m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00622)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
DAPK2
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
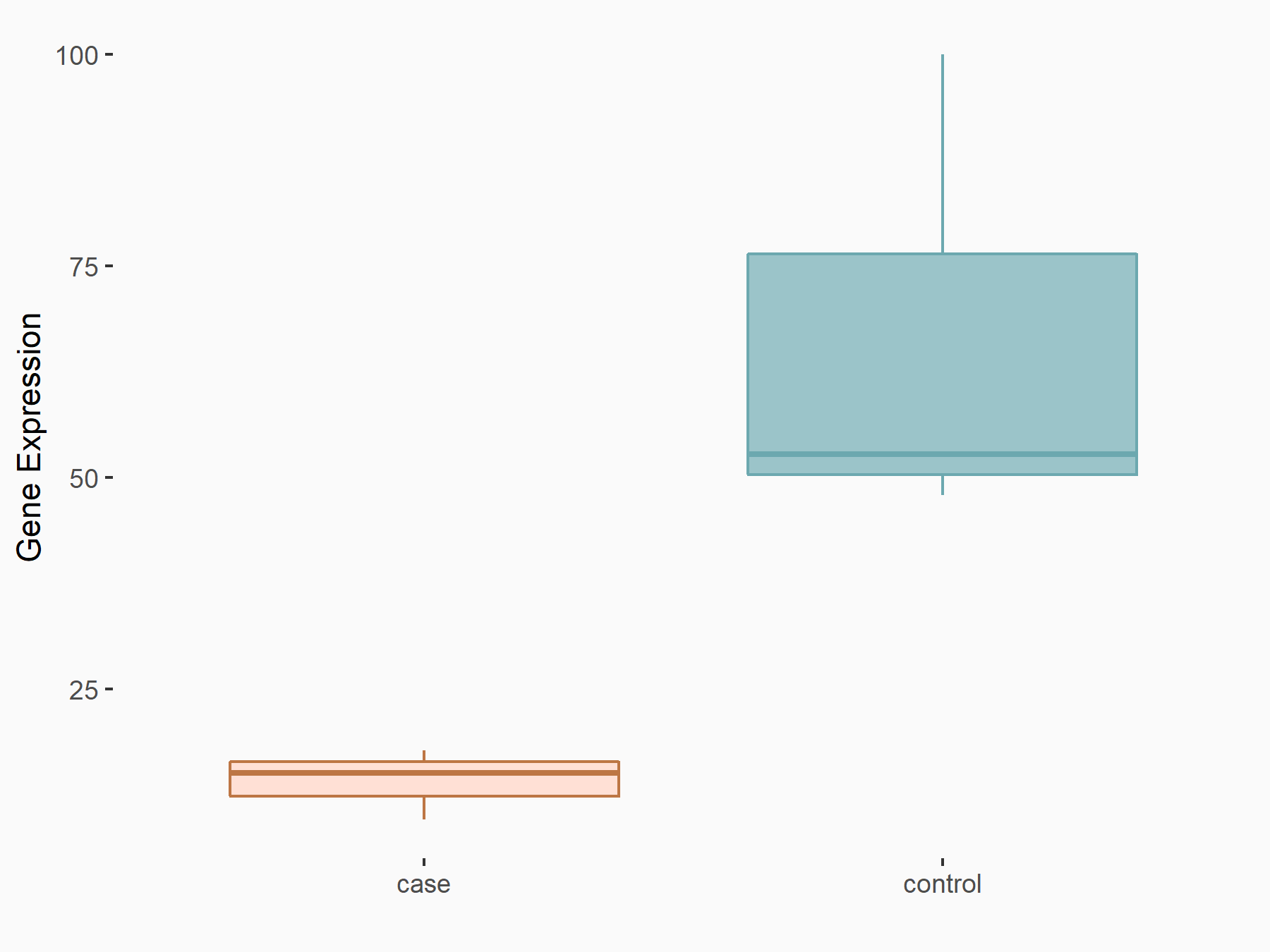  |
logFC: -2.25E+00 p-value: 1.26E-07 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between DAPK2 and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.83E+00 | GSE60213 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Cigarette smoking induced aberrant N6-methyladenosine modification of Death-associated protein kinase 2 (DAPK2), which resulted in decreased DAPK2 mRNA stability and expression of its mRNA and protein. This modification was mediated by the m6A "writer" METTL3 and the m6A "reader" YTHDF2. BAY 11-7085, a NF-Kappa-B signaling selective inhibitor, was shown to efficiently suppressed downregulation of DAPK2-induced oncogenic phenotypes of NSCLC cells. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| In-vitro Model | NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | The nude mice were maintained under pathogen-free conditions and kept under timed lighting conditions mandated by the committee with food and water provided ad libitum. For xenograft experiments, nude mice were injected subcutaneously with 5 × 106 cells resuspended in 0.1 mL PBS. When a tumor was palpable, it was measured every 3 days. | |||
YTH domain-containing family protein 2 (YTHDF2) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Human umbilical cord blood CD34+ cells | Homo sapiens |
|
Treatment: YTHDF2 knockdown UCB CD34+ cells
Control: Wild type UCB CD34+ cells
|
GSE107956 | |
| Regulation |
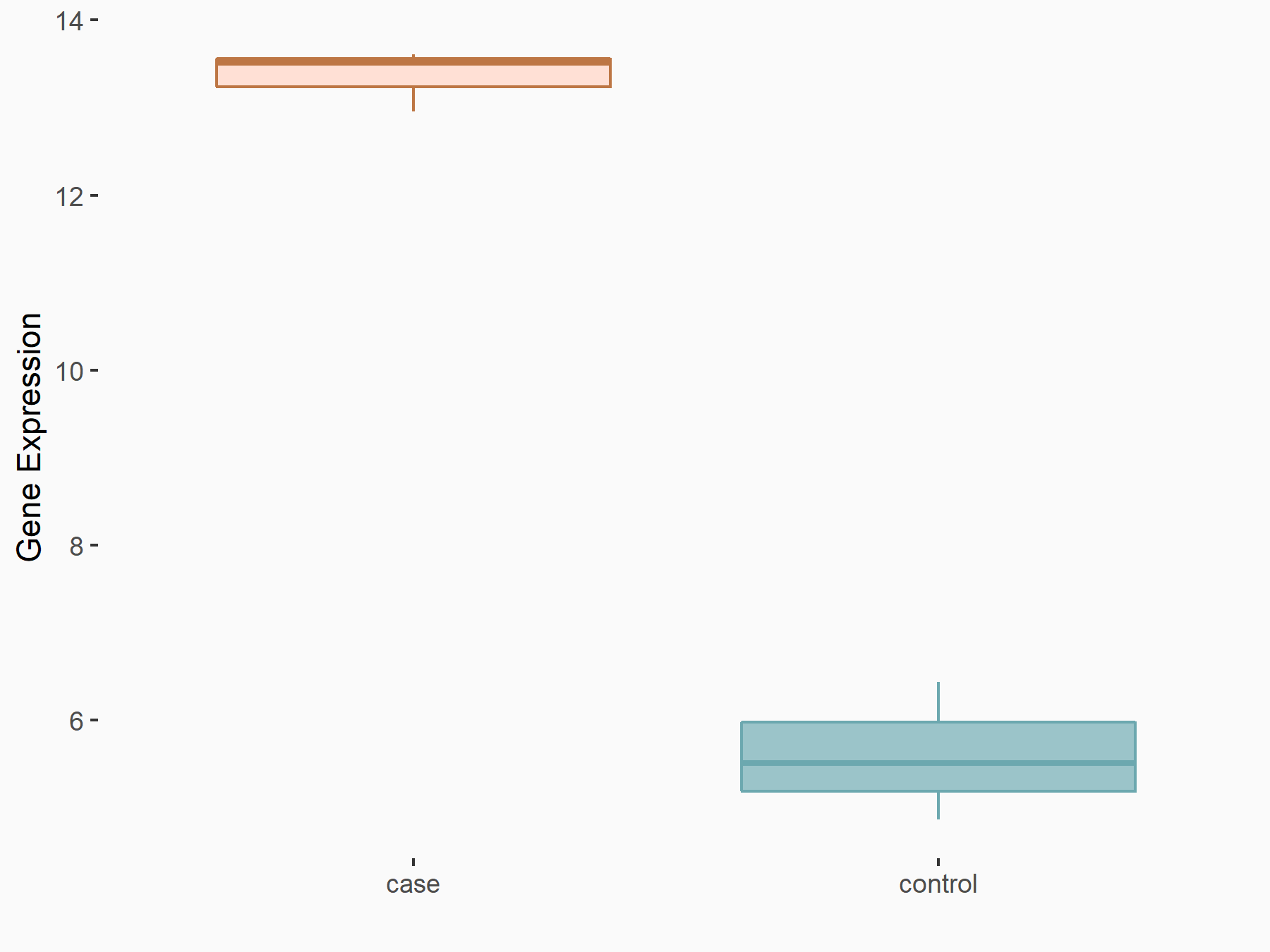  |
logFC: 1.13E+00 p-value: 1.77E-04 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Cigarette smoking induced aberrant N6-methyladenosine modification of Death-associated protein kinase 2 (DAPK2), which resulted in decreased DAPK2 mRNA stability and expression of its mRNA and protein. This modification was mediated by the m6A "writer" METTL3 and the m6A "reader" YTHDF2. BAY 11-7085, a NF-Kappa-B signaling selective inhibitor, was shown to efficiently suppressed downregulation of DAPK2-induced oncogenic phenotypes of NSCLC cells. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| In-vitro Model | NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | The nude mice were maintained under pathogen-free conditions and kept under timed lighting conditions mandated by the committee with food and water provided ad libitum. For xenograft experiments, nude mice were injected subcutaneously with 5 × 106 cells resuspended in 0.1 mL PBS. When a tumor was palpable, it was measured every 3 days. | |||
Lung cancer [ICD-11: 2C25]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Cigarette smoking induced aberrant N6-methyladenosine modification of Death-associated protein kinase 2 (DAPK2), which resulted in decreased DAPK2 mRNA stability and expression of its mRNA and protein. This modification was mediated by the m6A "writer" METTL3 and the m6A "reader" YTHDF2. BAY 11-7085, a NF-Kappa-B signaling selective inhibitor, was shown to efficiently suppressed downregulation of DAPK2-induced oncogenic phenotypes of NSCLC cells. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | The nude mice were maintained under pathogen-free conditions and kept under timed lighting conditions mandated by the committee with food and water provided ad libitum. For xenograft experiments, nude mice were injected subcutaneously with 5 × 106 cells resuspended in 0.1 mL PBS. When a tumor was palpable, it was measured every 3 days. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Cigarette smoking induced aberrant N6-methyladenosine modification of Death-associated protein kinase 2 (DAPK2), which resulted in decreased DAPK2 mRNA stability and expression of its mRNA and protein. This modification was mediated by the m6A "writer" METTL3 and the m6A "reader" YTHDF2. BAY 11-7085, a NF-Kappa-B signaling selective inhibitor, was shown to efficiently suppressed downregulation of DAPK2-induced oncogenic phenotypes of NSCLC cells. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | The nude mice were maintained under pathogen-free conditions and kept under timed lighting conditions mandated by the committee with food and water provided ad libitum. For xenograft experiments, nude mice were injected subcutaneously with 5 × 106 cells resuspended in 0.1 mL PBS. When a tumor was palpable, it was measured every 3 days. | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00622)
| In total 15 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE006920 | Click to Show/Hide the Full List | ||
| mod site | chr15:63909738-63909739:- | [2] | |
| Sequence | CACCATCTCAGCTCGCTGCAACCTCTGCCTCCCGGGTTCAA | ||
| Transcript ID List | ENST00000558064.1; ENST00000559007.5; ENST00000261891.7; ENST00000457488.5; ENST00000559731.5 | ||
| External Link | RMBase: RNA-editing_site_44273 | ||
| mod ID: A2ISITE006921 | Click to Show/Hide the Full List | ||
| mod site | chr15:63909793-63909794:- | [3] | |
| Sequence | TTTTTTTTTTTTTGAGACAGAGTCTCACTGTGTCTCCCAGG | ||
| Transcript ID List | ENST00000457488.5; ENST00000559007.5; ENST00000261891.7; ENST00000559731.5; ENST00000558064.1 | ||
| External Link | RMBase: RNA-editing_site_44274 | ||
| mod ID: A2ISITE006922 | Click to Show/Hide the Full List | ||
| mod site | chr15:63910618-63910619:- | [2] | |
| Sequence | GAGCCCAGGAGTTAAAGGTTACAGTGAATTATGATCACACT | ||
| Transcript ID List | rmsk_4360496; ENST00000457488.5; ENST00000559731.5; ENST00000558064.1; ENST00000559007.5; ENST00000261891.7 | ||
| External Link | RMBase: RNA-editing_site_44275 | ||
| mod ID: A2ISITE006923 | Click to Show/Hide the Full List | ||
| mod site | chr15:63910642-63910643:- | [2] | |
| Sequence | GGAGCTGAGGTGGGAGGATCACTTGAGCCCAGGAGTTAAAG | ||
| Transcript ID List | ENST00000559731.5; ENST00000558064.1; ENST00000457488.5; ENST00000559007.5; rmsk_4360496; ENST00000261891.7 | ||
| External Link | RMBase: RNA-editing_site_44276 | ||
| mod ID: A2ISITE006924 | Click to Show/Hide the Full List | ||
| mod site | chr15:63910678-63910679:- | [4] | |
| Sequence | GTATGGTGACAGGCACCTATAGTCCTAGCTACTCAGGGAGC | ||
| Transcript ID List | ENST00000559731.5; ENST00000457488.5; ENST00000558064.1; ENST00000261891.7; ENST00000559007.5; rmsk_4360496 | ||
| External Link | RMBase: RNA-editing_site_44277 | ||
| mod ID: A2ISITE006925 | Click to Show/Hide the Full List | ||
| mod site | chr15:63910680-63910681:- | [4] | |
| Sequence | AGGTATGGTGACAGGCACCTATAGTCCTAGCTACTCAGGGA | ||
| Transcript ID List | ENST00000558064.1; rmsk_4360496; ENST00000261891.7; ENST00000559731.5; ENST00000457488.5; ENST00000559007.5 | ||
| External Link | RMBase: RNA-editing_site_44278 | ||
| mod ID: A2ISITE006926 | Click to Show/Hide the Full List | ||
| mod site | chr15:63910755-63910756:- | [4] | |
| Sequence | AGACCAGCCCAGGCAACAACAGTGAGACCCTGACTCTACAA | ||
| Transcript ID List | rmsk_4360496; ENST00000558064.1; ENST00000457488.5; ENST00000261891.7; ENST00000559731.5; ENST00000559007.5 | ||
| External Link | RMBase: RNA-editing_site_44279 | ||
| mod ID: A2ISITE006927 | Click to Show/Hide the Full List | ||
| mod site | chr15:63910784-63910785:- | [2] | |
| Sequence | CGGGAGGTTCATTTAAGGCCAGGAGTTAAAGACCAGCCCAG | ||
| Transcript ID List | ENST00000559007.5; ENST00000457488.5; ENST00000558064.1; ENST00000559731.5; ENST00000261891.7; rmsk_4360496 | ||
| External Link | RMBase: RNA-editing_site_44280 | ||
| mod ID: A2ISITE006928 | Click to Show/Hide the Full List | ||
| mod site | chr15:63910789-63910790:- | [2] | |
| Sequence | TGAGGCGGGAGGTTCATTTAAGGCCAGGAGTTAAAGACCAG | ||
| Transcript ID List | ENST00000559731.5; ENST00000558064.1; rmsk_4360496; ENST00000261891.7; ENST00000559007.5; ENST00000457488.5 | ||
| External Link | RMBase: RNA-editing_site_44281 | ||
| mod ID: A2ISITE006929 | Click to Show/Hide the Full List | ||
| mod site | chr15:63911118-63911119:- | [2] | |
| Sequence | TCAAGCGATTCTCCTGCGTCAGCCTCCTGAGTAGCTGGGAT | ||
| Transcript ID List | ENST00000261891.7; ENST00000558064.1; ENST00000559007.5; ENST00000559731.5; ENST00000457488.5 | ||
| External Link | RMBase: RNA-editing_site_44282 | ||
| mod ID: A2ISITE006930 | Click to Show/Hide the Full List | ||
| mod site | chr15:63911179-63911180:- | [2] | |
| Sequence | ATTGCTTAGGTTGGAGTGCAATGGCGCGATCTCGGCTCACC | ||
| Transcript ID List | ENST00000558064.1; ENST00000457488.5; ENST00000559007.5; ENST00000559731.5; ENST00000261891.7 | ||
| External Link | RMBase: RNA-editing_site_44283 | ||
| mod ID: A2ISITE006931 | Click to Show/Hide the Full List | ||
| mod site | chr15:63911199-63911200:- | [2] | |
| Sequence | TTAGACAGAGTTTCACTCTTATTGCTTAGGTTGGAGTGCAA | ||
| Transcript ID List | ENST00000559731.5; ENST00000558064.1; ENST00000261891.7; ENST00000559007.5; ENST00000457488.5 | ||
| External Link | RMBase: RNA-editing_site_44284 | ||
| mod ID: A2ISITE006932 | Click to Show/Hide the Full List | ||
| mod site | chr15:63927927-63927928:- | [3] | |
| Sequence | GACAAAACCCCATCTCTACAAAAAACAAAACAAAAAACTAG | ||
| Transcript ID List | ENST00000559007.5; rmsk_4360527; ENST00000457488.5; ENST00000261891.7; ENST00000612884.4; ENST00000558069.5 | ||
| External Link | RMBase: RNA-editing_site_44285 | ||
| mod ID: A2ISITE006933 | Click to Show/Hide the Full List | ||
| mod site | chr15:63928188-63928189:- | [3] | |
| Sequence | ATATCACAATTGCTAGGGGTAGGGACTTAGTATCTGTATCT | ||
| Transcript ID List | ENST00000559007.5; ENST00000261891.7; ENST00000457488.5; rmsk_4360528; ENST00000558069.5; ENST00000612884.4 | ||
| External Link | RMBase: RNA-editing_site_44286 | ||
| mod ID: A2ISITE006934 | Click to Show/Hide the Full List | ||
| mod site | chr15:64003635-64003636:- | [3] | |
| Sequence | TCCAGCCTGGGTGACAGAGCAAGACTCTGTCAGAAGGAAAA | ||
| Transcript ID List | ENST00000558069.5; ENST00000561162.5; ENST00000559007.5; ENST00000559897.1; ENST00000261891.7; ENST00000612884.4; ENST00000559306.1; rmsk_4360695; ENST00000457488.5; ENST00000558482.5 | ||
| External Link | RMBase: RNA-editing_site_44287 | ||
N6-methyladenosine (m6A)
| In total 19 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE023546 | Click to Show/Hide the Full List | ||
| mod site | chr15:63907938-63907939:- | [5] | |
| Sequence | ATTTTGCTCATTTTTATTAAACTTCTGGTTTACCTGATGCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; A549; GM12878; peripheral-blood; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000559007.5; ENST00000261891.7; ENST00000457488.5 | ||
| External Link | RMBase: m6A_site_282358 | ||
| mod ID: M6ASITE023547 | Click to Show/Hide the Full List | ||
| mod site | chr15:63907997-63907998:- | [6] | |
| Sequence | CAGGCTGACCAACACCTCAGACCTTCTGAAGCAGCCCATTG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; A549; H1B; H1A; GM12878; LCLs; Huh7; CD4T; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000559007.5; ENST00000457488.5; ENST00000261891.7 | ||
| External Link | RMBase: m6A_site_282359 | ||
| mod ID: M6ASITE023548 | Click to Show/Hide the Full List | ||
| mod site | chr15:63908036-63908037:- | [6] | |
| Sequence | CAATCCTGTGACCTCCCAGAACCATGGAAGCCAGGACGTCA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; A549; H1A; H1B; GM12878; LCLs; Huh7; Jurkat; CD4T; peripheral-blood; MSC; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000559007.5; ENST00000261891.7; ENST00000457488.5 | ||
| External Link | RMBase: m6A_site_282360 | ||
| mod ID: M6ASITE023549 | Click to Show/Hide the Full List | ||
| mod site | chr15:63908058-63908059:- | [6] | |
| Sequence | GTGAACCTTGGGTGTGAGGGACCAATCCTGTGACCTCCCAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; H1A; H1B; GM12878; LCLs; Huh7; Jurkat; CD4T; peripheral-blood; MSC; TIME; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000457488.5; ENST00000559007.5; ENST00000261891.7 | ||
| External Link | RMBase: m6A_site_282361 | ||
| mod ID: M6ASITE023550 | Click to Show/Hide the Full List | ||
| mod site | chr15:63908074-63908075:- | [6] | |
| Sequence | AGGGTGATTGAGGAAAGTGAACCTTGGGTGTGAGGGACCAA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; H1A; H1B; GM12878; LCLs; Huh7; Jurkat; CD4T; peripheral-blood; MSC; TIME; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000261891.7; ENST00000457488.5; ENST00000559007.5 | ||
| External Link | RMBase: m6A_site_282362 | ||
| mod ID: M6ASITE023551 | Click to Show/Hide the Full List | ||
| mod site | chr15:63908096-63908097:- | [6] | |
| Sequence | CAGAGTCCTCCCCATTGGGAACAGGGTGATTGAGGAAAGTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; H1A; H1B; GM12878; LCLs; Huh7; Jurkat; peripheral-blood; MSC; TIME; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000457488.5; ENST00000559007.5; ENST00000261891.7 | ||
| External Link | RMBase: m6A_site_282363 | ||
| mod ID: M6ASITE023552 | Click to Show/Hide the Full List | ||
| mod site | chr15:63908117-63908118:- | [6] | |
| Sequence | CCAAAAATTACACCAGAGAGACAGAGTCCTCCCCATTGGGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; H1A; H1B; GM12878; LCLs; Huh7; peripheral-blood; MSC; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000457488.5; ENST00000261891.7; ENST00000559007.5 | ||
| External Link | RMBase: m6A_site_282364 | ||
| mod ID: M6ASITE023553 | Click to Show/Hide the Full List | ||
| mod site | chr15:63908147-63908148:- | [6] | |
| Sequence | ACAGGCTGAGGGGGTTCAGAACCAGCCTGGCCAAAAATTAC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; H1B; GM12878; LCLs; Huh7; MSC; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000457488.5; ENST00000261891.7; ENST00000559007.5 | ||
| External Link | RMBase: m6A_site_282365 | ||
| mod ID: M6ASITE023554 | Click to Show/Hide the Full List | ||
| mod site | chr15:63908185-63908186:- | [6] | |
| Sequence | CATCACGGGGTGAAGGTCAGACTAAGGCAGCCTTCTTCACA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; GM12878; Huh7; MSC | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000457488.5; ENST00000261891.7; ENST00000559007.5 | ||
| External Link | RMBase: m6A_site_282366 | ||
| mod ID: M6ASITE023555 | Click to Show/Hide the Full List | ||
| mod site | chr15:63908215-63908216:- | [6] | |
| Sequence | TTAGGAAAACAATATAAAGGACATCCTCATCATCACGGGGT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; GM12878; LCLs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000261891.7; ENST00000457488.5; ENST00000559007.5 | ||
| External Link | RMBase: m6A_site_282367 | ||
| mod ID: M6ASITE023556 | Click to Show/Hide the Full List | ||
| mod site | chr15:63908227-63908228:- | [6] | |
| Sequence | GGGAGTGTGGACTTAGGAAAACAATATAAAGGACATCCTCA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; hESC-HEK293T; GM12878; LCLs | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000457488.5; ENST00000559007.5; ENST00000261891.7 | ||
| External Link | RMBase: m6A_site_282368 | ||
| mod ID: M6ASITE023557 | Click to Show/Hide the Full List | ||
| mod site | chr15:63908237-63908238:- | [5] | |
| Sequence | AGTCTGGGGTGGGAGTGTGGACTTAGGAAAACAATATAAAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HEK293T; HeLa; A549; GM12878 | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000261891.7; ENST00000559007.5; ENST00000457488.5 | ||
| External Link | RMBase: m6A_site_282369 | ||
| mod ID: M6ASITE023558 | Click to Show/Hide the Full List | ||
| mod site | chr15:63908356-63908357:- | [6] | |
| Sequence | GCTTGCAGGCAAGCCAGGAGACCCTGGGAGCTGTGGCTGTC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; A549; U2OS; H1A; GM12878; peripheral-blood; endometrial; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000457488.5; ENST00000559731.5; ENST00000559007.5; ENST00000261891.7 | ||
| External Link | RMBase: m6A_site_282370 | ||
| mod ID: M6ASITE023559 | Click to Show/Hide the Full List | ||
| mod site | chr15:63908448-63908449:- | [6] | |
| Sequence | CCTTCTGTGCAGACTTTTGGACCCAGCTCAGCACCAGCACC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; U2OS; H1B; Jurkat; peripheral-blood; GSC-11; endometrial; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000559007.5; ENST00000261891.7; ENST00000457488.5; ENST00000559731.5 | ||
| External Link | RMBase: m6A_site_282371 | ||
| mod ID: M6ASITE023560 | Click to Show/Hide the Full List | ||
| mod site | chr15:63908456-63908457:- | [6] | |
| Sequence | CGGGGCTCCCTTCTGTGCAGACTTTTGGACCCAGCTCAGCA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; U2OS; H1B; Jurkat; peripheral-blood; GSC-11; endometrial; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000559731.5; ENST00000559007.5; ENST00000457488.5; ENST00000261891.7 | ||
| External Link | RMBase: m6A_site_282372 | ||
| mod ID: M6ASITE023561 | Click to Show/Hide the Full List | ||
| mod site | chr15:63908571-63908572:- | [6] | |
| Sequence | TGAGAGTGACACTGAGGAGGACATCGCCAGGAGGAAAGCCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; Jurkat; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000559007.5; ENST00000559731.5; ENST00000457488.5; ENST00000261891.7 | ||
| External Link | RMBase: m6A_site_282373 | ||
| mod ID: M6ASITE023562 | Click to Show/Hide the Full List | ||
| mod site | chr15:63922807-63922808:- | [7] | |
| Sequence | GGGCTCCACATTCTACCAGGACACCAGTGAATCCCTGTCAG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000558069.5; ENST00000559007.5; ENST00000557867.1; ENST00000261891.7; ENST00000457488.5; ENST00000612884.4 | ||
| External Link | RMBase: m6A_site_282374 | ||
| mod ID: M6ASITE023563 | Click to Show/Hide the Full List | ||
| mod site | chr15:63926049-63926050:- | [6] | |
| Sequence | TGGGAGACACGAAGCAGGAAACACTGGCAAATATCACAGCA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000261891.7; ENST00000558076.1; ENST00000558069.5; ENST00000457488.5; ENST00000612884.4; ENST00000559007.5; ENST00000557867.1 | ||
| External Link | RMBase: m6A_site_282375 | ||
| mod ID: M6ASITE023564 | Click to Show/Hide the Full List | ||
| mod site | chr15:63926063-63926064:- | [6] | |
| Sequence | AGCATCCCCTTTCCTGGGAGACACGAAGCAGGAAACACTGG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000558069.5; ENST00000557867.1; ENST00000261891.7; ENST00000559007.5; ENST00000612884.4; ENST00000558076.1; ENST00000457488.5 | ||
| External Link | RMBase: m6A_site_282376 | ||
References