m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00476)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
HMGA1
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Caco-2 cell line | Homo sapiens |
|
Treatment: shMETTL3 Caco-2 cells
Control: shNTC Caco-2 cells
|
GSE167075 | |
| Regulation |
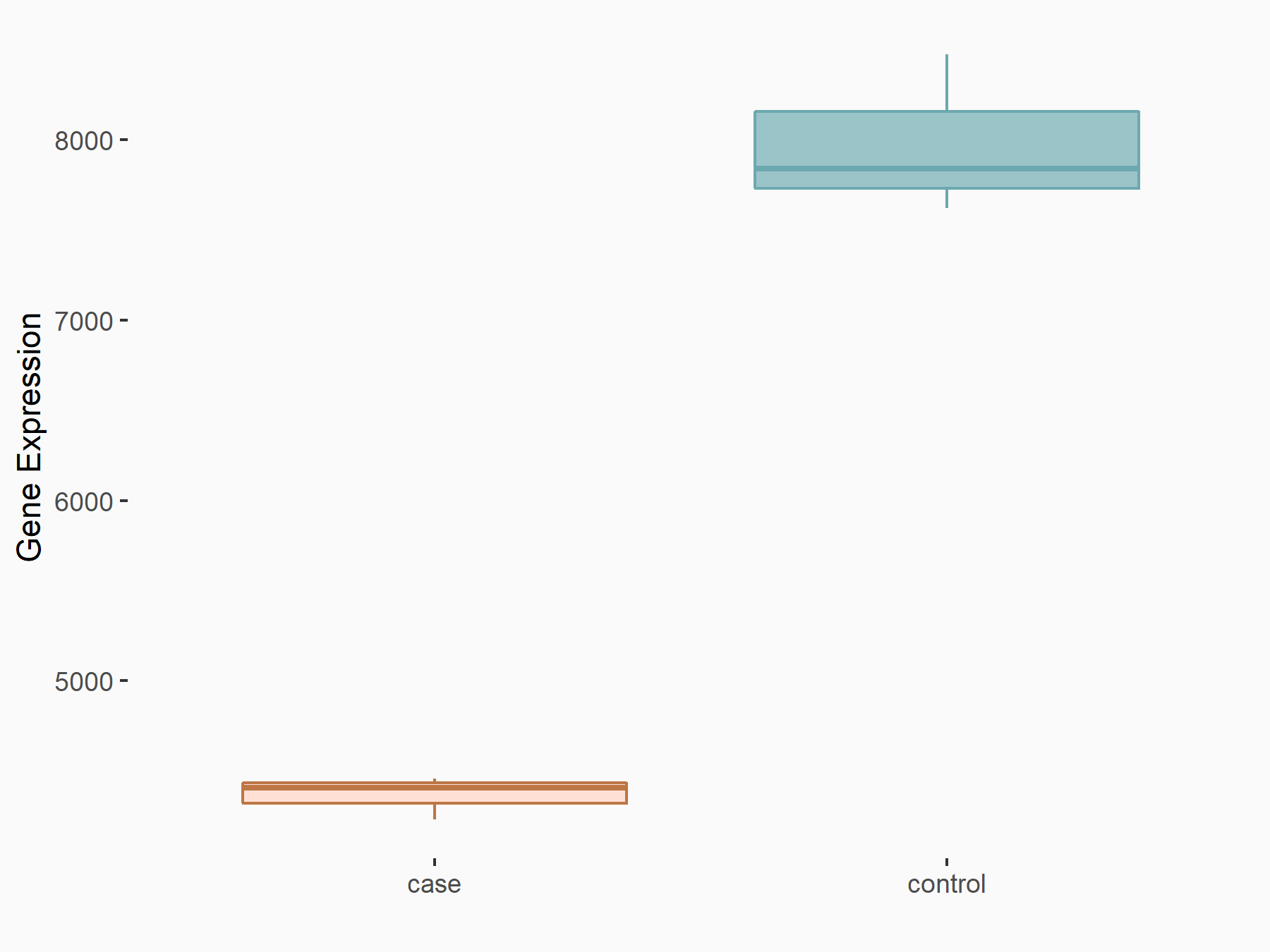  |
logFC: -8.70E-01 p-value: 2.47E-77 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | LINC00460 is a novel oncogene of colorectal cancer through interacting with IGF2BP2 and DHX9 and bind to the m6A modified High mobility group protein HMG-I/HMG-Y (HMGA1) mRNA to enhance the HMGA1 mRNA stability. The N6-methyladenosine (m6A) modification of HMGA1 mRNA by METTL3 enhanced HMGA1 expression in CRC. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| Cell Process | mRNA stability | |||
| Epithelial-mesenchymal transition | ||||
| In-vitro Model | DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| FHC | Normal | Homo sapiens | CVCL_3688 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | Groups of HCT116-Luc-shCtrl, HCT116-Luc-shLINC00460, and HCT116-Luc-shLINC00460 + HMGA1 cells (5 × 106) were injected subcutaneously into the flanks of mice correspondingly. | |||
Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) [READER]
| Representative RIP-seq result supporting the interaction between HMGA1 and the regulator | ||
| Cell Line | HEK293T | Homo sapiens |
| Regulation | logFC: 1.10E+00 | GSE90639 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | LINC00460 is a novel oncogene of colorectal cancer through interacting with IGF2BP2 and DHX9 and bind to the m6A modified High mobility group protein HMG-I/HMG-Y (HMGA1) mRNA to enhance the HMGA1 mRNA stability. The N6-methyladenosine (m6A) modification of HMGA1 mRNA by METTL3 enhanced HMGA1 expression in CRC. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| Cell Process | mRNA stability | |||
| Epithelial-mesenchymal transition | ||||
| In-vitro Model | DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| FHC | Normal | Homo sapiens | CVCL_3688 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | Groups of HCT116-Luc-shCtrl, HCT116-Luc-shLINC00460, and HCT116-Luc-shLINC00460 + HMGA1 cells (5 × 106) were injected subcutaneously into the flanks of mice correspondingly. | |||
Colorectal cancer [ICD-11: 2B91]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | LINC00460 is a novel oncogene of colorectal cancer through interacting with IGF2BP2 and DHX9 and bind to the m6A modified High mobility group protein HMG-I/HMG-Y (HMGA1) mRNA to enhance the HMGA1 mRNA stability. The N6-methyladenosine (m6A) modification of HMGA1 mRNA by METTL3 enhanced HMGA1 expression in CRC. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| Cell Process | mRNA stability | |||
| Epithelial-mesenchymal transition | ||||
| In-vitro Model | DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| FHC | Normal | Homo sapiens | CVCL_3688 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | Groups of HCT116-Luc-shCtrl, HCT116-Luc-shLINC00460, and HCT116-Luc-shLINC00460 + HMGA1 cells (5 × 106) were injected subcutaneously into the flanks of mice correspondingly. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | LINC00460 is a novel oncogene of colorectal cancer through interacting with IGF2BP2 and DHX9 and bind to the m6A modified High mobility group protein HMG-I/HMG-Y (HMGA1) mRNA to enhance the HMGA1 mRNA stability. The N6-methyladenosine (m6A) modification of HMGA1 mRNA by METTL3 enhanced HMGA1 expression in CRC. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| Cell Process | mRNA stability | |||
| Epithelial-mesenchymal transition | ||||
| In-vitro Model | DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| FHC | Normal | Homo sapiens | CVCL_3688 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | Groups of HCT116-Luc-shCtrl, HCT116-Luc-shLINC00460, and HCT116-Luc-shLINC00460 + HMGA1 cells (5 × 106) were injected subcutaneously into the flanks of mice correspondingly. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03552 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03597 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
Non-coding RNA
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05274 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 460 (LINC00460) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT05372 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 460 (LINC00460) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00476)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: AC4SITE000104 | Click to Show/Hide the Full List | ||
| mod site | chr6:34244855-34244856:+ | [3] | |
| Sequence | GGAGGAAGAGGAGGGCATCTCGCAGGAGTCCTCGGAGGAGG | ||
| Cell/Tissue List | H1 | ||
| Seq Type List | ac4C-seq | ||
| Transcript ID List | ENST00000401473.7; ENST00000347617.10; ENST00000374116.3; ENST00000478214.1; ENST00000311487.9; ENST00000447654.5 | ||
| External Link | RMBase: ac4C_site_1586 | ||
N1-methyladenosine (m1A)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: M1ASITE000107 | Click to Show/Hide the Full List | ||
| mod site | chr6:34246021-34246022:+ | [4] | |
| Sequence | CATGGCAGGCTGGGTGACCGACTACCCCAGTCCCAGGGAAG | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | m1A-seq | ||
| Transcript ID List | ENST00000374116.3; ENST00000347617.10; ENST00000311487.9; ENST00000401473.7; ENST00000447654.5 | ||
| External Link | RMBase: m1A_site_911 | ||
5-methylcytidine (m5C)
| In total 28 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE003890 | Click to Show/Hide the Full List | ||
| mod site | chr6:34236919-34236920:+ | [5] | |
| Sequence | GCGCTCCTCTAATTGGGACTCCGAGCCGGGGCTATTTCTGG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000347617.10; ENST00000401473.7; ENST00000311487.9; ENST00000447654.5 | ||
| External Link | RMBase: m5C_site_37804 | ||
| mod ID: M5CSITE003891 | Click to Show/Hide the Full List | ||
| mod site | chr6:34236920-34236921:+ | [5] | |
| Sequence | CGCTCCTCTAATTGGGACTCCGAGCCGGGGCTATTTCTGGC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000447654.5; ENST00000311487.9; ENST00000347617.10; ENST00000401473.7 | ||
| External Link | RMBase: m5C_site_37805 | ||
| mod ID: M5CSITE003892 | Click to Show/Hide the Full List | ||
| mod site | chr6:34236924-34236925:+ | [5] | |
| Sequence | CCTCTAATTGGGACTCCGAGCCGGGGCTATTTCTGGCGCTG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000447654.5; ENST00000347617.10; ENST00000311487.9; ENST00000401473.7 | ||
| External Link | RMBase: m5C_site_37806 | ||
| mod ID: M5CSITE003893 | Click to Show/Hide the Full List | ||
| mod site | chr6:34237228-34237229:+ | ||
| Sequence | CATTTGCTACCAGCGGCGGCCGCGGCGGAGCCAGGCCGGTC | ||
| Cell/Tissue List | muscle | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000401473.7; ENST00000478214.1; ENST00000311487.9; ENST00000447654.5; ENST00000347617.10 | ||
| External Link | RMBase: m5C_site_37807 | ||
| mod ID: M5CSITE003894 | Click to Show/Hide the Full List | ||
| mod site | chr6:34237534-34237535:+ | ||
| Sequence | GAGGACGCCCGCACCCCTTCCCCCGCGCCGCGGGCGCCCTG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000478214.1; ENST00000447654.5; ENST00000347617.10; ENST00000401473.7; ENST00000311487.9 | ||
| External Link | RMBase: m5C_site_37808 | ||
| mod ID: M5CSITE003895 | Click to Show/Hide the Full List | ||
| mod site | chr6:34237535-34237536:+ | ||
| Sequence | AGGACGCCCGCACCCCTTCCCCCGCGCCGCGGGCGCCCTGC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000447654.5; ENST00000478214.1; ENST00000347617.10; ENST00000311487.9; ENST00000401473.7 | ||
| External Link | RMBase: m5C_site_37809 | ||
| mod ID: M5CSITE003897 | Click to Show/Hide the Full List | ||
| mod site | chr6:34237536-34237537:+ | ||
| Sequence | GGACGCCCGCACCCCTTCCCCCGCGCCGCGGGCGCCCTGCG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000401473.7; ENST00000478214.1; ENST00000447654.5; ENST00000311487.9; ENST00000347617.10 | ||
| External Link | RMBase: m5C_site_37810 | ||
| mod ID: M5CSITE003898 | Click to Show/Hide the Full List | ||
| mod site | chr6:34237537-34237538:+ | ||
| Sequence | GACGCCCGCACCCCTTCCCCCGCGCCGCGGGCGCCCTGCGG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000311487.9; ENST00000478214.1; ENST00000401473.7; ENST00000347617.10; ENST00000447654.5 | ||
| External Link | RMBase: m5C_site_37811 | ||
| mod ID: M5CSITE003899 | Click to Show/Hide the Full List | ||
| mod site | chr6:34237539-34237540:+ | ||
| Sequence | CGCCCGCACCCCTTCCCCCGCGCCGCGGGCGCCCTGCGGGG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000447654.5; ENST00000311487.9; ENST00000478214.1; ENST00000347617.10; ENST00000401473.7 | ||
| External Link | RMBase: m5C_site_37812 | ||
| mod ID: M5CSITE003900 | Click to Show/Hide the Full List | ||
| mod site | chr6:34237541-34237542:+ | ||
| Sequence | CCCGCACCCCTTCCCCCGCGCCGCGGGCGCCCTGCGGGGGG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000347617.10; ENST00000311487.9; ENST00000478214.1; ENST00000447654.5; ENST00000401473.7 | ||
| External Link | RMBase: m5C_site_37813 | ||
| mod ID: M5CSITE003901 | Click to Show/Hide the Full List | ||
| mod site | chr6:34237542-34237543:+ | ||
| Sequence | CCGCACCCCTTCCCCCGCGCCGCGGGCGCCCTGCGGGGGGC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000347617.10; ENST00000447654.5; ENST00000478214.1; ENST00000311487.9; ENST00000401473.7 | ||
| External Link | RMBase: m5C_site_37814 | ||
| mod ID: M5CSITE003902 | Click to Show/Hide the Full List | ||
| mod site | chr6:34237544-34237545:+ | ||
| Sequence | GCACCCCTTCCCCCGCGCCGCGGGCGCCCTGCGGGGGGCGG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000447654.5; ENST00000401473.7; ENST00000347617.10; ENST00000311487.9; ENST00000478214.1 | ||
| External Link | RMBase: m5C_site_37815 | ||
| mod ID: M5CSITE003903 | Click to Show/Hide the Full List | ||
| mod site | chr6:34240725-34240726:+ | [5] | |
| Sequence | GTTTGTCTAAAAGCACTTTTCTGTCTCCCAGCATCCCAGCC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000374116.3; ENST00000401473.7; ENST00000621552.1; ENST00000478214.1; ENST00000447654.5; ENST00000311487.9; MIMAT0027571; ENST00000347617.10 | ||
| External Link | RMBase: m5C_site_37816 | ||
| mod ID: M5CSITE003904 | Click to Show/Hide the Full List | ||
| mod site | chr6:34241418-34241419:+ | ||
| Sequence | TCCCAGGTGAGAAACGGGTGCCTGCGTGGGCCAGCACCTGT | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000401473.7; ENST00000311487.9; ENST00000447654.5; ENST00000478214.1; ENST00000347617.10; ENST00000374116.3 | ||
| External Link | RMBase: m5C_site_37817 | ||
| mod ID: M5CSITE003905 | Click to Show/Hide the Full List | ||
| mod site | chr6:34244927-34244928:+ | [5] | |
| Sequence | ACTGGAGGAGCAGCTTCCTTCTGGGACTGGACAGCTTTGCT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000401473.7; ENST00000347617.10; ENST00000447654.5; ENST00000311487.9; ENST00000478214.1; ENST00000374116.3 | ||
| External Link | RMBase: m5C_site_37818 | ||
| mod ID: M5CSITE003906 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245099-34245100:+ | [5] | |
| Sequence | TGAGTGGGGAGCAGTTTTCCCCTGGCCTCAGTTCCCAGCTC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000447654.5; ENST00000311487.9; ENST00000347617.10; ENST00000401473.7; ENST00000374116.3; ENST00000478214.1 | ||
| External Link | RMBase: m5C_site_37819 | ||
| mod ID: M5CSITE003908 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245255-34245256:+ | [5] | |
| Sequence | GGGCTTTTGGTTTGGGGGCGCCCTCTCTGCTCCTTCACTGT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000447654.5; ENST00000311487.9; ENST00000374116.3; ENST00000347617.10; ENST00000401473.7 | ||
| External Link | RMBase: m5C_site_37820 | ||
| mod ID: M5CSITE003909 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245256-34245257:+ | [5] | |
| Sequence | GGCTTTTGGTTTGGGGGCGCCCTCTCTGCTCCTTCACTGTT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000374116.3; ENST00000311487.9; ENST00000401473.7; ENST00000347617.10; ENST00000447654.5 | ||
| External Link | RMBase: m5C_site_37821 | ||
| mod ID: M5CSITE003910 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245257-34245258:+ | [5] | |
| Sequence | GCTTTTGGTTTGGGGGCGCCCTCTCTGCTCCTTCACTGTTC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000347617.10; ENST00000401473.7; ENST00000311487.9; ENST00000374116.3; ENST00000447654.5 | ||
| External Link | RMBase: m5C_site_37822 | ||
| mod ID: M5CSITE003911 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245259-34245260:+ | [5] | |
| Sequence | TTTTGGTTTGGGGGCGCCCTCTCTGCTCCTTCACTGTTCCC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000347617.10; ENST00000311487.9; ENST00000447654.5; ENST00000374116.3; ENST00000401473.7 | ||
| External Link | RMBase: m5C_site_37823 | ||
| mod ID: M5CSITE003912 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245261-34245262:+ | [5] | |
| Sequence | TTGGTTTGGGGGCGCCCTCTCTGCTCCTTCACTGTTCCCTC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000347617.10; ENST00000401473.7; ENST00000447654.5; ENST00000311487.9; ENST00000374116.3 | ||
| External Link | RMBase: m5C_site_37824 | ||
| mod ID: M5CSITE003913 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245412-34245413:+ | [6] | |
| Sequence | CAATGGAGGGGGGTGCTGGCCCCCAGGATTCCCCCAGCCAA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000447654.5; ENST00000347617.10; ENST00000374116.3; ENST00000401473.7; ENST00000311487.9 | ||
| External Link | RMBase: m5C_site_37825 | ||
| mod ID: M5CSITE003914 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245413-34245414:+ | [6] | |
| Sequence | AATGGAGGGGGGTGCTGGCCCCCAGGATTCCCCCAGCCAAA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000447654.5; ENST00000374116.3; ENST00000401473.7; ENST00000347617.10; ENST00000311487.9 | ||
| External Link | RMBase: m5C_site_37826 | ||
| mod ID: M5CSITE003915 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245414-34245415:+ | [6] | |
| Sequence | ATGGAGGGGGGTGCTGGCCCCCAGGATTCCCCCAGCCAAAC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000447654.5; ENST00000347617.10; ENST00000401473.7; ENST00000374116.3; ENST00000311487.9 | ||
| External Link | RMBase: m5C_site_37827 | ||
| mod ID: M5CSITE003916 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245415-34245416:+ | [6] | |
| Sequence | TGGAGGGGGGTGCTGGCCCCCAGGATTCCCCCAGCCAAACT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000311487.9; ENST00000374116.3; ENST00000347617.10; ENST00000401473.7; ENST00000447654.5 | ||
| External Link | RMBase: m5C_site_37828 | ||
| mod ID: M5CSITE003917 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245458-34245459:+ | [5] | |
| Sequence | CTTTGTCACCACGTGGGGCTCACTTTTCATCCTTCCCCAAC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000401473.7; ENST00000374116.3; ENST00000447654.5; ENST00000311487.9; ENST00000347617.10 | ||
| External Link | RMBase: m5C_site_37829 | ||
| mod ID: M5CSITE003919 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245732-34245733:+ | [5] | |
| Sequence | CGCCACTCAGCCATTTCCCCCTCCTCAGATGGGGCACCAAT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000401473.7; ENST00000347617.10; ENST00000447654.5; ENST00000311487.9; ENST00000374116.3 | ||
| External Link | RMBase: m5C_site_37830 | ||
| mod ID: M5CSITE003920 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245961-34245962:+ | [5] | |
| Sequence | TCCTGTTCTGCCCACGATCCCCTCCCCCAAGATACTCTTTG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000347617.10; ENST00000401473.7; ENST00000311487.9; ENST00000374116.3; ENST00000447654.5 | ||
| External Link | RMBase: m5C_site_37831 | ||
N6-methyladenosine (m6A)
| In total 35 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE074653 | Click to Show/Hide the Full List | ||
| mod site | chr6:34236916-34236917:+ | [7] | |
| Sequence | GCTGCGCTCCTCTAATTGGGACTCCGAGCCGGGGCTATTTC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000447654.5; ENST00000347617.10; ENST00000401473.7; ENST00000311487.9 | ||
| External Link | RMBase: m6A_site_713498 | ||
| mod ID: M6ASITE074654 | Click to Show/Hide the Full List | ||
| mod site | chr6:34238848-34238849:+ | [8] | |
| Sequence | GTATGAGAGCTTCCAGGGAGACCCGCCAAGATCTCCAGGCA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; MT4; GSC-11; HEK293T; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000347617.10; ENST00000374116.3; ENST00000311487.9; ENST00000478214.1; ENST00000401473.7; ENST00000447654.5 | ||
| External Link | RMBase: m6A_site_713499 | ||
| mod ID: M6ASITE074656 | Click to Show/Hide the Full List | ||
| mod site | chr6:34238897-34238898:+ | [8] | |
| Sequence | CAGGCTCTCTGGGATTCCGGACTGGCTTCGCCCAACTGGAA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; MT4; HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374116.3; ENST00000447654.5; ENST00000478214.1; ENST00000311487.9; ENST00000347617.10; ENST00000401473.7 | ||
| External Link | RMBase: m6A_site_713500 | ||
| mod ID: M6ASITE074657 | Click to Show/Hide the Full List | ||
| mod site | chr6:34238917-34238918:+ | [8] | |
| Sequence | ACTGGCTTCGCCCAACTGGAACCCCCTCGATGAGGGGGCCC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; MT4; HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000347617.10; ENST00000401473.7; ENST00000311487.9; ENST00000447654.5; ENST00000478214.1; ENST00000374116.3 | ||
| External Link | RMBase: m6A_site_713501 | ||
| mod ID: M6ASITE074658 | Click to Show/Hide the Full List | ||
| mod site | chr6:34238945-34238946:+ | [8] | |
| Sequence | GATGAGGGGGCCCCAGGCAGACCTTATATGAGGTCAGCACT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; MT4; MM6; Huh7; peripheral-blood; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000447654.5; ENST00000347617.10; ENST00000401473.7; ENST00000478214.1; ENST00000374116.3; ENST00000311487.9 | ||
| External Link | RMBase: m6A_site_713502 | ||
| mod ID: M6ASITE074659 | Click to Show/Hide the Full List | ||
| mod site | chr6:34240894-34240895:+ | [8] | |
| Sequence | AGCCTCCGGTGAGTCCCGGGACAGCGCTGGTAGGGAGTCAG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; H1A; MT4; A549; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000311487.9; ENST00000401473.7; ENST00000374116.3; ENST00000447654.5; ENST00000347617.10; ENST00000478214.1 | ||
| External Link | RMBase: m6A_site_713503 | ||
| mod ID: M6ASITE074660 | Click to Show/Hide the Full List | ||
| mod site | chr6:34242732-34242733:+ | [9] | |
| Sequence | AGGAGCCCAGCGAAGTGCCAACACCTAAGAGACCTCGGGGC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000401473.7; ENST00000447654.5; ENST00000311487.9; ENST00000347617.10; ENST00000478214.1; ENST00000374116.3 | ||
| External Link | RMBase: m6A_site_713504 | ||
| mod ID: M6ASITE074661 | Click to Show/Hide the Full List | ||
| mod site | chr6:34242743-34242744:+ | [8] | |
| Sequence | GAAGTGCCAACACCTAAGAGACCTCGGGGCCGACCAAAGGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; H1A; MT4; A549; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000347617.10; ENST00000478214.1; ENST00000401473.7; ENST00000311487.9; ENST00000447654.5; ENST00000374116.3 | ||
| External Link | RMBase: m6A_site_713505 | ||
| mod ID: M6ASITE074662 | Click to Show/Hide the Full List | ||
| mod site | chr6:34242772-34242773:+ | [8] | |
| Sequence | CCGACCAAAGGGAAGCAAAAACAAGGGTGCTGCCAAGACCC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; U2OS; H1A; LCLs; MT4; A549; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000447654.5; ENST00000401473.7; ENST00000478214.1; ENST00000374116.3; ENST00000311487.9; ENST00000347617.10 | ||
| External Link | RMBase: m6A_site_713506 | ||
| mod ID: M6ASITE074663 | Click to Show/Hide the Full List | ||
| mod site | chr6:34242789-34242790:+ | [8] | |
| Sequence | AAAACAAGGGTGCTGCCAAGACCCGGGTGAGACTTGAGATG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; H1A; LCLs; MT4; A549; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000447654.5; ENST00000478214.1; ENST00000311487.9; ENST00000401473.7; ENST00000374116.3; ENST00000347617.10 | ||
| External Link | RMBase: m6A_site_713507 | ||
| mod ID: M6ASITE074664 | Click to Show/Hide the Full List | ||
| mod site | chr6:34243470-34243471:+ | [8] | |
| Sequence | TTTCTCTAACCCTCTAGAAAACCACCACAACTCCAGGAAGG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; H1A; LCLs; MT4; A549; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000447654.5; ENST00000374116.3; ENST00000311487.9; ENST00000478214.1; ENST00000401473.7; ENST00000347617.10 | ||
| External Link | RMBase: m6A_site_713508 | ||
| mod ID: M6ASITE074665 | Click to Show/Hide the Full List | ||
| mod site | chr6:34243476-34243477:+ | [9] | |
| Sequence | TAACCCTCTAGAAAACCACCACAACTCCAGGAAGGAAACCA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000374116.3; ENST00000347617.10; ENST00000311487.9; ENST00000401473.7; ENST00000447654.5; ENST00000478214.1 | ||
| External Link | RMBase: m6A_site_713509 | ||
| mod ID: M6ASITE074666 | Click to Show/Hide the Full List | ||
| mod site | chr6:34243493-34243494:+ | [8] | |
| Sequence | ACCACAACTCCAGGAAGGAAACCAAGGGGCAGACCCAAAAA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; H1A; LCLs; MT4; A549; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374116.3; ENST00000478214.1; ENST00000401473.7; ENST00000347617.10; ENST00000311487.9; ENST00000447654.5 | ||
| External Link | RMBase: m6A_site_713510 | ||
| mod ID: M6ASITE074667 | Click to Show/Hide the Full List | ||
| mod site | chr6:34243505-34243506:+ | [8] | |
| Sequence | GGAAGGAAACCAAGGGGCAGACCCAAAAAACTGGTAGGTGA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; H1A; LCLs; MT4; A549; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000347617.10; ENST00000401473.7; ENST00000374116.3; ENST00000478214.1; ENST00000311487.9; ENST00000447654.5 | ||
| External Link | RMBase: m6A_site_713511 | ||
| mod ID: M6ASITE074668 | Click to Show/Hide the Full List | ||
| mod site | chr6:34243514-34243515:+ | [8] | |
| Sequence | CCAAGGGGCAGACCCAAAAAACTGGTAGGTGAAGAAGCAGA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; H1A; LCLs; MT4; A549; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000478214.1; ENST00000374116.3; ENST00000447654.5; ENST00000347617.10; ENST00000311487.9; ENST00000401473.7 | ||
| External Link | RMBase: m6A_site_713512 | ||
| mod ID: M6ASITE074669 | Click to Show/Hide the Full List | ||
| mod site | chr6:34244932-34244933:+ | [8] | |
| Sequence | AGGAGCAGCTTCCTTCTGGGACTGGACAGCTTTGCTCCGCT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; H1A; LCLs; MT4; A549; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000347617.10; ENST00000311487.9; ENST00000374116.3; ENST00000447654.5; ENST00000401473.7; ENST00000478214.1 | ||
| External Link | RMBase: m6A_site_713513 | ||
| mod ID: M6ASITE074670 | Click to Show/Hide the Full List | ||
| mod site | chr6:34244937-34244938:+ | [8] | |
| Sequence | CAGCTTCCTTCTGGGACTGGACAGCTTTGCTCCGCTCCCAC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; U2OS; H1A; LCLs; MT4; A549; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; DART-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000347617.10; ENST00000311487.9; ENST00000447654.5; ENST00000374116.3; ENST00000401473.7; ENST00000478214.1 | ||
| External Link | RMBase: m6A_site_713514 | ||
| mod ID: M6ASITE074671 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245038-34245039:+ | [9] | |
| Sequence | CCACCTGTGCCCTCACCACCACACTACACAGCACACCAGCC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000401473.7; ENST00000347617.10; ENST00000311487.9; ENST00000374116.3; ENST00000447654.5; ENST00000478214.1 | ||
| External Link | RMBase: m6A_site_713515 | ||
| mod ID: M6ASITE074672 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245050-34245051:+ | [9] | |
| Sequence | TCACCACCACACTACACAGCACACCAGCCGCTGCAGGGCTC | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000447654.5; ENST00000478214.1; ENST00000374116.3; ENST00000347617.10; ENST00000311487.9; ENST00000401473.7 | ||
| External Link | RMBase: m6A_site_713516 | ||
| mod ID: M6ASITE074673 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245139-34245140:+ | [10] | |
| Sequence | CCCCCCGCCCACCCACGCATACACACATGCCCTCCTGGACA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000374116.3; ENST00000311487.9; ENST00000347617.10; ENST00000447654.5; ENST00000401473.7; ENST00000478214.1 | ||
| External Link | RMBase: m6A_site_713517 | ||
| mod ID: M6ASITE074674 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245157-34245158:+ | [8] | |
| Sequence | ATACACACATGCCCTCCTGGACAAGGCTAACATCCCACTTA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000374116.3; ENST00000447654.5; ENST00000311487.9; ENST00000347617.10; ENST00000401473.7 | ||
| External Link | RMBase: m6A_site_713518 | ||
| mod ID: M6ASITE074675 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245166-34245167:+ | [9] | |
| Sequence | TGCCCTCCTGGACAAGGCTAACATCCCACTTAGCCGCACCC | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000311487.9; ENST00000401473.7; ENST00000447654.5; ENST00000347617.10; ENST00000374116.3 | ||
| External Link | RMBase: m6A_site_713519 | ||
| mod ID: M6ASITE074676 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245223-34245224:+ | [8] | |
| Sequence | CCACTCCCTTGGTGGTGGGGACATTGCTCTCTGGGCTTTTG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000401473.7; ENST00000311487.9; ENST00000347617.10; ENST00000447654.5; ENST00000374116.3 | ||
| External Link | RMBase: m6A_site_713520 | ||
| mod ID: M6ASITE074677 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245433-34245434:+ | [8] | |
| Sequence | CCCAGGATTCCCCCAGCCAAACTGTCTTTGTCACCACGTGG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000401473.7; ENST00000311487.9; ENST00000447654.5; ENST00000374116.3; ENST00000347617.10 | ||
| External Link | RMBase: m6A_site_713521 | ||
| mod ID: M6ASITE074678 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245504-34245505:+ | [8] | |
| Sequence | TAGTCCCCGTACTAGGTTGGACAGCCCCCTTCGGTTACAGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; HepG2; Huh7; AML | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000311487.9; ENST00000401473.7; ENST00000374116.3; ENST00000347617.10; ENST00000447654.5 | ||
| External Link | RMBase: m6A_site_713522 | ||
| mod ID: M6ASITE074679 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245601-34245602:+ | [9] | |
| Sequence | CCTCCGCCTGGGATCTGAGTACATATTGTGGTGATGGAGAT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000311487.9; ENST00000401473.7; ENST00000347617.10; ENST00000374116.3; ENST00000447654.5 | ||
| External Link | RMBase: m6A_site_713523 | ||
| mod ID: M6ASITE074680 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245702-34245703:+ | [10] | |
| Sequence | GGGGCTGCTCCCCTAACCCTACTTTGCTTCCGCCACTCAGC | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000311487.9; ENST00000347617.10; ENST00000374116.3; ENST00000447654.5; ENST00000401473.7 | ||
| External Link | RMBase: m6A_site_713524 | ||
| mod ID: M6ASITE074681 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245754-34245755:+ | [9] | |
| Sequence | CCTCAGATGGGGCACCAATAACAAGGAGCTCACCCTGCCCG | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000447654.5; ENST00000311487.9; ENST00000374116.3; ENST00000347617.10; ENST00000401473.7 | ||
| External Link | RMBase: m6A_site_713525 | ||
| mod ID: M6ASITE074682 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245838-34245839:+ | [9] | |
| Sequence | GTTCCATTTTTCCTCTGTTCACAAACTACCTCTGGACAGTT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000374116.3; ENST00000401473.7; ENST00000311487.9; ENST00000347617.10; ENST00000447654.5 | ||
| External Link | RMBase: m6A_site_713526 | ||
| mod ID: M6ASITE074683 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245842-34245843:+ | [11] | |
| Sequence | CATTTTTCCTCTGTTCACAAACTACCTCTGGACAGTTGTGT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000401473.7; ENST00000347617.10; ENST00000447654.5; ENST00000374116.3; ENST00000311487.9 | ||
| External Link | RMBase: m6A_site_713527 | ||
| mod ID: M6ASITE074684 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245853-34245854:+ | [8] | |
| Sequence | TGTTCACAAACTACCTCTGGACAGTTGTGTTGTTTTTTGTT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; Huh7; AML | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000311487.9; ENST00000347617.10; ENST00000401473.7; ENST00000447654.5; ENST00000374116.3 | ||
| External Link | RMBase: m6A_site_713528 | ||
| mod ID: M6ASITE074685 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245891-34245892:+ | [10] | |
| Sequence | GTTCAATGTTCCATTCTTCGACATCCGTCATTGCTGCTGCT | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000347617.10; ENST00000401473.7; ENST00000374116.3; ENST00000447654.5; ENST00000311487.9 | ||
| External Link | RMBase: m6A_site_713529 | ||
| mod ID: M6ASITE074686 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245974-34245975:+ | [10] | |
| Sequence | ACGATCCCCTCCCCCAAGATACTCTTTGTGGGGAAGAGGGG | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000374116.3; ENST00000311487.9; ENST00000401473.7; ENST00000447654.5; ENST00000347617.10 | ||
| External Link | RMBase: m6A_site_713530 | ||
| mod ID: M6ASITE074687 | Click to Show/Hide the Full List | ||
| mod site | chr6:34246021-34246022:+ | [10] | |
| Sequence | CATGGCAGGCTGGGTGACCGACTACCCCAGTCCCAGGGAAG | ||
| Motif Score | 3.287565476 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000401473.7; ENST00000374116.3; ENST00000311487.9; ENST00000347617.10; ENST00000447654.5 | ||
| External Link | RMBase: m6A_site_713531 | ||
| mod ID: M6ASITE074688 | Click to Show/Hide the Full List | ||
| mod site | chr6:34246201-34246202:+ | [10] | |
| Sequence | CTGGTTTCCTATTTGCAGTTACTTGAATAAAAAAAATATCC | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000401473.7; ENST00000447654.5; ENST00000347617.10; ENST00000311487.9; ENST00000374116.3 | ||
| External Link | RMBase: m6A_site_713532 | ||
Pseudouridine (Pseudo)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: PSESITE000233 | Click to Show/Hide the Full List | ||
| mod site | chr6:34245631-34245632:+ | [12] | |
| Sequence | GTGATGGAGATGCAGTCACTTATTGTCCAGGTGAGGCCCAA | ||
| Transcript ID List | ENST00000311487.9; ENST00000374116.3; ENST00000347617.10; ENST00000447654.5; ENST00000401473.7 | ||
| External Link | RMBase: Pseudo_site_4234 | ||
| mod ID: PSESITE000234 | Click to Show/Hide the Full List | ||
| mod site | chr6:34246158-34246159:+ | [12] | |
| Sequence | GGGGAGGGGTAGCCATGATTTGTCCCAGCCTGGGGCTCCCT | ||
| Transcript ID List | ENST00000401473.7; ENST00000347617.10; ENST00000447654.5; ENST00000311487.9; ENST00000374116.3 | ||
| External Link | RMBase: Pseudo_site_4235 | ||
References