m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00388)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
SCD
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
|
Treatment: METTL3 knockdown MDA-MB-231 cells
Control: MDA-MB-231 cells
|
GSE70061 | |
| Regulation |
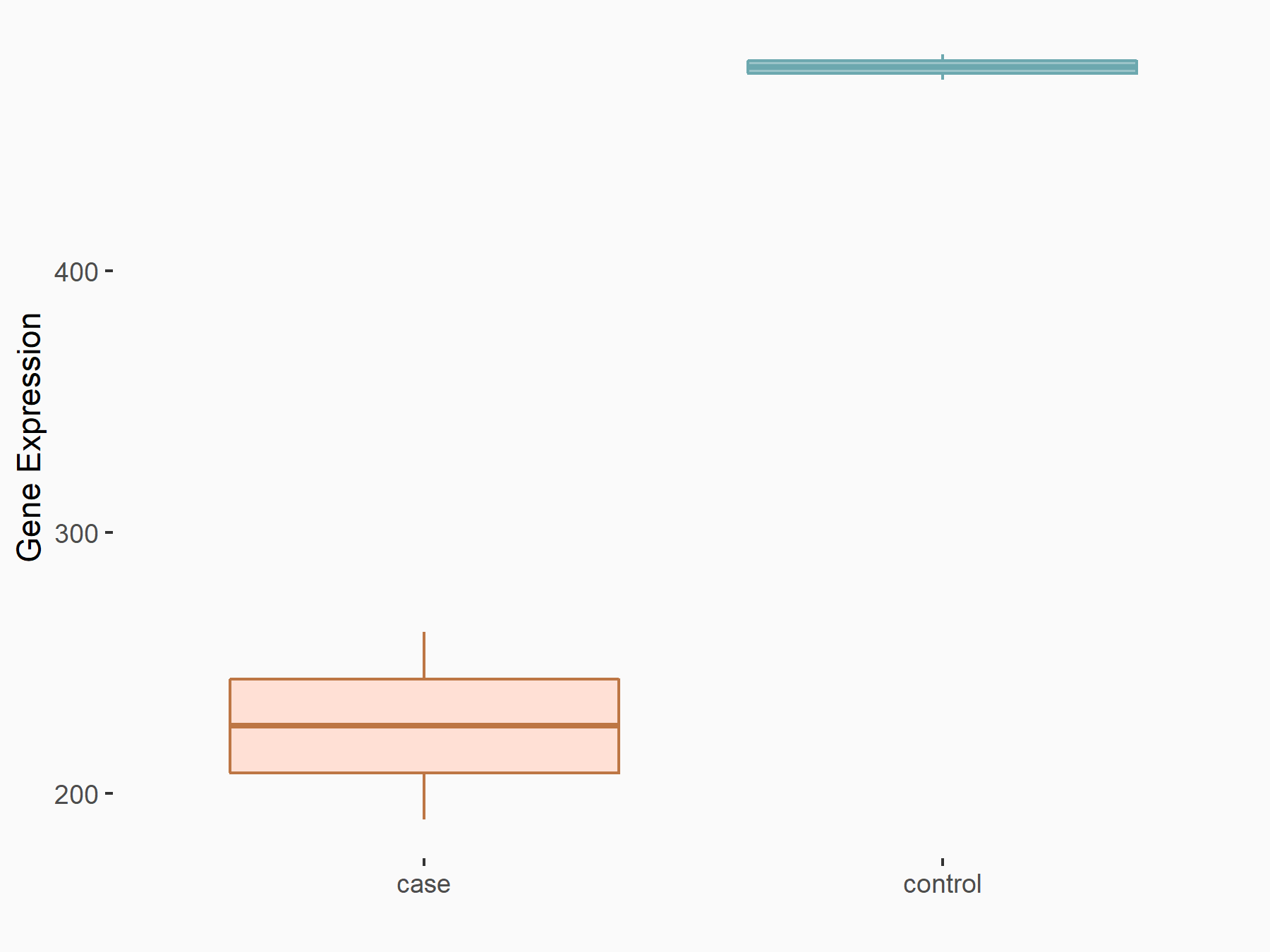  |
logFC: -1.10E+00 p-value: 2.11E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between SCD and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.17E+00 | GSE60213 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Targeting METTL3/14 in vitro increases protein level of ACLY and Stearoyl-CoA desaturase (SCD) as well as triglyceride and cholesterol production and accumulation of lipid droplets. These findings demonstrate a new NAFLD mouse model that provides a study platform for DM2-related NAFLD and reveals a unique epitranscriptional regulating mechanism for lipid metabolism via m6A-modified protein expression of ACLY and SCD1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-alcoholic fatty liver disease | ICD-11: DB92 | ||
| Pathway Response | Glycerolipid metabolism | hsa00561 | ||
| Cell Process | Lipid metabolism | |||
| In-vitro Model | LM3 | Malignant neoplasms | Mus musculus | CVCL_D269 |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| In-vivo Model | Mice with a Tmem30a deletion specifically in pancreatic beta cells were generated as previously described. Mice developed with NAFLD were named for Tmem30a-associated NAFLD (TAN) mice. The littermate mice with genotypes of Tmem30aloxP/loxP were used as controls. | |||
Methyltransferase-like 14 (METTL14) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL14 | ||
| Cell Line | HepG2 cell line | Homo sapiens |
|
Treatment: shMETTL14 HepG2 cells
Control: shCtrl HepG2 cells
|
GSE121949 | |
| Regulation |
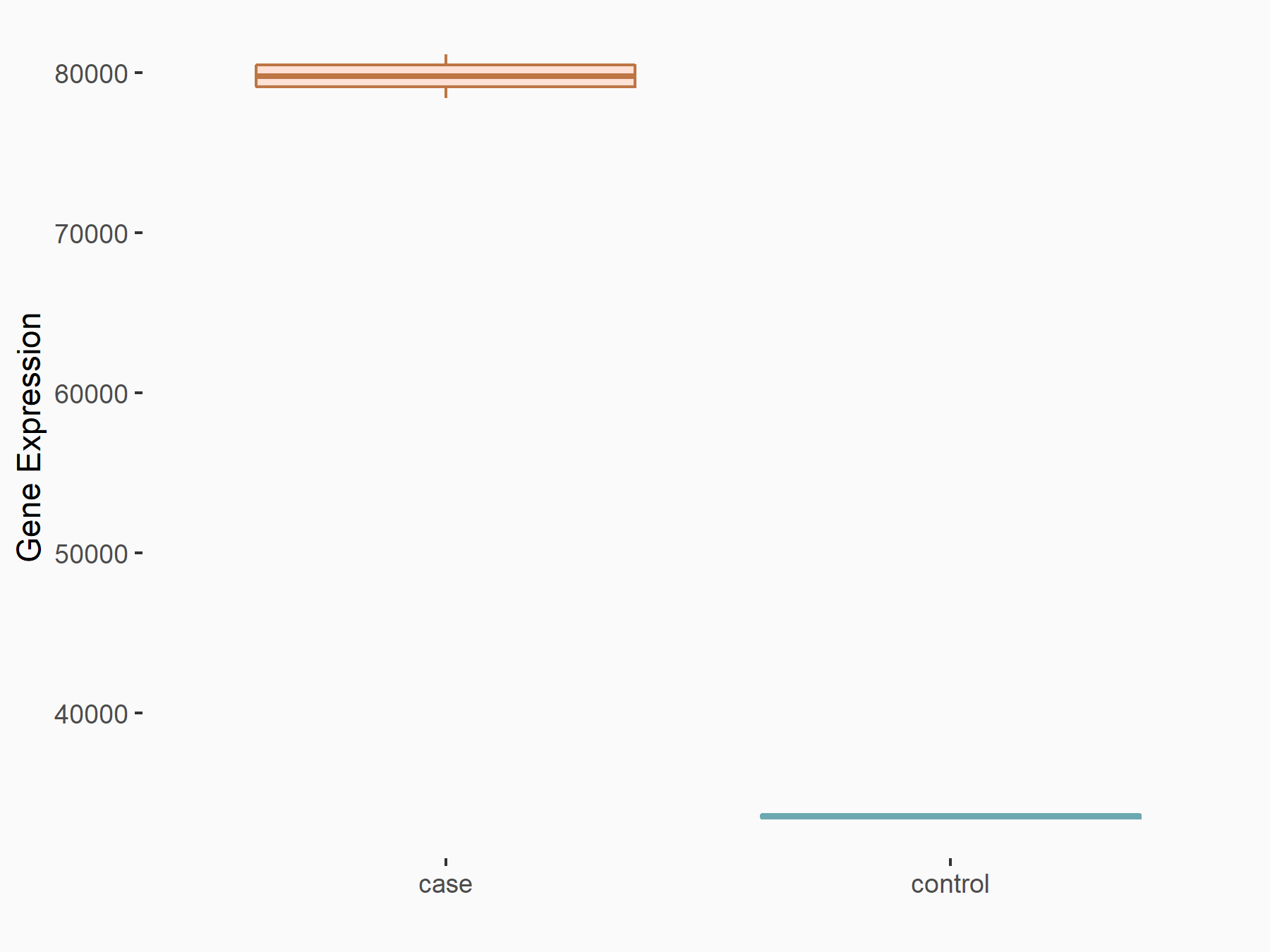  |
logFC: 1.25E+00 p-value: 7.56E-25 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Targeting METTL3/14 in vitro increases protein level of ACLY and Stearoyl-CoA desaturase (SCD) as well as triglyceride and cholesterol production and accumulation of lipid droplets. These findings demonstrate a new NAFLD mouse model that provides a study platform for DM2-related NAFLD and reveals a unique epitranscriptional regulating mechanism for lipid metabolism via m6A-modified protein expression of ACLY and SCD1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-alcoholic fatty liver disease | ICD-11: DB92 | ||
| Pathway Response | Glycerolipid metabolism | hsa00561 | ||
| Cell Process | Lipid metabolism | |||
| In-vitro Model | LM3 | Malignant neoplasms | Mus musculus | CVCL_D269 |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| In-vivo Model | Mice with a Tmem30a deletion specifically in pancreatic beta cells were generated as previously described. Mice developed with NAFLD were named for Tmem30a-associated NAFLD (TAN) mice. The littermate mice with genotypes of Tmem30aloxP/loxP were used as controls. | |||
YTH domain-containing protein 2 (YTHDC2) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | In nonalcoholic fatty liver disease, Ythdc2 could bind to mRNA of lipogenic genes, including sterol regulatory element-binding protein 1c, fatty acid synthase, Stearoyl-CoA desaturase (SCD), and acetyl-CoA carboxylase 1, to decrease their mRNA stability and inhibit gene expression. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Non-alcoholic fatty liver disease | ICD-11: DB92 | ||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vivo Model | All mice were housed at 21℃ ± 1℃ with a humidity of 55% ± 10% and a 12-hour light/dark cycle. The high-fat diets (HFDs), containing 60% kcal from fat, 20% kcal from carbohydrate, and 20% kcal from protein. | |||
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Targeting METTL3/14 in vitro increases protein level of ACLY and Stearoyl-CoA desaturase (SCD) as well as triglyceride and cholesterol production and accumulation of lipid droplets. These findings demonstrate a new NAFLD mouse model that provides a study platform for DM2-related NAFLD and reveals a unique epitranscriptional regulating mechanism for lipid metabolism via m6A-modified protein expression of ACLY and SCD1. | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycerolipid metabolism | hsa00561 | ||
| Cell Process | Lipid metabolism | |||
| In-vitro Model | LM3 | Malignant neoplasms | Mus musculus | CVCL_D269 |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| In-vivo Model | Mice with a Tmem30a deletion specifically in pancreatic beta cells were generated as previously described. Mice developed with NAFLD were named for Tmem30a-associated NAFLD (TAN) mice. The littermate mice with genotypes of Tmem30aloxP/loxP were used as controls. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [6] | |||
| Response Summary | In nonalcoholic fatty liver disease, Ythdc2 could bind to mRNA of lipogenic genes, including sterol regulatory element-binding protein 1c, fatty acid synthase, Stearoyl-CoA desaturase (SCD), and acetyl-CoA carboxylase 1, to decrease their mRNA stability and inhibit gene expression. | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulator | YTH domain-containing protein 2 (YTHDC2) | READER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vivo Model | All mice were housed at 21℃ ± 1℃ with a humidity of 55% ± 10% and a 12-hour light/dark cycle. The high-fat diets (HFDs), containing 60% kcal from fat, 20% kcal from carbohydrate, and 20% kcal from protein. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
DNA modification
m6A Regulator: Methyltransferase-like 16 (METTL16)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT02050 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase 16, RNA N6-adenosine (METTL16) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Papillary Thyroid Cancer | |
| Drug | Simvastatin | |
m6A Regulator: YTH domain-containing protein 2 (YTHDC2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT02051 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase 16, RNA N6-adenosine (METTL16) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Papillary Thyroid Cancer | |
| Drug | Simvastatin | |
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03134 | ||
| Epigenetic Regulator | Lactate dehydrogenase A (LDHA) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18lac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Nonalcoholic fatty liver disease | |
m6A Regulator: YTH domain-containing family protein 1 (YTHDF1)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03135 | ||
| Epigenetic Regulator | Lactate dehydrogenase A (LDHA) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18lac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Nonalcoholic fatty liver disease | |
Non-coding RNA
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05019 | ||
| Epigenetic Regulator | DARS1 antisense RNA 1 (DARS1-AS1) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 3 (IGF2BP3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Cervical cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00388)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: AC4SITE000144 | Click to Show/Hide the Full List | ||
| mod site | chr10:100364602-100364603:+ | [7] | |
| Sequence | ATTGGAGAAGCGGTGGATAACTAGCCAGACAAAATTTGAGA | ||
| Cell/Tissue List | H1 | ||
| Seq Type List | ac4C-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: ac4C_site_233 | ||
Adenosine-to-Inosine editing (A-to-I)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE004358 | Click to Show/Hide the Full List | ||
| mod site | chr10:100353971-100353972:+ | [8] | |
| Sequence | GTTGTATGAGCTCTTTTGCAAGGCACCAGAATGGTGCACCC | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: RNA-editing_site_18074 | ||
| mod ID: A2ISITE004359 | Click to Show/Hide the Full List | ||
| mod site | chr10:100361843-100361844:+ | [8] | |
| Sequence | TGGTAAAAACAGCAGCTCATAGAATTTTGAGTATTCCATGA | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: RNA-editing_site_18075 | ||
5-methylcytidine (m5C)
| In total 6 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE004741 | Click to Show/Hide the Full List | ||
| mod site | chr10:100348105-100348106:+ | [9] | |
| Sequence | CCACCACCATTACAGCGCCTCCCTCCAGGGTCCTGCAGAAT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m5C_site_5744 | ||
| mod ID: M5CSITE004742 | Click to Show/Hide the Full List | ||
| mod site | chr10:100348107-100348108:+ | [9] | |
| Sequence | ACCACCATTACAGCGCCTCCCTCCAGGGTCCTGCAGAATGG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m5C_site_5745 | ||
| mod ID: M5CSITE004743 | Click to Show/Hide the Full List | ||
| mod site | chr10:100354483-100354484:+ | ||
| Sequence | ACCACAAGTTTTCAGAAACACATGCTGATCCTCATAATTCC | ||
| Cell/Tissue List | lung | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m5C_site_5746 | ||
| mod ID: M5CSITE004744 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362130-100362131:+ | [9] | |
| Sequence | AGTTGGGATGCCAATTTCCTCTCCACTGCTGGACATGAGAT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m5C_site_5747 | ||
| mod ID: M5CSITE004745 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363382-100363383:+ | [9] | |
| Sequence | TGGAAGGGAGGAAGGCCTTTCTTCTGTGTTAATTGCGTAGA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m5C_site_5748 | ||
| mod ID: M5CSITE004746 | Click to Show/Hide the Full List | ||
| mod site | chr10:100364290-100364291:+ | ||
| Sequence | GGTGGTTAAGGCCAGGGCCTCTCCAACCACTGTGCCACTGA | ||
| Cell/Tissue List | liver | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m5C_site_5749 | ||
N6-methyladenosine (m6A)
| In total 115 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE001493 | Click to Show/Hide the Full List | ||
| mod site | chr10:100347295-100347296:+ | [10] | |
| Sequence | GGGGGCTGAGGAAATACCGGACACGGTCACCCGTTGCCAGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; A549; hESC-HEK293T; LCLs; MM6; TREX; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114339 | ||
| mod ID: M6ASITE001494 | Click to Show/Hide the Full List | ||
| mod site | chr10:100347343-100347344:+ | [10] | |
| Sequence | TTTAAATTCCCGGCTCGGGGACCTCCACGCACCGCGGCTAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; A549; LCLs; MM6; TREX; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114340 | ||
| mod ID: M6ASITE001495 | Click to Show/Hide the Full List | ||
| mod site | chr10:100347369-100347370:+ | [11] | |
| Sequence | ACGCACCGCGGCTAGCGCCGACAACCAGCTAGCGTGCAAGG | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114341 | ||
| mod ID: M6ASITE001496 | Click to Show/Hide the Full List | ||
| mod site | chr10:100347424-100347425:+ | [10] | |
| Sequence | GCGTACCGGCGGGCTTCGAAACCGCAGTCCTCCGGCGACCC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; CD34; A549; HepG2; MT4; MM6; TREX; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114342 | ||
| mod ID: M6ASITE001497 | Click to Show/Hide the Full List | ||
| mod site | chr10:100347448-100347449:+ | [10] | |
| Sequence | CAGTCCTCCGGCGACCCCGAACTCCGCTCCGGAGCCTCAGC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; CD34; A549; HepG2; MT4; MM6; Huh7; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114343 | ||
| mod ID: M6ASITE001498 | Click to Show/Hide the Full List | ||
| mod site | chr10:100348096-100348097:+ | [12] | |
| Sequence | ATACCACCACCACCACCATTACAGCGCCTCCCTCCAGGGTC | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114344 | ||
| mod ID: M6ASITE001499 | Click to Show/Hide the Full List | ||
| mod site | chr10:100348169-100348170:+ | [11] | |
| Sequence | GCCCCTCTACTTGGAAGACGACATTCGCCCTGATATAAAAG | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114345 | ||
| mod ID: M6ASITE001500 | Click to Show/Hide the Full List | ||
| mod site | chr10:100348202-100348203:+ | [13] | |
| Sequence | TATAAAAGATGATATATATGACCCCACCTACAAGGATAAGG | ||
| Motif Score | 2.839113095 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114346 | ||
| mod ID: M6ASITE001501 | Click to Show/Hide the Full List | ||
| mod site | chr10:100348211-100348212:+ | [11] | |
| Sequence | TGATATATATGACCCCACCTACAAGGATAAGGAAGGCCCAA | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114347 | ||
| mod ID: M6ASITE001502 | Click to Show/Hide the Full List | ||
| mod site | chr10:100348259-100348260:+ | [10] | |
| Sequence | GGTTGAATATGTCTGGAGAAACATCATCCTTATGTCTCTGC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; MT4 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114348 | ||
| mod ID: M6ASITE001503 | Click to Show/Hide the Full List | ||
| mod site | chr10:100348281-100348282:+ | [11] | |
| Sequence | ATCATCCTTATGTCTCTGCTACACTTGGGAGCCCTGTATGG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114349 | ||
| mod ID: M6ASITE001504 | Click to Show/Hide the Full List | ||
| mod site | chr10:100348331-100348332:+ | [14] | |
| Sequence | GATTCCTACCTGCAAGTTCTACACCTGGCTTTGGGGTAAGC | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114350 | ||
| mod ID: M6ASITE001505 | Click to Show/Hide the Full List | ||
| mod site | chr10:100352437-100352438:+ | [11] | |
| Sequence | TCTGTGGAGCCACCGCTCTTACAAAGCTCGGCTGCCCCTAC | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114351 | ||
| mod ID: M6ASITE001506 | Click to Show/Hide the Full List | ||
| mod site | chr10:100352479-100352480:+ | [11] | |
| Sequence | GCTCTTTCTGATCATTGCCAACACAATGGCATTCCAGGTAA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114352 | ||
| mod ID: M6ASITE001507 | Click to Show/Hide the Full List | ||
| mod site | chr10:100354451-100354452:+ | [13] | |
| Sequence | TGTCTATGAATGGGCTCGTGACCACCGTGCCCACCACAAGT | ||
| Motif Score | 2.839113095 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114353 | ||
| mod ID: M6ASITE001508 | Click to Show/Hide the Full List | ||
| mod site | chr10:100354466-100354467:+ | [11] | |
| Sequence | TCGTGACCACCGTGCCCACCACAAGTTTTCAGAAACACATG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114354 | ||
| mod ID: M6ASITE001509 | Click to Show/Hide the Full List | ||
| mod site | chr10:100354480-100354481:+ | [10] | |
| Sequence | CCCACCACAAGTTTTCAGAAACACATGCTGATCCTCATAAT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114355 | ||
| mod ID: M6ASITE001510 | Click to Show/Hide the Full List | ||
| mod site | chr10:100354506-100354507:+ | [14] | |
| Sequence | GCTGATCCTCATAATTCCCGACGTGGCTTTTTCTTCTCTCA | ||
| Motif Score | 2.839505952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114356 | ||
| mod ID: M6ASITE001511 | Click to Show/Hide the Full List | ||
| mod site | chr10:100354551-100354552:+ | [10] | |
| Sequence | GGTTGGCTGCTTGTGCGCAAACACCCAGCTGTCAAAGAGAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293; hESC-HEK293T; HepG2 | ||
| Seq Type List | m6A-seq; m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114357 | ||
| mod ID: M6ASITE001512 | Click to Show/Hide the Full List | ||
| mod site | chr10:100354586-100354587:+ | [10] | |
| Sequence | AGAGAAGGGGAGTACGCTAGACTTGTCTGACCTAGAAGCTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114358 | ||
| mod ID: M6ASITE001513 | Click to Show/Hide the Full List | ||
| mod site | chr10:100354611-100354612:+ | [10] | |
| Sequence | TCTGACCTAGAAGCTGAGAAACTGGTGATGTTCCAGAGGAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114359 | ||
| mod ID: M6ASITE001514 | Click to Show/Hide the Full List | ||
| mod site | chr10:100356536-100356537:+ | [11] | |
| Sequence | GGTCTGTCAATGTAGGTACTACAAACCTGGCTTGCTGATGA | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114360 | ||
| mod ID: M6ASITE001515 | Click to Show/Hide the Full List | ||
| mod site | chr10:100356540-100356541:+ | [10] | |
| Sequence | TGTCAATGTAGGTACTACAAACCTGGCTTGCTGATGATGTG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114361 | ||
| mod ID: M6ASITE001516 | Click to Show/Hide the Full List | ||
| mod site | chr10:100356604-100356605:+ | [10] | |
| Sequence | CCTGGTATTTCTGGGGTGAAACTTTTCAAAACAGTGTGTTC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114362 | ||
| mod ID: M6ASITE001517 | Click to Show/Hide the Full List | ||
| mod site | chr10:100356614-100356615:+ | [10] | |
| Sequence | CTGGGGTGAAACTTTTCAAAACAGTGTGTTCGTTGCCACTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114363 | ||
| mod ID: M6ASITE001518 | Click to Show/Hide the Full List | ||
| mod site | chr10:100356677-100356678:+ | [10] | |
| Sequence | TAATGCCACCTGGCTGGTGAACAGTGCTGCCCACCTCTTCG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114364 | ||
| mod ID: M6ASITE001519 | Click to Show/Hide the Full List | ||
| mod site | chr10:100356719-100356720:+ | [10] | |
| Sequence | ATATCGTCCTTATGACAAGAACATTAGCCCCCGGGAGAATA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114365 | ||
| mod ID: M6ASITE001520 | Click to Show/Hide the Full List | ||
| mod site | chr10:100356750-100356751:+ | [14] | |
| Sequence | CGGGAGAATATCCTGGTTTCACTTGGAGCTGTGGGTAAGTC | ||
| Motif Score | 2.469291667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114366 | ||
| mod ID: M6ASITE001521 | Click to Show/Hide the Full List | ||
| mod site | chr10:100360745-100360746:+ | [14] | |
| Sequence | TGTTTCAGGTGAGGGCTTCCACAACTACCACCACTCCTTTC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114367 | ||
| mod ID: M6ASITE001522 | Click to Show/Hide the Full List | ||
| mod site | chr10:100360799-100360800:+ | [11] | |
| Sequence | TGCCAGTGAGTACCGCTGGCACATCAACTTCACCACATTCT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114368 | ||
| mod ID: M6ASITE001523 | Click to Show/Hide the Full List | ||
| mod site | chr10:100360813-100360814:+ | [11] | |
| Sequence | GCTGGCACATCAACTTCACCACATTCTTCATTGATTGCATG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114369 | ||
| mod ID: M6ASITE001524 | Click to Show/Hide the Full List | ||
| mod site | chr10:100360903-100360904:+ | [10] | |
| Sequence | TCTTGGCCAGGATTAAAAGAACCGGAGATGGAAACTACAAG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114370 | ||
| mod ID: M6ASITE001525 | Click to Show/Hide the Full List | ||
| mod site | chr10:100360916-100360917:+ | [10] | |
| Sequence | TAAAAGAACCGGAGATGGAAACTACAAGAGTGGCTGAGTTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114371 | ||
| mod ID: M6ASITE001526 | Click to Show/Hide the Full List | ||
| mod site | chr10:100360919-100360920:+ | [11] | |
| Sequence | AAGAACCGGAGATGGAAACTACAAGAGTGGCTGAGTTTGGG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114372 | ||
| mod ID: M6ASITE001527 | Click to Show/Hide the Full List | ||
| mod site | chr10:100360964-100360965:+ | [10] | |
| Sequence | CTCAGGTTCCTTTTTCAAAAACCAGCCAGGCAGAGGTTTTA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114373 | ||
| mod ID: M6ASITE001528 | Click to Show/Hide the Full List | ||
| mod site | chr10:100361091-100361092:+ | [14] | |
| Sequence | TTCAAAAGTATTGAAAGCCAACAACTCTGCCTTTATGATGC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114374 | ||
| mod ID: M6ASITE001529 | Click to Show/Hide the Full List | ||
| mod site | chr10:100361262-100361263:+ | [10] | |
| Sequence | GTCCAGCTTCCAAAGCCTAGACAACCTTTCTGTAGCCTAAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T | ||
| Seq Type List | m6A-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114375 | ||
| mod ID: M6ASITE001530 | Click to Show/Hide the Full List | ||
| mod site | chr10:100361283-100361284:+ | [14] | |
| Sequence | CAACCTTTCTGTAGCCTAAAACGAATGGTCTTTGCTCCAGA | ||
| Motif Score | 2.179660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114376 | ||
| mod ID: M6ASITE001531 | Click to Show/Hide the Full List | ||
| mod site | chr10:100361445-100361446:+ | [14] | |
| Sequence | TGGAAAAGCAACTTCATTTGACACAAAGCTTCTAAAGCAGG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114377 | ||
| mod ID: M6ASITE001532 | Click to Show/Hide the Full List | ||
| mod site | chr10:100361530-100361531:+ | [10] | |
| Sequence | AGGGAAGCGAAGCAAGAGGAACCTCTCGCCATGATCAGACA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; AML | ||
| Seq Type List | m6A-seq; miCLIP | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114378 | ||
| mod ID: M6ASITE001533 | Click to Show/Hide the Full List | ||
| mod site | chr10:100361548-100361549:+ | [10] | |
| Sequence | GAACCTCTCGCCATGATCAGACATACAGCTGCCTACCTAAT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114379 | ||
| mod ID: M6ASITE001534 | Click to Show/Hide the Full List | ||
| mod site | chr10:100361573-100361574:+ | [10] | |
| Sequence | CAGCTGCCTACCTAATGAGGACTTCAAGCCCCACCACATAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114380 | ||
| mod ID: M6ASITE001535 | Click to Show/Hide the Full List | ||
| mod site | chr10:100361588-100361589:+ | [11] | |
| Sequence | TGAGGACTTCAAGCCCCACCACATAGCATGCTTCCTTTCTC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114381 | ||
| mod ID: M6ASITE001536 | Click to Show/Hide the Full List | ||
| mod site | chr10:100361666-100361667:+ | [11] | |
| Sequence | AATGCTAATTCAATGCCGCAACATATAGTTGAGGCCGAGGA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114382 | ||
| mod ID: M6ASITE001537 | Click to Show/Hide the Full List | ||
| mod site | chr10:100361697-100361698:+ | [10] | |
| Sequence | AGGCCGAGGATAAAGAAAAGACATTTTAAGTTTGTAGTAAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T | ||
| Seq Type List | m6A-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114383 | ||
| mod ID: M6ASITE001538 | Click to Show/Hide the Full List | ||
| mod site | chr10:100361765-100361766:+ | [14] | |
| Sequence | TTTTCTTTTTTTCTTTAATAACAAGGAGATTTCTTAGTTCA | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114384 | ||
| mod ID: M6ASITE001539 | Click to Show/Hide the Full List | ||
| mod site | chr10:100361831-100361832:+ | [10] | |
| Sequence | TGTTTCCAGAATTGGTAAAAACAGCAGCTCATAGAATTTTG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114385 | ||
| mod ID: M6ASITE001540 | Click to Show/Hide the Full List | ||
| mod site | chr10:100361958-100361959:+ | [10] | |
| Sequence | GATGGCTGTGGCATTTCCAAACATCCAAAAAAAGGGAAGGA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114386 | ||
| mod ID: M6ASITE001541 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362018-100362019:+ | [14] | |
| Sequence | GTCAAAAATAAAATATATATACATATATACATTGCTTAGAA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114387 | ||
| mod ID: M6ASITE001542 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362026-100362027:+ | [11] | |
| Sequence | TAAAATATATATACATATATACATTGCTTAGAACGTTAAAC | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114388 | ||
| mod ID: M6ASITE001543 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362038-100362039:+ | [14] | |
| Sequence | ACATATATACATTGCTTAGAACGTTAAACTATTAGAGTATT | ||
| Motif Score | 2.925321429 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114389 | ||
| mod ID: M6ASITE001544 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362045-100362046:+ | [10] | |
| Sequence | TACATTGCTTAGAACGTTAAACTATTAGAGTATTTCCCTTC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114390 | ||
| mod ID: M6ASITE001545 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362088-100362089:+ | [10] | |
| Sequence | AAGAGGGATGTTTGGAAAAAACTCTGAAGGAGAGGAGGAAT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114391 | ||
| mod ID: M6ASITE001546 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362142-100362143:+ | [10] | |
| Sequence | AATTTCCTCTCCACTGCTGGACATGAGATGGAGAGGCTGAG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; HepG2 | ||
| Seq Type List | m6A-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114392 | ||
| mod ID: M6ASITE001547 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362165-100362166:+ | [10] | |
| Sequence | TGAGATGGAGAGGCTGAGGGACAGGATCTATAGGCAGCTTC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2 | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114393 | ||
| mod ID: M6ASITE001548 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362195-100362196:+ | [15] | |
| Sequence | TAGGCAGCTTCTAAGAGCGAACTTCACATAGGAAGGGATCT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114394 | ||
| mod ID: M6ASITE001549 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362220-100362221:+ | [11] | |
| Sequence | ACATAGGAAGGGATCTGAGAACACGTTGCCAGGGGCTTGAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T; HepG2 | ||
| Seq Type List | MAZTER-seq; m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114395 | ||
| mod ID: M6ASITE001550 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362247-100362248:+ | [14] | |
| Sequence | GCCAGGGGCTTGAGAAGGTTACTGAGTGAGTTATTGGGAGT | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114396 | ||
| mod ID: M6ASITE001551 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362338-100362339:+ | [14] | |
| Sequence | TCTCCTTGAAATGAGTAAAAACTAGAAGGCTTCTCTCCACA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114397 | ||
| mod ID: M6ASITE001552 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362410-100362411:+ | [14] | |
| Sequence | GGAGAAGGGGGTCTCTGTTAACATCTAGCCTAAAGTATACA | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114398 | ||
| mod ID: M6ASITE001553 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362428-100362429:+ | [11] | |
| Sequence | TAACATCTAGCCTAAAGTATACAACTGCCTGGGGGGCAGGG | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114399 | ||
| mod ID: M6ASITE001554 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362463-100362464:+ | [14] | |
| Sequence | GCAGGGTTAGGAATCTCTTCACTACCCTGATTCTTGATTCC | ||
| Motif Score | 2.469291667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114400 | ||
| mod ID: M6ASITE001555 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362466-100362467:+ | [14] | |
| Sequence | GGGTTAGGAATCTCTTCACTACCCTGATTCTTGATTCCTGG | ||
| Motif Score | 2.05802381 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114401 | ||
| mod ID: M6ASITE001556 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362514-100362515:+ | [14] | |
| Sequence | CTGTCTGTCCCTTTTCTTTGACCAGATCTTTCTCTTCCCTG | ||
| Motif Score | 2.839113095 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114402 | ||
| mod ID: M6ASITE001557 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362556-100362557:+ | [11] | |
| Sequence | ACGTTTTCTTCTTTCCCTGGACAGGCAGCCTCCTTTGTGTG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T; AML | ||
| Seq Type List | MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114403 | ||
| mod ID: M6ASITE001558 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362595-100362596:+ | [14] | |
| Sequence | TGTATTCAGAGGCAGTGATGACTTGCTGTCCAGGCAGCTCC | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114404 | ||
| mod ID: M6ASITE001559 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362623-100362624:+ | [11] | |
| Sequence | TCCAGGCAGCTCCCTCCTGCACACAGAATGCTCAGGGTCAC | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114405 | ||
| mod ID: M6ASITE001560 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362642-100362643:+ | [14] | |
| Sequence | CACACAGAATGCTCAGGGTCACTGAACCACTGCTTCTCTTT | ||
| Motif Score | 2.469291667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114406 | ||
| mod ID: M6ASITE001561 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362647-100362648:+ | [15] | |
| Sequence | AGAATGCTCAGGGTCACTGAACCACTGCTTCTCTTTTGAAA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114407 | ||
| mod ID: M6ASITE001562 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362715-100362716:+ | [11] | |
| Sequence | CCTCCGCAGTGTCTCCACCTACACCCCTGTGCTCCCCTGCC | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114408 | ||
| mod ID: M6ASITE001563 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362752-100362753:+ | [15] | |
| Sequence | TGCCACACTGATGGCTCAAGACAAGGCTGGCAAACCCTCCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114409 | ||
| mod ID: M6ASITE001564 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362765-100362766:+ | [15] | |
| Sequence | GCTCAAGACAAGGCTGGCAAACCCTCCCAGAAACATCTCTG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114410 | ||
| mod ID: M6ASITE001565 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362777-100362778:+ | [11] | |
| Sequence | GCTGGCAAACCCTCCCAGAAACATCTCTGGCCCAGAAAGCC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T; HepG2 | ||
| Seq Type List | MAZTER-seq; m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114411 | ||
| mod ID: M6ASITE001566 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362825-100362826:+ | [12] | |
| Sequence | CCCTCCCTCTCTCATGAGGCACAGCCAAGCCAAGCGCTCAT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114412 | ||
| mod ID: M6ASITE001567 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362866-100362867:+ | [11] | |
| Sequence | GTTGAGCCAGTGGGCCAGCCACAGAGCAAAAGAGGGTTTAT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114413 | ||
| mod ID: M6ASITE001568 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362913-100362914:+ | [15] | |
| Sequence | TCCCCTCTCTCTGGGTCAGAACCAGAGGGCATGCTGAATGC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114414 | ||
| mod ID: M6ASITE001569 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363014-100363015:+ | [16] | |
| Sequence | GCTTGTAGAAGTAGGAGGAAACAGTTCTCACTGGGAAGAAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114415 | ||
| mod ID: M6ASITE001570 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363046-100363047:+ | [16] | |
| Sequence | GGGAAGAAGCAAGGGCAAGAACCCAAGTGCCTCACCTCGAA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HepG2; H1299; Huh7; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114416 | ||
| mod ID: M6ASITE001571 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363097-100363098:+ | [16] | |
| Sequence | GTTCCCTGGAGTCAGGGTGAACTGCAAAGCTTTGGCTGAGA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HepG2; H1299; Huh7; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114417 | ||
| mod ID: M6ASITE001572 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363117-100363118:+ | [16] | |
| Sequence | ACTGCAAAGCTTTGGCTGAGACCTGGGATTTGAGATACCAC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HepG2; H1299; Huh7; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114418 | ||
| mod ID: M6ASITE001573 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363136-100363137:+ | [11] | |
| Sequence | GACCTGGGATTTGAGATACCACAAACCCTGCTGAACACAGT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114419 | ||
| mod ID: M6ASITE001574 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363140-100363141:+ | [17] | |
| Sequence | TGGGATTTGAGATACCACAAACCCTGCTGAACACAGTGTCT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HepG2; BGC823; H1299; Huh7; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114420 | ||
| mod ID: M6ASITE001575 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363150-100363151:+ | [17] | |
| Sequence | GATACCACAAACCCTGCTGAACACAGTGTCTGTTCAGCAAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HepG2; BGC823; H1299; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114421 | ||
| mod ID: M6ASITE001576 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363170-100363171:+ | [17] | |
| Sequence | ACACAGTGTCTGTTCAGCAAACTAACCAGCATTCCCTACAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HepG2; BGC823; H1299; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114422 | ||
| mod ID: M6ASITE001577 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363201-100363202:+ | [11] | |
| Sequence | TTCCCTACAGCCTAGGGCAGACAATAGTATAGAAGTCTGGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T; BGC823; H1299; AML | ||
| Seq Type List | MAZTER-seq; m6A-seq; miCLIP | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114423 | ||
| mod ID: M6ASITE001578 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363227-100363228:+ | [14] | |
| Sequence | GTATAGAAGTCTGGAAAAAAACAAAAACAGAATTTGAGAAC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HEK293T; BGC823; H1299 | ||
| Seq Type List | DART-seq; m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114424 | ||
| mod ID: M6ASITE001579 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363233-100363234:+ | [18] | |
| Sequence | AAGTCTGGAAAAAAACAAAAACAGAATTTGAGAACCTTGGA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | BGC823; H1299 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114425 | ||
| mod ID: M6ASITE001580 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363246-100363247:+ | [18] | |
| Sequence | AACAAAAACAGAATTTGAGAACCTTGGACCACTCCTGTCCC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | BGC823; H1299 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114426 | ||
| mod ID: M6ASITE001581 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363253-100363254:+ | [18] | |
| Sequence | ACAGAATTTGAGAACCTTGGACCACTCCTGTCCCTGTAGCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | BGC823; H1299; AML | ||
| Seq Type List | m6A-seq; miCLIP | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114427 | ||
| mod ID: M6ASITE001582 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363323-100363324:+ | [11] | |
| Sequence | TCTATTAAGATTGGAAATGTACACTACCAAACACTCAGTCC | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114428 | ||
| mod ID: M6ASITE001583 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363333-100363334:+ | [11] | |
| Sequence | TTGGAAATGTACACTACCAAACACTCAGTCCACTGTTGAGC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T; BGC823 | ||
| Seq Type List | MAZTER-seq; m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114429 | ||
| mod ID: M6ASITE001584 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363423-100363424:+ | [14] | |
| Sequence | GGCTACAGGGGTTAGCCTGGACTAAAGGCATCCTTGTCTTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114430 | ||
| mod ID: M6ASITE001585 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363485-100363486:+ | [14] | |
| Sequence | AAAGGATCTAAGGGAAGATCACTGTAGTTTAGTTCTGTTGA | ||
| Motif Score | 2.469291667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114431 | ||
| mod ID: M6ASITE001586 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363586-100363587:+ | [14] | |
| Sequence | GCCTGCTTGATAATATATAAACAATAAAAACAACTTTCACT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114432 | ||
| mod ID: M6ASITE001587 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363595-100363596:+ | [11] | |
| Sequence | ATAATATATAAACAATAAAAACAACTTTCACTTCTTCCTAT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114433 | ||
| mod ID: M6ASITE001588 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363598-100363599:+ | [14] | |
| Sequence | ATATATAAACAATAAAAACAACTTTCACTTCTTCCTATTGT | ||
| Motif Score | 2.595904762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114434 | ||
| mod ID: M6ASITE001589 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363656-100363657:+ | [14] | |
| Sequence | CTGATCTGTACCATGACCCTACATAAGGCTGGATGGCACCT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114435 | ||
| mod ID: M6ASITE001590 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363743-100363744:+ | [10] | |
| Sequence | GTGTGTCTGCTGAGTAAGGAACACGATTTTCAAGATTCTAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114436 | ||
| mod ID: M6ASITE001591 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363794-100363795:+ | [10] | |
| Sequence | AAGTGACACATTAATGATAAACTCAGATCTGATCAAGAGTC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114437 | ||
| mod ID: M6ASITE001592 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363945-100363946:+ | [10] | |
| Sequence | AGAATCCCTTTGCACCTGAGACCCTACTGAAGTGGCTGGTA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114438 | ||
| mod ID: M6ASITE001593 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363950-100363951:+ | [14] | |
| Sequence | CCCTTTGCACCTGAGACCCTACTGAAGTGGCTGGTAGAAAA | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114439 | ||
| mod ID: M6ASITE001594 | Click to Show/Hide the Full List | ||
| mod site | chr10:100364070-100364071:+ | [11] | |
| Sequence | GCTGCTTTTCTTAAGTGCCCACATTTGATGGAGGGTGGAAA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114440 | ||
| mod ID: M6ASITE001595 | Click to Show/Hide the Full List | ||
| mod site | chr10:100364388-100364389:+ | [14] | |
| Sequence | GTTAAAATGGGGATAATAATACTGACCTACCTCAAAGGGCA | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | rmsk_3397884; ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114441 | ||
| mod ID: M6ASITE001596 | Click to Show/Hide the Full List | ||
| mod site | chr10:100364475-100364476:+ | [10] | |
| Sequence | TCAGCACAGGAATTCTCAAGACCTGAGTATTTTTTATAATA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; hESCs; fibroblasts; A549 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114442 | ||
| mod ID: M6ASITE001597 | Click to Show/Hide the Full List | ||
| mod site | chr10:100364512-100364513:+ | [10] | |
| Sequence | AATAGGAATGTCCACCATGAACTTGATACGTCCGTGTGTCC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; hNPCs; hESCs; fibroblasts; A549; AML | ||
| Seq Type List | m6A-seq; DART-seq; miCLIP | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114443 | ||
| mod ID: M6ASITE001598 | Click to Show/Hide the Full List | ||
| mod site | chr10:100364569-100364570:+ | [10] | |
| Sequence | TCTATATGGTTCTCCAAGAAACTGAATGAATCCATTGGAGA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; hNPCs; hESCs; fibroblasts; A549 | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114444 | ||
| mod ID: M6ASITE001599 | Click to Show/Hide the Full List | ||
| mod site | chr10:100364610-100364611:+ | [10] | |
| Sequence | AGCGGTGGATAACTAGCCAGACAAAATTTGAGAATACATAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; hNPCs; hESCs; fibroblasts; A549 | ||
| Seq Type List | m6A-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114445 | ||
| mod ID: M6ASITE001600 | Click to Show/Hide the Full List | ||
| mod site | chr10:100364631-100364632:+ | [10] | |
| Sequence | CAAAATTTGAGAATACATAAACAACGCATTGCCACGGAAAC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hNPCs; hESCs; fibroblasts; A549 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114446 | ||
| mod ID: M6ASITE001601 | Click to Show/Hide the Full List | ||
| mod site | chr10:100364634-100364635:+ | [14] | |
| Sequence | AATTTGAGAATACATAAACAACGCATTGCCACGGAAACATA | ||
| Motif Score | 2.147845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114447 | ||
| mod ID: M6ASITE001602 | Click to Show/Hide the Full List | ||
| mod site | chr10:100364650-100364651:+ | [10] | |
| Sequence | AACAACGCATTGCCACGGAAACATACAGAGGATGCCTTTTC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hNPCs; fibroblasts; A549 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114448 | ||
| mod ID: M6ASITE001603 | Click to Show/Hide the Full List | ||
| mod site | chr10:100364654-100364655:+ | [14] | |
| Sequence | ACGCATTGCCACGGAAACATACAGAGGATGCCTTTTCTGTG | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114449 | ||
| mod ID: M6ASITE001604 | Click to Show/Hide the Full List | ||
| mod site | chr10:100364718-100364719:+ | [14] | |
| Sequence | TTATGTGGGATATAGTAGTTACTTGTGACAAGAATAATTTT | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114450 | ||
| mod ID: M6ASITE001605 | Click to Show/Hide the Full List | ||
| mod site | chr10:100364725-100364726:+ | [14] | |
| Sequence | GGATATAGTAGTTACTTGTGACAAGAATAATTTTGGAATAA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114451 | ||
| mod ID: M6ASITE001606 | Click to Show/Hide the Full List | ||
| mod site | chr10:100364761-100364762:+ | [14] | |
| Sequence | AATAATTTCTATTAATATCAACTCTGAAGCTAATTGTACTA | ||
| Motif Score | 2.595904762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114452 | ||
| mod ID: M6ASITE001607 | Click to Show/Hide the Full List | ||
| mod site | chr10:100364778-100364779:+ | [14] | |
| Sequence | TCAACTCTGAAGCTAATTGTACTAATCTGAGATTGTGTTTG | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: m6A_site_114453 | ||
2'-O-Methylation (2'-O-Me)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: 2OMSITE000576 | Click to Show/Hide the Full List | ||
| mod site | chr10:100362092-100362093:+ | [19] | |
| Sequence | GGGATGTTTGGAAAAAACTCTGAAGGAGAGGAGGAATTAGT | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: Nm_site_810 | ||
| mod ID: 2OMSITE000577 | Click to Show/Hide the Full List | ||
| mod site | chr10:100363099-100363100:+ | [19] | |
| Sequence | TCCCTGGAGTCAGGGTGAACTGCAAAGCTTTGGCTGAGACC | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: Nm_site_811 | ||
Pseudouridine (Pseudo)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: PSESITE000306 | Click to Show/Hide the Full List | ||
| mod site | chr10:100347418-100347419:+ | [20] | |
| Sequence | CTCAGCGCGTACCGGCGGGCTTCGAAACCGCAGTCCTCCGG | ||
| Transcript ID List | ENST00000370355.3 | ||
| External Link | RMBase: Pseudo_site_749 | ||
References