m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00712)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
EGF
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
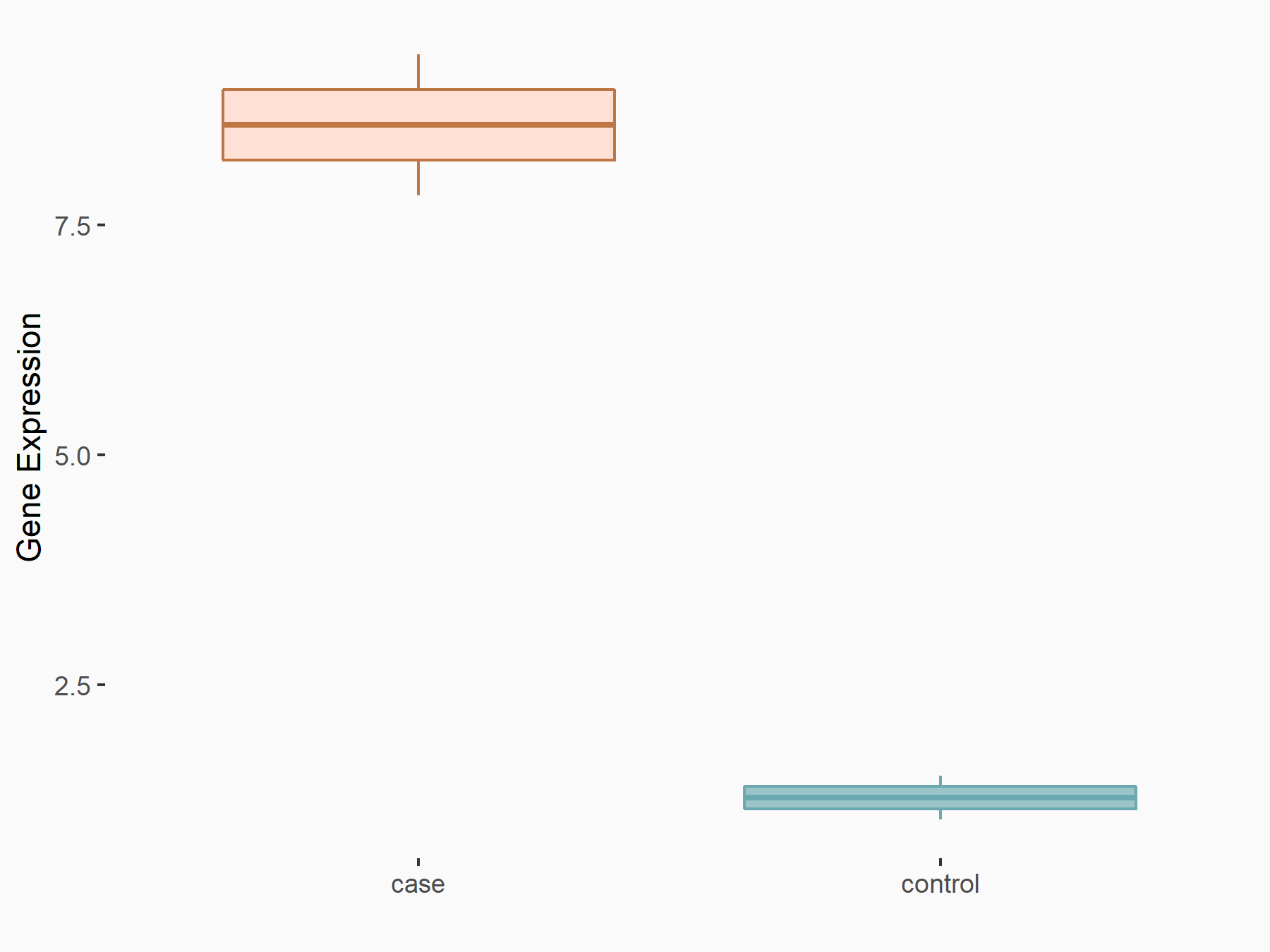 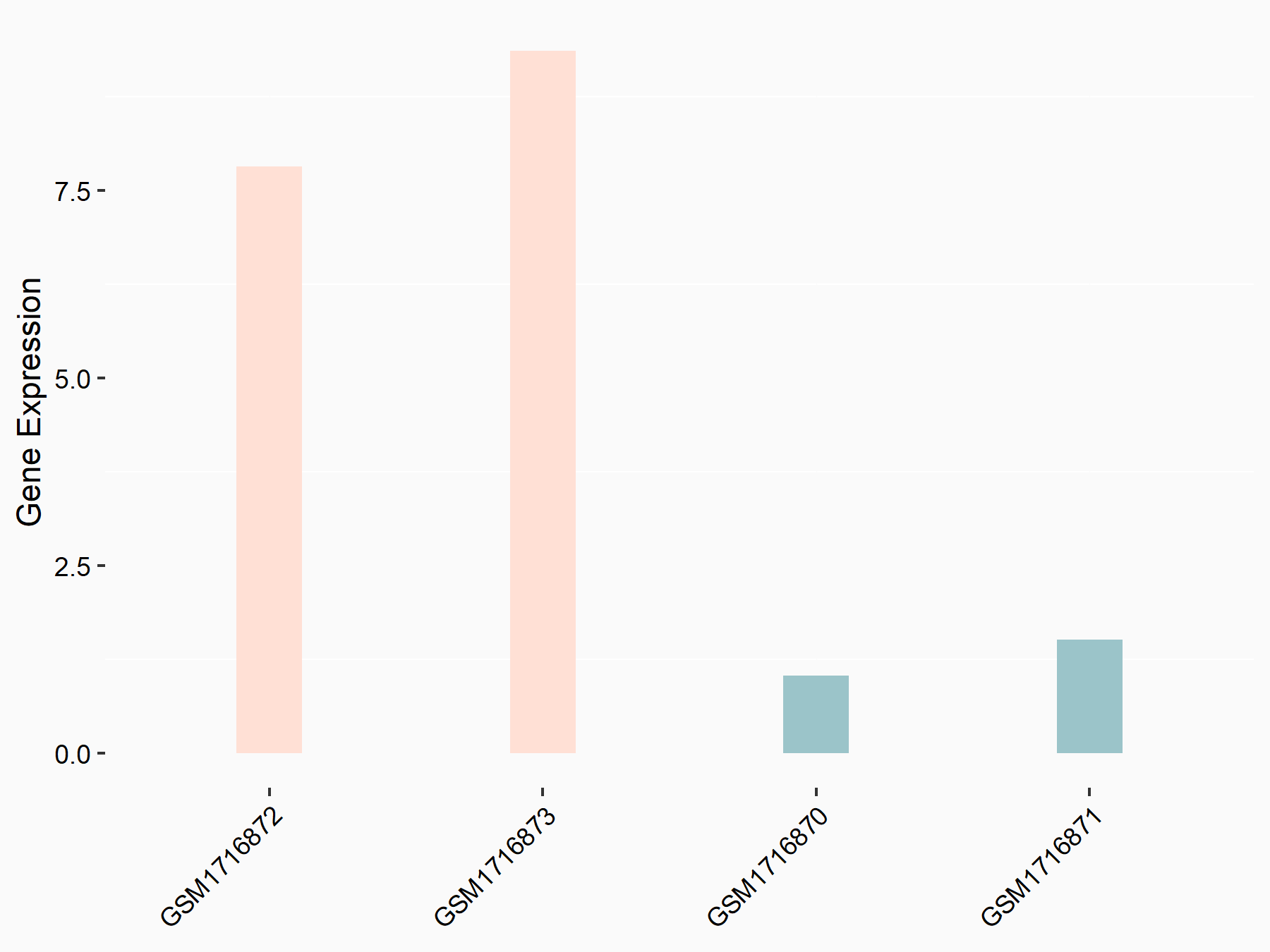 |
logFC: 2.08E+00 p-value: 3.08E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between EGF and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.98E+00 | GSE60213 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Knockdown of METTL3 sensitized these breast cancer cells to Adriamycin (ADR; also named as doxorubicin) treatment and increased accumulation of DNA damage. Mechanically, we demonstrated that inhibition of METTL3 impaired HR efficiency and increased ADR-induced DNA damage by regulating m6A modification of Pro-epidermal growth factor (EGF)/RAD51 axis. METTL3 promoted EGF expression through m6A modification, which further upregulated RAD51 expression, resulting in enhanced HR activity. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Doxil | Approved | ||
| Pathway Response | Homologous recombination | hsa03440 | ||
| Cell Process | Homologous recombination repair | |||
| In-vitro Model | MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | Cells were trypsinized and resuspended in DMEM at a consistence of 1 × 107 cells/ml. A total of 1 × 106 cells were injected into flank of mice. 27 days after injection, tumors were removed for paraffin-embedded sections. | |||
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Knockdown of METTL3 sensitized these breast cancer cells to Adriamycin (ADR; also named as doxorubicin) treatment and increased accumulation of DNA damage. Mechanically, we demonstrated that inhibition of METTL3 impaired HR efficiency and increased ADR-induced DNA damage by regulating m6A modification of Pro-epidermal growth factor (EGF)/RAD51 axis. METTL3 promoted EGF expression through m6A modification, which further upregulated RAD51 expression, resulting in enhanced HR activity. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Doxil | Approved | ||
| Pathway Response | Homologous recombination | hsa03440 | ||
| Cell Process | Homologous recombination repair | |||
| In-vitro Model | MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | Cells were trypsinized and resuspended in DMEM at a consistence of 1 × 107 cells/ml. A total of 1 × 106 cells were injected into flank of mice. 27 days after injection, tumors were removed for paraffin-embedded sections. | |||
Doxil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | Knockdown of METTL3 sensitized these breast cancer cells to Adriamycin (ADR; also named as doxorubicin) treatment and increased accumulation of DNA damage. Mechanically, we demonstrated that inhibition of METTL3 impaired HR efficiency and increased ADR-induced DNA damage by regulating m6A modification of Pro-epidermal growth factor (EGF)/RAD51 axis. METTL3 promoted EGF expression through m6A modification, which further upregulated RAD51 expression, resulting in enhanced HR activity. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Homologous recombination | hsa03440 | ||
| Cell Process | Homologous recombination repair | |||
| In-vitro Model | MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | Cells were trypsinized and resuspended in DMEM at a consistence of 1 × 107 cells/ml. A total of 1 × 106 cells were injected into flank of mice. 27 days after injection, tumors were removed for paraffin-embedded sections. | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00712)
| In total 6 m6A sequence/site(s) in this target gene | |||
| mod ID: 2OMSITE000345 | Click to Show/Hide the Full List | ||
| mod site | chr4:109992370-109992371:+ | [2] | |
| Sequence | AAAATTGGAATGATACAGAGAAGATTAGCATGGCCCCTGCG | ||
| Transcript ID List | ENST00000509996.1; ENST00000652245.1; ENST00000503392.1; ENST00000265171.9; ENST00000384530.1; ENST00000509793.5; rmsk_1406810 | ||
| External Link | RMBase: Nm_site_5260 | ||
| mod ID: 2OMSITE000346 | Click to Show/Hide the Full List | ||
| mod site | chr4:109992376-109992377:+ | [3] | |
| Sequence | GGAATGATACAGAGAAGATTAGCATGGCCCCTGCGCAAGGA | ||
| Cell/Tissue List | N.A.; HEK293 | ||
| Seq Type List | RiboMeth-seq | ||
| Transcript ID List | ENST00000652245.1; ENST00000384530.1; ENST00000265171.9; ENST00000503392.1; ENST00000509793.5; ENST00000509996.1; rmsk_1406810 | ||
| External Link | RMBase: Nm_site_5261 | ||
| mod ID: 2OMSITE000348 | Click to Show/Hide the Full List | ||
| mod site | chr4:109992383-109992384:+ | [3] | |
| Sequence | TACAGAGAAGATTAGCATGGCCCCTGCGCAAGGAGGACATG | ||
| Cell/Tissue List | N.A.; HEK293 | ||
| Seq Type List | RiboMeth-seq | ||
| Transcript ID List | ENST00000509793.5; ENST00000509996.1; ENST00000503392.1; ENST00000265171.9; ENST00000652245.1; rmsk_1406810; ENST00000384530.1 | ||
| External Link | RMBase: Nm_site_5263 | ||
| mod ID: 2OMSITE000349 | Click to Show/Hide the Full List | ||
| mod site | chr4:109992385-109992386:+ | [3] | |
| Sequence | CAGAGAAGATTAGCATGGCCCCTGCGCAAGGAGGACATGCA | ||
| Cell/Tissue List | N.A.; HEK293 | ||
| Seq Type List | RiboMeth-seq | ||
| Transcript ID List | rmsk_1406810; ENST00000509996.1; ENST00000652245.1; ENST00000384530.1; ENST00000265171.9; ENST00000503392.1; ENST00000509793.5 | ||
| External Link | RMBase: Nm_site_5264 | ||
| mod ID: 2OMSITE000350 | Click to Show/Hide the Full List | ||
| mod site | chr4:109992386-109992387:+ | [3] | |
| Sequence | AGAGAAGATTAGCATGGCCCCTGCGCAAGGAGGACATGCAA | ||
| Seq Type List | RiboMeth-seq | ||
| Transcript ID List | ENST00000503392.1; ENST00000652245.1; ENST00000509793.5; rmsk_1406810; ENST00000384530.1; ENST00000265171.9; ENST00000509996.1 | ||
| External Link | RMBase: Nm_site_5265 | ||
| mod ID: 2OMSITE000347 | Click to Show/Hide the Full List | ||
| mod site | chr4:109992377-109992378:+ | [3] | |
| Sequence | GAATGATACAGAGAAGATTAGCATGGCCCCTGCGCAAGGAG | ||
| Seq Type List | RiboMeth-seq | ||
| Transcript ID List | ENST00000384530.1; ENST00000652245.1; ENST00000509996.1; ENST00000509793.5; ENST00000265171.9; ENST00000503392.1; rmsk_1406810 | ||
| External Link | RMBase: Nm_site_5262 | ||
N6-methyladenosine (m6A)
| In total 12 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE067097 | Click to Show/Hide the Full List | ||
| mod site | chr4:109960917-109960918:+ | [4] | |
| Sequence | CTGTACTCTTGGGTGTAAAAACACCCCTGGATCCTATTACT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000652245.1; ENST00000509793.5; ENST00000504633.1; ENST00000265171.9; ENST00000503392.1 | ||
| External Link | RMBase: m6A_site_647467 | ||
| mod ID: M6ASITE067098 | Click to Show/Hide the Full List | ||
| mod site | chr4:109983450-109983451:+ | [5] | |
| Sequence | TTGATCTAAAGAACCAAGTAACACCATTGGACATCTTGTCC | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000652245.1; ENST00000509793.5; ENST00000265171.9; ENST00000509996.1; ENST00000511228.5; ENST00000503392.1 | ||
| External Link | RMBase: m6A_site_647468 | ||
| mod ID: M6ASITE067099 | Click to Show/Hide the Full List | ||
| mod site | chr4:110011344-110011345:+ | [6] | |
| Sequence | TTCATATGCCCTCCTATGGGACACAGACCCTTGAAGGGGGT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | fibroblasts | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000537316.5; ENST00000544918.1; ENST00000652245.1; ENST00000503392.1; ENST00000265171.9; ENST00000509996.1; ENST00000540840.1; ENST00000509793.5 | ||
| External Link | RMBase: m6A_site_647469 | ||
| mod ID: M6ASITE067100 | Click to Show/Hide the Full List | ||
| mod site | chr4:110011350-110011351:+ | [6] | |
| Sequence | TGCCCTCCTATGGGACACAGACCCTTGAAGGGGGTGTCGAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | fibroblasts | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000652245.1; ENST00000509793.5; ENST00000503392.1; ENST00000540840.1; ENST00000537316.5; ENST00000544918.1; ENST00000265171.9; ENST00000509996.1 | ||
| External Link | RMBase: m6A_site_647470 | ||
| mod ID: M6ASITE067101 | Click to Show/Hide the Full List | ||
| mod site | chr4:110011423-110011424:+ | [6] | |
| Sequence | ATGGCAACAAAGGGCCCTGGACCCACCACACCAAATGGAGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | fibroblasts; Huh7; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000509996.1; ENST00000537316.5; ENST00000540840.1; ENST00000503392.1; ENST00000509793.5; ENST00000544918.1; ENST00000265171.9; ENST00000652245.1 | ||
| External Link | RMBase: m6A_site_647471 | ||
| mod ID: M6ASITE067102 | Click to Show/Hide the Full List | ||
| mod site | chr4:110011457-110011458:+ | [6] | |
| Sequence | ATGGAGCTGACTCAGTGAAAACTGGAATTAAAAGGAAAGTC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | fibroblasts; Huh7; HEK293A-TOA; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000509996.1; ENST00000509793.5; ENST00000540840.1; ENST00000537316.5; ENST00000652245.1; ENST00000544918.1; ENST00000503392.1; ENST00000265171.9 | ||
| External Link | RMBase: m6A_site_647472 | ||
| mod ID: M6ASITE067103 | Click to Show/Hide the Full List | ||
| mod site | chr4:110011489-110011490:+ | [6] | |
| Sequence | AGGAAAGTCAAGAAGAATGAACTATGTCGATGCACAGTATC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | fibroblasts; Huh7; HEK293A-TOA; iSLK; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000540840.1; ENST00000509793.5; ENST00000265171.9; ENST00000503392.1; ENST00000544918.1; ENST00000652245.1; ENST00000509996.1; ENST00000537316.5 | ||
| External Link | RMBase: m6A_site_647473 | ||
| mod ID: M6ASITE067104 | Click to Show/Hide the Full List | ||
| mod site | chr4:110011533-110011534:+ | [6] | |
| Sequence | TCTTTCAAAAGTAGAGCAAAACTATAGGTTTTGGTTCCACA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | fibroblasts; Huh7; HEK293A-TOA; iSLK; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000509996.1; ENST00000509793.5; ENST00000652245.1; ENST00000265171.9; ENST00000540840.1; ENST00000544918.1 | ||
| External Link | RMBase: m6A_site_647474 | ||
| mod ID: M6ASITE067105 | Click to Show/Hide the Full List | ||
| mod site | chr4:110011589-110011590:+ | [7] | |
| Sequence | CACCTACTCAATGCCTGGAGACAGATACGTAGTTGTGCTTT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000265171.9; ENST00000540840.1; ENST00000544918.1; ENST00000652245.1; ENST00000509996.1; ENST00000509793.5 | ||
| External Link | RMBase: m6A_site_647475 | ||
| mod ID: M6ASITE067106 | Click to Show/Hide the Full List | ||
| mod site | chr4:110013579-110013580:+ | [7] | |
| Sequence | TAAAAATTCAAGTTTTAAGAACAGCATTTTCATGTAAAAAC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000652245.1 | ||
| External Link | RMBase: m6A_site_647476 | ||
| mod ID: M6ASITE067107 | Click to Show/Hide the Full List | ||
| mod site | chr4:110013598-110013599:+ | [7] | |
| Sequence | AACAGCATTTTCATGTAAAAACTTGATTTGTGTTTTTTCCA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000652245.1 | ||
| External Link | RMBase: m6A_site_647477 | ||
| mod ID: M6ASITE067108 | Click to Show/Hide the Full List | ||
| mod site | chr4:110013620-110013621:+ | [7] | |
| Sequence | TTGATTTGTGTTTTTTCCAGACTGAATACTTTTCCTCCCTA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000652245.1 | ||
| External Link | RMBase: m6A_site_647478 | ||
References