m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00271)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
HDGF
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
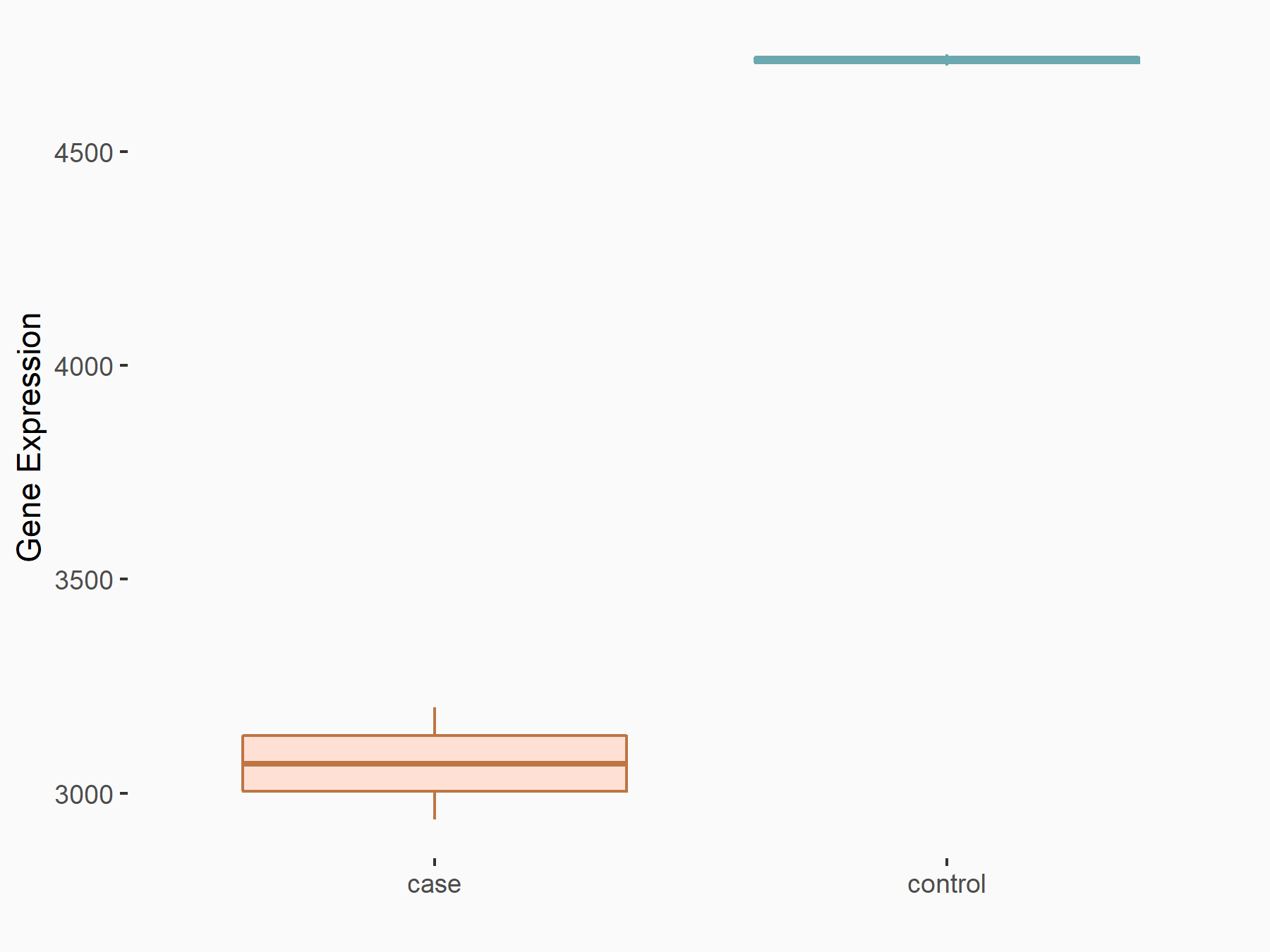  |
logFC: -6.20E-01 p-value: 6.85E-14 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between HDGF and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.03E+00 | GSE60213 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Elevated METTL3 expression promotes tumour angiogenesis and glycolysis in Gastric cancer. P300-mediated H3K27 acetylation activation in the promoter of METTL3 induced METTL3 transcription, which stimulated m6A modification of Hepatoma-derived growth factor (HDGF) mRNA, and the m6A reader IGF2BP3 then directly recognised and bound to the m6A site on HDGF mRNA and enhanced HDGF mRNA stability. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glycolysis | |||
| In-vitro Model | HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 |
| NCI-N87 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_1603 | |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| In-vivo Model | Mice 8 weeks after splenic portal vein injection of BGC823 cells with METTL3 overexpression or vector-transfected cells. | |||
Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by IGF2BP3 | ||
| Cell Line | ES-2 cell line | Homo sapiens |
|
Treatment: siIGF2BP3 ES-2 cells
Control: siControl ES-2 cells
|
GSE109604 | |
| Regulation |
  |
logFC: -6.81E-01 p-value: 1.47E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Elevated METTL3 expression promotes tumour angiogenesis and glycolysis in Gastric cancer. P300-mediated H3K27 acetylation activation in the promoter of METTL3 induced METTL3 transcription, which stimulated m6A modification of Hepatoma-derived growth factor (HDGF) mRNA, and the m6A reader IGF2BP3 then directly recognised and bound to the m6A site on HDGF mRNA and enhanced HDGF mRNA stability. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glycolysis | |||
| In-vitro Model | HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 |
| NCI-N87 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_1603 | |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| In-vivo Model | Mice 8 weeks after splenic portal vein injection of BGC823 cells with METTL3 overexpression or vector-transfected cells. | |||
Gastric cancer [ICD-11: 2B72]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Elevated METTL3 expression promotes tumour angiogenesis and glycolysis in Gastric cancer. P300-mediated H3K27 acetylation activation in the promoter of METTL3 induced METTL3 transcription, which stimulated m6A modification of Hepatoma-derived growth factor (HDGF) mRNA, and the m6A reader IGF2BP3 then directly recognised and bound to the m6A site on HDGF mRNA and enhanced HDGF mRNA stability. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glycolysis | |||
| In-vitro Model | HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 |
| NCI-N87 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_1603 | |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| In-vivo Model | Mice 8 weeks after splenic portal vein injection of BGC823 cells with METTL3 overexpression or vector-transfected cells. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Elevated METTL3 expression promotes tumour angiogenesis and glycolysis in Gastric cancer. P300-mediated H3K27 acetylation activation in the promoter of METTL3 induced METTL3 transcription, which stimulated m6A modification of Hepatoma-derived growth factor (HDGF) mRNA, and the m6A reader IGF2BP3 then directly recognised and bound to the m6A site on HDGF mRNA and enhanced HDGF mRNA stability. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glycolysis | |||
| In-vitro Model | HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 |
| NCI-N87 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_1603 | |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| In-vivo Model | Mice 8 weeks after splenic portal vein injection of BGC823 cells with METTL3 overexpression or vector-transfected cells. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
RNA modification
m6A Regulator: ELAV-like protein 1 (ELAVL1)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00073 | ||
| Epigenetic Regulator | Y-box-binding protein 1 (YBX1) | |
| Regulated Target | Hepatoma-derived growth factor (HDGF) | |
| Crosstalk relationship | m5C → m6A | |
| Disease | Bladder cancer | |
| Crosstalk ID: M6ACROT00074 | ||
| Epigenetic Regulator | RNA cytosine C(5)-methyltransferase NSUN2 (NSUN2) | |
| Regulated Target | Hepatoma-derived growth factor (HDGF) | |
| Crosstalk relationship | m5C → m6A | |
| Disease | Bladder cancer | |
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03645 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00271)
| In total 5 m6A sequence/site(s) in this target gene | |||
| mod ID: 2OMSITE000281 | Click to Show/Hide the Full List | ||
| mod site | chr1:156743862-156743863:- | [3] | |
| Sequence | CCTAGGACTCTCCTAAACGTCCCAAGGAGGCAGAAAACCCT | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000537739.5; ENST00000477306.1; ENST00000465180.5; ENST00000357325.10; ENST00000482651.5; ENST00000471377.5; ENST00000495212.1; ENST00000368206.5; ENST00000368209.9; ENST00000469145.5 | ||
| External Link | RMBase: Nm_site_399 | ||
| mod ID: 2OMSITE000283 | Click to Show/Hide the Full List | ||
| mod site | chr1:156743864-156743865:- | [3] | |
| Sequence | TGCCTAGGACTCTCCTAAACGTCCCAAGGAGGCAGAAAACC | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000357325.10; ENST00000495212.1; ENST00000368206.5; ENST00000482651.5; ENST00000469145.5; ENST00000477306.1; ENST00000537739.5; ENST00000471377.5; ENST00000465180.5; ENST00000368209.9 | ||
| External Link | RMBase: Nm_site_401 | ||
| mod ID: 2OMSITE000279 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742188-156742189:- | [3] | |
| Sequence | CTTGGGCTCTGATGAAAAATTGCTGACTGTAGCTTTGGAAG | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000368209.9; ENST00000537739.5; ENST00000357325.10 | ||
| External Link | RMBase: Nm_site_397 | ||
| mod ID: 2OMSITE000280 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742733-156742734:- | [3] | |
| Sequence | CTGAGACCTGAAGGGTTGACTTTACCCATTTGGGTGGGAGT | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000465180.5; ENST00000537739.5; ENST00000357325.10; ENST00000368209.9 | ||
| External Link | RMBase: Nm_site_398 | ||
| mod ID: 2OMSITE000282 | Click to Show/Hide the Full List | ||
| mod site | chr1:156743863-156743864:- | [3] | |
| Sequence | GCCTAGGACTCTCCTAAACGTCCCAAGGAGGCAGAAAACCC | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000477306.1; ENST00000368209.9; ENST00000465180.5; ENST00000357325.10; ENST00000537739.5; ENST00000469145.5; ENST00000471377.5; ENST00000482651.5; ENST00000368206.5; ENST00000495212.1 | ||
| External Link | RMBase: Nm_site_400 | ||
5-methylcytidine (m5C)
| In total 8 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE004198 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742963-156742964:- | [4] | |
| Sequence | GGGCAGCACAGGAGGGCGGCCTCCTTCTGAGCTCCTGTCCC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000357325.10; ENST00000368209.9; ENST00000465180.5; ENST00000537739.5 | ||
| External Link | RMBase: m5C_site_4010 | ||
| mod ID: M5CSITE004199 | Click to Show/Hide the Full List | ||
| mod site | chr1:156743365-156743366:- | [5] | |
| Sequence | AACTGGCCATGGCCTGCAAACTGGGAACCCCTTTCCCACCC | ||
| Cell/Tissue List | HEK293T; HeLa; T24 | ||
| Seq Type List | Bisulfite-seq; m5C-RIP-seq | ||
| Transcript ID List | ENST00000465180.5; ENST00000368209.9; ENST00000477306.1; ENST00000357325.10; ENST00000537739.5; ENST00000482651.5; ENST00000368206.5 | ||
| External Link | RMBase: m5C_site_4011 | ||
| mod ID: M5CSITE004200 | Click to Show/Hide the Full List | ||
| mod site | chr1:156744157-156744158:- | [4] | |
| Sequence | AGGGGACTTGCTGGAGGTAACTGCTGCCTGCCCAGGGCCTG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000368209.9; ENST00000495212.1; ENST00000471377.5; ENST00000465180.5; ENST00000537739.5; ENST00000477306.1; ENST00000482651.5; ENST00000368206.5; ENST00000357325.10; ENST00000469145.5 | ||
| External Link | RMBase: m5C_site_4012 | ||
| mod ID: M5CSITE004201 | Click to Show/Hide the Full List | ||
| mod site | chr1:156745104-156745105:- | [4] | |
| Sequence | CCTCTTCCCTTACGAGGAATCCAAGGAGAAGTTTGGCAAGC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000495212.1; ENST00000537739.5; ENST00000368206.5; ENST00000469145.5; ENST00000471377.5; ENST00000357325.10; ENST00000465180.5; ENST00000368209.9; ENST00000482651.5 | ||
| External Link | RMBase: m5C_site_4013 | ||
| mod ID: M5CSITE004202 | Click to Show/Hide the Full List | ||
| mod site | chr1:156751433-156751434:- | [5] | |
| Sequence | AGCCCGGGGCGCCCGGAGCCCCGCCATGTCGCGATCCAACC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000537739.5; ENST00000357325.10; ENST00000495212.1; ENST00000469145.5; ENST00000465180.5; ENST00000368206.5; ENST00000471377.5; ENST00000368209.9 | ||
| External Link | RMBase: m5C_site_4014 | ||
| mod ID: M5CSITE004203 | Click to Show/Hide the Full List | ||
| mod site | chr1:156751434-156751435:- | [5] | |
| Sequence | GAGCCCGGGGCGCCCGGAGCCCCGCCATGTCGCGATCCAAC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000471377.5; ENST00000537739.5; ENST00000465180.5; ENST00000368209.9; ENST00000495212.1; ENST00000357325.10; ENST00000469145.5; ENST00000368206.5 | ||
| External Link | RMBase: m5C_site_4015 | ||
| mod ID: M5CSITE004204 | Click to Show/Hide the Full List | ||
| mod site | chr1:156751435-156751436:- | [5] | |
| Sequence | GGAGCCCGGGGCGCCCGGAGCCCCGCCATGTCGCGATCCAA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000357325.10; ENST00000368209.9; ENST00000368206.5; ENST00000471377.5; ENST00000469145.5; ENST00000495212.1; ENST00000465180.5; ENST00000537739.5 | ||
| External Link | RMBase: m5C_site_4016 | ||
| mod ID: M5CSITE004205 | Click to Show/Hide the Full List | ||
| mod site | chr1:156751444-156751445:- | [5] | |
| Sequence | GCCCGGCGCGGAGCCCGGGGCGCCCGGAGCCCCGCCATGTC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000357325.10; ENST00000465180.5; ENST00000537739.5; ENST00000495212.1; ENST00000368206.5; ENST00000469145.5; ENST00000471377.5; ENST00000368209.9 | ||
| External Link | RMBase: m5C_site_4017 | ||
N6-methyladenosine (m6A)
| In total 52 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE061011 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742122-156742123:- | [6] | |
| Sequence | TTCAGTTCTAGGAAAATAAAACCCGTTGATTACTATATTGG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000537739.5; ENST00000368209.9; ENST00000357325.10 | ||
| External Link | RMBase: m6A_site_59031 | ||
| mod ID: M6ASITE061022 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742154-156742155:- | [6] | |
| Sequence | TTGGAAGTTTAGCTCTGAGAACCGTAGATGATTTCAGTTCT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000357325.10; ENST00000368209.9; ENST00000537739.5 | ||
| External Link | RMBase: m6A_site_59032 | ||
| mod ID: M6ASITE061033 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742183-156742184:- | [7] | |
| Sequence | GCTCTGATGAAAAATTGCTGACTGTAGCTTTGGAAGTTTAG | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000357325.10; ENST00000537739.5; ENST00000368209.9 | ||
| External Link | RMBase: m6A_site_59033 | ||
| mod ID: M6ASITE061044 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742300-156742301:- | [6] | |
| Sequence | ATTTGTAACCCACTGAGAGGACAGAGAGAAATAAGTGCCCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000357325.10; ENST00000368209.9; ENST00000537739.5 | ||
| External Link | RMBase: m6A_site_59034 | ||
| mod ID: M6ASITE061055 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742362-156742363:- | [7] | |
| Sequence | TCCAGTGTCATTTCTCATCCACATACCCTGACCTGGCCCCC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000357325.10; ENST00000537739.5; ENST00000368209.9 | ||
| External Link | RMBase: m6A_site_59035 | ||
| mod ID: M6ASITE061066 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742416-156742417:- | [7] | |
| Sequence | CTTCTATTTGGGGCTTGATGACTTTCTTTTTGTAGCTGGGG | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000368209.9; ENST00000537739.5; ENST00000357325.10 | ||
| External Link | RMBase: m6A_site_59036 | ||
| mod ID: M6ASITE061077 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742535-156742536:- | [7] | |
| Sequence | GTTTTTAATAAAAGAAAATTACAAAAAAAAATTTTAAAGAC | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000465180.5; ENST00000357325.10; ENST00000368209.9; ENST00000537739.5 | ||
| External Link | RMBase: m6A_site_59037 | ||
| mod ID: M6ASITE061086 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742571-156742572:- | [7] | |
| Sequence | CCAGACATAAAGGAAAAGACACATTTTTTAGGAAATGTTTT | ||
| Motif Score | 2.084928571 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000537739.5; ENST00000465180.5; ENST00000357325.10; ENST00000368209.9 | ||
| External Link | RMBase: m6A_site_59038 | ||
| mod ID: M6ASITE061087 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742573-156742574:- | [7] | |
| Sequence | ACCCAGACATAAAGGAAAAGACACATTTTTTAGGAAATGTT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000537739.5; ENST00000368209.9; ENST00000357325.10; ENST00000465180.5 | ||
| External Link | RMBase: m6A_site_59039 | ||
| mod ID: M6ASITE061088 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742587-156742588:- | [7] | |
| Sequence | TAGTCATATATATCACCCAGACATAAAGGAAAAGACACATT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000465180.5; ENST00000368209.9; ENST00000537739.5; ENST00000357325.10 | ||
| External Link | RMBase: m6A_site_59040 | ||
| mod ID: M6ASITE061089 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742630-156742631:- | [7] | |
| Sequence | AGCTGTCCTTTTTCCTCTCCACACAGTGCTCAAGGCCAGCT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000368209.9; ENST00000465180.5; ENST00000357325.10; ENST00000537739.5 | ||
| External Link | RMBase: m6A_site_59041 | ||
| mod ID: M6ASITE061090 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742677-156742678:- | [8] | |
| Sequence | CCCTTTAGATCTCTGAAGCCACAAATAGGATGCTTGGGAAG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | brain; liver; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000357325.10; ENST00000465180.5; ENST00000368209.9; ENST00000537739.5 | ||
| External Link | RMBase: m6A_site_59042 | ||
| mod ID: M6ASITE061091 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742735-156742736:- | [7] | |
| Sequence | ACCTGAGACCTGAAGGGTTGACTTTACCCATTTGGGTGGGA | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000357325.10; ENST00000368209.9; ENST00000465180.5; ENST00000537739.5 | ||
| External Link | RMBase: m6A_site_59043 | ||
| mod ID: M6ASITE061092 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742848-156742849:- | [9] | |
| Sequence | TAAATCCTTGATGATTGACAACACCCATTTTTCCTTTTGCC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000357325.10; ENST00000368209.9; ENST00000537739.5; ENST00000465180.5 | ||
| External Link | RMBase: m6A_site_59044 | ||
| mod ID: M6ASITE061093 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742851-156742852:- | [9] | |
| Sequence | CCATAAATCCTTGATGATTGACAACACCCATTTTTCCTTTT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000368209.9; ENST00000465180.5; ENST00000537739.5; ENST00000357325.10 | ||
| External Link | RMBase: m6A_site_59045 | ||
| mod ID: M6ASITE061094 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742897-156742898:- | [10] | |
| Sequence | CTAGATTCAGGGAAAGTGGGACAGCTTGTAGGGGAGGGGCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HepG2; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000357325.10; ENST00000465180.5; ENST00000368209.9; ENST00000537739.5 | ||
| External Link | RMBase: m6A_site_59046 | ||
| mod ID: M6ASITE061095 | Click to Show/Hide the Full List | ||
| mod site | chr1:156742937-156742938:- | [9] | |
| Sequence | CTGAGCTCCTGTCCCCTGCTACACCTATTATCCCAGCTGCC | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000537739.5; ENST00000368209.9; ENST00000357325.10; ENST00000465180.5 | ||
| External Link | RMBase: m6A_site_59047 | ||
| mod ID: M6ASITE061096 | Click to Show/Hide the Full List | ||
| mod site | chr1:156743084-156743085:- | [6] | |
| Sequence | CACTCTCCTAGGCATTCTGGACCTCTGGGTTGGGATCAGGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; MT4; A549; peripheral-blood; HEK293T; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000465180.5; ENST00000357325.10; ENST00000537739.5; ENST00000368209.9 | ||
| External Link | RMBase: m6A_site_59048 | ||
| mod ID: M6ASITE061097 | Click to Show/Hide the Full List | ||
| mod site | chr1:156743116-156743117:- | [8] | |
| Sequence | ACAACTGCTCCCACCTCCTGACACCCTTCTCCCACTCTCCT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | kidney; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000537739.5; ENST00000368209.9; ENST00000465180.5; ENST00000357325.10 | ||
| External Link | RMBase: m6A_site_59049 | ||
| mod ID: M6ASITE061098 | Click to Show/Hide the Full List | ||
| mod site | chr1:156743136-156743137:- | [6] | |
| Sequence | GGCTTCTGCTGGGGCCTGGGACAACTGCTCCCACCTCCTGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; HepG2; MT4; peripheral-blood; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000357325.10; ENST00000368209.9; ENST00000537739.5; ENST00000465180.5 | ||
| External Link | RMBase: m6A_site_59050 | ||
| mod ID: M6ASITE061099 | Click to Show/Hide the Full List | ||
| mod site | chr1:156743321-156743322:- | [7] | |
| Sequence | CCTGCTCTCCTCTTCTACTCACTTTTCCCACTCCAAGCCCA | ||
| Motif Score | 2.469291667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000537739.5; ENST00000368209.9; ENST00000482651.5; ENST00000357325.10; ENST00000477306.1; ENST00000465180.5 | ||
| External Link | RMBase: m6A_site_59051 | ||
| mod ID: M6ASITE061100 | Click to Show/Hide the Full List | ||
| mod site | chr1:156743359-156743360:- | [6] | |
| Sequence | CCATGGCCTGCAAACTGGGAACCCCTTTCCCACCCCAACCT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; MM6; Huh7; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000477306.1; ENST00000368209.9; ENST00000368206.5; ENST00000482651.5; ENST00000357325.10; ENST00000465180.5; ENST00000537739.5 | ||
| External Link | RMBase: m6A_site_59052 | ||
| mod ID: M6ASITE061101 | Click to Show/Hide the Full List | ||
| mod site | chr1:156743366-156743367:- | [6] | |
| Sequence | AAACTGGCCATGGCCTGCAAACTGGGAACCCCTTTCCCACC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; A549; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000537739.5; ENST00000465180.5; ENST00000368209.9; ENST00000357325.10; ENST00000477306.1; ENST00000368206.5; ENST00000482651.5 | ||
| External Link | RMBase: m6A_site_59053 | ||
| mod ID: M6ASITE061102 | Click to Show/Hide the Full List | ||
| mod site | chr1:156743384-156743385:- | [6] | |
| Sequence | GTCTGGGTGCTACTGGGGAAACTGGCCATGGCCTGCAAACT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; A549; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000368209.9; ENST00000477306.1; ENST00000357325.10; ENST00000482651.5; ENST00000537739.5; ENST00000465180.5; ENST00000368206.5 | ||
| External Link | RMBase: m6A_site_59054 | ||
| mod ID: M6ASITE061103 | Click to Show/Hide the Full List | ||
| mod site | chr1:156743393-156743394:- | [7] | |
| Sequence | CCTGCTGCTGTCTGGGTGCTACTGGGGAAACTGGCCATGGC | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000357325.10; ENST00000368209.9; ENST00000477306.1; ENST00000368206.5; ENST00000537739.5; ENST00000465180.5; ENST00000482651.5 | ||
| External Link | RMBase: m6A_site_59055 | ||
| mod ID: M6ASITE061104 | Click to Show/Hide the Full List | ||
| mod site | chr1:156743528-156743529:- | [6] | |
| Sequence | AGGGTGGGCTCTCCTAGGAGACCGGCACTGGCTGGGGCAAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; A549; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000537739.5; ENST00000465180.5; ENST00000368209.9; ENST00000482651.5; ENST00000368206.5; ENST00000477306.1; ENST00000357325.10 | ||
| External Link | RMBase: m6A_site_59056 | ||
| mod ID: M6ASITE061105 | Click to Show/Hide the Full List | ||
| mod site | chr1:156743608-156743609:- | [6] | |
| Sequence | TGGCAGCTGGACTAGGGGGGACCGGCCACTCTCGGAGAAGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; HEK293T; A549; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000368209.9; ENST00000477306.1; ENST00000465180.5; ENST00000482651.5; ENST00000368206.5; ENST00000537739.5; ENST00000357325.10 | ||
| External Link | RMBase: m6A_site_59057 | ||
| mod ID: M6ASITE061106 | Click to Show/Hide the Full List | ||
| mod site | chr1:156743618-156743619:- | [6] | |
| Sequence | CCTGACCCCTTGGCAGCTGGACTAGGGGGGACCGGCCACTC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; HEK293T; A549; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000477306.1; ENST00000368209.9; ENST00000368206.5; ENST00000537739.5; ENST00000357325.10; ENST00000465180.5; ENST00000482651.5 | ||
| External Link | RMBase: m6A_site_59058 | ||
| mod ID: M6ASITE061107 | Click to Show/Hide the Full List | ||
| mod site | chr1:156743846-156743847:- | [6] | |
| Sequence | ACGTCCCAAGGAGGCAGAAAACCCTGAAGGAGAGGAGAAGG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; U2OS; H1A; H1B; hNPCs; fibroblasts; A549; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000477306.1; ENST00000368209.9; ENST00000357325.10; ENST00000465180.5; ENST00000469145.5; ENST00000368206.5; ENST00000482651.5; ENST00000471377.5; ENST00000495212.1; ENST00000537739.5 | ||
| External Link | RMBase: m6A_site_59059 | ||
| mod ID: M6ASITE061108 | Click to Show/Hide the Full List | ||
| mod site | chr1:156744172-156744173:- | [6] | |
| Sequence | GTTGAAGAGGAGAGCAGGGGACTTGCTGGAGGTAACTGCTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; U2OS; H1A; H1B; hNPCs; fibroblasts; A549; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000368209.9; ENST00000471377.5; ENST00000482651.5; ENST00000469145.5; ENST00000465180.5; ENST00000537739.5; ENST00000477306.1; ENST00000357325.10; ENST00000495212.1; ENST00000368206.5 | ||
| External Link | RMBase: m6A_site_59060 | ||
| mod ID: M6ASITE061109 | Click to Show/Hide the Full List | ||
| mod site | chr1:156744250-156744251:- | [7] | |
| Sequence | GAATGCAGAGGGCAGCAGCGACGAGGAAGGGAAGCTGGTCA | ||
| Motif Score | 2.839505952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000471377.5; ENST00000368206.5; ENST00000368209.9; ENST00000357325.10; ENST00000537739.5; ENST00000495212.1; ENST00000465180.5; ENST00000482651.5; ENST00000477306.1; ENST00000469145.5 | ||
| External Link | RMBase: m6A_site_59061 | ||
| mod ID: M6ASITE061110 | Click to Show/Hide the Full List | ||
| mod site | chr1:156744312-156744313:- | [6] | |
| Sequence | AGCTGTGTGGAAGAGCCTGAACCAGAGCCCGAAGCTGCAGA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; U2OS; hNPCs; fibroblasts; A549; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000368209.9; ENST00000469145.5; ENST00000482651.5; ENST00000357325.10; ENST00000465180.5; ENST00000537739.5; ENST00000495212.1; ENST00000368206.5; ENST00000471377.5 | ||
| External Link | RMBase: m6A_site_59062 | ||
| mod ID: M6ASITE061111 | Click to Show/Hide the Full List | ||
| mod site | chr1:156745038-156745039:- | [6] | |
| Sequence | GGGGCTGTGGGAGATCGAGAACAACCCTACTGTCAAGGCTT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; hESC-HEK293T; U2OS; A549; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000465180.5; ENST00000368209.9; ENST00000482651.5; ENST00000469145.5; ENST00000537739.5; ENST00000471377.5; ENST00000495212.1; ENST00000368206.5; ENST00000357325.10 | ||
| External Link | RMBase: m6A_site_59063 | ||
| mod ID: M6ASITE061112 | Click to Show/Hide the Full List | ||
| mod site | chr1:156745080-156745081:- | [8] | |
| Sequence | GGAGAAGTTTGGCAAGCCCAACAAGAGGAAAGGGTTCAGCG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | brain; kidney; liver; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000368209.9; ENST00000482651.5; ENST00000368206.5; ENST00000495212.1; ENST00000357325.10; ENST00000465180.5; ENST00000469145.5; ENST00000537739.5; ENST00000471377.5 | ||
| External Link | RMBase: m6A_site_59064 | ||
| mod ID: M6ASITE061113 | Click to Show/Hide the Full List | ||
| mod site | chr1:156745125-156745126:- | [6] | |
| Sequence | GGCATTCCTGGGCCCCAAAGACCTCTTCCCTTACGAGGAAT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; U2OS; A549; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000465180.5; ENST00000537739.5; ENST00000357325.10; ENST00000368206.5; ENST00000482651.5; ENST00000368209.9; ENST00000471377.5; ENST00000495212.1; ENST00000469145.5 | ||
| External Link | RMBase: m6A_site_59065 | ||
| mod ID: M6ASITE061114 | Click to Show/Hide the Full List | ||
| mod site | chr1:156745250-156745251:- | [11] | |
| Sequence | GTCGGGAGGGCCCCGGGAGGACTACTCTGGGGAAGGTGAGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000482651.5; ENST00000537739.5; ENST00000495212.1; ENST00000368206.5; ENST00000465180.5; ENST00000357325.10; ENST00000368209.9; ENST00000469145.5; ENST00000471377.5 | ||
| External Link | RMBase: m6A_site_59066 | ||
| mod ID: M6ASITE061115 | Click to Show/Hide the Full List | ||
| mod site | chr1:156745306-156745307:- | [12] | |
| Sequence | ACCAAGTCTTTTTTTTCGGGACCCACGAGACGTGAGTGGAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | MT4; A549; MM6; Huh7 | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000368206.5; ENST00000471377.5; ENST00000469145.5; ENST00000357325.10; ENST00000368209.9; ENST00000495212.1; ENST00000482651.5; ENST00000537739.5; ENST00000465180.5 | ||
| External Link | RMBase: m6A_site_59067 | ||
| mod ID: M6ASITE061116 | Click to Show/Hide the Full List | ||
| mod site | chr1:156745332-156745333:- | [9] | |
| Sequence | TGCCGTGAAATCAACAGCCAACAAATACCAAGTCTTTTTTT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000495212.1; ENST00000465180.5; ENST00000482651.5; ENST00000537739.5; ENST00000357325.10; ENST00000471377.5; ENST00000368206.5; ENST00000469145.5; ENST00000368209.9 | ||
| External Link | RMBase: m6A_site_59068 | ||
| mod ID: M6ASITE061117 | Click to Show/Hide the Full List | ||
| mod site | chr1:156747458-156747459:- | [10] | |
| Sequence | CCCAGGCGTCCTGGCTGAGGACCGACTCCTCTGTCCCTGCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2; U2OS; MT4; A549; MM6; Huh7; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000357325.10; ENST00000368209.9; ENST00000465180.5; ENST00000471377.5; ENST00000482651.5; ENST00000368206.5; ENST00000495212.1; ENST00000469145.5; ENST00000537739.5 | ||
| External Link | RMBase: m6A_site_59069 | ||
| mod ID: M6ASITE061118 | Click to Show/Hide the Full List | ||
| mod site | chr1:156751355-156751356:- | [7] | |
| Sequence | CAAGATGAAGGGCTACCCACACTGGCCGGCCCGGGTGAGCA | ||
| Motif Score | 2.506922619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000471377.5; ENST00000357325.10; ENST00000495212.1; ENST00000368206.5; ENST00000368209.9; ENST00000537739.5; ENST00000469145.5; ENST00000465180.5 | ||
| External Link | RMBase: m6A_site_59070 | ||
| mod ID: M6ASITE061119 | Click to Show/Hide the Full List | ||
| mod site | chr1:156751357-156751358:- | [9] | |
| Sequence | GCCAAGATGAAGGGCTACCCACACTGGCCGGCCCGGGTGAG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000465180.5; ENST00000357325.10; ENST00000469145.5; ENST00000471377.5; ENST00000495212.1; ENST00000368206.5; ENST00000368209.9; ENST00000537739.5 | ||
| External Link | RMBase: m6A_site_59071 | ||
| mod ID: M6ASITE061120 | Click to Show/Hide the Full List | ||
| mod site | chr1:156751388-156751389:- | [13] | |
| Sequence | GAAGGAGTACAAATGCGGGGACCTGGTGTTCGCCAAGATGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | A549; HepG2; U2OS; MT4; MM6; Huh7; NB4 | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000537739.5; ENST00000469145.5; ENST00000368209.9; ENST00000368206.5; ENST00000357325.10; ENST00000471377.5; ENST00000465180.5; ENST00000495212.1 | ||
| External Link | RMBase: m6A_site_59072 | ||
| mod ID: M6ASITE061121 | Click to Show/Hide the Full List | ||
| mod site | chr1:156751400-156751401:- | [9] | |
| Sequence | ATCCAACCGGCAGAAGGAGTACAAATGCGGGGACCTGGTGT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000495212.1; ENST00000471377.5; ENST00000357325.10; ENST00000368206.5; ENST00000537739.5; ENST00000465180.5; ENST00000368209.9; ENST00000469145.5 | ||
| External Link | RMBase: m6A_site_59073 | ||
| mod ID: M6ASITE061122 | Click to Show/Hide the Full List | ||
| mod site | chr1:156751484-156751485:- | [6] | |
| Sequence | CGCCACCGCCGCACGCGCAAACTTGGGCTCGCGCTTCCCGG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; A549; U2OS; MM6; Jurkat; GSC-11; HEK293T; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000357325.10; ENST00000469145.5; ENST00000465180.5; ENST00000537739.5; ENST00000471377.5; ENST00000368206.5; ENST00000368209.9; ENST00000495212.1 | ||
| External Link | RMBase: m6A_site_59074 | ||
| mod ID: M6ASITE061123 | Click to Show/Hide the Full List | ||
| mod site | chr1:156751662-156751663:- | [14] | |
| Sequence | TTGAATTTCAAACACAAACAACTGCACGAGCGCGCACCCAC | ||
| Motif Score | 2.595904762 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000465180.5; ENST00000368206.5; ENST00000357325.10; ENST00000471377.5; ENST00000368209.9 | ||
| External Link | RMBase: m6A_site_59075 | ||
| mod ID: M6ASITE061124 | Click to Show/Hide the Full List | ||
| mod site | chr1:156751665-156751666:- | [6] | |
| Sequence | CAATTGAATTTCAAACACAAACAACTGCACGAGCGCGCACC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; A549; hESC-HEK293T; HepG2; U2OS; H1A; H1B; hESCs; fibroblasts; H1299; MM6; Huh7; Jurkat; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000368206.5; ENST00000465180.5; ENST00000368209.9; ENST00000357325.10; ENST00000471377.5 | ||
| External Link | RMBase: m6A_site_59076 | ||
| mod ID: M6ASITE061125 | Click to Show/Hide the Full List | ||
| mod site | chr1:156751671-156751672:- | [6] | |
| Sequence | AGGGAGCAATTGAATTTCAAACACAAACAACTGCACGAGCG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; A549; hESC-HEK293T; HepG2; U2OS; H1A; H1B; hESCs; fibroblasts; H1299; MM6; Huh7; Jurkat; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000368206.5; ENST00000357325.10; ENST00000471377.5; ENST00000465180.5; ENST00000368209.9 | ||
| External Link | RMBase: m6A_site_59077 | ||
| mod ID: M6ASITE061126 | Click to Show/Hide the Full List | ||
| mod site | chr1:156752080-156752081:- | [6] | |
| Sequence | GGAGCAGAGGGCAGGCGGAAACCGTGTACAGACGTCCACAC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; A549; HepG2; U2OS; H1A; H1B; hESCs; HEK293T; fibroblasts; H1299; MM6; Huh7; Jurkat; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000368209.9; ENST00000368206.5; ENST00000471377.5; ENST00000465180.5 | ||
| External Link | RMBase: m6A_site_59078 | ||
| mod ID: M6ASITE061127 | Click to Show/Hide the Full List | ||
| mod site | chr1:156752136-156752137:- | [6] | |
| Sequence | CGCCTAGCACGCTGCCAAGAACCGCGGGTGCCCAGTAAATA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; A549; H1B; H1A; H1299; peripheral-blood; GSC-11; HEK293A-TOA; MSC; iSLK; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | rmsk_276152; ENST00000471377.5; ENST00000465180.5; ENST00000368206.5; ENST00000368209.9 | ||
| External Link | RMBase: m6A_site_59079 | ||
| mod ID: M6ASITE061128 | Click to Show/Hide the Full List | ||
| mod site | chr1:156752202-156752203:- | [6] | |
| Sequence | CACGGCGAGGTCCAGAGAGGACCCCAGTGCGCGGAGCGGCA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; Huh7; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000471377.5; ENST00000368209.9; ENST00000368206.5; ENST00000465180.5 | ||
| External Link | RMBase: m6A_site_59080 | ||
| mod ID: M6ASITE061129 | Click to Show/Hide the Full List | ||
| mod site | chr1:156752303-156752304:- | [6] | |
| Sequence | GGCCATCTCCTGGCTACCAAACTCAAGCGTTTCCTCCTTAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000368206.5; ENST00000465180.5 | ||
| External Link | RMBase: m6A_site_59081 | ||
| mod ID: M6ASITE061130 | Click to Show/Hide the Full List | ||
| mod site | chr1:156752388-156752389:- | [6] | |
| Sequence | GTCCAAGTCTCCTTGGCAAGACAGTAGCTTAGAGAAACGAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000368206.5; ENST00000465180.5 | ||
| External Link | RMBase: m6A_site_59082 | ||
References