m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00389)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
SEC62
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | CT26 cell line | Mus musculus |
|
Treatment: METTL3 knockout CT26 cells
Control: CT26 cells
|
GSE142589 | |
| Regulation |
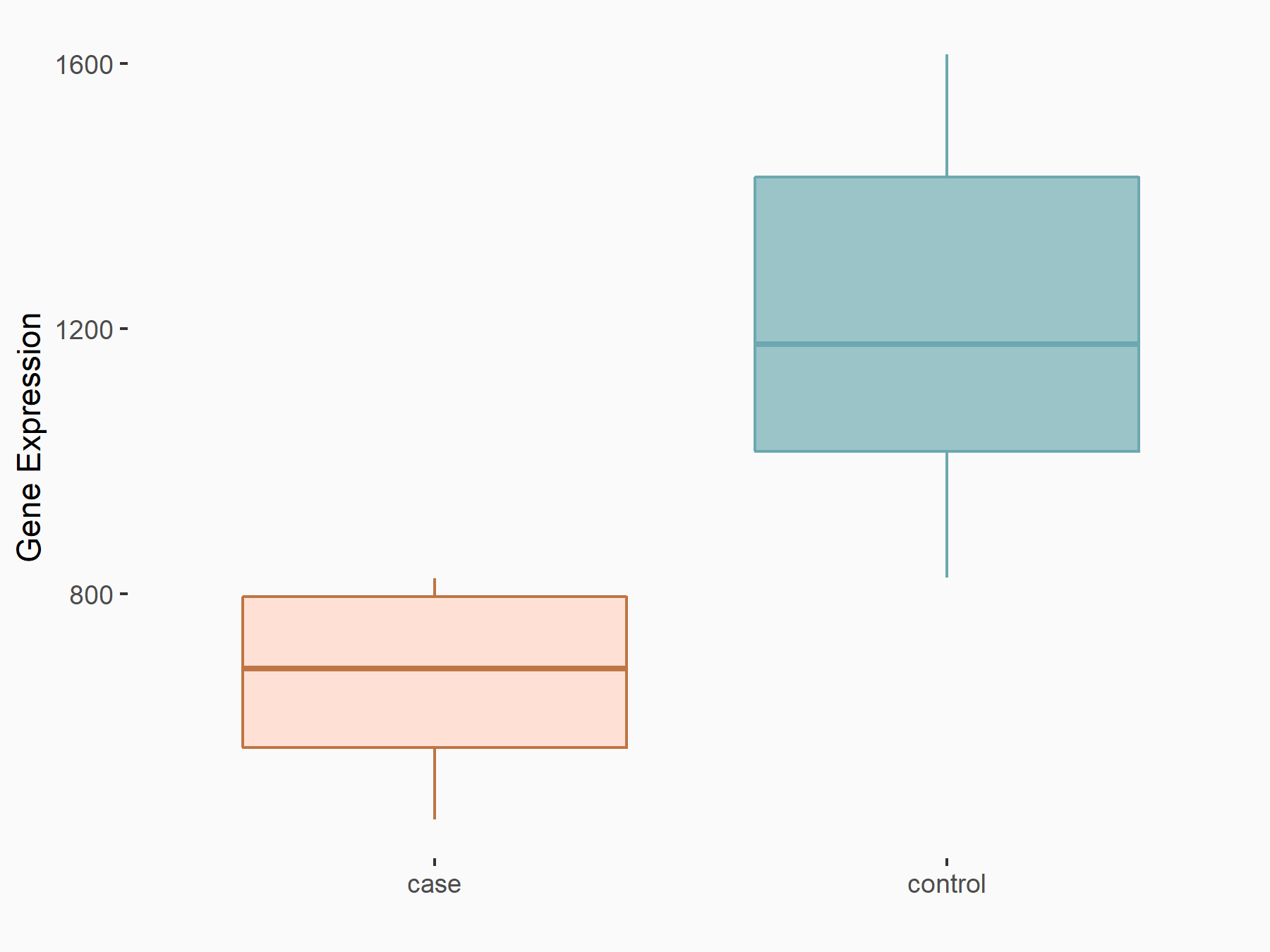 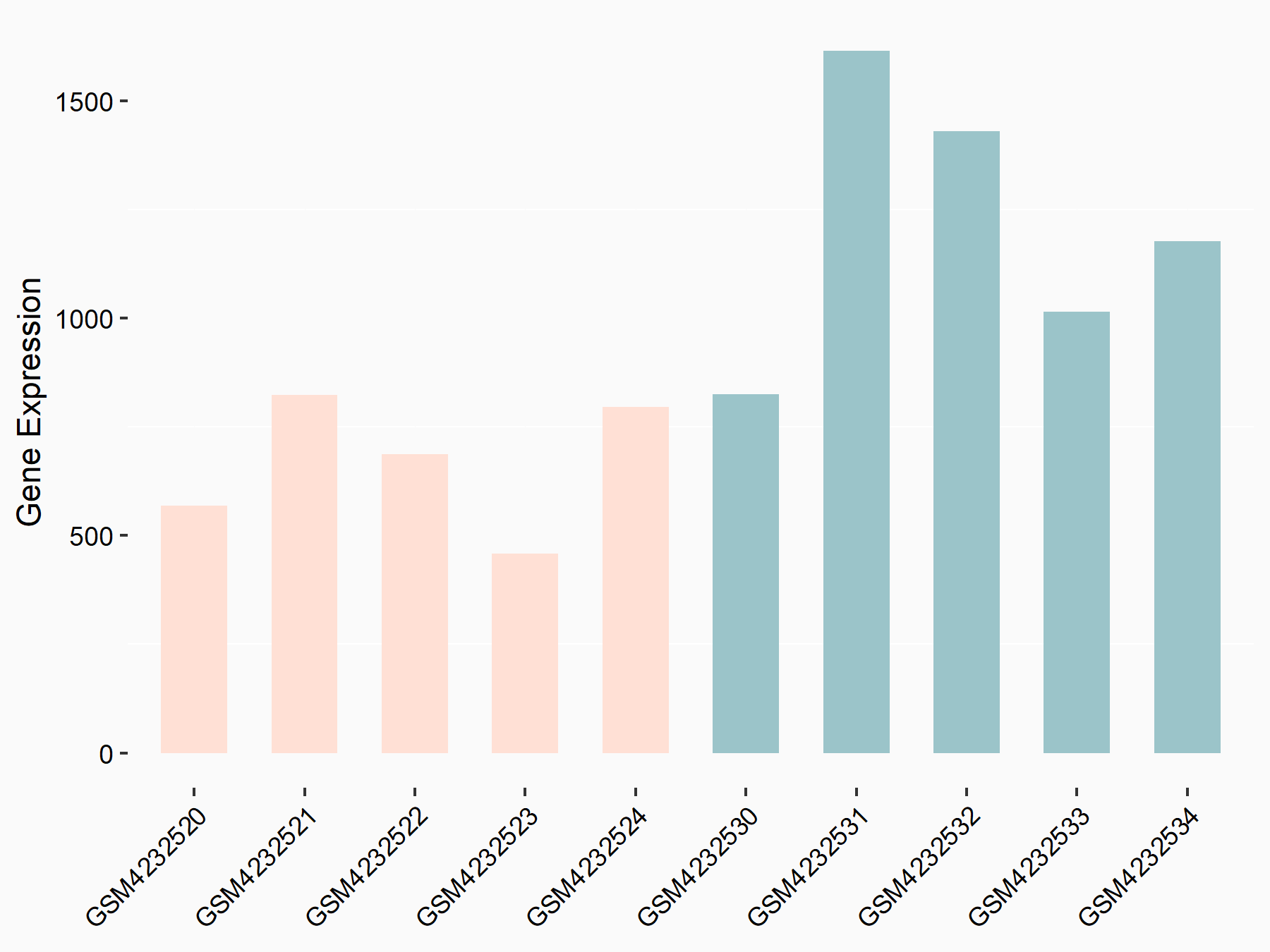 |
logFC: -8.61E-01 p-value: 1.21E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between SEC62 and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.99E+00 | GSE60213 |
| In total 3 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | MiR-4429 prevented gastric cancer progression through targeting METTL3 to inhibit m6A-caused stabilization of Translocation protein SEC62 (SEC62), indicating miR-4429 as a promising target for treatment improvement for Gastric cancer. METTL3 interacted with SEC62 to induce the m6A on SEC62 mRNA, therefore facilitated the stabilizing effect of IGF2BP1 on SEC62 mRNA. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Pathway Response | Protein processing in endoplasmic reticulum | hsa04141 | ||
| Cell Process | RNA stability | |||
| Cell apoptosis | ||||
| In-vitro Model | GES-1 | Normal | Homo sapiens | CVCL_EQ22 |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | Translocation protein SEC62 (SEC62) upregulated by the METTL3-mediated m6A modification promotes the stemness and chemoresistance of colorectal cancer by binding to beta-catenin and enhancing Wnt signalling. Depletion of Sec62 sensitized the CRC cells to 5-Fu or oxaliplatin treatment. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Responsed Drug | Fluorouracil | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Protein degradation | |||
| In-vitro Model | DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | DLD-1 cells were subcutaneously implanted into 4-6 weeks old female nude mice. When tumors reached a size of about 50 mm3, the nude mice were randomly divided into 6 groups. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | Translocation protein SEC62 (SEC62) upregulated by the METTL3-mediated m6A modification promotes the stemness and chemoresistance of colorectal cancer by binding to beta-catenin and enhancing Wnt signalling. Depletion of Sec62 sensitized the CRC cells to 5-Fu or oxaliplatin treatment. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Responsed Drug | Oxaliplatin | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Protein degradation | |||
| In-vitro Model | DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | DLD-1 cells were subcutaneously implanted into 4-6 weeks old female nude mice. When tumors reached a size of about 50 mm3, the nude mice were randomly divided into 6 groups. | |||
Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by IGF2BP1 | ||
| Cell Line | HepG2 cell line | Homo sapiens |
|
Treatment: siIGF2BP1 HepG2 cells
Control: siControl HepG2 cells
|
GSE161086 | |
| Regulation |
  |
logFC: -8.16E-01 p-value: 3.16E-03 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | MiR-4429 prevented gastric cancer progression through targeting METTL3 to inhibit m6A-caused stabilization of Translocation protein SEC62 (SEC62), indicating miR-4429 as a promising target for treatment improvement for Gastric cancer. METTL3 interacted with SEC62 to induce the m6A on SEC62 mRNA, therefore facilitated the stabilizing effect of IGF2BP1 on SEC62 mRNA. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Pathway Response | Protein processing in endoplasmic reticulum | hsa04141 | ||
| Cell Process | RNA stability | |||
| Cell apoptosis | ||||
| In-vitro Model | GES-1 | Normal | Homo sapiens | CVCL_EQ22 |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
Gastric cancer [ICD-11: 2B72]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | MiR-4429 prevented gastric cancer progression through targeting METTL3 to inhibit m6A-caused stabilization of Translocation protein SEC62 (SEC62), indicating miR-4429 as a promising target for treatment improvement for Gastric cancer. METTL3 interacted with SEC62 to induce the m6A on SEC62 mRNA, therefore facilitated the stabilizing effect of IGF2BP1 on SEC62 mRNA. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Protein processing in endoplasmic reticulum | hsa04141 | ||
| Cell Process | RNA stability | |||
| Cell apoptosis | ||||
| In-vitro Model | GES-1 | Normal | Homo sapiens | CVCL_EQ22 |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | MiR-4429 prevented gastric cancer progression through targeting METTL3 to inhibit m6A-caused stabilization of Translocation protein SEC62 (SEC62), indicating miR-4429 as a promising target for treatment improvement for Gastric cancer. METTL3 interacted with SEC62 to induce the m6A on SEC62 mRNA, therefore facilitated the stabilizing effect of IGF2BP1 on SEC62 mRNA. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Protein processing in endoplasmic reticulum | hsa04141 | ||
| Cell Process | RNA stability | |||
| Cell apoptosis | ||||
| In-vitro Model | GES-1 | Normal | Homo sapiens | CVCL_EQ22 |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
Colorectal cancer [ICD-11: 2B91]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | Translocation protein SEC62 (SEC62) upregulated by the METTL3-mediated m6A modification promotes the stemness and chemoresistance of colorectal cancer by binding to beta-catenin and enhancing Wnt signalling. Depletion of Sec62 sensitized the CRC cells to 5-Fu or oxaliplatin treatment. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Fluorouracil | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Protein degradation | |||
| In-vitro Model | DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | DLD-1 cells were subcutaneously implanted into 4-6 weeks old female nude mice. When tumors reached a size of about 50 mm3, the nude mice were randomly divided into 6 groups. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | Translocation protein SEC62 (SEC62) upregulated by the METTL3-mediated m6A modification promotes the stemness and chemoresistance of colorectal cancer by binding to beta-catenin and enhancing Wnt signalling. Depletion of Sec62 sensitized the CRC cells to 5-Fu or oxaliplatin treatment. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Oxaliplatin | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Protein degradation | |||
| In-vitro Model | DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | DLD-1 cells were subcutaneously implanted into 4-6 weeks old female nude mice. When tumors reached a size of about 50 mm3, the nude mice were randomly divided into 6 groups. | |||
Fluorouracil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [2] | |||
| Response Summary | Translocation protein SEC62 (SEC62) upregulated by the METTL3-mediated m6A modification promotes the stemness and chemoresistance of colorectal cancer by binding to beta-catenin and enhancing Wnt signalling. Depletion of Sec62 sensitized the CRC cells to 5-Fu or oxaliplatin treatment. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Protein degradation | |||
| In-vitro Model | DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | DLD-1 cells were subcutaneously implanted into 4-6 weeks old female nude mice. When tumors reached a size of about 50 mm3, the nude mice were randomly divided into 6 groups. | |||
Oxaliplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [2] | |||
| Response Summary | Translocation protein SEC62 (SEC62) upregulated by the METTL3-mediated m6A modification promotes the stemness and chemoresistance of colorectal cancer by binding to beta-catenin and enhancing Wnt signalling. Depletion of Sec62 sensitized the CRC cells to 5-Fu or oxaliplatin treatment. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Protein degradation | |||
| In-vitro Model | DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | DLD-1 cells were subcutaneously implanted into 4-6 weeks old female nude mice. When tumors reached a size of about 50 mm3, the nude mice were randomly divided into 6 groups. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 5 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03557 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Oxaliplatin | |
| Crosstalk ID: M6ACROT03558 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Fluorouracil | |
| Crosstalk ID: M6ACROT03602 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Oxaliplatin | |
| Crosstalk ID: M6ACROT03603 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Fluorouracil | |
| Crosstalk ID: M6ACROT03644 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
Non-coding RNA
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05312 | ||
| Epigenetic Regulator | hsa-miR-4429 | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05313 | ||
| Epigenetic Regulator | hsa-miR-4429 | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00389)
| In total 21 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE000097 | Click to Show/Hide the Full List | ||
| mod site | chr3:169969557-169969558:+ | [3] | |
| Sequence | AACATGAAAGCAGAGTGGTGATCCTACAGCTGACTGATCTC | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9; ENST00000469515.6; ENST00000460513.5; ENST00000481435.5; ENST00000487736.5 | ||
| External Link | RMBase: RNA-editing_site_101021 | ||
| mod ID: A2ISITE000098 | Click to Show/Hide the Full List | ||
| mod site | chr3:169970246-169970247:+ | [3] | |
| Sequence | TAGTGTTATTTGTGTAGCACATATTGTTACTAATGATTAAG | ||
| Transcript ID List | ENST00000481435.5; ENST00000480708.5; ENST00000337002.9; ENST00000460513.5; ENST00000487736.5; ENST00000469515.6 | ||
| External Link | RMBase: RNA-editing_site_101022 | ||
| mod ID: A2ISITE000099 | Click to Show/Hide the Full List | ||
| mod site | chr3:169971121-169971122:+ | [3] | |
| Sequence | ACTGTGGTGCCCAGGCTGGAATGCAGTGATGGCATCATAGC | ||
| Transcript ID List | ENST00000481435.5; ENST00000480708.5; ENST00000337002.9; ENST00000469515.6; ENST00000460513.5; ENST00000487736.5 | ||
| External Link | RMBase: RNA-editing_site_101023 | ||
| mod ID: A2ISITE000100 | Click to Show/Hide the Full List | ||
| mod site | chr3:169971147-169971148:+ | [3] | |
| Sequence | TGATGGCATCATAGCTCACTAGAGACTCAAACTCCTGGTCT | ||
| Transcript ID List | ENST00000481435.5; ENST00000480708.5; ENST00000487736.5; ENST00000337002.9; ENST00000460513.5; ENST00000469515.6 | ||
| External Link | RMBase: RNA-editing_site_101024 | ||
| mod ID: A2ISITE000101 | Click to Show/Hide the Full List | ||
| mod site | chr3:169971209-169971210:+ | [3] | |
| Sequence | CCTCCCAACTAGCTGGGACTACAAGTGTGTGCACCACCTCA | ||
| Transcript ID List | ENST00000337002.9; ENST00000481435.5; ENST00000480708.5; ENST00000487736.5; ENST00000460513.5; ENST00000469515.6 | ||
| External Link | RMBase: RNA-editing_site_101025 | ||
| mod ID: A2ISITE000102 | Click to Show/Hide the Full List | ||
| mod site | chr3:169971212-169971213:+ | [3] | |
| Sequence | CCCAACTAGCTGGGACTACAAGTGTGTGCACCACCTCACCT | ||
| Transcript ID List | ENST00000469515.6; ENST00000337002.9; ENST00000487736.5; ENST00000460513.5; ENST00000480708.5; ENST00000481435.5 | ||
| External Link | RMBase: RNA-editing_site_101026 | ||
| mod ID: A2ISITE000103 | Click to Show/Hide the Full List | ||
| mod site | chr3:169971294-169971295:+ | [3] | |
| Sequence | AAGTTACCTAGGCTGGTCTTAAACTCCTGGTCTCATGCGAT | ||
| Transcript ID List | ENST00000460513.5; ENST00000480708.5; ENST00000487736.5; ENST00000469515.6; ENST00000481435.5; ENST00000337002.9 | ||
| External Link | RMBase: RNA-editing_site_101027 | ||
| mod ID: A2ISITE000104 | Click to Show/Hide the Full List | ||
| mod site | chr3:169972514-169972515:+ | [4] | |
| Sequence | GCCTCAACCTACCAGGGCTCAGGTGGTCTTCCTACCTCAGC | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5; ENST00000469515.6; ENST00000460513.5; ENST00000487736.5; ENST00000481435.5 | ||
| External Link | RMBase: RNA-editing_site_101028 | ||
| mod ID: A2ISITE000105 | Click to Show/Hide the Full List | ||
| mod site | chr3:169972609-169972610:+ | [3] | |
| Sequence | TTTTTTTTTTTATGTAGAGAAGGAGTTTCACCATGTTGCCC | ||
| Transcript ID List | ENST00000337002.9; ENST00000487736.5; ENST00000480708.5; ENST00000469515.6; ENST00000481435.5; ENST00000460513.5 | ||
| External Link | RMBase: RNA-editing_site_101029 | ||
| mod ID: A2ISITE000106 | Click to Show/Hide the Full List | ||
| mod site | chr3:169972661-169972662:+ | [4] | |
| Sequence | AAGCCTGGTTTCCTGGGCTCAAGCAGTCTGCCTGCCTCAGC | ||
| Transcript ID List | ENST00000480708.5; ENST00000487736.5; ENST00000469515.6; ENST00000481435.5; ENST00000337002.9; ENST00000460513.5 | ||
| External Link | RMBase: RNA-editing_site_101030 | ||
| mod ID: A2ISITE000107 | Click to Show/Hide the Full List | ||
| mod site | chr3:169972863-169972864:+ | [3] | |
| Sequence | AACTGTAGCATCCAAAGATGATAGGGTGAGTACTCTGTAAG | ||
| Transcript ID List | ENST00000481435.5; ENST00000460513.5; ENST00000480708.5; ENST00000337002.9; ENST00000487736.5; ENST00000469515.6 | ||
| External Link | RMBase: RNA-editing_site_101031 | ||
| mod ID: A2ISITE000108 | Click to Show/Hide the Full List | ||
| mod site | chr3:169973482-169973483:+ | [5] | |
| Sequence | ACCAGCCTGGCTAACATGGTAAAATCCCATCCCTACAAAAA | ||
| Transcript ID List | ENST00000480708.5; ENST00000460513.5; ENST00000487736.5; ENST00000469515.6; rmsk_1162923; ENST00000337002.9; ENST00000481435.5 | ||
| External Link | RMBase: RNA-editing_site_101032 | ||
| mod ID: A2ISITE000109 | Click to Show/Hide the Full List | ||
| mod site | chr3:169974997-169974998:+ | [6] | |
| Sequence | GGCTTATAAAAAGAGACTATAGGCCGGTGACGATGGCTCAC | ||
| Transcript ID List | ENST00000487736.5; ENST00000460513.5; ENST00000337002.9; ENST00000469515.6; ENST00000481435.5; ENST00000480708.5 | ||
| External Link | RMBase: RNA-editing_site_101033 | ||
| mod ID: A2ISITE000110 | Click to Show/Hide the Full List | ||
| mod site | chr3:169975060-169975061:+ | [3] | |
| Sequence | GAGGCCTAGGCTGGCGGATCACTTGAGGTCAGGAGTTCAAG | ||
| Transcript ID List | ENST00000469515.6; rmsk_1162925; ENST00000487736.5; ENST00000460513.5; ENST00000337002.9; ENST00000481435.5; ENST00000480708.5 | ||
| External Link | RMBase: RNA-editing_site_101034 | ||
| mod ID: A2ISITE000111 | Click to Show/Hide the Full List | ||
| mod site | chr3:169986770-169986771:+ | [4] | |
| Sequence | GGGTCTCATTCCTGTCACCCAGGCTGGAATGCAATGGCACA | ||
| Transcript ID List | ENST00000337002.9; ENST00000469515.6; ENST00000480708.5; ENST00000470355.1 | ||
| External Link | RMBase: RNA-editing_site_101035 | ||
| mod ID: A2ISITE000112 | Click to Show/Hide the Full List | ||
| mod site | chr3:169986825-169986826:+ | [5] | |
| Sequence | CAGCCCGACTTGCTGGGGTCAGGTGATCTCCCACATCAGCC | ||
| Transcript ID List | ENST00000470355.1; ENST00000337002.9; ENST00000469515.6; ENST00000480708.5 | ||
| External Link | RMBase: RNA-editing_site_101036 | ||
| mod ID: A2ISITE000113 | Click to Show/Hide the Full List | ||
| mod site | chr3:169986864-169986865:+ | [4] | |
| Sequence | CCTACTGAGTAGCTGGAACTATAGGCACACTCCACTACACC | ||
| Transcript ID List | ENST00000470355.1; ENST00000469515.6; ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: RNA-editing_site_101037 | ||
| mod ID: A2ISITE000114 | Click to Show/Hide the Full List | ||
| mod site | chr3:169986866-169986867:+ | [4] | |
| Sequence | TACTGAGTAGCTGGAACTATAGGCACACTCCACTACACCCC | ||
| Transcript ID List | ENST00000480708.5; ENST00000469515.6; ENST00000470355.1; ENST00000337002.9 | ||
| External Link | RMBase: RNA-editing_site_101038 | ||
| mod ID: A2ISITE000115 | Click to Show/Hide the Full List | ||
| mod site | chr3:169987235-169987236:+ | [5] | |
| Sequence | CAAGGGTTCGAAACCAGCCTAGGCAACATGGCGAAACCTCG | ||
| Transcript ID List | ENST00000480708.5; ENST00000470355.1; ENST00000469515.6; ENST00000337002.9; rmsk_1162943 | ||
| External Link | RMBase: RNA-editing_site_101039 | ||
| mod ID: A2ISITE000116 | Click to Show/Hide the Full List | ||
| mod site | chr3:169991306-169991307:+ | [4] | |
| Sequence | TGACTCACAACTGCGATCCCAACTACTCGGGAGGCTGAGGT | ||
| Transcript ID List | rmsk_1162952; ENST00000337002.9; ENST00000480708.5; ENST00000470355.1 | ||
| External Link | RMBase: RNA-editing_site_101040 | ||
| mod ID: A2ISITE000117 | Click to Show/Hide the Full List | ||
| mod site | chr3:169991307-169991308:+ | [4] | |
| Sequence | GACTCACAACTGCGATCCCAACTACTCGGGAGGCTGAGGTG | ||
| Transcript ID List | ENST00000337002.9; rmsk_1162952; ENST00000480708.5; ENST00000470355.1 | ||
| External Link | RMBase: RNA-editing_site_101041 | ||
N6-methyladenosine (m6A)
| In total 108 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE062975 | Click to Show/Hide the Full List | ||
| mod site | chr3:169966803-169966804:+ | [7] | |
| Sequence | CTCGCTCCACGTCAGAGGGAACCGGGCGGAGCGGCCAACAT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; CD34; H1B; hESCs; HEK293A-TOA; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000460513.5; ENST00000461933.1 | ||
| External Link | RMBase: m6A_site_617680 | ||
| mod ID: M6ASITE062976 | Click to Show/Hide the Full List | ||
| mod site | chr3:169966839-169966840:+ | [7] | |
| Sequence | AACATGGCGGAACGCAGGAGACACAAGAAGCGGATCCAGGT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; H1B; hESCs; HEK293A-TOA; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000460513.5; ENST00000469515.6; ENST00000461933.1; ENST00000337002.9; ENST00000481435.5; ENST00000487736.5; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617681 | ||
| mod ID: M6ASITE062977 | Click to Show/Hide the Full List | ||
| mod site | chr3:169968421-169968422:+ | [7] | |
| Sequence | GACAGTGGCAGAGAGCTAAAACTTGAGCCAAACTGCCTGTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; CD34; H1B; hESCs; HEK293A-TOA; TIME; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000481435.5; ENST00000480708.5; ENST00000337002.9; rmsk_1162917; ENST00000487736.5; ENST00000460513.5; ENST00000469515.6; ENST00000461933.1 | ||
| External Link | RMBase: m6A_site_617682 | ||
| mod ID: M6ASITE062978 | Click to Show/Hide the Full List | ||
| mod site | chr3:169968432-169968433:+ | [7] | |
| Sequence | AGAGCTAAAACTTGAGCCAAACTGCCTGTTTGTACCCCACC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; CD34; HEK293A-TOA; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000461933.1; ENST00000480708.5; ENST00000337002.9; ENST00000481435.5; ENST00000469515.6; ENST00000487736.5; ENST00000460513.5; rmsk_1162917 | ||
| External Link | RMBase: m6A_site_617683 | ||
| mod ID: M6ASITE062979 | Click to Show/Hide the Full List | ||
| mod site | chr3:169975618-169975619:+ | [8] | |
| Sequence | ATGTTGCAGGAAGTTGGTGAACCATCTAAAGAAGAGAAGGC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | CD34; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000487736.5; ENST00000481435.5; ENST00000497277.1; ENST00000460513.5; ENST00000469515.6; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617684 | ||
| mod ID: M6ASITE062980 | Click to Show/Hide the Full List | ||
| mod site | chr3:169975662-169975663:+ | [9] | |
| Sequence | GGCCAAGTATCTTCGATTCAACTGTCCAACAAAGTCCACCA | ||
| Motif Score | 2.595904762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000469515.6; ENST00000337002.9; ENST00000497277.1; ENST00000487736.5; ENST00000481435.5; ENST00000460513.5 | ||
| External Link | RMBase: m6A_site_617685 | ||
| mod ID: M6ASITE062981 | Click to Show/Hide the Full List | ||
| mod site | chr3:169975670-169975671:+ | [10] | |
| Sequence | ATCTTCGATTCAACTGTCCAACAAAGTCCACCAATATGATG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000469515.6; ENST00000460513.5; ENST00000487736.5; ENST00000337002.9; ENST00000497277.1; ENST00000481435.5 | ||
| External Link | RMBase: m6A_site_617686 | ||
| mod ID: M6ASITE062982 | Click to Show/Hide the Full List | ||
| mod site | chr3:169976960-169976961:+ | [7] | |
| Sequence | TTTAGCTTCAAAAGCAGTGGACTGTCTTTTGGATTCAAAGT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; CD34; BGC823; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000497277.1; ENST00000487736.5; ENST00000469515.6; ENST00000460513.5; ENST00000481435.5; ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617687 | ||
| mod ID: M6ASITE062983 | Click to Show/Hide the Full List | ||
| mod site | chr3:169977016-169977017:+ | [10] | |
| Sequence | AAGGAGAGGAAGCTTTATTTACAACCAGGGAGTCTGTGGTT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000460513.5; ENST00000487736.5; ENST00000337002.9; ENST00000480708.5; ENST00000497277.1; ENST00000481435.5; ENST00000469515.6 | ||
| External Link | RMBase: m6A_site_617688 | ||
| mod ID: M6ASITE062984 | Click to Show/Hide the Full List | ||
| mod site | chr3:169977038-169977039:+ | [9] | |
| Sequence | AACCAGGGAGTCTGTGGTTGACTACTGCAACAGGTACTGTT | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000497277.1; ENST00000481435.5; ENST00000460513.5; ENST00000337002.9; ENST00000487736.5; ENST00000469515.6; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617689 | ||
| mod ID: M6ASITE062985 | Click to Show/Hide the Full List | ||
| mod site | chr3:169982768-169982769:+ | [7] | |
| Sequence | GAAAATGAAATATGATAAAGACATAAAGAAAGAAAAAGATA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; HEK293T; hESC-HEK293T; BGC823; hNPCs; hESCs; fibroblasts; A549; LCLs; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000481435.5; ENST00000469515.6; ENST00000460513.5; ENST00000487736.5; ENST00000337002.9; rmsk_1162935; ENST00000497277.1 | ||
| External Link | RMBase: m6A_site_617690 | ||
| mod ID: M6ASITE062986 | Click to Show/Hide the Full List | ||
| mod site | chr3:169982857-169982858:+ | [7] | |
| Sequence | AAAATATAAAGGATGAGAAGACAAAAAAAGAAAAAGAGAAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; hESC-HEK293T; BGC823; hNPCs; hESCs; fibroblasts; A549; LCLs; MM6; Huh7; Jurkat; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000497277.1; ENST00000460513.5; ENST00000469515.6; ENST00000337002.9; ENST00000480708.5; ENST00000481435.5; rmsk_1162935 | ||
| External Link | RMBase: m6A_site_617691 | ||
| mod ID: M6ASITE062987 | Click to Show/Hide the Full List | ||
| mod site | chr3:169983166-169983167:+ | [7] | |
| Sequence | TTGTGTTTTCATAGGAGGAAACTCCAGGAACTCCTAAAAAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; hNPCs; fibroblasts; A549; LCLs; MM6; Huh7; Jurkat; endometrial; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000460513.5; ENST00000480708.5; ENST00000481435.5; ENST00000469890.1; ENST00000469515.6; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617692 | ||
| mod ID: M6ASITE062988 | Click to Show/Hide the Full List | ||
| mod site | chr3:169983175-169983176:+ | [7] | |
| Sequence | CATAGGAGGAAACTCCAGGAACTCCTAAAAAGAAGGAAACT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; hNPCs; fibroblasts; A549; LCLs; MM6; Huh7; Jurkat; endometrial; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000337002.9; ENST00000481435.5; ENST00000469515.6; ENST00000480708.5; ENST00000460513.5; ENST00000469890.1 | ||
| External Link | RMBase: m6A_site_617693 | ||
| mod ID: M6ASITE062989 | Click to Show/Hide the Full List | ||
| mod site | chr3:169983193-169983194:+ | [11] | |
| Sequence | GAACTCCTAAAAAGAAGGAAACTAAGAAAAAATTCAAACTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HepG2; HEK293T; hNPCs; A549; LCLs; MM6; Huh7; Jurkat; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000469890.1; ENST00000481435.5; ENST00000469515.6; ENST00000337002.9; ENST00000480708.5; ENST00000460513.5 | ||
| External Link | RMBase: m6A_site_617694 | ||
| mod ID: M6ASITE062990 | Click to Show/Hide the Full List | ||
| mod site | chr3:169983210-169983211:+ | [11] | |
| Sequence | GAAACTAAGAAAAAATTCAAACTTGAGCCACATGATGATCA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HepG2; HEK293T; hNPCs; A549; LCLs; MM6; Huh7; Jurkat; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000460513.5; ENST00000469890.1; ENST00000481435.5; ENST00000469515.6; ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617695 | ||
| mod ID: M6ASITE062991 | Click to Show/Hide the Full List | ||
| mod site | chr3:169983219-169983220:+ | [10] | |
| Sequence | AAAAAATTCAAACTTGAGCCACATGATGATCAGGTTTTTCT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000469890.1; ENST00000480708.5; ENST00000337002.9; ENST00000460513.5; ENST00000481435.5; ENST00000469515.6 | ||
| External Link | RMBase: m6A_site_617696 | ||
| mod ID: M6ASITE062992 | Click to Show/Hide the Full List | ||
| mod site | chr3:169985158-169985159:+ | [7] | |
| Sequence | TAACTGCGCTTTTATCCCAGACTGGTTCATCATTACATTAG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; GM12878 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9; ENST00000469515.6; ENST00000481435.5; ENST00000470355.1 | ||
| External Link | RMBase: m6A_site_617697 | ||
| mod ID: M6ASITE062993 | Click to Show/Hide the Full List | ||
| mod site | chr3:169985181-169985182:+ | [7] | |
| Sequence | GGTTCATCATTACATTAGGGACAAAAAGGTTGGCAGTTGCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; GM12878 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000470355.1; ENST00000469515.6; ENST00000337002.9; ENST00000480708.5; ENST00000481435.5 | ||
| External Link | RMBase: m6A_site_617698 | ||
| mod ID: M6ASITE062994 | Click to Show/Hide the Full List | ||
| mod site | chr3:169985840-169985841:+ | [7] | |
| Sequence | ATGACCCAGTTCACTTTAAAACATTTGTCATGGGATTAATT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; GM12878; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000469515.6; ENST00000481435.5; ENST00000470355.1; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617699 | ||
| mod ID: M6ASITE062995 | Click to Show/Hide the Full List | ||
| mod site | chr3:169992667-169992668:+ | [12] | |
| Sequence | TTTGGTTCTTGCCAAATCTGACTGCTGATGTGGGCTTCATT | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5; ENST00000470355.1 | ||
| External Link | RMBase: m6A_site_617700 | ||
| mod ID: M6ASITE062996 | Click to Show/Hide the Full List | ||
| mod site | chr3:169992689-169992690:+ | [13] | |
| Sequence | TGCTGATGTGGGCTTCATTGACTCCTTCAGGCCTCTGTACA | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9; ENST00000470355.1 | ||
| External Link | RMBase: m6A_site_617701 | ||
| mod ID: M6ASITE062997 | Click to Show/Hide the Full List | ||
| mod site | chr3:169992719-169992720:+ | [10] | |
| Sequence | GCCTCTGTACACACATGAATACAAAGGACCAAAAGCAGACT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9; ENST00000470355.1 | ||
| External Link | RMBase: m6A_site_617702 | ||
| mod ID: M6ASITE062998 | Click to Show/Hide the Full List | ||
| mod site | chr3:169992726-169992727:+ | [7] | |
| Sequence | TACACACATGAATACAAAGGACCAAAAGCAGACTTAAAGAA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617703 | ||
| mod ID: M6ASITE062999 | Click to Show/Hide the Full List | ||
| mod site | chr3:169992737-169992738:+ | [7] | |
| Sequence | ATACAAAGGACCAAAAGCAGACTTAAAGAAAGATGAGAAGT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617704 | ||
| mod ID: M6ASITE063000 | Click to Show/Hide the Full List | ||
| mod site | chr3:169992763-169992764:+ | [7] | |
| Sequence | AGAAAGATGAGAAGTCTGAAACCAAAAAGCAACAGAAGTCC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617705 | ||
| mod ID: M6ASITE063001 | Click to Show/Hide the Full List | ||
| mod site | chr3:169992785-169992786:+ | [14] | |
| Sequence | CAAAAAGCAACAGAAGTCCGACAGTGAGGAAAAGTCAGACA | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | brain | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617706 | ||
| mod ID: M6ASITE063002 | Click to Show/Hide the Full List | ||
| mod site | chr3:169992803-169992804:+ | [7] | |
| Sequence | CGACAGTGAGGAAAAGTCAGACAGTGAGAAAAAGGAAGATG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; brain; liver; A549; hESC-HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617707 | ||
| mod ID: M6ASITE063003 | Click to Show/Hide the Full List | ||
| mod site | chr3:169992840-169992841:+ | [7] | |
| Sequence | GATGAGGAGGGGAAAGTAGGACCAGGAAATCATGGAACAGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617708 | ||
| mod ID: M6ASITE063004 | Click to Show/Hide the Full List | ||
| mod site | chr3:169992856-169992857:+ | [7] | |
| Sequence | TAGGACCAGGAAATCATGGAACAGAAGGCTCGGGGGGAGAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; brain; kidney; A549; hESC-HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-REF-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617709 | ||
| mod ID: M6ASITE063005 | Click to Show/Hide the Full List | ||
| mod site | chr3:169992887-169992888:+ | [7] | |
| Sequence | GGGGGGAGAACGGCATTCAGACACGGACAGTGACAGGAGGG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617710 | ||
| mod ID: M6ASITE063006 | Click to Show/Hide the Full List | ||
| mod site | chr3:169992893-169992894:+ | [7] | |
| Sequence | AGAACGGCATTCAGACACGGACAGTGACAGGAGGGAAGATG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617711 | ||
| mod ID: M6ASITE063007 | Click to Show/Hide the Full List | ||
| mod site | chr3:169992899-169992900:+ | [9] | |
| Sequence | GCATTCAGACACGGACAGTGACAGGAGGGAAGATGATCGAT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617712 | ||
| mod ID: M6ASITE063008 | Click to Show/Hide the Full List | ||
| mod site | chr3:169992926-169992927:+ | [9] | |
| Sequence | GGAAGATGATCGATCCCAGCACAGTAGTGGAAATGGAAATG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617713 | ||
| mod ID: M6ASITE063009 | Click to Show/Hide the Full List | ||
| mod site | chr3:169992961-169992962:+ | [10] | |
| Sequence | GAAATGATTTTGAAATGATAACAAAAGAGGAACTGGAACAG | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T; AML | ||
| Seq Type List | MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617714 | ||
| mod ID: M6ASITE063010 | Click to Show/Hide the Full List | ||
| mod site | chr3:169992972-169992973:+ | [7] | |
| Sequence | GAAATGATAACAAAAGAGGAACTGGAACAGCAAACAGATGG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617715 | ||
| mod ID: M6ASITE063011 | Click to Show/Hide the Full List | ||
| mod site | chr3:169992978-169992979:+ | [7] | |
| Sequence | ATAACAAAAGAGGAACTGGAACAGCAAACAGATGGGGATTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617716 | ||
| mod ID: M6ASITE063012 | Click to Show/Hide the Full List | ||
| mod site | chr3:169992985-169992986:+ | [7] | |
| Sequence | AAGAGGAACTGGAACAGCAAACAGATGGGGATTGTGAAGAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617717 | ||
| mod ID: M6ASITE063013 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993033-169993034:+ | [7] | |
| Sequence | AAGAGGAAAATGATGGAGAAACACCTAAATCTTCACATGAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617718 | ||
| mod ID: M6ASITE063014 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993047-169993048:+ | [10] | |
| Sequence | GGAGAAACACCTAAATCTTCACATGAAAAATCATAATCTGA | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617719 | ||
| mod ID: M6ASITE063015 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993067-169993068:+ | [12] | |
| Sequence | ACATGAAAAATCATAATCTGACTAATTTTGGGACTGAATGA | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617720 | ||
| mod ID: M6ASITE063016 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993079-169993080:+ | [7] | |
| Sequence | ATAATCTGACTAATTTTGGGACTGAATGAATAAGTACAAGA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617721 | ||
| mod ID: M6ASITE063017 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993094-169993095:+ | [9] | |
| Sequence | TTGGGACTGAATGAATAAGTACAAGAGGTTGGATTTTCTAT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617722 | ||
| mod ID: M6ASITE063018 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993136-169993137:+ | [7] | |
| Sequence | TTGGCTGATTACCATATTGAACACATGGCATTTGTAGCATT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617723 | ||
| mod ID: M6ASITE063019 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993171-169993172:+ | [9] | |
| Sequence | AGCATTCTTTAAATCTATCTACTGAAATGTATTTGACATTC | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617724 | ||
| mod ID: M6ASITE063020 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993186-169993187:+ | [9] | |
| Sequence | TATCTACTGAAATGTATTTGACATTCAAGCAGTTATATTCG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617725 | ||
| mod ID: M6ASITE063021 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993287-169993288:+ | [9] | |
| Sequence | TTTATCCATTTGATAATTTTACAGTAAGTAGGTCTCATTCA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617726 | ||
| mod ID: M6ASITE063022 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993313-169993314:+ | [9] | |
| Sequence | AGTAGGTCTCATTCATTTTGACAGTTATCAAAGATGTACTT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617727 | ||
| mod ID: M6ASITE063023 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993330-169993331:+ | [9] | |
| Sequence | TTGACAGTTATCAAAGATGTACTTTCCACAGTTAAATTTAC | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617728 | ||
| mod ID: M6ASITE063024 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993349-169993350:+ | [9] | |
| Sequence | TACTTTCCACAGTTAAATTTACATTAATGGCAATTTTTGAT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617729 | ||
| mod ID: M6ASITE063025 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993394-169993395:+ | [9] | |
| Sequence | TTATGGCTTTTTACTGTTAGACTAATCAAAAATAACTTTAA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617730 | ||
| mod ID: M6ASITE063026 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993420-169993421:+ | [9] | |
| Sequence | CAAAAATAACTTTAAAAGGAACAAAGAAACTCCAACATTTC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617731 | ||
| mod ID: M6ASITE063027 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993434-169993435:+ | [9] | |
| Sequence | AAAGGAACAAAGAAACTCCAACATTTCACATTATGCATAGT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617732 | ||
| mod ID: M6ASITE063028 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993491-169993492:+ | [9] | |
| Sequence | AGTTTCTTTAAGATGTGTAAACTCATTGTCCTTGATAGTTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617733 | ||
| mod ID: M6ASITE063029 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993591-169993592:+ | [7] | |
| Sequence | TTCAAAACTATTTTGTGGAAACAAGCCAACTAGTAACAATG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617734 | ||
| mod ID: M6ASITE063030 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993599-169993600:+ | [9] | |
| Sequence | TATTTTGTGGAAACAAGCCAACTAGTAACAATGCAGCAACA | ||
| Motif Score | 2.595904762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617735 | ||
| mod ID: M6ASITE063031 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993617-169993618:+ | [10] | |
| Sequence | CAACTAGTAACAATGCAGCAACACTTCTGGTTTAGCTAAAT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617736 | ||
| mod ID: M6ASITE063032 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993619-169993620:+ | [9] | |
| Sequence | ACTAGTAACAATGCAGCAACACTTCTGGTTTAGCTAAATTA | ||
| Motif Score | 2.506922619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617737 | ||
| mod ID: M6ASITE063033 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993661-169993662:+ | [10] | |
| Sequence | TTTTCCAATGTAGGAAATCCACACTGATTTGTACGTCTGAC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617738 | ||
| mod ID: M6ASITE063034 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993673-169993674:+ | [9] | |
| Sequence | GGAAATCCACACTGATTTGTACGTCTGACTGAGAGAAAGAT | ||
| Motif Score | 2.830077381 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617739 | ||
| mod ID: M6ASITE063035 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993680-169993681:+ | [9] | |
| Sequence | CACACTGATTTGTACGTCTGACTGAGAGAAAGATGGTCGTC | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617740 | ||
| mod ID: M6ASITE063036 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993718-169993719:+ | [7] | |
| Sequence | GTCTCCAGCAGAGAAAGTGAACAGCATTTGTTGGAAGGTGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: m6A_site_617741 | ||
| mod ID: M6ASITE063037 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993793-169993794:+ | [9] | |
| Sequence | TAACGTAAAGTGTATTCTGTACATAATTTACAAATAAAACA | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617742 | ||
| mod ID: M6ASITE063038 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993802-169993803:+ | [9] | |
| Sequence | GTGTATTCTGTACATAATTTACAAATAAAACATTTTATTTT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000480708.5; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617743 | ||
| mod ID: M6ASITE063039 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993855-169993856:+ | [9] | |
| Sequence | TTATTTAGATATTTCTCAACACTTAAATTCATAAAATTAAG | ||
| Motif Score | 2.506922619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617744 | ||
| mod ID: M6ASITE063040 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993876-169993877:+ | [9] | |
| Sequence | CTTAAATTCATAAAATTAAGACCATGTAAGGGTATGTTTTT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617745 | ||
| mod ID: M6ASITE063041 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993927-169993928:+ | [7] | |
| Sequence | AAGTTTGAGTAACCCACAGAACATCTGTGATCTTTCTACAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617746 | ||
| mod ID: M6ASITE063042 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993967-169993968:+ | [14] | |
| Sequence | GCAGCTTCAGTTTTGTGCCAACATTCCATGTATTTTGAATA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | brain; liver; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617747 | ||
| mod ID: M6ASITE063043 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993997-169993998:+ | [7] | |
| Sequence | TATTTTGAATATGAGCAAAAACTGATCTTAAGAGCAGACTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617748 | ||
| mod ID: M6ASITE063044 | Click to Show/Hide the Full List | ||
| mod site | chr3:169994014-169994015:+ | [7] | |
| Sequence | AAAACTGATCTTAAGAGCAGACTTAAAGTAGCTTTGTACGC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617749 | ||
| mod ID: M6ASITE063045 | Click to Show/Hide the Full List | ||
| mod site | chr3:169994031-169994032:+ | [9] | |
| Sequence | CAGACTTAAAGTAGCTTTGTACGCCTTAATGTTCATTTTGA | ||
| Motif Score | 2.830077381 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617750 | ||
| mod ID: M6ASITE063046 | Click to Show/Hide the Full List | ||
| mod site | chr3:169994068-169994069:+ | [9] | |
| Sequence | TTGATTTATTTTAAATCTTTACATTCAGAAATGAGATACTG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617751 | ||
| mod ID: M6ASITE063047 | Click to Show/Hide the Full List | ||
| mod site | chr3:169994172-169994173:+ | [10] | |
| Sequence | AATAAAGATAATGAAAGAAAACATAAGAGAACTATTGGACT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | rmsk_1162958; ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617752 | ||
| mod ID: M6ASITE063048 | Click to Show/Hide the Full List | ||
| mod site | chr3:169994237-169994238:+ | [10] | |
| Sequence | AGTTTGACTTTACGTCTTATACATCTTAGTTACAAGTCTTT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617753 | ||
| mod ID: M6ASITE063049 | Click to Show/Hide the Full List | ||
| mod site | chr3:169994307-169994308:+ | [9] | |
| Sequence | CATTCCTTGGAGTTATACTAACTAATAATGAATATTAAATG | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617754 | ||
| mod ID: M6ASITE063050 | Click to Show/Hide the Full List | ||
| mod site | chr3:169994490-169994491:+ | [9] | |
| Sequence | GTATCGACACATTTTATTAAACTTGGCTTGATTGCTTCTTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617755 | ||
| mod ID: M6ASITE063051 | Click to Show/Hide the Full List | ||
| mod site | chr3:169994531-169994532:+ | [14] | |
| Sequence | ATAGTAGTCCCTGTAAGTGGACATAAATGTGGCTATTTGAT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | brain | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617756 | ||
| mod ID: M6ASITE063052 | Click to Show/Hide the Full List | ||
| mod site | chr3:169994581-169994582:+ | [9] | |
| Sequence | ACTATGTCAAGGCTGCAATCACTTTAATAGTTTCTTTTTAG | ||
| Motif Score | 2.469291667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617757 | ||
| mod ID: M6ASITE063053 | Click to Show/Hide the Full List | ||
| mod site | chr3:169994661-169994662:+ | [9] | |
| Sequence | CGTCCCATTTGCAGAGCTCTACTACTTCATTTCCACAGTAA | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617758 | ||
| mod ID: M6ASITE063054 | Click to Show/Hide the Full List | ||
| mod site | chr3:169994664-169994665:+ | [9] | |
| Sequence | CCCATTTGCAGAGCTCTACTACTTCATTTCCACAGTAAGCA | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617759 | ||
| mod ID: M6ASITE063055 | Click to Show/Hide the Full List | ||
| mod site | chr3:169994747-169994748:+ | [10] | |
| Sequence | ATACCTATTTTGGGGTATAAACATGGTGTTTTTCAGAAATA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617760 | ||
| mod ID: M6ASITE063056 | Click to Show/Hide the Full List | ||
| mod site | chr3:169994870-169994871:+ | [10] | |
| Sequence | TAATTTGTCTAGTCTTAAAGACATTTGGAGCATGGCCAACA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617761 | ||
| mod ID: M6ASITE063057 | Click to Show/Hide the Full List | ||
| mod site | chr3:169994888-169994889:+ | [9] | |
| Sequence | AGACATTTGGAGCATGGCCAACAGGAAGGTCAGCTTCTTAC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617762 | ||
| mod ID: M6ASITE063058 | Click to Show/Hide the Full List | ||
| mod site | chr3:169994907-169994908:+ | [9] | |
| Sequence | AACAGGAAGGTCAGCTTCTTACTGATGTGTGTGGATTCTTA | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617763 | ||
| mod ID: M6ASITE063059 | Click to Show/Hide the Full List | ||
| mod site | chr3:169994927-169994928:+ | [9] | |
| Sequence | ACTGATGTGTGTGGATTCTTACAGAATGTTAGTACAAATTT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617764 | ||
| mod ID: M6ASITE063060 | Click to Show/Hide the Full List | ||
| mod site | chr3:169994940-169994941:+ | [9] | |
| Sequence | GATTCTTACAGAATGTTAGTACAAATTTTTATAAATATTTG | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617765 | ||
| mod ID: M6ASITE063061 | Click to Show/Hide the Full List | ||
| mod site | chr3:169995059-169995060:+ | [7] | |
| Sequence | ATTCAAGTTGTGTACTTTAAACAAATATAAAAATCAAAAGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617766 | ||
| mod ID: M6ASITE063062 | Click to Show/Hide the Full List | ||
| mod site | chr3:169995091-169995092:+ | [9] | |
| Sequence | ATCAAAAGGCTGCCTCTATTACAATCCTCAGTGGAATAGAT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617767 | ||
| mod ID: M6ASITE063063 | Click to Show/Hide the Full List | ||
| mod site | chr3:169995204-169995205:+ | [7] | |
| Sequence | ATGGCGATTTTTTTTTAAGAACTTCAGATTTGCTATGCTGC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617768 | ||
| mod ID: M6ASITE063064 | Click to Show/Hide the Full List | ||
| mod site | chr3:169995402-169995403:+ | [9] | |
| Sequence | TATTTTTCCTTTTCCCTCTTACTGGATTTTTCAATTTTCAA | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617769 | ||
| mod ID: M6ASITE063065 | Click to Show/Hide the Full List | ||
| mod site | chr3:169995440-169995441:+ | [9] | |
| Sequence | CAAACCATATGGCCTAGGTAACTTTAATTGAAATAATATAG | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617770 | ||
| mod ID: M6ASITE063066 | Click to Show/Hide the Full List | ||
| mod site | chr3:169995505-169995506:+ | [10] | |
| Sequence | GTAATAAATTTTAATATATCACATTGAAATTATGCAGTGGT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617771 | ||
| mod ID: M6ASITE063067 | Click to Show/Hide the Full List | ||
| mod site | chr3:169995562-169995563:+ | [10] | |
| Sequence | TCCCCCCTCTTTCTGGTTTTACATCATGAGGATCAGTTAGC | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617772 | ||
| mod ID: M6ASITE063068 | Click to Show/Hide the Full List | ||
| mod site | chr3:169995588-169995589:+ | [10] | |
| Sequence | TGAGGATCAGTTAGCTGCTGACAAGAAAAGTCATCTTGTGT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617773 | ||
| mod ID: M6ASITE063069 | Click to Show/Hide the Full List | ||
| mod site | chr3:169995647-169995648:+ | [9] | |
| Sequence | TTTTTTTAAATCTATTAGGAACTGGAAAACAGCAAGTATGG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617774 | ||
| mod ID: M6ASITE063070 | Click to Show/Hide the Full List | ||
| mod site | chr3:169995823-169995824:+ | [7] | |
| Sequence | TGTAAATTTCTGTTGTAAAGACACCTTATTTAATATAATCG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617775 | ||
| mod ID: M6ASITE063071 | Click to Show/Hide the Full List | ||
| mod site | chr3:169995907-169995908:+ | [9] | |
| Sequence | GCCTGAATGAAGCTTATCTGACACATAAGCTTCATAAGGCA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617776 | ||
| mod ID: M6ASITE063072 | Click to Show/Hide the Full List | ||
| mod site | chr3:169995953-169995954:+ | [7] | |
| Sequence | CAGCCTTTTTACCCTTAGGAACAGCTTTTTGCCCTTAGCAC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617777 | ||
| mod ID: M6ASITE063073 | Click to Show/Hide the Full List | ||
| mod site | chr3:169995995-169995996:+ | [7] | |
| Sequence | ATGCTTGGGGGCCATTTTAAACAGTGAAATCAAAAAAAGAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617778 | ||
| mod ID: M6ASITE063074 | Click to Show/Hide the Full List | ||
| mod site | chr3:169996015-169996016:+ | [7] | |
| Sequence | ACAGTGAAATCAAAAAAAGAACAGAAATGCAAAAACCATGG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617779 | ||
| mod ID: M6ASITE063075 | Click to Show/Hide the Full List | ||
| mod site | chr3:169996029-169996030:+ | [7] | |
| Sequence | AAAAGAACAGAAATGCAAAAACCATGGCATTAGCGTAAAAA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617780 | ||
| mod ID: M6ASITE063076 | Click to Show/Hide the Full List | ||
| mod site | chr3:169996052-169996053:+ | [7] | |
| Sequence | ATGGCATTAGCGTAAAAAGGACACGTTTATAGTATGAAAGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617781 | ||
| mod ID: M6ASITE063077 | Click to Show/Hide the Full List | ||
| mod site | chr3:169996077-169996078:+ | [7] | |
| Sequence | TTTATAGTATGAAAGCTGGAACATGGCCTTGTTCAACCTCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617782 | ||
| mod ID: M6ASITE063078 | Click to Show/Hide the Full List | ||
| mod site | chr3:169996160-169996161:+ | [10] | |
| Sequence | ATTTTAGGTTAGGCAAATTCACAAATACGGAATCCATGAAT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617783 | ||
| mod ID: M6ASITE063079 | Click to Show/Hide the Full List | ||
| mod site | chr3:169996192-169996193:+ | [9] | |
| Sequence | TCCATGAATAACGAGGATCAACTGTACTTAACTATATTTTA | ||
| Motif Score | 2.595904762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617784 | ||
| mod ID: M6ASITE063080 | Click to Show/Hide the Full List | ||
| mod site | chr3:169996221-169996222:+ | [9] | |
| Sequence | AACTATATTTTAAAGCTGTGACATTTATCTACCTGGTTTAT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617785 | ||
| mod ID: M6ASITE063081 | Click to Show/Hide the Full List | ||
| mod site | chr3:169996344-169996345:+ | [9] | |
| Sequence | TTGTGTGGTAATTTGCTTTTACTGTTTCCACAGTATTTCAG | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617786 | ||
| mod ID: M6ASITE063082 | Click to Show/Hide the Full List | ||
| mod site | chr3:169996379-169996380:+ | [9] | |
| Sequence | TTTCAGGTCTTTGCTCATTCACTCAGTAATACTGAGCACTT | ||
| Motif Score | 2.469291667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: m6A_site_617787 | ||
Pseudouridine (Pseudo)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: PSESITE000191 | Click to Show/Hide the Full List | ||
| mod site | chr3:169993109-169993110:+ | [15] | |
| Sequence | TAAGTACAAGAGGTTGGATTTTCTATGTTGGCTGATTACCA | ||
| Transcript ID List | ENST00000337002.9; ENST00000480708.5 | ||
| External Link | RMBase: Pseudo_site_3636 | ||
| mod ID: PSESITE000192 | Click to Show/Hide the Full List | ||
| mod site | chr3:169994312-169994313:+ | [15] | |
| Sequence | CTTGGAGTTATACTAACTAATAATGAATATTAAATGATTTT | ||
| Transcript ID List | ENST00000337002.9 | ||
| External Link | RMBase: Pseudo_site_3637 | ||
References