m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00331)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
MAP1LC3B
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
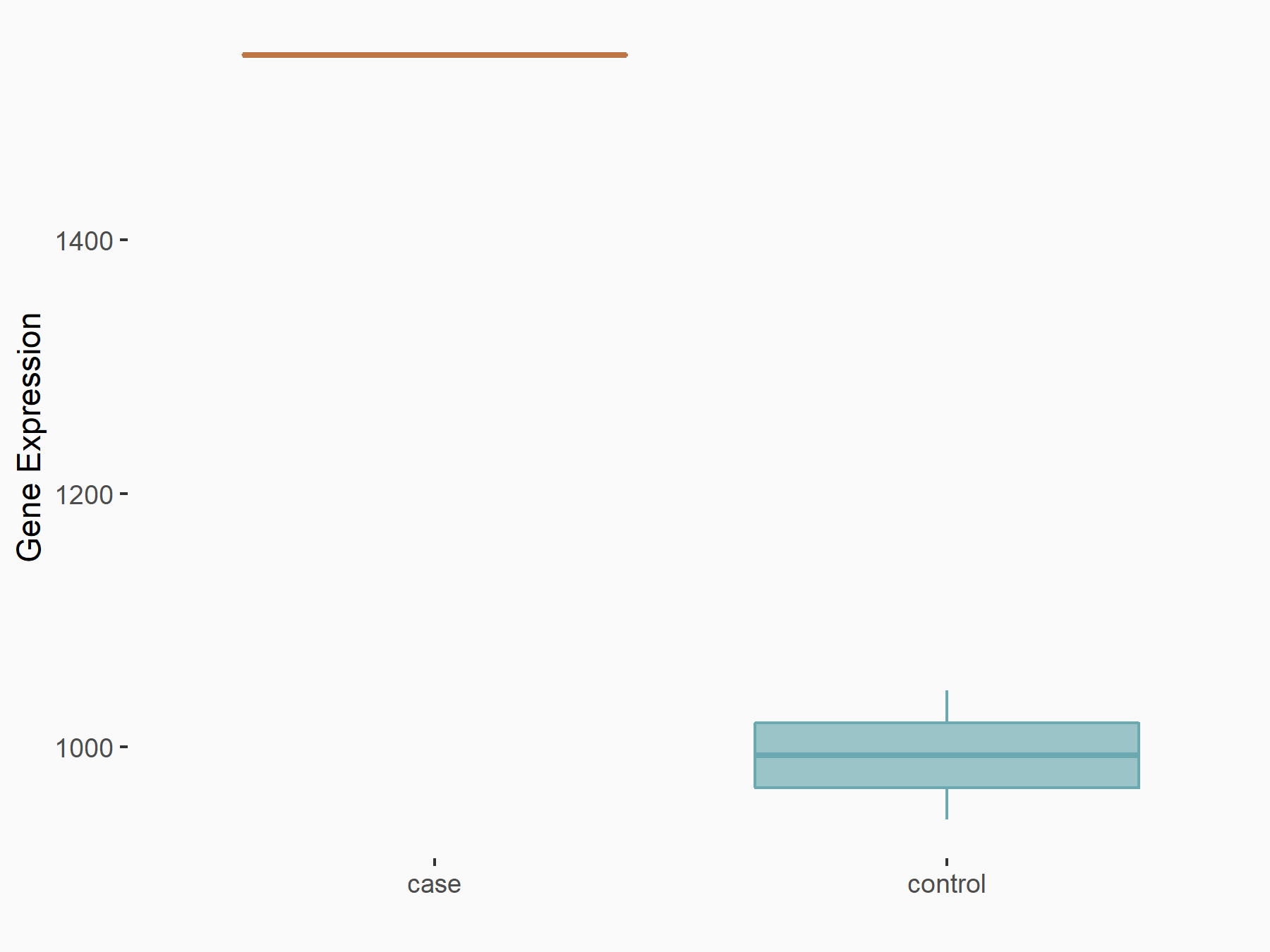  |
logFC: 6.38E-01 p-value: 4.93E-09 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between MAP1LC3B and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.34E+00 | GSE60213 |
| In total 4 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Knocking down METTL3 prevented Enterovirus 71-induced cell death and suppressed Enterovirus 71-induced expression of Bax while rescuing Bcl-2 expression after Enterovirus 71 infection. Knocking down METTL3 inhibited Enterovirus 71-induced expression of Atg5, Atg7 and Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II). Knocking down METTL3 inhibited Enterovirus 71-induced apoptosis and autophagy. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Enterovirus | ICD-11: 1A2Y | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| Cell autophagy | ||||
| In-vitro Model | Schwann cells (A type of glial cell that surrounds neurons) | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II). beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Responsed Drug | Chloroquine | Approved | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II). beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Responsed Drug | Gefitinib | Approved | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Experiment 4 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II). beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Responsed Drug | Beta-Elemen | Phase 3 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
Enterovirus [ICD-11: 1A2Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Knocking down METTL3 prevented Enterovirus 71-induced cell death and suppressed Enterovirus 71-induced expression of Bax while rescuing Bcl-2 expression after Enterovirus 71 infection. Knocking down METTL3 inhibited Enterovirus 71-induced expression of Atg5, Atg7 and Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II). Knocking down METTL3 inhibited Enterovirus 71-induced apoptosis and autophagy. | |||
| Responsed Disease | Enterovirus [ICD-11: 1A2Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| Cell autophagy | ||||
| In-vitro Model | Schwann cells (A type of glial cell that surrounds neurons) | |||
Lung cancer [ICD-11: 2C25]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II). beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Chloroquine | Approved | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II). beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Gefitinib | Approved | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II). beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Beta-Elemen | Phase 3 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
Chloroquine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [2] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II). beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
Gefitinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [2] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II). beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
Beta-Elemen
[Phase 3]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [2] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and Microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B/LC3B-II). beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00331)
| In total 39 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE007568 | Click to Show/Hide the Full List | ||
| mod site | chr16:87394522-87394523:+ | [4] | |
| Sequence | CAGGATTGACAGGGTTCTCAAAGTTGAAATGGATTTTAAGG | ||
| Transcript ID List | ENST00000268607.9; ENST00000650688.1; ENST00000564638.1; ENST00000564844.1; ENST00000565788.1; ENST00000570189.5 | ||
| External Link | RMBase: RNA-editing_site_53352 | ||
| mod ID: A2ISITE007569 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396181-87396182:+ | [5] | |
| Sequence | AACCTAGTTTCCTATATTAAAAAAATGTTTCCAGCCGGGTG | ||
| Transcript ID List | ENST00000564638.1; ENST00000564844.1; ENST00000650688.1; ENST00000268607.9; ENST00000570189.5; ENST00000565788.1 | ||
| External Link | RMBase: RNA-editing_site_53353 | ||
| mod ID: A2ISITE007570 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396219-87396220:+ | [5] | |
| Sequence | GTGTGGTGGCACATGCCTGTAATCCCAACATTTTGGTAGGC | ||
| Transcript ID List | ENST00000564844.1; ENST00000565788.1; ENST00000564638.1; rmsk_4605686; ENST00000650688.1; ENST00000268607.9; ENST00000570189.5 | ||
| External Link | RMBase: RNA-editing_site_53354 | ||
| mod ID: A2ISITE007571 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396220-87396221:+ | [5] | |
| Sequence | TGTGGTGGCACATGCCTGTAATCCCAACATTTTGGTAGGCC | ||
| Transcript ID List | ENST00000565788.1; ENST00000570189.5; ENST00000564638.1; rmsk_4605686; ENST00000650688.1; ENST00000564844.1; ENST00000268607.9 | ||
| External Link | RMBase: RNA-editing_site_53355 | ||
| mod ID: A2ISITE007572 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396226-87396227:+ | [5] | |
| Sequence | GGCACATGCCTGTAATCCCAACATTTTGGTAGGCCAAGGTG | ||
| Transcript ID List | ENST00000564638.1; ENST00000650688.1; ENST00000268607.9; rmsk_4605686; ENST00000564844.1; ENST00000570189.5; ENST00000565788.1 | ||
| External Link | RMBase: RNA-editing_site_53356 | ||
| mod ID: A2ISITE007573 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396236-87396237:+ | [5] | |
| Sequence | TGTAATCCCAACATTTTGGTAGGCCAAGGTGGGCGGATCAC | ||
| Transcript ID List | rmsk_4605686; ENST00000650688.1; ENST00000564638.1; ENST00000268607.9; ENST00000565788.1; ENST00000570189.5; ENST00000564844.1 | ||
| External Link | RMBase: RNA-editing_site_53357 | ||
| mod ID: A2ISITE007574 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396257-87396258:+ | [5] | |
| Sequence | GGCCAAGGTGGGCGGATCACAAGGTCAGGAGATTGAGACCA | ||
| Transcript ID List | ENST00000268607.9; ENST00000564844.1; ENST00000570189.5; ENST00000564638.1; ENST00000650688.1; ENST00000565788.1; rmsk_4605686 | ||
| External Link | RMBase: RNA-editing_site_53358 | ||
| mod ID: A2ISITE007575 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396258-87396259:+ | [5] | |
| Sequence | GCCAAGGTGGGCGGATCACAAGGTCAGGAGATTGAGACCAT | ||
| Transcript ID List | ENST00000564844.1; ENST00000564638.1; ENST00000650688.1; rmsk_4605686; ENST00000570189.5; ENST00000565788.1; ENST00000268607.9 | ||
| External Link | RMBase: RNA-editing_site_53359 | ||
| mod ID: A2ISITE007576 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396263-87396264:+ | [5] | |
| Sequence | GGTGGGCGGATCACAAGGTCAGGAGATTGAGACCATCCTGG | ||
| Transcript ID List | ENST00000564638.1; ENST00000650688.1; rmsk_4605686; ENST00000564844.1; ENST00000268607.9; ENST00000565788.1; ENST00000570189.5 | ||
| External Link | RMBase: RNA-editing_site_53360 | ||
| mod ID: A2ISITE007577 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396266-87396267:+ | [5] | |
| Sequence | GGGCGGATCACAAGGTCAGGAGATTGAGACCATCCTGGCTA | ||
| Transcript ID List | ENST00000564638.1; ENST00000564844.1; ENST00000650688.1; ENST00000570189.5; ENST00000268607.9; ENST00000565788.1; rmsk_4605686 | ||
| External Link | RMBase: RNA-editing_site_53361 | ||
| mod ID: A2ISITE007578 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396268-87396269:+ | [5] | |
| Sequence | GCGGATCACAAGGTCAGGAGATTGAGACCATCCTGGCTAAC | ||
| Transcript ID List | ENST00000268607.9; rmsk_4605686; ENST00000570189.5; ENST00000564638.1; ENST00000564844.1; ENST00000565788.1; ENST00000650688.1 | ||
| External Link | RMBase: RNA-editing_site_53362 | ||
| mod ID: A2ISITE007579 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396274-87396275:+ | [5] | |
| Sequence | CACAAGGTCAGGAGATTGAGACCATCCTGGCTAACATGATG | ||
| Transcript ID List | rmsk_4605686; ENST00000564844.1; ENST00000650688.1; ENST00000570189.5; ENST00000268607.9; ENST00000564638.1; ENST00000565788.1 | ||
| External Link | RMBase: RNA-editing_site_53363 | ||
| mod ID: A2ISITE007580 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396286-87396287:+ | [5] | |
| Sequence | AGATTGAGACCATCCTGGCTAACATGATGAAACCCCGTCTT | ||
| Transcript ID List | ENST00000565788.1; ENST00000650688.1; ENST00000570189.5; ENST00000564844.1; ENST00000564638.1; ENST00000268607.9; rmsk_4605686 | ||
| External Link | RMBase: RNA-editing_site_53364 | ||
| mod ID: A2ISITE007581 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396287-87396288:+ | [5] | |
| Sequence | GATTGAGACCATCCTGGCTAACATGATGAAACCCCGTCTTT | ||
| Transcript ID List | rmsk_4605686; ENST00000564638.1; ENST00000564844.1; ENST00000565788.1; ENST00000268607.9; ENST00000570189.5; ENST00000650688.1 | ||
| External Link | RMBase: RNA-editing_site_53365 | ||
| mod ID: A2ISITE007582 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396292-87396293:+ | [5] | |
| Sequence | AGACCATCCTGGCTAACATGATGAAACCCCGTCTTTACTAA | ||
| Transcript ID List | ENST00000565788.1; ENST00000268607.9; ENST00000564638.1; ENST00000564844.1; rmsk_4605686; ENST00000650688.1; ENST00000570189.5 | ||
| External Link | RMBase: RNA-editing_site_53366 | ||
| mod ID: A2ISITE007583 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396295-87396296:+ | [5] | |
| Sequence | CCATCCTGGCTAACATGATGAAACCCCGTCTTTACTAAAAA | ||
| Transcript ID List | rmsk_4605686; ENST00000268607.9; ENST00000570189.5; ENST00000565788.1; ENST00000564844.1; ENST00000650688.1; ENST00000564638.1 | ||
| External Link | RMBase: RNA-editing_site_53367 | ||
| mod ID: A2ISITE007584 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396297-87396298:+ | [5] | |
| Sequence | ATCCTGGCTAACATGATGAAACCCCGTCTTTACTAAAAATA | ||
| Transcript ID List | ENST00000268607.9; rmsk_4605686; ENST00000564638.1; ENST00000570189.5; ENST00000564844.1; ENST00000565788.1; ENST00000650688.1 | ||
| External Link | RMBase: RNA-editing_site_53368 | ||
| mod ID: A2ISITE007585 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396314-87396315:+ | [5] | |
| Sequence | GAAACCCCGTCTTTACTAAAAATACAAAAAATTAGCCGGGC | ||
| Transcript ID List | ENST00000650688.1; ENST00000564638.1; ENST00000565788.1; ENST00000564844.1; ENST00000268607.9; ENST00000570189.5; rmsk_4605686 | ||
| External Link | RMBase: RNA-editing_site_53369 | ||
| mod ID: A2ISITE007586 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396315-87396316:+ | [5] | |
| Sequence | AAACCCCGTCTTTACTAAAAATACAAAAAATTAGCCGGGCG | ||
| Transcript ID List | ENST00000564844.1; ENST00000650688.1; ENST00000268607.9; ENST00000570189.5; ENST00000564638.1; ENST00000565788.1; rmsk_4605686 | ||
| External Link | RMBase: RNA-editing_site_53370 | ||
| mod ID: A2ISITE007587 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396317-87396318:+ | [5] | |
| Sequence | ACCCCGTCTTTACTAAAAATACAAAAAATTAGCCGGGCGTG | ||
| Transcript ID List | ENST00000565788.1; ENST00000570189.5; ENST00000650688.1; ENST00000268607.9; rmsk_4605686; ENST00000564844.1; ENST00000564638.1 | ||
| External Link | RMBase: RNA-editing_site_53371 | ||
| mod ID: A2ISITE007588 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396322-87396323:+ | [5] | |
| Sequence | GTCTTTACTAAAAATACAAAAAATTAGCCGGGCGTGGTGGC | ||
| Transcript ID List | ENST00000570189.5; rmsk_4605686; ENST00000565788.1; ENST00000268607.9; ENST00000564844.1; ENST00000564638.1; ENST00000650688.1 | ||
| External Link | RMBase: RNA-editing_site_53372 | ||
| mod ID: A2ISITE007589 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396323-87396324:+ | [5] | |
| Sequence | TCTTTACTAAAAATACAAAAAATTAGCCGGGCGTGGTGGCG | ||
| Transcript ID List | ENST00000565788.1; ENST00000564638.1; rmsk_4605686; ENST00000570189.5; ENST00000268607.9; ENST00000650688.1; ENST00000564844.1 | ||
| External Link | RMBase: RNA-editing_site_53373 | ||
| mod ID: A2ISITE007590 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396327-87396328:+ | [5] | |
| Sequence | TACTAAAAATACAAAAAATTAGCCGGGCGTGGTGGCGGGCG | ||
| Transcript ID List | rmsk_4605686; ENST00000564844.1; ENST00000565788.1; ENST00000570189.5; ENST00000268607.9; ENST00000650688.1; ENST00000564638.1 | ||
| External Link | RMBase: RNA-editing_site_53374 | ||
| mod ID: A2ISITE007591 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396353-87396354:+ | [5] | |
| Sequence | GCGTGGTGGCGGGCGCCTGTAGTCCCAGCTACTTGGGATGC | ||
| Transcript ID List | ENST00000570189.5; ENST00000564638.1; ENST00000564844.1; ENST00000268607.9; ENST00000650688.1; ENST00000565788.1; rmsk_4605686 | ||
| External Link | RMBase: RNA-editing_site_53375 | ||
| mod ID: A2ISITE007592 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396363-87396364:+ | [5] | |
| Sequence | GGGCGCCTGTAGTCCCAGCTACTTGGGATGCTGAGGGAGGA | ||
| Transcript ID List | ENST00000570189.5; rmsk_4605686; ENST00000650688.1; ENST00000564844.1; ENST00000565788.1; ENST00000268607.9; ENST00000564638.1 | ||
| External Link | RMBase: RNA-editing_site_53376 | ||
| mod ID: A2ISITE007593 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396380-87396381:+ | [5] | |
| Sequence | GCTACTTGGGATGCTGAGGGAGGAGAATGGCATGAACCTGG | ||
| Transcript ID List | ENST00000268607.9; ENST00000565788.1; ENST00000564844.1; ENST00000650688.1; rmsk_4605686; ENST00000564638.1; ENST00000570189.5 | ||
| External Link | RMBase: RNA-editing_site_53377 | ||
| mod ID: A2ISITE007594 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396394-87396395:+ | [5] | |
| Sequence | TGAGGGAGGAGAATGGCATGAACCTGGGAGGCGTAGCTTGC | ||
| Transcript ID List | rmsk_4605686; ENST00000268607.9; ENST00000564638.1; ENST00000650688.1; ENST00000570189.5; ENST00000565788.1; ENST00000564844.1 | ||
| External Link | RMBase: RNA-editing_site_53378 | ||
| mod ID: A2ISITE007595 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396395-87396396:+ | [5] | |
| Sequence | GAGGGAGGAGAATGGCATGAACCTGGGAGGCGTAGCTTGCA | ||
| Transcript ID List | ENST00000650688.1; ENST00000564638.1; ENST00000564844.1; rmsk_4605686; ENST00000565788.1; ENST00000570189.5; ENST00000268607.9 | ||
| External Link | RMBase: RNA-editing_site_53379 | ||
| mod ID: A2ISITE007596 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396408-87396409:+ | [5] | |
| Sequence | GGCATGAACCTGGGAGGCGTAGCTTGCAGTGAGCAGAGATC | ||
| Transcript ID List | ENST00000564638.1; ENST00000650688.1; ENST00000570189.5; ENST00000268607.9; rmsk_4605686; ENST00000564844.1; ENST00000565788.1 | ||
| External Link | RMBase: RNA-editing_site_53380 | ||
| mod ID: A2ISITE007597 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396419-87396420:+ | [5] | |
| Sequence | GGGAGGCGTAGCTTGCAGTGAGCAGAGATCGCGCCACTGCA | ||
| Transcript ID List | ENST00000268607.9; ENST00000565788.1; ENST00000570189.5; rmsk_4605686; ENST00000564638.1; ENST00000564844.1; ENST00000650688.1 | ||
| External Link | RMBase: RNA-editing_site_53381 | ||
| mod ID: A2ISITE007598 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396422-87396423:+ | [5] | |
| Sequence | AGGCGTAGCTTGCAGTGAGCAGAGATCGCGCCACTGCACTC | ||
| Transcript ID List | rmsk_4605686; ENST00000268607.9; ENST00000565788.1; ENST00000564638.1; ENST00000650688.1; ENST00000564844.1; ENST00000570189.5 | ||
| External Link | RMBase: RNA-editing_site_53382 | ||
| mod ID: A2ISITE007599 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396434-87396435:+ | [5] | |
| Sequence | CAGTGAGCAGAGATCGCGCCACTGCACTCCAGCCTGAGTGA | ||
| Transcript ID List | ENST00000564638.1; ENST00000565788.1; rmsk_4605686; ENST00000268607.9; ENST00000650688.1; ENST00000570189.5; ENST00000564844.1 | ||
| External Link | RMBase: RNA-editing_site_53383 | ||
| mod ID: A2ISITE007600 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396450-87396451:+ | [5] | |
| Sequence | CGCCACTGCACTCCAGCCTGAGTGACAGAGCAAGACTCCAT | ||
| Transcript ID List | ENST00000650688.1; ENST00000268607.9; ENST00000564844.1; ENST00000565788.1; ENST00000564638.1; rmsk_4605686; ENST00000570189.5 | ||
| External Link | RMBase: RNA-editing_site_53384 | ||
| mod ID: A2ISITE007601 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396456-87396457:+ | [5] | |
| Sequence | TGCACTCCAGCCTGAGTGACAGAGCAAGACTCCATCTCAAA | ||
| Transcript ID List | ENST00000564638.1; ENST00000650688.1; ENST00000565788.1; ENST00000564844.1; ENST00000570189.5; ENST00000268607.9; rmsk_4605686 | ||
| External Link | RMBase: RNA-editing_site_53385 | ||
| mod ID: A2ISITE007602 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396461-87396462:+ | [5] | |
| Sequence | TCCAGCCTGAGTGACAGAGCAAGACTCCATCTCAAAAAAAA | ||
| Transcript ID List | rmsk_4605686; ENST00000565788.1; ENST00000650688.1; ENST00000564638.1; ENST00000570189.5; ENST00000564844.1; ENST00000268607.9 | ||
| External Link | RMBase: RNA-editing_site_53386 | ||
| mod ID: A2ISITE007603 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396462-87396463:+ | [5] | |
| Sequence | CCAGCCTGAGTGACAGAGCAAGACTCCATCTCAAAAAAAAA | ||
| Transcript ID List | ENST00000650688.1; rmsk_4605686; ENST00000564638.1; ENST00000564844.1; ENST00000570189.5; ENST00000565788.1; ENST00000268607.9 | ||
| External Link | RMBase: RNA-editing_site_53387 | ||
| mod ID: A2ISITE007604 | Click to Show/Hide the Full List | ||
| mod site | chr16:87396464-87396465:+ | [5] | |
| Sequence | AGCCTGAGTGACAGAGCAAGACTCCATCTCAAAAAAAAAAA | ||
| Transcript ID List | ENST00000564638.1; ENST00000570189.5; ENST00000268607.9; rmsk_4605686; ENST00000565788.1; ENST00000564844.1; ENST00000650688.1 | ||
| External Link | RMBase: RNA-editing_site_53388 | ||
| mod ID: A2ISITE007605 | Click to Show/Hide the Full List | ||
| mod site | chr16:87399145-87399146:+ | [6] | |
| Sequence | CCTCCTGAGTAGCTCTGACTAGAGGCACACACCACTACACC | ||
| Transcript ID List | ENST00000650688.1; ENST00000564638.1; ENST00000564844.1; ENST00000565788.1; ENST00000570189.5; ENST00000268607.9 | ||
| External Link | RMBase: RNA-editing_site_53389 | ||
| mod ID: A2ISITE007606 | Click to Show/Hide the Full List | ||
| mod site | chr16:87401248-87401249:+ | [4] | |
| Sequence | CCAGCCTCTCTGCCTCGGTAAGGCTTGAGGTCTTTTTCTGA | ||
| Transcript ID List | ENST00000268607.9; ENST00000570189.5; ENST00000650688.1; ENST00000564844.1; ENST00000565788.1 | ||
| External Link | RMBase: RNA-editing_site_53390 | ||
5-methylcytidine (m5C)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE000804 | Click to Show/Hide the Full List | ||
| mod site | chr16:87402955-87402956:+ | [7] | |
| Sequence | CTCAATGCTAATCAGGCCTTCTTCCTGTTGGTGAACGGACA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000268607.9; ENST00000570189.5; ENST00000564844.1 | ||
| External Link | RMBase: m5C_site_16978 | ||
| mod ID: M5CSITE000805 | Click to Show/Hide the Full List | ||
| mod site | chr16:87402959-87402960:+ | [7] | |
| Sequence | ATGCTAATCAGGCCTTCTTCCTGTTGGTGAACGGACACAGC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000564844.1; ENST00000570189.5; ENST00000650688.1; ENST00000268607.9 | ||
| External Link | RMBase: m5C_site_16979 | ||
N6-methyladenosine (m6A)
| In total 55 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE028933 | Click to Show/Hide the Full List | ||
| mod site | chr16:87384019-87384020:+ | [8] | |
| Sequence | TGCCGAGTGCAAAACGTTAGACTGCCCCGTGTGAGGCTCGA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | TREX | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000650688.1 | ||
| External Link | RMBase: m6A_site_335431 | ||
| mod ID: M6ASITE028934 | Click to Show/Hide the Full List | ||
| mod site | chr16:87384040-87384041:+ | [8] | |
| Sequence | CTGCCCCGTGTGAGGCTCGAACTCACGACCTTCAGATTATG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | TREX | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000650688.1 | ||
| External Link | RMBase: m6A_site_335432 | ||
| mod ID: M6ASITE028935 | Click to Show/Hide the Full List | ||
| mod site | chr16:87391968-87391969:+ | [9] | |
| Sequence | TCCCTCAAGAGTGCCCCGGGACACCCCGCCTGTGGCTCAGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335435 | ||
| mod ID: M6ASITE028936 | Click to Show/Hide the Full List | ||
| mod site | chr16:87392050-87392051:+ | [9] | |
| Sequence | AGGCTCCCGAGCGCCCACAGACCCGGGGTGCGGCCCAGCCC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000268607.9; ENST00000650688.1 | ||
| External Link | RMBase: m6A_site_335436 | ||
| mod ID: M6ASITE028937 | Click to Show/Hide the Full List | ||
| mod site | chr16:87392314-87392315:+ | [10] | |
| Sequence | AGGCTGCGGGCTGAGGAGATACAAGGGAAGTGGCTATCGCC | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000268607.9; ENST00000650688.1 | ||
| External Link | RMBase: m6A_site_335437 | ||
| mod ID: M6ASITE028938 | Click to Show/Hide the Full List | ||
| mod site | chr16:87392384-87392385:+ | [9] | |
| Sequence | CGCCCCCGGGAGCCGCCGGGACCCTCGCGTCGTCGCCGCCG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; fibroblasts; LCLs; MT4; Huh7; Jurkat; CD4T; GSC-11; HEK293A-TOA; MSC; iSLK; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | rmsk_4605680; ENST00000564844.1; ENST00000268607.9; ENST00000650688.1; ENST00000565788.1; ENST00000570189.5 | ||
| External Link | RMBase: m6A_site_335438 | ||
| mod ID: M6ASITE028939 | Click to Show/Hide the Full List | ||
| mod site | chr16:87392442-87392443:+ | [9] | |
| Sequence | GCACCATGCCGTCGGAGAAGACCTTCAAGCAGCGCCGCACC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; LCLs; MT4; Huh7; Jurkat; CD4T; GSC-11; HEK293A-TOA; MSC; iSLK; TIME; TREX; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000565788.1; ENST00000268607.9; ENST00000564638.1; ENST00000564844.1; ENST00000570189.5; ENST00000650688.1 | ||
| External Link | RMBase: m6A_site_335439 | ||
| mod ID: M6ASITE028941 | Click to Show/Hide the Full List | ||
| mod site | chr16:87398815-87398816:+ | [10] | |
| Sequence | TCTCTTCATTTCATTGCAGAACAAAGAGTAGAAGATGTCCG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000565788.1; ENST00000564638.1; ENST00000268607.9; ENST00000564844.1; ENST00000650688.1; ENST00000570189.5 | ||
| External Link | RMBase: m6A_site_335440 | ||
| mod ID: M6ASITE028942 | Click to Show/Hide the Full List | ||
| mod site | chr16:87398967-87398968:+ | [11] | |
| Sequence | GTTTTTACAGGAATCACCAGACAGCCAAACCCTGGGTGTCA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HepG2; A549; Huh7; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000268607.9; ENST00000564638.1; ENST00000564844.1; ENST00000565788.1; ENST00000650688.1; ENST00000570189.5 | ||
| External Link | RMBase: m6A_site_335441 | ||
| mod ID: M6ASITE028943 | Click to Show/Hide the Full List | ||
| mod site | chr16:87402190-87402191:+ | [10] | |
| Sequence | CCAGGTGATAATAGAACGATACAAGGGTGAGAAGCAGCTTC | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000570189.5; ENST00000650688.1; ENST00000564844.1; ENST00000268607.9; ENST00000565788.1 | ||
| External Link | RMBase: m6A_site_335442 | ||
| mod ID: M6ASITE028944 | Click to Show/Hide the Full List | ||
| mod site | chr16:87402973-87402974:+ | [10] | |
| Sequence | TTCTTCCTGTTGGTGAACGGACACAGCATGGTCAGCGTCTC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000564844.1; ENST00000570189.5; ENST00000268607.9; ENST00000650688.1 | ||
| External Link | RMBase: m6A_site_335443 | ||
| mod ID: M6ASITE028945 | Click to Show/Hide the Full List | ||
| mod site | chr16:87402995-87402996:+ | [10] | |
| Sequence | ACAGCATGGTCAGCGTCTCCACACCAATCTCAGAGGTGTAT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000564844.1; ENST00000570189.5; ENST00000650688.1; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335444 | ||
| mod ID: M6ASITE028946 | Click to Show/Hide the Full List | ||
| mod site | chr16:87403047-87403048:+ | [10] | |
| Sequence | AGATGAAGATGGATTCCTGTACATGGTCTATGCCTCCCAGG | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000570189.5; ENST00000564844.1; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335445 | ||
| mod ID: M6ASITE028947 | Click to Show/Hide the Full List | ||
| mod site | chr16:87403098-87403099:+ | [9] | |
| Sequence | GATGAAATTGTCAGTGTAAAACCAGAAAAAATGCAGCTCTT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000564844.1; ENST00000268607.9; ENST00000570189.5 | ||
| External Link | RMBase: m6A_site_335446 | ||
| mod ID: M6ASITE028948 | Click to Show/Hide the Full List | ||
| mod site | chr16:87403133-87403134:+ | [9] | |
| Sequence | GCTCTTCTAGAATTGTTTAAACCCTTACCAAGGAAAAAAAA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000268607.9; ENST00000650688.1; ENST00000564844.1; ENST00000570189.5 | ||
| External Link | RMBase: m6A_site_335447 | ||
| mod ID: M6ASITE028949 | Click to Show/Hide the Full List | ||
| mod site | chr16:87403204-87403205:+ | [9] | |
| Sequence | ATCCAATCACAGATCATGAAACAGTAGTGTTCCCACCTAGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000268607.9; ENST00000570189.5; ENST00000564844.1; ENST00000650688.1 | ||
| External Link | RMBase: m6A_site_335448 | ||
| mod ID: M6ASITE028950 | Click to Show/Hide the Full List | ||
| mod site | chr16:87403218-87403219:+ | [12] | |
| Sequence | CATGAAACAGTAGTGTTCCCACCTAGGAGTGTTAGGAAGTT | ||
| Motif Score | 2.032470238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000268607.9; ENST00000570189.5; ENST00000564844.1 | ||
| External Link | RMBase: m6A_site_335449 | ||
| mod ID: M6ASITE028952 | Click to Show/Hide the Full List | ||
| mod site | chr16:87403280-87403281:+ | [10] | |
| Sequence | AAACTGAGCTCCAAGTGAGCACATTCAGCTTTGGAAACTAT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000570189.5; ENST00000564844.1; ENST00000268607.9; ENST00000650688.1 | ||
| External Link | RMBase: m6A_site_335450 | ||
| mod ID: M6ASITE028953 | Click to Show/Hide the Full List | ||
| mod site | chr16:87403603-87403604:+ | [12] | |
| Sequence | CGCAAAACGCATTTGCCATCACAGTTGGCACAAACGCAGGG | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000570189.5; ENST00000564844.1; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335451 | ||
| mod ID: M6ASITE028954 | Click to Show/Hide the Full List | ||
| mod site | chr16:87403612-87403613:+ | [13] | |
| Sequence | CATTTGCCATCACAGTTGGCACAAACGCAGGGTAAACGGGC | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | kidney; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000570189.5; ENST00000650688.1; ENST00000564844.1; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335452 | ||
| mod ID: M6ASITE028955 | Click to Show/Hide the Full List | ||
| mod site | chr16:87403677-87403678:+ | [12] | |
| Sequence | TAAACTGCTGAAGGTCCCTGACTCCTAAGAGAACCACACCC | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000268607.9; ENST00000564844.1; ENST00000650688.1; ENST00000570189.5 | ||
| External Link | RMBase: m6A_site_335453 | ||
| mod ID: M6ASITE028956 | Click to Show/Hide the Full List | ||
| mod site | chr16:87403692-87403693:+ | [10] | |
| Sequence | CCCTGACTCCTAAGAGAACCACACCCAAAGTCCTCACTCTT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000564844.1; ENST00000570189.5; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335454 | ||
| mod ID: M6ASITE028957 | Click to Show/Hide the Full List | ||
| mod site | chr16:87403760-87403761:+ | [12] | |
| Sequence | TGTTCTCTAGATAGTTACACACATAAAGACACCACTCAAAA | ||
| Motif Score | 2.084928571 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000564844.1; ENST00000268607.9; ENST00000650688.1; ENST00000570189.5 | ||
| External Link | RMBase: m6A_site_335455 | ||
| mod ID: M6ASITE028958 | Click to Show/Hide the Full List | ||
| mod site | chr16:87403768-87403769:+ | [10] | |
| Sequence | AGATAGTTACACACATAAAGACACCACTCAAAAGGAAACTT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000570189.5; ENST00000564844.1; ENST00000650688.1; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335456 | ||
| mod ID: M6ASITE028959 | Click to Show/Hide the Full List | ||
| mod site | chr16:87403824-87403825:+ | [9] | |
| Sequence | TTGATCGAGTTTCTTAAAAGACCCTGGAGAAAGAGTGGCAT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000564844.1; ENST00000268607.9; ENST00000650688.1 | ||
| External Link | RMBase: m6A_site_335457 | ||
| mod ID: M6ASITE028960 | Click to Show/Hide the Full List | ||
| mod site | chr16:87403876-87403877:+ | [9] | |
| Sequence | CAGGTTTTGTCTGAGTTCAAACTAGTGCCTGTGTTGTTACG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000564844.1; ENST00000268607.9; ENST00000650688.1 | ||
| External Link | RMBase: m6A_site_335458 | ||
| mod ID: M6ASITE028961 | Click to Show/Hide the Full List | ||
| mod site | chr16:87403942-87403943:+ | [9] | |
| Sequence | TCTGGAGTACAGCGGGAGAAACACAAAATAGTATAACTGAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000564844.1; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335459 | ||
| mod ID: M6ASITE028963 | Click to Show/Hide the Full List | ||
| mod site | chr16:87403964-87403965:+ | [9] | |
| Sequence | ACAAAATAGTATAACTGAAAACATTAACATTCAGACACACT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000564844.1; ENST00000268607.9; ENST00000650688.1 | ||
| External Link | RMBase: m6A_site_335460 | ||
| mod ID: M6ASITE028964 | Click to Show/Hide the Full List | ||
| mod site | chr16:87403978-87403979:+ | [9] | |
| Sequence | CTGAAAACATTAACATTCAGACACACTCCCTTCTGCCTTCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000564844.1; ENST00000650688.1; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335461 | ||
| mod ID: M6ASITE028965 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404052-87404053:+ | [12] | |
| Sequence | TTTTTAATGTTAAATGTGTAACTCAGTATTACTGAAAAGGT | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000564844.1; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335462 | ||
| mod ID: M6ASITE028966 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404062-87404063:+ | [12] | |
| Sequence | TAAATGTGTAACTCAGTATTACTGAAAAGGTACCCACATTT | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000564844.1; ENST00000268607.9; ENST00000650688.1 | ||
| External Link | RMBase: m6A_site_335463 | ||
| mod ID: M6ASITE028967 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404073-87404074:+ | [12] | |
| Sequence | CTCAGTATTACTGAAAAGGTACCCACATTTTGAATAGTAGT | ||
| Motif Score | 2.8355 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000564844.1; ENST00000268607.9; ENST00000650688.1 | ||
| External Link | RMBase: m6A_site_335464 | ||
| mod ID: M6ASITE028968 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404077-87404078:+ | [10] | |
| Sequence | GTATTACTGAAAAGGTACCCACATTTTGAATAGTAGTTATC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000564844.1; ENST00000650688.1; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335465 | ||
| mod ID: M6ASITE028969 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404111-87404112:+ | [9] | |
| Sequence | AGTTATCACTCTTAGGTCAGACAGCCATCAGAATTCTCCCA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000268607.9; ENST00000564844.1 | ||
| External Link | RMBase: m6A_site_335466 | ||
| mod ID: M6ASITE028970 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404160-87404161:+ | [9] | |
| Sequence | GCATGTCAGTTGTGGAGAAAACATAGCAAAAAGAGCCGTAC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000564844.1; ENST00000650688.1; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335467 | ||
| mod ID: M6ASITE028971 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404194-87404195:+ | [12] | |
| Sequence | GCCGTACGCTCTTTACAGATACTAATGTCAAGAGTTAAACC | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000268607.9; ENST00000564844.1 | ||
| External Link | RMBase: m6A_site_335468 | ||
| mod ID: M6ASITE028972 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404212-87404213:+ | [9] | |
| Sequence | ATACTAATGTCAAGAGTTAAACCTCCTCAGGTTCAACCTGT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000268607.9; ENST00000564844.1 | ||
| External Link | RMBase: m6A_site_335469 | ||
| mod ID: M6ASITE028974 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404241-87404242:+ | [9] | |
| Sequence | GGTTCAACCTGTGATAAAAGACTAGTGCTTCCCAGTACTTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000564844.1; ENST00000268607.9; ENST00000650688.1 | ||
| External Link | RMBase: m6A_site_335470 | ||
| mod ID: M6ASITE028975 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404257-87404258:+ | [12] | |
| Sequence | AAAGACTAGTGCTTCCCAGTACTTGCATGGGGTTCACTATT | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000268607.9; ENST00000564844.1; ENST00000650688.1 | ||
| External Link | RMBase: m6A_site_335471 | ||
| mod ID: M6ASITE028976 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404316-87404317:+ | [12] | |
| Sequence | ATCACAGGAAAATCACAATTACACCACTTTAGACCCTATGT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000564844.1; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335472 | ||
| mod ID: M6ASITE028977 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404328-87404329:+ | [9] | |
| Sequence | TCACAATTACACCACTTTAGACCCTATGTGTAGCAGGTCAC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000268607.9; ENST00000564844.1 | ||
| External Link | RMBase: m6A_site_335473 | ||
| mod ID: M6ASITE028978 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404347-87404348:+ | [10] | |
| Sequence | GACCCTATGTGTAGCAGGTCACAACTTACCCTTGTGTGTTT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000268607.9; ENST00000564844.1 | ||
| External Link | RMBase: m6A_site_335474 | ||
| mod ID: M6ASITE028979 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404391-87404392:+ | [12] | |
| Sequence | TGTGTATGAAATACCTGTATACGTTAGTGAAAGCTGTTTAC | ||
| Motif Score | 2.084416667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000268607.9; ENST00000564844.1 | ||
| External Link | RMBase: m6A_site_335475 | ||
| mod ID: M6ASITE028980 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404410-87404411:+ | [12] | |
| Sequence | TACGTTAGTGAAAGCTGTTTACTGTAACGGGGAAAACCAGA | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000268607.9; ENST00000650688.1; ENST00000564844.1 | ||
| External Link | RMBase: m6A_site_335476 | ||
| mod ID: M6ASITE028981 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404425-87404426:+ | [9] | |
| Sequence | TGTTTACTGTAACGGGGAAAACCAGATTCTTTGCATCTGGG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; MT4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000564844.1; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335477 | ||
| mod ID: M6ASITE028982 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404510-87404511:+ | [12] | |
| Sequence | ACCCCCGCATGCGTCTGTCCACTTGGCTAACTTTTAATATG | ||
| Motif Score | 2.475107143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000564844.1; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335478 | ||
| mod ID: M6ASITE028983 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404519-87404520:+ | [12] | |
| Sequence | TGCGTCTGTCCACTTGGCTAACTTTTAATATGTGTATTTTT | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000268607.9; ENST00000564844.1; ENST00000650688.1 | ||
| External Link | RMBase: m6A_site_335479 | ||
| mod ID: M6ASITE028985 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404540-87404541:+ | [12] | |
| Sequence | CTTTTAATATGTGTATTTTTACATTATGTATATTCTTAACT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000268607.9; ENST00000564844.1 | ||
| External Link | RMBase: m6A_site_335480 | ||
| mod ID: M6ASITE028986 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404558-87404559:+ | [12] | |
| Sequence | TTACATTATGTATATTCTTAACTGGACTGTCTCGTTTAGAC | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000268607.9; ENST00000564844.1; ENST00000650688.1 | ||
| External Link | RMBase: m6A_site_335481 | ||
| mod ID: M6ASITE028987 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404563-87404564:+ | [9] | |
| Sequence | TTATGTATATTCTTAACTGGACTGTCTCGTTTAGACTGTAT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000564844.1; ENST00000650688.1; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335482 | ||
| mod ID: M6ASITE028988 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404577-87404578:+ | [9] | |
| Sequence | AACTGGACTGTCTCGTTTAGACTGTATACATCATATCTGAC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000564844.1; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335483 | ||
| mod ID: M6ASITE028989 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404596-87404597:+ | [12] | |
| Sequence | GACTGTATACATCATATCTGACATTATTGTAACTACCGTGT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000268607.9; ENST00000650688.1; ENST00000564844.1 | ||
| External Link | RMBase: m6A_site_335484 | ||
| mod ID: M6ASITE028990 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404665-87404666:+ | [10] | |
| Sequence | GCTTTTTAAGAAAAAAAATAACATGCTGAGGGGTGACCTAT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000268607.9; ENST00000650688.1; ENST00000564844.1 | ||
| External Link | RMBase: m6A_site_335485 | ||
| mod ID: M6ASITE028991 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404726-87404727:+ | [9] | |
| Sequence | TTATTTATAGGATCTTTAAAACATTTTTAATGAACTAAGTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000650688.1; ENST00000564844.1; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335486 | ||
| mod ID: M6ASITE028992 | Click to Show/Hide the Full List | ||
| mod site | chr16:87404739-87404740:+ | [9] | |
| Sequence | CTTTAAAACATTTTTAATGAACTAAGTTGAATAAAGGCACA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000564844.1; ENST00000650688.1; ENST00000268607.9 | ||
| External Link | RMBase: m6A_site_335487 | ||
References