m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00283)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
HK2
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
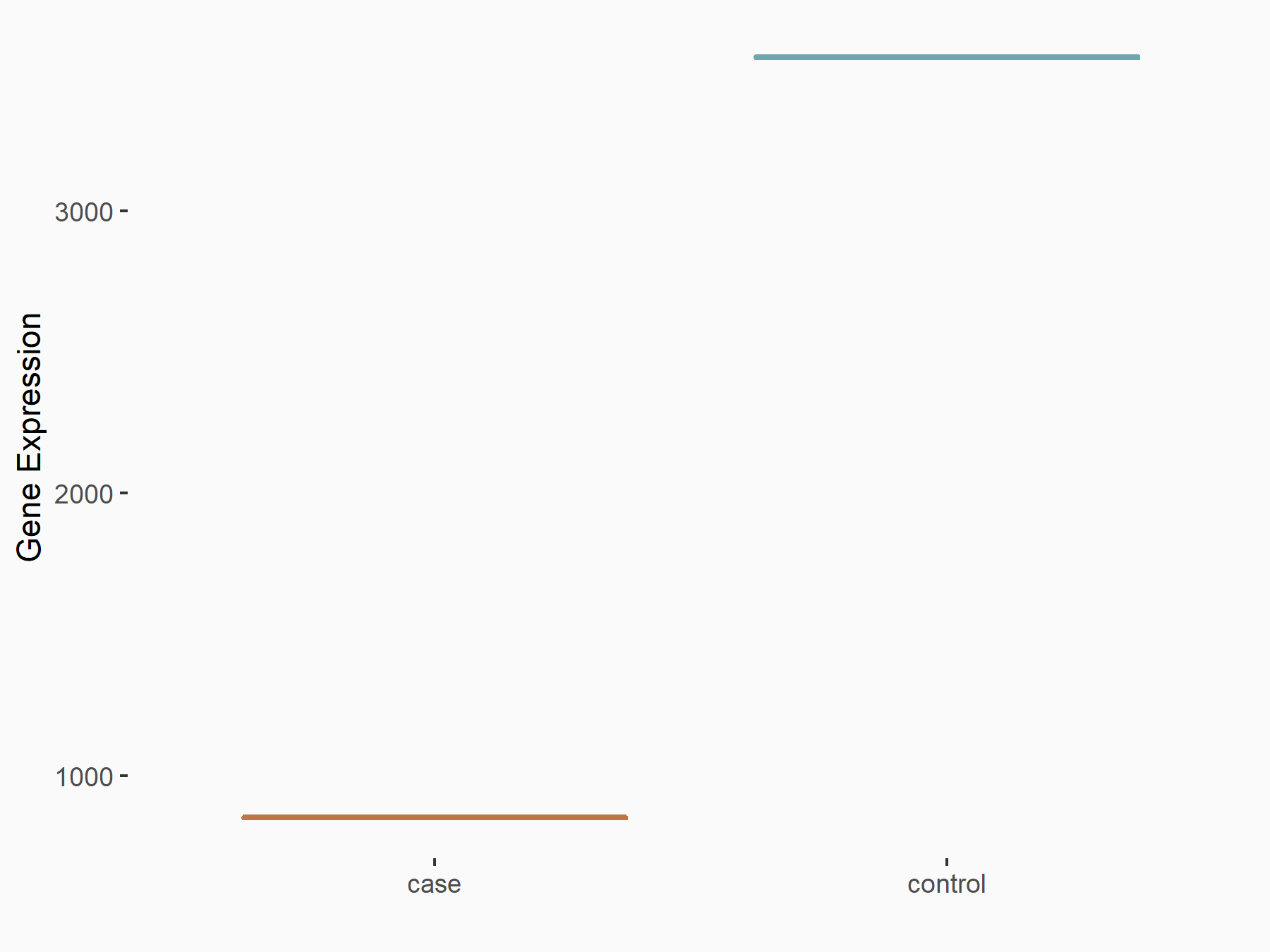 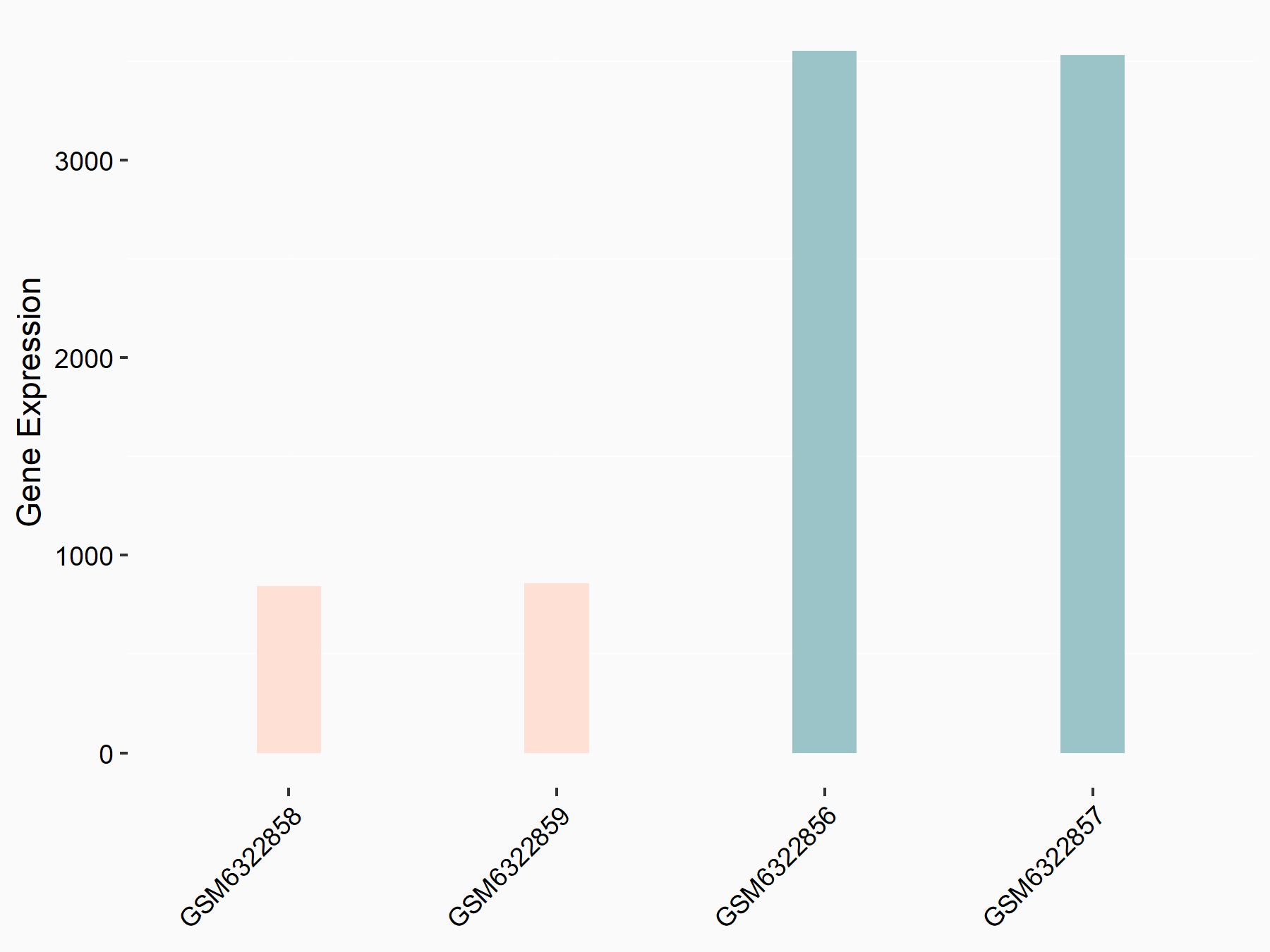 |
logFC: -2.06E+00 p-value: 3.79E-111 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between HK2 and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.17E+00 | GSE60213 |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 stabilizes Hexokinase-2 (HK2) and SLC2A1 (GLUT1) expression in colorectal cancer through an m6A-IGF2BP2/3- dependent mechanism. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glucose metabolism | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 enhanced the Hexokinase-2 (HK2) stability through YTHDF1-mediated m6A modification, thereby promoting the Warburg effect of CC, which promotes a novel insight for the CC treatment. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Cervical cancer | ICD-11: 2C77 | ||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010), Central carbon metabolism in cancer | ||
| Cell Process | Glycolysis | |||
| In-vitro Model | SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| HT-3 | Cervical carcinoma | Homo sapiens | CVCL_1293 | |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| In-vivo Model | Five-week-old male nude BALB/C mice were applied for this animal studies and fed with certified standard diet and tap water ad libitum in a light/dark cycle of 12 h on/12 h off.The assay was performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Stable transfection of METTL3 knockdown (sh-METTL3) or negative control (sh-blank) in SiHa cells (5 × 106 cells per 0.1 mL) were injected into the flank of mice. | |||
YTH domain-containing family protein 1 (YTHDF1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
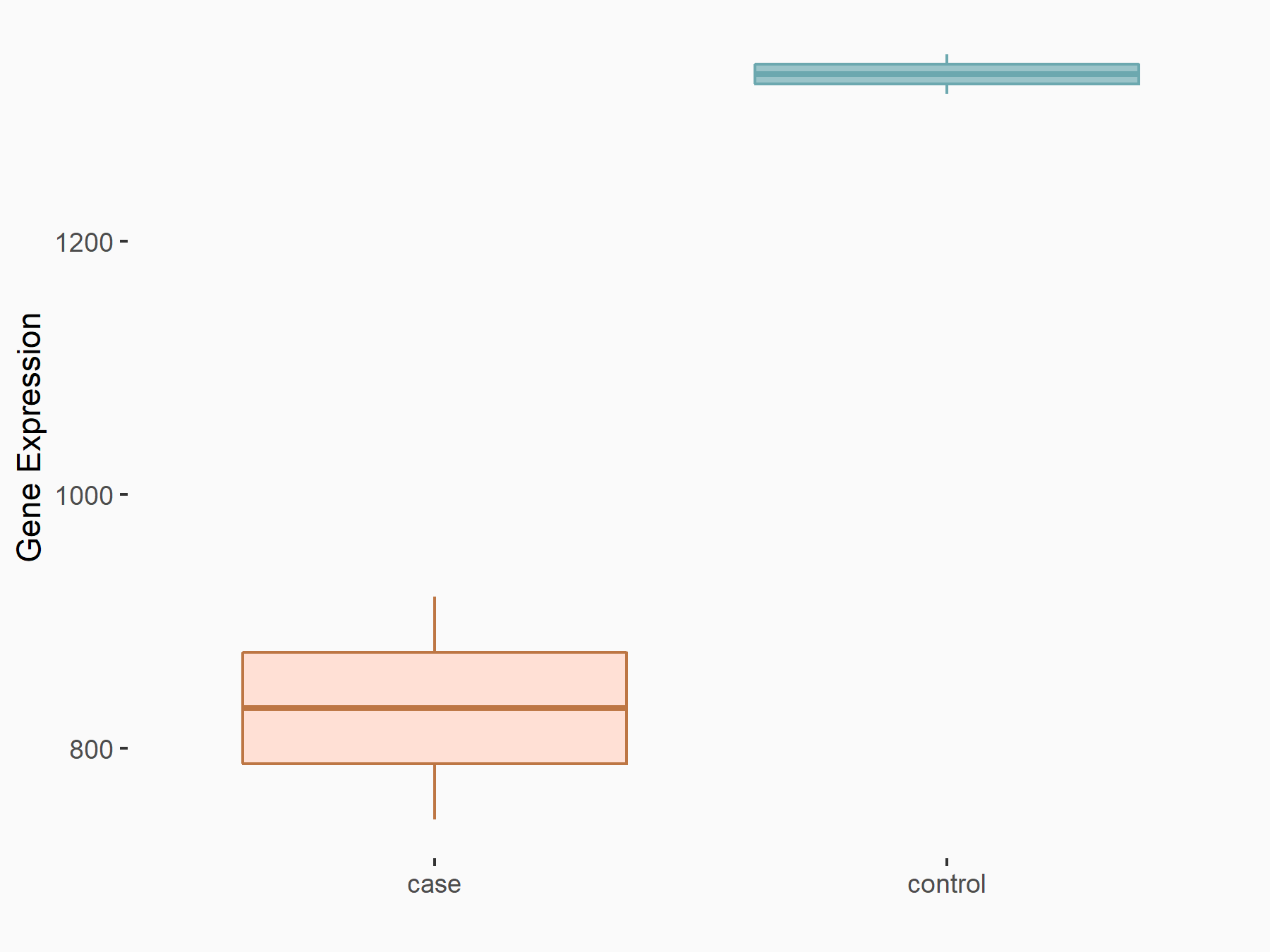 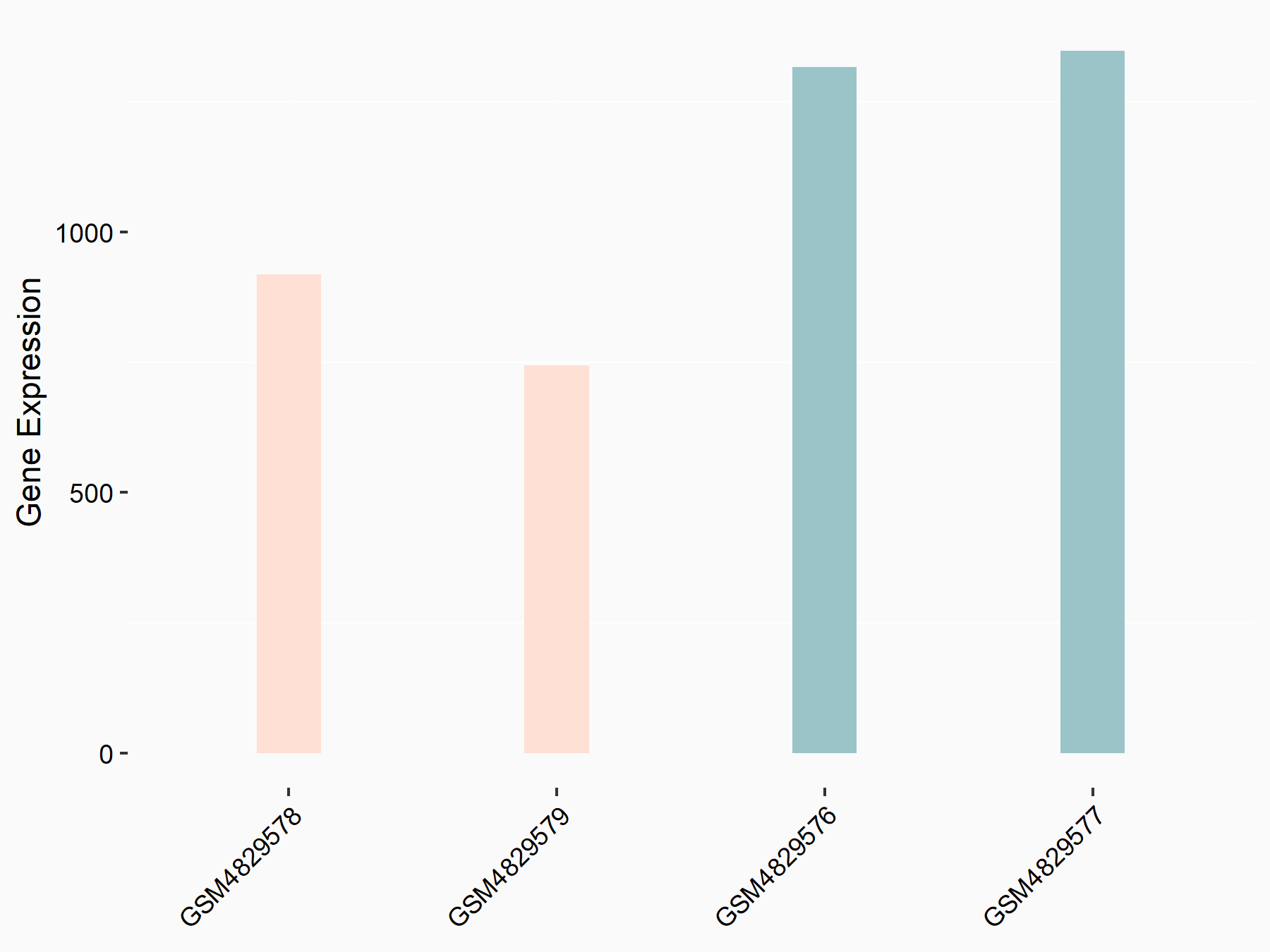 |
logFC: -6.80E-01 p-value: 6.28E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between HK2 and the regulator | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.46E+00 | GSE63591 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 enhanced the Hexokinase-2 (HK2) stability through YTHDF1-mediated m6A modification, thereby promoting the Warburg effect of CC, which promotes a novel insight for the CC treatment. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Cervical cancer | ICD-11: 2C77 | ||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010), Central carbon metabolism in cancer | ||
| Cell Process | Glycolysis | |||
| In-vitro Model | SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| HT-3 | Cervical carcinoma | Homo sapiens | CVCL_1293 | |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| In-vivo Model | Five-week-old male nude BALB/C mice were applied for this animal studies and fed with certified standard diet and tap water ad libitum in a light/dark cycle of 12 h on/12 h off.The assay was performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Stable transfection of METTL3 knockdown (sh-METTL3) or negative control (sh-blank) in SiHa cells (5 × 106 cells per 0.1 mL) were injected into the flank of mice. | |||
Wilms tumor 1-associating protein (WTAP) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by WTAP | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: WTAP-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
  |
logFC: -8.37E-01 p-value: 2.23E-65 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | Oncogenic role of WTAP and its m6A-mediated regulation on gastric cancer Warburg effect, providing a novel approach and therapeutic target in gastric cancer. WTAP enhanced the stability of Hexokinase-2 (HK2) mRNA through binding with the 3'-UTR m6A site. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glycolysis | |||
| In-vitro Model | SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 stabilizes Hexokinase-2 (HK2) and SLC2A1 (GLUT1) expression in colorectal cancer through an m6A-IGF2BP2/3- dependent mechanism. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glucose metabolism | |||
Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 stabilizes Hexokinase-2 (HK2) and SLC2A1 (GLUT1) expression in colorectal cancer through an m6A-IGF2BP2/3- dependent mechanism. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glucose metabolism | |||
Protein virilizer homolog (VIRMA) [WRITER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | KIAA1429 has the potential to promote CRC carcinogenesis by targeting Hexokinase-2 (HK2) via m6A-independent manner, providing insight into the critical roles of m6A in CRC. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | Metabolic pathways | hsa01100 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glycolysis | |||
| In-vitro Model | SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| FHC | Normal | Homo sapiens | CVCL_3688 | |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | Oncogenic role of WTAP and its m6A-mediated regulation on gastric cancer Warburg effect, providing a novel approach and therapeutic target in gastric cancer. WTAP enhanced the stability of Hexokinase-2 (HK2) mRNA through binding with the 3'-UTR m6A site. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Wilms tumor 1-associating protein (WTAP) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glycolysis | |||
| In-vitro Model | SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
Colorectal cancer [ICD-11: 2B91]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 stabilizes Hexokinase-2 (HK2) and SLC2A1 (GLUT1) expression in colorectal cancer through an m6A-IGF2BP2/3- dependent mechanism. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glucose metabolism | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 stabilizes Hexokinase-2 (HK2) and SLC2A1 (GLUT1) expression in colorectal cancer through an m6A-IGF2BP2/3- dependent mechanism. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glucose metabolism | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [6] | |||
| Response Summary | KIAA1429 has the potential to promote CRC carcinogenesis by targeting Hexokinase-2 (HK2) via m6A-independent manner, providing insight into the critical roles of m6A in CRC. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Protein virilizer homolog (VIRMA) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Metabolic pathways | hsa01100 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glycolysis | |||
| In-vitro Model | SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| FHC | Normal | Homo sapiens | CVCL_3688 | |
Cervical cancer [ICD-11: 2C77]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 enhanced the Hexokinase-2 (HK2) stability through YTHDF1-mediated m6A modification, thereby promoting the Warburg effect of CC, which promotes a novel insight for the CC treatment. | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010), Central carbon metabolism in cancer | ||
| Cell Process | Glycolysis | |||
| In-vitro Model | SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| HT-3 | Cervical carcinoma | Homo sapiens | CVCL_1293 | |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| In-vivo Model | Five-week-old male nude BALB/C mice were applied for this animal studies and fed with certified standard diet and tap water ad libitum in a light/dark cycle of 12 h on/12 h off.The assay was performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Stable transfection of METTL3 knockdown (sh-METTL3) or negative control (sh-blank) in SiHa cells (5 × 106 cells per 0.1 mL) were injected into the flank of mice. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 enhanced the Hexokinase-2 (HK2) stability through YTHDF1-mediated m6A modification, thereby promoting the Warburg effect of CC, which promotes a novel insight for the CC treatment. | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010), Central carbon metabolism in cancer | ||
| Cell Process | Glycolysis | |||
| In-vitro Model | SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| HT-3 | Cervical carcinoma | Homo sapiens | CVCL_1293 | |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| In-vivo Model | Five-week-old male nude BALB/C mice were applied for this animal studies and fed with certified standard diet and tap water ad libitum in a light/dark cycle of 12 h on/12 h off.The assay was performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Stable transfection of METTL3 knockdown (sh-METTL3) or negative control (sh-blank) in SiHa cells (5 × 106 cells per 0.1 mL) were injected into the flank of mice. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: RNA demethylase ALKBH5 (ALKBH5)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03364 | ||
| Epigenetic Regulator | Histone deacetylase 2 (HDAC2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03460 | ||
| Epigenetic Regulator | Histone deacetylase 1 (HDAC1) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03367 | ||
| Epigenetic Regulator | Histone deacetylase 2 (HDAC2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03368 | ||
| Epigenetic Regulator | Histone deacetylase 2 (HDAC2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03369 | ||
| Epigenetic Regulator | Histone deacetylase 2 (HDAC2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Regulator: Fat mass and obesity-associated protein (FTO)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03450 | ||
| Epigenetic Regulator | Histone deacetylase 1 (HDAC1) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 4 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03507 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03520 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03548 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03593 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
Non-coding RNA
m6A Regulator: YTH domain-containing family protein 1 (YTHDF1)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05116 | ||
| Epigenetic Regulator | HLA complex P5 (HCP5) | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Esophageal Squamous Cell Carcinoma | |
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05158 | ||
| Epigenetic Regulator | MIR4458 host gene (MIR4458HG) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Regulator: Wilms tumor 1-associating protein (WTAP)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05328 | ||
| Epigenetic Regulator | piR-30473 | |
| Regulated Target | Pre-mRNA-splicing regulator WTAP (WTAP) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Diffuse large B-cell lymphomas | |
m6A Regulator: RNA-binding motif protein 15 (RBM15)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05347 | ||
| Epigenetic Regulator | Circ_CTNNB1 | |
| Regulated Target | RNA binding motif protein 15 (RBM15) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Osteosarcoma | |
m6A Regulator: YTH domain-containing protein 1 (YTHDC1)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05900 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 294 (LINC00294) | |
| Regulated Target | YTH N6-methyladenosine RNA binding protein C1 (YTHDC1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00283)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: AC4SITE000027 | Click to Show/Hide the Full List | ||
| mod site | chr2:74854418-74854419:+ | [10] | |
| Sequence | CTACTGCAGCAGTGAAGATGCTGCCCACCTTTGTGAGGTCC | ||
| Cell/Tissue List | H1 | ||
| Seq Type List | ac4C-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000472302.1; ENST00000409174.1 | ||
| External Link | RMBase: ac4C_site_1122 | ||
Adenosine-to-Inosine editing (A-to-I)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE009786 | Click to Show/Hide the Full List | ||
| mod site | chr2:74841053-74841054:+ | [11] | |
| Sequence | GTTTTCTCAGCCTTGGTGCTATTGACATTTTGGGACAGATA | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: RNA-editing_site_77780 | ||
5-methylcytidine (m5C)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE002366 | Click to Show/Hide the Full List | ||
| mod site | chr2:74873890-74873891:+ | [12] | |
| Sequence | GTGAATGACACAGTTGGGACCATGATGACCTGTGGTTATGA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m5C_site_26759 | ||
| mod ID: M5CSITE002367 | Click to Show/Hide the Full List | ||
| mod site | chr2:74881707-74881708:+ | [12] | |
| Sequence | ACTTCCCTGGGCTTATTTTCCAGAGAAAGGGGACTTCTTGG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m5C_site_26760 | ||
N6-methyladenosine (m6A)
| In total 129 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE045851 | Click to Show/Hide the Full List | ||
| mod site | chr2:74834126-74834127:+ | [13] | |
| Sequence | CGGCAGCCCCTCAATAAGCCACATTGTTGCATGAAACTCCG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480474 | ||
| mod ID: M6ASITE045852 | Click to Show/Hide the Full List | ||
| mod site | chr2:74834141-74834142:+ | [14] | |
| Sequence | AAGCCACATTGTTGCATGAAACTCCGGCGCAGGAGTCCCGG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; H1299; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480475 | ||
| mod ID: M6ASITE045853 | Click to Show/Hide the Full List | ||
| mod site | chr2:74834175-74834176:+ | [13] | |
| Sequence | GTCCCGGGCTGCCGCTGGCAACATCGTGTCACCCAGCTAAG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480476 | ||
| mod ID: M6ASITE045854 | Click to Show/Hide the Full List | ||
| mod site | chr2:74834325-74834326:+ | [14] | |
| Sequence | AGTTAGCGCCTTGACGTGGGACAACCGGACACGTCGCCAGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; hESC-HEK293T; H1B; MM6; Jurkat; peripheral-blood; HEK293A-TOA; iSLK; TIME; TREX; MSC | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480477 | ||
| mod ID: M6ASITE045855 | Click to Show/Hide the Full List | ||
| mod site | chr2:74834333-74834334:+ | [14] | |
| Sequence | CCTTGACGTGGGACAACCGGACACGTCGCCAGGAGAGAACT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1B; MM6; Jurkat; peripheral-blood; HEK293A-TOA; iSLK; TIME; TREX; MSC | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480478 | ||
| mod ID: M6ASITE045856 | Click to Show/Hide the Full List | ||
| mod site | chr2:74834351-74834352:+ | [14] | |
| Sequence | GGACACGTCGCCAGGAGAGAACTGAGGCGCCTTCTAGCAGT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1B; MM6; Jurkat; peripheral-blood; HEK293A-TOA; TIME; TREX; iSLK; MSC; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480479 | ||
| mod ID: M6ASITE045857 | Click to Show/Hide the Full List | ||
| mod site | chr2:74834399-74834400:+ | [14] | |
| Sequence | CCAAAATCACGTCTCCGGAGACCCGCGCCCTCCGCCAGCCG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1B; MM6; Jurkat; peripheral-blood; HEK293A-TOA; TREX; iSLK; MSC; TIME; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480480 | ||
| mod ID: M6ASITE045858 | Click to Show/Hide the Full List | ||
| mod site | chr2:74834460-74834461:+ | [14] | |
| Sequence | CTTCTTTGTGCGCCGTCCGGACTCCCAGCTCCCGGCCCGGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1A; H1B; LCLs; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480481 | ||
| mod ID: M6ASITE045859 | Click to Show/Hide the Full List | ||
| mod site | chr2:74834508-74834509:+ | [14] | |
| Sequence | CCCCAGCACAAAGCAGTCGGACCGCGCCGCCCGCCTCCCCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1A; H1B; LCLs; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480482 | ||
| mod ID: M6ASITE045860 | Click to Show/Hide the Full List | ||
| mod site | chr2:74835200-74835201:+ | [14] | |
| Sequence | AGTAGGAAAGGGGCAGAGGAACTCCGCCCGCCGCGCCCCCT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1A; H1B; MM6; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; TIME; TREX; MSC; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480483 | ||
| mod ID: M6ASITE045861 | Click to Show/Hide the Full List | ||
| mod site | chr2:74835228-74835229:+ | [14] | |
| Sequence | CGCCGCGCCCCCTCCCCGGAACTGTGGCGCCCGGAACTGAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1A; H1B; MM6; HEK293A-TOA; iSLK; TREX; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480484 | ||
| mod ID: M6ASITE045862 | Click to Show/Hide the Full List | ||
| mod site | chr2:74835243-74835244:+ | [14] | |
| Sequence | CCGGAACTGTGGCGCCCGGAACTGAGGCTCGGCGAGGGGCG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1A; H1B; MM6; HEK293A-TOA; iSLK; TREX; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480485 | ||
| mod ID: M6ASITE045863 | Click to Show/Hide the Full List | ||
| mod site | chr2:74835302-74835303:+ | [14] | |
| Sequence | CCCTCCAAATCAGCCTCGGGACCGCCCAGCCTCGTGTCCTC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480486 | ||
| mod ID: M6ASITE045864 | Click to Show/Hide the Full List | ||
| mod site | chr2:74854311-74854312:+ | [13] | |
| Sequence | GGTTGACCAGTATCTCTACCACATGCGCCTCTCTGATGAGA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480487 | ||
| mod ID: M6ASITE045865 | Click to Show/Hide the Full List | ||
| mod site | chr2:74854331-74854332:+ | [14] | |
| Sequence | ACATGCGCCTCTCTGATGAGACCCTCTTGGAGATCTCTAAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480488 | ||
| mod ID: M6ASITE045866 | Click to Show/Hide the Full List | ||
| mod site | chr2:74854451-74854452:+ | [14] | |
| Sequence | TGAGGTCCACTCCAGATGGGACAGGTACTGCATCTGGGGGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000472302.1; ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480489 | ||
| mod ID: M6ASITE045867 | Click to Show/Hide the Full List | ||
| mod site | chr2:74867670-74867671:+ | [14] | |
| Sequence | TGGCTCTGGATCTTGGAGGGACCAACTTCCGTGTGCTTTGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000472302.1; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480490 | ||
| mod ID: M6ASITE045868 | Click to Show/Hide the Full List | ||
| mod site | chr2:74867704-74867705:+ | [14] | |
| Sequence | GCTTTGGGTGAAAGTAACGGACAATGGGCTCCAGAAGGTGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000472302.1; ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480491 | ||
| mod ID: M6ASITE045869 | Click to Show/Hide the Full List | ||
| mod site | chr2:74867758-74867759:+ | [14] | |
| Sequence | GATCTATGCCATCCCTGAGGACATCATGCGAGGCAGTGGCA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6; ENST00000472302.1 | ||
| External Link | RMBase: m6A_site_480492 | ||
| mod ID: M6ASITE045870 | Click to Show/Hide the Full List | ||
| mod site | chr2:74867823-74867824:+ | [14] | |
| Sequence | GCAGCCCCTGGTGACAAAAAACTTCCTTGGCATGTTCTTTC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000472302.1; ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480493 | ||
| mod ID: M6ASITE045871 | Click to Show/Hide the Full List | ||
| mod site | chr2:74867892-74867893:+ | [14] | |
| Sequence | TGGATAGGCAGTCACCCAAGACACTCATGGATGCCAGCACA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000472302.1; ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480494 | ||
| mod ID: M6ASITE045872 | Click to Show/Hide the Full List | ||
| mod site | chr2:74872357-74872358:+ | [14] | |
| Sequence | GGATAAGCTACAAATCAAAGACAAGAAGCTCCCACTGGGTT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480495 | ||
| mod ID: M6ASITE045873 | Click to Show/Hide the Full List | ||
| mod site | chr2:74872404-74872405:+ | [14] | |
| Sequence | TCTCGTTCCCCTGCCACCAGACTAAACTAGACGAGGTAAGA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480496 | ||
| mod ID: M6ASITE045874 | Click to Show/Hide the Full List | ||
| mod site | chr2:74872409-74872410:+ | [14] | |
| Sequence | TTCCCCTGCCACCAGACTAAACTAGACGAGGTAAGATGGGC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480497 | ||
| mod ID: M6ASITE045875 | Click to Show/Hide the Full List | ||
| mod site | chr2:74873293-74873294:+ | [14] | |
| Sequence | AGAGTTTCCTGGTCTCATGGACCAAGGGATTCAAGTCCAGT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480498 | ||
| mod ID: M6ASITE045876 | Click to Show/Hide the Full List | ||
| mod site | chr2:74873856-74873857:+ | [13] | |
| Sequence | TCCACAGGACTTTGATATCGACATTGTGGCTGTGGTGAATG | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480499 | ||
| mod ID: M6ASITE045877 | Click to Show/Hide the Full List | ||
| mod site | chr2:74873888-74873889:+ | [14] | |
| Sequence | TGGTGAATGACACAGTTGGGACCATGATGACCTGTGGTTAT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; MT4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480500 | ||
| mod ID: M6ASITE045878 | Click to Show/Hide the Full List | ||
| mod site | chr2:74874310-74874311:+ | [13] | |
| Sequence | GGAAGAGATGCGCCACATCGACATGGTGGAAGGCGATGAGG | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480501 | ||
| mod ID: M6ASITE045879 | Click to Show/Hide the Full List | ||
| mod site | chr2:74874388-74874389:+ | [13] | |
| Sequence | GGACGATGGCTCGCTCAACGACATTCGCACTGAGTTTGACC | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480502 | ||
| mod ID: M6ASITE045880 | Click to Show/Hide the Full List | ||
| mod site | chr2:74874418-74874419:+ | [13] | |
| Sequence | TGAGTTTGACCAGGAGATTGACATGGGCTCACTGAACCCGG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480503 | ||
| mod ID: M6ASITE045881 | Click to Show/Hide the Full List | ||
| mod site | chr2:74874433-74874434:+ | [14] | |
| Sequence | GATTGACATGGGCTCACTGAACCCGGGAAAGCAACTGTGAG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; MT4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480504 | ||
| mod ID: M6ASITE045882 | Click to Show/Hide the Full List | ||
| mod site | chr2:74877191-74877192:+ | [13] | |
| Sequence | GAAGATGATCAGTGGGATGTACATGGGGGAGCTGGTGAGGC | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480505 | ||
| mod ID: M6ASITE045883 | Click to Show/Hide the Full List | ||
| mod site | chr2:74877278-74877279:+ | [13] | |
| Sequence | GCTCAGCCCAGAGCTTCTCAACACCGGTCGCTTTGAGACCA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480506 | ||
| mod ID: M6ASITE045884 | Click to Show/Hide the Full List | ||
| mod site | chr2:74877295-74877296:+ | [14] | |
| Sequence | TCAACACCGGTCGCTTTGAGACCAAAGACATCTCAGACATT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; MT4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480507 | ||
| mod ID: M6ASITE045885 | Click to Show/Hide the Full List | ||
| mod site | chr2:74877302-74877303:+ | [14] | |
| Sequence | CGGTCGCTTTGAGACCAAAGACATCTCAGACATTGAAGGGT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; MT4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480508 | ||
| mod ID: M6ASITE045886 | Click to Show/Hide the Full List | ||
| mod site | chr2:74877311-74877312:+ | [14] | |
| Sequence | TGAGACCAAAGACATCTCAGACATTGAAGGGTGAGCTTCTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; MT4 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480509 | ||
| mod ID: M6ASITE045887 | Click to Show/Hide the Full List | ||
| mod site | chr2:74878740-74878741:+ | [14] | |
| Sequence | CCTGATGCGGTTGGGCCTGGACCCGACTCAGGAGGACTGCG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480510 | ||
| mod ID: M6ASITE045888 | Click to Show/Hide the Full List | ||
| mod site | chr2:74878755-74878756:+ | [14] | |
| Sequence | CCTGGACCCGACTCAGGAGGACTGCGTGGCCACTCACCGGA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480511 | ||
| mod ID: M6ASITE045889 | Click to Show/Hide the Full List | ||
| mod site | chr2:74878793-74878794:+ | [13] | |
| Sequence | GGATCTGCCAGATCGTGTCCACACGCTCCGCCAGCCTGTGC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480512 | ||
| mod ID: M6ASITE045890 | Click to Show/Hide the Full List | ||
| mod site | chr2:74878854-74878855:+ | [14] | |
| Sequence | GCTGCAGCGCATCAAGGAGAACAAAGGCGAGGAGCGGCTGC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480513 | ||
| mod ID: M6ASITE045891 | Click to Show/Hide the Full List | ||
| mod site | chr2:74878905-74878906:+ | [13] | |
| Sequence | TGGGGTCGACGGTTCCGTCTACAAGAAACACCCCCAGTGAG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480514 | ||
| mod ID: M6ASITE045892 | Click to Show/Hide the Full List | ||
| mod site | chr2:74878912-74878913:+ | [14] | |
| Sequence | GACGGTTCCGTCTACAAGAAACACCCCCAGTGAGTCAGTGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480515 | ||
| mod ID: M6ASITE045893 | Click to Show/Hide the Full List | ||
| mod site | chr2:74880279-74880280:+ | [13] | |
| Sequence | TGCAGTTTTGCCAAGCGTCTACATAAGACCGTGCGGCGGCT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480516 | ||
| mod ID: M6ASITE045894 | Click to Show/Hide the Full List | ||
| mod site | chr2:74880286-74880287:+ | [14] | |
| Sequence | TTGCCAAGCGTCTACATAAGACCGTGCGGCGGCTGGTGCCC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480517 | ||
| mod ID: M6ASITE045895 | Click to Show/Hide the Full List | ||
| mod site | chr2:74880415-74880416:+ | [14] | |
| Sequence | AACACCGTGCCCGCCAGAAGACATTAGAGCATCTGCAGCTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; MT4 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480518 | ||
| mod ID: M6ASITE045896 | Click to Show/Hide the Full List | ||
| mod site | chr2:74880505-74880506:+ | [14] | |
| Sequence | AGCGAGGTCTGAGCAAGGAGACTCATGCCAGTGCCCCCGTC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; peripheral-blood | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480519 | ||
| mod ID: M6ASITE045897 | Click to Show/Hide the Full List | ||
| mod site | chr2:74881719-74881720:+ | [14] | |
| Sequence | TTATTTTCCAGAGAAAGGGGACTTCTTGGCCTTGGACCTTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480520 | ||
| mod ID: M6ASITE045898 | Click to Show/Hide the Full List | ||
| mod site | chr2:74881734-74881735:+ | [14] | |
| Sequence | AGGGGACTTCTTGGCCTTGGACCTTGGAGGAACAAATTTCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480521 | ||
| mod ID: M6ASITE045899 | Click to Show/Hide the Full List | ||
| mod site | chr2:74881745-74881746:+ | [14] | |
| Sequence | TGGCCTTGGACCTTGGAGGAACAAATTTCCGGGTCCTGCTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; peripheral-blood | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480522 | ||
| mod ID: M6ASITE045900 | Click to Show/Hide the Full List | ||
| mod site | chr2:74881806-74881807:+ | [13] | |
| Sequence | GTGGGGTGGAGTGGAGATGCACAACAAGATCTACGCCATCC | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480523 | ||
| mod ID: M6ASITE045901 | Click to Show/Hide the Full List | ||
| mod site | chr2:74881809-74881810:+ | [13] | |
| Sequence | GGGTGGAGTGGAGATGCACAACAAGATCTACGCCATCCCGC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480524 | ||
| mod ID: M6ASITE045902 | Click to Show/Hide the Full List | ||
| mod site | chr2:74882129-74882130:+ | [13] | |
| Sequence | CTCCCTGCAGCTCTTTGACCACATTGTCCAGTGCATCGCGG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480525 | ||
| mod ID: M6ASITE045903 | Click to Show/Hide the Full List | ||
| mod site | chr2:74882150-74882151:+ | [14] | |
| Sequence | CATTGTCCAGTGCATCGCGGACTTCCTCGAGTACATGGGCA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480526 | ||
| mod ID: M6ASITE045904 | Click to Show/Hide the Full List | ||
| mod site | chr2:74882162-74882163:+ | [13] | |
| Sequence | CATCGCGGACTTCCTCGAGTACATGGGCATGAAGGGCGTGT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480527 | ||
| mod ID: M6ASITE045905 | Click to Show/Hide the Full List | ||
| mod site | chr2:74882225-74882226:+ | [14] | |
| Sequence | CTCCTTCCCCTGCCAGCAGAACAGCCTGGACGAGGTAACAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480528 | ||
| mod ID: M6ASITE045906 | Click to Show/Hide the Full List | ||
| mod site | chr2:74885511-74885512:+ | [14] | |
| Sequence | AGAGCATCCTCCTCAAGTGGACAAAAGGCTTCAAGGCATCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480529 | ||
| mod ID: M6ASITE045907 | Click to Show/Hide the Full List | ||
| mod site | chr2:74886327-74886328:+ | [13] | |
| Sequence | TGTGGTTGCTGTGGTGAACGACACAGTCGGAACTATGATGA | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480530 | ||
| mod ID: M6ASITE045908 | Click to Show/Hide the Full List | ||
| mod site | chr2:74886338-74886339:+ | [14] | |
| Sequence | TGGTGAACGACACAGTCGGAACTATGATGACCTGTGGCTTT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; Huh7; peripheral-blood; endometrial; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480531 | ||
| mod ID: M6ASITE045909 | Click to Show/Hide the Full List | ||
| mod site | chr2:74886363-74886364:+ | [14] | |
| Sequence | GATGACCTGTGGCTTTGAAGACCCTCACTGTGAAGTTGGCC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; Huh7; peripheral-blood | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480532 | ||
| mod ID: M6ASITE045910 | Click to Show/Hide the Full List | ||
| mod site | chr2:74886510-74886511:+ | [13] | |
| Sequence | CACGGGCAGCAATGCCTGCTACATGGAGGAGATGCGCAACG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480533 | ||
| mod ID: M6ASITE045911 | Click to Show/Hide the Full List | ||
| mod site | chr2:74886535-74886536:+ | [14] | |
| Sequence | GAGGAGATGCGCAACGTGGAACTGGTGGAAGGAGAAGAGGG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; MT4; peripheral-blood | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480534 | ||
| mod ID: M6ASITE045912 | Click to Show/Hide the Full List | ||
| mod site | chr2:74886570-74886571:+ | [14] | |
| Sequence | AGAGGGGCGGATGTGTGTGAACATGGAATGGGGGGCCTTCG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; MT4; peripheral-blood | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480535 | ||
| mod ID: M6ASITE045913 | Click to Show/Hide the Full List | ||
| mod site | chr2:74886594-74886595:+ | [14] | |
| Sequence | GGAATGGGGGGCCTTCGGGGACAATGGATGCCTAGATGACT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; peripheral-blood | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480536 | ||
| mod ID: M6ASITE045914 | Click to Show/Hide the Full List | ||
| mod site | chr2:74887949-74887950:+ | [13] | |
| Sequence | CCTGGGTGAGATTGTCCGTAACATTCTCATCGATTTCACCA | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480537 | ||
| mod ID: M6ASITE045915 | Click to Show/Hide the Full List | ||
| mod site | chr2:74887977-74887978:+ | [14] | |
| Sequence | ATCGATTTCACCAAGCGTGGACTACTCTTCCGAGGCCGCAT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; Jurkat | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480538 | ||
| mod ID: M6ASITE045916 | Click to Show/Hide the Full List | ||
| mod site | chr2:74888014-74888015:+ | [14] | |
| Sequence | GCATCTCAGAGCGGCTCAAGACAAGGGGCATCTTTGAAACC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; Jurkat | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480539 | ||
| mod ID: M6ASITE045917 | Click to Show/Hide the Full List | ||
| mod site | chr2:74888032-74888033:+ | [14] | |
| Sequence | AGACAAGGGGCATCTTTGAAACCAAGTTCTTGTCTCAGATT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; Jurkat | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480540 | ||
| mod ID: M6ASITE045918 | Click to Show/Hide the Full List | ||
| mod site | chr2:74889283-74889284:+ | [13] | |
| Sequence | CAAGTCCGAGCCATCCTGCAACACTTAGGGCTTGAGAGCAC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480541 | ||
| mod ID: M6ASITE045919 | Click to Show/Hide the Full List | ||
| mod site | chr2:74889396-74889397:+ | [15] | |
| Sequence | AGGCATGGCCGCTGTGGTGGACAGGATACGAGAAAACCGTG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | Jurkat; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480542 | ||
| mod ID: M6ASITE045920 | Click to Show/Hide the Full List | ||
| mod site | chr2:74889411-74889412:+ | [15] | |
| Sequence | GGTGGACAGGATACGAGAAAACCGTGGGCTGGACGCTCTCA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | Jurkat; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480543 | ||
| mod ID: M6ASITE045921 | Click to Show/Hide the Full List | ||
| mod site | chr2:74889437-74889438:+ | [16] | |
| Sequence | GGCTGGACGCTCTCAAAGTGACAGTGGGTGTGGATGGGACC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293 | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480544 | ||
| mod ID: M6ASITE045922 | Click to Show/Hide the Full List | ||
| mod site | chr2:74889455-74889456:+ | [14] | |
| Sequence | TGACAGTGGGTGTGGATGGGACCCTCTACAAGCTACATCCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; U2OS; MM6; Jurkat; peripheral-blood | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480545 | ||
| mod ID: M6ASITE045923 | Click to Show/Hide the Full List | ||
| mod site | chr2:74889462-74889463:+ | [16] | |
| Sequence | GGGTGTGGATGGGACCCTCTACAAGCTACATCCTCAGTGAG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | HEK293 | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480546 | ||
| mod ID: M6ASITE045924 | Click to Show/Hide the Full List | ||
| mod site | chr2:74890818-74890819:+ | [14] | |
| Sequence | TTGCCAAAGTCATGCATGAGACAGTGAAGGACCTGGCTCCG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; U2OS; MM6; Jurkat; peripheral-blood | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480547 | ||
| mod ID: M6ASITE045925 | Click to Show/Hide the Full List | ||
| mod site | chr2:74890828-74890829:+ | [14] | |
| Sequence | CATGCATGAGACAGTGAAGGACCTGGCTCCGAAATGTGATG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; U2OS; GM12878; MM6; Huh7; Jurkat; peripheral-blood; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480548 | ||
| mod ID: M6ASITE045926 | Click to Show/Hide the Full List | ||
| mod site | chr2:74890931-74890932:+ | [14] | |
| Sequence | TGCCGCATCCGTGAGGCTGGACAGCGATAGAACCCCTGAAA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; BGC823; U2OS; H1B; H1A; hESCs; fibroblasts; GM12878; LCLs; MM6; Huh7; Jurkat; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480549 | ||
| mod ID: M6ASITE045927 | Click to Show/Hide the Full List | ||
| mod site | chr2:74890942-74890943:+ | [14] | |
| Sequence | TGAGGCTGGACAGCGATAGAACCCCTGAAATCGGAAGGGAC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; BGC823; U2OS; H1B; H1A; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480550 | ||
| mod ID: M6ASITE045928 | Click to Show/Hide the Full List | ||
| mod site | chr2:74890961-74890962:+ | [14] | |
| Sequence | AACCCCTGAAATCGGAAGGGACTTCCTCTTTCTCTCCTTCT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; BGC823; U2OS; H1B; H1A; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480551 | ||
| mod ID: M6ASITE045929 | Click to Show/Hide the Full List | ||
| mod site | chr2:74891024-74891025:+ | [14] | |
| Sequence | GTCATCCCCTTGTGTCAGAGACAGACCCCTTGGCTTTTGCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480552 | ||
| mod ID: M6ASITE045930 | Click to Show/Hide the Full List | ||
| mod site | chr2:74891056-74891057:+ | [14] | |
| Sequence | GCTTTTGCTTGGCAGAGAGGACCCCACTGGACTGGGTTTTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1B; H1A; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480553 | ||
| mod ID: M6ASITE045931 | Click to Show/Hide the Full List | ||
| mod site | chr2:74891066-74891067:+ | [14] | |
| Sequence | GGCAGAGAGGACCCCACTGGACTGGGTTTTGTCTCTGCATC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1B; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480554 | ||
| mod ID: M6ASITE045932 | Click to Show/Hide the Full List | ||
| mod site | chr2:74891156-74891157:+ | [13] | |
| Sequence | TTCCAAATAAGGATTTGTTCACATGCATCATAACCATTCCC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480555 | ||
| mod ID: M6ASITE045933 | Click to Show/Hide the Full List | ||
| mod site | chr2:74891192-74891193:+ | [14] | |
| Sequence | TTCCCATTGGTTCTCCTAAAACATGAAAATTATCTCCCTTA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1B; hESCs; fibroblasts; GM12878; LCLs; H1299; Huh7; Jurkat; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480556 | ||
| mod ID: M6ASITE045934 | Click to Show/Hide the Full List | ||
| mod site | chr2:74891264-74891265:+ | [14] | |
| Sequence | TATAATTCTACAGGATGGGGACACTAATGAAGATACGGTTG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; U2OS; H1B; hESCs; GM12878; LCLs; Huh7; Jurkat; HEK293A-TOA; iSLK; MSC; TREX; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480557 | ||
| mod ID: M6ASITE045935 | Click to Show/Hide the Full List | ||
| mod site | chr2:74891303-74891304:+ | [14] | |
| Sequence | TGCTTCACCTTGGAGCCTGAACATGACATTTCTAAGTGGGG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; U2OS; H1B; H1A; hESCs; GM12878; LCLs; Huh7; Jurkat; HEK293A-TOA; MSC; TREX; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480558 | ||
| mod ID: M6ASITE045936 | Click to Show/Hide the Full List | ||
| mod site | chr2:74891308-74891309:+ | [13] | |
| Sequence | CACCTTGGAGCCTGAACATGACATTTCTAAGTGGGGTGCAT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480559 | ||
| mod ID: M6ASITE045937 | Click to Show/Hide the Full List | ||
| mod site | chr2:74891494-74891495:+ | [13] | |
| Sequence | GCCCCTCTTTAACCTCATCTACAAGCATTTGCCCTGTGGAT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480560 | ||
| mod ID: M6ASITE045938 | Click to Show/Hide the Full List | ||
| mod site | chr2:74891566-74891567:+ | ||
| Sequence | TCCTGAGTCTGGGCAATGAAACCAAAGCCAGGAGTTGACGC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480561 | ||
| mod ID: M6ASITE045939 | Click to Show/Hide the Full List | ||
| mod site | chr2:74891627-74891628:+ | [13] | |
| Sequence | GTCGCATCTCAGCGGGGCGCACATGTTATCCACAAGCAATG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480562 | ||
| mod ID: M6ASITE045940 | Click to Show/Hide the Full List | ||
| mod site | chr2:74891638-74891639:+ | [13] | |
| Sequence | GCGGGGCGCACATGTTATCCACAAGCAATGGACCTTTGGGG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480563 | ||
| mod ID: M6ASITE045941 | Click to Show/Hide the Full List | ||
| mod site | chr2:74891649-74891650:+ | [17] | |
| Sequence | ATGTTATCCACAAGCAATGGACCTTTGGGGAAGGGGGAGTT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HEK293T; MT4; AML | ||
| Seq Type List | MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480564 | ||
| mod ID: M6ASITE045942 | Click to Show/Hide the Full List | ||
| mod site | chr2:74891683-74891684:+ | [13] | |
| Sequence | GGGAGTTTTTAGTTTGTTTTACAAATTTTTCCTGCAAAAGT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480565 | ||
| mod ID: M6ASITE045943 | Click to Show/Hide the Full List | ||
| mod site | chr2:74891780-74891781:+ | [13] | |
| Sequence | AGATCTGCTTCTTCATCTTGACATGTAATGAATGGTCAGTT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480566 | ||
| mod ID: M6ASITE045944 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892031-74892032:+ | [14] | |
| Sequence | GAAGTGTGTGGAAGGCAGGGACAGAGATGGAGGCCGAGCCA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; LCLs; HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480567 | ||
| mod ID: M6ASITE045945 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892056-74892057:+ | [14] | |
| Sequence | GATGGAGGCCGAGCCAATAGACTGAAGAGACCACAGCAATT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; LCLs; HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480568 | ||
| mod ID: M6ASITE045946 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892065-74892066:+ | [14] | |
| Sequence | CGAGCCAATAGACTGAAGAGACCACAGCAATTGGCTCCTCC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; LCLs; HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480569 | ||
| mod ID: M6ASITE045947 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892134-74892135:+ | [14] | |
| Sequence | TGGGATGTTAAGCAAAGGAAACCAAAGGAATCGTTTCAAAT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; LCLs; HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480570 | ||
| mod ID: M6ASITE045948 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892157-74892158:+ | [14] | |
| Sequence | AAAGGAATCGTTTCAAATGGACTCATGGCTTAGAAATCTTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; LCLs; HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480571 | ||
| mod ID: M6ASITE045949 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892215-74892216:+ | [13] | |
| Sequence | TAGTATTCTAAAGCTTTCTGACAAGATAAAGGAAGTCACCA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480572 | ||
| mod ID: M6ASITE045950 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892232-74892233:+ | [18] | |
| Sequence | CTGACAAGATAAAGGAAGTCACCAAAATTTCTTTTTTTAAA | ||
| Motif Score | 2.026654762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480573 | ||
| mod ID: M6ASITE045951 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892269-74892270:+ | [13] | |
| Sequence | TAAATTGTATCTAATCCTCAACAACAAACCAAAACAGAACA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480574 | ||
| mod ID: M6ASITE045952 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892276-74892277:+ | [14] | |
| Sequence | TATCTAATCCTCAACAACAAACCAAAACAGAACAATTAAAC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480575 | ||
| mod ID: M6ASITE045953 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892282-74892283:+ | [14] | |
| Sequence | ATCCTCAACAACAAACCAAAACAGAACAATTAAACAGCCAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480576 | ||
| mod ID: M6ASITE045954 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892287-74892288:+ | [14] | |
| Sequence | CAACAACAAACCAAAACAGAACAATTAAACAGCCAAATAAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480577 | ||
| mod ID: M6ASITE045955 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892295-74892296:+ | [14] | |
| Sequence | AACCAAAACAGAACAATTAAACAGCCAAATAAAACCTCAGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; Huh7 | ||
| Seq Type List | m6A-seq; DART-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480578 | ||
| mod ID: M6ASITE045956 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892308-74892309:+ | [14] | |
| Sequence | CAATTAAACAGCCAAATAAAACCTCAGGGACAACATTTTTG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480579 | ||
| mod ID: M6ASITE045957 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892317-74892318:+ | [14] | |
| Sequence | AGCCAAATAAAACCTCAGGGACAACATTTTTGGTGTATTTG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480580 | ||
| mod ID: M6ASITE045958 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892384-74892385:+ | [13] | |
| Sequence | GTTTGTATTTTAAATGTTTTACAAGAATTGTCCATGTGCTT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480581 | ||
| mod ID: M6ASITE045959 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892463-74892464:+ | [18] | |
| Sequence | TGTGTTTTTCTTTTTTTTATACACAACATTTATTTCAAACT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480582 | ||
| mod ID: M6ASITE045960 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892468-74892469:+ | [13] | |
| Sequence | TTTTCTTTTTTTTATACACAACATTTATTTCAAACTATTGG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480583 | ||
| mod ID: M6ASITE045961 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892578-74892579:+ | [13] | |
| Sequence | GCCCGCCTCTGTCCTAAAATACAAACAAGCACAGACATTAA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480584 | ||
| mod ID: M6ASITE045962 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892582-74892583:+ | [14] | |
| Sequence | GCCTCTGTCCTAAAATACAAACAAGCACAGACATTAAACCT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480585 | ||
| mod ID: M6ASITE045963 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892592-74892593:+ | [14] | |
| Sequence | TAAAATACAAACAAGCACAGACATTAAACCTGGATACTATA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T | ||
| Seq Type List | m6A-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480586 | ||
| mod ID: M6ASITE045964 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892599-74892600:+ | [14] | |
| Sequence | CAAACAAGCACAGACATTAAACCTGGATACTATATGATAAA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480587 | ||
| mod ID: M6ASITE045965 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892645-74892646:+ | [18] | |
| Sequence | ATGTAACTATTGAATTGGATACAAGGATCAGAATGGAAAGA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480588 | ||
| mod ID: M6ASITE045966 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892667-74892668:+ | [14] | |
| Sequence | AAGGATCAGAATGGAAAGAAACTCACGATGAAATTGAACCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480589 | ||
| mod ID: M6ASITE045967 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892684-74892685:+ | [14] | |
| Sequence | GAAACTCACGATGAAATTGAACCTGGTTTTTGTATATTTAT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480590 | ||
| mod ID: M6ASITE045968 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892708-74892709:+ | [18] | |
| Sequence | GGTTTTTGTATATTTATCAAACTTGTGCTGAGAATAGTGTC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480591 | ||
| mod ID: M6ASITE045969 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892736-74892737:+ | [18] | |
| Sequence | TGAGAATAGTGTCTGATTATACGACTTTTAAGCAAAGTTGG | ||
| Motif Score | 2.084416667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480592 | ||
| mod ID: M6ASITE045970 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892739-74892740:+ | [18] | |
| Sequence | GAATAGTGTCTGATTATACGACTTTTAAGCAAAGTTGGGTG | ||
| Motif Score | 3.287565476 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480593 | ||
| mod ID: M6ASITE045971 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892880-74892881:+ | [18] | |
| Sequence | CCTCTGCTGCTCCCATCTGCACTAATTGACCCAAAACGTGG | ||
| Motif Score | 3.252583333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480594 | ||
| mod ID: M6ASITE045972 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892915-74892916:+ | [18] | |
| Sequence | ACGTGGGTATTTCCTGCTACACAAAAGCCAAAAGGTTTCAT | ||
| Motif Score | 2.084928571 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480595 | ||
| mod ID: M6ASITE045973 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892950-74892951:+ | [18] | |
| Sequence | TTTCATGTAGATTTTAGTTCACTAAAGGGTGCCCACAAAAT | ||
| Motif Score | 2.469291667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480596 | ||
| mod ID: M6ASITE045974 | Click to Show/Hide the Full List | ||
| mod site | chr2:74892964-74892965:+ | [13] | |
| Sequence | TAGTTCACTAAAGGGTGCCCACAAAATAGAGATTAATTTTA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000409174.1; ENST00000290573.6 | ||
| External Link | RMBase: m6A_site_480597 | ||
| mod ID: M6ASITE045975 | Click to Show/Hide the Full List | ||
| mod site | chr2:74893028-74893029:+ | [13] | |
| Sequence | TAGGTACTATCTGTGAAGTTACACTTTTTTTTTTTTTTTTA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480598 | ||
| mod ID: M6ASITE045976 | Click to Show/Hide the Full List | ||
| mod site | chr2:74893108-74893109:+ | [14] | |
| Sequence | TTGTTGGTTTCCAAAAAGGAACACTGGAAAATAAATTTTGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480599 | ||
| mod ID: M6ASITE045977 | Click to Show/Hide the Full List | ||
| mod site | chr2:74893110-74893111:+ | [18] | |
| Sequence | GTTGGTTTCCAAAAAGGAACACTGGAAAATAAATTTTGAAT | ||
| Motif Score | 2.506922619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480600 | ||
| mod ID: M6ASITE045978 | Click to Show/Hide the Full List | ||
| mod site | chr2:74893169-74893170:+ | [18] | |
| Sequence | AGGTTGACAGTCCCTTGCTGACATGGCTTTGCTTTGTGTAA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480601 | ||
| mod ID: M6ASITE045979 | Click to Show/Hide the Full List | ||
| mod site | chr2:74893281-74893282:+ | [13] | |
| Sequence | TGAAGATGTTGTCATTATGTACAATACATAACTAGTTTAAT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000290573.6; ENST00000409174.1 | ||
| External Link | RMBase: m6A_site_480602 | ||
References