m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00197)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
BECN1
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
RNA demethylase ALKBH5 (ALKBH5) [ERASER]
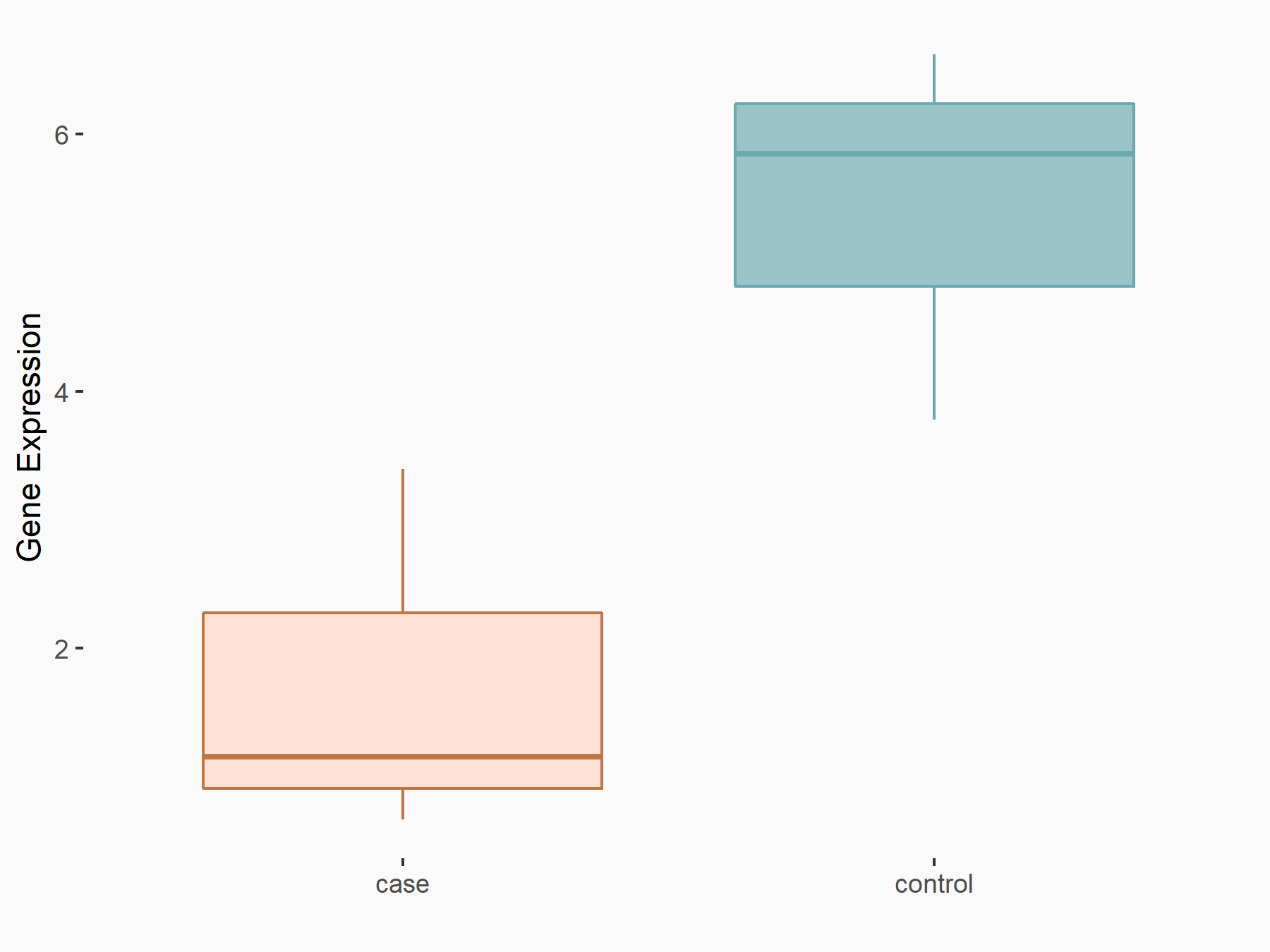
| Representative RNA-seq result indicating the expression of this target gene regulated by ALKBH5 | ||
| Cell Line | 143B cell line | Homo sapiens |
|
Treatment: siALKBH5 transfected 143B cells
Control: siControl 143B cells
|
GSE154528 | |
| Regulation |
  |
logFC: -1.52E+00 p-value: 9.98E-13 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | ALKBH5 is a tumor-promoting gene in epithelial ovarian cancer, which is involved in the mTOR pathway and BCL-2-Beclin-1 (BECN1) complex. ALKBH5 activated EGFR-PIK3CA-AKT-mTOR signaling pathway. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Ovarian cancer | ICD-11: 2C73 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Autophagy | hsa04140 | |||
| In-vitro Model | A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 |
| CoC1 | Ovarian adenocarcinoma | Homo sapiens | CVCL_6891 | |
| OVCAR-3 | Ovarian serous adenocarcinoma | Homo sapiens | CVCL_0465 | |
| SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 | |
| In-vivo Model | SKOV3 or A2780 cells were infected with the indicated lentiviral vectors and injected (5 × 106 cells/mouse in 200 uL volume) subcutaneously into the left armpit of 6-week-old BALB/c nude mice. After 21 days, the animals were sacrificed to confirm the presence of tumors and weigh the established tumors. | |||
YTH domain-containing family protein 1 (YTHDF1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
 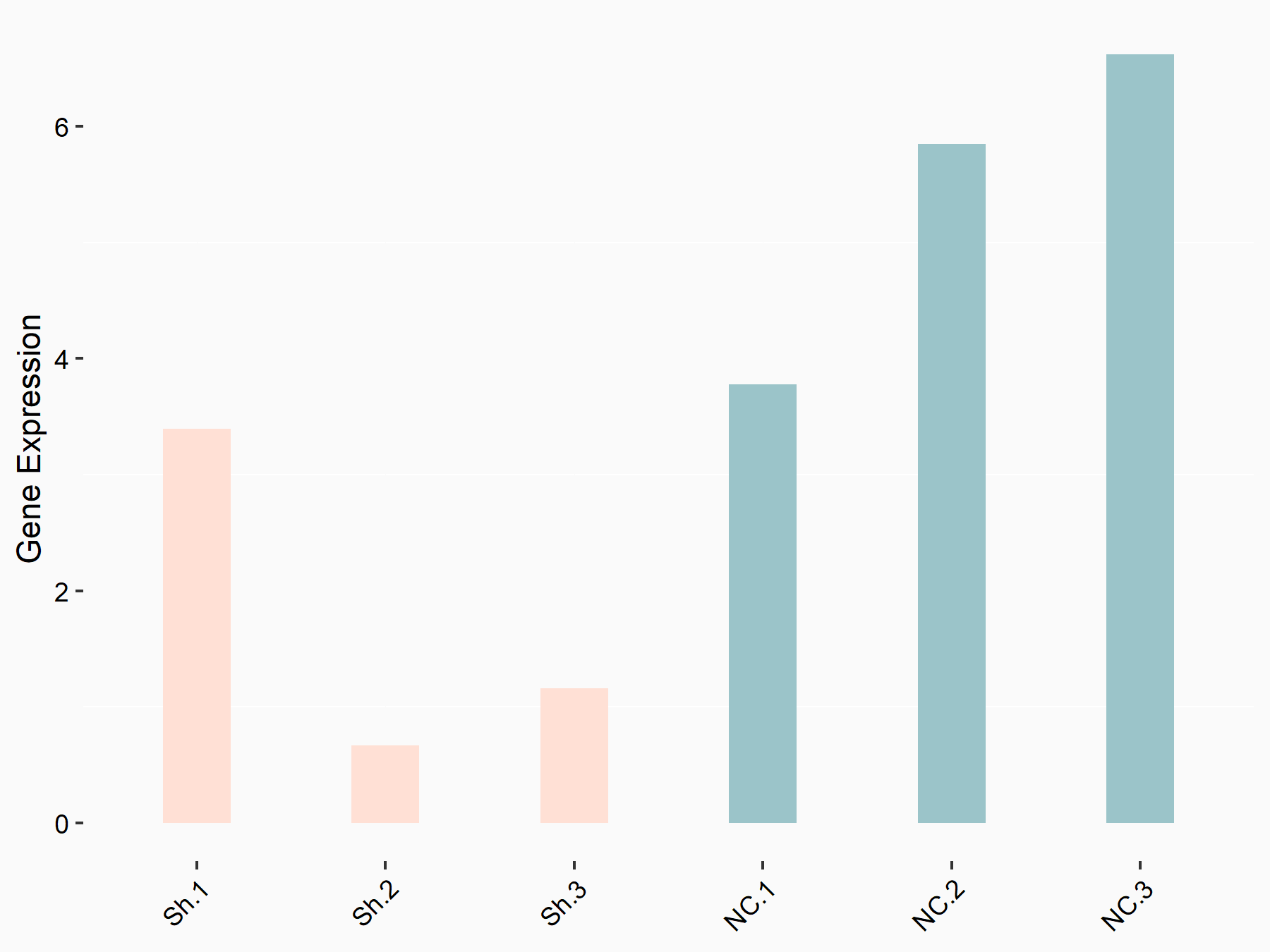 |
logFC: -1.33E+00 p-value: 2.29E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | Analyzed the effect of sorafenib on HSC ferroptosis and m6A modification in advanced fibrotic patients with hepatocellular carcinoma receiving sorafenib monotherapy. YTHDF1 promotes Beclin-1 (BECN1) mRNA stability and autophagy activation via recognizing the m6A binding site within BECN1 coding regions. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Responsed Drug | Sorafenib | Approved | ||
| Pathway Response | Ferroptosis | hsa04216 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Ferroptosis | |||
| Cell autophagy | ||||
| In-vitro Model | HSC (Hematopoietic stem cell) | |||
| In-vivo Model | VA-Lip-Mettl4-shRNA, VA-Lip-Fto-Plasmid and VA-Lip-Ythdf1-shRNA (0.75 mg/kg) were injected intravenously 3 times a week. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | Analyzed the effect of sorafenib on HSC ferroptosis and m6A modification in advanced fibrotic patients with hepatocellular carcinoma receiving sorafenib monotherapy. YTHDF1 promotes Beclin-1 (BECN1) mRNA stability and autophagy activation via recognizing the m6A binding site within BECN1 coding regions. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Sorafenib | Approved | ||
| Pathway Response | Ferroptosis | hsa04216 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Ferroptosis | |||
| Cell autophagy | ||||
| In-vitro Model | HSC (Hematopoietic stem cell) | |||
| In-vivo Model | VA-Lip-Mettl4-shRNA, VA-Lip-Fto-Plasmid and VA-Lip-Ythdf1-shRNA (0.75 mg/kg) were injected intravenously 3 times a week. | |||
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | ALKBH5 is a tumor-promoting gene in epithelial ovarian cancer, which is involved in the mTOR pathway and BCL-2-Beclin-1 (BECN1) complex. ALKBH5 activated EGFR-PIK3CA-AKT-mTOR signaling pathway. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Autophagy | hsa04140 | |||
| In-vitro Model | A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 |
| CoC1 | Ovarian adenocarcinoma | Homo sapiens | CVCL_6891 | |
| OVCAR-3 | Ovarian serous adenocarcinoma | Homo sapiens | CVCL_0465 | |
| SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 | |
| In-vivo Model | SKOV3 or A2780 cells were infected with the indicated lentiviral vectors and injected (5 × 106 cells/mouse in 200 uL volume) subcutaneously into the left armpit of 6-week-old BALB/c nude mice. After 21 days, the animals were sacrificed to confirm the presence of tumors and weigh the established tumors. | |||
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [2] | |||
| Response Summary | Analyzed the effect of sorafenib on HSC ferroptosis and m6A modification in advanced fibrotic patients with hepatocellular carcinoma receiving sorafenib monotherapy. YTHDF1 promotes Beclin-1 (BECN1) mRNA stability and autophagy activation via recognizing the m6A binding site within BECN1 coding regions. | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Pathway Response | Ferroptosis | hsa04216 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Ferroptosis | |||
| Cell autophagy | ||||
| In-vitro Model | HSC (Hematopoietic stem cell) | |||
| In-vivo Model | VA-Lip-Mettl4-shRNA, VA-Lip-Fto-Plasmid and VA-Lip-Ythdf1-shRNA (0.75 mg/kg) were injected intravenously 3 times a week. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03143 | ||
| Epigenetic Regulator | Histone acetyltransferase KAT2A (KAT2A) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00197)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: AC4SITE000224 | Click to Show/Hide the Full List | ||
| mod site | chr17:42820822-42820823:- | [6] | |
| Sequence | AGCTCCATTACTTACCACAGCCCAGGCGAAACCAGGAGAGA | ||
| Cell/Tissue List | H1 | ||
| Seq Type List | ac4C-seq | ||
| Transcript ID List | ENST00000593205.5; ENST00000361523.8; ENST00000593112.1; ENST00000590099.6; ENST00000543382.6; ENST00000438274.7; ENST00000585515.5; ENST00000591307.5; ENST00000589636.5; ENST00000591085.1; ENST00000617806.4 | ||
| External Link | RMBase: ac4C_site_798 | ||
Adenosine-to-Inosine editing (A-to-I)
| In total 24 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE008053 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812127-42812128:- | [7] | |
| Sequence | CCTCCCAAAGTGCTGGGATTACAGGCGTGAGCCACTGCTCC | ||
| Transcript ID List | ENST00000586754.2; ENST00000543382.6; ENST00000617806.4; ENST00000590764.1; ENST00000590099.6; ENST00000589663.5; ENST00000587880.1; ENST00000588276.5; ENST00000586589.5; ENST00000438274.7; ENST00000361523.8 | ||
| External Link | RMBase: RNA-editing_site_57660 | ||
| mod ID: A2ISITE008054 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812139-42812140:- | [7] | |
| Sequence | TGCCCACCTTGGCCTCCCAAAGTGCTGGGATTACAGGCGTG | ||
| Transcript ID List | ENST00000586589.5; ENST00000617806.4; ENST00000587880.1; ENST00000590099.6; ENST00000589663.5; ENST00000361523.8; ENST00000590764.1; ENST00000438274.7; ENST00000543382.6; ENST00000588276.5; ENST00000586754.2 | ||
| External Link | RMBase: RNA-editing_site_57661 | ||
| mod ID: A2ISITE008055 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812172-42812173:- | [7] | |
| Sequence | AGGCTAGTCTCAAACTCCCAACCTCAGGTGATCTGCCCACC | ||
| Transcript ID List | ENST00000361523.8; ENST00000589663.5; ENST00000590764.1; ENST00000586754.2; ENST00000587880.1; ENST00000586589.5; ENST00000438274.7; ENST00000590099.6; ENST00000617806.4; ENST00000588276.5; ENST00000543382.6 | ||
| External Link | RMBase: RNA-editing_site_57662 | ||
| mod ID: A2ISITE008056 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812179-42812180:- | [7] | |
| Sequence | GTTGGTCAGGCTAGTCTCAAACTCCCAACCTCAGGTGATCT | ||
| Transcript ID List | ENST00000587880.1; ENST00000438274.7; ENST00000361523.8; ENST00000590764.1; ENST00000586589.5; ENST00000589663.5; ENST00000617806.4; ENST00000586754.2; ENST00000588276.5; ENST00000543382.6; ENST00000590099.6 | ||
| External Link | RMBase: RNA-editing_site_57663 | ||
| mod ID: A2ISITE008057 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812180-42812181:- | [7] | |
| Sequence | TGTTGGTCAGGCTAGTCTCAAACTCCCAACCTCAGGTGATC | ||
| Transcript ID List | ENST00000438274.7; ENST00000589663.5; ENST00000586589.5; ENST00000586754.2; ENST00000617806.4; ENST00000590099.6; ENST00000361523.8; ENST00000587880.1; ENST00000543382.6; ENST00000590764.1; ENST00000588276.5 | ||
| External Link | RMBase: RNA-editing_site_57664 | ||
| mod ID: A2ISITE008058 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812187-42812188:- | [7] | |
| Sequence | TTCTCCATGTTGGTCAGGCTAGTCTCAAACTCCCAACCTCA | ||
| Transcript ID List | ENST00000586589.5; ENST00000543382.6; ENST00000589663.5; ENST00000587880.1; ENST00000617806.4; ENST00000588276.5; ENST00000361523.8; ENST00000438274.7; ENST00000590099.6; ENST00000590764.1; ENST00000586754.2 | ||
| External Link | RMBase: RNA-editing_site_57665 | ||
| mod ID: A2ISITE008059 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812201-42812202:- | [7] | |
| Sequence | AGTAGAGACGGGGTTTCTCCATGTTGGTCAGGCTAGTCTCA | ||
| Transcript ID List | ENST00000589663.5; ENST00000586589.5; ENST00000617806.4; ENST00000587880.1; ENST00000543382.6; ENST00000590099.6; ENST00000586754.2; ENST00000590764.1; ENST00000588276.5; ENST00000438274.7; ENST00000361523.8 | ||
| External Link | RMBase: RNA-editing_site_57666 | ||
| mod ID: A2ISITE008060 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812251-42812252:- | [7] | |
| Sequence | ATCTGGGATTACAGGCATGCACCACCACGCCCGGCTAATTA | ||
| Transcript ID List | ENST00000590099.6; ENST00000589663.5; ENST00000590764.1; ENST00000588276.5; ENST00000543382.6; ENST00000586754.2; ENST00000438274.7; ENST00000617806.4; ENST00000361523.8; ENST00000586589.5; ENST00000587880.1 | ||
| External Link | RMBase: RNA-editing_site_57667 | ||
| mod ID: A2ISITE008061 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812259-42812260:- | [7] | |
| Sequence | TCCTGAGTATCTGGGATTACAGGCATGCACCACCACGCCCG | ||
| Transcript ID List | ENST00000543382.6; ENST00000589663.5; ENST00000590764.1; ENST00000438274.7; ENST00000590099.6; ENST00000586754.2; ENST00000617806.4; ENST00000587880.1; ENST00000586589.5; ENST00000588276.5; ENST00000361523.8 | ||
| External Link | RMBase: RNA-editing_site_57668 | ||
| mod ID: A2ISITE008062 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812296-42812297:- | [7] | |
| Sequence | CCGCCTCCCGGGTTCAAGCAATTCCCCTGCCTCAGCCTCCT | ||
| Transcript ID List | ENST00000438274.7; ENST00000586754.2; ENST00000543382.6; ENST00000617806.4; ENST00000587880.1; ENST00000589663.5; ENST00000590099.6; ENST00000590764.1; ENST00000586589.5; ENST00000588276.5; ENST00000361523.8 | ||
| External Link | RMBase: RNA-editing_site_57669 | ||
| mod ID: A2ISITE008063 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812326-42812327:- | [7] | |
| Sequence | CAATGGCATGATCTCGGCTTACTGCAACCTCCGCCTCCCGG | ||
| Transcript ID List | ENST00000438274.7; ENST00000543382.6; ENST00000588276.5; ENST00000617806.4; ENST00000590099.6; ENST00000361523.8; ENST00000589663.5; ENST00000586754.2; ENST00000586589.5; ENST00000587880.1; ENST00000590764.1 | ||
| External Link | RMBase: RNA-editing_site_57670 | ||
| mod ID: A2ISITE008064 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812339-42812340:- | [7] | |
| Sequence | CCAGGCTGAAGTGCAATGGCATGATCTCGGCTTACTGCAAC | ||
| Transcript ID List | ENST00000590099.6; ENST00000543382.6; ENST00000586589.5; ENST00000589663.5; ENST00000586754.2; ENST00000361523.8; ENST00000588276.5; ENST00000617806.4; ENST00000587880.1; ENST00000590764.1; ENST00000438274.7 | ||
| External Link | RMBase: RNA-editing_site_57671 | ||
| mod ID: A2ISITE008065 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812344-42812345:- | [7] | |
| Sequence | GTTGCCCAGGCTGAAGTGCAATGGCATGATCTCGGCTTACT | ||
| Transcript ID List | ENST00000586589.5; ENST00000590099.6; ENST00000588276.5; ENST00000361523.8; ENST00000587880.1; ENST00000590764.1; ENST00000438274.7; ENST00000617806.4; ENST00000586754.2; ENST00000543382.6; ENST00000589663.5 | ||
| External Link | RMBase: RNA-editing_site_57672 | ||
| mod ID: A2ISITE008066 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812456-42812457:- | [7] | |
| Sequence | CCTCCCAAAGTGCTGGGATTACAGACGTGAGCCACTGTGCC | ||
| Transcript ID List | ENST00000361523.8; ENST00000586589.5; ENST00000590764.1; ENST00000543382.6; ENST00000438274.7; ENST00000586754.2; ENST00000617806.4; ENST00000589663.5; ENST00000587880.1; ENST00000590099.6; ENST00000588276.5 | ||
| External Link | RMBase: RNA-editing_site_57673 | ||
| mod ID: A2ISITE008067 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812468-42812469:- | [7] | |
| Sequence | CACCCGCCTTGGCCTCCCAAAGTGCTGGGATTACAGACGTG | ||
| Transcript ID List | ENST00000590099.6; ENST00000543382.6; ENST00000586589.5; ENST00000438274.7; ENST00000590764.1; ENST00000586754.2; ENST00000587880.1; ENST00000588276.5; ENST00000617806.4; ENST00000589663.5; ENST00000361523.8 | ||
| External Link | RMBase: RNA-editing_site_57674 | ||
| mod ID: A2ISITE008068 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812494-42812495:- | [7] | |
| Sequence | GGTCTTGATCTCCTGACCTCATGATCCACCCGCCTTGGCCT | ||
| Transcript ID List | ENST00000438274.7; ENST00000587880.1; ENST00000543382.6; ENST00000589663.5; ENST00000586754.2; ENST00000590764.1; ENST00000586589.5; ENST00000588276.5; ENST00000590099.6; ENST00000361523.8; ENST00000617806.4 | ||
| External Link | RMBase: RNA-editing_site_57675 | ||
| mod ID: A2ISITE008069 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812528-42812529:- | [7] | |
| Sequence | GGTAGTGACGGGGTTTCACCATGTTATCCATGATGGTCTTG | ||
| Transcript ID List | ENST00000586754.2; ENST00000587880.1; ENST00000543382.6; ENST00000438274.7; ENST00000589663.5; ENST00000590099.6; ENST00000586589.5; ENST00000361523.8; ENST00000617806.4; ENST00000590764.1; ENST00000588276.5 | ||
| External Link | RMBase: RNA-editing_site_57676 | ||
| mod ID: A2ISITE008070 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812658-42812659:- | [7] | |
| Sequence | GCGTGATCTCGGCTCATTGCAGGCTCTGCCTCCCAGGTTCA | ||
| Transcript ID List | ENST00000586589.5; ENST00000588276.5; ENST00000586754.2; ENST00000617806.4; ENST00000590099.6; ENST00000543382.6; ENST00000361523.8; ENST00000590764.1; ENST00000589663.5; ENST00000587880.1; ENST00000438274.7 | ||
| External Link | RMBase: RNA-editing_site_57677 | ||
| mod ID: A2ISITE008071 | Click to Show/Hide the Full List | ||
| mod site | chr17:42812698-42812699:- | [7] | |
| Sequence | AGACGGAGTCTCACTCTGTCACCCAGGCTGGAGTGCAGTGG | ||
| Transcript ID List | ENST00000617806.4; ENST00000586754.2; ENST00000543382.6; ENST00000361523.8; ENST00000586589.5; ENST00000438274.7; ENST00000587880.1; ENST00000590099.6; ENST00000589663.5; ENST00000588276.5; ENST00000590764.1 | ||
| External Link | RMBase: RNA-editing_site_57678 | ||
| mod ID: A2ISITE008072 | Click to Show/Hide the Full List | ||
| mod site | chr17:42813208-42813209:- | [8] | |
| Sequence | GTAGTCTGAGGTAATTTCATACAATATTTTAAATAATTTTG | ||
| Transcript ID List | ENST00000438274.7; ENST00000589663.5; ENST00000586754.2; ENST00000590764.1; ENST00000361523.8; ENST00000543382.6; ENST00000587880.1; ENST00000586589.5; ENST00000588276.5; ENST00000617806.4; ENST00000590099.6 | ||
| External Link | RMBase: RNA-editing_site_57679 | ||
| mod ID: A2ISITE008073 | Click to Show/Hide the Full List | ||
| mod site | chr17:42816434-42816435:- | [8] | |
| Sequence | GCTCTCGAACTCCTGACCTCAAGTTATCCACCCGTCTTGGC | ||
| Transcript ID List | ENST00000586589.5; ENST00000438274.7; ENST00000543382.6; ENST00000593205.5; ENST00000589663.5; ENST00000617806.4; ENST00000589493.1; ENST00000361523.8; ENST00000590099.6 | ||
| External Link | RMBase: RNA-editing_site_57680 | ||
| mod ID: A2ISITE008074 | Click to Show/Hide the Full List | ||
| mod site | chr17:42818643-42818644:- | [7] | |
| Sequence | TTGACATCATGTCGGGCCAGACAGATGTGGATCACCCACTC | ||
| Transcript ID List | ENST00000617806.4; ENST00000361523.8; ENST00000543382.6; ENST00000589493.1; ENST00000593205.5; ENST00000438274.7; ENST00000586589.5; ENST00000590099.6 | ||
| External Link | RMBase: RNA-editing_site_57681 | ||
| mod ID: A2ISITE008075 | Click to Show/Hide the Full List | ||
| mod site | chr17:42819383-42819384:- | [7] | |
| Sequence | TACCTGGCCCGTCACCTTCCAGTATGGATTTTTTCCTGTAG | ||
| Transcript ID List | ENST00000543382.6; ENST00000590099.6; ENST00000585515.5; ENST00000361523.8; ENST00000591085.1; ENST00000593205.5; ENST00000438274.7; ENST00000617806.4 | ||
| External Link | RMBase: RNA-editing_site_57682 | ||
| mod ID: A2ISITE008076 | Click to Show/Hide the Full List | ||
| mod site | chr17:42823032-42823033:- | [8] | |
| Sequence | ACAGAGAGGTTAAGTGACTTACCTAAGGTCCAACAGCTGGT | ||
| Transcript ID List | ENST00000585515.5; ENST00000589636.5; ENST00000438274.7; ENST00000591307.5; ENST00000593112.1; ENST00000590099.6; ENST00000591085.1; ENST00000593205.5; ENST00000543382.6; ENST00000617806.4; ENST00000361523.8 | ||
| External Link | RMBase: RNA-editing_site_57683 | ||
N6-methyladenosine (m6A)
| In total 68 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE032944 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810189-42810190:- | [9] | |
| Sequence | GGTGTTTGATACTGTTTGAGACATTATGGAGAGATTTAATT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361523.8; ENST00000590099.6; ENST00000617806.4 | ||
| External Link | RMBase: m6A_site_364419 | ||
| mod ID: M6ASITE032945 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810199-42810200:- | [10] | |
| Sequence | GATATGAAATGGTGTTTGATACTGTTTGAGACATTATGGAG | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000590099.6; ENST00000617806.4; ENST00000361523.8 | ||
| External Link | RMBase: m6A_site_364421 | ||
| mod ID: M6ASITE032946 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810237-42810238:- | [10] | |
| Sequence | GGTTCTTATACTGAATGCCGACTCTGCTCTGTGTTAGAGAT | ||
| Motif Score | 3.287565476 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000590099.6; ENST00000617806.4; ENST00000361523.8 | ||
| External Link | RMBase: m6A_site_364423 | ||
| mod ID: M6ASITE032947 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810268-42810269:- | [10] | |
| Sequence | TCAGATGCTTGAAATTTAACACTTTTCACTTGGTTCTTATA | ||
| Motif Score | 2.506922619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000361523.8; ENST00000590099.6; ENST00000617806.4 | ||
| External Link | RMBase: m6A_site_364425 | ||
| mod ID: M6ASITE032948 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810270-42810271:- | [9] | |
| Sequence | GATCAGATGCTTGAAATTTAACACTTTTCACTTGGTTCTTA | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361523.8; ENST00000590099.6; ENST00000617806.4 | ||
| External Link | RMBase: m6A_site_364426 | ||
| mod ID: M6ASITE032949 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810323-42810324:- | [10] | |
| Sequence | TTGGATAATGTGGGAAAATGACATCTAAGCTTTACCTGGTC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000438274.7; ENST00000617806.4; ENST00000588276.5; ENST00000361523.8; ENST00000590764.1; ENST00000590099.6 | ||
| External Link | RMBase: m6A_site_364427 | ||
| mod ID: M6ASITE032950 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810347-42810348:- | [10] | |
| Sequence | TGAGGATATGTTTGAGATGCACAGTTGGATAATGTGGGAAA | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000590764.1; ENST00000590099.6; ENST00000361523.8; ENST00000588276.5; ENST00000617806.4; ENST00000438274.7 | ||
| External Link | RMBase: m6A_site_364428 | ||
| mod ID: M6ASITE032951 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810374-42810375:- | [10] | |
| Sequence | AAATAGAAACTTTTGTCTTTACTGAGATGAGGATATGTTTG | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000438274.7; ENST00000590099.6; ENST00000361523.8; ENST00000617806.4; ENST00000588276.5; ENST00000590764.1 | ||
| External Link | RMBase: m6A_site_364430 | ||
| mod ID: M6ASITE032952 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810386-42810387:- | [11] | |
| Sequence | CTTAACAATATGAAATAGAAACTTTTGTCTTTACTGAGATG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000588276.5; ENST00000590764.1; ENST00000590099.6; ENST00000617806.4; ENST00000361523.8; ENST00000438274.7 | ||
| External Link | RMBase: m6A_site_364432 | ||
| mod ID: M6ASITE032953 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810402-42810403:- | [9] | |
| Sequence | CTCTCAACTTCCTGCACTTAACAATATGAAATAGAAACTTT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000590099.6; ENST00000588276.5; ENST00000590764.1; ENST00000438274.7; ENST00000361523.8; ENST00000617806.4 | ||
| External Link | RMBase: m6A_site_364433 | ||
| mod ID: M6ASITE032954 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810407-42810408:- | [10] | |
| Sequence | TATCTCTCTCAACTTCCTGCACTTAACAATATGAAATAGAA | ||
| Motif Score | 3.252583333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000588276.5; ENST00000361523.8; ENST00000590764.1; ENST00000590099.6; ENST00000438274.7; ENST00000617806.4 | ||
| External Link | RMBase: m6A_site_364434 | ||
| mod ID: M6ASITE032955 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810416-42810417:- | [10] | |
| Sequence | CTGGATTTTTATCTCTCTCAACTTCCTGCACTTAACAATAT | ||
| Motif Score | 2.595904762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000590099.6; ENST00000361523.8; ENST00000543382.6; ENST00000590764.1; ENST00000438274.7; ENST00000588276.5; ENST00000617806.4 | ||
| External Link | RMBase: m6A_site_364435 | ||
| mod ID: M6ASITE032956 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810437-42810438:- | [10] | |
| Sequence | ATTGAGATTGATGTGTTTTCACTGGATTTTTATCTCTCTCA | ||
| Motif Score | 2.469291667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000617806.4; ENST00000590764.1; ENST00000590099.6; ENST00000361523.8; ENST00000438274.7; ENST00000543382.6; ENST00000588276.5 | ||
| External Link | RMBase: m6A_site_364436 | ||
| mod ID: M6ASITE032957 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810467-42810468:- | [10] | |
| Sequence | ATTTGCTTTTCTTTTTTTTAACTGAGTTGAATTGAGATTGA | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000590764.1; ENST00000438274.7; ENST00000588276.5; ENST00000590099.6; ENST00000543382.6; ENST00000617806.4; ENST00000361523.8 | ||
| External Link | RMBase: m6A_site_364437 | ||
| mod ID: M6ASITE032958 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810497-42810498:- | [12] | |
| Sequence | CCCTTTCTTCTCTGAAAAAAACTAATTTAAATTTGCTTTTC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000590764.1; ENST00000543382.6; ENST00000617806.4; ENST00000590099.6; ENST00000361523.8; ENST00000438274.7; ENST00000588276.5 | ||
| External Link | RMBase: m6A_site_364438 | ||
| mod ID: M6ASITE032959 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810525-42810526:- | [12] | |
| Sequence | TTTGATTCTTAACCCCATGGACTCCTTTCCCTTTCTTCTCT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000543382.6; ENST00000588276.5; ENST00000361523.8; ENST00000617806.4; ENST00000590764.1; ENST00000590099.6; ENST00000438274.7 | ||
| External Link | RMBase: m6A_site_364439 | ||
| mod ID: M6ASITE032960 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810578-42810579:- | [9] | |
| Sequence | AGATACTTTCCAGAGCTACAACATGCCATCTATAGTTGCCA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361523.8; ENST00000590099.6; ENST00000438274.7; ENST00000590764.1; ENST00000617806.4; ENST00000543382.6; ENST00000588276.5 | ||
| External Link | RMBase: m6A_site_364441 | ||
| mod ID: M6ASITE032961 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810600-42810601:- | [10] | |
| Sequence | TTATTTTAAAATATCATGTGACAGATACTTTCCAGAGCTAC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000543382.6; ENST00000588276.5; ENST00000438274.7; ENST00000617806.4; ENST00000590764.1; ENST00000590099.6; ENST00000361523.8 | ||
| External Link | RMBase: m6A_site_364442 | ||
| mod ID: M6ASITE032962 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810623-42810624:- | [10] | |
| Sequence | AGAAAAAATCCACAAAAGCCACTTTATTTTAAAATATCATG | ||
| Motif Score | 2.475107143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000438274.7; ENST00000590099.6; ENST00000543382.6; ENST00000361523.8; ENST00000588276.5; ENST00000590764.1; ENST00000617806.4 | ||
| External Link | RMBase: m6A_site_364443 | ||
| mod ID: M6ASITE032963 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810632-42810633:- | [10] | |
| Sequence | TACCAAAAAAGAAAAAATCCACAAAAGCCACTTTATTTTAA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000617806.4; ENST00000361523.8; ENST00000588276.5; ENST00000438274.7; ENST00000590099.6; ENST00000543382.6; ENST00000590764.1 | ||
| External Link | RMBase: m6A_site_364444 | ||
| mod ID: M6ASITE032964 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810651-42810652:- | [10] | |
| Sequence | AAACAGTACATGTTTACAATACCAAAAAAGAAAAAATCCAC | ||
| Motif Score | 2.089839286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000438274.7; ENST00000617806.4; ENST00000590764.1; ENST00000543382.6; ENST00000590099.6; ENST00000361523.8; ENST00000588276.5 | ||
| External Link | RMBase: m6A_site_364445 | ||
| mod ID: M6ASITE032965 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810669-42810670:- | [10] | |
| Sequence | TTAAATTCGGGTAATATTAAACAGTACATGTTTACAATACC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000438274.7; ENST00000590764.1; ENST00000617806.4; ENST00000590099.6; ENST00000588276.5; ENST00000543382.6; ENST00000361523.8; ENST00000589663.5 | ||
| External Link | RMBase: m6A_site_364446 | ||
| mod ID: M6ASITE032966 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810701-42810702:- | [9] | |
| Sequence | TAATTTTGTTTTGTTTGCAAACATGTTTTAAATTAAATTCG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000617806.4; ENST00000588276.5; ENST00000438274.7; ENST00000590099.6; ENST00000590764.1; ENST00000361523.8; ENST00000589663.5; ENST00000543382.6 | ||
| External Link | RMBase: m6A_site_364447 | ||
| mod ID: M6ASITE032967 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810759-42810760:- | [10] | |
| Sequence | TCACAATTTTATAACAAATGACTTTTTTCCTTAGGGGGAGG | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000590764.1; ENST00000587880.1; ENST00000361523.8; ENST00000617806.4; ENST00000438274.7; ENST00000588276.5; ENST00000590099.6; ENST00000543382.6; ENST00000589663.5 | ||
| External Link | RMBase: m6A_site_364448 | ||
| mod ID: M6ASITE032968 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810766-42810767:- | [10] | |
| Sequence | GGTGTCCTCACAATTTTATAACAAATGACTTTTTTCCTTAG | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000543382.6; ENST00000617806.4; ENST00000587880.1; ENST00000590099.6; ENST00000588276.5; ENST00000438274.7; ENST00000589663.5; ENST00000590764.1; ENST00000361523.8 | ||
| External Link | RMBase: m6A_site_364449 | ||
| mod ID: M6ASITE032969 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810777-42810778:- | [13] | |
| Sequence | GGTCTTGCTTGGGTGTCCTCACAATTTTATAACAAATGACT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | HEK293; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000587880.1; ENST00000590764.1; ENST00000617806.4; ENST00000588276.5; ENST00000590099.6; ENST00000543382.6; ENST00000589663.5; ENST00000361523.8; ENST00000438274.7 | ||
| External Link | RMBase: m6A_site_364450 | ||
| mod ID: M6ASITE032970 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810812-42810813:- | [10] | |
| Sequence | AAGCTCTCAAGTTCATGCTGACGAATCTTAAGTGGGGTCTT | ||
| Motif Score | 2.833690476 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000590185.1; ENST00000590764.1; ENST00000589663.5; ENST00000617806.4; ENST00000438274.7; ENST00000590099.6; ENST00000543382.6; ENST00000588276.5; ENST00000587880.1; ENST00000361523.8 | ||
| External Link | RMBase: m6A_site_364451 | ||
| mod ID: M6ASITE032971 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810836-42810837:- | [10] | |
| Sequence | TTAACTCTGAGGAGCAGTGGACAAAAGCTCTCAAGTTCATG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000590185.1; ENST00000361523.8; ENST00000590764.1; ENST00000587880.1; ENST00000438274.7; ENST00000590099.6; ENST00000617806.4; ENST00000543382.6; ENST00000588276.5; ENST00000589663.5 | ||
| External Link | RMBase: m6A_site_364452 | ||
| mod ID: M6ASITE032972 | Click to Show/Hide the Full List | ||
| mod site | chr17:42810898-42810899:- | [14] | |
| Sequence | GGAGAAAGGCAAGATTGAAGACACAGGAGGCAGTGGCGGCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | MT4 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000590764.1; ENST00000587880.1; ENST00000589663.5; ENST00000617806.4; ENST00000586589.5; ENST00000590185.1; ENST00000438274.7; ENST00000361523.8; ENST00000588276.5; ENST00000543382.6; ENST00000590099.6 | ||
| External Link | RMBase: m6A_site_364453 | ||
| mod ID: M6ASITE032973 | Click to Show/Hide the Full List | ||
| mod site | chr17:42811676-42811677:- | [9] | |
| Sequence | AAGAGGTTGAGAAAGGCGAGACACGTTTTTGTCTTCCCTAC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T; MT4 | ||
| Seq Type List | MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000589663.5; ENST00000617806.4; ENST00000586589.5; ENST00000361523.8; ENST00000588276.5; ENST00000587880.1; ENST00000590099.6; ENST00000438274.7; ENST00000590185.1; ENST00000590764.1; ENST00000543382.6 | ||
| External Link | RMBase: m6A_site_364458 | ||
| mod ID: M6ASITE032974 | Click to Show/Hide the Full List | ||
| mod site | chr17:42811717-42811718:- | [14] | |
| Sequence | TGCAATGGTGGCTTTCCTGGACTGTGTGCAGCAGTTCAAAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | MT4 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000543382.6; ENST00000588276.5; ENST00000590099.6; ENST00000587880.1; ENST00000361523.8; ENST00000590764.1; ENST00000590185.1; ENST00000617806.4; ENST00000589663.5; ENST00000438274.7; ENST00000586589.5 | ||
| External Link | RMBase: m6A_site_364459 | ||
| mod ID: M6ASITE032975 | Click to Show/Hide the Full List | ||
| mod site | chr17:42811750-42811751:- | [9] | |
| Sequence | GTTGCGGTTTTTCTGGGACAACAAGTTTGACCATGCAATGG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000587880.1; ENST00000586754.2; ENST00000590099.6; ENST00000589663.5; ENST00000586589.5; ENST00000590764.1; ENST00000438274.7; ENST00000590185.1; ENST00000543382.6; ENST00000588276.5; ENST00000617806.4; ENST00000361523.8 | ||
| External Link | RMBase: m6A_site_364460 | ||
| mod ID: M6ASITE032976 | Click to Show/Hide the Full List | ||
| mod site | chr17:42811753-42811754:- | [9] | |
| Sequence | GGGGTTGCGGTTTTTCTGGGACAACAAGTTTGACCATGCAA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000617806.4; ENST00000589663.5; ENST00000587880.1; ENST00000590185.1; ENST00000543382.6; ENST00000586754.2; ENST00000590764.1; ENST00000586589.5; ENST00000438274.7; ENST00000588276.5; ENST00000361523.8; ENST00000590099.6 | ||
| External Link | RMBase: m6A_site_364461 | ||
| mod ID: M6ASITE032977 | Click to Show/Hide the Full List | ||
| mod site | chr17:42813957-42813958:- | [9] | |
| Sequence | ATATCTGGAGTCTCTGACAGACAAATCTAAGGTACTCTGTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000590099.6; ENST00000589663.5; ENST00000586754.2; ENST00000617806.4; ENST00000589492.1; ENST00000590764.1; ENST00000588276.5; ENST00000586589.5; ENST00000543382.6; ENST00000361523.8; ENST00000587880.1; ENST00000438274.7 | ||
| External Link | RMBase: m6A_site_364462 | ||
| mod ID: M6ASITE032978 | Click to Show/Hide the Full List | ||
| mod site | chr17:42813990-42813991:- | [10] | |
| Sequence | TAGATACCGACTTGTTCCTTACGGAAACCATTCATATCTGG | ||
| Motif Score | 2.046785714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000588276.5; ENST00000361523.8; ENST00000589663.5; ENST00000590099.6; ENST00000543382.6; ENST00000590764.1; ENST00000589492.1; ENST00000586754.2; ENST00000438274.7; ENST00000617806.4; ENST00000586589.5 | ||
| External Link | RMBase: m6A_site_364463 | ||
| mod ID: M6ASITE032979 | Click to Show/Hide the Full List | ||
| mod site | chr17:42814653-42814654:- | [9] | |
| Sequence | GGCACAGTGGACAGTTTGGCACAATCAATAACTTCAGGCTG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000593205.5; ENST00000586589.5; ENST00000588276.5; ENST00000590852.1; ENST00000543382.6; ENST00000586754.2; ENST00000590099.6; ENST00000589663.5; ENST00000589492.1; ENST00000438274.7; ENST00000617806.4; ENST00000361523.8 | ||
| External Link | RMBase: m6A_site_364464 | ||
| mod ID: M6ASITE032980 | Click to Show/Hide the Full List | ||
| mod site | chr17:42815913-42815914:- | [13] | |
| Sequence | CGTCTTTAATGCAACCTTCCACATCTGGTAATGAGAGCTAG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | brain; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000590099.6; ENST00000593205.5; ENST00000586754.2; ENST00000586589.5; ENST00000438274.7; ENST00000543382.6; ENST00000590852.1; ENST00000589663.5; ENST00000361523.8; ENST00000589493.1; ENST00000617806.4 | ||
| External Link | RMBase: m6A_site_364465 | ||
| mod ID: M6ASITE032981 | Click to Show/Hide the Full List | ||
| mod site | chr17:42815938-42815939:- | [11] | |
| Sequence | AGCTGGATAAGCTGAAGAAAACCAACGTCTTTAATGCAACC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000590099.6; ENST00000543382.6; ENST00000593205.5; ENST00000617806.4; ENST00000586589.5; ENST00000590852.1; ENST00000361523.8; ENST00000586754.2; ENST00000438274.7; ENST00000589493.1; ENST00000589663.5 | ||
| External Link | RMBase: m6A_site_364466 | ||
| mod ID: M6ASITE032982 | Click to Show/Hide the Full List | ||
| mod site | chr17:42815982-42815983:- | [11] | |
| Sequence | TGAGCTGAAGAGTGTTGAAAACCAGATGCGTTATGCCCAGA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; MM6; Jurkat; CD4T; peripheral-blood; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361523.8; ENST00000589663.5; ENST00000586589.5; ENST00000590099.6; ENST00000617806.4; ENST00000586754.2; ENST00000543382.6; ENST00000589493.1; ENST00000438274.7; ENST00000593205.5 | ||
| External Link | RMBase: m6A_site_364467 | ||
| mod ID: M6ASITE032983 | Click to Show/Hide the Full List | ||
| mod site | chr17:42818240-42818241:- | [11] | |
| Sequence | GTCCAGGCTGAGGCTGAGAGACTGGATCAGGAGGAAGCTCA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; hNPCs; LCLs; MM6; Jurkat; CD4T; peripheral-blood; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000617806.4; ENST00000590099.6; ENST00000361523.8; ENST00000593205.5; ENST00000543382.6; ENST00000438274.7; ENST00000589663.5; ENST00000589493.1; ENST00000586589.5 | ||
| External Link | RMBase: m6A_site_364468 | ||
| mod ID: M6ASITE032984 | Click to Show/Hide the Full List | ||
| mod site | chr17:42818292-42818293:- | [11] | |
| Sequence | GCTGGAAGACGTGGAAAAGAACCGCAAGATAGTGGCAGAAA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; hNPCs; A549; MM6; Jurkat; CD4T; peripheral-blood; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000617806.4; ENST00000590099.6; ENST00000361523.8; ENST00000586589.5; ENST00000589663.5; ENST00000438274.7; ENST00000543382.6; ENST00000593205.5; ENST00000589493.1 | ||
| External Link | RMBase: m6A_site_364469 | ||
| mod ID: M6ASITE032985 | Click to Show/Hide the Full List | ||
| mod site | chr17:42818369-42818370:- | [11] | |
| Sequence | ATGAATGAGGATGACAGTGAACAGTTACAGATGGAGCTAAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; hNPCs; HEK293T; A549; Jurkat; CD4T; peripheral-blood; endometrial; HEC-1-A; NB4; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000617806.4; ENST00000590099.6; ENST00000586589.5; ENST00000593205.5; ENST00000438274.7; ENST00000589663.5; ENST00000589493.1; ENST00000543382.6; ENST00000361523.8 | ||
| External Link | RMBase: m6A_site_364470 | ||
| mod ID: M6ASITE032986 | Click to Show/Hide the Full List | ||
| mod site | chr17:42818546-42818547:- | [9] | |
| Sequence | TGAAAATGAGTGTCAGAACTACAAGTGAGTGTCTCCAGAGC | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000590099.6; ENST00000617806.4; ENST00000438274.7; ENST00000361523.8; ENST00000593205.5; ENST00000589493.1; ENST00000589663.5; ENST00000586589.5; ENST00000543382.6 | ||
| External Link | RMBase: m6A_site_364471 | ||
| mod ID: M6ASITE032987 | Click to Show/Hide the Full List | ||
| mod site | chr17:42818549-42818550:- | [15] | |
| Sequence | CACTGAAAATGAGTGTCAGAACTACAAGTGAGTGTCTCCAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HepG2; hNPCs; Jurkat; CD4T; peripheral-blood; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000589663.5; ENST00000589493.1; ENST00000617806.4; ENST00000438274.7; ENST00000543382.6; ENST00000361523.8; ENST00000586589.5; ENST00000590099.6; ENST00000593205.5 | ||
| External Link | RMBase: m6A_site_364472 | ||
| mod ID: M6ASITE032988 | Click to Show/Hide the Full List | ||
| mod site | chr17:42818585-42818586:- | [15] | |
| Sequence | TACTCTTTTAGACCAGCTGGACACTCAGCTCAACGTCACTG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HepG2; Jurkat; CD4T; peripheral-blood; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000543382.6; ENST00000593205.5; ENST00000589493.1; ENST00000438274.7; ENST00000361523.8; ENST00000586589.5; ENST00000590099.6; ENST00000617806.4 | ||
| External Link | RMBase: m6A_site_364473 | ||
| mod ID: M6ASITE032989 | Click to Show/Hide the Full List | ||
| mod site | chr17:42818594-42818595:- | [15] | |
| Sequence | ATGCACAGATACTCTTTTAGACCAGCTGGACACTCAGCTCA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HepG2; Jurkat; CD4T; peripheral-blood; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000438274.7; ENST00000593205.5; ENST00000589493.1; ENST00000543382.6; ENST00000586589.5; ENST00000590099.6; ENST00000617806.4; ENST00000361523.8 | ||
| External Link | RMBase: m6A_site_364474 | ||
| mod ID: M6ASITE032990 | Click to Show/Hide the Full List | ||
| mod site | chr17:42818643-42818644:- | [16] | |
| Sequence | TTGACATCATGTCGGGCCAGACAGATGTGGATCACCCACTC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HepG2; HeLa; Jurkat; CD4T; peripheral-blood; HEK293T; endometrial; HEC-1-A; NB4; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000617806.4; ENST00000361523.8; ENST00000593205.5; ENST00000590099.6; ENST00000543382.6; ENST00000589493.1; ENST00000586589.5; ENST00000438274.7 | ||
| External Link | RMBase: m6A_site_364475 | ||
| mod ID: M6ASITE032991 | Click to Show/Hide the Full List | ||
| mod site | chr17:42818669-42818670:- | [11] | |
| Sequence | ACACTTCCAGGTCACTGGGGACCTTTTTGACATCATGTCGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; Jurkat; CD4T; peripheral-blood; HEK293T; endometrial; HEC-1-A; NB4; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361523.8; ENST00000543382.6; ENST00000617806.4; ENST00000438274.7; ENST00000589493.1; ENST00000591085.1; ENST00000593205.5; ENST00000590099.6 | ||
| External Link | RMBase: m6A_site_364476 | ||
| mod ID: M6ASITE032992 | Click to Show/Hide the Full List | ||
| mod site | chr17:42818792-42818793:- | [11] | |
| Sequence | ATGGAGAACCTCAGCCGAAGACTGAAGGCAAGTCGGCCCCA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361523.8; ENST00000617806.4; ENST00000590099.6; ENST00000438274.7; ENST00000543382.6; ENST00000591085.1; ENST00000593205.5 | ||
| External Link | RMBase: m6A_site_364477 | ||
| mod ID: M6ASITE032993 | Click to Show/Hide the Full List | ||
| mod site | chr17:42818805-42818806:- | [16] | |
| Sequence | TGATGGCGGCACCATGGAGAACCTCAGCCGAAGACTGAAGG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HepG2; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000590099.6; ENST00000438274.7; ENST00000617806.4; ENST00000361523.8; ENST00000591085.1; ENST00000543382.6; ENST00000593205.5 | ||
| External Link | RMBase: m6A_site_364478 | ||
| mod ID: M6ASITE032994 | Click to Show/Hide the Full List | ||
| mod site | chr17:42819138-42819139:- | [10] | |
| Sequence | CTGGTTCTCCCTTCTTTTCCACTGTAGACATATGAATTTTT | ||
| Motif Score | 2.475107143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000438274.7; ENST00000617806.4; ENST00000591085.1; ENST00000593205.5; ENST00000590099.6; ENST00000543382.6; ENST00000361523.8 | ||
| External Link | RMBase: m6A_site_364479 | ||
| mod ID: M6ASITE032995 | Click to Show/Hide the Full List | ||
| mod site | chr17:42819593-42819594:- | [14] | |
| Sequence | CCCAGGAGCCATTTATTGAAACTCCTCGCCAGGATGGTGTC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | MT4 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000593205.5; ENST00000590099.6; ENST00000617806.4; ENST00000585515.5; ENST00000591085.1; ENST00000438274.7; ENST00000361523.8; ENST00000543382.6 | ||
| External Link | RMBase: m6A_site_364480 | ||
| mod ID: M6ASITE032996 | Click to Show/Hide the Full List | ||
| mod site | chr17:42820372-42820373:- | [14] | |
| Sequence | TCAAAAGCATCCTGGTTAGGACAGTGAATTGTATGATTGCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | MT4 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000590099.6; ENST00000591307.5; ENST00000591085.1; ENST00000543382.6; ENST00000617806.4; ENST00000593205.5; ENST00000438274.7; ENST00000361523.8; ENST00000585515.5; rmsk_4695860 | ||
| External Link | RMBase: m6A_site_364481 | ||
| mod ID: M6ASITE032997 | Click to Show/Hide the Full List | ||
| mod site | chr17:42820787-42820788:- | [11] | |
| Sequence | GAGAGACCCAGGAGGAAGAGACTAACTCAGGAGAGGTAATA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; MT4; Huh7; peripheral-blood | ||
| Seq Type List | m6A-seq; DART-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000617806.4; ENST00000361523.8; ENST00000590099.6; ENST00000591085.1; ENST00000543382.6; ENST00000591307.5; ENST00000593112.1; ENST00000585515.5; ENST00000593205.5; ENST00000438274.7 | ||
| External Link | RMBase: m6A_site_364482 | ||
| mod ID: M6ASITE032998 | Click to Show/Hide the Full List | ||
| mod site | chr17:42820802-42820803:- | [11] | |
| Sequence | CCCAGGCGAAACCAGGAGAGACCCAGGAGGAAGAGACTAAC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; MT4; Huh7; peripheral-blood | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000593205.5; ENST00000591085.1; ENST00000590099.6; ENST00000438274.7; ENST00000585515.5; ENST00000593112.1; ENST00000591307.5; ENST00000617806.4; ENST00000589636.5; ENST00000543382.6; ENST00000361523.8 | ||
| External Link | RMBase: m6A_site_364483 | ||
| mod ID: M6ASITE032999 | Click to Show/Hide the Full List | ||
| mod site | chr17:42820812-42820813:- | [11] | |
| Sequence | CTTACCACAGCCCAGGCGAAACCAGGAGAGACCCAGGAGGA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; MT4; Huh7; peripheral-blood | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000591307.5; ENST00000589636.5; ENST00000591085.1; ENST00000438274.7; ENST00000593112.1; ENST00000617806.4; ENST00000543382.6; ENST00000590099.6; ENST00000593205.5; ENST00000585515.5; ENST00000361523.8 | ||
| External Link | RMBase: m6A_site_364484 | ||
| mod ID: M6ASITE033000 | Click to Show/Hide the Full List | ||
| mod site | chr17:42823737-42823738:- | [15] | |
| Sequence | GGAACTCACAGGTCAGCGAGACCCTTGGAAAGAGGGTGGTC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HepG2; MT4; Huh7; peripheral-blood; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000543382.6; ENST00000617806.4; ENST00000593112.1; ENST00000593205.5; ENST00000591085.1; ENST00000591307.5; ENST00000438274.7; ENST00000361523.8; ENST00000585515.5; ENST00000590099.6; ENST00000589636.5 | ||
| External Link | RMBase: m6A_site_364485 | ||
| mod ID: M6ASITE033001 | Click to Show/Hide the Full List | ||
| mod site | chr17:42823754-42823755:- | [15] | |
| Sequence | GACCGTGTCACCATCCAGGAACTCACAGGTCAGCGAGACCC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HepG2; MT4; Huh7; peripheral-blood; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000617806.4; ENST00000361523.8; ENST00000543382.6; ENST00000593205.5; ENST00000589636.5; ENST00000438274.7; ENST00000590099.6; ENST00000593112.1; ENST00000591307.5; ENST00000585515.5; ENST00000591085.1 | ||
| External Link | RMBase: m6A_site_364486 | ||
| mod ID: M6ASITE033002 | Click to Show/Hide the Full List | ||
| mod site | chr17:42823773-42823774:- | [15] | |
| Sequence | CACGAGTTTCAAGATCCTGGACCGTGTCACCATCCAGGAAC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2; MT4; Huh7; peripheral-blood; HEK293T; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000591307.5; ENST00000589636.5; ENST00000543382.6; ENST00000591085.1; ENST00000593112.1; ENST00000590099.6; ENST00000617806.4; ENST00000593205.5; ENST00000361523.8; ENST00000438274.7; ENST00000585515.5 | ||
| External Link | RMBase: m6A_site_364487 | ||
| mod ID: M6ASITE033003 | Click to Show/Hide the Full List | ||
| mod site | chr17:42823794-42823795:- | [16] | |
| Sequence | CAGCCAGCCCCTGAAACTGGACACGAGTTTCAAGATCCTGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HepG2; HeLa; A549; hESC-HEK293T; peripheral-blood; GSC-11; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000438274.7; ENST00000589636.5; ENST00000590099.6; ENST00000543382.6; ENST00000591307.5; ENST00000593112.1; ENST00000591085.1; ENST00000617806.4; ENST00000585515.5; ENST00000593205.5; ENST00000361523.8 | ||
| External Link | RMBase: m6A_site_364488 | ||
| mod ID: M6ASITE033004 | Click to Show/Hide the Full List | ||
| mod site | chr17:42823799-42823800:- | [16] | |
| Sequence | CGCTGCAGCCAGCCCCTGAAACTGGACACGAGTTTCAAGAT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HepG2; HeLa; A549; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000438274.7; ENST00000590099.6; ENST00000617806.4; ENST00000543382.6; ENST00000593112.1; ENST00000589636.5; ENST00000361523.8; ENST00000585515.5; ENST00000591085.1; ENST00000593205.5; ENST00000591307.5 | ||
| External Link | RMBase: m6A_site_364489 | ||
| mod ID: M6ASITE033005 | Click to Show/Hide the Full List | ||
| mod site | chr17:42823854-42823855:- | [9] | |
| Sequence | GGAAGGGTCTAAGACGTCCAACAACAGCACCATGCAGGTGA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000590099.6; ENST00000593205.5; ENST00000589636.5; ENST00000591085.1; ENST00000361523.8; ENST00000593112.1; ENST00000543382.6; ENST00000612631.1; ENST00000591307.5; ENST00000438274.7; ENST00000585515.5; ENST00000617806.4 | ||
| External Link | RMBase: m6A_site_364490 | ||
| mod ID: M6ASITE033006 | Click to Show/Hide the Full List | ||
| mod site | chr17:42824182-42824183:- | [11] | |
| Sequence | GTGCCTCCGCGGTAGACCGGACTTGGGTGACGGGCTCCGGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; A549; peripheral-blood; GSC-11; HEK293T; HEK293A-TOA; MSC; TIME; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361523.8; ENST00000591085.1; ENST00000589636.5; ENST00000593112.1; ENST00000590099.6; ENST00000438274.7; ENST00000617806.4; ENST00000593205.5; ENST00000543382.6; ENST00000591307.5; ENST00000612631.1; ENST00000585515.5 | ||
| External Link | RMBase: m6A_site_364491 | ||
| mod ID: M6ASITE033007 | Click to Show/Hide the Full List | ||
| mod site | chr17:42824187-42824188:- | [11] | |
| Sequence | CCACAGTGCCTCCGCGGTAGACCGGACTTGGGTGACGGGCT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; A549; peripheral-blood; GSC-11; HEK293T; HEK293A-TOA; MSC; TIME; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000543382.6; ENST00000590099.6; ENST00000591085.1; ENST00000593205.5; ENST00000593112.1; ENST00000589636.5; ENST00000591307.5; ENST00000438274.7; ENST00000585515.5; ENST00000617806.4; ENST00000361523.8; ENST00000612631.1 | ||
| External Link | RMBase: m6A_site_364492 | ||
| mod ID: M6ASITE033008 | Click to Show/Hide the Full List | ||
| mod site | chr17:42824255-42824256:- | [11] | |
| Sequence | CTACCGGGAAGTCGCTGAAGACAGAGCGATGGTAGTTCTGG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; HEK293A-TOA; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000617806.4; ENST00000438274.7; ENST00000590099.6; ENST00000612631.1; ENST00000543382.6; ENST00000591085.1; ENST00000593112.1; ENST00000589636.5; ENST00000585515.5; ENST00000361523.8 | ||
| External Link | RMBase: m6A_site_364493 | ||
| mod ID: M6ASITE033009 | Click to Show/Hide the Full List | ||
| mod site | chr17:42824337-42824338:- | [11] | |
| Sequence | CCGGGCCTGTGAGCCTGTGGACCAGGAGCTCCTGCTGCCGT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000612631.1; ENST00000589636.5 | ||
| External Link | RMBase: m6A_site_364494 | ||
| mod ID: M6ASITE033010 | Click to Show/Hide the Full List | ||
| mod site | chr17:42833217-42833218:- | [11] | |
| Sequence | GAGAACTGAGCCTCCGAGAGACTCGGAAACGGTGCGGCCCT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000589636.5; ENST00000612631.1 | ||
| External Link | RMBase: m6A_site_364495 | ||
| mod ID: M6ASITE033011 | Click to Show/Hide the Full List | ||
| mod site | chr17:42833233-42833234:- | [11] | |
| Sequence | TGGGGAATGGTGCGCTGAGAACTGAGCCTCCGAGAGACTCG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000589636.5; ENST00000612631.1 | ||
| External Link | RMBase: m6A_site_364496 | ||
References