m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00375)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
PTEN
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
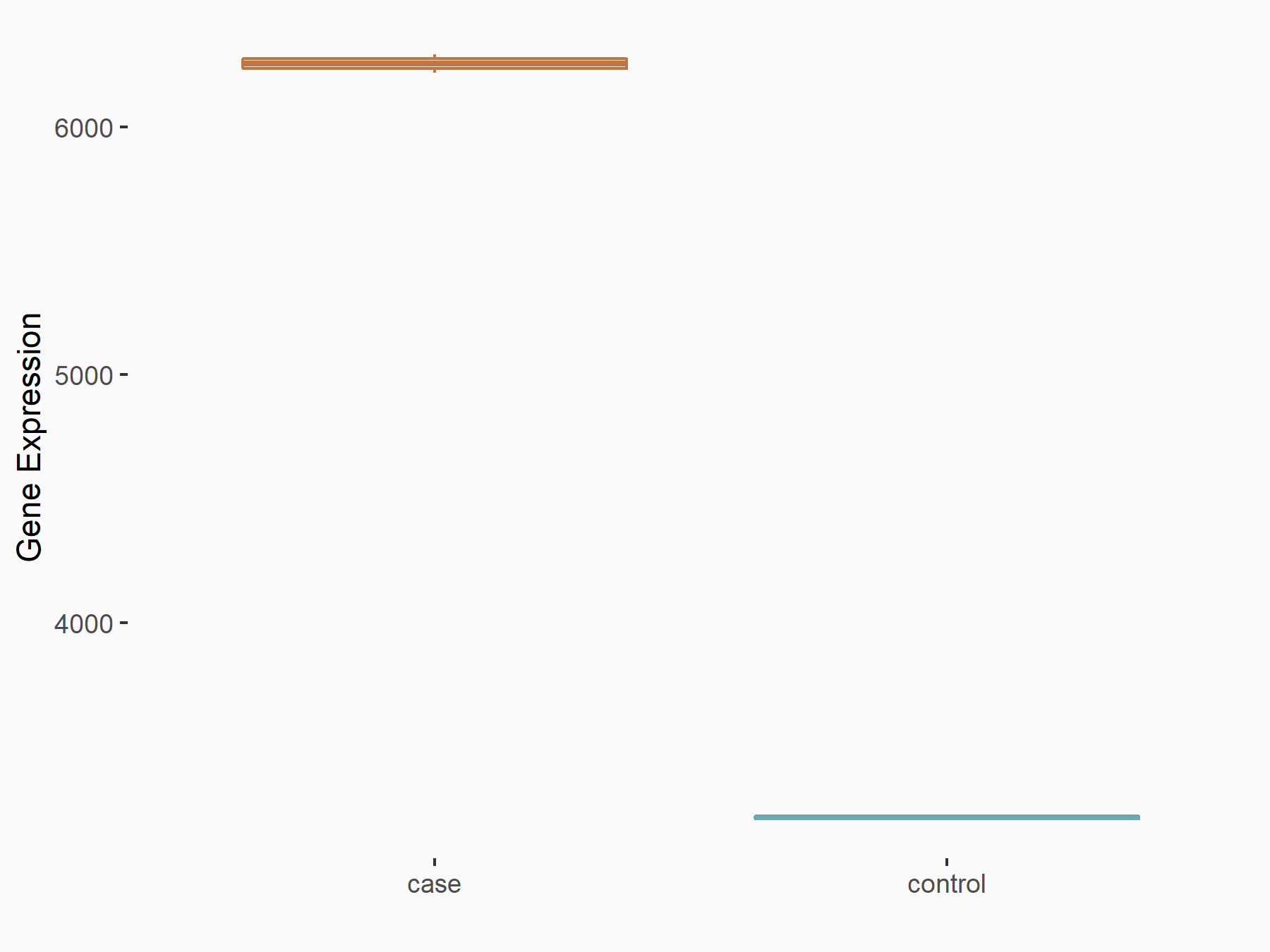  |
logFC: 9.60E-01 p-value: 1.46E-37 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between PTEN and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.25E+00 | GSE60213 |
| In total 4 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 depletion in human myeloid leukemia cell lines induces cell differentiation and apoptosis and delays leukemia progression in recipient mice in vivo. Single-nucleotide-resolution mapping of m6A coupled with ribosome profiling reveals that m6A promotes the translation of c-MYC, BCL2 and Mutated in multiple advanced cancers 1 (PTEN) mRNAs in the human acute myeloid leukemia MOLM-13 cell line. Moreover, loss of METTL3 leads to increased levels of phosphorylated AKT. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Acute myeloid leukaemia | ICD-11: 2A60 | ||
| Cell Process | Cell differentiation and apoptosis | |||
| Apoptosis (hsa04210) | ||||
| In-vitro Model | HSPC (Human hematopoietic stem cell) | |||
| In-vivo Model | 500,000 selected cells were injected via tail vein or retro-orbital route into female NSG (6-8 week old) recipient mice that had been sublethally irradiated with 475 cGy one day before transplantation. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | The molecular mechanism underlying the effect of LINC00470 on chronic myelocytic leukaemia by reducing the Mutated in multiple advanced cancers 1 (PTEN) stability via RNA methyltransferase METTL3, thus leading to the inhibition of cell autophagy while promoting chemoresistance in CML. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Chronic myeloid leukaemia | ICD-11: 2B33.2 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | RNA stability | |||
| Cell autophagy | ||||
| In-vitro Model | K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 |
| KCL-22 | Chronic myelogenous leukemia | Homo sapiens | CVCL_2091 | |
| In-vivo Model | In the control mice or ADR mice group, the parental or chemo-resistant K562 cells were infected with LV-shCtrl. In the ADR + shLINC00470 group, the chemo-resistant K562 cells were infected with LV- shLINC00470. These cells were injected, respectively, into these 5-week-old mice subcutaneously. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | Bete-elemene exerted the restrictive impacts on the cell growth of lung cancer in vivo and in vitro through targeting METTL3. Bete-elemene contributed to the augmented PTEN expression via suppressing its m6A modification. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Lung cancer | ICD-11: 2C25 | ||
| In-vitro Model | NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | Four-week-old BALB/c nude mice were randomly divided into three groups: (1) vector group, (2) vector + Bete-elemene group, and (3) Bete-elemene + METTL3 group. Nude mice were raised in an SPF level animal house and were free to eat and drink. Mice in the vector group were subcutaneously injected with lung cancer cells transfected with empty vector and did not receive Bete-elemene administration, and this group was implemented as the negative control. Following establishing orthotopic xenografts by using A549 or H1299 cells transfected with empty vector, mice in the vector + Bete-elemene group underwent intraperitoneal injection with Bete-elemene once a day. For the subcutaneous transplanted model, A549 or H1299 cells transfected with METTL3-overexpressing vector were inoculated into mice from the Bete-elemene + METTL3 group. Then, mice were intraperitoneally administrated with Bete-elemene once a day. Three weeks later, all the animals were euthanized with CO2. Xenografts were removed and weighted after mice were euthanatized. | |||
| Experiment 4 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | METTL3/YTHDF2/Mutated in multiple advanced cancers 1 (PTEN) axis exerts a significant role in hypoxia induced PASMCs proliferation, providing a novel therapeutic target for hypoxic pulmonary hypertension. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Pulmonary hypertension due to lung disease or hypoxia | ICD-11: BB01.2 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | PASMC cell line (Pulmonary artery smooth muscle cell) | |||
| In-vivo Model | 10 rats were divided into control and HPH group. In detail, 5 rats of the hypoxia groups were exposed to hypoxia (10%O2) chamber (AiPu XBS-02B, China) for 4 weeks. In addition, 5 rats of control group were kept under normoxic conditions (21% O2) for 4 weeks. Rats were housed in standard polypropylene cages under controlled photocycle (12 h light/12 h dark) under 22-25 ℃ temperature. | |||
Methyltransferase-like 14 (METTL14) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL14 | ||
| Cell Line | HepG2 cell line | Homo sapiens |
|
Treatment: shMETTL14 HepG2 cells
Control: shCtrl HepG2 cells
|
GSE121949 | |
| Regulation |
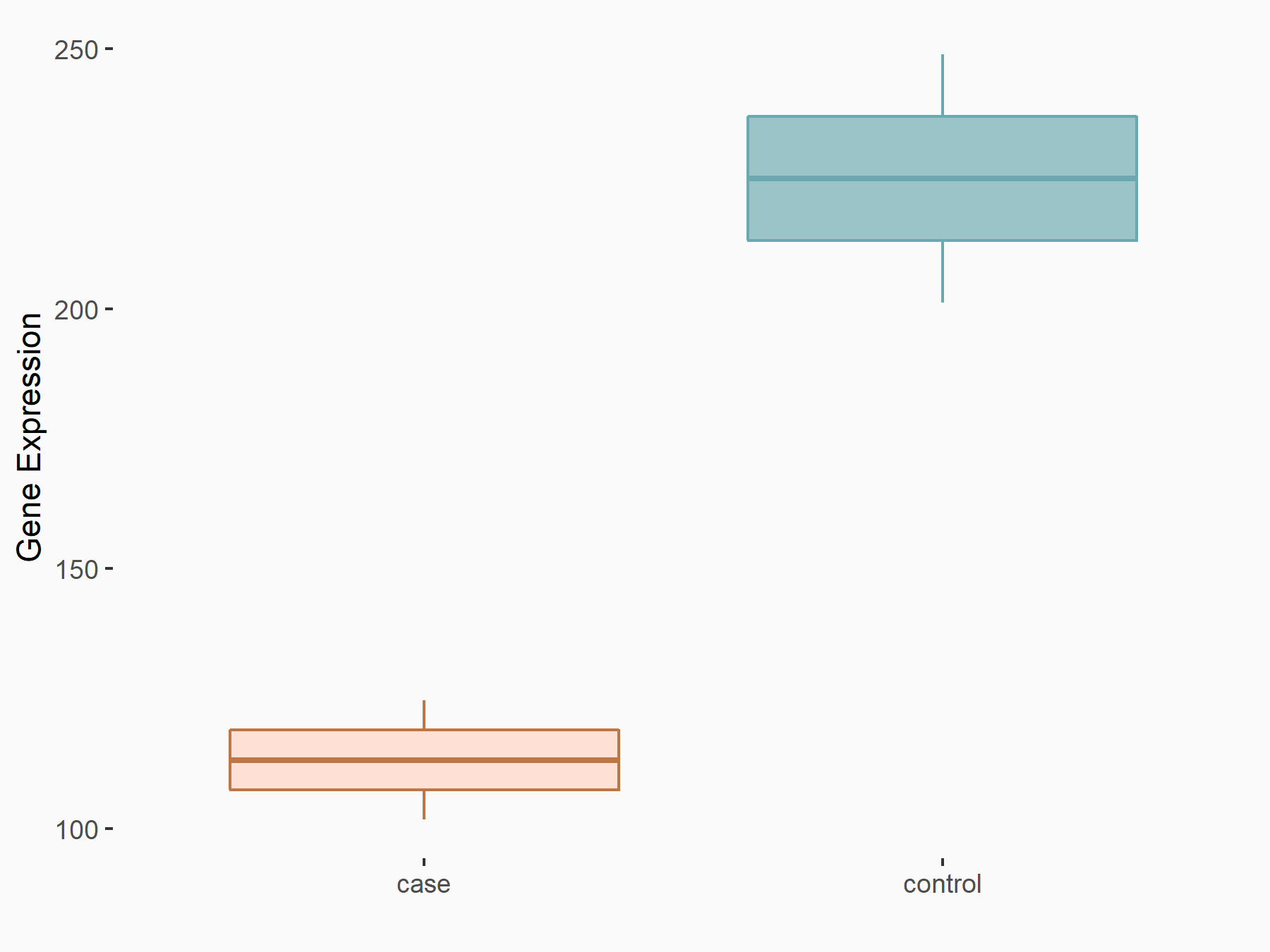  |
logFC: -9.93E-01 p-value: 1.46E-04 |
| More Results | Click to View More RNA-seq Results | |
| In total 3 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [5] | |||
| Response Summary | METTL14 inhibits tumor growth and metastasis of Stomach Adenocarcinoma via stabilization of Mutated in multiple advanced cancers 1 (PTEN) mRNA expression. Therefore, METTL14 is a potential biomarker of prognosis and therapeutic targets for Stomach Adenocarcinoma. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | RGM1 | Normal | Rattus norvegicus | CVCL_0499 |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | For the purpose of enhancing the overall randomization of the experiment, a random comparison table had been employed. Accordingly, 5-wk-old male nude athymic BALB/c nu/nu mice (Slack, Shanghai, China) were randomly divided into two parts including a control group (NC) and the experimental group METTL14-OE. For developing subcutaneous xeno transplantation model, 5 × 106 HGC-27 cells stably transfected with NC or METTL14 overexpression were subcutaneously incorporated for 5-week-old BALB/c nude mice. The mice experienced euthanasia after 27 days of inoculation and obtained xenografts's mass was obtained. Tumor volume over three days was obtained. To create mouse STAD liver metastasis orthotopic tumor model, 1 × 106 HGC-27 cells under stable transfection with NC or METTL14 overexpression were added to subserosal gastric wall of BALB/c nude mice. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | Upregulation of METTL14 inhibited ccRCC cells proliferation and migration in vitro. Overexpression of METTL14 increased the m6A enrichment of Mutated in multiple advanced cancers 1 (PTEN), and promoted Pten expression. METTL14-enhanced Pten mRNA stability was dependent on YTHDF1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Renal cell carcinoma of kidney | ICD-11: 2C90.0 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [7] | |||
| Response Summary | METTL14-regulated PI3K/Akt signaling pathway via Mutated in multiple advanced cancers 1 (PTEN) affected HDAC5-mediated EMT of renal tubular cells in diabetic kidney disease. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Chronic kidney disease | ICD-11: GB61.Z | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | HK-2 [Human kidney] | Normal | Homo sapiens | CVCL_0302 |
| In-vivo Model | Twenty mice were randomly divided into three groups: normal mice group (N), diabetic mice group (DM), and diabetic mice administrated with TSA group (DM + TSA). | |||
RNA demethylase ALKBH5 (ALKBH5) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by ALKBH5 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shALKBH5 MOLM-13 cells
Control: shNS MOLM-13 cells
|
GSE144968 | |
| Regulation |
  |
logFC: -2.10E+00 p-value: 1.89E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [8] | |||
| Response Summary | ALKBH5-mediated m6A demethylation enhanced the stability of KCNK15-AS1. In pancreatic cancer, KCNK15-AS1 bound to KCNK15 to inhibit its translation, and interacted with MDM2 to induce REST ubiquitination, which eventually facilitated Mutated in multiple advanced cancers 1 (PTEN) transcription to inactivate AKT pathway. | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Epithelial-mesenchymal transition | ||||
| Cell apoptosis | ||||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
YTH domain-containing protein 1 (YTHDC1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDC1 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shYTHDC1 MOLM13 cells
Control: shControl MOLM13 cells
|
GSE168565 | |
| Regulation |
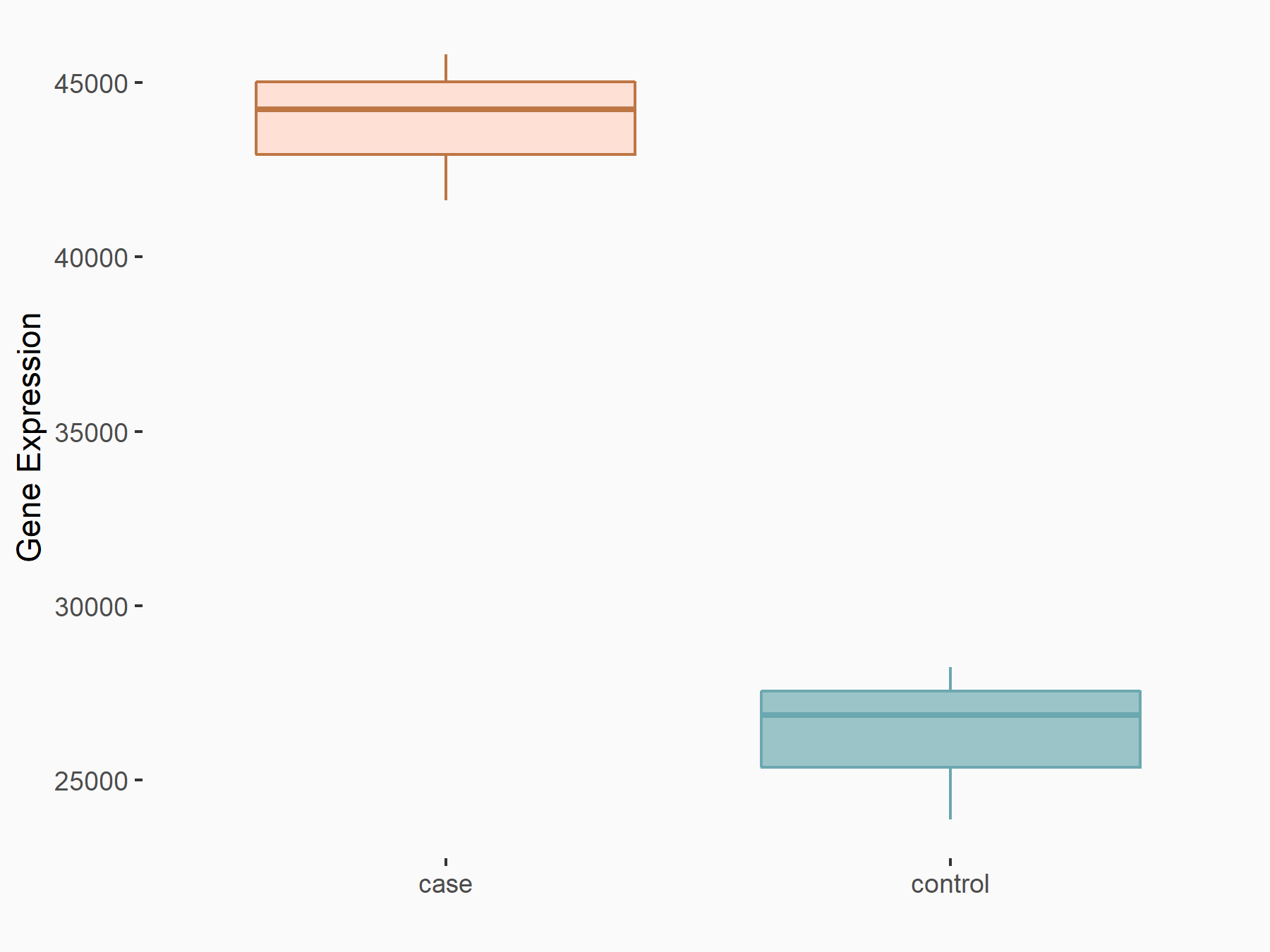  |
logFC: 7.38E-01 p-value: 4.67E-08 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [9] | |||
| Response Summary | YTHDC1 promoted Mutated in multiple advanced cancers 1 (PTEN) mRNA degradation to increase Akt phosphorylation, thus facilitating neuronal survival in particular after ischemia, modulating m6A reader YTHDC1 provide a potential therapeutic target for ischemic stroke. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Acute ischemic stroke | ICD-11: 8B11 | ||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| In-vivo Model | Male SD rats weighing 200-250 g were anesthetized using 4% isoflurane in 70% N2O and 30% O2 with a mask. A midline incision was made in the neck, the left external carotid artery (ECA) was carefully exposed and dissected, a monofilament nylon suture with a diameter of about 0.22 mm was inserted from the ECA into the internal carotid artery, and the left middle cerebral artery (MCA) was blocked. After occlusion for 90 minutes, the suture was removed for reperfusion, and ECA was ligated to close the wound. Sham-operated rats underwent the same surgery except for suture insertion. Rats were maintained on top of a warming pad (RWD, 69003) during the above procedures. The breathing machine was used to monitor the respiration of rats. The rats were returned to a heated cage during the recovery phase with free access to food and water. | |||
YTH domain-containing family protein 1 (YTHDF1) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | Upregulation of METTL14 inhibited ccRCC cells proliferation and migration in vitro. Overexpression of METTL14 increased the m6A enrichment of Mutated in multiple advanced cancers 1 (PTEN), and promoted Pten expression. METTL14-enhanced Pten mRNA stability was dependent on YTHDF1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Renal cell carcinoma of kidney | ICD-11: 2C90.0 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
YTH domain-containing family protein 2 (YTHDF2) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | METTL3/YTHDF2/Mutated in multiple advanced cancers 1 (PTEN) axis exerts a significant role in hypoxia induced PASMCs proliferation, providing a novel therapeutic target for hypoxic pulmonary hypertension. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Pulmonary hypertension due to lung disease or hypoxia | ICD-11: BB01.2 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | PASMC cell line (Pulmonary artery smooth muscle cell) | |||
| In-vivo Model | 10 rats were divided into control and HPH group. In detail, 5 rats of the hypoxia groups were exposed to hypoxia (10%O2) chamber (AiPu XBS-02B, China) for 4 weeks. In addition, 5 rats of control group were kept under normoxic conditions (21% O2) for 4 weeks. Rats were housed in standard polypropylene cages under controlled photocycle (12 h light/12 h dark) under 22-25 ℃ temperature. | |||
Solid tumour/cancer [ICD-11: 2A00-2F9Z]
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 depletion in human myeloid leukemia cell lines induces cell differentiation and apoptosis and delays leukemia progression in recipient mice in vivo. Single-nucleotide-resolution mapping of m6A coupled with ribosome profiling reveals that m6A promotes the translation of c-MYC, BCL2 and Mutated in multiple advanced cancers 1 (PTEN) mRNAs in the human acute myeloid leukemia MOLM-13 cell line. Moreover, loss of METTL3 leads to increased levels of phosphorylated AKT. | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell differentiation and apoptosis | |||
| Apoptosis (hsa04210) | ||||
| In-vitro Model | HSPC (Human hematopoietic stem cell) | |||
| In-vivo Model | 500,000 selected cells were injected via tail vein or retro-orbital route into female NSG (6-8 week old) recipient mice that had been sublethally irradiated with 475 cGy one day before transplantation. | |||
Malignant haematopoietic neoplasm [ICD-11: 2B33]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | The molecular mechanism underlying the effect of LINC00470 on chronic myelocytic leukaemia by reducing the Mutated in multiple advanced cancers 1 (PTEN) stability via RNA methyltransferase METTL3, thus leading to the inhibition of cell autophagy while promoting chemoresistance in CML. | |||
| Responsed Disease | Chronic myeloid leukaemia [ICD-11: 2B33.2] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | RNA stability | |||
| Cell autophagy | ||||
| In-vitro Model | K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 |
| KCL-22 | Chronic myelogenous leukemia | Homo sapiens | CVCL_2091 | |
| In-vivo Model | In the control mice or ADR mice group, the parental or chemo-resistant K562 cells were infected with LV-shCtrl. In the ADR + shLINC00470 group, the chemo-resistant K562 cells were infected with LV- shLINC00470. These cells were injected, respectively, into these 5-week-old mice subcutaneously. | |||
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [5] | |||
| Response Summary | METTL14 inhibits tumor growth and metastasis of Stomach Adenocarcinoma via stabilization of Mutated in multiple advanced cancers 1 (PTEN) mRNA expression. Therefore, METTL14 is a potential biomarker of prognosis and therapeutic targets for Stomach Adenocarcinoma. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | RGM1 | Normal | Rattus norvegicus | CVCL_0499 |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | For the purpose of enhancing the overall randomization of the experiment, a random comparison table had been employed. Accordingly, 5-wk-old male nude athymic BALB/c nu/nu mice (Slack, Shanghai, China) were randomly divided into two parts including a control group (NC) and the experimental group METTL14-OE. For developing subcutaneous xeno transplantation model, 5 × 106 HGC-27 cells stably transfected with NC or METTL14 overexpression were subcutaneously incorporated for 5-week-old BALB/c nude mice. The mice experienced euthanasia after 27 days of inoculation and obtained xenografts's mass was obtained. Tumor volume over three days was obtained. To create mouse STAD liver metastasis orthotopic tumor model, 1 × 106 HGC-27 cells under stable transfection with NC or METTL14 overexpression were added to subserosal gastric wall of BALB/c nude mice. | |||
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [8] | |||
| Response Summary | ALKBH5-mediated m6A demethylation enhanced the stability of KCNK15-AS1. In pancreatic cancer, KCNK15-AS1 bound to KCNK15 to inhibit its translation, and interacted with MDM2 to induce REST ubiquitination, which eventually facilitated Mutated in multiple advanced cancers 1 (PTEN) transcription to inactivate AKT pathway. | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Epithelial-mesenchymal transition | ||||
| Cell apoptosis | ||||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | Bete-elemene exerted the restrictive impacts on the cell growth of lung cancer in vivo and in vitro through targeting METTL3. Bete-elemene contributed to the augmented PTEN expression via suppressing its m6A modification. | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | Four-week-old BALB/c nude mice were randomly divided into three groups: (1) vector group, (2) vector + Bete-elemene group, and (3) Bete-elemene + METTL3 group. Nude mice were raised in an SPF level animal house and were free to eat and drink. Mice in the vector group were subcutaneously injected with lung cancer cells transfected with empty vector and did not receive Bete-elemene administration, and this group was implemented as the negative control. Following establishing orthotopic xenografts by using A549 or H1299 cells transfected with empty vector, mice in the vector + Bete-elemene group underwent intraperitoneal injection with Bete-elemene once a day. For the subcutaneous transplanted model, A549 or H1299 cells transfected with METTL3-overexpressing vector were inoculated into mice from the Bete-elemene + METTL3 group. Then, mice were intraperitoneally administrated with Bete-elemene once a day. Three weeks later, all the animals were euthanized with CO2. Xenografts were removed and weighted after mice were euthanatized. | |||
Renal cell carcinoma [ICD-11: 2C90]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [6] | |||
| Response Summary | Upregulation of METTL14 inhibited ccRCC cells proliferation and migration in vitro. Overexpression of METTL14 increased the m6A enrichment of Mutated in multiple advanced cancers 1 (PTEN), and promoted Pten expression. METTL14-enhanced Pten mRNA stability was dependent on YTHDF1. | |||
| Responsed Disease | Renal cell carcinoma of kidney [ICD-11: 2C90.0] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Experiment 2 Reporting the m6A-centered Disease Response | [6] | |||
| Response Summary | Upregulation of METTL14 inhibited ccRCC cells proliferation and migration in vitro. Overexpression of METTL14 increased the m6A enrichment of Mutated in multiple advanced cancers 1 (PTEN), and promoted Pten expression. METTL14-enhanced Pten mRNA stability was dependent on YTHDF1. | |||
| Responsed Disease | Renal cell carcinoma of kidney [ICD-11: 2C90.0] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
Acute ischemic stroke [ICD-11: 8B11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [9] | |||
| Response Summary | YTHDC1 promoted Mutated in multiple advanced cancers 1 (PTEN) mRNA degradation to increase Akt phosphorylation, thus facilitating neuronal survival in particular after ischemia, modulating m6A reader YTHDC1 provide a potential therapeutic target for ischemic stroke. | |||
| Responsed Disease | Acute ischemic stroke [ICD-11: 8B11] | |||
| Target Regulator | YTH domain-containing protein 1 (YTHDC1) | READER | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| In-vivo Model | Male SD rats weighing 200-250 g were anesthetized using 4% isoflurane in 70% N2O and 30% O2 with a mask. A midline incision was made in the neck, the left external carotid artery (ECA) was carefully exposed and dissected, a monofilament nylon suture with a diameter of about 0.22 mm was inserted from the ECA into the internal carotid artery, and the left middle cerebral artery (MCA) was blocked. After occlusion for 90 minutes, the suture was removed for reperfusion, and ECA was ligated to close the wound. Sham-operated rats underwent the same surgery except for suture insertion. Rats were maintained on top of a warming pad (RWD, 69003) during the above procedures. The breathing machine was used to monitor the respiration of rats. The rats were returned to a heated cage during the recovery phase with free access to food and water. | |||
Pulmonary hypertension [ICD-11: BB01]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | METTL3/YTHDF2/Mutated in multiple advanced cancers 1 (PTEN) axis exerts a significant role in hypoxia induced PASMCs proliferation, providing a novel therapeutic target for hypoxic pulmonary hypertension. | |||
| Responsed Disease | Pulmonary hypertension due to lung disease or hypoxia [ICD-11: BB01.2] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | PASMC cell line (Pulmonary artery smooth muscle cell) | |||
| In-vivo Model | 10 rats were divided into control and HPH group. In detail, 5 rats of the hypoxia groups were exposed to hypoxia (10%O2) chamber (AiPu XBS-02B, China) for 4 weeks. In addition, 5 rats of control group were kept under normoxic conditions (21% O2) for 4 weeks. Rats were housed in standard polypropylene cages under controlled photocycle (12 h light/12 h dark) under 22-25 ℃ temperature. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | METTL3/YTHDF2/Mutated in multiple advanced cancers 1 (PTEN) axis exerts a significant role in hypoxia induced PASMCs proliferation, providing a novel therapeutic target for hypoxic pulmonary hypertension. | |||
| Responsed Disease | Pulmonary hypertension due to lung disease or hypoxia [ICD-11: BB01.2] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | PASMC cell line (Pulmonary artery smooth muscle cell) | |||
| In-vivo Model | 10 rats were divided into control and HPH group. In detail, 5 rats of the hypoxia groups were exposed to hypoxia (10%O2) chamber (AiPu XBS-02B, China) for 4 weeks. In addition, 5 rats of control group were kept under normoxic conditions (21% O2) for 4 weeks. Rats were housed in standard polypropylene cages under controlled photocycle (12 h light/12 h dark) under 22-25 ℃ temperature. | |||
Chronic kidney disease [ICD-11: GB61]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [7] | |||
| Response Summary | METTL14-regulated PI3K/Akt signaling pathway via Mutated in multiple advanced cancers 1 (PTEN) affected HDAC5-mediated EMT of renal tubular cells in diabetic kidney disease. | |||
| Responsed Disease | Chronic kidney disease [ICD-11: GB61.Z] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | HK-2 [Human kidney] | Normal | Homo sapiens | CVCL_0302 |
| In-vivo Model | Twenty mice were randomly divided into three groups: normal mice group (N), diabetic mice group (DM), and diabetic mice administrated with TSA group (DM + TSA). | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
RNA modification
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1)
| In total 4 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00409 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00410 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00446 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 214 (MIR214) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00510 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | hsa-mir-18a | |
| Crosstalk relationship | A-to-I → m6A | |
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 4 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00411 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00412 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00447 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 214 (MIR214) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00511 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | hsa-mir-18a | |
| Crosstalk relationship | A-to-I → m6A | |
m6A Regulator: Methyltransferase-like 14 (METTL14)
| In total 4 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00413 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00414 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00448 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 214 (MIR214) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00512 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | hsa-mir-18a | |
| Crosstalk relationship | A-to-I → m6A | |
m6A Regulator: RNA demethylase ALKBH5 (ALKBH5)
| In total 4 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00415 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00416 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00449 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 214 (MIR214) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00513 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | hsa-mir-18a | |
| Crosstalk relationship | A-to-I → m6A | |
DNA modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 4 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT02148 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02161 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02174 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT02187 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
Histone modification
m6A Regulator: YTH domain-containing protein 2 (YTHDC2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03043 | ||
| Epigenetic Regulator | Polycomb Repressive Complex 2 (PRC2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Skin cancer | |
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03221 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03384 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03397 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
m6A Regulator: Methyltransferase-like 14 (METTL14)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03501 | ||
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
Non-coding RNA
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05287 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 470 (LINC00470) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
| Crosstalk ID: M6ACROT05364 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 470 (LINC00470) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Chronic myeloid leukaemia | |
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05288 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 470 (LINC00470) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00375)
| In total 28 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE004325 | Click to Show/Hide the Full List | ||
| mod site | chr10:87871594-87871595:+ | [18] | |
| Sequence | GCCTCCGCCTCCTGGGTTCAAGCAATTCTCTTGCCGCAGCC | ||
| Transcript ID List | ENST00000371953.8; ENST00000462694.1 | ||
| External Link | RMBase: RNA-editing_site_17372 | ||
| mod ID: A2ISITE004326 | Click to Show/Hide the Full List | ||
| mod site | chr10:87871598-87871599:+ | [18] | |
| Sequence | CCGCCTCCTGGGTTCAAGCAATTCTCTTGCCGCAGCCTCCC | ||
| Transcript ID List | ENST00000371953.8; ENST00000462694.1 | ||
| External Link | RMBase: RNA-editing_site_17373 | ||
| mod ID: A2ISITE004327 | Click to Show/Hide the Full List | ||
| mod site | chr10:87871633-87871634:+ | [19] | |
| Sequence | CCTCCCAAGTAGCTGGAATTACAGGCATGCGCCACCACGCC | ||
| Transcript ID List | ENST00000462694.1; ENST00000371953.8 | ||
| External Link | RMBase: RNA-editing_site_17374 | ||
| mod ID: A2ISITE004328 | Click to Show/Hide the Full List | ||
| mod site | chr10:87871763-87871764:+ | [18] | |
| Sequence | TGCCTGTCTCAGCCTGCTAAAGTGCTGAGATTACAGGCATG | ||
| Transcript ID List | ENST00000462694.1; ENST00000371953.8 | ||
| External Link | RMBase: RNA-editing_site_17375 | ||
| mod ID: A2ISITE004329 | Click to Show/Hide the Full List | ||
| mod site | chr10:87873061-87873062:+ | [19] | |
| Sequence | CAGCGTTGGCAGATTGCTTGAGTCTGGAAGTTCGAGACCAG | ||
| Transcript ID List | ENST00000371953.8; rmsk_3372605; ENST00000462694.1 | ||
| External Link | RMBase: RNA-editing_site_17376 | ||
| mod ID: A2ISITE004330 | Click to Show/Hide the Full List | ||
| mod site | chr10:87873094-87873095:+ | [18] | |
| Sequence | GAGACCAGTCTGGGCAACATAGGCAGACCCTGTCTCTACAA | ||
| Transcript ID List | ENST00000462694.1; ENST00000371953.8; rmsk_3372605 | ||
| External Link | RMBase: RNA-editing_site_17377 | ||
| mod ID: A2ISITE004331 | Click to Show/Hide the Full List | ||
| mod site | chr10:87873117-87873118:+ | [18] | |
| Sequence | CAGACCCTGTCTCTACAAAAAAAAATACAAAAATTAGTCGG | ||
| Transcript ID List | rmsk_3372605; ENST00000371953.8; ENST00000462694.1 | ||
| External Link | RMBase: RNA-editing_site_17378 | ||
| mod ID: A2ISITE004332 | Click to Show/Hide the Full List | ||
| mod site | chr10:87873120-87873121:+ | [18] | |
| Sequence | ACCCTGTCTCTACAAAAAAAAATACAAAAATTAGTCGGGTG | ||
| Transcript ID List | ENST00000371953.8; rmsk_3372605; ENST00000462694.1 | ||
| External Link | RMBase: RNA-editing_site_17379 | ||
| mod ID: A2ISITE004333 | Click to Show/Hide the Full List | ||
| mod site | chr10:87873158-87873159:+ | [19] | |
| Sequence | GTGTTATAGTGCGCATTGGTAGTCCCAGCTACTGAGGAGGC | ||
| Transcript ID List | rmsk_3372605; ENST00000371953.8; ENST00000462694.1 | ||
| External Link | RMBase: RNA-editing_site_17380 | ||
| mod ID: A2ISITE004334 | Click to Show/Hide the Full List | ||
| mod site | chr10:87873172-87873173:+ | [18] | |
| Sequence | ATTGGTAGTCCCAGCTACTGAGGAGGCTGAGGTGGGATCAC | ||
| Transcript ID List | ENST00000462694.1; ENST00000371953.8; rmsk_3372605 | ||
| External Link | RMBase: RNA-editing_site_17381 | ||
| mod ID: A2ISITE004335 | Click to Show/Hide the Full List | ||
| mod site | chr10:87873196-87873197:+ | [18] | |
| Sequence | GGCTGAGGTGGGATCACCTGAGACTGGGACTTTGAGGCTGC | ||
| Transcript ID List | ENST00000371953.8; rmsk_3372605; ENST00000462694.1 | ||
| External Link | RMBase: RNA-editing_site_17382 | ||
| mod ID: A2ISITE004336 | Click to Show/Hide the Full List | ||
| mod site | chr10:87887714-87887715:+ | [20] | |
| Sequence | CACAGGCCCAGAGCTCTAGGAGGGCAGAATGGTTTGTGGCT | ||
| Transcript ID List | ENST00000371953.8; rmsk_3372624; ENST00000462694.1 | ||
| External Link | RMBase: RNA-editing_site_17383 | ||
| mod ID: A2ISITE004337 | Click to Show/Hide the Full List | ||
| mod site | chr10:87890927-87890928:+ | [20] | |
| Sequence | TATTGCCCAGCCTAGCCTCAAACTCCTGGGCTGAAGGAATC | ||
| Transcript ID List | ENST00000462694.1; ENST00000371953.8 | ||
| External Link | RMBase: RNA-editing_site_17384 | ||
| mod ID: A2ISITE004338 | Click to Show/Hide the Full List | ||
| mod site | chr10:87890953-87890954:+ | [20] | |
| Sequence | TGGGCTGAAGGAATCCACCCATCTCAGCCTCCCAAAGTGCT | ||
| Transcript ID List | ENST00000462694.1; ENST00000371953.8 | ||
| External Link | RMBase: RNA-editing_site_17385 | ||
| mod ID: A2ISITE004339 | Click to Show/Hide the Full List | ||
| mod site | chr10:87891199-87891200:+ | [21] | |
| Sequence | CTCCTTCTCTTGGAATGAAAAGGTATCCTAGGCTCACCTGT | ||
| Transcript ID List | ENST00000462694.1; ENST00000371953.8 | ||
| External Link | RMBase: RNA-editing_site_17386 | ||
| mod ID: A2ISITE004340 | Click to Show/Hide the Full List | ||
| mod site | chr10:87891611-87891612:+ | [20] | |
| Sequence | TTGCTTGAGCTCAGGAGTTCAAGACCAACCTGGGCAACATG | ||
| Transcript ID List | ENST00000371953.8; ENST00000462694.1; rmsk_3372630 | ||
| External Link | RMBase: RNA-editing_site_17387 | ||
| mod ID: A2ISITE004341 | Click to Show/Hide the Full List | ||
| mod site | chr10:87891629-87891630:+ | [20] | |
| Sequence | TCAAGACCAACCTGGGCAACATGATGAAACCTTGTCTTTAC | ||
| Transcript ID List | ENST00000371953.8; rmsk_3372630; ENST00000462694.1 | ||
| External Link | RMBase: RNA-editing_site_17388 | ||
| mod ID: A2ISITE004342 | Click to Show/Hide the Full List | ||
| mod site | chr10:87891635-87891636:+ | [20] | |
| Sequence | CCAACCTGGGCAACATGATGAAACCTTGTCTTTACAAAAAA | ||
| Transcript ID List | ENST00000371953.8; rmsk_3372630; ENST00000462694.1 | ||
| External Link | RMBase: RNA-editing_site_17389 | ||
| mod ID: A2ISITE004343 | Click to Show/Hide the Full List | ||
| mod site | chr10:87891637-87891638:+ | [20] | |
| Sequence | AACCTGGGCAACATGATGAAACCTTGTCTTTACAAAAAATT | ||
| Transcript ID List | ENST00000371953.8; ENST00000462694.1; rmsk_3372630 | ||
| External Link | RMBase: RNA-editing_site_17390 | ||
| mod ID: A2ISITE004344 | Click to Show/Hide the Full List | ||
| mod site | chr10:87891684-87891685:+ | [20] | |
| Sequence | GCATGGTGGCATGTGCTTGTATTCCCAGCTACTCAGGAGGC | ||
| Transcript ID List | ENST00000462694.1; ENST00000371953.8; rmsk_3372630 | ||
| External Link | RMBase: RNA-editing_site_17391 | ||
| mod ID: A2ISITE004345 | Click to Show/Hide the Full List | ||
| mod site | chr10:87891694-87891695:+ | [20] | |
| Sequence | ATGTGCTTGTATTCCCAGCTACTCAGGAGGCTGAGGTGGGA | ||
| Transcript ID List | ENST00000462694.1; ENST00000371953.8; rmsk_3372630 | ||
| External Link | RMBase: RNA-editing_site_17392 | ||
| mod ID: A2ISITE004346 | Click to Show/Hide the Full List | ||
| mod site | chr10:87891698-87891699:+ | [20] | |
| Sequence | GCTTGTATTCCCAGCTACTCAGGAGGCTGAGGTGGGAGGAT | ||
| Transcript ID List | ENST00000462694.1; ENST00000371953.8; rmsk_3372630 | ||
| External Link | RMBase: RNA-editing_site_17393 | ||
| mod ID: A2ISITE004347 | Click to Show/Hide the Full List | ||
| mod site | chr10:87891720-87891721:+ | [20] | |
| Sequence | GAGGCTGAGGTGGGAGGATCACCTGAGCCCAGGAGGTCAAG | ||
| Transcript ID List | rmsk_3372630; ENST00000462694.1; ENST00000371953.8 | ||
| External Link | RMBase: RNA-editing_site_17394 | ||
| mod ID: A2ISITE004348 | Click to Show/Hide the Full List | ||
| mod site | chr10:87907473-87907474:+ | [21] | |
| Sequence | GTAATTTTGTTACTGTGATAATATATTTTATATATATGTGT | ||
| Transcript ID List | rmsk_3372651; ENST00000371953.8 | ||
| External Link | RMBase: RNA-editing_site_17395 | ||
| mod ID: A2ISITE004349 | Click to Show/Hide the Full List | ||
| mod site | chr10:87909322-87909323:+ | [19] | |
| Sequence | TTTATTTACTTATTTTTTGAAATAGCATCTGGCTCTGTTGC | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: RNA-editing_site_17396 | ||
| mod ID: A2ISITE004350 | Click to Show/Hide the Full List | ||
| mod site | chr10:87930808-87930809:+ | [21] | |
| Sequence | ACAAGTTTTTAAGCAAATGTATTAGCTCTAATTGCATACAC | ||
| Transcript ID List | ENST00000498703.1; ENST00000371953.8 | ||
| External Link | RMBase: RNA-editing_site_17397 | ||
| mod ID: A2ISITE004351 | Click to Show/Hide the Full List | ||
| mod site | chr10:87940181-87940182:+ | [18] | |
| Sequence | GGTTGGGCACGGTGGCTCATACCTATATTCCCAGCACTTTG | ||
| Transcript ID List | ENST00000371953.8; rmsk_3372703 | ||
| External Link | RMBase: RNA-editing_site_17398 | ||
| mod ID: A2ISITE004352 | Click to Show/Hide the Full List | ||
| mod site | chr10:87947839-87947840:+ | [21] | |
| Sequence | GCGTAGGTTGCAGTGAGCCGAGATCGTGCCACTGCACTCCA | ||
| Transcript ID List | ENST00000371953.8; rmsk_3372713 | ||
| External Link | RMBase: RNA-editing_site_17399 | ||
5-methylcytidine (m5C)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE004732 | Click to Show/Hide the Full List | ||
| mod site | chr10:87864050-87864051:+ | [22] | |
| Sequence | ATTTCCAGGGCTGGGAACGCCGGAGAGTTGGTCTCTCCCCT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m5C_site_5620 | ||
N6-methyladenosine (m6A)
| In total 50 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE001224 | Click to Show/Hide the Full List | ||
| mod site | chr10:87864117-87864118:+ | [1] | |
| Sequence | GGCGGCTGGCACATCCAGGGACCCGGGCCGGTTTTAAACCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109375 | ||
| mod ID: M6ASITE001225 | Click to Show/Hide the Full List | ||
| mod site | chr10:87864467-87864468:+ | [23] | |
| Sequence | CTTCAGCCACAGGCTCCCAGACATGACAGCCATCATCAAAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T; AML | ||
| Seq Type List | MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000371953.8; ENST00000487939.1; ENST00000462694.1 | ||
| External Link | RMBase: m6A_site_109376 | ||
| mod ID: M6ASITE001226 | Click to Show/Hide the Full List | ||
| mod site | chr10:87864503-87864504:+ | [23] | |
| Sequence | CAAAGAGATCGTTAGCAGAAACAAAAGGAGATATCAAGAGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000462694.1; ENST00000487939.1; ENST00000371953.8; ENST00000618586.1 | ||
| External Link | RMBase: m6A_site_109377 | ||
| mod ID: M6ASITE001227 | Click to Show/Hide the Full List | ||
| mod site | chr10:87864544-87864545:+ | [24] | |
| Sequence | ATGGATTCGACTTAGACTTGACCTGTATCCATTTCTGCGGC | ||
| Motif Score | 2.839113095 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000487939.1; ENST00000371953.8; ENST00000462694.1; ENST00000618586.1 | ||
| External Link | RMBase: m6A_site_109378 | ||
| mod ID: M6ASITE001228 | Click to Show/Hide the Full List | ||
| mod site | chr10:87876527-87876528:+ | [1] | |
| Sequence | TAAGAATTTAGTATTACAGGACTCAATCAGGGAACTGATTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000462694.1; ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109379 | ||
| mod ID: M6ASITE001229 | Click to Show/Hide the Full List | ||
| mod site | chr10:87894036-87894037:+ | [23] | |
| Sequence | GTACTCAGATATTTATCCAAACATTATTGCTATGGGATTTC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000462694.1; ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109380 | ||
| mod ID: M6ASITE001230 | Click to Show/Hide the Full List | ||
| mod site | chr10:87894087-87894088:+ | [23] | |
| Sequence | ACTTGAAGGCGTATACAGGAACAATATTGATGATGTAGTAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8; ENST00000462694.1 | ||
| External Link | RMBase: m6A_site_109381 | ||
| mod ID: M6ASITE001231 | Click to Show/Hide the Full List | ||
| mod site | chr10:87925541-87925542:+ | [23] | |
| Sequence | TTCAAAGCATAAAAACCATTACAAGATATACAATCTGTAAG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8; ENST00000498703.1 | ||
| External Link | RMBase: m6A_site_109382 | ||
| mod ID: M6ASITE001232 | Click to Show/Hide the Full List | ||
| mod site | chr10:87933105-87933106:+ | [23] | |
| Sequence | CCAATGGCTAAGTGAAGATGACAATCATGTTGCAGCAATTC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8; ENST00000498703.1 | ||
| External Link | RMBase: m6A_site_109383 | ||
| mod ID: M6ASITE001233 | Click to Show/Hide the Full List | ||
| mod site | chr10:87933149-87933150:+ | [1] | |
| Sequence | GTAAAGCTGGAAAGGGACGAACTGGTGTAATGATATGTGCA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000371953.8; ENST00000498703.1 | ||
| External Link | RMBase: m6A_site_109384 | ||
| mod ID: M6ASITE001234 | Click to Show/Hide the Full List | ||
| mod site | chr10:87933178-87933179:+ | [23] | |
| Sequence | ATGATATGTGCATATTTATTACATCGGGGCAAATTTTTAAA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000498703.1; ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109385 | ||
| mod ID: M6ASITE001235 | Click to Show/Hide the Full List | ||
| mod site | chr10:87933202-87933203:+ | [23] | |
| Sequence | CGGGGCAAATTTTTAAAGGCACAAGAGGCCCTAGATTTCTA | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000498703.1; ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109386 | ||
| mod ID: M6ASITE001236 | Click to Show/Hide the Full List | ||
| mod site | chr10:87933243-87933244:+ | [23] | |
| Sequence | TGGGGAAGTAAGGACCAGAGACAAAAAGGTAAGTTATTTTT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8; ENST00000498703.1 | ||
| External Link | RMBase: m6A_site_109387 | ||
| mod ID: M6ASITE001237 | Click to Show/Hide the Full List | ||
| mod site | chr10:87945821-87945822:+ | [25] | |
| Sequence | CTGGAAACTTTGGTTTTGGGACCCAGGAACACATTGATCTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000371953.8; ENST00000395256.2 | ||
| External Link | RMBase: m6A_site_109388 | ||
| mod ID: M6ASITE001238 | Click to Show/Hide the Full List | ||
| mod site | chr10:87952211-87952212:+ | [23] | |
| Sequence | ACCAGTGGCACTGTTGTTTCACAAGATGATGTTTGAAACTA | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000472832.2; ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109389 | ||
| mod ID: M6ASITE001239 | Click to Show/Hide the Full List | ||
| mod site | chr10:87957911-87957912:+ | [23] | |
| Sequence | ATTCCTCCAATTCAGGACCCACACGACGGGAAGACAAGTTC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8; ENST00000472832.2 | ||
| External Link | RMBase: m6A_site_109390 | ||
| mod ID: M6ASITE001240 | Click to Show/Hide the Full List | ||
| mod site | chr10:87957924-87957925:+ | [23] | |
| Sequence | AGGACCCACACGACGGGAAGACAAGTTCATGTACTTTGAGT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000472832.2; ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109391 | ||
| mod ID: M6ASITE001241 | Click to Show/Hide the Full List | ||
| mod site | chr10:87957993-87957994:+ | [23] | |
| Sequence | TATCAAAGTAGAGTTCTTCCACAAACAGAACAAGATGCTAA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000472832.2; ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109392 | ||
| mod ID: M6ASITE001242 | Click to Show/Hide the Full List | ||
| mod site | chr10:87960920-87960921:+ | [23] | |
| Sequence | TGTTTCACTTTTGGGTAAATACATTCTTCATACCAGGACCA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8; ENST00000472832.2 | ||
| External Link | RMBase: m6A_site_109393 | ||
| mod ID: M6ASITE001243 | Click to Show/Hide the Full List | ||
| mod site | chr10:87961026-87961027:+ | [23] | |
| Sequence | TATAGAGCGTGCAGATAATGACAAGGAATATCTAGTACTTA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8; ENST00000472832.2 | ||
| External Link | RMBase: m6A_site_109394 | ||
| mod ID: M6ASITE001244 | Click to Show/Hide the Full List | ||
| mod site | chr10:87961083-87961084:+ | [23] | |
| Sequence | TCTTGACAAAGCAAATAAAGACAAAGCCAACCGATACTTTT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000472832.2; ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109395 | ||
| mod ID: M6ASITE001245 | Click to Show/Hide the Full List | ||
| mod site | chr10:87965301-87965302:+ | [23] | |
| Sequence | TCTAGGTGAAGCTGTACTTCACAAAAACAGTAGAGGAGCCG | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109396 | ||
| mod ID: M6ASITE001246 | Click to Show/Hide the Full List | ||
| mod site | chr10:87965355-87965356:+ | [23] | |
| Sequence | CTAGCAGTTCAACTTCTGTAACACCAGATGTTAGTGACAAT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109397 | ||
| mod ID: M6ASITE001247 | Click to Show/Hide the Full List | ||
| mod site | chr10:87965497-87965498:+ | [23] | |
| Sequence | TTTTTATCAAGAGGGATAAAACACCATGAAAATAAACTTGA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109398 | ||
| mod ID: M6ASITE001248 | Click to Show/Hide the Full List | ||
| mod site | chr10:87965512-87965513:+ | [24] | |
| Sequence | ATAAAACACCATGAAAATAAACTTGAATAAACTGAAAATGG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | CD8T; A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109399 | ||
| mod ID: M6ASITE001249 | Click to Show/Hide the Full List | ||
| mod site | chr10:87965522-87965523:+ | [1] | |
| Sequence | ATGAAAATAAACTTGAATAAACTGAAAATGGACCTTTTTTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109400 | ||
| mod ID: M6ASITE001250 | Click to Show/Hide the Full List | ||
| mod site | chr10:87965559-87965560:+ | [23] | |
| Sequence | TTTTTTTTAATGGCAATAGGACATTGTGTCAGATTACCAGT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109401 | ||
| mod ID: M6ASITE001251 | Click to Show/Hide the Full List | ||
| mod site | chr10:87965642-87965643:+ | [23] | |
| Sequence | TATACATCCACAGGGTTTTGACACTTGTTGTCCAGTTGAAA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109402 | ||
| mod ID: M6ASITE001252 | Click to Show/Hide the Full List | ||
| mod site | chr10:87965710-87965711:+ | [23] | |
| Sequence | TACCTTTTTGTGTCAAAAGGACATTTAAAATTCAATTAGGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109403 | ||
| mod ID: M6ASITE001253 | Click to Show/Hide the Full List | ||
| mod site | chr10:87965897-87965898:+ | [26] | |
| Sequence | ACCCCTTTGCACTTGTGGCAACAGATAAGTTTGCAGTTGGC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | brain | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109404 | ||
| mod ID: M6ASITE001254 | Click to Show/Hide the Full List | ||
| mod site | chr10:87965945-87965946:+ | [23] | |
| Sequence | GGTTTCCGAAGGGTTTTGCTACATTCTAATGCATGTATTCG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109405 | ||
| mod ID: M6ASITE001255 | Click to Show/Hide the Full List | ||
| mod site | chr10:87966585-87966586:+ | [26] | |
| Sequence | AATTTTCAATTTGAGATTCTACAGTAAGCGTTTTTTTTCTT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109406 | ||
| mod ID: M6ASITE001256 | Click to Show/Hide the Full List | ||
| mod site | chr10:87966836-87966837:+ | [25] | |
| Sequence | ATGCTGCACACAAAAAAAAGACATTTGATTTTTCAGTAGAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109407 | ||
| mod ID: M6ASITE001257 | Click to Show/Hide the Full List | ||
| mod site | chr10:87966865-87966866:+ | [25] | |
| Sequence | TTTTCAGTAGAAATTGTCCTACATGTGCTTTATTGATTTGC | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109408 | ||
| mod ID: M6ASITE001258 | Click to Show/Hide the Full List | ||
| mod site | chr10:87967059-87967060:+ | [23] | |
| Sequence | CATACGATTTTAAGCGGAGTACAACTACTATTGTAAAGCTA | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109409 | ||
| mod ID: M6ASITE001259 | Click to Show/Hide the Full List | ||
| mod site | chr10:87967150-87967151:+ | [23] | |
| Sequence | CAAATTATACCTTCACCTTGACATTTGAATATCCAGCCATT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T; AML | ||
| Seq Type List | MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109410 | ||
| mod ID: M6ASITE001260 | Click to Show/Hide the Full List | ||
| mod site | chr10:87967257-87967258:+ | [23] | |
| Sequence | TGGTCTGACCTAGTTAATTTACAAATACAGATTGAATAGGA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109411 | ||
| mod ID: M6ASITE001261 | Click to Show/Hide the Full List | ||
| mod site | chr10:87967502-87967503:+ | [23] | |
| Sequence | TTAAATTGGTAAAGTTAGAGACAACTATTCTAACACCTCAC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109412 | ||
| mod ID: M6ASITE001262 | Click to Show/Hide the Full List | ||
| mod site | chr10:87967514-87967515:+ | [23] | |
| Sequence | AGTTAGAGACAACTATTCTAACACCTCACCATTGAAATTTA | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T; AML | ||
| Seq Type List | MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109413 | ||
| mod ID: M6ASITE001263 | Click to Show/Hide the Full List | ||
| mod site | chr10:87967630-87967631:+ | [23] | |
| Sequence | TATAAATATTGTTCTTTGTTACAATTTCGGGCACCGCATAT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109414 | ||
| mod ID: M6ASITE001264 | Click to Show/Hide the Full List | ||
| mod site | chr10:87967681-87967682:+ | [23] | |
| Sequence | CTTTATTGTTCCAATATGTAACATGGAGGGCCAGGTCATAA | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109415 | ||
| mod ID: M6ASITE001265 | Click to Show/Hide the Full List | ||
| mod site | chr10:87967708-87967709:+ | [23] | |
| Sequence | GGGCCAGGTCATAAATAATGACATTATAATGGGCTTTTGCA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109416 | ||
| mod ID: M6ASITE001266 | Click to Show/Hide the Full List | ||
| mod site | chr10:87967814-87967815:+ | [23] | |
| Sequence | GTTTTTGAGAAGCCTTGCTTACATTTTATGGTGTAGTCATT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109417 | ||
| mod ID: M6ASITE001267 | Click to Show/Hide the Full List | ||
| mod site | chr10:87967946-87967947:+ | [25] | |
| Sequence | ATTTGACAAGAATTGCTATGACTGAAAGGTTTTCGAGTCCT | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109418 | ||
| mod ID: M6ASITE001268 | Click to Show/Hide the Full List | ||
| mod site | chr10:87968077-87968078:+ | [26] | |
| Sequence | TTTTGGATGTGCAGCAGCTTACATGTCTGAAGTTACTTGAA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109419 | ||
| mod ID: M6ASITE001269 | Click to Show/Hide the Full List | ||
| mod site | chr10:87968229-87968230:+ | [26] | |
| Sequence | TCACCATTCTTTGCTGTGGCACAGGTTATAAACTTAAGTGG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | brain; kidney; liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109420 | ||
| mod ID: M6ASITE001270 | Click to Show/Hide the Full List | ||
| mod site | chr10:87968240-87968241:+ | [25] | |
| Sequence | TGCTGTGGCACAGGTTATAAACTTAAGTGGAGTTTACCGGC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109421 | ||
| mod ID: M6ASITE001271 | Click to Show/Hide the Full List | ||
| mod site | chr10:87968711-87968712:+ | [23] | |
| Sequence | ATTTGTTATTGTGTTTGTTAACAACCCTTTATCTCTTAGTG | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000371953.8 | ||
| External Link | RMBase: m6A_site_109422 | ||
| mod ID: M6ASITE094741 | Click to Show/Hide the Full List | ||
| mod site | KQ090021.1:79940-79941:+ | [27] | |
| Sequence | CGGCGGCGGCACATCCAGGGACCCGGGCCGGTTTTAAACCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; CD34; HEK293T; HepG2; H1A; H1B; A549; LCLs; MM6; Jurkat; HEK293A-TOA; TREX; iSLK; MSC; TIME; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000645317.1; ENST00000644628.1 | ||
| External Link | RMBase: m6A_site_881922 | ||
| mod ID: M6ASITE094742 | Click to Show/Hide the Full List | ||
| mod site | KQ090021.1:79957-79958:+ | [27] | |
| Sequence | GGGACCCGGGCCGGTTTTAAACCTCCCGTCCGCCGCCGCCG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; CD34; HEK293T; HepG2; H1A; H1B; A549; LCLs; MM6; Jurkat; HEK293A-TOA; TREX; iSLK; MSC; TIME; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000644628.1; ENST00000645317.1 | ||
| External Link | RMBase: m6A_site_881923 | ||
References