m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00370)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
PLK1
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
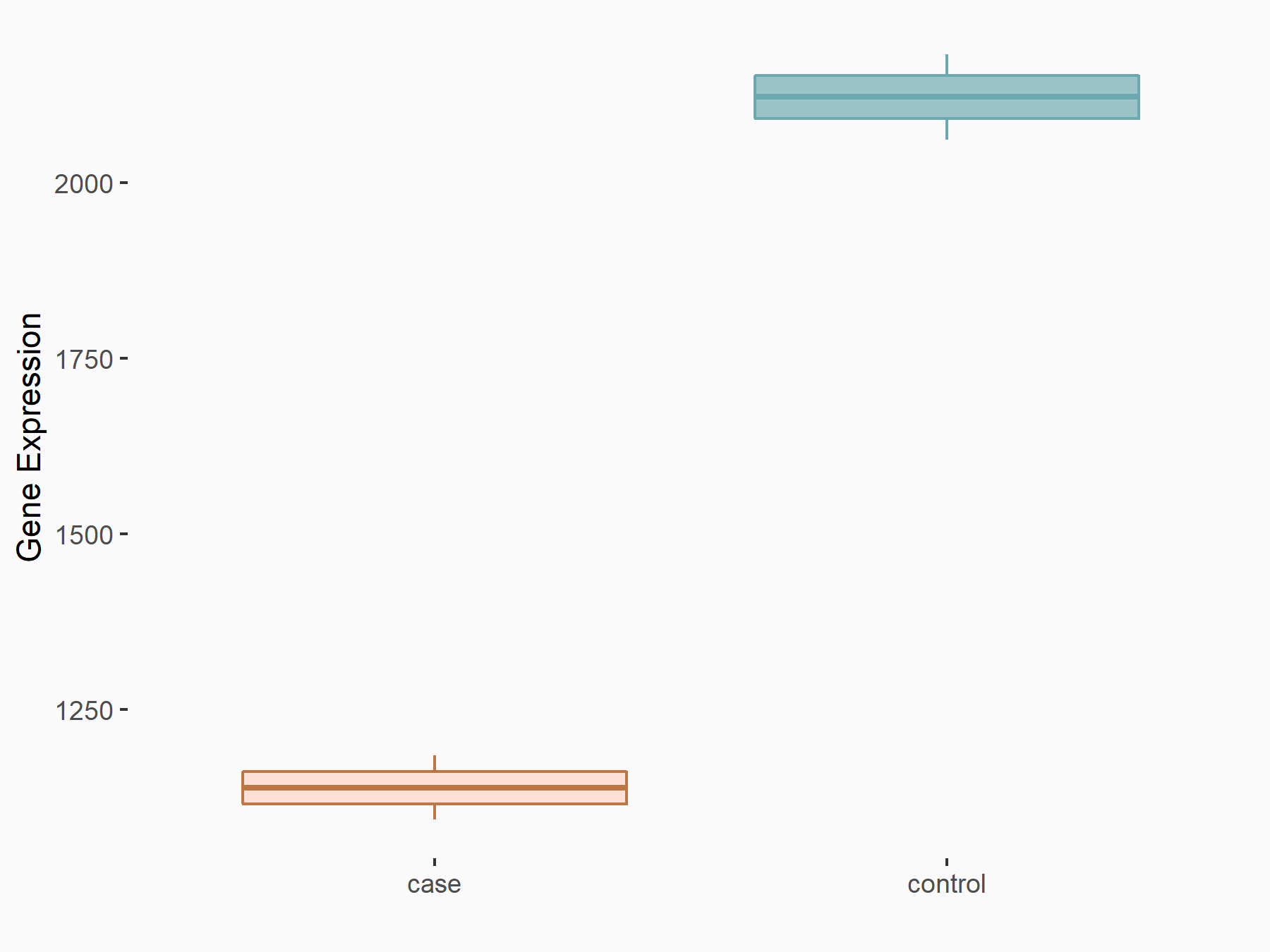  |
logFC: -8.98E-01 p-value: 1.15E-18 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Impaired METTL3 expression in dental pulp stem cells led to increasing cell senescence and apoptosis by interfering with the mitotic cell cycle in a m6A-dependent manner. The protein interaction network of differentially expressed genes identified Serine/threonine-protein kinase PLK1 (PLK1), a critical cycle modulator, as the target of METTL3-mediated m6A methylation in DPSCs. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Pulpitis | ICD-11: DA09 | ||
| Cell Process | Cell senescence | |||
| In-vitro Model | DPSC | Normal | Homo sapiens | CVCL_AV90 |
Pulpitis [ICD-11: DA09]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Impaired METTL3 expression in dental pulp stem cells led to increasing cell senescence and apoptosis by interfering with the mitotic cell cycle in a m6A-dependent manner. The protein interaction network of differentially expressed genes identified Serine/threonine-protein kinase PLK1 (PLK1), a critical cycle modulator, as the target of METTL3-mediated m6A methylation in DPSCs. | |||
| Responsed Disease | Pulpitis [ICD-11: DA09] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Cell Process | Cell senescence | |||
| In-vitro Model | DPSC | Normal | Homo sapiens | CVCL_AV90 |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00370)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: 2OMSITE000131 | Click to Show/Hide the Full List | ||
| mod site | chr16:23679149-23679150:+ | [2] | |
| Sequence | GTGCTTCGAGATCTCGGACGCGGACACCAAGGAGGTGTTCG | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000570220.5; ENST00000562272.5; ENST00000564202.1; ENST00000300093.9 | ||
| External Link | RMBase: Nm_site_2058 | ||
| mod ID: 2OMSITE000132 | Click to Show/Hide the Full List | ||
| mod site | chr16:23680096-23680097:+ | [2] | |
| Sequence | CCCACAGTCTCTCCTGGAGCTGCACAAGAGGAGGAAAGCCC | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000568568.1; ENST00000562272.5; ENST00000570220.5; ENST00000567897.6; ENST00000300093.9; ENST00000564202.1 | ||
| External Link | RMBase: Nm_site_2059 | ||
5-methylcytidine (m5C)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE000620 | Click to Show/Hide the Full List | ||
| mod site | chr16:23680910-23680911:+ | [3] | |
| Sequence | AACTCTTCCTCCCTCTGTCCCAGGGGATTTTGGACTGGCAA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000570220.5; ENST00000562272.5; ENST00000567897.6; ENST00000300093.9; ENST00000564202.1; ENST00000568568.1 | ||
| External Link | RMBase: m5C_site_15611 | ||
N6-methyladenosine (m6A)
| In total 44 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE026201 | Click to Show/Hide the Full List | ||
| mod site | chr16:23679068-23679069:+ | [4] | |
| Sequence | GATCCCGGAGGTCCTAGTGGACCCACGCAGCCGGCGGCGCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; A549; H1A; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; TREX; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000570220.5; ENST00000300093.9; ENST00000562272.5; ENST00000564202.1 | ||
| External Link | RMBase: m6A_site_312875 | ||
| mod ID: M6ASITE026202 | Click to Show/Hide the Full List | ||
| mod site | chr16:23679152-23679153:+ | [4] | |
| Sequence | CTTCGAGATCTCGGACGCGGACACCAAGGAGGTGTTCGCGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; HEK293T; NB4; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000570220.5; ENST00000300093.9; ENST00000564202.1; ENST00000562272.5 | ||
| External Link | RMBase: m6A_site_312876 | ||
| mod ID: M6ASITE026203 | Click to Show/Hide the Full List | ||
| mod site | chr16:23679296-23679297:+ | [4] | |
| Sequence | ATTCCACGGCTTTTTCGAGGACAACGACTTCGTGTTCGTGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; MT4; peripheral-blood | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000562272.5; ENST00000564202.1; ENST00000300093.9; ENST00000570220.5 | ||
| External Link | RMBase: m6A_site_312877 | ||
| mod ID: M6ASITE026204 | Click to Show/Hide the Full List | ||
| mod site | chr16:23679360-23679361:+ | [4] | |
| Sequence | GTGAGTGTCGCTGCTGGGGAACTGGAACTGCCTGCGGGGCA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000562272.5; ENST00000570220.5; ENST00000300093.9; ENST00000564202.1 | ||
| External Link | RMBase: m6A_site_312878 | ||
| mod ID: M6ASITE026205 | Click to Show/Hide the Full List | ||
| mod site | chr16:23679366-23679367:+ | [4] | |
| Sequence | GTCGCTGCTGGGGAACTGGAACTGCCTGCGGGGCAGTTGGA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000570220.5; ENST00000300093.9; ENST00000562272.5; ENST00000564202.1 | ||
| External Link | RMBase: m6A_site_312879 | ||
| mod ID: M6ASITE026206 | Click to Show/Hide the Full List | ||
| mod site | chr16:23679395-23679396:+ | [4] | |
| Sequence | GGGGCAGTTGGAGCGCCCAGACCTGGAGCTGCTGGAAAGAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000562272.5; ENST00000570220.5; ENST00000564202.1; ENST00000300093.9 | ||
| External Link | RMBase: m6A_site_312880 | ||
| mod ID: M6ASITE026207 | Click to Show/Hide the Full List | ||
| mod site | chr16:23679439-23679440:+ | [4] | |
| Sequence | CCAGCAAGGGAGAGCCTGGGACTTGGAGCTGCTAGAGAAGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000570220.5; ENST00000564202.1; ENST00000562272.5; ENST00000300093.9 | ||
| External Link | RMBase: m6A_site_312881 | ||
| mod ID: M6ASITE026208 | Click to Show/Hide the Full List | ||
| mod site | chr16:23679733-23679734:+ | [4] | |
| Sequence | GGGGGGGTAGTTGGAGCCAGACTTCGGGCCTTCCGGGGGAG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000568568.1; ENST00000564202.1; ENST00000562272.5; ENST00000300093.9; ENST00000570220.5; ENST00000567897.6 | ||
| External Link | RMBase: m6A_site_312882 | ||
| mod ID: M6ASITE026209 | Click to Show/Hide the Full List | ||
| mod site | chr16:23679786-23679787:+ | [4] | |
| Sequence | AAAGGGATCTGGTGCTGGGGACTCGGAGCTTCCGGAGTGGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000562272.5; ENST00000568568.1; ENST00000570220.5; ENST00000564202.1; ENST00000300093.9; ENST00000567897.6 | ||
| External Link | RMBase: m6A_site_312883 | ||
| mod ID: M6ASITE026210 | Click to Show/Hide the Full List | ||
| mod site | chr16:23679818-23679819:+ | [4] | |
| Sequence | CGGAGTGGGGCTGAGAGAGGACCCCCAGGTAAGGGGGGAAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000568568.1; ENST00000562272.5; ENST00000567897.6; ENST00000300093.9; ENST00000570220.5; ENST00000564202.1 | ||
| External Link | RMBase: m6A_site_312884 | ||
| mod ID: M6ASITE026211 | Click to Show/Hide the Full List | ||
| mod site | chr16:23679873-23679874:+ | [4] | |
| Sequence | GGTGCTGCGAATGGTTGTGGACAGTGTTAAGGCAGGGTCCA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000568568.1; ENST00000564202.1; ENST00000567897.6; ENST00000570220.5; ENST00000562272.5; ENST00000300093.9 | ||
| External Link | RMBase: m6A_site_312885 | ||
| mod ID: M6ASITE026212 | Click to Show/Hide the Full List | ||
| mod site | chr16:23679946-23679947:+ | [4] | |
| Sequence | GGGCATAGGGAAGGAGAGAAACCCACCAAGACCCCTCTTTC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000567897.6; ENST00000300093.9; ENST00000568568.1; ENST00000562272.5; ENST00000570220.5; ENST00000564202.1 | ||
| External Link | RMBase: m6A_site_312886 | ||
| mod ID: M6ASITE026213 | Click to Show/Hide the Full List | ||
| mod site | chr16:23679956-23679957:+ | [4] | |
| Sequence | AAGGAGAGAAACCCACCAAGACCCCTCTTTCATCCCTTGGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000300093.9; ENST00000570220.5; ENST00000567897.6; ENST00000568568.1; ENST00000564202.1; ENST00000562272.5 | ||
| External Link | RMBase: m6A_site_312887 | ||
| mod ID: M6ASITE026214 | Click to Show/Hide the Full List | ||
| mod site | chr16:23680099-23680100:+ | [5] | |
| Sequence | ACAGTCTCTCCTGGAGCTGCACAAGAGGAGGAAAGCCCTGA | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000564202.1; ENST00000562272.5; ENST00000300093.9; ENST00000567897.6; ENST00000568568.1; ENST00000570220.5 | ||
| External Link | RMBase: m6A_site_312888 | ||
| mod ID: M6ASITE026215 | Click to Show/Hide the Full List | ||
| mod site | chr16:23680183-23680184:+ | [4] | |
| Sequence | CTGCCAGTACCTGCACCGAAACCGAGTTATTCATCGAGACC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; MM6; Huh7; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000562272.5; ENST00000568568.1; ENST00000567897.6; ENST00000564202.1; ENST00000570220.5; ENST00000300093.9 | ||
| External Link | RMBase: m6A_site_312889 | ||
| mod ID: M6ASITE026216 | Click to Show/Hide the Full List | ||
| mod site | chr16:23680201-23680202:+ | [4] | |
| Sequence | AAACCGAGTTATTCATCGAGACCTCAAGCTGGGCAACCTTT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; MM6; Huh7; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000300093.9; ENST00000570220.5; ENST00000568568.1; ENST00000564202.1; ENST00000562272.5; ENST00000567897.6 | ||
| External Link | RMBase: m6A_site_312890 | ||
| mod ID: M6ASITE026217 | Click to Show/Hide the Full List | ||
| mod site | chr16:23680923-23680924:+ | [4] | |
| Sequence | TCTGTCCCAGGGGATTTTGGACTGGCAACCAAAGTCGAATA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000568568.1; ENST00000567897.6; ENST00000564202.1; ENST00000562272.5; ENST00000570220.5; ENST00000300093.9 | ||
| External Link | RMBase: m6A_site_312891 | ||
| mod ID: M6ASITE026218 | Click to Show/Hide the Full List | ||
| mod site | chr16:23680963-23680964:+ | [4] | |
| Sequence | ATGACGGGGAGAGGAAGAAGACCCTGTGTGGGACTCCTAAT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000568568.1; ENST00000570220.5; ENST00000300093.9; ENST00000564202.1; ENST00000567897.6; ENST00000562272.5 | ||
| External Link | RMBase: m6A_site_312892 | ||
| mod ID: M6ASITE026219 | Click to Show/Hide the Full List | ||
| mod site | chr16:23680975-23680976:+ | [4] | |
| Sequence | GGAAGAAGACCCTGTGTGGGACTCCTAATTACATAGCTCCC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000568568.1; ENST00000564202.1; ENST00000300093.9; ENST00000567897.6; ENST00000570220.5; ENST00000562272.5 | ||
| External Link | RMBase: m6A_site_312893 | ||
| mod ID: M6ASITE026220 | Click to Show/Hide the Full List | ||
| mod site | chr16:23680985-23680986:+ | [5] | |
| Sequence | CCTGTGTGGGACTCCTAATTACATAGCTCCCGAGGTGCTGA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000568568.1; ENST00000567897.6; ENST00000564202.1; ENST00000300093.9; ENST00000562272.5; ENST00000570220.5 | ||
| External Link | RMBase: m6A_site_312894 | ||
| mod ID: M6ASITE026221 | Click to Show/Hide the Full List | ||
| mod site | chr16:23682084-23682085:+ | [4] | |
| Sequence | TATACCTTGTTAGTGGGCAAACCACCTTTTGAGACTTCTTG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000562272.5; ENST00000568568.1; ENST00000567897.6; ENST00000300093.9 | ||
| External Link | RMBase: m6A_site_312895 | ||
| mod ID: M6ASITE026222 | Click to Show/Hide the Full List | ||
| mod site | chr16:23682097-23682098:+ | [4] | |
| Sequence | TGGGCAAACCACCTTTTGAGACTTCTTGCCTAAAAGAGACC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000300093.9; ENST00000562272.5; ENST00000567897.6; ENST00000568568.1 | ||
| External Link | RMBase: m6A_site_312896 | ||
| mod ID: M6ASITE026223 | Click to Show/Hide the Full List | ||
| mod site | chr16:23682115-23682116:+ | [4] | |
| Sequence | AGACTTCTTGCCTAAAAGAGACCTACCTCCGGATCAAGAAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000567897.6; ENST00000562272.5; ENST00000300093.9; ENST00000568568.1 | ||
| External Link | RMBase: m6A_site_312897 | ||
| mod ID: M6ASITE026224 | Click to Show/Hide the Full List | ||
| mod site | chr16:23683870-23683871:+ | [5] | |
| Sequence | GTCCCTCTCTCTGCCCCAGCACATCAACCCCGTGGCCGCCT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000562272.5; ENST00000568568.1; ENST00000567897.6; ENST00000300093.9 | ||
| External Link | RMBase: m6A_site_312898 | ||
| mod ID: M6ASITE026225 | Click to Show/Hide the Full List | ||
| mod site | chr16:23683914-23683915:+ | [4] | |
| Sequence | TCATCCAGAAGATGCTTCAGACAGATCCCACTGCCCGCCCA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000567897.6; ENST00000300093.9; ENST00000562272.5; ENST00000562407.1; ENST00000568568.1 | ||
| External Link | RMBase: m6A_site_312899 | ||
| mod ID: M6ASITE026226 | Click to Show/Hide the Full List | ||
| mod site | chr16:23684050-23684051:+ | [4] | |
| Sequence | GATTGCTCCCAGCAGCCTGGACCCCAGCAACCGGAAGCCCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000567897.6; ENST00000562272.5; ENST00000300093.9; ENST00000562407.1 | ||
| External Link | RMBase: m6A_site_312900 | ||
| mod ID: M6ASITE026227 | Click to Show/Hide the Full List | ||
| mod site | chr16:23687477-23687478:+ | [4] | |
| Sequence | CACCTTCCTAGGCTTGGAGAACCCCCTGCCTGAGCGTCCCC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000562272.5; ENST00000564947.1; ENST00000562407.1; ENST00000300093.9 | ||
| External Link | RMBase: m6A_site_312902 | ||
| mod ID: M6ASITE026228 | Click to Show/Hide the Full List | ||
| mod site | chr16:23687511-23687512:+ | [4] | |
| Sequence | CGTCCCCGGGAAAAAGAAGAACCAGTGGTTCGAGAGACAGG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000562407.1; ENST00000562272.5; ENST00000564947.1; ENST00000300093.9 | ||
| External Link | RMBase: m6A_site_312903 | ||
| mod ID: M6ASITE026229 | Click to Show/Hide the Full List | ||
| mod site | chr16:23687527-23687528:+ | [4] | |
| Sequence | AAGAACCAGTGGTTCGAGAGACAGGTGAGGTGGTCGACTGC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; endometrial | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000562272.5; ENST00000562407.1; ENST00000300093.9; ENST00000564947.1 | ||
| External Link | RMBase: m6A_site_312905 | ||
| mod ID: M6ASITE026230 | Click to Show/Hide the Full List | ||
| mod site | chr16:23687737-23687738:+ | [4] | |
| Sequence | TCAGGCCCTGTCTGCATAGGACCATTGGTTTCCCCATTTCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000564947.1; ENST00000300093.9; ENST00000562407.1; ENST00000562272.5 | ||
| External Link | RMBase: m6A_site_312906 | ||
| mod ID: M6ASITE026231 | Click to Show/Hide the Full List | ||
| mod site | chr16:23688730-23688731:+ | [5] | |
| Sequence | CAAGTGGGTGGACTATTCGGACAAGTACGGCCTTGGTAGGT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000300093.9; ENST00000564794.1; ENST00000562272.5; ENST00000564947.1 | ||
| External Link | RMBase: m6A_site_312907 | ||
| mod ID: M6ASITE026232 | Click to Show/Hide the Full List | ||
| mod site | chr16:23689324-23689325:+ | [5] | |
| Sequence | TGATGGTGACAGCCTGCAGTACATAGAGCGTGACGGCACTG | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000562272.5; ENST00000300093.9; ENST00000564794.1; rmsk_4493764 | ||
| External Link | RMBase: m6A_site_312908 | ||
| mod ID: M6ASITE026233 | Click to Show/Hide the Full List | ||
| mod site | chr16:23689521-23689522:+ | [5] | |
| Sequence | CCTTAAATATTTCCGCAATTACATGAGCGAGCACTTGCTGA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000564794.1; ENST00000562272.5; ENST00000300093.9 | ||
| External Link | RMBase: m6A_site_312909 | ||
| mod ID: M6ASITE026234 | Click to Show/Hide the Full List | ||
| mod site | chr16:23689554-23689555:+ | [5] | |
| Sequence | CTTGCTGAAGGCAGGTGCCAACATCACGCCGCGCGAAGGTG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000562272.5; ENST00000564794.1; ENST00000300093.9 | ||
| External Link | RMBase: m6A_site_312910 | ||
| mod ID: M6ASITE026235 | Click to Show/Hide the Full List | ||
| mod site | chr16:23689604-23689605:+ | [6] | |
| Sequence | CCCGGCTGCCCTACCTACGGACCTGGTTCCGCACCCGCAGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000562272.5; ENST00000564794.1; ENST00000300093.9 | ||
| External Link | RMBase: m6A_site_312911 | ||
| mod ID: M6ASITE026236 | Click to Show/Hide the Full List | ||
| mod site | chr16:23689863-23689864:+ | [5] | |
| Sequence | TGCTCTTCTCTTGCAGGATCACACCAAGCTCATCTTGTGCC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000562272.5; ENST00000300093.9; ENST00000564794.1 | ||
| External Link | RMBase: m6A_site_312912 | ||
| mod ID: M6ASITE026237 | Click to Show/Hide the Full List | ||
| mod site | chr16:23689905-23689906:+ | [5] | |
| Sequence | ACTGATGGCAGCCGTGACCTACATCGACGAGAAGCGGGACT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000562272.5; ENST00000300093.9; ENST00000564794.1 | ||
| External Link | RMBase: m6A_site_312913 | ||
| mod ID: M6ASITE026238 | Click to Show/Hide the Full List | ||
| mod site | chr16:23689923-23689924:+ | [6] | |
| Sequence | CTACATCGACGAGAAGCGGGACTTCCGCACATACCGCCTGA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HepG2; HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000300093.9; ENST00000564794.1; ENST00000562272.5 | ||
| External Link | RMBase: m6A_site_312914 | ||
| mod ID: M6ASITE026239 | Click to Show/Hide the Full List | ||
| mod site | chr16:23689931-23689932:+ | [5] | |
| Sequence | ACGAGAAGCGGGACTTCCGCACATACCGCCTGAGTCTCCTG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000300093.9; ENST00000564794.1; ENST00000562272.5 | ||
| External Link | RMBase: m6A_site_312915 | ||
| mod ID: M6ASITE026240 | Click to Show/Hide the Full List | ||
| mod site | chr16:23690013-23690014:+ | [5] | |
| Sequence | CTACGCCCGCACTATGGTGGACAAGCTGCTGAGCTCACGCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T; HepG2; HeLa; hESCs; A549; MM6 | ||
| Seq Type List | MAZTER-seq; m6A-seq | ||
| Transcript ID List | ENST00000564794.1; ENST00000562272.5; ENST00000300093.9 | ||
| External Link | RMBase: m6A_site_312916 | ||
| mod ID: M6ASITE026241 | Click to Show/Hide the Full List | ||
| mod site | chr16:23690082-23690083:+ | [7] | |
| Sequence | ATAGCTGCCCTCCCCTCCGGACTGGTGCCCTCCTCACTCCC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HepG2; HeLa; hESCs; A549; MM6; AML | ||
| Seq Type List | m6A-seq; miCLIP | ||
| Transcript ID List | ENST00000562272.5; ENST00000300093.9 | ||
| External Link | RMBase: m6A_site_312917 | ||
| mod ID: M6ASITE026242 | Click to Show/Hide the Full List | ||
| mod site | chr16:23690122-23690123:+ | [8] | |
| Sequence | CACCTGCATCTGGGGCCCATACTGGTTGGCTCCCGCGGTGC | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000300093.9; ENST00000562272.5 | ||
| External Link | RMBase: m6A_site_312918 | ||
| mod ID: M6ASITE026243 | Click to Show/Hide the Full List | ||
| mod site | chr16:23690231-23690232:+ | [5] | |
| Sequence | GCTGTATAAGTTATTTTTGTACATGTTCGGGTGTGGGTTCT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000562272.5; ENST00000300093.9 | ||
| External Link | RMBase: m6A_site_312919 | ||
| mod ID: M6ASITE026244 | Click to Show/Hide the Full List | ||
| mod site | chr16:23690344-23690345:+ | [5] | |
| Sequence | TCCTTTCCTTGGCTTTATGCACATTAAACAGATGTGAATAT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000562272.5; ENST00000300093.9 | ||
| External Link | RMBase: m6A_site_312921 | ||
References