m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00551)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
PARP1
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shMETTL3 MOLM13 cells
Control: MOLM13 cells
|
GSE98623 | |
| Regulation |
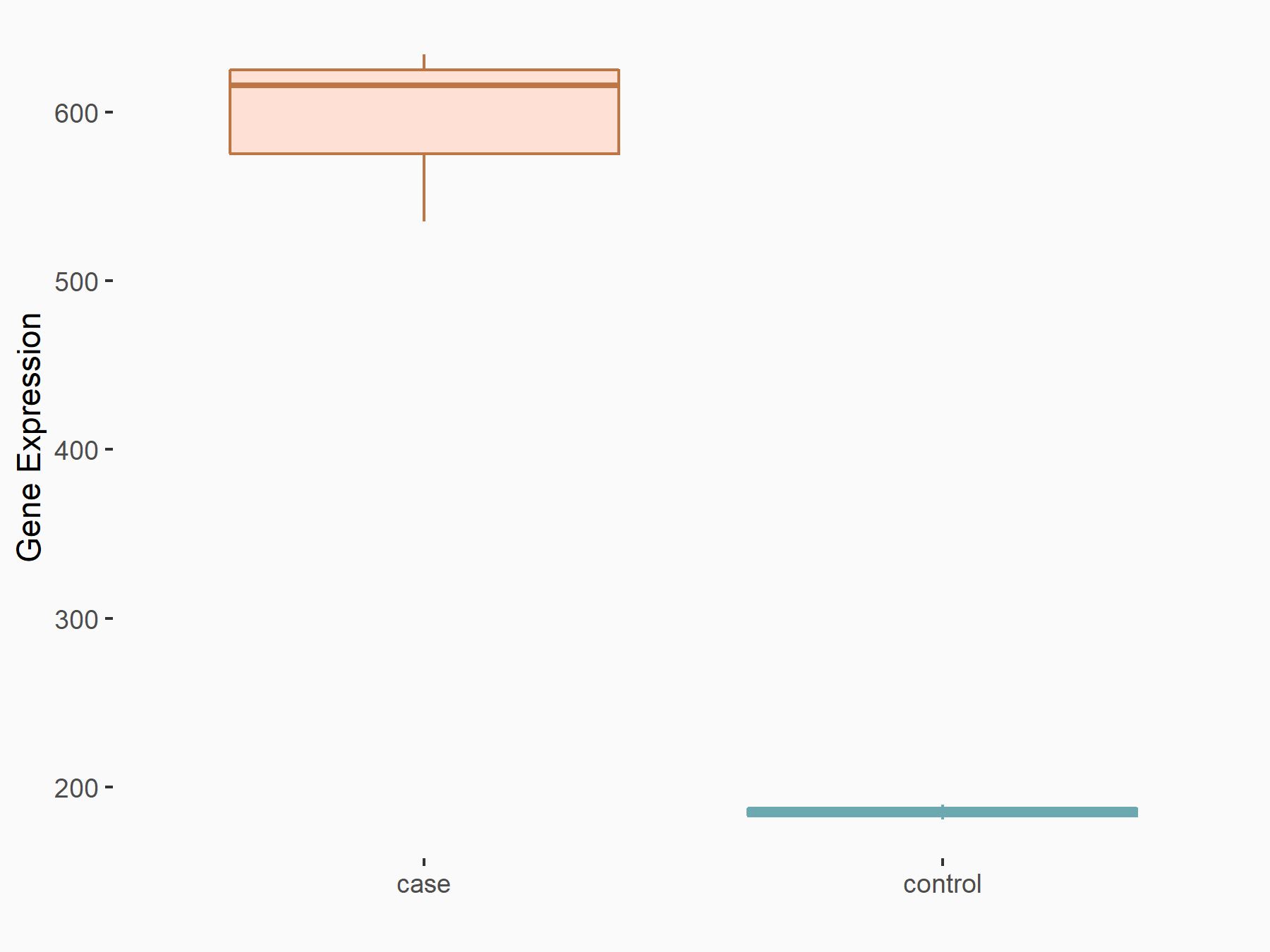  |
logFC: 1.69E+00 p-value: 1.05E-54 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | m6A methyltransferase METTL3 facilitates oxaliplatin resistance in CD133+ gastric cancer stem cells by Promoting Poly [ADP-ribose] polymerase 1 (PARP1) mRNA stability which increases base excision repair pathway activity. METTTL3 enhances the stability of PARP1 by recruiting YTHDF1 to target the 3'-untranslated Region (3'-UTR) of PARP1 mRNA. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Responsed Drug | Oxaliplatin | Approved | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | |||
| Cell Process | RNA stability | |||
| Excision repair | ||||
| In-vitro Model | SNU-719 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_5086 |
| MKN74 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_2791 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | 100,000 pLKO and PARP1-sh1 (PT1 and PT2) cells were mixed with matrix gel and inoculate into BALB/C nude mice, respectively. After 25 days, 6 organoid transplanted tumor mice were treated with oxaliplatin (Sellekchem, s1224) twice a week for 4 weeks at a dose of 5 mg/kg. | |||
YTH domain-containing family protein 1 (YTHDF1) [READER]
| Representative RIP-seq result supporting the interaction between PARP1 and the regulator | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.06E+00 | GSE63591 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | m6A methyltransferase METTL3 facilitates oxaliplatin resistance in CD133+ gastric cancer stem cells by Promoting PARP1 mRNA stability which increases base excision repair pathway activity. METTTL3 enhances the stability of PARP1 by recruiting Poly [ADP-ribose] polymerase 1 (PARP1) to target the 3'-untranslated Region (3'-UTR) of PARP1 mRNA. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Responsed Drug | Oxaliplatin | Approved | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | |||
| Cell Process | RNA stability | |||
| Excision repair | ||||
| In-vitro Model | SNU-719 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_5086 |
| MKN74 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_2791 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | 100,000 pLKO and PARP1-sh1 (PT1 and PT2) cells were mixed with matrix gel and inoculate into BALB/C nude mice, respectively. After 25 days, 6 organoid transplanted tumor mice were treated with oxaliplatin (Sellekchem, s1224) twice a week for 4 weeks at a dose of 5 mg/kg. | |||
Fat mass and obesity-associated protein (FTO) [ERASER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | Targeting FTO significantly suppresses cancer cell growth and enhances chemotherapy sensitivity, which not only mediating the balance of intracellular ROS by regulating G6PD expression, but also maintaining genome instability by regulating Poly [ADP-ribose] polymerase 1 (PARP1) expression. These findings shed light on new molecular mechanisms of CRC development and treatments mediated by m6A modification. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | Glutathione metabolism | hsa00480 | ||
| In-vitro Model | LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | For CDX model, nude mice (female, 4-6-week-old) were subcutaneously injected with 5 × 106 HCT116 cells on the both flank. For PDX model, the patient tumors were divided into small pieces and then inoculated on both flank of nude mice. For knockdown FTO mice model, FTO mice model, two weeks after inoculation, the shFTO#3 lenti-virus injected into the tumor for three consecutive days. For combined medication mice model, intraperitoneal injection of Rhein and Olaparib was started one week after inoculation. | |||
Gastric cancer [ICD-11: 2B72]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | m6A methyltransferase METTL3 facilitates oxaliplatin resistance in CD133+ gastric cancer stem cells by Promoting Poly [ADP-ribose] polymerase 1 (PARP1) mRNA stability which increases base excision repair pathway activity. METTTL3 enhances the stability of PARP1 by recruiting YTHDF1 to target the 3'-untranslated Region (3'-UTR) of PARP1 mRNA. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Oxaliplatin | Approved | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | |||
| Cell Process | RNA stability | |||
| Excision repair | ||||
| In-vitro Model | SNU-719 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_5086 |
| MKN74 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_2791 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | 100,000 pLKO and PARP1-sh1 (PT1 and PT2) cells were mixed with matrix gel and inoculate into BALB/C nude mice, respectively. After 25 days, 6 organoid transplanted tumor mice were treated with oxaliplatin (Sellekchem, s1224) twice a week for 4 weeks at a dose of 5 mg/kg. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | m6A methyltransferase METTL3 facilitates oxaliplatin resistance in CD133+ gastric cancer stem cells by Promoting PARP1 mRNA stability which increases base excision repair pathway activity. METTTL3 enhances the stability of PARP1 by recruiting Poly [ADP-ribose] polymerase 1 (PARP1) to target the 3'-untranslated Region (3'-UTR) of PARP1 mRNA. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Oxaliplatin | Approved | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | |||
| Cell Process | RNA stability | |||
| Excision repair | ||||
| In-vitro Model | SNU-719 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_5086 |
| MKN74 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_2791 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | 100,000 pLKO and PARP1-sh1 (PT1 and PT2) cells were mixed with matrix gel and inoculate into BALB/C nude mice, respectively. After 25 days, 6 organoid transplanted tumor mice were treated with oxaliplatin (Sellekchem, s1224) twice a week for 4 weeks at a dose of 5 mg/kg. | |||
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | Targeting FTO significantly suppresses cancer cell growth and enhances chemotherapy sensitivity, which not only mediating the balance of intracellular ROS by regulating G6PD expression, but also maintaining genome instability by regulating Poly [ADP-ribose] polymerase 1 (PARP1) expression. These findings shed light on new molecular mechanisms of CRC development and treatments mediated by m6A modification. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Glutathione metabolism | hsa00480 | ||
| In-vitro Model | LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | For CDX model, nude mice (female, 4-6-week-old) were subcutaneously injected with 5 × 106 HCT116 cells on the both flank. For PDX model, the patient tumors were divided into small pieces and then inoculated on both flank of nude mice. For knockdown FTO mice model, FTO mice model, two weeks after inoculation, the shFTO#3 lenti-virus injected into the tumor for three consecutive days. For combined medication mice model, intraperitoneal injection of Rhein and Olaparib was started one week after inoculation. | |||
Oxaliplatin
[Approved]
| In total 2 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | m6A methyltransferase METTL3 facilitates oxaliplatin resistance in CD133+ gastric cancer stem cells by Promoting Poly [ADP-ribose] polymerase 1 (PARP1) mRNA stability which increases base excision repair pathway activity. METTTL3 enhances the stability of PARP1 by recruiting YTHDF1 to target the 3'-untranslated Region (3'-UTR) of PARP1 mRNA. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | |||
| Cell Process | RNA stability | |||
| Excision repair | ||||
| In-vitro Model | SNU-719 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_5086 |
| MKN74 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_2791 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | 100,000 pLKO and PARP1-sh1 (PT1 and PT2) cells were mixed with matrix gel and inoculate into BALB/C nude mice, respectively. After 25 days, 6 organoid transplanted tumor mice were treated with oxaliplatin (Sellekchem, s1224) twice a week for 4 weeks at a dose of 5 mg/kg. | |||
| Experiment 2 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | m6A methyltransferase METTL3 facilitates oxaliplatin resistance in CD133+ gastric cancer stem cells by Promoting PARP1 mRNA stability which increases base excision repair pathway activity. METTTL3 enhances the stability of PARP1 by recruiting Poly [ADP-ribose] polymerase 1 (PARP1) to target the 3'-untranslated Region (3'-UTR) of PARP1 mRNA. | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | |||
| Cell Process | RNA stability | |||
| Excision repair | ||||
| In-vitro Model | SNU-719 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_5086 |
| MKN74 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_2791 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | 100,000 pLKO and PARP1-sh1 (PT1 and PT2) cells were mixed with matrix gel and inoculate into BALB/C nude mice, respectively. After 25 days, 6 organoid transplanted tumor mice were treated with oxaliplatin (Sellekchem, s1224) twice a week for 4 weeks at a dose of 5 mg/kg. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: Fat mass and obesity-associated protein (FTO)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03445 | ||
| Epigenetic Regulator | Histone deacetylase 1 (HDAC1) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03656 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
| Drug | Oxaliplatin | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00551)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE004673 | Click to Show/Hide the Full List | ||
| mod site | chr1:226383118-226383119:- | [3] | |
| Sequence | ACAGGACCGTATATTCCCCCCAGAAACCAGCGCCTCCGTGG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m5C_site_4827 | ||
| mod ID: M5CSITE004674 | Click to Show/Hide the Full List | ||
| mod site | chr1:226408075-226408076:- | [3] | |
| Sequence | ATCAGCAATCTATCAGGGAACGGCGGTGGCCGGTGCGGCGT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000366794.10; ENST00000629232.1 | ||
| External Link | RMBase: m5C_site_4828 | ||
N6-methyladenosine (m6A)
| In total 87 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE087903 | Click to Show/Hide the Full List | ||
| mod site | chr1:226360719-226360720:- | [4] | |
| Sequence | ACCCAAGGGCTAATAGTAATACTCGATTAAAAATGCAGATG | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82067 | ||
| mod ID: M6ASITE087904 | Click to Show/Hide the Full List | ||
| mod site | chr1:226360751-226360752:- | [5] | |
| Sequence | TTCCAAATTAAAACTCTACCACAAATATACTTACCCAAGGG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82068 | ||
| mod ID: M6ASITE087905 | Click to Show/Hide the Full List | ||
| mod site | chr1:226360759-226360760:- | [4] | |
| Sequence | CTTTTGTCTTCCAAATTAAAACTCTACCACAAATATACTTA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82069 | ||
| mod ID: M6ASITE087906 | Click to Show/Hide the Full List | ||
| mod site | chr1:226360794-226360795:- | [4] | |
| Sequence | AGTTGCATTTGAAATTCTTGACTTTCTTATGGGCACTTTTG | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82070 | ||
| mod ID: M6ASITE087907 | Click to Show/Hide the Full List | ||
| mod site | chr1:226360846-226360847:- | [6] | |
| Sequence | TCCTTCTCCAGGAATACTGAACATGGGAGCTCTTGAAATAT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | brain; HEK293T; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82071 | ||
| mod ID: M6ASITE087908 | Click to Show/Hide the Full List | ||
| mod site | chr1:226360887-226360888:- | [4] | |
| Sequence | GAACGCTAACAATTTCTCATACTTAGAAACAAAAAGAGCTT | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82072 | ||
| mod ID: M6ASITE087909 | Click to Show/Hide the Full List | ||
| mod site | chr1:226360977-226360978:- | [4] | |
| Sequence | GATTTTCTTTTTTATCTTGCACTTATTGTCCCCTTTTTAGT | ||
| Motif Score | 3.252583333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82073 | ||
| mod ID: M6ASITE087910 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361005-226361006:- | [4] | |
| Sequence | AAAATAAAAACTAATTTCATACTATTTAGATTTTCTTTTTT | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82074 | ||
| mod ID: M6ASITE087911 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361016-226361017:- | [4] | |
| Sequence | ATTTTTAGTTAAAAATAAAAACTAATTTCATACTATTTAGA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82075 | ||
| mod ID: M6ASITE087912 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361101-226361102:- | [4] | |
| Sequence | TTCCCCAGGGAAGGAAAAATACACTTCCACCCTTTTTTCTA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82076 | ||
| mod ID: M6ASITE087913 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361127-226361128:- | [4] | |
| Sequence | TTCGTTAGAATGTCTGCCTTACTGGTTTCCCCAGGGAAGGA | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82077 | ||
| mod ID: M6ASITE087919 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361154-226361155:- | [7] | |
| Sequence | AGACTAGTCCTATGGAAAAAACCAAGCTTCGTTAGAATGTC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82078 | ||
| mod ID: M6ASITE087920 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361172-226361173:- | [7] | |
| Sequence | AGAGAGATTCTGTTGCATAGACTAGTCCTATGGAAAAAACC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82079 | ||
| mod ID: M6ASITE087921 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361210-226361211:- | [4] | |
| Sequence | TTGCCAGGTAGATAAAACTGACATAGAGAAAAGGCTGGAGA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82080 | ||
| mod ID: M6ASITE087922 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361214-226361215:- | [7] | |
| Sequence | TTTCTTGCCAGGTAGATAAAACTGACATAGAGAAAAGGCTG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82081 | ||
| mod ID: M6ASITE087923 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361300-226361301:- | [4] | |
| Sequence | CGTGTTAAAGGTTTTCTCTAACTTCTCAAGTCCCTTGTTTT | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000366794.10; ENST00000490921.5 | ||
| External Link | RMBase: m6A_site_82082 | ||
| mod ID: M6ASITE087924 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361321-226361322:- | [4] | |
| Sequence | TCAGAAAGGATTTTACAGAAACGTGTTAAAGGTTTTCTCTA | ||
| Motif Score | 2.179660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000490921.5; ENST00000366794.10; ENST00000463968.5 | ||
| External Link | RMBase: m6A_site_82083 | ||
| mod ID: M6ASITE087925 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361327-226361328:- | [4] | |
| Sequence | ACCACCTCAGAAAGGATTTTACAGAAACGTGTTAAAGGTTT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000366794.10; ENST00000463968.5; ENST00000490921.5 | ||
| External Link | RMBase: m6A_site_82084 | ||
| mod ID: M6ASITE087926 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361352-226361353:- | [4] | |
| Sequence | TGCTGATGGGTAGTACCTGTACTAAACCACCTCAGAAAGGA | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000463968.5; ENST00000366794.10; ENST00000490921.5 | ||
| External Link | RMBase: m6A_site_82085 | ||
| mod ID: M6ASITE087927 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361387-226361388:- | [4] | |
| Sequence | AAGCGCTTCTGCACCAACTCACCTGGCCGCTAAGTTGCTGA | ||
| Motif Score | 2.026654762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000490921.5; ENST00000366794.10; ENST00000463968.5 | ||
| External Link | RMBase: m6A_site_82086 | ||
| mod ID: M6ASITE087928 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361438-226361439:- | [5] | |
| Sequence | TTGGGAGAGGTAGCCGAGTCACACCCGGTGGCTCTGGTATG | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000490921.5; ENST00000366794.10; ENST00000491816.1; ENST00000463968.5 | ||
| External Link | RMBase: m6A_site_82087 | ||
| mod ID: M6ASITE087929 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361473-226361474:- | [7] | |
| Sequence | AACTGAAATTCAATTTTAAGACCTCCCTGTGGTAATTGGGA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000366794.10; ENST00000490921.5; ENST00000491816.1; ENST00000463968.5 | ||
| External Link | RMBase: m6A_site_82088 | ||
| mod ID: M6ASITE087930 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361492-226361493:- | [7] | |
| Sequence | AATCTGAAGTATCTGCTGAAACTGAAATTCAATTTTAAGAC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000463968.5; ENST00000366794.10; ENST00000490921.5; ENST00000491816.1 | ||
| External Link | RMBase: m6A_site_82089 | ||
| mod ID: M6ASITE087931 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361538-226361539:- | [5] | |
| Sequence | TAACAAGCTTCCCCTCAGGTACATTGTCTATGATATTGCTC | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000490921.5; ENST00000366794.10; ENST00000463968.5; ENST00000491816.1 | ||
| External Link | RMBase: m6A_site_82090 | ||
| mod ID: M6ASITE087932 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361732-226361733:- | [7] | |
| Sequence | CCTCAGTCTGCCTGAAGAAGACTTAGAGTAACTTTCAGGCT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000366794.10; ENST00000463968.5; ENST00000490921.5; ENST00000468608.1; ENST00000491816.1 | ||
| External Link | RMBase: m6A_site_82091 | ||
| mod ID: M6ASITE087933 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361979-226361980:- | [4] | |
| Sequence | GGTGTGAATGACACCTCTCTACTATATAACGAGTATCCTTT | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000366794.10; ENST00000468608.1; ENST00000491816.1; ENST00000490921.5; ENST00000463968.5 | ||
| External Link | RMBase: m6A_site_82092 | ||
| mod ID: M6ASITE087934 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361989-226361990:- | [5] | |
| Sequence | GATTTCATCTGGTGTGAATGACACCTCTCTACTATATAACG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000463968.5; ENST00000491816.1; ENST00000468608.1; ENST00000490921.5; ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82093 | ||
| mod ID: M6ASITE087935 | Click to Show/Hide the Full List | ||
| mod site | chr1:226362028-226362029:- | [4] | |
| Sequence | CATTAGTCTGGATGGTGTAGACGTTCCTCTTGGGACCGGGA | ||
| Motif Score | 2.871321429 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000463968.5; ENST00000366794.10; ENST00000491816.1; ENST00000468608.1; ENST00000490921.5 | ||
| External Link | RMBase: m6A_site_82094 | ||
| mod ID: M6ASITE087936 | Click to Show/Hide the Full List | ||
| mod site | chr1:226362049-226362050:- | [4] | |
| Sequence | TACCCCTGATCCTTCAGCTAACATTAGTCTGGATGGTGTAG | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000366794.10; ENST00000463968.5; ENST00000490921.5; ENST00000491816.1; ENST00000468608.1 | ||
| External Link | RMBase: m6A_site_82095 | ||
| mod ID: M6ASITE087937 | Click to Show/Hide the Full List | ||
| mod site | chr1:226363153-226363154:- | [7] | |
| Sequence | CCTCTTTTGAATAGGTATGAACTGAAGCACGCTTCACATAT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000490921.5; ENST00000468608.1; ENST00000366794.10; ENST00000463968.5 | ||
| External Link | RMBase: m6A_site_82096 | ||
| mod ID: M6ASITE087938 | Click to Show/Hide the Full List | ||
| mod site | chr1:226363945-226363946:- | [7] | |
| Sequence | GGGAGAAGTTGCCCTTGGAAACATGTGAGTAAACCTGGCTC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000366794.10; ENST00000490921.5; ENST00000463968.5 | ||
| External Link | RMBase: m6A_site_82097 | ||
| mod ID: M6ASITE087939 | Click to Show/Hide the Full List | ||
| mod site | chr1:226363987-226363988:- | [7] | |
| Sequence | CTGCCATACGTCTCAGGGAGACCCAATAGGCTTAATCCTGT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000366794.10; ENST00000463968.5; ENST00000490921.5 | ||
| External Link | RMBase: m6A_site_82098 | ||
| mod ID: M6ASITE087940 | Click to Show/Hide the Full List | ||
| mod site | chr1:226364032-226364033:- | [5] | |
| Sequence | TAAAGGGATCTATTTCGCTGACATGGTCTCCAAGAGTGCCA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000366794.10; ENST00000463968.5; ENST00000490921.5 | ||
| External Link | RMBase: m6A_site_82099 | ||
| mod ID: M6ASITE087941 | Click to Show/Hide the Full List | ||
| mod site | chr1:226364069-226364070:- | [7] | |
| Sequence | TCCCTTTTCCGACCTTCCAGACAGGCTACATGTTTGGTAAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000463968.5; ENST00000366794.10; ENST00000490921.5 | ||
| External Link | RMBase: m6A_site_82100 | ||
| mod ID: M6ASITE087942 | Click to Show/Hide the Full List | ||
| mod site | chr1:226364128-226364129:- | [7] | |
| Sequence | CTCAAGTTGGCACTCAGTGAACAGCTGCTCCTAATTTCCCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000366794.10; rmsk_401830; ENST00000463968.5; ENST00000490921.5 | ||
| External Link | RMBase: m6A_site_82101 | ||
| mod ID: M6ASITE087943 | Click to Show/Hide the Full List | ||
| mod site | chr1:226365063-226365064:- | [7] | |
| Sequence | TGCTGTGGCACGGGTCCAGGACCACCAACTTTGCTGGGATC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000366794.10; ENST00000490921.5 | ||
| External Link | RMBase: m6A_site_82102 | ||
| mod ID: M6ASITE087944 | Click to Show/Hide the Full List | ||
| mod site | chr1:226365116-226365117:- | [5] | |
| Sequence | TGAAGGCGAATGCCAGCGTTACAAGCCCTTTAAGCAGCTTC | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000366794.10; ENST00000498787.1; ENST00000490921.5 | ||
| External Link | RMBase: m6A_site_82103 | ||
| mod ID: M6ASITE087945 | Click to Show/Hide the Full List | ||
| mod site | chr1:226365981-226365982:- | [5] | |
| Sequence | GAACACTCATGCAACCACACACAATGCGTATGACTTGGAAG | ||
| Motif Score | 2.084928571 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000366794.10; ENST00000498787.1; ENST00000490921.5 | ||
| External Link | RMBase: m6A_site_82104 | ||
| mod ID: M6ASITE087946 | Click to Show/Hide the Full List | ||
| mod site | chr1:226365999-226366000:- | [7] | |
| Sequence | CATCAGGAAGTATGTTAAGAACACTCATGCAACCACACACA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000490921.5; ENST00000366794.10; ENST00000498787.1 | ||
| External Link | RMBase: m6A_site_82105 | ||
| mod ID: M6ASITE087947 | Click to Show/Hide the Full List | ||
| mod site | chr1:226366044-226366045:- | [4] | |
| Sequence | TCTGTTTATCCAGGTGGTTGACAGAGATTCTGAAGAAGCCG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000498787.1; ENST00000366794.10; ENST00000490921.5 | ||
| External Link | RMBase: m6A_site_82106 | ||
| mod ID: M6ASITE087948 | Click to Show/Hide the Full List | ||
| mod site | chr1:226366186-226366187:- | [7] | |
| Sequence | GCCCAGCCCACAGGCCTGGGACACAGGCTCTCGGTCATGCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000498787.1; ENST00000490921.5; ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82107 | ||
| mod ID: M6ASITE087949 | Click to Show/Hide the Full List | ||
| mod site | chr1:226367486-226367487:- | [4] | |
| Sequence | CTATGAGAAGCTCAAAACTGACATTAAGGTAACAGGGTCAG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000498787.1; ENST00000490921.5; ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82108 | ||
| mod ID: M6ASITE087950 | Click to Show/Hide the Full List | ||
| mod site | chr1:226367490-226367491:- | [7] | |
| Sequence | TCAACTATGAGAAGCTCAAAACTGACATTAAGGTAACAGGG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000366794.10; ENST00000490921.5; ENST00000498787.1 | ||
| External Link | RMBase: m6A_site_82109 | ||
| mod ID: M6ASITE087951 | Click to Show/Hide the Full List | ||
| mod site | chr1:226367576-226367577:- | [7] | |
| Sequence | AATGCTTGACAACCTGCTGGACATCGAGGTGGCCTACAGTC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; CD34 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000490921.5; ENST00000366794.10; ENST00000498787.1 | ||
| External Link | RMBase: m6A_site_82110 | ||
| mod ID: M6ASITE087952 | Click to Show/Hide the Full List | ||
| mod site | chr1:226368208-226368209:- | [7] | |
| Sequence | TCCGCTCCTGAACAATGCAGACAGTGTGCAGGTAGCGCAAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000490921.5; ENST00000366794.10; ENST00000498787.1 | ||
| External Link | RMBase: m6A_site_82111 | ||
| mod ID: M6ASITE087953 | Click to Show/Hide the Full List | ||
| mod site | chr1:226368217-226368218:- | [7] | |
| Sequence | GAAGAAGCCTCCGCTCCTGAACAATGCAGACAGTGTGCAGG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; HepG2 | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000498787.1; ENST00000366794.10; ENST00000490921.5 | ||
| External Link | RMBase: m6A_site_82112 | ||
| mod ID: M6ASITE087954 | Click to Show/Hide the Full List | ||
| mod site | chr1:226368247-226368248:- | [4] | |
| Sequence | TTACACCCTGATCCCCCACGACTTTGGGATGAAGAAGCCTC | ||
| Motif Score | 3.287565476 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000498787.1; ENST00000366794.10; ENST00000490921.5 | ||
| External Link | RMBase: m6A_site_82113 | ||
| mod ID: M6ASITE087955 | Click to Show/Hide the Full List | ||
| mod site | chr1:226368265-226368266:- | [5] | |
| Sequence | GGATCTCTCAAATCGCTTTTACACCCTGATCCCCCACGACT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000490921.5; ENST00000498787.1; ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82114 | ||
| mod ID: M6ASITE087956 | Click to Show/Hide the Full List | ||
| mod site | chr1:226370669-226370670:- | [7] | |
| Sequence | ATGGTGGGGTGGGGCTACAGACTCTGGATTTGGGTCACTGG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000490921.5; ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82115 | ||
| mod ID: M6ASITE087957 | Click to Show/Hide the Full List | ||
| mod site | chr1:226374283-226374284:- | [7] | |
| Sequence | GCTCCCCAAGCCAGTTCAGGACCTCATCAAGATGATCTTTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82116 | ||
| mod ID: M6ASITE087958 | Click to Show/Hide the Full List | ||
| mod site | chr1:226377117-226377118:- | [4] | |
| Sequence | GTTCTACCCCCTGGAGATTGACTATGGCCAGGTAACCACAT | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82117 | ||
| mod ID: M6ASITE087959 | Click to Show/Hide the Full List | ||
| mod site | chr1:226377184-226377185:- | [7] | |
| Sequence | TGAAATTATATGAAGAAAAAACCGGGAACGCTTGGCACTCC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82118 | ||
| mod ID: M6ASITE087960 | Click to Show/Hide the Full List | ||
| mod site | chr1:226377242-226377243:- | [7] | |
| Sequence | ATCGGTAGCAACAAACTGGAACAGATGCCGTCCAAGGAGGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82119 | ||
| mod ID: M6ASITE087961 | Click to Show/Hide the Full List | ||
| mod site | chr1:226377248-226377249:- | [7] | |
| Sequence | ACGGTGATCGGTAGCAACAAACTGGAACAGATGCCGTCCAA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82120 | ||
| mod ID: M6ASITE087962 | Click to Show/Hide the Full List | ||
| mod site | chr1:226377252-226377253:- | [5] | |
| Sequence | GGGTACGGTGATCGGTAGCAACAAACTGGAACAGATGCCGT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82121 | ||
| mod ID: M6ASITE087963 | Click to Show/Hide the Full List | ||
| mod site | chr1:226379144-226379145:- | [7] | |
| Sequence | TCTGGAGGACGACAAGGAAAACAGGTGAGTTCTGCAGGTGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82122 | ||
| mod ID: M6ASITE087964 | Click to Show/Hide the Full List | ||
| mod site | chr1:226379153-226379154:- | [4] | |
| Sequence | GCTGCAGCTTCTGGAGGACGACAAGGAAAACAGGTGAGTTC | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82123 | ||
| mod ID: M6ASITE087965 | Click to Show/Hide the Full List | ||
| mod site | chr1:226379190-226379191:- | [7] | |
| Sequence | TGGTGGACATCGTTAAAGGAACCAACTCCTACTACAAGCTG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82124 | ||
| mod ID: M6ASITE087966 | Click to Show/Hide the Full List | ||
| mod site | chr1:226379204-226379205:- | [7] | |
| Sequence | TGCCACCCTTGGCCTGGTGGACATCGTTAAAGGAACCAACT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; HepG2; MT4 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82125 | ||
| mod ID: M6ASITE087967 | Click to Show/Hide the Full List | ||
| mod site | chr1:226379266-226379267:- | [7] | |
| Sequence | TCCTCTGGGACAGGACTGGAACACTCTGCGCATGTCCTGGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; HepG2; MT4 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82126 | ||
| mod ID: M6ASITE087968 | Click to Show/Hide the Full List | ||
| mod site | chr1:226379634-226379635:- | [5] | |
| Sequence | TGTTCTCCTCCAAGGTATCAACAAATCTGAAAAGAGAATGA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82127 | ||
| mod ID: M6ASITE087969 | Click to Show/Hide the Full List | ||
| mod site | chr1:226380037-226380038:- | [5] | |
| Sequence | TCAGGAGTTGTTCTTAGCGCACATCTTGTCCCCTTGGGGGG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82128 | ||
| mod ID: M6ASITE087970 | Click to Show/Hide the Full List | ||
| mod site | chr1:226380094-226380095:- | [7] | |
| Sequence | CATCCGAGTTGTGTCTGAGGACTTCCTCCAGGACGTCTCCG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; MT4; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82129 | ||
| mod ID: M6ASITE087971 | Click to Show/Hide the Full List | ||
| mod site | chr1:226380115-226380116:- | [5] | |
| Sequence | GGAGGAAGTAAAGGAAGCCAACATCCGAGTTGTGTCTGAGG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82130 | ||
| mod ID: M6ASITE087972 | Click to Show/Hide the Full List | ||
| mod site | chr1:226381096-226381097:- | [5] | |
| Sequence | GAAGTTGACGGGGACGGCCAACAAGGCTTCCCTGTGCATCA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82131 | ||
| mod ID: M6ASITE087973 | Click to Show/Hide the Full List | ||
| mod site | chr1:226381125-226381126:- | [7] | |
| Sequence | GTGAAGGCCATGATTGAGAAACTCGGGGGGAAGTTGACGGG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; MT4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82132 | ||
| mod ID: M6ASITE087974 | Click to Show/Hide the Full List | ||
| mod site | chr1:226381156-226381157:- | [7] | |
| Sequence | TCTCGGGAAGCTGTCCCGGAACAAGGATGAAGTGAAGGCCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; HepG2; MT4 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82133 | ||
| mod ID: M6ASITE087975 | Click to Show/Hide the Full List | ||
| mod site | chr1:226381192-226381193:- | [5] | |
| Sequence | CCCAGATAAGCCATTATCCAACATGAAGATCCTGACTCTCG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82134 | ||
| mod ID: M6ASITE087981 | Click to Show/Hide the Full List | ||
| mod site | chr1:226383052-226383053:- | [7] | |
| Sequence | CTCGGCTCCTGCTGCTGTGAACTCCTCTGCTTCAGCAGGTA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; MT4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82135 | ||
| mod ID: M6ASITE087992 | Click to Show/Hide the Full List | ||
| mod site | chr1:226383113-226383114:- | [7] | |
| Sequence | ACCGTATATTCCCCCCAGAAACCAGCGCCTCCGTGGCGGCC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82136 | ||
| mod ID: M6ASITE088003 | Click to Show/Hide the Full List | ||
| mod site | chr1:226383133-226383134:- | [7] | |
| Sequence | ATTGAAGGTTAAAAAACAGGACCGTATATTCCCCCCAGAAA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82137 | ||
| mod ID: M6ASITE088007 | Click to Show/Hide the Full List | ||
| mod site | chr1:226383138-226383139:- | [7] | |
| Sequence | AAGAAATTGAAGGTTAAAAAACAGGACCGTATATTCCCCCC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82138 | ||
| mod ID: M6ASITE088008 | Click to Show/Hide the Full List | ||
| mod site | chr1:226385535-226385536:- | [7] | |
| Sequence | AGTGTATGGTCAAGACACAGACACCCAACCGGAAGGAGTGG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; hESC-HEK293T; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82139 | ||
| mod ID: M6ASITE088009 | Click to Show/Hide the Full List | ||
| mod site | chr1:226385541-226385542:- | [7] | |
| Sequence | GGACCAAGTGTATGGTCAAGACACAGACACCCAACCGGAAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82140 | ||
| mod ID: M6ASITE088010 | Click to Show/Hide the Full List | ||
| mod site | chr1:226385559-226385560:- | [7] | |
| Sequence | CTGGGGACGTCACTGCCTGGACCAAGTGTATGGTCAAGACA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; CD34 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82141 | ||
| mod ID: M6ASITE088011 | Click to Show/Hide the Full List | ||
| mod site | chr1:226385672-226385673:- | [7] | |
| Sequence | CCACTGGCTGCAGATCTTGGACCGAGTAGCTGATGGCATGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; Huh7; peripheral-blood; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82142 | ||
| mod ID: M6ASITE088012 | Click to Show/Hide the Full List | ||
| mod site | chr1:226386356-226386357:- | [5] | |
| Sequence | GAAGGAGCTACTCATCTTCAACAAGCAGCAAGTGCCTTCTG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82143 | ||
| mod ID: M6ASITE088013 | Click to Show/Hide the Full List | ||
| mod site | chr1:226386419-226386420:- | [7] | |
| Sequence | TCAGAACGACCTGATCTGGAACATCAAGGACGAGCTAAAGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; hNPCs; LCLs; A549; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82144 | ||
| mod ID: M6ASITE088014 | Click to Show/Hide the Full List | ||
| mod site | chr1:226388686-226388687:- | [7] | |
| Sequence | GAAATCTAAAAAAGAAAAAGACAAGGATAGTAAGCTTGAAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; U2OS; hNPCs; fibroblasts; A549; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82145 | ||
| mod ID: M6ASITE088015 | Click to Show/Hide the Full List | ||
| mod site | chr1:226390529-226390530:- | [7] | |
| Sequence | TCCAGGCTGCTTTGTCAAGAACAGGGAGGAGCTGGGTTTCC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; hNPCs; HEK293T; fibroblasts; A549; LCLs; H1299; MM6; Huh7; peripheral-blood; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000366794.10; ENST00000469663.1 | ||
| External Link | RMBase: m6A_site_82146 | ||
| mod ID: M6ASITE088016 | Click to Show/Hide the Full List | ||
| mod site | chr1:226390592-226390593:- | [7] | |
| Sequence | CCTGTCCAAGAAGATGGTGGACCCGGAGAAGCCACAGCTAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; hNPCs; HEK293T; fibroblasts; LCLs; MT4; A549; H1299; MM6; Huh7; peripheral-blood; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000469663.1; ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82147 | ||
| mod ID: M6ASITE088017 | Click to Show/Hide the Full List | ||
| mod site | chr1:226392230-226392231:- | [4] | |
| Sequence | ATGCCAAGTCCAACAGAAGTACGTGCAAGGGGTGTATGGAG | ||
| Motif Score | 2.830077381 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000366794.10; ENST00000469663.1 | ||
| External Link | RMBase: m6A_site_82148 | ||
| mod ID: M6ASITE088018 | Click to Show/Hide the Full List | ||
| mod site | chr1:226392275-226392276:- | [7] | |
| Sequence | TTGGTAGCAAGGCAGAGAAGACTCTGGGTGACTTTGCAGCA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; hNPCs; HEK293T; fibroblasts; LCLs; MT4; H1299; MM6; Huh7; peripheral-blood; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000366794.10; ENST00000366792.3; ENST00000469663.1 | ||
| External Link | RMBase: m6A_site_82149 | ||
| mod ID: M6ASITE088019 | Click to Show/Hide the Full List | ||
| mod site | chr1:226402216-226402217:- | [6] | |
| Sequence | CAGCGGAAGCTGGAGGAGTGACAGGTGTGTACATACTGGGG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | brain | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000366790.3; ENST00000629232.1; ENST00000366792.3; ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82150 | ||
| mod ID: M6ASITE088020 | Click to Show/Hide the Full List | ||
| mod site | chr1:226402237-226402238:- | [7] | |
| Sequence | ACCAGCAGAAAGTCAAGAAGACAGCGGAAGCTGGAGGAGTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000629232.1; ENST00000366792.3; ENST00000366794.10; ENST00000366790.3 | ||
| External Link | RMBase: m6A_site_82151 | ||
| mod ID: M6ASITE088021 | Click to Show/Hide the Full List | ||
| mod site | chr1:226402352-226402353:- | [5] | |
| Sequence | ATGTTTGATGGAAAAGTCCCACACTGGTACCACTTCTCCTG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000366792.3; ENST00000366790.3; ENST00000629232.1; ENST00000366794.10 | ||
| External Link | RMBase: m6A_site_82152 | ||
| mod ID: M6ASITE088022 | Click to Show/Hide the Full List | ||
| mod site | chr1:226407837-226407838:- | [7] | |
| Sequence | CAGCGAGAGCATCCCCAAGGACTCGCTCCGGATGGCCATCA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; A549; U2OS; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; iSLK; MSC; TIME; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000366790.3; ENST00000629232.1; ENST00000366794.10; ENST00000366792.3 | ||
| External Link | RMBase: m6A_site_82153 | ||
2'-O-Methylation (2'-O-Me)
| In total 3 m6A sequence/site(s) in this target gene | |||
| mod ID: 2OMSITE000339 | Click to Show/Hide the Full List | ||
| mod site | chr1:226361350-226361351:- | [8] | |
| Sequence | CTGATGGGTAGTACCTGTACTAAACCACCTCAGAAAGGATT | ||
| Cell/Tissue List | HEK293 | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000463968.5; ENST00000490921.5; ENST00000366794.10 | ||
| External Link | RMBase: Nm_site_524 | ||
| mod ID: 2OMSITE000340 | Click to Show/Hide the Full List | ||
| mod site | chr1:226385666-226385667:- | [8] | |
| Sequence | GCTGCAGATCTTGGACCGAGTAGCTGATGGCATGGTGTTCG | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: Nm_site_525 | ||
| mod ID: 2OMSITE000344 | Click to Show/Hide the Full List | ||
| mod site | chr1:226386423-226386424:- | [8] | |
| Sequence | AGGCTCAGAACGACCTGATCTGGAACATCAAGGACGAGCTA | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000366794.10 | ||
| External Link | RMBase: Nm_site_526 | ||
References