m6A Regulator Information
General Information of the m6A Regulator (ID: REG00022)
| Regulator Name | YTH domain-containing protein 1 (YTHDC1) | ||||
|---|---|---|---|---|---|
| Synonyms |
Splicing factor YT521; YT521-B; KIAA1966; YT521
Click to Show/Hide
|
||||
| Gene Name | YTHDC1 | ||||
| Sequence |
MAADSREEKDGELNVLDDILTEVPEQDDELYNPESEQDKNEKKGSKRKSDRMESTDTKRQ
KPSVHSRQLVSKPLSSSVSNNKRIVSTKGKSATEYKNEEYQRSERNKRLDADRKIRLSSS ASREPYKNQPEKTCVRKRDPERRAKSPTPDGSERIGLEVDRRASRSSQSSKEEVNSEEYG SDHETGSSGSSDEQGNNTENEEEGVEEDVEEDEEVEEDAEEDEEVDEDGEEEEEEEEEEE EEEEEEEEEYEQDERDQKEEGNDYDTRSEASDSGSESVSFTDGSVRSGSGTDGSDEKKKE RKRARGISPIVFDRSGSSASESYAGSEKKHEKLSSSVRAVRKDQTSKLKYVLQDARFFLI KSNNHENVSLAKAKGVWSTLPVNEKKLNLAFRSARSVILIFSVRESGKFQGFARLSSESH HGGSPIHWVLPAGMSAKMLGGVFKIDWICRRELPFTKSAHLTNPWNEHKPVKIGRDGQEI ELECGTQLCLLFPPDESIDLYQVIHKMRHKRRMHSQPRSRGRPSRREPVRDVGRRRPEDY DIHNSRKKPRIDYPPEFHQRPGYLKDPRYQEVDRRFSGVRRDVFLNGSYNDYVREFHNMG PPPPWQGMPPYPGMEQPPHHPYYQHHAPPPQAHPPYSGHHPVPHEARYRDKRVHDYDMRV DDFLRRTQAVVSGRRSRPRERDRERERDRPRDNRRDRERDRGRDRERERERLCDRDRDRG ERGRYRR Click to Show/Hide
|
||||
| Function |
Regulator of alternative splicing that specifically recognizes and binds N6-methyladenosine (m6A)-containing RNAs. M6A is a modification present at internal sites of mRNAs and some non-coding RNAs and plays a role in the efficiency of mRNA splicing, processing and stability. Acts as a key regulator of exon-inclusion or exon-skipping during alternative splicing via interaction with mRNA splicing factors SRSF3 and SRSF10. Specifically binds m6A-containing mRNAs and promotes recruitment of SRSF3 to its mRNA-binding elements adjacent to m6A sites, leading to exon-inclusion during alternative splicing. In contrast, interaction with SRSF3 prevents interaction with SRSF10, a splicing factor that promotes exon skipping: this prevents SRSF10 from binding to its mRNA-binding sites close to m6A-containing regions, leading to inhibit exon skipping during alternative splicing. May also regulate alternative splice site selection. Also involved in nuclear export of m6A-containing mRNAs via interaction with SRSF3: interaction with SRSF3 facilitates m6A-containing mRNA-binding to both SRSF3 and NXF1, promoting mRNA nuclear export. Involved in S-adenosyl-L-methionine homeostasis by regulating expression of MAT2A transcripts, probably by binding m6A-containing MAT2A mRNAs (By similarity). Also recognizes and binds m6A on other RNA molecules. Involved in random X inactivation mediated by Xist RNA: recognizes and binds m6A-containing Xist and promotes transcription repression activity of Xist. Also recognizes and binds m6A-containing single-stranded DNA. Involved in germline development: required for spermatogonial development in males and oocyte growth and maturation in females, probably via its role in alternative splicing (By similarity).
Click to Show/Hide
|
||||
| Gene ID | 91746 | ||||
| Uniprot ID | |||||
| Regulator Type | WRITER ERASER READER | ||||
| Mechanism Diagram | Click to View the Original Diagram | ||||
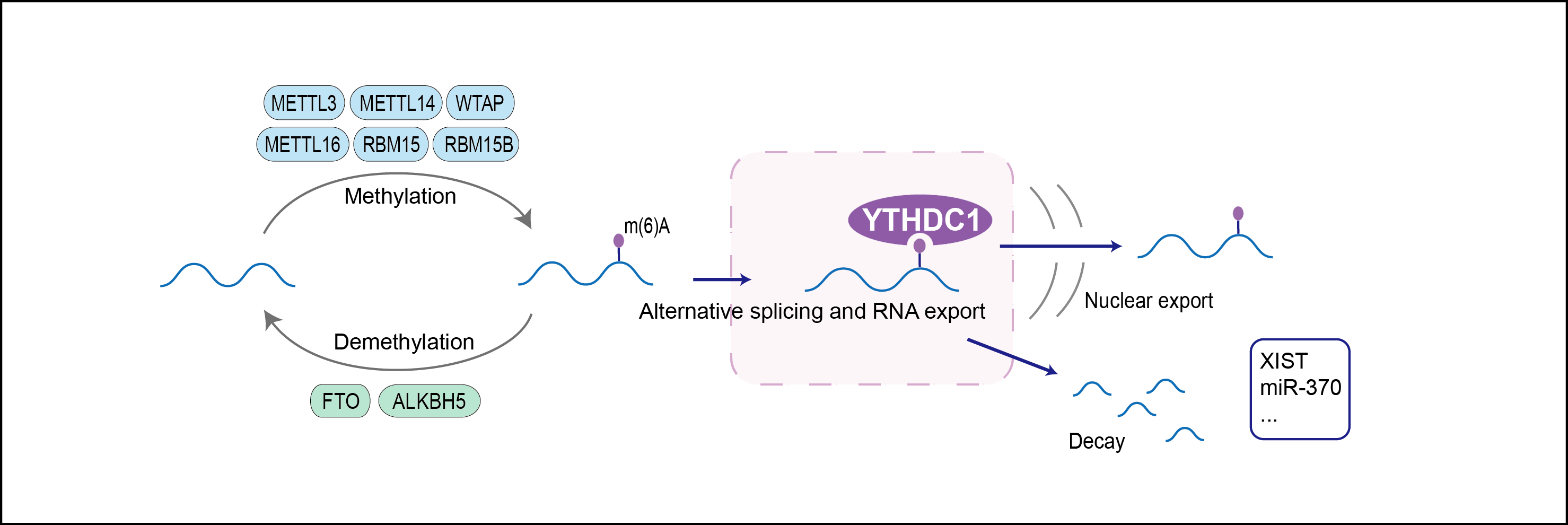
|
|||||
| Target Genes | Click to View Potential Target Genes of This Regulator | ||||
Full List of Target Gene(s) of This m6A Regulator and Corresponding Disease/Drug Response(s)
YTHDC1 can regulate the m6A methylation of following target genes, and result in corresponding disease/drug response(s). You can browse corresponding disease or drug response(s) resulted from the regulation of certain target gene.
Browse Target Gene related Disease
Browse Target Gene related Drug
DNA replication licensing factor MCM4 (MCM4)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDC1 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shYTHDC1 MOLM13 cells
Control: shControl MOLM13 cells
|
GSE168565 | |
| Regulation |
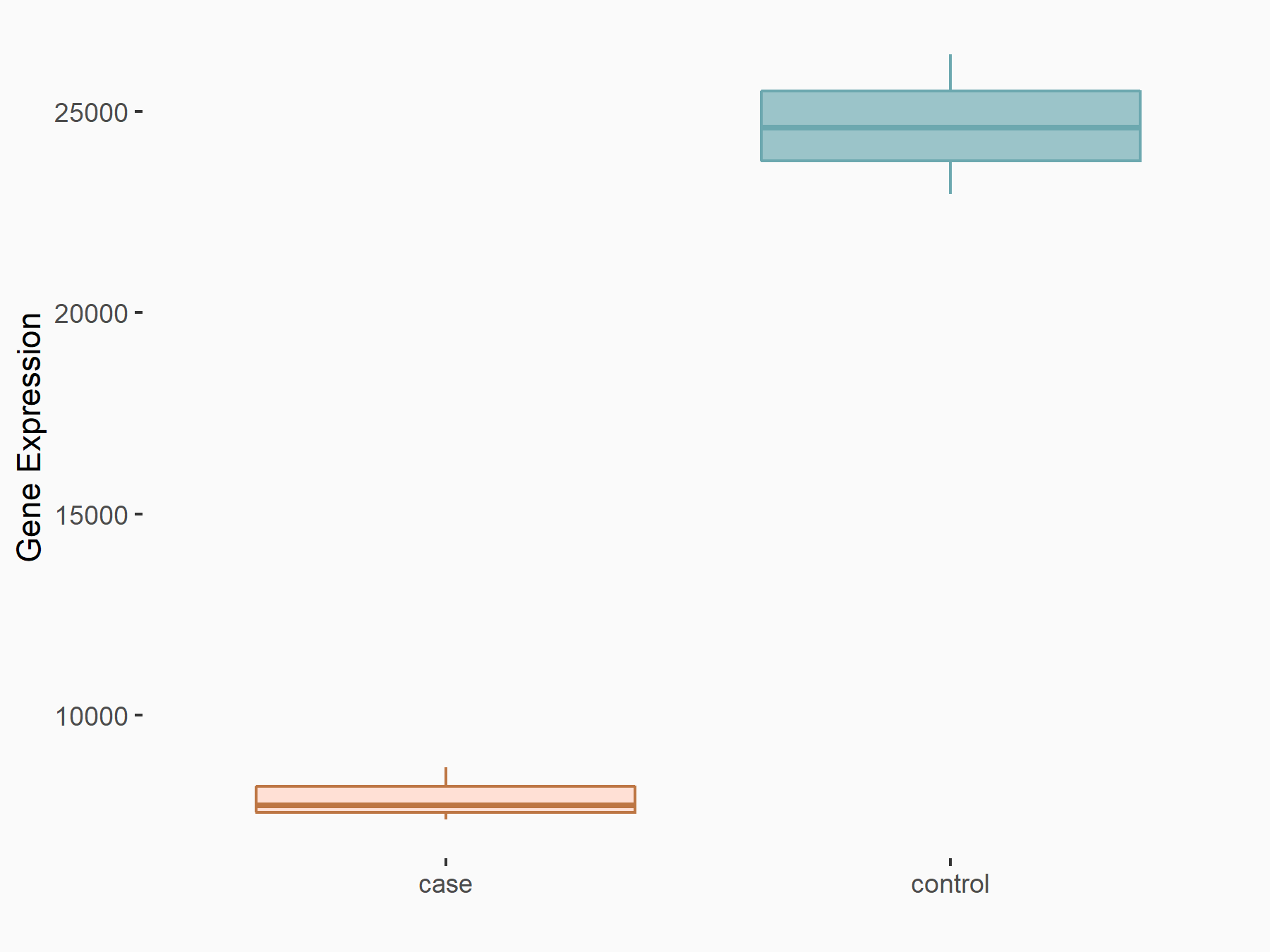  |
logFC: -1.63E+00 p-value: 3.74E-32 |
| More Results | Click to View More RNA-seq Results | |
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Pathway Response | DNA replication | hsa03030 | ||
| Cell Process | DNA replication | |||
| Response Summary | YTHDC1 knockdown has a strong inhibitory effect on proliferation of primary AML cells. YTHDC1 regulates leukemogenesis through DNA replication licensing factor MCM4 (MCM4), which is a critical regulator of DNA replication. | |||
Heat shock 70 kDa protein 1A (HSPA1A)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDC1 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shYTHDC1 MOLM13 cells
Control: shControl MOLM13 cells
|
GSE178859 | |
| Regulation |
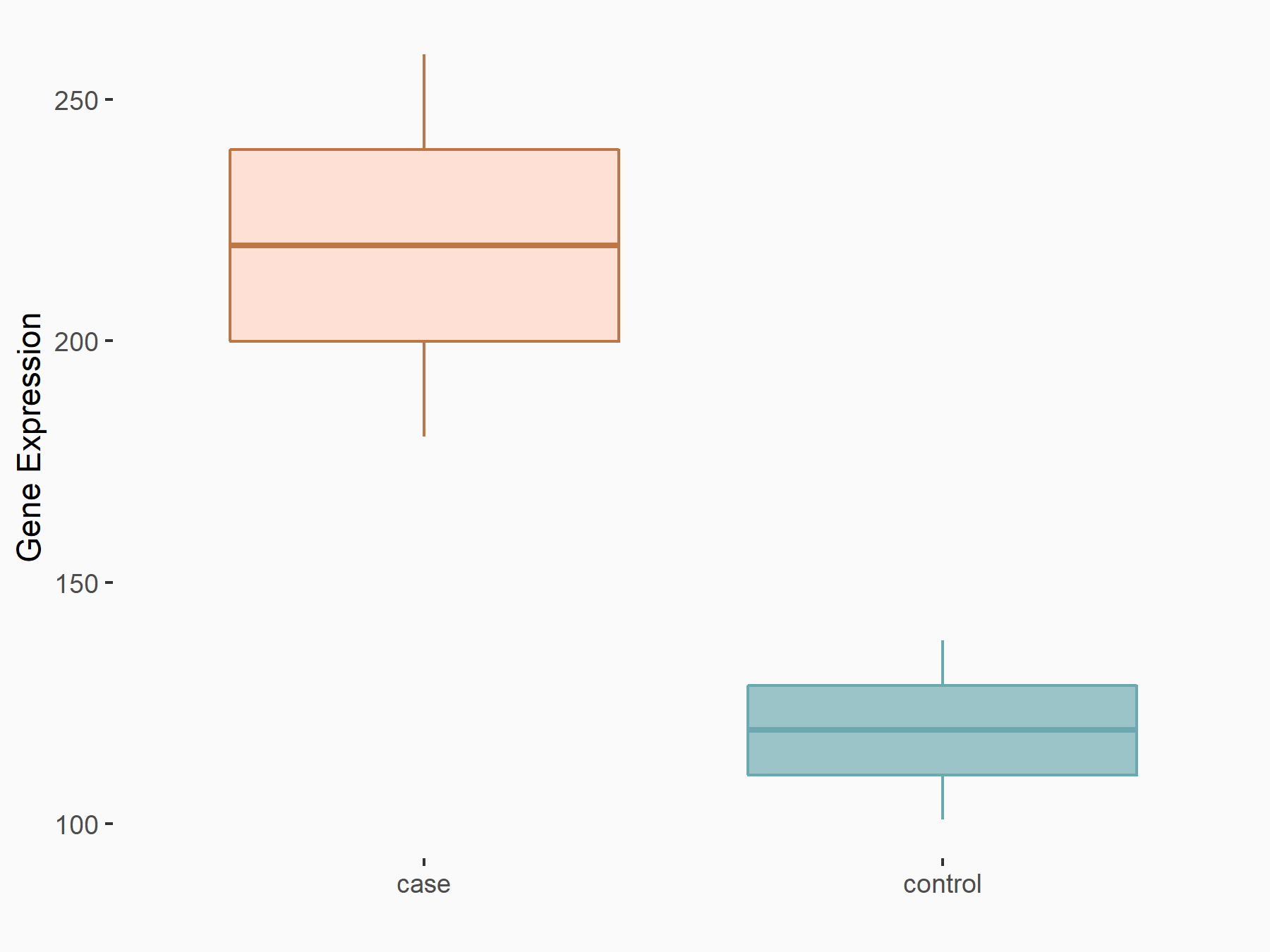 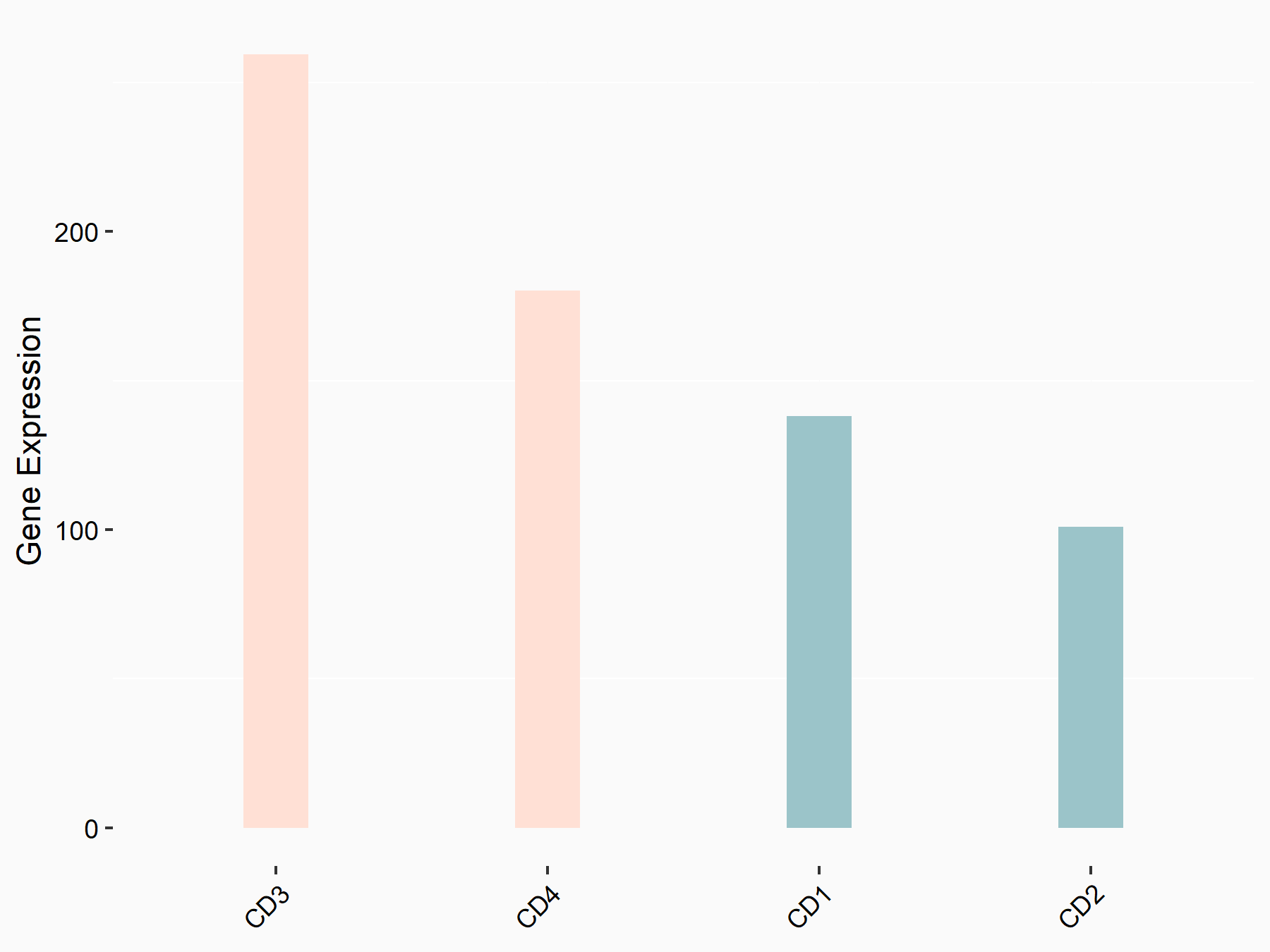 |
logFC: 8.78E-01 p-value: 3.66E-03 |
| More Results | Click to View More RNA-seq Results | |
Effects of heat [ICD-11: NF01]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [2] | |||
| Responsed Disease | Effects of heat [ICD-11: NF01] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
In-vitro Model |
HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| MEF (Mouse embryonic fibroblasts) | ||||
| Response Summary | Heat directly binds to Heat shock 70 kDa protein 1A (HSPA1A), thereby targeting stress genes in a trans-acting manner. Intriguingly, Heat is heavily methylated in the form of m6A. Heat mediates these effects via the nuclear m6A reader YTHDC1, forming a transcriptional silencing complex for stress genes. Reveals a crucial role of nuclear epitranscriptome in the transcriptional regulation of heat shock response. | |||
Mutated in multiple advanced cancers 1 (PTEN)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDC1 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shYTHDC1 MOLM13 cells
Control: shControl MOLM13 cells
|
GSE168565 | |
| Regulation |
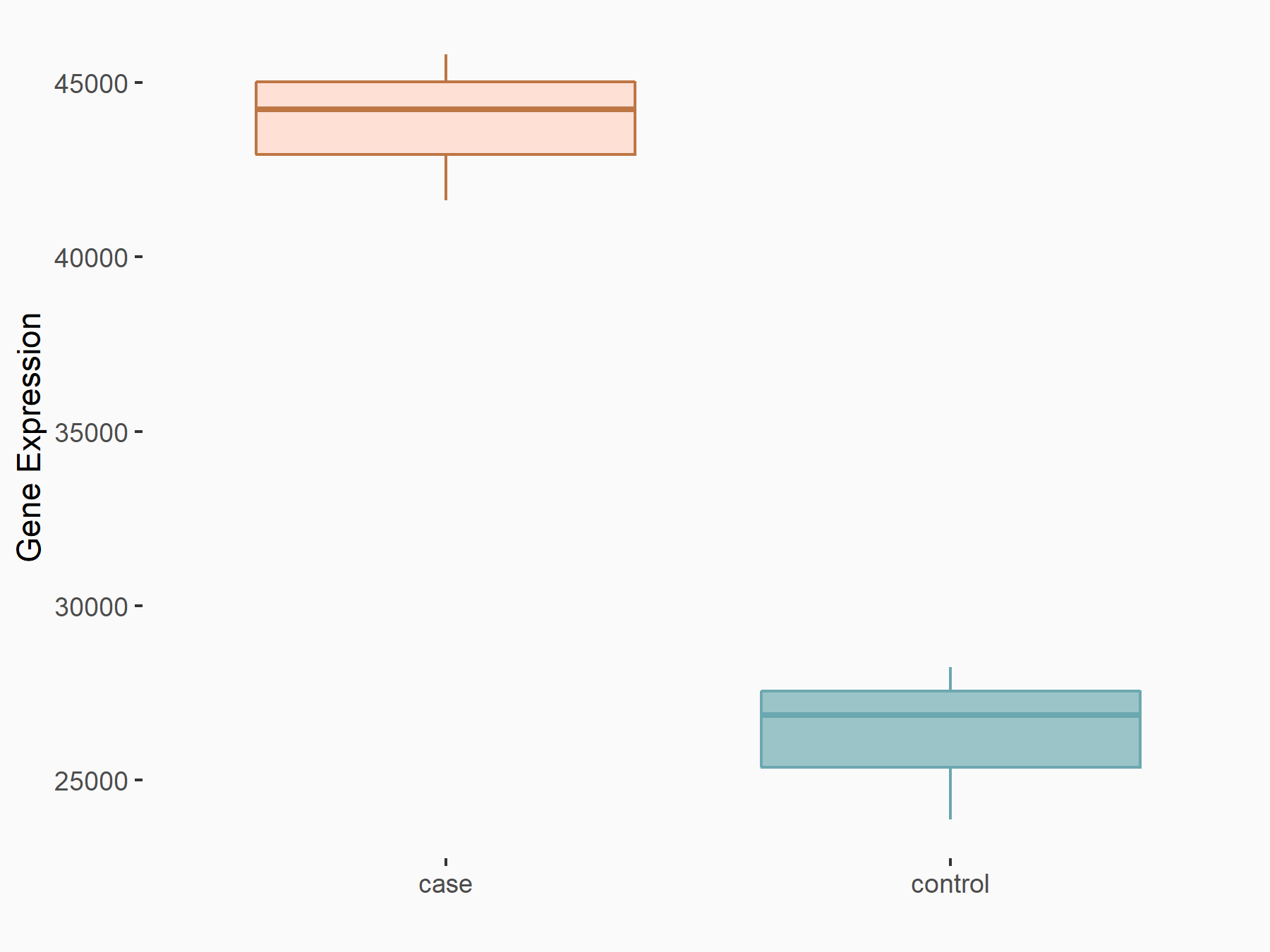 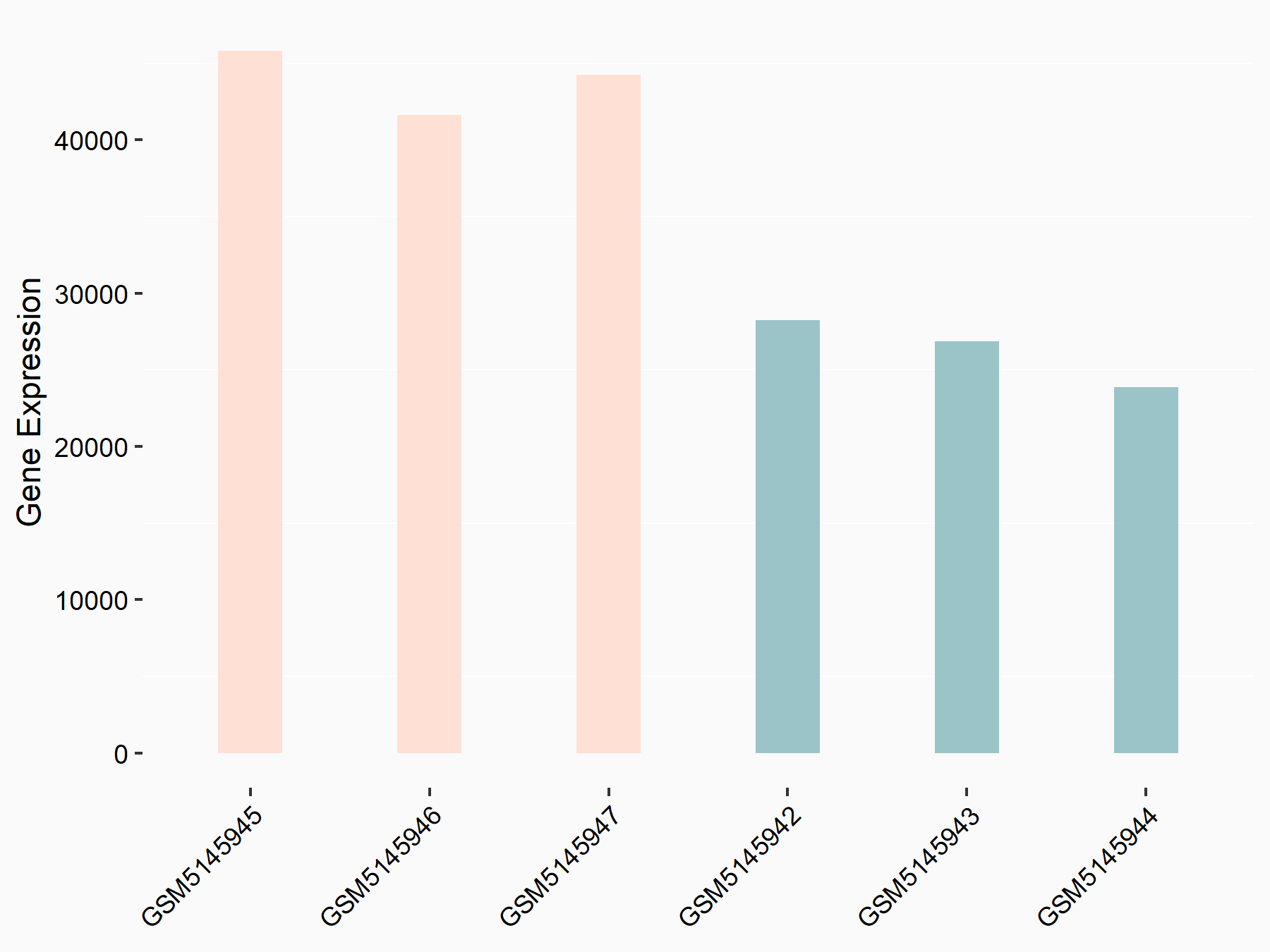 |
logFC: 7.38E-01 p-value: 4.67E-08 |
| More Results | Click to View More RNA-seq Results | |
Acute ischemic stroke [ICD-11: 8B11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [3] | |||
| Responsed Disease | Acute ischemic stroke [ICD-11: 8B11] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| In-vivo Model | Male SD rats weighing 200-250 g were anesthetized using 4% isoflurane in 70% N2O and 30% O2 with a mask. A midline incision was made in the neck, the left external carotid artery (ECA) was carefully exposed and dissected, a monofilament nylon suture with a diameter of about 0.22 mm was inserted from the ECA into the internal carotid artery, and the left middle cerebral artery (MCA) was blocked. After occlusion for 90 minutes, the suture was removed for reperfusion, and ECA was ligated to close the wound. Sham-operated rats underwent the same surgery except for suture insertion. Rats were maintained on top of a warming pad (RWD, 69003) during the above procedures. The breathing machine was used to monitor the respiration of rats. The rats were returned to a heated cage during the recovery phase with free access to food and water. | |||
| Response Summary | YTHDC1 promoted Mutated in multiple advanced cancers 1 (PTEN) mRNA degradation to increase Akt phosphorylation, thus facilitating neuronal survival in particular after ischemia, modulating m6A reader YTHDC1 provide a potential therapeutic target for ischemic stroke. | |||
Sequestosome-1 (SQSTM1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDC1 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shYTHDC1 MOLM13 cells
Control: shControl MOLM13 cells
|
GSE168565 | |
| Regulation |
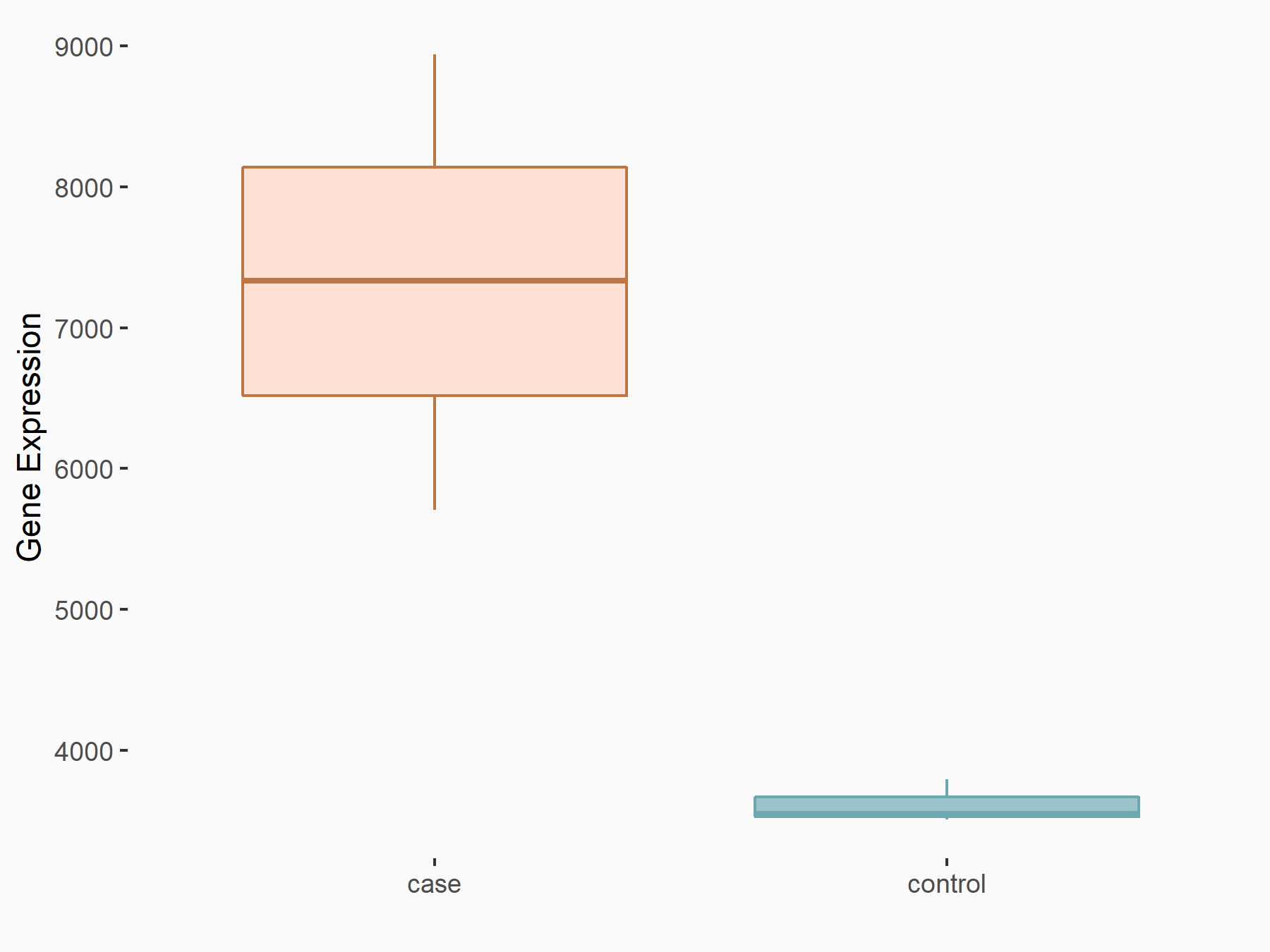 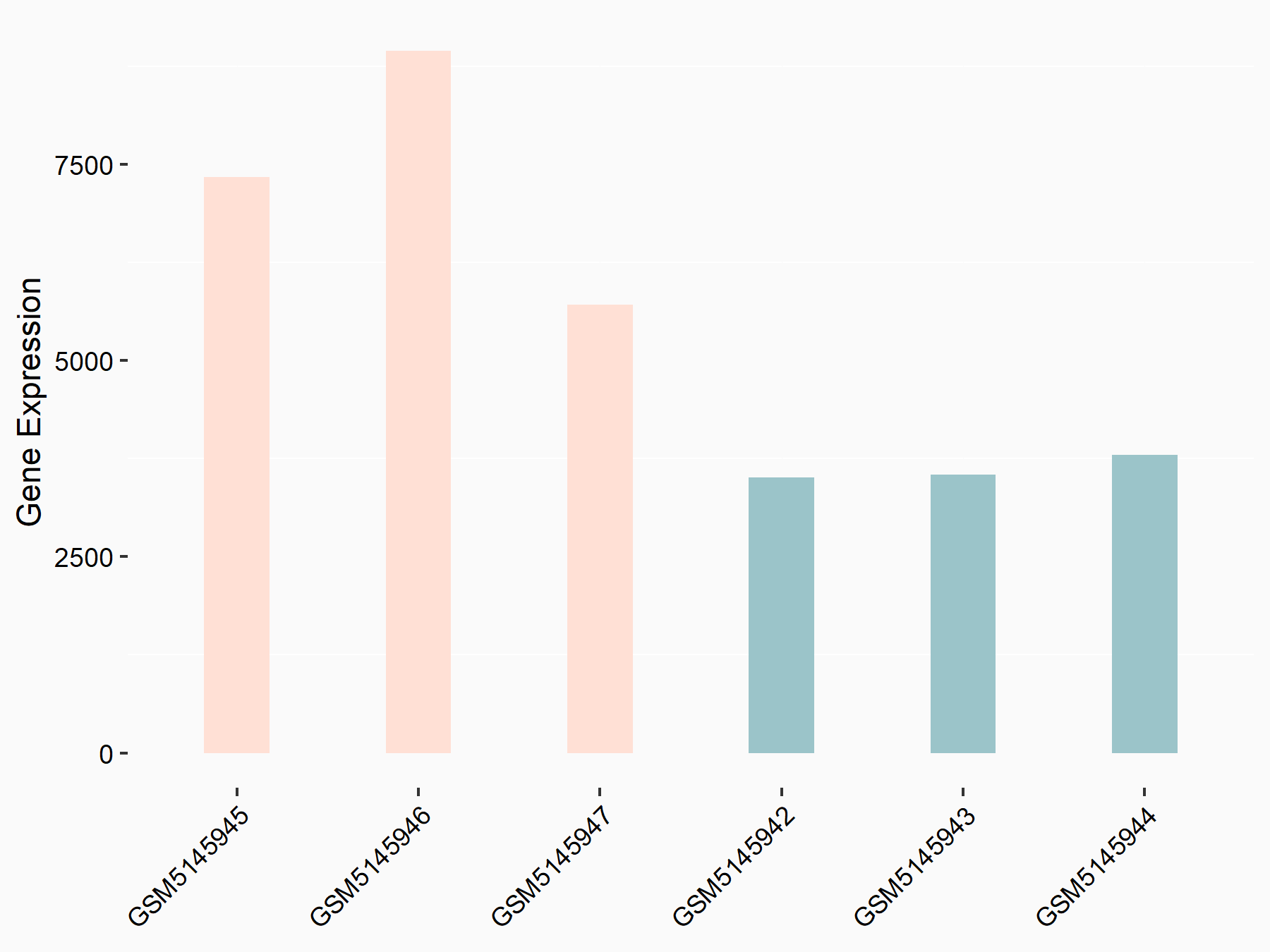 |
logFC: 1.02E+00 p-value: 3.33E-09 |
| More Results | Click to View More RNA-seq Results | |
Diabetes [ICD-11: 5A10-5A14]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Diabetes [ICD-11: 5A10-5A14] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular Processes | |||
| Cellular Transport | ||||
| Cellular catabolism | ||||
| Cell apoptosis | ||||
| Cell autophagy | ||||
In-vitro Model |
HaCaT | Normal | Homo sapiens | CVCL_0038 |
| NHEK (Normal human epithelial keratinocytes) | ||||
| In-vivo Model | The WT-si-NC, WT-si-Ythdc1 and WT-si-Sqstm1 groups were intracutaneously injected with corresponding siRNAs (si-NC, si-Ythdc1, or si-Sqstm1, 2.5 nmol) on the circle. | |||
| Response Summary | In diabetes/diabetic skin, YTHDC1 interacted and cooperated with ELAVL1/HuR (ELAV like RNA binding protein 1) in modulating the expression of Sequestosome-1 (SQSTM1). | |||
Diabetic skin lesions [ICD-11: EB90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Diabetic skin lesions [ICD-11: EB90.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular Processes | |||
| Cellular Transport | ||||
| Cellular catabolism | ||||
| Cell apoptosis | ||||
| Cell autophagy | ||||
In-vitro Model |
HaCaT | Normal | Homo sapiens | CVCL_0038 |
| NHEK (Normal human epithelial keratinocytes) | ||||
| In-vivo Model | The WT-si-NC, WT-si-Ythdc1 and WT-si-Sqstm1 groups were intracutaneously injected with corresponding siRNAs (si-NC, si-Ythdc1, or si-Sqstm1, 2.5 nmol) on the circle. | |||
| Response Summary | In diabetes/diabetic skin, YTHDC1 interacted and cooperated with ELAVL1/HuR (ELAV like RNA binding protein 1) in modulating the expression of Sequestosome-1 (SQSTM1). | |||
Serine/arginine-rich splicing factor 10 (SRSF10)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDC1 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shYTHDC1 MOLM13 cells
Control: shControl MOLM13 cells
|
GSE168565 | |
| Regulation |
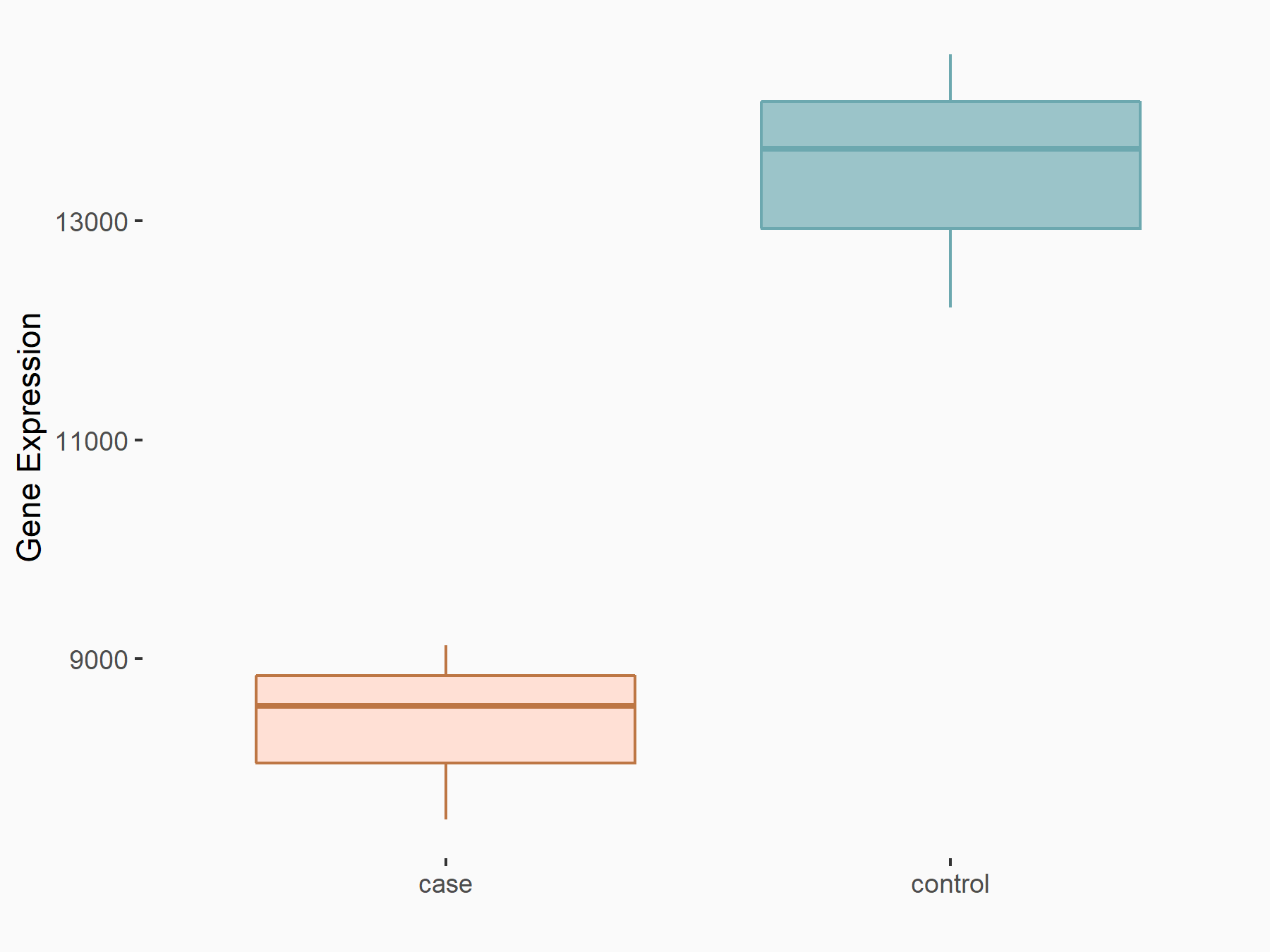 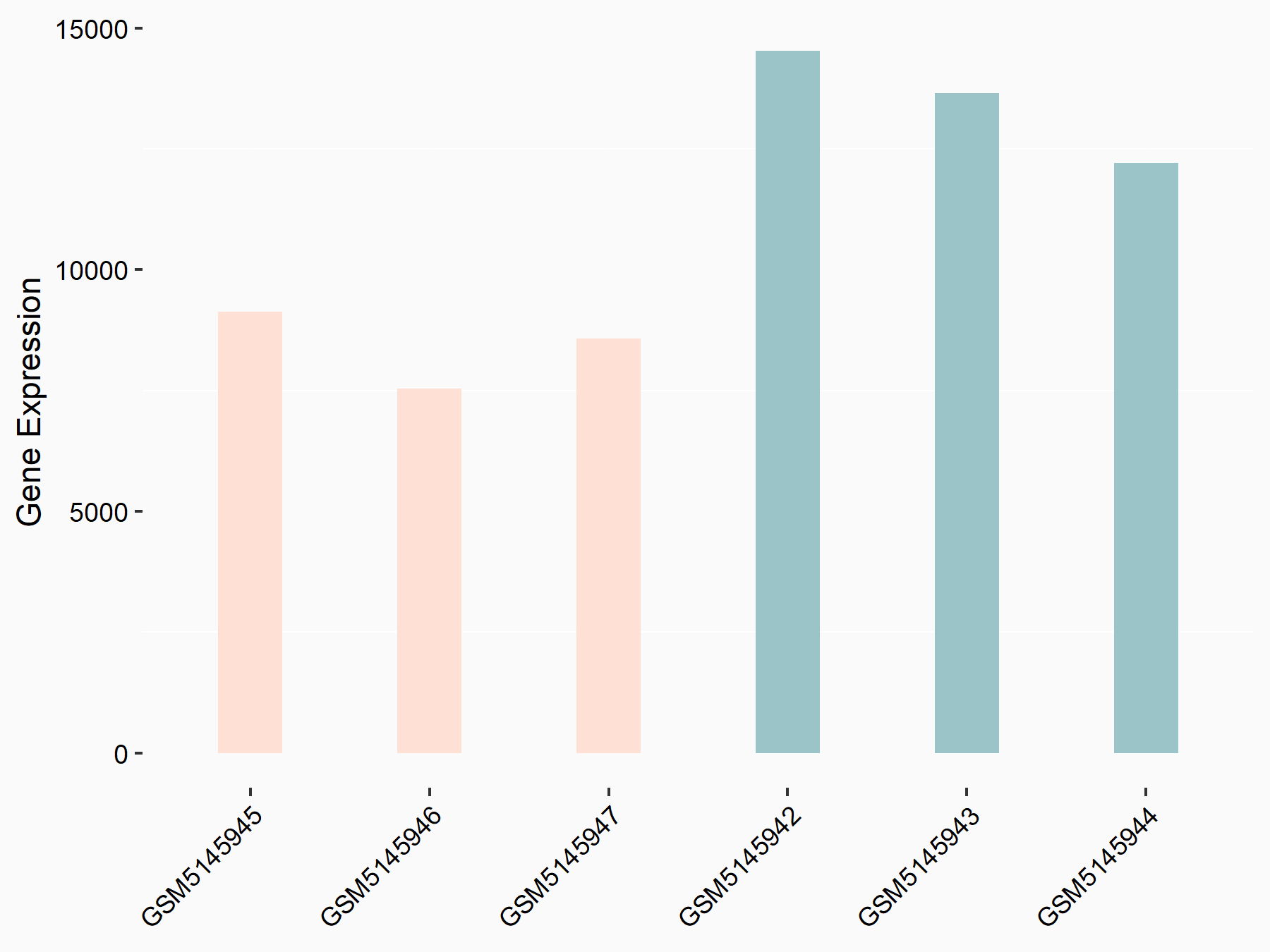 |
logFC: -6.79E-01 p-value: 2.34E-06 |
| More Results | Click to View More RNA-seq Results | |
Kaposi's sarcoma [ICD-11: 2B57]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [5] | |||
| Responsed Disease | Kaposi's sarcoma [ICD-11: 2B57] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Spliceosome | hsa03040 | ||
In-vitro Model |
TIVE-KSHV (KSHV-infected telomerase-immortalized human umbilical vein endothelial cells (TIVE-KSHV cells)) | |||
| iSLK-BAC16 cells (Kaposi's sarcoma cells carrying the recombinant KSHV bacterial artificial chromosome 16 (BAC16) (iSLK-BAC16 cells)) | ||||
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
| Response Summary | Kaposi's sarcoma-associated herpesvirus(KSHV) productive lytic replication plays a pivotal role in the initiation and progression of Kaposi's sarcoma tumors. m6A sites in RTA pre-mRNA crucial for splicing through interactions with YTHDC1, SRSF3 and Serine/arginine-rich splicing factor 10 (SRSF10). m6A in regulating RTA pre-mRNA splicing but also suggest that KSHV has evolved a mechanism to manipulate the host m6A machinery to its advantage in promoting lytic replication. | |||
Serine/arginine-rich splicing factor 11 (SRSF11)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDC1 | ||
| Cell Line | ICM cell line | Mus musculus |
|
Treatment: YTHDC1 knockout ICM cells
Control: Wild type ICM cells
|
GSE157267 | |
| Regulation |
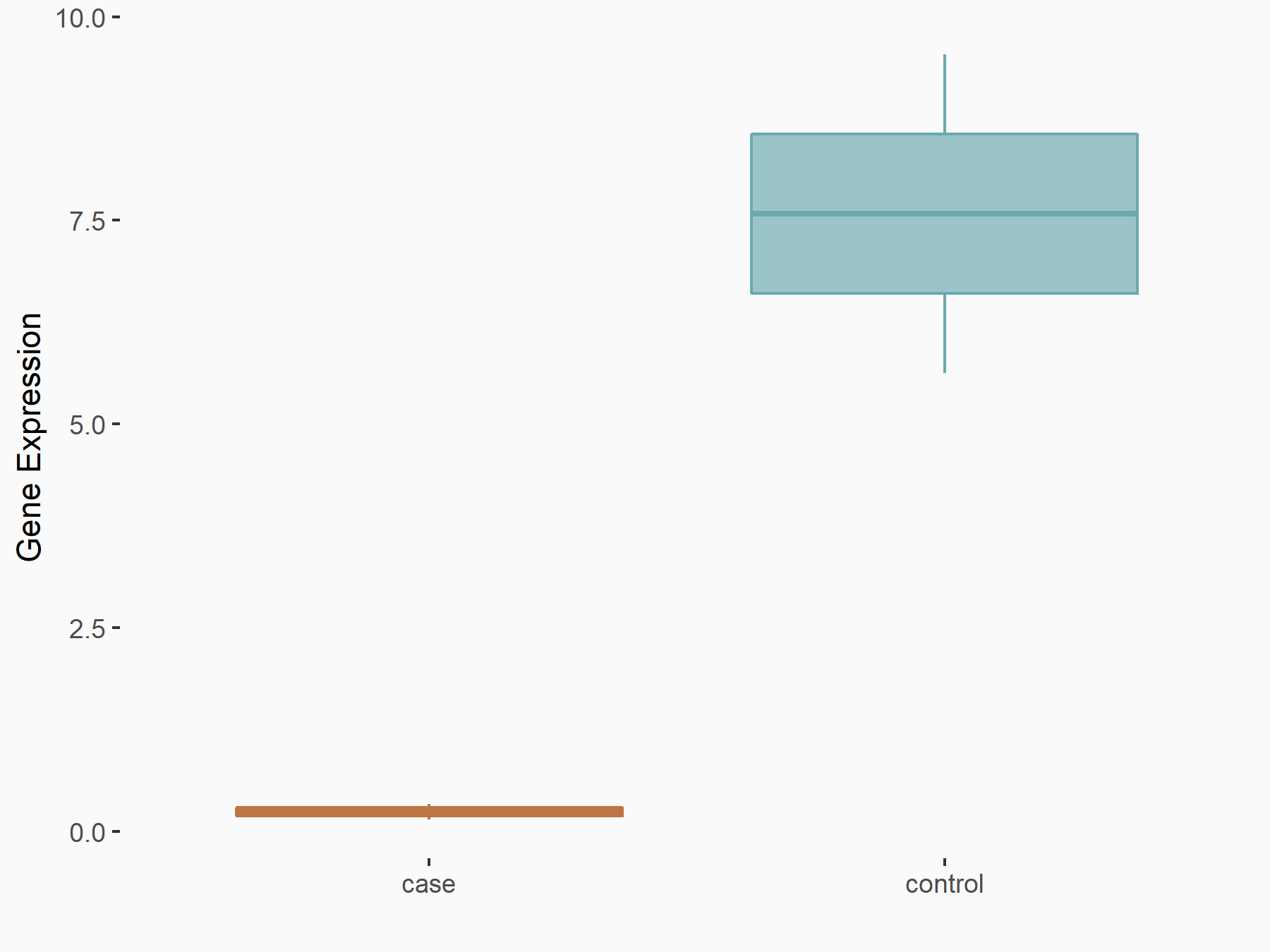 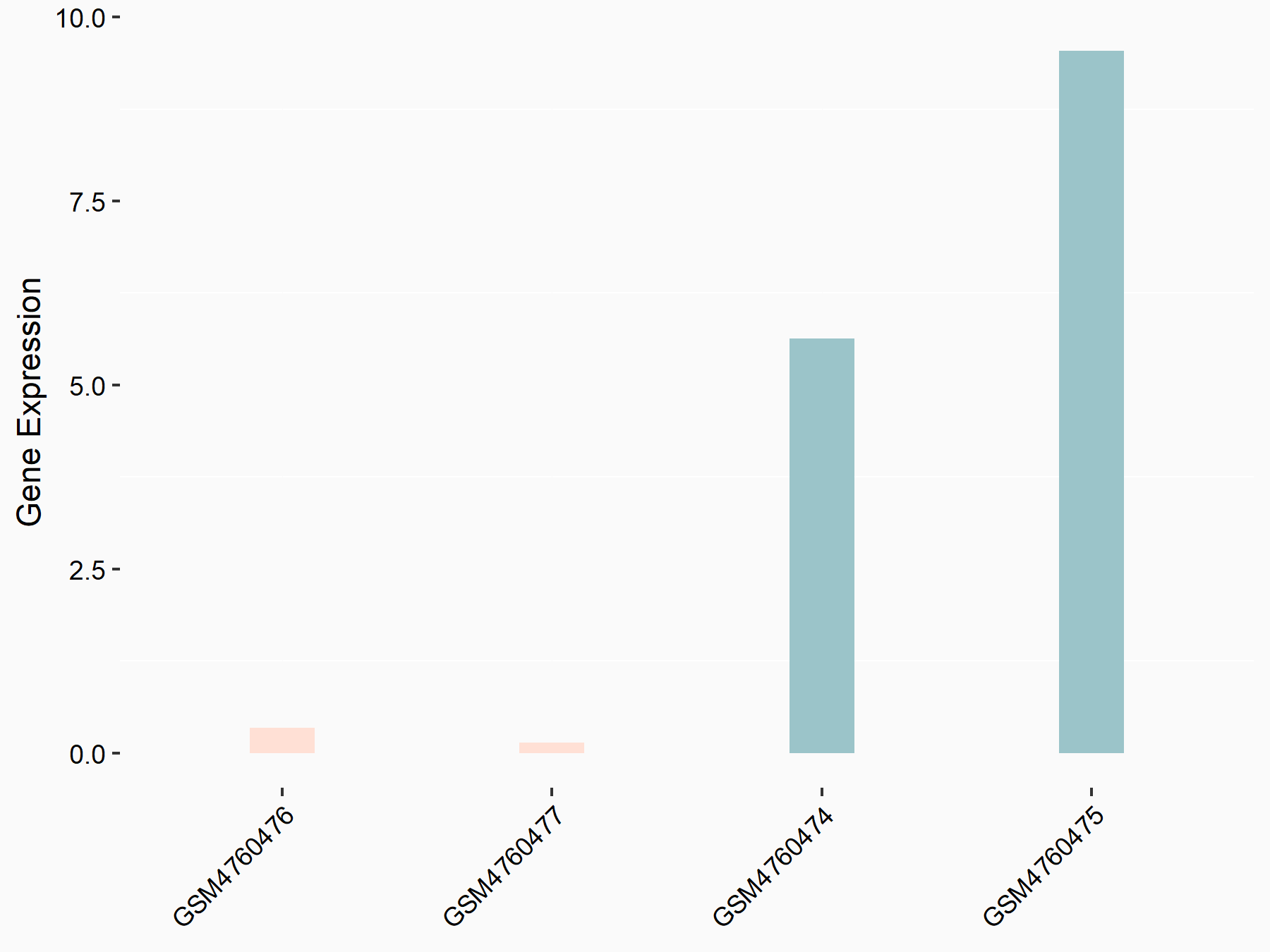 |
logFC: -2.75E+00 p-value: 7.07E-03 |
| More Results | Click to View More RNA-seq Results | |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | mRNA decay | |||
In-vitro Model |
U251 (Fibroblasts or fibroblast like cells) | |||
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| In-vivo Model | For subcutaneous tumor model, each mouse was injected subcutaneously in the right flank with 2 × 106 U87MG cells (METTL3-KD or control) in 100 uL PBS. | |||
| Response Summary | Silencing METTL3 or overexpressing dominant-negative mutant METTL3 suppressed the growth and self-renewal of Glioblastoma cells. Integrated transcriptome and MeRIP-seq analyses revealed that downregulating the expression of METTL3 decreased m6A modification levels of Serine/arginine-rich splicing factor 11 (SRSF11), which led to YTHDC1-dependent NMD of SRSF transcripts and decreased SRSF protein expression. | |||
Serine/arginine-rich splicing factor 3 (SRSF3)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDC1 | ||
| Cell Line | ICM cell line | Mus musculus |
|
Treatment: YTHDC1 knockout ICM cells
Control: Wild type ICM cells
|
GSE157267 | |
| Regulation |
 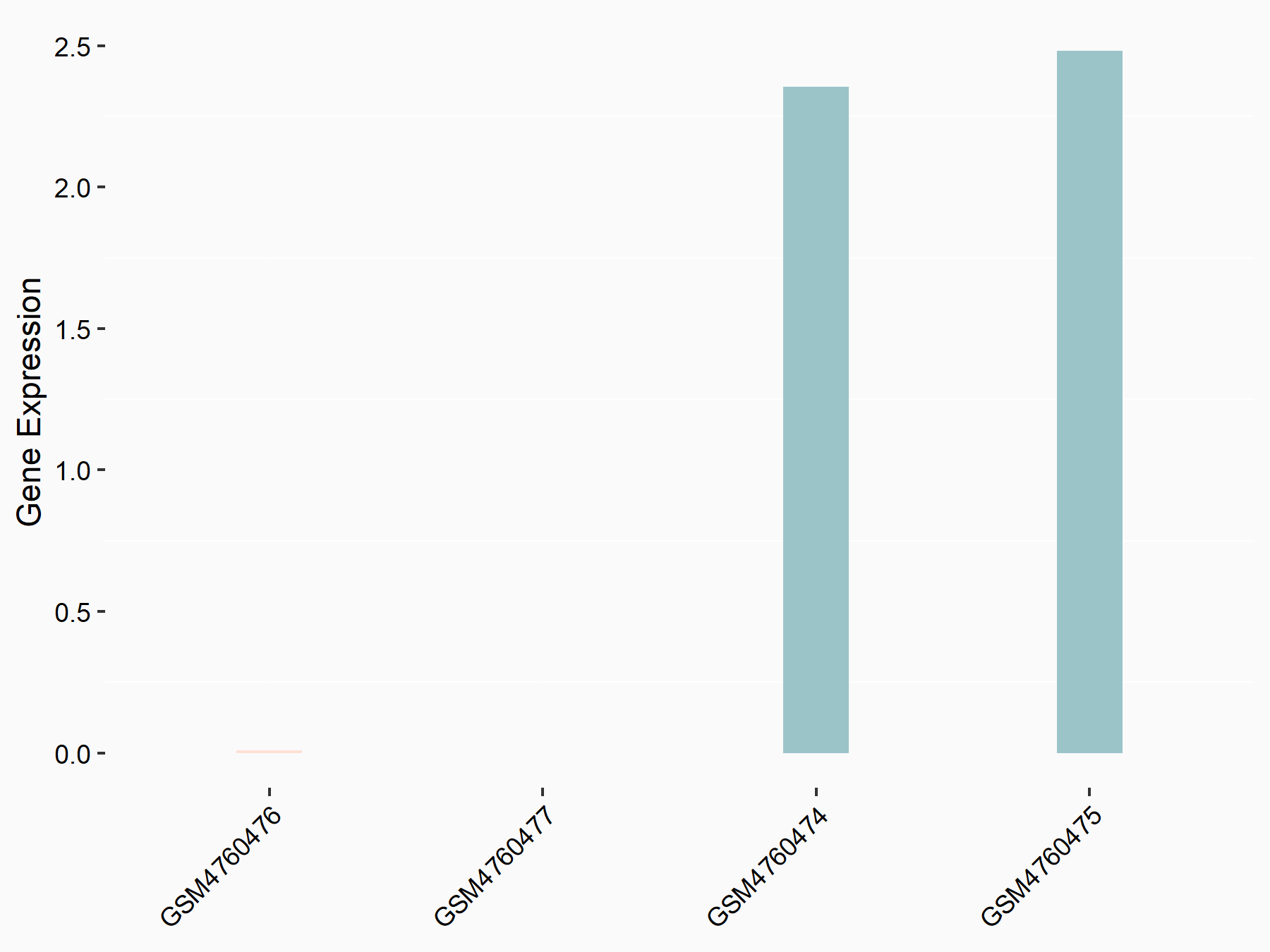 |
logFC: -1.77E+00 p-value: 4.66E-05 |
| More Results | Click to View More RNA-seq Results | |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | mRNA decay | |||
In-vitro Model |
U251 (Fibroblasts or fibroblast like cells) | |||
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| In-vivo Model | For subcutaneous tumor model, each mouse was injected subcutaneously in the right flank with 2 × 106 U87MG cells (METTL3-KD or control) in 100 uL PBS. | |||
| Response Summary | Silencing METTL3 or overexpressing dominant-negative mutant METTL3 suppressed the growth and self-renewal of Glioblastoma cells. Integrated transcriptome and MeRIP-seq analyses revealed that downregulating the expression of METTL3 decreased m6A modification levels of Serine/arginine-rich splicing factor 3 (SRSF3), which led to YTHDC1-dependent NMD of SRSF transcripts and decreased SRSF protein expression. | |||
Kaposi's sarcoma [ICD-11: 2B57]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [5] | |||
| Responsed Disease | Kaposi's sarcoma [ICD-11: 2B57] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Spliceosome | hsa03040 | ||
In-vitro Model |
TIVE-KSHV (KSHV-infected telomerase-immortalized human umbilical vein endothelial cells (TIVE-KSHV cells)) | |||
| iSLK-BAC16 cells (Kaposi's sarcoma cells carrying the recombinant KSHV bacterial artificial chromosome 16 (BAC16) (iSLK-BAC16 cells)) | ||||
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
| Response Summary | Kaposi's sarcoma-associated herpesvirus(KSHV) productive lytic replication plays a pivotal role in the initiation and progression of Kaposi's sarcoma tumors. m6A sites in RTA pre-mRNA crucial for splicing through interactions with YTHDC1, Serine/arginine-rich splicing factor 3 (SRSF3) and SRSF10. m6A in regulating RTA pre-mRNA splicing but also suggest that KSHV has evolved a mechanism to manipulate the host m6A machinery to its advantage in promoting lytic replication. | |||
Type 2 diabetes mellitus [ICD-11: 5A11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [12] | |||
| Responsed Disease | Type 2 diabetes mellitus [ICD-11: 5A11] | |||
In-vitro Model |
MIN6 | Mouse insulinoma | Mus musculus | CVCL_0431 |
Titin
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDC1 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shYTHDC1 MOLM13 cells
Control: shControl MOLM13 cells
|
GSE168565 | |
| Regulation |
 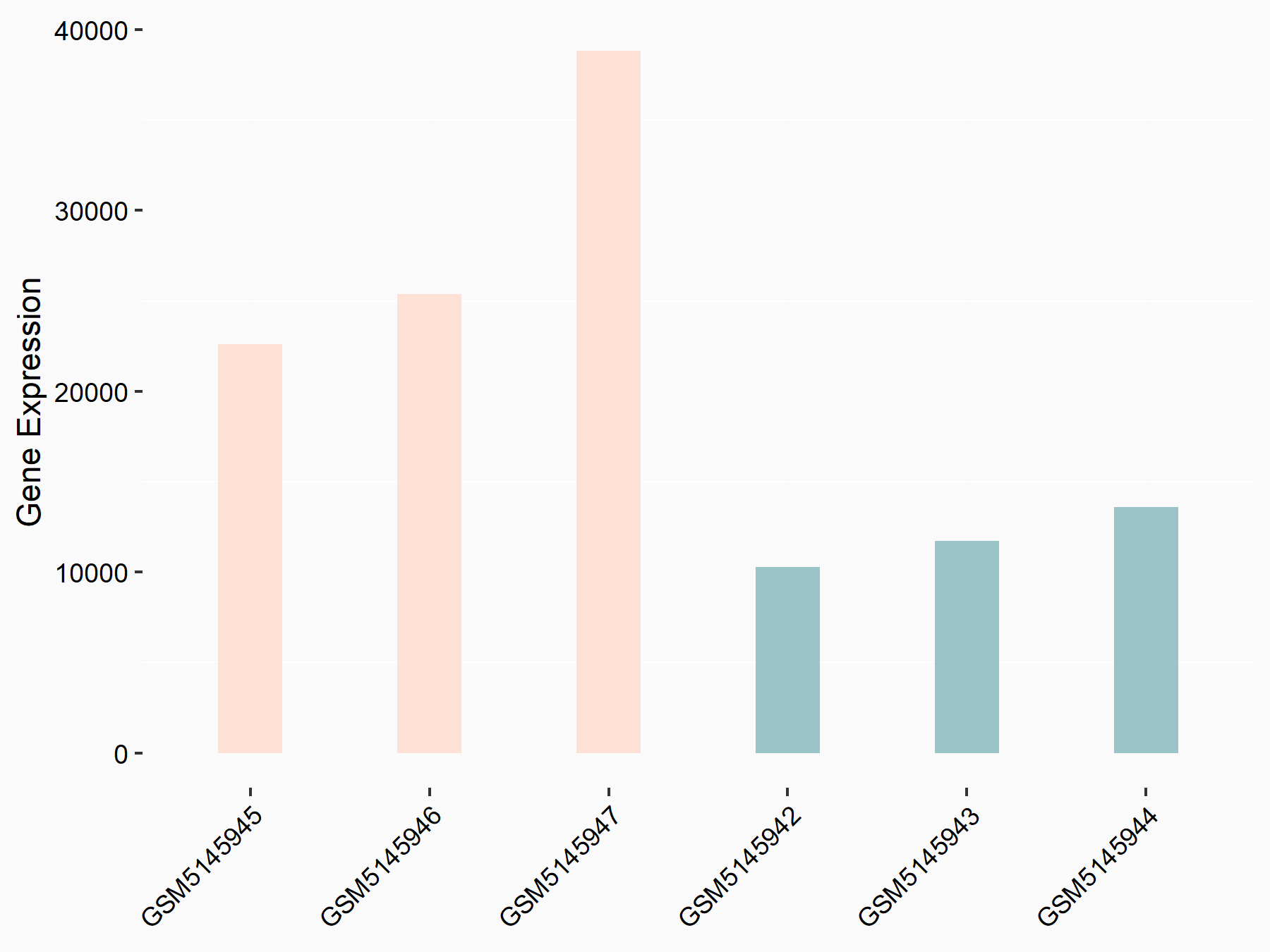 |
logFC: 1.29E+00 p-value: 9.48E-11 |
| More Results | Click to View More RNA-seq Results | |
Cardiomyopathy [ICD-11: BC43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Dilated cardiomyopathy [ICD-11: BC43.0] | |||
In-vitro Model |
Neonatal rat ventricular cardiomyocytes (Primary myocyte cells) | |||
| Response Summary | This study suggests that YTHDC1 plays crucial role in regulating the normal contractile function and the development of dilated cardiomyopathy. | |||
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDC1 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shYTHDC1 MOLM13 cells
Control: shControl MOLM13 cells
|
GSE168565 | |
| Regulation |
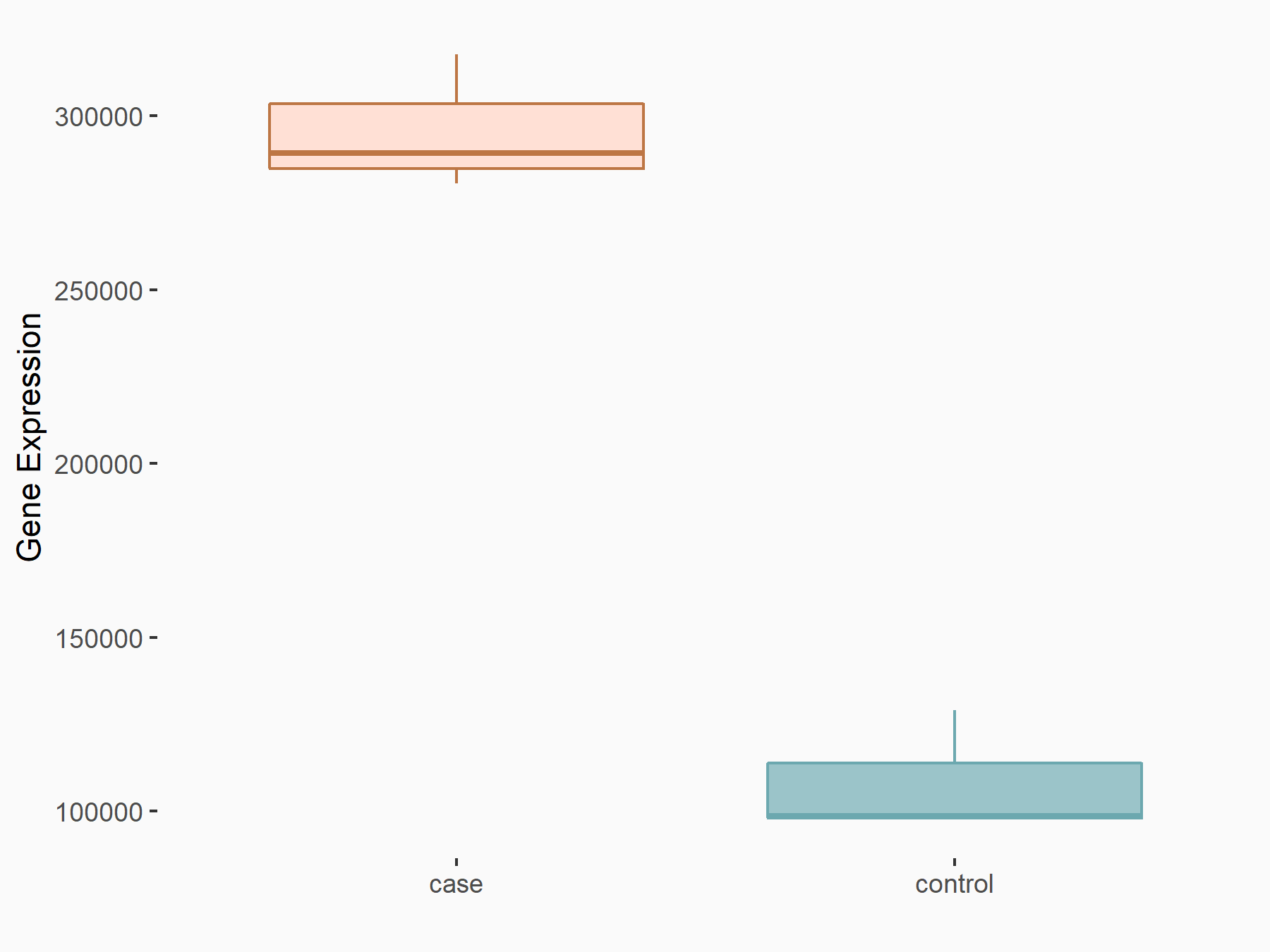 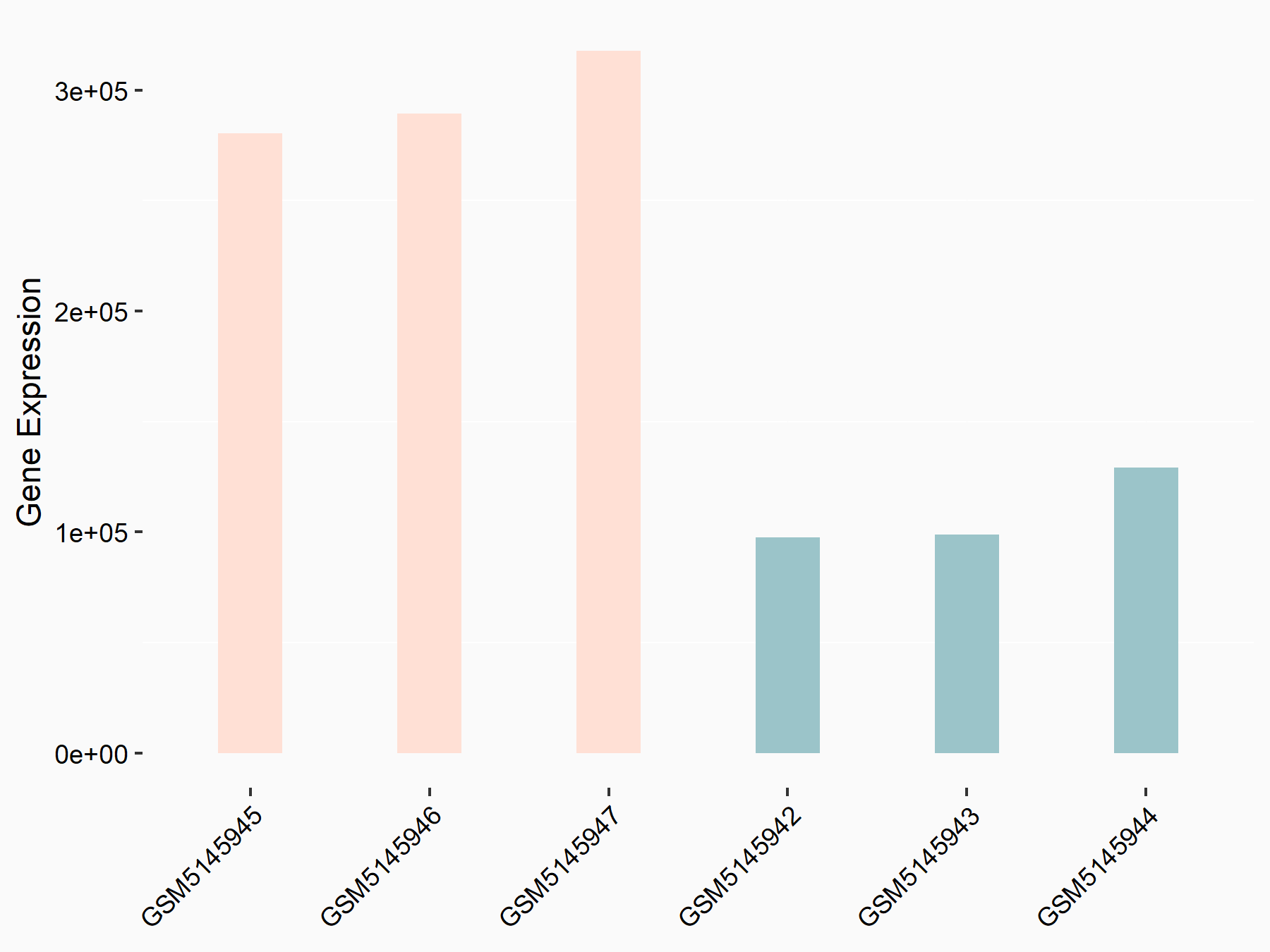 |
logFC: 1.45E+00 p-value: 5.75E-21 |
| More Results | Click to View More RNA-seq Results | |
Malignant haematopoietic neoplasm [ICD-11: 2B33]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [8] | |||
| Responsed Disease | Blood malignancies [ICD-11: 2B33.Y] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Oncogenic fusion protein expression | |||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 | |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| In-vivo Model | The NOD-SCID mice were intravenously (tail vein) implanted with sh-RNA-established NB4 cells. Direct injection of 5 × 106 shRNA-transformed NB4 cells into 150 uL of PBS was performed to establish intravenous (tail vein) leukemia. | |||
| Response Summary | MALAT1 hijacks both chimeric mRNAs and fusion protein in nuclear speckles during chromosomal translocation and mediates colocalization with METTL14 in an oncogenic fusion protein such as PML-RARalpha. Reducing MALAT1 or m6A methyltransferases and the 'reader' YTHDC1 result in the universal retention of distinct oncogenic gene (PML-RARalpha) mRNAs in nucleus. Targeting the lncRNA-triggered autoregulatory loop to disrupt chimeric mRNA transport represents a new common paradigm for treating blood malignancies. | |||
Epithelial membrane protein 3 (EMP3)
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [9] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Doxil | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| mTOR signaling pathway | hsa04150 | |||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | DNA repair | |||
In-vitro Model |
BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| Response Summary | Epithelial membrane protein 3 (EMP3) blocks Akt-mTOR signaling activation and induces autophagy. EMP3 downregulates YTHDC1, which at least in part mediates the effects of EMP3 on breast cancer cells. EMP3 sensitizes breast cancer cells to the DNA-damaging drug Adriamycin. EMP3 downregulation can be responsible for breast cancer chemoresistance. | |||
High mobility group protein HMGI-C (HMGA2)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [10] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Transcriptional misregulation in cancer | hsa05202 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell autophagy | ||||
In-vitro Model |
Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| In-vivo Model | To create the xenograft neoplasm system, 40 male BALB/c nude mice aged 5 weeks were randomly separated into sh-NC, sh-circHPS5, sh-circHPS5+CTRL, and sh-circHPS5+SAH groups (n = 5 for each group). HCC cells were subcutaneously injected into the axilla of the nude mice. | |||
| Response Summary | In hepatocellular carcinoma, METTL3 could direct the formation of circHPS5, and specific m6A controlled the accumulation of circHPS5. YTHDC1 facilitated the cytoplasmic output of circHPS5 under m6A modification. CircHPS5 can act as a miR-370 sponge to regulate the expression of High mobility group protein HMGI-C (HMGA2) and further accelerate hepatocellular carcinoma cell tumorigenesis. | |||
Programmed cell death 1 ligand 1 (CD274/PD-L1)
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [11] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
RWPE-1 | Normal | Homo sapiens | CVCL_3791 |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 | |
Serine/arginine-rich splicing factor 6 (SRSF6)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | mRNA decay | |||
In-vitro Model |
U251 (Fibroblasts or fibroblast like cells) | |||
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| In-vivo Model | For subcutaneous tumor model, each mouse was injected subcutaneously in the right flank with 2 × 106 U87MG cells (METTL3-KD or control) in 100 uL PBS. | |||
| Response Summary | Silencing METTL3 or overexpressing dominant-negative mutant METTL3 suppressed the growth and self-renewal of Glioblastoma cells. Integrated transcriptome and MeRIP-seq analyses revealed that downregulating the expression of METTL3 decreased m6A modification levels of Serine/arginine-rich splicing factor 6 (SRSF6), which led to YTHDC1-dependent NMD of SRSF transcripts and decreased SRSF protein expression. | |||
Vacuolar protein-sorting-associated protein 25 (VPS25)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [13] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
| Cell Process | Arrest cell cycle at G0/G1 phase | |||
| Cell apoptosis | ||||
In-vitro Model |
U251 (Fibroblasts or fibroblast like cells) | |||
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| In-vivo Model | The U87MG cells (1 × 107 cells in 0.1 ml PBS) were injected subcutaneously into BALB/c nude mice. Tumor width and length were recorded every 5 days. | |||
| Response Summary | YTHDC1 inhibited glioma proliferation by reducing the expression of Vacuolar protein-sorting-associated protein 25 (VPS25). | |||
Vascular endothelial growth factor A (VEGFA)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [14] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| In-vivo Model | The cells were then washed with PBS and digested with trypsin. After counting, 1 × 106 cells were injected subcutaneously into the flank regions of female BALB/c nude mice (4-6 weeks) to establish a subcutaneous implantation model. After 4 weeks, the volume and weight of the tumors were measured after the mice were sacrificed by CO2 inhalation and cervical dislocation. | |||
FOXF1 adjacent non-coding developmental regulatory RNA (FENDRR)
Pulmonary hypertension [ICD-11: BB01]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [15] | |||
| Responsed Disease | Hypoxic pulmonary hypertension [ICD-11: BB01.2] | |||
| Target Regulation | Down regulation | |||
| In-vivo Model | An aliquot of the vector at 1011genome equivalents was prepared in 20-30 μL of HBSS and isoflurane anesthesia followed by nasal drops. Mice were randomly divided into five groups as follows: normoxic environment plus control vector group (NOR + NC, n = 20), hypoxic environment plus control vector group (HYP + NC, n = 10), hypoxic environment plus FENDRR TFO2 adenovirus group (HYP + FENDRR TFO2, n = 10), normoxic environment plus FENDRR TFO2 adenovirus group (NOR + FENDRR TFO2, n = 10). | |||
Maternally expressed 3 (MEG3)
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [16] | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
In-vitro Model |
SCC-9 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1685 |
| SCC-25 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1682 | |
| CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 | |
Abortion [ICD-11: JA00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [17] | |||
| Responsed Disease | Abortion [ICD-11: JA00.0] | |||
| In-vivo Model | Sexually mature ICR mice, aged at least 10 weeks, were paired as one female and one male per cage to obtain pregnancies. The time of the vaginal plug sighting was denoted as day 0.5 of the pregnancy. The mice were divided into an FTO protein group and control group, with 10 pairs of mice in each group. Starting on day 0.5 of the pregnancy, mice in the FTO protein group were intraperitoneally injected daily with FTO protein (3.5 ng/kg), whereas those in the control group had daily intraperitoneal injections of equal volumes of saline. Female mice were euthanised on day 13.5 of the pregnancy (day 12.5-14.5), and the total numbers of normal and resorbed embryos were determined, along with the rate of embryo resorption. | |||
Nuclear paraspeckle assembly transcript 1 (NEAT1)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Target Regulation | Up regulation | |||
Coronary atherosclerosis [ICD-11: BA52]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [19] | |||
| Responsed Disease | Coronary atherosclerosis [ICD-11: BA52.0] | |||
| Target Regulation | Up regulation | |||
Atherosclerosis [ICD-11: BD40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [19] | |||
| Responsed Disease | Atherosclerosis [ICD-11: BD40] | |||
| Target Regulation | Up regulation | |||
Pvt1 oncogene (PVT1)
Chronic respiratory disease originating in the perinatal period [ICD-11: KB29]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [20] | |||
| Responsed Disease | Chronic respiratory disease originating in the perinatal period [ICD-11: KB29.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
MLE-12
|
N.A. | Mus musculus | CVCL_3751 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | Before hyperoxia treatment, mice in the BPD/PVT1 KO group were intratracheally instilled with 5 μL adenovirus vector expressing sh-PVT1 at a titer of 1 × 109 pfu/100 μL. Mice in the BPD/PVT1 KO + IL-33 group were intratracheally instilled with 5 μL adenovirus vector expressing sh-PVT1 and 5 μL adenovirus vector expressing IL-33. | |||
microRNA 370 (MIR370)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [10] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Pathway Response | Transcriptional misregulation in cancer | hsa05202 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell autophagy | ||||
In-vitro Model |
Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| In-vivo Model | To create the xenograft neoplasm system, 40 male BALB/c nude mice aged 5 weeks were randomly separated into sh-NC, sh-circHPS5, sh-circHPS5+CTRL, and sh-circHPS5+SAH groups (n = 5 for each group). HCC cells were subcutaneously injected into the axilla of the nude mice. | |||
| Response Summary | In hepatocellular carcinoma, METTL3 could direct the formation of circHPS5, and specific m6A controlled the accumulation of circHPS5. YTHDC1 facilitated the cytoplasmic output of circHPS5 under m6A modification. CircHPS5 can act as a microRNA 370 (MIR370) sponge to regulate the expression of HMGA2 and further accelerate hepatocellular carcinoma cell tumorigenesis. | |||
hsa-miR-30d
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [21] | |||
| Responsed Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Pathway Response | Central carbon metabolism in cancer | hsa05230 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glycolysis | |||
In-vitro Model |
SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| HPNE (Pancreatic ductal adenocarcinoma cell line HPNE were a gift from Dr. Lingye Tao from Renji hospital) | ||||
| HPAC | Pancreatic adenocarcinoma | Homo sapiens | CVCL_3517 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| Capan-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0026 | |
| Capan-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0237 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| In-vivo Model | To study the effect of miR-30d on liver metastasis of PDACs, 5 × 106 cells in 50 uL of PBS were injected into the spleens of nude mice (6 mice per group). Anesthetized mice were injected intraperitoneally with D-luciferin (150 mg/kg) every other week and imaged using an IVIS 100 imaging system (Xenogen, CA, USA) 10 min after the injection. The mice were sacrificed and their liver metastases were checked by standard histological examination 8-9 weeks after injection. | |||
| Response Summary | MiR-30d is a novel target for YTHDC1 through m6A modification, and hsa-miR-30d represses pancreatic ductal adenocarcinoma tumorigenesis via suppressing aerobic glycolysis. | |||
hsa_circ_0021427 (circ_HPS5)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [10] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Transcriptional misregulation in cancer | hsa05202 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell autophagy | ||||
In-vitro Model |
Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| In-vivo Model | To create the xenograft neoplasm system, 40 male BALB/c nude mice aged 5 weeks were randomly separated into sh-NC, sh-circHPS5, sh-circHPS5+CTRL, and sh-circHPS5+SAH groups (n = 5 for each group). HCC cells were subcutaneously injected into the axilla of the nude mice. | |||
| Response Summary | In hepatocellular carcinoma, METTL3 could direct the formation of circHPS5, and specific m6A controlled the accumulation of circHPS5. YTHDC1 facilitated the cytoplasmic output of hsa_circ_0021427 (circHPS5) under m6A modification. CircHPS5 can act as a miR-370 sponge to regulate the expression of HMGA2 and further accelerate hepatocellular carcinoma cell tumorigenesis. | |||
hsa_circ_0058493 (Circ_RHBDD1)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [22] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Hepatocellular carcinoma | hsa05225 | ||
| Cell Process | Cell growth and metastasis | |||
In-vitro Model |
BEL-7404 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6568 |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| SK-HEP-1 | Liver and intrahepatic bile duct epithelial neoplasm | Homo sapiens | CVCL_0525 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| In-vivo Model | Groups of HCT116-Luc-shCtrl, HCT116-Luc-shLINC00460, and HCT116-Luc-shLINC00460 + HMGA1 cells (5 × 106) were injected subcutaneously into the flanks of mice correspondingly. | |||
| Response Summary | In hepatocellular carcinoma, hsa_circ_0058493 contained m6A methylation sites and that METTL3 mediated the degree of methylation modification of hsa_circ_0058493. YTHDC1 could bind to hsa_circ_0058493 and promote its intracellular localization from the nucleus to the cytoplasm. | |||
hsa_circ_0092493 (circ_ARL3)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [23] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Reverse splicing and biogenesis | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | Mice were randomly divided into three groups, and subcutaneously injected with control, circ-ARL3-silenced and circ-ARL3&miR-1305-silenced HepG2.2.15 cells. | |||
| Response Summary | hsa_circ_0092493 (circ_ARL3) is a critical regulator in HBV-related HCC, targeting the axis of circ-ARL3/miR-1305 can be a promising treatment for HBV+ HCC patients. HBx protein upregulated N6 -methyladenosine (m6A) methyltransferases METTL3 expression, increasing the m6A modification of circ-ARL3; then, m6A reader YTHDC1 bound to m6A-modified of circ-ARL3 and favored its reverse splicing and biogenesis. Furthermore, circ-ARL3 was able to sponge miR-1305, antagonizing the inhibitory effects of miR-1305 on a cohort of target oncogenes, thereby promoting HBV+ HCC progression. | |||
Annexin A1 (ANXA1)
Clear cell renal cell carcinoma [ICD-11: XH46F1]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [24] | |||
| Responsed Disease | Clear cell renal cell carcinoma [ICD-11: XH46F1] | |||
| Target Regulation | Down regulation | |||
ATP8B1 antisense RNA 1 (ATP8B1-AS1)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [25] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
THLE-2 | Normal | Homo sapiens | CVCL_3803 |
| SNU-398 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0077 | |
| In-vivo Model | Indicated HCC cells were subcutaneously inoculated into the flank of 5-week-old male BALB/C athymic mice, which were purchased from Shanghai SLAC Laboratory Animal Co. and fed in specific pathogen-free condition. Subcutaneous tumor volumes were measured every week and calculated using the formula 0.5×length×width2. At the 28th day after inoculation, subcutaneous tumors were resected, weighed, and photographed. This study was approved by Affiliated Hospital of Youjiang Medical University for Nationalities Institutional Review Board. | |||
Chromatin-associated RNAs (caRNAs)
Malignant haematopoietic neoplasm [ICD-11: 2B33]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [26] | |||
| Responsed Disease | Myeloid leukaemia [ICD-11: 2B33.1] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell survival | |||
| Cell proliferation | ||||
| Cell differentiation | ||||
| Cell invasion | ||||
| Apoptosis | ||||
Circ_EPHB4
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [27] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulation | Up regulation | |||
Circ_IGF2BP3
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [28] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
| Cell Process | Immunity | |||
In-vitro Model |
SW900 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1731 |
| SK-MES-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0630 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1703 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1490 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| 16HBE14o- | Normal | Homo sapiens | CVCL_0112 | |
| In-vivo Model | A total of 106 LLC cells transfected with the following constructs were injected subcutaneously into C57BL/6 mice. | |||
| Response Summary | In non-small-cell lung carcinoma, METTL3 mediates the N6-methyladenosine (m6A) modification of Circ_IGF2BP3 and promotes its circularization in a manner dependent on the m6A reader protein YTHDC1. | |||
Circ_MPP1
Disorders of newborn related to slow fetal growth or fetal malnutrition [ICD-11: KA20]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [29] | |||
| Responsed Disease | Disorders of newborn related to slow fetal growth or fetal malnutrition [ICD-11: KA20.1] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | NF-kappa B signaling pathway | hsa04064 | ||
| MAPK signaling pathway | hsa04010 | |||
In-vitro Model |
JEG-3 | Gestational choriocarcinoma | Homo sapiens | CVCL_0363 |
Circ_TET2
Leukemogenesis [ICD-11: 2A82]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [30] | |||
| Responsed Disease | Chronic lymphocytic leukemia [ICD-11: 2A82.0] | |||
| Responsed Drug | CP028 | Investigative | ||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [30] | |||
| Responsed Disease | Chronic lymphocytic leukemia [ICD-11: 2A82.0] | |||
| Responsed Drug | Dactolisib | Investigative | ||
| Experiment 3 Reporting the m6A-centered Disease Response of This Target Gene | [30] | |||
| Responsed Disease | Chronic lymphocytic leukemia [ICD-11: 2A82.0] | |||
| Responsed Drug | Perhexiline | Approved | ||
Cleavage and polyadenylation specificity factor subunit 6 (CPSF6)
Type 2 diabetes mellitus [ICD-11: 5A11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [12] | |||
| Responsed Disease | Type 2 diabetes mellitus [ICD-11: 5A11] | |||
In-vitro Model |
MIN6 | Mouse insulinoma | Mus musculus | CVCL_0431 |
| In-vivo Model | Ythdc1flox/flox mice were maintained on a C57BL/6 background and normal chow. Ythdc1flox/flox mice were then crossed to a transgenic mouse line with MIP-CreERT [33]. MIP-CreERT mice have been described previously [15]. At the age of 10 weeks, both male and female mice were intraperitoneally injected with 100 mg/kg tamoxifen (T5648, Sigma) every other day for 3 times to delete Ythdc1 in beta-cells. Intraperitoneal glucose tolerance tests were performed on mice at age of 15 weeks after a 5-hour fast (2 g/kg dextrose). Insulin levels were measured at 0, 15 and 30 min after glucose challenge. Insulin tolerance tests were performed after a 5-hour fast by administering human recombinant insulin (0.75 U/kg). | |||
Elongation factor Ts, mitochondrial (TSFM)
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [31] | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 |
| Caov-3 | Ovarian serous adenocarcinoma | Homo sapiens | CVCL_0201 | |
| HO-8910 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6868 | |
| OVCAR-3 | Ovarian serous adenocarcinoma | Homo sapiens | CVCL_0465 | |
| In-vivo Model | In order to develop a xenograft model, 12 nude mice (1 × 107cells per 200 μL of FBS-free medium) were injected subcutaneously with CAOV3 cells. | |||
FAM111A divergent transcript (FAM111A-DT)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [32] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SK-HEP-1 | Liver and intrahepatic bile duct epithelial neoplasm | Homo sapiens | CVCL_0525 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| SNU-398 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0077 | |
| THLE-2 | Normal | Homo sapiens | CVCL_3803 | |
| THLE-3 | Normal | Homo sapiens | CVCL_3804 | |
Guanine nucleotide exchange factor C9orf72 (C9ORF72)
Frontotemporal dementia [ICD-11: 6D83]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [33] | |||
| Responsed Disease | Frontotemporal dementia [ICD-11: 6D83] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Motor neuron disease [ICD-11: 8B60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [33] | |||
| Responsed Disease | Amyotrophic lateral sclerosis [ICD-11: 8B60.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Histone-lysine N-methyltransferase 2C (KMT2C)
B-cell acute lymphoblastic leukemia [ICD-11: XH8NN2]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [34] | |||
| Responsed Disease | B-cell acute lymphoblastic leukemia [ICD-11: XH8NN2] | |||
| Target Regulation | Up regulation | |||
Interleukin-33 (IL33)
Chronic respiratory disease originating in the perinatal period [ICD-11: KB29]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [20] | |||
| Responsed Disease | Chronic respiratory disease originating in the perinatal period [ICD-11: KB29.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
MLE-12
|
N.A. | Mus musculus | CVCL_3751 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | Before hyperoxia treatment, mice in the BPD/PVT1 KO group were intratracheally instilled with 5 μL adenovirus vector expressing sh-PVT1 at a titer of 1 × 109 pfu/100 μL. Mice in the BPD/PVT1 KO + IL-33 group were intratracheally instilled with 5 μL adenovirus vector expressing sh-PVT1 and 5 μL adenovirus vector expressing IL-33. | |||
Lnc668
Silicosis [ICD-11: CA60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [35] | |||
| Responsed Disease | Pulmonary fibrosis [ICD-11: CA60] | |||
| Target Regulation | Up regulation | |||
long intergenic non-protein coding RNA 294 (LINC00294)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [36] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
long intergenic non-protein coding RNA 641 (LINC00641)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [37] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
|
HBE1
|
N.A. | Homo sapiens | CVCL_0287 | |
| In-vivo Model | A549 cells (5 × 106) with LINC00641 stable knockdown or control were suspended in 200 μL of 50% Matrigel (Corning), then injected subcutaneously into the flanks of the nude mice. For the metastatic tumor model, the male BALB/c-nude mice weighing 18-23 g were randomly divided into two groups. A549 cells with LINC00641 stable knockdown or control were injected into the tail veins of the nude mice (2 × 106 cells per mice). | |||
Long non-coding RNA epigenetically activating Wnt/beta-catenin signalling in HCC (LEAWBIH)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [38] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
In-vitro Model |
THLE-2 | Normal | Homo sapiens | CVCL_3803 |
| SK-HEP-1 | Liver and intrahepatic bile duct epithelial neoplasm | Homo sapiens | CVCL_0525 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
Long-chain-fatty-acid--CoA ligase 4 (ACSL4)
Injury of blood vessels of thorax [ICD-11: NB30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [39] | |||
| Responsed Disease | Sepsis-associated lung injury [ICD-11: NB30.4] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
MLE-12
|
N.A. | Mus musculus | CVCL_3751 |
miR-181a
Low bone mass disorder [ICD-11: FB83]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [40] | |||
| Responsed Disease | Osteoporosis [ICD-11: FB83.1] | |||
| Target Regulation | Up regulation | |||
miR-181c
Low bone mass disorder [ICD-11: FB83]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [40] | |||
| Responsed Disease | Osteoporosis [ICD-11: FB83.1] | |||
| Target Regulation | Up regulation | |||
Paired box protein Pax-8 (PAX8)
Thyroid Cancer [ICD-11: 2D10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [41] | |||
| Responsed Disease | Thyroid Cancer [ICD-11: 2D10] | |||
| Responsed Drug | Sodium iodide I 131 | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 |
| KTC-1 | Thyroid carcinoma | Homo sapiens | CVCL_6300 | |
| TPC-1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_6298 | |
| K1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_2537 | |
| CAL-62 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_1112 | |
| C-643 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_5969 | |
| Nthy-ori 3-1 | Normal | Homo sapiens | CVCL_2659 | |
| In-vivo Model | A concentration of 1 × 107 CAL62 cells stably transfected with OE METTL3 or OE METTL3/shPAX8-transfected CAL-62 cells were injected subcutaneously into the dorsal flanks of the mice (five mice per group) to establish a xenograft model. | |||
Phosphatidylinositol 3-kinase regulatory subunit alpha (PIK3R1)
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [42] | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 | |
| OVCAR-3 | Ovarian serous adenocarcinoma | Homo sapiens | CVCL_0465 | |
| A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 | |
PncRNA-D
Liposarcoma [ICD-11: 2B59]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [43] | |||
| Responsed Disease | Liposarcoma [ICD-11: 2B59] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Cell cycle | |||
In-vitro Model |
HAP1 | Chronic myelogenous leukemia | Homo sapiens | CVCL_Y019 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Response Summary | Knockdown of METTL3 prolonged the half-life of PncRNA-D, and among the known m6A recognition proteins, YTHDC1 was responsible for binding m6A of pncRNA-D Knockdown of METTL3 or YTHDC1 also enhanced the interaction of pncRNA-D with TLS, and results from RNA pulldown assays implicated YTHDC1 in the inhibitory effect on the TLS-pncRNA-D interaction. | |||
Protein E6 (E6)
Malignant neoplasms of tonsil [ICD-11: 2B69]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [44] | |||
| Responsed Disease | Malignant neoplasms of tonsil [ICD-11: 2B69] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
WSU-HN26 | Squamous cell carcinoma of the oral cavity | Homo sapiens | CVCL_5523 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| C33A2 (The C33A2 cell line originates from C33A and has the subgenomic HPV16 plasmid pBELsLuc stably integrated into the genome) | ||||
| Response Summary | Overexpression of the ALKBH5 promoted production of intron retention on the human papillomavirus type 16 (HPV16) E6 mRNAs thereby promoting Protein E6 (E6) mRNA production. METLL3 induced production of intron-containing HPV16 E1 mRNAs over spliced E2 mRNAs and altered HPV16 L1 mRNA splicing in a manner opposite to ALKBH5. Overexpression of YTHDC1, enhanced retention of the E6-encoding intron and promoted E6 mRNA production. HPV16 mRNAs are m6A-methylated in tonsillar cancer cells. | |||
Retinoic Acid Receptor Alpha-Retinoic Acid Receptor Alpha (PML-RARalpha)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [8] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell differentiation | |||
RNA, U1 small nuclear 8, pseudogene (RNU1-8P)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [45] | |||
| Responsed Disease | Intrahepatic cholangiocarcinoma [ICD-11: 2C12.10] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HuCC-T1 | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_0324 |
Ubiquitin-like-conjugating enzyme ATG10 (ATG10)
Acute ischemic stroke [ICD-11: 8B11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [47] | |||
| Responsed Disease | Acute ischemic stroke [ICD-11: 8B11] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
Unspecific Target Gene
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [48] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Cell Process | Cell migration | |||
| Cell invasion | ||||
In-vitro Model |
PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| Response Summary | Knockdown of HNRNPA2B1 or FTO prominently inhibited prostate cancer cells migration and invasion in vitro experiment. Determined CBLL1, FTO, YTHDC1, HNRNPA2B1 as crucial m6A regulators of prostate cancer. | |||
Epilepsy due to structural or metabolic conditions or diseases [ICD-11: 8A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [49] | |||
| Responsed Disease | Epilepsy due to structural or metabolic conditions or diseases [ICD-11: 8A60.5] | |||
| Responsed Drug | Betaine | Approved | ||
| Target Regulation | Down regulation | |||
| In-vivo Model | Mice were treated with a single i.p. injection of pentylenetetrazol (PTZ, Sigma-Aldrich, P6500) at a dose of 50 mg/kg to establish the animal model of acute seizures [16]. Betaine (Sigma-Aldrich, B2629) was dissolved in normal saline, and administrated at a dose of 200 mg/kg or 600 mg/kg, i.p., to mice for 14 days before PTZ injection in reference to a previous study. | |||
Acute ischemic stroke [ICD-11: 8B11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [50] | |||
| Responsed Disease | Acute ischemic stroke [ICD-11: 8B11] | |||
| In-vivo Model | 36 male C57BL/6J mice (20-22 g, 2-month-old) were purchased from SPF (Beijing) biotechnology co., Ltd (Beijing, China) and maintained in the specific pathogen-free (SPF) animal laboratory with a 12/12 h light/dark cycle with free access to food and water. The mice were randomly assigned into six groups (n = 6 per group): (1) sham-operated group (Sham), (2) MCAO 6 h, (3) MCAO 12 h, (4) MCAO 1 d, (5) MCAO 3 d, and (6) MCAO 7 d. | |||
Asthma [ICD-11: CA23]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [51] | |||
| Responsed Disease | Asthma [ICD-11: CA23] | |||
| In-vivo Model | Modelled two groups of female 6-week-old BALB/C mice: severe asthma group and blank control group (n = 3 per group). They had the same feeding conditions and growth environment. Immunization solution: Dissolve 20 mg ovalbumin (OVA) in 1 ml normal saline (NS), after OVA is completely dissolved, dilute 0.4-10 ml and mix well, then it was mixed with the same volume of liquid aluminium adjuvant and placed on a shaking table at 4℃ for 30 min. Challenge solution: Add 0.5 g OVA into 10 ml NS, fully dissolve it, and shake it on a shaking table at 4℃ for 30 min. Immunization: Mice were injected intraperitoneally on days 0 and 12, each with 0.2 ml; the control group was treated with equal volume of normal saline. Challenge: On days 18-23, the mice were atomized by ultrasound in a closed container at a dose of 10 ml once a day for 20 min. Lung tissue was taken 24 h after the last atomization and immediately stored in liquid nitrogen. | |||
| Response Summary | m6A(YTHDF3 and YTHDC1) modification plays a key role in severe asthma, and is able to guide the future strategy of immunotherapy. | |||
Epithelial membrane protein 3 (EMP3)
Doxil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [9] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| mTOR signaling pathway | hsa04150 | |||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | DNA repair | |||
| In-vitro Model | BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| Response Summary | Epithelial membrane protein 3 (EMP3) blocks Akt-mTOR signaling activation and induces autophagy. EMP3 downregulates YTHDC1, which at least in part mediates the effects of EMP3 on breast cancer cells. EMP3 sensitizes breast cancer cells to the DNA-damaging drug Adriamycin. EMP3 downregulation can be responsible for breast cancer chemoresistance. | |||
Circ_TET2
CP028
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [30] | |||
| Responsed Disease | Chronic lymphocytic leukemia | ICD-11: 2A82.0 | ||
Dactolisib
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [30] | |||
| Responsed Disease | Chronic lymphocytic leukemia | ICD-11: 2A82.0 | ||
Perhexiline
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [30] | |||
| Responsed Disease | Chronic lymphocytic leukemia | ICD-11: 2A82.0 | ||
Paired box protein Pax-8 (PAX8)
Sodium iodide I 131
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [41] | |||
| Responsed Disease | Thyroid Cancer | ICD-11: 2D10 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 |
| KTC-1 | Thyroid carcinoma | Homo sapiens | CVCL_6300 | |
| TPC-1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_6298 | |
| K1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_2537 | |
| CAL-62 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_1112 | |
| C-643 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_5969 | |
| Nthy-ori 3-1 | Normal | Homo sapiens | CVCL_2659 | |
| In-vivo Model | A concentration of 1 × 107 CAL62 cells stably transfected with OE METTL3 or OE METTL3/shPAX8-transfected CAL-62 cells were injected subcutaneously into the dorsal flanks of the mice (five mice per group) to establish a xenograft model. | |||
S-mu-GLT (SugLT)
Tamoxifen
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [46] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | DNA repair | |||
| In-vitro Model | CH12F3 | Mouse lymphoma | Mus musculus | CVCL_E067 |
| CH12F3 | Mouse lymphoma | Mus musculus | CVCL_E067 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | 6- to 12-week-old mice were used for experiments. | |||
| Response Summary | Direct suppression of m6A modification of S-mu-GLT (SugLT) or of m6A reader YTHDC1 reduces CSR. METTL3 enzyme-catalyzed N6-methyladenosine (m6A) RNA modification drives recognition and 3' end processing of S-mu-GLT by the RNA exosome, promoting class switch recombination and suppressing chromosomal translocations. Tamoxifen affects the role of METTL3 in B cell development. | |||
Unspecific Target Gene
Betaine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [49] | |||
| Responsed Disease | Epilepsy due to structural or metabolic conditions or diseases | ICD-11: 8A60.5 | ||
| Target Regulation | Down regulation | |||
| In-vivo Model | Mice were treated with a single i.p. injection of pentylenetetrazol (PTZ, Sigma-Aldrich, P6500) at a dose of 50 mg/kg to establish the animal model of acute seizures [16]. Betaine (Sigma-Aldrich, B2629) was dissolved in normal saline, and administrated at a dose of 200 mg/kg or 600 mg/kg, i.p., to mice for 14 days before PTZ injection in reference to a previous study. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
DNA modification
m6A Target: FOXF1 adjacent non-coding developmental regulatory RNA (FENDRR)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02013 | ||
| Regulated Target | Dynamin-1-like protein (DRP1) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Hypoxic pulmonary hypertension | |
Histone modification
m6A Target: Long-chain-fatty-acid--CoA ligase 4 (ACSL4)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03060 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Sepsis-associated lung injury | |
| Crosstalk ID: M6ACROT03061 | ||
| Epigenetic Regulator | CREB-binding protein (CREBBP) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Sepsis-associated lung injury | |
m6A Target: Maternally expressed 3 (MEG3)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03062 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Oral squamous cell carcinoma | |
| Crosstalk ID: M6ACROT05952 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Abortion | |
m6A Target: FAM111A divergent transcript (FAM111A-DT)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03067 | ||
| Epigenetic Regulator | Lysine-specific demethylase 3B (KDM3B) | |
| Regulated Target | Histone H3 lysine 9 dimethylation (H3K9me2) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT03489 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: Endogenous Retrovirus-K (ERVK)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03093 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase SETDB1 (SETDB1) | |
| Regulated Target | Histone H3 lysine 9 trimethylation (H3K9me3) | |
| Crosstalk relationship | m6A → Histone modification | |
m6A Target: LINE-1 retrotransposable element ORF1 protein (LINE1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03094 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase SETDB1 (SETDB1) | |
| Regulated Target | Histone H3 lysine 9 trimethylation (H3K9me3) | |
| Crosstalk relationship | m6A → Histone modification | |
m6A Target: Annexin A1 (ANXA1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03121 | ||
| Epigenetic Regulator | Histone deacetylase 2 (HDAC2) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Clear cell renal cell carcinoma | |
| Drug | Sunitinib | |
m6A Target: Nuclear paraspeckle assembly transcript 1 (NEAT1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03136 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT03137 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Liver cancer | |
m6A Target: Histone-lysine N-methyltransferase 2C (KMT2C)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03162 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase 2C (KMT2C) | |
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | B-cell acute lymphoblastic leukemia | |
| Crosstalk ID: M6ACROT03163 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase 2C (KMT2C) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | B-cell acute lymphoblastic leukemia | |
m6A Target: Polycomb protein EED (EED)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03178 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | m6A → Histone modification | |
m6A Target: ATP8B1 antisense RNA 1 (ATP8B1-AS1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03192 | ||
| Epigenetic Regulator | Lysine-specific demethylase 3B (KDM3B) | |
| Regulated Target | Histone H3 lysine 9 dimethylation (H3K9me2) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT03488 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: long non-coding RNA epigenetically activating Wnt/beta-catenin signalling in HCC (LEAWBIH)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03214 | ||
| Epigenetic Regulator | Lysine-specific demethylase 3B (KDM3B) | |
| Regulated Target | Histone H3 lysine 9 dimethylation (H3K9me2) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT03487 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: Lnc668
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03226 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 9 lactylation (H3K9la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pulmonary fibrosis | |
m6A Target: hsa_circ_0092493 (circ_ARL3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03482 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: hsa_circ_0021427 (circ_HPS5)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03483 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: microRNA 370 (MIR370)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03484 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: High mobility group protein HMGI-C (HMGA2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03485 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: hsa_circ_0058493 (Circ_RHBDD1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03486 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: long intergenic non-protein coding RNA 294 (LINC00294)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03490 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: Chromatin-associated RNAs (caRNAs)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05955 | ||
| Epigenetic Regulator | Polycomb Repressive Complex 2 (PRC2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Myeloid leukaemia | |
Non-coding RNA
m6A Target: Ubiquitin-like-conjugating enzyme ATG10 (ATG10)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05025 | ||
| Epigenetic Regulator | Colorectal neoplasia differentially expressed (CRNDE) | |
| Regulated Target | YTH N6-methyladenosine RNA binding protein C1 (YTHDC1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Acute ischemic stroke | |
m6A Target: Elongation factor Ts, mitochondrial (TSFM)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05106 | ||
| Epigenetic Regulator | piR-26441 | |
| Regulated Target | YTH N6-methyladenosine RNA binding protein C1 (YTHDC1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Ovarian cancer | |
m6A Target: Angiopoietin-related protein 4 (ANGPTL4)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05124 | ||
| Epigenetic Regulator | MIR22 host gene (MIR22HG) | |
| Regulated Target | YTH N6-methyladenosine RNA binding protein C1 (YTHDC1) | |
| Crosstalk relationship | ncRNA → m6A | |
m6A Target: Paired box protein Pax-8 (PAX8)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05170 | ||
| Epigenetic Regulator | hsa-miR-493-5p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Thyroid Cancer | |
| Drug | Sodium iodide I 131 | |
m6A Target: PncRNA-D
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05422 | ||
| Epigenetic Regulator | PncRNA-D | |
| Regulated Target | G1/S-specific cyclin-D1 (CCND1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liposarcoma | |
m6A Target: X inactive specific transcript (XIST)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05456 | ||
| Epigenetic Regulator | X inactive specific transcript (XIST) | |
| Regulated Target | Glypican-4 (GPC4) | |
| Crosstalk relationship | m6A → ncRNA | |
| Crosstalk ID: M6ACROT06024 | ||
| Epigenetic Regulator | X inactive specific transcript (XIST) | |
| Regulated Target | Transcriptional regulator ATRX (ATRX) | |
| Crosstalk relationship | m6A → ncRNA | |
m6A Target: hsa_circ_0092493 (circ_ARL3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05462 | ||
| Epigenetic Regulator | hsa_circ_0092493 (Circ_ARL3) | |
| Regulated Target | microRNA 1305 (MIR1305) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
m6A Target: hsa_circ_0021427 (circ_HPS5)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05533 | ||
| Epigenetic Regulator | hsa_circ_0021427 (Circ_HPS5) | |
| Regulated Target | hsa-miR-370-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
m6A Target: microRNA 370 (MIR370)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05535 | ||
| Epigenetic Regulator | MicroRNA 370 (MIR370) | |
| Regulated Target | High mobility group protein HMGI-C (HMGA2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
m6A Target: hsa_circ_0058493 (Circ_RHBDD1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05548 | ||
| Epigenetic Regulator | hsa_circ_0058493 (Circ_RHBDD1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
m6A Target: S-mu-GLT (SugLT)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05553 | ||
| Epigenetic Regulator | S-mu-GLT (SugLT) | |
| Crosstalk relationship | m6A → ncRNA | |
| Drug | Tamoxifen* | |
m6A Target: Circ_IGF2BP3
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05577 | ||
| Epigenetic Regulator | Circ_IGF2BP3 | |
| Regulated Target | hsa-miR-3173-5p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
m6A Target: hsa-miR-30d
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05634 | ||
| Epigenetic Regulator | hsa-miR-30d | |
| Regulated Target | Runt-related transcription factor 1 (RUNX1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Pancreatic ductal adenocarcinoma | |
m6A Target: Circ_MPP1
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05643 | ||
| Epigenetic Regulator | Circ_MPP1 | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Disorders of newborn related to slow fetal growth or fetal malnutrition | |
m6A Target: Pvt1 oncogene (PVT1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05647 | ||
| Epigenetic Regulator | Pvt1 oncogene (PVT1) | |
| Regulated Target | Interleukin-33 (IL33) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Chronic respiratory disease originating in the perinatal period | |
m6A Target: ATP8B1 antisense RNA 1 (ATP8B1-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05663 | ||
| Epigenetic Regulator | ATP8B1 antisense RNA 1 (ATP8B1-AS1) | |
| Regulated Target | Myc proto-oncogene protein (MYC) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
m6A Target: RNA, U1 small nuclear 8, pseudogene (RNU1-8P)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05673 | ||
| Epigenetic Regulator | RNA, U1 small nuclear 8, pseudogene (RNU1-8P) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Intrahepatic cholangiocarcinoma | |
m6A Target: Circ_EPHB4
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05709 | ||
| Epigenetic Regulator | Circ_EPHB4 | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Brain cancer | |
m6A Target: FAM111A divergent transcript (FAM111A-DT)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05711 | ||
| Epigenetic Regulator | FAM111A divergent transcript (FAM111A-DT) | |
| Regulated Target | YTH N6-methyladenosine RNA binding protein C1 (YTHDC1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
m6A Target: Circ_TET2
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05719 | ||
| Epigenetic Regulator | Circ_TET2 | |
| Regulated Target | Heterogeneous nuclear ribonucleoprotein C (HNRNPC) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Chronic lymphocytic leukemia | |
| Drug | CP028 | |
| Crosstalk ID: M6ACROT05720 | ||
| Epigenetic Regulator | Circ_TET2 | |
| Regulated Target | Heterogeneous nuclear ribonucleoprotein C (HNRNPC) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Chronic lymphocytic leukemia | |
| Drug | dactolisib | |
| Crosstalk ID: M6ACROT05721 | ||
| Epigenetic Regulator | Circ_TET2 | |
| Regulated Target | Heterogeneous nuclear ribonucleoprotein C (HNRNPC) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Chronic lymphocytic leukemia | |
| Drug | perhexiline | |
m6A Target: Nuclear paraspeckle assembly transcript 1 (NEAT1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05730 | ||
| Epigenetic Regulator | Nuclear paraspeckle assembly transcript 1 (NEAT1) | |
| Regulated Target | Krueppel-like factor 4 (KLF4) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Coronary atherosclerosis | |
m6A Target: miR-181a
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05754 | ||
| Epigenetic Regulator | miR-181a | |
| Regulated Target | Secreted frizzled related protein 1 (SFRP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Osteoporosis | |
m6A Target: miR-181c
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05755 | ||
| Epigenetic Regulator | miR-181c | |
| Regulated Target | Secreted frizzled related protein 1 (SFRP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Osteoporosis | |
m6A Target: Long intergenic non-protein coding RNA 294 (LINC00294)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05757 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 294 (LINC00294) | |
| Regulated Target | YTH N6-methyladenosine RNA binding protein C1 (YTHDC1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
m6A Target: Long intergenic non-protein coding RNA 641 (LINC00641)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05767 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 641 (LINC00641) | |
| Regulated Target | ELAV-like protein 1 (HuR/ELAVL1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Lung cancer | |
m6A Target: Maternally expressed 3 (MEG3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05831 | ||
| Epigenetic Regulator | Maternally expressed 3 (MEG3) | |
| Regulated Target | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Abortion | |
m6A Target: Lnc668
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05871 | ||
| Epigenetic Regulator | Lnc668 | |
| Regulated Target | Phosphatidylinositol-binding clathrin assembly protein (PICALM) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Pulmonary fibrosis | |
m6A Target: Retinoic Acid Receptor Alpha-Retinoic Acid Receptor Alpha (PML-RARalpha)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05881 | ||
| Epigenetic Regulator | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Regulated Target | YTH N6-methyladenosine RNA binding protein C1 (YTHDC1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Glioblastoma | |
m6A Target: Hexokinase-2 (HK2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05900 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 294 (LINC00294) | |
| Regulated Target | YTH N6-methyladenosine RNA binding protein C1 (YTHDC1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: Glucose transporter type 1 (GLUT1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05901 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 294 (LINC00294) | |
| Regulated Target | YTH N6-methyladenosine RNA binding protein C1 (YTHDC1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
Xenobiotics Compound(s) Regulating the m6A Methylation Regulator
| Compound Name | Tamoxifen | Approved |
|---|---|---|
| Synonyms |
tamoxifen; 10540-29-1; trans-Tamoxifen; Crisafeno; Soltamox; Tamoxifene; Diemon; Tamoxifenum; Tamoxifeno; Tamizam; Istubol; Tamoxen; Citofen; Oncomox; Valodex; Retaxim; Tamoxifene [INN-French]; Tamoxifenum [INN-Latin]; Tamoxifeno [INN-Spanish]; Tamoxifen (Z); Tamoxifen and its salts; Tamoxifen [INN:BAN]; ICI-46474; ICI 47699; TRANS FORM OF TAMOXIFEN; CCRIS 3275; UNII-094ZI81Y45; HSDB 6782; CHEMBL83; EINECS 234-118-0; 1-p-beta-Dimethylaminoethoxyphenyl-trans-1,2-diphenylbut-1-ene; Citofen; Nourytam; Novaldex; Tamone; Tamoxifeno;Tamoxifenum; Tomaxithen; Gen-Tamoxifen; Istubal (TN); Nolvadex (TN); Nolvadex-D; Novo-Tamoxifen; Pms-Tamoxifen; Tamoplex (TN); Tamoxifen (INN); Tamoxifen (TN); Trans-Tamoxifen; Valodex (TN); TAMOXIFEN (TAMOXIFEN CITRATE (54965-24-1)); Trans-2-[4-(1,2-Diphenyl-1-butenyl)phenoxy]-N,N-dimethylethylamine; (Z)-1-(p-Dimethylaminoethoxyphenyl)-1,2-diphenyl-1-butene; (Z)-2-(4-(1,2-Diphenyl-1-butenyl)phenoxy)-N,N-dimethylethanamine; (Z)-2-(4-(1,2-diphenylbut-1-enyl)phenoxy)-N,N-dimethylethanamine; (Z)-2-(para-(1,2-Diphenyl-1-butenyl)phenoxy)-N,N-dimethylamine (IUPAC); (Z)-2-[4-(1,2)-DIPHENYL-1-BUTENYL)-PHENOXY]-N,N-DIMETHYLETHANAMINE; (Z)-2-[p-(1,2-Diphenyl-1-butenyl)phenoxy]-N,N-dimethylethylamine; 1-p-beta-Dimethylamino-ethoxyphenyl-trans-1,2-diphenylbut-1-ene; 1-para-beta-Dimethylaminoethoxyphenyl-trans-1,2-diphenylbut-1-ene; 2-[4-[(Z)-1,2-diphenylbut-1-enyl]phenoxy]-N,N-dimethylethanamine; 2-{4-[(1Z)-1,2-diphenylbut-1-en-1-yl]phenoxy}-N,N-dimethylethanamine; Tamoxifen (Hormonal therapy); [3H]tamoxifen
Click to Show/Hide
|
|
| External link | ||
| Description |
Direct suppression of m6A modification ofS-mu-GLT or of m6A reader YTHDC1 reduces CSR. METTL3 enzyme-catalyzed N6-methyladenosine (m6A) RNA modification drives recognition and 3' end processing of S-mu-GLT by the RNA exosome, promotingclass switch recombination and suppressing chromosomal translocations. Tamoxifen affects the role of METTL3 in B cell development.
|
[46] |
| Compound Name | Beta-Elemene | Investigative |
| Synonyms |
BETA-ELEMENE; 515-13-9; beta-Elemen; (-)-beta-Elemene; Levo-beta-elemene; b-elemene; Levo-b-elemene; UNII-2QG8CX6LXD; (1S,2S,4R)-1-methyl-2,4-di(prop-1-en-2-yl)-1-vinylcyclohexane; 2,4-Diisopropenyl-1-methyl-1-vinylcyclohexane; (-)-b-Elemene; CHEBI:62855; 2QG8CX6LXD; (1S,2S,4R)-2,4-diisopropenyl-1-methyl-1-vinylcyclohexane; (1S,2S,4R)-(-)-1-methyl-1-vinyl-2,4-diisopropenylcyclohexane; (1S,2S,4R)-1-ethenyl-1-methyl-2,4-bis(prop-1-en-2-yl)cyclohexane; Cyclohexane, 1-ethenyl-1-methyl-2,4-bis(1-methylethenyl)-, (1S,2S,4R)-; 33880-83-0; (1S,2S,4R)-1-ethenyl-1-methyl-2,4-di(prop-1-en-2-yl)cyclohexane; beta-Elemene, (-)-; b-Elemen; E- .beta.-Elemene; Epitope ID:153551; Levo-b-elemene(-)-b-Elemene; CHEMBL448502; DTXSID60881211; SDP-111; 8064AH; s6957; ZINC14096289; AKOS028108977; (-)-beta-Elemene, analytical standard; Cyclohexane, 1-ethenyl-1-methyl-2,4-bis(1-methylethenyl)-, (1S-(1-alpha,2-beta,4-beta))-; Cyclohexane, 1-ethenyl-1-methyl-2,4-bis(1-methylethenyl)-, [1S-(1alpha,2beta,4beta)]-; AS-82909; HY-107324; CS-0028143; C17094; E79113; 880E830; Q27132237; 1-ethenyl-1-methyl-2,4-bis(1-methylethenyl)-cyclohexane; Cyclohexane, 2,4-diisopropenyl-1-methyl-1-vinyl-, (1S,2S,4R)-
Click to Show/Hide
|
|
| External link | ||
| Description |
Indicated that Beta-elemene administration led to the augment of YTHDC1 level was upregulated
|
[52] |
References