m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00325)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
MET
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
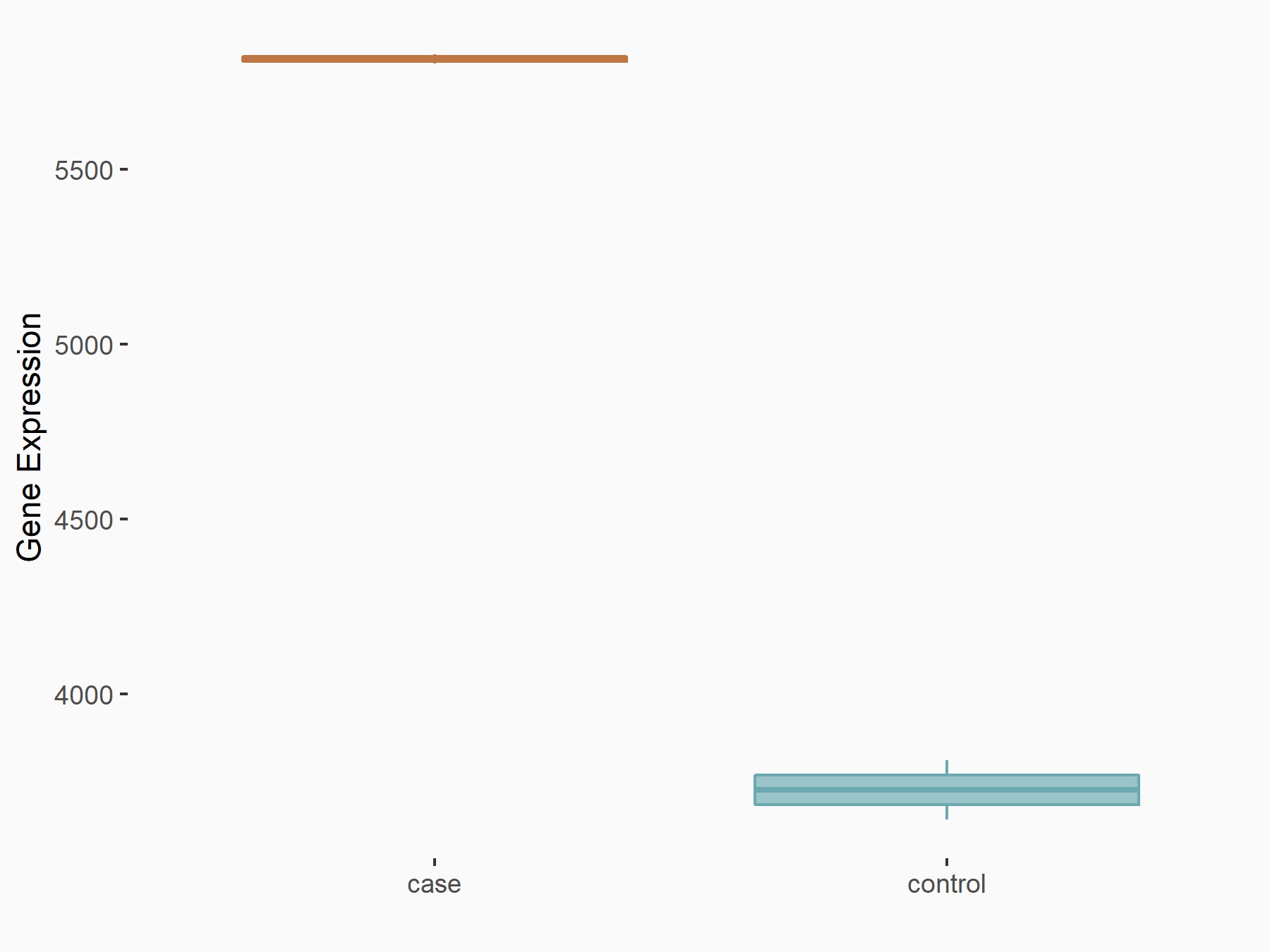  |
logFC: 6.42E-01 p-value: 3.45E-17 |
| More Results | Click to View More RNA-seq Results | |
| In total 3 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Chidamide could decrease Hepatocyte growth factor receptor (c-Met/MET) expression by inhibiting mRNA N6-methyladenosine (m6A) modification through the downregulation of METTL3 and WTAP expression, subsequently increasing the crizotinib sensitivity of NSCLC cells in a c-MET-/HGF-dependent manner. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Responsed Drug | Crizotinib | Approved | ||
| Pathway Response | EGFR tyrosine kinase inhibitor resistance | hsa01521 | ||
| In-vitro Model | HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 |
| NCI-H661 | Lung large cell carcinoma | Homo sapiens | CVCL_1577 | |
| NCI-H596 | Lung adenosquamous carcinoma | Homo sapiens | CVCL_1571 | |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| NCI-H358 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1559 | |
| NCI-H292 | Lung mucoepidermoid carcinoma | Homo sapiens | CVCL_0455 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1395 | Lung adenocarcinoma | Homo sapiens | CVCL_1467 | |
| EBC-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_2891 | |
| Calu-3 | Lung adenocarcinoma | Homo sapiens | CVCL_0609 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | HCC827 (3×106) cells suspended in 100 uL of PBS were injected into the left inguen of female Balb/c nude mice (body weight 18-20 g; age 6 weeks; Beijing Huafukang Bioscience Co., Inc.). When the tumor volumes reached 50-100 mm3 on the 10th posttransplantation day, the mice were randomized into four groups (10 mice per group) and were intragastrically administered vehicle (normal saline), crizotinib (25 mg/kg body weight), chidamide (5 mg/kg), or the combination of the two drugs daily for 21 days. The tumor volumes and body weights of the mice were measured every 3 days. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 combines with Hepatocyte growth factor receptor (c-Met/MET) and causes the PI3K/AKT signalling pathway to be manipulated, which affects the sensitivity of lung cancer cells to gefitinib. METTL3 knockdown promotes apoptosis and inhibits proliferation of lung cancer cells. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Lung cancer | ICD-11: 2C25 | ||
| Responsed Drug | Gefitinib | Approved | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| In-vitro Model | PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| NCI-H3255 | Lung adenocarcinoma | Homo sapiens | CVCL_6831 | |
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | METTL3-mediated m6A RNA methylation modulates uveal melanoma cell proliferation, migration, and invasion by targeting Hepatocyte growth factor receptor (c-Met/MET). Cycloleucine (Cyc) was used to block m6 A methylation in UM cells. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Melanoma of uvea | ICD-11: 2D0Y | ||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Arrest cell cycle at G1 phase | ||||
| In-vitro Model | M17 (Neuroblastoma cells) | |||
| M21 | Melanoma | Homo sapiens | CVCL_D031 | |
| M23 | Cutaneous melanoma | Homo sapiens | CVCL_RT32 | |
| SP-6.5 | Uveal melanoma | Homo sapiens | CVCL_7997 | |
Wilms tumor 1-associating protein (WTAP) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by WTAP | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: WTAP-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
  |
logFC: -2.39E+00 p-value: 1.81E-38 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Chidamide could decrease Hepatocyte growth factor receptor (c-Met/MET) expression by inhibiting mRNA N6-methyladenosine (m6A) modification through the downregulation of METTL3 and WTAP expression, subsequently increasing the crizotinib sensitivity of NSCLC cells in a c-MET-/HGF-dependent manner. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Responsed Drug | Crizotinib | Approved | ||
| Pathway Response | EGFR tyrosine kinase inhibitor resistance | hsa01521 | ||
| In-vitro Model | HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 |
| NCI-H661 | Lung large cell carcinoma | Homo sapiens | CVCL_1577 | |
| NCI-H596 | Lung adenosquamous carcinoma | Homo sapiens | CVCL_1571 | |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| NCI-H358 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1559 | |
| NCI-H292 | Lung mucoepidermoid carcinoma | Homo sapiens | CVCL_0455 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1395 | Lung adenocarcinoma | Homo sapiens | CVCL_1467 | |
| EBC-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_2891 | |
| Calu-3 | Lung adenocarcinoma | Homo sapiens | CVCL_0609 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | HCC827 (3×106) cells suspended in 100 uL of PBS were injected into the left inguen of female Balb/c nude mice (body weight 18-20 g; age 6 weeks; Beijing Huafukang Bioscience Co., Inc.). When the tumor volumes reached 50-100 mm3 on the 10th posttransplantation day, the mice were randomized into four groups (10 mice per group) and were intragastrically administered vehicle (normal saline), crizotinib (25 mg/kg body weight), chidamide (5 mg/kg), or the combination of the two drugs daily for 21 days. The tumor volumes and body weights of the mice were measured every 3 days. | |||
Lung cancer [ICD-11: 2C25]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Chidamide could decrease Hepatocyte growth factor receptor (c-Met/MET) expression by inhibiting mRNA N6-methyladenosine (m6A) modification through the downregulation of METTL3 and WTAP expression, subsequently increasing the crizotinib sensitivity of NSCLC cells in a c-MET-/HGF-dependent manner. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Crizotinib | Approved | ||
| Pathway Response | EGFR tyrosine kinase inhibitor resistance | hsa01521 | ||
| In-vitro Model | HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 |
| NCI-H661 | Lung large cell carcinoma | Homo sapiens | CVCL_1577 | |
| NCI-H596 | Lung adenosquamous carcinoma | Homo sapiens | CVCL_1571 | |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| NCI-H358 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1559 | |
| NCI-H292 | Lung mucoepidermoid carcinoma | Homo sapiens | CVCL_0455 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1395 | Lung adenocarcinoma | Homo sapiens | CVCL_1467 | |
| EBC-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_2891 | |
| Calu-3 | Lung adenocarcinoma | Homo sapiens | CVCL_0609 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | HCC827 (3×106) cells suspended in 100 uL of PBS were injected into the left inguen of female Balb/c nude mice (body weight 18-20 g; age 6 weeks; Beijing Huafukang Bioscience Co., Inc.). When the tumor volumes reached 50-100 mm3 on the 10th posttransplantation day, the mice were randomized into four groups (10 mice per group) and were intragastrically administered vehicle (normal saline), crizotinib (25 mg/kg body weight), chidamide (5 mg/kg), or the combination of the two drugs daily for 21 days. The tumor volumes and body weights of the mice were measured every 3 days. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 combines with Hepatocyte growth factor receptor (c-Met/MET) and causes the PI3K/AKT signalling pathway to be manipulated, which affects the sensitivity of lung cancer cells to gefitinib. METTL3 knockdown promotes apoptosis and inhibits proliferation of lung cancer cells. | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Gefitinib | Approved | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| In-vitro Model | PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| NCI-H3255 | Lung adenocarcinoma | Homo sapiens | CVCL_6831 | |
| Experiment 3 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Chidamide could decrease Hepatocyte growth factor receptor (c-Met/MET) expression by inhibiting mRNA N6-methyladenosine (m6A) modification through the downregulation of METTL3 and WTAP expression, subsequently increasing the crizotinib sensitivity of NSCLC cells in a c-MET-/HGF-dependent manner. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Wilms tumor 1-associating protein (WTAP) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Crizotinib | Approved | ||
| Pathway Response | EGFR tyrosine kinase inhibitor resistance | hsa01521 | ||
| In-vitro Model | HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 |
| NCI-H661 | Lung large cell carcinoma | Homo sapiens | CVCL_1577 | |
| NCI-H596 | Lung adenosquamous carcinoma | Homo sapiens | CVCL_1571 | |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| NCI-H358 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1559 | |
| NCI-H292 | Lung mucoepidermoid carcinoma | Homo sapiens | CVCL_0455 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1395 | Lung adenocarcinoma | Homo sapiens | CVCL_1467 | |
| EBC-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_2891 | |
| Calu-3 | Lung adenocarcinoma | Homo sapiens | CVCL_0609 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | HCC827 (3×106) cells suspended in 100 uL of PBS were injected into the left inguen of female Balb/c nude mice (body weight 18-20 g; age 6 weeks; Beijing Huafukang Bioscience Co., Inc.). When the tumor volumes reached 50-100 mm3 on the 10th posttransplantation day, the mice were randomized into four groups (10 mice per group) and were intragastrically administered vehicle (normal saline), crizotinib (25 mg/kg body weight), chidamide (5 mg/kg), or the combination of the two drugs daily for 21 days. The tumor volumes and body weights of the mice were measured every 3 days. | |||
Melanoma of uvea [ICD-11: 2D0Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | METTL3-mediated m6A RNA methylation modulates uveal melanoma cell proliferation, migration, and invasion by targeting Hepatocyte growth factor receptor (c-Met/MET). Cycloleucine (Cyc) was used to block m6 A methylation in UM cells. | |||
| Responsed Disease | Melanoma of uvea [ICD-11: 2D0Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Arrest cell cycle at G1 phase | ||||
| In-vitro Model | M17 (Neuroblastoma cells) | |||
| M21 | Melanoma | Homo sapiens | CVCL_D031 | |
| M23 | Cutaneous melanoma | Homo sapiens | CVCL_RT32 | |
| SP-6.5 | Uveal melanoma | Homo sapiens | CVCL_7997 | |
Crizotinib
[Approved]
| In total 2 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | Chidamide could decrease Hepatocyte growth factor receptor (c-Met/MET) expression by inhibiting mRNA N6-methyladenosine (m6A) modification through the downregulation of METTL3 and WTAP expression, subsequently increasing the crizotinib sensitivity of NSCLC cells in a c-MET-/HGF-dependent manner. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | EGFR tyrosine kinase inhibitor resistance | hsa01521 | ||
| In-vitro Model | HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 |
| NCI-H661 | Lung large cell carcinoma | Homo sapiens | CVCL_1577 | |
| NCI-H596 | Lung adenosquamous carcinoma | Homo sapiens | CVCL_1571 | |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| NCI-H358 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1559 | |
| NCI-H292 | Lung mucoepidermoid carcinoma | Homo sapiens | CVCL_0455 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1395 | Lung adenocarcinoma | Homo sapiens | CVCL_1467 | |
| EBC-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_2891 | |
| Calu-3 | Lung adenocarcinoma | Homo sapiens | CVCL_0609 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | HCC827 (3×106) cells suspended in 100 uL of PBS were injected into the left inguen of female Balb/c nude mice (body weight 18-20 g; age 6 weeks; Beijing Huafukang Bioscience Co., Inc.). When the tumor volumes reached 50-100 mm3 on the 10th posttransplantation day, the mice were randomized into four groups (10 mice per group) and were intragastrically administered vehicle (normal saline), crizotinib (25 mg/kg body weight), chidamide (5 mg/kg), or the combination of the two drugs daily for 21 days. The tumor volumes and body weights of the mice were measured every 3 days. | |||
| Experiment 2 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | Chidamide could decrease Hepatocyte growth factor receptor (c-Met/MET) expression by inhibiting mRNA N6-methyladenosine (m6A) modification through the downregulation of METTL3 and WTAP expression, subsequently increasing the crizotinib sensitivity of NSCLC cells in a c-MET-/HGF-dependent manner. | |||
| Target Regulator | Wilms tumor 1-associating protein (WTAP) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | EGFR tyrosine kinase inhibitor resistance | hsa01521 | ||
| In-vitro Model | HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 |
| NCI-H661 | Lung large cell carcinoma | Homo sapiens | CVCL_1577 | |
| NCI-H596 | Lung adenosquamous carcinoma | Homo sapiens | CVCL_1571 | |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| NCI-H358 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1559 | |
| NCI-H292 | Lung mucoepidermoid carcinoma | Homo sapiens | CVCL_0455 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1395 | Lung adenocarcinoma | Homo sapiens | CVCL_1467 | |
| EBC-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_2891 | |
| Calu-3 | Lung adenocarcinoma | Homo sapiens | CVCL_0609 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | HCC827 (3×106) cells suspended in 100 uL of PBS were injected into the left inguen of female Balb/c nude mice (body weight 18-20 g; age 6 weeks; Beijing Huafukang Bioscience Co., Inc.). When the tumor volumes reached 50-100 mm3 on the 10th posttransplantation day, the mice were randomized into four groups (10 mice per group) and were intragastrically administered vehicle (normal saline), crizotinib (25 mg/kg body weight), chidamide (5 mg/kg), or the combination of the two drugs daily for 21 days. The tumor volumes and body weights of the mice were measured every 3 days. | |||
Gefitinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [2] | |||
| Response Summary | METTL3 combines with Hepatocyte growth factor receptor (c-Met/MET) and causes the PI3K/AKT signalling pathway to be manipulated, which affects the sensitivity of lung cancer cells to gefitinib. METTL3 knockdown promotes apoptosis and inhibits proliferation of lung cancer cells. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Lung cancer | ICD-11: 2C25 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| In-vitro Model | PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| NCI-H3255 | Lung adenocarcinoma | Homo sapiens | CVCL_6831 | |
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
DNA modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 6 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT02146 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02159 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02172 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT02185 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT05855 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT05857 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03382 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03395 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00325)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE002306 | Click to Show/Hide the Full List | ||
| mod site | chr7:116753928-116753929:+ | [5] | |
| Sequence | AGGAGGTGCTTTATTAAGAAATAATTAATGAAACCTGGGCA | ||
| Transcript ID List | ENST00000495962.1; ENST00000436117.2; ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: RNA-editing_site_126692 | ||
5-methylcytidine (m5C)
| In total 3 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE004210 | Click to Show/Hide the Full List | ||
| mod site | chr7:116699926-116699927:+ | [6] | |
| Sequence | CACACAAGAATAATCAGGTTCTGTTCCATAAACTCTGGATT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000436117.2; ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m5C_site_40324 | ||
| mod ID: M5CSITE004211 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796912-116796913:+ | [6] | |
| Sequence | ACAGGCATGAGCCACTGCGCCCAGCCCTTATAAATTTTTGT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m5C_site_40325 | ||
| mod ID: M5CSITE004212 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796917-116796918:+ | [6] | |
| Sequence | CATGAGCCACTGCGCCCAGCCCTTATAAATTTTTGTATAGA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m5C_site_40326 | ||
N6-methyladenosine (m6A)
| In total 145 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE081926 | Click to Show/Hide the Full List | ||
| mod site | chr7:116672197-116672198:+ | [7] | |
| Sequence | ACAGCACGCGAGGCAGACAGACACGTGCTGGGGCGGGCAGG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HepG2; A549; HEK293T; iSLK; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773696 | ||
| mod ID: M6ASITE081927 | Click to Show/Hide the Full List | ||
| mod site | chr7:116699074-116699075:+ | [8] | |
| Sequence | GAACCTGTTTTGGCAGATAAACCTCTCATAATGAAGGCCCC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000456159.1; ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773697 | ||
| mod ID: M6ASITE081928 | Click to Show/Hide the Full List | ||
| mod site | chr7:116699231-116699232:+ | [8] | |
| Sequence | TTCCCAACTTCACCGCGGAAACACCCATCCAGAATGTCATT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000456159.1; ENST00000318493.11; ENST00000397752.7; ENST00000436117.2 | ||
| External Link | RMBase: m6A_site_773698 | ||
| mod ID: M6ASITE081929 | Click to Show/Hide the Full List | ||
| mod site | chr7:116699313-116699314:+ | [8] | |
| Sequence | TTATGTTTTAAATGAGGAAGACCTTCAGAAGGTTGCTGAGT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; fibroblasts; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000436117.2; ENST00000456159.1; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773699 | ||
| mod ID: M6ASITE081930 | Click to Show/Hide the Full List | ||
| mod site | chr7:116699339-116699340:+ | [8] | |
| Sequence | AGAAGGTTGCTGAGTACAAGACTGGGCCTGTGCTGGAACAC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; fibroblasts; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000456159.1; ENST00000436117.2; ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773700 | ||
| mod ID: M6ASITE081931 | Click to Show/Hide the Full List | ||
| mod site | chr7:116699356-116699357:+ | [8] | |
| Sequence | AAGACTGGGCCTGTGCTGGAACACCCAGATTGTTTCCCATG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; fibroblasts; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000436117.2; ENST00000456159.1; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773701 | ||
| mod ID: M6ASITE081932 | Click to Show/Hide the Full List | ||
| mod site | chr7:116699382-116699383:+ | [8] | |
| Sequence | AGATTGTTTCCCATGTCAGGACTGCAGCAGCAAAGCCAATT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; hESCs; fibroblasts; H1299; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000436117.2; ENST00000456159.1; ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773702 | ||
| mod ID: M6ASITE081933 | Click to Show/Hide the Full List | ||
| mod site | chr7:116699501-116699502:+ | [8] | |
| Sequence | GTGGCAGCGTCAACAGAGGGACCTGCCAGCGACATGTCTTT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; hESCs; fibroblasts; H1299; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000436117.2; ENST00000318493.11; ENST00000456159.1 | ||
| External Link | RMBase: m6A_site_773703 | ||
| mod ID: M6ASITE081934 | Click to Show/Hide the Full List | ||
| mod site | chr7:116699652-116699653:+ | [8] | |
| Sequence | AGTCCTTTCATCTGTAAAGGACCGGTTCATCAACTTCTTTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; fibroblasts; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7; ENST00000436117.2 | ||
| External Link | RMBase: m6A_site_773704 | ||
| mod ID: M6ASITE081935 | Click to Show/Hide the Full List | ||
| mod site | chr7:116699775-116699776:+ | [8] | |
| Sequence | TGGTTTTATGTTTTTGACGGACCAGTCCTACATTGATGTTT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; fibroblasts; A549; Huh7; HEK293A-TOA; iSLK; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000318493.11; ENST00000436117.2; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773705 | ||
| mod ID: M6ASITE081936 | Click to Show/Hide the Full List | ||
| mod site | chr7:116699850-116699851:+ | [9] | |
| Sequence | TGTCCATGCCTTTGAAAGCAACAATTTTATTTACTTCTTGA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T; A549 | ||
| Seq Type List | MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000436117.2; ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773706 | ||
| mod ID: M6ASITE081937 | Click to Show/Hide the Full List | ||
| mod site | chr7:116699885-116699886:+ | [8] | |
| Sequence | TCTTGACGGTCCAAAGGGAAACTCTAGATGCTCAGACTTTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; fibroblasts; A549; Huh7; HEK293A-TOA; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7; ENST00000436117.2 | ||
| External Link | RMBase: m6A_site_773707 | ||
| mod ID: M6ASITE081938 | Click to Show/Hide the Full List | ||
| mod site | chr7:116699900-116699901:+ | [8] | |
| Sequence | GGGAAACTCTAGATGCTCAGACTTTTCACACAAGAATAATC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; fibroblasts; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000397752.7; ENST00000436117.2; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773708 | ||
| mod ID: M6ASITE081939 | Click to Show/Hide the Full List | ||
| mod site | chr7:116699937-116699938:+ | [8] | |
| Sequence | AATCAGGTTCTGTTCCATAAACTCTGGATTGCATTCCTACA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; fibroblasts; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000436117.2; ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773709 | ||
| mod ID: M6ASITE081940 | Click to Show/Hide the Full List | ||
| mod site | chr7:116699955-116699956:+ | [9] | |
| Sequence | AAACTCTGGATTGCATTCCTACATGGAAATGCCTCTGGAGT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000436117.2; ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773710 | ||
| mod ID: M6ASITE081941 | Click to Show/Hide the Full List | ||
| mod site | chr7:116700008-116700009:+ | [9] | |
| Sequence | AAAAGAGAAAAAAGAGATCCACAAAGAAGGAAGTGTTTAAT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11; ENST00000436117.2 | ||
| External Link | RMBase: m6A_site_773711 | ||
| mod ID: M6ASITE081942 | Click to Show/Hide the Full List | ||
| mod site | chr7:116700076-116700077:+ | [8] | |
| Sequence | CCTGGGGCCCAGCTTGCTAGACAAATAGGAGCCAGCCTGAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; hESC-HEK293T; HepG2; fibroblasts; iSLK; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11; ENST00000436117.2 | ||
| External Link | RMBase: m6A_site_773712 | ||
| mod ID: M6ASITE081943 | Click to Show/Hide the Full List | ||
| mod site | chr7:116700102-116700103:+ | [9] | |
| Sequence | AGGAGCCAGCCTGAATGATGACATTCTTTTCGGGGTGTTCG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T; A549 | ||
| Seq Type List | MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000436117.2; ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773713 | ||
| mod ID: M6ASITE081944 | Click to Show/Hide the Full List | ||
| mod site | chr7:116700148-116700149:+ | [8] | |
| Sequence | AGCAAGCCAGATTCTGCCGAACCAATGGATCGATCTGCCAT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; fibroblasts; H1299; iSLK; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000436117.2; ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773714 | ||
| mod ID: M6ASITE081945 | Click to Show/Hide the Full List | ||
| mod site | chr7:116700225-116700226:+ | [8] | |
| Sequence | CAACAAGATCGTCAACAAAAACAATGTGAGATGTCTCCAGC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; A549; H1299; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7; ENST00000436117.2 | ||
| External Link | RMBase: m6A_site_773715 | ||
| mod ID: M6ASITE081946 | Click to Show/Hide the Full List | ||
| mod site | chr7:116700256-116700257:+ | [8] | |
| Sequence | TGTCTCCAGCATTTTTACGGACCCAATCATGAGCACTGCTT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; A549; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000436117.2; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773716 | ||
| mod ID: M6ASITE081947 | Click to Show/Hide the Full List | ||
| mod site | chr7:116724150-116724151:+ | [8] | |
| Sequence | CGGGACTCCGTGGGCGTAGGACCCTCCGAGCCAGGTGAGGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; A549 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7; ENST00000436117.2; ENST00000495962.1 | ||
| External Link | RMBase: m6A_site_773717 | ||
| mod ID: M6ASITE081948 | Click to Show/Hide the Full List | ||
| mod site | chr7:116724800-116724801:+ | [8] | |
| Sequence | GTGCTAGATTGGAGGTGAAGACCCTGGAGCCAGAGAGCCTA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | rmsk_2348879; ENST00000436117.2; ENST00000495962.1; ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773718 | ||
| mod ID: M6ASITE081949 | Click to Show/Hide the Full List | ||
| mod site | chr7:116731718-116731719:+ | [8] | |
| Sequence | CGCGCCGTGATGAATATCGAACAGAGTTTACCACAGCTTTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000436117.2; ENST00000318493.11; ENST00000495962.1; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773719 | ||
| mod ID: M6ASITE081950 | Click to Show/Hide the Full List | ||
| mod site | chr7:116731784-116731785:+ | [9] | |
| Sequence | AATTCAGCGAAGTCCTCTTAACATCTATATCCACCTTCATT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000436117.2; ENST00000397752.7; ENST00000495962.1 | ||
| External Link | RMBase: m6A_site_773720 | ||
| mod ID: M6ASITE081951 | Click to Show/Hide the Full List | ||
| mod site | chr7:116731812-116731813:+ | [8] | |
| Sequence | ATCCACCTTCATTAAAGGAGACCTCACCATAGCTAATCTTG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000495962.1; ENST00000436117.2; ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773721 | ||
| mod ID: M6ASITE081952 | Click to Show/Hide the Full List | ||
| mod site | chr7:116731835-116731836:+ | [8] | |
| Sequence | TCACCATAGCTAATCTTGGGACATCAGAGGGTCGCTTCATG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000436117.2; ENST00000397752.7; ENST00000495962.1; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773722 | ||
| mod ID: M6ASITE081953 | Click to Show/Hide the Full List | ||
| mod site | chr7:116739969-116739970:+ | [8] | |
| Sequence | GTTGTGGTTTCTCGATCAGGACCATCAACCCCTCATGTGAA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11; ENST00000436117.2; ENST00000495962.1 | ||
| External Link | RMBase: m6A_site_773723 | ||
| mod ID: M6ASITE081954 | Click to Show/Hide the Full List | ||
| mod site | chr7:116740001-116740002:+ | [8] | |
| Sequence | TCATGTGAATTTTCTCCTGGACTCCCATCCAGTGTCTCCAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000495962.1; ENST00000318493.11; ENST00000397752.7; ENST00000436117.2 | ||
| External Link | RMBase: m6A_site_773724 | ||
| mod ID: M6ASITE081955 | Click to Show/Hide the Full List | ||
| mod site | chr7:116740039-116740040:+ | [9] | |
| Sequence | CAGAAGTGATTGTGGAGCATACATTAAACCAAAATGGCTAC | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000436117.2; ENST00000318493.11; ENST00000495962.1 | ||
| External Link | RMBase: m6A_site_773725 | ||
| mod ID: M6ASITE081956 | Click to Show/Hide the Full List | ||
| mod site | chr7:116740046-116740047:+ | [8] | |
| Sequence | GATTGTGGAGCATACATTAAACCAAAATGGCTACACACTGG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000495962.1; ENST00000436117.2; ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773726 | ||
| mod ID: M6ASITE081957 | Click to Show/Hide the Full List | ||
| mod site | chr7:116740886-116740887:+ | [8] | |
| Sequence | TTGAATGGCTTGGGCTGCAGACATTTCCAGTCCTGCAGTCA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000436117.2; ENST00000397752.7; ENST00000495962.1; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773727 | ||
| mod ID: M6ASITE081958 | Click to Show/Hide the Full List | ||
| mod site | chr7:116740951-116740952:+ | [9] | |
| Sequence | TCAGTGTGGCTGGTGCCACGACAAATGTGTGCGATCGGAGG | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000495962.1; ENST00000397752.7; ENST00000436117.2 | ||
| External Link | RMBase: m6A_site_773728 | ||
| mod ID: M6ASITE081959 | Click to Show/Hide the Full List | ||
| mod site | chr7:116740986-116740987:+ | [8] | |
| Sequence | CGGAGGAATGCCTGAGCGGGACATGGACTCAACAGATCTGT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; liver | ||
| Seq Type List | m6A-seq; m6A-REF-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000495962.1; ENST00000397752.7; ENST00000436117.2 | ||
| External Link | RMBase: m6A_site_773729 | ||
| mod ID: M6ASITE081960 | Click to Show/Hide the Full List | ||
| mod site | chr7:116740992-116740993:+ | [8] | |
| Sequence | AATGCCTGAGCGGGACATGGACTCAACAGATCTGTCTGCCT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000495962.1; ENST00000436117.2; ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773730 | ||
| mod ID: M6ASITE081961 | Click to Show/Hide the Full List | ||
| mod site | chr7:116740997-116740998:+ | [10] | |
| Sequence | CTGAGCGGGACATGGACTCAACAGATCTGTCTGCCTGCAAT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000436117.2; ENST00000495962.1; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773731 | ||
| mod ID: M6ASITE081962 | Click to Show/Hide the Full List | ||
| mod site | chr7:116755387-116755388:+ | [8] | |
| Sequence | GTGCACCCCTTGAAGGAGGGACAAGGCTGACCATATGTGGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000495962.1; ENST00000397752.7; ENST00000436117.2; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773732 | ||
| mod ID: M6ASITE081963 | Click to Show/Hide the Full List | ||
| mod site | chr7:116755412-116755413:+ | [8] | |
| Sequence | GCTGACCATATGTGGCTGGGACTTTGGATTTCGGAGGAATA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000495962.1; ENST00000436117.2; ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773733 | ||
| mod ID: M6ASITE081964 | Click to Show/Hide the Full List | ||
| mod site | chr7:116755453-116755454:+ | [8] | |
| Sequence | ATAAATTTGATTTAAAGAAAACTAGAGTTCTCCTTGGAAAT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000436117.2; ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773734 | ||
| mod ID: M6ASITE081965 | Click to Show/Hide the Full List | ||
| mod site | chr7:116755513-116755514:+ | [9] | |
| Sequence | TAAGTGAGAGCACGATGAATACGTAAGGATCTTAAAATGCT | ||
| Motif Score | 2.084416667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000436117.2; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773735 | ||
| mod ID: M6ASITE081966 | Click to Show/Hide the Full List | ||
| mod site | chr7:116757509-116757510:+ | [8] | |
| Sequence | TTATTTCAAATGGCCACGGGACAACACAATACAGTACATTC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000436117.2; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773736 | ||
| mod ID: M6ASITE081967 | Click to Show/Hide the Full List | ||
| mod site | chr7:116757649-116757650:+ | [9] | |
| Sequence | CCCTCCAGGATCCTGTAATAACAAGTATTTCGCCGAAATAC | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000436117.2; ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773737 | ||
| mod ID: M6ASITE081968 | Click to Show/Hide the Full List | ||
| mod site | chr7:116757719-116757720:+ | [8] | |
| Sequence | TTTAACTGGAAATTACCTAAACAGTGGGAATTCTAGACACA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000436117.2; ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773738 | ||
| mod ID: M6ASITE081969 | Click to Show/Hide the Full List | ||
| mod site | chr7:116757735-116757736:+ | [8] | |
| Sequence | CTAAACAGTGGGAATTCTAGACACATTTCAATTGGTGGAAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000436117.2; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773739 | ||
| mod ID: M6ASITE081970 | Click to Show/Hide the Full List | ||
| mod site | chr7:116757757-116757758:+ | [8] | |
| Sequence | ACATTTCAATTGGTGGAAAAACATGTACTTTAAAAAGGTGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11; ENST00000436117.2 | ||
| External Link | RMBase: m6A_site_773740 | ||
| mod ID: M6ASITE081971 | Click to Show/Hide the Full List | ||
| mod site | chr7:116758466-116758467:+ | [8] | |
| Sequence | TTTTTTGTTCAGTGTGTCAAACAGTATTCTTGAATGTTATA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000436117.2; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773741 | ||
| mod ID: M6ASITE081972 | Click to Show/Hide the Full List | ||
| mod site | chr7:116758498-116758499:+ | [8] | |
| Sequence | AATGTTATACCCCAGCCCAAACCATTTCAACTGAGTTTGCT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000436117.2; ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773742 | ||
| mod ID: M6ASITE081973 | Click to Show/Hide the Full List | ||
| mod site | chr7:116758552-116758553:+ | [8] | |
| Sequence | TTGACTTAGCCAACCGAGAGACAAGCATCTTCAGTTACCGT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000422097.1; ENST00000397752.7; ENST00000318493.11; ENST00000436117.2 | ||
| External Link | RMBase: m6A_site_773743 | ||
| mod ID: M6ASITE081974 | Click to Show/Hide the Full List | ||
| mod site | chr7:116759351-116759352:+ | [8] | |
| Sequence | TACAGTACTTGGTGGAAAGAACCTCTCAACATTGTCAGTTT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000422097.1; ENST00000436117.2; ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773744 | ||
| mod ID: M6ASITE081975 | Click to Show/Hide the Full List | ||
| mod site | chr7:116759400-116759401:+ | [9] | |
| Sequence | GCTTTGCCAGTGGTGGGAGCACAATAACAGGTGTTGGGAAA | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000436117.2; ENST00000422097.1; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773745 | ||
| mod ID: M6ASITE081976 | Click to Show/Hide the Full List | ||
| mod site | chr7:116759422-116759423:+ | [8] | |
| Sequence | AATAACAGGTGTTGGGAAAAACCTGAATTCAGTTAGTGTCC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000436117.2; ENST00000318493.11; ENST00000422097.1; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773746 | ||
| mod ID: M6ASITE081977 | Click to Show/Hide the Full List | ||
| mod site | chr7:116759479-116759480:+ | [8] | |
| Sequence | TGTGCATGAAGCAGGAAGGAACTTTACAGTGGTAAGTCCTT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000422097.1; ENST00000318493.11; ENST00000436117.2; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773747 | ||
| mod ID: M6ASITE081978 | Click to Show/Hide the Full List | ||
| mod site | chr7:116763057-116763058:+ | [9] | |
| Sequence | GTCTTTCTCTAGGCATGTCAACATCGCTCTAATTCAGAGAT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000422097.1; ENST00000436117.2; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773748 | ||
| mod ID: M6ASITE081979 | Click to Show/Hide the Full List | ||
| mod site | chr7:116763133-116763134:+ | [8] | |
| Sequence | ATCTGCAACTCCCCCTGAAAACCAAAGCCTTTTTCATGTTA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000422097.1; ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773749 | ||
| mod ID: M6ASITE081980 | Click to Show/Hide the Full List | ||
| mod site | chr7:116763192-116763193:+ | [9] | |
| Sequence | TACTTTGATCTCATTTATGTACATAATCCTGTGTTTAAGCC | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000422097.1; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773750 | ||
| mod ID: M6ASITE081981 | Click to Show/Hide the Full List | ||
| mod site | chr7:116769715-116769716:+ | [9] | |
| Sequence | AATAAGAGCTGTGAGAATATACACTTACATTCTGAAGCCGT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000422097.1; ENST00000318493.11; ENST00000397752.7; ENST00000454623.1 | ||
| External Link | RMBase: m6A_site_773751 | ||
| mod ID: M6ASITE081982 | Click to Show/Hide the Full List | ||
| mod site | chr7:116769771-116769772:+ | [8] | |
| Sequence | CAATGACCTGCTGAAATTGAACAGCGAGCTAAATATAGAGG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000454623.1; ENST00000422097.1; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773752 | ||
| mod ID: M6ASITE081983 | Click to Show/Hide the Full List | ||
| mod site | chr7:116771965-116771966:+ | [8] | |
| Sequence | GGTTTCAAATGAATCTGTAGACTACCGAGCTACTTTTCCAG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000454623.1; ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773753 | ||
| mod ID: M6ASITE081984 | Click to Show/Hide the Full List | ||
| mod site | chr7:116774917-116774918:+ | [10] | |
| Sequence | TCTCAGAACGGTTCATGCCGACAAGTGCAGTATCCTCTGAC | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | HEK293; kidney; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000454623.1; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773754 | ||
| mod ID: M6ASITE081985 | Click to Show/Hide the Full List | ||
| mod site | chr7:116774940-116774941:+ | [8] | |
| Sequence | AGTGCAGTATCCTCTGACAGACATGTCCCCCATCCTAACTA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000454623.1; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773755 | ||
| mod ID: M6ASITE081986 | Click to Show/Hide the Full List | ||
| mod site | chr7:116774957-116774958:+ | [11] | |
| Sequence | CAGACATGTCCCCCATCCTAACTAGTGGGGACTCTGATATA | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000454623.1; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773756 | ||
| mod ID: M6ASITE081987 | Click to Show/Hide the Full List | ||
| mod site | chr7:116774967-116774968:+ | [8] | |
| Sequence | CCCCATCCTAACTAGTGGGGACTCTGATATATCCAGTCCAT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000454623.1; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773757 | ||
| mod ID: M6ASITE081988 | Click to Show/Hide the Full List | ||
| mod site | chr7:116777414-116777415:+ | [8] | |
| Sequence | TTGGTTGTGTATATCATGGGACTTTGTTGGACAATGATGGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773758 | ||
| mod ID: M6ASITE081989 | Click to Show/Hide the Full List | ||
| mod site | chr7:116777424-116777425:+ | [8] | |
| Sequence | ATATCATGGGACTTTGTTGGACAATGATGGCAAGAAAATTC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773759 | ||
| mod ID: M6ASITE081990 | Click to Show/Hide the Full List | ||
| mod site | chr7:116777466-116777467:+ | [8] | |
| Sequence | CTGTGCTGTGAAATCCTTGAACAGTAAGTGGCATTTTATTT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773760 | ||
| mod ID: M6ASITE081991 | Click to Show/Hide the Full List | ||
| mod site | chr7:116778784-116778785:+ | [9] | |
| Sequence | GGATTTCTCAGGAATCACTGACATAGGAGAAGTTTCCCAAT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773761 | ||
| mod ID: M6ASITE081992 | Click to Show/Hide the Full List | ||
| mod site | chr7:116778910-116778911:+ | [9] | |
| Sequence | TCCGCTGGTGGTCCTACCATACATGAAACATGGAGATCTTC | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773762 | ||
| mod ID: M6ASITE081993 | Click to Show/Hide the Full List | ||
| mod site | chr7:116778917-116778918:+ | [8] | |
| Sequence | GTGGTCCTACCATACATGAAACATGGAGATCTTCGAAATTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773763 | ||
| mod ID: M6ASITE081994 | Click to Show/Hide the Full List | ||
| mod site | chr7:116778951-116778952:+ | [8] | |
| Sequence | GAAATTTCATTCGAAATGAGACTCATGTAAGTTGACTGCCA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773764 | ||
| mod ID: M6ASITE081995 | Click to Show/Hide the Full List | ||
| mod site | chr7:116782069-116782070:+ | [11] | |
| Sequence | TGCAAGCAAAAAGTTTGTCCACAGAGACTTGGCTGCAAGAA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773765 | ||
| mod ID: M6ASITE081996 | Click to Show/Hide the Full List | ||
| mod site | chr7:116782075-116782076:+ | [8] | |
| Sequence | CAAAAAGTTTGTCCACAGAGACTTGGCTGCAAGAAACTGTA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773766 | ||
| mod ID: M6ASITE081997 | Click to Show/Hide the Full List | ||
| mod site | chr7:116782090-116782091:+ | [8] | |
| Sequence | CAGAGACTTGGCTGCAAGAAACTGTATGTAAGTATCAGAAT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773767 | ||
| mod ID: M6ASITE081998 | Click to Show/Hide the Full List | ||
| mod site | chr7:116783353-116783354:+ | [8] | |
| Sequence | TGATTTTGGTCTTGCCAGAGACATGTATGATAAAGAATACT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773768 | ||
| mod ID: M6ASITE081999 | Click to Show/Hide the Full List | ||
| mod site | chr7:116783391-116783392:+ | [8] | |
| Sequence | ACTATAGTGTACACAACAAAACAGGTGCAAAGCTGCCAGTG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773769 | ||
| mod ID: M6ASITE082000 | Click to Show/Hide the Full List | ||
| mod site | chr7:116783439-116783440:+ | [8] | |
| Sequence | TGGCTTTGGAAAGTCTGCAAACTCAAAAGTTTACCACCAAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773770 | ||
| mod ID: M6ASITE082001 | Click to Show/Hide the Full List | ||
| mod site | chr7:116795687-116795688:+ | [9] | |
| Sequence | TGCTCCTCTGGGAGCTGATGACAAGAGGAGCCCCACCTTAT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773771 | ||
| mod ID: M6ASITE082002 | Click to Show/Hide the Full List | ||
| mod site | chr7:116795718-116795719:+ | [8] | |
| Sequence | CCCACCTTATCCTGACGTAAACACCTTTGATATAACTGTTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773772 | ||
| mod ID: M6ASITE082003 | Click to Show/Hide the Full List | ||
| mod site | chr7:116795739-116795740:+ | [11] | |
| Sequence | CACCTTTGATATAACTGTTTACTTGTTGCAAGGGAGAAGAC | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773773 | ||
| mod ID: M6ASITE082004 | Click to Show/Hide the Full List | ||
| mod site | chr7:116795758-116795759:+ | [8] | |
| Sequence | TACTTGTTGCAAGGGAGAAGACTCCTACAACCCGAATACTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773774 | ||
| mod ID: M6ASITE082005 | Click to Show/Hide the Full List | ||
| mod site | chr7:116795784-116795785:+ | [8] | |
| Sequence | ACAACCCGAATACTGCCCAGACCCCTTGTAAGTAGTCTTTC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773775 | ||
| mod ID: M6ASITE082006 | Click to Show/Hide the Full List | ||
| mod site | chr7:116795946-116795947:+ | [8] | |
| Sequence | ATGCGCCCATCCTTTTCTGAACTGGTGTCCCGGATATCAGC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; U2OS; A549; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773776 | ||
| mod ID: M6ASITE082007 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796089-116796090:+ | [8] | |
| Sequence | TAACGCTGATGATGAGGTGGACACACGACCAGCCTCCTTCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; HepG2; U2OS; A549; HEK293A-TOA; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773777 | ||
| mod ID: M6ASITE082008 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796115-116796116:+ | [8] | |
| Sequence | GACCAGCCTCCTTCTGGGAGACATCATAGTGCTAGTACTAT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; A549; HEK293A-TOA; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773778 | ||
| mod ID: M6ASITE082009 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796154-116796155:+ | [11] | |
| Sequence | ATGTCAAAGCAACAGTCCACACTTTGTCCAATGGTTTTTTC | ||
| Motif Score | 2.506922619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773779 | ||
| mod ID: M6ASITE082010 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796227-116796228:+ | [11] | |
| Sequence | TTTGCTCTTGCCAAAATTGCACTATTATAGGACTTGTATTG | ||
| Motif Score | 3.252583333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773780 | ||
| mod ID: M6ASITE082011 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796238-116796239:+ | [8] | |
| Sequence | CAAAATTGCACTATTATAGGACTTGTATTGTTATTTAAATT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; fibroblasts; Huh7; HEK293A-TOA; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773781 | ||
| mod ID: M6ASITE082012 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796286-116796287:+ | [9] | |
| Sequence | TCTAAGGAATTTCTTATCTGACAGAGCATCAGAACCAGAGG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773782 | ||
| mod ID: M6ASITE082013 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796299-116796300:+ | [12] | |
| Sequence | TTATCTGACAGAGCATCAGAACCAGAGGCTTGGTCCCACAG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HEK293T; HeLa; A549; HepG2; fibroblasts; Huh7; HEK293A-TOA; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773783 | ||
| mod ID: M6ASITE082014 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796327-116796328:+ | [12] | |
| Sequence | CTTGGTCCCACAGGCCACGGACCAATGGCCTGCAGCCGTGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HEK293T; HeLa; A549; HepG2; fibroblasts; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773784 | ||
| mod ID: M6ASITE082015 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796350-116796351:+ | [9] | |
| Sequence | AATGGCCTGCAGCCGTGACAACACTCCTGTCATATTGGAGT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773785 | ||
| mod ID: M6ASITE082016 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796376-116796377:+ | [12] | |
| Sequence | CTGTCATATTGGAGTCCAAAACTTGAATTCTGGGTTGAATT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T; HeLa; A549; HepG2; fibroblasts; HEK293A-TOA; iSLK; TIME; endometrial; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773786 | ||
| mod ID: M6ASITE082017 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796474-116796475:+ | [8] | |
| Sequence | GGGCTTCTTGATCACAGAAAACTCAGAAGAGATAGTAATGC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773787 | ||
| mod ID: M6ASITE082018 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796500-116796501:+ | [8] | |
| Sequence | AAGAGATAGTAATGCTCAGGACAGGAGCGGCAGCCCCAGAA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773788 | ||
| mod ID: M6ASITE082019 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796520-116796521:+ | [8] | |
| Sequence | ACAGGAGCGGCAGCCCCAGAACAGGCCACTCATTTAGAATT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773789 | ||
| mod ID: M6ASITE082020 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796554-116796555:+ | [8] | |
| Sequence | TAGAATTCTAGTGTTTCAAAACACTTTTGTGTGTTGTATGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773790 | ||
| mod ID: M6ASITE082021 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796622-116796623:+ | [8] | |
| Sequence | GTCATTCACCCATTAGGTAAACATTCCCTTTTAAATGTTTG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773791 | ||
| mod ID: M6ASITE082022 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796656-116796657:+ | [8] | |
| Sequence | ATGTTTGTTTGTTTTTTGAGACAGGATCTCACTCTGTTGCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; kidney | ||
| Seq Type List | m6A-seq; m6A-REF-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773792 | ||
| mod ID: M6ASITE082023 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796754-116796755:+ | [8] | |
| Sequence | AAGCCTCCCGAATAGCTGGGACTACAGGCGCACACCACCAT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; A549 | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773793 | ||
| mod ID: M6ASITE082024 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796840-116796841:+ | [8] | |
| Sequence | GTTGCCAAGGCTGGTTTCAAACTCCTGGACTCAAGAAATCC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; A549 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773794 | ||
| mod ID: M6ASITE082025 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796848-116796849:+ | [8] | |
| Sequence | GGCTGGTTTCAAACTCCTGGACTCAAGAAATCCACCCACCT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; A549 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773795 | ||
| mod ID: M6ASITE082026 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796937-116796938:+ | [8] | |
| Sequence | CCTTATAAATTTTTGTATAGACATTCCTTTGGTTGGAAGAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773796 | ||
| mod ID: M6ASITE082027 | Click to Show/Hide the Full List | ||
| mod site | chr7:116796972-116796973:+ | [11] | |
| Sequence | GAAGAATATTTATAGGCAATACAGTCAAAGTTTCAAAATAG | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773797 | ||
| mod ID: M6ASITE082028 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797004-116797005:+ | [8] | |
| Sequence | TCAAAATAGCATCACACAAAACATGTTTATAAATGAACAGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773798 | ||
| mod ID: M6ASITE082029 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797020-116797021:+ | [8] | |
| Sequence | CAAAACATGTTTATAAATGAACAGGATGTAATGTACATAGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773799 | ||
| mod ID: M6ASITE082030 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797128-116797129:+ | [11] | |
| Sequence | CACTGTTTGAGAATGATGCTACTCTGATCTAATGAATGTGA | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773800 | ||
| mod ID: M6ASITE082031 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797149-116797150:+ | [8] | |
| Sequence | CTCTGATCTAATGAATGTGAACATGTAGATGTTTTGTGTGT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773801 | ||
| mod ID: M6ASITE082032 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797186-116797187:+ | [8] | |
| Sequence | GTGTATTTTTTTAAATGAAAACTCAAAATAAGACAAGTAAT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773802 | ||
| mod ID: M6ASITE082033 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797198-116797199:+ | [8] | |
| Sequence | AAATGAAAACTCAAAATAAGACAAGTAATTTGTTGATAAAT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T | ||
| Seq Type List | m6A-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773803 | ||
| mod ID: M6ASITE082034 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797232-116797233:+ | [11] | |
| Sequence | GATAAATATTTTTAAAGATAACTCAGCATGTTTGTAAAGCA | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773804 | ||
| mod ID: M6ASITE082035 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797381-116797382:+ | [8] | |
| Sequence | CACCTCGCAAGCAATTGGAAACAAAACTTTTGGGGAGTTTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773805 | ||
| mod ID: M6ASITE082036 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797386-116797387:+ | [8] | |
| Sequence | CGCAAGCAATTGGAAACAAAACTTTTGGGGAGTTTTATTTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773806 | ||
| mod ID: M6ASITE082037 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797436-116797437:+ | [8] | |
| Sequence | TGTGTTTTATGTTAAGCAAAACATACTTTAGAAACAAATGA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773807 | ||
| mod ID: M6ASITE082038 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797449-116797450:+ | [8] | |
| Sequence | AAGCAAAACATACTTTAGAAACAAATGAAAAAGGCAATTGA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773808 | ||
| mod ID: M6ASITE082039 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797515-116797516:+ | [8] | |
| Sequence | GAATAGCCACCCTGAGCAGAACTTTGTGATGCTTCATTCTG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773809 | ||
| mod ID: M6ASITE082040 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797554-116797555:+ | [11] | |
| Sequence | TGTGGAATTTTGTGCTTGCTACTGTATAGTGCATGTGGTGT | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773810 | ||
| mod ID: M6ASITE082041 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797605-116797606:+ | [8] | |
| Sequence | AACTGGTTTTGTCGACGTAAACATTTAAAGTGTTATATTTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773811 | ||
| mod ID: M6ASITE082042 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797680-116797681:+ | [8] | |
| Sequence | AATTTTGTTAGGCCACAAAAACACTGCACTGTGAACATTTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; fibroblasts; A549; Huh7; HEK293A-TOA; iSLK; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773812 | ||
| mod ID: M6ASITE082043 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797687-116797688:+ | [11] | |
| Sequence | TTAGGCCACAAAAACACTGCACTGTGAACATTTTAGAAAAG | ||
| Motif Score | 3.252583333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773813 | ||
| mod ID: M6ASITE082044 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797694-116797695:+ | [8] | |
| Sequence | ACAAAAACACTGCACTGTGAACATTTTAGAAAAGGTATGTC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; HepG2; fibroblasts; A549; Huh7; HEK293A-TOA; iSLK; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773814 | ||
| mod ID: M6ASITE082045 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797717-116797718:+ | [8] | |
| Sequence | TTTTAGAAAAGGTATGTCAGACTGGGATTAATGACAGCATG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; fibroblasts; A549; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773815 | ||
| mod ID: M6ASITE082046 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797730-116797731:+ | [9] | |
| Sequence | ATGTCAGACTGGGATTAATGACAGCATGATTTTCAATGACT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773816 | ||
| mod ID: M6ASITE082047 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797748-116797749:+ | [11] | |
| Sequence | TGACAGCATGATTTTCAATGACTGTAAATTGCGATAAGGAA | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T; A549 | ||
| Seq Type List | DART-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773817 | ||
| mod ID: M6ASITE082048 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797786-116797787:+ | [9] | |
| Sequence | GAAATGTACTGATTGCCAATACACCCCACCCTCATTACATC | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773818 | ||
| mod ID: M6ASITE082049 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797802-116797803:+ | [9] | |
| Sequence | CAATACACCCCACCCTCATTACATCATCAGGACTTGAAGCC | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773819 | ||
| mod ID: M6ASITE082050 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797813-116797814:+ | [8] | |
| Sequence | ACCCTCATTACATCATCAGGACTTGAAGCCAAGGGTTAACC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESCs; fibroblasts; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773820 | ||
| mod ID: M6ASITE082051 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797843-116797844:+ | [9] | |
| Sequence | AAGGGTTAACCCAGCAAGCTACAAAGAGGGTGTGTCACACT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773821 | ||
| mod ID: M6ASITE082052 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797859-116797860:+ | [9] | |
| Sequence | AGCTACAAAGAGGGTGTGTCACACTGAAACTCAATAGTTGA | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773822 | ||
| mod ID: M6ASITE082053 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797861-116797862:+ | [11] | |
| Sequence | CTACAAAGAGGGTGTGTCACACTGAAACTCAATAGTTGAGT | ||
| Motif Score | 2.506922619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773823 | ||
| mod ID: M6ASITE082054 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797867-116797868:+ | [8] | |
| Sequence | AGAGGGTGTGTCACACTGAAACTCAATAGTTGAGTTTGGCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; hESCs; fibroblasts; Huh7; HEK293A-TOA; iSLK; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773824 | ||
| mod ID: M6ASITE082055 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797911-116797912:+ | [13] | |
| Sequence | GTTGCAGGAAAATGATTATAACTAAAAGCTCTCTGATAGTG | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773825 | ||
| mod ID: M6ASITE082056 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797937-116797938:+ | [8] | |
| Sequence | AGCTCTCTGATAGTGCAGAGACTTACCAGAAGACACAAGGA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; fibroblasts; Huh7; HEK293A-TOA; iSLK; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773826 | ||
| mod ID: M6ASITE082057 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797949-116797950:+ | [8] | |
| Sequence | GTGCAGAGACTTACCAGAAGACACAAGGAATTGTACTGAAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; hESC-HEK293T; HepG2; fibroblasts; Huh7; HEK293A-TOA; iSLK; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773827 | ||
| mod ID: M6ASITE082058 | Click to Show/Hide the Full List | ||
| mod site | chr7:116797963-116797964:+ | [11] | |
| Sequence | CAGAAGACACAAGGAATTGTACTGAAGAGCTATTACAATCC | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773828 | ||
| mod ID: M6ASITE082059 | Click to Show/Hide the Full List | ||
| mod site | chr7:116798017-116798018:+ | [11] | |
| Sequence | TCATAAATGTAATAAGTAATACTAATTCACAGAGTATTGTA | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773829 | ||
| mod ID: M6ASITE082060 | Click to Show/Hide the Full List | ||
| mod site | chr7:116798049-116798050:+ | [9] | |
| Sequence | AGTATTGTAAATGGTGGATGACAAAAGAAAATCTGCTCTGT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773830 | ||
| mod ID: M6ASITE082061 | Click to Show/Hide the Full List | ||
| mod site | chr7:116798081-116798082:+ | [8] | |
| Sequence | CTGCTCTGTGGAAAGAAAGAACTGTCTCTACCAGGGTCAAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; H1B; hESCs; fibroblasts; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773831 | ||
| mod ID: M6ASITE082062 | Click to Show/Hide the Full List | ||
| mod site | chr7:116798126-116798127:+ | [8] | |
| Sequence | TGAACGCATCAATAGAAAGAACTCGGGGAAACATCCCATCA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1B; hESCs; fibroblasts; H1299; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773832 | ||
| mod ID: M6ASITE082063 | Click to Show/Hide the Full List | ||
| mod site | chr7:116798136-116798137:+ | [8] | |
| Sequence | AATAGAAAGAACTCGGGGAAACATCCCATCAACAGGACTAC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; brain; kidney; A549; hESC-HEK293T; U2OS; H1B; hESCs; fibroblasts; H1299; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773833 | ||
| mod ID: M6ASITE082064 | Click to Show/Hide the Full List | ||
| mod site | chr7:116798147-116798148:+ | [10] | |
| Sequence | CTCGGGGAAACATCCCATCAACAGGACTACACACTTGTATA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | brain; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773834 | ||
| mod ID: M6ASITE082065 | Click to Show/Hide the Full List | ||
| mod site | chr7:116798152-116798153:+ | [8] | |
| Sequence | GGAAACATCCCATCAACAGGACTACACACTTGTATATACAT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1B; hESCs; fibroblasts; H1299; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773835 | ||
| mod ID: M6ASITE082066 | Click to Show/Hide the Full List | ||
| mod site | chr7:116798181-116798182:+ | [8] | |
| Sequence | TTGTATATACATTCTTGAGAACACTGCAATGTGAAAATCAC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1B; hESCs; fibroblasts; H1299; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773836 | ||
| mod ID: M6ASITE082067 | Click to Show/Hide the Full List | ||
| mod site | chr7:116798217-116798218:+ | [8] | |
| Sequence | ATCACGTTTGCTATTTATAAACTTGTCCTTAGATTAATGTG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; hESCs; fibroblasts; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: m6A_site_773837 | ||
| mod ID: M6ASITE082068 | Click to Show/Hide the Full List | ||
| mod site | chr7:116798243-116798244:+ | [8] | |
| Sequence | CCTTAGATTAATGTGTCTGGACAGATTGTGGGAGTAAGTGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; hESC-HEK293T; HepG2; fibroblasts; Huh7; HEK293A-TOA; iSLK; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773838 | ||
| mod ID: M6ASITE082069 | Click to Show/Hide the Full List | ||
| mod site | chr7:116798310-116798311:+ | [8] | |
| Sequence | CTGCCTATACCTGCAGCTGAACTGAATGGTACTTCGTATGT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; Huh7; iSLK; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773839 | ||
| mod ID: M6ASITE082070 | Click to Show/Hide the Full List | ||
| mod site | chr7:116798320-116798321:+ | [11] | |
| Sequence | CTGCAGCTGAACTGAATGGTACTTCGTATGTTAATAGTTGT | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000318493.11; ENST00000397752.7 | ||
| External Link | RMBase: m6A_site_773840 | ||
2'-O-Methylation (2'-O-Me)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: 2OMSITE000464 | Click to Show/Hide the Full List | ||
| mod site | chr7:116782094-116782095:+ | [14] | |
| Sequence | GACTTGGCTGCAAGAAACTGTATGTAAGTATCAGAATCTCT | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000397752.7; ENST00000318493.11 | ||
| External Link | RMBase: Nm_site_6221 | ||
References