60213 |
Rapamycin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [44] | |||
| Responsed Disease | Retinoblastoma | ICD-11: 2D02.2 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| In-vitro Model | WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through Phosphatidylinositol 3-kinase regulatory subunit beta (PI3K-p85/PIK3R2)/AKT/mTOR pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-P70S6K/4EBP1 pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
Pro-epidermal growth factor (EGF)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
 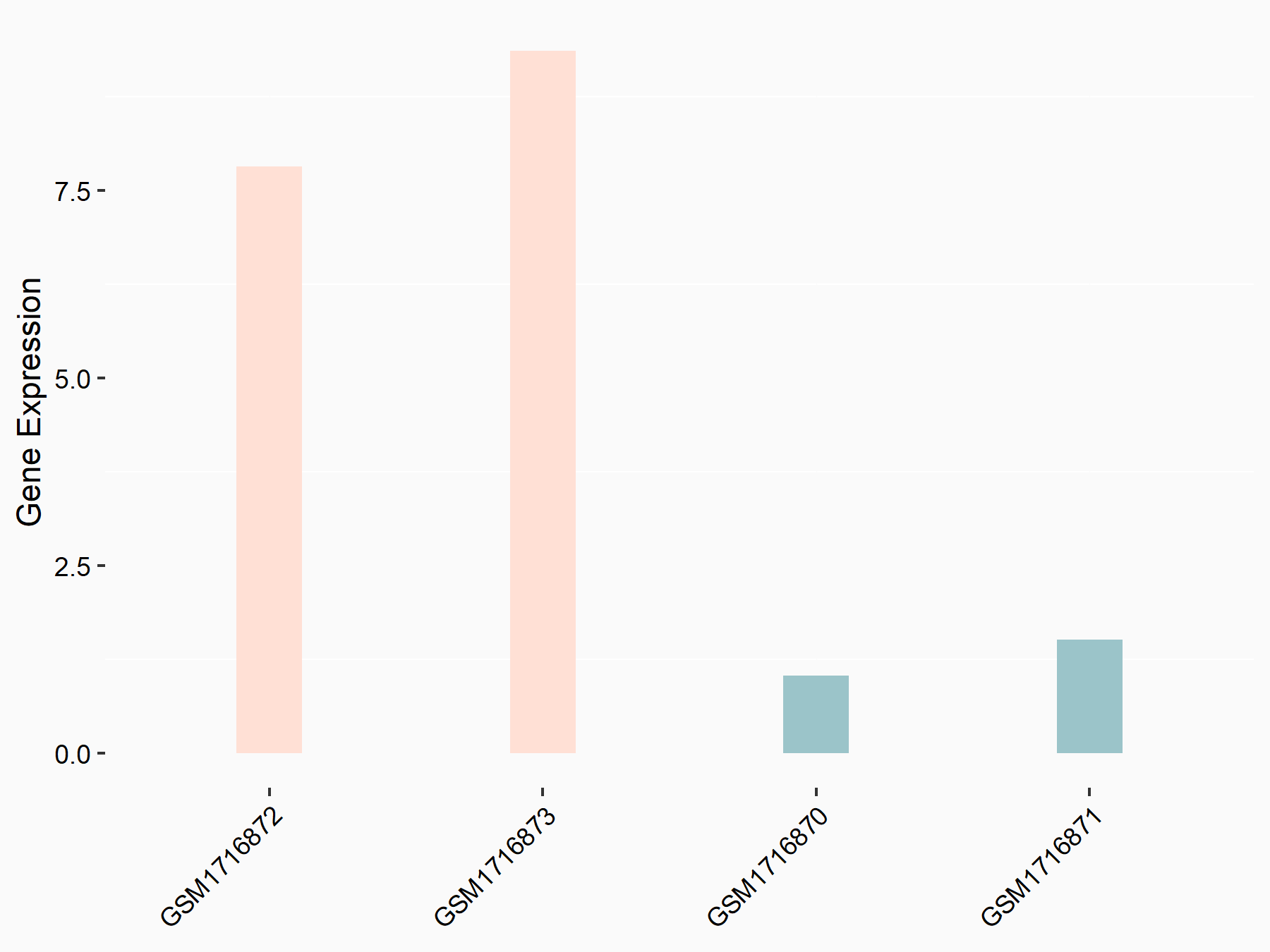 |
logFC: 2.08E+00 p-value: 3.08E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.98E+00 | GSE60213 |
Doxil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [98] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Homologous recombination | hsa03440 | ||
| Cell Process | Homologous recombination repair | |||
| In-vitro Model | MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | Cells were trypsinized and resuspended in DMEM at a consistence of 1 × 107 cells/ml. A total of 1 × 106 cells were injected into flank of mice. 27 days after injection, tumors were removed for paraffin-embedded sections. | |||
| Response Summary | Knockdown of METTL3 sensitized these breast cancer cells to Adriamycin (ADR; also named as doxorubicin) treatment and increased accumulation of DNA damage. Mechanically, we demonstrated that inhibition of METTL3 impaired HR efficiency and increased ADR-induced DNA damage by regulating m6A modification of Pro-epidermal growth factor (EGF)/RAD51 axis. METTL3 promoted EGF expression through m6A modification, which further upregulated RAD51 expression, resulting in enhanced HR activity. | |||
Ribosomal protein S6 kinase beta-1 (RPS6KB1/p70S6K)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | CT26 cell line | Mus musculus |
|
Treatment: METTL3 knockout CT26 cells
Control: CT26 cells
|
GSE142589 | |
| Regulation |
 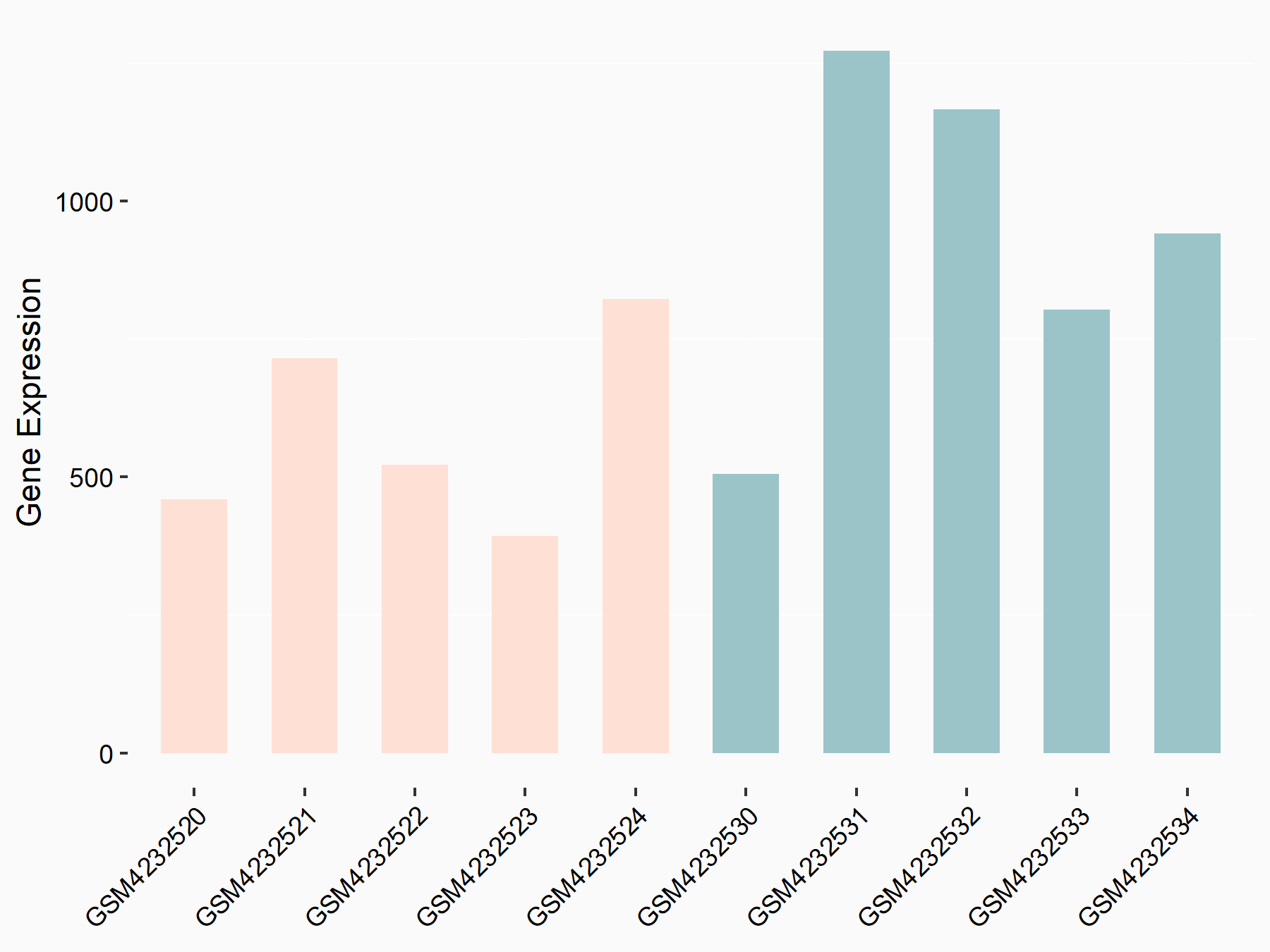 |
logFC: -6.86E-01 p-value: 2.67E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.70E+00 | GSE60213 |
Rapamycin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [44] | |||
| Responsed Disease | Retinoblastoma | ICD-11: 2D02.2 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| In-vitro Model | WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through PI3K/AKT/mTOR pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-Ribosomal protein S6 kinase beta-1 (RPS6KB1/p70S6K)/4EBP1 pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
Sequestosome-1 (SQSTM1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HUVEC cell line | Homo sapiens |
|
Treatment: shMETTL3 HUVEC cells
Control: shScramble HUVEC cells
|
GSE157544 | |
| Regulation |
 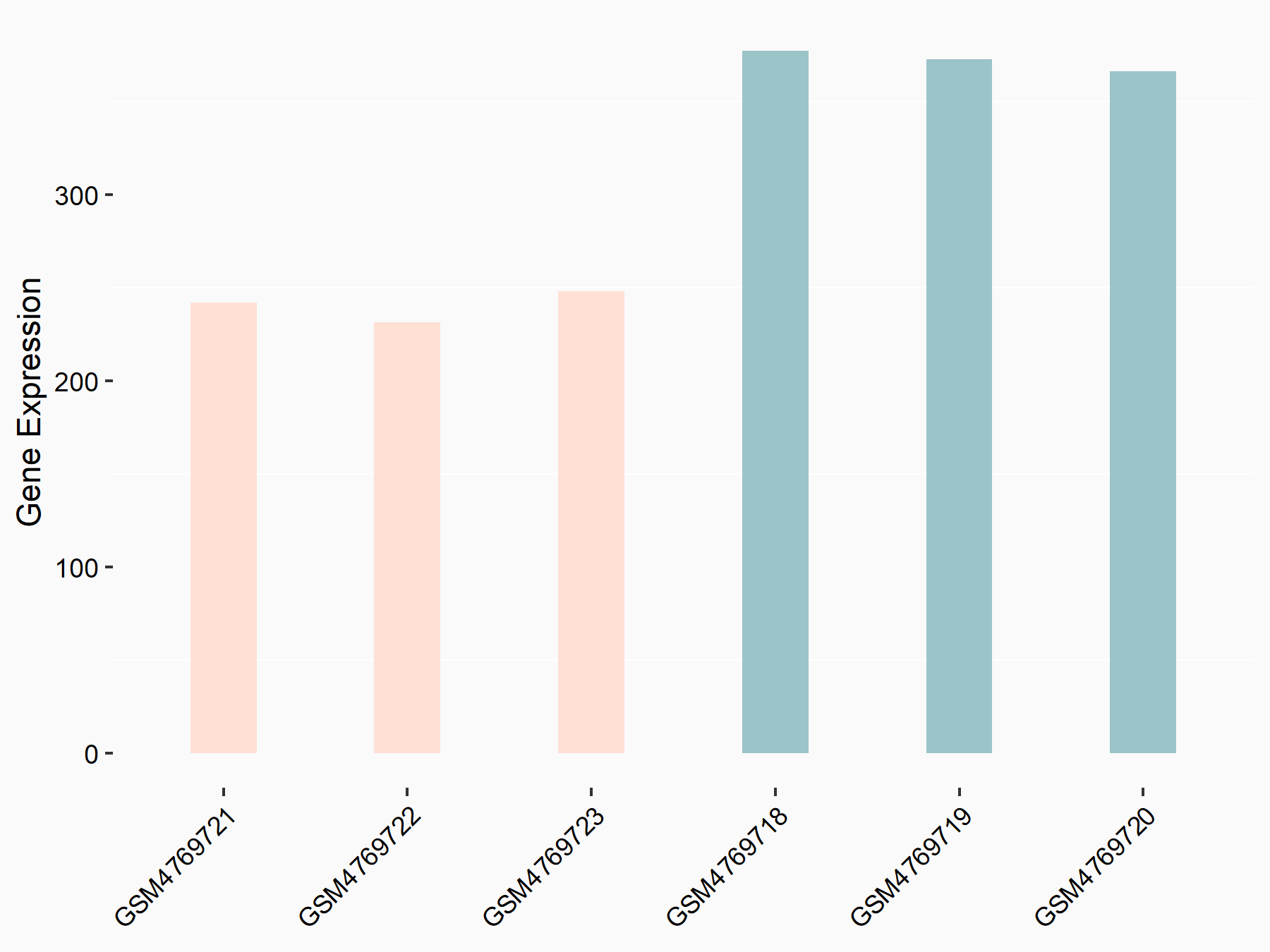 |
logFC: -6.28E-01 p-value: 1.94E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.03E+00 | GSE60213 |
Chloroquine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and Sequestosome-1 (SQSTM1). beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
Gefitinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and Sequestosome-1 (SQSTM1). beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
Beta-Elemen
[Phase 3]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and Sequestosome-1 (SQSTM1). beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
Paclitaxel
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [331] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | The experimental mice were housed in an SPF-grade animal laboratory at a temperature of approximately 25 °C, a humidity range of 50%, and an average daylight duration of 12 h. BALB/C female mice at 5-6 weeks of age were taken and prepared for tumor-bearing. After the mice were sacrificed, their tissues were collected, weighed and immediately snap-frozen in liquid nitrogen. | |||
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [331] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | The experimental mice were housed in an SPF-grade animal laboratory at a temperature of approximately 25 °C, a humidity range of 50%, and an average daylight duration of 12 h. BALB/C female mice at 5-6 weeks of age were taken and prepared for tumor-bearing. After the mice were sacrificed, their tissues were collected, weighed and immediately snap-frozen in liquid nitrogen. | |||
Serine/threonine-protein kinase mTOR (MTOR)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3 knockout liver
Control: Wild type liver cells
|
GSE198512 | |
| Regulation |
 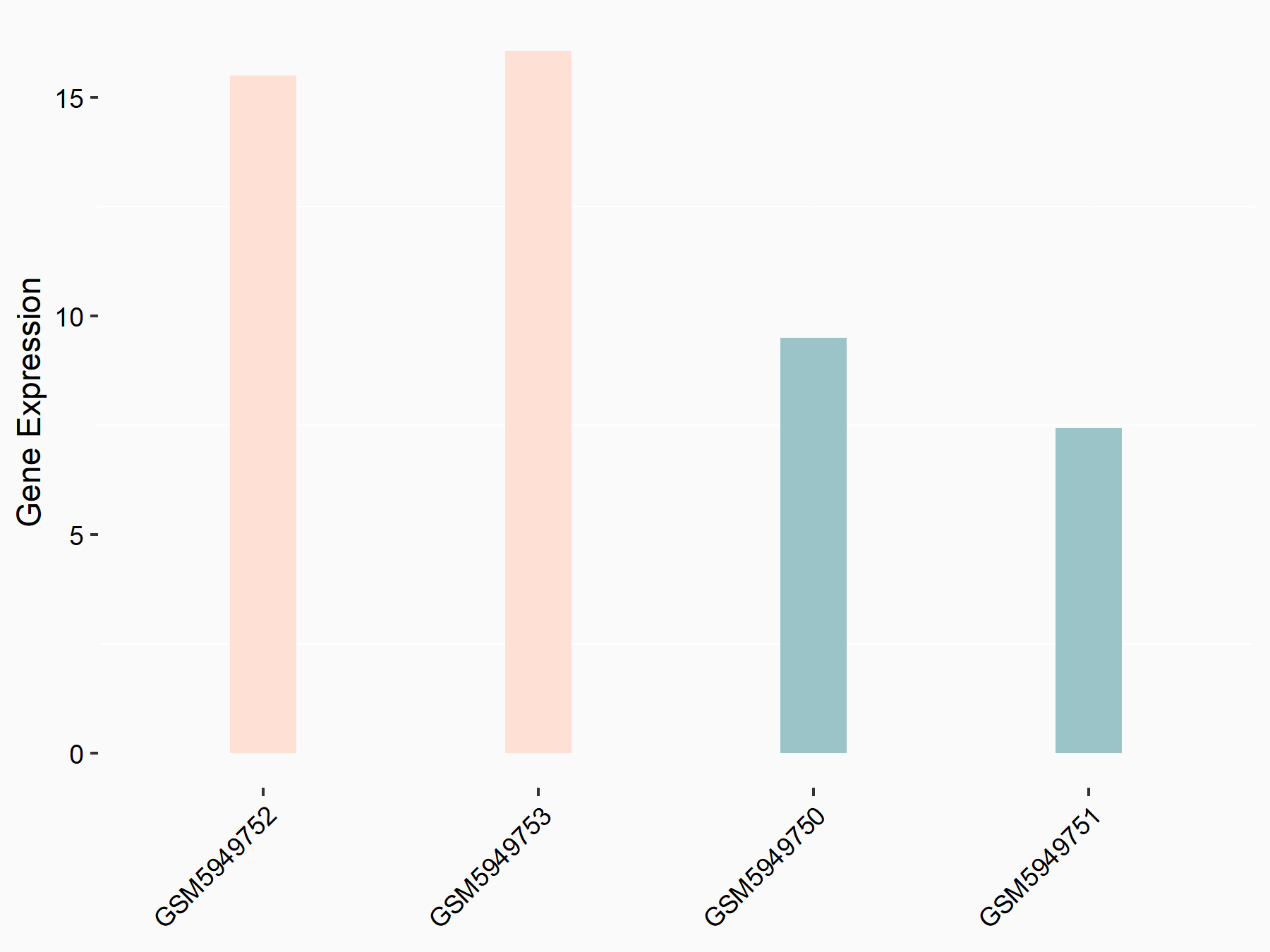 |
logFC: 8.34E-01 p-value: 1.58E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.47E+00 | GSE60213 |
Rapamycin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [44] | |||
| Responsed Disease | Retinoblastoma | ICD-11: 2D02.2 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| In-vitro Model | WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through PI3K/AKT/Serine/threonine-protein kinase mTOR (MTOR) pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-P70S6K/4EBP1 pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
TNF receptor-associated factor 5 (TRAF5)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 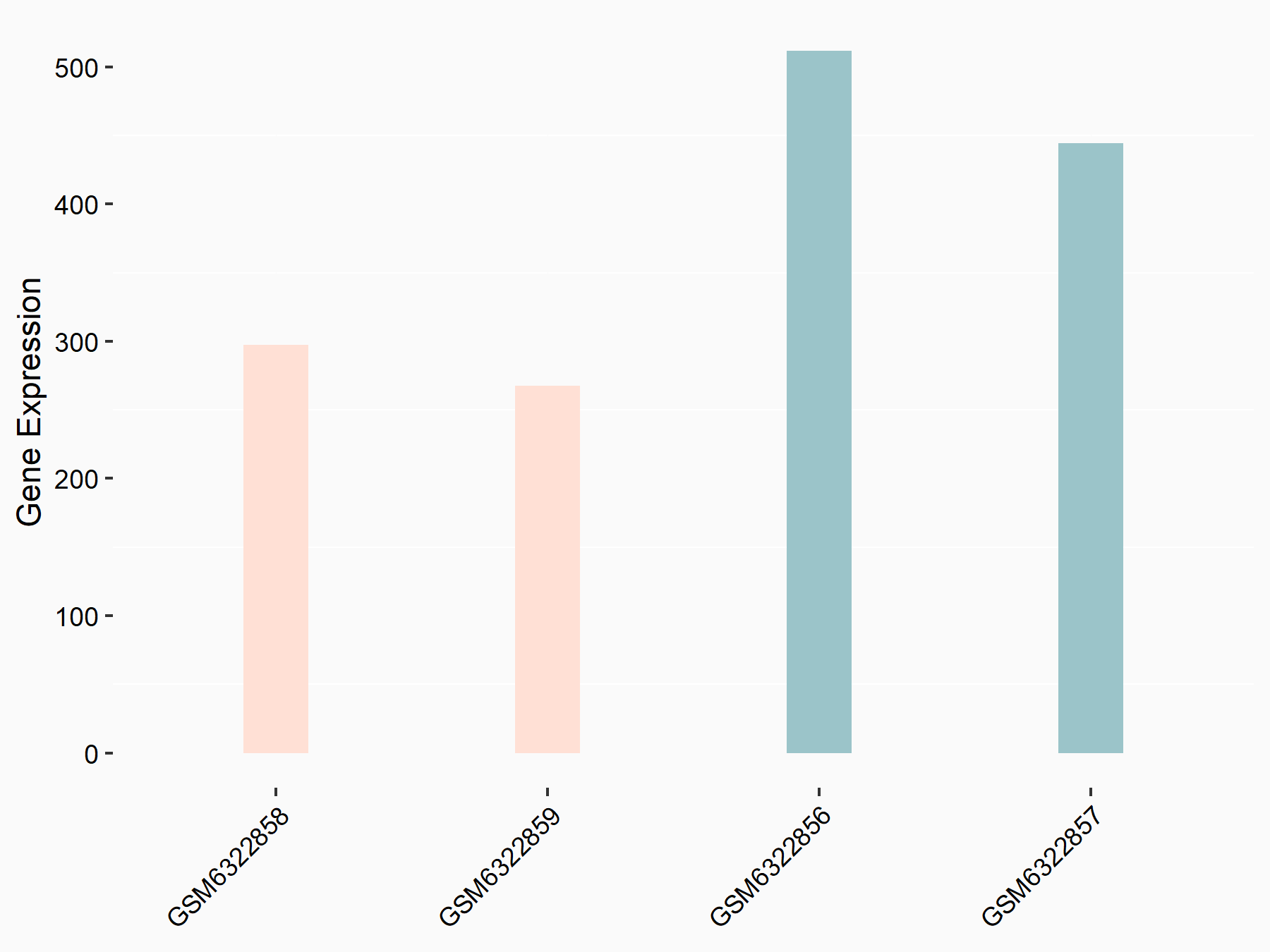 |
logFC: -7.59E-01 p-value: 1.50E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.24E+00 | GSE60213 |
Oxaliplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [113] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| In-vivo Model | HCT-116 cells (3 × 105 cells in 200 uL of saline) were subcutaneously injected into the nude mice to establish xenograft tumors. After 10 days, 10 mg/kg OX or saline was intraperitoneally injected (n = 5 for each group). Si-METTL3 or si-TRAF5 (10 nmol/20 g body weight) was injected twice intratumorally before the start of OX treatment. The mice were examined every 2 days and sacrificed 4 weeks after the OX treatment. | |||
| Response Summary | 2-polarized tumor-associated macrophages enabled the oxaliplatin resistance via the elevation of METTL3-mediated m6A modification in Colorectal Cancer cells. Furthermore, they found that TNF receptor-associated factor 5 (TRAF5) contributes to the METTL3-triggered OX resistance in CRC cells. | |||
Transcription factor AP-2 gamma (TFAP2C)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- mESCs
Control: Wild type ESCs
|
GSE147849 | |
| Regulation |
 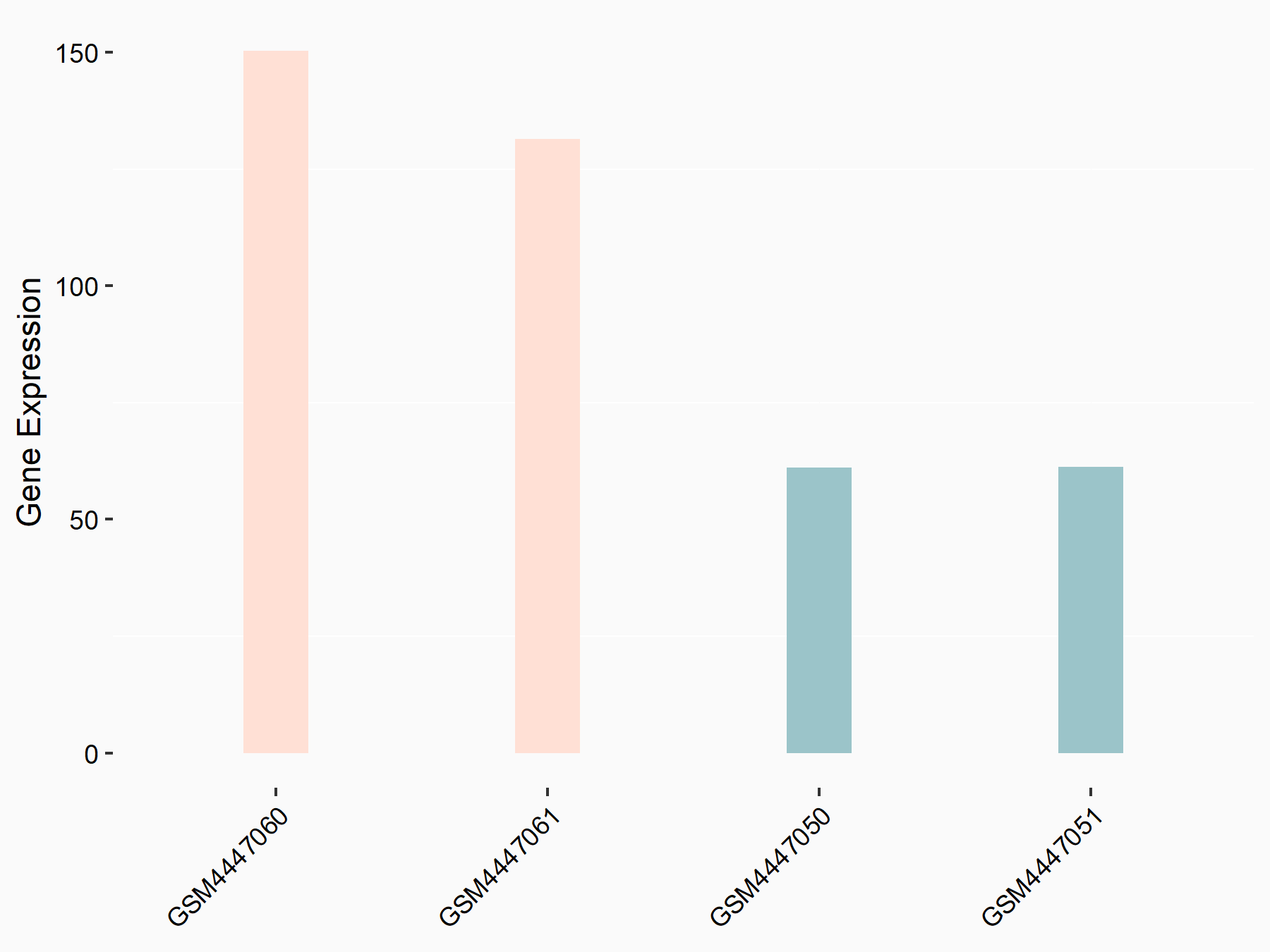 |
logFC: 1.20E+00 p-value: 4.47E-08 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.28E+00 | GSE60213 |
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [115] | |||
| Responsed Disease | Testicular cancer | ICD-11: 2C80 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | DNA repair | |||
| In-vitro Model | TCam-2 | Testicular seminoma | Homo sapiens | CVCL_T012 |
| In-vivo Model | Male mice were subcutaneously injected with tumour cells near the limbs to establish xenografts (1 × 106/mouse, 0.2 mL for each injection site; METTL3-overexpressing TCam-2/CDDP cells were inoculated once at the initial time and IGF2BP1-inhibited TCam-2/CDDP cells were inoculated every 3 days). | |||
| Response Summary | METTL3 potentiates resistance to cisplatin through m6A modification of Transcription factor AP-2 gamma (TFAP2C) in seminoma. Enhanced stability of TFAP2C mRNA promoted seminoma cell survival under cisplatin treatment burden probably through up-regulation of DNA repair-related genes. IGF2BP1 binds to TFAP2C and enhances TFAP2C mRNA stability. | |||
Transcription factor E2F1 (E2F1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
 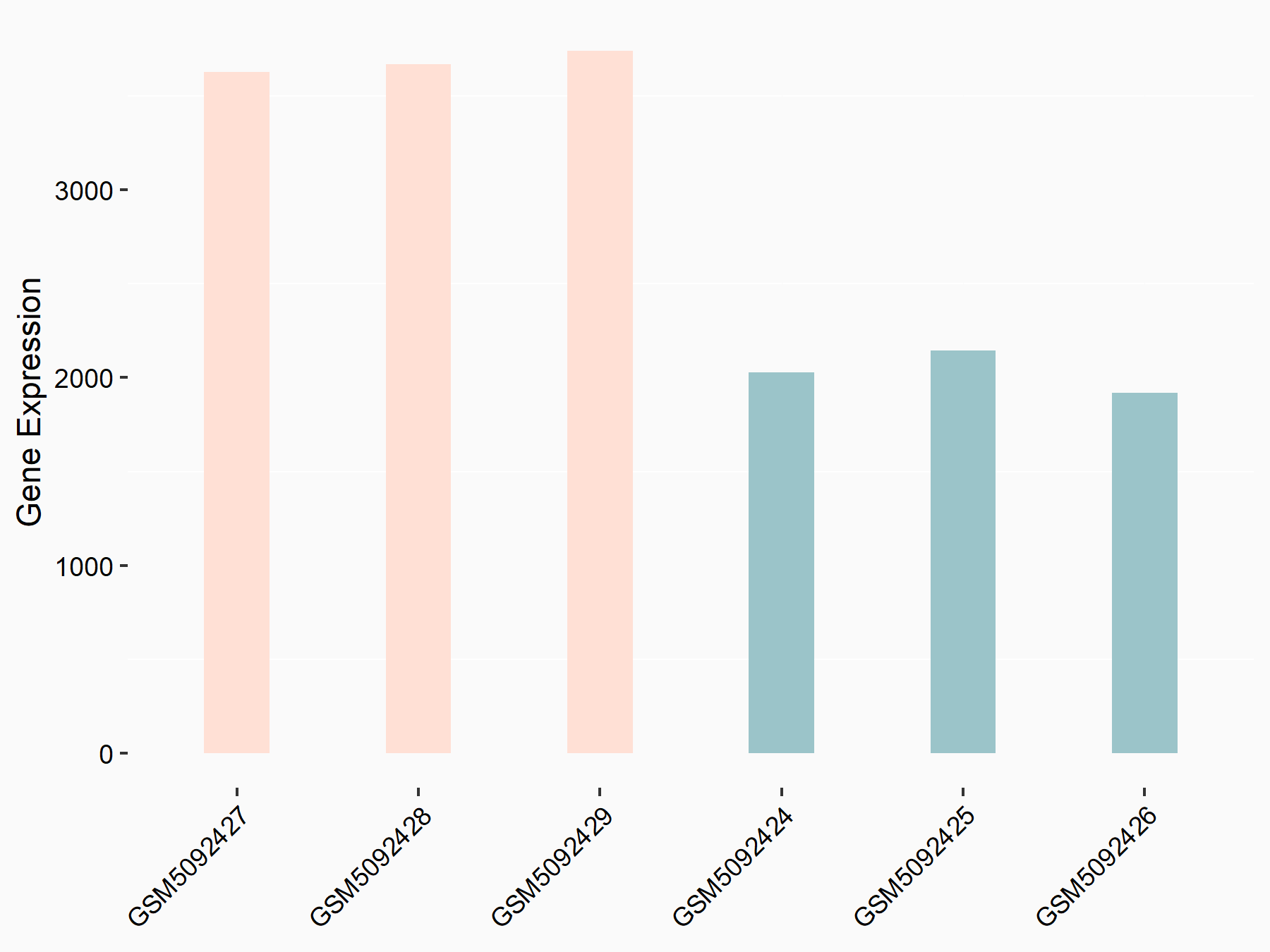 |
logFC: 8.59E-01 p-value: 2.73E-57 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.01E+00 | GSE60213 |
Doxil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [116] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | MCF7-DoxR (Adriamycin-resistant cell line MCF7-DoxR) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| In-vivo Model | Once the tumor volume increased to about 1 cm3, six groups of MCF7 bearing mice (n = 10 in each group) were injected with PBS (0.1 ml, caudal vein) and adriamycin (0.1 ml, 10 mg/kg), respectively. When the tumor reached 1.5 cm in any direction (defined as event-free survival analysis), 10 mice in each group were selected to measure the tumor size and weight on the 12th day after adriamycin injection. | |||
| Response Summary | METTL3 can regulate the expression of MALAT1 through m6A, mediate the Transcription factor E2F1 (E2F1)/AGR2 axis, and promote the adriamycin resistance of breast cancer. | |||
Transcriptional coactivator YAP1 (YAP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LNCaP cell line | Homo sapiens |
|
Treatment: shMETTL3 LNCaP cells
Control: shControl LNCaP cells
|
GSE147884 | |
| Regulation |
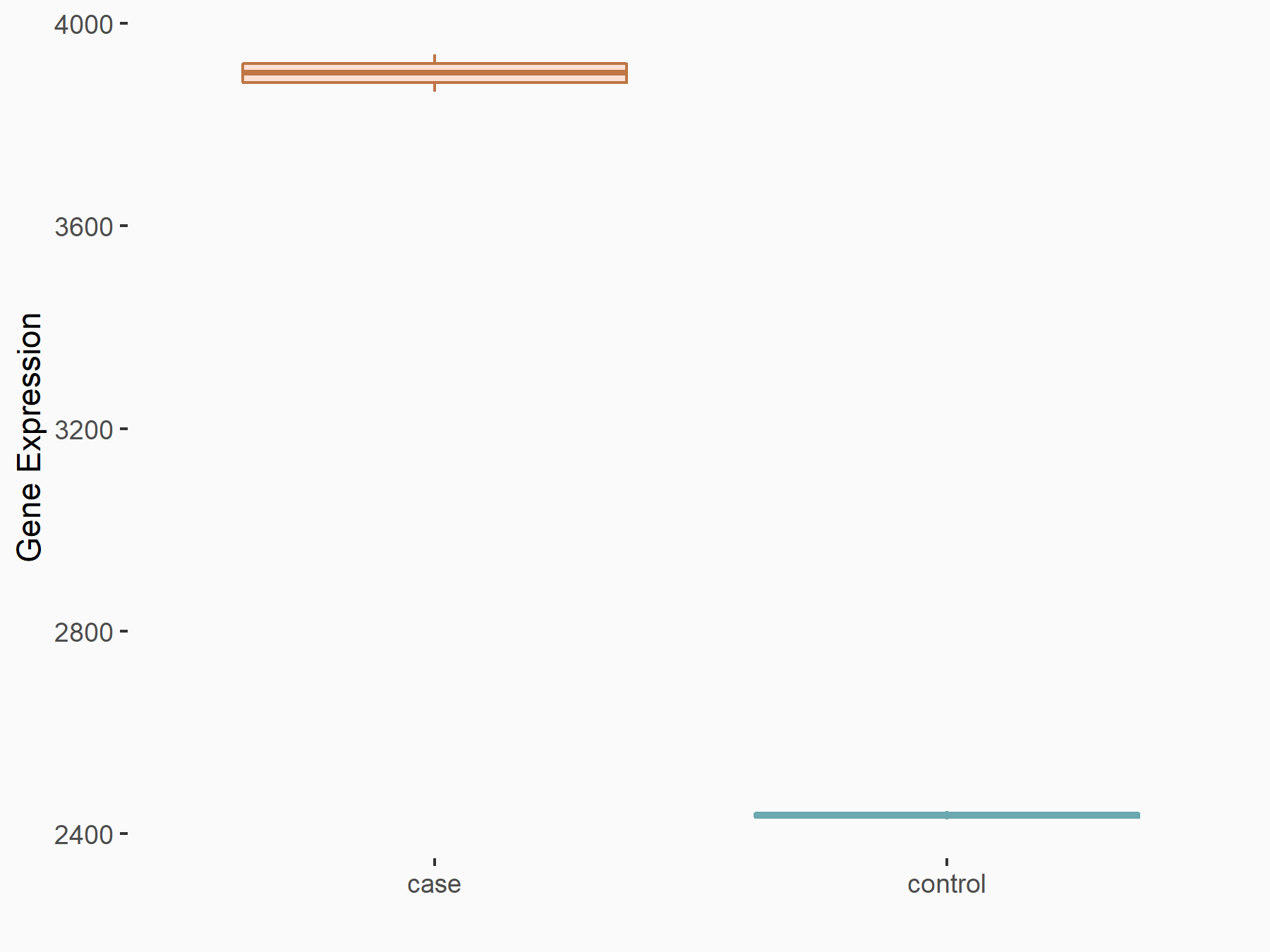 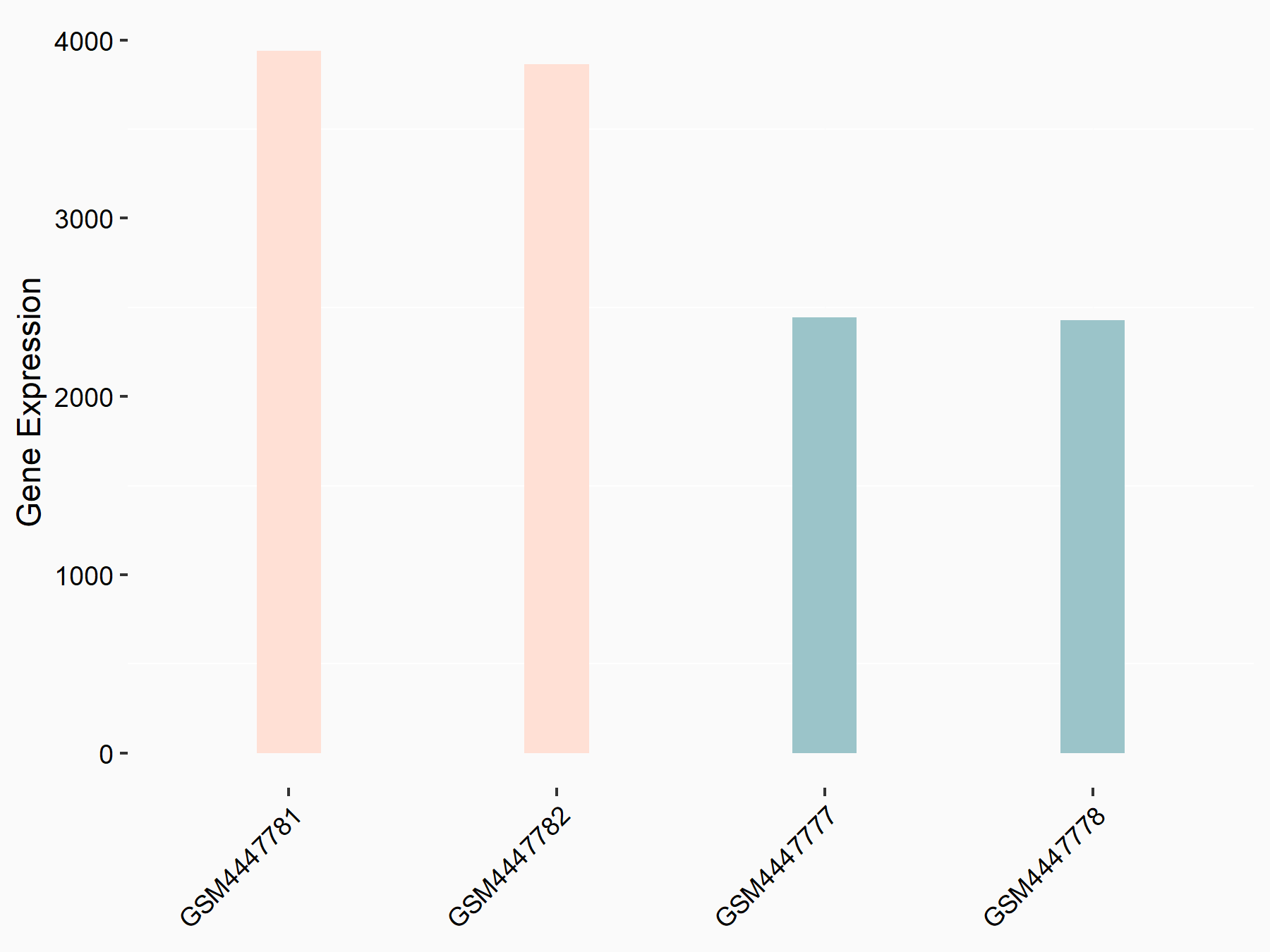 |
logFC: 6.80E-01 p-value: 6.99E-58 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.22E+00 | GSE60213 |
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [126] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Metabolic | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-6 | Lung adenocarcinoma | Homo sapiens | CVCL_0236 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | Mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then administrated with PBS or DDP (3 mg/kg per day) for three times a week, and tumor volume were measured every second day. | |||
| Response Summary | METTL3, YTHDF3, YTHDF1, and eIF3b directly promoted YAP translation through an interaction with the translation initiation machinery. METTL3 knockdown inhibits tumor growth and enhances sensitivity to DDP in vivo.m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-Transcriptional coactivator YAP1 (YAP1) axis to induce Non-small cell lung cancer drug resistance and metastasis. | |||
Translocation protein SEC62 (SEC62)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | CT26 cell line | Mus musculus |
|
Treatment: METTL3 knockout CT26 cells
Control: CT26 cells
|
GSE142589 | |
| Regulation |
 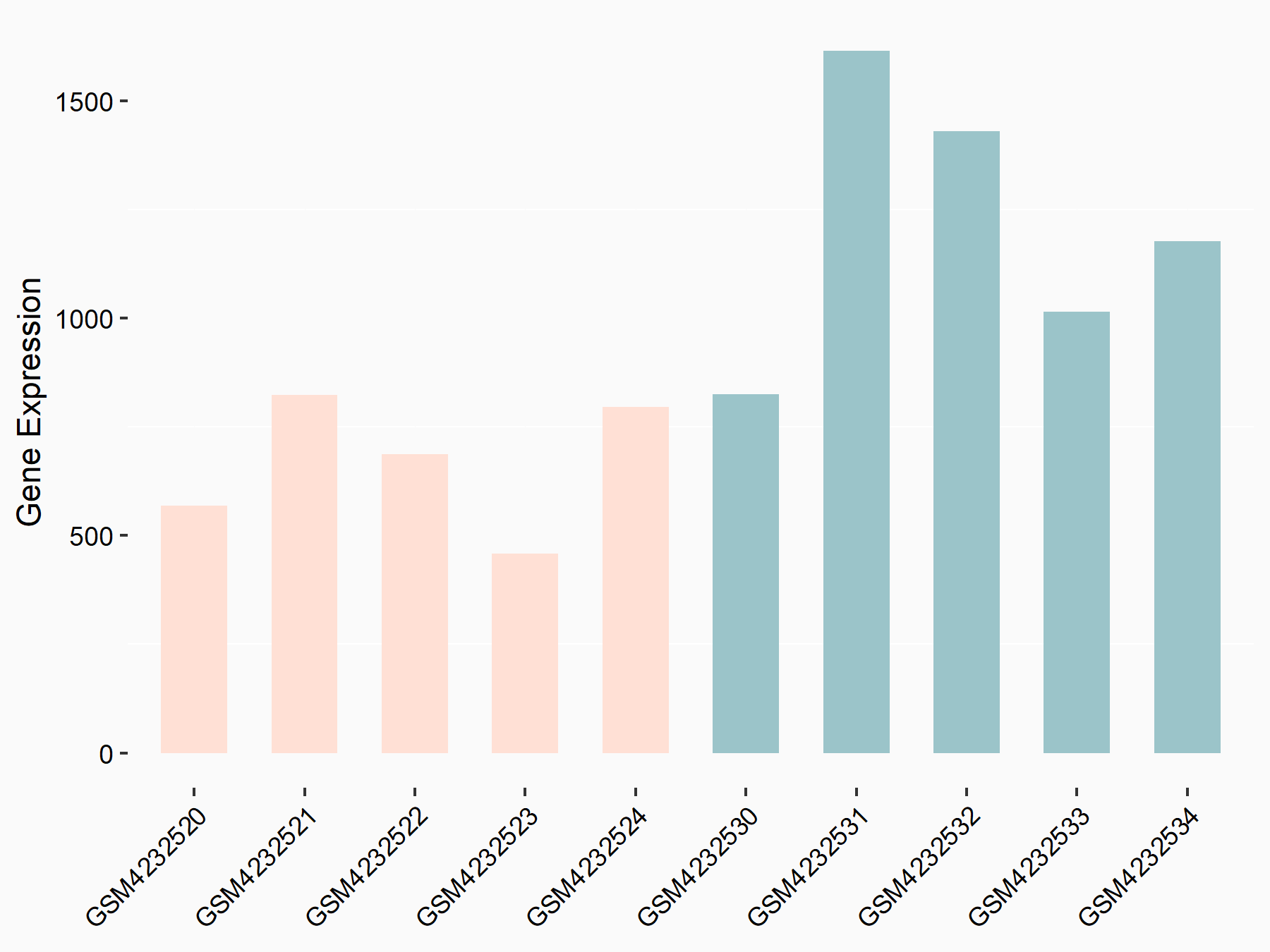 |
logFC: -8.61E-01 p-value: 1.21E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.99E+00 | GSE60213 |
Fluorouracil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [128] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Protein degradation | |||
| In-vitro Model | DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | DLD-1 cells were subcutaneously implanted into 4-6 weeks old female nude mice. When tumors reached a size of about 50 mm3, the nude mice were randomly divided into 6 groups. | |||
| Response Summary | Translocation protein SEC62 (SEC62) upregulated by the METTL3-mediated m6A modification promotes the stemness and chemoresistance of colorectal cancer by binding to beta-catenin and enhancing Wnt signalling. Depletion of Sec62 sensitized the CRC cells to 5-Fu or oxaliplatin treatment. | |||
Oxaliplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [128] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Protein degradation | |||
| In-vitro Model | DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | DLD-1 cells were subcutaneously implanted into 4-6 weeks old female nude mice. When tumors reached a size of about 50 mm3, the nude mice were randomly divided into 6 groups. | |||
| Response Summary | Translocation protein SEC62 (SEC62) upregulated by the METTL3-mediated m6A modification promotes the stemness and chemoresistance of colorectal cancer by binding to beta-catenin and enhancing Wnt signalling. Depletion of Sec62 sensitized the CRC cells to 5-Fu or oxaliplatin treatment. | |||
Ubiquitin-like modifier-activating enzyme ATG7 (ATG7)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 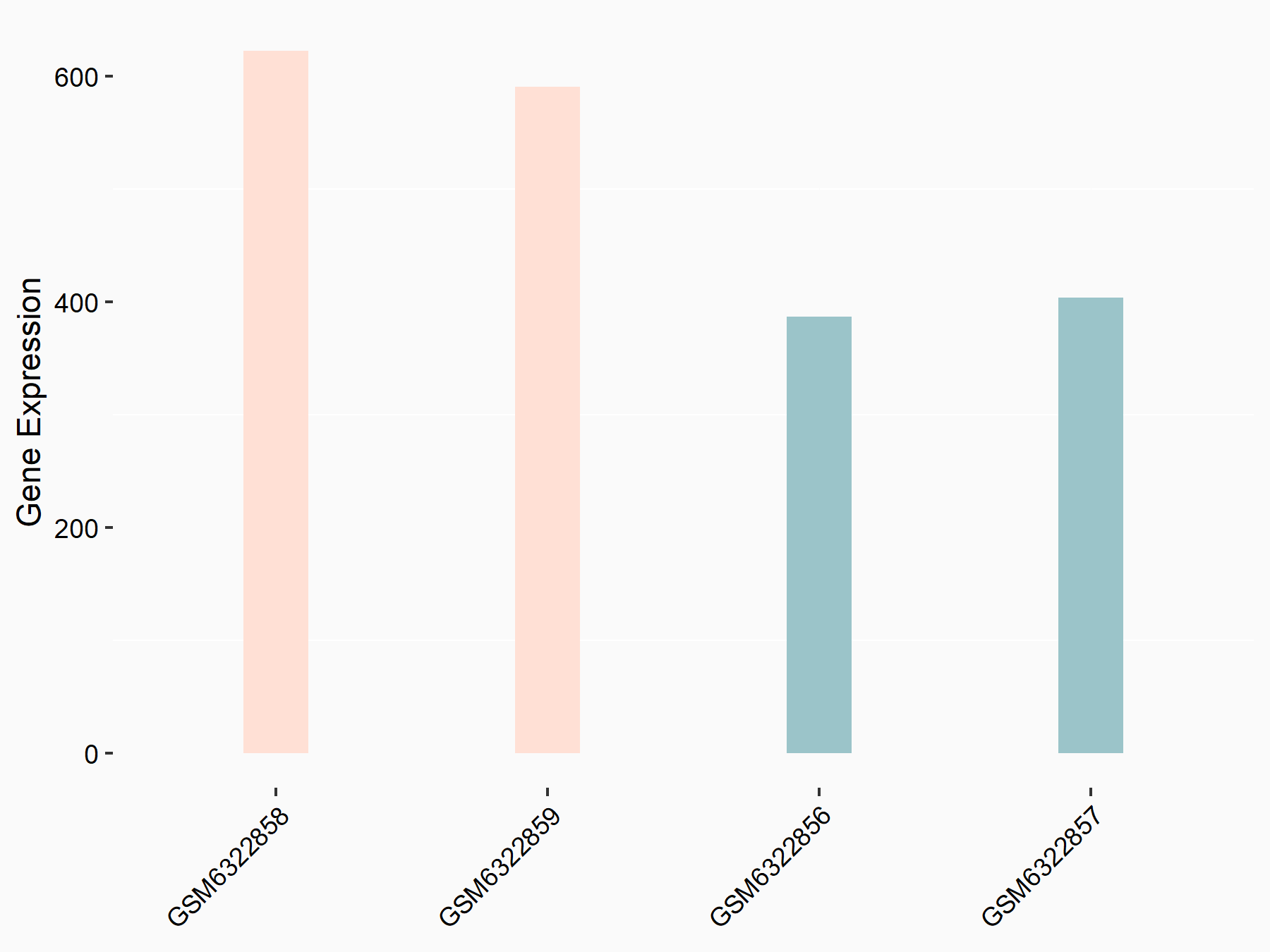 |
logFC: 6.17E-01 p-value: 3.67E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 4.77E+00 | GSE60213 |
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [17] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), ATG12, and ATG16L1. | |||
Chloroquine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
Gefitinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
Beta-Elemen
[Phase 3]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [18] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
Ubiquitin-like protein ATG12 (ATG12)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Mouse testis | Mus musculus |
|
Treatment: Mettl3 knockout mouse testis
Control: Mouse testis
|
GSE99771 | |
| Regulation |
 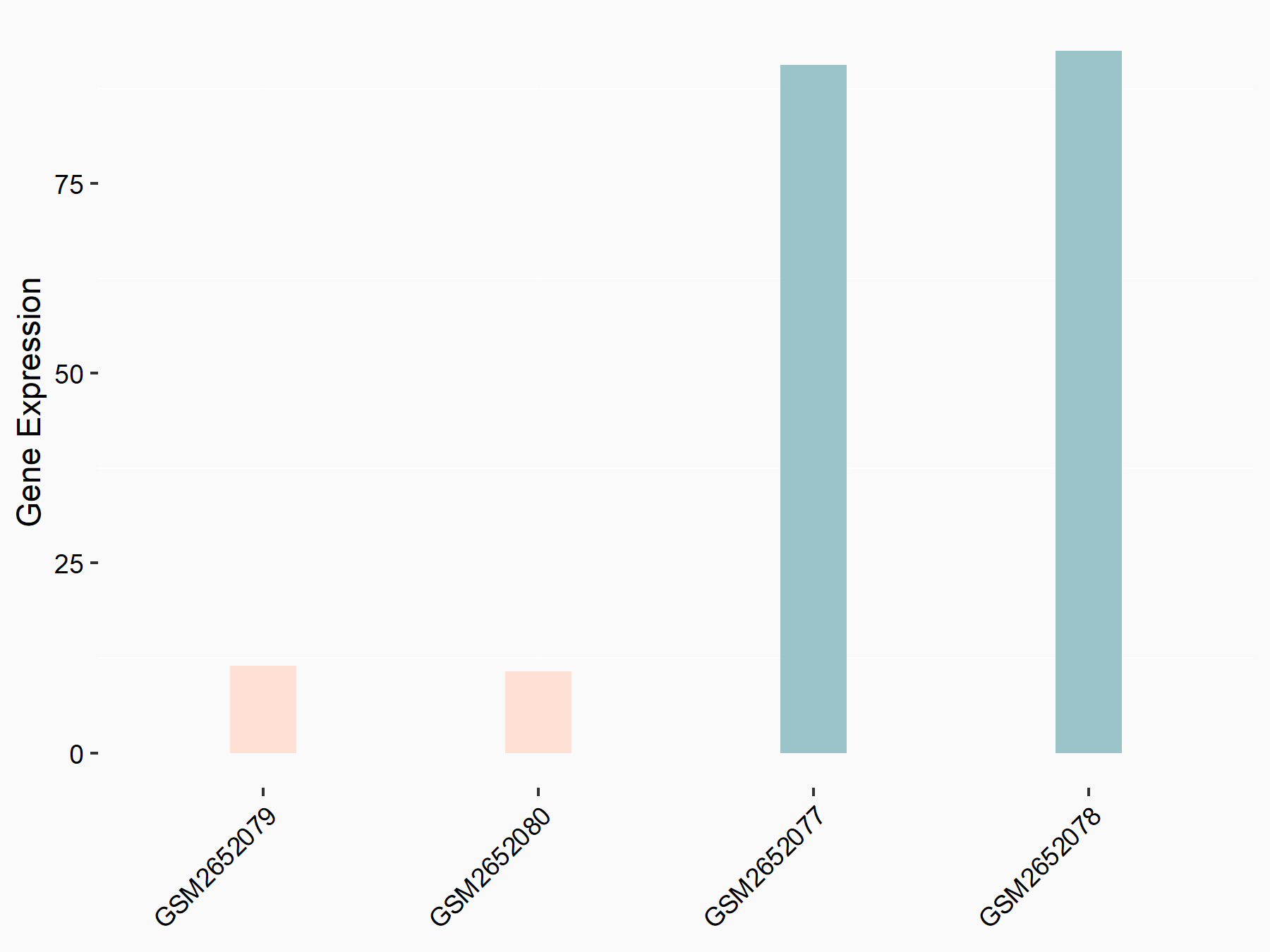 |
logFC: -2.94E+00 p-value: 6.14E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.85E+00 | GSE60213 |
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [17] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, ATG7, Ubiquitin-like protein ATG12 (ATG12), and ATG16L1. | |||
LBX2 antisense RNA 1 (LBX2-AS1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
 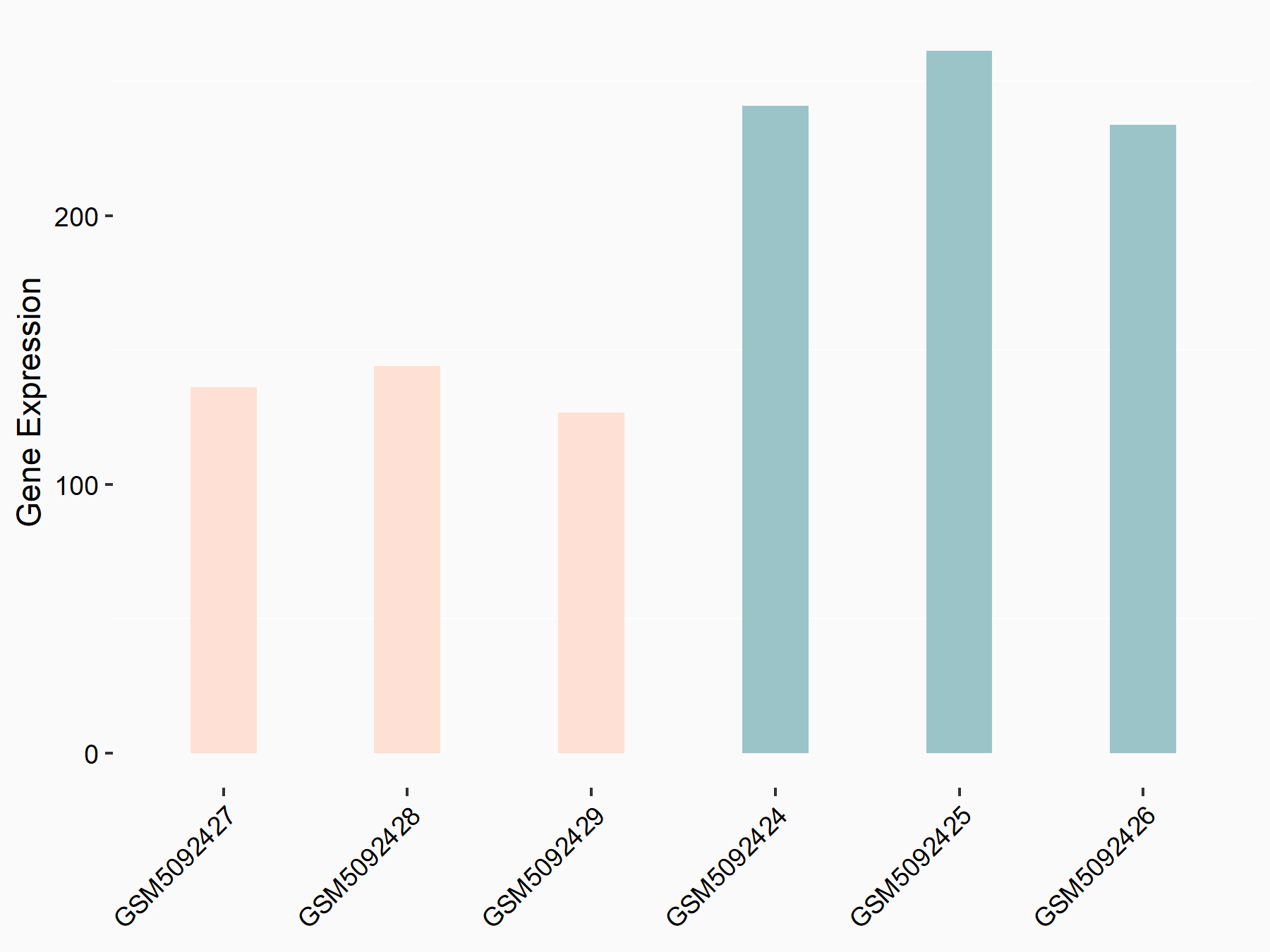 |
logFC: -8.54E-01 p-value: 2.13E-07 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.38E+00 | GSE60213 |
Fluorouracil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [138] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | The increased LBX2 antisense RNA 1 (LBX2-AS1) in CRC was mediated by METTL3-dependent m6A methylation. LBX2-AS1 serves as a therapeutic target and predictor of 5-FU benefit in colorectal cancer patients. | |||
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | DKO-1 cell line | Homo sapiens |
|
Treatment: METTL3 knockdown DKO-1 cell
Control: DKO-1 cell
|
GSE182382 | |
| Regulation |
  |
logFC: 8.20E-01 p-value: 5.93E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.28E+00 | GSE60213 |
Cisplatin
[Approved]
| In total 2 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [126] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Metabolic | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-6 | Lung adenocarcinoma | Homo sapiens | CVCL_0236 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | Mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then administrated with PBS or DDP (3 mg/kg per day) for three times a week, and tumor volume were measured every second day. | |||
| Response Summary | METTL3, YTHDF3, YTHDF1, and eIF3b directly promoted YAP translation through an interaction with the translation initiation machinery. METTL3 knockdown inhibits tumor growth and enhances sensitivity to DDP in vivo.m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)-miR-1914-3p-YAP axis to induce Non-small cell lung cancer drug resistance and metastasis. | |||
| Experiment 2 Reporting the m6A-centered Drug Response of This Target Gene | [144] | |||
| Responsed Disease | Thymic epithelial tumors | ICD-11: 2C27.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Cellular senescence | hsa04218 | ||
| Cell Process | Cell viability and proliferation | |||
| In-vitro Model | T1889 | Thymic undifferentiated carcinoma | Homo sapiens | CVCL_D024 |
| Response Summary | This study highlighted METTL3 as a tumor promoter in Thymic tumors and c-MYC as a promising target to be exploited for the treatment of TET. High expression of c-MYC protein is enabled by lncRNA Metastasis associated lung adenocarcinoma transcript 1 (MALAT1), which is methylated and delocalized by METTL3. Silencing of METTL3 combined with cisplatin or c-MYC inhibitor induces cell death in TET cells. Blocking of c-MYC by using JQ1 inhibitor cooperates with METTL3 depletion in the inhibition of proliferation and induction of cell death. | |||
JQ-1
[Phase 1]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [144] | |||
| Responsed Disease | Thymic epithelial tumors | ICD-11: 2C27.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Cellular senescence | hsa04218 | ||
| Cell Process | Cell viability and proliferation | |||
| In-vitro Model | T1889 | Thymic undifferentiated carcinoma | Homo sapiens | CVCL_D024 |
| Response Summary | This study highlighted METTL3 as a tumor promoter in Thymic tumors and c-MYC as a promising target to be exploited for the treatment of TET. High expression of c-MYC protein is enabled by lncRNA Metastasis associated lung adenocarcinoma transcript 1 (MALAT1), which is methylated and delocalized by METTL3. Silencing of METTL3 combined with cisplatin or c-MYC inhibitor induces cell death in TET cells. Blocking of c-MYC by using JQ1 inhibitor cooperates with METTL3 depletion in the inhibition of proliferation and induction of cell death. | |||
Doxil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [116] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | MCF7-DoxR (Adriamycin-resistant cell line MCF7-DoxR) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| In-vivo Model | Once the tumor volume increased to about 1 cm3, six groups of MCF7 bearing mice (n = 10 in each group) were injected with PBS (0.1 ml, caudal vein) and adriamycin (0.1 ml, 10 mg/kg), respectively. When the tumor reached 1.5 cm in any direction (defined as event-free survival analysis), 10 mice in each group were selected to measure the tumor size and weight on the 12th day after adriamycin injection. | |||
| Response Summary | METTL3 can regulate the expression of Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) through m6A, mediate the E2F1/AGR2 axis, and promote the adriamycin resistance of breast cancer. | |||
Artenimol
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [145] | |||
| Responsed Disease | Chronic kidney disease | ICD-11: GB61 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | HK-2 [Human kidney] | Normal | Homo sapiens | CVCL_0302 |
| HK2 | Normal | Acipenser baerii | CVCL_YE28 | |
| In-vivo Model | For the unilateral ureteral obstruction (UUO) model, male C57BL/6J mice at 8 weeks of age (20-22 g body weight) were first anaesthetized with pentobarbital sodium (50 mg/kg) via intraperitoneal injection. Then, the left ureter was ligated using 3-0 silk and a left lateral incision. | |||
| Response Summary | Renal fibrosis is a key factor in chronic kidney disease (CKD). Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)/miR-145/FAK pathway was involved in the effect of dihydroartemisinin (DHA) on TGF-beta1-induced renal fibrosis in vitro and in vivo. | |||
Simvastatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [367] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | Healthy adult ICR male (aged: 8 weeks old; weighing: 40- 70 g) and female mice (aged: 8 weeks old; weighing: 40-60 g) were obtained from Hunan SJA Laboratory Animal Co., Ltd. (Hunan, China). These mice were housed in the SPF laboratory at 25°C and 55% humidity under a 12-h light/dark cycle, with ad libitum access to food and water for acclimatization. | |||
Adenylate kinase 4, mitochondrial (AK4)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
 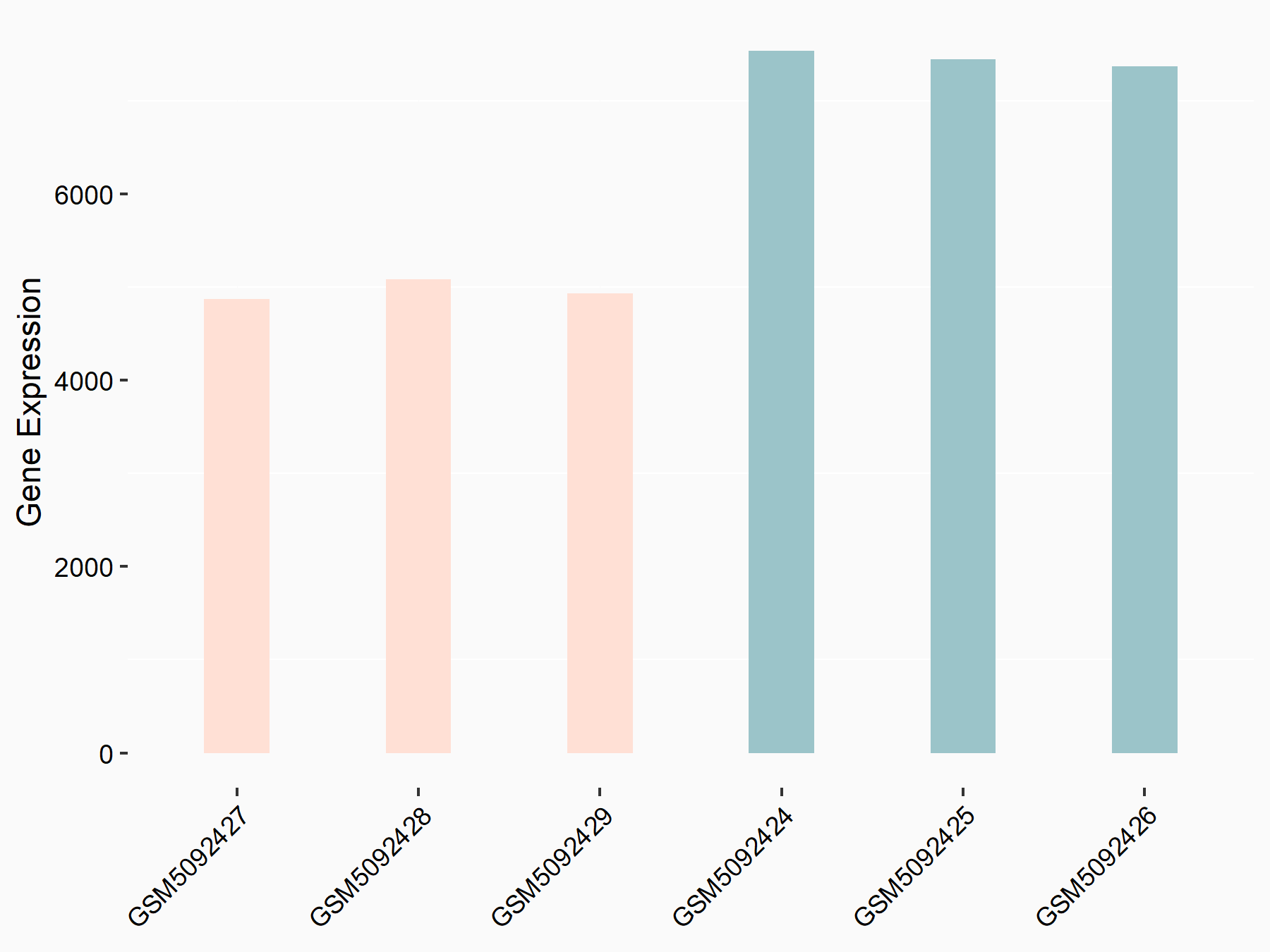 |
logFC: -5.87E-01 p-value: 2.40E-51 |
| More Results | Click to View More RNA-seq Results | |
Tamoxifen
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [152] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| Cell Process | Mitochondrial apoptosis | |||
| In-vitro Model | MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| MCF7-TamR | Invasive breast carcinoma | Homo sapiens | CVCL_EG55 | |
| Response Summary | Adenylate kinase 4 modulates the resistance of breast cancer cells to tamoxifen through an m6A-based epitranscriptomic mechanism. Genetic depletion of METTL3 in TamR MCF-7 cells led to a diminished Adenylate kinase 4, mitochondrial (AK4) protein level and attenuated resistance to tamoxifen. | |||
Anterior gradient protein 2 homolog (AGR2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
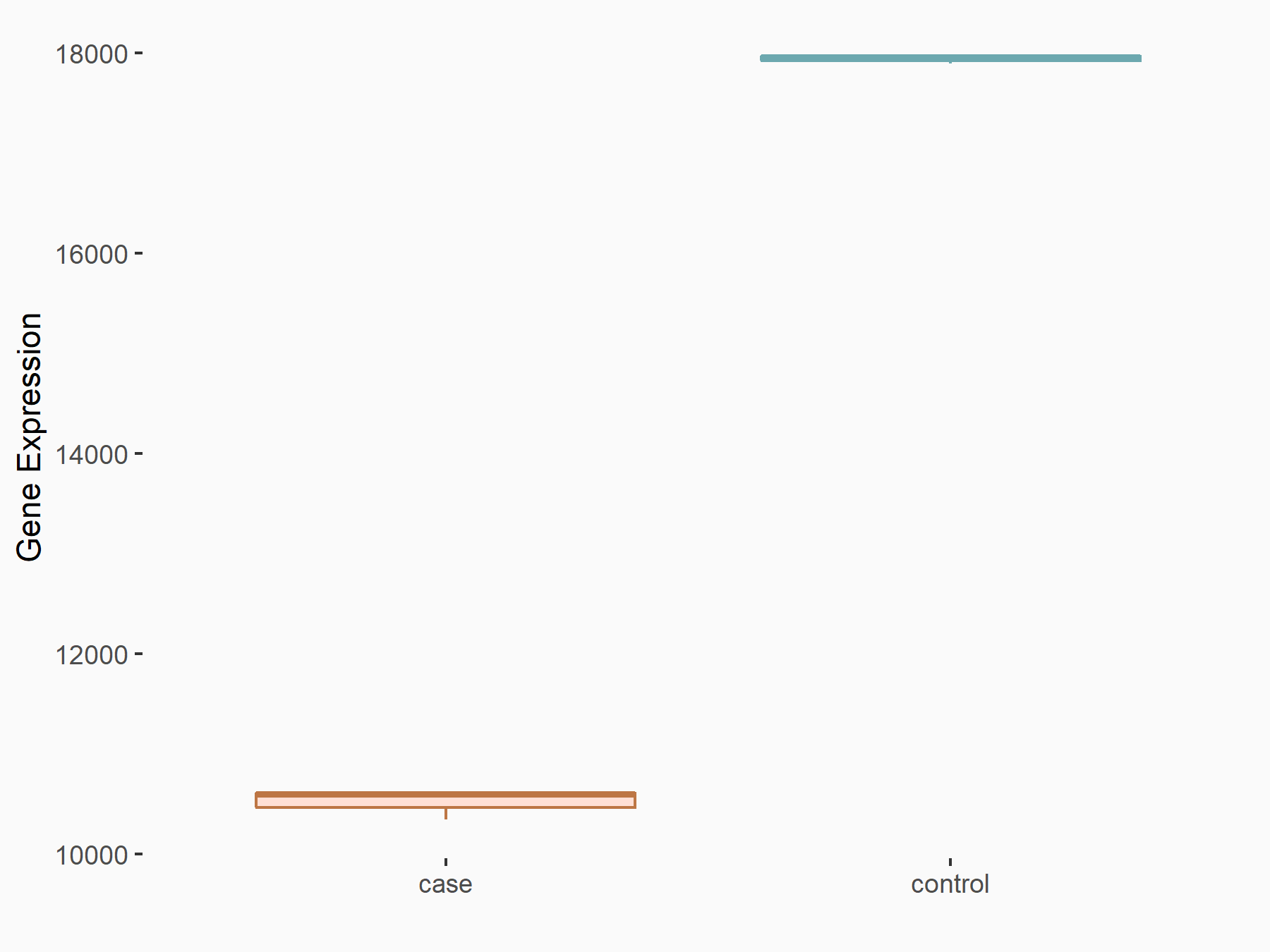 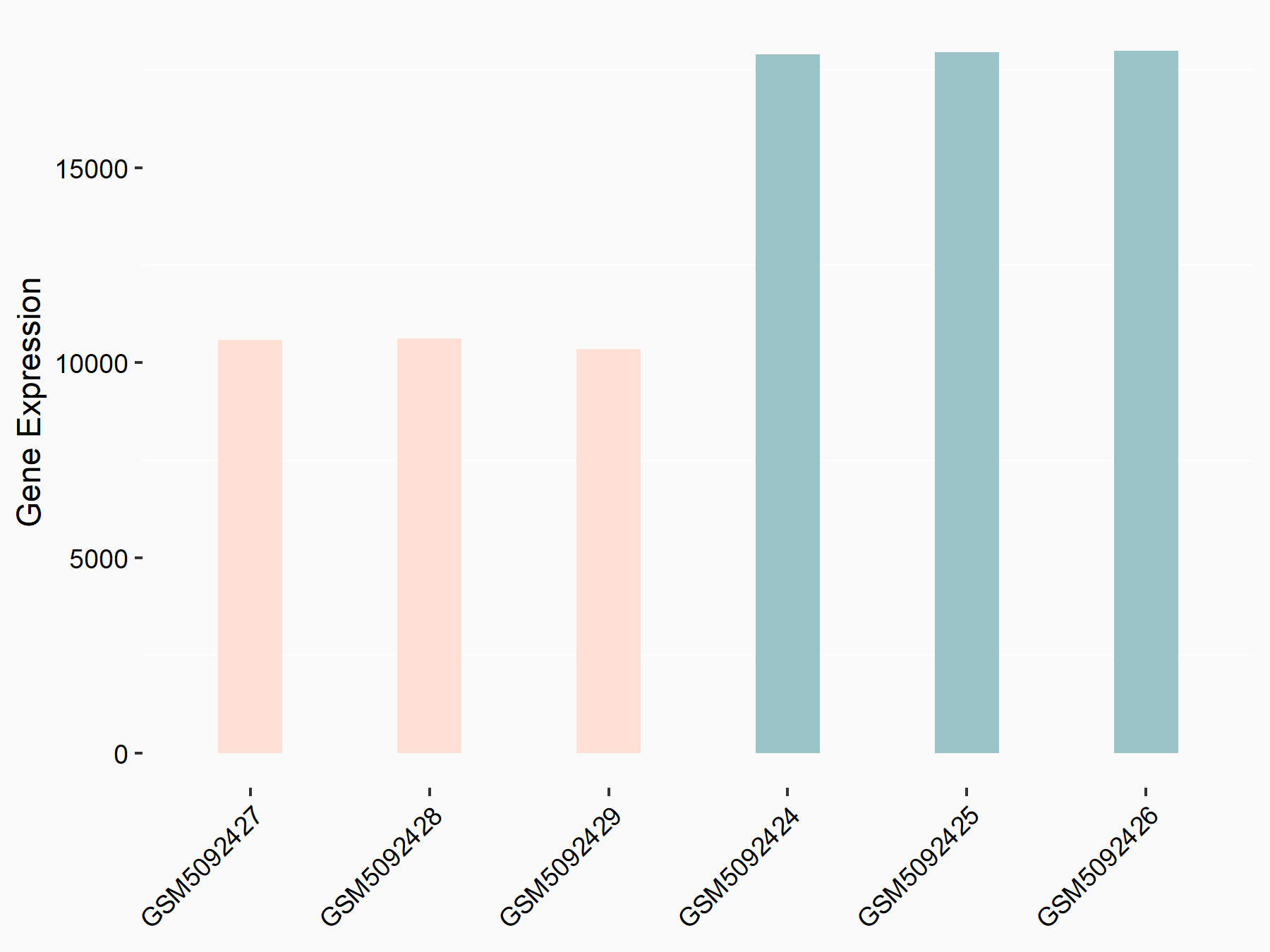 |
logFC: -7.71E-01 p-value: 1.08E-130 |
| More Results | Click to View More RNA-seq Results | |
Doxil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [116] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | MCF7-DoxR (Adriamycin-resistant cell line MCF7-DoxR) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| In-vivo Model | Once the tumor volume increased to about 1 cm3, six groups of MCF7 bearing mice (n = 10 in each group) were injected with PBS (0.1 ml, caudal vein) and adriamycin (0.1 ml, 10 mg/kg), respectively. When the tumor reached 1.5 cm in any direction (defined as event-free survival analysis), 10 mice in each group were selected to measure the tumor size and weight on the 12th day after adriamycin injection. | |||
| Response Summary | METTL3 can regulate the expression of MALAT1 through m6A, mediate the E2F1/Anterior gradient protein 2 homolog (AGR2) axis, and promote the adriamycin resistance of breast cancer. | |||
ATP-dependent translocase ABCB1 (ABCB1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
 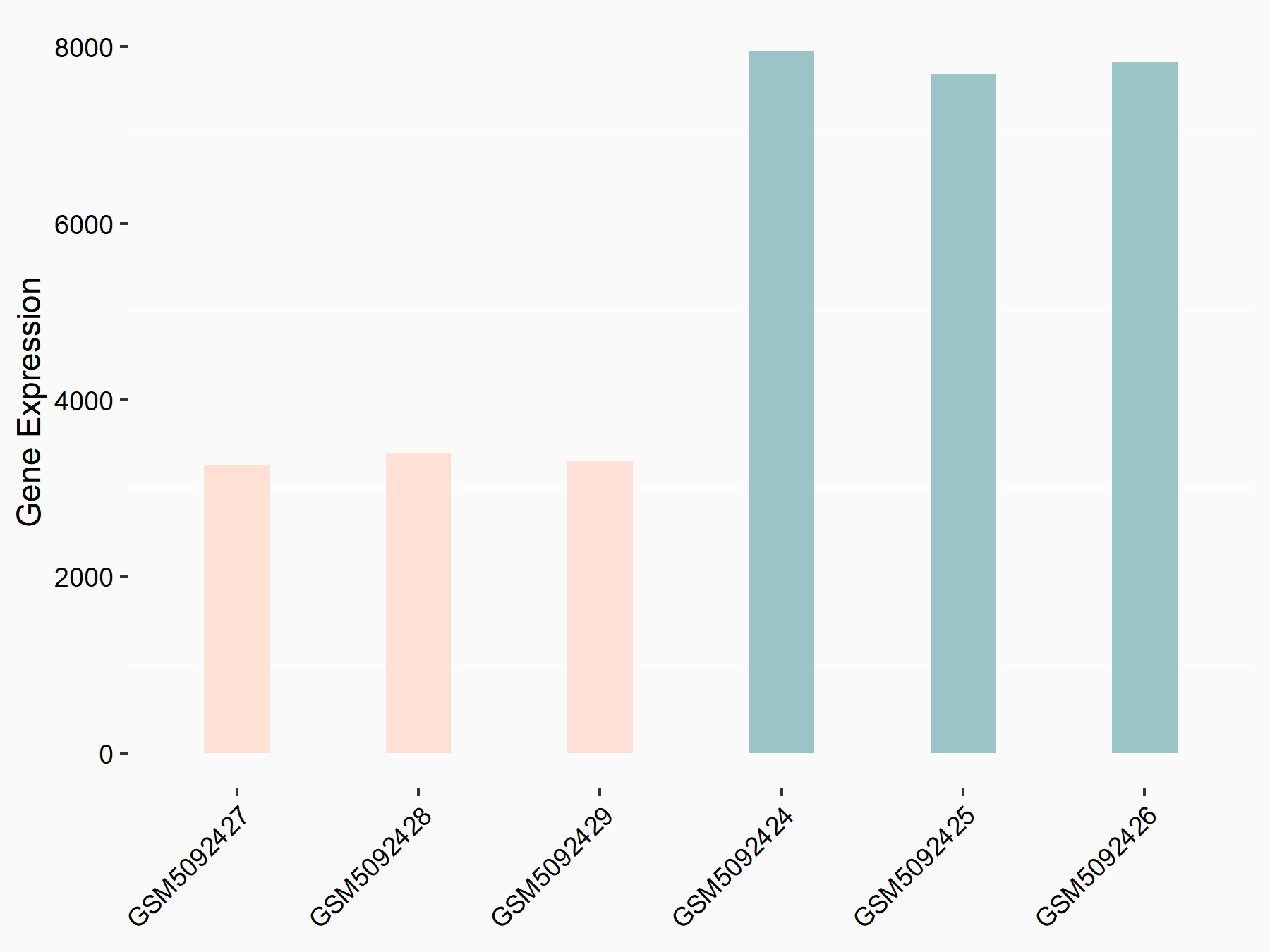 |
logFC: -1.24E+00 p-value: 2.14E-202 |
| More Results | Click to View More RNA-seq Results | |
Doxil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [22] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell growth and death | |||
| Cell apoptosis | ||||
| In-vitro Model | ADR-resistant MCF-7 (MCF-7/ADR) cells (Human breast cancer doxorubicin-resistant cell line) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| In-vivo Model | Cell suspensions (2 × 106 cells/mL) made with MCF-7/ADR cells stably expressing METTL3 and/or miR-221-3p inhibitor were subcutaneously implanted into each mouse. One week later, xenografted mice were injected with 0.1 mL ADR (25 mg/kg, intraperitoneal injection) twice a week. | |||
| Response Summary | METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. METTL3 knockdown was shown to reduce the expression of miR-221-3p by reducing pri-miR-221-3p m6A mRNA methylation, reducing the expression of ATP-dependent translocase ABCB1 (ABCB1) and BCRP, and inducing apoptosis. Identified the METTL3/miR-221-3p/HIPK2/Che-1 axis as a novel signaling event that will be responsible for resistance of BC cells to ADR. | |||
DNA repair protein RAD51 homolog 1 (RAD51)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
 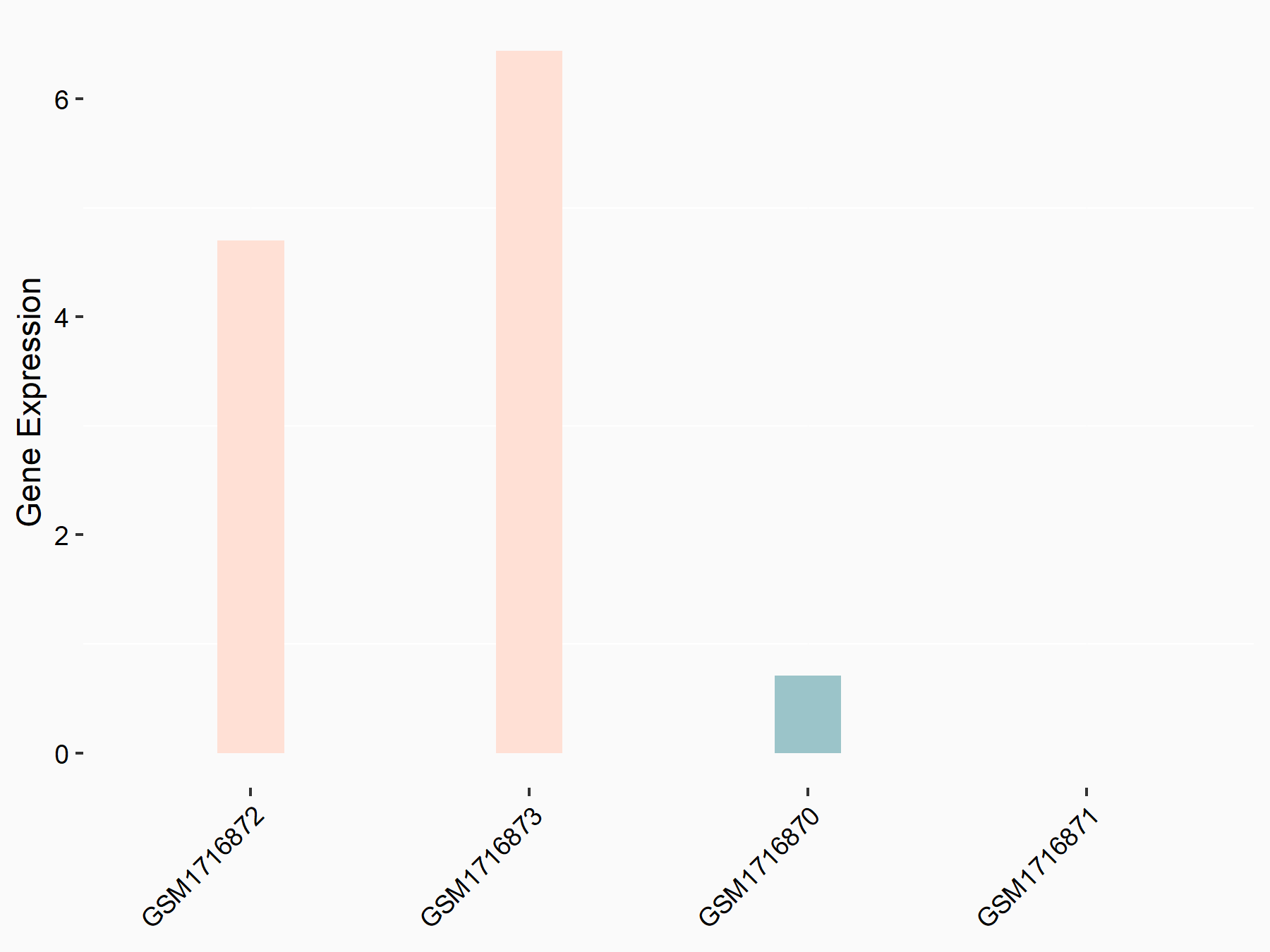 |
logFC: 2.32E+00 p-value: 1.62E-02 |
| More Results | Click to View More RNA-seq Results | |
Doxil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [98] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Homologous recombination | hsa03440 | ||
| Cell Process | Homologous recombination repair | |||
| In-vitro Model | MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | Cells were trypsinized and resuspended in DMEM at a consistence of 1 × 107 cells/ml. A total of 1 × 106 cells were injected into flank of mice. 27 days after injection, tumors were removed for paraffin-embedded sections. | |||
| Response Summary | Knockdown of METTL3 sensitized these breast cancer cells to Adriamycin (ADR; also named as doxorubicin) treatment and increased accumulation of DNA damage. Mechanically, we demonstrated that inhibition of METTL3 impaired HR efficiency and increased ADR-induced DNA damage by regulating m6A modification of EGF/RAD51 axis. METTL3 promoted EGF expression through m6A modification, which further upregulated DNA repair protein RAD51 homolog 1 (RAD51) expression, resulting in enhanced HR activity. | |||
DNA-3-methyladenine glycosylase (ANPG/MPG)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
  |
logFC: -6.00E-01 p-value: 3.04E-08 |
| More Results | Click to View More RNA-seq Results | |
Temozolomide
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [69] | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | DNA repair | |||
| In-vitro Model | U251 (Fibroblasts or fibroblast like cells) | |||
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| In-vivo Model | Subcutaneously injected shMETTL3 or shNC-expressing U87-MG-TMZ cells into BALB/c NOD mice. After confirmation of GBM implantation, mice were treated with TMZ (66 mg/kg/d, 5 d per week, for 3 cycles). | |||
| Response Summary | Two critical DNA repair genes (MGMT and DNA-3-methyladenine glycosylase (ANPG/MPG)) were m6A-modified by METTL3, whereas inhibited by METTL3 silencing or DAA-mediated total methylation inhibition, which is crucial for METTL3-improved temozolomide resistance in glioblastoma cells. | |||
Epidermal growth factor receptor (EGFR)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
 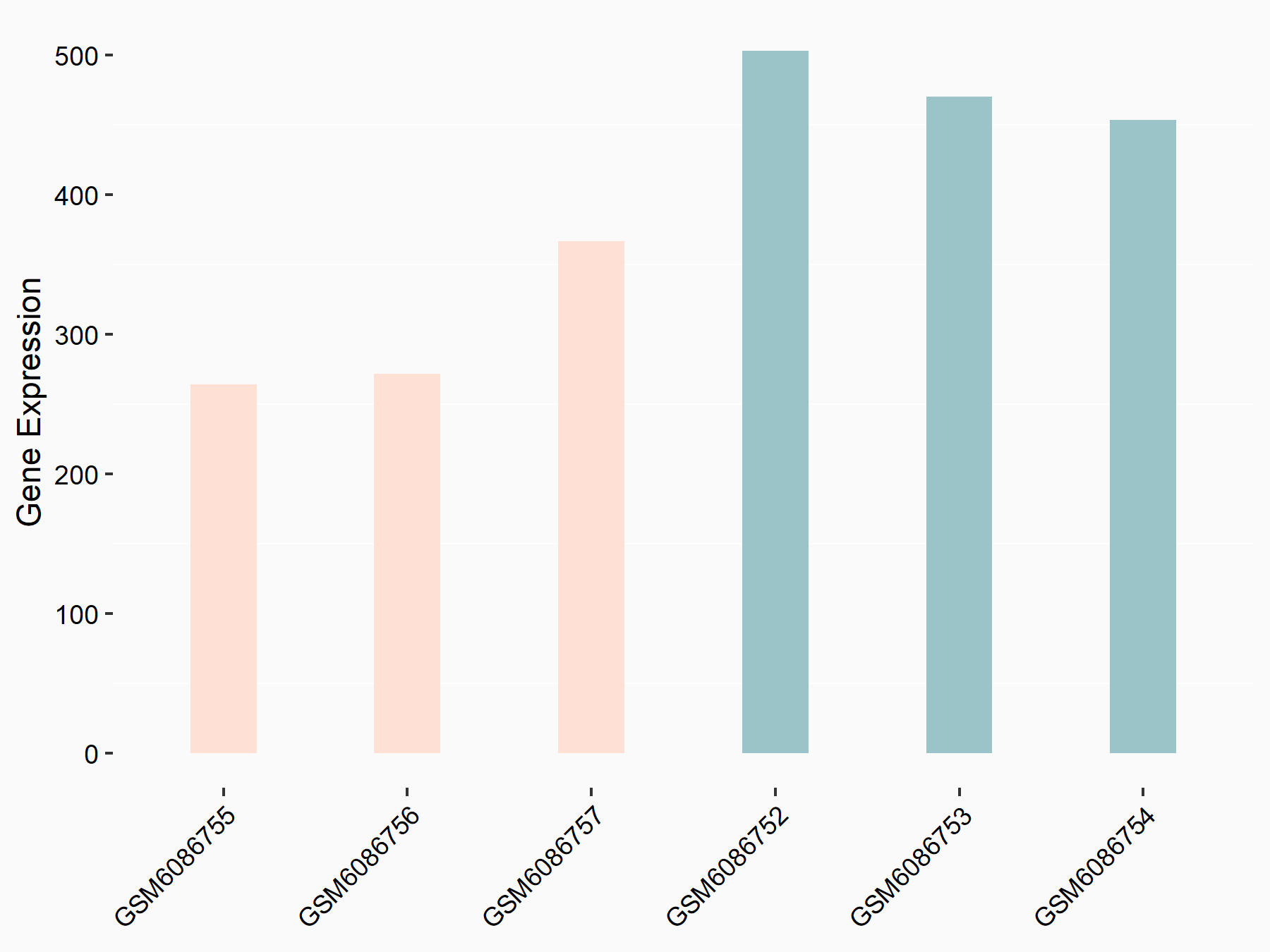 |
logFC: -6.74E-01 p-value: 1.53E-03 |
| More Results | Click to View More RNA-seq Results | |
PLX4032
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [164] | |||
| Responsed Disease | Melanoma | ICD-11: 2C30 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | EGFR tyrosine kinase inhibitor resistance | hsa01521 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | A375-R | Amelanotic melanoma | Homo sapiens | CVCL_6234 |
| Response Summary | METTL3 increased the m6A modification of Epidermal growth factor receptor (EGFR) mRNA in A375R cells, which promoted its translation efficiency. Inhibiting METTL3 function to restore PLX4032 sensitivity in patients with melanoma. | |||
Lenvatinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [290] | |||
| Responsed Disease | Liver hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 |
| In-vivo Model | Tumor cells derived xenograft (MHCC97H cells) from subcutaneous mouse model were cut into 1 mm3 pieces and implanted into the left lobe of the livers of six-week-old male NCG mice. Once the tumors were established and reached approximately 100 mm3, mice were randomly assigned to daily treatment with vehicle, lenvatinib (4 mg/kg, oral gavage), STM2457 (50 mg/kg, intraperitoneal injection) or a drug combination in which each compound was administered at the same dose and duration as the single agent. | |||
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [290] | |||
| Responsed Disease | Liver hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
Forkhead box protein O3 (FOXO3)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
 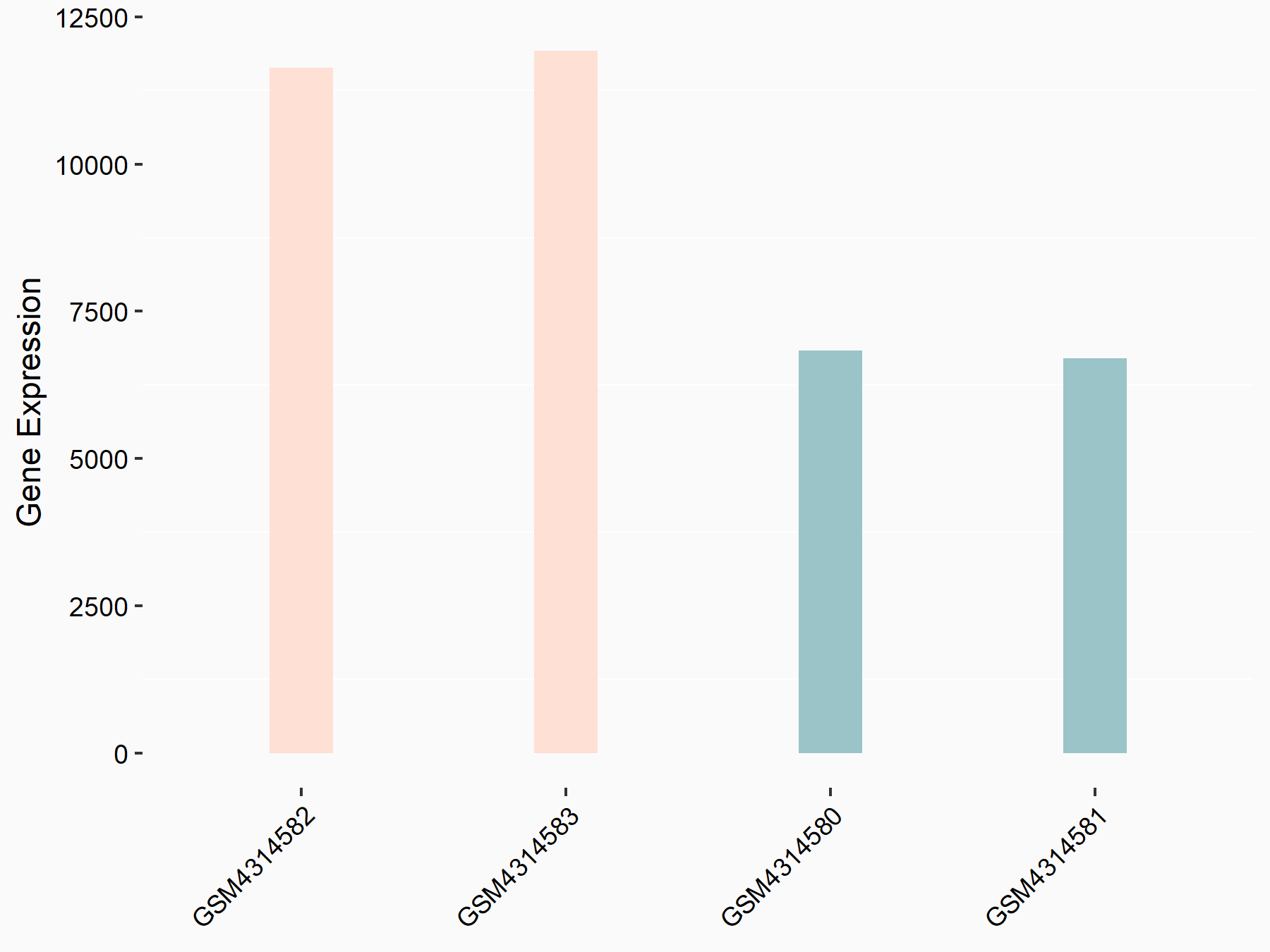 |
logFC: 8.00E-01 p-value: 1.28E-68 |
| More Results | Click to View More RNA-seq Results | |
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [167] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Cell Process | Cell Transport | |||
| Cell catabolism | ||||
| Cell autophagy | ||||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| Hepa 1-6 | Hepatocellular carcinoma of the mouse | Mus musculus | CVCL_0327 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
| WRL 68 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0581 | |
| In-vivo Model | For the drug-resistant subcutaneous tumor models, drug administration was adopted when the tumors reached about 50 mm3 in size, at which point mice were randomized for treatment with DMSO(intraperitoneally) or sorafenib (50 mg/kg/every 2 days, intraperitoneally). For the patient-derived tumor xenograft model, drug administration began 4 weeks after tumors reached about 100 mm3 in size with sorafenib (50 mg/kg/every 3 days, intraperitoneally) or siCtrl/siMETTL3 intratumor injection. | |||
| Response Summary | METTL3 and Forkhead box protein O3 (FOXO3) levels are tightly correlated in hepatocellular carcinoma patients. In mouse xenograft models, METTL3 depletion significantly enhances sorafenib resistance of HCC by abolishing the identified METTL3-mediated FOXO3 mRNA stabilization, and overexpression of FOXO3 restores m6 A-dependent sorafenib sensitivity. | |||
Hepatocyte growth factor receptor (c-Met/MET)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 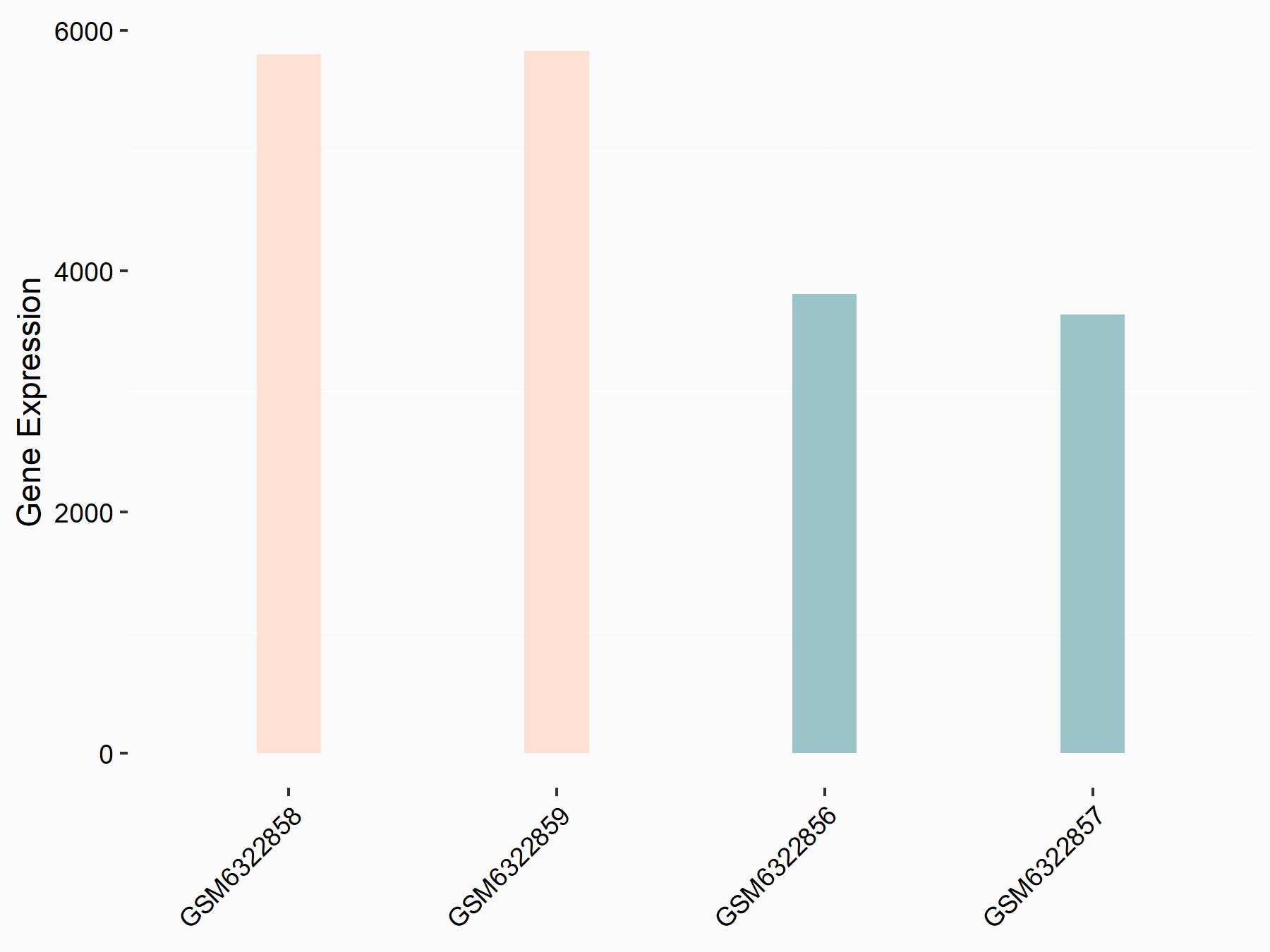 |
logFC: 6.42E-01 p-value: 3.45E-17 |
| More Results | Click to View More RNA-seq Results | |
Crizotinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [173] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | EGFR tyrosine kinase inhibitor resistance | hsa01521 | ||
| In-vitro Model | HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 |
| NCI-H661 | Lung large cell carcinoma | Homo sapiens | CVCL_1577 | |
| NCI-H596 | Lung adenosquamous carcinoma | Homo sapiens | CVCL_1571 | |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| NCI-H358 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1559 | |
| NCI-H292 | Lung mucoepidermoid carcinoma | Homo sapiens | CVCL_0455 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1395 | Lung adenocarcinoma | Homo sapiens | CVCL_1467 | |
| EBC-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_2891 | |
| Calu-3 | Lung adenocarcinoma | Homo sapiens | CVCL_0609 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | HCC827 (3×106) cells suspended in 100 uL of PBS were injected into the left inguen of female Balb/c nude mice (body weight 18-20 g; age 6 weeks; Beijing Huafukang Bioscience Co., Inc.). When the tumor volumes reached 50-100 mm3 on the 10th posttransplantation day, the mice were randomized into four groups (10 mice per group) and were intragastrically administered vehicle (normal saline), crizotinib (25 mg/kg body weight), chidamide (5 mg/kg), or the combination of the two drugs daily for 21 days. The tumor volumes and body weights of the mice were measured every 3 days. | |||
| Response Summary | Chidamide could decrease Hepatocyte growth factor receptor (c-Met/MET) expression by inhibiting mRNA N6-methyladenosine (m6A) modification through the downregulation of METTL3 and WTAP expression, subsequently increasing the crizotinib sensitivity of NSCLC cells in a c-MET-/HGF-dependent manner. | |||
Gefitinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [174] | |||
| Responsed Disease | Lung cancer | ICD-11: 2C25 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| In-vitro Model | PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| NCI-H3255 | Lung adenocarcinoma | Homo sapiens | CVCL_6831 | |
| Response Summary | METTL3 combines with Hepatocyte growth factor receptor (c-Met/MET) and causes the PI3K/AKT signalling pathway to be manipulated, which affects the sensitivity of lung cancer cells to gefitinib. METTL3 knockdown promotes apoptosis and inhibits proliferation of lung cancer cells. | |||
Meltrin-beta (ADAM19)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HUVEC cell line | Homo sapiens |
|
Treatment: shMETTL3 HUVEC cells
Control: shScramble HUVEC cells
|
GSE157544 | |
| Regulation |
 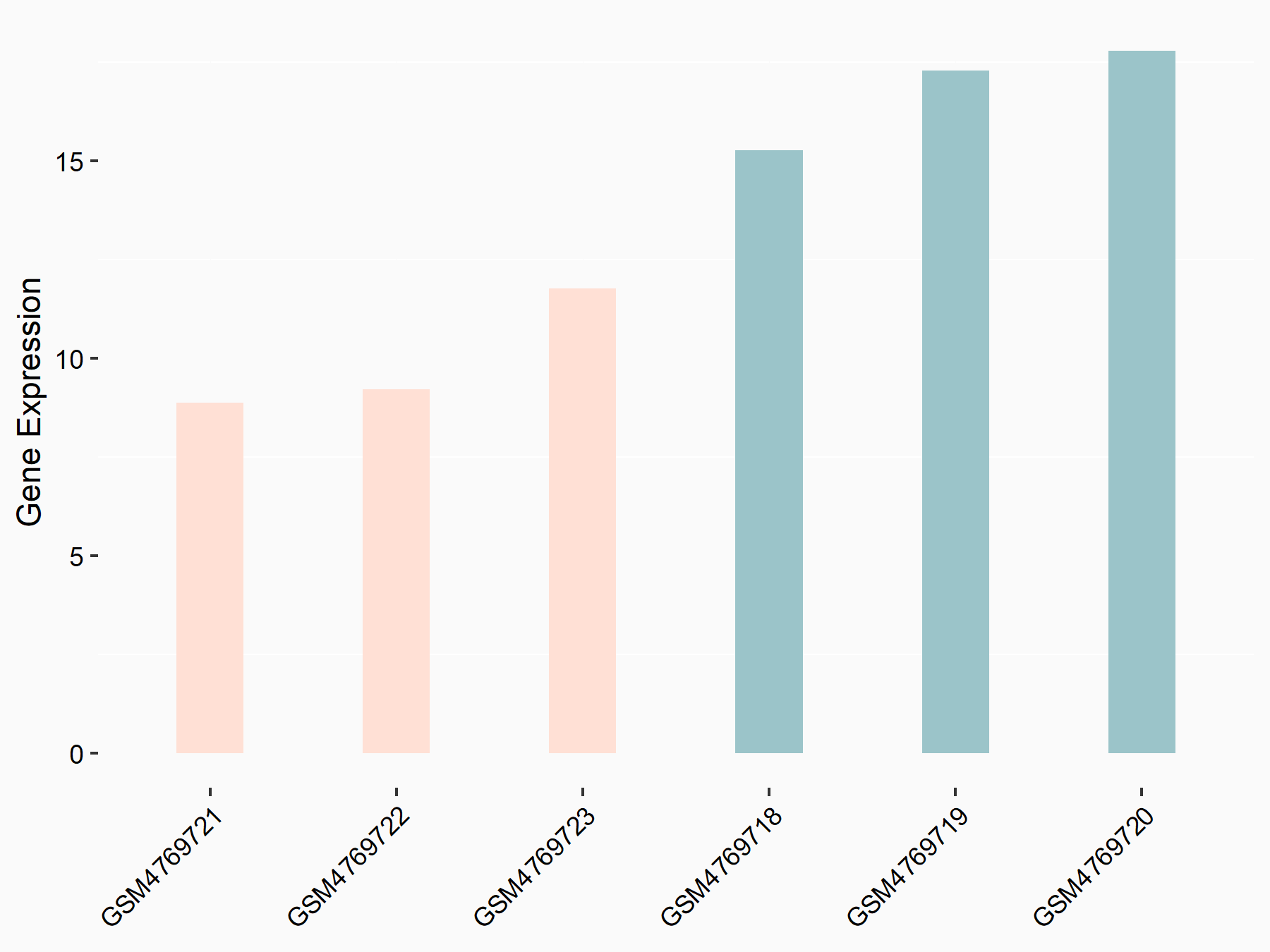 |
logFC: -7.06E-01 p-value: 4.35E-03 |
| More Results | Click to View More RNA-seq Results | |
Ethyl ester form of meclofenamic acid
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [194] | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| Target Regulation | Down regulation | |||
| Cell Process | Cells growth | |||
| Cells self-renewal | ||||
| Tumorigenesis | ||||
| MicroRNAs in cancer (hsa05206) | ||||
| In-vitro Model | GSC | Glioma | Epinephelus akaara | CVCL_M752 |
| In-vivo Model | 2 × 105 dissociated cells in 2 uL PBS were injected into the following site (anteroposterior [AP] +0.6 mm, mediolateral [ML] +1.6 mm, and dorsoventricular [DV] 2.6 mm) with a rate of 1 uL/min. | |||
| Response Summary | Knockdown of METTL3 or METTL14 induced changes in mRNA m6A enrichment and altered mRNA expression of genes (e.g., Meltrin-beta (ADAM19)) with critical biological functions in GSCs. Treatment with MA2, a chemical inhibitor of FTO, dramatically suppressed GSC-induced tumorigenesis and prolonged lifespan in GSC-grafted animals. | |||
Neurogenic locus notch homolog protein 2 (NOTCH2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
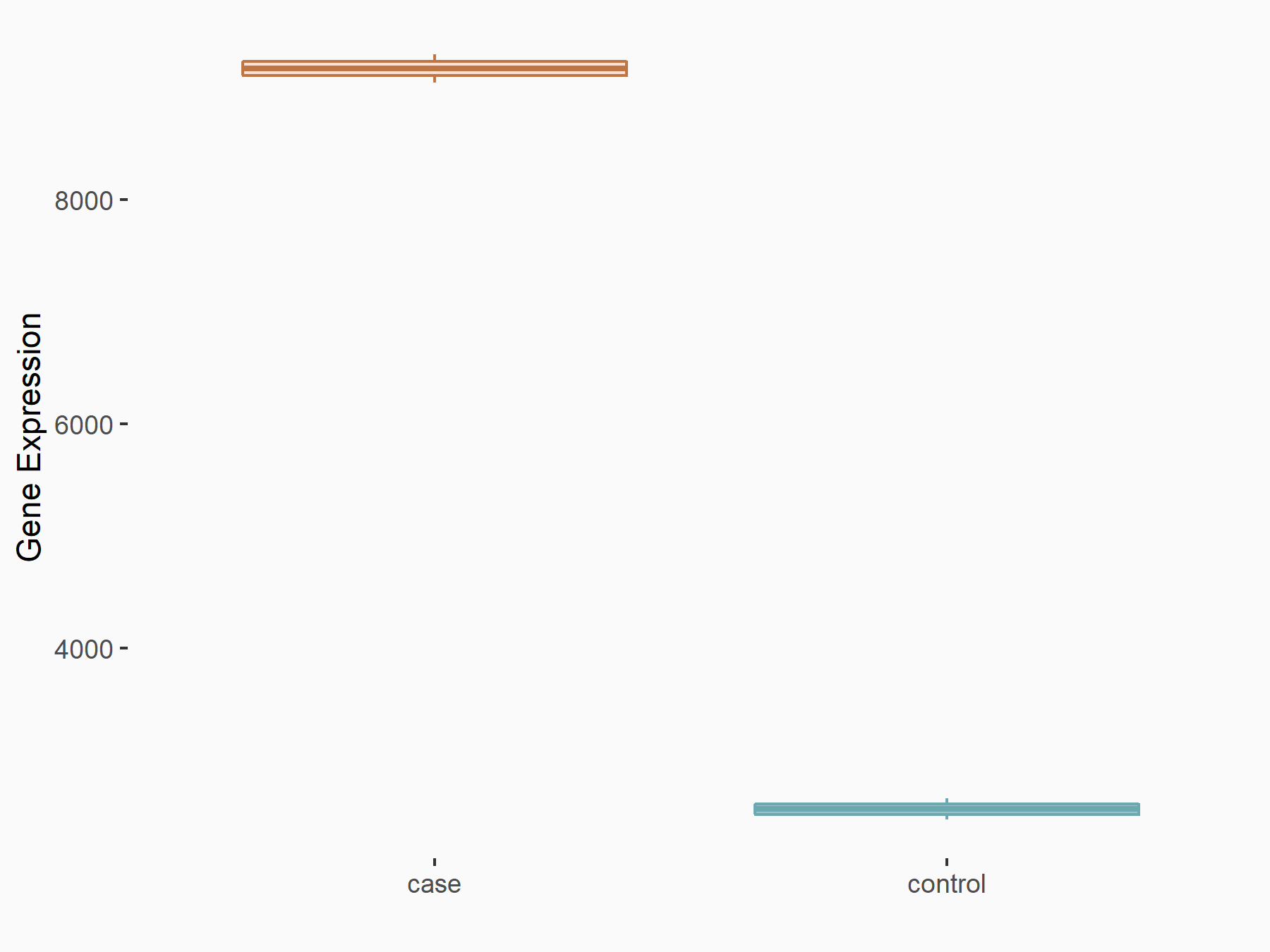 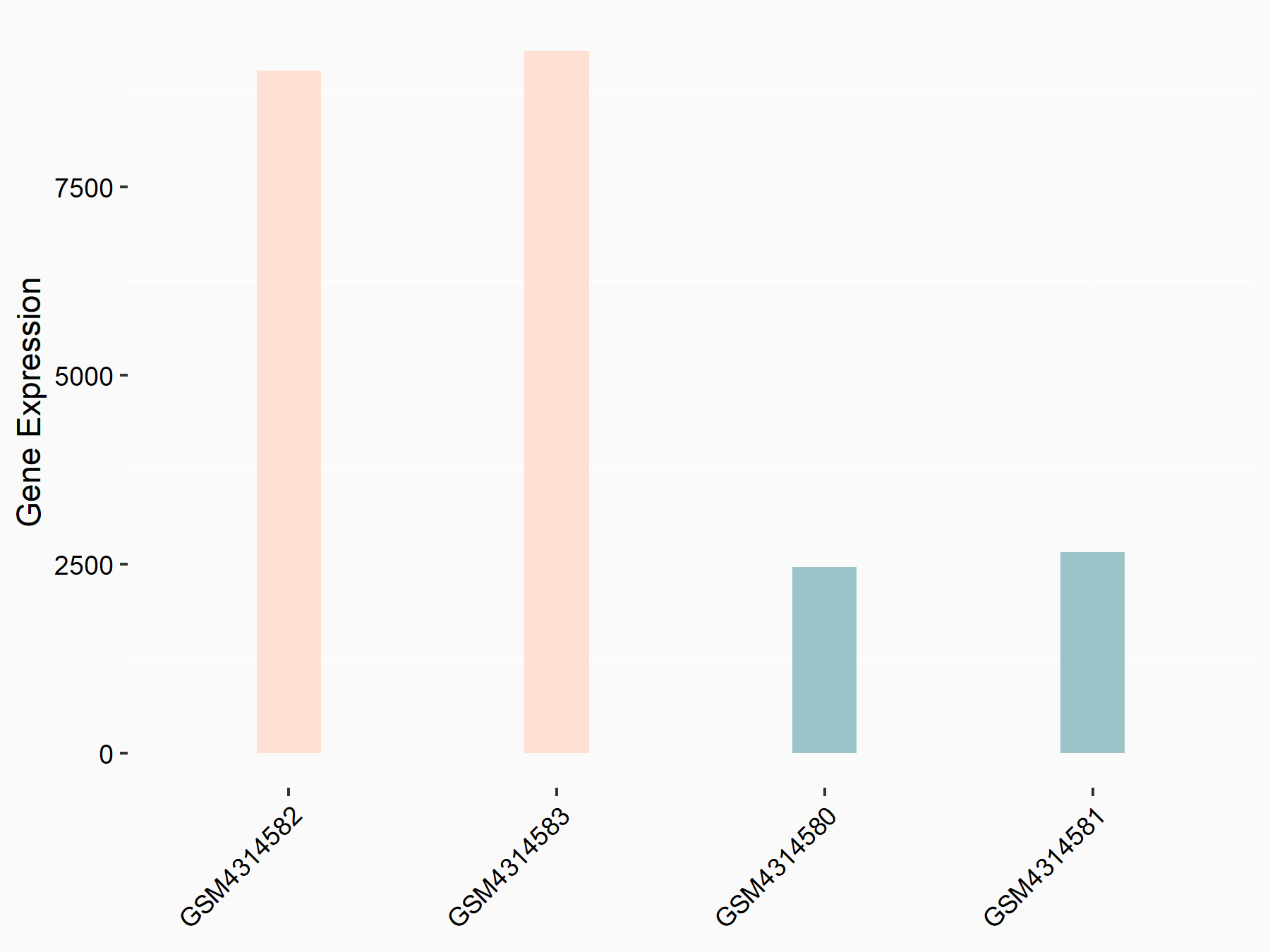 |
logFC: 1.84E+00 p-value: 2.61E-238 |
| More Results | Click to View More RNA-seq Results | |
Melittin
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [201] | |||
| Responsed Disease | Bladder cancer | ICD-11: 2C94 | ||
| Target Regulation | Down regulation | |||
| Cell Process | miRNA maturation | |||
| Cell apoptosis | ||||
| In-vitro Model | T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| EJ (Human bladder cancer cells) | ||||
| BIU-87 | Human bladder cancer cells | Homo sapiens | CVCL_6881 | |
| In-vivo Model | For melittin treatment study, 4-week-old female BALB/c nude mice were subcutaneously injected with 1 × 107 T24 or BIU87 cells. | |||
| Response Summary | METTL3 acts as a fate determinant that controls the sensitivity of bladder cancer cells to melittin treatment. Moreover, METTL3/miR-146a-5p/NUMB/Neurogenic locus notch homolog protein 2 (NOTCH2) axis plays an oncogenic role in bladder cancer pathogenesis and could be a potential therapeutic target for recurrent bladder cancer treatment. | |||
Nuclear factor erythroid 2-related factor 2 (NFE2L2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
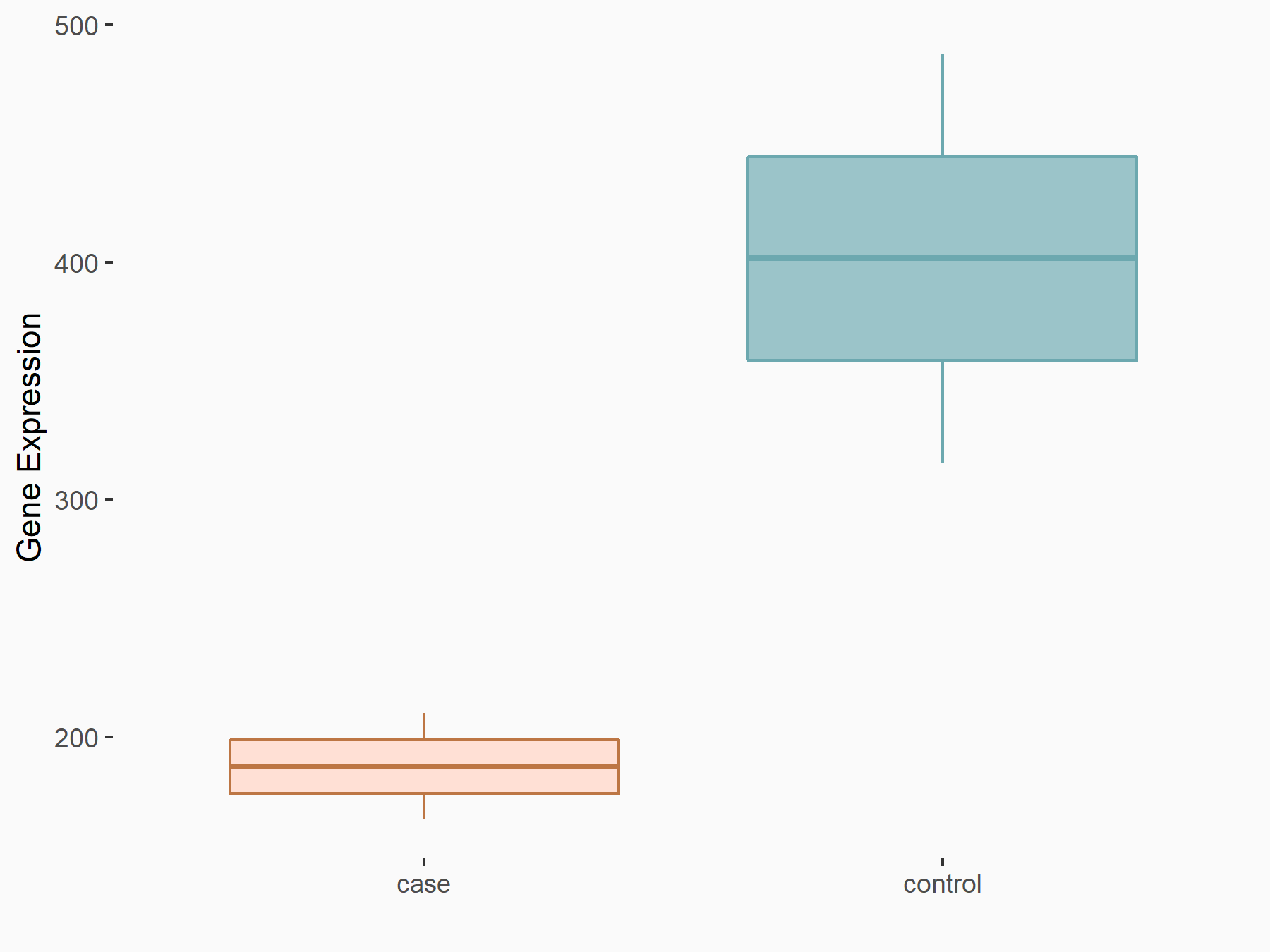 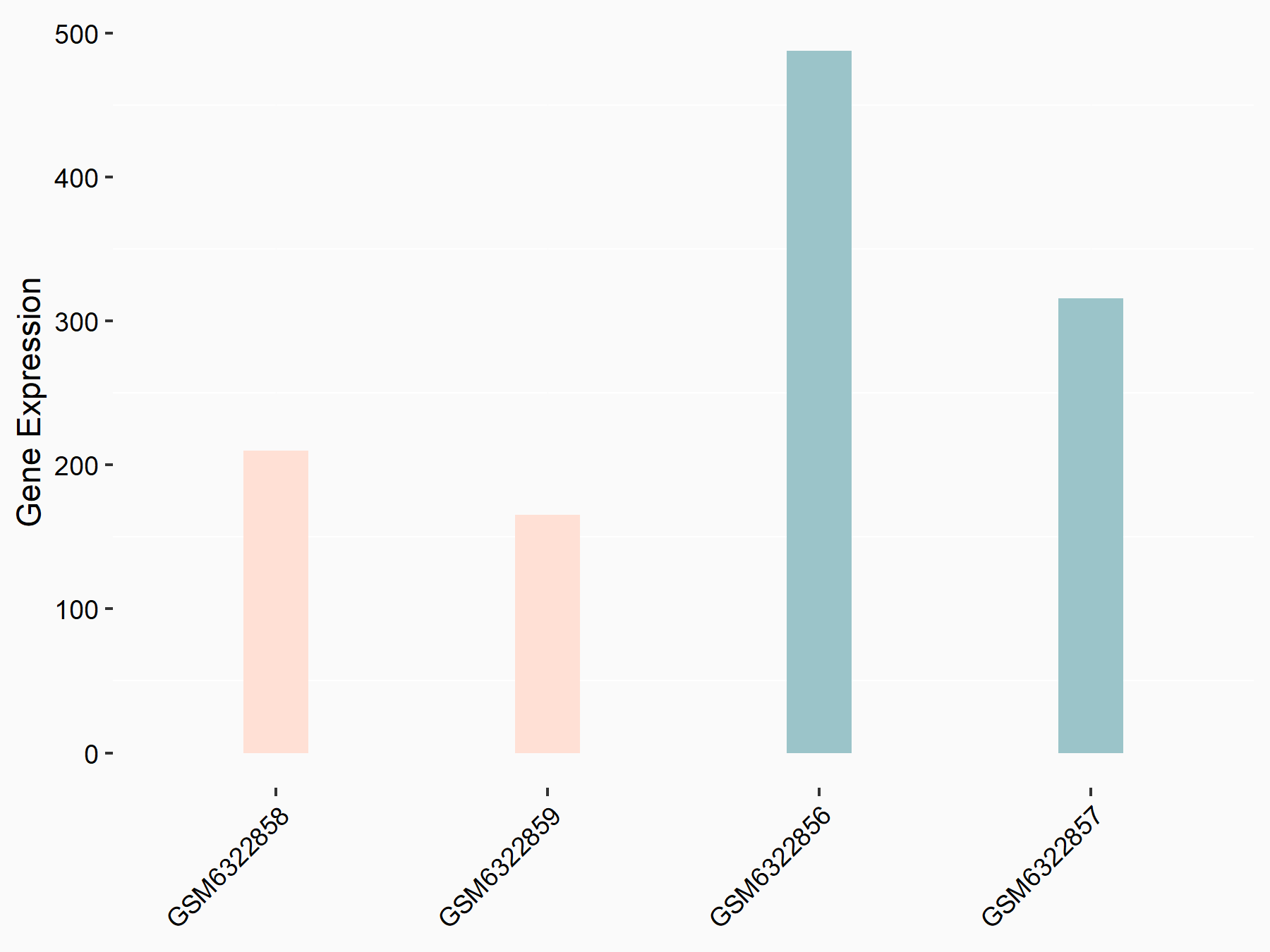 |
logFC: -1.10E+00 p-value: 4.72E-06 |
| More Results | Click to View More RNA-seq Results | |
Colistin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [65] | |||
| Responsed Disease | Diseases of the urinary system | ICD-11: GC2Z | ||
| Target Regulation | Up regulation | |||
| Cell Process | Oxidative stress | |||
| Cell apoptosis | ||||
| In-vivo Model | The 60 female Kunming mice were divided into two groups (n = 30): control group (injection of physiological saline through the caudal vein) and colistin group (injection of 15 mg/kg colistin, twice a day, with an eight-hour interval). | |||
| Response Summary | m6A methylation was involved in oxidative stress-mediated apoptosis in the mechanism of colistin nephrotoxicity. METTL3-mediated m6A methylation modification is involved in colistin-induced nephrotoxicity through apoptosis mediated by Keap1/Nuclear factor erythroid 2-related factor 2 (NFE2L2) signaling pathway. | |||
Nucleobindin-1 (NUCB1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | DKO-1 cell line | Homo sapiens |
|
Treatment: METTL3 knockdown DKO-1 cell
Control: DKO-1 cell
|
GSE182382 | |
| Regulation |
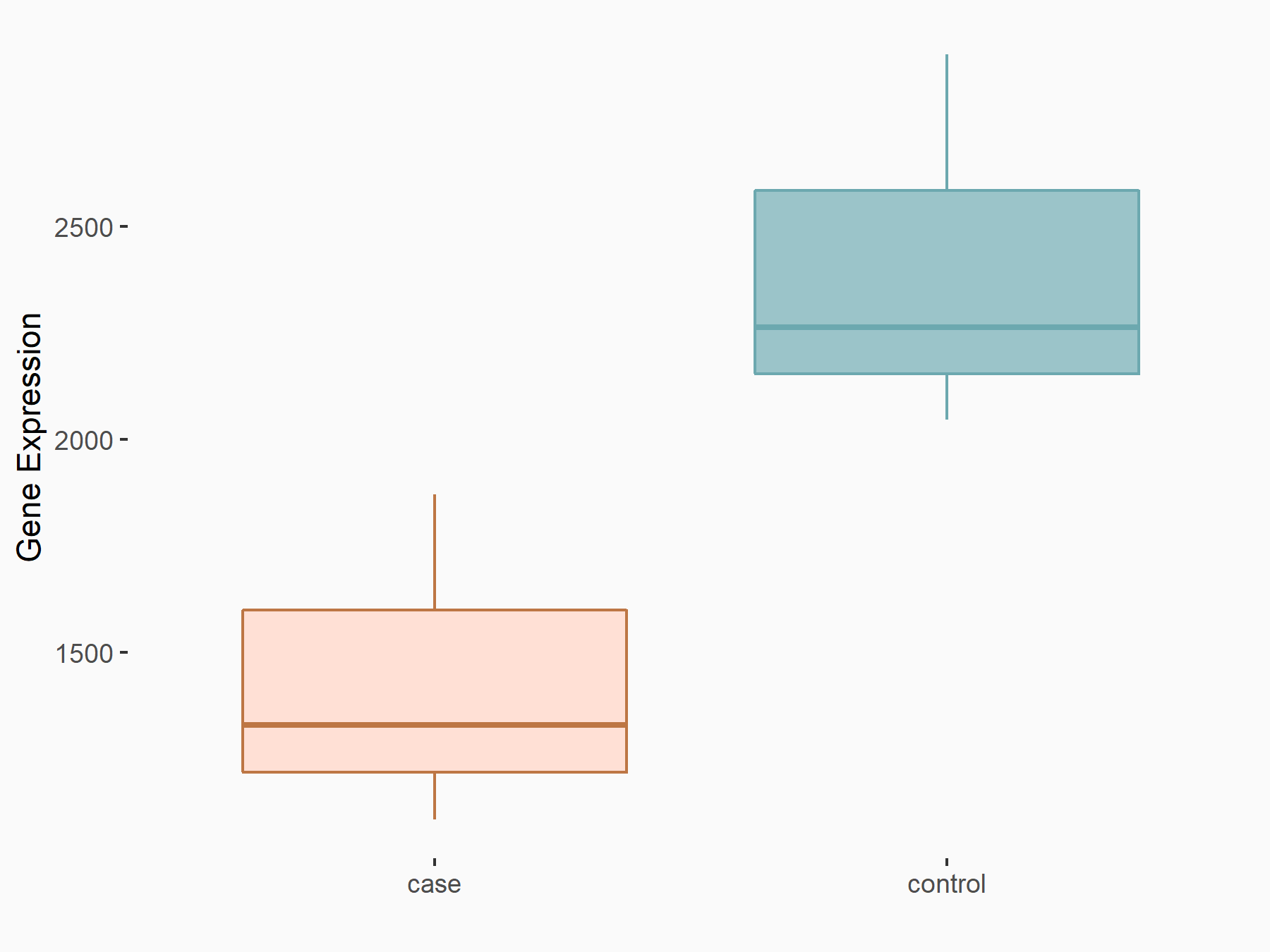 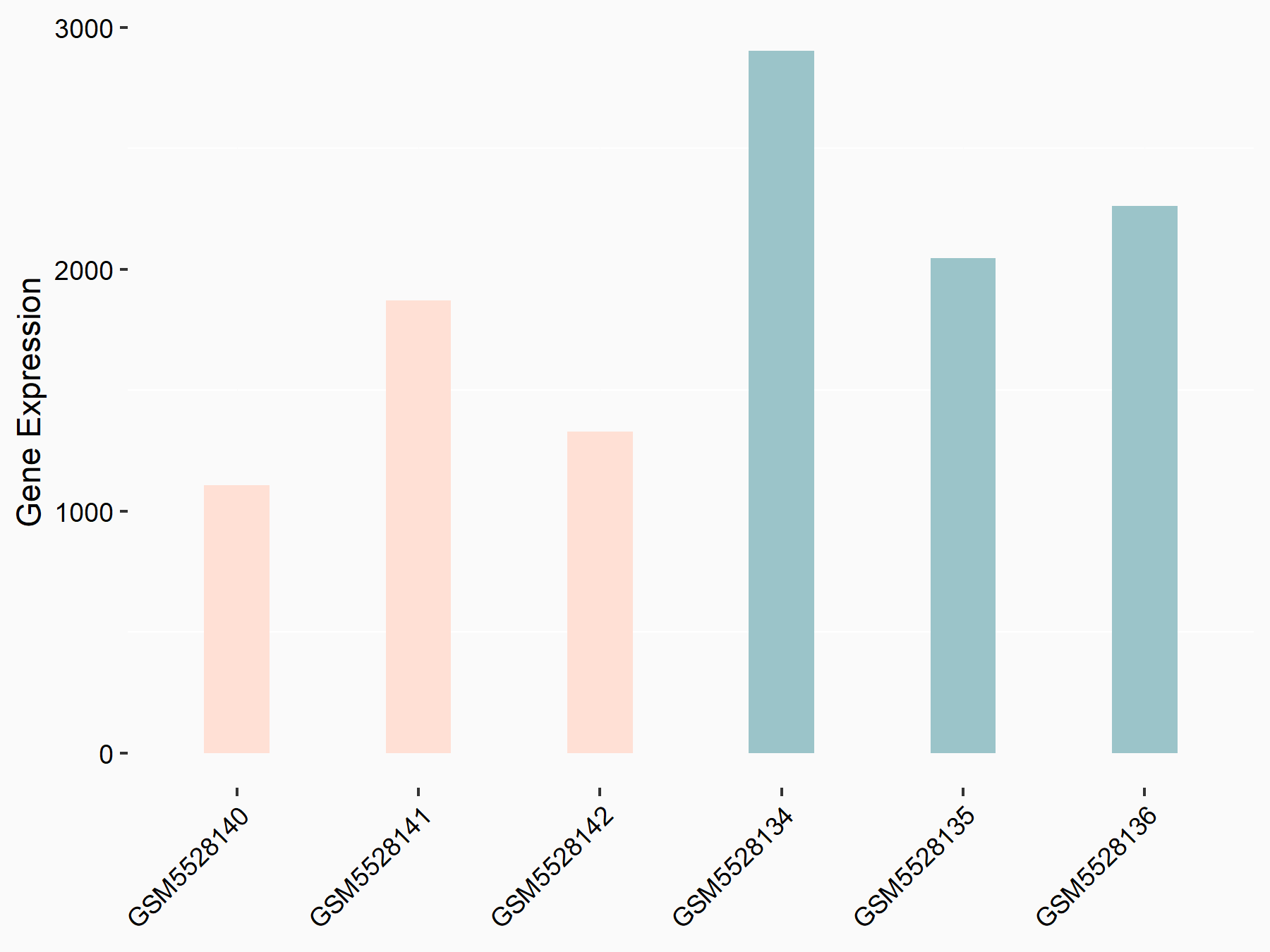 |
logFC: -7.45E-01 p-value: 2.41E-03 |
| More Results | Click to View More RNA-seq Results | |
Gemcitabine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [204] | |||
| Responsed Disease | Pancreatic ductal adenocarcinoma | ICD-11: 2C10.0 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell proliferation | |||
| Cell autophagy | ||||
| In-vitro Model | SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| In-vivo Model | 5 × 106 SW1990 cells expressing NUCB1 (oeNUCB1) or control vector (oeNC) were injected subcutaneously. | |||
| Response Summary | METTL3-mediated m6A modification on Nucleobindin-1 (NUCB1) 5'UTR via the reader YTHDF2 as a mechanism for NUCB1 downregulation in PDAC. This study revealed crucial functions of NUCB1 in suppressing proliferation and enhancing the effects of gemcitabine in pancreatic cancer cells. | |||
Poly [ADP-ribose] polymerase 1 (PARP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shMETTL3 MOLM13 cells
Control: MOLM13 cells
|
GSE98623 | |
| Regulation |
 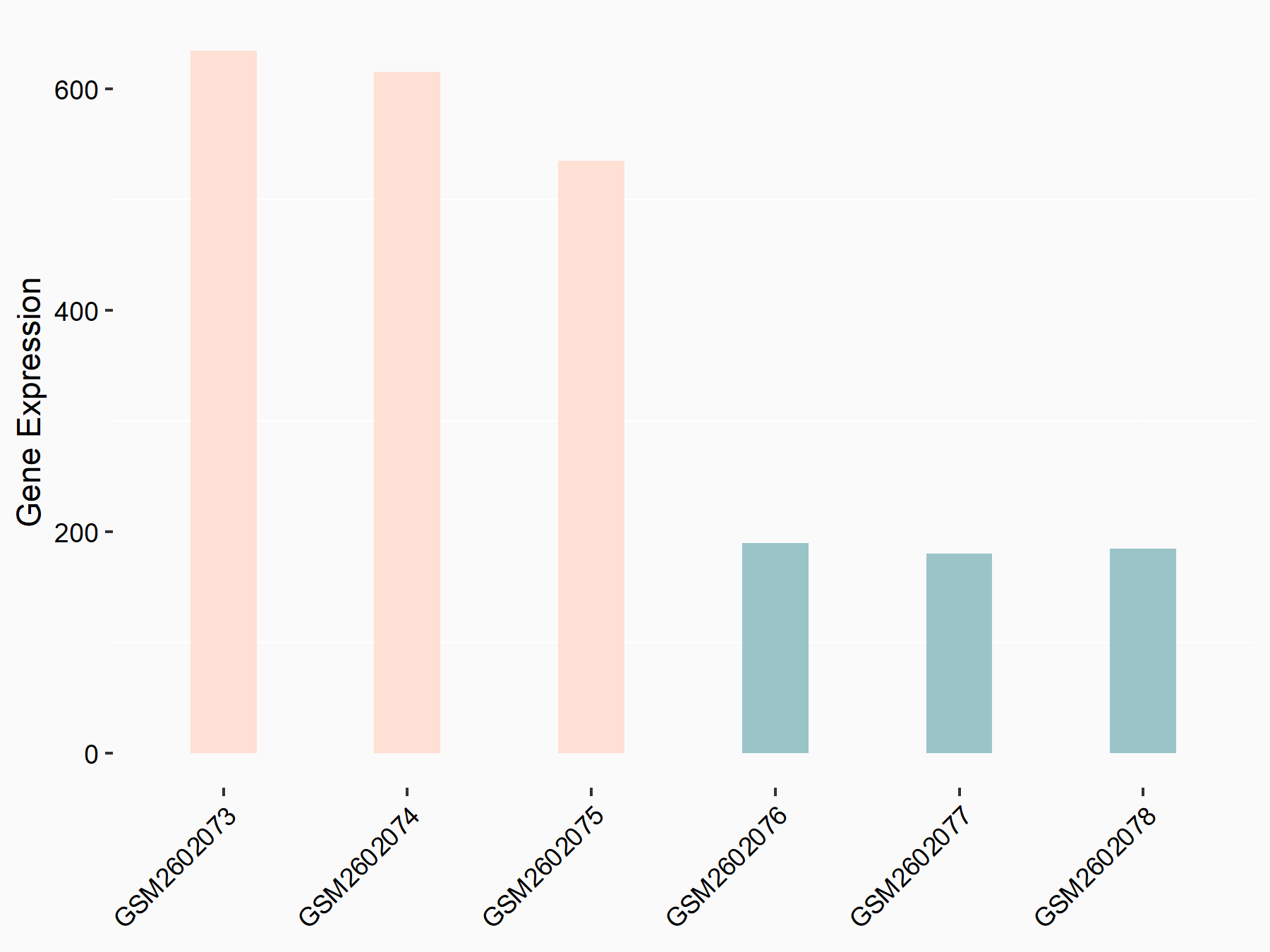 |
logFC: 1.69E+00 p-value: 1.05E-54 |
| More Results | Click to View More RNA-seq Results | |
Oxaliplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [208] | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | |||
| Cell Process | RNA stability | |||
| Excision repair | ||||
| In-vitro Model | SNU-719 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_5086 |
| MKN74 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_2791 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | 100,000 pLKO and PARP1-sh1 (PT1 and PT2) cells were mixed with matrix gel and inoculate into BALB/C nude mice, respectively. After 25 days, 6 organoid transplanted tumor mice were treated with oxaliplatin (Sellekchem, s1224) twice a week for 4 weeks at a dose of 5 mg/kg. | |||
| Response Summary | m6A methyltransferase METTL3 facilitates oxaliplatin resistance in CD133+ gastric cancer stem cells by Promoting Poly [ADP-ribose] polymerase 1 (PARP1) mRNA stability which increases base excision repair pathway activity. METTTL3 enhances the stability of PARP1 by recruiting YTHDF1 to target the 3'-untranslated Region (3'-UTR) of PARP1 mRNA. | |||
PR domain zinc finger protein 2 (PRDM2)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Pancreatic islets | Mus musculus |
|
Treatment: Mettl3 knockout mice
Control: Mettl3 flox/flox mice
|
GSE155612 | |
| Regulation |
 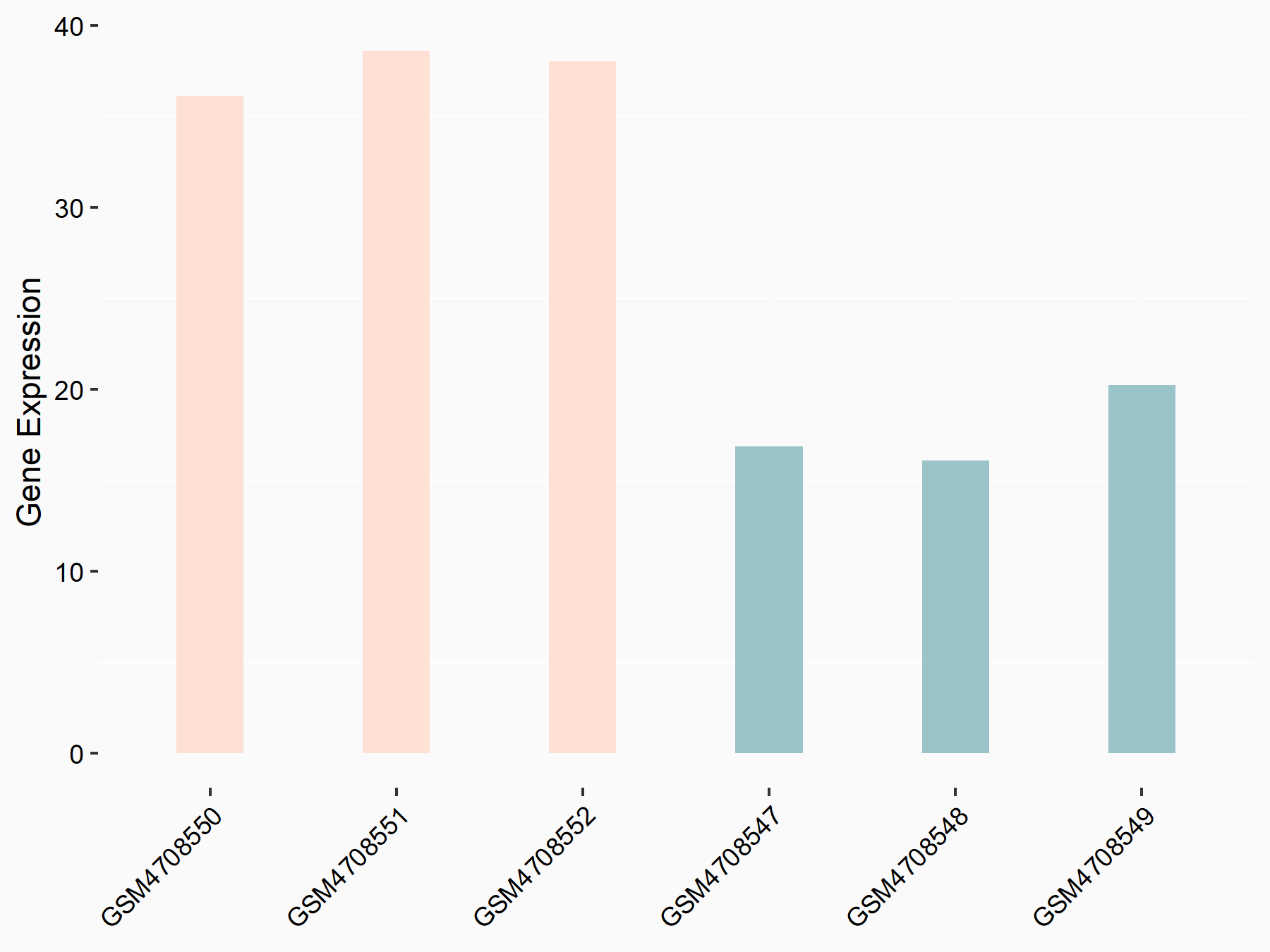 |
logFC: 1.05E+00 p-value: 2.68E-04 |
| More Results | Click to View More RNA-seq Results | |
Arsenite
[Phase 2]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [27] | |||
| Responsed Disease | Solid tumour/cancer | ICD-11: 2A00-2F9Z | ||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
| Response Summary | METTL3 significantly decreased m6A level, restoring p53 activation and inhibiting cellular transformation phenotypes in the arsenite-transformed cells. m6A downregulated the expression of the positive p53 regulator, PR domain zinc finger protein 2 (PRDM2), through the YTHDF2-promoted decay of PRDM2 mRNAs. m6A upregulated the expression of the negative p53 regulator, YY1 and MDM2 through YTHDF1-stimulated translation of YY1 and MDM2 mRNA. This study further sheds light on the mechanisms of arsenic carcinogenesis via RNA epigenetics. | |||
Protein AATF (AATF/CHE1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3 knockout liver
Control: Wild type liver cells
|
GSE198513 | |
| Regulation |
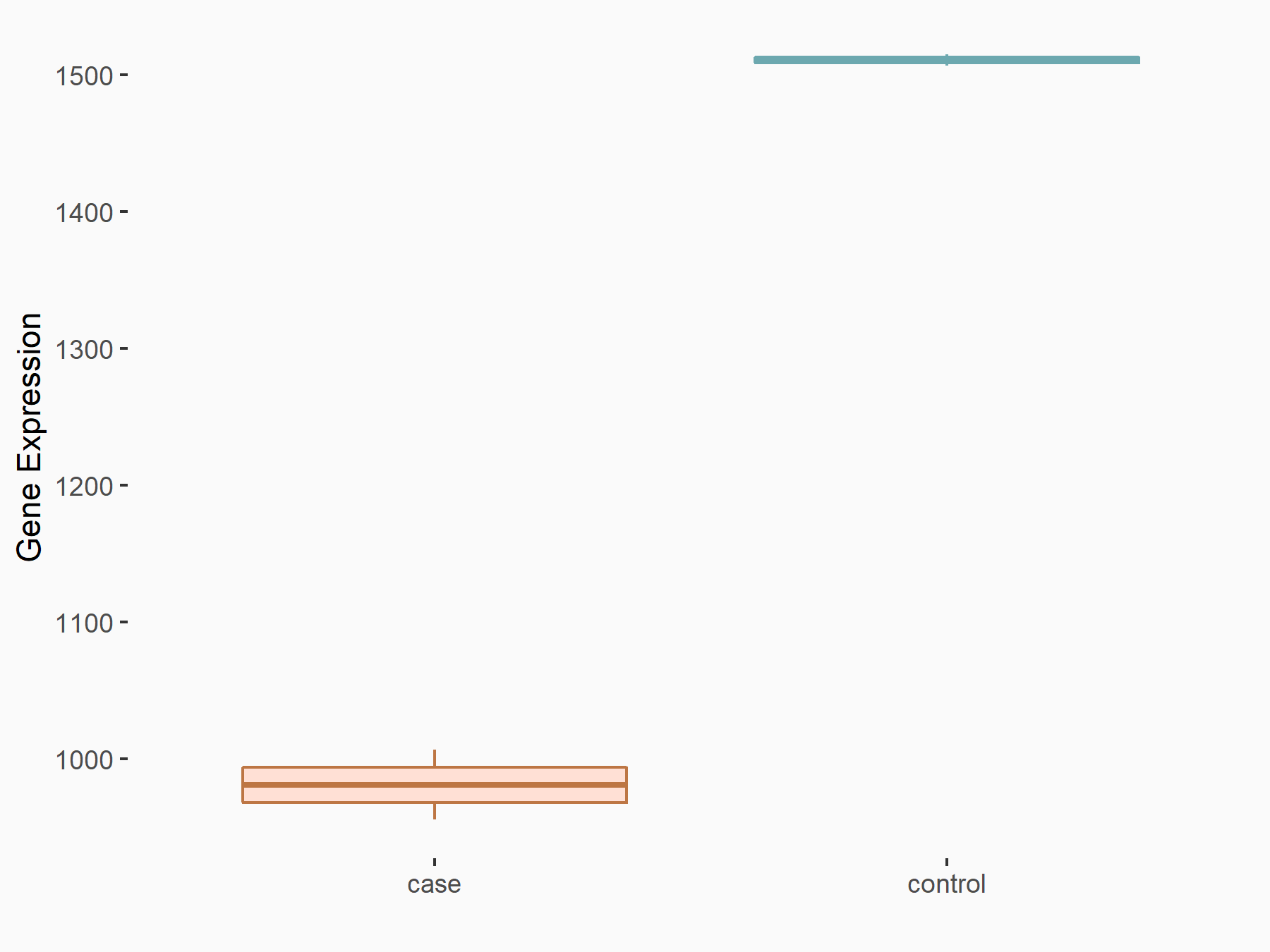 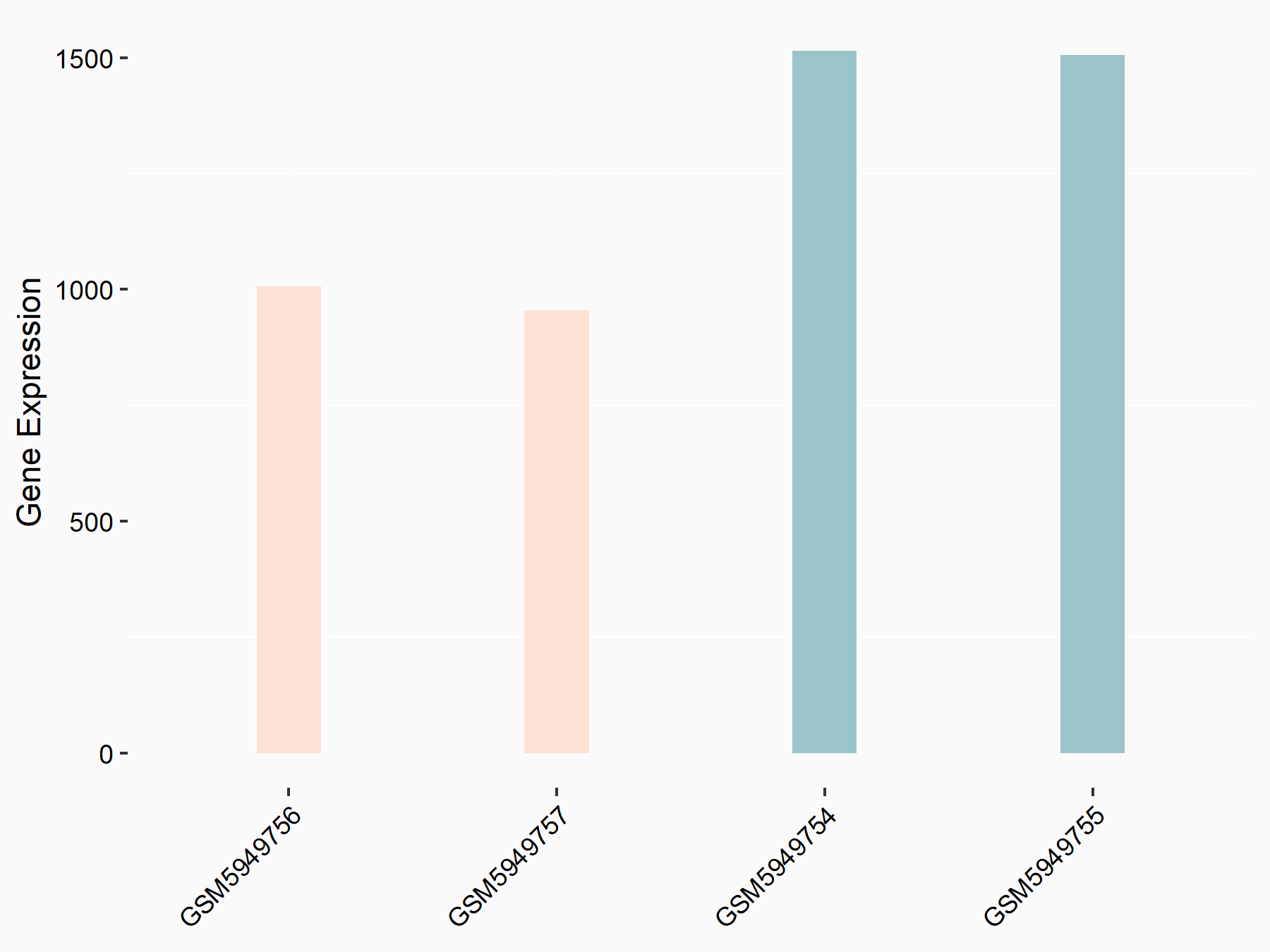 |
logFC: -6.23E-01 p-value: 2.02E-10 |
| More Results | Click to View More RNA-seq Results | |
Doxil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [22] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell growth and death | |||
| Cell apoptosis | ||||
| In-vitro Model | ADR-resistant MCF-7 (MCF-7/ADR) cells (Human breast cancer doxorubicin-resistant cell line) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| In-vivo Model | Cell suspensions (2 × 106 cells/mL) made with MCF-7/ADR cells stably expressing METTL3 and/or miR-221-3p inhibitor were subcutaneously implanted into each mouse. One week later, xenografted mice were injected with 0.1 mL ADR (25 mg/kg, intraperitoneal injection) twice a week. | |||
| Response Summary | METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. METTL3 knockdown was shown to reduce the expression of miR-221-3p by reducing pri-miR-221-3p m6A mRNA methylation, reducing the expression of MDR1 and BCRP, and inducing apoptosis. Identified the METTL3/miR-221-3p/HIPK2/Protein AATF (AATF/CHE1) axis as a novel signaling event that will be responsible for resistance of BC cells to ADR. | |||
Protein numb homolog (NUMB)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
|
Treatment: METTL3 knockdown MDA-MB-231 cells
Control: MDA-MB-231 cells
|
GSE70061 | |
| Regulation |
 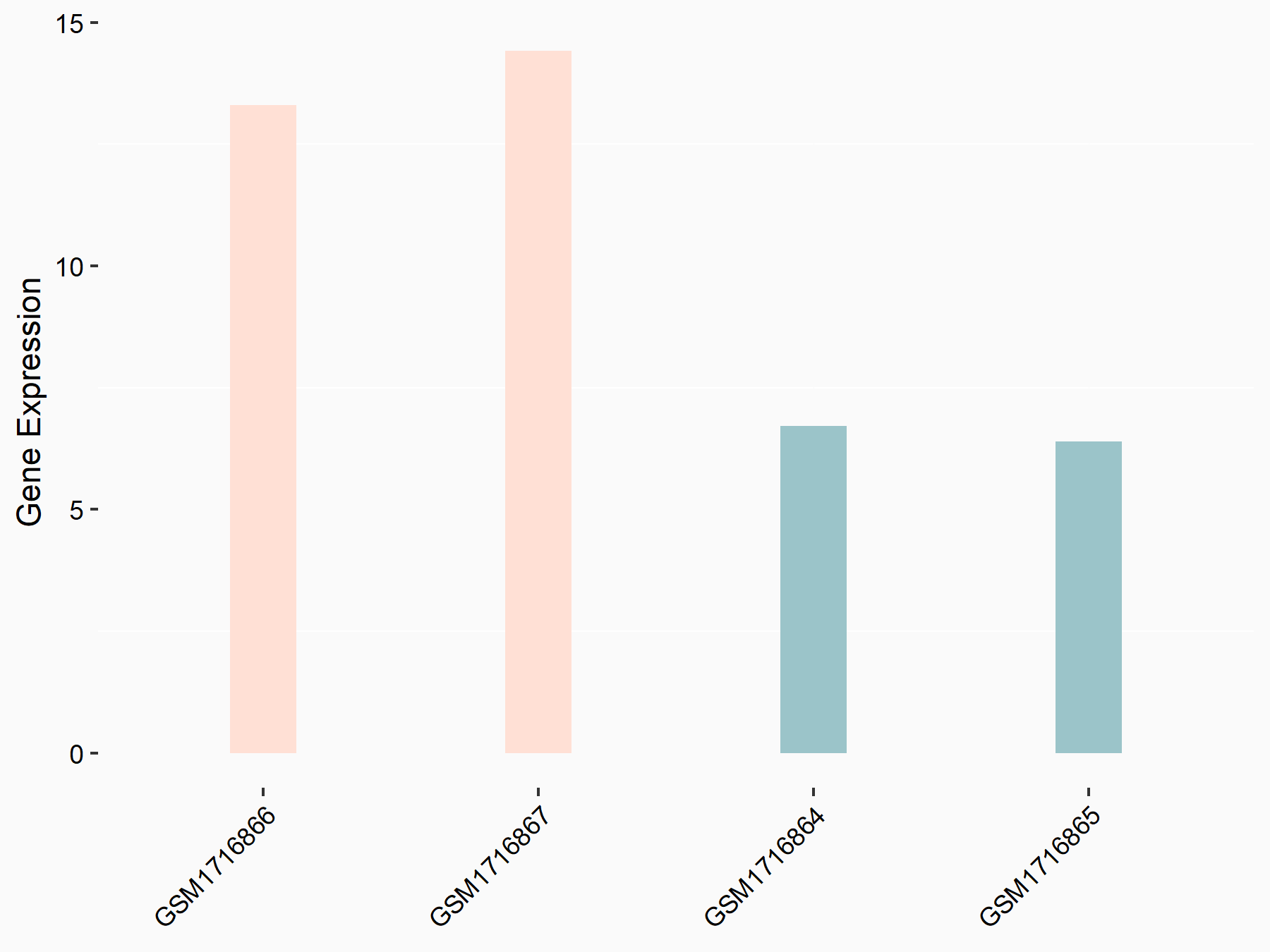 |
logFC: 9.75E-01 p-value: 1.24E-03 |
| More Results | Click to View More RNA-seq Results | |
Melittin
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [201] | |||
| Responsed Disease | Bladder cancer | ICD-11: 2C94 | ||
| Target Regulation | Down regulation | |||
| Cell Process | miRNA maturation | |||
| Cell apoptosis | ||||
| In-vitro Model | T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| EJ (Human bladder cancer cells) | ||||
| BIU-87 | Human bladder cancer cells | Homo sapiens | CVCL_6881 | |
| In-vivo Model | For melittin treatment study, 4-week-old female BALB/c nude mice were subcutaneously injected with 1 × 107 T24 or BIU87 cells. | |||
| Response Summary | METTL3 acts as a fate determinant that controls the sensitivity of bladder cancer cells to melittin treatment. Moreover, METTL3/miR-146a-5p/Protein numb homolog (NUMB)/NOTCH2 axis plays an oncogenic role in bladder cancer pathogenesis and could be a potential therapeutic target for recurrent bladder cancer treatment. | |||
RAC-alpha serine/threonine-protein kinase (AKT1)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3 knockout liver
Control: Wild type liver cells
|
GSE198512 | |
| Regulation |
 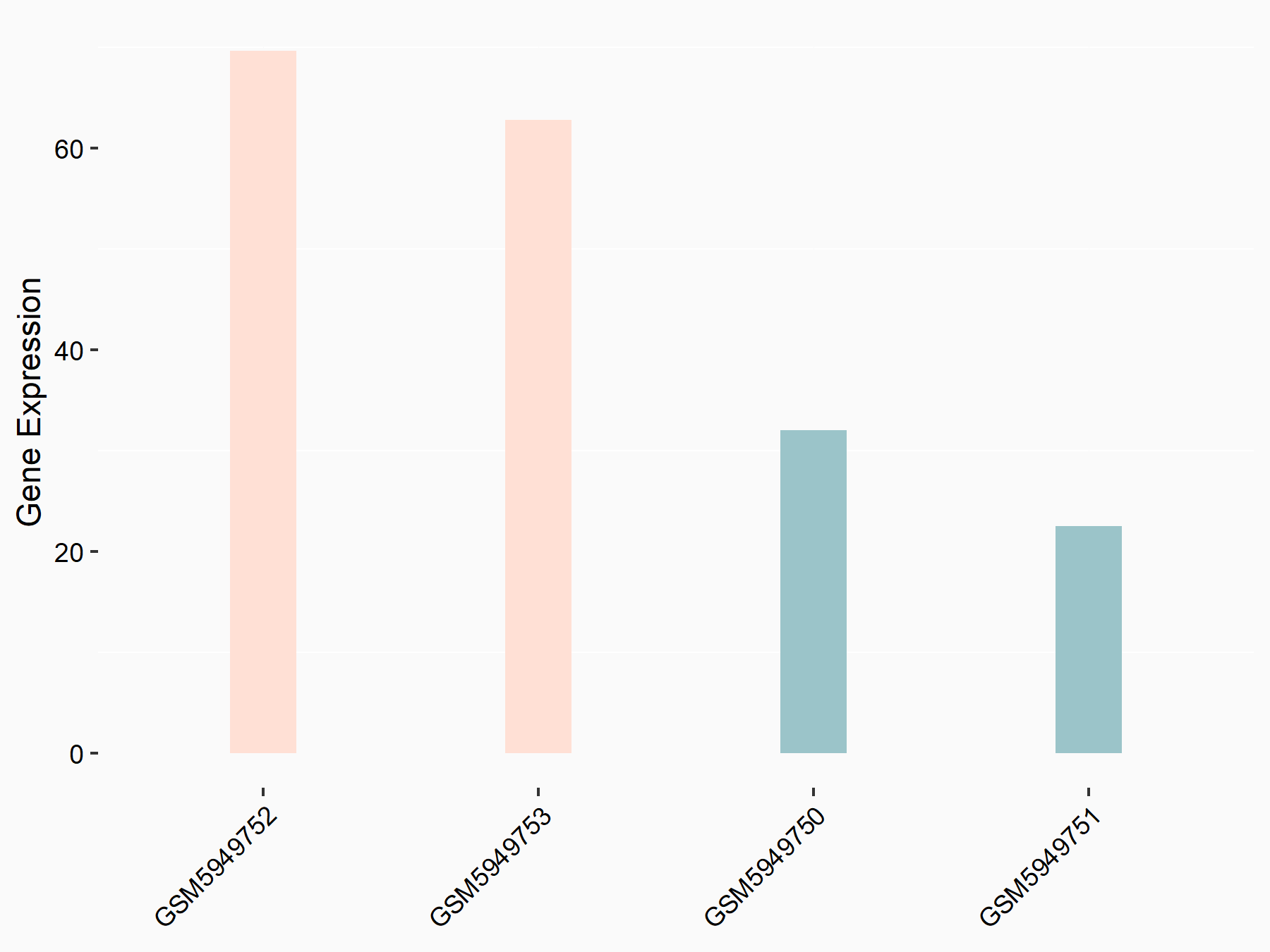 |
logFC: 1.27E+00 p-value: 1.77E-02 |
| More Results | Click to View More RNA-seq Results | |
Rapamycin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [44] | |||
| Responsed Disease | Retinoblastoma | ICD-11: 2D02.2 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| In-vitro Model | WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through PI3K/RAC-alpha serine/threonine-protein kinase (AKT1)/mTOR pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-P70S6K/4EBP1 pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
Rho GTPase activating protein 5 (ARHGAP5)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Pancreatic islets | Mus musculus |
|
Treatment: Mettl3 knockout mice
Control: Mettl3 flox/flox mice
|
GSE155612 | |
| Regulation |
 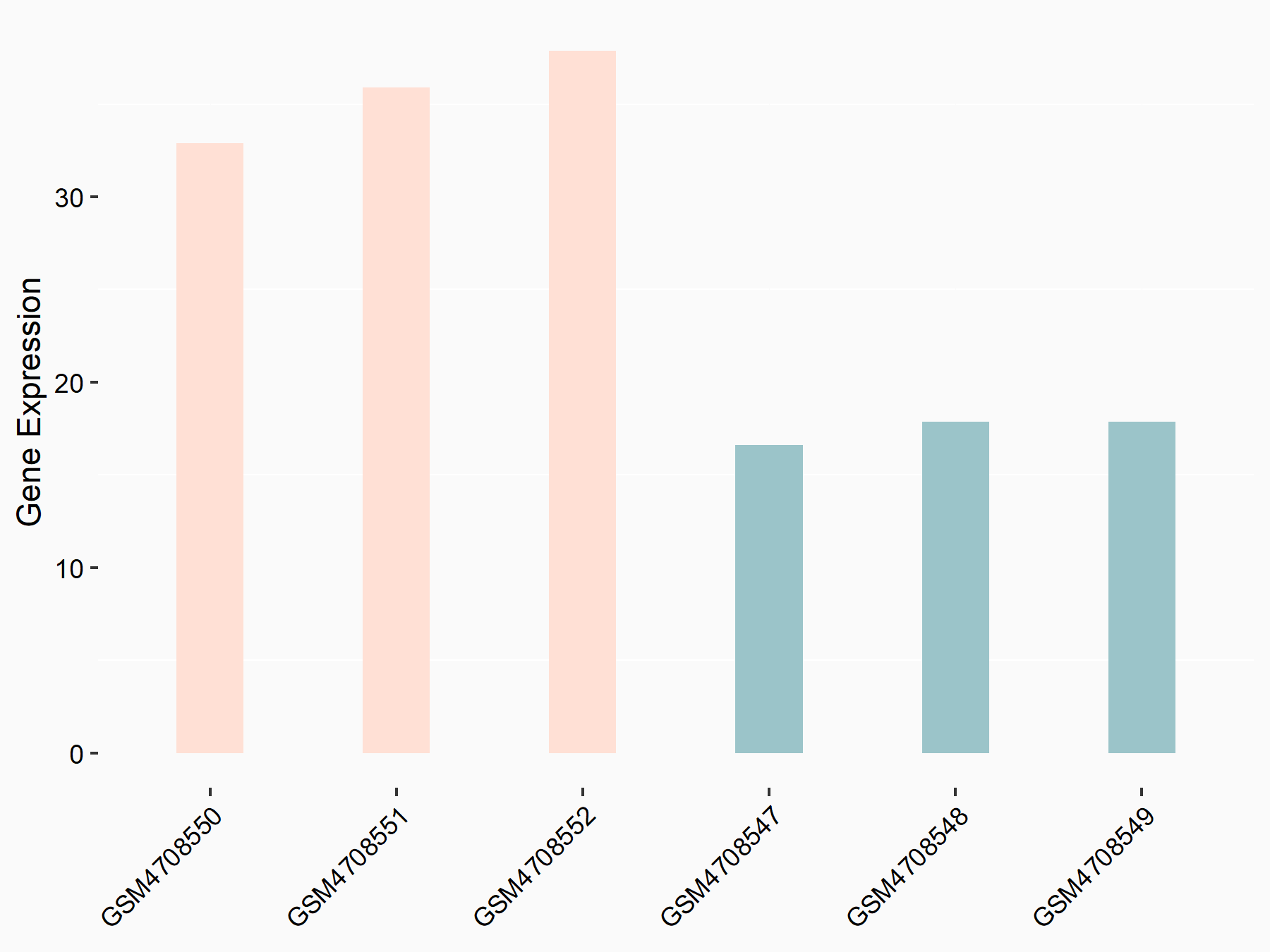 |
logFC: 9.85E-01 p-value: 6.23E-05 |
| More Results | Click to View More RNA-seq Results | |
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [215] | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular Processes | |||
| Transport and catabolism | ||||
| In-vitro Model | BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| Response Summary | ARHGAP5-AS1 also stabilized ARHGAP5 mRNA in the cytoplasm by recruiting METTL3 to stimulate m6A modification of Rho GTPase activating protein 5 (ARHGAP5) mRNA. As a result, ARHGAP5 was upregulated to promote chemoresistance and its upregulation was also associated with poor prognosis in gastric cancer. downregulation of ARHGAP5-AS1 in resistant cells evidently reversed the resistance to chemotherapeutic drugs including cisplatin (DDP), ADM, and 5-FU. | |||
Fluorouracil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [215] | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular processes | |||
| Cellular transport | ||||
| Cellular catabolism | ||||
| In-vitro Model | BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| Response Summary | ARHGAP5-AS1 also stabilized ARHGAP5 mRNA in the cytoplasm by recruiting METTL3 to stimulate m6A modification of Rho GTPase activating protein 5 (ARHGAP5) mRNA. As a result, ARHGAP5 was upregulated to promote chemoresistance and its upregulation was also associated with poor prognosis in gastric cancer. downregulation of ARHGAP5-AS1 in resistant cells evidently reversed the resistance to chemotherapeutic drugs including cisplatin (DDP), ADM, and 5-FU. | |||
Adriamycin
[Phase 3]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [215] | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular processes | |||
| Cellular transport | ||||
| Cellular catabolism | ||||
| In-vitro Model | BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| Response Summary | ARHGAP5-AS1 also stabilized ARHGAP5 mRNA in the cytoplasm by recruiting METTL3 to stimulate m6A modification of Rho GTPase activating protein 5 (ARHGAP5) mRNA. As a result, ARHGAP5 was upregulated to promote chemoresistance and its upregulation was also associated with poor prognosis in gastric cancer. downregulation of ARHGAP5-AS1 in resistant cells evidently reversed the resistance to chemotherapeutic drugs including cisplatin (DDP), ADM, and 5-FU. | |||
Ubiquitin-like-conjugating enzyme ATG3 (ATG3)
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3-deficient liver
Control: Wild type liver cells
|
GSE197800 | |
| Regulation |
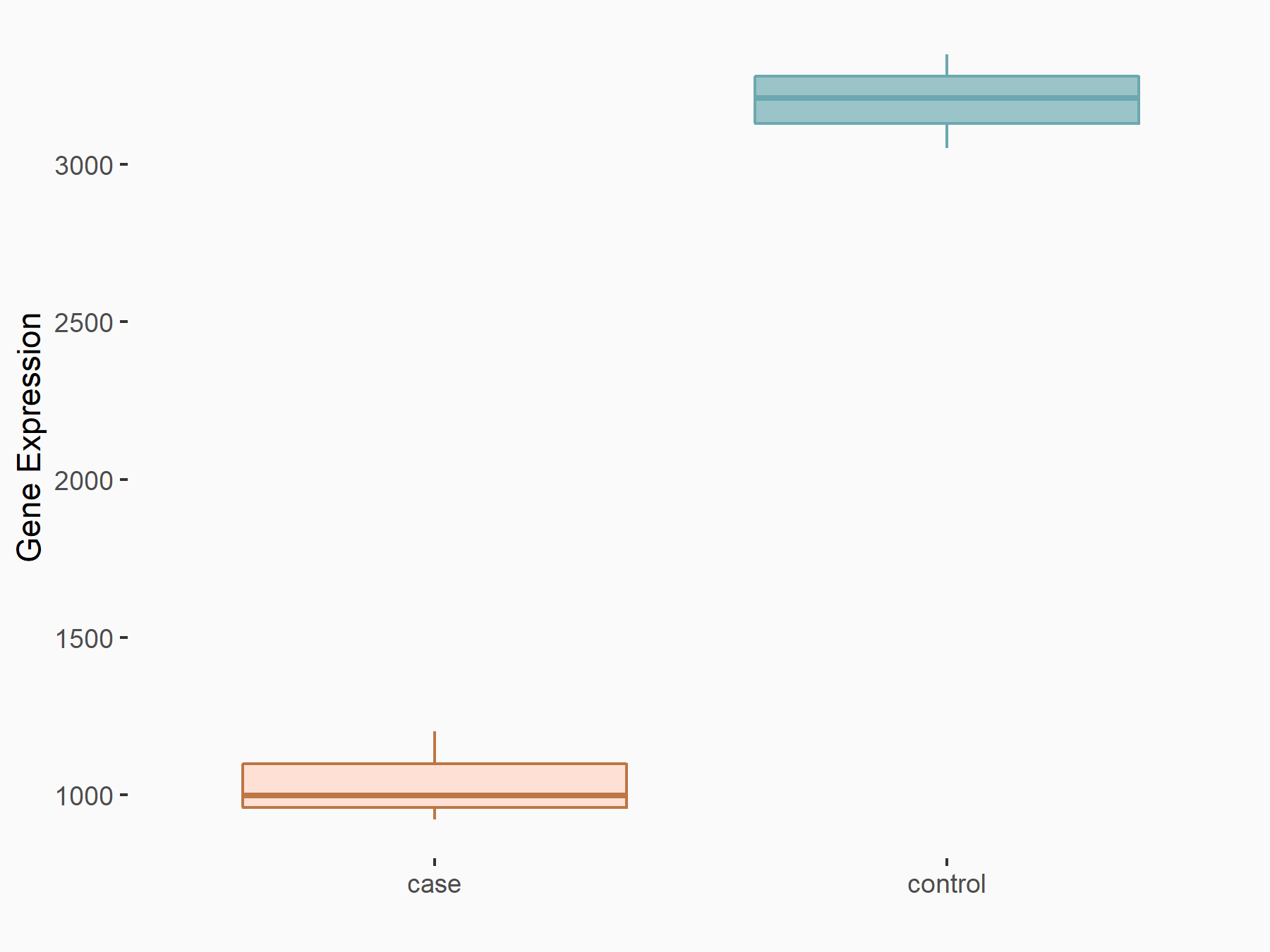 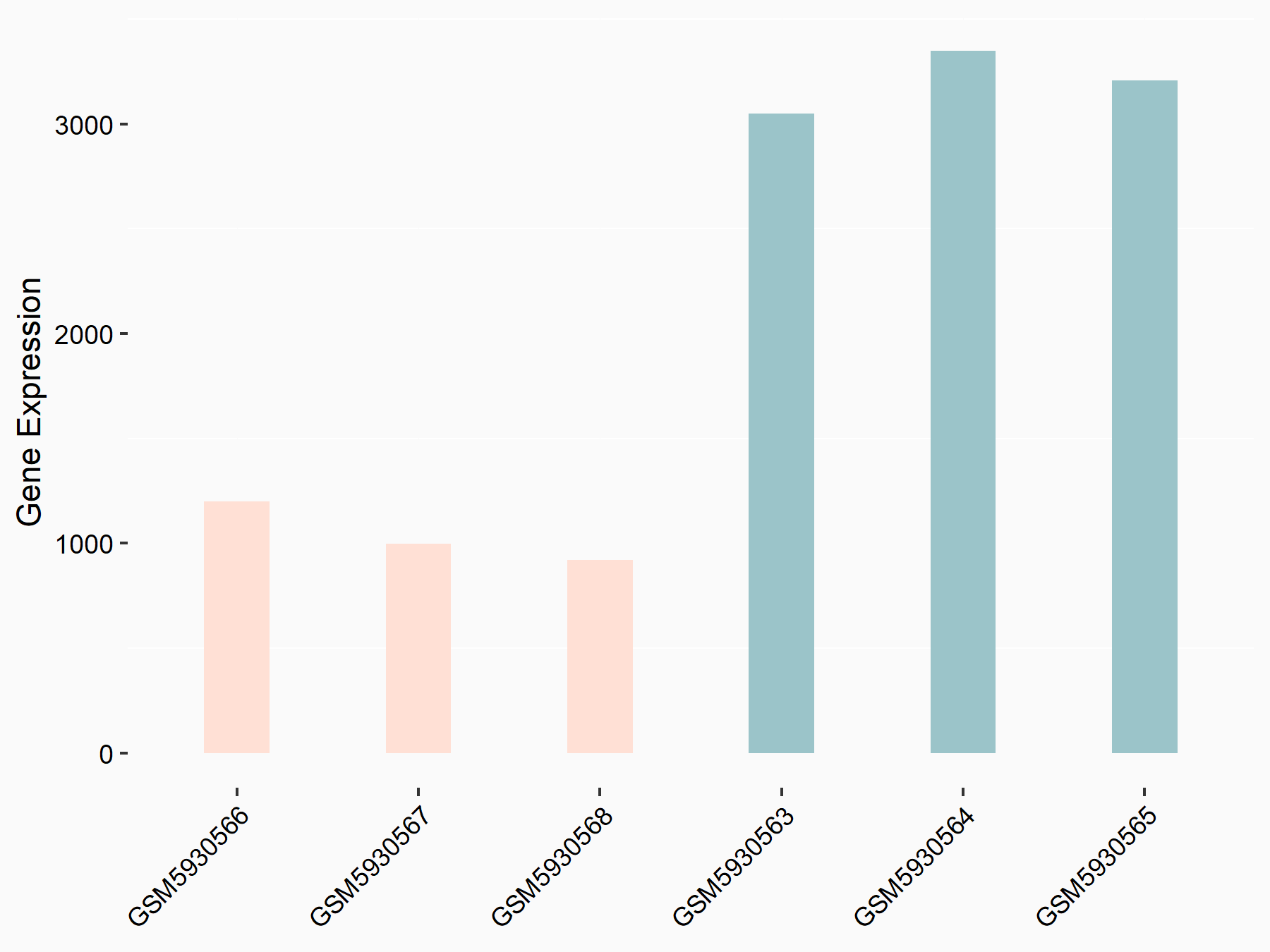 |
logFC: -1.62E+00 p-value: 2.55E-18 |
| More Results | Click to View More RNA-seq Results | |
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [17] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including Ubiquitin-like-conjugating enzyme ATG3 (ATG3), ATG5, ATG7, ATG12, and ATG16L1. | |||
Multidrug resistance-associated protein 1 (MRP1/ABCC1)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.47E+00 | GSE60213 |
Imatinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [252] | |||
| Responsed Disease | Gastrointestinal stromal tumour | ICD-11: 2B5B | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| In-vivo Model | For tumor growth assay, 4 × 106 logarithmically growing GIST cells were transfected with T1S-vector, T1S-METTL3, 882S-vector, or 882S-METTL3 constructs, and subcutaneously injected in 100 ul of PBS into the flank of female nude mice(4-week-old). Mice were then randomly divided into 8 groups (n = 5 in each group): (1) injected with T1S-vector-harboring cells, and treated with imatinib (600 mg/l in drinking water); (2) injected with T1S-vector-harboring cells, and treated with imatinib (600 mg/l in drinking water) and MRP1 inhibitor (100 mg/l in drinking water); (3) injected with T1S-METTL3-harboring cells, and treated with imatinib (600 mg/l in drinking water); (4) injected with T1S-METTL3-harboring cells, and treated with imatinib (600 mg/l in drinking water) and MRP1 inhibitor (100 mg/l in drinking water); (5) injected with 882S-vector-harboring cells and treated with imatinib (600 mg/l in drinking water); (6) injected with 882S-vector-harboring cells, and treated with imatinib (600 mg/l in drinking water) and MRP1 inhibitor (100 mg/l in drinking water); (7) injected with 882S-METTL3-harboring cells, and treated with imatinib (600 mg/l in drinking water); and (8) injected with 882S-METTL3-harboring cells, and treated with imatinib (600 mg/l in drinking water) and MRP1 inhibitor (100 mg/l in drinking water). | |||
| Response Summary | METTL3, and the YTHDF1/eEF-1 complex mediate the translation of Multidrug resistance-associated protein 1 (MRP1/ABCC1) mRNA in an m6A-dependent manner to regulate the intracellular concentration of imatinib and drug resistance of gastrointestinal stromal tumor (GIST). | |||
Long intergenic non-protein coding RNA 1273 (LINC01273)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.26E+00 | GSE60213 |
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [256] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Response Summary | Long intergenic non-protein coding RNA 1273 (LINC01273) was modified with m6A, METTL3 increased LINC01273 m6A modification, followed by LINC01273 decay in the presence of YTHDF2, a m6A 'reader'. And LINC01273 plays a key role in sorafenib resistant HCC cells. | |||
NIFK antisense RNA 1 (NIFK-AS1)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.43E+00 | GSE60213 |
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [258] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| THLE-3 | Normal | Homo sapiens | CVCL_3804 | |
| In-vivo Model | For the PDX model, fresh patient HCC tissues were cut into fragments with a volume of 3 × 3 mm3 and then implanted subcutaneously into the flanks of nude mice. The mice were given sorafenib (30 mg/kg) or vehicle orally twice a week for 24 days. This procedure was approved by the Ethics Committee of Jinling Hospital. | |||
| Response Summary | Identified the lncRNA NIFK antisense RNA 1 (NIFK-AS1) as being highly expressed in hepatocellular carcinoma tissues and cells, promotes disease progression and sorafenib resistance, and showed this up-regulation resulted from METTL3-dependent m6A methylation. | |||
Small nucleolar RNA host gene 3 (SNHG3)
| Representative RIP-seq result supporting the interaction between the target gene and METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.17E+00 | GSE60213 |
Pt
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [259] | |||
| Responsed Disease | Esophageal cancer | ICD-11: 2B70 | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cellular Processes | |||
| Cell growth and death | ||||
| Cell apoptosis | ||||
| In-vitro Model | Eca-9706 (Esophageal carcinoma cell line) | |||
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| In-vivo Model | Used 1 × 106 SNHG3 knocked down KY-SE150 cells and NC lentivirus to inject into the right flank of mice to generate xenografts. | |||
| Response Summary | Platinum can increase the overall m6A level of esophageal cancer. Small nucleolar RNA host gene 3 (SNHG3)/miR-186-5p, induced by platinum, was involved in regulating m6A level by targeting METTL3. miR-186-5p binds to the 3'UTR of METTL3 to inhibit its expression. Our manuscript has provided clues that regulating m6A level was a novel way to enhance the platinum efficacy. | |||
Apoptosis regulator Bcl-2 (BCL2)
Celastrol
[Preclinical]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [267] | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| In-vivo Model | AsPC-1 cells suspended in 100 μl of PBS (2 × 106 cells/100 μl) were injected subcutaneously into the lateral flank of the mice, which were randomly divided into solvent group (n = 6), 1.0 mg/kg of celastrol group (n = 6) and 3.0 mg/kg of celastrol group (n = 6). The administration of celastrol was performed by intraperitoneal injection into tumor-bearing mice every 2 day after 10 day inoculation. The tumor sizes were monitored with calipers every 5 days, and the tumor volume was calculated with the formula: Volume (mm3) = 1/2 × length × width2. | |||
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [268] | |||
| Responsed Disease | Spinal cord injury | ICD-11: ND51.2 | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | PC12 | Rat adrenal gland pheochromocytoma | Rattus norvegicus | CVCL_0481 |
| In-vivo Model | Rats were anesthetized with intraperitoneal injection of 1% sodium pentobarbital (20 mg/kg). For constructing a rat spinal cord hemisection model, the spinal colon was marked on the T9 spinous process and the skin was incised until exposing the T9-10 spinous process. The spinal cord was completely exposed by biting the vertebral plate with biting forceps. The spinal cord was cut on one side with ophthalmic scissors centered on the central canal of the spinal cord. | |||
E3 ubiquitin-protein ligase TRIM11 (TRIM11)
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [14] | |||
| Responsed Disease | Nasopharyngeal carcinoma | ICD-11: 2B6B | ||
| Target Regulation | Down regulation | |||
| Pathway Response | ABC transporters | hsa02010 | ||
| Wnt signaling pathway | hsa04310 | |||
| Ubiquitin mediated proteolysis | hsa04120 | |||
| Cell Process | Ubiquitination degradation | |||
| In-vitro Model | CNE-1 | Normal | Homo sapiens | CVCL_6888 |
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| In-vivo Model | A total of 2 × 106 cells was mixed with 0.2 ml PBS (pH 7.4) and 30% (v/v) Matrigel matrix (BD Biosciences). | |||
| Response Summary | TRIM11 regulates nasopharyngeal carcinoma drug resistance by positively modulating the Daple/beta-catenin/E3 ubiquitin-protein ligase TRIM11 (TRIM11) signaling pathway. TRIM11 enhanced the multidrug resistance in NPC by inhibiting apoptosis in vitro and promoting cisplatin (DDP) resistance in vivo. METTL3-mediated m6A modification caused the upregulation of TRIM11 via IGF2BP2 in NPC drug-resistant cells. | |||
Histone-lysine N-methyltransferase EZH2 (EZH2)
Temozolomide
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [298] | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | mRNA decay | |||
| In-vitro Model | U251 (Fibroblasts or fibroblast like cells) | |||
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| In-vivo Model | Used to inject 10 uL of the U87 MG-TMZ_R-luc cell suspension in the striatum at a depth of 3 mm from the dural surface. One week after the injection of the tumor cells, 40 mg/kg/day of TMZ in saline was administered for over 2 weeks by intraperitoneal injection. | |||
| Response Summary | Uncover the fundamental mechanisms underlying the interplay of m6 A RNA modification and histone modification in Temozolomide resistance and emphasize the therapeutic potential of targeting the SOX4/EZH2/METTL3 axis in the treatment of TMZ-resistant glioblastoma. | |||
Lactate dehydrogenase A (LDHA)
MICROCYSTIN-LR
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [306] | |||
| Responsed Disease | Male infertility | ICD-11: GB04 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model |
TM4
|
N.A. | Mus musculus | CVCL_4327 |
Mothers against decapentaplegic homolog 3 (SMAD3)
Lenvatinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [309] | |||
| Responsed Disease | Liver hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| In-vitro Model | Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| In-vivo Model | When the tumors in the CDX and PDX mice grew to about 80-150 mm3, forty tumor-bearing mice were randomly divided into four groups (ten mice in each group) according to the size of the transplanted tumor and the principle of consistency between groups. The four groups were: as follows: solvent control group, Len (3 mg kg-1) administration group, Res (0.375 mg kg-1) administration group, and Len (3 mg kg-1) plus Res (0.375 mg kg-1) administration group. The administration mode of all groups was intragastric and the volume was 0.2 ml/20 g. Mouse body weight and xenograft tumor volume were measured every other day. Tumor volume was measured weekly and calculated as V = (length × width2)/2. Three weeks after the onset of treatment, the tumors were excised and weighed. After the experiment, the heart, liver, spleen, lung and kidney tissues of the mice were completely removed and weighed. Then the organ index was calculated as organ index = weight of each organ/body weight × 100 %. | |||
Reserpine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [309] | |||
| Responsed Disease | Liver hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| In-vitro Model | Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| In-vivo Model | When the tumors in the CDX and PDX mice grew to about 80-150 mm3, forty tumor-bearing mice were randomly divided into four groups (ten mice in each group) according to the size of the transplanted tumor and the principle of consistency between groups. The four groups were: as follows: solvent control group, Len (3 mg kg-1) administration group, Res (0.375 mg kg-1) administration group, and Len (3 mg kg-1) plus Res (0.375 mg kg-1) administration group. The administration mode of all groups was intragastric and the volume was 0.2 ml/20 g. Mouse body weight and xenograft tumor volume were measured every other day. Tumor volume was measured weekly and calculated as V = (length × width2)/2. Three weeks after the onset of treatment, the tumors were excised and weighed. After the experiment, the heart, liver, spleen, lung and kidney tissues of the mice were completely removed and weighed. Then the organ index was calculated as organ index = weight of each organ/body weight × 100 %. | |||
NACHT, LRR and PYD domains-containing protein 3 (NLRP3)
Gefitinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [316] | |||
| Responsed Disease | Lung cancer | ICD-11: 2C25 | ||
| In-vitro Model | PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| PC-9/GR2 | Lung adenocarcinoma | Homo sapiens | CVCL_DI29 | |
|
HCC827/GR
|
N.A. | Homo sapiens | CVCL_E7R9 | |
| In-vivo Model | To establish xenograft models, five-week-old male mice were orthotopically injected with the same number of the PC9/GR cells. Tumours had developed after 4 days, at which time the xenografted mice were randomly divided into the following two experimental groups (each group with 6 mice): (1) the control group and (2) the gefitinib group. | |||
RAD51-associated protein 1 (RAD51AP1)
Fluorouracil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [327] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 |
| Response Summary | METTL3 augmented 5 FU induced DNA damage and overcame 5 FU resistance in HCT 8R cells, which could be mimicked by inhibition of RAD51-associated protein 1 (RAD51AP1). The present study revealed that the METTL3/RAD51AP1 axis plays an important role in the acquisition of 5 FU resistance in CRC. | |||
Serum response factor (SRF)
Tislelizumab
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [333] | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| In-vitro Model | U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 |
Staphylococcal nuclease domain-containing protein 1 (SND1)
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [336] | |||
| Responsed Disease | Nasal-type natural killer/T-cell lymphoma | ICD-11: XH3400 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | NK-92 | Natural killer cell lymphoblastic leukemia/lymphoma | Homo sapiens | CVCL_2142 |
| YTS | Lymphoblastic leukemia/lymphoma | Homo sapiens | CVCL_D324 | |
| SNT-8 | Nasal type extranodal NK/T-cell lymphoma | Homo sapiens | CVCL_A677 | |
| SNK-6 | Nasal type extranodal NK/T-cell lymphoma | Homo sapiens | CVCL_A673 | |
| In-vivo Model | A total of 32 nude mice were randomly divided into four groups with eight mice in each group. Nude mice were subcutaneously injected with 2 × 106 cells (suspended in 100 μl of PBS) transfected with NC shRNA or SND1 shRNA into the back flank of mice. In DDP administration group, DDP (3 mg/kg per week for 2 weeks) was intraperitoneally injected into mice every 3 days. The control group received 200 μl of 0.1% DMSO. The tumor volume was measured every week for a total of 4 weeks. The mice were sacrificed after 4 weeks, then the tumors were harvested for weight measurement and other analysis. The tumor volume (mm3) was estimated with the formula (0.5 × length × width2). | |||
Superoxide dismutase [Mn], mitochondrial (SOD2)
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [339] | |||
| Target Regulation | Up regulation | |||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
Suppressor of cytokine signaling 3 (SOCS3)
Colivelin
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [341] | |||
| Responsed Disease | Liver cancer | ICD-11: 2C12 | ||
Cycloleucine
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [344] | |||
| Responsed Disease | Chronic obstructive pulmonary disease | ICD-11: CA22 | ||
| Target Regulation | Down regulation | |||
Arsenite
[Phase 2]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [345] | |||
| Responsed Disease | Human skin lesions | ICD-11: ME60 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
Thioredoxin (TXN/SASP)
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [339] | |||
| Target Regulation | Up regulation | |||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
Transcription factor SOX-2 (SOX2)
LPS
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [350] | |||
| Responsed Disease | Cognitive disorders | ICD-11: 6E0Z | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
Transferrin receptor protein 1 (TFRC)
Doxil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [353] | |||
| In-vitro Model | NRCMs (Primary neonatal rat cardiomyocytes (NRCMs)NRCMs were prepared from the hearts of 2- to 3-day-old SD rats according to the following protocol) | |||
| HL-1 | Normal | Mus musculus | CVCL_0303 | |
| In-vivo Model | Eight-week-old male and female mice were given DOX (5 mg/kg, Cat No. D1515, Sigma-Aldrich) or saline once weekly for 4 weeks via intraperitoneal injection [29]. For experiments with STM2457 (Cat No. SML3360, Sigma-Aldrich), WT mice received STM2457 (50 mg/kg dissolved in 10% DMSO and 90% saline, i. p.) once daily . Cardiac function was evaluated using echocardiography 1 week following the final DOX. Then, heart tissues were collected after echocardiography assessment for biochemical assays. | |||
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [353] | |||
| In-vitro Model | NRCMs (Primary neonatal rat cardiomyocytes (NRCMs)NRCMs were prepared from the hearts of 2- to 3-day-old SD rats according to the following protocol) | |||
| HL-1 | Normal | Mus musculus | CVCL_0303 | |
| In-vivo Model | Eight-week-old male and female mice were given DOX (5 mg/kg, Cat No. D1515, Sigma-Aldrich) or saline once weekly for 4 weeks via intraperitoneal injection [29]. For experiments with STM2457 (Cat No. SML3360, Sigma-Aldrich), WT mice received STM2457 (50 mg/kg dissolved in 10% DMSO and 90% saline, i. p.) once daily . Cardiac function was evaluated using echocardiography 1 week following the final DOX. Then, heart tissues were collected after echocardiography assessment for biochemical assays. | |||
Ubiquitin carboxyl-terminal hydrolase 4 (USP4)
Metformin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [356] | |||
| Responsed Disease | Multiple myeloma | ICD-11: 2A83.1 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | NCI-H929 | Plasma cell myeloma | Homo sapiens | CVCL_1600 |
| MM1.S | Plasma cell myeloma | Homo sapiens | CVCL_8792 | |
| In-vivo Model | 5-week-old male athymic BALB/C nude mice were obtained from Experimental animal center of Fujian Medical University, and subsequently randomly divided into four groups (5 mice per group), including control, metformin, pcDNA-METTL3 and pcDNA-METTL3 + metformin. For the control or metformin group, to induce tumors, 5 × 105 H929 cells were suspended in 0.2 ml phosphate buffered saline and inoculated subcutaneously into the peritoneum of mice. Once the subcutaneous nodules grown to a rice grain size (required approximately a week), the subcutaneous xenograft model of MM in nude mice was successfully constructed. One week later, PBS or Metformin was diluted in drinking water (2 mg/ml) and administered orally throughout the indicated time periods. For the pcDNA-METTL3 group and pcDNA-METTL3 + metformin group, firstly, we transfected pcDNA-METTL3 into H929 cells to overexpress METTL3, and then 5 × 105 H929 cells were suspended in 0.2 ml phosphate-buffered saline and inoculated subcutaneously into the peritoneum of mice. One week later, PBS or Metformin was diluted in drinking water (2 mg/ml) and administered orally throughout the indicated time periods. All nude mice were closely monitored for tumor growth, skin condition, and behavior daily and any tumor ulceration or irritation was noted. Tumor length and width were calculated with vernier calipers every 7 days. After 35 days, the mice were humanely sacrificed, and the subcutaneous tumors were excised and removed. | |||
DBH antisense RNA 1 (Lnc_DBH-AS1)
Gemcitabine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [359] | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| In-vitro Model | MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 |
| HPDE | Normal | Homo sapiens | CVCL_4376 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| Canpan-2 (Pancreatic cancer cell line) | ||||
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| Response Summary | DBH antisense RNA 1 (Lnc_DBH-AS1) expression in pancreatic cancer(PC) was found to be linked to the METTL3-dependent m6A methylation of the lncRNA. MechanisticallyDBH-AS1 was able to increase PC cell sensitivity to gemcitabine by sequestering miR-3163 and thus upregulating USP44 in these tumor cells. | |||
KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1)
Doxil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [362] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | MCF-10A | Normal | Homo sapiens | CVCL_0598 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
microRNA let-7b (MIRLET7B)
Osimertinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [384] | |||
| Responsed Disease | Lung cancer | ICD-11: 2C25 | ||
| Pathway Response | Notch signaling pathway | hsa04330 | ||
| In-vitro Model | NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| H1975OR (Osimertinib resistant H1975 cells) | ||||
| HCC827OR (Osimertinib resistant HCC827 cells) | ||||
| Response Summary | The participation of Metformin decreased the bindings of DNMT3a/b to the METTL3 promoter with the help of the readers of NKAP and HNRNPA2B1.the mediation of m6A formation on pri-Let-7b processing increased the mature microRNA let-7b (MIRLET7B), whose key role is to suppress the Notch signaling and to re-captivate the Osimertinib treatment.The findings open up future drug development, targeting this pathway for lung cancer patients. | |||
hsa-miR-146a-5p
Melittin
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [201] | |||
| Responsed Disease | Bladder cancer | ICD-11: 2C94 | ||
| Target Regulation | Up regulation | |||
| Cell Process | miRNA maturation | |||
| Cell apoptosis | ||||
| In-vitro Model | T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| EJ (Human bladder cancer cells) | ||||
| BIU-87 | Human bladder cancer cells | Homo sapiens | CVCL_6881 | |
| In-vivo Model | For melittin treatment study, 4-week-old female BALB/c nude mice were subcutaneously injected with 1 × 107 T24 or BIU87 cells. | |||
| Response Summary | METTL3 acts as a fate determinant that controls the sensitivity of bladder cancer cells to melittin treatment. Moreover, METTL3/hsa-miR-146a-5p/NUMB/NOTCH2 axis plays an oncogenic role in bladder cancer pathogenesis and could be a potential therapeutic target for recurrent bladder cancer treatment. | |||
hsa-miR-181d-5p
Fluorouracil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [88] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | HT29 | Colon cancer | Mus musculus | CVCL_A8EZ |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| In-vivo Model | A tumor-bearing model was established by subcutaneously injecting 100 ul HT29 cells (5×106) followed by an intravenous injection of CAFs-derived exosomes (50 ug/mouse every three days) into the tail vein of the mice. An intraperitoneal injection of 5-FU (50 mg/kg, every week) was administered on day 12. | |||
| Response Summary | METTL3 dependent m6A methylation was upregulated in CRC to promote the processing of miR 181d 5p by DGCR8. This led to increased hsa-miR-181d-5p expression, which inhibited the 5 FU sensitivity of CRC cells by targeting NCALD. | |||
hsa-miR-186-5p
Pt
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [259] | |||
| Responsed Disease | Esophageal cancer | ICD-11: 2B70 | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cellular Processes | |||
| Cell growth and death | ||||
| Cell apoptosis | ||||
| In-vitro Model | Eca-9706 (Esophageal carcinoma cell line) | |||
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| In-vivo Model | Used 1 × 106 SNHG3 knocked down KY-SE150 cells and NC lentivirus to inject into the right flank of mice to generate xenografts. | |||
| Response Summary | Platinum can increase the overall m6A level of esophageal cancer. SNHG3/hsa-miR-186-5p, induced by platinum, was involved in regulating m6A level by targeting METTL3. miR-186-5p binds to the 3'UTR of METTL3 to inhibit its expression. Our manuscript has provided clues that regulating m6A level was a novel way to enhance the platinum efficacy. | |||
hsa-miR-1914-3p
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [126] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Metabolic | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-6 | Lung adenocarcinoma | Homo sapiens | CVCL_0236 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | Mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then administrated with PBS or DDP (3 mg/kg per day) for three times a week, and tumor volume were measured every second day. | |||
| Response Summary | METTL3, YTHDF3, YTHDF1, and eIF3b directly promoted YAP translation through an interaction with the translation initiation machinery. METTL3 knockdown inhibits tumor growth and enhances sensitivity to DDP in vivo.m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-hsa-miR-1914-3p-YAP axis to induce Non-small cell lung cancer drug resistance and metastasis. | |||
hsa-miR-221-3p
Doxil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [22] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell growth and death | |||
| Cell apoptosis | ||||
| In-vitro Model | ADR-resistant MCF-7 (MCF-7/ADR) cells (Human breast cancer doxorubicin-resistant cell line) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| In-vivo Model | Cell suspensions (2 × 106 cells/mL) made with MCF-7/ADR cells stably expressing METTL3 and/or miR-221-3p inhibitor were subcutaneously implanted into each mouse. One week later, xenografted mice were injected with 0.1 mL ADR (25 mg/kg, intraperitoneal injection) twice a week. | |||
| Response Summary | METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating hsa-miR-221-3p maturation in a m6A-dependent manner. METTL3 knockdown was shown to reduce the expression of miR-221-3p by reducing pri-miR-221-3p m6A mRNA methylation, reducing the expression of MDR1 and BCRP, and inducing apoptosis. Identified the METTL3/miR-221-3p/HIPK2/Che-1 axis as a novel signaling event that will be responsible for resistance of BC cells to ADR. | |||
hsa_circ_0008399 (Circ_RBM3)
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [402] | |||
| Responsed Disease | Bladder cancer | ICD-11: 2C94 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Protein export | hsa03060 | ||
| Cell Process | Eukaryotic translation | |||
| Cell apoptosis | ||||
| In-vitro Model | 5637 | Bladder carcinoma | Homo sapiens | CVCL_0126 |
| RT-4 | Bladder carcinoma | Homo sapiens | CVCL_0036 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| In-vivo Model | Chose 4-week-old female BALB/c nude mice for tumor xenograft experiments, which randomly were divided into four groups (n = 5 per group). Bladder cancer cells (3 × 106) were subcutaneously injected into the right axilla of the nude mice. | |||
| Response Summary | Circ0008399 bound WTAP to promote formation of the WTAP/METTL3/METTL14 m6A methyltransferase complex, reduce cisplatin sensitivity in bladder cancer, implicating the potential therapeutic value of targeting this axis. | |||
2,4-dienoyl-CoA reductase [ (DECR1)
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [408] | |||
| Responsed Disease | Liver cancer | ICD-11: 2C12 | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| SNU-449 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0454 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
|
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 | |
| In-vivo Model | For subcutaneous HCC mice xenograft model, the cell suspension was injected into the right flank of the mice (n = 5 per group). | |||
Aryl hydrocarbon receptor (AHR)
Tislelizumab
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [333] | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| In-vitro Model | U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 |
Baculoviral IAP repeat-containing protein 5 (BIRC5)
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [420] | |||
| Responsed Disease | Ovarian cancer | ICD-11: 2C73 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 |
| A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 | |
| In-vivo Model | Four weeks old female BALB/c nude mice were purchased from SJA Laboratory Animal Co, Ltd (Changsha, China), and acclimated in a specific pathogen free environment for 1 week. SKOV3/DDP cells treated with shBIRC5, or shBIRC5 combined with pcDNA-IGF2BP1 plasmid (IGF2BP1) or corresponding negative control were suspended in 100 μL McCoy's 5A medium and implanted subcutaneously into the left frank of mice in 1 × 107 cells/mice. After 7 days incubation, three groups of mice received 10 mg/kg cisplatin respectively, sacrificing and tumor collecting after 30 days. | |||
Calcium-binding and coiled-coil domain-containing protein 1 (CALCOCO1)
Paclitaxel
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [331] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | The experimental mice were housed in an SPF-grade animal laboratory at a temperature of approximately 25 °C, a humidity range of 50%, and an average daylight duration of 12 h. BALB/C female mice at 5-6 weeks of age were taken and prepared for tumor-bearing. After the mice were sacrificed, their tissues were collected, weighed and immediately snap-frozen in liquid nitrogen. | |||
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [331] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | The experimental mice were housed in an SPF-grade animal laboratory at a temperature of approximately 25 °C, a humidity range of 50%, and an average daylight duration of 12 h. BALB/C female mice at 5-6 weeks of age were taken and prepared for tumor-bearing. After the mice were sacrificed, their tissues were collected, weighed and immediately snap-frozen in liquid nitrogen. | |||
Catalase (CAT)
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [339] | |||
| Target Regulation | Up regulation | |||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
CD70 antigen (CD70)
JNJ-74494550
[Phase 2]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [427] | |||
| Responsed Disease | Papillary thyroid cancer | ICD-11: 2D10.1 | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | Nthy-ori 3-1 | Normal | Homo sapiens | CVCL_2659 |
| B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 | |
| K1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_2537 | |
| C-643 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_5969 | |
| BHT-101 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_1085 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| In-vivo Model | NCG mice (female, 4-5 weeks old, purchased from Jiangsu GemPharmatech) were acclimated for 1 week and were selected randomly to subcutaneously (s.c.) injected with 100 μl suspensions of 1 × 106 thyroid cancer cell lines. All mice were housed in animal facility under specific pathogen-free conditions. After 7 day, 5 × 106 huPBMCs were intravenously (i.v.) inoculated into mice. Mice were treated intravenously with anti-PD-1 mAb (nivolumab, 200 μg per injection), Combination (nivolumab + anti-CD70 mAb cusatuzumab, 200 μg per injection each), or PBS once a week. The treatment with M2-TAMs EVs or PBS continued in mice at 2ug EVs per injection at the tumor site twice a week [24]. The body weight of mice and the length (mm) and width (mm) of tumors were monitored every 3 or 4 days. Tumor volume (mm3) was calculated as Length × Width2 × 1/2. Tumor growth analyses were limited to 3 to 4 weeks because following this period mice started to show signs of xenograft-versus-host disease (xGVHD). Therefore, after 14 days or 21 days of PBMCs implantation, the mice were euthanized. | |||
Circ_ASK1
Gefitinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [433] | |||
| Responsed Disease | Lung adenocarcinoma | ICD-11: 2C25.0 | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 |
| SK-LU-1 | Lung adenocarcinoma | Homo sapiens | CVCL_0629 | |
| NCI-H1993 | Lung adenocarcinoma | Homo sapiens | CVCL_1512 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| 16HBE14o- | Normal | Homo sapiens | CVCL_0112 | |
| In-vivo Model | Established a xenograft model in BALB/c nude mice by inoculating HCC827-GR cells transfected with the constructs for circASK1 silencing, ASK1-272a.a overexpression and ASK1-272a.a overexpression/circASK1 knockdown. | |||
| Response Summary | Increased YTHDF2-mediated endoribonucleolytic cleavage of m6A-modified Circ_ASK1 accounts for its downregulation in gefitinib-resistant cells. Either METTL3 silencing or YTHDF2 silencing suppressed the decay of circASK1 in HCC827-GR cells. This study provides a novel therapeutic target to overcome gefitinib resistance in LUAD patients. | |||
Claspin (CLSPN)
Celastrol
[Preclinical]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [267] | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| In-vivo Model | AsPC-1 cells suspended in 100 μl of PBS (2 × 106 cells/100 μl) were injected subcutaneously into the lateral flank of the mice, which were randomly divided into solvent group (n = 6), 1.0 mg/kg of celastrol group (n = 6) and 3.0 mg/kg of celastrol group (n = 6). The administration of celastrol was performed by intraperitoneal injection into tumor-bearing mice every 2 day after 10 day inoculation. The tumor sizes were monitored with calipers every 5 days, and the tumor volume was calculated with the formula: Volume (mm3) = 1/2 × length × width2. | |||
Complement C1q subcomponent subunit A (C1QA)
Rituximab
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [445] | |||
| Responsed Disease | Diffuse large B-cell lymphomas | ICD-11: 2A81 | ||
| Target Regulation | Down regulation | |||
| In-vivo Model | Approximately 2 × 106 Farage/R or Farage/S cells stably transfected with shNC, shC1qA or shMETTL3 were subcutaneously injected into the left flank of each mouse. When the tumor volume reached ~100 mm3, each mouse received an intraperitoneal injection of Rituximab (20 mg/kg) every 4 days for a total of 5 injections. The diameter of each tumor was examined every 4 days using a caliper, and tumor volume was calculated as follows: (length × width2)/2. At 28 days after xenograft, the mice were sacrificed and the tumors were weighed and collected. | |||
Cytochrome P450 2B6 (CYP2B6)
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [451] | |||
| Responsed Disease | Insulin resistance | ICD-11: 5A44 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | C57/BL6 male mice (eight per group) were randomly divided into four groups: basal CD group, HFD + 0.9% NaCl injection group, HFD + DMSO injection group, and HFD + STM2457 (Catalog No. 2499663-01-1, Selleck Chemicals, Houston, Texas) injection group. When the mean body weight of mice in HFD group was statistically different from that of mice in the control group [(mean body weight in HFD:mean body weight in CD)/mean body weight in CD > 30%], the treatment was started once a day for one or two weeks. STM2457 was administered at a dose of 50 mg/kg and weighed prior to each injection. DMSO or saline was also administered at an equal volume. Body weight was recorded every day. Injection of STM2457 was conducted daily and totally for one or two weeks. | |||
D-3-phosphoglycerate dehydrogenase (PHGDH)
Lenvatinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [452] | |||
| Responsed Disease | Liver hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| In-vitro Model |
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| HepG3 | Hepatoblastoma | Homo sapiens | CVCL_Y479 | |
| In-vivo Model | For the subcutaneous injection model, 3 × 106 MHCC97L cells were washed with PBS thrice and resuspended in 100 μL 1:1 Matrixgel and DMEM-HG. HDTVi was performed as previously described.44 Plasmids were extracted using an EndoFree Plasmid extraction kit (QIAGEN). Plasmid sequence was confirmed using Sanger sequencing. A 2 mL mixture of 20 μg sgTp53 plasmids, 10 μg Myc overexpression plasmids, and 2 μg SB13 plasmids was dissolved in saline of 10% mice body weight and was tail-vein injected into C57BL/6 mice weighing at least 25 g. Plasmids were co-injected in the lateral tail vein to induce spontaneous HCC formation. Three weeks after HDTVi, mice were randomly subjected to vehicle or STM2457 treatment. STM2457 (DC chemicals) was dissolved in 20% (w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) and injected intraperitoneally. Sorafenib powder (Selleckchem) was dissolved in distilled water and administrated via oral gavage. A 5 days on/3 days off treatment regimen was applied. | |||
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [452] | |||
| Responsed Disease | Liver hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| In-vitro Model |
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | For the subcutaneous injection model, 3 × 106 MHCC97L cells were washed with PBS thrice and resuspended in 100 μL 1:1 Matrixgel and DMEM-HG. HDTVi was performed as previously described.44 Plasmids were extracted using an EndoFree Plasmid extraction kit (QIAGEN). Plasmid sequence was confirmed using Sanger sequencing. A 2 mL mixture of 20 μg sgTp53 plasmids, 10 μg Myc overexpression plasmids, and 2 μg SB13 plasmids was dissolved in saline of 10% mice body weight and was tail-vein injected into C57BL/6 mice weighing at least 25 g. Plasmids were co-injected in the lateral tail vein to induce spontaneous HCC formation. Three weeks after HDTVi, mice were randomly subjected to vehicle or STM2457 treatment. STM2457 (DC chemicals) was dissolved in 20% (w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) and injected intraperitoneally. Sorafenib powder (Selleckchem) was dissolved in distilled water and administrated via oral gavage. A 5 days on/2 days off treatment regimen was applied. | |||
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [452] | |||
| Responsed Disease | Liver hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| In-vitro Model |
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | For the subcutaneous injection model, 3 × 106 MHCC97L cells were washed with PBS thrice and resuspended in 100 μL 1:1 Matrixgel and DMEM-HG. HDTVi was performed as previously described.44 Plasmids were extracted using an EndoFree Plasmid extraction kit (QIAGEN). Plasmid sequence was confirmed using Sanger sequencing. A 2 mL mixture of 20 μg sgTp53 plasmids, 10 μg Myc overexpression plasmids, and 2 μg SB13 plasmids was dissolved in saline of 10% mice body weight and was tail-vein injected into C57BL/6 mice weighing at least 25 g. Plasmids were co-injected in the lateral tail vein to induce spontaneous HCC formation. Three weeks after HDTVi, mice were randomly subjected to vehicle or STM2457 treatment. STM2457 (DC chemicals) was dissolved in 20% (w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) and injected intraperitoneally. Sorafenib powder (Selleckchem) was dissolved in distilled water and administrated via oral gavage. A 5 days on/4 days off treatment regimen was applied. | |||
Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2 (RPN2)
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [456] | |||
| Responsed Disease | Bladder cancer | ICD-11: 2C94 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| In-vitro Model | 5637 | Bladder carcinoma | Homo sapiens | CVCL_0126 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
Ena/VASP-like protein (EVL)
Isoforsythiaside
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [460] | |||
| Responsed Disease | Macroscopic changes of size of the kidney | ICD-11: MF54.0 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | HEK293 | Normal | Homo sapiens | CVCL_0045 |
| HK-2 [Human kidney] | Normal | Homo sapiens | CVCL_0302 | |
| In-vivo Model | For the unilateral ureteral obstruction (UUO)-induced ON model, mice were subjected to permanent ureteral ligation of the left kidney ureter under aseptic and anaesthetic conditions, whereas sham-operated mice merely lacked ligation under equivalent conditions. The mice were sacrificed under anaesthesia at 3, 7 and 14 days postoperatively. For the ischemia/reperfusion (I/R)-induced renal fibrosis model, mice were recovered with blood supply after 42 min of bilateral renal artery clamping under both aseptic and anaesthetic conditions. These mice were sacrificed under anaesthesia at 7 and 14 days postoperatively. For drug treatment, isoforsythiaside was injected intraperitoneally at concentrations of 10, 25 and 50 mg/kg per day when the model was established. Similarly, the sham-operated group received an equivalent volume of saline intraperitoneally as a control. The harvested kidneys have been primarily used for morphological and histopathological observations and molecular biology studies. | |||
Ferroptosis suppressor protein 1 (AIFM2)
iFSP1
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [463] | |||
| Responsed Disease | Glioma | ICD-11: 2A00.0 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | U-251MG | Astrocytoma | Homo sapiens | CVCL_0021 |
| In-vivo Model | Briefly, the mice were anesthetized and placed in a stereotactic frame. A burr hole was drilled into the skull (1.0 mm posterior and 3.0 mm lateral to the bregma). A 50 μL microinjector was used to inject 10 μL of 1 × 106 U251 cells suspended in the serum-free DMEM. The injection was performed slowly within 10 min and stayed for 10 min. After that, mice in cages and a cat were placed in the same room for fear stress induction . | |||
FGD5 antisense RNA 1 (FGD5-AS1)
Paclitaxel
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [465] | |||
| Responsed Disease | Endometrial cancer | ICD-11: 2C76 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | Ishikawa | Endometrial adenocarcinoma | Homo sapiens | CVCL_2529 |
| HHUA | Endometrial adenocarcinoma | Homo sapiens | CVCL_3866 | |
Fos-related antigen 2 (FOSL2)
Short-chain fatty acid-butyric acid
[Phase 4]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [467] | |||
| Responsed Disease | Polycystic ovary syndrome | ICD-11: 5A80.1 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | KGN | Ovarian granulosa cell tumor | Homo sapiens | CVCL_0375 |
| In-vivo Model | Specific-pathogen-free (SPF) grade C57/B6 mice (6-8 weeks old) were purchased from Weitong Lihua Co. (Beijing, China). The mice were housed at constant temperature and humidity, and 12 h light/dark cycle a day and provided with adequate food and water. | |||
Glutathione peroxidase 1 (GPX1)
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [339] | |||
| Target Regulation | Up regulation | |||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
Guanine nucleotide-binding protein G (GNAO1)
Temozolomide
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [472] | |||
| Responsed Disease | Glioma | ICD-11: 2A00.0 | ||
hsa-mir-19a
Everolimus
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [490] | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Target Regulation | Up regulation | |||
hsa-mir-19b-1
Everolimus
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [490] | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Target Regulation | Up regulation | |||
hsa-miR-27a-3p
Propranolol
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [491] | |||
| Responsed Disease | Infantile hemangioma | ICD-11: 2E81 | ||
Oxymatrine
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [491] | |||
| Responsed Disease | Infantile hemangioma | ICD-11: 2E81 | ||
Hypoxia up-regulated protein 1 (HYOU1)
Doxil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [496] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
Inter-alpha-trypsin inhibitor heavy chain H1 (ITIH1)
LY2157299
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [499] | |||
| Responsed Disease | Liver hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| In-vivo Model | HCC cells stably transfected with a firefly luciferase expression vector (1 × 106 cells 30 μL) were injected into the left liver lobe of 5-week-old male BALB/c nude mice. Luciferase bioluminescence in each mouse was measured at week 6 after injection. Mice in all the groups were sacrificed 8 weeks after injection. ii) For the lung metastasis model, we slowly injected HCC cells (1 × 106cells 100 μL) into 5-week-old male BALB/c nude mice via the tail vein. Six weeks after injection, luciferase bioluminescence was measured, and the mice were then sacrificed. iii) As described in the previous study, 6-8-week-old male wild-type C57BL/6J mice were used for hydrodynamic tail vein injection (HTVi). The pT3-EF1alpha-myr-AKT-HA (human), pT3-EF1alpha-N90-beta-catenin (human), and pT3-EF1alpha-ITIH1 (human) plasmids were constructed by using the pT3-EF1alpha vector. The pooled pT3 plasmids and the Sleeping Beauty transposon plasmid (pCMV-SB) were mixed at a ratio of 25:1 and 20 μg of each pT3 plasmid was used for each mouse. | |||
Krueppel-like factor 9 (KLF9)
Methyl piperidine-3-carboxylate
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [507] | |||
| Responsed Disease | Obesity | ICD-11: 5B81 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | 3T3-L1 | Normal | Mus musculus | CVCL_0123 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
|
AAV-293
|
N.A. | Homo sapiens | CVCL_6871 | |
| In-vivo Model | For compound treatment, mice were intraperitoneally injected with methyl piperidine-3-carboxylate (MP3C) (Shanghai Aladdin Biochemical Technology) at 2.5 mg/kg every 3 days for 2 weeks. | |||
Long intergenic non-protein coding RNA 426 (LINC00426)
Bleomycin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [511] | |||
| Responsed Disease | Cervical cancer | ICD-11: 2C77 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| In-vivo Model | HeLa cells were stably transfected with LINC00426 overexpression lentivirus. Subsequently, for subcutaneously implanted tumor model, mice were randomly divided into two groups, and cells (1 × 107/100 μl) mixed with the same volume of matrix gel were injected subcutaneously into the right abdomen of mice. One month later the mice were executed and the tumors were stripped and weighed, measured for volume, and used for further analysis. For tumor metastasis assay, mice were randomly grouped, and 5 × 105/100 μl HeLa cells transfected with NC and LINC00426 overexpression lentivirus were intravenously injected into the tail vein of BALB/c nude mice which were sacrificed after 1 month. | |||
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [511] | |||
| Responsed Disease | Cervical cancer | ICD-11: 2C77 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| In-vivo Model | HeLa cells were stably transfected with LINC00426 overexpression lentivirus. Subsequently, for subcutaneously implanted tumor model, mice were randomly divided into two groups, and cells (1 × 107/100 μl) mixed with the same volume of matrix gel were injected subcutaneously into the right abdomen of mice. One month later the mice were executed and the tumors were stripped and weighed, measured for volume, and used for further analysis. For tumor metastasis assay, mice were randomly grouped, and 5 × 105/100 μl HeLa cells transfected with NC and LINC00426 overexpression lentivirus were intravenously injected into the tail vein of BALB/c nude mice which were sacrificed after 1 month. | |||
Imatinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [511] | |||
| Responsed Disease | Cervical cancer | ICD-11: 2C77 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| In-vivo Model | HeLa cells were stably transfected with LINC00426 overexpression lentivirus. Subsequently, for subcutaneously implanted tumor model, mice were randomly divided into two groups, and cells (1 × 107/100 μl) mixed with the same volume of matrix gel were injected subcutaneously into the right abdomen of mice. One month later the mice were executed and the tumors were stripped and weighed, measured for volume, and used for further analysis. For tumor metastasis assay, mice were randomly grouped, and 5 × 105/100 μl HeLa cells transfected with NC and LINC00426 overexpression lentivirus were intravenously injected into the tail vein of BALB/c nude mice which were sacrificed after 1 month. | |||
Long intergenic non-protein coding RNA 662 (LINC00662)
Docetaxel
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [513] | |||
| Responsed Disease | Triple-negative breast cancer | ICD-11: 2C6Z | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
Long-chain-fatty-acid--CoA ligase 4 (ACSL4)
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [515] | |||
| Responsed Disease | Sepsis-associated lung injury | ICD-11: NB30.4 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model |
MLE-12
|
N.A. | Mus musculus | CVCL_3751 |
| In-vivo Model | To investigate the effect of lactate on sepsis-induced lung injury, sodium oxalate (0.75 g/kg body weight, Selleck, S6871) [18] or 2-DG (0.5 g/kg body weight, Selleck, S4701) [19] was injected intraperitoneally 3 h before CLP or sham operation to inhibit lactate production. To suppress METTL3 levels, mice were injected with STM2457 (30 mg/kg body weight, MedChemExpress, HY-134836) 12 h before CLP or sham surgery, with a supplemental injection of STM2457 before surgery. | |||
m7GpppN-mRNA hydrolase (DCP2)
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [517] | |||
| Responsed Disease | Small cell lung cancer | ICD-11: 2C25.1 | ||
| Target Regulation | Down regulation | |||
| In-vitro Model |
H-69
|
N.A. | Homo sapiens | CVCL_8121 |
| H69AR | Lung small cell carcinoma | Homo sapiens | CVCL_3513 | |
| H446 | Lung small cell carcinoma | Homo sapiens | CVCL_1562 | |
|
HBE1
|
N.A. | Homo sapiens | CVCL_0287 | |
| In-vivo Model | SCLC cells were collected, and cell suspensions were prepared at a concentration of 1 × 106 cells per 100 μL PBS and injected subcutaneously into nude mice. When the tumour volume reached 60 mm3, mice were randomly divided into control and treatment groups with five mice in each group. The mice in the treatment group were treated regularly, and chemotherapy drugs (CDDP 3 mg/kg and VP16 2 mg/kg) were administered via intraperitoneal injection. | |||
miR-17-92
Everolimus
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [490] | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | miRNA maturation | |||
| In-vitro Model | MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | For subcutaneous xenograft models, 0.1 mL of cell suspension containing 106 cells were injected subcutaneously into the right flank of mice (n = 6 for each group). | |||
| Response Summary | In gastric cancer, m6A facilitated processing of pri-miR-17-92 into the miR-17-92 cluster through an m6A/DGCR8-dependent mechanism. METTL3-high tumors showed preferred sensitivity to an mTOR inhibitor, everolimus. | |||
Mitogen-activated protein kinase 13 (MAPK13)
Rapamycin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [519] | |||
| Target Regulation | Down regulation | |||
| In-vitro Model | 621-101 | Lung lymphangioleiomyomatosis | Homo sapiens | CVCL_S879 |
|
HEK293-EBNA
|
N.A. | Homo sapiens | CVCL_6974 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| UMB1949 | Kidney angiomyolipoma | Homo sapiens | CVCL_C471 | |
Neuronal acetylcholine receptor subunit alpha-4 (CHRNA4)
Bisphenol A
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [525] | |||
| Responsed Disease | Cognitive disorders | ICD-11: 6E0Z | ||
| Target Regulation | Up regulation | |||
| In-vivo Model | Rats were tested in the Morris Water Maze (MWM) after PND 60. The experimental device was a circular tank with a diameter of 120 cm and depth of 40 cm, containing water held at a constant 23 ± 1 °C. Rats were allowed to swim to a settled platform (a round, clear acrylic panel) for no more than 90 s with its top surface submerged 1.5 cm below the water level. Caramel was dissolved in water to ensuring that rats cannot directly observe the platform position. Rats were released from different positions in the water maze, and each rat performed four trials daily for five days. The escape latency was automatically recorded. On the sixth day, the rats were given a 90-s retention trial in which the platform was removed, and the mean speed, relative time in zone 2 and relative distance in zone 2 were recorded. | |||
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [525] | |||
| Responsed Disease | Cognitive disorders | ICD-11: 6E0Z | ||
| Target Regulation | Up regulation | |||
| In-vivo Model | Rats were tested in the Morris Water Maze (MWM) after PND 60. The experimental device was a circular tank with a diameter of 120 cm and depth of 40 cm, containing water held at a constant 23 ± 1 °C. Rats were allowed to swim to a settled platform (a round, clear acrylic panel) for no more than 90 s with its top surface submerged 1.5 cm below the water level. Caramel was dissolved in water to ensuring that rats cannot directly observe the platform position. Rats were released from different positions in the water maze, and each rat performed four trials daily for five days. The escape latency was automatically recorded. On the sixth day, the rats were given a 90-s retention trial in which the platform was removed, and the mean speed, relative time in zone 2 and relative distance in zone 2 were recorded. | |||
Nuclear factor of activated T-cells 5 (NFAT5)
Gemcitabine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [528] | |||
| Responsed Disease | Intrahepatic cholangiocarcinoma | ICD-11: 2C12.10 | ||
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [528] | |||
| Responsed Disease | Intrahepatic cholangiocarcinoma | ICD-11: 2C12.10 | ||
Nuclear receptor subfamily 4 group A member 2 (NR4A2)
Celecoxib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [530] | |||
| Responsed Disease | Esophageal Squamous Cell Carcinoma | ICD-11: 2B70.1 | ||
| In-vitro Model | KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 |
| Eca-109 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_6898 | |
| KYSE-450 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1353 | |
| KYSE-30 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1351 | |
| HET-1A | Normal | Homo sapiens | CVCL_3702 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Oxidized low-density lipoprotein receptor 1 (OLR1)
Temozolomide
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [534] | |||
| Responsed Disease | Glioma | ICD-11: 2A00.0 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | U-343MG | Glioblastoma | Homo sapiens | CVCL_S471 |
| U-251MG | Astrocytoma | Homo sapiens | CVCL_0021 | |
Paired box protein Pax-8 (PAX8)
Sodium iodide I 131
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [535] | |||
| Responsed Disease | Thyroid Cancer | ICD-11: 2D10 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 |
| KTC-1 | Thyroid carcinoma | Homo sapiens | CVCL_6300 | |
| TPC-1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_6298 | |
| K1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_2537 | |
| CAL-62 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_1112 | |
| C-643 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_5969 | |
| Nthy-ori 3-1 | Normal | Homo sapiens | CVCL_2659 | |
| In-vivo Model | A concentration of 1 × 107 CAL62 cells stably transfected with OE METTL3 or OE METTL3/shPAX8-transfected CAL-62 cells were injected subcutaneously into the dorsal flanks of the mice (five mice per group) to establish a xenograft model. | |||
Phosphoserine aminotransferase (PSAT1)
Lenvatinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [452] | |||
| Responsed Disease | Liver hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| In-vitro Model |
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | For the subcutaneous injection model, 3 × 106 MHCC97L cells were washed with PBS thrice and resuspended in 100 μL 1:1 Matrixgel and DMEM-HG. HDTVi was performed as previously described.44 Plasmids were extracted using an EndoFree Plasmid extraction kit (QIAGEN). Plasmid sequence was confirmed using Sanger sequencing. A 2 mL mixture of 20 μg sgTp53 plasmids, 10 μg Myc overexpression plasmids, and 2 μg SB13 plasmids was dissolved in saline of 10% mice body weight and was tail-vein injected into C57BL/6 mice weighing at least 25 g. Plasmids were co-injected in the lateral tail vein to induce spontaneous HCC formation. Three weeks after HDTVi, mice were randomly subjected to vehicle or STM2457 treatment. STM2457 (DC chemicals) was dissolved in 20% (w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) and injected intraperitoneally. Sorafenib powder (Selleckchem) was dissolved in distilled water and administrated via oral gavage. A 5 days on/6 days off treatment regimen was applied. | |||
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [452] | |||
| Responsed Disease | Liver hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| In-vitro Model |
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | For the subcutaneous injection model, 3 × 106 MHCC97L cells were washed with PBS thrice and resuspended in 100 μL 1:1 Matrixgel and DMEM-HG. HDTVi was performed as previously described.44 Plasmids were extracted using an EndoFree Plasmid extraction kit (QIAGEN). Plasmid sequence was confirmed using Sanger sequencing. A 2 mL mixture of 20 μg sgTp53 plasmids, 10 μg Myc overexpression plasmids, and 2 μg SB13 plasmids was dissolved in saline of 10% mice body weight and was tail-vein injected into C57BL/6 mice weighing at least 25 g. Plasmids were co-injected in the lateral tail vein to induce spontaneous HCC formation. Three weeks after HDTVi, mice were randomly subjected to vehicle or STM2457 treatment. STM2457 (DC chemicals) was dissolved in 20% (w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) and injected intraperitoneally. Sorafenib powder (Selleckchem) was dissolved in distilled water and administrated via oral gavage. A 5 days on/5 days off treatment regimen was applied. | |||
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [452] | |||
| Responsed Disease | Liver hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| In-vitro Model |
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | For the subcutaneous injection model, 3 × 106 MHCC97L cells were washed with PBS thrice and resuspended in 100 μL 1:1 Matrixgel and DMEM-HG. HDTVi was performed as previously described.44 Plasmids were extracted using an EndoFree Plasmid extraction kit (QIAGEN). Plasmid sequence was confirmed using Sanger sequencing. A 2 mL mixture of 20 μg sgTp53 plasmids, 10 μg Myc overexpression plasmids, and 2 μg SB13 plasmids was dissolved in saline of 10% mice body weight and was tail-vein injected into C57BL/6 mice weighing at least 25 g. Plasmids were co-injected in the lateral tail vein to induce spontaneous HCC formation. Three weeks after HDTVi, mice were randomly subjected to vehicle or STM2457 treatment. STM2457 (DC chemicals) was dissolved in 20% (w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) and injected intraperitoneally. Sorafenib powder (Selleckchem) was dissolved in distilled water and administrated via oral gavage. A 5 days on/7 days off treatment regimen was applied. | |||
Phosphoserine phosphatase (PSPH)
Lenvatinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [452] | |||
| Responsed Disease | Liver hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| In-vitro Model |
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | For the subcutaneous injection model, 3 × 106 MHCC97L cells were washed with PBS thrice and resuspended in 100 μL 1:1 Matrixgel and DMEM-HG. HDTVi was performed as previously described.44 Plasmids were extracted using an EndoFree Plasmid extraction kit (QIAGEN). Plasmid sequence was confirmed using Sanger sequencing. A 2 mL mixture of 20 μg sgTp53 plasmids, 10 μg Myc overexpression plasmids, and 2 μg SB13 plasmids was dissolved in saline of 10% mice body weight and was tail-vein injected into C57BL/6 mice weighing at least 25 g. Plasmids were co-injected in the lateral tail vein to induce spontaneous HCC formation. Three weeks after HDTVi, mice were randomly subjected to vehicle or STM2457 treatment. STM2457 (DC chemicals) was dissolved in 20% (w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) and injected intraperitoneally. Sorafenib powder (Selleckchem) was dissolved in distilled water and administrated via oral gavage. A 5 days on/9 days off treatment regimen was applied. | |||
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [452] | |||
| Responsed Disease | Liver hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| In-vitro Model |
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | For the subcutaneous injection model, 3 × 106 MHCC97L cells were washed with PBS thrice and resuspended in 100 μL 1:1 Matrixgel and DMEM-HG. HDTVi was performed as previously described.44 Plasmids were extracted using an EndoFree Plasmid extraction kit (QIAGEN). Plasmid sequence was confirmed using Sanger sequencing. A 2 mL mixture of 20 μg sgTp53 plasmids, 10 μg Myc overexpression plasmids, and 2 μg SB13 plasmids was dissolved in saline of 10% mice body weight and was tail-vein injected into C57BL/6 mice weighing at least 25 g. Plasmids were co-injected in the lateral tail vein to induce spontaneous HCC formation. Three weeks after HDTVi, mice were randomly subjected to vehicle or STM2457 treatment. STM2457 (DC chemicals) was dissolved in 20% (w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) and injected intraperitoneally. Sorafenib powder (Selleckchem) was dissolved in distilled water and administrated via oral gavage. A 5 days on/8 days off treatment regimen was applied. | |||
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [452] | |||
| Responsed Disease | Liver hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| In-vitro Model |
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | For the subcutaneous injection model, 3 × 106 MHCC97L cells were washed with PBS thrice and resuspended in 100 μL 1:1 Matrixgel and DMEM-HG. HDTVi was performed as previously described.44 Plasmids were extracted using an EndoFree Plasmid extraction kit (QIAGEN). Plasmid sequence was confirmed using Sanger sequencing. A 2 mL mixture of 20 μg sgTp53 plasmids, 10 μg Myc overexpression plasmids, and 2 μg SB13 plasmids was dissolved in saline of 10% mice body weight and was tail-vein injected into C57BL/6 mice weighing at least 25 g. Plasmids were co-injected in the lateral tail vein to induce spontaneous HCC formation. Three weeks after HDTVi, mice were randomly subjected to vehicle or STM2457 treatment. STM2457 (DC chemicals) was dissolved in 20% (w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) and injected intraperitoneally. Sorafenib powder (Selleckchem) was dissolved in distilled water and administrated via oral gavage. A 5 days on/9 days off treatment regimen was applied. | |||
Polyunsaturated fatty acid lipoxygenase ALOX12 (ALOX12)
Erianin
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [277] | |||
| Responsed Disease | Renal cell carcinoma | ICD-11: 2C90 | ||
| Target Regulation | Down regulation | |||
Prostaglandin E2 receptor EP4 subtype (PTGER2)
Carboplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [546] | |||
| Responsed Disease | Ovarian cancer | ICD-11: 2C73 | ||
| Target Regulation | Up regulation | |||
Pseudorabies Virus (PRV)
3-deazidenosine
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [552] | |||
| Responsed Disease | Rabies | ICD-11: 1C82 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model |
PK-15
|
N.A. | Sus scrofa | CVCL_2160 |
Receptor-interacting serine/threonine-protein kinase 4 (RIPK4)
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [557] | |||
| Responsed Disease | Malignant mixed epithelial mesenchymal tumour of ovary | ICD-11: 2B5D.0 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | NF-kappa B signaling pathway | hsa04064 | ||
| In-vitro Model | SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 |
| A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 | |
| In-vivo Model | Animals were purchased from Changzhou Cavens Laboratory Animal Company (Changzhou, China) and maintained under specific pathogen-free (SPF) conditions. All animal experiments were performed in accordance with the guidelines of the Laboratory Animal Health Committee of Jiangsu University. Approximately 1 × 107 cells were injected subcutaneously into the dorsal surface of female BALB/c nude mice (n = 5 for each group, 4 weeks old, 15 ± 2 g). When the xenograft volume reached approximately 60 mm3 (after 1 week), DDP was intraperitoneally injected three times per week for 3 weeks. On Day 28, the mice were sacrificed and the tumors were frozen at -80 °C for follow-up experiments. | |||
RNA component of 7SK nuclear ribonucleoprotein (RN7SK)
Etoposide
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [560] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| In-vitro Model | BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| BEL-7404 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6568 | |
| BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | For the generation of PDX mouse models, fresh LUSC specimens (2-3 mm3), which were from Shanghai Chest Hospital, were implanted into 4- to 6-week-old athymic nude mice (Jiesijie, Shanghai, China). After successful tumor growth was confirmed, the tumor tissues were passaged and implanted into the next generation of mice. The third to fifth generations of PDX-bearing mice were used for drug administration. When tumors reached approximately 200 mm3, mice were injected daily with DMSO (no. ST038, Beyotime Biotechnology, Shanghai, China) with or without MIT (no. S2485, 5 mg/kg, Selleck, Houston, TX) or HYD (no. S1896, 20 mg/kg, Selleck) (n = 5 mice/group). Tumor growth was monitored, and sizes were calculated by 0.5 × L × W2 (L indicating length and W indicating width). The mice were euthanized at day 28 after implantation. All animal experiments were approved by the institutional ethics committee of Shanghai Chest Hospital (approval number KS(Y)21381). | |||
Hydroxyurea
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [560] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| In-vitro Model | BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| BEL-7404 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6568 | |
| BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | For the generation of PDX mouse models, fresh LUSC specimens (2-3 mm3), which were from Shanghai Chest Hospital, were implanted into 4- to 6-week-old athymic nude mice (Jiesijie, Shanghai, China). After successful tumor growth was confirmed, the tumor tissues were passaged and implanted into the next generation of mice. The third to fifth generations of PDX-bearing mice were used for drug administration. When tumors reached approximately 200 mm3, mice were injected daily with DMSO (no. ST038, Beyotime Biotechnology, Shanghai, China) with or without MIT (no. S2485, 5 mg/kg, Selleck, Houston, TX) or HYD (no. S1896, 20 mg/kg, Selleck) (n = 5 mice/group). Tumor growth was monitored, and sizes were calculated by 0.5 × L × W2 (L indicating length and W indicating width). The mice were euthanized at day 28 after implantation. All animal experiments were approved by the institutional ethics committee of Shanghai Chest Hospital (approval number KS(Y)21381). | |||
Mitoxantrone
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [560] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| In-vitro Model | BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| BEL-7404 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6568 | |
| BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | For the generation of PDX mouse models, fresh LUSC specimens (2-3 mm3), which were from Shanghai Chest Hospital, were implanted into 4- to 6-week-old athymic nude mice (Jiesijie, Shanghai, China). After successful tumor growth was confirmed, the tumor tissues were passaged and implanted into the next generation of mice. The third to fifth generations of PDX-bearing mice were used for drug administration. When tumors reached approximately 200 mm3, mice were injected daily with DMSO (no. ST038, Beyotime Biotechnology, Shanghai, China) with or without MIT (no. S2485, 5 mg/kg, Selleck, Houston, TX) or HYD (no. S1896, 20 mg/kg, Selleck) (n = 5 mice/group). Tumor growth was monitored, and sizes were calculated by 0.5 × L × W2 (L indicating length and W indicating width). The mice were euthanized at day 28 after implantation. All animal experiments were approved by the institutional ethics committee of Shanghai Chest Hospital (approval number KS(Y)21381). | |||
Oxaliplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [560] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| In-vitro Model | BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| BEL-7404 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6568 | |
| BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | For the generation of PDX mouse models, fresh LUSC specimens (2-3 mm3), which were from Shanghai Chest Hospital, were implanted into 4- to 6-week-old athymic nude mice (Jiesijie, Shanghai, China). After successful tumor growth was confirmed, the tumor tissues were passaged and implanted into the next generation of mice. The third to fifth generations of PDX-bearing mice were used for drug administration. When tumors reached approximately 200 mm3, mice were injected daily with DMSO (no. ST038, Beyotime Biotechnology, Shanghai, China) with or without MIT (no. S2485, 5 mg/kg, Selleck, Houston, TX) or HYD (no. S1896, 20 mg/kg, Selleck) (n = 5 mice/group). Tumor growth was monitored, and sizes were calculated by 0.5 × L × W2 (L indicating length and W indicating width). The mice were euthanized at day 28 after implantation. All animal experiments were approved by the institutional ethics committee of Shanghai Chest Hospital (approval number KS(Y)21381). | |||
Raltitrexed
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [560] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| In-vitro Model | BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| BEL-7404 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6568 | |
| BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | For the generation of PDX mouse models, fresh LUSC specimens (2-3 mm3), which were from Shanghai Chest Hospital, were implanted into 4- to 6-week-old athymic nude mice (Jiesijie, Shanghai, China). After successful tumor growth was confirmed, the tumor tissues were passaged and implanted into the next generation of mice. The third to fifth generations of PDX-bearing mice were used for drug administration. When tumors reached approximately 200 mm3, mice were injected daily with DMSO (no. ST038, Beyotime Biotechnology, Shanghai, China) with or without MIT (no. S2485, 5 mg/kg, Selleck, Houston, TX) or HYD (no. S1896, 20 mg/kg, Selleck) (n = 5 mice/group). Tumor growth was monitored, and sizes were calculated by 0.5 × L × W2 (L indicating length and W indicating width). The mice were euthanized at day 28 after implantation. All animal experiments were approved by the institutional ethics committee of Shanghai Chest Hospital (approval number KS(Y)21381). | |||
RNA-binding protein 25 (RBM25)
Metformin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [356] | |||
| Responsed Disease | Multiple myeloma | ICD-11: 2A83.1 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | NCI-H929 | Plasma cell myeloma | Homo sapiens | CVCL_1600 |
| MM1.S | Plasma cell myeloma | Homo sapiens | CVCL_8792 | |
| In-vivo Model | 5-week-old male athymic BALB/C nude mice were obtained from Experimental animal center of Fujian Medical University, and subsequently randomly divided into four groups (5 mice per group), including control, metformin, pcDNA-METTL3 and pcDNA-METTL3 + metformin. For the control or metformin group, to induce tumors, 5 × 105 H929 cells were suspended in 0.2 ml phosphate buffered saline and inoculated subcutaneously into the peritoneum of mice. Once the subcutaneous nodules grown to a rice grain size (required approximately a week), the subcutaneous xenograft model of MM in nude mice was successfully constructed. One week later, PBS or Metformin was diluted in drinking water (2 mg/ml) and administered orally throughout the indicated time periods. For the pcDNA-METTL3 group and pcDNA-METTL3 + metformin group, firstly, we transfected pcDNA-METTL3 into H929 cells to overexpress METTL3, and then 5 × 105 H929 cells were suspended in 0.2 ml phosphate-buffered saline and inoculated subcutaneously into the peritoneum of mice. One week later, PBS or Metformin was diluted in drinking water (2 mg/ml) and administered orally throughout the indicated time periods. All nude mice were closely monitored for tumor growth, skin condition, and behavior daily and any tumor ulceration or irritation was noted. Tumor length and width were calculated with vernier calipers every 7 days. After 35 days, the mice were humanely sacrificed, and the subcutaneous tumors were excised and removed. | |||
Serine/threonine-protein kinase Nek6 (NEK6)
Rapamycin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [568] | |||
| Responsed Disease | Traumatic brain injury induced by controlled cortical impact injury | ICD-11: NA07 | ||
| In-vitro Model | SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
SREBF2 antisense RNA 1 (SREBF2-AS1)
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [575] | |||
| Responsed Disease | Liver hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| SNU-398 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0077 | |
| THLE-2 | Normal | Homo sapiens | CVCL_3803 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
Superoxide dismutase 1 (SOD1)
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [339] | |||
| Target Regulation | Up regulation | |||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
Thyroid hormone receptor-associated protein 3 (THRAP3)
Metformin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [356] | |||
| Responsed Disease | Multiple myeloma | ICD-11: 2A83.1 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | NCI-H929 | Plasma cell myeloma | Homo sapiens | CVCL_1600 |
| MM1.S | Plasma cell myeloma | Homo sapiens | CVCL_8792 | |
| In-vivo Model | 5-week-old male athymic BALB/C nude mice were obtained from Experimental animal center of Fujian Medical University, and subsequently randomly divided into four groups (5 mice per group), including control, metformin, pcDNA-METTL3 and pcDNA-METTL3 + metformin. For the control or metformin group, to induce tumors, 5 × 105 H929 cells were suspended in 0.2 ml phosphate buffered saline and inoculated subcutaneously into the peritoneum of mice. Once the subcutaneous nodules grown to a rice grain size (required approximately a week), the subcutaneous xenograft model of MM in nude mice was successfully constructed. One week later, PBS or Metformin was diluted in drinking water (2 mg/ml) and administered orally throughout the indicated time periods. For the pcDNA-METTL3 group and pcDNA-METTL3 + metformin group, firstly, we transfected pcDNA-METTL3 into H929 cells to overexpress METTL3, and then 5 × 105 H929 cells were suspended in 0.2 ml phosphate-buffered saline and inoculated subcutaneously into the peritoneum of mice. One week later, PBS or Metformin was diluted in drinking water (2 mg/ml) and administered orally throughout the indicated time periods. All nude mice were closely monitored for tumor growth, skin condition, and behavior daily and any tumor ulceration or irritation was noted. Tumor length and width were calculated with vernier calipers every 7 days. After 35 days, the mice were humanely sacrificed, and the subcutaneous tumors were excised and removed. | |||
Transmembrane protein 127 (TMEM127)
Rapamycin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [584] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | Approximately 1 × 106 viable MDA-MB-231 breast cancer cells were resuspended in 1:1 ratio in 50 μl medium and 50 μl matrigel (Corning, 354234) and injected orthotopically into the fourth mammary fat pad of each mouse. After injection, tumor size was measured twice a week using an electronic caliper. Tumor volumes were calculated with the formula: volume = (L × W2)/2, where L is the tumor length and W is the tumor width measured in millimeters. | |||
Ubiquitin carboxyl-terminal hydrolase 13 (USP13)
3-Methyladenine
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [587] | |||
| Responsed Disease | Gastrointestinal stromal tumor | ICD-11: 2B5B | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 | |
| In-vivo Model | 6 × 106 logarithmically growing GIST cells resuspended in 100 μL PBS were injected subcutaneously into the flank of 6-week-old female nude mice for tumor growth assays. The following treatments were initiated when the tumor volume reached approximately 300 mm3. (1) IM-sensitive GIST cell lines and IM-resistant GIST cell lines were used to establish a subcutaneous xenograft model of GIST (n = 6 mice/group). When the tumors grew to the required size, mice were treated with IM via drinking water; (2) GIST cells were pretreated with USP13 lentivirus or USP13 control lentivirus. In addition, IM-resistant GIST cells were pretreated with USP13 inhibitors or control. These lentivirus-transfected cells were implanted into subcutaneous mice tumors to construct the USP13 positive, USP13-negative, and control groups (n = 6 mice/group). When the transplanted tumor grew to the required volume, mice in each group were treated with IM via drinking water. (3) IM-resistant GIST cells-xenografted mice and control mice were treated with USP13 inhibitors or with USP13 inhibitors and autophagy inhibitors in combination therapy to assess reversal of IM resistance (n = 6 mice/group). Mice were treated with IM (45 mg/kg/day) and with or without 3-MA (15 mg/kg/day) via drinking water when the tumors grew to the required size. The USP13 inhibitor Spautin-1 was used at 20 mg/kg/day when required. | |||
Spautin 1
[Preclinical]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [587] | |||
| Responsed Disease | Gastrointestinal stromal tumor | ICD-11: 2B5B | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 | |
| In-vivo Model | 6 × 106 logarithmically growing GIST cells resuspended in 100 μL PBS were injected subcutaneously into the flank of 6-week-old female nude mice for tumor growth assays. The following treatments were initiated when the tumor volume reached approximately 300 mm3. (1) IM-sensitive GIST cell lines and IM-resistant GIST cell lines were used to establish a subcutaneous xenograft model of GIST (n = 6 mice/group). When the tumors grew to the required size, mice were treated with IM via drinking water; (2) GIST cells were pretreated with USP13 lentivirus or USP13 control lentivirus. In addition, IM-resistant GIST cells were pretreated with USP13 inhibitors or control. These lentivirus-transfected cells were implanted into subcutaneous mice tumors to construct the USP13 positive, USP13-negative, and control groups (n = 6 mice/group). When the transplanted tumor grew to the required volume, mice in each group were treated with IM via drinking water. (3) IM-resistant GIST cells-xenografted mice and control mice were treated with USP13 inhibitors or with USP13 inhibitors and autophagy inhibitors in combination therapy to assess reversal of IM resistance (n = 6 mice/group). Mice were treated with IM (45 mg/kg/day) and with or without 3-MA (15 mg/kg/day) via drinking water when the tumors grew to the required size. The USP13 inhibitor Spautin-1 was used at 20 mg/kg/day when required. | |||
Zinc finger and BTB domain-containing protein 7C (ZBTB7C)
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [588] | |||
| Responsed Disease | Osteosarcoma | ICD-11: 2B51 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | MNNG/HOS Cl #5 | Osteosarcoma | Homo sapiens | CVCL_0439 |
| MG-63 | Osteosarcoma | Homo sapiens | CVCL_0426 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| hFOB 1.19 | Normal | Homo sapiens | CVCL_3708 | |
| In-vivo Model | (1) ZBTB7C functional assay in vivo, 2 × 106 MNNG/HOS cells were stably transfected with lentivirus (vector control and si-ZBTB7C, n = 5), and then subcutaneously incubated in BALB/c nude mice; (2) STM2457 intervention. MNNG/HOS cells were implanted subcutaneously first. When tumors reached approximately 80-100 mm3, PBS or STM2457 (30 mg/kg (STM-30), 60 mg/kg (STM-60), n = 4) were administered every 2 days; (3) ZBTB7C effects of STM2457 assay. Mice were assigned to 4 groups (n = 5 per group). Two groups had MNNG/HOS cells stably transfected with lentiviral vectors (Ctrl) and 2 with lentivirus-mediated ZBTB7C overexpression (ZBTB7C-OE), and then mice were treated with control vehicle or STM2457 (60 mg/kg), respectively. The experiments went on for 22 days, and the xenografted tumors were measured, weighted, and characterized by H&E and IHC staining. | |||
Unspecific Target Gene
Arsenite
[Phase 2]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [590] | |||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | HBE (Human bronchial epithelial cell line) | |||
| Response Summary | m6A modification on RNA was significantly increased in arsenite-transformed cells and this modification was synergistically regulated by METTL3, METTL14, WTAP and FTO. Demonstrated the significant role of m6A in the prevention of tumor occurrence and progression induced by arsenite. | |||
Gemcitabine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [599] | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Pathway Response | Adipocytokine signaling pathway | hsa04920 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| HDE-CT cell line (A normal human pancreatic cell line) | ||||
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| Response Summary | Lasso regression identified a six-m6A-regulator-signature prognostic model (KIAA1429, HNRNPC, METTL3, YTHDF1, IGF2BP2, and IGF2BP3). Gene set enrichment analysis revealed m6A regulators (KIAA1429, HNRNPC, and IGF2BP2) were related to multiple biological behaviors in pancreatic cancer, including adipocytokine signaling, the well vs. poorly differentiated tumor pathway, tumor metastasis pathway, epithelial mesenchymal transition pathway, gemcitabine resistance pathway, and stemness pathway. | |||
Lenvatinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [602] | |||
| Responsed Disease | Liver hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| In-vitro Model | Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | To increase the accuracy of the experimental results in vivo, we subcutaneously transplanted tumor tissues from HCC patients into BALB/c nude mice and then treated the tumor-bearing mice with lenvatinib to establish a Lenvatinib-resistant PDX mode. | |||
A939572
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [623] | |||
| Responsed Disease | Cardiomegaly | ICD-11: BC45 | ||
| In-vivo Model | Slack Laboratory Animal Co., Ltd. (Shanghai, China) supplied the six-week-old C57BL/6 male mice. Animals were reared at constant temperature (21 ± 1 °C) and maintained in a room with water and food during a 12 h dark/light cycle. After acclimatizing for a week, the animals were randomly assigned to two groups: model (TAC) and control (sham). In the TACgroup, the aortic arch of the mice was constricted using a 27 G needle and 6.0 sutures. The Sham group was operated in the same way as the TAC group without ligation of the aorta. All mice were housed for four weeks for further research. | |||
STM2457
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [627] | |||
| Responsed Disease | Rheumatoid arthritis | ICD-11: FA20 | ||
| In-vivo Model | DBA/1 mice were injected intradermally with 100 μg of bovine type II collagen (CII) emulsified in 50 μL of complete Freund's adjuvant at a 1:1 ratio (vol/vol) as primary immunization on day 0. On day 21, the mice were boosted with CII emulsified at a 1:1 ratio in incomplete Freund's adjuvant. The weight, arthritis score, and thickness of the paws were observed and recorded once every 3 days. Starting on day 21, the CIA mice were administered an equal volume of methotrexate (MTX, 1 mg/kg) and METTL3 inhibitor STM2457 (50 mg/kg), which were intraperitoneally injected with twice a week. All mice were sacrificed and specimens were harvested on day 63. | |||
Ethyl ester form of meclofenamic acid
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [630] | |||
| Responsed Disease | Male infertility | ICD-11: GB04 | ||
| Cell Process | Cell cycle | |||
| Cell proliferation | ||||
| In-vitro Model | GC-1 spg | Normal | Mus musculus | CVCL_8872 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | Mouse GC-1 spg cells were treated with the ester form of meclofenamic acid (MA2) to inhibit the demethylase activity of FTO. | |||
| Response Summary | METTL3, METTL14, ALKBH5 and YTHDC2 are involved in the regulation of spermatogenesis and oogenesis. MA2 affected CDKs expression through the m6A-dependent mRNA degradation pathway, and thus repressed spermatogonial proliferation. Additionally, mutation of the predicted m6A sites in the Cdk2-3'UTR could mitigated the degradation of CDK2 mRNA after MA2 treatment. | |||
BHBA
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [635] | |||
| Responsed Disease | Liver injury | ICD-11: NB91.1 | ||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
RNA modification
m6A Target: Cyclin-dependent kinase inhibitor 1 (CDKN1A)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00001 | ||
| Epigenetic Regulator | RNA cytosine C(5)-methyltransferase NSUN2 (NSUN2) | |
| Regulated Target | Cyclin-dependent kinase inhibitor 1 (CDKN1A) | |
| Crosstalk relationship | m5C → m6A | |
m6A Target: Tyrosine-protein kinase EIF2AK2 (eIF2AK2/p68)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00003 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Tyrosine-protein kinase EIF2AK2 (eIF2AK2/p68) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | US9650366, 12 | |
m6A Target: LIM domain-containing protein ajuba (AJUBA)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00004 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | LIM domain-containing protein ajuba (AJUBA) | |
| Crosstalk relationship | m6A → A-to-I | |
m6A Target: Small nuclear ribonucleoprotein Sm D3 (SNRPD3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00005 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Small nuclear ribonucleoprotein Sm D3 (SNRPD3) | |
| Crosstalk relationship | m6A → A-to-I | |
m6A Target: DNA replication complex GINS protein SLD5 (GINS4)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00006 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | DNA replication complex GINS protein SLD5 (GINS4) | |
| Crosstalk relationship | m6A → A-to-I | |
m6A Target: Mitochondrial import inner membrane translocase subunit TIM50 (TIMM50)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00007 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Mitochondrial import inner membrane translocase subunit TIM50 (TIMM50) | |
| Crosstalk relationship | m6A → A-to-I | |
m6A Target: Protein-S-isoprenylcysteine O-methyltransferase (ICMT)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00008 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Protein-S-isoprenylcysteine O-methyltransferase (ICMT) | |
| Crosstalk relationship | m6A → A-to-I | |
m6A Target: Interleukin-17 receptor D (IL17RD)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00009 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Interleukin-17 receptor D (IL17RD) | |
| Crosstalk relationship | m6A → A-to-I | |
m6A Target: Gap junction gamma-1 protein (GJC1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00010 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Gap junction gamma-1 protein (GJC1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | Octanol | |
m6A Target: Methyltransferase-like protein 1 (METTL1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00011 | ||
| Epigenetic Regulator | Methyltransferase-like protein 1 (METTL1) | |
| Regulated Target | Cyclin-dependent kinase 4 (CDK4) | |
| Crosstalk relationship | m6A → m7G | |
| Disease | Neck squamous cell carcinoma | |
| Drug | Palbociclib | |
m6A Target: hsa-miR-582-3p
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00012 | ||
| Epigenetic Regulator | Y-box-binding protein 1 (YBX1) | |
| Regulated Target | NF-kappaB interacting lncRNA (NKILA) | |
| Crosstalk relationship | m5C → m6A | |
| Disease | Intrahepatic cholangiocarcinoma | |
| Crosstalk ID: M6ACROT00013 | ||
| Epigenetic Regulator | RNA cytosine C(5)-methyltransferase NSUN2 (NSUN2) | |
| Regulated Target | NF-kappaB interacting lncRNA (NKILA) | |
| Crosstalk relationship | m5C → m6A | |
| Disease | Intrahepatic cholangiocarcinoma | |
m6A Target: Collagen alpha-1 (COL1A1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00050 | ||
| Epigenetic Regulator | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 3 (ALKBH3) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m1A → m6A | |
| Disease | Keloid | |
| Crosstalk ID: M6ACROT00052 | ||
| Epigenetic Regulator | YTH domain-containing family protein 2 (YTHDF2) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m1A → m6A | |
| Disease | Keloid | |
m6A Target: Fibronectin (FN1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00051 | ||
| Epigenetic Regulator | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 3 (ALKBH3) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m1A → m6A | |
| Disease | Keloid | |
| Drug | Ocriplasmin | |
| Crosstalk ID: M6ACROT00053 | ||
| Epigenetic Regulator | YTH domain-containing family protein 2 (YTHDF2) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m1A → m6A | |
| Disease | Keloid | |
| Drug | Ocriplasmin | |
m6A Target: Interferon-inducible protein 4 (ADAR1)
| In total 81 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00063 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Cyclin-dependent kinase 2 (CDK2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Disease | Brain cancer | |
| Drug | PHA848125 | |
| Crosstalk ID: M6ACROT00076 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | UPF1 RNA helicase and ATPase (UPF1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00077 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Death associated protein 3 (DAP3) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00078 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Phosphatidylserine decarboxylase (PISD) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00079 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Thioredoxin reductase 2 (TXNRD2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00080 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | BLCAP apoptosis inducing factor (BLCAP) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00087 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Tyrosine-protein kinase EIF2AK2 (eIF2AK2/p68) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | US9650366, 12 | |
| Crosstalk ID: M6ACROT00088 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Eukaryotic translation initiation factor 2 subunit 1 (EIF2S1/eIF2-Alpha) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00091 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Antizyme inhibitor 1 (AZIN1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00092 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Zinc finger protein GLI1 (GLI1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | PMID26666870-Compound-16 | |
| Crosstalk ID: M6ACROT00097 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Rho GTPase activating protein 26 (ARHGAP26) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00110 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Prune homolog 2 with BCH domain (PRUNE2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00111 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | microRNA 222 (MIR222) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00112 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Aryl hydrocarbon receptor (AHR) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00113 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | microRNA 381 (MIR381) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00114 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Inhibitor of nuclear factor kappa-B kinase subunit beta (IKBKB) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | Arsenic trioxide | |
| Crosstalk ID: M6ACROT00115 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Actin, aortic smooth muscle (ACTA2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00116 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Collagen alpha-1 (COL1A1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00117 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Podocalyxin like (PODXL) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00120 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Cathepsin S (CTSS) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | RG7625 | |
| Crosstalk ID: M6ACROT00123 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Dihydrofolate reductase (DHFR) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | Trimethoprim | |
| Crosstalk ID: M6ACROT00124 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Signal transducer and activator of transcription 3 (STAT3) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | Acitretin | |
| Crosstalk ID: M6ACROT00125 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Intercellular adhesion molecule 1 (ICAM1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | ISIS 1570 | |
| Crosstalk ID: M6ACROT00126 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Vascular cell adhesion protein 1 (VCAM1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | ISIS 3792 | |
| Crosstalk ID: M6ACROT00127 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | E3 ubiquitin-protein ligase Mdm2 (Mdm2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | RG7388 | |
| Crosstalk ID: M6ACROT00128 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | X-linked inhibitor of apoptosis (XIAP) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | ASTX660 | |
| Crosstalk ID: M6ACROT00129 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | G1/S-specific cyclin-D1 (CCND1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | ABT-263 | |
| Crosstalk ID: M6ACROT00130 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | F11 receptor (F11R) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00131 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Protein tyrosine kinase 2 (PTK2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | VS-6063 | |
| Crosstalk ID: M6ACROT00132 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Thiol methyltransferase 1A (TMT1A) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00135 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Interferon induced with helicase C domain 1 (IFIH1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00136 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | microRNA 21 (MIR21) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00137 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Pellino E3 ubiquitin protein ligase 1 (PELI1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00138 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Protein sprouty homolog 2 (SPRY2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00143 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Heterogeneous nuclear ribonucleoprotein L like (HNRNPLL) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00146 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Paired box protein Pax-6 (PAX6) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00149 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Cyclin dependent kinase 13 (CDK13) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | THZ531 | |
| Crosstalk ID: M6ACROT00154 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | microRNA 26a-1 (MIR26A1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00155 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Cellular tumor antigen p53 (TP53/p53) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | Contusugene ladenovec | |
| Crosstalk ID: M6ACROT00156 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Integrin subunit alpha 2 (ITGA2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | E7820 | |
| Crosstalk ID: M6ACROT00157 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | HIF1A antisense RNA 2 (HIF1A-AS2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00158 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Cytochrome P450 2B6 (CYP2B6) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00159 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Cytochrome P450 2C8 (CYP2C8) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00160 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Cytochrome P450 family 3 subfamily A member 4 (CYP3A4) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | Curcumin | |
| Crosstalk ID: M6ACROT00161 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Hepatocyte nuclear factor 4 alpha (HNF4A) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | BI6015 | |
| Crosstalk ID: M6ACROT00168 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | CXADR cell adhesion molecule (CXADR) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00171 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Endoribonuclease Dicer (DICER1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00172 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | microRNA 155 (MIR155) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00173 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | microRNA 181a-1 (MIR181A1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00174 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | hsa-mir-18a | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00175 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | microRNA 19a (MIR19A) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00176 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | microRNA 210 (MIR210) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00177 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | POU domain, class 5, transcription factor 1 (POU5F1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00178 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Transcription factor SOX-2 (SOX2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00179 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | G1/S-specific cyclin-D2 (CCND2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00180 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Catenin beta-1 (CTNNB1/Beta-catenin) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | Recombinant human endostatin | |
| Crosstalk ID: M6ACROT00181 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Myc proto-oncogene protein (MYC) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | AVI-5126 | |
| Crosstalk ID: M6ACROT00182 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Transcription factor 4 (TCF4) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00183 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | microRNA 149 (MIR149) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00186 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | microRNA 200b (MIR200B) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00187 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Interferon induced protein with tetratricopeptide repeats 3 (IFIT3) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00188 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Interferon regulatory factor 9 (IRF9) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00189 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | microRNA 302a (MIR302A) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00190 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | microRNA 302b (MIR302B) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00191 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | microRNA 302c (MIR302C) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00192 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Transcription factor ISGF-3 components p91/p84 (Stat1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | Peptide analog 8 | |
| Crosstalk ID: M6ACROT00193 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Mitogen-activated protein kinase 1 (MAPK/ERK2/MAPK1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | BVD-523 | |
| Crosstalk ID: M6ACROT00194 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | APCDD1L divergent transcript (APCDD1L-DT) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00195 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | H1-10 antisense RNA 1 (H1-10-AS1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00196 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Long intergenic non-protein coding RNA 944 (LINC00944) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00197 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Long intergenic non-protein coding RNA 1003 (LINC01003) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00198 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Neurotrophic factor BDNF precursor form (BDNF) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00199 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | microRNA 432 (MIR432) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00200 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Nuclear receptor subfamily 1 group I member 2 (NR1I2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | Pregnenolone-16alpha-carbonitrile | |
| Crosstalk ID: M6ACROT00203 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | G1/S-specific cyclin-E1 (CCNE1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | PD-0183812 | |
| Crosstalk ID: M6ACROT00204 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Cyclin-dependent kinase 2 (CDK2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | PHA848125 | |
| Crosstalk ID: M6ACROT00205 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Cyclin-dependent kinase 4 (CDK4) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | Palbociclib | |
| Crosstalk ID: M6ACROT00206 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Cyclin-dependent kinase 6 (CDK6) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | Palbociclib | |
| Crosstalk ID: M6ACROT00207 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Nuclear paraspeckle assembly transcript 1 (NEAT1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00208 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | C-X3-C motif chemokine ligand 1 (CX3CL1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00209 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | hsa_circ_0000418 (Circ_CCT2) | |
| Crosstalk relationship | m6A → A-to-I | |
m6A Target: Rho GTPase activating protein 5 (ARHGAP5)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00065 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | A-to-I → m6A | |
| Disease | Breast cancer | |
m6A Target: Transcription factor SOX-9 (SOX9)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00067 | ||
| Epigenetic Regulator | 5-methylcytosine rRNA methyltransferase NSUN4 (NSUN4) | |
| Regulated Target | Transcription factor SOX-9 (SOX9) | |
| Crosstalk relationship | m5C → m6A | |
m6A Target: Cellular tumor antigen p53 (TP53/p53)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00069 | ||
| Epigenetic Regulator | tRNA (cytosine(72)-C(5))-methyltransferase NSUN6 (NSUN6) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m5C → m6A | |
| Disease | Colon cancer | |
| Drug | Contusugene ladenovec | |
| Crosstalk ID: M6ACROT00432 | ||
| Epigenetic Regulator | Methyltransferase-like protein 1 (METTL1) | |
| Regulated Target | MicroRNA 125a (MIR125A) | |
| Crosstalk relationship | m7G → m6A | |
| Crosstalk ID: M6ACROT00437 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 214 (MIR214) | |
| Crosstalk relationship | A-to-I → m6A | |
m6A Target: Suppressor of cytokine signaling 2 (SOCS2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00070 | ||
| Epigenetic Regulator | tRNA (cytosine(72)-C(5))-methyltransferase NSUN6 (NSUN6) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m5C → m6A | |
| Disease | Colon cancer | |
m6A Target: Cell division cycle-associated protein 7 (CDCA7)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00071 | ||
| Epigenetic Regulator | tRNA (cytosine(72)-C(5))-methyltransferase NSUN6 (NSUN6) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m5C → m6A | |
| Disease | Colon cancer | |
m6A Target: Double-stranded RNA-specific editase 1 (ADARB1)
| In total 27 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00081 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Filamin-A (FLNA) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | PTI-125 | |
| Crosstalk ID: M6ACROT00083 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Glutamate ionotropic receptor AMPA type subunit 2 (GRIA2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | E-2007 | |
| Crosstalk ID: M6ACROT00085 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Cytoplasmic FMR1 interacting protein 2 (CYFIP2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00089 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Cell division cycle 14B (CDC14B) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00093 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Zinc finger protein GLI1 (GLI1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | PMID26666870-Compound-16 | |
| Crosstalk ID: M6ACROT00095 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | microRNA 16-1 (MIR16-1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00098 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | microRNA 122 (MIR122) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00100 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | microRNA 214 (MIR214) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00102 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Oligophrenin 1 (OPHN1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00104 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | microRNA 21 (MIR21) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00106 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | microRNA 221 (MIR221) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00108 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | microRNA 222 (MIR222) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00118 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Podocalyxin like (PODXL) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00121 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Insulin-like growth factor-binding protein 7 (IGFBP7) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00133 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Thiol methyltransferase 1A (TMT1A) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00139 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Pellino E3 ubiquitin protein ligase 1 (PELI1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00141 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Protein sprouty homolog 2 (SPRY2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00144 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Heterogeneous nuclear ribonucleoprotein L like (HNRNPLL) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00147 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | microRNA 142 (MIR142) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00150 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Protein SON | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00152 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Glutamate ionotropic receptor kainate type subunit 2 (GRIK2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | 2,4-epi-neodysiherbaine | |
| Crosstalk ID: M6ACROT00162 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Cytochrome P450 2B6 (CYP2B6) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00164 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Cytochrome P450 2C8 (CYP2C8) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00166 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Hepatocyte nuclear factor 4 alpha (HNF4A) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | BI6015 | |
| Crosstalk ID: M6ACROT00169 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | CXADR cell adhesion molecule (CXADR) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00184 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Farnesyltransferase, CAAX box, subunit alpha (FNTA) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00201 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Nuclear receptor subfamily 1 group I member 2 (NR1I2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | Pregnenolone-16alpha-carbonitrile | |
m6A Target: Mutated in multiple advanced cancers 1 (PTEN)
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00411 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00412 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00447 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 214 (MIR214) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00511 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | hsa-mir-18a | |
| Crosstalk relationship | A-to-I → m6A | |
m6A Target: X inactive specific transcript (XIST)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00421 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00422 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | m6A → A-to-I | |
m6A Target: Maternally expressed 3 (MEG3)
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00429 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00430 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00481 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 155 (MIR155) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00515 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | hsa-mir-18a | |
| Crosstalk relationship | m6A → A-to-I | |
m6A Target: Epidermal growth factor receptor (EGFR)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00441 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 214 (MIR214) | |
| Crosstalk relationship | A-to-I → m6A | |
m6A Target: Macrophage colony-stimulating factor 1 (CSF1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00461 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 149 (MIR149) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00462 | ||
| Epigenetic Regulator | Methyltransferase-like protein 1 (METTL1) | |
| Regulated Target | MicroRNA 149 (MIR149) | |
| Crosstalk relationship | m7G → m6A | |
m6A Target: Stromal cell-derived factor 1 (CXCL12)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00469 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 149 (MIR149) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00470 | ||
| Epigenetic Regulator | Methyltransferase-like protein 1 (METTL1) | |
| Regulated Target | MicroRNA 149 (MIR149) | |
| Crosstalk relationship | m7G → m6A | |
m6A Target: Zinc finger protein SNAI1 (SNAI1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00486 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | hsa-mir-22 | |
| Crosstalk relationship | m6Am → m6A | |
m6A Target: H19 imprinted maternally expressed transcript (H19)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00488 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | hsa-mir-22 | |
| Crosstalk relationship | m6A → m6Am | |
| Crosstalk ID: M6ACROT00531 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 19a (MIR19A) | |
| Crosstalk relationship | m6A → A-to-I | |
m6A Target: Glucose transporter type 1 (GLUT1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00491 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | hsa-mir-22 | |
| Crosstalk relationship | m6Am → m6A | |
m6A Target: Cyclin-dependent kinase inhibitor 1B (CDKN1B/p27)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00499 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 222 (MIR222) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00500 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 222 (MIR222) | |
| Crosstalk relationship | A-to-I → m6A | |
m6A Target: Forkhead box protein O3 (FOXO3)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00521 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 221 (MIR221) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00556 | ||
| Epigenetic Regulator | Y-box-binding protein 1 (YBX1) | |
| Regulated Target | Growth arrest specific 5 (GAS5) | |
| Crosstalk relationship | m5C → m6A | |
m6A Target: Hypoxia-inducible factor 1-alpha (HIF-1-Alpha/HIF1A)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00524 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 142 (MIR142) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00528 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | MicroRNA 576 (MIR576) | |
| Crosstalk relationship | m6Am → m6A | |
m6A Target: Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00533 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 26a-1 (MIR26A1) | |
| Crosstalk relationship | m6A → A-to-I | |
m6A Target: Fascin (FSCN1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00540 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 200b (MIR200B) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00541 | ||
| Epigenetic Regulator | Y-box-binding protein 1 (YBX1) | |
| Regulated Target | MicroRNA 200b (MIR200B) | |
| Crosstalk relationship | m5C → m6A | |
m6A Target: Transcription factor SOX-6 (SOX6)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00553 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 122 (MIR122) | |
| Crosstalk relationship | A-to-I → m6A | |
m6A Target: pri-miR-143
| In total 5 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00566 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Signal transducer and activator of transcription 3 (STAT3) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00569 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | Signal transducer and activator of transcription 3 (STAT3) | |
| Crosstalk relationship | m6A → m6Am | |
| Crosstalk ID: M6ACROT00572 | ||
| Epigenetic Regulator | Y-box-binding protein 1 (YBX1) | |
| Regulated Target | Hypoxia-inducible factor 1-alpha (HIF-1-Alpha/HIF1A) | |
| Crosstalk relationship | m6A → m5C | |
| Crosstalk ID: M6ACROT00575 | ||
| Epigenetic Regulator | H/ACA ribonucleoprotein complex subunit DKC1 (DKC1) | |
| Regulated Target | Hypoxia-inducible factor 1-alpha (HIF-1-Alpha/HIF1A) | |
| Crosstalk relationship | m6A → Pseudouridine | |
| Crosstalk ID: M6ACROT00578 | ||
| Epigenetic Regulator | RNA cytosine C(5)-methyltransferase NSUN2 (NSUN2) | |
| Regulated Target | H19 imprinted maternally expressed transcript (H19) | |
| Crosstalk relationship | m5C → m6A | |
m6A Target: hsa-miR-193b
| In total 5 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00584 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | G1/S-specific cyclin-D1 (CCND1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00585 | ||
| Epigenetic Regulator | Y-box-binding protein 1 (YBX1) | |
| Regulated Target | G1/S-specific cyclin-D1 (CCND1) | |
| Crosstalk relationship | m6A → m5C | |
| Crosstalk ID: M6ACROT00586 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | G1/S-specific cyclin-D1 (CCND1) | |
| Crosstalk relationship | m6A → m5C | |
| Crosstalk ID: M6ACROT00587 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | G1/S-specific cyclin-D1 (CCND1) | |
| Crosstalk relationship | m6A → m6Am | |
| Crosstalk ID: M6ACROT00588 | ||
| Epigenetic Regulator | Methyltransferase-like protein 1 (METTL1) | |
| Regulated Target | G1/S-specific cyclin-D1 (CCND1) | |
| Crosstalk relationship | m6A → m7G | |
m6A Target: microRNA 21 (MIR21)
| In total 7 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00590 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Protein sprouty homolog 2 (SPRY2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00592 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Protein sprouty homolog 2 (SPRY2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00594 | ||
| Epigenetic Regulator | Y-box-binding protein 1 (YBX1) | |
| Regulated Target | Growth arrest specific 5 (GAS5) | |
| Crosstalk relationship | m5C → m6A | |
| Crosstalk ID: M6ACROT00596 | ||
| Epigenetic Regulator | Putative methyltransferase NSUN7 (NSUN7) | |
| Regulated Target | Phosphofructokinase, muscle (PFKM) | |
| Crosstalk relationship | m6A → m5C | |
| Crosstalk ID: M6ACROT00598 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → m5C | |
| Crosstalk ID: M6ACROT00600 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET2 (TET2) | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → m5C | |
| Crosstalk ID: M6ACROT00602 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET3 (TET3) | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → m5C | |
m6A Target: hsa-mir-320a
| In total 8 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00603 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Catenin beta-1 (CTNNB1/Beta-catenin) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00604 | ||
| Epigenetic Regulator | Y-box-binding protein 1 (YBX1) | |
| Regulated Target | Catenin beta-1 (CTNNB1/Beta-catenin) | |
| Crosstalk relationship | m6A → m5C | |
| Crosstalk ID: M6ACROT00605 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | Catenin beta-1 (CTNNB1/Beta-catenin) | |
| Crosstalk relationship | m6A → m6Am | |
| Crosstalk ID: M6ACROT00606 | ||
| Epigenetic Regulator | Y-box-binding protein 1 (YBX1) | |
| Regulated Target | Homeobox protein NANOG (NANOG) | |
| Crosstalk relationship | m6A → m5C | |
| Crosstalk ID: M6ACROT00607 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | Homeobox protein NANOG (NANOG) | |
| Crosstalk relationship | m6A → m6Am | |
| Crosstalk ID: M6ACROT00608 | ||
| Epigenetic Regulator | Methyltransferase-like protein 1 (METTL1) | |
| Regulated Target | Homeobox protein NANOG (NANOG) | |
| Crosstalk relationship | m6A → m7G | |
| Crosstalk ID: M6ACROT00609 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | Transcription factor E2F1 (E2F1) | |
| Crosstalk relationship | m6A → m6Am | |
| Crosstalk ID: M6ACROT00610 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Crosstalk relationship | m6Am → m6A | |
m6A Target: microRNA 222 (MIR222)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00636 | ||
| Epigenetic Regulator | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 3 (ALKBH3) | |
| Regulated Target | Cyclin-dependent kinase inhibitor 1B (CDKN1B/p27) | |
| Crosstalk relationship | m6A → m1A | |
| Crosstalk ID: M6ACROT00637 | ||
| Epigenetic Regulator | RNA cytosine C(5)-methyltransferase NSUN2 (NSUN2) | |
| Regulated Target | Cyclin-dependent kinase inhibitor 1B (CDKN1B/p27) | |
| Crosstalk relationship | m6A → m5C | |
| Crosstalk ID: M6ACROT00638 | ||
| Epigenetic Regulator | Methyltransferase-like protein 1 (METTL1) | |
| Regulated Target | Cyclin-dependent kinase inhibitor 1B (CDKN1B/p27) | |
| Crosstalk relationship | m6A → m7G | |
m6A Target: microRNA 25 (MIR25)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00639 | ||
| Epigenetic Regulator | Y-box-binding protein 1 (YBX1) | |
| Regulated Target | Zinc finger protein SNAI1 (SNAI1) | |
| Crosstalk relationship | m6A → m5C | |
m6A Target: hsa-mir-18a
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00640 | ||
| Epigenetic Regulator | Y-box-binding protein 1 (YBX1) | |
| Regulated Target | Growth arrest specific 5 (GAS5) | |
| Crosstalk relationship | m5C → m6A | |
| Crosstalk ID: M6ACROT00642 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → m5C | |
| Crosstalk ID: M6ACROT00644 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET2 (TET2) | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → m5C | |
| Crosstalk ID: M6ACROT00646 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET3 (TET3) | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → m5C | |
m6A Target: microRNA 221 (MIR221)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00648 | ||
| Epigenetic Regulator | Y-box-binding protein 1 (YBX1) | |
| Regulated Target | Growth arrest specific 5 (GAS5) | |
| Crosstalk relationship | m5C → m6A | |
| Crosstalk ID: M6ACROT00649 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | NAD-dependent protein deacetylase sirtuin-1 (SIRT1) | |
| Crosstalk relationship | m6A → m6Am | |
| Crosstalk ID: M6ACROT00650 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | Protein c-Fos (FOS) | |
| Crosstalk relationship | m6A → m6Am | |
m6A Target: microRNA 1246 (MIR1246)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00663 | ||
| Epigenetic Regulator | Y-box-binding protein 1 (YBX1) | |
| Regulated Target | Glycogen synthase kinase-3 beta (GSK3Beta/GSK3B) | |
| Crosstalk relationship | m6A → m5C | |
| Crosstalk ID: M6ACROT00664 | ||
| Epigenetic Regulator | tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4 (WDR4) | |
| Regulated Target | Interleukin 6 (IL6) | |
| Crosstalk relationship | m6A → m7G | |
m6A Target: hsa-mir-20a
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00665 | ||
| Epigenetic Regulator | Y-box-binding protein 1 (YBX1) | |
| Regulated Target | Ribonucleotide reductase regulatory subunit M2 (RRM2) | |
| Crosstalk relationship | m6A → m5C | |
| Crosstalk ID: M6ACROT00667 | ||
| Epigenetic Regulator | Y-box-binding protein 1 (YBX1) | |
| Regulated Target | 72 kDa type IV collagenase (MMP2) | |
| Crosstalk relationship | m6A → m5C | |
| Crosstalk ID: M6ACROT00669 | ||
| Epigenetic Regulator | H/ACA ribonucleoprotein complex subunit DKC1 (DKC1) | |
| Regulated Target | 72 kDa type IV collagenase (MMP2) | |
| Crosstalk relationship | m6A → Pseudouridine | |
| Crosstalk ID: M6ACROT00671 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Crosstalk relationship | m6Am → m6A | |
m6A Target: hsa-mir-19a
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00674 | ||
| Epigenetic Regulator | RNA cytosine C(5)-methyltransferase NSUN2 (NSUN2) | |
| Regulated Target | H19 imprinted maternally expressed transcript (H19) | |
| Crosstalk relationship | m5C → m6A | |
m6A Target: hsa-mir-19b-1
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00675 | ||
| Epigenetic Regulator | RNA cytosine C(5)-methyltransferase NSUN2 (NSUN2) | |
| Regulated Target | H19 imprinted maternally expressed transcript (H19) | |
| Crosstalk relationship | m5C → m6A | |
| Crosstalk ID: M6ACROT00676 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | NAD-dependent protein deacetylase sirtuin-1 (SIRT1) | |
| Crosstalk relationship | m6A → m6Am | |
m6A Target: microRNA-92a
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00679 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | Krueppel-like factor 4 (KLF4) | |
| Crosstalk relationship | m6A → m6Am | |
| Crosstalk ID: M6ACROT00680 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | Integrin alpha-6 (ITGA6) | |
| Crosstalk relationship | m6A → m6Am | |
DNA modification
m6A Target: ANKRD13C divergent transcript (ANKRD13C-DT)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02007 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | Long Terminal Repeat 7 (LTR7) | |
| Crosstalk relationship | m6A → DNA modification | |
m6A Target: DNA-binding protein SATB2 (SATB2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02008 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | DNA-binding protein SATB2 (SATB2) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Esophageal Squamous Cell Carcinoma | |
m6A Target: Methylcytosine dioxygenase TET1 (TET1)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02010 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m6A → DNA modification | |
| Crosstalk ID: M6ACROT02012 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Crosstalk ID: M6ACROT02094 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | Signal transducer and activator of transcription 3 (STAT3) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Inflammatory pain | |
m6A Target: Semaphorin-3F (SEMA3F)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02014 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Semaphorin-3F (SEMA3F) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Prostate cancer | |
| Drug | Docetaxel | |
m6A Target: microRNA 25 (MIR25)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02030 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT02031 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
m6A Target: Insulin-like Growth Factor Binding Protein 7 Overlapping Transcript
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02040 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Insulin-like growth factor-binding protein 7 (IGFBP7) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Oosteoarthritis | |
| Crosstalk ID: M6ACROT02041 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Insulin-like growth factor-binding protein 7 (IGFBP7) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Oosteoarthritis | |
m6A Target: Protein mono-ADP-ribosyltransferase PARP10 (PARP10)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02042 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Cardiomegaly | |
| Drug | Ang II | |
m6A Target: DNA (cytosine-5)-methyltransferase 1 (DNMT1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02043 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Forkhead box protein O3 (FOXO3) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT06013 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Eomesodermin homolog (EOMES) | |
| Crosstalk relationship | m6A → DNA modification | |
m6A Target: Neurogenic locus notch homolog protein 2 (NOTCH2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02044 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Neurogenic locus notch homolog protein 2 (NOTCH2) | |
| Crosstalk relationship | m6A → DNA modification | |
m6A Target: Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02047 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Secreted frizzled-related protein 2 (SFRP2) | |
| Crosstalk relationship | m6A → DNA modification | |
| Drug | Simvastatin | |
| Crosstalk ID: M6ACROT02048 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Secreted frizzled-related protein 2 (SFRP2) | |
| Crosstalk relationship | m6A → DNA modification | |
| Drug | Simvastatin | |
| Crosstalk ID: M6ACROT02049 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Secreted frizzled-related protein 2 (SFRP2) | |
| Crosstalk relationship | m6A → DNA modification | |
| Drug | Simvastatin | |
m6A Target: microRNA let-7b (MIRLET7B)
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02058 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02061 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02064 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT02067 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
m6A Target: Cysteine methyltransferase DNMT3A (DNMT3A)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02076 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Alpha tubulin acetyltransferase 1 (ATAT1) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Alzheimer disease | |
| Crosstalk ID: M6ACROT02106 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Gastric cancer | |
| Drug | Decitabine | |
m6A Target: Glucocorticoid receptor (NR3C1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02086 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Major depressive disorder | |
| Crosstalk ID: M6ACROT02090 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Major depressive disorder | |
m6A Target: Cyclic AMP-responsive element-binding protein 1 (CREB1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02087 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Major depressive disorder | |
| Crosstalk ID: M6ACROT02091 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Major depressive disorder | |
m6A Target: BDNF/NT-3 growth factors receptor (NTRK2)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02088 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Major depressive disorder | |
| Crosstalk ID: M6ACROT02092 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Major depressive disorder | |
m6A Target: Neurotrophic factor BDNF precursor form (BDNF)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02089 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Major depressive disorder | |
| Crosstalk ID: M6ACROT02093 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Major depressive disorder | |
m6A Target: SREBF2 antisense RNA 1 (SREBF2-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02098 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | Sterol regulatory element binding transcription factor 2 (SREBF2) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Liver cancer | |
m6A Target: Phosphoenolpyruvate carboxykinase [GTP], mitochondrial (PCK2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02113 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET2 (TET2) | |
| Regulated Target | Phosphoenolpyruvate carboxykinase [GTP], mitochondrial (PCK2) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Hepatic inflammation | |
m6A Target: LIFR antisense RNA 1 (LIFR-AS1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02116 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT02125 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
m6A Target: hsa-miR-380-3p
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02117 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT02126 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
m6A Target: DBH antisense RNA 1 (Lnc_DBH-AS1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02118 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
| Drug | Gemcitabine | |
| Crosstalk ID: M6ACROT02127 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
| Drug | Gemcitabine | |
m6A Target: DNA-binding protein inhibitor ID-2 (ID2)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02119 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT02128 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
m6A Target: BRAF-activated non-protein coding RNA (BANCR)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02120 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT02129 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
m6A Target: long intergenic non-protein coding RNA 662 (LINC00662)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02121 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT02130 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
m6A Target: Claspin (CLSPN)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02122 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
| Drug | Celastrol | |
| Crosstalk ID: M6ACROT02131 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
| Drug | Celastrol | |
m6A Target: Apoptosis regulator Bcl-2 (BCL2)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02123 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
| Drug | Celastrol | |
| Crosstalk ID: M6ACROT02132 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
| Drug | Celastrol | |
m6A Target: Integrin beta-1 (ITGB1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02124 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT02133 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
m6A Target: PI3-kinase subunit alpha (PI3k/PIK3CA)
| In total 8 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02141 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02149 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02154 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02162 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02167 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT02175 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT02180 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT02188 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
m6A Target: hsa-miR-143-3p
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02142 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02155 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02168 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT02181 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
m6A Target: Transcription factor JunB (JUNB)
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02143 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02156 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02169 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT02182 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
m6A Target: Histone-lysine N-methyltransferase EZH2 (EZH2)
| In total 6 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02145 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02158 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02171 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT02184 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT05854 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Simvastatin | |
| Crosstalk ID: M6ACROT05856 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Simvastatin | |
m6A Target: Hepatocyte growth factor receptor (c-Met/MET)
| In total 6 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02146 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02159 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02172 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT02185 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT05855 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT05857 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
m6A Target: Zinc finger and BTB domain-containing protein 4 (ZBTB4)
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02147 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02160 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02173 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT02186 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
m6A Target: Mutated in multiple advanced cancers 1 (PTEN)
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02148 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02161 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02174 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT02187 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
m6A Target: RAC-alpha serine/threonine-protein kinase (AKT1)
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02150 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02163 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02176 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT02189 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
m6A Target: Serine/threonine-protein kinase mTOR (MTOR)
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02151 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02164 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02177 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT02190 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
m6A Target: SVIL antisense RNA 1 (SVIL-AS1)
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02152 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02165 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02178 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT02191 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
m6A Target: SH3 domain-binding protein 5 (SH3BP5)
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02153 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02166 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02179 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT02192 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
m6A Target: Ribose-5-phosphate isomerase (RPIA)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT06003 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | Ribose-5-phosphate isomerase (RPIA) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Esophageal Squamous Cell Carcinoma | |
m6A Target: Wnt family member 7B (WNT7B)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT06005 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | Wnt family member 7B (WNT7B) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Esophageal Squamous Cell Carcinoma | |
m6A Target: B-cell lymphoma 6 protein (BCL6)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT06007 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | B-cell lymphoma 6 protein (BCL6) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Esophageal Squamous Cell Carcinoma | |
m6A Target: Protocadherin Fat 4 (FAT4)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT06009 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | Protocadherin Fat 4 (FAT4) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Esophageal Squamous Cell Carcinoma | |
m6A Target: Sterile alpha motif domain-containing protein 9-like (SAMD9L)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT06011 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | Sterile alpha motif domain-containing protein 9-like (SAMD9L) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Esophageal Squamous Cell Carcinoma | |
m6A Target: Insulin receptor (INSR)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT06015 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Insulin receptor (INSR) | |
| Crosstalk relationship | m6A → DNA modification | |
m6A Target: Mothers against decapentaplegic homolog 3 (SMAD3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT06017 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Mothers against decapentaplegic homolog 3 (SMAD3) | |
| Crosstalk relationship | m6A → DNA modification | |
Histone modification
m6A Target: Histone-lysine N-methyltransferase EZH2 (EZH2)
| In total 5 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03013 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Crosstalk ID: M6ACROT03038 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Brain cancer | |
| Drug | Temozolomide | |
| Crosstalk ID: M6ACROT03039 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
| Drug | Temozolomide | |
| Crosstalk ID: M6ACROT03381 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Simvastatin* | |
| Crosstalk ID: M6ACROT03394 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Simvastatin* | |
m6A Target: Lysine-specific demethylase 6B (KDM6B)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03015 | ||
| Epigenetic Regulator | Lysine-specific demethylase 6B (KDM6B) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Inflammatory response | |
| Crosstalk ID: M6ACROT03239 | ||
| Epigenetic Regulator | Lysine-specific demethylase 6B (KDM6B) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Crosstalk ID: M6ACROT05996 | ||
| Epigenetic Regulator | Lysine-specific demethylase 6B (KDM6B) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Inflammatory response | |
m6A Target: Interleukin-6 (IL-6)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03018 | ||
| Epigenetic Regulator | Lysine-specific demethylase 6B (KDM6B) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Inflammatory response | |
m6A Target: Histone-lysine N-methyltransferase SETD1A (SETD1A)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03030 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase SETD1A (SETD1A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | m6A → Histone modification | |
m6A Target: Histone-lysine N-methyltransferase SETD1B (SETD1B)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03031 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase SETD1B (SETD1B) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | m6A → Histone modification | |
m6A Target: Histone-lysine N-methyltransferase 2D (KMT2D)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03032 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase 2D (KMT2D) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | m6A → Histone modification | |
m6A Target: Histone-lysine N-methyltransferase SETD2 (SETD2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03042 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase SETD2 (SETD2) | |
| Regulated Target | Histone H3 lysine 36 trimethylation (H3K36me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Lung cancer | |
m6A Target: Bone morphogenetic protein 2 (BMP2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03044 | ||
| Epigenetic Regulator | Lactate dehydrogenase A (LDHA) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Ossification of spinal ligaments | |
m6A Target: Long-chain-fatty-acid--CoA ligase 4 (ACSL4)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03058 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Sepsis-associated lung injury | |
| Crosstalk ID: M6ACROT03059 | ||
| Epigenetic Regulator | CREB-binding protein (CREBBP) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Sepsis-associated lung injury | |
m6A Target: miR-503
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03071 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase 2A (KMT2A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Myocardial injury | |
| Drug | C646 | |
m6A Target: Suppressor of cytokine signaling 2 (SOCS2)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03072 | ||
| Epigenetic Regulator | Probable JmjC domain-containing histone demethylation protein 2C (JMJD1C) | |
| Regulated Target | Histone H3 lysine 9 monomethylation (H3K9me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
| Crosstalk ID: M6ACROT03659 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Acyl-CoA synthetase short-chain family member 3, mitochondrial (ACSS3)
| In total 9 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03077 | ||
| Regulated Target | Histone H3 lysine 14 propionylation (H3K14pr) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Nonalcoholic fatty liver disease | |
| Crosstalk ID: M6ACROT03078 | ||
| Regulated Target | Histone H3 lysine 23 propionylation (H3k23pr) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Nonalcoholic fatty liver disease | |
| Crosstalk ID: M6ACROT03481 | ||
| Epigenetic Regulator | Lactate dehydrogenase A (LDHA) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18lac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Nonalcoholic fatty liver disease | |
| Crosstalk ID: M6ACROT05838 | ||
| Regulated Target | Histone H3 lysine 14 propionylation (H3K14pr) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Nonalcoholic fatty liver disease | |
| Crosstalk ID: M6ACROT05839 | ||
| Regulated Target | Histone H3 lysine 14 propionylation (H3K14pr) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Nonalcoholic fatty liver disease | |
| Crosstalk ID: M6ACROT05840 | ||
| Regulated Target | Histone H3 lysine 14 propionylation (H3K14pr) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Nonalcoholic fatty liver disease | |
| Crosstalk ID: M6ACROT05841 | ||
| Regulated Target | Histone H3 lysine 23 propionylation (H3k23pr) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Nonalcoholic fatty liver disease | |
| Crosstalk ID: M6ACROT05842 | ||
| Regulated Target | Histone H3 lysine 23 propionylation (H3k23pr) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Nonalcoholic fatty liver disease | |
| Crosstalk ID: M6ACROT05843 | ||
| Regulated Target | Histone H3 lysine 23 propionylation (H3k23pr) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Nonalcoholic fatty liver disease | |
m6A Target: pri-rRNA
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03096 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase SUV39H1 (SUV39H1) | |
| Regulated Target | Histone H3 lysine 9 trimethylation (H3K9me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Crosstalk ID: M6ACROT03097 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase SUV39H2 (SUV39H2) | |
| Regulated Target | Histone H3 lysine 9 trimethylation (H3K9me3) | |
| Crosstalk relationship | m6A → Histone modification | |
m6A Target: Myc proto-oncogene protein (MYC)
| In total 6 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03106 | ||
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Esophageal Squamous Cell Carcinoma | |
| Crosstalk ID: M6ACROT03550 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03595 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03648 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
| Crosstalk ID: M6ACROT03651 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
| Crosstalk ID: M6ACROT03660 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: NACHT, LRR and PYD domains-containing protein 3 (NLRP3)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03107 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03109 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
m6A Target: Histone-lysine N-methyltransferase SETMAR (SETMAR)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03111 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase SETMAR (SETMAR) | |
| Regulated Target | Histone H3 lysine 36 dimethylation (H3K36me2) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Thyroid Cancer | |
| Crosstalk ID: M6ACROT03113 | ||
| Epigenetic Regulator | Probable global transcription activator SNF2L2 (SMARCA2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Thyroid Cancer | |
| Crosstalk ID: M6ACROT05848 | ||
| Epigenetic Regulator | Probable global transcription activator SNF2L2 (SMARCA2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Thyroid Cancer | |
m6A Target: Stearoyl-CoA desaturase (SCD)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03134 | ||
| Epigenetic Regulator | Lactate dehydrogenase A (LDHA) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18lac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Nonalcoholic fatty liver disease | |
m6A Target: Nuclear paraspeckle assembly transcript 1 (NEAT1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03141 | ||
| Epigenetic Regulator | Chromodomain-helicase-DNA-binding protein 4 (CHD4) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | m6A → Histone modification | |
m6A Target: Beclin-1 (BECN1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03143 | ||
| Epigenetic Regulator | Histone acetyltransferase KAT2A (KAT2A) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
m6A Target: Histone deacetylase 1 (HDAC1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03144 | ||
| Epigenetic Regulator | Histone deacetylase 1 (HDAC1) | |
| Crosstalk relationship | m6A → Histone modification | |
m6A Target: Histone acetyltransferase KAT2A (KAT2A)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03147 | ||
| Epigenetic Regulator | Histone acetyltransferase KAT2A (KAT2A) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Non-small cell lung cancer | |
m6A Target: Protein deacetylase HDAC6 (HDAC6)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03148 | ||
| Epigenetic Regulator | Protein deacetylase HDAC6 (HDAC6) | |
| Regulated Target | Histone acetylation | |
| Crosstalk relationship | m6A → Histone modification | |
m6A Target: Programmed cell death 1 ligand 1 (CD274/PD-L1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03149 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EHMT2 (EHMT2) | |
| Regulated Target | Histone H3 lysine 9 dimethylation (H3K9me2) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Inflammatory response | |
m6A Target: CX3C chemokine receptor 1 (CX3CR1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03150 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EHMT2 (EHMT2) | |
| Regulated Target | Histone H3 lysine 9 dimethylation (H3K9me2) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Inflammatory response | |
m6A Target: Caspase-1 (CASP1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03151 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EHMT2 (EHMT2) | |
| Regulated Target | Histone H3 lysine 9 dimethylation (H3K9me2) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Inflammatory response | |
m6A Target: Cysteine protease ATG4A (ATG4A)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03160 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase 2A (KMT2A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Intervertebral disc degeneration | |
m6A Target: Polycomb protein EED (EED)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03165 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | m6A → Histone modification | |
m6A Target: Thioredoxin domain-containing protein 5 (TXNDC5)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03169 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03170 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
m6A Target: E3 ubiquitin-protein ligase CHIP (STUB1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03177 | ||
| Epigenetic Regulator | Lysine-specific histone demethylase 1A (KDM1A) | |
| Regulated Target | Histone H3 lysine 9 trimethylation (H3K9me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Alzheimer disease | |
m6A Target: Methylcytosine dioxygenase TET1 (TET1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03182 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Crosstalk ID: M6ACROT03183 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
m6A Target: Histone deacetylase 9 (HDAC9)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03187 | ||
| Epigenetic Regulator | Histone deacetylase 9 (HDAC9) | |
| Regulated Target | Histone H4 lysine 12 acetylation (H4K12ac) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Acute laryngitis or tracheitis | |
m6A Target: Histone-lysine N-methyltransferase SUV39H2 (SUV39H2)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03190 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase SUV39H2 (SUV39H2) | |
| Regulated Target | Histone H3 lysine 9 trimethylation (H3K9me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Gastric cancer | |
| Drug | Cisplatin | |
| Crosstalk ID: M6ACROT03664 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Histone-lysine N-methyltransferase ASH1L (ASH1L)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03204 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase ASH1L (ASH1L) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Uveitis | |
| Crosstalk ID: M6ACROT05851 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase ASH1L (ASH1L) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Uveitis | |
m6A Target: Histone-lysine N-methyltransferase NSD2 (NSD2)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03209 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase NSD2 (NSD2) | |
| Regulated Target | Histone H3 lysine 36 dimethylation (H3K36me2) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Diabetic nephropathy | |
| Crosstalk ID: M6ACROT03211 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase NSD2 (NSD2) | |
| Regulated Target | Histone H3 lysine 36 trimethylation (H3K36me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Diabetic nephropathy | |
m6A Target: Spectrin beta, non-erythrocytic 2 (SPTBN2)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03222 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03223 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Lnc668
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03225 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 9 lactylation (H3K9la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pulmonary fibrosis | |
m6A Target: CUB domain-containing protein 1 (CDCP1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03228 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Pirin (PIR)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03233 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Melanoma of uvea | |
m6A Target: Chromobox protein homolog 8 (CBX8)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03249 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase 2B (KMT2B) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Colon cancer | |
m6A Target: NUTM2B antisense RNA 1 (NUTM2B-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03251 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase 2A (KMT2A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Liver cancer | |
m6A Target: Histone deacetylase 5 (HDAC5)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03253 | ||
| Epigenetic Regulator | Histone deacetylase 5 (HDAC5) | |
| Regulated Target | Histone H3 lysine 9 acetylation (H3K9Ac) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Osteosarcoma | |
| Crosstalk ID: M6ACROT03254 | ||
| Epigenetic Regulator | Histone deacetylase 5 (HDAC5) | |
| Regulated Target | Histone H3 lysine 14 acetylation (H3K14ac) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Osteosarcoma | |
m6A Target: Transcription factor SOX-2 (SOX2)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03293 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
| Crosstalk ID: M6ACROT03542 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03587 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Serine/arginine-rich splicing factor 3 (SRSF3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03294 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: Serine/arginine-rich splicing factor 6 (SRSF6)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03295 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: Serine/arginine-rich splicing factor 11 (SRSF11)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03296 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: Methylated-DNA--protein-cysteine methyltransferase (MGMT)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03297 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
| Drug | Temozolomide | |
| Crosstalk ID: M6ACROT03301 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: DNA-3-methyladenine glycosylase (ANPG/MPG)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03298 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
| Drug | Temozolomide | |
| Crosstalk ID: M6ACROT03302 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: Meltrin-beta (ADAM19)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03299 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
| Drug | Ethyl ester form of meclofenamic acid | |
m6A Target: Interferon-inducible protein 4 (ADAR1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03300 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: Optineurin (OPTN)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03303 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: Aryl hydrocarbon receptor (AHR)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03304 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
| Drug | Tislelizumab | |
m6A Target: Serum response factor (SRF)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03305 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
| Drug | Tislelizumab | |
m6A Target: X inactive specific transcript (XIST)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03306 | ||
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Ossification of spinal ligaments | |
m6A Target: pri-miR-143
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03311 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase 2A (KMT2A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Acute myocardial infarction | |
m6A Target: Tenascin (TNC)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03312 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase 2A (KMT2A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Acute myocardial infarction | |
m6A Target: TNF receptor-associated factor 6 (TRAF6)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03313 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase 2A (KMT2A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Acute myocardial infarction | |
m6A Target: Dynamin-1-like protein (DRP1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03314 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase 2A (KMT2A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Acute myocardial infarction | |
m6A Target: Heat shock protein HSP 90-alpha (HSP90/HSP90AA1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03315 | ||
| Epigenetic Regulator | Probable JmjC domain-containing histone demethylation protein 2C (JMJD1C) | |
| Regulated Target | Histone H3 lysine 9 monomethylation (H3K9me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03316 | ||
| Epigenetic Regulator | Probable JmjC domain-containing histone demethylation protein 2C (JMJD1C) | |
| Regulated Target | Histone H3 lysine 9 monomethylation (H3K9me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: Circ_DLC1
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03317 | ||
| Epigenetic Regulator | Probable JmjC domain-containing histone demethylation protein 2C (JMJD1C) | |
| Regulated Target | Histone H3 lysine 9 monomethylation (H3K9me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: hsa-miR-671-5p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03318 | ||
| Epigenetic Regulator | Probable JmjC domain-containing histone demethylation protein 2C (JMJD1C) | |
| Regulated Target | Histone H3 lysine 9 monomethylation (H3K9me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: Beta-catenin-interacting protein 1 (CTNNBIP1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03319 | ||
| Epigenetic Regulator | Probable JmjC domain-containing histone demethylation protein 2C (JMJD1C) | |
| Regulated Target | Histone H3 lysine 9 monomethylation (H3K9me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: UBX domain-containing protein 1 (UBXN1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03320 | ||
| Epigenetic Regulator | Probable JmjC domain-containing histone demethylation protein 2C (JMJD1C) | |
| Regulated Target | Histone H3 lysine 9 monomethylation (H3K9me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: Nuclear factor NF-kappa-B p105 subunit (NF-Kappa-B/NFKB1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03321 | ||
| Epigenetic Regulator | Probable JmjC domain-containing histone demethylation protein 2C (JMJD1C) | |
| Regulated Target | Histone H3 lysine 9 monomethylation (H3K9me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: HOXA transcript antisense RNA, myeloid-specific 1 (HOTAIRM1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03322 | ||
| Epigenetic Regulator | Probable JmjC domain-containing histone demethylation protein 2C (JMJD1C) | |
| Regulated Target | Histone H3 lysine 9 monomethylation (H3K9me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: Circ_EPHB4
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03323 | ||
| Epigenetic Regulator | Probable JmjC domain-containing histone demethylation protein 2C (JMJD1C) | |
| Regulated Target | Histone H3 lysine 9 monomethylation (H3K9me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: Ferroptosis suppressor protein 1 (AIFM2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03324 | ||
| Epigenetic Regulator | Probable JmjC domain-containing histone demethylation protein 2C (JMJD1C) | |
| Regulated Target | Histone H3 lysine 9 monomethylation (H3K9me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
| Drug | iFSP1 | |
m6A Target: Oxidized low-density lipoprotein receptor 1 (OLR1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03325 | ||
| Epigenetic Regulator | Probable JmjC domain-containing histone demethylation protein 2C (JMJD1C) | |
| Regulated Target | Histone H3 lysine 9 monomethylation (H3K9me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
| Drug | Temozolomide | |
m6A Target: Guanine nucleotide-binding protein G (GNAO1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03326 | ||
| Epigenetic Regulator | Probable JmjC domain-containing histone demethylation protein 2C (JMJD1C) | |
| Regulated Target | Histone H3 lysine 9 monomethylation (H3K9me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
| Drug | Temozolomide | |
m6A Target: L-glutamine amidohydrolase (GLS2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03371 | ||
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Esophageal Squamous Cell Carcinoma | |
m6A Target: Cyclin-dependent kinase inhibitor 1 (CDKN1A)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03372 | ||
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Esophageal Squamous Cell Carcinoma | |
m6A Target: Neurogenic locus notch homolog protein 1 (NOTCH1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03373 | ||
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Esophageal Squamous Cell Carcinoma | |
m6A Target: Microprocessor complex subunit DGCR8 (DGCR8)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03374 | ||
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Esophageal Squamous Cell Carcinoma | |
m6A Target: hsa-miR-320b
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03375 | ||
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Esophageal Squamous Cell Carcinoma | |
m6A Target: Nuclear receptor subfamily 4 group A member 2 (NR4A2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03376 | ||
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Esophageal Squamous Cell Carcinoma | |
| Drug | celecoxib | |
m6A Target: PI3-kinase subunit alpha (PI3k/PIK3CA)
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03377 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03385 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03390 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03398 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
m6A Target: hsa-miR-143-3p
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03378 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03391 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
m6A Target: Transcription factor JunB (JUNB)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03379 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03392 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
m6A Target: microRNA let-7b (MIRLET7B)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03380 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT03393 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
m6A Target: Hepatocyte growth factor receptor (c-Met/MET)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03382 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03395 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
m6A Target: Zinc finger and BTB domain-containing protein 4 (ZBTB4)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03383 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03396 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
m6A Target: Mutated in multiple advanced cancers 1 (PTEN)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03384 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03397 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
m6A Target: RAC-alpha serine/threonine-protein kinase (AKT1)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03386 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03399 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03637 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Serine/threonine-protein kinase mTOR (MTOR)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03387 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03400 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
m6A Target: SVIL antisense RNA 1 (SVIL-AS1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03388 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03401 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
m6A Target: SH3 domain-binding protein 5 (SH3BP5)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03389 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03402 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
m6A Target: Protein SON
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03491 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EHMT2 (EHMT2) | |
| Regulated Target | Histone H3 lysine 9 dimethylation (H3K9me2) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Inflammatory response | |
m6A Target: Protein-tyrosine kinase 2-beta (PYK2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03492 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EHMT2 (EHMT2) | |
| Regulated Target | Histone H3 lysine 9 dimethylation (H3K9me2) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Inflammatory response | |
m6A Target: Toll-like receptor 2 (TLR2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03503 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase 2A (KMT2A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Intervertebral disc degeneration | |
m6A Target: ZNFX1 antisense RNA 1 (ZFAS1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03504 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03517 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
m6A Target: FOXD2 adjacent opposite strand RNA 1 (FOXD2-AS1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03505 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03518 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
m6A Target: Apoptotic chromatin condensation inducer in the nucleus (ACIN1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03506 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03519 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
m6A Target: Hexokinase-2 (HK2)
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03507 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03520 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03548 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03593 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: hsa-miR-193b
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03508 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03521 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
m6A Target: G1/S-specific cyclin-D1 (CCND1)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03509 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03522 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03639 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Aspartate--tRNA ligase, cytoplasmic (DARS)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03510 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03523 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
m6A Target: Procathepsin L (CTSL)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03511 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03524 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
m6A Target: Circ_RNF13
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03512 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03525 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
m6A Target: long intergenic non-protein coding RNA 426 (LINC00426)
| In total 6 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03513 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Drug | Cisplatin | |
| Crosstalk ID: M6ACROT03514 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Drug | Bleomycin | |
| Crosstalk ID: M6ACROT03515 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Drug | Imatinib | |
| Crosstalk ID: M6ACROT03526 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Drug | Cisplatin | |
| Crosstalk ID: M6ACROT03527 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Drug | Bleomycin | |
| Crosstalk ID: M6ACROT03528 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Drug | Imatinib | |
m6A Target: Serine/threonine-protein kinase Nek2 (NEK2)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03516 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03529 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
m6A Target: Nucleolar protein 3 (ARC)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03530 | ||
| Epigenetic Regulator | Lysine-specific histone demethylase 1A (KDM1A) | |
| Regulated Target | Histone H3 lysine 9 trimethylation (H3K9me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Alzheimer disease | |
m6A Target: Cysteine methyltransferase DNMT3A (DNMT3A)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03531 | ||
| Epigenetic Regulator | Lysine-specific histone demethylase 1A (KDM1A) | |
| Regulated Target | Histone H3 lysine 9 trimethylation (H3K9me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Alzheimer disease | |
m6A Target: Mitogen-activated protein kinase 14 (p38/MAPK14)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03543 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03588 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Ephrin type-B receptor 2 (ERK/EPHB2)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03544 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03589 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Sprouty-related, EVH1 domain-containing protein 2 (SPRED2)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03545 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03590 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: microRNA 1246 (MIR1246)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03546 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03591 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: G1/S-specific cyclin-E1 (CCNE1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03547 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03592 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Glucose transporter type 1 (SLC2A1)
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03549 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03571 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03594 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03616 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: U4/U6 small nuclear ribonucleoprotein Prp31 (PRPF31/RP11)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03551 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03596 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: High mobility group protein HMG-I/HMG-Y (HMGA1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03552 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03597 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Long intergenic non-protein coding RNA 460 (LINC00460)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03553 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03598 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Circ_1662
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03554 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03599 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Transcriptional coactivator YAP1 (YAP1)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03555 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03600 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03658 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Mothers against decapentaplegic homolog 3 (SMAD3)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03556 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03601 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Translocation protein SEC62 (SEC62)
| In total 5 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03557 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Oxaliplatin | |
| Crosstalk ID: M6ACROT03558 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Fluorouracil | |
| Crosstalk ID: M6ACROT03602 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Oxaliplatin | |
| Crosstalk ID: M6ACROT03603 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Fluorouracil | |
| Crosstalk ID: M6ACROT03644 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: LBX2 antisense RNA 1 (LBX2-AS1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03559 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Fluorouracil | |
| Crosstalk ID: M6ACROT03604 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Fluorouracil | |
m6A Target: Putative pituitary tumor-transforming gene 3 protein (PTTG3P)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03560 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03605 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Class E basic helix-loop-helix protein 41 (BHLHE41)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03561 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03606 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Protein crumbs homolog 3 (CRB3)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03562 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03607 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Lactate dehydrogenase A (LDHA)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03563 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03608 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: RAD51-associated protein 1 (RAD51AP1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03564 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Fluorouracil | |
| Crosstalk ID: M6ACROT03609 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Fluorouracil | |
m6A Target: hsa_circ_0000677 (Circ_ABCC1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03565 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03610 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Interferon gamma (IFN-gamma)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03566 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03611 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: C-X-C motif chemokine 9 (Cxcl9)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03567 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03612 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: C-X-C motif chemokine 10 (Cxcl10)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03568 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03613 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Transcription factor ISGF-3 components p91/p84 (Stat1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03569 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03614 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Interferon regulatory factor 1 (Irf1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03570 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03615 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Zinc finger protein SNAI1 (SNAI1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03572 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03617 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Urokinase-type plasminogen activator (PLAU)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03573 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03618 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: hsa-miR-181d-5p
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03574 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Fluorouracil | |
| Crosstalk ID: M6ACROT03619 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Fluorouracil | |
m6A Target: Neurocalcin-delta (NCALD)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03575 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Fluorouracil | |
| Crosstalk ID: M6ACROT03620 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Fluorouracil | |
m6A Target: TNF receptor-associated factor 5 (TRAF5)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03576 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Oxaliplatin | |
| Crosstalk ID: M6ACROT03621 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Oxaliplatin | |
m6A Target: Protein yippee-like 5 (YPEL5)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03577 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03622 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Ephrin type-A receptor 2 (EphA2)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03578 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03623 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Vascular endothelial growth factor A (VEGFA)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03579 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03624 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Lithostathine-1-alpha (REG1A)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03580 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03625 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Polyamine-transporting ATPase 13A3 (ATP13A3)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03581 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03626 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Small nucleolar RNA host gene 1 (SNHG1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03582 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03627 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Circ_UHRF2
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03583 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03628 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Cohesin subunit SA-3 (STAG3)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03584 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03629 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: pri-miR-196b
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03585 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03630 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: hsa_circ_0124554
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03586 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03631 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Potassium voltage-gated channel subfamily H member 6 (KCNH6)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03632 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 9 lactylation (H3K9la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pulmonary fibrosis | |
m6A Target: Cadherin-1 (CDH1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03633 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 9 lactylation (H3K9la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pulmonary fibrosis | |
m6A Target: Apoptosis regulator Bcl-2 (BCL2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03634 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Apoptosis regulator BAX (BAX)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03635 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Caspase-3 (CASP3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03636 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Ribosomal protein S6 kinase beta-1 (RPS6KB1/p70S6K)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03638 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Rho GTPase activating protein 5 (ARHGAP5)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03640 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
| Drug | Doxil | |
| Crosstalk ID: M6ACROT03641 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
| Drug | Cisplatin | |
| Crosstalk ID: M6ACROT03642 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
| Drug | Fluorouracil | |
m6A Target: Actin, aortic smooth muscle (ACTA2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03643 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Hepatoma-derived growth factor (HDGF)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03645 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Zinc finger MYM-type protein 1 (ZMYM1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03646 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Long intergenic non-protein coding RNA 470 (LINC00470)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03647 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: DNA replication licensing factor MCM5 (MCM5)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03649 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: DNA replication licensing factor MCM6 (MCM6)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03650 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: THAP7 antisense RNA 1 (THAP7-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03652 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Krueppel-like factor 2 (KLF2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03653 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Sphingosine kinase 2 (SPHK2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03654 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: A disintegrin and metalloproteinase with thrombospondin motifs 9 (ADAMTS9)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03655 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Poly [ADP-ribose] polymerase 1 (PARP1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03656 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
| Drug | Oxaliplatin | |
m6A Target: miR-17-92
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03657 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
| Drug | Everolimus | |
m6A Target: Basic leucine zipper transcriptional factor ATF-like 2 (BATF2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03661 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Small nucleolar RNA host gene 3 (SNHG3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03662 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Small nucleolar RNA host gene (SNHG7)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03663 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Signal transducer and activator of transcription 3 (STAT3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03665 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Centromere protein F (CENPF)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03666 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Kelch-like protein 5 (KLHL5)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03667 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Target: Interleukin-12 subunit beta (IL12B)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05999 | ||
| Epigenetic Regulator | Lysine-specific demethylase 6B (KDM6B) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Inflammatory response | |
Non-coding RNA
m6A Target: LINC00035 (ABHD11-AS1)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05026 | ||
| Epigenetic Regulator | LINC00035 (ABHD11-AS1) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT05618 | ||
| Epigenetic Regulator | LINC00035 (ABHD11-AS1) | |
| Regulated Target | Krueppel-like factor 4 (KLF4) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05690 | ||
| Epigenetic Regulator | LINC00035 (ABHD11-AS1) | |
| Regulated Target | hsa-miR-1301-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute ischemic stroke | |
m6A Target: Integral membrane protein DGCR2/IDD (DGCR2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05028 | ||
| Epigenetic Regulator | hsa-miR-423-3p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
m6A Target: Tubulin beta-4B chain (TUBB4B)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05029 | ||
| Epigenetic Regulator | hsa-miR-1226-3p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
m6A Target: Transcription factor 4 (TCF4)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05030 | ||
| Epigenetic Regulator | hsa-miR-330-5p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
m6A Target: Small ribosomal subunit protein uS15 (RPS13)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05031 | ||
| Epigenetic Regulator | mmu-miR-1981-5p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
m6A Target: Epidermal growth factor receptor (EGFR)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05032 | ||
| Epigenetic Regulator | hsa-miR-33a | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
m6A Target: Tafazzin (TAFAZZIN)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05033 | ||
| Epigenetic Regulator | hsa-miR-33a | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
m6A Target: MAP kinase-activated protein kinase 2 (MAPKAPK2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05034 | ||
| Epigenetic Regulator | hsa-miR-33a | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
m6A Target: Cysteine methyltransferase DNMT3A (DNMT3A)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05035 | ||
| Epigenetic Regulator | hsa-miR-33a | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05961 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 942 (LINC00942) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
| Drug | Decitabine | |
m6A Target: Ragulator complex protein LAMTOR5 (LAMTOR5/HBXIP)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05036 | ||
| Epigenetic Regulator | MicroRNA let-7g (MIRLET7G) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Breast cancer | |
| Crosstalk ID: M6ACROT05375 | ||
| Epigenetic Regulator | MicroRNA let-7g (MIRLET7G) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Breast cancer | |
m6A Target: Rho GTPase activating protein 5 (ARHGAP5)
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05051 | ||
| Epigenetic Regulator | ARHGAP5 antisense RNA 1 (head to head) (ARHGAP5-AS1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
| Drug | Cisplatin | |
| Crosstalk ID: M6ACROT05218 | ||
| Epigenetic Regulator | hsa-miR-532-5p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Breast cancer | |
| Crosstalk ID: M6ACROT05982 | ||
| Epigenetic Regulator | ARHGAP5 antisense RNA 1 (head to head) (ARHGAP5-AS1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
| Drug | Adriamycin | |
| Crosstalk ID: M6ACROT05983 | ||
| Epigenetic Regulator | ARHGAP5 antisense RNA 1 (head to head) (ARHGAP5-AS1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
| Drug | Fluorouracil | |
m6A Target: Catenin beta-1 (CTNNB1/Beta-catenin)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05052 | ||
| Epigenetic Regulator | MicroRNA 186 (MIR186) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Hepatoblastoma | |
m6A Target: microRNA 186 (MIR186)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05053 | ||
| Epigenetic Regulator | MicroRNA 186 (MIR186) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Hepatoblastoma | |
| Crosstalk ID: M6ACROT05416 | ||
| Epigenetic Regulator | MicroRNA 186 (MIR186) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Hepatoblastoma | |
m6A Target: Cystine/glutamate transporter (SLC7A11)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05054 | ||
| Epigenetic Regulator | MicroRNA 186 (MIR186) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Hepatoblastoma | |
m6A Target: OIP5 antisense RNA 1 (OIP5-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05065 | ||
| Epigenetic Regulator | OIP5 antisense RNA 1 (OIP5-AS1) | |
| Regulated Target | Tripartite motif containing 5 (TRIM5) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Gastric cancer | |
m6A Target: GTPase KRas (KRAS)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05073 | ||
| Epigenetic Regulator | hsa-miR-30c-5p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Cervical cancer | |
m6A Target: Protein mono-ADP-ribosyltransferase PARP10 (PARP10)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05086 | ||
| Epigenetic Regulator | Cardiac-hypertrophy-associated piRNA | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Cardiomegaly | |
m6A Target: Transcriptional coactivator YAP1 (YAP1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05092 | ||
| Epigenetic Regulator | piR31115 | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Triple-negative breast cancer | |
m6A Target: Cytochrome c oxidase subunit 4 isoform 1, mitochondrial (COX4I1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05095 | ||
| Epigenetic Regulator | PM2.5-associated exosomal transcript PAET | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Asthma | |
m6A Target: Myc proto-oncogene protein (MYC)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05103 | ||
| Epigenetic Regulator | hsa-miR-493-5p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT05104 | ||
| Epigenetic Regulator | Maternally expressed 3 (MEG3) | |
| Regulated Target | hsa-miR-493-5p | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT05327 | ||
| Epigenetic Regulator | hsa-miR-338-5p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Lung cancer | |
m6A Target: Integrin alpha-6 (ITGA6)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05111 | ||
| Epigenetic Regulator | hsa-miR-502-3p | |
| Regulated Target | Ragulator complex protein LAMTOR5 (LAMTOR5/HBXIP) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver hepatocellular carcinoma | |
| Crosstalk ID: M6ACROT05112 | ||
| Epigenetic Regulator | Small nucleolar RNA host gene 3 (SNHG3) | |
| Regulated Target | hsa-miR-502-3p | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver hepatocellular carcinoma | |
m6A Target: NACHT, LRR and PYD domains-containing protein 3 (NLRP3)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05161 | ||
| Epigenetic Regulator | hsa-miR-1208 | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Osteoarthritis | |
| Crosstalk ID: M6ACROT05223 | ||
| Epigenetic Regulator | MIR570 host gene (MIR570HG) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
m6A Target: Tumor necrosis factor receptor superfamily member 6 (FAS)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05162 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 1410 (LINC01410) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Preeclampsia | |
m6A Target: Nuclear pore complex protein Nup214 (NUP214)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05164 | ||
| Epigenetic Regulator | hsa-miR-1208 | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Brain cancer | |
m6A Target: long intergenic non-protein coding RNA 662 (LINC00662)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05167 | ||
| Epigenetic Regulator | hsa-miR-186-5p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Triple-negative breast cancer | |
| Drug | Docetaxel | |
| Crosstalk ID: M6ACROT05168 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 662 (LINC00662) | |
| Regulated Target | hsa-miR-186-5p | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Triple-negative breast cancer | |
| Drug | Docetaxel | |
m6A Target: Paired box protein Pax-8 (PAX8)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05169 | ||
| Epigenetic Regulator | hsa-miR-493-5p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Thyroid Cancer | |
| Drug | Sodium iodide I 131 | |
m6A Target: Intraflagellar transport protein 80 homolog (IFT80)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05177 | ||
| Epigenetic Regulator | H2AZ2 pseudogene 1 (H2AZ2P1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Head and neck squamous carcinoma | |
m6A Target: Interleukin-1 beta (IL1B)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05180 | ||
| Epigenetic Regulator | Long noncoding RNA HZ06 (Lnc-HZ06) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Abortion | |
| Drug | Benzo[a]pyrene | |
m6A Target: CCN family member 2 (CTGF)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05184 | ||
| Epigenetic Regulator | AI662270 | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Chronic kidney disease | |
m6A Target: Collagen alpha-2(I) chain (COL1A2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05188 | ||
| Epigenetic Regulator | METTL3 binding lncRNA (MetBil) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Vascular disorders of the liver | |
m6A Target: Metalloproteinase inhibitor 1 (TIMP-1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05189 | ||
| Epigenetic Regulator | METTL3 binding lncRNA (MetBil) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Vascular disorders of the liver | |
m6A Target: Amphiregulin (AREG)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05214 | ||
| Epigenetic Regulator | hsa-miR-33a-3p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Pancreatic cancer | |
m6A Target: 2,4-dienoyl-CoA reductase [ (DECR1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05227 | ||
| Epigenetic Regulator | HNF4A antisense RNA 1 (HNF4A-AS1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
| Drug | Sorafenib | |
m6A Target: Transcription factor SOX-2 (SOX2)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05271 | ||
| Epigenetic Regulator | Circ_VMP1 | |
| Regulated Target | hsa-miR-524-5p | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
| Drug | Cisplatin | |
| Crosstalk ID: M6ACROT05272 | ||
| Epigenetic Regulator | hsa-miR-524-5p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
| Drug | Cisplatin | |
m6A Target: Heat shock factor protein 1 (HSF1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05277 | ||
| Epigenetic Regulator | hsa-miR-455-3p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Heme oxygenase 1 (HMOX1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05279 | ||
| Epigenetic Regulator | hsa-miR-340-3p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Diseases of the female genital system | |
m6A Target: Bone morphogenetic protein 2 (BMP2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05282 | ||
| Epigenetic Regulator | piR-36741 | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Osteoporosis | |
m6A Target: Mutated in multiple advanced cancers 1 (PTEN)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05287 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 470 (LINC00470) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
| Crosstalk ID: M6ACROT05364 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 470 (LINC00470) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Chronic myeloid leukaemia | |
m6A Target: CUB domain-containing protein 1 (CDCP1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05293 | ||
| Epigenetic Regulator | hsa-miR-338-5p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
m6A Target: Ferroptosis suppressor protein 1 (AIFM2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05301 | ||
| Epigenetic Regulator | hsa-miR-4443 | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
| Drug | Cisplatin | |
m6A Target: Wnt inhibitory factor 1 (WIF1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05304 | ||
| Epigenetic Regulator | hsa-miR-21-5p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Endometriosis | |
m6A Target: Interleukin enhancer-binding factor 3 (ILF3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05305 | ||
| Epigenetic Regulator | ILF3 divergent transcript (ILF3-DT) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: Translocation protein SEC62 (SEC62)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05312 | ||
| Epigenetic Regulator | hsa-miR-4429 | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
m6A Target: Signal transducer and activator of transcription 3 (STAT3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05324 | ||
| Epigenetic Regulator | LOC10013.776 (AGAP2-AS1) | |
| Regulated Target | Pre-mRNA-splicing regulator WTAP (WTAP) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
m6A Target: Neurotrophic factor BDNF precursor form (BDNF)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05336 | ||
| Epigenetic Regulator | SnoRNA-ended lncRNA Enhances pre-Ribosomal RNA Transcription (SLERT) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Endometrial cancer | |
m6A Target: Signal transducer and activator of transcription 2 (STAT2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05342 | ||
| Epigenetic Regulator | Small nucleolar RNA host gene 4 (SNHG4) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Congenital pneumonia | |
m6A Target: Kinesin-like protein KIF3C (KIF3C)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05343 | ||
| Epigenetic Regulator | hsa-miR-320d | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Prostate cancer | |
m6A Target: Homeobox protein NANOG (NANOG)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05351 | ||
| Epigenetic Regulator | MiR-135 family | |
| Regulated Target | Zinc finger protein 217 (ZNF217) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Breast cancer | |
m6A Target: Long intergenic non-protein coding RNA 1273 (LINC01273)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05354 | ||
| Epigenetic Regulator | hsa-miR-600 | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
| Drug | Sorafenib | |
| Crosstalk ID: M6ACROT05355 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 1273 (LINC01273) | |
| Regulated Target | hsa-miR-600 | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
| Drug | Sorafenib | |
| Crosstalk ID: M6ACROT05611 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 1273 (LINC01273) | |
| Regulated Target | hsa-miR-600 | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
| Drug | Sorafenib | |
m6A Target: Aspartate--tRNA ligase, cytoplasmic (DARS)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05356 | ||
| Epigenetic Regulator | DARS1 antisense RNA 1 (DARS1-AS1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Cervical cancer | |
m6A Target: Oxidized low-density lipoprotein receptor 1 (OLR1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05358 | ||
| Epigenetic Regulator | Circ_TTLL13 | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Brain cancer | |
| Drug | Temozolomide | |
m6A Target: Suppressor of cytokine signaling 2 (SOCS2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05360 | ||
| Epigenetic Regulator | hsa-miR-302a | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Brain cancer | |
m6A Target: H/ACA ribonucleoprotein complex subunit DKC1 (DKC1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05363 | ||
| Epigenetic Regulator | Circ_MEG3 | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: Transcription factor SOX-9 (SOX9)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05370 | ||
| Epigenetic Regulator | Circ_FTO | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Rheumatoid arthritis | |
m6A Target: microRNA 25 (MIR25)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05379 | ||
| Epigenetic Regulator | MicroRNA 25 (MIR25) | |
| Regulated Target | PHLPP-like (PHLPP2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Pancreatic cancer | |
m6A Target: hsa-miR-873-5p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05380 | ||
| Epigenetic Regulator | hsa-miR-873-5p | |
| Regulated Target | Kelch-like ECH-associated protein 1 (KEAP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Diseases of the urinary system | |
m6A Target: microRNA 221 (MIR221)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05381 | ||
| Epigenetic Regulator | MicroRNA 221 (MIR221) | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Bladder cancer | |
| Crosstalk ID: M6ACROT05561 | ||
| Epigenetic Regulator | MicroRNA 221 (MIR221) | |
| Regulated Target | Dickkopf-related protein 2 (DKK2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Cardiomegaly | |
m6A Target: microRNA 222 (MIR222)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05382 | ||
| Epigenetic Regulator | MicroRNA 222 (MIR222) | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Bladder cancer | |
| Crosstalk ID: M6ACROT05562 | ||
| Epigenetic Regulator | MicroRNA 222 (MIR222) | |
| Regulated Target | Dickkopf-related protein 2 (DKK2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Cardiomegaly | |
m6A Target: Family with sequence similarity 225 member A (FAM225A)
| In total 9 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05396 | ||
| Epigenetic Regulator | Family with sequence similarity 225 member A (FAM225A) | |
| Regulated Target | Integrin subunit beta 3 (ITGB3) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Nasopharyngeal carcinoma | |
| Crosstalk ID: M6ACROT05942 | ||
| Epigenetic Regulator | Family with sequence similarity 225 member A (FAM225A) | |
| Regulated Target | hsa-miR-590-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Cholangiocarcinoma | |
| Crosstalk ID: M6ACROT05943 | ||
| Epigenetic Regulator | Family with sequence similarity 225 member A (FAM225A) | |
| Regulated Target | MicroRNA 1275 (MIR1275) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Nasopharyngeal carcinoma | |
| Crosstalk ID: M6ACROT05944 | ||
| Epigenetic Regulator | Family with sequence similarity 225 member A (FAM225A) | |
| Regulated Target | hsa-miR-590-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Hepatocellular carcinoma | |
| Crosstalk ID: M6ACROT05945 | ||
| Epigenetic Regulator | Family with sequence similarity 225 member A (FAM225A) | |
| Regulated Target | MicroRNA 1275 (MIR1275) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Hepatocellular carcinoma | |
| Crosstalk ID: M6ACROT05946 | ||
| Epigenetic Regulator | hsa-miR-590-3p | |
| Regulated Target | Integrin subunit beta 3 (ITGB3) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Cholangiocarcinoma | |
| Crosstalk ID: M6ACROT05947 | ||
| Epigenetic Regulator | MicroRNA 1275 (MIR1275) | |
| Regulated Target | Integrin subunit beta 3 (ITGB3) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Nasopharyngeal carcinoma | |
| Crosstalk ID: M6ACROT05948 | ||
| Epigenetic Regulator | hsa-miR-590-3p | |
| Regulated Target | Integrin subunit beta 3 (ITGB3) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Hepatocellular carcinoma | |
| Crosstalk ID: M6ACROT05949 | ||
| Epigenetic Regulator | MicroRNA 1275 (MIR1275) | |
| Regulated Target | Integrin subunit beta 3 (ITGB3) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Hepatocellular carcinoma | |
m6A Target: microRNA 1246 (MIR1246)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05397 | ||
| Epigenetic Regulator | MicroRNA 1246 (MIR1246) | |
| Regulated Target | Sprouty-related, EVH1 domain-containing protein 2 (SPRED2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT05490 | ||
| Epigenetic Regulator | MicroRNA 1246 (MIR1246) | |
| Regulated Target | Paternally-expressed gene 3 protein (PEG3) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
m6A Target: Long intergenic non-protein coding RNA 470 (LINC00470)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05402 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 470 (LINC00470) | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Gastric cancer | |
| Crosstalk ID: M6ACROT05487 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 470 (LINC00470) | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Chronic myeloid leukaemia | |
m6A Target: Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)
| In total 14 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05404 | ||
| Epigenetic Regulator | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Regulated Target | Transcriptional coactivator YAP1 (YAP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
| Drug | Cisplatin | |
| Crosstalk ID: M6ACROT05424 | ||
| Epigenetic Regulator | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Regulated Target | MicroRNA 145 (MIR145) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Chronic kidney disease | |
| Drug | Dihydroartemisinin | |
| Crosstalk ID: M6ACROT05479 | ||
| Epigenetic Regulator | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Kidney failure | |
| Crosstalk ID: M6ACROT05492 | ||
| Epigenetic Regulator | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Regulated Target | Transcription factor p65 (RELA) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Brain cancer | |
| Crosstalk ID: M6ACROT05510 | ||
| Epigenetic Regulator | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Regulated Target | hsa-miR-26b | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Breast cancer | |
| Crosstalk ID: M6ACROT05515 | ||
| Epigenetic Regulator | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Regulated Target | Myc proto-oncogene protein (MYC) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Thymic epithelial tumors | |
| Drug | JQ1 | |
| Crosstalk ID: M6ACROT05516 | ||
| Epigenetic Regulator | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Regulated Target | Myc proto-oncogene protein (MYC) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Thymic epithelial tumors | |
| Drug | Cisplatin | |
| Crosstalk ID: M6ACROT05608 | ||
| Epigenetic Regulator | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Regulated Target | hsa-miR-26b | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Breast cancer | |
| Crosstalk ID: M6ACROT05613 | ||
| Epigenetic Regulator | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Regulated Target | Transcription factor E2F1 (E2F1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Breast cancer | |
| Drug | Adriamycin | |
| Crosstalk ID: M6ACROT05644 | ||
| Epigenetic Regulator | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Osteosarcoma | |
| Crosstalk ID: M6ACROT05694 | ||
| Epigenetic Regulator | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Regulated Target | hsa-miR-124-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Ewing's sarcoma | |
| Crosstalk ID: M6ACROT05728 | ||
| Epigenetic Regulator | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05822 | ||
| Epigenetic Regulator | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Osteosarcoma | |
| Crosstalk ID: M6ACROT05930 | ||
| Epigenetic Regulator | MicroRNA 145 (MIR145) | |
| Regulated Target | Focal adhesion kinase 1 (FAK) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Chronic kidney disease | |
| Drug | Dihydroartemisinin | |
m6A Target: hsa-miR-1914-3p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05406 | ||
| Epigenetic Regulator | hsa-miR-1914-3p | |
| Regulated Target | Transcriptional coactivator YAP1 (YAP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
| Drug | Cisplatin | |
m6A Target: hsa-miR-143-3p
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05409 | ||
| Epigenetic Regulator | hsa-miR-143-3p | |
| Regulated Target | Vasohibin 1 (VASH1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Lung cancer | |
| Crosstalk ID: M6ACROT05554 | ||
| Epigenetic Regulator | hsa-miR-143-3p | |
| Regulated Target | Protein kinase C epsilon (PRKCE) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Ischemic heart disease | |
| Crosstalk ID: M6ACROT05760 | ||
| Epigenetic Regulator | hsa-miR-143-3p | |
| Regulated Target | Mothers against decapentaplegic homolog 3 (SMAD3) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Asthma | |
m6A Target: mmu-miR-365-3p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05411 | ||
| Epigenetic Regulator | mmu-miR-365-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Chronic pain | |
m6A Target: Long intergenic non-protein coding RNA 958 (LINC00958)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05412 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 958 (LINC00958) | |
| Regulated Target | Hepatoma-derived growth factor (HDGF) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT05471 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 958 (LINC00958) | |
| Regulated Target | hsa-miR-378a-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Breast cancer | |
m6A Target: PncRNA-D
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05423 | ||
| Epigenetic Regulator | PncRNA-D | |
| Regulated Target | G1/S-specific cyclin-D1 (CCND1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liposarcoma | |
m6A Target: LncRNA activating regulator of DKK1 (LNCAROD)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05425 | ||
| Epigenetic Regulator | LncRNA activating regulator of DKK1 (LNCAROD) | |
| Regulated Target | Y-box-binding protein 1 (YBX1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Head and neck squamous carcinoma | |
m6A Target: mmu-miR-7212-5p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05430 | ||
| Epigenetic Regulator | mmu-miR-7212-5p | |
| Regulated Target | Fibroblast growth factor receptor 3 (FGFR3) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Unspecific body region injury | |
m6A Target: hsa-miR-25-3p
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05432 | ||
| Epigenetic Regulator | hsa-miR-25-3p | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Diabetic retinopathy | |
| Crosstalk ID: M6ACROT05433 | ||
| Epigenetic Regulator | hsa-miR-25-3p | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Diabetes | |
m6A Target: microRNA 335 (MIR335)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05434 | ||
| Epigenetic Regulator | MicroRNA 335 (MIR335) | |
| Regulated Target | ZFP36 ring finger protein like 1 (ZFP36L1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute ischemic stroke | |
m6A Target: hsa_circ_0029589 (Circ_CHFR)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05436 | ||
| Epigenetic Regulator | hsa_circ_0029589 (Circ_CHFR) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute coronary syndrome | |
m6A Target: Testis associated oncogenic lncRNA (THORLNC)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05441 | ||
| Epigenetic Regulator | Testis associated oncogenic lncRNA (THORLNC) | |
| Crosstalk relationship | m6A → ncRNA | |
m6A Target: hsa-miR-126-5p
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05445 | ||
| Epigenetic Regulator | hsa-miR-126-5p | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Ovarian cancer | |
| Crosstalk ID: M6ACROT05449 | ||
| Epigenetic Regulator | hsa-miR-126-5p | |
| Regulated Target | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3K-p85/PIK3R2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Chondropathies | |
m6A Target: Lnc_D63785
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05446 | ||
| Epigenetic Regulator | Lnc_D63785 | |
| Regulated Target | hsa-miR-422a | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Unspecific body region injury | |
| Crosstalk ID: M6ACROT05447 | ||
| Epigenetic Regulator | hsa-miR-422a | |
| Regulated Target | Mitogen-activated protein kinase kinase 6 (MAP2K6) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Unspecific body region injury | |
| Crosstalk ID: M6ACROT05940 | ||
| Epigenetic Regulator | hsa-miR-422a | |
| Regulated Target | Myocyte enhancer factor 2D (MEF2D) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Unspecific body region injury | |
m6A Target: H19 imprinted maternally expressed transcript (H19)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05450 | ||
| Epigenetic Regulator | H19 imprinted maternally expressed transcript (H19) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Abnormalities of breathing | |
m6A Target: ZNFX1 antisense RNA 1 (ZFAS1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05452 | ||
| Epigenetic Regulator | ZNFX1 antisense RNA 1 (ZFAS1) | |
| Regulated Target | MicroRNA 647 (MIR647) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT05627 | ||
| Epigenetic Regulator | ZNFX1 antisense RNA 1 (ZFAS1) | |
| Regulated Target | hsa-miR-100-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Nasopharyngeal carcinoma | |
m6A Target: Long intergenic non-protein coding RNA 2598 (LINC02598/RP11)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05461 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 2598 (LINC02598/RP11) | |
| Regulated Target | F-box/SPRY domain-containing protein 1 (FBXO45) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Colorectal cancer | |
m6A Target: hsa_circ_0092493 (circ_ARL3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05463 | ||
| Epigenetic Regulator | hsa_circ_0092493 (Circ_ARL3) | |
| Regulated Target | microRNA 1305 (MIR1305) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
m6A Target: hsa-miR-221-3p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05465 | ||
| Epigenetic Regulator | hsa-miR-221-3p | |
| Regulated Target | Homeodomain-interacting protein kinase 2 (HIPK2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Melanoma | |
| Drug | RSL3 | |
m6A Target: Long intergenic non-protein coding RNA 460 (LINC00460)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05470 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 460 (LINC00460) | |
| Regulated Target | ATP-dependent RNA helicase A (DHX9) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Colorectal cancer | |
m6A Target: hsa-miR-186-5p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05473 | ||
| Epigenetic Regulator | hsa-miR-186-5p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Melanoma | |
| Drug | ML162 | |
m6A Target: Small nucleolar RNA host gene 3 (SNHG3)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05474 | ||
| Epigenetic Regulator | Small nucleolar RNA host gene 3 (SNHG3) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Melanoma | |
| Drug | ML-210 | |
| Crosstalk ID: M6ACROT05662 | ||
| Epigenetic Regulator | Small nucleolar RNA host gene 3 (SNHG3) | |
| Regulated Target | hsa-miR-186-5p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Gastric cancer | |
| Crosstalk ID: M6ACROT05688 | ||
| Epigenetic Regulator | Small nucleolar RNA host gene 3 (SNHG3) | |
| Regulated Target | hsa-miR-330-5p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Melanoma | |
m6A Target: Pvt1 oncogene (PVT1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05480 | ||
| Epigenetic Regulator | Pvt1 oncogene (PVT1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Kidney failure | |
m6A Target: microRNA 93 (MIR93)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05481 | ||
| Epigenetic Regulator | MicroRNA 93 (MIR93) | |
| Regulated Target | Dual specificity protein phosphatase 2 (DUSP2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Emphysema | |
m6A Target: hsa_circ_0081609 (circ_CUX1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05484 | ||
| Epigenetic Regulator | hsa_circ_0081609 (Circ_CUX1) | |
| Regulated Target | Caspase-1 (CASP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Hypopharyngeal cancer | |
m6A Target: X inactive specific transcript (XIST)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05486 | ||
| Epigenetic Regulator | X inactive specific transcript (XIST) | |
| Regulated Target | hsa-miR-302a-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Ossification of spinal ligaments | |
m6A Target: Circ_1662
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05488 | ||
| Epigenetic Regulator | Circ_1662 | |
| Regulated Target | Transcriptional coactivator YAP1 (YAP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Colorectal cancer | |
m6A Target: Maternally expressed 3 (MEG3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05494 | ||
| Epigenetic Regulator | Maternally expressed 3 (MEG3) | |
| Regulated Target | MicroRNA 544b (MIR544B) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
m6A Target: hsa_circ_0004771 (circ_NRIP1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05496 | ||
| Epigenetic Regulator | hsa_circ_0004771 (Circ_NRIP1) | |
| Regulated Target | hsa-miR-199a-5p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Osteosarcoma | |
m6A Target: miR-199a
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05497 | ||
| Epigenetic Regulator | hsa-miR-199a-5p | |
| Regulated Target | Forkhead box protein C2 (FOXC2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Osteosarcoma | |
m6A Target: LCAT3
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05498 | ||
| Epigenetic Regulator | LCAT3 | |
| Regulated Target | Far upstream element-binding protein 1 (FUBP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Lung cancer | |
m6A Target: Nuclear paraspeckle assembly transcript 1 (NEAT1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05502 | ||
| Epigenetic Regulator | Nuclear paraspeckle assembly transcript 1 (NEAT1) | |
| Regulated Target | hsa-miR-766-5p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Chronic myeloid leukaemia | |
| Crosstalk ID: M6ACROT05712 | ||
| Epigenetic Regulator | Nuclear paraspeckle assembly transcript 1 (NEAT1) | |
| Regulated Target | hsa-miR-361-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
m6A Target: hsa-miR-766-5p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05503 | ||
| Epigenetic Regulator | hsa-miR-766-5p | |
| Regulated Target | Cyclin-dependent kinase inhibitor 1 (CDKN1A) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Chronic myeloid leukaemia | |
m6A Target: NIFK antisense RNA 1 (NIFK-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05504 | ||
| Epigenetic Regulator | NIFK antisense RNA 1 (NIFK-AS1) | |
| Regulated Target | MicroRNA 637 (MIR637) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
| Drug | Sorafenib | |
m6A Target: LBX2 antisense RNA 1 (LBX2-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05517 | ||
| Epigenetic Regulator | LBX2 antisense RNA 1 (LBX2-AS1) | |
| Regulated Target | hsa-miR-422a | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Colorectal cancer | |
| Drug | 5-FU | |
m6A Target: FOXD2 adjacent opposite strand RNA 1 (FOXD2-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05521 | ||
| Epigenetic Regulator | FOXD2 adjacent opposite strand RNA 1 (FOXD2-AS1) | |
| Regulated Target | Cyclin-dependent kinase inhibitor 1 (CDKN1A) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Cervical cancer | |
m6A Target: THAP7 antisense RNA 1 (THAP7-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05522 | ||
| Epigenetic Regulator | THAP7 antisense RNA 1 (THAP7-AS1) | |
| Regulated Target | Cullin 4B (CUL4B) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Gastric cancer | |
m6A Target: LIFR antisense RNA 1 (LIFR-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05526 | ||
| Epigenetic Regulator | LIFR antisense RNA 1 (LIFR-AS1) | |
| Regulated Target | hsa-miR-150-5p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Pancreatic cancer | |
m6A Target: hsa_circ_0008399 (Circ_RBM3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05530 | ||
| Epigenetic Regulator | hsa_circ_0008399 (Circ_RBM3) | |
| Regulated Target | Pre-mRNA-splicing regulator WTAP (WTAP) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Bladder cancer | |
| Drug | Cisplatin | |
m6A Target: hsa_circ_0021427 (circ_HPS5)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05532 | ||
| Epigenetic Regulator | hsa_circ_0021427 (Circ_HPS5) | |
| Regulated Target | hsa-miR-370-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
m6A Target: microRNA 370 (MIR370)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05534 | ||
| Epigenetic Regulator | MicroRNA 370 (MIR370) | |
| Regulated Target | High mobility group protein HMGI-C (HMGA2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
m6A Target: hsa-miR-140-3p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05544 | ||
| Epigenetic Regulator | hsa-miR-140-3p | |
| Regulated Target | Ubiquitin-conjugating enzyme E2 C (UBE2C) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
m6A Target: Small nucleolar RNA host gene 1 (SNHG1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05545 | ||
| Epigenetic Regulator | Small nucleolar RNA host gene 1 (SNHG1) | |
| Regulated Target | hsa-miR-140-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05701 | ||
| Epigenetic Regulator | Small nucleolar RNA host gene 1 (SNHG1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Colorectal cancer | |
m6A Target: hsa_circ_0058493 (Circ_RHBDD1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05547 | ||
| Epigenetic Regulator | hsa_circ_0058493 (Circ_RHBDD1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
m6A Target: microRNA let-7b (MIRLET7B)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05549 | ||
| Epigenetic Regulator | MicroRNA let-7b (MIRLET7B) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
m6A Target: hsa-miR-21-5p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05555 | ||
| Epigenetic Regulator | hsa-miR-21-5p | |
| Regulated Target | Sprouty RTK signaling antagonist 1 (SPRY1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Kidney disorders | |
m6A Target: hsa-miR-375-3p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05556 | ||
| Epigenetic Regulator | hsa-miR-375-3p | |
| Regulated Target | PDH kinase 1 (PDK1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Atherosclerosis | |
m6A Target: microRNA 150 (MIR150)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05558 | ||
| Epigenetic Regulator | MicroRNA 150 (MIR150) | |
| Regulated Target | Neurotrophic factor BDNF precursor form (BDNF) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Neuropathic pain | |
m6A Target: LOC10013.776 (AGAP2-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05560 | ||
| Epigenetic Regulator | LOC10013.776 (AGAP2-AS1) | |
| Regulated Target | hsa-miR-424-5p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Psoriasis | |
m6A Target: hsa-miR-380-3p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05563 | ||
| Epigenetic Regulator | hsa-miR-380-3p | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Pancreatic cancer | |
m6A Target: KRT7-AS
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05565 | ||
| Epigenetic Regulator | KRT7-AS | |
| Regulated Target | Keratin, type II cytoskeletal 7 (KRT7) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Breast cancer | |
m6A Target: hsa_circ_0000677
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05566 | ||
| Epigenetic Regulator | hsa_circ_0000677 (Circ_ABCC1) | |
| Regulated Target | ATP binding cassette subfamily C member 1 (ABCC1 blood group) (ABCC1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Colorectal cancer | |
m6A Target: hsa-miR-582-3p
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05568 | ||
| Epigenetic Regulator | hsa-miR-582-3p | |
| Regulated Target | Transcriptional coactivator YAP1 (YAP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Intrahepatic cholangiocarcinoma | |
| Crosstalk ID: M6ACROT05640 | ||
| Epigenetic Regulator | hsa-miR-582-3p | |
| Regulated Target | Transcriptional coactivator YAP1 (YAP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Intrahepatic cholangiocarcinoma | |
m6A Target: hsa_circ_0008542 (Circ_PHF12)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05569 | ||
| Epigenetic Regulator | hsa_circ_0008542 (Circ_PHF12) | |
| Regulated Target | hsa-miR-185-5p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Diseases of the musculoskeletal system | |
m6A Target: Prostate cancer associated transcript 6 (PCAT6)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05571 | ||
| Epigenetic Regulator | Prostate cancer associated transcript 6 (PCAT6) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Prostate cancer | |
m6A Target: Circ_IGF2BP3
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05576 | ||
| Epigenetic Regulator | Circ_IGF2BP3 | |
| Regulated Target | hsa-miR-328-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
m6A Target: hsa-miR-29a-3p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05580 | ||
| Epigenetic Regulator | hsa-miR-29a-3p | |
| Regulated Target | Tumor necrosis factor receptor superfamily member 1A (TNFRSF1A) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute respiratory distress syndrome | |
m6A Target: miR-17-92
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05582 | ||
| Epigenetic Regulator | hsa-miR-17-92 | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Gastric cancer | |
| Drug | Everolimus | |
| Crosstalk ID: M6ACROT05941 | ||
| Epigenetic Regulator | hsa-miR-17-92 | |
| Regulated Target | Transmembrane protein 127 (TMEM127) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Gastric cancer | |
| Drug | Everolimus | |
m6A Target: hsa-miR-146a-5p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05583 | ||
| Epigenetic Regulator | hsa-miR-146a-5p | |
| Regulated Target | Protein numb homolog (NUMB) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Bladder cancer | |
| Drug | Melittin | |
m6A Target: Long intergenic non-protein coding RNA 680 (LINC00680)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05584 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 680 (LINC00680) | |
| Regulated Target | NAD-dependent protein deacetylase sirtuin-1 (SIRT1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Osteoarthritis | |
m6A Target: Circ_ASK1
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05586 | ||
| Epigenetic Regulator | Circ_ASK1 | |
| Regulated Target | Mitogen-activated protein kinase kinase kinase 5 (MAP3K5) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
m6A Target: miR-182
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05587 | ||
| Epigenetic Regulator | MiR-182 | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Prostate cancer | |
m6A Target: hsa-miR-193b
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05593 | ||
| Epigenetic Regulator | hsa-miR-193b | |
| Regulated Target | G1/S-specific cyclin-D1 (CCND1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Cervical cancer | |
m6A Target: microRNA 126 (MIR126)
| In total 8 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05595 | ||
| Epigenetic Regulator | MicroRNA 126 (MIR126) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Endometriosis | |
| Crosstalk ID: M6ACROT05681 | ||
| Epigenetic Regulator | MicroRNA 126 (MIR126) | |
| Regulated Target | ADAM metallopeptidase domain 9 (ADAM9) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT05924 | ||
| Epigenetic Regulator | MicroRNA 126 (MIR126) | |
| Regulated Target | Scaffold protein ILK (ILK) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT05926 | ||
| Epigenetic Regulator | MicroRNA 126 (MIR126) | |
| Regulated Target | Cyclin-dependent kinase 3 (CDK3) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT05932 | ||
| Epigenetic Regulator | MicroRNA 126 (MIR126) | |
| Regulated Target | Target of Myb1 membrane trafficking protein (TOM1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT05934 | ||
| Epigenetic Regulator | MicroRNA 126 (MIR126) | |
| Regulated Target | High affinity immunoglobulin gamma Fc receptor I (FCGR1A) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT05936 | ||
| Epigenetic Regulator | MicroRNA 126 (MIR126) | |
| Regulated Target | Ficolin-1 (FCN1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT05938 | ||
| Epigenetic Regulator | MicroRNA 126 (MIR126) | |
| Regulated Target | Leucine-rich repeat and calponin homology domain-containing protein 4 (LRCH4) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute myeloid leukaemia | |
m6A Target: Circ_DLC1
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05596 | ||
| Epigenetic Regulator | Circ_DLC1 | |
| Regulated Target | hsa-miR-671-5p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Brain cancer | |
m6A Target: hsa-miR-671-5p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05597 | ||
| Epigenetic Regulator | hsa-miR-671-5p | |
| Regulated Target | Beta-catenin-interacting protein 1 (CTNNBIP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Brain cancer | |
m6A Target: pri-miR-143
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05600 | ||
| Epigenetic Regulator | pri-miR-143 | |
| Regulated Target | YY1-associated protein 1 (YY1AP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute myocardial infarction | |
| Crosstalk ID: M6ACROT06027 | ||
| Epigenetic Regulator | pri-miR-143 | |
| Regulated Target | Catenin delta-1 (CTNND1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute myocardial infarction | |
m6A Target: pri-miR-27
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05604 | ||
| Epigenetic Regulator | MicroRNA MIR27 family | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Multiple myeloma | |
m6A Target: hsa-miR-1915-3p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05605 | ||
| Epigenetic Regulator | hsa-miR-1915-3p | |
| Regulated Target | Protein SET (SET) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
m6A Target: hsa-miR-34a
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05607 | ||
| Epigenetic Regulator | hsa-miR-34a | |
| Regulated Target | NAD-dependent protein deacetylase sirtuin-1 (SIRT1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Abdominal aortic aneurysm | |
m6A Target: hsa-miR-26b
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05610 | ||
| Epigenetic Regulator | hsa-miR-26b | |
| Regulated Target | High mobility group protein HMGI-C (HMGA2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Breast cancer | |
m6A Target: hsa-miR-181d-5p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05615 | ||
| Epigenetic Regulator | hsa-miR-181d-5p | |
| Regulated Target | Neurocalcin-delta (NCALD) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Colorectal cancer | |
| Drug | 5-FU | |
m6A Target: SVIL antisense RNA 1 (SVIL-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05617 | ||
| Epigenetic Regulator | SVIL antisense RNA 1 (SVIL-AS1) | |
| Regulated Target | Transcription factor E2F1 (E2F1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Lung cancer | |
m6A Target: Long intergenic non-protein coding RNA 1833 (LINC01833)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05620 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 1833 (LINC01833) | |
| Regulated Target | Heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
m6A Target: Small nucleolar RNA host gene (SNHG7)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05623 | ||
| Epigenetic Regulator | Small nucleolar RNA host gene (SNHG7) | |
| Regulated Target | Myc proto-oncogene protein (MYC) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Prostate cancer | |
| Crosstalk ID: M6ACROT05698 | ||
| Epigenetic Regulator | Small nucleolar RNA host gene (SNHG7) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Gastric cancer | |
m6A Target: DBH antisense RNA 1 (Lnc_DBH-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05625 | ||
| Epigenetic Regulator | DBH antisense RNA 1 (Lnc_DBH-AS1) | |
| Regulated Target | MicroRNA 3163 (MIR3163) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Pancreatic cancer | |
| Drug | Gemcitabine | |
m6A Target: miR-503
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05626 | ||
| Epigenetic Regulator | MiR-503 | |
| Regulated Target | PPARG coactivator 1 beta (PPARGC1B) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Injury of heart | |
m6A Target: hsa-miR-222-3p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05629 | ||
| Epigenetic Regulator | hsa-miR-222-3p | |
| Regulated Target | Serine/threonine-protein kinase 4 (STK4) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Thyroid Cancer | |
m6A Target: hsa-miR-589-5p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05630 | ||
| Epigenetic Regulator | hsa-miR-589-5p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
m6A Target: hsa-miR-320b
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05635 | ||
| Epigenetic Regulator | hsa-miR-320b | |
| Regulated Target | Programmed cell death 4 (PDCD4) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Esophageal Squamous Cell Carcinoma | |
m6A Target: RNA component of 7SK nuclear ribonucleoprotein (RN7SK)
| In total 6 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05648 | ||
| Epigenetic Regulator | RNA component of 7SK nuclear ribonucleoprotein (RN7SK) | |
| Regulated Target | The positive transcription elongation factor b complex (P-TEFb) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05660 | ||
| Epigenetic Regulator | RNA component of 7SK nuclear ribonucleoprotein (RN7SK) | |
| Regulated Target | Cullin 1 (CUL1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Drug | mitoxantrone (MIT) | |
| Crosstalk ID: M6ACROT06030 | ||
| Epigenetic Regulator | RNA component of 7SK nuclear ribonucleoprotein (RN7SK) | |
| Regulated Target | Cullin 1 (CUL1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Drug | hydroxyurea (HYD) | |
| Crosstalk ID: M6ACROT06033 | ||
| Epigenetic Regulator | RNA component of 7SK nuclear ribonucleoprotein (RN7SK) | |
| Regulated Target | Cullin 1 (CUL1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Drug | raltitrexed (RAL) | |
| Crosstalk ID: M6ACROT06036 | ||
| Epigenetic Regulator | RNA component of 7SK nuclear ribonucleoprotein (RN7SK) | |
| Regulated Target | Cullin 1 (CUL1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Drug | oxaliplatin (OXA) | |
| Crosstalk ID: M6ACROT06039 | ||
| Epigenetic Regulator | RNA component of 7SK nuclear ribonucleoprotein (RN7SK) | |
| Regulated Target | Cullin 1 (CUL1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Drug | etoposide (ETO) | |
m6A Target: Long intergenic non-protein coding RNA 1638 (LINC01638)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05654 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 1638 (LINC01638) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Periodontitis | |
m6A Target: AC026356.1
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05656 | ||
| Epigenetic Regulator | AC026356.1 | |
| Regulated Target | Interleukin-11 (IL11) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
m6A Target: Circ_SETD2
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05657 | ||
| Epigenetic Regulator | Circ_SETD2 | |
| Regulated Target | hsa-miR-181a-5p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Pre-eclampsia | |
m6A Target: FGD5 antisense RNA 1 (FGD5-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05665 | ||
| Epigenetic Regulator | FGD5 antisense RNA 1 (FGD5-AS1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Endometrial cancer | |
| Drug | paclitaxel (PTX) | |
m6A Target: EN_42575
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05675 | ||
| Epigenetic Regulator | EN_42575 | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Change in bowel habit | |
m6A Target: chromosome 2 open reading frame 92 (C2orf92)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05678 | ||
| Epigenetic Regulator | Chromosome 2 open reading frame 92 (C2ORF92) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Papillary thyroid cancer | |
m6A Target: hsa_circ_0007905 (Circ_STX6)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05680 | ||
| Epigenetic Regulator | Circ_STX6 | |
| Regulated Target | hsa-miR-6749-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Age-related cataracts | |
m6A Target: Circ_MYO1C
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05686 | ||
| Epigenetic Regulator | Circ_MYO1C | |
| Regulated Target | CD274 molecule (CD274) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Pancreatic ductal adenocarcinoma | |
m6A Target: hsa-miR-181a-5p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05687 | ||
| Epigenetic Regulator | hsa-miR-181a-5p | |
| Regulated Target | NFKB activating protein (NKAP) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Ageing-related disease | |
m6A Target: BRAF-activated non-protein coding RNA (BANCR)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05689 | ||
| Epigenetic Regulator | BRAF-activated non-protein coding RNA (BANCR) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Pancreatic cancer | |
m6A Target: hsa-miR-148a-3p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05693 | ||
| Epigenetic Regulator | hsa-miR-148a-3p | |
| Regulated Target | Thioredoxin-interacting protein (TXNIP) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Prostate cancer | |
m6A Target: Long intergenic non-protein coding RNA 662 (LINC00662)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05699 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 662 (LINC00662) | |
| Regulated Target | Integrin subunit alpha 1 (ITGA1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Pancreatic cancer | |
m6A Target: KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05702 | ||
| Epigenetic Regulator | KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1) | |
| Regulated Target | hsa-miR-103a-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Breast cancer | |
| Drug | DOX | |
m6A Target: hsa-miR-1908-5p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05703 | ||
| Epigenetic Regulator | hsa-miR-1908-5p | |
| Regulated Target | HOP homeobox (HOPX) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Nasopharyngeal carcinoma | |
m6A Target: Circ_RNF13
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05706 | ||
| Epigenetic Regulator | Circ_RNF13 | |
| Regulated Target | C-X-C motif chemokine ligand 1 (CXCL1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Cervical cancer | |
m6A Target: Circ_EPHB4
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05708 | ||
| Epigenetic Regulator | Circ_EPHB4 | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Brain cancer | |
m6A Target: Long intergenic non-protein coding RNA 426 (LINC00426)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05713 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 426 (LINC00426) | |
| Regulated Target | hsa-miR-200a-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Cervical cancer | |
| Drug | Cisplatin | |
| Crosstalk ID: M6ACROT05714 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 426 (LINC00426) | |
| Regulated Target | hsa-miR-200a-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Cervical cancer | |
| Drug | Bleomycin | |
| Crosstalk ID: M6ACROT05715 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 426 (LINC00426) | |
| Regulated Target | hsa-miR-200a-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Cervical cancer | |
| Drug | imatinib | |
m6A Target: Circ_UHRF2
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05716 | ||
| Epigenetic Regulator | Circ_UHRF2 | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Colorectal cancer | |
m6A Target: Growth arrest specific 5 (GAS5)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05717 | ||
| Epigenetic Regulator | Growth arrest specific 5 (GAS5) | |
| Regulated Target | Dynamin-1-like protein (DRP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Vascular disorders of the liver | |
m6A Target: pri-miR-196b
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05731 | ||
| Epigenetic Regulator | pri-miR-196b | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Colorectal cancer | |
m6A Target: microRNA 675 (MIR675)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05732 | ||
| Epigenetic Regulator | MicroRNA 675 (MIR675) | |
| Regulated Target | myosin phosphatase targeting protein 1 (MYPT1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Gastrointestinal stromal tumor | |
m6A Target: hsa-miR-99a-5p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05737 | ||
| Epigenetic Regulator | hsa-miR-99a-5p | |
| Regulated Target | Beta-transducin repeat containing E3 ubiquitin protein ligase pseudogene 1 (BTRCP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Oral squamous cell carcinoma | |
m6A Target: HOXD antisense growth-associated long non-coding RNA (HAGLR)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05738 | ||
| Epigenetic Regulator | HOXD antisense growth-associated long non-coding RNA (HAGLR) | |
| Regulated Target | MicroRNA 135a-1 (MIR135A1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Pre-eclampsia | |
m6A Target: Circ_MIRLET7BHG
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05739 | ||
| Epigenetic Regulator | Circ_MIRLET7BHG | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Allergic rhinitis | |
m6A Target: pri-miR-935
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05743 | ||
| Epigenetic Regulator | pri-miR-935 | |
| Regulated Target | Gap junction alpha-1 protein (GJA1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Intrahepatic cholangiocarcinoma | |
m6A Target: Circ_ABCC4
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05744 | ||
| Epigenetic Regulator | Circ_ABCC4 | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Prostate cancer | |
m6A Target: hsa-miR-27a-3p
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05745 | ||
| Epigenetic Regulator | hsa-miR-27a-3p | |
| Regulated Target | Peroxisome proliferator-activated receptor gamma (PPARG) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Infantile hemangioma | |
| Drug | Oxymatrine | |
| Crosstalk ID: M6ACROT05746 | ||
| Epigenetic Regulator | hsa-miR-27a-3p | |
| Regulated Target | Peroxisome proliferator-activated receptor gamma (PPARG) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Infantile hemangioma | |
| Drug | propranolol (PPNL) | |
m6A Target: RNF32 divergent transcript (RNF32-DT)
| In total 16 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05750 | ||
| Epigenetic Regulator | RNF32 divergent transcript (RNF32-DT) | |
| Regulated Target | MicroRNA 2682 (MIR2682) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05902 | ||
| Epigenetic Regulator | RNF32 divergent transcript (RNF32-DT) | |
| Regulated Target | MicroRNA 34a (MIR34A) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05903 | ||
| Epigenetic Regulator | RNF32 divergent transcript (RNF32-DT) | |
| Regulated Target | MicroRNA 34b (MIR34B) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05904 | ||
| Epigenetic Regulator | RNF32 divergent transcript (RNF32-DT) | |
| Regulated Target | MicroRNA 34c (MIR34C) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05905 | ||
| Epigenetic Regulator | MicroRNA 2682 (MIR2682) | |
| Regulated Target | Myc proto-oncogene protein (MYC) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05906 | ||
| Epigenetic Regulator | hsa-miR-34a | |
| Regulated Target | Myc proto-oncogene protein (MYC) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05907 | ||
| Epigenetic Regulator | MicroRNA 34b (MIR34B) | |
| Regulated Target | Myc proto-oncogene protein (MYC) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05908 | ||
| Epigenetic Regulator | MicroRNA 34c (MIR34C) | |
| Regulated Target | Myc proto-oncogene protein (MYC) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05909 | ||
| Epigenetic Regulator | RNF32 divergent transcript (RNF32-DT) | |
| Regulated Target | MicroRNA 2682 (MIR2682) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05910 | ||
| Epigenetic Regulator | RNF32 divergent transcript (RNF32-DT) | |
| Regulated Target | MicroRNA 34a (MIR34A) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05911 | ||
| Epigenetic Regulator | RNF32 divergent transcript (RNF32-DT) | |
| Regulated Target | MicroRNA 34b (MIR34B) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05912 | ||
| Epigenetic Regulator | RNF32 divergent transcript (RNF32-DT) | |
| Regulated Target | MicroRNA 34c (MIR34C) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05913 | ||
| Epigenetic Regulator | MicroRNA 2682 (MIR2682) | |
| Regulated Target | Myc proto-oncogene protein (MYC) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05914 | ||
| Epigenetic Regulator | hsa-miR-34a | |
| Regulated Target | Myc proto-oncogene protein (MYC) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05915 | ||
| Epigenetic Regulator | MicroRNA 34b (MIR34B) | |
| Regulated Target | Myc proto-oncogene protein (MYC) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05916 | ||
| Epigenetic Regulator | MicroRNA 34c (MIR34C) | |
| Regulated Target | Myc proto-oncogene protein (MYC) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
m6A Target: hsa-miR-196b-5p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05751 | ||
| Epigenetic Regulator | hsa-miR-196b-5p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Structural developmental anomalies of teeth and periodontal tissues | |
m6A Target: Long intergenic non-protein coding RNA 294 (LINC00294)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05756 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 294 (LINC00294) | |
| Regulated Target | YTH N6-methyladenosine RNA binding protein C1 (YTHDC1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
m6A Target: SLC7A11 antisense RNA 1 (SLC7A11-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05761 | ||
| Epigenetic Regulator | SLC7A11 antisense RNA 1 (SLC7A11-AS1) | |
| Regulated Target | E3 ubiquitin-protein ligase CHIP (STUB1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
m6A Target: hsa_circ_0124554
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05763 | ||
| Epigenetic Regulator | hsa_circ_0124554 | |
| Regulated Target | MicroRNA 1184-1 (MIR1184-1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT05898 | ||
| Epigenetic Regulator | MicroRNA 1184 (MIR1184) | |
| Regulated Target | LIM and SH3 protein 1 (LASP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Colorectal cancer | |
m6A Target: Circ_ASXL1
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05764 | ||
| Epigenetic Regulator | Circ_ASXL1 | |
| Regulated Target | hsa-miR-320d | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Ovarian cancer | |
| Crosstalk ID: M6ACROT05896 | ||
| Epigenetic Regulator | hsa-miR-320d | |
| Regulated Target | Rac GTPase-activating protein 1 (RACGAP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Ovarian cancer | |
m6A Target: Circ_PRKAR1B
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05769 | ||
| Epigenetic Regulator | Circ_PRKAR1B | |
| Regulated Target | Spectrin beta, non-erythrocytic 1 (SPTBN1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Crohn disease | |
m6A Target: hsa-mir-320a
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05788 | ||
| Epigenetic Regulator | hsa-mir-320a | |
| Regulated Target | Runt-related transcription factor 2 (Runx2) | |
| Crosstalk relationship | m6A → ncRNA | |
m6A Target: MIRLET7E
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05805 | ||
| Epigenetic Regulator | MIRLET7E | |
| Regulated Target | Thrombospondin 1 (THBS1) | |
| Crosstalk relationship | m6A → ncRNA | |
m6A Target: hsa-miR-20a-5p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05814 | ||
| Epigenetic Regulator | hsa-miR-20a-5p | |
| Regulated Target | Nuclear factor 1 C-type (NFIC) | |
| Crosstalk relationship | m6A → ncRNA | |
m6A Target: Urothelial cancer associated 1 (UCA1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05815 | ||
| Epigenetic Regulator | Urothelial cancer associated 1 (UCA1) | |
| Crosstalk relationship | m6A → ncRNA | |
m6A Target: hsa_circ_0072380 (Circ_ZNF131)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05820 | ||
| Epigenetic Regulator | hsa_circ_0072380 (Circ_ZNF131) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Gestational diabetes mellitus | |
m6A Target: hsa_circ_0136959
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05824 | ||
| Epigenetic Regulator | hsa_circ_0136959 | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Papillary thyroid cancer | |
m6A Target: pre-miR-665
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05828 | ||
| Epigenetic Regulator | hsa-miR-665 | |
| Regulated Target | Homeobox protein DLX-3 (DLX3) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Structural developmental anomalies of teeth and periodontal tissues | |
m6A Target: SREBF2 antisense RNA 1 (SREBF2-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05833 | ||
| Epigenetic Regulator | SREBF2 antisense RNA 1 (SREBF2-AS1) | |
| Regulated Target | FMR1 autosomal homolog 1 (FXR1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute myeloid leukaemia | |
| Drug | Chidamide | |
m6A Target: Lnc668
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05870 | ||
| Epigenetic Regulator | Lnc668 | |
| Regulated Target | Phosphatidylinositol-binding clathrin assembly protein (PICALM) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Pulmonary fibrosis | |
m6A Target: Retinoic Acid Receptor Alpha-Retinoic Acid Receptor Alpha (PML-RARalpha)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05878 | ||
| Epigenetic Regulator | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Blood malignancies | |
m6A Target: Spectrin beta, non-erythrocytic 2 (SPTBN2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05929 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 1605 | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
Xenobiotics Compound(s) Regulating the m6A Methylation Regulator
| Compound Name | MA2 | Approved |
|---|---|---|
| Synonyms |
Ethyl ester form of meclofenamic acid (MA)
Click to Show/Hide
|
|
| Description |
Knockdown of METTL3 or METTL14 induced changes in mRNA m6A enrichment and altered mRNA expression of genes (e.g.,ADAM19) with critical biological functions inGSCs. Treatment with MA2, a chemical inhibitor of FTO, dramatically suppressed GSC-induced tumorigenesis and prolonged lifespan in GSC-grafted animals.
|
[194] |
| Compound Name | Simovil | Approved |
| Synonyms |
Cholestat; Coledis; Colemin; Corolin; Denan; Labistatin; Lipex; Lipovas; Lodales; Medipo; Nivelipol; Pantok; Rendapid; Simovil; Simvastatina; Simvastatine; Simvastatinum; Sinvacor; Sivastin; Synvinolin; Vasotenal; Zocor; Zocord; Simvast CR; Simvastatina [Spanish]; Simvastatine [French]; Simvastatinum [Latin]; MK 0733; MK 733; MK733; TNP00259; DRG-0320; KS-1113; L 644128-000U; MK-0733; MK-733; Simcard (TN); Simlup (TN); Simvacor (TN); Simvastatin & Primycin; Simvastatin, Compactin; Zocor (TN); Simvastatin [USAN:INN:BAN]; Simvastatin (JAN/USP/INN); Zocor, Simlup, Simcard, Simvacor, Simvoget, Zorced, Simvastatin; [(1S,3R,7S,8S,8aR)-8-[2-[(2R,4R)-4-hydroxy-6-oxooxan-2-yl]ethyl]-3,7-dimethyl-1,2,3,7,8,8a-hexahydronaphthalen-1-yl] 2,2-dimethylbutanoate; Butanoic acid, 2,2-dimethyl-, (1S,3R,7S,8S,8aR)-1,2,3,7,8,8a-hexahydro-3,7-dimethyl-8-[2-[(2R,4R)-tetrahydro-4-hydroxy-6-oxo-2H-pyran-2-yl]ethyl]-1-naphthalenyl ester; Butanoic acid, 2,2-dimethyl-, (1S,3R,7S,8S,8aR)-1,2,3,7,8,8a-hexahydro-3,7-dimethyl-8-(2-((2R,4R)-tetrahydro-4-hydroxy-6-oxo-2H-pyran-2-yl)ethyl)-1-naphthalenyl ester; Butanoic acid, 2,2-dimethyl-, (1S,3R,7S,8S,*aR)-1,2,3,7,8,8a-hexahydro-3,7-dimethyl-8-(2-((2R,4R)-tetrahydro-4-hydroxy-6-oxo-2H-pyran-2-yl)ethyl)-1-naphthalenyl ester; (1S,3R,7S,8S,8aR)-8-{2-[(2R,4R)-4-hydroxy-6-oxotetrahydro-2H-pyran-2-yl]ethyl}-3,7-dimethyl-1,2,3,7,8,8a-hexahydronaphthalen-1-yl 2,2-dimethylbutanoate; Butanoic acid, 2,2-dimethyl-,1,2,3,7,8,8a-hexahydro-3,7-dimethyl-8-[2-(tetrahydro-4-hydroxy-6-oxo-2H-pyran-2-yl)-ethyl]-1-naphthalenyl ester, [1S-[1 alpha,3 alpha,7 beta,8 beta(2S*,4S*),-8a beta; 2,2-Dimethylbutanoic acid (1S,3R,7S,8S,8aR)-1,2,3,7,8,8a-hexahydro-3,7-dimethyl-8-[2-[(2R,4R)-tetrahydro-4-hydroxy-6-oxo-2H-pyran-2-yl]ethyl]-1-naphthalenyl ester; 2,2-Dimethylbutyric acid, 8-ester with (4R,6R)-6-(2-((1S,2S,6R,8S,8aR)-1,2,6,7,8,8a-hexahydro-8-hydroxy-2,6-dimethyl-1-naphthyl)ethyl)tetrahydro-4-hydroxy-2H-pyran-2-one
Click to Show/Hide
|
|
| External link | ||
| Description |
Simvastatin induces METTL3 down-regulation in lung cancer tissues, which further influences EMT via m6A modification onEZH2 mRNA and thus inhibits the malignant progression oflung cancer.
|
[59] |
| Compound Name | Rapamycin | Approved |
| Synonyms |
Rapamune; Rapamycin (Sirolimus); AY-22989; Rapammune; sirolimusum; WY-090217; RAPA; Antibiotic AY 22989; AY 22989; UNII-W36ZG6FT64; CCRIS 9024; CHEBI:9168; SILA 9268A; W36ZG6FT64; HSDB 7284; C51H79NO13; NSC 226080; DE-109; NCGC00021305-05; DSSTox_CID_3582; DSSTox_RID_77091; DSSTox_GSID_23582; Cypher; Supralimus; Wy 090217; Perceiva; RAP; RPM; Rapamycin from Streptomyces hygroscopicus; SIIA 9268A; LCP-Siro; MS-R001; Rapamune (TN); Rapamycin (TN); Sirolimus (RAPAMUNE); Rapamycin C-7, analog 4; Sirolimus (USAN/INN); Sirolimus [USAN:BAN:INN]; Sirolimus, Rapamune,Rapamycin; Heptadecahydro-9,27-dihydroxy-3-[(1R)-2-[(1S,3R,4R)-4-hydroxy; 23,27-Epoxy-3H-pyrido(2,1-c)(1,4)oxaazacyclohentriacontine; 23,27-Epoxy-3H-pyrido[2,1-c][1,4]oxaazacyclohentriacontine; 23,27-epoxy-3H-pyrido[2,1-c][1,4]oxaazacyclohentriacontine-1,5,11,28,29; 3H-pyrido(2,1-c)(1,4)oxaazacyclohentriacontine-1,5,11,28,29(4H,6H,31H)-pentone; Sirolimus (MTOR inhibitor)
Click to Show/Hide
|
|
| External link | ||
| Description |
METTL3 promotes the progression ofretinoblastoma throughPI3K/AKT/mTOR pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-P70S6K/4EBP1 pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment.
|
[44] |
| Compound Name | BEZ235 | Phase 2 |
| Synonyms |
BEZ-235; S14-0511; NVP-BEZ-235; NVP-BEZ235, BEZ235; 2-(4-(2,3-dihydro-3-methyl-2-oxo-8-(quinolin-3-yl)imidazo[4,5-c]quinolin-1-yl)phenyl)-2-methylpropanenitrile
Click to Show/Hide
|
|
| External link | ||
| Description |
METTL3 plays a carcinogenic role in human EC progression partially throughAKT signaling pathways, suggesting that METTL3 serves as a potential therapeutic target foresophageal cancer therapy. A double-effect inhibitor (BEZ235) inhibited AKT and mTOR phosphorylation and hindered the effect of METTL3 overexpression on the proliferation and migration of Eca-109 and KY-SE150 cells.
|
[212] |
| Compound Name | STM2457 | Preclinical |
| Synonyms |
STM-2457; SCHEMBL22499068; GTPL11529; CHEBI:172325; EX-A5018; STM 2457; s9870; HY-134836; N-[(6-{[(cyclohexylmethyl)amino]methyl}imidazo[1,2-a]pyridin-2-yl)methyl]-4-oxo-4H-pyrido[1,2-a]pyrimidine-2-carboxamide; N-[[6-[(cyclohexylmethylamino)methyl]imidazo[1,2-a]pyridin-2-yl]methyl]-4-oxopyrido[1,2-a]pyrimidine-2-carboxamide
Click to Show/Hide
|
|
| External link | ||
| Description |
inhibition of METTL3 by STM2457 targets key stem cell populations ofacute myeloid leukaemia and reverses the AML phenotype, preventing or slowing the development of AML in re-transplantation experiments.
|
[595] |
| Compound Name | N~2~-hexyl-6,7-dimethoxy-N~4~-(1-methylpiperidin-4-yl)quinazoline-2,4-diamine | Investigative |
| Synonyms |
CHEMBL4086403; SCHEMBL23260478; MS012; BDBM50501525; MS 012; MS-012; N~2~-hexyl-6,7-dimethoxy-N~4~-(1-methylpiperidin-4-yl)quinazoline-2,4-diamine; 2089617-83-2
Click to Show/Hide
|
|
| External link | ||
| Activity |
IC50=7±2 nM
|
[638] |
| Compound Name | 6,7-Dimethoxy-N-(1-Methylpiperidin-4-Yl)-2-(Morpholin-4-Yl)quinazolin-4-Amine | Investigative |
| Synonyms |
CHEMBL571717; 6,7-Dimethoxy-N-(1-Methylpiperidin-4-Yl)-2-(Morpholin-4-Yl)quinazolin-4-Amine; 7L6; SCHEMBL15280520; BDBM50300032; NCGC00185861-01; 6,7-dimethoxy-N-(1-methylpiperidin-4-yl)-2-morpholinoquinazolin-4-amine
Click to Show/Hide
|
|
| External link | ||
| Activity |
IC50=13 ± 4 nM
|
[638] |
| Compound Name | Cycloleucine | Investigative |
| Synonyms |
1-Aminocyclopentanecarboxylic acid; 52-52-8; 1-Aminocyclopentane-1-carboxylic acid; Cycloleucin; 1-Amino-1-cyclopentanecarboxylic acid; 1-Amino-1-carboxycyclopentane; 1-Amino-cyclopentanecarboxylic acid; CYCLO-LEUCINE; Cyclopentanecarboxylic acid, 1-amino-; NSC 1026; CB 1639; X 201; UNII-0TQU7668EI; 1-Aminocyclopentanecarboxylate; HSDB 5195; WR 14,997; NSC1026; 1-amino cyclopentane carboxylic acid; EINECS 200-144-6; BRN 0636626; aminocyclopentanecarboxylic acid; Cyclopentanecarboxylic acid, 1-amino-, L-; AI3-26442
Click to Show/Hide
|
|
| External link | ||
| Description |
METTL3-mediated m6A RNA methylation modulatesuveal melanoma cell proliferation, migration, and invasion by targetingc-Met. Cycloleucine (Cyc) was used to block m6 A methylation in UM cells.
|
[175] |
| Compound Name | AdoHcy | Investigative |
| Synonyms |
S-Adenosyl-L-homocysteine; S-adenosylhomocysteine; S-adenosyl-L-homocysteine; 979-92-0; AdoHcy; S-(5'-adenosyl)-L-homocysteine; adenosylhomocysteine; Formycinylhomocysteine; Adenosyl-L-homocysteine; S-(5'-deoxyadenosin-5'-yl)-L-homocysteine; 2-S-adenosyl-L-homocysteine; 5'-Deoxy-S-adenosyl-L-homocysteine; S-adenosyl-homocysteine; S-Adenosyl Homocysteine; L-S-Adenosylhomocysteine; L-Homocysteine, S-(5'-deoxyadenosin-5'-yl)-; adenosylhomo-cys; adenosyl-homo-cys; UNII-8K31Q2S66S; (S)-5'-(S)-(3-Amino-3-carboxypropyl)-5'-thioadenosine; BRN 5166233; SAH; S-Adenosylhomocysteine; S-Adenosyl-L-Homocysteine
Click to Show/Hide
|
|
| External link | ||
| Description |
Mettl3 inhibitor, S-adenosylhomocysteine promoted the apoptosis and autophagy of chondrocytes with inflammation in vitro and aggravated the degeneration of chondrocytes and subchondral bone in monosodium iodoacetate (MIA) inducedtemporomandibular joint osteoarthritis mice in vivo. Bcl2 protein interacted with Beclin1 protein in chondrocytes induced by TNF-alpha stimulation. Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through m6A/Ythdf1/Bcl2 signal axis which provides promising therapeutic strategy fortemporomandibular joint osteoarthritis.
|
[10] |
References