m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00386)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
RUBCN
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
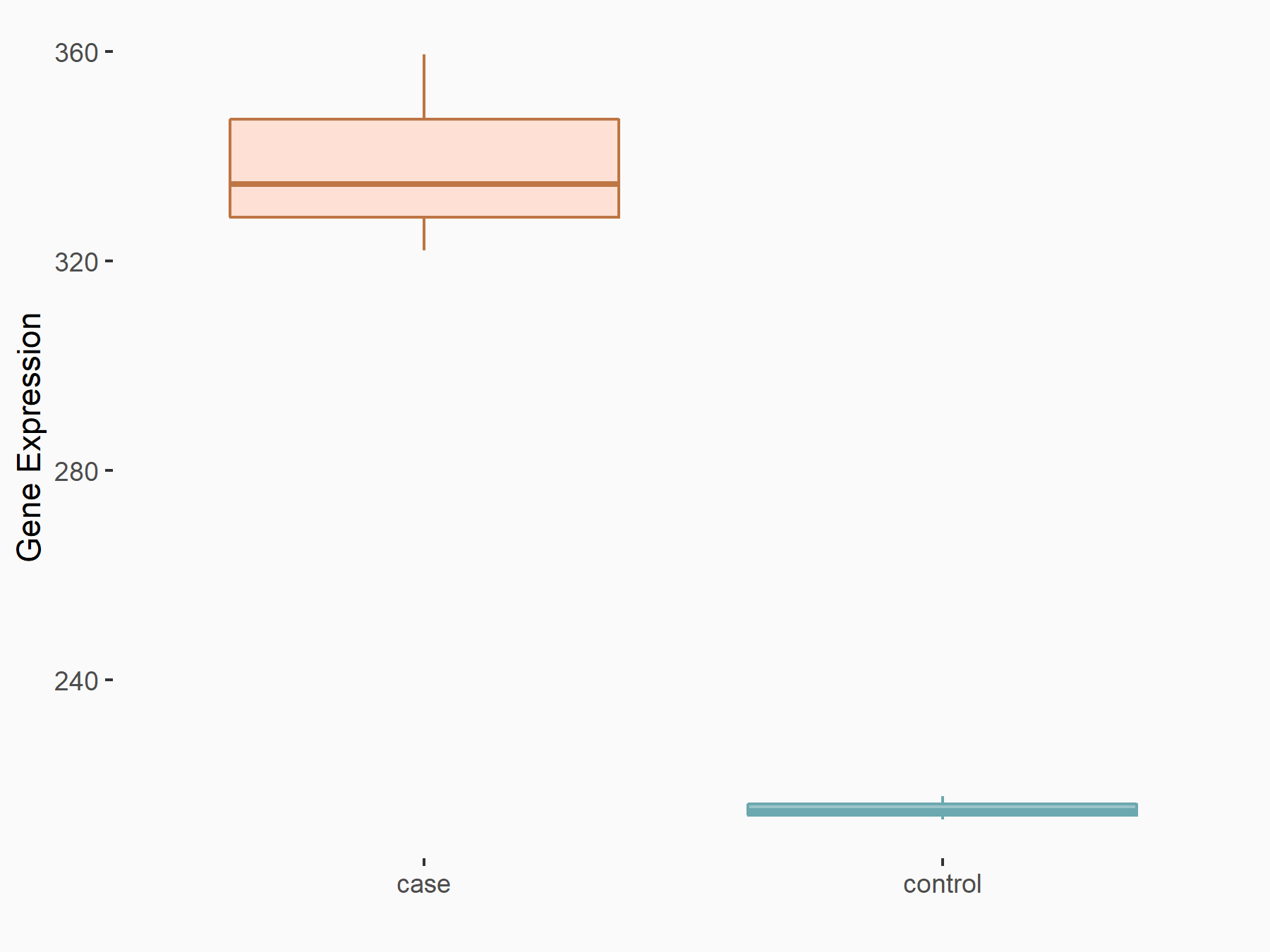 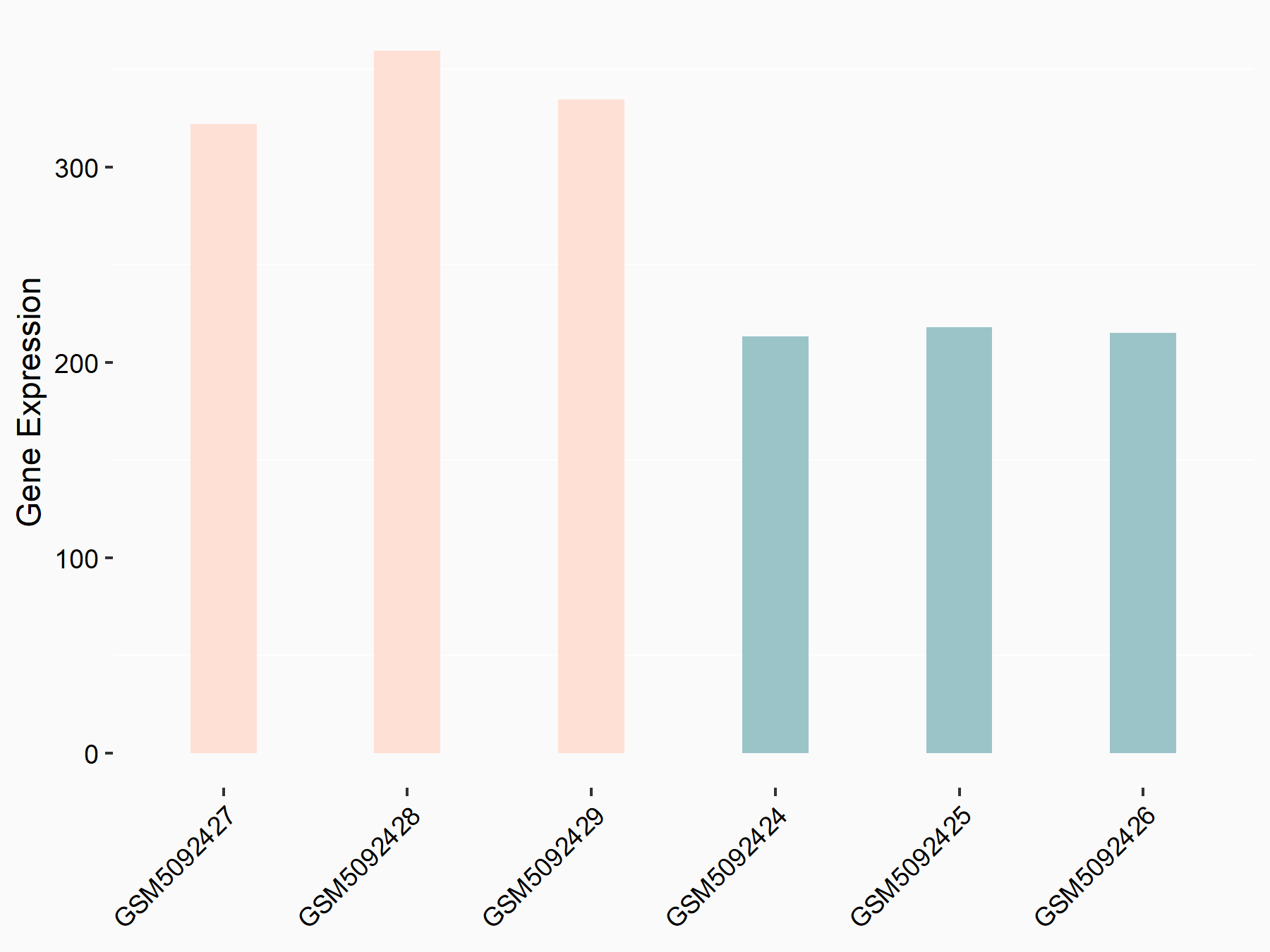 |
logFC: 6.54E-01 p-value: 1.80E-06 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | The upregulation of METTL3 and YTHDF1 induced by lipotoxicity contributes to the elevated expression level of Beclin-1 associated RUN domain containing protein (RUBCN/Rubicon) in an m6A-dependent manner, which inhibits the fusion of autophagosomes and lysosomes and further suppresses the clearance of LDs via lysosomes in nonalcoholic fatty liver disease. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-alcoholic fatty liver disease | ICD-11: DB92 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell autophagy | |||
| In-vitro Model | AML12 | Normal | Mus musculus | CVCL_0140 |
| Hepa 1-6 | Hepatocellular carcinoma of the mouse | Mus musculus | CVCL_0327 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | All mice were housed in specific pathogen-free conditions in the animal facility with constant temperature and humidity under a 12-h light/12-h dark cycle, with free access to water and food. After a week of adaptation, they were then divided into two groups randomly and fed with a HFD (60 kcal% fat, D12492, Research Diets) or standard normal chow diet (CD) for 16 weeks, respectively. | |||
YTH domain-containing family protein 1 (YTHDF1) [READER]
| Representative RIP-seq result supporting the interaction between RUBCN and the regulator | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.78E+00 | GSE63591 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | The upregulation of METTL3 and YTHDF1 induced by lipotoxicity contributes to the elevated expression level of Beclin-1 associated RUN domain containing protein (RUBCN/Rubicon) in an m6A-dependent manner, which inhibits the fusion of autophagosomes and lysosomes and further suppresses the clearance of LDs via lysosomes in nonalcoholic fatty liver disease. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-alcoholic fatty liver disease | ICD-11: DB92 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell autophagy | |||
| In-vitro Model | AML12 | Normal | Mus musculus | CVCL_0140 |
| Hepa 1-6 | Hepatocellular carcinoma of the mouse | Mus musculus | CVCL_0327 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | All mice were housed in specific pathogen-free conditions in the animal facility with constant temperature and humidity under a 12-h light/12-h dark cycle, with free access to water and food. After a week of adaptation, they were then divided into two groups randomly and fed with a HFD (60 kcal% fat, D12492, Research Diets) or standard normal chow diet (CD) for 16 weeks, respectively. | |||
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | The upregulation of METTL3 and YTHDF1 induced by lipotoxicity contributes to the elevated expression level of Beclin-1 associated RUN domain containing protein (RUBCN/Rubicon) in an m6A-dependent manner, which inhibits the fusion of autophagosomes and lysosomes and further suppresses the clearance of LDs via lysosomes in nonalcoholic fatty liver disease. | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell autophagy | |||
| In-vitro Model | AML12 | Normal | Mus musculus | CVCL_0140 |
| Hepa 1-6 | Hepatocellular carcinoma of the mouse | Mus musculus | CVCL_0327 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | All mice were housed in specific pathogen-free conditions in the animal facility with constant temperature and humidity under a 12-h light/12-h dark cycle, with free access to water and food. After a week of adaptation, they were then divided into two groups randomly and fed with a HFD (60 kcal% fat, D12492, Research Diets) or standard normal chow diet (CD) for 16 weeks, respectively. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | The upregulation of METTL3 and YTHDF1 induced by lipotoxicity contributes to the elevated expression level of Beclin-1 associated RUN domain containing protein (RUBCN/Rubicon) in an m6A-dependent manner, which inhibits the fusion of autophagosomes and lysosomes and further suppresses the clearance of LDs via lysosomes in nonalcoholic fatty liver disease. | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell autophagy | |||
| In-vitro Model | AML12 | Normal | Mus musculus | CVCL_0140 |
| Hepa 1-6 | Hepatocellular carcinoma of the mouse | Mus musculus | CVCL_0327 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | All mice were housed in specific pathogen-free conditions in the animal facility with constant temperature and humidity under a 12-h light/12-h dark cycle, with free access to water and food. After a week of adaptation, they were then divided into two groups randomly and fed with a HFD (60 kcal% fat, D12492, Research Diets) or standard normal chow diet (CD) for 16 weeks, respectively. | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00386)
| In total 5 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE000247 | Click to Show/Hide the Full List | ||
| mod site | chr3:197689055-197689056:- | [2] | |
| Sequence | TTCATCCTGGGTGACAGAGCAAGACCCTGTCTCAAAAAAAA | ||
| Transcript ID List | ENST00000471364.1; ENST00000413360.5; ENST00000273582.9; ENST00000415452.5; ENST00000296343.10; rmsk_1216476 | ||
| External Link | RMBase: RNA-editing_site_102366 | ||
| mod ID: A2ISITE000248 | Click to Show/Hide the Full List | ||
| mod site | chr3:197723384-197723385:- | [2] | |
| Sequence | GAGACTGGCCTGGCCAACATAGTGAAACCCCGTCTCTACTA | ||
| Transcript ID List | ENST00000449205.1; ENST00000474214.2; ENST00000296343.10; ENST00000467303.5; rmsk_1216552; ENST00000273582.9 | ||
| External Link | RMBase: RNA-editing_site_102367 | ||
| mod ID: A2ISITE000249 | Click to Show/Hide the Full List | ||
| mod site | chr3:197725621-197725622:- | [2] | |
| Sequence | GTCTGTAATCCTAGCACTTTAGGAGGCCGAGGCAGATGGTT | ||
| Transcript ID List | ENST00000273582.9; rmsk_1216556; ENST00000474214.2; ENST00000467303.5; ENST00000296343.10; ENST00000449205.1 | ||
| External Link | RMBase: RNA-editing_site_102368 | ||
| mod ID: A2ISITE000250 | Click to Show/Hide the Full List | ||
| mod site | chr3:197738387-197738388:- | [3] | |
| Sequence | TGAAAATACCTAAAAGCCATATATTATTTTAATATAGAGTC | ||
| Transcript ID List | ENST00000467303.5; ENST00000273582.9 | ||
| External Link | RMBase: RNA-editing_site_102369 | ||
| mod ID: A2ISITE000251 | Click to Show/Hide the Full List | ||
| mod site | chr3:197745914-197745915:- | [3] | |
| Sequence | CCTCCCCTATAGCTGAGATTACAGGTGCATGGCACCACGCC | ||
| Transcript ID List | ENST00000273582.9; ENST00000467303.5 | ||
| External Link | RMBase: RNA-editing_site_102370 | ||
5-methylcytidine (m5C)
N6-methyladenosine (m6A)
| In total 78 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE064550 | Click to Show/Hide the Full List | ||
| mod site | chr3:197669267-197669268:- | [4] | |
| Sequence | GAAAGGATATTAGTGAAAAAACTGGCAAAATCTGAATACAA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | rmsk_1216457; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625265 | ||
| mod ID: M6ASITE064551 | Click to Show/Hide the Full List | ||
| mod site | chr3:197669346-197669347:- | [4] | |
| Sequence | GCCGTCACGGACGAGAGGAGACTAAGCAGATACGATGACTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296343.10; rmsk_1216457 | ||
| External Link | RMBase: m6A_site_625266 | ||
| mod ID: M6ASITE064552 | Click to Show/Hide the Full List | ||
| mod site | chr3:197671561-197671562:- | [5] | |
| Sequence | TAAATTCCGTATTTCCAAAAACCTAACCATTAGTCACCCAG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625267 | ||
| mod ID: M6ASITE064553 | Click to Show/Hide the Full List | ||
| mod site | chr3:197671588-197671589:- | [5] | |
| Sequence | CTCCAACCAGTGTGTCTAAAACCAGCATAAATTCCGTATTT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000296343.10; rmsk_1216461; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625268 | ||
| mod ID: M6ASITE064554 | Click to Show/Hide the Full List | ||
| mod site | chr3:197672073-197672074:- | [4] | |
| Sequence | GTGTGCCGCCCGTGCTCTGAACTGCGTTTCCACCTGCTGTG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625269 | ||
| mod ID: M6ASITE064556 | Click to Show/Hide the Full List | ||
| mod site | chr3:197672203-197672204:- | [4] | |
| Sequence | ACATAGCAGAAGAGTGGAAGACAAATATTTTCCACTTAAAT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625270 | ||
| mod ID: M6ASITE064557 | Click to Show/Hide the Full List | ||
| mod site | chr3:197672259-197672260:- | [6] | |
| Sequence | AGTAAAGGGAAAGAAAAAAAACATAGTTTGCTTACCAGGTG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | CD34 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625271 | ||
| mod ID: M6ASITE064558 | Click to Show/Hide the Full List | ||
| mod site | chr3:197672330-197672331:- | [7] | |
| Sequence | CGGGGAGAAGCTTTGAATGCACTGAAGAGAATTCCTTCTAA | ||
| Motif Score | 3.252583333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625272 | ||
| mod ID: M6ASITE064559 | Click to Show/Hide the Full List | ||
| mod site | chr3:197672474-197672475:- | [7] | |
| Sequence | CGTCTGTTTTAGTGACTCTGACACGATGCCGTCCTCACCTT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625273 | ||
| mod ID: M6ASITE064560 | Click to Show/Hide the Full List | ||
| mod site | chr3:197672551-197672552:- | [7] | |
| Sequence | AACAGTCTCATAGGCTCCTCACTTGTCACTTATTTTTCCCT | ||
| Motif Score | 2.469291667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625274 | ||
| mod ID: M6ASITE064561 | Click to Show/Hide the Full List | ||
| mod site | chr3:197672570-197672571:- | [7] | |
| Sequence | AGCTTCTGAAGCCGAACTCAACAGTCTCATAGGCTCCTCAC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625275 | ||
| mod ID: M6ASITE064562 | Click to Show/Hide the Full List | ||
| mod site | chr3:197672575-197672576:- | [7] | |
| Sequence | CTGACAGCTTCTGAAGCCGAACTCAACAGTCTCATAGGCTC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625276 | ||
| mod ID: M6ASITE064563 | Click to Show/Hide the Full List | ||
| mod site | chr3:197672996-197672997:- | [7] | |
| Sequence | CCAGTTTTGGTTTAAATTACACATCCAGGCCAGGCACAGTG | ||
| Motif Score | 2.084928571 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625277 | ||
| mod ID: M6ASITE064564 | Click to Show/Hide the Full List | ||
| mod site | chr3:197672998-197672999:- | [7] | |
| Sequence | TTCCAGTTTTGGTTTAAATTACACATCCAGGCCAGGCACAG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625278 | ||
| mod ID: M6ASITE064565 | Click to Show/Hide the Full List | ||
| mod site | chr3:197673020-197673021:- | [7] | |
| Sequence | AGTTTATAAAAATGAGCTCTACTTCCAGTTTTGGTTTAAAT | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625279 | ||
| mod ID: M6ASITE064567 | Click to Show/Hide the Full List | ||
| mod site | chr3:197673271-197673272:- | [4] | |
| Sequence | GTGAGAGGGAACATGTGGGAACTGGCCCCTGCCCTTGATTC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625280 | ||
| mod ID: M6ASITE064568 | Click to Show/Hide the Full List | ||
| mod site | chr3:197673281-197673282:- | [4] | |
| Sequence | CTCCACCAGCGTGAGAGGGAACATGTGGGAACTGGCCCCTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625281 | ||
| mod ID: M6ASITE064569 | Click to Show/Hide the Full List | ||
| mod site | chr3:197673523-197673524:- | [4] | |
| Sequence | AAGAGCTGGCAGCTCTGTGAACTAAAGCCTGCATCATTTTT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625282 | ||
| mod ID: M6ASITE064570 | Click to Show/Hide the Full List | ||
| mod site | chr3:197673564-197673565:- | [7] | |
| Sequence | CTTCTAACCTCTGTGAAAACACTGGTCAGAGTGGCTTCTCC | ||
| Motif Score | 2.506922619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625283 | ||
| mod ID: M6ASITE064571 | Click to Show/Hide the Full List | ||
| mod site | chr3:197673566-197673567:- | [4] | |
| Sequence | AGCTTCTAACCTCTGTGAAAACACTGGTCAGAGTGGCTTCT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625284 | ||
| mod ID: M6ASITE064572 | Click to Show/Hide the Full List | ||
| mod site | chr3:197673644-197673645:- | [7] | |
| Sequence | CCCCGTTAATGTGTGTGTTGACAGTGAAGTCCTTGGGTGGG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625285 | ||
| mod ID: M6ASITE064573 | Click to Show/Hide the Full List | ||
| mod site | chr3:197673834-197673835:- | [8] | |
| Sequence | GTCCCTGTGTTCTTGGGATAACAAGAGCCTTTATGCCAATT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625286 | ||
| mod ID: M6ASITE064574 | Click to Show/Hide the Full List | ||
| mod site | chr3:197673879-197673880:- | [7] | |
| Sequence | TAGTGCTGTTACAGAACTTGACGGTTTGTGGATGTGAGTGT | ||
| Motif Score | 2.833690476 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625287 | ||
| mod ID: M6ASITE064575 | Click to Show/Hide the Full List | ||
| mod site | chr3:197673884-197673885:- | [4] | |
| Sequence | GCTATTAGTGCTGTTACAGAACTTGACGGTTTGTGGATGTG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625288 | ||
| mod ID: M6ASITE064576 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674092-197674093:- | [4] | |
| Sequence | ATCCTAAGGTGAGGACAGGAACCTCACCCACCATCTTCTTA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000413360.5; ENST00000296343.10; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625289 | ||
| mod ID: M6ASITE064578 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674098-197674099:- | [4] | |
| Sequence | GATGCCATCCTAAGGTGAGGACAGGAACCTCACCCACCATC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000413360.5; ENST00000296343.10; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625290 | ||
| mod ID: M6ASITE064579 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674215-197674216:- | [7] | |
| Sequence | CTGTCGCTGAAGGAGCGCTTACTCAGAGGAGCGGTCGGCCC | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000413360.5; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625291 | ||
| mod ID: M6ASITE064580 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674264-197674265:- | [7] | |
| Sequence | CCAGGCTCACCTGTAGCCTTACGCCGCAAGCATACGTGAGG | ||
| Motif Score | 2.046785714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000413360.5; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625292 | ||
| mod ID: M6ASITE064581 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674276-197674277:- | [7] | |
| Sequence | CTGATGCTCAGGCCAGGCTCACCTGTAGCCTTACGCCGCAA | ||
| Motif Score | 2.026654762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000413360.5; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625293 | ||
| mod ID: M6ASITE064582 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674297-197674298:- | [4] | |
| Sequence | ATCTTGAAGCAAAGAACAGAACTGATGCTCAGGCCAGGCTC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000413360.5; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625294 | ||
| mod ID: M6ASITE064583 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674302-197674303:- | [4] | |
| Sequence | TGGTGATCTTGAAGCAAAGAACAGAACTGATGCTCAGGCCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000413360.5; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625295 | ||
| mod ID: M6ASITE064584 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674422-197674423:- | [4] | |
| Sequence | CTATCCTAGACTCCTCAGAGACCTCAGAGGCAGCTGTGAAT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1B; H1A; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000296343.10; ENST00000413360.5; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625296 | ||
| mod ID: M6ASITE064585 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674433-197674434:- | [4] | |
| Sequence | AACAATCCTGACTATCCTAGACTCCTCAGAGACCTCAGAGG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1B; H1A; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000413360.5; ENST00000273582.9; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625297 | ||
| mod ID: M6ASITE064586 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674443-197674444:- | [9] | |
| Sequence | GGGGCCACAGAACAATCCTGACTATCCTAGACTCCTCAGAG | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000273582.9; ENST00000413360.5; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625298 | ||
| mod ID: M6ASITE064587 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674452-197674453:- | [4] | |
| Sequence | GGTATTTTGGGGGCCACAGAACAATCCTGACTATCCTAGAC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000296343.10; ENST00000273582.9; ENST00000413360.5 | ||
| External Link | RMBase: m6A_site_625299 | ||
| mod ID: M6ASITE064589 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674504-197674505:- | [4] | |
| Sequence | GAGAAGCAGCAGAGAATAGGACTACGTCATGGGCATTTCGT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000401223.1; MIMAT0004972; ENST00000273582.9; ENST00000296343.10; ENST00000413360.5 | ||
| External Link | RMBase: m6A_site_625300 | ||
| mod ID: M6ASITE064590 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674537-197674538:- | [4] | |
| Sequence | CTGTCCTGGACTGGGGTCAGACTGTGCCCCGAGGAGAAGCA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1B; H1A; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000296343.10; ENST00000413360.5; ENST00000273582.9; ENST00000401223.1 | ||
| External Link | RMBase: m6A_site_625301 | ||
| mod ID: M6ASITE064591 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674548-197674549:- | [4] | |
| Sequence | TTTCCCTCTCCCTGTCCTGGACTGGGGTCAGACTGTGCCCC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1B; H1A; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000296343.10; ENST00000413360.5; ENST00000273582.9; ENST00000401223.1 | ||
| External Link | RMBase: m6A_site_625302 | ||
| mod ID: M6ASITE064592 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674716-197674717:- | [4] | |
| Sequence | TCAGTGGAAGAGACAGCAAGACCAAGTTCTTCCAGCGTCTG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; A549; GM12878; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000413360.5; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625303 | ||
| mod ID: M6ASITE064593 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674724-197674725:- | [4] | |
| Sequence | TGCCTCTGTCAGTGGAAGAGACAGCAAGACCAAGTTCTTCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; A549; GM12878; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000273582.9; ENST00000413360.5 | ||
| External Link | RMBase: m6A_site_625304 | ||
| mod ID: M6ASITE064594 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674777-197674778:- | [4] | |
| Sequence | CACCAGGTGTTTCCATCAGAACTGACATGCGGGTCCTTCAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; U2OS; A549; GM12878; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000273582.9; ENST00000413360.5 | ||
| External Link | RMBase: m6A_site_625305 | ||
| mod ID: M6ASITE064595 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674798-197674799:- | [4] | |
| Sequence | TGGTGACAGATAATTTGTAAACACCAGGTGTTTCCATCAGA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; U2OS; A549; GM12878 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000413360.5; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625306 | ||
| mod ID: M6ASITE064596 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674813-197674814:- | [7] | |
| Sequence | TGGGTCCAGAGTCTGTGGTGACAGATAATTTGTAAACACCA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000296343.10; ENST00000413360.5 | ||
| External Link | RMBase: m6A_site_625307 | ||
| mod ID: M6ASITE064597 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674947-197674948:- | [4] | |
| Sequence | ACTGAGCCACTCTTTGAAGGACTCGCCCCACCTGGGGCTTC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; H1A; H1B; GM12878; LCLs; MM6; Jurkat; CD4T; GSC-11; HEK293A-TOA; iSLK; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000296343.10; ENST00000413360.5 | ||
| External Link | RMBase: m6A_site_625308 | ||
| mod ID: M6ASITE064598 | Click to Show/Hide the Full List | ||
| mod site | chr3:197674967-197674968:- | [4] | |
| Sequence | CGGGTCACACCTGTTGCAGAACTGAGCCACTCTTTGAAGGA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; H1A; H1B; GM12878; LCLs; MM6; Jurkat; CD4T; GSC-11; HEK293A-TOA; iSLK; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000413360.5; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625309 | ||
| mod ID: M6ASITE064600 | Click to Show/Hide the Full List | ||
| mod site | chr3:197675078-197675079:- | [4] | |
| Sequence | CCTGGAGTCTTACCTGTCAGACTACGAGGAGGAGCCCGCGG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HEK293A-TOA; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000296343.10; ENST00000413360.5 | ||
| External Link | RMBase: m6A_site_625310 | ||
| mod ID: M6ASITE064601 | Click to Show/Hide the Full List | ||
| mod site | chr3:197676993-197676994:- | [10] | |
| Sequence | CCCAGGCCACCTGACAGAGGACCTCCACCTGTACTCACTGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000413360.5; ENST00000273582.9; ENST00000296343.10; ENST00000415452.5 | ||
| External Link | RMBase: m6A_site_625311 | ||
| mod ID: M6ASITE064602 | Click to Show/Hide the Full List | ||
| mod site | chr3:197681222-197681223:- | [4] | |
| Sequence | CGTCAGCAACTTCTCCAAGGACCTGCTCATTAAGATCTGGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000413360.5; ENST00000273582.9; ENST00000415452.5; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625312 | ||
| mod ID: M6ASITE064603 | Click to Show/Hide the Full List | ||
| mod site | chr3:197681258-197681259:- | [4] | |
| Sequence | CCGGGTTCTGCGCAAGTGGGACTTCAGCAAGTACTACGTCA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000413360.5; ENST00000415452.5; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625313 | ||
| mod ID: M6ASITE064604 | Click to Show/Hide the Full List | ||
| mod site | chr3:197684198-197684199:- | [4] | |
| Sequence | AGATGCTGACATCAGAAGGAACACAGCCTCAAGCAGCAAAT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000471364.1; ENST00000413360.5; ENST00000415452.5; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625314 | ||
| mod ID: M6ASITE064605 | Click to Show/Hide the Full List | ||
| mod site | chr3:197691104-197691105:- | [4] | |
| Sequence | ACTCACATATCAAAGAACGGACTGTCAGTGTCACTGGCCTC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000296343.10; ENST00000415452.5; ENST00000413360.5; ENST00000471364.1 | ||
| External Link | RMBase: m6A_site_625315 | ||
| mod ID: M6ASITE064606 | Click to Show/Hide the Full List | ||
| mod site | chr3:197695893-197695894:- | [4] | |
| Sequence | CAGCTACCTCTCTGAGCAAGACTTCGGCAGCTGTGCCGACC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000415452.5; ENST00000447048.1; ENST00000273582.9; ENST00000413360.5; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625316 | ||
| mod ID: M6ASITE064607 | Click to Show/Hide the Full List | ||
| mod site | chr3:197695925-197695926:- | [4] | |
| Sequence | TTCCGAAGACCATCAGAAGGACAGTCCCTCATCAGCTACCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000447048.1; ENST00000413360.5; ENST00000273582.9; ENST00000415452.5; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625317 | ||
| mod ID: M6ASITE064608 | Click to Show/Hide the Full List | ||
| mod site | chr3:197695937-197695938:- | [4] | |
| Sequence | GGGGAAGGCATGTTCCGAAGACCATCAGAAGGACAGTCCCT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000447048.1; ENST00000413360.5; ENST00000273582.9; ENST00000415452.5 | ||
| External Link | RMBase: m6A_site_625318 | ||
| mod ID: M6ASITE064609 | Click to Show/Hide the Full List | ||
| mod site | chr3:197696964-197696965:- | [8] | |
| Sequence | CAGCACACCCAGCTCTCTGTACATGGAATATGGTGAGTACT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000415452.5; ENST00000413360.5; ENST00000447048.1; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625319 | ||
| mod ID: M6ASITE064611 | Click to Show/Hide the Full List | ||
| mod site | chr3:197700693-197700694:- | [4] | |
| Sequence | CCAGCTTCTCAGAGGGGCAGACACTCACTGTCACCAGTGGG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000296343.10; ENST00000273582.9; ENST00000413360.5; ENST00000447048.1; ENST00000415452.5; ENST00000449205.1 | ||
| External Link | RMBase: m6A_site_625320 | ||
| mod ID: M6ASITE064612 | Click to Show/Hide the Full List | ||
| mod site | chr3:197700761-197700762:- | [4] | |
| Sequence | TGCTGCCTCTTCCTTAGGGGACCAGGAAGGAGGTGGGGAGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; Jurkat; CD4T; peripheral-blood; MSC; iSLK; endometrial; HEC-1-A; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000415452.5; ENST00000413360.5; ENST00000296343.10; ENST00000449205.1; ENST00000447048.1 | ||
| External Link | RMBase: m6A_site_625321 | ||
| mod ID: M6ASITE064613 | Click to Show/Hide the Full List | ||
| mod site | chr3:197700867-197700868:- | [4] | |
| Sequence | ACTTGGACCCCCGAGGCCGGACTGCCAGCTGTCAGAGTCAC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; H1B; fibroblasts; A549; GM12878; LCLs; Jurkat; CD4T; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000296343.10; ENST00000415452.5; ENST00000449205.1; ENST00000413360.5; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625322 | ||
| mod ID: M6ASITE064614 | Click to Show/Hide the Full List | ||
| mod site | chr3:197700881-197700882:- | [4] | |
| Sequence | CCTGGCCATTGGCAACTTGGACCCCCGAGGCCGGACTGCCA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; H1B; fibroblasts; A549; GM12878; LCLs; Jurkat; CD4T; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000415452.5; ENST00000273582.9; ENST00000413360.5; ENST00000296343.10; ENST00000449205.1 | ||
| External Link | RMBase: m6A_site_625323 | ||
| mod ID: M6ASITE064615 | Click to Show/Hide the Full List | ||
| mod site | chr3:197701059-197701060:- | [4] | |
| Sequence | TCTCACCAGCAGAGGATCAAACCATCCAAGCCCCCCCAGTT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HEK293T; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000413360.5; ENST00000449205.1; ENST00000273582.9; ENST00000296343.10; ENST00000415452.5 | ||
| External Link | RMBase: m6A_site_625324 | ||
| mod ID: M6ASITE064616 | Click to Show/Hide the Full List | ||
| mod site | chr3:197701102-197701103:- | [4] | |
| Sequence | CTCTCTGGCCCTCCCCGGAAACCTCAAGAAAGCAGAGGGCA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000449205.1; ENST00000413360.5; ENST00000415452.5; ENST00000296343.10; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625325 | ||
| mod ID: M6ASITE064617 | Click to Show/Hide the Full List | ||
| mod site | chr3:197701778-197701779:- | [4] | |
| Sequence | TTCCACATACACCCCTCCAAACAGCTATGCTCAGCATTCCT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000449205.1; ENST00000413360.5; ENST00000467303.5; ENST00000296343.10 | ||
| External Link | RMBase: m6A_site_625326 | ||
| mod ID: M6ASITE064618 | Click to Show/Hide the Full List | ||
| mod site | chr3:197703584-197703585:- | [4] | |
| Sequence | CCTGGAAGCAGTGGAACAGAACAACCCCCGCCTCCTGGCTC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000449205.1; ENST00000467303.5; ENST00000413360.5; ENST00000296343.10; ENST00000273582.9; ENST00000474214.2 | ||
| External Link | RMBase: m6A_site_625327 | ||
| mod ID: M6ASITE064619 | Click to Show/Hide the Full List | ||
| mod site | chr3:197703589-197703590:- | [4] | |
| Sequence | CAGTGCCTGGAAGCAGTGGAACAGAACAACCCCCGCCTCCT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000449205.1; ENST00000296343.10; ENST00000273582.9; ENST00000413360.5; ENST00000474214.2; ENST00000467303.5 | ||
| External Link | RMBase: m6A_site_625328 | ||
| mod ID: M6ASITE064620 | Click to Show/Hide the Full List | ||
| mod site | chr3:197718016-197718017:- | [4] | |
| Sequence | CTTGGAGCGGCTTTGCAGGGACATGCAGAGCATCCTCTATC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000449205.1; ENST00000413360.5; ENST00000296343.10; ENST00000273582.9; ENST00000474214.2; ENST00000467303.5 | ||
| External Link | RMBase: m6A_site_625329 | ||
| mod ID: M6ASITE064622 | Click to Show/Hide the Full List | ||
| mod site | chr3:197748234-197748235:- | [11] | |
| Sequence | GACTGCTAAAAACTAGAGAAACTGGCTCACGCCTGTCATCC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | GM12878; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000472149.1; ENST00000467303.5; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625331 | ||
| mod ID: M6ASITE064623 | Click to Show/Hide the Full List | ||
| mod site | chr3:197748243-197748244:- | [11] | |
| Sequence | ATTTGTCTGGACTGCTAAAAACTAGAGAAACTGGCTCACGC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | GM12878; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000472149.1; ENST00000467303.5; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625332 | ||
| mod ID: M6ASITE064624 | Click to Show/Hide the Full List | ||
| mod site | chr3:197748253-197748254:- | [11] | |
| Sequence | AAGCCCCTTCATTTGTCTGGACTGCTAAAAACTAGAGAAAC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | GM12878; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000467303.5; ENST00000472149.1; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625333 | ||
| mod ID: M6ASITE064625 | Click to Show/Hide the Full List | ||
| mod site | chr3:197748351-197748352:- | [12] | |
| Sequence | TTGGACTCGGAAACTCAAGAACAATATTTGGCATGTCAAGG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; GM12878 | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000472149.1; ENST00000467303.5 | ||
| External Link | RMBase: m6A_site_625334 | ||
| mod ID: M6ASITE064626 | Click to Show/Hide the Full List | ||
| mod site | chr3:197748359-197748360:- | [12] | |
| Sequence | AGGAAAGTTTGGACTCGGAAACTCAAGAACAATATTTGGCA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; GM12878 | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000472149.1; ENST00000467303.5 | ||
| External Link | RMBase: m6A_site_625335 | ||
| mod ID: M6ASITE064627 | Click to Show/Hide the Full List | ||
| mod site | chr3:197748367-197748368:- | [12] | |
| Sequence | AATAGTGGAGGAAAGTTTGGACTCGGAAACTCAAGAACAAT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; GM12878 | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000467303.5; ENST00000472149.1 | ||
| External Link | RMBase: m6A_site_625336 | ||
| mod ID: M6ASITE064628 | Click to Show/Hide the Full List | ||
| mod site | chr3:197748392-197748393:- | [12] | |
| Sequence | CCTATCAAGGGGCACAAGAAACTTTAATAGTGGAGGAAAGT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; GM12878 | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000467303.5; ENST00000472149.1; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625337 | ||
| mod ID: M6ASITE064629 | Click to Show/Hide the Full List | ||
| mod site | chr3:197748420-197748421:- | [11] | |
| Sequence | TAACAATATAGGAAGAGTGAACCACACACCTATCAAGGGGC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | GM12878 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000467303.5; ENST00000273582.9; ENST00000472149.1 | ||
| External Link | RMBase: m6A_site_625338 | ||
| mod ID: M6ASITE064630 | Click to Show/Hide the Full List | ||
| mod site | chr3:197749289-197749290:- | [11] | |
| Sequence | GTGTTCACTACCCTAATAAGACCTCAGCTTCCCTCGGGAGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | GM12878 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000472149.1; ENST00000467303.5; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625339 | ||
| mod ID: M6ASITE064632 | Click to Show/Hide the Full List | ||
| mod site | chr3:197749321-197749322:- | [11] | |
| Sequence | TCTCAATCGGCACTCTCAAAACATCTGGCACGGTGTTCACT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | GM12878 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000472149.1; ENST00000467303.5 | ||
| External Link | RMBase: m6A_site_625340 | ||
| mod ID: M6ASITE064633 | Click to Show/Hide the Full List | ||
| mod site | chr3:197749519-197749520:- | [11] | |
| Sequence | ACGCAGCCTCGGGAGGCAGGACAGACCCCTGCGAGTGTAGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | GM12878 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000467303.5 | ||
| External Link | RMBase: m6A_site_625341 | ||
| mod ID: M6ASITE064634 | Click to Show/Hide the Full List | ||
| mod site | chr3:197749587-197749588:- | [11] | |
| Sequence | TCGAGTCCCTCCTGCTGCAGACAGGTCCCCTATACTTCGGT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | GM12878 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000467303.5; ENST00000273582.9 | ||
| External Link | RMBase: m6A_site_625342 | ||
| mod ID: M6ASITE064635 | Click to Show/Hide the Full List | ||
| mod site | chr3:197749660-197749661:- | [11] | |
| Sequence | CCCAAACTGGGGGATGCTAGACCGCGGGCACCTCTGAATCT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | GM12878; Jurkat | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000273582.9; ENST00000467303.5 | ||
| External Link | RMBase: m6A_site_625343 | ||
References