m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00484)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
SLC7A11
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by IGF2BP1 | ||
| Cell Line | MV3 cell line | Homo sapiens |
|
Treatment: siIGF2BP1 MV3 cells
Control: siControl MV3 cells
|
GSE146803 | |
| Regulation |
  |
logFC: -1.30E+00 p-value: 3.62E-06 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3-mediated Cystine/glutamate transporter (SLC7A11) m6A modification enhances hepatoblastoma ferroptosis resistance. The METTL3/IGF2BP1/m6A modification promotes SLC7A11 mRNA stability and upregulates its expression by inhibiting the deadenylation process. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatoblastoma | ICD-11: 2C12.01 | ||
| Pathway Response | Ferroptosis | hsa04216 | ||
| Cell Process | Ferroptosis | |||
| In-vitro Model | HuH-6 | Hepatoblastoma | Homo sapiens | CVCL_4381 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Methyltransferase-like 14 (METTL14) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL14 | ||
| Cell Line | HepG2 cell line | Homo sapiens |
|
Treatment: shMETTL14 HepG2 cells
Control: shCtrl HepG2 cells
|
GSE121949 | |
| Regulation |
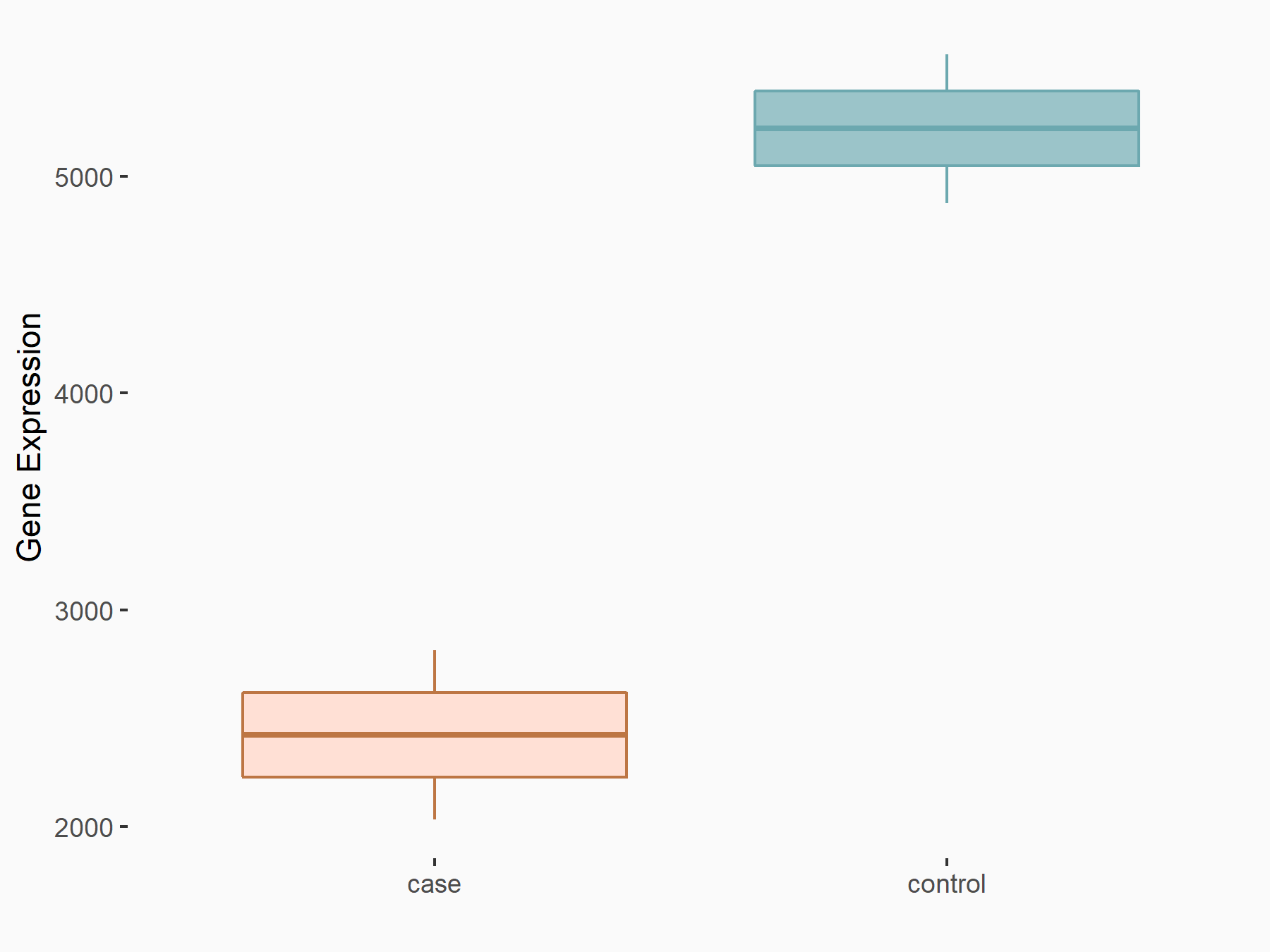  |
logFC: -1.11E+00 p-value: 3.96E-11 |
| More Results | Click to View More RNA-seq Results | |
| In total 4 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL14 induced m6A modification at 5'UTR of Cystine/glutamate transporter (SLC7A11) mRNA, which in turn underwent degradation relied on the YTHDF2-dependent pathway. Identify the HIF-1alpha /METTL14/YTHDF2/SLC7A11 axis as a potential therapeutic target for the HCC interventional embolization treatment. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Liver cancer | ICD-11: 2C12 | ||
| Pathway Response | HIF-1 signaling pathway | hsa04066 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 | |
| 7721 (Human hepatic malignant cell line) | ||||
| In-vivo Model | For the subcutaneous implantation model, 5 × 105 stable SLC7A11-knockdown HCCLM3 cells or SLC7A11-vector cells were injected subcutaneously into BALB/C nude mice. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates GSK-3beta and activates beta-catenin/TCF4-Cystine/glutamate transporter (SLC7A11)/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Pertuzumab | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates GSK-3beta and activates beta-catenin/TCF4-Cystine/glutamate transporter (SLC7A11)/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Trastuzumab | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
| Experiment 4 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates GSK-3beta and activates beta-catenin/TCF4-Cystine/glutamate transporter (SLC7A11)/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Tucatinib | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shMETTL3 MOLM13 cells
Control: MOLM13 cells
|
GSE98623 | |
| Regulation |
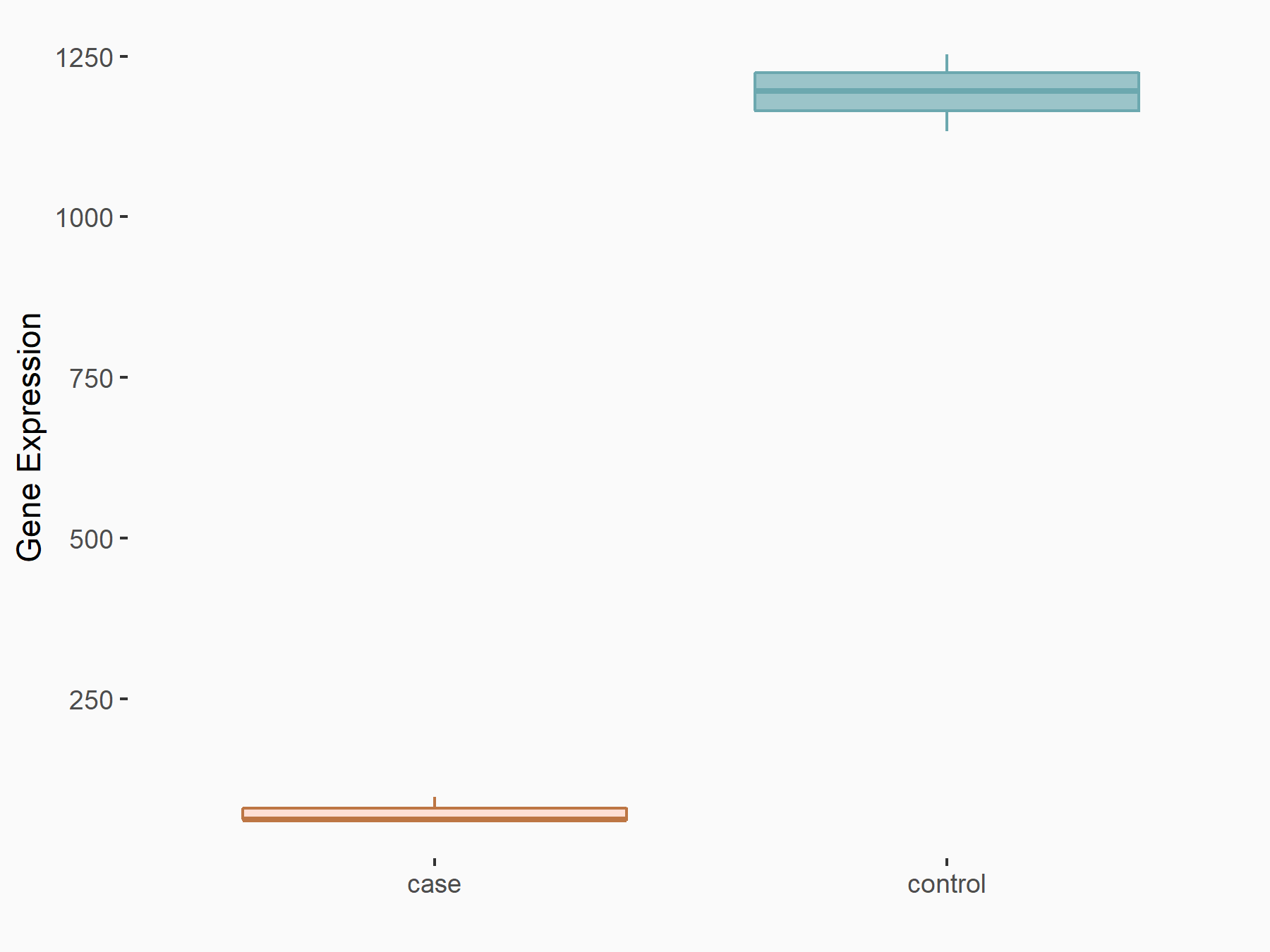  |
logFC: -4.02E+00 p-value: 2.26E-108 |
| More Results | Click to View More RNA-seq Results | |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3-mediated Cystine/glutamate transporter (SLC7A11) m6A modification enhances hepatoblastoma ferroptosis resistance. The METTL3/IGF2BP1/m6A modification promotes SLC7A11 mRNA stability and upregulates its expression by inhibiting the deadenylation process. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatoblastoma | ICD-11: 2C12.01 | ||
| Pathway Response | Ferroptosis | hsa04216 | ||
| Cell Process | Ferroptosis | |||
| In-vitro Model | HuH-6 | Hepatoblastoma | Homo sapiens | CVCL_4381 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | METTL3-mediated m-6A modification could stabilize Cystine/glutamate transporter (SLC7A11) mRNA and promote its translation, thus promoting LUAD cell proliferation and inhibiting cell ferroptosis, a novel form of programmed cell death. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Lung adenocarcinoma | ICD-11: 2C25.0 | ||
| Pathway Response | Ferroptosis | hsa04216 | ||
| Cell Process | Ferroptosis | |||
| In-vitro Model | SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| NCI-H322 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1556 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | For the subcutaneous xenograft model, PC9 cells stably transfected with METTL3 knockdown (shMETTL3) or negative control (shNC) shRNA (5 × 106 cells per mouse, n = 6) were suspended in 200 uL PBS with 50% Matrigel matrix (Corning, USA, 354234) and then injected into one side of the axilla of nude mice. | |||
NF-kappa-B-activating protein (NKAP) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [9] | |||
| Response Summary | This study NKAP knockdown induced cell death in glioblastoma cells. NKAP acted as a new ferroptosis suppressor by binding to m6A and then promoting Cystine/glutamate transporter (SLC7A11) mRNA splicing and maturation. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| Pathway Response | Ferroptosis | hsa04216 | ||
| Cell Process | Ferroptosis | |||
| In-vitro Model | U87MG (Astroblastoma cells from human brain) | |||
| U251 (Fibroblasts or fibroblast like cells) | ||||
| In-vivo Model | The male BALB/c nude mice were randomized divide into two groups, each group including six 4 weeks old nude mice. Investigators were blinded to the treatment groups during data collection and subsequent data analysis. In the subcutaneous xenograft model, 5 × 105 cells were subcutaneously injected in the right flanks of nude mice. In the orthotopic intracranial mouse model, each mouse was intracranially injected with 1 × 105 luciferase transfected U87MG cells in 10 uL PBS solution. | |||
YTH domain-containing protein 2 (YTHDC2) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [14] | |||
| Response Summary | YTHDC2 destabilized Cystine/glutamate transporter (SLC7A11) mRNA in an m6A-dependent manner because YTHDC2 preferentially bound to m6A-modified SLC7A11 mRNA and thereafter promoted its decay. the promotion of cystine uptake via the suppression of YTHDC2 is critical for LUAD tumorigenesis. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Lung adenocarcinoma | ICD-11: 2C25.0 | ||
| In-vitro Model | PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| NCI-H441 | Lung papillary adenocarcinoma | Homo sapiens | CVCL_1561 | |
| NCI-H292 | Lung mucoepidermoid carcinoma | Homo sapiens | CVCL_0455 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| Calu-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0608 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [9] | |||
| Response Summary | This study NKAP knockdown induced cell death in glioblastoma cells. NKAP acted as a new ferroptosis suppressor by binding to m6A and then promoting Cystine/glutamate transporter (SLC7A11) mRNA splicing and maturation. | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulator | NF-kappa-B-activating protein (NKAP) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Ferroptosis | hsa04216 | ||
| Cell Process | Ferroptosis | |||
| In-vitro Model | U87MG (Astroblastoma cells from human brain) | |||
| U251 (Fibroblasts or fibroblast like cells) | ||||
| In-vivo Model | The male BALB/c nude mice were randomized divide into two groups, each group including six 4 weeks old nude mice. Investigators were blinded to the treatment groups during data collection and subsequent data analysis. In the subcutaneous xenograft model, 5 × 105 cells were subcutaneously injected in the right flanks of nude mice. In the orthotopic intracranial mouse model, each mouse was intracranially injected with 1 × 105 luciferase transfected U87MG cells in 10 uL PBS solution. | |||
Liver cancer [ICD-11: 2C12]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3-mediated Cystine/glutamate transporter (SLC7A11) m6A modification enhances hepatoblastoma ferroptosis resistance. The METTL3/IGF2BP1/m6A modification promotes SLC7A11 mRNA stability and upregulates its expression by inhibiting the deadenylation process. | |||
| Responsed Disease | Hepatoblastoma [ICD-11: 2C12.01] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Ferroptosis | hsa04216 | ||
| Cell Process | Ferroptosis | |||
| In-vitro Model | HuH-6 | Hepatoblastoma | Homo sapiens | CVCL_4381 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| Experiment 2 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL14 induced m6A modification at 5'UTR of Cystine/glutamate transporter (SLC7A11) mRNA, which in turn underwent degradation relied on the YTHDF2-dependent pathway. Identify the HIF-1alpha /METTL14/YTHDF2/SLC7A11 axis as a potential therapeutic target for the HCC interventional embolization treatment. | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | HIF-1 signaling pathway | hsa04066 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 | |
| 7721 (Human hepatic malignant cell line) | ||||
| In-vivo Model | For the subcutaneous implantation model, 5 × 105 stable SLC7A11-knockdown HCCLM3 cells or SLC7A11-vector cells were injected subcutaneously into BALB/C nude mice. | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3-mediated Cystine/glutamate transporter (SLC7A11) m6A modification enhances hepatoblastoma ferroptosis resistance. The METTL3/IGF2BP1/m6A modification promotes SLC7A11 mRNA stability and upregulates its expression by inhibiting the deadenylation process. | |||
| Responsed Disease | Hepatoblastoma [ICD-11: 2C12.01] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Ferroptosis | hsa04216 | ||
| Cell Process | Ferroptosis | |||
| In-vitro Model | HuH-6 | Hepatoblastoma | Homo sapiens | CVCL_4381 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Lung cancer [ICD-11: 2C25]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | METTL3-mediated m-6A modification could stabilize Cystine/glutamate transporter (SLC7A11) mRNA and promote its translation, thus promoting LUAD cell proliferation and inhibiting cell ferroptosis, a novel form of programmed cell death. | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Ferroptosis | hsa04216 | ||
| Cell Process | Ferroptosis | |||
| In-vitro Model | SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| NCI-H322 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1556 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | For the subcutaneous xenograft model, PC9 cells stably transfected with METTL3 knockdown (shMETTL3) or negative control (shNC) shRNA (5 × 106 cells per mouse, n = 6) were suspended in 200 uL PBS with 50% Matrigel matrix (Corning, USA, 354234) and then injected into one side of the axilla of nude mice. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [14] | |||
| Response Summary | YTHDC2 destabilized Cystine/glutamate transporter (SLC7A11) mRNA in an m6A-dependent manner because YTHDC2 preferentially bound to m6A-modified SLC7A11 mRNA and thereafter promoted its decay. the promotion of cystine uptake via the suppression of YTHDC2 is critical for LUAD tumorigenesis. | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulator | YTH domain-containing protein 2 (YTHDC2) | READER | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| NCI-H441 | Lung papillary adenocarcinoma | Homo sapiens | CVCL_1561 | |
| NCI-H292 | Lung mucoepidermoid carcinoma | Homo sapiens | CVCL_0455 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| Calu-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0608 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
Breast cancer [ICD-11: 2C60]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates GSK-3beta and activates beta-catenin/TCF4-Cystine/glutamate transporter (SLC7A11)/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Pertuzumab | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates GSK-3beta and activates beta-catenin/TCF4-Cystine/glutamate transporter (SLC7A11)/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Trastuzumab | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates GSK-3beta and activates beta-catenin/TCF4-Cystine/glutamate transporter (SLC7A11)/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Tucatinib | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Pertuzumab
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [3] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates GSK-3beta and activates beta-catenin/TCF4-Cystine/glutamate transporter (SLC7A11)/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Trastuzumab
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [3] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates GSK-3beta and activates beta-catenin/TCF4-Cystine/glutamate transporter (SLC7A11)/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Tucatinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [3] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates GSK-3beta and activates beta-catenin/TCF4-Cystine/glutamate transporter (SLC7A11)/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
DNA modification
m6A Regulator: Methyltransferase-like 14 (METTL14)
| In total 9 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT02207 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Trastuzumab | |
| Crosstalk ID: M6ACROT02212 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Pertuzumab | |
| Crosstalk ID: M6ACROT02220 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Tucatinib | |
| Crosstalk ID: M6ACROT02231 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Trastuzumab | |
| Crosstalk ID: M6ACROT02236 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Pertuzumab | |
| Crosstalk ID: M6ACROT02244 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Tucatinib | |
| Crosstalk ID: M6ACROT02255 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Trastuzumab | |
| Crosstalk ID: M6ACROT02260 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Pertuzumab | |
| Crosstalk ID: M6ACROT02268 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Tucatinib | |
Histone modification
m6A Regulator: RNA demethylase ALKBH5 (ALKBH5)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03360 | ||
| Epigenetic Regulator | Histone deacetylase 2 (HDAC2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03456 | ||
| Epigenetic Regulator | Histone deacetylase 1 (HDAC1) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
Non-coding RNA
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05054 | ||
| Epigenetic Regulator | MicroRNA 186 (MIR186) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Hepatoblastoma | |
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05273 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 942 (LINC00942) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 3 (IGF2BP3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Regulator: Staphylococcal nuclease domain-containing protein 1 (SND1)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05311 | ||
| Epigenetic Regulator | Small nucleolar RNA host gene 1 (SNHG1) | |
| Regulated Target | Staphylococcal nuclease domain-containing protein 1 (SND1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
| Drug | Sorafenib | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00484)
| In total 33 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE000538 | Click to Show/Hide the Full List | ||
| mod site | chr4:138188216-138188217:- | [15] | |
| Sequence | TAAAAAAATTAGCTGGGTATAGTGATGGGTGCCTATAATCC | ||
| Transcript ID List | ENST00000280612.9; ENST00000509248.1; rmsk_1451296 | ||
| External Link | RMBase: RNA-editing_site_106610 | ||
| mod ID: A2ISITE000539 | Click to Show/Hide the Full List | ||
| mod site | chr4:138192339-138192340:- | [16] | |
| Sequence | CAACTAGCTCACTCAGGAAAAGAATGTCACAAGGTGACACA | ||
| Transcript ID List | ENST00000509248.1; ENST00000280612.9 | ||
| External Link | RMBase: RNA-editing_site_106611 | ||
| mod ID: A2ISITE000540 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196696-138196697:- | [17] | |
| Sequence | ATTGCACTCCCGCCCGGGTGACAAGAGTGAGGCTCTGTCTA | ||
| Transcript ID List | ENST00000280612.9; ENST00000509248.1; rmsk_1451306 | ||
| External Link | RMBase: RNA-editing_site_106612 | ||
| mod ID: A2ISITE000541 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196731-138196732:- | [17] | |
| Sequence | AGGAGGTGAAGGTTGCAGTGAGCCGAGATTGTGCCATTGCA | ||
| Transcript ID List | rmsk_1451306; ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: RNA-editing_site_106613 | ||
| mod ID: A2ISITE000542 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196742-138196743:- | [17] | |
| Sequence | TGCCTGAACCCAGGAGGTGAAGGTTGCAGTGAGCCGAGATT | ||
| Transcript ID List | rmsk_1451306; ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: RNA-editing_site_106614 | ||
| mod ID: A2ISITE000543 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196751-138196752:- | [17] | |
| Sequence | CAGAAGAATTGCCTGAACCCAGGAGGTGAAGGTTGCAGTGA | ||
| Transcript ID List | ENST00000280612.9; rmsk_1451306; ENST00000509248.1 | ||
| External Link | RMBase: RNA-editing_site_106615 | ||
| mod ID: A2ISITE000544 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196755-138196756:- | [17] | |
| Sequence | GAAGCAGAAGAATTGCCTGAACCCAGGAGGTGAAGGTTGCA | ||
| Transcript ID List | rmsk_1451306; ENST00000509248.1; ENST00000280612.9 | ||
| External Link | RMBase: RNA-editing_site_106616 | ||
| mod ID: A2ISITE000545 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196764-138196765:- | [17] | |
| Sequence | CGGGAGGCTGAAGCAGAAGAATTGCCTGAACCCAGGAGGTG | ||
| Transcript ID List | rmsk_1451306; ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: RNA-editing_site_106617 | ||
| mod ID: A2ISITE000546 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196765-138196766:- | [17] | |
| Sequence | TCGGGAGGCTGAAGCAGAAGAATTGCCTGAACCCAGGAGGT | ||
| Transcript ID List | rmsk_1451306; ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: RNA-editing_site_106618 | ||
| mod ID: A2ISITE000547 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196767-138196768:- | [17] | |
| Sequence | ACTCGGGAGGCTGAAGCAGAAGAATTGCCTGAACCCAGGAG | ||
| Transcript ID List | ENST00000509248.1; ENST00000280612.9; rmsk_1451306 | ||
| External Link | RMBase: RNA-editing_site_106619 | ||
| mod ID: A2ISITE000549 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196768-138196769:- | [17] | |
| Sequence | TACTCGGGAGGCTGAAGCAGAAGAATTGCCTGAACCCAGGA | ||
| Transcript ID List | rmsk_1451306; ENST00000509248.1; ENST00000280612.9 | ||
| External Link | RMBase: RNA-editing_site_106620 | ||
| mod ID: A2ISITE000550 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196770-138196771:- | [17] | |
| Sequence | GCTACTCGGGAGGCTGAAGCAGAAGAATTGCCTGAACCCAG | ||
| Transcript ID List | rmsk_1451306; ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: RNA-editing_site_106621 | ||
| mod ID: A2ISITE000551 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196773-138196774:- | [17] | |
| Sequence | CCAGCTACTCGGGAGGCTGAAGCAGAAGAATTGCCTGAACC | ||
| Transcript ID List | rmsk_1451306; ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: RNA-editing_site_106622 | ||
| mod ID: A2ISITE000552 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196780-138196781:- | [17] | |
| Sequence | TGTAATCCCAGCTACTCGGGAGGCTGAAGCAGAAGAATTGC | ||
| Transcript ID List | rmsk_1451306; ENST00000509248.1; ENST00000280612.9 | ||
| External Link | RMBase: RNA-editing_site_106623 | ||
| mod ID: A2ISITE000553 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196787-138196788:- | [17] | |
| Sequence | AGTTGCCTGTAATCCCAGCTACTCGGGAGGCTGAAGCAGAA | ||
| Transcript ID List | rmsk_1451306; ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: RNA-editing_site_106624 | ||
| mod ID: A2ISITE000554 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196791-138196792:- | [17] | |
| Sequence | TGGCAGTTGCCTGTAATCCCAGCTACTCGGGAGGCTGAAGC | ||
| Transcript ID List | ENST00000280612.9; ENST00000509248.1; rmsk_1451306 | ||
| External Link | RMBase: RNA-editing_site_106625 | ||
| mod ID: A2ISITE000555 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196796-138196797:- | [17] | |
| Sequence | TGTGGTGGCAGTTGCCTGTAATCCCAGCTACTCGGGAGGCT | ||
| Transcript ID List | rmsk_1451306; ENST00000509248.1; ENST00000280612.9 | ||
| External Link | RMBase: RNA-editing_site_106626 | ||
| mod ID: A2ISITE000556 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196797-138196798:- | [17] | |
| Sequence | GTGTGGTGGCAGTTGCCTGTAATCCCAGCTACTCGGGAGGC | ||
| Transcript ID List | ENST00000280612.9; ENST00000509248.1; rmsk_1451306 | ||
| External Link | RMBase: RNA-editing_site_106627 | ||
| mod ID: A2ISITE000557 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196807-138196808:- | [17] | |
| Sequence | AATAAGCTGAGTGTGGTGGCAGTTGCCTGTAATCCCAGCTA | ||
| Transcript ID List | ENST00000509248.1; rmsk_1451306; ENST00000280612.9 | ||
| External Link | RMBase: RNA-editing_site_106628 | ||
| mod ID: A2ISITE000558 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196827-138196828:- | [17] | |
| Sequence | GTCTCTACTAAAAATACAAAAATAAGCTGAGTGTGGTGGCA | ||
| Transcript ID List | rmsk_1451306; ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: RNA-editing_site_106629 | ||
| mod ID: A2ISITE000560 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196828-138196829:- | [17] | |
| Sequence | TGTCTCTACTAAAAATACAAAAATAAGCTGAGTGTGGTGGC | ||
| Transcript ID List | rmsk_1451306; ENST00000509248.1; ENST00000280612.9 | ||
| External Link | RMBase: RNA-editing_site_106630 | ||
| mod ID: A2ISITE000561 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196832-138196833:- | [17] | |
| Sequence | ACCCTGTCTCTACTAAAAATACAAAAATAAGCTGAGTGTGG | ||
| Transcript ID List | ENST00000509248.1; ENST00000280612.9; rmsk_1451306 | ||
| External Link | RMBase: RNA-editing_site_106631 | ||
| mod ID: A2ISITE000562 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196834-138196835:- | [17] | |
| Sequence | AAACCCTGTCTCTACTAAAAATACAAAAATAAGCTGAGTGT | ||
| Transcript ID List | ENST00000509248.1; rmsk_1451306; ENST00000280612.9 | ||
| External Link | RMBase: RNA-editing_site_106632 | ||
| mod ID: A2ISITE000563 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196852-138196853:- | [17] | |
| Sequence | AGCCTGGCCAACATGGTGAAACCCTGTCTCTACTAAAAATA | ||
| Transcript ID List | ENST00000280612.9; ENST00000509248.1; rmsk_1451306 | ||
| External Link | RMBase: RNA-editing_site_106633 | ||
| mod ID: A2ISITE000564 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196853-138196854:- | [17] | |
| Sequence | CAGCCTGGCCAACATGGTGAAACCCTGTCTCTACTAAAAAT | ||
| Transcript ID List | ENST00000280612.9; rmsk_1451306; ENST00000509248.1 | ||
| External Link | RMBase: RNA-editing_site_106634 | ||
| mod ID: A2ISITE000565 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196860-138196861:- | [17] | |
| Sequence | TTGAGACCAGCCTGGCCAACATGGTGAAACCCTGTCTCTAC | ||
| Transcript ID List | ENST00000509248.1; ENST00000280612.9; rmsk_1451306 | ||
| External Link | RMBase: RNA-editing_site_106635 | ||
| mod ID: A2ISITE000566 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196862-138196863:- | [17] | |
| Sequence | GCTTGAGACCAGCCTGGCCAACATGGTGAAACCCTGTCTCT | ||
| Transcript ID List | ENST00000509248.1; ENST00000280612.9; rmsk_1451306 | ||
| External Link | RMBase: RNA-editing_site_106636 | ||
| mod ID: A2ISITE000567 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196863-138196864:- | [17] | |
| Sequence | AGCTTGAGACCAGCCTGGCCAACATGGTGAAACCCTGTCTC | ||
| Transcript ID List | rmsk_1451306; ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: RNA-editing_site_106637 | ||
| mod ID: A2ISITE000568 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196872-138196873:- | [17] | |
| Sequence | GAGGTCAAGAGCTTGAGACCAGCCTGGCCAACATGGTGAAA | ||
| Transcript ID List | rmsk_1451306; ENST00000509248.1; ENST00000280612.9 | ||
| External Link | RMBase: RNA-editing_site_106638 | ||
| mod ID: A2ISITE000569 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196877-138196878:- | [17] | |
| Sequence | CACCAGAGGTCAAGAGCTTGAGACCAGCCTGGCCAACATGG | ||
| Transcript ID List | ENST00000280612.9; rmsk_1451306; ENST00000509248.1 | ||
| External Link | RMBase: RNA-editing_site_106639 | ||
| mod ID: A2ISITE000571 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196885-138196886:- | [17] | |
| Sequence | GGGCGTATCACCAGAGGTCAAGAGCTTGAGACCAGCCTGGC | ||
| Transcript ID List | rmsk_1451306; ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: RNA-editing_site_106640 | ||
| mod ID: A2ISITE000572 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196891-138196892:- | [17] | |
| Sequence | CAAGGTGGGCGTATCACCAGAGGTCAAGAGCTTGAGACCAG | ||
| Transcript ID List | ENST00000280612.9; ENST00000509248.1; rmsk_1451306 | ||
| External Link | RMBase: RNA-editing_site_106641 | ||
| mod ID: A2ISITE000573 | Click to Show/Hide the Full List | ||
| mod site | chr4:138196893-138196894:- | [17] | |
| Sequence | GCCAAGGTGGGCGTATCACCAGAGGTCAAGAGCTTGAGACC | ||
| Transcript ID List | rmsk_1451306; ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: RNA-editing_site_106642 | ||
5-methylcytidine (m5C)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE003334 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171131-138171132:- | [18] | |
| Sequence | ATTAGAACAACCACCTGTTTCACTAATAACTTACCCCTGAT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m5C_site_34338 | ||
| mod ID: M5CSITE003335 | Click to Show/Hide the Full List | ||
| mod site | chr4:138223279-138223280:- | [18] | |
| Sequence | TGAGTGTCAGCTGGAGCGCCCGGATCCAGATTTTCTTAACC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m5C_site_34339 | ||
N6-methyladenosine (m6A)
| In total 108 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE067671 | Click to Show/Hide the Full List | ||
| mod site | chr4:138164148-138164149:- | [19] | |
| Sequence | ACCATTTTTGTACATATTTTACTTGAAAATATTTTAAATGG | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651120 | ||
| mod ID: M6ASITE067672 | Click to Show/Hide the Full List | ||
| mod site | chr4:138164197-138164198:- | [19] | |
| Sequence | TTATATGTATTATACCTGTCACGCTTCTAGTTGCTTCAACC | ||
| Motif Score | 2.021232143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651121 | ||
| mod ID: M6ASITE067673 | Click to Show/Hide the Full List | ||
| mod site | chr4:138164392-138164393:- | [20] | |
| Sequence | AGAGTTCTGGTACTGCAATCACAATGCCAGATGGTGTTTAT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651122 | ||
| mod ID: M6ASITE067674 | Click to Show/Hide the Full List | ||
| mod site | chr4:138164474-138164475:- | [20] | |
| Sequence | TTCCTAGCACTGATGCCTGCACAAGCATGTGATATGTGAAA | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651123 | ||
| mod ID: M6ASITE067675 | Click to Show/Hide the Full List | ||
| mod site | chr4:138164527-138164528:- | [21] | |
| Sequence | GATATAAATGGTGCAGAGAGACTTTCATCTGTGGATTGCGT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651124 | ||
| mod ID: M6ASITE067676 | Click to Show/Hide the Full List | ||
| mod site | chr4:138164657-138164658:- | [20] | |
| Sequence | GGGGATTGTGTGTATTTTATACAAATTTAATACAATGTCTT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651125 | ||
| mod ID: M6ASITE067677 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165021-138165022:- | [19] | |
| Sequence | AATATTTTTCATATTCTGTGACAAGCATTTATAATTTGCAA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651126 | ||
| mod ID: M6ASITE067678 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165191-138165192:- | [19] | |
| Sequence | TTGTAAGCATATCTGAATTTACTTTATAAAGATGGTTTTAG | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651127 | ||
| mod ID: M6ASITE067679 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165279-138165280:- | [19] | |
| Sequence | GGATTACATGGTAGTGATGCACTGGTAGAAATGGTTTTTAG | ||
| Motif Score | 3.252583333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651128 | ||
| mod ID: M6ASITE067680 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165294-138165295:- | [19] | |
| Sequence | GAGCTTGCGTAGAATGGATTACATGGTAGTGATGCACTGGT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651129 | ||
| mod ID: M6ASITE067681 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165338-138165339:- | [22] | |
| Sequence | GGGTCTTCATTTTCTGACAGACAGGATTTGACTCAATATTT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651130 | ||
| mod ID: M6ASITE067682 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165365-138165366:- | [22] | |
| Sequence | TGTACTGAATACTTCGGCAAACTTATTGGGTCTTCATTTTC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651131 | ||
| mod ID: M6ASITE067683 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165382-138165383:- | [19] | |
| Sequence | CAAGACAAATGGCCTCTTGTACTGAATACTTCGGCAAACTT | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651132 | ||
| mod ID: M6ASITE067684 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165398-138165399:- | [22] | |
| Sequence | ACATGTCAGAAAAAGGCAAGACAAATGGCCTCTTGTACTGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651133 | ||
| mod ID: M6ASITE067685 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165418-138165419:- | [20] | |
| Sequence | CCTGTGATTTTATAGTATGCACATGTCAGAAAAAGGCAAGA | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651134 | ||
| mod ID: M6ASITE067686 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165478-138165479:- | [22] | |
| Sequence | TCCCTATGCCAAACAGGTGAACAAACGTAGTTGTTTTTTAC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651135 | ||
| mod ID: M6ASITE067687 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165486-138165487:- | [22] | |
| Sequence | TTGTTTTCTCCCTATGCCAAACAGGTGAACAAACGTAGTTG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651136 | ||
| mod ID: M6ASITE067688 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165509-138165510:- | [20] | |
| Sequence | ATCATTTAATGCTATGAGATACATTGTTTTCTCCCTATGCC | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651137 | ||
| mod ID: M6ASITE067689 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165576-138165577:- | [20] | |
| Sequence | CTTTTCACTAGAACTTCTCAACATTTGGGAACTTTGCAAAT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651138 | ||
| mod ID: M6ASITE067690 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165635-138165636:- | [19] | |
| Sequence | TTTGGTTATTTTGAATACAGACATTGGCTCCAAATTTTCAT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651139 | ||
| mod ID: M6ASITE067691 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165639-138165640:- | [19] | |
| Sequence | ATATTTTGGTTATTTTGAATACAGACATTGGCTCCAAATTT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651140 | ||
| mod ID: M6ASITE067692 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165708-138165709:- | [19] | |
| Sequence | CAACACTTTACTCTTTTAGGACAATTCCTAGAATCTATAGT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651141 | ||
| mod ID: M6ASITE067693 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165729-138165730:- | [20] | |
| Sequence | TATTCATGTTCAGAAAGGAAACAACACTTTACTCTTTTAGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651142 | ||
| mod ID: M6ASITE067694 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165785-138165786:- | [19] | |
| Sequence | CAAATTTCTATAAATTTCCTACTTAAGTCTTAAGAACTGGG | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651143 | ||
| mod ID: M6ASITE067695 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165817-138165818:- | [19] | |
| Sequence | TCCTTATGAATGTTATTACTACTGGTATAAATCAAATTTCT | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651144 | ||
| mod ID: M6ASITE067696 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165820-138165821:- | [19] | |
| Sequence | TTTTCCTTATGAATGTTATTACTACTGGTATAAATCAAATT | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651145 | ||
| mod ID: M6ASITE067697 | Click to Show/Hide the Full List | ||
| mod site | chr4:138165913-138165914:- | [20] | |
| Sequence | TAAATCACTATGATTATTGCACAAACAACCAGAATTCTCCA | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651146 | ||
| mod ID: M6ASITE067698 | Click to Show/Hide the Full List | ||
| mod site | chr4:138166086-138166087:- | [20] | |
| Sequence | TTTCTCCTTCATTCCAAAGTACAAACATACTTTGAAGAATG | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651147 | ||
| mod ID: M6ASITE067699 | Click to Show/Hide the Full List | ||
| mod site | chr4:138166194-138166195:- | [20] | |
| Sequence | CTTGGAAGCACAGTCATATCACACTGGGAGGCAATGCAATG | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651148 | ||
| mod ID: M6ASITE067700 | Click to Show/Hide the Full List | ||
| mod site | chr4:138166239-138166240:- | [19] | |
| Sequence | TTTAGACATCAAATTTTCCTACTAACTAACTTTATTAGATG | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651149 | ||
| mod ID: M6ASITE067701 | Click to Show/Hide the Full List | ||
| mod site | chr4:138166254-138166255:- | [22] | |
| Sequence | ATGTGTATTATAAAATTTAGACATCAAATTTTCCTACTAAC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651150 | ||
| mod ID: M6ASITE067702 | Click to Show/Hide the Full List | ||
| mod site | chr4:138166498-138166499:- | [19] | |
| Sequence | TTAATGTATTGGTTAGGAGAACTGCTTGCTAAGTCCTTATT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651151 | ||
| mod ID: M6ASITE067703 | Click to Show/Hide the Full List | ||
| mod site | chr4:138166582-138166583:- | [20] | |
| Sequence | CCAACTTTGTTCCAGAGTGAACATGCTTTTTTTCCTCAACA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651152 | ||
| mod ID: M6ASITE067704 | Click to Show/Hide the Full List | ||
| mod site | chr4:138166638-138166639:- | [22] | |
| Sequence | GTTAACAATTCTCTCACAAAACTGTAGAGCATTAGGCATCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651153 | ||
| mod ID: M6ASITE067705 | Click to Show/Hide the Full List | ||
| mod site | chr4:138166643-138166644:- | [20] | |
| Sequence | TTACAGTTAACAATTCTCTCACAAAACTGTAGAGCATTAGG | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651154 | ||
| mod ID: M6ASITE067706 | Click to Show/Hide the Full List | ||
| mod site | chr4:138166766-138166767:- | [20] | |
| Sequence | GTTACTATTGGCCTTAAAATACACAGAGGACGGTTACAGTG | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651155 | ||
| mod ID: M6ASITE067707 | Click to Show/Hide the Full List | ||
| mod site | chr4:138166858-138166859:- | [19] | |
| Sequence | AACAATGAATTTATATAATTACCTAGATTTTCTTAGTGTGA | ||
| Motif Score | 2.052208333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651156 | ||
| mod ID: M6ASITE067708 | Click to Show/Hide the Full List | ||
| mod site | chr4:138166888-138166889:- | [20] | |
| Sequence | GTGAGATAACTGTCCTTTCTACAACCTCATAACAATGAATT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651157 | ||
| mod ID: M6ASITE067709 | Click to Show/Hide the Full List | ||
| mod site | chr4:138166926-138166927:- | [19] | |
| Sequence | CATAGTCTCACAGGAAATTCACCAATTTTCCATATGTCGTG | ||
| Motif Score | 2.026654762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651158 | ||
| mod ID: M6ASITE067710 | Click to Show/Hide the Full List | ||
| mod site | chr4:138167472-138167473:- | [19] | |
| Sequence | CCTTAGTTTATTAGTACTGTACTTCAAAAAGATTTTTAAAT | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651159 | ||
| mod ID: M6ASITE067711 | Click to Show/Hide the Full List | ||
| mod site | chr4:138167612-138167613:- | [19] | |
| Sequence | TGATAGCCTCTGTTTACATTACTTGTATATGGGCAAAATAA | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651160 | ||
| mod ID: M6ASITE067712 | Click to Show/Hide the Full List | ||
| mod site | chr4:138167617-138167618:- | [20] | |
| Sequence | CACCTTGATAGCCTCTGTTTACATTACTTGTATATGGGCAA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651161 | ||
| mod ID: M6ASITE067713 | Click to Show/Hide the Full List | ||
| mod site | chr4:138167687-138167688:- | [20] | |
| Sequence | TTAAAATATGGCATTGTATAACAACTGGGAAGAAGCTCATA | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651162 | ||
| mod ID: M6ASITE067714 | Click to Show/Hide the Full List | ||
| mod site | chr4:138168125-138168126:- | [19] | |
| Sequence | GCAAAGTTGTGTACACTTCTACCCCCACAAAATCTGCATTG | ||
| Motif Score | 2.05802381 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | rmsk_1451273; ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651163 | ||
| mod ID: M6ASITE067715 | Click to Show/Hide the Full List | ||
| mod site | chr4:138168133-138168134:- | [20] | |
| Sequence | GAAGACCAGCAAAGTTGTGTACACTTCTACCCCCACAAAAT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | rmsk_1451273; ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651164 | ||
| mod ID: M6ASITE067716 | Click to Show/Hide the Full List | ||
| mod site | chr4:138168217-138168218:- | [20] | |
| Sequence | CTTCAGTGCCTCTCACTTAGACATGTTCCATTCGAGGTCCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | rmsk_1451273; ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651165 | ||
| mod ID: M6ASITE067717 | Click to Show/Hide the Full List | ||
| mod site | chr4:138168345-138168346:- | [22] | |
| Sequence | GGAAATGAAAGATAGTATGGACTGAAGGTAACAATATTTTA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651166 | ||
| mod ID: M6ASITE067718 | Click to Show/Hide the Full List | ||
| mod site | chr4:138168384-138168385:- | [22] | |
| Sequence | AAGGTATGAAAAGATAAAAAACCGAAGGCCAGAGAATCAGG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651167 | ||
| mod ID: M6ASITE067719 | Click to Show/Hide the Full List | ||
| mod site | chr4:138168520-138168521:- | [22] | |
| Sequence | CATAACAGAATTCCTCATGAACTGTAATCAGTCTACAGGAA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651168 | ||
| mod ID: M6ASITE067720 | Click to Show/Hide the Full List | ||
| mod site | chr4:138168574-138168575:- | [19] | |
| Sequence | GTTTCCCCAATTACAATTTGACATATCAATAGAGGGTTAAC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651169 | ||
| mod ID: M6ASITE067721 | Click to Show/Hide the Full List | ||
| mod site | chr4:138168582-138168583:- | [20] | |
| Sequence | GTAAAATTGTTTCCCCAATTACAATTTGACATATCAATAGA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651170 | ||
| mod ID: M6ASITE067722 | Click to Show/Hide the Full List | ||
| mod site | chr4:138168665-138168666:- | [19] | |
| Sequence | GGTTACGTTAGCAATGCATGACGGTTTCTCCAACACTAAGA | ||
| Motif Score | 2.833690476 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651171 | ||
| mod ID: M6ASITE067723 | Click to Show/Hide the Full List | ||
| mod site | chr4:138168709-138168710:- | [20] | |
| Sequence | GTAGCTCCTCAGCCCTGTGGACACAAAATTTGGACAGCTTA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651172 | ||
| mod ID: M6ASITE067724 | Click to Show/Hide the Full List | ||
| mod site | chr4:138168750-138168751:- | [20] | |
| Sequence | ATGGCTGACTTCGGTTCCCAACATGCGTATGCATTTAGACT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651173 | ||
| mod ID: M6ASITE067725 | Click to Show/Hide the Full List | ||
| mod site | chr4:138169254-138169255:- | [20] | |
| Sequence | ATTCTCAAAAACTCTCCTGAACACTTATTTATATATATGTT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651174 | ||
| mod ID: M6ASITE067726 | Click to Show/Hide the Full List | ||
| mod site | chr4:138169341-138169342:- | [20] | |
| Sequence | TTAGGACTCATCATGAGAAGACACAGTTCCTTTAATCAGGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651175 | ||
| mod ID: M6ASITE067727 | Click to Show/Hide the Full List | ||
| mod site | chr4:138169697-138169698:- | [20] | |
| Sequence | ACTAGGCTCATAGAATTTAAACAAGCAAATTTTCCTGAGCT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651176 | ||
| mod ID: M6ASITE067728 | Click to Show/Hide the Full List | ||
| mod site | chr4:138169717-138169718:- | [22] | |
| Sequence | TATTTTTTTTTTCCCCATAGACTAGGCTCATAGAATTTAAA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651177 | ||
| mod ID: M6ASITE067729 | Click to Show/Hide the Full List | ||
| mod site | chr4:138169788-138169789:- | [20] | |
| Sequence | TAAATATTGTTCATTTGTCCACATCTCTCCCTATGATATGT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651178 | ||
| mod ID: M6ASITE067730 | Click to Show/Hide the Full List | ||
| mod site | chr4:138169941-138169942:- | [20] | |
| Sequence | TTTCAATTTTGCTCTTGAAAACATTTAATATATATCCAAGA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651179 | ||
| mod ID: M6ASITE067731 | Click to Show/Hide the Full List | ||
| mod site | chr4:138170033-138170034:- | [20] | |
| Sequence | ATTAAAATTATTAAAGGCAAACATAGAATTCATAAAAAATT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651180 | ||
| mod ID: M6ASITE067732 | Click to Show/Hide the Full List | ||
| mod site | chr4:138170508-138170509:- | [22] | |
| Sequence | AGCCTGACCAACATGGAGAAACCCCATCTCTACTAAAAATA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | rmsk_1451278; ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651181 | ||
| mod ID: M6ASITE067733 | Click to Show/Hide the Full List | ||
| mod site | chr4:138170531-138170532:- | [22] | |
| Sequence | CCTGAGGTCGGGAGTTCTAGACCAGCCTGACCAACATGGAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9; rmsk_1451278 | ||
| External Link | RMBase: m6A_site_651182 | ||
| mod ID: M6ASITE067734 | Click to Show/Hide the Full List | ||
| mod site | chr4:138170595-138170596:- | [20] | |
| Sequence | TGAGCCGGGCATGGTGGCTTACATCTGTAATCCCAGCACTT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | rmsk_1451278; ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651183 | ||
| mod ID: M6ASITE067735 | Click to Show/Hide the Full List | ||
| mod site | chr4:138170704-138170705:- | [22] | |
| Sequence | TCATTCAACTTGCAAAAGAGACAACTGATAAGAAGAAAATT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651184 | ||
| mod ID: M6ASITE067736 | Click to Show/Hide the Full List | ||
| mod site | chr4:138170736-138170737:- | [20] | |
| Sequence | AGGCTTTGTCAGTAATTTCCACACCTTAATTATCATTCAAC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651185 | ||
| mod ID: M6ASITE067737 | Click to Show/Hide the Full List | ||
| mod site | chr4:138170794-138170795:- | [20] | |
| Sequence | TGTCGCTGTAAATAAGATTTACAACTGATGTTTCTAGAAAA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651186 | ||
| mod ID: M6ASITE067738 | Click to Show/Hide the Full List | ||
| mod site | chr4:138170835-138170836:- | [20] | |
| Sequence | TTAGGAAAAGGCATATATTAACATAAAAATTGCAAAAGAAA | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651187 | ||
| mod ID: M6ASITE067739 | Click to Show/Hide the Full List | ||
| mod site | chr4:138170949-138170950:- | [20] | |
| Sequence | TTAAATTTATGGCTATTTTTACACGATGATGAATTTTGACA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651188 | ||
| mod ID: M6ASITE067740 | Click to Show/Hide the Full List | ||
| mod site | chr4:138170998-138170999:- | [20] | |
| Sequence | CCATATCTGTAATCATATCTACATGCAATGTTAGTAATTCT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651189 | ||
| mod ID: M6ASITE067741 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171098-138171099:- | [22] | |
| Sequence | CCCCTGATGAGTCTATCTAAACATATGCATTTTAAGCCTTC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651190 | ||
| mod ID: M6ASITE067742 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171145-138171146:- | [22] | |
| Sequence | GGTATTAAATCCTCATTAGAACAACCACCTGTTTCACTAAT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651191 | ||
| mod ID: M6ASITE067743 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171325-138171326:- | [22] | |
| Sequence | TAGGGGCTACTGTTTATGAGACACATCCAGGAGTTATGTTT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651192 | ||
| mod ID: M6ASITE067744 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171337-138171338:- | [19] | |
| Sequence | AAGAAGAGTTTCTAGGGGCTACTGTTTATGAGACACATCCA | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651193 | ||
| mod ID: M6ASITE067745 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171389-138171390:- | [22] | |
| Sequence | TTATACCCAGAGCACTTTGAACAAAGGTCAGTGGGGATTGT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651194 | ||
| mod ID: M6ASITE067746 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171428-138171429:- | [22] | |
| Sequence | GATACCAATTTAGATAACAAACACTCATGCTTTAATGGATT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651195 | ||
| mod ID: M6ASITE067747 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171572-138171573:- | [20] | |
| Sequence | GGTTTTGTAAAGATGGTTTTACACACTATAGATGTCTATAC | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651196 | ||
| mod ID: M6ASITE067750 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171615-138171616:- | [22] | |
| Sequence | AAAGAACCTTCAAATTGAAGACTGAGATTTTTCTGTATATA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651199 | ||
| mod ID: M6ASITE067752 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171630-138171631:- | [22] | |
| Sequence | ATATGTTAGCACGGCAAAGAACCTTCAAATTGAAGACTGAG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651200 | ||
| mod ID: M6ASITE067753 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171679-138171680:- | [22] | |
| Sequence | GGGTTAGGAGAAAAGACTAGACAATTACTATGTGGTCATTC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651201 | ||
| mod ID: M6ASITE067754 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171684-138171685:- | [22] | |
| Sequence | TATTGGGGTTAGGAGAAAAGACTAGACAATTACTATGTGGT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651202 | ||
| mod ID: M6ASITE067755 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171805-138171806:- | [23] | |
| Sequence | ATATATTTTAGCATATTCGAACTAATTTCTAAGAAATTTAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | A549; HEK293T; Huh7; TIME | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651203 | ||
| mod ID: M6ASITE067756 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171851-138171852:- | [22] | |
| Sequence | AATTACAACTTTGGTGATAAACAAAAGGAGTCAGTTATTTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; Huh7; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000509248.1; ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651204 | ||
| mod ID: M6ASITE067757 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171867-138171868:- | [20] | |
| Sequence | TTCTGAAAGTCTAGAGAATTACAACTTTGGTGATAAACAAA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: m6A_site_651205 | ||
| mod ID: M6ASITE067758 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171914-138171915:- | [22] | |
| Sequence | GGCAATCTGCCCAAGGGGAGACACAAAATAGGGATTTTTAC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; hESC-HEK293T; hESCs; fibroblasts; MM6; Huh7; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: m6A_site_651206 | ||
| mod ID: M6ASITE067759 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171946-138171947:- | [22] | |
| Sequence | GATAAGTTATGAACTAATGGACTTGAGATCTTGGCAATCTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; hESCs; fibroblasts; MM6; Huh7; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: m6A_site_651207 | ||
| mod ID: M6ASITE067760 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171954-138171955:- | [22] | |
| Sequence | CAGAAGAAGATAAGTTATGAACTAATGGACTTGAGATCTTG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; hESCs; fibroblasts; MM6; Huh7; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000509248.1; ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651208 | ||
| mod ID: M6ASITE067761 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171997-138171998:- | [20] | |
| Sequence | GAGAAAATAACCAGAACATTACAAATAATACTGGAAGTTGT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000509248.1; ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651209 | ||
| mod ID: M6ASITE067762 | Click to Show/Hide the Full List | ||
| mod site | chr4:138172002-138172003:- | [22] | |
| Sequence | TTGTAGAGAAAATAACCAGAACATTACAAATAATACTGGAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESCs; fibroblasts; A549; Huh7; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000509248.1; ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651210 | ||
| mod ID: M6ASITE067763 | Click to Show/Hide the Full List | ||
| mod site | chr4:138179241-138179242:- | [22] | |
| Sequence | TTTATTATATGGGACAAGAAACCCAGGTGGTTTAGAATAAT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HEK293T; A549 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000509248.1; ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651211 | ||
| mod ID: M6ASITE067764 | Click to Show/Hide the Full List | ||
| mod site | chr4:138179248-138179249:- | [22] | |
| Sequence | TTATCTCTTTATTATATGGGACAAGAAACCCAGGTGGTTTA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000509248.1; ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651212 | ||
| mod ID: M6ASITE067765 | Click to Show/Hide the Full List | ||
| mod site | chr4:138179323-138179324:- | [22] | |
| Sequence | TGCCCTTTCCCTCTATTCGGACCCATTTAGTACAGGGATTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; A549 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: m6A_site_651213 | ||
| mod ID: M6ASITE067766 | Click to Show/Hide the Full List | ||
| mod site | chr4:138179360-138179361:- | [20] | |
| Sequence | TCCCAGCTTTGTTTTCCTTCACATGCCTCTTCATGGTTGCC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000509248.1; ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651214 | ||
| mod ID: M6ASITE067767 | Click to Show/Hide the Full List | ||
| mod site | chr4:138180671-138180672:- | [20] | |
| Sequence | TGGGCTGATTTATCTTCGATACAAATGCCCAGATATGCATC | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000509248.1; ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651215 | ||
| mod ID: M6ASITE067768 | Click to Show/Hide the Full List | ||
| mod site | chr4:138180755-138180756:- | [22] | |
| Sequence | GATAATGCTCTTCTCTGGAGACCTCGACAGTCTTTTGAATT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: m6A_site_651216 | ||
| mod ID: M6ASITE067769 | Click to Show/Hide the Full List | ||
| mod site | chr4:138185185-138185186:- | [20] | |
| Sequence | TCACCATTGGCTATGTGCTGACAAATGTGGCCTACTTTACG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: m6A_site_651217 | ||
| mod ID: M6ASITE067770 | Click to Show/Hide the Full List | ||
| mod site | chr4:138214593-138214594:- | [22] | |
| Sequence | TGTTACTGAAGAAGTAGAAAACCCTGAAAAGTAAGTAGCCA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: m6A_site_651218 | ||
| mod ID: M6ASITE067771 | Click to Show/Hide the Full List | ||
| mod site | chr4:138219352-138219353:- | [22] | |
| Sequence | AATTACAGGTCAAACGCAGAACTTTAAAGACGCCTTTTCAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9; ENST00000509248.1 | ||
| External Link | RMBase: m6A_site_651219 | ||
| mod ID: M6ASITE067772 | Click to Show/Hide the Full List | ||
| mod site | chr4:138232300-138232301:- | [22] | |
| Sequence | ATTCAATGTGAAATCCCTGAACTTGCGATCAAGCTCATTAC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651220 | ||
| mod ID: M6ASITE067773 | Click to Show/Hide the Full List | ||
| mod site | chr4:138232330-138232331:- | [22] | |
| Sequence | TTTGGACGCTACATTCTGGAACCATTTTTTATTCAATGTGA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651221 | ||
| mod ID: M6ASITE067774 | Click to Show/Hide the Full List | ||
| mod site | chr4:138236338-138236339:- | [22] | |
| Sequence | TTTGTACGAGTCTGGGTGGAACTCCTCATAATACGGTAAGA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651222 | ||
| mod ID: M6ASITE067775 | Click to Show/Hide the Full List | ||
| mod site | chr4:138236424-138236425:- | [22] | |
| Sequence | TGTCTTATGCTGAATTGGGAACAACTATAAAGAAATCTGGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651223 | ||
| mod ID: M6ASITE067776 | Click to Show/Hide the Full List | ||
| mod site | chr4:138241854-138241855:- | [22] | |
| Sequence | TCCTAAGGGCGTGCTCCAGAACACGGGCAGCGTGGGCATGT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651224 | ||
| mod ID: M6ASITE067777 | Click to Show/Hide the Full List | ||
| mod site | chr4:138241983-138241984:- | [20] | |
| Sequence | GAGGCTGCCTTCCCTGGGCAACAAGGAGCCACCTGGGCAGG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651225 | ||
| mod ID: M6ASITE067778 | Click to Show/Hide the Full List | ||
| mod site | chr4:138242132-138242133:- | [22] | |
| Sequence | TCGATCAGTAACACCAAGAGACACCAAAGTTGAAAGTTTTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; MM6; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651226 | ||
| mod ID: M6ASITE067779 | Click to Show/Hide the Full List | ||
| mod site | chr4:138242155-138242156:- | [22] | |
| Sequence | CAGTCTGAAAGCAGAGGAAGACATCGATCAGTAACACCAAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; GM12878; MM6; iSLK; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651227 | ||
| mod ID: M6ASITE067780 | Click to Show/Hide the Full List | ||
| mod site | chr4:138242176-138242177:- | [22] | |
| Sequence | ACACTATAGCGCTGAGAGAGACAGTCTGAAAGCAGAGGAAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; GM12878; MM6; iSLK; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651228 | ||
| mod ID: M6ASITE067781 | Click to Show/Hide the Full List | ||
| mod site | chr4:138242196-138242197:- | [22] | |
| Sequence | CTTTGAGTTTTCACCTGTGAACACTATAGCGCTGAGAGAGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; GM12878; MM6; iSLK; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: m6A_site_651229 | ||
Pseudouridine (Pseudo)
| In total 3 m6A sequence/site(s) in this target gene | |||
| mod ID: PSESITE000204 | Click to Show/Hide the Full List | ||
| mod site | chr4:138166302-138166303:- | [24] | |
| Sequence | GCATGTTAAAGCTGTATAATTTGTTGGGTTGTGAGTGGTCT | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: Pseudo_site_3796 | ||
| mod ID: PSESITE000205 | Click to Show/Hide the Full List | ||
| mod site | chr4:138166893-138166894:- | [24] | |
| Sequence | ATGTCGTGAGATAACTGTCCTTTCTACAACCTCATAACAAT | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: Pseudo_site_3797 | ||
| mod ID: PSESITE000206 | Click to Show/Hide the Full List | ||
| mod site | chr4:138171134-138171135:- | [24] | |
| Sequence | CTCATTAGAACAACCACCTGTTTCACTAATAACTTACCCCT | ||
| Transcript ID List | ENST00000280612.9 | ||
| External Link | RMBase: Pseudo_site_3798 | ||
References