m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00624)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
LHPP
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
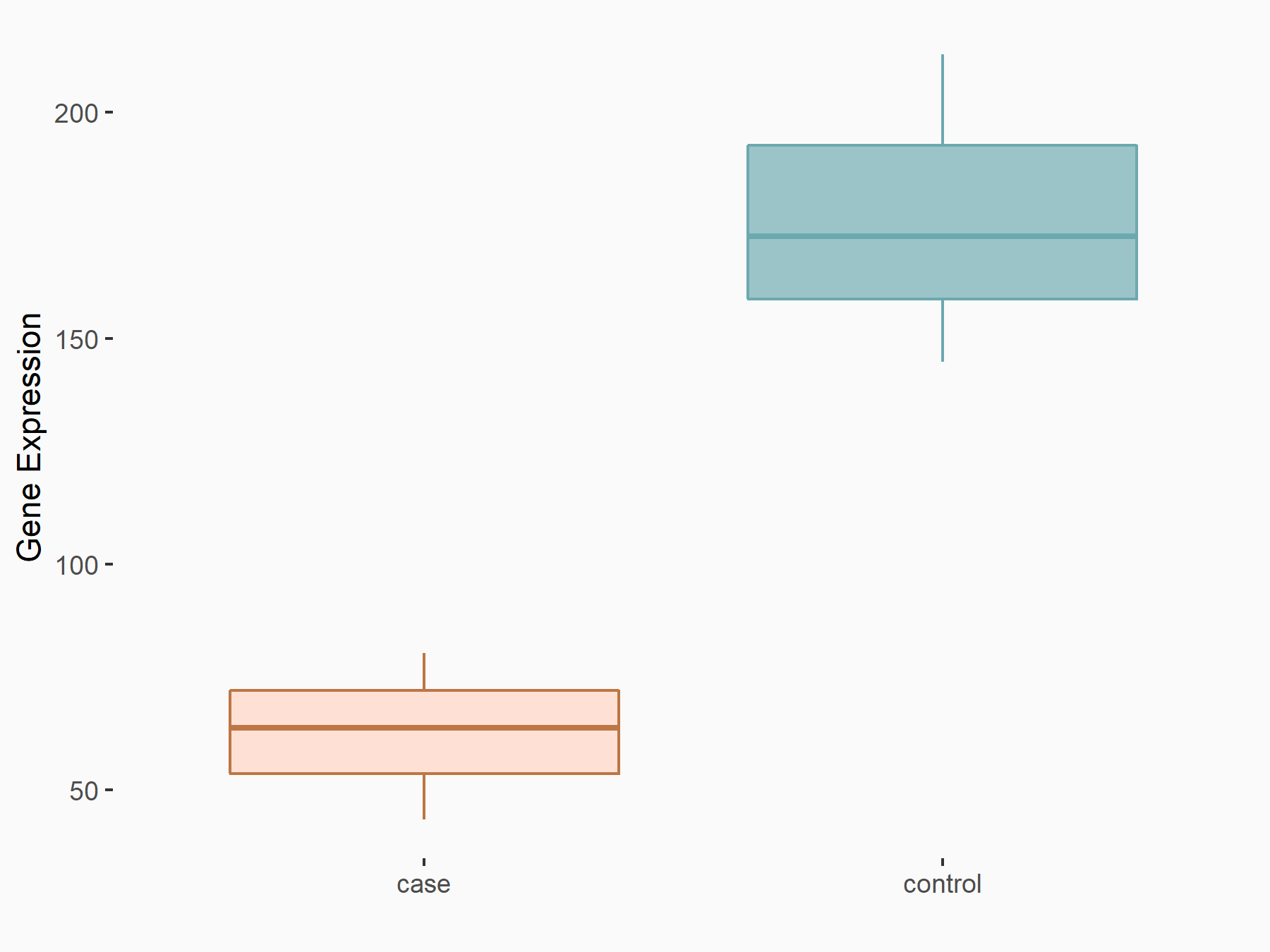  |
logFC: -1.51E+00 p-value: 1.12E-09 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between LHPP and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 8.41E+00 | GSE60213 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Knock-down of YTHDF2 or METTL3 significantly induced the expression of Phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP) and NKX3-1 at both mRNA and protein level with inhibited phosphorylated AKT. YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent way to regulate AKT phosphorylation-induced tumor progression in prostate cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Prostate cancer | ICD-11: 2C82 | ||
| Pathway Response | Oxidative phosphorylation | hsa00190 | ||
| In-vitro Model | VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| In-vivo Model | Approximately 2 × 106 PCa cells (PC-3 shNC, shYTHDF2, shMETTL3 cell lines) per mouse suspended in 100 uL PBS were injected in the flank of male BALB/c nude mice (4 weeks old). During the 40-day observation, the tumor size (V = (width2×length ×0.52)) was measured with vernier caliper. Approximately 1.5 × 106 PCa cells suspended in 100 uL of PBS (PC-3 shNC, shYTHDF2, and shMETTL3 cell lines) per mouse were injected into the tail vein of male BALB/c nude mice (4 weeks old). The IVIS Spectrum animal imaging system (PerkinElmer) was used to evaluate the tumor growth (40 days) and whole metastasis conditions (4 weeks and 6 weeks) with 100 uL XenoLight D-luciferin Potassium Salt (15 mg/ml, Perkin Elmer) per mouse. Mice were anesthetized and then sacrificed for tumors and metastases which were sent for further organ-localized imaging as above, IHC staining and hematoxylin-eosin (H&E) staining. | |||
YTH domain-containing family protein 2 (YTHDF2) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | B18-hi B cell line | Mus musculus |
|
Treatment: YTHDF2 knockout B18-hi B cells
Control: Wild type B18-hi B cells
|
GSE189819 | |
| Regulation |
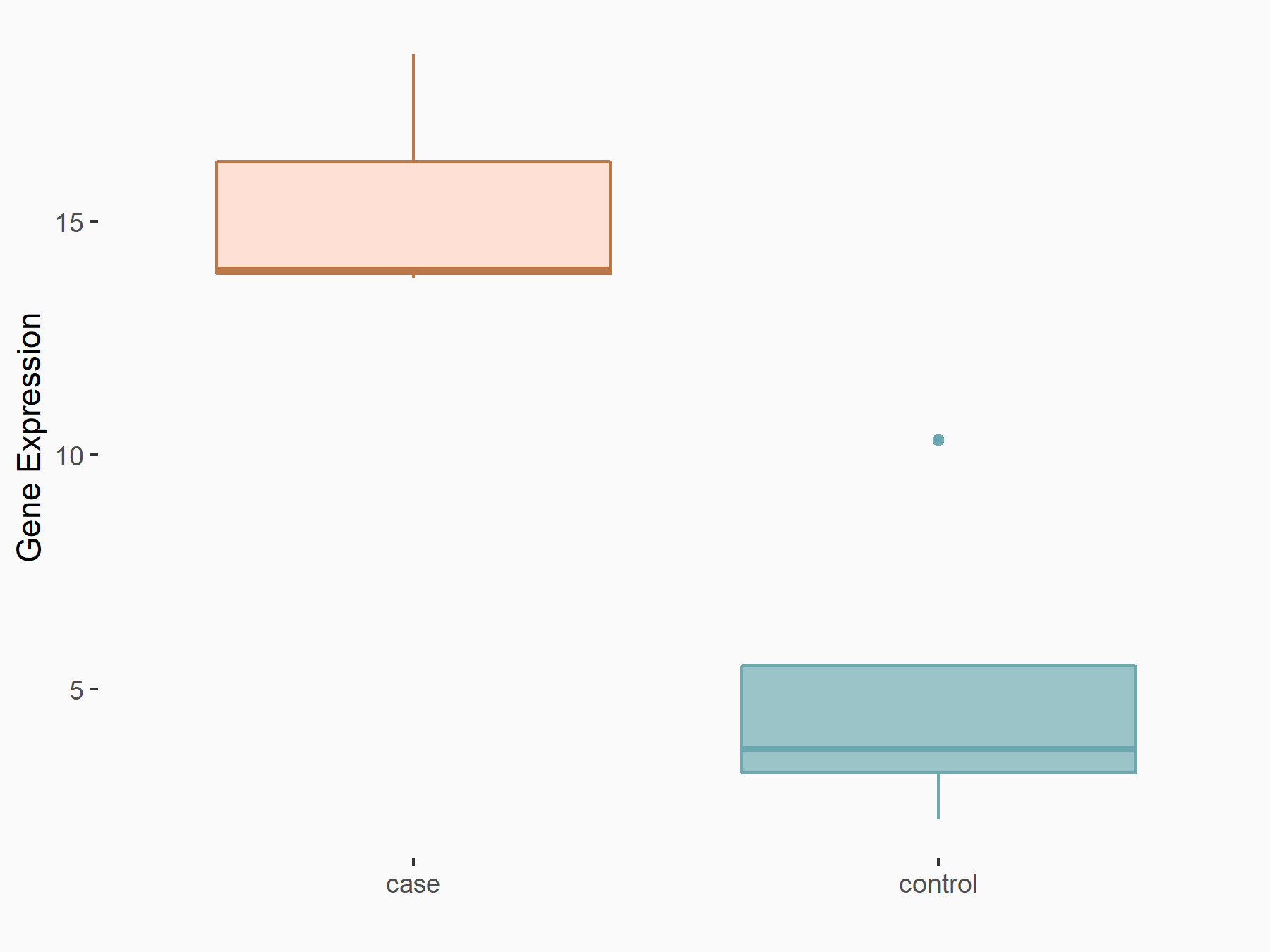  |
logFC: 1.54E+00 p-value: 1.87E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Knock-down of YTHDF2 or METTL3 significantly induced the expression of Phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP) and NKX3-1 at both mRNA and protein level with inhibited phosphorylated AKT. YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent way to regulate AKT phosphorylation-induced tumor progression in prostate cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Prostate cancer | ICD-11: 2C82 | ||
| Pathway Response | Oxidative phosphorylation | hsa00190 | ||
| In-vitro Model | VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| In-vivo Model | Approximately 2 × 106 PCa cells (PC-3 shNC, shYTHDF2, shMETTL3 cell lines) per mouse suspended in 100 uL PBS were injected in the flank of male BALB/c nude mice (4 weeks old). During the 40-day observation, the tumor size (V = (width2×length ×0.52)) was measured with vernier caliper. Approximately 1.5 × 106 PCa cells suspended in 100 uL of PBS (PC-3 shNC, shYTHDF2, and shMETTL3 cell lines) per mouse were injected into the tail vein of male BALB/c nude mice (4 weeks old). The IVIS Spectrum animal imaging system (PerkinElmer) was used to evaluate the tumor growth (40 days) and whole metastasis conditions (4 weeks and 6 weeks) with 100 uL XenoLight D-luciferin Potassium Salt (15 mg/ml, Perkin Elmer) per mouse. Mice were anesthetized and then sacrificed for tumors and metastases which were sent for further organ-localized imaging as above, IHC staining and hematoxylin-eosin (H&E) staining. | |||
Prostate cancer [ICD-11: 2C82]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Knock-down of YTHDF2 or METTL3 significantly induced the expression of Phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP) and NKX3-1 at both mRNA and protein level with inhibited phosphorylated AKT. YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent way to regulate AKT phosphorylation-induced tumor progression in prostate cancer. | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Oxidative phosphorylation | hsa00190 | ||
| In-vitro Model | VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| In-vivo Model | Approximately 2 × 106 PCa cells (PC-3 shNC, shYTHDF2, shMETTL3 cell lines) per mouse suspended in 100 uL PBS were injected in the flank of male BALB/c nude mice (4 weeks old). During the 40-day observation, the tumor size (V = (width2×length ×0.52)) was measured with vernier caliper. Approximately 1.5 × 106 PCa cells suspended in 100 uL of PBS (PC-3 shNC, shYTHDF2, and shMETTL3 cell lines) per mouse were injected into the tail vein of male BALB/c nude mice (4 weeks old). The IVIS Spectrum animal imaging system (PerkinElmer) was used to evaluate the tumor growth (40 days) and whole metastasis conditions (4 weeks and 6 weeks) with 100 uL XenoLight D-luciferin Potassium Salt (15 mg/ml, Perkin Elmer) per mouse. Mice were anesthetized and then sacrificed for tumors and metastases which were sent for further organ-localized imaging as above, IHC staining and hematoxylin-eosin (H&E) staining. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Knock-down of YTHDF2 or METTL3 significantly induced the expression of Phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP) and NKX3-1 at both mRNA and protein level with inhibited phosphorylated AKT. YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent way to regulate AKT phosphorylation-induced tumor progression in prostate cancer. | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Oxidative phosphorylation | hsa00190 | ||
| In-vitro Model | VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| In-vivo Model | Approximately 2 × 106 PCa cells (PC-3 shNC, shYTHDF2, shMETTL3 cell lines) per mouse suspended in 100 uL PBS were injected in the flank of male BALB/c nude mice (4 weeks old). During the 40-day observation, the tumor size (V = (width2×length ×0.52)) was measured with vernier caliper. Approximately 1.5 × 106 PCa cells suspended in 100 uL of PBS (PC-3 shNC, shYTHDF2, and shMETTL3 cell lines) per mouse were injected into the tail vein of male BALB/c nude mice (4 weeks old). The IVIS Spectrum animal imaging system (PerkinElmer) was used to evaluate the tumor growth (40 days) and whole metastasis conditions (4 weeks and 6 weeks) with 100 uL XenoLight D-luciferin Potassium Salt (15 mg/ml, Perkin Elmer) per mouse. Mice were anesthetized and then sacrificed for tumors and metastases which were sent for further organ-localized imaging as above, IHC staining and hematoxylin-eosin (H&E) staining. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03329 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5A (KDM5A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Prostate cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00624)
| In total 35 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE004433 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474170-124474171:+ | [2] | |
| Sequence | TTTTTTTGAGACAGGGTCTCACTTTGTCACCTAGGCTGGAG | ||
| Transcript ID List | ENST00000392757.8; ENST00000368842.10; ENST00000368839.1 | ||
| External Link | RMBase: RNA-editing_site_19158 | ||
| mod ID: A2ISITE004434 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474178-124474179:+ | [2] | |
| Sequence | AGACAGGGTCTCACTTTGTCACCTAGGCTGGAGTGCAGTGG | ||
| Transcript ID List | ENST00000392757.8; ENST00000368839.1; ENST00000368842.10 | ||
| External Link | RMBase: RNA-editing_site_19159 | ||
| mod ID: A2ISITE004435 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474194-124474195:+ | [2] | |
| Sequence | TGTCACCTAGGCTGGAGTGCAGTGGCACAATCTTGGCTCAC | ||
| Transcript ID List | ENST00000368842.10; ENST00000392757.8; ENST00000368839.1 | ||
| External Link | RMBase: RNA-editing_site_19160 | ||
| mod ID: A2ISITE004436 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474200-124474201:+ | [2] | |
| Sequence | CTAGGCTGGAGTGCAGTGGCACAATCTTGGCTCACTGCAGC | ||
| Transcript ID List | ENST00000368842.10; ENST00000392757.8; ENST00000368839.1 | ||
| External Link | RMBase: RNA-editing_site_19161 | ||
| mod ID: A2ISITE004437 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474203-124474204:+ | [2] | |
| Sequence | GGCTGGAGTGCAGTGGCACAATCTTGGCTCACTGCAGCCTC | ||
| Transcript ID List | ENST00000368839.1; ENST00000392757.8; ENST00000368842.10 | ||
| External Link | RMBase: RNA-editing_site_19162 | ||
| mod ID: A2ISITE004438 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474213-124474214:+ | [2] | |
| Sequence | CAGTGGCACAATCTTGGCTCACTGCAGCCTCGACCTCCTGG | ||
| Transcript ID List | ENST00000368839.1; ENST00000368842.10; ENST00000392757.8 | ||
| External Link | RMBase: RNA-editing_site_19163 | ||
| mod ID: A2ISITE004439 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474218-124474219:+ | [2] | |
| Sequence | GCACAATCTTGGCTCACTGCAGCCTCGACCTCCTGGGCTCA | ||
| Transcript ID List | ENST00000368842.10; ENST00000392757.8; ENST00000368839.1 | ||
| External Link | RMBase: RNA-editing_site_19164 | ||
| mod ID: A2ISITE004440 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474225-124474226:+ | [2] | |
| Sequence | CTTGGCTCACTGCAGCCTCGACCTCCTGGGCTCAAGCGATC | ||
| Transcript ID List | ENST00000368842.10; ENST00000392757.8; ENST00000368839.1 | ||
| External Link | RMBase: RNA-editing_site_19165 | ||
| mod ID: A2ISITE004441 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474238-124474239:+ | [2] | |
| Sequence | AGCCTCGACCTCCTGGGCTCAAGCGATCCTTTCACCTCAGC | ||
| Transcript ID List | ENST00000368839.1; ENST00000368842.10; ENST00000392757.8 | ||
| External Link | RMBase: RNA-editing_site_19166 | ||
| mod ID: A2ISITE004442 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474239-124474240:+ | [2] | |
| Sequence | GCCTCGACCTCCTGGGCTCAAGCGATCCTTTCACCTCAGCC | ||
| Transcript ID List | ENST00000368842.10; ENST00000392757.8; ENST00000368839.1 | ||
| External Link | RMBase: RNA-editing_site_19167 | ||
| mod ID: A2ISITE004443 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474251-124474252:+ | [2] | |
| Sequence | TGGGCTCAAGCGATCCTTTCACCTCAGCCCCCAAAGTAGCT | ||
| Transcript ID List | ENST00000392757.8; ENST00000368839.1; ENST00000368842.10 | ||
| External Link | RMBase: RNA-editing_site_19168 | ||
| mod ID: A2ISITE004444 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474256-124474257:+ | [2] | |
| Sequence | TCAAGCGATCCTTTCACCTCAGCCCCCAAAGTAGCTGAGAC | ||
| Transcript ID List | ENST00000392757.8; ENST00000368842.10; ENST00000368839.1 | ||
| External Link | RMBase: RNA-editing_site_19169 | ||
| mod ID: A2ISITE004445 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474264-124474265:+ | [2] | |
| Sequence | TCCTTTCACCTCAGCCCCCAAAGTAGCTGAGACTACAAGTG | ||
| Transcript ID List | ENST00000368842.10; ENST00000392757.8; ENST00000368839.1 | ||
| External Link | RMBase: RNA-editing_site_19170 | ||
| mod ID: A2ISITE004446 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474268-124474269:+ | [2] | |
| Sequence | TTCACCTCAGCCCCCAAAGTAGCTGAGACTACAAGTGCACA | ||
| Transcript ID List | ENST00000368839.1; ENST00000368842.10; ENST00000392757.8 | ||
| External Link | RMBase: RNA-editing_site_19171 | ||
| mod ID: A2ISITE004447 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474273-124474274:+ | [2] | |
| Sequence | CTCAGCCCCCAAAGTAGCTGAGACTACAAGTGCACACCACT | ||
| Transcript ID List | ENST00000392757.8; ENST00000368842.10; ENST00000368839.1 | ||
| External Link | RMBase: RNA-editing_site_19172 | ||
| mod ID: A2ISITE004448 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474278-124474279:+ | [2] | |
| Sequence | CCCCCAAAGTAGCTGAGACTACAAGTGCACACCACTGCACC | ||
| Transcript ID List | ENST00000392757.8; ENST00000368839.1; ENST00000368842.10 | ||
| External Link | RMBase: RNA-editing_site_19173 | ||
| mod ID: A2ISITE004449 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474281-124474282:+ | [2] | |
| Sequence | CCAAAGTAGCTGAGACTACAAGTGCACACCACTGCACCTGG | ||
| Transcript ID List | ENST00000368839.1; ENST00000368842.10; ENST00000392757.8 | ||
| External Link | RMBase: RNA-editing_site_19174 | ||
| mod ID: A2ISITE004450 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474319-124474320:+ | [2] | |
| Sequence | TGGCCAATTTTTGTATTTTTAGTACAGACAAGGTTTTGCCA | ||
| Transcript ID List | ENST00000368842.10; ENST00000368839.1; ENST00000392757.8 | ||
| External Link | RMBase: RNA-editing_site_19175 | ||
| mod ID: A2ISITE004451 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474324-124474325:+ | [2] | |
| Sequence | AATTTTTGTATTTTTAGTACAGACAAGGTTTTGCCACGTTG | ||
| Transcript ID List | ENST00000368842.10; ENST00000392757.8; ENST00000368839.1 | ||
| External Link | RMBase: RNA-editing_site_19176 | ||
| mod ID: A2ISITE004452 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474326-124474327:+ | [2] | |
| Sequence | TTTTTGTATTTTTAGTACAGACAAGGTTTTGCCACGTTGCC | ||
| Transcript ID List | ENST00000392757.8; ENST00000368839.1; ENST00000368842.10 | ||
| External Link | RMBase: RNA-editing_site_19177 | ||
| mod ID: A2ISITE004453 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474328-124474329:+ | [2] | |
| Sequence | TTTGTATTTTTAGTACAGACAAGGTTTTGCCACGTTGCCCA | ||
| Transcript ID List | ENST00000368839.1; ENST00000368842.10; ENST00000392757.8 | ||
| External Link | RMBase: RNA-editing_site_19178 | ||
| mod ID: A2ISITE004454 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474329-124474330:+ | [2] | |
| Sequence | TTGTATTTTTAGTACAGACAAGGTTTTGCCACGTTGCCCAG | ||
| Transcript ID List | ENST00000392757.8; ENST00000368842.10; ENST00000368839.1 | ||
| External Link | RMBase: RNA-editing_site_19179 | ||
| mod ID: A2ISITE004455 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474339-124474340:+ | [2] | |
| Sequence | AGTACAGACAAGGTTTTGCCACGTTGCCCAGGCTGGTCTTG | ||
| Transcript ID List | ENST00000392757.8; ENST00000368842.10; ENST00000368839.1 | ||
| External Link | RMBase: RNA-editing_site_19180 | ||
| mod ID: A2ISITE004456 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474348-124474349:+ | [2] | |
| Sequence | AAGGTTTTGCCACGTTGCCCAGGCTGGTCTTGAACTCCAGA | ||
| Transcript ID List | ENST00000368842.10; ENST00000368839.1; ENST00000392757.8 | ||
| External Link | RMBase: RNA-editing_site_19181 | ||
| mod ID: A2ISITE004457 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474373-124474374:+ | [2] | |
| Sequence | GGTCTTGAACTCCAGAGCTCAACGGATCTGCCCACCTCAGT | ||
| Transcript ID List | ENST00000368842.10; ENST00000392757.8; ENST00000368839.1 | ||
| External Link | RMBase: RNA-editing_site_19182 | ||
| mod ID: A2ISITE004458 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474391-124474392:+ | [2] | |
| Sequence | TCAACGGATCTGCCCACCTCAGTCTCCCAAAGTGTTGGGAT | ||
| Transcript ID List | ENST00000392757.8; ENST00000368839.1; ENST00000368842.10 | ||
| External Link | RMBase: RNA-editing_site_19183 | ||
| mod ID: A2ISITE004459 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474413-124474414:+ | [2] | |
| Sequence | TCTCCCAAAGTGTTGGGATTACAAGTATGAACCACCGTGCC | ||
| Transcript ID List | ENST00000368839.1; ENST00000392757.8; ENST00000368842.10 | ||
| External Link | RMBase: RNA-editing_site_19184 | ||
| mod ID: A2ISITE004460 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474416-124474417:+ | [2] | |
| Sequence | CCCAAAGTGTTGGGATTACAAGTATGAACCACCGTGCCCGG | ||
| Transcript ID List | ENST00000392757.8; ENST00000368842.10; ENST00000368839.1 | ||
| External Link | RMBase: RNA-editing_site_19185 | ||
| mod ID: A2ISITE004461 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474439-124474440:+ | [2] | |
| Sequence | ATGAACCACCGTGCCCGGCCATATGAAGTCTTGTAGTTGTT | ||
| Transcript ID List | ENST00000368839.1; ENST00000392757.8; ENST00000368842.10 | ||
| External Link | RMBase: RNA-editing_site_19186 | ||
| mod ID: A2ISITE004462 | Click to Show/Hide the Full List | ||
| mod site | chr10:124474444-124474445:+ | [2] | |
| Sequence | CCACCGTGCCCGGCCATATGAAGTCTTGTAGTTGTTGAACT | ||
| Transcript ID List | ENST00000368842.10; ENST00000392757.8; ENST00000368839.1 | ||
| External Link | RMBase: RNA-editing_site_19187 | ||
| mod ID: A2ISITE004463 | Click to Show/Hide the Full List | ||
| mod site | chr10:124501454-124501455:+ | [3] | |
| Sequence | CCACTAAAAATACAAACATTAGCCAGACGTGGTGGTGTGCA | ||
| Transcript ID List | ENST00000368842.10; rmsk_3443287; ENST00000481452.1; ENST00000368839.1 | ||
| External Link | RMBase: RNA-editing_site_19188 | ||
| mod ID: A2ISITE004464 | Click to Show/Hide the Full List | ||
| mod site | chr10:124554094-124554095:+ | [3] | |
| Sequence | AGCCTGGGCTGCCTGGATTCAGCTCCTGCTCTGACATTTAC | ||
| Transcript ID List | ENST00000493240.1; ENST00000368839.1; ENST00000482963.1; rmsk_3443376; ENST00000368842.10 | ||
| External Link | RMBase: RNA-editing_site_19189 | ||
| mod ID: A2ISITE004465 | Click to Show/Hide the Full List | ||
| mod site | chr10:124554113-124554114:+ | [3] | |
| Sequence | CAGCTCCTGCTCTGACATTTACAGCTGGGACGACCTGGGCC | ||
| Transcript ID List | ENST00000368842.10; rmsk_3443376; ENST00000368839.1; ENST00000482963.1; ENST00000493240.1 | ||
| External Link | RMBase: RNA-editing_site_19190 | ||
| mod ID: A2ISITE004466 | Click to Show/Hide the Full List | ||
| mod site | chr10:124554550-124554551:+ | [3] | |
| Sequence | ATAATGTGCATGCAGTGCTTAGCCTAGTGCCCGGTGCAGAG | ||
| Transcript ID List | ENST00000493240.1; ENST00000368842.10; ENST00000482963.1; ENST00000368839.1 | ||
| External Link | RMBase: RNA-editing_site_19191 | ||
| mod ID: A2ISITE004467 | Click to Show/Hide the Full List | ||
| mod site | chr10:124600534-124600535:+ | [3] | |
| Sequence | GCAGTTGGTCTCCAGAAGGAAATTCTCTGTATCTGGGTGCT | ||
| Transcript ID List | ENST00000493240.1; ENST00000482963.1; ENST00000368839.1; ENST00000368842.10 | ||
| External Link | RMBase: RNA-editing_site_19192 | ||
5-methylcytidine (m5C)
| In total 8 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE004797 | Click to Show/Hide the Full List | ||
| mod site | chr10:124461956-124461957:+ | [4] | |
| Sequence | CGGCGCGGGCGGCGGCACGGCCATCGCCGGCTCGGTGGAGG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000392757.8; ENST00000368842.10; ENST00000368839.1 | ||
| External Link | RMBase: m5C_site_6085 | ||
| mod ID: M5CSITE004798 | Click to Show/Hide the Full List | ||
| mod site | chr10:124461957-124461958:+ | [4] | |
| Sequence | GGCGCGGGCGGCGGCACGGCCATCGCCGGCTCGGTGGAGGC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000368842.10; ENST00000392757.8; ENST00000368839.1 | ||
| External Link | RMBase: m5C_site_6086 | ||
| mod ID: M5CSITE004799 | Click to Show/Hide the Full List | ||
| mod site | chr10:124461960-124461961:+ | [4] | |
| Sequence | GCGGGCGGCGGCACGGCCATCGCCGGCTCGGTGGAGGCGGT | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000368842.10; ENST00000368839.1; ENST00000392757.8 | ||
| External Link | RMBase: m5C_site_6087 | ||
| mod ID: M5CSITE004800 | Click to Show/Hide the Full List | ||
| mod site | chr10:124461962-124461963:+ | [4] | |
| Sequence | GGGCGGCGGCACGGCCATCGCCGGCTCGGTGGAGGCGGTGG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000368839.1; ENST00000392757.8; ENST00000368842.10 | ||
| External Link | RMBase: m5C_site_6088 | ||
| mod ID: M5CSITE004801 | Click to Show/Hide the Full List | ||
| mod site | chr10:124461963-124461964:+ | [4] | |
| Sequence | GGCGGCGGCACGGCCATCGCCGGCTCGGTGGAGGCGGTGGC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000392757.8; ENST00000368842.10; ENST00000368839.1 | ||
| External Link | RMBase: m5C_site_6089 | ||
| mod ID: M5CSITE004802 | Click to Show/Hide the Full List | ||
| mod site | chr10:124461977-124461978:+ | [4] | |
| Sequence | CATCGCCGGCTCGGTGGAGGCGGTGGCCAGGTGAGTGGGCC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000392757.8; ENST00000368839.1; ENST00000368842.10 | ||
| External Link | RMBase: m5C_site_6090 | ||
| mod ID: M5CSITE004803 | Click to Show/Hide the Full List | ||
| mod site | chr10:124461983-124461984:+ | [4] | |
| Sequence | CGGCTCGGTGGAGGCGGTGGCCAGGTGAGTGGGCCCCGGGA | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000368842.10; ENST00000368839.1; ENST00000392757.8 | ||
| External Link | RMBase: m5C_site_6091 | ||
| mod ID: M5CSITE004804 | Click to Show/Hide the Full List | ||
| mod site | chr10:124461984-124461985:+ | [4] | |
| Sequence | GGCTCGGTGGAGGCGGTGGCCAGGTGAGTGGGCCCCGGGAC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000368842.10; ENST00000392757.8; ENST00000368839.1 | ||
| External Link | RMBase: m5C_site_6092 | ||
N6-methyladenosine (m6A)
| In total 35 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE002282 | Click to Show/Hide the Full List | ||
| mod site | chr10:124488508-124488509:+ | [5] | |
| Sequence | AGAAAGCTTTTCTTATCAAAACATGAATAACGCCTTCCAGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000392757.8; ENST00000368842.10; ENST00000368839.1; ENST00000487190.1; ENST00000481452.1 | ||
| External Link | RMBase: m6A_site_122178 | ||
| mod ID: M6ASITE002283 | Click to Show/Hide the Full List | ||
| mod site | chr10:124496968-124496969:+ | [5] | |
| Sequence | GCTCTCTCCTAGGCGTTACTACAAGGAGACCTCTGGCCTGA | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000368839.1; ENST00000392757.8; ENST00000368842.10; ENST00000481452.1 | ||
| External Link | RMBase: m6A_site_122179 | ||
| mod ID: M6ASITE002284 | Click to Show/Hide the Full List | ||
| mod site | chr10:124613310-124613311:+ | [6] | |
| Sequence | GAAGGCTGATGGGTACGTGGACAACCTCGCAGAGGCAGTGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000368842.10; ENST00000482963.1; ENST00000486535.1; ENST00000368839.1; ENST00000493240.1; ENST00000492187.1 | ||
| External Link | RMBase: m6A_site_122180 | ||
| mod ID: M6ASITE002285 | Click to Show/Hide the Full List | ||
| mod site | chr10:124613331-124613332:+ | [6] | |
| Sequence | CAACCTCGCAGAGGCAGTGGACCTGCTGCTGCAGCACGCCG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000368842.10; ENST00000482963.1; ENST00000493240.1; ENST00000492187.1; ENST00000368839.1; ENST00000486535.1 | ||
| External Link | RMBase: m6A_site_122181 | ||
| mod ID: M6ASITE002286 | Click to Show/Hide the Full List | ||
| mod site | chr10:124613520-124613521:+ | [7] | |
| Sequence | CCACCTGCCCCAGTGCCCAGACCAACCAAGGCCCTGACAGC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HepG2; H1B; Jurkat; CD4T; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000368842.10; ENST00000368839.1; ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122182 | ||
| mod ID: M6ASITE002287 | Click to Show/Hide the Full List | ||
| mod site | chr10:124613655-124613656:+ | [8] | |
| Sequence | GTGAAGTCCAGGAGGGTGGGACAGGCCTGTCAGGCCTCTGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000482963.1; ENST00000368842.10; ENST00000368839.1 | ||
| External Link | RMBase: m6A_site_122183 | ||
| mod ID: M6ASITE002288 | Click to Show/Hide the Full List | ||
| mod site | chr10:124613695-124613696:+ | [8] | |
| Sequence | GGAATCTCCCAAATCCCAGAACTCACCACTCACCATGGGCC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000368842.10; ENST00000368839.1; ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122184 | ||
| mod ID: M6ASITE002289 | Click to Show/Hide the Full List | ||
| mod site | chr10:124613730-124613731:+ | [8] | |
| Sequence | TGGGCCTTTAAATGCAGTAAACTCCACCTAACCAGATTCAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000368839.1; ENST00000368842.10; ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122185 | ||
| mod ID: M6ASITE002290 | Click to Show/Hide the Full List | ||
| mod site | chr10:124613780-124613781:+ | [8] | |
| Sequence | GCCCACTGCCTCCTCTTCAGACTCTTTGCATTTCAGTGAAG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000368839.1; ENST00000482963.1; ENST00000368842.10 | ||
| External Link | RMBase: m6A_site_122186 | ||
| mod ID: M6ASITE002291 | Click to Show/Hide the Full List | ||
| mod site | chr10:124613813-124613814:+ | [8] | |
| Sequence | CAGTGAAGAGCCTGGAAGAAACCCAGGGGCCTCCTATGCAC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000482963.1; ENST00000368842.10; ENST00000368839.1 | ||
| External Link | RMBase: m6A_site_122187 | ||
| mod ID: M6ASITE002292 | Click to Show/Hide the Full List | ||
| mod site | chr10:124613851-124613852:+ | [8] | |
| Sequence | CACAGATCTTGCAGCCCAGAACCAAGTCAGCCTCCCTGCGA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000368839.1; ENST00000482963.1; ENST00000368842.10 | ||
| External Link | RMBase: m6A_site_122188 | ||
| mod ID: M6ASITE002293 | Click to Show/Hide the Full List | ||
| mod site | chr10:124613908-124613909:+ | [6] | |
| Sequence | CCCACCACCCCACCCCCGAAACAATGCCAGCCCGCTGCTTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; Huh7; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000368839.1; ENST00000482963.1; ENST00000368842.10 | ||
| External Link | RMBase: m6A_site_122189 | ||
| mod ID: M6ASITE002294 | Click to Show/Hide the Full List | ||
| mod site | chr10:124613955-124613956:+ | [6] | |
| Sequence | CCTCCCAGTCACCTTTGCAGACAAAGACCAGGGGCAGCTCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; Huh7; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000368839.1; ENST00000482963.1; ENST00000368842.10 | ||
| External Link | RMBase: m6A_site_122190 | ||
| mod ID: M6ASITE002295 | Click to Show/Hide the Full List | ||
| mod site | chr10:124613961-124613962:+ | [6] | |
| Sequence | AGTCACCTTTGCAGACAAAGACCAGGGGCAGCTCCCGAGGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; Huh7; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000368839.1; ENST00000482963.1; ENST00000368842.10 | ||
| External Link | RMBase: m6A_site_122191 | ||
| mod ID: M6ASITE002296 | Click to Show/Hide the Full List | ||
| mod site | chr10:124614003-124614004:+ | [5] | |
| Sequence | ACTGTGAAGGCTCCCATGCCACACAGTGAGAACTGTAGCCT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000482963.1; ENST00000368839.1; ENST00000368842.10 | ||
| External Link | RMBase: m6A_site_122192 | ||
| mod ID: M6ASITE002297 | Click to Show/Hide the Full List | ||
| mod site | chr10:124614014-124614015:+ | [6] | |
| Sequence | TCCCATGCCACACAGTGAGAACTGTAGCCTCTGCGTCCAAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; A549; Huh7; Jurkat; CD4T; HEK293A-TOA; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000368842.10; ENST00000482963.1; ENST00000368839.1 | ||
| External Link | RMBase: m6A_site_122193 | ||
| mod ID: M6ASITE002298 | Click to Show/Hide the Full List | ||
| mod site | chr10:124614055-124614056:+ | [6] | |
| Sequence | GCACACAGGGTACTTTCTGGACCCACTGCTGGACAGACTTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; Jurkat; CD4T; HEK293A-TOA; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000368842.10; ENST00000368839.1; ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122194 | ||
| mod ID: M6ASITE002299 | Click to Show/Hide the Full List | ||
| mod site | chr10:124614067-124614068:+ | [6] | |
| Sequence | CTTTCTGGACCCACTGCTGGACAGACTTGAAGGTGTCATGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; Jurkat; CD4T; HEK293A-TOA; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000482963.1; ENST00000368842.10; ENST00000368839.1 | ||
| External Link | RMBase: m6A_site_122195 | ||
| mod ID: M6ASITE002300 | Click to Show/Hide the Full List | ||
| mod site | chr10:124614107-124614108:+ | [6] | |
| Sequence | CCCGGTGTGTGCAGGAGGAAACTAACAGTTCAGTAAACTCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; Jurkat; CD4T; HEK293A-TOA; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000368842.10; ENST00000368839.1; ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122196 | ||
| mod ID: M6ASITE002301 | Click to Show/Hide the Full List | ||
| mod site | chr10:124614123-124614124:+ | [6] | |
| Sequence | GGAAACTAACAGTTCAGTAAACTCTGCCTTGACCAGCAGCC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; Jurkat; CD4T; HEK293A-TOA; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000482963.1; ENST00000368842.10; ENST00000368839.1 | ||
| External Link | RMBase: m6A_site_122197 | ||
| mod ID: M6ASITE002302 | Click to Show/Hide the Full List | ||
| mod site | chr10:124616722-124616723:+ | [6] | |
| Sequence | CCTGGCAGAGACGGTGTGGAACCGCCGGCCGCATGCGGGGC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; H1B | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122198 | ||
| mod ID: M6ASITE002303 | Click to Show/Hide the Full List | ||
| mod site | chr10:124616797-124616798:+ | [6] | |
| Sequence | TGTTCAACACATGTCGCGGAACTGTCGTGAAAAAAAGACTC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; H1A; H1B | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122199 | ||
| mod ID: M6ASITE002304 | Click to Show/Hide the Full List | ||
| mod site | chr10:124616814-124616815:+ | [6] | |
| Sequence | GGAACTGTCGTGAAAAAAAGACTCAGGACCACAGGTCCGGG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; H1A; H1B; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122200 | ||
| mod ID: M6ASITE002305 | Click to Show/Hide the Full List | ||
| mod site | chr10:124616821-124616822:+ | [6] | |
| Sequence | TCGTGAAAAAAAGACTCAGGACCACAGGTCCGGGGTGCTCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; H1A; H1B; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122201 | ||
| mod ID: M6ASITE002306 | Click to Show/Hide the Full List | ||
| mod site | chr10:124616965-124616966:+ | [9] | |
| Sequence | GGTGGGGGGTGGAGGGGAGGACAGGGAGAGCAGGCCTCTCG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | H1A; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122202 | ||
| mod ID: M6ASITE002307 | Click to Show/Hide the Full List | ||
| mod site | chr10:124617256-124617257:+ | [6] | |
| Sequence | CTGCAGAAAAGGCCCACGAGACTCAGACTGGCAGAGACTTA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; H1A; H1B; HEK293A-TOA; TREX; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122204 | ||
| mod ID: M6ASITE002308 | Click to Show/Hide the Full List | ||
| mod site | chr10:124617262-124617263:+ | [6] | |
| Sequence | AAAAGGCCCACGAGACTCAGACTGGCAGAGACTTAGGCGGA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; HEK293A-TOA; MSC; TREX; iSLK; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122205 | ||
| mod ID: M6ASITE002309 | Click to Show/Hide the Full List | ||
| mod site | chr10:124617272-124617273:+ | [6] | |
| Sequence | CGAGACTCAGACTGGCAGAGACTTAGGCGGACCAGGAACAG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; HEK293A-TOA; MSC; TREX; iSLK; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122206 | ||
| mod ID: M6ASITE002310 | Click to Show/Hide the Full List | ||
| mod site | chr10:124617282-124617283:+ | [6] | |
| Sequence | ACTGGCAGAGACTTAGGCGGACCAGGAACAGGGGCGCAGTC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; HEK293A-TOA; MSC; TREX; iSLK; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122207 | ||
| mod ID: M6ASITE002311 | Click to Show/Hide the Full List | ||
| mod site | chr10:124617289-124617290:+ | [6] | |
| Sequence | GAGACTTAGGCGGACCAGGAACAGGGGCGCAGTCTCCGTCC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; HEK293A-TOA; TREX; iSLK; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122208 | ||
| mod ID: M6ASITE002312 | Click to Show/Hide the Full List | ||
| mod site | chr10:124617317-124617318:+ | [6] | |
| Sequence | GCAGTCTCCGTCCCACCCAAACCCTAACCAGAGAGAACACG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; HEK293T; HEK293A-TOA; TREX; iSLK; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122210 | ||
| mod ID: M6ASITE002313 | Click to Show/Hide the Full List | ||
| mod site | chr10:124617333-124617334:+ | [6] | |
| Sequence | CCAAACCCTAACCAGAGAGAACACGGCACGTTGTGCCAGAC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; CD4T; HEK293T; HEK293A-TOA; TREX; iSLK; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122211 | ||
| mod ID: M6ASITE002314 | Click to Show/Hide the Full List | ||
| mod site | chr10:124617406-124617407:+ | [6] | |
| Sequence | CACTGCCGACAAGGCTGGGAACTGGGCCAAGTGAAGCAGAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; TREX; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122214 | ||
| mod ID: M6ASITE002315 | Click to Show/Hide the Full List | ||
| mod site | chr10:124617650-124617651:+ | [10] | |
| Sequence | ACCTCAGGGCAGAGGTGGGGACACAGGGCGGGACGGGCGAG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122220 | ||
| mod ID: M6ASITE002316 | Click to Show/Hide the Full List | ||
| mod site | chr10:124617753-124617754:+ | [10] | |
| Sequence | AGTTCCAGCTGCCCCTCAGAACTGTCCTGGTTTAGGAGGTG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000482963.1 | ||
| External Link | RMBase: m6A_site_122223 | ||
References