m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00426)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
TFEB
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LNCaP cell line | Homo sapiens |
|
Treatment: shMETTL3 LNCaP cells
Control: shControl LNCaP cells
|
GSE147884 | |
| Regulation |
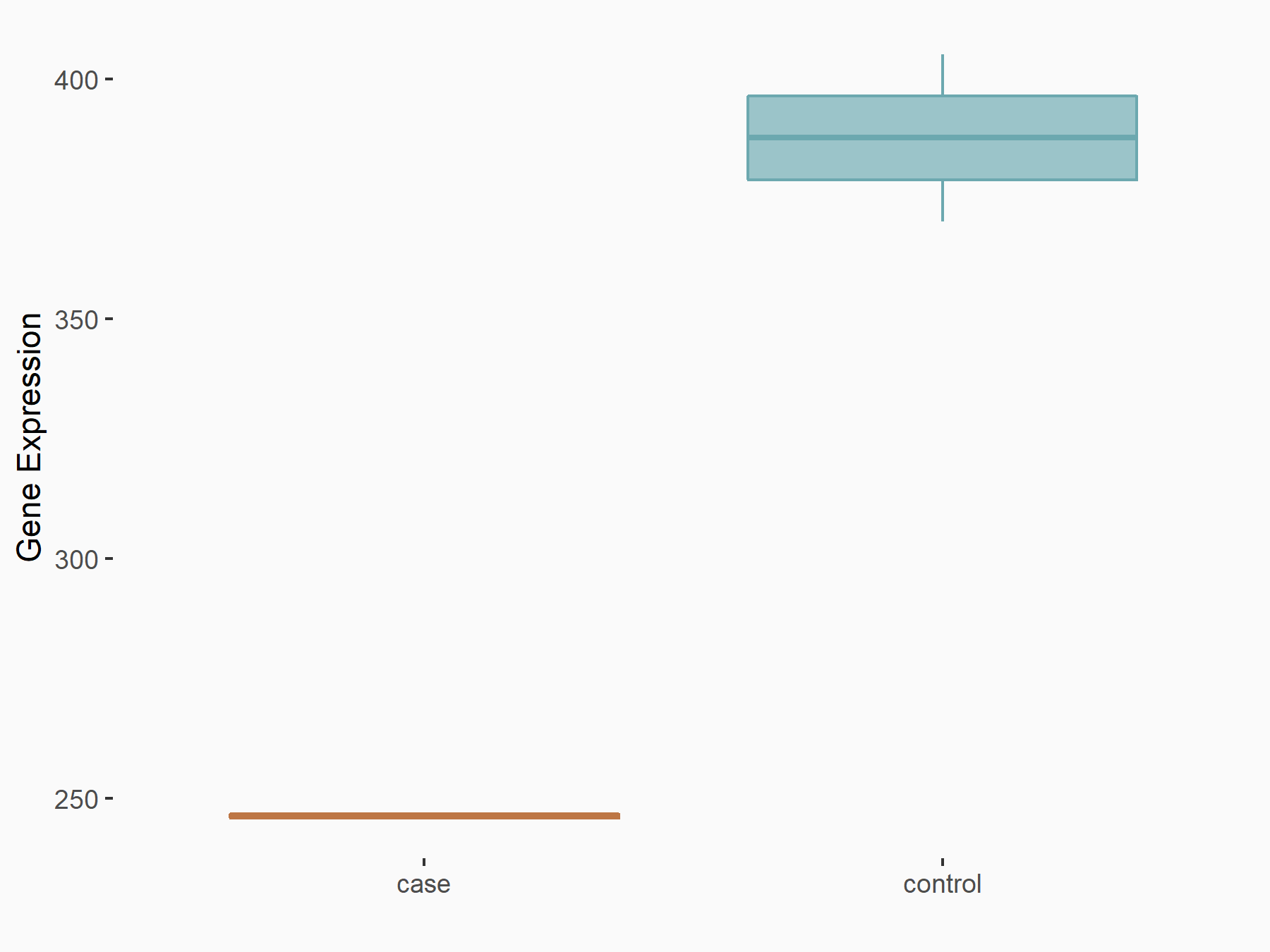  |
logFC: -6.54E-01 p-value: 8.08E-07 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between TFEB and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 8.22E+00 | GSE60213 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 methylates Transcription factor EB (TFEB), a master regulator of lysosomal biogenesis and autophagy genes, at two m6A residues in the 3'-UTR, which promotes the association of the RNA-binding protein HNRNPD with TFEB pre-mRNA and subsequently decreases the expression levels of TFEB. METTL3-ALKBH5 and autophagy, providing insight into the functional importance of the reversible mRNA m6A methylation and its modulators in ischemic heart disease. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Ischemic heart disease | ICD-11: BA40-BA6Z | ||
| Pathway Response | Apoptosis | hsa04210) | ||
| Cell Process | Cell proliferation | |||
| In-vitro Model | H9c2(2-1) | Normal | Rattus norvegicus | CVCL_0286 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | To cause I/R injury, mice were subjected to 30 min of LAD ischemia followed by 60 min of reperfusion. | |||
RNA demethylase ALKBH5 (ALKBH5) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by ALKBH5 | ||
| Cell Line | THP1 cell line | Homo sapiens |
|
Treatment: ALKBH5 knockdown THP1 cells
Control: Wild type THP1 cells
|
GSE128574 | |
| Regulation |
  |
logFC: -1.28E+00 p-value: 2.60E-04 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 methylates Transcription factor EB (TFEB), a master regulator of lysosomal biogenesis and autophagy genes, at two m6A residues in the 3'-UTR, which promotes the association of the RNA-binding protein HNRNPD with TFEB pre-mRNA and subsequently decreases the expression levels of TFEB. METTL3-ALKBH5 and autophagy, providing insight into the functional importance of the reversible mRNA m6A methylation and its modulators in ischemic heart disease. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Ischemic heart disease | ICD-11: BA40-BA6Z | ||
| Pathway Response | Apoptosis | hsa04210) | ||
| Cell Process | Cell proliferation | |||
| In-vitro Model | H9c2(2-1) | Normal | Rattus norvegicus | CVCL_0286 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | To cause I/R injury, mice were subjected to 30 min of LAD ischemia followed by 60 min of reperfusion. | |||
Ischemic heart disease [ICD-11: BA40-BA6Z]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 methylates Transcription factor EB (TFEB), a master regulator of lysosomal biogenesis and autophagy genes, at two m6A residues in the 3'-UTR, which promotes the association of the RNA-binding protein HNRNPD with TFEB pre-mRNA and subsequently decreases the expression levels of TFEB. METTL3-ALKBH5 and autophagy, providing insight into the functional importance of the reversible mRNA m6A methylation and its modulators in ischemic heart disease. | |||
| Responsed Disease | Ischemic heart disease [ICD-11: BA40-BA6Z] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Apoptosis | hsa04210) | ||
| Cell Process | Cell proliferation | |||
| In-vitro Model | H9c2(2-1) | Normal | Rattus norvegicus | CVCL_0286 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | To cause I/R injury, mice were subjected to 30 min of LAD ischemia followed by 60 min of reperfusion. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 methylates Transcription factor EB (TFEB), a master regulator of lysosomal biogenesis and autophagy genes, at two m6A residues in the 3'-UTR, which promotes the association of the RNA-binding protein HNRNPD with TFEB pre-mRNA and subsequently decreases the expression levels of TFEB. METTL3-ALKBH5 and autophagy, providing insight into the functional importance of the reversible mRNA m6A methylation and its modulators in ischemic heart disease. | |||
| Responsed Disease | Ischemic heart disease [ICD-11: BA40-BA6Z] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210) | ||
| Cell Process | Cell proliferation | |||
| In-vitro Model | H9c2(2-1) | Normal | Rattus norvegicus | CVCL_0286 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | To cause I/R injury, mice were subjected to 30 min of LAD ischemia followed by 60 min of reperfusion. | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00426)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE001448 | Click to Show/Hide the Full List | ||
| mod site | chr6:41707456-41707457:- | [2] | |
| Sequence | GGCCACAGGCAGGCACGCCAAGTCACAGCCCCTCTGACTTC | ||
| Transcript ID List | ENST00000416140.5; ENST00000394283.5; ENST00000420312.5; ENST00000358871.6; ENST00000373033.6; ENST00000445700.5; ENST00000425401.1; ENST00000230323.8; ENST00000403298.8; ENST00000419396.5; ENST00000424495.1; ENST00000433032.1 | ||
| External Link | RMBase: RNA-editing_site_116025 | ||
| mod ID: A2ISITE001449 | Click to Show/Hide the Full List | ||
| mod site | chr6:41715258-41715259:- | [2] | |
| Sequence | AGGGCCAGGGCTGTGGTCCTAGCTCTCCCTAAGCTGTGTGG | ||
| Transcript ID List | ENST00000358871.6; ENST00000424495.1; ENST00000394283.5; rmsk_1929789; ENST00000433032.1; ENST00000419396.5; ENST00000420312.5; ENST00000416140.5; ENST00000373033.6; ENST00000403298.8; ENST00000445700.5; ENST00000425401.1; ENST00000230323.8 | ||
| External Link | RMBase: RNA-editing_site_116026 | ||
5-methylcytidine (m5C)
N6-methyladenosine (m6A)
| In total 24 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE075051 | Click to Show/Hide the Full List | ||
| mod site | chr6:41683991-41683992:- | [3] | |
| Sequence | TCACTTTGTGCCTTTAGTAAACACTGTGCTTTGTACTTGCT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; GM12878; peripheral-blood; MSC; TIME; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000403298.8; ENST00000420312.5; ENST00000230323.8; ENST00000373033.6; ENST00000343317.10; ENST00000358871.6; ENST00000406563.6 | ||
| External Link | RMBase: m6A_site_716537 | ||
| mod ID: M6ASITE075052 | Click to Show/Hide the Full List | ||
| mod site | chr6:41684173-41684174:- | [3] | |
| Sequence | CTCAGGCCTGCCTTTTTGGGACTCAGATGGCAGGAGGTCCA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; GM12878; LCLs; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000358871.6; ENST00000343317.10; ENST00000420312.5; ENST00000406563.6; ENST00000373033.6; ENST00000230323.8; ENST00000403298.8 | ||
| External Link | RMBase: m6A_site_716538 | ||
| mod ID: M6ASITE075053 | Click to Show/Hide the Full List | ||
| mod site | chr6:41684248-41684249:- | [3] | |
| Sequence | CCCCTCATTACCAGTGAAGGACATGCTTGAGGGGTTCGGGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; fibroblasts; GM12878; LCLs; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000403298.8; ENST00000343317.10; ENST00000230323.8; ENST00000373033.6; ENST00000406563.6; ENST00000420312.5; ENST00000358871.6 | ||
| External Link | RMBase: m6A_site_716539 | ||
| mod ID: M6ASITE075054 | Click to Show/Hide the Full List | ||
| mod site | chr6:41684454-41684455:- | [3] | |
| Sequence | CCCCGTTGGCCCCTGTTTGGACTTAGTGCCTGTCTGGCAGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; GM12878; LCLs; MM6; CD4T; peripheral-blood; HEK293A-TOA; MSC; TIME; TREX; iSLK; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000403298.8; ENST00000420312.5; ENST00000230323.8; ENST00000406563.6; ENST00000358871.6; ENST00000343317.10; ENST00000373033.6 | ||
| External Link | RMBase: m6A_site_716540 | ||
| mod ID: M6ASITE075055 | Click to Show/Hide the Full List | ||
| mod site | chr6:41684573-41684574:- | [3] | |
| Sequence | GGCTGCCCCTGTGCCAGGGAACAGGGGCCGGCCTGGGGGCT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; LCLs; MM6; CD4T; peripheral-blood; MSC; TIME; TREX; iSLK; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000403298.8; ENST00000343317.10; ENST00000230323.8; ENST00000420312.5; ENST00000406563.6; ENST00000373033.6; ENST00000358871.6 | ||
| External Link | RMBase: m6A_site_716541 | ||
| mod ID: M6ASITE075056 | Click to Show/Hide the Full List | ||
| mod site | chr6:41684728-41684729:- | [3] | |
| Sequence | CCTGTCCAAGAAGGATCTGGACCTCATGCTCCTGGACGACT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1A; H1B; LCLs; A549; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000358871.6; ENST00000343317.10; ENST00000406563.6; ENST00000230323.8; ENST00000420312.5; ENST00000373033.6; ENST00000403298.8 | ||
| External Link | RMBase: m6A_site_716542 | ||
| mod ID: M6ASITE075057 | Click to Show/Hide the Full List | ||
| mod site | chr6:41684784-41684785:- | [3] | |
| Sequence | GGTCCCCCGGGCTACCCCGAACCCCTGGCGCCGGGGCATGG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1B; LCLs; A549; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000343317.10; ENST00000373033.6; ENST00000406563.6; ENST00000358871.6; ENST00000230323.8; ENST00000403298.8; ENST00000420312.5 | ||
| External Link | RMBase: m6A_site_716543 | ||
| mod ID: M6ASITE075058 | Click to Show/Hide the Full List | ||
| mod site | chr6:41684845-41684846:- | [3] | |
| Sequence | ATCCCCATTCCATCACCTGGACTTCAGCCACAGCCTGAGCT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; LCLs; A549; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; MSC; TIME; TREX; iSLK; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000406563.6; ENST00000343317.10; ENST00000358871.6; ENST00000420312.5; ENST00000403298.8; ENST00000230323.8; ENST00000373033.6 | ||
| External Link | RMBase: m6A_site_716544 | ||
| mod ID: M6ASITE075059 | Click to Show/Hide the Full List | ||
| mod site | chr6:41685019-41685020:- | [3] | |
| Sequence | CACCTCCCCGTCCGGCATGAACATGGCTGAGCTGGCCCAGC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000403298.8; ENST00000373033.6; ENST00000420312.5; ENST00000230323.8; ENST00000406563.6; ENST00000358871.6; ENST00000343317.10 | ||
| External Link | RMBase: m6A_site_716545 | ||
| mod ID: M6ASITE075060 | Click to Show/Hide the Full List | ||
| mod site | chr6:41686141-41686142:- | [4] | |
| Sequence | AAAGTCCAGGGAGCTGGAGAACCACTCTCGCCGCCTGGAGA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000358871.6; ENST00000343317.10; ENST00000373033.6; ENST00000230323.8; ENST00000403298.8; ENST00000420312.5; ENST00000406563.6 | ||
| External Link | RMBase: m6A_site_716546 | ||
| mod ID: M6ASITE075061 | Click to Show/Hide the Full List | ||
| mod site | chr6:41686168-41686169:- | [4] | |
| Sequence | CATCCGGAGGATGCAGAAGGACCTGCAAAAGTCCAGGGAGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000406563.6; ENST00000230323.8; ENST00000403298.8; ENST00000373033.6; ENST00000343317.10; ENST00000420312.5; ENST00000358871.6 | ||
| External Link | RMBase: m6A_site_716547 | ||
| mod ID: M6ASITE075062 | Click to Show/Hide the Full List | ||
| mod site | chr6:41686222-41686223:- | [5] | |
| Sequence | CTGCAGGGACGTGCGCTGGAACAAGGGCACCATCCTCAAGG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | brain; kidney; endometrial | ||
| Seq Type List | m6A-REF-seq; m6A-seq | ||
| Transcript ID List | ENST00000403298.8; ENST00000230323.8; ENST00000358871.6; ENST00000343317.10; ENST00000406563.6; ENST00000373033.6; ENST00000420312.5 | ||
| External Link | RMBase: m6A_site_716548 | ||
| mod ID: M6ASITE075063 | Click to Show/Hide the Full List | ||
| mod site | chr6:41686537-41686538:- | [6] | |
| Sequence | AGCCTGGGCAACAGAACGAGACTCCATCTCAAAAAAAAAAA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000373033.6; ENST00000358871.6; ENST00000343317.10; ENST00000403298.8; rmsk_1929735; ENST00000230323.8; ENST00000394283.5; ENST00000406563.6; ENST00000420312.5 | ||
| External Link | RMBase: m6A_site_716549 | ||
| mod ID: M6ASITE075064 | Click to Show/Hide the Full List | ||
| mod site | chr6:41687766-41687767:- | [3] | |
| Sequence | CAAGGAGCGGCAGAAGAAAGACAATCACAACTTAAGTGAGT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; LCLs | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000230323.8; ENST00000358871.6; ENST00000343317.10; ENST00000403298.8; ENST00000406563.6; ENST00000416140.5; ENST00000419396.5; ENST00000494822.1; ENST00000373033.6; ENST00000394283.5; ENST00000420312.5 | ||
| External Link | RMBase: m6A_site_716550 | ||
| mod ID: M6ASITE075065 | Click to Show/Hide the Full List | ||
| mod site | chr6:41688112-41688113:- | [3] | |
| Sequence | TCCAGGTGTCCCCAGGAAGGACTGCTCCTGGGATGATAGGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000406563.6; ENST00000343317.10; ENST00000403298.8; ENST00000416140.5; ENST00000494822.1; ENST00000373033.6; ENST00000358871.6; ENST00000230323.8; ENST00000394283.5; ENST00000419396.5; ENST00000419574.5; ENST00000420312.5 | ||
| External Link | RMBase: m6A_site_716551 | ||
| mod ID: M6ASITE075066 | Click to Show/Hide the Full List | ||
| mod site | chr6:41688182-41688183:- | [3] | |
| Sequence | AATAGCTCTGGACTCTGCAGACCTTCAGGTTGAATTGCTGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000358871.6; ENST00000343317.10; ENST00000373033.6; ENST00000494822.1; ENST00000403298.8; ENST00000406563.6; ENST00000394283.5; ENST00000419396.5; ENST00000230323.8; ENST00000420312.5; ENST00000416140.5; ENST00000419574.5 | ||
| External Link | RMBase: m6A_site_716552 | ||
| mod ID: M6ASITE075067 | Click to Show/Hide the Full List | ||
| mod site | chr6:41688191-41688192:- | [3] | |
| Sequence | ACATCTTATAATAGCTCTGGACTCTGCAGACCTTCAGGTTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000494822.1; ENST00000419396.5; ENST00000420312.5; ENST00000394283.5; ENST00000373033.6; ENST00000343317.10; ENST00000419574.5; ENST00000403298.8; ENST00000406563.6; ENST00000416140.5; ENST00000230323.8; ENST00000358871.6 | ||
| External Link | RMBase: m6A_site_716553 | ||
| mod ID: M6ASITE075068 | Click to Show/Hide the Full List | ||
| mod site | chr6:41690746-41690747:- | [3] | |
| Sequence | TCCCCAGGGGTGCGAGCTGGACACGTGCTGTCCTCCTCCGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000403298.8; ENST00000343317.10; ENST00000419574.5; ENST00000373033.6; ENST00000420312.5; ENST00000230323.8; ENST00000406563.6; ENST00000416140.5; ENST00000419396.5; ENST00000394283.5; ENST00000358871.6 | ||
| External Link | RMBase: m6A_site_716554 | ||
| mod ID: M6ASITE075069 | Click to Show/Hide the Full List | ||
| mod site | chr6:41691274-41691275:- | [3] | |
| Sequence | GCCCCTGTGCTTTGAGGAGGACACGTGCCTCATATACCTGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; CD4T; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000419574.5; ENST00000445214.1; ENST00000420312.5; ENST00000358871.6; ENST00000419396.5; ENST00000230323.8; ENST00000373033.6; ENST00000394283.5; rmsk_1929743; ENST00000424495.1; ENST00000343317.10; ENST00000425401.1; ENST00000416140.5; ENST00000403298.8; ENST00000445700.5; ENST00000433032.1 | ||
| External Link | RMBase: m6A_site_716555 | ||
| mod ID: M6ASITE075070 | Click to Show/Hide the Full List | ||
| mod site | chr6:41733157-41733158:- | [3] | |
| Sequence | TGAGAAGTAGGTGGAAGGGGACTCCCACCCCTCAAGTCTGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000433032.1; ENST00000230323.8; ENST00000358871.6; ENST00000419396.5; ENST00000373033.6; ENST00000425401.1; ENST00000394283.5; ENST00000445700.5; ENST00000403298.8; ENST00000424495.1; ENST00000420312.5 | ||
| External Link | RMBase: m6A_site_716556 | ||
| mod ID: M6ASITE075071 | Click to Show/Hide the Full List | ||
| mod site | chr6:41733755-41733756:- | [3] | |
| Sequence | AGCTCACTGTCCGGGGAGGGACAGCCAGCAATAGTTCCTGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000373033.6; ENST00000394283.5; ENST00000445700.5; ENST00000420312.5; ENST00000358871.6; ENST00000425401.1; ENST00000230323.8; ENST00000433032.1; ENST00000419396.5; ENST00000403298.8 | ||
| External Link | RMBase: m6A_site_716557 | ||
| mod ID: M6ASITE075072 | Click to Show/Hide the Full List | ||
| mod site | chr6:41733801-41733802:- | [3] | |
| Sequence | CTTGGTGCCACGACTGGGAGACAGTCCTCAGGCCCTCCCCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000419396.5; ENST00000230323.8; ENST00000373033.6; ENST00000433032.1; ENST00000425401.1; ENST00000403298.8; ENST00000358871.6; ENST00000420312.5; ENST00000445700.5; ENST00000394283.5 | ||
| External Link | RMBase: m6A_site_716558 | ||
| mod ID: M6ASITE075073 | Click to Show/Hide the Full List | ||
| mod site | chr6:41735370-41735371:- | [3] | |
| Sequence | CCGCGGGGCGCGGGCGGCGGACAGATTGACCTTCAGAGCGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000358871.6; ENST00000373033.6; ENST00000420312.5 | ||
| External Link | RMBase: m6A_site_716559 | ||
| mod ID: M6ASITE075074 | Click to Show/Hide the Full List | ||
| mod site | chr6:41735411-41735412:- | [3] | |
| Sequence | CTTGCGGGCCAGGCACCCGAACTTGCGACAAGTTGCCGGAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; GSC-11; HEK293T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000373033.6; ENST00000358871.6; ENST00000420312.5 | ||
| External Link | RMBase: m6A_site_716560 | ||
References