m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00802)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
SPTBN2
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
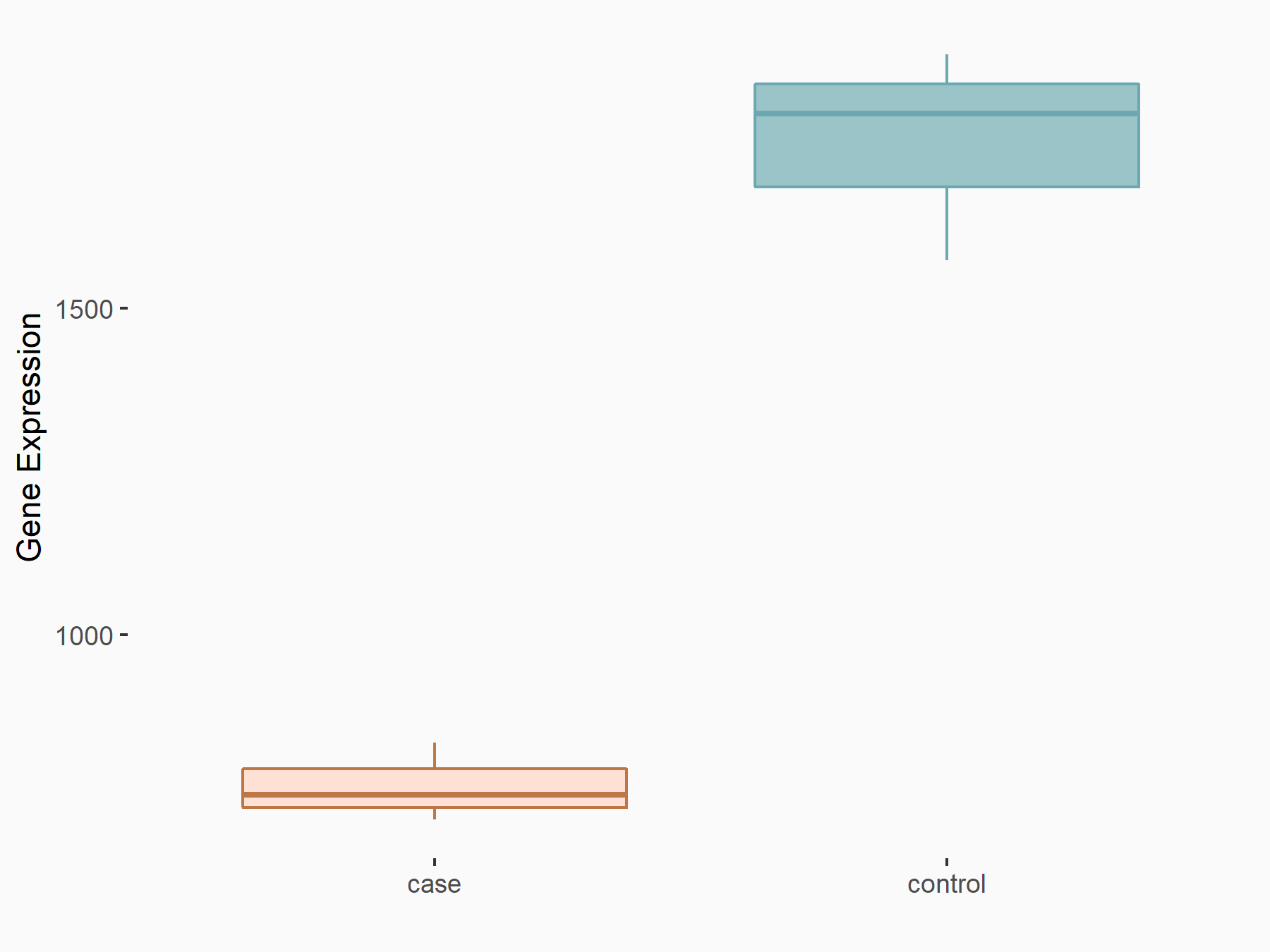 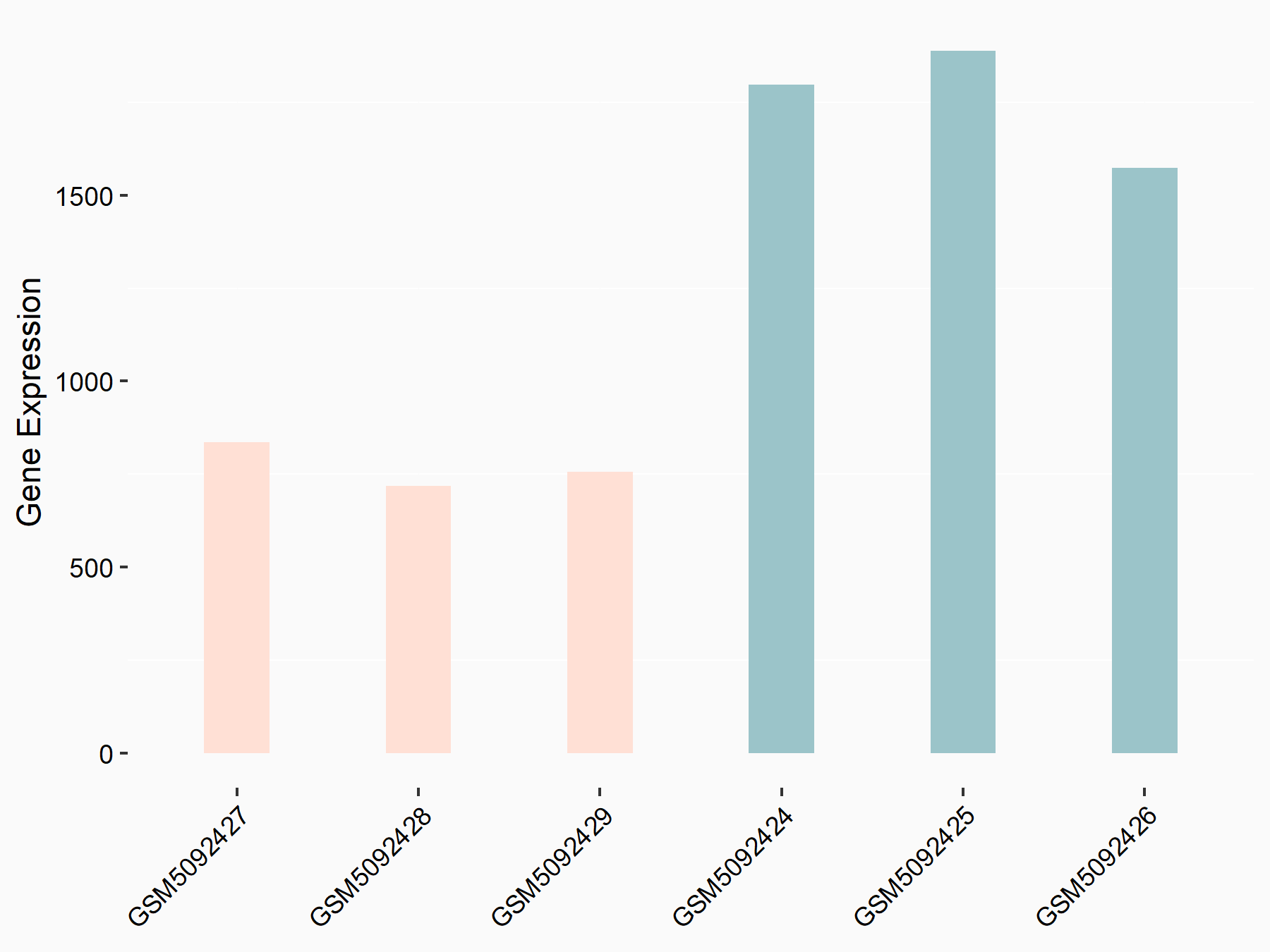 |
logFC: -1.19E+00 p-value: 2.92E-45 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Overexpression of LINC01605, regulated by SMYD2-EP300-mediated H3K27ac and H3K4me3 modifications, bound to METTL3 protein to promote m6A modification of Spectrin beta, non-erythrocytic 2 (SPTBN2) mRNA, leading to the development of colorectal cancer. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| In-vitro Model | Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| In-vivo Model | The growth of CC cells in vivo was examined by subcutaneous injection of LoVo cells or Caco-2 cells into NSG mice, while the metastatic ability of cells in vivo by intracardiac injection into mice. | |||
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Overexpression of LINC01605, regulated by SMYD2-EP300-mediated H3K27ac and H3K4me3 modifications, bound to METTL3 protein to promote m6A modification of Spectrin beta, non-erythrocytic 2 (SPTBN2) mRNA, leading to the development of colorectal cancer. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| In-vitro Model | Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| In-vivo Model | The growth of CC cells in vivo was examined by subcutaneous injection of LoVo cells or Caco-2 cells into NSG mice, while the metastatic ability of cells in vivo by intracardiac injection into mice. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03222 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03223 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
Non-coding RNA
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05929 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 1605 | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00802)
| In total 31 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE004997 | Click to Show/Hide the Full List | ||
| mod site | chr11:66712942-66712943:- | [2] | |
| Sequence | TGAGGCGAGAGGATCACTTGAGCCCAGGAGTTCTAGGCTGC | ||
| Transcript ID List | ENST00000473849.3; rmsk_3572549; ENST00000647510.1; ENST00000617502.4; ENST00000533211.5; ENST00000611817.4; ENST00000309996.6; ENST00000529997.5 | ||
| External Link | RMBase: RNA-editing_site_23077 | ||
| mod ID: A2ISITE004998 | Click to Show/Hide the Full List | ||
| mod site | chr11:66712947-66712948:- | [2] | |
| Sequence | GAGGCTGAGGCGAGAGGATCACTTGAGCCCAGGAGTTCTAG | ||
| Transcript ID List | ENST00000529997.5; ENST00000617502.4; ENST00000309996.6; ENST00000611817.4; ENST00000473849.3; ENST00000647510.1; ENST00000533211.5; rmsk_3572549 | ||
| External Link | RMBase: RNA-editing_site_23078 | ||
| mod ID: A2ISITE004999 | Click to Show/Hide the Full List | ||
| mod site | chr11:66712983-66712984:- | [2] | |
| Sequence | CCACGGTGGTACCTGCCTATAGTCCTAGCTGCTAGTGAGGC | ||
| Transcript ID List | ENST00000533211.5; ENST00000617502.4; ENST00000473849.3; ENST00000529997.5; rmsk_3572549; ENST00000611817.4; ENST00000309996.6; ENST00000647510.1 | ||
| External Link | RMBase: RNA-editing_site_23079 | ||
| mod ID: A2ISITE005000 | Click to Show/Hide the Full List | ||
| mod site | chr11:66712985-66712986:- | [2] | |
| Sequence | GGCCACGGTGGTACCTGCCTATAGTCCTAGCTGCTAGTGAG | ||
| Transcript ID List | ENST00000533211.5; rmsk_3572549; ENST00000611817.4; ENST00000529997.5; ENST00000473849.3; ENST00000647510.1; ENST00000617502.4; ENST00000309996.6 | ||
| External Link | RMBase: RNA-editing_site_23080 | ||
| mod ID: A2ISITE005001 | Click to Show/Hide the Full List | ||
| mod site | chr11:66719422-66719423:- | [2] | |
| Sequence | AATAAACACTGAATGAATGAAGATGAGGAAGGGCAAGTGTA | ||
| Transcript ID List | ENST00000611817.4; ENST00000527010.1; ENST00000309996.6; ENST00000533211.5; ENST00000529997.5; ENST00000617502.4; rmsk_3572558; ENST00000647510.1 | ||
| External Link | RMBase: RNA-editing_site_23081 | ||
| mod ID: A2ISITE005002 | Click to Show/Hide the Full List | ||
| mod site | chr11:66723313-66723314:- | [2] | |
| Sequence | GTAGGGCTAGGAGAAATGCAAAATTGGGGGTTTTGTTAAAG | ||
| Transcript ID List | ENST00000527010.1; ENST00000533211.5; ENST00000647510.1 | ||
| External Link | RMBase: RNA-editing_site_23082 | ||
| mod ID: A2ISITE005003 | Click to Show/Hide the Full List | ||
| mod site | chr11:66723325-66723326:- | [2] | |
| Sequence | TGGTCCTAGGAAGTAGGGCTAGGAGAAATGCAAAATTGGGG | ||
| Transcript ID List | ENST00000527010.1; ENST00000533211.5; ENST00000647510.1 | ||
| External Link | RMBase: RNA-editing_site_23083 | ||
| mod ID: A2ISITE005004 | Click to Show/Hide the Full List | ||
| mod site | chr11:66723331-66723332:- | [2] | |
| Sequence | CAGCCCTGGTCCTAGGAAGTAGGGCTAGGAGAAATGCAAAA | ||
| Transcript ID List | ENST00000533211.5; ENST00000647510.1; ENST00000527010.1 | ||
| External Link | RMBase: RNA-editing_site_23084 | ||
| mod ID: A2ISITE005005 | Click to Show/Hide the Full List | ||
| mod site | chr11:66723334-66723335:- | [2] | |
| Sequence | CTACAGCCCTGGTCCTAGGAAGTAGGGCTAGGAGAAATGCA | ||
| Transcript ID List | ENST00000527010.1; ENST00000647510.1; ENST00000533211.5 | ||
| External Link | RMBase: RNA-editing_site_23085 | ||
| mod ID: A2ISITE005006 | Click to Show/Hide the Full List | ||
| mod site | chr11:66723361-66723362:- | [2] | |
| Sequence | TCCTTCCAGAACCCGAGACTACCGAGACTACAGCCCTGGTC | ||
| Transcript ID List | ENST00000527010.1; ENST00000647510.1; ENST00000533211.5 | ||
| External Link | RMBase: RNA-editing_site_23086 | ||
| mod ID: A2ISITE005007 | Click to Show/Hide the Full List | ||
| mod site | chr11:66723371-66723372:- | [2] | |
| Sequence | CAGACCTCCTTCCTTCCAGAACCCGAGACTACCGAGACTAC | ||
| Transcript ID List | ENST00000527010.1; ENST00000647510.1; ENST00000533211.5 | ||
| External Link | RMBase: RNA-editing_site_23087 | ||
| mod ID: A2ISITE005008 | Click to Show/Hide the Full List | ||
| mod site | chr11:66723388-66723389:- | [2] | |
| Sequence | AGCACTTCTCTATTCTCCAGACCTCCTTCCTTCCAGAACCC | ||
| Transcript ID List | ENST00000647510.1; ENST00000527010.1; ENST00000533211.5 | ||
| External Link | RMBase: RNA-editing_site_23088 | ||
| mod ID: A2ISITE005009 | Click to Show/Hide the Full List | ||
| mod site | chr11:66723426-66723427:- | [2] | |
| Sequence | GCCCATGAGTCTCACCACCTACCTGAGTCTCACCACCCAGC | ||
| Transcript ID List | ENST00000527010.1; ENST00000647510.1; ENST00000533211.5 | ||
| External Link | RMBase: RNA-editing_site_23089 | ||
| mod ID: A2ISITE005010 | Click to Show/Hide the Full List | ||
| mod site | chr11:66723430-66723431:- | [2] | |
| Sequence | GTGTGCCCATGAGTCTCACCACCTACCTGAGTCTCACCACC | ||
| Transcript ID List | ENST00000533211.5; ENST00000647510.1; ENST00000527010.1 | ||
| External Link | RMBase: RNA-editing_site_23090 | ||
| mod ID: A2ISITE005011 | Click to Show/Hide the Full List | ||
| mod site | chr11:66723467-66723468:- | [2] | |
| Sequence | TGGAATCAGTTAATGATGGAAGCCAGGTTCTGCCTTTGTGT | ||
| Transcript ID List | ENST00000533211.5; ENST00000647510.1; ENST00000527010.1 | ||
| External Link | RMBase: RNA-editing_site_23091 | ||
| mod ID: A2ISITE005012 | Click to Show/Hide the Full List | ||
| mod site | chr11:66723472-66723473:- | [2] | |
| Sequence | CTTCATGGAATCAGTTAATGATGGAAGCCAGGTTCTGCCTT | ||
| Transcript ID List | ENST00000527010.1; ENST00000647510.1; ENST00000533211.5 | ||
| External Link | RMBase: RNA-editing_site_23092 | ||
| mod ID: A2ISITE005013 | Click to Show/Hide the Full List | ||
| mod site | chr11:66723494-66723495:- | [2] | |
| Sequence | CCTTCAAGTAGGTCAGGTTCAGCTTCATGGAATCAGTTAAT | ||
| Transcript ID List | ENST00000647510.1; ENST00000533211.5; ENST00000527010.1 | ||
| External Link | RMBase: RNA-editing_site_23093 | ||
| mod ID: A2ISITE005014 | Click to Show/Hide the Full List | ||
| mod site | chr11:66723508-66723509:- | [2] | |
| Sequence | GCAGACAACCCTGACCTTCAAGTAGGTCAGGTTCAGCTTCA | ||
| Transcript ID List | ENST00000527010.1; ENST00000647510.1; ENST00000533211.5 | ||
| External Link | RMBase: RNA-editing_site_23094 | ||
| mod ID: A2ISITE005015 | Click to Show/Hide the Full List | ||
| mod site | chr11:66724714-66724715:- | [2] | |
| Sequence | GCTGAACCTGACCTGCTTGAAGGCCAGGGTTGTCTGCCCCT | ||
| Transcript ID List | ENST00000647510.1; ENST00000533211.5; ENST00000527010.1 | ||
| External Link | RMBase: RNA-editing_site_23095 | ||
| mod ID: A2ISITE005016 | Click to Show/Hide the Full List | ||
| mod site | chr11:66724729-66724730:- | [2] | |
| Sequence | AACTGGTTCCATGAAGCTGAACCTGACCTGCTTGAAGGCCA | ||
| Transcript ID List | ENST00000647510.1; ENST00000527010.1; ENST00000533211.5 | ||
| External Link | RMBase: RNA-editing_site_23096 | ||
| mod ID: A2ISITE005017 | Click to Show/Hide the Full List | ||
| mod site | chr11:66724735-66724736:- | [2] | |
| Sequence | ATCATTAACTGGTTCCATGAAGCTGAACCTGACCTGCTTGA | ||
| Transcript ID List | ENST00000533211.5; ENST00000647510.1; ENST00000527010.1 | ||
| External Link | RMBase: RNA-editing_site_23097 | ||
| mod ID: A2ISITE005018 | Click to Show/Hide the Full List | ||
| mod site | chr11:66724736-66724737:- | [2] | |
| Sequence | CATCATTAACTGGTTCCATGAAGCTGAACCTGACCTGCTTG | ||
| Transcript ID List | ENST00000647510.1; ENST00000527010.1; ENST00000533211.5 | ||
| External Link | RMBase: RNA-editing_site_23098 | ||
| mod ID: A2ISITE005019 | Click to Show/Hide the Full List | ||
| mod site | chr11:66724748-66724749:- | [2] | |
| Sequence | CTTTCTGGCTTCCATCATTAACTGGTTCCATGAAGCTGAAC | ||
| Transcript ID List | ENST00000527010.1; ENST00000647510.1; ENST00000533211.5 | ||
| External Link | RMBase: RNA-editing_site_23099 | ||
| mod ID: A2ISITE005020 | Click to Show/Hide the Full List | ||
| mod site | chr11:66724749-66724750:- | [2] | |
| Sequence | CCTTTCTGGCTTCCATCATTAACTGGTTCCATGAAGCTGAA | ||
| Transcript ID List | ENST00000647510.1; ENST00000533211.5; ENST00000527010.1 | ||
| External Link | RMBase: RNA-editing_site_23100 | ||
| mod ID: A2ISITE005021 | Click to Show/Hide the Full List | ||
| mod site | chr11:66724805-66724806:- | [2] | |
| Sequence | GTGGTGAGACTCGGGTAGGTAGGGAGGTGTGTGCCCATGGA | ||
| Transcript ID List | ENST00000533211.5; ENST00000647510.1; ENST00000527010.1 | ||
| External Link | RMBase: RNA-editing_site_23101 | ||
| mod ID: A2ISITE005022 | Click to Show/Hide the Full List | ||
| mod site | chr11:66724809-66724810:- | [2] | |
| Sequence | ATGGGTGGTGAGACTCGGGTAGGTAGGGAGGTGTGTGCCCA | ||
| Transcript ID List | ENST00000533211.5; ENST00000647510.1; ENST00000527010.1 | ||
| External Link | RMBase: RNA-editing_site_23102 | ||
| mod ID: A2ISITE005023 | Click to Show/Hide the Full List | ||
| mod site | chr11:66724833-66724834:- | [2] | |
| Sequence | GGAGGTATGGAGAAGAGAGAAGTGATGGGTGGTGAGACTCG | ||
| Transcript ID List | ENST00000527010.1; ENST00000647510.1; ENST00000533211.5 | ||
| External Link | RMBase: RNA-editing_site_23103 | ||
| mod ID: A2ISITE005024 | Click to Show/Hide the Full List | ||
| mod site | chr11:66724875-66724876:- | [2] | |
| Sequence | ACCAGGGCTGTGGTCTTGGCAGTCTCTGGTTCTGGAAAGGA | ||
| Transcript ID List | ENST00000647510.1; ENST00000533211.5; ENST00000527010.1 | ||
| External Link | RMBase: RNA-editing_site_23104 | ||
| mod ID: A2ISITE005025 | Click to Show/Hide the Full List | ||
| mod site | chr11:66724905-66724906:- | [2] | |
| Sequence | CTGCATTTCTCCTGAGCCCTACTTCCTACCACCAGGGCTGT | ||
| Transcript ID List | ENST00000647510.1; ENST00000527010.1; ENST00000533211.5 | ||
| External Link | RMBase: RNA-editing_site_23105 | ||
| mod ID: A2ISITE005026 | Click to Show/Hide the Full List | ||
| mod site | chr11:66724941-66724942:- | [2] | |
| Sequence | TTGGGCCCCTTTGTAACTTTAACAAAACCCCCAATTCTGCA | ||
| Transcript ID List | ENST00000647510.1; ENST00000527010.1; ENST00000533211.5 | ||
| External Link | RMBase: RNA-editing_site_23106 | ||
| mod ID: A2ISITE005027 | Click to Show/Hide the Full List | ||
| mod site | chr11:66724947-66724948:- | [2] | |
| Sequence | TCCCAGTTGGGCCCCTTTGTAACTTTAACAAAACCCCCAAT | ||
| Transcript ID List | ENST00000527010.1; ENST00000647510.1; ENST00000533211.5 | ||
| External Link | RMBase: RNA-editing_site_23107 | ||
5-methylcytidine (m5C)
| In total 13 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE005229 | Click to Show/Hide the Full List | ||
| mod site | chr11:66685447-66685448:- | ||
| Sequence | CAGACCAGCCAGGAACCTCTCAGAGCTGAAGCAGGCCCTGG | ||
| Cell/Tissue List | testis; HeLa | ||
| Seq Type List | Bisulfite-seq; m5C-RIP-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000617502.4; ENST00000529997.5; ENST00000533211.5; ENST00000309996.6 | ||
| External Link | RMBase: m5C_site_8115 | ||
| mod ID: M5CSITE005230 | Click to Show/Hide the Full List | ||
| mod site | chr11:66685856-66685857:- | [3] | |
| Sequence | CAAGTAGTTGGGGGCAAGGTCCCAGGCCAACTCCCTCCCTC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000528051.1; ENST00000611817.4; ENST00000647510.1; ENST00000529997.5; ENST00000533211.5; ENST00000309996.6; ENST00000617502.4 | ||
| External Link | RMBase: m5C_site_8116 | ||
| mod ID: M5CSITE005231 | Click to Show/Hide the Full List | ||
| mod site | chr11:66687582-66687583:- | [3] | |
| Sequence | GCAGACCCGGACTCGGGGCCCGGCCCCATCTGCAATGCCCC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000309996.6; ENST00000533211.5; ENST00000647510.1; ENST00000529997.5; ENST00000611817.4 | ||
| External Link | RMBase: m5C_site_8117 | ||
| mod ID: M5CSITE005232 | Click to Show/Hide the Full List | ||
| mod site | chr11:66687583-66687584:- | [3] | |
| Sequence | GGCAGACCCGGACTCGGGGCCCGGCCCCATCTGCAATGCCC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000529997.5; ENST00000617502.4; ENST00000533211.5; ENST00000309996.6; ENST00000647510.1; ENST00000611817.4 | ||
| External Link | RMBase: m5C_site_8118 | ||
| mod ID: M5CSITE005233 | Click to Show/Hide the Full List | ||
| mod site | chr11:66687584-66687585:- | [3] | |
| Sequence | AGGCAGACCCGGACTCGGGGCCCGGCCCCATCTGCAATGCC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000533211.5; ENST00000529997.5; ENST00000617502.4; ENST00000611817.4; ENST00000647510.1 | ||
| External Link | RMBase: m5C_site_8119 | ||
| mod ID: M5CSITE005234 | Click to Show/Hide the Full List | ||
| mod site | chr11:66687589-66687590:- | [3] | |
| Sequence | GAGAGAGGCAGACCCGGACTCGGGGCCCGGCCCCATCTGCA | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m5C-RIP-seq; Bisulfite-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000647510.1; ENST00000617502.4; ENST00000611817.4; ENST00000529997.5; ENST00000309996.6 | ||
| External Link | RMBase: m5C_site_8120 | ||
| mod ID: M5CSITE005235 | Click to Show/Hide the Full List | ||
| mod site | chr11:66687613-66687614:- | [3] | |
| Sequence | GGGACGAAGCCAATGGGCCCCGGGGAGAGAGGCAGACCCGG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000611817.4; ENST00000532650.1; ENST00000529997.5; ENST00000647510.1; ENST00000309996.6; ENST00000533211.5 | ||
| External Link | RMBase: m5C_site_8121 | ||
| mod ID: M5CSITE005236 | Click to Show/Hide the Full List | ||
| mod site | chr11:66687614-66687615:- | [3] | |
| Sequence | GGGGACGAAGCCAATGGGCCCCGGGGAGAGAGGCAGACCCG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000529997.5; ENST00000647510.1; ENST00000611817.4; ENST00000309996.6; ENST00000533211.5; ENST00000617502.4; ENST00000532650.1 | ||
| External Link | RMBase: m5C_site_8122 | ||
| mod ID: M5CSITE005237 | Click to Show/Hide the Full List | ||
| mod site | chr11:66687615-66687616:- | [3] | |
| Sequence | AGGGGACGAAGCCAATGGGCCCCGGGGAGAGAGGCAGACCC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000532650.1; ENST00000617502.4; ENST00000611817.4; ENST00000647510.1; ENST00000529997.5; ENST00000533211.5 | ||
| External Link | RMBase: m5C_site_8123 | ||
| mod ID: M5CSITE005238 | Click to Show/Hide the Full List | ||
| mod site | chr11:66687616-66687617:- | [3] | |
| Sequence | CAGGGGACGAAGCCAATGGGCCCCGGGGAGAGAGGCAGACC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000611817.4; ENST00000617502.4; ENST00000532650.1; ENST00000529997.5; ENST00000647510.1; ENST00000309996.6 | ||
| External Link | RMBase: m5C_site_8124 | ||
| mod ID: M5CSITE005239 | Click to Show/Hide the Full List | ||
| mod site | chr11:66691316-66691317:- | ||
| Sequence | CTGCAGCGCCGACACTGTGCCTACGAGCATGACATTCAGGC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000533211.5; ENST00000529997.5; ENST00000611817.4; ENST00000309996.6; ENST00000647510.1 | ||
| External Link | RMBase: m5C_site_8126 | ||
| mod ID: M5CSITE005240 | Click to Show/Hide the Full List | ||
| mod site | chr11:66692586-66692587:- | ||
| Sequence | GAACAGTGGATCCAGGAGCGCGAGGTGGTGGCGGCCTCCCA | ||
| Cell/Tissue List | testis | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000533211.5; ENST00000647510.1; ENST00000529997.5; ENST00000617502.4; ENST00000611817.4 | ||
| External Link | RMBase: m5C_site_8127 | ||
| mod ID: M5CSITE005241 | Click to Show/Hide the Full List | ||
| mod site | chr11:66707749-66707750:- | ||
| Sequence | CACGAAGCCATTGAGACGGACATCGTGGCCTACAGCGGCCG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000611817.4; ENST00000533211.5; ENST00000617502.4; ENST00000647510.1; ENST00000529997.5 | ||
| External Link | RMBase: m5C_site_8131 | ||
N6-methyladenosine (m6A)
| In total 134 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE007341 | Click to Show/Hide the Full List | ||
| mod site | chr11:66685265-66685266:- | [4] | |
| Sequence | CTGGCCAAGGAGGTGATTAAACAGCTCAGCTTCTCTCACCG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000529997.5; ENST00000647510.1; ENST00000617502.4; ENST00000309996.6; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150339 | ||
| mod ID: M6ASITE007342 | Click to Show/Hide the Full List | ||
| mod site | chr11:66685303-66685304:- | [4] | |
| Sequence | TCCAGGTGTGTTGTTCGGGGACAGCCACCGCCTTGAGTCTG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000529997.5; ENST00000647510.1; ENST00000309996.6; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150340 | ||
| mod ID: M6ASITE007343 | Click to Show/Hide the Full List | ||
| mod site | chr11:66685453-66685454:- | [4] | |
| Sequence | ACCCCTCAGACCAGCCAGGAACCTCTCAGAGCTGAAGCAGG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000533211.5; ENST00000617502.4; ENST00000309996.6; ENST00000529997.5 | ||
| External Link | RMBase: m6A_site_150341 | ||
| mod ID: M6ASITE007344 | Click to Show/Hide the Full List | ||
| mod site | chr11:66685464-66685465:- | [4] | |
| Sequence | TCTGGTTCCTCACCCCTCAGACCAGCCAGGAACCTCTCAGA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000529997.5; ENST00000647510.1; ENST00000533211.5; ENST00000309996.6 | ||
| External Link | RMBase: m6A_site_150342 | ||
| mod ID: M6ASITE007345 | Click to Show/Hide the Full List | ||
| mod site | chr11:66685578-66685579:- | [4] | |
| Sequence | TGTCCCTCACCACGGTGTGGACAGTGCCGCACCCTCAACAT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; Huh7; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000617502.4; ENST00000529997.5; ENST00000647510.1; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150343 | ||
| mod ID: M6ASITE007346 | Click to Show/Hide the Full List | ||
| mod site | chr11:66685628-66685629:- | [5] | |
| Sequence | CCCCTCTCCCACCCCCGCCGACTTCTCCCCTTTCCCCAGCC | ||
| Motif Score | 3.287565476 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000617502.4; ENST00000309996.6; ENST00000533211.5; ENST00000529997.5 | ||
| External Link | RMBase: m6A_site_150344 | ||
| mod ID: M6ASITE007347 | Click to Show/Hide the Full List | ||
| mod site | chr11:66685659-66685660:- | [4] | |
| Sequence | AATTCCAGACTGGTGGTGGGACCCAGGTAACCCCCTCTCCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; H1A; H1B; A549; Huh7; HEK293A-TOA; iSLK; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000529997.5; ENST00000309996.6; ENST00000528051.1; ENST00000617502.4; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150345 | ||
| mod ID: M6ASITE007348 | Click to Show/Hide the Full List | ||
| mod site | chr11:66685671-66685672:- | [4] | |
| Sequence | CTTCTGCTGTTTAATTCCAGACTGGTGGTGGGACCCAGGTA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1A; H1B; A549; Huh7; Jurkat; HEK293A-TOA; iSLK; TIME; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000617502.4; ENST00000528051.1; ENST00000529997.5; ENST00000647510.1; ENST00000309996.6 | ||
| External Link | RMBase: m6A_site_150346 | ||
| mod ID: M6ASITE007349 | Click to Show/Hide the Full List | ||
| mod site | chr11:66685735-66685736:- | [4] | |
| Sequence | AGGTCAACCCATCACCAGGAACTGTCACTGGGGACGAGTCC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; hESCs; fibroblasts; MM6; Huh7; Jurkat; GSC-11; HEK293A-TOA; iSLK; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000529997.5; ENST00000617502.4; ENST00000533211.5; ENST00000309996.6; ENST00000528051.1; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150347 | ||
| mod ID: M6ASITE007350 | Click to Show/Hide the Full List | ||
| mod site | chr11:66685785-66685786:- | [4] | |
| Sequence | GGGACCGCCCTCTTGTCAGGACAACTGCCTGCTGCTAGGGT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; H1A; H1B; hESCs; fibroblasts; MM6; Huh7; Jurkat; HEK293A-TOA; iSLK; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000528051.1; ENST00000533211.5; ENST00000617502.4; ENST00000529997.5; ENST00000309996.6; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150348 | ||
| mod ID: M6ASITE007351 | Click to Show/Hide the Full List | ||
| mod site | chr11:66685802-66685803:- | [4] | |
| Sequence | GCCAGGGACAGTCGACAGGGACCGCCCTCTTGTCAGGACAA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1A; H1B; hNPCs; fibroblasts; A549; Huh7; Jurkat; HEK293A-TOA; iSLK; TIME; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000528051.1; ENST00000309996.6; ENST00000647510.1; ENST00000529997.5; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150349 | ||
| mod ID: M6ASITE007352 | Click to Show/Hide the Full List | ||
| mod site | chr11:66685815-66685816:- | [4] | |
| Sequence | CGTTCAGGAAACTGCCAGGGACAGTCGACAGGGACCGCCCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; hESC-HEK293T; H1A; H1B; hNPCs; fibroblasts; A549; Huh7; Jurkat; HEK293A-TOA; iSLK; TIME; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000529997.5; ENST00000528051.1; ENST00000647510.1; ENST00000617502.4; ENST00000309996.6 | ||
| External Link | RMBase: m6A_site_150350 | ||
| mod ID: M6ASITE007353 | Click to Show/Hide the Full List | ||
| mod site | chr11:66685825-66685826:- | [4] | |
| Sequence | TCCCTCCCTCCGTTCAGGAAACTGCCAGGGACAGTCGACAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1A; H1B; hNPCs; fibroblasts; A549; Huh7; Jurkat; HEK293A-TOA; iSLK; TIME; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000528051.1; ENST00000309996.6; ENST00000647510.1; ENST00000529997.5; ENST00000617502.4; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150351 | ||
| mod ID: M6ASITE007354 | Click to Show/Hide the Full List | ||
| mod site | chr11:66685877-66685878:- | [4] | |
| Sequence | CTTCAGCTTCTTTAAGAAGAACAAGTAGTTGGGGGCAAGGT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; HEK293A-TOA; iSLK; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000529997.5; ENST00000309996.6; ENST00000533211.5; ENST00000647510.1; ENST00000528051.1; ENST00000611817.4; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150352 | ||
| mod ID: M6ASITE007355 | Click to Show/Hide the Full List | ||
| mod site | chr11:66686209-66686210:- | [6] | |
| Sequence | CATTCTCCTAATAGCATGAAACAGTCAGCTCACTTTCTGCC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | Huh7; iSLK | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000611817.4; ENST00000528051.1; ENST00000533211.5; ENST00000529997.5; ENST00000309996.6; ENST00000617502.4; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150353 | ||
| mod ID: M6ASITE007356 | Click to Show/Hide the Full List | ||
| mod site | chr11:66686282-66686283:- | [4] | |
| Sequence | GTCTTGTCCTTTGTGGTAAGACTGGATGTGTGCGACGGCCG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; Huh7; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000611817.4; ENST00000309996.6; ENST00000529997.5; ENST00000647510.1; ENST00000528051.1; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150354 | ||
| mod ID: M6ASITE007357 | Click to Show/Hide the Full List | ||
| mod site | chr11:66686338-66686339:- | [4] | |
| Sequence | CCAGAAGCTTCCAGCTGCAGACTCCCCTTCTTTCCCTGTCC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; Huh7; iSLK; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000309996.6; ENST00000533211.5; ENST00000611817.4; ENST00000528051.1; ENST00000529997.5; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150355 | ||
| mod ID: M6ASITE007358 | Click to Show/Hide the Full List | ||
| mod site | chr11:66687116-66687117:- | [7] | |
| Sequence | GCGTGGGAGCCTCGGCTTTTACAAGGATGCCAAGGCAGCCA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | brain | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000309996.6; ENST00000529997.5; ENST00000611817.4; ENST00000647510.1; ENST00000617502.4; ENST00000530775.1 | ||
| External Link | RMBase: m6A_site_150356 | ||
| mod ID: M6ASITE007359 | Click to Show/Hide the Full List | ||
| mod site | chr11:66687592-66687593:- | [4] | |
| Sequence | GGGGAGAGAGGCAGACCCGGACTCGGGGCCCGGCCCCATCT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; H1A; Jurkat; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000309996.6; ENST00000533211.5; ENST00000611817.4; ENST00000529997.5; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150357 | ||
| mod ID: M6ASITE007360 | Click to Show/Hide the Full List | ||
| mod site | chr11:66687598-66687599:- | [4] | |
| Sequence | GGCCCCGGGGAGAGAGGCAGACCCGGACTCGGGGCCCGGCC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; H1A; Jurkat; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000309996.6; ENST00000617502.4; ENST00000647510.1; ENST00000529997.5; ENST00000611817.4 | ||
| External Link | RMBase: m6A_site_150358 | ||
| mod ID: M6ASITE007361 | Click to Show/Hide the Full List | ||
| mod site | chr11:66687644-66687645:- | [4] | |
| Sequence | ACACTGATTCCCCCCCAGGGACCTGGCTCAGGGGACGAAGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; Jurkat; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000532650.1; ENST00000533211.5; ENST00000611817.4; ENST00000309996.6; ENST00000529997.5; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150359 | ||
| mod ID: M6ASITE007362 | Click to Show/Hide the Full List | ||
| mod site | chr11:66687897-66687898:- | [4] | |
| Sequence | CCCCTGCTGGGACAACAGAGACTTGAGCACAGCAGCTTCCC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; Jurkat; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000533211.5; ENST00000529997.5; ENST00000611817.4; ENST00000617502.4; ENST00000532650.1; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150360 | ||
| mod ID: M6ASITE007363 | Click to Show/Hide the Full List | ||
| mod site | chr11:66687906-66687907:- | [4] | |
| Sequence | TCCTTGCAGCCCCTGCTGGGACAACAGAGACTTGAGCACAG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; Jurkat; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000309996.6; ENST00000529997.5; ENST00000533211.5; ENST00000617502.4; ENST00000611817.4; ENST00000532650.1 | ||
| External Link | RMBase: m6A_site_150361 | ||
| mod ID: M6ASITE007364 | Click to Show/Hide the Full List | ||
| mod site | chr11:66688006-66688007:- | [7] | |
| Sequence | TGCACAGATGGAGAGCCCTCACAGGTGACCCCACTGTCCCT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | brain | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000611817.4; ENST00000617502.4; ENST00000529997.5; ENST00000533211.5; ENST00000532650.1; ENST00000309996.6; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150362 | ||
| mod ID: M6ASITE007365 | Click to Show/Hide the Full List | ||
| mod site | chr11:66688193-66688194:- | [4] | |
| Sequence | GGGACCTGGTGGGCGGCCAGACAGCTTCTGACACCACCTGG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; Huh7; Jurkat; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000611817.4; ENST00000529997.5; ENST00000617502.4; ENST00000532650.1; ENST00000309996.6; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150363 | ||
| mod ID: M6ASITE007366 | Click to Show/Hide the Full List | ||
| mod site | chr11:66688210-66688211:- | [4] | |
| Sequence | AGCCAGTGTGCCTCCAGGGGACCTGGTGGGCGGCCAGACAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; Huh7; Jurkat; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000529997.5; ENST00000611817.4; ENST00000309996.6; ENST00000533211.5; ENST00000617502.4; ENST00000532650.1 | ||
| External Link | RMBase: m6A_site_150364 | ||
| mod ID: M6ASITE007367 | Click to Show/Hide the Full List | ||
| mod site | chr11:66688236-66688237:- | [4] | |
| Sequence | AAACAGCCGCCTGCTCCCGAACCCACAGCCAGTGTGCCTCC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; H1B; Huh7; Jurkat; HEK293T; endometrial; HEC-1-A; GSCs; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000529997.5; ENST00000611817.4; ENST00000532650.1; ENST00000647510.1; ENST00000617502.4; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150365 | ||
| mod ID: M6ASITE007368 | Click to Show/Hide the Full List | ||
| mod site | chr11:66688254-66688255:- | [4] | |
| Sequence | GAGGAGGAGGAGCGGCGGAAACAGCCGCCTGCTCCCGAACC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; H1B; MT4; Huh7; Jurkat; GSC-11; HEK293T; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000529997.5; ENST00000611817.4; ENST00000617502.4; ENST00000309996.6; ENST00000532650.1; ENST00000533211.5; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150366 | ||
| mod ID: M6ASITE007369 | Click to Show/Hide the Full List | ||
| mod site | chr11:66689109-66689110:- | [4] | |
| Sequence | GAAGTGGCAGGAGAAGATGGACTGGCTTCAGCTGGGTGAGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; Jurkat; GSC-11; HEK293T; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000532902.1; ENST00000529997.5; ENST00000533211.5; ENST00000611817.4; ENST00000617502.4; ENST00000309996.6 | ||
| External Link | RMBase: m6A_site_150367 | ||
| mod ID: M6ASITE007370 | Click to Show/Hide the Full List | ||
| mod site | chr11:66689137-66689138:- | [4] | |
| Sequence | TGCAGGCACGGCGCCAGGAGACAGCTGAGAAGTGGCAGGAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; Jurkat; GSC-11; HEK293T; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000611817.4; ENST00000647510.1; ENST00000533211.5; ENST00000309996.6; ENST00000529997.5; ENST00000617502.4; ENST00000532902.1 | ||
| External Link | RMBase: m6A_site_150368 | ||
| mod ID: M6ASITE007371 | Click to Show/Hide the Full List | ||
| mod site | chr11:66689871-66689872:- | [4] | |
| Sequence | AGAGATAGAGGCCCGGGCAGACCGCTTCTCCTCCTGCATCG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; Huh7; Jurkat; HEK293T; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000529997.5; ENST00000611817.4; ENST00000309996.6; ENST00000617502.4; ENST00000532902.1; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150369 | ||
| mod ID: M6ASITE007372 | Click to Show/Hide the Full List | ||
| mod site | chr11:66689910-66689911:- | [4] | |
| Sequence | CGCGGATCTAGTCATCAAGAACCAGCAAGGCATCAAGGCAG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; Huh7; Jurkat; HEK293T; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000529997.5; ENST00000647510.1; ENST00000309996.6; ENST00000611817.4; ENST00000532902.1; ENST00000617502.4; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150370 | ||
| mod ID: M6ASITE007373 | Click to Show/Hide the Full List | ||
| mod site | chr11:66690094-66690095:- | [4] | |
| Sequence | TTCTTCAAGGCTGTCCGGGAACTGATGCTCTGGATGGATGA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000532902.1; ENST00000647510.1; ENST00000533211.5; ENST00000529997.5; ENST00000617502.4; ENST00000611817.4; ENST00000309996.6 | ||
| External Link | RMBase: m6A_site_150371 | ||
| mod ID: M6ASITE007374 | Click to Show/Hide the Full List | ||
| mod site | chr11:66690125-66690126:- | [4] | |
| Sequence | GCTGCTGCTGGACACCACAGACAAGTTCCGCTTCTTCAAGG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; GSC-11; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000611817.4; ENST00000533211.5; ENST00000529997.5; ENST00000532902.1; ENST00000617502.4; ENST00000309996.6; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150372 | ||
| mod ID: M6ASITE007375 | Click to Show/Hide the Full List | ||
| mod site | chr11:66690134-66690135:- | [4] | |
| Sequence | CCGCCGGCAGCTGCTGCTGGACACCACAGACAAGTTCCGCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000532902.1; ENST00000533211.5; ENST00000647510.1; ENST00000529997.5; ENST00000611817.4; ENST00000309996.6; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150373 | ||
| mod ID: M6ASITE007376 | Click to Show/Hide the Full List | ||
| mod site | chr11:66690230-66690231:- | [4] | |
| Sequence | CCAGAAGGCCTACGCTGGAGACAAGGCTGAGGAGATCGGCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; Jurkat; HEK293T; iSLK; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000611817.4; ENST00000533211.5; ENST00000532902.1; ENST00000529997.5; ENST00000617502.4; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150374 | ||
| mod ID: M6ASITE007377 | Click to Show/Hide the Full List | ||
| mod site | chr11:66690347-66690348:- | [4] | |
| Sequence | AGGTGAGAGTGGCAGCCAGGACTGAGGCAGGCTGCGGCTCC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; Jurkat; GSC-11; HEK293T; iSLK; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000529997.5; ENST00000532902.1; ENST00000309996.6; ENST00000647510.1; ENST00000617502.4; ENST00000611817.4 | ||
| External Link | RMBase: m6A_site_150375 | ||
| mod ID: M6ASITE007378 | Click to Show/Hide the Full List | ||
| mod site | chr11:66691366-66691367:- | [4] | |
| Sequence | AGCAGCAGCTTCCGGACGGGACTGGCCGCGACCTCAACGCT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1A; H1B; MM6; Jurkat; GSC-11; HEK293A-TOA; iSLK; TIME; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000611817.4; ENST00000647510.1; ENST00000309996.6; ENST00000617502.4; ENST00000529997.5 | ||
| External Link | RMBase: m6A_site_150376 | ||
| mod ID: M6ASITE007379 | Click to Show/Hide the Full List | ||
| mod site | chr11:66691476-66691477:- | [4] | |
| Sequence | TGACCTGCTTGAGCTGCTGGACACACGGGGTCAGGTGCTGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; H1A; H1B; A549; Jurkat; HEK293A-TOA; iSLK; TIME; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000611817.4; ENST00000617502.4; ENST00000533211.5; ENST00000647510.1; ENST00000309996.6; ENST00000529997.5 | ||
| External Link | RMBase: m6A_site_150377 | ||
| mod ID: M6ASITE007380 | Click to Show/Hide the Full List | ||
| mod site | chr11:66691518-66691519:- | [4] | |
| Sequence | CACCGTGGCCGAGTGGAAGGACAGTCTCAACGAGGCCTGGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; brain; HepG2; H1A; H1B; A549; Jurkat; HEK293A-TOA; iSLK; TIME; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-REF-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000309996.6; ENST00000533211.5; ENST00000611817.4; ENST00000529997.5; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150378 | ||
| mod ID: M6ASITE007381 | Click to Show/Hide the Full List | ||
| mod site | chr11:66691623-66691624:- | [4] | |
| Sequence | ATTCCGAGAGTTCTCCCGGGACACAAGCACCATCGGTCAGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; Jurkat; HEK293T; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000529997.5; ENST00000611817.4; ENST00000647510.1; ENST00000533211.5; ENST00000309996.6 | ||
| External Link | RMBase: m6A_site_150379 | ||
| mod ID: M6ASITE007382 | Click to Show/Hide the Full List | ||
| mod site | chr11:66691647-66691648:- | [4] | |
| Sequence | CCTCTACCAGATGCTCCGAGACAAATTCCGAGAGTTCTCCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; Jurkat; HEK293T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000611817.4; ENST00000529997.5; ENST00000533211.5; ENST00000309996.6; ENST00000617502.4; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150380 | ||
| mod ID: M6ASITE007383 | Click to Show/Hide the Full List | ||
| mod site | chr11:66692551-66692552:- | [4] | |
| Sequence | CTCCCACGAGCTGGGCCAGGACTACGAGCATGTGACTGTGA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; Jurkat; HEK293T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000533211.5; ENST00000529997.5; ENST00000647510.1; ENST00000617502.4; ENST00000611817.4 | ||
| External Link | RMBase: m6A_site_150381 | ||
| mod ID: M6ASITE007384 | Click to Show/Hide the Full List | ||
| mod site | chr11:66692604-66692605:- | [4] | |
| Sequence | CGCGAGCTGGATGACCTGGAACAGTGGATCCAGGAGCGCGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; Jurkat; HEK293T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000529997.5; ENST00000611817.4; ENST00000647510.1; ENST00000309996.6; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150382 | ||
| mod ID: M6ASITE007385 | Click to Show/Hide the Full List | ||
| mod site | chr11:66692707-66692708:- | [4] | |
| Sequence | CATCCGCCAAGCCCAGGTGGACAAGCTGTATGCCGGCCTGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; MT4; Jurkat; GSC-11; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000529997.5; ENST00000647510.1; ENST00000617502.4; ENST00000309996.6; ENST00000611817.4; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150383 | ||
| mod ID: M6ASITE007386 | Click to Show/Hide the Full List | ||
| mod site | chr11:66692996-66692997:- | [4] | |
| Sequence | GCTGGCGGCCAGCAGCCAGGACATGATTGACCACGAGCACC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; Jurkat; GSC-11; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000611817.4; ENST00000647510.1; ENST00000529997.5; ENST00000617502.4; ENST00000309996.6; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150384 | ||
| mod ID: M6ASITE007387 | Click to Show/Hide the Full List | ||
| mod site | chr11:66693027-66693028:- | [4] | |
| Sequence | CCCTGGCCGACTACGCGCAGACCATCCACCAGCTGGCGGCC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; Jurkat; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000611817.4; ENST00000309996.6; ENST00000647510.1; ENST00000533211.5; ENST00000529997.5; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150385 | ||
| mod ID: M6ASITE007388 | Click to Show/Hide the Full List | ||
| mod site | chr11:66693305-66693306:- | [4] | |
| Sequence | CGCCTGGGCCACGAGCTGGAACTTCGAGGGAAGCGACTGGA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; MM6; Huh7; Jurkat; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000533211.5; ENST00000611817.4; ENST00000309996.6; ENST00000529997.5; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150386 | ||
| mod ID: M6ASITE007389 | Click to Show/Hide the Full List | ||
| mod site | chr11:66693402-66693403:- | [4] | |
| Sequence | CCATGAGCCCCGGATCGCGGACCTGAGGGAGCGGCAGCGTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; Huh7; HEK293T; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000309996.6; ENST00000647510.1; ENST00000617502.4; ENST00000611817.4; ENST00000529997.5 | ||
| External Link | RMBase: m6A_site_150387 | ||
| mod ID: M6ASITE007390 | Click to Show/Hide the Full List | ||
| mod site | chr11:66693775-66693776:- | [4] | |
| Sequence | CCAGCTTCTCATGAAGAAAAACCAGGTGAGGCAGAGGCTGA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; Huh7; HEK293T; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000533211.5; ENST00000529997.5; ENST00000611817.4; ENST00000309996.6; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150388 | ||
| mod ID: M6ASITE007391 | Click to Show/Hide the Full List | ||
| mod site | chr11:66693808-66693809:- | [4] | |
| Sequence | CTCCATGGAGCATGGCAAGGACCTGCCCAGCGTCCAGCTTC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; Huh7; HEK293T; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000611817.4; ENST00000647510.1; ENST00000309996.6; ENST00000529997.5; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150389 | ||
| mod ID: M6ASITE007392 | Click to Show/Hide the Full List | ||
| mod site | chr11:66694257-66694258:- | [4] | |
| Sequence | GTGCAGGGGAGGTGGAGAGAACCTCGAGGGCCGTGGAGGAG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; MM6; Huh7; Jurkat; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000617502.4; ENST00000647510.1; ENST00000611817.4; ENST00000309996.6; ENST00000529997.5 | ||
| External Link | RMBase: m6A_site_150390 | ||
| mod ID: M6ASITE007393 | Click to Show/Hide the Full List | ||
| mod site | chr11:66694283-66694284:- | [4] | |
| Sequence | CAAAGCACTGGCCCAGGAGGACCAGGGTGCAGGGGAGGTGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; MM6; Huh7; Jurkat; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000529997.5; ENST00000647510.1; ENST00000617502.4; ENST00000611817.4; ENST00000309996.6 | ||
| External Link | RMBase: m6A_site_150391 | ||
| mod ID: M6ASITE007394 | Click to Show/Hide the Full List | ||
| mod site | chr11:66696313-66696314:- | [4] | |
| Sequence | CTCGGATGACTACGGCAAGGACCTCACCAGCGTCAACATCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; A549; Huh7; Jurkat; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000529997.5; ENST00000617502.4; ENST00000533211.5; ENST00000530665.1; ENST00000309996.6; ENST00000647510.1; ENST00000611817.4 | ||
| External Link | RMBase: m6A_site_150392 | ||
| mod ID: M6ASITE007395 | Click to Show/Hide the Full List | ||
| mod site | chr11:66696446-66696447:- | [4] | |
| Sequence | GGCGCTGGGACGAGCTGGAGACCACCACCCAAGCCAAGGCC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; Jurkat; HEK293T; endometrial; HEC-1-A; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000611817.4; ENST00000529997.5; ENST00000533211.5; ENST00000530665.1; ENST00000617502.4; ENST00000309996.6 | ||
| External Link | RMBase: m6A_site_150393 | ||
| mod ID: M6ASITE007396 | Click to Show/Hide the Full List | ||
| mod site | chr11:66696469-66696470:- | [7] | |
| Sequence | GGAGAAGCTGAGAGACCTGCACAGGCGCTGGGACGAGCTGG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | brain | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000530665.1; ENST00000529997.5; ENST00000533211.5; ENST00000309996.6; ENST00000647510.1; ENST00000611817.4 | ||
| External Link | RMBase: m6A_site_150394 | ||
| mod ID: M6ASITE007397 | Click to Show/Hide the Full List | ||
| mod site | chr11:66696475-66696476:- | [4] | |
| Sequence | GGTGTCGGAGAAGCTGAGAGACCTGCACAGGCGCTGGGACG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; Jurkat; HEK293T; endometrial; HEC-1-A; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000611817.4; ENST00000309996.6; ENST00000530665.1; ENST00000647510.1; ENST00000529997.5; ENST00000617502.4; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150395 | ||
| mod ID: M6ASITE007398 | Click to Show/Hide the Full List | ||
| mod site | chr11:66698642-66698643:- | [4] | |
| Sequence | AGACTGGCTGGACAAGGTGGACAAGGTGAGCAGTGCTGTGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; Huh7; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000529997.5; ENST00000309996.6; ENST00000617502.4; ENST00000533211.5; ENST00000647510.1; ENST00000530665.1; ENST00000611817.4 | ||
| External Link | RMBase: m6A_site_150396 | ||
| mod ID: M6ASITE007399 | Click to Show/Hide the Full List | ||
| mod site | chr11:66698651-66698652:- | [4] | |
| Sequence | TGCCAACAAAGACTGGCTGGACAAGGTGGACAAGGTGAGCA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; Huh7; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000611817.4; ENST00000309996.6; ENST00000617502.4; ENST00000533211.5; ENST00000530665.1; ENST00000529997.5; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150397 | ||
| mod ID: M6ASITE007400 | Click to Show/Hide the Full List | ||
| mod site | chr11:66698660-66698661:- | [4] | |
| Sequence | CGAGCTGGCTGCCAACAAAGACTGGCTGGACAAGGTGGACA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; Huh7; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000309996.6; ENST00000617502.4; ENST00000533211.5; ENST00000529997.5; ENST00000611817.4; ENST00000530665.1 | ||
| External Link | RMBase: m6A_site_150398 | ||
| mod ID: M6ASITE007401 | Click to Show/Hide the Full List | ||
| mod site | chr11:66699031-66699032:- | [4] | |
| Sequence | ATTTCTGGGCCGTCTTCGGGACAACCGGGAGCAGCAGCATT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000611817.4; ENST00000617502.4; ENST00000529997.5; ENST00000309996.6; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150399 | ||
| mod ID: M6ASITE007402 | Click to Show/Hide the Full List | ||
| mod site | chr11:66699420-66699421:- | [4] | |
| Sequence | CAAGATTCGGGAAAAGGCAGACTCCATTGAGAGGAGGTCTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000611817.4; ENST00000529997.5; ENST00000309996.6; ENST00000647510.1; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150400 | ||
| mod ID: M6ASITE007403 | Click to Show/Hide the Full List | ||
| mod site | chr11:66699534-66699535:- | [4] | |
| Sequence | TGCCATTAAAAAACTGGAGGACTTCATGAGCACCATGGACG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000533211.5; ENST00000309996.6; ENST00000647510.1; ENST00000611817.4; ENST00000529997.5 | ||
| External Link | RMBase: m6A_site_150401 | ||
| mod ID: M6ASITE007404 | Click to Show/Hide the Full List | ||
| mod site | chr11:66699542-66699543:- | [4] | |
| Sequence | GCTGATGCTGCCATTAAAAAACTGGAGGACTTCATGAGCAC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000533211.5; ENST00000529997.5; ENST00000611817.4; ENST00000647510.1; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150402 | ||
| mod ID: M6ASITE007405 | Click to Show/Hide the Full List | ||
| mod site | chr11:66699574-66699575:- | [4] | |
| Sequence | CTCACACGGAGATGCCAGGGACACTCCAGGCTGCTGATGCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; H1B; Jurkat; HEK293T; iSLK; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000611817.4; ENST00000529997.5; ENST00000533211.5; ENST00000617502.4; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150403 | ||
| mod ID: M6ASITE007406 | Click to Show/Hide the Full List | ||
| mod site | chr11:66700644-66700645:- | [4] | |
| Sequence | AGCGACTGGAGGCCCTGGGAACTGGCTGGGAGGAGCTGGGC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; A549; HEK293T; H1A; H1B; MM6; Jurkat; GSC-11; HEK293A-TOA; iSLK; TIME; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000617502.4; ENST00000533211.5; ENST00000611817.4; ENST00000309996.6; ENST00000529997.5 | ||
| External Link | RMBase: m6A_site_150404 | ||
| mod ID: M6ASITE007407 | Click to Show/Hide the Full List | ||
| mod site | chr11:66700697-66700698:- | [4] | |
| Sequence | GGGCGAGGAGGTGACCCGGGACCAGGCTGACCCCCAGTGCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; MM6; Jurkat; GSC-11; HEK293A-TOA; iSLK; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000529997.5; ENST00000533211.5; ENST00000611817.4; ENST00000617502.4; ENST00000309996.6 | ||
| External Link | RMBase: m6A_site_150405 | ||
| mod ID: M6ASITE007408 | Click to Show/Hide the Full List | ||
| mod site | chr11:66700842-66700843:- | [4] | |
| Sequence | CCTGGCTAGGCCGCACTCAGACTGCTGTGGCCTCTGAAGAA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; MM6; Jurkat; GSC-11; HEK293A-TOA; iSLK; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000533211.5; ENST00000309996.6; ENST00000611817.4; ENST00000529997.5; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150406 | ||
| mod ID: M6ASITE007409 | Click to Show/Hide the Full List | ||
| mod site | chr11:66700871-66700872:- | [5] | |
| Sequence | CTTCTTGCGCAGCTTGGATGACTTCCAGGCCTGGCTAGGCC | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000529997.5; ENST00000647510.1; ENST00000611817.4; ENST00000309996.6; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150407 | ||
| mod ID: M6ASITE007410 | Click to Show/Hide the Full List | ||
| mod site | chr11:66700892-66700893:- | [4] | |
| Sequence | GGAGGCGCGGCGGCTGCAGGACTTCTTGCGCAGCTTGGATG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; MM6; Jurkat; GSC-11; HEK293A-TOA; iSLK; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000611817.4; ENST00000647510.1; ENST00000533211.5; ENST00000529997.5; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150408 | ||
| mod ID: M6ASITE007411 | Click to Show/Hide the Full List | ||
| mod site | chr11:66700952-66700953:- | [4] | |
| Sequence | GGTGCAGACCGGCTGGGAGGACCTCAGGGCCACCATGCGGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; MM6; Jurkat; GSC-11; HEK293A-TOA; iSLK; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000529997.5; ENST00000533211.5; ENST00000647510.1; ENST00000611817.4; ENST00000309996.6 | ||
| External Link | RMBase: m6A_site_150409 | ||
| mod ID: M6ASITE007412 | Click to Show/Hide the Full List | ||
| mod site | chr11:66700965-66700966:- | [4] | |
| Sequence | CCCGGCTGAGAGAGGTGCAGACCGGCTGGGAGGACCTCAGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; MM6; Jurkat; GSC-11; HEK293A-TOA; iSLK; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000617502.4; ENST00000611817.4; ENST00000309996.6; ENST00000533211.5; ENST00000529997.5 | ||
| External Link | RMBase: m6A_site_150410 | ||
| mod ID: M6ASITE007413 | Click to Show/Hide the Full List | ||
| mod site | chr11:66701047-66701048:- | [4] | |
| Sequence | ATCGCCGCCCGGGTGGGCGAACTGACTCGAGAGGCAAATGC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; A549; HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; MM6; Jurkat; GSC-11; HEK293A-TOA; iSLK; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000309996.6; ENST00000611817.4; ENST00000647510.1; ENST00000529997.5; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150411 | ||
| mod ID: M6ASITE007414 | Click to Show/Hide the Full List | ||
| mod site | chr11:66701078-66701079:- | [4] | |
| Sequence | GCTGGCCGGCACGGAGCGGGACCTGGAGGCCATCGCCGCCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; A549; HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; MM6; Jurkat; GSC-11; HEK293A-TOA; iSLK; TIME; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000611817.4; ENST00000529997.5; ENST00000647510.1; ENST00000617502.4; ENST00000309996.6; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150412 | ||
| mod ID: M6ASITE007415 | Click to Show/Hide the Full List | ||
| mod site | chr11:66701166-66701167:- | [4] | |
| Sequence | AGGCCTGGATGAGAGAGAAGACCAAAGTCATCGAGTCCACC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1B; Huh7; Jurkat; GSC-11; HEK293A-TOA; iSLK; endometrial; HEC-1-A; GSCs; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000647510.1; ENST00000529997.5; ENST00000611817.4; ENST00000309996.6; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150413 | ||
| mod ID: M6ASITE007416 | Click to Show/Hide the Full List | ||
| mod site | chr11:66701190-66701191:- | [4] | |
| Sequence | ACCACTTAGAGTGCACGGAGACCCAGGCCTGGATGAGAGAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1B; Huh7; Jurkat; GSC-11; HEK293A-TOA; iSLK; endometrial; HEC-1-A; GSCs; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000647510.1; ENST00000533211.5; ENST00000309996.6; ENST00000529997.5; ENST00000611817.4 | ||
| External Link | RMBase: m6A_site_150414 | ||
| mod ID: M6ASITE007417 | Click to Show/Hide the Full List | ||
| mod site | chr11:66701213-66701214:- | [4] | |
| Sequence | CTCAGCCCTGAGCATCCAGAACTACCACTTAGAGTGCACGG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; Huh7; Jurkat; GSC-11; iSLK; endometrial; HEC-1-A; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000611817.4; ENST00000647510.1; ENST00000309996.6; ENST00000617502.4; ENST00000533211.5; ENST00000529997.5 | ||
| External Link | RMBase: m6A_site_150415 | ||
| mod ID: M6ASITE007418 | Click to Show/Hide the Full List | ||
| mod site | chr11:66701607-66701608:- | [8] | |
| Sequence | AGGCAAAGACCGCATTGTCAACACCCAGGAGCAGCTCAACC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000529997.5; ENST00000617502.4; ENST00000647510.1; ENST00000533211.5; ENST00000611817.4 | ||
| External Link | RMBase: m6A_site_150416 | ||
| mod ID: M6ASITE007419 | Click to Show/Hide the Full List | ||
| mod site | chr11:66701619-66701620:- | [4] | |
| Sequence | GGCCAACCCCCCAGGCAAAGACCGCATTGTCAACACCCAGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; Huh7; Jurkat; GSC-11; HEK293T; iSLK; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000529997.5; ENST00000611817.4; ENST00000533211.5; ENST00000647510.1; ENST00000309996.6 | ||
| External Link | RMBase: m6A_site_150417 | ||
| mod ID: M6ASITE007420 | Click to Show/Hide the Full List | ||
| mod site | chr11:66701661-66701662:- | [8] | |
| Sequence | ACAAATCACCGCGGTGAATGACATTGCCGAGCAGTTACTGA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000647510.1; ENST00000309996.6; ENST00000529997.5; ENST00000611817.4; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150418 | ||
| mod ID: M6ASITE007421 | Click to Show/Hide the Full List | ||
| mod site | chr11:66701694-66701695:- | [4] | |
| Sequence | GACCCTGGAGCCTGAAATGAACACCCTTGCAGCACAAATCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; MM6; Huh7; Jurkat; GSC-11; HEK293T; iSLK; TIME; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000647510.1; ENST00000611817.4; ENST00000309996.6; ENST00000617502.4; ENST00000529997.5 | ||
| External Link | RMBase: m6A_site_150419 | ||
| mod ID: M6ASITE007422 | Click to Show/Hide the Full List | ||
| mod site | chr11:66701713-66701714:- | [4] | |
| Sequence | TCCACCCTCTCAGGTTCGAGACCCTGGAGCCTGAAATGAAC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; MM6; Huh7; Jurkat; GSC-11; HEK293T; iSLK; TIME; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000617502.4; ENST00000611817.4; ENST00000647510.1; ENST00000529997.5; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150420 | ||
| mod ID: M6ASITE007423 | Click to Show/Hide the Full List | ||
| mod site | chr11:66704618-66704619:- | [4] | |
| Sequence | CCTGCCTGAACGCCTGGAGGACCTGGAGGTCGTGCAGCAGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; A549; MM6; Huh7; Jurkat; GSC-11; HEK293T; iSLK; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000529997.5; ENST00000647510.1; ENST00000617502.4; ENST00000533211.5; ENST00000611817.4 | ||
| External Link | RMBase: m6A_site_150421 | ||
| mod ID: M6ASITE007424 | Click to Show/Hide the Full List | ||
| mod site | chr11:66704680-66704681:- | [4] | |
| Sequence | AGCGAGGCCGGGGCCTGTGGACTCTGGGTGGAGGAGAAGGA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1A; H1B; A549; MM6; Jurkat; GSC-11; HEK293A-TOA; iSLK; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000529997.5; ENST00000647510.1; ENST00000309996.6; ENST00000617502.4; ENST00000533211.5; ENST00000611817.4 | ||
| External Link | RMBase: m6A_site_150422 | ||
| mod ID: M6ASITE007425 | Click to Show/Hide the Full List | ||
| mod site | chr11:66704851-66704852:- | [4] | |
| Sequence | ACCCTGGACGCCTTGAGGGAACAGGCAGCAGCCCTGCCCCC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; H1B; MM6; Jurkat; GSC-11; HEK293T; HEK293A-TOA; iSLK; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000647510.1; ENST00000617502.4; ENST00000611817.4; ENST00000529997.5; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150423 | ||
| mod ID: M6ASITE007426 | Click to Show/Hide the Full List | ||
| mod site | chr11:66705103-66705104:- | [4] | |
| Sequence | GCCTCTGCCCGTGCAGCTGAACTCCAAGCCCAGTGGGAGCG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; MM6; Jurkat; HEK293T; HEK293A-TOA; iSLK; TIME; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000529997.5; ENST00000309996.6; ENST00000611817.4; ENST00000533211.5; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150424 | ||
| mod ID: M6ASITE007427 | Click to Show/Hide the Full List | ||
| mod site | chr11:66705260-66705261:- | [4] | |
| Sequence | CTCAGCCGACACGGGCCGAGACCTGACCGGTGCCCTCCGCC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; iSLK; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000611817.4; ENST00000533211.5; ENST00000647510.1; ENST00000529997.5; ENST00000309996.6; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150425 | ||
| mod ID: M6ASITE007428 | Click to Show/Hide the Full List | ||
| mod site | chr11:66705460-66705461:- | [4] | |
| Sequence | GTGACTTTCTCAGAGTATAGACCTTGCGACCCGCAGCTGGT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; Jurkat; HEK293T; TIME; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000611817.4; ENST00000309996.6; ENST00000647510.1; ENST00000617502.4; ENST00000529997.5; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150426 | ||
| mod ID: M6ASITE007429 | Click to Show/Hide the Full List | ||
| mod site | chr11:66705754-66705755:- | [4] | |
| Sequence | GCACGAGCTGGTGGAGGCAGACATCGCCGTGCAGGCCGAGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; H1B; Jurkat; HEK293T; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000309996.6; ENST00000533211.5; ENST00000529997.5; ENST00000611817.4; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150427 | ||
| mod ID: M6ASITE007430 | Click to Show/Hide the Full List | ||
| mod site | chr11:66705787-66705788:- | [4] | |
| Sequence | GCACCTAGCAGGAGTGGAGGACCTGCTGCAGCTGCACGAGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; H1B; Jurkat; HEK293T; endometrial; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000533211.5; ENST00000647510.1; ENST00000611817.4; ENST00000529997.5; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150428 | ||
| mod ID: M6ASITE007431 | Click to Show/Hide the Full List | ||
| mod site | chr11:66705817-66705818:- | [4] | |
| Sequence | GGGCCGGCTGCAGTCTCAGGACCTGGGCAGGCACCTAGCAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; H1B; Jurkat; HEK293T; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000611817.4; ENST00000533211.5; ENST00000529997.5; ENST00000647510.1; ENST00000309996.6; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150429 | ||
| mod ID: M6ASITE007432 | Click to Show/Hide the Full List | ||
| mod site | chr11:66707534-66707535:- | [4] | |
| Sequence | GGACCTGCTCTACCTCATGGACTGGATGGAAGAGATGAAGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; H1B; Jurkat; HEK293T; endometrial; GSCs; NB4; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000529997.5; ENST00000611817.4; ENST00000617502.4; ENST00000533211.5; ENST00000309996.6 | ||
| External Link | RMBase: m6A_site_150430 | ||
| mod ID: M6ASITE007433 | Click to Show/Hide the Full List | ||
| mod site | chr11:66707552-66707553:- | [4] | |
| Sequence | GCTGCAGAAGGTGTTCCAGGACCTGCTCTACCTCATGGACT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; H1B; Jurkat; HEK293T; endometrial; GSCs; NB4; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000611817.4; ENST00000647510.1; ENST00000617502.4; ENST00000533211.5; ENST00000529997.5 | ||
| External Link | RMBase: m6A_site_150431 | ||
| mod ID: M6ASITE007434 | Click to Show/Hide the Full List | ||
| mod site | chr11:66707627-66707628:- | [4] | |
| Sequence | CAACGTGGCACGGCTCTGGGACTTCTTGCGGCAGATGGTGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; Jurkat; HEK293T; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000617502.4; ENST00000611817.4; ENST00000533211.5; ENST00000647510.1; ENST00000529997.5 | ||
| External Link | RMBase: m6A_site_150432 | ||
| mod ID: M6ASITE007435 | Click to Show/Hide the Full List | ||
| mod site | chr11:66707750-66707751:- | [4] | |
| Sequence | GCACGAAGCCATTGAGACGGACATCGTGGCCTACAGCGGCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000617502.4; ENST00000611817.4; ENST00000309996.6; ENST00000529997.5; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150433 | ||
| mod ID: M6ASITE007436 | Click to Show/Hide the Full List | ||
| mod site | chr11:66707816-66707817:- | [8] | |
| Sequence | CTCCCTGCCCCCGACACAGGACAACTTTGGGCTGGAGCTGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000309996.6; ENST00000529997.5; ENST00000533211.5; ENST00000611817.4; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150434 | ||
| mod ID: M6ASITE007437 | Click to Show/Hide the Full List | ||
| mod site | chr11:66708159-66708160:- | [4] | |
| Sequence | GGAGACCTGGCTCAGCGAGAACCAGCGCCTCGTGTCCCAGG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000611817.4; ENST00000309996.6; ENST00000533211.5; ENST00000529997.5; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150435 | ||
| mod ID: M6ASITE007438 | Click to Show/Hide the Full List | ||
| mod site | chr11:66708175-66708176:- | [4] | |
| Sequence | GCAAGGCTGCCATGCGGGAGACCTGGCTCAGCGAGAACCAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000529997.5; ENST00000533211.5; ENST00000611817.4; ENST00000647510.1; ENST00000309996.6 | ||
| External Link | RMBase: m6A_site_150436 | ||
| mod ID: M6ASITE007439 | Click to Show/Hide the Full List | ||
| mod site | chr11:66708911-66708912:- | [4] | |
| Sequence | CGAGGGCCGGCTCATCTCGGACATCAACAAGGTCCGTGGCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; Huh7; HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000611817.4; ENST00000529997.5; ENST00000617502.4; ENST00000309996.6; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150437 | ||
| mod ID: M6ASITE007440 | Click to Show/Hide the Full List | ||
| mod site | chr11:66709001-66709002:- | [4] | |
| Sequence | CAGGTTTACCGAGAAAGGGAACTTGGAAGTGCTGCTCTTCA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000529997.5; ENST00000611817.4; ENST00000533211.5; ENST00000617502.4; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150438 | ||
| mod ID: M6ASITE007441 | Click to Show/Hide the Full List | ||
| mod site | chr11:66710629-66710630:- | [4] | |
| Sequence | CTCCCTTAGCGGGGTCCAGAACCAGCTGCAGTCCTTCAACT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000529997.5; ENST00000617502.4; ENST00000611817.4; ENST00000647510.1; ENST00000309996.6; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150439 | ||
| mod ID: M6ASITE007442 | Click to Show/Hide the Full List | ||
| mod site | chr11:66710761-66710762:- | [4] | |
| Sequence | TTCTGCCTCCCAGGTGCTGGACCATGCCATGGAGGCAGAGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000529997.5; ENST00000647510.1; ENST00000611817.4; ENST00000309996.6; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150440 | ||
| mod ID: M6ASITE007443 | Click to Show/Hide the Full List | ||
| mod site | chr11:66711016-66711017:- | [4] | |
| Sequence | TCTTGCAGACGTGAATGTGGACCAGCCAGATGAGAAGTCAA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000611817.4; ENST00000529997.5; ENST00000533211.5; ENST00000309996.6; ENST00000647510.1; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150441 | ||
| mod ID: M6ASITE007444 | Click to Show/Hide the Full List | ||
| mod site | chr11:66713655-66713656:- | [4] | |
| Sequence | CTGGCTGAAAAGGAACTGGGACTTACCAAGCTGCTGGATCC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000309996.6; ENST00000529997.5; ENST00000617502.4; ENST00000533211.5; ENST00000611817.4 | ||
| External Link | RMBase: m6A_site_150442 | ||
| mod ID: M6ASITE007445 | Click to Show/Hide the Full List | ||
| mod site | chr11:66713661-66713662:- | [4] | |
| Sequence | TTCAATCTGGCTGAAAAGGAACTGGGACTTACCAAGCTGCT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000533211.5; ENST00000529997.5; ENST00000611817.4; ENST00000647510.1; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150443 | ||
| mod ID: M6ASITE007446 | Click to Show/Hide the Full List | ||
| mod site | chr11:66713703-66713704:- | [8] | |
| Sequence | TCTCTGAAGAAGTGTAATGCACACTATAATCTGCAGAATGC | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000529997.5; ENST00000617502.4; ENST00000309996.6; ENST00000611817.4; ENST00000647510.1; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150444 | ||
| mod ID: M6ASITE007447 | Click to Show/Hide the Full List | ||
| mod site | chr11:66713740-66713741:- | [4] | |
| Sequence | TGCCTCCCCACCCAGGCCAGACCTGCTGGATTTTGAGTCTC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000529997.5; ENST00000647510.1; ENST00000617502.4; ENST00000611817.4; ENST00000309996.6; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150445 | ||
| mod ID: M6ASITE007448 | Click to Show/Hide the Full List | ||
| mod site | chr11:66714095-66714096:- | [4] | |
| Sequence | TTCAACGCCATCGTGCATAAACACCGGTGAGAAGATGGGGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000309996.6; ENST00000529997.5; ENST00000647510.1; ENST00000533211.5; ENST00000611817.4 | ||
| External Link | RMBase: m6A_site_150446 | ||
| mod ID: M6ASITE007449 | Click to Show/Hide the Full List | ||
| mod site | chr11:66714122-66714123:- | [4] | |
| Sequence | ACCACCAGCTGGAGAGATGGACTAGCTTTCAACGCCATCGT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000309996.6; ENST00000611817.4; ENST00000529997.5; ENST00000617502.4; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150447 | ||
| mod ID: M6ASITE007450 | Click to Show/Hide the Full List | ||
| mod site | chr11:66714152-66714153:- | [8] | |
| Sequence | AGTTATCCCAACGTCAATGTACACAACTTCACCACCAGCTG | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000309996.6; ENST00000529997.5; ENST00000611817.4; ENST00000617502.4; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150448 | ||
| mod ID: M6ASITE007451 | Click to Show/Hide the Full List | ||
| mod site | chr11:66714322-66714323:- | [4] | |
| Sequence | TTCTGTGGTGCCAGATGAAGACTGCAGGGTGAGGACACCCT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000529997.5; ENST00000647510.1; ENST00000611817.4; ENST00000309996.6; ENST00000533211.5; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150449 | ||
| mod ID: M6ASITE007452 | Click to Show/Hide the Full List | ||
| mod site | chr11:66714375-66714376:- | [8] | |
| Sequence | CAGTGTGGAGACAGAAGACAACAAGGAGAAGAAGTCAGCCA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000533211.5; ENST00000647510.1; ENST00000529997.5; ENST00000617502.4; ENST00000611817.4 | ||
| External Link | RMBase: m6A_site_150450 | ||
| mod ID: M6ASITE007453 | Click to Show/Hide the Full List | ||
| mod site | chr11:66714378-66714379:- | [4] | |
| Sequence | CATCAGTGTGGAGACAGAAGACAACAAGGAGAAGAAGTCAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000529997.5; ENST00000647510.1; ENST00000611817.4; ENST00000533211.5; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150451 | ||
| mod ID: M6ASITE007454 | Click to Show/Hide the Full List | ||
| mod site | chr11:66714385-66714386:- | [4] | |
| Sequence | TCCAAGACATCAGTGTGGAGACAGAAGACAACAAGGAGAAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000533211.5; ENST00000647510.1; ENST00000309996.6; ENST00000611817.4; ENST00000529997.5 | ||
| External Link | RMBase: m6A_site_150452 | ||
| mod ID: M6ASITE007455 | Click to Show/Hide the Full List | ||
| mod site | chr11:66714399-66714400:- | [4] | |
| Sequence | GCTTCCTTCCTAGATCCAAGACATCAGTGTGGAGACAGAAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; endometrial | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000529997.5; ENST00000617502.4; ENST00000647510.1; ENST00000533211.5; ENST00000611817.4 | ||
| External Link | RMBase: m6A_site_150453 | ||
| mod ID: M6ASITE007456 | Click to Show/Hide the Full List | ||
| mod site | chr11:66715241-66715242:- | [4] | |
| Sequence | TGACCCTTGGGCTGGTCTGGACCATCATCCTTCGATTCCAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000617502.4; ENST00000529997.5; ENST00000611817.4; ENST00000533211.5; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150454 | ||
| mod ID: M6ASITE007457 | Click to Show/Hide the Full List | ||
| mod site | chr11:66715270-66715271:- | [4] | |
| Sequence | CCATGACATTGTGGACGGAAACCACCGACTGACCCTTGGGC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HEK293T; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000533211.5; ENST00000617502.4; ENST00000529997.5; ENST00000611817.4; ENST00000309996.6 | ||
| External Link | RMBase: m6A_site_150455 | ||
| mod ID: M6ASITE007458 | Click to Show/Hide the Full List | ||
| mod site | chr11:66715285-66715286:- | [8] | |
| Sequence | GGAAAACATGGGCTCCCATGACATTGTGGACGGAAACCACC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000647510.1; ENST00000533211.5; ENST00000611817.4; ENST00000617502.4; ENST00000529997.5 | ||
| External Link | RMBase: m6A_site_150456 | ||
| mod ID: M6ASITE007459 | Click to Show/Hide the Full List | ||
| mod site | chr11:66715300-66715301:- | [4] | |
| Sequence | GCAGAAAGTGCACTTGGAAAACATGGGCTCCCATGACATTG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000617502.4; ENST00000309996.6; ENST00000529997.5; ENST00000533211.5; ENST00000611817.4 | ||
| External Link | RMBase: m6A_site_150457 | ||
| mod ID: M6ASITE007460 | Click to Show/Hide the Full List | ||
| mod site | chr11:66715345-66715346:- | [4] | |
| Sequence | CCACTGCCTGGAGAACGTGGACAAGGCACTGCAGTTCCTCA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000617502.4; ENST00000611817.4; ENST00000647510.1; ENST00000529997.5; ENST00000309996.6; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150458 | ||
| mod ID: M6ASITE007461 | Click to Show/Hide the Full List | ||
| mod site | chr11:66715899-66715900:- | [4] | |
| Sequence | GGTCACGTGCCGGGTGGGGGACCTGTACAGCGACCTCCGGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000611817.4; ENST00000529997.5; ENST00000309996.6; ENST00000647510.1; ENST00000527010.1; ENST00000617502.4; ENST00000651232.1 | ||
| External Link | RMBase: m6A_site_150459 | ||
| mod ID: M6ASITE007462 | Click to Show/Hide the Full List | ||
| mod site | chr11:66715935-66715936:- | [4] | |
| Sequence | AACCTTCACCAAGTGGGTAAACTCGCACCTGGCCCGGGTCA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000533211.5; ENST00000529997.5; ENST00000651232.1; ENST00000611817.4; ENST00000309996.6; ENST00000617502.4; ENST00000527010.1 | ||
| External Link | RMBase: m6A_site_150460 | ||
| mod ID: M6ASITE007463 | Click to Show/Hide the Full List | ||
| mod site | chr11:66715954-66715955:- | [4] | |
| Sequence | GAGAAGCTGTGCAGAAGAAAACCTTCACCAAGTGGGTAAAC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000651232.1; ENST00000529997.5; ENST00000611817.4; ENST00000647510.1; ENST00000309996.6; ENST00000527010.1; ENST00000617502.4; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150461 | ||
| mod ID: M6ASITE007464 | Click to Show/Hide the Full List | ||
| mod site | chr11:66718452-66718453:- | [4] | |
| Sequence | GTGCTGGAAGGACGCTTCAAACAGCTGCAAGGTGAGGCCCG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000611817.4; ENST00000651232.1; ENST00000529997.5; ENST00000309996.6; ENST00000527010.1; ENST00000647510.1; ENST00000617502.4; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150462 | ||
| mod ID: M6ASITE007465 | Click to Show/Hide the Full List | ||
| mod site | chr11:66718657-66718658:- | [9] | |
| Sequence | CCTGCCAGCGGGGAGCATGGACTCTGGGCCCTGGGCTCTGT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000533211.5; ENST00000611817.4; ENST00000527010.1; ENST00000529997.5; ENST00000309996.6; ENST00000617502.4; ENST00000651232.1 | ||
| External Link | RMBase: m6A_site_150463 | ||
| mod ID: M6ASITE007466 | Click to Show/Hide the Full List | ||
| mod site | chr11:66718685-66718686:- | [9] | |
| Sequence | GGTTCCACTGGGGAACAGAAACTGGCCCCCTGCCAGCGGGG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000611817.4; ENST00000529997.5; ENST00000617502.4; ENST00000647510.1; ENST00000651232.1; ENST00000527010.1; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150464 | ||
| mod ID: M6ASITE007467 | Click to Show/Hide the Full List | ||
| mod site | chr11:66718691-66718692:- | [9] | |
| Sequence | GCCTGAGGTTCCACTGGGGAACAGAAACTGGCCCCCTGCCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000527010.1; ENST00000611817.4; ENST00000309996.6; ENST00000651232.1; ENST00000647510.1; ENST00000533211.5; ENST00000529997.5; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150465 | ||
| mod ID: M6ASITE007468 | Click to Show/Hide the Full List | ||
| mod site | chr11:66718717-66718718:- | [9] | |
| Sequence | CTTGAATCCCCTGACGCTGGACTCCAGCCTGAGGTTCCACT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000611817.4; ENST00000617502.4; ENST00000529997.5; ENST00000647510.1; ENST00000527010.1; ENST00000651232.1; ENST00000533211.5; ENST00000309996.6 | ||
| External Link | RMBase: m6A_site_150466 | ||
| mod ID: M6ASITE007469 | Click to Show/Hide the Full List | ||
| mod site | chr11:66721139-66721140:- | [4] | |
| Sequence | CCTTCCTGACTCGGACTGGGACAATGACAGCAGCTCGGCCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; kidney | ||
| Seq Type List | m6A-seq; m6A-REF-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000611817.4; ENST00000647510.1; ENST00000529997.5; ENST00000309996.6; ENST00000527010.1; ENST00000617502.4 | ||
| External Link | RMBase: m6A_site_150467 | ||
| mod ID: M6ASITE007470 | Click to Show/Hide the Full List | ||
| mod site | chr11:66721145-66721146:- | [4] | |
| Sequence | CTGGGACCTTCCTGACTCGGACTGGGACAATGACAGCAGCT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000527010.1; ENST00000533211.5; ENST00000617502.4; ENST00000529997.5; ENST00000611817.4; ENST00000309996.6; ENST00000647510.1 | ||
| External Link | RMBase: m6A_site_150468 | ||
| mod ID: M6ASITE007471 | Click to Show/Hide the Full List | ||
| mod site | chr11:66721160-66721161:- | [4] | |
| Sequence | TGACATCAACAACCGCTGGGACCTTCCTGACTCGGACTGGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000647510.1; ENST00000309996.6; ENST00000533211.5; ENST00000617502.4; ENST00000527010.1; ENST00000611817.4; ENST00000529997.5 | ||
| External Link | RMBase: m6A_site_150469 | ||
| mod ID: M6ASITE007472 | Click to Show/Hide the Full List | ||
| mod site | chr11:66721214-66721215:- | [4] | |
| Sequence | CAGCACGCTGTCACCCACAGACTTTGACAGCTTGGAAATCC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000527010.1; ENST00000309996.6; ENST00000617502.4; ENST00000611817.4; ENST00000529997.5; ENST00000647510.1; ENST00000533211.5 | ||
| External Link | RMBase: m6A_site_150470 | ||
| mod ID: M6ASITE007473 | Click to Show/Hide the Full List | ||
| mod site | chr11:66728821-66728822:- | [10] | |
| Sequence | CGGAGGCTGGATCTCGGGAAACCTCGTCGGCAGTGTCATTA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | H1B; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000527010.1 | ||
| External Link | RMBase: m6A_site_150471 | ||
| mod ID: M6ASITE007474 | Click to Show/Hide the Full List | ||
| mod site | chr11:66728870-66728871:- | [10] | |
| Sequence | GGAATGCTTAGCGGAGACGGACTGGAGGTGCCCGGAGAGGA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | H1B; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000533211.5; ENST00000527010.1 | ||
| External Link | RMBase: m6A_site_150472 | ||
N7-methylguanosine (m7G)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: m7GSITE000011 | Click to Show/Hide the Full List | ||
| mod site | chr11:66687639-66687640:- | [11] | |
| Sequence | GATTCCCCCCCAGGGACCTGGCTCAGGGGACGAAGCCAATG | ||
| Cell/Tissue List | A549; HeLa; HepG2 | ||
| Seq Type List | BoRed-seq&m7G-RIP-seq; m7G-seq | ||
| Transcript ID List | ENST00000309996.6; ENST00000617502.4; ENST00000647510.1; ENST00000533211.5; ENST00000611817.4; ENST00000532650.1; ENST00000529997.5 | ||
| External Link | RMBase: m7G_site_172 | ||
References