m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00606)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
ARC
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Embryonic stem cells | Mus musculus |
|
Treatment: METTL3 knockout ESCs
Control: Wild type ESCs
|
GSE146466 | |
| Regulation |
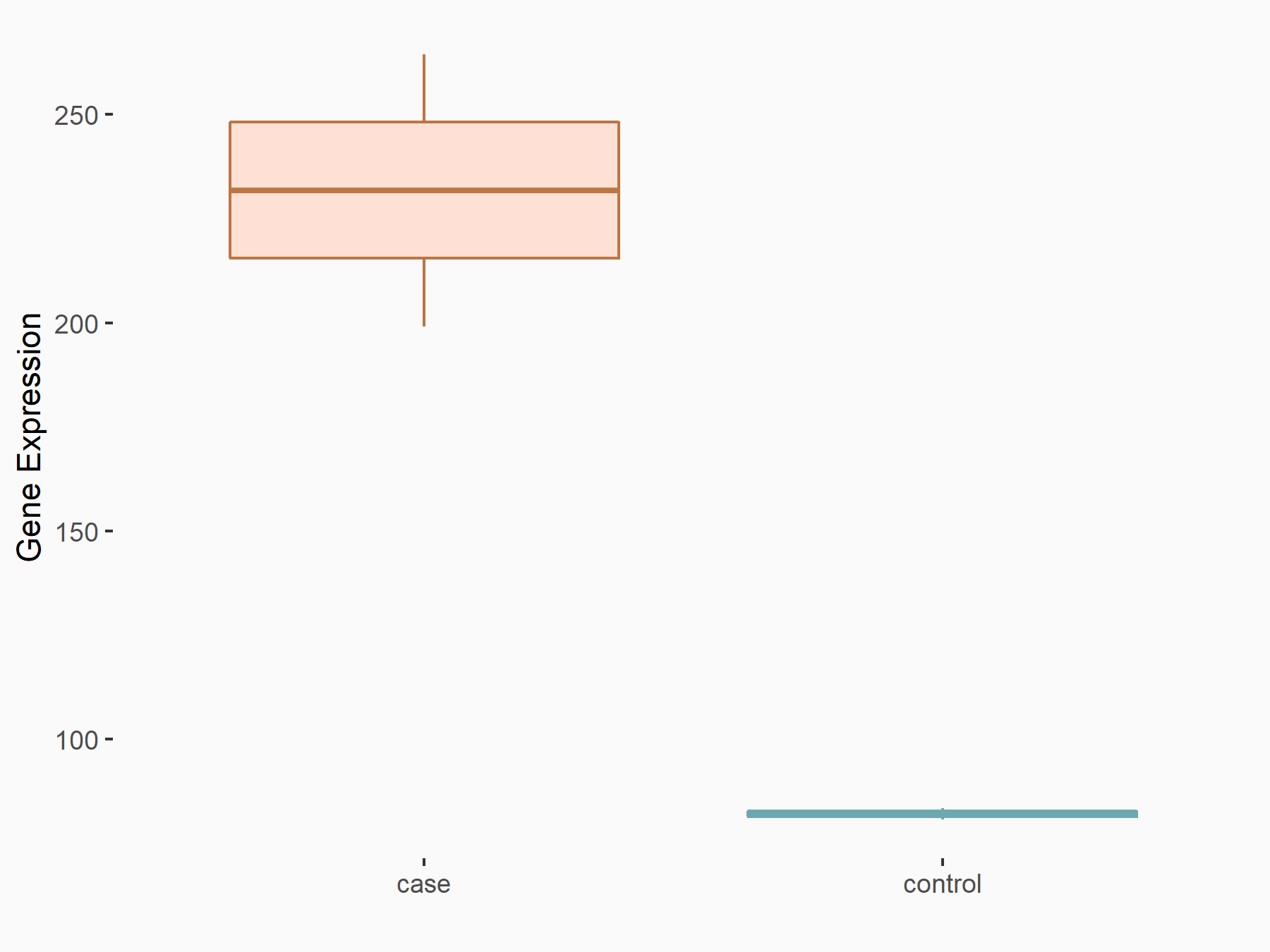 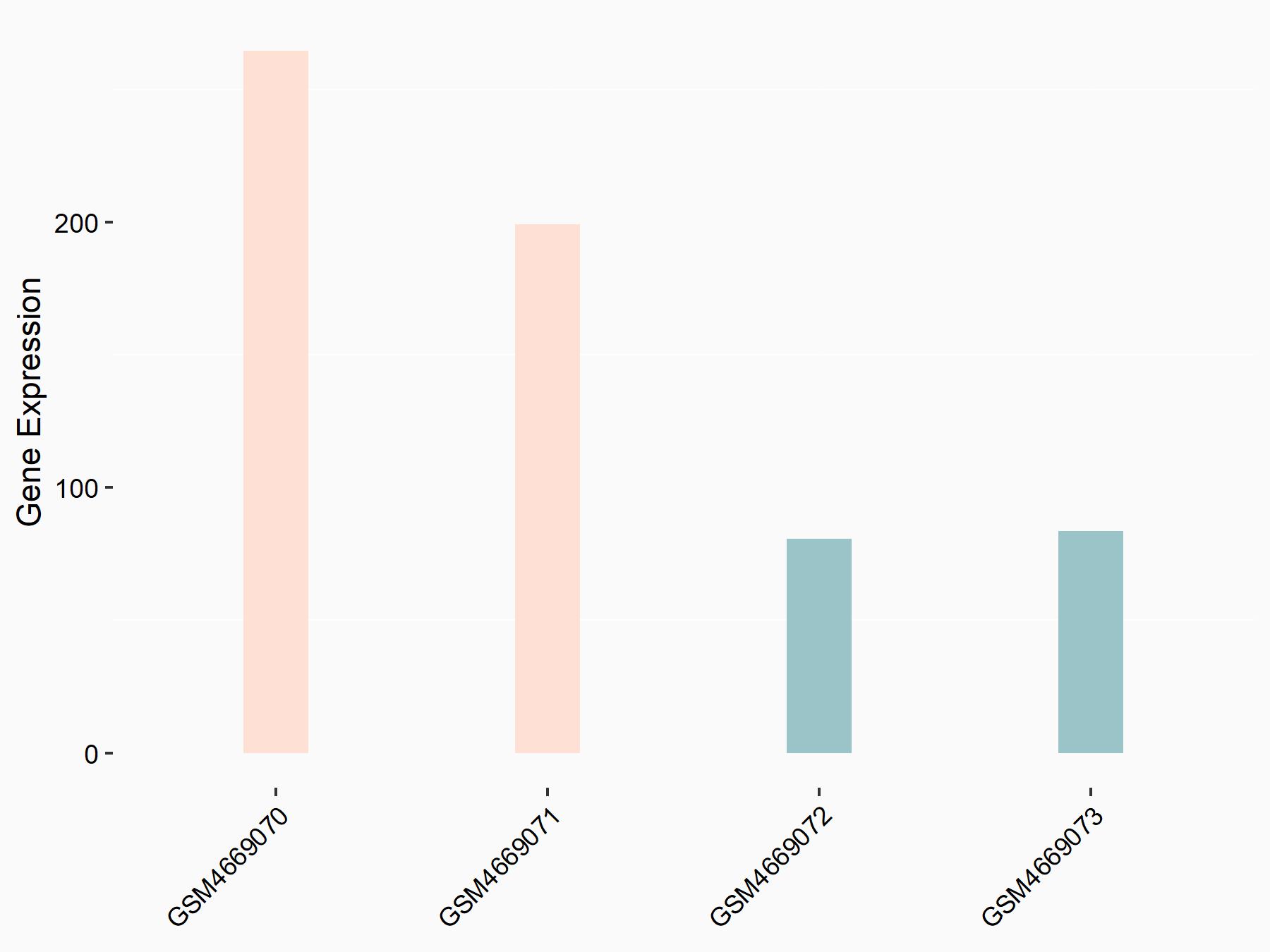 |
logFC: 1.50E+00 p-value: 1.12E-06 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 rescues the A-Bete-induced reduction of Nucleolar protein 3 (ARC) expression via YTHDF1-Dependent m6A modification, which suggests an important mechanism of epigenetic alteration in AD. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Alzheimer disease | ICD-11: 8A20 | ||
| In-vitro Model | SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
YTH domain-containing family protein 1 (YTHDF1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
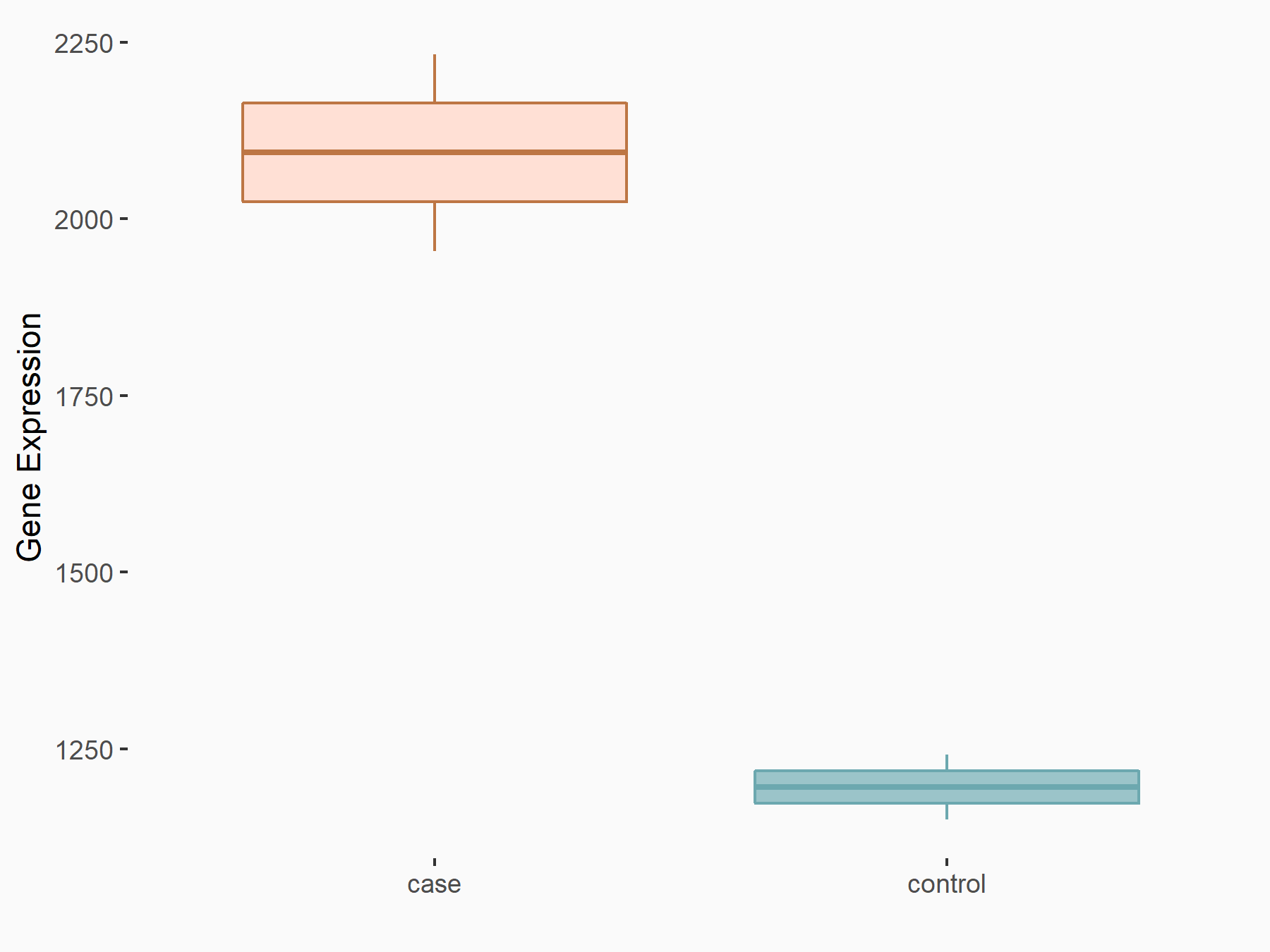 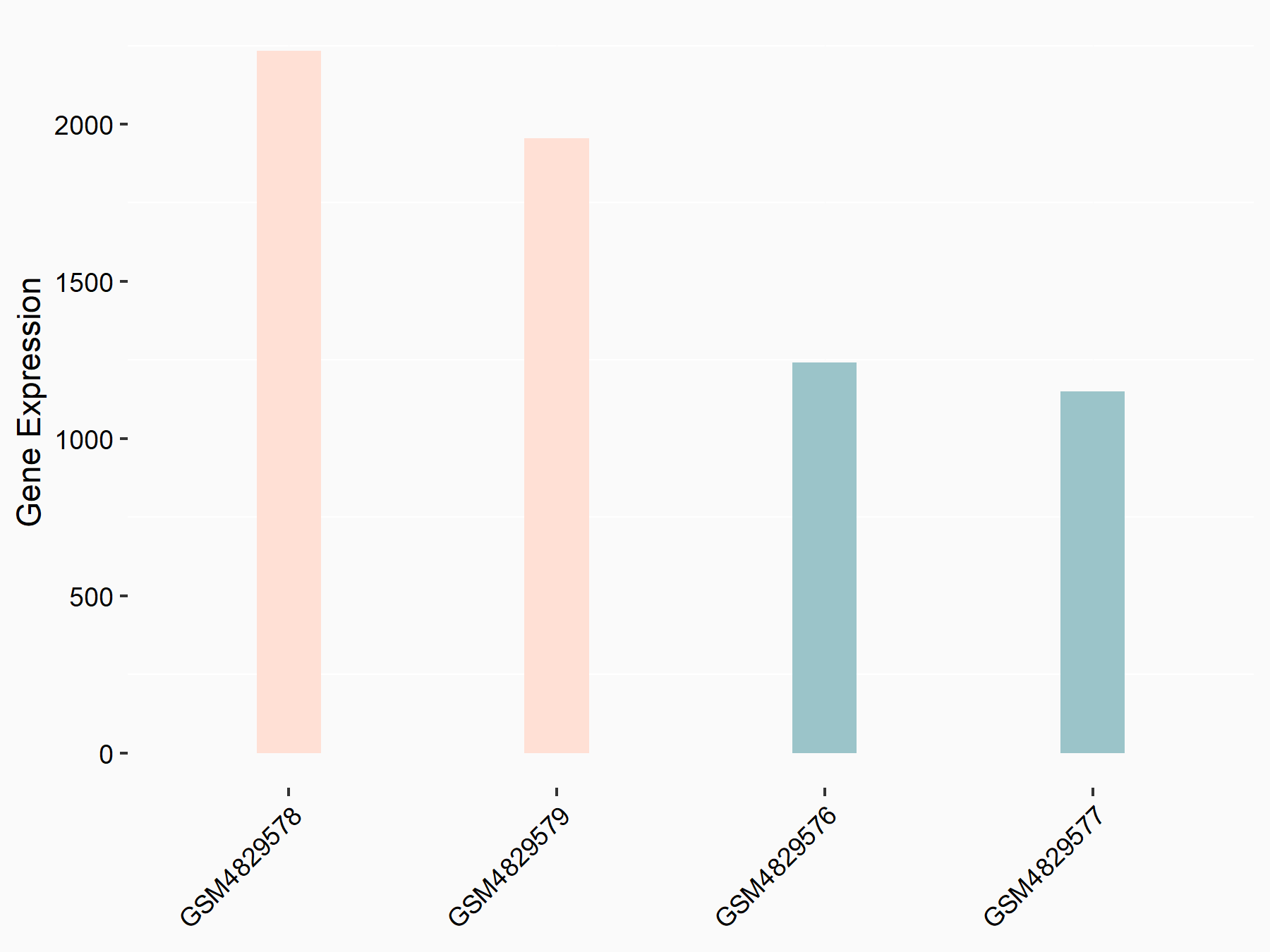 |
logFC: 8.08E-01 p-value: 1.08E-05 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 rescues the A-Bete-induced reduction of Nucleolar protein 3 (ARC) expression via YTHDF1-Dependent m6A modification, which suggests an important mechanism of epigenetic alteration in AD. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Alzheimer disease | ICD-11: 8A20 | ||
| In-vitro Model | SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
Alzheimer disease [ICD-11: 8A20]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 rescues the A-Bete-induced reduction of Nucleolar protein 3 (ARC) expression via YTHDF1-Dependent m6A modification, which suggests an important mechanism of epigenetic alteration in AD. | |||
| Responsed Disease | Alzheimer disease [ICD-11: 8A20] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 rescues the A-Bete-induced reduction of Nucleolar protein 3 (ARC) expression via YTHDF1-Dependent m6A modification, which suggests an important mechanism of epigenetic alteration in AD. | |||
| Responsed Disease | Alzheimer disease [ICD-11: 8A20] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03530 | ||
| Epigenetic Regulator | Lysine-specific histone demethylase 1A (KDM1A) | |
| Regulated Target | Histone H3 lysine 9 trimethylation (H3K9me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Alzheimer disease | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00606)
| In total 34 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE087282 | Click to Show/Hide the Full List | ||
| mod site | chr8:142611313-142611314:- | [2] | |
| Sequence | CTGGGGGACCAGCCCTGCAGACCACTCTGACAAGTCTTCAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | TREX | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811501 | ||
| mod ID: M6ASITE087283 | Click to Show/Hide the Full List | ||
| mod site | chr8:142611326-142611327:- | [2] | |
| Sequence | TCTGCCTTGTCCACTGGGGGACCAGCCCTGCAGACCACTCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | TREX | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811502 | ||
| mod ID: M6ASITE087284 | Click to Show/Hide the Full List | ||
| mod site | chr8:142611410-142611411:- | [3] | |
| Sequence | TGCCCTGCCCCGACACCAGAACCCTGGTGCTGGTGGGCCAC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | GSC-11; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811503 | ||
| mod ID: M6ASITE087285 | Click to Show/Hide the Full List | ||
| mod site | chr8:142611472-142611473:- | [3] | |
| Sequence | TGAGCTCAGCTGCAGGAAGGACATCCCAGAAGCCATGGCTC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | GSC-11; TREX; endometrial; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811504 | ||
| mod ID: M6ASITE087286 | Click to Show/Hide the Full List | ||
| mod site | chr8:142611582-142611583:- | [4] | |
| Sequence | GCCTCCCTCCCAGCAGGGAAACCCCGCCCGCTGCCAGGCCA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | Jurkat; GSC-11; endometrial; GSCs | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811505 | ||
| mod ID: M6ASITE087287 | Click to Show/Hide the Full List | ||
| mod site | chr8:142611681-142611682:- | [4] | |
| Sequence | CCCTGAGCGCCAGCGCGAGGACCAGGGCCTGCCACTAAGCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | Jurkat; GSC-11; TREX; endometrial; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811506 | ||
| mod ID: M6ASITE087288 | Click to Show/Hide the Full List | ||
| mod site | chr8:142611764-142611765:- | [3] | |
| Sequence | GTGGGAGGCGAAGTCCCAGGACAGCCCAGAGGGGGGCTACA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | GSC-11; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811507 | ||
| mod ID: M6ASITE087289 | Click to Show/Hide the Full List | ||
| mod site | chr8:142612079-142612080:- | [3] | |
| Sequence | AGCACCACCCTTCCACCCAGACCCCACCATCAGCTGTCCCG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | GSC-11; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000356613.4; ENST00000581404.1 | ||
| External Link | RMBase: m6A_site_811508 | ||
| mod ID: M6ASITE087290 | Click to Show/Hide the Full List | ||
| mod site | chr8:142612514-142612515:- | [3] | |
| Sequence | TCGCACACGCAGCAGAGCAGACCTGATGTCCCGGTGCTTCC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | GSC-11; TREX; endometrial; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4; ENST00000581404.1 | ||
| External Link | RMBase: m6A_site_811509 | ||
| mod ID: M6ASITE087291 | Click to Show/Hide the Full List | ||
| mod site | chr8:142612701-142612702:- | [5] | |
| Sequence | GCGGCCACCAAGCTCATGGGACACCCAGAGCAGAAGCTAGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; H1A; H1B; hESCs; GSC-11; HEK293A-TOA; MSC; TREX; endometrial; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4; ENST00000581404.1 | ||
| External Link | RMBase: m6A_site_811510 | ||
| mod ID: M6ASITE087292 | Click to Show/Hide the Full List | ||
| mod site | chr8:142612747-142612748:- | [6] | |
| Sequence | GCCCTCTGGACATCCAGAAAACCAAGTGTCCGGATGGCAGG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GSC-11; HEK293A-TOA; MSC; TREX; endometrial; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811511 | ||
| mod ID: M6ASITE087293 | Click to Show/Hide the Full List | ||
| mod site | chr8:142612758-142612759:- | [6] | |
| Sequence | ACAACACCCAGGCCCTCTGGACATCCAGAAAACCAAGTGTC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GSC-11; HEK293A-TOA; MSC; TREX; endometrial; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811512 | ||
| mod ID: M6ASITE087294 | Click to Show/Hide the Full List | ||
| mod site | chr8:142612775-142612776:- | [7] | |
| Sequence | AGCTTCCTCATTCTGCTACAACACCCAGGCCCTCTGGACAT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811513 | ||
| mod ID: M6ASITE087295 | Click to Show/Hide the Full List | ||
| mod site | chr8:142612802-142612803:- | [6] | |
| Sequence | CACTCTCAGGCCATCACAGAACACCCCAGCTTCCTCATTCT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HEK293T; U2OS; H1A; H1B; hESCs; GSC-11; HEK293A-TOA; MSC; TREX; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811514 | ||
| mod ID: M6ASITE087296 | Click to Show/Hide the Full List | ||
| mod site | chr8:142612835-142612836:- | [6] | |
| Sequence | CGTCTGTCTTTCCGGCCTGGACCCCACCCTCCACACTCTCA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HEK293T; U2OS; H1A; H1B; hESCs; Jurkat; GSC-11; HEK293A-TOA; MSC; TREX; endometrial; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811515 | ||
| mod ID: M6ASITE087297 | Click to Show/Hide the Full List | ||
| mod site | chr8:142612947-142612948:- | [5] | |
| Sequence | CCCCCTCGAGCTCCGGGCGGACTCGCACACACTCGTGTCAT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; U2OS; H1A; H1B; Jurkat; GSC-11; HEK293A-TOA; MSC; TREX; endometrial; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811516 | ||
| mod ID: M6ASITE087298 | Click to Show/Hide the Full List | ||
| mod site | chr8:142613019-142613020:- | [5] | |
| Sequence | CTGTGGCTTTGCCCACCAGGACTTTTGAGCTGGGGCTGACT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; U2OS; H1A; H1B; hESCs; Jurkat; GSC-11; HEK293A-TOA; MSC; TREX; endometrial; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811517 | ||
| mod ID: M6ASITE087299 | Click to Show/Hide the Full List | ||
| mod site | chr8:142613094-142613095:- | [5] | |
| Sequence | AGTCCGTGGCCAGTGACCGGACCCAGCCCGAGTAGAGGGCA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; U2OS; H1B; H1A; Jurkat; GSC-11; HEK293A-TOA; TREX; endometrial; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811518 | ||
| mod ID: M6ASITE087300 | Click to Show/Hide the Full List | ||
| mod site | chr8:142613139-142613140:- | [5] | |
| Sequence | CGGTGGAGGATGAGGCGGAGACCCTCACGCCCGCCCCCAAC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; U2OS; Jurkat; GSC-11; HEK293A-TOA; TREX; endometrial; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811519 | ||
| mod ID: M6ASITE087301 | Click to Show/Hide the Full List | ||
| mod site | chr8:142613241-142613242:- | [3] | |
| Sequence | TGCGCCACCCCCTGCCCAAGACCCTGGAGCAGCTCATCCAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | GSC-11; HEK293A-TOA; TREX; endometrial; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811520 | ||
| mod ID: M6ASITE087302 | Click to Show/Hide the Full List | ||
| mod site | chr8:142613351-142613352:- | [3] | |
| Sequence | GTTCCTGTGGCGCAAGCGGGACCTGTACCAGACGCTCTACG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | GSC-11; TREX; endometrial; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811521 | ||
| mod ID: M6ASITE087303 | Click to Show/Hide the Full List | ||
| mod site | chr8:142613375-142613376:- | [8] | |
| Sequence | GAAGCAGGGCGAGCCGCTGGACCAGTTCCTGTGGCGCAAGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HEK293T; GSC-11; TREX; endometrial; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811522 | ||
| mod ID: M6ASITE087304 | Click to Show/Hide the Full List | ||
| mod site | chr8:142613405-142613406:- | [8] | |
| Sequence | GGCCATCCAGCGCGAGCTGGACCTGCCGCAGAAGCAGGGCG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HEK293T; GSC-11; TREX; endometrial; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811523 | ||
| mod ID: M6ASITE087305 | Click to Show/Hide the Full List | ||
| mod site | chr8:142613483-142613484:- | [8] | |
| Sequence | CAAGCAGGGCTCCGTGAAGAACTGGGTGGAGTTCAAGAAGG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HEK293T; GSC-11; TREX; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811524 | ||
| mod ID: M6ASITE087306 | Click to Show/Hide the Full List | ||
| mod site | chr8:142613624-142613625:- | [3] | |
| Sequence | GGACACGCAGATCTTCGAGGACCCTCGAGAGTTCCTGAGCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HEK293T; GSC-11; HEK293A-TOA; TREX; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811525 | ||
| mod ID: M6ASITE087307 | Click to Show/Hide the Full List | ||
| mod site | chr8:142613642-142613643:- | [4] | |
| Sequence | GCAGCCCAGCCCCGGCGTGGACACGCAGATCTTCGAGGACC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HEK293T; Jurkat; GSC-11; HEK293A-TOA; TREX; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811526 | ||
| mod ID: M6ASITE087308 | Click to Show/Hide the Full List | ||
| mod site | chr8:142613970-142613971:- | [3] | |
| Sequence | GCCTGTGCCGCTGCCAGGAGACCATCGCCAACCTGGAGCGC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | GSC-11; TREX; endometrial; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811527 | ||
| mod ID: M6ASITE087309 | Click to Show/Hide the Full List | ||
| mod site | chr8:142614252-142614253:- | [4] | |
| Sequence | AGATGGAGCTGGACCACCGGACCAGCGGCGGGCTCCACGCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | Jurkat; GSC-11; TREX; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811528 | ||
| mod ID: M6ASITE087310 | Click to Show/Hide the Full List | ||
| mod site | chr8:142614260-142614261:- | [4] | |
| Sequence | CTGCGCACAGATGGAGCTGGACCACCGGACCAGCGGCGGGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | Jurkat; GSC-11; TREX; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811529 | ||
| mod ID: M6ASITE087311 | Click to Show/Hide the Full List | ||
| mod site | chr8:142614325-142614326:- | [4] | |
| Sequence | CCGCCGGACCCCAGCGCCGGACCACCCTCTGTCCGCCCCGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | Jurkat; GSC-11; HEK293T; TREX; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811530 | ||
| mod ID: M6ASITE087312 | Click to Show/Hide the Full List | ||
| mod site | chr8:142614338-142614339:- | [4] | |
| Sequence | CACGGGACCCAGGCCGCCGGACCCCAGCGCCGGACCACCCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | Jurkat; GSC-11; HEK293T; TREX; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811531 | ||
| mod ID: M6ASITE087313 | Click to Show/Hide the Full List | ||
| mod site | chr8:142614352-142614353:- | [4] | |
| Sequence | TCCCCGCAGCTCCGCACGGGACCCAGGCCGCCGGACCCCAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | Jurkat; GSC-11; HEK293T; TREX; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811532 | ||
| mod ID: M6ASITE087314 | Click to Show/Hide the Full List | ||
| mod site | chr8:142614380-142614381:- | [6] | |
| Sequence | CCGCCGGCCCGCAGCGACAGACAGGCGCTCCCCGCAGCTCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | U2OS; GSC-11; HEK293T; TREX; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811533 | ||
| mod ID: M6ASITE087315 | Click to Show/Hide the Full List | ||
| mod site | chr8:142614433-142614434:- | [3] | |
| Sequence | CTCCGCAGCAGCCGAGCCGGACCTGCCTCCCCGGGCGTGCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | GSC-11; HEK293T; TREX; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000356613.4 | ||
| External Link | RMBase: m6A_site_811534 | ||
References