m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00569)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
CDCP1
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
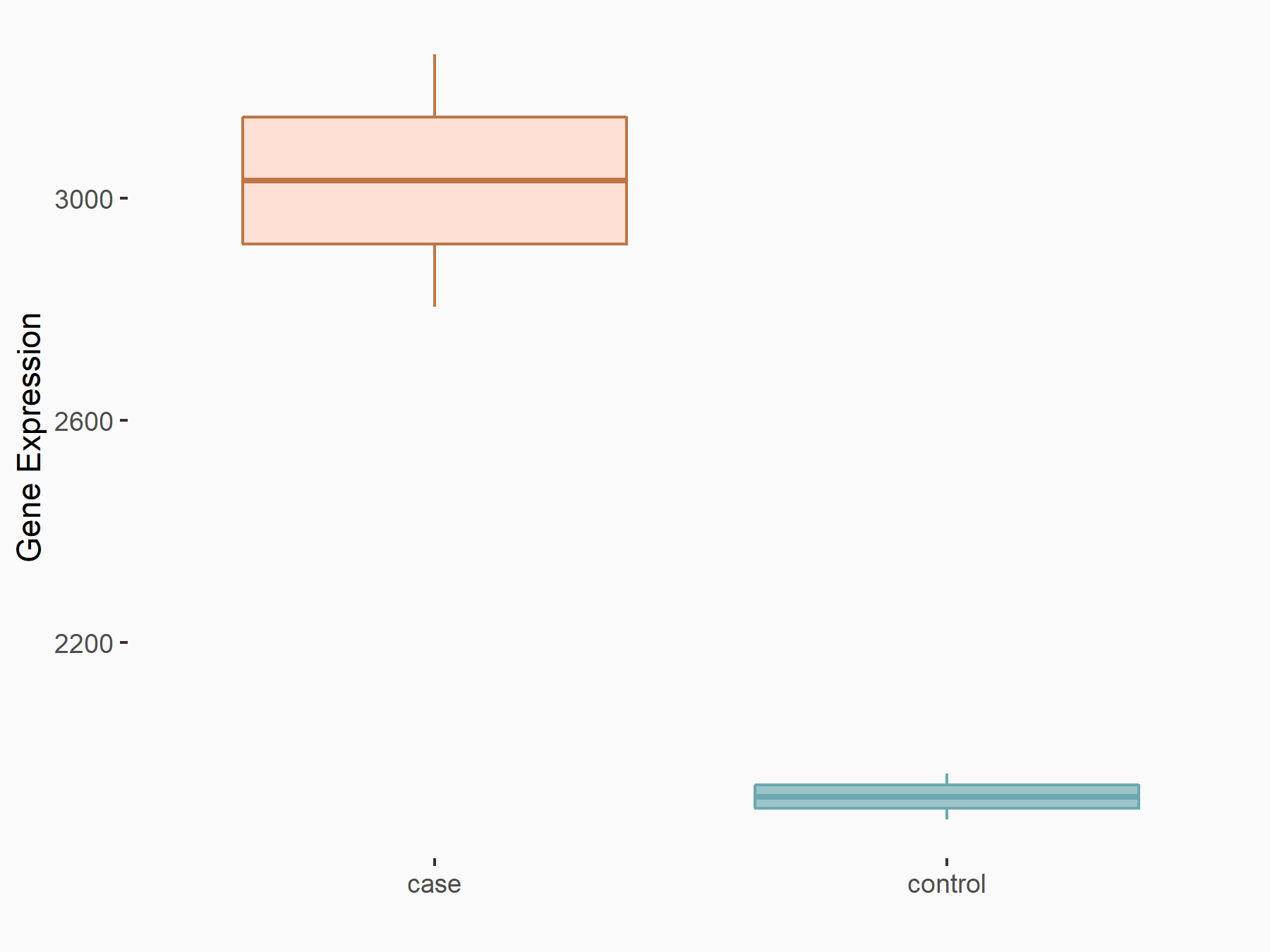 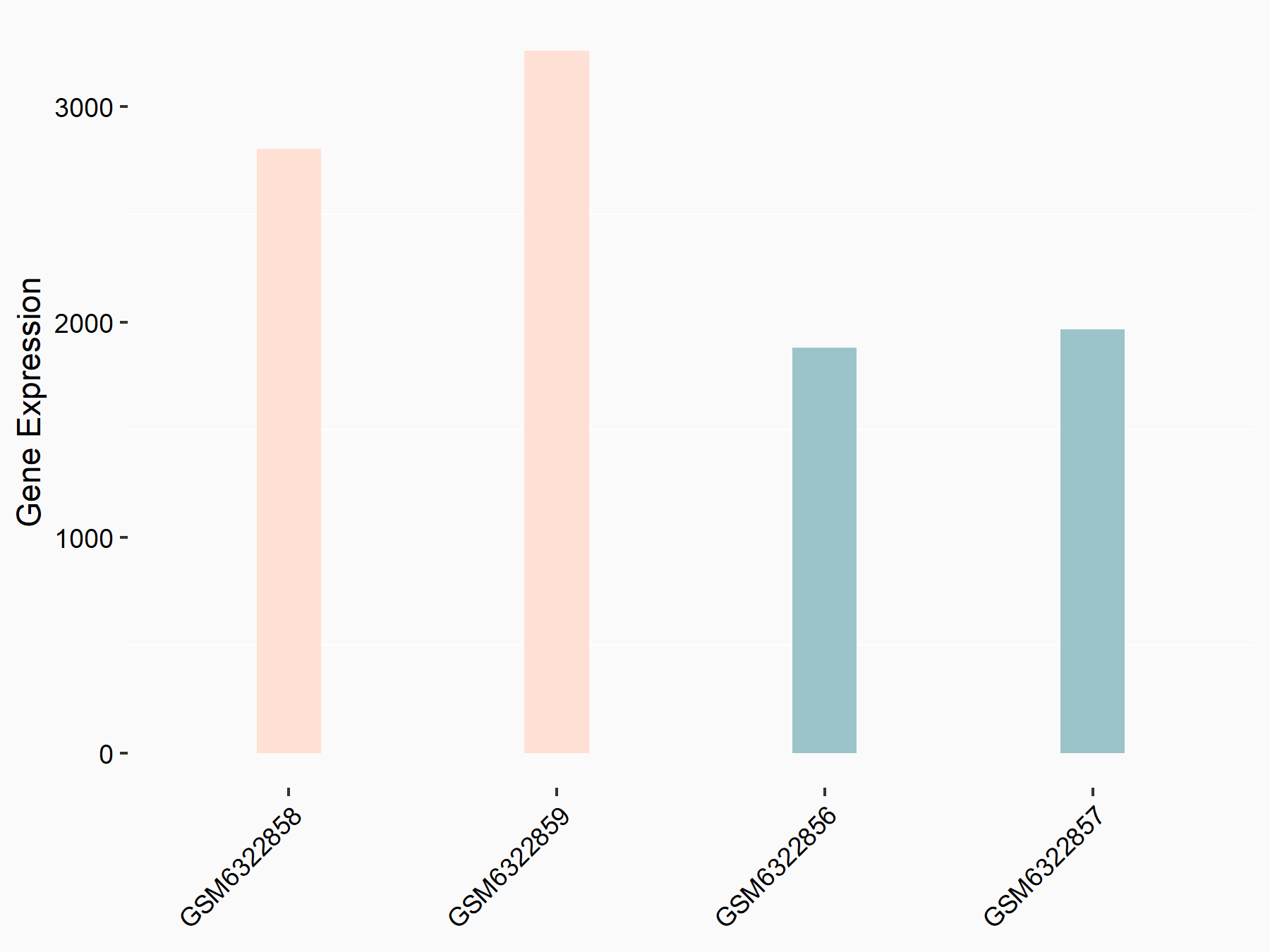 |
logFC: 6.56E-01 p-value: 5.99E-11 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between CDCP1 and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.49E+00 | GSE60213 |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | m6A methyltransferase METTL3 and demethylases ALKBH5 mediate the m6A modification in 3'-UTR of CDCP1 mRNA. METTL3 and CUB domain-containing protein 1 (CDCP1) are upregulated in the bladder cancer patient samples and the expression of METTL3 and CDCP1 is correlated with the progression status of the bladder cancers. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Bladder cancer | ICD-11: 2C94 | ||
| In-vitro Model | UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| NSTC2 (Nickel-induced transformation of human cells) | ||||
| MC-SV-HUC T-2 | Ureteral tumor cell | Homo sapiens | CVCL_6418 | |
| 16HBE14o- | Normal | Homo sapiens | CVCL_0112 | |
| In-vivo Model | To test for malignant transformation, 1×107 cells were inoculated subcutaneously in the dorsal thoracic midline of ten NOD/SCID mice (Weitong Lihua Experimental Animal Technology Co. Ltd). Tumor formation and growth were assessed every 3 days. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | The RCas9-METTL3 system mediates efficient sitespecific m6A installation on CUB domain-containing protein 1 (CDCP1) mRNA and promotes bladder cancer development. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Bladder cancer | ICD-11: 2C94 | ||
| In-vitro Model | T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
RNA demethylase ALKBH5 (ALKBH5) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by ALKBH5 | ||
| Cell Line | 143B cell line | Homo sapiens |
|
Treatment: siALKBH5 transfected 143B cells
Control: siControl 143B cells
|
GSE154528 | |
| Regulation |
  |
logFC: 1.57E+00 p-value: 8.38E-14 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | m6A methyltransferase METTL3 and demethylases ALKBH5 mediate the m6A modification in 3'-UTR of CDCP1 mRNA. METTL3 and CUB domain-containing protein 1 (CDCP1) are upregulated in the bladder cancer patient samples and the expression of METTL3 and CDCP1 is correlated with the progression status of the bladder cancers. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Bladder cancer | ICD-11: 2C94 | ||
| In-vitro Model | UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| NSTC2 (Nickel-induced transformation of human cells) | ||||
| MC-SV-HUC T-2 | Ureteral tumor cell | Homo sapiens | CVCL_6418 | |
| 16HBE14o- | Normal | Homo sapiens | CVCL_0112 | |
| In-vivo Model | To test for malignant transformation, 1×107 cells were inoculated subcutaneously in the dorsal thoracic midline of ten NOD/SCID mice (Weitong Lihua Experimental Animal Technology Co. Ltd). Tumor formation and growth were assessed every 3 days. | |||
Bladder cancer [ICD-11: 2C94]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | m6A methyltransferase METTL3 and demethylases ALKBH5 mediate the m6A modification in 3'-UTR of CDCP1 mRNA. METTL3 and CUB domain-containing protein 1 (CDCP1) are upregulated in the bladder cancer patient samples and the expression of METTL3 and CDCP1 is correlated with the progression status of the bladder cancers. | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| NSTC2 (Nickel-induced transformation of human cells) | ||||
| MC-SV-HUC T-2 | Ureteral tumor cell | Homo sapiens | CVCL_6418 | |
| 16HBE14o- | Normal | Homo sapiens | CVCL_0112 | |
| In-vivo Model | To test for malignant transformation, 1×107 cells were inoculated subcutaneously in the dorsal thoracic midline of ten NOD/SCID mice (Weitong Lihua Experimental Animal Technology Co. Ltd). Tumor formation and growth were assessed every 3 days. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | The RCas9-METTL3 system mediates efficient sitespecific m6A installation on CUB domain-containing protein 1 (CDCP1) mRNA and promotes bladder cancer development. | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Experiment 3 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | m6A methyltransferase METTL3 and demethylases ALKBH5 mediate the m6A modification in 3'-UTR of CDCP1 mRNA. METTL3 and CUB domain-containing protein 1 (CDCP1) are upregulated in the bladder cancer patient samples and the expression of METTL3 and CDCP1 is correlated with the progression status of the bladder cancers. | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| NSTC2 (Nickel-induced transformation of human cells) | ||||
| MC-SV-HUC T-2 | Ureteral tumor cell | Homo sapiens | CVCL_6418 | |
| 16HBE14o- | Normal | Homo sapiens | CVCL_0112 | |
| In-vivo Model | To test for malignant transformation, 1×107 cells were inoculated subcutaneously in the dorsal thoracic midline of ten NOD/SCID mice (Weitong Lihua Experimental Animal Technology Co. Ltd). Tumor formation and growth were assessed every 3 days. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03228 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
Non-coding RNA
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05293 | ||
| Epigenetic Regulator | hsa-miR-338-5p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00569)
| In total 10 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE003068 | Click to Show/Hide the Full List | ||
| mod site | chr3:45083660-45083661:- | ||
| Sequence | TGCCTCGGCCTCCCAAAGTGCTGGGATTACAGATGTGAGCC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m5C_site_31923 | ||
| mod ID: M5CSITE003069 | Click to Show/Hide the Full List | ||
| mod site | chr3:45083667-45083668:- | ||
| Sequence | ATCCTCCTGCCTCGGCCTCCCAAAGTGCTGGGATTACAGAT | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m5C_site_31924 | ||
| mod ID: M5CSITE003070 | Click to Show/Hide the Full List | ||
| mod site | chr3:45083668-45083669:- | ||
| Sequence | AATCCTCCTGCCTCGGCCTCCCAAAGTGCTGGGATTACAGA | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m5C_site_31925 | ||
| mod ID: M5CSITE003071 | Click to Show/Hide the Full List | ||
| mod site | chr3:45083669-45083670:- | ||
| Sequence | CAATCCTCCTGCCTCGGCCTCCCAAAGTGCTGGGATTACAG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m5C_site_31926 | ||
| mod ID: M5CSITE003072 | Click to Show/Hide the Full List | ||
| mod site | chr3:45083671-45083672:- | ||
| Sequence | AGCAATCCTCCTGCCTCGGCCTCCCAAAGTGCTGGGATTAC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m5C_site_31927 | ||
| mod ID: M5CSITE003073 | Click to Show/Hide the Full List | ||
| mod site | chr3:45083672-45083673:- | ||
| Sequence | AAGCAATCCTCCTGCCTCGGCCTCCCAAAGTGCTGGGATTA | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m5C_site_31928 | ||
| mod ID: M5CSITE003074 | Click to Show/Hide the Full List | ||
| mod site | chr3:45083675-45083676:- | ||
| Sequence | CTCAAGCAATCCTCCTGCCTCGGCCTCCCAAAGTGCTGGGA | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m5C_site_31929 | ||
| mod ID: M5CSITE003075 | Click to Show/Hide the Full List | ||
| mod site | chr3:45083677-45083678:- | ||
| Sequence | GGCTCAAGCAATCCTCCTGCCTCGGCCTCCCAAAGTGCTGG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m5C_site_31930 | ||
| mod ID: M5CSITE003076 | Click to Show/Hide the Full List | ||
| mod site | chr3:45083678-45083679:- | ||
| Sequence | GGGCTCAAGCAATCCTCCTGCCTCGGCCTCCCAAAGTGCTG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m5C_site_31931 | ||
| mod ID: M5CSITE003077 | Click to Show/Hide the Full List | ||
| mod site | chr3:45110683-45110684:- | [4] | |
| Sequence | TCCTTCCTCAACTTCAACCTCTCCAACTGTGAGAGGAAGGA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000425231.2; ENST00000296129.6 | ||
| External Link | RMBase: m5C_site_31932 | ||
N6-methyladenosine (m6A)
| In total 72 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE060370 | Click to Show/Hide the Full List | ||
| mod site | chr3:45082517-45082518:- | [5] | |
| Sequence | ATAAACAAATGTACCAAAAAACAAGTATCAAGCTGTTTAAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | A549; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585901 | ||
| mod ID: M6ASITE060371 | Click to Show/Hide the Full List | ||
| mod site | chr3:45082533-45082534:- | [5] | |
| Sequence | ATTTCCTGTGCTTTAAATAAACAAATGTACCAAAAAACAAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | A549; MSC; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585902 | ||
| mod ID: M6ASITE060372 | Click to Show/Hide the Full List | ||
| mod site | chr3:45082631-45082632:- | [5] | |
| Sequence | GCCTTGGTAAAGGTTAGCAGACTGGTGTGTGTGGATCTGCA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | A549; HepG2; H1B; H1A; hESCs; fibroblasts; H1299; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585903 | ||
| mod ID: M6ASITE060373 | Click to Show/Hide the Full List | ||
| mod site | chr3:45082656-45082657:- | [5] | |
| Sequence | AACAAGCACAAGCCCACCGGACATGGCCTTGGTAAAGGTTA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | A549; HepG2; H1B; H1A; hESCs; fibroblasts; H1299; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585904 | ||
| mod ID: M6ASITE060374 | Click to Show/Hide the Full List | ||
| mod site | chr3:45082675-45082676:- | [5] | |
| Sequence | TCAAGGAAGAGTCAACTGGAACAAGCACAAGCCCACCGGAC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | A549; HepG2; H1A; H1B; hESCs; fibroblasts; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585905 | ||
| mod ID: M6ASITE060375 | Click to Show/Hide the Full List | ||
| mod site | chr3:45082738-45082739:- | [5] | |
| Sequence | GGAAGGCCAGCGCATGCAGGACTGGTCTCTAATGCTGTGGT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | A549; HepG2; H1A; H1B; hESCs; fibroblasts; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585906 | ||
| mod ID: M6ASITE060376 | Click to Show/Hide the Full List | ||
| mod site | chr3:45082833-45082834:- | [5] | |
| Sequence | ACATTTCAGGGTCACTGGAAACAGTGAAGTGCCATTTGTTG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | A549; HepG2; H1A; H1B; fibroblasts; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585907 | ||
| mod ID: M6ASITE060377 | Click to Show/Hide the Full List | ||
| mod site | chr3:45082853-45082854:- | [5] | |
| Sequence | CCAGGCCCCACTTACGTAAAACATTTCAGGGTCACTGGAAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | A549; HepG2; H1A; H1B; fibroblasts; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585908 | ||
| mod ID: M6ASITE060378 | Click to Show/Hide the Full List | ||
| mod site | chr3:45082896-45082897:- | [5] | |
| Sequence | GAAGGGGCCCAGCCTGAGGAACCCTGGCTCTTTTCTTTAAA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | A549; HepG2; H1A; H1B; fibroblasts; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585909 | ||
| mod ID: M6ASITE060379 | Click to Show/Hide the Full List | ||
| mod site | chr3:45082945-45082946:- | [5] | |
| Sequence | ACAAGTTGCTGGCTCCTGAGACCAGTATTTCCTGGAGCTGT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | A549; HepG2; H1A; H1B; hESCs; fibroblasts; iSLK; TIME; MSC; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585910 | ||
| mod ID: M6ASITE060380 | Click to Show/Hide the Full List | ||
| mod site | chr3:45082965-45082966:- | [5] | |
| Sequence | TGAGGGCTCAAAACTCCTGGACAAGTTGCTGGCTCCTGAGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | A549; HepG2; H1A; H1B; hESCs; fibroblasts; iSLK; TIME; MSC; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585911 | ||
| mod ID: M6ASITE060381 | Click to Show/Hide the Full List | ||
| mod site | chr3:45082973-45082974:- | [5] | |
| Sequence | CCTTCAACTGAGGGCTCAAAACTCCTGGACAAGTTGCTGGC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | A549; HepG2; H1A; H1B; hESCs; fibroblasts; iSLK; TIME; MSC; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585912 | ||
| mod ID: M6ASITE060382 | Click to Show/Hide the Full List | ||
| mod site | chr3:45083011-45083012:- | [5] | |
| Sequence | CATCACTGTTGCCTGCAAGGACACCACGTGGCCATTTTCCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | A549; HepG2; H1A; H1B; hESCs; fibroblasts; iSLK; TIME; MSC; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585913 | ||
| mod ID: M6ASITE060383 | Click to Show/Hide the Full List | ||
| mod site | chr3:45083032-45083033:- | [6] | |
| Sequence | GCTACTTGTCACCATCACCGACATCACTGTTGCCTGCAAGG | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | hESC-HEK293T; A549 | ||
| Seq Type List | MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585914 | ||
| mod ID: M6ASITE060384 | Click to Show/Hide the Full List | ||
| mod site | chr3:45083102-45083103:- | [5] | |
| Sequence | CCCAGCCCCTTCCAAGCAGGACTAGGTGCCCTGCATTCCAC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | A549; HepG2; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585915 | ||
| mod ID: M6ASITE060385 | Click to Show/Hide the Full List | ||
| mod site | chr3:45083584-45083585:- | [7] | |
| Sequence | TACCTGCCCAGTAACTGTGGACTTTTGCTTCCTCACCCCTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | fibroblasts; A549 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585916 | ||
| mod ID: M6ASITE060386 | Click to Show/Hide the Full List | ||
| mod site | chr3:45083904-45083905:- | [8] | |
| Sequence | CTTTTTTTTTTTAATGTGAGACAGGATCTCATTCTGTTGCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585917 | ||
| mod ID: M6ASITE060387 | Click to Show/Hide the Full List | ||
| mod site | chr3:45083973-45083974:- | [8] | |
| Sequence | TCAGGTGAGTAGACCATGAGACCAATGTGTGCTCACATTAC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585918 | ||
| mod ID: M6ASITE060388 | Click to Show/Hide the Full List | ||
| mod site | chr3:45083981-45083982:- | [8] | |
| Sequence | ATCTCTAATCAGGTGAGTAGACCATGAGACCAATGTGTGCT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585919 | ||
| mod ID: M6ASITE060389 | Click to Show/Hide the Full List | ||
| mod site | chr3:45084011-45084012:- | [8] | |
| Sequence | TATTGTAATGAAAAAGAAAGACTGGGATTAATCTCTAATCA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585920 | ||
| mod ID: M6ASITE060390 | Click to Show/Hide the Full List | ||
| mod site | chr3:45084227-45084228:- | [5] | |
| Sequence | ACTCTAACACTTAAAAAAAAACCCAGATCAGAAGATCTGGC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | A549; fibroblasts; iSLK; MSC; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585921 | ||
| mod ID: M6ASITE060391 | Click to Show/Hide the Full List | ||
| mod site | chr3:45084247-45084248:- | [5] | |
| Sequence | ATGTTTTGTTTAGCTTGCGGACTCTAACACTTAAAAAAAAA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | A549; fibroblasts; iSLK; MSC; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585922 | ||
| mod ID: M6ASITE060392 | Click to Show/Hide the Full List | ||
| mod site | chr3:45084356-45084357:- | [5] | |
| Sequence | AGGTGGGGATTAGGACCAGAACATCTTTGGGGTGCTGTTAT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | A549; HepG2; fibroblasts; iSLK; MSC; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585923 | ||
| mod ID: M6ASITE060393 | Click to Show/Hide the Full List | ||
| mod site | chr3:45084362-45084363:- | [5] | |
| Sequence | AGCCACAGGTGGGGATTAGGACCAGAACATCTTTGGGGTGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | A549; HepG2; fibroblasts; iSLK; MSC; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585924 | ||
| mod ID: M6ASITE060394 | Click to Show/Hide the Full List | ||
| mod site | chr3:45084389-45084390:- | [5] | |
| Sequence | AGAGCCTTTTGCCATGCAAGACAACATAGCCACAGGTGGGG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | A549; HepG2; fibroblasts; iSLK; MSC; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585925 | ||
| mod ID: M6ASITE060395 | Click to Show/Hide the Full List | ||
| mod site | chr3:45084963-45084964:- | [9] | |
| Sequence | CATATCTGCTTTGATAAGAGACTTCCTGATTCTCTAGGTCG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HepG2; A549; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585926 | ||
| mod ID: M6ASITE060396 | Click to Show/Hide the Full List | ||
| mod site | chr3:45085123-45085124:- | [10] | |
| Sequence | TTTCACCTGACTTAGTAATAACTCATACTAACTGGTTTGGA | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585927 | ||
| mod ID: M6ASITE060397 | Click to Show/Hide the Full List | ||
| mod site | chr3:45085289-45085290:- | [5] | |
| Sequence | AGAGGCTTGCCCTCTTCAGGACAACAGTTCCAATTCCAAGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | A549; HepG2; H1B; fibroblasts; H1299; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585928 | ||
| mod ID: M6ASITE060398 | Click to Show/Hide the Full List | ||
| mod site | chr3:45085311-45085312:- | [5] | |
| Sequence | CCCTGGATTCAGAGTGTTAAACAGAGGCTTGCCCTCTTCAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | A549; HepG2; H1B; fibroblasts; H1299; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585929 | ||
| mod ID: M6ASITE060399 | Click to Show/Hide the Full List | ||
| mod site | chr3:45085464-45085465:- | [5] | |
| Sequence | GGACTCATTCTAAGGGCAAGACATTGAAAATGATGAATTCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | A549; HepG2; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; H1299; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585930 | ||
| mod ID: M6ASITE060400 | Click to Show/Hide the Full List | ||
| mod site | chr3:45085482-45085483:- | [5] | |
| Sequence | AACTTCACATTGCTCAGTGGACTCATTCTAAGGGCAAGACA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | A549; HepG2; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; H1299; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585931 | ||
| mod ID: M6ASITE060401 | Click to Show/Hide the Full List | ||
| mod site | chr3:45085510-45085511:- | [5] | |
| Sequence | ACAGCAGGAGGTTTTCCTGGACACCGCCAACTTCACATTGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | A549; HepG2; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; H1299; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585932 | ||
| mod ID: M6ASITE060402 | Click to Show/Hide the Full List | ||
| mod site | chr3:45085530-45085531:- | [5] | |
| Sequence | AGAGGAATTATACAGAAGGAACAGCAGGAGGTTTTCCTGGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | A549; HepG2; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; H1299; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585933 | ||
| mod ID: M6ASITE060403 | Click to Show/Hide the Full List | ||
| mod site | chr3:45085586-45085587:- | [5] | |
| Sequence | TCATAAAGCAGGGCACTGAGACACCCGTCCGTGTTCCTAAC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | A549; HepG2; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; H1299; Huh7; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585934 | ||
| mod ID: M6ASITE060404 | Click to Show/Hide the Full List | ||
| mod site | chr3:45085668-45085669:- | [5] | |
| Sequence | CACAGACATTCCCTTACTGAACACTCAGGAGCCCATGGAGC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | A549; hESC-HEK293T; HepG2; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; H1299; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; MAZTER-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585935 | ||
| mod ID: M6ASITE060405 | Click to Show/Hide the Full List | ||
| mod site | chr3:45085683-45085684:- | [5] | |
| Sequence | TGTAAGCAGCAAGGACACAGACATTCCCTTACTGAACACTC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | A549; hESC-HEK293T; HepG2; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; H1299; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; MAZTER-seq; m6A-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585936 | ||
| mod ID: M6ASITE060406 | Click to Show/Hide the Full List | ||
| mod site | chr3:45085689-45085690:- | [5] | |
| Sequence | TGGGGATGTAAGCAGCAAGGACACAGACATTCCCTTACTGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | A549; HepG2; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; H1299; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585937 | ||
| mod ID: M6ASITE060407 | Click to Show/Hide the Full List | ||
| mod site | chr3:45085713-45085714:- | [6] | |
| Sequence | GTACACCTTCTCCCATCCCAACAATGGGGATGTAAGCAGCA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T; A549 | ||
| Seq Type List | MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585938 | ||
| mod ID: M6ASITE060408 | Click to Show/Hide the Full List | ||
| mod site | chr3:45085736-45085737:- | [5] | |
| Sequence | CCTCCTGAGTCTGAGAGTGAACCGTACACCTTCTCCCATCC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | A549; HepG2; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; H1299; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585939 | ||
| mod ID: M6ASITE060409 | Click to Show/Hide the Full List | ||
| mod site | chr3:45085869-45085870:- | [5] | |
| Sequence | CTTCCTGCAGCCAGAGGTGGACACCTACCGGCCGTTCCAGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | A549; HepG2; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; H1299; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585940 | ||
| mod ID: M6ASITE060410 | Click to Show/Hide the Full List | ||
| mod site | chr3:45085932-45085933:- | [5] | |
| Sequence | TGTGTATGCAGTCATCGAGGACACCATGGTATATGGGCATC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | A549; HepG2; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; H1299; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585941 | ||
| mod ID: M6ASITE060411 | Click to Show/Hide the Full List | ||
| mod site | chr3:45085965-45085966:- | [5] | |
| Sequence | GTTTCAGAAAGGGCGAAAGGACAATGACTCCCATGTGTATG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | A549; hESC-HEK293T; HepG2; H1B; H1A; hESCs; fibroblasts; LCLs; H1299; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | MeRIP-seq; MAZTER-seq; m6A-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585942 | ||
| mod ID: M6ASITE060412 | Click to Show/Hide the Full List | ||
| mod site | chr3:45086052-45086053:- | [5] | |
| Sequence | AAATAGGAAAAAGAAGACAAACAAGGGCCCCGCTGTGGGTA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | A549; hESCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585943 | ||
| mod ID: M6ASITE060413 | Click to Show/Hide the Full List | ||
| mod site | chr3:45086056-45086057:- | [7] | |
| Sequence | TTCCAAATAGGAAAAAGAAGACAAACAAGGGCCCCGCTGTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | fibroblasts; MSC; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585944 | ||
| mod ID: M6ASITE060414 | Click to Show/Hide the Full List | ||
| mod site | chr3:45091178-45091179:- | [8] | |
| Sequence | CGGTGACACTTACCCCAAGGACTGTGGGTAAGGGCCTTCAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585945 | ||
| mod ID: M6ASITE060415 | Click to Show/Hide the Full List | ||
| mod site | chr3:45091210-45091211:- | [9] | |
| Sequence | CACGAGCGGCAAGCAGCTAGACCTGCTCTTCTCGGTGACAC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HepG2; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585946 | ||
| mod ID: M6ASITE060416 | Click to Show/Hide the Full List | ||
| mod site | chr3:45091328-45091329:- | [8] | |
| Sequence | TGATCATCCAGGAGCAGCGGACCCGGGCTGAGGAGATCTTC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585947 | ||
| mod ID: M6ASITE060417 | Click to Show/Hide the Full List | ||
| mod site | chr3:45091364-45091365:- | [11] | |
| Sequence | GGAGCGGCGTGGTCTGCCAGACAGGGCGCGCATTCATGATC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | TIME; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585948 | ||
| mod ID: M6ASITE060418 | Click to Show/Hide the Full List | ||
| mod site | chr3:45091417-45091418:- | [8] | |
| Sequence | GAACATCAGCGTGCCCAGAGACCAGGTGGCCTGCCTGACTT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585949 | ||
| mod ID: M6ASITE060419 | Click to Show/Hide the Full List | ||
| mod site | chr3:45091435-45091436:- | [8] | |
| Sequence | CCTCACCTCTGTGTCCTGGAACATCAGCGTGCCCAGAGACC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585950 | ||
| mod ID: M6ASITE060420 | Click to Show/Hide the Full List | ||
| mod site | chr3:45091471-45091472:- | [8] | |
| Sequence | CCTGAGGACCCCCAACTGGGACCGGGGCCTGCCATCCCTCA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585951 | ||
| mod ID: M6ASITE060421 | Click to Show/Hide the Full List | ||
| mod site | chr3:45091484-45091485:- | [8] | |
| Sequence | AAAGCAAGGTCTACCTGAGGACCCCCAACTGGGACCGGGGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585952 | ||
| mod ID: M6ASITE060422 | Click to Show/Hide the Full List | ||
| mod site | chr3:45093368-45093369:- | [8] | |
| Sequence | GCAGATCCAGGTGAAGCAGAACATCTCGGTGACCCTTCGCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585953 | ||
| mod ID: M6ASITE060423 | Click to Show/Hide the Full List | ||
| mod site | chr3:45093428-45093429:- | [8] | |
| Sequence | CAGTGCCATACCCAGCCAGGACCTGTACTTCGGCTCCTTCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585954 | ||
| mod ID: M6ASITE060424 | Click to Show/Hide the Full List | ||
| mod site | chr3:45093539-45093540:- | [8] | |
| Sequence | GAAGCTGCTGGTGCCCAAGGACAGGCTCAGCCTGGTGCTGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585955 | ||
| mod ID: M6ASITE060425 | Click to Show/Hide the Full List | ||
| mod site | chr3:45093647-45093648:- | [8] | |
| Sequence | ACTTCTTCCAGGCTGCACAGACCACCGGTACTGCCAAAGGA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585956 | ||
| mod ID: M6ASITE060426 | Click to Show/Hide the Full List | ||
| mod site | chr3:45095352-45095353:- | [8] | |
| Sequence | TGTGGATGAATGTGGAAAAAACCATAAGTACGCCCCTGAAT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585957 | ||
| mod ID: M6ASITE060427 | Click to Show/Hide the Full List | ||
| mod site | chr3:45095410-45095411:- | [8] | |
| Sequence | ACCCTGACATCTGGCTCCAAACACAAAATCTCCTTCCTTTG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585958 | ||
| mod ID: M6ASITE060428 | Click to Show/Hide the Full List | ||
| mod site | chr3:45095448-45095449:- | [8] | |
| Sequence | TCGTGTGTCTAGAATCTCGGACCTGCAGTAGCAACCTCACC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585959 | ||
| mod ID: M6ASITE060429 | Click to Show/Hide the Full List | ||
| mod site | chr3:45095497-45095498:- | [8] | |
| Sequence | ATCGAGCCACGGCCCGTCAAACAGAGCCGCAAGTTTGTCCC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585960 | ||
| mod ID: M6ASITE060430 | Click to Show/Hide the Full List | ||
| mod site | chr3:45110573-45110574:- | [8] | |
| Sequence | GCCTGGGAACATGGCGGGGAACTTCAACCTCTCTCTGCAAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000425231.2; ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585961 | ||
| mod ID: M6ASITE060431 | Click to Show/Hide the Full List | ||
| mod site | chr3:45110585-45110586:- | [8] | |
| Sequence | GGAGGACAAGCAGCCTGGGAACATGGCGGGGAACTTCAACC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6; ENST00000425231.2 | ||
| External Link | RMBase: m6A_site_585962 | ||
| mod ID: M6ASITE060432 | Click to Show/Hide the Full List | ||
| mod site | chr3:45110600-45110601:- | [8] | |
| Sequence | CGAGGTGTTCAAGCTGGAGGACAAGCAGCCTGGGAACATGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6; ENST00000425231.2 | ||
| External Link | RMBase: m6A_site_585963 | ||
| mod ID: M6ASITE060433 | Click to Show/Hide the Full List | ||
| mod site | chr3:45112099-45112100:- | [8] | |
| Sequence | CTCCGGCTTCAGCATTGCAAACCGCTCATCTATAAAACGTG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6; ENST00000425231.2 | ||
| External Link | RMBase: m6A_site_585964 | ||
| mod ID: M6ASITE060434 | Click to Show/Hide the Full List | ||
| mod site | chr3:45112211-45112212:- | [8] | |
| Sequence | CCACCGTGGTCAGGATCGGAACCTTCTGCAGCAATGGCACT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000425231.2; ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585965 | ||
| mod ID: M6ASITE060435 | Click to Show/Hide the Full List | ||
| mod site | chr3:45112367-45112368:- | [8] | |
| Sequence | TGTTGCCTACCCTCAACAGAACTTTCATCTGGGATGTCAAA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6; ENST00000490471.1; ENST00000425231.2 | ||
| External Link | RMBase: m6A_site_585966 | ||
| mod ID: M6ASITE060436 | Click to Show/Hide the Full List | ||
| mod site | chr3:45118514-45118515:- | [8] | |
| Sequence | TACATCGTCATTTCTAAAAGACATATAACCATGTTGTCCAT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000425231.2; ENST00000296129.6; ENST00000490471.1 | ||
| External Link | RMBase: m6A_site_585967 | ||
| mod ID: M6ASITE060437 | Click to Show/Hide the Full List | ||
| mod site | chr3:45118541-45118542:- | [8] | |
| Sequence | ACCCCGACTCTGCTGGCAAAACCCTGTTACATCGTCATTTC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000490471.1; ENST00000425231.2; ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585968 | ||
| mod ID: M6ASITE060438 | Click to Show/Hide the Full List | ||
| mod site | chr3:45118561-45118562:- | [8] | |
| Sequence | CAGTTCTCATAAAGCTGGGGACCCCGACTCTGCTGGCAAAA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6; ENST00000490471.1; ENST00000425231.2 | ||
| External Link | RMBase: m6A_site_585969 | ||
| mod ID: M6ASITE060439 | Click to Show/Hide the Full List | ||
| mod site | chr3:45139410-45139411:- | [8] | |
| Sequence | TTGGAGACAGCCTCATTCAGACAAGCTCTGGAGTTGGTCAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296129.6; ENST00000490471.1; rmsk_949942; ENST00000425231.2 | ||
| External Link | RMBase: m6A_site_585970 | ||
| mod ID: M6ASITE060440 | Click to Show/Hide the Full List | ||
| mod site | chr3:45139548-45139549:- | [8] | |
| Sequence | TACATTGCCCGTGTCTTCAGACTGCACATCCCAAAGGGCTT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000490471.1; ENST00000425231.2; ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585971 | ||
| mod ID: M6ASITE060441 | Click to Show/Hide the Full List | ||
| mod site | chr3:45146273-45146274:- | [8] | |
| Sequence | CGGAGTCATGGCCGGCCTGAACTGCGGGGTCTCTATCGCAC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000425231.2; ENST00000296129.6 | ||
| External Link | RMBase: m6A_site_585972 | ||
References