m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00416)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
SRSF3
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LNCaP cell line | Homo sapiens |
|
Treatment: shMETTL3 LNCaP cells
Control: shControl LNCaP cells
|
GSE147884 | |
| Regulation |
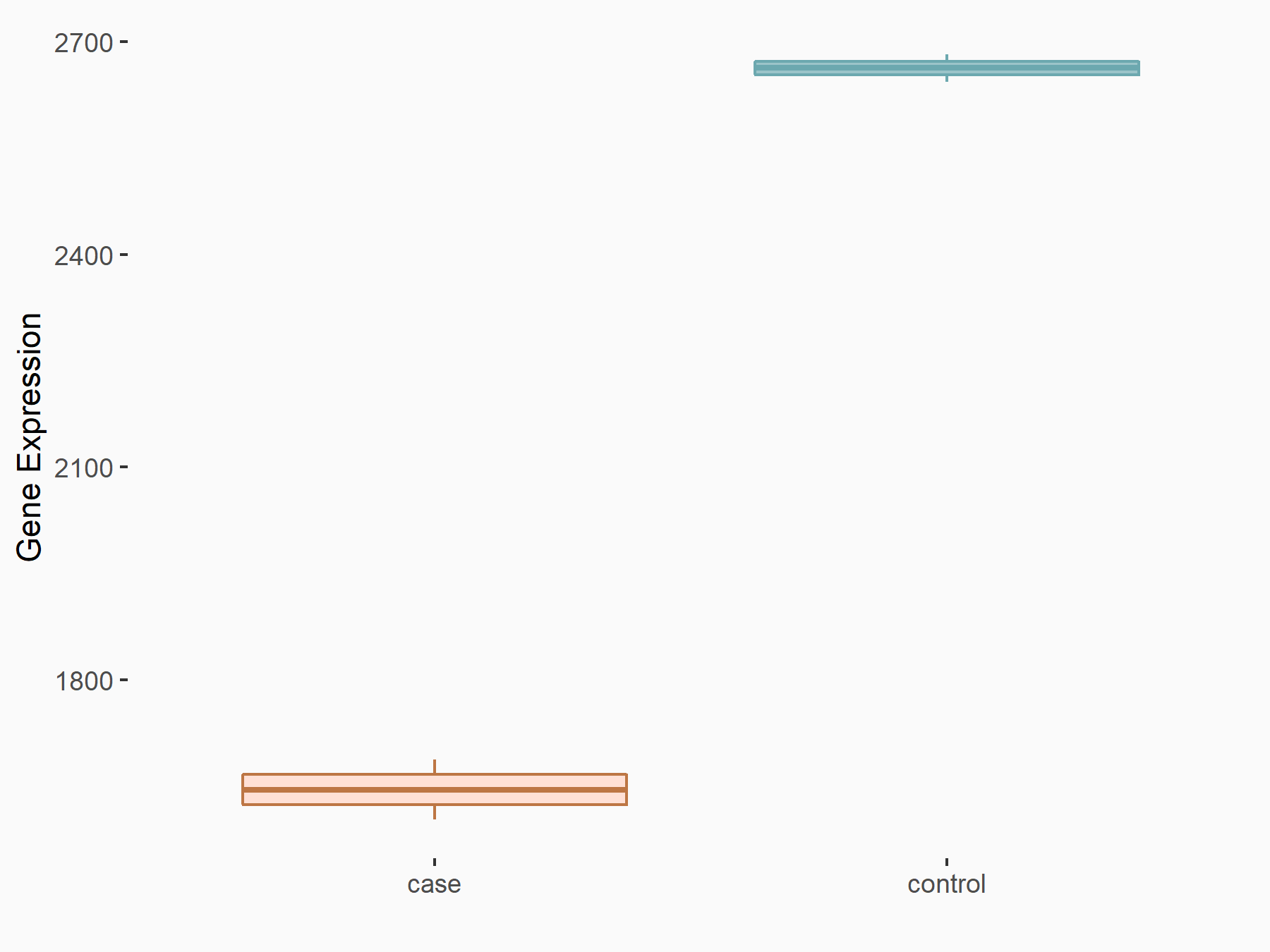  |
logFC: -6.96E-01 p-value: 4.00E-41 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Silencing METTL3 or overexpressing dominant-negative mutant METTL3 suppressed the growth and self-renewal of Glioblastoma cells. Integrated transcriptome and MeRIP-seq analyses revealed that downregulating the expression of METTL3 decreased m6A modification levels of Serine/arginine-rich splicing factor 3 (SRSF3), which led to YTHDC1-dependent NMD of SRSF transcripts and decreased SRSF protein expression. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | mRNA decay | |||
| In-vitro Model | U251 (Fibroblasts or fibroblast like cells) | |||
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| In-vivo Model | For subcutaneous tumor model, each mouse was injected subcutaneously in the right flank with 2 × 106 U87MG cells (METTL3-KD or control) in 100 uL PBS. | |||
YTH domain-containing protein 1 (YTHDC1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDC1 | ||
| Cell Line | ICM cell line | Mus musculus |
|
Treatment: YTHDC1 knockout ICM cells
Control: Wild type ICM cells
|
GSE157267 | |
| Regulation |
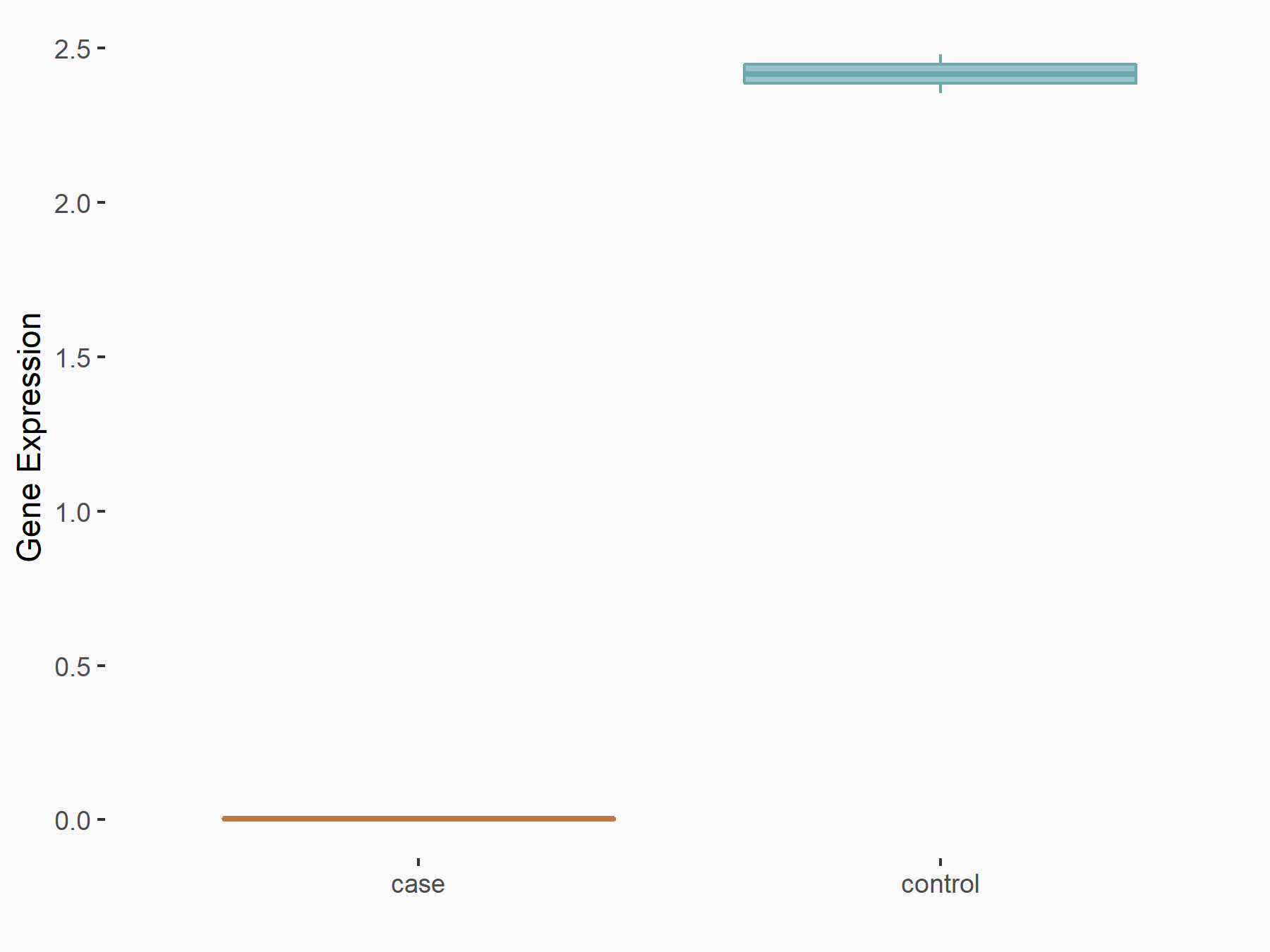  |
logFC: -1.77E+00 p-value: 4.66E-05 |
| More Results | Click to View More RNA-seq Results | |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Silencing METTL3 or overexpressing dominant-negative mutant METTL3 suppressed the growth and self-renewal of Glioblastoma cells. Integrated transcriptome and MeRIP-seq analyses revealed that downregulating the expression of METTL3 decreased m6A modification levels of Serine/arginine-rich splicing factor 3 (SRSF3), which led to YTHDC1-dependent NMD of SRSF transcripts and decreased SRSF protein expression. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | mRNA decay | |||
| In-vitro Model | U251 (Fibroblasts or fibroblast like cells) | |||
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| In-vivo Model | For subcutaneous tumor model, each mouse was injected subcutaneously in the right flank with 2 × 106 U87MG cells (METTL3-KD or control) in 100 uL PBS. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | Kaposi's sarcoma-associated herpesvirus(KSHV) productive lytic replication plays a pivotal role in the initiation and progression of Kaposi's sarcoma tumors. m6A sites in RTA pre-mRNA crucial for splicing through interactions with YTHDC1, Serine/arginine-rich splicing factor 3 (SRSF3) and SRSF10. m6A in regulating RTA pre-mRNA splicing but also suggest that KSHV has evolved a mechanism to manipulate the host m6A machinery to its advantage in promoting lytic replication. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Kaposi's sarcoma | ICD-11: 2B57 | ||
| Pathway Response | Spliceosome | hsa03040 | ||
| In-vitro Model | TIVE-KSHV (KSHV-infected telomerase-immortalized human umbilical vein endothelial cells (TIVE-KSHV cells)) | |||
| iSLK-BAC16 cells (Kaposi's sarcoma cells carrying the recombinant KSHV bacterial artificial chromosome 16 (BAC16) (iSLK-BAC16 cells)) | ||||
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
Brain cancer [ICD-11: 2A00]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Silencing METTL3 or overexpressing dominant-negative mutant METTL3 suppressed the growth and self-renewal of Glioblastoma cells. Integrated transcriptome and MeRIP-seq analyses revealed that downregulating the expression of METTL3 decreased m6A modification levels of Serine/arginine-rich splicing factor 3 (SRSF3), which led to YTHDC1-dependent NMD of SRSF transcripts and decreased SRSF protein expression. | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | mRNA decay | |||
| In-vitro Model | U251 (Fibroblasts or fibroblast like cells) | |||
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| In-vivo Model | For subcutaneous tumor model, each mouse was injected subcutaneously in the right flank with 2 × 106 U87MG cells (METTL3-KD or control) in 100 uL PBS. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Silencing METTL3 or overexpressing dominant-negative mutant METTL3 suppressed the growth and self-renewal of Glioblastoma cells. Integrated transcriptome and MeRIP-seq analyses revealed that downregulating the expression of METTL3 decreased m6A modification levels of Serine/arginine-rich splicing factor 3 (SRSF3), which led to YTHDC1-dependent NMD of SRSF transcripts and decreased SRSF protein expression. | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulator | YTH domain-containing protein 1 (YTHDC1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | mRNA decay | |||
| In-vitro Model | U251 (Fibroblasts or fibroblast like cells) | |||
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| In-vivo Model | For subcutaneous tumor model, each mouse was injected subcutaneously in the right flank with 2 × 106 U87MG cells (METTL3-KD or control) in 100 uL PBS. | |||
Kaposi's sarcoma [ICD-11: 2B57]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | Kaposi's sarcoma-associated herpesvirus(KSHV) productive lytic replication plays a pivotal role in the initiation and progression of Kaposi's sarcoma tumors. m6A sites in RTA pre-mRNA crucial for splicing through interactions with YTHDC1, Serine/arginine-rich splicing factor 3 (SRSF3) and SRSF10. m6A in regulating RTA pre-mRNA splicing but also suggest that KSHV has evolved a mechanism to manipulate the host m6A machinery to its advantage in promoting lytic replication. | |||
| Responsed Disease | Kaposi's sarcoma [ICD-11: 2B57] | |||
| Target Regulator | YTH domain-containing protein 1 (YTHDC1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Spliceosome | hsa03040 | ||
| In-vitro Model | TIVE-KSHV (KSHV-infected telomerase-immortalized human umbilical vein endothelial cells (TIVE-KSHV cells)) | |||
| iSLK-BAC16 cells (Kaposi's sarcoma cells carrying the recombinant KSHV bacterial artificial chromosome 16 (BAC16) (iSLK-BAC16 cells)) | ||||
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
Pancreatic cancer [ICD-11: 2C10]
Gemcitabine
[Approved]
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03294 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00416)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: AC4SITE000105 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602440-36602441:+ | [6] | |
| Sequence | TGCTGTGAAACACAGGCCATCAGGGAAAACGAAATGCTGCA | ||
| Cell/Tissue List | H1 | ||
| Seq Type List | ac4C-seq | ||
| Transcript ID List | ENST00000373715.11; ENST00000620389.1; ENST00000614136.1 | ||
| External Link | RMBase: ac4C_site_1592 | ||
5-methylcytidine (m5C)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE003933 | Click to Show/Hide the Full List | ||
| mod site | chr6:36596757-36596758:+ | [7] | |
| Sequence | ATTTTTGGTATTTTTCATTACAGAAATGCATCGTGATTCCT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000613941.4; ENST00000620941.1; ENST00000620242.1; ENST00000339436.11; ENST00000477442.6; ENST00000373715.11 | ||
| External Link | RMBase: m5C_site_37932 | ||
| mod ID: M5CSITE003934 | Click to Show/Hide the Full List | ||
| mod site | chr6:36598925-36598926:+ | [7] | |
| Sequence | AAATCGTGGCCCACCTCCCTCTTGGGGTCGTCGCCCTCGAG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000373715.11; ENST00000613941.4; ENST00000477442.6; ENST00000620941.1; ENST00000339436.11 | ||
| External Link | RMBase: m5C_site_37933 | ||
N6-methyladenosine (m6A)
| In total 57 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE074805 | Click to Show/Hide the Full List | ||
| mod site | chr6:36594428-36594429:+ | [8] | |
| Sequence | GGCCGGAGGAAAGCGGGAAGACTCATCGGAGCGTGTGGATT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; A549; H1B; MM6; CD4T; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000477442.6; ENST00000620242.1; ENST00000613941.4; ENST00000339436.11; ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715072 | ||
| mod ID: M6ASITE074806 | Click to Show/Hide the Full List | ||
| mod site | chr6:36596787-36596788:+ | [9] | |
| Sequence | TCGTGATTCCTGTCCATTGGACTGTAAGGTTTATGTAGGCA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | BGC823 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000339436.11; ENST00000620941.1; ENST00000613941.4; ENST00000620242.1; ENST00000477442.6; ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715073 | ||
| mod ID: M6ASITE074807 | Click to Show/Hide the Full List | ||
| mod site | chr6:36596817-36596818:+ | [10] | |
| Sequence | TTATGTAGGCAATCTTGGAAACAATGGCAACAAGACGGAAT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T; BGC823; HepG2; AML | ||
| Seq Type List | DART-seq; MAZTER-seq; m6A-seq; miCLIP | ||
| Transcript ID List | ENST00000477442.6; ENST00000620941.1; ENST00000339436.11; ENST00000620242.1; ENST00000613941.4; ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715074 | ||
| mod ID: M6ASITE074808 | Click to Show/Hide the Full List | ||
| mod site | chr6:36596826-36596827:+ | [10] | |
| Sequence | CAATCTTGGAAACAATGGCAACAAGACGGAATTGGAACGGG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000620941.1; ENST00000373715.11; ENST00000477442.6; ENST00000613941.4; ENST00000339436.11; ENST00000620242.1 | ||
| External Link | RMBase: m6A_site_715075 | ||
| mod ID: M6ASITE074809 | Click to Show/Hide the Full List | ||
| mod site | chr6:36596863-36596864:+ | [9] | |
| Sequence | CGGGCTTTTGGCTACTATGGACCACTCCGAAGTGTGTGGGT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | BGC823; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000620941.1; ENST00000477442.6; ENST00000613941.4; ENST00000339436.11; ENST00000373715.11; ENST00000620242.1 | ||
| External Link | RMBase: m6A_site_715076 | ||
| mod ID: M6ASITE074810 | Click to Show/Hide the Full List | ||
| mod site | chr6:36596892-36596893:+ | [9] | |
| Sequence | AAGTGTGTGGGTTGCTAGAAACCCACCCGGCTTTGCTTTTG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | BGC823; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000620941.1; ENST00000477442.6; ENST00000373715.11; ENST00000613941.4; ENST00000339436.11; ENST00000620242.1 | ||
| External Link | RMBase: m6A_site_715077 | ||
| mod ID: M6ASITE074811 | Click to Show/Hide the Full List | ||
| mod site | chr6:36598849-36598850:+ | [11] | |
| Sequence | AATATCTTTGCCCCCTCAGAACACTATGTGGCTGCCGTGTA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000373715.11; ENST00000613941.4; ENST00000477442.6; ENST00000620941.1; ENST00000339436.11 | ||
| External Link | RMBase: m6A_site_715078 | ||
| mod ID: M6ASITE074812 | Click to Show/Hide the Full List | ||
| mod site | chr6:36599808-36599809:+ | [8] | |
| Sequence | CCATCTATTAATAAAAATGAACCCCGTTACAGAGTCACCAT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000373715.11; ENST00000339436.11; ENST00000620389.1; ENST00000613941.4; ENST00000620941.1; ENST00000477442.6 | ||
| External Link | RMBase: m6A_site_715079 | ||
| mod ID: M6ASITE074813 | Click to Show/Hide the Full List | ||
| mod site | chr6:36600197-36600198:+ | [12] | |
| Sequence | CCAACGCAACATCTGGCAAAACCTTTTCAGCAAATTCTTCC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000339436.11; ENST00000620941.1; ENST00000620389.1; ENST00000373715.11; ENST00000477442.6 | ||
| External Link | RMBase: m6A_site_715080 | ||
| mod ID: M6ASITE074814 | Click to Show/Hide the Full List | ||
| mod site | chr6:36600265-36600266:+ | [12] | |
| Sequence | ACCATTTCTAGCTTGTTGAAACCCAAAACTAGTAAGTTTTT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000373715.11; ENST00000620389.1; ENST00000339436.11; ENST00000620941.1; ENST00000477442.6 | ||
| External Link | RMBase: m6A_site_715081 | ||
| mod ID: M6ASITE074815 | Click to Show/Hide the Full List | ||
| mod site | chr6:36600272-36600273:+ | [12] | |
| Sequence | CTAGCTTGTTGAAACCCAAAACTAGTAAGTTTTTCCTGCTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000339436.11; ENST00000620941.1; ENST00000620389.1; ENST00000373715.11; ENST00000477442.6 | ||
| External Link | RMBase: m6A_site_715082 | ||
| mod ID: M6ASITE074816 | Click to Show/Hide the Full List | ||
| mod site | chr6:36601250-36601251:+ | [13] | |
| Sequence | AGTGTTTCTGCTATTCCTAAACTTTTCCAGGTGGCTTAGTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000614136.1; ENST00000620389.1; ENST00000620941.1; ENST00000477442.6; ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715083 | ||
| mod ID: M6ASITE074817 | Click to Show/Hide the Full List | ||
| mod site | chr6:36601760-36601761:+ | [11] | |
| Sequence | GCTGTCTCGGGAGAGAAATCACAAGCCGTCCCGATCCTTCT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000620389.1; ENST00000620941.1; ENST00000614136.1; ENST00000477442.6; ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715084 | ||
| mod ID: M6ASITE074818 | Click to Show/Hide the Full List | ||
| mod site | chr6:36601822-36601823:+ | [14] | |
| Sequence | TTTGATAACTTGTATTTAAGACTTTGCATACATAGTATGCT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | MT4 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000373715.11; ENST00000620389.1; ENST00000620941.1; ENST00000477442.6; ENST00000614136.1 | ||
| External Link | RMBase: m6A_site_715085 | ||
| mod ID: M6ASITE074819 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602015-36602016:+ | [15] | |
| Sequence | GTTTGCAAGAGAAGTGGTGTACAGGAAATTACTTCATTTGA | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | kidney; liver; HEK293T | ||
| Seq Type List | m6A-REF-seq; DART-seq | ||
| Transcript ID List | ENST00000620941.1; ENST00000614136.1; ENST00000373715.11; ENST00000477442.6; ENST00000620389.1 | ||
| External Link | RMBase: m6A_site_715086 | ||
| mod ID: M6ASITE074820 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602025-36602026:+ | [10] | |
| Sequence | GAAGTGGTGTACAGGAAATTACTTCATTTGACAGGAGTATG | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000477442.6; ENST00000373715.11; ENST00000620389.1; ENST00000614136.1; ENST00000620941.1 | ||
| External Link | RMBase: m6A_site_715087 | ||
| mod ID: M6ASITE074821 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602035-36602036:+ | [10] | |
| Sequence | ACAGGAAATTACTTCATTTGACAGGAGTATGTACAGAAAAT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000373715.11; ENST00000620389.1; ENST00000614136.1; ENST00000477442.6; ENST00000620941.1 | ||
| External Link | RMBase: m6A_site_715088 | ||
| mod ID: M6ASITE074822 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602047-36602048:+ | [10] | |
| Sequence | TTCATTTGACAGGAGTATGTACAGAAAATTCAAGTTTTGTT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000373715.11; ENST00000614136.1; ENST00000620941.1; ENST00000620389.1 | ||
| External Link | RMBase: m6A_site_715089 | ||
| mod ID: M6ASITE074823 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602072-36602073:+ | [8] | |
| Sequence | AAATTCAAGTTTTGTTTGAGACTTCATAAGCTTGGTGCATT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000373715.11; ENST00000620389.1; ENST00000620941.1; ENST00000614136.1 | ||
| External Link | RMBase: m6A_site_715090 | ||
| mod ID: M6ASITE074824 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602134-36602135:+ | [8] | |
| Sequence | AAATCTGTTTGTCTCTTGAAACAGTGACACAAAGGTGTAAT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000373715.11; ENST00000620941.1; ENST00000620389.1; ENST00000614136.1 | ||
| External Link | RMBase: m6A_site_715091 | ||
| mod ID: M6ASITE074825 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602140-36602141:+ | [10] | |
| Sequence | GTTTGTCTCTTGAAACAGTGACACAAAGGTGTAATTCTCTA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000614136.1; ENST00000620389.1; ENST00000620941.1; ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715092 | ||
| mod ID: M6ASITE074826 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602206-36602207:+ | [10] | |
| Sequence | ATGTAATACCAAGAATTGTTACTTTACAATGTTCCCTTAAG | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000373715.11; ENST00000620389.1; ENST00000614136.1 | ||
| External Link | RMBase: m6A_site_715093 | ||
| mod ID: M6ASITE074827 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602211-36602212:+ | [11] | |
| Sequence | ATACCAAGAATTGTTACTTTACAATGTTCCCTTAAGCAAAA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000373715.11; ENST00000614136.1; ENST00000620389.1 | ||
| External Link | RMBase: m6A_site_715094 | ||
| mod ID: M6ASITE074828 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602247-36602248:+ | [8] | |
| Sequence | CAAAATTGAATTTGCTTTGAACTTTTAGTTATGCACAGACT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000614136.1; ENST00000373715.11; ENST00000620389.1 | ||
| External Link | RMBase: m6A_site_715095 | ||
| mod ID: M6ASITE074829 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602261-36602262:+ | [15] | |
| Sequence | CTTTGAACTTTTAGTTATGCACAGACTGATAATAAACCTCT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | liver; HEK293T | ||
| Seq Type List | m6A-REF-seq; DART-seq | ||
| Transcript ID List | ENST00000620389.1; ENST00000373715.11; ENST00000614136.1 | ||
| External Link | RMBase: m6A_site_715096 | ||
| mod ID: M6ASITE074830 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602265-36602266:+ | [8] | |
| Sequence | GAACTTTTAGTTATGCACAGACTGATAATAAACCTCTAAAC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000373715.11; ENST00000614136.1; ENST00000620389.1 | ||
| External Link | RMBase: m6A_site_715097 | ||
| mod ID: M6ASITE074831 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602276-36602277:+ | [8] | |
| Sequence | TATGCACAGACTGATAATAAACCTCTAAACCTGCCCAGCGG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000620389.1; ENST00000373715.11; ENST00000614136.1 | ||
| External Link | RMBase: m6A_site_715098 | ||
| mod ID: M6ASITE074832 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602284-36602285:+ | [8] | |
| Sequence | GACTGATAATAAACCTCTAAACCTGCCCAGCGGAAGTGTGT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000620389.1; ENST00000373715.11; ENST00000614136.1 | ||
| External Link | RMBase: m6A_site_715099 | ||
| mod ID: M6ASITE074834 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602323-36602324:+ | [10] | |
| Sequence | GTTTTTTTTTAAATTTAAATACAGAAACAACTGGCAAAAAT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000614136.1; ENST00000373715.11; ENST00000620389.1 | ||
| External Link | RMBase: m6A_site_715100 | ||
| mod ID: M6ASITE074835 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602329-36602330:+ | [8] | |
| Sequence | TTTTAAATTTAAATACAGAAACAACTGGCAAAAATTGAACT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000614136.1; ENST00000620389.1; ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715101 | ||
| mod ID: M6ASITE074836 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602332-36602333:+ | [10] | |
| Sequence | TAAATTTAAATACAGAAACAACTGGCAAAAATTGAACTAAG | ||
| Motif Score | 2.595904762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000620389.1; ENST00000373715.11; ENST00000614136.1 | ||
| External Link | RMBase: m6A_site_715102 | ||
| mod ID: M6ASITE074837 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602347-36602348:+ | [8] | |
| Sequence | AAACAACTGGCAAAAATTGAACTAAGATTTACTTTTTTTTC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000614136.1; ENST00000373715.11; ENST00000620389.1 | ||
| External Link | RMBase: m6A_site_715103 | ||
| mod ID: M6ASITE074838 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602401-36602402:+ | [8] | |
| Sequence | TAGGCTGCAGCTATAGTTGAACAAGCAGTCTTTAAAAACTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000620389.1; ENST00000373715.11; ENST00000614136.1 | ||
| External Link | RMBase: m6A_site_715104 | ||
| mod ID: M6ASITE074839 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602418-36602419:+ | [8] | |
| Sequence | TGAACAAGCAGTCTTTAAAAACTGCTGTGAAACACAGGCCA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000620389.1; ENST00000373715.11; ENST00000614136.1 | ||
| External Link | RMBase: m6A_site_715105 | ||
| mod ID: M6ASITE074840 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602429-36602430:+ | [8] | |
| Sequence | TCTTTAAAAACTGCTGTGAAACACAGGCCATCAGGGAAAAC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T | ||
| Seq Type List | m6A-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000614136.1; ENST00000373715.11; ENST00000620389.1 | ||
| External Link | RMBase: m6A_site_715106 | ||
| mod ID: M6ASITE074841 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602599-36602600:+ | [11] | |
| Sequence | TTTGATTGCTGTATATGGATACATGGCTGTTCGTGACATTC | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000620389.1; ENST00000373715.11; ENST00000614136.1 | ||
| External Link | RMBase: m6A_site_715107 | ||
| mod ID: M6ASITE074842 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602614-36602615:+ | [10] | |
| Sequence | TGGATACATGGCTGTTCGTGACATTCTTTATGTGCAAATTT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000614136.1; ENST00000373715.11; ENST00000620389.1 | ||
| External Link | RMBase: m6A_site_715108 | ||
| mod ID: M6ASITE074843 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602668-36602669:+ | [11] | |
| Sequence | TGTCCTGCCAGTTTAAGGGTACATTGTAGAGCCGAACTTTG | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000614136.1; ENST00000620389.1; ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715109 | ||
| mod ID: M6ASITE074844 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602683-36602684:+ | [8] | |
| Sequence | AGGGTACATTGTAGAGCCGAACTTTGAGTTACTGTGCAAGA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000373715.11; ENST00000620389.1; ENST00000614136.1 | ||
| External Link | RMBase: m6A_site_715110 | ||
| mod ID: M6ASITE074845 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602693-36602694:+ | [10] | |
| Sequence | GTAGAGCCGAACTTTGAGTTACTGTGCAAGATTTTTTTTTC | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000373715.11; ENST00000614136.1; ENST00000620389.1 | ||
| External Link | RMBase: m6A_site_715111 | ||
| mod ID: M6ASITE074846 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602768-36602769:+ | [11] | |
| Sequence | TTGGGATTAAAGTTTTGGTTACAAATTGTTCTTTAACTTGA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000614136.1; ENST00000373715.11; ENST00000620389.1 | ||
| External Link | RMBase: m6A_site_715112 | ||
| mod ID: M6ASITE074847 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602783-36602784:+ | [10] | |
| Sequence | TGGTTACAAATTGTTCTTTAACTTGAAAGCCTGTTTTTCCT | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715113 | ||
| mod ID: M6ASITE074848 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602809-36602810:+ | [8] | |
| Sequence | AAGCCTGTTTTTCCTTGCAAACTCAAATCTGTGAGCTTGGT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715114 | ||
| mod ID: M6ASITE074849 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602846-36602847:+ | [11] | |
| Sequence | TGGTACCAAGTCCAGGTATAACATTCCTATTGGAAGCCATA | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715115 | ||
| mod ID: M6ASITE074850 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602866-36602867:+ | [10] | |
| Sequence | ACATTCCTATTGGAAGCCATACTTATATTTTCTTGTAAAGT | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715116 | ||
| mod ID: M6ASITE074851 | Click to Show/Hide the Full List | ||
| mod site | chr6:36602978-36602979:+ | [11] | |
| Sequence | CCCATGGGAAGCAGTTGGTTACACGATTCTTATTTTATAAG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715117 | ||
| mod ID: M6ASITE074852 | Click to Show/Hide the Full List | ||
| mod site | chr6:36603001-36603002:+ | [8] | |
| Sequence | CGATTCTTATTTTATAAGAAACAGCTGAGAGGCACTATGGA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715118 | ||
| mod ID: M6ASITE074853 | Click to Show/Hide the Full List | ||
| mod site | chr6:36603132-36603133:+ | [11] | |
| Sequence | TCCATAATATTTAGTGACCAACATTTTAAAGTATAGCAGCA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715119 | ||
| mod ID: M6ASITE074854 | Click to Show/Hide the Full List | ||
| mod site | chr6:36603166-36603167:+ | [8] | |
| Sequence | AGCAGCAACCTGGTTCTTAAACACAAAGTAAGTTGCCCATT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715120 | ||
| mod ID: M6ASITE074855 | Click to Show/Hide the Full List | ||
| mod site | chr6:36603216-36603217:+ | [8] | |
| Sequence | CTTTTATCTTTAGCATGAAAACTTTCCACAGGTCTAAAAAT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715121 | ||
| mod ID: M6ASITE074856 | Click to Show/Hide the Full List | ||
| mod site | chr6:36603305-36603306:+ | [11] | |
| Sequence | CCATCATGATGTAAAAGTTCACAATATGGTTCAAATGTAAC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715122 | ||
| mod ID: M6ASITE074857 | Click to Show/Hide the Full List | ||
| mod site | chr6:36603324-36603325:+ | [11] | |
| Sequence | CACAATATGGTTCAAATGTAACAGTGCAGAATTGAATATGG | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T; AML | ||
| Seq Type List | MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715123 | ||
| mod ID: M6ASITE074858 | Click to Show/Hide the Full List | ||
| mod site | chr6:36603654-36603655:+ | [11] | |
| Sequence | GTTAAGCATGTTCAAGAAAGACACTTTTCAGACAGCCTGTT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715124 | ||
| mod ID: M6ASITE074859 | Click to Show/Hide the Full List | ||
| mod site | chr6:36603722-36603723:+ | [11] | |
| Sequence | ATTTTGTCTTTAGTATTCTCACATAGCCTACAACCTGTTCC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715125 | ||
| mod ID: M6ASITE074860 | Click to Show/Hide the Full List | ||
| mod site | chr6:36603751-36603752:+ | [11] | |
| Sequence | ACAACCTGTTCCTGTTAGGAACAAGCCAAGATAGCTAAGAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715126 | ||
| mod ID: M6ASITE074861 | Click to Show/Hide the Full List | ||
| mod site | chr6:36603800-36603801:+ | [11] | |
| Sequence | AGCCATGCAATATGTCAATTACATTGCTTTTTATAAATGGA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715127 | ||
| mod ID: M6ASITE074862 | Click to Show/Hide the Full List | ||
| mod site | chr6:36604198-36604199:+ | [11] | |
| Sequence | GTGTAAAAATGAAAGGAATCACAAAACTGAACTTAGACCTG | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000373715.11 | ||
| External Link | RMBase: m6A_site_715128 | ||
References