m6A-centered Crosstalk Information
Mechanism of Crosstalk between m6A Modification and Epigenetic Regulation
| Crosstalk ID |
M6ACROT03159
|
[1] | |||
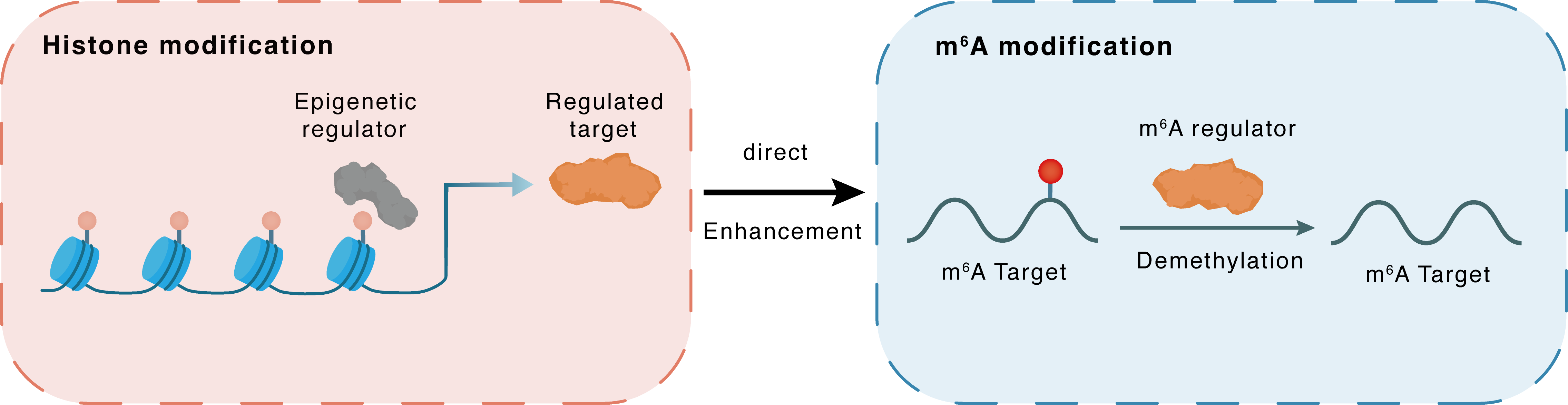 Histone modification
H3K9me3
KDM4C
Downstream Gene
Direct
Enhancement
m6A modification
SNAI1
SNAI1
ALKBH5
Demethylation
Histone modification
H3K9me3
KDM4C
Downstream Gene
Direct
Enhancement
m6A modification
SNAI1
SNAI1
ALKBH5
Demethylation
 : m6A sites : m6A sites
|
|||||
| m6A Modification: | |||||
|---|---|---|---|---|---|
| m6A Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | |||
| m6A Target | Zinc finger protein SNAI1 (SNAI1) | ||||
| Epigenetic Regulation that have Cross-talk with This m6A Modification: | |||||
| Epigenetic Regulation Type | Histone modification (HistMod) | ||||
| Epigenetic Regulator | Lysine-specific demethylase 4C (KDM4C) | ERASER | View Details | ||
| Regulated Target | Histone H3 lysine 9 trimethylation (H3K9me3) | View Details | |||
| Crosstalk Relationship | Histone modification → m6A | Enhancement | |||
| Crosstalk Mechanism | histone modification directly impacts m6A modification through modulating the level of m6A regulator | ||||
| Crosstalk Summary | KDM4C decreases Histone H3 lysine 9 trimethylation (H3K9me3) methylation to upregulate ALKBH5 and subsequently inhibits Zinc finger protein SNAI1 (SNAI1), ultimately impeding liver fibrosis. | ||||
| Responsed Disease | Hepatic fibrosis/cirrhosis | ICD-11: DB93 | |||
In-vitro Model |
HSC-T6
|
N.A. | Rattus norvegicus | CVCL_0315 | |
| In-vivo Model | The mice were divided into several groups: (a) NC; (b) liver fibrosis model; (c) sh-NC; (d) sh-snail1; (e) adeno-associated virus (AAV)-NC1; (f) AAV-KDM4C; (g) AAV-KDM4C + AAV-NC2; and (h) AAV-KDM4C + AAV-snail1 groups; there were 10 mice in each group. | ||||
Full List of Potential Compound(s) Related to This m6A-centered Crosstalk
| Lysine-specific demethylase 4C (KDM4C) | 14 Compound(s) Regulating the Target | Click to Show/Hide the Full List | ||
| Pyrido[1,2-a]indole-1.-carboxylic acid analog 4 | Patented | [2] | ||
| Synonyms |
PMID25468267-Compound-37
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| External Link | ||||
| Pyrido[1,2-a]indole-1.-carboxylic acid analog 1 | Patented | [2] | ||
| Synonyms |
PMID25468267-Compound-34
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| Activity | IC50 < 500 nM | |||
| External Link | ||||
| PMID25468267-Compound-56 | Patented | [2] | ||
| MOA | Inhibitor | |||
| Activity | IC50 < 1000 nM | |||
| External Link | ||||
| Pyrido[1,2-a]indole-1.-carboxylic acid analog 5 | Patented | [2] | ||
| Synonyms |
PMID25468267-Compound-38
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| Activity | IC50 < 500 nM | |||
| External Link | ||||
| PMID25468267-Compound-47 | Patented | [2] | ||
| MOA | Inhibitor | |||
| External Link | ||||
| Pyrido[1,2-a]indole-1.-carboxylic acid analog 7 | Patented | [2] | ||
| Synonyms |
PMID25468267-Compound-40
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| External Link | ||||
| Pyrido[1,2-a]indole-1.-carboxylic acid analog 3 | Patented | [2] | ||
| Synonyms |
PMID25468267-Compound-36
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| External Link | ||||
| Flavonoid derivative 6 | Patented | [2] | ||
| Synonyms |
PMID25468267-Compound-1
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| External Link | ||||
| PMID25468267-Compound-55 | Patented | [2] | ||
| MOA | Inhibitor | |||
| Activity | IC50 < 1000 nM | |||
| External Link | ||||
| Pyrido[1,2-a]indole-1.-carboxylic acid analog 2 | Patented | [2] | ||
| Synonyms |
PMID25468267-Compound-35
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| External Link | ||||
| Pyrido[1,2-a]indole-1.-carboxylic acid analog 6 | Patented | [2] | ||
| Synonyms |
PMID25468267-Compound-39
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| External Link | ||||
| Hydroxamate analog 1 | Patented | [2] | ||
| Synonyms |
PMID25468267-Compound-31
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| Activity | IC50 = 1000 nM | |||
| External Link | ||||
| PMID25468267-Compound-46 | Patented | [2] | ||
| MOA | Inhibitor | |||
| Activity | IC50 = 3400 nM | |||
| External Link | ||||
| IOX1 | Investigative | [3] | ||
| Synonyms |
5852-78-8; 8-Hydroxyquinoline-5-Carboxylic Acid; 8-Hydroxy-5-quinolinecarboxylic acid; 5-Carboxy-8-hydroxyquinoline; IOX 1; UNII-JM015YQC1C; IOX-1; 5-carboxy-8HQ; 5-Quinolinecarboxylic acid, 8-hydroxy-; JM015YQC1C; CHEMBL1230640; 4bio; 4jht; 8XQ; 4ie4; AC1LA0UV; MLS002729056; GTPL8230; SCHEMBL6068195; KS-00000PPH; CHEBI:93239; CTK1E0142; DTXSID20207236; AOB6499; JGRPKOGHYBAVMW-UHFFFAOYSA-N; MolPort-006-673-354; HMS3653E21; ZINC5933707; BCP16996; s7234; BDBM50396018; 2184AH; IOX1, > AKOS016371793
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| Activity | IC50 = 600 nM | |||
| External Link | ||||
| DB93: Hepatic fibrosis/cirrhosis | 5 Compound(s) Regulating the Disease | Click to Show/Hide the Full List | ||
| KB174 | Clinical Trial | [4] | ||
| External Link | ||||
| Quinazoline derivative 12 | Patented | [5] | ||
| Synonyms |
PMID26936077-Compound-23
Click to Show/Hide
|
|||
| External Link | ||||
| Quinazoline derivative 10 | Patented | [5] | ||
| Synonyms |
PMID26936077-Compound-21
Click to Show/Hide
|
|||
| External Link | ||||
| Quinazoline derivative 11 | Patented | [5] | ||
| Synonyms |
PMID26936077-Compound-22
Click to Show/Hide
|
|||
| External Link | ||||
| PMID27998201-Compound-5 | Patented | [6] | ||
| External Link | ||||
References