m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00714)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
KLF2
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Fat mass and obesity-associated protein (FTO) [ERASER]
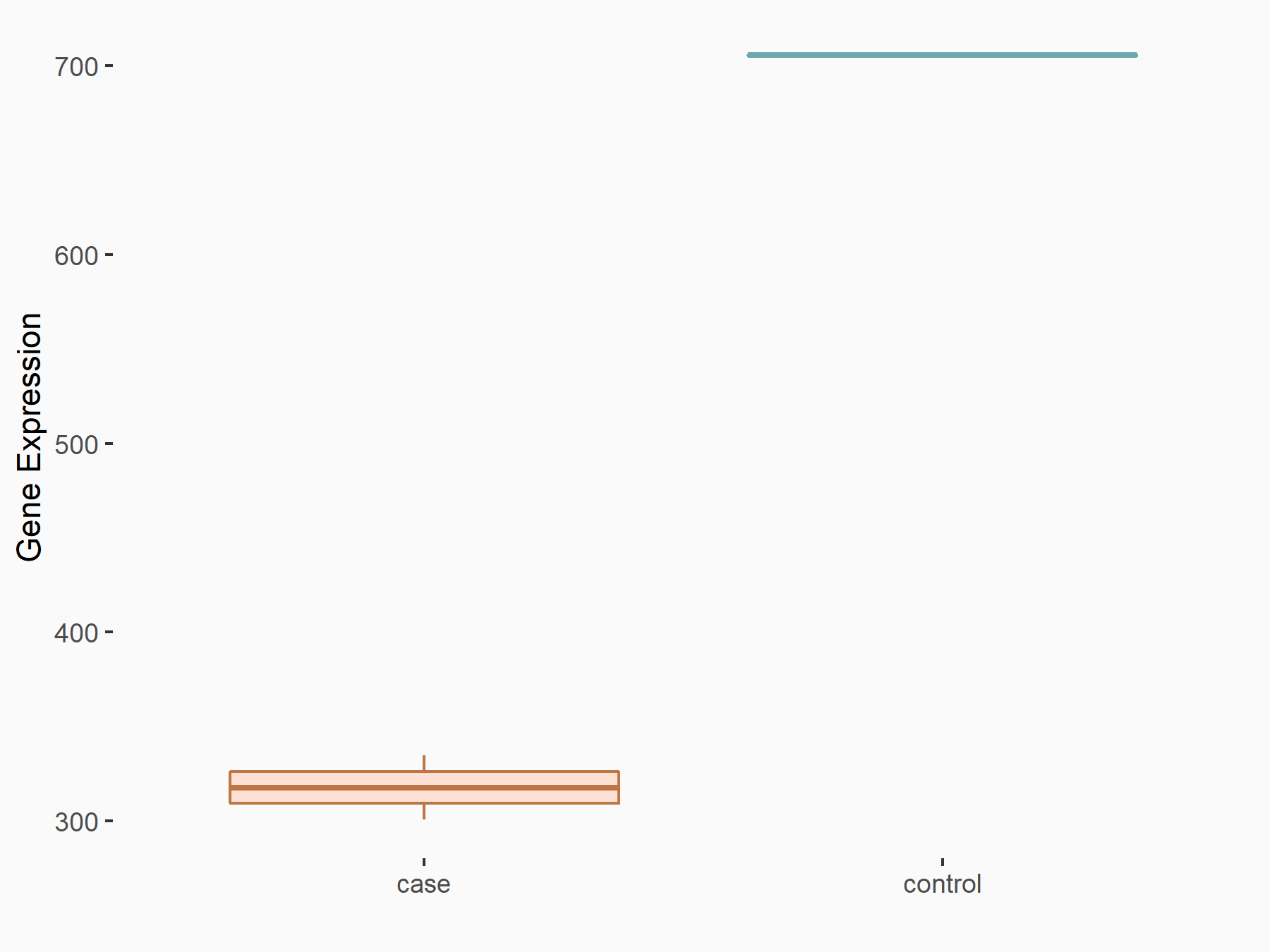
| Representative RNA-seq result indicating the expression of this target gene regulated by FTO | ||
| Cell Line | B16F10 cell line | Mus musculus |
|
Treatment: FTO knockout B16F10 cells
Control: B16F10 cells
|
GSE134388 | |
| Regulation |
  |
logFC: -1.68E+00 p-value: 3.25E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | FTO overexpression significantly upregulated the mRNA and protein levels of VCAM-1 and ICAM-1, downregulated those of Krueppel-like factor 2 (KLF2) and eNOS, and strongly attenuated the atorvastatin-mediated induction of KLF2 and eNOS expression. FTO could serve as a novel molecular target to modulate endothelial function in vascular diseases. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Vascular diseases | ICD-11: BE2Z | ||
| Responsed Drug | Atorvastatin | Approved | ||
| In-vitro Model | THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 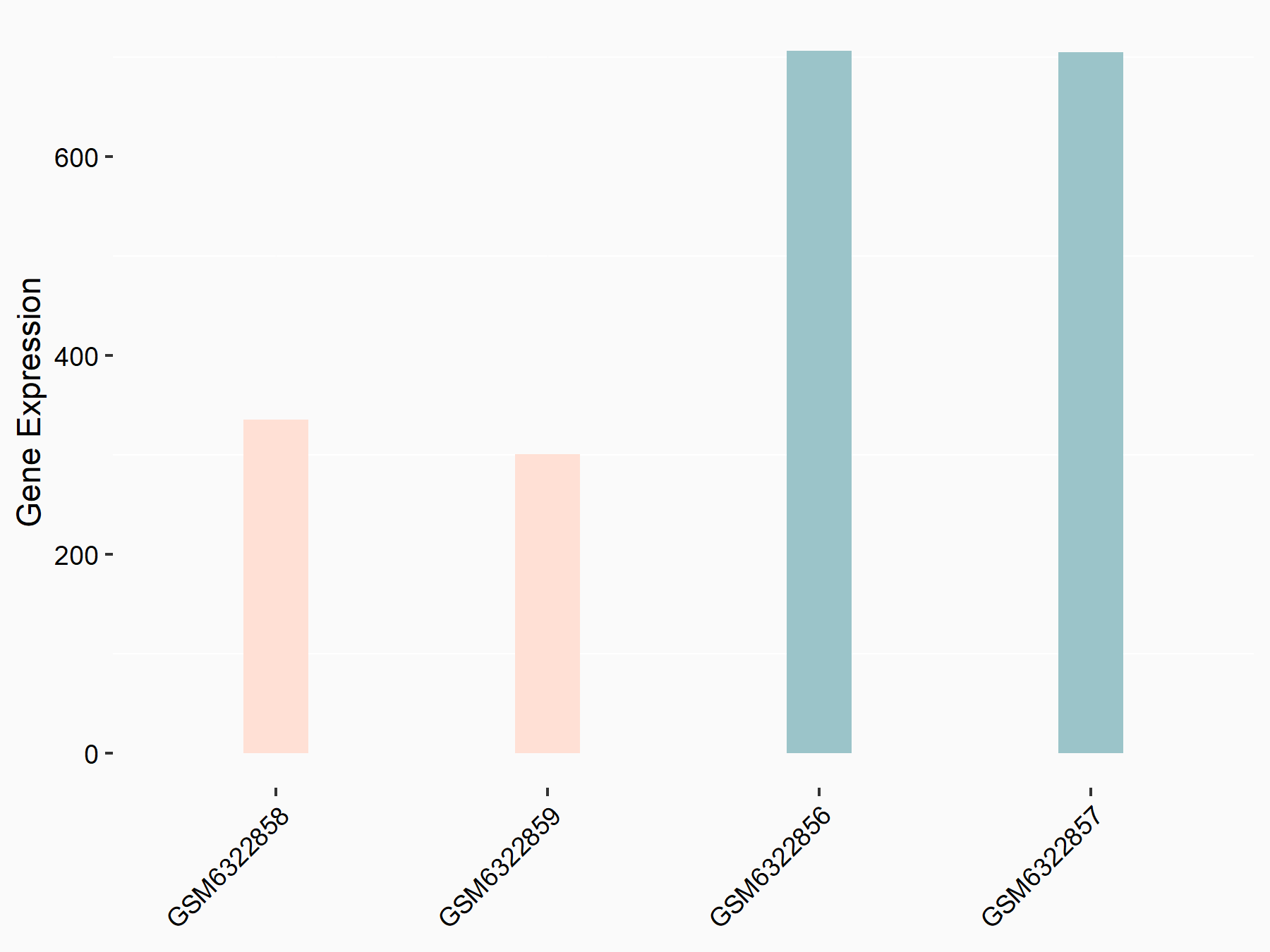 |
logFC: -1.15E+00 p-value: 3.23E-14 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 promotes translation of SPHK2 mRNA via an m6A-YTHDF1-dependent manner. Functionally, SPHK2 facilitates GC cell proliferation, migration and invasion by inhibiting Krueppel-like factor 2 (KLF2) expression. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 promotes translation of SPHK2 mRNA via an m6A-YTHDF1-dependent manner. Functionally, SPHK2 facilitates GC cell proliferation, migration and invasion by inhibiting Krueppel-like factor 2 (KLF2) expression. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
Diseases of the circulatory system [ICD-11: BE2Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | FTO overexpression significantly upregulated the mRNA and protein levels of VCAM-1 and ICAM-1, downregulated those of Krueppel-like factor 2 (KLF2) and eNOS, and strongly attenuated the atorvastatin-mediated induction of KLF2 and eNOS expression. FTO could serve as a novel molecular target to modulate endothelial function in vascular diseases. | |||
| Responsed Disease | Vascular diseases [ICD-11: BE2Z] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Atorvastatin | Approved | ||
| In-vitro Model | THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Atorvastatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | FTO overexpression significantly upregulated the mRNA and protein levels of VCAM-1 and ICAM-1, downregulated those of Krueppel-like factor 2 (KLF2) and eNOS, and strongly attenuated the atorvastatin-mediated induction of KLF2 and eNOS expression. FTO could serve as a novel molecular target to modulate endothelial function in vascular diseases. | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Vascular diseases | ICD-11: BE2Z | ||
| In-vitro Model | THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03653 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00714)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE009082 | Click to Show/Hide the Full List | ||
| mod site | chr19:16325053-16325054:+ | [3] | |
| Sequence | GTGGGCGGCGGGCTCGGGGTAGTAGAACGTGGGCTGCGGGG | ||
| Transcript ID List | ENST00000592003.1; ENST00000248071.5 | ||
| External Link | RMBase: RNA-editing_site_67126 | ||
5-methylcytidine (m5C)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE001844 | Click to Show/Hide the Full List | ||
| mod site | chr19:16325189-16325190:+ | ||
| Sequence | CCCGAGCCCCGCCGCCCGCTCACGCCCATTGCCCTGTCGCC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000248071.5; ENST00000592003.1; rmsk_4952862 | ||
| External Link | RMBase: m5C_site_23348 | ||
| mod ID: M5CSITE001845 | Click to Show/Hide the Full List | ||
| mod site | chr19:16325635-16325636:+ | [4] | |
| Sequence | GCATGCGAGGTCCCGGGGGCCGCCCCCCGCCGCCGCCCGAC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000248071.5; ENST00000592003.1 | ||
| External Link | RMBase: m5C_site_23349 | ||
N6-methyladenosine (m6A)
| In total 14 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE040444 | Click to Show/Hide the Full List | ||
| mod site | chr19:16324937-16324938:+ | [5] | |
| Sequence | CCGGCCATGGCGCTGAGTGAACCCATCCTGCCGTCCTTCTC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; A549; U2OS; Jurkat; CD4T; GSC-11; HEK293A-TOA; iSLK; TIME; TREX; MSC; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000248071.5; ENST00000592003.1 | ||
| External Link | RMBase: m6A_site_426233 | ||
| mod ID: M6ASITE040445 | Click to Show/Hide the Full List | ||
| mod site | chr19:16325232-16325233:+ | [6] | |
| Sequence | CAGCGCTGGCCGCGCGCCGAACCCGAGTCCGGCGGCACCGA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | A549; U2OS; Jurkat; CD4T; GSC-11; TREX; iSLK; TIME; MSC; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000248071.5; ENST00000592003.1 | ||
| External Link | RMBase: m6A_site_426234 | ||
| mod ID: M6ASITE040446 | Click to Show/Hide the Full List | ||
| mod site | chr19:16325276-16325277:+ | [6] | |
| Sequence | CGACCTCAACAGCGTGCTGGACTTCATCCTGTCCATGGGGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | A549; U2OS; Jurkat; CD4T; GSC-11; HEK293T; TREX; iSLK; TIME; MSC; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000248071.5; ENST00000592003.1 | ||
| External Link | RMBase: m6A_site_426235 | ||
| mod ID: M6ASITE040447 | Click to Show/Hide the Full List | ||
| mod site | chr19:16325370-16325371:+ | [6] | |
| Sequence | CCTGCGTTCTATTACCCCGAACCCGGCGCGCCCCCGCCCTA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | A549; U2OS; Jurkat; GSC-11; iSLK; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000592003.1; ENST00000248071.5 | ||
| External Link | RMBase: m6A_site_426236 | ||
| mod ID: M6ASITE040448 | Click to Show/Hide the Full List | ||
| mod site | chr19:16325983-16325984:+ | [7] | |
| Sequence | GCTACGCGGGCTGCGGCAAGACCTACACCAAGAGTTCGCAT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | CD34 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000592003.1; ENST00000248071.5 | ||
| External Link | RMBase: m6A_site_426237 | ||
| mod ID: M6ASITE040449 | Click to Show/Hide the Full List | ||
| mod site | chr19:16327100-16327101:+ | [5] | |
| Sequence | CGGGCGCGGCCCCCTCCCAAACTGTGACTGGTATTTATTGG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; fibroblasts; GM12878; LCLs; Jurkat; CD4T; HEK293A-TOA; iSLK; TIME; TREX; MSC; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000592003.1; ENST00000248071.5 | ||
| External Link | RMBase: m6A_site_426238 | ||
| mod ID: M6ASITE040450 | Click to Show/Hide the Full List | ||
| mod site | chr19:16327106-16327107:+ | [8] | |
| Sequence | CGGCCCCCTCCCAAACTGTGACTGGTATTTATTGGACCCAG | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000592003.1; ENST00000248071.5 | ||
| External Link | RMBase: m6A_site_426239 | ||
| mod ID: M6ASITE040451 | Click to Show/Hide the Full List | ||
| mod site | chr19:16327121-16327122:+ | [5] | |
| Sequence | CTGTGACTGGTATTTATTGGACCCAGAGAACCGGGCCGGGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; fibroblasts; GM12878; LCLs; H1299; MM6; Jurkat; CD4T; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000592003.1; ENST00000248071.5 | ||
| External Link | RMBase: m6A_site_426240 | ||
| mod ID: M6ASITE040452 | Click to Show/Hide the Full List | ||
| mod site | chr19:16327130-16327131:+ | [5] | |
| Sequence | GTATTTATTGGACCCAGAGAACCGGGCCGGGCACAGCGTGG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; fibroblasts; GM12878; LCLs; H1299; MM6; Jurkat; CD4T; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000592003.1; ENST00000248071.5 | ||
| External Link | RMBase: m6A_site_426241 | ||
| mod ID: M6ASITE040453 | Click to Show/Hide the Full List | ||
| mod site | chr19:16327212-16327213:+ | [8] | |
| Sequence | CACCCCAGCCCCCGTCTGTGACTGAAGGCCCGGTGGGAAAA | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000248071.5; ENST00000592003.1 | ||
| External Link | RMBase: m6A_site_426242 | ||
| mod ID: M6ASITE040454 | Click to Show/Hide the Full List | ||
| mod site | chr19:16327234-16327235:+ | [5] | |
| Sequence | TGAAGGCCCGGTGGGAAAAGACCACGATCCTCCTTGACGAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; H1B; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000248071.5; ENST00000592003.1 | ||
| External Link | RMBase: m6A_site_426243 | ||
| mod ID: M6ASITE040455 | Click to Show/Hide the Full List | ||
| mod site | chr19:16327312-16327313:+ | [9] | |
| Sequence | TCTTCTCTCCCACCGGGTCTACACTAGAGGATCGAGGCTTG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000248071.5 | ||
| External Link | RMBase: m6A_site_426244 | ||
| mod ID: M6ASITE040456 | Click to Show/Hide the Full List | ||
| mod site | chr19:16327408-16327409:+ | [9] | |
| Sequence | AATATTGTATATAGTGACTGACAAATATTGTATTACTGTAC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000248071.5 | ||
| External Link | RMBase: m6A_site_426245 | ||
| mod ID: M6ASITE040457 | Click to Show/Hide the Full List | ||
| mod site | chr19:16327437-16327438:+ | [10] | |
| Sequence | GTATTACTGTACATAGAGAGACAGGTGGGCATTTTTGGGCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | Huh7; TIME; TREX | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000248071.5 | ||
| External Link | RMBase: m6A_site_426246 | ||
References