m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00159)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
EIF4EBP1
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shMETTL3 MOLM13 cells
Control: MOLM13 cells
|
GSE98623 | |
| Regulation |
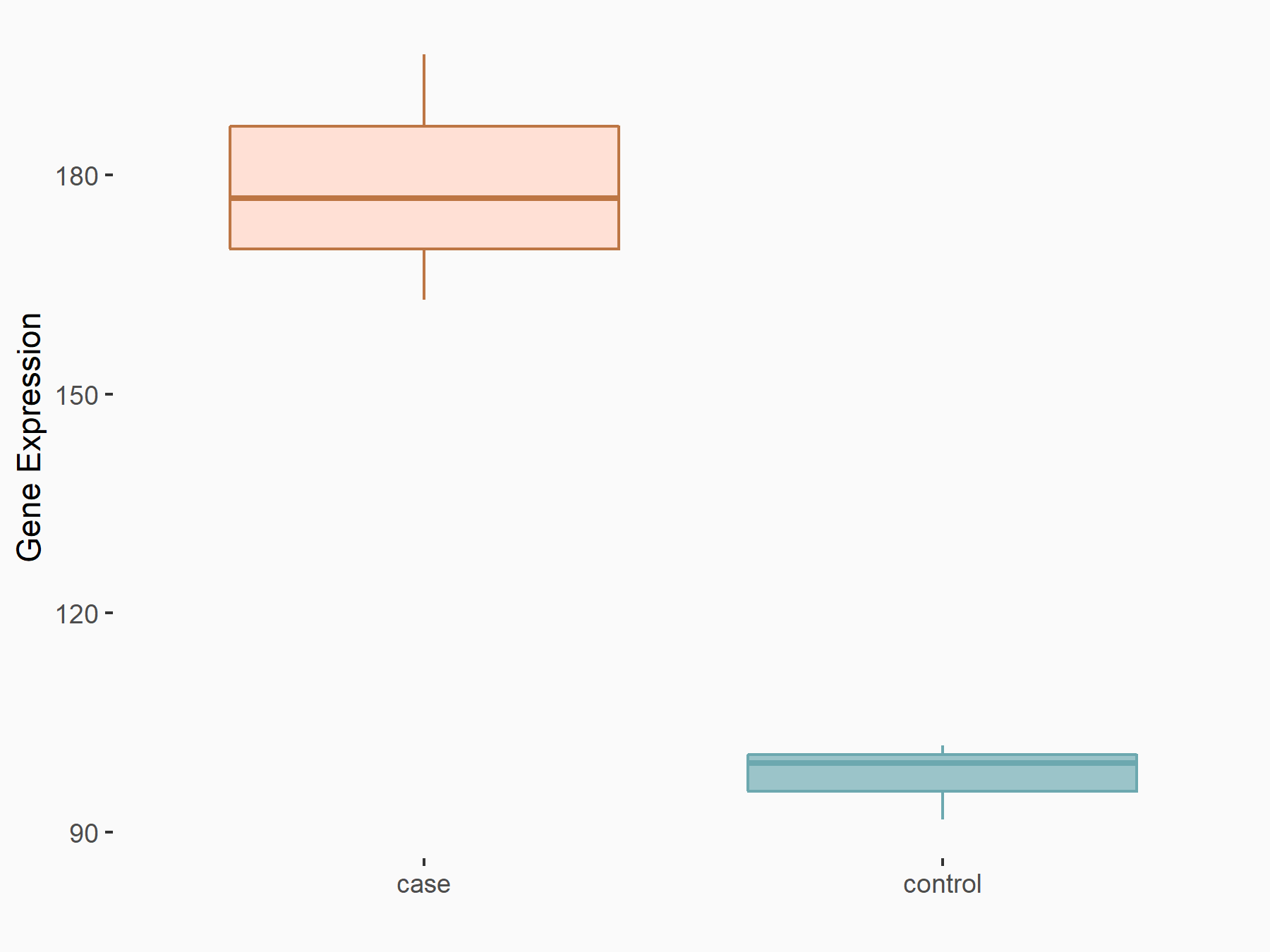 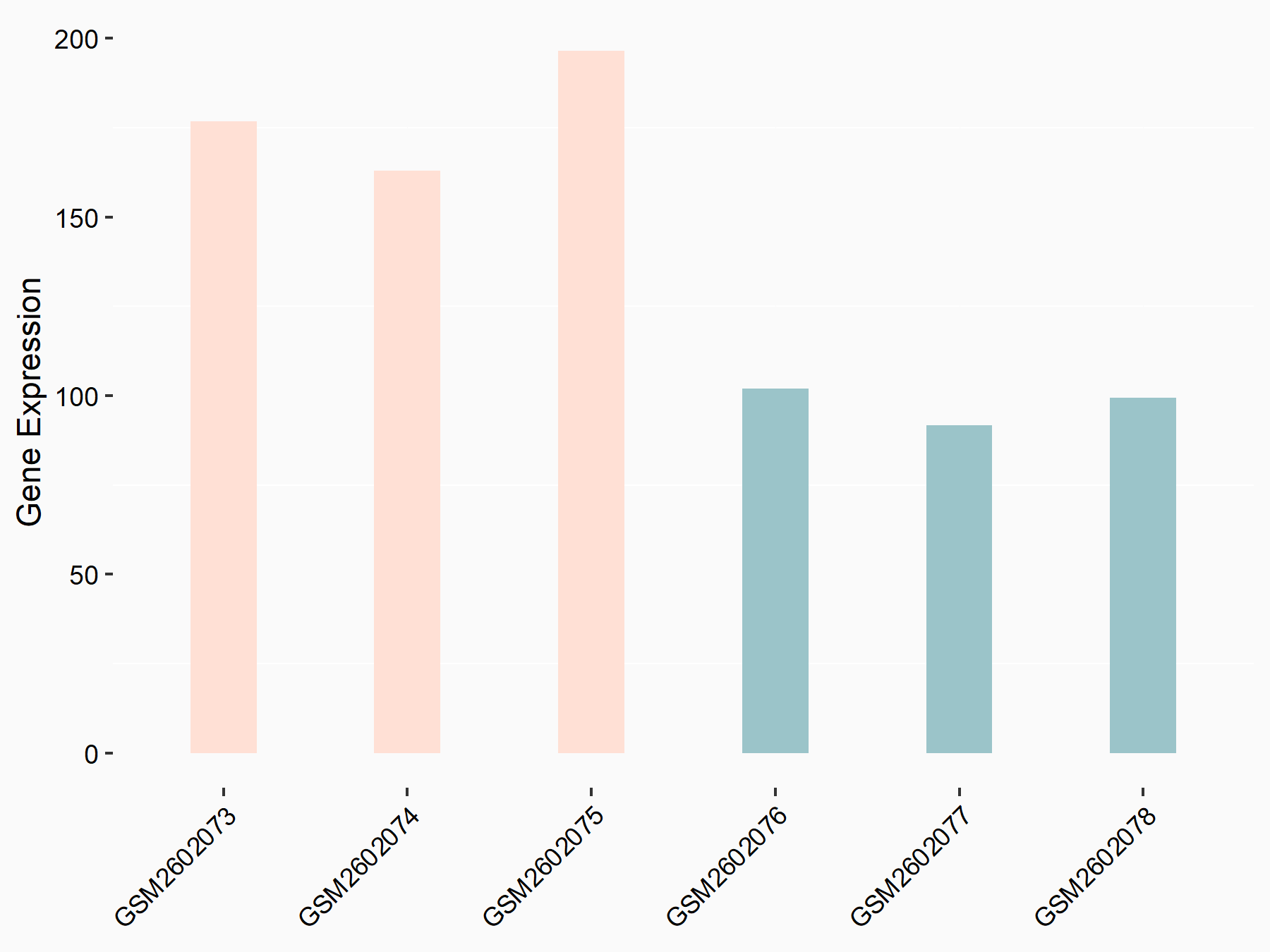 |
logFC: 8.71E-01 p-value: 3.32E-08 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between EIF4EBP1 and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 9.71E+00 | GSE60213 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through PI3K/AKT/mTOR pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-P70S6K/eIF4E-binding protein 1 (4EBP1/EIF4EBP1) pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Retinoblastoma | ICD-11: 2D02.2 | ||
| Responsed Drug | Rapamycin | Approved | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| In-vitro Model | WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
RNA demethylase ALKBH5 (ALKBH5) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by ALKBH5 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shALKBH5 MOLM-13 cells
Control: shNS MOLM-13 cells
|
GSE144968 | |
| Regulation |
  |
logFC: -7.12E-01 p-value: 2.44E-03 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | Bioactive peptides can inhibit acute myeloid leukemia cell proliferation by downregulating ALKBH5-mediated m6A demethylation of eIF4E-binding protein 1 (4EBP1/EIF4EBP1) and MLST8 mRNAs, which have potential to prevent and treat this disease. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Acute myeloid leukaemia | ICD-11: 2A60 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 |
| HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | HL-60 cells (1 × 107) suspended in 0.1 ml PBS containing 50% Matrigel were subcutaneously injected into the flanks of the mice. When tumor sizes reached 200 mm3, the mice were randomly distributed into four groups with the indicated dosages of saline, cytarabine and BP alone or in combination. For BP injections, the solution was delivered intraperitoneally at 106 ug/kg body weight for the first 8 consecutive days. For cytarabine injections, the solution was delivered intraperitoneally at 100 mg/kg body weight three times (once every three days). The combination group was administered intraperitoneally three times (once every three days) with the same dosages as described above. The control group was treated with an equivalent amount of saline. | |||
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | Bioactive peptides can inhibit acute myeloid leukemia cell proliferation by downregulating ALKBH5-mediated m6A demethylation of eIF4E-binding protein 1 (4EBP1/EIF4EBP1) and MLST8 mRNAs, which have potential to prevent and treat this disease. | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 |
| HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | HL-60 cells (1 × 107) suspended in 0.1 ml PBS containing 50% Matrigel were subcutaneously injected into the flanks of the mice. When tumor sizes reached 200 mm3, the mice were randomly distributed into four groups with the indicated dosages of saline, cytarabine and BP alone or in combination. For BP injections, the solution was delivered intraperitoneally at 106 ug/kg body weight for the first 8 consecutive days. For cytarabine injections, the solution was delivered intraperitoneally at 100 mg/kg body weight three times (once every three days). The combination group was administered intraperitoneally three times (once every three days) with the same dosages as described above. The control group was treated with an equivalent amount of saline. | |||
Retina cancer [ICD-11: 2D02]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through PI3K/AKT/mTOR pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-P70S6K/eIF4E-binding protein 1 (4EBP1/EIF4EBP1) pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
| Responsed Disease | Retinoblastoma [ICD-11: 2D02.2] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Rapamycin | Approved | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| In-vitro Model | WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
Rapamycin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through PI3K/AKT/mTOR pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-P70S6K/eIF4E-binding protein 1 (4EBP1/EIF4EBP1) pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Retinoblastoma | ICD-11: 2D02.2 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| In-vitro Model | WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: RNA demethylase ALKBH5 (ALKBH5)
| In total 5 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03257 | ||
| Epigenetic Regulator | Lysine-specific demethylase 4C (KDM4C) | |
| Regulated Target | Histone H3 lysine 9 trimethylation (H3K9me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT03261 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase 2A (KMT2A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT03265 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase 2C (KMT2C) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT03269 | ||
| Epigenetic Regulator | Lysine-specific demethylase 4B (KDM4B) | |
| Regulated Target | Histone H3 lysine 9 trimethylation (H3K9me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT03273 | ||
| Epigenetic Regulator | Probable JmjC domain-containing histone demethylation protein 2C (JMJD1C) | |
| Regulated Target | Histone H3 lysine 9 trimethylation (H3K9me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Acute myeloid leukaemia | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00159)
| In total 4 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE002517 | Click to Show/Hide the Full List | ||
| mod site | chr8:38041330-38041331:+ | [3] | |
| Sequence | CAGGCTGGTCTCGAACTTCTAACCTCAGGCGATCTGCCTGC | ||
| Transcript ID List | ENST00000338825.5 | ||
| External Link | RMBase: RNA-editing_site_130352 | ||
| mod ID: A2ISITE002518 | Click to Show/Hide the Full List | ||
| mod site | chr8:38041331-38041332:+ | [3] | |
| Sequence | AGGCTGGTCTCGAACTTCTAACCTCAGGCGATCTGCCTGCC | ||
| Transcript ID List | ENST00000338825.5 | ||
| External Link | RMBase: RNA-editing_site_130353 | ||
| mod ID: A2ISITE002519 | Click to Show/Hide the Full List | ||
| mod site | chr8:38042848-38042849:+ | [3] | |
| Sequence | CAAGGCGGGCAGATCGCTTGAGGTCAGGAGTTCAAGACCAG | ||
| Transcript ID List | ENST00000338825.5; rmsk_2492715 | ||
| External Link | RMBase: RNA-editing_site_130354 | ||
| mod ID: A2ISITE002520 | Click to Show/Hide the Full List | ||
| mod site | chr8:38053342-38053343:+ | [3] | |
| Sequence | TTTTGTTTTGTTTTGTTTTGAGACAGGGTCTTGTTCTGTCG | ||
| Transcript ID List | ENST00000338825.5; ENST00000520657.1 | ||
| External Link | RMBase: RNA-editing_site_130355 | ||
N6-methyladenosine (m6A)
| In total 16 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE084562 | Click to Show/Hide the Full List | ||
| mod site | chr8:38030570-38030571:+ | [4] | |
| Sequence | GCGGGTGCAGCGCACAGGAGACCATGTCCGGGGGCAGCAGC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; A549; H1B; H1A; hESCs; fibroblasts; H1299; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000338825.5 | ||
| External Link | RMBase: m6A_site_793852 | ||
| mod ID: M6ASITE084563 | Click to Show/Hide the Full List | ||
| mod site | chr8:38030600-38030601:+ | [4] | |
| Sequence | GGGGCAGCAGCTGCAGCCAGACCCCAAGCCGGGCCATCCCC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; A549; hESCs; fibroblasts; H1299; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000338825.5 | ||
| External Link | RMBase: m6A_site_793853 | ||
| mod ID: M6ASITE084564 | Click to Show/Hide the Full List | ||
| mod site | chr8:38030670-38030671:+ | [4] | |
| Sequence | CGTGCAGCTCCCGCCCGGGGACTACAGCACGACCCCCGGCG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; A549; fibroblasts; H1299; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000338825.5 | ||
| External Link | RMBase: m6A_site_793854 | ||
| mod ID: M6ASITE084565 | Click to Show/Hide the Full List | ||
| mod site | chr8:38047375-38047376:+ | [4] | |
| Sequence | GAAAAAAGGATTGAGCAAAGACCGGCAAATATGTGGGTAAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; A549; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; MSC; TREX; iSLK; TIME; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000520657.1; ENST00000338825.5 | ||
| External Link | RMBase: m6A_site_793855 | ||
| mod ID: M6ASITE084566 | Click to Show/Hide the Full List | ||
| mod site | chr8:38057125-38057126:+ | [4] | |
| Sequence | ATTCCTGATGGAGTGTCGGAACTCACCTGTGACCAAAACAC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; A549; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; TREX; iSLK; MSC; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000338825.5; ENST00000520657.1 | ||
| External Link | RMBase: m6A_site_793856 | ||
| mod ID: M6ASITE084567 | Click to Show/Hide the Full List | ||
| mod site | chr8:38057142-38057143:+ | [4] | |
| Sequence | GGAACTCACCTGTGACCAAAACACCCCCAAGGGATCTGCCC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; A549; hESC-HEK293T; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; TREX; iSLK; MSC; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000338825.5; ENST00000520657.1 | ||
| External Link | RMBase: m6A_site_793857 | ||
| mod ID: M6ASITE084568 | Click to Show/Hide the Full List | ||
| mod site | chr8:38057224-38057225:+ | [5] | |
| Sequence | CATGGAAGCCAGCCAGAGCCACCTGCGCAATAGCCCAGAAG | ||
| Motif Score | 2.032470238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000520657.1; ENST00000338825.5 | ||
| External Link | RMBase: m6A_site_793858 | ||
| mod ID: M6ASITE084569 | Click to Show/Hide the Full List | ||
| mod site | chr8:38059927-38059928:+ | [4] | |
| Sequence | AGAGTCACAGTTTGAGATGGACATTTAAAGCACCAGCCATC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; hESC-HEK293T; HepG2; hNPCs; hESCs; fibroblasts; LCLs; H1299; MM6; Huh7; peripheral-blood; TREX; endometrial; HEC-1-A; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-REF-seq; MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000338825.5; ENST00000520657.1 | ||
| External Link | RMBase: m6A_site_793859 | ||
| mod ID: M6ASITE084570 | Click to Show/Hide the Full List | ||
| mod site | chr8:38060053-38060054:+ | [4] | |
| Sequence | AGGCGTTGGCGTGGGGTCGGACACCCCAGCCCTTTCTCCCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; kidney; hESC-HEK293T; hESCs; A549; H1299; MM6; Huh7; peripheral-blood; iSLK; TIME; TREX; endometrial; HEC-1-A; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-REF-seq; DART-seq; MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000338825.5; ENST00000520657.1 | ||
| External Link | RMBase: m6A_site_793860 | ||
| mod ID: M6ASITE084571 | Click to Show/Hide the Full List | ||
| mod site | chr8:38060075-38060076:+ | [5] | |
| Sequence | ACCCCAGCCCTTTCTCCCTCACTCAGGGCACCTGCCCCCTC | ||
| Motif Score | 2.469291667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000520657.1; ENST00000338825.5 | ||
| External Link | RMBase: m6A_site_793861 | ||
| mod ID: M6ASITE084572 | Click to Show/Hide the Full List | ||
| mod site | chr8:38060106-38060107:+ | [4] | |
| Sequence | CTGCCCCCTCCTCTTCGTGAACACCAGCAGATACCTCCTTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; hESCs; fibroblasts; H1299; MM6; Huh7; peripheral-blood; iSLK; TIME; TREX; MSC; endometrial; HEC-1-A; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000338825.5; ENST00000520657.1 | ||
| External Link | RMBase: m6A_site_793862 | ||
| mod ID: M6ASITE084573 | Click to Show/Hide the Full List | ||
| mod site | chr8:38060118-38060119:+ | [5] | |
| Sequence | CTTCGTGAACACCAGCAGATACCTCCTTGTGCCTCCACTGA | ||
| Motif Score | 2.089839286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000338825.5; ENST00000520657.1 | ||
| External Link | RMBase: m6A_site_793863 | ||
| mod ID: M6ASITE084574 | Click to Show/Hide the Full List | ||
| mod site | chr8:38060179-38060180:+ | [6] | |
| Sequence | GGGGAGTGACCCCTGCCAGCACACCCTGCAGCCAAGGGCCA | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000338825.5; ENST00000520657.1 | ||
| External Link | RMBase: m6A_site_793864 | ||
| mod ID: M6ASITE084575 | Click to Show/Hide the Full List | ||
| mod site | chr8:38060208-38060209:+ | [4] | |
| Sequence | AGCCAAGGGCCAGGAAGTGGACAAGAACGAACCCTTCCTTC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; H1A; hESCs; fibroblasts; LCLs; H1299; MM6; Huh7; CD4T; peripheral-blood; iSLK; TIME; TREX; MSC; endometrial; HEC-1-A; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-REF-seq; MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000338825.5; ENST00000520657.1 | ||
| External Link | RMBase: m6A_site_793865 | ||
| mod ID: M6ASITE084576 | Click to Show/Hide the Full List | ||
| mod site | chr8:38060214-38060215:+ | [5] | |
| Sequence | GGGCCAGGAAGTGGACAAGAACGAACCCTTCCTTCCGAATG | ||
| Motif Score | 2.925321429 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000520657.1; ENST00000338825.5 | ||
| External Link | RMBase: m6A_site_793866 | ||
| mod ID: M6ASITE084577 | Click to Show/Hide the Full List | ||
| mod site | chr8:38060218-38060219:+ | [4] | |
| Sequence | CAGGAAGTGGACAAGAACGAACCCTTCCTTCCGAATGATCA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; hESCs; fibroblasts; LCLs; H1299; MM6; Huh7; CD4T; peripheral-blood; iSLK; TIME; TREX; MSC; endometrial; HEC-1-A; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000338825.5; ENST00000520657.1 | ||
| External Link | RMBase: m6A_site_793867 | ||
References