m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00234)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
E2F1
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
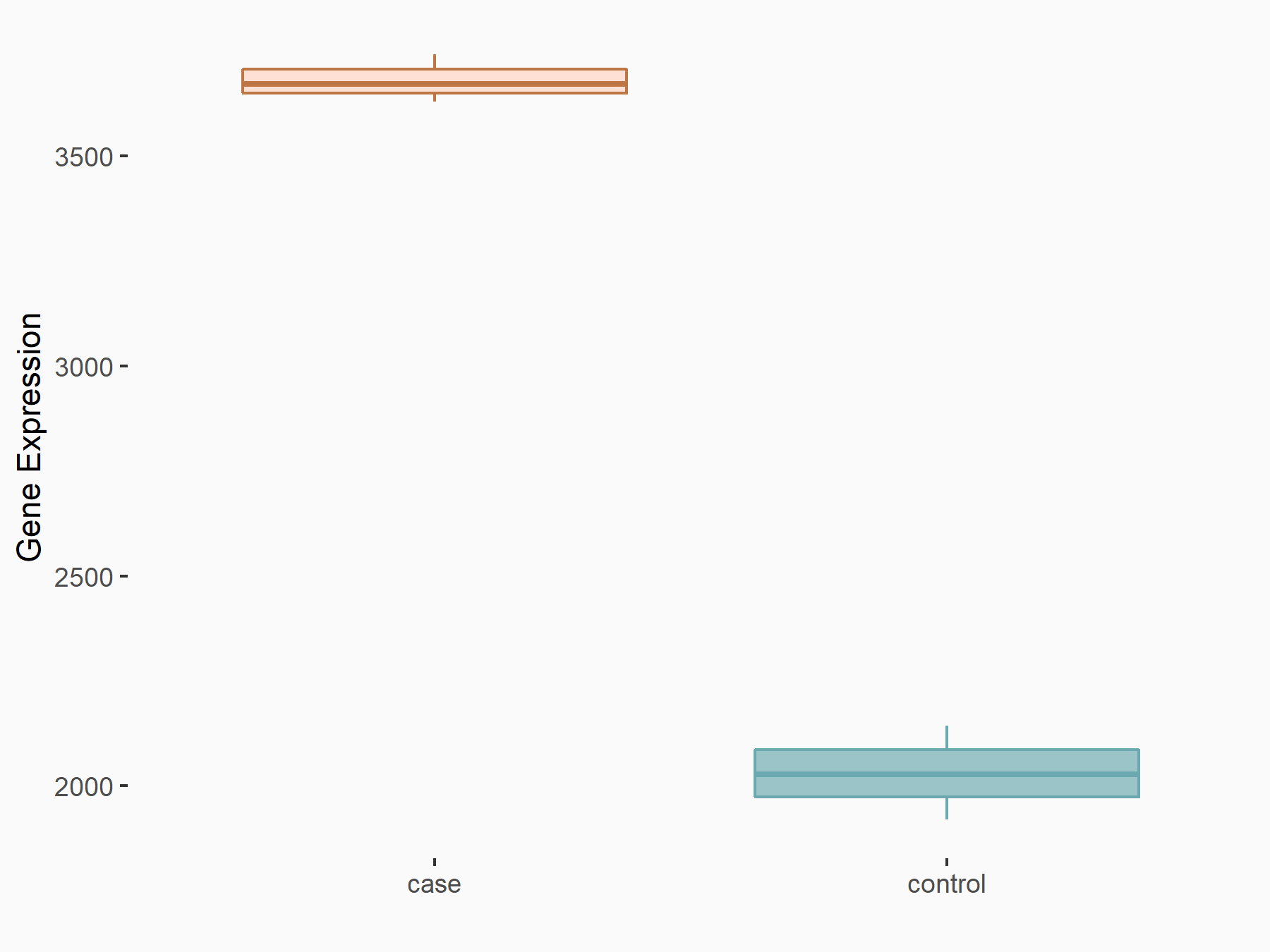  |
logFC: 8.59E-01 p-value: 2.73E-57 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between E2F1 and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.01E+00 | GSE60213 |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | This study revealed that m6A methylation is closely related to the poor prognosis of non-small cell lung cancer patients via interference with the TIME, which suggests that m6A plays a role in optimizing individualized immunotherapy management and improving prognosis. The expression levels of METTL3, FTO and YTHDF1 in non-small cell lung cancer were changed. Patients in Cluster 1 had lower immunoscores, higher programmed death-ligand 1 (PD-L1) expression, and shorter overall survival compared to patients in Cluster 2. The hallmarks of the Myelocytomatosis viral oncogene (MYC) targets, Transcription factor E2F1 (E2F1) targets were significantly enriched. | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 can regulate the expression of MALAT1 through m6A, mediate the Transcription factor E2F1 (E2F1)/AGR2 axis, and promote the adriamycin resistance of breast cancer. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Doxil | Approved | ||
| In-vitro Model | MCF7-DoxR (Adriamycin-resistant cell line MCF7-DoxR) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| In-vivo Model | Once the tumor volume increased to about 1 cm3, six groups of MCF7 bearing mice (n = 10 in each group) were injected with PBS (0.1 ml, caudal vein) and adriamycin (0.1 ml, 10 mg/kg), respectively. When the tumor reached 1.5 cm in any direction (defined as event-free survival analysis), 10 mice in each group were selected to measure the tumor size and weight on the 12th day after adriamycin injection. | |||
YTH domain-containing family protein 1 (YTHDF1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
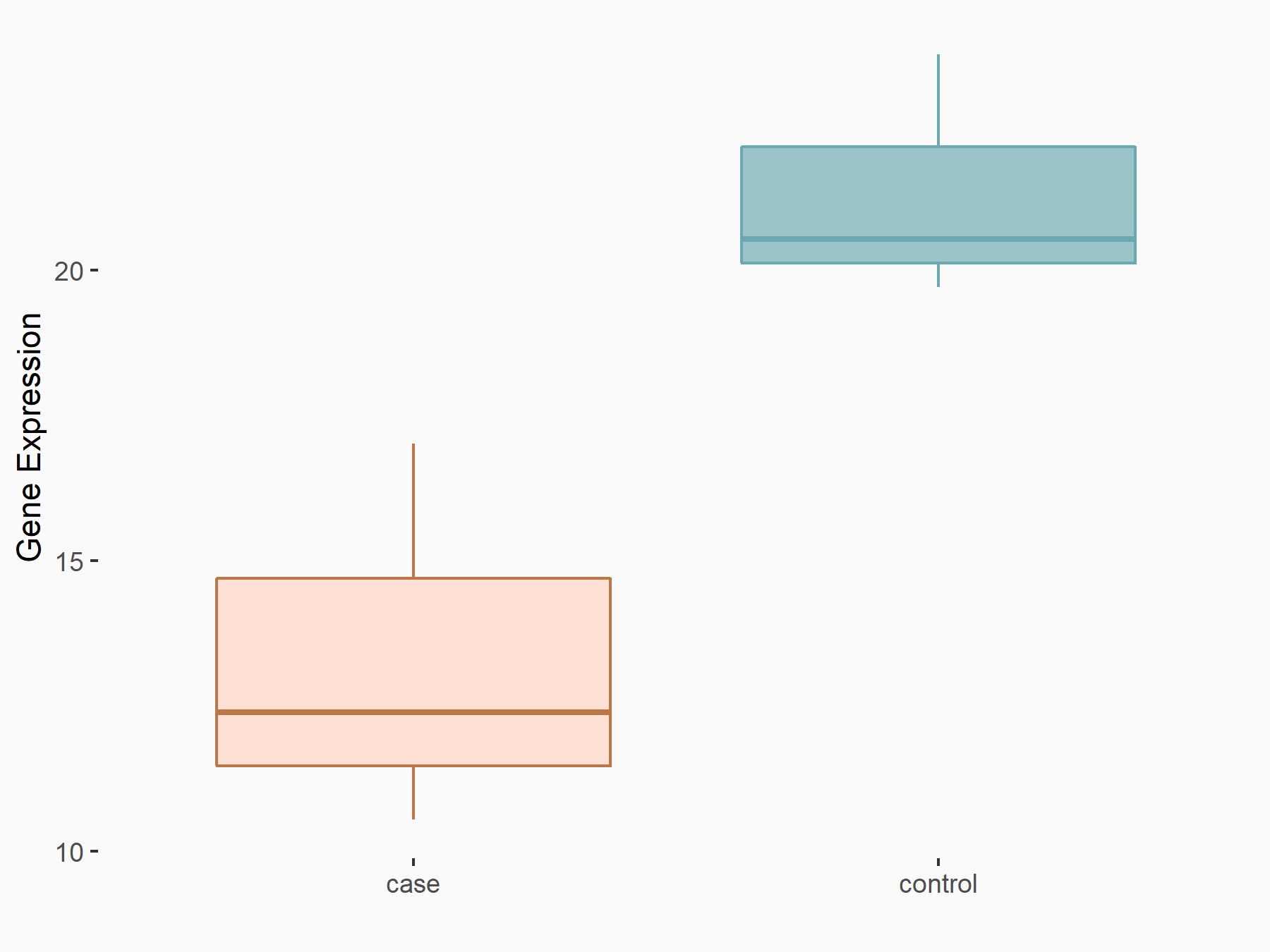  |
logFC: -6.62E-01 p-value: 3.39E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | This study revealed that m6A methylation is closely related to the poor prognosis of non-small cell lung cancer patients via interference with the TIME, which suggests that m6A plays a role in optimizing individualized immunotherapy management and improving prognosis. The expression levels of METTL3, FTO and YTHDF1 in non-small cell lung cancer were changed. Patients in Cluster 1 had lower immunoscores, higher programmed death-ligand 1 (PD-L1) expression, and shorter overall survival compared to patients in Cluster 2. The hallmarks of the Myelocytomatosis viral oncogene (MYC) targets, Transcription factor E2F1 (E2F1) targets were significantly enriched. | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | |||
Fat mass and obesity-associated protein (FTO) [ERASER]
| In total 3 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | This study revealed that m6A methylation is closely related to the poor prognosis of non-small cell lung cancer patients via interference with the TIME, which suggests that m6A plays a role in optimizing individualized immunotherapy management and improving prognosis. The expression levels of METTL3, FTO and YTHDF1 in non-small cell lung cancer were changed. Patients in Cluster 1 had lower immunoscores, higher programmed death-ligand 1 (PD-L1) expression, and shorter overall survival compared to patients in Cluster 2. The hallmarks of the Myelocytomatosis viral oncogene (MYC) targets, Transcription factor E2F1 (E2F1) targets were significantly enriched. | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | FTO upregulated the expression of Transcription factor E2F1 (E2F1) by inhibiting the m6A modification of E2F1. The FTO/E2F1/NELL2 axis modulated NSCLC cell viability, migration, and invasion in vitro as well as affected NSCLC tumor growth and metastasis in vivo. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Cell Process | Cell viability | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 |
| NCI-H23 | Lung adenocarcinoma | Homo sapiens | CVCL_1547 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| 16HBE14o- | Normal | Homo sapiens | CVCL_0112 | |
| In-vivo Model | The nude mice were maintained under pathogen-free conditions and kept under timed lighting conditions mandated by the committee with food and water provided ad libitum. For xenograft experiments, nude mice were injected subcutaneously with 5 × 106 cells resuspended in 0.1 mL PBS. When a tumor was palpable, it was measured every 3 days. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | FTO interacts with transcripts of Transcription factor E2F1 (E2F1) and Myc, inhibition of FTO significantly impairs the translation efficiency of E2F1 and Myc.FTO plays important oncogenic role in regulating cervical cancer cells' proliferation. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Cervical cancer | ICD-11: 2C77 | ||
| Cell Process | Cell proliferation and migration | |||
| In-vitro Model | HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
Lung cancer [ICD-11: 2C25]
| In total 4 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | This study revealed that m6A methylation is closely related to the poor prognosis of non-small cell lung cancer patients via interference with the TIME, which suggests that m6A plays a role in optimizing individualized immunotherapy management and improving prognosis. The expression levels of METTL3, FTO and YTHDF1 in non-small cell lung cancer were changed. Patients in Cluster 1 had lower immunoscores, higher programmed death-ligand 1 (PD-L1) expression, and shorter overall survival compared to patients in Cluster 2. The hallmarks of the Myelocytomatosis viral oncogene (MYC) targets, Transcription factor E2F1 (E2F1) targets were significantly enriched. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | FTO upregulated the expression of Transcription factor E2F1 (E2F1) by inhibiting the m6A modification of E2F1. The FTO/E2F1/NELL2 axis modulated NSCLC cell viability, migration, and invasion in vitro as well as affected NSCLC tumor growth and metastasis in vivo. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell viability | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 |
| NCI-H23 | Lung adenocarcinoma | Homo sapiens | CVCL_1547 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| 16HBE14o- | Normal | Homo sapiens | CVCL_0112 | |
| In-vivo Model | The nude mice were maintained under pathogen-free conditions and kept under timed lighting conditions mandated by the committee with food and water provided ad libitum. For xenograft experiments, nude mice were injected subcutaneously with 5 × 106 cells resuspended in 0.1 mL PBS. When a tumor was palpable, it was measured every 3 days. | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | This study revealed that m6A methylation is closely related to the poor prognosis of non-small cell lung cancer patients via interference with the TIME, which suggests that m6A plays a role in optimizing individualized immunotherapy management and improving prognosis. The expression levels of METTL3, FTO and YTHDF1 in non-small cell lung cancer were changed. Patients in Cluster 1 had lower immunoscores, higher programmed death-ligand 1 (PD-L1) expression, and shorter overall survival compared to patients in Cluster 2. The hallmarks of the Myelocytomatosis viral oncogene (MYC) targets, Transcription factor E2F1 (E2F1) targets were significantly enriched. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | |||
| Experiment 4 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | This study revealed that m6A methylation is closely related to the poor prognosis of non-small cell lung cancer patients via interference with the TIME, which suggests that m6A plays a role in optimizing individualized immunotherapy management and improving prognosis. The expression levels of METTL3, FTO and YTHDF1 in non-small cell lung cancer were changed. Patients in Cluster 1 had lower immunoscores, higher programmed death-ligand 1 (PD-L1) expression, and shorter overall survival compared to patients in Cluster 2. The hallmarks of the Myelocytomatosis viral oncogene (MYC) targets, Transcription factor E2F1 (E2F1) targets were significantly enriched. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | |||
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 can regulate the expression of MALAT1 through m6A, mediate the Transcription factor E2F1 (E2F1)/AGR2 axis, and promote the adriamycin resistance of breast cancer. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Doxil | Approved | ||
| In-vitro Model | MCF7-DoxR (Adriamycin-resistant cell line MCF7-DoxR) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| In-vivo Model | Once the tumor volume increased to about 1 cm3, six groups of MCF7 bearing mice (n = 10 in each group) were injected with PBS (0.1 ml, caudal vein) and adriamycin (0.1 ml, 10 mg/kg), respectively. When the tumor reached 1.5 cm in any direction (defined as event-free survival analysis), 10 mice in each group were selected to measure the tumor size and weight on the 12th day after adriamycin injection. | |||
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | FTO interacts with transcripts of Transcription factor E2F1 (E2F1) and Myc, inhibition of FTO significantly impairs the translation efficiency of E2F1 and Myc.FTO plays important oncogenic role in regulating cervical cancer cells' proliferation. | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation and migration | |||
| In-vitro Model | HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
Doxil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [2] | |||
| Response Summary | METTL3 can regulate the expression of MALAT1 through m6A, mediate the Transcription factor E2F1 (E2F1)/AGR2 axis, and promote the adriamycin resistance of breast cancer. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| In-vitro Model | MCF7-DoxR (Adriamycin-resistant cell line MCF7-DoxR) | |||
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| In-vivo Model | Once the tumor volume increased to about 1 cm3, six groups of MCF7 bearing mice (n = 10 in each group) were injected with PBS (0.1 ml, caudal vein) and adriamycin (0.1 ml, 10 mg/kg), respectively. When the tumor reached 1.5 cm in any direction (defined as event-free survival analysis), 10 mice in each group were selected to measure the tumor size and weight on the 12th day after adriamycin injection. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Non-coding RNA
m6A Regulator: Fat mass and obesity-associated protein (FTO)
| In total 4 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05238 | ||
| Epigenetic Regulator | hsa-miR-607 | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05242 | ||
| Epigenetic Regulator | hsa-miR-607 | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05252 | ||
| Epigenetic Regulator | hsa_circ_0072309 (Circ_LIFR) | |
| Regulated Target | hsa-miR-607 | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05256 | ||
| Epigenetic Regulator | hsa_circ_0072309 (Circ_LIFR) | |
| Regulated Target | hsa-miR-607 | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00234)
| In total 5 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE002667 | Click to Show/Hide the Full List | ||
| mod site | chr20:33675915-33675916:- | ||
| Sequence | AGGGGGCCAGTTCAGGGCCCCAGCTGCCCCCCAGGATGGAT | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m5C_site_28691 | ||
| mod ID: M5CSITE002668 | Click to Show/Hide the Full List | ||
| mod site | chr20:33675916-33675917:- | ||
| Sequence | CAGGGGGCCAGTTCAGGGCCCCAGCTGCCCCCCAGGATGGA | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m5C_site_28692 | ||
| mod ID: M5CSITE002669 | Click to Show/Hide the Full List | ||
| mod site | chr20:33675917-33675918:- | ||
| Sequence | CCAGGGGGCCAGTTCAGGGCCCCAGCTGCCCCCCAGGATGG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m5C_site_28693 | ||
| mod ID: M5CSITE002670 | Click to Show/Hide the Full List | ||
| mod site | chr20:33675918-33675919:- | [9] | |
| Sequence | TCCAGGGGGCCAGTTCAGGGCCCCAGCTGCCCCCCAGGATG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m5C_site_28694 | ||
| mod ID: M5CSITE002671 | Click to Show/Hide the Full List | ||
| mod site | chr20:33675928-33675929:- | [9] | |
| Sequence | GTGGGCCCTCTCCAGGGGGCCAGTTCAGGGCCCCAGCTGCC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m5C_site_28695 | ||
N6-methyladenosine (m6A)
| In total 43 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE052199 | Click to Show/Hide the Full List | ||
| mod site | chr20:33675653-33675654:- | [10] | |
| Sequence | GTTTTGGGACTCTGTTGGGAACATTTCGGGGCGGGAGAGGC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527693 | ||
| mod ID: M6ASITE052200 | Click to Show/Hide the Full List | ||
| mod site | chr20:33675665-33675666:- | [11] | |
| Sequence | CGTTTGGGCCGGGTTTTGGGACTCTGTTGGGAACATTTCGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527694 | ||
| mod ID: M6ASITE052201 | Click to Show/Hide the Full List | ||
| mod site | chr20:33675819-33675820:- | [11] | |
| Sequence | GAAGGCCTCCCCCAGCCCAGACCCTGTGGTCCCTCCTGCAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527695 | ||
| mod ID: M6ASITE052202 | Click to Show/Hide the Full List | ||
| mod site | chr20:33675869-33675870:- | [11] | |
| Sequence | ATGGGAGAGGTGAGTGGGGGACCTTCACTGATGTGGGCAGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; H1B; MT4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527696 | ||
| mod ID: M6ASITE052203 | Click to Show/Hide the Full List | ||
| mod site | chr20:33675974-33675975:- | [12] | |
| Sequence | GGAGGGAGACAGACTGACTGACAGCCATGGGTGGTCAGATG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527697 | ||
| mod ID: M6ASITE052204 | Click to Show/Hide the Full List | ||
| mod site | chr20:33675978-33675979:- | [12] | |
| Sequence | TGAGGGAGGGAGACAGACTGACTGACAGCCATGGGTGGTCA | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527698 | ||
| mod ID: M6ASITE052205 | Click to Show/Hide the Full List | ||
| mod site | chr20:33675982-33675983:- | [11] | |
| Sequence | AAAGTGAGGGAGGGAGACAGACTGACTGACAGCCATGGGTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; A549; MT4; Huh7 | ||
| Seq Type List | m6A-seq; m6A-CLIP/IP; MeRIP-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527699 | ||
| mod ID: M6ASITE052206 | Click to Show/Hide the Full List | ||
| mod site | chr20:33675986-33675987:- | [11] | |
| Sequence | TGGGAAAGTGAGGGAGGGAGACAGACTGACTGACAGCCATG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527700 | ||
| mod ID: M6ASITE052207 | Click to Show/Hide the Full List | ||
| mod site | chr20:33676031-33676032:- | [11] | |
| Sequence | TTCTGCCCCATACTGAAGGAACTGAGGCCTGGGTGATTTAT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527701 | ||
| mod ID: M6ASITE052208 | Click to Show/Hide the Full List | ||
| mod site | chr20:33676040-33676041:- | [12] | |
| Sequence | GAGCTGTTCTTCTGCCCCATACTGAAGGAACTGAGGCCTGG | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527702 | ||
| mod ID: M6ASITE052209 | Click to Show/Hide the Full List | ||
| mod site | chr20:33676228-33676229:- | [12] | |
| Sequence | TGGCTGGCTGGGCGTGTAGGACGGTGAGAGCACTTCTGTCT | ||
| Motif Score | 3.616982143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527703 | ||
| mod ID: M6ASITE052210 | Click to Show/Hide the Full List | ||
| mod site | chr20:33676270-33676271:- | [12] | |
| Sequence | TCCCAGAATCTGGTGCTCTGACCAGGCCAGGTGGGGAGGCT | ||
| Motif Score | 2.839113095 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527704 | ||
| mod ID: M6ASITE052211 | Click to Show/Hide the Full List | ||
| mod site | chr20:33676347-33676348:- | [12] | |
| Sequence | GTGCGCGTGGGGGGGCTCTAACTGCACTTTCGGCCCTTTTG | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527705 | ||
| mod ID: M6ASITE052212 | Click to Show/Hide the Full List | ||
| mod site | chr20:33676436-33676437:- | [12] | |
| Sequence | GTACCGGGGAATGAAGGTGAACATACACCTCTGTGTGTGCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527706 | ||
| mod ID: M6ASITE052213 | Click to Show/Hide the Full List | ||
| mod site | chr20:33676503-33676504:- | [13] | |
| Sequence | TGTGCATGCAGCCTACACCCACACGTGTGTACCGGGGGTGA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527707 | ||
| mod ID: M6ASITE052214 | Click to Show/Hide the Full List | ||
| mod site | chr20:33676509-33676510:- | [13] | |
| Sequence | TGTGTATGTGCATGCAGCCTACACCCACACGTGTGTACCGG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527708 | ||
| mod ID: M6ASITE052215 | Click to Show/Hide the Full List | ||
| mod site | chr20:33676575-33676576:- | [12] | |
| Sequence | TCTAGCTCTGGGGTCTGGCTACCGCTAGGAGGCTGAGCAAG | ||
| Motif Score | 2.05802381 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527709 | ||
| mod ID: M6ASITE052216 | Click to Show/Hide the Full List | ||
| mod site | chr20:33676634-33676635:- | [14] | |
| Sequence | TCCTCCCAGCCTGTTTGGAAACATTTAATTTATACCCCTCT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HepG2; HEK293T; HeLa; A549; U2OS; H1A; H1B; GM12878; H1299; MM6; Jurkat; HEK293A-TOA; iSLK; MSC; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527710 | ||
| mod ID: M6ASITE052217 | Click to Show/Hide the Full List | ||
| mod site | chr20:33676716-33676717:- | [11] | |
| Sequence | TTCTGACAGGGCTTGGAGGGACCAGGGTTTCCAGAGATGCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hESCs; GM12878; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527711 | ||
| mod ID: M6ASITE052218 | Click to Show/Hide the Full List | ||
| mod site | chr20:33676753-33676754:- | [11] | |
| Sequence | CTTCGACTGTGACTTTGGGGACCTCACCCCCCTGGATTTCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hESCs; GM12878; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527712 | ||
| mod ID: M6ASITE052219 | Click to Show/Hide the Full List | ||
| mod site | chr20:33676768-33676769:- | [12] | |
| Sequence | GGGCATCAGAGACCTCTTCGACTGTGACTTTGGGGACCTCA | ||
| Motif Score | 3.287565476 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527713 | ||
| mod ID: M6ASITE052220 | Click to Show/Hide the Full List | ||
| mod site | chr20:33676777-33676778:- | [11] | |
| Sequence | GGAGGGCGAGGGCATCAGAGACCTCTTCGACTGTGACTTTG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; GM12878; H1299; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527714 | ||
| mod ID: M6ASITE052221 | Click to Show/Hide the Full List | ||
| mod site | chr20:33676816-33676817:- | [12] | |
| Sequence | CCCACCCCACGAGGCCCTCGACTACCACTTCGGCCTCGAGG | ||
| Motif Score | 3.287565476 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527715 | ||
| mod ID: M6ASITE052222 | Click to Show/Hide the Full List | ||
| mod site | chr20:33676876-33676877:- | [11] | |
| Sequence | CCTGGAGCATGTGCGGGAGGACTTCTCCGGCCTCCTCCCTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hESCs; H1299; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527716 | ||
| mod ID: M6ASITE052223 | Click to Show/Hide the Full List | ||
| mod site | chr20:33676930-33676931:- | [11] | |
| Sequence | GCGGGCTCCCGTGGACGAGGACCGCCTGTCCCCGCTGGTGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; MM6; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527717 | ||
| mod ID: M6ASITE052224 | Click to Show/Hide the Full List | ||
| mod site | chr20:33677205-33677206:- | [11] | |
| Sequence | GGTCACTTCTGAGGAGGAGAACAGGGCCACTGACTCTGCCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; MT4; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527718 | ||
| mod ID: M6ASITE052225 | Click to Show/Hide the Full List | ||
| mod site | chr20:33677239-33677240:- | [11] | |
| Sequence | GTGGGATCAGCCCTGGGAAGACCCCATCCCAGGAGGTCACT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; MT4; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527719 | ||
| mod ID: M6ASITE052226 | Click to Show/Hide the Full List | ||
| mod site | chr20:33677266-33677267:- | [15] | |
| Sequence | TTTTCCTGTGCCCTGAGGAGACCGTAGGTGGGATCAGCCCT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; MT4; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527720 | ||
| mod ID: M6ASITE052227 | Click to Show/Hide the Full List | ||
| mod site | chr20:33677303-33677304:- | [13] | |
| Sequence | CAGATCTCCCTTAAGAGCAAACAAGGCCCGATCGATGTTTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T; HEK293A-TOA | ||
| Seq Type List | MAZTER-seq; m6A-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527721 | ||
| mod ID: M6ASITE052228 | Click to Show/Hide the Full List | ||
| mod site | chr20:33677498-33677499:- | [16] | |
| Sequence | GGACCTTCGTAGCATTGCAGACCCTGCAGAGCAGATGGTTA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527722 | ||
| mod ID: M6ASITE052229 | Click to Show/Hide the Full List | ||
| mod site | chr20:33677516-33677517:- | [16] | |
| Sequence | GGCCTACGTGACGTGTCAGGACCTTCGTAGCATTGCAGACC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527723 | ||
| mod ID: M6ASITE052230 | Click to Show/Hide the Full List | ||
| mod site | chr20:33678215-33678216:- | [11] | |
| Sequence | GCTGCGCCTGCTCTCCGAGGACACTGACAGCCAGCGATATC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; CD4T; peripheral-blood; GSC-11; HEC-1-A | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527724 | ||
| mod ID: M6ASITE052231 | Click to Show/Hide the Full List | ||
| mod site | chr20:33678243-33678244:- | [12] | |
| Sequence | ACCACCTGATGAATATCTGTACTACGCAGCTGCGCCTGCTC | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527725 | ||
| mod ID: M6ASITE052232 | Click to Show/Hide the Full List | ||
| mod site | chr20:33678263-33678264:- | [11] | |
| Sequence | GGAGAGCGAGCAGCAGCTGGACCACCTGATGAATATCTGTA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; CD4T; peripheral-blood; GSC-11; HEK293T; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527726 | ||
| mod ID: M6ASITE052233 | Click to Show/Hide the Full List | ||
| mod site | chr20:33678299-33678300:- | [11] | |
| Sequence | GCTTGAGGGGTTGACCCAGGACCTCCGACAGCTGCAGGAGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527727 | ||
| mod ID: M6ASITE052234 | Click to Show/Hide the Full List | ||
| mod site | chr20:33679766-33679767:- | [17] | |
| Sequence | TGCCAAGAAGTCCAAGAACCACATCCAGTGGCTGTAGGTAC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | HEK293; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527728 | ||
| mod ID: M6ASITE052235 | Click to Show/Hide the Full List | ||
| mod site | chr20:33679769-33679770:- | [11] | |
| Sequence | CATTGCCAAGAAGTCCAAGAACCACATCCAGTGGCTGTAGG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; HEK293T; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527729 | ||
| mod ID: M6ASITE052236 | Click to Show/Hide the Full List | ||
| mod site | chr20:33679820-33679821:- | [13] | |
| Sequence | GCAGAAGCGGCGCATCTATGACATCACCAACGTCCTTGAGG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527730 | ||
| mod ID: M6ASITE052237 | Click to Show/Hide the Full List | ||
| mod site | chr20:33679865-33679866:- | [11] | |
| Sequence | TGACGGTGTCGTCGACCTGAACTGGGCTGCCGAGGTGCTGA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527731 | ||
| mod ID: M6ASITE052238 | Click to Show/Hide the Full List | ||
| mod site | chr20:33679938-33679939:- | [11] | |
| Sequence | GGGAGAAGTCACGCTATGAGACCTCACTGAATCTGACCACC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527732 | ||
| mod ID: M6ASITE052239 | Click to Show/Hide the Full List | ||
| mod site | chr20:33680391-33680392:- | [11] | |
| Sequence | AGCGGAGGCTGGACCTGGAAACTGACCATCAGTACCTGGCC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; MT4; MM6; HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527733 | ||
| mod ID: M6ASITE052240 | Click to Show/Hide the Full List | ||
| mod site | chr20:33680399-33680400:- | [11] | |
| Sequence | ACAGGTGAAGCGGAGGCTGGACCTGGAAACTGACCATCAGT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; MT4; MM6; HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527734 | ||
| mod ID: M6ASITE052241 | Click to Show/Hide the Full List | ||
| mod site | chr20:33686381-33686382:- | [18] | |
| Sequence | GGCGCGTAAAAGTGGCCGGGACTTTGCAGGCAGCGGCGGCC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | A549; HEK293A-TOA; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000343380.6 | ||
| External Link | RMBase: m6A_site_527735 | ||
References