m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00520)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
ENO1
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
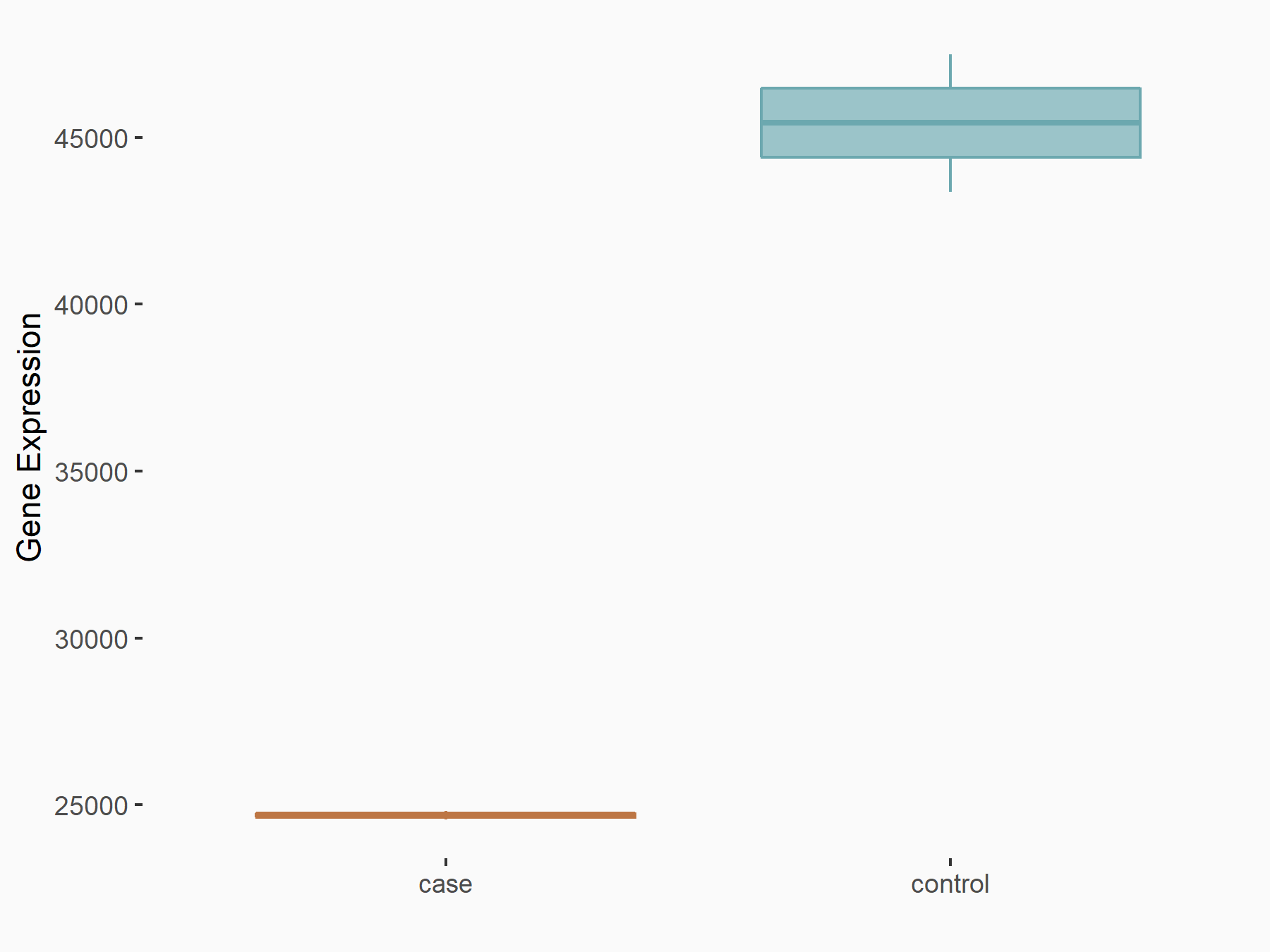 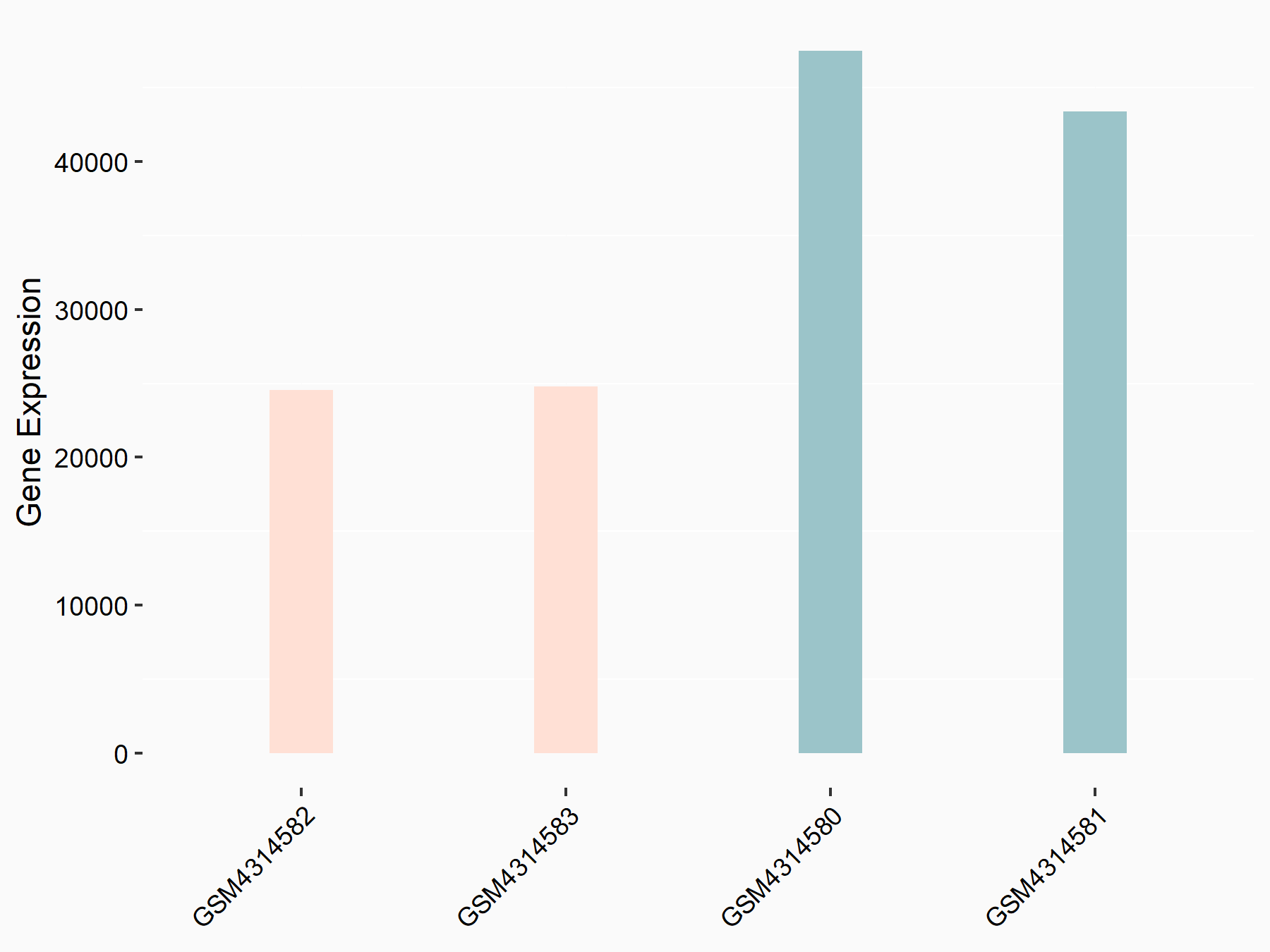 |
logFC: -8.80E-01 p-value: 3.03E-81 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Alpha-enolase (ENO1) positively correlated with METTL3 and global m6A levels, and negatively correlated with ALKBH5 in human Lung adenocarcinoma(LUAD). In addition, m6A-dependent elevation of ENO1 was associated with LUAD progression. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Lung adenocarcinoma | ICD-11: 2C25.0 | ||
| Pathway Response | Carbon metabolism | hsa01200 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glycolysis | |||
| In-vitro Model | PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| NCI-H441 | Lung papillary adenocarcinoma | Homo sapiens | CVCL_1561 | |
| NCI-H292 | Lung mucoepidermoid carcinoma | Homo sapiens | CVCL_0455 | |
| NCI-H2030 | Lung adenocarcinoma | Homo sapiens | CVCL_1517 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| 3LL | Malignant tumors of the mouse pulmonary system | Mus musculus | CVCL_5653 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| Calu-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0608 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| A-427 | Lung adenocarcinoma | Homo sapiens | CVCL_1055 | |
| 16HBE14o- | Normal | Homo sapiens | CVCL_0112 | |
| In-vivo Model | KP and Mettl3-/- mice were bred to generate KPM-/- mice. Afterwards, the KP and KPM-/- mice were intranasally infected under anesthesia with adeno-associated virus type 5 (AAV5) expressing Cre to initiate lung tumorigenesis along with ALKBH5-expressing AAV5 or Empty AAV5 to generate KPE, KPA, KPEM-/- and KPAM-/- spontaneous LUAD mouse models. For generation of LLC-based intra-pulmonary tumor mouse models, 1 × 107 LLC cells were injected into C57BL/6 mice via the tail vein.For cell-derived xenograft (CDX) mouse models, 1.0 × 107 H1299 or 1.5 × 107 H1975 cells were subcutaneously injected into 4-6-week-old athymic nude mice. The tumors were monitored at indicated time points and isolated for further analysis after sacrifice. | |||
RNA demethylase ALKBH5 (ALKBH5) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by ALKBH5 | ||
| Cell Line | human pluripotent stem cells | Homo sapiens |
|
Treatment: hILO ALKBH5knockout cells
Control: hILO wild type cells
|
GSE163945 | |
| Regulation |
  |
logFC: -9.52E-01 p-value: 3.98E-04 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Alpha-enolase (ENO1) positively correlated with METTL3 and global m6A levels, and negatively correlated with ALKBH5 in human Lung adenocarcinoma(LUAD). In addition, m6A-dependent elevation of ENO1 was associated with LUAD progression. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Lung adenocarcinoma | ICD-11: 2C25.0 | ||
| Pathway Response | Carbon metabolism | hsa01200 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glycolysis | |||
| In-vitro Model | PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| NCI-H441 | Lung papillary adenocarcinoma | Homo sapiens | CVCL_1561 | |
| NCI-H292 | Lung mucoepidermoid carcinoma | Homo sapiens | CVCL_0455 | |
| NCI-H2030 | Lung adenocarcinoma | Homo sapiens | CVCL_1517 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| 3LL | Malignant tumors of the mouse pulmonary system | Mus musculus | CVCL_5653 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| Calu-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0608 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| A-427 | Lung adenocarcinoma | Homo sapiens | CVCL_1055 | |
| 16HBE14o- | Normal | Homo sapiens | CVCL_0112 | |
| In-vivo Model | KP and Mettl3-/- mice were bred to generate KPM-/- mice. Afterwards, the KP and KPM-/- mice were intranasally infected under anesthesia with adeno-associated virus type 5 (AAV5) expressing Cre to initiate lung tumorigenesis along with ALKBH5-expressing AAV5 or Empty AAV5 to generate KPE, KPA, KPEM-/- and KPAM-/- spontaneous LUAD mouse models. For generation of LLC-based intra-pulmonary tumor mouse models, 1 × 107 LLC cells were injected into C57BL/6 mice via the tail vein.For cell-derived xenograft (CDX) mouse models, 1.0 × 107 H1299 or 1.5 × 107 H1975 cells were subcutaneously injected into 4-6-week-old athymic nude mice. The tumors were monitored at indicated time points and isolated for further analysis after sacrifice. | |||
Wilms tumor 1-associating protein (WTAP) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by WTAP | ||
| Cell Line | Human umbilical vein endothelial cells | Homo sapiens |
|
Treatment: siWTAP HUVECs
Control: siControl HUVECs
|
GSE167067 | |
| Regulation |
  |
logFC: -6.99E-01 p-value: 1.98E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | The stabilization of WTAP further promotes RNA m6A methylation of Alpha-enolase (ENO1), impacting the glycolytic activity of BC cells. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glycolysis | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | A total of 1 × 106 luciferase-labeled BC cells transfected with shWTAP or shNC were injected subcutaneously with or without C5aR1 neutrophils (tumor cells:neutrophils, 10:1). | |||
Lung cancer [ICD-11: 2C25]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Alpha-enolase (ENO1) positively correlated with METTL3 and global m6A levels, and negatively correlated with ALKBH5 in human Lung adenocarcinoma(LUAD). In addition, m6A-dependent elevation of ENO1 was associated with LUAD progression. | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Carbon metabolism | hsa01200 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glycolysis | |||
| In-vitro Model | PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| NCI-H441 | Lung papillary adenocarcinoma | Homo sapiens | CVCL_1561 | |
| NCI-H292 | Lung mucoepidermoid carcinoma | Homo sapiens | CVCL_0455 | |
| NCI-H2030 | Lung adenocarcinoma | Homo sapiens | CVCL_1517 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| 3LL | Malignant tumors of the mouse pulmonary system | Mus musculus | CVCL_5653 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| Calu-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0608 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| A-427 | Lung adenocarcinoma | Homo sapiens | CVCL_1055 | |
| 16HBE14o- | Normal | Homo sapiens | CVCL_0112 | |
| In-vivo Model | KP and Mettl3-/- mice were bred to generate KPM-/- mice. Afterwards, the KP and KPM-/- mice were intranasally infected under anesthesia with adeno-associated virus type 5 (AAV5) expressing Cre to initiate lung tumorigenesis along with ALKBH5-expressing AAV5 or Empty AAV5 to generate KPE, KPA, KPEM-/- and KPAM-/- spontaneous LUAD mouse models. For generation of LLC-based intra-pulmonary tumor mouse models, 1 × 107 LLC cells were injected into C57BL/6 mice via the tail vein.For cell-derived xenograft (CDX) mouse models, 1.0 × 107 H1299 or 1.5 × 107 H1975 cells were subcutaneously injected into 4-6-week-old athymic nude mice. The tumors were monitored at indicated time points and isolated for further analysis after sacrifice. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Alpha-enolase (ENO1) positively correlated with METTL3 and global m6A levels, and negatively correlated with ALKBH5 in human Lung adenocarcinoma(LUAD). In addition, m6A-dependent elevation of ENO1 was associated with LUAD progression. | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Carbon metabolism | hsa01200 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glycolysis | |||
| In-vitro Model | PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| NCI-H441 | Lung papillary adenocarcinoma | Homo sapiens | CVCL_1561 | |
| NCI-H292 | Lung mucoepidermoid carcinoma | Homo sapiens | CVCL_0455 | |
| NCI-H2030 | Lung adenocarcinoma | Homo sapiens | CVCL_1517 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| 3LL | Malignant tumors of the mouse pulmonary system | Mus musculus | CVCL_5653 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| Calu-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0608 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| A-427 | Lung adenocarcinoma | Homo sapiens | CVCL_1055 | |
| 16HBE14o- | Normal | Homo sapiens | CVCL_0112 | |
| In-vivo Model | KP and Mettl3-/- mice were bred to generate KPM-/- mice. Afterwards, the KP and KPM-/- mice were intranasally infected under anesthesia with adeno-associated virus type 5 (AAV5) expressing Cre to initiate lung tumorigenesis along with ALKBH5-expressing AAV5 or Empty AAV5 to generate KPE, KPA, KPEM-/- and KPAM-/- spontaneous LUAD mouse models. For generation of LLC-based intra-pulmonary tumor mouse models, 1 × 107 LLC cells were injected into C57BL/6 mice via the tail vein.For cell-derived xenograft (CDX) mouse models, 1.0 × 107 H1299 or 1.5 × 107 H1975 cells were subcutaneously injected into 4-6-week-old athymic nude mice. The tumors were monitored at indicated time points and isolated for further analysis after sacrifice. | |||
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | The stabilization of WTAP further promotes RNA m6A methylation of Alpha-enolase (ENO1), impacting the glycolytic activity of BC cells. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Wilms tumor 1-associating protein (WTAP) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glycolysis | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | A total of 1 × 106 luciferase-labeled BC cells transfected with shWTAP or shNC were injected subcutaneously with or without C5aR1 neutrophils (tumor cells:neutrophils, 10:1). | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00520)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE010498 | Click to Show/Hide the Full List | ||
| mod site | chr1:8876911-8876912:- | [5] | |
| Sequence | GCTGAGGTTGCAGTGAGCCAAAATGGCGCCATTGCACTCCA | ||
| Transcript ID List | ENST00000646906.1; ENST00000643438.1; ENST00000414948.1; ENST00000234590.10; ENST00000497492.1; rmsk_15815; ENST00000489867.2; ENST00000645609.1; ENST00000646660.1; ENST00000486051.5; ENST00000492343.2; ENST00000647408.1; ENST00000646156.1; ENST00000646539.1; ENST00000646680.1; ENST00000646370.2 | ||
| External Link | RMBase: RNA-editing_site_860 | ||
5-methylcytidine (m5C)
| In total 14 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE005072 | Click to Show/Hide the Full List | ||
| mod site | chr1:8861308-8861309:- | [6] | |
| Sequence | GTTGGCTACACAGACCCCTCCCCTCGTGTCAGCTCAGGCAG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000234590.10; ENST00000464920.2; ENST00000647408.1; ENST00000646370.2; ENST00000646539.1 | ||
| External Link | RMBase: m5C_site_699 | ||
| mod ID: M5CSITE005081 | Click to Show/Hide the Full List | ||
| mod site | chr1:8862948-8862949:- | ||
| Sequence | CAAATGGCCTTTCTTTTCTCCTAGATCAAGACTGGTGCCCC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000647408.1; ENST00000646539.1; ENST00000646370.2; ENST00000234590.10; ENST00000464920.2 | ||
| External Link | RMBase: m5C_site_700 | ||
| mod ID: M5CSITE005092 | Click to Show/Hide the Full List | ||
| mod site | chr1:8864097-8864098:- | [6] | |
| Sequence | ATGAAGCGTGTCCCTCCTTTCCTTAGTGGTGTCTATCGAAG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000646370.2; ENST00000645609.1; ENST00000646539.1; ENST00000234590.10; ENST00000647408.1; ENST00000464920.2 | ||
| External Link | RMBase: m5C_site_701 | ||
| mod ID: M5CSITE005094 | Click to Show/Hide the Full List | ||
| mod site | chr1:8864102-8864103:- | [6] | |
| Sequence | CACTGATGAAGCGTGTCCCTCCTTTCCTTAGTGGTGTCTAT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000234590.10; ENST00000646370.2; ENST00000464920.2; ENST00000645609.1; ENST00000646539.1; ENST00000647408.1 | ||
| External Link | RMBase: m5C_site_702 | ||
| mod ID: M5CSITE005095 | Click to Show/Hide the Full List | ||
| mod site | chr1:8866508-8866509:- | [6] | |
| Sequence | CCATGCTTCTCTGCTCTGCTCTCCCCAGGCGTTCAATGTCA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000645609.1; ENST00000646370.2; ENST00000646680.1; ENST00000234590.10; MIMAT0027358; ENST00000497492.1; ENST00000647408.1; ENST00000646539.1; ENST00000618792.1; ENST00000645600.1; ENST00000646660.1; ENST00000464920.2 | ||
| External Link | RMBase: m5C_site_703 | ||
| mod ID: M5CSITE005096 | Click to Show/Hide the Full List | ||
| mod site | chr1:8877314-8877315:- | ||
| Sequence | TCCTCAGGATCACCTGAGGTCGGGAGTTAGAGACCAGCCTG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000492343.2; ENST00000646660.1; ENST00000497492.1; ENST00000643438.1; ENST00000646539.1; ENST00000234590.10; ENST00000647408.1; ENST00000486051.5; ENST00000646156.1; ENST00000646680.1; ENST00000489867.2; ENST00000646906.1; ENST00000645609.1; ENST00000414948.1; rmsk_15817; ENST00000646370.2 | ||
| External Link | RMBase: m5C_site_704 | ||
| mod ID: M5CSITE005097 | Click to Show/Hide the Full List | ||
| mod site | chr1:8877321-8877322:- | ||
| Sequence | GTGGGCCTCCTCAGGATCACCTGAGGTCGGGAGTTAGAGAC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000486051.5; ENST00000234590.10; ENST00000647408.1; ENST00000492343.2; ENST00000646906.1; ENST00000646156.1; ENST00000643438.1; ENST00000497492.1; ENST00000489867.2; ENST00000414948.1; ENST00000646660.1; ENST00000646539.1; ENST00000646680.1; rmsk_15817; ENST00000646370.2; ENST00000645609.1 | ||
| External Link | RMBase: m5C_site_705 | ||
| mod ID: M5CSITE005099 | Click to Show/Hide the Full List | ||
| mod site | chr1:8877324-8877325:- | ||
| Sequence | GAGGTGGGCCTCCTCAGGATCACCTGAGGTCGGGAGTTAGA | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000643438.1; rmsk_15817; ENST00000489867.2; ENST00000414948.1; ENST00000497492.1; ENST00000646539.1; ENST00000646906.1; ENST00000646660.1; ENST00000646680.1; ENST00000486051.5; ENST00000234590.10; ENST00000647408.1; ENST00000646370.2; ENST00000645609.1; ENST00000492343.2; ENST00000646156.1 | ||
| External Link | RMBase: m5C_site_706 | ||
| mod ID: M5CSITE005102 | Click to Show/Hide the Full List | ||
| mod site | chr1:8877586-8877587:- | ||
| Sequence | CTGGGATTACAGGCGTGAGCCACCGCGCTAGGCCAAGAAGG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000414948.1; ENST00000489867.2; ENST00000234590.10; ENST00000645609.1; ENST00000646906.1; ENST00000646539.1; ENST00000646370.2; ENST00000646156.1; ENST00000646660.1; ENST00000647408.1; ENST00000497492.1; ENST00000492343.2; ENST00000643438.1; ENST00000646680.1; ENST00000486051.5 | ||
| External Link | RMBase: m5C_site_707 | ||
| mod ID: M5CSITE005111 | Click to Show/Hide the Full List | ||
| mod site | chr1:8877587-8877588:- | ||
| Sequence | GCTGGGATTACAGGCGTGAGCCACCGCGCTAGGCCAAGAAG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000234590.10; ENST00000492343.2; ENST00000646539.1; ENST00000646370.2; ENST00000486051.5; ENST00000646680.1; ENST00000414948.1; ENST00000645609.1; ENST00000497492.1; ENST00000643438.1; ENST00000646906.1; ENST00000489867.2; ENST00000647408.1; ENST00000646156.1; ENST00000646660.1 | ||
| External Link | RMBase: m5C_site_708 | ||
| mod ID: M5CSITE005113 | Click to Show/Hide the Full List | ||
| mod site | chr1:8877593-8877594:- | ||
| Sequence | CAAAGTGCTGGGATTACAGGCGTGAGCCACCGCGCTAGGCC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000234590.10; ENST00000492343.2; ENST00000646906.1; ENST00000646680.1; ENST00000414948.1; ENST00000497492.1; ENST00000646660.1; ENST00000647408.1; ENST00000646370.2; ENST00000489867.2; ENST00000645609.1; ENST00000646539.1; ENST00000643438.1; ENST00000646156.1; ENST00000486051.5 | ||
| External Link | RMBase: m5C_site_709 | ||
| mod ID: M5CSITE005114 | Click to Show/Hide the Full List | ||
| mod site | chr1:8877597-8877598:- | ||
| Sequence | CTCCCAAAGTGCTGGGATTACAGGCGTGAGCCACCGCGCTA | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000489867.2; ENST00000234590.10; ENST00000497492.1; ENST00000646680.1; ENST00000492343.2; ENST00000646906.1; ENST00000646660.1; ENST00000643438.1; ENST00000646370.2; ENST00000646539.1; ENST00000647408.1; ENST00000486051.5; ENST00000414948.1; ENST00000646156.1; ENST00000645609.1 | ||
| External Link | RMBase: m5C_site_710 | ||
| mod ID: M5CSITE005115 | Click to Show/Hide the Full List | ||
| mod site | chr1:8877639-8877640:- | ||
| Sequence | TGGTCTTGAGCTCCCGACCTCAGGTGATCCGCCCGCCTCGG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000492343.2; ENST00000645609.1; ENST00000234590.10; ENST00000646156.1; ENST00000497492.1; ENST00000646539.1; ENST00000646680.1; ENST00000646906.1; ENST00000646660.1; ENST00000489867.2; ENST00000643438.1; ENST00000647408.1; ENST00000486051.5; ENST00000414948.1; ENST00000646370.2 | ||
| External Link | RMBase: m5C_site_711 | ||
| mod ID: M5CSITE005116 | Click to Show/Hide the Full List | ||
| mod site | chr1:8877641-8877642:- | ||
| Sequence | GCTGGTCTTGAGCTCCCGACCTCAGGTGATCCGCCCGCCTC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000234590.10; ENST00000489867.2; ENST00000492343.2; ENST00000486051.5; ENST00000647408.1; ENST00000643438.1; ENST00000646906.1; ENST00000497492.1; ENST00000646539.1; ENST00000646680.1; ENST00000646660.1; ENST00000645609.1; ENST00000414948.1; ENST00000646156.1; ENST00000646370.2 | ||
| External Link | RMBase: m5C_site_712 | ||
N6-methyladenosine (m6A)
| In total 47 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE053300 | Click to Show/Hide the Full List | ||
| mod site | chr1:8861060-8861061:- | [7] | |
| Sequence | GTGTCATCTCCGGGGTGGCCACAGGCTAGATCCCCGGTGGT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000647408.1; ENST00000646539.1; ENST00000464920.2; ENST00000234590.10; ENST00000646370.2 | ||
| External Link | RMBase: m6A_site_5351 | ||
| mod ID: M6ASITE053301 | Click to Show/Hide the Full List | ||
| mod site | chr1:8861206-8861207:- | [8] | |
| Sequence | GTTCGTACCGCTTCCTTAGAACTTCTACAGAAGCCAAGCTC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; AML | ||
| Seq Type List | m6A-seq; DART-seq; miCLIP | ||
| Transcript ID List | ENST00000234590.10; ENST00000646539.1; ENST00000646370.2; ENST00000647408.1; ENST00000464920.2 | ||
| External Link | RMBase: m6A_site_5352 | ||
| mod ID: M6ASITE053302 | Click to Show/Hide the Full List | ||
| mod site | chr1:8861270-8861271:- | [7] | |
| Sequence | CAGCTCGAGGCCCCCGACCAACACTTGCAGGGGTCCCTGCT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000464920.2; ENST00000234590.10; ENST00000646539.1; ENST00000646370.2; ENST00000647408.1 | ||
| External Link | RMBase: m6A_site_5353 | ||
| mod ID: M6ASITE053303 | Click to Show/Hide the Full List | ||
| mod site | chr1:8861315-8861316:- | [8] | |
| Sequence | GTCACCTGTTGGCTACACAGACCCCTCCCCTCGTGTCAGCT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; Huh7; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000647408.1; ENST00000464920.2; ENST00000646370.2; ENST00000234590.10; ENST00000646539.1 | ||
| External Link | RMBase: m6A_site_5354 | ||
| mod ID: M6ASITE053344 | Click to Show/Hide the Full List | ||
| mod site | chr1:8861321-8861322:- | [7] | |
| Sequence | CCTTCGGTCACCTGTTGGCTACACAGACCCCTCCCCTCGTG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000464920.2; ENST00000646539.1; ENST00000647408.1; ENST00000646370.2; ENST00000234590.10 | ||
| External Link | RMBase: m6A_site_5355 | ||
| mod ID: M6ASITE053445 | Click to Show/Hide the Full List | ||
| mod site | chr1:8861375-8861376:- | [8] | |
| Sequence | TGCCGGCAGGAACTTCAGAAACCCCTTGGCCAAGTAAGCTG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; Huh7; endometrial; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000234590.10; ENST00000646370.2; ENST00000647408.1; ENST00000646539.1; ENST00000464920.2 | ||
| External Link | RMBase: m6A_site_5356 | ||
| mod ID: M6ASITE053459 | Click to Show/Hide the Full List | ||
| mod site | chr1:8861384-8861385:- | [8] | |
| Sequence | GGCTAAGTTTGCCGGCAGGAACTTCAGAAACCCCTTGGCCA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; Huh7; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000646539.1; ENST00000464920.2; ENST00000646370.2; ENST00000647408.1; ENST00000234590.10 | ||
| External Link | RMBase: m6A_site_5357 | ||
| mod ID: M6ASITE053481 | Click to Show/Hide the Full List | ||
| mod site | chr1:8862901-8862902:- | [7] | |
| Sequence | ATCTGAGCGCTTGGCCAAGTACAACCAGCTCCTCAGGTAAG | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000646539.1; ENST00000646370.2; ENST00000647408.1; ENST00000234590.10; ENST00000464920.2 | ||
| External Link | RMBase: m6A_site_5358 | ||
| mod ID: M6ASITE053511 | Click to Show/Hide the Full List | ||
| mod site | chr1:8862938-8862939:- | [8] | |
| Sequence | TTCTTTTCTCCTAGATCAAGACTGGTGCCCCTTGCCGATCT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; Huh7; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000646370.2; ENST00000647408.1; ENST00000234590.10; ENST00000464920.2; ENST00000646539.1 | ||
| External Link | RMBase: m6A_site_5359 | ||
| mod ID: M6ASITE053512 | Click to Show/Hide the Full List | ||
| mod site | chr1:8863175-8863176:- | [8] | |
| Sequence | ACTTCCTAGGAAGACCCAAAACCAGTGAGGCTCCATGTCTG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000464920.2; ENST00000646370.2; ENST00000647408.1; ENST00000646539.1; ENST00000645609.1; ENST00000234590.10 | ||
| External Link | RMBase: m6A_site_5360 | ||
| mod ID: M6ASITE053513 | Click to Show/Hide the Full List | ||
| mod site | chr1:8863182-8863183:- | [8] | |
| Sequence | GACCTCAACTTCCTAGGAAGACCCAAAACCAGTGAGGCTCC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000464920.2; ENST00000647408.1; ENST00000646539.1; ENST00000646370.2; ENST00000645609.1; ENST00000234590.10 | ||
| External Link | RMBase: m6A_site_5361 | ||
| mod ID: M6ASITE053529 | Click to Show/Hide the Full List | ||
| mod site | chr1:8863284-8863285:- | [8] | |
| Sequence | TGTCTCATCGTTCGGGGGAGACTGAAGATACCTTCATCGCT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000647408.1; ENST00000646370.2; ENST00000646539.1; ENST00000234590.10; ENST00000464920.2; ENST00000645609.1 | ||
| External Link | RMBase: m6A_site_5362 | ||
| mod ID: M6ASITE053540 | Click to Show/Hide the Full List | ||
| mod site | chr1:8865292-8865293:- | [8] | |
| Sequence | GTACAAGTCCTTCATCAAGGACTACCCAGGTGAGTGTTCCC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000464920.2; ENST00000646370.2; ENST00000647408.1; ENST00000646539.1; ENST00000645609.1; ENST00000234590.10 | ||
| External Link | RMBase: m6A_site_5363 | ||
| mod ID: M6ASITE053541 | Click to Show/Hide the Full List | ||
| mod site | chr1:8865310-8865311:- | [7] | |
| Sequence | TGACCAGCTGGCTGACCTGTACAAGTCCTTCATCAAGGACT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000464920.2; ENST00000646539.1; ENST00000234590.10; ENST00000647408.1; ENST00000646370.2; ENST00000645609.1 | ||
| External Link | RMBase: m6A_site_5364 | ||
| mod ID: M6ASITE053542 | Click to Show/Hide the Full List | ||
| mod site | chr1:8865328-8865329:- | [9] | |
| Sequence | CAGCAGGTACATCTCGCCTGACCAGCTGGCTGACCTGTACA | ||
| Motif Score | 2.839113095 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000464920.2; ENST00000647408.1; ENST00000646370.2; ENST00000646539.1; ENST00000234590.10; ENST00000645609.1 | ||
| External Link | RMBase: m6A_site_5365 | ||
| mod ID: M6ASITE053543 | Click to Show/Hide the Full List | ||
| mod site | chr1:8865340-8865341:- | [7] | |
| Sequence | TCCCGATGACCCCAGCAGGTACATCTCGCCTGACCAGCTGG | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000646370.2; ENST00000646539.1; ENST00000464920.2; ENST00000645609.1; ENST00000234590.10; ENST00000647408.1 | ||
| External Link | RMBase: m6A_site_5366 | ||
| mod ID: M6ASITE053544 | Click to Show/Hide the Full List | ||
| mod site | chr1:8865370-8865371:- | [8] | |
| Sequence | GTCTGGGAAGTATGACCTGGACTTCAAGTCTCCCGATGACC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000646539.1; ENST00000234590.10; ENST00000645609.1; ENST00000647408.1; ENST00000464920.2; ENST00000646370.2 | ||
| External Link | RMBase: m6A_site_5367 | ||
| mod ID: M6ASITE053545 | Click to Show/Hide the Full List | ||
| mod site | chr1:8865442-8865443:- | [7] | |
| Sequence | TGCTATTGGGAAAGCTGGCTACACTGATAAGGTGGTCATCG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000464920.2; ENST00000645609.1; ENST00000647408.1; ENST00000234590.10; ENST00000646539.1; ENST00000646370.2 | ||
| External Link | RMBase: m6A_site_5368 | ||
| mod ID: M6ASITE053546 | Click to Show/Hide the Full List | ||
| mod site | chr1:8865464-8865465:- | [8] | |
| Sequence | CAGGCCTGGAGCTGCTGAAGACTGCTATTGGGAAAGCTGGC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000646370.2; ENST00000645609.1; ENST00000646539.1; ENST00000464920.2; ENST00000234590.10; ENST00000647408.1 | ||
| External Link | RMBase: m6A_site_5369 | ||
| mod ID: M6ASITE053547 | Click to Show/Hide the Full List | ||
| mod site | chr1:8866298-8866299:- | [7] | |
| Sequence | TGAAGGCGGGTTTGCTCCCAACATCCTGGAGAATAAAGAAG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000647408.1; ENST00000645609.1; ENST00000645600.1; ENST00000646539.1; ENST00000234590.10; ENST00000646370.2; ENST00000464920.2 | ||
| External Link | RMBase: m6A_site_5370 | ||
| mod ID: M6ASITE053582 | Click to Show/Hide the Full List | ||
| mod site | chr1:8866376-8866377:- | [7] | |
| Sequence | CATTGGAGCAGAGGTTTACCACAACCTGAAGAATGTCATCA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000646370.2; ENST00000645609.1; ENST00000464920.2; ENST00000645600.1; ENST00000647408.1; ENST00000646539.1; ENST00000497492.1; ENST00000234590.10 | ||
| External Link | RMBase: m6A_site_5371 | ||
| mod ID: M6ASITE053621 | Click to Show/Hide the Full List | ||
| mod site | chr1:8866415-8866416:- | [8] | |
| Sequence | CCTCCCAGTCGGTGCAGCAAACTTCAGGGAAGCCATGCGCA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000645609.1; ENST00000464920.2; ENST00000234590.10; ENST00000497492.1; ENST00000645600.1; ENST00000646370.2; ENST00000647408.1; ENST00000646539.1 | ||
| External Link | RMBase: m6A_site_5372 | ||
| mod ID: M6ASITE053622 | Click to Show/Hide the Full List | ||
| mod site | chr1:8866463-8866464:- | [10] | |
| Sequence | TGGCGGTTCTCATGCTGGCAACAAGCTGGCCATGCAGGAGT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | brain; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000647408.1; ENST00000645609.1; ENST00000646370.2; ENST00000464920.2; ENST00000234590.10; ENST00000645600.1; ENST00000646660.1; ENST00000497492.1; ENST00000646539.1 | ||
| External Link | RMBase: m6A_site_5373 | ||
| mod ID: M6ASITE053659 | Click to Show/Hide the Full List | ||
| mod site | chr1:8867162-8867163:- | [7] | |
| Sequence | GGGGGTCCCCCTGTACCGCCACATCGCTGACTTGGCTGGCA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000647408.1; ENST00000645600.1; ENST00000645609.1; ENST00000464920.2; ENST00000646680.1; ENST00000646539.1; ENST00000497492.1; ENST00000646660.1; ENST00000234590.10; ENST00000646370.2 | ||
| External Link | RMBase: m6A_site_5374 | ||
| mod ID: M6ASITE053724 | Click to Show/Hide the Full List | ||
| mod site | chr1:8867999-8868000:- | [8] | |
| Sequence | TGATGATCGAGATGGATGGAACAGAAAATAAATGTGAGTGG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000646680.1; ENST00000646370.2; ENST00000234590.10; ENST00000645600.1; ENST00000645609.1; ENST00000646906.1; ENST00000646539.1; ENST00000647408.1; ENST00000497492.1; ENST00000464920.2; ENST00000646660.1 | ||
| External Link | RMBase: m6A_site_5375 | ||
| mod ID: M6ASITE053725 | Click to Show/Hide the Full List | ||
| mod site | chr1:8868021-8868022:- | [8] | |
| Sequence | GAACAAGAGAAGATTGACAAACTGATGATCGAGATGGATGG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; AML | ||
| Seq Type List | m6A-seq; miCLIP | ||
| Transcript ID List | ENST00000464920.2; ENST00000646906.1; ENST00000645609.1; ENST00000646539.1; ENST00000646660.1; ENST00000645600.1; ENST00000646680.1; ENST00000234590.10; ENST00000646370.2; ENST00000497492.1; ENST00000647408.1 | ||
| External Link | RMBase: m6A_site_5376 | ||
| mod ID: M6ASITE053762 | Click to Show/Hide the Full List | ||
| mod site | chr1:8868025-8868026:- | [11] | |
| Sequence | CACAGAACAAGAGAAGATTGACAAACTGATGATCGAGATGG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T; AML | ||
| Seq Type List | DART-seq; MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000464920.2; ENST00000645609.1; ENST00000646539.1; ENST00000645600.1; ENST00000646370.2; ENST00000646906.1; ENST00000234590.10; ENST00000646660.1; ENST00000647408.1; ENST00000497492.1; ENST00000646680.1 | ||
| External Link | RMBase: m6A_site_5377 | ||
| mod ID: M6ASITE053763 | Click to Show/Hide the Full List | ||
| mod site | chr1:8868039-8868040:- | [8] | |
| Sequence | TAGAAACTGAACGTCACAGAACAAGAGAAGATTGACAAACT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000646660.1; ENST00000645609.1; ENST00000646680.1; ENST00000497492.1; ENST00000464920.2; ENST00000646370.2; ENST00000646539.1; ENST00000647408.1; ENST00000234590.10; ENST00000646156.1; ENST00000646906.1; ENST00000645600.1 | ||
| External Link | RMBase: m6A_site_5378 | ||
| mod ID: M6ASITE053801 | Click to Show/Hide the Full List | ||
| mod site | chr1:8868054-8868055:- | [8] | |
| Sequence | CTCCCCTCTCCCTCGTAGAAACTGAACGTCACAGAACAAGA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000646680.1; ENST00000646370.2; ENST00000646156.1; ENST00000646660.1; ENST00000645609.1; ENST00000643438.1; ENST00000646539.1; ENST00000497492.1; ENST00000234590.10; ENST00000464920.2; ENST00000647408.1; ENST00000489867.2; ENST00000645600.1; ENST00000646906.1 | ||
| External Link | RMBase: m6A_site_5379 | ||
| mod ID: M6ASITE053802 | Click to Show/Hide the Full List | ||
| mod site | chr1:8870189-8870190:- | [8] | |
| Sequence | TCCCCCCTGCAACTCTCTGGACCTTTGAAGAAATTAAGATC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000489867.2; ENST00000646680.1; ENST00000234590.10; ENST00000497492.1; ENST00000645600.1; ENST00000464920.2; ENST00000646370.2; ENST00000486051.5; ENST00000646156.1; ENST00000643438.1; ENST00000646539.1; ENST00000646660.1; ENST00000646906.1; ENST00000645609.1; ENST00000647408.1 | ||
| External Link | RMBase: m6A_site_5380 | ||
| mod ID: M6ASITE053829 | Click to Show/Hide the Full List | ||
| mod site | chr1:8870232-8870233:- | [8] | |
| Sequence | AATTGCATCAGAAGAATGAGACACTTCGAGCCCTGAGGTTT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000486051.5; ENST00000647408.1; ENST00000234590.10; ENST00000646156.1; ENST00000464920.2; ENST00000497492.1; ENST00000643438.1; ENST00000646660.1; ENST00000646539.1; ENST00000645609.1; ENST00000646906.1; ENST00000645600.1; ENST00000646370.2; ENST00000489867.2; ENST00000646680.1 | ||
| External Link | RMBase: m6A_site_5381 | ||
| mod ID: M6ASITE053863 | Click to Show/Hide the Full List | ||
| mod site | chr1:8870316-8870317:- | [8] | |
| Sequence | ATTCCCTGTCACGCACGAGAACAATCCATGCATGACATAAT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000645600.1; ENST00000645609.1; ENST00000489867.2; ENST00000486051.5; ENST00000647408.1; ENST00000646370.2; ENST00000646680.1; ENST00000464920.2; ENST00000643438.1; ENST00000234590.10; ENST00000646660.1; ENST00000646539.1; ENST00000646156.1; ENST00000497492.1; ENST00000646906.1 | ||
| External Link | RMBase: m6A_site_5382 | ||
| mod ID: M6ASITE053928 | Click to Show/Hide the Full List | ||
| mod site | chr1:8870477-8870478:- | [8] | |
| Sequence | CTGTTGAGCACATCAATAAAACTATTGCGCCTGCCCTGGTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2 | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000646539.1; ENST00000646156.1; ENST00000464920.2; ENST00000234590.10; ENST00000646660.1; ENST00000643438.1; ENST00000646370.2; ENST00000647408.1; ENST00000646680.1; ENST00000645600.1; ENST00000646906.1; ENST00000645609.1; ENST00000497492.1; ENST00000489867.2; ENST00000486051.5 | ||
| External Link | RMBase: m6A_site_5383 | ||
| mod ID: M6ASITE053929 | Click to Show/Hide the Full List | ||
| mod site | chr1:8870488-8870489:- | [10] | |
| Sequence | TGTCTCAAAGGCTGTTGAGCACATCAATAAAACTATTGCGC | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | liver; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000645609.1; ENST00000234590.10; ENST00000486051.5; ENST00000646370.2; ENST00000647408.1; ENST00000645600.1; ENST00000489867.2; ENST00000646660.1; ENST00000497492.1; ENST00000464920.2; ENST00000646906.1; ENST00000646680.1; ENST00000646156.1; ENST00000646539.1; ENST00000643438.1 | ||
| External Link | RMBase: m6A_site_5384 | ||
| mod ID: M6ASITE053983 | Click to Show/Hide the Full List | ||
| mod site | chr1:8870872-8870873:- | [8] | |
| Sequence | TGCTGGGCAGGCGTCTCCAGACCCATTAAGTATATTAATGA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000646370.2; ENST00000646906.1; ENST00000645600.1; ENST00000643438.1; ENST00000646539.1; ENST00000645609.1; ENST00000497492.1; ENST00000646660.1; ENST00000464920.2; ENST00000489867.2; ENST00000234590.10; ENST00000646156.1; ENST00000646680.1; ENST00000486051.5; ENST00000647408.1 | ||
| External Link | RMBase: m6A_site_5385 | ||
| mod ID: M6ASITE053984 | Click to Show/Hide the Full List | ||
| mod site | chr1:8871877-8871878:- | [8] | |
| Sequence | GGGGAAGGGTAAGCCTTAGAACCCACAGCCCATGGCCTCCC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; Huh7; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000234590.10; ENST00000646906.1; ENST00000643438.1; ENST00000486051.5; ENST00000489867.2; ENST00000645600.1; ENST00000645609.1; ENST00000647408.1; ENST00000646680.1; ENST00000646156.1; ENST00000492343.2; ENST00000646660.1; ENST00000646539.1; ENST00000646370.2; ENST00000497492.1 | ||
| External Link | RMBase: m6A_site_5386 | ||
| mod ID: M6ASITE054044 | Click to Show/Hide the Full List | ||
| mod site | chr1:8871908-8871909:- | [8] | |
| Sequence | AGCTCCGGGACAATGATAAGACTCGCTATATGGGGAAGGGT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; Huh7; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; DART-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000643438.1; ENST00000645609.1; ENST00000234590.10; ENST00000646680.1; ENST00000486051.5; ENST00000497492.1; ENST00000646370.2; ENST00000646156.1; ENST00000646660.1; ENST00000645600.1; ENST00000647408.1; ENST00000489867.2; ENST00000492343.2; ENST00000646906.1; ENST00000646539.1 | ||
| External Link | RMBase: m6A_site_5387 | ||
| mod ID: M6ASITE054123 | Click to Show/Hide the Full List | ||
| mod site | chr1:8871919-8871920:- | [8] | |
| Sequence | TGAGGCCCTAGAGCTCCGGGACAATGATAAGACTCGCTATA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; HepG2; Huh7; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; DART-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000645600.1; ENST00000234590.10; ENST00000647408.1; ENST00000646660.1; ENST00000492343.2; ENST00000646906.1; ENST00000645609.1; ENST00000646539.1; ENST00000646370.2; ENST00000646680.1; ENST00000486051.5; ENST00000489867.2; ENST00000643438.1; ENST00000646156.1; ENST00000497492.1 | ||
| External Link | RMBase: m6A_site_5388 | ||
| mod ID: M6ASITE054124 | Click to Show/Hide the Full List | ||
| mod site | chr1:8873799-8873800:- | [8] | |
| Sequence | CGGTGCTAAACTCACAGAGAACTGCCAGTCGGAGCTGAGTC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000486051.5; ENST00000646539.1; ENST00000645609.1; ENST00000497492.1; ENST00000646680.1; ENST00000489867.2; ENST00000646370.2; ENST00000492343.2; ENST00000646906.1; ENST00000643438.1; ENST00000646156.1; ENST00000647408.1; ENST00000234590.10; ENST00000646660.1 | ||
| External Link | RMBase: m6A_site_5389 | ||
| mod ID: M6ASITE054125 | Click to Show/Hide the Full List | ||
| mod site | chr1:8873810-8873811:- | [8] | |
| Sequence | GAATCACTCAGCGGTGCTAAACTCACAGAGAACTGCCAGTC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000646906.1; ENST00000492343.2; ENST00000489867.2; ENST00000645609.1; ENST00000234590.10; ENST00000643438.1; ENST00000647408.1; ENST00000486051.5; ENST00000497492.1; ENST00000646660.1; ENST00000646539.1; ENST00000646680.1; ENST00000646370.2; ENST00000646156.1 | ||
| External Link | RMBase: m6A_site_5390 | ||
| mod ID: M6ASITE054126 | Click to Show/Hide the Full List | ||
| mod site | chr1:8873866-8873867:- | [8] | |
| Sequence | TCCAGCACATGGCGCGATGGACAGAGTGGGTGAGGTTGGCA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000646680.1; ENST00000643438.1; ENST00000646906.1; ENST00000646370.2; ENST00000486051.5; ENST00000647408.1; ENST00000492343.2; ENST00000234590.10; ENST00000489867.2; ENST00000646156.1; ENST00000645609.1; ENST00000646660.1; ENST00000497492.1; ENST00000646539.1 | ||
| External Link | RMBase: m6A_site_5391 | ||
| mod ID: M6ASITE054127 | Click to Show/Hide the Full List | ||
| mod site | chr1:8873936-8873937:- | [8] | |
| Sequence | AAAAATTAGAGAATTGTGAAACTCCTTCCTTTAAGAAGAAA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; A549; HepG2; LCLs; H1299; Huh7; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000646660.1; ENST00000647408.1; ENST00000643438.1; ENST00000486051.5; ENST00000492343.2; ENST00000646156.1; ENST00000489867.2; ENST00000646370.2; ENST00000646539.1; ENST00000234590.10; ENST00000497492.1; ENST00000646906.1; ENST00000645609.1; ENST00000646680.1 | ||
| External Link | RMBase: m6A_site_5392 | ||
| mod ID: M6ASITE054128 | Click to Show/Hide the Full List | ||
| mod site | chr1:8874870-8874871:- | [9] | |
| Sequence | CCATGCCAGGGAGATCTTTGACTCTCGCGGGAATCCCACTG | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000646156.1; ENST00000646370.2; ENST00000646539.1; ENST00000646906.1; ENST00000645609.1; ENST00000647408.1; ENST00000489867.2; ENST00000486051.5; ENST00000234590.10; ENST00000492343.2; ENST00000643438.1; ENST00000646660.1; ENST00000497492.1; ENST00000646680.1 | ||
| External Link | RMBase: m6A_site_5393 | ||
| mod ID: M6ASITE054148 | Click to Show/Hide the Full List | ||
| mod site | chr1:8875806-8875807:- | [8] | |
| Sequence | AGCCTGGGCAGCATAGTGAGACCGCCATCTCTTAAAAAAGT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; A549; HepG2; fibroblasts; LCLs; H1299; MM6; Huh7; CD4T; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000647408.1; ENST00000486051.5; ENST00000646660.1; ENST00000234590.10; ENST00000646370.2; ENST00000414948.1; ENST00000646156.1; ENST00000492343.2; ENST00000643438.1; rmsk_15811; ENST00000497492.1; ENST00000646680.1; ENST00000489867.2; ENST00000645609.1; ENST00000646539.1; ENST00000646906.1 | ||
| External Link | RMBase: m6A_site_5394 | ||
| mod ID: M6ASITE054149 | Click to Show/Hide the Full List | ||
| mod site | chr1:8878591-8878592:- | [8] | |
| Sequence | TTCCTCTCCTAGGCGACGAGACCCAGTGGCTAGGTAATGAT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; A549; H1B; H1A; HEK293T; fibroblasts; LCLs; MT4; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000643438.1; ENST00000486051.5; ENST00000646370.2; ENST00000646680.1; ENST00000489867.2; ENST00000492343.2; ENST00000647408.1; ENST00000646156.1; ENST00000646539.1; ENST00000645609.1; ENST00000234590.10 | ||
| External Link | RMBase: m6A_site_5395 | ||
| mod ID: M6ASITE054150 | Click to Show/Hide the Full List | ||
| mod site | chr1:8878687-8878688:- | [8] | |
| Sequence | AGTCGGCGCGGGCGGCGCGGACAGTATCTGTGGGTACCCGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; A549; H1B; H1A; hESCs; HEK293T; fibroblasts; LCLs; MT4; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000646539.1; ENST00000647408.1; ENST00000646156.1; ENST00000492343.2; ENST00000489867.2; ENST00000646680.1; ENST00000486051.5; ENST00000645609.1; ENST00000646370.2 | ||
| External Link | RMBase: m6A_site_5396 | ||
| mod ID: M6ASITE054312 | Click to Show/Hide the Full List | ||
| mod site | chr1:8879057-8879058:- | [8] | |
| Sequence | CGACGCTGAGTGCGTGCGGGACTCGGAGTACGTGACGGAGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; H1B; A549; GSC-11; HEK293T; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000645609.1; ENST00000489867.2; ENST00000646539.1 | ||
| External Link | RMBase: m6A_site_5400 | ||
References