m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00244)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
EPHB2
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
YTH domain-containing family protein 1 (YTHDF1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
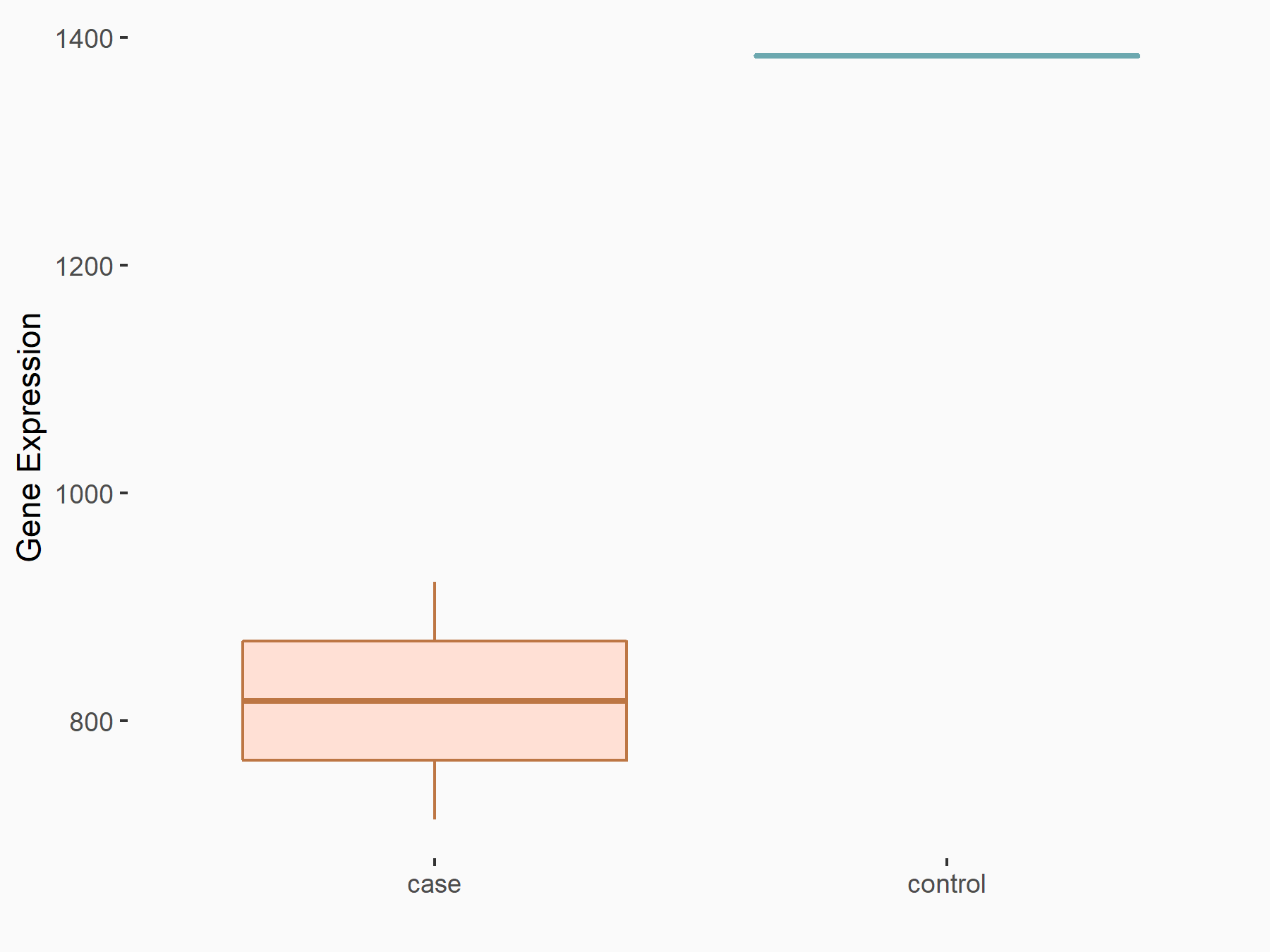  |
logFC: -7.59E-01 p-value: 2.20E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between EPHB2 and the regulator | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.91E+00 | GSE63591 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | m6A writers METTL3/METTL14 and the m6A reader YTHDF1 orchestrate MNK2 expression posttranscriptionally and thus control Ephrin type-B receptor 2 (ERK/EPHB2) signaling, which is required for the maintenance of muscle myogenesis and contribute to regeneration. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Muscular dystrophies | ICD-11: 8C70 | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| C2C12 | Normal | Mus musculus | CVCL_0188 | |
| In-vivo Model | For mouse muscle injury and regeneration experiment, tibialis anterior (TA) muscles of 6-week-old male mice were injected with 25 uL of 10 uM cardiotoxin (CTX, Merck Millipore, 217503), 0.9% normal saline (Saline) were used as control. The regenerated muscles were collected at day 1, 3, 5, and 10 post-injection. TA muscles were isolated for Hematoxylin and eosin staining or frozen in liquid nitrogen for RNA and protein extraction. | |||
Fat mass and obesity-associated protein (FTO) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by FTO | ||
| Cell Line | NB4 cell line | Homo sapiens |
|
Treatment: shFTO NB4 cells
Control: shNS NB4 cells
|
GSE103494 | |
| Regulation |
  |
logFC: 1.37E+00 p-value: 3.20E-04 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | FTO depended on its m6A RNA demethylase activity to activate PDGFRB/Ephrin type-B receptor 2 (ERK/EPHB2) signaling axis. FTO-mediated m6A demethylation plays an oncogenic role in NPM1-mutated Acute myeloid leukemia(AML). | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Acute myeloid leukaemia | ICD-11: 2A60 | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | OCI-AML-3 | Adult acute myeloid leukemia | Homo sapiens | CVCL_1844 |
| OCI-AML-2 | Adult acute myeloid leukemia | Homo sapiens | CVCL_1619 | |
Heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by HNRNPA2B1 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
|
Treatment: HNRNPA2B1 knockdown MDA-MB-231 cells
Control: MDA-MB-231 cells
|
GSE70061 | |
| Regulation |
  |
logFC: 6.38E-01 p-value: 4.72E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | hnRNPA2B1 promotes colon cancer progression via the MAPK pathway. hnRNPA2B1 is an upstream regulator of the Ephrin type-B receptor 2 (ERK/EPHB2)/MAPK pathway and inhibition of MAPK signaling blocked the effects of hnRNPA2B1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colon cancer | ICD-11: 2B90 | ||
| Pathway Response | MAPK signaling | hsa04010 | ||
| Apoptosis | hsa04210 | |||
| Cell Process | Arrest cell cycle at G0/G1 phase | |||
| Cell apoptosis | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| In-vivo Model | Four-week-old male BALB/c nude mice (purchased from Lingchang company) were randomly divided into three groups, each group has five mice. Each of the mice was injected subcutaneously on the right lateral back with 1 × 106 of each lentivirus infected SW480 cells in which hnRNPA2B1 was knocked out or negative control cells. Mice were killed at day 29, and tumors were then isolated, photographed. | |||
Methyltransferase-like 14 (METTL14) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL14 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
|
Treatment: siMETTL14 MDA-MB-231 cells
Control: MDA-MB-231 cells
|
GSE81164 | |
| Regulation |
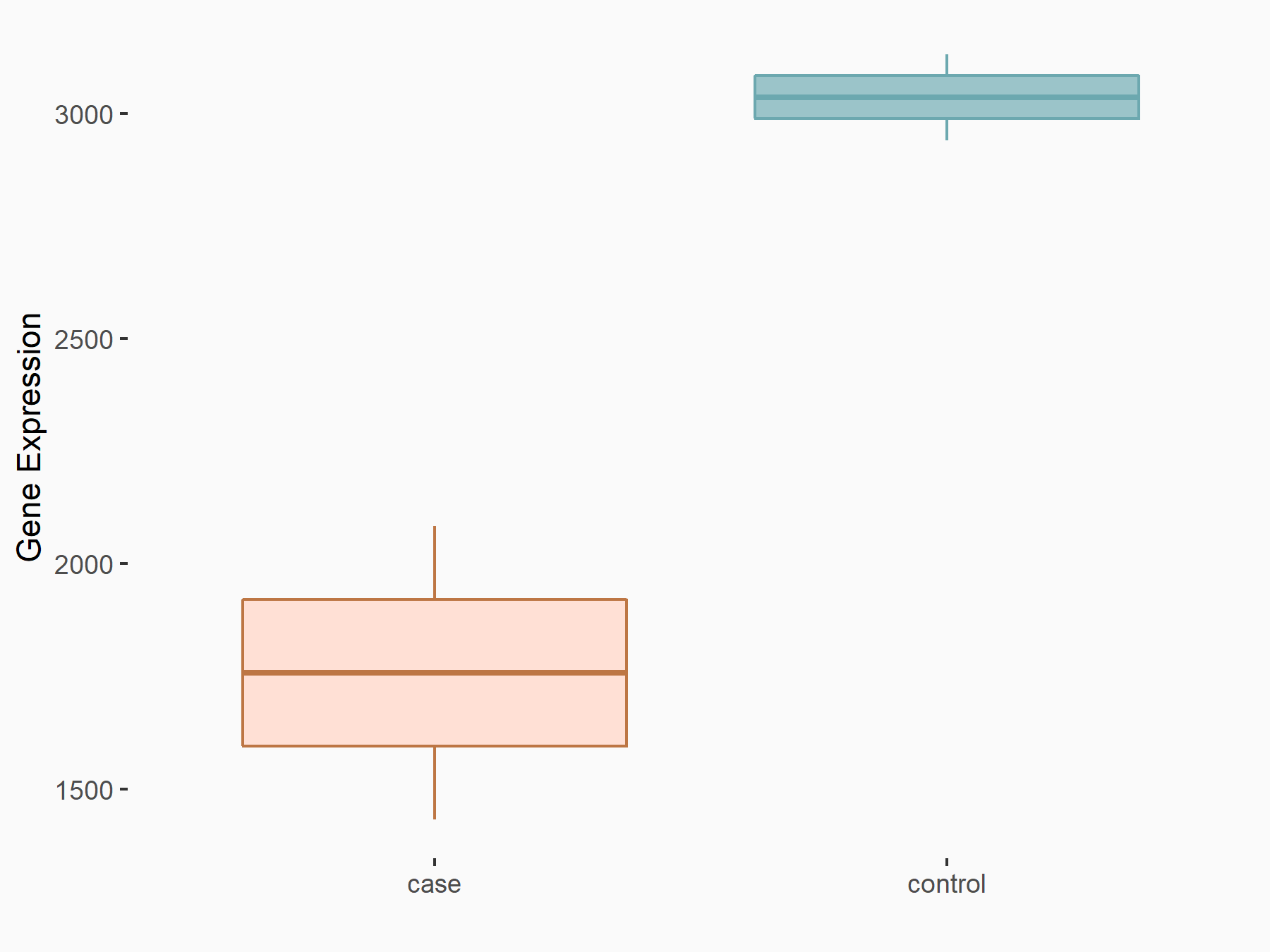  |
logFC: -7.89E-01 p-value: 4.97E-05 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | m6A writers METTL3/METTL14 and the m6A reader YTHDF1 orchestrate MNK2 expression posttranscriptionally and thus control Ephrin type-B receptor 2 (ERK/EPHB2) signaling, which is required for the maintenance of muscle myogenesis and contributes to regeneration. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Muscular dystrophies | ICD-11: 8C70 | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| C2C12 | Normal | Mus musculus | CVCL_0188 | |
| In-vivo Model | For mouse muscle injury and regeneration experiment, tibialis anterior (TA) muscles of 6-week-old male mice were injected with 25 uL of 10 uM cardiotoxin (CTX, Merck Millipore, 217503), 0.9% normal saline (Saline) were used as control. The regenerated muscles were collected at day 1, 3, 5, and 10 post-injection. TA muscles were isolated for Hematoxylin and eosin staining or frozen in liquid nitrogen for RNA and protein extraction. | |||
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
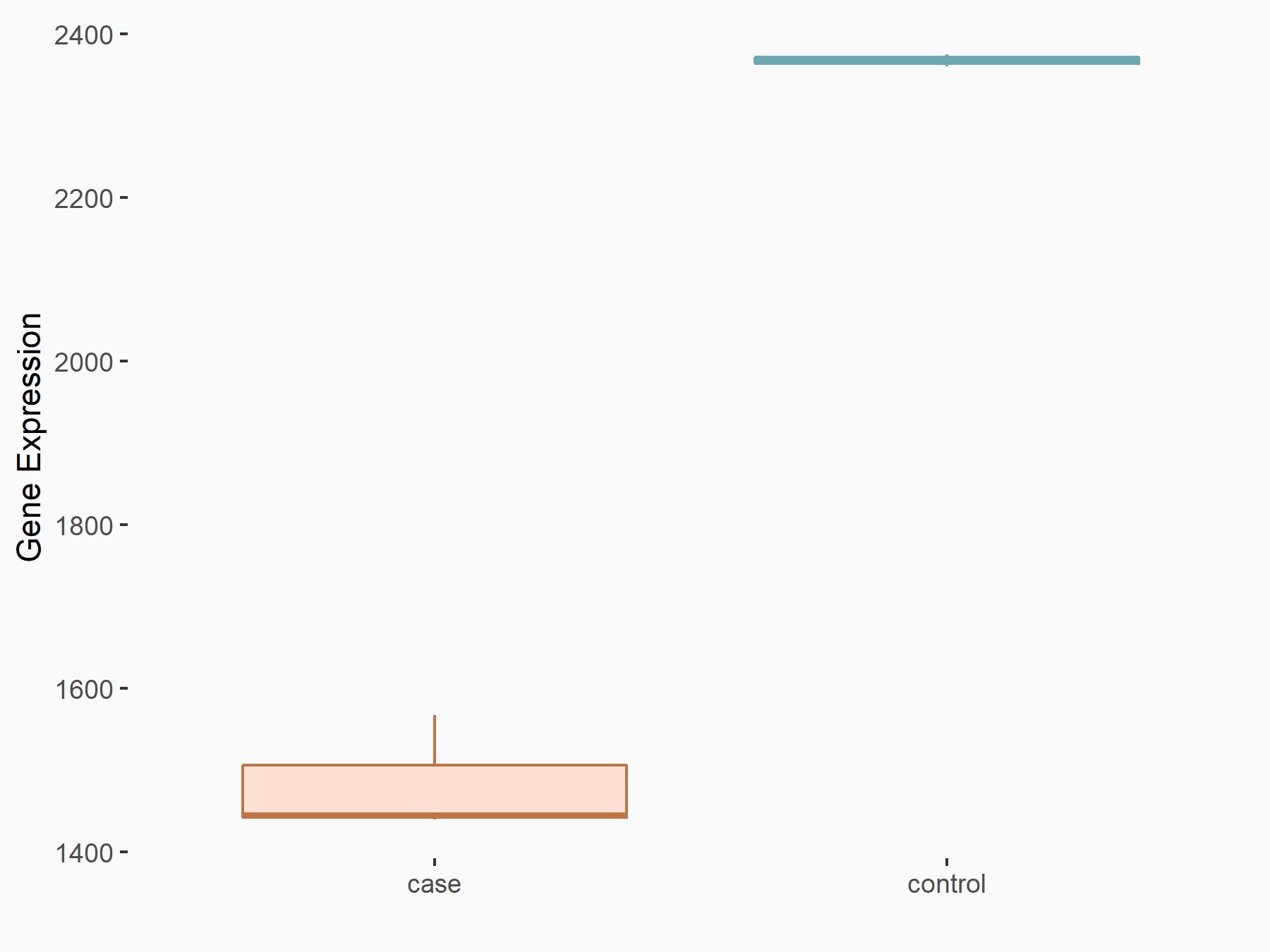 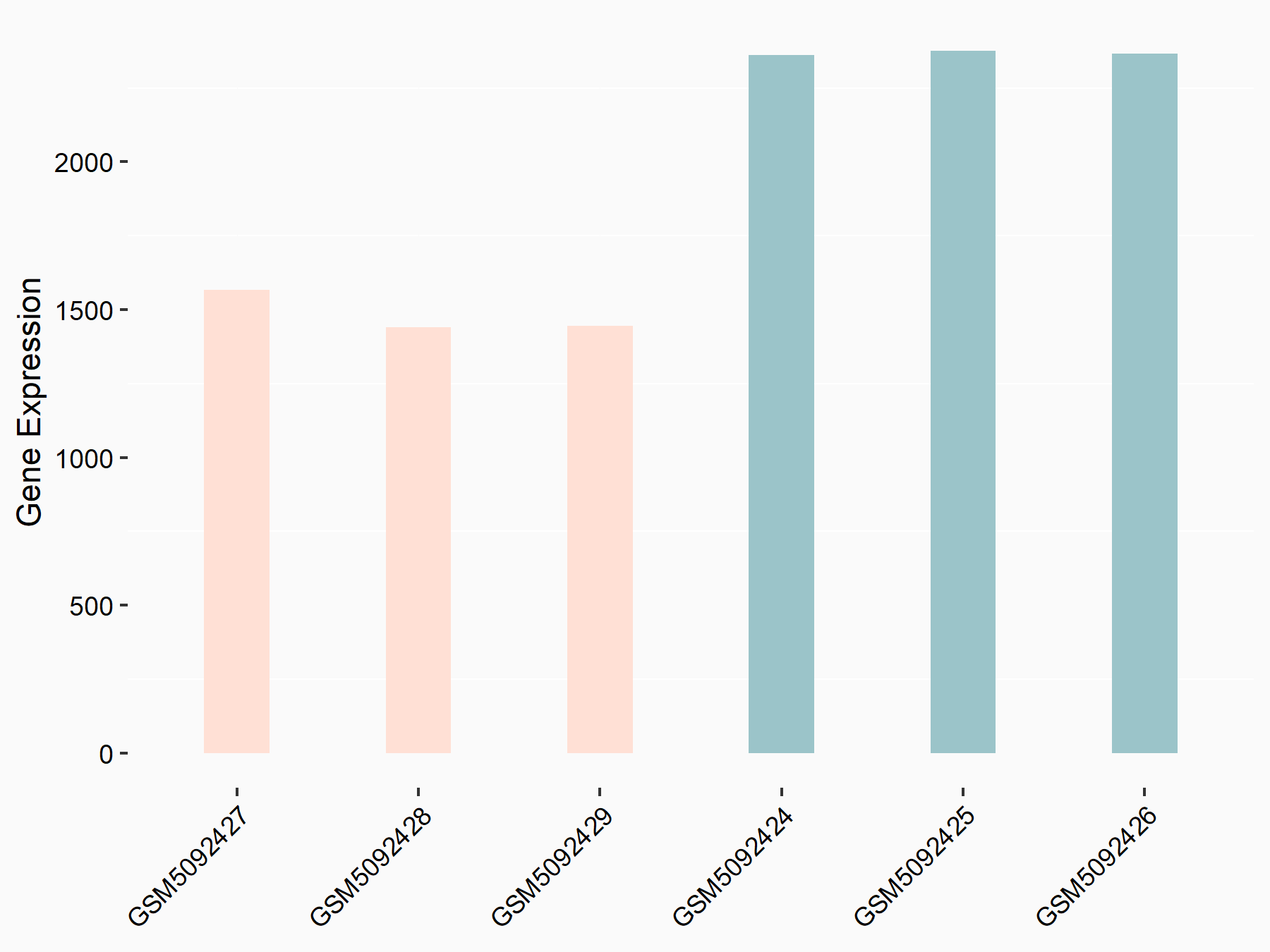 |
logFC: -6.74E-01 p-value: 5.11E-30 |
| More Results | Click to View More RNA-seq Results | |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | METTL3 played a tumor-suppressive role in Colorectal cancer cell proliferation, migration and invasion through p38/Ephrin type-B receptor 2 (ERK/EPHB2) pathways, which indicated that METTL3 was a novel marker for CRC carcinogenesis, progression and survival. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| KM12 | Colon carcinoma | Homo sapiens | CVCL_1331 | |
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | m6A writers METTL3/METTL14 and the m6A reader YTHDF1 orchestrate MNK2 expression posttranscriptionally and thus control Ephrin type-B receptor 2 (ERK/EPHB2) signaling, which is required for the maintenance of muscle myogenesis and contributes to regeneration. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Muscular dystrophies | ICD-11: 8C70 | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| C2C12 | Normal | Mus musculus | CVCL_0188 | |
| In-vivo Model | For mouse muscle injury and regeneration experiment, tibialis anterior (TA) muscles of 6-week-old male mice were injected with 25 uL of 10 uM cardiotoxin (CTX, Merck Millipore, 217503), 0.9% normal saline (Saline) were used as control. The regenerated muscles were collected at day 1, 3, 5, and 10 post-injection. TA muscles were isolated for Hematoxylin and eosin staining or frozen in liquid nitrogen for RNA and protein extraction. | |||
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | FTO depended on its m6A RNA demethylase activity to activate PDGFRB/Ephrin type-B receptor 2 (ERK/EPHB2) signaling axis. FTO-mediated m6A demethylation plays an oncogenic role in NPM1-mutated Acute myeloid leukemia(AML). | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | OCI-AML-3 | Adult acute myeloid leukemia | Homo sapiens | CVCL_1844 |
| OCI-AML-2 | Adult acute myeloid leukemia | Homo sapiens | CVCL_1619 | |
Colon cancer [ICD-11: 2B90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | hnRNPA2B1 promotes colon cancer progression via the MAPK pathway. hnRNPA2B1 is an upstream regulator of the Ephrin type-B receptor 2 (ERK/EPHB2)/MAPK pathway and inhibition of MAPK signaling blocked the effects of hnRNPA2B1. | |||
| Responsed Disease | Colon cancer [ICD-11: 2B90] | |||
| Target Regulator | Heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling | hsa04010 | ||
| Apoptosis | hsa04210 | |||
| Cell Process | Arrest cell cycle at G0/G1 phase | |||
| Cell apoptosis | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| In-vivo Model | Four-week-old male BALB/c nude mice (purchased from Lingchang company) were randomly divided into three groups, each group has five mice. Each of the mice was injected subcutaneously on the right lateral back with 1 × 106 of each lentivirus infected SW480 cells in which hnRNPA2B1 was knocked out or negative control cells. Mice were killed at day 29, and tumors were then isolated, photographed. | |||
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | METTL3 played a tumor-suppressive role in Colorectal cancer cell proliferation, migration and invasion through p38/Ephrin type-B receptor 2 (ERK/EPHB2) pathways, which indicated that METTL3 was a novel marker for CRC carcinogenesis, progression and survival. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| KM12 | Colon carcinoma | Homo sapiens | CVCL_1331 | |
Muscular dystrophies [ICD-11: 8C70]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | m6A writers METTL3/METTL14 and the m6A reader YTHDF1 orchestrate MNK2 expression posttranscriptionally and thus control Ephrin type-B receptor 2 (ERK/EPHB2) signaling, which is required for the maintenance of muscle myogenesis and contributes to regeneration. | |||
| Responsed Disease | Muscular dystrophies [ICD-11: 8C70] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| C2C12 | Normal | Mus musculus | CVCL_0188 | |
| In-vivo Model | For mouse muscle injury and regeneration experiment, tibialis anterior (TA) muscles of 6-week-old male mice were injected with 25 uL of 10 uM cardiotoxin (CTX, Merck Millipore, 217503), 0.9% normal saline (Saline) were used as control. The regenerated muscles were collected at day 1, 3, 5, and 10 post-injection. TA muscles were isolated for Hematoxylin and eosin staining or frozen in liquid nitrogen for RNA and protein extraction. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | m6A writers METTL3/METTL14 and the m6A reader YTHDF1 orchestrate MNK2 expression posttranscriptionally and thus control Ephrin type-B receptor 2 (ERK/EPHB2) signaling, which is required for the maintenance of muscle myogenesis and contributes to regeneration. | |||
| Responsed Disease | Muscular dystrophies [ICD-11: 8C70] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| C2C12 | Normal | Mus musculus | CVCL_0188 | |
| In-vivo Model | For mouse muscle injury and regeneration experiment, tibialis anterior (TA) muscles of 6-week-old male mice were injected with 25 uL of 10 uM cardiotoxin (CTX, Merck Millipore, 217503), 0.9% normal saline (Saline) were used as control. The regenerated muscles were collected at day 1, 3, 5, and 10 post-injection. TA muscles were isolated for Hematoxylin and eosin staining or frozen in liquid nitrogen for RNA and protein extraction. | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | m6A writers METTL3/METTL14 and the m6A reader YTHDF1 orchestrate MNK2 expression posttranscriptionally and thus control Ephrin type-B receptor 2 (ERK/EPHB2) signaling, which is required for the maintenance of muscle myogenesis and contribute to regeneration. | |||
| Responsed Disease | Muscular dystrophies [ICD-11: 8C70] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| C2C12 | Normal | Mus musculus | CVCL_0188 | |
| In-vivo Model | For mouse muscle injury and regeneration experiment, tibialis anterior (TA) muscles of 6-week-old male mice were injected with 25 uL of 10 uM cardiotoxin (CTX, Merck Millipore, 217503), 0.9% normal saline (Saline) were used as control. The regenerated muscles were collected at day 1, 3, 5, and 10 post-injection. TA muscles were isolated for Hematoxylin and eosin staining or frozen in liquid nitrogen for RNA and protein extraction. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03544 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03589 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00244)
| In total 6 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE005256 | Click to Show/Hide the Full List | ||
| mod site | chr1:22717638-22717639:+ | [5] | |
| Sequence | GATTCAGTGAGATGCTGTGTACCATGACTGGCAGAACTGCC | ||
| Transcript ID List | ENST00000374632.7; rmsk_48767; ENST00000374630.7; ENST00000544305.5; ENST00000400191.7 | ||
| External Link | RMBase: RNA-editing_site_2522 | ||
| mod ID: A2ISITE005257 | Click to Show/Hide the Full List | ||
| mod site | chr1:22782186-22782187:+ | [5] | |
| Sequence | GCACTTCCTTAATCTCTCTGAGCTTCAGTTTCTTCATCTAT | ||
| Transcript ID List | ENST00000374632.7; ENST00000374630.7; ENST00000544305.5; rmsk_48944; ENST00000374627.1; ENST00000400191.7 | ||
| External Link | RMBase: RNA-editing_site_2523 | ||
| mod ID: A2ISITE005258 | Click to Show/Hide the Full List | ||
| mod site | chr1:22782205-22782206:+ | [5] | |
| Sequence | GAGCTTCAGTTTCTTCATCTATAAAATGGTGATGATGTTTT | ||
| Transcript ID List | ENST00000374630.7; ENST00000374632.7; ENST00000400191.7; ENST00000544305.5; rmsk_48944; ENST00000374627.1 | ||
| External Link | RMBase: RNA-editing_site_2524 | ||
| mod ID: A2ISITE005259 | Click to Show/Hide the Full List | ||
| mod site | chr1:22807957-22807958:+ | [5] | |
| Sequence | AGCCTGGCCAACATGGTGAAACCCTGTCTCTACTAAAAATG | ||
| Transcript ID List | ENST00000544305.5; ENST00000374632.7; rmsk_49007; ENST00000465676.1; ENST00000374630.7; ENST00000374627.1; ENST00000400191.7 | ||
| External Link | RMBase: RNA-editing_site_2525 | ||
| mod ID: A2ISITE005260 | Click to Show/Hide the Full List | ||
| mod site | chr1:22889855-22889856:+ | [5] | |
| Sequence | TTTTTAAAAATAAATGTTTTAGTACAAATATGTCCCAAATA | ||
| Transcript ID List | ENST00000374627.1; ENST00000465676.1; ENST00000400191.7; ENST00000374630.7; ENST00000544305.5; ENST00000374632.7 | ||
| External Link | RMBase: RNA-editing_site_2526 | ||
| mod ID: A2ISITE005261 | Click to Show/Hide the Full List | ||
| mod site | chr1:22901727-22901728:+ | [5] | |
| Sequence | TTGGCAGCCTGAGGGTTACAAGTGGAGACTCTGTAGCCGGC | ||
| Transcript ID List | ENST00000400191.7; ENST00000374627.1; ENST00000374632.7; rmsk_49246; ENST00000374630.7 | ||
| External Link | RMBase: RNA-editing_site_2527 | ||
N6-methyladenosine (m6A)
| In total 71 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE004381 | Click to Show/Hide the Full List | ||
| mod site | chr1:22781432-22781433:+ | [6] | |
| Sequence | CCCCACAGAAACGCTAATGGACTCCACTACAGCGACTGCTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; iSLK; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374632.7; ENST00000374627.1; ENST00000400191.7; ENST00000544305.5; ENST00000374630.7 | ||
| External Link | RMBase: m6A_site_13451 | ||
| mod ID: M6ASITE004382 | Click to Show/Hide the Full List | ||
| mod site | chr1:22784419-22784420:+ | [6] | |
| Sequence | GGTGAGTGGCTACGATGAGAACATGAACACGATCCGCACGT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; U2OS; H1B; H1A; fibroblasts; GSC-11; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000400191.7; ENST00000374627.1; ENST00000374632.7; ENST00000544305.5; ENST00000374630.7 | ||
| External Link | RMBase: m6A_site_13452 | ||
| mod ID: M6ASITE004383 | Click to Show/Hide the Full List | ||
| mod site | chr1:22784425-22784426:+ | [6] | |
| Sequence | TGGCTACGATGAGAACATGAACACGATCCGCACGTACCAGG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; U2OS; H1B; H1A; fibroblasts; GSC-11; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374632.7; ENST00000374627.1; ENST00000374630.7; ENST00000544305.5; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13453 | ||
| mod ID: M6ASITE004384 | Click to Show/Hide the Full List | ||
| mod site | chr1:22784473-22784474:+ | [6] | |
| Sequence | CGTGTTTGAGTCAAGCCAGAACAACTGGCTACGGACCAAGT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; U2OS; H1B; H1A; hNPCs; hESCs; fibroblasts; GSC-11; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000544305.5; ENST00000374632.7; ENST00000374627.1; ENST00000374630.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13454 | ||
| mod ID: M6ASITE004385 | Click to Show/Hide the Full List | ||
| mod site | chr1:22784487-22784488:+ | [6] | |
| Sequence | GCCAGAACAACTGGCTACGGACCAAGTTTATCCGGCGCCGT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; U2OS; H1B; H1A; hNPCs; hESCs; fibroblasts; GSC-11; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000400191.7; ENST00000374630.7; ENST00000374632.7; ENST00000544305.5; ENST00000374627.1 | ||
| External Link | RMBase: m6A_site_13455 | ||
| mod ID: M6ASITE004386 | Click to Show/Hide the Full List | ||
| mod site | chr1:22784551-22784552:+ | [7] | |
| Sequence | GATGAAGTTTTCGGTGCGTGACTGCAGCAGCATCCCCAGCG | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000374627.1; ENST00000544305.5; ENST00000374632.7; ENST00000400191.7; ENST00000374630.7 | ||
| External Link | RMBase: m6A_site_13456 | ||
| mod ID: M6ASITE004387 | Click to Show/Hide the Full List | ||
| mod site | chr1:22784592-22784593:+ | [6] | |
| Sequence | TGCCTGGCTCCTGCAAGGAGACCTTCAACCTCTATTACTAT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; GSC-11; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000400191.7; ENST00000544305.5; ENST00000374632.7; ENST00000374630.7; ENST00000374627.1 | ||
| External Link | RMBase: m6A_site_13457 | ||
| mod ID: M6ASITE004388 | Click to Show/Hide the Full List | ||
| mod site | chr1:22784626-22784627:+ | [8] | |
| Sequence | TTACTATGAGGCTGACTTTGACTCGGCCACCAAGACCTTCC | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000374627.1; ENST00000374632.7; ENST00000400191.7; ENST00000374630.7; ENST00000544305.5 | ||
| External Link | RMBase: m6A_site_13458 | ||
| mod ID: M6ASITE004389 | Click to Show/Hide the Full List | ||
| mod site | chr1:22784640-22784641:+ | [6] | |
| Sequence | ACTTTGACTCGGCCACCAAGACCTTCCCCAACTGGATGGAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; Jurkat; GSC-11; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000400191.7; ENST00000374630.7; ENST00000374627.1; ENST00000374632.7; ENST00000544305.5 | ||
| External Link | RMBase: m6A_site_13459 | ||
| mod ID: M6ASITE004390 | Click to Show/Hide the Full List | ||
| mod site | chr1:22784716-22784717:+ | [6] | |
| Sequence | CGAGAGCTTCTCCCAGGTGGACCTGGGTGGCCGCGTCATGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; Jurkat; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374630.7; ENST00000400191.7; ENST00000544305.5; ENST00000374627.1; ENST00000374632.7 | ||
| External Link | RMBase: m6A_site_13460 | ||
| mod ID: M6ASITE004391 | Click to Show/Hide the Full List | ||
| mod site | chr1:22784743-22784744:+ | [9] | |
| Sequence | TGGCCGCGTCATGAAAATCAACACCGAGGTGCGGAGCTTCG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000374627.1; ENST00000400191.7; ENST00000374632.7; ENST00000374630.7; ENST00000544305.5 | ||
| External Link | RMBase: m6A_site_13461 | ||
| mod ID: M6ASITE004392 | Click to Show/Hide the Full List | ||
| mod site | chr1:22784765-22784766:+ | [6] | |
| Sequence | ACCGAGGTGCGGAGCTTCGGACCTGTGTCCCGCAGCGGCTT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; Jurkat; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374630.7; ENST00000374627.1; ENST00000400191.7; ENST00000374632.7; ENST00000544305.5 | ||
| External Link | RMBase: m6A_site_13462 | ||
| mod ID: M6ASITE004393 | Click to Show/Hide the Full List | ||
| mod site | chr1:22784803-22784804:+ | [6] | |
| Sequence | CTTCTACCTGGCCTTCCAGGACTATGGCGGCTGCATGTCCC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; H1A; H1B; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374630.7; ENST00000374632.7; ENST00000374627.1; ENST00000400191.7; ENST00000544305.5 | ||
| External Link | RMBase: m6A_site_13463 | ||
| mod ID: M6ASITE004394 | Click to Show/Hide the Full List | ||
| mod site | chr1:22784892-22784893:+ | [6] | |
| Sequence | ATGGCGCCATCTTCCAGGAAACCCTGTCGGGGGCTGAGAGC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; A549; U2OS; GSC-11; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000400191.7; ENST00000374627.1; ENST00000374630.7; ENST00000374632.7; ENST00000544305.5 | ||
| External Link | RMBase: m6A_site_13464 | ||
| mod ID: M6ASITE004395 | Click to Show/Hide the Full List | ||
| mod site | chr1:22863050-22863051:+ | [6] | |
| Sequence | TCTCAGGTTGTCCATCTGGGACTTTCAAGGCCAACCAAGGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000400191.7; ENST00000465676.1; ENST00000544305.5; ENST00000374632.7; ENST00000374630.7; ENST00000374627.1 | ||
| External Link | RMBase: m6A_site_13465 | ||
| mod ID: M6ASITE004396 | Click to Show/Hide the Full List | ||
| mod site | chr1:22863107-22863108:+ | [6] | |
| Sequence | ACTGTCCCATCAACAGCCGGACCACTTCTGAAGGGGCCACC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000544305.5; ENST00000374630.7; ENST00000374632.7; ENST00000400191.7; ENST00000374627.1; ENST00000465676.1 | ||
| External Link | RMBase: m6A_site_13466 | ||
| mod ID: M6ASITE004397 | Click to Show/Hide the Full List | ||
| mod site | chr1:22863162-22863163:+ | [6] | |
| Sequence | CAATGGCTACTACAGAGCAGACCTGGACCCCCTGGACATGC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000374632.7; ENST00000374630.7; ENST00000465676.1; ENST00000544305.5; ENST00000374627.1; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13467 | ||
| mod ID: M6ASITE004398 | Click to Show/Hide the Full List | ||
| mod site | chr1:22863168-22863169:+ | [6] | |
| Sequence | CTACTACAGAGCAGACCTGGACCCCCTGGACATGCCCTGCA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000374630.7; ENST00000400191.7; ENST00000544305.5; ENST00000374632.7; ENST00000465676.1; ENST00000374627.1 | ||
| External Link | RMBase: m6A_site_13468 | ||
| mod ID: M6ASITE004399 | Click to Show/Hide the Full List | ||
| mod site | chr1:22863177-22863178:+ | [6] | |
| Sequence | AGCAGACCTGGACCCCCTGGACATGCCCTGCACAAGTAAGT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000465676.1; ENST00000544305.5; ENST00000374632.7; ENST00000400191.7; ENST00000374630.7; ENST00000374627.1 | ||
| External Link | RMBase: m6A_site_13469 | ||
| mod ID: M6ASITE004400 | Click to Show/Hide the Full List | ||
| mod site | chr1:22863188-22863189:+ | [9] | |
| Sequence | ACCCCCTGGACATGCCCTGCACAAGTAAGTCCTAGGGCCCC | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000400191.7; ENST00000544305.5; ENST00000374630.7; ENST00000465676.1; ENST00000374627.1; ENST00000374632.7 | ||
| External Link | RMBase: m6A_site_13470 | ||
| mod ID: M6ASITE004401 | Click to Show/Hide the Full List | ||
| mod site | chr1:22864920-22864921:+ | [6] | |
| Sequence | TGATTTCCAGTGTCAATGAGACCTCCCTCATGCTGGAGTGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374630.7; ENST00000400191.7; ENST00000374627.1; ENST00000374632.7; ENST00000544305.5; ENST00000465676.1 | ||
| External Link | RMBase: m6A_site_13471 | ||
| mod ID: M6ASITE004402 | Click to Show/Hide the Full List | ||
| mod site | chr1:22864941-22864942:+ | [6] | |
| Sequence | CCTCCCTCATGCTGGAGTGGACCCCTCCCCGCGACTCCGGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; A549; iSLK; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000465676.1; ENST00000374632.7; ENST00000544305.5; ENST00000374627.1; ENST00000374630.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13472 | ||
| mod ID: M6ASITE004403 | Click to Show/Hide the Full List | ||
| mod site | chr1:22864972-22864973:+ | [6] | |
| Sequence | CGACTCCGGAGGCCGAGAGGACCTCGTCTACAACATCATCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; A549; iSLK; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374630.7; ENST00000374627.1; ENST00000374632.7; ENST00000400191.7; ENST00000465676.1; ENST00000544305.5 | ||
| External Link | RMBase: m6A_site_13473 | ||
| mod ID: M6ASITE004404 | Click to Show/Hide the Full List | ||
| mod site | chr1:22864981-22864982:+ | [9] | |
| Sequence | AGGCCGAGAGGACCTCGTCTACAACATCATCTGCAAGAGCT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000374630.7; ENST00000544305.5; ENST00000465676.1; ENST00000374627.1; ENST00000374632.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13474 | ||
| mod ID: M6ASITE004405 | Click to Show/Hide the Full List | ||
| mod site | chr1:22864984-22864985:+ | [9] | |
| Sequence | CCGAGAGGACCTCGTCTACAACATCATCTGCAAGAGCTGTG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000544305.5; ENST00000400191.7; ENST00000465676.1; ENST00000374630.7; ENST00000374627.1; ENST00000374632.7 | ||
| External Link | RMBase: m6A_site_13475 | ||
| mod ID: M6ASITE004406 | Click to Show/Hide the Full List | ||
| mod site | chr1:22865038-22865039:+ | [6] | |
| Sequence | TGCCTGCACCCGCTGCGGGGACAATGTACAGTACGCACCAC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; A549; iSLK; TIME | ||
| Seq Type List | m6A-seq; MAZTER-seq; m6A-CLIP/IP; MeRIP-seq | ||
| Transcript ID List | ENST00000400191.7; ENST00000374632.7; ENST00000544305.5; ENST00000465676.1; ENST00000374627.1; ENST00000374630.7 | ||
| External Link | RMBase: m6A_site_13476 | ||
| mod ID: M6ASITE004407 | Click to Show/Hide the Full List | ||
| mod site | chr1:22865089-22865090:+ | [9] | |
| Sequence | CCTGACCGAGCCACGCATTTACATCAGTGACCTGCTGGCCC | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000374627.1; ENST00000374630.7; ENST00000544305.5; ENST00000465676.1; ENST00000374632.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13477 | ||
| mod ID: M6ASITE004408 | Click to Show/Hide the Full List | ||
| mod site | chr1:22865119-22865120:+ | [9] | |
| Sequence | CCTGCTGGCCCACACCCAGTACACCTTCGAGATCCAGGCTG | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000374630.7; ENST00000400191.7; ENST00000465676.1; ENST00000374627.1; ENST00000544305.5; ENST00000374632.7 | ||
| External Link | RMBase: m6A_site_13478 | ||
| mod ID: M6ASITE004409 | Click to Show/Hide the Full List | ||
| mod site | chr1:22865191-22865192:+ | [6] | |
| Sequence | GCCTCAGTTCGCCTCTGTGAACATCACCACCAACCAGGCAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; iSLK; TIME | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374627.1; ENST00000465676.1; ENST00000490436.1; ENST00000374632.7; ENST00000400191.7; ENST00000544305.5; ENST00000374630.7 | ||
| External Link | RMBase: m6A_site_13479 | ||
| mod ID: M6ASITE004410 | Click to Show/Hide the Full List | ||
| mod site | chr1:22882403-22882404:+ | [10] | |
| Sequence | TCAGGTGAGCCGCACCGTGGACAGCATTACCCTGTCGTGGT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | iSLK | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000465676.1; ENST00000544305.5; ENST00000374627.1; ENST00000490436.1; ENST00000400191.7; ENST00000374632.7; ENST00000374630.7 | ||
| External Link | RMBase: m6A_site_13480 | ||
| mod ID: M6ASITE004411 | Click to Show/Hide the Full List | ||
| mod site | chr1:22895482-22895483:+ | [9] | |
| Sequence | TCTCCCCAGCCGAGTACCAGACAAGCATCCAGGAGAAGTTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000374627.1; ENST00000465676.1; ENST00000374630.7; ENST00000374632.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13481 | ||
| mod ID: M6ASITE004412 | Click to Show/Hide the Full List | ||
| mod site | chr1:22906796-22906797:+ | [9] | |
| Sequence | CAAGACGCTCAAGTCGGGCTACACGGAGAAGCAGCGCCGGG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000374627.1; ENST00000374630.7; ENST00000400191.7; ENST00000374632.7 | ||
| External Link | RMBase: m6A_site_13482 | ||
| mod ID: M6ASITE004413 | Click to Show/Hide the Full List | ||
| mod site | chr1:22906897-22906898:+ | [9] | |
| Sequence | AGGGTGTCGTGACCAAGAGCACACCTGTGATGATCATCACC | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000400191.7; ENST00000374630.7; ENST00000374627.1; ENST00000374632.7 | ||
| External Link | RMBase: m6A_site_13483 | ||
| mod ID: M6ASITE004414 | Click to Show/Hide the Full List | ||
| mod site | chr1:22906943-22906944:+ | [6] | |
| Sequence | CATGGAGAATGGCTCCCTGGACTCCTTTCTCCGGGTAGGGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000374627.1; ENST00000374630.7; ENST00000374632.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13484 | ||
| mod ID: M6ASITE004415 | Click to Show/Hide the Full List | ||
| mod site | chr1:22908031-22908032:+ | [6] | |
| Sequence | TGGCATGAAGTACCTGGCAGACATGAACTATGTTCACCGTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000374632.7; ENST00000400191.7; ENST00000374627.1; ENST00000374630.7 | ||
| External Link | RMBase: m6A_site_13485 | ||
| mod ID: M6ASITE004416 | Click to Show/Hide the Full List | ||
| mod site | chr1:22908037-22908038:+ | [6] | |
| Sequence | GAAGTACCTGGCAGACATGAACTATGTTCACCGTGACCTGG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000374632.7; ENST00000374627.1; ENST00000400191.7; ENST00000374630.7 | ||
| External Link | RMBase: m6A_site_13486 | ||
| mod ID: M6ASITE004417 | Click to Show/Hide the Full List | ||
| mod site | chr1:22908106-22908107:+ | [6] | |
| Sequence | CCTGGTCTGCAAGGTGTCGGACTTTGGGCTCTCACGCTTTC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000374632.7; ENST00000374630.7; ENST00000400191.7; ENST00000374627.1 | ||
| External Link | RMBase: m6A_site_13487 | ||
| mod ID: M6ASITE004418 | Click to Show/Hide the Full List | ||
| mod site | chr1:22910427-22910428:+ | [6] | |
| Sequence | TCGGCTGCCACCGCCCATGGACTGCCCGAGCGCCCTGCACC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000374627.1; ENST00000374630.7; ENST00000374632.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13488 | ||
| mod ID: M6ASITE004419 | Click to Show/Hide the Full List | ||
| mod site | chr1:22910460-22910461:+ | [6] | |
| Sequence | CCTGCACCAACTCATGCTGGACTGTTGGCAGAAGGACCGCA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000400191.7; ENST00000374630.7; ENST00000374627.1; ENST00000374632.7 | ||
| External Link | RMBase: m6A_site_13489 | ||
| mod ID: M6ASITE004420 | Click to Show/Hide the Full List | ||
| mod site | chr1:22910475-22910476:+ | [6] | |
| Sequence | GCTGGACTGTTGGCAGAAGGACCGCAACCACCGGCCCAAGT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000374627.1; ENST00000400191.7; ENST00000374630.7; ENST00000374632.7 | ||
| External Link | RMBase: m6A_site_13490 | ||
| mod ID: M6ASITE004421 | Click to Show/Hide the Full List | ||
| mod site | chr1:22910520-22910521:+ | [6] | |
| Sequence | CCAAATTGTCAACACGCTAGACAAGATGATCCGCAATCCCA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000374630.7; ENST00000374632.7; ENST00000374627.1; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13491 | ||
| mod ID: M6ASITE004422 | Click to Show/Hide the Full List | ||
| mod site | chr1:22912463-22912464:+ | [6] | |
| Sequence | CATCAACCTGCCGCTGCTGGACCGCACGATCCCCGACTACA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000374630.7; ENST00000374627.1; ENST00000374632.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13492 | ||
| mod ID: M6ASITE004423 | Click to Show/Hide the Full List | ||
| mod site | chr1:22913514-22913515:+ | [6] | |
| Sequence | CCACCAGAAAAAAATCCTGAACAGTATCCAGGTGATGCGGG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374627.1; ENST00000374630.7; ENST00000400191.7; ENST00000374632.7 | ||
| External Link | RMBase: m6A_site_13493 | ||
| mod ID: M6ASITE004424 | Click to Show/Hide the Full List | ||
| mod site | chr1:22913544-22913545:+ | [6] | |
| Sequence | GGTGATGCGGGCGCAGATGAACCAGATTCAGTCTGTGGAGG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; iSLK; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374632.7; ENST00000400191.7; ENST00000374630.7; ENST00000374627.1 | ||
| External Link | RMBase: m6A_site_13494 | ||
| mod ID: M6ASITE004425 | Click to Show/Hide the Full List | ||
| mod site | chr1:22913696-22913697:+ | [6] | |
| Sequence | GGCCACGGGCCACGGGAAGAACCAAGCGGTGCCAGCCACGA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; H1A; H1B; hNPCs; hESCs; fibroblasts; GSC-11; HEK293A-TOA; iSLK; TIME; MSC; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000400191.7; ENST00000374632.7; ENST00000374627.1; ENST00000374630.7 | ||
| External Link | RMBase: m6A_site_13495 | ||
| mod ID: M6ASITE004426 | Click to Show/Hide the Full List | ||
| mod site | chr1:22913732-22913733:+ | [6] | |
| Sequence | CACGAGACGTCACCAAGAAAACATGCAACTCAAACGACGGA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000374632.7; ENST00000374627.1; ENST00000374630.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13496 | ||
| mod ID: M6ASITE004427 | Click to Show/Hide the Full List | ||
| mod site | chr1:22913780-22913781:+ | [6] | |
| Sequence | AGGGAATGGGAAAAAAGAAAACAGATCCTGGGAGGGGGCGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374632.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13497 | ||
| mod ID: M6ASITE004428 | Click to Show/Hide the Full List | ||
| mod site | chr1:22913850-22913851:+ | [8] | |
| Sequence | TTCTCATAAGGAAAGCAATGACTGTTCTTGCGGGGGATAAA | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000400191.7; ENST00000374632.7 | ||
| External Link | RMBase: m6A_site_13498 | ||
| mod ID: M6ASITE004429 | Click to Show/Hide the Full List | ||
| mod site | chr1:22913910-22913911:+ | [6] | |
| Sequence | TGCGATGTGTCCAATCGGAGACAAAAGCAGTTTCTCTCCAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374632.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13499 | ||
| mod ID: M6ASITE004430 | Click to Show/Hide the Full List | ||
| mod site | chr1:22913967-22913968:+ | [6] | |
| Sequence | GACCTGGCCAGAGCCAAGAAACACTTTCAGAAAAACAAATG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000400191.7; ENST00000374632.7 | ||
| External Link | RMBase: m6A_site_13500 | ||
| mod ID: M6ASITE004431 | Click to Show/Hide the Full List | ||
| mod site | chr1:22913981-22913982:+ | [6] | |
| Sequence | CAAGAAACACTTTCAGAAAAACAAATGTGAAGGGGAGAGAC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000400191.7; ENST00000374632.7 | ||
| External Link | RMBase: m6A_site_13501 | ||
| mod ID: M6ASITE004432 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914000-22914001:+ | [6] | |
| Sequence | AACAAATGTGAAGGGGAGAGACAGGGGCCGCCCTTGGCTCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374632.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13502 | ||
| mod ID: M6ASITE004433 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914051-22914052:+ | [9] | |
| Sequence | GCTCCTCTAGGCCTCACTCAACAACCAAGCGCCTGGAGGAC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T; A549 | ||
| Seq Type List | MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000374632.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13503 | ||
| mod ID: M6ASITE004434 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914070-22914071:+ | [8] | |
| Sequence | AACAACCAAGCGCCTGGAGGACGGGACAGATGGACAGACAG | ||
| Motif Score | 3.616982143 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000374632.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13504 | ||
| mod ID: M6ASITE004435 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914075-22914076:+ | [6] | |
| Sequence | CCAAGCGCCTGGAGGACGGGACAGATGGACAGACAGCCACC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000374632.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13505 | ||
| mod ID: M6ASITE004436 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914083-22914084:+ | [6] | |
| Sequence | CTGGAGGACGGGACAGATGGACAGACAGCCACCCTGAGAAC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374632.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13506 | ||
| mod ID: M6ASITE004437 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914102-22914103:+ | [6] | |
| Sequence | GACAGACAGCCACCCTGAGAACCCCTCTGGGAAAATCTATT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000400191.7; ENST00000374632.7 | ||
| External Link | RMBase: m6A_site_13507 | ||
| mod ID: M6ASITE004438 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914132-22914133:+ | [7] | |
| Sequence | GAAAATCTATTCCTGCCACCACTGGGCAAACAGAAGAATTT | ||
| Motif Score | 2.475107143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000374632.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13508 | ||
| mod ID: M6ASITE004439 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914141-22914142:+ | [6] | |
| Sequence | TTCCTGCCACCACTGGGCAAACAGAAGAATTTTTCTGTCTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374632.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13509 | ||
| mod ID: M6ASITE004440 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914179-22914180:+ | [6] | |
| Sequence | CTTTGGAGAGTATTTTAGAAACTCCAATGAAAGACACTGTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374632.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13510 | ||
| mod ID: M6ASITE004441 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914192-22914193:+ | [6] | |
| Sequence | TTTAGAAACTCCAATGAAAGACACTGTTTCTCCTGTTGGCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000400191.7; ENST00000374632.7 | ||
| External Link | RMBase: m6A_site_13511 | ||
| mod ID: M6ASITE004442 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914263-22914264:+ | [6] | |
| Sequence | GGGTCAGGGAGAACGCGGGGACCCCAGAAAGGTCAGCCTTC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; U2OS; H1A; H1B; hNPCs; fibroblasts; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374632.7; ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13512 | ||
| mod ID: M6ASITE004443 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914390-22914391:+ | [6] | |
| Sequence | CGCCAGCCCCTGCCTCGAGGACTGATACTGCAGTGACTGCC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; H1A; H1B; hESCs; fibroblasts; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13513 | ||
| mod ID: M6ASITE004451 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914396-22914397:+ | [7] | |
| Sequence | CCCCTGCCTCGAGGACTGATACTGCAGTGACTGCCGTCAGC | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13514 | ||
| mod ID: M6ASITE004462 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914421-22914422:+ | [7] | |
| Sequence | AGTGACTGCCGTCAGCTCCGACTGCCGCTGAGAAGGGTTGA | ||
| Motif Score | 3.287565476 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13515 | ||
| mod ID: M6ASITE004473 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914476-22914477:+ | [6] | |
| Sequence | TTGTTTACAGCAATTCCTGGACTCGGGGGTATTTTGGTCAC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; U2OS; H1A; H1B; hESCs; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13516 | ||
| mod ID: M6ASITE004484 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914601-22914602:+ | [6] | |
| Sequence | ACATTTCCTACCTTTTGAGGACTTGATCCTTCTCCAGGAAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; iSLK; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13517 | ||
| mod ID: M6ASITE004495 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914787-22914788:+ | [6] | |
| Sequence | GCCATCTGGGCCATTCAGAGACTGGAGTGAGATTTGGGTGT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; U2OS; H1B; H1A; hESCs; A549; GSC-11; HEK293A-TOA; iSLK; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13518 | ||
| mod ID: M6ASITE004504 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914849-22914850:+ | [6] | |
| Sequence | GAGGAGCTTCCCACTCCAGGACTGTTGATGAAAGGGACAGA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; H1B; H1A; hESCs; GSC-11; HEK293A-TOA; iSLK; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13519 | ||
| mod ID: M6ASITE004510 | Click to Show/Hide the Full List | ||
| mod site | chr1:22914865-22914866:+ | [6] | |
| Sequence | CAGGACTGTTGATGAAAGGGACAGATTGAGGAGGAAGTGGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; H1B; H1A; hESCs; GSC-11; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13520 | ||
| mod ID: M6ASITE004512 | Click to Show/Hide the Full List | ||
| mod site | chr1:22915005-22915006:+ | [11] | |
| Sequence | AGTGCCCTCAGATGGTCAAAACCCAGTTTTCCCTCTGGGAG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000400191.7 | ||
| External Link | RMBase: m6A_site_13521 | ||
References