m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00188)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
ATG3
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3-deficient liver
Control: Wild type liver cells
|
GSE197800 | |
| Regulation |
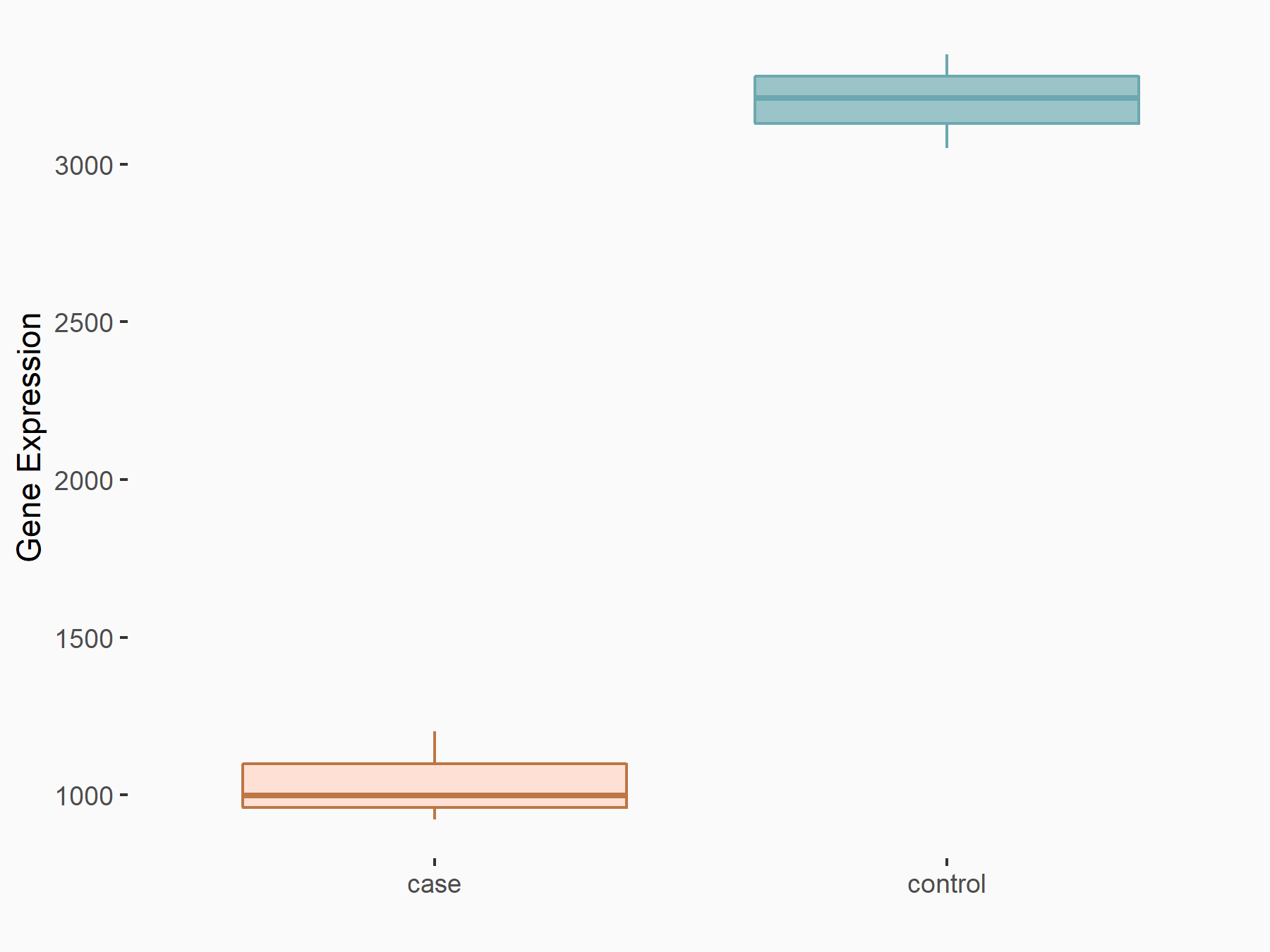  |
logFC: -1.62E+00 p-value: 2.55E-18 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including Ubiquitin-like-conjugating enzyme ATG3 (ATG3), ATG5, ATG7, ATG12, and ATG16L1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Responsed Drug | Sorafenib | Approved | ||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
YTH domain-containing family protein 1 (YTHDF1) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including Ubiquitin-like-conjugating enzyme ATG3 (ATG3), ATG5, ATG7, ATG12, and ATG16L1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Responsed Drug | Sorafenib | Approved | ||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
Liver cancer [ICD-11: 2C12]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including Ubiquitin-like-conjugating enzyme ATG3 (ATG3), ATG5, ATG7, ATG12, and ATG16L1. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Sorafenib | Approved | ||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including Ubiquitin-like-conjugating enzyme ATG3 (ATG3), ATG5, ATG7, ATG12, and ATG16L1. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Sorafenib | Approved | ||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
Sorafenib
[Approved]
| In total 2 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including Ubiquitin-like-conjugating enzyme ATG3 (ATG3), ATG5, ATG7, ATG12, and ATG16L1. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Experiment 2 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including Ubiquitin-like-conjugating enzyme ATG3 (ATG3), ATG5, ATG7, ATG12, and ATG16L1. | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00188)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE012014 | Click to Show/Hide the Full List | ||
| mod site | chr3:112556149-112556150:- | [2] | |
| Sequence | TAAAGATCCTATTCTGCCTTAGGTTTTATTACCTCAAATGC | ||
| Transcript ID List | ENST00000492886.5; rmsk_1062181; ENST00000283290.9; ENST00000465980.1; ENST00000496423.1; ENST00000402314.6; ENST00000495756.5 | ||
| External Link | RMBase: RNA-editing_site_98645 | ||
5-methylcytidine (m5C)
| In total 6 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE003134 | Click to Show/Hide the Full List | ||
| mod site | chr3:112561531-112561532:- | [3] | |
| Sequence | GCCGGCCGCTACTCCGGCCCCAGGATGCAGAATGTGATTAA | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000492886.5; ENST00000488910.1; ENST00000283290.9; ENST00000496423.1; ENST00000465980.1; ENST00000495756.5; ENST00000402314.6 | ||
| External Link | RMBase: m5C_site_32691 | ||
| mod ID: M5CSITE003135 | Click to Show/Hide the Full List | ||
| mod site | chr3:112561532-112561533:- | [3] | |
| Sequence | GGCCGGCCGCTACTCCGGCCCCAGGATGCAGAATGTGATTA | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000495756.5; ENST00000283290.9; ENST00000402314.6; ENST00000492886.5; ENST00000488910.1; ENST00000465980.1; ENST00000496423.1 | ||
| External Link | RMBase: m5C_site_32692 | ||
| mod ID: M5CSITE003136 | Click to Show/Hide the Full List | ||
| mod site | chr3:112561533-112561534:- | [3] | |
| Sequence | GGGCCGGCCGCTACTCCGGCCCCAGGATGCAGAATGTGATT | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000488910.1; ENST00000402314.6; ENST00000496423.1; ENST00000465980.1; ENST00000283290.9; ENST00000492886.5; ENST00000495756.5 | ||
| External Link | RMBase: m5C_site_32693 | ||
| mod ID: M5CSITE003137 | Click to Show/Hide the Full List | ||
| mod site | chr3:112561534-112561535:- | [3] | |
| Sequence | GGGGCCGGCCGCTACTCCGGCCCCAGGATGCAGAATGTGAT | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000492886.5; ENST00000496423.1; ENST00000402314.6; ENST00000465980.1; ENST00000283290.9; ENST00000488910.1; ENST00000495756.5 | ||
| External Link | RMBase: m5C_site_32694 | ||
| mod ID: M5CSITE003138 | Click to Show/Hide the Full List | ||
| mod site | chr3:112561549-112561550:- | [3] | |
| Sequence | TTTCCATCCCGTCGCGGGGCCGGCCGCTACTCCGGCCCCAG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000465980.1; ENST00000283290.9; ENST00000492886.5; ENST00000488910.1; ENST00000402314.6; ENST00000496423.1; ENST00000495756.5 | ||
| External Link | RMBase: m5C_site_32695 | ||
| mod ID: M5CSITE003139 | Click to Show/Hide the Full List | ||
| mod site | chr3:112561550-112561551:- | [3] | |
| Sequence | CTTTCCATCCCGTCGCGGGGCCGGCCGCTACTCCGGCCCCA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000492886.5; ENST00000496423.1; ENST00000402314.6; ENST00000488910.1; ENST00000283290.9; ENST00000495756.5; ENST00000465980.1 | ||
| External Link | RMBase: m5C_site_32696 | ||
N6-methyladenosine (m6A)
| In total 28 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE062126 | Click to Show/Hide the Full List | ||
| mod site | chr3:112532586-112532587:- | [4] | |
| Sequence | TACAGTTTCTCTAATAAGGGACTTATATGTTTATGCATTAA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000402314.6; ENST00000283290.9; ENST00000494571.1 | ||
| External Link | RMBase: m6A_site_602907 | ||
| mod ID: M6ASITE062127 | Click to Show/Hide the Full List | ||
| mod site | chr3:112532706-112532707:- | [5] | |
| Sequence | ATGACTACACAAGACACTTCACAATGTAATGAAGAGAGCAT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000283290.9; ENST00000402314.6; ENST00000494571.1 | ||
| External Link | RMBase: m6A_site_602908 | ||
| mod ID: M6ASITE062128 | Click to Show/Hide the Full List | ||
| mod site | chr3:112532713-112532714:- | [4] | |
| Sequence | ATAGAATATGACTACACAAGACACTTCACAATGTAATGAAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000402314.6; ENST00000494571.1; ENST00000283290.9 | ||
| External Link | RMBase: m6A_site_602909 | ||
| mod ID: M6ASITE062129 | Click to Show/Hide the Full List | ||
| mod site | chr3:112532720-112532721:- | [6] | |
| Sequence | TCCAACAATAGAATATGACTACACAAGACACTTCACAATGT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000402314.6; ENST00000283290.9; ENST00000494571.1 | ||
| External Link | RMBase: m6A_site_602910 | ||
| mod ID: M6ASITE062130 | Click to Show/Hide the Full List | ||
| mod site | chr3:112532752-112532753:- | [6] | |
| Sequence | CTTATTTTCTTGAAATTTGTACAAGCTGTCATTCCAACAAT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000402314.6; ENST00000494571.1; ENST00000283290.9 | ||
| External Link | RMBase: m6A_site_602911 | ||
| mod ID: M6ASITE062131 | Click to Show/Hide the Full List | ||
| mod site | chr3:112534305-112534306:- | [6] | |
| Sequence | TGATGAAGAAAATCATTGAGACTGTTGCAGAAGGAGGGGGA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000283290.9; ENST00000494571.1; ENST00000402314.6 | ||
| External Link | RMBase: m6A_site_602912 | ||
| mod ID: M6ASITE062132 | Click to Show/Hide the Full List | ||
| mod site | chr3:112536538-112536539:- | [4] | |
| Sequence | GTCAGGATCATGTGAAGAAAACAGTGACCATTGAAAATCAC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000495756.5; ENST00000283290.9; ENST00000402314.6; ENST00000494571.1 | ||
| External Link | RMBase: m6A_site_602913 | ||
| mod ID: M6ASITE062133 | Click to Show/Hide the Full List | ||
| mod site | chr3:112536564-112536565:- | [4] | |
| Sequence | AGTTGAGCACATGTATGAAGACATCAGTCAGGATCATGTGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000402314.6; ENST00000283290.9; ENST00000495756.5; ENST00000494571.1 | ||
| External Link | RMBase: m6A_site_602914 | ||
| mod ID: M6ASITE062134 | Click to Show/Hide the Full List | ||
| mod site | chr3:112537766-112537767:- | [4] | |
| Sequence | CTTATGATAAATATTACCAGACTCCACGATTATGGTTGTTT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000496423.1; ENST00000467275.1; ENST00000283290.9; ENST00000402314.6; ENST00000495756.5 | ||
| External Link | RMBase: m6A_site_602915 | ||
| mod ID: M6ASITE062135 | Click to Show/Hide the Full List | ||
| mod site | chr3:112537792-112537793:- | [6] | |
| Sequence | AACCAGAACTTATGACCTTTACATCACTTATGATAAATATT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000402314.6; ENST00000495756.5; ENST00000496423.1; ENST00000467275.1; ENST00000283290.9 | ||
| External Link | RMBase: m6A_site_602916 | ||
| mod ID: M6ASITE062136 | Click to Show/Hide the Full List | ||
| mod site | chr3:112537798-112537799:- | [6] | |
| Sequence | TTTGCAAACCAGAACTTATGACCTTTACATCACTTATGATA | ||
| Motif Score | 2.839113095 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000283290.9; ENST00000467275.1; ENST00000496423.1; ENST00000402314.6; ENST00000495756.5 | ||
| External Link | RMBase: m6A_site_602917 | ||
| mod ID: M6ASITE062137 | Click to Show/Hide the Full List | ||
| mod site | chr3:112537805-112537806:- | [4] | |
| Sequence | ATGCTATTTTGCAAACCAGAACTTATGACCTTTACATCACT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; Huh7 | ||
| Seq Type List | m6A-seq; DART-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000467275.1; ENST00000496423.1; ENST00000283290.9; ENST00000495756.5; ENST00000402314.6 | ||
| External Link | RMBase: m6A_site_602918 | ||
| mod ID: M6ASITE062138 | Click to Show/Hide the Full List | ||
| mod site | chr3:112537811-112537812:- | [4] | |
| Sequence | GTGAAGATGCTATTTTGCAAACCAGAACTTATGACCTTTAC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000283290.9; ENST00000467275.1; ENST00000496423.1; ENST00000495756.5; ENST00000402314.6 | ||
| External Link | RMBase: m6A_site_602919 | ||
| mod ID: M6ASITE062139 | Click to Show/Hide the Full List | ||
| mod site | chr3:112537844-112537845:- | [4] | |
| Sequence | TAGAAGCTTGTAAAGCCAAAACTGATGCTGGCGGTGAAGAT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; LCLs; Huh7 | ||
| Seq Type List | m6A-seq; DART-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000496423.1; ENST00000283290.9; ENST00000402314.6; ENST00000467275.1; ENST00000495756.5 | ||
| External Link | RMBase: m6A_site_602920 | ||
| mod ID: M6ASITE062140 | Click to Show/Hide the Full List | ||
| mod site | chr3:112537877-112537878:- | [5] | |
| Sequence | ATTTTCAGGCTACCCTAGATACAAGGAAAATAGTAGAAGCT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000283290.9; ENST00000467275.1; ENST00000402314.6; ENST00000495756.5; ENST00000496423.1 | ||
| External Link | RMBase: m6A_site_602921 | ||
| mod ID: M6ASITE062141 | Click to Show/Hide the Full List | ||
| mod site | chr3:112538153-112538154:- | [4] | |
| Sequence | AAGAGAGTGGATTGTTGGAAACAGATGAGGTTTGTAAATTG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; LCLs; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000467275.1; ENST00000496423.1; ENST00000283290.9; ENST00000495756.5; ENST00000402314.6; ENST00000492886.5 | ||
| External Link | RMBase: m6A_site_602922 | ||
| mod ID: M6ASITE062142 | Click to Show/Hide the Full List | ||
| mod site | chr3:112538213-112538214:- | [4] | |
| Sequence | GGGAATTTTCTTGAACTTGAACTTGATTAATCTTTTGTTGT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; hNPCs; A549; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000402314.6; ENST00000492886.5; ENST00000495756.5; ENST00000496423.1; ENST00000283290.9; ENST00000467275.1 | ||
| External Link | RMBase: m6A_site_602923 | ||
| mod ID: M6ASITE062143 | Click to Show/Hide the Full List | ||
| mod site | chr3:112538219-112538220:- | [4] | |
| Sequence | GGTTTAGGGAATTTTCTTGAACTTGAACTTGATTAATCTTT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; hNPCs; A549; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000283290.9; ENST00000402314.6; ENST00000496423.1; ENST00000467275.1; ENST00000495756.5; ENST00000492886.5 | ||
| External Link | RMBase: m6A_site_602924 | ||
| mod ID: M6ASITE062144 | Click to Show/Hide the Full List | ||
| mod site | chr3:112538280-112538281:- | [4] | |
| Sequence | CACTTAAAAACAAAGGCAAAACCCCAGTTTTTGAATGATAC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; hNPCs; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000495756.5; ENST00000283290.9; ENST00000496423.1; ENST00000402314.6; ENST00000467275.1; ENST00000492886.5 | ||
| External Link | RMBase: m6A_site_602925 | ||
| mod ID: M6ASITE062145 | Click to Show/Hide the Full List | ||
| mod site | chr3:112538291-112538292:- | [4] | |
| Sequence | TTCAATATCTTCACTTAAAAACAAAGGCAAAACCCCAGTTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hNPCs; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000495756.5; ENST00000492886.5; ENST00000496423.1; ENST00000467275.1; ENST00000283290.9; ENST00000402314.6 | ||
| External Link | RMBase: m6A_site_602926 | ||
| mod ID: M6ASITE062146 | Click to Show/Hide the Full List | ||
| mod site | chr3:112541882-112541883:- | [5] | |
| Sequence | AATATAATTAATTTCAAAGGACAATATAAGGCTTCAAGATT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000495756.5; ENST00000492886.5; ENST00000496423.1; ENST00000283290.9; ENST00000402314.6 | ||
| External Link | RMBase: m6A_site_602927 | ||
| mod ID: M6ASITE062147 | Click to Show/Hide the Full List | ||
| mod site | chr3:112548547-112548548:- | [5] | |
| Sequence | GTGATGGCGGATGGGTAGATACATATCACAACACAGGTAAG | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000402314.6; ENST00000465980.1; ENST00000495756.5; ENST00000283290.9; ENST00000492886.5; ENST00000496423.1 | ||
| External Link | RMBase: m6A_site_602928 | ||
| mod ID: M6ASITE062148 | Click to Show/Hide the Full List | ||
| mod site | chr3:112548617-112548618:- | [4] | |
| Sequence | CCGTGCTATAAGCGGTGCAAACAGATGGAATATTCAGATGA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; LCLs; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000465980.1; ENST00000495756.5; ENST00000283290.9; ENST00000492886.5; ENST00000496423.1; ENST00000402314.6 | ||
| External Link | RMBase: m6A_site_602929 | ||
| mod ID: M6ASITE062149 | Click to Show/Hide the Full List | ||
| mod site | chr3:112550213-112550214:- | [4] | |
| Sequence | GCATACCTACCAACAGGCAAACAATTTTTGGTAACCAAAAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; LCLs; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000496423.1; ENST00000495756.5; ENST00000402314.6; ENST00000492886.5; ENST00000283290.9; ENST00000465980.1 | ||
| External Link | RMBase: m6A_site_602930 | ||
| mod ID: M6ASITE062150 | Click to Show/Hide the Full List | ||
| mod site | chr3:112553289-112553290:- | [5] | |
| Sequence | ACCTAGTCCACCACTGTCCAACATGGCAATGGTAAGTAATA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000465980.1; ENST00000283290.9; ENST00000492886.5; ENST00000496423.1; ENST00000495756.5; ENST00000402314.6 | ||
| External Link | RMBase: m6A_site_602931 | ||
| mod ID: M6ASITE062151 | Click to Show/Hide the Full List | ||
| mod site | chr3:112561581-112561582:- | [5] | |
| Sequence | TTCTGACTCCCTGGCCCCTGACACGGCTGCACTTTCCATCC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000283290.9; ENST00000402314.6; ENST00000488910.1; ENST00000496423.1; ENST00000492886.5; ENST00000465980.1; ENST00000495756.5 | ||
| External Link | RMBase: m6A_site_602932 | ||
| mod ID: M6ASITE062152 | Click to Show/Hide the Full List | ||
| mod site | chr3:112561606-112561607:- | [4] | |
| Sequence | GCGAGTCGGTGGCAGCGAGGACATTTTCTGACTCCCTGGCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; A549; H1A; H1B; MM6; GSC-11; HEK293T; TREX; iSLK; MSC; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000488910.1; ENST00000495756.5; ENST00000283290.9; ENST00000465980.1; ENST00000402314.6; ENST00000496423.1; ENST00000492886.5 | ||
| External Link | RMBase: m6A_site_602933 | ||
| mod ID: M6ASITE062153 | Click to Show/Hide the Full List | ||
| mod site | chr3:112561869-112561870:- | [4] | |
| Sequence | GCGAGTGAAGCAAAGCGAGGACAGACAGCTCGCAGAGGGCG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; A549; HEK293T; H1A; H1B; hESCs; fibroblasts; H1299; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000492886.5; ENST00000402314.6; ENST00000495756.5; ENST00000283290.9 | ||
| External Link | RMBase: m6A_site_602937 | ||
References