m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00310)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
KLF4
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
YTH domain-containing family protein 2 (YTHDF2) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
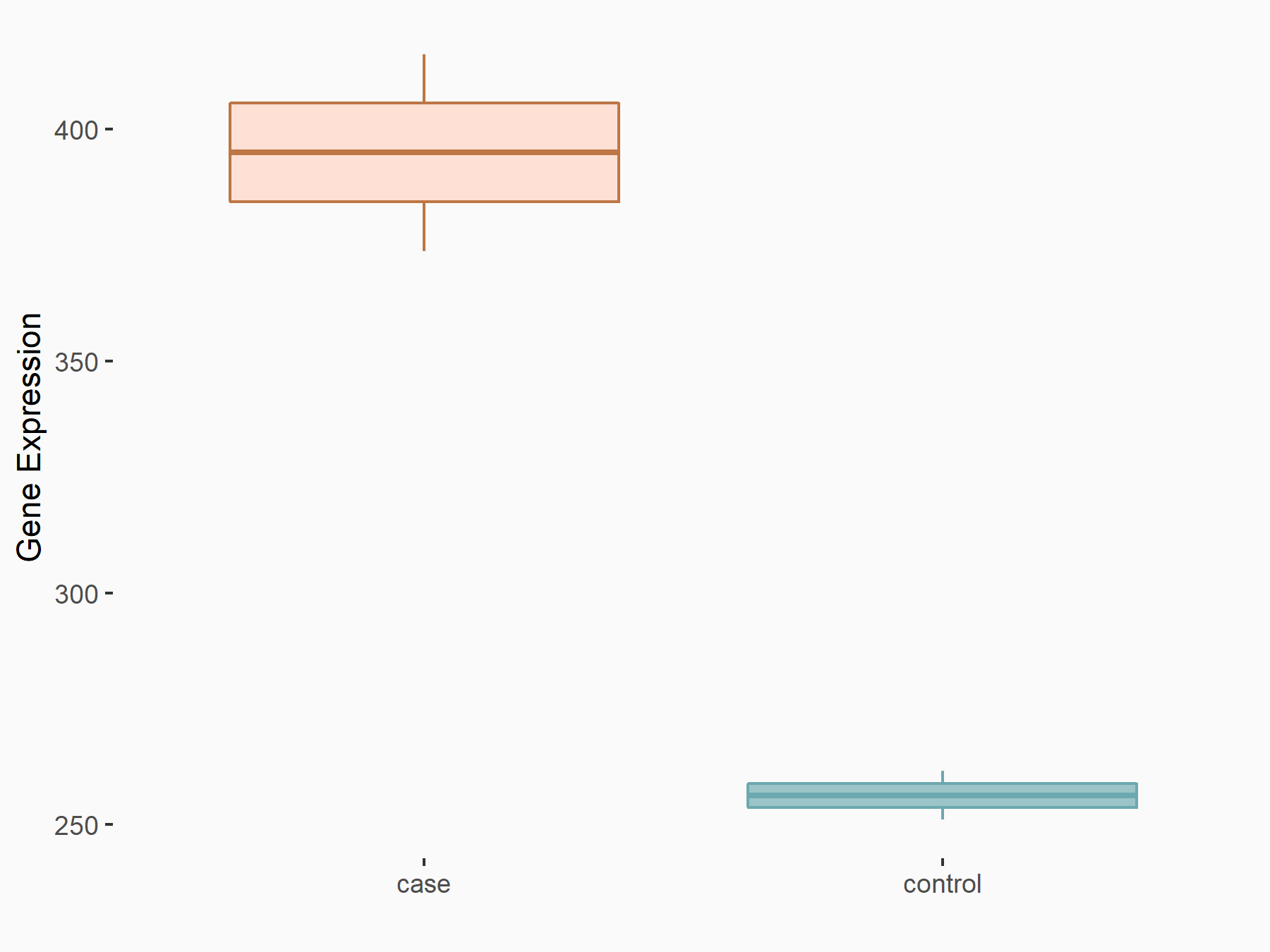  |
logFC: 6.24E-01 p-value: 8.92E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between KLF4 and the regulator | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.55E+00 | GSE49339 |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | MiR-1915-3p expression was regulated by METTL3/YTHDF2 m6A axis through transcription factor Krueppel-like factor 4 (KLF4). miR-1915-3p function as a tumor suppressor by targeting SET and has an anti-metastatic therapeutic potential for lung cancer treatment. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | TNF signaling pathway | hsa04668 | ||
| Cell Process | Cell migration | |||
| Cell invasion | ||||
| Epithelial-mesenchymal transition | ||||
| In-vitro Model | NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3/YTHDF2/SETD7/Krueppel-like factor 4 (KLF4) m6 A axis provide the insight into the underlying mechanism of carcinogenesis and highlight potential therapeutic targets for bladder cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Bladder cancer | ICD-11: 2C94 | ||
| Cell Process | Cancer proliferation | |||
| Cancer metastasis | ||||
| In-vitro Model | SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| In-vivo Model | For the subcutaneous implantation model, UM-UC-3 cells (2 × 106 cells per mouse) stably METTL3 knocked down (shMETTL3-1, shMETTL3-2) were injected into the flanks of mice. | |||
Methyltransferase-like 14 (METTL14) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL14 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
|
Treatment: siMETTL14 MDA-MB-231 cells
Control: MDA-MB-231 cells
|
GSE81164 | |
| Regulation |
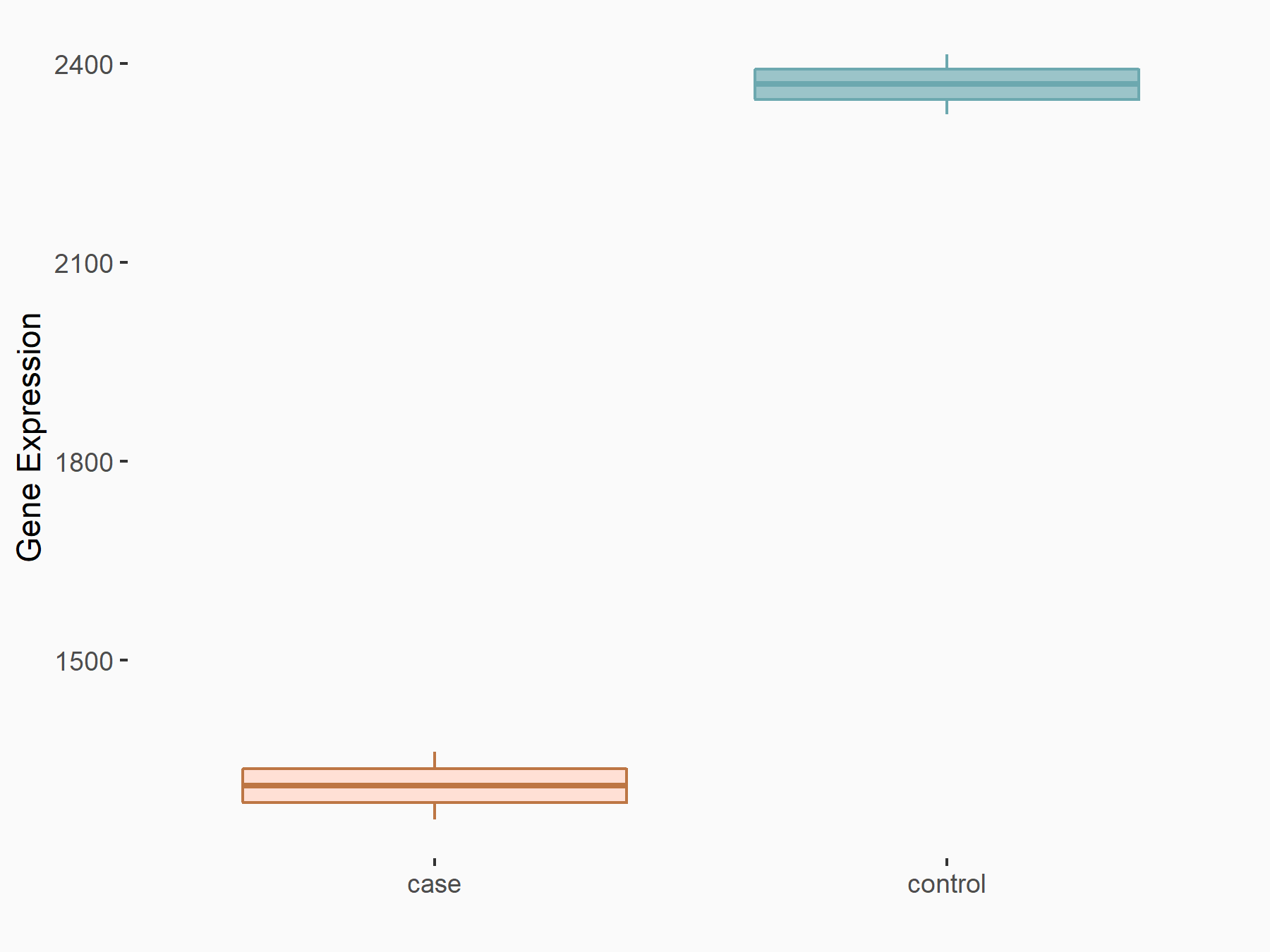  |
logFC: -8.55E-01 p-value: 7.89E-09 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | The expression of METTL14 was downregulated in CRC cells and METTL14 could inhibit the metastasis of CRC cells. MeCP2 could bind to METTL14 to coregulate tumor suppressor Krueppel-like factor 4 (KLF4) expression through changing m6 A methylation modification. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| In-vitro Model | SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| In-vivo Model | In vivo, the group of HCT116 cells labeled with luciferase were surgically injected into the spleen of nude mice. After 2 months, the nude mice were subjected to D-luciferin injection and the luciferase signals were monitored and quantified. | |||
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HUVEC cell line | Homo sapiens |
|
Treatment: shMETTL3 HUVEC cells
Control: shScramble HUVEC cells
|
GSE157544 | |
| Regulation |
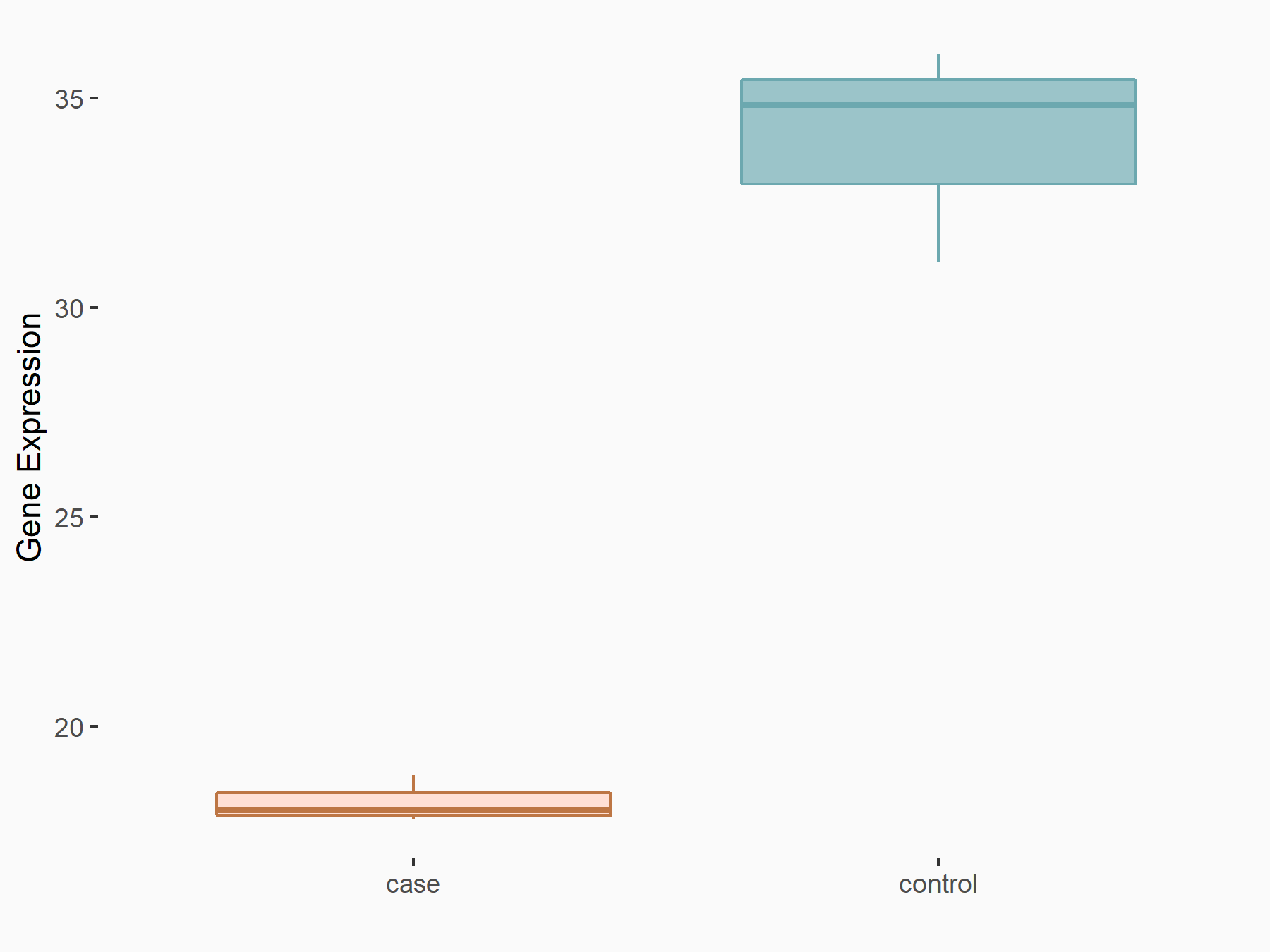 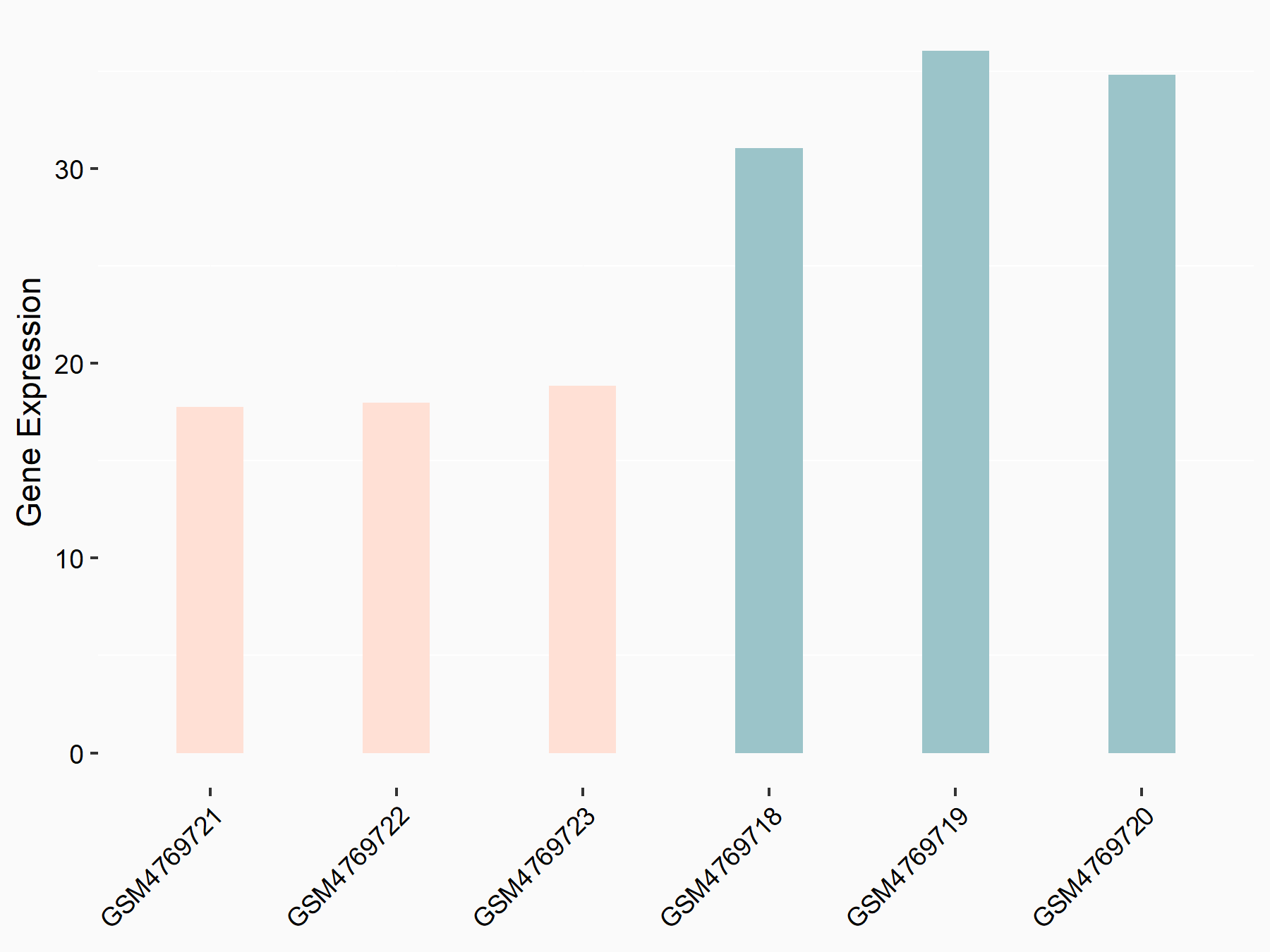 |
logFC: -8.63E-01 p-value: 1.22E-04 |
| More Results | Click to View More RNA-seq Results | |
| In total 3 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | MiR-1915-3p expression was regulated by METTL3/YTHDF2 m6A axis through transcription factor Krueppel-like factor 4 (KLF4). miR-1915-3p function as a tumor suppressor by targeting SET and has an anti-metastatic therapeutic potential for lung cancer treatment. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | TNF signaling pathway | hsa04668 | ||
| Cell Process | Cell migration | |||
| Cell invasion | ||||
| Epithelial-mesenchymal transition | ||||
| In-vitro Model | NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3/YTHDF2/SETD7/Krueppel-like factor 4 (KLF4) m6 A axis provide the insight into the underlying mechanism of carcinogenesis and highlight potential therapeutic targets for bladder cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Bladder cancer | ICD-11: 2C94 | ||
| Cell Process | Cancer proliferation | |||
| Cancer metastasis | ||||
| In-vitro Model | SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| In-vivo Model | For the subcutaneous implantation model, UM-UC-3 cells (2 × 106 cells per mouse) stably METTL3 knocked down (shMETTL3-1, shMETTL3-2) were injected into the flanks of mice. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | In the in vivo atherosclerosis model,partial ligation of the carotid artery led to plaque formation and up-regulation of METTL3 and NLRP1, with down-regulation of KLF4; knockdown of METTL3 via repetitive shRNA administration prevented the atherogenic process, NLRP1 up-regulation, and Krueppel-like factor 4 (KLF4) down-regulation. Collectively, it has demonstrated that METTL3 serves a central role in the atherogenesis induced by oscillatory stress and disturbed blood flow. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Atherosclerosis | ICD-11: BD40.Z | ||
| In-vitro Model | MAEC | Normal | Mus musculus | CVCL_U411 |
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | The expression of METTL14 was downregulated in CRC cells and METTL14 could inhibit the metastasis of CRC cells. MeCP2 could bind to METTL14 to coregulate tumor suppressor Krueppel-like factor 4 (KLF4) expression through changing m6 A methylation modification. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| In-vivo Model | In vivo, the group of HCT116 cells labeled with luciferase were surgically injected into the spleen of nude mice. After 2 months, the nude mice were subjected to D-luciferin injection and the luciferase signals were monitored and quantified. | |||
Lung cancer [ICD-11: 2C25]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | MiR-1915-3p expression was regulated by METTL3/YTHDF2 m6A axis through transcription factor Krueppel-like factor 4 (KLF4). miR-1915-3p function as a tumor suppressor by targeting SET and has an anti-metastatic therapeutic potential for lung cancer treatment. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | TNF signaling pathway | hsa04668 | ||
| Cell Process | Cell migration | |||
| Cell invasion | ||||
| Epithelial-mesenchymal transition | ||||
| In-vitro Model | NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | MiR-1915-3p expression was regulated by METTL3/YTHDF2 m6A axis through transcription factor Krueppel-like factor 4 (KLF4). miR-1915-3p function as a tumor suppressor by targeting SET and has an anti-metastatic therapeutic potential for lung cancer treatment. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | TNF signaling pathway | hsa04668 | ||
| Cell Process | Cell migration | |||
| Cell invasion | ||||
| Epithelial-mesenchymal transition | ||||
| In-vitro Model | NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
Bladder cancer [ICD-11: 2C94]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3/YTHDF2/SETD7/Krueppel-like factor 4 (KLF4) m6 A axis provide the insight into the underlying mechanism of carcinogenesis and highlight potential therapeutic targets for bladder cancer. | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Cell Process | Cancer proliferation | |||
| Cancer metastasis | ||||
| In-vitro Model | SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| In-vivo Model | For the subcutaneous implantation model, UM-UC-3 cells (2 × 106 cells per mouse) stably METTL3 knocked down (shMETTL3-1, shMETTL3-2) were injected into the flanks of mice. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3/YTHDF2/SETD7/Krueppel-like factor 4 (KLF4) m6 A axis provide the insight into the underlying mechanism of carcinogenesis and highlight potential therapeutic targets for bladder cancer. | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Down regulation | |||
| Cell Process | Cancer proliferation | |||
| Cancer metastasis | ||||
| In-vitro Model | SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| In-vivo Model | For the subcutaneous implantation model, UM-UC-3 cells (2 × 106 cells per mouse) stably METTL3 knocked down (shMETTL3-1, shMETTL3-2) were injected into the flanks of mice. | |||
Atherosclerosis [ICD-11: BD40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | In the in vivo atherosclerosis model,partial ligation of the carotid artery led to plaque formation and up-regulation of METTL3 and NLRP1, with down-regulation of KLF4; knockdown of METTL3 via repetitive shRNA administration prevented the atherogenic process, NLRP1 up-regulation, and Krueppel-like factor 4 (KLF4) down-regulation. Collectively, it has demonstrated that METTL3 serves a central role in the atherogenesis induced by oscillatory stress and disturbed blood flow. | |||
| Responsed Disease | Atherosclerosis [ICD-11: BD40.Z] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | MAEC | Normal | Mus musculus | CVCL_U411 |
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: Methyltransferase-like 14 (METTL14)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03284 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5C (KDM5C) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00310)
| In total 34 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE088979 | Click to Show/Hide the Full List | ||
| mod site | chr9:107485233-107485234:- | [5] | |
| Sequence | CTTGGTTCTAAAGGTACCAAACAAGGAAGCCAAAGTTTTCA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000374672.5; ENST00000610832.1; ENST00000493306.1; ENST00000497048.5 | ||
| External Link | RMBase: m6A_site_832383 | ||
| mod ID: M6ASITE088980 | Click to Show/Hide the Full List | ||
| mod site | chr9:107485416-107485417:- | [6] | |
| Sequence | CGTGAGTTGGGGGAGGGAAGACCAGAATTCCCTTGAATTGT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; MM6; Huh7; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000493306.1; ENST00000374672.5; ENST00000497048.5; ENST00000610832.1 | ||
| External Link | RMBase: m6A_site_832384 | ||
| mod ID: M6ASITE088981 | Click to Show/Hide the Full List | ||
| mod site | chr9:107485495-107485496:- | [6] | |
| Sequence | TCCGACTTGAATATTCCTGGACTTACAAAATGCCAAGGGGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; MM6; Huh7; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000374672.5; ENST00000493306.1; ENST00000610832.1; ENST00000497048.5 | ||
| External Link | RMBase: m6A_site_832385 | ||
| mod ID: M6ASITE088982 | Click to Show/Hide the Full List | ||
| mod site | chr9:107485511-107485512:- | [7] | |
| Sequence | TCATTCCAATTCTAAATCCGACTTGAATATTCCTGGACTTA | ||
| Motif Score | 3.287565476 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000374672.5; ENST00000610832.1; ENST00000497048.5; ENST00000493306.1 | ||
| External Link | RMBase: m6A_site_832386 | ||
| mod ID: M6ASITE088983 | Click to Show/Hide the Full List | ||
| mod site | chr9:107485560-107485561:- | [6] | |
| Sequence | CAAAAGACAAAAATCAAAGAACAGATGGGGTCTGTGACTGG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; MM6; Huh7; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000497048.5; ENST00000493306.1; ENST00000610832.1; ENST00000374672.5 | ||
| External Link | RMBase: m6A_site_832387 | ||
| mod ID: M6ASITE088984 | Click to Show/Hide the Full List | ||
| mod site | chr9:107485574-107485575:- | [6] | |
| Sequence | AAAAATGAGGAATCCAAAAGACAAAAATCAAAGAACAGATG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; MM6; Huh7; Jurkat; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000493306.1; ENST00000610832.1; ENST00000497048.5; ENST00000374672.5 | ||
| External Link | RMBase: m6A_site_832388 | ||
| mod ID: M6ASITE088985 | Click to Show/Hide the Full List | ||
| mod site | chr9:107485742-107485743:- | [6] | |
| Sequence | GAGGCATTTTTAAATCCCAGACAGTGGATATGACCCACACT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; fibroblasts; H1299; Huh7; HEK293A-TOA; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374672.5; ENST00000497048.5; ENST00000610832.1; ENST00000493306.1 | ||
| External Link | RMBase: m6A_site_832389 | ||
| mod ID: M6ASITE088986 | Click to Show/Hide the Full List | ||
| mod site | chr9:107485784-107485785:- | [6] | |
| Sequence | CCGAGCATTTTCCAGGTCGGACCACCTCGCCTTACACATGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; hESCs; fibroblasts; H1299; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000493306.1; ENST00000374672.5; ENST00000497048.5; ENST00000610832.1 | ||
| External Link | RMBase: m6A_site_832390 | ||
| mod ID: M6ASITE088987 | Click to Show/Hide the Full List | ||
| mod site | chr9:107485843-107485844:- | [6] | |
| Sequence | CTGACCAGGCACTACCGTAAACACACGGGGCACCGCCCGTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; fibroblasts; H1299; MM6; Huh7; Jurkat; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000493306.1; ENST00000374672.5; ENST00000610832.1; ENST00000497048.5 | ||
| External Link | RMBase: m6A_site_832391 | ||
| mod ID: M6ASITE088988 | Click to Show/Hide the Full List | ||
| mod site | chr9:107485864-107485865:- | [6] | |
| Sequence | AAATTCGCCCGCTCAGATGAACTGACCAGGCACTACCGTAA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; fibroblasts; H1299; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000497048.5; ENST00000493306.1; ENST00000610832.1; ENST00000374672.5 | ||
| External Link | RMBase: m6A_site_832392 | ||
| mod ID: M6ASITE088989 | Click to Show/Hide the Full List | ||
| mod site | chr9:107485898-107485899:- | [7] | |
| Sequence | ACCTTACCACTGTGACTGGGACGGCTGTGGATGGAAATTCG | ||
| Motif Score | 3.616982143 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000610832.1; ENST00000497048.5; ENST00000374672.5; ENST00000493306.1 | ||
| External Link | RMBase: m6A_site_832393 | ||
| mod ID: M6ASITE088990 | Click to Show/Hide the Full List | ||
| mod site | chr9:107485918-107485919:- | [6] | |
| Sequence | TTTACCTTTACAGGTGAGAAACCTTACCACTGTGACTGGGA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; fibroblasts; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000610832.1; ENST00000493306.1; ENST00000374672.5; ENST00000497048.5 | ||
| External Link | RMBase: m6A_site_832394 | ||
| mod ID: M6ASITE088991 | Click to Show/Hide the Full List | ||
| mod site | chr9:107487036-107487037:- | [6] | |
| Sequence | ATCTCAAGGCACACCTGCGAACCCACACAGGTAGGGGGACA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; fibroblasts; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000610832.1; ENST00000497048.5; ENST00000374672.5; ENST00000493306.1 | ||
| External Link | RMBase: m6A_site_832395 | ||
| mod ID: M6ASITE088992 | Click to Show/Hide the Full List | ||
| mod site | chr9:107487075-107487076:- | [6] | |
| Sequence | ATTACGCGGGCTGCGGCAAAACCTACACAAAGAGTTCCCAT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; fibroblasts; A549; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000610832.1; ENST00000497048.5; ENST00000493306.1; ENST00000374672.5 | ||
| External Link | RMBase: m6A_site_832396 | ||
| mod ID: M6ASITE088993 | Click to Show/Hide the Full List | ||
| mod site | chr9:107487114-107487115:- | [6] | |
| Sequence | GATCGTGGCCCCGGAAAAGGACCGCCACCCACACTTGTGAT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; fibroblasts; MM6; Huh7; iSLK; MSC; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000610832.1; ENST00000497048.5; ENST00000493306.1; ENST00000374672.5 | ||
| External Link | RMBase: m6A_site_832397 | ||
| mod ID: M6ASITE088994 | Click to Show/Hide the Full List | ||
| mod site | chr9:107487231-107487232:- | [6] | |
| Sequence | TGTGCTGGCCCCACTTCGGGACACACGGGATGATGCTCACC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; fibroblasts; Huh7; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000610832.1; ENST00000493306.1; ENST00000374672.5; ENST00000497048.5 | ||
| External Link | RMBase: m6A_site_832398 | ||
| mod ID: M6ASITE088995 | Click to Show/Hide the Full List | ||
| mod site | chr9:107487407-107487408:- | [6] | |
| Sequence | GGAAGTGCTGAGCAGCAGGGACTGTCACCCTGCCCTGCCGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; fibroblasts; MM6; Huh7; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000497048.5; ENST00000374672.5; ENST00000610832.1; ENST00000493306.1 | ||
| External Link | RMBase: m6A_site_832399 | ||
| mod ID: M6ASITE088996 | Click to Show/Hide the Full List | ||
| mod site | chr9:107487450-107487451:- | [6] | |
| Sequence | GGCGGCAGCTCCCCAGCAGGACTACCCCGACCCTGGGTCTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; H1A; H1B; fibroblasts; MM6; GSC-11; HEK293A-TOA; MSC; TIME; iSLK; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000497048.5; ENST00000610832.1; ENST00000374672.5; ENST00000493306.1 | ||
| External Link | RMBase: m6A_site_832400 | ||
| mod ID: M6ASITE088997 | Click to Show/Hide the Full List | ||
| mod site | chr9:107487520-107487521:- | [6] | |
| Sequence | TGCACCCACTTGGGCGCTGGACCCCCTCTCAGCAATGGCCA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; fibroblasts; MM6; GSC-11; HEK293A-TOA; MSC; iSLK; TIME; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374672.5; ENST00000610832.1; ENST00000493306.1; ENST00000497048.5 | ||
| External Link | RMBase: m6A_site_832401 | ||
| mod ID: M6ASITE088998 | Click to Show/Hide the Full List | ||
| mod site | chr9:107487767-107487768:- | [6] | |
| Sequence | GCTCCTGCGGCCAGAATTGGACCCGGTGTACATTCCGCCGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; H1A; H1B; fibroblasts; MM6; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000497048.5; ENST00000493306.1; ENST00000610832.1; ENST00000374672.5; ENST00000411706.1 | ||
| External Link | RMBase: m6A_site_832402 | ||
| mod ID: M6ASITE088999 | Click to Show/Hide the Full List | ||
| mod site | chr9:107487827-107487828:- | [6] | |
| Sequence | GGCTCCCTTCAACCTGGCGGACATCAACGACGTGAGCCCCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; CD34; HEK293T; A549; HepG2; fibroblasts; MM6; CD4T; GSC-11; HEK293A-TOA; MSC; iSLK; TIME; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000497048.5; ENST00000374672.5; ENST00000411706.1; ENST00000493306.1; ENST00000610832.1 | ||
| External Link | RMBase: m6A_site_832403 | ||
| mod ID: M6ASITE089000 | Click to Show/Hide the Full List | ||
| mod site | chr9:107488076-107488077:- | [6] | |
| Sequence | CAACGATCTCCTGGACCTGGACTTTATTCTCTCCAATTCGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; fibroblasts; MM6; HEK293A-TOA; iSLK; MSC; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000610832.1; ENST00000497048.5; ENST00000411706.1; ENST00000493306.1; ENST00000374672.5 | ||
| External Link | RMBase: m6A_site_832404 | ||
| mod ID: M6ASITE089001 | Click to Show/Hide the Full List | ||
| mod site | chr9:107488082-107488083:- | [6] | |
| Sequence | GGAGTTCAACGATCTCCTGGACCTGGACTTTATTCTCTCCA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; fibroblasts; MM6; HEK293A-TOA; iSLK; MSC; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000497048.5; ENST00000493306.1; ENST00000411706.1; ENST00000374672.5; ENST00000610832.1 | ||
| External Link | RMBase: m6A_site_832405 | ||
| mod ID: M6ASITE089002 | Click to Show/Hide the Full List | ||
| mod site | chr9:107488107-107488108:- | [6] | |
| Sequence | CGCCCCTACCTCGGAGAGAGACCGAGGAGTTCAACGATCTC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; fibroblasts; MM6; HEK293A-TOA; iSLK; MSC; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000610832.1; ENST00000497048.5; ENST00000493306.1; ENST00000411706.1; ENST00000374672.5 | ||
| External Link | RMBase: m6A_site_832406 | ||
| mod ID: M6ASITE089003 | Click to Show/Hide the Full List | ||
| mod site | chr9:107488175-107488176:- | [6] | |
| Sequence | GGCGGCGACCGTGGCCACAGACCTGGAGAGCGGCGGAGCCG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; fibroblasts; A549; MM6; HEK293A-TOA; iSLK; MSC; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374672.5; ENST00000497048.5; ENST00000411706.1; ENST00000610832.1; ENST00000493306.1 | ||
| External Link | RMBase: m6A_site_832407 | ||
| mod ID: M6ASITE089004 | Click to Show/Hide the Full List | ||
| mod site | chr9:107488304-107488305:- | [6] | |
| Sequence | ATGTCGGGTGGCAACAGGGGACCACCACTGACAGGTCTCTC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000497048.5; ENST00000493306.1; ENST00000610832.1; ENST00000411706.1; ENST00000374672.5 | ||
| External Link | RMBase: m6A_site_832408 | ||
| mod ID: M6ASITE089005 | Click to Show/Hide the Full List | ||
| mod site | chr9:107488958-107488959:- | [8] | |
| Sequence | GCCCGGCGGGAAGGGAGAAGACACTGCGTCAAGCAGGTGCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000610832.1; ENST00000411706.1; ENST00000493306.1; ENST00000374672.5 | ||
| External Link | RMBase: m6A_site_832409 | ||
| mod ID: M6ASITE089006 | Click to Show/Hide the Full List | ||
| mod site | chr9:107489125-107489126:- | [9] | |
| Sequence | GGGAGCAGGGAGGGGTGAGGACCGGTGGGACGCGCCGGAAC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000374672.5; ENST00000610832.1; ENST00000420475.1; ENST00000493306.1; ENST00000411706.1 | ||
| External Link | RMBase: m6A_site_832410 | ||
| mod ID: M6ASITE089007 | Click to Show/Hide the Full List | ||
| mod site | chr9:107489192-107489193:- | [6] | |
| Sequence | GCTGCTTCGGGCTGCCGAGGACCTTCTGGGCCCCCACATTA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000610832.1; ENST00000374672.5; ENST00000420475.1; ENST00000493306.1 | ||
| External Link | RMBase: m6A_site_832411 | ||
| mod ID: M6ASITE089008 | Click to Show/Hide the Full List | ||
| mod site | chr9:107489335-107489336:- | [6] | |
| Sequence | GAACTTTTGTATACAAAGGAACTTTTTAAAAAAGACGCTTC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000420475.1; ENST00000374672.5; ENST00000610832.1; ENST00000493306.1 | ||
| External Link | RMBase: m6A_site_832412 | ||
| mod ID: M6ASITE089009 | Click to Show/Hide the Full List | ||
| mod site | chr9:107489353-107489354:- | [6] | |
| Sequence | CGTTTACAACTTTTCTAAGAACTTTTGTATACAAAGGAACT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000420475.1; ENST00000374672.5; ENST00000493306.1; ENST00000610832.1 | ||
| External Link | RMBase: m6A_site_832413 | ||
| mod ID: M6ASITE089010 | Click to Show/Hide the Full List | ||
| mod site | chr9:107489408-107489409:- | [6] | |
| Sequence | GAGCGCAGCCCGGCCACCGGACCTACTTACTCGCCTTGCTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HEK293A-TOA; MSC | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000374672.5; ENST00000610832.1; ENST00000420475.1; ENST00000493306.1 | ||
| External Link | RMBase: m6A_site_832414 | ||
| mod ID: M6ASITE089011 | Click to Show/Hide the Full List | ||
| mod site | chr9:107490139-107490140:- | [6] | |
| Sequence | CCAGTCTTCGCGGGCTTCGAACCCAGGGAGCCGACAATGGC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; A549; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000420475.1 | ||
| External Link | RMBase: m6A_site_832415 | ||
| mod ID: M6ASITE089012 | Click to Show/Hide the Full List | ||
| mod site | chr9:107490224-107490225:- | [6] | |
| Sequence | CGCTCTTCTCCAGCGCGCAGACAGCGCGGCAAGCGCGTATG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000420475.1 | ||
| External Link | RMBase: m6A_site_832416 | ||
N7-methylguanosine (m7G)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: m7GSITE000119 | Click to Show/Hide the Full List | ||
| mod site | chr9:107489438-107489439:- | [10] | |
| Sequence | CAGTGGTGGGGGACGCTGCTGAGTGGAAGAGAGCGCAGCCC | ||
| Cell/Tissue List | A549; HeLa; HepG2 | ||
| Seq Type List | BoRed-seq&m7G-RIP-seq; m7G-seq | ||
| Transcript ID List | ENST00000374672.5; ENST00000610832.1; ENST00000420475.1 | ||
| External Link | RMBase: m7G_site_867 | ||
References