m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00299)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
JAK2
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Raw 264.7 cell line | Mus musculus |
|
Treatment: METTL3 knockout Raw 264.7 cells
Control: Wild type Raw 264.7 cells
|
GSE162248 | |
| Regulation |
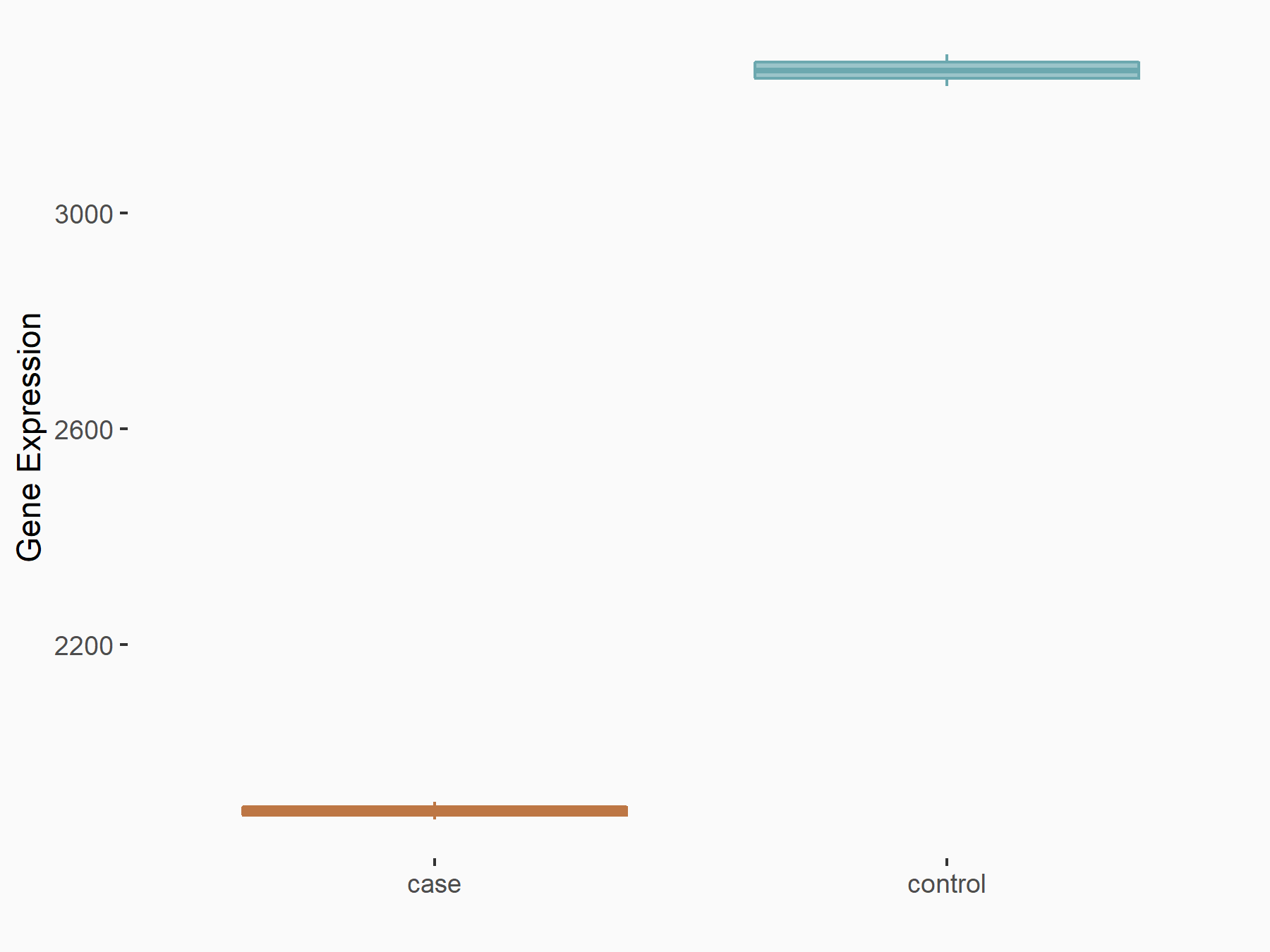  |
logFC: -7.87E-01 p-value: 4.26E-20 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between JAK2 and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 7.70E+00 | GSE60213 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 knockdown prevented Atherosclerosis progression by inhibiting Tyrosine-protein kinase JAK2 (JAK2)/STAT3 pathway via IGF2BP1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Atherosclerosis | ICD-11: BD40.Z | ||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
| Cell Process | Cell proliferation and migration | |||
| In-vitro Model | HUVEC-C | Normal | Homo sapiens | CVCL_2959 |
| In-vivo Model | The adeno-associated viruses (AAV) that could silence METTL3 (sh-METTL3) and the negative control adeno-associated viruses (sh-NC) were obtained from WZ Biosciences Inc. (Jinan, China). APOE-/- mice were randomly divided into AS + sh-NC and AS + sh-METTL3 groups. Each group contains five mice. Mice were fed with the standard diet for 1 week to acclimatize. After 1 week of acclimation, mice were challenged with a high-fat and high-cholesterol feed H10540 (Beijing HFK BIOSCIENCE Co., Ltd., Beijing, China). The formula of the H10540 feed was shown in Supplementary File S1. After 8 weeks of HFD feeding, sh-NC or sh-METTL3 adeno-associated virus serotype 9 (AAV9, 1012 viral genome copies per mouse) were respectively delivered into mice in AS + sh-NC or AS + sh-METTL3 group through tail vein injection. At 14 weeks after HDF feeding, mice fasted overnight. | |||
Fat mass and obesity-associated protein (FTO) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by FTO | ||
| Cell Line | 253J cell line | Homo sapiens |
|
Treatment: siFTO 253J cells
Control: 253J cells
|
GSE150239 | |
| Regulation |
  |
logFC: 9.68E-01 p-value: 1.29E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | FTO inhibits adipocytes apoptosis via reducing m6A epigenetic modification-mediated UPRmt.UPRmt-induced increase in PKR and eIF2-alpha phosphorylation, and ATF5-mediated upregulation of BAX expression promoted apoptosis. Furthermore, adipocytes apoptosis is inhibited by FTO-activated Tyrosine-protein kinase JAK2 (JAK2)/STAT3 signaling pathway . | |||
| Pathway Response | Adipocytokine signaling pathway | hsa04920 | ||
| JAK-STAT signaling pathway | hsa04630 | |||
| Cell Process | Mitochondria-dependent apoptosis | |||
| Cell apoptosis | ||||
| In-vitro Model | 3T3-L1 | Normal | Mus musculus | CVCL_0123 |
| In-vivo Model | Mice were provided adlibitum with water and a standard laboratory chow diet. The animal room was held a standard temperature, humidity and illumination. After intraperitoneal injection of FTO overexpression vector and NR to mice for 7 days, inguinal white adipose tissue (iWAT) were collected from mice. | |||
Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by IGF2BP1 | ||
| Cell Line | HepG2 cell line | Homo sapiens |
|
Treatment: siIGF2BP1 HepG2 cells
Control: siControl HepG2 cells
|
GSE161086 | |
| Regulation |
  |
logFC: -1.30E+00 p-value: 1.73E-03 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 knockdown prevented Atherosclerosis progression by inhibiting Tyrosine-protein kinase JAK2 (JAK2)/STAT3 pathway via IGF2BP1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Atherosclerosis | ICD-11: BD40.Z | ||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
| Cell Process | Cell proliferation and migration | |||
| In-vitro Model | HUVEC-C | Normal | Homo sapiens | CVCL_2959 |
| In-vivo Model | The adeno-associated viruses (AAV) that could silence METTL3 (sh-METTL3) and the negative control adeno-associated viruses (sh-NC) were obtained from WZ Biosciences Inc. (Jinan, China). APOE-/- mice were randomly divided into AS + sh-NC and AS + sh-METTL3 groups. Each group contains five mice. Mice were fed with the standard diet for 1 week to acclimatize. After 1 week of acclimation, mice were challenged with a high-fat and high-cholesterol feed H10540 (Beijing HFK BIOSCIENCE Co., Ltd., Beijing, China). The formula of the H10540 feed was shown in Supplementary File S1. After 8 weeks of HFD feeding, sh-NC or sh-METTL3 adeno-associated virus serotype 9 (AAV9, 1012 viral genome copies per mouse) were respectively delivered into mice in AS + sh-NC or AS + sh-METTL3 group through tail vein injection. At 14 weeks after HDF feeding, mice fasted overnight. | |||
RNA demethylase ALKBH5 (ALKBH5) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by ALKBH5 | ||
| Cell Line | NOMO-1 cell line | Homo sapiens |
|
Treatment: shALKBH5 NOMO-1 cells
Control: shNS NOMO-1 cells
|
GSE144968 | |
| Regulation |
  |
logFC: 1.17E+00 p-value: 2.74E-05 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | The ALKBH5-HOXA10 loop jointly activates the JAK2/STAT3 signaling pathway by mediating Tyrosine-protein kinase JAK2 (JAK2) m6A demethylation, promoting epithelial ovarian cancer resistance to cisplatin. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Malignant mixed epithelial mesenchymal tumour of ovary | ICD-11: 2B5D.0 | ||
| Responsed Drug | Cisplatin | Approved | ||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
| In-vitro Model | HO8910-DDP (HO8910 underwent continuous stepwise exposure to increasing concentrations of cisplatin to create the cisplatin-resistant cell lines HO8910-DDP) | |||
| HO-8910 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6868 | |
| A2780-DDP (A2780 underwent continuous stepwise exposure to increasing concentrations of cisplatin to create the cisplatin-resistant cell line) | ||||
| A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 | |
| In-vivo Model | About 5× 106 cells were injected subcutaneously into the axilla of the female athymic BALB/C nude mice (4 week-old, 18-20 g). When the average tumor size reached approximately 100mm3 (after 1 week), mice were then randomized into two groups and treated with cisplatin (5 mg/kg) or normal saline (NS) weekly. | |||
Malignant mixed epithelial mesenchymal tumour [ICD-11: 2B5D]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | The ALKBH5-HOXA10 loop jointly activates the JAK2/STAT3 signaling pathway by mediating Tyrosine-protein kinase JAK2 (JAK2) m6A demethylation, promoting epithelial ovarian cancer resistance to cisplatin. | |||
| Responsed Disease | Malignant mixed epithelial mesenchymal tumour of ovary [ICD-11: 2B5D.0] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Cisplatin | Approved | ||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
| In-vitro Model | HO8910-DDP (HO8910 underwent continuous stepwise exposure to increasing concentrations of cisplatin to create the cisplatin-resistant cell lines HO8910-DDP) | |||
| HO-8910 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6868 | |
| A2780-DDP (A2780 underwent continuous stepwise exposure to increasing concentrations of cisplatin to create the cisplatin-resistant cell line) | ||||
| A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 | |
| In-vivo Model | About 5× 106 cells were injected subcutaneously into the axilla of the female athymic BALB/C nude mice (4 week-old, 18-20 g). When the average tumor size reached approximately 100mm3 (after 1 week), mice were then randomized into two groups and treated with cisplatin (5 mg/kg) or normal saline (NS) weekly. | |||
Atherosclerosis [ICD-11: BD40]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 knockdown prevented Atherosclerosis progression by inhibiting Tyrosine-protein kinase JAK2 (JAK2)/STAT3 pathway via IGF2BP1. | |||
| Responsed Disease | Atherosclerosis [ICD-11: BD40.Z] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
| Cell Process | Cell proliferation and migration | |||
| In-vitro Model | HUVEC-C | Normal | Homo sapiens | CVCL_2959 |
| In-vivo Model | The adeno-associated viruses (AAV) that could silence METTL3 (sh-METTL3) and the negative control adeno-associated viruses (sh-NC) were obtained from WZ Biosciences Inc. (Jinan, China). APOE-/- mice were randomly divided into AS + sh-NC and AS + sh-METTL3 groups. Each group contains five mice. Mice were fed with the standard diet for 1 week to acclimatize. After 1 week of acclimation, mice were challenged with a high-fat and high-cholesterol feed H10540 (Beijing HFK BIOSCIENCE Co., Ltd., Beijing, China). The formula of the H10540 feed was shown in Supplementary File S1. After 8 weeks of HFD feeding, sh-NC or sh-METTL3 adeno-associated virus serotype 9 (AAV9, 1012 viral genome copies per mouse) were respectively delivered into mice in AS + sh-NC or AS + sh-METTL3 group through tail vein injection. At 14 weeks after HDF feeding, mice fasted overnight. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 knockdown prevented Atherosclerosis progression by inhibiting Tyrosine-protein kinase JAK2 (JAK2)/STAT3 pathway via IGF2BP1. | |||
| Responsed Disease | Atherosclerosis [ICD-11: BD40.Z] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
| Cell Process | Cell proliferation and migration | |||
| In-vitro Model | HUVEC-C | Normal | Homo sapiens | CVCL_2959 |
| In-vivo Model | The adeno-associated viruses (AAV) that could silence METTL3 (sh-METTL3) and the negative control adeno-associated viruses (sh-NC) were obtained from WZ Biosciences Inc. (Jinan, China). APOE-/- mice were randomly divided into AS + sh-NC and AS + sh-METTL3 groups. Each group contains five mice. Mice were fed with the standard diet for 1 week to acclimatize. After 1 week of acclimation, mice were challenged with a high-fat and high-cholesterol feed H10540 (Beijing HFK BIOSCIENCE Co., Ltd., Beijing, China). The formula of the H10540 feed was shown in Supplementary File S1. After 8 weeks of HFD feeding, sh-NC or sh-METTL3 adeno-associated virus serotype 9 (AAV9, 1012 viral genome copies per mouse) were respectively delivered into mice in AS + sh-NC or AS + sh-METTL3 group through tail vein injection. At 14 weeks after HDF feeding, mice fasted overnight. | |||
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [3] | |||
| Response Summary | The ALKBH5-HOXA10 loop jointly activates the JAK2/STAT3 signaling pathway by mediating Tyrosine-protein kinase JAK2 (JAK2) m6A demethylation, promoting epithelial ovarian cancer resistance to cisplatin. | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Malignant mixed epithelial mesenchymal tumour of ovary | ICD-11: 2B5D.0 | ||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
| In-vitro Model | HO8910-DDP (HO8910 underwent continuous stepwise exposure to increasing concentrations of cisplatin to create the cisplatin-resistant cell lines HO8910-DDP) | |||
| HO-8910 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6868 | |
| A2780-DDP (A2780 underwent continuous stepwise exposure to increasing concentrations of cisplatin to create the cisplatin-resistant cell line) | ||||
| A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 | |
| In-vivo Model | About 5× 106 cells were injected subcutaneously into the axilla of the female athymic BALB/C nude mice (4 week-old, 18-20 g). When the average tumor size reached approximately 100mm3 (after 1 week), mice were then randomized into two groups and treated with cisplatin (5 mg/kg) or normal saline (NS) weekly. | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00299)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE003070 | Click to Show/Hide the Full List | ||
| mod site | chr9:5120527-5120528:+ | [4] | |
| Sequence | CTAACAATATGGTTGACGGTAATAACTGGGGGAAAAAGTGC | ||
| Transcript ID List | ENST00000381652.3 | ||
| External Link | RMBase: RNA-editing_site_134209 | ||
5-methylcytidine (m5C)
| In total 3 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE004381 | Click to Show/Hide the Full List | ||
| mod site | chr9:5109394-5109395:+ | [5] | |
| Sequence | AAGAACTGCTAACTCATGCCCCCATGCCTAACAATATGGCT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000381652.3 | ||
| External Link | RMBase: m5C_site_42334 | ||
| mod ID: M5CSITE004382 | Click to Show/Hide the Full List | ||
| mod site | chr9:5109395-5109396:+ | [5] | |
| Sequence | AGAACTGCTAACTCATGCCCCCATGCCTAACAATATGGCTT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000381652.3 | ||
| External Link | RMBase: m5C_site_42335 | ||
| mod ID: M5CSITE004383 | Click to Show/Hide the Full List | ||
| mod site | chr9:5109396-5109397:+ | [5] | |
| Sequence | GAACTGCTAACTCATGCCCCCATGCCTAACAATATGGCTTT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000381652.3 | ||
| External Link | RMBase: m5C_site_42336 | ||
N6-methyladenosine (m6A)
| In total 49 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE087547 | Click to Show/Hide the Full List | ||
| mod site | chr9:4985095-4985096:+ | [6] | |
| Sequence | GTCTCCTGCCATTCGGGGAGACTGCAGGCCAACCGGGAGGC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; GSC-11; HEK293T; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000636127.1; ENST00000476574.5 | ||
| External Link | RMBase: m6A_site_815327 | ||
| mod ID: M6ASITE087548 | Click to Show/Hide the Full List | ||
| mod site | chr9:4985557-4985558:+ | [7] | |
| Sequence | CCCGTCCCGGGGCTTCGCAGACCTTGACCCGCCGGGTAGGA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000476574.5; ENST00000381652.3; ENST00000636127.1 | ||
| External Link | RMBase: m6A_site_815328 | ||
| mod ID: M6ASITE087549 | Click to Show/Hide the Full List | ||
| mod site | chr9:4985961-4985962:+ | [6] | |
| Sequence | TTCAGAAGCAGGCAACAGGAACAAGATGTGAACTGTTTCTC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000476574.5; ENST00000636127.1; ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815329 | ||
| mod ID: M6ASITE087550 | Click to Show/Hide the Full List | ||
| mod site | chr9:4985972-4985973:+ | [6] | |
| Sequence | GCAACAGGAACAAGATGTGAACTGTTTCTCTTCTGCAGAAA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000636127.1; ENST00000476574.5 | ||
| External Link | RMBase: m6A_site_815330 | ||
| mod ID: M6ASITE087551 | Click to Show/Hide the Full List | ||
| mod site | chr9:5021980-5021981:+ | [6] | |
| Sequence | AGGCAAATGTTCTGAAAAAGACTCTGCATGGGAATGGCCTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000636127.1; ENST00000381652.3; ENST00000476574.5 | ||
| External Link | RMBase: m6A_site_815331 | ||
| mod ID: M6ASITE087552 | Click to Show/Hide the Full List | ||
| mod site | chr9:5022026-5022027:+ | [6] | |
| Sequence | CGATGACAGAAATGGAGGGAACATCCACCTCTTCTATATAT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000476574.5; ENST00000381652.3; ENST00000636127.1 | ||
| External Link | RMBase: m6A_site_815332 | ||
| mod ID: M6ASITE087553 | Click to Show/Hide the Full List | ||
| mod site | chr9:5050736-5050737:+ | [6] | |
| Sequence | AAGTACCTGTGACTCATGAAACACAGGAAGAATGTCTTGGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000636127.1 | ||
| External Link | RMBase: m6A_site_815333 | ||
| mod ID: M6ASITE087554 | Click to Show/Hide the Full List | ||
| mod site | chr9:5050802-5050803:+ | [6] | |
| Sequence | TAGCCAAAGAAAACGATCAAACCCCACTGGCCATCTATAAC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000636127.1; ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815334 | ||
| mod ID: M6ASITE087555 | Click to Show/Hide the Full List | ||
| mod site | chr9:5054569-5054570:+ | [6] | |
| Sequence | TTTTTATCCCTAGCTACAAGACATTCTTACCAAAATGTATT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000636127.1; ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815335 | ||
| mod ID: M6ASITE087556 | Click to Show/Hide the Full List | ||
| mod site | chr9:5054606-5054607:+ | [6] | |
| Sequence | TATTCGAGCAAAGATCCAAGACTATCATATTTTGACAAGGA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000636127.1 | ||
| External Link | RMBase: m6A_site_815336 | ||
| mod ID: M6ASITE087557 | Click to Show/Hide the Full List | ||
| mod site | chr9:5054693-5054694:+ | [6] | |
| Sequence | ATGCAAAGCCACTGCCAGAAACTTGAAACTTAAGTATCTTA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000636127.1 | ||
| External Link | RMBase: m6A_site_815337 | ||
| mod ID: M6ASITE087558 | Click to Show/Hide the Full List | ||
| mod site | chr9:5054700-5054701:+ | [6] | |
| Sequence | GCCACTGCCAGAAACTTGAAACTTAAGTATCTTATAAATCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000636127.1 | ||
| External Link | RMBase: m6A_site_815338 | ||
| mod ID: M6ASITE087559 | Click to Show/Hide the Full List | ||
| mod site | chr9:5054725-5054726:+ | [6] | |
| Sequence | AGTATCTTATAAATCTGGAAACTCTGCAGTCTGCCTTCTAC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000636127.1 | ||
| External Link | RMBase: m6A_site_815339 | ||
| mod ID: M6ASITE087560 | Click to Show/Hide the Full List | ||
| mod site | chr9:5054769-5054770:+ | [6] | |
| Sequence | GAGAAATTTGAAGTAAAAGAACCTGGAAGTGGTCCTTCAGG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000636127.1; ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815340 | ||
| mod ID: M6ASITE087561 | Click to Show/Hide the Full List | ||
| mod site | chr9:5055726-5055727:+ | [6] | |
| Sequence | TGTCAGTATTAAGCAAGCAAACCAAGAGGGTTCAAATGAAA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000636127.1; ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815341 | ||
| mod ID: M6ASITE087562 | Click to Show/Hide the Full List | ||
| mod site | chr9:5064890-5064891:+ | [6] | |
| Sequence | TTCTCTGCTTAGGAAATTGAACTTAGCTCATTAAGGGAAGC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000636127.1; ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815342 | ||
| mod ID: M6ASITE087563 | Click to Show/Hide the Full List | ||
| mod site | chr9:5066698-5066699:+ | [6] | |
| Sequence | ATGGATTTTGCCATTAGTAAACTGAAGAAAGCAGGTAATCA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000636127.1; ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815343 | ||
| mod ID: M6ASITE087564 | Click to Show/Hide the Full List | ||
| mod site | chr9:5066720-5066721:+ | [6] | |
| Sequence | TGAAGAAAGCAGGTAATCAGACTGGACTGTATGTACTTCGA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000636127.1 | ||
| External Link | RMBase: m6A_site_815344 | ||
| mod ID: M6ASITE087565 | Click to Show/Hide the Full List | ||
| mod site | chr9:5066725-5066726:+ | [6] | |
| Sequence | AAAGCAGGTAATCAGACTGGACTGTATGTACTTCGATGCAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000636127.1 | ||
| External Link | RMBase: m6A_site_815345 | ||
| mod ID: M6ASITE087566 | Click to Show/Hide the Full List | ||
| mod site | chr9:5066754-5066755:+ | [6] | |
| Sequence | ACTTCGATGCAGTCCTAAGGACTTTAATAAATATTTTTTGA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000636127.1 | ||
| External Link | RMBase: m6A_site_815346 | ||
| mod ID: M6ASITE087567 | Click to Show/Hide the Full List | ||
| mod site | chr9:5069093-5069094:+ | [8] | |
| Sequence | AAGAGTACAACCTCAGTGGGACAAAGAAGAACTTCAGCAGT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000636127.1 | ||
| External Link | RMBase: m6A_site_815347 | ||
| mod ID: M6ASITE087568 | Click to Show/Hide the Full List | ||
| mod site | chr9:5069160-5069161:+ | [8] | |
| Sequence | GATGGAAACTGTTCGCTCAGACAATATAATTTTCCAGTTTA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000636127.1 | ||
| External Link | RMBase: m6A_site_815348 | ||
| mod ID: M6ASITE087569 | Click to Show/Hide the Full List | ||
| mod site | chr9:5069933-5069934:+ | [6] | |
| Sequence | TATTTTTTCAGATAAATCAAACCTTCTAGTCTTCAGAACGA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000636127.1 | ||
| External Link | RMBase: m6A_site_815349 | ||
| mod ID: M6ASITE087570 | Click to Show/Hide the Full List | ||
| mod site | chr9:5089782-5089783:+ | ||
| Sequence | TACTGAAGAGCACCTAAGAGACTTTGAAAGGGAAATTGAAA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815350 | ||
| mod ID: M6ASITE087571 | Click to Show/Hide the Full List | ||
| mod site | chr9:5090539-5090540:+ | [9] | |
| Sequence | GAACGGATAGATCACATAAAACTTCTGCAGTACACATCTCA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815351 | ||
| mod ID: M6ASITE087572 | Click to Show/Hide the Full List | ||
| mod site | chr9:5094889-5094890:+ | [9] | |
| Sequence | CACCGTTTTTATGAATCCGAACAGCATACCTCCAATTCCAC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000456661.1; ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815352 | ||
| mod ID: M6ASITE087573 | Click to Show/Hide the Full List | ||
| mod site | chr9:5111064-5111065:+ | [10] | |
| Sequence | CAGAGGTTCAGGTCTTGAGAACTGGCCCAGCTCAGGCTCCT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | GM12878 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000423021.2 | ||
| External Link | RMBase: m6A_site_815353 | ||
| mod ID: M6ASITE087574 | Click to Show/Hide the Full List | ||
| mod site | chr9:5111103-5111104:+ | [10] | |
| Sequence | CTGGGGCAGCGGCGACCAGAACAGCTCCTCCTTTGACCCCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | GM12878 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000423021.2 | ||
| External Link | RMBase: m6A_site_815354 | ||
| mod ID: M6ASITE087575 | Click to Show/Hide the Full List | ||
| mod site | chr9:5112413-5112414:+ | [6] | |
| Sequence | GGAGGAGGAGGATGAGGAGAACACGTCGGCGGCTGACCACT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000423021.2; ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815355 | ||
| mod ID: M6ASITE087576 | Click to Show/Hide the Full List | ||
| mod site | chr9:5112459-5112460:+ | [6] | |
| Sequence | GGAGAAGAAGGAGCTGAAGGACCCCCGGGACCGGACCAGCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000423021.2 | ||
| External Link | RMBase: m6A_site_815356 | ||
| mod ID: M6ASITE087577 | Click to Show/Hide the Full List | ||
| mod site | chr9:5112468-5112469:+ | [6] | |
| Sequence | GGAGCTGAAGGACCCCCGGGACCGGACCAGCCCAGACAAGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000423021.2; ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815357 | ||
| mod ID: M6ASITE087578 | Click to Show/Hide the Full List | ||
| mod site | chr9:5112473-5112474:+ | [6] | |
| Sequence | TGAAGGACCCCCGGGACCGGACCAGCCCAGACAAGGACGAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000423021.2 | ||
| External Link | RMBase: m6A_site_815358 | ||
| mod ID: M6ASITE087579 | Click to Show/Hide the Full List | ||
| mod site | chr9:5112483-5112484:+ | [6] | |
| Sequence | CCGGGACCGGACCAGCCCAGACAAGGACGAGGAGGAAGATG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000423021.2; ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815359 | ||
| mod ID: M6ASITE087580 | Click to Show/Hide the Full List | ||
| mod site | chr9:5114405-5114406:+ | [11] | |
| Sequence | CCTGCTGGTGGGCACCAGAGACTGGATCAAGGGGGAGACCT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000519308.1; ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815360 | ||
| mod ID: M6ASITE087581 | Click to Show/Hide the Full List | ||
| mod site | chr9:5114422-5114423:+ | [11] | |
| Sequence | GAGACTGGATCAAGGGGGAGACCTACCAGTGCAGGGTGACC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000519308.1 | ||
| External Link | RMBase: m6A_site_815361 | ||
| mod ID: M6ASITE087582 | Click to Show/Hide the Full List | ||
| mod site | chr9:5114531-5114532:+ | [11] | |
| Sequence | AACGCTAGAAGAGCCGAGGAACCAGGACAAGCGCACCCTCA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000519308.1 | ||
| External Link | RMBase: m6A_site_815362 | ||
| mod ID: M6ASITE087583 | Click to Show/Hide the Full List | ||
| mod site | chr9:5114537-5114538:+ | [11] | |
| Sequence | AGAAGAGCCGAGGAACCAGGACAAGCGCACCCTCACCTGCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000519308.1; ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815363 | ||
| mod ID: M6ASITE087584 | Click to Show/Hide the Full List | ||
| mod site | chr9:5114567-5114568:+ | [12] | |
| Sequence | CCTCACCTGCCTGATCCAGAACTTCTGGCCCAAGGACATCT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | iSLK; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000519308.1 | ||
| External Link | RMBase: m6A_site_815364 | ||
| mod ID: M6ASITE087585 | Click to Show/Hide the Full List | ||
| mod site | chr9:5114582-5114583:+ | [12] | |
| Sequence | CCAGAACTTCTGGCCCAAGGACATCTTGGTGCAGTGGCTGT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | iSLK; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000519308.1; ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815365 | ||
| mod ID: M6ASITE087586 | Click to Show/Hide the Full List | ||
| mod site | chr9:5114624-5114625:+ | [12] | |
| Sequence | CAACGAGGTGCAGCTCCCGGACACTTGGCACAGCATGACGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | iSLK | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000519308.1 | ||
| External Link | RMBase: m6A_site_815366 | ||
| mod ID: M6ASITE087587 | Click to Show/Hide the Full List | ||
| mod site | chr9:5114656-5114657:+ | [12] | |
| Sequence | GCATGACGCAGCCCCGCAAAACCAAGGGCTCTGGCGTCTTA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | iSLK | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000519308.1; ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815367 | ||
| mod ID: M6ASITE087588 | Click to Show/Hide the Full List | ||
| mod site | chr9:5123081-5123082:+ | [13] | |
| Sequence | TTTGGAGTGGTTCTGTATGAACTTTTCACATACATTGAGAA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | CD34 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815368 | ||
| mod ID: M6ASITE087589 | Click to Show/Hide the Full List | ||
| mod site | chr9:5126698-5126699:+ | [14] | |
| Sequence | TACAGATCTATATGATCATGACAGAATGCTGGAACAATAAT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000487310.1; ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815369 | ||
| mod ID: M6ASITE087590 | Click to Show/Hide the Full List | ||
| mod site | chr9:5126813-5126814:+ | ||
| Sequence | GAAATGACCTTCATTCTGAGACCAAAGTAGATTTACAGAAC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000487310.1; ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815370 | ||
| mod ID: M6ASITE087591 | Click to Show/Hide the Full List | ||
| mod site | chr9:5126832-5126833:+ | ||
| Sequence | GACCAAAGTAGATTTACAGAACAAAGTTTTATATTTCACAT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000487310.1; ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815371 | ||
| mod ID: M6ASITE087592 | Click to Show/Hide the Full List | ||
| mod site | chr9:5126861-5126862:+ | [15] | |
| Sequence | TATATTTCACATTGCTGTGGACTATTATTACATATATCATT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HEK293T; CD8T; HEK293A-TOA | ||
| Seq Type List | MeRIP-seq; m6A-CLIP/IP; m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000487310.1 | ||
| External Link | RMBase: m6A_site_815372 | ||
| mod ID: M6ASITE087593 | Click to Show/Hide the Full List | ||
| mod site | chr9:5126934-5126935:+ | [16] | |
| Sequence | TGTGAAAATATCTGCTCAAAACTTTCAAAGTTTAGTAAGTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T; Huh7; HEK293A-TOA; TIME; iSLK | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000381652.3; ENST00000487310.1 | ||
| External Link | RMBase: m6A_site_815373 | ||
| mod ID: M6ASITE087594 | Click to Show/Hide the Full List | ||
| mod site | chr9:5126981-5126982:+ | [16] | |
| Sequence | CATGAGGCCACCAGTAAAAGACATTAATGAGAATTCCTTAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HEK293T; Huh7; TIME | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815374 | ||
| mod ID: M6ASITE087595 | Click to Show/Hide the Full List | ||
| mod site | chr9:5127028-5127029:+ | [9] | |
| Sequence | TTTTGTAAGAAGTTTCTTAAACATTGTCAGTTAACATCACT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HEK293T; Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000381652.3 | ||
| External Link | RMBase: m6A_site_815375 | ||
Other RNA modification (t6A)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: RNASITE000004 | Click to Show/Hide the Full List | ||
| mod site | chr9:5109384-5109385:+ | [17] | |
| Sequence | GAAAGTATGCAAGAACTGCTAACTCATGCCCCCATGCCTAA | ||
| Cell/Tissue List | mitochondrial | ||
| Transcript ID List | ENST00000381652.3 | ||
| External Link | RMBase: otherMod_site_1147 | ||
References