m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00269)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
GLUT1
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
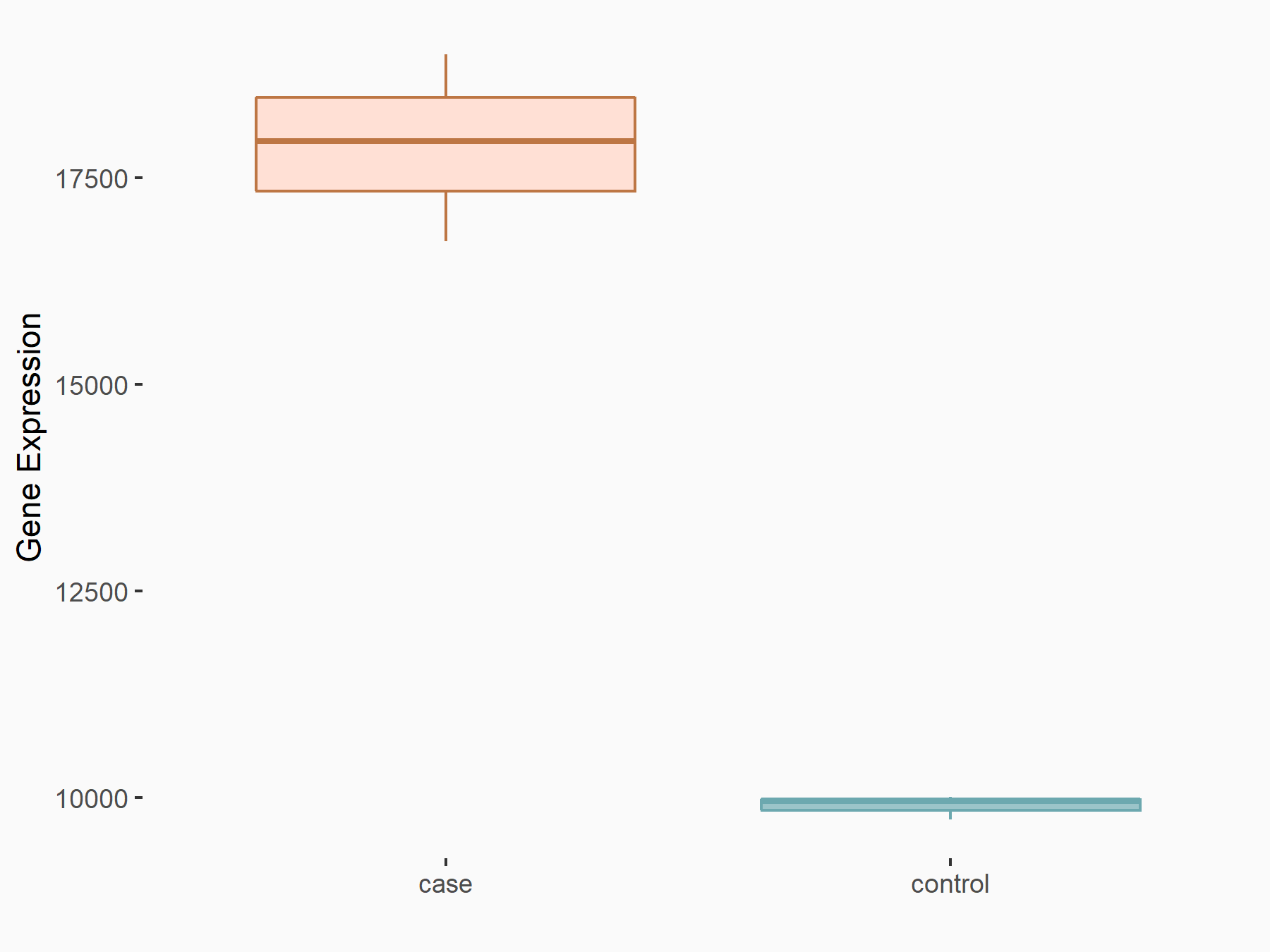 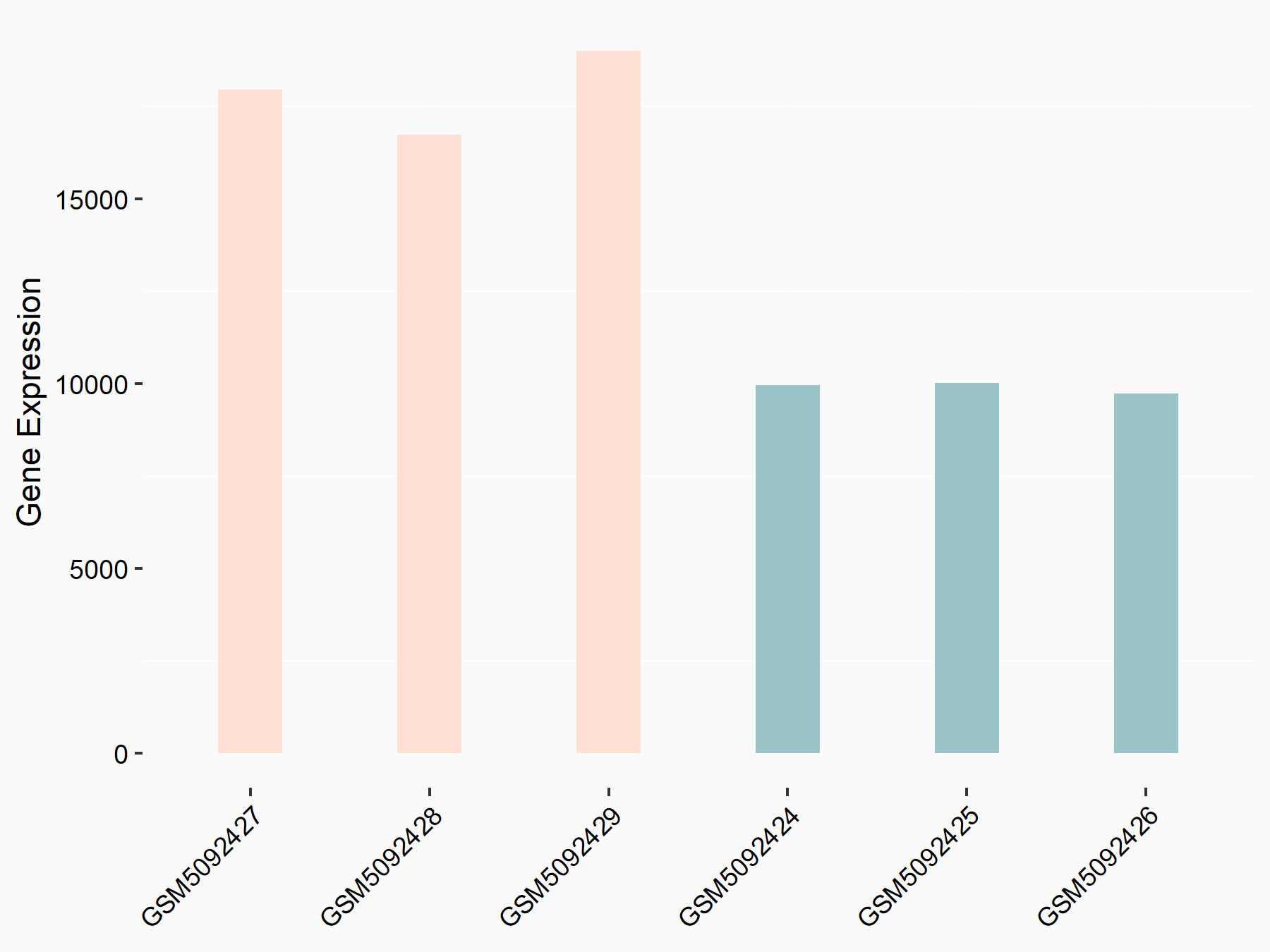 |
logFC: 8.54E-01 p-value: 1.08E-85 |
| More Results | Click to View More RNA-seq Results | |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 stabilizes HK2 and Glucose transporter type 1 (SLC2A1) (GLUT1) expression in colorectal cancer through an m6A-IGF2BP2/3- dependent mechanism. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glucose metabolism | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 induced Glucose transporter type 1 (SLC2A1) translation in an m6A-dependent manner, which subsequently promoted glucose uptake and lactate production, leading to the activation of mTORC1 signaling and CRC development. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| In-vivo Model | To generate the Mettl3+/- mice, 2 specific single-guide RNAs (sgRNAs) targeting exon 2 of Mettl3 were used, which resulted in a frameshift mutation and generated a premature stop codon. | |||
Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 stabilizes HK2 and Glucose transporter type 1 (SLC2A1) (GLUT1) expression in colorectal cancer through an m6A-IGF2BP2/3- dependent mechanism. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glucose metabolism | |||
Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 stabilizes HK2 and Glucose transporter type 1 (SLC2A1) (GLUT1) expression in colorectal cancer through an m6A-IGF2BP2/3- dependent mechanism. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glucose metabolism | |||
Colorectal cancer [ICD-11: 2B91]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 stabilizes HK2 and Glucose transporter type 1 (SLC2A1) (GLUT1) expression in colorectal cancer through an m6A-IGF2BP2/3- dependent mechanism. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glucose metabolism | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 stabilizes HK2 and Glucose transporter type 1 (SLC2A1) (GLUT1) expression in colorectal cancer through an m6A-IGF2BP2/3- dependent mechanism. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glucose metabolism | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 induced Glucose transporter type 1 (SLC2A1) translation in an m6A-dependent manner, which subsequently promoted glucose uptake and lactate production, leading to the activation of mTORC1 signaling and CRC development. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| In-vivo Model | To generate the Mettl3+/- mice, 2 specific single-guide RNAs (sgRNAs) targeting exon 2 of Mettl3 were used, which resulted in a frameshift mutation and generated a premature stop codon. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
RNA modification
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00489 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | hsa-mir-22 | |
| Crosstalk relationship | m6Am → m6A | |
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00490 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | hsa-mir-22 | |
| Crosstalk relationship | m6Am → m6A | |
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00491 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | hsa-mir-22 | |
| Crosstalk relationship | m6Am → m6A | |
m6A Regulator: Leucine-rich PPR motif-containing protein, mitochondrial (LRPPRC)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00492 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | hsa-mir-22 | |
| Crosstalk relationship | m6Am → m6A | |
Histone modification
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03417 | ||
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT03428 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 4 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03549 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03571 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03594 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03616 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
Non-coding RNA
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05045 | ||
| Epigenetic Regulator | MicroRNA 145 (MIR145) | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT05317 | ||
| Epigenetic Regulator | FTO intronic transcript 1 (FTO-IT1) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05159 | ||
| Epigenetic Regulator | MIR4458 host gene (MIR4458HG) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Regulator: Fat mass and obesity-associated protein (FTO)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05315 | ||
| Epigenetic Regulator | FTO intronic transcript 1 (FTO-IT1) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05341 | ||
| Epigenetic Regulator | Circ_FOXK2 | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 3 (IGF2BP3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Oral squamous cell carcinoma | |
m6A Regulator: YTH domain-containing protein 1 (YTHDC1)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05901 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 294 (LINC00294) | |
| Regulated Target | YTH N6-methyladenosine RNA binding protein C1 (YTHDC1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00269)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: AC4SITE000217 | Click to Show/Hide the Full List | ||
| mod site | chr1:42929735-42929736:- | [9] | |
| Sequence | CTGACGTGACCCATGACCTGCAGGAGATGAAGGAAGAGAGT | ||
| Cell/Tissue List | H1 | ||
| Seq Type List | ac4C-seq | ||
| Transcript ID List | ENST00000630287.2; ENST00000426263.8; ENST00000475162.3 | ||
| External Link | RMBase: ac4C_site_75 | ||
5-methylcytidine (m5C)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE001838 | Click to Show/Hide the Full List | ||
| mod site | chr1:42931121-42931122:- | [10] | |
| Sequence | CGCTCACCACGCTCTGGTCCCTCTCAGTGGCCATCTTTTCT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000415851.6; ENST00000372500.4; ENST00000625233.2; ENST00000475162.3; ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: m5C_site_2323 | ||
N6-methyladenosine (m6A)
| In total 53 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE021309 | Click to Show/Hide the Full List | ||
| mod site | chr1:42925688-42925689:- | [11] | |
| Sequence | TCCCCACACACACAAAATGAACCACGTTCTTTGTATGGGCC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26292 | ||
| mod ID: M6ASITE021315 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926155-42926156:- | [12] | |
| Sequence | TAAATGGCTGGTTTTTAGAAACATGGTTTTGAAATGCTTGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000630287.2; ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26293 | ||
| mod ID: M6ASITE021326 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926192-42926193:- | [13] | |
| Sequence | TACACCACCTCACTCCTGTTACTTACCTAAACAGATATAAA | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000630287.2; ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26294 | ||
| mod ID: M6ASITE021337 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926229-42926230:- | [13] | |
| Sequence | CTAAGTTATAGTATATCTGGACAAGCCAACTTGTAAATACA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26295 | ||
| mod ID: M6ASITE021348 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926252-42926253:- | [13] | |
| Sequence | GTTGTCAATATTAAATACAGACACTAAGTTATAGTATATCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000630287.2; ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26296 | ||
| mod ID: M6ASITE021359 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926256-42926257:- | [13] | |
| Sequence | TTTGGTTGTCAATATTAAATACAGACACTAAGTTATAGTAT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26297 | ||
| mod ID: M6ASITE021370 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926320-42926321:- | [12] | |
| Sequence | CTGCTCAAGAAGACATGGAGACTCCTGCCCTGTTGTGTATA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000630287.2; ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26298 | ||
| mod ID: M6ASITE021378 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926328-42926329:- | [12] | |
| Sequence | CAAACTCACTGCTCAAGAAGACATGGAGACTCCTGCCCTGT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000630287.2; ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26299 | ||
| mod ID: M6ASITE021379 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926345-42926346:- | [12] | |
| Sequence | GGGCCTGTGGGAGCCTGCAAACTCACTGCTCAAGAAGACAT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26300 | ||
| mod ID: M6ASITE021380 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926377-42926378:- | [13] | |
| Sequence | GCACTGAGGGCCACACTATTACCATGAGAAGAGGGCCTGTG | ||
| Motif Score | 2.052208333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26301 | ||
| mod ID: M6ASITE021381 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926383-42926384:- | [13] | |
| Sequence | TCCTTTGCACTGAGGGCCACACTATTACCATGAGAAGAGGG | ||
| Motif Score | 2.506922619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26302 | ||
| mod ID: M6ASITE021382 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926385-42926386:- | [14] | |
| Sequence | TCTCCTTTGCACTGAGGGCCACACTATTACCATGAGAAGAG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000630287.2; ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26303 | ||
| mod ID: M6ASITE021383 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926471-42926472:- | [12] | |
| Sequence | TTAATCTTTCTTTACCTGAGACCAGTTGGGAGCACTGGAGT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; fibroblasts; GM12878; LCLs; H1299; Huh7; iSLK; MSC; HEC-1-A; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000630287.2; ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26304 | ||
| mod ID: M6ASITE021384 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926555-42926556:- | [13] | |
| Sequence | CTGCTGGCCTGGATCTCCCCACTCTAGGGGTCAGGCTCCAT | ||
| Motif Score | 2.475107143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000630287.2; ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26305 | ||
| mod ID: M6ASITE021385 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926618-42926619:- | [12] | |
| Sequence | ACACACTAATCGAACTATGAACTACAAAGCTTCTATCCCAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26306 | ||
| mod ID: M6ASITE021386 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926625-42926626:- | [12] | |
| Sequence | CCTAAGGACACACTAATCGAACTATGAACTACAAAGCTTCT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26307 | ||
| mod ID: M6ASITE021387 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926634-42926635:- | [13] | |
| Sequence | GACCTATGTCCTAAGGACACACTAATCGAACTATGAACTAC | ||
| Motif Score | 2.506922619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000630287.2; ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26308 | ||
| mod ID: M6ASITE021388 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926638-42926639:- | [12] | |
| Sequence | GCTGGACCTATGTCCTAAGGACACACTAATCGAACTATGAA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; hESC-HEK293T; HepG2; U2OS; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26309 | ||
| mod ID: M6ASITE021389 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926653-42926654:- | [12] | |
| Sequence | CAGCTAATCTGTAGGGCTGGACCTATGTCCTAAGGACACAC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26310 | ||
| mod ID: M6ASITE021390 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926691-42926692:- | [12] | |
| Sequence | GGAGACTAAGCCCTGTCGAGACACTTGCCTTCTTCACCCAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; hESC-HEK293T; HepG2; U2OS; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000630287.2; ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26311 | ||
| mod ID: M6ASITE021391 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926707-42926708:- | [12] | |
| Sequence | TTCCATGCCTGAGGGTGGAGACTAAGCCCTGTCGAGACACT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26312 | ||
| mod ID: M6ASITE021392 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926752-42926753:- | [14] | |
| Sequence | AGCCTGAGTCTCCTGTGCCCACATCCCAGGCTTCACCCTGA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000630287.2; ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26313 | ||
| mod ID: M6ASITE021393 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926795-42926796:- | [13] | |
| Sequence | GGTTTTATAATTTTTTTATTACTGATTTTGTTATTTTTATA | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000475162.3; ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26314 | ||
| mod ID: M6ASITE021394 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926825-42926826:- | [12] | |
| Sequence | TGTTGCTCAAATCTATTCAGACAAGCAACAGGTTTTATAAT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; Huh7; Jurkat; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq | ||
| Transcript ID List | ENST00000630287.2; ENST00000426263.8; ENST00000475162.3 | ||
| External Link | RMBase: m6A_site_26315 | ||
| mod ID: M6ASITE021395 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926847-42926848:- | [12] | |
| Sequence | AGGATTTTAACAAAAGCAAGACTGTTGCTCAAATCTATTCA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000426263.8; ENST00000475162.3; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26316 | ||
| mod ID: M6ASITE021396 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926858-42926859:- | [13] | |
| Sequence | TTAACGGCTCCAGGATTTTAACAAAAGCAAGACTGTTGCTC | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000475162.3; ENST00000630287.2; ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26317 | ||
| mod ID: M6ASITE021397 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926875-42926876:- | [13] | |
| Sequence | AGAAGAATATTCAGGACTTAACGGCTCCAGGATTTTAACAA | ||
| Motif Score | 2.142029762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000475162.3; ENST00000630287.2; ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26318 | ||
| mod ID: M6ASITE021398 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926880-42926881:- | [12] | |
| Sequence | TGTCCAGAAGAATATTCAGGACTTAACGGCTCCAGGATTTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000630287.2; ENST00000475162.3; ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26319 | ||
| mod ID: M6ASITE021399 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926956-42926957:- | [12] | |
| Sequence | CAGCTGGATGAGACTTCCAAACCTGACAGATGTCAGCCGAG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000475162.3; ENST00000630287.2; ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26320 | ||
| mod ID: M6ASITE021400 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926964-42926965:- | [12] | |
| Sequence | AGCACAGGCAGCTGGATGAGACTTCCAAACCTGACAGATGT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000630287.2; ENST00000426263.8; ENST00000475162.3 | ||
| External Link | RMBase: m6A_site_26321 | ||
| mod ID: M6ASITE021401 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926981-42926982:- | [15] | |
| Sequence | CCTAAGGATCTCTCAGGAGCACAGGCAGCTGGATGAGACTT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | brain | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000475162.3; ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26322 | ||
| mod ID: M6ASITE021402 | Click to Show/Hide the Full List | ||
| mod site | chr1:42927026-42927027:- | [13] | |
| Sequence | AGTGTGAGTCGCCCCAGATCACCAGCCCGGCCTGCTCCCAG | ||
| Motif Score | 2.026654762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000475162.3; ENST00000630287.2; ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26323 | ||
| mod ID: M6ASITE021403 | Click to Show/Hide the Full List | ||
| mod site | chr1:42927087-42927088:- | [12] | |
| Sequence | GAGCCAGCCAAAGTGACAAGACACCCGAGGAGCTGTTCCAT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; U2OS; fibroblasts; A549; Huh7; GSC-11; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000475162.3; ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26324 | ||
| mod ID: M6ASITE021404 | Click to Show/Hide the Full List | ||
| mod site | chr1:42927092-42927093:- | [13] | |
| Sequence | GGGGGGAGCCAGCCAAAGTGACAAGACACCCGAGGAGCTGT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000475162.3; ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26325 | ||
| mod ID: M6ASITE021405 | Click to Show/Hide the Full List | ||
| mod site | chr1:42927144-42927145:- | [12] | |
| Sequence | TTCCTGAGACTAAAGGCCGGACCTTCGATGAGATCGCTTCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; Huh7; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000475162.3; ENST00000630287.2; ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26326 | ||
| mod ID: M6ASITE021412 | Click to Show/Hide the Full List | ||
| mod site | chr1:42927156-42927157:- | [12] | |
| Sequence | CCTACTTCAAAGTTCCTGAGACTAAAGGCCGGACCTTCGAT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; Huh7; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000426263.8; ENST00000475162.3; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26327 | ||
| mod ID: M6ASITE021423 | Click to Show/Hide the Full List | ||
| mod site | chr1:42927645-42927646:- | [11] | |
| Sequence | TTGCAGGCTTCTCCAACTGGACCTCAAATTTCATTGTGGGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000475162.3; ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26328 | ||
| mod ID: M6ASITE021434 | Click to Show/Hide the Full List | ||
| mod site | chr1:42927703-42927704:- | [11] | |
| Sequence | CCATGGTTCATCGTGGCTGAACTCTTCAGCCAGGGTCCACG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000426263.8; ENST00000475162.3; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26329 | ||
| mod ID: M6ASITE021445 | Click to Show/Hide the Full List | ||
| mod site | chr1:42929002-42929003:- | [16] | |
| Sequence | TGGAGCGAGCAGGCCGGCGGACCCTGCACCTCATAGGCCTC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2; HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000630287.2; ENST00000426263.8; ENST00000475162.3 | ||
| External Link | RMBase: m6A_site_26330 | ||
| mod ID: M6ASITE021456 | Click to Show/Hide the Full List | ||
| mod site | chr1:42929759-42929760:- | [12] | |
| Sequence | TGCTAAAGAAGCTGCGCGGGACAGCTGACGTGACCCATGAC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; MM6; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000630287.2; ENST00000475162.3; ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26331 | ||
| mod ID: M6ASITE021467 | Click to Show/Hide the Full List | ||
| mod site | chr1:42929886-42929887:- | [12] | |
| Sequence | CATCAACCGCAACGAGGAGAACCGGGCCAAGAGTGGTACGG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000426263.8; ENST00000630287.2; ENST00000475162.3 | ||
| External Link | RMBase: m6A_site_26332 | ||
| mod ID: M6ASITE021477 | Click to Show/Hide the Full List | ||
| mod site | chr1:42930000-42930001:- | [12] | |
| Sequence | CTCCATCATGGGCAACAAGGACCTGTGGCCCCTGCTGCTGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000475162.3; ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26333 | ||
| mod ID: M6ASITE021478 | Click to Show/Hide the Full List | ||
| mod site | chr1:42930021-42930022:- | [12] | |
| Sequence | CCCCCAGGTGTTCGGCCTGGACTCCATCATGGGCAACAAGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000426263.8; ENST00000475162.3; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26334 | ||
| mod ID: M6ASITE021479 | Click to Show/Hide the Full List | ||
| mod site | chr1:42930799-42930800:- | [12] | |
| Sequence | GTGCTCATGGGCTTCTCGAAACTGGGCAAGTCCTTTGAGAT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000372500.4; ENST00000475162.3; ENST00000426263.8; ENST00000625233.2; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26335 | ||
| mod ID: M6ASITE021480 | Click to Show/Hide the Full List | ||
| mod site | chr1:42930842-42930843:- | [12] | |
| Sequence | GAATTCAATGCTGATGATGAACCTGCTGGCCTTCGTGTCCG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000426263.8; ENST00000475162.3; ENST00000372500.4; ENST00000630287.2; ENST00000625233.2 | ||
| External Link | RMBase: m6A_site_26336 | ||
| mod ID: M6ASITE021481 | Click to Show/Hide the Full List | ||
| mod site | chr1:42931181-42931182:- | [12] | |
| Sequence | TCGAGGAGTTCTACAACCAGACATGGGTCCACCGCTATGGG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; A549; Jurkat; CD4T; peripheral-blood; GSC-11; endometrial; HEC-1-A; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000415851.6; ENST00000426263.8; ENST00000475162.3; ENST00000372500.4; ENST00000625233.2; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26337 | ||
| mod ID: M6ASITE021482 | Click to Show/Hide the Full List | ||
| mod site | chr1:42931189-42931190:- | [14] | |
| Sequence | GCAGGTGATCGAGGAGTTCTACAACCAGACATGGGTCCACC | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000475162.3; ENST00000426263.8; ENST00000372500.4; ENST00000625233.2; ENST00000630287.2; ENST00000415851.6 | ||
| External Link | RMBase: m6A_site_26338 | ||
| mod ID: M6ASITE021483 | Click to Show/Hide the Full List | ||
| mod site | chr1:42943064-42943065:- | [12] | |
| Sequence | TGGCCCTGATCATGTCAGAGACTCTTCTCTTGCTTTGGAAT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; A549; Jurkat; peripheral-blood; GSC-11; iSLK; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000475162.3; ENST00000630287.2; ENST00000630821.1; ENST00000426263.8; ENST00000372500.4; ENST00000625233.2; ENST00000415851.6 | ||
| External Link | RMBase: m6A_site_26339 | ||
| mod ID: M6ASITE021484 | Click to Show/Hide the Full List | ||
| mod site | chr1:42943195-42943196:- | [12] | |
| Sequence | GTGCCATCGTTGGCTTATGGACACCAGCCTGCTCTGTTGCA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; A549; Jurkat; CD4T; GSC-11; iSLK; endometrial; HEC-1-A; GSCs; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000475162.3; ENST00000372500.4; ENST00000630287.2; ENST00000415851.6; ENST00000630821.1; ENST00000426263.8; ENST00000625233.2 | ||
| External Link | RMBase: m6A_site_26340 | ||
| mod ID: M6ASITE021485 | Click to Show/Hide the Full List | ||
| mod site | chr1:42943253-42943254:- | [14] | |
| Sequence | CTCCCTGCAGTTTGGCTACAACACTGGAGTCATCAATGCCC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000630287.2; ENST00000426263.8; ENST00000372500.4; ENST00000415851.6; ENST00000625233.2; ENST00000630821.1 | ||
| External Link | RMBase: m6A_site_26341 | ||
| mod ID: M6ASITE021486 | Click to Show/Hide the Full List | ||
| mod site | chr1:42958743-42958744:- | [12] | |
| Sequence | GTCGCAGTGGGAGTCCCCGGACCGGAGCACGAGCCTGAGCG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; A549; U2OS; H1A; H1B; hESCs; fibroblasts; LCLs; MT4; H1299; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000625233.2; ENST00000372500.4; ENST00000630821.1; ENST00000630287.2; ENST00000426263.8; ENST00000460369.3; ENST00000415851.6 | ||
| External Link | RMBase: m6A_site_26342 | ||
| mod ID: M6ASITE021487 | Click to Show/Hide the Full List | ||
| mod site | chr1:42958789-42958790:- | [12] | |
| Sequence | AGCACGCCAGGGAGCAGGAGACCAAACGACGGGGGTCGGAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; A549; U2OS; H1A; H1B; hESCs; HEK293T; fibroblasts; LCLs; MT4; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000460369.3; ENST00000625233.2; ENST00000415851.6; ENST00000630821.1; ENST00000372500.4; ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: m6A_site_26343 | ||
| mod ID: M6ASITE021488 | Click to Show/Hide the Full List | ||
| mod site | chr1:42958981-42958982:- | [12] | |
| Sequence | CCCGCAGAGCGCCGCCCAGGACAGGCTGGGCCCCAGGCCCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000426263.8 | ||
| External Link | RMBase: m6A_site_26344 | ||
Pseudouridine (Pseudo)
| In total 4 m6A sequence/site(s) in this target gene | |||
| mod ID: PSESITE000036 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926243-42926244:- | [17] | |
| Sequence | ATTAAATACAGACACTAAGTTATAGTATATCTGGACAAGCC | ||
| Transcript ID List | ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: Pseudo_site_141 | ||
| mod ID: PSESITE000039 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926726-42926727:- | [18] | |
| Sequence | CAGGCTTCACCCTGAATGGTTCCATGCCTGAGGGTGGAGAC | ||
| Transcript ID List | ENST00000630287.2; ENST00000426263.8 | ||
| External Link | RMBase: Pseudo_site_142 | ||
| mod ID: PSESITE000041 | Click to Show/Hide the Full List | ||
| mod site | chr1:42926727-42926728:- | [18] | |
| Sequence | CCAGGCTTCACCCTGAATGGTTCCATGCCTGAGGGTGGAGA | ||
| Transcript ID List | ENST00000426263.8; ENST00000630287.2 | ||
| External Link | RMBase: Pseudo_site_143 | ||
| mod ID: PSESITE000044 | Click to Show/Hide the Full List | ||
| mod site | chr1:42931129-42931130:- | [17] | |
| Sequence | GCCCACCACGCTCACCACGCTCTGGTCCCTCTCAGTGGCCA | ||
| Transcript ID List | ENST00000625233.2; ENST00000426263.8; ENST00000630287.2; ENST00000415851.6; ENST00000475162.3; ENST00000372500.4 | ||
| External Link | RMBase: Pseudo_site_144 | ||
References