m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00330)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
MAPK14
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HeLa cell line | Homo sapiens |
|
Treatment: METTL3 knockdown HeLa cells
Control: HeLa cells
|
GSE70061 | |
| Regulation |
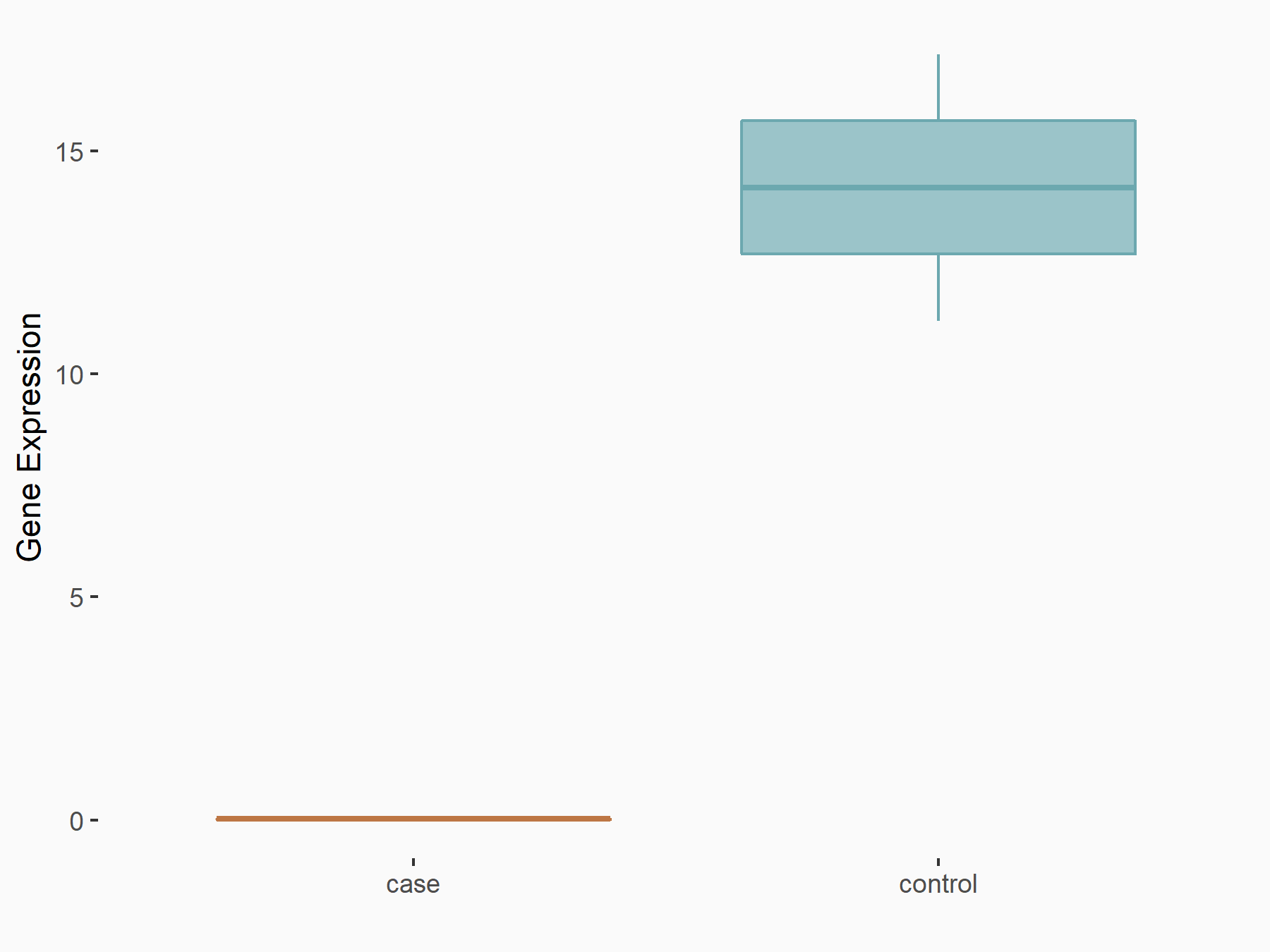  |
logFC: -3.87E+00 p-value: 1.84E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between MAPK14 and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.30E+00 | GSE60213 |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | High METTL3 expression was positively correlated with more severe clinical features of OSCC tumors. Furthermore, METTL3-KD and cycloleucine, a methylation inhibitor, decreased m6A levels and down-regulated Mitogen-activated protein kinase 14 (p38/MAPK14) expression in OSCC cells. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Oral squamous cell carcinoma | ICD-11: 2B6E.0 | ||
| In-vitro Model | SCC-25 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1682 |
| SCC-15 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1681 | |
| In-vivo Model | Male BALB/c-nu/nu mice, 9-10 weeks of age, were acclimatized for a week prior to the experiments. Nude mice (6 each group) were subcutaneously inoculated with 4 × 106 SCC-25 or SCC-15 cells through an injection into the center of the back, which consistently caused tumor formation within 1 week of inoculation. To monitor the initial tumor appearance, animals were observed every day. After tumor appeared, measurements were made every week with a caliper. After 28 days, mice were sacrificed and tumors were dissected out to be weighed. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 played a tumor-suppressive role in Colorectal cancer cell proliferation, migration and invasion through Mitogen-activated protein kinase 14 (p38/MAPK14)/ERK pathways, which indicated that METTL3 was a novel marker for CRC carcinogenesis, progression and survival. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| KM12 | Colon carcinoma | Homo sapiens | CVCL_1331 | |
Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated Mitogen-activated protein kinase 14 (p38/MAPK14), ERK1/2, AKT, and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, IL-1-beta and TNF-alpha secretion. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gangrene or necrosis of lung | ICD-11: CA43 | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
| In-vitro Model | BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
YTH domain-containing family protein 3 (YTHDF3) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated Mitogen-activated protein kinase 14 (p38/MAPK14), ERK1/2, AKT, and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, IL-1-beta and TNF-alpha secretion. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gangrene or necrosis of lung | ICD-11: CA43 | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
| In-vitro Model | BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | High METTL3 expression was positively correlated with more severe clinical features of OSCC tumors. Furthermore, METTL3-KD and cycloleucine, a methylation inhibitor, decreased m6A levels and down-regulated Mitogen-activated protein kinase 14 (p38/MAPK14) expression in OSCC cells. | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | SCC-25 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1682 |
| SCC-15 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1681 | |
| In-vivo Model | Male BALB/c-nu/nu mice, 9-10 weeks of age, were acclimatized for a week prior to the experiments. Nude mice (6 each group) were subcutaneously inoculated with 4 × 106 SCC-25 or SCC-15 cells through an injection into the center of the back, which consistently caused tumor formation within 1 week of inoculation. To monitor the initial tumor appearance, animals were observed every day. After tumor appeared, measurements were made every week with a caliper. After 28 days, mice were sacrificed and tumors were dissected out to be weighed. | |||
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 played a tumor-suppressive role in Colorectal cancer cell proliferation, migration and invasion through Mitogen-activated protein kinase 14 (p38/MAPK14)/ERK pathways, which indicated that METTL3 was a novel marker for CRC carcinogenesis, progression and survival. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| KM12 | Colon carcinoma | Homo sapiens | CVCL_1331 | |
Gangrene or necrosis of lung [ICD-11: CA43]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated Mitogen-activated protein kinase 14 (p38/MAPK14), ERK1/2, AKT, and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, IL-1-beta and TNF-alpha secretion. | |||
| Responsed Disease | Gangrene or necrosis of lung [ICD-11: CA43] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
| In-vitro Model | BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated Mitogen-activated protein kinase 14 (p38/MAPK14), ERK1/2, AKT, and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, IL-1-beta and TNF-alpha secretion. | |||
| Responsed Disease | Gangrene or necrosis of lung [ICD-11: CA43] | |||
| Target Regulator | YTH domain-containing family protein 3 (YTHDF3) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
| In-vitro Model | BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03543 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03588 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00330)
| In total 8 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE001427 | Click to Show/Hide the Full List | ||
| mod site | chr6:36027855-36027856:+ | [5] | |
| Sequence | GAACGGGAGTACTGCGACGCAGCCCGGAGTCGGCCTTGTAG | ||
| Transcript ID List | ENST00000468133.5; ENST00000229795.7; ENST00000229794.8; ENST00000622903.4 | ||
| External Link | RMBase: RNA-editing_site_115722 | ||
| mod ID: A2ISITE001428 | Click to Show/Hide the Full List | ||
| mod site | chr6:36036852-36036853:+ | [6] | |
| Sequence | CAGGTTCAAGCAATTCTCCCACCTTAGCCTCCCGAGTAGCT | ||
| Transcript ID List | ENST00000229794.8; ENST00000229795.7; ENST00000468133.5; ENST00000472333.1; ENST00000310795.8; ENST00000496250.5; ENST00000491957.5; ENST00000474429.5; ENST00000622903.4 | ||
| External Link | RMBase: RNA-editing_site_115723 | ||
| mod ID: A2ISITE001429 | Click to Show/Hide the Full List | ||
| mod site | chr6:36098400-36098401:+ | [7] | |
| Sequence | AGCAAAAGCAGACTGGGCACAGTGGCTCACATTTGTCATCA | ||
| Transcript ID List | ENST00000229794.8; rmsk_1918167; ENST00000310795.8; ENST00000468133.5; ENST00000229795.7; ENST00000474429.5 | ||
| External Link | RMBase: RNA-editing_site_115724 | ||
| mod ID: A2ISITE001430 | Click to Show/Hide the Full List | ||
| mod site | chr6:36098515-36098516:+ | [7] | |
| Sequence | CACTGCACTCCAGCCTGGGCAACAGAGCAAGACCTTGTCTT | ||
| Transcript ID List | ENST00000310795.8; ENST00000229794.8; ENST00000468133.5; ENST00000474429.5; rmsk_1918167; ENST00000229795.7 | ||
| External Link | RMBase: RNA-editing_site_115725 | ||
| mod ID: A2ISITE001431 | Click to Show/Hide the Full List | ||
| mod site | chr6:36101316-36101317:+ | [8] | |
| Sequence | GCATGATAGCACATGCCTGTAGTCCCAGCTACTTGGGAAGC | ||
| Transcript ID List | ENST00000474429.5; ENST00000229794.8; ENST00000310795.8; ENST00000229795.7; rmsk_1918168; ENST00000468133.5 | ||
| External Link | RMBase: RNA-editing_site_115726 | ||
| mod ID: A2ISITE001432 | Click to Show/Hide the Full List | ||
| mod site | chr6:36101368-36101369:+ | [8] | |
| Sequence | GGTTGCTTGAGTCAGGAGTTAGAGGCCATAGTGAGCTGTGA | ||
| Transcript ID List | ENST00000468133.5; ENST00000474429.5; rmsk_1918168; ENST00000310795.8; ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: RNA-editing_site_115727 | ||
| mod ID: A2ISITE001433 | Click to Show/Hide the Full List | ||
| mod site | chr6:36101529-36101530:+ | [8] | |
| Sequence | GATCTTAACTCTGTCACCCCAGCTGGAGTGCAGTGGCACGA | ||
| Transcript ID List | ENST00000468133.5; ENST00000474429.5; ENST00000229795.7; ENST00000310795.8; ENST00000229794.8 | ||
| External Link | RMBase: RNA-editing_site_115728 | ||
| mod ID: A2ISITE001434 | Click to Show/Hide the Full List | ||
| mod site | chr6:36101546-36101547:+ | [8] | |
| Sequence | CCCAGCTGGAGTGCAGTGGCACGATCTTAGCTCACTGGAAC | ||
| Transcript ID List | ENST00000229794.8; ENST00000310795.8; ENST00000474429.5; ENST00000468133.5; ENST00000229795.7 | ||
| External Link | RMBase: RNA-editing_site_115729 | ||
5-methylcytidine (m5C)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE003929 | Click to Show/Hide the Full List | ||
| mod site | chr6:36028046-36028047:+ | [9] | |
| Sequence | GCCGCACCTGCGCGGGCGACCAGCGCAAGGTCCCCGCCCGG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8; ENST00000622903.4; ENST00000468133.5 | ||
| External Link | RMBase: m5C_site_37918 | ||
N6-methyladenosine (m6A)
| In total 66 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE074700 | Click to Show/Hide the Full List | ||
| mod site | chr6:36027798-36027799:+ | [10] | |
| Sequence | CCCGTTTGCTGGCTCTTGGAACCGCGACCACTGGAGCCTTA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; A549; Jurkat; CD4T; HEK293A-TOA; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000468133.5; ENST00000229795.7; ENST00000229794.8; ENST00000622903.4 | ||
| External Link | RMBase: m6A_site_714667 | ||
| mod ID: M6ASITE074701 | Click to Show/Hide the Full List | ||
| mod site | chr6:36028197-36028198:+ | [10] | |
| Sequence | GTTCTACCGGCAGGAGCTGAACAAGACAATCTGGGAGGTGC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; CD34; HEK293T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000468133.5; ENST00000229795.7; ENST00000310795.8; ENST00000622903.4; ENST00000229794.8; ENST00000491957.5; ENST00000496250.5 | ||
| External Link | RMBase: m6A_site_714668 | ||
| mod ID: M6ASITE074702 | Click to Show/Hide the Full List | ||
| mod site | chr6:36028202-36028203:+ | [10] | |
| Sequence | ACCGGCAGGAGCTGAACAAGACAATCTGGGAGGTGCCCGAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000310795.8; ENST00000229794.8; ENST00000474429.5; ENST00000229795.7; ENST00000622903.4; ENST00000468133.5; ENST00000491957.5; ENST00000496250.5 | ||
| External Link | RMBase: m6A_site_714669 | ||
| mod ID: M6ASITE074703 | Click to Show/Hide the Full List | ||
| mod site | chr6:36028233-36028234:+ | [10] | |
| Sequence | GGTGCCCGAGCGTTACCAGAACCTGTCTCCAGTGGGCTCTG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000468133.5; ENST00000474429.5; ENST00000229795.7; ENST00000491957.5; ENST00000310795.8; ENST00000229794.8; ENST00000496250.5; ENST00000622903.4 | ||
| External Link | RMBase: m6A_site_714670 | ||
| mod ID: M6ASITE074704 | Click to Show/Hide the Full List | ||
| mod site | chr6:36052709-36052710:+ | [11] | |
| Sequence | TCTCCTTAGTGCTGCTTTTGACACAAAAACGGGGTTACGTG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000622903.4; ENST00000472333.1; ENST00000310795.8; ENST00000229795.7; ENST00000468133.5; ENST00000474429.5; ENST00000229794.8; ENST00000491957.5; ENST00000496250.5 | ||
| External Link | RMBase: m6A_site_714671 | ||
| mod ID: M6ASITE074705 | Click to Show/Hide the Full List | ||
| mod site | chr6:36059312-36059313:+ | [11] | |
| Sequence | TTGGTCTGTTGGACGTTTTTACACCTGCAAGGTCTCTGGAG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000474429.5; ENST00000491957.5; ENST00000229794.8; ENST00000472333.1; ENST00000468133.5; ENST00000496250.5; ENST00000622903.4; ENST00000310795.8 | ||
| External Link | RMBase: m6A_site_714672 | ||
| mod ID: M6ASITE074706 | Click to Show/Hide the Full List | ||
| mod site | chr6:36072907-36072908:+ | [11] | |
| Sequence | TCTCATGGGGGCAGATCTGAACAACATTGTGAAATGTCAGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000474429.5; ENST00000472333.1; ENST00000310795.8; ENST00000229795.7; ENST00000491957.5; ENST00000622903.4; ENST00000468133.5; ENST00000496250.5; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714673 | ||
| mod ID: M6ASITE074707 | Click to Show/Hide the Full List | ||
| mod site | chr6:36073695-36073696:+ | [11] | |
| Sequence | TTTTCTCTTTTGCAGTATATACATTCAGCTGACATAATTCA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000474429.5; ENST00000468133.5; ENST00000310795.8; ENST00000622903.4; ENST00000472333.1; ENST00000496250.5; ENST00000229794.8; ENST00000491957.5 | ||
| External Link | RMBase: m6A_site_714674 | ||
| mod ID: M6ASITE074708 | Click to Show/Hide the Full List | ||
| mod site | chr6:36074056-36074057:+ | [10] | |
| Sequence | TTTGCTTCCTAGGACCTAAAACCTAGTAATCTAGCTGTGAA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000310795.8; ENST00000491957.5; ENST00000474429.5; ENST00000229794.8; ENST00000468133.5; ENST00000472333.1; ENST00000496250.5; ENST00000622903.4; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714675 | ||
| mod ID: M6ASITE074709 | Click to Show/Hide the Full List | ||
| mod site | chr6:36074082-36074083:+ | [10] | |
| Sequence | TAATCTAGCTGTGAATGAAGACTGTGAGCTGAAGGTAAAAT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000622903.4; ENST00000310795.8; ENST00000472333.1; ENST00000496250.5; ENST00000229795.7; ENST00000474429.5; ENST00000468133.5; ENST00000229794.8; ENST00000491957.5 | ||
| External Link | RMBase: m6A_site_714676 | ||
| mod ID: M6ASITE074710 | Click to Show/Hide the Full List | ||
| mod site | chr6:36075861-36075862:+ | [10] | |
| Sequence | CTTTAGATTCTGGATTTTGGACTGGCTCGGCACACAGATGA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000468133.5; ENST00000496250.5; ENST00000474429.5; ENST00000310795.8; ENST00000472333.1; ENST00000229794.8; ENST00000491957.5 | ||
| External Link | RMBase: m6A_site_714677 | ||
| mod ID: M6ASITE074711 | Click to Show/Hide the Full List | ||
| mod site | chr6:36075872-36075873:+ | [11] | |
| Sequence | GGATTTTGGACTGGCTCGGCACACAGATGATGAAATGACAG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000310795.8; ENST00000229794.8; ENST00000496250.5; ENST00000491957.5; ENST00000229795.7; ENST00000468133.5; ENST00000472333.1; ENST00000474429.5; ENST00000490379.1 | ||
| External Link | RMBase: m6A_site_714678 | ||
| mod ID: M6ASITE074712 | Click to Show/Hide the Full List | ||
| mod site | chr6:36076586-36076587:+ | [11] | |
| Sequence | CCGAGCTGTTGACTGGAAGAACATTGTTTCCTGGTACAGAC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000468133.5; ENST00000491957.5; ENST00000496250.5; ENST00000229795.7; ENST00000310795.8; ENST00000490379.1; ENST00000229794.8; ENST00000474429.5 | ||
| External Link | RMBase: m6A_site_714679 | ||
| mod ID: M6ASITE074713 | Click to Show/Hide the Full List | ||
| mod site | chr6:36084651-36084652:+ | [12] | |
| Sequence | AATGAAAGGAAATGAACAAAACTTTTGAGAACTATGGGATT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000474429.5; ENST00000491957.5; rmsk_1918152; ENST00000229794.8; ENST00000229795.7; ENST00000496250.5; ENST00000468133.5; ENST00000310795.8 | ||
| External Link | RMBase: m6A_site_714680 | ||
| mod ID: M6ASITE074714 | Click to Show/Hide the Full List | ||
| mod site | chr6:36096205-36096206:+ | [13] | |
| Sequence | CCGGATGAGTGAGGTATAAGACAGTGTGAGTGTGTGTGTGC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | CD34 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000491957.5; ENST00000310795.8; ENST00000474429.5; ENST00000468133.5; ENST00000229795.7; ENST00000496250.5; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714681 | ||
| mod ID: M6ASITE074715 | Click to Show/Hide the Full List | ||
| mod site | chr6:36097967-36097968:+ | [13] | |
| Sequence | AGTGGATAAAAGTAGTTTAAACATCCTAAGGGGTAGACAAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | CD34 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8; ENST00000468133.5; ENST00000491957.5; ENST00000310795.8; ENST00000474429.5; ENST00000496250.5 | ||
| External Link | RMBase: m6A_site_714682 | ||
| mod ID: M6ASITE074716 | Click to Show/Hide the Full List | ||
| mod site | chr6:36097983-36097984:+ | [13] | |
| Sequence | TTAAACATCCTAAGGGGTAGACAAGATTAATTTGATGTTCG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | CD34 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000474429.5; ENST00000229795.7; ENST00000310795.8; ENST00000491957.5; ENST00000229794.8; ENST00000496250.5; ENST00000468133.5 | ||
| External Link | RMBase: m6A_site_714683 | ||
| mod ID: M6ASITE074717 | Click to Show/Hide the Full List | ||
| mod site | chr6:36102577-36102578:+ | [10] | |
| Sequence | CTTGTTTTTTCAGGCAAGAAACTATATTCAGTCTTTGACTC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000468133.5; ENST00000229794.8; ENST00000474429.5; ENST00000310795.8 | ||
| External Link | RMBase: m6A_site_714684 | ||
| mod ID: M6ASITE074718 | Click to Show/Hide the Full List | ||
| mod site | chr6:36102613-36102614:+ | [10] | |
| Sequence | GACTCAGATGCCGAAGATGAACTTTGCGAATGTATTTATTG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000474429.5; ENST00000310795.8; ENST00000229794.8; ENST00000468133.5 | ||
| External Link | RMBase: m6A_site_714685 | ||
| mod ID: M6ASITE074719 | Click to Show/Hide the Full List | ||
| mod site | chr6:36107487-36107488:+ | [10] | |
| Sequence | GGAGAAGATGCTTGTATTGGACTCAGATAAGAGAATTACAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000310795.8; ENST00000229794.8; ENST00000468133.5; ENST00000474429.5 | ||
| External Link | RMBase: m6A_site_714686 | ||
| mod ID: M6ASITE074720 | Click to Show/Hide the Full List | ||
| mod site | chr6:36107524-36107525:+ | [11] | |
| Sequence | ACAGCGGCCCAAGCCCTTGCACATGCCTACTTTGCTCAGTA | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000474429.5; ENST00000310795.8; ENST00000229794.8; ENST00000468133.5; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714687 | ||
| mod ID: M6ASITE074721 | Click to Show/Hide the Full List | ||
| mod site | chr6:36107563-36107564:+ | [10] | |
| Sequence | TACCACGATCCTGATGATGAACCAGTGGCCGATCCTTATGA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000468133.5; ENST00000310795.8; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714688 | ||
| mod ID: M6ASITE074722 | Click to Show/Hide the Full List | ||
| mod site | chr6:36108424-36108425:+ | [14] | |
| Sequence | CTTTGTGCCACCACCCCTTGACCAAGAAGAGATGGAGTCCT | ||
| Motif Score | 2.839113095 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000468133.5; ENST00000229794.8; ENST00000310795.8; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714689 | ||
| mod ID: M6ASITE074723 | Click to Show/Hide the Full List | ||
| mod site | chr6:36108507-36108508:+ | [15] | |
| Sequence | GGGAAGGCCTTTTCACGGGAACTCTCCAAATATTATTCAAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000468133.5; ENST00000310795.8; ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714690 | ||
| mod ID: M6ASITE074724 | Click to Show/Hide the Full List | ||
| mod site | chr6:36108634-36108635:+ | [13] | |
| Sequence | TGTCTTTGTGGGAGGGTAAGACAATATGAACAAACTATGAT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | CD34; hESC-HEK293T; U2OS; hESCs; A549 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000468133.5; ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714691 | ||
| mod ID: M6ASITE074725 | Click to Show/Hide the Full List | ||
| mod site | chr6:36108643-36108644:+ | [13] | |
| Sequence | GGGAGGGTAAGACAATATGAACAAACTATGATCACAGTGAC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | CD34; U2OS; hESCs; A549 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8; ENST00000468133.5 | ||
| External Link | RMBase: m6A_site_714692 | ||
| mod ID: M6ASITE074726 | Click to Show/Hide the Full List | ||
| mod site | chr6:36108662-36108663:+ | [14] | |
| Sequence | AACAAACTATGATCACAGTGACTTTACAGGAGGTTGTGGAT | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T; A549 | ||
| Seq Type List | DART-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000229795.7; ENST00000468133.5; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714693 | ||
| mod ID: M6ASITE074727 | Click to Show/Hide the Full List | ||
| mod site | chr6:36108667-36108668:+ | [16] | |
| Sequence | ACTATGATCACAGTGACTTTACAGGAGGTTGTGGATGCTCC | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8; ENST00000468133.5 | ||
| External Link | RMBase: m6A_site_714694 | ||
| mod ID: M6ASITE074728 | Click to Show/Hide the Full List | ||
| mod site | chr6:36108735-36108736:+ | [13] | |
| Sequence | CTGAGAGTTGGCTCAGGCAGACAAGAGCTGCTGTCCTTTTA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | CD34; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714695 | ||
| mod ID: M6ASITE074729 | Click to Show/Hide the Full List | ||
| mod site | chr6:36108848-36108849:+ | [14] | |
| Sequence | TCTCCTGTGACCCCACCTTGACGGTGGGGCGTAGACTTGAC | ||
| Motif Score | 2.833690476 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714696 | ||
| mod ID: M6ASITE074730 | Click to Show/Hide the Full List | ||
| mod site | chr6:36108862-36108863:+ | [10] | |
| Sequence | ACCTTGACGGTGGGGCGTAGACTTGACAACATCCCACAGTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000229794.8; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714697 | ||
| mod ID: M6ASITE074731 | Click to Show/Hide the Full List | ||
| mod site | chr6:36108867-36108868:+ | [11] | |
| Sequence | GACGGTGGGGCGTAGACTTGACAACATCCCACAGTGGCACG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714698 | ||
| mod ID: M6ASITE074732 | Click to Show/Hide the Full List | ||
| mod site | chr6:36108920-36108921:+ | [10] | |
| Sequence | ATACCTTCTGGTTGCTTCAGACCTGACACCGTCCCTCAGTG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714699 | ||
| mod ID: M6ASITE074734 | Click to Show/Hide the Full List | ||
| mod site | chr6:36108925-36108926:+ | [11] | |
| Sequence | TTCTGGTTGCTTCAGACCTGACACCGTCCCTCAGTGATACG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714700 | ||
| mod ID: M6ASITE074735 | Click to Show/Hide the Full List | ||
| mod site | chr6:36109035-36109036:+ | [14] | |
| Sequence | ATCTTGACAGTATATTTGAAACTGTAAATATGTTTGTGCCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000229794.8; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714701 | ||
| mod ID: M6ASITE074736 | Click to Show/Hide the Full List | ||
| mod site | chr6:36109138-36109139:+ | [13] | |
| Sequence | CAAGTCGAGAGGGCTGTTGGACAGCTGCTTGTGGGCCCGGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | CD34 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000229794.8; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714702 | ||
| mod ID: M6ASITE074737 | Click to Show/Hide the Full List | ||
| mod site | chr6:36109336-36109337:+ | [14] | |
| Sequence | GCAGGTTCTTGATGTCATGTACTTCCTGTGTACTCTTTATT | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714703 | ||
| mod ID: M6ASITE074738 | Click to Show/Hide the Full List | ||
| mod site | chr6:36109347-36109348:+ | [14] | |
| Sequence | ATGTCATGTACTTCCTGTGTACTCTTTATTTCTAGCAGAGT | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000229794.8; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714704 | ||
| mod ID: M6ASITE074739 | Click to Show/Hide the Full List | ||
| mod site | chr6:36109569-36109570:+ | [14] | |
| Sequence | CTGTGTTGAACACAATTGATACTTCAGGTGCTTTTGATGTG | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000229794.8; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714705 | ||
| mod ID: M6ASITE074740 | Click to Show/Hide the Full List | ||
| mod site | chr6:36109685-36109686:+ | [10] | |
| Sequence | AGGAATTATCATGGGAAAAGACCAGGGCTTTTCCCAGGAAT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000229794.8; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714706 | ||
| mod ID: M6ASITE074741 | Click to Show/Hide the Full List | ||
| mod site | chr6:36109713-36109714:+ | [10] | |
| Sequence | TTTTCCCAGGAATATCCCAAACTTCGGAAACAAGTTATTCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000229794.8; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714707 | ||
| mod ID: M6ASITE074742 | Click to Show/Hide the Full List | ||
| mod site | chr6:36109722-36109723:+ | [10] | |
| Sequence | GAATATCCCAAACTTCGGAAACAAGTTATTCTCTTCACTCC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714708 | ||
| mod ID: M6ASITE074743 | Click to Show/Hide the Full List | ||
| mod site | chr6:36109806-36109807:+ | [10] | |
| Sequence | TAAAGCCCATAAGGCCAGAAACTCCTTTTGCTGTCTTTCTC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000229794.8; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714709 | ||
| mod ID: M6ASITE074744 | Click to Show/Hide the Full List | ||
| mod site | chr6:36109857-36109858:+ | [11] | |
| Sequence | TACTTTAAAATAAAAAAGTAACAAGGTGTCTTTTCCACTCC | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714710 | ||
| mod ID: M6ASITE074745 | Click to Show/Hide the Full List | ||
| mod site | chr6:36109906-36109907:+ | [11] | |
| Sequence | AGGGTCTTCTTGGCAGCTTAACATTGACTTCTTGGTTTGGG | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000229794.8; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714711 | ||
| mod ID: M6ASITE074746 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110035-36110036:+ | [14] | |
| Sequence | TTTAATATTTTGTATTTTCAACTTTATAAAGATAAAATATC | ||
| Motif Score | 2.595904762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714712 | ||
| mod ID: M6ASITE074747 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110085-36110086:+ | [14] | |
| Sequence | GGAGAAGTGTCGTTTTCATAACTTGCTGAATTTCAGGCATT | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714713 | ||
| mod ID: M6ASITE074748 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110113-36110114:+ | [14] | |
| Sequence | AATTTCAGGCATTTTGTTCTACATGAGGACTCATATATTTA | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000229794.8; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714714 | ||
| mod ID: M6ASITE074749 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110121-36110122:+ | [10] | |
| Sequence | GCATTTTGTTCTACATGAGGACTCATATATTTAAGCCTTTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714715 | ||
| mod ID: M6ASITE074750 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110186-36110187:+ | [14] | |
| Sequence | CTTCCAGTGTTGGCTGTGTGACAGAATCTTGTATTTGGGCC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000229794.8; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714716 | ||
| mod ID: M6ASITE074751 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110240-36110241:+ | [11] | |
| Sequence | TTCTCAATCAGTGCAGTGATACATGTACTCCAGAGGGACAG | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000229794.8; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714717 | ||
| mod ID: M6ASITE074752 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110257-36110258:+ | [10] | |
| Sequence | GATACATGTACTCCAGAGGGACAGGGTGGACCCCCTGAGTC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714718 | ||
| mod ID: M6ASITE074753 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110266-36110267:+ | [10] | |
| Sequence | ACTCCAGAGGGACAGGGTGGACCCCCTGAGTCAACTGGAGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000229794.8; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714719 | ||
| mod ID: M6ASITE074754 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110304-36110305:+ | [10] | |
| Sequence | AGCAAGAAGGAAGGAGGCAGACTGATGGCGATTCCCTCTCA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000229794.8; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714720 | ||
| mod ID: M6ASITE074755 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110331-36110332:+ | [10] | |
| Sequence | GCGATTCCCTCTCACCCGGGACTCTCCCCCTTTCAAGGAAA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714721 | ||
| mod ID: M6ASITE074756 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110356-36110357:+ | [10] | |
| Sequence | CCCCCTTTCAAGGAAAGTGAACCTTTAAAGTAAAGGCCTCA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714722 | ||
| mod ID: M6ASITE074757 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110466-36110467:+ | [14] | |
| Sequence | AAATGCTCGTGTGATTTCCTACAGAAATACTGCTCTGAATA | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714723 | ||
| mod ID: M6ASITE074758 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110508-36110509:+ | [14] | |
| Sequence | TTTGTAATAAAGGTCTTTGCACATGTGACCACATACGTGTT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714724 | ||
| mod ID: M6ASITE074759 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110518-36110519:+ | [11] | |
| Sequence | AGGTCTTTGCACATGTGACCACATACGTGTTAGGAGGCTGC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714725 | ||
| mod ID: M6ASITE074760 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110522-36110523:+ | [14] | |
| Sequence | CTTTGCACATGTGACCACATACGTGTTAGGAGGCTGCATGC | ||
| Motif Score | 2.084416667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714726 | ||
| mod ID: M6ASITE074761 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110556-36110557:+ | [10] | |
| Sequence | TGCATGCTCTGGAAGCCTGGACTCTAAGCTGGAGCTCTTGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; hNPCs; fibroblasts; A549; HEK293A-TOA; MSC | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714727 | ||
| mod ID: M6ASITE074762 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110628-36110629:+ | [10] | |
| Sequence | CCATCTCCTGATTTCTCTGAACAGAAAACAAAAGAGAGAAT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; fibroblasts; A549; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq | ||
| Transcript ID List | ENST00000229794.8; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714728 | ||
| mod ID: M6ASITE074763 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110635-36110636:+ | [10] | |
| Sequence | CTGATTTCTCTGAACAGAAAACAAAAGAGAGAATGAGGGAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; HepG2; fibroblasts; A549; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714729 | ||
| mod ID: M6ASITE074764 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110681-36110682:+ | [10] | |
| Sequence | TATTTTATTTGTATTCATGAACTTGGCTGTAATCAGTTATG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; fibroblasts; A549; Huh7; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq | ||
| Transcript ID List | ENST00000229795.7; ENST00000229794.8 | ||
| External Link | RMBase: m6A_site_714730 | ||
| mod ID: M6ASITE074765 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110718-36110719:+ | [14] | |
| Sequence | TATGCCGTATAGGATGTCAGACAATACCACTGGTTAAAATA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T; HeLa; A549; Huh7; HEK293A-TOA; iSLK | ||
| Seq Type List | MeRIP-seq; DART-seq; MAZTER-seq; m6A-seq | ||
| Transcript ID List | ENST00000229794.8; ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714731 | ||
| mod ID: M6ASITE074766 | Click to Show/Hide the Full List | ||
| mod site | chr6:36110861-36110862:+ | [16] | |
| Sequence | TGACCTGGCATACTTGTCTGACAGATCTTAATACTACTCCT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000229795.7 | ||
| External Link | RMBase: m6A_site_714732 | ||
References