m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00399)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
SNAI1
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
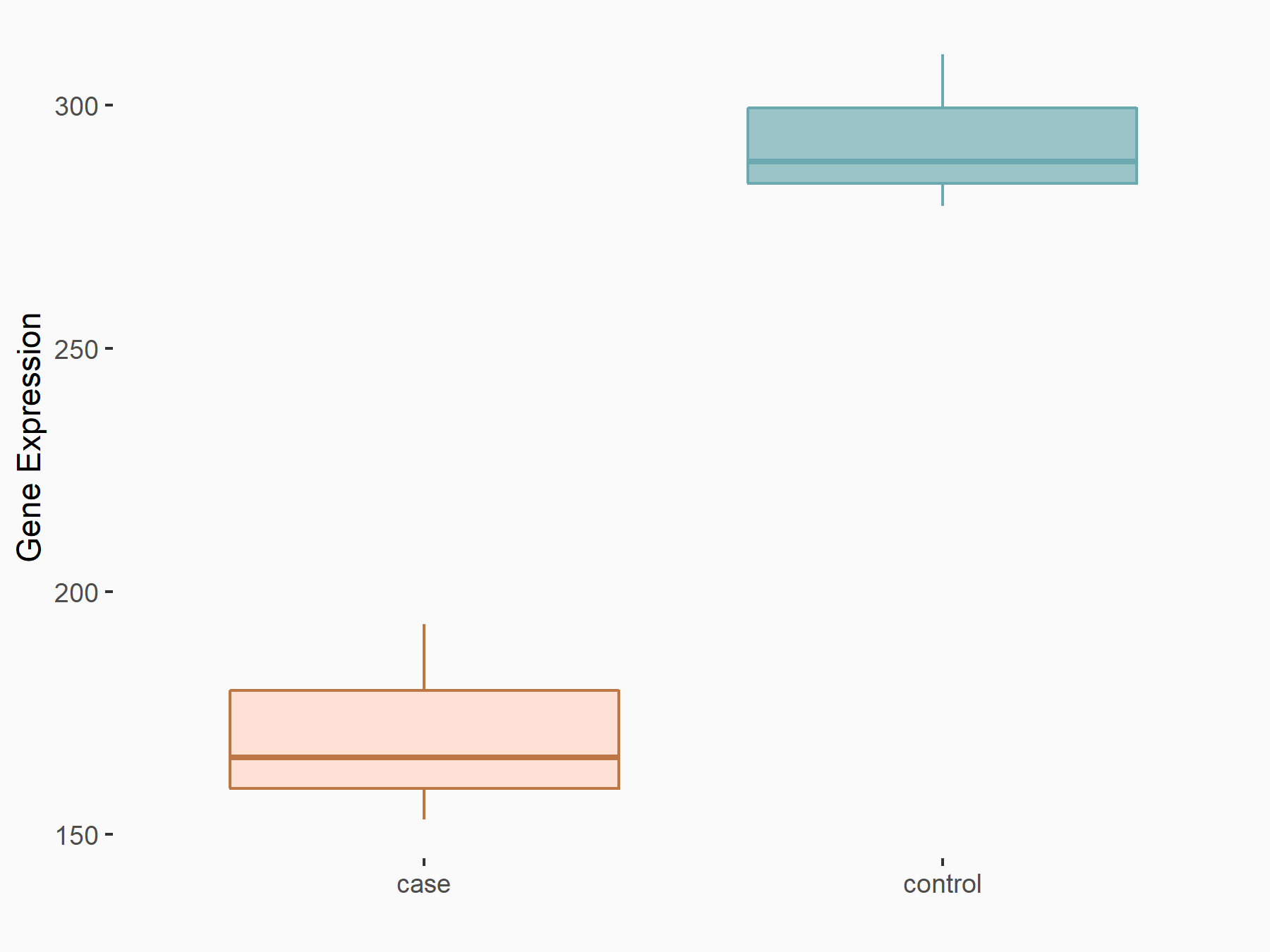 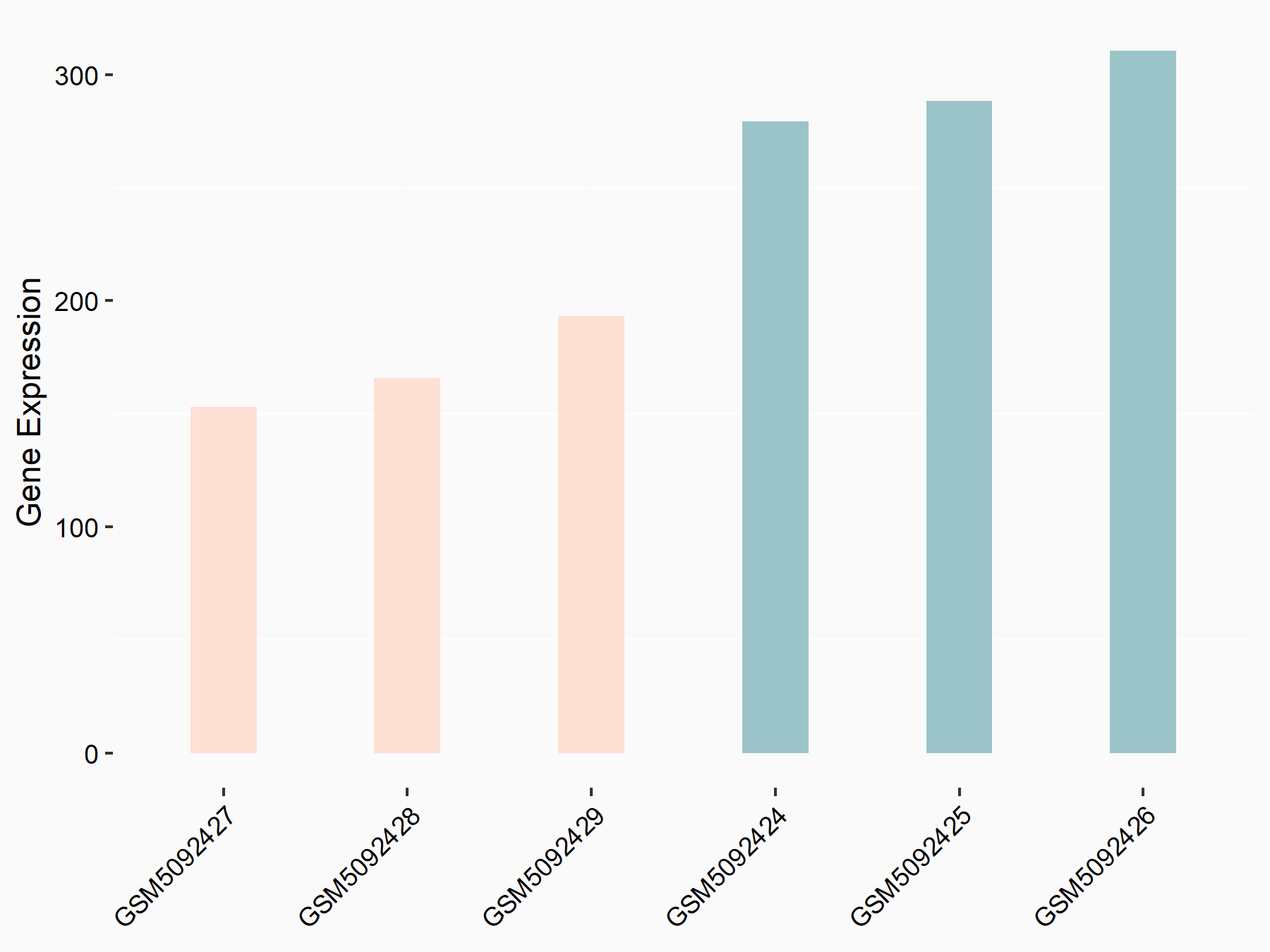 |
logFC: -7.78E-01 p-value: 5.01E-07 |
| More Results | Click to View More RNA-seq Results | |
| In total 5 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | The expression of TGFbeta1 was up-regulated, while self-stimulated expression of TGFbeta1 was suppressed in METTL3Mut/- cells. m6A performed multi-functional roles in TGFbeta1 expression and EMT modulation, suggesting the critical roles of m6A in cancer progression regulation. Zinc finger protein SNAI1 (SNAI1), which was down-regulated in Mettl3Mut/- cells, was a key factor responding to TGF-Beta-1-induced EMT. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Solid tumour/cancer | ICD-11: 2A00-2F9Z | ||
| Pathway Response | Adherens junction | hsa04520 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | Overexpression of Zinc finger protein SNAI1 (SNAI1) partially reversed the regulatory effects of METTL3 on EMT-related gene expressions and metastatic abilities in nasopharyngeal carcinoma. Knockdown of METTL3 decreased the enrichment abundance of Snail in anti-IGF2BP2. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Nasopharyngeal carcinoma | ICD-11: 2B6B | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | SUNE1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6946 |
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | METTL3 acts as a critical m6A methyltransferase capable of facilitating colorectal cancer(CRC) progression, and revealed a novel mechanism by which METTL3 promotes CRC cell proliferation and invasion via stabilizing Zinc finger protein SNAI1 (SNAI1) mRNA in an m6A-dependent manner. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | Adherens junction | hsa04520 | ||
| Cell Process | Cell proliferation | |||
| Cell invasion | ||||
| In-vitro Model | SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| HIEC-6 | Normal | Homo sapiens | CVCL_6C21 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| Experiment 4 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | The upregulation of METTL3 and YTHDF1 act as adverse prognosis factors for overall survival (OS) rate of liver cancer patients. m6A-sequencing and functional studies confirm that Zinc finger protein SNAI1 (SNAI1), a key transcription factor of EMT, is involved in m6A-regulated EMT. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| cell migration | ||||
| invasion | ||||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | All animal experiments complied with Zhongshan School of Medicine Policy on Care and Use of Laboratory Animals. For subcutaneous transplanted model, sh-control and sh-METTL3 Huh7 cells (5 × 106 per mouse, n = 5 for each group) were diluted in 200 uL of PBS + 200 uL Matrigel (BD Biosciences) and subcutaneously injected into immunodeficient female mice to investigate tumor growth. | |||
| Experiment 5 Reporting the m6A Methylation Regulator of This Target Gene | [5] | |||
| Response Summary | SUMOylation of Mettl3 was found to regulate hepatocellular carcinoma progression via controlling Zinc finger protein SNAI1 (SNAI1) mRNA homeostasis in an m6A methyltransferase activity-dependent manner. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| In-vivo Model | 1×106/mouse cells were suspended in 50 uL of serum-free DMEM and subcutaneously injected into the flank of each mouse. Tumor diameters and volume (length × width2 × 0.5236) were calculated every other day using calipers. | |||
RNA demethylase ALKBH5 (ALKBH5) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by ALKBH5 | ||
| Cell Line | CAG cell line | Homo sapiens |
|
Treatment: shALKBH5 CAG cells
Control: shNC CAG cells
|
GSE180214 | |
| Regulation |
  |
logFC: 1.03E+00 p-value: 7.72E-05 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | ALKBH5 overexpression incurred a significant reduction in iron-regulatory protein IRP2 and the modulator of epithelial-mesenchymal transition (EMT) Zinc finger protein SNAI1 (SNAI1). Owing to FBXL5-mediated degradation, ALKBH5 overexpression incurred a significant reduction in iron-regulatory protein IRP2 and the modulator of epithelial-mesenchymal transition (EMT) SNAI1. ALKBH5 in protecting against PDAC through modulating regulators of iron metabolism and underscore the multifaceted role of m6A in pancreatic cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Pathway Response | Adherens junction | hsa04520 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 |
Heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [9] | |||
| Response Summary | HNRNPA2B1, as an m6A reader, is critical in OSCC development. Its expression is significantly associated with the prognosis of Oral Squamous Cell Carcinoma(OSCC). m6A acts as a proto-oncogene that promotes the OSCC proliferation, migration, and invasion through the EMT progression via the LINE-1/TGF-beta1/Zinc finger protein SNAI1 (SNAI1)/Smad2 signaling pathway. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Oral squamous cell carcinoma | ICD-11: 2B6E.0 | ||
| Pathway Response | TGF-beta signaling pathway | hsa04350 | ||
| In-vitro Model | SCC-4 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1684 |
| CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 | |
Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | Overexpression of Zinc finger protein SNAI1 (SNAI1) partially reversed the regulatory effects of METTL3 on EMT-related gene expressions and metastatic abilities in nasopharyngeal carcinoma. Knockdown of METTL3 decreased the enrichment abundance of Snail in anti-IGF2BP2. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Nasopharyngeal carcinoma | ICD-11: 2B6B | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | SUNE1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6946 |
Protein virilizer homolog (VIRMA) [WRITER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [12] | |||
| Response Summary | KIAA1429 could significantly promote the migration and invasion of breast cancer cells. KIAA1429 could bind to the motif in the 3' UTR of SMC1A mRNA directly and enhance SMC1A mRNA stability. In conclusion, the study revealed a novel mechanism of the KIAA1429/SMC1A/Zinc finger protein SNAI1 (SNAI1) axis in the regulation of metastasis of breast cancer. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Adherens junction | hsa04520 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| SUM1315MO2 | Invasive breast carcinoma of no special type | Homo sapiens | CVCL_5589 | |
| In-vivo Model | For the mouse lung metastasis model, SUM-1315 cells (2 × 106/0.2 mL) expressing NC, shKIAA1429, SNAIL, or shKIAA1429+SNAIL were injected into the nude mice through the tail vein. | |||
YTH domain-containing family protein 1 (YTHDF1) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | The upregulation of METTL3 and YTHDF1 act as adverse prognosis factors for overall survival (OS) rate of liver cancer patients. m6A-sequencing and functional studies confirm that Zinc finger protein SNAI1 (SNAI1), a key transcription factor of EMT, is involved in m6A-regulated EMT. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | All animal experiments complied with Zhongshan School of Medicine Policy on Care and Use of Laboratory Animals. For subcutaneous transplanted model, sh-control and sh-METTL3 Huh7 cells (5 × 106 per mouse, n = 5 for each group) were diluted in 200 uL of PBS + 200 uL Matrigel (BD Biosciences) and subcutaneously injected into immunodeficient female mice to investigate tumor growth. | |||
Solid tumour/cancer [ICD-11: 2A00-2F9Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | The expression of TGFbeta1 was up-regulated, while self-stimulated expression of TGFbeta1 was suppressed in METTL3Mut/- cells. m6A performed multi-functional roles in TGFbeta1 expression and EMT modulation, suggesting the critical roles of m6A in cancer progression regulation. Zinc finger protein SNAI1 (SNAI1), which was down-regulated in Mettl3Mut/- cells, was a key factor responding to TGF-Beta-1-induced EMT. | |||
| Responsed Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Adherens junction | hsa04520 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | Overexpression of Zinc finger protein SNAI1 (SNAI1) partially reversed the regulatory effects of METTL3 on EMT-related gene expressions and metastatic abilities in nasopharyngeal carcinoma. Knockdown of METTL3 decreased the enrichment abundance of Snail in anti-IGF2BP2. | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) | READER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | SUNE1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6946 |
| Experiment 2 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | Overexpression of Zinc finger protein SNAI1 (SNAI1) partially reversed the regulatory effects of METTL3 on EMT-related gene expressions and metastatic abilities in nasopharyngeal carcinoma. Knockdown of METTL3 decreased the enrichment abundance of Snail in anti-IGF2BP2. | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | SUNE1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6946 |
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [9] | |||
| Response Summary | HNRNPA2B1, as an m6A reader, is critical in OSCC development. Its expression is significantly associated with the prognosis of Oral Squamous Cell Carcinoma(OSCC). m6A acts as a proto-oncogene that promotes the OSCC proliferation, migration, and invasion through the EMT progression via the LINE-1/TGF-beta1/Zinc finger protein SNAI1 (SNAI1)/Smad2 signaling pathway. | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Target Regulator | Heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | TGF-beta signaling pathway | hsa04350 | ||
| In-vitro Model | SCC-4 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1684 |
| CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | METTL3 acts as a critical m6A methyltransferase capable of facilitating colorectal cancer(CRC) progression, and revealed a novel mechanism by which METTL3 promotes CRC cell proliferation and invasion via stabilizing Zinc finger protein SNAI1 (SNAI1) mRNA in an m6A-dependent manner. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Adherens junction | hsa04520 | ||
| Cell Process | Cell proliferation | |||
| Cell invasion | ||||
| In-vitro Model | SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| HIEC-6 | Normal | Homo sapiens | CVCL_6C21 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [6] | |||
| Response Summary | ALKBH5 overexpression incurred a significant reduction in iron-regulatory protein IRP2 and the modulator of epithelial-mesenchymal transition (EMT) Zinc finger protein SNAI1 (SNAI1). Owing to FBXL5-mediated degradation, ALKBH5 overexpression incurred a significant reduction in iron-regulatory protein IRP2 and the modulator of epithelial-mesenchymal transition (EMT) SNAI1. ALKBH5 in protecting against PDAC through modulating regulators of iron metabolism and underscore the multifaceted role of m6A in pancreatic cancer. | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Adherens junction | hsa04520 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 |
Liver cancer [ICD-11: 2C12]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | The upregulation of METTL3 and YTHDF1 act as adverse prognosis factors for overall survival (OS) rate of liver cancer patients. m6A-sequencing and functional studies confirm that Zinc finger protein SNAI1 (SNAI1), a key transcription factor of EMT, is involved in m6A-regulated EMT. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Epithelial-mesenchymal transition | |||
| cell migration | ||||
| invasion | ||||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | All animal experiments complied with Zhongshan School of Medicine Policy on Care and Use of Laboratory Animals. For subcutaneous transplanted model, sh-control and sh-METTL3 Huh7 cells (5 × 106 per mouse, n = 5 for each group) were diluted in 200 uL of PBS + 200 uL Matrigel (BD Biosciences) and subcutaneously injected into immunodeficient female mice to investigate tumor growth. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [5] | |||
| Response Summary | SUMOylation of Mettl3 was found to regulate hepatocellular carcinoma progression via controlling Zinc finger protein SNAI1 (SNAI1) mRNA homeostasis in an m6A methyltransferase activity-dependent manner. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| In-vivo Model | 1×106/mouse cells were suspended in 50 uL of serum-free DMEM and subcutaneously injected into the flank of each mouse. Tumor diameters and volume (length × width2 × 0.5236) were calculated every other day using calipers. | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | The upregulation of METTL3 and YTHDF1 act as adverse prognosis factors for overall survival (OS) rate of liver cancer patients. m6A-sequencing and functional studies confirm that Zinc finger protein SNAI1 (SNAI1), a key transcription factor of EMT, is involved in m6A-regulated EMT. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | All animal experiments complied with Zhongshan School of Medicine Policy on Care and Use of Laboratory Animals. For subcutaneous transplanted model, sh-control and sh-METTL3 Huh7 cells (5 × 106 per mouse, n = 5 for each group) were diluted in 200 uL of PBS + 200 uL Matrigel (BD Biosciences) and subcutaneously injected into immunodeficient female mice to investigate tumor growth. | |||
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [12] | |||
| Response Summary | KIAA1429 could significantly promote the migration and invasion of breast cancer cells. KIAA1429 could bind to the motif in the 3' UTR of SMC1A mRNA directly and enhance SMC1A mRNA stability. In conclusion, the study revealed a novel mechanism of the KIAA1429/SMC1A/Zinc finger protein SNAI1 (SNAI1) axis in the regulation of metastasis of breast cancer. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Protein virilizer homolog (VIRMA) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Adherens junction | hsa04520 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| SUM1315MO2 | Invasive breast carcinoma of no special type | Homo sapiens | CVCL_5589 | |
| In-vivo Model | For the mouse lung metastasis model, SUM-1315 cells (2 × 106/0.2 mL) expressing NC, shKIAA1429, SNAIL, or shKIAA1429+SNAIL were injected into the nude mice through the tail vein. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
RNA modification
m6A Regulator: Heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00483 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | hsa-mir-22 | |
| Crosstalk relationship | m6Am → m6A | |
m6A Regulator: Zinc finger CCCH domain-containing protein 13 (ZC3H13)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00484 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | hsa-mir-22 | |
| Crosstalk relationship | m6Am → m6A | |
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00485 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | hsa-mir-22 | |
| Crosstalk relationship | m6Am → m6A | |
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00486 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | hsa-mir-22 | |
| Crosstalk relationship | m6Am → m6A | |
m6A Regulator: Leucine-rich PPR motif-containing protein, mitochondrial (LRPPRC)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00487 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | hsa-mir-22 | |
| Crosstalk relationship | m6Am → m6A | |
Histone modification
m6A Regulator: RNA demethylase ALKBH5 (ALKBH5)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03159 | ||
| Epigenetic Regulator | Lysine-specific demethylase 4C (KDM4C) | |
| Regulated Target | Histone H3 lysine 9 trimethylation (H3K9me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Hepatic fibrosis/cirrhosis | |
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03572 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03617 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
Non-coding RNA
m6A Regulator: Fat mass and obesity-associated protein (FTO)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05201 | ||
| Epigenetic Regulator | GATA6 antisense RNA 1 (head to head) (GATA6-AS1) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Pancreatic cancer | |
m6A Regulator: YTH domain-containing family protein 1 (YTHDF1)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05230 | ||
| Epigenetic Regulator | hsa-miR-145-5p | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00399)
| In total 15 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE053514 | Click to Show/Hide the Full List | ||
| mod site | chr20:49983129-49983130:+ | [16] | |
| Sequence | TAACTACAGCGAGCTGCAGGACTCTAATCCAGGTGCGTTGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; MT4; iSLK; MSC | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000244050.3 | ||
| External Link | RMBase: m6A_site_536116 | ||
| mod ID: M6ASITE053515 | Click to Show/Hide the Full List | ||
| mod site | chr20:49983925-49983926:+ | [16] | |
| Sequence | GCTGCCAATGCTCATCTGGGACTCTGTCCTGGCGCCCCAAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000244050.3 | ||
| External Link | RMBase: m6A_site_536117 | ||
| mod ID: M6ASITE053516 | Click to Show/Hide the Full List | ||
| mod site | chr20:49984024-49984025:+ | [16] | |
| Sequence | GACCTCCCTGTCAGATGAGGACAGTGGGAAAGGCTCCCAGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; Jurkat; peripheral-blood; GSC-11; TIME; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000244050.3 | ||
| External Link | RMBase: m6A_site_536118 | ||
| mod ID: M6ASITE053517 | Click to Show/Hide the Full List | ||
| mod site | chr20:49987945-49987946:+ | [16] | |
| Sequence | ACCTGCGGGCCCACCTCCAGACCCACTCAGATGTCAAGAAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000244050.3 | ||
| External Link | RMBase: m6A_site_536119 | ||
| mod ID: M6ASITE053518 | Click to Show/Hide the Full List | ||
| mod site | chr20:49987990-49987991:+ | [16] | |
| Sequence | AGTGCCAGGCGTGTGCTCGGACCTTCTCCCGAATGTCCCTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; U2OS | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000244050.3 | ||
| External Link | RMBase: m6A_site_536120 | ||
| mod ID: M6ASITE053519 | Click to Show/Hide the Full List | ||
| mod site | chr20:49988126-49988127:+ | [16] | |
| Sequence | CCCCAGCTCCAGCAGGAAGGACCCCACATCCTTCTCACTGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; U2OS; hESCs; HEK293A-TOA; MSC; TIME; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000244050.3 | ||
| External Link | RMBase: m6A_site_536121 | ||
| mod ID: M6ASITE053520 | Click to Show/Hide the Full List | ||
| mod site | chr20:49988198-49988199:+ | [16] | |
| Sequence | GGCCACATCAGCCCCACAGGACTTTGATGAAGACCATTTTC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; H1A; HEK293A-TOA; MSC; TIME; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000244050.3 | ||
| External Link | RMBase: m6A_site_536122 | ||
| mod ID: M6ASITE053521 | Click to Show/Hide the Full List | ||
| mod site | chr20:49988210-49988211:+ | [16] | |
| Sequence | CCCACAGGACTTTGATGAAGACCATTTTCTGGTTCTGTGTC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; H1A; HEK293A-TOA; MSC; TIME; iSLK; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000244050.3 | ||
| External Link | RMBase: m6A_site_536123 | ||
| mod ID: M6ASITE053522 | Click to Show/Hide the Full List | ||
| mod site | chr20:49988381-49988382:+ | [16] | |
| Sequence | CAGCTGCTTTGAGCTACAGGACAAAGGCTGACAGACTCACT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; H1A; H1B; hNPCs; hESCs; fibroblasts; MT4; Huh7; Jurkat; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000244050.3 | ||
| External Link | RMBase: m6A_site_536124 | ||
| mod ID: M6ASITE053523 | Click to Show/Hide the Full List | ||
| mod site | chr20:49988395-49988396:+ | [16] | |
| Sequence | TACAGGACAAAGGCTGACAGACTCACTGGGAAGCTCCCACC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; hNPCs; hESCs; fibroblasts; MT4; Huh7; Jurkat; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000244050.3 | ||
| External Link | RMBase: m6A_site_536125 | ||
| mod ID: M6ASITE053524 | Click to Show/Hide the Full List | ||
| mod site | chr20:49988427-49988428:+ | [16] | |
| Sequence | GCTCCCACCCCACTCAGGGGACCCCACTCCCCTCACACACA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; hNPCs; hESCs; fibroblasts; MT4; Huh7; Jurkat; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000244050.3 | ||
| External Link | RMBase: m6A_site_536126 | ||
| mod ID: M6ASITE053525 | Click to Show/Hide the Full List | ||
| mod site | chr20:49988462-49988463:+ | [16] | |
| Sequence | CACACACCCCCCCACAAGGAACCCTCAGGCCACCCTCCACG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; hNPCs; hESCs; fibroblasts; MT4; MM6; Huh7; Jurkat; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000244050.3 | ||
| External Link | RMBase: m6A_site_536127 | ||
| mod ID: M6ASITE053526 | Click to Show/Hide the Full List | ||
| mod site | chr20:49988550-49988551:+ | [16] | |
| Sequence | GCCTGCGGAGGCGGTGGCAGACTAGAGTCTGAGATGCCCCG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; H1A; H1B; Jurkat; HEK293A-TOA; iSLK; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000244050.3 | ||
| External Link | RMBase: m6A_site_536128 | ||
| mod ID: M6ASITE053527 | Click to Show/Hide the Full List | ||
| mod site | chr20:49988650-49988651:+ | [17] | |
| Sequence | TTAACAATGTCTGAAAAGGGACTGTGAGTAATGGCTGTCAC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | iSLK | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000244050.3 | ||
| External Link | RMBase: m6A_site_536129 | ||
| mod ID: M6ASITE053528 | Click to Show/Hide the Full List | ||
| mod site | chr20:49988722-49988723:+ | [17] | |
| Sequence | TGACCGATGTGTCTCCCAGAACTATTCTGGGGGCCCGACAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | iSLK | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000244050.3 | ||
| External Link | RMBase: m6A_site_536130 | ||
References