m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00356)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
TP53
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
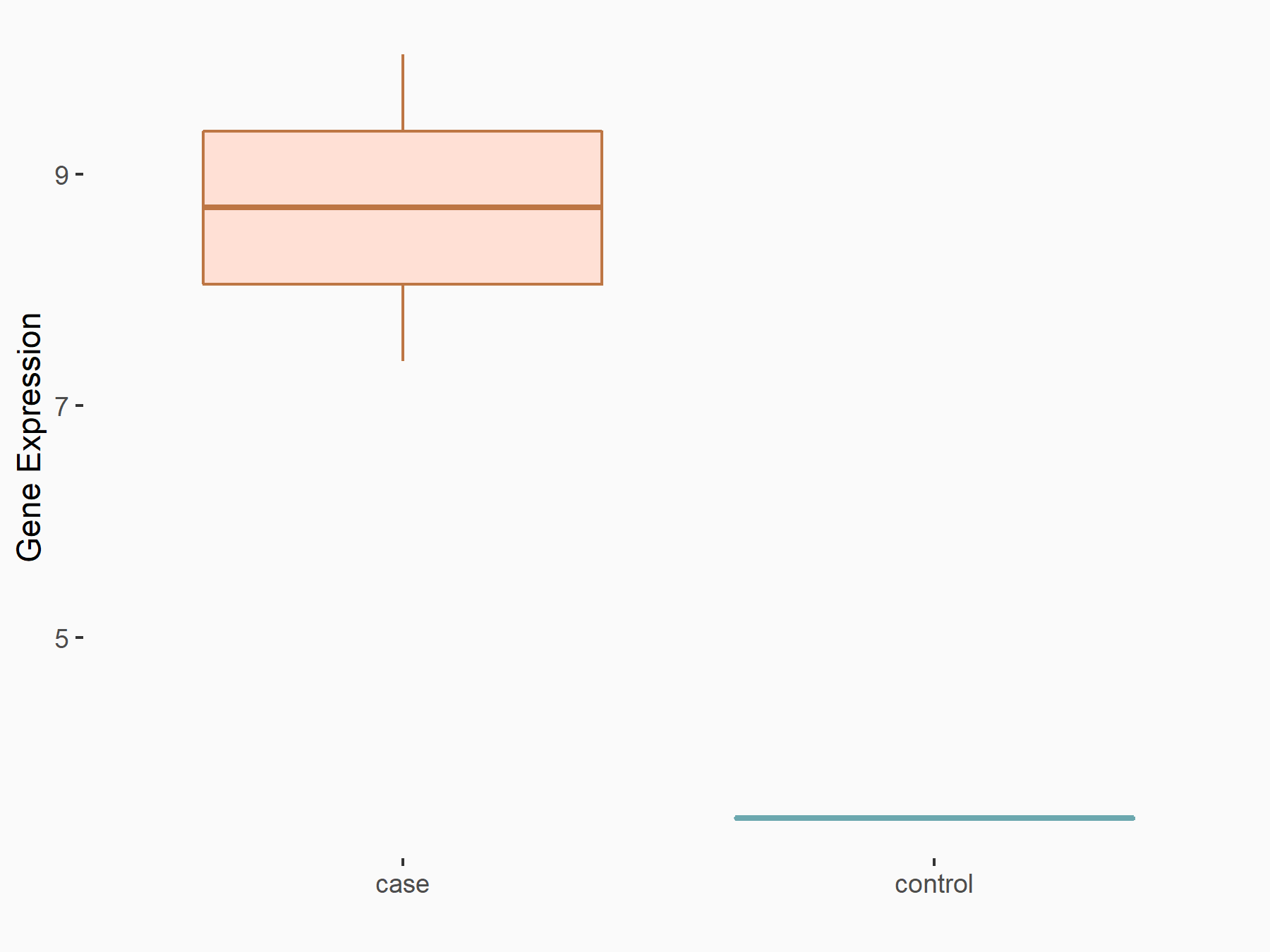  |
logFC: 1.12E+00 p-value: 1.48E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between TP53 and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.05E+00 | GSE60213 |
| In total 6 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Silencing the m(6)A methyltransferase significantly affects gene expression and alternative splicing patterns, resulting in modulation of the Cellular tumor antigen p53 (TP53/p53) (also known as TP53) signalling pathway and apoptosis. Modulation of p53 signalling through splicing is relevant to induction of apoptosis by silencing of METTL3. | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| p53 signaling pathway | hsa04115 | |||
| Cell Process | Gene expression | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 significantly decreased m6A level, restoring Cellular tumor antigen p53 (TP53/p53) activation and inhibiting cellular transformation phenotypes in the arsenite-transformed cells. m6A downregulated the expression of the positive p53 regulator, PRDM2, through the YTHDF2-promoted decay of PRDM2 mRNAs. m6A upregulated the expression of the negative p53 regulator, YY1 and MDM2 through YTHDF1-stimulated translation of YY1 and MDM2 mRNA. This study further sheds light on the mechanisms of arsenic carcinogenesis via RNA epigenetics. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Solid tumour/cancer | ICD-11: 2A00-2F9Z | ||
| Responsed Drug | Arsenite | Phase 2 | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | METTL3 and METTL14 play an oncogenic role in acute myeloid leukemia(AML) by targeting mdm2/Cellular tumor antigen p53 (TP53/p53) signal pathway. The knockdown of METTL3 and METTL14 in K562 cell line leads to several changes in the expression of p53 signal pathway, including the upregulation of p53, cyclin dependent kinase inhibitor 1A (CDKN1A/p21), and downregulation of mdm2. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Acute myeloid leukaemia | ICD-11: 2A60 | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Cell cycle | hsa04110 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell apoptosis | |||
| Cells in G3/M phase decreased | ||||
| In-vitro Model | THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| MOLT-4 | Adult T acute lymphoblastic leukemia | Homo sapiens | CVCL_0013 | |
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 | |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| CCRF-CEM C7 | T acute lymphoblastic leukemia | Homo sapiens | CVCL_6825 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| Experiment 4 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | The produced p53 R273H mutant protein resulted in acquired multidrug resistance in colon cancer cells. Either silencing METTL3 expression by using small interfering RNA (siRNA) or inhibiting RNA methylation with neplanocin A suppressed m6A formation in Cellular tumor antigen p53 (TP53/p53) pre-mRNA, and substantially increased the level of phosphorylated p53 protein (Ser15) and its function in cells heterozygously carrying the R273H mutation, thereby re-sensitizing these cells to anticancer drugs. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colon cancer | ICD-11: 2B90 | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Cell Process | Protein signaling | |||
| In-vitro Model | SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 |
| WiDr | Colon adenocarcinoma | Homo sapiens | CVCL_2760 | |
| Experiment 5 Reporting the m6A Methylation Regulator of This Target Gene | [5] | |||
| Response Summary | Cellular tumor antigen p53 (TP53/p53) n6-methyladenosine (m6A) played a decisive role in regulating Hepatocellular carcinoma(HCC) sensitivity to chemotherapy via the p53 activator RG7112 and the vascular endothelial growth factor receptor inhibitor apatinib. p53 mRNA m6A modification blockage induced by S-adenosyl homocysteine or siRNA-mediated METTL3 inhibition enhanced HCC sensitivity to chemotherapy. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Responsed Drug | Apatinib | Approved | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | QGY-7701 | Human papillomavirus-related endocervical adenocarcinoma | Homo sapiens | CVCL_6859 |
| HHL-5 | Normal | Homo sapiens | CVCL_S956 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | Nude mice (4-6 week-old) were administered sterile water and feed in a specific pathogen-free barrier. Using a 1-mL syringe, 1 × 107 HEPG2 cells were subcutaneously inoculated into the right axilla of nude mice to build the HCC xenograft model. When the tumor volume reached 50 mm3, the nude mice were randomly divided into 1 control (n = 4) and 3 treatment groups (n = 4 each). RG7112, apatinib, and RG7112 + apatinib were administered to the treatment groups and an equal volume of dimethyl sulfoxide to the control group by daily gavage for 14 d. The tumor length (L) and width (W) were measured on alternate days using vernier calipers. The following formula was used to calculate the tumor volume: volume (mm3) = 0.5 × L × W × W. At the end of the experiment, the nude mice were killed by CO2 overdose anesthesia. The tumors were dissected and weighed using a precision balance, and the tumor tissue was stored in liquid nitrogen for further analysis. | |||
| Experiment 6 Reporting the m6A Methylation Regulator of This Target Gene | [5] | |||
| Response Summary | Cellular tumor antigen p53 (TP53/p53) n6-methyladenosine (m6A) played a decisive role in regulating Hepatocellular carcinoma(HCC) sensitivity to chemotherapy via the p53 activator RG7112 and the vascular endothelial growth factor receptor inhibitor apatinib. p53 mRNA m6A modification blockage induced by S-adenosyl homocysteine or siRNA-mediated METTL3 inhibition enhanced HCC sensitivity to chemotherapy. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Responsed Drug | RG7112 | Phase 1 | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | QGY-7701 | Human papillomavirus-related endocervical adenocarcinoma | Homo sapiens | CVCL_6859 |
| HHL-5 | Normal | Homo sapiens | CVCL_S956 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | Nude mice (4-6 week-old) were administered sterile water and feed in a specific pathogen-free barrier. Using a 1-mL syringe, 1 × 107 HEPG2 cells were subcutaneously inoculated into the right axilla of nude mice to build the HCC xenograft model. When the tumor volume reached 50 mm3, the nude mice were randomly divided into 1 control (n = 4) and 3 treatment groups (n = 4 each). RG7112, apatinib, and RG7112 + apatinib were administered to the treatment groups and an equal volume of dimethyl sulfoxide to the control group by daily gavage for 14 d. The tumor length (L) and width (W) were measured on alternate days using vernier calipers. The following formula was used to calculate the tumor volume: volume (mm3) = 0.5 × L × W × W. At the end of the experiment, the nude mice were killed by CO2 overdose anesthesia. The tumors were dissected and weighed using a precision balance, and the tumor tissue was stored in liquid nitrogen for further analysis. | |||
Fat mass and obesity-associated protein (FTO) [ERASER]
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [7] | |||
| Response Summary | Meclofenamic acid increased Cellular tumor antigen p53 (TP53/p53) mRNA and protein levels in AKI both in vitro and in vivo, and FTO overexpression reduced p53 expression and reversed the MA-induced p53 increase in cisplatin-induced acute kidney injury Acute kidney injury. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Acute kidney failure | ICD-11: GB60 | ||
| Responsed Drug | Cisplatin | Approved | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | HK2 | Normal | Acipenser baerii | CVCL_YE28 |
| In-vivo Model | Induced AKI in c57BL/6 mice by intraperitoneal cisplatin injection and treated the animal with vehicle or an FTO inhibitor meclofenamic acid (MA) for 3 days. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [7] | |||
| Response Summary | Meclofenamic acid increased Cellular tumor antigen p53 (TP53/p53) mRNA and protein levels in AKI both in vitro and in vivo, and FTO overexpression reduced p53 expression and reversed the MA-induced p53 increase in cisplatin-induced Acute kidney injury. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Acute kidney failure | ICD-11: GB60 | ||
| Responsed Drug | Meclofenamic acid | Approved | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | HK2 | Normal | Acipenser baerii | CVCL_YE28 |
| In-vivo Model | Induced AKI in c57BL/6 mice by intraperitoneal cisplatin injection and treated the animal with vehicle or an FTO inhibitor meclofenamic acid (MA) for 3 days. | |||
Methyltransferase-like 14 (METTL14) [WRITER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | METTL3 and METTL14 play an oncogenic role in acute myeloid leukemia(AML) by targeting mdm2/Cellular tumor antigen p53 (TP53/p53) signal pathway. The knockdown of METTL3 and METTL14 in K562 cell line leads to several changes in the expression of p53 signal pathway, including the upregulation of p53, cyclin dependent kinase inhibitor 1A (CDKN1A/p21), and downregulation of mdm2. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Acute myeloid leukaemia | ICD-11: 2A60 | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Cell cycle | hsa04110 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell apoptosis | |||
| Cells in G6/M phase decreased | ||||
| In-vitro Model | THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| MOLT-4 | Adult T acute lymphoblastic leukemia | Homo sapiens | CVCL_0013 | |
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 | |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| CCRF-CEM C7 | T acute lymphoblastic leukemia | Homo sapiens | CVCL_6825 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Methyltransferase-like 16 (METTL16) [WRITER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [10] | |||
| Response Summary | Deletion of METTL16 or ALKBH5 predicted poor OS and DFS of hepatocellular carcinoma (HCC) patients. And this study found significant associations between the genetic alterations and clinicopathological features as well as Cellular tumor antigen p53 (TP53/p53) alteration. | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
RNA demethylase ALKBH5 (ALKBH5) [ERASER]
| In total 3 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [10] | |||
| Response Summary | Deletion of METTL16 or ALKBH5 predicted poor OS and DFS of hepatocellular carcinoma (HCC) patients. And this study found significant associations between the genetic alterations and clinicopathological features as well as Cellular tumor antigen p53 (TP53/p53) alteration. | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [14] | |||
| Response Summary | Knockdown of Cellular tumor antigen p53 (TP53/p53) or inhibition of p53's transcriptional activity by addition of its specific inhibitor PFT-Alpha decreased expression of ALKBH5 and Cancer stem cells' malignancies, the pivotal role of ALKBH5 in Cancer stem cells derived from nonsmall-cell lung cancer and highlight the regulatory function of the p53/ALKBH5 axis in modulating CSC progression. p53 transcriptionally regulates PRRX1, which is consistent with our previous report. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| In-vitro Model | PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | Cells at 1 × 106 were subcutaneously injected into the mice similarly to nude mice. Twenty-eight days later, grafted tumors were collected and morphologically analyzed. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [15] | |||
| Response Summary | ALKBH5 as another mammalian demethylase that oxidatively reverses m(6)A in mRNA in vitro and in vivo. Alkbh5-deficient male mice have increased m(6)A in mRNA and are characterized by impaired fertility resulting from apoptosis that affects meiotic metaphase-stage spermatocytes. Identified in mouse testes 1,551 differentially expressed genes that cover broad functional categories and include spermatogenesis-related mRNAs involved in the Cellular tumor antigen p53 (TP53/p53) functional interaction network. | |||
| Responsed Disease | Female reproductive system disorders | ICD-11: SC4Y | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| In-vitro Model | HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| In-vivo Model | Neo-resistant recombinant ESCs were analyzed for correct 3' and 5' targeting through hybridization by standard protocols, and we identified seven correctly targeted clones. Following ESC cell injection, several high-percentage chimera males were born. These were bred with Cre-expressing mice, and Cre-mediated excision of Alkbh5 exon 1 was tested in 26 agouti F1 pups. Two animals tested positive for the excised allele. These two mice, heterozygous for the constitutive allele (Alkbh5+/-), were analyzed by Southern blot analysis and used for further breeding to generate Alkbh5-/- mice. | |||
YTH domain-containing family protein 1 (YTHDF1) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 significantly decreased m6A level, restoring Cellular tumor antigen p53 (TP53/p53) activation and inhibiting cellular transformation phenotypes in the arsenite-transformed cells. m6A downregulated the expression of the positive p53 regulator, PRDM2, through the YTHDF2-promoted decay of PRDM2 mRNAs. m6A upregulated the expression of the negative p53 regulator, YY1 and MDM2 through YTHDF1-stimulated translation of YY1 and MDM2 mRNA. This study further sheds light on the mechanisms of arsenic carcinogenesis via RNA epigenetics. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Solid tumour/cancer | ICD-11: 2A00-2F9Z | ||
| Responsed Drug | Arsenite | Phase 2 | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
Solid tumour/cancer [ICD-11: 2A00-2F9Z]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 significantly decreased m6A level, restoring Cellular tumor antigen p53 (TP53/p53) activation and inhibiting cellular transformation phenotypes in the arsenite-transformed cells. m6A downregulated the expression of the positive p53 regulator, PRDM2, through the YTHDF2-promoted decay of PRDM2 mRNAs. m6A upregulated the expression of the negative p53 regulator, YY1 and MDM2 through YTHDF1-stimulated translation of YY1 and MDM2 mRNA. This study further sheds light on the mechanisms of arsenic carcinogenesis via RNA epigenetics. | |||
| Responsed Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Arsenite | Phase 2 | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
| Experiment 2 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 significantly decreased m6A level, restoring Cellular tumor antigen p53 (TP53/p53) activation and inhibiting cellular transformation phenotypes in the arsenite-transformed cells. m6A downregulated the expression of the positive p53 regulator, PRDM2, through the YTHDF2-promoted decay of PRDM2 mRNAs. m6A upregulated the expression of the negative p53 regulator, YY1 and MDM2 through YTHDF1-stimulated translation of YY1 and MDM2 mRNA. This study further sheds light on the mechanisms of arsenic carcinogenesis via RNA epigenetics. | |||
| Responsed Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Arsenite | Phase 2 | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | METTL3 and METTL14 play an oncogenic role in acute myeloid leukemia(AML) by targeting mdm2/Cellular tumor antigen p53 (TP53/p53) signal pathway. The knockdown of METTL3 and METTL14 in K562 cell line leads to several changes in the expression of p53 signal pathway, including the upregulation of p53, cyclin dependent kinase inhibitor 1A (CDKN1A/p21), and downregulation of mdm2. | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Cell cycle | hsa04110 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell apoptosis | |||
| Cells in G6/M phase decreased | ||||
| In-vitro Model | THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| MOLT-4 | Adult T acute lymphoblastic leukemia | Homo sapiens | CVCL_0013 | |
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 | |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| CCRF-CEM C7 | T acute lymphoblastic leukemia | Homo sapiens | CVCL_6825 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| Experiment 2 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | METTL3 and METTL14 play an oncogenic role in acute myeloid leukemia(AML) by targeting mdm2/Cellular tumor antigen p53 (TP53/p53) signal pathway. The knockdown of METTL3 and METTL14 in K562 cell line leads to several changes in the expression of p53 signal pathway, including the upregulation of p53, cyclin dependent kinase inhibitor 1A (CDKN1A/p21), and downregulation of mdm2. | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Cell cycle | hsa04110 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell apoptosis | |||
| Cells in G3/M phase decreased | ||||
| In-vitro Model | THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| MOLT-4 | Adult T acute lymphoblastic leukemia | Homo sapiens | CVCL_0013 | |
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 | |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| CCRF-CEM C7 | T acute lymphoblastic leukemia | Homo sapiens | CVCL_6825 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Colon cancer [ICD-11: 2B90]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [] | |||
| Response Summary | Colon cancer cell lines, either WiDr homozygous for missense-mutated Cellular tumor antigen p53 (TP53/p53) (R273H) or SW48/TP53-Dox bearing heterozygous TP53 mutant (R273H), display drug resistance with increased ceramide glycosylation. Increased Gb3-cSrc complex in GEMs of membranes in response to anticancer drug induced cell stress promotes expression of p53 mutant proteins and accordant cancer drug resistance. Genz-161 effectively inhibited GCS activity, and substantially suppressed the elevated Gb3 levels seen in GEMs of p53-mutant cells exposed to doxorubicin. | |||
| Responsed Disease | Colon cancer [ICD-11: 2B90] | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Wnt signaling pathway | hsa04310 | |||
| Cell Process | Cellular stress | |||
| Cell apoptosis | ||||
| In-vitro Model | SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 |
| In-vivo Model | Cell suspension of SW48 or SW48/TP53 (5-7 passages, 1 × 106 cells in 20 uL/mouse) was subcutaneously injected into the left flank of the mice. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | The produced p53 R273H mutant protein resulted in acquired multidrug resistance in colon cancer cells. Either silencing METTL3 expression by using small interfering RNA (siRNA) or inhibiting RNA methylation with neplanocin A suppressed m6A formation in Cellular tumor antigen p53 (TP53/p53) pre-mRNA, and substantially increased the level of phosphorylated p53 protein (Ser15) and its function in cells heterozygously carrying the R273H mutation, thereby re-sensitizing these cells to anticancer drugs. | |||
| Responsed Disease | Colon cancer [ICD-11: 2B90] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Cell Process | Protein signaling | |||
| In-vitro Model | SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 |
| WiDr | Colon adenocarcinoma | Homo sapiens | CVCL_2760 | |
Liver cancer [ICD-11: 2C12]
| In total 4 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [10] | |||
| Response Summary | Deletion of METTL16 or ALKBH5 predicted poor OS and DFS of hepatocellular carcinoma (HCC) patients. And this study found significant associations between the genetic alterations and clinicopathological features as well as Cellular tumor antigen p53 (TP53/p53) alteration. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | Methyltransferase-like 16 (METTL16) | WRITER | ||
| Experiment 2 Reporting the m6A-centered Disease Response | [5] | |||
| Response Summary | Cellular tumor antigen p53 (TP53/p53) n6-methyladenosine (m6A) played a decisive role in regulating Hepatocellular carcinoma(HCC) sensitivity to chemotherapy via the p53 activator RG7112 and the vascular endothelial growth factor receptor inhibitor apatinib. p53 mRNA m6A modification blockage induced by S-adenosyl homocysteine or siRNA-mediated METTL3 inhibition enhanced HCC sensitivity to chemotherapy. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Apatinib | Approved | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | QGY-7701 | Human papillomavirus-related endocervical adenocarcinoma | Homo sapiens | CVCL_6859 |
| HHL-5 | Normal | Homo sapiens | CVCL_S956 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | Nude mice (4-6 week-old) were administered sterile water and feed in a specific pathogen-free barrier. Using a 1-mL syringe, 1 × 107 HEPG2 cells were subcutaneously inoculated into the right axilla of nude mice to build the HCC xenograft model. When the tumor volume reached 50 mm3, the nude mice were randomly divided into 1 control (n = 4) and 3 treatment groups (n = 4 each). RG7112, apatinib, and RG7112 + apatinib were administered to the treatment groups and an equal volume of dimethyl sulfoxide to the control group by daily gavage for 14 d. The tumor length (L) and width (W) were measured on alternate days using vernier calipers. The following formula was used to calculate the tumor volume: volume (mm3) = 0.5 × L × W × W. At the end of the experiment, the nude mice were killed by CO2 overdose anesthesia. The tumors were dissected and weighed using a precision balance, and the tumor tissue was stored in liquid nitrogen for further analysis. | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [5] | |||
| Response Summary | Cellular tumor antigen p53 (TP53/p53) n6-methyladenosine (m6A) played a decisive role in regulating Hepatocellular carcinoma(HCC) sensitivity to chemotherapy via the p53 activator RG7112 and the vascular endothelial growth factor receptor inhibitor apatinib. p53 mRNA m6A modification blockage induced by S-adenosyl homocysteine or siRNA-mediated METTL3 inhibition enhanced HCC sensitivity to chemotherapy. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | RG7112 | Phase 1 | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | QGY-7701 | Human papillomavirus-related endocervical adenocarcinoma | Homo sapiens | CVCL_6859 |
| HHL-5 | Normal | Homo sapiens | CVCL_S956 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | Nude mice (4-6 week-old) were administered sterile water and feed in a specific pathogen-free barrier. Using a 1-mL syringe, 1 × 107 HEPG2 cells were subcutaneously inoculated into the right axilla of nude mice to build the HCC xenograft model. When the tumor volume reached 50 mm3, the nude mice were randomly divided into 1 control (n = 4) and 3 treatment groups (n = 4 each). RG7112, apatinib, and RG7112 + apatinib were administered to the treatment groups and an equal volume of dimethyl sulfoxide to the control group by daily gavage for 14 d. The tumor length (L) and width (W) were measured on alternate days using vernier calipers. The following formula was used to calculate the tumor volume: volume (mm3) = 0.5 × L × W × W. At the end of the experiment, the nude mice were killed by CO2 overdose anesthesia. The tumors were dissected and weighed using a precision balance, and the tumor tissue was stored in liquid nitrogen for further analysis. | |||
| Experiment 4 Reporting the m6A-centered Disease Response | [10] | |||
| Response Summary | Deletion of METTL16 or ALKBH5 predicted poor OS and DFS of hepatocellular carcinoma (HCC) patients. And this study found significant associations between the genetic alterations and clinicopathological features as well as Cellular tumor antigen p53 (TP53/p53) alteration. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [14] | |||
| Response Summary | Knockdown of Cellular tumor antigen p53 (TP53/p53) or inhibition of p53's transcriptional activity by addition of its specific inhibitor PFT-Alpha decreased expression of ALKBH5 and Cancer stem cells' malignancies, the pivotal role of ALKBH5 in Cancer stem cells derived from nonsmall-cell lung cancer and highlight the regulatory function of the p53/ALKBH5 axis in modulating CSC progression. p53 transcriptionally regulates PRRX1, which is consistent with our previous report. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| In-vitro Model | PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | Cells at 1 × 106 were subcutaneously injected into the mice similarly to nude mice. Twenty-eight days later, grafted tumors were collected and morphologically analyzed. | |||
Acute kidney failure [ICD-11: GB60]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [7] | |||
| Response Summary | Meclofenamic acid increased Cellular tumor antigen p53 (TP53/p53) mRNA and protein levels in AKI both in vitro and in vivo, and FTO overexpression reduced p53 expression and reversed the MA-induced p53 increase in cisplatin-induced acute kidney injury Acute kidney injury. | |||
| Responsed Disease | Acute kidney failure [ICD-11: GB60] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Cisplatin | Approved | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | HK2 | Normal | Acipenser baerii | CVCL_YE28 |
| In-vivo Model | Induced AKI in c57BL/6 mice by intraperitoneal cisplatin injection and treated the animal with vehicle or an FTO inhibitor meclofenamic acid (MA) for 3 days. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [7] | |||
| Response Summary | Meclofenamic acid increased Cellular tumor antigen p53 (TP53/p53) mRNA and protein levels in AKI both in vitro and in vivo, and FTO overexpression reduced p53 expression and reversed the MA-induced p53 increase in cisplatin-induced Acute kidney injury. | |||
| Responsed Disease | Acute kidney failure [ICD-11: GB60] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Meclofenamic acid | Approved | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | HK2 | Normal | Acipenser baerii | CVCL_YE28 |
| In-vivo Model | Induced AKI in c57BL/6 mice by intraperitoneal cisplatin injection and treated the animal with vehicle or an FTO inhibitor meclofenamic acid (MA) for 3 days. | |||
Cerebral development anomalies [ICD-11: LA05]
Female reproductive system disorders [ICD-11: SC4Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [15] | |||
| Response Summary | ALKBH5 as another mammalian demethylase that oxidatively reverses m(6)A in mRNA in vitro and in vivo. Alkbh5-deficient male mice have increased m(6)A in mRNA and are characterized by impaired fertility resulting from apoptosis that affects meiotic metaphase-stage spermatocytes. Identified in mouse testes 1,551 differentially expressed genes that cover broad functional categories and include spermatogenesis-related mRNAs involved in the Cellular tumor antigen p53 (TP53/p53) functional interaction network. | |||
| Responsed Disease | Female reproductive system disorders [ICD-11: SC4Y] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| In-vitro Model | HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| In-vivo Model | Neo-resistant recombinant ESCs were analyzed for correct 3' and 5' targeting through hybridization by standard protocols, and we identified seven correctly targeted clones. Following ESC cell injection, several high-percentage chimera males were born. These were bred with Cre-expressing mice, and Cre-mediated excision of Alkbh5 exon 1 was tested in 26 agouti F1 pups. Two animals tested positive for the excised allele. These two mice, heterozygous for the constitutive allele (Alkbh5+/-), were analyzed by Southern blot analysis and used for further breeding to generate Alkbh5-/- mice. | |||
Apatinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [5] | |||
| Response Summary | Cellular tumor antigen p53 (TP53/p53) n6-methyladenosine (m6A) played a decisive role in regulating Hepatocellular carcinoma(HCC) sensitivity to chemotherapy via the p53 activator RG7112 and the vascular endothelial growth factor receptor inhibitor apatinib. p53 mRNA m6A modification blockage induced by S-adenosyl homocysteine or siRNA-mediated METTL3 inhibition enhanced HCC sensitivity to chemotherapy. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | QGY-7701 | Human papillomavirus-related endocervical adenocarcinoma | Homo sapiens | CVCL_6859 |
| HHL-5 | Normal | Homo sapiens | CVCL_S956 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | Nude mice (4-6 week-old) were administered sterile water and feed in a specific pathogen-free barrier. Using a 1-mL syringe, 1 × 107 HEPG2 cells were subcutaneously inoculated into the right axilla of nude mice to build the HCC xenograft model. When the tumor volume reached 50 mm3, the nude mice were randomly divided into 1 control (n = 4) and 3 treatment groups (n = 4 each). RG7112, apatinib, and RG7112 + apatinib were administered to the treatment groups and an equal volume of dimethyl sulfoxide to the control group by daily gavage for 14 d. The tumor length (L) and width (W) were measured on alternate days using vernier calipers. The following formula was used to calculate the tumor volume: volume (mm3) = 0.5 × L × W × W. At the end of the experiment, the nude mice were killed by CO2 overdose anesthesia. The tumors were dissected and weighed using a precision balance, and the tumor tissue was stored in liquid nitrogen for further analysis. | |||
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [7] | |||
| Response Summary | Meclofenamic acid increased Cellular tumor antigen p53 (TP53/p53) mRNA and protein levels in AKI both in vitro and in vivo, and FTO overexpression reduced p53 expression and reversed the MA-induced p53 increase in cisplatin-induced acute kidney injury Acute kidney injury. | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Acute kidney failure | ICD-11: GB60 | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | HK2 | Normal | Acipenser baerii | CVCL_YE28 |
| In-vivo Model | Induced AKI in c57BL/6 mice by intraperitoneal cisplatin injection and treated the animal with vehicle or an FTO inhibitor meclofenamic acid (MA) for 3 days. | |||
Meclofenamic acid
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [7] | |||
| Response Summary | Meclofenamic acid increased Cellular tumor antigen p53 (TP53/p53) mRNA and protein levels in AKI both in vitro and in vivo, and FTO overexpression reduced p53 expression and reversed the MA-induced p53 increase in cisplatin-induced Acute kidney injury. | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Acute kidney failure | ICD-11: GB60 | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | HK2 | Normal | Acipenser baerii | CVCL_YE28 |
| In-vivo Model | Induced AKI in c57BL/6 mice by intraperitoneal cisplatin injection and treated the animal with vehicle or an FTO inhibitor meclofenamic acid (MA) for 3 days. | |||
Arsenite
[Phase 2]
| In total 2 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [2] | |||
| Response Summary | METTL3 significantly decreased m6A level, restoring Cellular tumor antigen p53 (TP53/p53) activation and inhibiting cellular transformation phenotypes in the arsenite-transformed cells. m6A downregulated the expression of the positive p53 regulator, PRDM2, through the YTHDF2-promoted decay of PRDM2 mRNAs. m6A upregulated the expression of the negative p53 regulator, YY1 and MDM2 through YTHDF1-stimulated translation of YY1 and MDM2 mRNA. This study further sheds light on the mechanisms of arsenic carcinogenesis via RNA epigenetics. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Solid tumour/cancer | ICD-11: 2A00-2F9Z | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
| Experiment 2 Reporting the m6A-centered Drug Response | [2] | |||
| Response Summary | METTL3 significantly decreased m6A level, restoring Cellular tumor antigen p53 (TP53/p53) activation and inhibiting cellular transformation phenotypes in the arsenite-transformed cells. m6A downregulated the expression of the positive p53 regulator, PRDM2, through the YTHDF2-promoted decay of PRDM2 mRNAs. m6A upregulated the expression of the negative p53 regulator, YY1 and MDM2 through YTHDF1-stimulated translation of YY1 and MDM2 mRNA. This study further sheds light on the mechanisms of arsenic carcinogenesis via RNA epigenetics. | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Solid tumour/cancer | ICD-11: 2A00-2F9Z | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
RG7112
[ Phase 1]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [5] | |||
| Response Summary | Cellular tumor antigen p53 (TP53/p53) n6-methyladenosine (m6A) played a decisive role in regulating Hepatocellular carcinoma(HCC) sensitivity to chemotherapy via the p53 activator RG7112 and the vascular endothelial growth factor receptor inhibitor apatinib. p53 mRNA m6A modification blockage induced by S-adenosyl homocysteine or siRNA-mediated METTL3 inhibition enhanced HCC sensitivity to chemotherapy. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | QGY-7701 | Human papillomavirus-related endocervical adenocarcinoma | Homo sapiens | CVCL_6859 |
| HHL-5 | Normal | Homo sapiens | CVCL_S956 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | Nude mice (4-6 week-old) were administered sterile water and feed in a specific pathogen-free barrier. Using a 1-mL syringe, 1 × 107 HEPG2 cells were subcutaneously inoculated into the right axilla of nude mice to build the HCC xenograft model. When the tumor volume reached 50 mm3, the nude mice were randomly divided into 1 control (n = 4) and 3 treatment groups (n = 4 each). RG7112, apatinib, and RG7112 + apatinib were administered to the treatment groups and an equal volume of dimethyl sulfoxide to the control group by daily gavage for 14 d. The tumor length (L) and width (W) were measured on alternate days using vernier calipers. The following formula was used to calculate the tumor volume: volume (mm3) = 0.5 × L × W × W. At the end of the experiment, the nude mice were killed by CO2 overdose anesthesia. The tumors were dissected and weighed using a precision balance, and the tumor tissue was stored in liquid nitrogen for further analysis. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
RNA modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 3 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00069 | ||
| Epigenetic Regulator | tRNA (cytosine(72)-C(5))-methyltransferase NSUN6 (NSUN6) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m5C → m6A | |
| Disease | Colon cancer | |
| Drug | Contusugene ladenovec | |
| Crosstalk ID: M6ACROT00432 | ||
| Epigenetic Regulator | Methyltransferase-like protein 1 (METTL1) | |
| Regulated Target | MicroRNA 125a (MIR125A) | |
| Crosstalk relationship | m7G → m6A | |
| Crosstalk ID: M6ACROT00437 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 214 (MIR214) | |
| Crosstalk relationship | A-to-I → m6A | |
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00431 | ||
| Epigenetic Regulator | Methyltransferase-like protein 1 (METTL1) | |
| Regulated Target | MicroRNA 125a (MIR125A) | |
| Crosstalk relationship | m7G → m6A | |
| Crosstalk ID: M6ACROT00436 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 214 (MIR214) | |
| Crosstalk relationship | A-to-I → m6A | |
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00433 | ||
| Epigenetic Regulator | Methyltransferase-like protein 1 (METTL1) | |
| Regulated Target | MicroRNA 125a (MIR125A) | |
| Crosstalk relationship | m7G → m6A | |
| Crosstalk ID: M6ACROT00438 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 214 (MIR214) | |
| Crosstalk relationship | A-to-I → m6A | |
Histone modification
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03049 | ||
| Epigenetic Regulator | Lactate dehydrogenase A (LDHA) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Melanoma of uvea | |
| Crosstalk ID: M6ACROT03050 | ||
| Epigenetic Regulator | L-lactate dehydrogenase B chain (LDHB) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Melanoma of uvea | |
m6A Regulator: RNA demethylase ALKBH5 (ALKBH5)
| In total 3 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03366 | ||
| Epigenetic Regulator | Histone deacetylase 2 (HDAC2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03463 | ||
| Epigenetic Regulator | Histone deacetylase 1 (HDAC1) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03533 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
Non-coding RNA
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05064 | ||
| Epigenetic Regulator | Antisense of IGF2R non-protein coding RNA (AIRN) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Vascular disorders of the liver | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00356)
| In total 8 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE007741 | Click to Show/Hide the Full List | ||
| mod site | chr17:7662895-7662896:- | [17] | |
| Sequence | CATGGTGGCTCACACCTGTAATCCCAGCACTTTGGGAGGCT | ||
| Transcript ID List | ENST00000413465.6; rmsk_4628373 | ||
| External Link | RMBase: RNA-editing_site_54881 | ||
| mod ID: A2ISITE007742 | Click to Show/Hide the Full List | ||
| mod site | chr17:7665877-7665878:- | [17] | |
| Sequence | ACAGCACATTGGGAGGCTAAAGCAGGAGGATAACTTGAGGC | ||
| Transcript ID List | ENST00000413465.6; ENST00000635293.1; rmsk_4628387 | ||
| External Link | RMBase: RNA-editing_site_54882 | ||
| mod ID: A2ISITE007743 | Click to Show/Hide the Full List | ||
| mod site | chr17:7666206-7666207:- | [17] | |
| Sequence | GCACGGTGGATCACACCTGTAATCCCAGCTACTCGGGAGGC | ||
| Transcript ID List | ENST00000359597.8; rmsk_4628388; ENST00000635293.1; ENST00000413465.6 | ||
| External Link | RMBase: RNA-editing_site_54883 | ||
| mod ID: A2ISITE007744 | Click to Show/Hide the Full List | ||
| mod site | chr17:7671228-7671229:- | [17] | |
| Sequence | AGGAGGATCTCTTGAGCCCAAGAGTTTGAGTCCAGCCTGAA | ||
| Transcript ID List | ENST00000620739.4; ENST00000619485.4; ENST00000359597.8; ENST00000504937.5; ENST00000504290.5; ENST00000269305.8; ENST00000617185.4; ENST00000576024.1; ENST00000510385.5; ENST00000610623.4; ENST00000413465.6; ENST00000610292.4; ENST00000445888.6; ENST00000635293.1; ENST00000455263.6; ENST00000420246.6; ENST00000619186.4; ENST00000622645.4; ENST00000610538.4; ENST00000618944.4; rmsk_4628398; ENST00000615910.4 | ||
| External Link | RMBase: RNA-editing_site_54884 | ||
| mod ID: A2ISITE007745 | Click to Show/Hide the Full List | ||
| mod site | chr17:7671925-7671926:- | [17] | |
| Sequence | TTTTTATTTTCTGTGAAGTCAAGGTCTTGCTACGTTGCCCA | ||
| Transcript ID List | ENST00000504937.5; ENST00000510385.5; ENST00000617185.4; ENST00000455263.6; ENST00000610623.4; ENST00000420246.6; ENST00000576024.1; ENST00000610538.4; ENST00000359597.8; ENST00000619186.4; ENST00000619485.4; ENST00000504290.5; ENST00000635293.1; ENST00000615910.4; ENST00000413465.6; ENST00000269305.8; ENST00000610292.4; ENST00000620739.4; ENST00000618944.4; ENST00000622645.4; ENST00000445888.6 | ||
| External Link | RMBase: RNA-editing_site_54885 | ||
| mod ID: A2ISITE007746 | Click to Show/Hide the Full List | ||
| mod site | chr17:7672745-7672746:- | [17] | |
| Sequence | GATTACAGTGATGTGATCATAGCTCATTATACCCTCCTGGG | ||
| Transcript ID List | ENST00000620739.4; ENST00000617185.4; ENST00000269305.8; ENST00000610292.4; ENST00000610538.4; ENST00000615910.4; ENST00000635293.1; ENST00000445888.6; ENST00000622645.4; ENST00000359597.8; ENST00000420246.6; ENST00000413465.6; ENST00000610623.4; ENST00000504290.5; ENST00000455263.6; ENST00000619485.4; ENST00000504937.5; ENST00000619186.4; ENST00000510385.5; ENST00000576024.1; ENST00000618944.4 | ||
| External Link | RMBase: RNA-editing_site_54886 | ||
| mod ID: A2ISITE007747 | Click to Show/Hide the Full List | ||
| mod site | chr17:7684206-7684207:- | [17] | |
| Sequence | AGGATCTCACTATGTTGCCCAGGCTGGTCTCAAACTGCTGG | ||
| Transcript ID List | ENST00000635293.1; ENST00000610538.4; ENST00000445888.6; ENST00000455263.6; ENST00000514944.5; ENST00000622645.4; ENST00000505014.5; ENST00000604348.5; ENST00000503591.1; ENST00000509690.5; ENST00000620739.4; ENST00000617185.4; ENST00000420246.6; ENST00000610292.4; ENST00000269305.8; ENST00000619485.4 | ||
| External Link | RMBase: RNA-editing_site_54887 | ||
| mod ID: A2ISITE007748 | Click to Show/Hide the Full List | ||
| mod site | chr17:7684352-7684353:- | [18] | |
| Sequence | ACTGTTGCCCAGGCTAGAGTACAATGGCACAATCAAGGCTT | ||
| Transcript ID List | ENST00000514944.5; ENST00000509690.5; ENST00000445888.6; ENST00000619485.4; ENST00000505014.5; ENST00000617185.4; ENST00000503591.1; ENST00000269305.8; ENST00000620739.4; ENST00000635293.1; ENST00000622645.4; ENST00000455263.6; ENST00000420246.6; ENST00000610538.4; ENST00000610292.4; ENST00000604348.5 | ||
| External Link | RMBase: RNA-editing_site_54888 | ||
N1-methyladenosine (m1A)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: M1ASITE000049 | Click to Show/Hide the Full List | ||
| mod site | chr17:7673779-7673780:- | [19] | |
| Sequence | GTTTGTGCCTGTCCTGGGAGAGACCGGCGCACAGAGGAAGA | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | m1A-MAP-seq | ||
| Transcript ID List | ENST00000504937.5; ENST00000610623.4; ENST00000510385.5; ENST00000618944.4; ENST00000619485.4; ENST00000635293.1; ENST00000455263.6; ENST00000610292.4; ENST00000610538.4; ENST00000445888.6; ENST00000504290.5; ENST00000420246.6; ENST00000622645.4; ENST00000413465.6; ENST00000619186.4; ENST00000359597.8; ENST00000617185.4; ENST00000509690.5; ENST00000620739.4; ENST00000269305.8; ENST00000615910.4 | ||
| External Link | RMBase: m1A_site_414 | ||
| mod ID: M1ASITE000050 | Click to Show/Hide the Full List | ||
| mod site | chr17:7674891-7674892:- | [20] | |
| Sequence | GATGACAGAAACACTTTTCGACATAGTGTGGTGGTGCCCTA | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | m1A-seq | ||
| Transcript ID List | ENST00000504290.5; ENST00000610292.4; ENST00000359597.8; ENST00000617185.4; ENST00000413465.6; ENST00000610538.4; ENST00000455263.6; ENST00000509690.5; ENST00000574684.1; ENST00000615910.4; ENST00000269305.8; ENST00000635293.1; ENST00000622645.4; ENST00000420246.6; ENST00000504937.5; ENST00000618944.4; ENST00000619186.4; ENST00000514944.5; ENST00000445888.6; ENST00000619485.4; ENST00000620739.4; ENST00000610623.4; ENST00000505014.5; ENST00000510385.5 | ||
| External Link | RMBase: m1A_site_415 | ||
5-methylcytidine (m5C)
| In total 3 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE000941 | Click to Show/Hide the Full List | ||
| mod site | chr17:7670712-7670713:- | [21] | |
| Sequence | CCTCCTCTGTTGCTGCAGATCCGTGGGCGTGAGCGCTTCGA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000504290.5; ENST00000620739.4; ENST00000504937.5; ENST00000619186.4; ENST00000617185.4; ENST00000576024.1; ENST00000359597.8; ENST00000455263.6; ENST00000618944.4; ENST00000445888.6; ENST00000622645.4; ENST00000510385.5; ENST00000610538.4; ENST00000269305.8; ENST00000635293.1; ENST00000610292.4; ENST00000615910.4; ENST00000420246.6; ENST00000610623.4; ENST00000619485.4; ENST00000413465.6 | ||
| External Link | RMBase: m5C_site_17915 | ||
| mod ID: M5CSITE000942 | Click to Show/Hide the Full List | ||
| mod site | chr17:7676583-7676584:- | [21] | |
| Sequence | GGTCACTGCCATGGAGGAGCCGCAGTCAGATCCTAGCGTCG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000420246.6; ENST00000619485.4; ENST00000455263.6; ENST00000514944.5; ENST00000617185.4; ENST00000509690.5; ENST00000505014.5; ENST00000269305.8; ENST00000359597.8; ENST00000604348.5; ENST00000610292.4; ENST00000615910.4; ENST00000620739.4; ENST00000622645.4; ENST00000635293.1; ENST00000413465.6; ENST00000508793.5; ENST00000503591.1; ENST00000445888.6; ENST00000610538.4 | ||
| External Link | RMBase: m5C_site_17916 | ||
| mod ID: M5CSITE000943 | Click to Show/Hide the Full List | ||
| mod site | chr17:7686881-7686882:- | ||
| Sequence | CCCAAGCTGCTAAGGTCCCACAACTTCCGGACCTTTGTCCT | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000505014.5; ENST00000610292.4; ENST00000509690.5; ENST00000445888.6; ENST00000622645.4; ENST00000269305.8; ENST00000420246.6; ENST00000620739.4; ENST00000619485.4; ENST00000617185.4; ENST00000455263.6; ENST00000604348.5; ENST00000610538.4; ENST00000635293.1; ENST00000514944.5; ENST00000503591.1 | ||
| External Link | RMBase: m5C_site_17917 | ||
N6-methyladenosine (m6A)
| In total 49 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE030086 | Click to Show/Hide the Full List | ||
| mod site | chr17:7667034-7667035:- | [22] | |
| Sequence | ACTCTGCAGATCTCAAGTAAACAGAAAATCTCCCTTTTTGA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000359597.8; ENST00000413465.6; ENST00000635293.1 | ||
| External Link | RMBase: m6A_site_346500 | ||
| mod ID: M6ASITE030087 | Click to Show/Hide the Full List | ||
| mod site | chr17:7667058-7667059:- | [22] | |
| Sequence | CCTACGACGAGGACACTTGGACCTACTCTGCAGATCTCAAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000413465.6; ENST00000359597.8; ENST00000635293.1 | ||
| External Link | RMBase: m6A_site_346501 | ||
| mod ID: M6ASITE030088 | Click to Show/Hide the Full List | ||
| mod site | chr17:7667066-7667067:- | [22] | |
| Sequence | TGTGTTCACCTACGACGAGGACACTTGGACCTACTCTGCAG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000413465.6; ENST00000359597.8; ENST00000635293.1 | ||
| External Link | RMBase: m6A_site_346502 | ||
| mod ID: M6ASITE030089 | Click to Show/Hide the Full List | ||
| mod site | chr17:7667392-7667393:- | [23] | |
| Sequence | ACTCTCCCCAGGAGATCCAGACCCGCCTCTTTCAGAGACTT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000359597.8; ENST00000635293.1; ENST00000413465.6 | ||
| External Link | RMBase: m6A_site_346503 | ||
| mod ID: M6ASITE030090 | Click to Show/Hide the Full List | ||
| mod site | chr17:7667414-7667415:- | [22] | |
| Sequence | TTCTTACTAGGGAATGCCAAACACTCTCCCCAGGAGATCCA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000413465.6; ENST00000359597.8; ENST00000635293.1 | ||
| External Link | RMBase: m6A_site_346504 | ||
| mod ID: M6ASITE030091 | Click to Show/Hide the Full List | ||
| mod site | chr17:7668431-7668432:- | [22] | |
| Sequence | ATCTCTTATTTTACAATAAAACTTTGCTGCCACCTGTGTGT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000619485.4; ENST00000445888.6; ENST00000269305.8; ENST00000620739.4; ENST00000420246.6; ENST00000504937.5; ENST00000622645.4; ENST00000618944.4; ENST00000359597.8; ENST00000617185.4; ENST00000610538.4; ENST00000610623.4; ENST00000455263.6; ENST00000610292.4; ENST00000504290.5; ENST00000635293.1; ENST00000619186.4; ENST00000413465.6; ENST00000510385.5 | ||
| External Link | RMBase: m6A_site_346505 | ||
| mod ID: M6ASITE030092 | Click to Show/Hide the Full List | ||
| mod site | chr17:7668624-7668625:- | [24] | |
| Sequence | GTTGCCCAGGCTGGTCTCAAACTCCTGGGCTCAGGCGATCC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | MT4; Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000455263.6; ENST00000622645.4; ENST00000619186.4; ENST00000504290.5; ENST00000610623.4; ENST00000619485.4; ENST00000635293.1; ENST00000413465.6; ENST00000504937.5; ENST00000359597.8; ENST00000618944.4; ENST00000510385.5; ENST00000617185.4; ENST00000445888.6; ENST00000620739.4; ENST00000610538.4; ENST00000610292.4; ENST00000269305.8; ENST00000420246.6 | ||
| External Link | RMBase: m6A_site_346506 | ||
| mod ID: M6ASITE030093 | Click to Show/Hide the Full List | ||
| mod site | chr17:7668710-7668711:- | [25] | |
| Sequence | CAGCCTCCGGAGTAGCTGGGACCACAGGTTCATGCCACCAT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000620739.4; ENST00000610292.4; ENST00000635293.1; ENST00000622645.4; ENST00000619485.4; ENST00000504937.5; ENST00000445888.6; ENST00000618944.4; ENST00000619186.4; ENST00000617185.4; ENST00000269305.8; ENST00000455263.6; ENST00000610623.4; ENST00000413465.6; ENST00000610538.4; ENST00000420246.6; ENST00000359597.8; ENST00000510385.5; ENST00000504290.5 | ||
| External Link | RMBase: m6A_site_346507 | ||
| mod ID: M6ASITE030094 | Click to Show/Hide the Full List | ||
| mod site | chr17:7669054-7669055:- | [26] | |
| Sequence | TCGGTGGGTTGGTAGTTTCTACAGTTGGGCAGCTGGTTAGG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000420246.6; ENST00000620739.4; ENST00000413465.6; ENST00000618944.4; ENST00000622645.4; ENST00000510385.5; ENST00000455263.6; ENST00000635293.1; ENST00000610538.4; ENST00000504937.5; ENST00000610292.4; ENST00000619186.4; ENST00000445888.6; ENST00000359597.8; ENST00000617185.4; ENST00000610623.4; ENST00000619485.4; ENST00000504290.5; ENST00000269305.8 | ||
| External Link | RMBase: m6A_site_346508 | ||
| mod ID: M6ASITE030095 | Click to Show/Hide the Full List | ||
| mod site | chr17:7669274-7669275:- | [27] | |
| Sequence | GGGCCCACTTCACCGTACTAACCAGGGAAGCTGTCCCTCAC | ||
| Motif Score | 2.147452381 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000504937.5; ENST00000619485.4; ENST00000635293.1; ENST00000359597.8; ENST00000269305.8; ENST00000618944.4; ENST00000619186.4; ENST00000617185.4; ENST00000610538.4; ENST00000610292.4; ENST00000510385.5; ENST00000622645.4; ENST00000504290.5; ENST00000610623.4; ENST00000620739.4; ENST00000445888.6; ENST00000455263.6; ENST00000413465.6; ENST00000420246.6 | ||
| External Link | RMBase: m6A_site_346509 | ||
| mod ID: M6ASITE030096 | Click to Show/Hide the Full List | ||
| mod site | chr17:7669288-7669289:- | [28] | |
| Sequence | CACATTCTAGGTAGGGGCCCACTTCACCGTACTAACCAGGG | ||
| Motif Score | 2.475107143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000455263.6; ENST00000420246.6; ENST00000504937.5; ENST00000622645.4; ENST00000617185.4; ENST00000635293.1; ENST00000510385.5; ENST00000610623.4; ENST00000269305.8; ENST00000359597.8; ENST00000619485.4; ENST00000620739.4; ENST00000504290.5; ENST00000413465.6; ENST00000610292.4; ENST00000618944.4; ENST00000610538.4; ENST00000619186.4; ENST00000445888.6 | ||
| External Link | RMBase: m6A_site_346510 | ||
| mod ID: M6ASITE030097 | Click to Show/Hide the Full List | ||
| mod site | chr17:7669401-7669402:- | [24] | |
| Sequence | GGAGGAGGATGGGGAGTAGGACATACCAGCTTAGATTTTAA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | MT4 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000455263.6; ENST00000504937.5; ENST00000269305.8; ENST00000445888.6; ENST00000610538.4; ENST00000635293.1; ENST00000420246.6; ENST00000622645.4; ENST00000510385.5; ENST00000619485.4; ENST00000619186.4; ENST00000413465.6; ENST00000610292.4; ENST00000610623.4; ENST00000359597.8; ENST00000617185.4; ENST00000504290.5; ENST00000620739.4; ENST00000618944.4 | ||
| External Link | RMBase: m6A_site_346511 | ||
| mod ID: M6ASITE030098 | Click to Show/Hide the Full List | ||
| mod site | chr17:7669453-7669454:- | [24] | |
| Sequence | TTGTCCCGGGGCTCCACTGAACAAGTTGGCCTGCACTGGTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | MT4 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000455263.6; ENST00000620739.4; ENST00000610538.4; ENST00000618944.4; ENST00000269305.8; ENST00000622645.4; ENST00000610292.4; ENST00000359597.8; ENST00000635293.1; ENST00000445888.6; ENST00000619186.4; ENST00000413465.6; ENST00000504937.5; ENST00000504290.5; ENST00000420246.6; ENST00000619485.4; ENST00000510385.5; ENST00000610623.4; ENST00000617185.4 | ||
| External Link | RMBase: m6A_site_346512 | ||
| mod ID: M6ASITE030099 | Click to Show/Hide the Full List | ||
| mod site | chr17:7669486-7669487:- | [24] | |
| Sequence | GTGCGTCAGAAGCACCCAGGACTTCCATTTGCTTTGTCCCG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | MT4 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000610292.4; ENST00000619186.4; ENST00000510385.5; ENST00000622645.4; ENST00000610538.4; ENST00000445888.6; ENST00000610623.4; ENST00000504937.5; ENST00000455263.6; ENST00000420246.6; ENST00000619485.4; ENST00000618944.4; ENST00000635293.1; ENST00000617185.4; ENST00000269305.8; ENST00000620739.4; ENST00000359597.8; ENST00000504290.5; ENST00000413465.6 | ||
| External Link | RMBase: m6A_site_346513 | ||
| mod ID: M6ASITE030100 | Click to Show/Hide the Full List | ||
| mod site | chr17:7669525-7669526:- | [24] | |
| Sequence | TTTGGGTTTTGGGTCTTTGAACCCTTGCTTGCAATAGGTGT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | MT4; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000617185.4; ENST00000610292.4; ENST00000610538.4; ENST00000618944.4; ENST00000504290.5; ENST00000622645.4; ENST00000504937.5; ENST00000635293.1; ENST00000445888.6; ENST00000620739.4; ENST00000359597.8; ENST00000619186.4; ENST00000413465.6; ENST00000455263.6; ENST00000619485.4; ENST00000269305.8; ENST00000610623.4; ENST00000510385.5; ENST00000420246.6 | ||
| External Link | RMBase: m6A_site_346514 | ||
| mod ID: M6ASITE030101 | Click to Show/Hide the Full List | ||
| mod site | chr17:7669581-7669582:- | [28] | |
| Sequence | CCACTTCTTGTTCCCCACTGACAGCCTCCCACCCCCATCTC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000413465.6; ENST00000619485.4; ENST00000618944.4; ENST00000455263.6; ENST00000619186.4; ENST00000420246.6; ENST00000445888.6; ENST00000635293.1; ENST00000510385.5; ENST00000610623.4; ENST00000610538.4; ENST00000620739.4; ENST00000359597.8; ENST00000610292.4; ENST00000269305.8; ENST00000504937.5; ENST00000504290.5; ENST00000622645.4; ENST00000576024.1; ENST00000617185.4 | ||
| External Link | RMBase: m6A_site_346515 | ||
| mod ID: M6ASITE030102 | Click to Show/Hide the Full List | ||
| mod site | chr17:7669612-7669613:- | [24] | |
| Sequence | GACAGAAGGGCCTGACTCAGACTGACATTCTCCACTTCTTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | MT4; Huh7; CD4T; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000445888.6; ENST00000610538.4; ENST00000610623.4; ENST00000510385.5; ENST00000413465.6; ENST00000504290.5; ENST00000420246.6; ENST00000359597.8; ENST00000269305.8; ENST00000618944.4; ENST00000455263.6; ENST00000617185.4; ENST00000576024.1; ENST00000620739.4; ENST00000619485.4; ENST00000622645.4; ENST00000504937.5; ENST00000615910.4; ENST00000635293.1; ENST00000610292.4; ENST00000619186.4 | ||
| External Link | RMBase: m6A_site_346516 | ||
| mod ID: M6ASITE030103 | Click to Show/Hide the Full List | ||
| mod site | chr17:7669631-7669632:- | [24] | |
| Sequence | ATAAAAAACTCATGTTCAAGACAGAAGGGCCTGACTCAGAC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | MT4; Huh7; CD4T; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000619485.4; ENST00000413465.6; ENST00000615910.4; ENST00000576024.1; ENST00000610623.4; ENST00000619186.4; ENST00000617185.4; ENST00000610292.4; ENST00000622645.4; ENST00000504290.5; ENST00000359597.8; ENST00000635293.1; ENST00000610538.4; ENST00000420246.6; ENST00000455263.6; ENST00000510385.5; ENST00000445888.6; ENST00000269305.8; ENST00000618944.4; ENST00000620739.4; ENST00000504937.5 | ||
| External Link | RMBase: m6A_site_346517 | ||
| mod ID: M6ASITE030104 | Click to Show/Hide the Full List | ||
| mod site | chr17:7669644-7669645:- | [24] | |
| Sequence | TCTACCTCCCGCCATAAAAAACTCATGTTCAAGACAGAAGG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | MT4; Huh7; CD4T; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000610623.4; ENST00000615910.4; ENST00000359597.8; ENST00000610292.4; ENST00000455263.6; ENST00000504290.5; ENST00000619485.4; ENST00000610538.4; ENST00000413465.6; ENST00000420246.6; ENST00000617185.4; ENST00000620739.4; ENST00000269305.8; ENST00000510385.5; ENST00000445888.6; ENST00000618944.4; ENST00000504937.5; ENST00000576024.1; ENST00000635293.1; ENST00000619186.4; ENST00000622645.4 | ||
| External Link | RMBase: m6A_site_346518 | ||
| mod ID: M6ASITE030105 | Click to Show/Hide the Full List | ||
| mod site | chr17:7670661-7670662:- | [23] | |
| Sequence | GAGCTGAATGAGGCCTTGGAACTCAAGGATGCCCAGGCTGG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; MT4; CD4T; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000619186.4; ENST00000413465.6; ENST00000359597.8; ENST00000620739.4; ENST00000576024.1; ENST00000635293.1; ENST00000615910.4; ENST00000504937.5; ENST00000269305.8; ENST00000622645.4; ENST00000510385.5; ENST00000617185.4; ENST00000504290.5; ENST00000619485.4; ENST00000610292.4; ENST00000618944.4; ENST00000455263.6; ENST00000610623.4; ENST00000445888.6; ENST00000420246.6; ENST00000610538.4 | ||
| External Link | RMBase: m6A_site_346519 | ||
| mod ID: M6ASITE030106 | Click to Show/Hide the Full List | ||
| mod site | chr17:7673228-7673229:- | [23] | |
| Sequence | ATGGTGTTACTTCCTGATAAACTCGTCGTAAGTTGAAAATA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; CD4T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000618944.4; ENST00000620739.4; ENST00000359597.8; ENST00000510385.5; ENST00000610292.4; ENST00000504290.5; ENST00000445888.6; ENST00000413465.6; ENST00000455263.6; ENST00000619485.4; ENST00000619186.4; ENST00000576024.1; ENST00000610623.4; ENST00000610538.4; ENST00000635293.1; ENST00000617185.4; ENST00000420246.6; ENST00000269305.8; ENST00000615910.4; ENST00000622645.4; ENST00000504937.5 | ||
| External Link | RMBase: m6A_site_346520 | ||
| mod ID: M6ASITE030107 | Click to Show/Hide the Full List | ||
| mod site | chr17:7673332-7673333:- | [29] | |
| Sequence | TATATATTTTAAAGGACCAGACCAGCTTTCAAAAAGAAAAT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | A549; CD4T; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000576024.1; ENST00000413465.6; ENST00000619186.4; ENST00000269305.8; ENST00000617185.4; ENST00000619485.4; ENST00000510385.5; ENST00000420246.6; ENST00000610292.4; ENST00000618944.4; ENST00000504290.5; ENST00000504937.5; ENST00000620739.4; ENST00000610538.4; ENST00000610623.4; ENST00000455263.6; ENST00000615910.4; ENST00000359597.8; ENST00000622645.4; ENST00000635293.1; ENST00000445888.6 | ||
| External Link | RMBase: m6A_site_346521 | ||
| mod ID: M6ASITE030108 | Click to Show/Hide the Full List | ||
| mod site | chr17:7673564-7673565:- | [30] | |
| Sequence | TCTCCCCAGCCAAAGAAGAAACCACTGGATGGAGAATATTT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HepG2; A549; CD4T; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000576024.1; ENST00000622645.4; ENST00000620739.4; ENST00000359597.8; ENST00000610292.4; ENST00000510385.5; ENST00000504290.5; ENST00000635293.1; ENST00000610623.4; ENST00000445888.6; ENST00000617185.4; ENST00000420246.6; ENST00000610538.4; ENST00000413465.6; ENST00000618944.4; ENST00000615910.4; ENST00000619186.4; ENST00000504937.5; ENST00000269305.8; ENST00000455263.6; ENST00000509690.5; ENST00000619485.4 | ||
| External Link | RMBase: m6A_site_346522 | ||
| mod ID: M6ASITE030109 | Click to Show/Hide the Full List | ||
| mod site | chr17:7673777-7673778:- | [29] | |
| Sequence | TTGTGCCTGTCCTGGGAGAGACCGGCGCACAGAGGAAGAGA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | A549; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000504290.5; ENST00000635293.1; ENST00000504937.5; ENST00000510385.5; ENST00000619485.4; ENST00000610538.4; ENST00000617185.4; ENST00000622645.4; ENST00000445888.6; ENST00000610623.4; ENST00000413465.6; ENST00000615910.4; ENST00000509690.5; ENST00000620739.4; ENST00000359597.8; ENST00000618944.4; ENST00000619186.4; ENST00000455263.6; ENST00000269305.8; ENST00000610292.4; ENST00000420246.6 | ||
| External Link | RMBase: m6A_site_346523 | ||
| mod ID: M6ASITE030110 | Click to Show/Hide the Full List | ||
| mod site | chr17:7674901-7674902:- | [23] | |
| Sequence | GGAGTATTTGGATGACAGAAACACTTTTCGACATAGTGTGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000504937.5; ENST00000622645.4; ENST00000610538.4; ENST00000445888.6; ENST00000574684.1; ENST00000618944.4; ENST00000509690.5; ENST00000455263.6; ENST00000617185.4; ENST00000269305.8; ENST00000510385.5; ENST00000420246.6; ENST00000514944.5; ENST00000505014.5; ENST00000413465.6; ENST00000619485.4; ENST00000610292.4; ENST00000615910.4; ENST00000610623.4; ENST00000620739.4; ENST00000359597.8; ENST00000619186.4; ENST00000504290.5; ENST00000635293.1 | ||
| External Link | RMBase: m6A_site_346524 | ||
| mod ID: M6ASITE030111 | Click to Show/Hide the Full List | ||
| mod site | chr17:7675193-7675194:- | [23] | |
| Sequence | TGTTTTGCCAACTGGCCAAGACCTGCCCTGTGCAGCTGTGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000610538.4; ENST00000622645.4; ENST00000610623.4; ENST00000618944.4; ENST00000508793.5; ENST00000509690.5; ENST00000514944.5; ENST00000455263.6; ENST00000610292.4; ENST00000505014.5; ENST00000615910.4; ENST00000269305.8; ENST00000510385.5; ENST00000617185.4; ENST00000620739.4; ENST00000413465.6; ENST00000504937.5; ENST00000504290.5; ENST00000359597.8; ENST00000420246.6; ENST00000445888.6; ENST00000619186.4; ENST00000604348.5; ENST00000619485.4; ENST00000635293.1 | ||
| External Link | RMBase: m6A_site_346525 | ||
| mod ID: M6ASITE030112 | Click to Show/Hide the Full List | ||
| mod site | chr17:7676016-7676017:- | [23] | |
| Sequence | TGGGCTTCTTGCATTCTGGGACAGCCAAGTCTGTGACTTGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000514944.5; ENST00000620739.4; ENST00000269305.8; ENST00000622645.4; ENST00000503591.1; ENST00000508793.5; ENST00000635293.1; ENST00000615910.4; ENST00000359597.8; ENST00000610292.4; ENST00000509690.5; ENST00000445888.6; ENST00000505014.5; ENST00000619485.4; ENST00000420246.6; ENST00000610538.4; ENST00000413465.6; ENST00000604348.5; ENST00000617185.4; ENST00000455263.6 | ||
| External Link | RMBase: m6A_site_346526 | ||
| mod ID: M6ASITE030113 | Click to Show/Hide the Full List | ||
| mod site | chr17:7676064-7676065:- | [23] | |
| Sequence | CTTCTGTCCCTTCCCAGAAAACCTACCAGGGCAGCTACGGT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000635293.1; ENST00000514944.5; ENST00000622645.4; ENST00000269305.8; ENST00000617185.4; ENST00000509690.5; ENST00000455263.6; ENST00000604348.5; ENST00000610292.4; ENST00000420246.6; ENST00000620739.4; ENST00000413465.6; ENST00000508793.5; ENST00000503591.1; ENST00000619485.4; ENST00000359597.8; ENST00000610538.4; ENST00000445888.6; ENST00000615910.4; ENST00000505014.5 | ||
| External Link | RMBase: m6A_site_346527 | ||
| mod ID: M6ASITE030114 | Click to Show/Hide the Full List | ||
| mod site | chr17:7676198-7676199:- | [23] | |
| Sequence | TGAACAATGGTTCACTGAAGACCCAGGTCCAGATGAAGCTC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000455263.6; ENST00000620739.4; ENST00000269305.8; ENST00000420246.6; ENST00000505014.5; ENST00000617185.4; ENST00000622645.4; ENST00000445888.6; ENST00000359597.8; ENST00000610538.4; ENST00000610292.4; ENST00000503591.1; ENST00000514944.5; ENST00000615910.4; ENST00000413465.6; ENST00000635293.1; ENST00000508793.5; ENST00000604348.5; ENST00000509690.5; ENST00000619485.4 | ||
| External Link | RMBase: m6A_site_346528 | ||
| mod ID: M6ASITE030115 | Click to Show/Hide the Full List | ||
| mod site | chr17:7676215-7676216:- | [23] | |
| Sequence | CTGTCCCCGGACGATATTGAACAATGGTTCACTGAAGACCC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000420246.6; ENST00000604348.5; ENST00000508793.5; ENST00000509690.5; ENST00000635293.1; ENST00000455263.6; ENST00000610292.4; ENST00000619485.4; ENST00000514944.5; ENST00000505014.5; ENST00000503591.1; ENST00000413465.6; ENST00000617185.4; ENST00000269305.8; ENST00000359597.8; ENST00000445888.6; ENST00000610538.4; ENST00000622645.4; ENST00000620739.4; ENST00000615910.4 | ||
| External Link | RMBase: m6A_site_346529 | ||
| mod ID: M6ASITE030116 | Click to Show/Hide the Full List | ||
| mod site | chr17:7676391-7676392:- | [23] | |
| Sequence | GTCTTTCAGACTTCCTGAAAACAACGTTCTGGTAAGGACAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000620739.4; ENST00000269305.8; ENST00000604348.5; ENST00000445888.6; ENST00000509690.5; ENST00000413465.6; ENST00000615910.4; ENST00000622645.4; ENST00000420246.6; ENST00000610292.4; ENST00000619485.4; ENST00000455263.6; ENST00000508793.5; ENST00000617185.4; ENST00000505014.5; ENST00000610538.4; ENST00000503591.1; ENST00000635293.1; ENST00000514944.5; ENST00000359597.8 | ||
| External Link | RMBase: m6A_site_346530 | ||
| mod ID: M6ASITE030117 | Click to Show/Hide the Full List | ||
| mod site | chr17:7676402-7676403:- | [23] | |
| Sequence | TTTCTGCTCTTGTCTTTCAGACTTCCTGAAAACAACGTTCT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000269305.8; ENST00000445888.6; ENST00000413465.6; ENST00000505014.5; ENST00000610538.4; ENST00000610292.4; ENST00000635293.1; ENST00000503591.1; ENST00000615910.4; ENST00000620739.4; ENST00000619485.4; ENST00000622645.4; ENST00000359597.8; ENST00000508793.5; ENST00000604348.5; ENST00000455263.6; ENST00000420246.6; ENST00000514944.5; ENST00000617185.4; ENST00000509690.5 | ||
| External Link | RMBase: m6A_site_346531 | ||
| mod ID: M6ASITE030118 | Click to Show/Hide the Full List | ||
| mod site | chr17:7676428-7676429:- | [23] | |
| Sequence | GGAAGCGAAAATTCCATGGGACTGACTTTCTGCTCTTGTCT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000514944.5; ENST00000505014.5; ENST00000622645.4; ENST00000619485.4; ENST00000610538.4; ENST00000455263.6; ENST00000445888.6; ENST00000617185.4; ENST00000359597.8; ENST00000503591.1; ENST00000610292.4; ENST00000269305.8; ENST00000413465.6; ENST00000615910.4; ENST00000620739.4; ENST00000420246.6; ENST00000509690.5; ENST00000508793.5; ENST00000604348.5; ENST00000635293.1 | ||
| External Link | RMBase: m6A_site_346532 | ||
| mod ID: M6ASITE030119 | Click to Show/Hide the Full List | ||
| mod site | chr17:7676455-7676456:- | [23] | |
| Sequence | ACCCCAGCCCCCTAGCAGAGACCTGTGGGAAGCGAAAATTC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000622645.4; ENST00000620739.4; ENST00000420246.6; ENST00000359597.8; ENST00000505014.5; ENST00000610538.4; ENST00000615910.4; ENST00000455263.6; ENST00000617185.4; ENST00000503591.1; ENST00000269305.8; ENST00000610292.4; ENST00000509690.5; ENST00000635293.1; ENST00000514944.5; ENST00000445888.6; ENST00000413465.6; ENST00000619485.4; ENST00000508793.5; ENST00000604348.5 | ||
| External Link | RMBase: m6A_site_346533 | ||
| mod ID: M6ASITE030120 | Click to Show/Hide the Full List | ||
| mod site | chr17:7676522-7676523:- | [23] | |
| Sequence | ACATTTTCAGACCTATGGAAACTGTGAGTGGATCCATTGGA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; A549; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; iSLK; TIME; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000455263.6; ENST00000604348.5; ENST00000503591.1; ENST00000509690.5; ENST00000610292.4; ENST00000269305.8; ENST00000413465.6; ENST00000610538.4; ENST00000420246.6; ENST00000619485.4; ENST00000620739.4; ENST00000508793.5; ENST00000635293.1; ENST00000622645.4; ENST00000505014.5; ENST00000617185.4; ENST00000514944.5; ENST00000445888.6; ENST00000615910.4; ENST00000359597.8 | ||
| External Link | RMBase: m6A_site_346534 | ||
| mod ID: M6ASITE030121 | Click to Show/Hide the Full List | ||
| mod site | chr17:7676532-7676533:- | [23] | |
| Sequence | GAGTCAGGAAACATTTTCAGACCTATGGAAACTGTGAGTGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; A549; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; iSLK; TIME; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000359597.8; ENST00000615910.4; ENST00000619485.4; ENST00000610538.4; ENST00000455263.6; ENST00000604348.5; ENST00000509690.5; ENST00000514944.5; ENST00000445888.6; ENST00000413465.6; ENST00000610292.4; ENST00000622645.4; ENST00000269305.8; ENST00000635293.1; ENST00000508793.5; ENST00000503591.1; ENST00000617185.4; ENST00000620739.4; ENST00000420246.6; ENST00000505014.5 | ||
| External Link | RMBase: m6A_site_346535 | ||
| mod ID: M6ASITE030122 | Click to Show/Hide the Full List | ||
| mod site | chr17:7676542-7676543:- | [23] | |
| Sequence | AGCCCCCTCTGAGTCAGGAAACATTTTCAGACCTATGGAAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; A549; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; iSLK; TIME; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000617185.4; ENST00000445888.6; ENST00000622645.4; ENST00000615910.4; ENST00000610292.4; ENST00000505014.5; ENST00000269305.8; ENST00000604348.5; ENST00000413465.6; ENST00000420246.6; ENST00000508793.5; ENST00000610538.4; ENST00000514944.5; ENST00000620739.4; ENST00000359597.8; ENST00000619485.4; ENST00000503591.1; ENST00000455263.6; ENST00000635293.1; ENST00000509690.5 | ||
| External Link | RMBase: m6A_site_346536 | ||
| mod ID: M6ASITE030123 | Click to Show/Hide the Full List | ||
| mod site | chr17:7676614-7676615:- | [23] | |
| Sequence | TTTCCTCTTGCAGCAGCCAGACTGCCTTCCGGGTCACTGCC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; A549; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; HEK293A-TOA; iSLK; TIME; MSC; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000508793.5; ENST00000604348.5; ENST00000503591.1; ENST00000514944.5; ENST00000635293.1; ENST00000610292.4; ENST00000620739.4; ENST00000509690.5; ENST00000619485.4; ENST00000622645.4; ENST00000269305.8; ENST00000420246.6; ENST00000445888.6; ENST00000505014.5; ENST00000610538.4; ENST00000455263.6; ENST00000617185.4 | ||
| External Link | RMBase: m6A_site_346537 | ||
| mod ID: M6ASITE030124 | Click to Show/Hide the Full List | ||
| mod site | chr17:7677384-7677385:- | [23] | |
| Sequence | ATGGGCAGGTGAGCGGTGAGACAGTTGTTCTTCCAGAAGCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; A549; CD4T; peripheral-blood; GSC-11; HEK293T; HEK293A-TOA; iSLK; TIME; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000620739.4; ENST00000269305.8; ENST00000455263.6; ENST00000508793.5; ENST00000509690.5; ENST00000505014.5; ENST00000514944.5; ENST00000617185.4; ENST00000445888.6; ENST00000622645.4; ENST00000610292.4; ENST00000420246.6; ENST00000635293.1; ENST00000503591.1; ENST00000604348.5; ENST00000610538.4; ENST00000619485.4 | ||
| External Link | RMBase: m6A_site_346538 | ||
| mod ID: M6ASITE030125 | Click to Show/Hide the Full List | ||
| mod site | chr17:7685261-7685262:- | [23] | |
| Sequence | ATGAAATAAAGCTGCTATAAACATTCTTGTACAATTCTTTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000420246.6; ENST00000269305.8; ENST00000610538.4; ENST00000604348.5; ENST00000505014.5; ENST00000622645.4; ENST00000445888.6; ENST00000617185.4; ENST00000619485.4; ENST00000455263.6; ENST00000571370.1; ENST00000610292.4; ENST00000503591.1; ENST00000514944.5; ENST00000620739.4; ENST00000635293.1; ENST00000509690.5 | ||
| External Link | RMBase: m6A_site_346539 | ||
| mod ID: M6ASITE030126 | Click to Show/Hide the Full List | ||
| mod site | chr17:7685473-7685474:- | [23] | |
| Sequence | GCCTGGGCGACAAGAACGAAACTCCGTCTCAAAAAAAAGGG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; Jurkat; CD4T; HEK293T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000420246.6; ENST00000610538.4; ENST00000635293.1; ENST00000620739.4; ENST00000610292.4; ENST00000617185.4; ENST00000514944.5; ENST00000619485.4; ENST00000445888.6; ENST00000509690.5; rmsk_4628448; ENST00000571370.1; ENST00000455263.6; ENST00000505014.5; ENST00000269305.8; ENST00000604348.5; ENST00000622645.4; ENST00000503591.1 | ||
| External Link | RMBase: m6A_site_346540 | ||
| mod ID: M6ASITE030127 | Click to Show/Hide the Full List | ||
| mod site | chr17:7685543-7685544:- | [23] | |
| Sequence | GAGGCAGGAGAATCGCTTGAACCCGGGAGGCAGAGGTTGCA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; Jurkat; HEK293T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000509690.5; ENST00000610292.4; ENST00000514944.5; ENST00000269305.8; ENST00000505014.5; ENST00000617185.4; ENST00000619485.4; ENST00000455263.6; ENST00000503591.1; rmsk_4628448; ENST00000620739.4; ENST00000622645.4; ENST00000571370.1; ENST00000420246.6; ENST00000610538.4; ENST00000445888.6; ENST00000604348.5; ENST00000635293.1 | ||
| External Link | RMBase: m6A_site_346541 | ||
| mod ID: M6ASITE030128 | Click to Show/Hide the Full List | ||
| mod site | chr17:7685661-7685662:- | [31] | |
| Sequence | CTTGAGGATAGGAGTTCCAGACCAGCGTGGCCAACGTGGTG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; H1B; A549; Jurkat; HEK293A-TOA; MSC; TREX | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000622645.4; ENST00000610538.4; ENST00000514944.5; ENST00000617185.4; ENST00000503591.1; ENST00000635293.1; ENST00000619485.4; ENST00000571370.1; ENST00000604348.5; ENST00000509690.5; ENST00000620739.4; rmsk_4628448; ENST00000505014.5; ENST00000420246.6; ENST00000445888.6; ENST00000269305.8; ENST00000455263.6; ENST00000610292.4 | ||
| External Link | RMBase: m6A_site_346542 | ||
| mod ID: M6ASITE030129 | Click to Show/Hide the Full List | ||
| mod site | chr17:7685756-7685757:- | [23] | |
| Sequence | ACCATTTTGCCTGTTGGAGAACTTCATATAGAATGGAATCA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; H1A; H1B; A549; HEK293A-TOA; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000604348.5; ENST00000505014.5; ENST00000509690.5; ENST00000420246.6; ENST00000619485.4; ENST00000503591.1; ENST00000622645.4; ENST00000571370.1; ENST00000610292.4; ENST00000617185.4; ENST00000610538.4; ENST00000514944.5; ENST00000445888.6; ENST00000269305.8; ENST00000635293.1; ENST00000455263.6; ENST00000620739.4 | ||
| External Link | RMBase: m6A_site_346543 | ||
| mod ID: M6ASITE030130 | Click to Show/Hide the Full List | ||
| mod site | chr17:7685857-7685858:- | [23] | |
| Sequence | TGATGCCTACCAAGTCACAGACCCTTTTCATCGTCCCAGAA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000617185.4; ENST00000610538.4; ENST00000619485.4; ENST00000505014.5; ENST00000269305.8; ENST00000604348.5; ENST00000620739.4; ENST00000610292.4; ENST00000445888.6; ENST00000635293.1; ENST00000514944.5; ENST00000571370.1; ENST00000503591.1; ENST00000622645.4; ENST00000509690.5; ENST00000455263.6; ENST00000420246.6 | ||
| External Link | RMBase: m6A_site_346544 | ||
| mod ID: M6ASITE030131 | Click to Show/Hide the Full List | ||
| mod site | chr17:7686080-7686081:- | [23] | |
| Sequence | TCTTCCTGGTAGGAGGCGGAACTCGAATTCATTTCTCCCGC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000610538.4; ENST00000514944.5; ENST00000445888.6; ENST00000571370.1; ENST00000503591.1; ENST00000619485.4; ENST00000610292.4; ENST00000617185.4; ENST00000505014.5; ENST00000620739.4; ENST00000604348.5; ENST00000420246.6; ENST00000635293.1; ENST00000455263.6; ENST00000622645.4; ENST00000269305.8; ENST00000509690.5 | ||
| External Link | RMBase: m6A_site_346545 | ||
| mod ID: M6ASITE030132 | Click to Show/Hide the Full List | ||
| mod site | chr17:7686232-7686233:- | [23] | |
| Sequence | TTTGATAATTTGTCGGAAAAACAATCTACCTGTTATCTAGC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000505014.5; ENST00000619485.4; ENST00000635293.1; ENST00000620739.4; ENST00000503591.1; ENST00000420246.6; ENST00000269305.8; ENST00000610292.4; ENST00000514944.5; ENST00000571370.1; ENST00000610538.4; ENST00000445888.6; ENST00000622645.4; ENST00000455263.6; ENST00000617185.4; ENST00000509690.5; ENST00000604348.5 | ||
| External Link | RMBase: m6A_site_346546 | ||
| mod ID: M6ASITE030133 | Click to Show/Hide the Full List | ||
| mod site | chr17:7687436-7687437:- | [23] | |
| Sequence | GTAGCTGCTGGGCTCCGGGGACACTTTGCGTTCGGGCTGGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; H1B; fibroblasts; A549; HEK293A-TOA; TIME; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000622645.4; ENST00000455263.6; ENST00000619485.4; ENST00000617185.4; ENST00000620739.4; ENST00000420246.6; ENST00000269305.8; ENST00000604348.5; ENST00000610538.4; ENST00000505014.5; ENST00000635293.1; ENST00000610292.4; ENST00000509690.5; ENST00000445888.6 | ||
| External Link | RMBase: m6A_site_346553 | ||
| mod ID: M6ASITE030134 | Click to Show/Hide the Full List | ||
| mod site | chr17:7687513-7687514:- | [23] | |
| Sequence | TCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; fibroblasts; A549; HEK293A-TOA; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000269305.8; ENST00000620739.4; ENST00000617185.4 | ||
| External Link | RMBase: m6A_site_346556 | ||
References