m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00401)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
SOCS2
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
YTH domain-containing family protein 2 (YTHDF2) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Human umbilical cord blood CD34+ cells | Homo sapiens |
|
Treatment: YTHDF2 knockdown UCB CD34+ cells
Control: Wild type UCB CD34+ cells
|
GSE107956 | |
| Regulation |
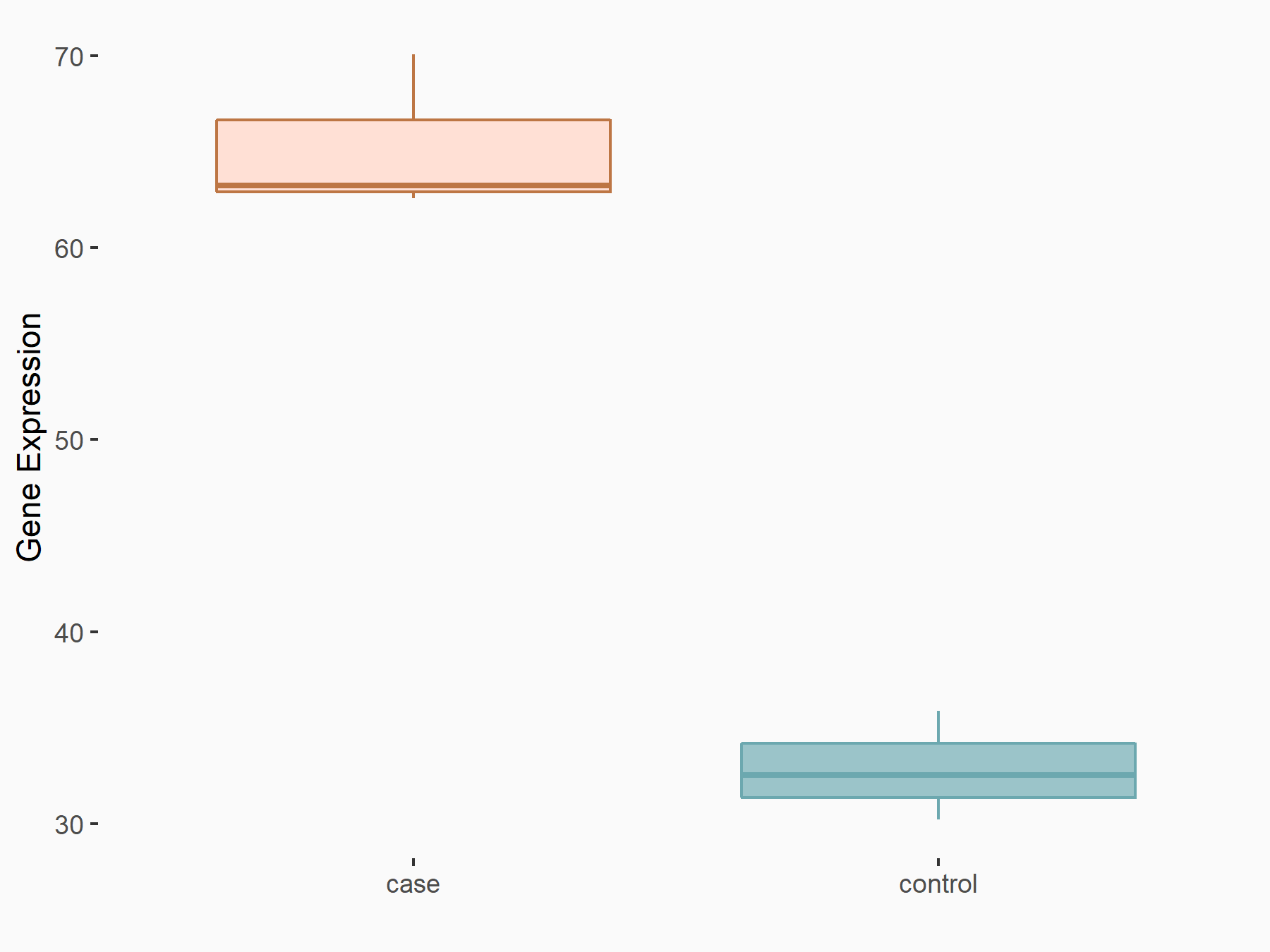  |
logFC: 9.71E-01 p-value: 1.71E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between SOCS2 and the regulator | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.81E+00 | GSE49339 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 is frequently up-regulated in human HCC and contributes to HCC progression. METTL3 represses Suppressor of cytokine signaling 2 (SOCS2) expression in HCC through an m6A-YTHDF2-dependent mechanism. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Liver cancer | ICD-11: 2C12 | ||
| Cell Process | Cells proliferation | |||
| Cells migration | ||||
| Cells invasion | ||||
| RNA degradation (hsa03018) | ||||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| In-vivo Model | For the subcutaneous implantation model, 2 × 106 METTL3 stable knockdown Huh-7 cells or METTL3 overexpression MHCC97L cells were injected subcutaneously into BABL/cAnN-nude mice. For orthotopic implantation, wild-type and METTL3 knockout Huh-7 cells were luciferase labelled, and 2 × 106 cells were then injected orthotopically into the left liver lobe of nude mice. | |||
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | ARPE-19 cell line | Homo sapiens |
|
Treatment: shMETTL3 ARPE-19 cells
Control: shControl ARPE-19 cells
|
GSE202017 | |
| Regulation |
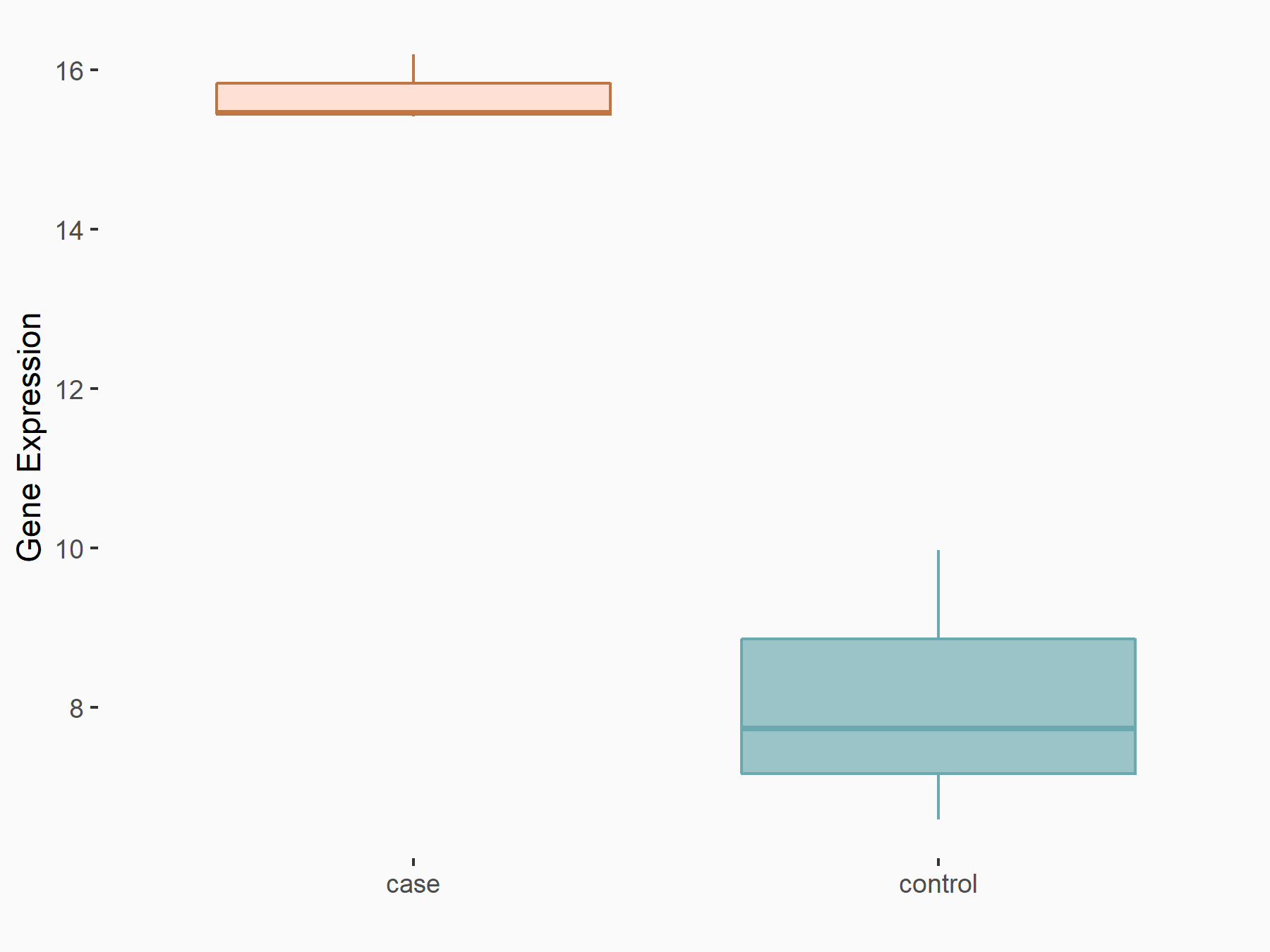  |
logFC: 8.91E-01 p-value: 2.94E-04 |
| More Results | Click to View More RNA-seq Results | |
| In total 5 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | MiR-302a was identified to target METTL3, which could inhibit Suppressor of cytokine signaling 2 (SOCS2) expression via m6A modification in glioma. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Glioma | ICD-11: 2A00.0 | ||
| Pathway Response | Transcriptional misregulation in cancer | hsa05202 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 |
| LN-18 | Glioblastoma | Homo sapiens | CVCL_0392 | |
| HEB (human normal glial cell line HEB were obtained from Tongpai (Shanghai) biotechnology co., LTD (Shanghai, China)) | ||||
| In-vivo Model | Mice were subcutaneously injected with 2.5 × 106 U251 cells stably infected with OE-NC, OE-JMJD1C, OE-NC and sh-NC, OE-JMJD1C and sh-NC or OE-JMJD1C and sh-SOCS2 (n = 10 in each group) in 0.1 ml PBS. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | METTL3-KO in gastric cancer cells resulted in the suppression of cell proliferation by inducing Suppressor of cytokine signaling 2 (SOCS2), suggesting a potential role of elevated METTL3 expression in gastric cancer progression. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| In-vitro Model | AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | An increased level of METTL3 may maintain the tumorigenicity of colon cancer cells by suppressing Suppressor of cytokine signaling 2 (SOCS2). | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Colon cancer | ICD-11: 2B90 | ||
| Cell Process | Cell proliferation | |||
| In-vitro Model | SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 |
| Experiment 4 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 is frequently up-regulated in human HCC and contributes to HCC progression. METTL3 represses Suppressor of cytokine signaling 2 (SOCS2) expression in HCC through an m6A-YTHDF2-dependent mechanism. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Liver cancer | ICD-11: 2C12 | ||
| Cell Process | Cells proliferation | |||
| Cells migration | ||||
| Cells invasion | ||||
| RNA degradation (hsa03018) | ||||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| In-vivo Model | For the subcutaneous implantation model, 2 × 106 METTL3 stable knockdown Huh-7 cells or METTL3 overexpression MHCC97L cells were injected subcutaneously into BABL/cAnN-nude mice. For orthotopic implantation, wild-type and METTL3 knockout Huh-7 cells were luciferase labelled, and 2 × 106 cells were then injected orthotopically into the left liver lobe of nude mice. | |||
| Experiment 5 Reporting the m6A Methylation Regulator of This Target Gene | [5] | |||
| Response Summary | METTL3 knock-down experiment revealed that expressions of SOCS family members SOCS1, Suppressor of cytokine signaling 2 (SOCS2), SOCS4, SOCS5, and SOCS6 were increased after METTL3 knock-down. It indicated that METTL3 is involved in the development of Graves' disease (GD) by inducing mRNA m6A methylation modification of SOCS family members. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Graves disease | ICD-11: 5A02.0 | ||
| In-vitro Model | PBMCs (Human peripheral blood mononuclear cells (PBMCs) are isolated from peripheral blood and identified as any blood cell with a round nucleus) | |||
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | MiR-302a was identified to target METTL3, which could inhibit Suppressor of cytokine signaling 2 (SOCS2) expression via m6A modification in glioma. | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Transcriptional misregulation in cancer | hsa05202 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 |
| LN-18 | Glioblastoma | Homo sapiens | CVCL_0392 | |
| HEB (human normal glial cell line HEB were obtained from Tongpai (Shanghai) biotechnology co., LTD (Shanghai, China)) | ||||
| In-vivo Model | Mice were subcutaneously injected with 2.5 × 106 U251 cells stably infected with OE-NC, OE-JMJD1C, OE-NC and sh-NC, OE-JMJD1C and sh-NC or OE-JMJD1C and sh-SOCS2 (n = 10 in each group) in 0.1 ml PBS. | |||
Solid tumour/cancer [ICD-11: 2A00-2F9Z]
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | METTL3-KO in gastric cancer cells resulted in the suppression of cell proliferation by inducing Suppressor of cytokine signaling 2 (SOCS2), suggesting a potential role of elevated METTL3 expression in gastric cancer progression. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
Colon cancer [ICD-11: 2B90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | An increased level of METTL3 may maintain the tumorigenicity of colon cancer cells by suppressing Suppressor of cytokine signaling 2 (SOCS2). | |||
| Responsed Disease | Colon cancer [ICD-11: 2B90] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Cell Process | Cell proliferation | |||
| In-vitro Model | SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 |
Liver cancer [ICD-11: 2C12]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 is frequently up-regulated in human HCC and contributes to HCC progression. METTL3 represses Suppressor of cytokine signaling 2 (SOCS2) expression in HCC through an m6A-YTHDF2-dependent mechanism. | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Cell Process | Cells proliferation | |||
| Cells migration | ||||
| Cells invasion | ||||
| RNA degradation (hsa03018) | ||||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| In-vivo Model | For the subcutaneous implantation model, 2 × 106 METTL3 stable knockdown Huh-7 cells or METTL3 overexpression MHCC97L cells were injected subcutaneously into BABL/cAnN-nude mice. For orthotopic implantation, wild-type and METTL3 knockout Huh-7 cells were luciferase labelled, and 2 × 106 cells were then injected orthotopically into the left liver lobe of nude mice. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 is frequently up-regulated in human HCC and contributes to HCC progression. METTL3 represses Suppressor of cytokine signaling 2 (SOCS2) expression in HCC through an m6A-YTHDF2-dependent mechanism. | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Down regulation | |||
| Cell Process | Cells proliferation | |||
| Cells migration | ||||
| Cells invasion | ||||
| RNA degradation (hsa03018) | ||||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| In-vivo Model | For the subcutaneous implantation model, 2 × 106 METTL3 stable knockdown Huh-7 cells or METTL3 overexpression MHCC97L cells were injected subcutaneously into BABL/cAnN-nude mice. For orthotopic implantation, wild-type and METTL3 knockout Huh-7 cells were luciferase labelled, and 2 × 106 cells were then injected orthotopically into the left liver lobe of nude mice. | |||
Thyrotoxicosis [ICD-11: 5A02]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [5] | |||
| Response Summary | METTL3 knock-down experiment revealed that expressions of SOCS family members SOCS1, Suppressor of cytokine signaling 2 (SOCS2), SOCS4, SOCS5, and SOCS6 were increased after METTL3 knock-down. It indicated that METTL3 is involved in the development of Graves' disease (GD) by inducing mRNA m6A methylation modification of SOCS family members. | |||
| Responsed Disease | Graves disease [ICD-11: 5A02.0] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | PBMCs (Human peripheral blood mononuclear cells (PBMCs) are isolated from peripheral blood and identified as any blood cell with a round nucleus) | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
RNA modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00070 | ||
| Epigenetic Regulator | tRNA (cytosine(72)-C(5))-methyltransferase NSUN6 (NSUN6) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m5C → m6A | |
| Disease | Colon cancer | |
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03072 | ||
| Epigenetic Regulator | Probable JmjC domain-containing histone demethylation protein 2C (JMJD1C) | |
| Regulated Target | Histone H3 lysine 9 monomethylation (H3K9me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
| Crosstalk ID: M6ACROT03659 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
Non-coding RNA
m6A Regulator: Wilms tumor 1-associating protein (WTAP)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05155 | ||
| Epigenetic Regulator | AC115619-22aa | |
| Regulated Target | Pre-mRNA-splicing regulator WTAP (WTAP) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05360 | ||
| Epigenetic Regulator | hsa-miR-302a | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Brain cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00401)
| In total 49 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE014545 | Click to Show/Hide the Full List | ||
| mod site | chr12:93570141-93570142:+ | [8] | |
| Sequence | CCCTGCAGGGAATGGCAGAGACACTCTCCGGACTGAGGGAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; endometrial; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000340600.6 | ||
| External Link | RMBase: m6A_site_202237 | ||
| mod ID: M6ASITE014546 | Click to Show/Hide the Full List | ||
| mod site | chr12:93570152-93570153:+ | [8] | |
| Sequence | ATGGCAGAGACACTCTCCGGACTGAGGGAACCGAGGCCAGT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; endometrial; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000340600.6 | ||
| External Link | RMBase: m6A_site_202238 | ||
| mod ID: M6ASITE014547 | Click to Show/Hide the Full List | ||
| mod site | chr12:93570161-93570162:+ | [8] | |
| Sequence | ACACTCTCCGGACTGAGGGAACCGAGGCCAGTCACCAAGCC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; endometrial; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000340600.6 | ||
| External Link | RMBase: m6A_site_202239 | ||
| mod ID: M6ASITE014548 | Click to Show/Hide the Full List | ||
| mod site | chr12:93570412-93570413:+ | [8] | |
| Sequence | TCCAGGACCTGATCAAGGGGACCGCCTCCGGTCCCCGGCCG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000549206.5; ENST00000340600.6 | ||
| External Link | RMBase: m6A_site_202240 | ||
| mod ID: M6ASITE014549 | Click to Show/Hide the Full List | ||
| mod site | chr12:93570642-93570643:+ | [8] | |
| Sequence | GGATCGCTATCCTTCCCTGAACCCGGGCGCTCAGCTGGCCC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000340600.6; ENST00000549206.5 | ||
| External Link | RMBase: m6A_site_202241 | ||
| mod ID: M6ASITE014550 | Click to Show/Hide the Full List | ||
| mod site | chr12:93571029-93571030:+ | [8] | |
| Sequence | GCGCACTCGCTGCTCCTGGGACCGACGTTTAACTCTTGCCA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000536696.6; ENST00000549206.5; ENST00000340600.6 | ||
| External Link | RMBase: m6A_site_202242 | ||
| mod ID: M6ASITE014551 | Click to Show/Hide the Full List | ||
| mod site | chr12:93571144-93571145:+ | [8] | |
| Sequence | CCTCACCGCGTGGCCCGCGGACAGTGCGCGCCGGGGTCCCG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; AML | ||
| Seq Type List | m6A-seq; miCLIP | ||
| Transcript ID List | ENST00000549206.5; ENST00000340600.6; ENST00000536696.6 | ||
| External Link | RMBase: m6A_site_202243 | ||
| mod ID: M6ASITE014552 | Click to Show/Hide the Full List | ||
| mod site | chr12:93571357-93571358:+ | [8] | |
| Sequence | CCGGGGAAGAGAGAGTCCGAACCGCGGCTCTGCCCCGCGGC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000536696.6; ENST00000549206.5; ENST00000340600.6; ENST00000548091.5 | ||
| External Link | RMBase: m6A_site_202245 | ||
| mod ID: M6ASITE014553 | Click to Show/Hide the Full List | ||
| mod site | chr12:93571457-93571458:+ | [8] | |
| Sequence | CTTTTTTTTATTTCCCCTAAACTGCCATTCAAATTAATAAT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000536696.6; ENST00000548091.5; ENST00000549206.5; ENST00000340600.6; ENST00000549122.5 | ||
| External Link | RMBase: m6A_site_202246 | ||
| mod ID: M6ASITE014554 | Click to Show/Hide the Full List | ||
| mod site | chr12:93571590-93571591:+ | [8] | |
| Sequence | AGGGTCGCAGCCGGAGGGAAACCCGGCAGCAGTCCGAGAGT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000549122.5; ENST00000548091.5; ENST00000536696.6; ENST00000549206.5; ENST00000340600.6 | ||
| External Link | RMBase: m6A_site_202247 | ||
| mod ID: M6ASITE014555 | Click to Show/Hide the Full List | ||
| mod site | chr12:93571706-93571707:+ | [8] | |
| Sequence | GTGTGCCCTCTGCGCACGGAACTCCGACCGCCGCCTCCGAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000549122.5; ENST00000340600.6; ENST00000536696.6; ENST00000548537.1; ENST00000622746.4; ENST00000548091.5; ENST00000549206.5 | ||
| External Link | RMBase: m6A_site_202250 | ||
| mod ID: M6ASITE014556 | Click to Show/Hide the Full List | ||
| mod site | chr12:93572047-93572048:+ | [8] | |
| Sequence | CAGCGCCCGCGGCGGCGAGAACCACCGCAGCCATCCTGGTC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000548091.5; ENST00000548537.1; ENST00000549206.5; ENST00000549122.5; ENST00000622746.4; ENST00000536696.6; ENST00000340600.6; ENST00000549887.1 | ||
| External Link | RMBase: m6A_site_202251 | ||
| mod ID: M6ASITE014557 | Click to Show/Hide the Full List | ||
| mod site | chr12:93572225-93572226:+ | [8] | |
| Sequence | GCAGCGCTTTAGGGCGGAGAACCAAGTTTGTGTGGGTGCTT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000549887.1; ENST00000548537.1; ENST00000549122.5; ENST00000536696.6; ENST00000549206.5; ENST00000622746.4; ENST00000548091.5; ENST00000340600.6; ENST00000547229.1 | ||
| External Link | RMBase: m6A_site_202252 | ||
| mod ID: M6ASITE014558 | Click to Show/Hide the Full List | ||
| mod site | chr12:93572714-93572715:+ | [8] | |
| Sequence | GGTGACCGCGGCTGCGAGGGACTTTGTCATCCGTCCTCCAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; A549; H1A; hESCs; MM6; GSC-11; HEK293T; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000549206.5; ENST00000549122.5; ENST00000551556.2; ENST00000549887.1; ENST00000547229.1; ENST00000622746.4; ENST00000340600.6; ENST00000536696.6; ENST00000548537.1; ENST00000548091.5 | ||
| External Link | RMBase: m6A_site_202253 | ||
| mod ID: M6ASITE014559 | Click to Show/Hide the Full List | ||
| mod site | chr12:93572784-93572785:+ | [8] | |
| Sequence | TCTCTCTGCCACCATTTCGGACACCCCGCAGGGACTCGTTT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1B; H1A; hESCs; GM12878; MM6; Jurkat; CD4T; GSC-11; HEK293A-TOA; MSC; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000622746.4; ENST00000548537.1; ENST00000549887.1; ENST00000340600.6; ENST00000549122.5; ENST00000551556.2; ENST00000547229.1; ENST00000549206.5; ENST00000536696.6; ENST00000548091.5 | ||
| External Link | RMBase: m6A_site_202254 | ||
| mod ID: M6ASITE014560 | Click to Show/Hide the Full List | ||
| mod site | chr12:93572797-93572798:+ | [8] | |
| Sequence | ATTTCGGACACCCCGCAGGGACTCGTTTTGGGATTCGCACT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1B; H1A; hESCs; GM12878; MM6; Jurkat; CD4T; GSC-11; HEK293A-TOA; MSC; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000548537.1; ENST00000340600.6; ENST00000622746.4; ENST00000549122.5; ENST00000549206.5; ENST00000551556.2; ENST00000536696.6; ENST00000549887.1; ENST00000547229.1; ENST00000548091.5 | ||
| External Link | RMBase: m6A_site_202255 | ||
| mod ID: M6ASITE014561 | Click to Show/Hide the Full List | ||
| mod site | chr12:93572838-93572839:+ | [8] | |
| Sequence | GACTTCAAGGAAGGACGCGAACCCTTCTCTGACCCCAGCTC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; H1B; H1A; hESCs; GM12878; MM6; Jurkat; CD4T; GSC-11; HEK293A-TOA; TREX; MSC; endometrial; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000551556.2; ENST00000548537.1; ENST00000536696.6; ENST00000549122.5; ENST00000548091.5; ENST00000547229.1; ENST00000622746.4; ENST00000549206.5; ENST00000549887.1; ENST00000340600.6 | ||
| External Link | RMBase: m6A_site_202256 | ||
| mod ID: M6ASITE014562 | Click to Show/Hide the Full List | ||
| mod site | chr12:93572960-93572961:+ | [8] | |
| Sequence | GGACGCGGAGCCAGTGGGGGACCGCGGGGTCGGCGGAGGAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; H1A; GM12878; MM6; Jurkat; CD4T; GSC-11; HEK293A-TOA; TREX; endometrial; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000549887.1; ENST00000551556.2; ENST00000548091.5; ENST00000536696.6; ENST00000549122.5; ENST00000549206.5; ENST00000622746.4; ENST00000548537.1; ENST00000340600.6 | ||
| External Link | RMBase: m6A_site_202257 | ||
| mod ID: M6ASITE014563 | Click to Show/Hide the Full List | ||
| mod site | chr12:93573032-93573033:+ | [8] | |
| Sequence | CCCTGCGGGAGCTCGGTCAGACAGGTAGGGAGCCGATCGGC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; H1A; MM6; Jurkat; GSC-11; HEK293A-TOA; TREX; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000551556.2; ENST00000549122.5; ENST00000536696.6; ENST00000549206.5; ENST00000549887.1; ENST00000622746.4; ENST00000340600.6; ENST00000548091.5; ENST00000548537.1 | ||
| External Link | RMBase: m6A_site_202258 | ||
| mod ID: M6ASITE014564 | Click to Show/Hide the Full List | ||
| mod site | chr12:93573158-93573159:+ | [8] | |
| Sequence | ACGTCCCTTGCTTCCAAGGGACCGCGGAGAGCACGCTCGCA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; CD34; HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000548091.5; ENST00000340600.6; ENST00000536696.6; ENST00000549206.5; ENST00000548537.1; ENST00000551556.2; ENST00000622746.4; ENST00000549887.1; ENST00000549122.5 | ||
| External Link | RMBase: m6A_site_202259 | ||
| mod ID: M6ASITE014565 | Click to Show/Hide the Full List | ||
| mod site | chr12:93573485-93573486:+ | [9] | |
| Sequence | CTTGGCGGCCAAGTTCAGGGACTGACACTGCCGCGAGGGCG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000536696.6; ENST00000340600.6; ENST00000622746.4; ENST00000549206.5; ENST00000549887.1; ENST00000548537.1; ENST00000548091.5; ENST00000551556.2; ENST00000549122.5 | ||
| External Link | RMBase: m6A_site_202260 | ||
| mod ID: M6ASITE014566 | Click to Show/Hide the Full List | ||
| mod site | chr12:93574040-93574041:+ | [8] | |
| Sequence | GAGTTTAGGGAAGATTGTAAACTTGAAGTAGTAATTATCAC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000340600.6; ENST00000551556.2; ENST00000549887.1; ENST00000548091.5; ENST00000536696.6; ENST00000549122.5; ENST00000622746.4; ENST00000548537.1; ENST00000549206.5 | ||
| External Link | RMBase: m6A_site_202261 | ||
| mod ID: M6ASITE014567 | Click to Show/Hide the Full List | ||
| mod site | chr12:93574708-93574709:+ | [8] | |
| Sequence | TCTTTTCTTTTTCTTTTTGAACAAAAACCCCAGGATGGTAC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; CD34; HEK293T; A549; hESCs; fibroblasts; GM12878; MM6; Huh7; MSC; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000536696.6; ENST00000622746.4; ENST00000548091.5; ENST00000549887.1; ENST00000549122.5; ENST00000551556.2; ENST00000549206.5; ENST00000340600.6; ENST00000548537.1 | ||
| External Link | RMBase: m6A_site_202262 | ||
| mod ID: M6ASITE014568 | Click to Show/Hide the Full List | ||
| mod site | chr12:93574714-93574715:+ | [8] | |
| Sequence | CTTTTTCTTTTTGAACAAAAACCCCAGGATGGTACTGGGGA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; CD34; HEK293T; A549; hESCs; fibroblasts; GM12878; MM6; Huh7; MSC; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000548537.1; ENST00000536696.6; ENST00000340600.6; ENST00000549122.5; ENST00000548091.5; ENST00000549206.5; ENST00000549887.1; ENST00000551556.2; ENST00000622746.4 | ||
| External Link | RMBase: m6A_site_202263 | ||
| mod ID: M6ASITE014569 | Click to Show/Hide the Full List | ||
| mod site | chr12:93574741-93574742:+ | [10] | |
| Sequence | GATGGTACTGGGGAAGTATGACTGTTAATGAAGCCAAAGAG | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000549887.1; ENST00000548537.1; ENST00000549122.5; ENST00000548091.5; ENST00000549206.5; ENST00000536696.6; ENST00000622746.4; ENST00000551556.2; ENST00000340600.6 | ||
| External Link | RMBase: m6A_site_202264 | ||
| mod ID: M6ASITE014570 | Click to Show/Hide the Full List | ||
| mod site | chr12:93574786-93574787:+ | [8] | |
| Sequence | TAAAAGAGGCACCAGAAGGAACTTTCTTGATTAGAGATAGC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESCs; fibroblasts; GM12878; LCLs; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000622746.4; ENST00000536696.6; ENST00000340600.6; ENST00000549122.5; ENST00000549887.1; ENST00000551556.2; ENST00000548091.5; ENST00000549206.5; ENST00000548537.1 | ||
| External Link | RMBase: m6A_site_202265 | ||
| mod ID: M6ASITE014571 | Click to Show/Hide the Full List | ||
| mod site | chr12:93574817-93574818:+ | [8] | |
| Sequence | TAGAGATAGCTCGCATTCAGACTACCTACTAACAATATCTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESCs; fibroblasts; GM12878; LCLs; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000536696.6; ENST00000340600.6; ENST00000549887.1; ENST00000622746.4; ENST00000551556.2; ENST00000548091.5; ENST00000549206.5; ENST00000549122.5; ENST00000548537.1 | ||
| External Link | RMBase: m6A_site_202266 | ||
| mod ID: M6ASITE014572 | Click to Show/Hide the Full List | ||
| mod site | chr12:93574843-93574844:+ | [8] | |
| Sequence | TACTAACAATATCTGTTAAAACATCAGCTGGACCAACTAAT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESCs; fibroblasts; GM12878; LCLs; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000549122.5; ENST00000548091.5; ENST00000536696.6; ENST00000549887.1; ENST00000340600.6; ENST00000548537.1; ENST00000549206.5; ENST00000551556.2; ENST00000622746.4 | ||
| External Link | RMBase: m6A_site_202267 | ||
| mod ID: M6ASITE014573 | Click to Show/Hide the Full List | ||
| mod site | chr12:93574854-93574855:+ | [8] | |
| Sequence | TCTGTTAAAACATCAGCTGGACCAACTAATCTTCGAATCGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESCs; fibroblasts; GM12878; LCLs; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000548537.1; ENST00000549206.5; ENST00000548091.5; ENST00000536696.6; ENST00000551556.2; ENST00000549887.1; ENST00000549122.5; ENST00000622746.4; ENST00000340600.6 | ||
| External Link | RMBase: m6A_site_202268 | ||
| mod ID: M6ASITE014574 | Click to Show/Hide the Full List | ||
| mod site | chr12:93574901-93574902:+ | [8] | |
| Sequence | AGACGGAAAATTCAGATTGGACTCTATCATATGTGTCAAAT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESCs; fibroblasts; GM12878; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000536696.6; ENST00000549887.1; ENST00000549206.5; ENST00000340600.6; ENST00000548091.5; ENST00000551556.2; ENST00000549122.5; ENST00000548537.1; ENST00000622746.4 | ||
| External Link | RMBase: m6A_site_202269 | ||
| mod ID: M6ASITE014575 | Click to Show/Hide the Full List | ||
| mod site | chr12:93574932-93574933:+ | [8] | |
| Sequence | TGTGTCAAATCCAAGCTTAAACAATTTGACAGTGTGGTTCA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESCs; fibroblasts; GM12878; MM6; Huh7; Jurkat; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000551556.2; ENST00000549887.1; ENST00000622746.4; ENST00000549122.5; ENST00000536696.6; ENST00000549206.5; ENST00000548537.1; ENST00000340600.6; ENST00000548091.5 | ||
| External Link | RMBase: m6A_site_202270 | ||
| mod ID: M6ASITE014576 | Click to Show/Hide the Full List | ||
| mod site | chr12:93574993-93574994:+ | [8] | |
| Sequence | AGATGTGCAAGGATAAGCGGACAGGTCCAGAAGCCCCCCGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESCs; fibroblasts; GM12878; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000549122.5; ENST00000551556.2; ENST00000548091.5; ENST00000549887.1; ENST00000340600.6; ENST00000536696.6; ENST00000549206.5; ENST00000551883.1; ENST00000548537.1; ENST00000622746.4 | ||
| External Link | RMBase: m6A_site_202271 | ||
| mod ID: M6ASITE014577 | Click to Show/Hide the Full List | ||
| mod site | chr12:93575043-93575044:+ | [8] | |
| Sequence | GTTCACCTTTATCTGACCAAACCGCTCTACACGTCAGCACC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESCs; fibroblasts; GM12878; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000551883.1; ENST00000549122.5; ENST00000340600.6; ENST00000548537.1; ENST00000549206.5; ENST00000551556.2; ENST00000536696.6; ENST00000622746.4; ENST00000549887.1 | ||
| External Link | RMBase: m6A_site_202272 | ||
| mod ID: M6ASITE014578 | Click to Show/Hide the Full List | ||
| mod site | chr12:93575051-93575052:+ | [11] | |
| Sequence | TTATCTGACCAAACCGCTCTACACGTCAGCACCATCTCTGC | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000549206.5; ENST00000548537.1; ENST00000549122.5; ENST00000549887.1; ENST00000622746.4; ENST00000551556.2; ENST00000536696.6; ENST00000551883.1; ENST00000340600.6 | ||
| External Link | RMBase: m6A_site_202273 | ||
| mod ID: M6ASITE014579 | Click to Show/Hide the Full List | ||
| mod site | chr12:93575096-93575097:+ | [10] | |
| Sequence | TCTCTGTAGGCTCACCATTAACAAATGTACCGGTGCCATCT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000551556.2; ENST00000551883.1; ENST00000340600.6; ENST00000536696.6; ENST00000622746.4; ENST00000549510.1; ENST00000549206.5; ENST00000549122.5; ENST00000548537.1 | ||
| External Link | RMBase: m6A_site_202274 | ||
| mod ID: M6ASITE014580 | Click to Show/Hide the Full List | ||
| mod site | chr12:93575121-93575122:+ | [8] | |
| Sequence | TGTACCGGTGCCATCTGGGGACTGCCTTTACCAACAAGACT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; hESCs; fibroblasts; GM12878; H1299; Huh7; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000340600.6; ENST00000622746.4; ENST00000548537.1; ENST00000551883.1; ENST00000536696.6; ENST00000549122.5; ENST00000551556.2; ENST00000549206.5; ENST00000549510.1 | ||
| External Link | RMBase: m6A_site_202275 | ||
| mod ID: M6ASITE014581 | Click to Show/Hide the Full List | ||
| mod site | chr12:93575139-93575140:+ | [8] | |
| Sequence | GGACTGCCTTTACCAACAAGACTAAAAGATTACTTGGAAGA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; hESCs; fibroblasts; GM12878; H1299; Huh7; iSLK; MSC; TIME; TREX; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; miCLIP | ||
| Transcript ID List | ENST00000551883.1; ENST00000622746.4; ENST00000549122.5; ENST00000549510.1; ENST00000548537.1; ENST00000536696.6; ENST00000551556.2; ENST00000549206.5; ENST00000340600.6 | ||
| External Link | RMBase: m6A_site_202276 | ||
| mod ID: M6ASITE014582 | Click to Show/Hide the Full List | ||
| mod site | chr12:93575197-93575198:+ | [8] | |
| Sequence | AAATGTTTCTCTTTTTTTAAACATGTCTCACATAGAGTATC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; hESCs; fibroblasts; GM12878; LCLs; H1299; Huh7; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000551883.1; ENST00000549510.1; ENST00000340600.6; ENST00000549206.5; ENST00000549122.5; ENST00000548537.1; ENST00000622746.4; ENST00000551556.2 | ||
| External Link | RMBase: m6A_site_202277 | ||
| mod ID: M6ASITE014583 | Click to Show/Hide the Full List | ||
| mod site | chr12:93575206-93575207:+ | [11] | |
| Sequence | TCTTTTTTTAAACATGTCTCACATAGAGTATCTCCGAATGC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000551556.2; ENST00000549206.5; ENST00000548537.1; ENST00000549510.1; ENST00000340600.6; ENST00000549122.5; ENST00000551883.1; ENST00000622746.4 | ||
| External Link | RMBase: m6A_site_202278 | ||
| mod ID: M6ASITE014584 | Click to Show/Hide the Full List | ||
| mod site | chr12:93575243-93575244:+ | [8] | |
| Sequence | ATGCAGCTATGTAAAAGAGAACCAAAACTTGAGTGCTCTGG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; LCLs; Huh7; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000549510.1; ENST00000622746.4; ENST00000548537.1; ENST00000551883.1; ENST00000549122.5; ENST00000340600.6; ENST00000549206.5; ENST00000551556.2 | ||
| External Link | RMBase: m6A_site_202279 | ||
| mod ID: M6ASITE014585 | Click to Show/Hide the Full List | ||
| mod site | chr12:93575249-93575250:+ | [8] | |
| Sequence | CTATGTAAAAGAGAACCAAAACTTGAGTGCTCTGGATAACT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; LCLs; Huh7; iSLK; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; miCLIP | ||
| Transcript ID List | ENST00000622746.4; ENST00000549206.5; ENST00000549510.1; ENST00000551883.1; ENST00000551556.2; ENST00000549122.5; ENST00000548537.1; ENST00000340600.6 | ||
| External Link | RMBase: m6A_site_202280 | ||
| mod ID: M6ASITE014586 | Click to Show/Hide the Full List | ||
| mod site | chr12:93575290-93575291:+ | [8] | |
| Sequence | ATATGGAATGCTTTCTAAGAACAGCTGAAGCTAATCTAATT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; Huh7; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000549206.5; ENST00000551883.1; ENST00000549122.5; ENST00000622746.4; ENST00000549510.1; ENST00000340600.6; ENST00000551556.2; ENST00000548537.1 | ||
| External Link | RMBase: m6A_site_202281 | ||
| mod ID: M6ASITE014587 | Click to Show/Hide the Full List | ||
| mod site | chr12:93575403-93575404:+ | [8] | |
| Sequence | CCTTCCTAAGGCTGACCAAGACCTGTTGATCCTTTTAGATT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000548537.1; ENST00000551556.2; ENST00000549122.5; ENST00000549206.5; ENST00000551883.1; ENST00000549510.1; ENST00000622746.4; ENST00000340600.6 | ||
| External Link | RMBase: m6A_site_202282 | ||
| mod ID: M6ASITE014588 | Click to Show/Hide the Full List | ||
| mod site | chr12:93575535-93575536:+ | [8] | |
| Sequence | GCTCCAGTAGGAGAGAAAGAACTCCTCATAGGAATACTGAA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000549510.1; ENST00000551556.2; ENST00000548537.1; ENST00000622746.4; ENST00000549122.5; ENST00000340600.6; ENST00000549206.5; ENST00000551883.1 | ||
| External Link | RMBase: m6A_site_202283 | ||
| mod ID: M6ASITE014589 | Click to Show/Hide the Full List | ||
| mod site | chr12:93575569-93575570:+ | [8] | |
| Sequence | TACTGAAGAAGTGGGAAGGAACCAAGCTGACACAGGCCTCA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000549206.5; ENST00000551556.2; ENST00000548537.1; ENST00000549510.1; ENST00000340600.6; ENST00000549122.5; ENST00000622746.4; ENST00000551883.1 | ||
| External Link | RMBase: m6A_site_202284 | ||
| mod ID: M6ASITE014590 | Click to Show/Hide the Full List | ||
| mod site | chr12:93575840-93575841:+ | [8] | |
| Sequence | TTATTTCAGAAATATTTCAAACTGGTGCAAATGGAAAAGAC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000549122.5; ENST00000551556.2; ENST00000622746.4; ENST00000340600.6; ENST00000549206.5; ENST00000548537.1; ENST00000549510.1; ENST00000551883.1 | ||
| External Link | RMBase: m6A_site_202285 | ||
| mod ID: M6ASITE014591 | Click to Show/Hide the Full List | ||
| mod site | chr12:93575859-93575860:+ | [8] | |
| Sequence | AACTGGTGCAAATGGAAAAGACTTTCTCTTTTCCTTTAAAG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000622746.4; ENST00000551556.2; ENST00000549206.5; ENST00000548537.1; ENST00000340600.6; ENST00000549510.1; ENST00000551883.1; ENST00000549122.5 | ||
| External Link | RMBase: m6A_site_202286 | ||
| mod ID: M6ASITE014592 | Click to Show/Hide the Full List | ||
| mod site | chr12:93575886-93575887:+ | [8] | |
| Sequence | CTTTTCCTTTAAAGCTAAAGACAAGAATATCATGCTATACA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000340600.6; ENST00000551556.2; ENST00000548537.1; ENST00000549510.1; ENST00000549122.5; ENST00000622746.4; ENST00000551883.1; ENST00000549206.5 | ||
| External Link | RMBase: m6A_site_202287 | ||
| mod ID: M6ASITE014593 | Click to Show/Hide the Full List | ||
| mod site | chr12:93575934-93575935:+ | [8] | |
| Sequence | CTCAATCCCCGTTAATAAAAACCAATGTAGGTATAGGCATT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000551556.2; ENST00000340600.6; ENST00000622746.4; ENST00000549510.1; ENST00000549122.5; ENST00000551883.1; ENST00000549206.5; ENST00000548537.1 | ||
| External Link | RMBase: m6A_site_202288 | ||
References