m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00157)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
LINC00470
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
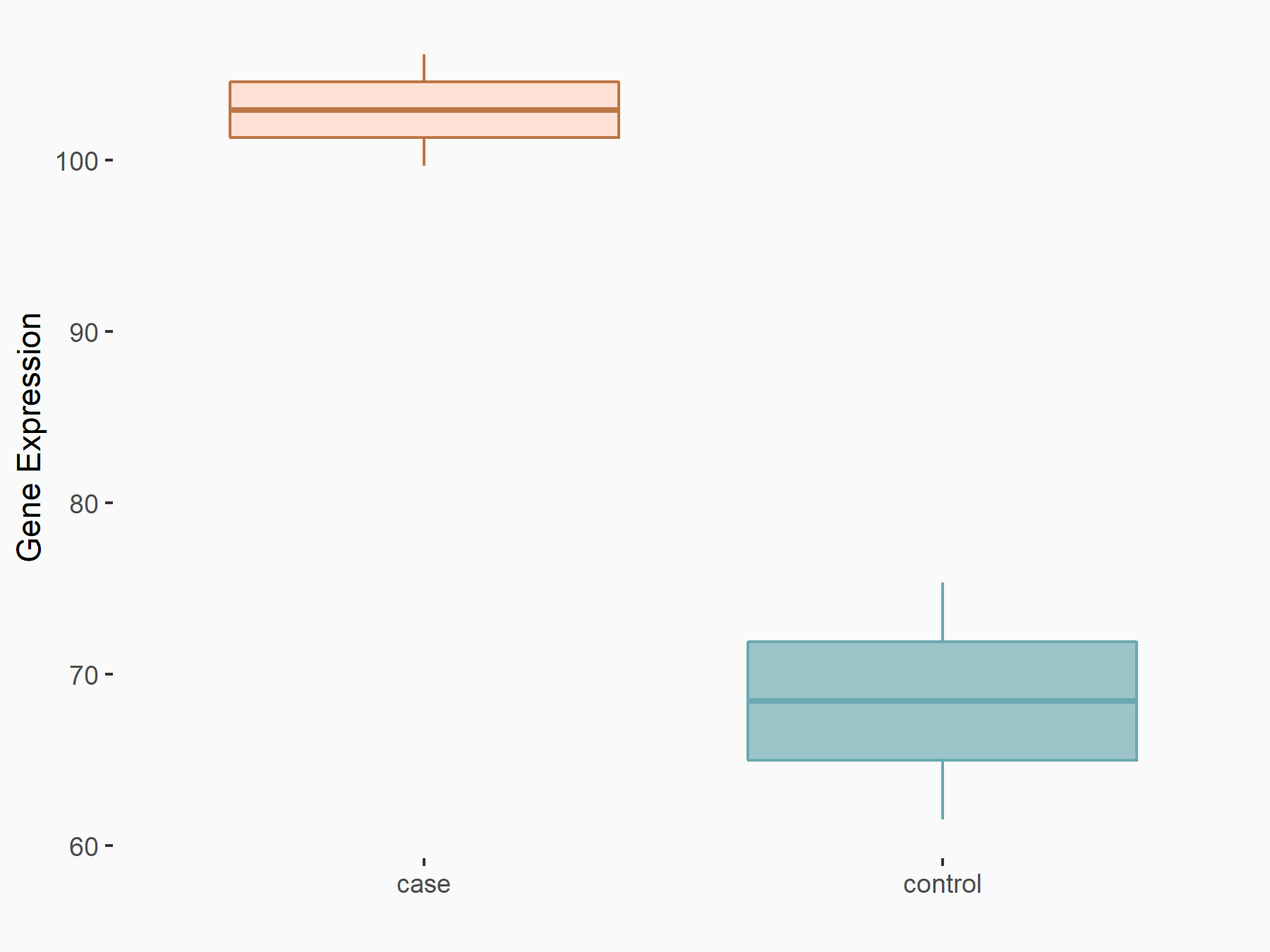  |
logFC: 5.89E-01 p-value: 3.48E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | The molecular mechanism underlying the effect of Long intergenic non-protein coding RNA 470 (LINC00470) on chronic myelocytic leukaemia by reducing the PTEN stability via RNA methyltransferase METTL3, thus leading to the inhibition of cell autophagy while promoting chemoresistance in CML. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Chronic myeloid leukaemia | ICD-11: 2B33.2 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | RNA stability | |||
| Cell autophagy | ||||
| In-vitro Model | K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 |
| KCL-22 | Chronic myelogenous leukemia | Homo sapiens | CVCL_2091 | |
| In-vivo Model | In the control mice or ADR mice group, the parental or chemo-resistant K562 cells were infected with LV-shCtrl. In the ADR + shLINC00470 group, the chemo-resistant K562 cells were infected with LV- shLINC00470. These cells were injected, respectively, into these 5-week-old mice subcutaneously. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | Long intergenic non-protein coding RNA 470 (LINC00470)-METTL3-mediated PTEN mRNA degradation relied on the m6A reader protein YTHDF2-dependent pathway.LINC00470 served as a therapeutic target for Gastric cancer patients. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Pathway Response | Gastric cancer | hsa05226 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
YTH domain-containing family protein 2 (YTHDF2) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | Long intergenic non-protein coding RNA 470 (LINC00470)-METTL3-mediated PTEN mRNA degradation relied on the m6A reader protein YTHDF2-dependent pathway. LINC00470 served as a therapeutic target for Gastric cancer patients. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Pathway Response | Gastric cancer | hsa05226 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
Malignant haematopoietic neoplasm [ICD-11: 2B33]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | The molecular mechanism underlying the effect of Long intergenic non-protein coding RNA 470 (LINC00470) on chronic myelocytic leukaemia by reducing the PTEN stability via RNA methyltransferase METTL3, thus leading to the inhibition of cell autophagy while promoting chemoresistance in CML. | |||
| Responsed Disease | Chronic myeloid leukaemia [ICD-11: 2B33.2] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | RNA stability | |||
| Cell autophagy | ||||
| In-vitro Model | K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 |
| KCL-22 | Chronic myelogenous leukemia | Homo sapiens | CVCL_2091 | |
| In-vivo Model | In the control mice or ADR mice group, the parental or chemo-resistant K562 cells were infected with LV-shCtrl. In the ADR + shLINC00470 group, the chemo-resistant K562 cells were infected with LV- shLINC00470. These cells were injected, respectively, into these 5-week-old mice subcutaneously. | |||
Gastric cancer [ICD-11: 2B72]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | Long intergenic non-protein coding RNA 470 (LINC00470)-METTL3-mediated PTEN mRNA degradation relied on the m6A reader protein YTHDF2-dependent pathway.LINC00470 served as a therapeutic target for Gastric cancer patients. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Gastric cancer | hsa05226 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| Experiment 2 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | Long intergenic non-protein coding RNA 470 (LINC00470)-METTL3-mediated PTEN mRNA degradation relied on the m6A reader protein YTHDF2-dependent pathway. LINC00470 served as a therapeutic target for Gastric cancer patients. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Gastric cancer | hsa05226 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03647 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
Non-coding RNA
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05401 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 470 (LINC00470) | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Gastric cancer | |
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05402 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 470 (LINC00470) | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Gastric cancer | |
| Crosstalk ID: M6ACROT05487 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 470 (LINC00470) | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Chronic myeloid leukaemia | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00157)
| In total 15 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE036613 | Click to Show/Hide the Full List | ||
| mod site | chr18:1362553-1362554:- | [3] | |
| Sequence | TCGGCCTGTTCAACAGCAGGACCTGCATCGGGCAATTGAAA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | TREX | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000581430.5; ENST00000412816.7; ENST00000579735.1 | ||
| External Link | RMBase: m6A_site_392124 | ||
| mod ID: M6ASITE036614 | Click to Show/Hide the Full List | ||
| mod site | chr18:1362662-1362663:- | [3] | |
| Sequence | TGATGCGTTTTCAGGAAGGGACCTAAATGCGATGTGTTGAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | TREX | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000579735.1; ENST00000581430.5; ENST00000412816.7 | ||
| External Link | RMBase: m6A_site_392125 | ||
| mod ID: M6ASITE036615 | Click to Show/Hide the Full List | ||
| mod site | chr18:1362684-1362685:- | [3] | |
| Sequence | TGCTAGAAGTTGCCCAGGAAACTGATGCGTTTTCAGGAAGG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | TREX | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000412816.7; ENST00000579735.1; ENST00000581430.5 | ||
| External Link | RMBase: m6A_site_392126 | ||
| mod ID: M6ASITE036616 | Click to Show/Hide the Full List | ||
| mod site | chr18:1362707-1362708:- | [3] | |
| Sequence | AAATGTGGATAGGCATGTAGACCTGCTAGAAGTTGCCCAGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | TREX | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000579735.1; ENST00000412816.7; ENST00000581430.5 | ||
| External Link | RMBase: m6A_site_392127 | ||
| mod ID: M6ASITE036617 | Click to Show/Hide the Full List | ||
| mod site | chr18:1362745-1362746:- | [4] | |
| Sequence | CAGAAAGAAGCAATCCTGAAACTCATCTTGAAAAATGAAAA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; U2OS; TREX | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000579735.1; ENST00000581430.5; ENST00000412816.7 | ||
| External Link | RMBase: m6A_site_392128 | ||
| mod ID: M6ASITE036618 | Click to Show/Hide the Full List | ||
| mod site | chr18:1362766-1362767:- | [4] | |
| Sequence | ATCAACCAGCCTGCTTTAAAACAGAAAGAAGCAATCCTGAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; U2OS; TREX | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000579735.1; ENST00000581430.5; ENST00000412816.7 | ||
| External Link | RMBase: m6A_site_392129 | ||
| mod ID: M6ASITE036619 | Click to Show/Hide the Full List | ||
| mod site | chr18:1362788-1362789:- | [5] | |
| Sequence | AAGAATTCCTACAAGATTTCACATCAACCAGCCTGCTTTAA | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000581430.5; ENST00000579735.1; ENST00000412816.7 | ||
| External Link | RMBase: m6A_site_392130 | ||
| mod ID: M6ASITE036620 | Click to Show/Hide the Full List | ||
| mod site | chr18:1362830-1362831:- | [4] | |
| Sequence | AGCTACCATTCATCCTCAGGACATTGACACGGCTATAACGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; U2OS; TREX | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000579735.1; ENST00000412816.7; ENST00000581430.5 | ||
| External Link | RMBase: m6A_site_392131 | ||
| mod ID: M6ASITE036621 | Click to Show/Hide the Full List | ||
| mod site | chr18:1362884-1362885:- | [4] | |
| Sequence | GAGTCTCTGGGATGGACTGGACACTGATCACCGCTGCCAGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; U2OS; TREX | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000579735.1; ENST00000581430.5; ENST00000412816.7 | ||
| External Link | RMBase: m6A_site_392132 | ||
| mod ID: M6ASITE036622 | Click to Show/Hide the Full List | ||
| mod site | chr18:1362889-1362890:- | [4] | |
| Sequence | TTTATGAGTCTCTGGGATGGACTGGACACTGATCACCGCTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; U2OS; TREX | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000579735.1; ENST00000412816.7; ENST00000581430.5 | ||
| External Link | RMBase: m6A_site_392133 | ||
| mod ID: M6ASITE036623 | Click to Show/Hide the Full List | ||
| mod site | chr18:1362957-1362958:- | [4] | |
| Sequence | AATAGACTCCTTTGTACGAAACCGTTCAGAGTTCTGACCAT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; U2OS; TREX | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000579735.1; ENST00000581430.5; ENST00000412816.7 | ||
| External Link | RMBase: m6A_site_392134 | ||
| mod ID: M6ASITE036624 | Click to Show/Hide the Full List | ||
| mod site | chr18:1362972-1362973:- | [4] | |
| Sequence | CATCATTATAGACGAAATAGACTCCTTTGTACGAAACCGTT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; U2OS; TREX | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000579735.1; ENST00000412816.7; ENST00000581430.5 | ||
| External Link | RMBase: m6A_site_392135 | ||
| mod ID: M6ASITE036625 | Click to Show/Hide the Full List | ||
| mod site | chr18:1362981-1362982:- | [5] | |
| Sequence | ACTGTCCATCATCATTATAGACGAAATAGACTCCTTTGTAC | ||
| Motif Score | 2.871321429 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000581430.5; ENST00000412816.7; ENST00000579735.1 | ||
| External Link | RMBase: m6A_site_392136 | ||
| mod ID: M6ASITE036626 | Click to Show/Hide the Full List | ||
| mod site | chr18:1363042-1363043:- | [6] | |
| Sequence | AGTGGTATGGAGAATCTCAAACACTGGCTGCTGCTGTCTTC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | U2OS | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000581430.5; ENST00000412816.7; ENST00000579735.1 | ||
| External Link | RMBase: m6A_site_392137 | ||
| mod ID: M6ASITE036627 | Click to Show/Hide the Full List | ||
| mod site | chr18:1363292-1363293:- | [5] | |
| Sequence | ACCCTCTTAATATGCATGTTACTTGGAAGTGATATAGCAGG | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000579735.1; ENST00000581430.5; ENST00000412816.7 | ||
| External Link | RMBase: m6A_site_392138 | ||
References