m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00175)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
AKT1
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 14 (METTL14) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL14 | ||
| Cell Line | Neural progenitor cell line | Mus musculus |
|
Treatment: METTL14 knockout NPCs
Control: Wild type NPCs
|
GSE158985 | |
| Regulation |
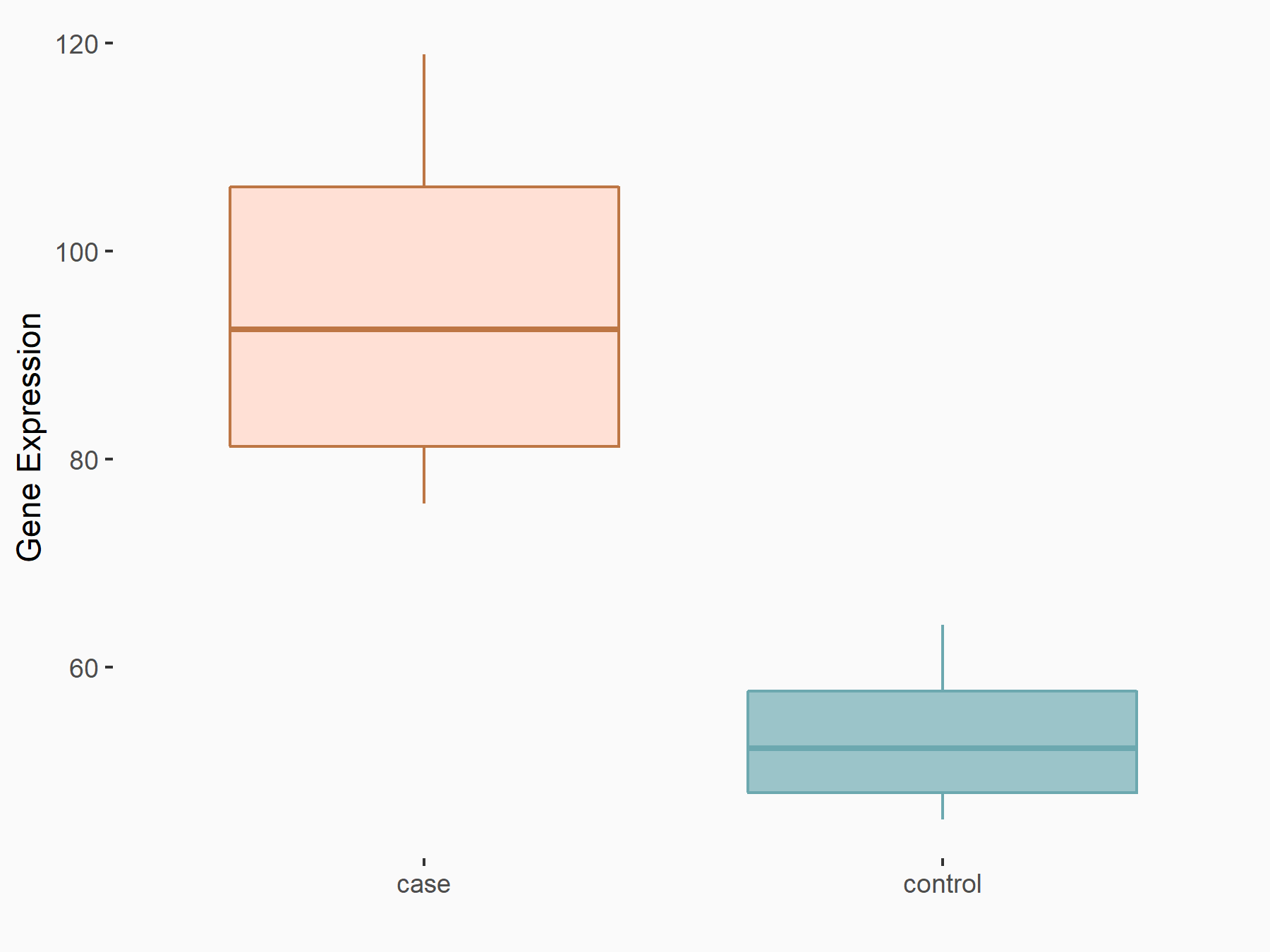  |
logFC: 8.08E-01 p-value: 3.19E-03 |
| More Results | Click to View More RNA-seq Results | |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | The m6A modification level was decreased in GC and METTL14 was a key regulator resulting in m6A disorder in GC. METTL14 overexpression suppressed GC cell proliferation and aggression by deactivating the PI3K/RAC-alpha serine/threonine-protein kinase (AKT1)/mTOR pathway and the EMT pathway, respectively. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| In-vitro Model | SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL14 was found to inhibit HCC cell migration, invasion, and EMT through modulating EGFR/PI3K/RAC-alpha serine/threonine-protein kinase (AKT1) signaling pathway in an m6A-dependent manner. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | YY-8103 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_WY40 |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| In-vivo Model | For the lung metastasis model, stably transfected HepG2 cells (1 × 106/0.1 mL DMEM) were injected into each nude mouse through the tail vein. Five weeks later, mice were euthanized, and the lung tissues were collected. | |||
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3 knockout liver
Control: Wild type liver cells
|
GSE198512 | |
| Regulation |
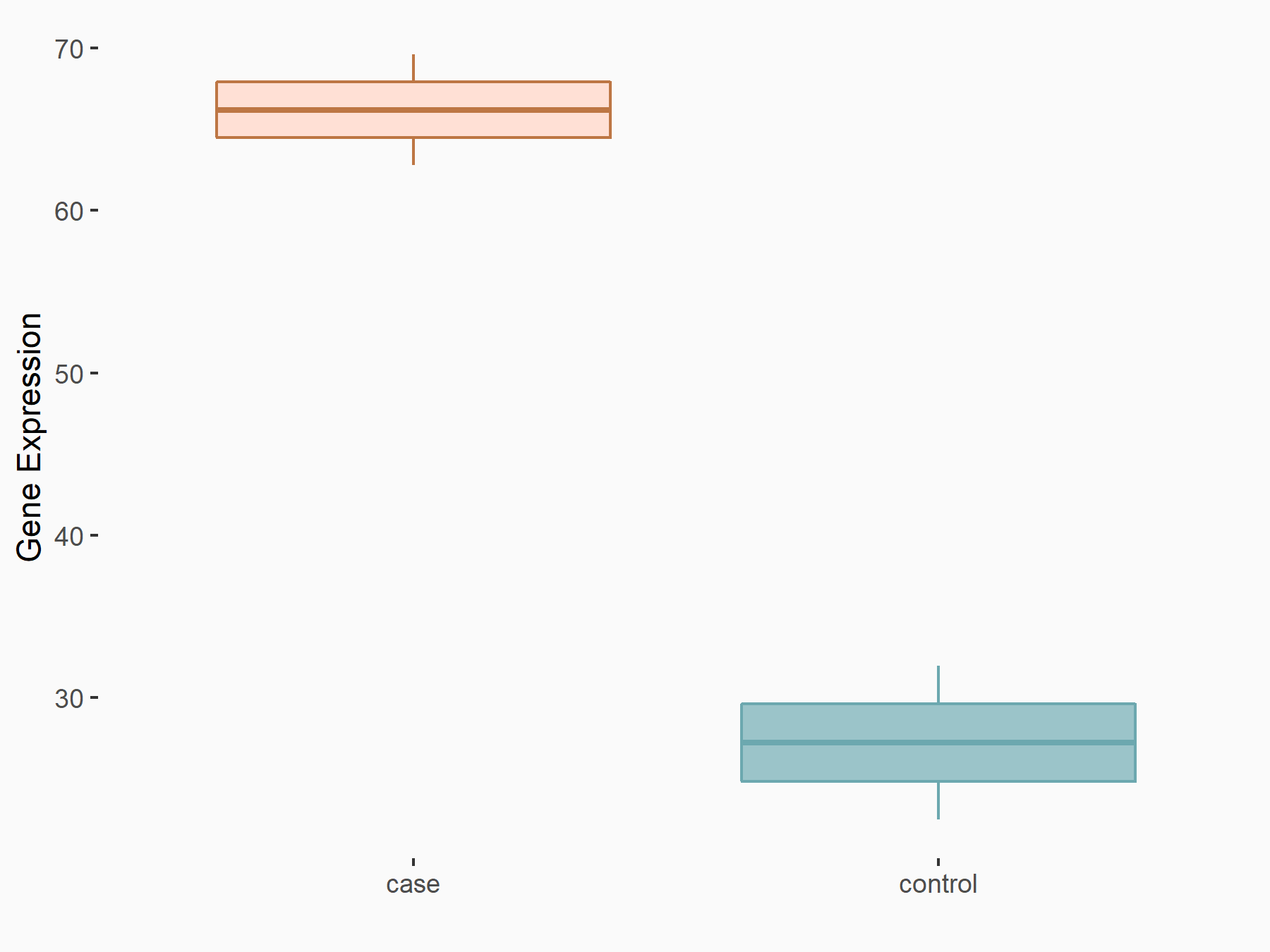  |
logFC: 1.27E+00 p-value: 1.77E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 10 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | METTL3 depletion in human myeloid leukemia cell lines induces cell differentiation and apoptosis and delays leukemia progression in recipient mice in vivo. Single-nucleotide-resolution mapping of m6A coupled with ribosome profiling reveals that m6A promotes the translation of c-MYC, BCL2 and PTEN mRNAs in the human acute myeloid leukemia MOLM-13 cell line. Moreover, loss of METTL3 leads to increased levels of phosphorylated RAC-alpha serine/threonine-protein kinase (AKT1). | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Acute myeloid leukaemia | ICD-11: 2A60 | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell differentiation and apoptosis | |||
| In-vitro Model | HSPC (Human hematopoietic stem cell) | |||
| In-vivo Model | 500,000 selected cells were injected via tail vein or retro-orbital route into female NSG (6-8 week old) recipient mice that had been sublethally irradiated with 475 cGy one day before transplantation. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | Downregulated METTL3 expression in AML-MSCs induced an increase in RAC-alpha serine/threonine-protein kinase (AKT1) protein, resulting in enhanced MSC adipogenesis, thereby contributing to chemoresistance in acute myeloid leukaemia (AML) cells. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Acute myeloid leukaemia | ICD-11: 2A60 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Adipogenesis | |||
| In-vitro Model | HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 |
| U-937 | Adult acute monocytic leukemia | Homo sapiens | CVCL_0007 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [5] | |||
| Response Summary | METTL3 plays a carcinogenic role in human EC progression partially through RAC-alpha serine/threonine-protein kinase (AKT1) signaling pathways, suggesting that METTL3 serves as a potential therapeutic target for esophageal cancer therapy. A double-effect inhibitor (BEZ235) inhibited AKT and mTOR phosphorylation and hindered the effect of METTL3 overexpression on the proliferation and migration of Eca-109 and KY-SE150 cells. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Esophageal cancer | ICD-11: 2B70 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Wnt signaling pathway | hsa04310 | |||
| mTOR signaling pathway | hsa04150 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell proliferation and invasion | |||
| Cell apoptosis | ||||
| In-vitro Model | Eca-109 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_6898 |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| Normal esophageal epithelial cell line (HEEC) (Isolated from the human esophagus) | ||||
| Experiment 4 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | Down-regulation of METTL3 inhibits the proliferation and mobility of human gastric cancer cells and leads to inactivation of the AKT signaling pathway, suggesting that METTL3 is a potential target for the treatment of human gastric cancer. METTL3 knockdown decreased Bcl2 and increased Bax and active Caspase-3 in gastric cancer cells, which suggested the apoptotic pathway was activated. METTL3 led to inactivation of the AKT signaling pathway in human gastric cancer cells, including decreased phosphorylation levels of RAC-alpha serine/threonine-protein kinase (AKT1) and expression of down-stream effectors p70S6K and Cyclin D1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| Experiment 5 Reporting the m6A Methylation Regulator of This Target Gene | [7] | |||
| Response Summary | METTL3 contributes to the progression and chemoresistance of NSCLC by promoting RAC-alpha serine/threonine-protein kinase (AKT1) protein expression through regulating AKT1 mRNA m6A levels, and provides an efficient therapeutic intervention target for overcoming chemoresistance in NSCLC. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Experiment 6 Reporting the m6A Methylation Regulator of This Target Gene | [8] | |||
| Response Summary | METTL3-mediated m6 A methylation promotes lung cancer progression via activating PI3K/RAC-alpha serine/threonine-protein kinase (AKT1)/mTOR pathway. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Lung cancer | ICD-11: 2C25 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| In-vivo Model | 5 × 106 A549 cells overexpressing METTL3 (Lv-METTL3) or control (Lv-Ctrl) were suspended in 100 uL phosphate-buffered saline (PBS), and were subcutaneously injected into mouse lower right flank. Drug treatment started in the Lv-METTL3 group when the tumour volume reached around 100 mm3. Mice were randomly divided into three groups to receive vehicle, GSK2536771 (30 mg/kg) or rapamycin (1 mg/kg). Drugs were administrated daily through intraperitoneal injection for 18 days. Treatment conditions were chosen as previously reported. | |||
| Experiment 7 Reporting the m6A Methylation Regulator of This Target Gene | [9] | |||
| Response Summary | METTL3 knockdown downregulated the phosphorylation levels of RAC-alpha serine/threonine-protein kinase (AKT1) and the expression of the downstream effector Cyclin D1 in ovarian cancer. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Ovarian cancer | ICD-11: 2C73 | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell cycle | |||
| Cell apoptosis | ||||
| In-vitro Model | OVCAR-3 | Ovarian serous adenocarcinoma | Homo sapiens | CVCL_0465 |
| SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 | |
| Experiment 8 Reporting the m6A Methylation Regulator of This Target Gene | [10] | |||
| Response Summary | Knock-down of YTHDF2 or METTL3 significantly induced the expression of LHPP and NKX3-1 at both mRNA and protein level with inhibited phosphorylated RAC-alpha serine/threonine-protein kinase (AKT1). YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent way to regulate AKT phosphorylation-induced tumor progression in prostate cancer. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Prostate cancer | ICD-11: 2C82 | ||
| Pathway Response | Oxidative phosphorylation | hsa00190 | ||
| In-vitro Model | VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| In-vivo Model | Approximately 2 × 106 PCa cells (PC-3 shNC, shYTHDF2, shMETTL3 cell lines) per mouse suspended in 100 uL PBS were injected in the flank of male BALB/c nude mice (4 weeks old). During the 40-day observation, the tumor size (V = (width2×length ×0.52)) was measured with vernier caliper. Approximately 1.5 × 106 PCa cells suspended in 100 uL of PBS (PC-3 shNC, shYTHDF2, and shMETTL3 cell lines) per mouse were injected into the tail vein of male BALB/c nude mice (4 weeks old). The IVIS Spectrum animal imaging system (PerkinElmer) was used to evaluate the tumor growth (40 days) and whole metastasis conditions (4 weeks and 6 weeks) with 100 uL XenoLight D-luciferin Potassium Salt (15 mg/ml, Perkin Elmer) per mouse. Mice were anesthetized and then sacrificed for tumors and metastases which were sent for further organ-localized imaging as above, IHC staining and hematoxylin-eosin (H&E) staining. | |||
| Experiment 9 Reporting the m6A Methylation Regulator of This Target Gene | [11] | |||
| Response Summary | Knockdown of METTL3 could obviously promote cell proliferation, migration and invasion function, and induce G0/G1 arrest,METTL3 acts as a novel marker for tumorigenesis, development and survival of RCC. Knockdown of METTL3 promoted changes in pI3K/RAC-alpha serine/threonine-protein kinase (AKT1)/mTOR markers' expression with a gain in p-PI3k, p-AKT, p-mTOR and p-p70, and a loss of p-4EBP1. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Renal cell carcinoma | ICD-11: 2C90 | ||
| Cell Process | Epithelial-to-mesenchymal transition | |||
| Arrest cell cycle at G0/G1 phase | ||||
| In-vitro Model | ACHN | Papillary renal cell carcinoma | Homo sapiens | CVCL_1067 |
| Caki-1 | Clear cell renal cell carcinoma | Homo sapiens | CVCL_0234 | |
| Caki-2 | Papillary renal cell carcinoma | Homo sapiens | CVCL_0235 | |
| HK2 | Normal | Acipenser baerii | CVCL_YE28 | |
| In-vivo Model | Cells (5×106 cells in 200 uL) were suspended with 100 uL PBS and 100 uL Matrigel Matrix, and injected subcutaneously into the left armpit of each mouse. | |||
| Experiment 10 Reporting the m6A Methylation Regulator of This Target Gene | [12] | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through PI3K/RAC-alpha serine/threonine-protein kinase (AKT1)/mTOR pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-P70S6K/4EBP1 pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Retinoblastoma | ICD-11: 2D02.2 | ||
| Responsed Drug | Rapamycin | Approved | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| In-vitro Model | WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
RNA demethylase ALKBH5 (ALKBH5) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by ALKBH5 | ||
| Cell Line | 143B cell line | Homo sapiens |
|
Treatment: siALKBH5 transfected 143B cells
Control: siControl 143B cells
|
GSE154528 | |
| Regulation |
  |
logFC: -9.73E-01 p-value: 3.10E-07 |
| More Results | Click to View More RNA-seq Results | |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [13] | |||
| Response Summary | ALKBH5-mediated m6A demethylation enhanced the stability of KCNK15-AS1. In pancreatic cancer, KCNK15-AS1 bound to KCNK15 to inhibit its translation, and interacted with MDM2 to induce REST ubiquitination, which eventually facilitated PTEN transcription to inactivate RAC-alpha serine/threonine-protein kinase (AKT1) pathway. | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Epithelial-mesenchymal transition | ||||
| Cell apoptosis | ||||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [14] | |||
| Response Summary | The overexpression of ALKBH5 led to the activation of RAC-alpha serine/threonine-protein kinase (AKT1), and BMP2 was regulated by ALKBH5 through the AKT signaling pathway. ALKBH5 promoted the osteogenesis of the ligamentum flavum cells through BMP2 demethylation and AKT activation. MK22606 is an AKT inhibitor. Moreover, when ALKBH5 was knocked down in the ligamentum flavum cells, p-AKT was inhibited when compared with that in the overexpressed ALKBH5 and control groups. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Ossification of spinal ligaments | ICD-11: FA83 | ||
| Responsed Drug | MK22606 | Investigative | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Ossification | |||
| In-vitro Model | Ligamentum flavum cells (Ligamentum flavum cells) | |||
YTH domain-containing family protein 1 (YTHDF1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
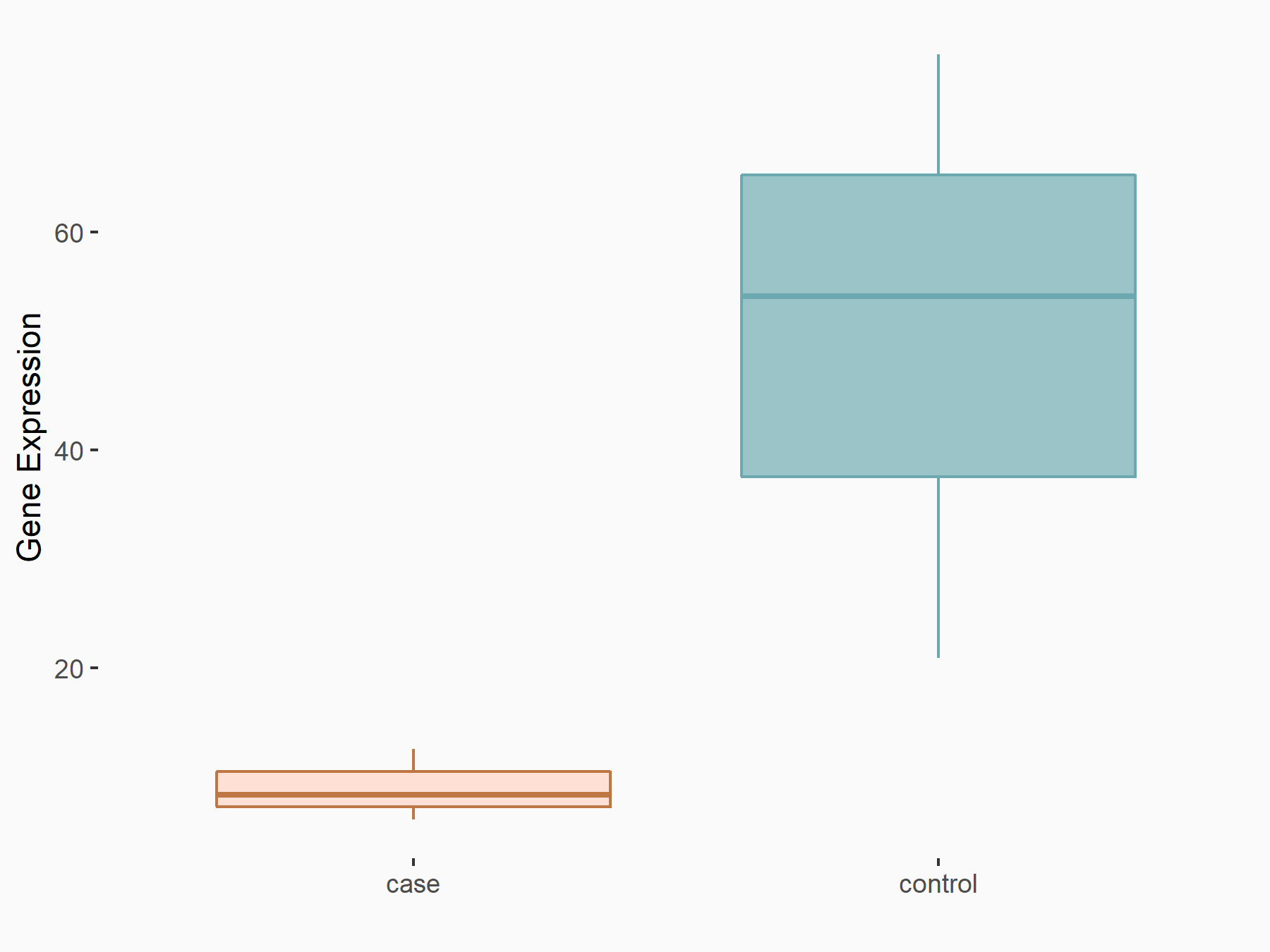  |
logFC: -2.24E+00 p-value: 7.14E-03 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [15] | |||
| Response Summary | YTHDF1 contributes to the progression of HCC by activating PI3K/RAC-alpha serine/threonine-protein kinase (AKT1)/mTOR signaling pathway and inducing EMT. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell proliferation | ||||
| In-vitro Model | SNU-398 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0077 |
| SK-HEP-1 | Liver and intrahepatic bile duct epithelial neoplasm | Homo sapiens | CVCL_0525 | |
| PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| In-vivo Model | Ten four-week-old BALB/c male nude mice (GemPharmatech, Jiangsu, China) were subcutaneously injected with control Huh7 cells 2 × 106 (left-back) and stable knockdown of YTHDF1 Huh7 cells 2 × 106 (right-back). These cells were respectively premixed with 50 ul Matrigel (Corning, 354,234) in 100 ul PBS. | |||
Heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1) [READER]
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [17] | |||
| Response Summary | In breast cancer, modest stable overexpression of A2B1 in MCF-7 cells (MCF-7-A2B1 cells) resulted in tamoxifen and fulvestrant- resistance whereas knockdown of A2B1 in LCC9 and LY2 cells restored tamoxifen and fulvestrant, endocrine-sensitivity. MCF-7-A2B1 cells have increased ER-alpha and reduced miR-222-3p that targets ER-alpha. MCF-7-A2B1 have activated RAC-alpha serine/threonine-protein kinase (AKT1) and MAPK that depend on A2B1 expression and are growth inhibited by inhibitors of these pathways. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Fulvestrant | Approved | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell migration and invasion | |||
| In-vitro Model | HCC1806 | Breast squamous cell carcinoma | Homo sapiens | CVCL_1258 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [17] | |||
| Response Summary | In breast cancer, modest stable overexpression of A2B1 in MCF-7 cells (MCF-7-A2B1 cells) resulted in tamoxifen and fulvestrant - resistance whereas knockdown of A2B1 in LCC9 and LY2 cells restored tamoxifen and fulvestrant, endocrine-sensitivity. MCF-7-A2B1 cells have increased ER-alpha and reduced miR-222-3p that targets ER-alpha. MCF-7-A2B1 have activated RAC-alpha serine/threonine-protein kinase (AKT1) and MAPK that depend on A2B1 expression and are growth inhibited by inhibitors of these pathways. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Tamoxifen | Approved | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell migration and invasion | |||
| In-vitro Model | HCC1806 | Breast squamous cell carcinoma | Homo sapiens | CVCL_1258 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [20] | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated p38, ERK1/2, RAC-alpha serine/threonine-protein kinase (AKT1), and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, IL-1-beta and TNF-alpha secretion. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gangrene or necrosis of lung | ICD-11: CA43 | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
| In-vitro Model | BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
YTH domain-containing family protein 2 (YTHDF2) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [10] | |||
| Response Summary | Knock-down of YTHDF2 or METTL3 significantly induced the expression of LHPP and NKX3-1 at both mRNA and protein level with inhibited phosphorylated RAC-alpha serine/threonine-protein kinase (AKT1). YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent way to regulate AKT phosphorylation-induced tumor progression in prostate cancer. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Prostate cancer | ICD-11: 2C82 | ||
| Pathway Response | Oxidative phosphorylation | hsa00190 | ||
| In-vitro Model | VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
YTH domain-containing family protein 3 (YTHDF3) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [20] | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated p38, ERK1/2, RAC-alpha serine/threonine-protein kinase (AKT1), and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, IL-1-beta and TNF-alpha secretion. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gangrene or necrosis of lung | ICD-11: CA43 | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
| In-vitro Model | BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
YTH domain-containing protein 2 (YTHDC2) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [21] | |||
| Response Summary | YTHDC2 promotes radiotherapy resistance of NPC cells by activating the IGF1R/RAC-alpha serine/threonine-protein kinase (AKT1)/S6 signaling axis and serves as a potential therapeutic target in radiosensitization of NPC cells. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Nasopharyngeal carcinoma | ICD-11: 2B6B | ||
| In-vitro Model | HK1-IRR (HK1-IRR (HK1-ionizing radiation radioresistent cell line) was derived from HK1 after a prolonged exposure of irradiation.HK1, a generous gift from Prof. Ya Cao (Cancer Research Institute, Central South University), was established from a recurrent nasopharynx carcinoma of a Chinese 17-year-old male patient) | |||
| NPC/HK1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_7084 | |
| CNE2-IRR (CNE2-IRR (CNE2-ionizing radiation radioresistent cell line) was derived from CNE2 after a prolonged exposure of irradiation) | ||||
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| In-vivo Model | 2 × 106 cells resuspended in 50 uL of Matrigel (Corning) were subcutaneously injected into 4-6 weeks old male nude mice. When tumor volumes reached 150-200 mm3, animals were divided into control group and radiotherapy group. In the radiotherapy group, tumors were treated with a single irradiation (4 Gy) when tumor volumes reached approximately 150-200 mm3. The tumor stopped growing in the next few days and then restarted growth. | |||
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | METTL3 depletion in human myeloid leukemia cell lines induces cell differentiation and apoptosis and delays leukemia progression in recipient mice in vivo. Single-nucleotide-resolution mapping of m6A coupled with ribosome profiling reveals that m6A promotes the translation of c-MYC, BCL2 and PTEN mRNAs in the human acute myeloid leukemia MOLM-13 cell line. Moreover, loss of METTL3 leads to increased levels of phosphorylated RAC-alpha serine/threonine-protein kinase (AKT1). | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell differentiation and apoptosis | |||
| In-vitro Model | HSPC (Human hematopoietic stem cell) | |||
| In-vivo Model | 500,000 selected cells were injected via tail vein or retro-orbital route into female NSG (6-8 week old) recipient mice that had been sublethally irradiated with 475 cGy one day before transplantation. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | Downregulated METTL3 expression in AML-MSCs induced an increase in RAC-alpha serine/threonine-protein kinase (AKT1) protein, resulting in enhanced MSC adipogenesis, thereby contributing to chemoresistance in acute myeloid leukaemia (AML) cells. | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Adipogenesis | |||
| In-vitro Model | HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 |
| U-937 | Adult acute monocytic leukemia | Homo sapiens | CVCL_0007 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [21] | |||
| Response Summary | YTHDC2 promotes radiotherapy resistance of NPC cells by activating the IGF1R/RAC-alpha serine/threonine-protein kinase (AKT1)/S6 signaling axis and serves as a potential therapeutic target in radiosensitization of NPC cells. | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulator | YTH domain-containing protein 2 (YTHDC2) | READER | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | HK1-IRR (HK1-IRR (HK1-ionizing radiation radioresistent cell line) was derived from HK1 after a prolonged exposure of irradiation.HK1, a generous gift from Prof. Ya Cao (Cancer Research Institute, Central South University), was established from a recurrent nasopharynx carcinoma of a Chinese 17-year-old male patient) | |||
| NPC/HK1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_7084 | |
| CNE2-IRR (CNE2-IRR (CNE2-ionizing radiation radioresistent cell line) was derived from CNE2 after a prolonged exposure of irradiation) | ||||
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| In-vivo Model | 2 × 106 cells resuspended in 50 uL of Matrigel (Corning) were subcutaneously injected into 4-6 weeks old male nude mice. When tumor volumes reached 150-200 mm3, animals were divided into control group and radiotherapy group. In the radiotherapy group, tumors were treated with a single irradiation (4 Gy) when tumor volumes reached approximately 150-200 mm3. The tumor stopped growing in the next few days and then restarted growth. | |||
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [5] | |||
| Response Summary | METTL3 plays a carcinogenic role in human EC progression partially through RAC-alpha serine/threonine-protein kinase (AKT1) signaling pathways, suggesting that METTL3 serves as a potential therapeutic target for esophageal cancer therapy. A double-effect inhibitor (BEZ235) inhibited AKT and mTOR phosphorylation and hindered the effect of METTL3 overexpression on the proliferation and migration of Eca-109 and KY-SE150 cells. | |||
| Responsed Disease | Esophageal cancer [ICD-11: 2B70] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Wnt signaling pathway | hsa04310 | |||
| mTOR signaling pathway | hsa04150 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell proliferation and invasion | |||
| Cell apoptosis | ||||
| In-vitro Model | Eca-109 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_6898 |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| Normal esophageal epithelial cell line (HEEC) (Isolated from the human esophagus) | ||||
Gastric cancer [ICD-11: 2B72]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | The m6A modification level was decreased in GC and METTL14 was a key regulator resulting in m6A disorder in GC. METTL14 overexpression suppressed GC cell proliferation and aggression by deactivating the PI3K/RAC-alpha serine/threonine-protein kinase (AKT1)/mTOR pathway and the EMT pathway, respectively. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| In-vitro Model | SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| Experiment 2 Reporting the m6A-centered Disease Response | [6] | |||
| Response Summary | Down-regulation of METTL3 inhibits the proliferation and mobility of human gastric cancer cells and leads to inactivation of the AKT signaling pathway, suggesting that METTL3 is a potential target for the treatment of human gastric cancer. METTL3 knockdown decreased Bcl2 and increased Bax and active Caspase-3 in gastric cancer cells, which suggested the apoptotic pathway was activated. METTL3 led to inactivation of the AKT signaling pathway in human gastric cancer cells, including decreased phosphorylation levels of RAC-alpha serine/threonine-protein kinase (AKT1) and expression of down-stream effectors p70S6K and Cyclin D1. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [13] | |||
| Response Summary | ALKBH5-mediated m6A demethylation enhanced the stability of KCNK15-AS1. In pancreatic cancer, KCNK15-AS1 bound to KCNK15 to inhibit its translation, and interacted with MDM2 to induce REST ubiquitination, which eventually facilitated PTEN transcription to inactivate RAC-alpha serine/threonine-protein kinase (AKT1) pathway. | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Epithelial-mesenchymal transition | ||||
| Cell apoptosis | ||||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
Gastrointestinal cancer [ICD-11: 2C11]
Liver cancer [ICD-11: 2C12]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL14 was found to inhibit HCC cell migration, invasion, and EMT through modulating EGFR/PI3K/RAC-alpha serine/threonine-protein kinase (AKT1) signaling pathway in an m6A-dependent manner. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | YY-8103 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_WY40 |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| In-vivo Model | For the lung metastasis model, stably transfected HepG2 cells (1 × 106/0.1 mL DMEM) were injected into each nude mouse through the tail vein. Five weeks later, mice were euthanized, and the lung tissues were collected. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [15] | |||
| Response Summary | YTHDF1 contributes to the progression of HCC by activating PI3K/RAC-alpha serine/threonine-protein kinase (AKT1)/mTOR signaling pathway and inducing EMT. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell proliferation | ||||
| In-vitro Model | SNU-398 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0077 |
| SK-HEP-1 | Liver and intrahepatic bile duct epithelial neoplasm | Homo sapiens | CVCL_0525 | |
| PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| In-vivo Model | Ten four-week-old BALB/c male nude mice (GemPharmatech, Jiangsu, China) were subcutaneously injected with control Huh7 cells 2 × 106 (left-back) and stable knockdown of YTHDF1 Huh7 cells 2 × 106 (right-back). These cells were respectively premixed with 50 ul Matrigel (Corning, 354,234) in 100 ul PBS. | |||
Lung cancer [ICD-11: 2C25]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [7] | |||
| Response Summary | METTL3 contributes to the progression and chemoresistance of NSCLC by promoting RAC-alpha serine/threonine-protein kinase (AKT1) protein expression through regulating AKT1 mRNA m6A levels, and provides an efficient therapeutic intervention target for overcoming chemoresistance in NSCLC. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [8] | |||
| Response Summary | METTL3-mediated m6 A methylation promotes lung cancer progression via activating PI3K/RAC-alpha serine/threonine-protein kinase (AKT1)/mTOR pathway. | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| In-vivo Model | 5 × 106 A549 cells overexpressing METTL3 (Lv-METTL3) or control (Lv-Ctrl) were suspended in 100 uL phosphate-buffered saline (PBS), and were subcutaneously injected into mouse lower right flank. Drug treatment started in the Lv-METTL3 group when the tumour volume reached around 100 mm3. Mice were randomly divided into three groups to receive vehicle, GSK2536771 (30 mg/kg) or rapamycin (1 mg/kg). Drugs were administrated daily through intraperitoneal injection for 18 days. Treatment conditions were chosen as previously reported. | |||
Breast cancer [ICD-11: 2C60]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [17] | |||
| Response Summary | In breast cancer, modest stable overexpression of A2B1 in MCF-7 cells (MCF-7-A2B1 cells) resulted in tamoxifen and fulvestrant- resistance whereas knockdown of A2B1 in LCC9 and LY2 cells restored tamoxifen and fulvestrant, endocrine-sensitivity. MCF-7-A2B1 cells have increased ER-alpha and reduced miR-222-3p that targets ER-alpha. MCF-7-A2B1 have activated RAC-alpha serine/threonine-protein kinase (AKT1) and MAPK that depend on A2B1 expression and are growth inhibited by inhibitors of these pathways. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Fulvestrant | Approved | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell migration and invasion | |||
| In-vitro Model | HCC1806 | Breast squamous cell carcinoma | Homo sapiens | CVCL_1258 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| Experiment 2 Reporting the m6A-centered Disease Response | [17] | |||
| Response Summary | In breast cancer, modest stable overexpression of A2B1 in MCF-7 cells (MCF-7-A2B1 cells) resulted in tamoxifen and fulvestrant - resistance whereas knockdown of A2B1 in LCC9 and LY2 cells restored tamoxifen and fulvestrant, endocrine-sensitivity. MCF-7-A2B1 cells have increased ER-alpha and reduced miR-222-3p that targets ER-alpha. MCF-7-A2B1 have activated RAC-alpha serine/threonine-protein kinase (AKT1) and MAPK that depend on A2B1 expression and are growth inhibited by inhibitors of these pathways. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Tamoxifen | Approved | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell migration and invasion | |||
| In-vitro Model | HCC1806 | Breast squamous cell carcinoma | Homo sapiens | CVCL_1258 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [9] | |||
| Response Summary | METTL3 knockdown downregulated the phosphorylation levels of RAC-alpha serine/threonine-protein kinase (AKT1) and the expression of the downstream effector Cyclin D1 in ovarian cancer. | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell cycle | |||
| Cell apoptosis | ||||
| In-vitro Model | OVCAR-3 | Ovarian serous adenocarcinoma | Homo sapiens | CVCL_0465 |
| SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 | |
Prostate cancer [ICD-11: 2C82]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [10] | |||
| Response Summary | Knock-down of YTHDF2 or METTL3 significantly induced the expression of LHPP and NKX3-1 at both mRNA and protein level with inhibited phosphorylated RAC-alpha serine/threonine-protein kinase (AKT1). YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent way to regulate AKT phosphorylation-induced tumor progression in prostate cancer. | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Oxidative phosphorylation | hsa00190 | ||
| In-vitro Model | VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| In-vivo Model | Approximately 2 × 106 PCa cells (PC-3 shNC, shYTHDF2, shMETTL3 cell lines) per mouse suspended in 100 uL PBS were injected in the flank of male BALB/c nude mice (4 weeks old). During the 40-day observation, the tumor size (V = (width2×length ×0.52)) was measured with vernier caliper. Approximately 1.5 × 106 PCa cells suspended in 100 uL of PBS (PC-3 shNC, shYTHDF2, and shMETTL3 cell lines) per mouse were injected into the tail vein of male BALB/c nude mice (4 weeks old). The IVIS Spectrum animal imaging system (PerkinElmer) was used to evaluate the tumor growth (40 days) and whole metastasis conditions (4 weeks and 6 weeks) with 100 uL XenoLight D-luciferin Potassium Salt (15 mg/ml, Perkin Elmer) per mouse. Mice were anesthetized and then sacrificed for tumors and metastases which were sent for further organ-localized imaging as above, IHC staining and hematoxylin-eosin (H&E) staining. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [10] | |||
| Response Summary | Knock-down of YTHDF2 or METTL3 significantly induced the expression of LHPP and NKX3-1 at both mRNA and protein level with inhibited phosphorylated RAC-alpha serine/threonine-protein kinase (AKT1). YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent way to regulate AKT phosphorylation-induced tumor progression in prostate cancer. | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Oxidative phosphorylation | hsa00190 | ||
| In-vitro Model | VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
Renal cell carcinoma [ICD-11: 2C90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [11] | |||
| Response Summary | Knockdown of METTL3 could obviously promote cell proliferation, migration and invasion function, and induce G0/G1 arrest,METTL3 acts as a novel marker for tumorigenesis, development and survival of RCC. Knockdown of METTL3 promoted changes in pI3K/RAC-alpha serine/threonine-protein kinase (AKT1)/mTOR markers' expression with a gain in p-PI3k, p-AKT, p-mTOR and p-p70, and a loss of p-4EBP1. | |||
| Responsed Disease | Renal cell carcinoma [ICD-11: 2C90] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Cell Process | Epithelial-to-mesenchymal transition | |||
| Arrest cell cycle at G0/G1 phase | ||||
| In-vitro Model | ACHN | Papillary renal cell carcinoma | Homo sapiens | CVCL_1067 |
| Caki-1 | Clear cell renal cell carcinoma | Homo sapiens | CVCL_0234 | |
| Caki-2 | Papillary renal cell carcinoma | Homo sapiens | CVCL_0235 | |
| HK2 | Normal | Acipenser baerii | CVCL_YE28 | |
| In-vivo Model | Cells (5×106 cells in 200 uL) were suspended with 100 uL PBS and 100 uL Matrigel Matrix, and injected subcutaneously into the left armpit of each mouse. | |||
Retina cancer [ICD-11: 2D02]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [12] | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through PI3K/RAC-alpha serine/threonine-protein kinase (AKT1)/mTOR pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-P70S6K/4EBP1 pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
| Responsed Disease | Retinoblastoma [ICD-11: 2D02.2] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Rapamycin | Approved | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| In-vitro Model | WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
Gangrene or necrosis of lung [ICD-11: CA43]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [20] | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated p38, ERK1/2, RAC-alpha serine/threonine-protein kinase (AKT1), and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, IL-1-beta and TNF-alpha secretion. | |||
| Responsed Disease | Gangrene or necrosis of lung [ICD-11: CA43] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
| In-vitro Model | BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [20] | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated p38, ERK1/2, RAC-alpha serine/threonine-protein kinase (AKT1), and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, IL-1-beta and TNF-alpha secretion. | |||
| Responsed Disease | Gangrene or necrosis of lung [ICD-11: CA43] | |||
| Target Regulator | YTH domain-containing family protein 3 (YTHDF3) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
| In-vitro Model | BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
Ossification of spinal ligaments [ICD-11: FA83]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [14] | |||
| Response Summary | The overexpression of ALKBH5 led to the activation of RAC-alpha serine/threonine-protein kinase (AKT1), and BMP2 was regulated by ALKBH5 through the AKT signaling pathway. ALKBH5 promoted the osteogenesis of the ligamentum flavum cells through BMP2 demethylation and AKT activation. MK22606 is an AKT inhibitor. Moreover, when ALKBH5 was knocked down in the ligamentum flavum cells, p-AKT was inhibited when compared with that in the overexpressed ALKBH5 and control groups. | |||
| Responsed Disease | Ossification of spinal ligaments [ICD-11: FA83] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | MK22606 | Investigative | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Ossification | |||
| In-vitro Model | Ligamentum flavum cells (Ligamentum flavum cells) | |||
Fulvestrant
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [17] | |||
| Response Summary | In breast cancer, modest stable overexpression of A2B1 in MCF-7 cells (MCF-7-A2B1 cells) resulted in tamoxifen and fulvestrant- resistance whereas knockdown of A2B1 in LCC9 and LY2 cells restored tamoxifen and fulvestrant, endocrine-sensitivity. MCF-7-A2B1 cells have increased ER-alpha and reduced miR-222-3p that targets ER-alpha. MCF-7-A2B1 have activated RAC-alpha serine/threonine-protein kinase (AKT1) and MAPK that depend on A2B1 expression and are growth inhibited by inhibitors of these pathways. | |||
| Target Regulator | Heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell migration and invasion | |||
| In-vitro Model | HCC1806 | Breast squamous cell carcinoma | Homo sapiens | CVCL_1258 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
Rapamycin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [12] | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through PI3K/RAC-alpha serine/threonine-protein kinase (AKT1)/mTOR pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-P70S6K/4EBP1 pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Retinoblastoma | ICD-11: 2D02.2 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| In-vitro Model | WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
Tamoxifen
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [17] | |||
| Response Summary | In breast cancer, modest stable overexpression of A2B1 in MCF-7 cells (MCF-7-A2B1 cells) resulted in tamoxifen and fulvestrant - resistance whereas knockdown of A2B1 in LCC9 and LY2 cells restored tamoxifen and fulvestrant, endocrine-sensitivity. MCF-7-A2B1 cells have increased ER-alpha and reduced miR-222-3p that targets ER-alpha. MCF-7-A2B1 have activated RAC-alpha serine/threonine-protein kinase (AKT1) and MAPK that depend on A2B1 expression and are growth inhibited by inhibitors of these pathways. | |||
| Target Regulator | Heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell migration and invasion | |||
| In-vitro Model | HCC1806 | Breast squamous cell carcinoma | Homo sapiens | CVCL_1258 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
MK22606
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [14] | |||
| Response Summary | The overexpression of ALKBH5 led to the activation of RAC-alpha serine/threonine-protein kinase (AKT1), and BMP2 was regulated by ALKBH5 through the AKT signaling pathway. ALKBH5 promoted the osteogenesis of the ligamentum flavum cells through BMP2 demethylation and AKT activation. MK22606 is an AKT inhibitor. Moreover, when ALKBH5 was knocked down in the ligamentum flavum cells, p-AKT was inhibited when compared with that in the overexpressed ALKBH5 and control groups. | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Ossification of spinal ligaments | ICD-11: FA83 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Ossification | |||
| In-vitro Model | Ligamentum flavum cells (Ligamentum flavum cells) | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
DNA modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 4 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT02150 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02163 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02176 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT02189 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
Histone modification
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03328 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5A (KDM5A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Prostate cancer | |
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 3 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03386 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03399 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03637 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Regulator: Methyltransferase-like 14 (METTL14)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03499 | ||
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
Non-coding RNA
m6A Regulator: Heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05143 | ||
| Epigenetic Regulator | Prostate cancer associated transcript 6 (PCAT6) | |
| Regulated Target | Heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Breast cancer | |
| Drug | Tamoxifen | |
| Crosstalk ID: M6ACROT05145 | ||
| Epigenetic Regulator | Prostate cancer associated transcript 6 (PCAT6) | |
| Regulated Target | Heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Breast cancer | |
| Drug | Fulvestrant | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00175)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: 2OMSITE000095 | Click to Show/Hide the Full List | ||
| mod site | chr14:104770802-104770803:- | [22] | |
| Sequence | GTCACGTCGGAGACTGACACCAGGTATTTTGATGAGGAGTT | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000553506.5; ENST00000555528.5; ENST00000554192.5; ENST00000554585.5; ENST00000544168.5; ENST00000402615.6; ENST00000649815.1; ENST00000610370.1; ENST00000555458.5; ENST00000557552.1; ENST00000554581.5; ENST00000554848.5; ENST00000349310.7; ENST00000407796.6 | ||
| External Link | RMBase: Nm_site_1824 | ||
| mod ID: 2OMSITE000096 | Click to Show/Hide the Full List | ||
| mod site | chr14:104770805-104770806:- | [22] | |
| Sequence | CAGGTCACGTCGGAGACTGACACCAGGTATTTTGATGAGGA | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000407796.6; ENST00000553506.5; ENST00000544168.5; ENST00000554581.5; ENST00000610370.1; ENST00000649815.1; ENST00000554192.5; ENST00000554848.5; ENST00000555528.5; ENST00000557552.1; ENST00000402615.6; ENST00000349310.7; ENST00000555458.5; ENST00000554585.5 | ||
| External Link | RMBase: Nm_site_1825 | ||
5-methylcytidine (m5C)
N6-methyladenosine (m6A)
| In total 104 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE021750 | Click to Show/Hide the Full List | ||
| mod site | chr14:104769422-104769423:- | [23] | |
| Sequence | GCGTTTTTTTACAACATTCAACTTTAGTATTTTTACTATTA | ||
| Motif Score | 2.595904762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000407796.6; ENST00000555458.5; ENST00000555528.5; ENST00000649815.1; ENST00000554581.5; ENST00000557552.1; ENST00000402615.6; ENST00000349310.7; ENST00000610370.1; ENST00000553506.5 | ||
| External Link | RMBase: m6A_site_266107 | ||
| mod ID: M6ASITE021751 | Click to Show/Hide the Full List | ||
| mod site | chr14:104769432-104769433:- | [24] | |
| Sequence | CTGCACCACGGCGTTTTTTTACAACATTCAACTTTAGTATT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000349310.7; ENST00000407796.6; ENST00000610370.1; ENST00000649815.1; ENST00000557552.1; ENST00000402615.6; ENST00000553506.5; ENST00000555528.5; ENST00000555458.5; ENST00000554581.5 | ||
| External Link | RMBase: m6A_site_266108 | ||
| mod ID: M6ASITE021752 | Click to Show/Hide the Full List | ||
| mod site | chr14:104769555-104769556:- | [23] | |
| Sequence | GGTAGCACTTGACCTTTTCGACGCTTAACCTTTCCGCTGTC | ||
| Motif Score | 2.839505952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000555528.5; ENST00000402615.6; ENST00000555458.5; ENST00000649815.1; ENST00000610370.1; ENST00000553506.5; ENST00000557552.1; ENST00000349310.7; ENST00000554581.5; ENST00000407796.6 | ||
| External Link | RMBase: m6A_site_266109 | ||
| mod ID: M6ASITE021753 | Click to Show/Hide the Full List | ||
| mod site | chr14:104769629-104769630:- | [25] | |
| Sequence | CCCCCTTCCCCTTCTGTGTCACAGTTCTTGGTGACTGTCCC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000555528.5; ENST00000554581.5; ENST00000553506.5; ENST00000557552.1; ENST00000555458.5; ENST00000649815.1; ENST00000402615.6; ENST00000349310.7; ENST00000407796.6; ENST00000610370.1 | ||
| External Link | RMBase: m6A_site_266110 | ||
| mod ID: M6ASITE021754 | Click to Show/Hide the Full List | ||
| mod site | chr14:104769662-104769663:- | [23] | |
| Sequence | GGGGGTTTTTAATCTTTGTGACAGGAAAGCCCTCCCCCTTC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000554581.5; ENST00000553506.5; ENST00000402615.6; ENST00000407796.6; ENST00000557552.1; ENST00000555528.5; ENST00000649815.1; ENST00000610370.1; ENST00000555458.5; ENST00000349310.7 | ||
| External Link | RMBase: m6A_site_266111 | ||
| mod ID: M6ASITE021755 | Click to Show/Hide the Full List | ||
| mod site | chr14:104769738-104769739:- | [23] | |
| Sequence | GCCAGGGTTTACCCAGTGGGACAGAGGAGCAAGGTTTAAAT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000610370.1; ENST00000407796.6; ENST00000554581.5; ENST00000555528.5; ENST00000349310.7; ENST00000555458.5; ENST00000553506.5; ENST00000557552.1; ENST00000402615.6; ENST00000649815.1 | ||
| External Link | RMBase: m6A_site_266112 | ||
| mod ID: M6ASITE021756 | Click to Show/Hide the Full List | ||
| mod site | chr14:104769748-104769749:- | [23] | |
| Sequence | TGGGGGATGGGCCAGGGTTTACCCAGTGGGACAGAGGAGCA | ||
| Motif Score | 2.052208333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000557552.1; ENST00000555458.5; ENST00000553506.5; ENST00000555528.5; ENST00000407796.6; ENST00000402615.6; ENST00000554581.5; ENST00000610370.1; ENST00000649815.1; ENST00000349310.7 | ||
| External Link | RMBase: m6A_site_266113 | ||
| mod ID: M6ASITE021757 | Click to Show/Hide the Full List | ||
| mod site | chr14:104770054-104770055:- | [26] | |
| Sequence | CACGTAGGGAAATGTTAAGGACTTCTGCAGCTATGCGCAAT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; A549; CD8T; iSLK | ||
| Seq Type List | m6A-seq; m6A-CLIP/IP; MeRIP-seq | ||
| Transcript ID List | ENST00000554192.5; ENST00000555458.5; ENST00000649815.1; ENST00000553506.5; ENST00000557552.1; ENST00000610370.1; ENST00000554581.5; ENST00000402615.6; ENST00000555528.5; ENST00000407796.6; ENST00000349310.7 | ||
| External Link | RMBase: m6A_site_266118 | ||
| mod ID: M6ASITE021758 | Click to Show/Hide the Full List | ||
| mod site | chr14:104770073-104770074:- | [23] | |
| Sequence | CCCTCAGAACAATCCGATTCACGTAGGGAAATGTTAAGGAC | ||
| Motif Score | 2.021232143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000554581.5; ENST00000649815.1; ENST00000407796.6; ENST00000402615.6; ENST00000610370.1; ENST00000555528.5; ENST00000349310.7; ENST00000555458.5; ENST00000553506.5; ENST00000557552.1; ENST00000554192.5 | ||
| External Link | RMBase: m6A_site_266119 | ||
| mod ID: M6ASITE021759 | Click to Show/Hide the Full List | ||
| mod site | chr14:104770085-104770086:- | [24] | |
| Sequence | GTGTGGCCTCAGCCCTCAGAACAATCCGATTCACGTAGGGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T; HeLa; A549; iSLK | ||
| Seq Type List | MAZTER-seq; m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000349310.7; ENST00000553506.5; ENST00000407796.6; ENST00000555458.5; ENST00000649815.1; ENST00000554192.5; ENST00000402615.6; ENST00000554581.5; ENST00000610370.1; ENST00000555528.5; ENST00000557552.1 | ||
| External Link | RMBase: m6A_site_266120 | ||
| mod ID: M6ASITE021760 | Click to Show/Hide the Full List | ||
| mod site | chr14:104770150-104770151:- | [26] | |
| Sequence | GCAGCGGGGTAGGGAAGAAAACTATCCTGCGGGTTTTAATT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000554192.5; ENST00000407796.6; ENST00000554581.5; ENST00000402615.6; ENST00000610370.1; ENST00000649815.1; ENST00000555528.5; ENST00000557552.1; ENST00000553506.5; ENST00000555458.5; ENST00000349310.7 | ||
| External Link | RMBase: m6A_site_266123 | ||
| mod ID: M6ASITE021761 | Click to Show/Hide the Full List | ||
| mod site | chr14:104770202-104770203:- | [27] | |
| Sequence | TCTCGAGCCCAGATGGAAAGACGTTTTTGTGCTGTGGGCAG | ||
| Motif Score | 2.871321429 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000610370.1; ENST00000553506.5; ENST00000555458.5; ENST00000554192.5; ENST00000554581.5; ENST00000555528.5; ENST00000649815.1; ENST00000554585.5; ENST00000407796.6; ENST00000557552.1; ENST00000544168.5; ENST00000349310.7; ENST00000402615.6 | ||
| External Link | RMBase: m6A_site_266124 | ||
| mod ID: M6ASITE021762 | Click to Show/Hide the Full List | ||
| mod site | chr14:104770328-104770329:- | [28] | |
| Sequence | ACGGCCTGAGGCGGCGGTGGACTGCGCTGGACGATAGCTTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HepG2; HEK293T; MM6; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000554585.5; ENST00000407796.6; ENST00000349310.7; ENST00000402615.6; ENST00000554581.5; ENST00000610370.1; ENST00000544168.5; ENST00000555528.5; ENST00000554192.5; ENST00000553506.5; ENST00000555458.5; ENST00000557552.1; ENST00000649815.1 | ||
| External Link | RMBase: m6A_site_266127 | ||
| mod ID: M6ASITE021763 | Click to Show/Hide the Full List | ||
| mod site | chr14:104770398-104770399:- | [29] | |
| Sequence | TGACAGCATGGAGTGTGTGGACAGCGAGCGCAGGCCCCACT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; MM6; peripheral-blood; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000553506.5; ENST00000554192.5; ENST00000554848.5; ENST00000402615.6; ENST00000554581.5; ENST00000544168.5; ENST00000557552.1; ENST00000610370.1; ENST00000649815.1; ENST00000555528.5; ENST00000554585.5; ENST00000555458.5; ENST00000407796.6; ENST00000349310.7 | ||
| External Link | RMBase: m6A_site_266128 | ||
| mod ID: M6ASITE021764 | Click to Show/Hide the Full List | ||
| mod site | chr14:104770759-104770760:- | [25] | |
| Sequence | CGGCCCAGATGATCACCATCACACCACCTGACCAAGGTGAG | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | kidney; liver; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000555458.5; ENST00000407796.6; ENST00000610370.1; ENST00000649815.1; ENST00000553506.5; ENST00000554581.5; ENST00000554585.5; ENST00000554848.5; ENST00000554192.5; ENST00000557552.1; ENST00000349310.7; ENST00000555528.5; ENST00000402615.6; ENST00000544168.5 | ||
| External Link | RMBase: m6A_site_266129 | ||
| mod ID: M6ASITE021765 | Click to Show/Hide the Full List | ||
| mod site | chr14:104770806-104770807:- | [24] | |
| Sequence | CCAGGTCACGTCGGAGACTGACACCAGGTATTTTGATGAGG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000407796.6; ENST00000554585.5; ENST00000557552.1; ENST00000649815.1; ENST00000554192.5; ENST00000349310.7; ENST00000402615.6; ENST00000554581.5; ENST00000553506.5; ENST00000555458.5; ENST00000554848.5; ENST00000555528.5; ENST00000544168.5; ENST00000610370.1 | ||
| External Link | RMBase: m6A_site_266130 | ||
| mod ID: M6ASITE021766 | Click to Show/Hide the Full List | ||
| mod site | chr14:104770810-104770811:- | [30] | |
| Sequence | AGCCCCAGGTCACGTCGGAGACTGACACCAGGTATTTTGAT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HepG2; Huh7; peripheral-blood; HEK293T; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000554581.5; ENST00000555528.5; ENST00000555458.5; ENST00000554848.5; ENST00000402615.6; ENST00000554192.5; ENST00000349310.7; ENST00000557552.1; ENST00000407796.6; ENST00000553506.5; ENST00000554585.5; ENST00000610370.1; ENST00000649815.1; ENST00000544168.5 | ||
| External Link | RMBase: m6A_site_266131 | ||
| mod ID: M6ASITE021767 | Click to Show/Hide the Full List | ||
| mod site | chr14:104770949-104770950:- | [31] | |
| Sequence | GACATCAGTCCTGCCTGGAGACCCCTTGGAGATCCAGGTGC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000553506.5; ENST00000554192.5; ENST00000554848.5; ENST00000554581.5; ENST00000555458.5; ENST00000610370.1; ENST00000554585.5; ENST00000649815.1; ENST00000407796.6; ENST00000402615.6; ENST00000349310.7; ENST00000544168.5; ENST00000557552.1; ENST00000555528.5 | ||
| External Link | RMBase: m6A_site_266132 | ||
| mod ID: M6ASITE021768 | Click to Show/Hide the Full List | ||
| mod site | chr14:104771399-104771400:- | [29] | |
| Sequence | AGGGGGTCCTTGACTGTGGGACCTGAGGGGCTCTCTGGCAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; H1A; H1B; GM12878; HEK293A-TOA; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000557552.1; ENST00000554581.5; ENST00000554585.5; ENST00000544168.5; ENST00000349310.7; ENST00000402615.6; ENST00000555458.5; ENST00000554192.5; ENST00000554848.5; ENST00000555528.5; ENST00000407796.6; ENST00000649815.1; ENST00000553506.5; ENST00000610370.1 | ||
| External Link | RMBase: m6A_site_266133 | ||
| mod ID: M6ASITE021769 | Click to Show/Hide the Full List | ||
| mod site | chr14:104771442-104771443:- | [29] | |
| Sequence | AATGCTGGTGCCCAGCCCAGACTCTGGGGTGGGGCAGTGCT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; H1A; H1B; GM12878; HEK293A-TOA; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000349310.7; ENST00000407796.6; ENST00000544168.5; ENST00000555458.5; ENST00000554581.5; ENST00000554192.5; ENST00000610370.1; ENST00000553506.5; ENST00000555528.5; ENST00000554585.5; ENST00000554848.5; ENST00000649815.1; ENST00000557552.1; ENST00000402615.6 | ||
| External Link | RMBase: m6A_site_266134 | ||
| mod ID: M6ASITE021770 | Click to Show/Hide the Full List | ||
| mod site | chr14:104771464-104771465:- | [29] | |
| Sequence | ACCTTGCTAATTGACAATGGACAATGCTGGTGCCCAGCCCA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; H1A; H1B; GM12878; HEK293A-TOA; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000553506.5; ENST00000402615.6; ENST00000554848.5; ENST00000555528.5; ENST00000544168.5; ENST00000649815.1; ENST00000555458.5; ENST00000557552.1; ENST00000554192.5; ENST00000407796.6; ENST00000554585.5; ENST00000554581.5; ENST00000349310.7; ENST00000610370.1 | ||
| External Link | RMBase: m6A_site_266135 | ||
| mod ID: M6ASITE021771 | Click to Show/Hide the Full List | ||
| mod site | chr14:104771484-104771485:- | [29] | |
| Sequence | CTGCCCAGCCTTGACCAGAGACCTTGCTAATTGACAATGGA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; H1A; HEK293A-TOA; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000349310.7; ENST00000554585.5; ENST00000649815.1; ENST00000544168.5; ENST00000610370.1; ENST00000555528.5; ENST00000554581.5; ENST00000554192.5; ENST00000554848.5; ENST00000555458.5; ENST00000402615.6; ENST00000407796.6; ENST00000553506.5; ENST00000557552.1 | ||
| External Link | RMBase: m6A_site_266136 | ||
| mod ID: M6ASITE021772 | Click to Show/Hide the Full List | ||
| mod site | chr14:104771633-104771634:- | [29] | |
| Sequence | TCGGGCTGGGCAGCCCTGGGACCCAGCTGGGCACCTGTTCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; HEK293A-TOA; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000544168.5; ENST00000554581.5; ENST00000557552.1; ENST00000407796.6; ENST00000349310.7; ENST00000554848.5; ENST00000402615.6; ENST00000555528.5; ENST00000554585.5; ENST00000555458.5; ENST00000553506.5; ENST00000554192.5; ENST00000610370.1; ENST00000649815.1 | ||
| External Link | RMBase: m6A_site_266137 | ||
| mod ID: M6ASITE021773 | Click to Show/Hide the Full List | ||
| mod site | chr14:104771813-104771814:- | [32] | |
| Sequence | TGGGTGCTCATCTTTCAGGGACCCTAGGAGCCCTGGCCATC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000544168.5; ENST00000555528.5; ENST00000349310.7; ENST00000554581.5; ENST00000554585.5; ENST00000553506.5; ENST00000402615.6; ENST00000649815.1; ENST00000555458.5; ENST00000407796.6; ENST00000557552.1; ENST00000554192.5; ENST00000554848.5; ENST00000610370.1 | ||
| External Link | RMBase: m6A_site_266138 | ||
| mod ID: M6ASITE021774 | Click to Show/Hide the Full List | ||
| mod site | chr14:104771908-104771909:- | [29] | |
| Sequence | CTGGCTGTGCAGGGACAGGGACAGCACCTACTTTTCTGGCA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000649815.1; ENST00000555458.5; ENST00000554585.5; ENST00000544168.5; ENST00000554192.5; ENST00000553506.5; ENST00000402615.6; ENST00000554581.5; ENST00000610370.1; ENST00000555528.5; ENST00000557552.1; ENST00000407796.6; ENST00000554848.5; ENST00000349310.7 | ||
| External Link | RMBase: m6A_site_266139 | ||
| mod ID: M6ASITE021775 | Click to Show/Hide the Full List | ||
| mod site | chr14:104771914-104771915:- | [29] | |
| Sequence | CTATTTCTGGCTGTGCAGGGACAGGGACAGCACCTACTTTT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000349310.7; ENST00000554581.5; ENST00000553506.5; ENST00000554848.5; ENST00000544168.5; ENST00000557552.1; ENST00000555528.5; ENST00000610370.1; ENST00000554192.5; ENST00000407796.6; ENST00000649815.1; ENST00000402615.6; ENST00000555458.5; ENST00000554585.5 | ||
| External Link | RMBase: m6A_site_266140 | ||
| mod ID: M6ASITE021776 | Click to Show/Hide the Full List | ||
| mod site | chr14:104772013-104772014:- | [29] | |
| Sequence | TGAAGGCTGGCTGGTGGTGGACCAGGGGTGCTGCCCCTTGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000554581.5; ENST00000349310.7; ENST00000555528.5; ENST00000553506.5; ENST00000407796.6; ENST00000557552.1; ENST00000544168.5; ENST00000555458.5; ENST00000649815.1; ENST00000554848.5; ENST00000402615.6; ENST00000610370.1; ENST00000554192.5; ENST00000554585.5 | ||
| External Link | RMBase: m6A_site_266141 | ||
| mod ID: M6ASITE021777 | Click to Show/Hide the Full List | ||
| mod site | chr14:104772889-104772890:- | [29] | |
| Sequence | TTCAGGGCTGCTCAAGAAGGACCCCAAGCAGAGGTGAGGGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; peripheral-blood; HEK293T; iSLK; MSC; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000554585.5; ENST00000553506.5; ENST00000554192.5; ENST00000554581.5; ENST00000610370.1; ENST00000649815.1; ENST00000544168.5; ENST00000555458.5; ENST00000402615.6; ENST00000554848.5; ENST00000349310.7; ENST00000407796.6; ENST00000555528.5; ENST00000557552.1 | ||
| External Link | RMBase: m6A_site_266142 | ||
| mod ID: M6ASITE021778 | Click to Show/Hide the Full List | ||
| mod site | chr14:104772991-104772992:- | [29] | |
| Sequence | CCTGCCCTTCTACAACCAGGACCATGAGAAGCTTTTTGAGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; LCLs; A549; H1299; MM6; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000554848.5; ENST00000553506.5; ENST00000544168.5; ENST00000557552.1; ENST00000349310.7; ENST00000554192.5; ENST00000554581.5; ENST00000402615.6; ENST00000555458.5; ENST00000407796.6; ENST00000649815.1; ENST00000554585.5; ENST00000555528.5 | ||
| External Link | RMBase: m6A_site_266143 | ||
| mod ID: M6ASITE021779 | Click to Show/Hide the Full List | ||
| mod site | chr14:104773000-104773001:- | [24] | |
| Sequence | GTGCGGTCGCCTGCCCTTCTACAACCAGGACCATGAGAAGC | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000554848.5; ENST00000553506.5; ENST00000554581.5; ENST00000554192.5; ENST00000349310.7; ENST00000555458.5; ENST00000407796.6; ENST00000554585.5; ENST00000557552.1; ENST00000402615.6; ENST00000544168.5; ENST00000555528.5; ENST00000649815.1 | ||
| External Link | RMBase: m6A_site_266144 | ||
| mod ID: M6ASITE021780 | Click to Show/Hide the Full List | ||
| mod site | chr14:104773057-104773058:- | [29] | |
| Sequence | TGACTACGGCCGTGCAGTGGACTGGTGGGGGCTGGGCGTGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; HEK293T; A549; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000553506.5; ENST00000407796.6; ENST00000554848.5; ENST00000555528.5; ENST00000554192.5; ENST00000349310.7; ENST00000555458.5; ENST00000649815.1; ENST00000554585.5; ENST00000544168.5; ENST00000557552.1; ENST00000402615.6; ENST00000554581.5 | ||
| External Link | RMBase: m6A_site_266145 | ||
| mod ID: M6ASITE021781 | Click to Show/Hide the Full List | ||
| mod site | chr14:104773081-104773082:- | [29] | |
| Sequence | ACCCGTGCAGGTGCTGGAGGACAATGACTACGGCCGTGCAG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; H1A; H1B; A549; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000555528.5; ENST00000553506.5; ENST00000554192.5; ENST00000557552.1; ENST00000407796.6; ENST00000402615.6; ENST00000554585.5; ENST00000554848.5; ENST00000544168.5; ENST00000649815.1; ENST00000554581.5; ENST00000349310.7; ENST00000555458.5 | ||
| External Link | RMBase: m6A_site_266146 | ||
| mod ID: M6ASITE021782 | Click to Show/Hide the Full List | ||
| mod site | chr14:104773134-104773135:- | [29] | |
| Sequence | CTCCCCCGGCAGTGTCCCGGACACCCCTTGATGCCGAGTCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; HEK293T; A549; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000555458.5; ENST00000554581.5; ENST00000349310.7; ENST00000554848.5; ENST00000553506.5; ENST00000554585.5; ENST00000402615.6; ENST00000554192.5; ENST00000557552.1; ENST00000649815.1; ENST00000555528.5; ENST00000544168.5; ENST00000407796.6 | ||
| External Link | RMBase: m6A_site_266147 | ||
| mod ID: M6ASITE021783 | Click to Show/Hide the Full List | ||
| mod site | chr14:104773273-104773274:- | [24] | |
| Sequence | CCATGAAGACCTTTTGCGGCACACCTGAGTACCTGGCCCCC | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000349310.7; ENST00000554848.5; ENST00000544168.5; ENST00000555528.5; ENST00000557552.1; ENST00000649815.1; ENST00000555458.5; ENST00000554585.5; ENST00000407796.6; ENST00000553506.5; ENST00000554192.5; ENST00000554581.5; ENST00000402615.6 | ||
| External Link | RMBase: m6A_site_266148 | ||
| mod ID: M6ASITE021784 | Click to Show/Hide the Full List | ||
| mod site | chr14:104773285-104773286:- | [30] | |
| Sequence | AGGACGGTGCCACCATGAAGACCTTTTGCGGCACACCTGAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HepG2; Huh7; peripheral-blood; HEK293A-TOA; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000554581.5; ENST00000544168.5; ENST00000553506.5; ENST00000349310.7; ENST00000554848.5; ENST00000557552.1; ENST00000554585.5; ENST00000407796.6; ENST00000554192.5; ENST00000402615.6; ENST00000649815.1; ENST00000555528.5; ENST00000555458.5 | ||
| External Link | RMBase: m6A_site_266149 | ||
| mod ID: M6ASITE021785 | Click to Show/Hide the Full List | ||
| mod site | chr14:104773332-104773333:- | [30] | |
| Sequence | CGGGCACATTAAGATCACAGACTTCGGGCTGTGCAAGGAGG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HepG2; Huh7; peripheral-blood; HEK293A-TOA; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000349310.7; ENST00000553506.5; ENST00000407796.6; ENST00000554848.5; ENST00000544168.5; ENST00000557552.1; ENST00000554585.5; ENST00000649815.1; ENST00000402615.6; ENST00000554192.5; ENST00000555528.5; ENST00000554581.5 | ||
| External Link | RMBase: m6A_site_266150 | ||
| mod ID: M6ASITE021786 | Click to Show/Hide the Full List | ||
| mod site | chr14:104773359-104773360:- | [28] | |
| Sequence | GCTGGAGAACCTCATGCTGGACAAGGACGGGCACATTAAGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HepG2; hESC-HEK293T; Huh7; peripheral-blood; HEK293A-TOA; endometrial; AML | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000555528.5; ENST00000349310.7; ENST00000407796.6; ENST00000554848.5; ENST00000557552.1; ENST00000402615.6; ENST00000554192.5; ENST00000544168.5; ENST00000554581.5; ENST00000649815.1; ENST00000554585.5; ENST00000553506.5 | ||
| External Link | RMBase: m6A_site_266151 | ||
| mod ID: M6ASITE021787 | Click to Show/Hide the Full List | ||
| mod site | chr14:104773371-104773372:- | [30] | |
| Sequence | ACCTTTCCCCTAGCTGGAGAACCTCATGCTGGACAAGGACG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HepG2; Huh7; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000349310.7; ENST00000402615.6; ENST00000555528.5; ENST00000407796.6; ENST00000557552.1; ENST00000554848.5; ENST00000553506.5; ENST00000554585.5; ENST00000649815.1; ENST00000554581.5; ENST00000554192.5; ENST00000544168.5 | ||
| External Link | RMBase: m6A_site_266152 | ||
| mod ID: M6ASITE021788 | Click to Show/Hide the Full List | ||
| mod site | chr14:104773461-104773462:- | [30] | |
| Sequence | GAAGAACGTGGTGTACCGGGACCTCAAGGTGCGCTGGCGGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2; Huh7; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000554192.5; ENST00000554581.5; ENST00000557552.1; ENST00000555528.5; ENST00000554848.5; ENST00000349310.7; ENST00000553506.5; ENST00000407796.6; ENST00000649815.1; ENST00000402615.6; ENST00000544168.5 | ||
| External Link | RMBase: m6A_site_266153 | ||
| mod ID: M6ASITE021789 | Click to Show/Hide the Full List | ||
| mod site | chr14:104773497-104773498:- | [30] | |
| Sequence | TGAGATTGTGTCAGCCCTGGACTACCTGCACTCGGAGAAGA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HepG2; Huh7; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000553506.5; ENST00000349310.7; ENST00000557552.1; ENST00000649815.1; ENST00000554848.5; ENST00000544168.5; ENST00000407796.6; ENST00000555528.5; ENST00000402615.6; ENST00000554581.5 | ||
| External Link | RMBase: m6A_site_266154 | ||
| mod ID: M6ASITE021790 | Click to Show/Hide the Full List | ||
| mod site | chr14:104773539-104773540:- | [30] | |
| Sequence | GGAGCGTGTGTTCTCCGAGGACCGGGCCCGCTTCTATGGCG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000553506.5; ENST00000554848.5; ENST00000349310.7; ENST00000649815.1; ENST00000554581.5; ENST00000407796.6; ENST00000557552.1; ENST00000402615.6; ENST00000555528.5; ENST00000544168.5 | ||
| External Link | RMBase: m6A_site_266155 | ||
| mod ID: M6ASITE021791 | Click to Show/Hide the Full List | ||
| mod site | chr14:104773649-104773650:- | [29] | |
| Sequence | GCCGAGACCCGGTCTGAGAAACCCCAGCCCTGCAGGCCATG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000557552.1; ENST00000555528.5; ENST00000349310.7; ENST00000554848.5; ENST00000554581.5; ENST00000553506.5; ENST00000402615.6; ENST00000544168.5; ENST00000649815.1; ENST00000407796.6 | ||
| External Link | RMBase: m6A_site_266156 | ||
| mod ID: M6ASITE021792 | Click to Show/Hide the Full List | ||
| mod site | chr14:104773663-104773664:- | [29] | |
| Sequence | CTTCCCTGTGCAATGCCGAGACCCGGTCTGAGAAACCCCAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000557552.1; ENST00000349310.7; ENST00000649815.1; ENST00000554581.5; ENST00000544168.5; ENST00000553506.5; ENST00000407796.6; ENST00000555528.5; ENST00000402615.6; ENST00000554848.5 | ||
| External Link | RMBase: m6A_site_266157 | ||
| mod ID: M6ASITE021793 | Click to Show/Hide the Full List | ||
| mod site | chr14:104773719-104773720:- | [29] | |
| Sequence | GCTGAGTTAGGGCTTCTGAGACTTTCCAGGCCCCCGTGCCC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000649815.1; ENST00000349310.7; ENST00000402615.6; ENST00000544168.5; ENST00000553506.5; ENST00000554581.5; ENST00000554848.5; ENST00000407796.6; ENST00000555528.5; ENST00000557552.1 | ||
| External Link | RMBase: m6A_site_266158 | ||
| mod ID: M6ASITE021794 | Click to Show/Hide the Full List | ||
| mod site | chr14:104773884-104773885:- | [29] | |
| Sequence | GCTGGGGCTGCGGGGGATGGACTTCGCGGCCTGTGGGCCGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; CD4T; peripheral-blood; GSC-11; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000554848.5; ENST00000544168.5; ENST00000554581.5; ENST00000349310.7; ENST00000554826.1; ENST00000402615.6; ENST00000557552.1; ENST00000555528.5; ENST00000553506.5; ENST00000407796.6; ENST00000649815.1 | ||
| External Link | RMBase: m6A_site_266159 | ||
| mod ID: M6ASITE021795 | Click to Show/Hide the Full List | ||
| mod site | chr14:104773958-104773959:- | [29] | |
| Sequence | CCCTGAAGTACTCTTTCCAGACCCACGACCGCCTCTGCTTT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; A549; MM6; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000554848.5; ENST00000544168.5; ENST00000557552.1; ENST00000402615.6; ENST00000649815.1; ENST00000554826.1; ENST00000407796.6; ENST00000555528.5; ENST00000553506.5; ENST00000349310.7; ENST00000554581.5 | ||
| External Link | RMBase: m6A_site_266160 | ||
| mod ID: M6ASITE021796 | Click to Show/Hide the Full List | ||
| mod site | chr14:104774804-104774805:- | [29] | |
| Sequence | TGGACTCCGCCGCTGGTGAGACAGGCCTGCCCTGGAGGGTC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000544168.5; ENST00000557552.1; ENST00000649815.1; ENST00000407796.6; ENST00000555528.5; ENST00000554581.5; ENST00000554848.5; ENST00000349310.7; ENST00000554826.1; ENST00000556836.1; ENST00000402615.6 | ||
| External Link | RMBase: m6A_site_266161 | ||
| mod ID: M6ASITE021797 | Click to Show/Hide the Full List | ||
| mod site | chr14:104774821-104774822:- | [29] | |
| Sequence | GAGGGCTTTACACACCGTGGACTCCGCCGCTGGTGAGACAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000555528.5; ENST00000649815.1; ENST00000556836.1; ENST00000349310.7; ENST00000557552.1; ENST00000554581.5; ENST00000544168.5; ENST00000407796.6; ENST00000554826.1; ENST00000554848.5; ENST00000402615.6 | ||
| External Link | RMBase: m6A_site_266162 | ||
| mod ID: M6ASITE021798 | Click to Show/Hide the Full List | ||
| mod site | chr14:104774939-104774940:- | [25] | |
| Sequence | ACTCCAGGCACCCCTTCCTCACAGTGAGTGGGAGCCCAGAT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000557552.1; ENST00000349310.7; ENST00000407796.6; ENST00000402615.6; ENST00000554581.5; ENST00000556836.1; ENST00000554826.1; ENST00000544168.5; ENST00000555528.5; ENST00000649815.1; ENST00000554848.5 | ||
| External Link | RMBase: m6A_site_266163 | ||
| mod ID: M6ASITE021799 | Click to Show/Hide the Full List | ||
| mod site | chr14:104774959-104774960:- | [29] | |
| Sequence | CGAGAACCGCGTCCTGCAGAACTCCAGGCACCCCTTCCTCA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; A549; MM6; CD4T; peripheral-blood; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000555528.5; ENST00000554581.5; ENST00000349310.7; ENST00000649815.1; ENST00000407796.6; ENST00000402615.6; ENST00000557552.1; ENST00000554848.5; ENST00000556836.1; ENST00000544168.5; ENST00000554826.1 | ||
| External Link | RMBase: m6A_site_266164 | ||
| mod ID: M6ASITE021800 | Click to Show/Hide the Full List | ||
| mod site | chr14:104774974-104774975:- | [29] | |
| Sequence | GGCCCACACACTCACCGAGAACCGCGTCCTGCAGAACTCCA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; A549; MM6; CD4T; peripheral-blood; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000554581.5; ENST00000554848.5; ENST00000407796.6; ENST00000649815.1; ENST00000349310.7; ENST00000402615.6; ENST00000544168.5; ENST00000554826.1; ENST00000555528.5; ENST00000556836.1; ENST00000557552.1 | ||
| External Link | RMBase: m6A_site_266165 | ||
| mod ID: M6ASITE021801 | Click to Show/Hide the Full List | ||
| mod site | chr14:104774989-104774990:- | [24] | |
| Sequence | TTTACAGGACGAGGTGGCCCACACACTCACCGAGAACCGCG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000557552.1; ENST00000544168.5; ENST00000556836.1; ENST00000555528.5; ENST00000402615.6; ENST00000554848.5; ENST00000554581.5; ENST00000349310.7; ENST00000407796.6; ENST00000649815.1; ENST00000554826.1 | ||
| External Link | RMBase: m6A_site_266166 | ||
| mod ID: M6ASITE021802 | Click to Show/Hide the Full List | ||
| mod site | chr14:104775206-104775207:- | [29] | |
| Sequence | ACCCTGGTGCCTGCCCATAGACCATGAACGAGTTTGAGTAC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; A549; H1299; MM6; CD4T; peripheral-blood; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000402615.6; ENST00000554581.5; ENST00000557552.1; ENST00000349310.7; ENST00000554848.5; ENST00000555528.5; ENST00000407796.6; ENST00000555380.1; ENST00000544168.5; ENST00000556836.1; ENST00000649815.1; ENST00000554826.1 | ||
| External Link | RMBase: m6A_site_266167 | ||
| mod ID: M6ASITE021803 | Click to Show/Hide the Full List | ||
| mod site | chr14:104775359-104775360:- | [29] | |
| Sequence | TGTGTTCCCTGTAAGCCTGGACTCCCCGCTGAGCCCAGTAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000649815.1; ENST00000554581.5; ENST00000555380.1; ENST00000555528.5; ENST00000554826.1; ENST00000407796.6; ENST00000402615.6; ENST00000544168.5; ENST00000349310.7; ENST00000554848.5; ENST00000556836.1; ENST00000557552.1 | ||
| External Link | RMBase: m6A_site_266168 | ||
| mod ID: M6ASITE021804 | Click to Show/Hide the Full List | ||
| mod site | chr14:104775559-104775560:- | [33] | |
| Sequence | GTCCCAGCTCGGCCCAGGAGACACTCAGCACTCCACGGCAT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | Jurkat; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000649815.1; ENST00000554826.1; ENST00000407796.6; ENST00000402615.6; ENST00000554848.5; ENST00000349310.7; ENST00000555380.1; ENST00000557552.1; ENST00000544168.5; ENST00000555528.5; ENST00000554581.5 | ||
| External Link | RMBase: m6A_site_266169 | ||
| mod ID: M6ASITE021805 | Click to Show/Hide the Full List | ||
| mod site | chr14:104775606-104775607:- | [29] | |
| Sequence | TGTGCCTGGGGCTGCCTTGGACTGTGGAGGGCTGGGTGGGT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; NB4; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000402615.6; ENST00000649815.1; ENST00000555380.1; ENST00000407796.6; ENST00000349310.7; ENST00000554848.5; ENST00000554581.5; ENST00000555528.5; ENST00000554826.1; ENST00000557552.1; ENST00000544168.5 | ||
| External Link | RMBase: m6A_site_266170 | ||
| mod ID: M6ASITE021806 | Click to Show/Hide the Full List | ||
| mod site | chr14:104775706-104775707:- | [24] | |
| Sequence | CCGGTCGGGCTCACCCAGTGACAACTCAGGGGCTGAAGAGA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000544168.5; ENST00000555528.5; ENST00000554848.5; ENST00000554581.5; ENST00000554826.1; ENST00000407796.6; ENST00000402615.6; ENST00000557552.1; ENST00000649815.1; ENST00000555380.1; ENST00000349310.7 | ||
| External Link | RMBase: m6A_site_266171 | ||
| mod ID: M6ASITE021807 | Click to Show/Hide the Full List | ||
| mod site | chr14:104775730-104775731:- | [29] | |
| Sequence | GCAGGAGGAGGAGGAGATGGACTTCCGGTCGGGCTCACCCA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000557552.1; ENST00000349310.7; ENST00000554581.5; ENST00000407796.6; ENST00000555528.5; ENST00000544168.5; ENST00000554826.1; ENST00000649815.1; ENST00000554848.5; ENST00000402615.6; ENST00000555380.1 | ||
| External Link | RMBase: m6A_site_266172 | ||
| mod ID: M6ASITE021808 | Click to Show/Hide the Full List | ||
| mod site | chr14:104775773-104775774:- | [29] | |
| Sequence | AGTGGACAACCGCCATCCAGACTGTGGCTGACGGCCTCAAG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; MM6; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000555528.5; ENST00000349310.7; ENST00000554848.5; ENST00000649815.1; ENST00000402615.6; ENST00000554581.5; ENST00000407796.6; ENST00000544168.5; ENST00000555380.1; ENST00000557552.1; ENST00000554826.1 | ||
| External Link | RMBase: m6A_site_266173 | ||
| mod ID: M6ASITE021809 | Click to Show/Hide the Full List | ||
| mod site | chr14:104775788-104775789:- | [29] | |
| Sequence | CCTCCTGCAGGGAGGAGTGGACAACCGCCATCCAGACTGTG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; MM6; CD4T; peripheral-blood; GSC-11; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000544168.5; ENST00000555528.5; ENST00000555380.1; ENST00000554581.5; ENST00000349310.7; ENST00000554848.5; ENST00000402615.6; ENST00000557552.1; ENST00000554826.1; ENST00000649815.1; ENST00000407796.6 | ||
| External Link | RMBase: m6A_site_266174 | ||
| mod ID: M6ASITE021810 | Click to Show/Hide the Full List | ||
| mod site | chr14:104775889-104775890:- | [29] | |
| Sequence | GGAACCCCGACAGCTGTGGAACCACGCTTGTGAGGCTGGCG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; HEK293T; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000554581.5; ENST00000649815.1; ENST00000402615.6; ENST00000557552.1; ENST00000555528.5; ENST00000554848.5; ENST00000349310.7; ENST00000555380.1; ENST00000407796.6; ENST00000554826.1; ENST00000544168.5 | ||
| External Link | RMBase: m6A_site_266175 | ||
| mod ID: M6ASITE021811 | Click to Show/Hide the Full List | ||
| mod site | chr14:104775906-104775907:- | [29] | |
| Sequence | CCTCAGGGCTGTCTCTGGGAACCCCGACAGCTGTGGAACCA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000554581.5; ENST00000402615.6; ENST00000544168.5; ENST00000554848.5; ENST00000555380.1; ENST00000407796.6; ENST00000349310.7; ENST00000557552.1; ENST00000554826.1; ENST00000555528.5; ENST00000649815.1 | ||
| External Link | RMBase: m6A_site_266176 | ||
| mod ID: M6ASITE021812 | Click to Show/Hide the Full List | ||
| mod site | chr14:104775946-104775947:- | [29] | |
| Sequence | CCCTGAGTGTATGTGGCCAGACCAGGTCAAGTGGTGGCCTC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000557552.1; ENST00000407796.6; ENST00000649815.1; ENST00000402615.6; ENST00000555380.1; ENST00000349310.7; ENST00000554826.1; ENST00000544168.5; ENST00000554581.5; ENST00000555528.5; ENST00000554848.5 | ||
| External Link | RMBase: m6A_site_266177 | ||
| mod ID: M6ASITE021813 | Click to Show/Hide the Full List | ||
| mod site | chr14:104776025-104776026:- | [32] | |
| Sequence | GTTGGAGTCCGGTTGTGGGGACAGAGTCCTGCTGGGGGGCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000349310.7; ENST00000554826.1; ENST00000555528.5; ENST00000554581.5; ENST00000407796.6; ENST00000649815.1; ENST00000554848.5; ENST00000402615.6; ENST00000544168.5; ENST00000557552.1; ENST00000555380.1 | ||
| External Link | RMBase: m6A_site_266178 | ||
| mod ID: M6ASITE021814 | Click to Show/Hide the Full List | ||
| mod site | chr14:104776152-104776153:- | [29] | |
| Sequence | TCCGTCTCAAAAAAAGAAAAACCCGGTGGAGAGGGCCAGTC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000407796.6; ENST00000554848.5; ENST00000555528.5; ENST00000402615.6; ENST00000544168.5; ENST00000554826.1; ENST00000649815.1; ENST00000557552.1; ENST00000349310.7; ENST00000555380.1; rmsk_4276828; ENST00000554581.5 | ||
| External Link | RMBase: m6A_site_266179 | ||
| mod ID: M6ASITE021815 | Click to Show/Hide the Full List | ||
| mod site | chr14:104776174-104776175:- | [29] | |
| Sequence | AGCCAGGGCGATAGAGCAAGACTCCGTCTCAAAAAAAGAAA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000349310.7; rmsk_4276828; ENST00000555380.1; ENST00000555528.5; ENST00000554581.5; ENST00000402615.6; ENST00000649815.1; ENST00000407796.6; ENST00000554848.5; ENST00000544168.5; ENST00000554826.1; ENST00000557552.1 | ||
| External Link | RMBase: m6A_site_266180 | ||
| mod ID: M6ASITE021816 | Click to Show/Hide the Full List | ||
| mod site | chr14:104776242-104776243:- | [29] | |
| Sequence | GAGGCACAAGAATCGCTTGAACCCGAGAGGCAGAGGCTGCA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000557552.1; ENST00000554581.5; ENST00000554826.1; ENST00000555528.5; ENST00000555380.1; ENST00000349310.7; ENST00000407796.6; rmsk_4276828; ENST00000544168.5; ENST00000554848.5; ENST00000649815.1; ENST00000402615.6 | ||
| External Link | RMBase: m6A_site_266181 | ||
| mod ID: M6ASITE021817 | Click to Show/Hide the Full List | ||
| mod site | chr14:104776444-104776445:- | [29] | |
| Sequence | GAGATAAAAACTGGCTAGGGACCGGGTGCAGTGGCTCACGT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000544168.5; ENST00000402615.6; ENST00000557552.1; rmsk_4276828; ENST00000407796.6; ENST00000554848.5; ENST00000554826.1; ENST00000554581.5; ENST00000555528.5; ENST00000649815.1; ENST00000349310.7; ENST00000555380.1 | ||
| External Link | RMBase: m6A_site_266182 | ||
| mod ID: M6ASITE021818 | Click to Show/Hide the Full List | ||
| mod site | chr14:104776455-104776456:- | [29] | |
| Sequence | AACTGAGGCTGGAGATAAAAACTGGCTAGGGACCGGGTGCA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000544168.5; ENST00000649815.1; ENST00000349310.7; ENST00000555528.5; ENST00000554826.1; ENST00000554581.5; ENST00000555380.1; ENST00000557552.1; ENST00000402615.6; ENST00000554848.5; ENST00000407796.6 | ||
| External Link | RMBase: m6A_site_266183 | ||
| mod ID: M6ASITE021819 | Click to Show/Hide the Full List | ||
| mod site | chr14:104776474-104776475:- | [29] | |
| Sequence | TGTGGGGTTTGGGTGGGGAAACTGAGGCTGGAGATAAAAAC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000407796.6; ENST00000554581.5; ENST00000555528.5; ENST00000402615.6; ENST00000649815.1; ENST00000349310.7; ENST00000557552.1; ENST00000555380.1; ENST00000544168.5; ENST00000554826.1; ENST00000554848.5 | ||
| External Link | RMBase: m6A_site_266184 | ||
| mod ID: M6ASITE021820 | Click to Show/Hide the Full List | ||
| mod site | chr14:104776511-104776512:- | [29] | |
| Sequence | CCTCAGAGCCCAGTTTGGAAACTGGCCCAGTCCTGCCTGTG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000649815.1; ENST00000555380.1; ENST00000544168.5; ENST00000555528.5; ENST00000349310.7; ENST00000554848.5; ENST00000554581.5; ENST00000402615.6; ENST00000554826.1; ENST00000407796.6; ENST00000557552.1 | ||
| External Link | RMBase: m6A_site_266185 | ||
| mod ID: M6ASITE021821 | Click to Show/Hide the Full List | ||
| mod site | chr14:104776600-104776601:- | [29] | |
| Sequence | AGCCCCAGCGCTTAGGAGGGACACGGGGATGGCGGGGTGCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000554826.1; ENST00000544168.5; ENST00000349310.7; ENST00000555528.5; ENST00000555380.1; ENST00000554581.5; ENST00000554848.5; ENST00000649815.1; ENST00000402615.6; ENST00000407796.6; ENST00000557552.1 | ||
| External Link | RMBase: m6A_site_266186 | ||
| mod ID: M6ASITE021822 | Click to Show/Hide the Full List | ||
| mod site | chr14:104776671-104776672:- | [29] | |
| Sequence | AACGCACCTTCCATGTGGAGACTCCTGAGGAGCGGTACGTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000407796.6; ENST00000544168.5; ENST00000402615.6; ENST00000555380.1; ENST00000557552.1; ENST00000649815.1; ENST00000554581.5; ENST00000555528.5; ENST00000554826.1; ENST00000554848.5; ENST00000555926.1; ENST00000349310.7 | ||
| External Link | RMBase: m6A_site_266187 | ||
| mod ID: M6ASITE021823 | Click to Show/Hide the Full List | ||
| mod site | chr14:104776704-104776705:- | [29] | |
| Sequence | TCATCCGCTGCCTGCAGTGGACCACTGTCATCGAACGCACC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000402615.6; ENST00000544168.5; ENST00000555528.5; ENST00000557552.1; ENST00000407796.6; ENST00000649815.1; ENST00000349310.7; ENST00000555926.1; ENST00000554848.5; ENST00000555380.1; ENST00000554581.5 | ||
| External Link | RMBase: m6A_site_266188 | ||
| mod ID: M6ASITE021824 | Click to Show/Hide the Full List | ||
| mod site | chr14:104776733-104776734:- | [24] | |
| Sequence | GACGGAGCGGCCCCGGCCCAACACCTTCATCATCCGCTGCC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000407796.6; ENST00000349310.7; ENST00000555380.1; ENST00000555528.5; ENST00000649815.1; ENST00000555926.1; ENST00000544168.5; ENST00000554848.5; ENST00000557552.1; ENST00000402615.6; ENST00000554581.5 | ||
| External Link | RMBase: m6A_site_266189 | ||
| mod ID: M6ASITE021825 | Click to Show/Hide the Full List | ||
| mod site | chr14:104776792-104776793:- | [29] | |
| Sequence | GGGCAGGGGGATGCACGCAGACAGAGGCTCTGCCTGCCCGC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000557552.1; ENST00000554581.5; ENST00000554848.5; ENST00000649815.1; ENST00000544168.5; ENST00000349310.7; ENST00000407796.6; ENST00000555926.1; ENST00000555528.5; ENST00000402615.6; ENST00000555380.1 | ||
| External Link | RMBase: m6A_site_266190 | ||
| mod ID: M6ASITE021826 | Click to Show/Hide the Full List | ||
| mod site | chr14:104776878-104776879:- | [29] | |
| Sequence | AAGAGATGGGGCTTCCCAGGACCTGGTGGGTGGTATGCAAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000349310.7; ENST00000402615.6; ENST00000554581.5; ENST00000555528.5; ENST00000544168.5; ENST00000557552.1; ENST00000555380.1; ENST00000407796.6; ENST00000649815.1; ENST00000555926.1; ENST00000554848.5 | ||
| External Link | RMBase: m6A_site_266191 | ||
| mod ID: M6ASITE021827 | Click to Show/Hide the Full List | ||
| mod site | chr14:104777154-104777155:- | [29] | |
| Sequence | ACATTTCTACACTAGGTGGGACAAGCAGGTTTCTGGGGTGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000407796.6; ENST00000649815.1; ENST00000555926.1; ENST00000555380.1; ENST00000554848.5; ENST00000554581.5; ENST00000349310.7; ENST00000544168.5; ENST00000557552.1; ENST00000555528.5; ENST00000402615.6 | ||
| External Link | RMBase: m6A_site_266192 | ||
| mod ID: M6ASITE021828 | Click to Show/Hide the Full List | ||
| mod site | chr14:104777174-104777175:- | [29] | |
| Sequence | GAGGTTGGGAAAAAAGCAGGACATTTCTACACTAGGTGGGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000544168.5; ENST00000649815.1; ENST00000402615.6; ENST00000555926.1; ENST00000554581.5; ENST00000555528.5; ENST00000349310.7; ENST00000555380.1; ENST00000407796.6; ENST00000554848.5; ENST00000557552.1 | ||
| External Link | RMBase: m6A_site_266193 | ||
| mod ID: M6ASITE021829 | Click to Show/Hide the Full List | ||
| mod site | chr14:104778244-104778245:- | [29] | |
| Sequence | CGTGCCTGGTGGCACCTGGGACAGGGCCTTGCTGGGGGTGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000557552.1; ENST00000554581.5; ENST00000555528.5; ENST00000555926.1; ENST00000349310.7; ENST00000402615.6; ENST00000555380.1; ENST00000649815.1; ENST00000407796.6; ENST00000554848.5 | ||
| External Link | RMBase: m6A_site_266194 | ||
| mod ID: M6ASITE021830 | Click to Show/Hide the Full List | ||
| mod site | chr14:104780104-104780105:- | [24] | |
| Sequence | CCAACGTGAGGCTCCCCTCAACAACTTCTCTGTGGCGCGTA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000555926.1; ENST00000407796.6; ENST00000555380.1; ENST00000649815.1; ENST00000554581.5; ENST00000554848.5; ENST00000555528.5; ENST00000349310.7; ENST00000402615.6 | ||
| External Link | RMBase: m6A_site_266195 | ||
| mod ID: M6ASITE021831 | Click to Show/Hide the Full List | ||
| mod site | chr14:104780125-104780126:- | [29] | |
| Sequence | GGAGCGGCCGCAGGATGTGGACCAACGTGAGGCTCCCCTCA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; MT4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000554581.5; ENST00000349310.7; ENST00000649815.1; ENST00000402615.6; ENST00000554848.5; ENST00000555528.5; ENST00000555926.1; ENST00000407796.6; ENST00000555380.1 | ||
| External Link | RMBase: m6A_site_266196 | ||
| mod ID: M6ASITE021832 | Click to Show/Hide the Full List | ||
| mod site | chr14:104780149-104780150:- | [24] | |
| Sequence | TGATGGCACCTTCATTGGCTACAAGGAGCGGCCGCAGGATG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000349310.7; ENST00000555528.5; ENST00000555380.1; ENST00000554581.5; ENST00000554848.5; ENST00000649815.1; ENST00000407796.6; ENST00000402615.6; ENST00000555926.1 | ||
| External Link | RMBase: m6A_site_266197 | ||
| mod ID: M6ASITE021833 | Click to Show/Hide the Full List | ||
| mod site | chr14:104780201-104780202:- | [29] | |
| Sequence | CTGTAGGGGAGTACATCAAGACCTGGCGGCCACGCTACTTC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; MT4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000402615.6; ENST00000555380.1; ENST00000554581.5; ENST00000349310.7; ENST00000554848.5; ENST00000407796.6; ENST00000555926.1; ENST00000555528.5; ENST00000649815.1 | ||
| External Link | RMBase: m6A_site_266198 | ||
| mod ID: M6ASITE021834 | Click to Show/Hide the Full List | ||
| mod site | chr14:104780209-104780210:- | [24] | |
| Sequence | CCGCACGTCTGTAGGGGAGTACATCAAGACCTGGCGGCCAC | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000555380.1; ENST00000649815.1; ENST00000555528.5; ENST00000402615.6; ENST00000554581.5; ENST00000407796.6; ENST00000554848.5; ENST00000349310.7; ENST00000555926.1 | ||
| External Link | RMBase: m6A_site_266199 | ||
| mod ID: M6ASITE021835 | Click to Show/Hide the Full List | ||
| mod site | chr14:104792951-104792952:- | [29] | |
| Sequence | GAGAGGCCCAGGTCCAGGGAACTGTGTTCAATTACATGGAT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; MT4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000554848.5; ENST00000402615.6; ENST00000349310.7; ENST00000649815.1; ENST00000555926.1; ENST00000554581.5; ENST00000555528.5; ENST00000407796.6 | ||
| External Link | RMBase: m6A_site_266200 | ||
| mod ID: M6ASITE021836 | Click to Show/Hide the Full List | ||
| mod site | chr14:104793242-104793243:- | [29] | |
| Sequence | CATGGGTAGGAACACCATGGACAGGGAGAGCAAACGGGGCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000554848.5; ENST00000554581.5; ENST00000349310.7; ENST00000557494.1; ENST00000555528.5; ENST00000402615.6; ENST00000555926.1; ENST00000407796.6; ENST00000649815.1 | ||
| External Link | RMBase: m6A_site_266201 | ||
| mod ID: M6ASITE021837 | Click to Show/Hide the Full List | ||
| mod site | chr14:104793251-104793252:- | [29] | |
| Sequence | CCCAGGATCCATGGGTAGGAACACCATGGACAGGGAGAGCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000557494.1; ENST00000554581.5; ENST00000402615.6; ENST00000555528.5; ENST00000407796.6; ENST00000554848.5; ENST00000555926.1; ENST00000349310.7; ENST00000649815.1 | ||
| External Link | RMBase: m6A_site_266202 | ||
| mod ID: M6ASITE021838 | Click to Show/Hide the Full List | ||
| mod site | chr14:104793374-104793375:- | [29] | |
| Sequence | CTTGCGCTGGAGGGCTCTGGACTCCCGTTTGCGCCAGTGGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000402615.6; ENST00000407796.6; ENST00000557494.1; ENST00000555528.5; ENST00000554581.5; ENST00000555926.1; ENST00000349310.7; ENST00000649815.1; ENST00000554848.5 | ||
| External Link | RMBase: m6A_site_266203 | ||
| mod ID: M6ASITE021839 | Click to Show/Hide the Full List | ||
| mod site | chr14:104793453-104793454:- | [29] | |
| Sequence | AATGAATGAACCAGATTCAGACCGGCAGGGGCGCTGTGGTT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000349310.7; ENST00000555528.5; ENST00000407796.6; ENST00000557494.1; ENST00000554848.5; ENST00000649815.1; ENST00000554581.5; ENST00000555926.1; ENST00000402615.6 | ||
| External Link | RMBase: m6A_site_266204 | ||
| mod ID: M6ASITE021840 | Click to Show/Hide the Full List | ||
| mod site | chr14:104793464-104793465:- | [29] | |
| Sequence | CATGTTGGCCAAATGAATGAACCAGATTCAGACCGGCAGGG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000407796.6; ENST00000554848.5; ENST00000557494.1; ENST00000402615.6; rmsk_4276839; ENST00000349310.7; ENST00000649815.1; ENST00000554581.5; ENST00000555926.1; ENST00000555528.5 | ||
| External Link | RMBase: m6A_site_266205 | ||
| mod ID: M6ASITE021841 | Click to Show/Hide the Full List | ||
| mod site | chr14:104793501-104793502:- | [29] | |
| Sequence | CCAGCACTGGGCCTGGCAAAACCTGAGACGCCCGGTACATG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000555926.1; ENST00000554848.5; ENST00000649815.1; ENST00000557494.1; ENST00000349310.7; ENST00000555528.5; ENST00000407796.6; ENST00000402615.6; ENST00000554581.5; rmsk_4276839 | ||
| External Link | RMBase: m6A_site_266206 | ||
| mod ID: M6ASITE021842 | Click to Show/Hide the Full List | ||
| mod site | chr14:104793542-104793543:- | [29] | |
| Sequence | TCACTGACGGACTTGTCTGAACCTCTCTTTGTCTCCAGCGC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | rmsk_4276839; ENST00000557494.1; ENST00000407796.6; ENST00000555926.1; ENST00000553797.1; ENST00000555528.5; ENST00000402615.6; ENST00000349310.7; ENST00000649815.1; ENST00000554581.5; ENST00000554848.5 | ||
| External Link | RMBase: m6A_site_266207 | ||
| mod ID: M6ASITE021843 | Click to Show/Hide the Full List | ||
| mod site | chr14:104793552-104793553:- | [29] | |
| Sequence | TGGGTGCTCCTCACTGACGGACTTGTCTGAACCTCTCTTTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000555528.5; ENST00000554848.5; ENST00000402615.6; ENST00000407796.6; ENST00000555926.1; ENST00000554581.5; ENST00000553797.1; ENST00000557494.1; ENST00000349310.7; ENST00000649815.1; rmsk_4276839 | ||
| External Link | RMBase: m6A_site_266208 | ||
| mod ID: M6ASITE021844 | Click to Show/Hide the Full List | ||
| mod site | chr14:104793574-104793575:- | [29] | |
| Sequence | TGGGTCTGTAACCACCCTGGACTGGGTGCTCCTCACTGACG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; iSLK; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000555528.5; ENST00000649815.1; ENST00000407796.6; ENST00000402615.6; ENST00000554848.5; ENST00000555926.1; rmsk_4276839; ENST00000557494.1; ENST00000553797.1; ENST00000554581.5; ENST00000349310.7 | ||
| External Link | RMBase: m6A_site_266209 | ||
| mod ID: M6ASITE021845 | Click to Show/Hide the Full List | ||
| mod site | chr14:104793766-104793767:- | [29] | |
| Sequence | CTGTAGTTGGTGATGTGGGAACTGGGTTTTGAGGGATGCCT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000553797.1; ENST00000649815.1; ENST00000555926.1; ENST00000557494.1; ENST00000554848.5; ENST00000407796.6; ENST00000402615.6; ENST00000349310.7; ENST00000554581.5 | ||
| External Link | RMBase: m6A_site_266210 | ||
| mod ID: M6ASITE021846 | Click to Show/Hide the Full List | ||
| mod site | chr14:104793830-104793831:- | [29] | |
| Sequence | ATCTGGAAGGGTAGCAGAGGACTGCTCACAGGAAGAGCATG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000407796.6; ENST00000554581.5; ENST00000555926.1; ENST00000557494.1; ENST00000402615.6; ENST00000349310.7; ENST00000553797.1; ENST00000554848.5; ENST00000649815.1 | ||
| External Link | RMBase: m6A_site_266211 | ||
| mod ID: M6ASITE021847 | Click to Show/Hide the Full List | ||
| mod site | chr14:104793874-104793875:- | [29] | |
| Sequence | GCCTGGAGGTCCCAGAGAGGACCCTCCTCCCCTCAGGAAGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000349310.7; ENST00000557494.1; ENST00000402615.6; ENST00000649815.1; ENST00000554581.5; ENST00000407796.6; ENST00000553797.1; ENST00000554848.5; ENST00000555926.1 | ||
| External Link | RMBase: m6A_site_266212 | ||
| mod ID: M6ASITE021848 | Click to Show/Hide the Full List | ||
| mod site | chr14:104793942-104793943:- | [29] | |
| Sequence | TCTAAGCTGGCTGTGGAAAGACCCATGTTGGGATCCATTCC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000349310.7; ENST00000554848.5; ENST00000554581.5; ENST00000407796.6; ENST00000402615.6; ENST00000557494.1; ENST00000555926.1; ENST00000553797.1; ENST00000649815.1 | ||
| External Link | RMBase: m6A_site_266213 | ||
| mod ID: M6ASITE021849 | Click to Show/Hide the Full List | ||
| mod site | chr14:104794085-104794086:- | [29] | |
| Sequence | CTTCCCTGTCCTGGCATTAGACACAAGGCAGTGGGCCTTCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000649815.1; ENST00000555926.1; ENST00000554581.5; ENST00000402615.6; ENST00000554848.5; ENST00000553797.1; ENST00000407796.6; ENST00000349310.7; ENST00000557494.1 | ||
| External Link | RMBase: m6A_site_266214 | ||
| mod ID: M6ASITE021850 | Click to Show/Hide the Full List | ||
| mod site | chr14:104794298-104794299:- | [29] | |
| Sequence | GCCCGTGTGGGGGTGGGGGGACATCCAGAGGTCTTTGAGTC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000349310.7; ENST00000649815.1; ENST00000407796.6; ENST00000555926.1; ENST00000553797.1; ENST00000557494.1; ENST00000402615.6 | ||
| External Link | RMBase: m6A_site_266215 | ||
| mod ID: M6ASITE021851 | Click to Show/Hide the Full List | ||
| mod site | chr14:104794362-104794363:- | [29] | |
| Sequence | ATTTTTACACATGGCTCTGGACAGCTGTCTGACTCTGTCAG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000557494.1; ENST00000555926.1; ENST00000349310.7; ENST00000649815.1; ENST00000553797.1; ENST00000407796.6; ENST00000402615.6 | ||
| External Link | RMBase: m6A_site_266216 | ||
| mod ID: M6ASITE021852 | Click to Show/Hide the Full List | ||
| mod site | chr14:104795035-104795036:- | [29] | |
| Sequence | TGGGAGGGCCCCGGAAGGAGACTTCGTTTCCCACGGACGAA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; MT4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000349310.7; ENST00000557494.1; ENST00000407796.6; ENST00000649815.1; ENST00000553797.1; ENST00000555926.1 | ||
| External Link | RMBase: m6A_site_266217 | ||
| mod ID: M6ASITE021853 | Click to Show/Hide the Full List | ||
| mod site | chr14:104795735-104795736:- | [29] | |
| Sequence | GGGCCGGCGGCGGCGGCAGGACCGAGCGCGGCAGGCGGCTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; A549; GSC-11; HEK293T; HEK293A-TOA; MSC; iSLK; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000349310.7; ENST00000557494.1 | ||
| External Link | RMBase: m6A_site_266218 | ||
Pseudouridine (Pseudo)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: PSESITE000054 | Click to Show/Hide the Full List | ||
| mod site | chr14:104770043-104770044:- | [34] | |
| Sequence | ATGTTAAGGACTTCTGCAGCTATGCGCAATGTGGCATTGGG | ||
| Transcript ID List | ENST00000610370.1; ENST00000555528.5; ENST00000407796.6; ENST00000349310.7; ENST00000554581.5; ENST00000402615.6; ENST00000554192.5; ENST00000555458.5; ENST00000557552.1; ENST00000649815.1; ENST00000553506.5 | ||
| External Link | RMBase: Pseudo_site_1550 | ||
References