m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00155)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
ITGB3
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
 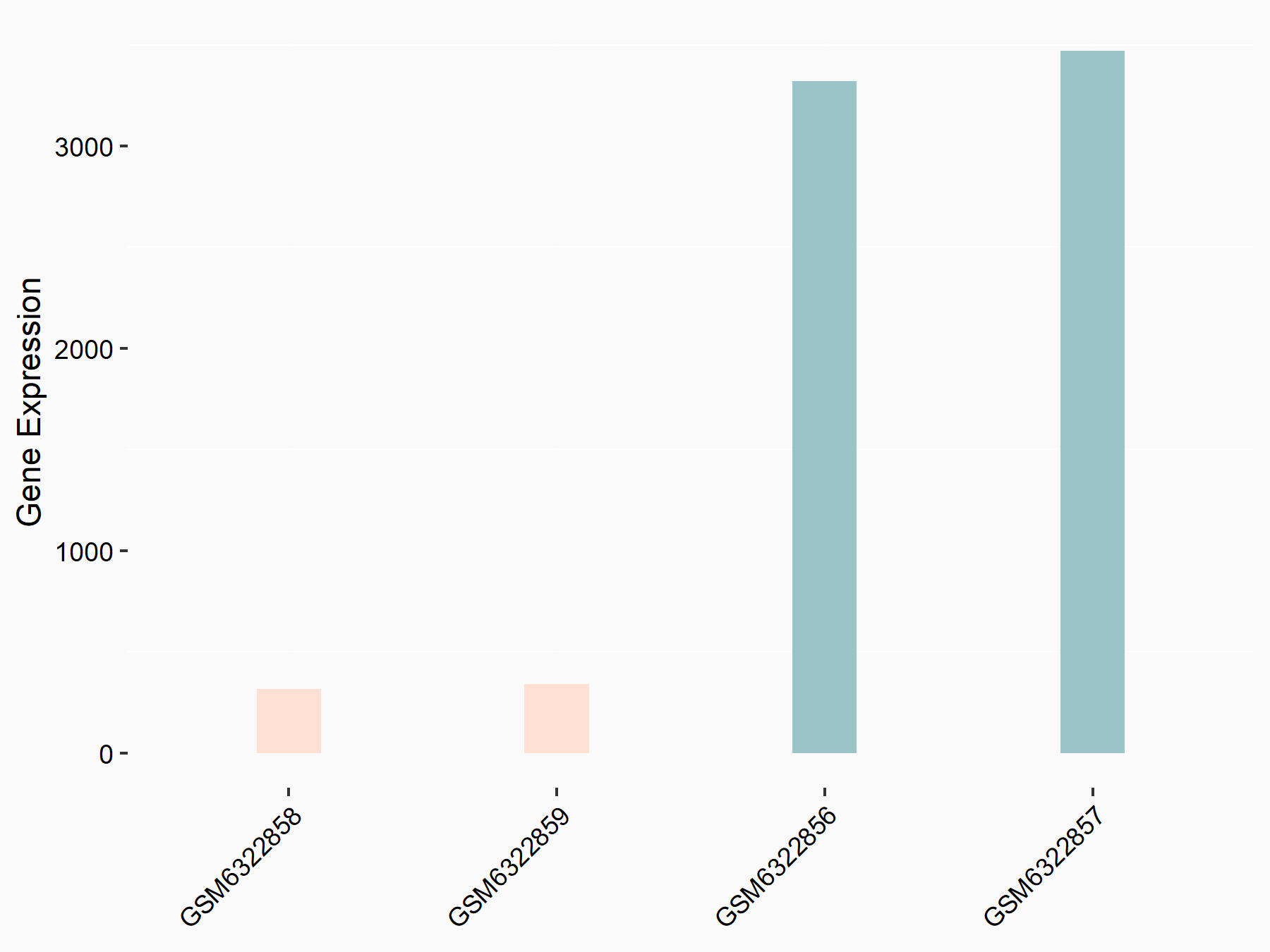 |
logFC: -3.38E+00 p-value: 2.43E-215 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between ITGB3 and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.57E+00 | GSE60213 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | FAM225A functioned as a competing endogenous RNA (ceRNA) for sponging miR-590-3p and miR-1275, leading to the upregulation of their target Integrin subunit beta 3 (ITGB3), and the activation of FAK/PI3K/Akt signaling to promote Nasopharyngeal carcinoma cell proliferation and invasion. FAM225A showed lower RNA stability after silencing of METTL3. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Nasopharyngeal carcinoma | ICD-11: 2B6B | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell proliferation and metastasis | |||
| In-vitro Model | 5-8F | Nasopharyngeal carcinoma | Homo sapiens | CVCL_C528 |
| 6-10B | Nasopharyngeal carcinoma | Homo sapiens | CVCL_C529 | |
| C666-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_7949 | |
| CNE-1 | Normal | Homo sapiens | CVCL_6888 | |
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| HK1 | Nasopharyngeal carcinoma | Acipenser baerii | CVCL_YE27 | |
| HNE-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_0308 | |
| HONE-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_8706 | |
| N2Tert (The human immortalized nasopharyngeal epithelial cell lines) | ||||
| NP69 (A human immortalized nasopharyngeal epithelial) | ||||
| S18 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_B0U9 | |
| S26 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_B0UB | |
| SUNE1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6946 | |
| In-vivo Model | For the tumor growth model, 1 × 106 HNE1-Scrambled or HNE1-shFAM2225A 2# cells were injected into the axilla of the mice, and the tumor size was measured every 3 days. | |||
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | FAM225A functioned as a competing endogenous RNA (ceRNA) for sponging miR-590-3p and miR-1275, leading to the upregulation of their target Integrin subunit beta 3 (ITGB3), and the activation of FAK/PI3K/Akt signaling to promote Nasopharyngeal carcinoma cell proliferation and invasion. FAM225A showed lower RNA stability after silencing of METTL3. | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell proliferation and metastasis | |||
| In-vitro Model | 5-8F | Nasopharyngeal carcinoma | Homo sapiens | CVCL_C528 |
| 6-10B | Nasopharyngeal carcinoma | Homo sapiens | CVCL_C529 | |
| C666-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_7949 | |
| CNE-1 | Normal | Homo sapiens | CVCL_6888 | |
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| HK1 | Nasopharyngeal carcinoma | Acipenser baerii | CVCL_YE27 | |
| HNE-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_0308 | |
| HONE-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_8706 | |
| N2Tert (The human immortalized nasopharyngeal epithelial cell lines) | ||||
| NP69 (A human immortalized nasopharyngeal epithelial) | ||||
| S18 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_B0U9 | |
| S26 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_B0UB | |
| SUNE1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6946 | |
| In-vivo Model | For the tumor growth model, 1 × 106 HNE1-Scrambled or HNE1-shFAM2225A 2# cells were injected into the axilla of the mice, and the tumor size was measured every 3 days. | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00155)
| In total 5 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE008218 | Click to Show/Hide the Full List | ||
| mod site | chr17:47299049-47299050:+ | [2] | |
| Sequence | AAAGGGCAAGAGAGAGGGCAAGAGGCCCAGGAGACTTAGTT | ||
| Transcript ID List | ENST00000560629.1; ENST00000559488.5 | ||
| External Link | RMBase: RNA-editing_site_58674 | ||
| mod ID: A2ISITE008219 | Click to Show/Hide the Full List | ||
| mod site | chr17:47309271-47309272:+ | [3] | |
| Sequence | TCTGTTACCCAGGCTGGTCTAGAACTCCTGGGCTCAAGTGA | ||
| Transcript ID List | ENST00000560629.1; ENST00000559488.5 | ||
| External Link | RMBase: RNA-editing_site_58675 | ||
| mod ID: A2ISITE008220 | Click to Show/Hide the Full List | ||
| mod site | chr17:47309299-47309300:+ | [3] | |
| Sequence | TGGGCTCAAGTGATCTTCCTACCTTGGCCCCTAAAAGTGCT | ||
| Transcript ID List | ENST00000560629.1; ENST00000559488.5 | ||
| External Link | RMBase: RNA-editing_site_58676 | ||
| mod ID: A2ISITE008221 | Click to Show/Hide the Full List | ||
| mod site | chr17:47309722-47309723:+ | [3] | |
| Sequence | GGGCAACATGAAGAAACCCCATCTCTACAAAAAAATGCAAA | ||
| Transcript ID List | ENST00000560629.1; ENST00000559488.5; rmsk_4706758 | ||
| External Link | RMBase: RNA-editing_site_58677 | ||
| mod ID: A2ISITE008222 | Click to Show/Hide the Full List | ||
| mod site | chr17:47309839-47309840:+ | [3] | |
| Sequence | GGGGAGGTGGAGGTTTCAGTAAGCCGAGATTGTGCCACTGC | ||
| Transcript ID List | rmsk_4706758; ENST00000560629.1; ENST00000559488.5 | ||
| External Link | RMBase: RNA-editing_site_58678 | ||
5-methylcytidine (m5C)
N6-methyladenosine (m6A)
| In total 14 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE033476 | Click to Show/Hide the Full List | ||
| mod site | chr17:47283462-47283463:+ | [4] | |
| Sequence | TGAGGCCCGAGTACTAGAGGACAGGCCCCTCAGCGACAAGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | kidney; endometrial | ||
| Seq Type List | m6A-REF-seq; m6A-seq | ||
| Transcript ID List | ENST00000559488.5; ENST00000571680.1; ENST00000560629.1 | ||
| External Link | RMBase: m6A_site_368416 | ||
| mod ID: M6ASITE033477 | Click to Show/Hide the Full List | ||
| mod site | chr17:47283492-47283493:+ | [5] | |
| Sequence | CAGCGACAAGGGCTCTGGAGACAGCTCCCAGGTCACTCAAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000559488.5; ENST00000560629.1; ENST00000571680.1 | ||
| External Link | RMBase: m6A_site_368417 | ||
| mod ID: M6ASITE033478 | Click to Show/Hide the Full List | ||
| mod site | chr17:47284496-47284497:+ | [5] | |
| Sequence | GGTGGAGGATTACCCTGTGGACATCTACTACTTGATGGACC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000571680.1; ENST00000559488.5; ENST00000560629.1 | ||
| External Link | RMBase: m6A_site_368418 | ||
| mod ID: M6ASITE033479 | Click to Show/Hide the Full List | ||
| mod site | chr17:47284514-47284515:+ | [5] | |
| Sequence | GGACATCTACTACTTGATGGACCTGTCTTACTCCATGAAGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000560629.1; ENST00000559488.5; ENST00000571680.1 | ||
| External Link | RMBase: m6A_site_368419 | ||
| mod ID: M6ASITE033480 | Click to Show/Hide the Full List | ||
| mod site | chr17:47292223-47292224:+ | [6] | |
| Sequence | AAAGCCCGTGGGCTTCAAGGACAGCCTGATCGTCCAGGTCA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | GM12878; HEK293A-TOA; MSC; TIME; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000560629.1; ENST00000559488.5 | ||
| External Link | RMBase: m6A_site_368420 | ||
| mod ID: M6ASITE033481 | Click to Show/Hide the Full List | ||
| mod site | chr17:47292281-47292282:+ | [7] | |
| Sequence | GCCTGCCAGGCCCAAGCTGAACCTAATAGCCATCGCTGCAA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | U2OS; fibroblasts; GM12878; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000560629.1; ENST00000559488.5 | ||
| External Link | RMBase: m6A_site_368421 | ||
| mod ID: M6ASITE033482 | Click to Show/Hide the Full List | ||
| mod site | chr17:47292315-47292316:+ | [8] | |
| Sequence | GCTGCAACAATGGCAATGGGACCTTTGAGTGTGGGGTATGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; U2OS; fibroblasts; GM12878; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000560629.1; ENST00000559488.5 | ||
| External Link | RMBase: m6A_site_368422 | ||
| mod ID: M6ASITE033483 | Click to Show/Hide the Full List | ||
| mod site | chr17:47292385-47292386:+ | [8] | |
| Sequence | GTGTGAGTGCTCAGAGGAGGACTATCGCCCTTCCCAGCAGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; A549; U2OS; fibroblasts; GM12878; CD4T; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000560629.1; ENST00000559488.5 | ||
| External Link | RMBase: m6A_site_368423 | ||
| mod ID: M6ASITE033484 | Click to Show/Hide the Full List | ||
| mod site | chr17:47292547-47292548:+ | [4] | |
| Sequence | CGACTTCTCCTGTGTCCGCTACAAGGGGGAGATGTGCTCAG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000559488.5; ENST00000560629.1 | ||
| External Link | RMBase: m6A_site_368424 | ||
| mod ID: M6ASITE033485 | Click to Show/Hide the Full List | ||
| mod site | chr17:47299331-47299332:+ | [9] | |
| Sequence | TGGCCAGTGCAGCTGTGGGGACTGCCTGTGTGACTCCGACT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | GSC-11; iSLK; MSC; TIME; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000559488.5; ENST00000560629.1 | ||
| External Link | RMBase: m6A_site_368425 | ||
| mod ID: M6ASITE033486 | Click to Show/Hide the Full List | ||
| mod site | chr17:47299354-47299355:+ | [9] | |
| Sequence | GCCTGTGTGACTCCGACTGGACCGGCTACTACTGCAACTGT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | GSC-11; iSLK; MSC; TIME; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000559488.5; ENST00000560629.1 | ||
| External Link | RMBase: m6A_site_368426 | ||
| mod ID: M6ASITE033487 | Click to Show/Hide the Full List | ||
| mod site | chr17:47307561-47307562:+ | [10] | |
| Sequence | GCCGCCCTGCTCATCTGGAAACTCCTCATCACCATCCACGA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | MT4 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000560629.1; ENST00000559488.5 | ||
| External Link | RMBase: m6A_site_368427 | ||
| mod ID: M6ASITE033488 | Click to Show/Hide the Full List | ||
| mod site | chr17:47307632-47307633:+ | [10] | |
| Sequence | ACGCGCCAGAGCAAAATGGGACACAGTAAGAGACGGGGCTG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | MT4 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000560629.1; ENST00000559488.5 | ||
| External Link | RMBase: m6A_site_368428 | ||
| mod ID: M6ASITE033489 | Click to Show/Hide the Full List | ||
| mod site | chr17:47310280-47310281:+ | [11] | |
| Sequence | TGCCATCATGTTTACAGAGGACAGTATTTGTGGGGAGGGAT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | fibroblasts | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000559488.5; ENST00000560629.1 | ||
| External Link | RMBase: m6A_site_368429 | ||
References