m6A-centered Crosstalk Information
Mechanism of Crosstalk between m6A Modification and Epigenetic Regulation
| Crosstalk ID |
M6ACROT05942
|
[1] | |||
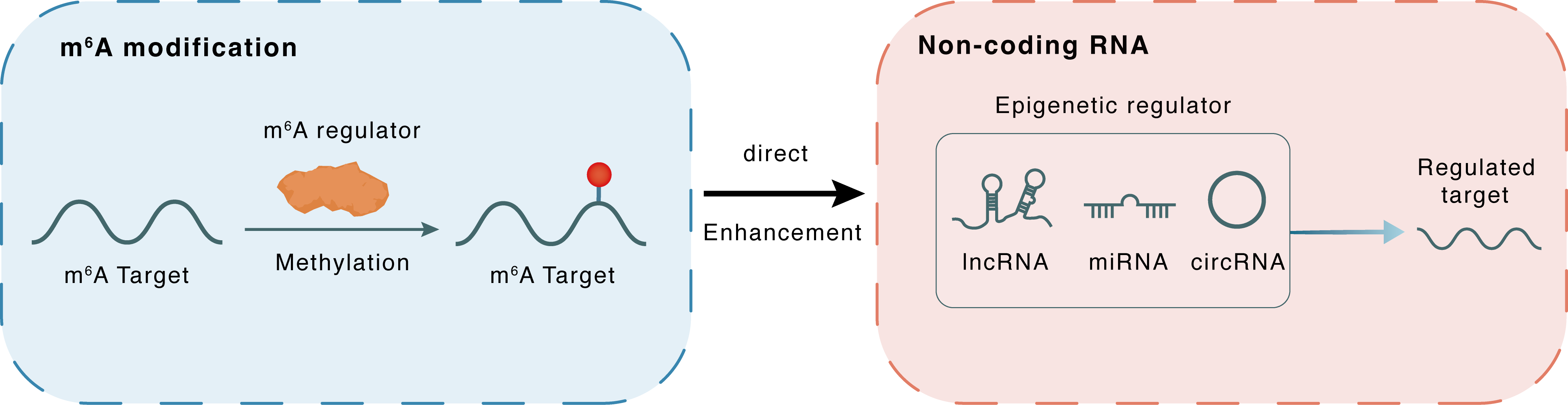 m6A modification
FAM225A
FAM225A
METTL3
Methylation
m6A modification
FAM225A
FAM225A
METTL3
Methylation
 : m6A sites
Direct
Enhancement
Non-coding RNA
FAM225A
miR-590-3p
lncRNA miRNA circRNA : m6A sites
Direct
Enhancement
Non-coding RNA
FAM225A
miR-590-3p
lncRNA miRNA circRNA
|
|||||
| m6A Modification: | |||||
|---|---|---|---|---|---|
| m6A Regulator | Methyltransferase-like 3 (METTL3) | WRITER | |||
| m6A Target | Family with sequence similarity 225 member A (FAM225A) | ||||
| Epigenetic Regulation that have Cross-talk with This m6A Modification: | |||||
| Epigenetic Regulation Type | Non-coding RNA (ncRNA) | ||||
| Epigenetic Regulator | Family with sequence similarity 225 member A (FAM225A) | LncRNA | View Details | ||
| Regulated Target | hsa-miR-590-3p | View Details | |||
| Crosstalk Relationship | m6A → ncRNA | Enhancement | |||
| Crosstalk Mechanism | m6A regulators directly modulate the functionality of ncRNAs through specific targeting ncRNA | ||||
| Crosstalk Summary | Silencing METTL3 decreases Family with sequence similarity 225 member A (FAM225A) RNA stability, which serves as the ceRNA for sponging both hsa-miR-590-3p and miR-1275, increasing the levels of their target integrin beta3 (ITGB3), finally stimulating FAK/PI3K/Akt signaling. miR-590-3p has been reported as a tumor suppressor in cholangiocarcinoma and hepatocellular carcinoma and miR-1275 can inhibit NPC cell growth and suppress hepatocellular carcinoma cell proliferation. | ||||
| Responsed Disease | Cholangiocarcinoma | ICD-11: XH7M15 | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | mRNA stability | ||||
| Cell proliferation | |||||
| Cell invasion | |||||