m6A-centered Crosstalk Information
Mechanism of Crosstalk between m6A Modification and Epigenetic Regulation
| Crosstalk ID |
M6ACROT05871
|
[1] | |||
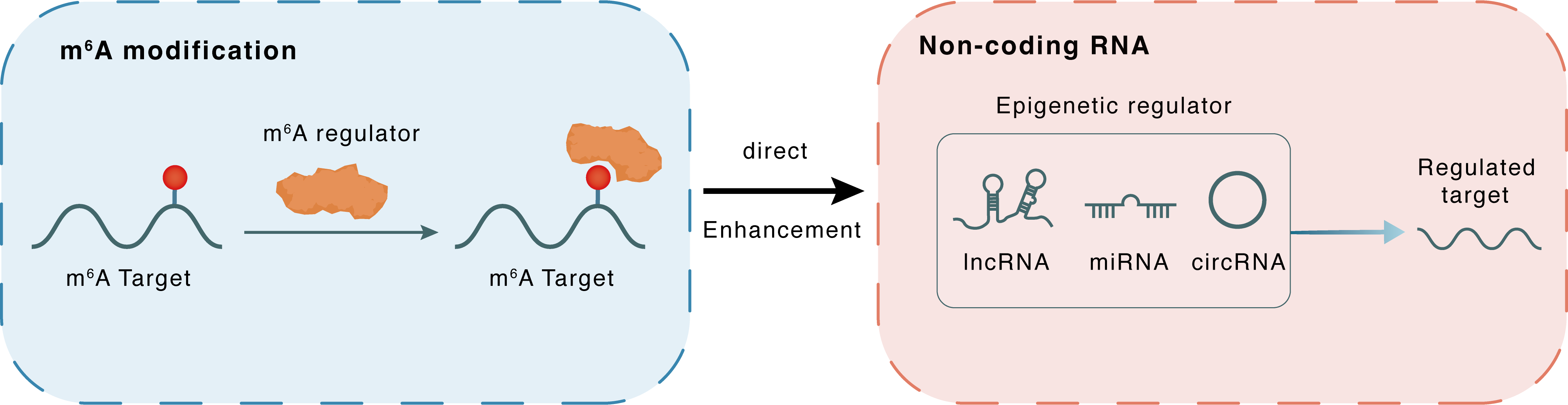 m6A modification
Lnc668
Lnc668
YTHDC1
m6A modification
Lnc668
Lnc668
YTHDC1
 : m6A sites
Direct
Enhancement
Non-coding RNA
lnc668
PICALM
lncRNA miRNA circRNA : m6A sites
Direct
Enhancement
Non-coding RNA
lnc668
PICALM
lncRNA miRNA circRNA
|
|||||
| m6A Modification: | |||||
|---|---|---|---|---|---|
| m6A Regulator | YTH domain-containing protein 1 (YTHDC1) | READER | |||
| m6A Target | Lnc668 | ||||
| Epigenetic Regulation that have Cross-talk with This m6A Modification: | |||||
| Epigenetic Regulation Type | Non-coding RNA (ncRNA) | ||||
| Epigenetic Regulator | Lnc668 | LncRNA | View Details | ||
| Regulated Target | Phosphatidylinositol-binding clathrin assembly protein (PICALM) | View Details | |||
| Crosstalk Relationship | m6A → ncRNA | Enhancement | |||
| Crosstalk Mechanism | m6A regulators directly modulate the functionality of ncRNAs through specific targeting ncRNA | ||||
| Crosstalk Summary | The m6A reader YTHDC1 recognized m6A-modified Lnc668 and elevated the METTL3-mediated lnc668 modification. lnc668 enhanced Phosphatidylinositol-binding clathrin assembly protein (PICALM) mRNA stability to promote pulmonary fibrogenesis depending on YTHDC1 phase separation | ||||
| Responsed Disease | Pulmonary fibrosis | ICD-11: CA60 | |||
| Cell Process | Cell differentiation | ||||
In-vitro Model |
MRC-5 | Normal | Homo sapiens | CVCL_0440 | |
| In-vivo Model | Eight-week-old C57BL/6 mice were provided by Jinan Pengyue Company and divided into the following groups according to experimental requirements: sham operation group, BLM model group, BLM + si-lnc668/NC group, BLM + si-lnc668 group, overexpressed lnc668/NC group, and overexpressed lnc668 treatment group, with 10 mice in each group. Anesthesia was induced by intraperitoneal injection of 4% chloral hydrate (10 mg/kg). The sham operation group received an equivalent volume of saline as a control. | ||||