m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00196)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
BCL2
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Th1 cell line | Mus musculus |
|
Treatment: METTL3 knockout splenic Th1 cells
Control: Wild type splenic Th1 cells
|
GSE129648 | |
| Regulation |
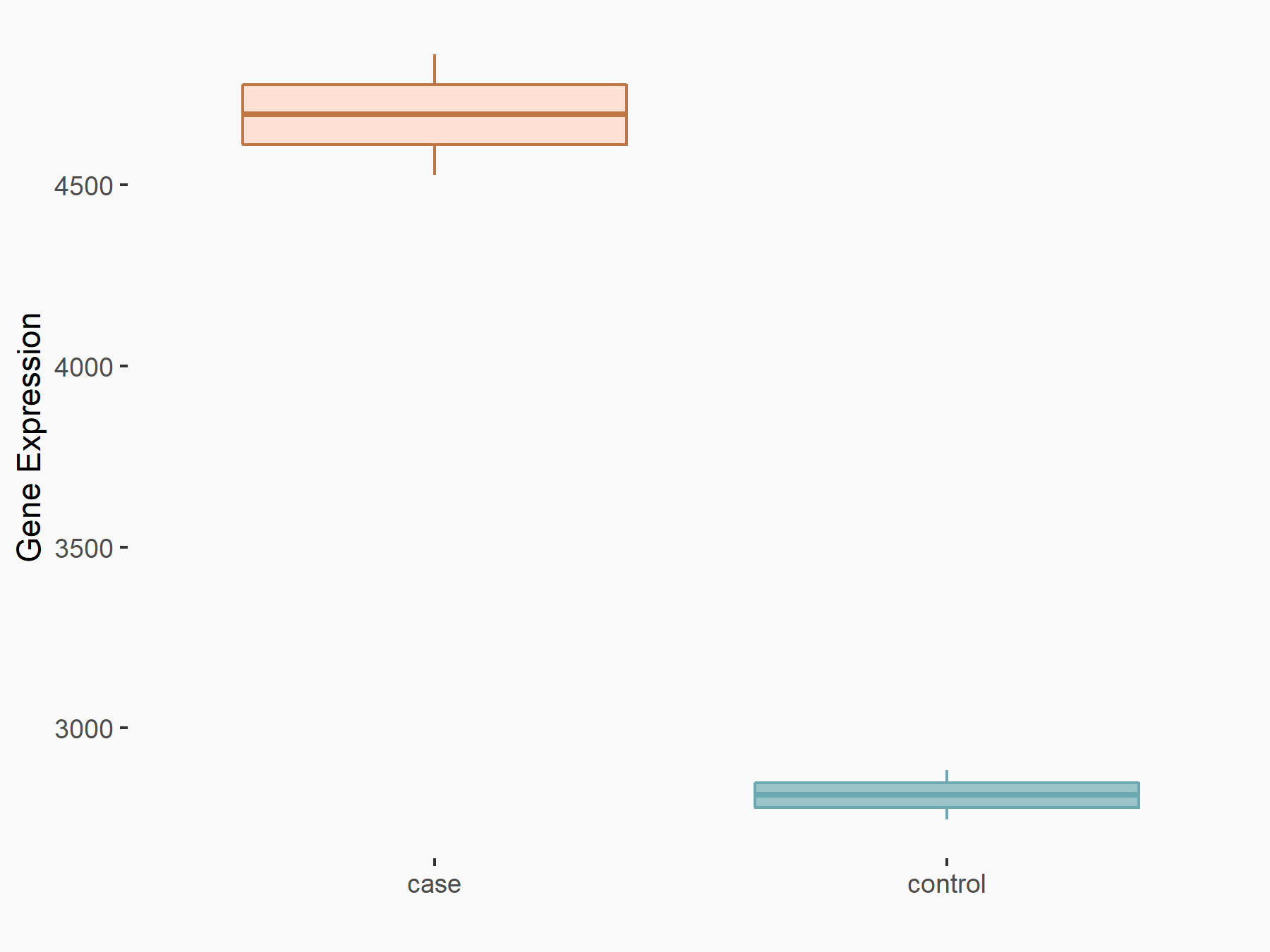  |
logFC: 7.38E-01 p-value: 3.55E-06 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between BCL2 and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 6.70E+00 | GSE60213 |
| In total 6 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Knocking down METTL3 prevented Enterovirus 71-induced cell death and suppressed Enterovirus 71-induced expression of BAX while rescuing Apoptosis regulator Bcl-2 (BCL2) expression after Enterovirus 71 infection. Knocking down METTL3 inhibited Enterovirus 71-induced expression of Atg5, Atg7 and LC3 II. Knocking down METTL3 inhibited Enterovirus 71-induced apoptosis and autophagy. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Enterovirus | ICD-11: 1A2Y | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| Cell autophagy | ||||
| In-vitro Model | Schwann cells (A type of glial cell that surrounds neurons) | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 depletion in human myeloid leukemia cell lines induces cell differentiation and apoptosis and delays leukemia progression in recipient mice in vivo. Single-nucleotide-resolution mapping of m6A coupled with ribosome profiling reveals that m6A promotes the translation of c-MYC, Apoptosis regulator Bcl-2 (BCL2) and PTEN mRNAs in the human acute myeloid leukemia MOLM-13 cell line. Moreover, loss of METTL3 leads to increased levels of phosphorylated AKT. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Acute myeloid leukaemia | ICD-11: 2A60 | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell differentiation and apoptosis | |||
| In-vitro Model | HSPC (Human hematopoietic stem cell) | |||
| In-vivo Model | 500,000 selected cells were injected via tail vein or retro-orbital route into female NSG (6-8 week old) recipient mice that had been sublethally irradiated with 475 cGy one day before transplantation. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | Mettl3 inhibitor, S-adenosylhomocysteine promoted the apoptosis and autophagy of chondrocytes with inflammation in vitro and aggravated the degeneration of chondrocytes and subchondral bone in monosodium iodoacetate (MIA) induced temporomandibular joint osteoarthritis mice in vivo. Bcl2 protein interacted with Beclin1 protein in chondrocytes induced by TNF-alpha stimulation. Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through m6A/Ythdf1/Apoptosis regulator Bcl-2 (BCL2) signal axis which provides promising therapeutic strategy for temporomandibular joint osteoarthritis. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | B-cell lymphomas | ICD-11: 2A86 | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| In-vitro Model | ATDC-5 | Mouse teratocarcinoma | Mus musculus | CVCL_3894 |
| In-vivo Model | For MIA + SAH control, S-adenosylhomocysteine (SAH), Mettl3 inhibitor (10 mg/kg) (MCE, NJ, USA) was injected intraperitoneally before MIA injection and maintained twice a week until mice were sacrificed. | |||
| Experiment 4 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | Down-regulation of METTL3 inhibits the proliferation and mobility of human gastric cancer cells and leads to inactivation of the AKT signaling pathway, suggesting that METTL3 is a potential target for the treatment of human gastric cancer. METTL3 knockdown decreased Apoptosis regulator Bcl-2 (BCL2) and increased Bax and active Caspase-3 in gastric cancer cells, which suggested the apoptotic pathway was activated. METTL3 led to inactivation of the AKT signaling pathway in human gastric cancer cells, including decreased phosphorylation levels of AKT and expression of down-stream effectors p70S6K and Cyclin D1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| Experiment 5 Reporting the m6A Methylation Regulator of This Target Gene | [5] | |||
| Response Summary | METTL3 regulated cellular growth, survival and migration in non-small cell lung cancer. METTL3 promoted non-small cell lung cancer progression by modulating the level of Apoptosis regulator Bcl-2 (BCL2). | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | The mice were housed with filtered air, 12 h light/dark cycle, constant temperature (25℃), relative humidity (50±5%) and free access to food and water. In order to establish a human NSCLC xenograft model, 5×106 H1299, sh-METTL3-H1299 and METTL3 stably overexpressed H1299 cells (2×106 per mouse) were subcutaneously injected into mice. Tumor growth was observed daily. Tumor volume was calculated as follows: 0.5× (length × width2). At 24 days post-inoculation, the maximum diameter exhibited by a single subcutaneous tumor was 15 mm and mice were anesthetized by intraperitoneal administration of sodium pentobarbital (50 mg/kg), then sacrificed by cervical dislocation. | |||
| Experiment 6 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | Apoptosis regulator Bcl-2 (BCL2) acted as the target of METTL3, thereby regulating the proliferation and apoptosis of breast cancer. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | MCF-10A | Normal | Homo sapiens | CVCL_0598 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| In-vivo Model | Mice were maintained at 22 ± 2 ℃ with a humidity of 35 ± 5% under a 12 h light and 12 h dark cycle, with free access to water and food. For the HFD experiment, female control (Ftoflox/flox) and adipose-selective fto knockout (Fabp4-Cre Ftoflox/flox, fto-AKO) mice were fed with high-fat diet (60% fat in calories; Research Diets, D12492) for the desired periods of time, and food intake and body weight were measured every week after weaning (at 3 weeks of age). | |||
Fat mass and obesity-associated protein (FTO) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by FTO | ||
| Cell Line | NB4 cell line | Homo sapiens |
|
Treatment: shFTO NB4 cells
Control: shNS NB4 cells
|
GSE103494 | |
| Regulation |
  |
logFC: -6.55E-01 p-value: 1.57E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [7] | |||
| Response Summary | Studies of the aberrant expression of m6A mediators in breast cancer revealed that they were associated with different BC subtypes and functions, such as proliferation, apoptosis, stemness, the cell cycle, migration, and metastasis, through several factors and signaling pathways, such as Apoptosis regulator Bcl-2 (BCL2) and the PI3K/Akt pathway, among others. Fat mass and obesity-associated protein (FTO) was identified as the first m6A demethylase, and a series of inhibitors that target FTO were reported to have potential for the treatment of BC by inhibiting cell proliferation and promoting apoptosis. | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Meclofenamic acid | Approved | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell proliferation | |||
| Cell apoptosis | ||||
Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by IGF2BP1 | ||
| Cell Line | ES-2 cell line | Homo sapiens |
|
Treatment: siIGF2BP1 ES-2 cells
Control: siControl ES-2 cells
|
GSE109604 | |
| Regulation |
  |
logFC: -7.97E-01 p-value: 9.99E-03 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [8] | |||
| Response Summary | YBX1 selectively functions in regulating survival of myeloid leukemia cells. YBX1 interacts with insulin-like growth factor 2 messenger RNA (mRNA)-binding proteins (IGF2BPs) and stabilizes m6A-tagged RNA. YBX1 deficiency dysregulates the expression of apoptosis-related genes and promotes mRNA decay of MYC and Apoptosis regulator Bcl-2 (BCL2) in an m6A-dependent manner, which contributes to the defective survival that results from deletion of YBX1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Myeloid leukaemia | ICD-11: 2B33.1 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | Leukemia stem cell line (Leukemia stem cell line) | |||
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| BV-173 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0181 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| KG-1a | Adult acute myeloid leukemia | Homo sapiens | CVCL_1824 | |
Methyltransferase-like 14 (METTL14) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL14 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
|
Treatment: siMETTL14 MDA-MB-231 cells
Control: MDA-MB-231 cells
|
GSE81164 | |
| Regulation |
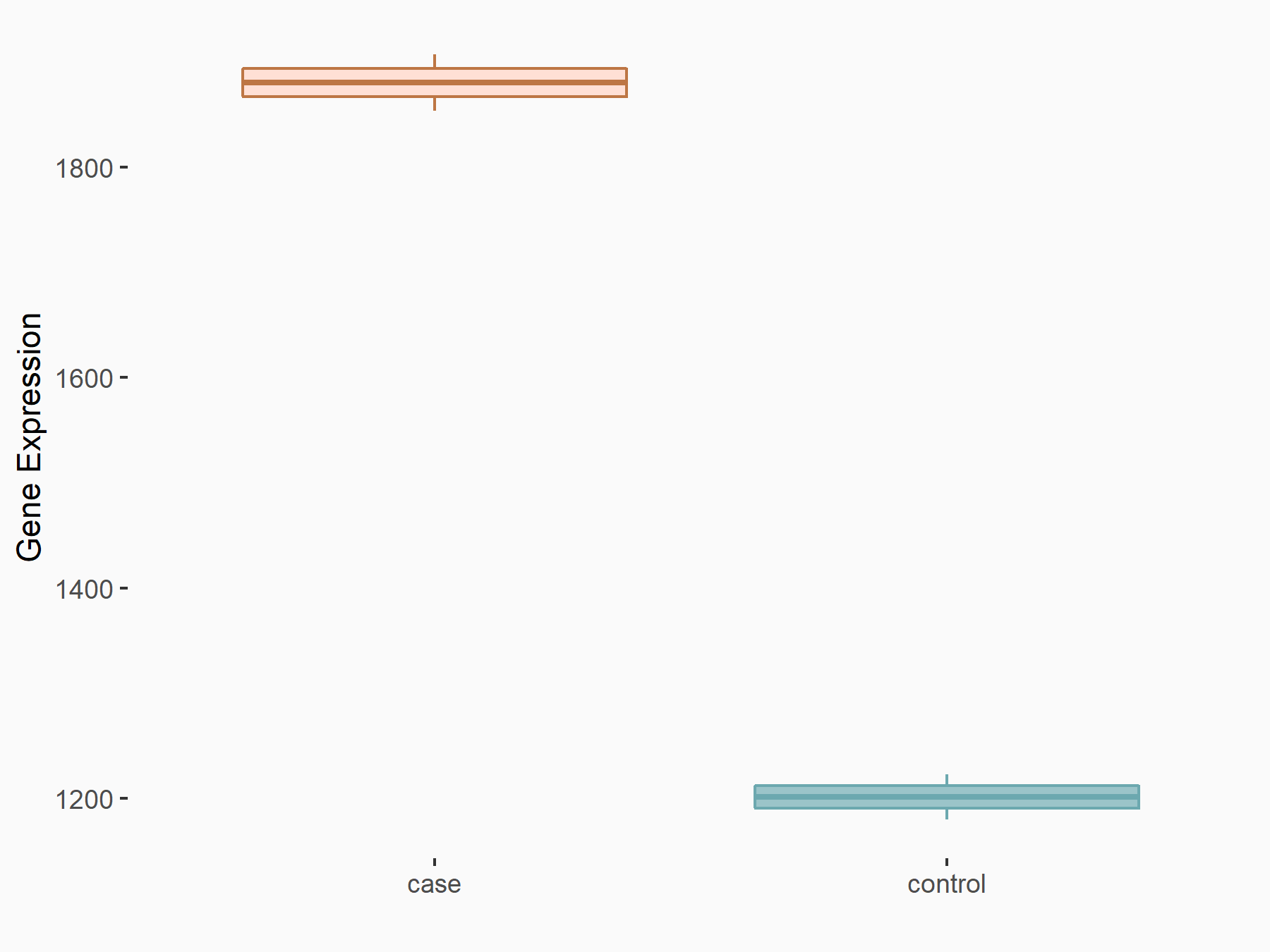  |
logFC: 6.46E-01 p-value: 1.74E-05 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [9] | |||
| Response Summary | Knocking down METTL14 could inhibit the development of atherosclerosis in high-fat diet-treated APOE mice. After transfection with si-METTL14, the Apoptosis regulator Bcl-2 (BCL2) expression level and the viability of ox-LDL-incubated cells increased, whereas the apoptosis rate and the expressions of Bax and cleaved caspase-3 decreased. However, the effect of METTL14 knockdown was reversed by p65 overexpression. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Atherosclerosis | ICD-11: BD40.Z | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | HUVEC-C | Normal | Homo sapiens | CVCL_2959 |
| EA.hy 926 | Normal | Homo sapiens | CVCL_3901 | |
| In-vivo Model | The mice were randomly divided into control, Ad-sh-NC, and Ad-sh-METTL14 groups (10 mice per group). The mice in the control group were fed a normal diet, while the Ad-sh-NC and Ad-sh-METTL14 groups were fed a high-fat diet (20% fat and 0.25% cholesterol). Furthermore, 300 uL of constructed sh-NC or sh-METTL14 adenovirus was injected every 3 weeks into the caudal veins of mice from the Ad-sh-NC or Ad-sh-METTL14 groups, respectively. The constructed vectors were obtained from HanBio Technology Co., Ltd. (Shanghai, China). All mice were sacrificed after 24 weeks and the aortas were separated for further experiments. | |||
RNA demethylase ALKBH5 (ALKBH5) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by ALKBH5 | ||
| Cell Line | CAG cell line | Homo sapiens |
|
Treatment: shALKBH5 CAG cells
Control: shNC CAG cells
|
GSE180214 | |
| Regulation |
  |
logFC: -7.25E-01 p-value: 1.10E-04 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [10] | |||
| Response Summary | ALKBH5 is a tumor-promoting gene in epithelial ovarian cancer, which is involved in the mTOR pathway and Apoptosis regulator Bcl-2 (BCL2)-Beclin1 complex. ALKBH5 activated EGFR-PIK3CA-AKT-mTOR signaling pathway. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Ovarian cancer | ICD-11: 2C73 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Autophagy | hsa04140 | |||
| In-vitro Model | A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 |
| CoC1 | Ovarian adenocarcinoma | Homo sapiens | CVCL_6891 | |
| OVCAR-3 | Ovarian serous adenocarcinoma | Homo sapiens | CVCL_0465 | |
| SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 | |
| In-vivo Model | SKOV3 or A2780 cells were infected with the indicated lentiviral vectors and injected (5 × 106 cells/mouse in 200 uL volume) subcutaneously into the left armpit of 6-week-old BALB/c nude mice. After 21 days, the animals were sacrificed to confirm the presence of tumors and weigh the established tumors. | |||
YTH domain-containing family protein 1 (YTHDF1) [READER]
| Representative RIP-seq result supporting the interaction between BCL2 and the regulator | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.25E+00 | GSE63591 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | Mettl3 inhibitor, S-adenosylhomocysteine promoted the apoptosis and autophagy of chondrocytes with inflammation in vitro and aggravated the degeneration of chondrocytes and subchondral bone in monosodium iodoacetate (MIA) induced temporomandibular joint osteoarthritis mice in vivo. Bcl2 protein interacted with Beclin1 protein in chondrocytes induced by TNF-alpha stimulation. Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through m6A/Ythdf1/Apoptosis regulator Bcl-2 (BCL2) signal axis which provides promising therapeutic strategy for temporomandibular joint osteoarthritis. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | B-cell lymphomas | ICD-11: 2A86 | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| In-vitro Model | ATDC-5 | Mouse teratocarcinoma | Mus musculus | CVCL_3894 |
| In-vivo Model | For MIA + SAH control, S-adenosylhomocysteine (SAH), Mettl3 inhibitor (10 mg/kg) (MCE, NJ, USA) was injected intraperitoneally before MIA injection and maintained twice a week until mice were sacrificed. | |||
Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [8] | |||
| Response Summary | YBX1 selectively functions in regulating survival of myeloid leukemia cells. YBX1 interacts with insulin-like growth factor 2 messenger RNA (mRNA)-binding proteins (IGF2BPs) and stabilizes m6A-tagged RNA. YBX1 deficiency dysregulates the expression of apoptosis-related genes and promotes mRNA decay of MYC and Apoptosis regulator Bcl-2 (BCL2) in an m6A-dependent manner, which contributes to the defective survival that results from deletion of YBX1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Myeloid leukaemia | ICD-11: 2B33.1 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | Leukemia stem cell line (Leukemia stem cell line) | |||
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| BV-173 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0181 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| KG-1a | Adult acute myeloid leukemia | Homo sapiens | CVCL_1824 | |
Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [8] | |||
| Response Summary | YBX1 selectively functions in regulating survival of myeloid leukemia cells. YBX1 interacts with insulin-like growth factor 2 messenger RNA (mRNA)-binding proteins (IGF2BPs) and stabilizes m6A-tagged RNA. YBX1 deficiency dysregulates the expression of apoptosis-related genes and promotes mRNA decay of MYC and Apoptosis regulator Bcl-2 (BCL2) in an m6A-dependent manner, which contributes to the defective survival that results from deletion of YBX1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Myeloid leukaemia | ICD-11: 2B33.1 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | Leukemia stem cell line (Leukemia stem cell line) | |||
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| BV-173 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0181 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| KG-1a | Adult acute myeloid leukemia | Homo sapiens | CVCL_1824 | |
Enterovirus [ICD-11: 1A2Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Knocking down METTL3 prevented Enterovirus 71-induced cell death and suppressed Enterovirus 71-induced expression of BAX while rescuing Apoptosis regulator Bcl-2 (BCL2) expression after Enterovirus 71 infection. Knocking down METTL3 inhibited Enterovirus 71-induced expression of Atg5, Atg7 and LC3 II. Knocking down METTL3 inhibited Enterovirus 71-induced apoptosis and autophagy. | |||
| Responsed Disease | Enterovirus [ICD-11: 1A2Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Apoptosis | hsa04210 | |||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| Cell autophagy | ||||
| In-vitro Model | Schwann cells (A type of glial cell that surrounds neurons) | |||
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 depletion in human myeloid leukemia cell lines induces cell differentiation and apoptosis and delays leukemia progression in recipient mice in vivo. Single-nucleotide-resolution mapping of m6A coupled with ribosome profiling reveals that m6A promotes the translation of c-MYC, Apoptosis regulator Bcl-2 (BCL2) and PTEN mRNAs in the human acute myeloid leukemia MOLM-13 cell line. Moreover, loss of METTL3 leads to increased levels of phosphorylated AKT. | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell differentiation and apoptosis | |||
| In-vitro Model | HSPC (Human hematopoietic stem cell) | |||
| In-vivo Model | 500,000 selected cells were injected via tail vein or retro-orbital route into female NSG (6-8 week old) recipient mice that had been sublethally irradiated with 475 cGy one day before transplantation. | |||
B-cell lymphomas [ICD-11: 2A86]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | Mettl3 inhibitor, S-adenosylhomocysteine promoted the apoptosis and autophagy of chondrocytes with inflammation in vitro and aggravated the degeneration of chondrocytes and subchondral bone in monosodium iodoacetate (MIA) induced temporomandibular joint osteoarthritis mice in vivo. Bcl2 protein interacted with Beclin1 protein in chondrocytes induced by TNF-alpha stimulation. Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through m6A/Ythdf1/Apoptosis regulator Bcl-2 (BCL2) signal axis which provides promising therapeutic strategy for temporomandibular joint osteoarthritis. | |||
| Responsed Disease | B-cell lymphomas [ICD-11: 2A86] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| In-vitro Model | ATDC-5 | Mouse teratocarcinoma | Mus musculus | CVCL_3894 |
| In-vivo Model | For MIA + SAH control, S-adenosylhomocysteine (SAH), Mettl3 inhibitor (10 mg/kg) (MCE, NJ, USA) was injected intraperitoneally before MIA injection and maintained twice a week until mice were sacrificed. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | Mettl3 inhibitor, S-adenosylhomocysteine promoted the apoptosis and autophagy of chondrocytes with inflammation in vitro and aggravated the degeneration of chondrocytes and subchondral bone in monosodium iodoacetate (MIA) induced temporomandibular joint osteoarthritis mice in vivo. Bcl2 protein interacted with Beclin1 protein in chondrocytes induced by TNF-alpha stimulation. Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through m6A/Ythdf1/Apoptosis regulator Bcl-2 (BCL2) signal axis which provides promising therapeutic strategy for temporomandibular joint osteoarthritis. | |||
| Responsed Disease | B-cell lymphomas [ICD-11: 2A86] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| In-vitro Model | ATDC-5 | Mouse teratocarcinoma | Mus musculus | CVCL_3894 |
| In-vivo Model | For MIA + SAH control, S-adenosylhomocysteine (SAH), Mettl3 inhibitor (10 mg/kg) (MCE, NJ, USA) was injected intraperitoneally before MIA injection and maintained twice a week until mice were sacrificed. | |||
Malignant haematopoietic neoplasm [ICD-11: 2B33]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [8] | |||
| Response Summary | YBX1 selectively functions in regulating survival of myeloid leukemia cells. YBX1 interacts with insulin-like growth factor 2 messenger RNA (mRNA)-binding proteins (IGF2BPs) and stabilizes m6A-tagged RNA. YBX1 deficiency dysregulates the expression of apoptosis-related genes and promotes mRNA decay of MYC and Apoptosis regulator Bcl-2 (BCL2) in an m6A-dependent manner, which contributes to the defective survival that results from deletion of YBX1. | |||
| Responsed Disease | Myeloid leukaemia [ICD-11: 2B33.1] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) | READER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | Leukemia stem cell line (Leukemia stem cell line) | |||
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| BV-173 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0181 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| KG-1a | Adult acute myeloid leukemia | Homo sapiens | CVCL_1824 | |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | Down-regulation of METTL3 inhibits the proliferation and mobility of human gastric cancer cells and leads to inactivation of the AKT signaling pathway, suggesting that METTL3 is a potential target for the treatment of human gastric cancer. METTL3 knockdown decreased Apoptosis regulator Bcl-2 (BCL2) and increased Bax and active Caspase-3 in gastric cancer cells, which suggested the apoptotic pathway was activated. METTL3 led to inactivation of the AKT signaling pathway in human gastric cancer cells, including decreased phosphorylation levels of AKT and expression of down-stream effectors p70S6K and Cyclin D1. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [5] | |||
| Response Summary | METTL3 regulated cellular growth, survival and migration in non-small cell lung cancer. METTL3 promoted non-small cell lung cancer progression by modulating the level of Apoptosis regulator Bcl-2 (BCL2). | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | The mice were housed with filtered air, 12 h light/dark cycle, constant temperature (25℃), relative humidity (50±5%) and free access to food and water. In order to establish a human NSCLC xenograft model, 5×106 H1299, sh-METTL3-H1299 and METTL3 stably overexpressed H1299 cells (2×106 per mouse) were subcutaneously injected into mice. Tumor growth was observed daily. Tumor volume was calculated as follows: 0.5× (length × width2). At 24 days post-inoculation, the maximum diameter exhibited by a single subcutaneous tumor was 15 mm and mice were anesthetized by intraperitoneal administration of sodium pentobarbital (50 mg/kg), then sacrificed by cervical dislocation. | |||
Breast cancer [ICD-11: 2C60]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [7] | |||
| Response Summary | Studies of the aberrant expression of m6A mediators in breast cancer revealed that they were associated with different BC subtypes and functions, such as proliferation, apoptosis, stemness, the cell cycle, migration, and metastasis, through several factors and signaling pathways, such as Apoptosis regulator Bcl-2 (BCL2) and the PI3K/Akt pathway, among others. Fat mass and obesity-associated protein (FTO) was identified as the first m6A demethylase, and a series of inhibitors that target FTO were reported to have potential for the treatment of BC by inhibiting cell proliferation and promoting apoptosis. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Responsed Drug | Meclofenamic acid | Approved | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell proliferation | |||
| Cell apoptosis | ||||
| Experiment 2 Reporting the m6A-centered Disease Response | [6] | |||
| Response Summary | Apoptosis regulator Bcl-2 (BCL2) acted as the target of METTL3, thereby regulating the proliferation and apoptosis of breast cancer. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | MCF-10A | Normal | Homo sapiens | CVCL_0598 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| In-vivo Model | Mice were maintained at 22 ± 2 ℃ with a humidity of 35 ± 5% under a 12 h light and 12 h dark cycle, with free access to water and food. For the HFD experiment, female control (Ftoflox/flox) and adipose-selective fto knockout (Fabp4-Cre Ftoflox/flox, fto-AKO) mice were fed with high-fat diet (60% fat in calories; Research Diets, D12492) for the desired periods of time, and food intake and body weight were measured every week after weaning (at 3 weeks of age). | |||
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [10] | |||
| Response Summary | ALKBH5 is a tumor-promoting gene in epithelial ovarian cancer, which is involved in the mTOR pathway and Apoptosis regulator Bcl-2 (BCL2)-Beclin1 complex. ALKBH5 activated EGFR-PIK3CA-AKT-mTOR signaling pathway. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Autophagy | hsa04140 | |||
| In-vitro Model | A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 |
| CoC1 | Ovarian adenocarcinoma | Homo sapiens | CVCL_6891 | |
| OVCAR-3 | Ovarian serous adenocarcinoma | Homo sapiens | CVCL_0465 | |
| SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 | |
| In-vivo Model | SKOV3 or A2780 cells were infected with the indicated lentiviral vectors and injected (5 × 106 cells/mouse in 200 uL volume) subcutaneously into the left armpit of 6-week-old BALB/c nude mice. After 21 days, the animals were sacrificed to confirm the presence of tumors and weigh the established tumors. | |||
Atherosclerosis [ICD-11: BD40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [9] | |||
| Response Summary | Knocking down METTL14 could inhibit the development of atherosclerosis in high-fat diet-treated APOE mice. After transfection with si-METTL14, the Apoptosis regulator Bcl-2 (BCL2) expression level and the viability of ox-LDL-incubated cells increased, whereas the apoptosis rate and the expressions of Bax and cleaved caspase-3 decreased. However, the effect of METTL14 knockdown was reversed by p65 overexpression. | |||
| Responsed Disease | Atherosclerosis [ICD-11: BD40.Z] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | HUVEC-C | Normal | Homo sapiens | CVCL_2959 |
| EA.hy 926 | Normal | Homo sapiens | CVCL_3901 | |
| In-vivo Model | The mice were randomly divided into control, Ad-sh-NC, and Ad-sh-METTL14 groups (10 mice per group). The mice in the control group were fed a normal diet, while the Ad-sh-NC and Ad-sh-METTL14 groups were fed a high-fat diet (20% fat and 0.25% cholesterol). Furthermore, 300 uL of constructed sh-NC or sh-METTL14 adenovirus was injected every 3 weeks into the caudal veins of mice from the Ad-sh-NC or Ad-sh-METTL14 groups, respectively. The constructed vectors were obtained from HanBio Technology Co., Ltd. (Shanghai, China). All mice were sacrificed after 24 weeks and the aortas were separated for further experiments. | |||
Dentofacial anomalies [ICD-11: DA0E]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | Mettl3 inhibitor, S-adenosylhomocysteine promoted the apoptosis and autophagy of chondrocytes with inflammation in vitro and aggravated the degeneration of chondrocytes and subchondral bone in monosodium iodoacetate (MIA) induced temporomandibular joint osteoarthritis mice in vivo. Bcl2 protein interacted with Beclin1 protein in chondrocytes induced by TNF-alpha stimulation. Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through m6A/Ythdf1/Apoptosis regulator Bcl-2 (BCL2) signal axis which provides promising therapeutic strategy for temporomandibular joint osteoarthritis. | |||
| Responsed Disease | Temporomandibular joint disorders [ICD-11: DA0E.8] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| In-vitro Model | ATDC-5 | Mouse teratocarcinoma | Mus musculus | CVCL_3894 |
| In-vivo Model | For MIA + SAH control, S-adenosylhomocysteine (SAH), Mettl3 inhibitor (10 mg/kg) (MCE, NJ, USA) was injected intraperitoneally before MIA injection and maintained twice a week until mice were sacrificed. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | Mettl3 inhibitor, S-adenosylhomocysteine promoted the apoptosis and autophagy of chondrocytes with inflammation in vitro and aggravated the degeneration of chondrocytes and subchondral bone in monosodium iodoacetate (MIA) induced temporomandibular joint osteoarthritis mice in vivo. Bcl2 protein interacted with Beclin1 protein in chondrocytes induced by TNF-alpha stimulation. Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through m6A/Ythdf1/Apoptosis regulator Bcl-2 (BCL2) signal axis which provides promising therapeutic strategy for temporomandibular joint osteoarthritis. | |||
| Responsed Disease | Temporomandibular joint disorders [ICD-11: DA0E.8] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| In-vitro Model | ATDC-5 | Mouse teratocarcinoma | Mus musculus | CVCL_3894 |
| In-vivo Model | For MIA + SAH control, S-adenosylhomocysteine (SAH), Mettl3 inhibitor (10 mg/kg) (MCE, NJ, USA) was injected intraperitoneally before MIA injection and maintained twice a week until mice were sacrificed. | |||
Meclofenamic acid
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [7] | |||
| Response Summary | Studies of the aberrant expression of m6A mediators in breast cancer revealed that they were associated with different BC subtypes and functions, such as proliferation, apoptosis, stemness, the cell cycle, migration, and metastasis, through several factors and signaling pathways, such as Apoptosis regulator Bcl-2 (BCL2) and the PI3K/Akt pathway, among others. Fat mass and obesity-associated protein (FTO) was identified as the first m6A demethylase, and a series of inhibitors that target FTO were reported to have potential for the treatment of BC by inhibiting cell proliferation and promoting apoptosis. | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell proliferation | |||
| Cell apoptosis | ||||
R-2HG
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [7] | |||
| Response Summary | Studies of the aberrant expression of m6A mediators in breast cancer revealed that they were associated with different BC subtypes and functions, such as proliferation, apoptosis, stemness, the cell cycle, migration, and metastasis, through several factors and signaling pathways, such as Apoptosis regulator Bcl-2 (BCL2) and the PI3K/Akt pathway, among others. Fat mass and obesity-associated protein (FTO) was identified as the first m6A demethylase, and a series of inhibitors that target FTO were reported to have potential for the treatment of BC by inhibiting cell proliferation and promoting apoptosis. | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell proliferation | |||
| Cell apoptosis | ||||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
DNA modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT02123 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
| Drug | Celastrol | |
| Crosstalk ID: M6ACROT02132 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
| Drug | Celastrol | |
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03634 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00196)
| In total 23 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE008682 | Click to Show/Hide the Full List | ||
| mod site | chr18:63192681-63192682:- | [19] | |
| Sequence | TTGGAGCTGATGCAATGGCAAGTGCTGTGGCCGTGGTATCC | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: RNA-editing_site_63837 | ||
| mod ID: A2ISITE008683 | Click to Show/Hide the Full List | ||
| mod site | chr18:63211745-63211746:- | [19] | |
| Sequence | ACACAGCTTGCAGCCACAGTAGGACAATGCCAAGTCCAATG | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: RNA-editing_site_63838 | ||
| mod ID: A2ISITE008684 | Click to Show/Hide the Full List | ||
| mod site | chr18:63234217-63234218:- | [20] | |
| Sequence | GGGAGAGCATCAGAATAAATAGCTAATGCATACGGAGCTTA | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5; rmsk_4883120 | ||
| External Link | RMBase: RNA-editing_site_63839 | ||
| mod ID: A2ISITE008685 | Click to Show/Hide the Full List | ||
| mod site | chr18:63240081-63240082:- | [20] | |
| Sequence | TGGGAGGATTGCTTGAGCCCAGGAGGGAGAGATTGTAGTGA | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5; rmsk_4883129 | ||
| External Link | RMBase: RNA-editing_site_63840 | ||
| mod ID: A2ISITE008686 | Click to Show/Hide the Full List | ||
| mod site | chr18:63246507-63246508:- | [19] | |
| Sequence | TCATGAGACCTGGTGGTTTTATAAAGGGGAGTTTGCCTGCA | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1; rmsk_4883138 | ||
| External Link | RMBase: RNA-editing_site_63841 | ||
| mod ID: A2ISITE008687 | Click to Show/Hide the Full List | ||
| mod site | chr18:63246532-63246533:- | [19] | |
| Sequence | GTGCTGTTGTGATAATGAATAAGTCTCATGAGACCTGGTGG | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1; rmsk_4883138 | ||
| External Link | RMBase: RNA-editing_site_63842 | ||
| mod ID: A2ISITE008688 | Click to Show/Hide the Full List | ||
| mod site | chr18:63246587-63246588:- | [19] | |
| Sequence | TGTCATGGGAGGGAGCGGGTAGGATGTAATTGAATCATGGG | ||
| Transcript ID List | ENST00000398117.1; rmsk_4883138; ENST00000333681.5 | ||
| External Link | RMBase: RNA-editing_site_63843 | ||
| mod ID: A2ISITE008689 | Click to Show/Hide the Full List | ||
| mod site | chr18:63246626-63246627:- | [19] | |
| Sequence | CAAATTTCATCTTGGATCGTAGCTCCCACAATTCCCACATG | ||
| Transcript ID List | rmsk_4883138; ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: RNA-editing_site_63844 | ||
| mod ID: A2ISITE008690 | Click to Show/Hide the Full List | ||
| mod site | chr18:63252485-63252486:- | [19] | |
| Sequence | TCCCACCGGGTTCCTCCCACAGCATGTGGGAATTGTGGGAG | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: RNA-editing_site_63845 | ||
| mod ID: A2ISITE008691 | Click to Show/Hide the Full List | ||
| mod site | chr18:63252517-63252518:- | [19] | |
| Sequence | GCAGAAAGACCTACCACCATAATTCAATTGCCTCCCACCGG | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: RNA-editing_site_63846 | ||
| mod ID: A2ISITE008692 | Click to Show/Hide the Full List | ||
| mod site | chr18:63252580-63252581:- | [20] | |
| Sequence | AGGGAACTCCCATTTTTAAAACCATCACATCTTGTGAGACT | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: RNA-editing_site_63847 | ||
| mod ID: A2ISITE008693 | Click to Show/Hide the Full List | ||
| mod site | chr18:63252581-63252582:- | [20] | |
| Sequence | CAGGGAACTCCCATTTTTAAAACCATCACATCTTGTGAGAC | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: RNA-editing_site_63848 | ||
| mod ID: A2ISITE008694 | Click to Show/Hide the Full List | ||
| mod site | chr18:63252804-63252805:- | [19] | |
| Sequence | TCAAATTTACAAAGTGTATTAGTCTGTTTTCACGCTGTTGA | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: RNA-editing_site_63849 | ||
| mod ID: A2ISITE008695 | Click to Show/Hide the Full List | ||
| mod site | chr18:63277421-63277422:- | [20] | |
| Sequence | CAGGCTGGTCTCAAATGCCTAACCTTGAGTGATTTTCCTTC | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: RNA-editing_site_63850 | ||
| mod ID: A2ISITE008696 | Click to Show/Hide the Full List | ||
| mod site | chr18:63296330-63296331:- | [20] | |
| Sequence | CACTCCAGCCTGGATGACAGAGTGAGACCTTGTCTCAAAAA | ||
| Transcript ID List | rmsk_4883223; ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: RNA-editing_site_63851 | ||
| mod ID: A2ISITE008697 | Click to Show/Hide the Full List | ||
| mod site | chr18:63299836-63299837:- | [20] | |
| Sequence | CTGGGCTTGAAGGATACATAAGAGTTTGTTCAAAAAAAAAA | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: RNA-editing_site_63852 | ||
| mod ID: A2ISITE008698 | Click to Show/Hide the Full List | ||
| mod site | chr18:63299837-63299838:- | [19] | |
| Sequence | CCTGGGCTTGAAGGATACATAAGAGTTTGTTCAAAAAAAAA | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: RNA-editing_site_63853 | ||
| mod ID: A2ISITE008699 | Click to Show/Hide the Full List | ||
| mod site | chr18:63299839-63299840:- | [20] | |
| Sequence | GGCCTGGGCTTGAAGGATACATAAGAGTTTGTTCAAAAAAA | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: RNA-editing_site_63854 | ||
| mod ID: A2ISITE008700 | Click to Show/Hide the Full List | ||
| mod site | chr18:63299841-63299842:- | [19] | |
| Sequence | TTGGCCTGGGCTTGAAGGATACATAAGAGTTTGTTCAAAAA | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: RNA-editing_site_63855 | ||
| mod ID: A2ISITE008701 | Click to Show/Hide the Full List | ||
| mod site | chr18:63299870-63299871:- | [20] | |
| Sequence | TAAGGATGGTTTCACAGAGAAGGTGCCGTTTGGCCTGGGCT | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: RNA-editing_site_63856 | ||
| mod ID: A2ISITE008702 | Click to Show/Hide the Full List | ||
| mod site | chr18:63299875-63299876:- | [19] | |
| Sequence | AAAGTTAAGGATGGTTTCACAGAGAAGGTGCCGTTTGGCCT | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: RNA-editing_site_63857 | ||
| mod ID: A2ISITE008703 | Click to Show/Hide the Full List | ||
| mod site | chr18:63300391-63300392:- | [19] | |
| Sequence | TTTTTTGAACAAACTCTTACATATCCTTCAAGCCCAGGCCA | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5; rmsk_4883235 | ||
| External Link | RMBase: RNA-editing_site_63858 | ||
| mod ID: A2ISITE008704 | Click to Show/Hide the Full List | ||
| mod site | chr18:63300393-63300394:- | [19] | |
| Sequence | TTTTTTTTGAACAAACTCTTACATATCCTTCAAGCCCAGGC | ||
| Transcript ID List | rmsk_4883235; ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: RNA-editing_site_63859 | ||
N6-methyladenosine (m6A)
| In total 92 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE037566 | Click to Show/Hide the Full List | ||
| mod site | chr18:63123857-63123858:- | [21] | |
| Sequence | GAGTAAATCCATGCACCTAAACCTTTTGGAAAATCTGCCGT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HEK293T; hNPCs; GM12878; LCLs; MM6; CD4T; peripheral-blood; HEK293A-TOA; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403695 | ||
| mod ID: M6ASITE037567 | Click to Show/Hide the Full List | ||
| mod site | chr18:63123894-63123895:- | [21] | |
| Sequence | TGAGACTATTAATAAATAAGACTGTAGTGTAGATACTGAGT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; hNPCs; GM12878; LCLs; CD8T; MM6; CD4T; peripheral-blood; HEK293A-TOA; iSLK; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403696 | ||
| mod ID: M6ASITE037568 | Click to Show/Hide the Full List | ||
| mod site | chr18:63123910-63123911:- | [21] | |
| Sequence | TCAATCAAGAAAATTCTGAGACTATTAATAAATAAGACTGT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; hNPCs; GM12878; LCLs; MM6; CD4T; peripheral-blood; HEK293A-TOA; iSLK; endometrial; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403697 | ||
| mod ID: M6ASITE037569 | Click to Show/Hide the Full List | ||
| mod site | chr18:63123957-63123958:- | [21] | |
| Sequence | GCTGAGCACAGAAGATGGGAACACTGGTGGAGGATGGAAAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hNPCs; GM12878; LCLs; MM6; CD4T; HEK293A-TOA; iSLK; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403698 | ||
| mod ID: M6ASITE037570 | Click to Show/Hide the Full List | ||
| mod site | chr18:63124013-63124014:- | [21] | |
| Sequence | AGAAAAATAACTTCAAGCAAACATCCTATCAACAACAAGGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; hNPCs; GM12878; LCLs; MM6; HEK293A-TOA; iSLK; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1; rmsk_4882956 | ||
| External Link | RMBase: m6A_site_403699 | ||
| mod ID: M6ASITE037571 | Click to Show/Hide the Full List | ||
| mod site | chr18:63124041-63124042:- | [21] | |
| Sequence | TACTGTACCGGTTCATCTGGACTGCCCCAGAAAAATAACTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; hNPCs; GM12878; LCLs; CD8T; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | rmsk_4882956; ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403700 | ||
| mod ID: M6ASITE037572 | Click to Show/Hide the Full List | ||
| mod site | chr18:63124119-63124120:- | [21] | |
| Sequence | TTGGACAACCATGACCTTGGACAATCATGAAATATGCATCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; hNPCs; GM12878; CD8T | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | rmsk_4882956; ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403701 | ||
| mod ID: M6ASITE037573 | Click to Show/Hide the Full List | ||
| mod site | chr18:63124135-63124136:- | [21] | |
| Sequence | ATTCTGTTCCATGTCTTTGGACAACCATGACCTTGGACAAT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; GM12878 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | rmsk_4882956; ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403702 | ||
| mod ID: M6ASITE037574 | Click to Show/Hide the Full List | ||
| mod site | chr18:63124205-63124206:- | ||
| Sequence | ATAAAATTAAGCAATGTGAAACTGAATTGGAGAGTGATAAT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403703 | ||
| mod ID: M6ASITE037575 | Click to Show/Hide the Full List | ||
| mod site | chr18:63124475-63124476:- | [21] | |
| Sequence | ATGTGATGCCTCTGCGAAGAACCTTGTGTGACAAATGAGAA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403704 | ||
| mod ID: M6ASITE037576 | Click to Show/Hide the Full List | ||
| mod site | chr18:63124511-63124512:- | [21] | |
| Sequence | CAGAACTAAGGGTATGAAGGACCTGTATTGGGGTCGATGTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403705 | ||
| mod ID: M6ASITE037577 | Click to Show/Hide the Full List | ||
| mod site | chr18:63124527-63124528:- | [21] | |
| Sequence | AAGTTTTCAGAATAACCAGAACTAAGGGTATGAAGGACCTG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403706 | ||
| mod ID: M6ASITE037578 | Click to Show/Hide the Full List | ||
| mod site | chr18:63125266-63125267:- | [22] | |
| Sequence | AGCACCTCAATTTAGTTCAAACAAGACGCCAACATTCTCTC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | MM6; CD4T; peripheral-blood; endometrial; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403707 | ||
| mod ID: M6ASITE037579 | Click to Show/Hide the Full List | ||
| mod site | chr18:63125331-63125332:- | [21] | |
| Sequence | AGATTTGGCAGGGGCAGAAAACTCTGGCAGGCTTAAGATTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; MM6; CD4T; peripheral-blood; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403708 | ||
| mod ID: M6ASITE037580 | Click to Show/Hide the Full List | ||
| mod site | chr18:63125448-63125449:- | [21] | |
| Sequence | AAGGAGAAGAACATCTGAGAACCTCCTCGGCCCTCCCAGTC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; MM6; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403709 | ||
| mod ID: M6ASITE037581 | Click to Show/Hide the Full List | ||
| mod site | chr18:63125458-63125459:- | [21] | |
| Sequence | AAAGCCCCAAAAGGAGAAGAACATCTGAGAACCTCCTCGGC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; MM6; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403710 | ||
| mod ID: M6ASITE037582 | Click to Show/Hide the Full List | ||
| mod site | chr18:63125493-63125494:- | [21] | |
| Sequence | ATGCACTTTCCAAATTGGGGACAAGGGCTCTAAAAAAAGCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; MM6; endometrial; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403711 | ||
| mod ID: M6ASITE037583 | Click to Show/Hide the Full List | ||
| mod site | chr18:63125607-63125608:- | [23] | |
| Sequence | TTTGTATTGAAGAGGGATTCACATCTGCATCTTAACTGCTC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403712 | ||
| mod ID: M6ASITE037584 | Click to Show/Hide the Full List | ||
| mod site | chr18:63126810-63126811:- | [24] | |
| Sequence | GAAAGCTGCTTTAAAAAAATACATGCATCTCAGCGTTTTTT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403713 | ||
| mod ID: M6ASITE037585 | Click to Show/Hide the Full List | ||
| mod site | chr18:63126934-63126935:- | [24] | |
| Sequence | AGTTTAGAATCAGCCTTGAAACATTGATGGAATAACTCTGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T; GM12878 | ||
| Seq Type List | MeRIP-seq; MAZTER-seq; m6A-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403714 | ||
| mod ID: M6ASITE037586 | Click to Show/Hide the Full List | ||
| mod site | chr18:63126977-63126978:- | [21] | |
| Sequence | GTGACATCTTCAGCAAATAAACTAGGAAATTTTTTTTTCTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; hNPCs; GM12878; LCLs; HEK293A-TOA; iSLK; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403715 | ||
| mod ID: M6ASITE037587 | Click to Show/Hide the Full List | ||
| mod site | chr18:63126994-63126995:- | [24] | |
| Sequence | AAGAAGTAACAAAAGAAGTGACATCTTCAGCAAATAAACTA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T; AML | ||
| Seq Type List | MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403716 | ||
| mod ID: M6ASITE037588 | Click to Show/Hide the Full List | ||
| mod site | chr18:63127020-63127021:- | [21] | |
| Sequence | AAAAAAATCAATGGTGGGGAACTATAAAGAAGTAACAAAAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; hNPCs; GM12878; LCLs; CD8T; MM6; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403717 | ||
| mod ID: M6ASITE037589 | Click to Show/Hide the Full List | ||
| mod site | chr18:63127105-63127106:- | [21] | |
| Sequence | TAGTTTGGTTTTATTTGAAAACCTGACAAAAAAAAAGTTCC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HEK293T; hNPCs; GM12878; LCLs; MM6; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403718 | ||
| mod ID: M6ASITE037590 | Click to Show/Hide the Full List | ||
| mod site | chr18:63127142-63127143:- | [21] | |
| Sequence | TCTATGGAATTACATGTAAAACATTATCTTGTCACTGTAGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; HepG2; hNPCs; GM12878; LCLs; MM6; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403719 | ||
| mod ID: M6ASITE037591 | Click to Show/Hide the Full List | ||
| mod site | chr18:63127169-63127170:- | [25] | |
| Sequence | GAGGTGTCATGGATTAATTGACCCCTGTCTATGGAATTACA | ||
| Motif Score | 2.839113095 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403720 | ||
| mod ID: M6ASITE037592 | Click to Show/Hide the Full List | ||
| mod site | chr18:63127351-63127352:- | [21] | |
| Sequence | GGGCCCTTCCTATCAGAAGGACATGGTGAAGGCTGGGAACG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; HepG2; H1A; H1B; hNPCs; hESCs; GM12878; LCLs; MM6; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403721 | ||
| mod ID: M6ASITE037593 | Click to Show/Hide the Full List | ||
| mod site | chr18:63127382-63127383:- | [21] | |
| Sequence | TATCACTGTAGAGGGAAGGAACAGAGGCCCTGGGCCCTTCC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; HepG2; H1A; H1B; hNPCs; hESCs; GM12878; LCLs; CD8T; MM6; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403722 | ||
| mod ID: M6ASITE037594 | Click to Show/Hide the Full List | ||
| mod site | chr18:63127407-63127408:- | [21] | |
| Sequence | GAGCCACGACCCTTCTTAAGACATGTATCACTGTAGAGGGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; HepG2; H1A; H1B; hNPCs; GM12878; LCLs; CD8T; MM6; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403723 | ||
| mod ID: M6ASITE037595 | Click to Show/Hide the Full List | ||
| mod site | chr18:63127606-63127607:- | [21] | |
| Sequence | CTTGGAGGCCTGGTCCTGGAACTGAGCCGGGGCCCTCACTG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; H1B; hNPCs; GM12878; LCLs; MM6; CD4T; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; endometrial; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403724 | ||
| mod ID: M6ASITE037596 | Click to Show/Hide the Full List | ||
| mod site | chr18:63127643-63127644:- | [21] | |
| Sequence | AGGAGACCGAAGTCCGCAGAACCTGCCTGTGTCCCAGCTTG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; H1A; H1B; hNPCs; hESCs; GM12878; LCLs; MM6; CD4T; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403725 | ||
| mod ID: M6ASITE037597 | Click to Show/Hide the Full List | ||
| mod site | chr18:63127658-63127659:- | [21] | |
| Sequence | CAAGTGCCTGCTTTTAGGAGACCGAAGTCCGCAGAACCTGC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; H1B; hNPCs; hESCs; GM12878; LCLs; MM6; CD4T; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403726 | ||
| mod ID: M6ASITE037598 | Click to Show/Hide the Full List | ||
| mod site | chr18:63127736-63127737:- | [21] | |
| Sequence | TCTGGAACTTGAGGAAGTGAACATTTCGGTGACTTCCGCAT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; HepG2; H1B; hNPCs; hESCs; GM12878; LCLs; CD8T; MM6; CD4T; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; endometrial; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403727 | ||
| mod ID: M6ASITE037599 | Click to Show/Hide the Full List | ||
| mod site | chr18:63127750-63127751:- | [21] | |
| Sequence | AAGAAACAGAATCCTCTGGAACTTGAGGAAGTGAACATTTC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; H1B; hNPCs; hESCs; GM12878; LCLs; MM6; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403728 | ||
| mod ID: M6ASITE037600 | Click to Show/Hide the Full List | ||
| mod site | chr18:63127765-63127766:- | [21] | |
| Sequence | TGGCTCTGTCTGAGTAAGAAACAGAATCCTCTGGAACTTGA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; H1B; hNPCs; hESCs; GM12878; LCLs; MM6; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403729 | ||
| mod ID: M6ASITE037601 | Click to Show/Hide the Full List | ||
| mod site | chr18:63127949-63127950:- | [21] | |
| Sequence | TGAGATTTCCACGCCGAAGGACAGCGATGGGAAAAATGCCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; H1A; H1B; hNPCs; GM12878; LCLs; MM6; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403730 | ||
| mod ID: M6ASITE037602 | Click to Show/Hide the Full List | ||
| mod site | chr18:63127982-63127983:- | [21] | |
| Sequence | GAGTTGGGAACTTCAGATGGACCTAGTACCCACTGAGATTT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; H1A; H1B; hNPCs; hESCs; GM12878; LCLs; CD8T; MM6; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403731 | ||
| mod ID: M6ASITE037603 | Click to Show/Hide the Full List | ||
| mod site | chr18:63127993-63127994:- | [21] | |
| Sequence | TGGGAGGAAAAGAGTTGGGAACTTCAGATGGACCTAGTACC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; H1A; H1B; hNPCs; hESCs; GM12878; LCLs; CD8T; MM6; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403732 | ||
| mod ID: M6ASITE037604 | Click to Show/Hide the Full List | ||
| mod site | chr18:63128048-63128049:- | [21] | |
| Sequence | CTCCCTGGCCTGAAGAAGAGACTCTTTGCATATGACTCACA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; H1A; H1B; hNPCs; hESCs; GM12878; LCLs; MM6; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403733 | ||
| mod ID: M6ASITE037605 | Click to Show/Hide the Full List | ||
| mod site | chr18:63128103-63128104:- | [24] | |
| Sequence | CCCTGGGGGCTGTGATATTAACAGAGGGAGGGTTCCTGTGG | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T; AML | ||
| Seq Type List | MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403734 | ||
| mod ID: M6ASITE037606 | Click to Show/Hide the Full List | ||
| mod site | chr18:63128199-63128200:- | [21] | |
| Sequence | AGCAGACGGATGGAAAAAGGACCTGATCATTGGGGAAGCTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; H1A; H1B; GM12878; LCLs; MM6; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403735 | ||
| mod ID: M6ASITE037607 | Click to Show/Hide the Full List | ||
| mod site | chr18:63128214-63128215:- | [25] | |
| Sequence | GATCACCATCTGAAGAGCAGACGGATGGAAAAAGGACCTGA | ||
| Motif Score | 2.871321429 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000333681.5; ENST00000590515.1; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403736 | ||
| mod ID: M6ASITE037608 | Click to Show/Hide the Full List | ||
| mod site | chr18:63128264-63128265:- | [21] | |
| Sequence | TGGATGTTCTGTGCCTGTAAACATAGATTCGCTTTCCATGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; GM12878; LCLs; MM6; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000590515.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403737 | ||
| mod ID: M6ASITE037609 | Click to Show/Hide the Full List | ||
| mod site | chr18:63128343-63128344:- | [26] | |
| Sequence | TTTAAGACAGTCCCATCAAAACTCCTGTCTTTGGAAATCCG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T; GM12878; LCLs; HEK293A-TOA | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000590515.1; ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403738 | ||
| mod ID: M6ASITE037610 | Click to Show/Hide the Full List | ||
| mod site | chr18:63128357-63128358:- | [21] | |
| Sequence | AAAAGATTTATTTATTTAAGACAGTCCCATCAAAACTCCTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; GM12878; LCLs; HEK293A-TOA; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1; ENST00000590515.1 | ||
| External Link | RMBase: m6A_site_403739 | ||
| mod ID: M6ASITE037611 | Click to Show/Hide the Full List | ||
| mod site | chr18:63128481-63128482:- | [21] | |
| Sequence | TATAAACATCACACACACAGACAGACACACACACACACAAC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hNPCs; GM12878; LCLs; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000590515.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403740 | ||
| mod ID: M6ASITE037612 | Click to Show/Hide the Full List | ||
| mod site | chr18:63128496-63128497:- | [21] | |
| Sequence | AAAAAATAACACACATATAAACATCACACACACAGACAGAC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; CD34; HEK293T; hNPCs; GM12878; LCLs; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000590515.1; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403741 | ||
| mod ID: M6ASITE037613 | Click to Show/Hide the Full List | ||
| mod site | chr18:63128538-63128539:- | [27] | |
| Sequence | TGTCAGTGATGTACCATGAAACAAAGCTGCAGGCTGTTTAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | CD34; HEK293T; hNPCs; GM12878; LCLs; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000590515.1; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403742 | ||
| mod ID: M6ASITE037614 | Click to Show/Hide the Full List | ||
| mod site | chr18:63128603-63128604:- | [21] | |
| Sequence | AGTCAACATGCCTGCCCCAAACAAATATGCAAAAGGTTCAC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; CD34; HEK293T; GM12878; LCLs; MM6; peripheral-blood; iSLK; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1; ENST00000590515.1 | ||
| External Link | RMBase: m6A_site_403743 | ||
| mod ID: M6ASITE037615 | Click to Show/Hide the Full List | ||
| mod site | chr18:63128689-63128690:- | [21] | |
| Sequence | TCTCCTGGCTGTCTCTGAAGACTCTGCTCAGTTTGGCCCTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; CD34; HEK293T; HepG2; H1A; H1B; GM12878; LCLs; MM6; CD4T; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000590515.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403744 | ||
| mod ID: M6ASITE037616 | Click to Show/Hide the Full List | ||
| mod site | chr18:63128744-63128745:- | [21] | |
| Sequence | CTGCAGGATGCCTTTGTGGAACTGTACGGCCCCAGCATGCG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; H1A; H1B; GM12878; LCLs; MM6; CD4T; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1; ENST00000590515.1 | ||
| External Link | RMBase: m6A_site_403745 | ||
| mod ID: M6ASITE037617 | Click to Show/Hide the Full List | ||
| mod site | chr18:63216101-63216102:- | [2] | |
| Sequence | CACCAAAAACAAACAAACAAACAAAACCACAGCCCTATTGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403746 | ||
| mod ID: M6ASITE037618 | Click to Show/Hide the Full List | ||
| mod site | chr18:63281093-63281094:- | [2] | |
| Sequence | TAGGTACCTCGCATAAGTGGACTCACACTTGTGACTGGCTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403747 | ||
| mod ID: M6ASITE037619 | Click to Show/Hide the Full List | ||
| mod site | chr18:63313896-63313897:- | [21] | |
| Sequence | TTTTAGCAATCTTTGATTGAACTGGTACATTTCAGATTACT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000589955.2; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403748 | ||
| mod ID: M6ASITE037620 | Click to Show/Hide the Full List | ||
| mod site | chr18:63314614-63314615:- | [27] | |
| Sequence | AGTGACCCTCTAAACTGGGAACTGTGACGACATTCAGCTTC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | CD34 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000589955.2; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403749 | ||
| mod ID: M6ASITE037621 | Click to Show/Hide the Full List | ||
| mod site | chr18:63314621-63314622:- | [27] | |
| Sequence | GATGGAGAGTGACCCTCTAAACTGGGAACTGTGACGACATT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | CD34 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000589955.2; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403750 | ||
| mod ID: M6ASITE037622 | Click to Show/Hide the Full List | ||
| mod site | chr18:63314676-63314677:- | [27] | |
| Sequence | AGCTTCTTTAAGACAAAGGGACTCTTGCAGGCAAGGGATTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | CD34 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000589955.2; ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403751 | ||
| mod ID: M6ASITE037623 | Click to Show/Hide the Full List | ||
| mod site | chr18:63317459-63317460:- | [21] | |
| Sequence | TGACATTCCAAAGTCCTTGGACTGGAATCTCCAGTGTGTGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; GM12878; LCLs; peripheral-blood; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5; ENST00000589955.2 | ||
| External Link | RMBase: m6A_site_403752 | ||
| mod ID: M6ASITE037624 | Click to Show/Hide the Full List | ||
| mod site | chr18:63317507-63317508:- | [21] | |
| Sequence | TAAGTCTTTTTCCCTGAAAAACTTAGTTGTGTTTGTAAAGA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; GM12878; LCLs; peripheral-blood; HEK293A-TOA; iSLK; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5; ENST00000589955.2 | ||
| External Link | RMBase: m6A_site_403753 | ||
| mod ID: M6ASITE037625 | Click to Show/Hide the Full List | ||
| mod site | chr18:63317534-63317535:- | [21] | |
| Sequence | AAATTTTCCAAAAGAGGAAAACCAATGTAAGTCTTTTTCCC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; GM12878; LCLs; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000589955.2; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403754 | ||
| mod ID: M6ASITE037626 | Click to Show/Hide the Full List | ||
| mod site | chr18:63317577-63317578:- | [21] | |
| Sequence | AAGAATAGACTCCACAGAAGACCAAGGGCATCATCAATCGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; hNPCs; GM12878; LCLs; MM6; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000589955.2; ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403755 | ||
| mod ID: M6ASITE037627 | Click to Show/Hide the Full List | ||
| mod site | chr18:63317589-63317590:- | [21] | |
| Sequence | CAACCCGATTAGAAGAATAGACTCCACAGAAGACCAAGGGC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; H1A; H1B; hNPCs; GM12878; LCLs; MM6; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000589955.2; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403756 | ||
| mod ID: M6ASITE037628 | Click to Show/Hide the Full List | ||
| mod site | chr18:63317610-63317611:- | [21] | |
| Sequence | TTGTACACTAAAATTGGAGAACAACCCGATTAGAAGAATAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; H1A; H1B; hNPCs; GM12878; LCLs; CD8T; MM6; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000589955.2; ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403757 | ||
| mod ID: M6ASITE037629 | Click to Show/Hide the Full List | ||
| mod site | chr18:63317639-63317640:- | [21] | |
| Sequence | CCCAAACAAGCTCCCCTGAAACAAGCCCGTTGTACACTAAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; H1A; H1B; GM12878; LCLs; MM6; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5; ENST00000589955.2 | ||
| External Link | RMBase: m6A_site_403758 | ||
| mod ID: M6ASITE037630 | Click to Show/Hide the Full List | ||
| mod site | chr18:63317654-63317655:- | [21] | |
| Sequence | CTTGACAGTCTGCATCCCAAACAAGCTCCCCTGAAACAAGC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; H1A; H1B; GM12878; LCLs; MM6; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000589955.2; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403759 | ||
| mod ID: M6ASITE037631 | Click to Show/Hide the Full List | ||
| mod site | chr18:63317693-63317694:- | [21] | |
| Sequence | GTTTCAGGGACTGCTTATGAACCAGATACCAGGTTGGGTCT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; H1A; H1B; GM12878; LCLs; MM6; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000589955.2; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403760 | ||
| mod ID: M6ASITE037632 | Click to Show/Hide the Full List | ||
| mod site | chr18:63317704-63317705:- | [21] | |
| Sequence | ACCGGAGGGAGGTTTCAGGGACTGCTTATGAACCAGATACC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; H1A; H1B; GM12878; LCLs; peripheral-blood; HEK293A-TOA; iSLK; TIME; MSC; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000589955.2; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403761 | ||
| mod ID: M6ASITE037633 | Click to Show/Hide the Full List | ||
| mod site | chr18:63317724-63317725:- | [21] | |
| Sequence | GGCTTGAGCAGCTTGTTGGAACCGGAGGGAGGTTTCAGGGA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; H1A; H1B; GM12878; LCLs; peripheral-blood; HEK293A-TOA; iSLK; TIME; MSC; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5; ENST00000589955.2 | ||
| External Link | RMBase: m6A_site_403762 | ||
| mod ID: M6ASITE037634 | Click to Show/Hide the Full List | ||
| mod site | chr18:63317757-63317758:- | [21] | |
| Sequence | AAGGTCCAGGAGTCCCCAGGACCACCGTTTCCTGGCTTGAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; H1A; H1B; GM12878; LCLs; peripheral-blood; HEK293A-TOA; iSLK; TIME; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5; ENST00000589955.2 | ||
| External Link | RMBase: m6A_site_403763 | ||
| mod ID: M6ASITE037635 | Click to Show/Hide the Full List | ||
| mod site | chr18:63317925-63317926:- | [21] | |
| Sequence | GTCCCTACCTCCTCCTCTGGACAAAGCGTTCACTCCCAACC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; H1B; H1A; hESCs; GM12878; LCLs; CD8T; MT4; peripheral-blood; HEK293A-TOA; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000589955.2; ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403764 | ||
| mod ID: M6ASITE037636 | Click to Show/Hide the Full List | ||
| mod site | chr18:63318121-63318122:- | [21] | |
| Sequence | GTGGATGACTGAGTACCTGAACCGGCACCTGCACACCTGGA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; H1A; H1B; GM12878; LCLs; MT4; MM6; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5; ENST00000589955.2 | ||
| External Link | RMBase: m6A_site_403765 | ||
| mod ID: M6ASITE037637 | Click to Show/Hide the Full List | ||
| mod site | chr18:63318154-63318155:- | [21] | |
| Sequence | GGAGATGTCGCCCCTGGTGGACAACATCGCCCTGTGGATGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; GM12878; LCLs; CD8T; MT4; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5; ENST00000589955.2 | ||
| External Link | RMBase: m6A_site_403766 | ||
| mod ID: M6ASITE037638 | Click to Show/Hide the Full List | ||
| mod site | chr18:63318238-63318239:- | [21] | |
| Sequence | GCTCTTCAGGGACGGGGTGAACTGGGGGAGGATTGTGGCCT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; U2OS; H1A; H1B; hESCs; GM12878; LCLs; CD8T; MT4; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000589955.2; ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403767 | ||
| mod ID: M6ASITE037639 | Click to Show/Hide the Full List | ||
| mod site | chr18:63318282-63318283:- | [25] | |
| Sequence | ACGCCCTTCACCGCGCGGGGACGCTTTGCCACGGTGGTGGA | ||
| Motif Score | 3.616982143 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000589955.2; ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403768 | ||
| mod ID: M6ASITE037640 | Click to Show/Hide the Full List | ||
| mod site | chr18:63318446-63318447:- | [21] | |
| Sequence | CCAGGACCTCGCCGCTGCAGACCCCGGCTGCCCCCGGCGCC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; U2OS; GM12878; LCLs; MT4; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000589955.2; ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403769 | ||
| mod ID: M6ASITE037641 | Click to Show/Hide the Full List | ||
| mod site | chr18:63318461-63318462:- | [21] | |
| Sequence | CCCGGGACCCGGTCGCCAGGACCTCGCCGCTGCAGACCCCG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; U2OS; GM12878; LCLs; MT4; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000589955.2; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403770 | ||
| mod ID: M6ASITE037642 | Click to Show/Hide the Full List | ||
| mod site | chr18:63318475-63318476:- | [21] | |
| Sequence | CCATCCAGCCGCATCCCGGGACCCGGTCGCCAGGACCTCGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; U2OS; GM12878; LCLs; MT4; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000589955.2; ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403771 | ||
| mod ID: M6ASITE037643 | Click to Show/Hide the Full List | ||
| mod site | chr18:63318647-63318648:- | [21] | |
| Sequence | GGATGGCGCACGCTGGGAGAACAGGGTACGATAACCGGGAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; CD34; HEK293T; hESC-HEK293T; HepG2; GM12878; LCLs; MT4; MM6; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; endometrial; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000333681.5; ENST00000589955.2; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403772 | ||
| mod ID: M6ASITE037644 | Click to Show/Hide the Full List | ||
| mod site | chr18:63318749-63318750:- | [2] | |
| Sequence | AATAAAATAGCTGGATTATAACTCCTCTTCTTTCTCTGGGG | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1; ENST00000589955.2 | ||
| External Link | RMBase: m6A_site_403773 | ||
| mod ID: M6ASITE037645 | Click to Show/Hide the Full List | ||
| mod site | chr18:63318802-63318803:- | [21] | |
| Sequence | AGTGTTCCGCGTGATTGAAGACACCCCCTCGTCCAAGAATG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; hNPCs; GM12878; LCLs; MT4; HEK293A-TOA; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000589955.2; ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403774 | ||
| mod ID: M6ASITE037646 | Click to Show/Hide the Full List | ||
| mod site | chr18:63318834-63318835:- | [21] | |
| Sequence | CATCTCATGCCAAGGGGGAAACACCAGAATCAAGTGTTCCG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; hNPCs; GM12878; LCLs; HEK293A-TOA; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403775 | ||
| mod ID: M6ASITE037647 | Click to Show/Hide the Full List | ||
| mod site | chr18:63318934-63318935:- | [21] | |
| Sequence | CAGCATCACAGAGGAAGTAGACTGATATTAACAATACTTAC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; hNPCs; GM12878; LCLs; HEK293A-TOA; iSLK; MSC; TIME; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403776 | ||
| mod ID: M6ASITE037648 | Click to Show/Hide the Full List | ||
| mod site | chr18:63318960-63318961:- | [25] | |
| Sequence | ATTAGTTTGTTTTTTCTTTAACCTTTCAGCATCACAGAGGA | ||
| Motif Score | 2.147452381 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403777 | ||
| mod ID: M6ASITE037649 | Click to Show/Hide the Full List | ||
| mod site | chr18:63319125-63319126:- | [26] | |
| Sequence | TAATGTAACTTTCAATGGAAACCTTTGAGATTTTTTACTTA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HEK293T; GM12878; LCLs; iSLK | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403778 | ||
| mod ID: M6ASITE037650 | Click to Show/Hide the Full List | ||
| mod site | chr18:63319209-63319210:- | [21] | |
| Sequence | TTTGGTTTTACAAAAAGGAAACTTGACAGAGGATCATGCTG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; GM12878; LCLs; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000333681.5; ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403779 | ||
| mod ID: M6ASITE037651 | Click to Show/Hide the Full List | ||
| mod site | chr18:63319382-63319383:- | [2] | |
| Sequence | GTTGGGATTCCTGCGGATTGACATTTCTGTGAAGCAGAAGT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403780 | ||
| mod ID: M6ASITE037652 | Click to Show/Hide the Full List | ||
| mod site | chr18:63319469-63319470:- | [21] | |
| Sequence | AGGACTTCTGCGAATACCGGACTGAAAATTGTAATTCATCT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; hNPCs; A549; GM12878; LCLs; CD8T; MT4; MM6; CD4T; peripheral-blood; HEK293A-TOA; iSLK; MSC; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403781 | ||
| mod ID: M6ASITE037653 | Click to Show/Hide the Full List | ||
| mod site | chr18:63319486-63319487:- | [21] | |
| Sequence | GAAGGGGAAAACATCACAGGACTTCTGCGAATACCGGACTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; hNPCs; A549; GM12878; LCLs; CD8T; MT4; MM6; CD4T; peripheral-blood; HEK293A-TOA; iSLK; MSC; endometrial; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403782 | ||
| mod ID: M6ASITE037654 | Click to Show/Hide the Full List | ||
| mod site | chr18:63319496-63319497:- | [21] | |
| Sequence | AAATTGCCGAGAAGGGGAAAACATCACAGGACTTCTGCGAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; hNPCs; A549; GM12878; LCLs; MT4; MM6; CD4T; peripheral-blood; HEK293A-TOA; iSLK; MSC; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1; ENST00000333681.5 | ||
| External Link | RMBase: m6A_site_403783 | ||
| mod ID: M6ASITE037655 | Click to Show/Hide the Full List | ||
| mod site | chr18:63319795-63319796:- | [21] | |
| Sequence | GAAAAGAGGGAAAAAATAAAACCCTCCCCCACCACCTCCTT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; hNPCs; LCLs; MT4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403784 | ||
| mod ID: M6ASITE037656 | Click to Show/Hide the Full List | ||
| mod site | chr18:63319840-63319841:- | [21] | |
| Sequence | GAGGGGGTCCAGGGGGGAGAACTTCGTAGCAGTCATCCTTT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; H1B; hNPCs; LCLs; MT4; MM6; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403785 | ||
| mod ID: M6ASITE037657 | Click to Show/Hide the Full List | ||
| mod site | chr18:63319923-63319924:- | [21] | |
| Sequence | AGCCAGCGCCGCCGCGCAGGACCAGGAGGAGGAGAAAGGGT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; CD34; hNPCs; LCLs; MT4; HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000398117.1 | ||
| External Link | RMBase: m6A_site_403786 | ||
References