m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00625)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
NKX3-1
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
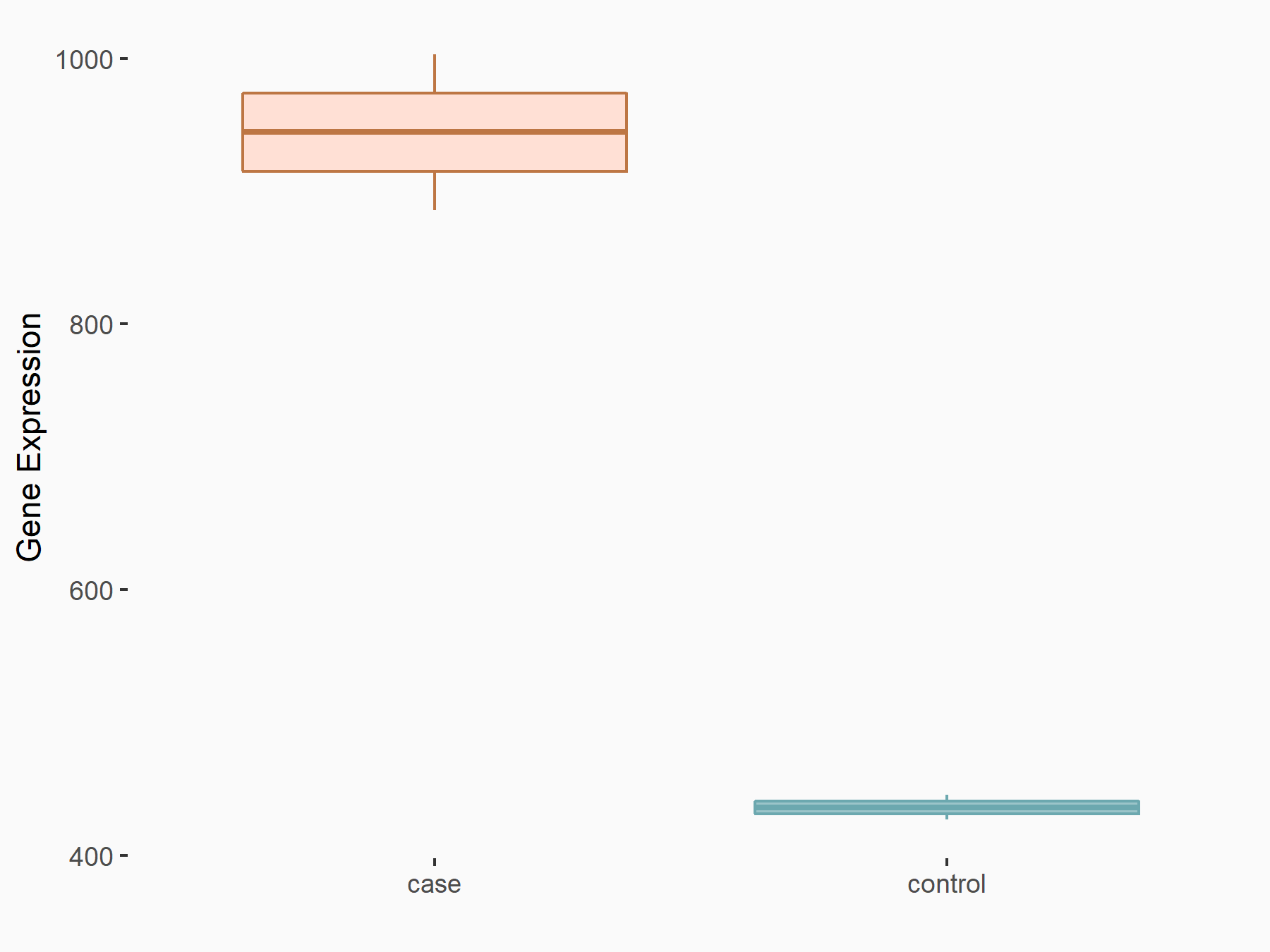 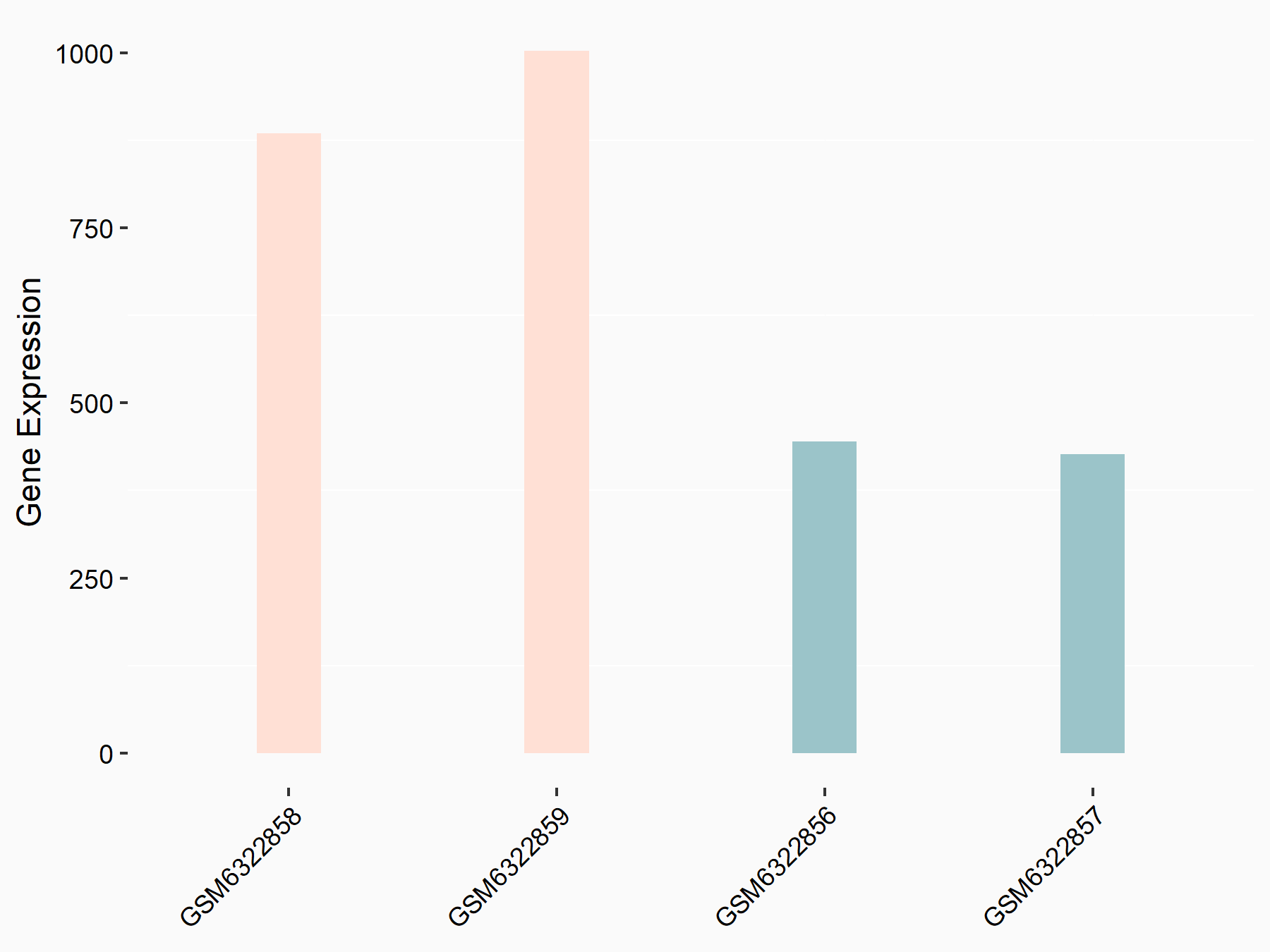 |
logFC: 1.12E+00 p-value: 7.44E-16 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between NKX3-1 and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.11E+00 | GSE60213 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Knock-down of YTHDF2 or METTL3 significantly induced the expression of LHPP and Homeobox protein Nkx-3.1 (NKX3-1) at both mRNA and protein level with inhibited phosphorylated AKT. YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent way to regulate AKT phosphorylation-induced tumor progression in prostate cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Prostate cancer | ICD-11: 2C82 | ||
| Pathway Response | Oxidative phosphorylation | hsa00190 | ||
| In-vitro Model | VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| In-vivo Model | Approximately 2 × 106 PCa cells (PC-3 shNC, shYTHDF2, shMETTL3 cell lines) per mouse suspended in 100 uL PBS were injected in the flank of male BALB/c nude mice (4 weeks old). During the 40-day observation, the tumor size (V = (width2×length ×0.52)) was measured with vernier caliper. Approximately 1.5 × 106 PCa cells suspended in 100 uL of PBS (PC-3 shNC, shYTHDF2, and shMETTL3 cell lines) per mouse were injected into the tail vein of male BALB/c nude mice (4 weeks old). The IVIS Spectrum animal imaging system (PerkinElmer) was used to evaluate the tumor growth (40 days) and whole metastasis conditions (4 weeks and 6 weeks) with 100 uL XenoLight D-luciferin Potassium Salt (15 mg/ml, Perkin Elmer) per mouse. Mice were anesthetized and then sacrificed for tumors and metastases which were sent for further organ-localized imaging as above, IHC staining and hematoxylin-eosin (H&E) staining. | |||
YTH domain-containing family protein 2 (YTHDF2) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
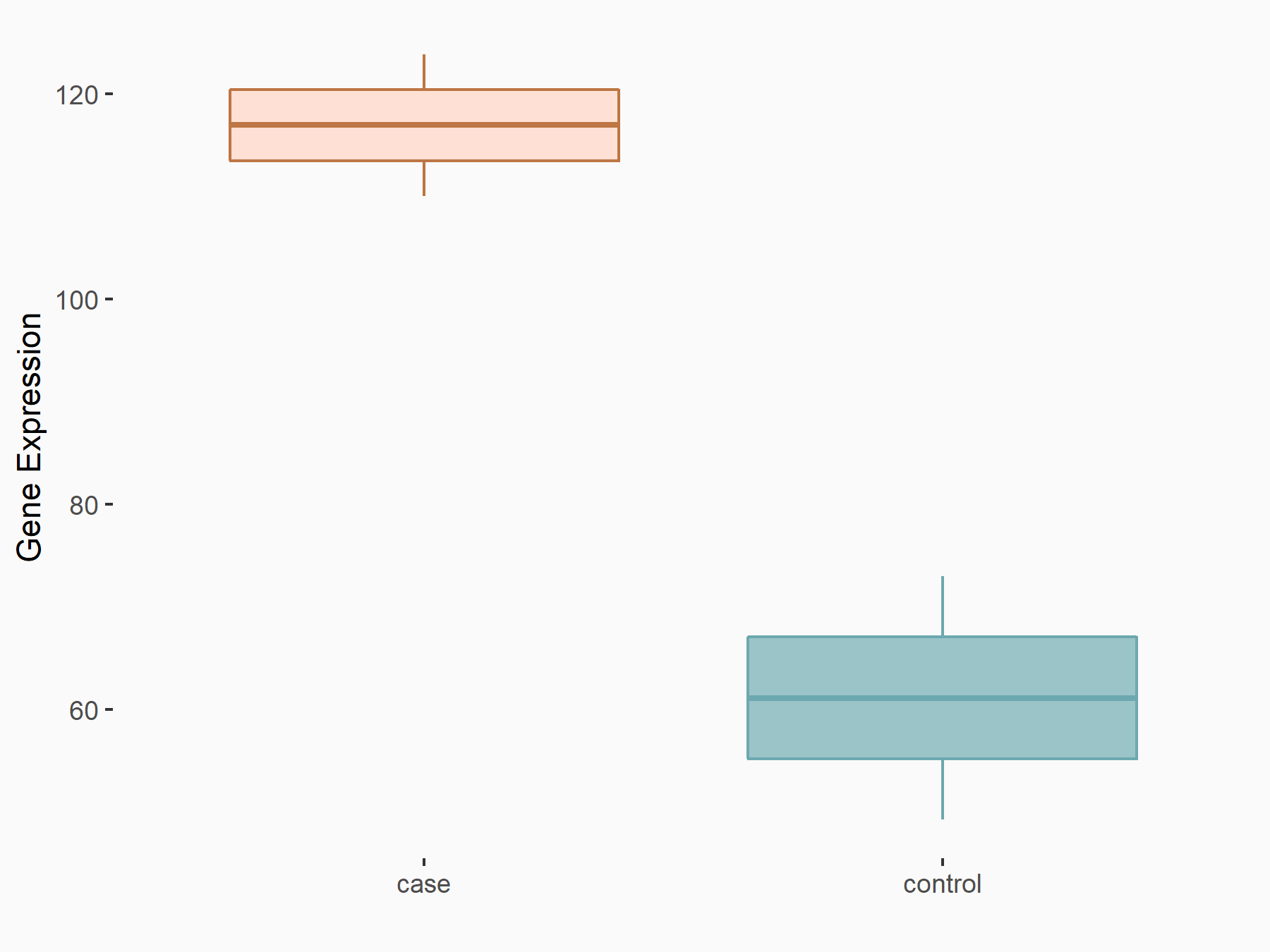 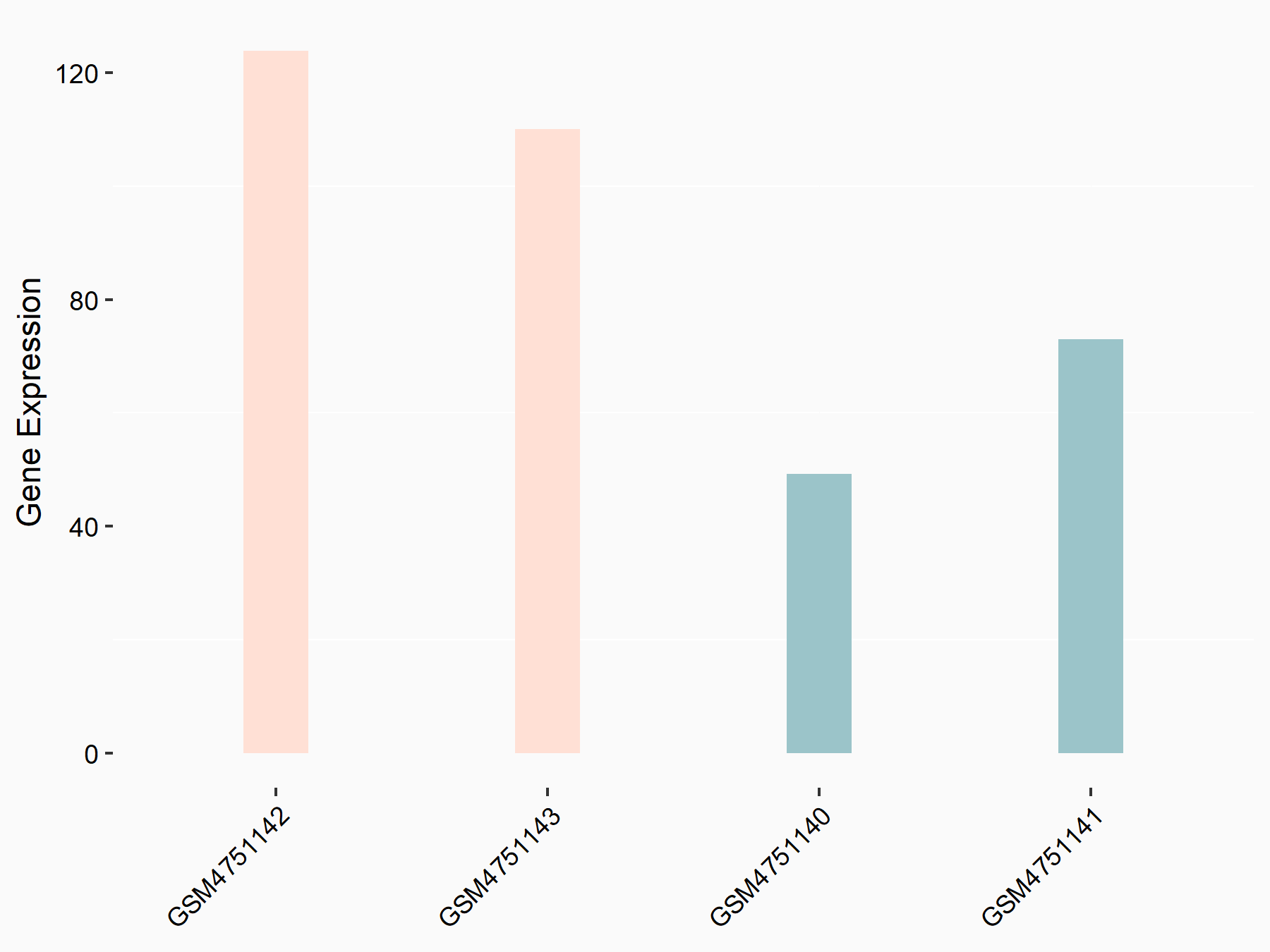 |
logFC: 9.36E-01 p-value: 4.17E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between NKX3-1 and the regulator | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.20E+00 | GSE49339 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Knock-down of YTHDF2 or METTL3 significantly induced the expression of LHPP and Homeobox protein Nkx-3.1 (NKX3-1) at both mRNA and protein level with inhibited phosphorylated AKT. YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent way to regulate AKT phosphorylation-induced tumor progression in prostate cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Prostate cancer | ICD-11: 2C82 | ||
| Pathway Response | Oxidative phosphorylation | hsa00190 | ||
| In-vitro Model | VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| In-vivo Model | Approximately 2 × 106 PCa cells (PC-3 shNC, shYTHDF2, shMETTL3 cell lines) per mouse suspended in 100 uL PBS were injected in the flank of male BALB/c nude mice (4 weeks old). During the 40-day observation, the tumor size (V = (width2×length ×0.52)) was measured with vernier caliper. Approximately 1.5 × 106 PCa cells suspended in 100 uL of PBS (PC-3 shNC, shYTHDF2, and shMETTL3 cell lines) per mouse were injected into the tail vein of male BALB/c nude mice (4 weeks old). The IVIS Spectrum animal imaging system (PerkinElmer) was used to evaluate the tumor growth (40 days) and whole metastasis conditions (4 weeks and 6 weeks) with 100 uL XenoLight D-luciferin Potassium Salt (15 mg/ml, Perkin Elmer) per mouse. Mice were anesthetized and then sacrificed for tumors and metastases which were sent for further organ-localized imaging as above, IHC staining and hematoxylin-eosin (H&E) staining. | |||
Prostate cancer [ICD-11: 2C82]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Knock-down of YTHDF2 or METTL3 significantly induced the expression of LHPP and Homeobox protein Nkx-3.1 (NKX3-1) at both mRNA and protein level with inhibited phosphorylated AKT. YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent way to regulate AKT phosphorylation-induced tumor progression in prostate cancer. | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Oxidative phosphorylation | hsa00190 | ||
| In-vitro Model | VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| In-vivo Model | Approximately 2 × 106 PCa cells (PC-3 shNC, shYTHDF2, shMETTL3 cell lines) per mouse suspended in 100 uL PBS were injected in the flank of male BALB/c nude mice (4 weeks old). During the 40-day observation, the tumor size (V = (width2×length ×0.52)) was measured with vernier caliper. Approximately 1.5 × 106 PCa cells suspended in 100 uL of PBS (PC-3 shNC, shYTHDF2, and shMETTL3 cell lines) per mouse were injected into the tail vein of male BALB/c nude mice (4 weeks old). The IVIS Spectrum animal imaging system (PerkinElmer) was used to evaluate the tumor growth (40 days) and whole metastasis conditions (4 weeks and 6 weeks) with 100 uL XenoLight D-luciferin Potassium Salt (15 mg/ml, Perkin Elmer) per mouse. Mice were anesthetized and then sacrificed for tumors and metastases which were sent for further organ-localized imaging as above, IHC staining and hematoxylin-eosin (H&E) staining. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Knock-down of YTHDF2 or METTL3 significantly induced the expression of LHPP and Homeobox protein Nkx-3.1 (NKX3-1) at both mRNA and protein level with inhibited phosphorylated AKT. YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent way to regulate AKT phosphorylation-induced tumor progression in prostate cancer. | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Oxidative phosphorylation | hsa00190 | ||
| In-vitro Model | VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| In-vivo Model | Approximately 2 × 106 PCa cells (PC-3 shNC, shYTHDF2, shMETTL3 cell lines) per mouse suspended in 100 uL PBS were injected in the flank of male BALB/c nude mice (4 weeks old). During the 40-day observation, the tumor size (V = (width2×length ×0.52)) was measured with vernier caliper. Approximately 1.5 × 106 PCa cells suspended in 100 uL of PBS (PC-3 shNC, shYTHDF2, and shMETTL3 cell lines) per mouse were injected into the tail vein of male BALB/c nude mice (4 weeks old). The IVIS Spectrum animal imaging system (PerkinElmer) was used to evaluate the tumor growth (40 days) and whole metastasis conditions (4 weeks and 6 weeks) with 100 uL XenoLight D-luciferin Potassium Salt (15 mg/ml, Perkin Elmer) per mouse. Mice were anesthetized and then sacrificed for tumors and metastases which were sent for further organ-localized imaging as above, IHC staining and hematoxylin-eosin (H&E) staining. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03330 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5A (KDM5A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Prostate cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00625)
| In total 41 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE084189 | Click to Show/Hide the Full List | ||
| mod site | chr8:23679194-23679195:- | [2] | |
| Sequence | TGAATTGCTTTCTGCTCTTTACATTTCTTTTAAAATAAGCA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790389 | ||
| mod ID: M6ASITE084190 | Click to Show/Hide the Full List | ||
| mod site | chr8:23679256-23679257:- | [2] | |
| Sequence | TTCAATGCCACGTGCTGCTGACACCGACCGGAGTACTAGCC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790390 | ||
| mod ID: M6ASITE084191 | Click to Show/Hide the Full List | ||
| mod site | chr8:23679313-23679314:- | [2] | |
| Sequence | ATAGAAAGTTGGCCAATTTCACCCCATTTTCTGTGGTTTGG | ||
| Motif Score | 2.026654762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790391 | ||
| mod ID: M6ASITE084192 | Click to Show/Hide the Full List | ||
| mod site | chr8:23679422-23679423:- | [2] | |
| Sequence | GGGAGTCTCTTGACTCCACTACTTAATTCCGTTTAGTGAGA | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790392 | ||
| mod ID: M6ASITE084193 | Click to Show/Hide the Full List | ||
| mod site | chr8:23679479-23679480:- | [2] | |
| Sequence | AGACATTAGAAAAAAATGAAACAACAAAACAATTACTAATG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790393 | ||
| mod ID: M6ASITE084194 | Click to Show/Hide the Full List | ||
| mod site | chr8:23679497-23679498:- | [2] | |
| Sequence | GGTGAGCTGGTAGAGGGGAGACATTAGAAAAAAATGAAACA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790394 | ||
| mod ID: M6ASITE084195 | Click to Show/Hide the Full List | ||
| mod site | chr8:23679632-23679633:- | [2] | |
| Sequence | ACTGTCCAAATGCTTTGGGAACTGTGTTTATTGCCTATAAT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HEK293T; Jurkat | ||
| Seq Type List | DART-seq; m6A-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790395 | ||
| mod ID: M6ASITE084196 | Click to Show/Hide the Full List | ||
| mod site | chr8:23679652-23679653:- | [2] | |
| Sequence | ATTCTAAAACACAACAAGAAACTGTCCAAATGCTTTGGGAA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T; Jurkat | ||
| Seq Type List | DART-seq; m6A-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790396 | ||
| mod ID: M6ASITE084197 | Click to Show/Hide the Full List | ||
| mod site | chr8:23679662-23679663:- | [2] | |
| Sequence | AAGGAAAACCATTCTAAAACACAACAAGAAACTGTCCAAAT | ||
| Motif Score | 2.084928571 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790397 | ||
| mod ID: M6ASITE084198 | Click to Show/Hide the Full List | ||
| mod site | chr8:23679664-23679665:- | [3] | |
| Sequence | AAAAGGAAAACCATTCTAAAACACAACAAGAAACTGTCCAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | Jurkat | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790398 | ||
| mod ID: M6ASITE084199 | Click to Show/Hide the Full List | ||
| mod site | chr8:23679675-23679676:- | [2] | |
| Sequence | AAAGGCTAAGAAAAAGGAAAACCATTCTAAAACACAACAAG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HEK293T; Jurkat | ||
| Seq Type List | DART-seq; m6A-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790399 | ||
| mod ID: M6ASITE084200 | Click to Show/Hide the Full List | ||
| mod site | chr8:23679717-23679718:- | [4] | |
| Sequence | GTCCACTGAGCAAGCAAGGGACTGAGTGAGCCTTTTGCAGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; Jurkat | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790400 | ||
| mod ID: M6ASITE084201 | Click to Show/Hide the Full List | ||
| mod site | chr8:23679775-23679776:- | [4] | |
| Sequence | GTGGAGGGCTCATGGGTGGGACATGGAAAAGAAGGCAGCCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790401 | ||
| mod ID: M6ASITE084202 | Click to Show/Hide the Full List | ||
| mod site | chr8:23679836-23679837:- | [2] | |
| Sequence | TGGATTTTCACAGAGGAAGAACACAGCGCAGAATGAAGGGC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790402 | ||
| mod ID: M6ASITE084203 | Click to Show/Hide the Full List | ||
| mod site | chr8:23679899-23679900:- | [2] | |
| Sequence | GCTGTTACGTTTGAAGTCTGACAATCCTTGAGAATCTTTGC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790403 | ||
| mod ID: M6ASITE084204 | Click to Show/Hide the Full List | ||
| mod site | chr8:23679913-23679914:- | [2] | |
| Sequence | CTTCTGGTGGTTCTGCTGTTACGTTTGAAGTCTGACAATCC | ||
| Motif Score | 2.046785714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790404 | ||
| mod ID: M6ASITE084205 | Click to Show/Hide the Full List | ||
| mod site | chr8:23679949-23679950:- | [2] | |
| Sequence | GACAGCTTGAGAAGGTCACTACTGCATTTATAGGACCTTCT | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790405 | ||
| mod ID: M6ASITE084206 | Click to Show/Hide the Full List | ||
| mod site | chr8:23680005-23680006:- | [2] | |
| Sequence | TATTTCTTTCCTTTAAAAATACATAGCATTAAATCCCAAAT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790406 | ||
| mod ID: M6ASITE084207 | Click to Show/Hide the Full List | ||
| mod site | chr8:23680030-23680031:- | [2] | |
| Sequence | ATGCATTTTTAAAACTAGCAACTCTTATTTCTTTCCTTTAA | ||
| Motif Score | 2.595904762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790407 | ||
| mod ID: M6ASITE084208 | Click to Show/Hide the Full List | ||
| mod site | chr8:23680037-23680038:- | [2] | |
| Sequence | AAAAGACATGCATTTTTAAAACTAGCAACTCTTATTTCTTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790408 | ||
| mod ID: M6ASITE084209 | Click to Show/Hide the Full List | ||
| mod site | chr8:23680052-23680053:- | [2] | |
| Sequence | GATTCTGAATTGGCTAAAAGACATGCATTTTTAAAACTAGC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790409 | ||
| mod ID: M6ASITE084210 | Click to Show/Hide the Full List | ||
| mod site | chr8:23680096-23680097:- | [2] | |
| Sequence | CCAAAGCTTTATCTGTCTTGACTTTTTAAAAAAGTTTGGGG | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790410 | ||
| mod ID: M6ASITE084211 | Click to Show/Hide the Full List | ||
| mod site | chr8:23680790-23680791:- | [4] | |
| Sequence | GATGTCACCAACTGAATTAAACTTAAGTCCAGAAGCCTCCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; Huh7; Jurkat; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790411 | ||
| mod ID: M6ASITE084212 | Click to Show/Hide the Full List | ||
| mod site | chr8:23680824-23680825:- | [4] | |
| Sequence | CAAACCTAACAGGAGAAAGGACAACCAGGATGAGGATGTCA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; hESC-HEK293T; CD8T; Huh7; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790412 | ||
| mod ID: M6ASITE084213 | Click to Show/Hide the Full List | ||
| mod site | chr8:23680841-23680842:- | [4] | |
| Sequence | TATCAACTCTGAAAGAGCAAACCTAACAGGAGAAAGGACAA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; A549; CD8T; Huh7; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790413 | ||
| mod ID: M6ASITE084214 | Click to Show/Hide the Full List | ||
| mod site | chr8:23681050-23681051:- | [4] | |
| Sequence | GAGACAATGAAACAACAGAGACAGTGAAAGTTTTAATACCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; hESC-HEK293T; U2OS; Huh7; Jurkat; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000380871.5; ENST00000523261.1 | ||
| External Link | RMBase: m6A_site_790414 | ||
| mod ID: M6ASITE084215 | Click to Show/Hide the Full List | ||
| mod site | chr8:23681059-23681060:- | [4] | |
| Sequence | TTCACTGGTGAGACAATGAAACAACAGAGACAGTGAAAGTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; hESC-HEK293T; U2OS; fibroblasts; CD8T; Huh7; Jurkat; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000523261.1; ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790415 | ||
| mod ID: M6ASITE084216 | Click to Show/Hide the Full List | ||
| mod site | chr8:23681067-23681068:- | [4] | |
| Sequence | TCAGATTCTTCACTGGTGAGACAATGAAACAACAGAGACAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; U2OS; fibroblasts; Huh7; Jurkat; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000523261.1; ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790416 | ||
| mod ID: M6ASITE084217 | Click to Show/Hide the Full List | ||
| mod site | chr8:23681186-23681187:- | [4] | |
| Sequence | GACAACCATTATGATCAAAAACTGCCTTCCCCAGGGTGTCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; fibroblasts; GM12878; Huh7; Jurkat; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000523261.1; ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790417 | ||
| mod ID: M6ASITE084218 | Click to Show/Hide the Full List | ||
| mod site | chr8:23681356-23681357:- | [4] | |
| Sequence | GCTCTCCTCGGAGCTGGGAGACTTGGAGAAGCACTCCTCTT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1B; H1A; hESCs; fibroblasts; GM12878; Huh7; Jurkat; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000523261.1; ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790418 | ||
| mod ID: M6ASITE084219 | Click to Show/Hide the Full List | ||
| mod site | chr8:23681390-23681391:- | [4] | |
| Sequence | TCCAGAACAGACGCTATAAGACTAAGCGAAAGCAGCTCTCC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1B; H1A; hESCs; fibroblasts; GM12878; Jurkat; HEK293A-TOA; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000523261.1; ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790419 | ||
| mod ID: M6ASITE084220 | Click to Show/Hide the Full List | ||
| mod site | chr8:23681400-23681401:- | [5] | |
| Sequence | AAGATATGGTTCCAGAACAGACGCTATAAGACTAAGCGAAA | ||
| Motif Score | 2.871321429 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000380871.5; ENST00000523261.1 | ||
| External Link | RMBase: m6A_site_790420 | ||
| mod ID: M6ASITE084221 | Click to Show/Hide the Full List | ||
| mod site | chr8:23681404-23681405:- | [4] | |
| Sequence | AGTGAAGATATGGTTCCAGAACAGACGCTATAAGACTAAGC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1B; H1A; hESCs; fibroblasts; GM12878; CD8T; Jurkat; HEK293A-TOA; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000523261.1; ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790421 | ||
| mod ID: M6ASITE084222 | Click to Show/Hide the Full List | ||
| mod site | chr8:23681429-23681430:- | [4] | |
| Sequence | AGAACCTCAAGCTCACGGAGACCCAAGTGAAGATATGGTTC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1B; H1A; hESCs; fibroblasts; GM12878; Jurkat; HEK293A-TOA; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000380871.5; ENST00000523261.1 | ||
| External Link | RMBase: m6A_site_790422 | ||
| mod ID: M6ASITE084223 | Click to Show/Hide the Full List | ||
| mod site | chr8:23681446-23681447:- | [4] | |
| Sequence | ACGGGCCCACCTGGCCAAGAACCTCAAGCTCACGGAGACCC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; hESCs; Jurkat; HEK293A-TOA; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000523261.1; ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790423 | ||
| mod ID: M6ASITE084224 | Click to Show/Hide the Full List | ||
| mod site | chr8:23681570-23681571:- | [4] | |
| Sequence | CCCTTCCAAGGCTTCCCCAAACCCCTAAGCAGCCGCAGAAG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; A549; U2OS; Jurkat; GSC-11; HEK293T; HEK293A-TOA; iSLK; MSC; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000523261.1; ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790424 | ||
| mod ID: M6ASITE084225 | Click to Show/Hide the Full List | ||
| mod site | chr8:23681602-23681603:- | [6] | |
| Sequence | TTATCTGTTGGACTCTGAAAACACTTCAGGCGCCCTTCCAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | A549; U2OS; Jurkat; GSC-11; HEK293T; HEK293A-TOA; iSLK; MSC; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000523261.1; ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790425 | ||
| mod ID: M6ASITE084226 | Click to Show/Hide the Full List | ||
| mod site | chr8:23681611-23681612:- | [6] | |
| Sequence | CTTGGGGTCTTATCTGTTGGACTCTGAAAACACTTCAGGCG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | A549; U2OS; Jurkat; GSC-11; HEK293T; HEK293A-TOA; iSLK; MSC; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000523261.1; ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790426 | ||
| mod ID: M6ASITE084227 | Click to Show/Hide the Full List | ||
| mod site | chr8:23682612-23682613:- | [4] | |
| Sequence | AGGCCGAGACGCTGGCAGAGACCGAGCCAGGTAAGCGGCGA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; A549; HEK293T; Jurkat; GSC-11; HEK293A-TOA; iSLK; MSC; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000523261.1; ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790427 | ||
| mod ID: M6ASITE084228 | Click to Show/Hide the Full List | ||
| mod site | chr8:23682739-23682740:- | [4] | |
| Sequence | GGCCGCACGAGCAGCCAGAGACAGCGCGACCCGGAGCCGGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; A549; U2OS; Jurkat; GSC-11; HEK293T; HEK293A-TOA; iSLK; MSC; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000523261.1; ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790428 | ||
| mod ID: M6ASITE084229 | Click to Show/Hide the Full List | ||
| mod site | chr8:23682791-23682792:- | [4] | |
| Sequence | CACGTCCTTCCTCATCCAGGACATCCTGCGGGACGGCGCGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; A549; U2OS; Jurkat; GSC-11; HEK293T; iSLK; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000523261.1; ENST00000380871.5 | ||
| External Link | RMBase: m6A_site_790429 | ||
References