m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00620)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
PDGFRA
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
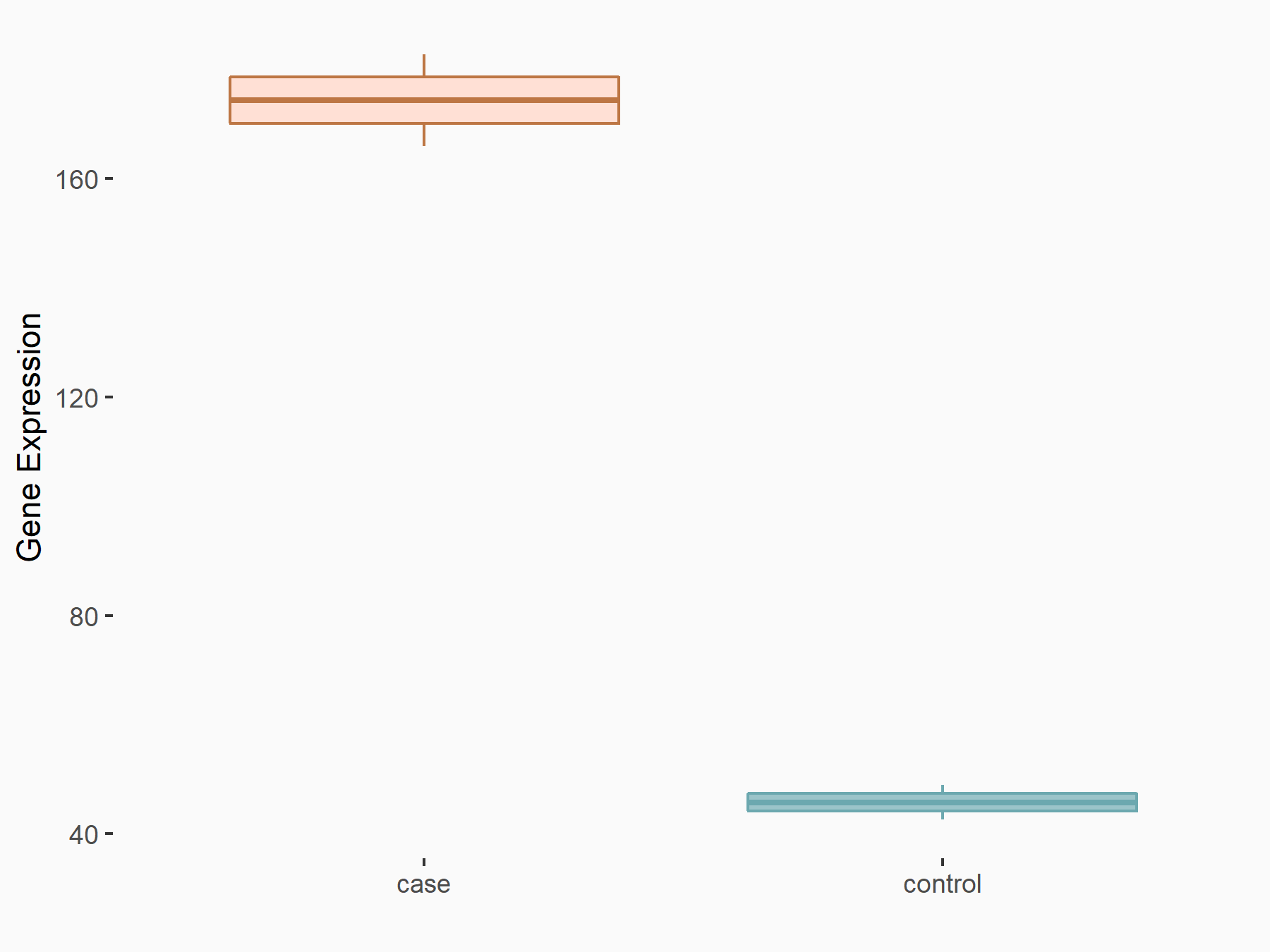 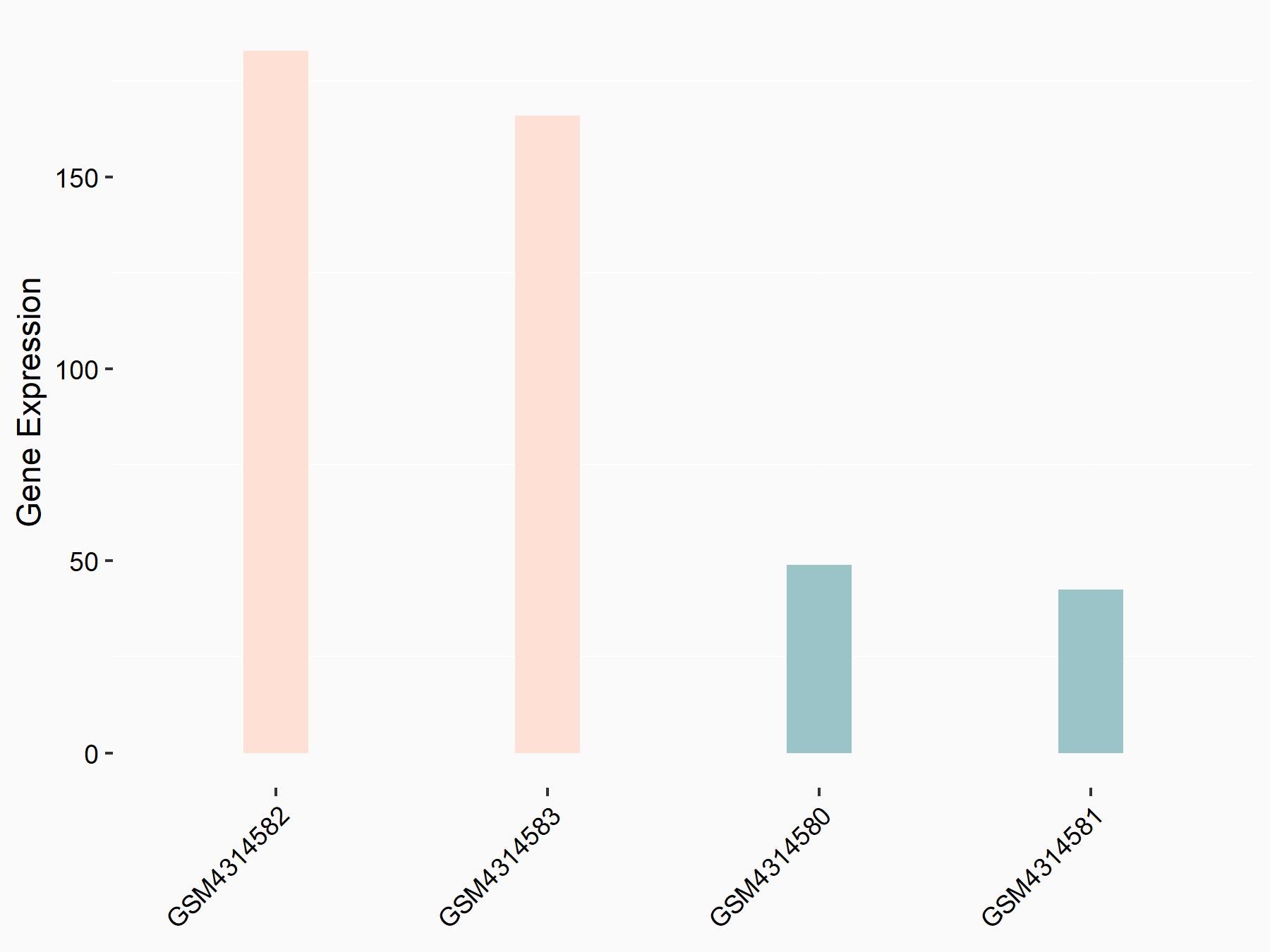 |
logFC: 1.93E+00 p-value: 8.84E-11 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between PDGFRA and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 4.83E+00 | GSE60213 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Specific depletion of METTL3 in pericytes suppressed diabetes-induced pericyte dysfunction and Microvascular complication in vivo. METTL3 overexpression impaired pericyte function by repressing PKC-Eta, FAT4, and Platelet-derived growth factor receptor alpha (PDGFRA) expression, which was mediated by YTHDF2-dependent mRNA decay. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Diseases of arteries or arterioles | ICD-11: BD5Y | ||
| In-vitro Model | ACBRI-183 (Human retinal pericytes (ACBRI-183) was obtained from Cell Systems Corp. (CSC, USA)) | |||
| In-vivo Model | Mettl3 floxed mice were purchased from GemPharmatech Co. Ltd (Nanjing, China). Pdgfr-Beta-Cre mice were purchased from Beijing Biocytogen Co. Ltd (Beijing, China) generated on C57BL/6J background. Mettl3 flox/flox mice were crossed with Pdgfr-Beta-Cre mice to generate pericyte-specific Mettl3 knockout mice. All mice were bred under the specific-pathogen free condition with free access to diet and water or their nursing mothers with alternating 12/12 light-dark cycle (lights on at 08:00 and off at 20:00). | |||
Diseases of arteries or arterioles [ICD-11: BD5Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Specific depletion of METTL3 in pericytes suppressed diabetes-induced pericyte dysfunction and Microvascular complication in vivo. METTL3 overexpression impaired pericyte function by repressing PKC-Eta, FAT4, and Platelet-derived growth factor receptor alpha (PDGFRA) expression, which was mediated by YTHDF2-dependent mRNA decay. | |||
| Responsed Disease | Diseases of arteries or arterioles [ICD-11: BD5Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | ACBRI-183 (Human retinal pericytes (ACBRI-183) was obtained from Cell Systems Corp. (CSC, USA)) | |||
| In-vivo Model | Mettl3 floxed mice were purchased from GemPharmatech Co. Ltd (Nanjing, China). Pdgfr-Beta-Cre mice were purchased from Beijing Biocytogen Co. Ltd (Beijing, China) generated on C57BL/6J background. Mettl3 flox/flox mice were crossed with Pdgfr-Beta-Cre mice to generate pericyte-specific Mettl3 knockout mice. All mice were bred under the specific-pathogen free condition with free access to diet and water or their nursing mothers with alternating 12/12 light-dark cycle (lights on at 08:00 and off at 20:00). | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00620)
| In total 50 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE065726 | Click to Show/Hide the Full List | ||
| mod site | chr4:54229319-54229320:+ | [2] | |
| Sequence | TTGGAGCTACAGGGAGAGAAACAGAGGAGGAGACTGCAAGA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | MSC; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000512143.1; ENST00000507166.5; ENST00000257290.10; ENST00000509490.5; ENST00000508170.5; ENST00000509092.5 | ||
| External Link | RMBase: m6A_site_637821 | ||
| mod ID: M6ASITE065727 | Click to Show/Hide the Full List | ||
| mod site | chr4:54229331-54229332:+ | [2] | |
| Sequence | GGAGAGAAACAGAGGAGGAGACTGCAAGAGATCATTGGAGG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | MSC; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000512143.1; ENST00000508170.5; ENST00000509092.5; ENST00000507166.5; ENST00000509490.5; ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637822 | ||
| mod ID: M6ASITE065728 | Click to Show/Hide the Full List | ||
| mod site | chr4:54229383-54229384:+ | [2] | |
| Sequence | CTCTTTACTCCATGTGTGGGACATTCATTGCGGAATAACAT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | MSC; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10; ENST00000509490.5; ENST00000512143.1; ENST00000509092.5; ENST00000507166.5; ENST00000508170.5 | ||
| External Link | RMBase: m6A_site_637823 | ||
| mod ID: M6ASITE065729 | Click to Show/Hide the Full List | ||
| mod site | chr4:54229890-54229891:+ | [2] | |
| Sequence | CTCGCCTGCGGTTTGGCGGGACAGCGCTGCAGCCCATGGTG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | MSC | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000257290.10; ENST00000512143.1; ENST00000509092.5; ENST00000508170.5; ENST00000507166.5; ENST00000503856.5; ENST00000509490.5 | ||
| External Link | RMBase: m6A_site_637824 | ||
| mod ID: M6ASITE065730 | Click to Show/Hide the Full List | ||
| mod site | chr4:54272445-54272446:+ | [3] | |
| Sequence | CACCATGGCTCAACTGGGGGACAGACGGTGAGGTGCACAGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000509490.5; ENST00000257290.10; ENST00000509092.5; ENST00000507166.5 | ||
| External Link | RMBase: m6A_site_637825 | ||
| mod ID: M6ASITE065731 | Click to Show/Hide the Full List | ||
| mod site | chr4:54274962-54274963:+ | [4] | |
| Sequence | TGGGAGTTTCCAAGAGATGGACTAGTGCTTGGTAAGTTCCA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000507166.5; ENST00000509092.5; ENST00000257290.10; ENST00000509490.5 | ||
| External Link | RMBase: m6A_site_637826 | ||
| mod ID: M6ASITE065732 | Click to Show/Hide the Full List | ||
| mod site | chr4:54277917-54277918:+ | [5] | |
| Sequence | ACGGCCAGATCCAGTGAAAAACAAGCTCTCATGTCTGAACT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000507166.5; ENST00000461294.2; ENST00000509092.5; ENST00000507536.1; ENST00000257290.10; ENST00000509490.5 | ||
| External Link | RMBase: m6A_site_637827 | ||
| mod ID: M6ASITE065733 | Click to Show/Hide the Full List | ||
| mod site | chr4:54277935-54277936:+ | [5] | |
| Sequence | AAACAAGCTCTCATGTCTGAACTGAAGATAATGACTCACCT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000509092.5; ENST00000461294.2; ENST00000507166.5; ENST00000257290.10; ENST00000507536.1; ENST00000509490.5 | ||
| External Link | RMBase: m6A_site_637828 | ||
| mod ID: M6ASITE065734 | Click to Show/Hide the Full List | ||
| mod site | chr4:54277970-54277971:+ | [5] | |
| Sequence | TCACCTGGGGCCACATTTGAACATTGTAAACTTGCTGGGAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000507536.1; ENST00000507166.5; ENST00000257290.10; ENST00000509092.5; ENST00000509490.5; ENST00000461294.2 | ||
| External Link | RMBase: m6A_site_637829 | ||
| mod ID: M6ASITE065735 | Click to Show/Hide the Full List | ||
| mod site | chr4:54277979-54277980:+ | [5] | |
| Sequence | GCCACATTTGAACATTGTAAACTTGCTGGGAGCCTGCACCA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000507166.5; ENST00000257290.10; ENST00000509490.5; ENST00000461294.2; ENST00000509092.5; ENST00000507536.1 | ||
| External Link | RMBase: m6A_site_637830 | ||
| mod ID: M6ASITE065736 | Click to Show/Hide the Full List | ||
| mod site | chr4:54281729-54281730:+ | [6] | |
| Sequence | AGCTACTCTTCACTTGCTGAACATTTTCAAAAAGAATTGAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | fibroblasts | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000257290.10; ENST00000507166.5; ENST00000507536.1; ENST00000509490.5 | ||
| External Link | RMBase: m6A_site_637831 | ||
| mod ID: M6ASITE065737 | Click to Show/Hide the Full List | ||
| mod site | chr4:54281751-54281752:+ | [6] | |
| Sequence | ATTTTCAAAAAGAATTGAGAACTTCTGGATTAAATTGCCTT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | fibroblasts; MSC; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000507536.1; ENST00000509490.5; ENST00000507166.5; ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637832 | ||
| mod ID: M6ASITE065738 | Click to Show/Hide the Full List | ||
| mod site | chr4:54281783-54281784:+ | [7] | |
| Sequence | AATTGCCTTCTTCCTCGAAAACCCTGGGACCCTTCCAGATG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | H1B; H1A; fibroblasts; Huh7; MSC; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000507166.5; ENST00000509490.5; ENST00000257290.10; ENST00000507536.1 | ||
| External Link | RMBase: m6A_site_637833 | ||
| mod ID: M6ASITE065739 | Click to Show/Hide the Full List | ||
| mod site | chr4:54281791-54281792:+ | [7] | |
| Sequence | TCTTCCTCGAAAACCCTGGGACCCTTCCAGATGGGACTAAC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | H1B; H1A; fibroblasts; Huh7; MSC; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000509490.5; ENST00000507536.1; ENST00000507166.5; ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637834 | ||
| mod ID: M6ASITE065740 | Click to Show/Hide the Full List | ||
| mod site | chr4:54281806-54281807:+ | [7] | |
| Sequence | CTGGGACCCTTCCAGATGGGACTAACTGGGGAAAGTGGACA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | H1B; H1A; fibroblasts; Huh7; MSC; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000257290.10; ENST00000509490.5; ENST00000507166.5; ENST00000507536.1 | ||
| External Link | RMBase: m6A_site_637835 | ||
| mod ID: M6ASITE065741 | Click to Show/Hide the Full List | ||
| mod site | chr4:54281824-54281825:+ | [7] | |
| Sequence | GGACTAACTGGGGAAAGTGGACAAGTTACAAACAAAGAAAC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | H1B; H1A; fibroblasts; Huh7; MSC; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000507536.1; ENST00000507166.5; ENST00000509490.5; ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637836 | ||
| mod ID: M6ASITE065742 | Click to Show/Hide the Full List | ||
| mod site | chr4:54281835-54281836:+ | [7] | |
| Sequence | GGAAAGTGGACAAGTTACAAACAAAGAAACTCAAAGGAAAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | H1B; H1A; fibroblasts; Huh7; MSC; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000509490.5; ENST00000257290.10; ENST00000507536.1; ENST00000507166.5 | ||
| External Link | RMBase: m6A_site_637837 | ||
| mod ID: M6ASITE065743 | Click to Show/Hide the Full List | ||
| mod site | chr4:54281843-54281844:+ | [7] | |
| Sequence | GACAAGTTACAAACAAAGAAACTCAAAGGAAAGTCATTGGC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | H1B; H1A; fibroblasts; Huh7; MSC; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000509490.5; ENST00000507536.1; ENST00000507166.5; ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637838 | ||
| mod ID: M6ASITE065744 | Click to Show/Hide the Full List | ||
| mod site | chr4:54281974-54281975:+ | [4] | |
| Sequence | AAATAAAGCAATTCACAGAAACTACTTTTTCATGTAGCTTG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000509490.5; ENST00000507536.1; ENST00000257290.10; ENST00000507166.5 | ||
| External Link | RMBase: m6A_site_637839 | ||
| mod ID: M6ASITE065745 | Click to Show/Hide the Full List | ||
| mod site | chr4:54285385-54285386:+ | [4] | |
| Sequence | TGCAGACTCAGAAGTCAAAAACCTCCTTTCAGATGATAACT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000257290.10; ENST00000507166.5 | ||
| External Link | RMBase: m6A_site_637840 | ||
| mod ID: M6ASITE065746 | Click to Show/Hide the Full List | ||
| mod site | chr4:54289101-54289102:+ | [8] | |
| Sequence | GTGGAGAATCTGCTGCCTGGACAATATAAAAAGGTGTGTTT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | U2OS; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10; ENST00000507166.5 | ||
| External Link | RMBase: m6A_site_637841 | ||
| mod ID: M6ASITE065747 | Click to Show/Hide the Full List | ||
| mod site | chr4:54290334-54290335:+ | [8] | |
| Sequence | TTATGAAAAAATTCACCTGGACTTCCTGAAGAGTGACCATC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | U2OS; MSC; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10; ENST00000507166.5 | ||
| External Link | RMBase: m6A_site_637842 | ||
| mod ID: M6ASITE065748 | Click to Show/Hide the Full List | ||
| mod site | chr4:54290379-54290380:+ | [8] | |
| Sequence | TGTGGCACGCATGCGTGTGGACTCAGACAATGCATACATTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | U2OS; H1A; H1B; GSC-11; MSC; endometrial; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10; ENST00000507166.5 | ||
| External Link | RMBase: m6A_site_637843 | ||
| mod ID: M6ASITE065749 | Click to Show/Hide the Full List | ||
| mod site | chr4:54290385-54290386:+ | [8] | |
| Sequence | ACGCATGCGTGTGGACTCAGACAATGCATACATTGGTGTCA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | U2OS; H1A; H1B; GSC-11; MSC; TREX; endometrial; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10; ENST00000507166.5 | ||
| External Link | RMBase: m6A_site_637844 | ||
| mod ID: M6ASITE065750 | Click to Show/Hide the Full List | ||
| mod site | chr4:54290424-54290425:+ | [8] | |
| Sequence | CACCTACAAAAACGAGGAAGACAAGCTGAAGGACTGGGAGG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | U2OS; H1A; H1B; Huh7; GSC-11; MSC; TREX; endometrial; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10; ENST00000507166.5 | ||
| External Link | RMBase: m6A_site_637845 | ||
| mod ID: M6ASITE065751 | Click to Show/Hide the Full List | ||
| mod site | chr4:54290436-54290437:+ | [8] | |
| Sequence | CGAGGAAGACAAGCTGAAGGACTGGGAGGGTGGTCTGGATG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | U2OS; H1A; H1B; Huh7; GSC-11; MSC; TREX; endometrial; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10; ENST00000507166.5 | ||
| External Link | RMBase: m6A_site_637846 | ||
| mod ID: M6ASITE065752 | Click to Show/Hide the Full List | ||
| mod site | chr4:54290464-54290465:+ | [8] | |
| Sequence | GGTGGTCTGGATGAGCAGAGACTGAGCGCTGACAGTGGCTA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | U2OS; H1A; H1B; hNPCs; Huh7; GSC-11; MSC; TREX; endometrial; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10; ENST00000507166.5 | ||
| External Link | RMBase: m6A_site_637847 | ||
| mod ID: M6ASITE065753 | Click to Show/Hide the Full List | ||
| mod site | chr4:54290529-54290530:+ | [8] | |
| Sequence | CCCTGTCCCTGAGGAGGAGGACCTGGGCAAGAGGAACAGAC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | U2OS; H1A; H1B; hNPCs; Huh7; GSC-11; MSC; TREX; endometrial; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10; ENST00000507166.5 | ||
| External Link | RMBase: m6A_site_637848 | ||
| mod ID: M6ASITE065754 | Click to Show/Hide the Full List | ||
| mod site | chr4:54290544-54290545:+ | [8] | |
| Sequence | GGAGGACCTGGGCAAGAGGAACAGACACAGGTAGCTGTGGG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | U2OS; H1A; H1B; hNPCs; Huh7; GSC-11; MSC; endometrial; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000507166.5; ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637849 | ||
| mod ID: M6ASITE065755 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295131-54295132:+ | [8] | |
| Sequence | CTCCCTCCTCCAGCTCGCAGACCTCTGAAGAGAGTGCCATT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | U2OS; H1A; H1B; Huh7; GSC-11; MSC; endometrial; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10; ENST00000507166.5 | ||
| External Link | RMBase: m6A_site_637850 | ||
| mod ID: M6ASITE065756 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295197-54295198:+ | [8] | |
| Sequence | TCATCAAGAGAGAGGACGAGACCATTGAAGACATCGACATG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | U2OS; H1B; Huh7; GSC-11; HEK293T; MSC; endometrial; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000507166.5; ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637851 | ||
| mod ID: M6ASITE065757 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295207-54295208:+ | [8] | |
| Sequence | AGAGGACGAGACCATTGAAGACATCGACATGATGGATGACA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | U2OS; H1B; Huh7; GSC-11; HEK293T; MSC; endometrial; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10; ENST00000507166.5 | ||
| External Link | RMBase: m6A_site_637852 | ||
| mod ID: M6ASITE065758 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295237-54295238:+ | [8] | |
| Sequence | GATGGATGACATCGGCATAGACTCTTCAGACCTGGTGGAAG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | U2OS; H1B; Huh7; GSC-11; HEK293T; MSC; endometrial; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000507166.5; ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637853 | ||
| mod ID: M6ASITE065759 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295246-54295247:+ | [8] | |
| Sequence | CATCGGCATAGACTCTTCAGACCTGGTGGAAGACAGCTTCC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | U2OS; H1B; Huh7; GSC-11; HEK293T; MSC; endometrial; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10; ENST00000507166.5 | ||
| External Link | RMBase: m6A_site_637854 | ||
| mod ID: M6ASITE065760 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295258-54295259:+ | [3] | |
| Sequence | CTCTTCAGACCTGGTGGAAGACAGCTTCCTGTAACTGGCGG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | brain; liver; U2OS; H1B; Huh7; GSC-11; HEK293T; MSC; endometrial; GSCs | ||
| Seq Type List | m6A-REF-seq; MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000507166.5; ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637855 | ||
| mod ID: M6ASITE065761 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295331-54295332:+ | [8] | |
| Sequence | CTCTGGATCCCGTTCAGAAAACCACTTTATTGCAATGCAGA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | U2OS; H1B; Huh7; GSC-11; MSC; TREX; endometrial; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637856 | ||
| mod ID: M6ASITE065762 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295365-54295366:+ | [8] | |
| Sequence | ATGCAGAGGTTGAGAGGAGGACTTGGTTGATGTTTAAAGAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | U2OS; H1B; fibroblasts; Huh7; GSC-11; MSC; TREX; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637857 | ||
| mod ID: M6ASITE065763 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295450-54295451:+ | [8] | |
| Sequence | AATGGGATATTTTGAAATGAACTTTGTCAGTGTTGCCTCTT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HEK293T; U2OS; fibroblasts; Huh7; GSC-11; MSC; TREX; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637858 | ||
| mod ID: M6ASITE065764 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295550-54295551:+ | [8] | |
| Sequence | TAATAGGCCACAGAAGGTGAACTTTGTGCTTCAAGGACATT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | U2OS; hNPCs; fibroblasts; Huh7; GSC-11; MSC; TREX; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637859 | ||
| mod ID: M6ASITE065765 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295566-54295567:+ | [8] | |
| Sequence | GTGAACTTTGTGCTTCAAGGACATTGGTGAGAGTCCAACAG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | U2OS; hNPCs; fibroblasts; Huh7; GSC-11; MSC; TREX; endometrial; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637860 | ||
| mod ID: M6ASITE065766 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295587-54295588:+ | [8] | |
| Sequence | CATTGGTGAGAGTCCAACAGACACAATTTATACTGCGACAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | U2OS; fibroblasts; Huh7; GSC-11; MSC; TREX; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637861 | ||
| mod ID: M6ASITE065767 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295609-54295610:+ | [8] | |
| Sequence | ACAATTTATACTGCGACAGAACTTCAGCATTGTAATTATGT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | U2OS; fibroblasts; Huh7; GSC-11; MSC; TREX; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637862 | ||
| mod ID: M6ASITE065768 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295681-54295682:+ | [8] | |
| Sequence | GTATTAACTATCTTCTTTGGACTTCTGAAGAGACCACTCAA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | U2OS; fibroblasts; Huh7; HEK293A-TOA; MSC; TREX; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637863 | ||
| mod ID: M6ASITE065769 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295693-54295694:+ | [8] | |
| Sequence | TTCTTTGGACTTCTGAAGAGACCACTCAATCCATCCATGTA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | U2OS; fibroblasts; Huh7; HEK293A-TOA; MSC; TREX; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637864 | ||
| mod ID: M6ASITE065770 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295727-54295728:+ | [8] | |
| Sequence | CCATGTACTTCCCTCTTGAAACCTGATGTCAGCTGCTGTTG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | U2OS; fibroblasts; Huh7; HEK293A-TOA; MSC; TREX | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637865 | ||
| mod ID: M6ASITE065771 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295749-54295750:+ | [8] | |
| Sequence | CTGATGTCAGCTGCTGTTGAACTTTTTAAAGAAGTGCATGA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | U2OS; fibroblasts; Huh7; MSC; TREX | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637866 | ||
| mod ID: M6ASITE065772 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295773-54295774:+ | [8] | |
| Sequence | TTTAAAGAAGTGCATGAAAAACCATTTTTGAACCTTAAAAG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | U2OS; fibroblasts; Huh7; MSC; TREX | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637867 | ||
| mod ID: M6ASITE065773 | Click to Show/Hide the Full List | ||
| mod site | chr4:54295784-54295785:+ | [8] | |
| Sequence | GCATGAAAAACCATTTTTGAACCTTAAAAGGTACTGGTACT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | U2OS; fibroblasts; Huh7; MSC; TREX | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637868 | ||
| mod ID: M6ASITE065774 | Click to Show/Hide the Full List | ||
| mod site | chr4:54297475-54297476:+ | [3] | |
| Sequence | ACGTTTGTGTTTCTAGAATCACAGCTCAAGCATTCTGTTTA | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | brain; liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637869 | ||
| mod ID: M6ASITE065775 | Click to Show/Hide the Full List | ||
| mod site | chr4:54298096-54298097:+ | [3] | |
| Sequence | GATACTTACATGTTCCCAAAACAATGGTGTGGTGAATGTGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000257290.10 | ||
| External Link | RMBase: m6A_site_637870 | ||
References