m6A-centered Crosstalk Information
Mechanism of Crosstalk between m6A Modification and Epigenetic Regulation
| Crosstalk ID |
M6ACROT03182
|
[1] | |||
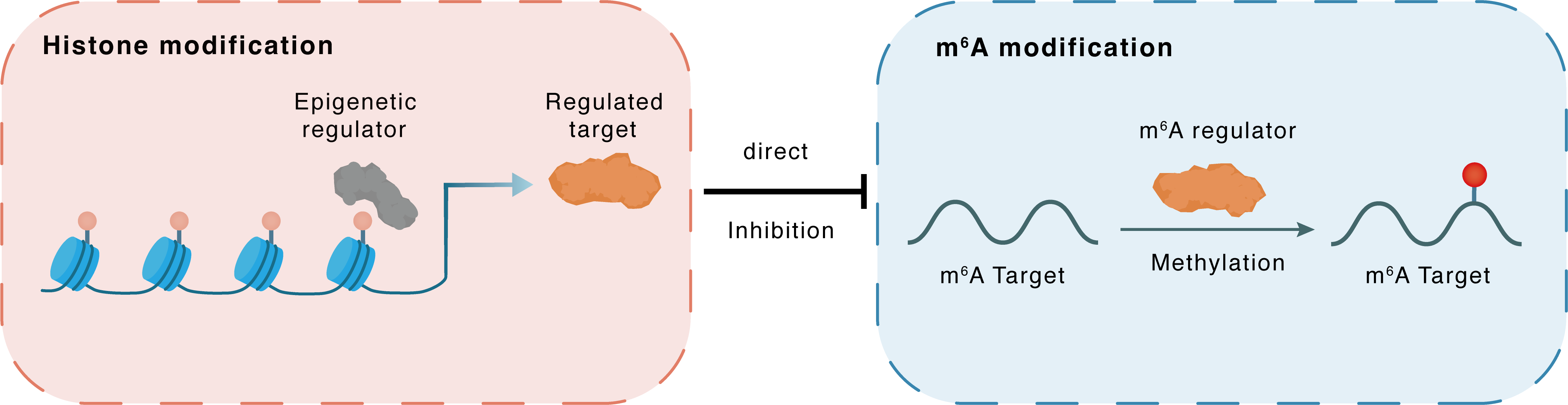 Histone modification
H3K4me3
TET1
METTL3
Direct
Inhibition
m6A modification
TET1
TET1
METTL3
Methylation
Histone modification
H3K4me3
TET1
METTL3
Direct
Inhibition
m6A modification
TET1
TET1
METTL3
Methylation
 : m6A sites : m6A sites
|
|||||
| m6A Modification: | |||||
|---|---|---|---|---|---|
| m6A Regulator | Methyltransferase-like 3 (METTL3) | WRITER | |||
| m6A Target | Methylcytosine dioxygenase TET1 (TET1) | ||||
| Epigenetic Regulation that have Cross-talk with This m6A Modification: | |||||
| Epigenetic Regulation Type | Histone modification (HistMod) | ||||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | ERASER | View Details | ||
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | View Details | |||
| Downstream Gene | METTL3 | View Details | |||
| Crosstalk Relationship | Histone modification → m6A | Inhibition | |||
| Crosstalk Mechanism | histone modification directly impacts m6A modification through modulating the level of m6A regulator | ||||
| Crosstalk Summary | Methylcytosine dioxygenase TET1 (TET1) knockdown contributed to the binding of Histone H3 lysine 4 trimethylation (H3K4me3) and H3K27me3 to METTL3 DNA. Our results revealed a negative feedback regulatory loop between TET1 and METTL3/YTHDF2 in myoblast differentiation, which unveiled the interplay among DNA methylation, RNA methylation and histone methylation in skeletal myogenesis. | ||||