m6A-centered Crosstalk Information
Mechanism of Crosstalk between m6A Modification and Epigenetic Regulation
| Crosstalk ID |
M6ACROT03150
|
[1] | |||
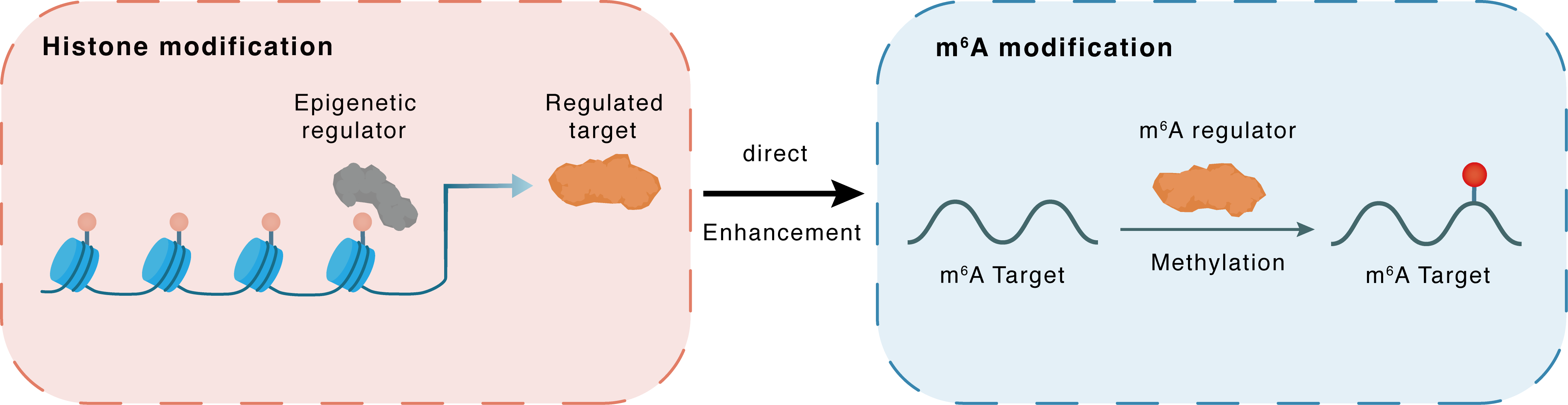 Histone modification
H3K9me2
G9a
METTL3
Direct
Enhancement
m6A modification
CX3CR1
CX3CR1
METTL3
Methylation
Histone modification
H3K9me2
G9a
METTL3
Direct
Enhancement
m6A modification
CX3CR1
CX3CR1
METTL3
Methylation
 : m6A sites : m6A sites
|
|||||
| m6A Modification: | |||||
|---|---|---|---|---|---|
| m6A Regulator | Methyltransferase-like 3 (METTL3) | WRITER | |||
| m6A Target | CX3C chemokine receptor 1 (CX3CR1) | ||||
| Epigenetic Regulation that have Cross-talk with This m6A Modification: | |||||
| Epigenetic Regulation Type | Histone modification (HistMod) | ||||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EHMT2 (EHMT2) | WRITER | View Details | ||
| Regulated Target | Histone H3 lysine 9 dimethylation (H3K9me2) | View Details | |||
| Downstream Gene | METTL3 | View Details | |||
| Crosstalk Relationship | Histone modification → m6A | Enhancement | |||
| Crosstalk Mechanism | histone modification directly impacts m6A modification through modulating the level of m6A regulator | ||||
| Crosstalk Summary | EHMT2 promotes m6A methyltransferase activity of METTL3, which upregulates PD-L1, CX3C chemokine receptor 1 (CX3CR1) and CASP1, at translational/post-translational level by regulating Histone H3 lysine 9 dimethylation (H3K9me2) level during ET. | ||||
| Responsed Disease | Inflammatory response | ICD-11: MG46 | |||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| RAW 264.7 | Mouse leukemia | Mus musculus | CVCL_0493 | ||
Full List of Potential Compound(s) Related to This m6A-centered Crosstalk
| Histone-lysine N-methyltransferase EHMT2 (EHMT2) | 7 Compound(s) Regulating the Target | Click to Show/Hide the Full List | ||
| MS012 | Preclinical | [2] | ||
| Synonyms |
CHEMBL4086403; 2089617-83-2 (free base); N2-hexyl-6,7-dimethoxy-N4-(1-methylpiperidin-4-yl)quinazoline-2,4-diamine; BDBM50501525; N~2~-hexyl-6,7-dimethoxy-N~4~-(1-methylpiperidin-4-yl)quinazoline-2,4-diamine
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| External Link | ||||
| BIX-01294 | Preclinical | [3] | ||
| Synonyms |
BIX01294; BIX 01294
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| Activity | IC50 = 67 nM | |||
| External Link | ||||
| A-366 | Preclinical | [4] | ||
| Synonyms |
A 366; A366
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| Activity | IC50 = 3 nM | |||
| External Link | ||||
| UNC0321 | Investigative | [5] | ||
| Synonyms |
UNC-0321; UNC 0321
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| Activity | IC50 = 9000 nM | |||
| External Link | ||||
| BRD9539 | Investigative | [6] | ||
| Synonyms |
BRD-9539; BRD 9539
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| Activity | IC50 = 1500 nM | |||
| External Link | ||||
| UNC0642 | Investigative | [7] | ||
| Synonyms |
1481677-78-4; UNC 0642; UNC-0642; CHEMBL2441082; 2-(4,4-Difluoro-1-piperidinyl)-6-methoxy-N-[1-(1-methylethyl)-4-piperidinyl]-7-[3-(1-pyrrolidinyl)propoxy]-4-quinazolinamine; Barrett; GTPL7017; SCHEMBL17372593; AOB2595; MolPort-035-765-953; EX-A2241; BCP08266; ZINC96285772; BDBM50442103; AKOS024458509; SB19046; CS-5269; NCGC00189140-01; NCGC00189140-02; AS-16721; HY-13980; BC600721; AK547424; UNC0642, > KB-146019; J-008448
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| Activity | IC50 < 2.5 nM | |||
| External Link | ||||
| UNC0638 | Investigative | [8] | ||
| Synonyms |
1255580-76-7; UNC-0638; UNC 0638; UNII-26A103L2FO; 2-Cyclohexyl-N-(1-isopropylpiperidin-4-yl)-6-methoxy-7-(3-(pyrrolidin-1-yl)propoxy)quinazolin-4-amine; CHEMBL1231795; 26A103L2FO; 2-Cyclohexyl-6-methoxy-N-[1-(1-methylethyl)-4-piperidinyl]-7-[3-(1-pyrrolidinyl)propoxy]-4-quinazolinamine; 1255517-77-1; 2-cyclohexyl-6-methoxy-N-[1-(propan-2-yl)piperidin-4-yl]-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-4-amine; 2-Cyclohexyl-N-(1-isopropyl-4-piperidinyl)-6-methoxy-7-[3-(1-pyrrolidinyl)propoxy]-4-quinazolinamine
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| Activity | Ki = 3.7 nM | |||
| External Link | ||||
References