m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00227)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
DGAT2
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Fat mass and obesity-associated protein (FTO) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by FTO | ||
| Cell Line | B16-OVA cell line | Mus musculus |
|
Treatment: shFTO B16-OVA cells
Control: shNC B16-OVA cells
|
GSE154952 | |
| Regulation |
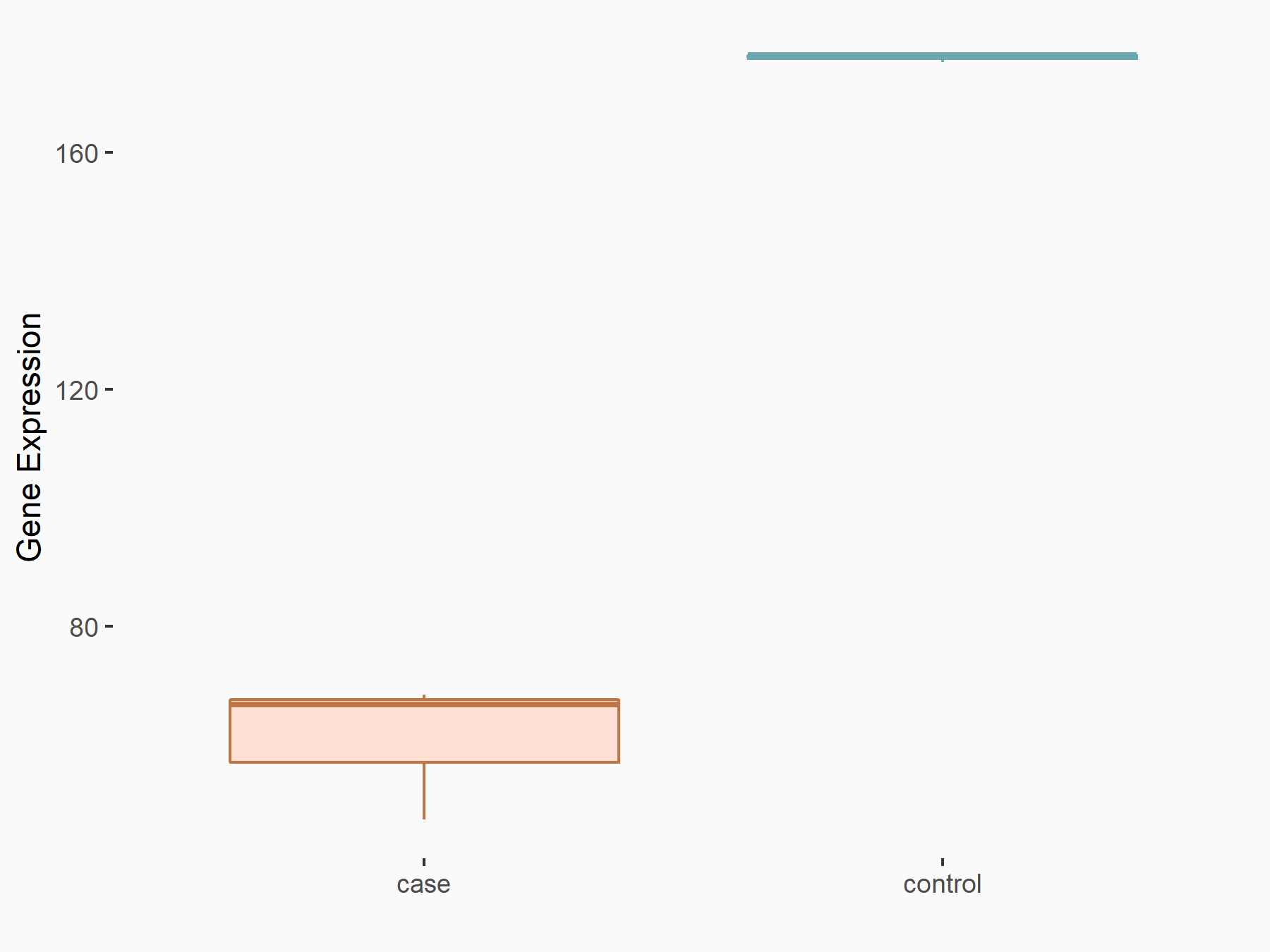
  |
logFC: -6.69E-01 p-value: 1.09E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Glucose Is Involved in the Dynamic Regulation of m6A in Patients With Type 2 Diabetes.high-glucose stimulation enhances FTO expression, which leads to decreased m6A, and the lower m6A induces methyltransferase upregulation; FTO then triggers the mRNA expression of FOXO1, FASN, G6PC, and Diacylglycerol O-acyltransferase 2 (DGAT2), and these four genes were correlated with glucose and lipid metabolism. | |||
| Responsed Disease | Diabetes | ICD-11: 5A10-5A14 | ||
| Pathway Response | Metabolic pathways | hsa01100 | ||
| Cell Process | Lipid metabolism | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shMETTL3 MOLM13 cells
Control: MOLM13 cells
|
GSE98623 | |
| Regulation |
 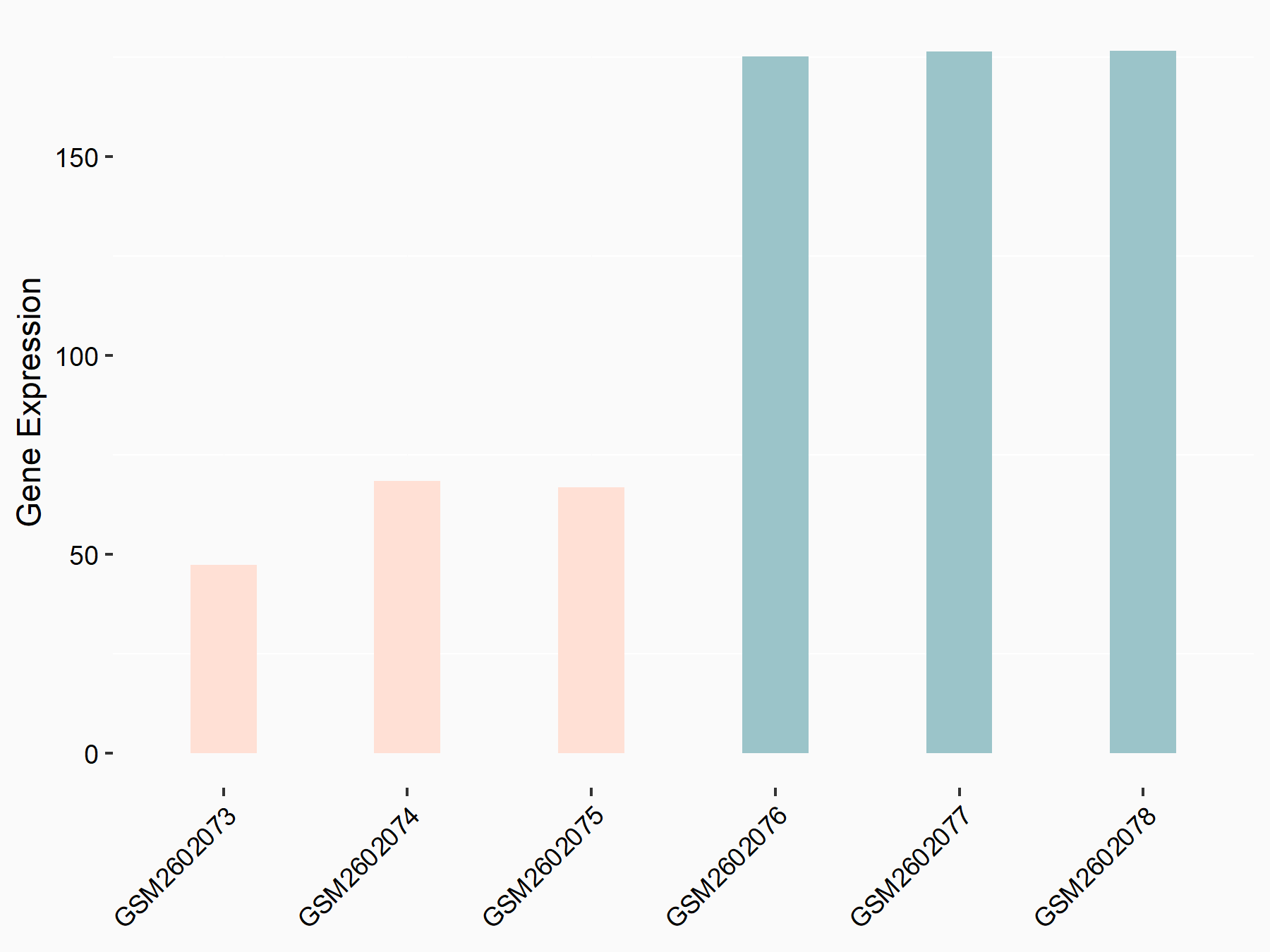 |
logFC: -1.53E+00 p-value: 7.36E-18 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | Type 2 diabetes (T2D) is characterized by lack of insulin, insulin resistance and high blood sugar. METTL3 silence decreased the m6A methylated and total mRNA level of Fatty acid synthase (Fasn), subsequently inhibited fatty acid metabolism. The expression of Acc1, Acly, Diacylglycerol O-acyltransferase 2 (DGAT2), Ehhadh, Fasn, Foxo, Pgc1a and Sirt1, which are critical to the regulation of fatty acid synthesis and oxidation were dramatically decreased in livers of hepatocyte-specific METTL3 knockout mice. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Type 2 diabetes mellitus | ICD-11: 5A11 | ||
| Pathway Response | Insulin resistance | hsa04931 | ||
| Cell Process | Lipid metabolism | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | Hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were generated by crossing mice with TBG-Cre Tg mice. METTL3 flox (METTL3 fl/fl) and hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were used for experiments. | |||
Diabetes [ICD-11: 5A10-5A14]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Glucose Is Involved in the Dynamic Regulation of m6A in Patients With Type 2 Diabetes.high-glucose stimulation enhances FTO expression, which leads to decreased m6A, and the lower m6A induces methyltransferase upregulation; FTO then triggers the mRNA expression of FOXO1, FASN, G6PC, and Diacylglycerol O-acyltransferase 2 (DGAT2), and these four genes were correlated with glucose and lipid metabolism. | |||
| Responsed Disease | Diabetes [ICD-11: 5A10-5A14] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Pathway Response | Metabolic pathways | hsa01100 | ||
| Cell Process | Lipid metabolism | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
Type 2 diabetes mellitus [ICD-11: 5A11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | Type 2 diabetes (T2D) is characterized by lack of insulin, insulin resistance and high blood sugar. METTL3 silence decreased the m6A methylated and total mRNA level of Fatty acid synthase (Fasn), subsequently inhibited fatty acid metabolism. The expression of Acc1, Acly, Diacylglycerol O-acyltransferase 2 (DGAT2), Ehhadh, Fasn, Foxo, Pgc1a and Sirt1, which are critical to the regulation of fatty acid synthesis and oxidation were dramatically decreased in livers of hepatocyte-specific METTL3 knockout mice. | |||
| Responsed Disease | Type 2 diabetes mellitus [ICD-11: 5A11] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Insulin resistance | hsa04931 | ||
| Cell Process | Lipid metabolism | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | Hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were generated by crossing mice with TBG-Cre Tg mice. METTL3 flox (METTL3 fl/fl) and hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were used for experiments. | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00227)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE005120 | Click to Show/Hide the Full List | ||
| mod site | chr11:75781605-75781606:+ | [3] | |
| Sequence | AGAGCTCACCACCTTGCAGCAGTCCCGGTACAGTTGCCCAG | ||
| Transcript ID List | ENST00000604733.5; ENST00000604935.5; ENST00000228027.12; ENST00000603276.5; ENST00000376262.7 | ||
| External Link | RMBase: RNA-editing_site_24162 | ||
N6-methyladenosine (m6A)
| In total 13 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE007867 | Click to Show/Hide the Full List | ||
| mod site | chr11:75768997-75768998:+ | [4] | |
| Sequence | GGCGGGCTTCAGCCATGAAGACCCTCATAGCCGCCTACTCC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000376262.7; ENST00000604935.5; ENST00000603276.5; ENST00000228027.12; ENST00000605608.1 | ||
| External Link | RMBase: m6A_site_156188 | ||
| mod ID: M6ASITE007868 | Click to Show/Hide the Full List | ||
| mod site | chr11:75769077-75769078:+ | [4] | |
| Sequence | AGCCAGCGCTCTCACGGAGGACCTGCGCTGTCGCGCGAGGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000603276.5; ENST00000604935.5; ENST00000228027.12; ENST00000605608.1; ENST00000604733.5; ENST00000376262.7 | ||
| External Link | RMBase: m6A_site_156189 | ||
| mod ID: M6ASITE007869 | Click to Show/Hide the Full List | ||
| mod site | chr11:75773749-75773750:+ | [4] | |
| Sequence | CCTGCGTTTGAGGAAGTGAGACCAGGGGGATGTACACAAGA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000604733.5; ENST00000603276.5; ENST00000604935.5; ENST00000376262.7; ENST00000605608.1; ENST00000228027.12 | ||
| External Link | RMBase: m6A_site_156190 | ||
| mod ID: M6ASITE007870 | Click to Show/Hide the Full List | ||
| mod site | chr11:75796423-75796424:+ | [5] | |
| Sequence | GTGCCTTCTGCAACTTCAGCACAGAGGCCACAGAAGTGAGC | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000604733.5; ENST00000605099.1; ENST00000603276.5; ENST00000604935.5; ENST00000603363.5; ENST00000376262.7; ENST00000603865.1; ENST00000228027.12 | ||
| External Link | RMBase: m6A_site_156191 | ||
| mod ID: M6ASITE007871 | Click to Show/Hide the Full List | ||
| mod site | chr11:75798270-75798271:+ | [6] | |
| Sequence | CTTTGGAGAGAATGAAGTGTACAAGCAGGTGATCTTCGAGG | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000603363.5; ENST00000376262.7; ENST00000603865.1; ENST00000604733.5; ENST00000228027.12 | ||
| External Link | RMBase: m6A_site_156192 | ||
| mod ID: M6ASITE007872 | Click to Show/Hide the Full List | ||
| mod site | chr11:75798333-75798334:+ | [6] | |
| Sequence | CCAGAAGAAGTTCCAGAAATACATTGGTTTCGCCCCATGCA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000604733.5; ENST00000376262.7; ENST00000603865.1; ENST00000603363.5; ENST00000228027.12 | ||
| External Link | RMBase: m6A_site_156193 | ||
| mod ID: M6ASITE007873 | Click to Show/Hide the Full List | ||
| mod site | chr11:75800463-75800464:+ | [4] | |
| Sequence | AGCTCTTCGACAAGCACAAGACCAAGTTCGGCCTCCCGGAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000603865.1; ENST00000228027.12; ENST00000376262.7; ENST00000603363.5 | ||
| External Link | RMBase: m6A_site_156194 | ||
| mod ID: M6ASITE007874 | Click to Show/Hide the Full List | ||
| mod site | chr11:75800484-75800485:+ | [4] | |
| Sequence | CCAAGTTCGGCCTCCCGGAGACTGAGGTCCTGGAGGTGAAC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000376262.7; ENST00000603865.1; ENST00000603363.5; ENST00000228027.12 | ||
| External Link | RMBase: m6A_site_156195 | ||
| mod ID: M6ASITE007875 | Click to Show/Hide the Full List | ||
| mod site | chr11:75800503-75800504:+ | [4] | |
| Sequence | GACTGAGGTCCTGGAGGTGAACTGAGCCAGCCTTCGGGGCC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000603865.1; ENST00000603363.5; ENST00000228027.12; ENST00000376262.7 | ||
| External Link | RMBase: m6A_site_156196 | ||
| mod ID: M6ASITE007876 | Click to Show/Hide the Full List | ||
| mod site | chr11:75800538-75800539:+ | [4] | |
| Sequence | GGGGCCAATTCCCTGGAGGAACCAGCTGCAAATCACTTTTT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000228027.12; ENST00000376262.7; ENST00000603865.1; ENST00000603363.5 | ||
| External Link | RMBase: m6A_site_156197 | ||
| mod ID: M6ASITE007877 | Click to Show/Hide the Full List | ||
| mod site | chr11:75801081-75801082:+ | [4] | |
| Sequence | TCACTCATATCGGAGCCTGGACTGGCCTCCAGGATGAGGAT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; MM6; HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000228027.12; ENST00000376262.7; ENST00000603363.5 | ||
| External Link | RMBase: m6A_site_156200 | ||
| mod ID: M6ASITE007878 | Click to Show/Hide the Full List | ||
| mod site | chr11:75801134-75801135:+ | [4] | |
| Sequence | GACACCCTGCAGGGGAAAGGACTGCCCCCCATGCACCATTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; MM6; HEK293A-TOA; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000376262.7; ENST00000228027.12; ENST00000603363.5 | ||
| External Link | RMBase: m6A_site_156201 | ||
| mod ID: M6ASITE007879 | Click to Show/Hide the Full List | ||
| mod site | chr11:75801233-75801234:+ | [4] | |
| Sequence | ATGACATGGATGCAGCACAGACTCAGCCTTGGCCTGGAGCA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; GM12878; MM6; HEK293A-TOA; MSC; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000603363.5; ENST00000228027.12; ENST00000376262.7 | ||
| External Link | RMBase: m6A_site_156203 | ||
References