m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00729)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
DARS
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3 knockout liver
Control: Wild type liver cells
|
GSE198513 | |
| Regulation |
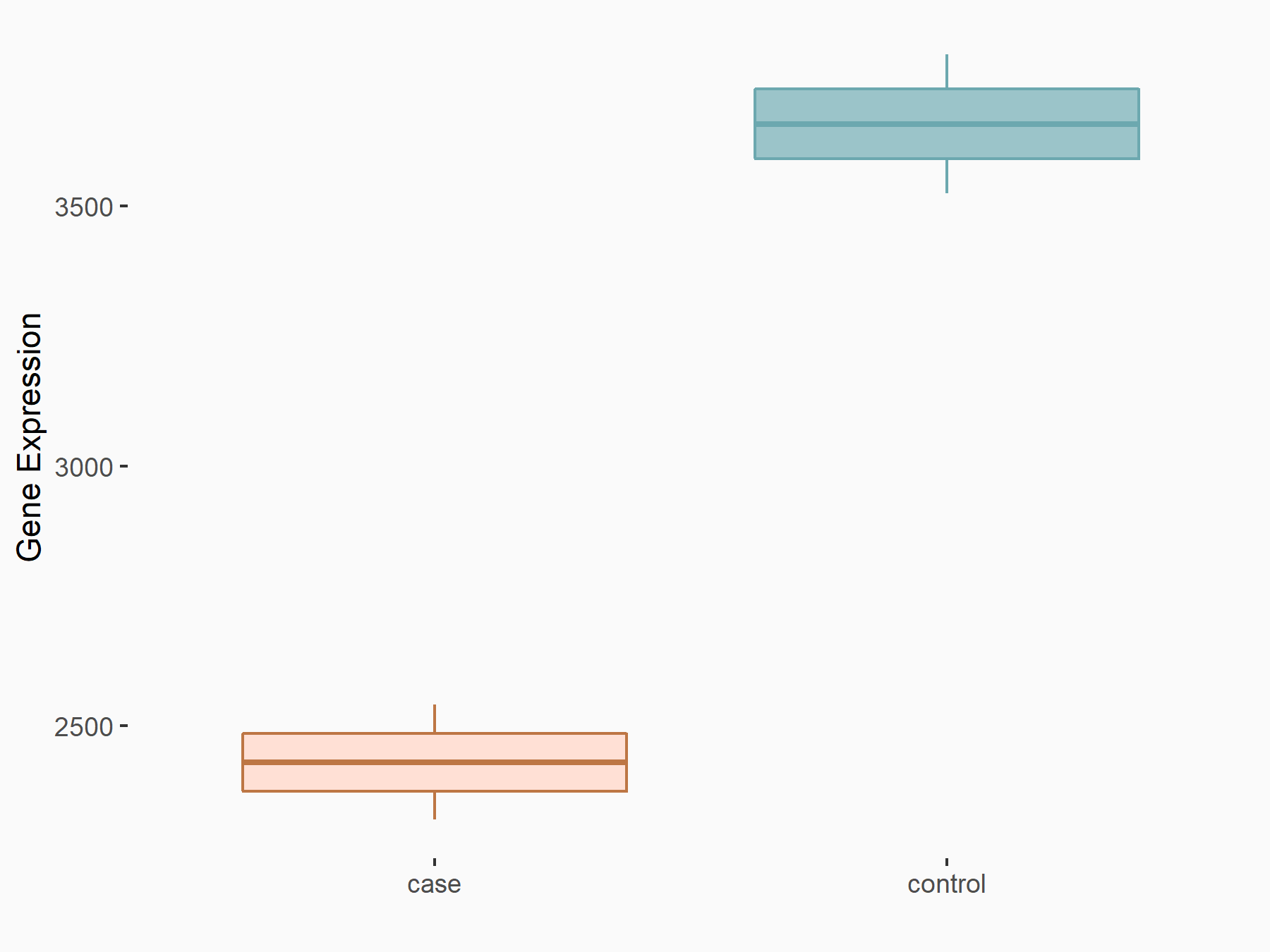  |
logFC: -5.90E-01 p-value: 7.45E-13 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between DARS and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.22E+00 | GSE60213 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | DARS-AS1 was validated to facilitate DARS translation via recruiting METTL3 and METTL14, which bound with DARS mRNA Aspartate--tRNA ligase, cytoplasmic (DARS) mRNA 5' untranslated region (5'UTR) and promoting its translation. The present study demonstrated that the 'HIF1-Alpha/DARS-AS1/DARS/ATG5/ATG3' pathway regulated the hypoxia-induced cytoprotective autophagy of cervical cancer(CC) and is a promising target of therapeutic strategies for patients afflicted with CC. | |||
| Responsed Disease | Cervical cancer | ICD-11: 2C77 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell autophagy | |||
| In-vitro Model | SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| End1/E6E7 | Normal | Homo sapiens | CVCL_3684 | |
| DoTc2 4510 | Cervical carcinoma | Homo sapiens | CVCL_1181 | |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
Methyltransferase-like 14 (METTL14) [WRITER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | DARS-AS1 was validated to facilitate DARS translation via recruiting METTL3 and METTL14, which bound with DARS mRNA Aspartate--tRNA ligase, cytoplasmic (DARS) mRNA 5' untranslated region (5'UTR) and promoting its translation. The present study demonstrated that the 'HIF1-Alpha/DARS-AS1/DARS/ATG5/ATG3' pathway regulated the hypoxia-induced cytoprotective autophagy of cervical cancer(CC) and is a promising target of therapeutic strategies for patients afflicted with CC. | |||
| Responsed Disease | Cervical cancer | ICD-11: 2C77 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell autophagy | |||
| In-vitro Model | SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| End1/E6E7 | Normal | Homo sapiens | CVCL_3684 | |
| DoTc2 4510 | Cervical carcinoma | Homo sapiens | CVCL_1181 | |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
Cervical cancer [ICD-11: 2C77]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | DARS-AS1 was validated to facilitate DARS translation via recruiting METTL3 and METTL14, which bound with DARS mRNA Aspartate--tRNA ligase, cytoplasmic (DARS) mRNA 5' untranslated region (5'UTR) and promoting its translation. The present study demonstrated that the 'HIF1-Alpha/DARS-AS1/DARS/ATG5/ATG3' pathway regulated the hypoxia-induced cytoprotective autophagy of cervical cancer(CC) and is a promising target of therapeutic strategies for patients afflicted with CC. | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell autophagy | |||
| In-vitro Model | SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| End1/E6E7 | Normal | Homo sapiens | CVCL_3684 | |
| DoTc2 4510 | Cervical carcinoma | Homo sapiens | CVCL_1181 | |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | DARS-AS1 was validated to facilitate DARS translation via recruiting METTL3 and METTL14, which bound with DARS mRNA Aspartate--tRNA ligase, cytoplasmic (DARS) mRNA 5' untranslated region (5'UTR) and promoting its translation. The present study demonstrated that the 'HIF1-Alpha/DARS-AS1/DARS/ATG5/ATG3' pathway regulated the hypoxia-induced cytoprotective autophagy of cervical cancer(CC) and is a promising target of therapeutic strategies for patients afflicted with CC. | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell autophagy | |||
| In-vitro Model | SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| End1/E6E7 | Normal | Homo sapiens | CVCL_3684 | |
| DoTc2 4510 | Cervical carcinoma | Homo sapiens | CVCL_1181 | |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03510 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03523 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
Non-coding RNA
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05356 | ||
| Epigenetic Regulator | DARS1 antisense RNA 1 (DARS1-AS1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Cervical cancer | |
m6A Regulator: Methyltransferase-like 14 (METTL14)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05357 | ||
| Epigenetic Regulator | DARS1 antisense RNA 1 (DARS1-AS1) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Cervical cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00729)
| In total 11 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE010028 | Click to Show/Hide the Full List | ||
| mod site | chr2:135908908-135908909:- | [2] | |
| Sequence | ATATGTACTACGTTTACTTTATCCAGTCTATCATTGATGGG | ||
| Transcript ID List | ENST00000478212.5; ENST00000489964.5; ENST00000422708.3; ENST00000264161.9 | ||
| External Link | RMBase: RNA-editing_site_80411 | ||
| mod ID: A2ISITE010029 | Click to Show/Hide the Full List | ||
| mod site | chr2:135909622-135909623:- | [2] | |
| Sequence | CTTTTAAAAGTTGAACCCATAGACAAAGAGTAGAATGGTTG | ||
| Transcript ID List | rmsk_683690; ENST00000264161.9; ENST00000422708.3; ENST00000489964.5; ENST00000478212.5 | ||
| External Link | RMBase: RNA-editing_site_80412 | ||
| mod ID: A2ISITE010030 | Click to Show/Hide the Full List | ||
| mod site | chr2:135909679-135909680:- | [2] | |
| Sequence | ATTAGCACATCAGGCACAGAAAGACAAATACTGTGTGATCT | ||
| Transcript ID List | rmsk_683690; ENST00000489964.5; ENST00000264161.9; ENST00000422708.3; ENST00000478212.5 | ||
| External Link | RMBase: RNA-editing_site_80413 | ||
| mod ID: A2ISITE010031 | Click to Show/Hide the Full List | ||
| mod site | chr2:135909720-135909721:- | [2] | |
| Sequence | GTAGCAATGTAGATGAACATAGAGGACATCATGCTAAACAA | ||
| Transcript ID List | rmsk_683690; ENST00000264161.9; ENST00000422708.3; ENST00000478212.5; ENST00000489964.5 | ||
| External Link | RMBase: RNA-editing_site_80414 | ||
| mod ID: A2ISITE010032 | Click to Show/Hide the Full List | ||
| mod site | chr2:135910119-135910120:- | [3] | |
| Sequence | ATTTGTGCACTCTGATGTTCAATGCAGCATTATTTACAATA | ||
| Transcript ID List | ENST00000489964.5; ENST00000478212.5; rmsk_683692; ENST00000264161.9; ENST00000422708.3 | ||
| External Link | RMBase: RNA-editing_site_80415 | ||
| mod ID: A2ISITE010033 | Click to Show/Hide the Full List | ||
| mod site | chr2:135912971-135912972:- | [4] | |
| Sequence | GCACTTTGGGAGGCCAAGGTAGGAGGATCATTTGAGGCCAG | ||
| Transcript ID List | ENST00000489964.5; ENST00000264161.9; ENST00000422708.3; rmsk_683697 | ||
| External Link | RMBase: RNA-editing_site_80416 | ||
| mod ID: A2ISITE010034 | Click to Show/Hide the Full List | ||
| mod site | chr2:135917076-135917077:- | [4] | |
| Sequence | CCTTCACCTCTCAGGCCCAAACAATTCTCCTGCCTCAGCCT | ||
| Transcript ID List | ENST00000422708.3; ENST00000264161.9 | ||
| External Link | RMBase: RNA-editing_site_80417 | ||
| mod ID: A2ISITE010035 | Click to Show/Hide the Full List | ||
| mod site | chr2:135917156-135917157:- | [4] | |
| Sequence | TTTTCTTTTTCTTTTTTTTGAGACGAGTCTTGCTCTGTCGC | ||
| Transcript ID List | ENST00000264161.9; ENST00000422708.3 | ||
| External Link | RMBase: RNA-editing_site_80418 | ||
| mod ID: A2ISITE010036 | Click to Show/Hide the Full List | ||
| mod site | chr2:135918278-135918279:- | [2] | |
| Sequence | ATTTTTGTACTACCATTGATAGACATGATGAATGGGAGTTG | ||
| Transcript ID List | ENST00000264161.9; ENST00000422708.3 | ||
| External Link | RMBase: RNA-editing_site_80419 | ||
| mod ID: A2ISITE010037 | Click to Show/Hide the Full List | ||
| mod site | chr2:135919248-135919249:- | [2] | |
| Sequence | CACTTGAGGTCAGAAGTTCAAGACCGACCCGGCCAACTTAG | ||
| Transcript ID List | ENST00000422708.3; rmsk_683710; ENST00000264161.9 | ||
| External Link | RMBase: RNA-editing_site_80420 | ||
| mod ID: A2ISITE010038 | Click to Show/Hide the Full List | ||
| mod site | chr2:135929999-135930000:- | [3] | |
| Sequence | CATGTACTTGCCTTGACTTAAGTTACCAGTCTGTCCAAGAG | ||
| Transcript ID List | ENST00000264161.9; ENST00000456565.5; ENST00000441323.5 | ||
| External Link | RMBase: RNA-editing_site_80421 | ||
N6-methyladenosine (m6A)
| In total 54 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE047547 | Click to Show/Hide the Full List | ||
| mod site | chr2:135906710-135906711:- | [5] | |
| Sequence | ATTAATTTGTTAAATTCTAAACTTGAATTTCAATAAAATTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494453 | ||
| mod ID: M6ASITE047548 | Click to Show/Hide the Full List | ||
| mod site | chr2:135906761-135906762:- | [5] | |
| Sequence | AAATTATGTAAAGCTAAACTACTGGTTAGAAAGTATTCAGT | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494454 | ||
| mod ID: M6ASITE047549 | Click to Show/Hide the Full List | ||
| mod site | chr2:135906910-135906911:- | [6] | |
| Sequence | CTTAGATTTTCAGTATTTGAACTTATTTTTTTAAATTCTGT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000478212.5; ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494455 | ||
| mod ID: M6ASITE047550 | Click to Show/Hide the Full List | ||
| mod site | chr2:135906952-135906953:- | [5] | |
| Sequence | TTTTTCTTCTTTTCTATATTACAAGGGCCCCAGTGTTAATG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000478212.5; ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494456 | ||
| mod ID: M6ASITE047551 | Click to Show/Hide the Full List | ||
| mod site | chr2:135906976-135906977:- | [5] | |
| Sequence | TTTTAAACAGTTAAGATTGTACAATTTTTCTTCTTTTCTAT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000264161.9; ENST00000478212.5 | ||
| External Link | RMBase: m6A_site_494457 | ||
| mod ID: M6ASITE047552 | Click to Show/Hide the Full List | ||
| mod site | chr2:135906990-135906991:- | [6] | |
| Sequence | GTAAACGTTAAGAGTTTTAAACAGTTAAGATTGTACAATTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; AML | ||
| Seq Type List | m6A-seq; MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000264161.9; ENST00000478212.5 | ||
| External Link | RMBase: m6A_site_494458 | ||
| mod ID: M6ASITE047553 | Click to Show/Hide the Full List | ||
| mod site | chr2:135907006-135907007:- | [5] | |
| Sequence | ATATTCTGTTACTACAGTAAACGTTAAGAGTTTTAAACAGT | ||
| Motif Score | 2.179660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000478212.5; ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494459 | ||
| mod ID: M6ASITE047554 | Click to Show/Hide the Full List | ||
| mod site | chr2:135907016-135907017:- | [5] | |
| Sequence | TTACAAATTCATATTCTGTTACTACAGTAAACGTTAAGAGT | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000264161.9; ENST00000478212.5 | ||
| External Link | RMBase: m6A_site_494460 | ||
| mod ID: M6ASITE047555 | Click to Show/Hide the Full List | ||
| mod site | chr2:135907034-135907035:- | [5] | |
| Sequence | GAAATTCATATGGTTATGTTACAAATTCATATTCTGTTACT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000478212.5; ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494461 | ||
| mod ID: M6ASITE047556 | Click to Show/Hide the Full List | ||
| mod site | chr2:135907060-135907061:- | [5] | |
| Sequence | ACATGCATCAGTAGGAAATAACTTGAGAAATTCATATGGTT | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000264161.9; ENST00000478212.5 | ||
| External Link | RMBase: m6A_site_494462 | ||
| mod ID: M6ASITE047557 | Click to Show/Hide the Full List | ||
| mod site | chr2:135907080-135907081:- | [6] | |
| Sequence | AGAGATTTTTATAACTTCAGACATGCATCAGTAGGAAATAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T | ||
| Seq Type List | m6A-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000478212.5; ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494463 | ||
| mod ID: M6ASITE047558 | Click to Show/Hide the Full List | ||
| mod site | chr2:135907087-135907088:- | [5] | |
| Sequence | GTTTAAAAGAGATTTTTATAACTTCAGACATGCATCAGTAG | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000478212.5; ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494464 | ||
| mod ID: M6ASITE047559 | Click to Show/Hide the Full List | ||
| mod site | chr2:135907140-135907141:- | [5] | |
| Sequence | TTGCAAGGCCCTAAAATATCACTGTTATTTTTGGAGTAATT | ||
| Motif Score | 2.469291667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000478212.5; ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494465 | ||
| mod ID: M6ASITE047560 | Click to Show/Hide the Full List | ||
| mod site | chr2:135907310-135907311:- | [7] | |
| Sequence | ACGACTCACTCCTTAAATTCACACTTTGCCACTTAACTCCA | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000489964.5; ENST00000478212.5; ENST00000422708.3; ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494466 | ||
| mod ID: M6ASITE047561 | Click to Show/Hide the Full List | ||
| mod site | chr2:135911188-135911189:- | [7] | |
| Sequence | TTTGGAGAAAATTAAGGCTTACATTGATTCCTTCCGCTTTG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000422708.3; ENST00000489964.5; ENST00000491481.1; ENST00000264161.9; ENST00000478212.5 | ||
| External Link | RMBase: m6A_site_494467 | ||
| mod ID: M6ASITE047562 | Click to Show/Hide the Full List | ||
| mod site | chr2:135911427-135911428:- | [7] | |
| Sequence | TTGTCAGGAGCTCAAAGAATACATGATCCTCAACTGCTAAC | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000422708.3; ENST00000264161.9; ENST00000489964.5; ENST00000478212.5; ENST00000491481.1 | ||
| External Link | RMBase: m6A_site_494468 | ||
| mod ID: M6ASITE047563 | Click to Show/Hide the Full List | ||
| mod site | chr2:135911490-135911491:- | [6] | |
| Sequence | GTGTTTTTTCTCTTGCAGAAACAGTCCAACTCTTACGATAT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264161.9; ENST00000489964.5; ENST00000491481.1; ENST00000478212.5; ENST00000422708.3 | ||
| External Link | RMBase: m6A_site_494469 | ||
| mod ID: M6ASITE047564 | Click to Show/Hide the Full List | ||
| mod site | chr2:135911514-135911515:- | [6] | |
| Sequence | AGGATGGTAGTCACTTGAGGACTGGTGTTTTTTCTCTTGCA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000491481.1; ENST00000489964.5; ENST00000478212.5; ENST00000264161.9; ENST00000422708.3 | ||
| External Link | RMBase: m6A_site_494470 | ||
| mod ID: M6ASITE047565 | Click to Show/Hide the Full List | ||
| mod site | chr2:135914509-135914510:- | [7] | |
| Sequence | TTTCACGTTTCTTTTTCAGCACACCAAATGAAAAGCTGTTG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000422708.3; ENST00000489964.5; ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494471 | ||
| mod ID: M6ASITE047566 | Click to Show/Hide the Full List | ||
| mod site | chr2:135914536-135914537:- | [6] | |
| Sequence | AGTTGCCATTTGATCTTCAAACACTGATTTCACGTTTCTTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000489964.5; ENST00000264161.9; ENST00000422708.3 | ||
| External Link | RMBase: m6A_site_494472 | ||
| mod ID: M6ASITE047567 | Click to Show/Hide the Full List | ||
| mod site | chr2:135914591-135914592:- | [6] | |
| Sequence | ATTTTGTAATCCCTTGAGAAACTCAAATAGAAGAAGTTCCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264161.9; ENST00000422708.3; ENST00000489964.5 | ||
| External Link | RMBase: m6A_site_494473 | ||
| mod ID: M6ASITE047568 | Click to Show/Hide the Full List | ||
| mod site | chr2:135916296-135916297:- | [6] | |
| Sequence | TTTTTGGAGCCAACTCTAAGACTAGAATATTGTGAAGCATT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264161.9; ENST00000422708.3 | ||
| External Link | RMBase: m6A_site_494474 | ||
| mod ID: M6ASITE047569 | Click to Show/Hide the Full List | ||
| mod site | chr2:135916341-135916342:- | [6] | |
| Sequence | GAAATTCAAACAGTGAATAAACAGTTCCCATGTGAGCCATT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264161.9; ENST00000422708.3 | ||
| External Link | RMBase: m6A_site_494475 | ||
| mod ID: M6ASITE047570 | Click to Show/Hide the Full List | ||
| mod site | chr2:135916352-135916353:- | [6] | |
| Sequence | GGTTTCAGACTGAAATTCAAACAGTGAATAAACAGTTCCCA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; kidney | ||
| Seq Type List | m6A-seq; m6A-REF-seq | ||
| Transcript ID List | ENST00000264161.9; ENST00000422708.3 | ||
| External Link | RMBase: m6A_site_494476 | ||
| mod ID: M6ASITE047571 | Click to Show/Hide the Full List | ||
| mod site | chr2:135916364-135916365:- | [6] | |
| Sequence | ATTCTGCAAATAGGTTTCAGACTGAAATTCAAACAGTGAAT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264161.9; ENST00000422708.3 | ||
| External Link | RMBase: m6A_site_494477 | ||
| mod ID: M6ASITE047572 | Click to Show/Hide the Full List | ||
| mod site | chr2:135920463-135920464:- | [6] | |
| Sequence | ATGGTACAAATATTCAAAGGACTTCAAGAAAGGTAAAAAGT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000422708.3; ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494478 | ||
| mod ID: M6ASITE047573 | Click to Show/Hide the Full List | ||
| mod site | chr2:135920478-135920479:- | [7] | |
| Sequence | GAAATTGCTGACACCATGGTACAAATATTCAAAGGACTTCA | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000422708.3; ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494479 | ||
| mod ID: M6ASITE047574 | Click to Show/Hide the Full List | ||
| mod site | chr2:135920488-135920489:- | [5] | |
| Sequence | AGTTATGGAAGAAATTGCTGACACCATGGTACAAATATTCA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000264161.9; ENST00000422708.3 | ||
| External Link | RMBase: m6A_site_494480 | ||
| mod ID: M6ASITE047575 | Click to Show/Hide the Full List | ||
| mod site | chr2:135920542-135920543:- | [6] | |
| Sequence | AACTGAGTTTGTTGGTTTGGACATTGAAATGGCTTTTAATT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T | ||
| Seq Type List | m6A-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000264161.9; ENST00000422708.3 | ||
| External Link | RMBase: m6A_site_494481 | ||
| mod ID: M6ASITE047576 | Click to Show/Hide the Full List | ||
| mod site | chr2:135920561-135920562:- | [5] | |
| Sequence | CTAATACCCATAGACATCTAACTGAGTTTGTTGGTTTGGAC | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494482 | ||
| mod ID: M6ASITE047577 | Click to Show/Hide the Full List | ||
| mod site | chr2:135920568-135920569:- | [6] | |
| Sequence | GAAGACTCTAATACCCATAGACATCTAACTGAGTTTGTTGG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T | ||
| Seq Type List | m6A-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494483 | ||
| mod ID: M6ASITE047578 | Click to Show/Hide the Full List | ||
| mod site | chr2:135920576-135920577:- | [5] | |
| Sequence | TCAGAGCGGAAGACTCTAATACCCATAGACATCTAACTGAG | ||
| Motif Score | 2.089839286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494484 | ||
| mod ID: M6ASITE047579 | Click to Show/Hide the Full List | ||
| mod site | chr2:135920584-135920585:- | [6] | |
| Sequence | CACAGTATTCAGAGCGGAAGACTCTAATACCCATAGACATC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494485 | ||
| mod ID: M6ASITE047580 | Click to Show/Hide the Full List | ||
| mod site | chr2:135922787-135922788:- | [6] | |
| Sequence | GAGAAGGTTTTCTCTATTGGACCAGGTAAGATTTTGGCAGT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494486 | ||
| mod ID: M6ASITE047581 | Click to Show/Hide the Full List | ||
| mod site | chr2:135922888-135922889:- | [5] | |
| Sequence | AAGGAGGAGCCAATGTTTTTACTGTGTCATATTTTAAAAAT | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494487 | ||
| mod ID: M6ASITE047582 | Click to Show/Hide the Full List | ||
| mod site | chr2:135924427-135924428:- | [7] | |
| Sequence | CTTCCGAGAAACTTTAATTAACAAAGGTTTTGTGGAAATCC | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264161.9; ENST00000441323.5 | ||
| External Link | RMBase: m6A_site_494488 | ||
| mod ID: M6ASITE047583 | Click to Show/Hide the Full List | ||
| mod site | chr2:135924497-135924498:- | [7] | |
| Sequence | ATTTTATTACTGTCTTCTAGACATCAACTAGTCAGGCAGTC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000441323.5; ENST00000456565.5; ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494489 | ||
| mod ID: M6ASITE047584 | Click to Show/Hide the Full List | ||
| mod site | chr2:135932801-135932802:- | [5] | |
| Sequence | CCAGGATACAAGATTAGACAACAGAGTCATTGATCTTAGGG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000441323.5; ENST00000456565.5; ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494490 | ||
| mod ID: M6ASITE047585 | Click to Show/Hide the Full List | ||
| mod site | chr2:135932804-135932805:- | [8] | |
| Sequence | TAACCAGGATACAAGATTAGACAACAGAGTCATTGATCTTA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000456565.5; ENST00000441323.5; ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494491 | ||
| mod ID: M6ASITE047586 | Click to Show/Hide the Full List | ||
| mod site | chr2:135932814-135932815:- | [7] | |
| Sequence | GAGCTACTGTTAACCAGGATACAAGATTAGACAACAGAGTC | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000441323.5; ENST00000264161.9; ENST00000456565.5 | ||
| External Link | RMBase: m6A_site_494492 | ||
| mod ID: M6ASITE047587 | Click to Show/Hide the Full List | ||
| mod site | chr2:135943389-135943390:- | [7] | |
| Sequence | ACACAGCAAGACGTTGAGTTACATGTTCAGAAGGTAAGTTT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000449218.5; ENST00000463008.1; ENST00000435076.1; ENST00000456565.5; ENST00000441323.5; ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494493 | ||
| mod ID: M6ASITE047588 | Click to Show/Hide the Full List | ||
| mod site | chr2:135943409-135943410:- | [7] | |
| Sequence | ATCAGAAAATTGGAAGCTGTACACAGCAAGACGTTGAGTTA | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000456565.5; ENST00000264161.9; ENST00000463008.1; ENST00000435076.1; ENST00000441323.5; ENST00000449218.5 | ||
| External Link | RMBase: m6A_site_494494 | ||
| mod ID: M6ASITE047589 | Click to Show/Hide the Full List | ||
| mod site | chr2:135943474-135943475:- | [7] | |
| Sequence | TTTTTCTCTTGACAGCATCAACAAAGAGAGCATTGTGGATG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000435076.1; ENST00000441323.5; ENST00000264161.9; ENST00000449218.5; ENST00000456565.5; ENST00000463008.1 | ||
| External Link | RMBase: m6A_site_494495 | ||
| mod ID: M6ASITE047590 | Click to Show/Hide the Full List | ||
| mod site | chr2:135961395-135961396:- | [7] | |
| Sequence | GATGGTTAAATTTGCTGCCAAGTAAGTAAGCAATTATATCC | ||
| Motif Score | 1.724672619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264161.9; ENST00000463008.1; ENST00000449218.5; ENST00000441323.5; ENST00000456565.5; ENST00000435076.1 | ||
| External Link | RMBase: m6A_site_494496 | ||
| mod ID: M6ASITE047591 | Click to Show/Hide the Full List | ||
| mod site | chr2:135979288-135979289:- | [7] | |
| Sequence | GGGTACGTGCAAGAGTTCATACAAGCAGAGCTAAAGGTAGG | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000435076.1; ENST00000441323.5; ENST00000264161.9; ENST00000456565.5; ENST00000474184.1; ENST00000449218.5; ENST00000463008.1 | ||
| External Link | RMBase: m6A_site_494497 | ||
| mod ID: M6ASITE047592 | Click to Show/Hide the Full List | ||
| mod site | chr2:135979331-135979332:- | [7] | |
| Sequence | CGGGTTAGAGACTTGACAATACAAAAAGCTGATGAAGTTGT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000463008.1; ENST00000435076.1; ENST00000456565.5; ENST00000441323.5; ENST00000264161.9; ENST00000449218.5; ENST00000474184.1 | ||
| External Link | RMBase: m6A_site_494498 | ||
| mod ID: M6ASITE047593 | Click to Show/Hide the Full List | ||
| mod site | chr2:135979336-135979337:- | [5] | |
| Sequence | TGGTTCGGGTTAGAGACTTGACAATACAAAAAGCTGATGAA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000435076.1; ENST00000449218.5; ENST00000463008.1; ENST00000456565.5; ENST00000264161.9; ENST00000441323.5; ENST00000474184.1 | ||
| External Link | RMBase: m6A_site_494499 | ||
| mod ID: M6ASITE047594 | Click to Show/Hide the Full List | ||
| mod site | chr2:135979341-135979342:- | [6] | |
| Sequence | AGTTTTGGTTCGGGTTAGAGACTTGACAATACAAAAAGCTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000435076.1; ENST00000456565.5; ENST00000463008.1; ENST00000449218.5; ENST00000441323.5; ENST00000474184.1; ENST00000264161.9 | ||
| External Link | RMBase: m6A_site_494500 | ||
| mod ID: M6ASITE047595 | Click to Show/Hide the Full List | ||
| mod site | chr2:135980521-135980522:- | [9] | |
| Sequence | CTTTGCTTTTGGCTATGAGAACCGTGTTATTTATTCTCCTG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | A549; HEC-1-A | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000441323.5; ENST00000435076.1; ENST00000463008.1; ENST00000456565.5; ENST00000264161.9; ENST00000474184.1; ENST00000449218.5 | ||
| External Link | RMBase: m6A_site_494501 | ||
| mod ID: M6ASITE047596 | Click to Show/Hide the Full List | ||
| mod site | chr2:135983400-135983401:- | [6] | |
| Sequence | ATGATACAATCACAAGAAAAACCAGGTTAGTAGCTATGTGA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; A549; H1299; MM6; CD4T; peripheral-blood; MSC; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000474184.1; ENST00000449218.5; ENST00000456565.5; ENST00000435076.1; ENST00000264161.9; ENST00000441323.5 | ||
| External Link | RMBase: m6A_site_494502 | ||
| mod ID: M6ASITE047597 | Click to Show/Hide the Full List | ||
| mod site | chr2:135983409-135983410:- | [7] | |
| Sequence | ATATCTTCAATGATACAATCACAAGAAAAACCAGGTTAGTA | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000456565.5; ENST00000474184.1; ENST00000449218.5; ENST00000264161.9; ENST00000435076.1; ENST00000441323.5 | ||
| External Link | RMBase: m6A_site_494503 | ||
| mod ID: M6ASITE047598 | Click to Show/Hide the Full List | ||
| mod site | chr2:135983415-135983416:- | [5] | |
| Sequence | TATGGAATATCTTCAATGATACAATCACAAGAAAAACCAGG | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000474184.1; ENST00000435076.1; ENST00000456565.5; ENST00000441323.5; ENST00000264161.9; ENST00000449218.5 | ||
| External Link | RMBase: m6A_site_494504 | ||
| mod ID: M6ASITE047606 | Click to Show/Hide the Full List | ||
| mod site | chr2:135985553-135985554:- | [6] | |
| Sequence | CCAGGGTCGGGAGGGCGGAGACTGGGAGGGAGGGAGAAGCC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; A549; H1A; H1B; MT4; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; HEK293A-TOA; MSC; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000435076.1; ENST00000456565.5; ENST00000264161.9; ENST00000441323.5; ENST00000449218.5 | ||
| External Link | RMBase: m6A_site_494512 | ||
| mod ID: M6ASITE047607 | Click to Show/Hide the Full List | ||
| mod site | chr2:135985576-135985577:- | [6] | |
| Sequence | GTCCGAGCGCGTGTGCTGAGACCCCAGGGTCGGGAGGGCGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; A549; H1A; H1B; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; HEK293A-TOA; MSC; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000264161.9; ENST00000456565.5; ENST00000449218.5; ENST00000441323.5 | ||
| External Link | RMBase: m6A_site_494513 | ||
Pseudouridine (Pseudo)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: PSESITE000142 | Click to Show/Hide the Full List | ||
| mod site | chr2:135920539-135920540:- | [10] | |
| Sequence | TGAGTTTGTTGGTTTGGACATTGAAATGGCTTTTAATTACC | ||
| Transcript ID List | ENST00000264161.9; ENST00000422708.3 | ||
| External Link | RMBase: Pseudo_site_2724 | ||
| mod ID: PSESITE000143 | Click to Show/Hide the Full List | ||
| mod site | chr2:135979305-135979306:- | [11] | |
| Sequence | AGCTGATGAAGTTGTTTGGGTACGTGCAAGAGTTCATACAA | ||
| Transcript ID List | ENST00000474184.1; ENST00000435076.1; ENST00000456565.5; ENST00000441323.5; ENST00000463008.1; ENST00000449218.5; ENST00000264161.9 | ||
| External Link | RMBase: Pseudo_site_2725 | ||
References