m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00213)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
CDKN1A
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
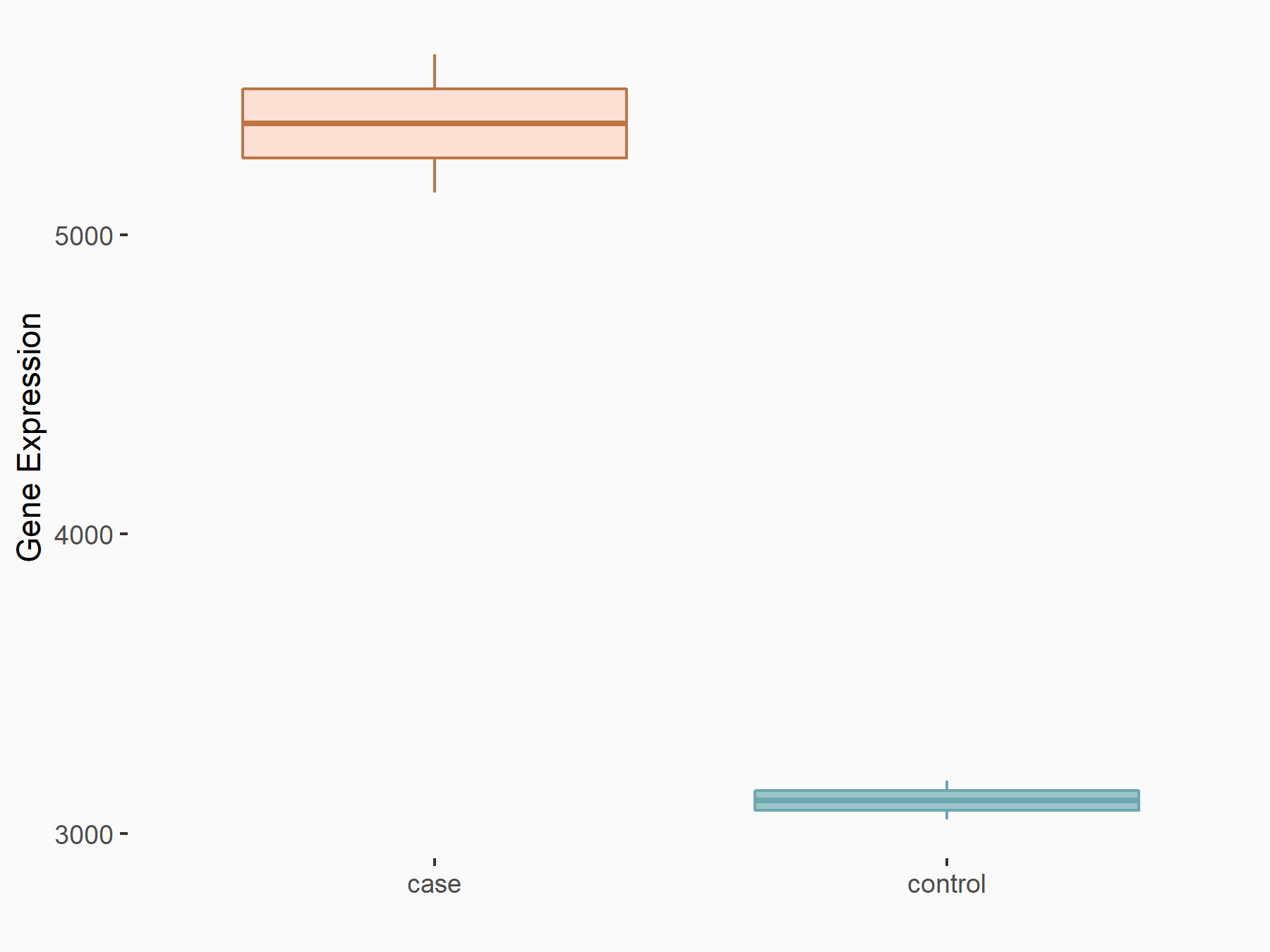  |
logFC: 7.88E-01 p-value: 3.07E-21 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between CDKN1A and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 9.39E+00 | GSE60213 |
| In total 3 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 and METTL14 play an oncogenic role in acute myeloid leukemia(AML) by targeting mdm2/p53 signal pathway. The knockdown of METTL3 and METTL14 in K562 cell line leads to several changes in the expression of p53 signal pathway, including the upregulation of p53, cyclin dependent kinase inhibitor 1A (CDKN1A/Cyclin-dependent kinase inhibitor 1 (CDKN1A)), and downregulation of mdm2. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Acute myeloid leukaemia | ICD-11: 2A60 | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Cell apoptosis | |||
| Cells in G4/M phase decreased | ||||
| In-vitro Model | THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| MOLT-4 | Adult T acute lymphoblastic leukemia | Homo sapiens | CVCL_0013 | |
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 | |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| CCRF-CEM C7 | T acute lymphoblastic leukemia | Homo sapiens | CVCL_6825 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 modulates the cell cycle of Esophageal squamous cell carcinoma(ESCC) cells through a Cyclin-dependent kinase inhibitor 1 (CDKN1A)-dependent pattern. METTL3-guided m6A modification contributes to the progression of ESCC via the p21-axis. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Esophageal squamous cell carcinoma | ICD-11: 2B70.1 | ||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Arrest cell cycle at G2/M phase | |||
| In-vitro Model | TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| In-vivo Model | Totally 5 × 106 ESCC cells after treatment were administered to randomized animals by the subcutaneous route (right flank) in 200 uL DMEM. Tumor measurement was performed every other day. At study end (at least 3 weeks), the animals underwent euthanasia, and the tumors were extracted for histology. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | METTL3 is able to promote breast cancer cell proliferation by regulating the Cyclin-dependent kinase inhibitor 1 (CDKN1A) expression by an m6A-dependent manner. Metformin can take p21 as the main target to inhibit such effect. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| SUM1315MO2 | Invasive breast carcinoma of no special type | Homo sapiens | CVCL_5589 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HBL-100 | Normal | Homo sapiens | CVCL_4362 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| In-vivo Model | About 1 × 107 stable METTL3 overexpression and negative control SUM-1315 cells were injected subcutaneously into the axilla of the female BALB/C nude mice (4-6 weeks old, 18-20 g, 10 mice/group). One week after injection, the two groups, METTL3 OE and NC, were then randomly allocated into the control group and experimental group (5 mice/group), which were treated with PBS or metformin (250 mg/kg/dose, respectively). PBS or metformin was administered every 2 days via intraperitoneal injection. | |||
Fat mass and obesity-associated protein (FTO) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by FTO | ||
| Cell Line | 253J cell line | Homo sapiens |
|
Treatment: siFTO 253J cells
Control: 253J cells
|
GSE150239 | |
| Regulation |
  |
logFC: 9.72E-01 p-value: 1.24E-24 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | The elevated FTO in esophageal squamous cell carcinoma decreased m6A methylation of LINC00022 transcript, leading to the inhibition of LINC00022 decay via the m6A reader YTHDF2. LINC00022 directly binds to Cyclin-dependent kinase inhibitor 1 (CDKN1A) protein and promotes its ubiquitination-mediated degradation, thereby facilitating cell-cycle progression and proliferation. | |||
| Responsed Disease | Esophageal squamous cell carcinoma | ICD-11: 2B70.1 | ||
| Pathway Response | Cell cycle | hsa04110 | ||
| Ubiquitin mediated proteolysis | hsa04120 | |||
| Cell Process | Ubiquitination degradation | |||
| Cell apoptosis | ||||
| Decreased G0/G1 phase | ||||
| In-vitro Model | HET-1A | Normal | Homo sapiens | CVCL_3702 |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| KYSE-450 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1353 | |
| KYSE-70 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1356 | |
| TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 | |
| In-vivo Model | The number of cells inoculated in each mouse was 4 × 106, 1 × 106, 2 × 106 and 1 × 106, respectively. | |||
Methyltransferase-like 14 (METTL14) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL14 | ||
| Cell Line | Neural progenitor cell line | Mus musculus |
|
Treatment: METTL14 knockout NPCs
Control: Wild type NPCs
|
GSE158985 | |
| Regulation |
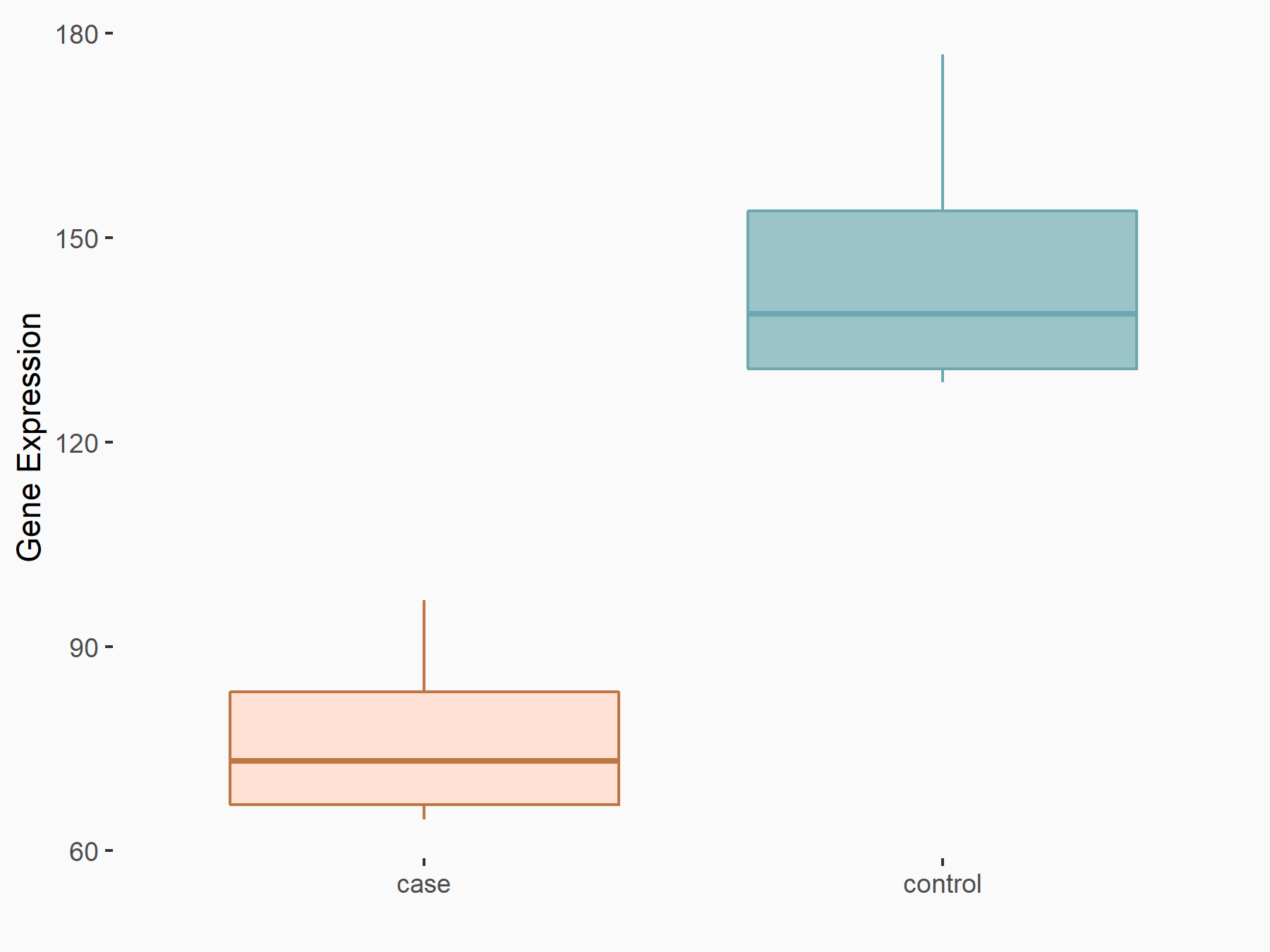  |
logFC: -9.21E-01 p-value: 1.04E-03 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 and METTL14 play an oncogenic role in acute myeloid leukemia(AML) by targeting mdm2/p53 signal pathway. The knockdown of METTL3 and METTL14 in K562 cell line leads to several changes in the expression of p53 signal pathway, including the upregulation of p53, cyclin dependent kinase inhibitor 1A (CDKN1A/Cyclin-dependent kinase inhibitor 1 (CDKN1A)), and downregulation of mdm2. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Acute myeloid leukaemia | ICD-11: 2A60 | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Cell apoptosis | |||
| Cells in G7/M phase decreased | ||||
| In-vitro Model | THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| MOLT-4 | Adult T acute lymphoblastic leukemia | Homo sapiens | CVCL_0013 | |
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 | |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| CCRF-CEM C7 | T acute lymphoblastic leukemia | Homo sapiens | CVCL_6825 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
RNA demethylase ALKBH5 (ALKBH5) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by ALKBH5 | ||
| Cell Line | CAG cell line | Homo sapiens |
|
Treatment: shALKBH5 CAG cells
Control: shNC CAG cells
|
GSE180214 | |
| Regulation |
  |
logFC: -1.90E+00 p-value: 1.35E-05 |
| More Results | Click to View More RNA-seq Results | |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [5] | |||
| Response Summary | Expression of CDKN1A (p21) was significantly up-regulated in ALKBH5-depleted cells, and m6 A modification and stability of Cyclin-dependent kinase inhibitor 1 (CDKN1A) mRNA were increased by ALKBH5 knockdown. Identify ALKBH5 as the first m6 A demethylase that accelerates cell cycle progression and promotes cell proliferation of ESCC cells, which is associated with poor prognosis of esophageal squamous cell carcinoma patients. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Esophageal squamous cell carcinoma | ICD-11: 2B70.1 | ||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Cell cycle | |||
| Cell apoptosis | ||||
| In-vitro Model | OE21 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_2661 |
| TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 | |
| TE-5 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1764 | |
| TE-9 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1767 | |
| In-vivo Model | The OE21 cells were stably transfected with sh-ALKBH5 (#1), sh-ALKBH5 (#2) or sh-scramble as negative control and injected (2 × 106 cells/mouse in 100 uL volume) subcutaneously into the back of male athymic BALB/c nude mice (6 weeks old, Japan SLC). | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | The expression of Cyclin-dependent kinase inhibitor 1 (CDKN1A) or TIMP3 was increased by ALKBH5 knockdown. In conclusions, the ALKBH5-IGF2BPs axis promotes cell proliferation and tumorigenicity, which in turn causes the unfavorable prognosis of NSCLC. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Arrest cell cycle at G1 phase | |||
| In-vitro Model | NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| NCI-H2087 | Lung adenocarcinoma | Homo sapiens | CVCL_1524 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| ABC-1 | Lung adenocarcinoma | Homo sapiens | CVCL_1066 | |
| NCI-H358 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1559 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| HEK293 | Normal | Homo sapiens | CVCL_0045 | |
| PC-3 [Human lung carcinoma] | Lung adenocarcinoma | Homo sapiens | CVCL_S982 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| RERF-LC-MS | Lung adenocarcinoma | Homo sapiens | CVCL_1655 | |
| HLC-1 | Lung adenocarcinoma | Homo sapiens | CVCL_5529 | |
| LC-2/ad | Lung adenocarcinoma | Homo sapiens | CVCL_1373 | |
YTH domain-containing protein 2 (YTHDC2) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [12] | |||
| Response Summary | Knockdown of YTHDC2 substantially promoted the proliferation rate of esophageal squamous cell carcinoma cells by affecting several cancer-related signaling pathways. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Esophageal squamous cell carcinoma | ICD-11: 2B70.1 | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| NF-kappa B signaling pathway | hsa04064 | |||
| JAK-STAT signaling pathway | hsa04630 | |||
| Cell Process | Genetic variants | |||
| In-vitro Model | KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 |
| KYSE-30 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1351 | |
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 and METTL14 play an oncogenic role in acute myeloid leukemia(AML) by targeting mdm2/p53 signal pathway. The knockdown of METTL3 and METTL14 in K562 cell line leads to several changes in the expression of p53 signal pathway, including the upregulation of p53, cyclin dependent kinase inhibitor 1A (CDKN1A/Cyclin-dependent kinase inhibitor 1 (CDKN1A)), and downregulation of mdm2. | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Cell apoptosis | |||
| Cells in G7/M phase decreased | ||||
| In-vitro Model | THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| MOLT-4 | Adult T acute lymphoblastic leukemia | Homo sapiens | CVCL_0013 | |
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 | |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| CCRF-CEM C7 | T acute lymphoblastic leukemia | Homo sapiens | CVCL_6825 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 and METTL14 play an oncogenic role in acute myeloid leukemia(AML) by targeting mdm2/p53 signal pathway. The knockdown of METTL3 and METTL14 in K562 cell line leads to several changes in the expression of p53 signal pathway, including the upregulation of p53, cyclin dependent kinase inhibitor 1A (CDKN1A/Cyclin-dependent kinase inhibitor 1 (CDKN1A)), and downregulation of mdm2. | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Cell apoptosis | |||
| Cells in G4/M phase decreased | ||||
| In-vitro Model | THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| MOLT-4 | Adult T acute lymphoblastic leukemia | Homo sapiens | CVCL_0013 | |
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 | |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| CCRF-CEM C7 | T acute lymphoblastic leukemia | Homo sapiens | CVCL_6825 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Esophageal cancer [ICD-11: 2B70]
| In total 4 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | The elevated FTO in esophageal squamous cell carcinoma decreased m6A methylation of LINC00022 transcript, leading to the inhibition of LINC00022 decay via the m6A reader YTHDF2. LINC00022 directly binds to Cyclin-dependent kinase inhibitor 1 (CDKN1A) protein and promotes its ubiquitination-mediated degradation, thereby facilitating cell-cycle progression and proliferation. | |||
| Responsed Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.1] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Pathway Response | Cell cycle | hsa04110 | ||
| Ubiquitin mediated proteolysis | hsa04120 | |||
| Cell Process | Ubiquitination degradation | |||
| Cell apoptosis | ||||
| Decreased G0/G1 phase | ||||
| In-vitro Model | HET-1A | Normal | Homo sapiens | CVCL_3702 |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| KYSE-450 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1353 | |
| KYSE-70 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1356 | |
| TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 | |
| In-vivo Model | The number of cells inoculated in each mouse was 4 × 106, 1 × 106, 2 × 106 and 1 × 106, respectively. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 modulates the cell cycle of Esophageal squamous cell carcinoma(ESCC) cells through a Cyclin-dependent kinase inhibitor 1 (CDKN1A)-dependent pattern. METTL3-guided m6A modification contributes to the progression of ESCC via the p21-axis. | |||
| Responsed Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.1] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Arrest cell cycle at G2/M phase | |||
| In-vitro Model | TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| In-vivo Model | Totally 5 × 106 ESCC cells after treatment were administered to randomized animals by the subcutaneous route (right flank) in 200 uL DMEM. Tumor measurement was performed every other day. At study end (at least 3 weeks), the animals underwent euthanasia, and the tumors were extracted for histology. | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [5] | |||
| Response Summary | Expression of CDKN1A (p21) was significantly up-regulated in ALKBH5-depleted cells, and m6 A modification and stability of Cyclin-dependent kinase inhibitor 1 (CDKN1A) mRNA were increased by ALKBH5 knockdown. Identify ALKBH5 as the first m6 A demethylase that accelerates cell cycle progression and promotes cell proliferation of ESCC cells, which is associated with poor prognosis of esophageal squamous cell carcinoma patients. | |||
| Responsed Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.1] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Cell cycle | |||
| Cell apoptosis | ||||
| In-vitro Model | OE21 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_2661 |
| TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 | |
| TE-5 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1764 | |
| TE-9 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1767 | |
| In-vivo Model | The OE21 cells were stably transfected with sh-ALKBH5 (#1), sh-ALKBH5 (#2) or sh-scramble as negative control and injected (2 × 106 cells/mouse in 100 uL volume) subcutaneously into the back of male athymic BALB/c nude mice (6 weeks old, Japan SLC). | |||
| Experiment 4 Reporting the m6A-centered Disease Response | [12] | |||
| Response Summary | Knockdown of YTHDC2 substantially promoted the proliferation rate of esophageal squamous cell carcinoma cells by affecting several cancer-related signaling pathways. | |||
| Responsed Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.1] | |||
| Target Regulator | YTH domain-containing protein 2 (YTHDC2) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| NF-kappa B signaling pathway | hsa04064 | |||
| JAK-STAT signaling pathway | hsa04630 | |||
| Cell Process | Genetic variants | |||
| In-vitro Model | KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 |
| KYSE-30 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1351 | |
Lung cancer [ICD-11: 2C25]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [] | |||
| Response Summary | In lung squamous cell carcinoma, seven m6A-related autophagy genes were screened to construct a prognostic model: CASP4, Cyclin-dependent kinase inhibitor 1 (CDKN1A), DLC1, ITGB1, PINK1, TP63, and EIF4EBP1. | |||
| Responsed Disease | Lung squamous cell carcinoma [ICD-11: 2C25.2] | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell proliferation and metastasis | |||
| Cell autophagy | ||||
| Experiment 2 Reporting the m6A-centered Disease Response | [6] | |||
| Response Summary | The expression of Cyclin-dependent kinase inhibitor 1 (CDKN1A) or TIMP3 was increased by ALKBH5 knockdown. In conclusions, the ALKBH5-IGF2BPs axis promotes cell proliferation and tumorigenicity, which in turn causes the unfavorable prognosis of NSCLC. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Arrest cell cycle at G1 phase | |||
| In-vitro Model | NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| NCI-H2087 | Lung adenocarcinoma | Homo sapiens | CVCL_1524 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| ABC-1 | Lung adenocarcinoma | Homo sapiens | CVCL_1066 | |
| NCI-H358 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1559 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| HEK293 | Normal | Homo sapiens | CVCL_0045 | |
| PC-3 [Human lung carcinoma] | Lung adenocarcinoma | Homo sapiens | CVCL_S982 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| RERF-LC-MS | Lung adenocarcinoma | Homo sapiens | CVCL_1655 | |
| HLC-1 | Lung adenocarcinoma | Homo sapiens | CVCL_5529 | |
| LC-2/ad | Lung adenocarcinoma | Homo sapiens | CVCL_1373 | |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | METTL3 is able to promote breast cancer cell proliferation by regulating the Cyclin-dependent kinase inhibitor 1 (CDKN1A) expression by an m6A-dependent manner. Metformin can take p21 as the main target to inhibit such effect. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| SUM1315MO2 | Invasive breast carcinoma of no special type | Homo sapiens | CVCL_5589 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HBL-100 | Normal | Homo sapiens | CVCL_4362 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| In-vivo Model | About 1 × 107 stable METTL3 overexpression and negative control SUM-1315 cells were injected subcutaneously into the axilla of the female BALB/C nude mice (4-6 weeks old, 18-20 g, 10 mice/group). One week after injection, the two groups, METTL3 OE and NC, were then randomly allocated into the control group and experimental group (5 mice/group), which were treated with PBS or metformin (250 mg/kg/dose, respectively). PBS or metformin was administered every 2 days via intraperitoneal injection. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
RNA modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00001 | ||
| Epigenetic Regulator | RNA cytosine C(5)-methyltransferase NSUN2 (NSUN2) | |
| Regulated Target | Cyclin-dependent kinase inhibitor 1 (CDKN1A) | |
| Crosstalk relationship | m5C → m6A | |
m6A Regulator: Methyltransferase-like 14 (METTL14)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00002 | ||
| Epigenetic Regulator | RNA cytosine C(5)-methyltransferase NSUN2 (NSUN2) | |
| Regulated Target | Cyclin-dependent kinase inhibitor 1 (CDKN1A) | |
| Crosstalk relationship | m5C → m6A | |
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03372 | ||
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Esophageal Squamous Cell Carcinoma | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00213)
| In total 6 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE001436 | Click to Show/Hide the Full List | ||
| mod site | chr6:36682188-36682189:+ | [13] | |
| Sequence | CTTATGAACTGTGTGATGTTAGGCAAGTTACTTAACTTATC | ||
| Transcript ID List | ENST00000405375.5; ENST00000459970.1; ENST00000615513.4; ENST00000244741.9; ENST00000373711.3; ENST00000448526.6; ENST00000478800.1; rmsk_1919667 | ||
| External Link | RMBase: RNA-editing_site_115776 | ||
| mod ID: A2ISITE001437 | Click to Show/Hide the Full List | ||
| mod site | chr6:36682197-36682198:+ | [13] | |
| Sequence | TGTGTGATGTTAGGCAAGTTACTTAACTTATCTGAGCCTTG | ||
| Transcript ID List | ENST00000615513.4; ENST00000244741.9; ENST00000448526.6; ENST00000459970.1; rmsk_1919667; ENST00000373711.3; ENST00000478800.1; ENST00000405375.5 | ||
| External Link | RMBase: RNA-editing_site_115777 | ||
| mod ID: A2ISITE001438 | Click to Show/Hide the Full List | ||
| mod site | chr6:36682206-36682207:+ | [13] | |
| Sequence | TTAGGCAAGTTACTTAACTTATCTGAGCCTTGGTTGCCTCT | ||
| Transcript ID List | ENST00000448526.6; ENST00000478800.1; ENST00000459970.1; ENST00000244741.9; ENST00000405375.5; ENST00000615513.4; ENST00000373711.3; rmsk_1919667 | ||
| External Link | RMBase: RNA-editing_site_115778 | ||
| mod ID: A2ISITE001439 | Click to Show/Hide the Full List | ||
| mod site | chr6:36683265-36683266:+ | [13] | |
| Sequence | GAGGAAACTAGAGCTCAGACAAGTTAAGTTGCTTGCCCAGG | ||
| Transcript ID List | ENST00000373711.3; ENST00000405375.5; ENST00000615513.4; ENST00000478800.1; ENST00000459970.1; ENST00000448526.6; ENST00000244741.9 | ||
| External Link | RMBase: RNA-editing_site_115779 | ||
| mod ID: A2ISITE001440 | Click to Show/Hide the Full List | ||
| mod site | chr6:36683266-36683267:+ | [13] | |
| Sequence | AGGAAACTAGAGCTCAGACAAGTTAAGTTGCTTGCCCAGGG | ||
| Transcript ID List | ENST00000478800.1; ENST00000459970.1; ENST00000615513.4; ENST00000448526.6; ENST00000244741.9; ENST00000373711.3; ENST00000405375.5 | ||
| External Link | RMBase: RNA-editing_site_115780 | ||
| mod ID: A2ISITE001441 | Click to Show/Hide the Full List | ||
| mod site | chr6:36684876-36684877:+ | [13] | |
| Sequence | GTCTTAACTCTGTTGGCCAGACTGGAGTGCAGTGATACGAT | ||
| Transcript ID List | ENST00000448526.6; ENST00000373711.3; ENST00000615513.4; ENST00000405375.5; ENST00000244741.9 | ||
| External Link | RMBase: RNA-editing_site_115781 | ||
N6-methyladenosine (m6A)
| In total 46 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE074863 | Click to Show/Hide the Full List | ||
| mod site | chr6:36676546-36676547:+ | [14] | |
| Sequence | GAGGAAGGGGATGGTAGGAGACAGGAGACCTCTAAAGACCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000448526.6; ENST00000615513.4; ENST00000459970.1 | ||
| External Link | RMBase: m6A_site_715130 | ||
| mod ID: M6ASITE074864 | Click to Show/Hide the Full List | ||
| mod site | chr6:36676553-36676554:+ | [14] | |
| Sequence | GGGATGGTAGGAGACAGGAGACCTCTAAAGACCCCAGGTAA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000459970.1; ENST00000615513.4; ENST00000448526.6 | ||
| External Link | RMBase: m6A_site_715131 | ||
| mod ID: M6ASITE074865 | Click to Show/Hide the Full List | ||
| mod site | chr6:36676563-36676564:+ | [14] | |
| Sequence | GAGACAGGAGACCTCTAAAGACCCCAGGTAAACCTTAGCCT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000615513.4; ENST00000448526.6; ENST00000459970.1 | ||
| External Link | RMBase: m6A_site_715132 | ||
| mod ID: M6ASITE074866 | Click to Show/Hide the Full List | ||
| mod site | chr6:36677795-36677796:+ | [14] | |
| Sequence | GATCTGAGTTAGGTCACCAGACTTCTCTGAGCCCCAGTTTC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000459970.1; rmsk_1919658; ENST00000448526.6; ENST00000615513.4 | ||
| External Link | RMBase: m6A_site_715133 | ||
| mod ID: M6ASITE074867 | Click to Show/Hide the Full List | ||
| mod site | chr6:36677856-36677857:+ | [14] | |
| Sequence | ATGTGGGGAGTATTCAGGAGACAGACAACTCACTCGTCAAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000448526.6; ENST00000459970.1; ENST00000615513.4 | ||
| External Link | RMBase: m6A_site_715134 | ||
| mod ID: M6ASITE074868 | Click to Show/Hide the Full List | ||
| mod site | chr6:36678816-36678817:+ | [14] | |
| Sequence | AGGTGAGAGAGCGGCGGCAGACAACAGGGGACCCCGGGCCG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; A549; MT4; CD4T; peripheral-blood; GSC-11; HEK293T; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000478800.1; ENST00000459970.1; ENST00000615513.4; ENST00000373711.3; ENST00000405375.5; ENST00000462537.3; ENST00000448526.6; ENST00000244741.9 | ||
| External Link | RMBase: m6A_site_715135 | ||
| mod ID: M6ASITE074869 | Click to Show/Hide the Full List | ||
| mod site | chr6:36678826-36678827:+ | [14] | |
| Sequence | GCGGCGGCAGACAACAGGGGACCCCGGGCCGGCGGCCCAGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; A549; MT4; CD4T; peripheral-blood; GSC-11; HEK293T; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000615513.4; ENST00000478800.1; ENST00000373711.3; ENST00000459970.1; ENST00000462537.3; ENST00000405375.5; ENST00000448526.6; ENST00000244741.9 | ||
| External Link | RMBase: m6A_site_715136 | ||
| mod ID: M6ASITE074870 | Click to Show/Hide the Full List | ||
| mod site | chr6:36680530-36680531:+ | [14] | |
| Sequence | CGAGCAGCGCCTTGCAGGAAACTGACTCATCACTACTCCCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; GSC-11 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000615513.4; ENST00000405375.5; ENST00000244741.9; ENST00000459970.1; ENST00000462537.3; ENST00000373711.3; ENST00000448526.6; ENST00000478800.1 | ||
| External Link | RMBase: m6A_site_715137 | ||
| mod ID: M6ASITE074871 | Click to Show/Hide the Full List | ||
| mod site | chr6:36684109-36684110:+ | [14] | |
| Sequence | GCTGCAGGCGCCATGTCAGAACCGGCTGGGGATGTCCGTCA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; A549; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000615513.4; ENST00000459970.1; ENST00000448526.6; ENST00000405375.5; ENST00000373711.3; ENST00000244741.9; ENST00000478800.1 | ||
| External Link | RMBase: m6A_site_715138 | ||
| mod ID: M6ASITE074872 | Click to Show/Hide the Full List | ||
| mod site | chr6:36684132-36684133:+ | [14] | |
| Sequence | GGCTGGGGATGTCCGTCAGAACCCATGCGGCAGCAAGGCCT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; A549; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000373711.3; ENST00000478800.1; ENST00000459970.1; ENST00000615513.4; ENST00000448526.6; ENST00000405375.5; ENST00000244741.9 | ||
| External Link | RMBase: m6A_site_715139 | ||
| mod ID: M6ASITE074873 | Click to Show/Hide the Full List | ||
| mod site | chr6:36684177-36684178:+ | [14] | |
| Sequence | CCGCCTCTTCGGCCCAGTGGACAGCGAGCAGCTGAGCCGCG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; A549; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; TIME; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000244741.9; ENST00000373711.3; ENST00000459970.1; ENST00000478800.1; ENST00000615513.4; ENST00000405375.5; ENST00000448526.6 | ||
| External Link | RMBase: m6A_site_715140 | ||
| mod ID: M6ASITE074874 | Click to Show/Hide the Full List | ||
| mod site | chr6:36684249-36684250:+ | [14] | |
| Sequence | GGAGGCCCGTGAGCGATGGAACTTCGACTTTGTCACCGAGA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; A549; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; TIME; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000244741.9; ENST00000615513.4; ENST00000405375.5; ENST00000459970.1; ENST00000478800.1; ENST00000448526.6; ENST00000373711.3 | ||
| External Link | RMBase: m6A_site_715141 | ||
| mod ID: M6ASITE074875 | Click to Show/Hide the Full List | ||
| mod site | chr6:36684269-36684270:+ | [14] | |
| Sequence | ACTTCGACTTTGTCACCGAGACACCACTGGAGGGTGACTTC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; MT4; A549; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; TIME; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000478800.1; ENST00000448526.6; ENST00000615513.4; ENST00000459970.1; ENST00000405375.5; ENST00000373711.3; ENST00000244741.9 | ||
| External Link | RMBase: m6A_site_715142 | ||
| mod ID: M6ASITE074876 | Click to Show/Hide the Full List | ||
| mod site | chr6:36684285-36684286:+ | [15] | |
| Sequence | CGAGACACCACTGGAGGGTGACTTCGCCTGGGAGCGTGTGC | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000615513.4; ENST00000478800.1; ENST00000373711.3; ENST00000459970.1; ENST00000448526.6; ENST00000405375.5; ENST00000244741.9 | ||
| External Link | RMBase: m6A_site_715143 | ||
| mod ID: M6ASITE074877 | Click to Show/Hide the Full List | ||
| mod site | chr6:36684413-36684414:+ | [14] | |
| Sequence | CACCTGCTCTGCTGCAGGGGACAGCAGAGGAAGACCATGTG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; A549; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; HEK293A-TOA; TIME; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000405375.5; ENST00000373711.3; ENST00000615513.4; ENST00000459970.1; ENST00000448526.6; ENST00000244741.9; ENST00000478800.1 | ||
| External Link | RMBase: m6A_site_715144 | ||
| mod ID: M6ASITE074878 | Click to Show/Hide the Full List | ||
| mod site | chr6:36684426-36684427:+ | [14] | |
| Sequence | GCAGGGGACAGCAGAGGAAGACCATGTGGACCTGTCACTGT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; A549; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; HEK293A-TOA; TIME; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000448526.6; ENST00000405375.5; ENST00000478800.1; ENST00000244741.9; ENST00000615513.4; ENST00000373711.3; ENST00000459970.1 | ||
| External Link | RMBase: m6A_site_715145 | ||
| mod ID: M6ASITE074879 | Click to Show/Hide the Full List | ||
| mod site | chr6:36684435-36684436:+ | [14] | |
| Sequence | AGCAGAGGAAGACCATGTGGACCTGTCACTGTCTTGTACCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; A549; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; HEK293A-TOA; TIME; endometrial; HEC-1-A; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000373711.3; ENST00000244741.9; ENST00000615513.4; ENST00000478800.1; ENST00000459970.1; ENST00000405375.5; ENST00000448526.6 | ||
| External Link | RMBase: m6A_site_715146 | ||
| mod ID: M6ASITE074880 | Click to Show/Hide the Full List | ||
| mod site | chr6:36684499-36684500:+ | [14] | |
| Sequence | GCTGAAGGGTCCCCAGGTGGACCTGGAGACTCTCAGGGTCG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; A549; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; TIME; endometrial; HEC-1-A; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000244741.9; ENST00000459970.1; ENST00000405375.5; ENST00000373711.3; ENST00000615513.4; ENST00000448526.6 | ||
| External Link | RMBase: m6A_site_715147 | ||
| mod ID: M6ASITE074881 | Click to Show/Hide the Full List | ||
| mod site | chr6:36684507-36684508:+ | [14] | |
| Sequence | GTCCCCAGGTGGACCTGGAGACTCTCAGGGTCGAAAACGGC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; A549; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; TIME; endometrial; HEC-1-A; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000615513.4; ENST00000448526.6; ENST00000405375.5; ENST00000459970.1; ENST00000373711.3; ENST00000244741.9 | ||
| External Link | RMBase: m6A_site_715148 | ||
| mod ID: M6ASITE074882 | Click to Show/Hide the Full List | ||
| mod site | chr6:36684533-36684534:+ | [14] | |
| Sequence | AGGGTCGAAAACGGCGGCAGACCAGCATGACAGGTGCGGAC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; A549; CD4T; peripheral-blood; GSC-11; HEK293T; TIME; endometrial; HEC-1-A; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000405375.5; ENST00000448526.6; ENST00000244741.9; ENST00000373711.3; ENST00000615513.4 | ||
| External Link | RMBase: m6A_site_715149 | ||
| mod ID: M6ASITE074883 | Click to Show/Hide the Full List | ||
| mod site | chr6:36685856-36685857:+ | [16] | |
| Sequence | GGGCCTCAAAGGCCCGCTCTACATCTTCTGCCTTAGTCTCA | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000615513.4; ENST00000244741.9; ENST00000405375.5; ENST00000373711.3; ENST00000448526.6 | ||
| External Link | RMBase: m6A_site_715150 | ||
| mod ID: M6ASITE074884 | Click to Show/Hide the Full List | ||
| mod site | chr6:36685915-36685916:+ | [17] | |
| Sequence | TTATTTGTGTTTTAATTTAAACACCTCCTCATGTACATACC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000405375.5; ENST00000448526.6; ENST00000615513.4; ENST00000373711.3; ENST00000244741.9 | ||
| External Link | RMBase: m6A_site_715151 | ||
| mod ID: M6ASITE074885 | Click to Show/Hide the Full List | ||
| mod site | chr6:36685929-36685930:+ | [17] | |
| Sequence | ATTTAAACACCTCCTCATGTACATACCCTGGCCGCCCCCTG | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000615513.4; ENST00000448526.6; ENST00000244741.9; ENST00000405375.5 | ||
| External Link | RMBase: m6A_site_715152 | ||
| mod ID: M6ASITE074886 | Click to Show/Hide the Full List | ||
| mod site | chr6:36685981-36685982:+ | [14] | |
| Sequence | CTGGCATTAGAATTATTTAAACAAAAACTAGGCGGTTGAAT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T | ||
| Seq Type List | m6A-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000244741.9; ENST00000405375.5; ENST00000615513.4; ENST00000448526.6 | ||
| External Link | RMBase: m6A_site_715153 | ||
| mod ID: M6ASITE074887 | Click to Show/Hide the Full List | ||
| mod site | chr6:36685987-36685988:+ | [14] | |
| Sequence | TTAGAATTATTTAAACAAAAACTAGGCGGTTGAATGAGAGG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000405375.5; ENST00000448526.6; ENST00000615513.4; ENST00000244741.9 | ||
| External Link | RMBase: m6A_site_715154 | ||
| mod ID: M6ASITE074888 | Click to Show/Hide the Full List | ||
| mod site | chr6:36686044-36686045:+ | [17] | |
| Sequence | GCATTTTTATTTTATGAAATACTATTTAAAGCCTCCTCATC | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000448526.6; ENST00000244741.9; ENST00000405375.5; ENST00000615513.4 | ||
| External Link | RMBase: m6A_site_715155 | ||
| mod ID: M6ASITE074889 | Click to Show/Hide the Full List | ||
| mod site | chr6:36686153-36686154:+ | [17] | |
| Sequence | CCACTTGTCCGCTGGGTGGTACCCTCTGGAGGGGTGTGGCT | ||
| Motif Score | 2.8355 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000615513.4; ENST00000244741.9; ENST00000405375.5; ENST00000448526.6 | ||
| External Link | RMBase: m6A_site_715156 | ||
| mod ID: M6ASITE074890 | Click to Show/Hide the Full List | ||
| mod site | chr6:36686190-36686191:+ | [17] | |
| Sequence | GGCTCCTTCCCATCGCTGTCACAGGCGGTTATGAAATTCAC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000244741.9; ENST00000615513.4; ENST00000448526.6; ENST00000405375.5 | ||
| External Link | RMBase: m6A_site_715157 | ||
| mod ID: M6ASITE074891 | Click to Show/Hide the Full List | ||
| mod site | chr6:36686260-36686261:+ | [17] | |
| Sequence | CTTTTTCATTTGAGAAGTAAACAGATGGCACTTTGAAGGGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000244741.9; ENST00000615513.4; ENST00000405375.5; ENST00000448526.6 | ||
| External Link | RMBase: m6A_site_715158 | ||
| mod ID: M6ASITE074892 | Click to Show/Hide the Full List | ||
| mod site | chr6:36686269-36686270:+ | [17] | |
| Sequence | TTGAGAAGTAAACAGATGGCACTTTGAAGGGGCCTCACCGA | ||
| Motif Score | 3.252583333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000244741.9; ENST00000615513.4; ENST00000405375.5; ENST00000448526.6 | ||
| External Link | RMBase: m6A_site_715159 | ||
| mod ID: M6ASITE074893 | Click to Show/Hide the Full List | ||
| mod site | chr6:36686308-36686309:+ | [14] | |
| Sequence | GAGTGGGGGCATCATCAAAAACTTTGGAGTCCCCTCACCTC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000615513.4; ENST00000448526.6; ENST00000405375.5; ENST00000244741.9 | ||
| External Link | RMBase: m6A_site_715160 | ||
| mod ID: M6ASITE074894 | Click to Show/Hide the Full List | ||
| mod site | chr6:36686385-36686386:+ | [14] | |
| Sequence | AGCCTAGGGCTGAGCTGGGGACCTGGTACCCTCCTGGCTCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000448526.6; ENST00000405375.5; ENST00000244741.9; ENST00000615513.4 | ||
| External Link | RMBase: m6A_site_715161 | ||
| mod ID: M6ASITE074895 | Click to Show/Hide the Full List | ||
| mod site | chr6:36686459-36686460:+ | [14] | |
| Sequence | AAGGTGGGGTCCTGGAGCAGACCACCCCGCCTGCCCTCATG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000405375.5; ENST00000244741.9; ENST00000615513.4; ENST00000448526.6 | ||
| External Link | RMBase: m6A_site_715162 | ||
| mod ID: M6ASITE074896 | Click to Show/Hide the Full List | ||
| mod site | chr6:36686699-36686700:+ | [18] | |
| Sequence | CCCCACCCCCAGCTCAATGGACTGGAAGGGGAAGGGACACA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | MT4 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000405375.5; ENST00000615513.4; ENST00000448526.6; ENST00000244741.9 | ||
| External Link | RMBase: m6A_site_715163 | ||
| mod ID: M6ASITE074897 | Click to Show/Hide the Full List | ||
| mod site | chr6:36686715-36686716:+ | [18] | |
| Sequence | ATGGACTGGAAGGGGAAGGGACACACAAGAAGAAGGGCACC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | MT4 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000244741.9; ENST00000615513.4; ENST00000405375.5; ENST00000448526.6 | ||
| External Link | RMBase: m6A_site_715164 | ||
| mod ID: M6ASITE074898 | Click to Show/Hide the Full List | ||
| mod site | chr6:36686719-36686720:+ | [16] | |
| Sequence | ACTGGAAGGGGAAGGGACACACAAGAAGAAGGGCACCCTAG | ||
| Motif Score | 2.084928571 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000448526.6; ENST00000244741.9; ENST00000615513.4; ENST00000405375.5 | ||
| External Link | RMBase: m6A_site_715165 | ||
| mod ID: M6ASITE074899 | Click to Show/Hide the Full List | ||
| mod site | chr6:36686885-36686886:+ | [17] | |
| Sequence | TAGATCTTTCTAGGAGGGAGACACTGGCCCCTCAAATCGTC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000405375.5; ENST00000244741.9; ENST00000448526.6; ENST00000615513.4 | ||
| External Link | RMBase: m6A_site_715166 | ||
| mod ID: M6ASITE074900 | Click to Show/Hide the Full List | ||
| mod site | chr6:36686949-36686950:+ | [17] | |
| Sequence | ATCCCTCCCCAGTTCATTGCACTTTGATTAGCAGCGGAACA | ||
| Motif Score | 3.252583333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000244741.9; ENST00000615513.4; ENST00000448526.6; ENST00000405375.5 | ||
| External Link | RMBase: m6A_site_715167 | ||
| mod ID: M6ASITE074901 | Click to Show/Hide the Full List | ||
| mod site | chr6:36686967-36686968:+ | [16] | |
| Sequence | GCACTTTGATTAGCAGCGGAACAAGGAGTCAGACATTTTAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000615513.4; ENST00000405375.5; ENST00000448526.6; ENST00000244741.9 | ||
| External Link | RMBase: m6A_site_715168 | ||
| mod ID: M6ASITE074902 | Click to Show/Hide the Full List | ||
| mod site | chr6:36687132-36687133:+ | [16] | |
| Sequence | GTATTGGGGTCTGACCCCAAACACCTTCCAGCTCCTGTAAC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T; HeLa; peripheral-blood | ||
| Seq Type List | MAZTER-seq; m6A-seq | ||
| Transcript ID List | ENST00000405375.5; ENST00000244741.9; ENST00000448526.6; ENST00000615513.4 | ||
| External Link | RMBase: m6A_site_715169 | ||
| mod ID: M6ASITE074903 | Click to Show/Hide the Full List | ||
| mod site | chr6:36687151-36687152:+ | [16] | |
| Sequence | AACACCTTCCAGCTCCTGTAACATACTGGCCTGGACTGTTT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000244741.9; ENST00000405375.5; ENST00000448526.6; ENST00000615513.4 | ||
| External Link | RMBase: m6A_site_715170 | ||
| mod ID: M6ASITE074904 | Click to Show/Hide the Full List | ||
| mod site | chr6:36687165-36687166:+ | [19] | |
| Sequence | CCTGTAACATACTGGCCTGGACTGTTTTCTCTCGGCTCCCC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000615513.4; ENST00000405375.5; ENST00000448526.6; ENST00000244741.9 | ||
| External Link | RMBase: m6A_site_715171 | ||
| mod ID: M6ASITE074905 | Click to Show/Hide the Full List | ||
| mod site | chr6:36687216-36687217:+ | [19] | |
| Sequence | GTTCCCGTTTCTCCACCTAGACTGTAAACCTCTCGAGGGCA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000244741.9; rmsk_1919675; ENST00000448526.6; ENST00000615513.4; ENST00000405375.5 | ||
| External Link | RMBase: m6A_site_715172 | ||
| mod ID: M6ASITE074906 | Click to Show/Hide the Full List | ||
| mod site | chr6:36687223-36687224:+ | [14] | |
| Sequence | TTTCTCCACCTAGACTGTAAACCTCTCGAGGGCAGGGACCA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000448526.6; rmsk_1919675; ENST00000615513.4; ENST00000405375.5; ENST00000244741.9 | ||
| External Link | RMBase: m6A_site_715173 | ||
| mod ID: M6ASITE074907 | Click to Show/Hide the Full List | ||
| mod site | chr6:36687240-36687241:+ | [14] | |
| Sequence | TAAACCTCTCGAGGGCAGGGACCACACCCTGTACTGTTCTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000615513.4; ENST00000405375.5; ENST00000448526.6; ENST00000244741.9; rmsk_1919675 | ||
| External Link | RMBase: m6A_site_715174 | ||
| mod ID: M6ASITE074908 | Click to Show/Hide the Full List | ||
| mod site | chr6:36687243-36687244:+ | [16] | |
| Sequence | ACCTCTCGAGGGCAGGGACCACACCCTGTACTGTTCTGTGT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | rmsk_1919675; ENST00000405375.5; ENST00000615513.4; ENST00000244741.9; ENST00000448526.6 | ||
| External Link | RMBase: m6A_site_715175 | ||
References